Abstract
Bacterial infections are one of the main health concerns humanity faces today and bacterial resistances and protection mechanisms are set to aggravate the issue in the coming years. An increasing number of bacterial strains evades antibiotic treatment by hiding inside cells. Conventional antimicrobial agents are unable to penetrate or be retained in the infected mammalian cells. Recent approaches to overcome these limitations have focused on load-carrier systems, requiring a triggered discharge leading to complex release kinetics. The unison of potent antimicrobial activity with high mammalian cell compatibility is a prerequisite for intracellular activity, which is not well-met by otherwise well-established inorganic systems, such as silver-based nanoparticles. In this work, load and carrier are combined into one functional inorganic nanoparticle system, which unites antimicrobial activity with mammalian cell compatibility. These multicomponent nanohybrids based on cerium oxide are produced in one step, yet unite complex materials. The nanoparticles form suprastructures of similar size and surface charge as bacteria, therefore facilitating the uptake into the same subcellular compartments, where they unleash their antibacterial effect. Such intrinsically antibacterial nanohybrids significantly reduce bacterial survival inside macrophages without harming the latter. Furthermore, blocking of nanoparticle endocytosis and subcellular electron microscopy elucidate the mechanism of action. Taken together, this work presents the first demonstration of antibacterial activity of ceria-based nanoparticles inside of mammalian cells and offers a route to straightforward and robust intracellular antibacterial agents that do not depend on payload delivery or biological constituents.
Ceria/bioglass nanohybrids significantly reduce bacterial survival inside human cells without harming the latter and overcome major shortcomings of conventional antibiotics.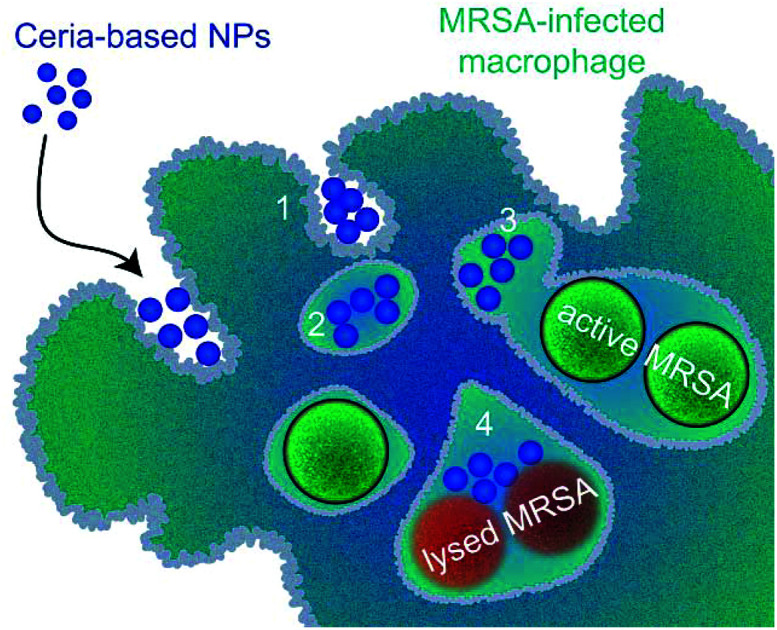
1. Introduction
Relapse of bacterial infection after well-conducted antibiotic treatment remains a major health concern.1 Apart from direct antibiotic resistance, invasion into host cells is the biggest obstacle to effective antibiotic treatment.2 Whereas most microorganisms are detected and destroyed by the host organism through processes including phagocytosis, some microbes are capable of evading destruction by residing intracellularly. The environment inside host cells provides a safe haven where certain pathogens can avoid detection and subsequent elimination by the immune system.3 Clinical significance of intracellular pathogens is especially high because the direct contact between host cells allows intracellular bacteria to spread within the body without being attacked by the immune system.3 In addition to well-known intracellular pathogens, such as Salmonella enterica and Listeria monocytogenes, there is growing evidence for the intracellular survival of Staphylococcus aureus (S. aureus).4,5 Staphylococci are adaptable pathogens and have the ability to infect, invade, persist, and replicate in any human tissue, especially skin, bone, and visceral organs.6 When S. aureus infections disseminate into the bloodstream, mortality rates of up to 30% have been reported,7,8 making S. aureus one of the most lethal infections worldwide. Intracellular survival of S. aureus has been observed in a diversity of host cells including endothelial cells, epithelial cells, fibroblasts, osteoblasts, keratinocytes,4 and professional phagocytes such as macrophages and neutrophils.9–11 Especially the intracellular survival of methicillin-resistant Staphylococcus aureus (MRSA) has raised great concerns for public health.12 A recent in vivo study has shown how MRSA survives antibiotic therapy by hiding in Kupffer cells (macrophages) and ultimately escapes to colonize other tissues.13
The intracellular survival of bacteria may result from the lack of access of the antibiotics to the intracellular niche as well as the ability of S. aureus to neutralize the phagolysosome (Scheme 1). For example, it has been described that S. aureus can resist the lytic effect of lysozymes on the peptidoglycan in the cell wall14 and is able to block the action of antimicrobial peptides. S. aureus has also been shown to express urease, which catalyzes the hydrolysis of urea to form ammonia, resulting in the pH neutralization of the phagosome.15 Additionally, the degradation of reactive oxygen species (ROS) by expression of catalase, superoxide dismutase and peroxiredoxins has been observed in S. aureus.16–19 Research has made tremendous efforts to provide a solution for the intracellular killing of pathogens. Most small-molecule antimicrobial agents have poor cellular penetration, are not well-distributed within the cell and thus show reduced intracellular activity. For example, the widely used antibiotic gentamicin has a very low permeability across non-polar cell membranes due to its high polarity, rendering the intracellular killing of pathogens inefficient.20,21
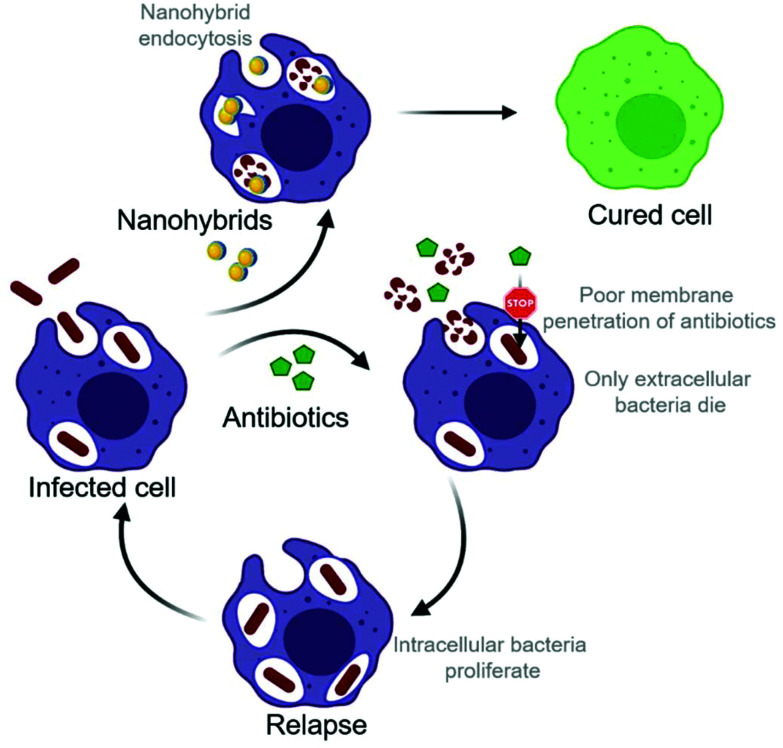
Scheme 1. Intracellular infections are hard to treat with conventional antibiotics due to their poor membrane permeability. Nanohybrids with intrinsic antimicrobial activity tend end up in the same compartment as bacteria hiding inside the cells because of their similar size and surface charge. Treating intracellular infections with endocytosed nanohybrids has the prospect to bring significant benefit.
