Summary
Many bacterial and archaeal organisms use CRISPR-Cas (clustered regularly interspaced short palindromic repeats-CRISPR associated) systems to defend themselves from mobile genetic elements. These CRISPR-Cas systems are classified into six types based on their composition and mechanism. CRISPR-Cas enzymes are widely used for genome editing and offer immense therapeutic opportunity to treat genetic diseases. To realize their full potential, it is important to control the timing, duration, efficiency and specificity of CRISPR-Cas enzyme activities. In this review we discuss the mechanisms of natural CRISPR-Cas regulatory biomolecules that enhance or inhibit CRISPR-Cas immunity by altering enzyme function. We also discuss the potential applications of these CRISPR regulators and highlight unanswered questions about their evolution and purpose in nature.
Introduction
CRISPR-Cas systems provide diverse bacterial and archaeal species with adaptive immunity against invading mobile genetic elements (MGEs). This naturally occurring immune system has been repurposed into a revolutionary genome engineering technology for research and clinical applications. In its natural form, each CRISPR genomic locus is organized as an array of repeat sequences interspersed with variable sequences corresponding to a segment of an invasive MGE. These variable sequences, termed “spacers”, provide a genetic record of past infections and the basis for protection from future infections by the same MGE1,2. CRISPR arrays are flanked by multiple conserved protein-coding genes that function at different stages of CRISPR-mediated immunity and are generally prefixed as “CRISPR-associated” (Cas). CRISPR-Cas immunity operates in three stages: adaptation, biogenesis and interference. At the adaptation stage, a Cas integrase incorporates MGE-derived DNA sequences as spacers into the CRISPR array. During biogenesis, a precursor transcript produced from the CRISPR array is cleaved within the repeat to form processed CRISPR RNAs (crRNAs) containing a spacer and at least one portion of the repeat sequence. During the interference stage, crRNA-guided Cas complexes recognize a specific site on target nucleic acids through complementary base-pairing, triggering Cas enzyme-catalyzed target cleavage. Based on the composition of CRISPR loci, CRISPR systems can be classified into six types with each CRISPR type further classified into subtypes. The observed diversity of CRISPR locus organization and functional complexity can be attributed to the physiological environment of the host and the MGEs that they are exposed to. For more detailed information on CRISPR evolution and classification, refer to3.
Although CRISPR-Cas adaptive immune systems can ensure bacterial survival during phage infection, they can also trigger autoimmunity and present a substantial metabolic burden to the host4. To maintain CRISPR immunity while limiting fitness costs, host regulatory factors control the operation of the CRISPR immune pathways. As a result of the arms race, MGEs also encode CRISPR-regulators called anti-CRISPRs (Acr) that operate through multiple distinct mechanisms to inhibit CRISPR-cas activity. Advances in the field of CRISPR regulation have advanced our understanding of CRISPR biology and improved genome engineering tools that are being widely adopted. In this review we describe the mechanisms underlying regulation of CRISPR systems and how these enhance control of CRISPR enzymes both in nature and in the laboratory.
Regulation of CRISPR-Cas gene expression
In many bacterial genomes, CRISPR-Cas systems are subject to control by transcriptional and post-transcriptional modes of gene regulation (Fig. 1, table 1). Transposon mutagenesis experiments uncovered multiple transcription factors that are involved in controlling the expression of CRISPR-Cas components. Regulators identified in this way from Escherichia coli (E.coli) and Salmonella typhi include nucleoid associated protein, H-NS, and Leucine responsive regulatory proteins, LRP and leuO. H-NS and LRP negatively regulate CRISPR-Cas expression, while leuO positively regulates expression, by binding to the Cas promoter5,6. Similarly, in archaeon S. islandicus, two transcriptional regulators, Csa3a and Csa3b, containing CRISPR-associated rossman fold (CARF) and helix-turn-helix (HTH) domains have been identified that regulate the expression of type I-A CRISPR locus. Csa3a transcriptionally activates expression of the CRISPR array and adaptation genes, while Csa3b represses the expression of interference genes in the type I-A CRISPR locus7,8. Interestingly, in the absence of viral infection, Csa3b requires co-binding by the cascade complex to exert complete repression of the CRISPR loci. However, during viral infection, redistribution of the cascade complex to the invading viral genome leads to reactivation of CRISPR loci8. Bacteria can vary the expression of their CRISPR-Cas genes based on external stimuli like nutrient availability, extracytoplasmic stress and growth lifestyle9–14. In P. atrosepticum, the availability of glucose can reduce Cas gene expression through a decrease in the levels of the transcriptional activator, CRP-cAMP. The CRP-cAMP complex consists of a dimer of CRP (cAMP regulatory protein) associated with cAMP (cyclic adenosine 3,5-monophosphate), produced by adenylate cyclase. In the presence of high glucose levels, the activity of the adenylate cyclase gene, cya1, is low and thus the level of CRP-cAMP complex is also low. This loss of CRP-cAMP complex leads to lower Cas gene expression9. Similarly, under iron deprivation, type I-F CRISPR-cas genes show heightened expression in P. aeruginosa mediated by Extracytoplasmic Function (ECF) sigma factor, PvdS, which promotes transcription of the CRISPR locus by interacting with the Cas gene promoter15. These responses to limiting levels of glucose and iron could be a bacterial strategy to adapt to increased metabolic stress induced during phage infection9. In contrast, bacteria respond to extracytoplasmic stress by promoting Cas gene expression through the activity of the BaeSR signalling system10. It is unclear whether Cas genes function in a non-defense stress response pathway or whether induction occurs as a result of common signalling networks between stress response and phage defense10. Consistent with the role of CRISPR systems in defense against MGEs, bacteria regulate Cas gene expression in correspondence to the risk of phage infection. Susceptibility to phage infection heavily depends on the growth lifestyle of the bacteria. At high cell density where the risk of phage infection is high, Cas gene expression is positively regulated by components of the quorum sensing pathway. In P. aeruginosa, quorum sensing autoinducer-receptor pairs LasIR, RhIIR and SmaIR promote expression of type I-E and I-F Cas genes11,12. In contrast, alginate regulator proteins that are involved in biofilm production– AlgU, AlgR and AmrZ - repress expression of type I-F Cas genes13. By regulating the expression of multiple Cas genes, these transcriptional regulatory mechanisms not only affect CRISPR-Cas immunity at the interference stage but also the adaptation stage9,11. Considering the high cost of maintaining defense systems, additional transcriptional regulatory mechanisms are likely to exist. Similar to CRP-cAMP regulatory activity, other DNA-binding proteins fused to small-molecule sensor domains might exist that can function as allosteric transcription factors. One potential mechanism could involve the function of allosteric transcription factors containing cyclic oligoadenylate (cOA) sensing, CARF domain, and helix-turn-helix DNA binding domain3,16.
Figure 1. Transcriptional modulation of a CRISPR locus under diverse environments.
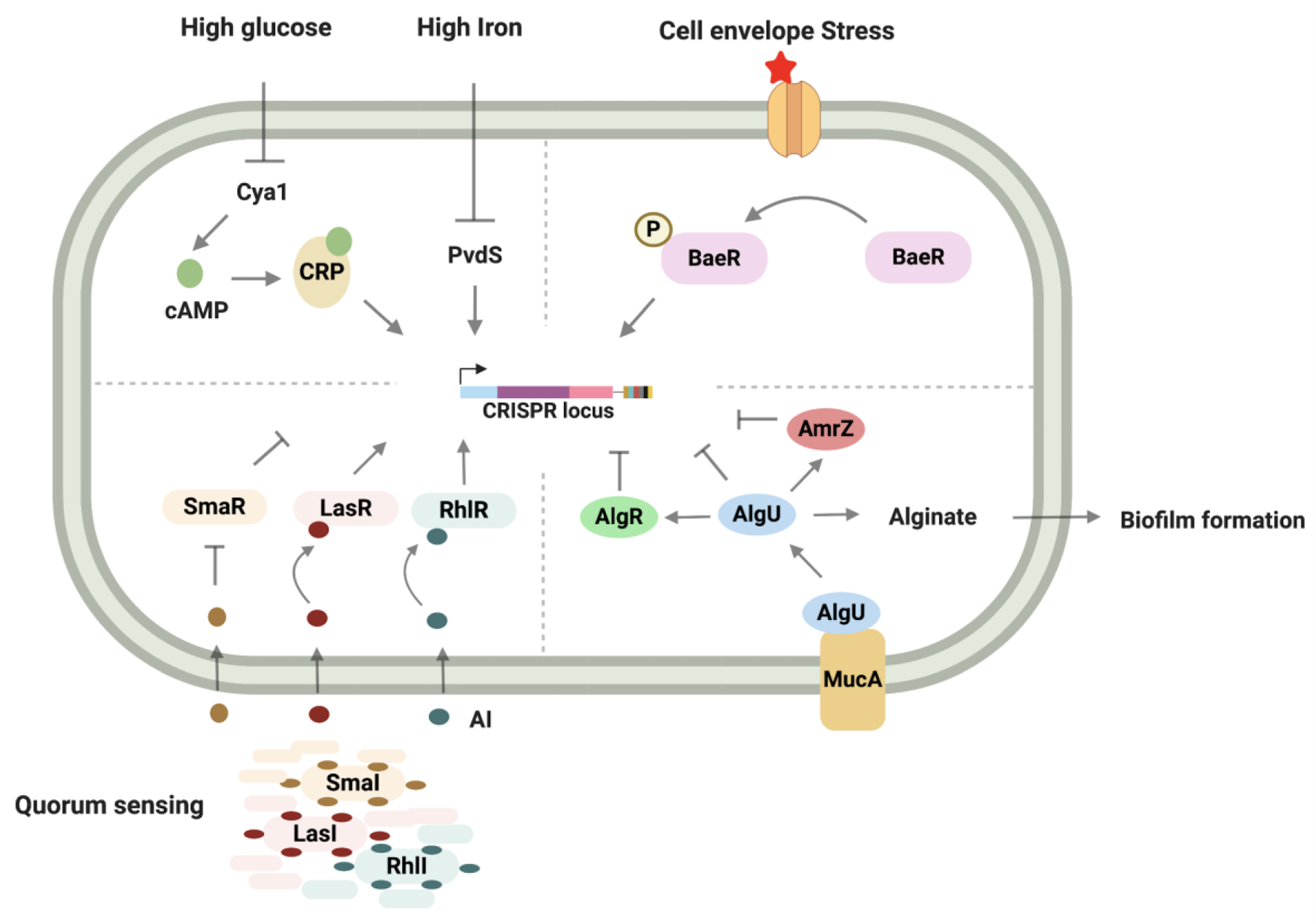
Schematic showing activation of CRISPR expression in bacteria in response to diverse external stimuli like glucose and iron levels (Top, left), extracytoplasmic/envelope stress (Top right), quorum sensing (Bottom left) and alginate biosynthesis (Bottom right). Created with BioRender.com
Table 1.
