Abstract
Ion mobility-mass spectrometry (IM-MS) combines complementary size- and mass-selective separations into a single analytical platform. This chapter provides context for both the instrumental arrangements and key application areas that are commonly encountered in bioanalytical settings. New advances in these high-throughput strategies are described with description of complementary informatics tools to effectively utilize these data-intensive measurements. Rapid separations such as these are especially important in systems, synthetic, and chemical biology in which many small molecules are transient and correspond to various biological classes for integrated omics measurements. This chapter highlights the fundamentals of IM-MS and its applications toward biomolecular separations and discusses methods currently being used in the fields of proteomics, lipidomics, and metabolomics.
Keywords: Ion mobility, Ion mobility-mass spectrometry, Biomolecules, Omics
1. Origins of Biomolecular Analysis by Ion Mobility-Mass Spectrometry
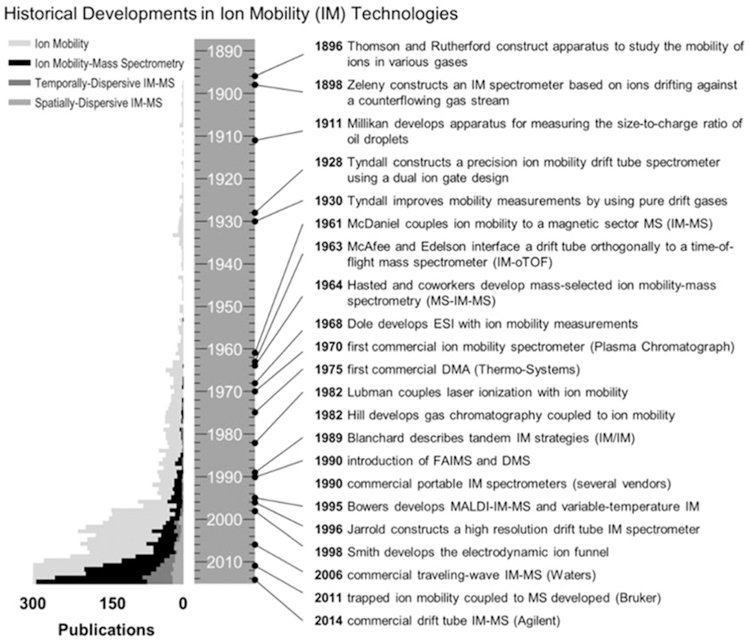
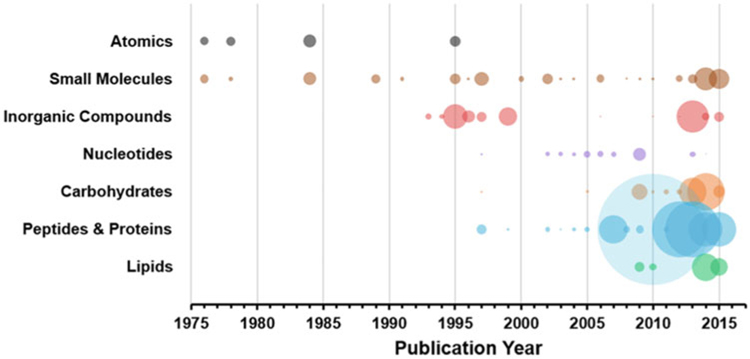
Ion mobility-mass spectrometry (IM-MS) is an integrated chemical separation technique which combines complementary size- and mass-selective separations into a single analytical platform. IM-MS is capable of separating individual chemical compounds on the millisecond time scale and as such is considered a high-throughput technique, and IM-MS is currently used in numerous chemical analysis applications including those of biological origin. A timeline of historical developments in IM and IM-MS is provided in Fig. 1. Ion mobility traces its roots back to experiments by Rutherford and Thomson in the late 1890s [1] and was further improved by Tyndall in the 1920s using pure drift gases [2, 3], before being paired with mass spectrometry in the 1960s [4, 5]. Electrospray ionization (ESI) and laser ionization were first used with IM in 1968 [6, 7] and 1982 [8], respectively, with Bowers demonstrating matrix-assisted laser desorption/ionization (MALDI) with IM-MS in the mid-1990s [9]. In the late 1990s onward, both MALDI and ESI ionization techniques were utilized with custom- and home-built IM-MS instruments by several laboratories, establishing IM-MS as highly sensitive technique for biomolecular analysis [10–14]. These soft ionization techniques were readily paired with IM-MS and enabled the analysis of large, intact molecules [15], and the growth in the application of IM-MS to biomolecules is illustrated in the timeline in Fig. 2 [16]. In 2006, a commercially available ion mobility-mass spectrometer well suited for biomolecular analysis was developed by Waters Corporation, named the Synapt HDMS (“High Definition” MS), which combined the soft ionization of ESI and liquid chromatography-ESI with the rapid separation of traveling wave-based IM technology and time-of-flight MS. The traveling wave IM technique was improved significantly in 2011 and the platform subsequently supported other optional analytical capabilities including MALDI and MS imaging, as well as integration with a variety of other ion sources and accessories [17–19]. This first commercial offering enabled a larger scientific community access to IM-MS which otherwise was only available to researchers with the capability to build their own instruments. Subsequently, other vendors entered the market with their own commercial IM-MS offerings, including Agilent Technologies in 2014 (drift tube IM) [20] and Bruker Corporation in 2016 (trapped IM) [21]. With the availability of versatile commercial instrumentation, biomolecular analysis using IM-MS flourished and continues to be a rapidly growing field providing fundamental insight into some of our most important biological questions.
Fig. 1.
A timeline and histogram summarizing the number of publications published between 1890 and 2014 as pertaining to ion mobility and ion mobility-mass spectrometry. Specific historical milestones in the development of ion mobility and IM-MS instrumentation are summarized at the right of the timeline (Figure from May et al. [5])
Fig. 2.
A bubble plot summarizing the number of CCS values reported over time for seven chemical compound classes. The size of each bubble represents the relative number of CCS values in each respective year (Figure from May et al. [16])
2. Biomolecular Separation and Analytical Utility of IM-MS
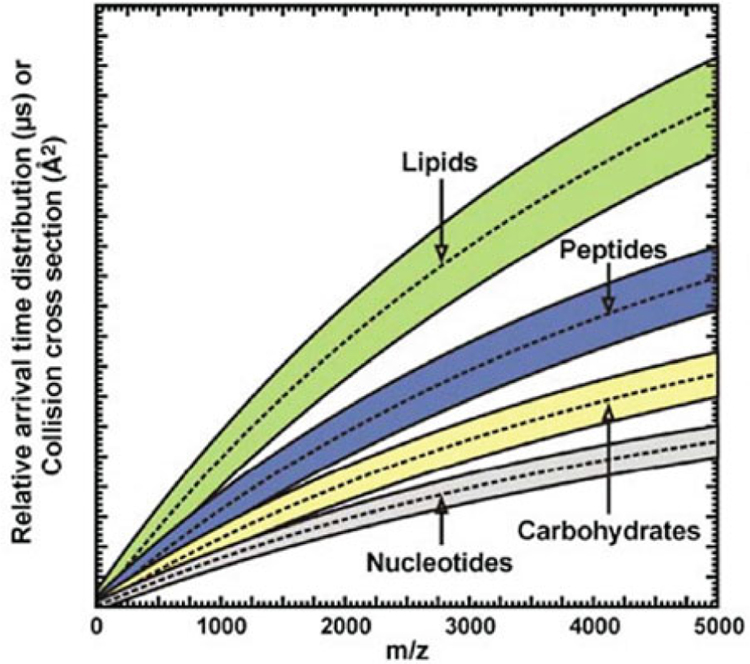
IM-MS has found great utility for biomolecular separation due to its broad sample compatibility, resolution, and sensitivity. It has been used to separate and analyze a wide range of masses, from small molecules to large protein complexes [22–29]. The analytical importance of IM is summarized in Table 1 [16]. IM provides a broad range of analytically valuable enhancements, including increasing peak capacity when coupled with other separation stages (chromatography, MS, etc.), enhancing low abundance signals of importance by reducing interference from chemical noise, and allowing for structural characterization of analytes through the measurement of their gas-phase cross-sectional area. Furthermore, it has been demonstrated that biomolecular classes maintain different molecular packing efficiencies in the gas phase, which results in class-specific correlations in 2-dimensional IM-MS spectra (Fig. 3) [20, 30, 31]. Notably, the canonical biochemical classes (lipids, peptides, carbohydrates, and nucleotides) align themselves into unique regions of IM-MS analytical space, which reflect their conformational preferences. Within each of these broad biochemical class correlations, subclass trends may also exist. As a prominent example, lipids have a characteristic behavior in IM-MS indicative of their primary class and structure [32]. Specifically, lipid packing efficiency is affected by both headgroup identity and modifications within their acyl tail(s), forming different correlations of gas-phase collision cross section (CCS) versus mass. Current research is considering the effects of degree of unsaturation and carbon chain length on lipid packing efficiency and correlation behavior in IM-MS [33–35]. Novel approaches such as ozonolysis paired with IM-MS are now also being used in lipidomics to allow for determination of double-bond location in lipids [36, 37]. For broad-scale identification of unknowns, empirically derived databases of mass and CCS are being assembled for a variety of biomolecular compounds [16, 24, 29, 38–44]. In addition to specific chemical class information, IM can address the challenge of resolving and identifying isobaric compounds that are unresolvable by conventional MS. In many cases, isobaric species including isomers and conformers can be distinguished in the IM dimension due to the different conformations they adopt in the gas phase, whether peptides, carbohydrates, or lipids [45–50]. Collectively, the capability to rapidly resolve biochemicals based on both structure and mass has made IM-MS an invaluable analytical tool for biomolecular analysis.
Table 1.
Three analytical uses of ion mobility
| Analytical use of ion mobility | Description | Additional requirements | Example application areas |
|---|---|---|---|
| 1. Chemical separation | Partition signal from chemical noise and increase peak capacity of the analysis | None | Detection of illicit compounds (e.g.., drugs and explosives) and screening of exogenous metabolites (e.g., pesticides and industrial chemicals) |
| 2. Analyte identification and characterization | Use CCS measurement to characterize unknown by correlation | Reference values from databases and libraries incorporating normalized drift times, reduced mobilities, and/or CCS | Emerging omics and small molecule discovery initiatives |
| 3. Structural analysis | Utilize the experimental CCS to infer structural information | Computational methods to link theoretical structure(s) to the experimental CCS | Insights into protein complex arrangements and structure |
Fig. 3.
A conceptual depiction of where various biomolecular classes are observed in IM–MS conformational space (Figure from Fenn et al. [30])
3. Instrumentation Considerations
3.1. Chromatography
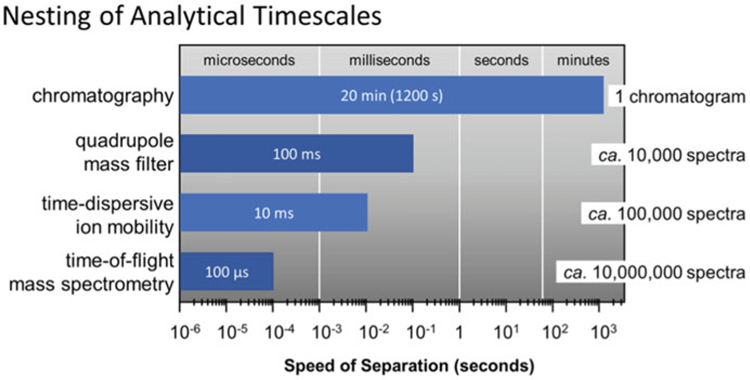
The inclusion of a chromatographic separation prior to IM-MS provides added peak capacity while enhancing the analytical sensitivity of both the IM and MS stages by reducing ion suppression resulting from the simultaneous infusion of multiple analytes. Liquid chromatography (LC) is commonly implemented with IM-MS due to its ability to use a wide variety of flow rates, column choices, and solvents that pair well with ESI ion generation [51, 52]. Since the IM separation occurs postionization, all of the LC conditions which work in LC-MS are also compatible with IM-MS, including both normal- and reverse-phase LC columns and numerous solvent systems [53]. Autosampler systems are also commonly utilized with LC to facilitate automated, high-throughput LC-IM-MS workflows [54]. Other chromatographic separation techniques such gas chromatography (GC) and supercritical fluid chromatography (SFC) have also been demonstrated with IM-MS [55–57], although these are less common due to the more limited analytical space that these techniques encompass (e.g., volatile and nonpolar analytes, respectively). Figure 4 illustrates the compatibility of timescales for chromatography with IM-MS [5]. While chromatography operates on a timescale of minutes, the downstream analytical strategies such as IM and MS operate on the order of milliseconds to microseconds, allowing further separation of components eluting from the chromatographic dimension without affecting the overall sample throughput.