Nanoparticles have been considered promising vehicles for the intracellular delivery of antibiotics.22,23 In recent work, inorganic nanoparticles have been functionalized with gentamicin in order to eliminate intracellular bacteria.24,25 Lipid and polymer-based capsules have been investigated for their function as carriers, where the core of the nanoparticle serves as a vehicle to carry active molecules.26 Fascinating results have been achieved in a study using antibiotic-loaded liposomes for the delivery of vancomycin to Kupffer cells containing S. aureus by harnessing their intrinsic ability to catch foreign particles.13 In this way, vancomycin could be delivered to the previously inaccessible intracellular reservoir. However, the potentially limited intracellular antibiotic activity, antibiotic release prior to reaching the target site, and systemic toxicity remain major issues of all these load-carrier systems.
Here, inorganic nanoparticles with direct antibiotic activity are proposed instead, thus effectively uniting load and carrier. Various inorganic nanoparticles with antimicrobial properties have been developed. Most prominently, silver-based systems have shown high antibiotic activity countless times. However, the toxicity of released silver ions towards mammalian cells has raised concerns. Toxicity of silver-based nanoparticles especially becomes an issue following nanoparticle internalization into phagocytes and exposure times of several hours.27,28 Therefore, few inorganic systems have been explored for the elimination of intracellular pathogens. Nonetheless, antimicrobial nanoparticles deserve further consideration due to their unique tissue homing, intratissural and intracellular distribution. Various antimicrobial mechanisms of metal oxide nanoparticles have been proposed, including the induction of oxidative stress,29 binding to proteins,30 and interruption of electron transport processes.31 Interestingly, due to the mostly unspecific nature of those mechanisms, bacteria are reported to be less like to develop resistance against nanoparticles compared to conventional antibiotics.32
Apart from silver-based systems, materials of interest include the biodegradable material bioglass (a blend of SiO2, CaO, Na2O, and P2O5). Bioglass has been widely used in the biomedical field for soft (especially with the incorporation of Sr into the bioglass matrix) and hard tissue applications33–35 and has exhibited antimicrobial properties in nanoparticulate form.36 Similarly, cerium oxide (ceria, CeO2) nanoparticles have documented antimicrobial properties, which strongly depend on material characteristics, including the Ce3+/Ce4+ surface ratio, and environmental factors, such as pH and the presence of phosphate.37–39 The antibacterial activity of cerium oxide has been attributed to the reversible conversion between two valence states of Ce3+/Ce4+.40 On this basis, several mechanisms of action have been proposed.41 To the best of our knowledge, however, the antibacterial activity of nanoceria inside mammalian cells has not yet been demonstrated.
In this work, we present the development of inorganic hybrid metal oxide nanoparticles that unify direct antimicrobial activity with low mammalian cell toxicity. The study is focused on metal–oxide nanoparticles based on bioglass (BG), ceria, hybrids of the bioglass and cerium oxide (BG/ceria), and 2% Zn-doped Sr-substituted hybrids (Zn2-SrBG/ceria), building on their high biocompatibility and potential for wound healing.33 Despite their sophisticated formulation, all nanoparticles were produced in a single step using liquid-feed flame spray pyrolysis (LF-FSP), a highly scalable nanoparticle production method.42 Additionally, silver-based systems based on X% Ag-doped bioglass (AgX-BG, X = 0.1, 0.5, 2%) have been included as a nanoparticle-based reference material with well documented antimicrobial activity.43 Gentamicin, an established conventional small molecule antibiotic was used as a control. The antimicrobial activity of the ceria-based nanoparticles against planktonic MRSA is compared to the silver-based systems used for reference. After confirming mammalian cell compatibility of the metal oxide hybrid nanoparticles, potent intracellular bacterial growth inhibition is demonstrated, especially for Zn2-SrBG/ceria. The inhibition of nanoparticle uptake into cells by blocking actin polymerization provides evidence for the direct antimicrobial effect of inorganic nanoparticles on intracellular MRSA. Scanning transmission electron microscopy (STEM) coupled with elemental mapping further illustrates the uptake of the nanoparticles into the same compartment as the bacteria where they selectively disrupt their membranes.
2. Results and discussion
2.1. Physicochemical properties
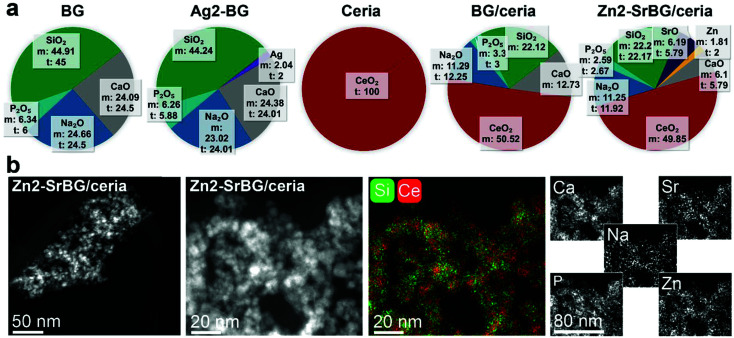
The physicochemical characteristics of the nanoparticles are displayed in Fig. 1. Elemental composition analysis by inductively coupled plasma optical emission spectrometry (Fig. 1a) indicates excellent agreement between the measured and the theoretical composition (according to the stoichiometry of the LF-FSP precursor; maximum deviation <10%). This agreement shows the aptitude of LF-FSP for the one-step and sterile production of sophisticated metal oxide nanoparticles (here with up to 7 different components in one particle). The LF-FSP process is highly scalable with production rates of 5 g h−1 achieved in the lab-scale reactor operating in continuous mode. In pilot scale reactors, production rates of 10–100 g h−1 are achievable for a great variety of metal oxide nanoparticles, including CeO2.44
Fig. 1. (a) Elemental composition of the different nanoparticles measured by inductively coupled plasma optical emission spectrometry (m: measured, t: theoretical). (b) Dark-field scanning transmission electron micrographs of the Zn2-SrBG/ceria nanoparticles including energy-dispersive X-ray spectroscopy mapping.
