Abstract
The zoonotic SARS-CoV-2 virus that causes COVID-19 continues to spread worldwide, with devastating consequences. While the medical community has gained insight into the epidemiology of COVID-19, important questions remain about the clinical complexities and underlying mechanisms of disease phenotypes. Severe COVID-19 most commonly involves respiratory manifestations, although other systems are also affected, and acute disease is often followed by protracted complications. Such complex manifestations suggest that SARS-CoV-2 dysregulates the host response, triggering wide-ranging immuno-inflammatory, thrombotic, and parenchymal derangements. We review the intricacies of COVID-19 pathophysiology, its various phenotypes, and the anti-SARS-CoV-2 host response at the humoral and cellular levels. Some similarities exist between COVID-19 and respiratory failure of other origins, but evidence for many distinctive mechanistic features indicates that COVID-19 constitutes a new disease entity, with emerging data suggesting involvement of an endotheliopathy-centred pathophysiology. Further research, combining basic and clinical studies, is needed to advance understanding of pathophysiological mechanisms and to characterise immuno-inflammatory derangements across the range of phenotypes to enable optimum care for patients with COVID-19.
This is the first in a Series of four papers about COVID-19
Introduction
The emergence of SARS-CoV-2 has resulted in a health crisis not witnessed since the 1918–19 Spanish influenza pandemic. The most plausible origin of SARS-CoV-2 is natural selection of the virus in an animal host followed by zoonotic transfer.1 After the first cases of COVID-19 were identified in Wuhan, China, in December, 2019, the virus spread rapidly and had been reported in 220 countries as of April 25, 2021.2 Among the countries most affected by COVID-19 so far are the USA, Brazil, Mexico, and India (totalling >1·3 million deaths and >65 million infections by April 25, 2021).3 Despite the markedly lower fatality rate of COVID-19 compared with the previous severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS) coronavirus epidemics, its dissemination has had devastating effects on health systems and national economies worldwide.
We are only beginning to understand the dynamics of SARS-CoV-2 infectivity and transmissibility—a pressing goal with the emergence of new variants of concern—and the clinical intricacies of multiple COVID-19 phenotypes. Although acute respiratory manifestations are the most common feature of severe COVID-19, many non-respiratory effects have been reported in the acute phase of the disease, and emerging evidence points to various long-lasting complications after SARS-CoV-2 infection (the post-COVID syndrome or long COVID).4, 5, 6 Such a complex clinical picture suggests that SARS-CoV-2 generates a dysregulated host response to infection, including wide-ranging immuno-inflammatory derangements. Understanding of the pathophysiology and phenotypes of COVID-19, including the host response to SARS-CoV-2, will be key to developing personalised management strategies for patients.
The response of the medical and scientific communities has been unprecedented. An extraordinary number of research reports related to SARS-CoV-2 and COVID-19 has been published over the past year, including impressive advances in understanding but also information of dubious quality.7 With a desire to help patients quickly and to curb the pandemic, some decisions have been politicised or made hastily on the basis of anecdotal or poorly verified evidence (eg, on the use of ibuprofen, hydroxychloroquine, angiotensin-converting enzyme [ACE] inhibitors, angiotensin receptor blockers, and lopinavir–ritonavir), provoking confusion among medical personnel and public mistrust.8, 9, 10, 11 In the research context, these challenges have potentially been aggravated by viewing COVID-19 in terms of pre-existing pathophysiological concepts, which could lead to the generation of biased hypotheses, misleading findings, and unjustified clinical decision making that does not benefit and, at worst, harms patients with COVID-19. For example, controversy about the pathophysiology of SARS-CoV-2-associated lung failure—and the use of anti-inflammatory treatments in the face of a presumed cytokine storm—has been fuelled by the sepsis research legacy.
Key messages.
-
•
Understanding of the pathophysiology and the clinical and mechanistic phenotypes of COVID-19 will be key to developing personalised management strategies and improving outcomes for patients
-
•
Presentations of SARS-CoV-2 infection range from asymptomatic, to mild or moderate respiratory and non-respiratory symptoms, to severe COVID-19 pneumonia and ARDS with multiorgan failure; emerging evidence also points to various long-lasting complications after SARS-CoV-2 infection (the post-COVID syndrome or long COVID)
-
•
The complex clinical picture suggest that SARS-CoV-2 elicits a host response that triggers wide-ranging immuno-inflammatory, thrombotic, and parenchymal derangements
-
•
SARS-CoV-2-induced endotheliitis might be a common factor in the respiratory and non-respiratory manifestations of COVID-19
-
•
The systemic inflammatory response to SARS-CoV-2 infection seems to be relatively mild compared with that of non-COVID-19-associated ARDS or severe bacterial infections
-
•
The association between viral load and disease severity suggests that poor viral infection control by the immune system is a major pathogenic contributor to severe COVID-19
-
•
COVID-19 is a new disease entity with a pathophysiology distinct from that of influenza, non-COVID-19-related ARDS, and other coronavirus infections
ARDS=acute respiratory distress syndrome.
To implement optimum management strategies and improve outcomes for patients, recognition of the similarities in pathophysiology and phenotypes—where these exist—between COVID-19 and other, better known conditions is needed, along with understanding of the distinctive features that set this new disease apart from other entities. In this Series paper, we aim to provide a comprehensive and critical summary of available evidence on the pathophysiology of COVID-19 for clinicians and clinical scientists. We review disease mechanisms that underpin the respiratory manifestations of COVID-19, with a consideration of the similarities and differences between COVID-19 and respiratory diseases of other causes. We discuss the potential role of a distinctive endotheliopathy-centred pathophysiology in the respiratory and non-respiratory manifestations of COVID-19, and consider features of the host response to SARS-CoV-2 at the humoral and cellular levels. A complex array of immuno-inflammatory mechanisms underlies the range of phenotypes in COVID-19, but many uncertainties remain. We conclude by identifying key questions for future research.
Clinical pathology of COVID-19
Comparison of SARS, MERS, and COVID-19
SARS-CoV-2 belongs to a large family of coronaviruses that includes seven human pathogens, four of which cause common colds. SARS-CoV-2 has many similarities to SARS-CoV, which was responsible for the 2003 SARS outbreak that began in China,12 and MERS-CoV, responsible for the 2012 MERS outbreak that was first reported in Saudi Arabia.13 A comparison of key features of these three coronaviruses is presented in the table . SARS-CoV and SARS-CoV-2 both use the human ACE2 receptor for viral entry and cell tropism for infectivity. Although SARS-CoV-2 primarily targets the lung epithelial cells, the intestinal and other epithelia can be also be infected, with active replication and de-novo production of infective virus.17, 18 The role of enteric replication in COVID-19 is not fully understood, but it has been suggested to exacerbate the inflammatory response.19 In MERS, blood immune cells can be infected, enabling further viral replication,20 whereas in SARS, infection of circulating immune cells is abortive (ie, no replication and cell death).21, 22 The details of infection and lytic replication mechanisms of SARS-CoV-2 in immune cells are currently unclear, with two reports describing direct monocyte infection.23, 24
Table.
Comparison of key features of SARS-CoV, MERS-CoV, and SARS-CoV-2
| SARS-CoV | MERS-CoV | SARS-CoV-2 | |
|---|---|---|---|
| Year of outbreak | 2003 | 2012 | 2019 |
| Number of countries affected | 29 | 27 | 220 as of April 25, 20212 |
| Confirmed cases | 8096 | 2538 | >149 million as of April 25, 20213 |
| Confirmed deaths (mortality) | 744 (9·2%) | 871 (34%) | >3·1 million (2·2%)* as of April 25, 20213 |
| Receptor for viral entry | ACE2 | DPP4 | ACE2 |
| Cells susceptible to infection | Respiratory and intestinal epithelial cells; abortive infection in haematopoietic cells | Respiratory epithelial cells, activated T cells, monocytes, macrophages, and dendritic cells | Respiratory epithelial and intestinal epithelial cells, alveolar macrophages, cardiocytes, olfactory sustentacular cells, bile duct cells, and testicular Sertoli cells; abortive infection in haematopoietic cells |
| Upper respiratory tract viral replication | No | No | Yes |
| Lower respiratory tract viral replication | Yes | Yes | Yes |
| Lymphopenia | Yes: CD4+ and CD8+ T cells, B cells, natural killer cells | Yes: CD4+ and CD8+ T cells | Yes: CD4+ and CD8+ T cells, B cells, natural killer cells |
| Neutrophilia | Yes | Yes | Yes: elevated levels of NETosis |
| Monocytes and Macrophages | High levels | High levels | Counts decreased (but not significantly), especially for CD86+ HLA-DR+ monocytes |
| Systemically elevated cytokines | IL-6, IL-8, CXCL10 (IP10), IFN-γ, MIP-1α (CCL3), MCP1 (CCL2), IL-1, IL-12, CXCL9, TGF-β | IL-6, IL-8, CXCL10, IFN-γ, MIP-1α, MCP1, CCL5, IL-12 | IL-6, IL-8, CXCL10, IFN-γ, MIP-1α, MCP1, IL-2, IL-15, IL-1RA, IL-4, G-CSF, TNF, IL-10 |
| Presence of viral RNA in plasma | Yes; associated with critical illness and fatal outcome | Yes; associated with critical illness and fatal outcome | Yes; associated with critical illness and fatal outcome |
| Interferon response† | Persistent type I and type II interferon responses; ISG response in the most severe cases; defective expression of MHC class II and immunoglobulin-related genes | Infection induces repressive histone modifications that downregulate expression of specific ISGs | Delayed and dysregulated type I interferon response in severe cases; autoantibodies against type I interferons in a subset of patients |
| Viral load peak, days after symptom onset | ∼10 days | ∼10 days | 3–5 days |
| Humoral response: time to antibody detection, days (IQR) | 12 (8–15·2) | 16 (13–19) | 12 (10–15) |
| Duration of immunity | Long-lasting memory T-cell response; IgG levels decrease over time but have been detected 13 years after infection; association with severity inconclusive | Long-lasting memory T-cell response; IgG levels decrease over time but have been detected up to 3 years after infection; higher peak levels, seroconversion rates, and time to seroconversion in more severe disease | Unknown |
| Cross-reactivity of patient antibodies with other coronaviruses | Cross-reactivity with hCoV and MERS-CoV | Low level of cross-reactivity with hCoV and SARS-CoV | Cross-reactivity with SARS-CoV, but only rare neutralisation of live virus |
ACE2=angiotensin-converting enzyme 2. CCL=C-C motif chemokine. CXCL=C-X-C motif chemokine. DPP4=dipeptidyl peptidase 4. G-CSF=granulocyte colony-stimulating factor. hCoV=human coronaviruses. IFN-γ=interferon-γ. IL=interleukin. IL-1RA=interleukin-1 receptor antagonist. IP10=10 kDa interferon gamma-induced protein. ISG=interferon-stimulated gene. MCP1=monocyte chemotactic protein 1. MIP-1α=macrophage inflammatory protein-1α. NET=neutrophil extracellular trap. TGF-β=transforming growth factor-β. TNF=tumour necrosis factor.
Mortality for SARS-CoV-2 might be inaccurate because of insufficient recording of all cases, particularly asymptomatic cases.
Antagonism of the antiviral type I interferon response through virally encoded proteins is an immune evasion strategy used by all three coronavirus infections, although mediated by different mechanisms. Virally encoded proteins strongly suppress antiviral type I and type III interferon expression through shutdown of the translational machinery.14, 15 The underlying evasion strategies targeting the type III interferon pathway are currently unknown but appear to be strongest for SARS-CoV-2.16
The relatively wide transmissibility of SARS-CoV-2 is probably related to active viral replication in the upper airways in the pre-symptomatic and symptomatic phases.25, 26 In COVID-19, viral loads in respiratory samples peak earlier (3–5 days after symptom onset) than in MERS and SARS (table).25, 27, 28, 29, 30, 31 The quality and duration of protective immunity to SARS-CoV-2 is unclear. However, early re-infection is rare and protective immunity in rhesus macaques re-challenged with SARS-CoV-2 inoculation has been shown.32 Observational studies have demonstrated robust antibody and T-cell responses in a substantial proportion of patients after infection with SARS-CoV-2.33 However, the humoral response to SARS-CoV-2 appears to be proportional to COVID-19 severity: the highest antibody titres are detected in severe and protracted cases, compared with low or undetectable titres in patients with mild or asymptomatic COVID-19.34 The magnitude of the antibody response often correlates with T-cell responses, although uncoupled responses with strong specific T-cell production and no antibody production have been described.35 SARS, MERS, and COVID-19 have a similarly short-lived humoral antibody response.36, 37 In two studies of SARS survivors, anti-SARS-CoV antibody titres remained high for the first 2 years after infection, only 55% of patients had IgG antibodies after 3 years, and there was no detectable peripheral memory B-cell response after 6 years,38, 39 although a low IgG titre has been detected in some survivors 13 years after SARS infection.40 Regarding SARS-CoV-2, immunological memory has been shown to persist for more than 6 months.41 The precise duration of protective immunity against SARS-CoV-2 is difficult to predict, but T-cell immunity might be more durable: Le Bert and colleagues showed that T-cell reactivity for SARS-CoV persisted for 17 years after infection.42
The same authors also demonstrated cross-reactivity in T-cell immunity between SARS-CoV and SARS-CoV-2.42 Approximately 20–50% of tested populations show pre-existing T-cell responses to SARS-CoV-2 that are probably related to exposure to endemic human coronaviruses.43 A range of memory CD4+ T-cell clones cross-reacted with similar affinity to SARS-CoV-2 and four common cold coronaviruses (hCoV-OC43, hCoV-229E, hCOV-NL63, and hCoV-HKU1).44 Cross-reactive antibody-binding responses among several coronaviruses are known to exist; in patients with SARS, antibodies reactive to human coronaviruses, MERS-CoV, and SARS-CoV have been found.45, 46, 47 However, antibody cross-neutralisation of live viruses appears to be rare.48, 49 It will be crucial to define the role of pre-existing reactive T cells in COVID-19 by undertaking large-scale population studies and preclinical tests.
Pneumonia and lung damage
The leading symptom of COVID-19 pneumonia is hypoxaemia, which can worsen and progress to various stages of acute respiratory distress syndrome (ARDS), defined as an impairment of oxygenation—ie, a ratio of partial pressure of arterial oxygen (PaO2) to fractional concentration of oxygen in inspired air (FiO2) of 300 mm Hg or less. COVID-19 pneumonia in adults is characterised by clinical signs (fever, cough) and a respiratory rate of more than 30 breaths per min or respiratory distress evidenced by either impaired PaO2 relative to FiO2 or oxygen saturation (SpO2) <93%.50 About 80% of patients with COVID-19 develop mild-to-moderate disease, 15% progress to severe stages requiring oxygen support, and 5% develop critical disease including ARDS, septic shock, or multiorgan failure.51 Age, various comorbidities (eg, diabetes, obesity, and muco-obstructive lung and cardiovascular diseases), and some genetic polymorphisms correlate with a higher risk of respiratory failure.52, 53, 54
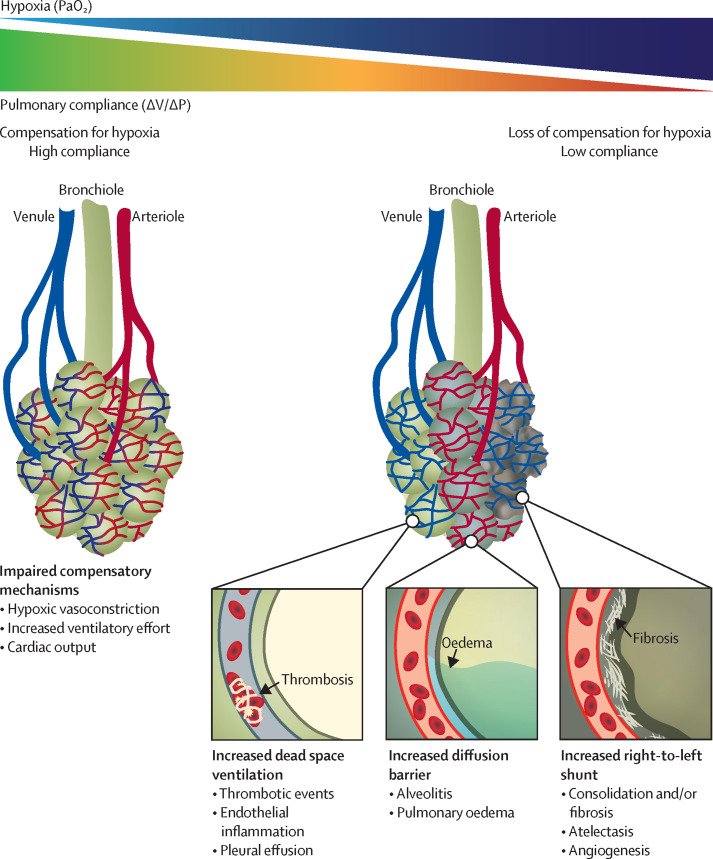
An unusual phenomenon in COVID-19 compared with other causes of respiratory failure is so-called silent hypoxaemia, characterised by critically low PaO2 but only mild respiratory discomfort and dyspnoea.55, 56 One study demonstrated shortness of breath in only about 19% of patients who had critical PaO2/FiO2 ratios.57 Pathophysiologically, hypoxaemia has only a limited role in the sensation of breathlessness,58 but the response to low PaO2 (<60 mm Hg) increases the respiratory drive, defined as the intensity of the neural stimulus to breathe via regulation of respiratory rate and depth (tidal volume, Vt). Therefore, tachypnoea and high Vt (but not necessarily dyspnoea) are signs of hypoxia, especially in COVID-19 pneumonia.57, 59 Understanding this concept is fundamental for clinical management; an excessive respiratory drive could lead to further deterioration of lung function in a vicious cycle of patient-inflicted lung injury, although this concept remains controversial.60 Several hypotheses have been proposed to explain hypoxia, including SARS-CoV-2-specific effects on oxygen receptor chemosensitivity,61 reduced diffusion capacity,62 and loss of hypoxic vasoconstrictive mechanisms.63 Indeed, many pathophysiological events in COVID-19 affect either lung perfusion (Q) or ventilation (V; dead space ventilation or right-to-left shunt formation), all of which could lead to a V/Q mismatch. Altered lung mechanics due to progressive lung oedema related to sustained pulmonary inflammation, alveolar collapse, atelectasis, and fibrosis further impair global lung function, resulting in progressive tissue hypoxia (Figure 1, Figure 2 ). Other hallmarks of severe COVID-19 are endothelial inflammation, neovascularisation, and thrombotic events (see below).
