Abstract
Background
Mosquitoes host and transmit numerous arthropod-borne viruses (arboviruses) that cause disease in both humans and animals. Effective surveillance of virome profiles in mosquitoes is vital to the prevention and control of mosquito-borne diseases in northwestern China, where epidemics occur frequently.
Methods
Mosquitoes were collected in the Shaanxi-Gansu-Ningxia region (Shaanxi Province, Gansu Province, and Ningxia Hui Autonomous Region) of China from June to August 2019. Morphological methods were used for taxonomic identification of mosquito species. High-throughput sequencing and metagenomic analysis were used to characterize mosquito viromes.
Results
A total of 22,959 mosquitoes were collected, including Culex pipiens (45.7%), Culex tritaeniorhynchus (40.6%), Anopheles sinensis (8.4%), Aedes (5.2%), and Armigeres subalbatus (0.1%). In total, 3,014,183 (0.95% of clean reads) viral sequences were identified and assigned to 116 viral species (including pathogens such as Japanese encephalitis virus and Getah virus) in 31 viral families, including Flaviviridae, Togaviridae, Phasmaviridae, Phenuiviridae, and some unclassified viruses. Mosquitoes collected in July (86 species in 26 families) showed greater viral diversity than those from June and August. Culex pipiens (69 species in 25 families) and Culex tritaeniorhynchus (73 species in 24 families) carried more viral species than Anopheles sinensis (50 species in 19 families) or Aedes (38 species in 20 families) mosquitoes.
Conclusion
Viral diversity and abundance were affected by mosquito species and collection time. The present study elucidates the virome compositions of various mosquito species in northwestern China, improving the understanding of virus transmission dynamics for comparison with those of disease outbreaks.
Author summary
Most traditional viral research methods are limited to screening for a few types of viruses with low throughput. Here, we relied on metagenomic sequencing, which is a broadly non-specific screen for any virus, for monitoring of mosquito-borne viruses in the Shaanxi-Gansu-Ningxia region of China in 2019. The baseline of viral abundance and diversity in local Culex pipiens, Culex tritaeniorhynchus, Anopheles sinensis, and Aedes mosquitoes were investigated, with 116 viral species identified in 31 viral families, including Flaviviridae, Togaviridae, Phasmaviridae, Phenuiviridae, and some unclassified viruses. This study provides a baseline survey of the viromes of mosquitoes in the study area, highlighting the necessity of strengthened monitoring of mosquito-borne pathogens.
Introduction
Arthropod-borne viruses (arboviruses) are viruses that are maintained in natural reservoirs and transmitted by blood-sucking arthropods between susceptible vertebrate hosts. More than 500 arboviruses have been identified around the world, of which approximately 130 are capable of causing disease in human and animals [1]. In recent years, researchers in China have successfully isolated 29 viral species belonging to 7 families and 13 genera [2]. Mosquitoes are major vectors that transmit numerous arboviruses to various hosts, causing disease in both humans and animals [3, 4]. For example, the Aedes aegypti is the primary vector for Dengue virus, Zika virus, and Chikungunya virus, and Culex is a primary vector for Japanese Encephalitis Virus (JEV) and West Nile Virus (WNV) [5, 6]. Mosquitoes thus pose an immense threat to public health due to their role in transmitting viral diseases.
Viromes are structurally and functionally diverse, with differences among habitats in hosts and environments; prior to the development of high-throughput sequencing, little was known about the viromes of invertebrates and vertebrates [7–9]. Viral profiling through metagenomics is a relatively new technique that takes advantage of high-throughput sequencing and is broadly non-specific to identify any viruses present in a given sample [10–12]. Metagenomics methods have provided new insights into the substantial complexity and diversity of viruses carried by mosquitoes, which has significance for dynamic surveillance of pathogens and the response to emerging and remerging infectious diseases [13–15].
The Shaanxi-Gansu-Ningxia geographic region (Shaanxi Province, Gansu Province, and Ningxia Hui Autonomous Region) is located in northwestern China. This area mainly contains hills and gully areas of the Loess Plateau, a typical agricultural and pastoral zone of northern China. In recent years, accompanying the rapid development of the region’s economy and livestock husbandry, frequent population movement, and accompanying climate and environmental changes, the risk of mosquito-borne viral disease transmission in the area has increased [16–19]. The Shaanxi-Gansu-Ningxia region is traditionally a low epidemic area for JEV, but endured a significant outbreak of this disease in 2018, with approximately 833 cases reported [20, 21]. Characterization of the baseline mosquito virome in the Shaanxi-Gansu-Ningxia region is of great importance for assessments of mosquito-borne disease transmission risk and vector competence, and will provide scientific data for the prevention and control of arbovirus disease transmission [22–24]. However, few data have been reported from this region. In the present study, we investigated the virome compositions of various mosquito species in the Shaanxi-Gansu-Ningxia region using a viral metagenomics approach to provide additional insights into the viruses circulating in this region.
Materials and methods
Mosquito collection
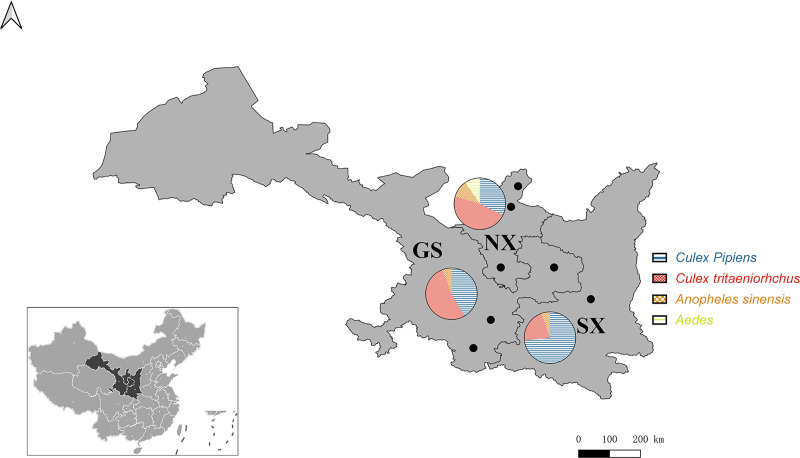
Mosquitoes were collected between June to August of 2019 from seven cities in the Shaanxi-Gansu-Ningxia region (Fig 1). Farms, pigpens, sheep pens, and residential areas were sampled. A trapping lamp was placed outdoors at 18:00–20:00 and retrieved in the early morning of the next day. On an ice bath, male mosquitoes were removed and the species of female mosquitoes were determined based on morphological characteristics. The female mosquitoes were pooled by species, time of collection, and location (30–50 mosquitoes/pool), and then stored at −80°C for subsequent use. The collection sites distribution of mosquito and location of the Shaanxi-Gansu-Ningxia region were presented using ArcGIS (v.10.0; ESRI, Redlands, CA, USA).
