Abstract
Proper Golgi complex function depends on the activity of Arf1, a GTPase whose effectors assemble and transport outgoing vesicles. Phosphatidylinositol 4-phosphate (PI4P) generated at the Golgi by the conserved PI 4-kinase Pik1 (PI4KIIIβ) is also essential for Golgi function, although its precise roles in vesicle formation are less clear. Arf1 has been reported to regulate PI4P production, but whether Pik1 is a direct Arf1 effector is not established. Using a combination of live-cell time-lapse imaging analyses, acute PI4P depletion experiments, and in vitro protein–protein interaction assays on Golgi-mimetic membranes, we present evidence for a model in which Arf1 initiates the final stages of Golgi maturation by tightly controlling PI4P production through direct recruitment of the Pik1-Frq1 PI4-kinase complex. This PI4P serves as a critical signal for AP-1 and secretory vesicle formation, the final events at maturing Golgi compartments. This work therefore establishes the regulatory and temporal context surrounding Golgi PI4P production and its precise roles in Golgi maturation.
INTRODUCTION
Eukaryotes employ intricate networks of trafficking pathways among subcellular compartments to maintain protein and lipid homeostasis. Many of these networks converge at the Golgi complex, where lipids and membrane proteins are sorted and prepared for delivery to the endolysosomal system, to the plasma membrane, or to the endoplasmic reticulum (ER) via membrane-bound vesicles and tubules. Formation of these transport carriers is subject to tight spatial and temporal control, much of which is exerted by Arf GTPases.
Arf GTPases are, in essence, signaling molecules: when GTP-bound, or “active,” Arfs bind Golgi membranes and recruit effector proteins that assemble transport carriers (reviewed in D’Souza-Schorey and Chavrier, 2006). Intrinsic nucleotide exchange by Arfs is quite slow, so guanine nucleotide exchange factors (Arf-GEFs) accelerate GDP-for-GTP exchange to ensure timely, appropriate Arf activation. In humans, four Arf paralogues are activated throughout the Golgi by the Arf-GEFs GBF1, which functions at the early and medial Golgi, and BIG1/2, which function at the late Golgi/trans-Golgi network (TGN) and at endosomes. In budding yeast (Saccharomyces cerevisiae), the redundant Arf paralogues Arf1/2 are activated by three Arf-GEFs: Gea1 (GBF1 homologue) at the early Golgi, Gea2 (GBF1 homologue) at the medial Golgi, and Sec7 (BIG1/2 homologue) at the late Golgi/TGN (Bui et al., 2009).
Specific lipids also regulate transport carrier formation. Like Arf, localized enrichment of certain lipids in the Golgi membrane can contribute to membrane deformation and recruit proteins that form and move transport carriers. One particularly noteworthy Golgi lipid is phosphatidylinositol-4-phosphate (PI4P), which is essential for Golgi function and cell viability (reviewed in Tan and Brill, 2014). In S. cerevisiae, the essential conserved PI 4-kinase (PI4K) Pik1 generates PI4P at the Golgi (Flanagan et al., 1993; Walch-Solimena and Novick, 1999). In humans, the responsibility is shared by the Pik1 homologue PI4KIIIβ, which functions throughout the Golgi and TGN (de Graaf et al., 2004; Weixel et al., 2005), and PI4KIIα, which is concentrated at the TGN and at endosomes (Wang et al., 2003; Minogue et al., 2010) but is found only at the vacuole and plasma membrane in S. cerevisiae (Han et al., 2002). Pik1 and its homologues form heterodimers with frequenin (Frq1 in S. cerevisiae; NCS-1 in humans), which promotes their association with Golgi membranes (Hendricks et al., 1999; Ames et al., 2000; Taverna et al., 2002; Rajebhosale et al., 2003; Strahl et al., 2003, 2005).
Studies in mammalian systems and in S. cerevisiae have shown that PI4P production is tied to Arf activity (Godi et al., 1998; Walch-Solimena and Novick, 1999; Haynes et al., 2005; Sciorra et al., 2005; Saila et al., 2020), but it is unclear whether yeast Pik1 or metazoan PI4KIIIβ is a direct Arf effector (Boura and Nencka, 2015). Mammalian ARF enhances PI4KIIIβ membrane association and catalytic activity on isolated Golgi membranes in vitro (Godi et al., 1998), and it was reported that ARF1 binds to PI4KIIIβ, but this interaction appeared to be weak and was lost in the presence of NCS-1 (Haynes et al., 2005). Therefore, whether Arf binds to PI4KIIIβ directly—and whether frequenin/NCS-1 is involved in the interaction—has not been clearly established.
Furthermore, although the general importance of Golgi PI4P is well-appreciated, its precise roles in Golgi function are less well-defined. Loss of Pik1 activity halts secretion (Hama et al., 1999; Walch-Solimena and Novick, 1999; Audhya et al., 2000; Sciorra et al., 2005), slows Golgi-to-endosome transport (Audhya et al., 2000), and alters Golgi maturation kinetics (Daboussi et al., 2012), but the extent to which PI4P directly participates in these pathways is unclear. Most published pik1 conditional mutant-based analyses of PI4P function have relied on lengthy Pik1 inactivation times, which increases the risk of observing indirect trafficking defects that arise from compromised Golgi trafficking.
Here, using S. cerevisiae as a model system, we show that activated Arf1 directly recruits the Pik1-Frq1 PI4K complex to the late Golgi/TGN and present evidence of an unexpected role for lipid packing defects in facilitating binding of the complex to membranes. We also use an unbiased, high-specificity PI4P probe to establish the timing of PI4P production by Pik1 in the context of Golgi maturation. Consistent with this timing, we confirm previous reports that PI4P is critical for secretory trafficking and for localization of the AP-1 clathrin adaptor. Contrary to other reports, we find that PI4P is not required for localization of the GGA clathrin adaptor Gga2. Our data reveal that the production of PI4P regulates a discrete set of late events at the TGN, serving as the final signal to conclude the Golgi maturation process.
RESULTS
Arf1 directly binds the Pik1-Frq1 complex
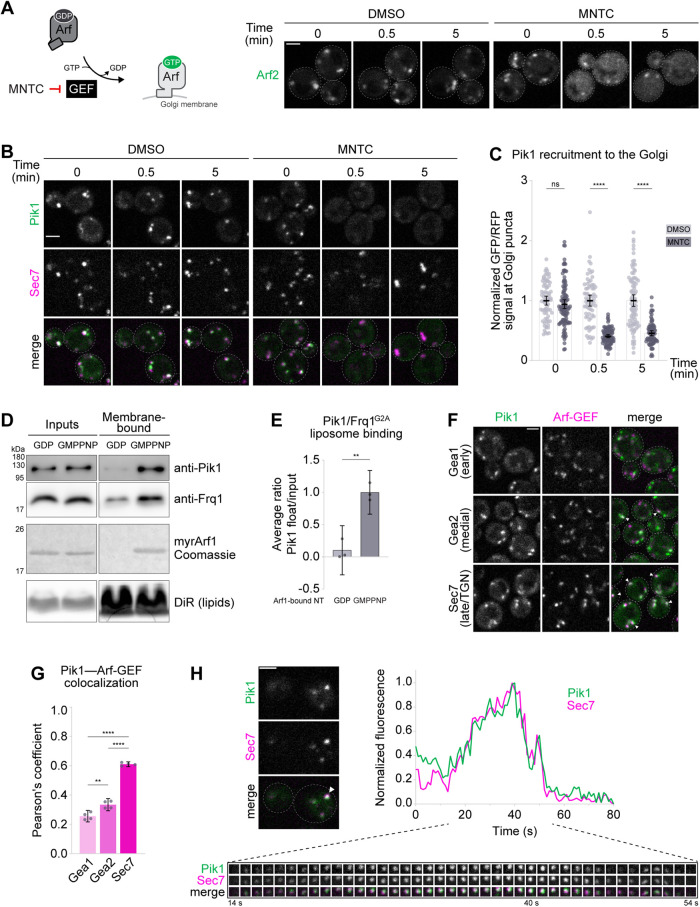
To determine the role of Arf1/2, herein referred to collectively as Arf1, in Pik1 localization in vivo, we first examined Pik1 localization in S. cerevisiae cells lacking active Arf1. To do so, we inhibited Arf1 activation throughout the Golgi with MNTC, a Golgi Arf-GEF inhibitor (Lee et al., 2014; Thomas and Fromme, 2016) (Figure 1A), and found that Pik1 became largely mislocalized from the Golgi (labeled by the late Golgi Arf-GEF Sec7) within 30 s of MNTC treatment (Figure 1, B and C). Similarly, we measured an ∼50% reduction in Pik1 Golgi localization in an arf1Δ mutant (Supplemental Figure S1, A and B), which expresses only ∼10% of the total Arf1/2 present in wild-type cells (Stearns et al., 1990). This indicates that robust Pik1 localization is dependent on Arf1 activation.
FIGURE 1:
Arf1 recruits Pik1-Frq1 to the late Golgi/TGN. (A) Left: Schematic of MNTC Arf-GEF inactivation. Right: Live-cell fluorescence microscopy of Arf2-GFP in cells treated with MNTC or with solvent alone (DMSO) for the indicated times. (B) Live-cell fluorescence microscopy of iGFP-Pik1 and Sec7-6xdsRed in cells treated with MNTC or DMSO for the indicated times. (C) Quantification of Pik1 Golgi localization for the experiment shown in B. Bars represent normalized average ratios of GFP/RFP signal measured at individual Golgi compartments; dots show individual intensity ratio measurements. Error bars represent 95% confidence intervals for n ≥ 70 Golgi puncta from ≥45 cells. (D) Liposome floatation assay to assess binding between purified Pik1-Frq1G2A loaded with AMP-PNP, a nonhydrolyzable analogue of ATP, and purified myrArf1 loaded with GDP (inactive) or with GMP-PNP, a nonhydrolyzable analogue of GTP (active), on 100-nm TGN-like liposomes. myrArf1 visualized by Coomassie stain (the brightness and contrast of the image shown was altered to enhance band visibility); lipids in gel visualized by DiR; Pik1 and Frq1G2A visualized by Western blot. “Inputs” are reaction inputs; “Membrane-bound” are lipids and associated proteins following separation from free protein via a sucrose gradient. (E) Quantification of Pik1 recruitment for n = 3 independent myrArf1-binding assays (as shown in D). Dots show individual Membrane-bound/Input Coomassie band intensity ratios; error bars represent 95% confidence intervals. (F) Live-cell fluorescence microscopy of iGFP-Pik1 alongside the three Golgi Arf-GEFs: Gea1-3xmars, Gea2-3xmars, and Sec7-6xdsRed. Arrowheads indicate examples of Pik1-GEF colocalization. (G) Quantification of Pik1-GEF colocalization for experiment shown in F. Bars represent average Pearson correlation coefficients; dots show individual Pearson correlation coefficient values. Error bars represent 95% confidence intervals for n = 4 micrographs (containing a total of ≥92 cells per strain) analyzed. (H) Representative time-lapse imaging data for iGFP-Pik1 and Sec7-6xdsRed at a single Golgi punctum (top left; arrowhead indicates the region of interest). Images taken at 1-s intervals (bottom). Top right: Relative GFP and RFP intensity values in the region of interest over time. Micrograph scale bars measure 2 μm. Single focal planes are shown. ns, not significant; ** p < 0.01, **** p < 0.0001.
There is an established interaction between human PI4KIIIβ and the Rab GTPase Rab11 (Burke et al., 2014), so we also tested whether Ypt31/32, the S. cerevisiae Rab11 homologues, are important for Pik1 localization. It was demonstrated previously that Ypt31/32 activation at the Golgi is abolished in cells carrying a temperature-sensitive allele of TRS130 (trs130Δ33ts), which encodes a subunit of the Ypt31/32 Rab-GEF complex TRAPPII (Sacher et al., 2001; Thomas and Fromme, 2016). In trs130Δ33ts cells, we found only a minor mislocalization of Pik1 that was not detectably exacerbated upon shift to the restrictive temperature (Supplemental Figure S1, C and D), suggesting that Ypt31/32 activation has little to no direct role in Pik1 localization at the Golgi.
Because Pik1 was rapidly mislocalized from the Golgi upon MNTC-induced Arf1 inactivation, we hypothesized that Pik1 might bind Arf1 directly, rather than being recruited to the Golgi by interaction with another Arf1 effector. Previous studies (Godi et al., 1998; Haynes et al., 2005) have proposed that Arf1 may directly recruit PI4KIIIβ, but there are no reports that clearly demonstrate specific, GTP-dependent PI4K-Arf binding. This is likely because it is difficult to purify appreciable quantities of full-length Pik1 or its homologues for stringent in vitro protein–protein interaction studies—we have found that full-length Pik1 is not stably expressed in Escherichia coli, even when coexpressed with Frq1. Furthermore, our attempts to purify endogenous or overexpressed Pik1 from S. cerevisiae yielded very small amounts of pure protein, likely because Pik1 is not particularly abundant (Ho et al., 2018) and strong PIK1 overexpression causes severe trafficking defects (Schorr et al., 2001) that essentially halt cell proliferation.
We therefore employed Pichia pastoris, a methylotrophic yeast frequently used in industrial-scale protein production (Ahmad et al., 2014), to establish a system for expressing and purifying a stable, stoichiometric, enzymatically active S. cerevisiae Pik1-Frq1 complex (Supplemental Figure S1, E and F). We found that expressing Pik1 alone in P. pastoris yielded mostly Pik1 degradation products, but when coexpressed with Frq1, Pik1 remained intact. Additionally, we used a nonmyristoylated Frq1 mutant (Frq1G2A) to avoid potential aggregation and solubility problems introduced by the solvent-exposed myristoyl group (Hendricks et al., 1999; Ames et al., 2000). Importantly, myristoylation of Frq1 is not essential, as the frq1G2A mutant complements a frq1Δ mutant (Huttner et al., 2003).
