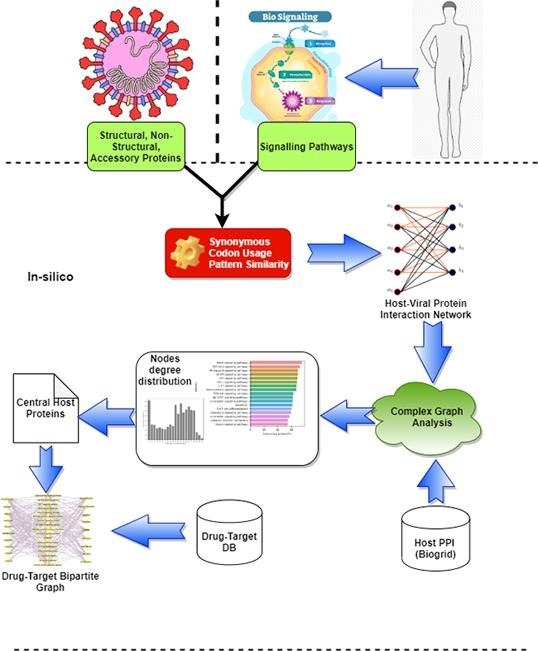
Graphical abstract
Keywords: Protein interaction network, Codon usage bias, Bipartite graph, Cell signaling, Relative synonymous codon usage, Centrality, Drugs
Abstract
Understanding the molecular mechanism of COVID-19 pathogenesis helps in the rapid therapeutic target identification. Usually, viral protein targets host proteins in an organized fashion. The expression of any viral gene depends mostly on the host translational machinery. Recent studies report the great significance of codon usage biases in establishing host-viral protein–protein interactions (PPI). Exploring the codon usage patterns between a pair of co-evolved host and viral proteins may present novel insight into the host-viral protein interactomes during disease pathogenesis. Leveraging the similarity in codon usage patterns, we propose a computational scheme to recreate the host-viral protein–protein interaction network. We use host proteins from seventeen (17) essential signaling pathways for our current work towards understanding the possible targeting mechanism of SARS-CoV-2 proteins. We infer both negatively and positively interacting edges in the network. Further, extensive analysis is performed to understand the host PPI network topologically and the attacking behavior of the viral proteins. Our study reveals that viral proteins mostly utilize codons, rare in the targeted host proteins (negatively correlated interaction). Among them, non-structural proteins, NSP3 and structural protein, Spike (S), are the most influential proteins in interacting with multiple host proteins. While ranking the most affected pathways, MAPK pathways observe to be the worst affected during the SARS-CoV-2 infection. Several proteins participating in multiple pathways are highly central in host PPI and mostly targeted by multiple viral proteins. We observe many potential targets (host proteins) from the affected pathways associated with the various drug molecules, including Arsenic trioxide, Dexamethasone, Hydroxychloroquine, Ritonavir, and Interferon beta, which are either under clinical trial or in use during COVID-19.
1. Introduction
The entire world is passing through an unprecedented pandemic situation due to massive outbreak of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) infected viral disease, COVID-19. SARS-CoV-2 belongs to the Coronaviridae family, and members of this family are enveloped, non-segmented, and have single-stranded, positive-sense large RNA genomes [1]. This virus rapidly spreads from person to person through respiratory droplets during close physical contact. Other than respiratory system, it is reported to attack the immune system, and other vital cellular machinery leading to multi-organ failure [2]. The need for the hour is utmost crucial for the scientific community to understand the disease pathogenesis of SARS-CoV-2 at genomic and proteomic levels for the rapid development of effective drugs or vaccines to control the COVID-19. Many recent works use host-viral protein–protein interaction network as an input to elucidate potential drug targets or repurposed drug molecules [3], [4], [5]. Host-pathogen protein interactions provide essential insights into the molecular mechanisms of pathogenesis [6].
The host defense mechanism activates signal transduction molecules that initiate signals, which activate immune effector mechanisms to protect the host from any pathogenic infections. Studies show that viral immune modulators perturb the human PPI network by targeting signaling pathways [7] to suppress the immunity in mammalian hosts [8]. To understand the molecular mechanism of pathogenicity of SARS-CoV-2 during COVID-19 disease, investigation of the host-viral protein interactions is important. Knowledge gained through understanding the interactions among viral and host proteins involved in signaling pathways may translate into effective therapies and vaccines. There are four (04) basic categories of chemical signaling found in multi-cellular organisms namely; paracrine signaling, autocrine signaling, endocrine signaling, and signaling by direct contact. Various regulatory signaling pathways are involved in signaling transduction and cellular interactions, many of which are playing an important role during COVID-19. Signal transduction focuses on molecular and functional aspects of viral interactions with host cell signaling, important for the anti-viral response, the viral life cycle, viral pathogenesis, and cell transformation [9]. We aim to study the interaction pattern of SARS-CoV-2 with its target host proteins involved in signaling pathways [10], [11], [12], [13], [14], [15] (see Materials and Method section). Working with them can help in deciphering the possible involvement of pathways and key genes during COVID-19 pathogenesis.
The virus-host interactome is essential for understanding virulence factors influencing SARS-CoV-2 pathogenesis [16]. Recent studies reported the use of SARS-CoV-2 and host PPI networks to study the pathogenesis of SARS-CoV-2 and identify repurposed drugs [17], [4], [18]. Several studies have shown that viral proteins interact with hubs in complex host PPI networks [19], [20]. Considering different features of proteins such as sequence homology, gene co-expression, or phylogenetic profiles [21], [22], [23], the pairwise similarity is computed between a pair of proteins to predict a possible interaction between them. In addition to non-structural information, structural data about a pair of proteins appears to be more effective in improving prediction [24], [25], [4].
Several computational methods have been developed to predict PPIs by focusing on protein sequence features [26], [27], [28]. In reality, predicting whether two given proteins are physically interacting or not based on the similarity of different structural and non-structural features is challenging and not feasible due to the expensive experimental setup. In the case of viral genome study, codon usage biases play an important role [29], [30], [31]. Viral gene depends largely on the host translational machinery for their expression [32]. Viral proteins are co-evolved with host proteins. Several studies have reported that physically interacting or functionally associated protein pairs have similar codon usage bias [33], [34], [35], [36], [37], [38], [39]. Therefore, codon usage bias can be utilized in establishing host-viral protein interactions [40], [41]. State-of-the-art methods for inferring interactions do not consider the co-expression or co-adaptation between a pair of virus and host proteins for drawing possible attacking mechanisms of a virus [42], [43]. According to the genome hypothesis proposed by Grantham et al. [44], the pattern of codon usage is species-specific and in some way unique. Interestingly, even in the same genome, the codon usage varies significantly among genes with different expression levels [39], functions [45], and tissue-specific patterns [46]. Few works hypothesized that viral proteins enrich with few codons that are rare in their target host proteins [47], [48].
In this work, we explore host-viral interactions by leveraging the inherent correlation (co-expression or co-adaptation) between viral and host proteins’ codon usage patterns, which applies to any nucleotide (CDS) sequences. To the best of our knowledge, no prior work explored the codon usage similarity to infer host-viral PPI network. We capture both positive and negative interactions in the host-viral PPI. We use host proteins involved in different human cellular signaling pathways that might be affected during COVID-19 disease pathogenesis. Topologically, we try to establish the relevance of the host proteins and highlight a few essential proteins in the network, which are also useful as potential drug targets for certain reported drugs during COVID-19.
2. Materials and method
This section discusses the proposed scheme for constructing a host-viral PPI network using pair-wise codon usage patterns of host and viral proteins. To analyze the interaction mechanism of SARS-CoV-2 viral proteins in host signaling pathways, we select all the genes involved in 17 candidate signaling pathways. We calculate the RSCU similarity score for all pairs of host-viral candidate proteins to build the network. We further analyze the host PPI network to list highly connected host proteins and highlighted a few potential drugs targeting those proteins for possible repairing of affected pathways during COVID-19.
2.1. Data acquisition and processing
Structurally, SARS-CoV-2 consists of three categories of proteins, structural, non-structural, and accessory proteins. We select four (04) structural proteins, sixteen (16) non-structural proteins, and six (06) accessory proteins reported in [49], [50]. The details of the viral proteins are listed in Table 1 (NCBI accession numbers for SARS-CoV-2 proteins: MN908947.3, NC_045512).
Table 1.
SARS-CoV-2 proteins considered for host-viral PPI construction.
| Protein category | Count | Protein Name |
|---|---|---|
| Structural | 4 | Spike (S), Envelope (E), Membrane (M), Nucleocapsid (N) |
| Non-structural | 16 | Nsp1,Nsp2,,Nsp16 |
| Accessory | 6 | Orf3a, Orf6, Orf7a, Orf7b, Orf8, Orf10 |
As discussed before, we consider seventeen (17) signaling pathways, namely TGF-β, JAK-STAT, PI3K-Akt, MAPK, HIF-1, TNF, NF-κ B, Cytokine-cytokine receptor interaction, Apoptosis, Th17 cell differentiation, Chemokine, Toll-like receptor, RIG-like receptor, IL-17, Insulin Signaling, mTOR, and Adipocytokine, which are reported to associate with COVID-19 and other viral diseases [10], [11], [12], [13], [14], [15], [51], [52], [53]. We search the Kyoto Encyclopedia of Genes and Genomes (KEGG) database 1 for the set of human host proteins (genes) that participated in our selected candidate pathways. We observe a total of 2600 genes involved in the above pathways (Supplementary-A,Table S1). We use 1313 unique genes participating exclusively in our 17 target pathways (Supplementary-B). Our proposed scheme is relying on the codon usage pattern, for which the nucleotide sequence (coding region) of each host protein is obtained from the NCBI database. A good number of genes (total 1274) are also involved in more than one pathways. We summarize our target pathways and the number of genes involved in each pathway in the Table 2 .
Table 2.
