Abstract
Background & Aims
Hepatoblastoma (HB), the most common pediatric liver cancer, often bears β-catenin mutations and deregulates the Hippo tumor suppressor pathway. Murine HBs can be generated by co-expressing β-catenin mutants and the constitutively active Hippo effector YAPS127A. Some HBs and other cancers also express mutants of NFE2L2/NRF2 (NFE2L2), a transcription factor that tempers oxidative and electrophilic stress. In doing so, NFE2L2 either suppresses or facilitates tumorigenesis.
Methods
We evaluated NFE2L2’s role in HB pathogenesis by co-expressing all combinations of mutant β-catenin, YAPS127A, and the patient-derived NFE2L2 mutants L30P and R34P in murine livers. We evaluated growth, biochemical and metabolic profiles, and transcriptomes of the ensuing tumors.
Results
In association with β-catenin+YAPS127A, L30P and R34P markedly accelerated HB growth and generated widespread cyst formation and necrosis, which are otherwise uncommon features. Surprisingly, any 2 members of the mutant β-catenin-YAPS127A-L30P/R34P triad were tumorigenic, thus directly establishing NFE2L2’s oncogenicity. Each tumor group displayed distinct features but shared 22 similarly deregulated transcripts, 10 of which perfectly correlated with survival in human HBs and 17 of which correlated with survival in multiple adult cancers. One highly up-regulated transcript encoded serpin E1, a serine protease inhibitor that regulates fibrinolysis, growth, and extracellular matrix. Although the combination of mutant β-catenin, YAPS127A, and serpin E1 did not accelerate cystogenic tumor growth, it did promote the widespread necrosis associated with mutant β-catenin-YAPS127A-L30P/R34P tumors.
Conclusions
Our findings establish the direct oncogenicity of NFE2L2 mutants and key transcripts, including serpin E1, that drive specific HB features.
Keywords: KEAP1, Hepatocellular Carcinoma, Plasminogen Activator Inhibitor, Warburg Effect
Abbreviations used in this paper: ADP, adenosine diphosphate; ARE, antioxidant response element; CNV, copy number variant; FAO, fatty acid oxidase; GFP, green fluorescent protein; HB, hepatoblastoma; HCC, hepatocellular carcinoma; HDTVI, hydrodynamic tail vein injection; IPA, Ingenuity Pathway Analysis; KEAP1, Kelch-like ECH-associated protein 1; OCR, oxygen consumption rate; PC, pyruvate carboxylase; pPDHα1, phosphorylated pyruvate dehydrogenase α1 catalytic subunit; PFK-1, phosphofructokinase; PKM-2, pyruvate kinase isozymes M1/M2; ROS, reactive oxygen species; SB, Sleeping Beauty; TCA, tricarboxylic acid; TCGA, The Cancer Genome Atlas; WT, wild-type; YAP, yes-associated protein
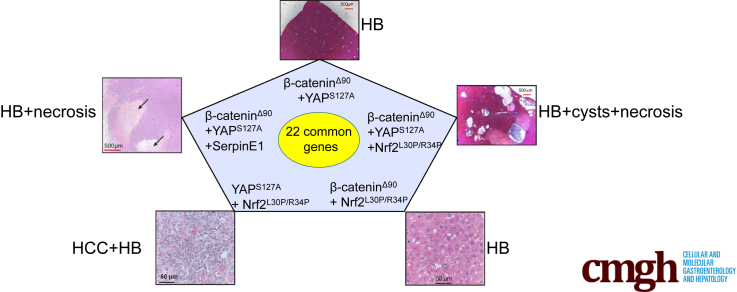
Graphical abstract
Summary.
The consequences of NFE2L2 dysregulation in hepatoblastoma are unclear. The co-expression of NFE2L2 mutants along with β-catenin and YAP mutants markedly accelerates hepatoblastoma growth. A core set of 22 transcripts appears responsible for this, and NFE2L2 is demonstrated to be a bona fide oncogene.
Hepatoblastoma (HB), the most common pediatric liver cancer, usually arises before 3 years of age. Factors impacting survival include age, α-fetoprotein levels, histologic subtype, and transcriptional profiles.1 Heterogeneous mutations in the β-catenin transcription factor occur in ∼80% of HBs in association with β-catenin’s nuclear accumulation.2 Most HBs also deregulate the Hippo pathway and also aberrantly accumulate its terminal effector yes-associated protein (YAP) in the nucleus.2,3
Mutations impair β-catenin’s interaction with the adenomatous polyposis coli complex that normally phosphorylates β-catenin and licenses its proteasome-mediated degradation.1,3,4 Stabilized β-catenin then enters the nucleus, associates the Tcf/Lef family of transcriptional co-regulators, up-regulates oncogenic drivers such as c-Myc and Cyclin D1, and initiates tumorigenesis.1,3,4. Hydrodynamic tail vein injection (HDTVI) of Sleeping Beauty (SB) plasmids encoding a patient-derived 90 base pair in-frame N-terminal deletion of β-catenin [Δ(90)] and YAPS127A, a nuclearly localized YAP mutant, efficiently promotes HB tumorigenesis, whereas neither individual factor is tumorigenic.4
Different β-catenin mutants uniquely influence HB features.3 For example, mice with Δ(90)-driven HBs survive ∼11–13 weeks. Their tumors display the “crowded fetal” histology of the most common human HB subtype and differentially express ∼5300 transcripts relative to the liver.3,4 In contrast, tumors generated by the Δ(36-53) mutant grow slower, demonstrate greater histologic heterogeneity, and differentially express >6400 transcripts.3 The causes of these differences are complex and determined by each mutant’s stability, nuclear localization, and transcriptional potency.3
HBs are less genetically complex than other cancers.5,6 Nevertheless, ∼5%–10% of HBs also harbor recurrent missense mutations in the NFE2L2/NRF2 (NFE2L2) gene, and up to 50% have copy number increases.7 Similar changes occur in adult cancers and correlate with shorter survival.8 HBs with NFE2L2 mutations are associated with shorter survival, although how β-catenin heterogeneity affects this is unknown.7 Our profiling of 45 murine HBs generated by 8 patient-derived β-catenin mutants identified one NFE2L2 point mutation (L30P).3 These findings present largely anecdotal reckonings of NFE2L2’s role in HB, which cannot be further evaluated because of the small case numbers and β-catenin’s functional heterogeneity.
NFE2L2, a “Cap ’n’ Collar” bZIP transcription factor, mediates adaptations to oxidative, electrophilic, and xenobiotic stresses.9,10 NFE2L2 normally forms a cytoplasmic complex with Kelch-like ECH-associated protein 1 (KEAP1) via short NFE2L2 segments known as the ETGE and DLG domains. NFE2L2-KEAP1 complexes interact with Cullin 3, an E3 ubiquitin ligase that targets NFE2L2’s proteosomal degradation and ensures low basal expression.11 The aforementioned stresses prompt the oxidation of multiple KEAP1 cysteine thiol groups that maintain complex integrity.10,11 Dissociated and stabilized NFE2L2 thus translocates to the nucleus, heterodimerizes with members of the Maf family of bZIP transcription factors, and binds to antioxidant response elements in numerous target genes. Their products restore redox balance, metabolize xenobiotics, counter stress and apoptosis, regulate metabolic pathways, and maintain genomic and mitochondrial integrity.9, 10, 11, 12 Most NFE2L2 mutations reside within or near the ETGE or DLG domains and abrogate KEAP1 association, thereby leading to constitutive NFE2L2 nuclear translocation.12,13 Alternatively, copy number variants (CNVs) in NFE2L2 and KEAP1 create stoichiometric imbalance and allow nuclear accumulation of wild-type (WT) NFE2L2.
NFE2L2s suppress reactive oxygen species (ROS)-mediated and electrophile-mediated genotoxicity in cancer susceptible nfe2l2-/- mice.14 ROS-scavenging NFE2L2 target gene products include peroxiredoxins, thioredoxin, and thioredoxin reductase, whereas others detoxify and/or promote the excretion of α,β-unsaturated carbonyl-, epoxide-, and quinone-containing moieties.15,16 In contrast, NFE2L2 deregulation may facilitate tumor growth by increasing oxidative stress tolerance and allowing previously unattainable levels of oncogene-stimulated proliferation.17,18 NFE2L2 target gene products unrelated to redox regulation also benefit tumor growth, angiogenesis, and metastasis.19,20
We show here that 2 patient-derived NFE2L2 missense mutants, L30P and R34P, dramatically accelerate hepatic tumorigenesis by β-catenin mutants and YAPS127A. The tumors also possessed large necrotic areas and innumerable fluid-filled cysts, an otherwise rare HB feature.21 The exceedingly rapid growth of β-catenin+YAPS127A+L30P/R34P tumors was associated with a more robust antioxidant response. When co-expressed with either Δ(90) or YAPS127A individually, L30P and R34P were also transforming, thus establishing them as actual oncoproteins. HBs expressing each combination of Δ(90), YAPS127A, and L30P/R34P shared a “core” set of 22 similarly regulated transcripts whose expression correlated with long-term survival in HBs and other cancers. Although not affecting tumor growth or cyst formation when co-expressed with Δ(90) and YAPS127A, one transcript encoding the serine protease inhibitor serpin E1 generated highly necrotic tumors, thereby supporting the idea that these 22 common transcripts cooperatively contribute to different tumor phenotypes.
Results
NFE2L2 Mutants Accelerate Tumorigenesis
β-catenin mutations and WT β-catenin itself are tumorigenic when co-expressed with YAPS127A,2, 3, 4 with specific tumor features and transcriptomes determined by the β-catenin mutant’s identity.3 Despite these differences, endogenous NFE2L2 transcripts were up-regulated approximately 2-fold in all tumor groups relative to control livers (Figure 1A). Similar induction was observed in slower growing Δ(90)+YAPS127A tumors lacking Myc, ChREBP, or Myc+ChREBP.22 Like Myc, ChREBP is a transcription factor that regulates target genes involved in ribosomal biogenesis and carbohydrate and lipid metabolism.22 These results indicate that regardless of growth rates or other features, NFE2L2 is induced equally in murine HBs, whereas KEAP1 expression is unchanged (Figure 1B).
Figure 1.
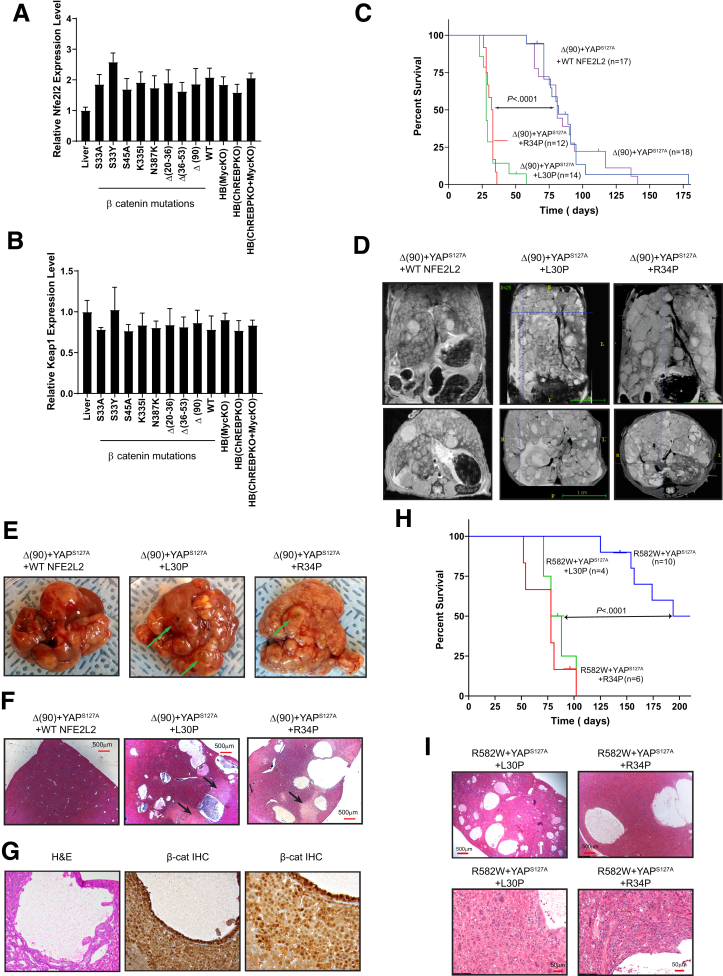
NFE2L2 mutants L30P and R34P accelerate HB growth. (A) NFE2L2 transcript levels were quantified from previously reported RNA-seq data obtained from murine HBs generated by the enforced hepatic overexpression of YAPS127A and 8 missense or in-frame deletion mutants of β-catenin or WT β-catenin.3 Transcripts were also quantified in Δ(90) β-catenin-generated HBs arising in myc-/-, chrebp-/-, and myc-/- x chrebp-/- hepatocyte backgrounds.22,25 All transcript levels are expressed relative to those in normal liver (n = 5 samples/group). (B) KEAP1 transcript levels in the tissues shown in (A). (C) Kaplan-Meier survival curves of the indicated cohorts, n = 10–12 mice/group. (D) Magnetic resonance images of comparably sized tumors just before death. (E) Gross appearance of typical tumors from each of the groups, with examples of typical fluid-filled cysts indicated by arrows. (F) H&E-stained sections of the tumors shown in (C) showing multiple cysts, with areas of prominent adjacent necrosis indicated by arrows. (G) Higher power magnification of H&E- and β-catenin immunohistochemistry-stained sections showing the lumens of cysts lined with cells resembling tumor cells that stain strongly for nuclearly localized β-catenin. (H) Kaplan-Meier survival curves of mice expressing β-catenin missense mutant R582W, YAPS127A, and the indicated NFE2L2 proteins. (I) H&E stained sections of the indicated tumors from (H).
NFE2L2 missense mutations occur in 5%–10% of HBs and at higher frequencies in other cancers.7,23,24 To test the impact of these on survival, we injected SB vectors encoding Δ(90)+YAPS127A with or without WT-NFE2L2, L30P, or R34P. As expected, Δ(90)+YAPS127A tumor-bearing mice had median survivals of ∼11–13 weeks (Figure 1C),4,25 which were unaltered by coinjection with WT-NFE2L2. In contrast, L30P and R34P co-expression significantly shortened survival (Figure 1C).
L30P/R34P-expressing tumors displayed the crowded fetal pattern of Δ(90)+YAPS127A HBs.4,25 In addition, they contained extensive areas of necrosis and innumerable fluid-filled cysts (Figure 1D–F). Cells lining the cysts were indistinguishable from tumor cells and stained intensely for nuclearly localized β-catenin but not for the endothelial marker CD34 (Figure 1G), thus confirming their derivation from epithelium (Figure 1G).26 Δ(90)+YAPS127A tumors contained neither cysts nor significant necrosis.
Together with YAPS127A, the R582W β-catenin mutant generates slowly growing tumors that more closely resemble hepatocellular carcinomas (HCCs).3 L30P and R34P also accelerated these tumors’ growth and generated numerous cysts (Figure 1H and I).
