Abstract
Separated by a century, the influenza pandemic of 1918 and the COVID-19 pandemic of 2019–2021 are among the most disastrous infectious disease emergences of modern times. Although caused by unrelated viruses, the two pandemics are nevertheless similar in their clinical, pathological, and epidemiological features, and in the civic, public health, and medical responses to combat them. Comparing and contrasting the two pandemics, we consider what lessons we have learned over the span of a century and how we are applying those lessons to the challenges of COVID-19.
It was the best of times when renowned artist Marilee Shapiro Asher finally left the hospital, in April 2020, after five days of struggling with COVID-19 (caused by severe acute respiratory syndrome coronavirus 2 [SARS-CoV-2]).1 We know that Asher was glad to be home, looking forward to returning to her studio and her exhibitions, because she had already written about what it was like surviving an eerily similar respiratory illness as a 6-year-old girl: her most vivid memory was not the days in bed, but finally being allowed to get up one morning and join her family at the breakfast table, a joyous event signaling recovery.
That joyous breakfast was in 1918, when Marilee survived the so-called “Spanish” influenza, estimated to have killed at least 50 million people worldwide, one of the deadliest single events in all of human history.2 Recovering from COVID-19 last year, Asher, then in her 108th year, was among a dwindling cohort of 1918 pandemic survivors who not only still remembered it but who also were now facing another lethal pandemic: COVID-19. Childhood memories like Asher’s were supplemented by an enormous body of medical, scientific, public heath, and societal information concerning that earlier pandemic. It is worth reflecting on this body of collective memory as we travel through the dark uncertainty of another pandemic that threatens and impacts millions of lives. From the vantage point of an additional century of medical and social progress, it is hoped that we are mastering history’s lessons.
THE 1918 INFLUENZA PANDEMIC
Before 1918, influenza was a poorly understood disease of unknown cause. The 1918 pandemic appeared suddenly in a few populous cities including in China in June3 and in Northern Europe in July‒August 1918.4 It rebounded over most of the world (in both the Northern and Southern Hemispheres) in September‒November 1918, featuring from one to several additional recurrences beginning in late 1918‒early 1919.2,5,6 In the United States, an estimated 675 000 people died in the first year, equivalent to about 2.16 million deaths in today’s much larger population, an approximate 1% case‒fatality ratio.2 The explosivity of the pandemic was staggering. Bodies were sometimes “stacked like cord wood” in hospitals, or by roads outside of cemeteries; coffins had to be mass produced on a large scale (Figure 1).
FIGURE 1—
Both the (a) 1918 and the (b) 2020 Pandemics Featured Hastily Assembled Cemeteries, Mass Graves, and Collections of Unburied Bodies
Note. Photo by Willy Kurniawan, courtesy of Reuters. Printed with permission.
Over a few years, the 1918 pandemic settled into a pattern of less fatal annual seasonality. Human influenza A viruses were first isolated in 1933.7 At that time, isolation materials from the 1918 pandemic were thought not to exist; however, decades later (1996–2005) the viral genome was fully sequenced from RNA fragments in pathological materials of 1918‒1919 pandemic victims; soon thereafter, it was reconstructed as a fully infectious virus and studied experimentally.7 Viral descendants of the 1918 “founder” virus are still circulating today as seasonal influenza A viruses; subsequent pandemics in 1957, 1968, and 2009 all resulted from genetic updating of the 1918 virus via a mutational mechanism called gene segment reassortment.8 Over the period of a century, viral descendants of this single emergent virus have caused tens of millions of additional deaths, adding to the tragic losses of 1918. Fortunately, to date, there is evidence that public health restrictions to control COVID-19 (e.g., social distancing, mask wearing, business closures) are controlling influenza as well. As we are in the early second year of the COVID-19 pandemic, we cannot predict with certainty whether the virus will persist as the 1918 influenza virus did, or die out in the face of growing population immunity associated with natural infection and new COVID-19 vaccines.
CLINICAL AND PATHOLOGICAL COMPARISONS
Although caused by unrelated viruses, the two diseases are similar in their clinical features (Figure 2). Both are respiratory viruses transmitted and acquired via respiratory inoculation, and both emerged in global populations with little or no preexisting immunity. Typical signs and symptoms of both full-blown diseases include fever, chills, fatigue, muscle aches, nasal congestion or rhinorrhea, headache, and cough, with variable sore throat, dyspnea, and nausea, vomiting, or diarrhea. Both diseases feature many mild, atypical, and asymptomatic infections, but also complicating, sometimes fatal, pneumonias in about 2% of those clinically ill. In the case of COVID-19, unusual as well as late complications are being noted with increasing frequency, including tissue and organ damage, neurological complications, and inflammatory syndromes. It is not clear to what extent, if any, such complications occurred with 1918 influenza, although, curiously, neurological complications were said to be prominent in the 1889 influenza pandemic.