To directly test whether Arf1 recruits Pik1-Frq1G2A to membranes, we assayed binding between Pik1-Frq1G2A and Arf1, which we prepared in its native myristoylated form (myrArf1), on artificial liposomes that mimic the membrane lipid composition of the late Golgi/TGN (Klemm et al., 2009) but lack PI4P (Supplemental Table S3). Pik1-Frq1G2A exhibited some intrinsic affinity for these liposomes but bound most robustly in the presence of activated, membrane-bound myrArf1 (Figure 1, D and E). This indicates that activated Arf1 binds directly to the Pik1-Frq1G2A complex.
There are reports that mammalian Arf1 and NCS-1 can bind to each other independently of PI4KIIIβ (Haynes et al., 2005, 2006), but we did not observe an interaction between myrArf1 and Frq1, even when Frq1 was bound to a peptide fragment of Pik1 that is necessary for Pik1-Frq1 complex formation (Strahl et al., 2007) (‘Pik1FB’) (Supplemental Figure S1, G and H). These in vitro experiments, together with our in vivo data demonstrating that Pik1 localization at the Golgi depends on Arf activation, establish that the Pik1-Frq1 PI4K complex is a direct Arf1 effector.
Pik1 localizes to the late Golgi/TGN
Arf1 controls trafficking events across the entire Golgi and is activated by distinct Arf-GEFs throughout: Gea1 at the early Golgi, Gea2 at the medial Golgi, and Sec7 at the late Golgi/TGN (Spang et al., 2001; Bui et al., 2009; Gustafson and Fromme, 2017). Pik1 and Frq1, however, are generally found at more mature regions of the Golgi (Walch-Solimena and Novick, 1999; Sciorra et al., 2005; Strahl et al., 2005; Gloor et al., 2010). Indeed, we found that punctate Pik1 colocalized almost exclusively with Sec7 (Figure 1, F and G). Further, in sec7-4ts cells, which suffer impaired Arf1 activation at the late Golgi (Jones et al., 1999; Deitz et al., 2000; McDonold and Fromme, 2014), Pik1 was largely cytosolic at the restrictive temperature. Normal Pik1 localization was restored upon expression of wild-type SEC7 (Supplemental Figure S1I).
We next examined the temporal dynamics of Pik1 localization by measuring relative fluorescence intensities of Pik1 and Sec7 at individual maturing Golgi compartments over time. On average, Pik1 signal peaked simultaneously with that of Sec7 (Figure 1H), consistent with a previous report (Daboussi et al., 2012). We conclude that Sec7-dependent activation of Arf1 recruits Pik1 to the late Golgi/TGN.
Diacylglycerol is important for Arf1 recruitment of Pik1-Frq1 in vitro
We next aimed to understand why the Arf1-Pik1-Frq1 interaction is confined to the late Golgi/TGN, despite the broader localization of Arf1 across the entire Golgi complex. This behavior suggested that another factor, acting in conjunction with Arf1, ensures that Pik1-Frq1 is recruited to the Golgi at the appropriate time. Although modest, the intrinsic affinity of Pik1-Frq1G2A for TGN-like liposomes in vitro (Figure 1D) raised the possibility that some property of the late Golgi/TGN membrane might serve as such a factor. Membrane curvature, packing defects, and electrostatics attributable to the local lipid environment commonly play roles in peripheral membrane protein localization (reviewed in Bigay and Antonny, 2012; Jackson et al., 2016).
To explore this possibility, we altered the composition of our TGN-like liposomes (Figure 2A) and retested the capacity of activated myrArf1 to recruit Pik1-Frq1G2A. We found that myrArf1 did not recruit Pik1-Frq1G2A to liposomes composed of only PC (Figure 2B). This observation suggested that some feature unique to the TGN-like liposomes acted in synergy with myrArf1 to recruit Pik1-Frq1G2A to the membrane.
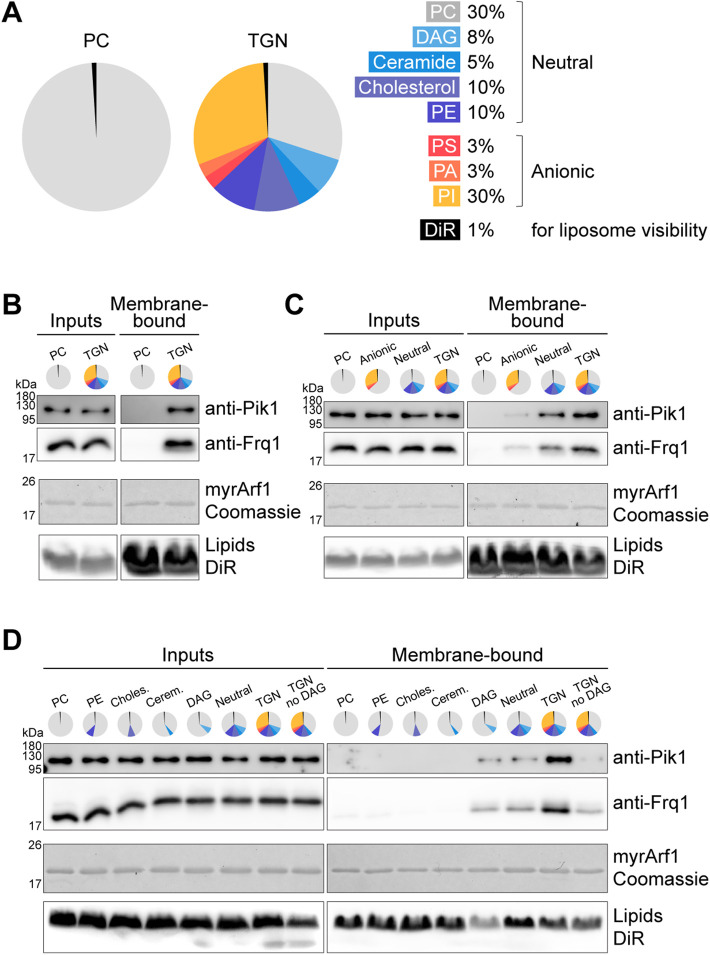
FIGURE 2:
DAG facilitates direct recruitment of Pik1-Frq1G2A to membranes by Arf1 in vitro. (A) Baseline compositions of liposomes used for biochemical assays. PC, phosphatidylcholine; DAG, diacylglycerol; PE, phosphatidylethanolamine; PS, phosphatidylserine; PA, phosphatidic acid; PI, phosphatidylinositol; DiR, 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindotricarbocyanine iodide. See Supplemental Table S3 for details. (B–D) Liposome floatation assay to assess binding between purified GMP-PNP–loaded (activated) myrArf1 and purified AMP-PNP–loaded Pik1-Frq1G2A on 100-nm liposomes of the indicated compositions. myrArf1 visualized by Coomassie stain; lipids in gel visualized by DiR; Pik1 and Frq1G2A visualized by Western blot. “Inputs” are reaction inputs; “Membrane-bound” are lipids and associated proteins following separation from free protein via a sucrose gradient. Concentrations of all individual lipid components are the same as in the TGN-like mixture shown in A, except for PC, which was increased to replace omitted lipids. See Supplemental Table S3 for details.
We then repeated the binding assay with PC liposomes containing neutral TGN lipids (diacylglycerol, DAG; ceramide; cholesterol; and phosphatidylethanolamine, PE) or anionic TGN lipids (phosphatidylinositol, PI; phosphatidylserine, PS; and phosphatidic acid, PA). Intriguingly, far more Pik1-Frq1G2A was recruited to neutral liposomes, despite the presence of its substrate, PI, in the anionic liposomes (Figure 2C). In addition, myrArf1 did not recruit Pik1-Frq1G2A to liposomes containing PC and PI alone (Supplemental Figure S2A). These results indicated that some component(s) of the neutral TGN lipid mix might be important for the ability of myrArf1 to recruit Pik1-Frq1G2A to membranes.
Therefore, we then tested myrArf1 recruitment of Pik1-Frq1G2A to liposomes containing PC supplemented with individual neutral TGN lipids and found that DAG was sufficient to enable Pik1-Frq1G2A recruitment in this context (Figure 2D). Importantly, Pik1-Frq1G2A recruitment was dramatically reduced on TGN-like liposomes lacking DAG, indicating that the Pik1-Frq1 complex has a strong preference for DAG-containing membranes. In cells, a small Golgi pool of cytidine diphosphate–linked diacylglycerol (CDP-DAG) serves as a branch point for several lipid synthesis pathways (Carman and Han, 2009), but we found CDP-DAG to be unimportant for myrArf1-Pik1-Frq1G2A binding in vitro (Supplemental Figure S2B).
In an attempt to assess the potential importance of DAG for Pik1 localization in vivo, we deleted LPP1, which encodes an integral membrane PA phosphatase that produces DAG at the Golgi (Toke et al., 1998; Huh et al., 2003). Localization of Pik1 was unperturbed, and the morphology of the late Golgi/TGN appeared typical (Supplemental Figure S2, C and D), indicating that DAG production by Lpp1 does not directly influence Pik1 localization. We were unable to delete PAH1, which encodes an ER-localized PA phosphatase that produces the bulk of cellular DAG (Han et al., 2006), in haploids of our strain background, but found that Pik1 localized normally in PAH1/pah1Δ heterozygous diploid cells (unpublished data). Additionally, overexpression of the ER-localized DAG kinase DGK1 (Han et al., 2008) in an effort to decrease overall DAG levels in favor of phosphatidic acid production had no impact on Pik1 localization or Golgi morphology (unpublished data). We note that existing in vivo DAG probes do not label the Golgi in S. cerevisiae (Ganesan et al., 2015), so we have been unable to verify Golgi-specific changes in DAG levels in these experiments.
These results suggest that there is a general membrane property that can be conferred by DAG in vitro that promotes Pik1-Frq1 recruitment in vivo. DAG has a small headgroup relative to many other lipid species in the membrane, which enables it to significantly alter membrane organization and facilitate membrane curvature (Goñi and Alonso, 1999). As such, we considered that Pik1-Frq1 may have an affinity for curved membranes. However, when we tested myrArf1 recruitment of Pik1-Frq1G2A to smaller, more highly curved PC liposomes, we did not detect an appreciable interaction (Supplemental Figure S2E). Thus, perhaps some other property of DAG-containing membranes—possibly lipid packing defects induced by DAG (Soulages et al., 1995; Vanni et al., 2014)—makes the surface contours, overall fluidity, or other features of the membrane more conducive to Pik1-Frq1 membrane association.
PI4P is generated at the late Golgi/TGN upstream of secretory vesicle formation and AP-1 localization
We next sought to characterize the distribution of PI4P, the product of Pik1-Frq1 activity, throughout the Golgi. Most in vivo probes previously used to visualize PI4P at the Golgi (e.g., PH domain–based probes derived from FAPP or OSBP proteins) are inherently Golgi-biased because they also bind to Arf1 and thus label only regions of the Golgi enriched in both PI4P and Arf1 (Levine and Munro, 1998, 2002; Godi et al., 2004; Weixel et al., 2005). Although we can reasonably assume, on the basis of our Pik1-Arf1 binding assays and Pik1 localization measurements (Figures 1 and 2), that regions of the Golgi enriched in PI4P are also enriched in Arf1, we wanted to evaluate PI4P distribution using a probe without bias for any additional factors.