L30P and R34P Are Stabilized, Nuclearly Localized, and Alter Metabolic and Redox States
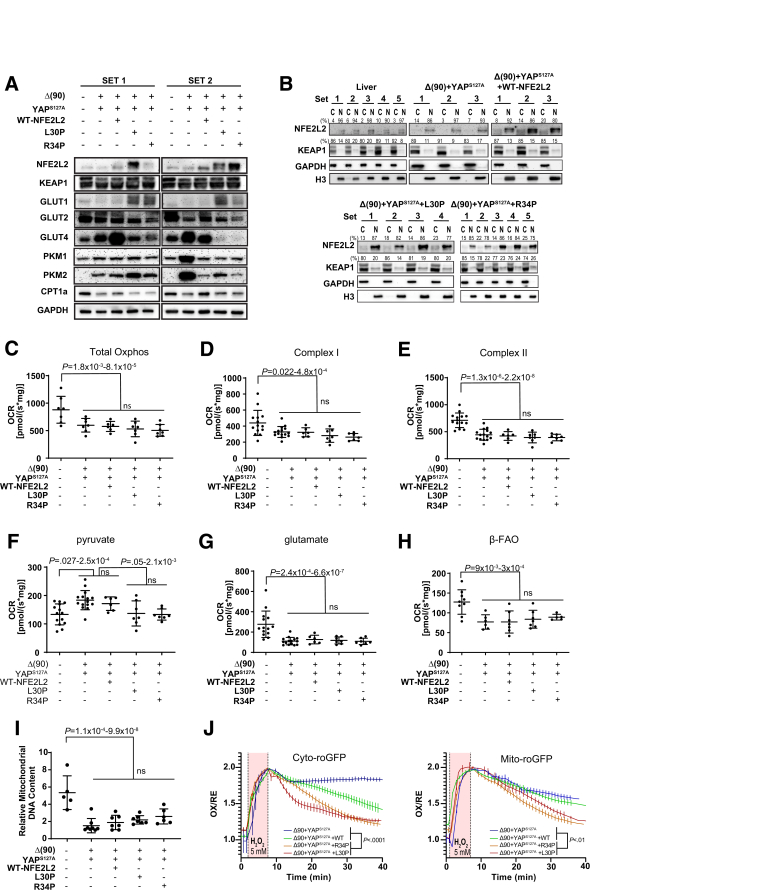
L30P and R34P were modestly overexpressed relative to WT or endogenous NFE2L2 protein in livers and Δ(90)+YAPS127A tumors, and KEAP1 was expressed equally across cohorts (Figure 2A, Figure 1B). Relative to control livers, tumor cohorts expressed distinct patterns of glucose transporters. For example, L30P/R34P tumors up-regulated ubiquitously expressed Glut1, consistent with their rapid growth and greater reliance on Warburg-type respiration (Figure 2A).3,22,27 They also tended to down-regulate liver-specific Glut2 and adipose- and muscle-specific Glut4. In contrast, Δ(90)+YAPS127A tumors and Δ(90)+YAPS127A+WT-NFE2L2 tumors both up-regulated Glut4 and modestly down-regulated Glut2. All tumors markedly up-regulated the M2 isoform of pyruvate kinase, which favors tumor growth because of its lower Km for phosphoenolpyruvate, leading to the accumulation of glycolytically derived anabolic substrates.28 Finally, all tumors expressed lower levels of carnitine palmitoyltransferase I, fatty acid beta oxidation's (β-FAO) rate-limiting enzyme.28 KEAP 1 was cytoplasmically localized (Figure 2B), whereas WT NFE2L2, despite being cytoplasmic in most cell lines, was nuclear in livers and HBs, as was Δ(90). WT-NFE2L2 nuclear localization may reflect the liver’s heavy engagement in xenobiotic metabolism.10,12
Figure 2.
Distribution and metabolic consequences of L30P/R34P expression in HBs. (A) Expression of NFE2L2, KEAP1, GLUT1, GLUT2, GLUT4, PKM-1, PKM-2, and Cpt1a in 2 representative sets of total lysates from the indicated tissues. (B) Nuclear (N)/cytoplasmic (C) fractionation of the indicated tissues, n = 3–5 samples/group. GAPDH and histone H3 (H3) immunoblots were performed as controls for protein loading and the purity of each fraction. Numbers above the NFE2L2 and KEAP1 panels indicate the fraction of protein associated with each compartment as determined by densitometric scanning of bands. (C) Total OCRs of mitochondria from the indicated tissues in the presence of malate, ADP, pyruvate, glutamate, and succinate. (D) Complex I responses, calculated after addition of rotenone to the reactions in (C) without succinate. (E) Complex II responses as determined from residual activity after addition of rotenone. (F) Responses to pyruvate. (G) Responses to glutamate. (H) β-FAO responses after addition of malate, L-carnitine, and palmitoyl-CoA. (I) Quantification of mitochondrial DNA (mtDNA) in representative tissues. TaqMan reactions amplified a segment of the mt D-loop region.3,25Each point represents the mean of triplicate TaqMan reactions after normalizing to a control TaqMan reaction for the ApoE nuclear gene. (J) In vitro recovery from oxidative stress. Monolayer cultures of the indicated tumor cells expressing cyto-roGFP or mito-roGFP were exposed to 5 mmol/L hydrogen peroxide (bar) while being monitored by live cell confocal microscope.
To assess L30P/R34P’s influence on metabolic functions, we examined mitochondrial oxygen consumption rates (OCRs) in response to tricarboxylic acid (TCA) cycle substrates. Consistent with the switch from oxidative phosphorylation (Oxphos) to Warburg respiration, total OCRs and individual Complex I and Complex II contributions were reduced across all tumor groups relative to livers (Figure 2C–E). Further consistent with previous observations,3,22,29 all tumors increased their pyruvate response while suppressing their glutamate response (Figure 2F and G).
HBs suppress β-FAO as they shift to glycolysis and hepatic lipid is mobilized for de novo membrane synthesis.3,22,29 Regardless of NFE2L2 status, all tumors also down-regulated β-FAO (Figure 2H), in keeping with lower carnitine palmitoyltransferase I expression (Figure 2A) and a reduction in mitochondrial DNA content as previously described (Figure 2I).22
We next generated tumors expressing redox-sensitive forms of cytoplasmic- or mitochondrial-localized green fluorescent protein (GFP) (cyto-roGFP and mito-roGFP, respectively).30 Tumor cells were cultured for several days and briefly exposed to H2O2 while monitoring recovery by live-confocal microscopy. Δ(90)+YAPS127+WT-NFE2L2 cells recovered somewhat more rapidly than Δ(90)+YAPS127A cells, but this was significantly accelerated in tumor cells expressing L30P/R34P (Figure 2J).
L30P/R34P Are Tumorigenic in Combination With Either Δ(90) or YAPS127A
Rapid tumorigenesis mediated by L30P/R34P (Figure 1C) raised questions regarding the latter’s role(s) in transformation. Although no individual factor was oncogenic (Figure 3A),4,22 L30P and R34P each generated tumors when co-expressed with either Δ(90) or YAPS127A, with survivals being comparable with those of the R582W+YAPS127A cohort (Figure 1H, Figure 3A).4 Thus, any pairwise combination of Δ(90), YAPS127A, and L30P/R34P is oncogenic. Δ(90)+L30P/R34P tumors were highly differentiated, whereas YAPS127A+L30P/R34P tumors resembled HCCs with HB-like features (Figure 3B).3,25
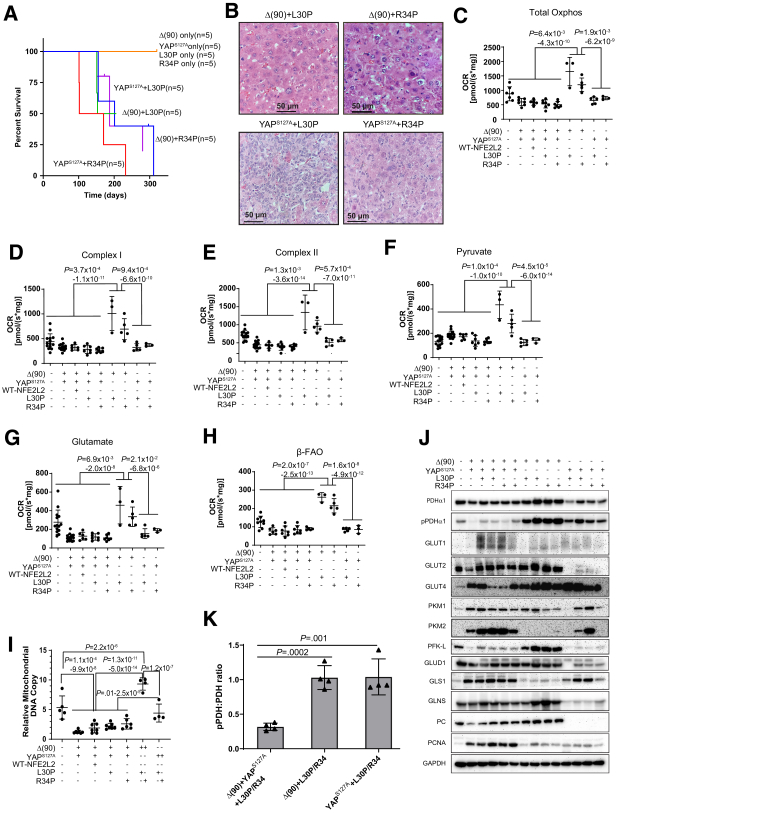
Figure 3.
Characteristics of tumors generated by L30P and R34P co-expressed with Δ(90) or YAPS127A. (A) Kaplan-Meier survival curves. Survival was determined as described in Figure 1C. (B) Histopathologic features of representative tumors from the indicated cohorts. (C–H) OCRs performed as described in Figure 2C–H. (C) Total Oxphos; (D) Complex I; (E) Complex II; (F) pyruvate response; (G) glutamate response; (H) β-FAO. (I) Mitochondrial DNA content of representative tumors from the indicated cohorts performed as described in Figure 2I. (J) Immunoblots from representative tissues of the indicated cohorts. GAPDH was used as a loading control. (K) Quantification of PDH and pPDH immunoblot results from (J).
Metabolically, YAPS127A+L30P/R34P tumors were similar to those previously described (Figure 3C–G),3,25 whereas Δ(90)+L30P/R34P tumors were distinct, with higher total OCRs and Complex I and II activities. Rather than the reciprocal relationship between β-FAO and pyruvate consumption and lower glutamate consumption documented previously (Figure 2F-H),3,25 all activities and mitochondrial DNA content were increased in Δ(90)+L30P/R34P and exceeded even those of livers (Figure 3F–I).
Components of the above pathways were again expressed in complex and cohort-specific ways. For example, pyruvate dehydrogenase α1 catalytic subunit (PDHα1) expression generally reflected each cohort’s mitochondrial DNA content (Figure 3J). However, Ser293-phosphorylated PDHα1 (pPDHα1) was lower in Δ(90)+YAPS127A tumors, irrespective of L30P/R34P status, indicating PDH activation and likely explaining the higher OCRs in response to pyruvate (Figure 3F).3,25,30 Most notable were the higher pPDHα1 levels in Δ(90)+L30P/R34P and YAPS127A+L30P/R34P tumors, whose relative levels of PDH inactivation were similar (Figure 3J and K).
Compared with Δ(90)+YAPS127A and Δ(90)+YAPS127A+L30P/R34P tumors, Δ(90)+L30P/R34P and YAPS127A+L30P/R34P tumors down-regulated Glut1 and up-regulated Glut4. However, most notable was the marked down-regulation of Glut2 expression by the latter cohort (Figure 2A, Figure 3J). Δ(90)+L30P/R34P and YAPS127A+L30P/R34P tumors also tended to down-regulate pyruvate kinase isozymes M1/M2 (PKM-2), suggesting that it was not necessarily associated with transformation but rather reflected rapid growth rates.
The higher pyruvate utilization and greater mitochondrial activity of Δ(90)+L30P/R34P tumors were further accompanied by increased expression of the rate-limiting liver-type phosphofructokinase (PFK-L) (Figure 3J). Together with reduced PKM-2 expression, this suggested that the more robust Oxphos of these tumors (Figure 3C–H) was associated with increased glycolytic flow into the TCA cycle rather than into anabolic pathways that support rapid growth. Pyruvate production and its use for Oxphos in these different cohorts were thus under complex but cooperative control by factors that coordinated glucose uptake and its fate.
The complex Oxphos dependencies were further underscored by the glutamate response, which was reduced in most tumor groups relative to liver but up-regulated in Δ(90)+L30P/R34P tumors (Figure 3G). Yet, levels of mitochondrial glutamate dehydrogenase 1, which catalyzes glutamate’s conversion to α-ketoglutarate, were unchanged, and those of mitochondrial glutaminase 1, which catalyzes glutamine conversion to glutamate, were actually reduced (Figure 3J). Levels of glutamine synthase, which converts cytoplasmic glutamate to glutamine, were somewhat higher, suggesting that intratumoral glutamine availability poses a barrier to more extensive glutaminolysis. The extremely low levels of glutamate dehydrogenase 1 in YAPS127A+L30P/R34P tumors, which might limit the supply of α-ketoglutarate, were accompanied by a parallel marked decline of pyruvate carboxylase (PC), which anaplerotically furnishes oxaloacetate from pyruvate. As already demonstrated, the expected increase in pyruvate availability was not associated with greater pyruvate utilization. Instead, pyruvate might now prove a more abundant source of α-ketoglutarate via alanine transaminase. Finally, proliferating cell nuclear antigen levels closely matched tumor growth rates and survival (Figure 1C, Figure 3A).
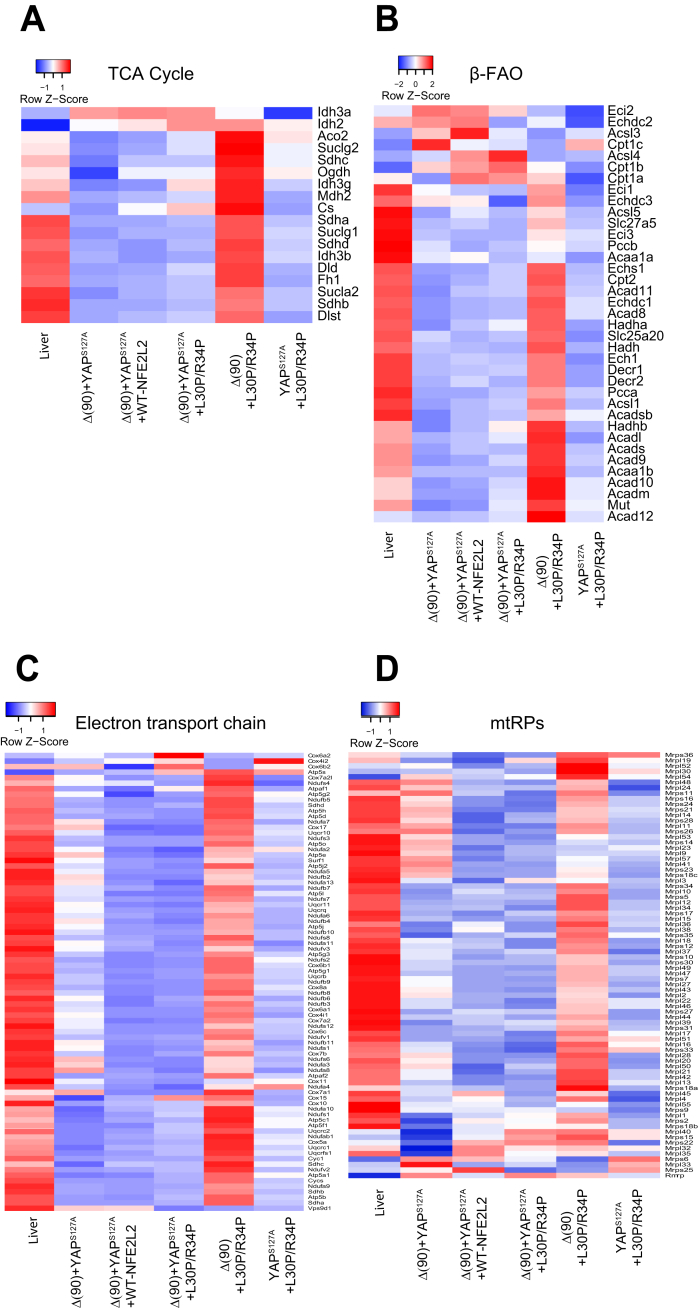
RNAseq analysis on the above cohorts demonstrated that most tumors reduced expression of transcripts encoding components of the TCA cycle, β-FAO, the ETC, and ribosomes (Figure 4). The sole exception was seen in Δ(90)+L30P/R34P HBs, which up-regulated these pathways as expected.
