Graphical abstract
Abbreviations: AES, atomic emission spectroscopy; ASAXS, anomalous small-angle X-ray scattering; BE, buffer equilibration; ICP-MS, inductively coupled plasma mass spectrometry; NMR, nuclear magnetic resonance; PRE, paramagnetic relaxation enhancement; SAXS, small-angle X-ray scattering
Keywords: Dynamics, Electrostatic interactions, Ion atmosphere, Ionic diffusion, Spatial distribution
Abstract
Ionic interactions are crucial to biological functions of DNA, RNA, and proteins. Experimental research on how ions behave around biological macromolecules has lagged behind corresponding theoretical and computational research. In the 21st century, quantitative experimental approaches for investigating ionic interactions of biomolecules have become available and greatly facilitated examinations of theoretical electrostatic models. These approaches utilize anomalous small-angle X-ray scattering, atomic emission spectroscopy, mass spectrometry, or nuclear magnetic resonance (NMR) spectroscopy. We provide an overview on the experimental methodologies that can quantify and characterize ions within the ion atmospheres around nucleic acids, proteins, and their complexes.
1. Introduction
Ions are essential for life. Cellular fluids contain various inorganic and organic ions. These ions strongly influence thermodynamics and kinetics of macromolecular interactions [1], [2], [3], [4]. Such influences on DNA, RNA, and proteins depend not only on ionic strength, but also on ionic species, as well-known for Hofmeister series of ions that affect solubility and stability of proteins [3], [5]. While ions are important as essential constituents of biological systems, it is not well understood how these ions interact with biological macromolecules and impact their functions.
Largely due to their mobile nature, ions around nucleic acids and proteins are difficult to directly observe. Protein Data Bank (PDB) has collected > 170,000 structures of biological macromolecules and their complexes over the past five decades [6]. Multivalent ions tightly bound to particular sites are seen in many PDB structures. However, the vast majority of monovalent cations and anions undergo territorial binding (as opposed to site binding) and are unresolved even in high-resolution crystal structures of biological macromolecules. The spatial distribution of mobile ions and their interactions with macromolecules remains elusive in structural biology.
On the other hand, the ion atmosphere where counterions are condensed around nucleic acids was proposed even prior to the inception of PDB. In 1950–60 s, studies using electrophoresis, volumetry, and dialysis equilibrium approaches had suggested that nucleic acids are surrounded by counterions [7], [8], [9], [10]. As three-dimensional structures of DNA and RNA were revealed, researchers developed theoretical models of ionic distribution around nucleic acids and its relevance to the thermodynamics of nucleic acid-protein interactions [11], [12], [13]. In 1970–90 s, the theoretical models for the ion atmosphere were used to explain thermodynamic and 23Na nuclear magnetic resonance (NMR) data [14], [15], [16]. However, the validity range of the theoretical models remained to be addressed. The ion atmosphere can be convoluted with solvation or conformational dynamics, which may limit the validity of simple models. It could be even more challenging to theoretically describe the ion atmosphere around proteins because their overall charges are smaller and the charge distribution on the molecular surfaces is more uneven. Validations through experiments are essential for any theoretical models. However, in the 20th century, experimental research on the ion atmosphere lagged far behind theoretical and computational research.
In the first two decades of the 21st century, there has been remarkable progress in experimental research on the ion atmosphere around DNA, RNA, and proteins. Some newly developed approaches have greatly facilitated examination and validation of theoretical models and yielded deeper insight into how ions behave around biological macromolecules. In this mini-review, we provide an overview on the recent progress in the experimental approaches for investigating the ion atmosphere around nucleic acids and proteins. Introducing some relevant fundamental concepts, we explain the basic principles of these experimental approaches and their applications.
2. Theoretical aspects of ion accumulation around biological macromolecules
2.1. The concept of ion atmosphere
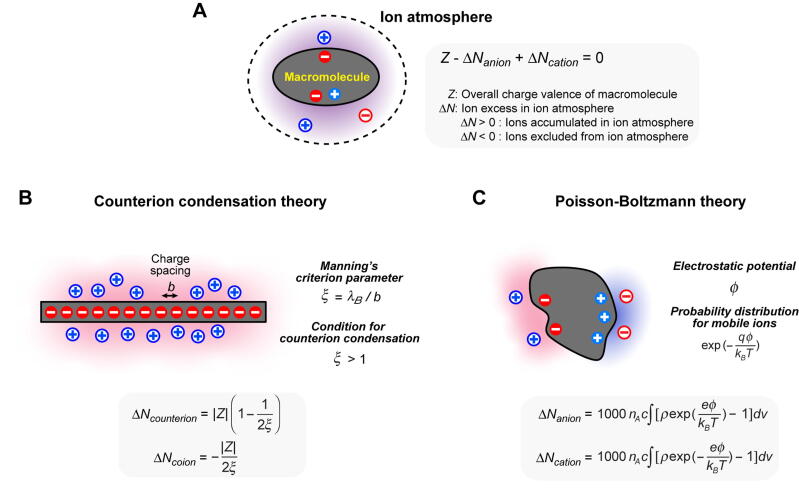
The ion atmosphere of a macromolecule is a zone near the macromolecular surface where the probability distribution of mobile ions differs from the background due to their electrostatic interactions with the macromolecule. For nucleic acids, cations acting as counterions are accumulated in the ion atmosphere and anions acting as coions are excluded from the ion atmosphere. It is important to note that the charge neutralization of a macromolecule occurs not only via accumulation of counterions in the ion atmosphere but also via exclusion of coions from the ion atmosphere (Fig. 1A). The space comprising of the macromolecule and its ion atmosphere should satisfy Z – ΔNanion + ΔNcation = 0, where Z is the overall charge valence of the macromolecule; ΔN is a parameter referred to as ‘ion excess’ and represents the number of ions accumulated in (ΔN > 0) or excluded from (ΔN < 0) the ion atmosphere per macromolecule. An ion excess ΔN represents the difference between the number of ions in the ion atmosphere and the number of ions in the same volume outside the ion atmosphere (i.e., the background).
Fig. 1.