Scanning transmission electron micrographs (Fig. 1b and Fig. S1†) and dynamic light scattering measurements (DLS, Table S1†) show comparable agglomerate sizes for the different types of particles, respectively. The discrepancy between the particle size observed in electron microscopy and the hydrodynamic size measured by DLS originates from partial agglomeration as well as overestimation of the average hydrodynamic size of fractal agglomerates and is well in line with literature.45,46 Note the similar hydrodynamic size and the remarkably comparable surface charge of the nanoparticle agglomerates (range −35 to −25 mV) with S. aureus (−32 mV).47 The similarity in surface charge and size of the nanoparticles and the bacteria facilitates their uptake into the same cellular compartment, thus allowing the nanoparticles to unfold their activity in close proximity to the bacteria. Energy-dispersive spectroscopy mappings of the Zn2-SrBG/ceria hybrid nanoparticles (Fig. 1b) demonstrate a uniform distribution of the dopants within the bioglass as well as ceria rich regions. X-ray diffraction measurements further confirm the crystalline nature of the ceria component and the amorphous structure of the bioglass (Fig. S2†). X-ray photoelectron spectroscopy (XPS, Tables S2, S3†) and Raman (Fig. S2c and d†) measurements indicate the presence of Ce(iii) in the ceria-based nanoparticles. Ce(iii) and the associated oxygen vacancies are catalytically active, which is demonstrated by the H2O2 scavenging, catalase and superoxide dismutase mimetic activity of the nanoparticles (Fig. S3†). Such catalytic activity is important for their antibacterial activity.48
2.2. Effects on human macrophages and planktonic MRSA
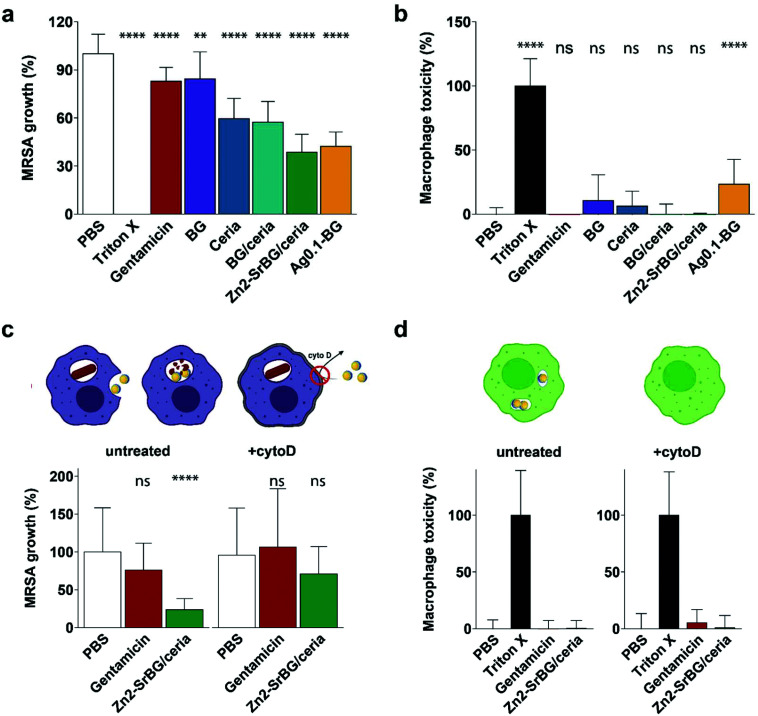
The direct effects of different nanoparticles on MRSA were assessed in a planktonic growth assay. Growth curves of treated and untreated MRSA were recorded by repeated optical measurements at 600 nm (Fig. S4†). The bacteria reach their highest growth phase (log phase) after approx. 2.5 h. Because this phase marks the highest proliferation of bacteria, the 2.5 h time-point was chosen for all subsequent bacterial assays (compare Fig. S4c and d†). After 2.5 h, the bacterial inhibition was assessed by ATP generation quantified by a luciferase assay (Fig. S4a†), whereas growth was measured by counting colony-forming units (CFU, Fig. S4b†). The Ag2-BG nanoparticles show the strongest inhibition on MRSA growth, with values comparable to the treatment of the bacteria with high-dose gentamicin (100 μg ml−1). Furthermore, ceria and Zn2-SrBG/ceria show substantial bacterial inhibition (Fig. S4a†). Although differences between treatments are less pronounced, also the CFU counts show growth inhibition by silver and ceria containing particles (Fig. S4b†). Lower antimicrobial activity was observed for Ag0.1-BG as well as BG nanoparticles (Fig. S4†). Taken together, ceria, Zn2-SrBG/ceria, Ag0.5-BG, and Ag2-BG consistently exhibit strong antimicrobial properties against planktonic MRSA.
2.3. Effects against MRSA hiding inside human macrophages
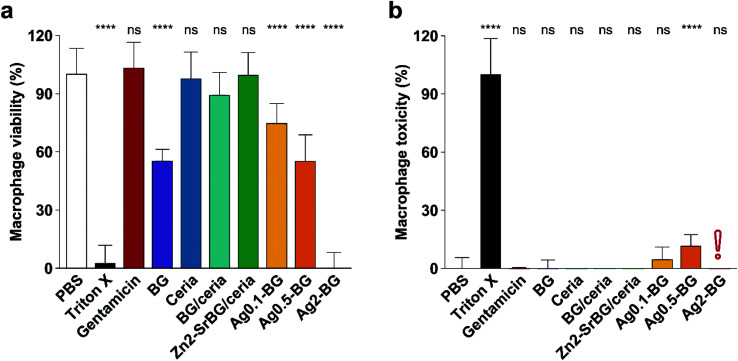
After confirming the antimicrobial activity of the different nanoparticles against planktonic MRSA, their activity towards mammalian cells was assessed in human monocyte-derived macrophages. The viability of the cells (Fig. 2a) and the toxicity of the nanoparticles (Fig. 2b) were assessed by measuring mitochondrial ATP and released lactate dehydrogenase (LDH, indicator for membrane damage) respectively. Significant effects on macrophage viability were found for all silver-containing nanoparticles and bioglass following 2.5 h incubation of macrophages with 0.5 mg ml−1 nanoparticles (Fig. 2a). Viability was also compromised by silver-containing nanoparticles at higher concentrations and incubation times (for 24 h data see ESI Fig. S4e†). In contrast, BG/ceria and especially Zn2-SrBG/ceria nanoparticles were well tolerated at all time points and concentrations up to 1 mg ml−1. Assessment of macrophage cytotoxicity indicates that already after 2.5 h, the membrane integrity is severely compromised by the presence of silver (Fig. 2b and Fig. S4f). The silver-containing nanoparticles partially interfered with the LDH cytotoxicity assay,49 falsely showing low cytotoxicity despite high cell death (Fig. S5a and b†). Taken together, ceria, BG/ceria, and Zn2-SrBG/ceria nanoparticles are well compatible with human macrophages, in contrast to silver-containing nanoparticles. Note that at the concentrations where no significant mammalian cytotoxicity was observed (e.g. 0.1 mg ml−1, Fig. S4e†) also no antimicrobial effects could be detected for the silver-containing nanoparticles (Fig. S4c†). Due to their high toxicity towards macrophages, Ag0.5-BG and Ag2-BG were excluded from further experiments.
Fig. 2. (a) Macrophage viability after 2.5 h treatment (0.5 mg ml−1 NPs, 100 μg ml−1 gentamicin) quantified by ATP generation. BG and especially Ag compromise macrophage viability. (b) Macrophage toxicity quantified by LDH release, which indicates membrane damage. In line with viability measurements, Ag-containing particles express toxicity towards macrophages. The red exclamation point indicates the interference of silver with the LDH assay (compare Fig. S5†). Control PBS was set as 100% viability and 0% toxicity. Significance levels compared to PBS are shown, p < 0.0001. N = 3.
In order to quantify the intracellular antimicrobial effects of the cytocompatible ceria-based nanoparticles, human monocyte-derived macrophages were infected with MRSA (multiplicity of infection, MOI = 10). Extracellular bacteria were eliminated by the addition of gentamicin to the culture media. Following the treatment with the different nanoparticles, gentamicin, or vehicle control (PBS), the cell walls of the macrophages were perforated with saponin without affecting the bacteria50,51 and intracellular bacteria were counted. To ensure that counted bacteria were in fact released from an intracellular compartment, possibly present extracellular bacteria were also counted before saponin perforation (ESI, Fig. S5c and d†). CFU counts of the released bacteria showed that there was a substantial difference in intracellular MRSA survival between different nanoparticle treatments (Fig. 3a). Macrophages treated with Zn2-SrBG/ceria and Ag0.1-BG showed the lowest CFU counts and thus the highest antibacterial activity. Whereas ceria and BG/ceria treated macrophages also showed reduced intracellular bacteria survival, BG nanoparticles had no influence on the intracellular MRSA count. Cytotoxicity was monitored for the full duration of the experiment to exclude macrophage cell wall disruption (Fig. 3b). Significant toxicity was only found for Ag0.1-BG, whereas LDH release was comparable to healthy controls for all other experimental groups indicating that there was no effect of cytotoxicity confounding the experimental findings. These findings show that the sophisticated Zn2-SrBG/ceria nanoparticles unite strong intracellular antimicrobial properties with human cell compatibility.