Fig 1. Map of mosquito collection sites.
The Shanxi-Gansu-Ningxia region is labeled in light gray and cities where the collection sites were located are indicated with black dots. The pie charts on the map show the composition of mosquito species collected in each province, with each color representing a mosquito species. The location of the Shaanxi-Gansu-Ningxia region is shown in dark gray on the map of China in the lower left of corner (GS: Gansu Province, SX: Shaanxi Province, NX: Ningxia Hui Autonomous Region). The base layer of this modified map originated from National Earth System Science Data Center, National Science & Technology Infrastructure of China (http://www.geodata.cn).
Nucleic acid extraction and high-throughput sequencing
Each mosquito pool was homogenized on an ice bath. Total RNA was extracted with the QIAamp Viral RNA Minikit (QIAGEN) according to the manufacturer’s instructions. cDNA was synthesized using Superscript III reverse transcriptase (ThermoFisher) and random primers, followed by the addition of Klenow enzyme (New England BioLabs) to complete second-strand cDNA synthesis. The REPLI-g Mini Kit (QIAGEN) was used for pre-enrichment via multiple displacement amplification. The NexteraXT DNA Library Preparation Kit (Illumina) was used to construct sequencing libraries. After each library was quantified on the Qubit 4.0 fluorometer (ThermoFisher), sequencing was performed on the Illumina Novaseq (2×150 bp) or MiSeq (2×300 bp) high-throughput sequencing platform. The amount of sequencing data was adjusted according to the number of mosquitoes in each pool to balance the data available for all pools.
Metagenomics data analysis
The raw paired-end reads obtained from high-throughput sequencing were subjected to quality control using the Trimmomatic (v0.39), BBDUK (v38.79), and Fastp (v0.20.0) tools. Adapters, low-complexity reads, and low-quality bases (with scores less than 15) were removed to generate clean data. Then, the clean reads were aligned to genomes of Culex quinquefasciatus (CulPip1.0, RefSeq: GCF_000209185.1), Anopheles sinensis (AS2, GenBank: GCA_000441895.2), and Aedes albopictus (Aalbo_primary.1, RefSeq: GCF_006496715.1), respectively. All mosquito genomes were available at Genomes–NCBI Datasets (https://www.ncbi.nlm.nih.gov/datasets/genomes/). Unmapped reads were extracted and then paired-end aligned to the NCBI viral whole-genome database (https://www.ncbi.nlm.nih.gov/labs/virus/vssi/) using Bowtie 2 (v2.3.5) in end-to-end mode (with the parameter ‘very-sensitive’). Mapped reads were extracted and duplicate reads were removed using SAMtools (v1.9) to obtain unique virus-related reads. The Blastn alignment function in NCBI-Blast+ (v2.9.0) was used to align the unique reads with the local virus reference genome database in ‘best hit only’ mode. Alignment results with E-values lower than 1e-10 and identity higher than 90% were retained for the next step. Finally, MEGAN6 (v6.20) software was used for metagenomic annotation based on a genomic DNA taxonomic database (Jul2020) with the default lowest common ancestor parameter. Annotations with less than five reads to any reference genome were removed from further analysis. Data visualization was conducted using Graphpad, VIP [25] and TBtools [26]. Sequencing coverage and depth were analyzed with SAMtools. The length coverage percentage of each viral species was calculated using the following formula: total length of read covering the reference / number of bases in the reference. The consensus sequence identity with the reference genome was determined using Blastn.
Results
Mosquito collection in the Shaanxi-Gansu-Nanjing region from June to August 2019
In total, 22,959 mosquitoes were collected during the peak mosquito activity period from June to August in the Shaanxi-Gansu-Ningxia region of China (Fig 1 and S1 Table). Among them, Culex pipiens (Cx.p) accounted for 45.7% (10,498/22,959) of all mosquitoes, and was the most abundant mosquito species. Culex tritaeniorhynchus (Cx.t), Anopheles sinensis (An.), Aedes (Ae.), and Armigeres subalbatus accounted for 40.6% (9322/22,959), 8.4% (1940/22,959), 5.2% (1184/22,959), and 0.1% (15/22,959) of the total mosquitoes, respectively. Armigeres subalbatus were removed from further analysis, as the number of mosquitoes was insufficient for library construction.