Concepts of the ion atmosphere and relevant theories. (A) Charge neutralization for the space comprising of a macromolecule and its ion atmosphere. (B) View of the counterion condensation theory. λB is the Bjerrum length. For nucleic acids, ΔNcounterion = ΔNcation and ΔNcoion = ΔNanion. (C) View of the Poisson-Boltzmann theory. Symbols used in the ion-excess numbers ΔNcation and ΔNanion are as follows: nA is Avogadro’s number; c is the bulk ion concentration in mol/L units; ρ defines ion accessibility in the box with ρ = 1 for accessible regions and ρ = 0 for regions that are inaccessible due to macromolecular atoms; q, the ionic charge; e, the elementary charge; kB, the Boltzmann constant; T, the temperature; and v, the volume in m3. A factor of 1000 is for the conversion of the volume unit from L to m3. Subtraction of 1 in the integral corresponds to subtraction of the Boltzmann factor for the background with an electrostatic potential of zero. The shown equations for ion excess are for monovalent ions.
2.2. Counterion condensation theory
The counterion condensation theory is a relatively simple theory about condensation around linear polyelectrolytes such as nucleic acids. It was developed by Manning in 1969 [12], [17], [18], and applied to the thermodynamics of nucleic acid-protein interactions by Record in late 1970 s [13], [19], [20]. According to this theory, counterion condensation around a linear polyelectrolyte occurs when its effective mean charge spacing b is smaller than the Bjerrum length λB (=7.14 Å for water at 25 °C). The so-called Manning parameter ξ = λB / b is of key importance in this theory. ξ > 1 is the criterion for the counterion condensation. B-form DNA is predicted to condense counterions because b = 1.7 Å (i.e., two phosphates per 3.4 Å along the double-helical axis) and ξ = 4.2. The ion-excess numbers ΔNcation and ΔNanion can be estimated from ξ and the overall macromolecular charge valance Z. The total number of counterions in the condensation region is |Z|[1 − ξ-1], whereas the total number of excluded coions is |Z|(2ξ)-1 [12]. Manning’s theory separately considers ‘condensed counterions’ and ‘uncondensed counterions’ for the charge neutralization, but this distinction has been criticized as a nonphysical treatment [11], [21]. When screening over the entire atmosphere is considered for monovalent ions, the ion-excess numbers are ΔNcounterion = |Z|[1 − (2ξ)-1] and ΔNcoion = −|Z|(2ξ)-1 (Fig. 1B) [13]. For example, this theory predicts that a chemically synthesized 24-bp B-form DNA (Z = -46) accumulates 40.5 cations in the ion atmosphere and excludes 5.5 anions (i.e., ΔNcation = 40.5 and ΔNanion = -5.5).
The counterion condensation theory has been successful in explaining various experimental data. In particular, for DNA, this theory explained the salt concentration dependence of thermodynamics for many DNA-protein association processes [22], [23]. However, due to the assumption of linear charge distribution, the counterion condensation theory is not applicable for globular proteins and folded RNAs. Another limitation is that this theory does not predict the dependence of ΔN on ionic strength. Recent studies by ion-counting methods clearly showed that ΔNcation and ΔNanion significantly depend on ionic strength, especially when ionic strength is >100 mM [24], [25].
2.3. Poisson-Boltzmann theory
If the three-dimensional structure is known for a macromolecule, the spatial probability distribution of mobile ions around the macromolecular surface at a particular ionic strength can be predicted by the Poisson-Boltzmann theory [26], [27]. In this theory, the Poisson equation for electrostatics is combined with the assumption that the probability distribution of mobile ions is given by a Boltzmann factor with respect to the electrostatic potential. The Poisson-Boltzmann equation is a second-order differential equation and can be numerically solved using atomic coordinates of biomolecules. Software for this purpose such as APBS [28] and DelPhi [29], [30] has gained popularity in a wide variety of research areas. Using the calculated electrostatic potentials, the spatial distribution of mobile ions can be predicted from the Boltzmann factors. Based on the probability distribution of mobile ions, the ion-excess numbers ΔNcation and ΔNanion can also be predicted using the equations shown in Fig. 1C.
The Poisson-Boltzmann theory is approximate under assumptions that simplifies calculations [31]. The lack of consideration of correlations between ions can diminish accuracy in calculations of electrostatic potentials for systems at high ionic strength [32]. Solvation of ions and macromolecules is neglected and ions are treated as point charges. Due to the assumption of a dielectric continuum, the electrostatic potentials predicted with the Poisson-Boltzmann theory may be inaccurate for zones near the first hydration layer. Many Poisson-Boltzmann models assume a uniform dielectric constant for the interior or exterior of the macromolecule, creating a sharp dielectric jump at the boundary that could cause problems. Modified Poisson-Boltzmann models have been proposed for improvement, for example, by including corrections for finite ion sizes or solvent dielectric saturation effects [33], [34], [35], [36]. There are needs of examinations for the theoretical models through experiments.
3. Experimental methods for investigating ion atmosphere around biological macromolecules
Since Hofmeister discovered a series of ions that affect the solubility and stability of proteins in the late 19th century, there have been numerous investigations into ionic interactions of proteins and nucleic acids [2], [3], [4]. Here, we focus on recently developed methods that are truly quantitative in determining the ion-excess numbers or/and are incisive in characterizing ions in the ion atmosphere. These methods are summarized in Table 1.
Table 1.
Quantitative experimental methods suited for investigations of the ion atmosphere.
| Methods |
||||
|---|---|---|---|---|
| Investigations | AES | ICP-MS | ASAXS | NMR |
| Quantification of ions in the ion atmosphere | Yes [24], [37] | Yes [25], [38], [39] | Yes [40] | Yes [41] |
| Competition between ions for macromolecules | Yes [24] | Yes [42] | Yes [43], [44] | Yes [41], [45], [46] |
| Spatial distribution of ions around macromolecules | n.a. | n.a. | Yes [43], [47] | Yes [41], [48], [66] |
| Diffusional properties of ions interacting with macromolecules | n.a. | n.a. | n.a. | Yes [41], [46] |
| Ion release upon macromolecular association | p.a. | p.a. | p.a. | Yes [41], [46] |
Yes: The feasibility demonstrated in the cited references.
n.a.: Not applicable.
p.a.: Potentially applicable.