Figure 1.
Hypoxia and lung failure in COVID-19
Conceptual and simplified view of COVID-19 lung pathogenesis. In most patients, hypoxia is the principal and most severe COVID-19 symptom; although still debated, compensatory mechanisms to maintain oxygen delivery such as increased respiratory effort, hypoxic vasoconstriction, and cardiac output are thought to eventually lose efficacy with increasing COVID-19 severity. With a further reduction in functional lung capacity, hypoxia becomes life-threatening and frequently necessitates intensive care support. Mechanisms contributing to COVID-19 severity include increased dead space ventilation secondary to endothelial inflammation and microthrombi, an elevated diffusion barrier secondary to alveolitis and pulmonary oedema, and right-to-left shunt formation secondary to atelectasis, which is related to increased oedema and fibrosis in the long term; these mechanisms collectively reduce gas-exchange capacity. ΔP=change in pleural pressure. ΔV=change in volume. PaO2=partial pressure of arterial oxygen.
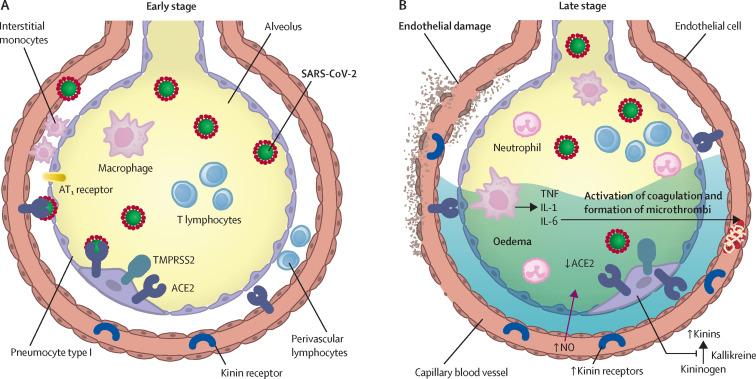
Figure 2.
Inflammatory mechanisms, alveolar epithelial and endothelial damage, and coagulopathy in COVID-19
(A) In the early stage of disease, SARS-CoV-2 infects the bronchial epithelial cells as well as type I and type II alveolar pneumocytes and capillary endothelial cells. The serine protease TMPRSS2 promotes viral uptake by cleaving ACE2 and activating the SARS-CoV-2 S-protein. During early infection, viral copy numbers can be high in the lower respiratory tract. Inflammatory signalling molecules are released by infected cells and alveolar macrophages, in addition to recruited T lymphocytes, monocytes, and neutrophils. (B) With disease progression, plasma and tissue kallikreins release vasoactive peptides known as kinins that activate kinin receptors on the lung endothelium, which in turn leads to vascular smooth muscle relaxation and increased vascular permeability. This process is controlled by the ACE2 receptor. Without ACE2 blocking the ligands of kinin receptor B1, the lungs are prone to vascular leakage, angioedema, and downstream activation of coagulation. Dysregulated proinflammatory cytokine (TNF, IL-1, IL-6) and NO release and signalling contribute to these processes. As a consequence, pulmonary oedema fills the alveolar spaces, followed by hyaline membrane formation, compatible with early-phase acute respiratory distress syndrome. Anomalous coagulation frequently results in the formation of microthrombi and subsequent thrombotic sequelae. ACE2=angiotensin-converting enzyme 2. AT1 receptor=type 1 angiotensin II receptor. IL=interleukin. NO=nitric oxide. TMPRSS2=transmembrane protease serine 2. TNF=tumour necrosis factor.
Mechanisms of cellular infection and dissemination
SARS-CoV-2 invades ciliated cells in the superficial epithelium of the nasal cavity.52 A disseminated viral infection with viraemia and high viral loads in the airways on hospital admission are both associated with severe outcomes,64, 65 although not all studies support this notion.31 In contrast to influenza viruses, which mainly infect airway cells and immune cells (alveolar and interstitial macrophages and natural killer cells), SARS-CoV-2 can infect a wider range of cells, including cardiocytes and endothelial, testicular, and bile duct cells.66 The viral spike (S) glycoprotein mediates viral entry by binding to ACE2 on the epithelial cell surface, a process supported by transmembrane protease serine 2 (TMPRSS2).67 ACE2 expression is high in the epithelial cells of the nasal cavity, supporting an initially localised infection by SARS-CoV-2.52, 67 How SARS-CoV-2 disseminates to the lower respiratory tract is unclear. Two theories predominate: first, (micro-)aspiration of SARS-CoV-2 particles causes spread from the oropharynx to the lungs;68 and second, airborne microparticles are transported directly into the lower respiratory tract by airflow, bypassing the upper airways.69, 70 Of note, involvement of other receptors (eg, neuropilin 1) acting as co-factors to SARS-CoV-2 cellular entry and tropism has been suggested.71, 72
Histopathology
In deceased patients with COVID-19, the average lung weight is typically increased with apparent oedema and congestion (figure 1).73 Microscopically, diffuse damage to the respiratory tract occurs with mucus and cell debris deposits in the bronchi, and the alveoli are typically filled with fluid, fibrin, and hyaluran.74 At the cellular level, type II pneumocytes frequently have disrupted cell membranes and the lung parenchyma undergoes remodelling (hyperplastic, metaplastic, and necrotic pneumocytes).73, 75 Microangiopathy predominates, leading to widespread platelet–fibrin microthrombi in alveolar capillaries.73 Several markers of angiogenesis are upregulated.76 Leukocyte infiltration is common, including perivascular T cells and macrophages in the alveolar lumina, and lymphocytes and monocytes in the interstitium (figure 2).73 A subset of patients shows haemophagocytosis in the pulmonary lymph nodes,77 as observed in SARS and H1N1 influenza,78 suggesting a potential cytokine storm syndrome-like response in these patients.
COVID-19-associated ARDS
COVID-19-associated ARDS shares some general characteristics of ARDS such as impaired gas exchange and characteristic CT findings. However, the combination of different pathological mechanisms in COVID-19-induced ARDS results in a more variable clinical appearance.
ARDS related to COVID-19 is often associated with an almost normal respiratory system compliance, unlike non-COVID-19-related ARDS (figure 1).79 However, compliance can vary depending on the predominant pathomechanism and time. Gattinoni and colleagues proposed two simplified phenotypes of SARS-CoV-2 ARDS: type H, dominated by low compliance (high elastance), high right-to-left shunt, high lung weight, and high recruitability (severe COVID-19-related ARDS, similar to classic ARDS); and type L, characterised by high compliance (low elastance), low ventilation-to-perfusion ratio, low lung weight, and low recruitability (mild COVID-19-related ARDS).80 However, these proposed descriptors are controversial, especially in view of a possible bias owing to early intubation of type L patients.81 The current consensus is that the type H and L phenotypes should not be used to guide clinical practice. Another phenotyping possibility for establishing early prognosis in COVID-19-associated ARDS includes the combination of static compliance and D-dimer concentrations. Accordingly, patients with low compliance and high D-dimer concentrations have the highest risk of mortality.82 Beyond standard lung-protective ventilation regimes, intermittent prone positioning appears to improve gas exchange by reducing the V/Q mismatch, but this has not been fully verified in clinical studies. If further deterioration occurs, extracorporeal membrane oxygenation should be considered early as a treatment option: indications include an oxygenation index of less than 80 mm Hg for more than 3 hours (FiO2 >90%) or an airway plateau pressure of at least 30 cm H2O (for patients with respiratory failure only).50, 79
The increased respiratory drive in ARDS related to COVID-19 (typically within the first few days from ARDS onset) differs from that in non-COVID-19 ARDS and is probably underestimated, potentially masking the true extent of hypoxaemia.83 From a conceptual viewpoint, COVID-19 provides a unique opportunity to decipher a specific cause of ARDS; it also emphasises the fact that ARDS is a syndrome that occurs in various critical care conditions and is not a discrete disease entity per se.
Lung fibrosis
Lung fibrosis is characterised by deterioration of lung function and respiratory failure, correlates with poor prognosis, and is irreversible.84 Approximately 50% of people with severe COVID-19 develop ARDS, for which lung fibrosis is a known complication. Lung fibrosis is driven by pro-fibrotic factors, predominantly transforming growth factor-β (TGF-β). Typically, secretion of TGF-β from the injured lung promotes repair and resolution of infection-induced damage,84 but in severe COVID-19 the infection can cause excessive TGF-β signalling.85, 86 Indeed, hallmarks of pro-fibrotic processes such as epithelial-to-mesenchymal and endothelial-to-mesenchymal transition have been observed in COVID-19.87 Follow-up of SARS and MERS survivors showed that older patients frequently developed residual pulmonary fibrosis.88, 89 In SARS, 36% of patients developed lung fibrosis approximately 3 months after diagnosis,89, 90 with a direct correlation between disease duration and extent of fibrosis.91, 92 Given the similarities between SARS, MERS, and COVID-19, it is likely that pulmonary fibrosis will be a common complication of COVID-19. Accordingly, autopsies have revealed a high proportion of COVID-19 cases with lung fibrosis.93, 94 In studies of follow-up chest CT scans, progression of so-called ground glass opacities was shown in a mixed pattern peaking at 10–11 days from symptom onset before persisting or gradually resolving as irregular fibrosis.95 Among individuals in recovery after COVID-19, those who were severely affected and had an increased inflammatory reaction were most likely to develop pulmonary fibrosis.96 The presence of interstitial thickening, irregular interface, thick reticular pattern, and a parenchymal band on CT imaging have been suggested as predictors of early COVID-19 pulmonary fibrosis (figure 1).96
Coagulopathy and endothelial damage
Coagulopathy and endothelial damage are key occurrences in severe COVID-19, with frequent reports of arterial and venous thromboembolism.97 Between 21% and 69% of critically ill patients with COVID-19 present with venous thromboembolism,98 far exceeding the 7·5% occurrence reported in surgical patients in the intensive care unit (ICU).99 Additionally, COVID-19 has a higher prevalence of thrombosis than does influenza.100 Available data suggest an association between deranged coagulation and severity of lung failure and mortality.57, 101, 102, 103, 104 Patients with severe COVID-19 frequently present with signs of hypercoagulability—ie, high circulating D-dimer concentrations (3–40 times normal concentrations), increased fibrinogen, elevated prothrombin time and activated partial thromboplastin time, and thrombocytopenia.57, 105, 106 D-dimer concentrations are consistently higher in patients with severe COVID-19 than in general ICU patients107 and patients with severe pneumonia not related to COVID-19.108
Although the exact pathomechanism of hypercoagulabilty in COVID-19 remains unclear, it is likely that direct virus-induced endothelial damage and ensuing inflammation (mediated by cytokines, reactive oxygen species, and acute-phase reactants) are key factors.109 Some data suggest that SARS-CoV-2 can infect vascular endothelial cells,76, 110, 111 but other studies have shown no evidence of the virus in these cells.73, 112, 113 The notion of pulmonary circulation thrombosis induced by endothelial injury is supported by evidence that alveolar damage in COVID-19 is frequently accompanied by thrombotic microangiopathy (Figure 1, Figure 2).114, 115 COVID-19-associated endotheliopathy with diffuse microcirculatory injury in the lungs appears to be a central feature of the severe disease phenotype. We henceforth refer to this endotheliopathy-centred condition as endotheliitis, following the first use of this term by Varga and colleagues;110 however, descriptions of the same or similar small-vessel COVID-19 pathology by the same authors and other investigators have included the terms angiocentric lymphocytic inflammation,76 mononuclear and neutrophilic inflammation of microvessels,110 lymphocytic endotheliitis,110 thrombotic microangiopathy,116 cytoplasmic vacuolisation,117 and (pulmonary) capillaritis.111 These findings of endotheliopathy have also been termed diffuse pulmonary intravascular coagulopathy118 and, in essense, they describe different variants of the same phenomenon: dysfunctional cross-talk between leukocytes and endothelial cells that manifests as vascular immunopathology predominantly confined to the lungs, which aggravates hypoxaemia.119 It is important to note that coagulopathy and endothelial damage, or endotheliitis, are not unique to COVID-19 but are a common feature of ARDS in general.
Endotheliitis might be partly responsible for refractory COVID-19-related ARDS with disturbed, hyperperfused intrapulmonary blood flow and loss of hypoxic vasoconstriction with alveolar damage, featuring oedema, haemorrhage, and intra-alveolar fibrin in the most severe cases.76, 117, 120, 121 Accordingly, several studies support the assumption that the combination of pulmonary vascular dysfunction, as signalled by a reduction in respiratory system compliance, and thrombosis, indicated by high D-dimer concentrations, represents the worst-case scenario, being associated with a substantially elevated risk of mortality in patients with COVID-19 and ARDS.82, 122, 123 SARS-CoV-2 infection is not restricted to the lungs; vascular disorders are often systemic and feature generalised vasodysregulation including stasis, disturbed endothelial barrier and permeability control, disrupted cell membranes, endothelium-localised inflammation, and an actively (clinically evident) prothrombotic endothelial cell state, with intracellular virus particles as a probable causative factor, localised to the lungs, brain, heart, kidneys, gut, and liver.76, 110, 113, 117, 124, 125, 126
Widespread endotheliitis is a common hallmark of severe infectious disease that is shared with viral sepsis and shock.127, 128 These perturbations largely stem from abnormal nitric oxide metabolism and upregulation of reactive oxygen species, with oxidative stress additionally aggravated by downregulation of endothelium-associated antioxidant defence mechanisms (figure 2).129 Moreover, endothelium-associated downstream effector programmes lead to activation of proteases, exposure of adhesion molecules, and induction of tissue factor.
Notably, COVID-19 has multiple extrapulmonary clinical manifestations (panel 1 ) that are likely to be related to COVID-19-associated widespread vascular pathology. Some of these manifestations are common to all critical illness states (eg, renal dysfunction) or are reminiscent of the complications of other viral pneumonias (eg, neurological sequelae) and are also commonly seen in critically ill patients with influenza. However, prominent pulmonary as well as systemic endotheliitis represents a distinguishable and distinct feature of COVID-19.
Panel 1. Extrapulmonary clinical manifestations of COVID-19.