Regulation of CRISPR-Cas gene expression
| Regulator | CRISPR system | Key Mechanism |
|---|---|---|
| LRP6 | IE - S. Typhi | Transcriptional repressor |
| H-NS5,6 | IE - S. Typhi, E.coli | Transcriptional repressor |
| leuO5,6 | IE - S. Typhi, E.coli | Relieves repression by LRP and H-NS |
| Csa3a7 | IA - S. Islandicus | Transcriptional activator |
| Csa3b8 | IA - S. Islandicus | Transcriptional repressor |
| CRP-cAMP9 | IF - P. atrosepticum | Glucose responsive transcriptional activator |
| PvdS15 | IF - P. aeruginosa | Iron responsive transcriptional activator |
| BaeSR10 | IE - E.coli | Transcriptional activation likely in response to extracellular stress |
| LasR12, RhlR12 | IF - P. aeruginosa | Quorum sensing mediated transcriptional regulation |
| SmaR11 | I-E, I-F, IIIA - Serratia | Quorum sensing mediated transcriptional regulation |
| AlgU, AlgR, AmrZ13 | IF - P. aeruginosa | Inhibition of CRISPR-Cas during planktonic growth |
| trL18 | IIA - S. pyogenes | Repurposes Cas9 for autoregulation |
In addition to transcription factors, RNA regulatory mechanisms promote expression of type I CRISPR loci and inhibit expression of type II CRISPR loci. PhrS, identified from an sRNA library screen in P. aeruginosa, promotes expression of the CRISPR array by blocking the binding of Rho protein to the leader sequence of the CRISPR array, thereby inhibiting Rho-dependent transcriptional termination17. A transposon-based screen in S. aureus showed that RNA produced from the CRISPR locus complexes with Cas9 and repurposed it to autoregulate its own expression18. In type II CRISPR systems, two classes of RNA, pre-crRNA and tracrRNA, are produced from the CRISPR locus. Pre-crRNA is transcribed from the CRISPR array into a long transcript containing multiple spacer-repeat segments. TracrRNA is produced from two promoters into a long and a short form. During biogenesis, the short form of the tracrRNA base pairs with the repeat regions of the pre-crRNA followed by processing of tracrRNA-crRNA hybrids into mature crRNA. The long form of the tracrRNA recruits Cas9 to the Cas gene promoter within the CRISPR locus to repress Cas gene expression18.
Controlling CRISPR effectors
Bacteria and phages undergo repeated cycles of adaptation and counter-adaptation that have likely resulted in diversification of CRISPR systems. As a result of this bacteria-phage arms race, phages have also evolved proteins (Acrs) that counteract the CRISPR defense system. Like CRISPR-Cas effectors, Acrs are highly diverse, with ~50 different families that are named based on the CRISPR-Cas type they inhibit and the order in which they were discovered19. Acrs can vary in strength of inhibition, specificity, mechanism, and a majority of them counteract specific Cas orthologs from specific CRISPR systems. Mechanistically, Acrs can be subdivided into three subclasses- competitive, allosteric and enzymatic inhibitors (Fig. 2 and Table 2). Among the known Acrs, competitive inhibitors are the most common and operate by interacting with critical residues on Cas effectors to sterically block the interaction with their target nucleic acid or recruitment of a Cas endonuclease20–27. A majority of the competitive Acrs have a negatively charged surface mimicking the DNA phosphate backbone, allowing them to compete with the same binding site on the Cas effector. As competitive inhibitors, the potency of inhibition is a function of the Acrs stoichiometric ratio with Cas effectors. As a result, these types of inhibitors are generally produced at concentrations matching or greater than that of the Cas effectors. Allosteric inhibitors, on the other hand, take advantage of the fact that multiple domains of Cas proteins are interconnected, and inhibition involves trapping Cas effectors in conformations in which they are incapable of binding or cleaving their targets23,28–30. In contrast to the other classes, enzymatic Acrs are highly potent due to their multi-turnover property, which gives them the ability to inactivate multiple Cas complexes31–33. Enzymatic Acrs are, however, less common compared to competitive inhibitors, probably due to their greater complexity and hence, increased evolutionary cost.
Figure 2. Summary of the different strategies utilized by Acrs to inhibit CRISPR activity.
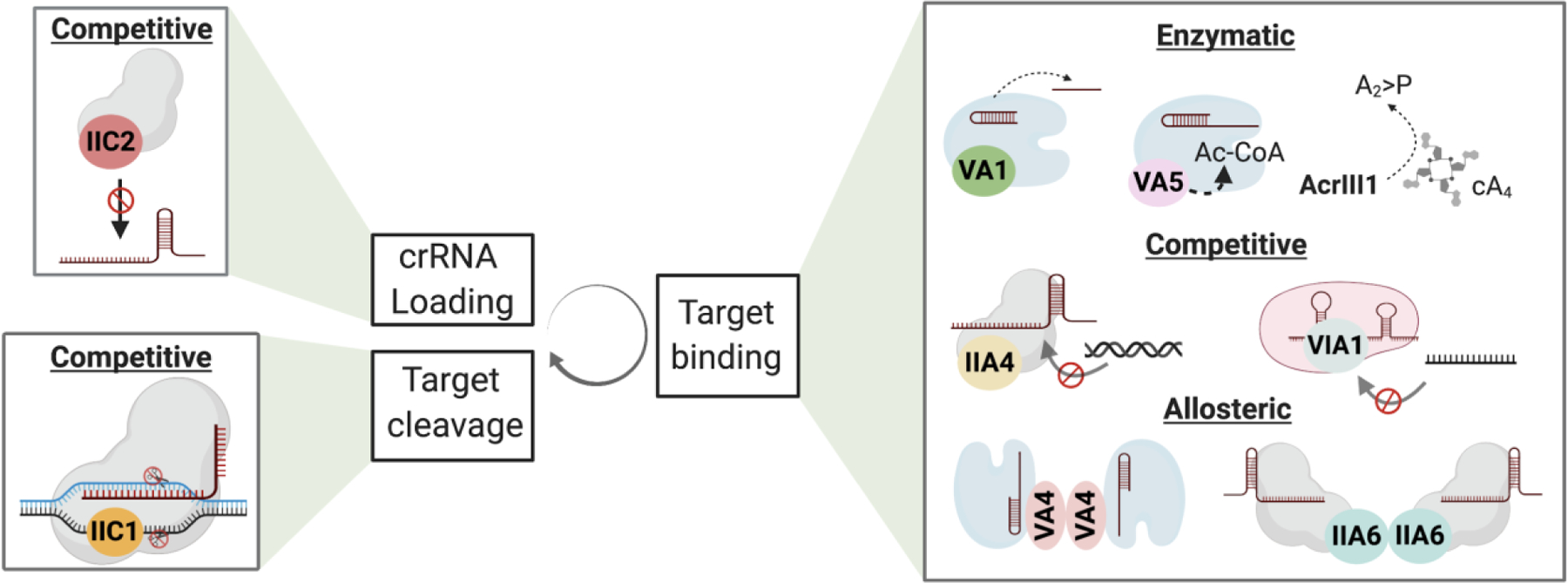
Acrs can inhibit CRISPR-Cas activity during biogenesis (crRNA loading), target recognition and target cleavage. Mechanistically they can be classified as competitive, allosteric or enzymatic inhibitors. Acrs with unique mechanisms under each category are shown as examples. Refer to able 1 for a complete list of Acrs with known mechanisms. Created with BioRender.com
Table 2.
