Abstract
Foot-and-mouth disease (FMD) is a highly contagious animal disease caused by an RNA virus subdivided into seven serotypes that are unevenly distributed in Asia, Africa, and South America. Despite the challenges of controlling FMD, since 1996 there have been only two outbreaks attributed to serotype C, in Brazil and in Kenya, in 2004. This article describes the historical distribution and origins of serotype C and its disappearance. The serotype was first described in Europe in the 1920s, where it mainly affected pigs and cattle but as a less common cause of outbreaks than serotypes O and A. No serotype C outbreaks have been reported in Europe since vaccination stopped in 1990. FMD virus is presumed to have been introduced into South America from Europe in the nineteenth century, although whether serotype C evolved there or in Europe is not known. As in Europe, this serotype was less widely distributed and caused fewer outbreaks than serotypes O and A. Since 1994, serotype C had not been reported from South America until four small outbreaks were detected in the Amazon region in 2004. Elsewhere, serotype C was introduced to Asia, in the 1950s to the 1970s, persisting and evolving for several decades in the Indian subcontinent and for eighteen years in the Philippines. Serotype C virus also circulated in East Africa between 1957 and 2004. Many serotype C viruses from European and Kenyan outbreaks were closely related to vaccine strains, including the most recently recovered Kenyan isolate from 2004. International surveillance has not confirmed any serotype C cases, worldwide, for over 15 years, despite more than 2,000 clinical submissions per year to reference laboratories. Serology provides limited evidence for absence of this serotype, as unequivocal interpretation is hampered by incomplete intra-serotype specificity of immunoassays and the continued use of this serotype in vaccines. It is recommended to continue strengthening surveillance in regions of FMD endemicity, to stop vaccination against serotype C and to reduce working with the virus in laboratories, since inadvertent escape of virus during such activities is now the biggest risk for its reappearance in the field.
Keywords: foot-and-mouth, serotype C, phylogeny, extinction
1. Introduction
Foot-and-mouth disease (FMD) is an economically important vesicular condition of cloven-hoofed artiodactyls, characterised by blisters in and around the mouth, feet, and udders and accompanied by fever, lameness, loss of condition, milk drop in dairy cattle, and sudden death in young stock due to myocarditis (Thomson and Bastos 2005). The disease is highly contagious and occurs in many African and Asian countries where the incidence of new cases is often cyclical, associated with waning immunity and changing opportunities for transmission.
FMD is caused by a virus (FMDV) in the genus Aphthovirus and the family Picornaviridae. The virus genome is a single strand of positive-sense RNA and viral replication is associated with copying errors that underlie rapid mutation rates and the emergence of new virus lineages and strains. FMDV has been divided into seven serotypes that do not cross-protect. Serotypes O and A were the first to be recognised (Vallée and Carré 1922), followed shortly after by serotype C (Waldmann and Trautwein 1926) and later by the Southern African Territories serotypes (SAT 1-3; Brooksby 1958) and Asia 1 (Brooksby and Rogers 1957). All the serotypes cause a similar disease, although there may be differences in host specificity and virulence between viral strains. Sequencing of the viral genome is used to trace the spread and emergence of new FMDV lineages, especially based on similarity of the sequences encoding the variable capsid protein (VP1) (Knowles et al. 2016). These data, together with geographical separation, are the basis for subdivision of the serotypes into geographical topotypes (Knowles and Samuel 2003). Five serological subtypes of FMDV were defined for serotype C (Davie 1962), but subtype classification has now been largely abandoned (Pereira 1977). Due to recombination, the division into serotypes only holds when sequences of the capsid encoding parts of the genome are compared and this means that when we talk of a serotype becoming extinct, we are primarily referring to these genetic elements (Bachanek-Bankowska et al. 2018).
FMD has been eradicated from North America, Australasia, Europe, and much of South America by zoosanitary measures supported in some cases by vaccination campaigns. Efforts directed towards wider global control and eventual eradication have made progress where economic development has underpinned investment in animal health services (http://www.gf-tads.org/fmd/fmd/en/; last accessed 29 Jan 2021). In view of the continued endemic circulation of FMD and the logistical and technical difficulties to control the disease, it is perhaps surprising that serotype C appears to have become extinct, having disappeared from global surveillance. Serotype C has not been recorded by the World Organisation for Animal Health (OIE)/Food and Agriculture Organization of the United Nations (FAO) FMD Laboratory Network (http://foot-and-mouth.org; last accessed 29 Jan 2021) since outbreaks were detected in Brazil and Kenya in 2004. In addition to South America and Africa, this serotype was also previously present in Europe and Asia. The geographical origin of FMDV is not known (although an African origin is suspected) but it is believed that Europe was ultimately the origin for all the FMDV serotypes reported in South America, including serotype C, and that either Europe or South America were the likely sources of characterised serotype C viruses from Africa and Asia (Fig. 1). Topotypes are defined within FMDV serotypes with a cut-off of <15 per cent genetic divergence in VP1 and Knowles and Samuel (2003) debated whether there should be one or seven topotypes for serotype C but the World Reference Laboratory for FMD (WRLFMD) currently describes three topotypes: EURO-SA, AFRICA, and ASIA, excluding the C/GER/c.32 and C/UK/149/34 isolates. This article considers the historical occurrence and connections between outbreaks of serotype C, evidence that the serotype has become extinct and steps needed to verify elimination and to ensure that it does not return.
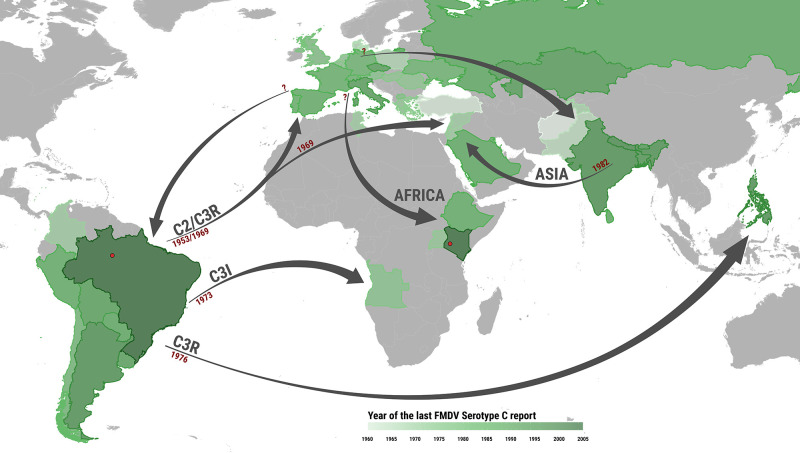
Figure 1.
Geographic and temporal distribution of historical FMDV serotype C events. The map provides the most likely timing and spatial directionality of the historical events attributed to serotype C by examining historical information on outbreaks, the genetic relationship between sampled isolates and the results generated by the phylogeographic analysis (Fig. 2, Table 1). Countries are coloured according to the last year when a serotype C case was reported. Arrows show the route of virus movements between continents for each of the topotypes, along with the suggested year of introduction. Geographic locations denoted in red are those of the 2004 outbreaks reported in Brazil and Kenya.
2. Materials and methods
Serotype C viruses have been archived at the FAO WRLFMD, The Pirbright Institute, UK, arising from samples collected between 1932 and 2004. Sequences of the VP1 coding region of serotype C FMDV were available for 100 isolates from public databases. An additional 49 VP1 sequences were either newly determined or obtained from collaborating organisations and submitted to GenBank (Supplementary Table S1). VP1 coding sequences were amplified from nucleic acid extracted from cell cultured virus preparations, using reverse transcription - polymerase chain reaction (RT-PCR) with the primers and methods described in Knowles et al. (2016).
A final dataset of 149 serotype C sequences, encoding the VP1 capsid region (630 nucleotides), were aligned using MAFFT 7.450 (Katoh and Standley 2013). Recombination analyses performed in GARD (Pond et al. 2006) and LDhat (Auton and McVean, 2007) using both the VP1 and the full-capsid region sequences did not find evidence of recombination breakpoints.