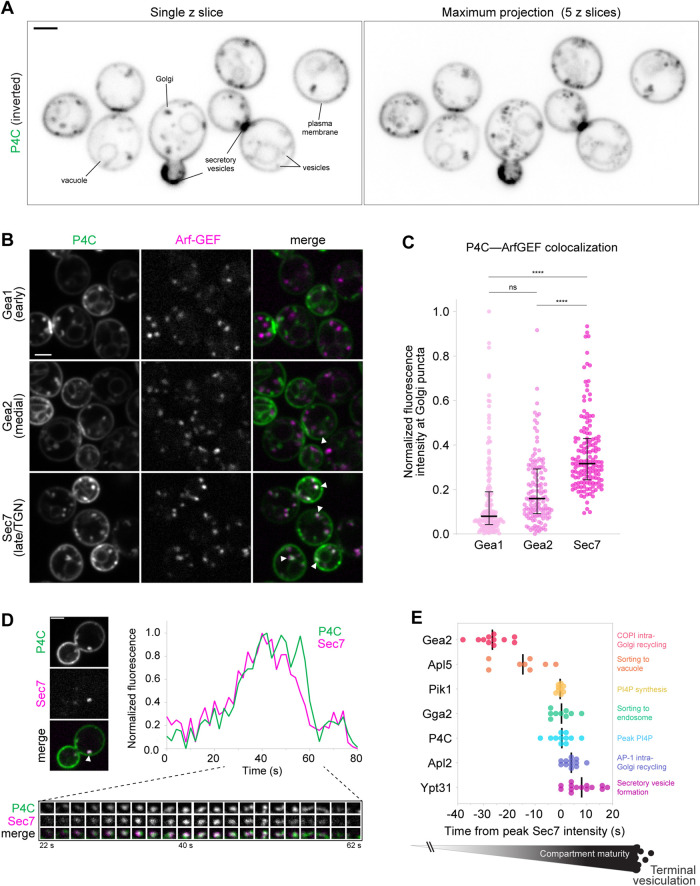
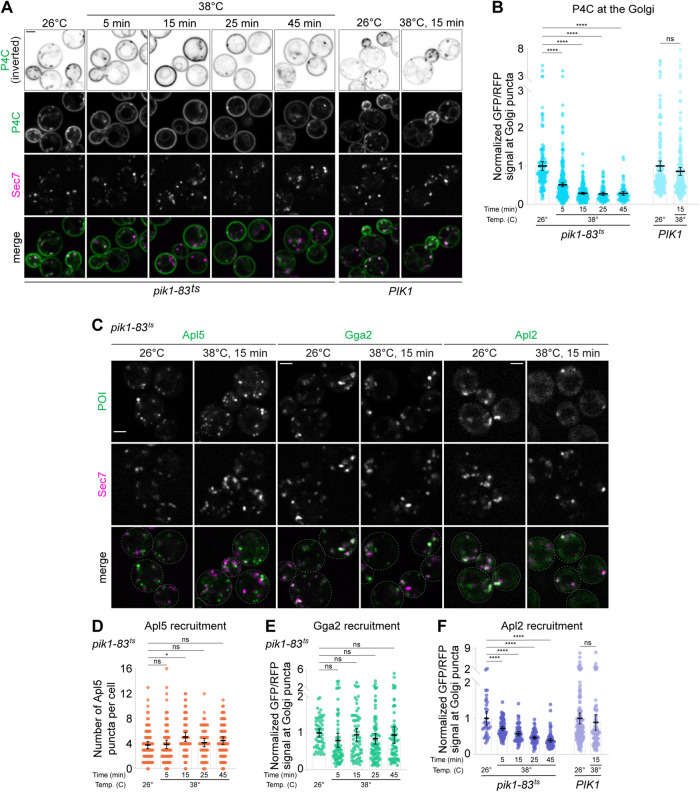
Therefore, to visualize PI4P in cells, we utilized the small PI4P-binding “P4C” domain from the Legionella pneumophilia virulence factor SidC (Luo et al., 2015). Like any probe, P4C has the potential to alter PI4P distribution and availability to effectors, but we found that expression of P4C did not hamper cell growth or alter cell morphology (Figure 3A; Supplemental Figure S3A). GFP-P4C labeled known PI4P pools throughout the cell–-at the Golgi, at the plasma membrane, and on the vacuole membrane (Figure 3A)–-and colocalized with the Rab GTPase Sec4 (Rab8 homologue) on secretory vesicles and at sites of polarized secretion (Supplemental Figure S3B). mScarlet-P4C did not label endosomes, marked by the Rab5 homologue Vps21 (Supplemental Figure S3C), which are known to be enriched in PI3P (Di Paolo and De Camilli, 2006), and purified P4C bound specifically and robustly to PI4P-containing liposomes in vitro (Supplemental Figure S3D) (Luo et al., 2015). Loss of Golgi PI4P production upon Pik1 inactivation in pik1-83ts temperature-sensitive catalytic mutants (Hendricks et al., 1999) resulted in a rapid and near-complete release of P4C from the Golgi (Supplemental Figure S3E), indicating that P4C can detect dynamic changes in cellular PI4P distribution.
FIGURE 3:
PI4P distribution and dynamics in vivo. (A) Live-cell fluorescence microscopy of GFP-P4C distribution. Left: Single focal plane. Right: Maximum projection of five focal planes spaced 0.4 μm apart. (B) Live-cell fluorescence microscopy of GFP-P4C alongside the three Golgi Arf-GEFs: Gea1-3xmars, Gea2-3xmars, and Sec7-6xdsRed. Arrowheads indicate examples of P4C-GEF colocalization. Single focal planes are shown. (C) Quantification of P4C-GEF colocalization for experiment shown in B. Horizonal lines and vertical bars represent median and interquartile ranges, respectively, of GFP/RFP signal measured at individual Golgi compartments; dots show individual intensity ratio measurements (n ≥70 Golgi puncta from ≥35 cells). ns, not significant; **** p < 0.0001. (D) Representative time-lapse imaging data for GFP-P4C and Sec7-6xdsRed at a single Golgi punctum (top left; arrowhead indicates ROI). Images taken at 2-s intervals (bottom). Top right: Relative GFP and RFP intensity values in the region of interest over time. A single focal plane is shown. (E) Summary of time-lapse imaging data for markers of various events at the medial-late Golgi/TGN. Each dot represents peak-to-peak time between maximum intensity of POI and maximum intensity of Sec7 at a single Golgi punctum, where peak Sec7 signal time = 0. Proteins of interest were GFP tagged, and Sec7 was 6xdsRed tagged (except for the Gea2 experiment, in which Gea2 was tagged with mNeonGreen and Sec7 was tagged with mars). Vertical black lines indicate average peak-to-peak times. See Figures 1H and 3D and Supplemental Figure S4 for representative data. Micrograph scale bars measure 2 μm.
We found that P4C intensity at Golgi puncta was reduced by ∼50% in arf1Δ cells (Supplemental Figure S3, F and G), which correlates well with the partial loss of Pik1 from Golgi compartments in arf1Δ cells (Supplemental Figure S1, A and B). In wild-type cells, punctate pools of P4C colocalized extensively with Sec7 and only minimally with Gea2 (Figure 3, B and C). In single-compartment intensity tracking analyses, Golgi P4C levels peaked alongside Sec7 (Figure 3D), a pattern similar to that of the Arf-binding PHOSH1 PI4P probe (Daboussi et al., 2012). In the context of the lifetime of a Golgi compartment, this occurs ∼20–30 s downstream of peak Gea2 localization (Figure 3E; Supplemental Figure S4A). Thus, Pik1 and PI4P are hallmarks of the late Golgi/TGN, and both share little overlap with markers of less mature Golgi compartments.
We next incorporated our analysis of PI4P localization dynamics into the context of the major trafficking events that occur at the late Golgi/TGN during Golgi maturation (Losev et al., 2006; Matsuura-Tokita et al., 2006): AP-1–mediated intra-Golgi trafficking (Day et al., 2018; Casler et al., 2019), AP-3–mediated Golgi-to-vacuole trafficking (Cowles et al., 1997), GGA-mediated Golgi-to-late endosome trafficking (Costaguta et al., 2001), and secretory vesicle formation. To do so, we measured fluorescence intensities of several vesicle biogenesis pathway markers relative to Sec7 at maturing Golgi compartments.
Consistent with recent reports (Day et al., 2018; Tojima et al., 2019; Casler and Glick, 2020), we observed distinct waves of vesicle formation events throughout the Sec7 compartment lifetime (Figure 3E; Supplemental Figure S4, B–E). First, Apl5, a subunit of the AP-3 adaptor complex, peaks as Sec7 first begins to appear at the Golgi (on average, ∼15 s upstream of peak Sec7 levels). Next, Gga2, one of two S. cerevisiae GGA clathrin adaptor paralogues (Zhdankina et al., 2001), peaks concurrently with Sec7, followed by Apl2, a subunit of the AP-1 clathrin adaptor complex (on average, ∼4 s downstream of Sec7). Finally, the machinery for secretory vesicle formation, marked by the Rab GTPase Ypt31, assembles ∼8 s downstream of Sec7 and remains through terminal vesiculation of the compartment.
These analyses position peak PI4P production roughly in the middle of the Sec7 compartment lifetime: upstream of AP-1–dependent intra-Golgi trafficking and secretory vesicle formation, downstream of AP-3–dependent trafficking to the vacuole, and in parallel with GGA-dependent trafficking to the late endosome.
Inhibition of Arf1 activation results in rapid loss of cargo adaptor localization
Studies in several eukaryotic model systems have shown that PI4P has roles in maintenance of overall Golgi morphology, cargo glycosylation, retention and distribution of resident protein populations, secretion, and some aspects of cargo adaptor localization kinetics, particularly at the late Golgi/TGN (reviewed in Tan and Brill, 2014). Our P4C localization dynamics data indicated that PI4P was present throughout the lifetime of the late Golgi/TGN compartment, but we wanted to systematically assess its importance for individual outbound late Golgi/TGN pathways.
Because Arf1 participates in virtually all trafficking events at the Golgi (D’Souza-Schorey and Chavrier, 2006), including the initiation of PI4P production by Pik1-Frq1, we began by first catalogueing the immediate effects of MNTC-induced Arf1 inactivation. Given the near-immediate dissociation of Pik1 from the Golgi upon MNTC treatment (Figure 1, B and C), we hypothesized that other late Golgi/TGN Arf1 effectors would also be rapidly mislocalized upon acute Arf1 inactivation.
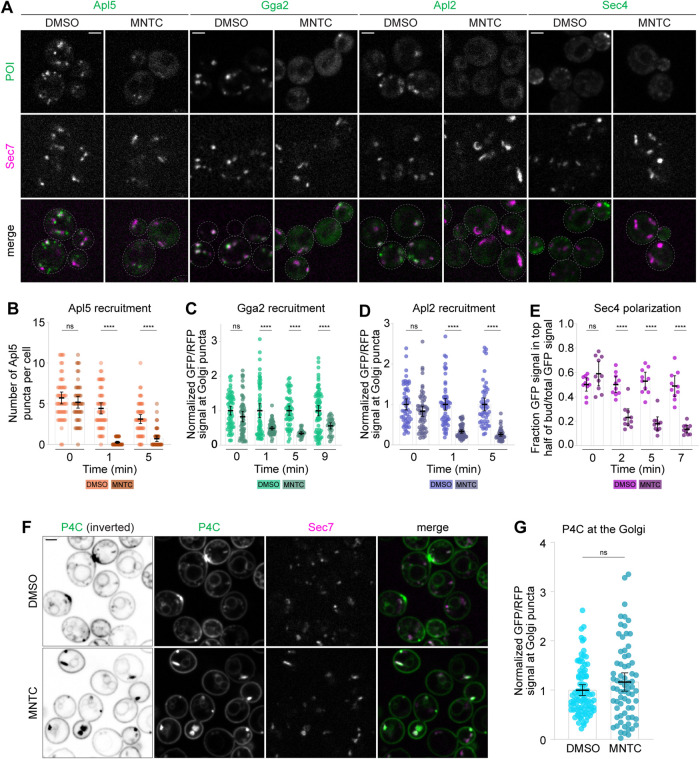
As anticipated, we measured significant mislocalization of the Arf-dependent cargo adaptors Gga2 and AP-1 (Apl2) within 1 min and of AP-3 (Apl5) within 2 min of MNTC treatment (Figure 4, A–D Supplemental Figure S5, A–C). We also observed that the Rab GTPase Sec4, which is normally present on secretory vesicles, became almost completely cytosolic within 2 min of MNTC treatment (Figure 4, A and E; Supplemental Figure S5D), suggesting that secretory vesicle formation was also abolished. This observation is consistent with the role of Arf1 in a GTPase cascade in which Arf1 recruits the TRAPPII Rab-GEF complex, which activates Ypt31/32 (Thomas and Fromme, 2016, 2020). Ypt31/32 then recruit Sec2, the Rab-GEF for Sec4, to secretory vesicles (Walch-Solimena et al., 1997; Ortiz et al., 2002).
FIGURE 4:
Pharmacological inhibition of Arf1 activation results in rapid cargo adaptor mislocalization and loss of new secretory vesicles. (A) Live-cell fluorescence microscopy of GFP-tagged vesicle biogenesis markers (Apl5 for vacuole-bound vesicles, Gga2 for endosome-bound vesicles, Apl2 for intra-Golgi recycling vesicles, Sec4 for secretory vesicles) and Sec7-6xdsRed in cells treated with MNTC or DMSO for 5 min. Full time courses are shown in Supplemental Figure S5. (B) Quantification of Apl5 Golgi localization for experiment shown in A and Supplemental Figure S5A. Dots show individual numbers of Apl5 puncta per cell; bars represent average puncta counts. Error bars represent 95% confidence intervals for n ≥ 47 cells. (C) Quantification of Gga2 Golgi localization for experiment shown in A and Supplemental Figure S5B. Bars represent normalized average ratios of GFP/RFP signal measured at individual Golgi compartments; dots show individual intensity ratio measurements. Error bars represent 95% confidence intervals for n ≥ 25 Golgi puncta from ≥23 cells. (D) Quantification of Apl2 Golgi localization for experiment shown in A and Supplemental Figure S5C. Bars represent normalized average ratios of GFP/RFP signal measured at individual Golgi compartments; dots show individual intensity ratio measurements. Error bars represent 95% confidence intervals for n ≥ 42 Golgi puncta from ≥50 cells. (E) Quantification of Sec4 polarization for experiment shown in A and Supplemental Figure S5D. Dots show GFP polarization values (i.e., the ratio of GFP signal in the top half of the daughter cell to total GFP signal in the mother + daughter cell). Bars represent average polarization values. Error bars represent 95% confidence intervals for n = 10 budding cells (representative of ≥35 budding cells) per condition; for each condition, GFP polarization was quantified for the same 10 cells. (F) Live-cell fluorescence microscopy of GFP-P4C and Sec7-6xdsRed in cells treated with MNTC or DMSO for 5 min. (G) Quantification of P4C Golgi localization for experiment shown in F. Bars represent normalized average ratios of GFP/RFP signal measured at individual Golgi compartments; dots show individual intensity ratio measurements. Error bars represent 95% confidence intervals for n ≥ 70 Golgi puncta from ≥40 cells. Micrograph scale bars measure 2 μm. Single focal planes are shown. ns, not significant; **** p < 0.0001.
In MNTC-treated cells, Golgi compartments quickly become enlarged (Supplemental Figure S6A). This phenotype likely arises from an accumulation of stalled cargo due to loss of Arf1-dependent cargo adaptor function. Strikingly, the medial Golgi Arf-GEF Gea2 colocalized nearly completely with Sec7 (Supplemental Figure S6, B and C), and P4C remained associated with the swollen Golgi compartments (Figure 4, F and G). We attribute the persistence of PI4P under these conditions to the dramatic and rapid block of trafficking and maturation at the Golgi in the absence of Arf1 activation.
