Abstract
Aim:
This study was designed to investigate the prevalence of Clostridioides difficile, its toxin-producing genes, and antibiotic resistance patterns in diarrheal samples from hospitalized patients in Hamadan, Iran.
Background:
Today, concerns over Clostridioides difficile infection (CDI) have significantly increased due to reduced susceptibility to antibiotics used for CDI treatment. Toxins produced by C. difficile strains are associated with disease severity and outcome.
Methods:
In this cross-sectional study, a total of 130 diarrheal samples of patients admitted to different wards of three hospitals in Hamadan from November 2018 to September 2019 were collected. C. difficile isolates were identified by culture on CCFA and PCR (Polymerase chain reaction). The presence of toxin-encoding genes (tcdA and tcdB) and binary toxin genes (cdtA and cdtB) was analyzed by PCR. Resistance of the isolates to metronidazole, vancomycin and clindamycin antibiotics was determined using agar dilution method.
Results:
Out of 130 diarrheal samples from hospitalized patients, 16 (12.3%) C. difficile isolates were obtained. PCR results were positive for two toxin-producing genes, tcdA and tcdB, in all (100%) C. difficile isolates, and the binary toxin genes cdtA and cdtB were detected in 6 (37.5%) and 8 (50%) isolates, respectively. The results of antibiotic susceptibility testing showed resistance to metronidazole, vancomycin, and clindamycin in 3 (18.7%), 3 (18.7%), and 2 (12.5%) isolates, respectively, and all isolates were resistant to rifampicin.
Conclusion:
The results of this study showed toxigenic C. difficile with tcdA+/tcdB+ profile is a major cause of nosocomial diarrhea in Hamadan, and clinical laboratories should routinely perform C. difficile diagnostic testing on diarrheal specimens of hospitalized patients. Resistance to conventional antibiotic therapy against C. difficile should be considered as a warning to prevent irrational administration of antibiotics.
Key Words: Clostridioides difficile, TcdA, TcdB, Binary toxin, Antibiotic resistance
Introduction
Clostridioides difficile is a Gram-positive, obligate anaerobic bacterium that forms spores and is considered an important human pathogen. Attention to this bacterium has increased since it was described in 1978 as the main cause of antibiotic-induced diarrhea and the development of almost all cases of pseudomembranous colitis and toxic megacolon (1). C. difficile was initially thought to be a hospital-acquired bacterium, but there are some reports of C. difficile infection (CDI) among people outside hospitals or people who had not taken antibiotics. However, increasing prevalence in the community compared to the hospital is reported to be 1300 times lower, which may be due to lower use of antibiotics in the community (2).
C. difficile causes colitis and diarrhea by producing two exotoxins: toxin A (enterotoxin) and toxin B (cytotoxin). In human studies, the level of toxins in the stool is related to the severity of the disease. Toxin A causes inflammation, which leads to the secretion of intestinal fluids and mucosal damage. Toxin B is approximately 10 times more involved in colonic mucosal damage than toxin A, suggesting that toxin B may be more critical than toxin A in the pathogenesis of C. difficile colitis. A few strains of C. difficile can produce another toxin called binary toxin, which consists of a cell binding component and an enzymatic component that shows an action-specific ADP ribosyltransferase activity leading to disorganization of the cytoskeleton (3-5).
Various risk factors affect the prevalence of C. difficile nosocomial infections, which include antibiotics, advanced age, hospitalization, debilitating diseases such as cancer or treatment with immunosuppressive drugs, abdominal surgery, chemotherapy, and prolonged stay in a healthcare setting. Two major roles for antibiotics in the pathogenesis of C. difficile have been described. In first, antibiotics destroy the normal intestinal flora and provide conditions for C. difficile to multiply and produce toxins. Second, the rapid growth of C. difficile resistance to clindamycin and fluoroquinolones seems to play an important role in increasing the prevalence and pathogenicity of this microbe (6).
Antibiotics that play a major role in predisposing the host to C. difficile-associated diarrhea include fluoroquinolones, clindamycin, a range of penicillins and cephalosporins. Any antibiotic, even metronidazole and vancomycin, used to treat C. difficile can cause antibiotic-dependent colitis. Although an increased resistance to metronidazole has been observed in clinical isolates of C. difficile, it is still the most effective agent in the treatment of infections caused by this bacterium (5, 7-9).
Currently, there is insufficient information about the prevalence of C. difficile in the west of Iran. Thus, the aim of the present study was to determine the frequency of C. difficile, its toxin genes, and resistance to metronidazole, vancomycin and clindamycin among isolates obtained from fecal samples of patients with diarrhea who were admitted to hospitals in Hamadan.
Methods
Patients and C. difficile clinical isolates
In this cross-sectional study, a total of 130 fecal specimens were collected from hospitalized patients with diarrhea in the hospitals of Hamadan from November 2018 to September 2019. This study was approved by the Ethics Committee of Hamadan University of Medical Sciences (IR.UMSHA. REC.1397.510). Diarrhea was defined as the passage of more than two loose or watery stools during a 24 h period or fewer hours (10). Fecal samples from patients were collected in a specific stool container and then immediately transferred to the microbiology laboratory in Hamadan University of Medical Sciences. Stool samples were treated as previously described (11, 12). To isolate C. difficile, the treated suspensions were cultured on cycloserine-cefoxitin fructose agar (CCFA; Mast Co, UK) supplemented with 5% fresh sheep blood and incubated anaerobically for 48 h at 37 °C using an anaerobic jar (MART Microbiology B.V. the Netherlands). Identification of the isolates was performed based on Gram staining, odor and colony characteristics on CCFA plates. For molecular confirmation of C. difficile isolates, the cdd3, as a housekeeping gene (C. difficile downstream 3), was targeted by PCR (Polymerase chain reaction) using specific primers as described previously by Cohen et al. (13). Samples confirmed as C. difficile were stored in cooked meat broth (Merck, Germany) at 4 °C, and were subjected to further molecular identification. A questionnaire containing demographic data and time of hospitalization, age and sex of patient was completed for patients.