Maximum-likelihood (ML) tree topology reconstruction was pipelined using PhyML 3.3 (Guindon et al. 2010) to obtain the initial BioNJ tree and optimise branch lengths and substitution rates, along with RAxML 8.2.12 (Stamatakis 2014) to search for the ML topology and to bootstrap for 200 runs. A similarity matrix was generated by estimating raw evolutionary distances between pairwise sets of VP1 sequences in R 4.0.3 (R Core Team 2020) using the ape package (Paradis and Schliep 2019). Phylogenies were plotted in R using the ggtree package (Yu et al. 2017).
The time-stamped phylogeny was obtained from BEAST 1.10.4 (Suchard et al. 2018) using the uncorrelated relaxed clock model based on a log-normal distribution (Drummond et al. 2006), along with the GTR + Γ4 model of nucleotide substitution and the Bayesian skygrid coalescent tree prior (Gill et al. 2013). Discrete trait phylogeographic analysis (Lemey et al. 2009) was further performed in BEAST to reconstruct FMDV serotype C movements between continents. Posterior estimates were obtained by running the Markov chain Monte Carlo for 200 million iterations, sampling every 20,000 states and setting the burn in at 10 per cent of the chain.
The sequence alignment and the BEAST .xml files used to perform the analyses have been uploaded to Dryad (https://doi.org/10.5061/dryad.v6wwpzgv6; last accessed 29 Jan 2021).
3. Results and discussion
3.1 Serotype C phylogeny
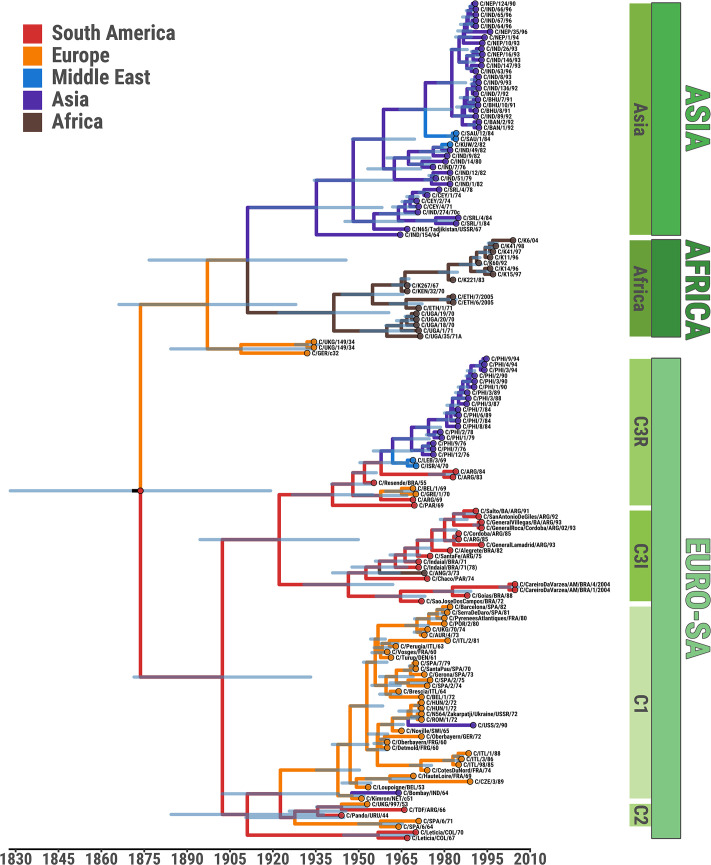
Figures 2 and 3 display inferred phylogenies from representative viruses of all the main geographical reservoirs where serotype C has been reported (Fig. 1), with the earliest virus being from Europe in 1932 and the most recent from Brazil and Kenya in 2004. Figure 3 shows a matrix of estimated pairwise evolutionary distances annotated to the reconstructed ML phylogeny. Molecular clock analyses in this study were performed using the widest possible collection of serotype C VP1 sequences available (n = 149) to minimise sampling bias and suggested the year of the most recent common ancestor (tMRCA) of serotype C to be approximately 1873 (95% Bayesian credible interval (BCI) 1825–1916) (Fig. 2). A previous estimate of the divergence times for FMDV serotypes put the origin of serotype C at around 1919 (95% BCI between 1856 and 1940) albeit using a smaller collection of VP1 sequences (Tully and Fares 2008). These dates overlap with the earliest recorded appearance of FMD in South America (i.e. 1870; Machado 1969). The molecular clock estimate for the rate of FMDV VP1 capsid protein evolution in serotype C (2.86 × 10−3 nt substitution/site/year, 95% BCI 1.82 × 10−3–3.99 ×10−3) is slightly higher than that reported by Tully and Fares (2008) but at the lower boundary of rates previously reported for other serotypes (Freimanis et al. 2016). From the beginning of the twentieth century, at least four major ancestors of subsequently circulating serotype C lineages are predicted to have been in existence (Fig. 2; Table 1).
Figure 2.
Space-time evolution of the FMDV serotype C based on sequences of the VP1 capsid encoding region generated from 149 isolates. Time-calibrated genetic relationships between VP1 sequences, with the 95% Bayesian credible interval (BCI) region of node timing represented in light blue; branches and nodes are coloured according to the most probable ancestral continent inferred from the discrete phylogeography analysis. Topotypes with their subtypes are denoted (green panels). The viruses labelled as C/ETH/6/2005 and C/ETH/7/2005 were submitted to the WRLFMD in 2005 from samples collected in 1983.
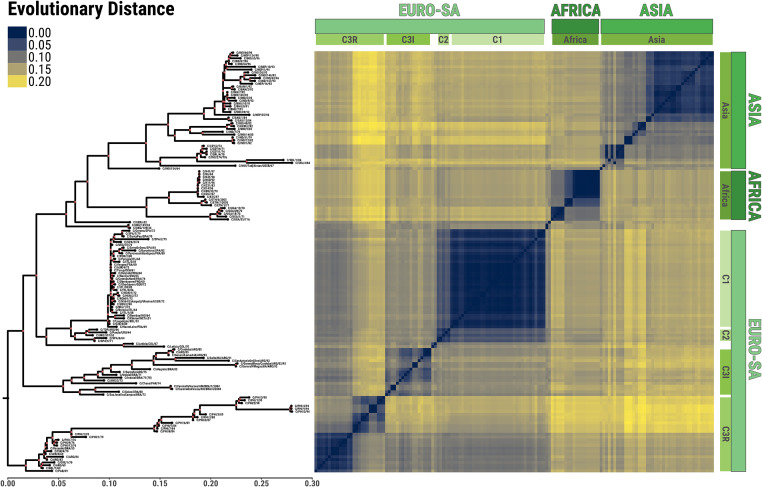
Figure 3.
Evolutionary signature of the FMDV serotype C generated using 149 partial FMDV genome sequences encoding the VP1 capsid region. Left panel, maximum-likelihood topology where internal nodes with bootstrap values of ≥75 are coloured in red; right panel, matrix of pairwise evolutionary distances. Topotypes with their subtypes are denoted (green panels). The viruses labelled as C/ETH/6/2005 and C/ETH/7/2005 were submitted to the WRLFMD in 2005 from samples collected in 1983. There are two slightly different viruses called C3/Indaial/Brazil/1971, which differ in cell culture passage history and by about 4% in VP1 (Ouldridge, July 1988, pers. comm.).
Table 1.