Candidate signaling pathways and the number of host proteins (or genes) participating in the pathway.
| Pathways | #Genes involved | Pathways | #Genes involved |
|---|---|---|---|
| NF-κ B signaling pathway | 105 | Th17 cell differentiationl | 108 |
| Cytokine-cytokine receptor interaction | 295 | TGF-β signaling pathway | 95 |
| TNF signaling pathway | 113 | Toll-like receptor signaling pathway | 105 |
| IL-17 signaling pathway | 95 | HIF-1 signaling pathway | 110 |
| RIG-I-like receptor signaling pathway | 70 | Apoptosis | 137 |
| MAPK signaling pathway | 295 | Insulin signaling pathway | 138 |
| Chemokine signaling pathway | 190 | mTOR signaling pathway | 156 |
| PI3K-Akt signaling pathway | 355 | Adipocytokine signaling pathway | 70 |
| JAK-STAT signaling pathway | 163 |
2.2. Computing Relative Synonymous Codon Usage (RSCU)
The genetic code describes how the 64-nucleotide triplets specify only twenty (20) different translated amino acids. These alternative codons for the same amino acids are termed as synonymous codons. However, most of the amino acids have at least two synonymous codons that are not used at the same frequencies in different genomes. Differences in the frequency of occurrence on synonymous codons in coding DNA is termed as synonymous codon usage bias [54].
Several indices are available to quantify the synonymous codon usage bias [55]. Effective Number of Codons (ENC) focuses on GC content, Rare Codon (RC) focuses on the abundance of low usage codon, and Codon Adaptation Index (CAI) calculates the frequency of the overall codons based on a reference set. They are either partially capturing the usage or generating values based on the relative whole reference set. We are looking for a normalized frequency of codon usage for comparing the variation of usage between host and viral proteins. RSCU is one of the indices for measuring codon bias used to examine synonymous codon usage without the confounding influence of the amino acid composition of different gene products [55]. It is widely used to estimate the codon usage bias [56], [57], [58], [59]. It can be used to quantify the similarity between any two gene sequences by applying any classical proximity measure between a pair of RSCU vectors. The similarity between RSCU vectors may reflect the possible interactions between a couple of proteins in the PPI [58], [60], [57].
RSCU is the ratio between the observed number of occurrences of codons and expected during uniform usage of synonymous codons and calculated as follows.
| (1) |
where, is the number of occurrences of the codon for the amino acid, which is encoded by synonymous codons. The RSCU score of a codon more than 1.0 indicates excess usage (biased) of the codon, and less than 1.0 marks low usage of that particular codon.
We generate a 59-dimensional RSCU feature vector for each coding protein. We consider the usage pattern of only 59 codons (out of 64 available codons). We ignore 03 stop codons and uniquely coded codons ATG and TGG coded for Met and Trp amino acids, respectively [61]. For RSCU calculation, we use the CAI package [62] available free at 2 . Using the feature vectors, we try to draw the similarity between host and viral proteins to form a network, as discussed next.
2.3. Inferring host-viral protein interaction network
Protein–Protein Interactions (PPI) are usually studied computationally from a graph-theoretic perspective [63]. Interactions among different organisms, such as a host and its pathogen, are primarily driven by interactions among the host proteins and pathogen proteins. These interactions can also be represented as host-pathogen PPI. Host-pathogen PPI is usually represented as a bipartite graph where any given interacting pair of nodes (proteins) does not belong to the same organism. This network essentially provides the known interactions of host proteins with pathogen proteins.
Pearson’s correlation coefficient () is used to calculate relationship between two variables with different magnitudes [64], [65]. Assume and are the RSCU vectors for a pair of viral and host proteins, respectively. Based on and can be calculated as follows.
| (2) |
where, and and are the mean of the vectors and respectively.
To determine significantly correlated pair of RSCU vectors, we use 2-tailed p measurement [66]. Two proteins are strongly connected if the p is less than certain cutoff threshold, , i.e. . We use SciPy version 1.5.0 (sipy.stats) 3 for calculating and p value.
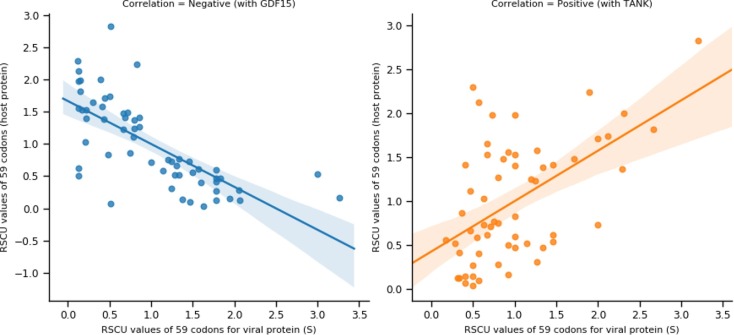
We consider two (02) kinds of interactions, positive and negative, between a host and viral proteins while inferring the network. Positive interaction indicates possible similar codon usage, whereas a negative score signifies possible rare codon usage by SARS-CoV-2 proteins compared to its interacting host proteins. The Pearson correlation coefficient () is used to determine the possible sign of the inferred edges. An example is shown in Fig. 1 for negative and positive correlations computed between two pairs of host-viral proteins.
Fig. 1.
The host-viral codon usage (RSCU) patterns. The scatter plot shows the RSCU value of 59 codons for viral (X-axis) and host (Y-axis) proteins. The regression line represents the trend of RSCU patterns. Viral protein S showing positive correlation () with host protein TANK and negative correlation () with host protein GDF15.
Given a set of viral proteins, and host proteins we can create a bipartite graph in the form of adjacency matrix using the above and p values as follows.
| (3) |
Next, we investigate the interaction mechanism of SARS-CoV-2 proteins with the proteins involved in certain signaling pathways given in Table 2.
3. Results and discussion
3.1. Benchmarking
To assess the effectiveness of codon usage bias measure in predicting possible viral and host protein interactions, the reported host-viral networks with physically verified interactions are considered. For example, 332 host protein interactions with 27 SARS-CoV-2 proteins [3] (network-1) are reported that utilized affinity purification mass spectrometry (AP-MS) based method to infer the physical interactions. It reports host-viral interactions forming star-like topology, where one host is exclusively interacting with one viral node. Few other similar studies report SARS-CoV-2 viral proteins interactions with more than 1100 [67](network-2) and 200 [16](network-3) host proteins. Altogether, there are total of 294 interactions (network-1), 1106 interactions (network-2), and 517 interactions (network-3) in the above networks, which are available in BioGRID database [68]. We apply our method to these three networks, and we observe approximately interactions (average) for the above three networks (Table 3 ). In addition, we use three (03) other viral-host networks, such as Epstein-Barr from Virhostome 4 , Hepatitis-C and Influenza-A from VirusMINT [69] for validation. We report the performance in Table 3. For more detailed results, one can refer to Supplementary-c.
Table 3.
Performance assessment of the proposed scheme on few virus-host interaction networks.
Unlike reported physical interactions methods, codon usage infers a possible co-expression between a pair of host and viral proteins computed quantitatively using pairwise RSCU score similarity. Possibly this might be a possible reason for inferring low matching interactions with the true networks. We consider for inferring the above networks.
3.2. Comparison of RSCU patterns among viral and host proteins
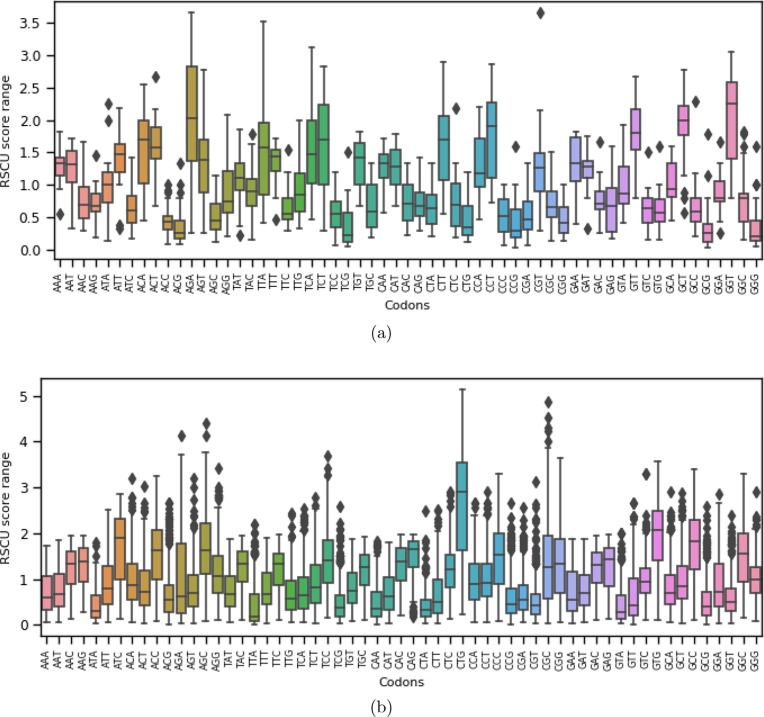
We report codon usage distribution of 59 codons across 26 viral proteins (SARS-CoV-2) and 1313 host proteins in Fig. 2 (a) and (b), respectively, involved in our candidate signaling pathways. We observe that GGT, AGA, GCT, CCT, GTT, TCT, ACA, CTT, TTA, ACT are the highly used (median RSCU score for each codon) codons in SARS-CoV-2 proteins. On the other hand, CGA, AGC, ACC, CGG, CTG, CCG, ACG, GCG, TCG, GGG rarely used codons. In the host proteins (from 17 signaling pathways), codons such as CTG, GTG, ATC, GCC, CAG, ACC, AGC, GGC, and CCC are highly used (median RSCU score for each codon). The distribution margins of RSCU values of those codons are relatively wider (Fig. 2 (b)). However, CCG, GTT, CGT, GCG, TCG, CAA, CTA, ATA, GTA, TTA rarely used codons in host proteins. It is worth mentioning that for SARS-CoV-2 proteins, highly used codons are ending (third position of codon) with T or A that shows similar characteristics with Nipah virus [54], SARS-CoV [70], and coronavirus N genes [71]. But for host proteins from candidate signaling pathways, the highly used codons are ending with G or C at the third position of the codons.
Fig. 2.
Distribution of RSCU scores for 59 codons for all (a) SARS-CoV-2 proteins; (b) Host proteins.