DNA extraction and PCR
Genomic DNA was extracted from freshly grown colonies using the boiling method. All C. difficile isolates were subjected for determination of toxin genes. The detection of toxin A gene (tcdA), toxin B gene (tcdB), and binary toxin genes (cdtA and cdtB) was performed by PCR described by Cohen et al. and Terhes et al., using specific primers (Metabion, Germany) shown in Table 1 (13, 14). The PCR reactions for the detection of tcdA and tcdB genes were done in a total volume of 25 μL. The reaction mixture contained 10 µl master mix (Amplicon, Denmark), 0.5 μM of each primer, 1 μL DNA template, and 10 μL distilled water. The PCR reactions consisted of an initial denaturation step at 95 °C for 5 min, followed by 30 cycles of 60 sec at 95 °C, annealing for 45 sec at 51 °C (for tcdA), 50 °C (for tcdB), 53 °C (for cdtA and cdtB), and extension for 50 sec at 72 °C. A final extension step was performed at 72 °C for 5 min. The PCR products were separated by electrophoresis in 1.2% agarose gels.
Table 1.
Primer sequences and fragment lengths used for amplification of cdd3, tcdA, tcdB, cdtA, and cdtB genes
| Primer (gene) | Nucleotide sequence | Fragment Length (bp) | References |
|---|---|---|---|
| cdd3 | F: 5´ TCC AAT ATA ATA AAT TAG CAT TCC A 3´ R: 5´ GGC TAT TAC ACG TAA TCC AGA TA 3´ |
622 | |
| tcdA | F: 5´ ATG ATA AGG CAA CTT CAG TGG 3´ R: 5´ TAA GTT CCT CCT GCT CCA TCA A 3´ |
624 | |
| tcdB | F: 5´ GAG CTG CTT CAA TTG GAG AGA 3´ R: 5´ GTA ACC TAC TTT CAT AAC ACC AG 3´ |
412 | |
| cdtA | F: 5ʹ TGA ACC TGG AAA AGG TGA TG 3ʹ R: 5ʹ AGG ATT ATT TAC TGG ACC ATT TG 3´ |
375 | |
| cdtB | F: 5ʹ CTTAATGCAAGTAAATACTGAG 3ʹ R: 5ʹ AACGGATCTCTTGCTTCAGTC 3ʹ |
512 |
Antimicrobial susceptibility testing
Antimicrobial susceptibility of C. difficile isolates to metronidazole and clindamycin and was determined using the breakpoints defined by the Clinical and Laboratory Standards Institute (document M11-A8) and to vancomycin by the European Committee for Antimicrobial Susceptibility Testing (EUCAST) criteria (15). The MIC breakpoints for vancomycin and rifampicin were used as previously described (16). The minimum inhibitory concentration (MIC) of these antibiotics (Sigma-Aldrich, St. Louis, Mo) was determined by the agar dilution method. The range of MIC value used for antimicrobial agents was 0.5 to 256 μg/ml. Media with different concentrations of each antibiotic were prepared by adding defined amounts of each antibiotic to cooled Brucella agar medium supplemented with hemin (5 μg/ml), vitamin K1 (10 μg/ml), and 5% sheep blood (5). The turbidity of each bacterial suspension was adjusted equivalent to a no.1 McFarland standard, and 20 μl of each suspension was inoculated on Brucella agar plates containing different concentrations of each antibiotic. Inoculated plates without antibiotics served as control.
Statistical analysis
The data was analyzed using SPSS software, version 21 for Windows (SPSS Inc., Chicago, IL, USA). A p-value <0.05 was considered statistically significant.
Results
Out of 130 collected stool specimens, 16 (12.36%) were C. difficile culture positive and confirmed with the cdd3 gene PCR. Of 130 stool samples, 70 (53.8%) were collected from women and 60 (46.1%) were from men. For men, 7 cases (11.6%) and for women, 9 cases (12.8%) were positive for the presence of C. difficile. The results indicate that C. difficile is more common in diarrhea specimens isolated from women; however, there was no significant difference according to the gender of patients (p = 0.38). Based on the length of hospital stay of the patients, 50% of C. difficile-positive patients were hospitalized for more than 7 days (longest time), the shortest time (12.5%) was at 1 to 3 days, and 37.5% of C. difficile-positive patients were hospitalized for 3 to 7 days. In this study, the age range of patients who showed C. difficile infection ranged from 28 to 89 years; 8 (50%) of the positive cases were in the age range of 50 to 70 years old, and 6 (37.5% ) and 2 (12.5%) positive cases were in 70-90 and 28-50 age ranges, respectively. Most (87.5%) C. difficile-positive samples were isolated from patients in the internal wards of the hospitals; only 2 (12.5%) positive samples were isolated from patients in the intensive care unit (ICU).
