Abstract
Hematological malignancies are a class of malignant neoplasms attributed to abnormal differentiation of hematopoietic stem cells (HSCs). The systemic involvement, poor prognosis, chemotherapy resistance, and recurrence common in hematological malignancies urge researchers to look for novel treatment targets and mechanisms. In recent years, epigenetic abnormalities have been shown to play a vital role in tumorigenesis and progression in hematological malignancies. In addition to DNA methylation and histone modifications, which are most studied, RNA methylation has become increasingly significant. In this review, we elaborate recent advances in the understanding of RNA modification in the pathogenesis, diagnosis and molecular targeted therapies of hematological malignancies and discuss its intricate interactions with other epigenetic modifications, including DNA methylation, histone modifications and noncoding RNAs.
Subject terms: Haematological cancer, Cancer epigenetics, Targeted therapies
Introduction
Hematological malignancies are a class of malignant neoplasms initiated from aberrant self-renewal, blocked differentiation, clonal expansion, and biological dysfunction of hematopoietic cells. Many aspects contribute to hematological malignancies, such as DNA damage response [1], epigenetic alteration [2], and dysregulation of the bone marrow niches [3]. Among them, epigenetics focuses on heritable variations in gene expression or cell phenotype that occur without altering the actual DNA sequence. Compared with genetic mechanisms, epigenetic pathways exhibit much greater flexibility, and tumors probably rely more on epigenetic alterations to escape immune surveillance and develop drug resistance [4]. Recently, protein drugs targeting epigenetic modifiers such as DNA methyltransferase inhibitors and histone deacetylase inhibitors have achieved significant therapeutic effects in hematological malignancies, which has made epigenetics a hot topic of research [5].
Epigenetics is divided into three levels of modifications and alterations: transcriptional, posttranscriptional, and posttranslational. Epigenetic transcriptional regulation mainly involves DNA methylation, histone modification and chromatin remodeling. Posttranscriptional regulation involves RNA modification and ncRNAs. Posttranslational modifications are covalent modifications of proteins during or after translation.
To date, more than 170 kinds of RNA modifications have been identified [6]. With the discovery of RNA demethylases and the application of methylated RNA immunoprecipitation sequencing (MeRIP-seq), the latest studies on methylation at N-6 adenosine (m6A) in RNA have attracted widespread interest due to the identification of m6A as the most abundant internal modification in higher eukaryotes [7]. M6A RNA regulates mRNA posttranscriptionally by affecting splicing [8, 9], export [10], stability [8, 11], and translation of transcripts [11]. M6A RNA is strongly associated with cancer initiation and progression, and m6A regulators are potential targets in cancer therapy [12].
Here, we elaborate recent discoveries of m6A RNA modification in the pathogenesis, diagnosis and molecular targeted therapies of hematological malignancies and discuss its intricate interplay with DNA methylation, histone modifications, and ncRNAs.
The mechanism of m6A RNA methylation
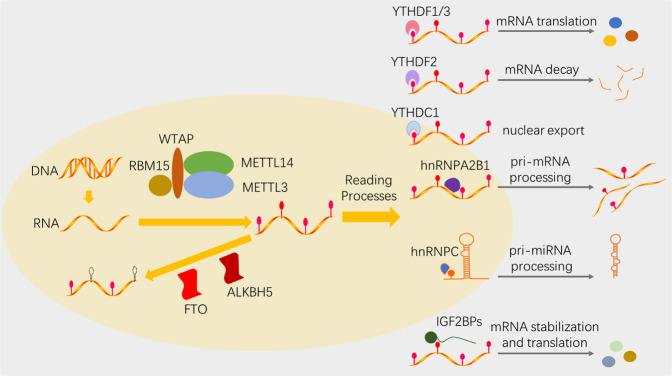
MeRIP-seq has been used to identify mRNAs with m6A modification from more than 7000 mammalian genes, and m6A peaks with the consensus motif RRACH (R = A/G, H = A/C/U) were enriched in the 5′ UTR, 3′ UTR, and near coding regions [13, 14]. A group of m6A regulators modulate m6A modification dynamically and reversibly: the “writers” install the m6A marks, the “erasers” selectively remove m6A marks, and the “readers” specifically recognize the m6A marks [7]. S-adenosylmethionine (SAM) serves as the methyl donor [7, 15] (Fig. 1).
Fig. 1. The mechanism of m6A RNA methylation.
The dynamic and reversible m6A formation is realized through different m6A regulators: the “writers” install m6A marks, the “erasers” selectively remove m6A marks, and the “readers” specifically recognize m6A marks.
m6A Writers
Methyltransferase-like 3 (METTL3), initially named MT-A70, was the first reported m6A RNA writer and has been acknowledged as the core m6A catalytic enzyme [16, 17]. Liu et al. found that methyltransferase-like 14 (METTL14) formed a stable heterodimer core complex with METTL3 and mediated mammalian nuclear RNA m6A methylation [18]. However, studies on the crystal structure of the METTL3/METTL14 complex suggested that METTL14 was not the catalytic core but played a structural role in facilitating RNA substrate recognition and binding [17, 19]. It is now generally considered that METTL14 works as a coregulator of the m6A methyltransferase complex (MTC) and assists m6A RNA methylation.
Other coregulators of MTC are still under investigation. WTAP (Wilms tumor 1-associated protein), which interacts with the N-terminal leader helix of METTL3, is required for METTL3/METTL14 localization to nuclear speckles [20, 21]. RNA-binding motif protein 15 (RBM15) and its paralog RBM15B bind to the MTC and recruit it to U-rich regions of RNA [22]. RBM15/15B mediates m6A formation on the long noncoding RNA X-inactive specific transcript (XIST) to regulate the transcriptional silencing of genes on the X chromosome [22, 23]. Methyltransferase-like 16 (METTL16) modulates the homeostasis of SAM by binding to a vertebrate-conserved hairpin in the 3′ UTR of the SAM synthetase MAT2A and regulating splicing and stability of MAT2A mRNA in a m6A-dependent manner [24]. In addition, Mettl16 is responsible for m6A methylation of human pre-mRNAs and various ncRNAs [25]. VIRMA (Vir-like m6A methyltransferase associated), also known as KIAA1429, regulates preferential mRNA methylation in the 3′ UTR and near the stop codon by recruiting an active METTL3/METTL14/WTAP MTC complex [26]. In eukaryotes, ZC3H13 [27] and HAKAI [28] also participate in m6A RNA formation.
m6A Erasers
The fat mass and obesity-associated (FTO) protein was the first discovered m6A RNA demethylase [29]. Another m6A demethylase, alkB homologue 5 human protein (ALKBH5), was subsequently found by the same team and associated with sperm infertility [30]. They are both homologues of the Fe2+ and alpha-ketoglutarate (α-KG)-dependent (AlkB) human dioxygenase family [31, 32].
However, the substrate preference of FTO remains debatable. Mauer et al. considered that FTO’s physiologic target was not m6A but N6,2′-O-dimethyladenosine (m6Am), one of the most prevalent and reversible modifications at the 5′ ends of capped mRNAs [33]. They discovered that in vitro, FTO demethylated nearly all m6Am marks on RNA substrates at low concentrations (100 nM), but m6A demethylation was readily detected only at higher FTO concentrations (200 nM) [33, 34]. Zhang et al. found that FTO displayed the same demethylation activity toward m6A and m6Am in the same RNA sequence, suggesting that the substrate specificity of FTO resulted from the interaction between residues in the FTO catalytic pocket and the nucleobases of RNA substrates [31]. They claimed that m6A was still the most favorable substrate of FTO because the total amount of m6A was tenfold higher than that of m6Am in mRNAs [31].
m6A Readers
Different m6A readers elicit distinct or even opposite effects by recruiting different functional enzymes. There are three m6A reader families: the YT521-B homologue (YTH) protein family, the heterogeneous nuclear ribonucleoprotein (hnRNP) family, and the common m6A RNA-binding protein family. YTH protein family members, including YTHDF1, YTHDF2, YTHDF3, YTHDC1, and YTHDC2, directly bind to m6A marks with their YTH domain and mediate mRNA alternative splicing [35], nuclear export [10], degradation [36], and translation [37]. The hnRNP family contains hnRNPC, hnRNPG, and hnRNPA2B1. They modulate pre-mRNA processing via an “m6A-switch mechanism”: m6A-dependent RNA structural alterations decrease the stability of Watson-Crick base pairing and thereby allow hnRNPs to bind m6A-modified RNAs but not direct m6A marks [9, 38, 39]. Common m6A RNA-binding protein family members, mainly insulin-like growth factor 2 mRNA-binding proteins (IGF2BP1, IGF2BP2, and IGF2BP3), utilize specific KH domains and their flanking regions to selectively bind m6A-containing RNAs in an RNA structure-independent manner, fortify targeted mRNA stability, and promote targeted mRNA translation, but the exact mechanisms have yet to be elucidated [11].
m6A RNA methylation in hematological malignancies
To date, m6A RNA modifications have been shown to govern normal hematopoiesis and leukemogenesis. Most m6A regulators have been affirmed to be dysregulated and play a critical role in hematological tumorigenesis, which holds great therapeutic potential.
m6A Writers
The role of METTL3 as an oncogene or tumor suppressor in human solid malignancies is still controversial [40]. In normal hematopoiesis, the expression of METTL3 in hematopoietic stem and progenitor cells (HSPCs) was higher than that in mature differentiated myeloid cells [41]. Mechanistically, Mettl3 blocked hematopoietic stem cell (HSC) differentiation by enhancing the mRNA stability of MYC [42, 43], which is involved in regulating the self-renewal and differentiation of HSCs [44]. HSCs from the bone marrow of METTL3-null adult mice showed increased cell counts, reduced self-renewal capacity and lower long-term reconstituting activity than controls [42, 45]. In summary, Mettl3 maintained HSCs in a quiescent state.