Summary of known Anti-CRISPR mechanisms
| Regulator | Acr type | Key Mechanism |
|---|---|---|
| AcrF120 | Competitive | - Blocks dsDNA binding by interacting with Cas7f |
| AcrF220 | - Blocks dsDNA binding by wedging between Cas8f and Cas7.6f | |
| AcrF322 | - Interacts with Cas3 and blocks its interaction with Csy complex | |
| AcrF10100 | - Blocks dsDNA binding by making contacts with Cas5f, 7f and 8f. | |
| AcrIIA225 | Competitive | - Blocks dsDNA by interacting with PAM interaction sites on Cas9 |
| AcrIIA426 | - Blocks dsDNA by interacting with PAM interaction sites on Cas9 | |
| AcrIIC123 | - Inhibits catalytic activity of Cas9 by interacting with the HNH domain | |
| AcrIIC224 | - Interacts with the bridge-helix region of Cas9 and blocks guide RNA binding | |
| AcrVIA127,72 | Competitive | - Blocks target RNA binding by interacting with the crRNA, Helical-1, NTD, HEPN-1, and Linker domains of Cas13a-crRNA complex |
| AcrIIA628 | Allosteric | - Interacts as a dimer with the Cas9-sgRNA complex formed by WED and PAM-interacting domain reducing target DNA binding. |
| AcrIIC323 | - Interacts as a dimer with non-catalytic sites of HNH and dimerizes Cas9 to prevent dsDNA binding | |
| AcrVA429 | Allosteric | - Interacts as a dimer with Cas12a at the WED and bridge-helix to prevent stable dsDNA interaction. |
| AcrIII133 | Enzymatic | - Degrades cOAs through its ring nuclease activity |
| AcrVA131 | Enzymatic | - Occupies PAM binding groove on Cas12a-crRNA complex and endoribonucleolytically cleaves crRNA |
| AcrVA532 | - Acetylates Lys635 on Cas12a and prevents dsDNA binding | |
Although a few Acrs, such as AcrIIA5, AcrIIA16, AcrIIA17 and AcrVA1, are broad-spectrum in the sense that they can inhibit multiple different CRISPR subtypes, none show inhibition across multiple CRISPR types34,35. This selectivity in inhibition can be attributed to (1) selective pressure from limited bacterial encounters, and (2) antagonistic interphage interaction within the phage community. In contrast to Cas effectors, Cas integrases involved in CRISPR adaptation are highly conserved and yet we still lack evidence for the existence of phage counter strategies against them. It also remains to be tested if Acrs known to inhibit the interference activity of some Cas effectors, can also inhibit their function during primed adaptation36. It is possible that the discovery of anti-adaptation mechanisms are limited by the lack of bioinformatic clues and an appropriate functional genomic screen. An alternative hypothesis could be that the anti-adaptation strategies are rare, evidenced by the lack of sequence divergence in the Cas adaptation machinery that otherwise would occur as a result of a host escape mechanism to combat the invading MGE.
Recent findings suggest that phages can deploy additional mechanisms to evade CRISPR defense by bacteria without directly interacting with their components. This phage strategy involves spatial evasion of CRISPR systems by housing their genomic DNA within a nucleus-like structure during infection. The nucleus structure is enclosed within a shell made of several phage proteins that is spatially organized with the help of phage-encoded tubulin spindle. This strategy not only provides phage resistance to CRISPR-Cas but also to the restriction-modifications systems37–39. This strategy of forming the nucleus-like barrier has been demonstrated only for jumbo phages, whose genomes are >200 kilobases37,38. Work by Banfield and colleagues published earlier this year revealed the existence of hundreds of such huge phages40. Many of these large phages contain homologs of proteins that make up the phage shell, suggesting that the phage-barrier strategy of evading bacterial defense could be a more prevalent phenomenon40.
In addition to Acrs encoded within prophage regions, bacterial and archaeal genomes also contain accessory regulators in CRISPR-Cas genomic neighborhoods. Bioinformatic analysis of CRISPR-Cas neighborhoods has helped identify proteins that form core components of these pathways and others that supplement the CRISPR interference machinery3,41. With increasing availability of genomes from cultivable and metagenomic samples, the list of proteins enriched in CRISPR genomic neighborhoods is growing. However, functional validation for many of these proteins is still lacking. Most bioinformatically predicted CRISPR-associated accessory proteins are associated with RNA-targeting type III and VI CRISPR systems41.
Enhancing type III CRISPR immunity with second messenger signalling
Type III CRISPR systems include A-F subtypes, with mechanistic understanding coming from studies of III-A and III-B systems. Both of these systems contain Cas10 (known as Csm1 in III-A and Cmr2 in III-B) in their multisubunit effector complexes. In addition to Cas10, the type III-A effector complex (Csm) contains Csm2–5 proteins while the III-B effector complex (Cmr) contains Cmr1, and Cmr3–5 proteins3. At the interference stage of type III CRISPR immunity, the effector complex targets ssRNA generated by transcription. This binding event results in cleavage of the target RNA by Csm3/Cmr4 proteins and activation of Cas1042,43. The Cas10 subunit of the effector complex consists of a histidine-aspartate (HD) nuclease domain and two palm domains that structurally resemble polymerase/cyclase domain where one of the palm domains contains a catalytic GGDD motif (Fig. 3). On activation, Cas10 cleaves the target DNA with its HD nuclease domain and initiates second messenger signaling through conversion of ATP into cyclic oligoadenylates (cOA)42,44,45. The cOA synthesis by Cas10 is catalyzed by the divalent metal-ion dependent GGDD motif of its palm domain. Although only one of the palm domains catalyzes the cOA synthesis, both palm domains are required for ATP binding46. These cOAs further enhance type III CRISPR immunity by activating a trans-acting ribonuclease known as Csm6 in III-A and Csx1 in III-B systems (Fig. 3)47–50. Csm6/Csx1 contains a CARF domain on the N-terminal end and a higher eukaryotic prokaryotic nucleotide binding (HEPN) domain on the C-terminal end. The CARF domains of the two monomers form a highly conserved positively charged cleft that serves as the cOA binding site (Fig. 3)51,52. The CARF domain and the HEPN domain are allosterically coordinated such that upon cOA binding, the HEPN domain adopts a conformation that orients the catalytic histidine residues for RNase activity49,51,52. The Csm6/Csx1 proteins vary in sequence, structure and ligand preference not only between different type III subtypes but also between different orthologs within the same subtype. For example, ToCsm6 and TtCsm6 show strongest activation by cA4 while MtCsm6 and EiCsm6 prefer cA652–54. The CARF domain of ToCsm6 also possesses an autoregulatory ring nuclease function where it sequentially degrades cA4 into linear products. This activity is absent from SisCsx1 attributable to the lack of conservation of the key residues involved in the ring nuclease activity of ToCsm6’s CARF domain53.
Figure 3. cOA signalling in type III CRISPR system.
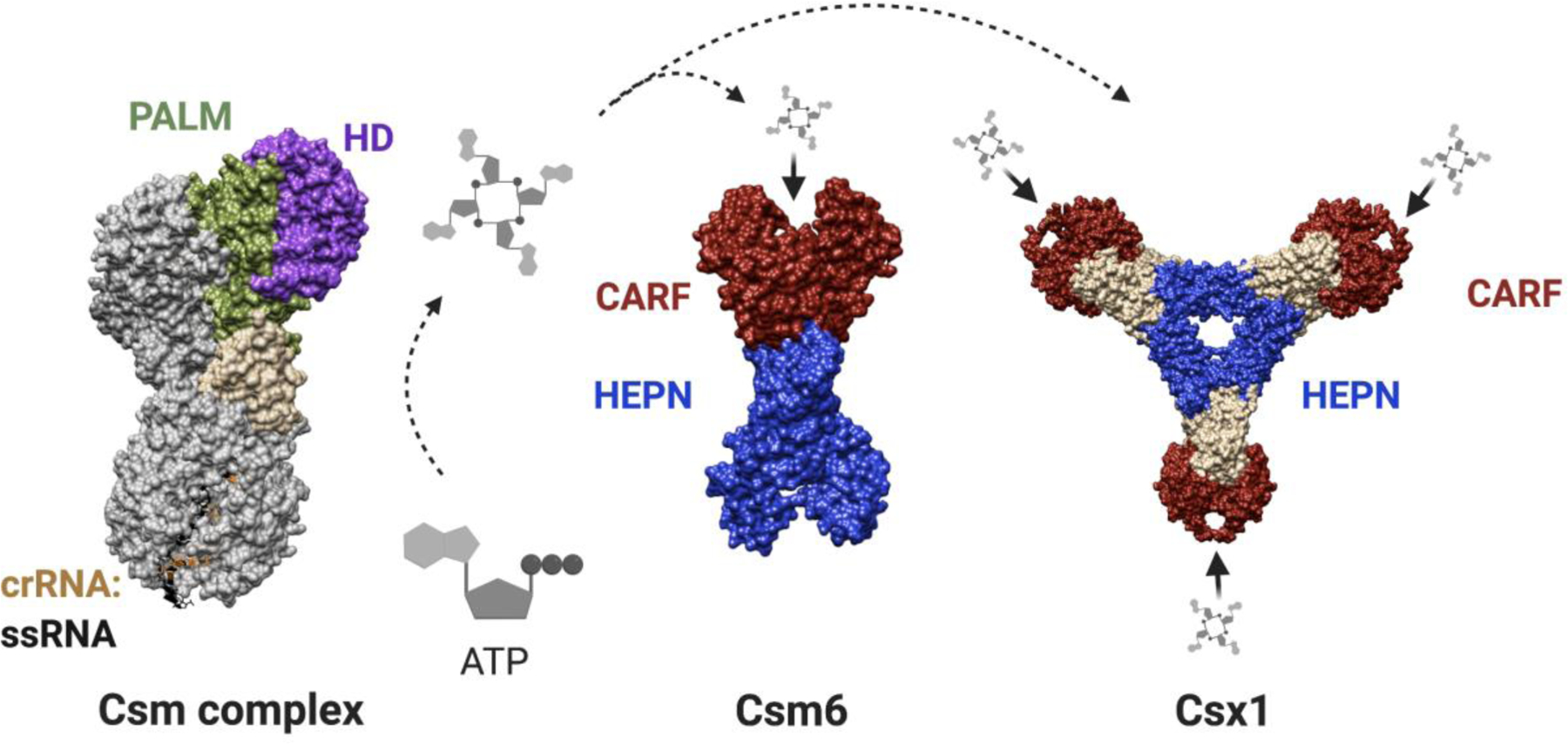
Solid surface representation of the Csm complex (PDB ID: 6IFY) highlighting the Cas 10 domains involved in DNA cleavage (HD domain) and cOA synthesis (Palm domain) followed by a schematic showing the docking of cOA onto Csm6 and Csx1. Surface representation of Csm6 (PDB ID- 5FSH) and Csx1 (PDB ID - 6QZT) with the CARF domain responsible for cOA recognition shown in red and the HEPN RNase domain colored blue. Created with BioRender.com
Surveying the genomic neighborhood of type III CRISPR systems has led to identification of many accessory proteins including Csm6/Csx1. Biochemical characterization of another CARF domain containing accessory protein, Can1 (CRISPR ancillary nuclease 1) showed that it may supplement type III CRISPR immunity by cleaving supercoiled DNA48. Can1 exists as a monomer and contains four domains- two CARF domains, a nuclease domain and another nuclease-like domain called “domain2” consisting of an HTH motif in addition to a segment that partly resembles the core domain of the PD-D/ExK nuclease family. Similar to Csm6/Csx1, the interface between the two CARF domains makes up the cA4 binding pocket. Upon binding of cA4, Can1 undergoes structural rearrangement, activating the metal-dependent non-specific nuclease domain, which then nicks supercoiled DNA. This activity of Can1 could be used to target phage DNA during replication or transcription when the DNA undergoes supercoiling48. Some type III CRISPR-Cas locus also contain homologs of NucC, the effector nuclease of the bacterial defense system, CBASS (cyclic oligonucleotide-based anti-phage signaling system). Upon binding of cA3, NucC forms a homohexameric complex and cleaves dsDNA non-specifically. It is possible that NucC also functions as an accessory nuclease as part of the type III CRISPR pathway in addition to its role in the CBASS defense system55.
