Abstract
Protein conformational diseases are characterized by misfolding and toxic aggregation of metastable proteins, often culminating in neurodegeneration. Enteric bacteria influence the pathogenesis of neurodegenerative diseases; however, the complexity of the human microbiome hinders our understanding of how individual microbes influence these diseases. Disruption of host protein homeostasis, or proteostasis, affects the onset and progression of these diseases. To investigate the effect of bacteria on host proteostasis, we used Caenorhabditis elegans expressing tissue-specific polyglutamine reporters that detect changes in the protein folding environment. We found that colonization of the C. elegans gut with enteric bacterial pathogens disrupted proteostasis in the intestine, muscle, neurons, and the gonad, while the presence of bacteria that conditionally synthesize butyrate, a molecule previously shown to be beneficial in neurodegenerative disease models, suppressed aggregation and the associated proteotoxicity. Co-colonization with this butyrogenic strain suppressed bacteria-induced protein aggregation, emphasizing the importance of microbial interaction and its impact on host proteostasis. Further experiments demonstrated that the beneficial effect of butyrate depended on the bacteria that colonized the gut and that this protective effect required SKN-1/Nrf2 and DAF-16/FOXO transcription factors. We also found that bacteria-derived protein aggregates contribute to the observed disruption of host proteostasis. Together, these results reveal the significance of enteric infection and gut dysbiosis on the pathogenesis of protein conformational diseases and demonstrate the potential of using butyrate-producing microbes as a preventative and treatment strategy for neurodegenerative disease.
Author summary
Protein conformational diseases are one of the leading causes of geriatric death and disability, worldwide. Individuals suffering from these ailments are limited to palliative care, as there are no cures or effective treatments. Correlational evidence suggests that the human gut microbiota is a culprit, but the effect of individual bacteria remains elusive, in part, due to the complexity of the microbiome. A single-bacterium approach can help to deconvolute the complexity of the microbiome and reveal the effect of individual bacterial species on organismal proteostasis. As such, we utilized the intestine of C. elegans as a “test tube” to identify the effect of bacteria on the host using tissue-specific polyglutamine repeats as protein folding sensors. We found that colonization of the C. elegans intestine with pathogenic gram-negative bacteria disrupted proteostasis in the intestine, muscle, neurons, and gonads. Furthermore, we demonstrated that butyrogenic bacteria enhanced proteostasis, which was evidenced by a decrease in polyglutamine aggregation and suppression of aggregate-dependent toxicity. Further experiments revealed that co-colonization with butyrogenic bacteria inhibited protein aggregation in C. elegans and the butyrate-mediated suppression of aggregation is dependent on SKN-1/Nrf2 and DAF-16/FOXO–two transcription factors involved in the regulation of oxidative stress responses. While the mechanism of bacteria-mediated induction of protein aggregation remains elusive, our results suggest that bacterial aggregates, in addition to the contribution of oxidative stress, are the contributing factor. These results are intriguing as they suggest that enteric bacteria directly contribute to the pathogenicity of protein conformational diseases.
Introduction
Neurodegenerative protein conformational diseases (PCDs), including amyotrophic lateral sclerosis (ALS), Alzheimer’s, Huntington’s, and Parkinson’s disease, are characterized by the misfolding and aggregation of metastable proteins that reside within the proteome, often resulting in loss of tissue function that manifests in disease progression [1]. Despite the high prevalence and enormous financial and social burdens imposed on afflicted individuals and their families [2], no effective treatment or cure has been found; moreover, the etiology of these diseases remains largely unknown [3]. Factors such as age, diet, stress, trauma, toxins, infections, or antibiotics, have been shown to increase the risk of PCDs [4–10]. Notably, these triggers are also associated with changes in the microbiome [11–13], suggesting that bacteria may contribute to the pathogenesis of PCDs, which may explain their sporadic onset. Nonetheless, the relationship between the microbiome and disease progression remains poorly defined.
The human gut microbiota (HGM) is a complex assembly of microorganisms capable of synthesizing molecules that can impact the host’s physiology [14]. A healthy commensal relationship between a balanced microbiome and the host is fostered by their cross-talk [15]. Conversely, gut dysbiosis results from the loss of beneficial bacteria accompanied by the overgrowth of opportunistic pathogens, a condition often induced by antibiotics [16,17]. Recent evidence has established a link between the HGM and PCDs, whereby gut dysbiosis or direct enteric infection exacerbate the disease [18–20]. Commensal residents of the human microbiota, specifically those involved in the synthesis of short-chain fatty acids (SCFAs) such as butyrate, were shown to be beneficial to the host [21]. Unfortunately, environmental factors and the complexity of the human microbiome often hinder the consistency of results from experiments that pertain to PCDs [22]. To eliminate such complexity, we employed Caenorhabditis elegans as a model to study the effect of enteric pathogens on host proteostasis and examined the benefits provided by butyrogenic bacteria.
Here, we show that colonization of the C. elegans gut with gram-negative enteric bacterial pathogens disrupts host proteostasis, leading to aggregation and proteotoxicity of polyglutamine (polyQ) tracts across multiple tissues, including intestine, muscle, neurons, and the gonads. Such tissue non-autonomous effects of bacteria on the host may explain the impact of the gut microbiota on the onset and progression of PCDs. While pathogenic bacteria contribute to aggregation, commensal strains suppress it. Additionally, we show that butyrate and butyrogenic bacteria enhance host proteostasis and lead to suppression of bacteria-mediated polyQ aggregation and proteotoxicity. Oxidative stress and bacteria-derived protein aggregates seem to contribute to the observed disruption of proteostasis. Collectively, our results suggest that dysbiosis between enteric pathogens and commensal butyrogenic bacteria contributes to the pathogenicity of PCDs.
Results
Colonization of the C. elegans intestine with human bacterial pathogens disrupts proteostasis and affects animal motility
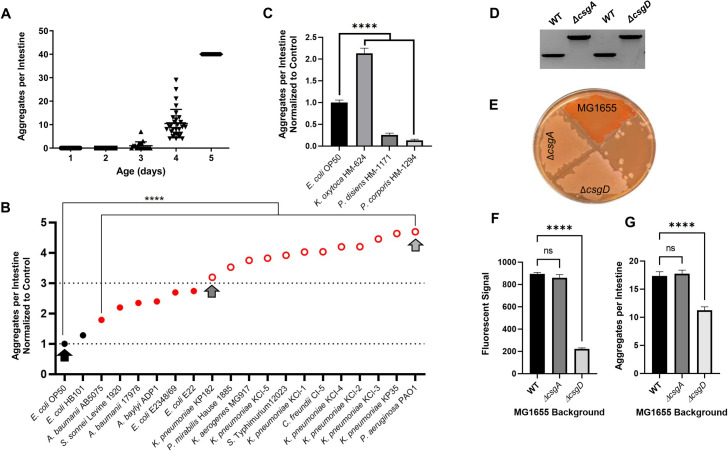
We tested the ability of select bacterial species from the Enterobacteriaceae family to affect the protein folding environment in C. elegans upon intestinal colonization. Select bacterial species were from the following genera: Escherichia, Klebsiella, Proteus, Citrobacter, Shigella, and Salmonella, as well as additional pathogenic bacteria that are associated with gut microbiota; these include gram-negative Pseudomonas and Acinetobacter. Although the physical manifestations of human proteopathies involve aggregate-induced dysfunction of neuronal and musculoskeletal tissue, we concluded that the intestinal tissue was best suited for our initial experiments because it is the immediate environment of bacterial colonization, presumably resulting in the most robust effect. As such, to assess the effect of these candidate strains on the protein folding environment in the C. elegans gut, we used animals that constitutively express intestine-specific polyQs fused to yellow fluorescent protein (polyQ44::YFP) [23]. Nematodes carrying this reporter exhibit age-dependent aggregation of intestinal polyQs (Fig 1A). These aggregates present as quantifiable fluorescent foci and were previously characterized as a proxy to assess the influence of bacteria on aggregation [23]. To determine the age at which nematodes harbor an intermediate range of aggregation for identifying bacteria that suppress or enhance aggregation, we quantified aggregates between days 1–5, post-hatching. Worms were grown at room temperature (~23°C) on the control bacteria, E. coli OP50. Because a high number of aggregates can introduce counting error, an aggregation threshold was set at 40 aggregates per intestine to ensure the reliability of counting. Aggregates increased each day, becoming visible after three days, and reaching our set threshold after five days (Fig 1A). Hence, to identify bacteria that either enhance or suppress aggregation, we chose to grow C. elegans on test bacteria for four days before assessing the ability of each strain to influence protein aggregation in the intestine. Out of 19 strains tested, one did not have any effect on aggregation, six strains significantly enhanced aggregation two- to three-fold, and 12 strains enhanced aggregation more than three-fold with respect to E. coli OP50 (Fig 1B). We found that the strongest enhancers were P. aeruginosa, K. pneumoniae, C. freundii, S. Typhimurium, K. aerogenes, and P. mirabilis. The only strain that did not significantly induce aggregation relative to E. coli OP50 was E. coli HB101, a non-pathogenic strain derived from E. coli K-12. To eliminate the possibility that bacteria change polyQ expression levels, using an antibody specific to YFP/GFP (S1A Fig), we confirmed that polyQ44:YFP soluble protein levels do not change between E. coli OP50 and P. aeruginosa PAO1 –two bacterial strains that elicited the lowest and highest intestinal aggregation, respectively (S1B Fig). Moreover, western blot analysis of the insoluble polyQ44::YFP fraction confirmed bacteria-mediated increase in aggregation (S2A Fig), as it parallels the aggregation profile of worms fed E. coli OP50 and P. aeruginosa PAO1 (Fig 1B). These results are intriguing as they demonstrate that common human enteric pathogens significantly affect polyQ aggregation, and therefore host proteostasis. We next asked whether gram-negative bacteria that are associated with commensal microflora would also enhance aggregation. We tested three strains known to be part of the commensal microbiome: K. oxytoca HM-624, Prevotella disiens HM-1171, and Prevotella corporis HM-1294. While K. oxytoca enhanced aggregation, as we have seen with the other Klebsiella spp., both Prevotella spp. suppressed aggregation relative to E. coli OP50 (Fig 1C). In fact, P. corporis almost completely suppressed aggregation.
Fig 1. Gram-negative bacteria found in the human gut affect C. elegans proteostasis and enhance protein aggregation in the intestine.
A) Age-dependent aggregation of intestinal polyQs (polyQ44) colonized by E. coli OP50. Data are represented as the number of aggregates per 20–30 intestines quantified in animals at day 1–5 post-hatching. B) The effect of select enteric bacteria on polyQ aggregation in the C. elegans intestine. Data are represented as the average number of aggregates per C. elegans intestine normalized to E. coli OP50 control strain (lower dotted line). Each data point is an average of a minimum of three independent experiments with a total of at least 100 animals. Black circles represent control bacteria and bacteria that did not have any significant effect on polyglutamine aggregation upon colonization of the C. elegans intestine. Red solid circles represent bacteria that significantly enhanced aggregation by <3-fold. Red open circles represent bacteria that significantly enhanced aggregation by >3-fold (arbitrary threshold). Arrows represent bacteria that were chosen for follow-up experiments. C) The effect of select commensal bacteria on polyQ aggregation in the C. elegans intestine. Data are represented as the average number of aggregates per C. elegans intestine normalized to E. coli OP50 control strain. Each data point is an average of a minimum of three independent experiments with a total of at least 100 animals. D) PCR confirmation of E. coli MG1655 curli mutant strains. WT, csgA::kan (left two bands) amplified with primers flanking the ΔcsgA locus. WT, csgD::kan (right two bans) amplified with primers flanking the ΔcsgD locus. E) Phenotypic confirmation of the curli-deficient ΔcsgA and ΔcsgD mutant strains using Congo Red plate assay. F) ProteoStat staining of total aggregates produced by E. coli MG1655 wild-type (WT), MG1655 ΔcsgA, and MG1655 ΔcsgD. Data are represented as the average fluorescent signal per bacterial strain stained with ProteoStat. Each data point is an average of two independent experiments with three replicates per run. G) The effect of E. coli MG1655 curli mutants on polyQ (polyQ44) aggregation in the C. elegans intestine. Data are represented as the average number of aggregates per C. elegans intestine. Each data point is an average of three independent experiments with a total of 90 animals. Error bars represent standard error of the mean (SEM). Statistical significance was calculated using one-way analysis of variance (ANOVA) followed by multiple comparison Dunnett’s post-hoc test (****p< 0.0001).
To begin understanding how bacteria could contribute to the disruption of host proteostasis, we used E. coli defective in the production of curli, which are aggregation-prone amyloids that are specific to Enterobacteriaceae [24]. We generated two mutant E. coli strains: one deficient in csgA, which encodes an amyloid structural subunit of curli fimbriae, and one deficient in csgD, a transcription factor that positively regulates the curli operon (Fig 1D). While staining with congo red confirmed a deficiency in curli production (Fig 1E), staining for bacterial protein aggregates using ProteoStat, which provides superior sensitivity in aggregate detection [25], revealed that only csgD mutant cells have significantly fewer aggregates (Fig 1F). In agreement with these results, we found that colonization of C. elegans only with the ΔcsgD E. coli strain led to a significant decrease in the number of polyQ aggregates per intestine (Fig 1G). These results suggest that bacteria-derived aggregates significantly contribute to the disruption of host proteostasis.