In hematological malignancies, METTL3 plays a vital role in acute myeloid leukemia (AML) pathogenesis. METTL3 was significantly more abundant in AML cells than in HSPCs [41]. Depletion of METTL3 in vitro resulted in cell cycle arrest, blocked cell growth, differentiation of leukemic cells, and cell apoptosis [41, 46]. METTL3 was found to enhance the translation of many oncogenic mRNAs by interacting with eIF3h [47]. It promoted the translation of the MYC and BCL-2 mRNAs in human AML MOLM13 cells, which are critical for regulating apoptosis and differentiation [41]. In addition, METTL3 is recruited by the CAATT-box binding protein CEBPZ to the promoters of leukemic active genes, such as the transcription factor SP1, and METTL3 at the associated mRNA transcripts induced m6A modification and enhanced translation by relieving ribosome stalling [46]. Recently, a promising small molecule inhibitor targeting METTL3 in the bone marrow and spleen of AML patients was identified, which is likely the first discovered in vivo RNA methyltransferase inhibitor [48].
METTL14, implicated as an adverse prognosis factor, has been shown to be expressed at low levels in several solid cancers, such as hepatocellular carcinoma (HCC) [49], bladder cancer (BCa) [50], and colorectal cancer (CRC) [51]. METTL14 was also highly expressed in normal HSPCs [45]. Silencing of METTL14 promoted terminal myeloid differentiation, but the functions of Mettl14 in m6A formation and HSC regulation were dependent on Mettl3 [45]. Another study uncovered that in AML carrying t(11q23), t(15;17), or t(8;21), upregulated Mettl14 acted as an oncogene via the SPI1-Mettl14-MYB/MYC signaling axis [52]. However, Mettl3 and Mettl14 have been found to be expressed at lower levels in ETV6/RUNX (E/R)1-positive acute lymphoblastic leukemia (ALL) children than in controls, and their expression in relapsed patients was lower than that in nonrelapsed patients [53].
In this decade, researchers have begun to focus on the oncogenic role of WTAP in leukemogenesis. The overexpression of WTAP led to abnormal proliferation and arrested differentiation of leukemic cells [54]. The expression of WTAP was positively correlated with the levels of various cell proliferation-related proteins (cyclins and HSP90), antiapoptotic proteins (BCL-2 and BAX), oncoproteins (FLI1), and proteins important for stem cell functions (MYC and Ash2L) [54]. WTAP knockdown markedly increased apoptosis following etoposide treatment, which suggests an association between the increased expression of WTAP and chemoresistance in AML [54]. Sorci et al. substantiated that the function of WTAP in cell proliferation was impaired if METTL3 was knocked down in HeLa cells and K562 cells, reconfirming WTAP as a coregulator in MTC [55].
RBM15 has exhibited specific functions in acute megakaryoblastic leukemia (AMKL), a rare type of AML that is more common in children and infants. The fusion of the RBM15 gene and megakaryoblastic leukemia-1 gene by the t(1;22)(p13.3;q13.1) translocation in AMKL was found to be related to recurrent aberrations [56]. RBM15 has been reported to modulate the homeostasis of long-term HSCs, and RBM15-deleted HSCs display decreased self-renewal and aspects associated with ageing through NF-κB activation and DNA damage [57, 58]. RBM15 also plays an important role in megakaryocytic differentiation [58, 59]. RBM15 knockdown in mice produces defective megakaryopoiesis [59]. At the molecular level, RBM15 promoted megakaryocyte terminal differentiation by binding to pre-mRNA intronic regions of genes important for megakaryopoiesis in MEG-01 cells, such as GATA1, RUNX1, TAL1, and c-MPL, and to the 3′ UTRs of genes involved in RNA splicing and metabolic regulation [60]. This process could be enhanced by antisense AS-RBM15 RNA, a lncRNA overlapped with the 5′ UTR of RBM15 [61].
m6A Erasers
Single-nucleotide polymorphisms (SNPs) of FTO are strongly associated with an increased risk of various cancers, including leukemia, myeloma, and lymphoma [62]. FTO plays an oncogenic role in the occurrence, progression and prognosis of many types of cancers in a m6A-dependent manner, especially AML [63], glioblastoma (GBM) [64], and breast cancer (BC) [65]. It is critical for regulating cancer stem cell function and promotes the growth, self-renewal, and metastasis of cancer cells [66].
FTO was found to be aberrantly overexpressed in certain AML subtypes carrying t(11q23)/MLL, t(15;17)/PML-RARA, FLT3-ITD, and NPM1 mutations and was associated with decreased global m6A levels [63]. Further experiments discovered that FTO promoted leukemogenesis and inhibited all-trans-retinoic acid (ATRA)-induced AML cell differentiation by regulating ankyrin repeat and SOCS box protein 2 (ASB2) and retinoic acid receptor alpha (RARα) by reducing their m6A levels [63]. In mice, xenografted leukemic cells with mRNA m6A hypomethylation induced by upregulated FTO had more tyrosine kinase inhibitor (TKI) tolerance and higher growth rates than control cells [67]. Hence, in leukemia, alteration of m6A levels by FTO might be a driver of drug resistance and is a promising target for diagnosis and treatment. In multiple myeloma, FTO was aberrantly upregulated in extramedullary myeloma and enhanced heat shock transcription factor 1 (HSF1) expression by reducing the m6A levels of its transcripts and inhibiting the degradation effect of YTHDF2 [68].
FTO inhibitors are currently under development for the treatment of hematological malignancies. Rhein, the first identified competitive inhibitor for FTO in vitro and in vivo, reversibly bound to the FTO catalytic domain and competitively prevented its recognition of m6A [69]. Meclofenamic acid (MA) is a highly selective inhibitor of FTO on single-stranded nucleic acids over ALKBH5 [70]. A recent study declared that R-2-hydroxyglutarate (R-2HG), an oncometabolite of mutant isocitrate dehydrogenases 1/2 (IDH1/2), downregulated FTO in R-2HG-sensitive leukemia cells, which in turn lessened the mRNA stability of MYC and CEBPA, exerting broad and intrinsic antitumor activity in leukemia [71]. In addition, a novel FTO inhibitor, FB23, designed based on the principle of MA selectivity, was reported to significantly suppress the progression of human leukemia cells and mouse AML models [72]. Su et al. developed two potent small molecule FTO inhibitors, CS1 and CS2, which efficiently suppressed the m6A demethylase activity of FTO with IC50 values in the nanomolar range and exhibited strong antitumor effects in multiple types of cancers, including AML [73]. These FTO inhibitors increase the likelihood of success in developing new therapeutic strategies for hematological malignancies.
Overexpressed ALKBH5 plays an oncogenic role in many solid tumors, such as BC [74], GBM [75], and pancreatic cancer (PC) [76], and is associated with poor outcomes. However, there have been only a few studies on ALKBH5 in hematological malignancies. Two recent studies simultaneously uncovered that ALKBH5 selectively promoted leukemogenesis and modulated self-renewal of leukemia stem cells (LSCs) but had little effect on normal hematopoiesis, suggesting ALKBH5 as a pivotal biotarget of AML [77, 78]. Increased expression of ALKBH5 was correlated with poor prognosis in AML patients, and it exerted tumor-promoting effects in AML via posttranscriptional modulation of TACC3 [77], a prognosis-associated oncogene in various cancers [79–81]. Currently, inhibitors specific to ALKBH5 are under development [82, 83].
m6A Readers
A remote study showed that low IGF2BP1/3 and high IGF2BP2 levels were observed in healthy donor bone marrow and peripheral blood [84], opening the avenue for exploring the oncofetal role of IGF2BPs in normal and malignant hematopoiesis.
IGF2BP1 was upregulated in many types of leukemia and other hematological malignancies. Lower IGF2BP1 expression in AML patients was associated with better survival [85], and depletion of IGPF2BP1 led to a reduction in proliferation of AML cell lines [86]. In ALL, excessive IGF2BP1 is associated with genetic alterations. IGF2BP1 was significantly overexpressed in t(12;21)(p13;q22) ALL carrying the E/R fusion gene compared with other ALL subtypes, and a meta-analysis identified risk SNPs in the IGF2BP1 gene for E/R-positive ALL [84, 87]. Some B-ALL patients carry recurrent t(14;17)(q32;q21) translocations resulting in an IGH-IGF2BP1 fusion, which probably has prognostic significance in B-ALL [88, 89]. In B-ALL REH cells, downregulation of IGF2BP1 reduces many key mRNAs (ETV6/RUNX1, ACTB, CTNNB1, and MYC) by impairing their stability, thus regulating the cell cycle, migration and an aggressive phenotype [90]. Depletion of IGF2BP1 in REH cells also lessens STAT3 mRNA, an attractive therapeutic outcome in leukemia and lymphoma, and augments the sensitivity of STAT3 to its selective inhibitor S3I-201 [90, 91].
IGF2BP2, overexpressed in AML patients, was not only positively associated with poor prognostic factors (FLT3-ITD mutation and IDH1 mutation) but also negatively correlated with a good prognosis indicator (CEBPA mutation status) [92]. IGF2BP2 knockdown significantly inhibited cell growth in AML cell lines, but the mechanism still requires investigation [92].
IGF2BP3 overexpression leads to an increase in the numbers of bone marrow hematopoietic progenitors and the proliferation rate, providing hematopoietic progenitors with a survival advantage [93]. In CML K562 cells, overexpressed IGF2BP3 promoted cell proliferation and survival through the IGF-II pathway [94]. In AML THP-1 cells, IGF2BP3 bound to the 3′ UTR of COX-2 mRNA and affected its stability [95], which had an antiapoptotic role in AML cells [96]. IGF2BP3 is specifically overexpressed in MLL-rearranged B-ALL, and knockdown of IGF2BP3 disrupted cell growth and accelerated apoptosis [93]. Mechanistically, IGF2BP3 positively targeted mRNAs of the quintessential oncogenes CDK6 and MYC in B-ALL RS4;11 cells and REH cells [93]. Additionally, IGF2BP3 was overexpressed in Hodgkin lymphoma cells against a nearly negative background by immunohistochemical staining and showed a relationship with cytoplasmic immunoreactivity to both CD15 and CD30, indicating IGF2BP3 as a novel biomarker in the diagnosis of Hodgkin lymphoma [97].