Virome profiles of mosquitoes
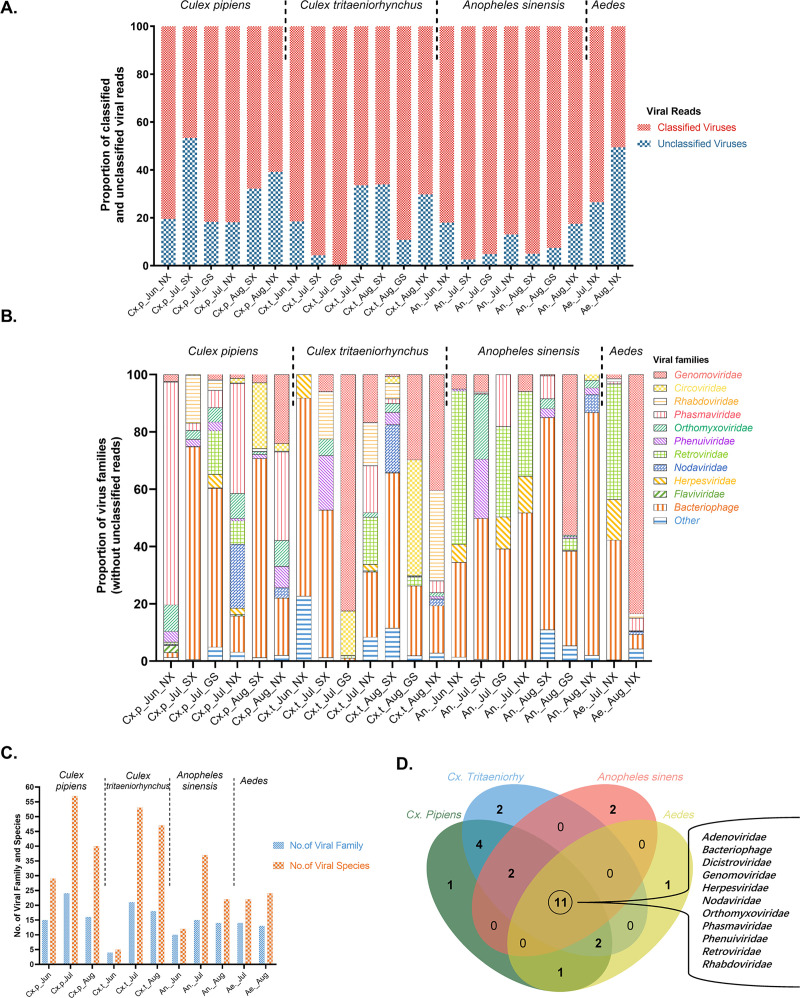
We characterized the viromes of 22,944 mosquitoes in 22 pools, representing four mosquito species from three provinces (Shaanxi, Gansu, and Ningxia) (Table 1). A total of 394,144,760 paired-end reads (55.4 G base pair) were generated through high-throughput sequencing, with an average of about 9.9 M reads per 500 mosquitoes (2.2–23.8 M reads/500 mosquitoes). After quality filtering, 15.8 G bases (77.3 M reads) were removed and 316,821,976 clean reads were retained for the subsequent analysis pipeline. Among those, 3,014,183 reads were identified as viral reads, accounting for 0.95% (3,014,183/316,821,976) of all clean reads, which suggests a rich diversity of viruses harbored by mosquitoes. The proportion of reads aligned to the viral genome database in each pool ranged from 0.01% to 5.96% of clean reads (Table 1). The three pools of Cx.t in July in Gansu, Cx.p in July in Shaanxi, and An. in July in Shaanxi achieved the highest proportions of viral reads, with 5.96%, 3.45%, and 2.99% of clean reads attributed to viruses, respectively. Meanwhile, the proportions of viral reads in all other pools were below 1%. Of the viral reads in each pool, 0.28–53.34% of reads were labeled as unclassified viral sequences, which could not be annotated to families or species (Fig 2A). Unclassified viral reads accounted for 27.74% (836,150/3,014,183) of total viral reads.
Table 1. Mosquito collection and sequencing data generation.
| Mosquito species | Month | Location | Pool name | No. of mosquitoes | Raw reads | Viral reads | Viral read proportion |
|---|---|---|---|---|---|---|---|
| Culex pipiens | June | Ningxia | Cx.p_Jun_NX | 1651 | 31,927,546 | 59,303 | 0.23% |
| July | Shaanxi | Cx.p_Jul_SX | 2104 | 47,610,058 | 1,282,927 | 3.45% | |
| July | Gansu | Cx.p_Jul_GS | 1647 | 22,426,930 | 56,294 | 0.30% | |
| July | Ningxia | Cx.p_Jul_NX | 2139 | 40,324,558 | 142,331 | 0.43% | |
| August | Shaanxi | Cx.p_Aug_SX | 2750 | 19,976,476 | 55,304 | 0.33% | |
| August | Ningxia | Cx.p_Aug_NX | 207 | 5,454,292 | 14,588 | 0.32% | |
| Culex tritaeniorhynchus | June | Ningxia | Cx.t_Jun_NX | 27 | 2,178,600 | 119 | 0.01% |
| July | Shaanxi | Cx.t_Jul_SX | 503 | 12,653,570 | 55,281 | 0.65% | |
| July | Gansu | Cx.t_Jul_GS | 782 | 15,096,738 | 769,383 | 5.96% | |
| July | Ningxia | Cx.t_Jul_NX | 1440 | 47,547,796 | 40,818 | 0.11% | |
| August | Shaanxi | Cx.t_Aug_SX | 900 | 15,298,094 | 8379 | 0.07% | |
| August | Gansu | Cx.t_Aug_GS | 1300 | 13,032,834 | 95,124 | 0.97% | |
| August | Ningxia | Cx.t_Aug_NX | 4370 | 24,813,632 | 31,030 | 0.15% | |
| Anopheles sinensis | June | Ningxia | An._Jun_NX | 39 | 7,935,140 | 1256 | 0.02% |
| July | Shaanxi | An._Jul_SX | 64 | 12,097,926 | 290,905 | 2.99% | |
| July | Gansu | An._Jul_GS | 33 | 15,104,164 | 6873 | 0.05% | |
| July | Ningxia | An._Jul_NX | 1034 | 9,425,030 | 1367 | 0.02% | |
| August | Shaanxi | An._Aug_SX | 300 | 8,799,222 | 2838 | 0.04% | |
| August | Gansu | An._Aug_GS | 200 | 9,246,162 | 14,362 | 0.18% | |
| August | Ningxia | An._Aug_NX | 270 | 8,313,570 | 366 | 0.01% | |
| Aedes | July | Ningxia | Ae._Jul_NX | 374 | 11,929,360 | 50,793 | 0.53% |
| August | Ningxia | Ae._Aug_NX | 810 | 12,953,062 | 34,542 | 0.32% | |
| Sum | 22,944 | 394,144,760 | 3,014,183 |
Fig 2. Virome of mosquitoes in the Shaanxi-Gansu-Ningxia region.