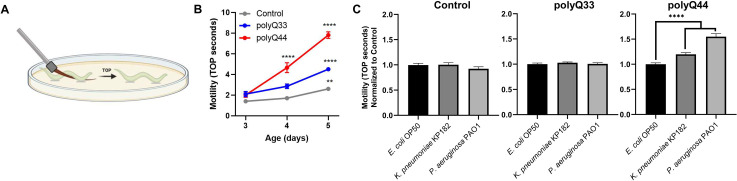
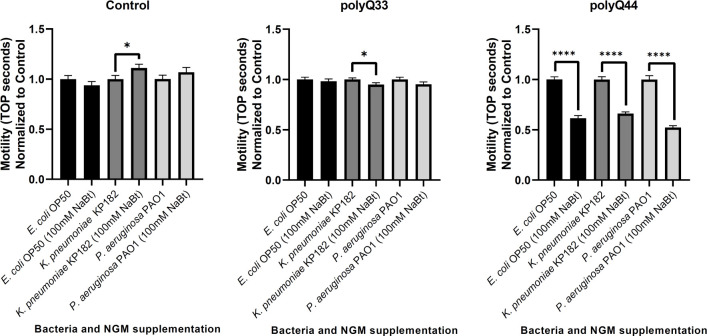
Tissue non-autonomous effects between C. elegans neurons and other somatic cells (i.e., intestine and epithelium) have been recently described where perturbation of proteostasis in one tissue impacts another [26–28]. As such, we postulated that bacteria-induced disruption of proteostasis in the intestine could affect other tissues. To begin assessing the impact of bacteria on muscle, we examined motility. The nematodes that express intestinal polyQ have an integrated rol-6 marker, which results in a roller phenotype and renders animals incompatible with known motility assays, such as thrashing. Thus, we developed a novel motility readout that entails sliding an eyebrow hair under the mid-section of the worm and counting the number of seconds until the worm completely crawls off—we refer to this phenotype as time-off-pick (TOP) (Fig 2A). We found that TOP increases with age and the decrease in motility is dependent on polyQ when compared to control roller worms that do not express polyQs, as well as those that express non-aggregating polyQs (Fig 2B). To ensure that the TOP method measures polyQ-specific defects and not an undesired effect related to the roller phenotype or bacterial pathogenicity, we used non-roller strains that express muscle-specific polyQ35 (unc-54p::Q35::YFP, AM140) and polyQ40 (unc-54p::Q40::YFP, AM141). Both of these strains are known to exhibit polyQ-length and age-dependent motility defects [29]. In support of the functionality of our assay, we found that worms expressing these constructs also exhibited a significant polyQ-length and age-dependent TOP motility defect that correlated with the enumeration of body bends, which is a well-established assessment of motility in C. elegans (S3A Fig). Additionally, colonization of these polyQ-expressing strains with P. aeruginosa PAO1 inhibited motility in both assays yet had little to no effect in wild-type N2 animals. These findings indicate that polyQ toxicity, and not bacterial pathogenicity, is the factor that is measured by TOP (S3B Fig). Therefore, we further used the TOP method to determine whether colonization of the C. elegans intestine with bacteria that induced polyQ aggregation aggravates this phenotype. We chose two bacterial strains: K. pneumoniae KP182 (medium inducer); P. aeruginosa PAO1 (high inducer) and found that the “time-off-pick” was significantly increased by both strains (Fig 2C), supporting the trend seen in their respective aggregation profiles (Fig 1B). We did not detect any effect on the TOP phenotype in roller controls or in roller nematodes expressing a non-aggregating and intestine-specific polyQ33, confirming that this motility defect was the result of bacteria-induced polyQ-dependent toxicity rather than general pathogenicity from a bacterial infection (Fig 2C). While it is known that worms expressing polyQ33 do not exhibit age-dependent aggregation, we wanted to ensure that aggregation does not occur when these animals are colonized by bacteria that induce polyQ44 aggregation. We did not detect polyQ33 aggregates in animals colonized with the strongest inducer, P. aeruginosa PAO1 (S4 Fig). Based on our results, we conclude that bacterial colonization of the C. elegans intestine affects protein folding in the immediate environment and that such localized intestinal protein aggregation influences motility, likely by acting on muscle or neuronal tissues.
Fig 2. Colonization of C. elegans expressing intestinal polyQ with human enteric pathogens influences motility.
A) A cartoon depicting time-off-pick (TOP) phenotype measure as the time (seconds) it takes a worm to crawl off a pick when pickup up by the midbody section. B) Age-dependent changes of motility in animals expressing intestinal polyQ (polyQ33 and polyQ44) grown on E. coli OP50 control strain. Higher motility measured in TOP seconds indicates a higher motility defect. Data are represented as the average TOP per worm. Each data point represents 20 worms. C) Motility of animals expressing intestinal polyQ grown on K. pneumoniae KP182 and P. aeruginosa PAO1. Data are represented as the average TOP per 60 worms over three independent experiments, normalized to E. coli OP50 control strain. Error bars represent SEM. Statistical significance was calculated using one-way ANOVA followed by multiple comparison Dunnett’s post-hoc test (**p<0.01, ****p <0.0001).
Bacterial colonization of the C. elegans intestine disrupts protein folding in distal tissues
Muscle tissue
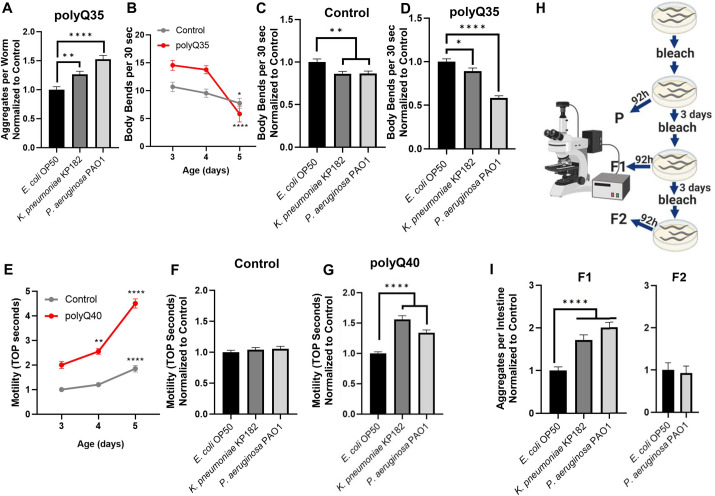
Because bacteria-induced polyQ aggregation in the intestine influenced C. elegans motility, an effect that likely involves distal tissues, we sought to determine whether the influence of bacteria on protein aggregation is confined to the intestine or if bacteria can impact protein folding across other tissues. We employed nematodes expressing muscle-specific polyQ35 (unc-54p::Q35::YFP) as a proxy to detect changes in proteostasis in that tissue. Different tissues have different thresholds for polyQ aggregation; therefore, while polyQ33 did not aggregate in the intestine, polyQ35 aggregates in the muscle. This reporter exhibits age-dependent and muscle-specific aggregation and the associated motility defect [29]. We colonized the intestines of these worms with two strains that enhanced intestinal polyQ aggregation: K. pneumoniae KP182 and P. aeruginosa PAO1. Animals expressing muscle polyQs exhibit a more robust aggregation profile compared to the intestinal polyQ model; thus, we had to quantify the aggregates on day three instead of day four. The results paralleled the trend seen in the aggregation profile of the intestinal polyQ model, where K. pneumoniae enhanced aggregation compared to control E. coli OP50 and P. aeruginosa PAO1 showed the most robust enhancement (Fig 3A). To determine whether the bacteria-induced proteostasis imbalance detected by the polyQ aggregation in the muscle also leads to tissue dysfunction, we employed a previously characterized motility readout to enumerate the body bends of polyQ-expressing worms fed these two bacterial strains. The animals expressing muscle polyQ exhibit a sharp and age-dependent decrease in body bends (Fig 3B). We found that colonization of the C. elegans intestine with enteric bacterial pathogens led to a significant defect in motility, particularly in worms fed the most potent enhancer of aggregation, P. aeruginosa, which decreased the number of body bends 2-fold when compared to the control E. coli OP50; a larger and more significant defect than what was observed in control wild-type animals (Fig 3C and 3D).
Fig 3. Colonization of the C. elegans intestine with human enteric pathogens influences proteostasis in a tissue non-autonomous manner.
A) The effect of select bacteria on protein folding in the muscle. Data are represented as the average number of aggregates per worm normalized to E. coli OP50 control strain. Each bar is an average of three independent experiments with a total of 100 animals. B) Age-dependent decline in motility assessed by enumerating body bends per 30 seconds in muscle-specific polyQ35 and N2 control worms. The data are represented as the average number of body bends quantified on day 3–5 post-hatching in a total of 15 animals per each day. The effect of bacteria on the motility of C) control animals and D) animals expressing muscle-specific polyQ35. Data are represented as the average number of body bends per worm normalized to worms fed E. coli OP50 control strain. Each bar is an average of three independent experiments with a total of 45 animals. E) Age-dependent decline in motility assessed by increased TOP. The effect of bacteria on the motility of F) control animals and G) animals expressing neuron-specific polyQ40. Data are represented as the average TOP per worm normalized to animals fed E. coli OP50 control strain. Each bar is an average of three independent experiments with a total of 60 animals. H) A cartoon depicting experiments which demonstrated that bacterial colonization of the C. elegans intestine affects the F1 generation. I) Quantification of intestinal aggregates in the F1 and F2 generations from parental animals that were fed select test and control bacteria. Each bar represents the average number of aggregates per intestine normalized to the control (E. coli OP50). Data are representative of three (F1) and one (F2) independent experiments with a total of 100 and 30 animals, respectively. Error bars represent SEM. Statistical significance was calculated using one-way ANOVA followed by multiple comparison Dunnett’s post-hoc test (*p<0.05, **p<0.01, ****p<0.0001).
Neuronal tissue
To determine whether proteostasis in neuronal tissue is also affected by bacterial colonization of the intestine, we utilized worms carrying neuronal polyQ40 (F25B3.3p::Q40::YFP) [30]. The small size and high concentration of neurons preclude us from accurately quantifying the exact number of aggregates in this tissue; as such, we measured toxicity by assessing TOP motility. Animals carrying this reporter exhibit an age-dependent decrease in motility compared to control N2 worms (Fig 3E). We found that colonization of the intestine with K. pneumoniae and P. aeruginosa led to a significant defect in motility when compared to the control E. coli OP50 (Fig 3F and 3G).
Gonad
Thus far, we demonstrated that colonization of the C. elegans gut with enteric bacterial pathogens leads to proteostasis imbalance and proteotoxicity in proximal tissues, including muscle and neurons. To further examine the extent to which gut bacteria influence host proteostasis, we asked whether colonization of the C. elegans intestine had any effect on the gonad. If the colonization of the intestine affects the muscle and neurons, we reasoned that bacteria might also affect proteostasis in the germline—if this is the case, then we should be able to detect changes in polyQ aggregation in the progeny of parents that were colonized by pathogenic bacteria. To test the effect of bacteria on the next generation, we fed C. elegans either control E. coli OP50, K. pneumoniae KP182, or P. aeruginosa PAO1 for three days, followed by “bleaching” adults, and age-synchronizing the progeny. C. elegans synchronization involves dissolving gravid adults by hypochlorite treatment leaving intact and sterile embryos which hatch into L1 larvae. Synchronized F1 animals were placed on fresh plates containing a lawn of control E. coli OP50 and intestinal aggregates were enumerated after 92 hours (h). The F2 generation was isolated and assessed using the same method (Fig 3H). Strikingly, we found that colonization of the parental intestines with the select bacteria enhanced polyQ aggregation in the F1 progeny, despite the fact that they never encountered the bacteria that were fed to the parental generation (with the exception of E. coli OP50) (S5 Fig). The extent of the enhancement in the number of polyQ aggregates per worm correlated with previously assessed phenotypes where P. aeruginosa showed the highest induction of aggregation (Fig 3I). Since P. aeruginosa induced the strongest effect, we looked at the F2 progeny from a parental generation fed these bacteria. We saw no enhancement of aggregation compared to control E. coli, indicating that only F1 generation is affected (Fig 3I). Using western blotting, we ruled out the possibility that these results were due to changes in polyQ44::YFP expression level (S1E Fig) and we confirmed an increase of the insoluble polyQ44::YFP (S2D Fig). Collectively, these results are intriguing as they demonstrate that colonization of the parental intestines affects proteostasis in the offspring.
Butyrate suppresses bacteria-induced polyglutamine aggregation and aggregate-dependent toxicity across tissues
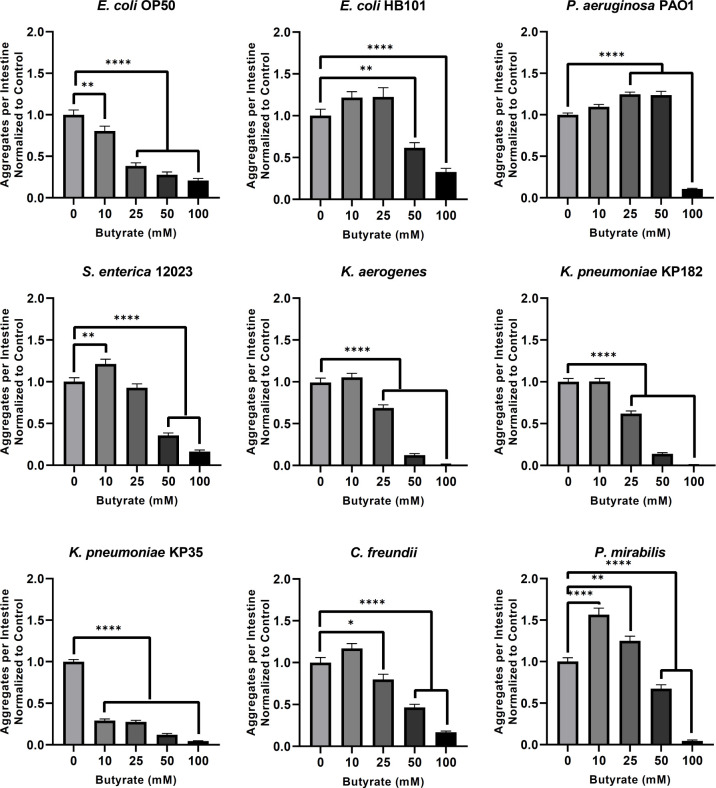
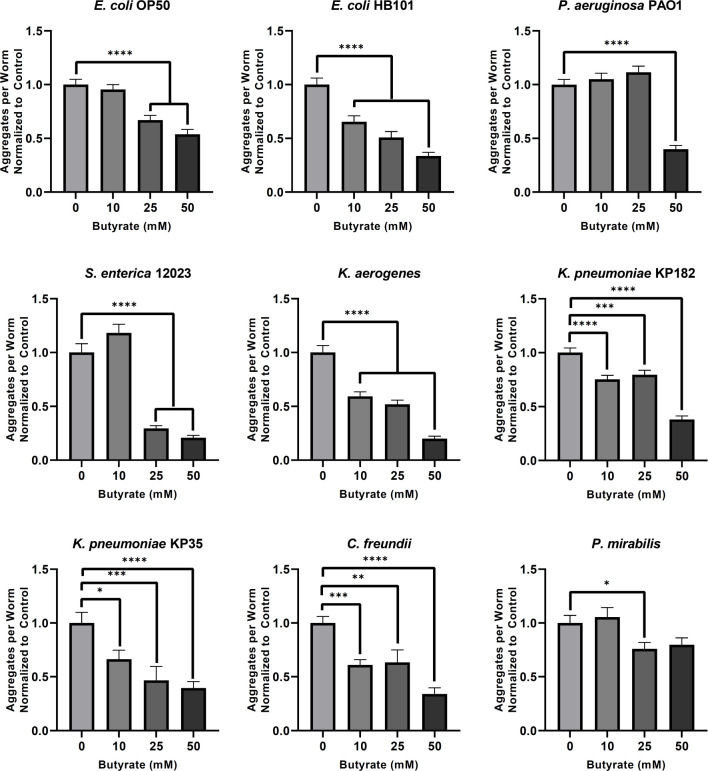
The depletion of butyrogenic bacteria in the gut establishes an environment in which pathogenic bacteria, particularly Enterobacteriaceae, can flourish [31]. In addition, butyrate itself has been found to suppress the growth of pathogenic bacteria and was shown to protect against neurodegeneration [32–34]. To assess the effect of exogenous butyrate on the observed bacteria-induced aggregation, we first tested its effect on intestinal polyQ aggregation in C. elegans. We used all bacterial genera that induced protein aggregation by three-fold or more (Fig 1B). The physiological concentration of human colonic butyrate is estimated to be 10–20 mM, and a therapeutic dose of 150 mM butyrate was safely delivered via enema directly into the human large intestine [35,36]. Therefore, we tested the effect of butyrate on bacteria-induced protein aggregation at concentrations ranging from 10–100 mM. Unexpectedly, we found that lower concentrations of butyrate differentially affected host proteostasis where enhancement of aggregation was dependent on the type of bacteria colonizing the intestine: 10 mM: S. enterica 12023, P. mirabilis; 25 mM: P. mirabilis, P. aeruginosa PAO1; 50 mM: P. aeruginosa PAO1. However, at higher butyrate concentrations (>25 mM), bacteria-induced aggregation was suppressed in a dose-dependent manner in worms colonized with all bacteria except P. aeruginosa. Supplementation with 100 mM butyrate suppressed bacteria-induced aggregation across all strains. (Fig 4). These results are unexpected, as they suggest that at low physiological concentrations (<25 mM), butyrate may contribute to the bacteria-mediated disruption of host proteostasis, as demonstrated by differentially enhanced polyQ aggregation; however, with the exception of P. aeruginosa PAO1, higher concentrations (50–100 mM) of butyrate suppressed aggregation induced by all other bacterial strains, which suggests therapeutic potential. We employed western blot analysis of polyQ44::YFP levels to eliminate the possibility that these results are due to changes in protein level (S1C and S1D Fig), and we confirmed these results by assessing insoluble fractions (S2B and S2C Fig).