3.1. ASAXS methods
Small-angle X-ray scattering (SAXS) is a powerful technique that can provide global structural information of biological macromolecules in solution. Anomalous SAXS (ASAXS) occurs at wavelengths near the X-ray absorption edge of an element [50]. When SAXS data are recorded using a wavelength close to the absorption edge of an element (resonant) and another wavelength slightly far from the absorption edge, only the resonant element exhibits differences between the two datasets. Subtraction of resonant from non-resonant scattering profile yields ASAXS data, which provide information about the resonant element (e.g., ions) correlated with a non-resonant structure (e.g., DNA). Although ASAXS for Na+, K+, and Mg2+ ions are difficult to detect due to interference by water [51], ASAXS can readily be measured for heavier ions such as Rb+, Sr2+ and Co3+.
In 2003, using ASAXS, Pollack and coworkers achieved the first direct confirmation of the physical presence of the ion atmosphere around DNA [47]. They measured ASAXS for Rb+ and Sr2+ ions in solutions of a 25-bp DNA duplex. Ionic competition between Co3+ and Rb+ ions for DNA phosphates was also studied by ASAXS [44]. In the charge neutralization of DNA, multivalent ions were found to occupy the ion atmosphere more favorably than monovalent ions. It was also demonstrated that the ASAXS-based methodology can quantify the number of ions around DNA [40]. ASAXS was used to study ion atmosphere around RNA as well. The anomalous profiles of Rb+ and Sr2+ ions around 25-bp DNA and RNA duplexes of the same sequence were compared [52]. The data show that the RNA duplex attracts both monovalent and divalent ions closer to its surface and hence have more effective charge screening as compared to the corresponding DNA duplex. The discrepancy was attributed to the topological difference between the A-form RNA and the B-form DNA.
ASAXS data agreed well with the predictions from the Poisson-Boltzmann theory with finite ion sizes taken into account [53], [54], [55]. Excellent agreement was also found between ASAXS profile of short double-stranded RNA and the results from explicit solvent molecular dynamics (MD) simulations [56]. However, for a RNA pseudoknot, both experimental ASAXS and atomistic MD results were significantly different from the Poisson-Boltzmann-based predictions from the crystal structure. These findings encourage further improvement of theoretical models for dynamic structures. Thus, ASAXS greatly facilitated the examination of theoretical models on spatial distribution of mobile ions around DNA and RNA.
3.2. BE-AES and BE-ICP-MS methods
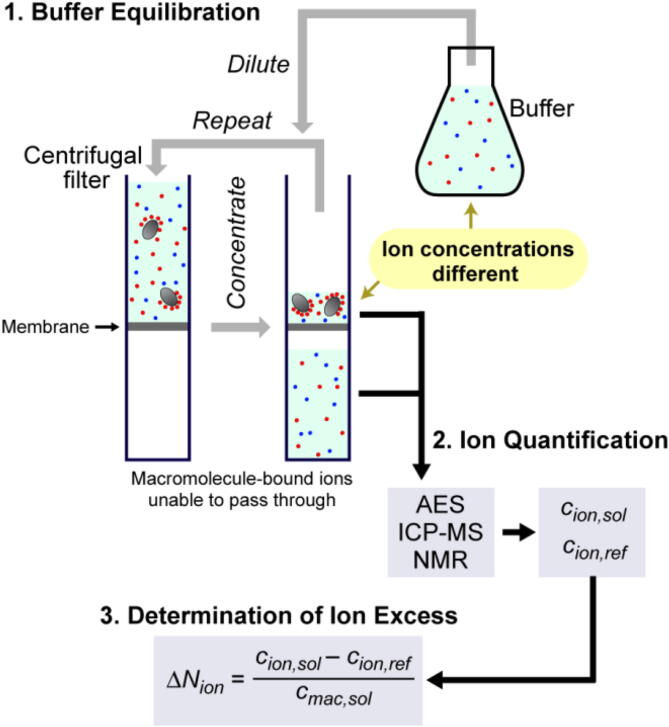
The buffer equilibration-atomic emission spectroscopy (BE-AES) method is an ion-counting method that allows for quantifying ions within the ion atmosphere [24], [37], [57]. The first step in ion-counting methods is buffer equilibration using centrifugal filters (Fig. 2) or dialysis membrane [58]. Ions in the final macromolecular solution and in the flow-through liquid are quantified using atomic emission spectroscopy (AES). The quantification of ions can also be performed by inductively coupled plasma mass spectroscopy (ICP-MS) [25], [38], [42] or NMR [41], [46]. The BE-AES and BE-ICP-MS methods can quantify various elements and analyze biologically relevant ions such as Na+, K+, and Mg2+ ions [57], [58].
Fig. 2.
Ion-counting methods for quantifying ions in the ion atmosphere. The ion excess per macromolecule (ΔNion) is determined from the ion concentrations in the final solution (cion,sol) and in the reference buffer used for the equilibration (cion,ref) as well as the macromolecular concentration in the final solution (cmac,sol).
The BE-AES ion-counting method also allows for investigations of competition between ions for nucleic acids [24]. Competition experiments using this method showed that Mg2+ and Ca2+ ions associate with DNA ~ 40-fold more strongly than Na+ and K+ ions. The BE-AES method was also used to investigate anion exclusion. The ion-excess numbers at high salt concentrations were found to depend strongly on the mean activity coefficients of the salts. The BE-AES or BE-ICP-MS methods have been used to study ion atmosphere around RNA as well [38]. As anticipated from the structural difference, the cation-excess number determined for a 24-bp RNA duplex was larger than that for the corresponding DNA duplex of the same sequence [38]. The BE-ICP-MS method has been applied to study the ion atmosphere around a nucleosome core particle [39]. Intriguingly, although the nucleosome formation reduces the overall charge by half, the ion-counting data suggested that a strong negative electrostatic field remains.