YTHDF2-mediated m6A mRNA decay regulates HSC protection, self-renewal and regeneration by targeting multiple critical genes, such as Wnt target genes (CyclinD1, C-MYC, and AXIN2), pro-survival genes (BCL-2 and MCL-1), anti-apoptosis genes (BAX, BAD, and BIM), and the key transcription factor TAL1 [98, 99]. Depletion of YTHDF2 in mouse HPSCs and human umbilical cord blood HSCs boosted HSC expansion without marked lineage bias or leukemic potential [98, 99]. However, long-term hematopoiesis-specific YTHDF2 deficiency in young mice led to a progressive myeloid bias, a loss of lymphoid potential, an expansion of HSCs and the failure of multilineage reconstitution and activated proinflammatory pathways by elevating key genes such as STAT1, IL6RA and GADD45G [100]. In addition, YTHDF2 was upregulated in AML, inhibition of which enhanced myeloid reconstitution in HSCs and specifically compromised the propagation of LSCs, suggesting YTHDF2 as a unique therapeutic AML target [101].
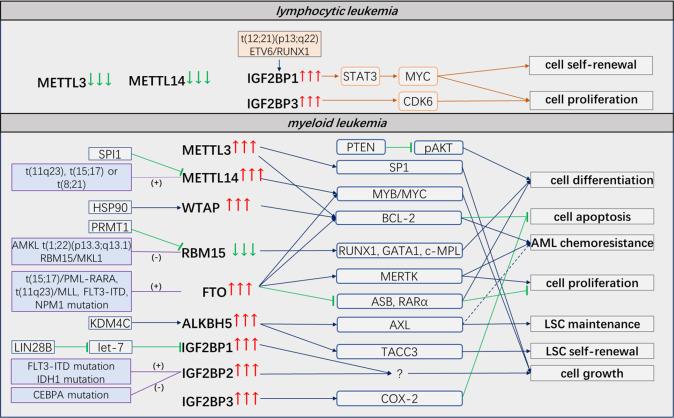
It seems contradictory that both m6A readers and erasers are overexpressed in AML. In fact, they modulate leukemogenesis through different pathways and downstream targets (Fig. 2). METTL3 and METTL14 appear to affect myeloid differentiation and regulate critical genes, such as MYC and MYB, which are essential for the initiation of leukemogenesis [41, 52]. FTO preferentially impacts cell proliferation and modulates drug resistance via RARα and ASB2 [63]. ALKBH5 promotes leukemogenesis and controls the self-renewal of LSCs via TACC3 [77]. Alternatively, m6A readers and erasers act on different m6A sites of the same downstream transcript, leading to identical results. For instance, METTL14 and FTO have both been reported to promote MYC gene expression in AML. The former mainly facilitates m6A formation on the 3′-terminal exon of MYC mRNA [52], and the latter removes m6A marks on the 5′-terminal and internal exons [71]. More importantly, the outcomes of methylation and demethylation, which promote or repress gene expression, are ultimately implemented through distinct reading processes. High expression levels of leukemic oncogenes are ascribed to either the IGF2BP1/2/3-mediated promotion of mRNA stabilization and translation after MTC deposition of m6A marks or the inhibition of YTHDF2/3-mediated mRNA decay by FTO/ALKBH5-induced m6A mark removal. Therefore, we speculate that it is the site where m6A marks are specifically located that enables the removal by m6A eraser and determines what kind of reader it appeals to.
Fig. 2. The functions of m6A regulators in leukemia.
Most m6A regulators are dysregulated and play a critical role in leukemia. They modulate leukemogenesis through different pathways and downstream targets.
The internal epigenetic modification network
The internal interactions among different epigenetic modifications are realized via various epigenetic modifiers. Taking DNA methylation and histone modification as examples, DNA and histones intertwine to form nucleosomes, and diverse epigenetic marks deposited on DNA/histones synergistically or antagonistically alter the regional chromatin structure and then enfold or expose double-stranded DNA buried by nucleosomes to control gene transcription. The crosstalk between DNA methylation and histone modifications has been extensively stated in the last two decades [102]. In hematological malignancies, the integration of AML DNA methylation profiles and HSPCs ChIP-seq data revealed that hypermethylated CpG islands in AML were related to a significant reduction in H3K4me3 with a concomitant increase in H3K4me0 levels, suggesting that H3K4me3 levels are strongly associated with DNA methylation patterns in AML [103].
Recently, increasing evidence has highlighted the interaction of RNA modification with other epigenetic modifications: histone modifications, DNA methylation, and ncRNAs.
Interaction of RNA methylation with histone modifications
RNA methylation selectively acts on the transcripts encoding epigenetic modifiers of histone modifications. Chen et al. discovered m6A modifications on the transcripts of the histone methyltransferase EZH2 in adult neural stem cells (NSCs) by MeRIP-seq, and knockdown of METTL3 resulted in both EZH2 protein reduction and a subsequent global H3K27me3 decrease [104]. In Mettl14-knockout NSCs, the stability of the mRNA transcripts of the H3K27 acetyltransferases CBP and P300 was distinctly strengthened compared to that in controls [105]. Increased m6A RNA methylation on mRNA transcripts of the H3K9me3 methyltransferase Suv39H1 after baicalin hydrate treatment was detected in nasopharyngeal carcinoma (NPC) [106]. In mouse embryonic stem cells and HEK293 cells, loss of METTL3 or METTL14 increased global H3K9me2 levels without altering the expression of H3K9me2-related epigenetic modifiers, and loss of FTO or ALKBH5 decreased H3K9me2 levels [107]. The mechanism involved YTHDC1 recruiting the H3K9me2 demethylase KDM3B to m6A-associated chromatin regions to regulate local H3K9me2 levels and gene expression [107]. In AML THP-1 cells, YTHDF2 increased the H3K27me3 marks on the promoters of proinflammatory cytokines by degrading transcripts of the H3K27me3 demethylase KDM6B in an m6A-dependent manner during bacterial infection [108]. In addition, m6A modification also regulates histone modifiers indirectly. ALKBH5 has been found to regulate EZH2 at the protein level rather than the mRNA level by depleting m6A modifications on the lncRNA NEAT1, which recognizes EZH2 as a downstream target, thus affecting the invasion and metastasis of gastric cancer [109].
In contrast, histone modifications guide m6A function. Suppressed Mettl14 was triggered through the H3K36me3 loss induced by methyltransferase SETD2 knockdown or demethylase KDM4A overexpression, resulting in global m6A demethylation [110]. Mechanistic research has implied that Mettl14 serves as a novel H3K36me3 reader and connects the MTC complex to RNAP II, thereby depositing m6A modifications on nascent RNAs during transcriptional elongation [110]. Wang et al. found that the H3K9me3 demethylase KDM4C was accumulated in the ALKBH5 genetic coding area, eliminating the repressive effect of H3K9me3 and recruiting transcription factors MYB and Pol II to upregulate the expression of ALKBH5 in AML [78].
In summary, m6A marks on transcripts encoding epigenetic modifiers of histone modifications regulate histone modifications, and histone modifications dynamically and specifically mediate m6A RNA methylation at both the posttranscriptional and transcriptional levels.
Interaction of RNA methylation with DNA methylation
DNA methylation directly controls the genetic expression of m6A regulators. For instance, in pancreatic duct epithelial cells, cigarette smoke condensate causes hypomethylation of the METTL3 promoter, induces METTL3 expression and promotes the development and progression of PC [111].
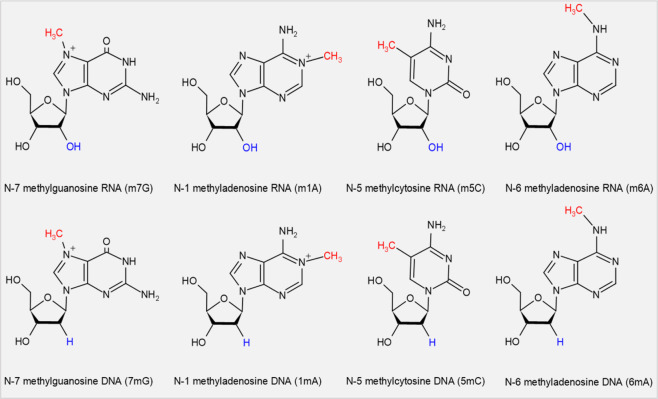
Although there is little direct evidence that RNA methylation modulates DNA methylation, they share the same methyl donor and identical epigenetic marks. Methyl marks on nucleic acids and histones are installed when the active methyl-group donor SAM turns into S-adenosylhomocysteine (SAH) after donating its methyl-group under the catalysis of relevant methyltransferases [15, 112]. Owing to their chemical structural similarities, DNA and RNA possess many identical epigenetic marks (Fig. 3).
Fig. 3. The identical methyl marks of DNA and RNA methylation.
DNA and RNA possess many identical epigenetic marks, such as N-7 methylguanosine, N-1 methyladenosine, N-5 methylcytosine, and N-6 methyladenosine.
Furthermore, in eukaryotes, most writers and erasers of these identical methyl marks are homologues with the same domain or region. The core RNA m6A methyltransferase METTL3 and the DNA 6 mA methyltransferase METTL4 [113] belong to the eukaryotic methyltransferase-like (METTL) protein family. METTLs deposit methylation modification through their SAM binding region largely onto RNAs, including mRNAs, tRNAs, rRNAs, and even ncRNAs, and less onto DNA and proteins (Table 1). The RNA m6A demethylase FTO and the DNA 6 mA demethylase ALKBH1 [134] are homologues of the AlkB dioxygenase superfamily with the same Fe2OG dioxygenase domain and α-KG binding region that recruits Fe2+/α-KG, and they serve to transfer methyl groups from methylated nucleobases to formaldehyde. Another m6A RNA demethylase, ALKBH5, is also an alkB homologue with the same α-KG binding region [135]. Here, we list the known epigenetic functions of the alkB homologues (Table 2).