Proteins involved in the regulation of CRISPR activity may not necessarily be limited to CRISPR-accessory proteins. To identify such regulators, proteins that co-purify with members of the type III CRISPR system were identified by mass spectrometry. Based on this approach, two components of the degradosome, PNPase and RNase J2, showed evidence of physical association with the type III CRISPR system. Both of these nucleases play a role in efficient clearance of pathogen-derived transcripts, supplementing the nuclease activity of the type III CRISPR components (Fig. 5)56.
Figure 5. Fine tuning type III CRISPR-Cas immunity.
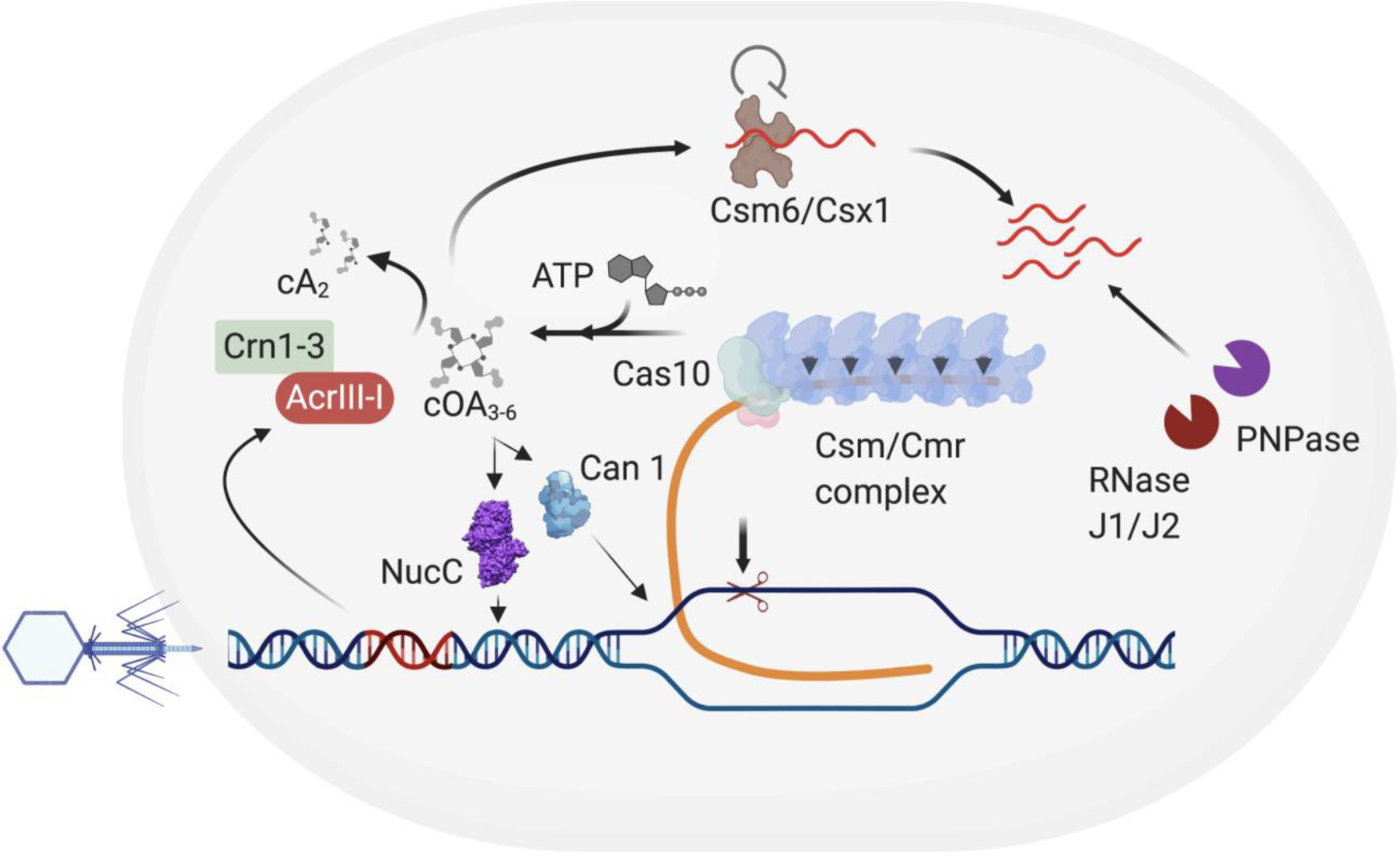
Schematic showing the type III CRISPR mechanism and the role of accessory nucleases in controlling type III immunity. The type III CRISPR-Cas pathway involves targeting of RNA by the Csm/Cmr complex during transcription followed by the activation of Cas10 that results in cleavage of DNA and the production of cOAs. The cOAs released by Cas10 then activates the accessory nuclease Csm6/Csx1 which then leads to non-specific cleavage of RNA. This second messenger pathway can further be supplemented by other accessory RNA-targeting nucleases, RNaseJ1/J2 and PNPase and DNA targeting nucleases including NucC and Can1 that can cleave dsDNA and ssDNA, respectively. The concentration of the cOA molecules can be controlled both by the host ring nucleases (Crn1–3) and MGE-derived ring nuclease (AcrIII-I)
Accessory regulators of type VI CRISPR system
Type VI CRISPR systems are RNA-guided RNA targeting systems that are further subclassified into type VI A-D. These systems contain at least two HEPN domains but vary in sequence and domain organization of their loci3. Type VI-B are unique in that they contain accessory transmembrane (TM) proteins (Csx27 and Csx28) in their locus. One of these TM proteins, Csx28 found in type VI B1 locus, also contains a HEPN domain. To explore their effect on Cas13b mediated immunity, Cas13b’s interference activity was assayed in the presence of Csx27 or Csx28 where they were found to inhibit and enhance Cas13b activity, respectively. Interestingly, irrespective of them containing transmembrane domains, neither of the accessory proteins showed membrane localization in E.coli57. The prevailing hypothesis for the Csx28 mechanism is that it acts as an additional transacting RNase57 (Fig. 4b, Left). Type VI-D also contains an accessory protein that enhances Cas13d activity. One of their orthologs found in Ruminococcus Sp, RspWYL1, comprises an N-terminal domain that consists of DNA binding ribbon-helix-helix motif, a WYL domain and an oligomerization C-terminal domain. Although the detailed mechanism is still under investigation, RspWYL1 has been shown to weakly interact with Cas13d and possess high affinity for ssRNA. Two likely hypotheses for its mechanism are that (1) it functions as an RNA sponge due to its high affinity for ssRNA and enhances Cas13d activity by localizing more RNA and (2) Even with weak interaction, RspWYL1 can allosterically enhance Cas13d cleavage activity58,59 (Fig. 4a).
Figure 4. Potential mechanisms of CRISPR-associated accessory proteins.