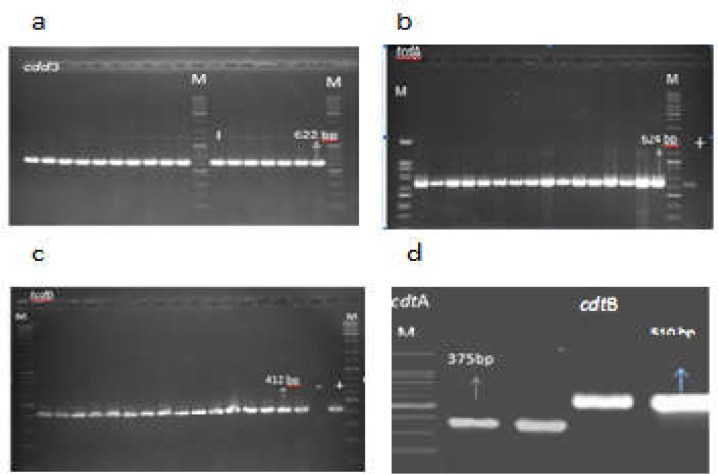
All C. difficile isolates (100%) carried both tcdA and tcdB genes, showed a tcdA+/tcdB+ profile, and were considered as toxigenic strains. The cdtA and cdtB genes were detected in 6 (37.5%) and 8 (50%) isolates, respectively. Two isolates were negative for both cdtA and cdtB genes. Simultaneous detection of binary toxin genes did not occur in any of the isolates. Therefore, tcdA/tcdB/cdtA and tcdA/tcdB/cdtB profiles were detected in 6 (37.5%) and 8 (50%) of the isolates, respectively (Figure 1). The details of C. difficile isolates obtained in this study are shown in Table 2.
Figure 1.
Gel electrophoresis of PCR products for C. difficile identification and toxin encoding genes in isolates from hospitalized patients in Hamadan. (a) cdd3; (b) tcdA; (c) tcdB; (d) cdtA and cdtB genes. Lane M, 100 bp DNA size marker
Table 2.
Characteristics of C. difficile isolates from hospitalized patients in Hamadan
| Isolates | Patient sex | Ward | tcdA | tcdB | cdtA | cdtB |
|---|---|---|---|---|---|---|
| 1 | M | Int | + | + | - | + |
| 2 | M | Int | + | + | - | + |
| 3 | F | Int | + | + | - | + |
| 4 | M | Int | + | + | + | - |
| 5 | F | Int | + | + | + | - |
| 6 | F | Int | + | + | - | + |
| 7 | M | Int | + | + | - | + |
| 8 | F | ICU | + | + | - | + |
| 9 | F | Int | + | + | + | - |
| 10 | F | Int | + | + | + | - |
| 11 | M | ICU | + | + | - | + |
| 12 | F | Int | + | + | + | - |
| 13 | M | Int | + | + | - | + |
| 14 | F | Int | + | + | + | - |
| 15 | F | Int | + | + | - | - |
| 16 | F | Int | + | + | - | - |
M, male; F, female; Int, internal ward; ICU, intensive care unit
According to antimicrobial susceptibility testing by agar dilution, resistance to metronidazole (MIC ≥32 μg/ml), clindamycin (MIC ≥8 μg/ml), and vancomycin (MIC ≥8 μg/ml) was observed in 3 (18.7%), 2 (12.5%), and 3 (18.7%) of the isolates, respectively. All isolates were resistant to rifampicin (MIC ≥4 μg/ml). Reduced susceptibility to vancomycin and clindamycin was observed in one (6.2 %) and 13 (81.2 %) of isolates, respectively, which was interpreted as intermediate phenotype (MIC = 4 μg/ml). The interpretive criteria of the MIC values for C. difficile isolates are shown in Table 3.
Table 3.
Interpretive criteria of the MIC values for C. difficile isolates
| Antibiotic agent | (μg/ml) |
No. of isolates with MIC of (µg/ml) |
Susceptibility profile |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S | I | R | 0.5 1 2 4 8 16 32 64 128 256 | S (%) | I (%) | R (%) | ||||||||||
| Metronidazole* | ≤8 | 16 | ≥32 | 12 | 1 | - | - | - | - | - | - | 3 | - | 81.3 | - | 18.7 |
| Vancomycin§ | ≤2 | 4 | ≥8 | 12 | - | - | 1 | 3 | - | - | - | - | 75 | 6.3 | 18.7 | |
| Rifampicin§ | - | - | ≥4 | - | - | - | - | 16 | - | - | - | - | - | - | - | 100 |
| Clindamycin* | ≤2 | 4 | ≥8 | 1 | - | - | 13 | - | 2 | - | - | - | - | 6.3 | 81.2 | 12.5 |
Breakpoints were defined as susceptible (S), intermediately resistant (I), or resistant (R) with reference to CLSI (*) or published data (§)
Discussion
This research is the first study of the prevalence of CDI in diarrheal samples of patients admitted to hospitals in Hamadan. The prevalence of CDI was detected to be 12% in this study. It was established that the incidence rate of CDI is significantly higher among hospitalized people than among those who acquired it from the community (17, 18). The rate of CDI in different countries has been reported as being from zero to 36% (19-22). Currently, there is limited data about the molecular epidemiology of CDI in Africa, Asia, and Latin America (23, 24). It is difficult to obtain accurate epidemiological information about the prevalence of CDI in developing countries, because their diagnosis is based on immunoassay (EIA) methods rather than culture (25-27). Limited laboratory capacity, inefficient infection management and control systems affect the accurate reporting of CDI prevalence in developing countries (28, 29). In recent years, several studies were performed on anaerobic bacteria due to the provision of facilities and optimizing of anaerobic culture and isolation methods. In a study by Borren et al., which included 51 studies from throughout Asia, the rate of CDI was 14.8% among all patients with diarrhea and showed a higher prevalence in East Asia (19.5%) compared with South Asia (10.5%) or the Middle East (11.1%) (30). Pooled prevalence of CDI in Persian Gulf countries ranged from 2.8% to 21.7% (31). In Saudi Arabia, the prevalence of CDI varied from 17% to 20% in suspected diarrheal samples (32, 33). The current findings also showed a lower prevalence of CDI compared to some studies from Saudi Arabia; however, the prevalence of CDI in hospitals in Kuwait and Qatar is lower than the current results (34, 35). Moreover, the current findings showed a prevalence rate of CDI similar to that in South and East Asia, such as India (10.9%), China (14%), and South-Korea (14.3%) (36-38). According to studies conducted from 2010 to 2020 in Iran, different prevalence rates of C. difficile ranging from 4.7% to 39% have been reported (5, 10, 12, 39-41). The prevalence of CDI in hospitals in Hamadan is lower than that in Tehran, Isfahan, and Kerman (10, 12, 42). This variation in the prevalence of CDI in different studies might be due to differences in geographical distribution, infection control policies, diagnostic methods, or the studied population (30).