Fig 4. The effect of butyrate on bacteria-induced aggregation in the intestine.
Data are represented as the average number of aggregates per intestine normalized to the control (0 mM butyrate). Each bar is an average of three independent experiments with a total of 100 animals. Error bars represent SEM. Statistical significance was calculated using one-way ANOVA followed by multiple comparison Dunnett’s post-hoc test (*p<0.05, **p<0.01, ****p<0.0001).
Furthermore, to rule out the possibility that butyrate decreased bacteria-mediated protein aggregation by merely killing bacteria, we tested the effect of butyrate on bacterial growth and viability in liquid medium and on solid NGM plates, respectively, at concentrations up to 100 mM. While butyrate did affect the growth and viability of some bacteria, all of the strains tested remained viable. We found that most overnight cultures supplemented with butyrate at concentrations that inhibited aggregation reached optical densities comparable to unsupplemented controls (S6 Fig). We also tested the effect of butyrate on bacteria cultured on solid NGM media and found that, while some strains were affected, all were viable across concentrations (S7 Fig). Additionally, feeding C. elegans dead bacteria resulted in higher aggregation profiles that are distinct from those treated with 100 mM butyrate, further suggesting that butyrate does not suppress bacteria-mediated aggregation by solely affecting bacterial viability (S8 Fig). Next, we asked whether butyrate could affect colonization of the intestine. We found that at 25 mM, butyrate significantly enhanced intestinal colonization by E. coli OP50, but exhibited no effect on colonization at 100 mM (S9 Fig). These results indicate that it is unlikely that the butyrate’s effect on bacterial colonization affects aggregation.
A trade-off between organismal resources available to maintain either proteostasis or fecundity has been proposed [37]. As such, we asked whether butyrate affects fecundity in our experiments, which may explain the suppression of aggregation. Indeed, we found that butyrate decreases the number of progeny per worm, but the effect seems to depend on the presence of polyQ (S10A and S10B Fig). Moreover, 25 mM butyrate significantly decreased the number of progeny in worms colonized by P. aeruginosa (S10C Fig). Yet, we found that under the same conditions, butyrate increased polyQ aggregation in animals colonized by P. aeruginosa (Figs 4 and S2C). These results indicate that a decrease in fecundity alone does not explain the observed suppression of polyQ aggregation.
To determine whether exogenous butyrate would also rescue the aggregate-dependent motility defect observed in the intestinal polyQ44 model, we measured the TOP phenotype in animals colonized by E. coli OP50, K. pneumoniae KP182, and P. aeruginosa PAO1 in the absence or presence of 100 mM butyrate. While little to no changes in TOP were observed in the roller controls, exogenous butyrate supplementation (100 mM) rescued motility defects in intestinal polyQ44 worms (Figs 2C and 5). We further explored the ability of butyrate to suppress aggregation in other tissues by repeating the above experiment using the muscle polyQ model and assessed the effect of butyrate on bacteria-induced protein aggregation at concentrations ranging from 0–50 mM. Unlike the intestinal polyQ44 model, worms expressing muscle polyQ35 did not experience a significant enhancement of aggregation at lower butyrate concentrations and exhibited a dose-response or near dose-response suppression of aggregation across all bacterial strains except for P. mirabilis and P. aeruginosa PAO1, wherein the only suppressive concentrations of butyrate were 25 and 50 mM, respectively (Fig 6). It is worth noting that the trends of butyrate-suppressed aggregation in worms expressing muscle polyQ parallel those seen in worms expressing intestinal polyQs (Fig 4). For example, animals cultured on plates containing 0–50 mM butyrate exhibited a significant dose-dependent decrease in intestinal and muscle aggregation when colonized by E. coli OP50 or Klebsiella spp. On the other hand, lower concentrations of butyrate did not significantly suppress aggregation in either tissue when animals were fed P. aeruginosa PAO1, S. enterica 12023, and P. mirabilis, suggesting that the beneficial effect of butyrate is, at least in part, facilitated by bacteria (Figs 4 and 6).
Fig 5. The effect of butyrate on toxicity associated with bacteria-mediated intestinal polyQ aggregation.
Aggregation-dependent toxicity is assessed by measuring motility (TOP phenotype) in the presence of 100 mM butyrate. Data are represented as the average number of TOP seconds per worm normalized to the control (0 mM butyrate). Two roller strains, Control (AM446, no polyQ) and polyQ33 (no aggregates), are controls. Each bar is an average of three independent experiments with a total of 60 animals. Error bars represent SEM. Statistical significance between each pair was calculated using Student’s t-test (*p<0.05, ****p<0.0001).
Fig 6. Butyrate affects bacteria-mediated protein aggregation in the C. elegans muscle.
Data are represented as the average number of aggregates of muscle-specific polyQ35 (AM140) per worm normalized to the control (0 mM butyrate). Each bar is an average of three independent experiments with a total of 100 animals and the error bars represent SEM. Statistical significance was calculated using one-way ANOVA followed by multiple comparison Dunnett’s post-hoc test (*p<0.05, **p<0.01, ***p<0.001, ****p<0.0001).
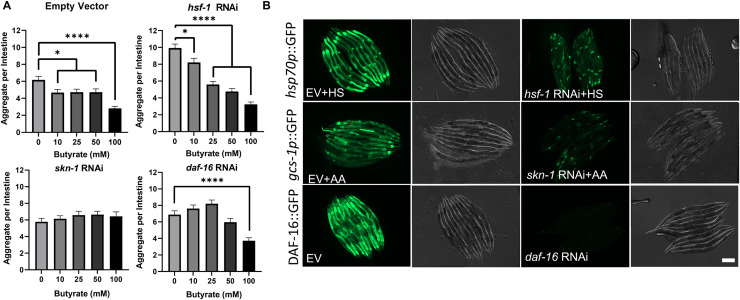
To gain a mechanistic understanding of how butyrate suppresses bacteria-mediated aggregation, we investigated the dependence of the observed response on three major evolutionary-conserved transcription factors, HSF-1, SKN-1/Nrf2, and DAF-16/FOXO. Each of these transcription factors was previously shown to provide protection against proteotoxicity [38–40]. Because butyrate suppressed aggregation upon colonization with multiple strains of bacteria, including E. coli OP50 (Fig 4), we investigated the dependence of butyrate-mediated suppression of aggregation on the abovementioned transcription factors by knocking-down each candidate using RNAi. C. elegans carrying intestinal polyQ44 that were fed E. coli HT115 (DE3) expressing empty vector control (L4440) RNAi, exhibited a significant suppression of aggregation in the presence of butyrate (10–100 mM) (Fig 7A). While butyrate-mediated suppression of aggregation was not affected when HSF-1 was knocked-down, the beneficial effect of butyrate was abolished upon downregulation of SKN-1/Nrf2 and DAF-16/FOXO transcription factors. As expected, HSF-1 knockdown enhanced aggregation in the absence of butyrate when compared to control animals (Fig 7A) [41]. To confirm that the knockdown is functional, we used fluorescent reporters to assess the expression of downstream target genes of HSF-1 (hsp70p::GFP also known as C12C8.1p::GFP) and SKN-1 (gcs-1p::GFP), and we used DAF-16 (DAF-16::GFP) fluorescent fusion (Fig 7B). The expression of all fluorescent reporters was attenuated upon knockdown of the respective target genes.
Fig 7. SKN-1 and DAF-16 are involved in butyrate-mediated suppression of aggregation.
A) The effect of hsf-1, skn-1, and daf-16 knockdown on intestine-specific polyQ44 aggregation in the presence of exogenous butyrate supplementation, compared to empty vector control (L4440). Data are represented as the average number of aggregates per C. elegans intestine. Each data point is an average of a minimum of three independent experiments with a total of at least 85 animals. Error bars represent SEM. Statistical significance was calculated using one-way ANOVA followed by multiple comparison Dunnett’s post-hoc test (*p<0.05, ****p<0.0001). B) Fluorescent and Nomarski images of worms expressing reporter constructs regulated by HSF-1, SKN-1, and DAF-16. Worms were fed E. coli expressing either the control empty vector (EV) shown in the left panels or specific RNAi shown in the right panels. To activate the reporters, animals expressing hsp70p::GFP and gcs-1p::GFP were either heat shocked (HS) or exposed to 5 mM acrylamide (AA), respectively. Functional knockdowns were confirmed by the attenuated GFP expression in animals fed RNAi versus EV. Scale bar = 200 μm.
Together, these results indicate that butyrate can suppress bacteria-induced polyQ aggregation across tissues and rescues aggregate-dependent toxicity in the host. Such protective effect of butyrate seems to be mediated by SKN-1 and DAF-16 transcription factors.
Colonization of the C. elegans intestine with E. coli engineered to overproduce butyrate suppresses protein aggregation across C. elegans tissues
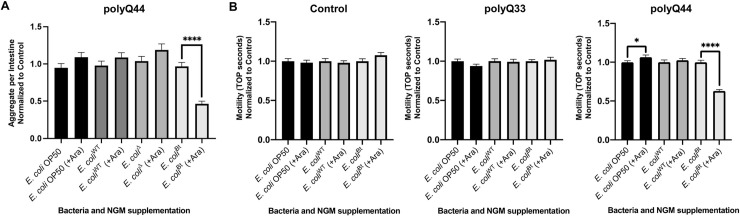
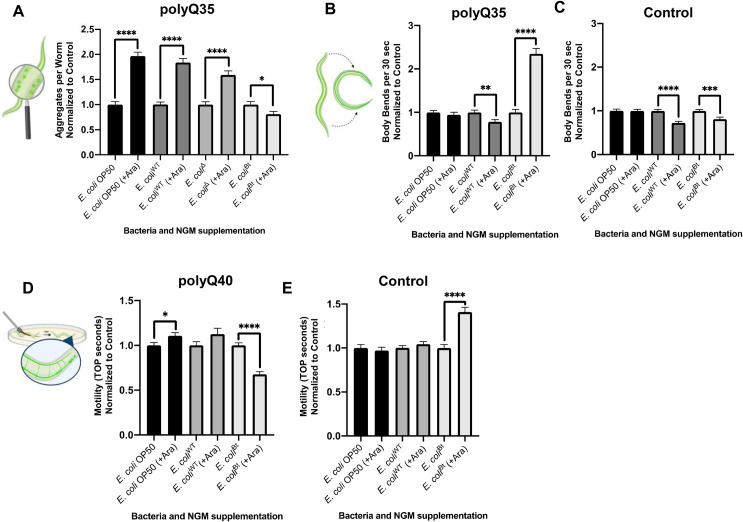
SCFA-producing bacteria are common residents of the human gut microbiota. Many of these bacteria, such as those belonging to the Firmicutes phylum, are known to produce butyrate [42]. Butyrate has been demonstrated to provide numerous health benefits against a variety of ailments, and neurodegenerative diseases are no exception [32]. To determine the effect that butyrogenic bacteria have on organismal proteostasis, we used an E. coli strain, LW393 (E. coliBt), engineered to conditionally overproduce butyrate [43]. Colonizing C. elegans intestines with this strain allows us to model simplified physiological, yet controlled, conditions. The strain was derived from E. coli W (E. coliWT), a non-pathogenic strain closely related to commensal bacteria by first creating a quadruple deletion mutant (ldhA, frdABCD, ackA, and adhE), E. coli BEM9 (E. coliΔ) strain, followed by overexpression of enzymes that channel intermediates into the pathway and which are essential for butyrate synthesis [43]. Therefore, we used E. coliWT and, in some instances, E. coliΔ, as controls. E. coliBt synthesizes butyrate in the presence of 5- and 6-carbon sugar substrates to levels up to three times higher than any other available commercial strain [43]. We first used this strain to colonize the intestine of C. elegans expressing the intestinal polyQ44 reporter over a period of four days. We demonstrated that, on days three and four, C. elegans intestines are colonized by viable bacteria (S11 Fig). While glucose is a suitable substrate for butyrate synthesis, we found that it affects polyQ aggregation and thus cannot be utilized in our experiments (S12 Fig); however, L-arabinose is another butyrate synthesis substrate and does not affect aggregation. To confirm that L-arabinose from the NGM plates is available to intestinal bacteria, we colonized C. elegans intestines with bacteria that express an arabinose-inducible fluorescent reporter and cultured them in the absence of the sugar. We then transferred these animals onto plates containing E. coli OP50 and 3% L-arabinose and detected a strong induction of fluorescence, indicating that L-arabinose is accessible to the intestinal bacteria (S13 Fig). We supplemented NGM plates with 3% L-arabinose and quantified the number of aggregates after four days feeding polyQ-expressing animals the three control (E. coli OP50, E. coliWT, and E. coliΔ) and the butyrate-producing (E. coliBt) strains. We did not detect any effect on polyQ aggregation in worms colonized with each of the control E. coli strains; however, supplementation of L-arabinose did result in significant suppression of polyQ aggregation in animals colonized by E. coliBt (Fig 8A). To determine whether the aggregate-suppressing effect of E. coliBt would also rescue polyQ-associated motility defects, we measured the TOP phenotype in worms expressing intestinal polyQ44. In agreement with the observed suppression of aggregation, supplementation of L-arabinose to animals colonized by E. coliBt also improved motility, indicating that bacteria-derived butyrate can rescue aggregation and aggregate-associate toxicity (Fig 8B).
Fig 8. Butyrate-producing E. coli suppresses aggregation and the associated toxicity.
Animals were fed four different strains of bacteria: non-butyrogenic controls (E. coli OP50, E. coliWT, E. coliΔ) and conditional butyrogenic E. coli (E. coliBt). A) The graphs represent the average number of intestinal polyQ44 aggregates per worm normalized to the control (no arabinose). Each bar is an average of three independent experiments with a total of 100 animals. B) Intestinal aggregate-dependent toxicity normalized to the control (no arabinose) assessed with the TOP phenotype. The left panel represents roller worms (Control), the middle panel represents polyQ33 worms, and the right panel represents worms expressing polyQ44. Each bar is an average of three independent experiments with a total of 60 animals. Error bars represent SEM. Statistical significance between each pair was calculated using Student’s t-test (*p<0.05, ****p<0.0001).