Fig. 4.
The relationship of various analytical timescales based on the respective speed of each separation. The total number of possible spectra obtained through nesting the subsequent analytical separation dimensions is summarized to the right (Figure from May et al. [5])
3.2. Ion Sources
Two of the most common methods for ionization of biomolecules are MALDI and ESI. These so called “soft” ionization techniques impart minimal energy to molecules during the ionization process, which helps to maintain the analyte’s structural integrity during transfer to the gas phase, which is crucial for the analysis of fragile biomolecules [9, 15]. MALDI is performed on solid samples using a laser to desorb and generate ions and tends to generate low charge states in a narrow distribution, which simplifies the mass spectra [58, 59]. The use of a laser also facilitates operating MALDI in an imaging mode, allowing spatially resolved mass information to be obtained [60]. MALDI-based MS imaging (MSI) has been utilized to determine analyte location in tissue samples [61], and MSI has also been coupled with IM-MS to provide a high-dimensional separation strategy that can simultaneously separate analytes based on spatial location, size, and mass [62, 63]. MALDI, however, is not conducive to direct analysis of liquid samples such as the effluent stream originating from LC, which requires offline sample fraction collection for MALDI analysis. Liquid sample analysis is, however, readily facilitated by ESI, which generates a continuous flow of ions from liquid-phase samples. ESI has enabled high-throughput LC-MS experiments and has been used to combine LC with IM-MS. In contrast to the low charge states observed in MALDI, ESI typically generates multiply charged ions. Since mass spectrometers measure ions as a mass-to-charge ratio, higher charge states effectively increase the practical mass range accessible to a mass spectrometer, facilitating analysis of proteins and other high mass analytes alongside small molecules in a complex biological sample [64, 65]. Multiply charged ions have also improved detection limits for Fourier transform MS (FTMS) techniques [66, 67], improved the ion fragment yields for traditional collisional activation experiments (collision-induced dissociation, CID) [68], and have enabled electron-induced fragmentation techniques such as electron capture dissociation (ECD) and electron transfer dissociation (ETD) to be implemented [69, 70]. Specifically for IM analysis, the various charge states generated from ESI are readily resolved into distinct correlations of cross section and mass, which facilitates the separation and identification of multiply charged species [10].
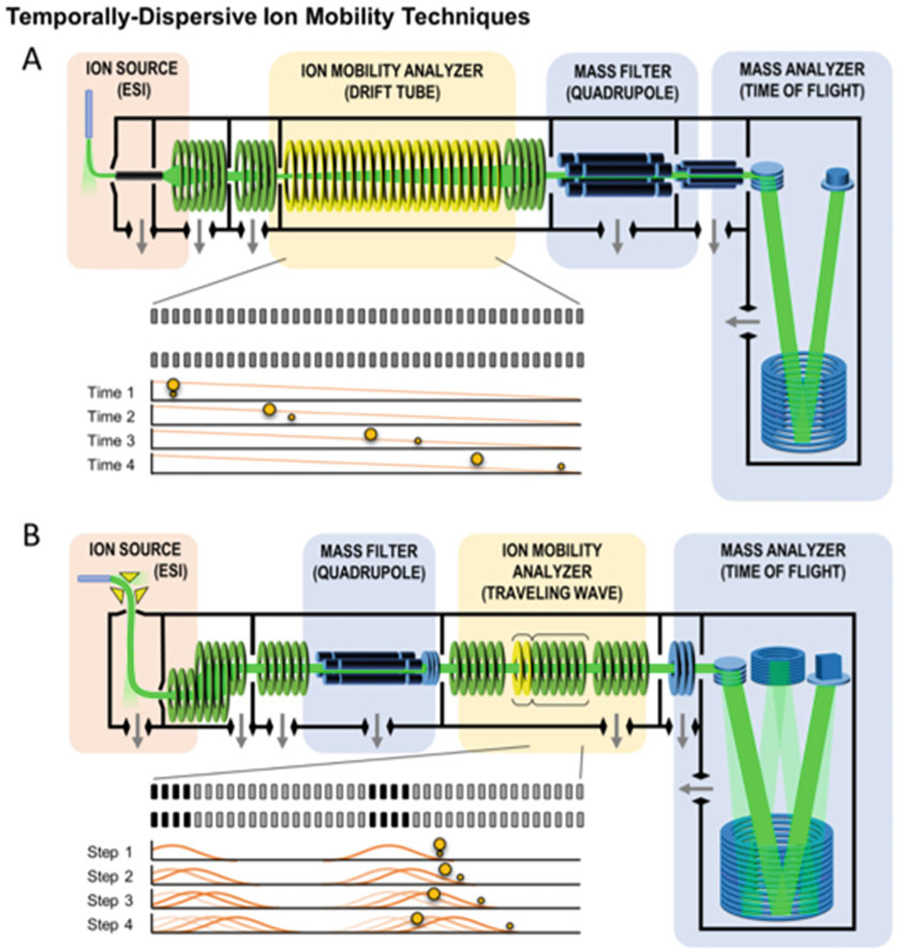
3.3. Time-Dispersive Ion Mobility Techniques
Two ion mobility techniques that are commonly used are drift tube ion mobility spectrometry (DTIMS) and traveling wave ion mobility spectrometry (TWIMS) (Fig. 5) [5]. TWIMS was one of the first IM techniques to be used in a commercially available IM-MS platform. In TWIMS, ion separations result from a series of dynamically pulsed voltages (an electrodynamic field) which creates a traveling wave potential that transfers ions through the drift region in a mobility-selective mode. TWIMS separations are similar to DTIMS separations and can access similar resolving powers [71]. Although DTIMS combined with MS was implemented commercially several years after TWIMS, it is an older technique and considered the standard and classically most accurate method for CCS determination. Unlike TWIMS, DTIMS separates ions using a constant voltage gradient (a uniform electric field) which allows the measured ion drift times to be related directly to CCS via the kinetic theory of gases, commonly referred to as the Mason–Schamp relationship [72, 73]. On the other hand, TWIMS-derived CCS values are determined via an empirical calibration relationship between TWIMS measured drift times and known CCS values obtained from DTIMS [27, 32, 74]. Due to the fact that TWIMS does not require high voltages for separations, it is readily scalable to longer path lengths, which fundamentally improves instrument resolution. In contrast, high operational voltages must be utilized in order to increase the path length in DTIMS. Next-generation TWIMS instruments are taking advantage of this scalability, producing high resolving power platforms based on serpentine and cyclic designs [45, 75].
Fig. 5.
Two conceptual schematic diagrams of commercially-available time-dispersive IM-MS instrumentation. (a) An electrostatic drift tube ion mobility spectrometer (DTIMS) coupled to a quadrupole-time-of-flight (QTOF) mass spectrometer, and (b) an electrodynamic traveling wave ion mobility spectrometer (TWIMS) coupled with a quadrupole mass filter and a QTOF. Conceptual experimental sequences are shown in the insets of each panel and illustrate the mechanism giving rise to the temporal separation of smaller and larger collision cross section ions experimentally observed in each technique (Figure from May et al. [5])
3.4. Mass Spectrometry Considerations
Mass spectrometry is routinely conducted after IM separations to provide mass measurement on ion mobility separated ions. In this IM-MS configuration, most of the chemical separation occurs in the mass dimension due to the high resolving power (>10,000) accessible by modern mass spectrometers; however, the added IM dimension provides improved peak capacity and the capability for resolving isomeric compounds based on structural differences [76]. IM is commonly coupled with time-of-flight (TOF) mass spectrometry due to its ability to rapidly analyze a wide mass range simultaneously and with high resolution. The timescale of the TOF is on the order of microseconds, pairing well with an upstream IM which is on the order of milliseconds per separation. Quadrupole MS is also commonly used with IM-MS as an added mass-filtering stage for tandem MS/MS experiments, which can further aid in analyte identification and characterization.
Fragmentation methods have been utilized with great success in tandem mass spectrometry and in IM-MS, where it can be initiated between the IM and MS stages (IM-MS/MS) or prior to IM-MS either with a front-end mass filter, or without, as is the case with in-source ion activation [77]. Collision-induced dissociation (CID) is commonly used to fragment ions as it is readily implemented with existing ion optics. CID is implemented through ion collisions with an inert background gas (such as nitrogen or argon) under elevated voltage conditions. CID is considered an ergodic process where energy is restributed within the molecule and the weakest bonds are preferentially cleaved, which leads to predictable and reproducible fragmentation of ions. The analysis of CID fragmentation spectra has been used with IM-MS to identify isobaric species which exhibit different bond energies. The CID method is commonly used in proteomics to determine the amino acid sequence of peptides. While highly predictable, CID does not preserve weak bonds typical of noncovalent complexes and post-translational modifications, and is less effective at fragmenting large molecules, such as intact proteins. To address these deficiencies, electron capture dissociation (ECD) and electron transfer dissociation (ETD) have been used, both of which fragment the ion in a nonergodic process and thus creating different but informative fragments compared to CID. ECD requires simultaneous trapping of ions and free electrons and has only been implemented in ion cyclotron resonance MS. ETD and CID have been used simultaneously with IM-MS for glycoproteomics [78–80]. This allows a variety of fragments to be determined for a given precursor ion, in particular for preserving informative labile bonds when obtaining fragmentation information. Combining the ion drift time, precursor mass, and fragment mass information can facilitate confident identification of the precursor molecular.
IM-MS holds an important and ever increasing role in biomolecular analyses. The ability of IM-MS to integrate with a wide variety of chromatographic techniques, ion generation methods, fragmentation methods, and mass analyzers results in a versatile and power technique for biomolecular analysis, particularly in the omic sciences (i.e., proteomics, lipidomics, metabolomics, etc.) where deriving biological answers requires high sensitivity and high-confidence identifications. Additionally, IM improves peak capacity compared to standalone MS and is compatible with other analytical stages such as LC separations and tandem ion fragmentation. In this context, IM can differentiate isobaric species and provides an additional descriptor (ion drift times or gas phase CCS) for the identification of unknowns. The following sections describe current trends in specific applications of IM-MS to biomolecular analyses.
4. Current Trends in IM-MS for Biomolecular Analyses
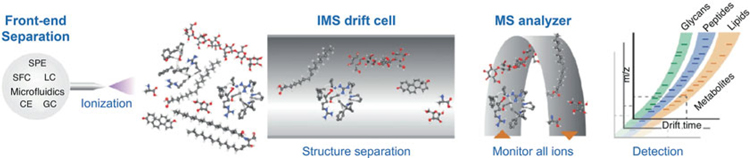
The confident identification of small molecules continues to be one of the most difficult challenges in omic studies. While advances in proteomics have streamlined IM-MS-based identification efforts for peptides and proteins [38, 81, 82], metabolomic and lipidomic identification capabilities have generally lagged behind [83, 84]. One reason for this is that while the structure of peptides consists of translationally assembled amino acid building blocks which can often be elucidated through ion fragmentation strategies, metabolites and lipids are less predictable and contain a wide array of substructural units. Additionally, the prevalence of isobaric species in lipidomics complicates feature identification, and metabolomics studies often encounter features of redundant mass that lack unique fragmentation patterns, which further confounds attempts at identification by MS alone. Therefore, a combination of analytical techniques is required for high-confidence lipidomics and metabolomics. As shown in Fig. 6, gas chromatography, liquid chromatography, or another front-end separations can be readily combined with IM and MS analysis to provide high-dimensional data sets which can be partitioned into specific omics workflows [53, 84]. The IM analysis provides charge state and chemical class-specific separations based on differences in intrinsic gas phase packing, as well as quantitative size information via CCS measurement which can be used as a reproducible measurement for identification purposes [84].
Fig. 6.