The BE-AES and BE-ICP-MS ion-counting data illuminated the strengths and limitations of the theoretical models on the ion atmosphere. The discrepancies between Poisson-Boltzmann predictions and experimental data were more significant for ions with higher valence or larger size. Based on the experimental data, new theoretical models that can better reproduce the experimental ion-counting data were developed [59], [60], [61]. The ion-counting methods helped advance knowledge about the ion atmosphere surrounding nucleic acids.
3.3. NMR methods for quantifying and characterizing ions in the ion atmosphere
NMR spectroscopy is a powerful technique for probing the structural and dynamic properties of biomolecules [62]. In 1970–80 s, 23Na NMR was used to study interactions between Na+ ions and DNA [45], [63], [64]. Later on, magnetic field-dependence of 23Na NMR relaxation was used to invetigate more details about the behavior of Na+ ions interacting with DNA [49]. In 1990s, NMR paramagnetic relaxation enhancement (PRE) arising from Mn2+ ions as well as nuclear Overhauser effects (NOEs) for NH4+ ions were also used to investigate ion-DNA interactions [48], [65], [66]. More recently, NMR-based ion-counting methods have been developed for more quantitative investigations of ions around nucleic acids and proteins [41], [46].
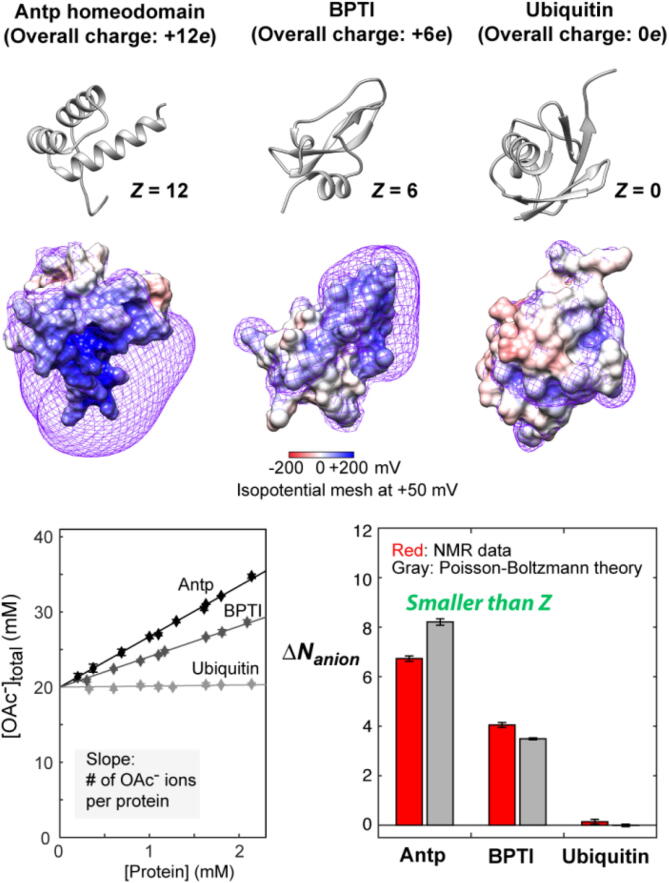
The NMR-based ion-counting method were applied to both DNA and proteins. The ion-excess number ΔNanion was measured for the Antp homeodomain, bovine pancreatic trypsin inhibitor (BPTI) and ubiquitin (Fig. 3) [41]. The NMR experiments clearly showed accumulation of anions around the positively charged proteins. The measured ΔNanion was significantly smaller than the overall charge valence (Z) of each protein, which suggest that the charge neutralization occurs via both the accumulation of counterions and the exclusion of coions [41]. The experimental results were consistent with the predication from the Poisson-Boltzmann theory (Fig. 3).
Fig. 3.
NMR-based quantification of anions accumulated around the Antp homeodomain, BPTI, and ubiquitin. Note that the measured ΔNanion was smaller than the overall charge valence Z. This means that the charge neutralization occurs not only via accumulation of anions, but also via exclusion of cations from the ion atmosphere. Adopted from Yu et al. [41]
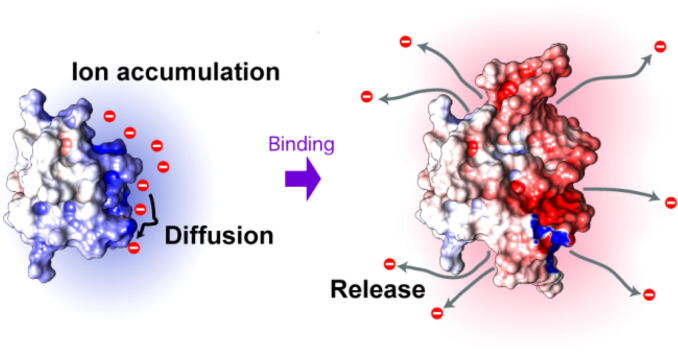
Unlike other ion-counting methods, the NMR-based method can also provide information about the diffusional properties of ions within and outside the ion atmosphere [41], [46]. NMR-based diffusion experiments showed that the acetate ions in the ion atmosphere are only loosely constrained by the proteins. By comparing the apparent diffusion coefficients of ions in solutions of free proteins, free nucleic acids, and the complex as well as those in the sample buffer alone, the number of ions released upon the formation of the protein-nucleic acid complex can also be determined. NMR-based diffusion data showed the release of 7 anions from the ion atmosphere around Antp homeodomain and 11 cations from the ion atmosphere around a 15-bp DNA duplex upon the formation of the protein-DNA complex [41], [46].