Conjectured timeline of FMDV type C events.
| Time | tMRCA [95% BCI] |
Ancestral virus location
[P] |
Event |
|---|---|---|---|
| Late 19th century | 1873 [1825–1916] | South America [0.87] | Serotype C evolves from type A, either in Europe or in South America (following the introduction of FMD into Argentina in c.1870). New ecological niche? |
| 1920s | 1922 [1890–1946] | South America [0.96] | C3 splits into two distinct lineages in South America, C3R (named after the Resende strain) and C3I (named after the Indaial strain) |
| 1920–30 | 1908 [1880–1929] | Europe [0.89] | C/GER/c.32 and C/UK/149/34 may have evolved in Europe either from indigenous viruses or from a virus introduced from South America |
| 1920–30 | 1935 [1908–1954] | Europe [0.42] | Serotype C introduced into Asia (possibly from Europe) |
| 1920–40 | 1941 [1919–1957] | Europe [0.42] | Serotype C introduced into Africa (probably from Europe or Asia). Independent lineages evolve in Uganda, Kenya and Ethiopia |
| 1930s | 1942 [1934–1949] | South America [0.89] | Serotype C introduced into Europe from South America which became the C1 lineage (dominant in Europe until 1989) |
| 1953 | 1927 [1907–1942] | South America [0.84] | Serotype C2 was introduced into the UK from South America. It is likely that this lineage did not persist |
| c.1964 | 1947 [1942–1950] | Europe [0.99] | C1 spreads to Bombay, India (or is introduced as a vaccine strain?) |
| 1969 | 1967 [1962–1969] | South America [0.88] | C3R introduced into the Middle East (Lebanon/Jordan/Syria) from South America with spread to Israel |
| 1969 | 1957 [1948–1967] | South America [0.94] | C5-like virus introduced into Europe from South America (Belgium in 1969; Greece in 1970) |
| 1973 | 1957 [1947–1966] | South America [0.99] | C3I-like virus introduced into Angola from South America |
| 1976 | 1968 [1961–1974] | South America [0.88] | C3R introduced into the Philippines from South America |
| 1982 | 1979 [1975–1981] | Asia [0.99] | Serotype C introduced into Kuwait from South Asia |
| 1983 | 1979 [1972–1982] | South America [1.0] | C3R reintroduced into the field in Argentina, persisting until 1984 |
| 1984 | 1982 [1980–1984] | Asia [0.99] | Serotype C introduced into Saudi Arabia from South Asia. It did not persist |
| 1990 | – | Serotype C disappears from Europe (last report in Italy in July 1989) | |
| 1994 | – | Serotype C disappears from South America (last reports in Argentina in 1994) | |
| 1996 | – | Serotype C disappears from the Philippines (following the introduction of type O in 1994) | |
| 1996 | 1986 [1983–1989] | Asia [1.0] | Last serotype C recorded on the Indian subcontinent (Nepal) |
| 2004 | 1996 [1994–1997] | Africa [1.0] | Last serotype C in Kenya |
| 2004 | 2002 [1998–2004] | South America [1.0] | Serotype C reappears in the Amazonas region of Brazil |
The tMRCA virus for different events has been estimated from the time-calibrated tree (Fig. 2) and is expressed as mean values with associated 95% BCI. Most likely location of ancestral virus with associated posterior probability (P) for each of event was estimated from the discrete phylogeographic diffusion model.
The first of these (tMRCA c.1911, 95% BCI 1882–1935) gave rise to a cluster of viruses isolated between 1944 and 1990 that correspond to the subtypes C1 (average evolutionary distance of 0.013, 95% probability interval (PI) 0–0.042) and C2 (average evolutionary distance of 0.032, 95% PI 0.014–0.051) (Fig. 2). These were mostly of European origin (C1) but included one Indian vaccine strain (C/Bombay/IND/64), two South American viruses from Uruguay (1944) and Argentina (1966), as well as a virus isolated in UK from 1953. As for other FMD outbreaks at the time, this UK isolate may have been introduced via imported South American meat, directly or via mainland Europe (Fogedby 1963). Two Colombian viruses (from 1967 and 1970) also clustered with the C1/C2 lineages but as a distinct outgroup.
The second progenitor (tMRCA c.1922, 95% BCI 1890–1946) was reconstructed to give rise to two subsequent virus clusters (Fig. 2). One of these, C3R (average evolutionary distance of 0.079, 95% PI 0.005–0.137), contained viruses from South America (Argentina 1969, 1983, and 1984; Paraguay 1969), the Philippines (1976–94), Europe (Belgium 1969 and Greece 1970), and the Middle East (Lebanon 1969 and Israel 1970). Its oldest representative was the vaccine strain C3/Resende/Brazil/1955. The other cluster, C3I (average evolutionary distance of 0.081, 95% PI 0.004–0.127), comprised almost entirely South American viruses from Argentina, Brazil, and Paraguay isolated between 1971 and 1993 along with a single isolate from Angola (1973). Once again, a vaccine strain was the oldest representative (C3/Indaial/Brazil/1971).
The third serotype C ancestor diverged around 1911 (95% BCI 1872–1941) into two clusters of viruses of Asian (n = 42, reported between 1964 and 1996; average evolutionary distance of 0.081, 95% PI 0.007–0.143) and East African origin (n = 18, collected during 1967–2004; average evolutionary distance of 0.052, 95% PI 0–0.100) (Fig. 2). Most of the Asian viruses were from the Indian subcontinent (n = 22 from India, 1964–93; n = 6 from Sri Lanka, 1971–84; n = 5 from Nepal, 1990–96; n = 3 from Bhutan, 1991; n = 2 from Bangladesh, 1992), but there were also representatives from Tajikistan (1967), Saudi Arabia (n = 2, 1984), and Kuwait (1982). Also, in this cluster, the oldest representative was a vaccine strain. The East African viruses were from Kenya (n = 10, 1967–2004), Uganda (n = 5, 1970–71), and Ethiopia (n = 3, 1971–83).
The fourth and final ancestral clade (tMRCA c.1908, 95% BCI 1880–1929) included only European viruses, one from Germany (c.1932; received from Insel-Reims in January 1933) and one from the United Kingdom (collected on a farm in May 1934) (average evolutionary distance of 0.068, 95% PI 0.007–0.102) (not assigned in Fig. 2).
3.2 Serotype C in Europe
The history of FMD in Europe in the mid-twentieth century has been reviewed by Fogedby (1963). He noted that prior to the 1930s, FMD outbreaks in Europe were often ascribed to incursions from the East (from Russia and Asia Minor through the Balkans) although the serotype divisions were not then recognised. After this time, serotypes O, A, and C were recovered from a series of FMD epizootics, until brought under control by systematic vaccination from the 1950s onwards, leading to eradication of FMD by the end of the 1980s, and the last prophylactic vaccination took place in 1990 (Sutmoller et al. 2003). Fogedby (1963) reported a tendency for clusters of disease to begin with viruses from serotypes O or A and then for serotype C to become involved and to spread more slowly, giving rise to fewer outbreaks. However, a serotype C virus of high virulence was responsible for a major epizootic in Northern Italy that persisted for three years from 1945 to 1948 and was associated with mortality in adult cattle. Serotype C was also the cause of a series of European outbreaks from 1960 to 1961 (e.g. Italy reported 18,722 outbreaks during 1961) affecting pigs that were not routinely vaccinated. An outbreak of serotype C in pigs in Normandy, France, in 1974 was considered the source of an airborne incursion on the Island of Jersey in the same year (Mowat 1987) and in experimental trials the virus C1/Noville/SWI/65 was found to be excreted at a very high level in the breath of infected pigs (Donaldson et al. 1982). The last recorded FMD outbreaks caused by serotype C in western Europe occurred in Italy in 1989 (Valarcher et al. 2008), affecting primarily pigs and cattle.
Martínez et al. (1992) compared VP1 sequences from 13 European isolates and found that 11 of them were almost identical, despite being sampled from seven countries over a time window of 36 years (1953–89). This lack of evolution was ascribed to a common origin as vaccine escapes. A subsequent comparison of 51 partial VP1 sequences for serotype C viruses has been reported by Knowles and Samuel (2003), including six European isolates. The European isolates belonged to the subtype C1, apart from: (i) a 1953 UK field isolate that was closely related to the Uruguayan C2 Pando vaccine strain, originally isolated in 1944 and (ii) C5/BEL/1/69, a C5 subtype virus related to C5/ARG/69. These C2 and C5 viruses may have been introduced into Europe from South America. Valarcher et al. (2008) reviewed European incursions since 1985, noting that the last serotype C occurrence in Italy was caused by a virus closely related to those causing earlier Italian outbreaks of the 1980s, which probably arose from the vaccine strain, C1/Brescia/ITL/64. Unlike serotypes O and A, serotype C viruses have not recurred in Europe after FMD freedom was attained in 1990 and vaccination ceased.