Figure 4.
Differential expression of transcripts involved in mitochondrial function. Control livers or tumors from each of the indicated cohorts (n = 5/group) were subjected to RNAseq and analyzed for expression of transcripts encoding proteins in the following pathways or structures. (A) TCA cycle, (B) β-FAO, (C) electron transport chain, (D) mitochondria ribosomal proteins (mtRPs).
RNAseq Reveals Common Transformation-Specific Transcripts
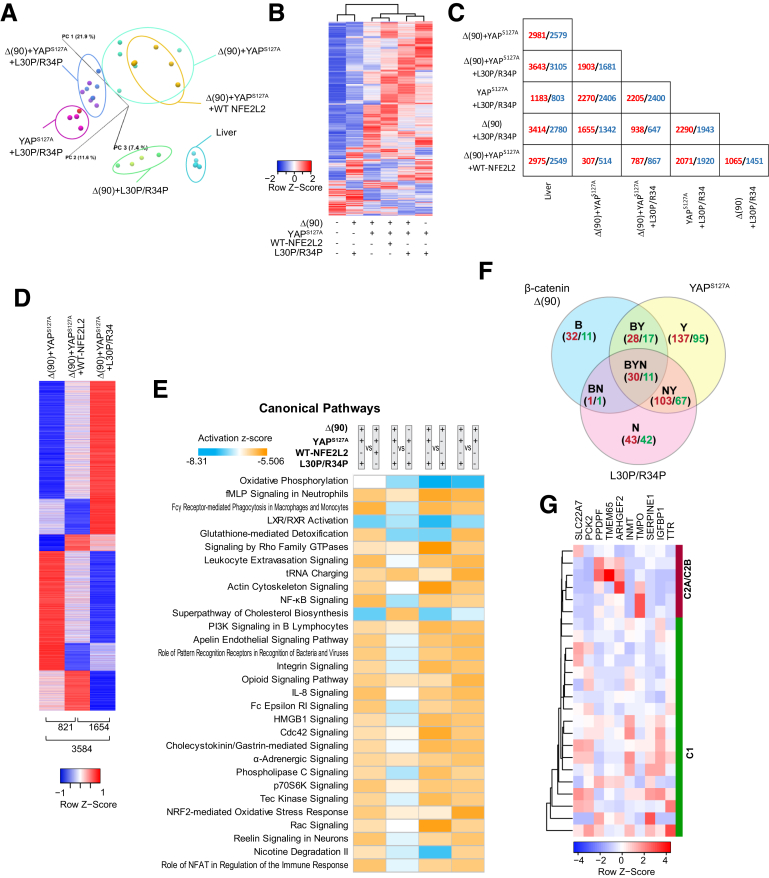
The finding that any 2 members of the Δ(90)+YAPS127A+L30P/R34P trio were oncogenic (Figure 1C, Figure 3A) suggested a model of tumorigenesis (and perhaps cystogenesis and necrosis) involving common transcripts that we sought to identify by RNAseq. Principal component analysis and hierarchical clustering readily distinguished tumor cohorts from liver (Figure 5A and B). Because Δ(90)+YAPS127A+L30P and Δ(90)+YAPS127A+R34P tumors differed in the expression of only one transcript, they were subsequently combined. All other cohorts were distinguishable (Figure 5B and C, Figure 4B and C). Δ(90)+YAPS127A tumors and Δ(90)+YAPS127A+WT-NFE2L2 tumors differed in the expression of 821 genes (Figure 5C and D), indicating that WT-NFE2L2 was not entirely silent as suspected from its nuclear localization and modest redox buffering (Figure 2B and J). Δ(90)+YAPS127A+L30P/R34P tumors showed more extensive transcriptional dysregulation, differing from Δ(90)+YAPS127A and Δ(90)+YAPS127A+WT-NFE2L2 tumors by 3584 transcripts and 1654 transcripts, respectively (Figure 5D).
Figure 5.
RNAseq analysis of tumors generated by combinations of Δ(90), YAPS127A, WT-NFE2L2, L30P, and R34P. (A) Principal components analysis of transcriptomic profiles of livers and tumors (n = 4–5 samples/group. (B) Heat maps of differentially expressed transcripts from the tissues depicted in (A) arranged by hierarchical clustering. Because only a single transcript difference was found between tumors expressing L30P and R34P, results were combined for this and subsequent analyses (L30P/R34P). (C) Pairwise comparisons showing the number of significant gene expression differences between any 2 of the tissues depicted in (B). Red and blue, up-regulated and down-regulated, respectively, in the tumors depicted at the left relative to those indicated at the bottom. (D) Distinct transcript patterns of Δ(90)+YAPS127A, Δ(90)+YAPS127A+WT-NFE2L2, and Δ(90)+YAPS127A+L30P/R34P tumor cohorts. Numbers at bottom of each column indicate the significant expression differences between each pairwise comparison. (E) Top IPA pathways among different tumor groups, expressed as z-scores. Orange, up-regulated; blue, down-regulated. (F) Shared gene expression subsets between and among the indicated cohorts. Red and green, number of transcripts up-regulated and down-regulated, respectively, relative to liver. (G) Hierarchical clustering of C1 and C2A/C2B subsets31,32 of human HBs using the 10 “BYN” transcripts from Table 2 that were dysregulated in human tumors.
The 821 gene expression differences between the Δ(90)+YAPS127A and Δ(90)+YAPS127A+WT-NFE2L2 cohorts were categorized by using Ingenuity Pathway Analysis (IPA). Among the 4 pathways with the most disparate z-scores (z> +2.8), only one (NRF2-mediated oxidative stress response) was considered NFE2L2-responsive (Figure 5E, Table 1, Supplementary Table 1). In contrast, the 3584 differences between the Δ(90)+YAPS127A and Δ(90)+YAPS127A+L30P/R34P cohorts involved 12 similarly dysregulated pathways, with 3 involving redox homeostasis. Finally, the 1654 differences between Δ(90)+YAPS127A+WT-NFE2L2 and Δ(90)+YAPS127A+L30P/R34P tumors showed up-regulation of the NRF2-mediated oxidative stress response and glutathione redox reactions I pathways in the latter. L30P/R34P thus dysregulated more redox target genes than WT-NFE2L2.
Table 1.
Top IPA Pathways Deregulated in HBs Expressing WT-NFE2L2 vs L30P/R34P
| Name of IPA pathway | Δ90+YAPS127A +L30P/R34P vs Δ90+YAPS127A Z-score | Name of IPA pathway | Δ90+YAPS127A +WT-NFE2L2 vs Δ90+YAPS127A Z-score | Name of IPA pathway | Δ90+YAPS127A +L30P/R34P vs Δ90+YAPS127A +WT-NFE2L2 Z-score |
|---|---|---|---|---|---|
| NRF2-mediated oxidative stress responsea | 3.888 | Oxidative phosphorylation | –4.359 | Fcγ receptor-mediated phagocytosis in macrophages and monocytes | 3.71 |
| tRNA charging | 3.771 | EIF2 signaling | –3.128 | NF-κB signaling | 3.266 |
| Leukocyte extravasation signaling | 3.43 | Sirtuin signaling pathway | 2.985 | Leukocyte extravasation signaling | 3.157 |
| Glutathione-mediated detoxificationa | 3.317 | NRF2-mediated oxidative stress responsea | 2.828 | IL-8 signaling | 3 |
| Role of pattern recognition receptors in recognition of bacteria and viruses | 3 | Superpathway of cholesterol biosynthesis | –3 | ||
| Ephrin receptor signaling | 3 | Role of NFAT in regulation of the immune response | 2.858 | ||
| RAN signaling | 3 | Reelin signaling in neurons | 2.84 | ||
| Apelin endothelial signaling pathway | 2.985 | LXR/RXR activation | –2.828 | ||
| Fcγ receptor-mediated phagocytosis in macrophages and monocytes | 2.982 | fMLP signaling in neutrophils | 2.673 | ||
| Opioid signaling pathway | 2.92 | Fc epsilon RI signaling | 2.668 | ||
| fMLP signaling in neutrophils | 2.828 | NRF2-mediated oxidative stress responsea | 2.558 | ||
| Glutathione redox reactions Ia | 2.828 | Apelin endothelial signaling pathway | 2.53 | ||
| TREM1 signaling | 2.53 | ||||
| ERK5 signaling | 2.53 | ||||
| Role of pattern recognition receptors in recognition of bacteria and viruses | 2.496 | ||||
| NER pathway | 2.449 | ||||
| Glutathione redox reactions Ia | 2.449 |
Indicates redox-regulated pathway.
ChIP-seq results from human HepG2 HB cells (https://www.encodeproject.org/data-standards/chip-seq/) indicated that 4.7%–5.5% of the transcripts in Figure 5D were orthologs of NFE2L2 direct target genes. Including data from 3 additional cell lines (A549, HeLa-S3, and IMR90) increased this to 13.8%–22.2%. IPA profiling identified additional variably deregulated pathways among the tumor cohorts, with the most prominent ones pertaining to Oxphos, mitochondrial dysfunction, cholesterol/bile acid synthesis, and cell signaling (Figure 5E).
Forty-one transcripts were deregulated in all tumors, with 29 always being deregulated in the same direction (Figure 5F). Twenty-two members of this core “BYN” subset were similarly deregulated in HBs driven by other β-catenin mutants and in slowly growing Δ(90)+YAPS127A HBs arising in myc-/-, chrebp-/-, and myc-/-+chrebp-/- hepatocytes (Table 2).3,22,25 Δ(90)+YAPS127A+L30P/R34P HBs also deregulated these transcripts more than occurred with any other pairwise combination of factors. Fourteen transcripts were also dysregulated in Δ(90)+YAPS127A+WT-NFE2L2 tumors, suggesting that they were less likely to be involved in the growth-accelerating and cystogenic properties of L30P/R34P. Together, these considerations indicated that the remaining 8 coordinately deregulated transcripts contributed to L30P/R34P’s cooperation with Δ(90) or YAPS127A. On average, these 8 transcripts were deregulated by 47.5-fold in the Δ(90)+YAPS127A+L30P/R34P cohort versus 13-fold in the next highest expressing cohort (P < .001).
Table 2.
Gene Responsiveness to the Indicated Combinations of Δ(90), YAPS127A, and L30P/R34P
| Name of gene | Mouse gene ID | Human gene ID | Expression in HBs: fold change vs liver |
Association with shortened survival in human cancersa, b, c | |||
|---|---|---|---|---|---|---|---|
| Δ(90)+ YAPS127A | Δ(90)+ L30P/R3P | YAPS127A+ L30P/R3P | Δ(90)+ YAPS127A+ L30P/R3P | ||||
| Slc22a7 | 108114 | 10864 | –25.69 | –7.41 | –13.99 | –109.62 | KIRC,b LIHCb |
| Inmt | 21743 | 11185 | –10.55 | –5.52 | –3.85 | –31.22 | LIHCb |
| Gas1 | 14451 | 2619 | –6.98 | –3.76 | –6.84 | –22.76 | THCA,a KIRC,a KIRP,a KICH,a COAD,a READ,a STADa |
| Atp5kd | 11958 | 521 | –2.28 | –1.89 | –5.73 | –11.40 | |
| Ttrd | 22139 | 7276 | –5.35 | –2.71 | –3.31 | –9.42 | |
| Cmtm8 | 70031 | 152189 | –2.67 | –1.95 | –1.98 | –6.14 | KIRC,b KIRPb |
| Ppdpfd | 66496 | 79144 | –2.16 | –1.89 | –2.51 | –3.74 | KIRP,b KICHb |
| Camkk2d | 207565 | 10645 | –1.69 | –1.66 | –1.79 | –3.51 | |
| Tmem65d | 74868 | 157378 | 2.08 | 1.82 | 1.99 | 2.95 | UCECa |
| Tpm1d | 22003 | 7168 | 2.69 | 2.05 | 2.52 | 3.50 | BLCAa |
| Tgfad | 21802 | 7039 | 2.12 | 2.33 | 2.62 | 3.98 | PAAD,a THCAa |
| Tmpod | 21917 | 7112 | 2.91 | 1.67 | 2.71 | 4.25 | KIRP,a KICHa |
| Garsd | 353172 | 2617 | 2.86 | 1.68 | 2.01 | 4.57 | KIRC,a KIRP,a KICH,a BLCA,a LIHC,a BRCAa |
| Rock2d | 19878 | 9475 | 2.77 | 2.83 | 3.08 | 5.22 | KIRC,b KIRP,a KICHa |
| Il22ra1d | 230828 | 58985 | 3.15 | 3.80 | 3.40 | 6.58 | |
| Arhgef2d | 16800 | 9181 | 4.93 | 1.86 | 4.45 | 8.72 | KIRC,a KICH,a LIHCa |
| Acot9d | 56360 | 23597 | 8.89 | 2.39 | 7.67 | 13.02 | LIHCa |
| Pck2 | 74551 | 5106 | 6.95 | 2.22 | 6.22 | 15.99 | LIHCa |
| Lgals3 | 16854 | 3958 | 7.06 | 2.34 | 8.42 | 18.40 | |
| Uap1l1d | 227620 | 91373 | 15.51 | 6.65 | 16.40 | 23.67 | LIHC,a PAADb |
| Serpine1 | 18787 | 5054 | 7.35 | 9.25 | 11.86 | 43.70 | KIRC,a KIRP,a KICH,a LIHC,a STAD,a HNSCa |
| Igfbp1 | 16006 | 3484 | 58.42 | 12.80 | 31.41 | 132.32 | KIRC,a LUAD,a STADa |
NOTE. The 22 transcripts listed here are a subset of the original 41 “BYN” groups depicted in Figure 5F that remained after eliminating those that were not regulated in the same direction in each case or that were not also deregulated in tumors driven by other types of β-catenin mutations or in tumors arising from myc-/- and/or chrebp-/- hepatocytes.22
Up-regulated.
Down-regulated.
See Figure 6 for Kaplan-Meier survival curves.
Also deregulated by Δ(90)+YAPS127A+WT-NFE2L2 tumors.