Clinical presentations are generally listed in order of clinical relevance. Further information on extrapulmonary manifestations is provided in a review by Gupta and colleagues.130
Brain, nervous system
-
•
Encephalitis
-
•
Headache
-
•
Ageusia, anosmia
-
•
Encephalopathy
-
•
Guillain-Barré syndrome
-
•
Stroke
-
•
Myalgia
Kidney
-
•
Acute kidney injury
-
•
Proteinuria
-
•
Haematuria
-
•
Metabolic acidosis
-
•
Electrolyte imbalances
Liver
-
•
Elevated aminotransferases
-
•
Elevated conjugated bilirubin
-
•
Low serum albumin
Gastrointestinal tract
-
•
Diarrhoea
-
•
Nausea, vomiting
-
•
Abdominal pain
-
•
Mesenteric ischaemia (rare)
-
•
Gastrointestinal bleeding (rare)
Heart, cardiovascular system
-
•
Myocardial injury, myocarditis
-
•
Thromboembolism, endotheliitis
-
•
Cardiac arrhythmias
-
•
Cardiogenic shock
-
•
Myocardial ischaemia, acute coronary syndrome
-
•
Cardiomyopathy (biventricular, left ventricular, or right ventricular)
Endocrine system
-
•
Hyperglycaemia
-
•
Diabetic ketoacidosis
Skin
-
•
Petechiae
-
•
Pernio-like skin lesions
-
•
Livedo reticularis
-
•
Erythematous rash
-
•
Urticaria
-
•
Vesicles
Host response to SARS-CoV-2
Cytokine response
Cytokine production in COVID-19 typically occurs via two pathways: upon direct viral recognition by immune cells through pattern-recognition receptors, prominently virus-specific Toll-like receptors (TLR3, TLR7, TLR8, and TLR9), and indirectly through the mediation of damage-associated molecular patterns (DAMPS) released from epithelial cells damaged by SARS-CoV-2 (eg, preformed cytokines, high mobility group protein B1 [HMGB1], or ATP).131, 132 In support of the direct pathway, TLR7 defects are associated with severe COVID-19 in young men,133 and up to 3·5% of patients with severe COVID-19 have been shown to have inborn defects in TLR3-dependent and interferon regulatory factor 7 (IRF7)-dependent type I interferon immunity.134 Upon injury, endothelial, epithelial, and other parenchymal cells release inflammatory mediators that activate immune cells. These events collectively trigger a release of multiple proinflammatory cytokines and chemokines.135 Circulating concentrations of proinflammatory mediators including tumour necrosis factor (TNF), monocyte chemotactic protein 1 (MCP1; C-C motif chemokine 2 [CCL2]), macrophage inflammatory protein-1α (MIP-1α; CCL3), and C-X-C motif chemokine 10 (CXCL10; 10 kDa interferon gamma-induced protein [IP10]) were increased in patients with COVID-19 who were admitted to the ICU compared with those not admitted,136 while conflicting data exist for IL-6.136, 137 Type I and type III interferon responses and the IL-1–IL-6 axis are likely to constitute biologically relevant signalling pathways in SARS-CoV-2 infection. For example, circulating IL-6 concentrations were correlated with COVID-19 severity, outcomes, or both, in several138, 139, 140, 141, 142 but not all143 studies. IL-1 production by the inflammasome is upregulated in cells from patients with COVID-19,144 and single-cell RNA sequencing of circulating cells from patients with COVID-19 has shown a greater abundance of classic CD14+ monocytes with high IL-1β expression.145
In a study of patients with respiratory failure associated with COVID-19, expression of the HLA DR isotype (HLA-DR; an MHC class II cell-surface molecule that is crucial for antigen presentation) on circulating monocytes was decreased but, in contrast to bacterial sepsis, monocytes retained high cytokine production capacity.146, 147 Similarly, children with post-COVID-19 multisystem inflammatory syndrome showed a severe systemic inflammatory syndrome affecting the cardiac, digestive, and haematopoietic systems with features of a septic cytokine storm.148, 149, 150 A type I interferon signature (ie, low concentrations) has been associated with mild COVID-19, whereas interferon signalling was defective in severe COVID-19.126, 134, 151, 152, 153, 154 Of note, patients with severe COVID-19 have restricted or delayed interferon type I and type II responses in contrast to patients with severe influenza, who have a dominant early interferon response.155, 156
There is controversy concerning the role of the so-called cytokine storm in COVID-19 pathophysiology. Although an increased systemic cytokine response in COVID-19 is undisputed, comparisons of TNF, IL-6, and IL-8 concentrations across acute conditions—COVID-19-induced ARDS and non-COVID-19 ARDS, sepsis, trauma, cardiac arrest, and cytokine storm syndrome—show that systemic inflammation during COVID-19 is less robust than in these other conditions.106, 138, 157, 158 The current evidence neither confirms nor refutes the role of the hyperinflammatory phenomenon in subsets of patients with COVID-19; it is possible that if a cytokine storm syndrome does occur in COVID-19, it might differ from that in other acute conditions in terms of scale of sensitivity or response characteristics (eg, different inflammatory mediators or profiles).
Reports on tissue-specific activation and secretion patterns of cytokines in patients with COVID-19 are only rudimentary and have mainly focused on the lungs. A meta-transcriptomic sequencing study comparing inflammatory gene expression in the bronchoalveolar lavage fluid (BALF) in patients with COVID-19, patients with community-acquired pneumonia, and healthy individuals showed a COVID-19-specific activation signature of proinflammatory genes (ie, IL1B, interleukin-1 receptor antagonist [IL1RN], CXCL17, CXCL8, and MCP1) and robust upregulation of numerous interferon-inducible genes.159 Small BALF-based studies have shown upregulation of TNF, IL1RN, IL-1β, IL-6, IL-8, IL-10, MCP1, CXCL10, MIP-1α, and MIP-1β (CCL4) in patients with COVID-19.160, 161, 162
A few small studies have directly examined the lungs for evidence of the cytokine response in COVID-19. Ackermann and colleagues compared lungs from seven patients who died of respiratory failure related to COVID-19 and seven patients who died of ARDS secondary to influenza A H1N1, with results showing increased RNA expression of CXCL8 and CXCL13 in lungs from patients with COVID-19 and increased IL6 expression in both groups.76 Another autopsy study reported varying mRNA expression levels of IL1B and IL6 in the lungs of four individuals who died of COVID-19.111 The above activation patterns have been reproduced in a lung organoid model derived from human pluripotent stem cells and infected with SARS-CoV-2.163 A multinational study showed contrasting expression patterns (high and low) in interferon-stimulated genes and cytokines in post-mortem lung tissue from 16 patients with COVID-19.164 The abundant NACHT, LRR, and PYD domains-containing protein 3 (NLRP3) inflammasome aggregates found in the lungs of deceased patients with COVID-19 are consistent with the above findings.165 Cytokine responses were not investigated in any other organs, which is a serious limitation given that ACE2 can be more robustly expressed in the small intestine, kidneys, and heart compared with the lungs.166
We caution against premature dismissal or acceptance of elevated cytokine responses (ie, a cytokine storm) as a cause of COVID-19 severity or mortality until more comprehensive profiles of circulating mediators are available. The need for caution is emphasised by the absence of reports of organ-dependent cytokine characteristics; a role for compartmentalised (versus systemic) inflammatory dynamics that influence outcomes might be plausible.
Non-cytokine mediators
With the exception of ferritin and C-reactive protein (CRP; both acute-phase proteins), non-cytokine inflammatory mediators have been under-investigated in COVID-19. Circulating ferritin is a recognisable marker of secondary haemophagocytic lymphohistiocytosis (HLH) and macrophage activation syndrome (MAS-HLH) in critically ill patients.167, 168 In severe COVID-19, ferritin concentrations of at least 4420 μg/L (a cutoff used for MAS in patients with bacterial sepsis)168 indicated a MAS-HLH-like phenotype and were associated with enhanced IL-1β production from ex vivo-stimulated circulating monocytes.147 Various studies and meta-analyses have shown that ferritin is both a good marker of disease severity and a predictor of in-hospital mortality.169, 170, 171, 172 High circulating ferritin might therefore constitute a potential marker to guide anti-inflammatory treatments (eg, an IL-1 receptor antagonist) in patients with severe COVID-19.173, 174, 175 Furthermore, the diagnosis of hyperinflammation, MAS-HLH, or HLH, confirmed by autopsy of lymph nodes and bone marrow of patients with COVID-19,77, 176 should not rely solely on circulating ferritin concentrations, but also on additional laboratory (eg, proinflammatory cytokines) and clinical parameters. A composite H-score has been proposed for the secondary diagnosis of HLH and a similar approach might be applicable in COVID-19.177
A meta-analysis of 51 225 patients identified CRP as an important predictor of COVID-19-related mortality in patients aged 60 years or older.169 A systematic review of 2591 patients showed that elevated CRP concentrations correlated with COVID-19 severity.178 In patients with severe COVID-19 who were admitted to the ICU, CRP kinetics were similar to those observed in bacterial sepsis: a high CRP concentration at admission followed by a gradual decline.179 Circulating ferritin, CRP, and D-dimers were reported to be expressed to a similar extent or more robustly expressed in COVID-19 compared with other acute states.106
The complement system is another key host-defence mechanism with capacity to exacerbate tissue injury through its proinflammatory effects (and via promotion of coagulation). Activation peptide of complement component 5a (C5a) and the membrane attack complex (MAC; C5b-9) were increased according to disease severity in the blood (and BALF for C5a) of patients with COVID-19.162, 180 Furthermore, circulating sC5b-9 and C4d concentrations in patients with COVID-19 at hospital admission were correlated with ferritin concentrations.181 Whether complement system activation contributes to COVID-19 tissue damage and organ failure is currently not clear.
Procalcitonin has proven useful in the early diagnosis of lower respiratory tract infections of bacterial origin and to guide antibiotic therapy.182, 183 In a meta-analysis including 5912 patients with COVID-19 (35 studies), procalcitonin concentrations were twice as high in those with severe disease versus non-severe disease.184 Two other meta-analyses showed no difference in circulating procalcitonin concentrations between patients with COVID-19 and those with non-COVID-19 pneumonia,185 and among patients with COVID-19 according to disease severity (in contrast to the procalcitonin spike seen in sepsis).106 In 66 patients with COVID-19 admitted to the ICU, procalcitonin concentrations predicted the occurrence of secondary infections with an area under the receiver operating characteristic curve of 0·80.186
Hormones and endocrine factors
Hypertension, type 2 diabetes, and obesity are comorbidities that are associated with risk of COVID-19 complications and mortality.57, 187 In patients with SARS, circulating glucose and diabetes independently predicted morbidity and mortality.188, 189 ACE2 is expressed in key metabolic tissues including the thyroid, endocrine pancreas, testes, ovaries, and adrenal and pituitary glands.190, 191 An infection of pancreatic β cells with SARS-CoV-2 contributed to glycaemic dysregulation,163 suggesting that coronavirus-dependent β-cell dysfunction might also occur in patients without pre-existing diabetes. Data confirm that glycaemic control affects outcomes in COVID-19 patients with diabetes.192 In a retrospective study of 952 patients with type 2 diabetes, COVID-19 severity and mortality were associated with glucose control: patients with well controlled glycaemia (variability within 3·9–10·0 mmol/L) had lower mortality than did patients with high glycaemic variability (>10·0 mmol/L).192 The mechanisms that underpin increased COVID-19 severity in patients with diabetes might include a higher cardiovascular disease prevalence, increased ACE2 expression, decreased viral clearance, and metabolic derangements.193 Evidence suggests that hyperglycaemia and insufficient glycaemic control could be associated with increased oxidative stress and hyperinflammation in severe infections, potentially promoting endothelial or organ damage.194, 195 Additionally, COVID-19 is often associated with hypokalaemia, which is known to affect glucose control in diabetes.196
The renin–angiotensin–aldosterone system (RAAS) is highly activated in patients with severe COVID-19.197 Although the effect of therapeutic RAAS blockade (ACE inhibitors and angiotensin receptor blockers) is unclear, current evidence does not support discontinuation of prescribed RAAS blockers.197, 198 Angiotensin II might promote hyperinflammation through IL-6 induction in endothelial and vascular smooth muscle cells via the type 1 angiotensin II receptor (AT1) receptor.199 Furthermore, by increasing aldosterone concentrations, angiotensin II triggers vasoconstriction and water reabsorption. Exogenous angiotensin II, which has been approved in the USA and the EU for use in patients with COVID-19, competes with SARS-CoV-2 to bind to the ACE2 receptor; angiotensin II receptor occupation causes internalisation and downregulation of ACE2 followed by lysosomal degradation.197 Another proposed consequence of ACE2 downregulation following SARS-CoV-2 infection is an imbalance between the ACE–angiotensin II–AT1 receptor axis and the ACE2–angiotensin (1–7)–Mas receptor axis, potentially favouring local proinflammatory and prothrombotic processes.200, 201
The expression of TMPRSS2 is controlled by androgenic hormones, which might partly explain the sex differences observed in critically ill patients with COVID-19.67, 202 Although SARS-CoV-2 equally affects men and women, the severity of COVID-19 and the number of deaths are up to twice as high in men.202 TMPRSS2 expression might be upregulated in prostate cancer; patients with prostate cancer treated with androgen-deprivation therapies have been reported to have a decreased risk for the severe COVID-19 phenotype.203 Sex hormone-induced differences might therefore be of particular relevance in SARS-CoV-2 infections.204
Emerging data demonstrate a profound association of endocrine and metabolic dysfunctions with clinical COVID-19 outcomes. Given that SARS-CoV-2 binding to the ACE2 receptor and RAAS activation are apparent in patients with severe COVID-19, modulation of the co-stimulatory molecules (eg, TMPRSS inhibition) should be investigated.
Cellular immune response
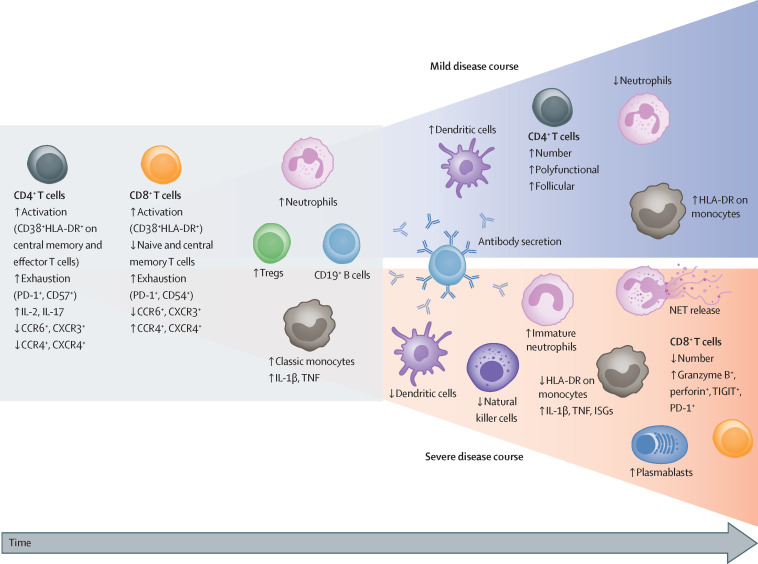
Patients with COVID-19 frequently have mild neutrophilia and T-cell lymphopenia resulting in an increased neutrophil-to-lymphocyte ratio,144, 205 which is a useful prognostic marker for COVID-19 severity.206 Other leukocyte subsets also undergo characteristic, albeit more heterogeneous, fluctuations and trajectories (figure 3 ).
Figure 3.