The similarity of many European serotype C isolates can be seen in Fig. 3. Two viruses from the former Czechoslovakia and Kazakhstan, respectively (C/CZE/3/89 and C/USS/2/90) are also ‘vaccine-like’ (Clade C1). The first was submitted in 1989 as ‘the Czech vaccine strain’ but its isolation date from the field is unknown. The second was from an outbreak in Kazakhstan in 1990. However, evolutionary progression consistent with a period of circulation in the field is evident for some of the viruses, for example, those Clade C1 viruses isolated in Spain in the 1970s and 1980s. Figure 3 also shows the similarity of both C/GRE/1/70 and C5/BEL/1/69 with C5/ARG/69 within the C3R clade, pointing to a likely South American source for these outbreaks.
3.3 Serotype C in South America
Cattle from Europe were first brought to South America in the sixteenth century, but FMDV is thought to have been introduced from later arrivals in 1870 (Machado 1969), at a time when serotypes had not been identified. Thereafter, most of the trade that might have transferred FMDV was in livestock products coming from South America to Europe, following the first refrigerated meat shipments in 1876. As supported by the ML phylogeny (Fig. 3), the genetic diversity of the South American serotype C viruses was found to be markedly higher than that of viruses isolated from Europe. The topology further supported evidence of at least three distinct lineages, structured as sister clades of the C3 Indaial subtype, and characterised by substantial within-lineage evolution. Serotype C was first recognised in South America during an epidemic which occurred in Brazil (Rio Grande do Sul), Uruguay and Argentina during 1943–45. Ultimately, four subtypes of serotype C were reported (C2–5) and, between 1968 and 2004, 8,519 outbreaks due to serotype C were recognised in eight countries (Sanchez-Vazquez et al., 2018). In December 1966, an outbreak of FMDV serotype C occurred in cattle on seven farms on the island of Tierra del Fuego at the southern tip of Argentina. The outbreak was unusual in its severity, with deaths occurring due to myocarditis even amongst older cattle (Gimeno 1967). The virus was characterised as a new antigenic subtype (C4); however, sequence analyses have shown a close relationship to C2/Pando/Uruguay/44 (Fig. 3). The decade with the most recorded serotype C outbreaks was the 1970s (5,095 outbreaks were recorded between 1972 and 1979), after which the numbers reduced substantially in the 1980s (1,635), whilst only 274 outbreaks were ascribed to this serotype in the 1990s. These years also saw a decline in other serotypes due to increasingly effective FMD control measures. Serotype C was less widely distributed than serotypes O and A, being present in Argentina, Bolivia, Brazil, Chile, Paraguay, Uruguay, and sporadically in Peru and in Colombia (interestingly, only on two occasions, when cases were reported in the south-east corner of the country from the city of Leticia, next to the Amazon river and close to the border with Brazil). Many Argentinean serotype C viruses circulating between 1975 and 1994 were more closely related to the vaccine strain C3/Indaial/Brazil/1971 than C3/Resende/Brazil/1955 but were distinct enough not to have been considered vaccine escapes (Konig et al. 2001). However, some outbreaks between 1982 and 1984 were probably caused by the C3 Resende vaccine strain (Bergmann et al. 1988; Konig et al. 2001; Knowles and Samuel 2003). A Peruvian outbreak due to serotype C in 1980 was also attributed to vaccine containing virus that had been incompletely inactivated by formalin and released before the full testing regimen had been completed (Rweyemamu et al. 2008).
After 1995, no serotype C outbreaks were reported until 2004, when the virus was detected in cattle from the Amazonas region near Manaus (Correa Melo 2004). A Neighbor-joining phylogenetic tree reconstructed at that time showed that the virus responsible (although genetically ‘South American’) was not closely related to the South American vaccine strains (C3 Resende or C3 Indaial) nor very similar to any previously characterised isolates (maximum VP1 identity of 89%). However, both the ML and time-stamped phylogenies here reconstructed identified this virus as descending from Brazilian strains previously circulating in 1972 and 1988 (with a divergence of ∼7%), and clustering within the C3I subtype clade (Figs. 2 and 3). The tMRCA of the 2004 outbreaks was estimated at c.2002 (95% BCI 1998–2004). Four outbreaks were officially declared, and the region was not a part of one of the OIE-recognised Brazilian FMD-free zones as it was said to have low disease detection capacity and vaccination levels. Presumably, the virus had been circulating undetected in this region over the previous nine or more years. A serological study performed to test the likelihood that the C3 Indaial vaccine would protect against the new field isolate showed that a two-course primary vaccination, but not a single vaccine dose, gave an acceptable expectancy of protection (Correa Melo 2004).
Since 2004, there have been no reports of serotype C anywhere in South America. Following a resolution of the OIE and the South American Commission for the Fight against FMD that recommended to withdraw the serotype C from the formulation of vaccines in use, Bolivia in 2018 and Brazil and Paraguay in 2019 have withdrawn the serotype C from their vaccination programs (PANAFTOSA-OPS/OMS 2019; OIE/FAO, 2019). However, Argentina still maintains the serotype C in the vaccines in use (PANAFTOSA-OPS/OMS 2019), and is the only country known to do so. Other serotypes (O and A) have also been absent from most countries of South America since 2012, apart from outbreaks occurring in Venezuela and Colombia (PANAFTOSA-OPS/OMS 2019). Thus, the last report for the serotype A took place in Venezuela in 2013, while for the serotype O, it was in the North of the Andean Region, caused by a particular lineage of serotype O circulating in that zone since 2004 (PANAFTOSA-OPS/OMS 2019).
In short, a further fifteen years have now elapsed since the last serotype C outbreaks in South America and the areas where FMD remains (only Venezuela, with spill over to northern Colombia) have never reported this serotype. Furthermore, the rest of the sub-continent has FMD-free status (with or without vaccination) validated by OIE as compliant with their surveillance requirements (OIE Terrestrial Animal Health Code 2019). Therefore, it must now be considered extremely unlikely that serotype C FMDV is still circulating anywhere in the continent. As in other regions, the main risk of a recurrence appears to be from continued manufacture, testing and use of serotype C vaccine or from a laboratory escape.
3.4 Serotype C in Asia
According to the WRLFMD serotyping records (WRLFMD, unpublished data; Ferris and Donaldson 1992), FMDV serotype C was first identified across the Asian continent in West Pakistan and Sri Lanka in 1954, India in 1955, Afghanistan in 1957, Turkey in 1959, and East Pakistan in 1960. Unfortunately, none of these virus isolates are available for genetic analyses.
FMDV serotype C was first recognised in India during 1955 and was recorded as present in 1956, 1963–64, 1970–88, and 1991–95. It has been the least prevalent of the four serotypes reported from India, causing only sporadic outbreaks (∼8% during 1971–90, and 1.6% for the period between 1991 and 1994), with the last case in 1995 (Hemadri et al. 2003; Bhattacharya et al. 2005). The published sequence of the Indian vaccine strain C/Bombay/IND/64 (GenBank Accession No: MK413704; Hemadri et al. 2003), isolated from a pig at the Aarey Milk colony, Mumbai, Maharashtra, is closely related to C1 viruses circulating at that time in Europe (Figs. 2 and 3; Hemadri et al. 2003). However, another virus purporting to be C/Bombay/IND/64 (Indian Ref. No. IND/154/64) was received by the WRLFMD in 1971 (WRLFMD Ref. No. IND/4/71) and is completely unrelated, sitting close to the root of the ASIA topotype. A later vaccine strain, C/IND/51/79 (collected in 1977; Nagendrakumar et al. 2005), along with field isolates from the 1970s, 1980s, and 1990s are clustered in a separate clade (Figs. 2 and 3). Despite its genetic distance from characterised Indian field strains, C/Bombay/IND/64 showed a good antigenic match with isolates from the 1990s (Hemadri et al. 2003) as was also shown for C/IND/51/79 (Nagendrakumar et al. 2005). Based on their close historical links, Britain and Europe represent likely sources of serotype C viruses found in India, with at least two distinct introductions having given rise to the C1-like/Bombay/IND/64 and the other ASIA lineages.