FIGURE 2—
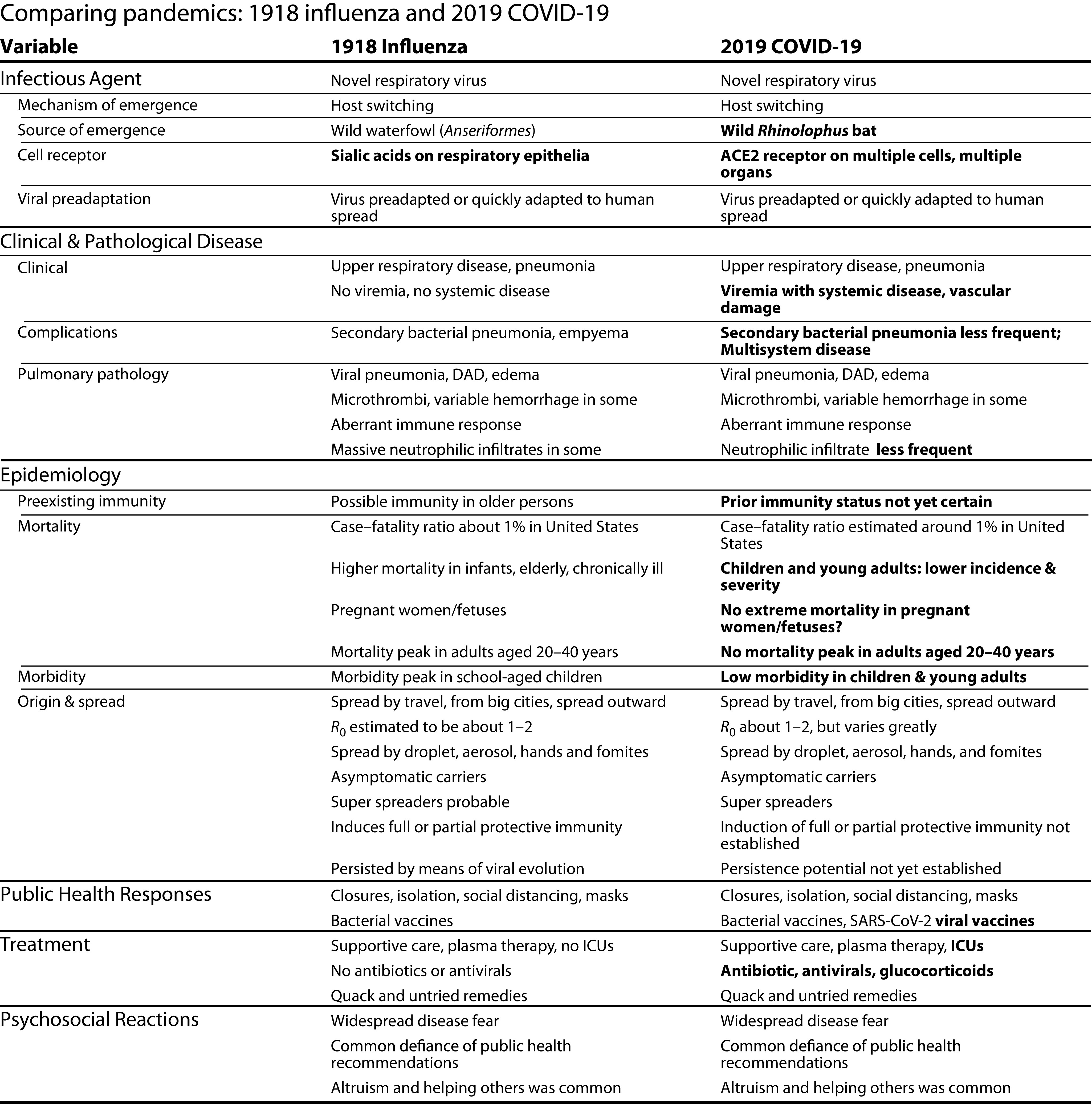
Comparing Pandemics: 1918 Influenza and 2019 COVID-19
Note. DAD = diffuse alveolar damage; ICU = intensive care unit; SARS-CoV-2 = severe acute respiratory syndrome-2.
Typical influenza pneumonia in 1918 occurred in a bronchopneumonic pattern associated with secondary bacterial pneumonias caused by pathogens carried silently in the upper respiratory tract, including Streptococcus pneumoniae, Streptococcus pyogenes, and Staphylococcus aureus.9,10 Initial autopsy data from COVID-19 patients suggest a similar histologic picture of viral pneumonic damage with, however, fewer secondary bacterial pneumonias,11 perhaps in part reflecting widespread use of broad-spectrum antibiotics not available in 1918.