Severity-dependent changes in peripheral blood immune cells during COVID-19
COVID-19 is associated with a marked decrease in circulating effector CD4+ and CD8+ T lymphocytes, leading to an increase in Tregs. In addition to a numerical loss, the high proportion of exhausted and suppressed T cells expressing CTLA-4, PD-1, or TIGIT suggest compromised T-cell immunity. Both CD4+ and CD8+ T cells show early upregulation of activation markers such as CD38 and HLA-DR concurrent with an altered chemokine receptor pattern (a decrease in CCR6 and CXCR3). Circulating CD4+ T cells are skewed towards an IL-2 and IL-17-positive profile. Early in the course of COVID-19, the numbers of B cells and monocytes remain unchanged, whereas mild neutrophilia is frequently observed. Blue area: patients with favourable outcomes have an increased number of circulating dendritic cells and intermediate monocytes that upregulate HLA-DR expression, suggesting recovery of immunocompetence. T-cell numbers recover with an increase in polyfunctional and follicular helper T cells. These changes are accompanied by a humoral antibody response produced by activated and expanded B cells. During recovery, neutrophil numbers normalise. Red area: a more severe disease course is characterised by a decrease in dendritic cells, natural killer cells, and monocytes; monocytes further downregulate HLA-DR expression but upregulate IL-1β, TNF, and ISGs. Although low in number, CD8+ T cells in severe COVID-19 show substantial upregulation of their effector molecules, such as granzyme B and perforin. Of note, the numbers of B cells decrease, whereas plasmablasts are increased in the blood; specific antibodies are also produced in patients with severe COVID-19. Profound changes occur among myeloid cells including egress of immature neutrophils and myeloid-derived suppressor cells from the bone marrow, and enhanced formation of NETs. NET formation is likely to be an important contributor to the development of organ and endothelial injury in conjunction with activation of the complement and coagulation cascades. CCR=C-C chemokine receptor. CTLA-4=cytotoxic T-lymphocyte protein 4. CXCR=C-X-C chemokine receptor. HLA-DR=HLA DR isotype. IL=interleukin. ISGs=interferon-stimulated genes. NETs=neutrophil extracellular traps. PD-1=programmed cell death 1. TIGIT=T-cell immunoglobulin and ITIM domain. TNF=tumour necrosis factor. Tregs=regulatory T cells.
Granulocytes
Although conflicting data exist on eosinophil count,207, 208 most studies have documented mild peripheral neutrophilia in patients with COVID-19 independent of disease severity.144, 205 Peripheral neutrophils typically include highly activated CD38+, CD11b+, and HLA-DR+ cells and myeloid-derived suppressor cells (MDSCs).207, 209 The increase in MDSCs is suggestive of an inflammation-related, emergency haematopoiesis.210, 211 One study reported an immunosuppressive MDSC phenotype in patients with severe COVID-19 upon hospital admission.212 Whether MDSC functionality in COVID-19 is stable or changes over time as observed in bacterial sepsis is not known.213 Consistent with accelerated myelopoiesis, patients with COVID-19 have an increase in peripheral immature (CD10−, CD16low) neutrophil granulocytes, cells that are commonly released under stress.207, 214 Concurrently, the serum of patients with COVID-19 features elevated markers of neutrophil extracellular trap (NET) formation and induces NET formation in healthy neutrophils,215, 216, 217 both typical features of neutrophil functionality. Notably, some BALF samples from patients with COVID-19 contained elevated levels of chemokines that are crucial for neutrophil recruitment (eg, CXCL1, CXCL2, CXCL8) and their concentrations correlated with the SARS-CoV-2 viral load.159 Simultaneously, autopsy reports have revealed neutrophilic tissue infiltrations in a subgroup of patients;112 whether those infiltrations were contingent on SARS-CoV-2 presence, chemokine production, or an exacerbated T-helper-17 (Th17) response in the affected tissues (eg, lungs) is not known.93, 112 Taken together, the evidence suggests that the aberrant recruitment and response of expanded immature neutrophils are likely to contribute to COVID-19 pathogenesis.
Monocytes and macrophages
Although monocyte counts are not substantially altered in COVID-19,205, 218 reports on monocyte–macrophage composition vary. Some studies have documented a shift from CD16+ monocytes towards classic CD14+ monocytes,214 whereas others have shown only marginal alterations.146, 208, 219, 220 Circulating monocytes show signs of activation such as increased CD169 expression,207 with elevated IL-1 and IL-6 production in severe COVID-19.145, 221 The observed cellular activation is associated with upregulation of interferon-stimulated genes attributed to bystander cytokine effects.145, 222 Activated monocytes have been shown to migrate to the lungs in patients with COVID-19 and in mouse models of SARS-CoV-2 infection.222, 223 A concomitant reduction in alveolar macrophages has been observed,222 perhaps reflecting direct infection and depletion of these cells by SARS-CoV-2.23, 24 In the lungs of patients with severe COVID-19, a vicious cycle between infected macrophages (and, secondarily, monocytes) and T cells develops, driving alveolar inflammation and damage.224 Thus, monocytes are aberrantly activated in COVID-19 and might contribute to a proinflammatory state in the infected lungs.
Dendritic cells
Scarce data indicate a depletion of myeloid and plasmacytoid dendritic cells in the blood of patients with COVID-19.214, 223 A single study reported reduced dendritic cell counts in the lungs of patients with severe COVID-19.222 Given the key function of dendritic cells in pathogen sensing and adaptive immune responses, dendritic cell depletion might compromise the development of a protective anti-viral T-cell response.
HLA-DR expression and antigen presentation
HLA-DR expressed on monocytes (mHLA-DR) is a well established marker of immune suppression—eg, in bacterial sepsis, trauma, and following major surgery.225, 226, 227, 228, 229, 230 mHLA-DR expression in patients with COVID-19 has been investigated with flow cytometry and single-cell RNA sequencing.147, 179, 214, 231, 232, 233, 234 An inverse relationship between mHLA-DR expression and COVID-19 disease severity and the extent of inflammation has been established, exemplified by lower expression of mHLA-DR in patients admitted to the ICU versus non-ICU patients,231 and in non-survivors versus survivors,232, 234 concurrent with a negative correlation between circulating IL-6 and mHLA-DR expression.147 mHLA-DR expression in patients not admitted to the ICU was in the normal range.235 Suppressed mHLA-DR expression in patients with COVID-19 admitted to the ICU appears to be closely associated with disease severity and ARDS occurrence. Notably, mHLA-DR downregulation might be additionally amplified by steroid administration.214, 236 mHLA-DR expression kinetics in patients with COVID-19 admitted to ICUs were consistent across studies with no changes during ICU admission to the end of follow-up.179, 231, 233, 234 Although more studies on mHLA-DR kinetics are needed, two distinct phases of mHLA-DR expression in COVID-19 progression can be suggested: first, normal-range HLA-DR expression during the pre-ICU stage; and second, a steep decrease in HLA-DR expression following transfer to the ICU. Investigations are needed into the putative associations between HLA-DR (also on dendritic cells and B cells) and the frequency of secondary infections, the magnitude of lymphopenia, SARS-CoV-2 viral load dynamics, functional testing (eg, leukocytic cytokine production capacity), and immunomodulatory interventions such as dexamethasone.
T cells
The cytotoxic T-cell response, a complex process that also involves antigen-activated CD4+ helper T cells, is crucial for virus eradication.237 The T-cell response must be tightly regulated; exaggerated activation could lead to host cell death, whereas insufficient activation facilitates viral spread. The destruction of the lung epithelium and endothelium in severe COVID-19 has been associated with the appearance of perivascular T lymphocytes.76 Several COVID-19 studies have described a profound decline in virtually all T-cell subtypes.145, 238, 239, 240, 241 Notably, in children with mild-to-moderate COVID-19, lymphopenia is rare and mild,148, 242 whereas in severe paediatric cases with multisystem inflammatory syndrome, it is frequent and pronounced.148, 149 CD8+ T-cell lymphopenia results from the loss of mainly naive and central memory cell subsets, whereas virtually all subpopulations are reduced in CD4+ T lymphopenia.109, 243 The extent of CD8+ T-cell (but not CD4+) lymphopenia inversely correlates with the degree of inflammation244 and is more profound in non-survivors than survivors, but it does not differ between patients admitted versus those not admitted to intensive care.109 T-cell loss has been attributed not only to overwhelming T-cell activation, but also to defects in IL-2–IL-2 receptor signalling.245, 246 Notably, normalisation of several parameters (eg, a late-onset increase in CD8+ T-cells) can occur in mild cases and in recovering patients.244 Although a cause–effect relationship remains to be proven, plummeting T-cell counts could serve as a prognostic marker of increased risk of mortality when considered with IL-6247, 248 and circulating neutrophil counts.249
Evidence suggests that a higher proportion of T cells from patients with COVID-19 express markers characteristic of activation and exhaustion than do those from healthy participants.243 However, functional analysis of T cells has produced contradictory observations supporting both preserved and compromised T-cell functionality.250, 251 Moreover, other investigators have observed a marked skewing of CD4+ T cells towards the Th17 phenotype.243 Several studies have reported the presence of SARS-CoV-2-reactive T cells in approximately half of tested patients with COVID-19,33, 252, 253 supporting the possibility that COVID-19 induces a T-cell response in the presence of lymphopenia and, perhaps, T-cell dysfunction.
Lung infection by respiratory viruses typically results in recruitment and local accumulation of distinct T-cell populations mediated by various chemoattractants. Emerging evidence shows that T cells of patients with COVID-19 have an altered chemokine receptor pattern.243 These findings are suggestive of altered migratory and homing behaviour of T cells. Accordingly, single-cell RNA sequencing of bronchoalveolar cells has provided evidence that in moderate COVID-19 cases, highly clonally expanded (possibly virus-specific) T lymphocytes proliferate in the lungs, whereas clonally heterogeneous T-cell pools populate the lungs of severely affected patients.222
The above data are consistent with a contribution of dysfunctional antiviral adaptive immunity to COVID-19 pathogenesis. Correspondingly, SARS-CoV-2 has been detected in the respiratory tracts of dying patients with COVID-19.254 High viral loads in respiratory samples and plasma are associated with disease severity, and with a deeper dysregulation of the host response, including the magnitude of lymphopenia.137, 255 These findings suggest that in severely affected patients, the immune system is incapable of eradicating the virus, which could further trigger the induction of non-homoeostatic responses to the infection. Consistent with this suggestion, in a European multicentre ICU study, the incidence of secondary nosocomial ventilator-associated lower respiratory tract infections was 50·5% in patients with SARS-COV-2,256 which was significantly higher than the incidence of such infections in patients with influenza pneumonia. Similarly, secondary nosocomial infections occurred in up to 50% of Chinese patients with COVID-19 who were admitted to hospital.121 Available evidence is therefore consistent with impaired T-cell immunity (both numerical and functional deficits) weakening infection control and increasing lung tissue damage in COVID-19.
Natural killer cells
Natural killer cells are commonly enriched in the lungs and respond rapidly to a broad collection of viral infections.257, 258 Consequently, patients with moderate-to-severe COVID-19 feature an accumulation of natural killer cells in the infected lungs,222, 244 whereas natural killer cells counts in the periphery decline.214, 259 Current evidence on natural killer cell immune function in COVID-19 is contradictory. Whereas some investigators have reported signs of natural killer cell hyporesponsiveness and exhaustion,214, 260 marked activation of these cells in the blood of patients with COVID-19 has also been documented.261
B cells and the humoral response
Several studies have reported selective B-cell plasmablast expansion in severe COVID-19, indicative of a strong SARS-CoV-2-specific humoral response (see below) concomitant with a decline in peripheral naive and memory B-cell counts.145, 148, 249, 262 In contrast to the fluctuations in the periphery, B-cell and plasma cell counts were unchanged in BALF from patients with COVID-19.222 Both naive and memory B-cell compartments are activated during COVID-19 infection,145, 208, 263, 264 as reflected by increased expression of proliferation markers.145 Thus, existing data indicate that SARS-CoV-2 infection exerts a pronounced, systemic, and multifaceted B-cell response including memory B-cell recall.
Activation of the B-cell compartment is accompanied by a humoral reaction; several studies have detected a robust antibody response varying from oligoclonal to highly multiclonal patterns. Clonality reportedly peaks during the early recovery phase and subsequently decreases.145 Most clones produce antibodies against the receptor-binding domain (RBD) of the SARS-CoV-2 spike protein, as observed in previous coronavirus-based infections.265, 266 Crystal structures of antibody–RBD–ACE2 complexes underscore the neutralising effect of these antibodies.267, 268 Most antibodies do not cross-react with the RBD from the related SARS-CoV and MERS capsids (table).268 Although the majority of studies reported convergent antibody responses against the spike protein RBD,262, 264 others have shown divergent responses.145, 208 This variability might be attributable to differences in COVID-19 patient cohorts.
The extent to which the ubiquitous antibody responses contribute to COVID-19 resolution is not yet clear. It has been suggested that vaccination protocols using the major RBD spike protein antigens with particularly high convergence of antibodies are promising. It is also puzzling that patients with COVID-19 with primary antibody deficiencies do not exhibit markedly worse disease course or outcomes.269 For example, patients with COVID-19 who have reduced or absent B-cell functionality due to agammaglobulinaemias of genetic origin (X-linked agammaglobulinaemia or autosomal recessive agammaglobulinaemia) or anti-B-cell therapies (eg, anti-CD20 monoclonal antibody ocrelizumab or tyrosine kinase inhibitor ibrutinib) have fully recovered without signs of severe disease.130, 270, 271, 272, 273 In contrast, there is a minority of patients with COVID-19 who have B lymphocyte clones that are able to produce autoantibodies against type I interferons, which could potentially compromise viral eradication.151
B-cell reactions to SARS-CoV-2 might be beneficial in cases of particularly efficient antibody responses. However, B-cell responses might be ineffective or harmful in other scenarios (eg, antibody-dependent enhancement).
Conclusions and future perspectives
An overview of current evidence regarding immuno-inflammatory reactions induced by SARS-CoV-2 and subsequent organ-based consequences suggests several conclusions. First, a new infectious profile is evident: low pathogenic Coronaviridae subspecies of human coronavirus, such as hCoV-229E, hCoV-OC43, and hCoV-NL63, infect the upper airways causing mild-to-moderate (common cold-like) respiratory disease, whereas highly pathogenic viruses settle in the lower respiratory tract, typically causing severe pneumonia and ARDS. SARS-CoV-2 shares features of low and high pathogenic coronavirus subspecies: after infecting the upper respiratory tract, it is capable of subsequently spreading to the lower respiratory tract. This duality, accompanied by the capacity for viral transmission during the incubation period, appears as an evolutionary advantage and a unique feature of this new coronavirus pathogen. Second, endothelial and epithelial infection appears to predominate, rather than alveolar-centred infection: the disruption of the alveolar epithelial–endothelial barrier is central to the development of severe pneumonia and ARDS. Compared with influenza and SARS, multiorgan involvement and thromboembolic events are more common in COVID-19,100, 130 implying that SARS-CoV-2 is an endotheliophilic virus. Third, the inflammatory response is atypical: although patients with COVID-19 have elevated circulating proinflammatory cytokines over a longer period of time than do patients with influenza,155 for example, the concentrations seem to be significantly lower than are typical in non-COVID-19-related ARDS.123 The inflammatory characteristics thus far observed in patients with COVID-19 suggest that either the systemic cytokine component is not a crucial contributor to COVID-19 severity or that the disease features its own unique, poorly understood, yet detrimental, inflammatory profile. Fourth, a maladaptive host response is unable to combat the virus: there is a marked association between high viral loads (local and systemic) and the phenotype and magnitude of the dysregulated host response, suggesting that poor control of SARS-CoV-2 virus by the immune response leads to severe COVID-19.
The concept of virus-induced pulmonary vasculitis is consistent with the frequently observed failure of simple ventilatory support in patients with COVID-19. Typically, a substantial V/Q mismatch in COVID-19 is more often a consequence of right-to-left shunt due to inflamed, hyperperfused lungs and failure of hypoxic vasoconstriction. This scenario might trigger a vicious cycle beginning with hypoxia and an increase in respiratory effort and oxygen consumption. If a patient with severe COVID-19 is not able to satisfy their oxygen demand by physiological adaptation of cardiac output and oxygen content, the outcome will be fatal. This concept could help to explain why older patients and patients with obesity who have reduced lung capacity, and patients with cardiovascular comorbidities have the highest risks for unfavourable outcomes.
In conclusion, we propose that COVID-19 should be perceived as a new entity with its own characteristic and distinct pathophysiology. However, the differences between ARDS related to COVID-19 and ARDS of other causes (and other critical illnesses) should not prompt abandonment of the existing consensus principles of critical care, as noted by others.274 Regardless of whether this notion is confirmed, it is advisable to study COVID-19 pathophysiology without preconceptions based on other critical diseases, which might be misleading for clinical care and treatments. Directions for future research into COVID-19 pathophysiology are proposed in panel 2 . An unbiased, gradual assembly of key pieces in the COVID-19 pathophysiological puzzle for different patient cohorts (eg, based on sex, age, ethnicity, pre-existing comorbidities), albeit not as rapid as desired, will eventually create a more accurate depiction of the disease. Armed with this knowledge, existing treatment guidelines could potentially be updated, enabling medical professionals to provide optimum care for patients with COVID-19.