The role of toxins A and B in the pathogenicity of C. difficile is well studied. In the current study, all isolates were toxigenic and showed a tcdA+/tcdB+/CDT- profile, because none of the isolates contained binary toxins simultaneously, but 6 (37.5%) and 8 (50%) of the isolates contained cdtA and cdtB genes, respectively. Due to the importance of the TcdB toxin in bacterial pathogenesis, the presence of the tcdB gene in all isolates is important and indicates that we are facing high virulence strains in the hospitals of Hamadan. Multiple studies have reported the tcdA+/tcdB+/CDT- profile as a main toxin profile. In a study done by Shoaei et al., 11.5% of clinical samples from inpatients in hospitals of Isfahan were toxigenic and showed a tcdA+/tcdB+/CDT- profile; only one (2.2%) isolate harbored all toxin-associated genes with tcdA+, tcdB+, cdtA+, and cdtB+ profiles (10).
Rezazadeh et al. reported different results in diarrheal samples from ICU patients in Kerman (43). In their study, the frequency of CDI was 41%, and tcdA+/tcdB+/CDT- profiles were detected in 20% of isolates; only one isolate with the tcdA+/tcdB+/CDT+ profile was detected. The dominance of the tcdA+/tcdB+/CDT- profile has also been reported by Heidari et al., Goudarzi et al., and Azimirad et al. from hospitals in Tehran and Shiraz (5, 12, 39). Moreover, the tcdA+/tcdB+/CDT- profile was reported in 18.8%, 92%, 8%, and 71% of C. difficile isolates from Japan, Czech Republic, Argentina, and China, respectively (4, 44, 45). Consistent with the current results, the presence of one of the two binary genes in the clinical strains of C. difficile has been reported in most studies from Asian countries such as Japan, Korea, China, and Thailand (10, 46). In European countries, however, the prevalence of binary toxins has been reported in 4% to 12% of C. difficile isolates, and these strains have been associated with higher mortality and CDI recurrence (47, 48).
C. difficile is the main infectious cause of antibiotic-associated diarrhea (AAD), and metronidazole and vancomycin remain as the first-line drugs in the treatment of CDI. One of the main goals of the current study was to determine the prevalence of resistance to metronidazole, vancomycin and clindamycin. Resistance to metronidazole, clindamycin and vancomycin was observed in this study. C. difficile isolates showed a geographically dispersed antibiotic resistance pattern due to the use of different standards and susceptibility testing methods. In European countries and the United States, E-test has been the most common method for testing antimicrobial susceptibility of C. difficile (45). Most results are reported according to CLSI criteria (49). Resistance to metronidazole has been reported in various parts of the world, being first reported in 2011-2012 (50). According to the results of various studies, the rate of resistance to metronidazole was reported to be from zero to 18.3% until 2018. The results also showed that resistance to metronidazole has decreased by about 0.8%, and this dramatic decrease observed in some countries may be due to the choice of test for antibiotic susceptibility testing (49). In recent years, vancomycin and metronidazole resistance has been reported in Iran and other countries. According to the results of previous studies, the proportions of vancomycin non-susceptibility varied from 0 to 87.7% in BI/NAP1/027 isolates (51-54). This data indicates an increase in vancomycin resistance over time. High rates of vancomycin resistance have been reported in the United States and then in Asia (7, 55). Vancomycin resistance averaged 0.4% before 2012 and reached 4% afterward, that shows a significant difference (7, 56). One of the reasons for the increase in vancomycin resistance is the extended use of vancomycin in US hospitals, after which resistance to vancomycin significantly increased worldwide (49). In 2013, the first report of vancomycin resistance was reported to be 1% among C. difficile isolates in Tehran (39). In the current study, resistance to vancomycin was detected in three isolates (18.7%), and a decreasing trend in the susceptibility of vancomycin (intermediate phenotype) was detected in 6.2% of the isolates. Three isolates were also found to be resistant to metronidazole. Resistance to clindamycin was found to be lower than resistance to vancomycin and metronidazole in this study. In various studies from European, Asian, and North American countries, resistance to clindamycin has been reported to be from 8.3% to 100% (46, 57, 58). There are different reports on resistance to vancomycin, metronidazole, and clindamycin. Based on recent studies in Iran by Shoaei et al. from Isfahan and Heidari et al. from Shiraz, the antimicrobial susceptibility determination by E-test showed that all toxicogenic C. difficile isolates were sensitive to vancomycin and metronidazole (5, 10), while Baghani et al. reported that 30% of C. difficile strains isolated from Tehran hospitals were resistant to vancomycin (59). Baghani et al. also reported that 81.5% of the isolates were resistant to metronidazole, which showed a high rate of resistance to this antibiotic and raised great concern about the treatment of patients with CDI (59). In a study conducted by Mohammadbeigi et al., all C. difficile isolates were susceptible to vancomycin, and 8.16% and 72.1% of toxigenic isolates were resistant to metronidazole and clindamycin, respectively, in Kerman hospitals (60). Some studies from China showed susceptibility to vancomycin in 100% and resistance to metronidazole in 15.6-35.3% of C. difficile isolates (61, 62). In Europe, resistance to therapeutic antibiotics of choice in the case of CDI, like metronidazole and vancomycin, showed low resistance rates of 0.1% and 2.3%, respectively (62, 63).