We further investigated whether the effect of endogenous butyrate produced by E. coliBt strain was limited to the intestine or was ubiquitous across tissues, as seen when the animals were exposed to exogenous butyrate. To accomplish this, we colonized the intestines of animals expressing muscle and neuronal polyQ with E. coliBt. While supplementation of 3% L-arabinose into NGM plates did not affect intestinal polyQ aggregation in animals colonized by control E. coli strains (Fig 8A), it did enhance aggregation in animals expressing muscle-specific polyQs (Fig 9A). Intriguingly, this enhancement of aggregation was significantly reduced in the presence of L-arabinose in animals colonized by E. coliBt, suggesting that endogenously produced butyrate suppresses protein aggregation in the muscle tissue as well (Fig 9A). We then assessed the effects of this strain on aggregate-dependent toxicity in this model utilizing the body bend assay. Our results show that worms expressing muscle-specific polyQ have 2.5x more body bends when E. coliBt is endogenously producing butyrate, compared to worms colonized with non-butyrogenic bacteria or butyrogenic bacteria in the absence of the sugar substrate (Fig 9B). The observed beneficial effect of endogenous butyrate is dependent on the presence of polyQ since endogenous butyrate did not increase the number of body bends in control wild-type worms (Fig 9C). In fact, the presence of endogenous butyrate was slight, but significantly detrimental to wild-type C. elegans. These results support the beneficial effect of butyrate in the suppression of aggregation and the associated proteotoxicity in the muscle tissues.
Fig 9. Colonization of the C. elegans intestine with butyrogenic E. coli suppresses aggregation and associated toxicity in distal tissues.
A) The effect of butyrogenic bacteria (E. coliBt) on the aggregation profile of worms expressing muscle-specific polyQ35. Data are represented as the average number of aggregates of muscle-specific polyQ35 (AM140) per worm normalized to the control (no arabinose). Animals were fed four different strains of bacteria: non-butyrogenic controls (E. coli OP50, E. coliWT, E. coliΔ) and conditional butyrogenic E. coli (E. coliBt). B) The effect of butyrogenic bacteria on the motility of animals expressing muscle-specific polyQ35 and C) control N2 worms. Data are represented as the average number of body bends per worm normalized to control (no arabinose). Each bar is an average of three independent experiments with a total of 45 animals. D) The effect of butyrogenic bacteria on the motility of animals expressing neuronal polyQ40 and on E) control N2 worms. Data are represented as the average TOP seconds per worm normalized to control animals (no arabinose). Error bars represent SEM. Statistical significance between each pair was calculated using Student’s t-test (*p<0.05, **p<0.01, ***p<0.0005 ****p<0.0001).
We expanded our investigation to animals carrying a neuronal polyQ40 reporter by measuring toxicity using the TOP phenotypic readout. We employed this method to assess the effect of endogenous butyrate on aggregate-dependent toxicity. The availability of L-arabinose to control E. coli strains (E. coli OP50 and E. coliWT) did not improve motility. Still, the butyrate produced by E. coliBt significantly enhanced motility in animals carrying the neuronal polyQ reporter (Fig 9D), but not in N2 control worms (Fig 9E). While further investigation is needed to elucidate the basis of this response, the result could be key to understanding the mechanism(s) by which butyrogenic bacteria affect host proteostasis.
Co-colonization of the C. elegans intestine with butyrogenic bacteria suppresses bacteria-induced aggregation
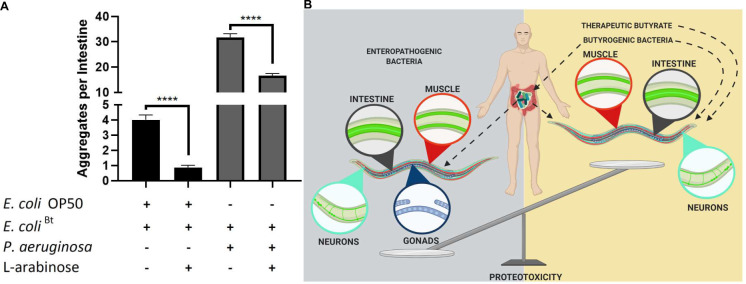
The human gut microbiota is a polymicrobial community and the effect of the interactions between individual strains and the host is hindered by the complexity and wealth of its composition. We co-colonized the C. elegans gut with pathogenic and butyrogenic bacteria as a strategy to begin deconvoluting complex polymicrobial interactions within the human microbiome and their effect on the host. We fed worms cultures of E. coliBt with either E. coli OP50 as a control or P. aeruginosa PAO1, which showed the most robust enhancement of aggregation. In the presence of L-arabinose, we found a significant decrease in aggregation of intestinal polyQs in co-cultures with both strains (Fig 10A). E. coliBt suppressed E. coli OP50- and P. aeruginosa PAO1-mediated aggregation by approximately 4- and 2-fold, respectively. These results indicate that endogenous butyrate synthesized by butyrogenic bacteria can enhance host proteostasis and suppress protein misfolding. Furthermore, our experiments demonstrate the potential benefits of butyrate and butyrogenic bacteria in the suppression of bacteria-induced proteotoxicity associated with metastable proteins and also emphasize the importance of having a balance between commensal butyrogenic and enteropathogenic microbes (Fig 10B).
Fig 10. Co-colonization of E. coliBt suppresses P. aeruginosa-mediated enhancement of polyQ aggregation.
A) The effect of butyrogenic bacteria (E. coliBt) on the aggregation of intestinal polyQ44 (AM738) in worms co-colonized with E. coli OP50 or P. aeruginosa in the presence and absence of 3% L-arabinose. Data are represented as the average number of aggregates per intestine. Each bar is an average of three independent experiments with a total of 100 animals. Error bars represent SEM. Statistical significance between each pair was calculated using Student’s t-test (****p<0.0001). B) Model figure. Enteropathogenic bacteria disrupt proteostasis across C. elegans tissues, including intestine, muscle, neurons, and gonads. The bacteria-mediated proteotoxicity is alleviated by butyrate and butyrogenic bacteria.
Discussion
We used a simple metazoan model, C. elegans, to study the effect of bacterial gut colonization on organismal proteostasis. We found that the colonization of the C. elegans intestine with enteric bacterial pathogens disrupts organismal proteostasis and enhances proteotoxicity associated with polyQ. Importantly, our results demonstrate that bacteria alone do not cause major toxicity in control animals that do not express polyQ, which would hinder the interpretation of our data (Figs 2B, 2C, 3C, and 3F). It is not surprising that the detrimental effect of bacteria on polyQ aggregation is observed in the most proximal tissue, the intestine, as similar results were previously observed [23]. However, to our knowledge, this is the first report of bacteria having detrimental, tissue non-autonomous effects on protein folding. Recently, Bacillus subtilis, a probiotic strain that is also part of the human commensal microbiota, was shown to inhibit α-synuclein aggregation in C. elegans muscle [44]. The fact that certain bacteria disrupt host proteostasis while others can enhance it emphasizes the importance of microbes in the pathogenesis of PCDs. Previous research suggests that increased presence of bacterial species such as K. pneumoniae and P. aeruginosa in the human gut is associated with an increased prevalence of PCDs [45,46]. Interestingly, among all of the strains that we tested, Klebsiella spp. and P. aeruginosa were the most potent inducers of polyQ aggregation–this was the case even for a commensal strain, K. oxytoca (Fig 1B and 1C). Both of these species are ubiquitous in the environment, and multidrug-resistant strains of these bacteria are often associated with nosocomial and opportunistic infections [47]. Due to their rapidly increasing resistance to antibiotics, they are classified by the Centers for Disease Control and Prevention as serious threats [48]. While these strains are often associated with infections, their enteric presence is also evident in healthy individuals and may possibly support their contribution to neurodegenerative diseases [49,50]. In fact, both of these microbes have recently been associated with neurodegenerative diseases in humans [45,46], a correlation that is in agreement with our results. The increasing prevalence of antibiotic resistance among Klebsiella spp. and P. aeruginosa strains may specifically enrich for their growth within the human microbiome upon an antibiotic treatment while decreasing the abundance of beneficial bacteria. In agreement with this hypothesis, population-based studies of patients with Parkinson’s and ALS revealed that a history of antibiotic treatment is associated with an elevated risk for these diseases [7,8].
Another notable strain that induced aggregation by more than three-fold is P. mirabilis. This gram-negative bacterium is part of human commensal microbiota but can also become an opportunistic pathogen, most often leading to urinary tract infections, and eventually, bacteremia [51]. Interestingly, in one study, blood cultures of PD septic patients had a five-fold higher prevalence of P. mirabilis compared to non-PD septic patients [52]. Recently, it was demonstrated in a mouse model that P. mirabilis contributes to PD pathogenesis, including the induction of α-synuclein in the mouse brain [53]. Furthermore, colonization of the gut with P. mirabilis and Enterobacteriaceae, in particular, K. pneumoniae, was shown to be accelerated by antibiotic treatment, leading to detrimental dysbiosis in mouse microbiota [54]. On the other side of the aggregation spectrum, the non-pathogenic E. coli HB101 did not significantly enhance aggregation in our C. elegans polyQ model. The E. coli HB101 strain is also a common laboratory food for C. elegans; hence, we did not expect any enhancement with these bacteria. However, both P. disiens HM-1171 and P. corporis HM-1294 suppressed aggregation relative to E. coli OP50 (Fig 1C). The mechanism by which these Prevotella spp. suppress aggregation does not depend on butyrate as both strains were deficient in its production [55,56]. Interestingly, Prevotella was the only bacterial genus whose abundance significantly decreased in patients suffering from constipation, a condition that is a hallmark in Parkinson’s disease and is associated with up to 80% of cases [57–59]. Further evidence that these two species could be beneficial to host proteostasis comes from a recent study that demonstrated a negative correlation between the abundance of Prevotella in the gut of Parkinson’s disease patients and the severity of symptoms [60]. A negative correlation between the abundance of Prevotella and pathogenesis of Alzheimer’s disease mouse model has also been observed [61]. Collectively, these results suggest that the members of the Prevotella genus could play a more direct role in enhancing host proteostasis, and that the mechanisms by which bacteria affect host proteostasis in our C. elegans model may be conserved in higher organisms.
Although P. aeruginosa was previously found to induce neurodegeneration in C. elegans [62], the results of decreased polyQ-dependent motility of animals colonized with P. aeruginosa and K. pneumoniae were unexpected and suggest that either bacteria, or host responses to these bacteria, affect the function of distal tissues (i.e., muscle or neurons), but only in animals that express metastable aggregation-prone proteins such as polyQs (Figs 2B, 2C, and 3A–3G). Recent studies show that colonization of the C. elegans intestine with pathogenic bacteria affects bacterial avoidance behavior [63] and consequently increases lifespan [64]. While it is possible that feeding behavior might have been affected in our experiments, it is unlikely that avoidance would contribute to an increase in polyQ aggregation and the associated motility defects. In fact, if the animals in our experiments experienced food avoidance behavior, we would expect to see enhanced proteostasis, as dietary restriction was previously shown to suppress polyQ aggregation [65].
It is known that bacterial colonization of the C. elegans intestine increases the production of reactive oxygen species (ROS), which in turn enhances aggregation in that tissue [23]. Oxidative stress is known to affect overall animal physiology, including motility [66]. Since our results show the bacteria-mediated motility defects only in the presence of aggregation-prone polyQ44, but not in non-aggregating polyQ33 or non-polyQ controls (Fig 2C), it is possible that ROS generated by the worm in response to bacterial colonization enhanced polyQ aggregation, which further affected the function of other tissues involved in motility. Such a mechanism likely contributes to the observed bacteria-mediated disruption of host proteostasis which is supported by our results demonstrating that butyrate rescued polyQ aggregation in a DAF-16 and SKN-1-dependent manner (Fig 7A). However, ROS is likely not the sole contributor since our results also indicate that bacterial aggregates affect host proteostasis and consequently increase aggregation of host polyQ (Fig 1E–1G).
Tissue non-autonomous regulation has been noted in C. elegans under various conditions [27,67]. Our results demonstrating that bacterial colonization of the C. elegans intestine affects proteostasis across tissues are intriguing, as they suggest that bacteria impose tissue non-autonomous effects on the host. In human and mouse studies, the effect of bacterial gut colonization on host proteostasis has been primarily attributed to non-specific mechanisms, such as systemic inflammation, which broadly affects many tissues [68]; however, C. elegans lacks an inflammatory response, indicating that other and possibly more direct mechanisms may be involved. Bacterial amyloids have also been shown to specifically contribute to pathogenesis by affecting metastable aggregation-prone proteins [69]. The introduction of metastable proteins is known to affect the stability and folding of other proteins within the proteome [70,71]. As such, it is likely that bacteria may be secreting factors that translocate from the intestine across other tissues and affect proteostasis. A cross-seeding between disease-associated proteins (i.e., Aβ) with bacterial amyloids, including those from E. coli and P. aeruginosa, was previously observed [72,73]. Also, an enhancement of alpha-synuclein aggregation mediated by bacterial curli was shown in mouse and C. elegans Parkinson’s disease models [74]. While our results indicate that E. coli curli fimbriae were not responsible for the enhancement of polyQ aggregation, E. coli aggregates did play a contributing factor (Fig 1G). It is possible that alpha-synuclein and polyQ are affected differently by bacterial amyloids. In addition to regulating the csg operon, CsgD also functions as a global transcriptional regulator and its target gene(s), other than csgA, could affect host proteostasis [75]. It is intriguing to speculate that the transmission of aggregation-prone proteins between tissues [67], may underlie how bacterial colonization of parental intestines affects protein aggregation in the progeny (Fig 3I).
A symbiosis between commensal and pathogenic bacteria can be disrupted by many factors, including infections, diet, and antibiotics [76]. While dysbiosis of the human gut microbiota has been associated with the pathogenesis of PCDs, the mechanisms that contribute to the disruption of host proteostasis and disease progression remain poorly understood. Our results demonstrate that butyrate, a common metabolite produced by commensal microbiota, can suppress aggregation and the associated toxicity when supplied exogenously (Figs 4 and 6) or produced by intestinal bacteria (Figs 8 and 9). We also showed that butyrate did not simply inhibit intestinal colonization (S9 Fig). In agreement with our results, the protective effects of butyrate have been previously demonstrated in a mouse model and in humans [21,32]. Our data indicate that the benefits of butyrate depend on SKN-1/Nrf2 and DAF-16/FOXO transcription factors, suggesting that butyrate may inhibit bacteria-induced aggregation by activating protective stress responses, in particular, oxidative stress responses. Indeed, bacteria are known to trigger oxidative stress, which can contribute to polyQ aggregation [23]. It is possible that various bacteria introduce different levels of oxidative stress and the strongest contributors affect host proteostasis, consequently leading to misfolding and aggregation of metastable proteins present within the proteome. In fact, oxidative stress is one of the major contributors to PCDs [77]. Recently, it was found that butyrate and β-hydroxybutyrate provide Nrf2- and FOXO-dependent protection against oxidative stress, respectively [78,79]. In addition, both SKN-1 and DAF-16 were shown to be involved in β-hydroxybutyrate-mediated lifespan extension [80]. These results, along with our data, collectively suggest that butyrate activates SKN-1/Nrf2 and DAF-16/FOXO, which attenuate bacteria-induced oxidative stress. The effect of butyrate on polyQ aggregation also seems to depend on the type of pathogenic bacteria present, suggesting that butyrate may contribute to host proteostasis by acting on bacteria in addition to the host. These results are consistent throughout the intestinal and muscle-specific polyQ models. Additionally, the co-colonization experiment further supports that the extent of the beneficial effect of butyrate depends on the bacteria present (Fig 10A). The fact that bacteria are viable in the presence of butyrate at concentrations that suppress aggregation (S6 and S7 Figs) and that dead bacteria exhibit a distinct effect on aggregation (S8 Fig) suggest that microbial processes which affect the host may be targeted by butyrate. Whether butyrate affects specific bacterial signals that contribute to PCDs remains to be determined and further studies are underway to identify bacterial factors that enhance aggregation of host metastable proteins. Whatever the mechanism may be, our results suggest a therapeutic potential of butyrate in the prevention or treatment of PCDs.