As for other FMDV serotypes, similar viruses are shared between India and its neighbours (Nepal, Bhutan, Sri Lanka, and Bangladesh), reflecting the close connections between these countries (Ranaweera et al. 2019). FMDV serotype C outbreaks reported both in Kuwait (1982) and Saudi Arabia (1984) appear to be independent introductions from the Indian subcontinent (Figs. 2 and 3). The link between the subcontinent and Tajikistan is uncertain (Figs. 2 and 3), with not enough resolution from sampling to draw substantiated conclusions; however, Sri Lanka and former Soviet Union countries have maintained trade ties for many years.
In 1969, FMDV serotype C was reported in Lebanon, Syria, and Jordan and subsequently in 1970 in Israel. Sequencing of VP1 showed that the viruses from Lebanon (C/LEB/3/69) and Israel (C/ISR/4/70) were closely related to C3/Resende/BRA/55, suggesting a likely South American origin (Figs. 2 and 3).
Following the introduction of a European/South American-like type A virus into the Philippines in 1975 and the subsequent use of a trivalent South American vaccine, serotype C was first recognised in the Philippines in 1976, after which major outbreaks occurred until the early 1980s. Then the serotype was detected only sporadically and was not detected at all between 1991 and 1993, although it was probably present. The last outbreak was confirmed in 1994 (Rweyemamu et al. 2008), shortly after the introduction of the O/CATHAY topotype (probably from Hong Kong), which went on to cause major outbreaks and took many years to eradicate (Di Nardo et al. 2014). The most likely ultimate source of the serotype C introduction was South America since the earliest Philippine virus was very closely related to the C3 Resende vaccine virus of Brazilian origin. Knowles and Samuel (2003) describe a steady evolution of Philippine isolates over time, consistent with diversification from a single source (Fig. 3). Interestingly, as judged by VP1 sequences, serotype C viruses in the Philippines evolved rapidly, resulting in codon deletions at positions 144 and 207 (loss of up to three or six nucleotides). These viruses became more and more difficult to identify in typing assays, suggesting antigenic evolution was also taking place.
3.5 Serotype C in Africa
FMDV type C was first reported from Africa, in Niger, in 1946 by the former Laboratoire Central Recherches Vétérinaire à Alfort, Maisons-Alfort, France (now the ANSES laboratory). According to Sangula et al. (2011), serotype C was first recognised in Kenya in 1957, with outbreaks reported on a regular basis thereafter, mainly in cattle, until 1985. After this time, the number of outbreaks were few and restricted to two districts only. A single vaccine strain, K267/67 produced by the Kenya Veterinary Vaccines Production Institute, has been used. Phylogenetic analysis of seven serotype C isolates obtained from outbreaks that occurred in Kenya between 1992 and 2004 indicated that all were extremely similar and probably represent vaccine escapes or laboratory contamination (Sangula et al. 2011).
Elsewhere in Africa, outbreaks due to serotype C were confirmed in Ethiopia in 1957, 1971, and 1983 (Ayelet et al. 2009); the 1957 isolate is no longer available. Serotype C was also identified in Uganda in 1970–71, but subsequent reports of outbreaks between 1972 and 1987, as far as we are aware, were not confirmed by laboratory isolation and serotyping. Serotype C outbreaks occurred in Tunisia in 1965, 1967, and 1969; however, these samples are no longer available for sequencing. Serotype C was also recorded in Angola in 1973 and sequencing studies showed this isolate as clustering with South American viruses, suggesting a likely South American origin despite its very low identity to the available South American viruses (Knowles and Samuel 2003; Fig. 3). Subsequent reported outbreaks in 1974–78 were not confirmed by laboratory serotyping.
A recent serological survey of Eritrean cattle found an unexpectedly high level of serotype C reactors, considering that the virus had never been isolated from outbreaks in that country (Tekleghiorghis et al. 2014). Possible explanations suggested were that undisclosed infection had occurred or else that the results reflected cross-serotype reactions that may have been exacerbated by infections with more than one serotype. Serotype C reactor African buffalo were also detected in Central and West Africa from samples collected from wildlife between 1993 and 2003, during the rinderpest eradication campaign, including a few animals with higher virus neutralising antibody titres to serotype C than other serotypes (Di Nardo et al. 2015). In another study Ouagal et al. (2018), failed to detect antibodies to serotypes C and Asia 1 (with the exception of marginal cross-reactivity) in blood samples collected in Chad in 2010, while the presence of antibodies to four other FMDV serotypes (O, A, SAT 1, and SAT 2) was described. Since serotype C virus has never been recorded in the countries concerned, undisclosed infection was considered a less likely explanation than cross-serotype serological reactivity.
3.6 Likely origins of serotype C
The concentration of serotype C viruses in Europe and South America versus the relative lack of occurrence and persistence in Africa and Asia points towards it originating in one of the former continents. If the serotype was best adapted to conditions in Europe or South America, then this would have contributed to its disappearance when these continents became wholly or nearly FMD-free, along with failure to persist in other parts of the world, where it was less suited, even those where control measures have been less rigorous. Even in Europe and South America, serotype C was less prevalent than serotypes O and A pointing to a relatively lower fitness. Conventional wisdom has been that serotype C was taken to South America along with serotypes O and A. However, dating of the tMRCA of serotype C viruses to the late 19th Century (∼1873) could mean that this serotype evolved either in Europe or in South America. Since most diversification is evident in South America, it cannot be excluded that type C viruses evolved initially there, following the introduction of serotype A, to which it is most closely related based on VP1 sequence data (Fig. 3, Table 1). If as a result of undersampling, other serotype C lineages that became extinct have not been represented in our analysis, then the dating of the tMRCA of serotype C viruses might be pushed further back in time, making a South American origin less likely. However, excluding two early clades that were sampled but went extinct (GER/c32 and UKG/149/34; Leticia/COL/67 and Leticia/COL/70), the result of the tMRCA is unchanged at 1870 (95% BCI 1807–1921). This is because the FMDV clock is strict and the evolution linear, so removing early extinct isolates does not change the slope of the root-to-tip versus time regression line, and the estimated molecular clocks are almost identical for these two scenarios (2.85 × 10−3 versus 2.86 × 10−3 nt substitutions/site/year for the truncated and full trees, respectively).
3.7 Risk of re-introduction from vaccines
The phylogenetic reconstruction of serotype C shows instances of outbreaks that appear to have been caused by vaccine viruses (Fig. 3). Beck and Strohmaier (1987) reported that most of the outbreaks in Europe in the 1980s were caused by viruses closely related to strains which had been circulating more than 20 years before and subsequently used for the production of vaccines. In 1985, speaking about the progress of FMD eradication in Western Europe, Bachrach (1985) stated that ‘the presence of live virus in some batches of vaccine and the escape of virus from vaccine from manufacturing facilities are now responsible for a large proportion of the outbreaks that still occur there’. This makes the point that once a pathogen is very rare, the risk–benefit of FMD vaccination changes markedly from when it was common.
Although the safety of FMD vaccine manufacture and vaccines have improved in many parts of the world, current vaccine manufacture still relies on in vitro culture of FMD virus on a large-scale, followed by inactivation prior to formulation and use. Biocontainment standards for vaccine production have been regularly revised and enhanced but local enforcement standards may vary. Despite the precautions taken, suspected or actual escapes of FMDV from facilities where vaccines were prepared (Germany 1987; Russia 1993 and 2016; UK 2007) suggest that this risk is hard to completely eliminate (Valarcher et al. 2008; Cottam et al. 2008; OIE/FAO Ref Lab Network Report 2016), and risks are higher if live virus challenge studies are undertaken to monitor the performance and potency of FMDV vaccines.
The relative contributions to past virus escape from vaccine manufacture versus use is not clear, although it has been noted that most vaccines connected with European outbreaks were prepared by formaldehyde inactivation, which can give rise to residual infection in finished vaccines (Brown 1991). The risk of residual FMDV infectivity in vaccines has been greatly reduced by the use of two cycles of inactivation with binary ethyleneimine instead of formaldehyde (Bahnemann 1990; Brown 1991). Efforts continue to develop safer production methods based on use of highly attenuated FMDV vaccine (Arzt et al. 2017) or recombinant technology that does not require live FMDV (Porta et al. 2013). A recent paper even described the creation of a recombinant serotype C vaccine with reduced virulence that could be used to produce vaccine more safely for vaccine bank reserves (You et al. 2019). Until such products are licensed and commercially available, the risk of continuing with production, let alone use of serotype C vaccines, is hard to justify, considering the low likelihood that animals will be challenged with serotype C FMDV in the field. In South American countries where there have been prolonged periods of FMD freedom with ongoing vaccination, there is already a desire to switch to a non-vaccination policy for all the serotypes.