To conclude, a relatively high frequency of C. difficile was detected in diarrheal samples collected from two hospitals in Hamadan. The data also indicated that the tcdA+/tcdB+/CDT- toxigenic pattern was predominant among C. difficile isolates, and clinical laboratories should routinely perform C. difficile diagnostic tests on diarrheal specimens of hospitalized patients in this area.
There were some limitations in our study, such as a small sample size, the lack of molecular typing information of the isolates by ribotyping method, and insufficient equipment for anaerobic culture of bacteria. The rate of resistance to conventional antibiotic therapy against C. difficile was alarming in Hamadan. Thus, further studies are needed to monitor the rate and pattern of antibiotic resistance of C. difficile isolates in this region.
Acknowledgment
This research was supported by the Vice Chancellor for Research & Technology of Hamadan University of Medical Sciences, Hamadan, IRAN (Grant no: 9804101833). The authors would like to thank all members of the Foodborne and Waterborne Diseases Research Center, Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences for providing the conditions to subculture our samples and verify the identification of C. difficile isolates. We also appreciate the efforts of the staffs of the microbiology laboratory of Shahid Beheshti and Sina hospitals in Hamadan.
Conflict of interests
The authors declare that they have no conflict of interest.
References
- 1.Riggs MM, Sethi AK, Zabarsky TF, Eckstein EC, Jump RL, Donskey CJ. Asymptomatic carriers are a potential source for transmission of epidemic and nonepidemic Clostridium difficile strains among long-term care facility residents. Clin infect Dis. 2007;45:992–8. doi: 10.1086/521854. [DOI] [PubMed] [Google Scholar]
- 2.Rodriguez-Palacios A, Borgmann S, Kline TR, LeJeune JT. Clostridium difficile in foods and animals: history and measures to reduce exposure. Anim Health Res Rev. 2013;14 doi: 10.1017/S1466252312000229. [DOI] [PubMed] [Google Scholar]
- 3.Azimirad M, Noukabadi FN, Lahmi F, Yadegar A. Prevalence of binary-toxin genes (cdtA and cdtB) among clinical strains of Clostridium difficile isolated from diarrheal patients in Iran. Gastroenterol Hepatol Bed Bench. 2018;11:59–65. [PMC free article] [PubMed] [Google Scholar]
- 4.Tokimatsu I, Shigemura K, Osawa K, Kinugawa S, Kitagawa K, Nakanishi N, et al. Molecular epidemiologic study of Clostridium difficile infections in university hospitals: Results of a nationwide study in Japan. J Infect Chemother 2018. 24:641–7. doi: 10.1016/j.jiac.2018.03.015. [DOI] [PubMed] [Google Scholar]
- 5.Heidari H, Ebrahim-Saraie HS, Amanati A, Motamedifar M, Hadi N, Bazargani A. Toxin profiles and antimicrobial resistance patterns among toxigenic clinical isolates of Clostridioides (Clostridium) difficile. Iran J Basic Med Sci. 2019;22:813–9. doi: 10.22038/ijbms.2019.35223.8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McDonald LC, Killgore GE, Thompson A, Owens Jr RC, Kazakova SV, Sambol SP, et al. An epidemic, toxin gene–variant strain of Clostridium difficile. N Engl J Med. 2005;353:2433–41. doi: 10.1056/NEJMoa051590. [DOI] [PubMed] [Google Scholar]
- 7.Peng Z, Jin D, Kim HB, Stratton CW, Wu B, Tang YW, et al. Update on Antimicrobial Resistance in Clostridium difficile: Resistance Mechanisms and Antimicrobial Susceptibility Testing. J Clin Microbiol. 2017;55:1998–2008. doi: 10.1128/JCM.02250-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang H, Weintraub A, Fang H, Nord CE. Antimicrobial resistance in Clostridium difficile. Int J Antimicrob Agents. 2009;34:516–22. doi: 10.1016/j.ijantimicag.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 9.Goudarzi M, Navidinia M. Overview Perspective of Bacterial Strategies of Resistance to Biocides and Antibiotics. Arch Clin Infect Dis. 2019;14:e65744. [Google Scholar]
- 10.Shoaei P, Shojaei H, Khorvash F, Hosseini SM, Ataei B, Tavakoli H, et al. Molecular epidemiology of Clostridium difficile infection in Iranian hospitals. Antimicrob Resist Infect Control. 2019;8:1–7. doi: 10.1186/s13756-018-0454-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Azimirad M, Krutova M, Yadegar A, Shahrokh S, Olfatifar M, Aghdaei HA, et al. Clostridioides difficile ribotypes 001 and 126 were predominant in Tehran healthcare settings from 2004 to 2018: A 14-year-long cross-sectional study. Emerg Microbes Infect. 2020;9:1432–43. doi: 10.1080/22221751.2020.1780949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Azimirad M, Krutova M, Balaii H, Kodori M, Shahrokh S, Azizi O, et al. Coexistence of Clostridioides difficile and Staphylococcus aureus in gut of Iranian outpatients with underlying inflammatory bowel disease. Anaerobe. 2020;61:102113. doi: 10.1016/j.anaerobe.2019.102113. [DOI] [PubMed] [Google Scholar]