Table 1.
The interacted epigenetic function of mammalian METTL protein family.
| METTLs | Function |
|---|---|
| METTL1 | tRNA, mRNA, and miRNA 7-methylguanosine (m7G) methyltransferase [114, 115] |
| METTL2 | tRNA 3-methylcytidine (m3C) methyltransferase [116] |
| METTL3 | mRNA 6-methyladenosine (m6A) methyltransferase[17] |
| METTL4 |
DNA 6-methyladenosine (6 mA) methyltransferase [117] snRNA N6 2′-O-dimethyladenosine (m6Am) methyltransferase [118] |
| METTL5 | 18S rRNA m6A methyltransferase [119] |
| METTL6 | tRNA m3C methyltransferase [116] |
| METTL8 | mRNA m3C methyltransferase [116] |
| METTL10 | Nonhistone lysine methyltransferase [120] |
| METTL11A | Histone lysine methyltransferase [121] |
| METTL11B | Proteins N-terminal mono-methytransferase [122] |
| METTL12 | Mitochondrial protein (citrate synthase) lysine methyltransferase [123] |
| METTL13 | Protein (eEF1A) lysine methyltransferase [124] |
| METTL14 | mRNA m6A methyltransferase [17, 18] |
| METTL15 | Mitochondrial 12S rRNA N4-methylcytidine (m4C) methyltransferase [125] |
| METTL16 | snRNA, lncRNA and pre-mRNA m6A methyltransferase [24, 25] |
| METTL17 | Mitochondrial 12S rRNA m4C and 5-methylcytidine (m5C) methyltransferase [126] |
| METTL20 | Mitochondrial lysine methyltransferase [127] |
| METTL21B | Nonhistone lysine methyltransferase [128] |
| METTL21C | Nonhistone lysine methyltransferase [129, 130] |
| METTL21D | Nonhistone lysine methyltransferase [131] |
| METTL22 | Nonhistone lysine methyltransferase [132] |
| METTL23 | Histone arginine methyltransferase (H3R17me2a) [133] |
Table 2.
The interacted epigenetic function of mammalian AlkB proteins.
| Enzymes | Function |
|---|---|
| ALKBH1 | DNA 6 mA demethylase [134] |
| tRNA N1-methyladenosine (m1A) demethylase [136] | |
| Mitochondrial DNA 3-methylcytidine (3mC) and RNA m3C demethylase [137] | |
| Mitochondrial tRNA m5C demethylase [138] | |
| ALKBH2 | DNA 3mC or 1 mA demethylase [139] |
| DNA and N4-ethenocytosine dioxygenase [140] | |
| DNA N3-ethylthymidine decarboxymethylase [141] | |
| ALKBH2/3 | DNA m5C and m3C demethylase [142] |
| ALKBH3 | tRNA m1A and m3C demethylase [143] |
| ALKBH5 | RNA m6A demethylase [30] |
| ALKBH8 | tRNA 5-methoxycarbonylmethyluridine (mcm5U) methyltransferase [144] |
| ALKBH9 (FTO) | RNA m6A demethylase [29] |
| (FTO) | DNA 6 mA demethylase [145] |
| Single-stranded DNA 3-methylthymine and single-stranded RNA 3-methyluracil demethylase [34] |
Interaction of RNA methylation with ncRNAs
Noncoding RNAs, previously called the “dark matter” of the genome, lack coding capacity but constitute more than 90% of the RNA from the human genome [146]. In recent decades, many ncRNAs, such as long noncoding RNAs (lncRNAs), miRNAs, PIWI-interacting RNAs (piRNAs), pseudogenes and circular RNAs (circRNAs), have been found to play crucial roles in both normal cellular function and diseases, including cancer. Recently, ncRNAs have also been reported to be m6A-modified.
lncRNAs
LncRNAs, composed of more than 200 nt in length, induce chromatin remodeling and histone modifications in the nucleus, modulate alternative splicing patterns and generate endo-siRNAs (miRNAs and piRNAs) at the transcriptional level, and alter the activity and localization of specific proteins directly or indirectly in the cytoplasm [147]. lncRNAs contribute to the aging, malfunction, and malignant transformation of blood cells by various mechanisms during normal and malignant hematopoiesis [148], and their aberrant expression in hematological malignancies has been proposed as a diagnostic biomarker, novel therapeutic target, and predictor of clinical outcomes [149].
The majority of lncRNAs are 5′-capped, polyadenylated and spliced like mRNAs. They can be N-6 adenosine-methylated reversibly by different RNA m6A regulators. In most cancers, METTL3 promotes tumorigenesis, progression and chemoresistance by augmenting oncogenic lncRNA expression and stabilizing the respective transcripts [150–153]. M6A modification triggered by overexpression of METTL3 and YTHDF3 in non-small cell lung cancer (NSCLC) increased lncRNA MALAT1 mRNA and enhanced its stability, inducing the invasion, metastasis and drug resistance of NSCLC via the MALAT1-miR-1914-3p-YAP axis [154]. In NSCLC H1299 cells, the lncRNA THOR was m6A-methylated by METTL3, which could be read and stabilized by YTHDF1 [155]. However, in HeLa cells, the m6A modification added by METTL3 to the lncRNA pncRNA-D shortened its half-life, and METTL3 together with YTHDC1 impaired the interaction of pncRNA-D with the RNA-binding protein TLS, thus relieving their inhibitory function on cyclinD1 gene expression and cell cycle progression [156].
METTL14 has been confirmed to be downregulated in CRC, and depletion of METTL14 abolished the m6A level of lncRNA XIST and augmented its expression by reducing the degradation function of YTHDF2, resulting in increased CRC cell growth and invasion [51]. In addition, m6A downregulation by VIRMA depletion suppressed oncogenic lncRNA CCAT1 expression, and higher VIRMA and lncRNA CCAT1 expression predicted poor prognosis in prostate cancer (PCa) patients [157]. In PC, upregulated IGF2BP2 served as an m6A reader for the lncRNA DANCR and stabilized its RNA; DANCR interacted with IGF2BP2 to promote cell proliferation and stemness-like properties [158].
ALKBH5 has been reported to positively control the expression of lncRNAs in tumor cells. Overexpressed ALKBH5 in gastric cancer (GC) upregulated the lncRNA NEAT1 by demethylating its transcripts, thus promoting invasion and metastasis of GC cells via targeting of downstream EZH2 [109]. ALKBH5 also promoted osteosarcoma cell proliferation in vitro and tumor growth in vivo by demethylating the lncRNA PVT1 and enhancing its transcripts by inhibiting the recruitment of the degradation-promoting reader YTHDF2 [159]. ALKBH5 and the lncRNA KCNK15-AS1 were both downregulated in PC. Overexpression of ALKBH5 decreased the m6A marks on KCNK15-AS1 but stabilized its mRNA. These outcomes might jointly inhibit the epithelial–mesenchymal transition of PC cells [160].
On the other hand, lncRNAs alter m6A modification of mRNAs by interacting with m6A RNA regulators. In GC, the oncogenic lncRNA LINC00470 binds to the mRNA of the classical tumor suppressor PTEN and recruits METTL3 to it to form m6A marks, eventually mediating PTEN mRNA degradation via the LINC00470-METTL3-YTHDF2 axis [161]. In BC cells, METTL14 is directly recruited by LNC942, promoting the stability and expression of its downstream targets, such as CXCR4 and CYP1B1 (mediated by m6A methylation levels), and subsequently accelerates cell proliferation and colony formation and reduces cell apoptosis in vitro and in vivo [162].The antisense lncRNA GATA3-AS functions as a cis-acting element for the preferential interaction of KIAA1429 with the pre-mRNA of the tumor suppressor gene GATA3, promotes m6A modification on the 3′ UTR of GATA3 pre-mRNA, and mediates tumor growth and metastasis in HCC [163].
In addition to m6A writers, overexpressed lncRNAs also interact with m6A erasers to control target mRNA expression and cell fate. In glioblastoma stem-like cells (GSCs), the antisense lncRNA FOXM1-AS promotes the interaction of ALKBH5 with nascent transcripts of the FOXM1 gene, increasing FOXM1 expression. Depletion of ALKBH5 and FOXM1-AS disrupts GSC tumorigenesis through the FOXM1 axis [75]. In GBM, the lncRNA SOX2OT upregulates SOX2 expression by recruiting ALKBH5 to its mRNA, inhibits cell apoptosis, and promotes cell proliferation and temozolomide resistance via the Wnt5a/β-catenin signaling pathway [164]. In addition, the antisense lncRNA GAS5-AS1 was found to be markedly decreased in cervical cancer. GAS5-AS1 bound to the tumor suppressor GAS5 and enhanced its stability by interacting with ALKBH5, while this process was inhibited by the YTHDF2-dependent degradation pathway [165].
miRNAs
MiRNAs, with lengths less than 22 nt, are highly conserved in evolution and act in transcriptional repression, mRNA cleavage and degradation. M6A RNA modification regulates miRNA synthesis, processing, and maturation, which are important in tumorigenesis and cancer progression.