A conceptual IM-MS analytical workflow incorporating various front-end separation techniques (SPE, solid phase extraction; SFC, super critical fluidic chromatography; LC, liquid chromatography; CE, capillary electrophoresis GC, gas chromatography). The resulting IM-MS analysis yields drift time vs. m/z spectra which can be interpreted for relative size-mass information intrinsic to different biochemical classes (Figure adapted from Zhang et al. [84])
5. Integrating Ion Mobility for Omics Analyses
IM-MS provides a fast separation of chemically unique biological groups with the added benefit of measuring collision cross section concurrently with mass-to-charge ratio [31, 82, 85]. This capability is important for omics studies utilizing complex biological samples which routinely require extensive sample purification strategies to isolate molecules of interest from undesirable compounds that would otherwise make MS analysis difficult (e.g., salts, detergents, and other types of chemical noise) [81, 86]. Sample preparation strategies have the potential to chemically alter molecules of interest, for example, by oxidation, reduction, conversion to a secondary metabolite, or loss of a post-translational modification in peptides. Integrating IM with MS can help offset some of the burden of chemical separation and alleviate the need for extensive sample handling. In certain cases where fragmentation occurs postmobility, either intrinsically or intentionally, IM-MS also allows the alignment of precursor and product ions which can further aid in identification [83, 87]. The following sections provide examples of how ion mobility has been integrated in proteomic, lipidomic, and metabolomic analyses. Although each analysis is discussed separately, it should be noted that ion mobility allows for simultaneous analysis of these individual omic classes via chemical class separation, providing a truly integrated multiomic experiment.
5.1. Proteomics
Proteomics, the large-scale study of proteins, has been a driving force in systems biology and has increased our understanding of diseases and human health. Proteins serve as the machines for all cellular processes; thus proteomic studies are one of the most crucial tasks in systems biology. MS-based proteomics is a major component to the advancements in the field [88, 89]. Proteins encompass a large dynamic range of concentrations, necessitating separation techniques to enhance lower abundance species prior to mass analysis. Common separation methods for proteomics include gel electrophoresis, liquid chromatography, and, in a growing number of instances, ion mobility [90–92].
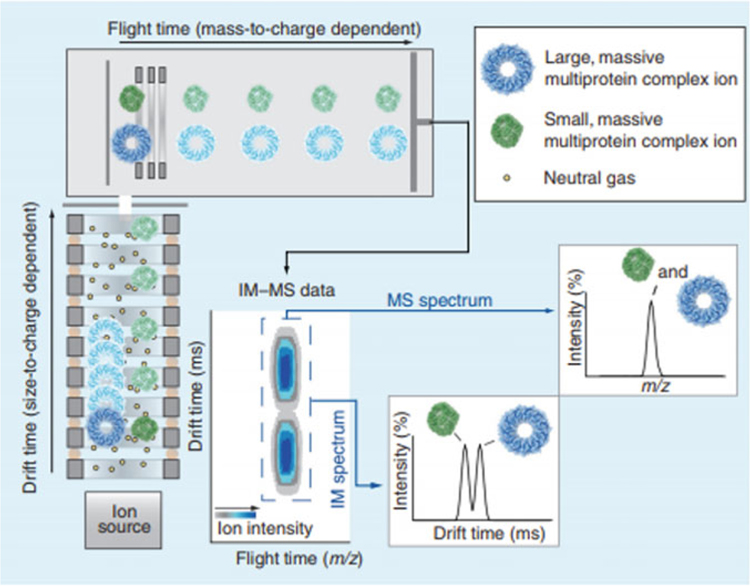
Ion mobility has been utilized extensively in structural proteomics [82, 93]. Proteins of similar or exact mass, such as protein conformers, can be separated by IM due to differences in their gas phase structure [94]. Figure 7 illustrates IM separations for ions of protein complexes of different sizes and configurations, but similar mass-to-charge ratio [93]. The majority of multiprotein complexes have been analyzed on TWIMS instruments, and it has been demonstrated that incorporation of TWIMS with MS analysis can increase proteome coverage by up to 60% [95].
Fig. 7.
Basic principles of IM-MS data acquisition and the resulting spectra. Ions generated in the ion source (lower left) are introduced into an IM spectrometer, which is effectively an ion guide consisting of a background of neutral gas molecules and a weak electric field. Following IM analysis, the ions are introduced into a mass analyzer operated under vacuum. The resulting IM and MS data are 3-dimensional, containing ion intensity, size, and mass information, and can be projected either as a contour plot (middle, bottom), or 2D selections in either drift time or m/z space (lower right) (Figure from Zhang et al. [93])
In addition to separating proteins, IM is used to probe structural information [93]. Temperature-controlled ESI sources and heated ion transfer capillaries have been used prior to IM-MS to rapidly heat proteins and monitor their controlled denaturation [11, 96, 97]. In addition, thermally induced protein conformational transformations as well as protein–ligand interactions are able to be observed. In these ways and others, IM-MS progresses from a separation strategy to a structural measurement technique to broaden our understanding of how protein clusters are formed and stabilized [96, 97].
5.2. Lipidomics
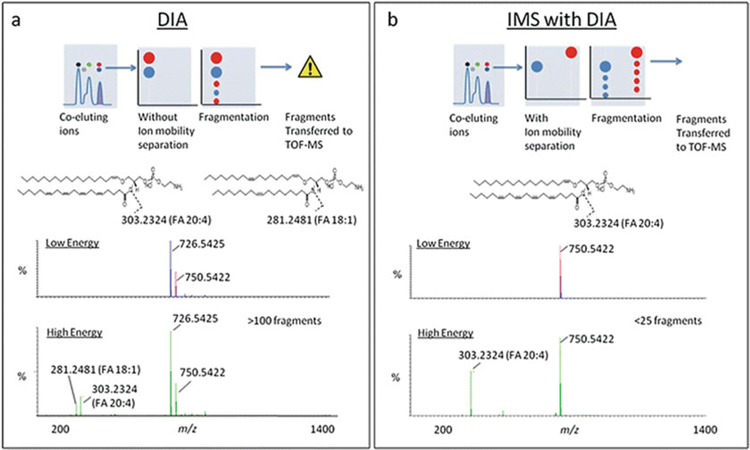
Lipids comprise a large portion of the small molecules extracted from organisms. They have three major functions in biological systems: energy storage, cellular signaling, and structural functions. Lipids can be divided into eight major categories (fatty acids, glycerolipids, glycerophospholipids, sphingolipids, sterols, prenols, saccharolipids, and polyketides) with many different structural motifs ranging from fused cyclic molecules to long-chain fatty acids. Lipids cover a large range of m/z ratios, and while mass spectrometry has been a powerful tool in lipidomics, the numerous isomeric lipid species which can exist make lipid structural characterization by MS challenging [98–100]. Contributing to lipid structural complexity are the numerous potential double-bond positions, geometric (cis/trans), constitutional (linear and branched), and stereochemical orientations that a lipid can adopt which are all isobaric in mass. Identifying complex lipid structures has been a struggle in the field of lipidomics, one that ion mobility is well-suited to address [99–102]. Lipid identifications in MS-based lipidomics utilize the exact mass measurement, as well as characteristic fragmentation patterns obtained from tandem MS/MS experiments to assign confidence to structural identifications. As many lipids are chemically and structurally similar, lipids can be challenging to separate using LC alone, making it difficult to correlate fragment ions with their precursor ion forms. As demonstrated by Paglia et al., ion mobility can be used to align fragmentation spectra with precursor parent ions to increase the confidence in lipid identification (Fig. 8). In this study, ion mobility was also demonstrated to be useful in separating coeluting lipid structures with the same m/z ratios [103].
Fig. 8.
Two examples of data-independent acquisition (DIA) using IM-MS instrumentation. (a) Conventional DIA where ions are comprehensively fragmented following chromatography, (MSE) and (b) MSE coupled with IM (HDMSE). DIA combined with IM separation enables mobility-aligned fragmentation spectra, which can increase the specificity and confidence in identifying complex lipids (Figure from Paglia et al. [103])
Ozone-induced dissociation has been recently demonstrated with IM-MS to elucidate the location of double bonds in the acyl tail region of lipids. Two separate strategies have been described: solution-phase ozonolysis of lipids prior to being introduced to the mass spectrometer [104], and gas-phase ozonolysis of lipid ions within the MS, the latter technique termed OzID. These ozonolysis strategies have been shown to be useful for locating the position of double bonds within lipids; however, one shortcoming with this approach is that it does not provide any information about the geometry (cis/trans) of the double bond prior to ozonolysis [104, 105]. This emphasizes the strength of IM-MS analysis, where ion mobility allows for the differentiation of some geometric lipid isomers, such as cis versus trans, even when present in a complex biological mixture. Groessl et al. demonstrated that CCS differences of 1% or more are sufficient to baseline separate lipids in DTIMS [101]. Although the possibility to use IM for identification purposes was also discussed, it was noted that high precision and accuracy are needed to create and populate reliable reference data libraries. Recently, Leaptrot et al. reported on an empirical CCS database of various sphingolipids and phospholipids incorporating 456 high-precision DTIMS measurements (global average RSD of 0.18%), which provides a quantitative means of identifying unknown lipids, including isomers, by IM-derived CCS information [33]. As these CCS libraries become more available, it is expected that lipidomics using IM-MS will continue to grow.
5.3. Metabolomics
Metabolomics is the measurement of the thousands of small molecules in a biological system. Unlike genomics and proteomics, metabolomics encompasses a large amount of chemical diversity as it consists of molecules from many different biological classes, such as carbohydrates, amino acids, hormones, and lipids [106]. There are generally two approaches to MS-based metabolomics: (1) targeted analysis, in which a panel of metabolites are selected for chemical analysis, and (2) untargeted analysis, in which all small molecules present in the sample are analyzed simultaneously [106]. Both approaches have their advantages. Targeted studies allows for semiquantitative analysis of small molecules on a curated list. Isotope standards can be analyzed concurrently with unknowns and calibrated against empirically measured response curves (calibration curves) in order to determine the concentration of specific metabolites within a sample. Since only a small number of m/z values are prioritized, targeted studies are commonly conducted on MS instrumentation with selective ion monitoring capabilities, such as triple quadrupole and ion trap instruments where a select m/z can be tuned and monitored in a continuous duration, greatly increasing the instrument sensitivity and response reproducibility, which improves quantitative results. While targeted approaches provide quantitative information regarding metabolites of interest, these studies do not provide detailed information for other small molecules present in the sample. On the other hand, untargeted approaches focus on separating and comprehensively measuring all of the small molecules present in the sample but lacks robust means of quantifying these signals. Also, untargeted studies generally utilize analytical methods and settings that attempt to measure a large breadth of molecules, and thus can be less sensitive to a specific class or pool of analytes [106–108]. While untargeted studies can detect a large number of molecules present in a sample, identifying the oftentimes thousands of metabolites observed in a single untargeted study can be an arduous task. For a single m/z feature, there can be hundreds of matches to known compounds in various metabolomic databases, making an absolute identification difficult [107–109]. Thus in order to improve confidence in metabolite identification, multiple dimensions of analytical information are generally obtained in untargeted experiments, which can include measurements from front-end chromatographic separations (analyte polarity and retention times), as well as post-ionization techniques such as ion mobility (likely chemical class and collision cross sections) and tandem MS (ion stability and fragmentation information). Ion mobility in particular can provide additional peak capacity, isomeric differentiation, and an additional molecular descriptor (CCS) which can help to alleviate some of the difficulties associated with confident metabolite identification [107–111].
As noted in the previous section, IM can improve fragmentation identification in lipidomics, and this advantage applies to metabolomics as well. With such chemical diversity found in the metabolome, ion isolation is complicated by coeluting species, and thus aligning precursor ions with their fragments originating from ion activation experiments is challenging. Using IM prior to ion fragmentation allows coeluting small molecules to be further resolved following chromatography [107–109]. For example, Wickramasekara et al. classified coeluting lipid species based on the differences in their IM drift times (Fig. 9) [112]. In addition, the drift time-extracted spectra can be used to align fragment ions to the corresponding parent ion, as these signals have identical drift times when conducting the fragmentation postmobility. This capability to mobility-align ion precursors and fragments can help to collate specific ion fragment information which can then be used with the accurate mass measurement to match unknowns to database entries, thus increasing the confidence in assigning metabolite identifications.