3.8 Evidence for absence
Although caution is needed in declaring a serotype extinct, it is now many years since the last reported cases of serotype C infection in each of the different continents where it has occurred: 1989 (Europe), 1996 (Asia) and 2004 (South America and Africa). Moreover, the most recent cases in Kenya have been attributed to either use of vaccine or misdiagnosis, rather than virus persistence in the field.
FMD endemicity, such as occurs in parts of Africa, is often linked to insensitive systems of surveillance (including lack of resources and incentives for investigation or reporting of cases). Under these conditions a specific serotype might be missed. However, over several years, even with highly sub-optimal sampling, the likelihood of detection for persisting viruses will increase substantially. Furthermore, in the absence of control measures, cycles of emerging strains occur, linked to the waxing and waning of immunity and associated with periodic epidemics that will increase the likelihood of investigation, sampling and characterisation of the virus(es) involved. Research is warranted to improve the technologies that support FMD surveillance in endemic regions, both to check for serotype C as well as to improve risk management for other serotypes. The coverage of virological testing could be broadened by non-invasive sampling, for example collection of pooled milk (Armson et al. 2019) or the swabbing of high-risk environments (Colenutt et al. 2018), followed by RT-PCR testing with primers developed to specifically detect serotype C. Random nasal swabbing and testing of animals at markets could also be performed (Klein et al. 2008).
Serological testing for serotype C has given equivocal results as the finding of seroreactors could be explained either by virus circulation, post-vaccination responses or by cross-serotype reactions. The latter may be predisposed by infection with multiple serotypes, as commonly occurs in Africa, boosting antibody responses to shared epitopes. There is a variable degree of antigenic diversity within serotypes that can give rise to incomplete cross-protection and five serological subtypes of FMDV were defined for serotype C (Davie 1962). Although less well studied than some other serotypes, later work mapped the main antigenic sites on the serotype C capsid surface and their contributions to antigenic diversity (Lea et al. 1994; Mateu et al. 1994; Grazioli et al. 2013). The relative strength of the test result for a given serotype may also be influenced by the antigenic match between the test viruses and the immunogens (field viruses or vaccines) that gave rise to the antibody response. Therefore, seroreactivity may need to be measured against more than one representative of a given serotype. Further research to better understand the basis for cross-serotype antibody reactions, may lead to more specific serosurveillance tools as well as being needed to improve the cross-protection afforded by vaccines. Modelling approaches can help in interpreting cross-reactivity findings from comparative serology where complex serological patterns are found in endemic settings (Casey-Bryars et al. 2018). As for other serosurveys, results can be better interpreted if there is good metadata collected at the time that animals are sampled, so that follow-up can assess spatial and temporal clustering of positive animals and epidemiological units and tracing and testing of contacts can be conducted.
FMDV may also persist undetected where there is partial control by vaccination, such that the disease is masked but the infection is not eradicated. Sequencing suggests that this situation may have pertained in South America, where the 2004 outbreaks of serotype C resulted from undisclosed virus circulation over a nine-year period under conditions of incomplete vaccination and weak surveillance associated with the undeveloped veterinary services at the time in the estates of the Amazon basin (Sanchez-Vazquez et al. 2018). There have been similar examples, with other FMD serotypes, of FMD recrudescence from within South America, for example the outbreaks of serotype O in Paraguay (2002, 2003, 2011, and 2012), Brazil in 2005 and Argentina 2006 (Naranjo and Cosivi 2013). However, since this time, Brazil has moved to complete eradication of FMD in its entire territory, including the Amazonas region, now recognised as FMD-free with vaccination by OIE. Moreover, continental South America has not reported outbreaks of FMD since January of 2012, apart from recent outbreaks due to serotype O in the North of the Andean region near the borders between Colombia and Venezuela. Meanwhile, the yearly sero-surveys to rule out the presence of FMD transmission contribute to demonstration of the absence of this and other serotypes in vaccinated herds.
Therefore, notwithstanding the need for caution, after fifteen years without a reported case, it appears that serotype C is no longer circulating in livestock and unlike the SAT serotypes, serotype C virus has never been shown to persist in non-domestic species. In view of this epidemiological situation, several specific recommendations were made concerning the management of FMDV serotype C (Roeder and Knowles 2008, the OIE/FAO FMD Laboratory Network 2015; the OIE General Session 2017) endorsed a resolution to highlight the importance of sharing laboratory and field data regarding serotype C and to recommend that the continued use of this serotype in FMDV vaccines is halted (apart for maintenance of stocks in antigen banks). Researchers were also encouraged to participate in surveillance activities for FMD serotype C at the international level, and this review was written to help understand the history of this serotype and frame research priorities for work that should now be undertaken, which mostly relate to better surveillance tools and safer vaccines.
Lessons on mitigating the possibility of iatrogenic release of an extinct virus may be learnt from the efforts taken following rinderpest eradication. In this case, facilities that meet stringent biosecurity requirements have been designated for holding residual stocks of rinderpest virus or rinderpest vaccine, whilst other stocks have been destroyed (Myers et al. 2018). It is therefore recommended to discontinue the manufacture, testing and use of serotype C vaccines, to take stock of global repositories and to only manipulate the virus in vitro where risk assessment provides sufficient justification and where facilities conform to a high level of biocontainment, such as EU BSL3+ (EuFMD Commission 2013).
Supplementary Material
Acknowledgements
The authors acknowledge the support of other members of the OIE/FAO Reference Laboratory Network for foot-and-mouth disease and the contributions of Aldo Dekker, Rossana Allende, Abraham Sangula and Graham Belsham at the Annual meeting of the Network in 2015. The purpose of the Network is to make available accurate and timely data to support global surveillance and control of FMD (https://www.foot-and-mouth.org/Ref-Lab-Network; last accessed 29 Jan 2021).
Funding
Work of the OIE/FAO Reference Laboratory Network for foot-and-mouth disease is supported by the European Union (via a contracted project granted from the European Commission for the control of Foot-and-Mouth Disease, EuFMD). The views expressed herein can in no way be taken to reflect the official opinion of the European Union. Sequencing work at The Pirbright Institute was funded by the Department for Environment, Food and Rural Affairs (Defra), United Kingdom [research grant SE2944]. The Pirbright Institute receives grant-aided support from the Biotechnology and Biological Sciences Research Council of the United Kingdom [projects BBS/E/I/00007035, BBS/E/I/00007036].
Conflict of interest
None declared.