3.3. Analysis of host-viral inferred networks
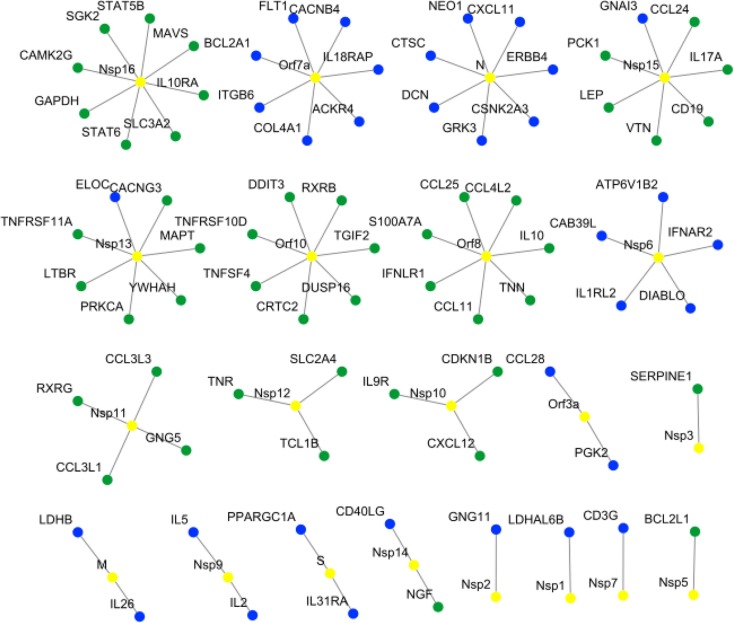
We predict the host-viral (SARS-CoV-2) interaction graph based on the Eq. 3 involving 26 SARS-CoV-2 proteins with 1313 host proteins participating in 17 signaling pathways. Out of 34138 () maximum possible interactions, our method infers 9412 ( 36%) strong interactions. In our network, 859 distinct host proteins ( 66%) are connected to at least one viral protein. We set for deciding the strong interaction (edge) between two proteins. Interestingly, our inferred network reveals that out of 859 host proteins, a total of 779 proteins is targeted by more than one viral proteins. A snapshot of isolated networks with one (viral) to many (host) interactions are shown in Fig. 3 between viral and host proteins.
Fig. 3.
The host-viral interactions network showing host proteins, which are connected to a single viral protein. In the network, the yellow-color represents viral nodes, whereas the blue and green colors represents host nodes, represent positive and negative interactions, respectively. As shown in the figure, 09 viral proteins (Nsp1, Nsp2, Nsp6, Nsp7, Nsp9, M, N, Orf3a, and Orf7a), 08 viral proteins (Nsp3, Nsp5, Nsp10, Nsp11, Nsp12, Nsp16, Orf8, and Orf10), and 03 viral proteins (Nsp13, Nsp14, and Nsp15) interactions with host proteins are positive, negative and both, respectively.
Similar researches on SARS-CoV-2 host protein interactions [3] shows viral protein oriented star-like topology only and unable to report any host protein oriented multiple interactions. We report a list of such highly connected host proteins with the viral proteins (total of 15) in Table 4 . Many (viral) to one (host) interactions are also reported (Supplementary-D).
Table 4.
The list of top few host proteins targeted by number of SARS-CoV-2 interacting viral proteins. For each host protein (Hp), number of interacting viral proteins (IVP) count and calculated average correlation value () are shown. There are total of 40 host proteins (20 for positive interactions and 20 for negative interactions).
| Sl. No. | Hp | IVP count | Avg. () | SARS-CoV-2 interacting viral proteins |
|---|---|---|---|---|
| 1 | COL4A5 | 21 | 0.62 | Nsp1, Nsp2, Nsp3, Nsp4, Nsp5, Nsp6, Nsp7, Nsp8, Nsp9, Nsp10, Nsp12, Nsp13, Nsp14, Nsp15, Nsp16, S, M, N, Orf3a, Orf7a, Orf8 |
| 2 | STAM2 | 21 | 0.63 | Nsp2, Nsp3, Nsp4, Nsp5, Nsp6, Nsp7, Nsp8, Nsp9, Nsp10, Nsp12, Nsp13, Nsp14, Nsp15, Nsp16, S, M, N, Orf3a, Orf6, Orf7a, Orf8 |
| 3 | LIFR | 21 | 0.64 | Nsp2, Nsp3, Nsp4, Nsp5, Nsp6, Nsp7, Nsp8, Nsp9, Nsp10, Nsp12, Nsp13, Nsp14, Nsp15, Nsp16, S, M, N, Orf3a, Orf6, Orf7a, Orf8 |
| 4 | IFNAR1 | 20 | 0.59 | Nsp2, Nsp3, Nsp4, Nsp5, Nsp6, Nsp7, Nsp8, Nsp9, Nsp10, Nsp12, Nsp13, Nsp14, Nsp15, Nsp16, S, M, N, Orf3a, Orf7a, Orf8 |
| 5 | PPM1B | 20 | 0.61 | Nsp2, Nsp3, Nsp4, Nsp5, Nsp6, Nsp7, Nsp8, Nsp9, Nsp10, Nsp12, Nsp13, Nsp14, Nsp15, Nsp16, S, M, N, Orf3a, Orf7a, Orf8 |
| 6 | RPS6KA6 | 20 | 0.62 | Nsp2, Nsp3, Nsp4, Nsp5, Nsp6, Nsp7, Nsp8, Nsp9, Nsp10, Nsp12, Nsp13, Nsp14, Nsp15, Nsp16, S, N, Orf3a, Orf6, Orf7a, Orf8 |
| 7 | SOS2 | 20 | 0.63 | Nsp2, Nsp3, Nsp4, Nsp5, Nsp6, Nsp7, Nsp8, Nsp9, Nsp10, Nsp12, Nsp13, Nsp14, Nsp15, Nsp16, S, M, N, Orf3a, Orf7a, Orf8 |
| 8 | PKN2 | 20 | 0.66 | Nsp2, Nsp3, Nsp4, Nsp5, Nsp6, Nsp7, Nsp8, Nsp9, Nsp10, Nsp12, Nsp13, Nsp14, Nsp15, Nsp16, S, M, N, Orf3a, Orf7a, Orf8 |
| 9 | IRAK4 | 20 | 0.69 | Nsp2, Nsp3, Nsp4, Nsp5, Nsp6, Nsp7, Nsp8, Nsp9, Nsp10, Nsp12, Nsp13, Nsp14, Nsp15, Nsp16, S, M, N, Orf3a, Orf7a, Orf8 |
| 10 | IL13RA2 | 19 | 0.61 | Nsp2, Nsp3, Nsp4, Nsp5, Nsp6, Nsp7, Nsp8, Nsp9, Nsp12, Nsp13, Nsp14, Nsp15, Nsp16, S, M, N, Orf3a, Orf7a, Orf8 |
| 11 | APAF1 | 19 | 0.61 | Nsp2, Nsp3, Nsp4, Nsp5, Nsp6, Nsp7, Nsp8, Nsp9, Nsp12, Nsp13, Nsp14, Nsp15, Nsp16, S, M, N, Orf3a, Orf7a, Orf8 |
| 12 | CUL2 | 19 | 0.61 | Nsp2, Nsp3, Nsp4, Nsp5, Nsp6, Nsp7, Nsp8, Nsp9, Nsp12, Nsp13, Nsp14, Nsp15, Nsp16, S, M, N, Orf3a, Orf7a, Orf8 |
| 13 | DNM1L | 19 | 0.63 | Nsp2, Nsp3, Nsp4, Nsp5, Nsp6, Nsp7, Nsp8, Nsp9, Nsp12, Nsp13, Nsp14, Nsp15, Nsp16, S, M, N, Orf3a, Orf7a, Orf8 |
| 14 | MIOS | 19 | 0.64 | Nsp2, Nsp3, Nsp4, Nsp5, Nsp6, Nsp8, Nsp9, Nsp10, Nsp12, Nsp13, Nsp14, Nsp15, Nsp16, S, M, N, Orf3a, Orf7a, Orf8 |
| 15 | BIRC2 | 19 | 0.65 | Nsp2, Nsp3, Nsp4, Nsp5, Nsp6, Nsp7, Nsp8, Nsp9, Nsp12, Nsp13, Nsp14, Nsp15, Nsp16, S, M, N, Orf3a, Orf7a, Orf8 |
| 16 | RPS6KA3 | 19 | 0.68 | Nsp2, Nsp3, Nsp4, Nsp5, Nsp6, Nsp8, Nsp9, Nsp10, Nsp12, Nsp13, Nsp14, Nsp15, Nsp16, S, M, N, Orf3a, Orf7a, Orf8 |
| 17 | PPP1R3A | 19 | 0.70 | Nsp2, Nsp3, Nsp4, Nsp5, Nsp6, Nsp7, Nsp8, Nsp9, Nsp10, Nsp12, Nsp13, Nsp14, Nsp15, Nsp16, S, N, Orf3a, Orf7a, Orf8 |
| 18 | SGK3 | 18 | 0.61 | Nsp2, Nsp3, Nsp4, Nsp5, Nsp6, Nsp8, Nsp9, Nsp12, Nsp13, Nsp14, Nsp15, Nsp16, S, M, N, Orf3a, Orf7a, Orf8 |
| 19 | PPP3CB | 18 | 0.62 | Nsp2, Nsp3, Nsp4, Nsp5, Nsp6, Nsp8, Nsp9, Nsp10, Nsp12, Nsp13, Nsp14, Nsp15, Nsp16, S, N, Orf3a, Orf7a, Orf8 |
| 20 | HIF1A | 18 | 0.62 | Nsp2, Nsp3, Nsp4, Nsp5, Nsp6, Nsp8, Nsp9, Nsp12, Nsp13, Nsp14, Nsp15, Nsp16, S, M, N, Orf3a, Orf7a, Orf8 |
| 1 | GDF15 | 19 | −0.62 | Nsp2, Nsp3, Nsp4, Nsp5, Nsp6, Nsp7, Nsp8, Nsp9, Nsp10, Nsp12, Nsp13, Nsp14, Nsp15, Nsp16, S, M, Orf3a, Orf6, Orf8 |
| 2 | FGF4 | 19 | −0.60 | Nsp2, Nsp3, Nsp4, Nsp5, Nsp6, Nsp7, Nsp8, Nsp9, Nsp10, Nsp12, Nsp13, Nsp14, Nsp15, Nsp16, S, M, Orf3a, Orf6, Orf8 |
| 3 | SHC2 | 19 | −0.58 | Nsp2, Nsp3, Nsp4, Nsp5, Nsp6, Nsp7, Nsp8, Nsp9, Nsp10, Nsp12, Nsp13, Nsp14, Nsp15, Nsp16, S, M, Orf3a, Orf6, Orf8 |