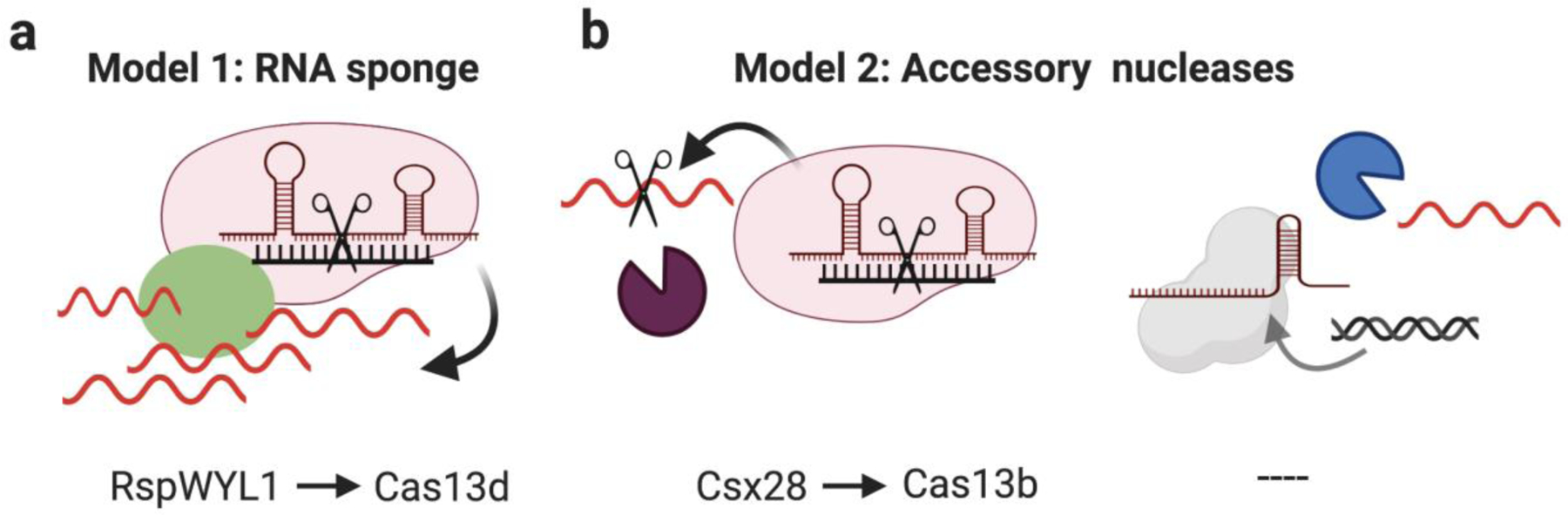
a, Accessory proteins (Green circle) can stimulate RNA-targeting activity of Cas enzymes by increasing the local concentration of RNA and/or by allosterically modulating the Cas enzyme activity. b Additional nucleases could also be deployed to clear residual MGE derived nucleic acids. Potential candidates that support the hypothesis are displayed under their respective models. Created with BioRender.com.
Research on the regulatory role of proteins functionally associated with CRISPR has focused on RNA-targeting CRISPR systems (type III and VI), in part because bacteria containing RNA-targeting CRISPR systems are more likely to require supplementary activity of accessory proteins to attain complete immunity from phages. In contrast to DNA, where a single cleavage event could lead to loss of phage replication, RNA-targeting systems need to target multiple copies of RNA molecules to achieve complete phage immunity. Based on existing evidence, two models can be proposed for how enhanced RNA targeting can be achieved. In model 1, the accessory protein increases the local concentration of RNA around the Cas protein and/or allosterically stimulates the Cas activity (Fig. 4a). In model 2, accessory nucleases are deployed that supplement the RNA targeting activity of Cas systems (Fig. 4b, left). That said, it is possible that DNA-targeting systems might recruit accessory proteins to clear residual phage-derived nucleic acids without their requirement for phage immunity (Fig. 4b, right). In addition to RNA-targeting systems, CARF and WYL domain containing proteins also exist proximal to DNA-targeting type I and V CRISPR systems suggesting that second messenger signaling might play a role in their regulation as well3. Furthermore, several accessory proteins have been computationally identified with predicted functions including helicase, protease and membrane association3,41. Future research on these proteins is likely to reveal unique mechanisms of CRISPR regulation.
Fine tuning the activity of CRISPR regulators
Just as bacteria regulate the expression of their Cas genes, phages also control the expression of their Acrs. Many Acr genes co-occur with genes containing helix-turn-helix (HTH) motifs and are thus called Acr-associated (Aca) genes. These Aca genes transcriptionally repress the expression of Acr genes by binding to the inverted repeat sequences in their operon’s promoter. Recent findings suggest that phages can also encode for Aca-Acr fusions that possess dual CRISPR inhibition and autoregulatory functions60–63. Just like the expression of Cas genes positively correlates with the risk of infection, expression of Acrs might also vary with the expression of Cas proteins.
Type III and VI RNA-targeting CRISPR systems use non-specific RNase activity for complete clearance of MGE. In the type III system, binding of the effector complex to an RNA target leads to the production of cOA molecules that then activates the accessory non-specific RNase, Csm6/Csx1. In type VI, the Cas13 effector itself possesses a non-specific RNase activity that is activated by the target RNA to cut both the target in cis and other RNA in trans64. For both these systems, uncontrolled RNase activity can lead to cell growth arrest which can be detrimental to the host once the infection has been cleared65,66. Bacteria and archaea with type III CRISPR-Cas systems circumvent this with the help of ring nucleases that degrade the cOA molecules required for the activation of Csm6/Csx1. In many cases, the CARF domain of Csm6/Csx1 possesses an autoregulatory ring nuclease activity that inactivates the HEPN domain resulting in loss of Csm6/Csx1’s RNase activity53,67. In addition, bacteria and archaea can also encode for accessory CARF-domain containing ring nucleases within their CRISPR locus68–70 (Fig. 5). In one instance, a fusion of Csx1 and a CRISPR-associated ring nuclease, Crn2 has also been discovered where the two enzymes compete for cOA binding and counteract each other resulting in reduced RNase activity71. Not surprisingly, phages and archaeal viruses have also adopted this strategy of controlling type III CRISPR immunity by encoding their own ring nuclease, AcrIII-I (homolog of Crn2), that degrades cA4 (Fig. 5). Since AcrIII-1 targets the small messenger molecule rather than a specific protein, it is likely that AcrIII-1 has broad spectrum inhibitory activity against type III systems that utilize cA433,68. Despite having a similar non-specific RNase component, such host-encoded control mechanisms are yet to be discovered for type VI CRISPR systems. However, Acrs against type VI-A systems were recently discovered that inhibit both cis and trans-cleavage activity of Cas1327,72. Like ring nucleases, future studies might reveal proteins that specifically inhibit the non-specific trans-cleavage activity of Cas13.
CRISPR Control Engineering and Applications
The emergence of CRISPR-Cas proteins as transformative therapeutic agents and research tools has demanded precise and tuneable control over their activity in both time and space. For example, CRISPR therapeutics deployed for in vivo editing could benefit from control elements that limit off-target editing stemming from sustained high level expression73 or that ensure editing activity is cell-, tissue-, and even organelle-specific. As CRISPR-based gene drives, a technology that allows a genomic change to spread through generations across a population at a higher rate, edge closer to real world deployment, externally controllable off-switches will be critical safeguards. While CRISPR-Cas systems and anti-CRISPR loci employ diverse regulatory mechanisms, these strategies have been shaped by evolutionary pressure to maintain fitness advantages in their natural settings and are not optimized for control of biotechnological and gene editing tools. The high scientific and biomedical value of these proteins has spurred engineering of diverse strategies better suited to enable precise spatiotemporal control over their expression and activity outside of prokaryotic cells (Fig. 6). In some cases, these engineered control schemes are analogous to their natural counterparts, while many employ alternative regulatory mechanisms that have been inspired from outside of CRISPR biology.
Figure 6. Engineered regulation of CRISPR-Cas biogenesis and interference.