- 13.Cohen SH, Tang YJ, Silva Jr J. Analysis of the pathogenicity locus in Clostridium difficile strains. J Infect Dis. 2000;181:659–63. doi: 10.1086/315248. [DOI] [PubMed] [Google Scholar]
- 14.Terhes G, Urbán E, Sóki J, Hamid KA, Nagy E. Community-acquired Clostridium difficile diarrhea caused by binary toxin, toxin A, and toxin B gene-positive isolates in Hungary. J Clin Microbiol. 2004;42:4316–8. doi: 10.1128/JCM.42.9.4316-4318.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.CLSI . Performance Standards for Antimicrobial Susceptibility Testing. 28th ed. CLSI supplement M100 Wayne, PA: Clinical and Laboratory Standards Institute; 2018. [Google Scholar]
- 16.Peng Z, Addisu A, Alrabaa S, Sun X. Antibiotic resistance and toxin production of Clostridium difficile isolates from the hospitalized patients in a large hospital in Florida. Front Microbiol. 2017;8:2584. doi: 10.3389/fmicb.2017.02584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bouza E. Consequences of Clostridium difficile infection: understanding the healthcare burden. Clin Microbiol Infect. 2012;18:5–12. doi: 10.1111/1469-0691.12064. [DOI] [PubMed] [Google Scholar]
- 18.Bauer MP, Notermans DW, Van Benthem BH, Brazier JS, Wilcox MH, Rupnik M, et al. Clostridium difficile infection in Europe: a hospital-based survey. Lancet. 2011;377:63–73. doi: 10.1016/S0140-6736(10)61266-4. [DOI] [PubMed] [Google Scholar]
- 19.Curcio D, Cané A, Fernández FA, Correa J. Clostridium difficile-associated diarrhea in developing countries: A systematic review and meta-analysis. Infect Dis Ther. 2019;8:87–103. doi: 10.1007/s40121-019-0231-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cohen SH, Gerding DN, Johnson S, Kelly CP, Loo VG, McDonald LC, et al. Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the society for healthcare epidemiology of America (SHEA) and the infectious diseases society of America (IDSA) Infect Control Hosp Epidemiol. 2010;31:431–55. doi: 10.1086/651706. [DOI] [PubMed] [Google Scholar]
- 21.Depestel DD, Aronoff DM. Epidemiology of Clostridium difficile infection. J Pharm Pract. 2013;26:464–75. doi: 10.1177/0897190013499521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forrester JD, Cai LZ, Mbanje C, Rinderknecht TN, Wren SM. Clostridium difficile infection in low‐and middle‐human development index countries: a systematic review. Trop Med Int Health. 2017;22:1223–32. doi: 10.1111/tmi.12937. [DOI] [PubMed] [Google Scholar]
- 23.Freeman J, Bauer MP, Baines SD, Corver J, Fawley WN, Goorhuis B, et al. The changing epidemiology of Clostridium difficile infections. Clin Microbiol Rev. 2010;23:529–49. doi: 10.1128/CMR.00082-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burke KE, Lamont JT. Clostridium difficile infection: a worldwide disease. Gut Liver. 2014;8:1–6. doi: 10.5009/gnl.2014.8.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crobach MJ, Dekkers OM, Wilcox MH, Kuijper EJ. European Society of Clinical Microbiology and Infectious Diseases (ESCMID): data review and recommendations for diagnosing Clostridium difficile-infection (CDI) Clin Microbiol Infect. 2009;15:1053–66. doi: 10.1111/j.1469-0691.2009.03098.x. [DOI] [PubMed] [Google Scholar]
- 26.Planche T, Aghaizu A, Holliman R, Riley P, Poloniecki J, Breathnach A, et al. Diagnosis of Clostridium difficile infection by toxin detection kits: a systematic review. Lancet Infect Dis. 2008;8:777–84. doi: 10.1016/S1473-3099(08)70233-0. [DOI] [PubMed] [Google Scholar]
- 27.Eastwood K, Else P, Charlett A, Wilcox M. Comparison of nine commercially available Clostridium difficile toxin detection assays, a real-time PCR assay for difficile tcdB, and a glutamate dehydrogenase detection assay to cytotoxin testing and cytotoxigenic culture methods. J Clin Microbiol. 2009;47:3211–7. doi: 10.1128/JCM.01082-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McDonald LC, Gerding DN, Johnson S, Bakken JS, Carroll KC, Coffin SE, et al. Clinical practice guidelines for Clostridium difficile infection in adults and children: 2017 update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA) Clin Infect Dis. 2018;66:e1–48. doi: 10.1093/cid/cix1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng JW, Xiao M, Kudinha T, Xu ZP, Sun LY, Hou X, et al. The role of glutamate dehydrogenase (GDH) testing assay in the diagnosis of Clostridium difficile infections: a high sensitive screening test and an essential step in the proposed laboratory diagnosis workflow for developing countries like China. PLoS one. 2015;10:e0144604. doi: 10.1371/journal.pone.0144604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borren NZ, Ghadermarzi S, Hutfless S, Ananthakrishnan AN. The emergence of Clostridium difficile infection in Asia: A systematic review and meta-analysis of incidence and impact. PloS one. 2017;12:e0176797. doi: 10.1371/journal.pone.0176797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malekzadegan Y, Halaji M, Hasannejad-Bibalan M, Jalalifar S, Fathi J, Ebrahim-Saraie HS. Burden of Clostridium (Clostridioides) difficile Infection among Patients in Western Asia: A Systematic Review and Meta-Analysis. Iran J Public Health. 2019;48:1589. [PMC free article] [PubMed] [Google Scholar]