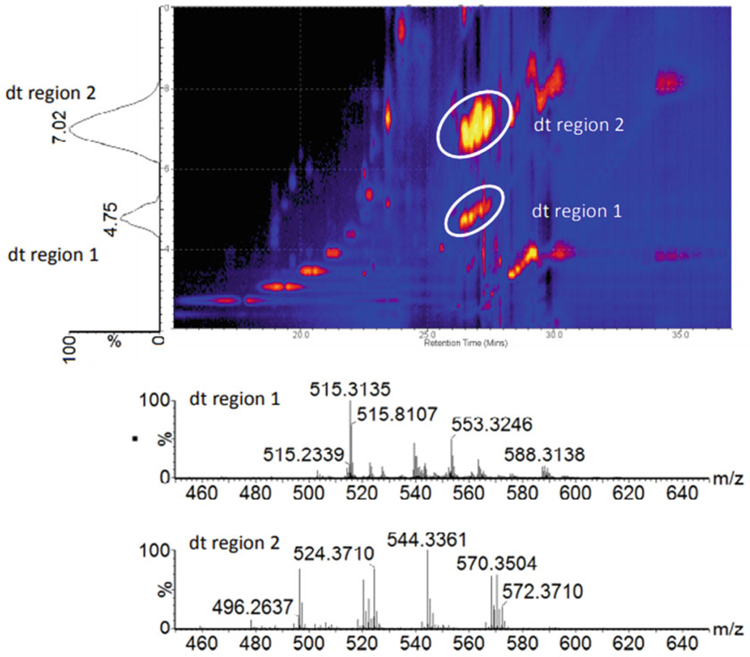
Fig. 9.
An example 2D LC-IM spectrum (drift time vs. retention time) showing IM separation of different compound classes observed in rat plasma. The encircled regions represent compound classes that elute at a similar retention time (26–28 min). Extracted IM spectra (bottom) reveal that these two regions of ion signal represent different lipid classes, namely Lyso-PC and SM lipids (sphingosine phosphocholines) (Figure from Wickramasekara et al. [112])
In addition to utilizing the enhanced separation and fragment alignment capabilities from IM, CCS measurements derived from IM experiments can improve metabolite validation [109]. CCS is linked to an intrinsic molecular property of the analyte (the microscopic molecular cross section) and thus is considered more robust than other conditional measurement parameters such as the chromatographic retention time. This property makes CCS useful as an additional molecular descriptor that can be used in metabolomic studies along with accurate mass and fragmentation information. Currently, there are several laboratories attempting to use CCS in metabolite identification workflows. For example, Paglia et al. have described a robust analytical workflow incorporating CCS for both metabolite and lipid identifications and report a ca. 2% interlaboratory reproducibility of the TWIMS-derived CCS [42, 83]. Stow et al. utilized standardized DTIMS instrumentation deployed across several international laboratories to achieve an interlaboratory CCS reproducibility of better than 0.5% [113], and recent work by Nichols et al. describe the utility of DTIMS CCS measurements as a molecular descriptor in untargeted studies of primary human metabolites [111]. As more research shifts toward incorporating CCS into metabolomic analysis, there is a need for database expansion to support bioinformatic strategies. Recent efforts for developing CCS databases to support metabolite identifications have included pesticides, pollutants, xenobiotics, and steroids [114–117]. Leveraging the standardization efforts for DTIMS, Picache et al. have recently described a “Unified CCS Compendium” which compiles over 3800 CCS measurements obtained from different studies into a single, self-consistent resource with a global average CCS precision of 0.25% RSD [43]. These and other efforts will allow rapid and reliable metabolite identification and quantification, which becomes increasingly important as the field shifts toward comprehensively characterizing individual metabolomic pathways.
6. Accelerating Advancements in IM-MS Technology
Innovation in IM-MS instrumentation continues at a rapid pace. Various improvements have been suggested and novel IM-MS technologies now being described allow for ingenious solutions for challenges encountered in biomolecular analysis. For example, a novel instrument design approach now being actively developed for IM-MS utilizes a scalable ion optical architecture consisting of electrode pads on a printed circuit board (PCB) and driven with electrodynamic (RF) fields that confine ions to a predefined ion optical path. This approach, named by R.D. Smith and colleagues as “structures for lossless ion manipulations” (SLIM), utilizes a 2-dimensional electrode geometry that is both modular and scalable such that various different experiments can be achieved on the same instrument platform [45, 118–121]. In SLIM, two PCBs with a mirrored electrode symmetry are placed above and below one another to create the ion path of travel in between the boards, and dynamic electric fields are used to both contain and guide the ions through the SLIM device [122]. SLIM allows high ion transfer transmission through elevated pressure regions, and SLIM-based ion mobility separations based on both DTIMS and TWIMS have been demonstrated [123, 124]. The ability to print electrodes on a two-dimensional surface allows for various ion manipulation modules to be fabricated, including modules to move ions at 90° angles (elbows and tees) [123, 125]. This facilitates cyclic racetrack and serpentine geometries to be fabricated for long path length, high-resolution ion mobility separations, and incorporation of “tee” junctions allows selection of a discreet ion mobility region for further tandem analysis by either IM or MS. An example of SLIM-based ion mobility instrumentation is shown in Fig. 10 [124]. Current designs have created instruments with some of the highest IM resolution currently available [71].
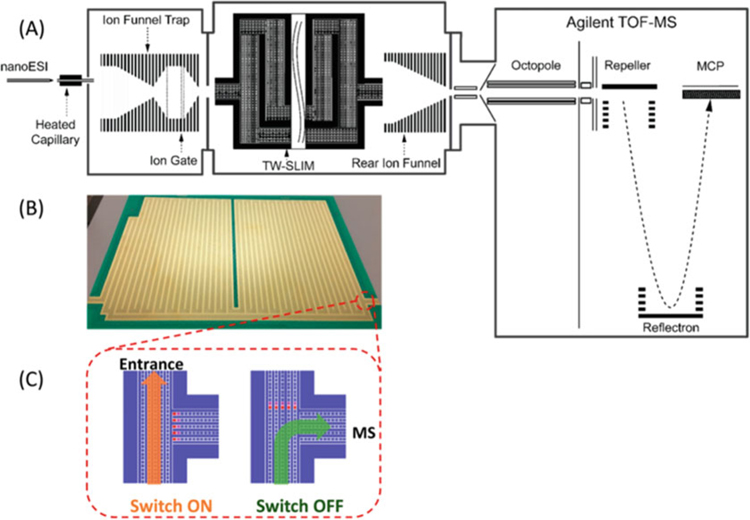
Fig. 10.
(a) A schematic of the multipass SLIM SUPER IM-MS instrument. (b) Photograph of one of the two SLIM surfaces which are stacked in mirrored symmetry in the instrumentation, and (c) an illustration of an ion switch which is used to direct ions through various stages of the experiment (Figure from Deng et al. [124])
In addition to TWIMS and DTIMS, a relatively new ion mobility technique called trapped ion mobility spectrometry (TIMS) is currently available in commercial instrumentation [126, 127]. TIMS performs ion mobility separations by selectively releasing ions trapped in a mobility “analyzer” cell combining a gas flow and an opposing electric field [128, 129]. The ions trapped in TIMS are released slowly by lowering the electric field barrier, which allows a mobility spectrum to be obtained and subsequent MS analysis to be performed. The rate at which the electric field is lowered corresponds with IM resolution, with slower scan rates leading to higher resolution. TIMS instruments are capable of high IM resolution and are very versatile due to its variable scan rate [130]. Either high-throughput or high-resolution scan rates may be chosen as needed, or a scan rate may be variable during analysis to allow high resolution only for a certain range of mobilities, enabling targeted high-resolution experiments to be conducted. This is in contrast to current high-resolution approaches based on cyclic or racetrack IM technologies, which relies on selecting a narrow population of mobilities for high-resolution analysis, at a cost of rejecting other ions outside of this mobility window.
While most efforts have focused on improving IM resolution, some approaches have sought to improve sensitivity and throughput. An example of such an approach is a multichannel IM spectrometer shown in Fig. 11 [5]. This instrument utilizes eight discrete ion optical paths to perform eight ion mobility separations in parallel [5, 131]. Each ion channel can act independently, analyzing eight unique samples at a time, improving throughput. Alternatively, the multichannel instrument can analyze the same sample simultaneously across the eight ion optical paths, increasing the sensitivity of the instrument. As the analytical community pushes for rapid extraction of more information from complex samples, advancements in high-throughput instrument designs remain crucial.
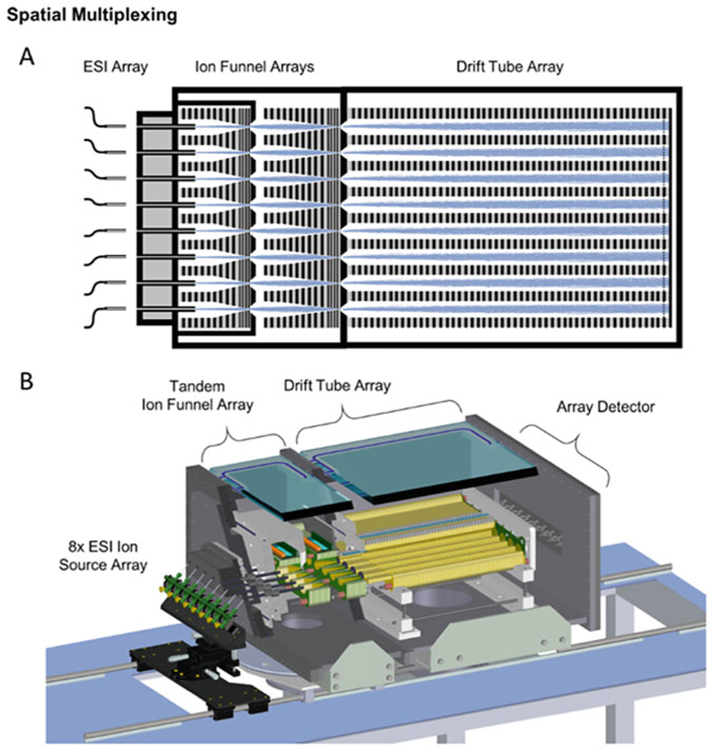
Fig. 11.
An example of a spatial multiplexing strategy for DTIMS using eight individual IM analysis channels: (a) simulation of ion trajectories through the interfacing ion funnels and the drift tube array, and (b) a cutaway showing component details of the spatially multiplexed IM instrument (Figure from May et al. [5])
Additional strategies for improving ion mobility separation have focused on increasing the chemical selectivity of existing IM instrumentation by using alternate drift gases [132]. The understanding and application of the effect of drift gas on IM separations and the associated CCS measurement are still in early development, but there is now mounting evidence that the use of more polarizable drift gases (e.g., CO2, N2O, NO2) can increase the resolution for certain ion species [132, 133]. While the hard-sphere interactions between the ion and drift gas tend to predominate the mobility of ions in the IM experiment [134], long-range interactions also play a role in the observed IM separations and are exploited by varying the drift gas polarizability. Improved selectivity can occur between certain ionic species depending upon their susceptibility to long-range interactions [135]. The effect on separation efficiency by varying the drift gas composition is similar to the effect of varying the solvent conditions in capillary electrophoresis to affect separation selectivity. It is common to alter the drift gas in some high-field IM techniques, such as high-field asymmetric waveform ion mobility spectrometry (FAIMS) and differential mobility spectrometry (DMS) [136–141]. In low-field IM techniques, this is less common, but there have been a number of significant examples [142, 143]. Using an ambient pressure drift tube, Hill and coworkers showed that varying the drift gas polarizability could improve selectivity for various small molecules including drugs, carbohydrates, and peptides [132, 133, 144, 145]. Using a reduced pressure drift tube, Yost and coworkers reported that carbon dioxide improved resolving power for several isobaric steroids [146]. Eberlin and coworkers demonstrated that replacing nitrogen with carbon dioxide in a TWIMS instrument could improve separation of a number of analytes such as carbohydrates and isomeric haloanilines [50, 147–149]. However, it is common for observations of more polarizable drift gases to report minimal improvement to overall IM peak capacity or resolution compared to nitrogen or helium [23, 135, 150–152]. For example, Fjeldsted and coworkers investigated the separation of various small molecule pesticides, isomeric carbohydrates, and fluoroalkyl phosphazenes in a wide variety of drift gases, including helium, nitrogen, argon, carbon dioxide, nitrous oxide, and sulfur hexafluoride [23]. Generally, it was observed that helium and nitrogen had the highest resolution and resolving power, with some of the more polarizable drift gases demonstrating better selectivity when comparing certain analyte pairs [23]. To fully evaluate drift gas effect, a broader range of masses and biological classes needs to be reported in a variety of drift gases across multiple platforms and laboratories. Recently, Morris et al. utilized DTIMS operated in a variety of drift gases (helium, nitrogen, argon, and carbon dioxide) to study several different classes of compounds (quaternary ammoniums, phosphazenes, polypeptides, and carbohydrates) and provided recommended experimental parameters that allow for comparable CCS values to be determined among multiple laboratories [153]. Currently, the majority of CCS measurements have been reported in either helium or nitrogen, hindering the evaluation of IM separation performance in other alternate drift gases [16]. As the methods and parameters to perform these variable drift gas experiments become standardized, normalized measurements such as CCS will be vital for allowing direct comparison of separations conducted on different platforms (e.g., drift tube or traveling wave) [16]. The potential analytical importance of drift gas composition on improving IM resolution and separation selectivity is significant, and currently this area of research is largely unexplored.