Previous analyses of 24 HBs by DNA microarrays and another set of 25 by RNAseq had identified 16 and 4 unrelated transcripts, respectively, whose expression correlated with survival.31,32 In the former, tumors associated with favorable, poor, and intermediate survival were designated C1, C2A, and C2B, respectively. Ten transcripts from Table 2 were differentially expressed by the C1 and C2A/C2B groups (q < 0.05), with hierarchical clustering perfectly segregating the C1 and C2A/C2B groups (Figure 5G). No transcripts in Table 2 were included in the previous panels.31,32
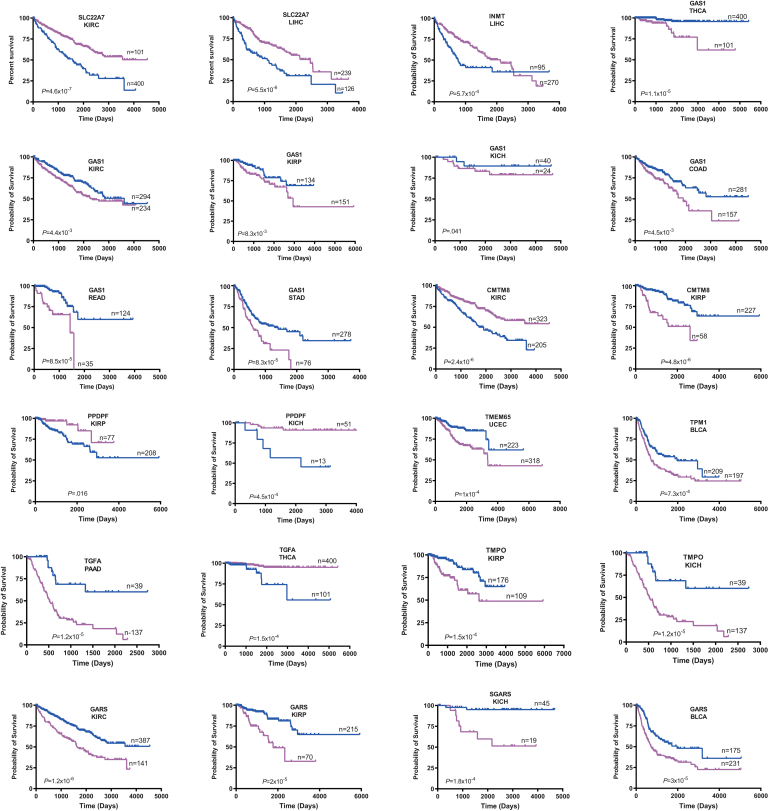
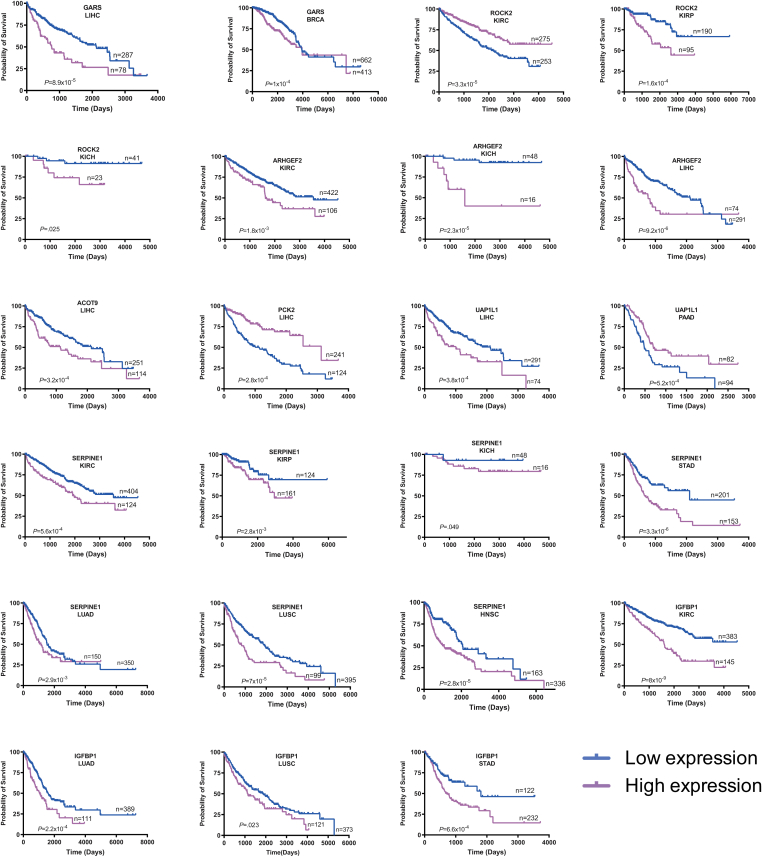
Seventeen of the above 22 transcripts also correlated with survival in 14 cancer types from The Cancer Genome Atlas (TCGA) (Figure 6). Recurrent associations were seen with the 3 most common kidney cancers and HCC, and some transcripts correlated with survival in multiple cancers. Of the 45 instances where expression correlated with survival, the direction of change relative to that of matched normal tissues was discordant in only 1 case and partially discordant in 2 others (Table 2).
Figure 6.
Correlation between expression levels of transcripts listed inTable 2and survival in select human cancers. Each depicted tumor type was divided into 2 groups displaying the highest and lowest expression of the indicated transcript. Standard Kaplan-Meir survival curves for each group were then generated, and P values were determined by a standard rank test.
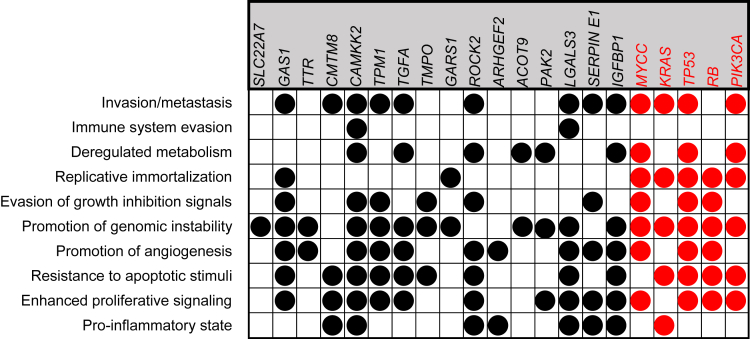
The Cancer Hallmarks Analytics Tool (http://chat.lionproject.net/) showed associations of the above 22 transcripts with known cancer-promoting/enabling features.33,34 Sixteen transcripts were associated with 1 or more hallmarks, and 8 were associated with 5–9 (Figure 7). By comparison, the associations of 5 well-known oncogenes and tumor suppressors with broad human cancer associations ranged from 5 to 8. Therefore, BYN transcript levels frequently correlate with the properties, behaviors, and lethality of multiple cancers, including HBs.
Figure 7.
Association of relevant “BYN” transcripts with The Hallmarks of Cancer. Each gene was queried in The Cancer Hallmarks Analytics Tool website (http://chat.lionproject.net/) to identify the hallmarks that have been previously associated with the gene’s mutation or dysregulation.33 Those shown in red were included as examples of genes that have been broadly implicated in the causation of multiple different mammalian cancers.
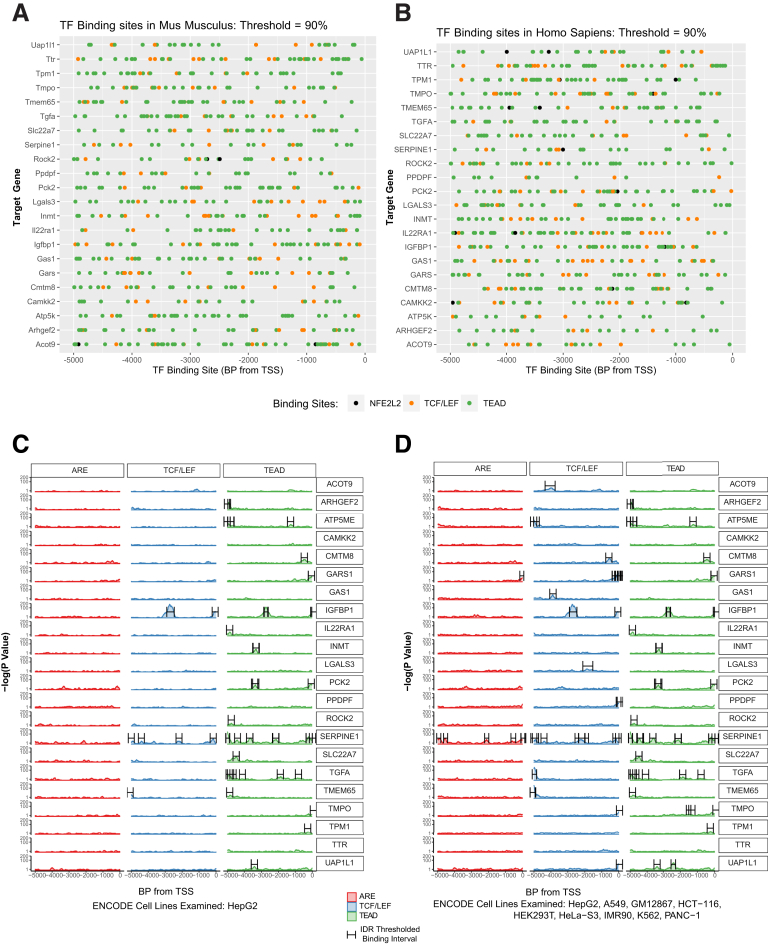
The promoters of all 22 BYN genes, and their human counterparts, contained multiple computationally-identified consensus binding sites for β-catenin/Tcf/Lef, YAP/TAZ (TEAD sites) and NFE2L2/Maf (ARE sites) (Figure 8A and B). However, using data from HepG2 HB cells and eight additional human cell lines in the ENCODE v.5 CHIP-seq data base, many fewer sites were identified. 20 genes contain at least one bona fide binding site for b-catenin/Tcf/Lef or YAP/TAZ but only two genes contained documented sites for NFE2L2/Maf (Figure 8C and D). Thus, many of the BYN genes are either indirect NFE2L2 targets or harbor biding sites elsewhere.
Figure 8.
In silico promoter analysis. Five kb of upstream promoter sequence for each of the genes listed in Table 2 were screened for the presence of consensus elements for Tcf/Lef, TEAD, and ARE binding sites. (A) Murine genes; (B) human genes. (C) True binding sites based on ChIP results for Tcf/Lef, TEAD, and ARE obtained from human HepG2 HB cells in the Encode v.5 database (https://www.encodeproject.org/datastandards/chip-seq/) were mapped to the promoter sequences from (B). (D) Binding sites for Tcf/Lef, TEAD, and ARE after the evaluation of Chip-Seq data from 8 other human cell lines (see Materials and Methods).
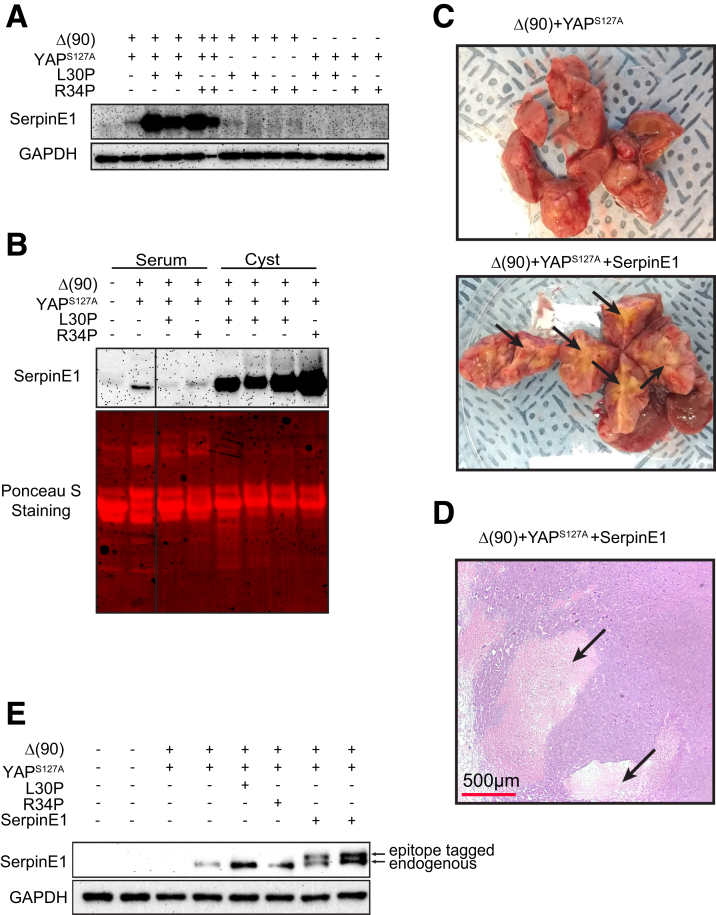
The serpine1 gene promoter contained the largest number of binding sites for all 3 transcription factors (Figure 8D). Serpin E1 protein levels were particularly high in Δ(90)+YAPS127A+L30P/R34P tumors (Figure 9A) and agreed with RNAseq results (Table 2). Cyst fluid and, to a lesser extent, plasma from tumor-bearing mice also tended to contain high levels of serpin E1, whereas plasma from mice bearing Δ(90)+YAPS127A tumors contained low-undetectable levels (Figure 9B).
Figure 9.
Serpin E1 deregulation recapitulates the extensive necrosis associated with L30P/R34P overexpression. (A) Serpin E1 levels in the indicated tumors. (B) Serpin E1 levels in plasma and cyst fluid from the indicated cohorts. Plasma and cyst fluids were diluted proportionately, and 40 μg of each was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis. To account for differences in the types of samples present, Ponceau acid red staining of the membrane was used to confirm protein concentrations. (C) Gross appearance of Δ(90)+YAPS127A+serpin E1 tumor sections showing extensive necrosis (arrows) relative to that of a typical Δ(90)+YAPS127A control. (D) Histologic appearance of typical section from the tumor shown in (C) with large pericystic necrotic areas indicated by arrows. (E) Immunoblots for serpin E1 expression in indicated tumor types. Note slower mobility of exogenously expressed, epitope-tagged serpin E1.
Serpin E1 co-expression by HDTVI neither affected the survival of Δ(90)+YAPS127A tumor-bearing mice nor promoted cysts (Figure 1C). However, the tumors were necrotic and expressed serpin E1 (Figure 9C–E). Serpin E1 deregulation thus recapitulated the extensive necrosis associated with L30P/R34P overexpression but neither the accelerated growth nor cystogenesis.
NFE2L2 Mutations, CNVs, and Overexpression in HBs and Other Cancers
Our RNAseq analyses of 45 murine tumors generated by 9 β-catenin variants identified only L30P, which was previously described in human HBs.3 We therefore queried RNAseq or exome-seq data from 194 previously reported primary HBs and cell lines (https://cancer.sanger.ac.uk/cosmic)7,32,35 and identified 9 NFE2L2 missense mutations (frequency = 3.5%) (Table 3).
Table 3.
Previously Identified NFE2L2 Point Mutations in Primary Human HBs and HB Cell Lines
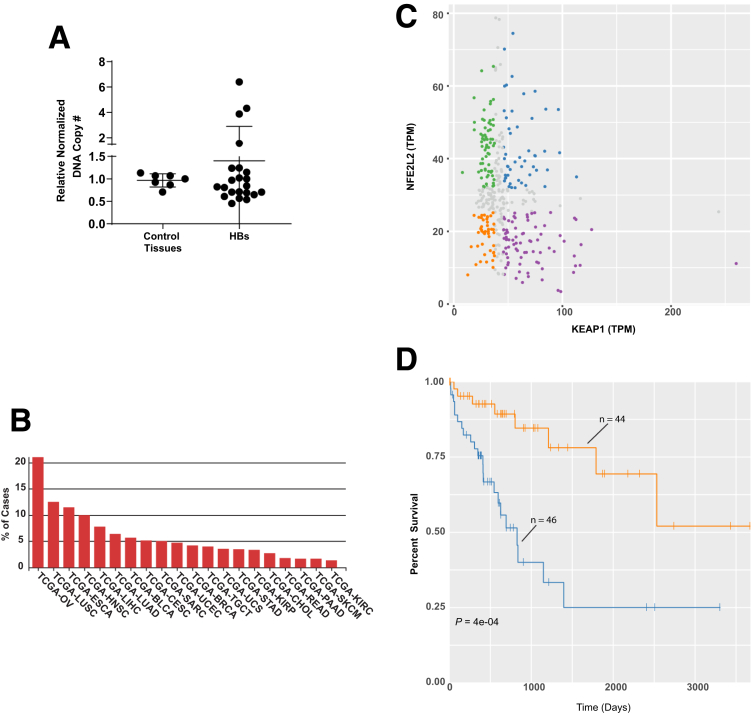
In contrast to missense mutations, up to 50% of HBs harbor NFE2L2 CNVs.35 We quantified NFE2L2 gene copy numbers in 22 primary HBs from our own institution and identified 3 (13.6%) with 4.0-fold to 6.4-fold increases in copy number (Figure 10A). As many as 10%-20% of other human cancers also showed NFE2L2 gene amplification (https://www.mycancergenome.org/content/gene/nfe2l2/) (Figure 10B). However, an association between low levels of NFE2L2 and KEAP1 transcripts and favorable survival was seen only in HCCs (Figure 10C and D).