- 32.Senok AC, Aldosari KM, Alowaisheq RA, Abid OA, Alsuhaibani KA, Khan MA, et al. Detection of Clostridium difficile antigen and toxin in stool specimens: comparison of the ifficile Quik Chek Complete enzyme immunoassay and GeneXpert C difficile polymerase chain reaction assay. Saudi J Gastroenterol. 2017;23:259. doi: 10.4103/sjg.SJG_80_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Al-Tawfiq JA, Rabaan AA, Bazzi AM, Raza S, Noureen M. Clostridioides (Clostridium) difficile-associated disease: Epidemiology among patients in a general hospital in Saudi Arabia. Am J Infect Control. 2020;48:1152–7. doi: 10.1016/j.ajic.2020.01.011. [DOI] [PubMed] [Google Scholar]
- 34.Jamal W, Pauline E, Rotimi V. A prospective study of community-associated Clostridium difficile infection in Kuwait: epidemiology and ribotypes. Anaerob. 2015;35:28–32. doi: 10.1016/j.anaerobe.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 35.Al-Thani AA, Hamdi WS, Al-Ansari NA, Doiphode SH, Wilson GJ. Erratum: Polymerase chain reaction ribotyping of Clostridium difficile isolates in Qatar: a hospital-based study. BMC Infect Dis. 2015;15:173. doi: 10.1186/s12879-015-0866-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vaishnavi C, Singh M, Mahmood S, Kochhar R. Prevalence and molecular types of Clostridium difficile isolates from faecal specimens of patients in a tertiary care centre. J Med Microbiol. 2015;64:1297–304. doi: 10.1099/jmm.0.000169. [DOI] [PubMed] [Google Scholar]
- 37.Tang C, Cui L, Xu Y, Xie L, Sun P, Liu C, et al. The incidence and drug resistance of Clostridium difficile infection in Mainland China: a systematic review and meta-analysis. Sci Rep. 2016;6:37865. doi: 10.1038/srep37865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kwon SS, Gim JL, Kim MS, Kim H, Choi JY, Yong D, et al. Clinical and molecular characteristics of community-acquired Clostridium difficile infections in comparison with those of hospital-acquired C. difficile. Anaerobe. 2017;48:42–6. doi: 10.1016/j.anaerobe.2017.06.014. [DOI] [PubMed] [Google Scholar]
- 39.Goudarzi M, Goudarzi H, Alebouyeh M, Rad MA, Mehr FSS, Zali MR, et al. Antimicrobial susceptibility of Clostridium difficile clinical isolates in Iran. Iran Red Crescent Med J. 2013;15:704–11. doi: 10.5812/ircmj.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hematyar Y, Pirzadeh T, Moaddab SR, Rezaee MA, Memar MY, Kafil HS. Clostridium difficile in patients with nosocomial diarrhea, Northwest of Iran. Health Promot Perspect. 2020;10:148–151. doi: 10.34172/hpp.2020.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gholam-Mostafaei FS, Yadegar A, Aghdaei HA, Azimirad M, Daryani NE, Zali MR. Anti-TNF containing regimens may be associated with increased risk of Clostridioides difficile infection in patients with underlying inflammatory bowel disease. Curr Res Transl Med. 2020;68:125–30. doi: 10.1016/j.retram.2020.03.002. [DOI] [PubMed] [Google Scholar]
- 42.Alimolaei M, Rahimi HR, Ezatkhah M, Bafti MS, Afzali S. Prevalence, characteristics and antimicrobial susceptibility patterns of Clostridioides difficile isolated from hospitals in Iran. J Glob Antimicrob Resist. 2019;19:22–7. doi: 10.1016/j.jgar.2019.02.013. [DOI] [PubMed] [Google Scholar]
- 43.Rezazadeh Zarandi E, Mansouri S, Nakhaee N, Sarafzadeh F, Iranmanesh Z, Moradi M. Frequency of antibiotic associated diarrhea caused by Clostridium difficile among hospitalized patients in intensive care unit, Kerman, Iran. Gastroenterol Hepatol Bed Bench. 2017;10:229–34. [PMC free article] [PubMed] [Google Scholar]
- 44.Krutova M, Nyc O, Matejkova J, Allerberger F, Wilcox MH, Kuijper EJ. Molecular characterisation of Czech Clostridium difficile isolates collected in 2013–2015. Int J Med Microbiol. 2016;306:479–85. doi: 10.1016/j.ijmm.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 45.Cejas D, Ríos Osorio NR, Quirós R, Sadorin R, Berger MA, Gutkind G, et al. Detection and molecular characterization of Clostridium difficile ST 1 in Buenos Aires, Argentina. Anaerobe. 2018;49:14–7. doi: 10.1016/j.anaerobe.2017.10.007. [DOI] [PubMed] [Google Scholar]