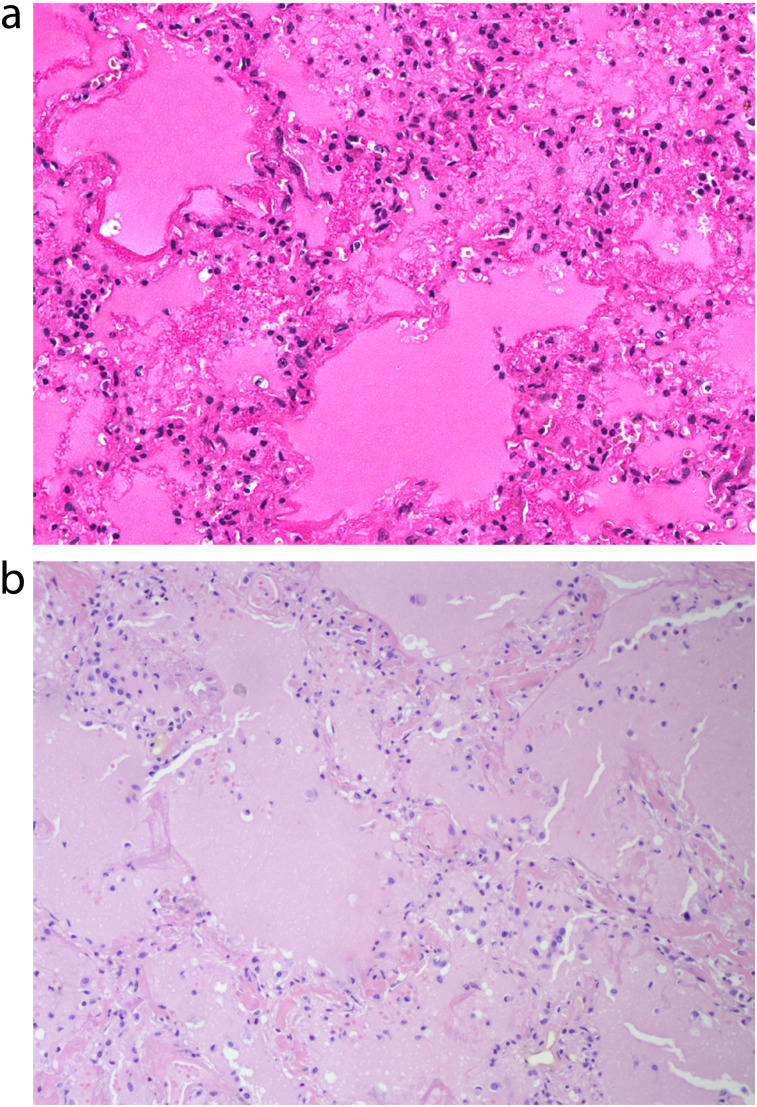
In both diseases, severe pneumonias have been associated histologically with diffuse alveolar damage, hyaline membrane formation, pulmonary edema, and, often, neutrophilic infiltrates11,12 (Figure 3). Autopsy studies of COVID-19 patients reveal widespread medium- and small-vessel thromboses13; pulmonary small-vessel thrombosis was prominent in 1918 influenza as well14,15; however, it has been less frequently observed in more recent influenza autopsies (e.g., during the 2009 H1N1 pandemic16,17). In contrast to 1918, in which tissue damage was mostly pulmonary, in COVID-19, tissue damage has been observed in tissues and organs systemically.18
FIGURE 3—
Representative Pulmonary Histopathology of (a) Fatal 1918 Influenza and (b) Fatal SARS-CoV-2 Infection Showing Acute Diffuse Alveolar Damage With Pulmonary Edema and Hyaline Membranes
Source. Sauter et al.11 and Sheng et al.12
Note. SARS-CoV-2 = severe acute respiratory syndrome coronavirus 2. The histologic patterns of acute diffuse alveolar damage are virtually indistinguishable.
Important pathological differences between the two infections (Figure 2) include the following: influenza infects primarily by binding to sialic acid receptors found on respiratory epithelial cells, whereas SARS-CoV-2 infects various cells of the respiratory tract, gastrointestinal enterocytes, and arterial and venous endothelial cells, as well as arterial smooth-muscle cells, presumably by binding to ACE2 receptors.19 As influenza caused by human-adapted influenza viruses is not associated with viremia, live influenza virus has little direct interaction with the systemic immune system, explaining in part why natural and vaccine-induced protective immunity against influenza is often imperfect. Preliminary data from COVID-19, however, suggest systemic infection of multiple organs,20–22 which can potentially elicit protective immunity more durable than that of influenza, although duration of COVID-19 protection remains to be determined, and reinfections have been documented.23
EPIDEMIOLOGICAL COMPARISONS
It is extremely difficult to know the exact origin of any pandemic disease, because emerging infectious agents arise via host switching from an animal to a human, after which successful adaptation associated with human-to-human transmission occurs.24–26 This process necessarily takes time: by the time the new disease is eventually recognized, its occult beginnings are unlikely to be discovered. In this regard, it is noteworthy that over many centuries, from the 1500s until the modern era, almost all influenza pandemics were first recognized in Asia or Southeast Asia, and then spread westward to Europe and, at some point after the 16th century, from Europe or Asia to the Western Hemisphere.27 As some of the earliest evidence of the existence of the 1918 pandemic came from China,28 this same historical pattern remains plausible, although the geographic origin of the 1918 pandemic remains unknown, with hypotheses ranging from China to Europe to the United States.28–30
When the 1918 pandemic was first recognized clinically and epidemiologically in July 1918, and again in September‒November 1918, it was robustly emerging almost simultaneously in large populous cities all over the globe, in both the Northern and Southern Hemispheres. This pattern indicates that rather than spreading from city to city along travel routes at the time of such explosive emergence, many regions of the world must have been seeded by the virus previously.2 Presumably, the relatively slow global spread of infections by ship, rail, and other means of human travel went undetected until international metropolitan mortality data began to show excess respiratory mortality increases. From these large cities, the disease spread outward to smaller towns and to rural areas, and also caused additional rounds of global spread by ships.