Figure 10.
CNVs in HB and select human tumors. (A) NFE2L2 CNVs determined from FFPE samples of 7 normal tissues and 22 primary HBs. TaqMan-based assay was used to quantify the copy number ratios of NFE2L2 versus 2 control genes (GAPDH and RPPH1). Each reaction was performed in triplicate, and each point represents the calculated mean of the empirically determined CNVs relative to those of the 2 control genes. (B) Primary human tumors with the highest frequency of NFE2L2 gene amplification. Data were obtained from the Genomic Data Commons site in TCGA. (C) Scatterplots of NFE2L2 and KEAP1 transcript expression in 371 HCC samples from the TCGA PANCAN data set. Sets are divided into quadrants that indicate those samples with the highest and lowest expression of NFE2L2 and KEAP1. (D) Kaplan-Meier survival of individuals from (C) whose tumors contained the highest and lowest levels of NFE2L2 and KEAP1 expression.
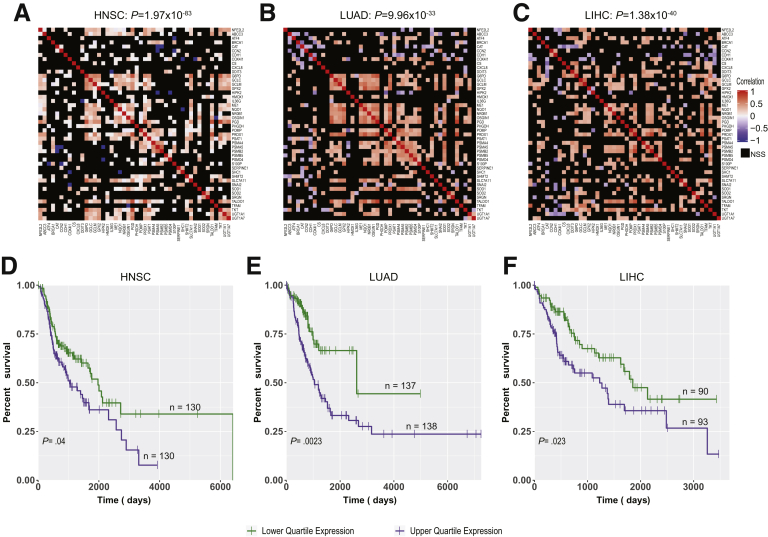
Transcript and protein levels often correlate poorly, particularly in cancer.36 This is especially true for NFE2L2, whose activation occurs post-translationally.10,11,23 Therefore, evaluating survival on the basis of transcript levels (Figure 10C and D) might not fully capture the magnitude of NFE2L2’s influence. A more meaningful way to identify NFE2L2 deregulation that reflects the protein’s redox-dependent function might be to survey tumors for NFE2L2 target gene expression. We did this for 45 direct targets from the IPA Knowledge Base (Table 4) in 3 cancer types from TCGA that preliminary analysis indicated as having high incidences of NFE2L2 CNV or mutation. NFE2L2 transcript levels in these tumors were similar to those in matched normal tissues and did not correlate with target gene transcript levels. However, as a group these transcripts were significantly up-regulated (1.36-fold to 1.63-fold) (Figure 11A–C), and the survival of individuals with tumors expressing the highest levels was significantly shorter (Figure 3, Figure 11D–F). This suggests that NFE2L2 target gene products suppress ROS and other electrophiles that compromise tumor aggressiveness.8,19,35
Table 4.
Forty-six NFE2L2 Target Gene Transcripts From IPA
| Symbol | Synonym(s) | Entrez gene name | Location | Family | Entrez gene ID |
||||
|---|---|---|---|---|---|---|---|---|---|
| Human | Mouse | Rat | |||||||
| ABCC3 | 1700019L09Rik, ABC31, ATP binding cassette subfamily C member 3, ATP-binding cassette C3, ATP-binding cassette, sub-family C (CFTR/MRP), member 3, cMOAT2, EST90757, MLP2, MOAT-D, MRP3, Multidrug Resistant Protein 3 | ATP binding cassette subfamily C member 3 | Plasma membrane | transporter | 8714 | 76408 | 140668 | ||
| ATF4 | activating transcription factor 4, C/ATF, CREB-2, TAXREB67, TXREB | Activating transcription factor 4 | Nucleus | transcription regulator | 468 | 11911 | 79255 | ||
| BRCA1 | BRCA1 DNA repair associated, BRCA1, DNA repair associated, BRCAI, BRCC1, breast cancer 1, early onset, BROVCA1, FANCS, PNCA4, PPP1R53, PSCP, RNF53 | BRCA1 DNA repair associated | Nucleus | transcription regulator | 672 | 12189 | 497672 | ||
| CAT | 2210418N07, ACATALASIA, Cas-1, Cat01, Catalase, Catalase1, Catl, CS1 | Catalase | Cytoplasm | enzyme | 847 | 12359 | 24248 | ||
| CCN2 | AMPHIROGULIN, cellular communication network factor 1, cellular communication network factor 2, CTGF, CTGF isoform 1, CTGRP, Fibroblast-inducible secreted, Fisp12, HCS24, IGFBP8, IGFBP-RP2, NOV2, Tissue growth factor | Cellular communication network factor 2 | Extracellular space | growth factor | 1490 | 14219 | 64032 | ||
| CDH1 | AA960649, ARC-1, BCDS1, cadherin 1, Cadherin E, CD324, CDHE, CSEIL, E-cadherin, ECAD, L-CAM, Um, UVO, uvomorulin | Cadherin 1 | Plasma membrane | other | 999 | 12550 | 83502 | ||
| COX4I1 | AL024441, CcO IVi1, COX, COX IV-1, COX4, COX4-1, COX4A, COX4I, COXIV, cytochrome c oxidase subunit 4I1, IV-1 | Cytochrome c oxidase subunit 4I1 | Cytoplasm | enzyme | 1327 | 12857 | 29445 | ||
| CS | 2610511A05Rik, 9030605P22Rik, Ahl4, BB234005, Cis, citrate synthase | Citrate synthase | Cytoplasm | enzyme | 1431 | 12974 | 170587 | ||
| CXCL8 | C-X-C motif chemokine ligand 8, GCP-1, IL8, LECT, LUCT, LYNAP, MDNCF, MONAP, Monocyte-derived neutrophil chemotactic factor, NAF, NAP-1 | C-X-C motif chemokine ligand 8 | Extracellular space | cytokine | 3576 | ||||
| DDIT3 | AC144852.1, AltDDIT3, C/EBP homology, C/EBP-homologous, C/EBPzeta, Cebp Zeta, Cebp ζ, CEBPZ, CHOP, CHOP-10, DNA DAMAGE-INDUCIBLE transcript, DNA-damage inducible transcript 3, GADD153, RM4 | DNA damage inducible transcript 3 | Nucleus | transcription regulator | 1649 | 13198 | 29467 | ||
| G6PD | G28A, G6PD1, G6PDX, Glucose-6-P Dehydrogenase, glucose-6-phosphate dehydrogenase, glucose-6-phosphate dehydrogenase X-linked, Gpdx | Glucose-6-phosphate dehydrogenase | Cytoplasm | enzyme | 2539 | 14381 | 24377 | ||
| GCLC | D9Wsu168e, gamma GCS HEAVY CHAIN, Gamma Glutamyl Cysteine Synthetase Light Subunit, Gamma Glutamylcysteine Synthetase, Gamma glutamylcysteine synthetase heavy subunit, GCL, GCS, GCS, Catalytic, GCS-HS, Ggcs-hs, GLCL, GLCL-H, GLCLC, glutamate-cysteine ligase catalytic subunit, Glutamate-Cysteine Ligase, Catalytic Subunit, γ Gcs, γ GCS HEAVY CHAIN, γ Glutamyl Cysteine Synthetase Light Subunit, γ Glutamylcysteine Synthetase, γ glutamylcysteine synthetase heavy subunit, γ-Gcsh | Glutamate-cysteine ligase catalytic subunit | Cytoplasm | enzyme | 2729 | 14629 | 25283 | ||
| GCLM | AI649393, Gamma gclm, gamma GCS LIGHT CHAIN, Gamma glutamylcysteine synthase (regulatory), gamma GLUTAMYLCYSTEINE SYNTHETASE, gamma-glutamylcysteine synthetase light (regulatory) subunit, Gcmc, Gcs, Regulatory, Gcs-ls, GLCLR, glutamat-cystein ligase, regulatory subunit, glutamate-cysteine ligase modifier subunit, Glutamate-Cysteine Ligase, Modifier Subunit, γ gclm, γ GCS LIGHT CHAIN, γ glutamylcysteine synthase (regulatory), γ GLUTAMYLCYSTEINE SYNTHETASE, γ-glutamylcysteine synthetase light (regulatory) subunit | Glutamate-cysteine ligase modifier subunit | Cytoplasm | enzyme | 2730 | 14630 | 29739 | ||
| GPX2 | GI-GPx, glutathione peroxidase 2, GPRP, GPRP-2, GPX-GI, GSHPx-2, GSHPX-GI | Glutathione peroxidase 2 | Cytoplasm | enzyme | 2877 | 14776 | 29326 | ||
| HIPK2 | 1110014O20RIK, B230339E18RIK, homeodomain interacting protein kinase 2, LOC100505582, LOC653052, PRO0593, Stank | Homeodomain interacting protein kinase 2 | Nucleus | kinase | 28996 | 15258 | 362342 | ||
| HMOX1 | bK286B10, D8Wsu38e, haemox, HEME OXYGENASE, HEME OXYGENASE (DECYCLIZING) 1, Heme oxygenase 1, Hemox, Heox, HEOXG, Hmox, HMOX1D, HO-1, HSP32 | Heme oxygenase 1 | Cytoplasm | enzyme | 3162 | 15368 | 24451 | ||
| IL36G | IL-1F9, IL-1H1, IL-1RP2, IL1E, interleukin 1 family, member 9, interleukin 36 gamma, interleukin 36 γ, interleukin 36, gamma, interleukin 36, γ, RGD1563019 | Interleukin 36 gamma | Extracellular space | cytokine | 56300 | 215257 | 499744 | ||
| ME1 | BRCAME, D9Ertd267e, HUMNDME, Malate Nadp Oxyreductase, Malic enzyme, malic enzyme 1, malic enzyme 1, NADP(+)-dependent, cytosolic, Mdh-1, MES, MOD1 | Malic enzyme 1 | Cytoplasm | enzyme | 4199 | 17436 | 24552 | ||
| NQO1 | AV001255, DHQU, DIA4, DT-diaphorase, DTD, NAD DT-diaphorase, NAD(P)H dehydrogenase, quinone 1, NAD(P)H quinone dehydrogenase 1, NAD(P)H:quinone oxidoreductase, Nadph dehydrogenase, Nadph diaphorase, Nadph Quinone Oxidoreductase-1, NMO1, NMOR, NMOR1, NMORI, Nqo, Ox-1, Qr, QR1, Quinone reductase | NAD(P)H quinone dehydrogenase 1 | Cytoplasm | enzyme | 1728 | 18104 | 24314 | ||
| NR0B1 | AHC, AHCH, AHX, DAX-1, DSS, GTD, HHG, NROB1, nuclear receptor subfamily 0 group B member 1, nuclear receptor subfamily 0, group B, member 1, SRXY2 | Nuclear receptor subfamily 0 group B member 1 | Nucleus | ligand-dependent nuclear receptor | 190 | 11614 | 58850 | ||
| OSGIN1 | 1700012B18Rik, BDGI, OKL38, oxidative stress induced growth inhibitor 1 | Oxidative stress induced growth inhibitor 1 | Other | growth factor | 29948 | 71839 | 171493 | ||
| PGD | 0610042A05Rik, 6PGD, 6PGDH, AU019875, C78335, Cc2-27, LOC100363662, phosphogluconate dehydrogenase | Phosphogluconate dehydrogenase | Cytoplasm | enzyme | 5226 | 110208 | 1E+08 | ||
| PHGDH | 3-PGDH, 3-phosphoglycerate dehydrogenase, 4930479N23, A10, HEL-S-113, NLS, NLS1, PDG, PGAD, PGD, PGDH, PGDH3, PHGDHD, phosphoglycerate dehydrogenase, SERA | Phosphoglycerate dehydrogenase | Cytoplasm | enzyme | 26227 | 236539 | 58835 | ||
| POMP | 2510048O06Rik, C13orf12, HSPC014, LOC100911238, PNAS-110, PRAAS2, proteasome maturation protein, proteasome maturation protein-like, RGD1305831, UMP1 | Proteasome maturation protein | Nucleus | other | 51371 | 66537 | 288455 100911238 |
||
| PRDX1 | ENHANCER, Enhancer protein, Hbp23, MSP23, NKEF-A, OSF-3, PAG, PAGA, PAGB, PEROXIREDOXIN 1, peroxiredoxin 1-like 1, PEROXYREDOXIN 1, Prdx1l1, PrdxI, PRX1, PRXI, TDPX2, TDX2, TPxA | Peroxiredoxin 1 | Cytoplasm | enzyme | 5052 | 18477 | 117254 100363379 |
||
| PSAT1 | D8Ertd814e, EPIP, NLS2, Phosphoserine Aminotransferase, phosphoserine aminotransferase 1, PSA, PSA1, PSAT, PSATD, Similar to phosphoserine aminotransferase | Phosphoserine aminotransferase 1 | Cytoplasm | enzyme | 29968 | 107272 | 293820 | ||
| PSMA4 | 20S PROTEASOME alpha 4 subunit, 20S PROTEASOME α 4 subunit, C9, HC9, HsT17706, Macropain subunit C9, proteasome (prosome, macropain) subunit, alpha type 4, proteasome (prosome, macropain) subunit, α type 4, proteasome subunit alpha 4, Proteasome subunit c9, proteasome subunit α 4, Proteosome subunit α 4, PSC9 | Proteasome subunit alpha 4 | Cytoplasm | peptidase | 5685 | 26441 | 29671 | ||
| PSMA5 | Aa409047, Macropain zeta chain, Macropain ζ chain, proteasome (prosome, macropain) subunit, alpha type 5, proteasome (prosome, macropain) subunit, α type 5, proteasome subunit alpha 5, proteasome subunit α 5, PSC5, ZETA, ζ | Proteasome subunit alpha 5 | Cytoplasm | peptidase | 5686 | 26442 | 29672 | ||
| PSMB2 | AU045357, AW108089, beta 2 PROTEASOME subunit, C7-I, D4Wsu33e, HC7-I, proteasome (prosome, macropain) subunit, beta type 2, proteasome (prosome, macropain) subunit, β type 2, proteasome subunit beta 2, proteasome subunit β 2, β 2 PROTEASOME subunit | Proteasome subunit beta 2 | Cytoplasm | peptidase | 5690 | 26445 | 29675 | ||
| PSMB5 | 26s Proteasome Beta 5, 26s Proteasome β 5, beta 5 PROTEASOME subunit, Lmp17, LMPX, MB1, proteasome (prosome, macropain) subunit, beta type 5, proteasome (prosome, macropain) subunit, β type 5, Proteasome 20S X, proteasome subunit beta 5, proteasome subunit β 5, X proteasome subunit, β 5 PROTEASOME subunit | Proteasome subunit beta 5 | Cytoplasm | peptidase | 5693 | 19173 | 29425 | ||
| PSMD4 | AF, AF-1, angiocidin, ASF, MCB1, proteasome (prosome, macropain) 26S subunit, non-ATPase, 4, Proteasome 26s subunit non-atpase 4, proteasome 26S subunit, non-ATPase 4, pUB-R5, Rpn10, S5A | Proteasome 26S subunit, non-ATPase 4 | Cytoplasm | other | 5710 | 19185 | 83499 | ||
| S100P | MIG9, PLACENTAL CALCIUM binding, S100 calcium binding protein P | S100 calcium binding protein P | Cytoplasm | other | 6286 | ||||
| SERPINE1 | beta MIGRATING PLAS ACTIVATOR, beta-MIGRATING PLASMINOGEN ACTIVATOR INHIBITOR I, PAI, PAI-1, PAI1A, Pai1aa, Planh, PLANH1, Plasminogen activator inhibitor 1, RATPAI1A, serine (or cysteine) peptidase inhibitor, clade E, member 1, serpin family E member 1, SERPINE, β MIGRATING PLAS ACTIVATOR, β-MIGRATING PLASMINOGEN ACTIVATOR INHIBITOR I | Serpin family E member 1 | Extracellular space | other | 5054 | 18787 | 24617 | ||
| SHC1 | p52SHC, p66, P66shc, SHC, Shc (46 kDa isoform), SHC adaptor protein 1, Shc p66 isoform, SHCA, src homology 2 domain-containing transforming protein C1 | SHC adaptor protein 1 | Cytoplasm | other | 6464 | 20416 | 85385 | ||
| SHMT2 | 2700043D08Rik, AA408223, AA986903, GLYA, HEL-S-51e, serine hydroxymethyltransferase 2 (mitochondrial), Serine Hydroxymethyltransferase2, SHMT | Serine hydroxymethyltransferase 2 | Cytoplasm | enzyme | 6472 | 108037 | 299857 | ||
| SLC7A11 | 9930009M05RIK, AI451155, CCBR1, CYSTEINE GLUATAMINE TRANSPORTER, solute carrier family 7 (cationic amino acid transporter, y+ system), member 11, solute carrier family 7 member 11, sut, xCT | Solute carrier family 7 member 11 | Plasma Membrane | transporter | 23657 | 26570 | 310392 | ||
| SNAI2 | SLUG, SLUGH, SLUGH1, snail family transcriptional repressor 2, snail family zinc finger 2, SNAIL2, WS2D | Snail family transcriptional repressor 2 | Nucleus | transcription regulator | 6591 | 20583 | 25554 | ||