Panel 2. Priorities for future research.
The proposed research aims will be achieved most effectively with a complementary combination of preclinical and clinical research.
-
•
Establish the molecular basis for lower pathogenicity of SARS-CoV-2 compared with SARS-CoV
-
•
Define the role of pre-existing and acquired T-cell immunity in COVID-19 development and progression
-
•
Establish precise predictive thresholds for known biomarkers of COVID-19 severity, outcomes, and complications
-
•
Develop novel prognostic biomarkers and risk predictors for COVID-19 pneumopathy, acute respiratory distress syndrome, and fibrosis
-
•
Elucidate compartmentalisation profiles of soluble inflammatory mediators and cell subsets (ie, in individual organs and systems)
-
•
Characterise immunological deficiencies secondary to ageing and comorbidities that impair efficient immunological responses against SARS-CoV-2
-
•
Characterise short-term and long-term COVID-19 vasculopathies and their sequelae
-
•
Develop a unified post-COVID-19 monitoring platform to characterise long-term outcomes and immune derangements after SARS-CoV-2 infection
-
•
Conduct high-quality, prospective clinical studies to identify optimum anti-coagulative and immunomodulatory strategies for patients with SARS-CoV-2 infection
Search strategy and selection criteria
We searched PubMed and MEDLINE for relevant peer-reviewed articles published in English from database inception to Feb 13, 2021, using a set of topic-specific search terms (eg, “pulmonary” OR “respiratory”) in combination with at least one of the following terms: “SARS-CoV-2”, “COVID-19”, “MERS-CoV”, “SARS-CoV”, “SARS-CoV-1”, and “coronavirus”. The choice of topic-specific search terms was not defined a priori but left to the discretion of the panel of experts responsible for each section to allow wide scrutiny of the literature. Similarly, the selection of articles for consideration and citation was done by the individual expert panels. This selection was subsequently scrutinised, and modified if needed, by the co-authors who internally reviewed the individual sections. The final list of cited articles was selected on the basis of their relevance to the aims of this Series paper, with a primary focus on peer-reviewed publications. Preprint publications (searched on the medRxiv, bioRxiv, Research Square, Preprints, and PeerJ servers) were referenced only as a justified exception (<2% of all citations in the final version) and with the approval of all section authors and internal reviewers.
Declaration of interests
MSW has received unrestricted funding from Sartorius. GL reports personal fees from Swedish Orphan Biovitrum. TSp and JCS disclose institutional funding (received by the University of Bern) from Orion Pharma, Abbott Nutrition International, B Braun Medical, CSEM, Edwards Lifesciences, Kenta Biotech, Maquet Critical Care, Omnicare Clinical Research, Nestlé, Pierre Fabre Pharma, Pfizer, Bard Medica, Abbott, Anandic Medical Systems, PanGas Healthcare, Bracco, Hamilton Medical, Fresenius Kabi, Getinge Group Maquet, Dräger, Teleflex Medical, GlaxoSmithKline, Merck Sharp and Dohme, Eli Lilly, Baxter, Astellas, AstraZeneca, CSL Behring, Novartis, Covidien, Nycomed, Phagenesis, and Hemotune. MGN reports grants from GlaxoSmithKline and ViiV Healthcare, and was a scientific founder of TTxD. All other authors declare no competing interests.
Acknowledgments
Acknowledgments
No funding was provided for this Series paper. TSk is supported by the Poland National Science Centre (UMO-2020/01/0/NZ6/00218). MS-H is funded by a UK National Institute for Health Research (NIHR) Clinician Scientist Award (CS-2016-16-011). GL is a participant of the Berlin Institute of Health (BIH) Charité Clinician Scientist Programme, which receives grants from the Charité–Universitätsmedizin Berlin and the Berlin Institute of Health. MB is supported by the Deutsche Forschungsgemeinschaft-funded Collaborative Research Centre PolyTarget (SFB 1278, Project ID 316213987). WJW is supported by the Netherlands Organisation for Scientific Research. AT and JFB-M acknowledge funding from CIBERES in the context of CIBERESUCICOVID (part of Instituto de Salud Carlos III). AC acknowledges funding from the Ministero della Salute, Bando Ricerca COVID-19 (2020–2021, grant number COVID-2020-12371808). The views expressed in this publication are those of the authors and not necessarily those of the UK National Health Service, the NIHR, or the Department of Health and Social Care. We thank Margit Leitner for technical help with figure design.
Contributors
Each author co-wrote at least one section of this Series paper as part of a panel composed of three or more authors who were selected on the basis of their expertise (biochemistry, immunology, critical care, respiratory medicine, or infectious diseases). The panels (and sections) were as follows: SC, RA, AdlF, and JFB-M (Comparison of SARS, MERS, and COVID-19); MB, MAW, CC, AT, RF, and MG (Pneumonia and lung damage); MFO, MSW, IM-L, and JFB-M (Coagulopathy and endothelial damage); MFO, SC, GL, SW, WJW, MGN, and TvdP (Cytokine response); MK, SBF, EJG-B, MG, and MGN (Non-cytokine mediators); FMB, CM, TSp, and JCS (Hormones and endocrine factors); MFO, TSk, GM, FV, AG-S, J-MC, FU, AC, and IR (Cellular immune response); MS-H, AC, and IR (B cells and the humoral response). All authors except for MS-H, FMB, AdlF, CC, RF, MG, and AC also served as internal reviewers for other sections. Each draft section was internally reviewed by at least two other authors; to ensure unbiased internal review, the section drafts were allocated by IR to reviewers who were blinded to section authors. MFO, MSW, TSk, WJW, and IR designed and created the figures. MFO, MSW, and IR wrote the abstract and the introduction and concluding sections, compiled all sections, and provided final editing of the manuscript. IR coordinated work on the Series paper. All authors participated in literature searches and critical analysis of published data, and revised their drafts and carried out a final update to include all recent relevant studies published during the writing of the manuscript. All authors read and approved the final manuscript. All authors except for AdlF, TSp, and CC are members of the European Group on Immunology of Sepsis.
References
- 1.Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS-CoV-2. Nat Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Worldometer COVID-19 coronavirus pandemic. April 25, 2021. https://www.worldometers.info/coronavirus/
- 3.Johns Hopkins University of Medicine COVID-19 dashboard by the Center for Systems Science and Engineering (CSS) at Johns Hopkins University (JHU) https://coronavirus.jhu.edu/map.html
- 4.Carfì A, Bernabei R, Landi F, Gemelli Against COVID-19 Post-Acute Care Study Group Persistent symptoms in patients after acute COVID-19. JAMA. 2020;324:603–605. doi: 10.1001/jama.2020.12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Helms J, Kremer S, Merdji H, et al. Neurologic features in severe SARS-CoV-2 infection. N Engl J Med. 2020;382:2268–2270. doi: 10.1056/NEJMc2008597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fraser E. Long term respiratory complications of covid-19. BMJ. 2020;370 doi: 10.1136/bmj.m3001. [DOI] [PubMed] [Google Scholar]
- 7.The Editors of the Lancet Group Learning from a retraction. Lancet. 2020;396 doi: 10.1016/S0140-6736(20)31958-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rochwerg B, Parke R, Murthy S, et al. Misinformation during the coronavirus disease 2019 outbreak: how knowledge emerges from noise. Crit Care Explor. 2020;2 doi: 10.1097/CCE.0000000000000098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steinberg I. The Conversation: Coronavirus research done too fast is testing publishing safeguards, bad science is getting through. April 9, 2020. https://theconversation.com/coronavirus-research-done-too-fast-is-testing-publishing-safeguards-bad-science-is-getting-through-134653
- 10.Weber B, CBC Scientists cut peer-review corners under pressure of COVID-19 pandemic. April 21, 2020. https://www.cbc.ca/news/canada/edmonton/scientists-covid-pandemic-research-misinformation-1.5539997
- 11.Osuchowski MF, Aletti F, Cavaillon JM, et al. SARS-CoV-2/COVID-19: evolving reality, global response, knowledge gaps, and opportunities. Shock. 2020;54:416–437. doi: 10.1097/SHK.0000000000001565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Centers for Disease Control and Prevention Frequently asked questions about SARS. https://www.cdc.gov/sars/about/faq.html
- 13.WHO Middle East respiratory syndrome coronavirus (MERS-CoV) – The Kingdom of Saudi Arabia. https://www.who.int/csr/don/08-april-2020-mers-saudi-arabia/en/
- 14.Thoms M, Buschauer R, Ameismeier M, et al. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science. 2020;369:1249–1255. doi: 10.1126/science.abc8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xia H, Cao Z, Xie X, et al. Evasion of type I interferon by SARS-CoV-2. Cell Rep. 2020;33 doi: 10.1016/j.celrep.2020.108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harrison AG, Lin T, Wang P. Mechanisms of SARS-CoV-2 transmission and pathogenesis. Trends Immunol. 2020;41:1100–1115. doi: 10.1016/j.it.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang W, Xu Y, Gao R, et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323:1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiao F, Tang M, Zheng X, Liu Y, Li X, Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158:1831–1833.e3. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stanifer ML, Kee C, Cortese M, et al. Critical role of type III interferon in controlling SARS-CoV-2 infection in human intestinal epithelial cells. Cell Rep. 2020;32 doi: 10.1016/j.celrep.2020.107863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chu H, Zhou J, Wong BH, et al. Productive replication of Middle East respiratory syndrome coronavirus in monocyte-derived dendritic cells modulates innate immune response. Virology. 2014;454–455:197–205. doi: 10.1016/j.virol.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheung CY, Poon LL, Ng IH, et al. Cytokine responses in severe acute respiratory syndrome coronavirus-infected macrophages in vitro: possible relevance to pathogenesis. J Virol. 2005;79:7819–7826. doi: 10.1128/JVI.79.12.7819-7826.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou J, Chu H, Li C, et al. Active replication of Middle East respiratory syndrome coronavirus and aberrant induction of inflammatory cytokines and chemokines in human macrophages: implications for pathogenesis. J Infect Dis. 2014;209:1331–1342. doi: 10.1093/infdis/jit504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Y, Feng Z, Diao B, et al. The novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) directly decimates human spleens and lymph nodes. medRxiv. 2020 doi: 10.1101/2020.03.27.20045427. published online March 31. (preprint). [DOI] [Google Scholar]
- 24.Wang C, Xie J, Zhao L, et al. Alveolar macrophage dysfunction and cytokine storm in the pathogenesis of two severe COVID-19 patients. EBioMedicine. 2020;57 doi: 10.1016/j.ebiom.2020.102833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wölfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 26.Rockx B, Kuiken T, Herfst S, et al. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science. 2020;368:1012–1015. doi: 10.1126/science.abb7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peiris JSM, Chu CM, Cheng VCC, et al. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pan Y, Zhang D, Yang P, Poon LLM, Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect Dis. 2020;20:411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He X, Lau EHY, Wu P, et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med. 2020;26:672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- 30.Corman VM, Albarrak AM, Omrani AS, et al. Viral shedding and antibody response in 37 patients with Middle East respiratory syndrome coronavirus infection. Clin Infect Dis. 2016;62:477–483. doi: 10.1093/cid/civ951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yilmaz A, Marklund E, Andersson M, et al. Upper respiratory tract levels of SARS-CoV-2 RNA and duration of viral RNA shedding do not differ between patients with mild and severe/critical COVID-19. J Infect Dis. 2021;223:15–18. doi: 10.1093/infdis/jiaa632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chandrashekar A, Liu J, Martinot AJ, et al. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science. 2020;369:812–817. doi: 10.1126/science.abc4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ni L, Ye F, Cheng M-L, et al. Detection of SARS-CoV-2-specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity. 2020;52:971–977.e3. doi: 10.1016/j.immuni.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weis S, Scherag A, Baier M, et al. Antibody response using six different serological assays in a completely PCR-tested community after a COVID-19 outbreak—the CoNAN study. Clin Microbiol Infect. 2021;27:470.e1–470.e9. doi: 10.1016/j.cmi.2020.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sekine T, Perez-Potti A, Rivera-Ballesteros O, et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020;183:158–168.e14. doi: 10.1016/j.cell.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ibarrondo FJ, Fulcher JA, Goodman-Meza D, et al. Rapid decay of anti-SARS-CoV-2 antibodies in persons with mild Covid-19. N Engl J Med. 2020;383:1085–1087. doi: 10.1056/NEJMc2025179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cao W-C, Liu W, Zhang P-H, Zhang F, Richardus JH. Disappearance of antibodies to SARS-associated coronavirus after recovery. N Engl J Med. 2007;357:1162–1163. doi: 10.1056/NEJMc070348. [DOI] [PubMed] [Google Scholar]
- 38.Tang F, Quan Y, Xin ZT, et al. Lack of peripheral memory B cell responses in recovered patients with severe acute respiratory syndrome: a six-year follow-up study. J Immunol. 2011;186:7264–7268. doi: 10.4049/jimmunol.0903490. [DOI] [PubMed] [Google Scholar]
- 39.Wu LP, Wang NC, Chang YH, et al. Duration of antibody responses after severe acute respiratory syndrome. Emerg Infect Dis. 2007;13:1562–1564. doi: 10.3201/eid1310.070576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo X, Guo Z, Duan C, et al. Long-term persistence of IgG antibodies in SARS-CoV infected healthcare workers. medRxiv. 2020 doi: 10.1101/2020.02.12.20021386. published online Feb 14. (preprint). [DOI] [Google Scholar]
- 41.Dan JM, Mateus J, Kato Y, et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021;371 doi: 10.1126/science.abf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Le Bert N, Tan AT, Kunasegaran K, et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 43.Altmann DM, Boyton RJ. SARS-CoV-2 T cell immunity: specificity, function, durability, and role in protection. Sci Immunol. 2020;5 doi: 10.1126/sciimmunol.abd6160. [DOI] [PubMed] [Google Scholar]
- 44.Mateus J, Grifoni A, Tarke A, et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;370:89–94. doi: 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Du L, Ma C, Jiang S. Antibodies induced by receptor-binding domain in spike protein of SARS-CoV do not cross-neutralize the novel human coronavirus hCoV-EMC. J Infect. 2013;67:348–350. doi: 10.1016/j.jinf.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Che XY, Qiu LW, Liao ZY, et al. Antigenic cross-reactivity between severe acute respiratory syndrome-associated coronavirus and human coronaviruses 229E and OC43. J Infect Dis. 2005;191:2033–2037. doi: 10.1086/430355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chan KH, Cheng VC, Woo PC, et al. Serological responses in patients with severe acute respiratory syndrome coronavirus infection and cross-reactivity with human coronaviruses 229E, OC43, and NL63. Clin Diagn Lab Immunol. 2005;12:1317–1321. doi: 10.1128/CDLI.12.11.1317-1321.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lv H, Wu NC, Tsang OT-Y, et al. Cross-reactive antibody response between SARS-CoV-2 and SARS-CoV infections. Cell Rep. 2020;31 doi: 10.1016/j.celrep.2020.107725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Premkumar L, Segovia-Chumbez B, Jadi R, et al. The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci Immunol. 2020;5 doi: 10.1126/sciimmunol.abc8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.vWei P-F, ed. Diagnosis and treatment protocol for novel coronavirus pneumonia (trial version 7). Chin Med J (Engl) 2020; 133: 1087–95. [DOI] [PMC free article] [PubMed]
- 51.Epidemiology Working Group for NCIP Epidemic Response. Chinese Center for Disease Control and Prevention The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19) in China. Zhonghua Liu Xing Bing Xue Za Zhi. 2020;41:145–151. doi: 10.3760/cma.j.issn.0254-6450.2020.02.003. [DOI] [PubMed] [Google Scholar]