Fig. 3. (a) Activity towards intracellular MRSA and (b) corresponding low mammalian cell toxicity. Gentamicin dose: 100 μg ml−1; NP dose: 500 μg ml−1. (c) Inhibition of the nanoparticle internalization by blocking of the actin polymerization with cytoD leads to suppression of the antimicrobial activity. (d) No macrophage toxicity was observed for any of the treatments (in absence of MRSA). Control PBS was set as 100% growth and 0% toxicity. Significance levels compared to PBS are shown, p < 0.0001. N = 3.
To confirm that the antimicrobial effect of the nanoparticles is due to the internalization of the nanoparticles and subsequent intracellular co-localization with the bacteria, an experiment was designed to block the uptake of nanoparticles following the infection with bacteria. Cytochalasin D (cytoD) blocks actin polymerization and is used to reduce nanoparticle uptake into macrophages.52 The above experiment was repeated with and without blocking using cytoD. In the blocked samples, no significant antimicrobial effect of the Zn2-SrBG/ceria nanoparticles against the intracellular MRSA was observed. This finding strongly suggests that a direct interaction between intracellular nanoparticles and MRSA is required in order to eradicate the intracellular bacteria (Fig. 3c). The dose of antimicrobial ions released from the nanoparticles in a comparable time frame was found to be orders of magnitude below their level of activity (Fig. S6†). Again, no toxicity was found for any of the treatment groups (Fig. 3d).
2.4. Electron microscopy investigation into the mechanism
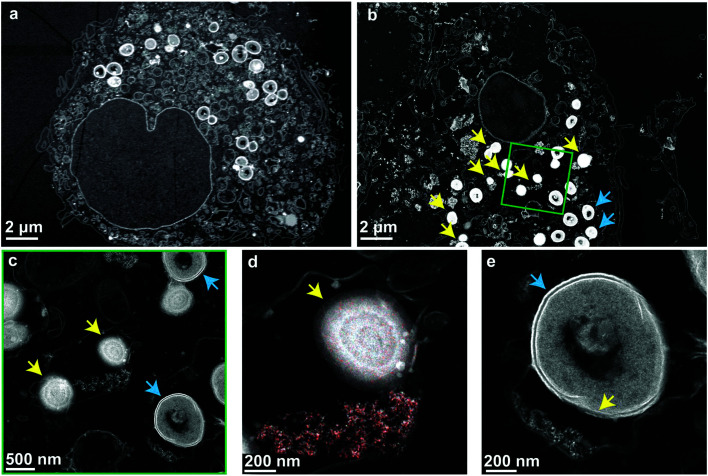
Finally, to verify that the nanoparticles and cells are taken up into the same compartments and to illustrate their interaction, high-angle annular dark-field (HAADF) scanning transmission electron micrographs of the nanoparticle-treated MRSA infected macrophages were collected (Fig. 4). The micrographs show high numbers of viable bacteria with intact membranes in untreated macrophages (Fig. 4a). In nanoparticle-treated macrophages, bacterial membrane disruption was observed at different stages wherever bacteria were co-localized with intracellular nanoparticles (Fig. 4b). This membrane disruption is observable as diffuse shells around bacteria and their decreased size (Fig. 4c, d, e and Fig. S7a, b†). The observed structural changes are comparable to experiments on planktonic bacteria interactions with aluminum oxide nanoparticles (cf. Fig. 5, Ansari et al.53). Intact bacterial cells were mostly observed in nanoparticle-free regions. Mammalian cell membranes remained largely unaffected by the nanoparticles, as indicated by the electron micrographs as well as the LDH values being comparable to nanoparticle-free macrophage samples (Fig. 3d).
Fig. 4. High-angle annular dark-field (HAADF) scanning transmission electron micrographs of macrophages infected with MRSA. Blue arrows indicate intact bacterial membranes and yellow arrows compromised ones. (a) Non-treated macrophages hosting intracellular bacteria with intact cell membranes. (b) Nanoparticle-treated macrophages with intact bacteria (blue arrows) and lysed bacteria (yellow arrows) where particles and bacteria are in the same cellular compartment. (c) Higher magnification of the region indicated by the green square. (d) Energy-dispersive X-ray spectroscopy mapping of Ce (red) confirms the co-localization of particles with the lysed bacteria. The particles are compressed to compact agglomerates in the phagosome. (e) A bacterium is partially lysed by contact with a small number of nanoparticles.
The antibacterial mechanism of neither ceria nor bioglass is fully understood.40,54 Several nanoceria systems including the one presented here have exhibited good tolerance by mammalian cells but toxicity towards planktonic microbial cells.40 While this ambiguity is not well understood, studies have shown that nanoceria is strongly attracted to bacterial cell walls and locally generates reactive oxygen species (ROS) through changing the Ce oxidation state from IV to III.55,56 Since the initial oxidation state is regenerated and due to their physical stability, ceria nanoparticles may stay active for prolonged times and can, therefore, express a more sustained effect.57 Supporting these findings, a strong dependence of the antimicrobial activity on environmental variables56,58 and nanoceria material properties (such as morphology59,60 or the initial oxidation state37,38,48,61) has been reported. Generally, catalytically active nanoceria tends to also be antimicrobial,48,61 which holds true for the nanoparticles discussed in this work (H2O2 decomposition and enzyme mimetic activity shown in Fig. S3†). Most literature indicates a mechanism mediated by direct contact of the nanoparticles with the bacterial membrane resulting in membrane disintegration, which is in line with the STEM investigations shown in Fig. 4.
Energy-dispersive X-ray spectroscopy mapping of ceria revealed the distribution of nanoparticles in the vesicles and around the bacteria (Fig. 4d and Fig. S7c, d). They were localized very close to partially or fully lysed cells. Fig. 4e shows a bacterium that is partially lysed due to limited contact with nanoparticles after vesicular co-localization of the two. The contact-based mechanism is further supported by the observation that the treatment of planktonic bacteria with nanoparticle supernatants showed unaffected growth and the comparatively poor solubility of ceria,62 indicating a minor role of dissolved ions in the observed intracellular antimicrobial activity (compare Fig. S7†).
3. Conclusions
This work describes a new strategy against microbial infections, which are a fast-growing problem due to rising antibiotic resistance and bacterial intracellular survival mechanisms. Inherently antimicrobial cerium oxide-based nanohybrids have been demonstrated to effectively reduce the intracellular spread of antibiotic-resistant bacteria. This is in stark contrast to more complex load-carrier systems that have been explored in the past and falls in line with the current push of simplifying bioactive agents. Flame-made metal oxide nanoparticles provide a versatile framework for biomedical applications that enables the tuning of chemical and physical properties. Here, this freedom has been utilized to unite mammalian cell compatibility with antimicrobial properties in a Zn-doped Sr-substituted bioglass-ceria hybrid system that is taken up into the same cellular compartments as bacteria. While this work is a solid base for further exploration, a more in-depth understanding of the particle–bacteria interaction and the involved mechanisms through advanced imaging and spectroscopy methods will allow increasing the efficacy of the nanoparticles by further tweaking of their composition and structure. While the (intracellular) antimicrobial properties of the produced nanoparticles have yet to be evaluated in vivo, infected wound in vivo models have numerous challenges.63,64 However, recent studies investigating the topical application of a ceria-based nanoparticle suspension to the subcutis of rats in a perforator flap model (see Fig. S8†) have shown a pronounced accumulation of nanoparticles in tissue macrophages, which indicates the feasibility of the approach.65 Also, long-term co-culture studies would be of high interest, however, cell viability of infected cells is limited in currently available models.28 While the stability of nanoceria is welcomed for sustainable treatments, a degradable particle might reduce long-term toxicity concerns – such a nanoparticle system can be engineered by harnessing the freedom of LF-FSP.66 Taken together, this work illustrates that ceria-based nanohybrids which unify mammalian cell compatibility and antimicrobial activity can be harnessed to eliminate intracellular pathogens, which hide within cells and are only poorly accessible by conventional antibiotics.