A. Proportions of classified and unclassified viral reads. Each bar represents a mosquito pool, as described in Table 1. B. Proportions of viral families among the classified viral reads. Each bar represents a mosquito pool, as described in Table 1. Read percentages lower than 0.1% are grouped as Others. C. Number of viral families and species among mosquitoes sorted by mosquito species and collection month. D. Venn diagram showing viral families found in the four mosquito groups. The four mosquito groups are presented in separate areas of the diagram. The numbers are representative of viral families found in mosquitos of each genus.
Viral diversity of classified viruses
The classified viral reads were distributed into 31 viral families and 116 viral species (S2 and S3 Tables). Among them, 90.66% of viral reads belonged to 11 viral families, including Genomoviridae (33.11%), Bacteriophage (containing six viral families, 34.25%), Circoviridae (7.62%), Rhabdoviridae (5.7%), Phasmaviridae (5.14%), and Orthomyxoviridae (4.83%). A further 20 viral families, including plant viruses and viroids, accounted for 9.34% of total classified viral reads (Fig 2B). The highest viral diversity was observed in the pool of Cx.p collected from Gansu in July (57 viral species in 24 viral families) (Fig 2C). Among mosquito species, Cx.p pools (69 species in 25 families) and Cx.t pools (73 species in 24 families) showed greater viral diversity than those of An. (50 species in 19 families) and Aedes (38 species in 20 families) (S1 Fig and S4 Table). The pools of Cx.p (57 species in 24 families) and Cx.t (53 species in 21 families) collected in July revealed the highest viral diversity (Fig 2C). Among collection months, we observed that mosquitoes collected in July showed greater viral diversity (86 species in 26 families) than those obtained in June (36 species in 18 families) or August (72 species in 25 families).
Furthermore, at the viral species level, 17 viral species show broad vector ranges encompassing all four mosquito groups, including Culex orthophasmavirus, Wuhan Mosquito Virus 6, Hubei mosquito virus 2, Yongsan picorna-like virus 3, Yongsan picorna-like virus 4, and Culex Hubei-like virus (S2 Fig). Fifty-three viral species showed narrow mosquito vector ranges, of which twenty-five (including JEV) were observed in Cx.t, fourteen in Cx.p, ten in An., and four in Aedes (S5 Table). In terms of viral families, eleven families, including Adenoviridae, Herpesviridae, Orthomyxoviridae, Phasmaviridae, Phenuiviridae, and Rhabdoviridae, were identified in all four mosquito groups (Fig 2D). Anelloviridae and Baculoviridae were only detected in Cx.t, Poxviridae and Xinmoviridae were only observed in An., and Togaviridae were found only in Aedes. The families Luteoviridae, Iflaviridae, and Flaviviridae were found in two Culex species (S5 Table).
Viral characterization of selected viruses
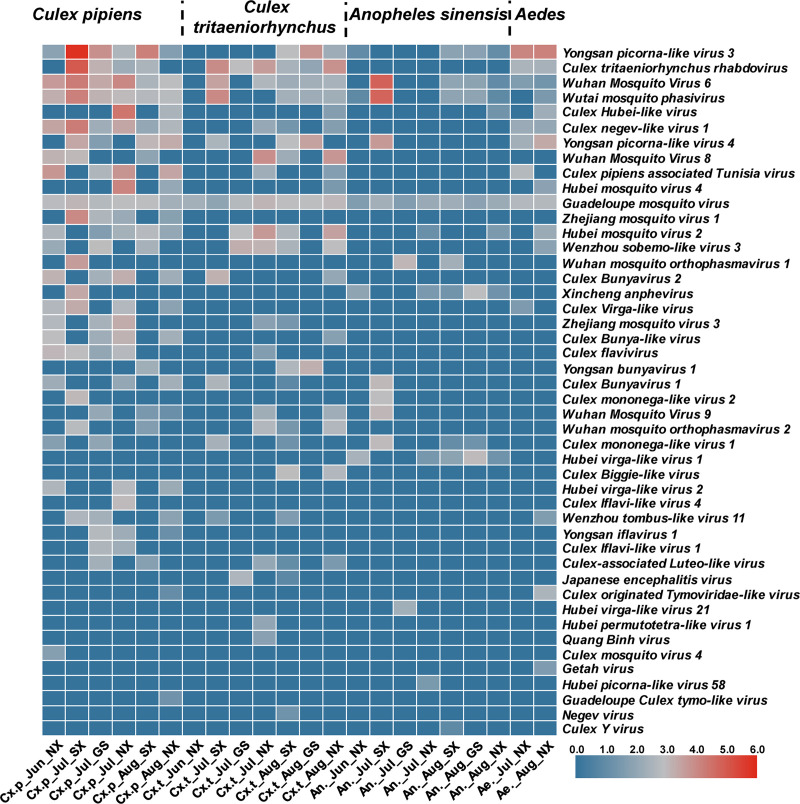
Forty-six species of mosquito-borne viruses belonging to 13 viral families were discovered in mosquitoes (Fig 3), with a minimum of six and a maximum of 668,502 reads, equivalent to 2.6–100.0% coverage and 1.3–3643.7× of the reference genome. The viral consensus sequences generated here had more than 93.9% nucleotide identity to the corresponding reference genomes (S6 Table). For example, we obtained the near full-length genome of Yongsan picorna-like virus 3 from Cx.p collected in Shaanxi in July, which showed 100% coverage and 98.4% identity with the reference genome (NC040584). Five other viruses also reached greater than 99% coverage and greater than 95% identity to their reference genomes, including Culex Bunyavirus 1 and Wutai mosquito Phasivirus in the family Phenuiviridae, as well as Cx.p associated Tunisia virus, Wuhan Mosquito Virus 8, and Hubei mosquito virus 4 in the family Tymoviridae.
Fig 3. Heatmap showing sequence reads of selected mosquito-borne viruses in each pool.
The names of the 46 viral species are listed in the right column. Mosquito species are shown at the top. Pool information is provided in the bottom row. The colors represent the base-10 logarithm of sequence read number, from blue (low read number) to red (high read number).