- 46.Luo Y, Zhang W, Cheng JW, Xiao M, Sun GR, Guo CJ, et al. Molecular epidemiology of Clostridium difficile in two tertiary care hospitals in Shandong Province China. Infect Drug Resist. 2018;11:489–500. doi: 10.2147/IDR.S152724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sandokji AM, Murshid KR, El-Badry AA, Al-Ali KH, Shalaby SA. Infectious Nosocomial Diarrhea in the Surgical Wards: Role of Parasites and Microbes Imply Stool Analysis. J Taibah Univ Sci. 2009;4:73–81. [Google Scholar]
- 48.Putsathit P, Kiratisin P, Ngamwongsatit P, Riley TV. Clostridium difficile infection in Thailand. Int J Antimicrob Agents. 2015;45:1–7. doi: 10.1016/j.ijantimicag.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 49.Saha S, Kapoor S, Tariq R, Schuetz AN, Tosh PK, Pardi DS, et al. Increasing antibiotic resistance in Clostridioides difficile: A systematic review and meta-analysis. Anaerobe. 2019;58:35–46. doi: 10.1016/j.anaerobe.2019.102072. [DOI] [PubMed] [Google Scholar]
- 50.Jin D, Luo Y, Huang C, Cai J, Ye J, Zheng Y, et al. Molecular epidemiology of Clostridium difficile infection in hospitalized patients in eastern China. J Clin Microbiol. 2017;55:801. doi: 10.1128/JCM.01898-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tkhawkho L, Nitzan O, Pastukh N, Brodsky D, Jackson K, Peretz A. Antimicrobial susceptibility of Clostridium difficile isolates in Israel. J Glob Antimicrob Resist. 2017;10:161–4. doi: 10.1016/j.jgar.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 52.Fraga EG, Nicodemo AC, Sampaio JL. Antimicrobial susceptibility of Brazilian Clostridium difficile strains determined by agar dilution and disk diffusion. Braz J Infect Dis. 2016;20:476–81. doi: 10.1016/j.bjid.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Banawas SS. Clostridium difficile Infections: A Global Overview of Drug Sensitivity and Resistance Mechanisms. Biomed Res Int. 2018;2018:8414257. doi: 10.1155/2018/8414257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sholeh M, Krutova M, Forouzesh M, Mironov S, Sadeghifard N, Molaeipour L, et al. Antimicrobial resistance in Clostridioides (Clostridium) difficile derived from humans: a systematic review and meta-analysis. Antimicrob Resist Infect Control. 2020;9:158. doi: 10.1186/s13756-020-00815-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Collins DA, Sohn KM, Wu Y, Ouchi K, Ishii Y, Elliott B, Riley TV, Tateda K. Clostridioides difficile Asia-Pacific Study Group Clostridioides difficile infection in the Asia-Pacific region. Emerg Microbes Infect. 2020;9:42–52. doi: 10.1080/22221751.2019.1702480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martin H, Willey B, Low DE, Staempfli HR, McGeer A, Boerlin P, et al. Characterization of Clostridium difficile strains isolated from patients in Ontario, Canada, from 2004 to 2006. J Clin Microbiol. 2008;46:2999–3004. doi: 10.1128/JCM.02437-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Spigaglia P. Recent advances in the understanding of antibiotic resistance in Clostridium difficile infection. Ther Adv Infect Dis. 2016;3:23–42. doi: 10.1177/2049936115622891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Slimings C, Riley TV. Antibiotics and hospital-acquired Clostridium difficile infection: update of systematic review and meta-analysis. J Antimicrob Chemother. 2014;69:881–91. doi: 10.1093/jac/dkt477. [DOI] [PubMed] [Google Scholar]
- 59.Baghani A, Mesdaghinia A, Kuijper EJ, Aliramezani A, Talebi M, Douraghi M. High prevalence of Clostridiodes diffiicle PCR ribotypes 001 and 126 in Iran. Sci Rep. 2020;10:4658. doi: 10.1038/s41598-020-61604-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mohammadbeigi M, Safayi Delouyi Z, Mohammadzadeh N, Alaalmohadesin A, Taheri K, Edalati E, et al. Prevalence and antimicrobial susceptibility pattern of toxigenic Clostridium difficilestrains isolated in Iran. Turk J Med Sci. 2019;49:384–91. doi: 10.3906/sag-1808-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jin D, Luo Y, Huang C, Cai J, Ye J, Zheng Y, et al. Molecular Epidemiology of Clostridium difficile Infection in Hospitalized Patients in Eastern China. J Clin Microbiol. 2017;55:801–10. doi: 10.1128/JCM.01898-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tilkorn F, Frickmann H, Simon IS, Schwanbeck J, Horn S, Zimmermann O, et al. Antimicrobial Resistance Patterns in Clostridioides difficile Strains Isolated from Neonates in Germany. Antibiotics. 2020;9:481. doi: 10.3390/antibiotics9080481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Freeman J, Vernon J, Morris K, Nicholson S, Todhunter S, Longshaw C, et al. Pan-European longitudinal surveillance of antibiotic resistance among prevalent Clostridium difficile ribotypes. Clin Microbiol Infect. 2015;21:248. doi: 10.1016/j.cmi.2014.09.017. [DOI] [PubMed] [Google Scholar]