Because of modern international air travel, COVID-19 spread slightly more rapidly than 1918 pandemic influenza; however, the patterns of spread were probably very similar: (1) local emergences and initial spread that went undetected because of low case‒fatality, followed by (2) local, national, and eventually international movement of infectious persons, leading to seeding of cases in crowded metropolitan areas, followed by (3) clusters of respiratory disease mortality that were eventually detected in sensitive metropolitan mortality data, followed quickly by (4) massive global emergence.31
SARS-CoV-2 was first detected in Wuhan, Hubei Province, China, and spread simultaneously outward within China and via international air routes. It is highly likely that SARS-CoV-2 emerged from within a tight phylogenetic cluster of Sarbecoviruses infecting Rhinolophus (horseshoe) bats found mostly in Southwest China and contiguous areas of Cambodia, the Lao PDR, Myanmar, and Vietnam.26,32,33 How the virus got to the place of its initial detection, at least 850 miles away in Wuhan, remains unknown; possible explanations include the mobility and long-distance ranges of various bat species, undetected cross-infection from Rhinolophus to other bat species, or infection and movement of secondarily infected animal hosts or of humans.
EMERGENCE VIA ANIMAL-TO-HUMAN VIRAL HOST SWITCHING
The 1918 pandemic “founder” virus was genetically and functionally very similar, in sequences of all eight genes, to avian viruses that then existed, and that still exist, in the global reservoir of wild waterfowl (Anseriformes).34 It is unknown whether an avian virus host-switched directly into humans or first switched into a different host, perhaps another mammal, and from there to humans.35 However, phylogenetic analysis of the human virus suggests that emergence must have occurred in or shortly before 1918.2
SARS-CoV-2 is very close genetically to numerous enzootic Sarbecoviruses of Rhinolophus bats found in Southwest China and contiguous areas, suggesting one of three possibilities32,36–38: (1) an as-yet-undiscovered enzootic Sarbecovirus identical to SARS-CoV-2 emerged into humans directly; (2) a different but closely related Sarbecovirus emerged directly into humans and spread silently for some period of time, accumulating new mutations as it adapted to human transmission; or (3) humans were infected via an intermediate animal host that had originally been infected by a Rhinolophus-transmitted Sarbecovirus.36,37,39 Thus, 1918 influenza and SARS-CoV-2 share the same origin mysteries of direct versus indirect emergence from a natural animal host, and of extent of postemergence genetic adaptation to humans.
Both 1918 influenza and COVID-19 are among the deadliest examples of viral emergences from the animal‒human interface.33 How this happens and what we can do to prevent it from happening are among the most important areas of research in the study of emerging infections.33,40 The host-switching ability of both viruses may be an established evolutionary mechanism: both 1918 influenza and SARS-CoV-2 are promiscuous in their ability to infect mammals, facilitating broad epidemicity and epizooticity. In 1918, the human virus was quickly transmitted to pigs,41 while housecats were sometimes infected by their owners (as seen in previous influenza pandemics). A century later, humans and pigs are still frequently exchanging their influenza viruses.42 Unexpected deaths of chimpanzees and gorillas in 1918 were thought to be attributable to influenza. Horses, dogs, seals, and other animals have also been involved in influenza virus exchanges.43 SARS-CoV-2 has infected not only Rhinolophus bats, their reservoir host, but also cats, dogs, minks, and other animals44; closely related SARS-like viruses have infected pangolins (Manis javanica, a species of anteater).32 Such efficient intra- and interspecies exchanges may have enhanced evolution and survival of both viruses.
VIRAL TRANSMISSION
Both viruses are transmitted by the respiratory route via large droplets, fine-particle (< 5 μm) aerosols, or by hands or fomites contaminated with respiratory secretions. Both viruses spread by silent transmission—that is, transmission by presymptomatic (incubating) people, by asymptomatic infected people, by people with mild or atypical symptoms who are not recognized as being potentially infectious, and, less commonly, by people who have recovered from illness but may still be excreting virus.39,45 Unlike influenza, SARS-CoV-2 infects enteric cells, but gastrointestinal transmission has not yet been shown to be important.
Preliminary evidence suggests roughly equivalent effects of environmental variables on spread of both viruses (e.g., effects of airflow, temperature, and humidity). This has important implications for COVID-19 public health control measures such as social distancing and controlling airflow in hospitals, nursing homes, workplaces, and recreational venues, such as restaurants and bars.
Regarding seasonality, 1918 pandemic influenza was first detected in the early summer of the Northern Hemisphere and did not spread globally until September‒October 1918. When it did so, it aggressively spread not only in the Northern Hemisphere but also in the Southern Hemisphere’s spring season (e.g., in South Africa46 and in New Zealand47). Five hundred years of observation48 suggest that influenza pandemics can appear at any time of year, but when they arrive in summer they are likely to be somewhat blunted until they rebound more forcefully in the fall; when pandemics arrive at other times of year, summers seem to temporarily slow viral spread.48 This pattern was seen in both the 1957 and 2009 influenza pandemics; in the United States, both pandemics arrived in the spring, slowed down in the summer, and then picked up in the fall. The presumed reasons for this pattern include physical effects of temperature and humidity on viral spread and more summer hours spent outdoors where airflow is optimal and crowding usually less extreme. To date, seasonal effects on COVID-19 spread have not been fully documented because few regions have been in the throes of COVID-19 for much more than a full calendar year. Moreover, the effects of season and of often-intermittent and incomplete public health control efforts are hard to disentangle.