Through metagenomics, we also identified viral reads assigned to JEV, Culex Flavivirus, and Quang Binh virus belonging to the family Flaviviridae genus Flavivirus (Table 2). JEV was detected in the pools of Cx.t from Gansu in July (396 reads, 41% coverage, 16.4× depth). The consensus sequences showed 98.1% of nucleotide identity to JEV Genotype-I (HM366552). Culex flavivirus was found in five Culex pools (S3 Table). Quang Binh virus was detected in Cx.t from Ningxia in July, with a coverage and identity of 35.2% and 98.1%, respectively, compared to the strain MH827524. Pathogenic flaviviruses such as Yellow fever virus, WNV, and Zika virus were not found. In addition, two species of Phasmaviridae and five species of Phenuiviridae, which belong to the Bunyavirales, were identified in mosquitoes. Getah virus (GETV), belonging to family Togaviridae genus Alphavirus, was detected in Aedes from Ningxia in August with coverage and identity of 19.4% and 98.1% compared to the strain NC_006558, respectively (Table 2).
Table 2. Characterization of selected species of mosquito-borne viruses.
| Viral family | Viral species | Reference strain | Coverage* (%) | Depth | Per. ident. (%) | Collection month | Collection location | Mosquito species |
|---|---|---|---|---|---|---|---|---|
| Flaviviridae | Culex flavivirus | NC_008604 | 98.1 | 12.8 | 96.8 | Jun | NX | Cx.p |
| Flaviviridae | JEV | HM366552 | 41.0 | 16.4 | 98.1 | Jul | GS | Cx.t |
| Flaviviridae | Quang Binh virus | MH827524 | 35.2 | 1.9 | 98.1 | Jul | NX | Cx.t |
| Iflaviridae | Yongsan picorna-like virus 3 | NC_040584 | 100.0 | 3643.7 | 98.4 | Jul | SX | Cx.p |
| Phasmaviridae | Wuhan mosquito orthophasmavirus 1 | NC_031307 | 96.8 | 62.2 | 96.6 | Jul | SX | Cx.p |
| Phasmaviridae | Wuhan mosquito orthophasmavirus 2 | NC_031312 | 69.4 | 14.3 | 97.8 | Jul | SX | Cx.p |
| Phenuiviridae | Culex Bunyavirus 1 | MH188051 | 99.3 | 550.0 | 95.2 | Jul | SX | An. |
| Phenuiviridae | Wutai mosquito phasivirus | NC_031317 | 99.2 | 549.8 | 97.7 | Jul | SX | An. |
| Phenuiviridae | Yongsan bunyavirus 1 | NC_040759 | 97.8 | 29.8 | 97.5 | Aug | GS | Cx.t |
| Phenuiviridae | Culex Bunyavirus 2 | MH188052 | 97.0 | 29.3 | 96.3 | Jul | NX | Cx.p |
| Phenuiviridae | Culex Bunya-like virus | MH188002 | 15.0 | 105.8 | 97.2 | Jul | NX | Cx.p |
| Togaviridae | GETV | NC_006558 | 19.4 | 1.8 | 98.1 | Aug | NX | Ae. |
| Tymoviridae | Culex pipiens associated Tunisia virus | NC_040723 | 99.9 | 59.5 | 96.7 | Jul | NX | Cx.p |
| Tymoviridae | Wuhan Mosquito Virus 8 | NC_028265 | 99.6 | 72.3 | 98.0 | Jul | NX | Cx.t |
| Tymoviridae | Hubei mosquito virus 4 | NC_032231 | 99.0 | 644.9 | 95.6 | Jul | NX | Cx.p |
* The coverage (%) indicates the percentage of bases on the reference sequence covered by the reads.
Cx.p (Culex pipiens), Cx.t (Culex tritaeniorhynchus), An. (Anopheles sinensis), SX (Shaanxi Province), GS (Gansu Province), NX (Ningxia Hui Autonomous Region)
Discussion
Mosquito-borne arboviruses and their associated diseases are a great threat to human health [2, 27]. The life cycle of arboviruses relies on infection of and transmission by arthropod vectors [28]. With changes in climate, urban ecology, high-speed railway and airline travel, and other economic and social activities, the reproduction of mosquito vectors and arboviruses is no longer limited to specific regions [20, 29]. Changes in the population structure of mosquitoes and adaptation by arboviruses to new vectors may increase their geographic distribution. Additionally, some adaptive mutations enhance virus transmissibility by various mosquito species [30]. Thus, some mosquito-borne diseases may become emerging or re-emerging infectious diseases [31]. From the perspective of disease control, tracking the genetic diversity of mutating virus populations with meta-genomic methods will facilitate comparison of virulence traits with available genomic databases, determine disease susceptibility, and enable prediction and prevention of future public health threats [32].
Numerous metagenomic analyses have been conducted in invertebrates such as mosquitoes, spiders, and ticks from many regions of China [9, 22, 33–35], but few efforts have been made to characterize mosquito viromes in northwestern China. In this study, we explored mosquito viromes from seven locations in the Shaanxi-Gansu-Ningxia region using high-throughput sequencing and metagenomics analysis, focusing on identifying the viruses and potential diseases present in mosquitoes. A total of 116 viral species in 31 viral families were observed, including mammalian viruses, arboviruses, and bacteriophages, as well as some virus-related sequences that have not yet been specifically classified. Thus, the spectrum of mosquito-borne viruses is extremely diverse and varies with the time of collection, as well as the type of mosquitoes. Overall, Culex mosquitoes play an important role in harboring viruses in the study area, as Cx.p and Cx.t are not only the predominant in mosquitoes in terms of population size but also these species harbor more diverse viruses than other mosquitoes, in accordance with previous reports [5, 6, 36]. Aedes is rare in this area, and a small population was observed. In this survey, Aedes mosquitoes were collected only in July and August from Ningxia, and they harbored fewer viruses than other mosquito groups (S1 Table). Our results also showed that mosquitoes collected in July harbored greater viral abundance than those from June and August in all Culex and Anopheles species, but not Aedes.