We also noticed a puzzling relationship between the late Golgi/TGN and endosomes, marked by Sec7 and Vps21, respectively: ordinarily, Sec7 and Vps21 do not colocalize, but upon a 5-min MNTC treatment, they colocalized extensively (Supplemental Figure S6, D and E). A recent study reported similar accumulation of Vps21 at large puncta upon treatment with brefeldin A (BFA) (Nagano et al., 2019), another chemical Arf-GEF inhibitor (Donaldson et al., 1992; Peyroche et al., 1999), though those puncta reportedly did not contain Sec7. Exactly which Arf-dependent factor(s) are responsible for maintaining the normal endosomal localization of active Vps21 is beyond the scope of this study, but one possibility is that blocking Arf-dependent trafficking out of the TGN results in ectopic activation of Vps21 at the Golgi.
These MNTC experiments emphasize the importance of Arf1 for Golgi function and the high sensitivity of several types of vesicle formation factors—a lipid-modifying enzyme, cargo adaptors, and other GTPases—to Arf1 activation state. They also provide important complementary context for the PI4P depletion experiments presented below.
PI4P is required for secretory trafficking and AP-1 localization, but not for AP-3 or GGA localization
We found that pik1-83ts growth defects cannot be fully suppressed by overexpression of ARF1 or SEC7 (Supplemental Figure S7A), underscoring the importance of PI4P for Golgi function. To determine which major late Golgi/TGN vesicle formation events are PI4P-dependent and which are PI4P-independent, we sought to measure the impact of acute PI4P depletion on these pathways. To do so, we imaged the same vesicle markers used in the MNTC experiments in pik1-83ts mutant cells over 5- to 45-min temperature shift time courses. In pik1-83tscells, more than 50% of P4C signal is lost from the Golgi within 5 min of temperature shift, and more than 70% is lost within 15 min (Figure 5, A and B); this mutant is therefore appropriate for analysis of both immediate and longer-term consequences of Pik1 inactivation. Notably, upon PI4P loss, Sec7 compartments do not swell as dramatically they do upon loss of Arf1 activation, suggesting that at least some cargo is able to exit the Golgi.
FIGURE 5:
PI4P is not required for AP-3 or Gga localization but is required for AP-1 localization. (A) Live-cell fluorescence microscopy of GFP-P4C and Sec7-6xdsRed in wild-type and pik1-83ts cells at the indicated temperatures and times. (B) Quantification of P4C-Sec7 colocalization for experiment shown in A. Bars represent normalized average ratios of GFP/RFP signal measured at individual Golgi compartments; dots show individual intensity ratio measurements. Error bars represent 95% confidence intervals for n ≥ 78 Golgi puncta from ≥47 cells. (C) Live-cell fluorescence microscopy of GFP-tagged vesicle biogenesis markers (Apl5 for vacuole-bound vesicles, Gga2 for endosome-bound vesicles, Apl2 for intra-Golgi recycling vesicles) and Sec7-6xdsRed in pik1-83ts cells at the indicated temperatures for the indicated times. Full time courses are shown in Supplemental Figure S7, B, C, and F. (D) Quantification of Apl5 Golgi localization for experiment shown in C and Supplemental Figure S7B. Dots show individual number of Apl5 puncta per cell; bars represent average puncta counts. Error bars represent 95% confidence intervals for n ≥ 43 cells. (E) Quantification of Gga2 Golgi localization for experiment shown in C and Supplemental Figure S7C. Bars represent normalized average ratios of GFP/RFP signal measured at individual Golgi compartments; dots show individual intensity ratio measurements. Error bars represent 95% confidence intervals for n ≥ 55 Golgi puncta from ≥25 cells. (F) Quantification of Apl2 Golgi localization for experiment shown in C and Supplemental Figure S7F. Bars represent normalized average ratios of GFP/RFP signal measured at individual Golgi compartments; dots show individual intensity ratio measurements. Error bars represent 95% confidence intervals for n ≥ 33 Golgi puncta from ≥46 cells. Micrograph scale bars measure 2 μm. Single focal planes are shown. ns, not significant; * p < 0.05, **** p < 0.0001.
We found that Apl5 localized normally across the entire time course (Figure 5, C and D; Supplemental Figure S7B), indicating that PI4P is not required for AP-3 localization. This is consistent with our observation that little PI4P is present at the Golgi during peak AP-3 localization (Figure 3E).
Gga2 localization was also essentially unperturbed (Figure 5, C and E; Supplemental Figure S7C), in contrast with some previous reports (Wang et al., 2007; Demmel et al., 2008) but in agreement with others (Daboussi et al., 2012; Aoh et al., 2013). We did notice a slight, albeit insignificant, decrease in Gga2 Golgi localization 5 min post–temperature shift, suggesting that PI4P may play a minor role in Gga2 localization, but that other signals likely exert greater influence. Vps21 distribution was normal (Supplemental Figure S7, D and E), in contrast to the unexpected Sec7-Vps21 colocalization we observed upon Arf1 inactivation (Supplemental Figure S6, D and E).
Conversely, we found that trafficking pathways occurring downstream of peak PI4P production were substantially disrupted upon PI4P depletion. We found that significantly less AP-1 was present at the Golgi within 5 min of temperature shift (Figure 5, C and F; Supplemental Figure S7F), in agreement with previous reports of mammalian AP-1 PI4P dependence (Wang et al., 2003; Heldwein et al., 2004). Thus, loss of PI4P causes a near-immediate impairment of AP-1 localization.
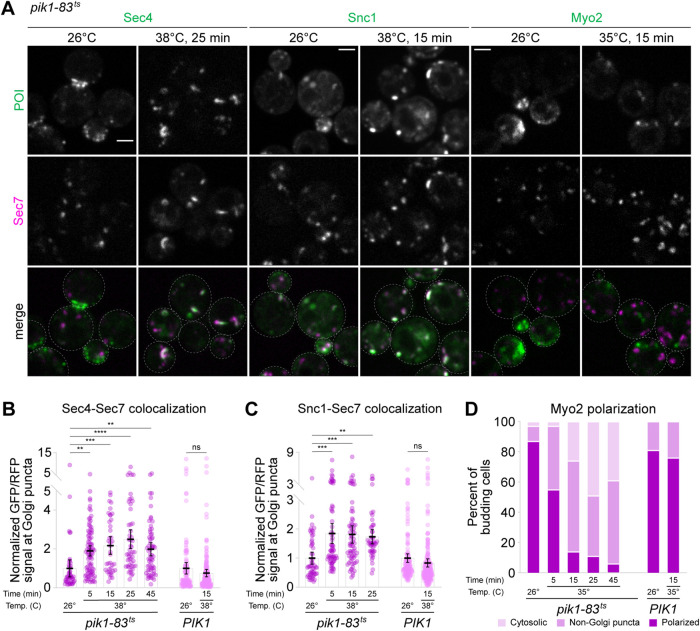
Likewise, secretory vesicle formation was perturbed in the absence of PI4P. Within 15 min of temperature shift, Sec4-labeled secretory vesicles disappeared, and Sec4 instead accumulated at the Golgi (Figure 6, A and B; Supplemental Figure S8A), as did Ypt31 (Supplemental Figure S8, B and C). Given that pik1ts mutants accumulate secretory cargos at restrictive temperatures (Audhya et al., 2000), we infer that our observations reflect decreased secretory vesicle formation upon loss of Pik1 function. In support of this interpretation, we found that Snc1, a secretory vesicle R-SNARE that, when overexpressed, localizes both to the late Golgi/TGN and to exocytic sites at the plasma membrane (Protopopov et al., 1993; Lewis et al., 2000), also accumulated at the Golgi at the restrictive temperature (Figure 6, A and C; Supplemental Figure S8D), in agreement with a previous report (Santiago-Tirado et al., 2011). Finally, Myo2, a myosin V motor protein that transports secretory vesicles from the Golgi to the plasma membrane (Johnston et al., 1991; Rossanese et al., 2001) and is normally most clearly visible at sites of polarized secretion, became less polarized but, unlike other secretory vesicle machinery, did not localize to the Golgi (Figure 6, A and D; Supplemental Figure S8E). PI4P is required for Myo2 transport of secretory vesicles (Santiago-Tirado et al., 2011), so we presume that the observed halt in secretory traffic is due, at least in part, to loss of Myo2 association with nascent secretory vesicles.
FIGURE 6:
PI4P ion results in ectopic activation of Sec4 at the TGN. (A) Live-cell fluorescence microscopy of GFP-Sec4, GFP-Snc1, and Myo2-3xGFP alongside Sec7-6xdsRed in pik1-83ts cells at the indicated temperatures for the indicated times. Full time courses are shown in Supplemental Figure S8, A, D, and E. (B) Quantification of Sec4 Golgi localization for experiment shown in A and Supplemental Figure S8A. Bars represent normalized average ratios of GFP/RFP signal measured at individual Golgi compartments; dots show individual intensity ratio measurements. Error bars represent 95% confidence intervals for n ≥ 39 Golgi puncta from ≥27 cells. (C) Quantification of Snc1 Golgi localization for experiment shown in A and Supplemental Figure S8D. Bars represent normalized average ratios of GFP/RFP signal measured at individual Golgi compartments; dots show individual intensity ratio measurements. Error bars represent 95% confidence intervals for n ≥ 47 Golgi puncta from ≥26 cells. (D) Quantification of Myo2 polarization for experiment shown in A and Supplemental Figure S8E. Cells with completely polarized Myo2 (darkest pink), punctate nonpolarized Myo2, (medium pink), and completely cytosolic Myo2 (lightest pink) were counted. Only budding cells with small and medium daughter cells (n ≥ 35) were analyzed. Micrograph scale bars measure 2 μm. Single focal planes are shown. ns, not significant; ** p < 0.01, *** p <0.001, **** p < 0.0001.
Collectively, these PI4P depletion experiments establish that PI4P production by the Pik1-Frq1 PI4K complex is required for the final stages of Golgi maturation in S. cerevisiae, namely AP-1 vesicle formation and formation of secretory vesicles. PI4P is not required for AP-3 recruitment, which occurs at less mature compartments, or for Gga2 recruitment. These experiments provide new insights and clarification about the physiological functions of PI4P at the late Golgi/TGN.
DISCUSSION
In this study, we definitively established that the Pik1-Frq1 PI4K complex is a direct Arf1 effector. We identified a potential role for membrane packing defects in this interaction, a discovery that may explain why the Pik1-Frq1 complex is constrained to late Golgi/TGN compartments despite the presence of activated Arf1 throughout the Golgi complex. We also systematically assessed the immediate impacts of PI4P depletion on vesicle formation at the late Golgi/TGN and found that PI4P is critical for AP-1 localization and for secretory trafficking but is not important for localization of either AP-3 or GGA. Central to all these pathways is Arf1 (Figure 7).
FIGURE 7:
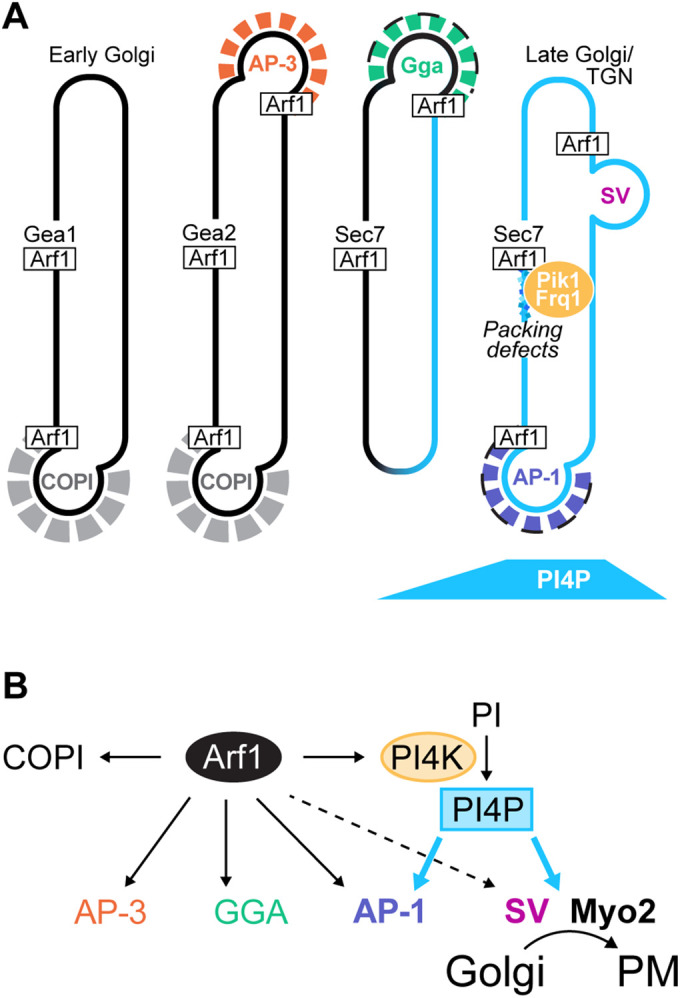
Model: Arf1 regulates PI4P synthesis at the Golgi by directly recruiting the Pik1-Frq1 PI4K complex. (A, B) Model for the role of Arf1 in controlling PI4P synthesis at the late Golgi/TGN by direct PI4K recruitment. Activated Arf1 recruits the Pik1-Frq1 complex, an interaction that is likely dependent on features of the late Golgi/TGN membrane, such as lipid packing defects. Once at the Golgi, Pik1-Frq1 generates PI4P, which is required for AP-1 recruitment and for secretory trafficking (“SV”) but is not required for AP-3 or GGA recruitment.