3.4. NMR methods for investigating spatial distribution of ions in the ion atmosphere
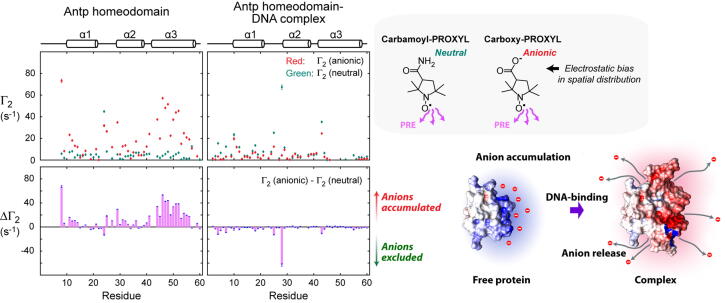
NMR PRE arising from paramagnetic cosolutes, carboxy-PROXYL (anionic) and carbamoyl-PROXYL (neutral), can be used to investigate anion distribution around a protein. The PRE rate for the transverse nuclear magnetizations, Г2, is sensitive to spatial distribution of paramagnetic cosolutes [67]. The difference (ΔГ2) between the PRE Г2rates for the anionic and neutral PROXYL derivatives at the same concentration reflect a bias in spatial distribution of anions due to electrostatic interactions with the protein. Fig. 4 shows examples of ΔГ2 data for the Antp homeodomain in the free state and those for the Antp homeodomain-DNA complex. Many residues of the Antp homeodomain in the free state exhibited large positive ΔГ2 values, suggesting that anions are accumulated around the positively charged surface of this protein. In contrast, most residues in the complex with DNA exhibited negative ΔГ2, suggesting that anions are excluded from the complex surface. This is reasonable since the overall charge for the Antp homeodomain-DNA complex is −16e. Moreover, the negative ΔГ2 is consistent with the NMR diffusion data indicating anion release from the Antp homeodomain upon binding to DNA. The NMR-based approaches for investigating the ion atmosphere can readily be applied to many other systems of proteins, nucleic acids, and their complexes.
Fig. 4.
NMR paramagnetic relaxation enhancement (PRE)-based approach for investigating the spatial distribution of anions around the Antp homeodomain and its complex with 15-bp DNA. The data were adopted from Yu et al. [41] Comparison of PRE arising from analogous anionic and neutral paramagnetic cosolutes provides site-specific information about ion accumulation or exclusion. The data shown on the left-hand side suggest that anions are accumulated around the Antp homeodomain in the free state but excluded upon formation of the protein-DNA complex.
4. Future perspectives
Many experimental data show that thermodynamics and kinetics of protein-nucleic acid or protein–protein association are strongly influenced by ions [16], [22], [68], [69]. Such influences may arise not only from the screening effect [70], but also from the entropic effects of the ion release upon the macromolecular complex formation [4], [16], [22]. Further research on the ion atmospheres around macromolecules may explain why activities of some proteins strongly depend on types of ions present in the same solutions. For instance, when glutamate (Glu-) ions are used instead of Cl- ions in biochemical experiments, some DNA-binding proteins exhibit substantially stronger (>100-fold for some cases) affinity for DNA [71], [72]. This effect may be related to differences in the behavior of Glu-and Cl- ions around positively charged proteins. Future applications of the aforementioned methodologies will likely provide mechanistic insight into how ions affect functions of proteins and nucleic acids. Since NMR methods for charged moieties of proteins are available [74], [75], more detailed investigations of interactions between ions and individual charged side chains of proteins will be feasible.
Further methodological progress can be made through an advancement in hardware. For instance, advances in synchrotron radiation sources and corresponding detectors would improve the precision of the ASAXS method and may extend its capability toward more proximal ionic interactions [73]. Advances in NMR instrumentation will facilitate diffusion coefficient measurements for various ions. Typical NMR probes can generate magnetic field gradients of up to ~55 gauss/cm. This magnitude of gradients is sufficient for diffusion experiments on 1H, 13C, 15N, 19F and 31P nuclei, but insufficient for diffusion experiments on 23Na and 35Cl nuclei. Because quadrupole nuclei exhibit rapid longitudinal relaxation and relatively small gyromagnetic ratios, diffusion NMR experiments for these nuclei require special broadband probe hardware that can generate far stronger field gradients (e.g., >200 gauss/cm). Such hardware will enable investigations of the diffusional properties of physiologically important Na+ and Cl- ions in the vicinity of nucleic acids and proteins.
5. Concluding remarks
In the past two decades, there have been great advances in experimental research on the ion atmospheres around biomolecules. ASAXS, BE-AE and BE-ICP-MS methods have provided significant insights into the ion atmosphere around DNA and RNA. More recently, NMR-based methods that are capable of quantifying, characterizing, and visualizing ions in the ion atmosphere have provided unprecedented information about the ion atmosphere around nucleic acids and proteins. These experimental approaches can readily generate mutually beneficial feedback loops between theoretical/computational and experimental studies. Further research on weak interactions between ions and macromolecules will likely advance our knowledge about how ions impact biological macromolecules and their functions in living systems.
CRediT authorship contribution statement
Binhan Yu: Writing - original draft. Junji Iwahara: Funding acquisition, Conceptualization, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by Grant R35-GM130326 (to J.I.) from the National Institutes of Health. We thank Channing Pletka for editing the manuscript.