References
- Armson B. et al. (2019) ‘Opportunities for Enhanced Surveillance of Foot-and-Mouth Disease in Endemic Settings Using Milk Samples’, Transboundary and Emerging Diseases, 66: 1405–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auton A., McVean G. (2007) ‘Recombination Rate Estimation in the Presence of Hotspots’, Genome Research, 17: 1219–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arzt J. et al. (2017) ‘Pathogenesis of Virulent and Attenuated Foot-and-Mouth Disease Virus in Cattle’, Virology Journal, 14: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayelet G. et al. (2009) ‘Genetic Characterization of Foot-and-Mouth Disease Viruses, Ethiopia, 1981-2007’, Emerging Infectious Diseases, 15: 1409–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachanek-Bankowska K. et al. (2018) ‘Reconstructing the Evolutionary History of Pandemic Foot-and-Mouth Disease Viruses: The Impact of Recombination within the Emerging O/ME-SA/Ind-2001 Lineage’, Scientific Reports, 8: 14693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahnemann H. B. (1990) ‘Inactivation of Viral Antigens for Vaccine Preparation with Particular Reference to the Application of Binary Ethylenimine’, Vaccine, 8: 299–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachrach H. L. (1985) ‘Foot-and-Mouth Disease and Its Antigens’, Advances in Experimental Medicine and Biology, 185: 27–46. [DOI] [PubMed] [Google Scholar]
- Bhattacharya S. et al. (2005) ‘Studies of the Outbreaks of Foot and Mouth Disease in West Bengal, India, between 1985 and 2002’, Revue Scientifique et Technique (International Office of Epizootics), 24: 945–52. [PubMed] [Google Scholar]
- Beck E., Strohmaier K. (1987) ‘Subtyping of European Foot-and-Mouth Disease Virus Strains by Nucleotide Sequence Determination’, Journal of Virology, 61: 1621–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann I. E. et al. (1988) ‘Serological and Biochemical Analysis of Foot-and-Mouth Disease Virus (Serotype C3) Isolated in Argentina between 1981 and 1986’, Vaccine, 6: 245–52. [DOI] [PubMed] [Google Scholar]
- Brooksby J. B. (1958) ‘The Virus of Foot-and-Mouth Disease’, Advances in Virus Research, 5: 1–37. [DOI] [PubMed] [Google Scholar]
- Brooksby J. B., Rogers J. (1957) ‘Methods Used in Typing the Virus of Foot-and-Mouth Disease at Pirbright, 1950–55’, in Methods of Typing and Cultivation of Foot-and-Mouth Disease Virus: Project No. 208. Paris: European Productivity Agency of the Organization of European Cooperation (OEEC).
- Brown F. (1991) ‘An Overview of the Inactivation of FMDV and the Implications When Residual Virus is Present in Vaccines’, Developments in Biological Standardization, 75: 37–41. [PubMed] [Google Scholar]
- Casey-Bryars M. et al. (2018) ‘Waves of Endemic Foot-and-Mouth Disease in Eastern Africa Suggest Feasibility of Proactive Vaccination Approaches’, Nature Ecology & Evolution, 2: 1449–57. [DOI] [PubMed] [Google Scholar]
- Colenutt C. et al. (2018) ‘Evaluation of Environmental Sampling as a Low Technology Method for Surveillance of Foot-and-Mouth Disease Virus in an Endemic Area’, Applied and Environmental Microbiology, 84: e00686–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottam E. M. et al. (2008) ‘Transmission Pathways of Foot-and-Mouth Disease Virus in the United Kingdom In’, PLoS Pathogens, 4: e1000050–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa Melo E. (2004) ‘Outbreaks of FMD Type C in Amazonas, Brazil’, in Organisation for European Economic Co-operation. EMPRES Transboundary Animal Disease Bulletin No 26, pp. 31-34. Food and Agriculture Organization of the United Nations (FAO), Rome, Italy. [Google Scholar]
- Davie J. (1962) ‘The Classification of Subtype Variants of the Virus of Foot-and-Mouth Disease’, Bulletin de L'Office International Des Epizooties, 57: 962–7. [Google Scholar]
- Di Nardo A. et al. (2014) ‘Phylodynamic Reconstruction of O CATHAY Topotype Foot-and-Mouth Disease Virus Epidemics in the Philippines’, Veterinary Research, 45: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Nardo A. et al. (2015) ‘Serological Profile of Foot-and-Mouth Disease in Wildlife Populations of West and Central Africa with Special Reference to Syncerus Caffer Subspecies’, Veterinary Research, 46: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson A. I., Ferris N. P., Gloster J. (1982) ‘Air Sampling of Pigs Infected with Foot-and-Mouth Disease Virus: Comparison of Litton and Cyclone Samplers’, Research in Veterinary Science, 33: 384–5. [PubMed] [Google Scholar]
- Drummond A. J. et al. (2006) ‘Relaxed Phylogenetics and Dating with Confidence’, PLoS Biology, 4: e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EuFMD Commission (2013) ‘Minimum biorisk management standards for laboratories working with foot-and-mouth disease virus’ in Proceedings of the 40th General Session of the EuFMD Commission, 22–24 April 2013, Rome, Italy. <http://www.fao.org/fileadmin/user_upload/eufmd/Lab_guidelines/FMD_Minimumstandards_2013_Final_version.pdf> accessed 3 Feb 2021.
- Ferris N. P., Donaldson A. I. (1992) ‘The World Reference Laboratory for Foot and Mouth Disease: A Review of Thirty-Three Years of Activity (1958-1991)’, Revue’, Revue Scientifique et Technique de l'OIE, 11: 657–84. [DOI] [PubMed] [Google Scholar]
- Fogedby E. (1963) ‘Review of epizootiology and control of foot-and-mouth disease in Europe 1937-1961’, in Proceedings of the 10th Session of the European Commission for the Control of FMD, 17-19 April 1963, Rome, Italy. <http://www.fao.org/3/ca8527en/ca8527en.pdf> accessed 3 Feb 2021.
- Freimanis G. L. et al. (2016) ‘Genomics and Outbreaks: Foot and Mouth Disease’, Revue Scientifique et Technique de l'OIE, 35: 175–89. [DOI] [PubMed] [Google Scholar]
- Gill M. S. et al. (2013) ‘Improving Bayesian Population Dynamics Inference: A Coalescent-Based Model for Multiple Loci’, Molecular Biology and Evolution, 30: 713–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno E. J. (1967) ‘Informe Sobre el Caso de Fiebre Aftosa Aparecido en Tierra Del Fuego en el Mes de Diciembre de 1966 [Study on a Case of Foot-and-Mouth Diseases Occurring in Tierra Del Fuego in the Month of December 1966]’, Bulletin de L'Office International Des Epizooties, 68: 555–64.[Spanish]. [PubMed] [Google Scholar]
- Grazioli S. et al. (2013) ‘Mapping of Antigenic Sites of Foot-and-Mouth Disease Virus Serotype Asia 1 and Relationships with Sites Described in Other Serotypes’, Journal of General Virology, 94: 559–69. [DOI] [PubMed] [Google Scholar]
- Guindon S. et al. (2010) ‘New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3’, Systematic Biology, 59: 307–21. [DOI] [PubMed] [Google Scholar]
- Hemadri D. et al. (2003) ‘Serotype C Foot-and-Mouth Disease Virus Isolates from India Belong to a Separate so Far Not Described Lineage’, Veterinary Microbiology, 92: 25–35. [DOI] [PubMed] [Google Scholar]
- Katoh K., Standley D. M. (2013) ‘MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability’, Molecular Biology and Evolution, 30: 772–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein J. et al. (2008) ‘Epidemiology of Foot-and-Mouth Disease in Landhi Dairy Colony, Pakistan, the World Largest Buffalo Colony’, Virology Journal, 5: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles N. J., Samuel A. R. (2003) ‘Molecular Epidemiology of Foot-and-Mouth Disease Virus’, Virus Research, 91: 65–80. [DOI] [PubMed] [Google Scholar]
- Knowles N. J. et al. (2016) ‘VP1 Sequencing Protocol for Foot and Mouth Disease Virus Molecular Epidemiology’, Revue Scientifique et Technique de l'OIE, 35: 741–55. [DOI] [PubMed] [Google Scholar]
- Konig G. et al. (2001) ‘Phylogenetic Analysis of Foot-and-Mouth Disease Viruses Isolated in Argentina’, Virus Genes, 23: 175–82. [DOI] [PubMed] [Google Scholar]
- Lea S. et al. (1994) ‘The Structure and Antigenicity of a Type C Foot-and-Mouth Disease Virus’, Structure, 2: 123–39. [DOI] [PubMed] [Google Scholar]
- Lemey P. et al. (2009) ‘Bayesian Phylogeography Finds Its Roots’, PLoS Computational Biology, 5: e1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado M. A. Jr. (1969) AFTOSA: A Historical Survey of Foot-and-Mouth Disease and Inter-American Relations. Albany: State University of New York Press. [Google Scholar]
- Martínez M. A. et al. (1992) ‘Evolution of the Capsid Protein Genes of Foot-and-Mouth Disease Virus: Antigenic Variation without Accumulation of Amino Acid Substitutions over Six Decades’, Journal of Virology, 66: 3557–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateu M. G. et al. (1994) ‘Antigenic Heterogeneity of a Foot-and-Mouth Disease Virus Serotype in the Field is Mediated by Very Limited Sequence Variation at Several Antigenic Sites’, Journal of Virology, 68: 1407–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowat G. N. (1987) ‘Advances in the Epidemiology of Foot-and-Mouth Disease in the United Kingdom since ’, State Veterinary Journal, 41: 121–41. [Google Scholar]
- Myers L. et al. (2018) Global Rinderpest Action Plan: Post-Eradication. Rome: Food and Agriculture Organization of the United Nations (FAO) and World Organisation for Animal Health (OIE). [Google Scholar]
- Nagendrakumar S. B. et al. (2005) ‘Molecular Characterization of Foot-and-Mouth Disease Virus Type C of Indian Origin’, Journal of Clinical Microbiology, 43: 966–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naranjo J., Cosivi O. (2013) ‘Elimination of Foot-and-Mouth Disease in South America: Lessons and Challenges’ Philosophical ’, Philosophical Transactions of the Royal Society B: Biological Sciences, 368: 20120381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OIE/FAO FMD Laboratory Network (2015) ‘Research priorities to provide evidence that serotype C is no longer circulating’, in King D.P. (ed.) Proceedings of the 10th OIE/FAO FMD Laboratory Network Meeting, 24–26 November, Brussels, Belgium. <https://www.wrlfmd.org/sites/world/files/quick_media/OIE-FAO%20FMD%20Ref%20Lab%20Network%20Report%202015.pdf> accessed 3 Feb 2021.