PATTERNS OF MORBIDITY AND MORTALITY
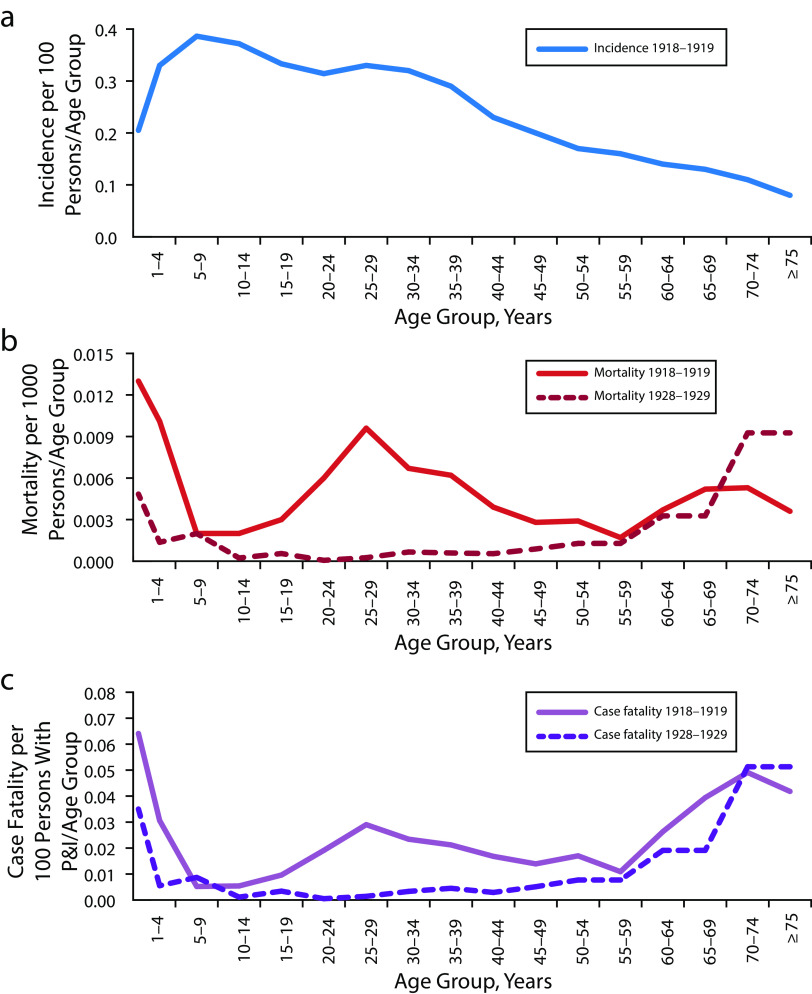
In all circumstances studied over the past 130 years, except in 1918–1920, patterns of age-specific morbidity, mortality, and case‒fatality for pandemic influenza have been similar. Because influenza pandemics emerge when all or most of the global population lacks immunity to the new pandemic virus, moderately high attack rates within the first year, usually between 30% and 60% of the population, are common. Age-specific morbidity patterns have been highly similar for known influenza pandemics, featuring peak morbidity rates in school-aged children and young adults, slightly lower rates in both very young children and in adults aged 30 to 55 years, and much lower rates at older ages (Figure 4). This pattern presumably reflects exposure risks related to school, work, and other congregating activities, as well as the possibility of prior exposure to related influenza viruses within the older age group.
FIGURE 4—
Age-Specific Morbidity and Mortality of Influenza in 1918–1919 and, for Comparison, in 1928–1929, as Determined by US “P and I” Data by (a) Incidence per 100 Persons Ill With Pneumonia and Influenza per Age Group; (b) Mortality per 1000 Persons per Age Group; and (c) Case‒Fatality
Source. Morens and Taubenberger.49
Note. P and I = pneumonia and influenza. Parts b and c compare the W-shaped curves of age-specific mortality and case‒fatality seen in 1918–1919 with more typical U-shaped curves from 1928 to 1929. Between 1889 and the present time, U-shaped curves have been seen in all pandemics and seasonal epidemics except for 1918 and the several years thereafter. Morbidity and mortality data reflecting diagnoses of pneumonia and influenza (so-called “P and I”) are still widely used today for epidemiological purposes (e.g., for estimating total influenza deaths during periods of influenza prevalence) because incomplete morbidity reporting and imperfect death certificate accuracy greatly underestimate infections and deaths from influenza and its secondary bacterial complications. National or large-population data permitting similar calculations for COVID-19 are not yet available, although preliminary data suggest that age-specific mortality is very low in infants and children, rising regularly with age thereafter.
Overall influenza mortality varies significantly, with some pandemic viruses being highly pathogenic (approximate 1% case‒fatality in the United States in the 1918 pandemic vs less than 0.05% case‒fatality in the 2009 pandemic). The elderly; people with serious respiratory, cardiac, metabolic, and other diseases; and pregnant women are always at elevated mortality risk from influenza.
With the exception of 1918–1920, pandemic and seasonal influenza exhibit a characteristic mortality pattern. Age-specific influenza mortality is classically U-shaped, with elevated mortality in infants and young toddlers and the elderly, but with very low mortality at all ages in between. A different pattern was seen in 1918–1920: a W-shaped pattern (Figure 4) featured a third mortality peak in those aged 20 to 40 years. This pattern, never seen before or since, disappeared entirely in the early 1920s.50,51 It remains unexplained, and, while likely not a signature of the 1918 virus, it may be related to preexisting age cohort‒specific, cross-protective immunity.