METTL3 has been reported to play a vital role in pri-miRNA processing and miRNA maturation in a global and non-cell-type-specific manner by recognizing DGCR8 of the microprocessor complex and binding it with pri-miRNAs [166]. In BCa cells, METTL3 interacted with DGCR8 to accelerate the maturation of pri-miR-221/222, resulting in a reduction in PTEN, which ultimately led to the growth of BCa [167]. In pancreatic cells, overexpressed METTL3 increased m6A formation on pri-miR-25 and facilitated the interaction of pri-miR-25 with DGCR8 by recruiting the RNA-binding protein NKAP, promoting the maturation of oncogenic miR-25-3p [111]. Excessive miR-25-3p targeted PHLPP2 mRNA and promoted the initiation and progression of pancreatic ductal adenocarcinoma via the METTL3-miR-25-3p-PHLPP2-AKT axis [111]. In CRC, METTL3 methylated pri-miR-1246 and promoted the maturation of the metastasis-related RNA miR-1246, which downregulated the anti-oncogenic protein SPRED2 and further inactivated the Raf/MEK/ERK pathway [168]. In addition, Mettl3 increased the splicing of precursor miR-143-3p and enhanced mature miR-143-3p in lung cancer cells, while miR-143-3p overexpression fostered brain metastasis, angiogenesis and tubulin depolymerization in lung cancer cells by repressing VASH1 [169].
Other m6A regulators participate in cancer bioprocesses by targeting miRNAs as well. MiRNA profiling in BC cells showed that METTL14 affected the migration and invasion of BC cells by regulating m6A modification of miRNAs such as miR-146a-5p [170]. However, the underlying mechanism remains to be explored. HNRNPA2B1 reads m6A marks on pri-miRNAs and interacts with DGCR8 to promote pri-miRNA processing [38]. In tamoxifen-sensitive MCF-7 BC cells, overexpression of HNRNPA2B1 reduced tamoxifen and fulvestrant responses via TGFβ signaling and other pathways, which resulted from alterations in miRNA expression (downregulation of miR-29a-3p, miR-29b-3p, and miR-222 and upregulation of miR-1266-5p, miR-1268a, and miR-671-3p) [171]. ALKBH5 inhibited the expression of miR-7 by interacting with HuR to regulate miR-7 processing in epithelial ovarian cancer cells, which promoted EGFR expression [172].
In contrast, miRNAs regulate m6A RNA abundance. The transcriptome-wide distribution of m6A modification in mouse ESCs and HeLa cells revealed that 75% of the m6A peaks with RRACH motifs were identified as potential targets of miRNAs [173]. Further experiments validated that miRNAs induced de novo m6A methylation in a sequence-dependent manner and miRNAs regulated the m6A methyltransferase activity of METTL3 by modulating its binding to mRNAs [173].
A novel mechanism is that miRNAs bind to the 3′ UTR of transcripts encoding m6A regulators and inhibit their expression. Mammalian hepatitis B X-interacting protein (HBXIP) increased METTL3 expression via decreased let-7g miRNA levels, which targeted the 3′ UTR of METTL3 and inhibited its expression, and METTL3 enhanced HBXIP through m6A modification. This HBXIP/let-7g/METTL3/HBXIP loop drove the aggressiveness of BC [174]. In hepatoblastoma, METTL3 was identified to be a direct target of miR-186, and the miR-186/METTL3 axis contributed to the progression of hepatoblastoma via the Wnt/β-catenin pathway [175]. The tumor-suppressive microRNA-1266 in CRC promoted the occurrence and progression of CRC by directly targeting FTO [176]. Fluorescent enzyme reporter gene experiments validated that miR-346 bound to the 3′ UTR of YTHDF1, which has an essential function in glioma diagnosis, treatment and prognosis [177]. MiR-145 decreased the luciferase activities of the 3′ UTR of YTHDF2 mRNA in HCC, and miR-145 inhibition strongly reduced m6A levels by targeting the 3′ UTR of YTHDF2 mRNA in HCC cells [178].
Other ncRNAs
NcRNAs containing the consensus motif RRACH could theoretically be m6A-modified. In addition to their effects on lncRNAs and miRNAs, many METTLs and AlkB homologues have been reported to install methyl marks on rRNAs, tRNAs and other ncRNAs.
CircRNAs, a class of covalently closed ncRNAs without the 5′ cap structure, have recently shown potential to encode proteins via m6A-mediated initiation of translation [179]. m6A modification was also found to regulate circRNA metabolism by guiding the back-splicing reaction, increasing export to the cytoplasm and facilitating degradation by m6A RNA regulators [180–182].
Discussion
Unlike DNA methylation and histone modifications that chiefly act on the chromatin structure, m6A RNA broadly targets nearly all transcripts [13, 14] and widely modulates their location, processing, stability, and translation [7], playing a critical role in normal hemopoiesis and hematological malignancies [45, 63, 77, 78].
As research continues, the actions of m6A readers gradually become explicit. The seemingly contradictory fact that some m6A writers and erasers are synchronously abundant in certain cancers, such as AML, may be further realized through distinct reading processes. More emphasis should be placed on the deeper investigation of mechanisms through which m6A writers and erasers selectively recruit or hinder m6A readers to or from specific m6A sites of downstream transcripts.
RNA epigenetic drugs targeting m6A regulators have caused immense concern. One METTL3 inhibitor is currently being developed [48] and is in sharp contrast to inhibitors targeting m6A erasers, especially FTO [69–72, 82, 83]. FTO acts on <10% of m6A-containing transcripts but dominates leukemogenesis and drug tolerance to ATRA and TKI treatment [63, 67], which partly explains why FTO inhibitors are expected to have a superior clinical future. However, the role of FTO in normal hemopoiesis remains unclear. On the other hand, ALKBH5 should be investigated as much as FTO, given that it is the only present prognosis-related biotarget among m6A writers and erasers in AML and has no or slight effects on normal hematopoiesis [77, 78]. Considering the few overlapping downstream transcripts of FTO and ALKBH5 (fewer than 10%) [78] and their identical effect on leukemogenesis, the combined application of FTO inhibitors and ALKBH5 inhibitors may hold great therapeutic potential in the future.
The emerging phenomenon of the interplay between m6A and histone modifications [78, 107] prompted us to review to previous related studies. The data that we have summarized powerfully demonstrates that RNA methylation strongly interacts with other epigenetic modifications, including DNA methylation, histone modifications and ncRNAs. Even though there is no direct evidence that RNA methylation guides DNA methylation, the intricate function of METTLs and ALKBHs may provide us answers in the future. Our presentation may enlighten researchers who are interested in RNA modifications on m6A-centered epigenetic interactions and future therapeutic development for all cancers that are not restricted to hematological malignancies.
Acknowledgements
This work is supported by the National Natural Science Foundation of PR China (no. 81570193 and no. 81770219, for QW).
Compliance with ethical standards
Conflict of interest
The author declares no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Lan Yao, Hua Yin, Mei Hong
References
- 1.Delia D, Mizutani S. The DNA damage response pathway in normal hematopoiesis and malignancies. Int J Hematol. 2017;106:328–34. doi: 10.1007/s12185-017-2300-7. [DOI] [PubMed] [Google Scholar]
- 2.Hu D, Shilatifard A. Epigenetics of hematopoiesis and hematological malignancies. Genes Dev. 2016;30:2021–41. doi: 10.1101/gad.284109.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mendez-Ferrer S, Bonnet D, Steensma DP, Hasserjian RP, Ghobrial IM, Gribben JG, et al. Bone marrow niches in haematological malignancies. Nat Rev Cancer. 2020;20:285–98. doi: 10.1038/s41568-020-0245-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones PA, Issa JP, Baylin S. Targeting the cancer epigenome for therapy. Nat Rev Genet. 2016;17:630–41. doi: 10.1038/nrg.2016.93. [DOI] [PubMed] [Google Scholar]
- 5.Dimopoulos K, Gronbaek K. Epigenetic therapy in hematological cancers. APMIS. 2019;127:316–28. doi: 10.1111/apm.12906. [DOI] [PubMed] [Google Scholar]
- 6.Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, et al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018;46:D303–7. doi: 10.1093/nar/gkx1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi H, Wei J, He C. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol Cell. 2019;74:640–50. doi: 10.1016/j.molcel.2019.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang C, Klukovich R, Peng H, Wang Z, Yu T, Zhang Y, et al. ALKBH5-dependent m6A demethylation controls splicing and stability of long 3’-UTR mRNAs in male germ cells. Proc Natl Acad Sci USA. 2018;115:E325–33. doi: 10.1073/pnas.1717794115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou KI, Shi H, Lyu R, Wylder AC, Matuszek Z, Pan JN, et al. Regulation of Co-transcriptional Pre-mRNA splicing by m(6)A through the low-complexity protein hnRNPG. Mol Cell. 2019;76:70–81.e9. doi: 10.1016/j.molcel.2019.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roundtree IA, Luo GZ, Zhang ZJ, Wang X, Zhou T, Cui YQ, et al. YTHDC1 mediates nuclear export of N-6 - methyladenosine methylated mRNAs. Elife 2017;6:e31311. [DOI] [PMC free article] [PubMed]