High metabolic rates of bacteria can lead to depletion of available oxygen and introduce a hypoxic environment on the plates. Low oxygen levels are known to disrupt cellular processes leading to increased polyQ aggregation [81]. One of the possible explanations regarding how butyrate may inhibit bacteria-mediated polyQ aggregation is by suppressing bacterial metabolism, which restores the normoxic environment. However, if this were true, we would expect to have seen the restoration of a normoxic environment when we fed worms heat-killed bacteria, which would result in polyQ aggregation similar to 100 mM butyrate (S8 Fig). Therefore, it is likely that butyrate could have some additional effects on bacteria in addition to the host. In fact, it is known that butyrate can affect bacterial pathogenicity [34,82,83]. Further research is underway to determine what changes butyrate might exert on bacteria and how that influences the host.
Methods
Bacterial strains
Detailed information about bacterial strains used and generated in this study is provided in Table 1.
Table 1. Reagents and resources used in this study and their associated sources and identifiers.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial Strains | ||
| Escherichia coli OP50 | Caenorhabditis Genetics Center | WB OP50; RRID: WB-STRAIN: OP50; NCBI TaxID: 637912; DC199 |
| Escherichia coli HB101 | Caenorhabditis Genetics Center | WormBase ID: WBStrain00041075; DC210 |
| Escherichia coli E22 | Shuman Lab, (University of Chicago) | E22, DC66 |
| Escherichia coli E2348/69 | Shuman Lab (University of Chicago) | E2348/69, DC67 |
| Klebsiella aerogenes | Shuman Lab (University of Chicago) | NCBI TaxID: 548; DC134; MG917 |
| Klebsiella pneumoniae KP35 | Prince Lab (Columbia University) | NCBI TaxID: 1460422; KP35/ST258; DC86 |
| Klebsiella pneumoniae KP182 | Shuman Lab (University of Chicago) | NCBI TaxID: 1352932; DC4 |
| Klebsiella pneumoniae KCI-1 | University of Chicago Clinical Microbiology | F50024/KCI-1, DC94 |
| Klebsiella pneumoniae KCI-2 | University of Chicago Clinical Microbiology | W70979/KCI-2, DC95 |
| Klebsiella pneumoniae KCI-3 | University of Chicago Clinical Microbiology | W71530/KCI-3, DC96 |
| Klebsiella pneumoniae KCI-4 | University of Chicago Clinical Microbiology | T32014/KCI-4, DC97 |
| Klebsiella pneumoniae KCI-5 | University of Chicago Clinical Microbiology | F53563/KCI-5, DC98 |
| Proteus mirabilis Hauser 1885 | ATCC | ATCC 35659 NCBI TaxID: 584; DC225 |
| Citrobacter freundii | NorthShore Research Institute | ACI-5, DC153 |
| Shigella sonnei Levine 1920 | ATCC | ATCC 9290, DC227 |
| Salmonella Typhimurium 12023 | ATCC | ATCC 14028; DC216 |
| Pseudomonas aeruginosa PAO1 | Shuman Lab (University of Chicago) | PAO1; DC3 |
| Acinetobacter baylyi ADP1 | ATCC | ATCC 33305; ADP1; DC167 |
| Acinetobacter baumannii AB5075 | Shuman Lab (University of Chicago) | AB5075; DC1 |
| Acinetobacter baumannii 17978 | Shuman Lab (University of Chicago) | 17978; DC2 |
| Escherichia coli W | Shanmugam Lab (University of Florida) | ATCC9637, E. coliW, DC222 |
| Escherichia coli LW393 | Shanmugam Lab (University of Florida) | E. coliBt, DC208 |
| Escherichia coli BEM9 | Shanmugam Lab (University of Florida) | E. coliΔ, DC223 |
| Escherichia coli DC228 (LW393-pMJG125.sfGFP) | This study | DC228 |
| Klebsiella oxytoca | BEI Resources | HM-624, MIT 10–5243 |
| Prevotella disiens | BEI Resources | HM-1171, DNF00882 |
| Prevotella corporis | BEI Resources | HM-1294, MJR7716 |
| Escherichia coli HT115 | Ahringer RNAi Library [84] | skn-1 (T19E7.2) RNAi |
| Escherichia coli HT115 | Ahringer RNAi Library [84] | daf-16 (R13H8.1) RNAi |
| Escherichia coli HT115 | Ahringer RNAi Library [84] | hsf-1 (Y53C10A.12) RNAi |
| Escherichia coli HT115 | Ahringer RNAi Library [84] | Empty vector, L4440 |
| Escherichia coli MG1655 | CGSC (no. 6300) | E. coli MG1655, F− λ− rph-1 |
| Escherichia coli BW25113 ΔcsgA | [85] | BW25113 ΔcsgA::kan |
| Escherichia coli BW25113 ΔcsgD | [85] | BW25113 ΔcsgD::kan |
| Escherichia coli MG1655 ΔcsgD | This study | E. coli MG1655 ΔcsgD::kan |
| Escherichia coli MG1655 ΔcsgA | This study | E. coli MG1655 ΔcsgA::kan |
| C. elegans Strains | ||
| C. elegans N2, Bristol | Caenorhabditis Genetics Center | N2 |
| C. elegans: AM738: rmIs297[vha-6p::q44::yfp; rol-6(su1006)] | Morimoto Lab (Northwestern University) | AM738, Q44::YFP |
| C. elegans: AM712 rm1s281[vha-6p::q33::yfp; rol-6(su1006)] | Morimoto Lab (Northwestern University) | AM712, Q33::YFP |
| C. elegans: AM446 rmIs223[C12C8.1p::gfp;rol-6(su1006)] | Morimoto Lab (Northwestern University) | AM446, hsp70p::GFP |
| C. elegans: AM140 rmIs132[unc-54p::q35::yfp] | Morimoto Lab (Northwestern University) | AM140, Q35::YFP |
| C. elegans AM141 rmls133[unc-54p::q40::yfp] | Morimoto Lab (Northwestern University) | AM141, Q40::YFP |
| C. elegans: AM101 rmIs110 [F25B3.3p::q40::yfp] | Morimoto Lab (Northwestern University) | AM101, Q40::YFP |
| C. elegans: DA597 phm-2(ad597) | Caenorhabditis Genetics Center | DA597 |
| C. elegans: LD1171 ldIs3[gcs-1p::gfp+rol-6(su1006)] | Caenorhabditis Genetics Center | LD1171, gcs-1p::GFP |
| C. elegans TJ356 zls356[daf-16p::daf-16a/b::gfp + rol-6(su1006)], | Caenorhabditis Genetics Center | TJ356, DAF-16::GFP |
| Chemicals & Commercial Assays | ||
| Levamisole | Fisher Scientific | Cat#ICN15522805 |
| Cholesterol | Fisher Scientific | Cat#ICN10138201 |
| Sodium butyrate | Fisher Scientific | Cat#A11079-22 |
| Thermo Scientific—AnaeroPack | Fisher Scientific | Cat#23-246-376 |
| Triton X-100, Molecular Biology Grade | Promega | Cat#H5141 |
| ProSignal Blotting Film | Prometheus | Cat#30-810L |
| Powdered nonfat milk | Research Products International | M17200-1000 |
| Tween-20 | Fisher BioReagents | Cat#C58H114O26 |
| Trans-Blot Turbo Midi-size Transfer Stacks | BioRad | Cat#1704273 |
| Trans-Blot Turbo Midi-size PDVF Membrane | BioRad | Cat#10026933 |
| Trans-Blot Turbo 5x transfer buffer | BioRad | Cat#10026938 |
| Criterion XT Precast Gel | BioRad | Cat#3450124 |
| XT 4x Sample Buffer | BioRad | Cat#1610791 |
| 20X Reducing Agent | BioRad | Cat#1610792 |
| XT MOPS | BioRad | Cat#1610788 |
| Clarity Western ECL | BioRad | |
| Isopropyl β-D-1-thiogalactopyranoside (IPTG) | GoldBio | Cat #12481C5 |
| Congo Red | Acros Organics | Cat#22962–0250 |
| Brilliant Blue G-250 | Fisher Biotech | CAS#6104-58-1 |
| Luria broth (Lennox) | Apex | Cat#11–125 |
| ProteoStat Protein aggregation assay | Enzo | Product #ENZ-51023-KP002 |
| Antibodies | ||
| Goat anti-mouse HRP secondary antibody | Thermo Scientific | Prod#31430 |
| Living Colors JL-8 primary monoclonal antibody | Takara Bio | Cat#632381 |
| Plasmids | ||
| pMJG125.sfGFP | This study | N/A |
| pBSK-sfGFP | Thomas Bernhardt (Harvard Medical School) | N/A |
| pKD46 | [86] | N/A |
| pXDC18.mCherry | [87] | N/A |
| P1vir | [88] | N/A |
| Software and Algorithms | ||
| GraphPad Prism v8.4.3 | GraphPad Software, Inc | https://www.graphpad.com |
| BioRender | BioRender | www.biorender.com |
| Oligonucleotides | ||
| ATAATAATTAATGTTATTGTCTCATGAGCGGATACA | IDT | MJG368 |
| ATCCATATGTTATAACCTCCTTAGAGC | IDT | MJG369 |
| ATTTATCATATGTCTAAAGGTGAAGAACTGTTCACCG | IDT | MJG1004 |
| TAAAATTCTAGATTATTTGTAGAGCTCATCCATGCCGTG | IDT | MJG1005 |
| CAGTATTTCGCAAGGTGCTTATG | IDT | csgA conf fwd |
| CCCTTGCTGGGTCGTATTAAA | IDT | csgA conf rev |
| GCAACATCTGTCAGTACTTCTGG | IDT | csgD conf fwd |
| CAGTATGGTCAGTTAGCAATCCC | IDT | csgD conf rev |
Caenorhabditis elegans maintenance and strains
Nematodes were maintained according to previously established protocols [89]. All experiments were performed at room temperature (RT, ~23 oC) unless described otherwise. All C. elegans strains used in this study can be found in Table 1.
Bacterial culture conditions
Culture conditions for bacteria used in endogenous and exogenous butyrate treatment can be found in “Endogenous Butyrate Production” and “Exogenous Butyrate Treatment”, respectively. DC228 was inoculated in the presence of 2 μg/mL gentamicin. Strains were plated onto Nematode Growth Media (NGM) +/- 3% arabinose and were left to dry overnight at RT prior to having worms plated on them. Curli-mutant bacteria and wild-type control (MG1655) were plated on NGM and kept at RT for 2 days prior to having worms plated on them. K. oxytoca (HM-624), Prevotella disiens (HM-1171), Prevotella corporis (HM-1294), and their E. coli OP50 control were grown in Reinforced Clostridial Broth (RCB). Unless otherwise indicated, all other bacteria were grown overnight in a 37°C incubator shaking at 220 revolutions per minute (RPM) in Lennox Luria Broth (LB), were seeded on NGM, left to dry overnight, and were kept at 4°C for another day prior to having worms plated on them.
Transduction and verification of curli (-) mutants
The bacterial strains used in this study are listed in Table 1. All solutions were prepared using ultrapure water, 18.2 MΩ/cm (Barnstead). Bacterial strains were grown in LB (5 g yeast extract, 10 g Bacto tryptone, and 10 g NaCl per liter of water), pH 7.4, at 37°C, with shaking (250 RPM), unless otherwise indicated. Transduction with P1vir was done to introduce gene deletions from E. coli donor strains from the Keio library [85]. Gene deletions were confirmed using PCR amplification of the target gene sequence using the primers listed in Table 1. The PCR products were visualized on an ethidium bromide-stained agarose gel.
Congo red assay
Curli production of each strain was determined by Congo Red plate assay. Congo Red plates were made using LB medium without salt (5 g yeast extract and 10 g Bacto tryptone per liter of ultrapure water), to which 50 μg/mL filter sterilized Congo Red and 1 μg/mL filter sterilized Brilliant Blue G250 were added after autoclaving. Overnight cultures were pelleted and washed once in LB without salt. Cells were then resuspended in LB without salt and cell density (measured by OD600) was normalized across all samples. Twenty-five microliters of each strain was spotted in a separate quadrant of Congo Red agar plate, which was then incubated at 26°C for 48 h. Plates were imaged after 48 h incubation at 4°C.
ProteoStat assay
All bacteria were grown in LB in a 37°C incubator shaking at 220 RPM overnight. Bacteria were diluted to OD600 of 0.5 and seeded in the center of a 96-well plate. Outer wells were filled with water and the plate was sealed with parafilm. For consistency, each bacterial strain was tested in a separate plate. Plates were incubated for ~44 h at 26°C. Following the incubation, LB was removed, and wells were washed twice with ddH20. Adhered biofilm was resuspended by pipetting with 100 μL 1x ProteoStat Assay Buffer. Two microliters of 1x ProteoStat Detection Reagent was dispensed into wells in a black, clear-bottom 96-well plate and 98 μL of resuspended biofilm was added and mixed. The plate was incubated for 15 minutes (min) at RT away from light. Following the incubation, fluorescence was read at excitation wavelength 550 nm and emission wavelength 600 nm, gain 100, in a Tecan Infinite 200 Pro plate reader. Assays were performed in triplicates.
Plasmid construction
pXDC18.mCherry was digested with EcoRI and AseI to remove lacIq and Ptac. A cassette harboring the araC gene and ParaBAD promoter from plasmid pKD46 was amplified with primers MJG368 and MJG369, digested with EcoRI & AseI and cloned into the pXDC18.mCherry backbone to yield plasmid pMJG125. The sfGFP cassette from plasmid pBSK-sfGFP was amplified with oligonucleotide primers MJG1004 and MJG1005. The resulting PCR product was digested with NdeI and XbaI and subsequently cloned into pMJG125 to yield pMJG125.sfGFP.
Generation of bacterial strains
The arabinose-inducible E. coli reporter strain (DC228) was generated by the electroporation of arabinose-inducible fluorescent reporter encoded on plasmid pMJG125.sfGFP into E. coli LW393 using the BioRad Gene Pulser Xcell electroporator according to the manufacturer’s instructions.