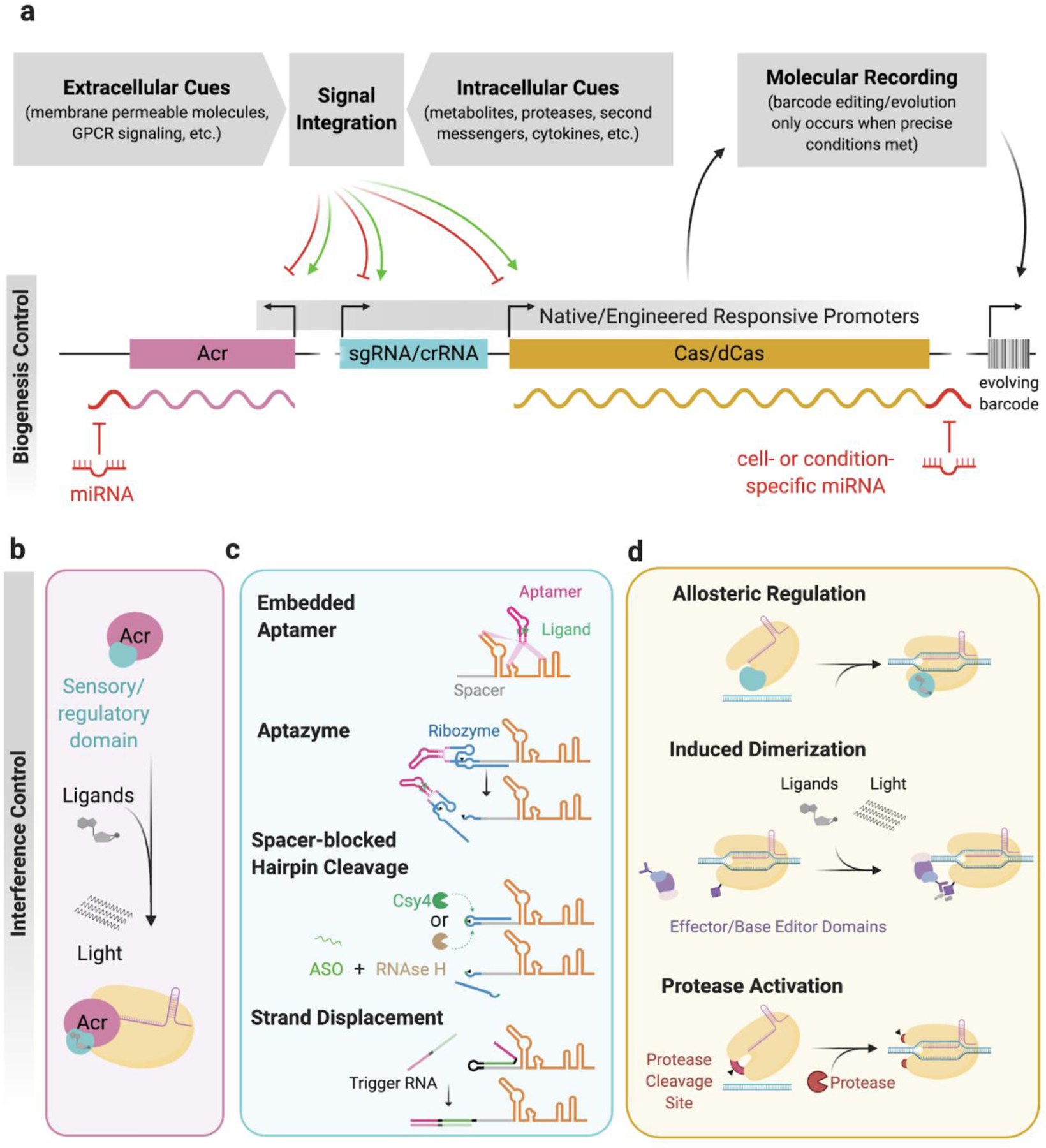
Customized transcriptional (a), post-transcriptional (a and c), and post-translational (b and d) control strategies enable precise spatiotemporal regulation of CRISPR-Cas activity. Engineering CRISPR-Cas elements to automatically and predictably sense and respond to a wide range of extra- and intracellular molecules could usher in a new era of biology, where these genetically-encoded tools illuminate the relationship between the current state of a cell and its past molecular history. a, describes how molecular signals can control expression/biogenesis of crispr components b-d, examples of how CRISPR-Cas systems have been engineered to achieve tight control of the CRISPR interference pathway. Created with BioRender.com
Just as environmental cues regulate the biogenesis of natural CRISPR systems, small molecule-inducible promoters are often employed to control the transcription of Cas proteins and crRNA/sgRNA for routine applications. This robust and simple strategy enables on-demand expression of genetically encoded CRISPR systems and is generally sufficient for most functions, unless the delay between induction and mature complex formation is a barrier to an application’s efficacy. Despite this subtle limitation, encoding Cas components under transcriptional control of endogenous stimulus-responsive promoters facilitates easy coupling of CRISPR expression to existing and engineered regulatory networks and signaling pathways74–76. Predictably linking expression of CRISPR components to spatiotemporally dynamic intra- and extracellular cues will be vital to the development of powerful CRISPR-based molecular recorders, or DNA editors that convert molecular signals into genetic mutations, thereby recording the history of a signal over the lifetime of a cell in manner that is retrievable by DNA sequencing. For example, a CRISPR-based molecular recorder implemented in human cells was used to track exposure to exogenous small molecule inducers doxycycline and isopropyl-b-D-thiogalactoside, and it was also utilized to record the transcriptional activation of an inflammatory response pathway over time75. This powerful automatic control mechanism will be integral to achieving the full potential of CRISPR-based molecular recorders, where the roles that various biomolecules play in development, pathogenesis, and oncogenesis can be probed in longitudinal animal studies with single cell resolution.
While implementing control over CRISPR biogenesis has unique advantages, customized transcriptional regulation of CRISPR components is unfavorable in certain applications due to the long timescale spanning stimulus, transcription, translation, and complex formation. Engineered regulation of the interference stage, or post-transcriptional/-translational control, is better suited for applications where a short timescale between sensing and responding events is required, such as rapid response to infection or other transient stimuli. Robust interference antagonism with Acr proteins has been widely adopted for a range of functions as recently reviewed elsewhere77, where the majority of reports simply demonstrate that various Acr proteins function as potent inhibitors of genome editing in eukaryotic cells. Notable applications include tissue-specific microRNA-mediated repression of AcrIIA4, AcrIIC1, and AcrIIC3 to restrict SpyCas9 and NmeCas9 activity to specific tissues78–80, and the interruption of Cas9-based gene drives by AcrIIA2 and AcrIIA4 in yeast81. Similarly, high-throughput chemical library screening has identified a submicromolar small molecule inhibitor of Cas9 DNA binding82. Given the notoriously long residence time of Cas9 and Cas12a on their DNA targets, the ability of allosteric Acr’s to dislodge nuclease-inactivated Cas proteins (dCas) from their targets offers a distinct kinetic advantage over enzymatic and competitive Acr’s and small molecule inhibitors that are unable to reverse Cas DNA-binding29,31, but applications exploiting this property have not yet been reported.
Akin to allosteric activation of Csm6 and Csx1 in type III CRISPR systems by second messengers, various protein engineering strategies have been applied to install post-translationally regulated on- and off- switches in Cas and Acr proteins83–85. Insertion of ligand-binding domains into allosterically sensitive sites in Cas9 has yielded variants that are conditionally activated in the presence of the small molecules 4-hydroxytamoxifen84 and trimethoprim86, while fusion of ligand-inducible degrons to Cas proteins facilitates rapid conditional degradation or stabilization in response to various small molecules87,88. While these post-translational Cas control strategies rely on addition of external inducers, more advanced strategies have enabled automatic post-translational control of genetically-encoded CRISPR systems. For example, viral protease-activated circularly permuted ProCas9s facilitate rapid altruistic cell death in response to viral infection85. Another strategy commonly utilized to engineer conditional CRISPR activity relies on fusion of small molecule-inducible heterodimerization domains to two halves of a split Cas protein or to a Cas protein and an effector domain76. Chimeric receptors have been engineered to use extracellular cues to drive dimerization and release of both catalytically dead Cas9 (dCas9) and split dCas9 to the nucleus74, and similar systems will likely be integral components of automatically regulated molecular recorders.
Furthermore, conditional crRNAs/sgRNAs that require specific small molecules, endogenous RNA molecules, targeted antisense oligonucleotides, or a sequence-specific endoribonuclease for activation or deactivation have been used to control Cas9 and Cas12a activities89–95. Ligand-regulated guide RNAs have been constructed by prepending a ligand-dependent aptazyme to the 5’ end of a Cas9 sgRNA91 and by embedding small molecule-binding RNA aptamers within various locations of gRNAs, such as the upper stem/tetraloop, nexus, and tracrRNA hairpins of a Cas9 sgRNA89,90. Spacer-blocking hairpin formation by extension of the spacer with a complementary sequence has been shown to be an effective method to reduce Cas activity, and targeting can be restored by cleaving the spacer-complementary strand with a sequence-specific endonuclease like Csy4 or by directing RNAse H activity to the hairpin loop with a targeted anti-sense oligo92. Additionally, custom guide RNAs have been constructed such that binding of strand-displacing, complementary RNA trigger molecules like small RNA or endogenous mRNA induces guide RNA-activating or -deactivating conformational changes93–95, although rules governing predictable redirection of specificity to alternative trigger RNA molecules require further investigation. Extending the ligand specificities of these engineered Cas systems to a wide range of intracellular metabolites and RNAs will facilitate direct interfacing of molecular recorders with host metabolic and transcriptional cues without relying on stimulus responsive promoters or extracellular signals.
Conclusion and future outlook
Research in the past couple of years has significantly advanced our understanding of how bacteria and MGEs regulate CRISPR activity. Non-coding RNAs thought to primarily play a role in crRNA biogenesis were shown to regulate expression of the CRISPR locus suggesting the possibility of finding additional functions of non-coding RNAs in CRISPR immunity17,18. Trans-factors that enhance the function of CRISPR effectors during interference have only been identified for RNA-targeting systems49,50,57,58. It remains to be seen if such regulators might exist for other CRISPR systems. In contrast to DNA-targeting systems, type VI CRISPR systems suffer from non-specific RNase (trans-cleavage) activity that could lead to cellular toxicity and limit its use for RNA editing applications. Mechanistic understanding of the recently discovered type VI Acrs could guide strategies to overcome this limitation72. Although rare, future studies could also reveal Acrs with additional enzymatic activities that modify the crRNA or Cas effector activity. Although Anti-Anti-CRISPR proteins that inhibit Acr expression have been identified, bacterial proteins that directly counteract Acrs, still remain to be found60,61.
The easily programmable, multiplexable, and functionalizable nature of CRISPR has positioned it as a revolutionary component in genetic circuits that perform computation inside living cells. Seminal work has demonstrated the capability of microbial96 and human cells97,98 to perform logical operations using layered CRISPR-based logic gates. An assortment of robust orthogonal Cas:Acr protein pairs will ultimately facilitate construction of more complex logic gates programmed to precisely and automatically control gene expression in response to numerous simultaneous inputs, as recently demonstrated in human and yeast cells99, but there is currently a dearth of Cas:Acr pairs that exhibit low crosstalk for this purpose. While bioinformatic mining and high-throughput characterization of Cas and Acr specificities will continue to expand this toolset, engineering Cas chimeras to alter Acr protein susceptibility is a proven strategy that will undoubtedly yield additional orthogonal sets28–30. Discovery of additional allosteric inhibitors like AcrVA4 that rapidly displace Cas proteins from their targets will be uniquely suited as off-switches in stable gene circuits that rely on reversibility of DNA binding components, such as circuits utilizing CRISPRi/a and CRISPR-mediated epigenome modification. CRISPR-Cas enhancer strategies will also be important for further development of CRISPR-based diagnostics where the implementation of novel nuclease enhancers could drastically improve on the current best limit of detection.
Acknowledgements
We thank Tina Y. Liu for helpful discussions and proofreading the manuscript. The authors acknowledge financial support from the Defense Advanced Research Projects Agency (DARPA) (award HR0011- 17-2-0043 to J.A.D.), the Paul G. Allen Frontiers Group and the National Science Foundation (MCB-1244557 to J.A.D.). B.F.C. is supported by an NIH/NIGMS postdoctoral fellowship (F32 GM131654). G.J.K is supported by an NHMRC Investigator Grant (ELI, 1175568). J.A.D. is an investigator of the Howard Hughes Medical Institute (HHMI), and this study was supported in part by HHMI; J.A.D is also a Paul Allen Distinguished Investigator.