- 11.Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20:285–95. doi: 10.1038/s41556-018-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He L, Li H, Wu A, Peng Y, Shu G, Yin G. Functions of N6-methyladenosine and its role in cancer. Mol Cancer. 2019;18:176. doi: 10.1186/s12943-019-1109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–6. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 14.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell. 2012;149:1635–46. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tuck MT. The formation of internal 6-methyladenine residues in eucaryotic messenger RNA. Int J Biochem. 1992;24:379–86. doi: 10.1016/0020-711x(92)90028-y. [DOI] [PubMed] [Google Scholar]
- 16.Bokar JA, Shambaugh ME, Polayes D, Matera AG, Rottman FM. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA. 1997;3:1233–47. [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X, Feng J, Xue Y, Guan Z, Zhang D, Liu Z, et al. Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature. 2016;534:575–8. doi: 10.1038/nature18298. [DOI] [PubMed] [Google Scholar]
- 18.Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10:93–5. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang P, Doxtader KA, Nam Y. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol Cell. 2016;63:306–17. doi: 10.1016/j.molcel.2016.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–89. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scholler E, Weichmann F, Treiber T, Ringle S, Treiber N, Flatley A, et al. Interactions, localization, and phosphorylation of the m(6)A generating METTL3-METTL14-WTAP complex. Rna. 2018;24:499–512. doi: 10.1261/rna.064063.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, et al. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537:369–73. doi: 10.1038/nature19342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moindrot B, Cerase A, Coker H, Masui O, Grijzenhout A, Pintacuda G, et al. A pooled shRNA screen identifies Rbm15, Spen, and Wtap as factors required for Xist RNA-mediated silencing. Cell Rep. 2015;12:562–72. doi: 10.1016/j.celrep.2015.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pendleton KE, Chen B, Liu K, Hunter OV, Xie Y, Tu BP, et al. The U6 snRNA m(6)A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell. 2017;169:824–35.e14. doi: 10.1016/j.cell.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Warda AS, Kretschmer J, Hackert P, Lenz C, Urlaub H, Hobartner C, et al. Human METTL16 is a N-6-methyladenosine (m(6)A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. Embo Rep. 2017;18:2004–14. doi: 10.15252/embr.201744940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yue Y, Liu J, Cui X, Cao J, Luo G, Zhang Z, et al. VIRMA mediates preferential m(6)A mRNA methylation in 3’UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018;4:10. doi: 10.1038/s41421-018-0019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wen J, Lv RT, Ma HH, Shen HJ, He CX, Wang JH, et al. Zc3h13 Regulates Nuclear RNA m(6)A Methylation and Mouse Embryonic Stem Cell Self-Renewal. Mol Cell. 2018;69:1028–38.e6. doi: 10.1016/j.molcel.2018.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruzicka K, Zhang M, Campilho A, Bodi Z, Kashif M, Saleh M, et al. Identification of factors required for m(6)A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. N Phytol. 2017;215:157–72. doi: 10.1111/nph.14586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885–7. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang X, Wei LH, Wang Y, Xiao Y, Liu J, Zhang W, et al. Structural insights into FTO’s catalytic mechanism for the demethylation of multiple RNA substrates. Proc Natl Acad Sci USA. 2019;116:2919–24. doi: 10.1073/pnas.1820574116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feng C, Liu Y, Wang G, Deng Z, Zhang Q, Wu W, et al. Crystal structures of the human RNA demethylase Alkbh5 reveal basis for substrate recognition. J Biol Chem. 2014;289:11571–83. doi: 10.1074/jbc.M113.546168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mauer J, Luo X, Blanjoie A, Jiao X, Grozhik AV, Patil DP, et al. Reversible methylation of m(6)Am in the 5’ cap controls mRNA stability. Nature. 2017;541:371–5. doi: 10.1038/nature21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mauer J, Jaffrey SR. FTO, m(6) Am, and the hypothesis of reversible epitranscriptomic mRNA modifications. FEBS Lett. 2018;592:2012–22. doi: 10.1002/1873-3468.13092. [DOI] [PubMed] [Google Scholar]
- 35.Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, et al. Nuclear m(6)A reader YTHDC1 Regulates mRNA splicing. Mol Cell. 2016;61:507–19. doi: 10.1016/j.molcel.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 36.Du H, Zhao Y, He JQ, Zhang Y, Xi HR, Liu MF, et al. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun. 2016;7:12626. [DOI] [PMC free article] [PubMed]
- 37.Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, et al. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 2017;27:315–28. doi: 10.1038/cr.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alarcon CR, Goodarzi H, Lee H, Liu XH, Tavazoie S, Tavazoie SF. HNRNPA2B1 is a mediator of m(6)A-dependent nuclear RNA processing events. Cell. 2015;162:1299–308. doi: 10.1016/j.cell.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu N, Dai Q, Zheng G, He C, Parisien M, Pan TN. (6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–4. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng W, Dong X, Zhao Y, Wang S, Jiang H, Zhang M, et al. Multiple functions and mechanisms underlying the role of METTL3 in human cancers. Front Oncol. 2019;9:1403. doi: 10.3389/fonc.2019.01403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vu LP, Pickering BF, Cheng Y, Zaccara S, Nguyen D, Minuesa G, et al. The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med. 2017;23:1369–76. doi: 10.1038/nm.4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee H, Bao SY, Qian YZ, Geula S, Leslie J, Zhang CL, et al. Stage-specific requirement for Mettl3-dependent m(6)A mRNA methylation during haematopoietic stem cell differentiation. Nat Cell Biol. 2019;21:700–9. doi: 10.1038/s41556-019-0318-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheng Y, Luo H, Izzo F, Pickering BF, Nguyen D, Myers R, et al. m(6)A RNA methylation maintains hematopoietic stem cell identity and symmetric commitment. Cell Rep. 2019;28:1703–16.e6. doi: 10.1016/j.celrep.2019.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sheng Y, Ma R, Yu C, Wu Q, Zhang S, Paulsen K, et al. Role of c-Myc haploinsufficiency in the maintenance of HSCs in mice. Blood 2021;137:610–23. [DOI] [PMC free article] [PubMed]
- 45.Yao QJ, Sang L, Lin M, Yin X, Dong W, Gong Y, et al. Mettl3-Mettl14 methyltransferase complex regulates the quiescence of adult hematopoietic stem cells. Cell Res. 2018;28:952–4. doi: 10.1038/s41422-018-0062-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barbieri I, Tzelepis K, Pandolfini L, Shi J, Millan-Zambrano G, Robson SC, et al. Promoter-bound METTL3 maintains myeloid leukaemia by m(6)A-dependent translation control. Nature. 2017;552:126–31. doi: 10.1038/nature24678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Choe J, Lin S, Zhang W, Liu Q, Wang L, Ramirez-Moya J, et al. mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature. 2018;561:556–60. doi: 10.1038/s41586-018-0538-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Albertella M, Blackaby W, Fosbeary R, Hendrick A, Leggate D, Ofir-Rosenfeld Y, et al. A small molecule inhibitor of the RNA m6A writer METTL3 inhibits the development of acute myeloid leukemia (AML) in vivo. Mol Cancer Ther 2019;18(12 Supplement):B126–B126.
- 49.Ma JZ, Yang F, Zhou CC, Liu F, Yuan JH, Wang F, et al. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N(6)-methyladenosine-dependent primary MicroRNA processing. Hepatology. 2017;65:529–43. doi: 10.1002/hep.28885. [DOI] [PubMed] [Google Scholar]
- 50.Gu C, Wang Z, Zhou N, Li G, Kou Y, Luo Y, et al. Mettl14 inhibits bladder TIC self-renewal and bladder tumorigenesis through N(6)-methyladenosine of Notch1. Mol Cancer. 2019;18:168. doi: 10.1186/s12943-019-1084-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang X, Zhang S, He C, Xue P, Zhang L, He Z, et al. METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol Cancer. 2020;19:46. doi: 10.1186/s12943-020-1146-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weng H, Huang H, Wu H, Qin X, Zhao BS, Dong L, et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m(6)A modification. Cell Stem Cell. 2018;22:191–205.e9. doi: 10.1016/j.stem.2017.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun C, Chang L, Liu C, Chen X, Zhu X. The study of METTL3 and METTL14 expressions in childhood ETV6/RUNX1-positive acute lymphoblastic leukemia. Mol Genet Genom Med. 2019;7:e00933. doi: 10.1002/mgg3.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bansal H, Yihua Q, Iyer SP, Ganapathy S, Proia D, Penalva LO, et al. WTAP is a novel oncogenic protein in acute myeloid leukemia. Leukemia. 2014;28:1171–4. doi: 10.1038/leu.2014.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sorci M, Ianniello Z, Cruciani S, Larivera S, Ginistrelli LC, Capuano E, et al. METTL3 regulates WTAP protein homeostasis. Cell Death Dis 2018;9:796. [DOI] [PMC free article] [PubMed]
- 56.Wiseman DH, Bonney DK, Wynn RF. Hemophagocytosis by leukemic megakaryoblasts in acute myeloid leukemia (Megakaryoblastic) With t(1;22)(p13;q13);RBM15-MKL1. J Pediatr Hematol Oncol. 2012;34:576–80. doi: 10.1097/MPH.0b013e318245a027. [DOI] [PubMed] [Google Scholar]