Exogenous butyrate treatment
All bacteria were grown aerobically in LB and seeded on NGM plates supplemented with indicated concentrations of sodium butyrate or controls lacking the compound. To ensure uniform coverage of the bacterial lawn, cultures were spread evenly across the plate. Plates were left to dry overnight at RT and were placed at 4°C one day prior to plating worms. Control experiments compared the aggregation profiles of worms fed heat-killed bacteria versus bacteria cultured in the presence of 100 mM sodium butyrate. Bacterial cultures were collected in 15 mL conical tubes and were killed by heating for 60 min at 70°C water bath. Bacterial viability was assessed by inoculating 10% of the culture in LB followed by overnight growth at 37°C shaking at 220 RPM. No growth confirmed bacteria to be dead. To bypass developmental delay that could result from feeding worms dead bacteria, animals were age synchronized, and cultured on plain NGM seeded with E. coli OP50 at 20°C for 48 h. After 48 h, animals were washed 6x with 5 mL M9 and transferred to NGM plates containing either heat-killed bacteria or 100 mM sodium butyrate with live bacteria. Worms were incubated at RT for three additional days and aggregates were quantified as described in “Aggregate Quantification”.
Endogenous butyrate production
Overnight cultures of E. coliBt and control strains, E. coliWT and E. coli OP50, were grown anaerobically. E. coli OP50 was grown anaerobically only in experiments where it was used as a control for E. coliBt. E. coliΔ was grown aerobically, because this strain is not capable of anaerobic growth. To ensure uniform coverage of the bacterial lawn, bacterial cultures were spread evenly across the plate. The NGM plates +/- 3% L-arabinose were seeded with the bacteria and dried overnight. E. coliBt and control strains, E. coliWT and E. coli OP50, were placed in air-tight containers with anaerobic gas packs, and all plates were stored at 4°C for one to two days prior to the experiment. Synchronized animals were plated and fed on the bacteria for 72 h (muscle polyQ and control) or 92 h (intestinal polyQ, neuronal polyQ and controls) prior to assessing aggregation or motility. Aggregates were quantified as described in “Aggregate Quantification”. The same method was followed when using 3% glucose rather than 3% L-arabinose.
Aggregate quantification
Worms were cultured and maintained as described in “Caenorhabditis elegans Maintenance and Strains”. Following age synchronization, animals were plated on NGM plates seeded with specific bacteria. Unless otherwise indicated, after 92 h (intestinal polyQ44) or 72 h (muscle polyQ35) of feeding, animals were picked from the plates and placed in 96-well plates containing M9/levamisole, and frozen at -20°C. Aggregates were quantified between 18–48 h post-freezing. PolyQ aggregates form rapidly, and their quantification is labor-intensive; therefore, the freezing step is essential to halt the formation of new aggregates during the quantification process. Worms expressing intestinal polyQ (Fig 1A and 1B) were imaged using a Nikon Eclipse 80i epifluorescence microscope equipped with a 4X Plan Apochromat objective (0.2 NA), metal halide lamp, and Ex450-490nm/Em500-550nm excitation filter. For all other aggregate quantification assays, worms were viewed using a Zeiss Axiovert S100 equipped with an Achrostigmat 10X Ph1 phase-contrast infinity microscope objective (0.25 NA), Chroma EGFP/FITC long-pass filter set (19002), and a mercury lamp.
Fecundity
Bacteria were grown as described in “Bacterial Culture Conditions”. Synchronized L1s were plated on NGM containing bacteria with indicated concentrations of sodium butyrate and incubated for 48 h. After 48 h, worms were replated every 24 h until they were 96 h old. The number of adult worms per plate was accounted for after each transfer. To count the sum of the eggs and L1 larvae laid by gravid adults between 48–72 and 72–96 h of age, entire plates were washed with M9, pipetting up and down meticulously to ensure removal of eggs and larvae. Plates were verified void of eggs and larvae by confirming their removal under the Zeiss Stemi 305 stereo microscope. Eggs and larvae were washed twice with 15 mL M9 and were resuspended in 2 mL M9. Ten or 100 μL samples of M9 were taken from the falcon tube and eggs + L1-stage worms were counted in replicates of six per run. The total number of offspring was calculated by factoring in the dilution and was expressed as the total number of progenies per worm.
RNAi knockdown
RNAi-mediated knockdown was carried out according to a previously published protocol [90]. Briefly, E. coli carrying DAF-16, SKN-1, and HSF-1 RNAi constructs and empty vector (L4440) were cultured overnight in a 37°C incubator shaking at 220 RPM in LB medium supplemented with 5 μg/mL tetracycline and 50 μg/mL ampicillin. Overnight cultures were induced for 4 h with 1 mM Isopropyl β-d-1-thiogalactopyranoside (IPTG) prior to seeding NGM plates supplemented with 12.5 μg/mL tetracycline, 100 μg/mL ampicillin, and 0.4 mM IPTG. Where specified, plates were also supplemented with 10, 25, 50, and 100 mM sodium butyrate. The seeded plates were allowed to dry overnight and were kept at 4°C for a maximum of two days. To bypass any effect on the development, L1 synchronized worms were placed on E. coli OP50 for 48 h at RT, washed three times with 15 mL M9, and plated on RNAi bacteria with or without butyrate. Aggregates were quantified after 48 h, as described under “Aggregate Quantification”. Knockdown was confirmed using fluorescent reporters. The induction of C12C8.1p::GFP (AM446) was used to assess HSF-1 knockdown. The expression of daf-16p::DAF-16::GFP (TJ365) was used to assess DAF-16 knockdown. The induction of gcs-1p::GFP (LD1171) was used to assess SKN-1 knockdown. Each reporter strain was synchronized, plated on E. coli OP50 for 48 h, washed, and replated on corresponding RNAi plates. After 24 h on RNAi plates, worms expressing C12C8.1p::GFP were heat-shocked for 1 h at 33°C and allowed to recover on RNAi plates for additional 24 h before imaging. The strain expressing daf-16p::DAF-16::GFP was imaged after 48 h on RNAi. To induce the expression of gcs-1p::GFP, the RNAi plates seeded with bacteria expressing SKN-1 RNAi were supplemented with 5 mM acrylamide. Worms were imaged after 48 h on RNAi/acrylamide plates. For each strain, 10 worms were picked from control L4440 and test RNAi plate and mounted in 2 mM levamisole on 3% agarose pads covered with a coverslip. Images were taken using a Zeiss Axio Observer 7 microscope as described under “Live Imaging”.
Colonizing the C. elegans intestine with polyculture
All strains were grown as described in “Bacterial Culture Conditions”, the cultures were normalized to OD600 of 1.0, and were plated in equal parts on NGM plates +/- 3% L-arabinose. Worms were cultured as described in “Caenorhabditis elegans Maintenance and Strains”, and aggregates were quantified as described in “Aggregate Quantification”.
Motility assays
All assays were performed at RT on age-synchronized animals. Unless otherwise indicated, worms were 72 h old at the time of body bend assays and 92 h old at the time of time-off-pick (TOP) assays. A body bend was defined as a change in the direction of the midline constituting approximately a 90° bend and a significant decrease in nose-to-tail distance. The rate was determined by placing animals in a drop of M9, allowing them to recover for 1 min, and counting the number of body bends in 30 s. TOP motility was defined by the time it took a worm to crawl completely off an eyebrow hair that was placed under its mid-section (Fig 2A). Worms were visualized using Zeiss Stemi 305 stereo microscopes.
PolyQ aggregation in the progeny
Worms were cultured and maintained as described in “Caenorhabditis elegans Maintenance and Strains”. Following synchronization, parental generation worms (P) were plated on NGM plates seeded with bacteria. After 72–78 h, animals (P) were bleached and the F1 progeny were age-synchronized, followed by plating on NGM plates containing control E. coli OP50 lawn. The F2 progeny were isolated by repeating the same method. F1 and F2 progeny were grown for 92 h at RT prior to aggregate quantification.
Confirmation of bacterial killing after embryo isolation and age-synchronization of progeny
Worms were cultured and maintained as described in “PolyQ Aggregation in the Progeny”. Following both the bleaching of the parental generations (P), and overnight synchronization of F1 L1s, 100 μL of M9 containing embryos and L1s, respectively, were spread across LB plates, put at 37°C overnight, and examined the following day for the presence or absence of bacterial growth.
Bacterial colonization assays
NGM plates were supplemented with indicated concentrations of sodium butyrate or 3% L-arabinose and seeded with bacteria. E. coli OP50 and P. aeruginosa PAO1 were grown under aerobic conditions and E. coliBt was grown anaerobically. The plates were then dried. Worms expressing intestinal polyQ44 were age-synchronized and plated on NGM seeded with the above bacteria, followed by a 3- and 4-day incubation. To remove any extracellular bacteria, 12 or more worms from the plate of interest were transferred onto a plain NGM plate for approximately 30 min. Ten worms were then transferred from each plain NGM plate into 1 mL of M9 supplemented with 80 μM levamisole and the tube was agitated to move worms around in solution. Once worms settled to the bottom, 950 μL of the supernatant was gently removed by pipetting. To remove any remaining external bacteria, worms were washed twice, each wash lasting 10 min, with M9 containing gentamicin (100 μg/mL) and levamisole (80 μM). Worms were then washed three times with M9/levamisole and the last wash was saved. To account for any external bacteria, the last wash was plated onto LB plates. Two hundred microliters of 1% triton X-100 was added to the worms sitting in 50 μL M9/levamisole. A handheld homogenizer (Bel-Art) was used to grind worms until no large remains were visible. The samples were then serially diluted with M9 and spotted onto LB plates. Plates were incubated at 37°C overnight and colonies were quantified the next day.
Butyrate dose response in axenic culture
Bacterial strains were cultured in LB. Strains were grown overnight in a 37°C incubator shaking at 220 RPM. Two-fold serial dilutions of sodium butyrate were prepared in LB in a 96-well plate. The concentrations ranged from 0 to 200 mM. Three microliters of culture adjusted to OD600 of 1.0 were added to 1.5 mL of LB media. An equal volume of this culture was added to the wells of the 96-well plate, which diluted the final concentrations of sodium butyrate by half (0 to 100 mM). The plate was incubated at 37°C in a Tecan Infinite 200 Pro plate reader for 24 h. Every 30 min, the plate was shaken for 5 s and OD600 readings were taken.
Bacterial viability on NGM supplemented with butyrate
All bacteria were grown aerobically in LB overnight in a 37°C incubator shaking at 220 RPM. The next day, cultures were adjusted to OD600 of 0.5 and 10 μL of a 10−1 dilution were spotted onto NGM plates supplemented with 0, 10, 25, 50, or 100 mM sodium butyrate. The plates were then placed in a 37°C incubator for 48 h. After incubation, the spots of bacteria were scraped with an inoculating loop, suspended in 1 mL LB, and 10 μL were spotted onto LB agar plates and placed in a 37°C incubator overnight. Images were taken the next day and viability was assessed by the presence or absence of bacterial lawns or distinguishable colonies.
Live imaging
Nematodes expressing intestinal polyQ33 or polyQ44 fed E. coli OP50 and P. aeruginosa PAO1 for four days were mounted in a drop of 1 mM levamisole on a 3% agarose pad and covered with a coverslip. GFP fluorescence and Nomarski images were taken using a Zeiss Axio Observer 7 microscope equipped with an Axiocam 503 mono camera, Solid-State Light Source Colibri 7, and a 5x Plan-Neofluar objective (0.16 NA). Pharyngeal mutant worms (DA597) were imaged using a 20x Plan-Neofluar objective (0.5 NA) and were processed using ZEN Tiles & Positions Module in Zeiss ZenPro software.
Arabinose availability assay
Bacteria expressing the arabinose reporter (E. coli DC228) were inoculated with 2 μg/mL gentamicin and were cultured as described in “Bacterial Culture Conditions”. Bacteria were seeded onto NGM plates +/- 3% L-arabinose and kept at RT for two days before worms were plated on them. Pharyngeal mutant animals (DA597) were age-synchronized and cultured for four days on E. coli bacteria (DC228) carrying an L-arabinose-inducible reporter (Ara-sfGFP) on NGM +/- 3% L-arabinose (positive and negative control, respectively). Animals used to test the availability of arabinose to the intestine were cultured for two days on E. coli bacteria carrying L-arabinose inducible reporter in the absence of L-arabinose followed by their transfer onto 3% L-arabinose plates containing E. coli OP50. Image acquisition is described in “Live Imaging”.
Western blotting and preparation
Worms were washed off NGM plates with M9, washed with 10 mL M9, and plated on unseeded NGM plates for ~30 min, or until M9 was dry to allow picking. To obtain soluble protein fractions, 20–50 (number was kept consistent between blots) worms were prepared and samples were processed as previously described [91]. To obtain the insoluble fraction of polyQ44::YFP aggregates, 50 worms were picked in 10 μL M9 with 1 mM phenylmethylsulfonyl fluoride (PMSF) and were flash-frozen three times in -80 oC ethanol bath. One glass bead was placed in each tube and tubes were vortexed for 1 min and placed on ice for 1 min for a total of five cycles. Tubes were spun down 1500 RPM for 10 min. Nine microliters of samples were transferred into a new tube and combined with 9 μL 2x XT loading buffer and reducing agent. Samples were heated at 98 oC for 7 min. Twelve microliters of samples were loaded into wells. Proteins were separated on 4–12% gradient sodium dodecyl sulfate-polyacrylamide gel (SDS-PAGE) gels by electrophoresis and transferred onto polyvinylidene difluoride (PVDF) membrane using Trans-Blot Turbo Transfer machine in the presence of Trans-Blot Turbo Transfer Buffer. Membranes were blocked with 5% nonfat milk in 0.1% PBS-Tween-20 (PBST) for 30 min and probed with Living Colors JL-8 monoclonal primary antibody (1:2000, soluble fraction; 1:800, insoluble fraction) for at least 24 h at 4 oC followed by (1:40,000) goat-anti-mouse horse-radish peroxidase (HRP) conjugated secondary antibody. Clarity Western ECL substrate was used for chemiluminescence. Image J (v1.52) was used to quantify western blot bands.
Quantification and statistical analysis
Data were considered statistically significant when p<0.05 was obtained by Student’s t-test or one-way ANOVA followed by multiple comparison Dunnett’s post-hoc test as indicated in figure legends. Asterisks denote corresponding statistical significance (*p<0.05, **p<0.01, ***p<0.001, ****p<0.0001). Where indicated, data are representative of the mean normalized to control strain E. coli OP50. Error bars represent standard error of the mean. Statistical analyses were performed using GraphPad Prism 8.4.3 software.
The numerical data used in all figures are included in S1 Data.
Supporting information
A) Western blot confirmation of antibody specificity. N2: no band; gcs-1p::GFP, polyQ33::YFP, polyQ44::YFP all show bands corresponding to their increasing molecular weight, respectively. B-E) Western blotting and image-J quantification of the soluble fraction of polyQ44::YFP in four-day-old C. elegans colonized with: B) E. coli OP50 and P. aeruginosa PAO1, C) E. coli OP50 with 0, 25, and 100 mM butyrate, D) P. aeruginosa PAO1 with 0, 25, 100 mM butyrate, E) F1 progeny from parental generations colonized with E. coli OP50 and P. aeruginosa PAO1. Band intensity is measured in arbitrary units (A.U). Data are representative of five independent experiments, B; four independent experiments, C and D; three independent experiments, E. Error bars represent SEM. Significance for C and D was calculated using one-way ANOVA followed by multiple comparison Dunnett’s post-hoc test. Significance for B and E was calculated using Student’s t-test (ns = not significant).