Arboviruses in the families Flaviviridae and Togaviridae, including JEV, Quang Binh virus, Culex Flavivirus, and GETV, were observed in this study, but no member of Reoviridae was found. JEV is a significant human pathogen and GETV causes disease in animals such as pigs and horses. In this study, JEV was found in Cx.t from pigpen environments in Longnan City (Gansu Province in July). The consensus sequences showed close genetic identity (98.1% nt identity) to JEV Genotype-I. As an outbreak of JEV occurred in adults from the Shaanxi-Gansu-Ningxia region in 2018, JEV is of great concern as the main arbovirus disease in the study area. Therefore, monitoring of Cx.t density and tracing the genetic variations of JEV is a vital step in routine surveillance of mosquitoes and mosquito-borne viruses in the study area. In this study, sequence matches to GETV were observed in Aedes specimens collected in August from Ningxia Hui Autonomous Region. GETV, a member of the genus Alphavirus in the family Togaviridae, is an important animal pathogen that causes disease in livestock such as pigs, cattle, and horses [37–39]. GETV was first isolated in Malaysia in 1955, and then spread to Japan, China, South Korea, Mongolia, Russia, and other regions [40]. In China, GETV has been isolated in Yunnan Province, Hebei Province, Shanghai City, Sichuan Province, and Inner Mongolia Autonomous Region [41, 42]. An outbreak was reported at a swine farm in Hunan Province in 2017[43]. Culex and Armigeres have been reported as the major vectors of GETV [44]. Quang Binh virus was first identified from Cx.t collected in Vietnam in 2002, and has since been reported in southern China, such as Yunnan Province, Guangdong Province, and Shanghai City, but this study was the first report of Quang Binh virus in northwestern China [41, 43, 45]. Routine surveillance of these important pathogens is essential to disease control, and will provide useful information to prevent future outbreaks in this region.
Furthermore, the near-complete genomes of six mosquito-borne viruses were found, including Yongsan picorna-like virus 3, Culex Bunyavirus 1, Culex pipiens associated Tunisia virus, Wuhan Mosquito Virus 8, Hubei mosquito virus 4, and Wutai mosquito phasivirus. These viruses showed high identity with viruses reported in a metagenomic study from China, indicating that they do not exhibit strict geographical or mosquito species restrictions [33, 46]. These viruses can potentially affect the transmission of viruses that are pathogenic to vertebrates, thus representing a potential tool for vector control strategies. However, whether they have any direct or indirect public health risk requires further investigation [47].
A limitation of this study is that the species of Aedes mosquitoes were not determined, and all Aedes were pooled together without species distinction. Another shortcoming is that this research mainly focused on the viruses present in mosquitoes of the Shaanxi-Gansu-Ningxia region of China, rather than new viral discovery. Our analysis pipeline was based on a known viral reference genome database. Next-generation sequencing may also detect sequences from endogenous viral elements, which have been incorporated into the mosquito genome [23, 24, 48]. To exclude possible endogenous viral elements, each dataset was aligned to the Cx.t, An., and Aedes albopictus genomes. However, no mosquito reference genome sequences were available for some mosquito genera; therefore, the effect of this step cannot be evaluated. Moreover, although fewer mosquitoes were collected in June, the differences in sequencing depth and number of mosquitoes among pools were calibrated during the library construction and sequencing processes to support balanced comparison. Mosquitoes collected from seven cities in three provinces were used in this study; in future works, a larger sample size of each mosquito species from each location and coverage of a broader geographic region including more environments are needed. Such information would help to more comprehensively determine the baseline level of mosquito-borne viruses in the study area.
In conclusion, this study provided baseline information on viruses circulating in various mosquito species in the Shaanxi-Gansu-Ningxia region of China, which is of great importance for prediction and prevention of future mosquito-borne disease outbreaks in this region.
Supporting information
A. Culex pipiens, B. Culex tritaeniorhynchus, C. Anopheles sinensis, D. Aedes.
(TIF)
The four mosquito groups are presented in separate areas of the diagram. The numbers are representative of viral families found in mosquitos of each genus.
(TIF)
(DOCX)
(XLSX)
(XLSX)
(DOCX)
(DOCX)
(XLSX)
Data Availability
The data that support the findings of this study have been deposited into CNGB Sequence Archive (CNSA: https://db.cngb.org/cnsa; accession number CNP0001261). Other data that support the findings of this study are available within the paper and its Supplementary Information files.