7. Accelerating Advancements in Integrated Omics Analyses
When utilized in various omics studies, IM has been used primarily to partition analytes of interest from chemical noise originating from complex samples, resolve ambiguities within coeluting features, and align precursor and fragmentation data acquired in data-independent strategies. While the majority of IM-MS applications in biomolecular analysis have focused on specific omic fields (e.g., proteomics, lipidomics, metabolomics), there is increasing interest in utilizing IM-MS for untargeted, multiomic studies which examine a large breadth of molecule types simultaneously [154]. A truly, multiomic analytical workflow will facilitate the development of system maps which connect relationships between molecule types and allow perturbed pathways to be elucidated. To illustrate this concept, Fig. 12 displays work from Paglia et al. on building pathway maps to track metabolites being shuttled between mitochondria and the cytosol [155]. An area that could significantly benefit from this type of analysis is the microbiome and chemical communications fields. The microbiome has experienced recent and significant attention aimed at understanding the integral role that commensal bacteria plays on human health [156–158]. This focus, in large part, is due to advancements in sequencing of bacterial communities allowing for whole populations to be analyzed simultaneously, facilitating the comparison of healthy versus disease states [157, 158]. One challenge that remains in microbiome research is the understanding of the mechanisms that lead to disease, which can be addressed at least in part by building biochemical inventories of small molecule metabolites observed within the samples. MS-based metabolomics can be applied to a variety of sample types, can measure thousands of metabolites, requires minute sample quantities, and thus has high potential for integration into multiomics investigations. In particular, the ability of IM-MS experiments to simultaneously separate and detect multiple biological classes and chemical motifs can allow perturbed metabolites to be observed along with changes in the bacterial community. Uncovering changes in the metabolomic profile can provide insight into the role that particular bacteria plays within the complex interplay of biomolecules associated with the microbiome and/or host organisms.
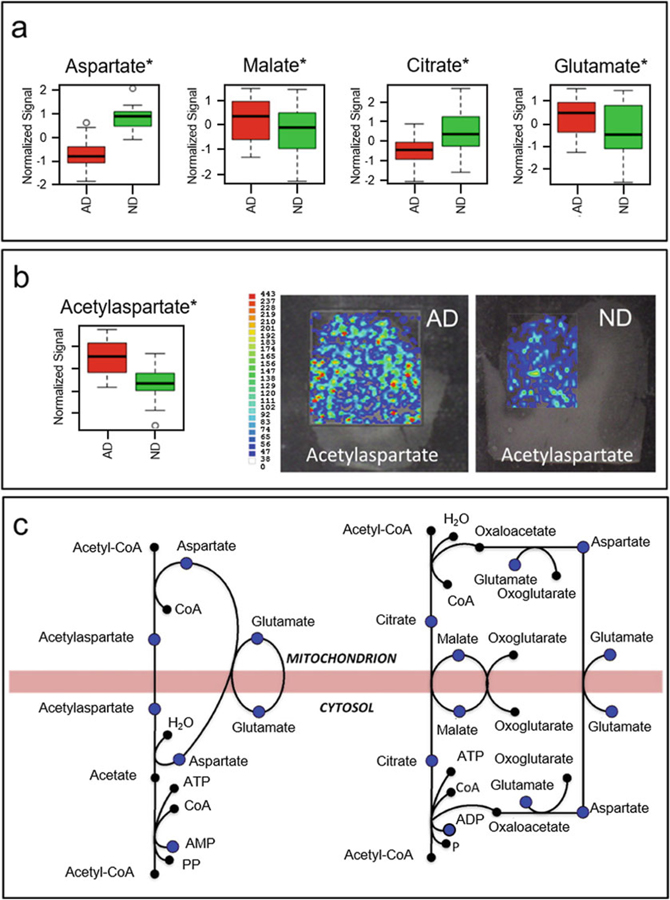
Fig. 12.
(a) Bar charts summarizing the normalized ion signal observed in IM-MS experiments for aspartate, malate, citrate, and glutamate obtained from AD (red) and ND control subjects (green). (b) N-acetylaspartate (NAA) levels between AD (red) and control subjects (green) and MS images obtained from brain sections of both AD and control subjects. (c) Mitochondrial shuttles describing the exchange of metabolites between the cytosol and mitochondria and the metabolites quantified in the IM-MS experiments (blue dots) of Paglia et al. *p < 0.05 (t test) (Figure from Paglia et al. [155])
Throughout this chapter, biomolecular analysis by various IM-MS technologies has been highlighted. Currently, IM is used primarily to enhance the separation of molecules present within complex samples. However, with higher quality IM measurements now being reported throughout the community [43], the role of IM for supporting biomolecular identifications is poised to make a significant impact in the omics sciences, particularly within the metabolomics and lipidomics communities. As technological and experimental advances are continuing at an accelerated pace, IM-MS will play an increasingly important role in providing deeper levels of information in large-scale biomolecular analysis initiatives including those in systems, synthetic, and chemical biology.
Acknowledgements
This work was supported in part using the resources of the Center for Innovative Technology at Vanderbilt University. The authors gratefully acknowledge financial support for this work provided by the National Institutes of Health (NIH NIGMS R01GM092218 and NCI R03CA222452) and the U.S. Environmental Protection Agency under Assistance Agreement No. 83573601. This work has not been formally reviewed by the EPA. The views expressed in this document are solely those of the authors and do not necessarily reflect those of the funding agencies and organizations. The U.S. Government does not endorse any products or commercial services mentioned in this publication.
References
- 1.Thomson JJ, Rutherford E (1896) On the passage of electricity through gases exposed to Roentgen rays. Phil Mag Ser 5 42 (258):392–407 [Google Scholar]
- 2.Tyndall AM, Powell CF (1930) The mobility of ions in pure gases. Proc R Soc Lond Ser A 129(809):162–180 [Google Scholar]
- 3.Tyndall AM, Starr LH, Powell CF (1928) The mobility of ions in air. Part IV. Investigations by two new methods. Proc R Soc Lond Ser A 121(787):172–184 [Google Scholar]
- 4.Barnes WS, Martin DW, McDaniel EW (1961) Mass spectrographic identification of the ion observed in hydrogen mobility experiments. Phys Rev Lett 6(3):110–111 [Google Scholar]
- 5.May JC, McLean JA (2015) Ion mobility-mass spectrometry: time-dispersive instrumentation. Anal Chem 87(3):1422–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dole M, Mack LL, Hines RL, Mobley RC, Ferguson LD, Alice MB (1968) Molecular beams of macroions. J Chem Phys 49 (5):2240–2249 [Google Scholar]
- 7.Dole M, Hines RL, Mack LL, Mobley RC, Ferguson LD, Alice MB (1968) Gas phase macroions. Macromolecules 1(1):96–97 [Google Scholar]
- 8.Lubman DM, Kronick MN (1982) Plasma chromatography with laser-produced ions. Anal Chem 54(9):1546–1551 [Google Scholar]
- 9.von Helden G, Wyttenbach T, Bowers MT (1995) Inclusion of a MALDI ion source in the ion chromatography technique: conformational information on polymer and biomolecular ions. Int J Mass Spectrom Ion Process 146–147:349–364 [Google Scholar]
- 10.Wittmer D, Chen YH, Luckenbill BK, Hill HH Jr (1994) Electrospray ionization ion mobility spectrometry. Anal Chem 66 (14):2348–2355 [Google Scholar]
- 11.Clemmer DE, Jarrold MF (1997) Ion mobility measurements and their applications to clusters and biomolecules. J Mass Spectrom 32(6):577–592 [Google Scholar]
- 12.Hoaglund CS, Valentine SJ, Sporleder CR, Reilly JP, Clemmer DE (1998) Three-dimensional ion mobility/TOFMS analysis of electrosprayed biomolecules. Anal Chem 70(11):2236–2242 [DOI] [PubMed] [Google Scholar]
- 13.Henderson SC, Valentine SJ, Counterman AE, Clemmer DE (1999) ESI/ion trap/ion mobility/time-of-flight mass spectrometry for rapid and sensitive analysis of biomolecular mixtures. Anal Chem 71(2):291–301 [DOI] [PubMed] [Google Scholar]
- 14.Gillig KJ, Ruotolo B, Stone EG, Russell DH, Fuhrer K, Gonin M, Schultz AJ (2000) Coupling high-pressure MALDI with ion mobility/orthogonal time-of-flight mass spectrometry. Anal Chem 72(17):3965–3971 [DOI] [PubMed] [Google Scholar]
- 15.Smith RD, Loo JA, Loo RRO, Busman M, Udseth HR (1991) Principles and practice of electrospray ionization–mass spectrometry for large polypeptides and proteins. Mass Spectrom Rev 10(5):359–452 [Google Scholar]
- 16.May JC, Morris CB, McLean JA (2017) Ion mobility collision cross section compendium. Anal Chem 89(2):1032–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giles K, Williams JP, Campuzano I (2011) Enhancements in travelling wave ion mobility resolution. Rapid Commun Mass Spectrom 25(11):1559–1566 [DOI] [PubMed] [Google Scholar]
- 18.Giles K, Pringle SD, Worthington KR, Little DLW, Bateman RH (2004) Applications of a traveling wave-based radio-frequency-only stacked ring ion guide. Rapid Commun Mass Spectrom 18(20):2401–2414 [DOI] [PubMed] [Google Scholar]
- 19.Pringle SD, Giles K, Wildgoose JL, Williams JP, Slade SE, Thalassinos K, Bateman RH, Bowers MT, Scrivens JH (2007) An investigation of the mobility separation of some peptide and protein ions using a new hybrid quadrupole/travelling wave IMS/oa-ToF instrument. Int J Mass Spectrom 261 (1):1–12 [Google Scholar]
- 20.May JC, Goodwin CR, Lareau NM, Leaptrot KL, Morris CB, Kurulugama RT, Mordehai A, Klein C, Barry W, Darland E, Overney G, Imatani K, Stafford GC, Fjeldsted JC, McLean JA (2014) Conformational ordering of biomolecules in the gas phase: nitrogen collision cross sections measured on a prototype high resolution drift tube ion mobility-mass spectrometer. Anal Chem 86 (4):2107–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Silveira JA, Ridgeway ME, Park MA (2014) High resolution trapped ion mobility spectrometry of peptides. Anal Chem 86 (12):5624–5627 [DOI] [PubMed] [Google Scholar]
- 22.Zhang F, Guo S, Zhang MY, Zhang ZX, Guo YL (2015) Characterizing ion mobility and collision cross section of fatty acids using electrospray ion mobility mass spectrometry. J Mass Spectrom 50(7):906–913 [DOI] [PubMed] [Google Scholar]
- 23.Kurulugama RT, Darland E, Kuhlmann F, Stafford G, Fjeldsted J (2015) Evaluation of drift gas selection in complex sample analyses using a high performance drift tube ion mobility-QTOF mass spectrometer. Analyst 14(20):6834–6844 [DOI] [PubMed] [Google Scholar]
- 24.Paglia G, Williams JP, Menikarachchi L, Thompson JW, Tyldesley-Worster R, Halldorsson S, Rolfsson O, Moseley A, Grant D, Langridge J, Palsson BO, Astarita G (2014) Ion mobility derived collision cross sections to support metabolomics applications. Anal Chem 86(8):3985–3993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allen SJ, Schwartz AM, Bush MF (2013) Effects of polarity on the structures and charge states of native-like proteins and protein complexes in the gas phase. Anal Chem 85(24):12055–12061 [DOI] [PubMed] [Google Scholar]
- 26.Campuzano I, Bush MF, Robinson CV, Beaumont C, Richardson K, Kim H, Kim HI (2012) Structural characterization of drug-like compounds by ion mobility mass spectrometry: comparison of theoretical and experimentally derived nitrogen collision cross sections. Anal Chem 84(2):1026–1033 [DOI] [PubMed] [Google Scholar]
- 27.Bush MF, Campuzano IDG, Robinson CV (2012) Ion mobility mass spectrometry of peptide ions: effects of drift gas and calibration strategies. Anal Chem 84 (16):7124–7130 [DOI] [PubMed] [Google Scholar]
- 28.Shah AR, Agarwal K, Baker ES, Singhal M, Mayampurath AM, Ibrahim YM, Kangas LJ, Monroe ME, Zhao R, Belov ME, Anderson GA, Smith RD (2010) Machine learning based prediction for peptide drift times in ion mobility spectrometry. Bioinformatics 26 (13):1601–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McLean JA (2009) The mass-mobility correlation redux: the conformational landscape of anhydrous biomolecules. J Am Soc Mass Spectrom 20(10):1775–1781 [DOI] [PubMed] [Google Scholar]
- 30.Fenn LS, McLean JA (2008) Biomolecular structural separations by ion-mobility-mass spectrometry. Anal Bioanal Chem 391 (3):905–909 [DOI] [PubMed] [Google Scholar]