In the early stages of the COVID-19 pandemic, morbidity and mortality patterns are still not fully established, in part because of the relatively high percentage of asymptomatic infections coupled with underdiagnosis of cases. Overall case‒ and infection‒fatality ratios, which are population structure‒dependent, have been estimated from as high as 3% to well below 1%.52 Speculative theories to explain low morbidity and mortality in the young include (1) protection afforded by prior and recent exposure to circulating endemic coronaviruses, two of which—HCoV-HKU1 and HCoV-OC43—are β-coronaviruses, albeit not closely related to SARS-CoV-2; (2) increased exposures to other infectious agents that stimulate generic innate immune responses; or (3) immune enhancement mechanisms.39
In contrast to influenza, which causes high mortality and high fetal loss, significant COVID-19 mortality in pregnant women and their fetuses is only now beginning to become better appreciated, although the extent of maternal and fetal risks remains to be fully established.53–56 In 1918, as in 2020,57 mortality was higher in the poor, in African Americans and Native Americans, in health care workers, and in workers in crowded occupations.50,58–60 These patterns, observed for most infectious diseases, reflect societal inequalities and inadequate occupational safety measures.
As descendants of the 1918 influenza virus persist to this day,8 a question arises about whether SARS-CoV-2 will do the same. Furthermore, a possibility to be considered is whether, similar to influenza, it will elicit a weakly protective immune response and then circumvent that response with further viral evolution by antigenic drift or other mechanisms such as viral genetic recombination. The recent (in late 2020) emergences of SARS-CoV-2 genetic variants, some apparently associated with increased transmissibility and immune escape,61 may be an early answer to this question, auguring future COVID-19 reemergences caused by antigenically drifting strains, in a manner analogous to the genetic drift of influenza A viruses. Descendants of the 1918 virus still circulate; we can only speculate whether SARS-CoV-2 or its descendants will still be circulating in 2120. (Continued in Part II.62)
ACKNOWLEDGMENTS
This work was supported by the intramural research program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
CONFLICTS OF INTEREST
The authors have no conflicts of interest.
Footnotes
See also Leavitt, p. 996.
REFERENCES
- 1.Dvorak P. At 107, she’s a force who’s conquered 2 pandemics. Washington Post. May 8, 2020. Available at: https://www.washingtonpost.com/local/at-107-this-artist-just-beat-covid-19-it-was-the-second-pandemic-she-survived/2020/05/07/2aba2e28-9084-11ea-a0bc-4e9ad4866d21_story.html. Accessed February 4, 2021.
- 2.Taubenberger JK, Kash JC, Morens DM. The 1918 influenza pandemic: 100 years of questions answered and unanswered. Sci Transl Med. 2019;11(502):eaau5485. doi: 10.1126/scitranslmed.aau5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cadbury WW. The 1918 pandemic of influenza in Canton. China Med J. 1920;34:1–17. [Google Scholar]
- 4.Low RB. The incidence of epidemic influenza during 1918‒19 in Europe and in the Western Hemisphere. In: Ministry of Health, ed. Reports on Public Health and Medical Subjects. No. 4. Reports on the Pandemic of Influenza, 1918‒19. Part II. Influenza in Foreign Countries, 1918‒19. Chapter 1. Europe and the Western Hemisphere. London, England: His Majesty’s Stationery Office; 1920:202‒348.
- 5.Taubenberger JK, Morens DM. 1918 influenza: the mother of all pandemics. Emerg Infect Dis. 2006;12(1):15–22. doi: 10.3201/eid1209.05-0979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morens DM, Taubenberger JK. Influenza cataclysm, 1918. N Engl J Med. 2018;379(24):2285–2287. doi: 10.1056/NEJMp1814447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taubenberger JK, Hultin JV, Morens DM. Discovery and characterization of the 1918 pandemic influenza virus in historical context. Antivir Ther. 2007;12(4 pt B):581–591. [PMC free article] [PubMed] [Google Scholar]
- 8.Morens DM, Taubenberger JK, Fauci AS. The persistent legacy of the 1918 influenza virus. N Engl J Med. 2009;361(3):225–229. doi: 10.1056/NEJMp0904819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winternitz MC, Wason IM, McNamara FP. The Pathology of Influenza. New Haven, CT: Yale University Press; 1920. [Google Scholar]
- 10.Morens DM, Taubenberger JK, Fauci AS. Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: implications for pandemic influenza preparedness. J Infect Dis. 2008;198(7):962–970. doi: 10.1086/591708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sauter JL, Baine MK, Butnor KJ et al. Insights into pathogenesis of fatal COVID-19 pneumonia from histopathology with immunohistochemical and viral RNA studies. Histopathology. 2020;77(6):915–925. doi: 10.1111/his.14201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sheng ZM, Chertow DS, Ambroggio X et al. Autopsy series of 68 cases dying before and during the 1918 influenza pandemic peak. Proc Natl Acad Sci U S A. 2011;108(39):16416–16421. doi: 10.1073/pnas.1111179108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fox SE, Akmatbekov A, Harbert JL, Li G, Quincy Brown J, Vander Heide RS. Pulmonary and cardiac pathology in African American patients with COVID-19: an autopsy series from New Orleans. Lancet Respir Med. 2020;8(7):681–686. doi: 10.1016/S2213-2600(20)30243-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.LeCount ER. Disseminated necrosis of the pulmonary capillaries in influenzal pneumonia. J Am Med Assoc. 1919;72(21):1519–1520. doi: 10.1001/jama.1919.02610210015003. [DOI] [Google Scholar]