(TIF)
A-C) Insoluble fraction of polyQ44::YFP aggregates in four-day-old C. elegans expressing intestinal polyQ44::YFP colonized with: A) E. coli OP50 and P. aeruginosa PAO1, B) E. coli OP50 in the presence of 0, 25, 100 mM butyrate, and C) P. aeruginosa PAO1 in the presence of 0, 25, 100 mM butyrate. D) insoluble polyQ44 extracted from F1 progeny whose parents were colonized with E. coli OP50 or P. aeruginosa PAO1.
(TIF)
A) Age-dependent decline in motility assessed by increased TOP (left) and decreased number of body bends per 30 seconds (right) in muscle-specific polyQ35 (AM140), muscle-specific polyQ40 (AM141), and control (N2) worms. The data are represented as the average TOP seconds or average number of body bends per worm normalized to day 3. Each data point represents the average of two independent experiments with a total of 30 worms. B) The effect of bacteria on the motility at day 3 assessed by TOP (left) and body bends (right) in muscle-specific polyQ35, muscle-specific polyQ40, and N2 control worms. Data are represented as the average TOP or average number of body bends per worm normalized to animals fed E. coli OP50. Each bar is an average of two independent experiments with a total of 30 worms. Error bars represent SEM. Significance was calculated using one-way ANOVA followed by multiple comparison Dunnett’s post-hoc test (*p<0.05, ****p<0.0001).
(TIF)
Fluorescent images represent worms expressing either polyQ33 or polyQ44 that were fed control E. coli OP50 or test strain P. aeruginosa PAO1 for a period of four days. Scale bar = 200 μm.
(TIF)
Images represent LB agar plates seeded with samples of M9 media from a 3rd wash of embryos post-bleaching and from overnight incubation of embryos that were allowed to hatch into L1 stage. Parental strains were colonized either with E. coli OP50 or P. aeruginosa PAO1. No colony-forming units were detected, indicating that all samples were void of bacteria.
(TIF)
The growth of bacterial cultures was assessed in the presence of butyrate (0–100 mM) by measuring optical density at 600 nm (OD600).
(TIF)
A) A cartoon depicting the procedure. B) Growth of bacteria collected from butyrate NGM plates and spotted on LB agar.
(TIF)
Data are represented as the average number of aggregates per worm. Each bar is an average of three independent experiments with a total of 100 animals. Error bars represent SEM. Statistical significance was calculated using Student’s t-test (ns: non-significant, ***p<0.0005, ****p<0.0001).
(TIF)
Bacterial load was enumerated by extracting intestinal E. coli OP50 on day four from animals expressing intestinal polyQ44 reporter (AM738). Data are represented as the average bacterial load per C. elegans intestine normalized to the control (0 mM butyrate). Each bar represents three independent experiments with a total of 30 animals. Error bars represent SEM. Significance was calculated using one-way ANOVA followed by multiple comparison Dunnett’s post-hoc test (ns: non-significant, ****p<0.0001).
(TIF)
The effect of 0, 25, 100 mM butyrate on fecundity, days three and four, in: A) N2 worms colonized with E. coli OP50, B) polyQ44 colonized with E. coli OP50, C) polyQ44 colonized with P. aeruginosa PAO1. Each bar represents the average of three (B) and two (A, C) independent experiments with a total of 93 and 62 worms, respectively. Error bars represent SEM. Significance was calculated using one-way ANOVA followed by multiple comparison Dunnett’s post-hoc test (ns: non-significant, *p<0.05, ****p<0.0001).
(TIF)
Bacterial load was enumerated by extracting intestinal bacteria (E. coli OP50, P. aeruginosa PAO1, E. coliBt +/- L-arabinose) on days three and four from animals expressing intestinal polyQ44 reporter (AM738). Each bar represents an average of three independent experiments with a total of 30 animals. Error bars represent SEM.
(TIF)
Data are represented as the average number of aggregates of intestine-specific polyQ44 (AM738) per worm normalized to the control (no glucose). Each bar is an average of two independent experiments with a total of 40 animals. Error bars represent SEM. Statistical significance between each pair was calculated using Student’s t-test (**p<0.01, ***p<0.001, ****p<0.0001).
(TIF)
A) An overlay of Nomarski and GFP images of phm-2 worms cultured for four days on E. coli bacteria (DC228) carrying an L-arabinose-inducible reporter (Ara-sfGFP). B) Phm-2 worms cultured on E. coli bacteria in the presence of 3% L-arabinose. C) phm-2 worms cultured for two days on E. coli bacteria carrying L-arabinose inducible reporter in the absence of L-arabinose followed by a transfer onto 3% L-arabinose plates containing E. coli OP50. Scale bar = 200 μm.
(TIF)
Excel spreadsheet containing, in separate worksheets, the underlying numerical data for Figure panels: 1A, 1B, 1C, 1F, 1G, 2B, 2C, 3A, 3B, 3C, 3D, 3E, 3F, 3G, 3I, 4, 5, 6, 7, 8A, 8B, 9A, 9B, 9C, 9D, 9E, 10A, S1, S3A, S3B, S6, S8, S9, S10, S11, S12.
(XLSX)
Acknowledgments
We thank Dr. Howard Shuman (University of Chicago), Dr. Keelnathan Shanmugam (University of Florida), Alice Prince (Columbia University), the University of Chicago Clinical Microbiology Laboratory, and the NorthShore Research Institute for sharing bacterial strains. Also, we want to thank Dr. Richard Morimoto and Sue Fox (Northwestern University) for generously providing C. elegans strains. The other C. elegans strains were provided by the Caenorhabditis Genetics Center, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). We want to thank Dr. Brent Christner (University of Florida) and Dr. Mariola Edelmann (University of Florida) for allowing us to use their laboratory space and equipment and Dr. Keith Choe (University of Florida) for advice on experiments. Finally, we want to thank Dr. Cindy Voisine, Mark Gorelik, and members of the Czyz Lab for reviewing our manuscript and providing insightful feedback. Cartoon figures were created using BioRender.com. The following reagents were obtained through BEI Resources, NIAID, NIH as part of the Human Microbiome Project: Klebsiella oxytoca Strain MIT 10–5243, HM-624; Prevotella disiens Strain DNF00882, HM-1171; Prevotella corporis Strain MJR7716 HM-1294.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was supported in part by the Infectious Diseases Society of America and Start-up Funding provided by the Microbiology & Cell Science Department at the University of Florida Institute of Food and Agricultural Sciences to DMC. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Soto C. Unfolding the role of protein misfolding in neurodegenerative diseases. Nature Reviews Neuroscience. 2003;4:49–60. 10.1038/nrn1007 [DOI] [PubMed] [Google Scholar]
- 2.El-Hayek YH, Wiley RE, Khoury CP, Daya RP, Ballard C, Evans AR, et al. Tip of the Iceberg: Assessing the Global Socioeconomic Costs of Alzheimer’s Disease and Related Dementias and Strategic Implications for Stakeholders. Journal of Alzheimer’s disease: JAD. 2019;70(2):323–41. 10.3233/JAD-190426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Durães F, Pinto M, Sousa E. Old Drugs as New Treatments for Neurodegenerative Diseases. Pharmaceuticals (Basel). 2018;11(2):44. 10.3390/ph11020044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beard JD, Kamel F. Military service, deployments, and exposures in relation to amyotrophic lateral sclerosis etiology and survival. Epidemiol Rev. 2015;37(1):55–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bianchi VE, Herrera PF, Laura R. Effect of nutrition on neurodegenerative diseases. A systematic review. Nutritional neuroscience. 2019:1–25. 10.1080/1028415X.2019.1681088 [DOI] [PubMed] [Google Scholar]
- 6.Esch T, Stefano GB, Fricchione GL, Benson H. The role of stress in neurodegenerative diseases and mental disorders. Neuro endocrinology letters. 2002;23(3):199–208. [PubMed] [Google Scholar]
- 7.Mertsalmi TH, Pekkonen E, Scheperjans F. Antibiotic Exposure and Risk of Parkinson’s Disease in Finland: A Nationwide Case-Control Study. Movement Disorders. 2020;35(3):431–42. 10.1002/mds.27924 [DOI] [PubMed] [Google Scholar]
- 8.Sun J, Zhan Y, Mariosa D, Larsson H, Almqvist C, Ingre C, et al. Antibiotics use and risk of amyotrophic lateral sclerosis in Sweden. 2019;26(11):1355–61. 10.1111/ene.13986 [DOI] [PubMed] [Google Scholar]
- 9.Dominy SS, Lynch C, Ermini F, Benedyk M, Marczyk A, Konradi A, et al. Porphyromonas gingivalis in Alzheimer’s disease brains: Evidence for disease causation and treatment with small-molecule inhibitors. Science Advances. 2019;5(1):eaau3333. 10.1126/sciadv.aau3333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Honjo K, van Reekum R, Verhoeff NPLG. Alzheimer’s disease and infection: Do infectious agents contribute to progression of Alzheimer’s disease? Alzheimer’s & Dementia. 2009;5(4):348–60. 10.1016/j.jalz.2008.12.001 [DOI] [PubMed] [Google Scholar]
- 11.Dethlefsen L, Huse S, Sogin ML, Relman DA. The Pervasive Effects of an Antibiotic on the Human Gut Microbiota, as Revealed by Deep 16S rRNA Sequencing. PLOS Biology. 2008;6(11):e280. 10.1371/journal.pbio.0060280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hemmings SMJ, Malan-Müller S, van den Heuvel LL, Demmitt BA, Stanislawski MA, Smith DG, et al. The Microbiome in Posttraumatic Stress Disorder and Trauma-Exposed Controls: An Exploratory Study. Psychosom Med. 2017;79(8):936–46. 10.1097/PSY.0000000000000512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin Y, Wu S, Zeng Z, Fu Z. Effects of environmental pollutants on gut microbiota. Environmental Pollution. 2017;222:1–9. 10.1016/j.envpol.2016.11.045 [DOI] [PubMed] [Google Scholar]
- 14.Glowacki RWP, Martens EC. In sickness and health: Effects of gut microbial metabolites on human physiology. PLoS Pathog. 2020;16(4):e1008370–e. 10.1371/journal.ppat.1008370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kho ZY, Lal SK. The Human Gut Microbiome—A Potential Controller of Wellness and Disease. Frontiers in microbiology. 2018;9:1835–. 10.3389/fmicb.2018.01835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toor D, Wsson MK, Kumar P, Karthikeyan G, Kaushik NK, Goel C, et al. Dysbiosis Disrupts Gut Immune Homeostasis and Promotes Gastric Diseases. International journal of molecular sciences. 2019;20(10). 10.3390/ijms20102432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Francino MP. Antibiotics and the Human Gut Microbiome: Dysbioses and Accumulation of Resistances. Frontiers in microbiology. 2016;6:1543–. 10.3389/fmicb.2015.01543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerhardt S, Mohajeri MH. Changes of Colonic Bacterial Composition in Parkinson’s Disease and Other Neurodegenerative Diseases. 2018;10(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mazzini L, Mogna L, De Marchi F, Amoruso A, Pane M, Aloisio I, et al. Potential Role of Gut Microbiota in ALS Pathogenesis and Possible Novel Therapeutic Strategies. Journal of Clinical Gastroenterology. 2018;52:S68–S70. 10.1097/MCG.0000000000001042 [DOI] [PubMed] [Google Scholar]
- 20.Scheperjans F, Aho V, Pereira PA, Koskinen K, Paulin L, Pekkonen E, et al. Gut microbiota are related to Parkinson’s disease and clinical phenotype. Mov Disord. 2015;30(3):350–8. 10.1002/mds.26069 [DOI] [PubMed] [Google Scholar]
- 21.Zhu S, Jiang Y, Xu K, Cui M, Ye W, Zhao G, et al. The progress of gut microbiome research related to brain disorders. Journal of Neuroinflammation. 2020;17(1):25. 10.1186/s12974-020-1705-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burberry A, Wells MF, Limone F, Couto A, Smith KS, Keaney J, et al. C9orf72 suppresses systemic and neural inflammation induced by gut bacteria. Nature. 2020. 10.1038/s41586-020-2288-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mohri-Shiomi A, Garsin DA. Insulin Signaling and the Heat Shock Response Modulate Protein Homeostasis in the Caenorhabditis elegans Intestine during Infection. Journal of Biological Chemistry. 2008;283(1):194–201. 10.1074/jbc.M707956200 [DOI] [PubMed] [Google Scholar]
- 24.Barnhart MM, Chapman MR. Curli biogenesis and function. Annual review of microbiology. 2006;60:131–47. 10.1146/annurev.micro.60.080805.142106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen D, Coleman J, Chan E, Nicholson TP, Dai L, Sheppard PW, et al. Novel cell- and tissue-based assays for detecting misfolded and aggregated protein accumulation within aggresomes and inclusion bodies. Cell biochemistry and biophysics. 2011;60(3):173–85. 10.1007/s12013-010-9138-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.E L, Zhou T, Koh S, Chuang M, Sharma R, Pujol N, et al. An Antimicrobial Peptide and Its Neuronal Receptor Regulate Dendrite Degeneration in Aging and Infection. Neuron. 2018;97(1):125–38.e5. 10.1016/j.neuron.2017.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chikka MR, Anbalagan C, Dvorak K, Dombeck K, Prahlad V. The Mitochondria-Regulated Immune Pathway Activated in the C. elegans Intestine Is Neuroprotective. Cell reports. 2016;16(9):2399–414. 10.1016/j.celrep.2016.07.077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sandhof CA, Hoppe SO, Druffel-Augustin S, Gallrein C, Kirstein J, Voisine C, et al. Reducing INS-IGF1 signaling protects against non-cell autonomous vesicle rupture caused by SNCA spreading. Autophagy. 2020;16(5):878–99. 10.1080/15548627.2019.1643657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morley JF, Brignull HR, Weyers JJ, Morimoto RI. The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans. Proceedings of the National Academy of Sciences. 2002;99(16):10417–22. 10.1073/pnas.152161099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brignull HR, Moore FE, Tang SJ, Morimoto RI. Polyglutamine Proteins at the Pathogenic Threshold Display Neuron-Specific Aggregation in a Pan-Neuronal Caenorhabditis elegans Model. The Journal of Neuroscience. 2006;26(29):7597–606. 10.1523/JNEUROSCI.0990-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Byndloss MX, Olsan EE, Rivera-Chávez F, Tiffany CR, Cevallos SA, Lokken KL, et al. Microbiota-activated PPAR-γ signaling inhibits dysbiotic Enterobacteriaceae expansion. Science. 2017;357(6351):570–5. 10.1126/science.aam9949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y-g, Wu S, Yi J, Xia Y, Jin D, Zhou J, et al. Target Intestinal Microbiota to Alleviate Disease Progression in Amyotrophic Lateral Sclerosis. Clinical Therapeutics. 2017;39(2):322–36. 10.1016/j.clinthera.2016.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haenen D, Zhang J, Souza da Silva C, Bosch G, van der Meer IM, van Arkel J, et al. A diet high in resistant starch modulates microbiota composition, SCFA concentrations, and gene expression in pig intestine. The Journal of nutrition. 2013;143(3):274–83. 10.3945/jn.112.169672 [DOI] [PubMed] [Google Scholar]