Funding Statement
This work was supported by the National Natural Science Foundation of China (31900156)(FL); Key Research Project of Ningxia Science and Technology in 2019 (2019BCG01003)(HW); National Science and Technology Major Project (2018ZX10711001)(HW, XM), (2017ZX10104001)(FL, XM) and (2018ZX10713-002)(XM), the Development Grant of State Key Laboratory of Infectious Disease Prevention and Control (2015SKLID505)(HW). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Xia H, Wang Y, Atoni E, Zhang B, Yuan Z. Mosquito-Associated Viruses in China. Virol Sin. 2018;1(33):5–20 10.1007/s12250-018-0002-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liang G, Li X, Gao X, Fu S, Wang H, Li M, et al. Arboviruses and their related infections in China: A comprehensive field and laboratory investigation over the last 3 decades. Rev Med Virol. 2018;1(28) 10.1002/rmv.1959 [DOI] [PubMed] [Google Scholar]
- 3.Aragao CF, Pinheiro V, Nunes NJ, Silva E, Pereira G, Nascimento B, et al. Natural Infection of Aedes aegypti by Chikungunya and Dengue type 2 Virus in a Transition Area of North-Northeast Brazil. Viruses. 2019;12(11) 10.3390/v11121126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fu S, Song S, Li X, Xu Z, He Y, Li F, et al. Comparisons of Zika Viruses Isolated from Culex Quinquefasciatus and Armigeres Subalbatus Mosquitoes. Chinese Journal of Virology. 2019;03(35):385–395 [Google Scholar]
- 5.Shi C, Beller L, Deboutte W, Yinda KC, Delang L, Vega-Rua A, et al. Stable distinct core eukaryotic viromes in different mosquito species from Guadeloupe, using single mosquito viral metagenomics. Microbiome. 2019;1(7):121 10.1186/s40168-019-0734-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nanfack Minkeu F, Vernick K. A Systematic Review of the Natural Virome of Anopheles Mosquitoes. Viruses. 2018;5(10):222 10.3390/v10050222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Temmam S, Monteil-Bouchard S, Robert C, Baudoin JP, Sambou M, Aubadie-Ladrix M, et al. Characterization of Viral Communities of Biting Midges and Identification of Novel Thogotovirus Species and Rhabdovirus Genus. Viruses. 2016;3(8):77 10.3390/v8030077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lim ES, Zhou Y, Zhao G, Bauer IK, Droit L, Ndao IM, et al. Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat Med. 2015;10(21):1228–1234 10.1038/nm.3950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao P, Han J, Zhang Y, Li C, Guo X, Wen S, et al. Metagenomic Analysis of Flaviviridae in Mosquito Viromes Isolated From Yunnan Province in China Reveals Genes From Dengue and Zika Viruses. Front Cell Infect Microbiol. 2018;(8):359 10.3389/fcimb.2018.00359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roux S, Chan LK, Egan R, Malmstrom RR, McMahon KD, Sullivan MB. Ecogenomics of virophages and their giant virus hosts assessed through time series metagenomics. Nat Commun. 2017;1(8):858 10.1038/s41467-017-01086-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zablocki O, van Zyl L, Adriaenssens EM, Rubagotti E, Tuffin M, Cary SC, et al. High-level diversity of tailed phages, eukaryote-associated viruses, and virophage-like elements in the metaviromes of antarctic soils. Appl Environ Microbiol. 2014;22(80):6888–6897 10.1128/AEM.01525-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohlund P, Hayer J, Lunden H, Hesson JC, Blomstrom AL. Viromics Reveal a Number of Novel RNA Viruses in Swedish Mosquitoes. Viruses. 2019;11(11) 10.3390/v11111027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cholleti H, Hayer J, Fafetine J, Berg M, Blomstrom AL. Genetic characterization of a novel picorna-like virus in Culex spp. mosquitoes from Mozambique. Virol J. 2018;1(15):71 10.1186/s12985-018-0981-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Atoni E, Wang Y, Karungu S, Waruhiu C, Zohaib A, Obanda V, et al. Metagenomic Virome Analysis of Culex Mosquitoes from Kenya and China. Viruses. 2018;1(10) 10.3390/v10010030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sadeghi M, Altan E, Deng X, Barker CM, Fang Y, Coffey LL, et al. Virome of >12 thousand Culex mosquitoes from throughout California. Virology. 2018;(523):74–88 10.1016/j.virol.2018.07.029 [DOI] [PubMed] [Google Scholar]
- 16.Wang X, Cai Y, Jia X, Chen P, Yang H. Correlation between Japanese encephalitis epidemic and mosquito vector surveillance results in Longnan, Gansu province, China. CHINESE JOURNAL OF VECTOR BIOLOGY AND CONTROL. 2019;05(30):502–505 [Google Scholar]
- 17.Zhang J, Han X, He X. A primary investigation on mosquito species and mosquito-carried virus in Shaanxi Province. Chinese Journal of Hygienic Insecticides & Equipments. 2014;05(20):488–489 [Google Scholar]
- 18.Li G, Fu H, Fu S, Shen M, Ding X. Investigation of mosquitoes and mosquito⁃borne viruses in the Yangtze River Basin within Gansu province. CHINESE JOURNAL OF VECTOR BIOLOGY AND CONTROL. 2010;04(21):303–305 [Google Scholar]
- 19.Yu D, Cui S, Fu S, Cao L, Cao Y, Chen S, et al. Investigation of mosquitoes and mosquito-borne arboviruses in Gansu Province, China. Journal of Microbes and Infections. 2016;03(11):153–158 [Google Scholar]
- 20.Danwu, Yin Z, Li J, Shi W, Wang H, Fu H, et al. Epidemiology of Japanese encephalitis in China, 2014-2018. Chinese journal of Vaccies and Immunization. 2020;01(26):1–4 [Google Scholar]
- 21.Liu W, Fu S, Ma X, Chen X, Wu D, Zhou L, et al. An outbreak of Japanese encephalitis caused by genotype Ib Japanese encephalitis virus in China, 2018: A laboratory and field investigation. PLoS Negl Trop Dis. 2020;5(14):e8312 10.1371/journal.pntd.0008312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Du J, Li F, Han Y, Fu S, Liu B, Shao N, et al. Characterization of viromes within mosquito species in China. Sci China Life Sci. 2020;7(63):1089–1092 10.1007/s11427-019-1583-9 [DOI] [PubMed] [Google Scholar]