Recruitment of the Pik1-Frq1 PI4K complex by Arf1
Before this work, it was clear that the Arf1 activation state influences the localization and activity of mammalian PI4KIIIβ: Arf1 activation enhances PI4KIIIβ association with isolated Golgi membranes in vitro, and PI4KIIIβ is mislocalized from the Golgi in cells treated with the Arf-GEF inhibitor BFA (Godi et al., 1998). However, whether PI4KIIIβ and Pik1 are direct Arf1 effectors—that is, whether Arf1 directly binds these PI-4-kinases to recruit them to the Golgi, rather than recruiting them through an intermediary factor—had not yet been determined. Using purified recombinant proteins, we have shown that activated Arf1 recruits the Pik1-Frq1 complex to Golgi-mimetic membranes.
It has been proposed that the late Golgi/TGN Arf-GEF Sec7, rather than its substrate Arf1, binds Pik1 to recruit it to Golgi membranes (Gloor et al., 2010), but our data do not support this idea: the catalytically defective Sec7-4ts mutant protein (Jones et al., 1999; Deitz et al., 2000; McDonold and Fromme, 2014) remains associated with the Golgi upon temperature shift, yet Pik1 becomes mislocalized.
Our data also challenge another model in which the Arf-dependent GGA adaptor proteins recruit Pik1 (Daboussi et al., 2012, 2017). Given the strong Arf1-Pik1-Frq1 interaction in the absence of other endogenous factors (Figure 1D) and previous evidence that Pik1 still localizes to the Golgi in gga1Δgga2Δ mutant cells (Daboussi et al., 2012), we propose that the primary determinant of Pik1 localization is direct recruitment by Arf1. Nonetheless, because Gga2 can bind Pik1 in vitro (Daboussi et al., 2012, 2017), we cannot rule out a potential role for GGA in helping to refine Pik1 localization.
Mammalian ARF1 and NCS-1 were reported to bind each other independently of PI4KIIIβ (Haynes et al., 2005, 2006), but we did not detect a direct interaction between Arf1 and Frq1 in the absence of Pik1. We also found that Pik1 expressed alone in P. pastoris was prone to extensive degradation, whereas coexpression of Pik1 and Frq1 together yielded a stable protein complex, indicating that association with Frq1 stabilizes Pik1. Further, as observed previously (Hendricks et al., 1999), we found that endogenous S. cerevisiae copurifies with Frq1. Given the high conservation of all three proteins, the conserved role of Frq1 in enhancing Pik1 activity and stability (Hendricks et al., 1999; Rajebhosale et al., 2003; Gromada et al., 2005; Strahl et al., 2007), and the ability of human NCS-1 to complement loss of Frq1 in S. cerevisiae (Strahl et al., 2003), we expect that human ARF1 also recruits a PI4KIIIβ/NCS-1 complex, rather than unaccompanied PI4KIIIβ, to the Golgi. However, although human NCS-1 can strongly bind a fragment of Pik1 in vitro (Strahl et al., 2003), PI4KIIIβ lacks an equivalent to one of the two helices in Pik1 involved in its binding to Frq1 (Strahl et al., 2003, 2007; Burke et al., 2014). As such, we cannot rule out the possibility that NCS-1 may be less important for PI4KIIIβ function than Frq1 is for Pik1 function.
In our in vitro liposome recruitment assays, we found that the presence of DAG was both necessary and sufficient to facilitate Pik1-Frq1 binding to Arf1 on TGN-like liposomes. The precise role of DAG is not clear: the best-known DAG-binding structural motif is the C1 domain (Mérida et al., 2010), yet neither Pik1/PI4KIIIβ nor Frq1/NCS-1 possesses a C1 domain (Ames et al., 2000; Bourne et al., 2001; Mérida et al., 2010; Burke et al., 2014). Intriguingly, human PI4KIIIβ is phosphorylated and consequently stimulated by the C1 domain–containing protein kinase D (PKD) (Maeda et al., 2001; Hausser et al., 2005), which itself binds DAG and ARF1 at the Golgi (Baron and Malhotra, 2002; Pusapati et al., 2010). However, there is no known PKD homologue in S. cerevisiae and, although nearly two dozen residues in Pik1 have been identified as phosphosites in various proteomic studies (Soulard et al., 2010; Swaney et al., 2013; MacGilvray et al., 2020), no stimulatory phosphorylation of Pik1 in S. cerevisiae has been reported.
DAG is known to induce lipid packing defects and membrane curvature that contribute to vesicle budding and fission (reviewed in Goñi and Alonso, 1999; Bigay and Antonny, 2012), but we did not identify a role for overall membrane curvature in Arf1-Pik1-Frq1 binding. Others have shown in vitro and in silico that DAG can induce significant perturbations to packing of nearby lipid molecules (Soulages et al., 1995; González-Rubio et al., 2011; Vanni et al., 2014), so we suspect that packing defects introduced by DAG are responsible for generating the biophysical state most conducive to Arf1-dependent docking of Pik1-Frq1 on the membrane. We propose that Pik1-Frq1 has an affinity for membranes with packing defects, which, alongside activated Arf, serve as a mechanism for concentrating Pik1-Frq1—and thus, PI4P production—at vesicle formation sites. This scenario would not be without precedent, as targeting of the S. cerevisiae ArfGAP Gcs1 to nascent vesicles at the late Golgi/TGN is dependent on its packing defect–sensing ALPS (ArfGAP1 lipid packing sensor) motif (Xu et al., 2013) and may be directly linked to DAG metabolism at the Golgi (Yanagisawa et al., 2002).
It is important to note, however, that this hypothesis does not explain the exclusion of Pik1 from earlier Golgi compartments, which are also rich in nascent Arf-dependent vesicles and are thought to contain significant overall packing defects (Bigay and Antonny, 2012). If packing defects alone are not responsible for the confinement of the Pik1-Frq1-Arf1 interaction to the late Golgi/TGN, another factor must contribute. After decades of study, it remains unresolved how several other Arf1 effectors, such as COPI and AP-3, achieve their localization to specific Golgi compartments, and future studies are needed to determine precisely how Pik1-Frq1 is targeted to the late-Golgi/TGN. One possibility is that the enrichment of anionic lipids at the late Golgi/TGN has a greater role in Pik1 localization in vivo than our in vitro assays suggest; perhaps the in vitro DAG requirement simply magnifies the most significant membrane properties required for Arf-Pik1-Frq1 binding.
PI4P marks the beginning of the end
We found that the emergence of PI4P at late Golgi/TGN compartments follows a consistent timeline: downstream of AP-3 vesicle formation, approximately in parallel with GGA vesicle formation, upstream of AP-1 vesicle formation, and further still upstream of secretory vesicle formation. Although Pik1 activity has been shown to be important for normal Golgi maturation (Hama et al., 1999; Walch-Solimena and Novick, 1999; Audhya et al., 2000; Daboussi et al., 2012) and regulates effectors such as the COPI adaptor Vps74 (Tu et al., 2008; Wood et al., 2009) and the lipid flippase Drs2 (Natarajan et al., 2009; Zhou et al., 2013), the immediate contributions of PI4P to vesicle formation at the late Golgi/TGN have not been entirely clear.
To address this, we imaged vesicle biogenesis markers in a pik1ts catalytic mutant (Hendricks et al., 1999), which we found is largely devoid of Golgi PI4P within 5–15 min of shift to a restrictive temperature. We reasoned that the most immediate effects of PI4P loss would be the most direct, so we imaged cells within 5 min of temperature shift and limited our experiments to 45-min time courses to avoid complicating our results by compromising overall Golgi integrity. From these analyses, we conclude that PI4P only appears to play critical, direct roles in AP-1 vesicle and secretory vesicle formation, both of which occur downstream of peak Pik1 recruitment to the Golgi.
Upon Pik1 inactivation, AP-1 recruitment was quickly diminished. Mammalian AP-1 has a known PI4P-binding site (Heldwein et al., 2004) that, alongside functional PI4KIIα, is required for AP-1 localization (Wang et al., 2003; Heldwein et al., 2004). This indicates a conserved role for PI4P in AP-1 function.
We and others (Santiago-Tirado et al., 2011) have found that transport of the secretory cargo Snc1 out of the Golgi is completely blocked and secretion of invertase is dramatically reduced in the absence of PI4P (Hama et al., 1999; Audhya et al., 2000). Nascent secretory vesicles appeared to stall at the Golgi upon PI4P loss, as we found that Sec4, which ordinarily localizes exclusively to secretory vesicles, rapidly accumulated at Sec7 compartments upon PI4P depletion. This was initially surprising because PI4P is thought to serve alongside Ypt31/32 as part of a dual signal for Sec2 recruitment and, thus, Sec4 activation (Mizuno-Yamasaki et al., 2010). However, we also found that Ypt31 accumulated at the Golgi, suggesting that PI4P binding may be dispensable for Sec2 recruitment in the presence of excess Ypt31/32.
The PI4P-binding factor that is responsible for secretory vesicle release remains unidentified, but we did find that Myo2, whose association with secretory vesicles is regulated by PI4P (Santiago-Tirado et al., 2011) and by direct binding to Sec4 (Jin et al., 2011) and to Ypt31/32 (Lipatova et al., 2008), became depolarized upon PI4P loss. Unlike Sec4 and Ypt31, however, Myo2 did not accumulate at the Golgi, which indicates that PI4P—and not Sec4 or Ypt31—is the crucial signal for Myo2 recruitment.
Notably, in myo2-12ts mutants, Sec4 becomes depolarized and accumulates at intracellular puncta (Santiago-Tirado et al., 2011), a phenotype remarkably similar to our observations of stalled secretory vesicle machinery in pik1-83ts mutants. This suggests that failure of Myo2 to associate with nascent secretory vesicles in the absence of PI4P results in blocked secretion. Further work is needed to determine whether Myo2 alone is the essential PI4P secretory effector, but our results provide further insight into the mechanism by which PI4P drives secretion.
Our observation that Gga2 localization is essentially independent of PI4P aligns with two other pik1ts mutant studies (Daboussi et al., 2012; Aoh et al., 2013) but differs from another (Demmel et al., 2008). We did notice a subtle, transient decrease in Gga2 Golgi localization upon initial PI4P depletion, suggesting that PI4P may provide some temporal regulation to S. cerevisiae GGA vesicle formation but that other signals, such as protein cargo, are more important. Indeed, maturation of carboxypeptidase Y (CPY), a GGA-dependent cargo that undergoes proteolytic processing as it traverses the Golgi and the late endosome before being delivered to the vacuole (Mullins and Bonifacino, 2001), is slowed, but not abolished, in pik1-83ts cells (Hama et al., 1999; Audhya et al., 2000). As such, although our data indicate that Gga2 localization is not PI4P-dependent, it is possible that PI4P may help ensure efficient GGA vesicle assembly or cargo sorting in S. cerevisiae. PI4P appears to be more crucial for GGA function in higher eukaryotes, as PI4P-binding sites in the three human GGA proteins have been identified and characterized (Wang et al., 2007), and rapamycin-induced recruitment of the PI 4-phosphatase Sac1 to the TGN in mammalian cell culture decreases Golgi localization of GGA1 and GGA2, but not GGA3, by ∼50% (Szentpetery et al., 2010).
Similarly, we found that GGA localization is not entirely dependent on Arf1. Although initially cytosolic upon Arf1 inactivation, some Gga2 eventually returned to the Golgi. Others have reported a similar GGA “partial sensitivity” to Arf1 inactivation via BFA (Fernandez and Payne, 2006; Hirst et al., 2007; Aoh et al., 2013). Our interpretation of this observation is that cargo binding plays a larger role in GGA localization than does its interaction with Arf1. This aligns with a report that S. cerevisiae Arf1-Gga2 binding is not required for timely CPY maturation (Boman et al., 2002).
In summary, our findings add a critical component to the substantial repertoire of factors under direct Arf1 control—regulation of PI4P synthesis by direct recruitment of the Pik1-Frq1 PI4K complex—and more clearly distinguish PI4P-dependent from PI4P-independent events during maturation of the late Golgi. We favor a model in which Arf1-directed PI4P synthesis primes the Golgi for secretory trafficking by recruiting AP-1 for a final wave of intra-Golgi resident cargo recycling and then recruits proteins needed for secretory vesicle release.
MATERIALS AND METHODS
Yeast strains and plasmids
Plasmids were constructed using conventional molecular cloning techniques and are described in Supplemental Table S1. Genes were driven by endogenous promoters and terminators unless otherwise noted.
Pichia pastoris protein expression strains were generated by homologous recombination of integrating plasmids in the commercially available KM71H strain (Invitrogen cat #C18200). Saccharomyces cerevisiae strains were generated in SEY6210 or SEY6211 by mating or by homologous recombination of PCR-amplified pFa6 cassettes (Longtine et al., 1998) or integrating plasmids and are described in Supplemental Table S2. All S. cerevisiae fluorescent fusion proteins were expressed from endogenous genomic loci as single copies, with the exceptions of GFP-Snc1, which was overexpressed under the TPI1 promoter, and GFP-Vps21 and GFP-Ypt31, which were each expressed as duplicate copies under their endogenous promoters. All S. cerevisiae fluorescent fusion proteins have been previously published, and we verified their functionality by monitoring cell growth and/or protein localization patterns.
Yeast growth assays
Yeast were grown to OD600 ∼2 and then diluted to OD600 = 1 in synthetic complete or selective media. Fivefold serial dilutions of the cells were then spot-plated on the appropriate medium and grown at the indicated temperature for 2 d.
Antibodies
Polyclonal anti-Pik1 and anti-Frq1 antibodies were gifts from Jeremy Thorner (University of California, Berkeley, Berkeley, CA) and were used at 1:1000 and 1:10,000 dilutions, respectively.