| 4 | CEBPB | 18 | −0.59 | Nsp2, Nsp3, Nsp4, Nsp5, Nsp6, Nsp7, Nsp8, Nsp9, Nsp10, Nsp12, Nsp13, Nsp14, Nsp15, Nsp16, S, Orf3a, Orf6, Orf8 |
| 5 | IRS2 | 18 | −0.59 | Nsp2, Nsp3, Nsp4, Nsp5, Nsp6, Nsp7, Nsp8, Nsp9, Nsp10, Nsp12, Nsp13, Nsp14, Nsp15, Nsp16, S, Orf3a, Orf6, Orf8 |
| 6 | JUN | 18 | −0.58 | Nsp2, Nsp3, Nsp4, Nsp5, Nsp6, Nsp8, Nsp9, Nsp10, Nsp12, Nsp13, Nsp14, Nsp15, Nsp16, S, Orf3a, Orf6, Orf817 |
| 7 | EFNA2 | 18 | −0.58 | Nsp2, Nsp3, Nsp4, Nsp5, Nsp6, Nsp7, Nsp8, Nsp9, Nsp10, Nsp12, Nsp13, Nsp14, Nsp15, Nsp16, S, Orf3a, Orf6, Orf8 |
| 8 | LPAR5 | 18 | −0.58 | Nsp2, Nsp3, Nsp4, Nsp5, Nsp6, Nsp7, Nsp8, Nsp9, Nsp10, Nsp12, Nsp13, Nsp14, Nsp15, Nsp16, S, Orf3a, Orf6, Orf8 |
| 9 | GDF7 | 18 | −0.58 | Nsp2, Nsp3, Nsp4, Nsp5, Nsp6, Nsp7, Nsp8, Nsp9, Nsp10, Nsp12, Nsp13, Nsp14, Nsp15, Nsp16, S, Orf3a, Orf6, Orf8 |
| 10 | FZD1 | 18 | −0.58 | Nsp2, Nsp3, Nsp4, Nsp5, Nsp6, Nsp7, Nsp8, Nsp9, Nsp10, Nsp12, Nsp13, Nsp14, Nsp15, Nsp16, S, Orf3a, Orf818 |
| 11 | FZD9 | 18 | −0.57 | Nsp2, Nsp3, Nsp4, Nsp5, Nsp6, Nsp7, Nsp8, Nsp9, Nsp10, Nsp12, Nsp13, Nsp14, Nsp15, Nsp16, S, Orf3a, Orf6, Orf8 |
| 12 | PPP2R3B | 18 | −0.57 | Nsp2, Nsp3, Nsp4, Nsp5, Nsp6, Nsp7, Nsp8, Nsp9, Nsp10, Nsp12, Nsp13, Nsp14, Nsp15, Nsp16, S, Orf3a, Orf6, Orf8 |
| 13 | MAPK8IP2 | 18 | −0.57 | Nsp2, Nsp3, Nsp4, Nsp5, Nsp6, Nsp7, Nsp8, Nsp9, Nsp10, Nsp12, Nsp13, Nsp14, Nsp15, Nsp16, S, Orf3a, Orf6, Orf8 |
| 14 | DDIT4 | 18 | −0.57 | Nsp2, Nsp3, Nsp4, Nsp5, Nsp6, Nsp7, Nsp8, Nsp9, Nsp10, Nsp12, Nsp13, Nsp14, Nsp15, Nsp16, S, Orf3a, Orf6, Orf8 |
| 15 | NOG | 18 | −0.57 | Nsp2, Nsp3, Nsp4, Nsp5, Nsp6, Nsp7, Nsp8, Nsp9, Nsp10, Nsp12, Nsp13, Nsp14, Nsp15, Nsp16, S, Orf3a, Orf6, Orf8 |
| 16 | SMAD6 | 18 | −0.57 | Nsp2, Nsp3, Nsp4, Nsp5, Nsp6, Nsp7, Nsp8, Nsp9, Nsp10, Nsp12, Nsp13, Nsp14, Nsp15, Nsp16, S, Orf3a, Orf6, Orf8 |
| 17 | WNT6 | 18 | −0.57 | Nsp2, Nsp3, Nsp4, Nsp5, Nsp6, Nsp7, Nsp8, Nsp9, Nsp10, Nsp12, Nsp13, Nsp14, Nsp15, Nsp16, S, Orf3a, Orf6, Orf8 |
| 18 | GREM2 | 18 | −0.57 | Nsp2, Nsp3, Nsp4, Nsp5, Nsp6, Nsp7, Nsp8, Nsp9, Nsp10, Nsp12, Nsp13, Nsp14, Nsp15, Nsp16, S, Orf3a, Orf6, Orf8 |
| 19 | BMP7 | 18 | −0.56 | Nsp2, Nsp3, Nsp4, Nsp5, Nsp6, Nsp7, Nsp8, Nsp9, Nsp10, Nsp12, Nsp13, Nsp14, Nsp15, Nsp16, S, Orf3a, Orf6, Orf8 |
| 20 | FZD8 | 18 | −0.56 | Nsp2, Nsp3, Nsp4, Nsp5, Nsp6, Nsp7, Nsp8, Nsp9, Nsp10, Nsp12, Nsp13, Nsp14, Nsp15, Nsp16, S, Orf3a, Orf6, Orf8 |
3.4. Distribution of correlation scores
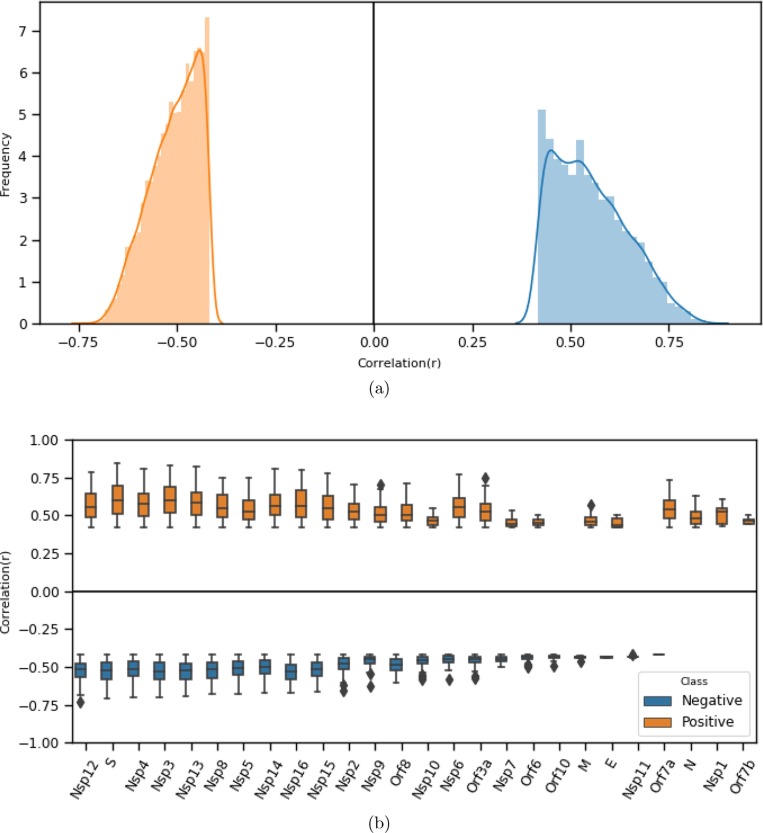
Statistically, it is also important to study the distribution of correlation values (both positive and negative) between pairs of proteins in terms of codon usage patterns. From the distribution plot given in Fig. 4 reveals that the host-viral codon usage pattern (edge correlation) shows non-normal distribution pattern (with for positive correlation, and for negative correlation based on normality test, performed using SciPy.stats.normaltest3) [72], [73]. The negative correlation is varied in the range [-.73, −4.18], which covers 6325 (67%) interactions, and a positive correlation is varied in the range [4.18, 8.44], which covers 3087 (33%) interactions. So, positive correlation exhibits a wider range of values than the negative range.
Fig. 4.
(a) Frequency distribution of positive (right) and negative (left) correlation scores for interacting proteins in terms of RSCU based codon usage similarity. (b) Box plot showing the range of correlation values for each viral protein while associated with its target proteins.
We further look into the correlation value distribution of a viral protein interacting with its target proteins. We report the correlation value range (both positive and negative) for 26 viral proteins in Fig. 4. While fixing at high (significance level) value, correlation values also appear to be significant which are ranging between ±.05 and above. Except few, most of the viral proteins are participated in the network, both positively and negatively. Viral proteins, Orf10 and Nsp10 are interacted with their targets, negatively. Similarly, viral proteins like N, Nsp1 and Orf7b are interacted, positively.
Based on correlation analysis, we may confirm that while a viral protein targets its host, it mimics similar codon usage of its target to uphold the expression of target host protein. Similarly, viral proteins use a set of codons that are rarely used in their targets to down-regulate the expression of its target. We observe in the case of host proteins, involved in signaling pathways, the majority of SARS-CoV-2 proteins aimed to break down the normal pathways by down regulating the key proteins involved in such pathways.
3.5. Degree distribution of host and viral proteins
In any interacting network, the node’s degree conveys essential information about the node’s influence within the network. In the case of host-viral PPI, a high degree viral protein (highly connected) may be a critical protein that influences the functional activities of a number of host proteins. Pharmacologically, identifying such (hub) proteins may help in designing a small molecule that may bind with it to inhibit its influence during disease pathogenesis. The same may be applicable to host proteins. If host protein have a high degree, it indicates that more viral proteins target the host protein. However, it may require further investigation about its importance in its own network, i.e., host–host protein network. If a host protein is significant concerning its degree, suitable repurposed drug molecules may be identified for the same.