References
- 1.Lipfert J., Doniach S., Das R., Herschlag D. Understanding nucleic acid-ion interactions. Annu Rev Biochem. 2014;83(1):813–841. doi: 10.1146/annurev-biochem-060409-092720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Okur H.I., Hladílková J., Rembert K.B., Cho Y., Heyda J., Dzubiella J. Beyond the Hofmeister Series: Ion-Specific Effects on Proteins and Their Biological Functions. J Phys Chem B. 2017;121(9):1997–2014. doi: 10.1021/acs.jpcb.6b10797. [DOI] [PubMed] [Google Scholar]
- 3.Record M.T., Jr., Zhang W., Anderson C.F. Analysis of effects of salts and uncharged solutes on protein and nucleic acid equilibria and processes: a practical guide to recognizing and interpreting polyelectrolyte effects, Hofmeister effects, and osmotic effects of salts. Adv Protein Chem. 1998;51:281–353. doi: 10.1016/s0065-3233(08)60655-5. [DOI] [PubMed] [Google Scholar]
- 4.Yu B., Pettitt B.M., Iwahara J. Dynamics of ionic interactions at protein–nucleic acid interfaces. Acc Chem Res. 2020;53(9):1802–1810. doi: 10.1021/acs.accounts.0c00212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y., Cremer P.S. Chemistry of Hofmeister anions and osmolytes. Annu Rev Phys Chem. 2010;61(1):63–83. doi: 10.1146/annurev.physchem.59.032607.093635. [DOI] [PubMed] [Google Scholar]
- 6.Burley S.K., Bhikadiya C., Bi C., Bittrich S., Chen L., Crichlow G.V. RCSB Protein Data Bank: powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering and energy sciences. Nucleic Acids Res. 2021;49:D437–D451. doi: 10.1093/nar/gkaa1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ross P.D., Scruggs R.L. Electrophoresis of DNA. II. Specific interactions of univalent and divalent cations with DNA. Biopolymers. 1964;2(1):79–89. [Google Scholar]
- 8.Shapiro J.T., Stannard B.S., Felsenfeld G. The binding of small cations to deoxyribonucleic acid. Nucleotide specificity. Biochemistry. 1969;8(8):3233–3241. doi: 10.1021/bi00836a015. [DOI] [PubMed] [Google Scholar]
- 9.Skerjanc J., Strauss U.P. Interactions of polyelectrolytes with simple electrolytes. III. The binding of magnesium ion by deoxyribonucleic acid. J Am Chem Soc. 1968;90(12):3081–3085. doi: 10.1021/ja01014a017. [DOI] [PubMed] [Google Scholar]
- 10.Zubay G., Doty P. Nucleic acid interactions with metal ions and amino acids. BBA - Biochimica et Biophysica Acta. 1958;29(1):47–58. doi: 10.1016/0006-3002(58)90145-8. [DOI] [PubMed] [Google Scholar]
- 11.Bret M.L., Zimm B.H. Distribution of counterions around a cylindrical polyelectrolyte and manning's condensation theory. Biopolymers. 1984;23:287–312. doi: 10.1002/bip.360230208. [DOI] [PubMed] [Google Scholar]
- 12.Manning G.S. Limiting Laws and Counterion Condensation in Polyelectrolyte Solutions. I. Colligative Properties. J Chem Phys. 1969;51:924–933. [Google Scholar]
- 13.Record M.T., Jr., Lohman M.L., De Haseth P. Ion effects on ligand-nucleic acid interactions. J Mol Biol. 1976;107:145–158. doi: 10.1016/s0022-2836(76)80023-x. [DOI] [PubMed] [Google Scholar]
- 14.Anderson C.F., Record M.T., Jr. Ion distributions around DNA and other cylindrical polyions: theoretical descriptions and physical implications. Annu Rev Biophys Biophys Chem. 1990;19:423–465. doi: 10.1146/annurev.bb.19.060190.002231. [DOI] [PubMed] [Google Scholar]
- 15.Manning G.S. Molecular theory of polyelectrolyte solutions with applications to electrostatic properties of polynucleotides. Q Rev Biophys. 1978;11:179–246. doi: 10.1017/s0033583500002031. [DOI] [PubMed] [Google Scholar]
- 16.Record M.T., Jr., Anderson C.F., Lohman T.M. Thermodynamic analysis of ion effects on the binding and conformational equilibria of proteins and nucleic acids: the roles of ion association or release, screening, and ion effects on water activity. Q Rev Biophys. 1978;11:103–178. doi: 10.1017/s003358350000202x. [DOI] [PubMed] [Google Scholar]
- 17.Manning G.S. Limiting Laws and Counterion Condensation in Polyelectrolyte Solutions. 3. An Analysis Based on Mayer Ionic Solution Theory. J Chem Phys. 1969;51:3249–4000. [Google Scholar]
- 18.Manning G.S. Limiting Laws and Counterion Condensation in Polyelectrolyte Solutions. 2. Self-Diffusion of Small Ions. J Chem Phys. 1969;51:934–1000. [Google Scholar]
- 19.Dehaseth P.L., Lohman T.M., Record M.T. Nonspecific Interaction of Lac Repressor with DNA - Association Reaction Driven by Counterion Release. Biochemistry. 1977;16:4783–4790. doi: 10.1021/bi00641a004. [DOI] [PubMed] [Google Scholar]
- 20.Record M.T., Jr., deHaseth P.L., Lohman T.M. Interpretation of monovalent and divalent cation effects on the lac repressor-operator interaction. Biochemistry. 1977;16:4791–4796. doi: 10.1021/bi00641a005. [DOI] [PubMed] [Google Scholar]
- 21.Stigter D. Evaluation of the counterion condensation theory of polyelectrolytes. Biophys J. 1995;69:380–388. doi: 10.1016/S0006-3495(95)79910-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Privalov P.L., Dragan A.I., Crane-Robinson C. Interpreting protein/DNA interactions: distinguishing specific from non-specific and electrostatic from non-electrostatic components. Nucleic Acids Res. 2011;39:2483–2491. doi: 10.1093/nar/gkq984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Record M.T., Jr., Ha J.H., Fisher M.A. Analysis of equilibrium and kinetic measurements to determine thermodynamic origins of stability and specificity and mechanism of formation of site-specific complexes between proteins and helical DNA. Methods Enzymol. 1991;208:291–343. doi: 10.1016/0076-6879(91)08018-d. [DOI] [PubMed] [Google Scholar]