4. Experimental section
4.1. Bacterial strains, cell lines, and growth conditions
Methicillin-resistant Staphylococcus aureus (MRSA, DSMZ 11729) was obtained from DSMZ German Collection of Microorganisms and Cell Cultures Gmbh (Braunschweig, Germany). Human monocytic cell line THP-1 from ATCC was used as a precursor for monocyte-derived macrophages. The THP-1 cells were cultured in RPMI-1640 according to the supplier's protocol and used from passage 6 up to 21.
4.2. Synthesis of nanoparticles
Nanoparticles were synthesized under controlled and sterile conditions as previously described.48 In brief, metal precursors were dissolved at 0.3 M in combustible solvents and dispersed into a flame, allowing the formation of nanoparticles. In this work, five types of nanoparticles were used: (i) Bioglass (BG: 40 wt% calcium acetylacetonate hydrate, 37 wt% sodium 2-ethylhexanoate, 6 wt% tributyl phosphate, and 17 wt% HMDSO in THF); (ii) Bioglass containing 0.1, 0.5 or 2% silver (Ag0.1-BG, Ag0.5-BG and Ag2-BG respectively by the addition of silver acetate to the BG precursor); (iii) Bioglass and ceria hybrid particles (BG/ceria, 1 : 1 mixture of the BG and ceria precursor); (iv) Bioglass and ceria hybrid, matrix substituted with strontium oxide (SrO) and doped with 2% zinc (Zn2-SrBG/ceria, 68 wt% cerium(iii) 2-ethylhexanoate, 5 wt% calcium acetylacetonate hydrate, 13 wt% sodium 2-ethylhexanoate, 2 wt% tributyl phosphate, 6 wt% HMDSO in THF, 4 wt% strontium acetylacetonate hydrate, and 2 wt% zinc acetylacetonate); (v) Cerium oxide (ceria: 100 wt% cerium(iii) 2-ethylhexanoate in THF). Prior to experimental usage, nanoparticles were suspended in Dulbecco's PBS (PBS) (56064C, Sigma Aldrich) and probe-sonicated for 2 min with an interval of 8 seconds sonication and 2 seconds pause at an amplitude of 30% on ice.
4.3. Electron microscopy of nanoparticles
Nanoparticles were suspended in 0.5 mL deionized water and a drop of 10 μL was placed on a clean parafilm surface. A grid with a formvar film (Electron Microscopy Science, Lucerna-Chem AG, Lucerne, Switzerland) was placed on top of the drop for 10 min. Any excess liquid was removed and the grid air-dried. After drying, the grid was imaged in FEI Tecnai F30 FEG transmission electron microscope (TEM). Scanning transmission electron microscopy (STEM) was performed on a Jeol 2200FS TEM/STEM microscope equipped with an energy dispersive X-ray detector (EDX) for elemental analysis and operated at 200 kV.
4.4. Elemental analysis
Similarly as described before,65 the as-prepared nanoparticles were weighted into PTFE tubes and mixed with 6 mL 65% HNO3 p.a. (Merck) and 1 mL 30% H2O2 p.a. (Merck). The samples were then digested in a microwave and elements were quantified with an ICP-OES (Agilent 5110, Santa Clara, CA, USA). Due to the high silicate content and thus incomplete Si dissolution, the Si percentage was underestimated in some samples. For these samples, the residual mass was assumed to be Si as well.
4.5. Powder X-ray diffraction
X-ray diffraction (XRD) patterns were recorded with a Bruker AXS D8 Advance diffractometer (40 kV, 40 mA, Cu Kα radiation) at 2θ = 15–70° with a step size of 0.03° as described before.48
4.6. Dynamic light scattering
As described in earlier work,48 particle suspensions of 0.13 mg mL−1 in physiological saline solution were probe-sonicated for 2 minutes. Three dynamic light scattering (DLS) measurements per particle were conducted with a Zetasizer instrument (Nano ZS90, Malvern Instruments). The suspensions were vortex-mixed between measurements to ensure homogeneity.
4.7. Raman spectroscopy
Similarly to described elsewhere,65,67 measurements were performed on a WITec alpha 300R confocal Raman microscope, equipped with a UHTS 300 Vis spectrometer and an Andor Newton EMCCD. A linearly polarized 532 nm laser was used for excitation. Spectra were acquired with an integration time of 2 s.
4.8. X-ray photoelectron spectroscopy
X-ray photoelectron spectroscopy (XPS) analyses were carried out by SuSoS AG, Dübendorf, Switzerland.
4.9. Catalytic activity
The catalytic activity was assessed as described before.48 An SOD assay kit (Sigma-Aldrich, 19160) was used to measure superoxide dismutase mimicry. The final nanoparticle concentration was 0.05 mg ml−1. Catalase activity was quantified with the Amplex™ Red Catalase Assay Kit (Thermofisher, A22180). The final nanoparticle concentration was 0.05 mg ml−1. Vitamin C (Sigma, 95209) at 0.05 mg ml−1 was used as a positive control.
H2O2 decomposition was evaluated by incubating 0.5 mg ml−1 nanoparticles at 37 °C in 50 mM H2O2. Supernatants were collected at different times and transferred to an UV-transparent 96-well plate (Greiner, 655801). The remaining H2O2 concentration in the samples was assessed by measuring the absorbance at 235 nm. A H2O2 standard curve was also recorded to quantify the remaining H2O2.
4.10. Cell culture
THP-1 was grown in T75 culture flasks (90075, TPP Switzerland) containing RPMI 1640 medium (R7388, Sigma-Aldrich) supplemented with 10% fetal calf serum (FCS, F9665, Sigma Aldrich), 1% l-glutamine (D7513, Sigma Aldrich) and 1% Penicillin Streptomycin Neomycin mix (P4083, Sigma Aldrich) at 37 °C in a humidified atmosphere of 5% CO2. For routine maintenance, THP-1 was subcultured and the medium was refreshed every 3–4 days. The Suspension cells were counted using an automated hemocytometer (Casy TT ® cell counter, Roche Innovatis AG). Subcultures were seeded at density 0.5–1 × 106 per 20 ml fresh medium at 37 °C in a humidified atmosphere of 5% CO2.
4.11. THP-1 differentiation
To obtain THP-1 derived macrophages, differentiation of monocytes was induced by addition of Phorbol-12-myristate-13-acetate (PMA, 200 nM, P1585, Sigma Aldrich) to monocytes in supplemented RPMI 1640 medium following a well-established protocol.68 Cells were seeded at a density of 3 × 106 per ml (60.000 cells per well, 1900 cells per mm2) in a 96-well plate and cultured for 72 h at 37 °C in a humidified atmosphere of 5% CO2. After incubation, non-attached cells were removed by aspiration and adherent monocyte-derived macrophage cells were washed three times with pre-warmed RPMI 1640 to remove PMA.
4.12. Nanoparticle mammalian cell toxicity
THP-1 derived macrophages seeded at a density of 1900 cells per mm2 in a 96-well plate were treated with nanoparticles at different concentrations: 0.1, 0.5 mg ml−1 and 1 mg ml−1 for 24 h, 37 °C in a humidified atmosphere of 5% CO2. Subsequently, the supernatant was collected and nanoparticles were removed by centrifugation at 5000g for 15 min at room temperature (RT). Then, lactate dehydrogenase (LDH) was quantified using a Cytotox 96 ® Non-radioactive Cytotoxicity assay (Promega). The substrate was added to the supernatant in a 1 : 1 ratio (50 μl) and incubated for 30 min on a shaker. For each sample, biological triplicates and two technical replicates were used. Negative and positive controls, as well as blank samples (medium only), were included. The negative control included THP-1 derived macrophages treated with an equal volume of PBS and the positive control included THP-1 derived macrophages treated with 1% Triton X-100 (93443, Sigma Aldrich). Absorbance was recorded at 490 nm with a Mithras-2 LB943 Multimode Reader (Berthold Technologies GmbH & Co. KG, Germany). Mitochondrial ATP was measured by adding Celltiter Glo (G7570, Promega) reagent in a 1 : 1 ratio (50 μl) to the treated macrophages for 10 min on a shaker in dark at RT. Luminescence was then recorded using the Mithras-2 LB943 Multimode Reader. For both assays, nanoparticle interference was measured using the same experimental procedure in the absence of macrophages (cell-free control).