- OIE/FAO FMD Laboratory Network. (2016). ‘Annual Report 2016’, in Donald King and Mark Henstock (eds), The Pirbright Institute, UK. <https://www.foot-and-mouth.org/publications/oiefao-fmd-laboratory-network-2016>accessed 3 Feb 2021.
- OIE Terrestrial Animal Health Code (2019) ‘Chapter 8.8. Infection with Foot and Mouth Disease Virus’. <https://www.oie.int/standard-setting/terrestrial-code/access-online/> accessed 3 Feb 2021.
- OIE Resolution (2017). ‘Resolutions Adopted by the World Assembly of OIE Delegates during Their 85th General Session, Resolution No. 30, Foot and Mouth Disease Serotype C’, p 50, 21–26 May 2017. <https://www.oie.int/fileadmin/Home/eng/About_us/docs/pdf/Session/2017/A_RESO_2017_Public.pdf> accessed 29 Jan 2021.
- Ouagal M. et al. (2018) ‘Study on Seroprevalence and Serotyping of Foot and Mouth Disease in Chad’, Revue Scientifique et Technique de l'OIE, 37: 937–47. [DOI] [PubMed] [Google Scholar]
- PANAFTOSA-OPS/OMS (2019) Informe de Situación de los Programas de Erradicación de la Fiebre Aftosa. Sudamerica y Panamá en 2018. Rio de Janeiro, Brazil. <https://iris.paho.org/handle/10665.2/51789> accessed 29 Jan 2021. [Google Scholar]
- Paradis E., Schliep K. (2019) ‘Ape 5.0: An Environment for Modern Phylogenetics and Evolutionary Analyses in R’, Bioinformatics, 35: 526–8. [DOI] [PubMed] [Google Scholar]
- Pereira H. G. (1977) ‘Subtyping of Foot-and-Mouth Disease Virus’, Developments in Biological Standardization, 35: 167–74. [PubMed] [Google Scholar]
- Pond S. L. K. et al. (2006) ‘GARD: A Genetic Algorithm for Recombination Detection’, Bioinformatics, 22: 3096–8. [DOI] [PubMed] [Google Scholar]
- Porta C. et al. (2013) ‘Rational Engineering of Recombinant Picornavirus Capsids to Produce Safe, Protective Vaccine Antigen’, PLoS Pathogens, 9: e1003255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core Team R., (2020). R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing. <https://www.R-project.org/> accessed 3 Feb 2021. [Google Scholar]
- Ranaweera L. T. et al. (2019) ‘Transboundary Movements of Foot-and-Mouth Disease from India to Sri Lanka: A Common Pattern is Shared by Serotypes O and C’, PLoS One, 14: e0227126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder P. L., Knowles N. J. (2008) ‘Foot-and-Mouth Disease Virus Type C Situation–The First Target for Eradication?’, in Proceedings of the Global Control of FMD–Tools, Ideas and Ideals, 14–17 Oct. Erice, Italy. <http://www.fao.org/ag/againfo/commissions/docs/research_group/erice/APPENDIX_07.pdf> accessed 29 Jan 2021.
- Rweyemamu M. et al. (2008) ‘Epidemiological Patterns of Foot-and-Mouth Disease Worldwide’, Transboundary and Emerging Diseases, 55: 57–72. [DOI] [PubMed] [Google Scholar]
- Sanchez-Vazquez M. J. et al. (2019) ‘Investigating the Temporal and Spatial Distribution of Foot-and-Mouth Disease Virus Serotype C in the Region of South America, 1968-2016’, Transboundary and Emerging Diseases, 66: 653–61. [DOI] [PubMed] [Google Scholar]
- Sangula A. K. et al. (2011) ‘Low Diversity of Foot-and-Mouth Disease Serotype C Virus in Kenya: Evidence for Probable Vaccine Strain Re-Introductions in the Field’, Epidemiology and Infection, 139: 189–96. [DOI] [PubMed] [Google Scholar]
- OIE/FAO FMD Laboratory Network Report (2019) ‘Annual Report 2019’ in Donald King, Antonello Di Nardo and Mark Henstock (eds), The Pirbright Institute, UK.
- Stamatakis A. (2014) ‘RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies’, Bioinformatics, 30: 1312–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchard M. A. et al. (2018) ‘Bayesian Phylogenetic and Phylodynamic Data Integration Using BEAST 1’, Virus Evolution, 4: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutmoller P. et al. (2003) ‘Control and Eradication of Foot-and-Mouth Disease. A Review’, Virus Research, 91: 101–44. [DOI] [PubMed] [Google Scholar]
- Tekleghiorghis T. et al. (2014) ‘Serological Evidence Indicates That Foot-and-Mouth DiseaseVirus Serotype O, C and SAT1 Are Most Dominant in Eritrea’, Transboundary and Emerging Diseases, 61: e83-8–e88. [DOI] [PubMed] [Google Scholar]
- Thomson G. R., Bastos A. D. S. (2005) ‘Foot-and-Mouth Disease’, in Coetzer J. A. W., Tustin R. C (eds.) Infectious Diseases of Livestock,Vol 2, pp.1324–65. Oxford: Oxford University Press. [Google Scholar]
- Tully D. C., Fares M. A. (2008) ‘The Tale of a Modern Animal Plague: Tracing the Evolutionary History and Determining the Time-Scale for Foot and Mouth Disease Virus’, Virology, 382: 250–6. [DOI] [PubMed] [Google Scholar]
- Valarcher J. F. et al. (2008) ‘Incursions of Foot-and-Mouth Disease Virus into Europe between 1985 and 2006’, Transboundary and Emerging Diseases, 55: 14–34. [DOI] [PubMed] [Google Scholar]
- Vallée H., Carré H. (1922) ‘Sur la Pluralité du Virus Aphteux’ Comptes Rendus de L'Académie Des Sciences’, French, 174: 1498–500. [Google Scholar]
- Waldmann O., Trautwein K. (1926) ‘Experimentalle Untersuchungen Ueber Die Pluralitet Des Maul-Und Klauenseuche Virus’, Berlin Tierarztl Wochenschr, 42: 569–71.[German]. [Google Scholar]
- You S. H. et al. (2019) ‘Evaluation of Novel Inactivated Vaccine for Type C Foot-and-Mouth Disease in Cattle and Pigs’, Veterinary Microbiology, 234: 44–50. [DOI] [PubMed] [Google Scholar]
- Yu G. et al. (2017) ‘Ggtree: An R Package for Visualization and Annotation of Phylogenetic Trees with Their Covariates and Other Associated Data’, Methods in Ecology and Evolution, 8: 28–36. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.