| SOD1 | ALS, ALS1, B430204E11Rik, Cu/Zn-SOD, CuZnSOD, czSOD, HEL-S-44, hSod1, Ipo-1, IPOA, SOD, SOD1L1, SODC, superoxide dismutase 1, superoxide dismutase 1, soluble | Superoxide dismutase 1 | Cytoplasm | enzyme | 6647 | 20655 | 24786 | ||
| SOD2 | IMAGE:4711494, IPO-B, MANGANESE DEPENDENT SOD, Manganese Superoxide Dismutase, Manganese Superoxide Dismutase 2, MGC5618, MITOCHONDRIAL SOD, Mn superoxide dismutase, MNSOD, mtSOD, MVCD6, superoxide dismutase 2, superoxide dismutase 2, mitochondrial | Superoxide dismutase 2 | Cytoplasm | enzyme | 6648 | 20656 | 24787 | ||
| SRGN | haematoPOETIC PROTEOGLYCAN CORE, HEMATOPOETIC PROTEOGLYCAN CORE, PGSG, PPG, PRG, PRG1, Serglycin, Sgc | Serglycin | Cytoplasm | other | 5552 | ||||
| TALDO1 | TAL, TAL-H, TALDOR, Transaldolase, transaldolase 1 | Transaldolase 1 | Cytoplasm | enzyme | 6888 | 21351 | 83688 | ||
| TFAM | AI661103, Hmgts, MTDPS15, MTTF1, MTTFA, TCF6, TCF6L1, TCF6L2, TCF6L3, Tfa, transcription factor A, mitochondrial, tsHMG | Transcription factor A, mitochondrial | Cytoplasm | transcription regulator | 7019 | 21780 | 83474 | ||
| TKT | HEL-S-48, HEL107, p68, SDDHD, TK, TKT1, TRANSKETOLASE | Transketolase | Cytoplasm | enzyme | 7086 | 21881 | 64524 | ||
| UGT1A1 | BILIQTL1, BILIRUBIN UDP-GLUCURONOSYLTRANSFERASE ISOENZYME1, GNT1, HUG-BR1, UDP glucuronosyltransferase 1 family, polypeptide A1, UDP glucuronosyltransferase family 1 member A1, Udp Glycosyltransferase 1, UDPGT, UDPGT 1-1, Udpgt-1a, UGT1, UGT1A, UGT1A01, Ugt1a2b, UgtBr1 | UDP glucuronosyltransferase family 1 member A1 | Cytoplasm | enzyme | 54658 | 394436 | 24861 | ||
| UGT1A7 (includes others) | A13, GNT1, HLUGP4, hUG-BR1, LOC100048744, LUGP4, mUGTBr/P, UDP glucuronosyltransferase 1 family, polypeptide A9, UDP glucuronosyltransferase family 1 member A10, UDP glucuronosyltransferase family 1 member A7, UDP glucuronosyltransferase family 1 member A8, UDP glucuronosyltransferase family 1 member A9, UDP glycosyltransferase 1 family, polypeptide A10, UDPGT, UDPGT 1-7, UDPGT 1-8, UDPGT 1-9, UGT-1A, UGT-1G, UGT-1H, UGT-1I, UGT-1J, UGT1, UGT1-01, UGT1-07, UGT1-08, UGT1-09, UGT1-10, UGT1-9, UGT1.1, UGT1.10, UGT1.8, UGT1.9, UGT1A1, UGT1A10, Ugt1a11, Ugt1a12, Ugt1a13, UGT1A8, UGT1A8S, UGT1A9, UGT1A9S, UGT1AI, UGTP4 | UDP glucuronosyltransferase family 1 member A9 | Cytoplasm | enzyme | 54577 54576 54575 54600 |
394434 394430 |
301595 | ||
Figure 11.
Correlation between NFE2L2 and its target genes in 3 human cancer types. (A–C) Correlation matrices of NFE2L2 and 45 NFE2L2 target gene transcripts (Table 4) from 3 of the tumor groups depicted in Figure 10B. (A) Head and neck squamous cell cancer (HNSC), (B) lung adenocarcinoma (LUAD), (C) HCC (LIHC). The preponderance of positive correlations is apparent and was assessed using a binomial test. P values are shown at top of each panel. (D–F) Long-term survival of patients whose tumors are profiled in (A–C). Expression levels of the 45 NFE2L2 target genes (Table 4) were averaged across all samples for each cancer type. Survival differences between the 2 quartiles of individuals whose tumors expressed the highest and lowest levels of these transcripts were determined by using Kaplan-Meier survival and were assessed using log-rank tests.
Discussion
L30P and R34P, but not WT-NFE2L2, markedly accelerate murine liver tumorigenesis, supporting the idea that NFE2L2-KEAP1 dysregulation suppresses the proliferative limitations imposed by oxidative, electrophilic, and metabolic stresses.10, 11, 12, 13 Depending on context and timing, this may have opposite outcomes.9 For example, it can reduce ROS-mediated oncogenic lesions that initiate tumorigenesis. At later times, it can increase tolerance to oncoproteins and facilitate tumor evolution, expansion, and therapy resistance. Contributing non-canonical functions of NFE2L2 targets might include the regulation of apoptosis, metabolism, angiogenesis, and chemotherapeutic drug detoxification.19,20,37 However, support for this model has derived largely from in vitro studies or from molecularly heterogeneous tumor xenografts.37, 38, 39 Our findings show that L30P/R34P not only accelerate Δ(90)+YAPS127A-mediated tumorigenesis but are directly transforming when co-expressed with either oncoprotein, thus indicating that some HBs could be NFE2L2-driven when only one arm of the β-catenin/Hippo axis is deregulated.2,4 Further examination of other cancers harboring NFE2L2 mutations or CNVs may reveal unrecognized codependencies with other oncoproteins or tumor suppressors.
A unique feature of Δ(90)+YAPS127A+L30P/R34P or R582W+YAPS127A+L30P/R34P HBs is widespread cystogenesis (Figure 1D–G, I). Tumor vasculature is sometimes composed of actual tumor cells or tumor cell-derived endothelium.40,41 However, the numerous, large, and bloodless cysts we observed do not appear to be of vascular origin. Peliosis hepatis is a rare condition associated with epithelial- or endothelial-lined cyst-like lesions, but these are typically blood-filled and less abundant. The cysts we observed are more reminiscent of those occasionally seen in human HBs,21 were independent of tumor growth rates, and arose only in the context of β-catenin+YAPS127A+L30P/R34P co-expression.
In contrast to its cytoplasmic location in cultured cells, nuclear WT-NFE2L2 (Figure 2B) may reflect the oxidized cytoplasm of tumor cells and/or the hepatocyte’s abundance of dietary electrophiles.34,42 These chronic stresses could maintain high levels of oxidized KEAP1, with any reduced residuum being responsible for lowering WT-NFE2L2 levels (Figure 2A). The 821 transcript differences between Δ(90)+YAPS127A tumors and Δ(90) +YAPS127A+WT-NFE2L2 tumors (Figure 5D, Supplementary Table 1) are testimony to the consequences of even small perturbations in the NFE2L2-KEAP1 balance. This likely explains why either mutations or CNVs increase the fractional nuclear concentration of NFE2L2 with similar consequences.8,38
Δ(90)+YAPS127A+L30P/R34P tumor metabolism resembled that of Δ(90)+YAPS127A HBs including lower Oxphos and mitochondrial mass and higher pyruvate consumption compared with livers (Figure 2C–I, Figure 3C–I).3,22 In contrast, they variably increased pPDHα1, Glut1, Glut2, PKM-2, and PFK-L (Figure 3F and J). Higher activity of PDH, which links glycolysis and Oxphos, may allow tumors to compensate for lower FAO rates by maximizing acetyl coenzyme A synthesis in the face of ongoing Warburg respiration.3,22 This dynamic behavior underscores the balancing of FAO and glycolysis via the Randle cycle,43 which allows for new membranes to be synthesized from preexisting fatty acids, thus minimizing the need for de novo synthesis from more primitive precursors.
Despite alterations of glutaminolysis pathway enzymes (Figure 3J), Δ(90)+YAPS127A+L30P/R34P tumors also decreased glutamine-driven Oxphos, which is used by some tumors to maintain α-ketoglutarate pools.43 Collectively, these studies indicate that metabolic demands of these tumors were addressed by reprogramming glycolysis and the TCA cycle.
Δ(90)+L30P/R34P HBs were unique in their down-regulation of Glut1 and PKM-2 and their up-regulation of pPDHα1, Glut4, and PFK-L, a rate-limiting glycolytic enzyme.44 This complex relationship suggested a potentially higher rate of glycolytic flux into mitochondria, while simultaneously reducing the accumulation of anabolic precursors as a result of slower tumor growth rates. However, the mitochondrial mass and Oxphos of these tumors exceeded even those of normal livers (Figure 3C–E, I, Figure 4). Together, these findings underscored a reduced reliance on Warburg respiration and a return to a more liver-like metabolic profile.
In further contrast, the metabolic behavior of YAPS127A+L30P/R34P tumors more closely resembled that of Δ(90)+YAPS127A tumors (Figure 3C–H), although with different expression patterns of the above proteins. Notable differences included the virtual absence of PC, the consequence of which might be the enhanced catalysis of pyruvate to acetyl coenzyme A.
Tumors with pairwise Δ(90), YAPS127A, and L30P/R34P combinations allowed for the assignment of distinct, albeit general, functions for each factor and the identification of key individual transcriptional repertoires. Thus, Δ(90), but not YAPS127A, appears to promote increased mitochondrial mass and a more metabolically active state in response to all tested substrates. Yet, the metabolic behaviors of Δ(90)+YAPS127A and YAPS127A+L30P/R34P tumors (Figure 3C–H) suggest that YAPS127A was dominant over Δ(90), which tended to suppress these more exaggerated responses.
Unbiased RNAseq permitted the identification of specific transcripts associated with each 2- to 3-member combination of Δ(90)/YAPS127A/L30P/R34P (Figure 5B–F). Importantly, it revealed a 22 transcript subset shared by all tumors, likely being the most important contributors to transformation and perhaps contributing to the accelerated growth rates, cystogenesis and necrosis of Δ(90)+YAPS127A+L30P/R34P tumors as well (Table 2). Further evaluation provided additional evidence for its importance to HBs and other cancers. First, all 22 transcripts were altered in the same direction in each HB cohort (Table 2, Figure 5F). Second, they were expressed in tumors with disparate growth rates generated by different β-catenin mutants.3,22 Third, they were more deregulated in Δ(90)+YAPS127A+L30P/R34P tumors (Table 2). Fourth, 10 of the transcripts perfectly identified human HB groups with different prognoses (Figure 5G).31,32 Last, 16 of the transcripts were associated with cancer-promoting or enabling functions (Figure 7).
Among the most intriguing of the 22 transcripts was that encoding serpin E1/plasminogen activator inhibitor-1, a serine protease inhibitor with roles in tumor growth, metastasis, angiogenesis, and matrix remodeling in addition to canonical functions in fibrinolysis.45 Serpin E1 transcripts were the second most highly up-regulated in β-catenin+YAPS127A+L30P/R34P tumors (Table 2), and the serpine1 promoter was unique in containing multiple bona fide response elements for each transcription factor (Figure 8). We also extended previous observations that serpin E1 levels correlate with unfavorable survival in human cancers (Figure 6).46,47 High levels of circulating serpin E1 are associated with polycystic ovary syndrome, and serpin E1’s enforced expression in murine ovaries is cystogenic.48,49 Serpin E1 co-expression in β-catenin+YAPS127A HBs did not accelerate growth rates or promote cystogenesis but did recapitulate the extensive necrosis of β-catenin+YAPS127A+L30P/R34P HBs (Figure 1F, Figure 9C and D). Therefore, other transcripts likely impact tumor growth rates and cystogenesis.
Our findings demonstrate that NFE2L2 mutants alter redox balance in β-catenin+YAPS127A HBs and increase growth, cystogenesis, and necrosis. The unanticipated oncogenicity of L30P/R34P when co-expressed with β-catenin or YAPS127A also demonstrated their direct role in transformation in vivo and unequivocally established NFE2L2 as an oncoprotein that can be activated by mutation, overexpression, or other factors that perturb the normal NFE2L2:KEAP1 balance. This work also identified key transcripts that likely mediate these unique features and directly contribute to tumorigenesis. The high rate of NFE2L2-KEAP1 axis dysregulation in cancer and its prognostic implications can now be better appreciated in light of these findings.
Materials and Methods
Plasmid DNAs
Plasmids used included SB vectors encoding human β-catenin mutants Δ(90) and R582W, YAPS127A, WT-NFE2L2, NFE2L2L30P (L30P), NFE2L2R34P (R34P), cyto-roGFP, mito-roGFP,22,29 or Myc-epitope-tagged murine Serpin E1. All plasmids were purified with Plasmid Plus Midi columns (Qiagen, Inc, Germantown, MD), and 10 μg each was administered to 6- to 8-week-old FVB mice via HDTVI.3,25 All inocula also contained 2 μg of a non-SB vector encoding SB transposase.
Tumor Induction in Mice
All animal studies were performed in accordance with the Public Health Service Policy on Humane Care and Use of Laboratory Animals Institute for Laboratory Animal Research Guide for Care and Use of Laboratory Animals. They were also approved by the Institutional Animal Care and Use Committee at the University of Pittsburgh. Animals were euthanized when tumors achieved maximal permissible size or if tumors caused obvious distress. Fresh tissues were used immediately for the indicated metabolic studies or were apportioned into small pieces, snap-frozen in liquid nitrogen, and immediately stored at −80°C.