Protein purification
GST-Pik1 and the GST-Pik1-Frq1G2A complex were produced in P. pastoris. Cells were grown, shaking, at 30°C in buffered complex glycerol medium (2% peptone, 1% yeast extract, 1.34% yeast nitrogen base, 0.2% histidine, 4 × 10–5% biotin, 100 mM KH2PO4, pH 6.0, 1% glycerol) to OD600 ∼5, and then collected by centrifugation and resuspended in buffered complex methanol medium (2% peptone, 1% yeast extract, 1.34% yeast nitrogen base, 0.2% histidine, 4 × 10–5% biotin, 100 mM KH2PO4, pH 6.0, 1% methanol) to induce protein expression. Importantly, following induction, cells were kept well-aerated by limiting culture volumes to <10% flask capacity and covering flasks with only cheesecloth. Cells were grown, shaking, at 30°C for 24 h. Fresh methanol was added to 1%. After an additional 24 h, cells were harvested by centrifugation and lysed in lysis buffer (25 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM MgCl2, 1 mM CaCl2, 10% glycerol, 1 mM ATP, 0.1% β-octylglucoside, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF], 1× Roche Protease Inhibitor Cocktail) (Roche cOmplete PIC SKU 11836145001) using a freezer mill (SPEX SamplePrep). Insoluble material was collected by centrifugation and discarded. GST-tagged protein was isolated from the remaining lysate by incubation with glutathione resin (G-Biosciences cat #786-310) for 4 h. The resin was washed thrice with a high-salt Triton X-100–containing buffer (25 mM Tris-HCl, pH 7.5, 500 mM NaCl, 1 mM MgCl2, 1 mM CaCl2, 10% glycerol, 1 mM ATP, 0.5% Triton X-100, 1 mM DTT) and then once with lysis buffer. Protein was then eluted from the resin (and separated from the GST tag) by treatment with PreScission protease for ∼16 h.
6xHis-Frq1 was produced in Rosetta 2 cells (Novagen cat #71402). Cells were grown, shaking, at 37°C in terrific broth to OD600 ∼ 3. Protein expression was then induced by the addition of 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) and continued at 18°C for ∼16 h. Cells were lysed by sonication in lysis buffer (40 mM Tris-HCl, pH 8.0, 300 mM NaCl, 10% glycerol, 1 mM CaCl2, 10 mM imidazole, 1 mM PMSF, 10 mM β-mercaptoethanol [β-ME]). Insoluble material was collected by centrifugation and discarded. 6xHis-Frq1 was isolated from the remaining lysate by incubation with Ni-NTA affinity resin (Qiagen cat #30210) for 1 h and then eluted from the resin with a high-imidazole buffer (40 mM Tris-HCl, pH 8.0, 300 mM NaCl, 10% glycerol, 1 mM CaCl2, 250 mM imidazole, 10 mM β-ME). 6xHis-Frq1 was further purified by gel filtration (Superdex 200 Increase 10/300 GL column) in 20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM CaCl2, 1 mM DTT). The 6xHis tag was removed via ∼16-h TEV protease treatment, and untagged Frq1 was collected by gel filtration (Superdex 200 Increase 10/300 GL column) in the same buffer.
The Frq1/Pik1(121-174) complex was prepared by incubating purified Frq1 with a 3× molar excess of the Pik1(121-174) peptide (custom synthesized by Biomatik) at 30°C for 30 min. The complex was separated from free Frq1 and Pik1(121-174) peptide by gel filtration (Superdex 200 Increase 10/300 GL column) in 20 mM Tris-HCl, pH 7.5, 1 mM CaCl2, 150 mM NaCl, 1 mM DTT.
Myristoylated Arf1 (myrArf1) was purified as previously described (Ha et al., 2005): BL21(DE3) cells (Novagen cat #69450) coexpressing Arf1 and Nmt1 were grown, shaking, at 37°C to OD600 ∼ 0.7 in lysogeny broth medium (LB)-Luria. Protein expression was then induced by the addition of 1 mM IPTG and myristate (Sigma-Aldrich cat #M3128) and continued for ∼16 h at 18°C. Cells were lysed by sonication in lysis buffer (20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM MgCl2, 10 mM β-ME, and 1 mM PMSF). Insoluble material was collected by centrifugation and discarded. The remaining lysate was then cleared of major contaminants by incubation with DEAE Sephacel (GE Healthcare cat #17050001) for 1.5 h. The clarified lysate was brought to 3 M NaCl by the addition of solid NaCl and then incubated with TOYOPEARL Phenyl-650M resin (Tosoh Bioscience cat #19818) for 1 h (DTT added to 1 mM) to isolate myrArf1. myrArf1 was then eluted with a low-salt buffer (20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM MgCl2, 1 mM DTT) and further purified by gel filtration (Superdex 200 Increase 10/300 GL column) in 20 mM HEPES, pH 7.4, 100 mM NaCl, 1 mM MgCl2, 1 mM DTT.
GST-CFP-P4C was produced in Rosetta2 cells (Novagen cat #71402). Cells were grown, shaking, at 37°C in LB-Miller to OD600 ∼2. Protein expression was then induced by the addition of 100 µM IPTG and continued at 18°C for ∼16 h. Cells were lysed by sonication in lysis buffer (40 mM Tris-HCl, pH 7.8, 150 mM NaCl, 1 mM PMSF, 10 mM DTT). Insoluble material was collected by centrifugation and discarded. GST-CFP-P4C was isolated from the remaining lysate by incubation with glutathione resin (G-Biosciences cat #786-310) for 2.5 h and then eluted from the resin (and separated from the GST tag) by treatment with PreScission protease for ∼16 h. CFP-P4C was collected by gel filtration (Superdex 200 Increase 10/300 GL column) in 20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM DTT.
Liposome preparation
Dehydrated lipids (Avanti Polar Lipids) were dissolved in chloroform and combined as specified in Supplemental Table S3. Lipids were dried under vacuum for ∼15 min and then rehydrated in HK buffer (20 mM HEPES, pH 7.4, 150 mM KOAc) at 37°C for ∼16 h. Lipids were extruded through 30- or 100-nm filters using a using a miniextruder (Avanti Polar Lipids).
32P kinase assay
Reactions (50 µl) containing 70 μM Pik1-Frq1G2A, 20 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 500 μM CaCl2, 10 μCi 32P-γ-ATP, 50 μM ATP, and 70 μM TGN liposomes were incubated at room temperature for 30 min. Lipids were extracted with 2:1 vol:vol chloroform:methanol and dried under a nitrogen stream. Dried lipids were then resuspended in 2:1 chloroform:methanol, separated by thin-layer chromatography (K2C2O4-treated silica thin-layer chromatography plate; Millipore part #1.05715.0001) with a water/acetic acid/methanol/acetone/chloroform (8:20:15:18:39 vol:vol:vol:vol:vol) solvent, and visualized by phosphorimaging.
Liposome floatation assays
For P4C lipid specificity experiments, 80-µl reactions containing 250 µM liposomes and 3 µg CFP-P4C in HK buffer (+1 mM DTT) were incubated at 30°C for 30 min. Lipids and bound proteins were separated from free protein via a discontinuous sucrose gradient as previously described (Richardson and Fromme, 2015): reactions were mixed with 2.5 M sucrose in HK buffer to a final sucrose concentration of 1 M (“Input” samples) and then transferred to 7 × 20 mm Polycarbonate centrifuge tubes (Beckman Coulter cat #343775). Mixtures were overlaid with a layer of 0.75 M sucrose in HK buffer followed by a final layer of HK buffer and then centrifuged at 390,000 × g/20°C for 20 min. Lipids were collected from the top layer of the gradient (“Membrane-bound” samples). Proteins were visualized by SDS–PAGE/Coomassie stain; lipids were visualized by DiR dye.
Arf1-Pik1-Frq1G2A–binding experiments were carried out as described above, but with the following modifications: first, 3 µg myrArf1 was loaded with GDP or GMP-PNP via EDTA-mediated nucleotide exchange as previously described (Richardson and Fromme, 2015) and incubated with liposomes at 30°C for 30 min before the addition of AMP-PNP–loaded Pik1-Frq1G2A and subsequent incubation at 30°C for 10 min. Second, the sucrose gradient was prepared with HK buffer containing 2 mM MgCl2 to preserve myrArf1-nucelotide binding. myrArf1 was visualized by SDS–PAGE/Coomassie stain; lipids were visualized in gel by DiR dye; Pik1 and Frq1G2A were visualized by Western blot. All liposome floatation assay results shown are representative of two to five independent experiments.
Fluorescence microscopy
Cells were grown to OD600 ∼ 0.5 in synthetic media at 26°C and then imaged at room temperature on glass slides or glass-bottomed dishes using a CSU-X spinning-disk confocal system (Intelligent Imaging Innovations) with a DMI6000 B microscope (Leica Microsystems), 100× 1.46 NA oil immersion objective, and an Evolve 512Delta EMCCD camera (Photometrics). Unless otherwise specified in the figure legends, exposures (≤500 ms, usually 200 ms) were taken at single focal planes. For time-lapse imaging experiments, 200-ms exposures were taken every 1 or 2 s for 4 min. All images were captured with Slidebook 6 software (Intelligent Imaging Innovations).
For temperature shift experiments, cultures were first imaged at 26°C and then transferred to water baths of the indicated temperatures for the indicated times. For all experiments except those involving Myo2-3xGFP, 38°C was used as the restrictive temperature. For those involving Myo2-3xGFP, 35°C was used, as we found that it appeared to aggregate in wild-type cells at 38°C.
For MNTC treatment experiments, cells were treated with 18 µM 6-methyl-5-nitro-2-(trifluoro-methyl)-4H-chromen-4-one (MNTC) (MolPort-000-729-160; dissolved in dimethyl sulfoxide [DMSO]) or with DMSO alone for the indicated times. In most cases, images were taken at 1-min intervals for up to 15 min, but only time points at which we observed significant changes in protein of interest (POI) localization are included in the figures.
Image analysis
Time-lapse imaging analysis was done in Slidebook 6 software (Intelligent Imaging Innovations). Individual Golgi compartments that remained in the same focal plane and general region of the cell throughout their lifetimes (∼50–60 s) were analyzed as regions of interest (ROIs). Green fluorescent protein (GFP) and red fluorescent protein (RFP) fluorescence intensities within ROIs were measured over time; intensity values were normalized to values between 0 and 1 for each channel, with the lowest value set to 0 and the highest value set to 1. “Peak-to-peak time” values denote the time between maximum intensities of GFP and RFP signals at individual ROIs.
All other image analysis and processing was done in ImageJ (FIJI). For Pik1-GEF colocalization experiments, Pearson correlation coefficients were measured using the JACoP plug-in. For Apl5 Arf inactivation and PI4P depletion experiments, Apl5-GFP puncta were defined via Otsu automatic thresholding and then manually counted. For Sec4 Arf inactivation experiments, GFP-Sec4 polarization in budding cells was measured by dividing the GFP signal in the top half of daughter cells by the total GFP signal in mother and daughter cells. For Myo2 PI4P depletion experiments, budding cells with polarized, punctate/nonpolarized, and cytosolic Myo2-3xGFP were manually counted. For all other imaging experiments, GFP/RFP intensity ratios at individual Golgi compartments were calculated by first defining Sec7 (RFP) ROIs via automatic Otsu thresholding and then measuring GFP and RFP fluorescence in each ROI using the Multimeasure tool.
Statistical analysis
All statistical tests were performed using GraphPad Prism 8 software. Significance was determined by unpaired two-tailed t test with Welch’s correction (Figure 1, C and E, Supplemental Figures S1, B and D, S2D, S3G, S6, C and E, and Figure 4, B–E, and G, and wild-type comparisons in Figures 5, B and F, and 6, B and C, and Supplemental Figure S8C) or by one-way analysis of variance with Tukey’s (Figures 1G and 3C) or Dunnett’s (pik1ts comparisons in Figures 5, B and D–F, and 6, B and C, and Supplemental Figures S7E and S8C) tests for multiple comparison.
Supplementary Material
Acknowledgments
We thank the laboratories of S. Emr, C. Stefan, J. Thorner, and A. Bretscher for advice, reagents, and equipment. We thank Fromme lab members for helpful discussions and for reagent preparation (R. Feathers prepared pPICK B; S. Wallace prepared pSW1). We thank S. Halaby for generating preliminary sec7-4 ts data. We thank J. Thorner and S. Emr for careful reading of and constructive comments on the manuscript. This study was supported by funds from National Institutes of Health/National Institute of General Medical Sciences Grants R01GM098621 and R35GM136258 to J.C.F. and by a National Science Foundation Graduate Research Fellowship (grant DGE-1650441) to C.M.H.
Abbreviations used:
- AMP-PNP
adenylyl-imidodiphosphate
- Arf-GEF
Arf guanine nucleotide exchange factor
- ATP
adenosine triphosphate
- BFA
brefeldin A
- DAG
diacylglycerol
- DiR
1,1′-dioctadecyl-3,3,3′,3′-tetramethylindotricarbocyanine iodide
- DMSO
dimethyl sulfoxide
- DTT
dithiothreitol
- ER
endoplasmic reticulum
- GDP
guanosine diphosphate
- GMP-PNP
5′-guanylyl imidodiphosphate
- GST
glutathione S-transferase
- GTP
guanosine-5′-triphosphate
- IPTG
isopropyl β- d-1-thiogalactopyranoside
- LB
lysogeny broth medium
- β-ME
β-mercaptoethanol
- OD 600
optical density at 600 nm
- PA
phosphatidic acid
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PI
phosphatidylinositol
- PI3P
phosphatidylinositol 3-phosphate
- PI4K
PI 4-kinase
- PI4P
phosphatidylinositol 4-phosphate
- PI4,5P 2
phosphatidylinositol 4,5-bisphosphate
- PMSF
phenylmethylsulfonyl fluoride
- POI
protein of interest
- PS
phosphatidylserine
- Rab-GEF
Rab guanine nucleotide exchange factor
- ROI
region of interest
- TGN
trans-Golgi network
- ts
temperature sensitive.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E21-02-0069) on March 31, 2021.