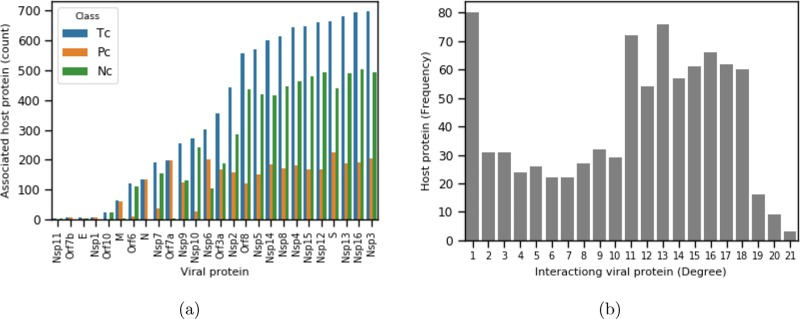
While focusing on highly interacting viral proteins, interestingly, we observe that the maximum number of highly interacting proteins belongs to the non-structural family. In the case of structural proteins, S is a highly interacting (more than 600) protein. Out of accessory proteins, Orf8 shows a maximum interaction count next to protein S.
We report the degree distribution for each of the viral proteins from our network in Fig. 5 (a). From the figure, it can be observed that majority of the viral proteins carrying a high node degree. Out of all the SARS-CoV-2 proteins, Nsp3 shows the maximum degree ( 700), which interacts with more than 80% of the candidate host proteins involved in 17 different signaling pathways. Concerning negative edges, Nsp3 is still on top, followed by Nsp16, Nsp13, and few others. While considering positive edges, S, Nsp6, and Orf7a are found to be highly interactive. Few viral proteins like Nsp11, Orf7b, E, Nsp1, Orf10, and Nsp11 are comparatively less interactive.
Fig. 5.
(a) The bar chart represents the host protein count for each viral protein based on correlation analysis (p-value ). Pc-positive count, Nc-negative count, positive and negative count are based on positive and negative correlations. (b) Degree distribution of 859 host proteins in terms of number of associated viral proteins (degree) count (x-axis) with host protein frequency (y-axis).
We show the degree distribution of 859 host proteins in Fig. 5 (b), interacting with 26 viral proteins. From the distribution plot, we can observe that majority (82) of the host proteins are connected with only one viral node. While considering host proteins targeted by multiple viral proteins, we observe less than ten (10) such proteins are highly connected proteins with the degree 21 (maximum within the network). Even though our network is a bipartite graph, we observe that the number of low-degree nodes is high, fav minor in number, which is somehow follows the scale-free properties [74] of a complex network. We observe relatively good host nodes possessing a degree within the range of 11 to 18.
3.6. Ranking highly targeted signaling pathways during COVID-19
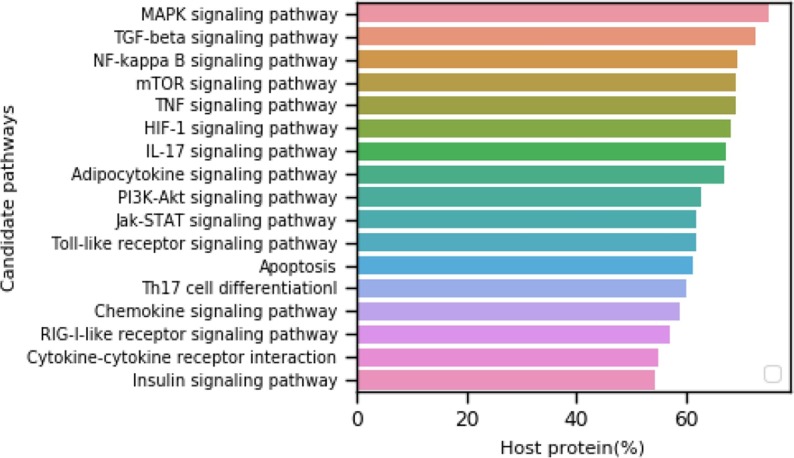
To study the most affected pathways in our 17 candidate set of pathways, we rank them based on the percentage of host proteins targeted by any SARS-CoV-2 viral protein (out of total proteins involved in those pathways) and report in Fig. 6 .
Fig. 6.
Ranking of 17 candidate signaling pathways. The pathway ranking is done by observing the host protein percentage from pathways that interact with any of the SARS-CoV-2 (26) proteins.
The topmost pathway is the Mitogen-Activated Protein Kinase (MAPK) signaling pathway. Different SARS-CoV-2 proteins target more than 50% of proteins from this pathway. This pathway is associated with the COVID-19 immune response [75] and involves in papain-like protease activation of promoter as observed in the SARS coronavirus [76]. MAPK proteins communicate signals from a receptor on the cell’s surface to the DNA in the cell’s nucleus, essential from a viral infection point of view. Further, MAPK proteins are involved in a series of vital signal transduction pathways that regulate processes such as cell proliferation, cell differentiation, and cell death in humans.
Besides MAPK, other ranked signaling pathways are significantly affected during COVID-19 infection. Under physiological conditions, adipokines act mainly in adipose tissue (paracrine or autocrine) or circulate through the blood to distant target organs, regulating their growth and development, metabolism, and tissue remodeling. However, adipokines’ synthesis and secretion are disordered under pathological conditions, leading to obesity, diabetes, heart disease, and other metabolic disorders. Our results show that the adipocytokine pathway is affected by COVID-19. It implicates that patients with comorbid conditions like diabetes and heart disease may show the worst disease aggression, which is already observed in various reports. The mTOR pathway [12] is a central regulator of mammalian metabolism and physiology, with essential roles in tissues’ function, including liver, muscle, white and brown adipose tissue, and the brain. It is dysregulated in human diseases, such as diabetes, obesity, depression, aging-related problems, and certain cancers. Our result corroborates with the same, and it has reported that aged patients are more prone to the infection due to the dysregulation of the mTOR pathway or some other unknown reasons.
It has been observed that some COVID-19 affected deaths are due to multiple organ failures. HIF1 [89], [90] and RIG1 [91] like receptor pathways are involved in normal immunoregulation and various organ functioning. Dysregulation may cause immune compromisation and multiple organ failure through ischaemic heart disease, acute lung injury, pulmonary hypertension, pulmonary fibrosis, chronic obstructive pulmonary disease (COPD), acute liver failure, liver fibrosis, and acute kidney injury, etc. Our result also supports these findings.
In our ranking, the fourth most affected pathway is the TGF- (Transforming growth factor-beta) [92], which is a multi-functional cytokine belonging to the transforming growth factor superfamily that includes three different mammalian isoforms (TGF- 1 to 3, HGNC symbols TGFB1, TGFB2, TGFB3) and many other signaling proteins. All-white blood cell lineages produce TGF-β proteins. This pathway activates different downstream substrates and regulatory proteins, inducing transcription of various target genes that function in differentiation, chemotaxis, proliferation, and activation of many immune cells.
3.7. Centrality analysis of targeted host proteins and candidate signaling pathways
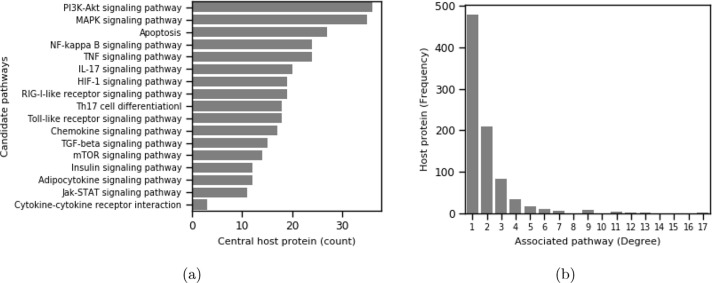
Studies on human host-viral protein interactions reveal that viruses tends to target- attacks towards host proteins [77], [19], [78] by interacting with key (central) host proteins. We consider a host protein important if it interacts with many other host proteins in host-host protein network. We use BioGRID [68] to calculate the centrality score of our candidate host proteins 5 . We report the top 100 central proteins in Supplementary-A (Table S2). We observe that a good number of interacting host proteins in our network are highly central in their own (host) PPI. A common set of viral proteins targets central genes, and such proteins are involved in multiple pathways. For instance, if we consider few top central proteins, MYC (2843), TRIM25 (2656), EGFR (2452), BRCA1 (2236), MDM2 (2219), NTRK1 (2030), KRAS (1944), ELAVL1 (1914) and HSP90AA1 (1734), they are found to be targeted jointly by the viral proteins such Nsp2, Nsp3, Nsp4, Nsp5, Nsp8, Nsp10, Nsp12, Nsp13.
If we consider the most central proteins in our candidate pathways, we observe that the PI3K-Akt signaling pathway (36 target proteins) and MAPK signaling pathway (35 target proteins) contain most of the central proteins targeted by the viral proteins. A pathway may be more crucial from the disease pathogenesis perspective if it contains highly central proteins targeted by viral proteins. Moving one step ahead, we may rank our 17 pathways based on the number of participating central proteins (out of the top 100 centrality list) in the above pathways and shown in Fig. 7 (a). More details about the top 100 central host proteins are listed in Supplementary-A (Table S2). Interestingly, in terms of the number of target proteins, which are also central in host-host PPI, the signaling pathway MAPK is one of the worst affected pathways among 17 candidate pathways. In addition to PPI centrality, we study the pathway centrality of the host proteins regarding our 17 signaling pathways. Prior researches also identified an exciting fact that viral proteins target host proteins that are pathway central, i.e., participating in multiple pathways [78]. The degree distribution of host proteins in terms of their density of participation in 17 pathways is reported in Fig. 7(b). We observe a nice power-law [74] like distribution where the majority of proteins are participating in only one pathway, and fewer numbers are having high participation in multiple pathways. We list a few top highly pathway-central proteins and few interesting facts in Table 5 . The table shows that the pathway-central proteins are also highly connected in their own PPI and mostly targeted by multiple viral proteins.
Fig. 7.
(a) Participation host protein count of central proteins in candidate pathways; (b) Degree distribution of 859 interacting host proteins in terms of number of associated signaling pathways (candidate).
Table 5.