- 57.Ma X, Renda MJ, Wang L, Cheng EC, Niu C, Morris SW, et al. Rbm15 modulates Notch-induced transcriptional activation and affects myeloid differentiation. Mol Cell Biol. 2007;27:3056–64. doi: 10.1128/MCB.01339-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schuschel K, Helwig M, Huttelmaier S, Heckl D, Klusmann JH, Hoell JI. RNA-Binding Proteins in Acute Leukemias. Int J Mol Sci 2020;21:3409. [DOI] [PMC free article] [PubMed]
- 59.Jin S, Mi Y, Song J, Zhang P, Liu Y. PRMT1-RBM15 axis regulates megakaryocytic differentiation of human umbilical cord blood CD34(+) cells. Exp Ther Med. 2018;15:2563–8. doi: 10.3892/etm.2018.5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang L, Tran NT, Su H, Wang R, Lu Y, Tang H, et al. Cross-talk between PRMT1-mediated methylation and ubiquitylation on RBM15 controls RNA splicing. Elife. 2015;4:e07938. [DOI] [PMC free article] [PubMed]
- 61.Tran NT, Su HR, Khodadadi-Jamayran A, Lin S, Zhang L, Zhou DW, et al. The AS-RBM15 lncRNA enhances RBM15 protein translation during megakaryocyte differentiation. Embo Rep. 2016;17:887–900. doi: 10.15252/embr.201541970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Deng XL, Su R, Stanford S, Chen JJ. Critical enzymatic functions of FTO in obesity and cancer. Front Endocrinol. 2018;9:396. [DOI] [PMC free article] [PubMed]
- 63.Li ZJ, Weng HY, Su R, Weng XC, Zuo ZX, Li CY, et al. FTO plays an oncogenic role in acute myeloid leukemia as a N-6-methyladenosine RNA demethylase. Cancer Cell. 2017;31:127–41. doi: 10.1016/j.ccell.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, et al. m(6)A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 2017;18:2622–34. doi: 10.1016/j.celrep.2017.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Niu Y, Lin Z, Wan A, Chen H, Liang H, Sun L, et al. RNA N6-methyladenosine demethylase FTO promotes breast tumor progression through inhibiting BNIP3. Mol Cancer. 2019;18:46. doi: 10.1186/s12943-019-1004-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen J, Du B. Novel positioning from obesity to cancer: FTO, an m(6)A RNA demethylase, regulates tumour progression. J Cancer Res Clin Oncol. 2019;145:19–29. doi: 10.1007/s00432-018-2796-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yan F, Al-Kali A, Zhang Z, Liu J, Pang J, Zhao N, et al. A dynamic N(6)-methyladenosine methylome regulates intrinsic and acquired resistance to tyrosine kinase inhibitors. Cell Res. 2018;28:1062–76. doi: 10.1038/s41422-018-0097-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu A, Zhang J, Zuo L, Xu J, Yin X, Cheng Q, et al. Fto promotes extramedullary progression of multiple myeloma by regulation of HSF1 through m6a RNA methylation. Blood. 2019;134:3063. [Google Scholar]
- 69.Chen B, Ye F, Yu L, Jia G, Huang X, Zhang X, et al. Development of cell-active N6-methyladenosine RNA demethylase FTO inhibitor. J Am Chem Soc. 2012;134:17963–71. doi: 10.1021/ja3064149. [DOI] [PubMed] [Google Scholar]
- 70.Huang Y, Yan J, Li Q, Li J, Gong S, Zhou H, et al. Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Res. 2015;43:373–84. doi: 10.1093/nar/gku1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Su R, Dong L, Li C, Nachtergaele S, Wunderlich M, Qing Y, et al. R-2HG exhibits anti-tumor activity by targeting FTO/m(6)A/MYC/CEBPA signaling. Cell. 2018;172:90–105.e123. doi: 10.1016/j.cell.2017.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang Y, Su R, Sheng Y, Dong L, Dong Z, Xu H, et al. Small-molecule targeting of oncogenic FTO demethylase in acute myeloid leukemia. Cancer Cell. 2019;35:677–91.e10. doi: 10.1016/j.ccell.2019.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Su R, Dong L, Li Y, Gao M, Han L, Wunderlich M, et al. Targeting FTO suppresses cancer stem cell maintenance and immune evasion. Cancer Cell. 2020;38:79–96.e11. doi: 10.1016/j.ccell.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang CZ, Samanta D, Lu HQ, Bullen JW, Zhang HM, Chen I, et al. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m(6)A-demethylation of NANOG mRNA. Proc Natl Acad Sci USA. 2016;113:E2047–56. doi: 10.1073/pnas.1602883113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, et al. m(6)A Demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31:591–606.e6. doi: 10.1016/j.ccell.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tang B, Yang Y, Kang M, Wang Y, Wang Y, Bi Y, et al. m(6)A demethylase ALKBH5 inhibits pancreatic cancer tumorigenesis by decreasing WIF-1 RNA methylation and mediating Wnt signaling. Mol Cancer. 2020;19:3. doi: 10.1186/s12943-019-1128-6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 77.Shen C, Sheng Y, Zhu AC, Robinson S, Jiang X, Dong L, et al. RNA demethylase ALKBH5 selectively promotes tumorigenesis and cancer stem cell self-renewal in acute myeloid leukemia. Cell Stem Cell. 2020;27:64–80.e9. doi: 10.1016/j.stem.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang J, Li Y, Wang P, Han G, Zhang T, Chang J, et al. Leukemogenic Chromatin Alterations Promote AML Leukemia Stem Cells via a KDM4C-ALKBH5-AXL Signaling Axis. Cell Stem Cell 2020;27:81–97 e88. [DOI] [PubMed]
- 79.Li Q, Ye L, Guo W, Wang M, Huang S, Peng X. Overexpression of TACC3 is correlated with tumor aggressiveness and poor prognosis in prostate cancer. Biochem Biophys Res Commun. 2017;486:872–8. doi: 10.1016/j.bbrc.2017.03.090. [DOI] [PubMed] [Google Scholar]
- 80.Lin ZR, Wang MY, He SY, Cai ZM, Huang WR. TACC3 transcriptionally upregulates E2F1 to promote cell growth and confer sensitivity to cisplatin in bladder cancer. Cell Death Dis. 2018;9:72. doi: 10.1038/s41419-017-0112-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Moritsubo M, Miyoshi H, Matsuda K, Yoshida N, Nakashima K, Yanagida E, et al. TACC3 expression as a prognostic factor in aggressive types of adult T-cell leukemia/lymphoma patients. Int J Lab Hematol 2020;42:842–48. [DOI] [PubMed]
- 82.Shen DD, Suo FZ, Song QM, Chang J, Zhang T, Hong JJ, et al. Development of formaldehyde dehydrogenase-coupled assay and antibody-based assays for ALKBH5 activity evaluation. J Pharm Biomed Anal. 2019;162:9–15. doi: 10.1016/j.jpba.2018.09.018. [DOI] [PubMed] [Google Scholar]
- 83.Malacrida A, Rivara M, Di Domizio A, Cislaghi G, Miloso M, Zuliani V, et al. 3D proteome-wide scale screening and activity evaluation of a new ALKBH5 inhibitor in U87 glioblastoma cell line. Bioorg Med Chem. 2020;28:115300. [DOI] [PubMed]
- 84.Stoskus M, Gineikiene E, Valceckiene V, Valatkaite B, Pileckyte R, Griskevicius L. Identification of characteristic IGF2BP expression patterns in distinct B-ALL entities. Blood Cells Mol Dis. 2011;46:321–6. doi: 10.1016/j.bcmd.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 85.Elcheva IA, Wood T, Chiarolanzio K, Chim B, Wong M, Singh V, et al. RNA-binding protein IGF2BP1 maintains leukemia stem cell properties by regulating HOXB4, MYB, and ALDH1A1. Leukemia. 2020;34:1354–63. doi: 10.1038/s41375-019-0656-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhou J, Bi C, Ching YQ, Chooi JY, Lu X, Quah JY, et al. Inhibition of LIN28B impairs leukemia cell growth and metabolism in acute myeloid leukemia. J Hematol Oncol. 2017;10:138. doi: 10.1186/s13045-017-0507-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vijayakrishnan J, Qian M, Studd JB, Yang W, Kinnersley B, Law PJ, et al. Identification of four novel associations for B-cell acute lymphoblastic leukaemia risk. Nat Commun. 2019;10:5348. doi: 10.1038/s41467-019-13069-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jeffries SJ, Jones L, Harrison CJ, Russell LJ. IGH@ translocations co-exist with other primary rearrangements in B-cell precursor acute lymphoblastic leukemia. Haematologica. 2014;99:1334–42. doi: 10.3324/haematol.2014.103820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gu G, Sederberg MC, Drachenberg MR, South ST. IGF2BP1: a novel IGH translocation partner in B acute lymphoblastic leukemia. Cancer Genet. 2014;207:332–4. doi: 10.1016/j.cancergen.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 90.Stoskus M, Eidukaite A, Griskevicius L. Defining the significance of IGF2BP1 overexpression in t(12;21)(p13;q22)-positive leukemia REH cells. Leuk Res. 2016;47:16–21. doi: 10.1016/j.leukres.2016.05.009. [DOI] [PubMed] [Google Scholar]
- 91.Bai L, Zhou H, Xu R, Zhao Y, Chinnaswamy K, McEachern D, et al. A potent and selective small-molecule degrader of STAT3 achieves complete tumor regression in vivo. Cancer Cell. 2019;36:498–511.e7. doi: 10.1016/j.ccell.2019.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.He X, Li W, Liang X, Zhu X, Zhang L, Huang Y, et al. IGF2BP2 overexpression indicates poor survival in patients with acute myelocytic leukemia. Cell Physiol Biochem. 2018;51:1945–56. doi: 10.1159/000495719. [DOI] [PubMed] [Google Scholar]
- 93.Palanichamy JK, Tran TM, Howard JM, Contreras JR, Fernando TR, Sterne-Weiler T, et al. RNA-binding protein IGF2BP3 targeting of oncogenic transcripts promotes hematopoietic progenitor proliferation. J Clin Investig. 2016;126:1495–511. doi: 10.1172/JCI80046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liao B, Hu Y, Brewer G. RNA-binding protein insulin-like growth factor mRNA-binding protein 3 (IMP-3) promotes cell survival via insulin-like growth factor II signaling after ionizing radiation. J Biol Chem. 2011;286:31145–52. doi: 10.1074/jbc.M111.263913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ko CY, Wang WL, Li CF, Jeng YM, Chu YY, Wang HY, et al. IL-18-induced interaction between IMP3 and HuR contributes to COX-2 mRNA stabilization in acute myeloid leukemia. J Leukoc Biol. 2016;99:131–41. doi: 10.1189/jlb.2A0414-228RR. [DOI] [PubMed] [Google Scholar]