- 52.Hou YJ, Okuda K, Edwards CE, et al. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell. 2020;182:429–446.e14. doi: 10.1016/j.cell.2020.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tang X, Du RH, Wang R, et al. Comparison of hospitalized patients with ARDS caused by COVID-19 and H1N1. Chest. 2020;158:195–205. doi: 10.1016/j.chest.2020.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ellinghaus D, Degenhardt F, Bujanda L, et al. Genomewide association study of severe Covid-19 with respiratory failure. N Engl J Med. 2020;383:1522–1534. doi: 10.1056/NEJMoa2020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Siswanto GM, Gani M, Fauzi AR, et al. Possible silent hypoxemia in a COVID-19 patient: a case report. Ann Med Surg (Lond) 2020;60:583–586. doi: 10.1016/j.amsu.2020.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dhont S, Derom E, Van Braeckel E, Depuydt P, Lambrecht BN. The pathophysiology of ‘happy’ hypoxemia in COVID-19. Respir Res. 2020;21:198. doi: 10.1186/s12931-020-01462-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guan WJ, Ni ZY, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Manning HL, Schwartzstein RM. Pathophysiology of dyspnea. N Engl J Med. 1995;333:1547–1553. doi: 10.1056/NEJM199512073332307. [DOI] [PubMed] [Google Scholar]
- 59.Duffin J. Measuring the ventilatory response to hypoxia. J Physiol. 2007;584:285–293. doi: 10.1113/jphysiol.2007.138883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brochard L, Slutsky A, Pesenti A. Mechanical ventilation to minimize progression of lung injury in acute respiratory failure. Am J Respir Crit Care Med. 2017;195:438–442. doi: 10.1164/rccm.201605-1081CP. [DOI] [PubMed] [Google Scholar]
- 61.Tobin MJ, Laghi F, Jubran A. Why COVID-19 silent hypoxemia is baffling to physicians. Am J Respir Crit Care Med. 2020;202:356–360. doi: 10.1164/rccm.202006-2157CP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mo X, Jian W, Su Z, et al. Abnormal pulmonary function in COVID-19 patients at time of hospital discharge. Eur Respir J. 2020;55 doi: 10.1183/13993003.01217-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lang M, Som A, Mendoza DP, et al. Hypoxaemia related to COVID-19: vascular and perfusion abnormalities on dual-energy CT. Lancet Infect Dis. 2020;20:1365–1366. doi: 10.1016/S1473-3099(20)30367-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Westblade LF, Brar G, Pinheiro LC, et al. SARS-CoV-2 viral load predicts mortality in patients with and without cancer who are hospitalized with COVID-19. Cancer Cell. 2020;38:661–671.e2. doi: 10.1016/j.ccell.2020.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Magleby R, Westblade LF, Trzebucki A, et al. Impact of SARS-CoV-2 viral load on risk of intubation and mortality among hospitalized patients with coronavirus disease 2019. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa851. published online June 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hikmet F, Méar L, Edvinsson Å, Micke P, Uhlén M, Lindskog C. The protein expression profile of ACE2 in human tissues. Mol Syst Biol. 2020;16 doi: 10.15252/msb.20209610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dickson RP, Erb-Downward JR, Martinez FJ, Huffnagle GB. The microbiome and the respiratory tract. Annu Rev Physiol. 2016;78:481–504. doi: 10.1146/annurev-physiol-021115-105238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Morawska L, Cao J. Airborne transmission of SARS-CoV-2: the world should face the reality. Environ Int. 2020;139 doi: 10.1016/j.envint.2020.105730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wilson NM, Norton A, Young FP, Collins DW. Airborne transmission of severe acute respiratory syndrome coronavirus-2 to healthcare workers: a narrative review. Anaesthesia. 2020;75:1086–1095. doi: 10.1111/anae.15093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cantuti-Castelvetri L, Ojha R, Pedro LD, et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370:856–860. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang S, Qiu Z, Hou Y, et al. AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res. 2021;31:126–140. doi: 10.1038/s41422-020-00460-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Carsana L, Sonzogni A, Nasr A, et al. Pulmonary post-mortem findings in a series of COVID-19 cases from northern Italy: a two-centre descriptive study. Lancet Infect Dis. 2020;20:1135–1140. doi: 10.1016/S1473-3099(20)30434-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hellman U, Karlsson MG, Engström-Laurent A, et al. Presence of hyaluronan in lung alveoli in severe Covid-19: an opening for new treatment options? J Biol Chem. 2020;295:15418–15422. doi: 10.1074/jbc.AC120.015967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yao XH, He ZC, Li TY, et al. Pathological evidence for residual SARS-CoV-2 in pulmonary tissues of a ready-for-discharge patient. Cell Res. 2020;30:541–543. doi: 10.1038/s41422-020-0318-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ackermann M, Verleden SE, Kuehnel M, et al. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in Covid-19. N Engl J Med. 2020;383:120–128. doi: 10.1056/NEJMoa2015432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Prilutskiy A, Kritselis M, Shevtsov A, et al. SARS-CoV-2 infection-associated hemophagocytic lymphohistiocytosis. Am J Clin Pathol. 2020;154:466–474. doi: 10.1093/ajcp/aqaa124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nicholls JM, Poon LL, Lee KC, et al. Lung pathology of fatal severe acute respiratory syndrome. Lancet. 2003;361:1773–1778. doi: 10.1016/S0140-6736(03)13413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sorbello M, El-Boghdadly K, Di Giacinto I, et al. The Italian coronavirus disease 2019 outbreak: recommendations from clinical practice. Anaesthesia. 2020;75:724–732. doi: 10.1111/anae.15049. [DOI] [PubMed] [Google Scholar]
- 80.Gattinoni L, Chiumello D, Caironi P, et al. COVID-19 pneumonia: different respiratory treatments for different phenotypes? Intensive Care Med. 2020;46:1099–1102. doi: 10.1007/s00134-020-06033-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tobin MJ, Laghi F, Jubran A. Caution about early intubation and mechanical ventilation in COVID-19. Ann Intensive Care. 2020;10:78. doi: 10.1186/s13613-020-00692-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Grasselli G, Tonetti T, Protti A, et al. Pathophysiology of COVID-19-associated acute respiratory distress syndrome: a multicentre prospective observational study. Lancet Respir Med. 2020;8:1201–1208. doi: 10.1016/S2213-2600(20)30370-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gattinoni L, Marini JJ, Camporota L. The respiratory drive: an overlooked tile of COVID-19 pathophysiology. Am J Respir Crit Care Med. 2020;202:1079–1080. doi: 10.1164/rccm.202008-3142ED. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wilson MS, Wynn TA. Pulmonary fibrosis: pathogenesis, etiology and regulation. Mucosal Immunol. 2009;2:103–121. doi: 10.1038/mi.2008.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Quartuccio L, Semerano L, Benucci M, Boissier MC, De Vita S. Urgent avenues in the treatment of COVID-19: targeting downstream inflammation to prevent catastrophic syndrome. Joint Bone Spine. 2020;87:191–193. doi: 10.1016/j.jbspin.2020.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen W. A potential treatment of COVID-19 with TGF-β blockade. Int J Biol Sci. 2020;16:1954–1955. doi: 10.7150/ijbs.46891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Eapen MS, Lu W, Gaikwad AV, et al. Endothelial to mesenchymal transition: a precursor to post-COVID-19 interstitial pulmonary fibrosis and vascular obliteration? Eur Respir J. 2020;56 doi: 10.1183/13993003.03167-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gu J, Korteweg C. Pathology and pathogenesis of severe acute respiratory syndrome. Am J Pathol. 2007;170:1136–1147. doi: 10.2353/ajpath.2007.061088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hui DS, Joynt GM, Wong KT, et al. Impact of severe acute respiratory syndrome (SARS) on pulmonary function, functional capacity and quality of life in a cohort of survivors. Thorax. 2005;60:401–409. doi: 10.1136/thx.2004.030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Herridge MS, Tansey CM, Matté A, et al. Functional disability 5 years after acute respiratory distress syndrome. N Engl J Med. 2011;364:1293–1304. doi: 10.1056/NEJMoa1011802. [DOI] [PubMed] [Google Scholar]
- 91.Hwang DM, Chamberlain DW, Poutanen SM, Low DE, Asa SL, Butany J. Pulmonary pathology of severe acute respiratory syndrome in Toronto. Mod Pathol. 2005;18:1–10. doi: 10.1038/modpathol.3800247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Guo Y, Korteweg C, McNutt MA, Gu J. Pathogenetic mechanisms of severe acute respiratory syndrome. Virus Res. 2008;133:4–12. doi: 10.1016/j.virusres.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Edler C, Schröder AS, Aepfelbacher M, et al. Dying with SARS-CoV-2 infection-an autopsy study of the first consecutive 80 cases in Hamburg, Germany. Int J Legal Med. 2020;134:1275–1284. doi: 10.1007/s00414-020-02317-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Barton LM, Duval EJ, Stroberg E, Ghosh S, Mukhopadhyay S. COVID-19 autopsies, Oklahoma, USA. Am J Clin Pathol. 2020;153:725–733. doi: 10.1093/ajcp/aqaa062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ojha V, Mani A, Pandey NN, Sharma S, Kumar S. CT in coronavirus disease 2019 (COVID-19): a systematic review of chest CT findings in 4410 adult patients. Eur Radiol. 2020;30:6129–6138. doi: 10.1007/s00330-020-06975-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yu M, Liu Y, Xu D, Zhang R, Lan L, Xu H. Prediction of the development of pulmonary fibrosis using serial thin-section CT and clinical features in patients discharged after treatment for COVID-19 pneumonia. Korean J Radiol. 2020;21:746–755. doi: 10.3348/kjr.2020.0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Klok FA, Kruip MJHA, van der Meer NJM, et al. Incidence of thrombotic complications in critically ill ICU patients with COVID-19. Thromb Res. 2020;191:145–147. doi: 10.1016/j.thromres.2020.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Obi AT, Barnes GD, Napolitano LM, Henke PK, Wakefield TW. Venous thrombosis epidemiology, pathophysiology, and anticoagulant therapies and trials in severe acute respiratory syndrome coronavirus 2 infection. J Vasc Surg Venous Lymphat Disord. 2021;9:23–35. doi: 10.1016/j.jvsv.2020.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Obi AT, Pannucci CJ, Nackashi A, et al. Validation of the Caprini venous thromboembolism risk assessment model in critically ill surgical patients. JAMA Surg. 2015;150:941–948. doi: 10.1001/jamasurg.2015.1841. [DOI] [PubMed] [Google Scholar]
- 100.Burkhard-Koren NM, Haberecker M, Maccio U, et al. Higher prevalence of pulmonary macrothrombi in SARS-CoV-2 than in influenza A: autopsy results from ‘Spanish flu’ 1918/1919 in Switzerland to Coronavirus disease 2019. J Pathol Clin Res. 2021;7:135–143. doi: 10.1002/cjp2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wu C, Chen X, Cai Y, et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med. 2020;180:934–943. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chen T, Wu D, Chen H, et al. Clinical characteristics of 113 deceased patients with coronavirus disease 2019: retrospective study. BMJ. 2020;368 doi: 10.1136/bmj.m1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Goyal P, Choi JJ, Pinheiro LC, et al. Clinical characteristics of Covid-19 in New York City. N Engl J Med. 2020;382:2372–2374. doi: 10.1056/NEJMc2010419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Thachil J, Tang N, Gando S, et al. ISTH interim guidance on recognition and management of coagulopathy in COVID-19. J Thromb Haemost. 2020;18:1023–1026. doi: 10.1111/jth.14810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Leisman DE, Ronner L, Pinotti R, et al. Cytokine elevation in severe and critical COVID-19: a rapid systematic review, meta-analysis, and comparison with other inflammatory syndromes. Lancet Respir Med. 2020;8:1233–1244. doi: 10.1016/S2213-2600(20)30404-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lippi G, Plebani M. Laboratory abnormalities in patients with COVID-2019 infection. Clin Chem Lab Med. 2020;58:1131–1134. doi: 10.1515/cclm-2020-0198. [DOI] [PubMed] [Google Scholar]
- 108.Jirak P, Larbig R, Shomanova Z, et al. Myocardial injury in severe COVID-19 is similar to pneumonias of other origin: results from a multicentre study. ESC Heart Fail. 2021;8:37–46. doi: 10.1002/ehf2.13136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mazzoni A, Salvati L, Maggi L, et al. Impaired immune cell cytotoxicity in severe COVID-19 is IL-6 dependent. J Clin Invest. 2020;130:4694–4703. doi: 10.1172/JCI138554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Varga Z, Flammer AJ, Steiger P, et al. Endothelial cell infection and endotheliitis in COVID-19. Lancet. 2020;395:1417–1418. doi: 10.1016/S0140-6736(20)30937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bösmüller H, Traxler S, Bitzer M, et al. The evolution of pulmonary pathology in fatal COVID-19 disease: an autopsy study with clinical correlation. Virchows Arch. 2020;477:349–357. doi: 10.1007/s00428-020-02881-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Schaefer IM, Padera RF, Solomon IH, et al. In situ detection of SARS-CoV-2 in lungs and airways of patients with COVID-19. Mod Pathol. 2020;33:2104–2114. doi: 10.1038/s41379-020-0595-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Su H, Yang M, Wan C, et al. Renal histopathological analysis of 26 postmortem findings of patients with COVID-19 in China. Kidney Int. 2020;98:219–227. doi: 10.1016/j.kint.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Buja LM, Wolf DA, Zhao B, et al. The emerging spectrum of cardiopulmonary pathology of the coronavirus disease 2019 (COVID-19): report of 3 autopsies from Houston, Texas, and review of autopsy findings from other United States cities. Cardiovasc Pathol. 2020;48 doi: 10.1016/j.carpath.2020.107233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sadegh Beigee F, Pourabdollah Toutkaboni M, Khalili N, et al. Diffuse alveolar damage and thrombotic microangiopathy are the main histopathological findings in lung tissue biopsy samples of COVID-19 patients. Pathol Res Pract. 2020;216 doi: 10.1016/j.prp.2020.153228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Iba T, Levy JH, Connors JM, Warkentin TE, Thachil J, Levi M. The unique characteristics of COVID-19 coagulopathy. Crit Care. 2020;24:360. doi: 10.1186/s13054-020-03077-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Copin MC, Parmentier E, Duburcq T, Poissy J, Mathieu D. Time to consider histologic pattern of lung injury to treat critically ill patients with COVID-19 infection. Intensive Care Med. 2020;46:1124–1126. doi: 10.1007/s00134-020-06057-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.McGonagle D, O'Donnell JS, Sharif K, Emery P, Bridgewood C. Immune mechanisms of pulmonary intravascular coagulopathy in COVID-19 pneumonia. Lancet Rheumatol. 2020;2:e437–e445. doi: 10.1016/S2665-9913(20)30121-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Arachchillage DJ, Stacey A, Akor F, Scotz M, Laffan M. Thrombolysis restores perfusion in COVID-19 hypoxia. Br J Haematol. 2020;190:e270–e274. doi: 10.1111/bjh.17050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang Y, Xiao M, Zhang S, et al. Coagulopathy and antiphospholipid antibodies in patients with Covid-19. N Engl J Med. 2020;382:e38. doi: 10.1056/NEJMc2007575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Botta M, Tsonas AM, Pillay J, et al. Ventilation management and clinical outcomes in invasively ventilated patients with COVID-19 (PRoVENT-COVID): a national, multicentre, observational cohort study. Lancet Respir Med. 2021;9:139–148. doi: 10.1016/S2213-2600(20)30459-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sinha P, Calfee CS, Cherian S, et al. Prevalence of phenotypes of acute respiratory distress syndrome in critically ill patients with COVID-19: a prospective observational study. Lancet Respir Med. 2020;8:1209–1218. doi: 10.1016/S2213-2600(20)30366-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Puelles VG, Lütgehetmann M, Lindenmeyer MT, et al. Multiorgan and renal tropism of SARS-CoV-2. N Engl J Med. 2020;383:590–592. doi: 10.1056/NEJMc2011400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wichmann D, Sperhake JP, Lütgehetmann M, et al. Autopsy findings and venous thromboembolism in patients with COVID-19: a prospective cohort study. Ann Intern Med. 2020;173:268–277. doi: 10.7326/M20-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gill SE, Dos Santos CC, O'Gorman DB, et al. Transcriptional profiling of leukocytes in critically ill COVID19 patients: implications for interferon response and coagulation. Intensive Care Med Exp. 2020;8:75. doi: 10.1186/s40635-020-00361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Deinhardt-Emmer S, Wittschieber D, Sanft J, et al. Early postmortem mapping of SARS-CoV-2 RNA in patients with COVID-19 and the correlation with tissue damage. Elife. 2021;10 doi: 10.7554/eLife.60361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.South AM, Diz DI, Chappell MC. COVID-19, ACE2, and the cardiovascular consequences. Am J Physiol Heart Circ Physiol. 2020;318:H1084–H1090. doi: 10.1152/ajpheart.00217.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lelubre C, Vincent JL. Mechanisms and treatment of organ failure in sepsis. Nat Rev Nephrol. 2018;14:417–427. doi: 10.1038/s41581-018-0005-7. [DOI] [PubMed] [Google Scholar]