- 31.Fenn LS, Kliman M, Mahsut A, Zhao SR, McLean JA (2009) Characterizing ion mobility-mass spectrometry conformation space for the analysis of complex biological samples. Anal Bioanal Chem 394 (1):235–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hines KM, May JC, McLean JA, Xu LB (2016) Evaluation of collision cross section calibrants for structural analysis of lipids by traveling wave ion mobility-mass spectrometry. Anal Chem 88(14):7329–7336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leaptrot KL, May JC, Dodds JN, McLean JA (2019) Ion mobility conformational lipid atlas for high confidence lipidomics. Nat Commun 10(1):985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baker ES, Burnum-Johnson KE, Jacobs JM, Diamond DL, Brown RN, Ibrahim YM, Orton DJ, Piehowski PD, Purdy DE, Moore RJ, Danielson WF, Monroe ME, Crowell KL, Slysz GW, Gritsenko MA, Sandoval JD, LaMarche BL, Matzke MM, Webb-Robertson B-JM, Simons BC, McMahon BJ, Bhattacharya R, Perkins JD, Carithers RL, Strom S, Self SG, Katze MG, Anderson GA, Smith RD (2014) Advancing the high throughput identification of liver fibrosis protein signatures using multiplexed ion mobility spectrometry. Mol Cell Proteomics 13 (4):1119–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu X, Plasencia M, Ragg S, Valentine SJ, Clemmer DE (2004) Development of high throughput dispersive LC-ion mobility-TOFMS techniques for analysing the human plasma proteome. Brief Funct Genomics Proteomics 3(2):177–186 [DOI] [PubMed] [Google Scholar]
- 36.Poad BLJ, Zheng XY, Mitchell TW, Smith RD, Baker ES, Blanksby SJ (2018) Online ozonolysis combined with ion mobility-mass spectrometry provides a new platform for lipid isomer analyses. Anal Chem 90(2):1292–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harris RA, May JC, Stinson CA, Xia Y, McLean JA (2018) Determining double bond position in lipids using online ozonolysis coupled to liquid chromatography and ion mobility-mass spectrometry. Anal Chem 90(3):1915–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valentine SJ, Counterman AE, Clemmer DE (1999) A database of 660 peptide ion cross sections: use of intrinsic size parameters for bona fide predictions of cross sections. J Am Soc Mass Spectrom 10(11):1188–1211 [DOI] [PubMed] [Google Scholar]
- 39.Tao L, McLean JR, McLean JA, Russell DH (2007) A collision cross-section database of singly-charged peptide ions. J Am Soc Mass Spectrom 18(9):1727–1728 [DOI] [PubMed] [Google Scholar]
- 40.Dilger JM, Valentine SJ, Glover MS, Ewing MA, Clemmer DE (2012) A database of alkali metal-containing peptide cross sections: influence of metals on size parameters for specific amino acids. Int J Mass Spectrom 330–332:35–45 [Google Scholar]
- 41.Dilger JM, Valentine SJ, Glover MS, Clemmer DE (2013) A database of alkaline-earth-coordinated peptide cross sections: insight into general aspects of structure. J Am Soc Mass Spectrom 24(5):768–779 [DOI] [PubMed] [Google Scholar]
- 42.Paglia G, Angel P, Williams JP, Richardson K, Olivos HJ, Thompson JW, Menikarachchi L, Lai S, Walsh C, Moseley A, Plumb RS, Grant DF, Palsson BO, Langridge J, Geromanos S, Astarite G (2015) Ion mobility-derived collision cross section as an additional measure for lipid fingerprinting and identification. Anal Chem 87(2):1137–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Picache JA, Rose BS, Balinski A, Leaptrot KL, Sherrod SD, May JC, McLean JA (2019) Collision cross section compendium to annotate and predict multi-omic compound identities. Chem Sci 10(4):983–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ellis HW, Thackston MG, McDaniel EW, Mason EA (1984) Transport properties of gaseous ions over a wide energy range. Part III. At Data Nucl Data Tables 31(1):113–151 [Google Scholar]
- 45.Deng LL, Ibrahim YM, Baker ES, Aly NA, Hamid AM, Zhang X, Zheng XY, Garimella SVB, Webb IK, Prost SA, Sandoval JA, Norheim RV, Anderson GA, Tolmachev AV, Smith RD (2016) Ion mobility separations of isomers based upon long path length structures for lossless ion manipulations combined with mass spectrometry. Chem Select 1 (10):2396–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dodds JN, May JC, McLean JA (2017) Investigation of the complete suite of the leucine and isoleucine isomers: toward prediction of ion mobility separation capabilities. Anal Chem 89(1):952–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dwivedi P, Bendiak B, Clowers BH, Hill HH Jr (2007) Rapid resolution of carbohydrate isomers by electrospray ionization ambient pressure ion mobility spectrometry-time-of-flight mass spectrometry (ESI-APIMS-TOFMS). J Am Soc Mass Spectrom 18(7):1163–1175 [DOI] [PubMed] [Google Scholar]
- 48.Fenn LS, McLean JA (2011) Structural resolution of carbohydrate positional and structural isomers based on gas-phase ion mobility-mass spectrometry. Phys Chem Chem Phys 13(6):2196–2205 [DOI] [PubMed] [Google Scholar]
- 49.Groessl M, Graf S, Knochenmuss R (2015) High resolution ion mobility-mass spectrometry for separation and identification of isomeric lipids. Analyst 14(20):6904–6911 [DOI] [PubMed] [Google Scholar]
- 50.Lalli PM, Corilo YE, Fasciotti M, Riccio MF, de Sa GF, Daroda RJ, Souza GHMF, McCullagh M, Bartberger MD, Eberlin MN, Campuzano IDG (2013) Baseline resolution of isomers by traveling wave ion mobility mass spectrometry: investigating the effects of polarizable drift gases and ionic charge distribution. J Mass Spectrom 48(9):989–997 [DOI] [PubMed] [Google Scholar]
- 51.Valentine SJ, Kulchania M, Barnes CAS, Clemmer DE (2001) Multidimensional separations of complex peptide mixtures: a combined high-performance liquid chromatography/ion mobility/time-of-flight mass spectrometry approach. Int J Mass Spectrom 212(1–3):97–109 [Google Scholar]
- 52.Matz LM, Dion HM, Hill HH Jr (2002) Evaluation of capillary liquid chromatography–electrospray ionization ion mobility spectrometry with mass spectrometry detection. J Chromatogr A 946(1–2):59–68 [DOI] [PubMed] [Google Scholar]
- 53.Lareau NM, May JC, McLean JA (2015) Non-derivatized glycan analysis by reverse phase liquid chromatography and ion mobility-mass spectrometry. Analyst 140 (10):3335–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nichols CM, May JC, Sherrod SD, McLean JA (2018) Automated flow injection method for the high precision determination of drift tube ion mobility collision cross sections. Analyst 143(7):1556–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gerhardt N, Schwolow S, Rohn S, Perez-Cacho PR, Galan-Soldevilla H, Arce L, Weller P (2019) Quality assessment of olive oils based on temperature-ramped HS-GC-IMS and sensory evaluation: comparison of different processing approaches by LDA, kNN, and SVM. Food Chem 278:720–728 [DOI] [PubMed] [Google Scholar]
- 56.Donato P, Giuffrida D, Oteri M, Inferrera V, Dugo P, Mondello L (2018) Supercritical fluid chromatography x ultra-high pressure liquid chromatography for red chilli pepper fingerprinting by photodiode array, quadrupole-time-of-flight and ion mobility mass spectrometry (SFC x RP-UHPLC-PDA-Q-ToF MS-IMS). Food Anal Methods 11 (12):3331–3341 [Google Scholar]
- 57.Hill HH, Stlouis RH, Morrissey MA, Shumate CB, Siems WF, McMinn DG (1992) A detection method for unified chromatography-ion mobility monitoring. J High Resol Chromatogr 15(7):417–422 [Google Scholar]
- 58.Hillenkamp F, Karas M, Beavis RC, Chait BT (1991) Matrix-assisted laser desorption/ionization mass spectrometry of biopolymers. Anal Chem 63(24):1193A–1203A [DOI] [PubMed] [Google Scholar]
- 59.Karas M, Bahr U, Gießmann U (1991) Matrix-assisted laser desorption ionization mass spectrometry. Mass Spectrom Rev 10 (5):335–357 [Google Scholar]
- 60.Stoeckli M, Chaurand P, Hallahan DE, Caprioli RM (2001) Imaging mass spectrometry: a new technology for the analysis of protein expression in mammalian tissues. Nat Med 7(4):493. [DOI] [PubMed] [Google Scholar]
- 61.Cornett DS, Reyzer ML, Chaurand P, Caprioli RM (2007) MALDI imaging mass spectrometry: molecular snapshots of biochemical systems. Nat Methods 4(10):828. [DOI] [PubMed] [Google Scholar]
- 62.McLean JA, Ridenour WB, Caprioli RM (2007) Profiling and imaging of tissues by imaging ion mobility-mass spectrometry. J Mass Spectrom 42(8):1099–1105 [DOI] [PubMed] [Google Scholar]
- 63.Jackson SN, Ugarov M, Egan T, Post JD, Langlais D, Albert Schultz J, Woods AS (2007) MALDI-ion mobility-TOFMS imaging of lipids in rat brain tissue. J Mass Spectrom 42(8):1093–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM (1989) Electrospray ionization for mass spectrometry of large biomolecules. Science 246(4926):64–71 [DOI] [PubMed] [Google Scholar]
- 65.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM (1990) Electrospray ionization–principles and practice. Mass Spectrom Rev 9(1):37–70 [Google Scholar]
- 66.Belov ME, Gorshkov MV, Udseth HR, Anderson GA, Smith RD (2000) Zeptomole-sensitivity electrospray ionization fourier transform ion cyclotron resonance mass spectrometry of proteins. Anal Chem 72(10):2271–2279 [DOI] [PubMed] [Google Scholar]
- 67.Hardman M, Makarov AA (2003) Interfacing the orbitrap mass analyzer to an electrospray ion source. Anal Chem 75(7):1699–1705 [DOI] [PubMed] [Google Scholar]
- 68.Tang XJ, Thibault P, Boyd RK (1993) Fragmentation reactions of multiply-protonated peptides and implications for sequencing by tandem mass spectrometry with low-energy collision-induced dissociation. Anal Chem 65(20):2824–2834 [DOI] [PubMed] [Google Scholar]
- 69.Zubarev RA, Kelleher NL, McLafferty FW (1998) Electron capture dissociation of multiply charged protein cations. A nonergodic process. J Am Chem Soc 120(13):3265–3266 [Google Scholar]
- 70.Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF (2004) Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Natl Acad Sci 101(26):9528–9533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dodds JN, May JC, McLean JA (2017) Correlating resolving power, resolution, and collision cross section: unifying cross-platform assessment of separation efficiency in ion mobility spectrometry. Anal Chem 89 (22):12176–12184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mason EA, McDaniel EW (1988) Transport properties of ions in gases. John Wiley & Sons, New York, NY, p 560 [Google Scholar]
- 73.Siems WF, Viehland LA, Hill HH Jr (2012) Improved momentum-transfer theory for ion mobility. 1. Derivation of the fundamental equation. Anal Chem 84(22):9782–9791 [DOI] [PubMed] [Google Scholar]
- 74.Bush MF, Hall Z, Giles K, Hoyes J, Robinson CV, Ruotolo BT (2010) Collision cross sections of proteins and their complexes: a calibration framework and database for gas-phase structural biology. Anal Chem 82 (22):9557–9565 [DOI] [PubMed] [Google Scholar]