Footnotes
Competing Interests
JAD is a co-founder of Caribou Biosciences, Editas Medicine, Intellia Therapeutics, Scribe Therapeutics, and Mammoth Biosciences. JAD is a scientific advisory board member of Caribou Biosciences, Intellia Therapeutics, eFFECTOR Therapeutics, Scribe Therapeutics, Synthego, Metagenomi, Mammoth Biosciences, and Inari. JAD is a Director at Johnson & Johnson and has sponsored research projects supported by Pfizer and Biogen.
References
- 1.Jansen R, van Embden JDA, Gaastra W & Schouls LM Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol 43, 1565–1575 (2002).This article reports the first use of the acronym “CRISPR”.
- 2.Mojica FJM, Díez-Villaseñor C, García-Martínez J & Soria E Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol 60, 174–182 (2005).First report showing that spacers within CRISPR arrays serve as memory of past infections.
- 3.Makarova KS et al. Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat. Rev. Microbiol 18, 67–83 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vale PF et al. Costs of CRISPR-Cas-mediated resistance in Streptococcus thermophilus. Proc. Biol. Sci 282, 20151270 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Westra ER et al. H-NS-mediated repression of CRISPR-based immunity in Escherichia coli K12 can be relieved by the transcription activator LeuO. Mol. Microbiol 77, 1380–1393 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Medina-Aparicio L et al. The CRISPR/Cas immune system is an operon regulated by LeuO, H-NS, and leucine-responsive regulatory protein in Salmonella enterica serovar Typhi. J. Bacteriol 193, 2396–2407 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu T et al. Coupling transcriptional activation of CRISPR-Cas system and DNA repair genes by Csa3a in Sulfolobus islandicus. Nucleic Acids Res 45, 8978–8992 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He F, Vestergaard G, Peng W, She Q & Peng X CRISPR-Cas type I-A Cascade complex couples viral infection surveillance to host transcriptional regulation in the dependence of Csa3b. Nucleic Acids Res 45, 1902–1913 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patterson AG, Chang JT, Taylor C & Fineran PC Regulation of the Type I-F CRISPR-Cas system by CRP-cAMP and GalM controls spacer acquisition and interference. Nucleic Acids Res 43, 6038–6048 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perez-Rodriguez R et al. Envelope stress is a trigger of CRISPR RNA-mediated DNA silencing in Escherichia coli. Mol. Microbiol 79, 584–599 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patterson AG et al. Quorum Sensing Controls Adaptive Immunity through the Regulation of Multiple CRISPR-Cas Systems. Mol. Cell 64, 1102–1108 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Høyland-Kroghsbo NM et al. Quorum sensing controls the Pseudomonas aeruginosa CRISPR-Cas adaptive immune system. Proc. Natl. Acad. Sci. U. S. A 114, 131–135 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borges AL et al. Bacterial alginate regulators and phage homologs repress CRISPR-Cas immunity. Nat Microbiol 5, 679–687 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Høyland-Kroghsbo NM, Muñoz KA & Bassler BL Temperature, by Controlling Growth Rate, Regulates CRISPR-Cas Activity in Pseudomonas aeruginosa. MBio 9, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahator SD, Jianhe W & Zhang L-H The ECF sigma factor PvdS regulates the type I-F CRISPR-Cas system in Pseudomonas aeruginosa. bioRxiv 2020.01.31.929752 (2020) doi: 10.1101/2020.01.31.929752. [DOI] [Google Scholar]
- 16.Koonin EV & Makarova KS Discovery of Oligonucleotide Signaling Mediated by CRISPR-Associated Polymerases Solves Two Puzzles but Leaves an Enigma. ACS Chem. Biol 13, 309–312 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin P et al. High-throughput screen reveals sRNAs regulating crRNA biogenesis by targeting CRISPR leader to repress Rho termination. Nat. Commun 10, 3728 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Workman RE et al. A natural single-guide RNA repurposes Cas9 to autoregulate CRISPR-Cas expression. bioRxiv 2020.05.21.102756 (2020) doi: 10.1101/2020.05.21.102756. [DOI] [PubMed] [Google Scholar]
- 19.Bondy-Denomy J et al. A Unified Resource for Tracking Anti-CRISPR Names. CRISPR J 1, 304–305 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chowdhury S et al. Structure Reveals Mechanisms of Viral Suppressors that Intercept a CRISPR RNA-Guided Surveillance Complex. Cell 169, 47–57.e11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rollins MF et al. Structure Reveals a Mechanism of CRISPR-RNA-Guided Nuclease Recruitment and Anti-CRISPR Viral Mimicry. Mol. Cell 74, 132–142.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X et al. Structural basis of Cas3 inhibition by the bacteriophage protein AcrF3. Nat. Struct. Mol. Biol 23, 868–870 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Harrington LB et al. A Broad-Spectrum Inhibitor of CRISPR-Cas9. Cell 170, 1224–1233.e15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thavalingam A et al. Inhibition of CRISPR-Cas9 ribonucleoprotein complex assembly by anti-CRISPR AcrIIC2. Nat. Commun 10, 2806 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu L, Yin M, Wang M & Wang Y Phage AcrIIA2 DNA Mimicry: Structural Basis of the CRISPR and Anti-CRISPR Arms Race. Mol. Cell 73, 611–620.e3 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Shin J et al. Disabling Cas9 by an anti-CRISPR DNA mimic. Sci Adv 3, e1701620 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meeske AJ et al. A phage-encoded anti-CRISPR enables complete evasion of type VI-A CRISPR-Cas immunity. Science (2020) doi: 10.1126/science.abb6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fuchsbauer O et al. Cas9 Allosteric Inhibition by the Anti-CRISPR Protein AcrIIA6. Mol. Cell 76, 922–937.e7 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Knott GJ et al. Structural basis for AcrVA4 inhibition of specific CRISPR-Cas12a. Elife 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu Y et al. Diverse Mechanisms of CRISPR-Cas9 Inhibition by Type IIC Anti-CRISPR Proteins. Mol. Cell 74, 296–309.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knott GJ et al. Broad-spectrum enzymatic inhibition of CRISPR-Cas12a. Nat. Struct. Mol. Biol 26, 315–321 (2019).One of the first reports showing that Acrs can possess enzymatic activity
- 32.Dong L et al. An anti-CRISPR protein disables type V Cas12a by acetylation. Nat. Struct. Mol. Biol 26, 308–314 (2019).One of the first reports showing that Acrs can possess enzymatic activity
- 33.Athukoralage JS et al. An anti-CRISPR viral ring nuclease subverts type III CRISPR immunity. Nature 577, 572–575 (2020).This reports the discovery of the first Acr against type III CRISPR system
- 34.Garcia B et al. Anti-CRISPR AcrIIA5 Potently Inhibits All Cas9 Homologs Used for Genome Editing. Cell Rep 29, 1739–1746.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mahendra C et al. Broad-spectrum anti-CRISPR proteins facilitate horizontal gene transfer. Nat Microbiol 5, 620–629 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei Y, Terns RM & Terns MP Cas9 function and host genome sampling in Type II-A CRISPR-Cas adaptation. Genes Dev 29, 356–361 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malone LM et al. A jumbo phage that forms a nucleus-like structure evades CRISPR-Cas DNA targeting but is vulnerable to type III RNA-based immunity. Nat Microbiol 5, 48–55 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Mendoza SD et al. A bacteriophage nucleus-like compartment shields DNA from CRISPR nucleases. Nature 577, 244–248 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chaikeeratisak V et al. Assembly of a nucleus-like structure during viral replication in bacteria. Science 355, 194–197 (2017).The first report showing that bacteriophages can form nucleus-like structure during infection in bacteria
- 40.Al-Shayeb B et al. Clades of huge phages from across Earth’s ecosystems. Nature 578, 425–431 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shmakov SA, Makarova KS, Wolf YI, Severinov KV & Koonin EV Systematic prediction of genes functionally linked to CRISPR-Cas systems by gene neighborhood analysis. Proc. Natl. Acad. Sci. U. S. A 115, E5307–E5316 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Samai P et al. Co-transcriptional DNA and RNA Cleavage during Type III CRISPR-Cas Immunity. Cell 161, 1164–1174 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu TY, Liu J-J, Aditham AJ, Nogales E & Doudna JA Target preference of Type III-A CRISPR-Cas complexes at the transcription bubble. Nat. Commun 10, 3001 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elmore JR et al. Bipartite recognition of target RNAs activates DNA cleavage by the Type III-B CRISPR-Cas system. Genes Dev 30, 447–459 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kazlauskiene M, Tamulaitis G, Kostiuk G, Venclovas Č & Siksnys V Spatiotemporal Control of Type III-A CRISPR-Cas Immunity: Coupling DNA Degradation with the Target RNA Recognition. Mol. Cell 62, 295–306 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Han W et al. A Type III-B Cmr effector complex catalyzes the synthesis of cyclic oligoadenylate second messengers by cooperative substrate binding. Nucleic Acids Res 46, 10319–10330 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mogila I et al. Genetic Dissection of the Type III-A CRISPR-Cas System Csm Complex Reveals Roles of Individual Subunits. Cell Rep 26, 2753–2765.e4 (2019). [DOI] [PubMed] [Google Scholar]