Oxphos Measurements
OCRs were determined with an Oroboros Oxygraph 2k instrument (Oroboros Instruments, Inc, Innsbruck, Austria) as described previously using partially purified mitochondrial suspensions in MiR05 buffer.22,29 (OCRs) Reactions were performed after the addition of cytochrome c (10 μmol/L), malate (2 mmol/L), pyruvate (5 mmol/L), adenosine diphosphate (ADP) (5 mmol/L), and glutamate (10 mmol/L) to initiate electron transport chain activity via Complex I. Succinate was added to a final concentration of 10 mmol/L to allow the sum of Complexes I + II activities to be measured. Rotenone (0.5 μmol/L final concentration) was added to inhibit Complex I and to allow the individual contributions of Complex I + Complex II to be confirmed. All activities were normalized to total protein. To measure β-FAO, palmitoyl-CoA and L-carnitine (3 μmol/L and 10 μmol/L, respectively) were added to reactions already primed with ADP and malate. Similar studies performed only in response to L-carnitine alone were used to assess the endogenous stores of potentially oxidizable fatty acids.
Response to Oxidative Stress
Subsets of mice were injected with the usual SB vectors and additional ones encoding cyto-roGFP or mito-roGFP, which are redox-sensitive forms of GFP that localize to the cytoplasm or mitochondrial matrix, respectively.30 Fresh tumors were finely minced in Dulbecco modified Eagle medium + 10% fetal bovine serum, and single cells were allowed to attach to the bottom of a 100-cm tissue culture plate until achieving approximately 80% confluency. The cells were then trypsinized, re-seeded into 6-well glass bottom tissue culture plates (MatTek, Corp, Ashland, MA), allowed to attach for 2–3 days, and then visualized in real time in a temperature- and CO2-controlled environment using a LSM710 laser scanning confocal microscope (Carl Zeiss, Munich, Germany). The ratio of emission signal intensities at 488/405 nm from regions of comparable brightness was used to determine the baseline redox state of each compartment.30 Cells were exposed to 5 mmol/L H2O2 for 5 minutes, followed by rapid replacement with fresh H2O2-free medium. Continuous monitoring was maintained for ∼40 minutes to allow redox normalization rates to be determined.
TaqMan Assays
To identify CNVs in human HBs, DNAs from stored, de-identified FFPE specimens of 8 normal tissues and 22 HBs were isolated by using a QIAmp DNA FFPE Tissue Kit (Qiagen). Ten nanograms of DNA was amplified in triplicate 12-μL reactions using a KAPA Probe Fast qPCR Kit (2×) (GeneWorks, Thebarton, Australia). Each primer’s final concentration was 250 nmol/L, and each TaqMan probe concentration was 300 nmol/L. NFE2L2 primer sequences were the following: (Forward): 5′-TCATCTACAAACGGGAATGTCT-3′, (Reverse): 5′-GTTGCCCACATTCCCAAATC-3′, TaqMan probe: 5′-56-FAM/AGATGCTTT/ZEN/GTACTTTGAT-3′. Two nuclear genes were used for controls, namely glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and ribonuclease P RNA component H1 (RPPH1). GAPDH primer sequences were the following: (Forward): 5′-CCTAGGGCTCACATATTC-3′, (Reverse): 5′-CGCCCAATACGACCAAATCTA-3′, TaqMan probe: 5′-/5Cy5/TCCTCATGC/TAO/CTTCTTGCCTCTTGT/3IABRQSP/3′. RPPH1 polymerase chain reaction primer sequences were the following: (Forward): 5′-TCTGGCCCTAGTCTCAGACCTT-3′, (Reverse): 5′-GAGCTGAGTGC GTCCTGTC-3′, TaqMan probe: 5′-56-FAM/CCAAGGGAC/ZEN/ATGGGAGTG-3′. Amplification conditions were 95°C for 10 seconds, followed by 40 cycles at 95°C for 15 seconds, and 60°C for 60 seconds.
Mitochondrial DNA content was quantified by using a TaqMan-based approach that amplified a 90 base pair segment of the mtD-loop region.3,25 The results were normalized to a control TaqMan reaction that amplified a 73 base pair region of the nuclear apolipoprotein B gene. Reactions contained 10 ng of total DNA and were performed on a CFX96 Touch real-time polymerase chain reaction detection system (Bio-Rad, Inc, Hercules, CA) using the conditions of 95°C for 10 seconds, 40 cycles at 95°C for 15 seconds, followed by 60°C for 60 seconds.
Immunoblotting
Tissue lysates were prepared in sodium dodecyl sulfate-lysis buffer as previously described.3,25,29 Immunoblots were developed by using SuperSignal West Pico Chemiluminescent Substrate kit (Thermo Fisher, Waltham, MA). Antibodies, the vendors from which they were obtained, and the conditions used are listed in Table 5.
Table 5.
Antibodies Used in the Current Study
| Name of antibody | Vendor | Catalog no. | Dilution used |
|---|---|---|---|
| CPT1A | Abcam | ab128568 | 1:1000 |
| GAPDH | Sigma-Aldrich | G8795 | 1:10,000 |
| GLUT1 | Abcam | ab115730 | 1:20,000 |
| Glutamine synthetase (GLNS) | GeneTex | GTX109121 | 1:5000 |
| Glutaminase (GLS) | Abcam | ab131554 | 1:1000 |
| β-Catenin | Abcam | ab16051 | 1:4000 |
| PAI1 (Serpine1) | R&D | AF3828sp | 1:1000 |
| PDHA1 | Santa Cruz | SC-377092 | 1:1000 |
| PDHA1(pSER293) | Calbiochem | AP1062 | 1:1000 |
| PC | Abcam | Ab128952 | 1:2000 |
| PKM1 | CST | 7067 | 1:1000 |
| PKM2 | CST | 3198 | 1:1000 |
| PFK-L | Aviva System Biology | ARP45774 | 1:1000 |
| Glut2 | ProteinTech | 20436-1-AP | 1:300 |
| Glut4 | CST | 2213 | 1:1000 |
| Histone H3 | CST | 9715 | 1:2000 |
| Glud1 | CST | 12793 | 1:1000 |
For subcellular fractionation nuclear and cytoplasmic fractionations were performed on ∼100 mg tissue fragments using a Subcellular Protein Fractionation Kit according to the supplier’s directions (Thermo Fisher).3
RNAseq
RNA purification was performed by using Qiagen RNAeasy columns.3 An Agilent 2100 Bioanalyzer (Agilent Technologies, Foster City, CA) was used to evaluate integrity, and only samples with RIN values >8.5 were used. Samples were prepared for paired-end sequencing by using an NEB NEBNext Ultra Directional RNA Library Prep kit (New England Biolab, Beverly, MA), and sequencing was performed on a NovaSeq 600 Instrument (Illumina, Inc, San Diego, CA) by Novagene, Inc (Sacramento, CA). Raw and processed original data are accessible through GEO (accession number: GSE157623) (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE157623). Differentially expressed transcripts were identified by using DeSEq2, CLC Genomic Workbench v. 12.0 (Qiagen) and EdgeR and were analyzed by using the Galaxy platform (https://galaxyproject.org/use/). For DeSeq2 and EdgeR, FASTQ file reads were mapped against the GRCm38.p6 mouse reference genome using STAR (https://github.com/alexdobin/STAR/releases) version 2.5.2. The output files were analyzed by featureCounts (http://bioinf.wehi.edu.au/featureCounts/) to quantify transcript abundance. Only transcripts whose differential expression was demonstrated by all 3 methods are reported and only after significance was adjusted for false discovery using the Bonferonni–Hochberg correction (q < 0.05). IPA (Qiagen) was used for pathway identification. The following parameters were recorded for each pathway: P value, ratios of dysregulated transcripts to all transcripts associated with that pathway (ratio), and predicted pathway activation, inhibition, and indeterminate (Z-score).
Target Gene Promoter Analysis
Human and mouse genomes were accessed through the BSgenome.Hsapiens.UCSC.hg38 and BSgenome.Mmusculus.UCSC.mm10 R packages, respectively. Gene position information was accessed for human and mouse using the TxDb.Hsapiens.UCSC.hg38.knownGene and TxDb.Mmusculus.UCSC.mm10.knownGene packages, respectively. The flank function was used to query the upstream 5000 base pair promoter regions for each gene, and the searchSeq function of the TFBSTools R package was used to assess the binding sites in these flanking regions using the aforementioned position frequency matrices. Binding position frequency matrices were queried in R from the JASPAR database using the package JASPAR2018 and converted to position weight matrices using the toPWM function of the TFBSTools R package. Sequences with binding scores >90% were accepted as putative binding sites.
Chip-Seq Data were accessed from the ENCODE v.5 database (https://www.encodeproject.org/) for the terminal transcription factors of the NFE2L2 (antioxidant response elements [ARE] sites), Wnt/β-Catenin (Tcf/Lcf sites), and Hippo/Yap (TEAD sites) pathways. Data were only included from cell lines with readily available IDR thresholded peaks identified, and P values derived from more than one experimental replicate. The cell lines from which data were obtained included HepG2, A549, Hela-S3, IMR90, GM12867, HCT-116, HEK293T, K562, and PANC-1. The RTrackLayer R package was used to process the data, and the R packages BSgenome.Hsapiens.UCSC.hg38 and TxDb.Hsapiens.UCSC.hg38.knownGene were used to align the Chip-Seq data to the promoters of the genes of interest. For figures summarizing the binding of more than one terminal transcription factor, the P value plotted was the maximum of any of the terminal transcription factors at that base pair.
Dysregulation of NFE2L2 and Its Direct Target Genes in Human Cancers
RNA-seq data from 141 HB samples7,32,35 were searched for NFE2L2 mutations using CLC Genomics Workbench 11. The data were uploaded, and a track from reference CDS annotation of gene NFE2L2 was generated from the “Homo_sapiens_refseq_GRCh38.p11_o_Genes” track. Amino acid changes within each sample were found by using Basic variant detection: “Toolbox>Resequencing Analysis>Functional Consequences>Variant detection>Basic variant detection” followed by “Amino Acid Changes”: “Toolbox>Resequencing Analysis>Functional Consequences>Amino Acid Changes”. NFE2L2 mutations from 116 human HBs from the COSMIC database were identified by performing a search using “Tissue Distribution>Liver>Sub-Histology> Hepatoblastoma”. NFE2L2 gene CNVs across multiple cancers were identified by analyzing TCGA genomic data in Genomic Data Commons (GDC).
NFE2L2 and KEAP1 transcript expression data were downloaded from the TCGA PANCAN dataset as FPKM, converted to TPM, and filtered to contain only data from HCCs. Samples were defined as having high (top 40%) or low (bottom 40%) levels of KEAP1 and NFE2L2, thus allowing data to be independently categorized according to the expression of each tumor’s transcripts. Survival analysis for each group was performed using the Survival package in R, displayed as Kaplan-Meier curves, and compared pairwise for log-rank P values. This procedure was repeated for OV, LUSC, ESCA, HNSC, SARC, and LUAD tumors.
To determine the relationship between NFE2L2 expression and its direct targets, we used IPA to identify 45 promoters that contained established NFE2L2-binding ARE elements. Expression data (TPM) for these genes and NFE2L2 itself were accessed from the GDC TCGA dataset available on the UCSC Xenabrowser (xena.ucsc.edu) from cohorts with the highest levels of NFE2L2 CNVs. Natural log-fold up-regulation or down-regulation for each transcript was computed relative to that in matched normal tissues from the same individual. For visualization purposes, the pairwise correlation matrices of differential values were plotted as a heat map for each cohort, first with transcripts in an arbitrary order that was the same across cohorts and then as hierarchically clustered heat maps. In all cases only significant correlations were expressed in color, with any remaining ones being blacked out. The default settings of the R “ComplexHeatmap” package were used for hierarchical clustering. The predominance of positive correlations was assessed by using a binomial test.
Statistical Analyses
Survival data for patients in TCGA were analyzed with GraphPad Prism 7 (GraphPad Software, Inc, San Diego, CA). Depending on the tumor type and “BYN” member transcript being analyzed (Figure 5F), different expression level cutoffs were used to define the intragroup subsets with the highest and lowest levels of expression (eg, highest vs lowest 50%, quartile, etc). Kaplan-Meier survival curves were generated by log-rank (Mantel-Cox) test. One-way analysis of variance was applied for multiple comparisons using Fisher least significant difference test. Student 2-tailed t test was used for comparing differences between 2 groups.
Acknowledgments
CRediT Authorship Contributions
Huabo Wang, PhD (Conceptualization: Equal; Formal analysis: Lead; Investigation: Lead; Methodology: Lead; Supervision: Supporting; Writing – original draft: Equal; Writing – review & editing: Equal)
Jie Lu, BA (Formal analysis: Supporting; Investigation: Equal; Project administration: Equal; Visualization: Equal; figure preparation: Equal)
Jordan A. Mandel, BA (Data curation: Equal; Methodology: Equal; Project administration: Equal)
Weiqi Zhang, MD (Investigation: Equal; figure preparation: Equal)
Marie Schwalbe, MD (Investigation: Equal; figure preparation: Equal)
Joanna Gorka, BA (Investigation: Equal)
Ying Liu, MD (Investigation: Equal; figure preparation: Supporting)
Brady Marburger, MD (Investigation: Equal; figure preparation: Equal)
Jinling Wang, MD (Investigation: Equal)
Sarangarajan Ranganathan, MD (Formal analysis: Equal; Methodology: Equal)
Edward V. Prochownik, MD, PhD (Conceptualization: Lead; Data curation: Lead; Formal analysis: Lead; Funding acquisition: Lead; Project administration: Lead; Resources: Lead; Supervision: Lead; Writing – original draft: Lead; Writing – review & editing: Lead)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding M.S., Y. L., and B. M. were supported by the University of PittsburghSchool of Medicine Dean’s Summer Research Program. W. Zhang and J. Wang were supported by joint visiting medical student research program between UPMC and Tsinghua UniversitySchool of Medicine, Beijing, China and Central South University Xiangya University Medical School, Changsha, People’s Republic of China, respectively. The work was also supported by NIH grant RO1 CA174713 to E.V.P. and by awards from The V Foundation and The Mellon Foundation (both to E.V.P.). Computational work was supported in part by the University of Pittsburgh Center for Research Computing.