- 75.Deng LL, Ibrahim YM, Hamid AM, Garimella SVB, Webb IK, Zheng XY, Prost SA, Sandoval JA, Norheim RV, Anderson GA, Tolmachev AV, Baker ES, Smith RD (2016) Ultra-high resolution ion mobility separations utilizing traveling waves in a 13 m serpentine path length structures for lossless ion manipulations module. Anal Chem 88 (18):8957–8964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.May JC, McLean JA (2016) Advanced multidimensional separations in mass spectrometry: navigating the big data deluge. Ann Rev Anal Chem 9:387–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.May JC, Goodwin CR, McLean JA (2011) Gas-phase ion mobility-mass spectrometry (IM-MS) and tandem IM-MS/MS strategies for metabolism studies and metabolomics. Encyclopedia Drug Metab Interact. 10.1002/9780470921920.edm099 [DOI]
- 78.Kolli V, Schumacher KN, Dodds ED (2017) Ion mobility-resolved collision-induced dissociation and electron transfer dissociation of N-glycopeptides: gathering orthogonal connectivity information from a single mass-selected precursor ion population. Analyst 142(24):4691–4702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lermyte F, Verschueren T, Brown JM, Williams JP, Valkenborg D, Sobott F (2015) Characterization of top-down ETD in a travelling-wave ion guide. Methods 89:22–29 [DOI] [PubMed] [Google Scholar]
- 80.Williams JP, Pringle S, Richardson K, Gethings L, Vissers JPC, De Cecco M, Houel S, Chakraborty AB, Yu YQ, Chen WB, Brown JM (2013) Characterisation of glycoproteins using a quadrupole time-of-flight mass spectrometer configured for electron transfer dissociation. Rapid Commun Mass Spectrom 27(21):2383–2390 [DOI] [PubMed] [Google Scholar]
- 81.Ruotolo BL, Benesch JLP, Sandercock AM, Hyung S, Robinson C (2008) Ion mobility-mass spectrometry analysis of large protein complexes. Nat Protoc 3:1139–1152 [DOI] [PubMed] [Google Scholar]
- 82.McLean JA, Ruotolo BT, Gillig KJ, Russel DH (2005) Ion mobility-mass spectrometry: a new paradigm for proteomics. Int J Mass Spectrom 240(3):301–315 [Google Scholar]
- 83.Paglia G, Astarita G (2017) Metabolomics and lipidomics using traveling-wave ion mobility mass spectrometry. Nat Protoc 12:797–813 [DOI] [PubMed] [Google Scholar]
- 84.Zhang X, Quinn K, Cruickshank-Quinn C, Reisdorph R, Reisdorph N (2018) The application of ion mobility mass spectrometry to metabolomics. Curr Opin Chem Biol 42:60–66 [DOI] [PubMed] [Google Scholar]
- 85.May JC, Goodwin CR, McLean JA (2015) Ion mobility mass spectrometry strategies for untargeted systems, synthetic, and chemical biology. Curr Opin Biotechnol 31:117–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dunn WB, Broadhurst D, Begley P, Zelena E, Francis-McIntyre S, Anderson N, Brown M, Knowles JD, Halsall A, Haselden JN, Nicholls AW, Wilson ID, Kell DB, Goodacre R, The Human Serum Metabolome Consortium (2011) Procedures for large-scale metabolic profiling of seruma and plasma using gas chromatography and liquid chromatraphy coupled to mass spectrometry. Nat Protoc 6:1060–1083 [DOI] [PubMed] [Google Scholar]
- 87.Hoaglund-Hyzer CS, Li J, Clemmer DE (2000) Mobility labeling for parallel CID of ion mixtures. Anal Chem 72(13):2737–2740 [DOI] [PubMed] [Google Scholar]
- 88.Taguchi F, Solomon B, Gregorc V, Roder H, Gray R, Kashara K, Nisho M, Brahmer J, Spreafico A, Ludovini V, Massion PP, Dziadziuszho R, Schiller J, Grigorieva J, Tsypin M, Hunsucker SW, Caprioli R, Duncan MW, Hirsch FR, Bunn PA, Carbone DP (2007) Mass spectrometry to classify non-small-lung cell lung cancer patients for clinical outcome after treatment with epidermal growth factor receptor tyrosine kinase inhibitors: a multicohort cross-institutional study. J Natl Cancer Instit 99(11):838–846 [DOI] [PubMed] [Google Scholar]
- 89.Mori H, Takio K, Ogawara M, Selkoe DJ (1992) Mass spectrometry of purified amyloid beta protein in Alzheimer’s disease. J Biochem Chem 267:17082–17086 [PubMed] [Google Scholar]
- 90.Djidja MC, Francese S, Loadman PM, Sutton CW, Scriven P, Claude E, Snel MF, Franck J, Salzet M, Clench MR (2009) Detergent addition to tryptic digests and ion mobility separation prior to MS/MS improves peptide yield and protein identification for in situ proteomics investigation of frozen and formalin-fixed paraffin-embedded adenocarcinoma tissue sections. Proteomics 9(10):2750–2763 [DOI] [PubMed] [Google Scholar]
- 91.Moon MH, Myung S, Plasencia M, Hilderbrand AE, Clemmer DE (2003) Nanoflow LC/Ion mobility/CID/TOF for proteomics: analysis of a human urinary proteome. J Proteome Res 2(6):589–597 [DOI] [PubMed] [Google Scholar]
- 92.Thalassinos K, Grabenauer M, Slade SE, Hilton GR, Bowers MT, Scrivens JH (2009) Characterization of phosphorylated peptides using traveling wave-based and drift cell ion mobility mass spectrometry. Anal Chem 81(1):248–254 [DOI] [PubMed] [Google Scholar]
- 93.Zhong Y, Hyung S, Ruotolo BT (2012) Ion mobility-mass spectrometry for structual proteomics. Expert Rev Proteomics 9(1):47–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.May JC, McLean JA (2015) A uniform field ion mobility study of melittin and implications of low-field mobility for resolving fine cross-sectional detail in peptide and protein experiments. Proteomics 15(16):2862–2871 [DOI] [PubMed] [Google Scholar]
- 95.Shliaha PV, Bond NJ, Gatto L, Liley KS (2013) Effects of traveling wave ion mobility separation of data independent acquistion in proteomics studies. J Proteome Res 12 (6):2323–2339 [DOI] [PubMed] [Google Scholar]
- 96.Wang G, Abzalimov RR, Kaltashov IA (2011) Direct monitoring of heat-stressed biopolymers with temperature-controlled electrospray ionization mass spectrometry. Anal Chem 83(8):2870–2876 [DOI] [PubMed] [Google Scholar]
- 97.El-Baba TJ, Woodall DW, Raab SA, Fuller DR, Langanowsky A, Russell DH, Clemmer DE (2017) Melting Proteins: evidence for multiple stable structures upon thermal denaturation of native ubiquitin from ion mobility spectrometry-mass spectrometry measurements. J Am Chem Soc 139(18):6306–6309 [DOI] [PubMed] [Google Scholar]
- 98.Han X, Yang K, Gross RW (2011) Multidimensional mass spectrometry-based shot-gun lipidomics and novel strategies for lipidomic analysis. Mass Spectrom Rev 31 (1):134–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kliman M, May JC, McLean JA (2011) Lipid analysis and lipidomics by structurally selective ion mobility-mass spectrometry. Biochim Biophys Acta 1811(11):935–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Paglia G, Kliman M, Claude E, Geromanos S, Astarita G (2015) Application of ion-mobility mass spectrometry forl lipids. Anal Bioanal Chem 407(17):4995–5007 [DOI] [PubMed] [Google Scholar]
- 101.Grossel M, Graf S, Knochenmuss R (2015) High resolution ion mobility-mass spectrometry for separation and identification of isomeric lipids. Analyst 140(20):6904–6911 [DOI] [PubMed] [Google Scholar]
- 102.Di Giovanni JP, Barkley RM, Jones DNM, Hankins JA, Murphy RC (2018) Tandem mass spectrometry and ion mobility reveals structural insight into eicosanoid product ion formation. J Am Soc Mass Spectrom 29 (6):1231–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Paglia G, Shrestha B, Astarita G (2017) Ion-mobility mass spectrometry for lipidomics applications. In: Wood P (ed) Lipidomics. Humana, New York, NY, pp 61–79 [Google Scholar]
- 104.Thomas MC, Mitchell TW, Harman DG, Deeley JM, Murphy RC, Blanksby SJ (2007) Elucidation of double bond position in unsaturated lipids by ozone electrospray ionization mass spectrometry. Anal Chem 79 (13):5013–5022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sun C, Zhao Y, Curtis JM (2014) Elucidation of phosphatidylcholine isomers using two dimensional liquid chromatography coupled inline with ozonolysis mass spectrometry. J Chromatogr A 1351:37–45 [DOI] [PubMed] [Google Scholar]
- 106.Dettmer K, Aronov PA, Hammock BD (2007) Mass spectrometry-based metabolomics. Mass Spectrom Rev 26(1):51–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhang X, Quinn K, Cruickshank-Quinn C, Reisdorph R, Reisdorph N (2018) Applications of ion mobility mass spectrometry to metabolomics. Curr Opin Chem Biol 42:60–66 [DOI] [PubMed] [Google Scholar]
- 108.Sinclair E, Hollywood KA, Yan C, Blankley R, Rainer B, Barran P (2018) Mobilising ion mobility mass spectrometry for metabolomics. Analyst 19:4783–4788 [DOI] [PubMed] [Google Scholar]
- 109.Schrimpe-Rutledge AC, Codreanu SG, Sherrod SD, McLean JA (2016) Untargeted metabolomics strategies—challenges and emerging directions. J Am Soc Mass Spectrom 27(12):1897–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.May JC, Gant-Branum RL, McLean JA (2016) Targeting the untargeted in molecular phenomics with structurally-selective ion mobility-mass spectrometry. Curr Opin Biotechnol 39:192–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nichols CM, Dodds JN, Rose BS, Picache JA, Morris CB, Codreanu SG, May JC, Sherrod SD, McLean JA (2018) Untargeted molecular discovery in primary metabolism: collision cross section as a molecular descriptor in ion mobility-mass spectrometry. Anal Chem 90 (24):14484–14492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wickramasekara SI, Zandkarimi F, Morré J, Kirkwood J, Legette L, Jiang Y, Gombart AF, Stevens JF, Maier CS (2013) Electrospray quadrupole travelling wave ion mobility time-of-flight mass spectrometry for the detection of plasma metabolome changes caused by xanthohumol in obese zucker (fa/fa) rats. Meta 3:701–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Stow MS, Causon TJ, Zheng X, Kurulugama RT, Mairinger T, May JC, Rennie EE, Smith RD, McLean JA, Hann S, Fjeldsted JC (2017) An interlaboratory evaluation of drift tube ion mobility-mass spectrometry collision cross section measurements. Anal Chem 89 (17):9048–9055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Regueiro J, Negreira N, Berntssen MH (2016) Ion-mobility-derived collision cross section as an additional identification point for multiresidue screening of pesticides in fish feed. Anal Chem 88(22):11169–11177 [DOI] [PubMed] [Google Scholar]
- 115.Stephan S, Hippler J, Köhler T, Deeb AA, Schmidt TC, Schmitz OJ (2016) Contaminant screening of wastewater with HPLC-IM-qTOF-MS and LC+ LC-IM-qTOF-MS using a CCS database. Anal Bioanal Chem 408(24):6545–6555 [DOI] [PubMed] [Google Scholar]
- 116.Zheng X, Aly NA, Zhou Y, Dupuis KT, Bilbao A, Paurus VL, Orton DJ, Wilson R, Payne SH, Smith RD, Baker ES (2017) A structural examination and collision cross section database for over 500 metabolites and xenobiotics using drift tube ion mobility spectrometry. Chem Sci 8(11):7724–7736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hernández-Mesa M, Le Bizec B, Monteau F, García-Campaña AM, Dervilly-Pinel G (2018) Collision cross section (CCS) database: an additional measure to characterize steroids. Anal Chem 90(7):4616–4625 [DOI] [PubMed] [Google Scholar]