- 24.Bai Y., Greenfeld M., Travers K.J., Chu V.B., Lipfert J., Doniach S. Quantitative and comprehensive decomposition of the ion atmosphere around nucleic acids. J Am Chem Soc. 2007;129:14981–14988. doi: 10.1021/ja075020g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gebala M., Giambasu G.M., Lipfert J., Bisaria N., Bonilla S., Li G. Cation-anion interactions within the nucleic acid ion atmosphere revealed by ion counting. J Am Chem Soc. 2015;137:14705–14715. doi: 10.1021/jacs.5b08395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fogolari F., Brigo A., Molinari H. The Poisson-Boltzmann equation for biomolecular electrostatics: a tool for structural biology. J Mol Recognit. 2002;15:377–392. doi: 10.1002/jmr.577. [DOI] [PubMed] [Google Scholar]
- 27.Honig B., Nicholls A. Classical electrostatics in biology and chemistry. Science. 1995;268:1144–1149. doi: 10.1126/science.7761829. [DOI] [PubMed] [Google Scholar]
- 28.Baker N.A., Sept D., Joseph S., Holst M.J., McCammon J.A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gilson M.K., Sharp K.A., Honig B.H. Calculating the electrostatic potential of molecules in solution: Method and error assessment. J Comput Chem. 1988;9:327–335. [Google Scholar]
- 30.Li C., Jia Z., Chakravorty A., Pahari S., Peng Y., Basu S. DelPhi Suite: New Developments and Review of Functionalities. J Comput Chem. 2019;40:2502–2508. doi: 10.1002/jcc.26006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grochowski P., Trylska J. Continuum molecular electrostatics, salt effects, and counterion binding—A review of the Poisson-Boltzmann theory and its modifications. Biopolymers. 2008;89:93–113. doi: 10.1002/bip.20877. [DOI] [PubMed] [Google Scholar]
- 32.Netz R.R., Orland H. Beyond Poisson-Boltzmann: Fluctuation effects and correlation functions. Eur Phys J E. 2000;1:203–214. [Google Scholar]
- 33.Chu V.B., Bai Y., Lipfert J., Herschlag D., Doniach S. Evaluation of ion binding to DNA duplexes using a size-modified Poisson-Boltzmann theory. Biophys J. 2007;93:3202–3209. doi: 10.1529/biophysj.106.099168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Granot J. Effect of finite ionic size on the solution of the Poisson-Boltzmann equation: Application to the binding of divalent metal ions to DNA. Biopolymers. 1983;22:1831–1841. doi: 10.1002/bip.360220715. [DOI] [PubMed] [Google Scholar]
- 35.Gavryushov S. Electrostatics of B-DNA in NaCl and CaCl2 Solutions: Ion Size, Interionic Correlation, and Solvent Dielectric Saturation Effects. J Phys Chem B. 2008;112:8955–8965. doi: 10.1021/jp711339d. [DOI] [PubMed] [Google Scholar]
- 36.Borukhov I., Andelman D., Orland H. Steric Effects in Electrolytes: A Modified Poisson-Boltzmann Equation. Phys Rev Lett. 1997;79:435–438. [Google Scholar]
- 37.Das R., Travers K.J., Bai Y., Herschlag D. Determining the Mg2+ Stoichiometry for Folding an RNA Metal Ion Core. J Am Chem Soc. 2005;127:8272–8273. doi: 10.1021/ja051422h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gebala M., Herschlag D. Quantitative studies of an RNA duplex electrostatics by ion counting. Biophys J. 2019;117:1116–1124. doi: 10.1016/j.bpj.2019.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gebala M., Johnson S.L., Narlikar G.J., Herschlag D. Ion counting demonstrates a high electrostatic field generated by the nucleosome. Elife. 2019;8 doi: 10.7554/eLife.44993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pabit S.A., Meisburger S.P., Li L., Blose J.M., Jones C.D., Pollack L. Counting Ions around DNA with Anomalous Small-Angle X-ray Scattering. J Am Chem Soc. 2010;132:16334–16336. doi: 10.1021/ja107259y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu B., Pletka C.C., Iwahara J. Quantifying and visualizing weak interactions between anions and proteins. Proc Natl Acad Sci U S A. 2021;118 doi: 10.1073/pnas.2015879118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gebala M., Bonilla S., Bisaria N., Herschlag D. Does cation size affect occupancy and electrostatic screening of the nucleic acid ion atmosphere? J Am Chem Soc. 2016;138:10925–10934. doi: 10.1021/jacs.6b04289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andresen K., Das R., Park H.Y., Smith H., Kwok L.W., Lamb J.S. Spatial Distribution of Competing Ions around DNA in Solution. Phys Rev Lett. 2004;93 doi: 10.1103/PhysRevLett.93.248103. [DOI] [PubMed] [Google Scholar]
- 44.Andresen K., Qiu X., Pabit S.A., Lamb J.S., Park H.Y., Kwok L.W. Mono- and Trivalent Ions around DNA: A Small-Angle Scattering Study of Competition and Interactions. Biophys J. 2008;95:287–295. doi: 10.1529/biophysj.107.123174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bleam M.L., Anderson C.F., Record M.T. Relative binding affinities of monovalent cations for double-stranded DNA. Proc Natl Acad Sci U S A. 1980;77:3085–3089. doi: 10.1073/pnas.77.6.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pletka C.C., Nepravishta R., Iwahara J. Detecting counterion dynamics in DNA-protein association. Angew Chem Int Ed Engl. 2020;59:1465–1468. doi: 10.1002/anie.201910960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Das R., Mills T.T., Kwok L.W., Maskel G.S., Millett I.S., Doniach S. Counterion distribution around DNA probed by solution X-ray scattering. Phys Rev Lett. 2003;90 doi: 10.1103/PhysRevLett.90.188103. [DOI] [PubMed] [Google Scholar]
- 48.Hud N.V., Feigon J. Localization of divalent metal ions in the minor groove of DNA A-tracts. J Am Chem Soc. 1997;119:5756–5757. [Google Scholar]
- 49.Denisov V.P., Halle B. Sequence-specific binding of counterions to B-DNA. Proc Natl Acad Sci U S A. 2000;97:629–633. doi: 10.1073/pnas.97.2.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pabit S.A., Finkelstein K.D., Chapter P.L. Using Anomalous Small Angle X-Ray Scattering to Probe the Ion Atmosphere Around Nucleic Acids. Methods in Enzymology: Academic Press. 2009;19:391–410. doi: 10.1016/S0076-6879(09)69019-4. [DOI] [PubMed] [Google Scholar]
- 51.Pollack L. SAXS studies of ion-nucleic acid interactions. Annu Rev Biophys. 2011;40:225–242. doi: 10.1146/annurev-biophys-042910-155349. [DOI] [PubMed] [Google Scholar]