4.13. Antimicrobial effect of nanoparticles on planktonic bacteria
MRSA DMSZ 11729 was grown overnight in Tryptic soy broth (TSB100) at 37 °C on a shaker at 160 rpm. Bacteria were then transferred onto a TSB agar plate to form colonies overnight at 37 °C, 5% CO2. For each experiment, one colony was inoculated in TSB100 and grown overnight. Then, bacteria were diluted to an optical density (OD) of 0.1 and grown for 1–1.5 h, to reassure that the bacteria were in log phase. After reaching log phase (around OD600 = 0.6), the culture was diluted to OD600 = 0.1 again and added to a 96-well plate (TPP, Sigma Aldrich) with nanoparticles in a 1 : 4 ratio to obtain final nanoparticle concentrations of 0.5 mg ml−1. The plate was sealed with a gas-permeable membrane Breathe-easy® (Z380059, Sigma Aldrich) and OD at 600 nm was measured every 15 min during 2.5 h under shaking conditions. Controls included equal volumes of PBS and gentamicin (150 μg ml−1) in PBS. The absorbance of nanoparticles at 600 nm was tested for interference in TSB100 in the absence of bacteria.
The metabolic activity of the planktonic bacteria was quantified with Bactiter Glo (G8230, Promega) according to the manufacturer's instruction. Briefly, bacterial suspensions were mixed in a 1 : 1 ratio (50 μl each) with Bactiter Glo in microplate wells and incubated in the dark for 5 min at RT on a shaker at 100 rpm. Luminescence was recorded in a plate reader (Synergy H1, Biotek) to quantify ATP presence of the bacteria indicative of the number of viable bacteria.
4.14. CFU counting
The samples containing MRSA were diluted in PBS to a concentration of around 1 × 103 CFU ml−1 and 100 μl was plated on LB agar plates (Plate count agar, 70152, Sigma Aldrich). After 18–20 h incubation at 37 °C, CFU were quantified with a SCAN® 300 instrument (Interscience, France) using the SCAN® software.
4.15. Antimicrobial effect of nanoparticles on internalized bacteria
THP-1 derived macrophages were obtained as described above. Macrophages were incubated with RPMI 1640 without PMA for 22 h in a 96-well plate. After this period, the medium was replaced with antibiotic-free RPMI without phenol red, supplemented with 10% FCS. Cells were infected with MRSA (DSMZ 11729) at a multiplicity of infection (MOI) of 10 for 1 h at 37 °C, 5% CO2. Then, wells were washed three times with cold PBS (4 °C) and incubated with 100 μg ml−1 gentamicin in RPMI 1640 for 1 h at 37 °C, 5% CO2 to eliminate extracellular bacteria in line with available protocols.69 The gentamicin-containing medium was then removed and all wells were rinsed three times with cold PBS and filled with 150 μl RPMI 1640 medium supplemented with 10% FCS and 0.5 mg ml−1 BG, Ceria, Zn2-SrBG/ceria, BG/Ceria, Ag0.1-BG, Ag0.5-BG, or Ag2-BG and subsequently incubated for 2.5 h at 37 °C, 5% CO2.
In order to assess the cytotoxicity of the nanoparticles, the release of LDH into the supernatant was measured using Cytotox 96 non-radioactive Cytotoxicity assay as described above. Triton X-100 (1%) in RPMI was used as a negative control and PBS in RPMI as a positive control.
After incubation with nanoparticles or control conditions (PBS and Triton X-100), cells were washed three times with cold PBS and macrophage cell walls were subsequently perforated with 0.1% saponin (47036, Sigma Aldrich) for 10 min at RT on a shaker to release internalized bacteria. The suspension containing the released bacteria (0.1 ml) was then 10–1000× diluted in 0.9% NaCl, plated in triplicates on a PC agar plate for 18–20 h at 37 °C and CFUs were counted as described before. For reference, CFU of the supernatants prior to saponin treatment were measured to account for potentially present extracellular bacteria.
4.16. Inhibition of nanoparticle endocytosis
THP-1 derived macrophages were infected with bacteria at an MOI of 10 for 1 h as described above. After washing the macrophages three times with PBS, one group was treated with 20 μM Cytochalasin-D (cytoD)70 to block endocytosis of nanoparticles as well as 100 μg ml−1 gentamicin to kill extracellular bacteria during 1 h at 37 °C, 5% CO2. The control group was incubated with 100 μg ml−1 gentamicin in absence of cytoD. After washing twice, RPMI + 10% FCS was added to the unblocked control and 10 μM cytoD in RPMI + 10% FCS was added to the inhibited group. Then 0.5 mg ml−1 Zn2-SrBG/ceria nanoparticles, 100 μg ml−1 gentamicin or an equal volume of PBS were added to both groups and left to incubate for 2.5 h at 37 °C, 5% CO2. After the incubation time, the supernatants were collected and LDH was measured following the aforementioned protocol. Then, samples were washed twice with RPMI + 10% FCS and 100 μg ml−1 gentamicin to remove potentially present extracellular bacteria and finally washed twice with PBS. Subsequently, all cells were permeabilized with 0.1% saponin for 10 min. Each sample was diluted 10–1000× and plated on PC-agar plates for CFU quantification.
4.17. Scanning transmission electron microscopy
For scanning transmission electron microscopy (STEM) examination, 1 × 107 THP-1 cells were seeded in RPMI into T75 cell culture flasks. Four groups were prepared: macrophages (control, 1), macrophages with 0.5 mg ml−1 Zn2-SrBG/ceria (2), Macrophages with internalized MRSA (3), macrophages with internalized MRSA and 0.5 mg ml−1 Zn2-SrBG/ceria (4) as described above. Cells were detached from the flask using trypsin-EDTA and a cell scraper, pelleted by centrifugation (500g, 5 min, 20 °C) and resuspended in 1 ml paraformaldehyde. After fixation at 4 °C overnight, the cells were spun down at 1500g for 5 min and stained with 2% osmium tetroxide in 0.1 M sodium cacodylate buffer. After drying in an ethanol gradient series, the cells were embedded in epoxy resin (EPON 812, Sigma-Aldrich). Ultrathin sections were either imaged on an FEI Helios 660 G3 UC FIB/SEM at 25 kV or contrasted with lead citrate (Reynolds 1963) before observation by TEM. Elemental distribution maps were recorded from these ultrathin sections in scanning transmission mode on a Talos F200X TEM (Super-X EDS, 4 detector configuration, FEI, USA) at an accelerating voltage of 200 kV. Samples were mounted on a double-tilt holder and fixed using a molybdenum ring and clamp. The data were processed using the software Velox 2.9 (FEI, USA).
4.18. Statistics
For comparison between treated and non-treated groups, a non-parametric Wilcoxon signed-rank test was performed.
Author contributions
Martin T. Matter: Conceptualization, methodology, investigation, writing – original draft preparation, analysis, visualization. Meagan Doppegieter: Investigation, writing – original draft preparation, analysis. Alexander Gogos, Kerda Keevend: Investigation. Qun Ren: Methodology. Inge K. Herrmann: Conceptualization, supervision, project administration, writing – reviewing and editing.