Few top pathway central proteins with the number of pathways they are participating (out of 17 pathways), PPI centrality score and number of viral proteins (Vp) targeting the proteins.
| Host protein | #Pathway centrality | PPI centrality | Interacting Vp count |
|---|---|---|---|
| IKBKB | 13 | 552 | 2 |
| CHUK | 12 | 462 | 11 |
| MAPK3 | 12 | 337 | 16 |
| RELA | 12 | 859 | 10 |
| AKT1 | 11 | 886 | 11 |
| AKT2 | 11 | 113 | 12 |
| AKT3 | 11 | 61 | 15 |
| IKBKG | 11 | 959 | 9 |
| TNF | 11 | 497 | 11 |
| MAPK8 | 9 | 444 | 12 |
| MAPK9 | 9 | 260 | 15 |
| NFKBIA | 9 | 501 | 8 |
| PIK3CA | 9 | 190 | 19 |
| PIK3CB | 9 | 82 | 17 |
| PIK3CD | 9 | 28 | 17 |
| PIK3R1 | 9 | 684 | 5 |
| PIK3R2 | 9 | 190 | 16 |
3.8. Quantitative association of key pathway proteins and drugs
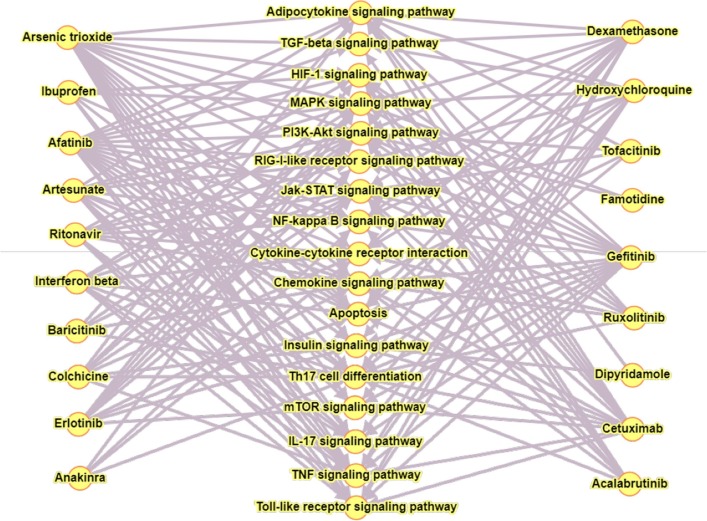
To investigate further the significance of key proteins in our network, we analyse protein-drug association. We primarily consider approved or under-trial drugs that are in use during COVID-19. We searched online drug target resource database 6 to count hits with different key proteins in our network (Supplementary-A, Table S3). We ranked those drugs based on their counts of protein targets in our network (Table 6 ). A good number of drugs are also observed to be associated with central proteins that are not reported so far used in COVID-19. The list of such drugs is given in Supplementary-A (Table S3). It can be observed from Table 6 that a single drug is having targets in multiple pathways forming a bipartite graph as shown in Fig. 8 .
Table 6.
Few COVID-19 drugs with their actual number of target host proteins from our inferred network, number of targets that are highly central and number of targets involved in candidate pathways.
| Drug Name | #Actual targets | #Targets in | #Targets (central) in | #Involved |
|---|---|---|---|---|
| inferred networks | top-100 host PPI | pathway | ||
| Arsenic trioxide | 25 | 11 | 8 | 16 |
| Dexamethasone | 64 | 10 | 5 | 12 |
| Hydroxychloroquine | 9 | 5 | 1 | 11 |
| Interferon beta | 5 | 4 | 0 | 9 |
| Ritonavir | 15 | 3 | 0 | 10 |
Fig. 8.
Bipartite graph showing 19 drugs linked with 17 signaling pathways. Left and right panel are showing drug name and middle panel is showing signaling pathway name.
We discuss below the drugs that are associated with COVID-19 and having potential target host proteins involved in our inferred host-viral networks.
Arsenic trioxide is a widely known chemical used for multiple disease conditions. The Ministry of Ayush 7 , Govt. Of India, advised for Arsenicum album 30 as a potential homeopath drug for COVID-19. Arsenic trioxide is a mother tincture of Arsenicum album 30, used as a homeopath medicine. Symptoms like severe respiratory adverse effects frequently occur in patients with promyelocytic leukemia. Arsenic trioxide could be used in consolidation therapy [79], [80]. If we consider central genes (present in the top 100 list) involved in MAPK pathways, we observe that six target proteins (RARA, FGFR1, IKBKB, CCND1, CDKN1A, JUN, MAPK3, AKT1) are the excellent target of this chemical. Interestingly, all such proteins are targeted negatively by viral proteins. In a comorbid situation where these signaling pathway genes are already perturbed, arsenic trioxide may play a protective role in boosting up the immunity and other unknown vital regulators that are yet to discover. In addition to MAPK, several targets are present in PI3K-Akt, TNF, and Apoptosis signaling pathways.
Dexamethasone is another most widely used COVID-19 drug with 64 target genes8 . This is the first drug to show life-saving efficacy in patients infected with COVID-19 [81] and widely utilized in a large trial in the UK [82] Our result shows that NTRK1, HSPA8, SMAD3, VCAM1, and RARA are the targets (central) for the drug involved in MAPK, PI3K-Akt, Th17 cell differentiation, TGF-β, and NF-κ B. In addition to that, Dexamethasone also targets a few other interacting host proteins (low centrality), JUNB, LIF, CD86, SLC2A4, and IRS2. Dexamethasone is predicted to maintain these signaling pathways’ normal functioning and shows protection against COVID-19 symptoms, as we assume from our results.
Hydroxychloroquine is another important drug, has been widely utilized for COVID-19 treatment [83], [84]. The only central target is TNF, which is present in several signaling pathways. It can rapidly be transcribed in various cell types following exposure to a broad range of pathogens and signals of inflammation and stress [85]. Other low centrality targets are TLR3, TLR7, PTGS2, and TLR9.
The other two important drugs recommended by WHO 9 are Ritonavir , and Interferon Alfa B observed in our list used for the COVID-19 clinical trial [86], [87], [88]. Interestingly, we found that ritonavir shows three target central genes (CXCL10, TLR4, IFNL3) in our study. These genes share NF κ B, HIF1, toll-like receptor, PI3K-AKT, JAK-STAT, cytokine-cytokine receptor, TNF, IL7, RIG1 receptor, chemokine signaling pathways. Interferon Alfa 1B is another option for solidarity trials with targets like IFNAR1, IFNAR2, and IL13 genes. These genes participate in PI3K-AKT, toll-like receptor, cytokine-cytokine receptor, JAK-STAT, Il17 pathways. These pathways are essential for maintaining normal immunological functioning, which is thought to dysregulate in COVID-19.
We believe that a pathway targeted by different SARS-CoV-2 proteins and involvement of highly central proteins in its own PPI is the most crucial (affected) pathway. It is worth mentioning that the above-highlighted drug molecules are based on quantitative analysis of host proteins from our inferred host-viral network and their hits with the existing drug target database. Hence, our proposed scheme is not a new drug repurposing methodology and needs due attention while designing therapeutic solutions pharmacologically.
4. Conclusion
This work introduced a novel effort into recreating host-viral PPI. Proposed work explored the codon usage pattern similarity between host proteins that are participating in a few major signaling pathways and SARS-CoV-2 viral proteins. Both positive and negative edges between interacting proteins were inferred, which depict an essential association between viral and host proteins. The inferred network was analyzed topologically, considering nodes’ degree distribution and node centrality. An interesting fact has been observed on how viral proteins are targeting their host proteins. Our analysis highlighted a few drugs already in use for COVID-19, having potential targets in some of the essential host proteins involved in important candidate signaling pathways such as MAPK and PI3K-Akt. Several central proteins were identified (AKT1, CCND1, CDKN1A, FGFR1, HSPA8, IKBKB, JUN, MAPK3, NTRK1, RARA, SMAD3, TNF, and VCAM1), which are involved in critical signaling pathways and targeted by few drug molecules. The topmost few drug molecules highlighted by this study are Arsenic trioxide, Dexamethasone, and Hydroxychloroquine, which might play an influential role in preventing COVID-19 mortality.
Our method is generic and useful to draw a more extensive network, covering genes from all critical pathways. Even the method can be applied to any set of host-viral proteins (other than SARS-CoV-2 or Human). Currently, we used a correlation score to measure the similarity between two RSCU vectors. However, other measures such as cosine similarity, mutual information, and ensemble approach might improve the result. One may consider a multi-layer network approach considering viral-viral and host–host networks that may shed better light on the possible viral-host interaction patterns.
Acknowledgement
This research is partly supported by the Department of Science & Technology (DST), Govt. of India under DST-ICPS Data Science program [DST/ICPS/Cluster/Data Science/General]. Thanks to Ms Arpita Roy, for reviewing and correcting the final draft of the manuscript.
CRediT authorship contribution statement
Jayanta Kumar Das: Conceptualization, Data curation, Methodology, Software, Formal analysis, Visualization, Writing - original draft, Writing - review & editing. Subhadip Chakraborty: Software, Validation, Visualization, Writing - original draft, Writing - review & editing. Swarup Roy: Conceptualization, Supervision, Visualization, Writing - original draft, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.jbi.2021.103801.