- 130.Gupta A, Madhavan MV, Sehgal K, et al. Extrapulmonary manifestations of COVID-19. Nat Med. 2020;26:1017–1032. doi: 10.1038/s41591-020-0968-3. [DOI] [PubMed] [Google Scholar]
- 131.Onofrio L, Caraglia M, Facchini G, Margherita V, Placido S, Buonerba C. Toll-like receptors and COVID-19: a two-faced story with an exciting ending. Future Sci OA. 2020;6 doi: 10.2144/fsoa-2020-0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Totura AL, Whitmore A, Agnihothram S, et al. Toll-like receptor 3 signaling via TRIF contributes to a protective innate immune response to severe acute respiratory syndrome coronavirus infection. MBio. 2015;6:e00638–e00715. doi: 10.1128/mBio.00638-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.van der Made CI, Simons A, Schuurs-Hoeijmakers J, et al. Presence of genetic variants among young men with severe COVID-19. JAMA. 2020;324:663–673. doi: 10.1001/jama.2020.13719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhang Q, Bastard P, Liu Z, et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370 doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Merad M, Martin JC. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat Rev Immunol. 2020;20:355–362. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bermejo-Martin JF, González-Rivera M, Almansa R, et al. Viral RNA load in plasma is associated with critical illness and a dysregulated host response in COVID-19. Crit Care. 2020;24:691. doi: 10.1186/s13054-020-03398-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sinha P, Matthay MA, Calfee CS. Is a “cytokine storm” relevant to COVID-19? JAMA Intern Med. 2020;180:1152–1154. doi: 10.1001/jamainternmed.2020.3313. [DOI] [PubMed] [Google Scholar]
- 139.Chen X, Zhao B, Qu Y, et al. Detectable serum severe acute respiratory syndrome coronavirus 2 viral load (RNAemia) is closely correlated with drastically elevated interleukin 6 level in critically ill patients with coronavirus disease 2019. Clin Infect Dis. 2020;71:1937–1942. doi: 10.1093/cid/ciaa449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wen W, Su W, Tang H, et al. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discov. 2020;6:31. doi: 10.1038/s41421-020-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gao Y, Li T, Han M, et al. Diagnostic utility of clinical laboratory data determinations for patients with the severe COVID-19. J Med Virol. 2020;92:791–796. doi: 10.1002/jmv.25770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ruan Q, Yang K, Wang W, Jiang L, Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020;46:846–848. doi: 10.1007/s00134-020-05991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Balnis J, Adam AP, Chopra A, et al. Unique inflammatory profile is associated with higher SARS-CoV-2 acute respiratory distress syndrome (ARDS) mortality. Am J Physiol Regul Integr Comp Physiol. 2021;320:R250–R257. doi: 10.1152/ajpregu.00324.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chen G, Wu D, Guo W, et al. Clinical and immunologic features in severe and moderate coronavirus disease 2019. J Clin Invest. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wen W, Su W, Tang H, et al. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discov. 2020;6:31. doi: 10.1038/s41421-020-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Payen D, Cravat M, Maadadi H, et al. A longitudinal study of immune cells in severe COVID-19 patients. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.580250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Giamarellos-Bourboulis EJ, Netea MG, Rovina N, et al. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe. 2020;27:992–1000.e3. doi: 10.1016/j.chom.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Carter MJ, Fish M, Jennings A, et al. Peripheral immunophenotypes in children with multisystem inflammatory syndrome associated with SARS-CoV-2 infection. Nat Med. 2020;26:1701–1707. doi: 10.1038/s41591-020-1054-6. [DOI] [PubMed] [Google Scholar]
- 149.Feldstein LR, Rose EB, Horwitz SM, et al. Multisystem inflammatory syndrome in U.S. children and adolescents. N Engl J Med. 2020;383:334–346. doi: 10.1056/NEJMoa2021680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Dufort EM, Koumans EH, Chow EJ, et al. Multisystem inflammatory syndrome in children in New York State. N Engl J Med. 2020;383:347–358. doi: 10.1056/NEJMoa2021756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bastard P, Rosen LB, Zhang Q, et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370 doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hadjadj J, Yatim N, Barnabei L, et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Blanco-Melo D, Nilsson-Payant BE, Liu WC, et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Trouillet-Assant S, Viel S, Gaymard A, et al. Type I IFN immunoprofiling in COVID-19 patients. J Allergy Clin Immunol. 2020;146:206–208.e2. doi: 10.1016/j.jaci.2020.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Galani IE, Rovina N, Lampropoulou V, et al. Untuned antiviral immunity in COVID-19 revealed by temporal type I/III interferon patterns and flu comparison. Nat Immunol. 2021;22:32–40. doi: 10.1038/s41590-020-00840-x. [DOI] [PubMed] [Google Scholar]
- 156.Lee JS, Park S, Jeong HW, et al. Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19. Sci Immunol. 2020;5 doi: 10.1126/sciimmunol.abd1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kox M, Waalders NJB, Kooistra EJ, Gerretsen J, Pickkers P. Cytokine levels in critically ill patients with COVID-19 and other conditions. JAMA. 2020;324:1565–1567. doi: 10.1001/jama.2020.17052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Monneret G, Benlyamani I, Gossez M, et al. COVID-19: What type of cytokine storm are we dealing with? J Med Virol. 2021;93:197–198. doi: 10.1002/jmv.26317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zhou Z, Ren L, Zhang L, et al. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe. 2020;27:883–890.e2. doi: 10.1016/j.chom.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Xiong Y, Liu Y, Cao L, et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg Microbes Infect. 2020;9:761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ronit A, Berg RMG, Bay JT, et al. Compartmental immunophenotyping in COVID-19 ARDS: a case series. J Allergy Clin Immunol. 2021;147:81–91. doi: 10.1016/j.jaci.2020.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Carvelli J, Demaria O, Vély F, et al. Association of COVID-19 inflammation with activation of the C5a-C5aR1 axis. Nature. 2020;588:146–150. doi: 10.1038/s41586-020-2600-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Yang L, Han Y, Nilsson-Payant BE, et al. A human pluripotent stem cell-based platform to study SARS-CoV-2 tropism and model virus infection in human cells and organoids. Cell Stem Cell. 2020;27:125–136.e7. doi: 10.1016/j.stem.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Nienhold R, Ciani Y, Koelzer VH, et al. Two distinct immunopathological profiles in autopsy lungs of COVID-19. Nat Commun. 2020;11 doi: 10.1038/s41467-020-18854-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Toldo S, Bussani R, Nuzzi V, et al. Inflammasome formation in the lungs of patients with fatal COVID-19. Inflamm Res. 2021;70:7–10. doi: 10.1007/s00011-020-01413-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Li MY, Li L, Zhang Y, Wang XS. Expression of the SARS-CoV-2 cell receptor gene ACE2 in a wide variety of human tissues. Infect Dis Poverty. 2020;9:45. doi: 10.1186/s40249-020-00662-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Lachmann G, Knaak C, Vorderwülbecke G, et al. Hyperferritinemia in critically ill patients. Crit Care Med. 2020;48:459–465. doi: 10.1097/CCM.0000000000004131. [DOI] [PubMed] [Google Scholar]
- 168.Kyriazopoulou E, Leventogiannis K, Norrby-Teglund A, et al. Macrophage activation-like syndrome: an immunological entity associated with rapid progression to death in sepsis. BMC Med. 2017;15:172. doi: 10.1186/s12916-017-0930-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Mesas AE, Cavero-Redondo I, Álvarez-Bueno C, et al. Predictors of in-hospital COVID-19 mortality: a comprehensive systematic review and meta-analysis exploring differences by age, sex and health conditions. PLoS One. 2020;15 doi: 10.1371/journal.pone.0241742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Cheng L, Li H, Li L, et al. Ferritin in the coronavirus disease 2019 (COVID-19): a systematic review and meta-analysis. J Clin Lab Anal. 2020;34 doi: 10.1002/jcla.23618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Masetti C, Generali E, Colapietro F, et al. High mortality in COVID-19 patients with mild respiratory disease. Eur J Clin Invest. 2020;50 doi: 10.1111/eci.13314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Al-Samkari H, Karp Leaf RS, Dzik WH, et al. COVID-19 and coagulation: bleeding and thrombotic manifestations of SARS-CoV-2 infection. Blood. 2020;136:489–500. doi: 10.1182/blood.2020006520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Huet T, Beaussier H, Voisin O, et al. Anakinra for severe forms of COVID-19: a cohort study. Lancet Rheumatol. 2020;2:e393–e400. doi: 10.1016/S2665-9913(20)30164-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Cavalli G, De Luca G, Campochiaro C, et al. Interleukin-1 blockade with high-dose anakinra in patients with COVID-19, acute respiratory distress syndrome, and hyperinflammation: a retrospective cohort study. Lancet Rheumatol. 2020;2:e325–e331. doi: 10.1016/S2665-9913(20)30127-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Dimopoulos G, de Mast Q, Markou N, et al. Favorable anakinra responses in severe covid-19 patients with secondary hemophagocytic lymphohistiocytosis. Cell Host Microbe. 2020;28:117–123.e1. doi: 10.1016/j.chom.2020.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Vasquez-Bonilla WO, Orozco R, Argueta V, et al. A review of the main histopathological findings in coronavirus disease 2019. Hum Pathol. 2020;105:74–83. doi: 10.1016/j.humpath.2020.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Fardet L, Galicier L, Lambotte O, et al. Development and validation of the HScore, a score for the diagnosis of reactive hemophagocytic syndrome. Arthritis Rheumatol. 2014;66:2613–2620. doi: 10.1002/art.38690. [DOI] [PubMed] [Google Scholar]
- 178.Feng X, Li S, Sun Q, et al. Immune-inflammatory parameters in COVID-19 cases: a systematic review and meta-analysis. Front Med (Lausanne) 2020;7:301. doi: 10.3389/fmed.2020.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Kox M, Frenzel T, Schouten J, van de Veerdonk FL, Koenen HJPM, Pickkers P. COVID-19 patients exhibit less pronounced immune suppression compared with bacterial septic shock patients. Crit Care. 2020;24:263. doi: 10.1186/s13054-020-02896-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Cugno M, Meroni PL, Gualtierotti R, et al. Complement activation in patients with COVID-19: a novel therapeutic target. J Allergy Clin Immunol. 2020;146:215–217. doi: 10.1016/j.jaci.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Holter JC, Pischke SE, de Boer E, et al. Systemic complement activation is associated with respiratory failure in COVID-19 hospitalized patients. Proc Natl Acad Sci USA. 2020;117:25018–25025. doi: 10.1073/pnas.2010540117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Schuetz P, Albrich W, Mueller B. Procalcitonin for diagnosis of infection and guide to antibiotic decisions: past, present and future. BMC Med. 2011;9:107. doi: 10.1186/1741-7015-9-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Tujula B, Hämäläinen S, Kokki H, Pulkki K, Kokki M. Review of clinical practice guidelines on the use of procalcitonin in infections. Infect Dis (Lond) 2020;52:227–234. doi: 10.1080/23744235.2019.1704860. [DOI] [PubMed] [Google Scholar]
- 184.Bao J, Li C, Zhang K, Kang H, Chen W, Gu B. Comparative analysis of laboratory indexes of severe and non-severe patients infected with COVID-19. Clin Chim Acta. 2020;509:180–194. doi: 10.1016/j.cca.2020.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Soraya GV, Ulhaq ZS. Crucial laboratory parameters in COVID-19 diagnosis and prognosis: an updated meta-analysis. Med Clin (Barc) 2020;155:143–151. doi: 10.1016/j.medcli.2020.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.van Berkel M, Kox M, Frenzel T, Pickkers P, Schouten J. Biomarkers for antimicrobial stewardship: a reappraisal in COVID-19 times? Crit Care. 2020;24:600. doi: 10.1186/s13054-020-03291-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Yang X, Yu Y, Xu J, et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Apicella M, Campopiano MC, Mantuano M, Mazoni L, Coppelli A, Del Prato S. COVID-19 in people with diabetes: understanding the reasons for worse outcomes. Lancet Diabetes Endocrinol. 2020;8:782–792. doi: 10.1016/S2213-8587(20)30238-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Yang JK, Feng Y, Yuan MY, et al. Plasma glucose levels and diabetes are independent predictors for mortality and morbidity in patients with SARS. Diabet Med. 2006;23:623–628. doi: 10.1111/j.1464-5491.2006.01861.x. [DOI] [PubMed] [Google Scholar]
- 190.Taneera J, El-Huneidi W, Hamad M, Mohammed AK, Elaraby E, Hachim MY. Expression profile of SARS-CoV-2 host receptors in human pancreatic islets revealed upregulation of ACE2 in diabetic donors. Biology (Basel) 2020;9:E215. doi: 10.3390/biology9080215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Pal R, Banerjee M. COVID-19 and the endocrine system: exploring the unexplored. J Endocrinol Invest. 2020;43:1027–1031. doi: 10.1007/s40618-020-01276-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Zhu L, She ZG, Cheng X, et al. Association of blood glucose control and outcomes in patients with COVID-19 and pre-existing type 2 diabetes. Cell Metab. 2020;31:1068–1077.e3. doi: 10.1016/j.cmet.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Rao S, Lau A, So HC. Exploring diseases/traits and blood proteins causally related to expression of ACE2, the putative receptor of SARS-CoV-2: a mendelian randomization analysis highlights tentative relevance of diabetes-related traits. Diabetes Care. 2020;43:1416–1426. doi: 10.2337/dc20-0643. [DOI] [PubMed] [Google Scholar]
- 194.Marshall RJ, Armart P, Hulme KD, et al. Glycemic variability in diabetes increases the severity of influenza. MBio. 2020;11:e02841–e02849. doi: 10.1128/mBio.02841-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Nielsen TB, Pantapalangkoor P, Yan J, et al. Diabetes exacerbates infection via hyperinflammation by signaling through TLR4 and RAGE. MBio. 2017;8:e00818–e00917. doi: 10.1128/mBio.00818-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Pal R, Bhansali A. COVID-19, diabetes mellitus and ACE2: the conundrum. Diabetes Res Clin Pract. 2020;162 doi: 10.1016/j.diabres.2020.108132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Busse LW, Chow JH, McCurdy MT, Khanna AK. COVID-19 and the RAAS-a potential role for angiotensin II? Crit Care. 2020;24:136. doi: 10.1186/s13054-020-02862-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Fosbøl EL, Butt JH, Østergaard L, et al. Association of angiotensin-converting enzyme inhibitor or angiotensin receptor blocker use with COVID-19 diagnosis and mortality. JAMA. 2020;324:168–177. doi: 10.1001/jama.2020.11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Han Y, Runge MS, Brasier AR. Angiotensin II induces interleukin-6 transcription in vascular smooth muscle cells through pleiotropic activation of nuclear factor-kappa B transcription factors. Circ Res. 1999;84:695–703. doi: 10.1161/01.res.84.6.695. [DOI] [PubMed] [Google Scholar]
- 200.Franco R, Rivas-Santisteban R, Serrano-Marín J, Rodríguez-Pérez AI, Labandeira-García JL, Navarro G. SARS-CoV-2 as a factor to disbalance the renin-angiotensin system: a suspect in the case of exacerbated IL-6 production. J Immunol. 2020;205:1198–1206. doi: 10.4049/jimmunol.2000642. [DOI] [PubMed] [Google Scholar]