REFERENCES
- Ahmad M, Hirz M, Pichler H, Schwab H (2014). Protein expression in Pichia pastoris: recent achievements and perspectives for heterologous protein production. Appl Microbiol Biotechnol 98, 5301–5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames JB, Hendricks KB, Strahl T, Huttner IG, Hamasaki N, Thorner J (2000). Structure and calcium-binding properties of Frq1, a novel calcium sensor in the yeast Saccharomyces cerevisiae. Biochemistry 39, 12149–12161. [DOI] [PubMed] [Google Scholar]
- Aoh QL, Hung C, Duncan MC (2013). Energy metabolism regulates clathrin adaptors at the trans-Golgi network and endosomes. Mol Biol Cell 24, 832–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audhya A, Foti M, Emr SD (2000). Distinct roles for the yeast phosphatidylinositol 4-kinases, Stt4p and Pik1p, in secretion, cell growth, and organelle membrane dynamics. Mol Biol Cell 11, 2673–2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron CL, Malhotra V (2002). Role of diacylglycerol in PKD recruitment to the TGN and protein transport to the plasma membrane. Science 295, 325–328. [DOI] [PubMed] [Google Scholar]
- Bigay J, Antonny B (2012). Curvature, lipid packing, and electrostatics of membrane organelles: defining cellular territories in determining specificity. Dev Cell 23, 886–895. [DOI] [PubMed] [Google Scholar]
- Boman AL, Salo PD, Hauglund MJ, Strand NL, Rensink SJ, Zhdankina O (2002). ADP-ribosylation factor (ARF) interaction is not sufficient for yeast GGA protein function or localization. Mol Biol Cell 13, 3078–3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boura E, Nencka R (2015). Phosphatidylinositol 4-kinases: function, structure, and inhibition. Exp Cell Res 337, 136–145. [DOI] [PubMed] [Google Scholar]
- Bourne Y, Dannenberg J, Pollmann V, Marchot P, Pongs O (2001). Immunocytochemical localization and crystal structure of human frequenin (Neuronal Calcium Sensor 1). J Biol Chem 276, 11949–11955. [DOI] [PubMed] [Google Scholar]
- Bui QT, Golinelli-Cohen M-P, Jackson CL (2009). Large Arf1 guanine nucleotide exchange factors: evolution, domain structure, and roles in membrane trafficking and human disease. Mol Genet Genomics 282, 329–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke JE, Inglis AJ, Perisic O, Masson GR, McLaughlin SH, Rutaganira F, Shokat KM, Williams RL (2014). Structures of PI4KIIIβ complexes show simultaneous recruitment of Rab11 and its effectors. Science 344, 1035–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman GM, Han GS (2009). Regulation of phospholipid synthesis in yeast. Annu Rev Biochem 50, 69–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casler JC, Glick BS (2020). A microscopy-based kinetic analysis of yeast vacuolar protein sorting. eLife 9, e56844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casler JC, Papanikou E, Barrero JJ, Glick BS (2019). Maturation-driven transport and AP-1–dependent recycling of a secretory cargo in the Golgi. J Cell Biol 218, 1582–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costaguta G, Stefan CJ, Bensen ES, Emr SD, Payne GS (2001). Yeast Gga coat proteins function with clathrin in Golgi to endosome transport. Mol Biol Cell 12, 1885–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles CR, Odorizzi G, Payne GS, Emr SD (1997). The AP-3 adaptor complex is essential for cargo-selective transport to the yeast vacuole. Cell 91, 109–118. [DOI] [PubMed] [Google Scholar]
- Daboussi L, Costaguta G, Ghukasyan R, Payne GS (2017). Conserved role for Gga proteins in phosphatidylinositol 4-kinase localization to the trans-Golgi network. Proc Natl Acad Sci USA 114, 3433–3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daboussi L, Costaguta G, Payne GS (2012). Phosphoinositide-mediated clathrin adaptor progression at the trans-Golgi network. Nat Cell Biol 14, 239–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day KJ, Casler JC, Glick BS (2018). Budding yeast has a minimal endomembrane system. Dev Cell 44, 56–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaf P, Zwart WT, van Dijken RAJ, Deneka M, Schulz TKF, Geijsen N, Coffer PJ, Gadella BM, Verkleij AJ, van der Sluijs P, et al. (2004). Phosphatidylinositol 4-kinaseβ is critical for functional association of rab11 with the Golgi complex. Mol Biol Cell 15, 2038–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deitz SB, Hambourg A, Képès F, Franzusoff A (2000). Sec7p directs the transitions required for yeast golgi biogenesis. Traffic 1, 172–183. [DOI] [PubMed] [Google Scholar]
- Demmel L, Gravert M, Ercan E, Habermann B, Müller-Reichert T, Kukhtina V, Haucke V, Baust T, Sohrmann M, Kalaidzidis Y, et al. (2008). The clathrin adaptor Gga2p is a phosphatidylinositol 4-phosphate effector at the Golgi exit. Mol Biol Cell 19, 1991–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paolo G, De Camilli P (2006). Phosphoinositides in cell regulation and membrane dynamics. Nat Rev 443, 651–657. [DOI] [PubMed] [Google Scholar]
- Donaldson JG, Cassel D, Kahn RA, Klausner RD (1992). ADP-ribosylation factor, a small GTP-binding protein, is required for binding of the coatomer protein beta-COP to Golgi membranes. Proc Natl Acad Sci USA 89, 6408–6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza-Schorey C, Chavrier P (2006). ARF proteins: roles in membrane traffic and beyond. Nat Rev Mol Cell Biol 7, 347–358. [DOI] [PubMed] [Google Scholar]
- Fernandez GE, Payne GS (2006). Laa1p, a conserved AP-1 accessory protein important for AP-1 localization in yeast. Mol Biol Cell 17, 3304–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan CA, Schnieders EA, Emerick AW, Kunisawa R, Admon A, Thorner J (1993). Phosphatidylinositol 4-kinase: gene structure and requirement for yeast cell viability. Science 262, 1444–1448. [DOI] [PubMed] [Google Scholar]
- Ganesan S, Shabits BN, Zaremberg V (2015). Tracking diacylglycerol and phosphatidic acid pools in budding yeast. Lipid Insights 8, 75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloor Y, Schöne M, Habermann B, Ercan E, Beck M, Weselek G, Müller-Reichert T, Walch-Solimena C (2010). Interaction between Sec7p and Pik1p: the first clue for the regulation of a coincidence detection signal. Eur J Cell Biol 89, 575–583. [DOI] [PubMed] [Google Scholar]
- Godi A, Di Campli A, Konstantakopoulos A, Di Tullio G, Alessi DR, Kular GS, Daniele T, Marra P, Lucocq JM, De Matteis MA (2004). FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4)P. Nat Cell Biol 6, 393–404. [DOI] [PubMed] [Google Scholar]
- Godi A, Pertile P, Meyers R, Marra P, Di Tullio G, Iurisci C, Luini A, Corda D, De Matteis MA (1998). ARF mediates recruitment of PtdIns-4-OH kinase-β and stimulates synthesis of PtdIns(4,5)P2 on the Golgi complex. Nat Cell Biol 1, 280–287. [DOI] [PubMed] [Google Scholar]
- Goñi FM, Alonso A (1999). Structure and functional properties of diacylglycerols in membranes. Prog Lipid Res 38, 1–48. [DOI] [PubMed] [Google Scholar]
- González-Rubio P, Gautier R, Etchebest C, Fuchs PFJ (2011). Amphipathic-Lipid-Packing-Sensor interactions with lipids assessed by atomistic molecular dynamics. Biochim Biophys Acta 1808, 2119–2127. [DOI] [PubMed] [Google Scholar]
- Gromada JC, Bark C, Smidt K, Efanov AM, Janson J, Mandic SA, Webb D-L, Zhang W, Meister B, Jeromin A, et al. (2005). Neuronal calcium sensor-1 potentiates glucose-dependent exocytosis in pancreatic β cells through activation of phosphatidylinositol 4-kinase β. Proc Natl Acad Sci USA 102, 10303–10308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson MA, Fromme JC (2017). Regulation of Arf activation occurs via distinct mechanisms at early and late Golgi compartments. Mol Biol Cell 28, 3660–3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha VL, Thomas GMH, Stauffer S, Randazzo PA (2005). Preparation of myristoylated Arf1 and Arf6. Methods Enzymol 404, 164–174. [DOI] [PubMed] [Google Scholar]
- Hama H, Schnieders EA, Thorner J, Takemoto JY, DeWald DB (1999). Direct involvement of phosphatidylinositol 4-phosphate in secretion in the yeast Saccharomyces cerevisiae. J Biol Chem 274, 34294–34300. [DOI] [PubMed] [Google Scholar]
- Han G-S, Audhya A, Markley DJ, Emr SD, Carman GM (2002). The Saccharomyces cerevisiae LSB6 gene encodes phosphatidylinositol 4-kinase activity. J Biol Chem 277, 47709–47718. [DOI] [PubMed] [Google Scholar]
- Han G-S, O’Hara L, Siniossoglou S, Carman GM (2008). Characterization of the yeast DGK1-encoded CTP-dependent diacylglycerol kinase. J Biol Chem 283, 20443–20453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G-S, Wu W-I, Carman GM (2006). The Saccharomyces cerevisiae lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J Biol Chem 281, 9210–9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausser A, Märtens S, Link G, Pfizenmaier K (2005). Protein kinase D regulates vesicular transport by phosphorylation and activation of phosphatidylinositol-4 kinase III β at the Golgi complex. Nat Cell Biol 7, 880–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes LP, Fitzgerald DJ, Wareing B, O’Callaghan DW, Morgan A, Burgoyne RD (2006). Analysis of the interacting partners of the neuronal calcium-binding proteins L-CaBP1, hippocalcin, NCS-1 and neurocalcin δ. Proteomics 6, 1822–1832. [DOI] [PubMed] [Google Scholar]
- Haynes LP, Thomas GMH, Burgoyne RD (2005). Interaction of neuronal calcium sensor-1 and ADP-ribosylation factor 1 allows bidirectional control of phosphatidylinositol 4-kinase β and trans-Golgi network-plasma membrane traffic. J Biol Chem 280, 6047–6054. [DOI] [PubMed] [Google Scholar]
- Heldwein EE, Macia E, Wang J, Yin HL, Kirchhausen T, Harrison SC (2004). Crystal structure of the clathrin adaptor protein 1 core. Proc Natl Acad Sci USA 101, 14108–14113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks KB, Wang BQ, Schnieders EA, Thorner J (1999). Yeast homologue of neuronal frequenin is a regulator of phosphatidylinositol-4-OH kinase. Nat Cell Biol 1, 234–241. [DOI] [PubMed] [Google Scholar]
- Hirst J, Seaman MNJ, Buschow SI, Robinson MS (2007). The role of cargo proteins in GGA recruitment. Traffic 8, 594–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho B, Baryshnikova A, Brown GW (2018). Unification of protein abundance datasets yields a quantitative Saccharomyces cerevisiae proteome. Cell Syst 6, 192–205. [DOI] [PubMed] [Google Scholar]
- Huh W-K, Falvo JV, Gerke LC, Carrol AS, Howson RW, Weissman JS, O’Shea EK (2003). Global analysis of protein localization in budding yeast. Nature 425, 686–691. [DOI] [PubMed] [Google Scholar]
- Huttner IG, Strahl T, Osawa M, King DS, Ames JB, Thorner J (2003). Molecular interactions of yeast frequenin (Frq1) with the phosphatidylinositol 4-kinase isoform, Pik1. J Biol Chem 278, 4862–4874. [DOI] [PubMed] [Google Scholar]
- Jackson CL, Walch L, Verbavatz J-M (2016). Lipids and their trafficking: an integral part of cellular organization. Dev Cell 39, 139–153. [DOI] [PubMed] [Google Scholar]
- Jin Y, Sultana A, Gandhi P, Franklin E, Hamamoto S, Khan AR, Munson M, Schekman R, Weisman LS (2011). Myosin V transports secretory vesicles via a Rab GTPase cascade and interaction with the exocyst complex. Dev Cell 21, 1156–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston GC, Prendergast JA, Singer RA (1991). The Saccharomyces cerevisiae MYO2 gene encodes an essential myosin for vectorial transport of vesicles. J Cell Biol 113, 539–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Jedd G, Kahn RA, Franzusoff A, Bartolini F, Segev N (1999). Genetic interactions in yeast between Ypt GTPases and Arf guanine nucleotide exchangers. Genetics 152, 1543–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm RW, Ejsing CS, Surma MA, Kaiser HJ, Gerl MJ, Sampaio JL, De Robillard Q, Ferguson C, Proszynski TJ, Shevchenko A, et al. (2009). Segregation of sphingolipids and sterols during formation of secretory vesicles at the trans-Golgi network. J Cell Biol 185, 601–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AY, St Onge RP, Proctor MJ, Wallace IM, Nile AH, Spagnuolo PA, Jitkova Y, Gronda M, Wu Y, Kim MK, et al. (2014). Mapping the cellular response to small molecules using chemogenomic fitness signatures. Science 344, 208–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine TP, Munro S (1998). The pleckstrin homology domain of oxysterol-binding protein recognises a determinant specific to Golgi membranes. Curr Biol 8, 729–739. [DOI] [PubMed] [Google Scholar]
- Levine TP, Munro S (2002). Targeting of Golgi-specific pleckstrin homology domains involves both Ptdlns 4-kinase-dependent and -independent components. Curr Biol 12, 695–704. [DOI] [PubMed] [Google Scholar]