Supplementary material
The following are the Supplementary data to this article:
References
- 1.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-em structure of the 2019-ncov spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jain U. Effect of covid-19 on the organs. Cureus. 2020;12 doi: 10.7759/cureus.9540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., et al. A sars-cov-2 protein interaction map reveals targets for drug repurposing. Nature. 2020:1–13. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F. Network-based drug repurposing for novel coronavirus 2019-ncov/sars-cov-2. Cell Discovery. 2020;6:1–18. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.B.R. Beck, B. Shin, Y. Choi, S. Park, K. Kang, Predicting commercially available antiviral drugs that may act on the novel coronavirus (sars-cov-2) through a drug-target interaction deep learning model, Comput. Struct. Biotechnol. J. (2020). [DOI] [PMC free article] [PubMed]
- 6.Memišević V., Zavaljevski N., Rajagopala S.V., Kwon K., Pieper R., DeShazer D., Reifman J., Wallqvist A. Mining host-pathogen protein interactions to characterize burkholderia mallei infectivity mechanisms. PLoS Comput. Biol. 2015;11:e1004088. doi: 10.1371/journal.pcbi.1004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pichlmair A., Kandasamy K., Alvisi G., Mulhern O., Sacco R., Habjan M., Binder M., Stefanovic A., Eberle C.-A., Goncalves A., et al. Viral immune modulators perturb the human molecular network by common and unique strategies. Nature. 2012;487:486–490. doi: 10.1038/nature11289. [DOI] [PubMed] [Google Scholar]
- 8.R. Draenert, J. Frater, J.G. Prado, Virus immune evasion: new mechanism and implications in disease outcome, 2012. [DOI] [PMC free article] [PubMed]
- 9.Kieser A. Signal transduction by viral factors: critical interface between the virus and its host cell with implications for the viral life cycle and disease development. Signal Transduction. 2007;7:3–4. [Google Scholar]
- 10.Seif F., Aazami H., Khoshmirsafa M., Kamali M., Mohsenzadegan M., Pornour M., Mansouri D. Jak inhibition as a new treatment strategy for patients with covid-19. Int. Arch. Allergy Immunol. 2020;181:467–475. doi: 10.1159/000508247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu D., Yang X.O. Th17 responses in cytokine storm of covid-19: An emerging target of jak2 inhibitor fedratinib. J. Microbiol., Immunol. Infect. 2020;53:368–370. doi: 10.1016/j.jmii.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ganesan H., Balasubramanian V., Iyer M., Venugopal A., Subramaniam M.D., Cho S.-G., Vellingiri B. mTOR signalling pathway-a root cause for idiopathic autism? BMB Reports. 2019;52:424. doi: 10.5483/BMBRep.2019.52.7.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo W., Li Y.-X., Jiang L.-J., Chen Q., Wang T., Ye D.-W. Targeting jak-stat signaling to control cytokine release syndrome in covid-19. Trends Pharmacol. Sci. 2020 doi: 10.1016/j.tips.2020.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shibabaw T. Inflammatory cytokine: Il-17a signaling pathway in patients present with covid-19 and current treatment strategy. J. Inflammat. Res. 2020;13:673. doi: 10.2147/JIR.S278335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grimes J.M., Grimes K.V. p38 mapk inhibition: A promising therapeutic approach for covid-19. J. Mol. Cell. Cardiol. 2020 doi: 10.1016/j.yjmcc.2020.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J., Guo M., Tian X., Wang X., Yang X., Wu P., Liu C., Xiao Z., Qu Y., Yin Y., et al. Virus-host interactome and proteomic survey of pbmcs from covid-19 patients reveal potential virulence factors influencing sars-cov-2 pathogenesis. Med. 2020 doi: 10.1016/j.medj.2020.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Messina F., Giombini E., Agrati C., Vairo F., Bartoli T.A., Al Moghazi S., Piacentini M., Locatelli F., Kobinger G., Maeurer M., et al. Covid-19: viral–host interactome analyzed by network based-approach model to study pathogenesis of sars-cov-2 infection. J. Translat. Med. 2020;18:1–10. doi: 10.1186/s12967-020-02405-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar Das J., Tradigo G., Veltri P., Guzzi P.H., Roy S. Data science in unveiling covid-19 pathogenesis and diagnosis: evolutionary origin to drug repurposing. Briefings Bioinformat. 2021;22:855–872. doi: 10.1093/bib/bbaa420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Navratil V., de Chassey B., Combe C.R., Lotteau V. When the human viral infectome and diseasome networks collide: towards a systems biology platform for the aetiology of human diseases. BMC Syste. Biol. 2011;5:13. doi: 10.1186/1752-0509-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dyer M.D., Murali T., Sobral B.W. The landscape of human proteins interacting with viruses and other pathogens. PLoS Pathog. 2008;4:e32. doi: 10.1371/journal.ppat.0040032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuchaiev O., Rašajski M., Higham D.J., Pržulj N. Geometric de-noising of protein-protein interaction networks. PLoS Comput. Biol. 2009;5:e1000454. doi: 10.1371/journal.pcbi.1000454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murakami Y., Mizuguchi K. Homology-based prediction of interactions between proteins using averaged one-dependence estimators. BMC Bioinformat. 2014;15:213. doi: 10.1186/1471-2105-15-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salwinski L., Eisenberg D. Computational methods of analysis of protein–protein interactions. Current Opin. Struct. Biol. 2003;13:377–382. doi: 10.1016/s0959-440x(03)00070-8. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Q.C., Petrey D., Deng L., Qiang L., Shi Y., Thu C.A., Bisikirska B., Lefebvre C., Accili D., Hunter T., et al. Structure-based prediction of protein–protein interactions on a genome-wide scale. Nature. 2012;490:556–560. doi: 10.1038/nature11503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh R., Park D., Xu J., Hosur R., Berger B. Struct2net: a web service to predict protein–protein interactions using a structure-based approach. Nucleic Acids Res. 2010;38:W508–W515. doi: 10.1093/nar/gkq481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jain S., Bader G.D. An improved method for scoring protein-protein interactions using semantic similarity within the gene ontology. BMC Bioinformat. 2010;11:1–14. doi: 10.1186/1471-2105-11-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang X., Yang S., Li Q., Wuchty S., Zhang Z. Prediction of human-virus protein-protein interactions through a sequence embedding-based machine learning method. Comput. Struct. Biotechnol. J. 2020;18:153–161. doi: 10.1016/j.csbj.2019.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alguwaizani S., Park B., Zhou X., Huang D.-S., Han K. Predicting interactions between virus and host proteins using repeat patterns and composition of amino acids. J. Healthcare Eng. 2018;2018 doi: 10.1155/2018/1391265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dilucca M., Forcelloni S., Georgakilas A.G., Giansanti A., Pavlopoulou A. Codon usage and phenotypic divergences of sars-cov-2 genes. Viruses. 2020;12:498. doi: 10.3390/v12050498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharp P.M., Emery L.R., Zeng K. Forces that influence the evolution of codon bias. Philosoph. Trans. Roy. Soc. B: Biol. Sci. 2010;365:1203–1212. doi: 10.1098/rstb.2009.0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jenkins G.M., Holmes E.C. The extent of codon usage bias in human rna viruses and its evolutionary origin. Virus Res. 2003;92:1–7. doi: 10.1016/s0168-1702(02)00309-x. [DOI] [PubMed] [Google Scholar]
- 32.Gale M., Tan S.-L., Katze M.G. Translational control of viral gene expression in eukaryotes. Microbiol. Molecular Biol. Rev. 2000;64:239–280. doi: 10.1128/mmbr.64.2.239-280.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lithwick G., Margalit H. Relative predicted protein levels of functionally associated proteins are conserved across organisms. Nucleic Acidsr Res. 2005;33:1051–1057. doi: 10.1093/nar/gki261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fraser H.B., Hirsh A.E., Wall D.P., Eisen M.B. Coevolution of gene expression among interacting proteins. Proc. Nat. Acad. Sci. 2004;101:9033–9038. doi: 10.1073/pnas.0402591101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bahir I., Fromer M., Prat Y., Linial M. Viral adaptation to host: a proteome-based analysis of codon usage and amino acid preferences. Molecular Syst. Biol. 2009;5:311. doi: 10.1038/msb.2009.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou Z., Dang Y., Zhou M., Li L., Yu C.-H., Fu J., Chen S., Liu Y. Codon usage is an important determinant of gene expression levels largely through its effects on transcription. Proc. Nat. Acad. Sci. 2016;113:E6117–E6125. doi: 10.1073/pnas.1606724113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jitobaom K., Phakaratsakul S., Sirihongthong T., Chotewutmontri S., Suriyaphol P., Suptawiwat O., Auewarakul P. Codon usage similarity between viral and some host genes suggests a codon-specific translational regulation. Heliyon. 2020;6:e03915. doi: 10.1016/j.heliyon.2020.e03915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song H., Gao H., Liu J., Tian P., Nan Z. Comprehensive analysis of correlations among codon usage bias, gene expression, and substitution rate in arachis duranensis and arachis ipaënsis orthologs. Sci. Rep. 2017;7:1–12. doi: 10.1038/s41598-017-13981-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dos Reis M., Wernisch L., Savva R. Unexpected correlations between gene expression and codon usage bias from microarray data for the whole escherichia coli k-12 genome. Nucleic Acids Res. 2003;31:6976–6985. doi: 10.1093/nar/gkg897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chaney J.L., Clark P.L. Roles for synonymous codon usage in protein biogenesis. Annual Rev. Biophys. 2015;44:143–166. doi: 10.1146/annurev-biophys-060414-034333. [DOI] [PubMed] [Google Scholar]
- 41.Zhou Y., Zhou Y.-S., He F., Song J., Zhang Z. Can simple codon pair usage predict protein–protein interaction? Mol. BioSyst. 2012;8:1396–1404. doi: 10.1039/c2mb05427b. [DOI] [PubMed] [Google Scholar]
- 42.Peng X., Wang J., Peng W., Wu F.-X., Pan Y. Protein–protein interactions: detection, reliability assessment and applications. Briefings Bioinformat. 2016;18:798–819. doi: 10.1093/bib/bbw066. [DOI] [PubMed] [Google Scholar]
- 43.Rao V.S., Srinivas K., Sujini G., Kumar G. Protein-protein interaction detection: methods and analysis. Int. J. Proteom. 2014;2014 doi: 10.1155/2014/147648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grantham R., Gautier C., Gouy M., Mercier R., Pave A. Codon catalog usage and the genome hypothesis. Nucleic Acids Res. 1980;8:197. doi: 10.1093/nar/8.1.197-c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Y.-S., Zhou J.-H., Chen H.-T., Ma L.-N., Pejsak Z., Ding Y.-Z., Zhang J. The characteristics of the synonymous codon usage in enterovirus 71 virus and the effects of host on the virus in codon usage pattern. Infection, Genetics Evol. 2011;11:1168–1173. doi: 10.1016/j.meegid.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Plotkin J.B., Robins H., Levine A.J. Tissue-specific codon usage and the expression of human genes. Proc. Nat. Acad. Sci. 2004;101:12588–12591. doi: 10.1073/pnas.0404957101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mueller S., Papamichail D., Coleman J.R., Skiena S., Wimmer E. Reduction of the rate of poliovirus protein synthesis through large-scale codon deoptimization causes attenuation of viral virulence by lowering specific infectivity. J. Virol. 2006;80:9687–9696. doi: 10.1128/JVI.00738-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tindle R.W. Immune evasion in human papillomavirus-associated cervical cancer. Nat. Rev. Cancer. 2002;2:59–64. doi: 10.1038/nrc700. [DOI] [PubMed] [Google Scholar]
- 49.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., et al. A new coronavirus associated with human respiratory disease in china. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Das J.K., Roy S. A study on non-synonymous mutational patterns in structural proteins of sars-cov-2. Genome. 2021 doi: 10.1139/gen-2020-0157. [DOI] [PubMed] [Google Scholar]
- 51.M.A.-A.-K. Khan, A.B.M.M.K. Islam, Sars-cov-2 proteins exploit host’s genetic and epigenetic mediators for the annexation of key host signaling pathways, Front. Mol. Biosci. 7 (2020). [DOI] [PMC free article] [PubMed]
- 52.Chen L., Deng H., Cui H., Fang J., Zuo Z., Deng J., Li Y., Wang X., Zhao L. Inflammatory responses and inflammation-associated diseases in organs. Oncotarget. 2018;9:7204. doi: 10.18632/oncotarget.23208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao Z., Xia J., Tastan O., Singh I., Kshirsagar M., Carbonell J., Klein-Seetharaman J. Virus interactions with human signal transduction pathways. Int. J. Comput. Biol. Drug Des. 2011;4:83–105. doi: 10.1504/IJCBDD.2011.038658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Khandia R., Singhal S., Kumar U., Ansari A., Tiwari R., Dhama K., Das J., Munjal A., Singh R.K. Analysis of nipah virus codon usage and adaptation to hosts. Front. Microbiol. 2019;10:886. doi: 10.3389/fmicb.2019.00886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sharp P.M., Tuohy T.M., Mosurski K.R. Codon usage in yeast: cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res. 1986;14:5125–5143. doi: 10.1093/nar/14.13.5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sharp P.M., Li W.-H. An evolutionary perspective on synonymous codon usage in unicellular organisms. J. Molecul. Evol. 1986;24:28–38. doi: 10.1007/BF02099948. [DOI] [PubMed] [Google Scholar]
- 57.M. Dilucca, S. Forcelloni, G. Cimini, A. Giansanti, Co-evolution between codon usage and protein-protein interaction networks in bacterial genomes, bioRxiv (2020). [DOI] [PubMed]
- 58.Najafabadi H.S., Salavati R. Sequence-based prediction of protein-protein interactions by means of codon usage. Genome Biol. 2008;9:1–9. doi: 10.1186/gb-2008-9-5-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.J. Das, S. Roy, Comparative analysis of human coronaviruses focusing on nucleotide variability and synonymous codon usage pattern, BioRxiv (2020). [DOI] [PMC free article] [PubMed]
- 60.Meintjes P.L., Rodrigo A.G. Evolution of relative synonymous codon usage in human immunodeficiency virus type-1. J. Bioinformatics Comput. Biol. 2005;3:157–168. doi: 10.1142/s0219720005000953. [DOI] [PubMed] [Google Scholar]
- 61.Chen Y., Xu Q., Yuan X., Li X., Zhu T., Ma Y., Chen J.-L. Analysis of the codon usage pattern in middle east respiratory syndrome coronavirus. Oncotarget. 2017;8:110337. doi: 10.18632/oncotarget.22738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee B.D. Python implementation of codon adaptation index. J. Open Source Softw. 2018;3:903. doi: 10.21105/joss.00905. [DOI] [Google Scholar]
- 63.P.H. Guzzi, S. Roy, Biological Network Analysis: Trends, Approaches, Graph Theory, and Algorithms, Academic Press, 2020.