Conflicts of interest
M. T. M and I. K. H. declare inventorship on a patent application related to the nanoparticle compositions (EP21168502). All other authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Supplementary Material
Acknowledgments
The Scientific Center for Optical and Electron Microscopy (ScopeM) of ETH Zürich is acknowledged for ultramicrotomy performed by A. G. Bittermann and access to their electron microscopes. We acknowledge financial support from the Gottfried and Julia Bangerter Rhyner Foundation, the Helmut Horten Foundation, OPO-Foundation and the Swiss National Science Foundation (SNSF, Eccellenza grant number 181290).
Electronic supplementary information (ESI) available. See DOI: 10.1039/d0nr08285f
References
- Rollin G. Tan X. Tros F. Dupuis M. Nassif X. Charbit A. Coureuil M. Front. Microbiol. 2017;8:1354. doi: 10.3389/fmicb.2017.01354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamaruzzaman N. F. Kendall S. Good L. Br. J. Pharmacol. 2017;174:2225–2236. doi: 10.1111/bph.13664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdonnay E. Henry T. eLife. 2016;5:e14721. doi: 10.7554/eLife.14721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzoni C. Kelley W. L. Trends Microbiol. 2009;17(2):59–65. doi: 10.1016/j.tim.2008.11.005. [DOI] [PubMed] [Google Scholar]
- Fraunholz M. Sinha B. Front. Cell. Infect. Microbiol. 2012;2:43. doi: 10.3389/fcimb.2012.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowy F. D. N. Engl. J. Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- van Hal S. J. Jensen S. O. Vaska V. L. Espedido B. A. Paterson D. L. Gosbell I. B. Clin. Microbiol. Rev. 2012;25:362–386. doi: 10.1128/CMR.05022-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomer L. Schneewind O. Missiakas D. Annu. Rev. Pathol. 2016;11:343–364. doi: 10.1146/annurev-pathol-012615-044351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresham H. D. Lowrance J. H. Caver T. E. Wilson B. S. Cheung A. L. Lindberg F. P. J. Immunol. 2000;164(7):3713–3722. doi: 10.4049/jimmunol.164.7.3713. [DOI] [PubMed] [Google Scholar]
- Kubica M. Guzik K. Koziel J. Zarebski M. Richter W. Gajkowska B. Golda A. Maciag-Gudowska A. Brix K. Shaw L. Foster T. Potempa J. PLoS One. 2008;3(1):e1409. doi: 10.1371/journal.pone.0001409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q. Fei Y. Hu L. Huang Z. Li L.-L. Wang H. Nano Lett. 2018;18:6229–6236. doi: 10.1021/acs.nanolett.8b02286. [DOI] [PubMed] [Google Scholar]
- Gordon R. J. Lowy F. D. Clin. Infect. Dis. 2008;46(5):S350–S359. doi: 10.1086/533591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surewaard B. G. J. Deniset J. F. Zemp F. J. Amrein M. Otto M. Conly J. Omri A. Yates R. M. Kubes P. J. Exp. Med. 2016;213:1141–1151. doi: 10.1084/jem.20160334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bera A. Herbert S. Jakob A. Vollmer W. Götz F. Mol. Microbiol. 2005;55:778–787. doi: 10.1111/j.1365-2958.2004.04446.x. [DOI] [PubMed] [Google Scholar]
- Uribe-Querol E. Rosales C. Front. Immunol. 2017;8:1368. doi: 10.3389/fimmu.2017.01368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clauditz A. Resch A. Wieland K.-P. Peschel A. Götz F. Infect. Immun. 2006;74(8):4950–4953. doi: 10.1128/IAI.00204-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove K. Coutts G. Jonsson I.-M. Tarkowski A. Kokai-Kun J. F. Mond J. J. Foster S. J. J. Bacteriol. 2007;189(3):1025–1035. doi: 10.1128/JB.01524-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karavolos M. H. Horsburgh M. J. Ingham E. Foster S. J. Microbiology. 2003;149(10):2749–2758. doi: 10.1099/mic.0.26353-0. [DOI] [PubMed] [Google Scholar]
- Liu G. Y. Essex A. Buchanan J. T. Datta V. Hoffman H. M. Bastian J. F. Fierer J. Nizet V. J. Exp. Med. 2005;202(2):209–215. doi: 10.1084/jem.20050846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristuccia A. M. Cunha B. A. Med. Clin. North Am. 1982;66(1):303–312. doi: 10.1016/s0025-7125(16)31462-6. [DOI] [PubMed] [Google Scholar]
- Lecaroz C. Gamazo C. Blanco-Prieto M. J. J. Nanosci. Nanotechnol. 2006;6:3296–3302. doi: 10.1166/jnn.2006.478. [DOI] [PubMed] [Google Scholar]
- Abed N. Saïd-Hassane F. Zouhiri F. Mougin J. Nicolas V. Desmaële D. Gref R. Couvreur P. Sci. Rep. 2015;5:13500. doi: 10.1038/srep13500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladavière C. Gref R. Nanomedicine. 2015;10:3033–3055. doi: 10.2217/nnm.15.128. [DOI] [PubMed] [Google Scholar]
- Mu H. Tang J. Liu Q. Sun C. Wang T. Duan J. Sci. Rep. 2016;6:18877. doi: 10.1038/srep18877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S. Han X. Yang Y. Qiao H. Yu Z. Liu Y. Wang J. Tang T. ACS Appl. Mater. Interfaces. 2018;10:14299–14311. doi: 10.1021/acsami.7b15678. [DOI] [PubMed] [Google Scholar]
- Flamant Q. Caravaca C. Meille S. Gremillard L. Chevalier J. Biotteau-Deheuvels K. Kuntz M. Chandrawati R. Herrmann I. K. Spicer C. D. Stevens M. M. Anglada M. Acta Biomater. 2016;46:308–322. doi: 10.1016/j.actbio.2016.09.017. [DOI] [PubMed] [Google Scholar]
- Pratsinis A. Hervella P. Leroux J.-C. Pratsinis S. E. Sotiriou G. A. Small. 2013;9:2576–2584. doi: 10.1002/smll.201202120. [DOI] [PubMed] [Google Scholar]
- Kang J. Dietz M. J. Hughes K. Xing M. Li B. J. Antimicrob. Chemother. 2019;74:1578–1585. doi: 10.1093/jac/dkz053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H. Ji Z. Xia T. Meng H. Low-Kam C. Liu R. Pokhrel S. Lin S. Wang X. Liao Y.-P. Wang M. Li L. Rallo R. Damoiseaux R. Telesca D. Mädler L. Cohen Y. Zink J. I. Nel A. E. ACS Nano. 2012;6:4349–4368. doi: 10.1021/nn3010087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold K. Slay B. Knackstedt M. Gaharwar A. K. Adv. Ther. 2018;1:1700033. [Google Scholar]
- Dizaj S. M. Lotfipour F. Barzegar-Jalali M. Zarrintan M. H. Adibkia K. Mater. Sci. Eng., C. 2014;44:278–284. doi: 10.1016/j.msec.2014.08.031. [DOI] [PubMed] [Google Scholar]
- Niño-Martínez N. Salas Orozco M. F. Martínez-Castañón G.-A. Torres Méndez F. Ruiz F. Int. J. Mol. Sci. 2019;20:2808. doi: 10.3390/ijms20112808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lese I. Graf D. A. Tsai C. Taddeo A. Matter M. T. Constantinescu M. A. Herrmann I. K. Olariu R. PLoS One. 2018;13:e0207802. doi: 10.1371/journal.pone.0207802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miguez-Pacheco V. Hench L. L. Boccaccini A. R. Acta Biomater. 2015;13:1–15. doi: 10.1016/j.actbio.2014.11.004. [DOI] [PubMed] [Google Scholar]