Supplementary Material
References
- 1.Czauderna P., Lopez-Terrada D., Hiyama E., Haberle B., Malogolowkin M.H., Meyers R.L. Hepatoblastoma state of the art: pathology, genetics, risk stratification, and chemotherapy. Curr Opin Pediatr. 2014;26:19–28. doi: 10.1097/MOP.0000000000000046. [DOI] [PubMed] [Google Scholar]
- 2.Bell D., Ranganathan S., Tao J., Monga S.P. Novel advances in understanding of molecular pathogenesis of hepatoblastoma: a Wnt/beta-catenin perspective. Gene Expr. 2017;17:141–154. doi: 10.3727/105221616X693639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang W., Meyfeldt J., Wang H., Kulkarni S., Lu J., Mandel J.A., Marburger B., Liu Y., Gorka J.E., Ranganathan S., Prochownik E.V. beta-Catenin mutations as determinants of hepatoblastoma phenotypes in mice. J Biol Chem. 2019;294:17524–17542. doi: 10.1074/jbc.RA119.009979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tao J., Calvisi D.F., Ranganathan S., Cigliano A., Zhou L., Singh S., Jiang L., Fan B., Terracciano L., Armeanu-Ebinger S., Ribback S., Dombrowski F., Evert M., Chen X., Monga S.P.S. Activation of beta-catenin and Yap1 in human hepatoblastoma and induction of hepatocarcinogenesis in mice. Gastroenterology. 2014;147:690–701. doi: 10.1053/j.gastro.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grobner S.N., Worst B.C., Weischenfeldt J., Buchhalter I., Kleinheinz K., Rudneva V.A., Johann P.D., Balasubramanian G.P., Segura-Wang M., Brabetz S., Bender S., Hutter B., Sturm D., Pfaff E., Hubschmann D., Zipprich G., Heinold M., Eils J., Lawerenz C., Erkek S., Lambo S., Waszak S., Blattmann C., Borkhardt A., Kuhlen M., Eggert A., Fulda S., Gessler M., Wegert J., Kappler R., Baumhoer D., Burdach S., Kirschner-Schwabe R., Kontny U., Kulozik A.E., Lohmann D., Hettmer S., Eckert C., Bielack S., Nathrath M., Niemeyer C., Richter G.H., Schulte J., Siebert R., Westermann F., Molenaar J.J., Vassal G., Witt H., IP-S Project, IM-S Project, Burkhardt B., Kratz C.P., Witt O., van Tilburg C.M., Kramm C.M., Fleischhack G., Dirksen U., Rutkowski S., Fruhwald M., von Hoff K., Wolf S., Klingebiel T., Koscielniak E., Landgraf P, Koster J., Resnick A.C., Zhang J., Liu Y., Zhou X., Waanders A.J., Zwijnenburg D.A., Raman P., Brors B., Weber U.D., Northcott P.A., Pajtler K.W., Kool M., Piro R.M., Korbel J.O., Schlesner M., Eils R., Jones D.T.W., Lichter P., Chavez L., Zapatka M., Pfister S.M. The landscape of genomic alterations across childhood cancers. Nature. 2018;555:321–327. doi: 10.1038/nature25480. [DOI] [PubMed] [Google Scholar]
- 6.Ma X., Liu Y., Liu Y., Alexandrov L.B., Edmonson M.N., Gawad C., Zhou X., Li Y., Rusch M.C., Easton J., Huether R., Gonzalez-Pena V., Wilkinson M.R., Hermida L.C., Davis S., Sioson E., Pounds S., Cao X., Ries R.E., Wang Z., Chen X., Dong L., Diskin S.J., Smith M.A., Guidry Auvil J.M., Meltzer P.S., Lau C.C., Perlman E.J., Maris J.M., Meshinchi S., Hunger S.P., Gerhard D.S., Zhang J. Pan-cancer genome and transcriptome analyses of 1,699 paediatric leukaemias and solid tumours. Nature. 2018;555:371–376. doi: 10.1038/nature25795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sumazin P., Chen Y., Trevino L.R., Sarabia S.F., Hampton O.A., Patel K., Mistretta T.A., Zorman B., Thompson P., Heczey A., Comerford S., Wheeler D.A., Chintagumpala M., Meyers R., Rakheja D., Finegold M.J., Tomlinson G., Parsons D.W., Lopez-Terrada D. Genomic analysis of hepatoblastoma identifies distinct molecular and prognostic subgroups. Hepatology. 2017;65:104–121. doi: 10.1002/hep.28888. [DOI] [PubMed] [Google Scholar]
- 8.Kerins M.J., Ooi A. A catalogue of somatic NRF2 gain-of-function mutations in cancer. Sci Rep. 2018;8:12846. doi: 10.1038/s41598-018-31281-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu S., Lu H., Bai Y. Nrf2 in cancers: a double-edged sword. Cancer Med. 2019;8:2252–2267. doi: 10.1002/cam4.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dodson M., de la Vega M.R., Cholanians A.B., Schmidlin C.J., Chapman E., Zhang D.D. Modulating NRF2 in disease: timing is everything. Annu Rev Pharmacol Toxicol. 2019;59:555–575. doi: 10.1146/annurev-pharmtox-010818-021856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobayashi A., Kang M.I., Watai Y., Tong K.I., Shibata T., Uchida K., Yamamoto M. Oxidative and electrophilic stresses activate Nrf2 through inhibition of ubiquitination activity of Keap1. Mol Cell Biol. 2006;26:221–229. doi: 10.1128/MCB.26.1.221-229.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dinkova-Kostova A.T., Holtzclaw W.D., Cole R.N., Itoh K., Wakabayashi N., Katoh Y., Yamamoto M., Talalay P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci U S A. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tong K.I., Padmanabhan B., Kobayashi A., Shang C., Hirotsu Y., Yokoyama S., Yamamoto M. Different electrostatic potentials define ETGE and DLG motifs as hinge and latch in oxidative stress response. Mol Cell Biol. 2007;27:7511–7521. doi: 10.1128/MCB.00753-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kitamura Y., Umemura T., Kanki K., Kodama Y., Kitamoto S., Saito K., Itoh K., Yamamoto M., Masegi T., Nishikawa A., Hirose M. Increased susceptibility to hepatocarcinogenicity of Nrf2-deficient mice exposed to 2-amino-3-methylimidazo[4,5-f]quinoline. Cancer Sci. 2007;98:19–24. doi: 10.1111/j.1349-7006.2006.00352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chorley B.N., Campbell M.R., Wang X., Karaca M., Sambandan D., Bangura F., Xue P., Pi J., Kleeberger S.R., Bell D.A. Identification of novel NRF2-regulated genes by ChIP-Seq: influence on retinoid X receptor alpha. Nucleic Acids Res. 2012;40:7416–7429. doi: 10.1093/nar/gks409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malhotra D., Portales-Casamar E., Singh A., Srivastava S., Arenillas D., Happel C., Shyr C., Wakabayashi N., Kensler T.W., Wasserman W.W., Biswal S. Global mapping of binding sites for Nrf2 identifies novel targets in cell survival response through ChIP-Seq profiling and network analysis. Nucleic Acids Res. 2010;38:5718–5734. doi: 10.1093/nar/gkq212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liou G.Y., Storz P. Reactive oxygen species in cancer. Free Radic Res. 2010;44:479–496. doi: 10.3109/10715761003667554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ogrunc M., Di Micco R., Liontos M., Bombardelli L., Mione M., Fumagalli M., Gorgoulis V.G., d’Adda di Fagagna F. Oncogene-induced reactive oxygen species fuel hyperproliferation and DNA damage response activation. Cell Death Differ. 2014;21:998–1012. doi: 10.1038/cdd.2014.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niture S.K., Jaiswal A.K. Nrf2 protein up-regulates antiapoptotic protein Bcl-2 and prevents cellular apoptosis. J Biol Chem. 2012;287:9873–9886. doi: 10.1074/jbc.M111.312694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kowaltowski A.J., Fenton R.G., Fiskum G. Bcl-2 family proteins regulate mitochondrial reactive oxygen production and protect against oxidative stress. Free Radic Biol Med. 2004;37:1845–1853. doi: 10.1016/j.freeradbiomed.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 21.Miller J.H. The ultrasonographic appearance of cystic hepatoblastoma. Radiology. 1981;138:141–143. doi: 10.1148/radiology.138.1.6256810. [DOI] [PubMed] [Google Scholar]
- 22.Wang H., Dolezal J.M., Kulkarni S., Lu J., Mandel J., Jackson L.E., Alencastro F., Duncan A.W., Prochownik E.V. Myc and ChREBP transcription factors cooperatively regulate normal and neoplastic hepatocyte proliferation in mice. J Biol Chem. 2018;293:14740–14757. doi: 10.1074/jbc.RA118.004099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rojo de la Vega M., Chapman E., Zhang D.D. NRF2 and the hallmarks of cancer. Cancer Cell. 2018;34:21–43. doi: 10.1016/j.ccell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Solis L.M., Behrens C., Dong W., Suraokar M., Ozburn N.C., Moran C.A., Corvalan A.H., Biswal S., Swisher S.G., Bekele B.N., Minna J.D., Stewart D.J., Wistuba Nrf2 and Keap1 abnormalities in non-small cell lung carcinoma and association with clinicopathologic features. Clin Cancer Res. 2010;16:3743–3753. doi: 10.1158/1078-0432.CCR-09-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang H., Lu J., Edmunds L.R., Kulkarni S., Dolezal J., Tao J., Ranganathan S., Jackson L., Fromherz M., Beer-Stolz D., Uppala R., Bharathi S., Monga S.P., Goetzman E.S., Prochownik E.V. Coordinated activities of nultiple Myc-dependent and Myc-independent biosynthetic pathways in hepatoblastoma. J Biol Chem. 2016;291:26241–26251. doi: 10.1074/jbc.M116.754218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rawla P., Sunkara T., Muralidharan P., Raj J.P. An updated review of cystic hepatic lesions. Clin Exp Hepatol. 2019;5:22–29. doi: 10.5114/ceh.2019.83153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vander Heiden M.G., Cantley L.C., Thompson C.B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qu Q., Zeng F., Liu X., Wang Q.J., Deng F. Fatty acid oxidation and carnitine palmitoyltransferase I: emerging therapeutic targets in cancer. Cell Death Dis. 2016;7:e2226. doi: 10.1038/cddis.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jackson L.E., Kulkarni S., Wang H., Lu J., Dolezal J.M., Bharathi S.S., Ranganathan S., Patel M.S., Deshpande R., Alencastro F., Wendell S.G., Goetzman E.S., Duncan A.W., Prochownik E.V. Genetic dissociation of glycolysis and the TCA cycle affects neither normal nor neoplastic proliferation. Cancer Res. 2017;77:5795–5807. doi: 10.1158/0008-5472.CAN-17-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanson G.T., Aggeler R., Oglesbee D., Cannon M., Capaldi R.A., Tsien R.Y., Remington S.J. Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. J Biol Chem. 2004;279:13044–13053. doi: 10.1074/jbc.M312846200. [DOI] [PubMed] [Google Scholar]
- 31.Cairo S., Armengol C., De Reynies A., Wei Y., Thomas E., Renard C.A., Goga A., Balakrishnan A., Semeraro M., Gresh L., Pontoglio M., Strick-Marchand H., Levillayer F., Nouet Y., Rickman D., Gauthier F., Branchereau S., Brugieres L., Laithier V., Bouvier R., Boman F., Basso G., Michiels J.F., Hofman P., Arbez-Gindre F., Jouan H., Rousselet-Chapeau M.C., Berrebi D., Marcellin L., Plenat F., Zachar D., Joubert M., Selves J., Pasquier D., Bioulac-Sage P., Grotzer M., Childs M., Fabre M., Buendia M.A. Hepatic stem-like phenotype and interplay of Wnt/beta-catenin and Myc signaling in aggressive childhood liver cancer. Cancer Cell. 2008;14:471–484. doi: 10.1016/j.ccr.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 32.Hooks K.B., Audoux J., Fazli H., Lesjean S., Ernault T., Dugot-Senant N., Leste-Lasserre T., Hagedorn M., Rousseau B., Danet C., Branchereau S., Brugieres L., Taque S., Guettier C., Fabre M., Rullier A., Buendia M.A., Commes T., Grosset C.F., Raymond A.A. New insights into diagnosis and therapeutic options for proliferative hepatoblastoma. Hepatology. 2018;68:89–102. doi: 10.1002/hep.29672. [DOI] [PubMed] [Google Scholar]
- 33.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 34.Weinberg F., Ramnath N., Nagrath D. Reactive oxygen species in the tumor microenvironment: an overview. Cancers (Basel) 2019;11:1191. doi: 10.3390/cancers11081191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eichenmuller M., Trippel F., Kreuder M., Beck A., Schwarzmayr T., Haberle B., Cairo S., Leuschner I., von Schweinitz D., Strom T.M., Kappler R. The genomic landscape of hepatoblastoma and their progenies with HCC-like features. J Hepatol. 2014;61:1312–1320. doi: 10.1016/j.jhep.2014.08.009. [DOI] [PubMed] [Google Scholar]
- 36.Liu Y., Beyer A., Aebersold R. On the dependency of cellular protein levels on mRNA abundance. Cell. 2016;165:535–550. doi: 10.1016/j.cell.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 37.Chio I.I.C., Tuveson D.A. ROS in cancer: the burning question. Trends Mol Med. 2017;23:411–429. doi: 10.1016/j.molmed.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frohlich D.A., McCabe M.T., Arnold R.S., Day M.L. The role of Nrf2 in increased reactive oxygen species and DNA damage in prostate tumorigenesis. Oncogene. 2008;27:4353–4362. doi: 10.1038/onc.2008.79. [DOI] [PubMed] [Google Scholar]
- 39.Sunamura M., Duda D.G., Ghattas M.H., Lozonschi L., Motoi F., Yamauchi J., Matsuno S., Shibahara S., Abraham N.G. Heme oxygenase-1 accelerates tumor angiogenesis of human pancreatic cancer. Angiogenesis. 2003;6:15–24. doi: 10.1023/a:1025803600840. [DOI] [PubMed] [Google Scholar]
- 40.Hendrix M.J., Seftor E.A., Hess A.R., Seftor R.E. Vasculogenic mimicry and tumour-cell plasticity: lessons from melanoma. Nat Rev Cancer. 2003;3:411–421. doi: 10.1038/nrc1092. [DOI] [PubMed] [Google Scholar]
- 41.McGuire T.F., Sajithlal G.B., Lu J., Nicholls R.D., Prochownik E.V. In vivo evolution of tumor-derived endothelial cells. PLoS One. 2012;7 doi: 10.1371/journal.pone.0037138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cook J.A., Gius D., Wink D.A., Krishna M.C., Russo A., Mitchell J.B. Oxidative stress, redox, and the tumor microenvironment. Semin Radiat Oncol. 2004;14:259–266. doi: 10.1016/j.semradonc.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 43.Goetzman E.S., Prochownik E.V. The role for Myc in coordinating glycolysis, oxidative phosphorylation, glutaminolysis, and fatty acid metabolism in normal and neoplastic tissues. Front Endocrinol (Lausanne) 2018;9:129. doi: 10.3389/fendo.2018.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Al Hasawi N., Alkandari M.F., Luqmani Y.A. Phosphofructokinase: a mediator of glycolytic flux in cancer progression. Crit Rev Oncol Hematol. 2014;92:312–321. doi: 10.1016/j.critrevonc.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 45.Placencio V.R., DeClerck Y.A. Plasminogen activator inhibitor-1 in cancer: rationale and insight for future therapeutic testing. Cancer Res. 2015;75:2969–2974. doi: 10.1158/0008-5472.CAN-15-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Binder B.R., Mihaly J. The plasminogen activator inhibitor "paradox" in cancer. Immunol Lett. 2008;118:116–124. doi: 10.1016/j.imlet.2008.03.017. [DOI] [PubMed] [Google Scholar]
- 47.Kubala M.H., DeClerck Y.A. The plasminogen activator inhibitor-1 paradox in cancer: a mechanistic understanding. Cancer Metastasis Rev. 2019;38:483–492. doi: 10.1007/s10555-019-09806-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Devin J.K., Johnson J.E., Eren M., Gleaves L.A., Bradham W.S., Bloodworth J.R., Jr., Vaughan D.E. Transgenic overexpression of plasminogen activator inhibitor-1 promotes the development of polycystic ovarian changes in female mice. J Mol Endocrinol. 2007;39:9–16. doi: 10.1677/JME-06-0057. [DOI] [PubMed] [Google Scholar]
- 49.Lin S., Yongmei G. Plasminogen activator and plasma activator inhibitor-1 in young women with polycystic ovary syndrome. Int J Gynaecol Obstet. 2008;100:285–286. doi: 10.1016/j.ijgo.2007.09.019. [DOI] [PubMed] [Google Scholar]
- 50.Valanejad L., Cast A., Wright M., Bissig K.D., Karns R., Weirauch M.T., Timchenko N. PARP1 activation increases expression of modified tumor suppressors and pathways underlying development of aggressive hepatoblastoma. Commun Biol. 2018;1:67. doi: 10.1038/s42003-018-0077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.