- 52.Pabit S.A., Qiu X., Lamb J.S., Li L., Meisburger S.P., Pollack L. Both helix topology and counterion distribution contribute to the more effective charge screening in dsRNA compared with dsDNA. Nucleic Acids Res. 2009;37:3887–3896. doi: 10.1093/nar/gkp257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rouzina I., Bloomfield V.A. Competitive electrostatic binding of charged ligands to polyelectrolytes: practical approach using the non-linear Poisson-Boltzmann equation. Biophys Chem. 1997;64:139–155. doi: 10.1016/s0301-4622(96)02231-4. [DOI] [PubMed] [Google Scholar]
- 54.Rouzina I., Bloomfield V.A. Competitive Electrostatic Binding of Charged Ligands to Polyelectrolytes: Planar and Cylindrical Geometries. J Phys Chem. 1996;100:4292–4304. doi: 10.1016/s0301-4622(96)02231-4. [DOI] [PubMed] [Google Scholar]
- 55.Rouzina I., Bloomfield V.A. Influence of Ligand Spatial Organization on Competitive Electrostatic Binding to DNA. J Phys Chem. 1996;100:4305–4313. [Google Scholar]
- 56.Kirmizialtin S., Pabit S.A., Meisburger S.P., Pollack L., Elber R. RNA and its ionic cloud: solution scattering experiments and atomically detailed simulations. Biophys J. 2012;102:819–828. doi: 10.1016/j.bpj.2012.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Greenfeld M, Herschlag D. Probing Nucleic Acid–Ion Interactions with Buffer Exchange-Atomic Emission Spectroscopy. Biophysical, Chemical, and Functional Probes of RNA Structure, Interactions and Folding: Part B2009. p. 375-89. [DOI] [PubMed]
- 58.Jacobson D.R., Saleh O.A. Counting the ions surrounding nucleic acids. Nucleic Acids Res. 2017;45:1596–1605. doi: 10.1093/nar/gkw1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Giambasu G.M., Gebala M.K., Panteva M.T., Luchko T., Case D.A., York D.M. Competitive interaction of monovalent cations with DNA from 3D-RISM. Nucleic Acids Res. 2015;43:8405–8415. doi: 10.1093/nar/gkv830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Giambasu G.M., Case D.A., York D.M. Predicting Site-Binding Modes of Ions and Water to Nucleic Acids Using Molecular Solvation Theory. J Am Chem Soc. 2019;141:2435–2445. doi: 10.1021/jacs.8b11474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Giambasu G.M., Luchko T., Herschlag D., York D.M., Case D.A. Ion counting from explicit-solvent simulations and 3D-RISM. Biophys J. 2014;106:883–894. doi: 10.1016/j.bpj.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cavanagh J., Fairbrother W.J., Palmer A.G., III, Rance M., Skelton N.J. 2 ed. Elsevier Academic Press; Burlignton: 2007. Protein NMR Sprectroscopy: Principles and Practice. [Google Scholar]
- 63.Anderson C.F., Record M.T., Jr., Hart P.A. Sodium-23 NMR studies of cation-DNA interactions. Biophys Chem. 1978;7:301–316. doi: 10.1016/0301-4622(78)85007-8. [DOI] [PubMed] [Google Scholar]
- 64.Reuben J., Shporer M., Gabbay E.J. The alkali ion-DNA interaction as reflected in the nuclear relaxation rates of Na and Rb. Proc Natl Acad Sci U S A. 1975;72:245–247. doi: 10.1073/pnas.72.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hud N.V., Schultze P., Sklenar V., Feigon J. Binding sites and dynamics of ammonium ions in a telomere repeat DNA quadruplex. J Mol Biol. 1999;285:233–243. doi: 10.1006/jmbi.1998.2327. [DOI] [PubMed] [Google Scholar]
- 66.Hud N.V., Sklenar V., Feigon J. Localization of ammonium ions in the minor groove of DNA duplexes in solution and the origin of DNA A-tract bending. J Mol Biol. 1999;286:651–660. doi: 10.1006/jmbi.1998.2513. [DOI] [PubMed] [Google Scholar]
- 67.Clore G.M., Iwahara J. Theory, practice, and applications of paramagnetic relaxation enhancement for the characterization of transient low-population states of biological macromolecules and their complexes. Chem Rev. 2009;109:4108–4139. doi: 10.1021/cr900033p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Iwahara J., Kolomeisky A.B. Discrete-state stochastic kinetic models for target DNA search by proteins: Theory and experimental applications. Biophys Chem. 2021;269 doi: 10.1016/j.bpc.2020.106521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schreiber G., Fersht A.R. Rapid, electrostatically assisted association of proteins. Nat Struct Biol. 1996;3:427–431. doi: 10.1038/nsb0596-427. [DOI] [PubMed] [Google Scholar]
- 70.Zhou H.X., Pang X. Electrostatic Interactions in Protein Structure, Folding, Binding, and Condensation. Chem Rev. 2018;118:1691–1741. doi: 10.1021/acs.chemrev.7b00305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ha J.H., Capp M.W., Hohenwalter M.D., Baskerville M., Record M.T., Jr. Thermodynamic stoichiometries of participation of water, cations and anions in specific and non-specific binding of lac repressor to DNA. Possible thermodynamic origins of the “glutamate effect” on protein-DNA interactions. J Mol Biol. 1992;228:252–264. doi: 10.1016/0022-2836(92)90504-d. [DOI] [PubMed] [Google Scholar]
- 72.Leirmo S., Harrison C., Cayley D.S., Burgess R.R., Record M.T., Jr. Replacement of potassium chloride by potassium glutamate dramatically enhances protein-DNA interactions in vitro. Biochemistry. 1987;26:2095–2101. doi: 10.1021/bi00382a006. [DOI] [PubMed] [Google Scholar]
- 73.Lombardo D., Calandra P., Kiselev M.A. Structural Characterization of Biomaterials by Means of Small Angle X-rays and Neutron Scattering (SAXS and SANS), and Light Scattering Experiments. Molecules. 2020;25 doi: 10.3390/molecules25235624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hass M.A., Mulder F.A. Contemporary NMR Studies of Protein Electrostatics. Annu Rev Biophys. 2015;44:53–75. doi: 10.1146/annurev-biophys-083012-130351. [DOI] [PubMed] [Google Scholar]
- 75.Nguyen D., Chen C., Pettitt B.M., Iwahara J. NMR methods for characterizing the basic side chains of proteins: electrostatic interactions, hydrogen bonds, and conformational dynamics. Methods Enzymol. 2019;615:285–332. doi: 10.1016/bs.mie.2018.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]