- 118.Chen TC, Ibrahim YM, Webb IK, Garimella SVB, Zhang X, Hamid AM, Deng LL, Karnesky WE, Prost SA, Sandoval JA, Norheim RV, Anderson GA, Tolmachev AV, Baker ES, Smith RD (2016) Mobility-selected ion trapping and enrichment using structures for lossless ion manipulations. Anal Chem 88(3):1728–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ibrahim YM, Hamid AM, Deng LL, Garimella SVB, Webb IK, Baker ES, Smith RD (2017) New frontiers for mass spectrometry based upon structures for lossless ion manipulations. Analyst 142(7):1010–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang XY, Garimella SVB, Prost SA, Webb IK, Chen TC, Tang KQ, Tolmachev AV, Norheim RV, Baker ES, Anderson GA, Ibrahim YM, Smith RD (2015) Ion trapping, storage, and ejection in structures for lossless ion manipulations. Anal Chem 87 (12):6010–6016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chouinard CD, Nagy G, Webb IK, Shi TJ, Baker ES, Prost SA, Liu T, Ibrahim YM, Smith RD (2018) Improved sensitivity and separations for phosphopeptides using online liquid chromatography coupled with structures for lossless ion manipulations ion mobility-mass spectrometry. Anal Chem 90 (18):10889–10896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Garimella SVB, Ibrahim YM, Webb IK, Tolmachev AV, Zhang XY, Prost SA, Anderson GA, Smith RD (2014) Simulation of electric potentials and ion motion in planar electrode structures for lossless ion manipulations (SLIM). J Am Soc Mass Spectrom 25 (11):1890–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Webb IK, Garimella SVB, Tolmachev AV, Chen TC, Zhang XY, Cox JT, Norheim RV, Prost SA, LaMarche B, Anderson GA, Ibrahim YM, Smith RD (2014) Mobility-resolved ion selection in uniform drift field ion mobility spectrometry/mass spectrometry: dynamic switching in structures for lossless ion manipulations. Anal Chem 86 (19):9632–9637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Deng LL, Webb IK, Garimella SVB, Hamid AM, Zheng XY, Norheim RV, Prost SA, Anderson GA, Sandoval JA, Baker ES, Ibrahim YM, Smith RD (2017) Serpentine ultralong path with extended routing (SUPER) high resolution traveling wave ion mobility-MS using structures for lossless ion manipulations. Anal Chem 89(8):4628–4634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Garimella SVB, Ibrahim YM, Webb IK, Ipsen AB, Chen TC, Tolmachev AV, Baker ES, Anderson GA, Smith RD (2015) Ion manipulations in structures for lossless ion manipulations (SLIM): computational evaluation of a 90 degrees turn and a switch. Analyst 14 (20):6845–6852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Fernandez-Lima FA, Kaplan DA, Suetering J, Park MA (2011) Gas-phase separation using a trapped ion mobility spectrometer. Int J Ion Mobil Spectrom 14(2–3):93–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Adams KJ, Montero D, Aga D, Fernandez-Lima F (2016) Isomer separation of polybrominated diphenyl ether metabolites using nanoESI-TIMS-MS. Int J Ion Mobil Spectrom 19(2–3):69–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Michelmann K, Silveira JA, Ridgeway ME, Park MA (2015) Fundamentals of trapped ion mobility spectrometry. J Am Soc Mass Spectrom 26(1):14–24 [DOI] [PubMed] [Google Scholar]
- 129.Hernandez DR, DeBord JD, Ridgeway ME, Kaplan DA, Park MA, Fernandez-Lima F (2014) Ion dynamics in a trapped ion mobility spectrometer. Analyst 139(8):1913–1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Silveira JA, Danielson W, Ridgeway ME, Park MA (2016) Altering the mobility-time continuum: nonlinear scan functions for targeted high resolution trapped ion mobility-mass spectrometry. Int J Ion Mobil Spectrom 19 (2–3):87–94 [Google Scholar]
- 131.May JC, Leaptrot KL, Sundarapandian S, McLean JA. In theoretical evaluation and performance characterization of an 8-channel spatially multiplexed ion mobility-mass spectrometer, 60th annual ASMS conference on mass spectrometry and allied topics, Vancouver, BC, May 2012 [Google Scholar]
- 132.Asbury GR, Hill HH (2000) Using different drift gases to change separation factors (alpha) in ion mobility spectrometry. Anal Chem 72 (3):580–584 [DOI] [PubMed] [Google Scholar]
- 133.Matz LM, Hill HH Jr, Beegle LW, Kanik I (2002) Investigation of drift gas selectivity in high resolution ion mobility spectrometry with mass spectrometry detection. J Am Soc Mass Spectrom 13(4):300–307 [DOI] [PubMed] [Google Scholar]
- 134.Berant Z, Karpas Z (1989) Mass-mobility correlation of ions in view of new mobility data. J Am Chem Soc 111(11):3819–3824 [Google Scholar]
- 135.Howdle MD, Eckers C, Laures AMF, Creaser CS (2010) The effect of drift gas on the separation of active pharmaceutical ingredients and impurities by ion mobility–mass spectrometry. Int J Mass Spectrom 298 (1–3):72–77 [Google Scholar]
- 136.Purves RW, Ozog AR, Ambrose SJ, Prasad S, Belford M, Dunyach JJ (2014) Using gas modifiers to significantly improve sensitivity and selectivity in a cylindrical FAIMS device. J Am Soc Mass Spectrom 25(7):1274–1284 [DOI] [PubMed] [Google Scholar]
- 137.Waraksa E, Perycz U, Namiesnik J, Sillanpaa M, Dymerski T, Wojtowicz M, Puton J (2016) Dopants and gas modifiers in ion mobility spectrometry. TRAC Trends Anal Chem 82:237–249 [Google Scholar]
- 138.Schneider BB, Covey TR, Nazarov EG (2013) Investigation of the chemical orthogonality effect of transport gas modifiers on DMS separations. Abstr Pap Am Chem Soc 246:1 [Google Scholar]
- 139.Kafle A, Coy SL, Wong BM, Fornace AJ, Glick JJ, Vouros P (2014) Understanding gas phase modifier interactions in rapid analysis by differential mobility-tandem mass spectrometry. J Am Soc Mass Spectrom 25 (7):1098–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Levin DS, Vouros P, Miller RA, Nazarov EG, Morris JC (2006) Characterization of gas-phase molecular interactions on differential mobility ion behavior utilizing an electrospray ionization-differential mobility-mass spectrometer system. Anal Chem 78 (1):96–106 [DOI] [PubMed] [Google Scholar]
- 141.Porta T, Varesio E, Hopfgartner G (2013) Gas-phase separation of drugs and metabolites using modifier-assisted differential ion mobility spectrometry hyphenated to liquid extraction surface analysis and mass spectrometry. Anal Chem 85(24):11771–11779 [DOI] [PubMed] [Google Scholar]
- 142.Fernandez-Maestre R, Wu C, Hill HH (2012) Buffer gas modifiers effect resolution in ion mobility spectrometry through selective ion-molecule clustering reactions. Rapid Commun Mass Spectrom 26(19):2211–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Garabedian A, Leng FF, Ridgeway ME, Park MA, Fernandez-Lima F (2018) Tailoring peptide conformational space with organic gas modifiers in TIMS-MS. Int J Ion Mobil Spectrom 21(1–2):43–48 [Google Scholar]
- 144.Beegle LW, Kanik I, Matz L, Hill HH (2001) Electrospray ionization nigh-resolution ion mobility spectrometry for the detection of organic compounds, 1. Amino acids. Anal Chem 73(13):3028–3034 [DOI] [PubMed] [Google Scholar]
- 145.Beegle LW, Kanik I, Matz L, Hill HH (2002) Effects of drift-gas polarizability on glycine peptides in ion mobility spectrometry. Int J Mass Spectrom 216(3):257–268 [Google Scholar]
- 146.Chouinard CD, Beekman CR, Kemperman RHJ, King HM, Yost RA (2017) Ion mobility-mass spectrometry separation of steroid structural isomers and epimers. Int J Ion Mobil Spectrom 20(1–2):31–39 [Google Scholar]
- 147.Fasciotti M, Lalli PM, Klitzke CF, Corilo YE, Pudenzi MA, Pereira RCL, Bastos W, Daroda RJ, Eberlin MN (2013) Petroleomics by traveling wave ion mobility–mass spectrometry using CO2 as a drift gas. Energy Fuel 27 (12):7277–7286 [Google Scholar]
- 148.Fasciotti M, Sanvido GB, Santos VG, Lalli PM, McCullagh M, de Sá GF, Daroda RJ, Peter MG, Eberlin MN (2012) Separation of isomeric disaccharides by traveling wave ion mobility mass spectrometry using CO2 as drift gas. J Mass Spectrom 47 (12):1643–1647 [DOI] [PubMed] [Google Scholar]
- 149.Bataglion GA, Souza G, Heerdt G, Morgon NH, Dutra JDL, Freire RO, Eberlin MN, Tata A (2015) Separation of glycosidic catiomers by TWIM-MS using CO2 as a drift gas. J Mass Spectrom 50(2):336–343 [DOI] [PubMed] [Google Scholar]
- 150.Ruotolo BT, McLean JA, Gillig KJ, Russell DH (2004) Peak capacity of ion mobility mass spectrometry: the utility of varying drift gas polarizability for the separation of tryptic peptides. J Mass Spectrom 39(4):361–367 [DOI] [PubMed] [Google Scholar]
- 151.Jurneczko E, Kalapothakis J, Campuzano IDG, Morris M, Barran PE (2012) Effects of drift gas on collision cross sections of a protein standard in linear drift tube and traveling wave ion mobility mass spectrometry. Anal Chem 84(20):8524–8531 [DOI] [PubMed] [Google Scholar]
- 152.Davidson KL, Bush MF (2017) Effects of drift gas selection on the ambient-temperature, ion mobility mass spectrometry analysis of amino acids. Anal Chem 89 (3):2017–2023 [DOI] [PubMed] [Google Scholar]
- 153.Morris CB, May JC, Leaptrot KL, McLean JA (2019) Evaluating separation selectivity and collision cross section measurement reproducibility in helium, nitrogen, argon, and carbon dioxide drift gases for drift tube ion mobility-mass spectrometry. J Am Soc Mass Spectrom 30(6):1059–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Norris JL, Farrow MA, Gutierrez DB, Palmer LD, Muszynski N, Sherrod SD, Pino JC, Allen JL, Spraggins JM, ALR L, Jordan A, Burns W, Poland JC, Romer C, Manier ML, Nei Y, Prentice BM, Rose KL, Hill S, Van de Plas R, Tsui T, Braman NM, Keller MR, Rutherford A, Lobdell N, Lopez CF, Lacy DB, JA ML, Wikswo JP, Skaar EP, Caprioli RM (2017) Integrated, high-throughput, multiomics platform enables data-driven construction of cellular responses and reveals global drug mechanisms of action. J Proteome Res 16(3):1364–1375 [DOI] [PubMed] [Google Scholar]
- 155.Paglia G, Stocchero M, Cacciato S, Lai S, Angel P, Alam MT, Keller M, Ralser M, Astarita G (2016) Unbiased metabolomic investigation of Alzheimer’s disease brain points to dysregulation of mitochondrial aspartate metabolism. J Proteome Res 15:608–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Morgan XC, Segata N, Huttenhower C (2013) Biodiversity and functional genomics in the human microbiome. Trends Genet 29 (1):51–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Weir TL, Manter DK, Brittany BA, Heuberger AL, Ryan EP (2013) Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS One 8(8):e70803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Raman M, Ahmed I, Gillevet PM, Probert CS, Ratcliffe NM, Smith S, Greenwood R, Sikaroodi M, Lam V, Crotty P, Bailey J, Meyes RP, Rious KP (2013) Fecal microbiome and volatile organic compound metabolome in obese humans with nonalcoholic fatty liver disease. Clin Gastroenterol Hepatol 11(7):868–875 [DOI] [PubMed] [Google Scholar]