- Lewis MJ, Nichols BJ, Prescianotto-Baschong C, Riezman H, Pelham HRB (2000). Specific retrieval of the exocytic SNARE Snc1p from early yeast endosomes. Mol Biol Cell 11, 23–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipatova Z, Tokarev AA, Jin Y, Mulholland J, Weisman LS, Segev N (2008). Direct interaction between a myosin V motor and the Rab GTPases Ypt31/32 is required for polarized secretion. Mol Biol Cell 19, 4177–4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961. [DOI] [PubMed] [Google Scholar]
- Losev E, Reinke CA, Jellen J, Strongin DE, Bevis BJ, Glick BS (2006). Golgi maturation visualized in living yeast. Nature 441, 1002–1006. [DOI] [PubMed] [Google Scholar]
- Luo X, Wasilko DJ, Liu Y, Sun J, Wu X, Luo ZQ, Mao Y (2015). Structure of the Legionella virulence factor, SidC reveals a unique PI(4)P-specific binding domain essential for its targeting to the bacterial phagosome. PLoS Pathog 11, 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGilvray ME, Shishkova E, Place M, Wagner ER, Coon JJ, Gasch AP (2020). Phosphoproteome response to dithiothreitol reveals unique versus shared features of Saccharomyces cerevisiae stress responses. J Proteome Res 19, 3405–3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda Y, Beznoussenko GV, Lint JV, Mironov AA, Malhotra V (2001). Recruitment of protein kinase D to the trans-Golgi network via the first cysteine-rich domain. EMBO J 20, 5982–5990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura-Tokita K, Takeuchi M, Ichihara A, Mikuriya K, Nakano A (2006). Live imaging of yeast Golgi cisternal maturation. Nature 441, 1007–1010. [DOI] [PubMed] [Google Scholar]
- McDonold CM, Fromme JC (2014). Four GTPases differentially regulate the Sec7 Arf-GEF to direct traffic at the trans-Golgi Network. Dev Cell 30, 759–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mérida I, Carrasco S, Avila-Flores A (2010). Diacylglycerol signaling: the C1 domain, generation of DAG, and termination of signals. In: Protein Kinase C in Cancer Signaling and Therapy, ed. Kazanietz MG, New York: Springer Science+Business Media, LLC, 55–78. [Google Scholar]
- Minogue S, Chu KME, Westover EJ, Covey DF, Hsuan JJ, Waugh MG (2010). Relationship between phosphatidylinositol 4-phosphate synthesis, membrane organization, and lateral diffusion of PI4KII at the trans-Golgi network. J Lipid Res 51, 2314–2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno-Yamasaki E, Medkova M, Coleman J, Novick P (2010). Phosphatidylinositol 4-phosphate controls both membrane recruitment and a regulatory switch of the Rab GEF Sec2p. Dev Cell 18, 828–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins C, Bonifacino JS (2001). Structural requirements for function of yeast GGAs in vacuolar protein sorting, α-factor maturation, and interactions with clathrin. Mol Cell Biol 21, 7981–7994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano M, Toshima JY, Siekhaus DE, Toshima J (2019). Rab5-mediated endosome formation is regulated at the trans-Golgi network. Commun Biol 2, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan P, Liu K, Patil DV, Sciorra VA, Jackson CL, Graham TR (2009). Regulation of a Golgi flippase by phosphoinositides and an ArfGEF. Nat Cell Biol 11, 1421–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz D, Medkova M, Walch-Solimena C, Novick P (2002). Ypt32 recruits the Sec4p guanine nucleotide exchange factor, Sec2p, to secretory vesicles; evidence for a Rab cascade in yeast. J Cell Biol 157, 1005–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyroche A, Antonny B, Robineau S, Acker J, Cherfils J, Jackson CL (1999). Brefeldin A acts to stabilize an abortive ARF-GDP-Sec7 domain protein complex: involvement of specific residues of the Sec7 domain. Mol Cell 3, 275–285. [DOI] [PubMed] [Google Scholar]
- Protopopov V, Govindan B, Novick P, Gerst JE (1993). Homologs of the synaptobrevin/VAMP family of synaptic vesicle proteins function on the late secretory pathway in S. cerevisiae. Cell 74, 855–861. [DOI] [PubMed] [Google Scholar]
- Pusapati GV, Krndija D, Armacki M, vin Wichert G, von Blume J, Malhotra V, Adler G, Seufferlein T (2010). Role of the second cysteine-rich domain and Pro275 in protein kinase D2 interaction with ADP-ribosylation factor 1, trans-Golgi network recruitment, and protein transport. Mol Biol Cell 21, 1011–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajebhosale M, Greenwood S, Vidugiriene J, Jeromin A, Hilfiker S (2003). Phosphatidylinositol 4-OH kinase is a downstream target of neuronal calcium sensor-1 in enhancing exocytosis in neuroendocrine cells. J Biol Chem 278, 6075–6084. [DOI] [PubMed] [Google Scholar]
- Richardson BC, Fromme JC (2015). Biochemical methods for studying kinetic regulation of Arf1 activation by Sec7. Methods Cell Biol 130, 101–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossanese OW, Reinke CA, Bevis BJ, Hammond AT, Sears IB, O’Connor J, Glick BS (2001). A role for actin, Cdc1p, and Myo2p in the inheritance of late Golgi elements in Saccharomyces cerevisiae. J Cell Biol 153, 47–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacher M, Barrowman J, Wang W, Horecka J, Zhang Y, Pypaert M, Ferro-Novick S (2001). TRAPP I implicated in the specificity of tethering in ER-to-Golgi transport. Mol Cell 7, 433–442. [DOI] [PubMed] [Google Scholar]
- Saila S, Gyanwali GC, Hussain M, Gianfelice A, Ireton K (2020). The host GTPase Arf1 and its effectors AP1 and PICK1 stimulate actin polymerization and exocytosis to promote entry of Listeria monocytogenes. Infect Immun 88, 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Tirado FH, Legesse-Miller A, Schott D, Bretscher A (2011). PI4P and Rab inputs collaborate in myosin-V-dependent transport of secretory compartments in yeast. Dev Cell 20, 47–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorr M, Then A, Tahirovic S, Hug N, Mayinger P (2001). The phosphoinositide phosphatase Sac1p controls trafficking of the yeast Chs3p chitin synthase. Curr Biol 11, 1421–1426. [DOI] [PubMed] [Google Scholar]
- Sciorra VA, Audhya A, Parsons AB, Segev N, Boone C, Emr SD (2005). Synthetic genetic array analysis of the PtdIns 4-kinase Pik1p identifies components in a Golgi-specific Ypt31/Rab-GTPase signaling pathway. Mol Biol Cell 16, 776–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulages JL, Salamon Z, Wells M, Tollin G (1995). Low concentrations of diacylglycerol promote the binding of apolipophorin III to a phospholipid bilayer: a surface plasmon resonance spectroscopy study. Proc Natl Acad Sci USA 92, 5650–5654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulard A, Cremonesi A, Moes S, Schütz F, Jenö P, Hall MN (2010). The rapamycin-sensitive phosphoproteome reveals that TOR controls protein kinase A toward some but not all substrates. Mol Biol Cell 21, 3475–3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Herrmann JM, Hamamoto S, Schekman R (2001). The ADP ribosylation factor-nucleotide exchange factors Gea1p and Gea2p have overlapping, but not redundant functions in retrograde transport from the Golgi to the endoplasmic reticulum. Mol Biol Cell 12, 1035–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns T, Kahn RA, Botstein D, Hoyt MA (1990). ADP ribosylation factor is an essential protein in Saccharomyces cerevisiae and is encoded by two genes. Mol Cell Biol 10, 6690–6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl T, Grafelmann B, Dannenberg J, Thorner J, Pongs O (2003). Conservation of regulatory function in calcium-binding proteins: human frequenin (neuronal calcium sensor-1) associates productively with yeast phosphatidylinositol 4-kinase isoform, Pik1. J Biol Chem 278, 49589–49599. [DOI] [PubMed] [Google Scholar]
- Strahl T, Hama H, DeWald DB, Thorner J (2005). Yeast phosphatidylinositol 4-kinase, Pik1, has essential roles at the Golgi and in the nucleus. J Cell Biol 171, 967–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl T, Huttner IG, Lusin JD, Osawa M, King D, Thorner J, Ames JB (2007). Structural insights into activation of phosphatidylinositol 4-kinase (Pik1) by yeast frequenin (Frq1). J Biol Chem 282, 30949–30959. [DOI] [PubMed] [Google Scholar]
- Swaney DL, Beltrao P, Starita L, Guo A, Rush J, Fields S, Krogan NJ, Villén J (2013). Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat Methods 10, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szentpetery Z, Várnai P, Balla T (2010). Acute manipulation of Golgi phosphoinositides to assess their importance in cellular trafficking and signaling. Proc Natl Acad Sci USA 107, 8225–8230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, Brill JA (2014). Cinderella story: PI4P goes from precursor to key signaling molecule. Crit Rev Biochem Mol Biol 49, 33–58. [DOI] [PubMed] [Google Scholar]
- Taverna E, Francolini M, Jeromin A, Hilfiker S, Roder J, Rosa P (2002). Neuronal calcium sensor 1 and phosphatidylinositol 4-OH kinase β interact in neuronal cells and are translocated to membranes during nucleotide-evoked exocytosis. J Cell Sci 115, 3909–3922. [DOI] [PubMed] [Google Scholar]
- Thomas LL, Fromme JC (2016). GTPase cross talk regulates TRAPPII activation of Rab11 homologues during vesicle biogenesis. J Cell Biol 215, 499–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas LL, Fromme JC (2020). Extensive GTPase crosstalk regulates Golgi trafficking and maturation. Curr Opin Cell Biol 65, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tojima T, Suda Y, Ishii M, Kurokawa K, Nakano A (2019). Spatiotemporal dissection of the trans-Golgi network in budding yeast. J Cell Sci 132, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toke, DA, Bennett LW, Dillon DA, Wu WI, Chen X, Ostrander DB, Oshiro J, Cremesti A, Voelker DR, Fischl AS, et al. (1998). Isolation and characterization of the Saccharomyces cerevisiae DPP1 gene encoding diacylglycerol pyrophosphate phosphatase. J Biol Chem 273, 3278–3284. [DOI] [PubMed] [Google Scholar]
- Tu L, Tai WCS, Chen L, Banfield DK (2008). Signal-mediated dynamic retention of glycosyltransferases in the Golgi. Science 321, 404–407. [DOI] [PubMed] [Google Scholar]
- Vanni S, Hirose H, Barelli H, Antonny B, Gautier R (2014). A sub-nanometre view of how membrane curvature and composition modulate lipid packing and protein recruitment. Nat Commun 5, 1–10. [DOI] [PubMed] [Google Scholar]
- Walch-Solimena C, Collins RN, Novick PJ (1997). Sec2p mediates nucleotide exchange on Sec4p and is involved in polarized delivery of post-Golgi vesicles. J Cell Biol 137, 1495–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walch-Solimena C, Novick P (1999). The yeast phosphatidylinositol-4-OH kinase Pik1 regulates secretion at the Golgi. Nat Cell Biol 1, 523–525. [DOI] [PubMed] [Google Scholar]
- Wang J, Sun H-Q, Macia E, Kirchhausen T, Watson H, Bonifacino JS, Yin HL (2007). PI4P promotes the recruitment of the GGA adaptor proteins to the trans-Golgi network and regulates their recognition of the ubiquitin sorting signal. Mol Biol Cell 18, 2646–2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YJ, Wang J, Sun HQ, Martinez M, Sun YX, Macia E, Kirchhausen T, Albanesi JP, Roth MG, Yin HL (2003). Phosphatidylinositol 4 phosphate regulates targeting of clathrin adaptor AP-1 complexes to the Golgi. Cell 114, 299–310. [DOI] [PubMed] [Google Scholar]
- Weixel KM, Blumental-Perry A, Watkins SC, Aridor M, Weisz OA (2005). Distinct Golgi populations of phosphatidylinositol 4-phosphate regulated by phosphatidylinositol 4-kinases. J Biol Chem 280, 10501–10508. [DOI] [PubMed] [Google Scholar]
- Wood CS, Schmitz KR, Bessman NJ, Setty TG, Ferguson KM, Burd CG (2009). PtdIns4P recognition by Vps74/GOLPH3 links PtdIns 4-kinase signaling to retrograde Golgi trafficking. J Cell Biol 187, 967–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Baldridge RD, Chi RJ, Burd CG, Graham TR (2013). Phosphatidylserine flipping enhances membrane curvature and negative charge required for vesicular transport. J Cell Biol 202, 875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa LL, Marchena J, Xie Z, Li X, Poon PP, Singer RA, Johnston GC, Randazzo PA, Bankaitis VA (2002). Activity of specific lipid-regulated ADP ribosylation factor-GTPase–activating proteins is required for Sec14p-dependent Golgi secretory function in yeast. Mol Biol Cell 13, 2193–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhdankina O, Strand NL, Redmond JM, Boman AL (2001). Yeast GGA proteins interact with GTP-bound Arf and facilitate transport through the Golgi. Yeast 18, 1–18. [DOI] [PubMed] [Google Scholar]
- Zhou X, Sebastian TT, Graham TR (2013). Auto-inhibition of Drs2p, a yeast phospholipid flippase, by its carboxyl-terminal tail. J Biol Chem 288, 31807–31815. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.