- 64.Häne B.G., Jäger K., Drexler H.G. The pearson product-moment correlation coefficient is better suited for identification of dna fingerprint profiles than band matching algorithms. Electrophoresis. 1993;14:967–972. doi: 10.1002/elps.11501401154. [DOI] [PubMed] [Google Scholar]
- 65.J. Benesty, J. Chen, Y. Huang, I. Cohen, Pearson correlation coefficient, in: Noise Reduction in Speech Processing, Springer, 2009, pp. 1–4.
- 66.Sedgwick P. Pearson’s correlation coefficient. Bmj. 2012;345:e4483. [Google Scholar]
- 67.A. Stukalov, V. Girault, V. Grass, V. Bergant, O. Karayel, C. Urban, D.A. Haas, Y. Huang, L. Oubraham, A. Wang, et al., Multi-level proteomics reveals host-perturbation strategies of sars-cov-2 and sars-cov, Biorxiv (2020). [DOI] [PubMed]
- 68.Stark C., Breitkreutz B.-J., Reguly T., Boucher L., Breitkreutz A., Tyers M. Biogrid: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chatr-Aryamontri A., Ceol A., Peluso D., Nardozza A., Panni S., Sacco F., Tinti M., Smolyar A., Castagnoli L., Vidal M., et al. Virusmint: a viral protein interaction database. Nucleic Acids Res. 2009;37:D669–D673. doi: 10.1093/nar/gkn739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gu W., Zhou T., Ma J., Sun X., Lu Z. Analysis of synonymous codon usage in sars coronavirus and other viruses in the nidovirales. Virus Res. 2004;101:155–161. doi: 10.1016/j.virusres.2004.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sheikh A., Al-Taher A., Al-Nazawi M., Al-Mubarak A.I., Kandeel M. Analysis of preferred codon usage in the coronavirus n genes and their implications for genome evolution and vaccine design. J. Virolog. Methods. 2020;277:113806. doi: 10.1016/j.jviromet.2019.113806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.d’Agostino R.B. An omnibus test of normality for moderate and large size samples. Biometrika. 1971;58:341–348. [Google Scholar]
- 73.D’Agostino R., Pearson E.S. Tests for departure from normality. empirical results for the distributions of b 2 andv b. Biometrika. 1973;60:613–622. [Google Scholar]
- 74.Albert R., Barabási A.-L. Statistical mechanics of complex networks. Reviews Modern Phys. 2002;74:47. [Google Scholar]
- 75.Huang L., Shi Y., Gong B., Jiang L., Liu X., Yang J., Tang J., You C., Jiang Q., Long B., et al. Blood single cell immune profiling reveals the interferon-mapk pathway mediated adaptive immune response for covid-19. MedRxiv. 2020 [Google Scholar]
- 76.Li S.-W., Wang C.-Y., Jou Y.-J., Yang T.-C., Huang S.-H., Wan L., Lin Y.-J., Lin C.-W. Sars coronavirus papain-like protease induces egr-1-dependent up-regulation of tgf-β1 via ros/p38 mapk/stat3 pathway. Sci. Reports. 2016;6:1–13. doi: 10.1038/srep25754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Albert R., Jeong H., Barabási A.-L. Error and attack tolerance of complex networks. Nature. 2000;406:378–382. doi: 10.1038/35019019. [DOI] [PubMed] [Google Scholar]
- 78.Halehalli R.R., Nagarajaram H.A. Molecular principles of human virus protein–protein interactions. Bioinformatics. 2015;31:1025–1033. doi: 10.1093/bioinformatics/btu763. [DOI] [PubMed] [Google Scholar]
- 79.Gavillet M., Klappert J.C., Spertini O., Blum S. Acute leukemia in the time of covid-19. Leukemia Res. 2020;92:106353. doi: 10.1016/j.leukres.2020.106353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ferrara F., Zappasodi P., Roncoroni E., Borlenghi E., Rossi G. Impact of covid-19 on the treatment of acute myeloid leukemia. Leukemia. 2020;34:2254–2256. doi: 10.1038/s41375-020-0925-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lammers T., Sofias A.M., van der Meel R., Schiffelers R., Storm G., Tacke F., Koschmieder S., Brümmendorf T.H., Kiessling F., Metselaar J.M. Dexamethasone nanomedicines for covid-19. Nature Nanotechnol. 2020;15:622–624. doi: 10.1038/s41565-020-0752-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Theoharides T., Conti P. Dexamethasone for covid-19? not so fast. J. Biol. Regul. Homeost. Agents. 2020;34:10–23812. doi: 10.23812/20-EDITORIAL_1-5. [DOI] [PubMed] [Google Scholar]
- 83.Chen Z., Hu J., Zhang Z., Jiang S., Han S., Yan D., Zhuang R., Hu B., Zhang Z. Efficacy of hydroxychloroquine in patients with covid-19: results of a randomized clinical trial. medrxiv. 2020 [Google Scholar]
- 84.Skipper C.P., Pastick K.A., Engen N.W., Bangdiwala A.S., Abassi M., Lofgren S.M., Williams D.A., Okafor E.C., Pullen M.F., Nicol M.R., et al. Hydroxychloroquine in nonhospitalized adults with early covid-19: a randomized trial. Annals Internal Med. 2020;173:623–631. doi: 10.7326/M20-4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Falvo J.V., Tsytsykova A.V., Goldfeld A.E. Transcriptional control of the tnf gene. TNF Pathophysiol. 2010;11:27–60. doi: 10.1159/000289196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.P. Dalerba, B. Levin, J.L. Thompson, A trial of lopinavir-ritonavir in covid-19, New Engl. J. Med. 382 (2020). [DOI] [PubMed]
- 87.Ye X., Luo Y., Xia S., Sun Q., Ding J., Zhou Y., Chen W., Wang X., Zhang W., Du W., et al. Clinical efficacy of lopinavir/ritonavir in the treatment of coronavirus disease 2019. Eur. Rev. Med. Pharmacol. Sci. 2020;24:3390–3396. doi: 10.26355/eurrev_202003_20706. [DOI] [PubMed] [Google Scholar]
- 88.Zha L., Li S., Pan L., Tefsen B., Li Y., French N., Chen L., Yang G., Villanueva E.V. Corticosteroid treatment of patients with coronavirus disease 2019 (covid-19) Med. J. Aust. 2019;212(2020):416–420. doi: 10.5694/mja2.50577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Maxwell P. HIF-1: an oxygen response system with special relevance to the kidney. J. Am. Soc. Nephrol. 2003;14(11):2712–2722. doi: 10.1097/01.asn.0000092792.97122.e0. [DOI] [PubMed] [Google Scholar]
- 90.Jahani M., Sadat D., Kamran M. Hypoxia: A key feature of COVID-19 launching activation of HIF-1 and cytokine storm. J. Inflamm. 2020;17(1):1–10. doi: 10.1186/s12950-020-00263-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kawai T., Shizuo A. Toll‐like receptor and RIG‐1‐like receptor signaling. Ann. NY. Acad. Sci. 2008;1143(1):1–20. doi: 10.1196/annals.1443.020. [DOI] [PubMed] [Google Scholar]
- 92.Larson C., et al. TGF-beta: a master immune regulator. Expert Opin. Ther. Targets. 2020;24(5):427–438. doi: 10.1080/14728222.2020.1744568. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.