- 201.Verdecchia P, Cavallini C, Spanevello A, Angeli F. The pivotal link between ACE2 deficiency and SARS-CoV-2 infection. Eur J Intern Med. 2020;76:14–20. doi: 10.1016/j.ejim.2020.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Gebhard C, Regitz-Zagrosek V, Neuhauser HK, Morgan R, Klein SL. Impact of sex and gender on COVID-19 outcomes in Europe. Biol Sex Differ. 2020;11:29. doi: 10.1186/s13293-020-00304-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Montopoli M, Zumerle S, Vettor R, et al. Androgen-deprivation therapies for prostate cancer and risk of infection by SARS-CoV-2: a population-based study (N = 4532) Ann Oncol. 2020;31:1040–1045. doi: 10.1016/j.annonc.2020.04.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Scully EP, Haverfield J, Ursin RL, Tannenbaum C, Klein SL. Considering how biological sex impacts immune responses and COVID-19 outcomes. Nat Rev Immunol. 2020;20:442–447. doi: 10.1038/s41577-020-0348-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Qin C, Zhou L, Hu Z, et al. Dysregulation of immune response in patients with Coronavirus 2019 (COVID-19) in Wuhan, China. Clin Infect Dis. 2020;71:762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Ma A, Cheng J, Yang J, Dong M, Liao X, Kang Y. Neutrophil-to-lymphocyte ratio as a predictive biomarker for moderate-severe ARDS in severe COVID-19 patients. Crit Care. 2020;24:288. doi: 10.1186/s13054-020-03007-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Carissimo G, Xu W, Kwok I, et al. Whole blood immunophenotyping uncovers immature neutrophil-to-VD2 T-cell ratio as an early marker for severe COVID-19. Nat Commun. 2020;11 doi: 10.1038/s41467-020-19080-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Kuri-Cervantes L, Pampena MB, Meng W, et al. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci Immunol. 2020;5 doi: 10.1126/sciimmunol.abd7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Bordoni V, Sacchi A, Cimini E, et al. An inflammatory profile correlates with decreased frequency of cytotoxic cells in COVID-19. Clin Infect Dis. 2020;71:2272–2275. doi: 10.1093/cid/ciaa577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Silvin A, Chapuis N, Dunsmore G, et al. Elevated calprotectin and abnormal myeloid cell subsets discriminate severe from mild COVID-19. Cell. 2020;182:1401–1418.e18. doi: 10.1016/j.cell.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Schulte-Schrepping J, Reusch N, Paclik D, et al. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell. 2020;182:1419–1440.e23. doi: 10.1016/j.cell.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Agrati C, Sacchi A, Bordoni V, et al. Expansion of myeloid-derived suppressor cells in patients with severe coronavirus disease (COVID-19) Cell Death Differ. 2020;27:3196–3207. doi: 10.1038/s41418-020-0572-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Brudecki L, Ferguson DA, McCall CE, El Gazzar M. Myeloid-derived suppressor cells evolve during sepsis and can enhance or attenuate the systemic inflammatory response. Infect Immun. 2012;80:2026–2034. doi: 10.1128/IAI.00239-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Wilk AJ, Rustagi A, Zhao NQ, et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat Med. 2020;26:1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Wang J, Jiang M, Chen X, Montaner LJ. Cytokine storm and leukocyte changes in mild versus severe SARS-CoV-2 infection: review of 3939 COVID-19 patients in China and emerging pathogenesis and therapy concepts. J Leukoc Biol. 2020;108:17–41. doi: 10.1002/JLB.3COVR0520-272R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Zuo Y, Yalavarthi S, Shi H, et al. Neutrophil extracellular traps in COVID-19. JCI Insight. 2020;5 doi: 10.1172/jci.insight.138999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Barnes BJ, Adrover JM, Baxter-Stoltzfus A, et al. Targeting potential drivers of COVID-19: Neutrophil extracellular traps. J Exp Med. 2020;217 doi: 10.1084/jem.20200652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Carter MJ, Fish M, Jennings A, et al. Immunophenotyping of circulating leukocytes reveal non-specific activation of innate and adaptive immune systems in multi-system inflammatory syndrome of childhood temporally associated with SARS-Cov-2 infection: descriptive cohort study. Preprints. 2020 doi: 10.20944/preprints202007.0252.v1. published online July 12. (preprint). [DOI] [Google Scholar]
- 219.Payen D, Cravat M, Maadadi H, et al. A longitudinal study of immune cells in severe COVID-19 patients. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.580250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Laing AG, Lorenc A, Del Molino Del Barrio I, et al. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat Med. 2020;26:1623–1635. doi: 10.1038/s41591-020-1038-6. [DOI] [PubMed] [Google Scholar]
- 221.Zhou Y, Fu B, Zheng X, et al. Pathogenic T cells and inflammatory monocytes incite inflammatory storm in severe COVID-19 patients. Natl Sci Rev. 2020;7:998–1002. doi: 10.1093/nsr/nwaa041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Liao M, Liu Y, Yuan J, et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 223.Sanchez-Cerrillo I, Landete P, Aldave B, et al. COVID-19 severity associates with pulmonary redistribution of CD1c+ DCs and inflammatory transitional and nonclassical monocytes. J Clin Invest. 2020;130:6290–6300. doi: 10.1172/JCI140335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Grant RA, Morales-Nebreda L, Markov NS, et al. Circuits between infected macrophages and T cells in SARS-CoV-2 pneumonia. Nature. 2021;590:635–641. doi: 10.1038/s41586-020-03148-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Leijte GP, Rimmelé T, Kox M, et al. Monocytic HLA-DR expression kinetics in septic shock patients with different pathogens, sites of infection and adverse outcomes. Crit Care. 2020;24:110. doi: 10.1186/s13054-020-2830-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Torrance HDT, Longbottom ER, Vivian ME, et al. Post-operative immune suppression is mediated via reversible, Interleukin-10 dependent pathways in circulating monocytes following major abdominal surgery. PLoS One. 2018;13 doi: 10.1371/journal.pone.0203795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Cheron A, Floccard B, Allaouchiche B, et al. Lack of recovery in monocyte human leukocyte antigen-DR expression is independently associated with the development of sepsis after major trauma. Crit Care. 2010;14:R208. doi: 10.1186/cc9331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Monneret G, Lepape A, Voirin N, et al. Persisting low monocyte human leukocyte antigen-DR expression predicts mortality in septic shock. Intensive Care Med. 2006;32:1175–1183. doi: 10.1007/s00134-006-0204-8. [DOI] [PubMed] [Google Scholar]
- 229.Kim OY, Monsel A, Bertrand M, Coriat P, Cavaillon JM, Adib-Conquy M. Differential down-regulation of HLA-DR on monocyte subpopulations during systemic inflammation. Crit Care. 2010;14:R61. doi: 10.1186/cc8959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Flohé S, Lendemans S, Schade FU, Kreuzfelder E, Waydhas C. Influence of surgical intervention in the immune response of severely injured patients. Intensive Care Med. 2004;30:96–102. doi: 10.1007/s00134-003-2041-3. [DOI] [PubMed] [Google Scholar]
- 231.Spinetti T, Hirzel C, Fux M, et al. Reduced monocytic human leukocyte antigen-DR expression indicates immunosuppression in critically ill COVID-19 patients. Anesth Analg. 2020;131:993–999. doi: 10.1213/ANE.0000000000005044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Wang F, Hou H, Yao Y, et al. Systemically comparing host immunity between survived and deceased COVID-19 patients. Cell Mol Immunol. 2020;17:875–877. doi: 10.1038/s41423-020-0483-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Jeannet R, Daix T, Formento R, Feuillard J, François B. Severe COVID-19 is associated with deep and sustained multifaceted cellular immunosuppression. Intensive Care Med. 2020;46:1769–1771. doi: 10.1007/s00134-020-06127-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Monneret G, Cour M, Viel S, Venet F, Argaud L. Coronavirus disease 2019 as a particular sepsis: a 2-week follow-up of standard immunological parameters in critically ill patients. Intensive Care Med. 2020;46:1764–1765. doi: 10.1007/s00134-020-06123-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Remy S, Gossez M, Belot A, et al. Massive increase in monocyte HLA-DR expression can be used to discriminate between septic shock and hemophagocytic lymphohistiocytosis-induced shock. Crit Care. 2018;22:213. doi: 10.1186/s13054-018-2146-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Le Tulzo Y, Pangault C, Amiot L, et al. Monocyte human leukocyte antigen-DR transcriptional downregulation by cortisol during septic shock. Am J Respir Crit Care Med. 2004;169:1144–1151. doi: 10.1164/rccm.200309-1329OC. [DOI] [PubMed] [Google Scholar]
- 237.Schmidt ME, Varga SM. The CD8 T cell response to respiratory virus infections. Front Immunol. 2018;9:678. doi: 10.3389/fimmu.2018.00678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Liu R, Wang Y, Li J, et al. Decreased T cell populations contribute to the increased severity of COVID-19. Clin Chim Acta. 2020;508:110–114. doi: 10.1016/j.cca.2020.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Travis CR. As plain as the nose on your face: the case for a nasal (mucosal) route of vaccine administration for Covid-19 disease prevention. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.591897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Chen R, Sang L, Jiang M, et al. Longitudinal hematologic and immunologic variations associated with the progression of COVID-19 patients in China. J Allergy Clin Immunol. 2020;146:89–100. doi: 10.1016/j.jaci.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Jiang M, Guo Y, Luo Q, et al. T-cell subset counts in peripheral blood can be used as discriminatory biomarkers for diagnosis and severity prediction of coronavirus disease 2019. J Infect Dis. 2020;222:198–202. doi: 10.1093/infdis/jiaa252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Wu H, Zhu H, Yuan C, et al. Clinical and immune features of hospitalized pediatric patients with coronavirus disease 2019 (COVID-19) in Wuhan, China. JAMA Netw Open. 2020;3 doi: 10.1001/jamanetworkopen.2020.10895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.De Biasi S, Meschiari M, Gibellini L, et al. Marked T cell activation, senescence, exhaustion and skewing towards TH17 in patients with COVID-19 pneumonia. Nat Commun. 2020;11 doi: 10.1038/s41467-020-17292-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Wang F, Nie J, Wang H, et al. Characteristics of peripheral lymphocyte subset alteration in COVID-19 pneumonia. J Infect Dis. 2020;221:1762–1769. doi: 10.1093/infdis/jiaa150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Shi H, Wang W, Yin J, et al. The inhibition of IL-2/IL-2R gives rise to CD8+ T cell and lymphocyte decrease through JAK1-STAT5 in critical patients with COVID-19 pneumonia. Cell Death Dis. 2020;11:429. doi: 10.1038/s41419-020-2636-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Hou H, Zhang B, Huang H, et al. Using IL-2R/lymphocytes for predicting the clinical progression of patients with COVID-19. Clin Exp Immunol. 2020;201:76–84. doi: 10.1111/cei.13450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Luo M, Liu J, Jiang W, Yue S, Liu H, Wei S. IL-6 and CD8+ T cell counts combined are an early predictor of in-hospital mortality of patients with COVID-19. JCI Insight. 2020;5 doi: 10.1172/jci.insight.139024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Urra JM, Cabrera CM, Porras L, Ródenas I. Selective CD8 cell reduction by SARS-CoV-2 is associated with a worse prognosis and systemic inflammation in COVID-19 patients. Clin Immunol. 2020;217 doi: 10.1016/j.clim.2020.108486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Liu J, Li S, Liu J, et al. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS-CoV-2 infected patients. EBioMedicine. 2020;55 doi: 10.1016/j.ebiom.2020.102763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Wang F, Hou H, Luo Y, et al. The laboratory tests and host immunity of COVID-19 patients with different severity of illness. JCI Insight. 2020;5 doi: 10.1172/jci.insight.137799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Remy KE, Mazer M, Striker DA, et al. Severe immunosuppression and not a cytokine storm characterizes COVID-19 infections. JCI Insight. 2020;5 doi: 10.1172/jci.insight.140329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Gimenez E, Albert E, Torres I, et al. SARS-CoV-2-reactive interferon-gamma-producing CD8+ T cells in patients hospitalized with coronavirus disease 2019. J Med Virol. 2021;93:375–382. doi: 10.1002/jmv.26213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Grifoni A, Weiskopf D, Ramirez SI, et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Schaller T, Hirschbühl K, Burkhardt K, et al. Postmortem examination of patients with COVID-19. JAMA. 2020;323:2518–2520. doi: 10.1001/jama.2020.8907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Fajnzylber J, Regan J, Coxen K, et al. SARS-CoV-2 viral load is associated with increased disease severity and mortality. Nat Commun. 2020;11 doi: 10.1038/s41467-020-19057-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Rouzé A, Martin-Loeches I, Povoa P, et al. Relationship between SARS-CoV-2 infection and the incidence of ventilator-associated lower respiratory tract infections: a European multicenter cohort study. Intensive Care Med. 2021;47:188–198. doi: 10.1007/s00134-020-06323-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Scharenberg M, Vangeti S, Kekäläinen E, et al. Influenza A virus infection induces hyperresponsiveness in human lung tissue-resident and peripheral blood NK cells. Front Immunol. 2019;10 doi: 10.3389/fimmu.2019.01116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Björkström NK, Ljunggren HG, Michaëlsson J. Emerging insights into natural killer cells in human peripheral tissues. Nat Rev Immunol. 2016;16:310–320. doi: 10.1038/nri.2016.34. [DOI] [PubMed] [Google Scholar]
- 259.Jiang Y, Wei X, Guan J, et al. COVID-19 pneumonia: CD8+ T and NK cells are decreased in number but compensatory increased in cytotoxic potential. Clin Immunol. 2020;218 doi: 10.1016/j.clim.2020.108516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Marquardt N, Kekäläinen E, Chen P, et al. Human lung natural killer cells are predominantly comprised of highly differentiated hypofunctional CD69−CD56dim cells. J Allergy Clin Immunol. 2017;139:1321–1330.e4. doi: 10.1016/j.jaci.2016.07.043. [DOI] [PubMed] [Google Scholar]
- 261.Maucourant C, Filipovic I, Ponzetta A, et al. Natural killer cell immunotypes related to COVID-19 disease severity. Sci Immunol. 2020;5 doi: 10.1126/sciimmunol.abd6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Nielsen SCA, Yang F, Jackson KJL, et al. Human B cell clonal expansion and convergent antibody responses to SARS-CoV-2. Cell Host Microbe. 2020;28:516. doi: 10.1016/j.chom.2020.09.002. 25.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Galson JD, Schaetzle S, Bashford-Rogers RJM, et al. Deep sequencing of B cell receptor repertoires from COVID-19 patients reveals strong convergent immune signatures. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.605170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Robbiani DF, Gaebler C, Muecksch F, et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Zhu Z, Chakraborti S, He Y, et al. Potent cross-reactive neutralization of SARS coronavirus isolates by human monoclonal antibodies. Proc Natl Acad Sci USA. 2007;104:12123–12128. doi: 10.1073/pnas.0701000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Zhang H, Wang G, Li J, et al. Identification of an antigenic determinant on the S2 domain of the severe acute respiratory syndrome coronavirus spike glycoprotein capable of inducing neutralizing antibodies. J Virol. 2004;78:6938–6945. doi: 10.1128/JVI.78.13.6938-6945.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Cao Y, Su B, Guo X, et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients' B cells. Cell. 2020;182:73–84.e16. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Ju B, Zhang Q, Ge J, et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- 269.Quinti I, Lougaris V, Milito C, et al. A possible role for B cells in COVID-19? Lesson from patients with agammaglobulinemia. J Allergy Clin Immunol. 2020;146:211–213.e4. doi: 10.1016/j.jaci.2020.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Soresina A, Moratto D, Chiarini M, et al. Two X-linked agammaglobulinemia patients develop pneumonia as COVID-19 manifestation but recover. Pediatr Allergy Immunol. 2020;31:565–569. doi: 10.1111/pai.13263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Montero-Escribano P, Matías-Guiu J, Gómez-Iglesias P, Porta-Etessam J, Pytel V, Matias-Guiu JA. Anti-CD20 and COVID-19 in multiple sclerosis and related disorders: a case series of 60 patients from Madrid, Spain. Mult Scler Relat Disord. 2020;42 doi: 10.1016/j.msard.2020.102185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Giovannoni G. Anti-CD20 immunosuppressive disease-modifying therapies and COVID-19. Mult Scler Relat Disord. 2020;41 doi: 10.1016/j.msard.2020.102135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Treon SP, Castillo JJ, Skarbnik AP, et al. The BTK inhibitor ibrutinib may protect against pulmonary injury in COVID-19-infected patients. Blood. 2020;135:1912–1915. doi: 10.1182/blood.2020006288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Rice TW, Janz DR. In defense of evidence-based medicine for the treatment of COVID-19 acute respiratory distress syndrome. Ann Am Thorac Soc. 2020;17:787–789. doi: 10.1513/AnnalsATS.202004-325IP. [DOI] [PMC free article] [PubMed] [Google Scholar]