- Matter M. T. Starsich F. Galli M. Hilber M. Schlegel A. A. Bertazzo S. Pratsinis S. E. Herrmann I. K. Nanoscale. 2017;9:8418–8426. doi: 10.1039/c7nr01176h. [DOI] [PubMed] [Google Scholar]
- Waltimo T. Brunner T. J. Vollenweider M. Stark W. J. Zehnder M. J. Dent. Res. 2007;86:754–757. doi: 10.1177/154405910708600813. [DOI] [PubMed] [Google Scholar]
- Szymanski C. J. Munusamy P. Mihai C. Xie Y. Hu D. Gilles M. K. Tyliszczak T. Thevuthasan S. Baer D. R. Orr G. Biomaterials. 2015;62:147–154. doi: 10.1016/j.biomaterials.2015.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C. Lin Y. Wang J. Wu L. Wei W. Ren J. Qu X. Adv. Healthcare Mater. 2013;2:1591–1599. doi: 10.1002/adhm.201200464. [DOI] [PubMed] [Google Scholar]
- Maria Magdalane C. Kaviyarasu K. Raja A. Arularasu M. V. Mola G. T. Isaev A. B. Al-Dhabi N. A. Arasu M. V. Jeyaraj B. Kennedy J. Maaza M. J. Photochem. Photobiol., B. 2018;185:275–282. doi: 10.1016/j.jphotobiol.2018.06.011. [DOI] [PubMed] [Google Scholar]
- Zhang M. Zhang C. Zhai X. Luo F. Du Y. Yan C. Sci. China Mater. 2019;62:1727–1739. [Google Scholar]
- Qi M. Li W. Zheng X. Li X. Sun Y. Wang Y. Li C. Wang L. Front. Mater. 2020;7:213. [Google Scholar]
- Gröhn A. J. Pratsinis S. E. Sánchez-Ferrer A. Mezzenga R. Wegner K. Ind. Eng. Chem. Res. 2014;53:10734–10742. [Google Scholar]
- Chernousova S. Epple M. Angew. Chem., Int. Ed. 2013;52:1636–1653. doi: 10.1002/anie.201205923. [DOI] [PubMed] [Google Scholar]
- Mueller R. Mädler L. Pratsinis S. E. Chem. Eng. Sci. 2003;58:1969–1976. [Google Scholar]
- Pratsinis A. Kelesidis G. A. Zuercher S. Krumeich F. Bolisetty S. Mezzenga R. Leroux J.-C. Sotiriou G. A. ACS Nano. 2017;11:12210–12218. doi: 10.1021/acsnano.7b05518. [DOI] [PubMed] [Google Scholar]
- Maguire C. M. Rösslein M. Wick P. Prina-Mello A. Sci. Technol. Adv. Mater. 2018;19:732–745. doi: 10.1080/14686996.2018.1517587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z. Wang X. Pang Y. Cheng S. Liu J. Nat. Commun. 2019;10:5783. doi: 10.1038/s41467-019-13727-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matter M. T. Furer L. A. Starsich F. H. L. Fortunato G. Pratsinis S. E. Herrmann I. K. ACS Appl. Mater. Interfaces. 2018;11:2830–2839. doi: 10.1021/acsami.8b18778. [DOI] [PubMed] [Google Scholar]
- Oh S.-J. Kim H. Liu Y. Han H.-K. Kwon K. Chang K.-H. Park K. Kim Y. Shim K. An S. S. A. Lee M.-Y. Toxicol. Lett. 2014;225:422–432. doi: 10.1016/j.toxlet.2014.01.015. [DOI] [PubMed] [Google Scholar]
- White R. C. Cianciotto N. P. Infect. Immun. 2016;84:3313–3327. doi: 10.1128/IAI.00750-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y. Lenz J. Arvidson C. G. Infect. Immun. 2005;73:4834–4845. doi: 10.1128/IAI.73.8.4834-4845.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iversen T.-G. Skotland T. Sandvig K. Nano Today. 2011;6:176–185. [Google Scholar]
- Ansari M. A. Khan H. M. Khan A. A. Pal R. Cameotra S. S. J. Nanopart. Res. 2013;15:1970. [Google Scholar]
- Drago L. Toscano M. Bottagisio M. Materials. 2018;11:326. doi: 10.3390/ma11020326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeyons O. Thill A. Chauvat F. Menguy N. Cassier-Chauvat C. Oréar C. Daraspe J. Auffan M. Rose J. Spalla O. Nanotoxicology. 2009;3:284–295. [Google Scholar]
- Alpaslan E. Geilich B. M. Yazici H. Webster T. J. Sci. Rep. 2017;7:srep45859. doi: 10.1038/srep45859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarnuzzer R. W. Colon J. Patil S. Seal S. Nano Lett. 2005;5:2573–2577. doi: 10.1021/nl052024f. [DOI] [PubMed] [Google Scholar]
- Sayle T. X. T. Molinari M. Das S. Bhatta U. M. Möbus G. Parker S. C. Seal S. Sayle D. C. Nanoscale. 2013;5:6063. doi: 10.1039/c3nr00917c. [DOI] [PubMed] [Google Scholar]
- Pelletier D. A. Suresh A. K. Holton G. A. McKeown C. K. Wang W. Gu B. Mortensen N. P. Allison D. P. Joy D. C. Allison M. R. Brown S. D. Phelps T. J. Doktycz M. J. Appl. Environ. Microbiol. 2010;76:7981–7989. doi: 10.1128/AEM.00650-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X. Li X. Feng Y. Yang S. RSC Adv. 2018;8:11764–11770. doi: 10.1039/c8ra00622a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farias I. A. P. dos Santos C. C. L. Sampaio F. C. BioMed Res. Int. 2018;2018:1–14. doi: 10.1155/2018/1923606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plakhova T. V. Romanchuk A. Yu. Yakunin S. N. Dumas T. Demir S. Wang S. Minasian S. G. Shuh D. K. Tyliszczak T. Shiryaev A. A. Egorov A. V. Ivanov V. K. Kalmykov S. N. J. Phys. Chem. C. 2016;120:22615–22626. [Google Scholar]
- Warren H. S. Fitting C. Hoff E. Adib-Conquy M. Beasley-Topliffe L. Tesini B. Liang X. Valentine C. Hellman J. Hayden D. Cavaillon J.-M. J. Infect. Dis. 2010;201:223–232. doi: 10.1086/649557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolker J. Nature. 2012;491:31–33. doi: 10.1038/491031a. [DOI] [PubMed] [Google Scholar]
- Matter M. T. Li J.-H. Lese I. Schreiner C. Bernard L. Scholder O. Hubeli J. Keevend K. Tsolaki E. Bertero E. Bertazzo S. Zboray R. Olariu R. Constantinescu M. A. Figi R. Herrmann I. K. Adv. Sci. 2020;7:2000912. doi: 10.1002/advs.202000912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rösslein M. Liptrott N. J. Owen A. Boisseau P. Wick P. Herrmann I. K. Nanotoxicology. 2017;11:147–149. doi: 10.1080/17435390.2017.1279361. [DOI] [PubMed] [Google Scholar]
- Saxena N. Keilhofer J. Maurya A. K. Fortunato G. Overbeck J. Müller-Buschbaum P. ACS Appl. Energy Mater. 2018;1:336–342. [Google Scholar]
- Daigneault M. Preston J. A. Marriott H. M. Whyte M. K. B. Dockrell D. H. PLoS One. 2010;5:e8668. doi: 10.1371/journal.pone.0008668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jubrail J. Morris P. Bewley M. A. Stoneham S. Johnston S. A. Foster S. J. Peden A. A. Read R. C. Marriott H. M. Dockrell D. H. Cell. Microbiol. 2016;18:80–96. doi: 10.1111/cmi.12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popēna I. Ābols A. Saulīte L. Pleiko K. Zandberga E. Jēkabsons K. Endzeliņš E. Llorente A. Linē A. Riekstiņa U. Cell Commun. Signaling. 2018;16:17–17. doi: 10.1186/s12964-018-0229-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.