- 15.Walters KA, D’Agnillo F, Sheng ZM et al. 1918 pandemic influenza virus and Streptococcus pneumoniae co-infection results in activation of coagulation and widespread pulmonary thrombosis in mice and humans. J Pathol. 2016;238(1):85–97. doi: 10.1002/path.4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taubenberger JK, Morens DM. The pathology of influenza virus infections. Annu Rev Pathol. 2008;3(1):499–522. doi: 10.1146/annurev.pathmechdis.3.121806.154316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gill JR, Sheng ZM, Ely SF et al. Pulmonary pathologic findings of fatal 2009 pandemic influenza A/H1N1 viral infections. Arch Pathol Lab Med. 2010;134(2):235–243. doi: 10.5858/134.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rapkiewicz AV, Mai X, Carsons SE et al. Megakaryocytes and platelet-fibrin thrombi characterize multi-organ thrombosis at autopsy in COVID-19: A case series. EClinicalMedicine. 2020;24:100434. doi: 10.1016/j.eclinm.2020.100434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Q, Zhang Y, Wu L et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181(4):894–904.e9. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Braun F, Lutgehetmann M, Pfefferle S et al. SARS-CoV-2 renal tropism associates with acute kidney injury. Lancet. 2020;396(10251):597–598. doi: 10.1016/S0140-6736(20)31759-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Puelles VG, Lutgehetmann M, Lindenmeyer MT et al. Multiorgan and renal tropism of SARS-CoV-2. N Engl J Med. 2020;383(6):590–592. doi: 10.1056/NEJMc2011400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fajnzylber J, Regan J, Coxen K et al. SARS-CoV-2 viral load is associated with increased disease severity and mortality. Nat Commun. 2020;11(1):5493. doi: 10.1038/s41467-020-19057-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Elslande J, Vermeersch P, Vandervoort K et al. Symptomatic SARS-CoV-2 reinfection by a phylogenetically distinct strain. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa1330. ciaa1330; epub ahead of print September 5, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuiken T, Holmes EC, McCauley J, Rimmelzwaan GF, Williams CS, Grenfell BT. Host species barriers to influenza virus infections. Science. 2006;312(5772):394–397. doi: 10.1126/science.1122818. [DOI] [PubMed] [Google Scholar]
- 25.Parrish CR, Holmes EC, Morens DM et al. Cross-species virus transmission and the emergence of new epidemic diseases. Microbiol Mol Biol Rev. 2008;72(3):457–470. doi: 10.1128/MMBR.00004-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Latinne A, Hu B, Olival KJ Origin and cross-species transmission of bat coronaviruses in China. Preprint. Posted online May 31, 2020. bioRxiv. [DOI] [PMC free article] [PubMed]
- 27.Patterson KD, Pyle GF. The geography and mortality of the 1918 influenza pandemic. Bull Hist Med. 1991;65(1):4–21. [PubMed] [Google Scholar]
- 28.Shortridge KF. The 1918 “Spanish” flu: pearls from swine? Nat Med. 1999;5(4):384–385. doi: 10.1038/7383. [DOI] [PubMed] [Google Scholar]
- 29.Oxford JS, Gill D. A possible European origin of the Spanish influenza and the first attempts to reduce mortality to combat superinfecting bacteria: an opinion from a virologist and a military historian. Hum Vaccin Immunother. 2019;15(9):2009–2012. doi: 10.1080/21645515.2019.1607711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barry JM. The site of origin of the 1918 influenza pandemic and its public health implications. J Transl Med. 2004;2(1):3. doi: 10.1186/1479-5876-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Worobey M, Pekar J, Larsen BB et al. The emergence of SARS-CoV-2 in Europe and North America. Science. 2020;370(6516):564–570. doi: 10.1126/science.abc8169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morens DM, Breman JG, Calisher CH et al. The origin of COVID-19 and why it matters. Am J Trop Med Hyg. 2020;103(3):955–959. doi: 10.4269/ajtmh.20-0849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morens DM, Fauci AS. Emerging pandemic diseases: how we got to COVID-19 [erratum in Cell. 2020;183(3):837] Cell. 2020;182(5):1077–1092. doi: 10.1016/j.cell.2020.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dugan VG, Chen R, Spiro DJ et al. The evolutionary genetics and emergence of avian influenza viruses in wild birds. PLoS Pathog. 2008;4(5):e1000076. doi: 10.1371/journal.ppat.1000076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wille M, Holmes EC. The ecology and evolution of influenza viruses. Cold Spring Harb Perspect Med. 2020;10(7):a038489. doi: 10.1101/cshperspect.a038489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang YZ, Holmes EC. A genomic perspective on the origin and emergence of SARS-CoV-2. Cell. 2020;181(2):223–227. doi: 10.1016/j.cell.2020.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS-CoV-2. Nat Med. 2020;26(4):450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu R, Zhao X, Li J et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morens DM, Daszak P, Taubenberger JK. Escaping Pandora’s Box—another novel coronavirus. N Engl J Med. 2020;382(14):1293–1295. doi: 10.1056/NEJMp2002106. [DOI] [PubMed] [Google Scholar]