- 34.Sorbara MT, Dubin K, Littmann ER, Moody TU, Fontana E. Inhibiting antibiotic-resistant Enterobacteriaceae by microbiota-mediated intracellular acidification. 2019;216(1):84–98. 10.1084/jem.20181639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cummings JH, Pomare EW, Branch WJ, Naylor CP, Macfarlane GT. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut. 1987;28(10):1221–7. 10.1136/gut.28.10.1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. Review article: the role of butyrate on colonic function. Alimentary pharmacology & therapeutics. 2008;27(2):104–19. 10.1111/j.1365-2036.2007.03562.x [DOI] [PubMed] [Google Scholar]
- 37.Labbadia J, Morimoto RI. Repression of the Heat Shock Response Is a Programmed Event at the Onset of Reproduction. Molecular cell. 2015;59(4):639–50. 10.1016/j.molcel.2015.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blackwell TK, Steinbaugh MJ, Hourihan JM, Ewald CY, Isik M. SKN-1/Nrf, stress responses, and aging in Caenorhabditis elegans. Free radical biology & medicine. 2015;88(Pt B):290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fujimoto M, Takaki E, Hayashi T, Kitaura Y, Tanaka Y, Inouye S, et al. Active HSF1 significantly suppresses polyglutamine aggregate formation in cellular and mouse models. The Journal of biological chemistry. 2005;280(41):34908–16. 10.1074/jbc.M506288200 [DOI] [PubMed] [Google Scholar]
- 40.Hesp K, Smant G, Kammenga JE. Caenorhabditis elegans DAF-16/FOXO transcription factor and its mammalian homologs associate with age-related disease. Experimental gerontology. 2015;72:1–7. 10.1016/j.exger.2015.09.006 [DOI] [PubMed] [Google Scholar]
- 41.Nollen EAA, Garcia SM, van Haaften G, Kim S, Chavez A, Morimoto RI, et al. Genome-wide RNA interference screen identifies previously undescribed regulators of polyglutamine aggregation. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(17):6403–8. 10.1073/pnas.0307697101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Louis P, Flint HJ. Formation of propionate and butyrate by the human colonic microbiota. Environmental Microbiology. 2017;19(1):29–41. 10.1111/1462-2920.13589 [DOI] [PubMed] [Google Scholar]
- 43.Wang L, Chauliac D, Moritz BE, Zhang G, Ingram LO, Shanmugam KT. Metabolic engineering of Escherichia coli for the production of butyric acid at high titer and productivity. Biotechnology for Biofuels. 2019;12(1):62. 10.1186/s13068-019-1408-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goya ME, Xue F, Sampedro-Torres-Quevedo C, Arnaouteli S, Riquelme-Dominguez L, Romanowski A, et al. Probiotic Bacillus subtilis Protects against α-Synuclein Aggregation in C. elegans. Cell reports. 2020;30(2):367–80.e7. 10.1016/j.celrep.2019.12.078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haran JP, Bhattarai SK, Foley SE, Dutta P, Ward DV, Bucci V, et al. Alzheimer’s Disease Microbiome Is Associated with Dysregulation of the Anti-Inflammatory P-Glycoprotein Pathway. mBio. 2019;10(3):e00632–19. 10.1128/mBio.00632-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steenblock DA IT, Antonio ASS, Wardaningsih E, Azizi MJ Amyotrophic Lateral Sclerosis (ALS) Linked to Intestinal Microbiota Dysbiosis & Systemic Microbial Infection in Human Patients: A Cross-Sectional Clinical Study. Int J Neurodegener Dis. 2018;1(1). 10.23937/IJND-2017/1710002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rice LB. Federal Funding for the Study of Antimicrobial Resistance in Nosocomial Pathogens: No ESKAPE. The Journal of infectious diseases. 2008;197(8):1079–81. 10.1086/533452 [DOI] [PubMed] [Google Scholar]
- 48.CDC. Antibiotic Resistance Threats in the United States, 2019. US Department of Health and Human Services. 2019. [Google Scholar]
- 49.Zou Y, Xue W, Luo G, Deng Z, Qin P, Guo R, et al. 1,520 reference genomes from cultivated human gut bacteria enable functional microbiome analyses. Nature biotechnology. 2019;37(2):179–85. 10.1038/s41587-018-0008-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Estepa V, Rojo-Bezares B, Torres C, Sáenz Y. Faecal carriage of Pseudomonas aeruginosa in healthy humans: antimicrobial susceptibility and global genetic lineages. FEMS microbiology ecology. 2014;89(1):15–9. 10.1111/1574-6941.12301 [DOI] [PubMed] [Google Scholar]
- 51.Chen C-Y, Chen Y-H, Lu P-L, Lin W-R, Chen T-C, Lin C-Y. Proteus mirabilis urinary tract infection and bacteremia: Risk factors, clinical presentation, and outcomes. Journal of Microbiology, Immunology and Infection. 2012;45(3):228–36. [DOI] [PubMed] [Google Scholar]
- 52.Su CM, Kung CT, Chen FC, Cheng HH, Hsiao SY, Lai YR, et al. Manifestations and Outcomes of Patients with Parkinson’s Disease and Serious Infection in the Emergency Department. BioMed research international. 2018;2018:6014896. 10.1155/2018/6014896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Choi JG, Kim N, Ju IG, Eo H, Lim S-M, Jang S-E, et al. Oral administration of Proteus mirabilis damages dopaminergic neurons and motor functions in mice. Scientific reports. 2018;8(1):1275. 10.1038/s41598-018-19646-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, et al. Enterobacteriaceae Act in Concert with the Gut Microbiota to Induce Spontaneous and Maternally Transmitted Colitis. Cell Host & Microbe. 2010;8(3):292–300. 10.1016/j.chom.2010.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yousefi-Mashouf R, Eley A, Duerden BI. Characterisation of non-pigmented species of the genus Prevotellaby polyacrylamide gel electrophoresis. Journal of medical microbiology. 1992;37(2):83–90. 10.1099/00222615-37-2-83 [DOI] [PubMed] [Google Scholar]
- 56.Harding GK, Sutter VL, Finegold SM, Bricknell KS. Characterization of bacteroides melaninogenicus. Journal of clinical microbiology. 1976;4(4):354–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu L, Liu W, Alkhouri R, Baker RD, Bard JE, Quigley EM, et al. Structural changes in the gut microbiome of constipated patients. Physiological genomics. 2014;46(18):679–86. 10.1152/physiolgenomics.00082.2014 [DOI] [PubMed] [Google Scholar]
- 58.Stocchi F, Torti M. Chapter Twenty-Seven—Constipation in Parkinson’s Disease. In: Chaudhuri KR, Titova N, editors. International review of neurobiology. 134: Academic Press; 2017. p. 811–26. [DOI] [PubMed] [Google Scholar]
- 59.Jost WH, Schimrigk K. Constipation in Parkinson’s disease. Klinische Wochenschrift. 1991;69(20):906–9. 10.1007/BF01798536 [DOI] [PubMed] [Google Scholar]
- 60.Jin M, Li J, Liu F, Lyu N, Wang K, Wang L, et al. Analysis of the Gut Microflora in Patients With Parkinson’s Disease. Frontiers in Neuroscience. 2019;13(1184). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shen L, Liu L, Ji HF. Alzheimer’s Disease Histological and Behavioral Manifestations in Transgenic Mice Correlate with Specific Gut Microbiome State. Journal of Alzheimer’s disease: JAD. 2017;56(1):385–90. 10.3233/JAD-160884 [DOI] [PubMed] [Google Scholar]
- 62.Wu Q, Cao X, Yan D, Wang D, Aballay A. Genetic Screen Reveals Link between the Maternal Effect Sterile Gene mes-1 and Pseudomonas aeruginosa-induced Neurodegeneration in Caenorhabditis elegans. Journal of Biological Chemistry. 2015;290(49):29231–9. 10.1074/jbc.M115.674259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Singh J, Aballay A. Intestinal infection regulates behavior and learning via neuroendocrine signaling. Elife. 2019;8:e50033. 10.7554/eLife.50033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kumar S, Egan BM, Kocsisova Z, Schneider DL, Murphy JT, Diwan A, et al. Lifespan Extension in C. elegans Caused by Bacterial Colonization of the Intestine and Subsequent Activation of an Innate Immune Response. Developmental cell. 2019;49(1):100–17.e6. 10.1016/j.devcel.2019.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Steinkraus KA, Smith ED, Davis C, Carr D, Pendergrass WR, Sutphin GL, et al. Dietary restriction suppresses proteotoxicity and enhances longevity by an hsf-1-dependent mechanism in Caenorhabditis elegans. Aging Cell. 2008;7(3):394–404. 10.1111/j.1474-9726.2008.00385.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kumsta C, Thamsen M, Jakob U. Effects of oxidative stress on behavior, physiology, and the redox thiol proteome of Caenorhabditis elegans. Antioxidants & redox signaling. 2011;14(6):1023–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nussbaum-Krammer CI, Morimoto RI. Caenorhabditis elegans as a model system for studying non-cell-autonomous mechanisms in protein-misfolding diseases. Disease models & mechanisms. 2014;7(1):31–9. 10.1242/dmm.013011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fehmida B, Muhammad Y, Sayed SS, Esam IA, Mohammed HA-Q, Adel MA, et al. Link Between Chronic Bacterial Inflammation and Alzheimer Disease. CNS & Neurological Disorders—Drug Targets. 2014;13(7):1140–7. 10.2174/1871527313666140917115741 [DOI] [PubMed] [Google Scholar]
- 69.Sampson TR, Challis C, Jain N, Moiseyenko A, Ladinsky MS, Shastri GG, et al. A gut bacterial amyloid promotes α-synuclein aggregation and motor impairment in mice. Elife. 2020;9:e53111. 10.7554/eLife.53111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gidalevitz T, Ben-Zvi A, Ho KH, Brignull HR, Morimoto RI. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science. 2006;311(5766):1471–4. 10.1126/science.1124514 [DOI] [PubMed] [Google Scholar]
- 71.Pace MC, Xu G, Fromholt S, Howard J, Giasson BI, Lewis J, et al. Differential induction of mutant SOD1 misfolding and aggregation by tau and α-synuclein pathology. Molecular Neurodegeneration. 2018;13(1):23. 10.1186/s13024-018-0253-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Javed I, Zhang Z, Adamcik J, Andrikopoulos N, Li Y, Otzen DE, et al. Accelerated Amyloid Beta Pathogenesis by Bacterial Amyloid FapC. Advanced Science. 2020;7(18):2001299. 10.1002/advs.202001299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hartman K, Brender JR, Monde K, Ono A, Evans Margery L, Popovych N, et al. Bacterial curli protein promotes the conversion of PAP248-286 into the amyloid SEVI: cross-seeding of dissimilar amyloid sequences. PeerJ. 2013;1:e5. 10.7717/peerj.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen SG, Stribinskis V, Rane MJ, Demuth DR, Gozal E, Roberts AM, et al. Exposure to the Functional Bacterial Amyloid Protein Curli Enhances Alpha-Synuclein Aggregation in Aged Fischer 344 Rats and Caenorhabditis elegans. Scientific reports. 2016;6(1):34477. 10.1038/srep34477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ogasawara H, Yamamoto K, Ishihama A. Role of the biofilm master regulator CsgD in cross-regulation between biofilm formation and flagellar synthesis. Journal of bacteriology. 2011;193(10):2587–97. 10.1128/JB.01468-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lazar V, Ditu L-M, Pircalabioru GG, Gheorghe I, Curutiu C, Holban AM, et al. Aspects of Gut Microbiota and Immune System Interactions in Infectious Diseases, Immunopathology, and Cancer. Frontiers in Immunology. 2018;9(1830). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Uttara B, Singh AV, Zamboni P, Mahajan RT. Oxidative stress and neurodegenerative diseases: a review of upstream and downstream antioxidant therapeutic options. Curr Neuropharmacol. 2009;7(1):65–74. 10.2174/157015909787602823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sun B, Jia Y, Yang S, Zhao N, Hu Y, Hong J, et al. Sodium butyrate protects against high-fat diet-induced oxidative stress in rat liver by promoting expression of nuclear factor E2-related factor 2. British Journal of Nutrition. 2019;122(4):400–10. 10.1017/S0007114519001399 [DOI] [PubMed] [Google Scholar]
- 79.Kim DH, Park MH, Ha S, Bang EJ, Lee Y, Lee AK, et al. Anti-inflammatory action of β-hydroxybutyrate via modulation of PGC-1α and FoxO1, mimicking calorie restriction. Aging (Albany NY). 2019;11(4):1283–304. 10.18632/aging.101838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Edwards C, Canfield J, Copes N, Rehan M, Lipps D, Bradshaw PC. D-beta-hydroxybutyrate extends lifespan in C. elegans. Aging (Albany NY). 2014;6(8):621–44. 10.18632/aging.100683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fawcett EM, Hoyt JM, Johnson JK, Miller DL. Hypoxia disrupts proteostasis in Caenorhabditis elegans. Aging Cell. 2015;14(1):92–101. 10.1111/acel.12301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gantois I, Ducatelle R, Pasmans F, Haesebrouck F, Hautefort I, Thompson A, et al. Butyrate Specifically Down-Regulates Salmonella Pathogenicity Island 1 Gene Expression. Applied and Environmental Microbiology. 2006;72(1):946–9. 10.1128/AEM.72.1.946-949.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tobe T, Nakanishi N, Sugimoto N. Activation of motility by sensing short-chain fatty acids via two steps in a flagellar gene regulatory cascade in enterohemorrhagic Escherichia coli. Infection and immunity. 2011;79(3):1016–24. 10.1128/IAI.00927-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421(6920):231–7. 10.1038/nature01278 [DOI] [PubMed] [Google Scholar]
- 85.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006.0008–2006.0008. 10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proceedings of the National Academy of Sciences. 2000;97(12):6640–5. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Charpentier X, Faucher SP, Kalachikov S, Shuman HA. Loss of RNase R induces competence development in Legionella pneumophila. Journal of bacteriology. 2008;190(24):8126–36. 10.1128/JB.01035-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Singer M, Baker TA, Schnitzler G, Deischel SM, Goel M, Dove W, et al. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989;53(1):1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77(1):71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Guisbert E, Czyz DM, Richter K, McMullen PD, Morimoto RI. Identification of a tissue-selective heat shock response regulatory network. PLoS genetics. 2013;9(4):e1003466. 10.1371/journal.pgen.1003466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Labbadia J, Brielmann RM, Neto MF, Lin Y-F, Haynes CM, Morimoto RI. Mitochondrial Stress Restores the Heat Shock Response and Prevents Proteostasis Collapse during Aging. Cell reports. 2017;21(6):1481–94. 10.1016/j.celrep.2017.10.038 [DOI] [PMC free article] [PubMed] [Google Scholar]