- 48.McMahon SA et al. Structure and mechanism of a Type III CRISPR defence DNA nuclease activated by cyclic oligoadenylate. Nat. Commun 11, 500 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kazlauskiene M, Kostiuk G, Venclovas Č, Tamulaitis G & Siksnys V A cyclic oligonucleotide signaling pathway in type III CRISPR-Cas systems. Science 357, 605–609 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Niewoehner O et al. Type III CRISPR-Cas systems produce cyclic oligoadenylate second messengers. Nature 548, 543–548 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Niewoehner O & Jinek M Structural basis for the endoribonuclease activity of the type III-A CRISPR-associated protein Csm6. RNA 22, 318–329 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Garcia-Doval C et al. Activation and self-inactivation mechanisms of the cyclic oligoadenylate-dependent CRISPR ribonuclease Csm6. Nat. Commun 11, 1596 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Molina R et al. Structure of Csx1-cOA4 complex reveals the basis of RNA decay in Type III-B CRISPR-Cas. Nat. Commun 10, 4302 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grüschow S, Athukoralage JS, Graham S, Hoogeboom T & White MF Cyclic oligoadenylate signalling mediates Mycobacterium tuberculosis CRISPR defence. Nucleic Acids Res 47, 9259–9270 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lau RK et al. Structure and Mechanism of a Cyclic Trinucleotide-Activated Bacterial Endonuclease Mediating Bacteriophage Immunity. Mol. Cell 77, 723–733.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chou-Zheng L & Hatoum-Aslan A A type III-A CRISPR-Cas system employs degradosome nucleases to ensure robust immunity. Elife 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smargon AA et al. Cas13b Is a Type VI-B CRISPR-Associated RNA-Guided RNase Differentially Regulated by Accessory Proteins Csx27 and Csx28. Mol. Cell 65, 618–630.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yan WX et al. Cas13d Is a Compact RNA-Targeting Type VI CRISPR Effector Positively Modulated by a WYL-Domain-Containing Accessory Protein. Mol. Cell 70, 327–339.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang H, Dong C, Li L, Wasney GA & Min J Structural insights into the modulatory role of the accessory protein WYL1 in the Type VI-D CRISPR-Cas system. Nucleic Acids Res 47, 5420–5428 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stanley SY et al. Anti-CRISPR-Associated Proteins Are Crucial Repressors of Anti-CRISPR Transcription. Cell 178, 1452–1464.e13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Birkholz N, Fagerlund RD, Smith LM, Jackson SA & Fineran PC The autoregulator Aca2 mediates anti-CRISPR repression. Nucleic Acids Res 47, 9658–9665 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Watters KE et al. Potent CRISPR-Cas9 inhibitors from Staphylococcus genomes. Proc. Natl. Acad. Sci. U. S. A 117, 6531–6539 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Osuna BA et al. Critical Anti-CRISPR Locus Repression by a Bi-functional Cas9 Inhibitor. Cell Host Microbe (2020) doi: 10.1016/j.chom.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.O’Connell MR Molecular Mechanisms of RNA Targeting by Cas13-containing Type VI CRISPR-Cas Systems. J. Mol. Biol 431, 66–87 (2019). [DOI] [PubMed] [Google Scholar]
- 65.Meeske AJ, Nakandakari-Higa S & Marraffini LA Cas13-induced cellular dormancy prevents the rise of CRISPR-resistant bacteriophage. Nature 570, 241–245 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rostøl JT & Marraffini LA Non-specific degradation of transcripts promotes plasmid clearance during type III-A CRISPR-Cas immunity. Nat Microbiol 4, 656–662 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jia N, Jones R, Yang G, Ouerfelli O & Patel DJ CRISPR-Cas III-A Csm6 CARF Domain Is a Ring Nuclease Triggering Stepwise cA4 Cleavage with ApA>p Formation Terminating RNase Activity. Mol. Cell 75, 944–956.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Athukoralage JS et al. The dynamic interplay of host and viral enzymes in type III CRISPR-mediated cyclic nucleotide signalling. Elife 9, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Athukoralage JS, Rouillon C, Graham S, Grüschow S & White MF Ring nucleases deactivate type III CRISPR ribonucleases by degrading cyclic oligoadenylate. Nature 562, 277–280 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Athukoralage JS et al. Tetramerisation of the CRISPR ring nuclease Crn3/Csx3 facilitates cyclic oligoadenylate cleavage. Elife 9, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Samolygo A, Athukoralage JS, Graham S & White MF Fuse to defuse: a self-limiting ribonuclease-ring nuclease fusion for type III CRISPR defence. Nucleic Acids Res (2020) doi: 10.1093/nar/gkaa298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lin P et al. CRISPR-Cas13 Inhibitors Block RNA Editing in Bacteria and Mammalian Cells. Mol. Cell (2020) doi: 10.1016/j.molcel.2020.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fu Y et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol 31, 822–826 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Baeumler TA, Ahmed AA & Fulga TA Engineering Synthetic Signaling Pathways with Programmable dCas9-Based Chimeric Receptors. Cell Rep 20, 2639–2653 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Perli SD, Cui CH & Lu TK Continuous genetic recording with self-targeting CRISPR-Cas in human cells. Science 353, (2016).Pioneering work demonstrating use of Cas9 as a genetic recorder of molecular events in mammalian cells and mice.
- 76.Kempton HR, Goudy LE, Love KS & Qi LS Multiple Input Sensing and Signal Integration Using a Split Cas12a System. Mol. Cell 78, 184–191.e3 (2020). [DOI] [PubMed] [Google Scholar]
- 77.Marino ND, Pinilla-Redondo R, Csörgő B & Bondy-Denomy J Anti-CRISPR protein applications: natural brakes for CRISPR-Cas technologies. Nat. Methods 17, 471–479 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hoffmann MD et al. Cell-specific CRISPR–Cas9 activation by microRNA-dependent expression of anti-CRISPR proteins. Nucleic Acids Res 47, e75–e75 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hirosawa M, Fujita Y & Saito H Cell-Type-Specific CRISPR Activation with MicroRNA-Responsive AcrllA4 Switch. ACS Synth. Biol 8, 1575–1582 (2019). [DOI] [PubMed] [Google Scholar]
- 80.Lee J et al. Tissue-restricted genome editing in vivo specified by microRNA-repressible anti-CRISPR proteins. RNA 25, 1421–1431 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Basgall EM et al. Gene drive inhibition by the anti-CRISPR proteins AcrIIA2 and AcrIIA4 in Saccharomyces cerevisiae. Microbiology 164, 464–474 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Maji B et al. A High-Throughput Platform to Identify Small-Molecule Inhibitors of CRISPR-Cas9. Cell 177, 1067–1079.e19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Polstein LR & Gersbach CA A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat. Chem. Biol 11, 198–200 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Oakes BL et al. Profiling of engineering hotspots identifies an allosteric CRISPR-Cas9 switch. Nat. Biotechnol 34, 646–651 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Oakes BL et al. CRISPR-Cas9 Circular Permutants as Programmable Scaffolds for Genome Modification. Cell 176, 254–267.e16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Manna D et al. A Singular System with Precise Dosing and Spatiotemporal Control of CRISPR-Cas9. Angew. Chem. Int. Ed 58, 6285–6289 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kleinjan DA, Wardrope C, Nga Sou S & Rosser SJ Drug-tunable multidimensional synthetic gene control using inducible degron-tagged dCas9 effectors. Nat. Commun 8, 1191 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Maji B et al. Multidimensional chemical control of CRISPR-Cas9. Nat. Chem. Biol 13, 9–11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Iwasaki RS, Ozdilek BA, Garst AD, Choudhury A & Batey RT Small molecule regulated sgRNAs enable control of genome editing in E. coli by Cas9. Nat. Commun 11, 1394 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kundert K et al. Controlling CRISPR-Cas9 with ligand-activated and ligand-deactivated sgRNAs. Nat. Commun 10, 2127 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tang W, Hu JH & Liu DR Aptazyme-embedded guide RNAs enable ligand-responsive genome editing and transcriptional activation. Nat. Commun 8, 15939 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ferry QRV, Lyutova R & Fulga TA Rational design of inducible CRISPR guide RNAs for de novo assembly of transcriptional programs. Nat. Commun 8, 14633 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Siu K-H & Chen W Riboregulated toehold-gated gRNA for programmable CRISPR–Cas9 function. Nat. Chem. Biol 15, 217–220 (2018). [DOI] [PubMed] [Google Scholar]
- 94.Oesinghaus L & Simmel FC Switching the activity of Cas12a using guide RNA strand displacement circuits. Nat. Commun 10, 2092 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hanewich-Hollatz MH, Chen Z, Hochrein LM, Huang J & Pierce NA Conditional Guide RNAs: Programmable Conditional Regulation of CRISPR/Cas Function in Bacterial and Mammalian Cells via Dynamic RNA Nanotechnology. ACS Cent Sci 5, 1241–1249 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nielsen AAK & Voigt CA Multi-input CRISPR/C as genetic circuits that interface host regulatory networks. Mol. Syst. Biol 10, 763 (2014).First implementation Cas9-based genetic circuits capable of performing logic operations in mammalian cells
- 97.Kiani S et al. CRISPR transcriptional repression devices and layered circuits in mammalian cells. Nat. Methods 11, 723–726 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nissim L, Perli SD, Fridkin A, Perez-Pinera P & Lu TK Multiplexed and programmable regulation of gene networks with an integrated RNA and CRISPR/Cas toolkit in human cells. Mol. Cell 54, 698–710 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nakamura M et al. Anti-CRISPR-mediated control of gene editing and synthetic circuits in eukaryotic cells. Nat. Commun 10, 194 (2019).Highlights the utility of Acr’s as gene circuit components in eukaryotic cells.
- 100.Guo TW et al. Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex. Cell 171, 414–426.e12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]