- 40.Allen T, Murray KA, Zambrana-Torrelio C et al. Global hotspots and correlates of emerging zoonotic diseases. Nat Commun. 2017;8(1):1124. doi: 10.1038/s41467-017-00923-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koen J. A practical method for field diagnosis of swine diseases. Am J Vet Med. 1918;14:468–470. [Google Scholar]
- 42.Nelson MI, Gramer MR, Vincent AL, Holmes EC. Global transmission of influenza viruses from humans to swine. J Gen Virol. 2012;93(pt 10):2195–2203. doi: 10.1099/vir.0.044974-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56(1):152–179. doi: 10.1128/MR.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haider N, Rothman-Ostrow P, Osman AY et al. COVID-19—zoonosis or emerging infectious disease? Front Public Health. 2020;8:596944. doi: 10.3389/fpubh.2020.596944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morens DM, Daszak P, Markel H, Taubenberger JK. Pandemic COVID-19 joins history’s pandemic legion. MBio. 2020;11(3):e00812–20. doi: 10.1128/mBio.00812-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patterson KD, Pyle GF. The diffusion of influenza in sub-Saharan Africa during the 1918‒1919 pandemic. Soc Sci Med. 1983;17(17):1299–1307. doi: 10.1016/0277-9536(83)90022-9. [DOI] [PubMed] [Google Scholar]
- 47.Maze MJ, Beckert L. The 1918‒1919 influenza epidemic in New Zealand: end of the century reflections. N Z Med J. 2019;132(1507):8–10. [PubMed] [Google Scholar]
- 48.Morens DM, Taubenberger JK. Pandemic influenza: certain uncertainties. Rev Med Virol. 2011;21(5):262–284. doi: 10.1002/rmv.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morens DM, Taubenberger JK. The 1918 influenza pandemic: a still-mysterious litmus test for pandemic prevention and control. In: Blackburn CC, editor. Preparing for Pandemics in the Modern World. College Station, TX: Texas A&M University Press; 2020. [Google Scholar]
- 50.Jordan EO. Epidemic Influenza: A Survey. Chicago, IL: American Medical Association; 1927. [Google Scholar]
- 51.Viboud C, Eisenstein J, Reid AH, Janczewski TA, Morens DM, Taubenberger JK. Age- and sex-specific mortality associated with the 1918‒1919 influenza pandemic in Kentucky. J Infect Dis. 2013;207(5):721–729. doi: 10.1093/infdis/jis745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O’Driscoll M, Ribeiro Dos Santos G, Wang L et al. Age-specific mortality and immunity patterns of SARS-CoV-2. Nature. 2021;590(7844):140‒145. doi: 10.1038/s41586-020-2918-0. [DOI] [PubMed] [Google Scholar]
- 53.Ellington S, Strid P, Tong VT et al. Characteristics of women of reproductive age with laboratory-confirmed SARS-CoV-2 infection by pregnancy status—United States, January 22‒June 7, 2020. MMWR Morb Mortal Wkly Rep. 2020;69(25):769–775. doi: 10.15585/mmwr.mm6925a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dashraath P, Wong JJL, Su LL, See KC, Fisher D. Coronavirus disease 2019 (COVID-19) and pregnancy: responding to a rapidly evolving situation. Obstet Gynecol. 2020;136(1):191–192. doi: 10.1097/AOG.0000000000003963. [DOI] [PubMed] [Google Scholar]
- 55.Memoli MJ, Harvey H, Morens DM, Taubenberger JK. Influenza in pregnancy. Influenza Other Respir Viruses. 2013;7(6):1033–1039. doi: 10.1111/irv.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de Souza Silva GA, da Silva SP, da Costa MAS et al. SARS-CoV, MERS-CoV and SARS-CoV-2 infections in pregnancy and fetal development. J Gynecol Obstet Hum Reprod. 2020;49(10):101846. doi: 10.1016/j.jogoh.2020.101846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alcendor DJ. Racial disparities-associated COVID-19 mortality among minority populations in the US. J Clin Med. 2020;9(8):2442. doi: 10.3390/jcm9082442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sattenspiel L. Regional patterns of mortality during the 1918 influenza pandemic in Newfoundland. Vaccine. 2011;29(suppl 2):B33–B37. doi: 10.1016/j.vaccine.2011.02.046. [DOI] [PubMed] [Google Scholar]
- 59.Økland H, Mamelund SE. Race and 1918 influenza pandemic in the United States: a review of the literature. Int J Environ Res Public Health. 2019;16(14):2487. doi: 10.3390/ijerph16142487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mamelund SE. A socially neutral disease? Individual social class, household wealth and mortality from Spanish influenza in two socially contrasting parishes in Kristiania 1918‒19. Soc Sci Med. 2006;62(4):923–940. doi: 10.1016/j.socscimed.2005.06.051. [DOI] [PubMed] [Google Scholar]
- 61.Centers for Disease Control and Prevention. Emerging SARS-CoV-2 variants. 2021. Available at: https://www.cdc.gov/coronavirus/2019-ncov/more/science-and-research/scientific-brief-emerging-variants.html. Accessed February 3, 2021. [PubMed]
- 62.Morens DM, Taubenberger JK, Fauci AS. A centenary tale of two pandemics: the 1918 influenza pandemic and COVID-19. Part II. Am J Public Health. 2021 doi: 10.2105/AJPH.2021.306310. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]