- 23.Frey KG, Biser T, Hamilton T, Santos CJ, Pimentel G, Mokashi VP, et al. Bioinformatic Characterization of Mosquito Viromes within the Eastern United States and Puerto Rico: Discovery of Novel Viruses. Evol Bioinform Online. 2016;Suppl 2(12):1–12 10.4137/EBO.S38518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zakrzewski M, Rašić G, Darbro J, Krause L, Poo YS, Filipović I, et al. Mapping the virome in wild-caught Aedes aegypti from Cairns and Bangkok. Scientific Reports. 2018;1(8) 10.1038/s41598-018-22945-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Wang H, Nie K, Zhang C, Zhang Y, Wang J, et al. VIP: an integrated pipeline for metagenomics of virus identification and discovery. Scientific Reports. 2016;1(6) 10.1038/srep23774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, et al. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol Plant. 2020; 10.1016/j.molp.2020.06.009 [DOI] [PubMed] [Google Scholar]
- 27.Liang G. Arbovirus-the very important pathogen of neglected tropical infectious disease in the world. China Tropical Medicine. 2018;01(18):1–5 [Google Scholar]
- 28.Sawchenko PE, Swanson LW, Vale WW. Corticotropin-releasing factor: co-expression within distinct subsets of oxytocin-, vasopressin-, and neurotensin-immunoreactive neurons in the hypothalamus of the male rat. J Neurosci. 1984;4(4):1118–1129 10.1523/JNEUROSCI.04-04-01118.1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Junglen S, Drosten C. Virus discovery and recent insights into virus diversity in arthropods. Curr Opin Microbiol. 2013;4(16):507–513 10.1016/j.mib.2013.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fischer C, de Lamballerie X, Drexler JF. Enhanced Molecular Surveillance of Chikungunya Virus. mSphere. 2019;4(4) 10.1128/mSphere.00295-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu M, Zheng Y, Wu J, Tan Y. Results and analysis of mosquito density monitoring in three districts of Yinchuan, Ningxia, China, 2016-2018.CHINESE JOURNAL OF VECTOR BIOLOGY AND CONTROL. 2020:1–3 [Google Scholar]
- 32.Christaki E. New technologies in predicting, preventing and controlling emerging infectious diseases. Virulence. 2015;6(6):558–565 10.1080/21505594.2015.1040975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi M, Lin XD, Tian JH, Chen LJ, Chen X, Li CX, et al. Redefining the invertebrate RNA virosphere. Nature. 2016;7634(540):539–543 10.1038/nature20167 [DOI] [PubMed] [Google Scholar]
- 34.Xia H, Hu C, Zhang D, Tang S, Zhang Z, Kou Z, et al. Metagenomic profile of the viral communities in Rhipicephalus spp. ticks from Yunnan, China. PLoS One. 2015;3(10):e121609 10.1371/journal.pone.0121609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi C, Liu Y, Hu X, Xiong J, Zhang B, Yuan Z. A metagenomic survey of viral abundance and diversity in mosquitoes from Hubei province. PLoS One. 2015;6(10):e129845 10.1371/journal.pone.0129845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xia H, Wang Y, Shi C, Atoni E, Zhao L, Yuan Z. Comparative Metagenomic Profiling of Viromes Associated with Four Common Mosquito Species in China. Virol Sin. 2018;1(33):59–66 10.1007/s12250-018-0015-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crabtree MB, Nga PT, Miller BR. Isolation and characterization of a new mosquito flavivirus, Quang Binh virus, from Vietnam. Arch Virol. 2009;5(154):857–860 10.1007/s00705-009-0373-1 [DOI] [PubMed] [Google Scholar]
- 38.Fang Y, Zhang Y, Zhou ZB, Shi WQ, Xia S, Li YY, et al. Co-circulation of Aedes flavivirus, Culex flavivirus, and Quang Binh virus in Shanghai, China. Infect Dis Poverty. 2018;1(7):75 10.1186/s40249-018-0457-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zuo S, Zhao Q, Guo X, Zhou H, Cao W, Zhang J. Detection of Quang Binh virus from mosquitoes in China. Virus Res. 2014;(180):31–38 10.1016/j.virusres.2013.12.005 [DOI] [PubMed] [Google Scholar]
- 40.Liu H, Zhang X, Li LX, Shi N, Sun XT, Liu Q, et al. First isolation and characterization of Getah virus from cattle in northeastern China. BMC Vet Res. 2019;1(15):320 10.1186/s12917-019-2061-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang T, Li R, Hu Y, Yang L, Zhao D, Du L, et al. An outbreak of Getah virus infection among pigs in China, 2017. Transbound Emerg Dis. 2018;3(65):632–637 10.1111/tbed.12867 [DOI] [PubMed] [Google Scholar]
- 42.Li YY, Fu SH, Guo XF, Lei WW, Li XL, Song JD, et al. Identification of a Newly Isolated Getah Virus in the China-Laos Border, China. Biomed Environ Sci. 2017;3(30):210–214 10.3967/bes2017.028 [DOI] [PubMed] [Google Scholar]
- 43.Li YY, Liu H, Fu SH, Li XL, Guo XF, Li MH, et al. From discovery to spread: The evolution and phylogeny of Getah virus. Infect Genet Evol. 2017;(55):48–55 10.1016/j.meegid.2017.08.016 [DOI] [PubMed] [Google Scholar]
- 44.Rui C, Xiaowei N, Na F, Shihong F, Xiaoyan S, Lin Z, et al. Emerging of Japanese encephalitis virus and Getah virus from specimen of mosquitoes in Inner Mongolia Autonomous Region. China Journal of Epidemiol. 2020;04:571–572 [DOI] [PubMed] [Google Scholar]
- 45.Wei L, Ming P, Xingyu Z, Shihua L, Xuecheng L, Shihong F, et al. First isolatio and identification of Getah virus SC1210 in Sichuan. China Joural of experiemnt and clinical virol. 2017;01(31):2–7 [Google Scholar]
- 46.Maia L, Pinto A, Carvalho MS, Melo FL, Ribeiro BM, Slhessarenko RD. Novel Viruses in Mosquitoes from Brazilian Pantanal. Viruses. 2019;10(11) 10.3390/v11100957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manni M, Zdobnov EM. A Novel Anphevirus in Aedes albopictus Mosquitoes Is Distributed Worldwide and Interacts with the Host RNA Interference Pathway. Viruses. 2020;11(12) 10.3390/v12111264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Katzourakis A, Gifford RJ. Endogenous viral elements in animal genomes. PLoS Genet. 2010;11(6):e1001191 10.1371/journal.pgen.1001191 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A. Culex pipiens, B. Culex tritaeniorhynchus, C. Anopheles sinensis, D. Aedes.
(TIF)
The four mosquito groups are presented in separate areas of the diagram. The numbers are representative of viral families found in mosquitos of each genus.
(TIF)
(DOCX)
(XLSX)
(XLSX)
(DOCX)
(DOCX)
(XLSX)
Data Availability Statement
The data that support the findings of this study have been deposited into CNGB Sequence Archive (CNSA: https://db.cngb.org/cnsa; accession number CNP0001261). Other data that support the findings of this study are available within the paper and its Supplementary Information files.