- 96.Carter BZ, Mak PY, Wang X, Tao W, Ruvolo V, Mak D, et al. An ARC-regulated IL1beta/Cox-2/PGE2/beta-Catenin/ARC circuit controls leukemia-microenvironment interactions and confers drug resistance in AML. Cancer Res. 2019;79:1165–77. doi: 10.1158/0008-5472.CAN-18-0921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Masoud R, Ibrahiem A, Tantawy D, Eldosoky I. The complementary role of insulin-like growth factor II mRNA-binding protein 3 (IMP3) in diagnosis of Hodgkin’s lymphoma. Ann Diagn Pathol. 2019;42:64–8. doi: 10.1016/j.anndiagpath.2019.06.006. [DOI] [PubMed] [Google Scholar]
- 98.Li ZR, Qian PX, Shao WQ, Shi HL, He XC, Gogol M, et al. Suppression of m(6)A reader Ythdf2 promotes hematopoietic stem cell expansion (vol 28, pg 904, 2018) Cell Res. 2018;28:1042. doi: 10.1038/s41422-018-0072-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang H, Zuo H, Liu J, Wen F, Gao Y, Zhu X, et al. Loss of YTHDF2-mediated m(6)A-dependent mRNA clearance facilitates hematopoietic stem cell regeneration. Cell Res. 2018;28:1035–8. doi: 10.1038/s41422-018-0082-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mapperley C, van de Lagemaat LN, Lawson H, Tavosanis A, Paris J, Campos J, et al. The mRNA m6A reader YTHDF2 suppresses proinflammatory pathways and sustains hematopoietic stem cell function. J Exp Med. 2021;218:e20200829. [DOI] [PMC free article] [PubMed]
- 101.Paris J, Morgan M, Campos J, Spencer GJ, Shmakova A, Ivanova I, et al. Targeting the RNA m(6)A reader YTHDF2 selectively compromises cancer stem cells in acute myeloid leukemia. Cell Stem Cell. 2019;25:137–48.e6. doi: 10.1016/j.stem.2019.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Furuya K, Ikura M, Ikura T. Epigenetic interplays between DNA demethylation and histone methylation for protecting oncogenesis. J Biochem. 2019;165:297–9. doi: 10.1093/jb/mvy124. [DOI] [PubMed] [Google Scholar]
- 103.Scalea S, Maresca C, Catalanotto C, Marino R, Cogoni C, Reale A, et al. Modifications of H3K4 methylation levels are associated with DNA hypermethylation in acute myeloid leukemia. Febs J. 2020;287:1155–75. doi: 10.1111/febs.15086. [DOI] [PubMed] [Google Scholar]
- 104.Chen J, Zhang YC, Huang C, Shen H, Sun B, Cheng X, et al. m(6)A Regulates neurogenesis and neuronal development by modulating histone methyltransferase Ezh2. Genom Proteom Bioinform. 2019;17:154–68. doi: 10.1016/j.gpb.2018.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang Y, Li Y, Yue MH, Wang J, Kumar S, Wechsler-Reya RJ, et al. N-6-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications. Nat Neurosci. 2018;21:195–206. doi: 10.1038/s41593-017-0057-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lai WW, Jia JT, Yan B, Jiang YQ, Shi Y, Chen L, et al. Baicalin hydrate inhibits cancer progression in nasopharyngeal carcinoma by affecting genome instability and splicing. Oncotarget. 2018;9:901–14. doi: 10.18632/oncotarget.22868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Li Y, Xia L, Tan K, Ye X, Zuo Z, Li M, et al. N(6)-Methyladenosine co-transcriptionally directs the demethylation of histone H3K9me2. Nat Genet. 2020;52:870–7. doi: 10.1038/s41588-020-0677-3. [DOI] [PubMed] [Google Scholar]
- 108.Wu C, Chen W, He J, Jin S, Liu Y, Yi Y, et al. Interplay of m(6)A and H3K27 trimethylation restrains inflammation during bacterial infection. Sci Adv. 2020;6:eaba0647. doi: 10.1126/sciadv.aba0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhang J, Guo S, Piao HY, Wang Y, Wu Y, Meng XY, et al. ALKBH5 promotes invasion and metastasis of gastric cancer by decreasing methylation of the lncRNA NEAT1. J Physiol Biochem. 2019;75:379–89. doi: 10.1007/s13105-019-00690-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Huang H, Weng H, Zhou K, Wu T, Zhao BS, Sun M, et al. Histone H3 trimethylation at lysine 36 guides m(6)A RNA modification co-transcriptionally. Nature. 2019;567:414–9. doi: 10.1038/s41586-019-1016-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhang JL, Bai RH, Li M, Ye HL, Wu C, Wang CF, et al. Excessive miR-25-3p maturation via N-6-methyladenosine stimulated by cigarette smoke promotes pancreatic cancer progression. Nat Commun. 2019;10:1858. [DOI] [PMC free article] [PubMed]
- 112.Zhang J, Zheng YG. SAM/SAH analogs as versatile tools for SAM-dependent methyltransferases. Acs Chem Biol. 2016;11:583–97. doi: 10.1021/acschembio.5b00812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Xiao CL, Zhu S, He M, Chen, Zhang Q, Chen Y, et al. N(6)-Methyladenine DNA modification in the human genome. Mol Cell. 2018;71:306–18.e7. doi: 10.1016/j.molcel.2018.06.015. [DOI] [PubMed] [Google Scholar]
- 114.Lin SB, Liu Q, Lelyveld VS, Choe J, Szostak JW, Gregory RI. Mettl1/Wdr4-mediated m(7)G tRNA methylome is required for normal mRNA translation and embryonic stem cell self-renewal and differentiation. Mol Cell. 2018;71:244–55.e5. doi: 10.1016/j.molcel.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Boulias K, Greer EL. Put the pedal to the METTL1: adding Internal m(7)G increases mRNA translation efficiency and augments miRNA processing. Mol Cell. 2019;74:1105–7. doi: 10.1016/j.molcel.2019.06.004. [DOI] [PubMed] [Google Scholar]
- 116.Xu L, Liu XY, Sheng N, Oo KS, Liang JX, Chionh YH, et al. Three distinct 3-methylcytidine (m(3)C) methyltransferases modify tRNA and mRNA in mice and humans. J Biol Chem. 2017;292:14695–703. doi: 10.1074/jbc.M117.798298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hao Z, Wu T, Cui X, Zhu P, Tan C, Dou X, et al. N(6)-Deoxyadenosine methylation in mammalian mitochondrial DNA. Mol Cell. 2020;78:382–95.e8. doi: 10.1016/j.molcel.2020.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chen H, Gu L, Orellana EA, Wang Y, Guo J, Liu Q, et al. METTL4 is an snRNA m(6)Am methyltransferase that regulates RNA splicing. Cell Res. 2020;30:544–47. [DOI] [PMC free article] [PubMed]
- 119.van Tran N, Ernst FGM, Hawley BR, Zorbas C, Ulryck N, Hackert P, et al. The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. 2019;47:7719–33. doi: 10.1093/nar/gkz619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Shimazu T, Barjau J, Sohtome Y, Sodeoka M, Shinkai Y. Selenium-based S-adenosylmethionine analog reveals the mammalian seven-beta-strand methyltransferase METTL10 to be an EF1A1 lysine methyltransferase. Plos ONE. 2014;9:e105394. [DOI] [PMC free article] [PubMed]
- 121.Richon VM, Johnston D, Sneeringer CJ, Jin L, Majer CR, Elliston K, et al. Chemogenetic analysis of human protein methyltransferases. Chem Biol Drug Des. 2011;78:199–210. doi: 10.1111/j.1747-0285.2011.01135.x. [DOI] [PubMed] [Google Scholar]
- 122.Petkowski JJ, Bonsignore LA, Tooley JG, Wilkey DW, Merchant ML, Macara IG, et al. NRMT2 is an N-terminal monomethylase that primes for its homologue NRMT1. Biochem J. 2013;456:453–62. doi: 10.1042/BJ20131163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Malecki J, Jakobsson ME, Ho AYY, Moen A, Rustan AC, Falnes PO. Uncovering human METTL12 as a mitochondrial methyltransferase that modulates citrate synthase activity through metabolite-sensitive lysine methylation. J Biol Chem. 2017;292:17950–62. doi: 10.1074/jbc.M117.808451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jakobsson ME, Malecki JM, Halabelian L, Nilges BS, Pinto R, Kudithipudi S, et al. The dual methyltransferase METTL13 targets N terminus and Lys55 of eEF1A and modulates codon-specific translation rates. Nat Commun. 2018;9:3411. [DOI] [PMC free article] [PubMed]
- 125.Van Haute L, Hendrick AG, D’Souza AR, Powell CA, Rebelo-Guiomar P, Harbour ME, et al. METTL15 introduces N4-methylcytidine into human mitochondrial 12S rRNA and is required for mitoribosome biogenesis. Nucleic Acids Res. 2019;47:10267–81. doi: 10.1093/nar/gkz735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Shi ZN, Xu SY, Xing SH, Yao K, Zhang L, Xue LX, et al. Mettl17, a regulator of mitochondrial ribosomal RNA modifications, is required for the translation of mitochondrial coding genes. Faseb J. 2019;33:13040–50. doi: 10.1096/fj.201901331R. [DOI] [PubMed] [Google Scholar]
- 127.Malecki J, Ho AYY, Moen A, Dahl HA, Falnes PO. Human METTL20 is a mitochondrial lysine methyltransferase that targets the beta subunit of electron transfer flavoprotein (ETF beta) and modulates its activity. J Biol Chem. 2015;290:423–34. doi: 10.1074/jbc.M114.614115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Malecki J, Aileni VK, Ho AYY, Schwarz J, Moen A, Sorensen V, et al. The novel lysine specific methyltransferase METTL21B affects mRNA translation through inducible and dynamic methylation of Lys-165 in human eukaryotic elongation factor 1 alpha (eEF1A) Nucleic Acids Res. 2017;45:4370–89. doi: 10.1093/nar/gkx002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wiederstein JL, Nolte H, Gunther S, Piller T, Baraldo M, Kostin S, et al. Skeletal muscle-specific methyltransferase METTL21C trimethylates p97 and regulates autophagy-associated protein breakdown. Cell Rep. 2018;23:1342–56. doi: 10.1016/j.celrep.2018.03.136. [DOI] [PubMed] [Google Scholar]
- 130.Wang C, Arrington J, Ratliff AC, Chen JJ, Horton HE, Nie YH, et al. Methyltransferase-like 21c methylates and stabilizes the heat shock protein Hspa8 in type I myofibers in mice. J Biol Chem. 2019;294:13718–28. doi: 10.1074/jbc.RA119.008430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kernstock S, Davydova E, Jakobsson M, Moen A, Pettersen S, Maelandsmo GM, et al. Lysine methylation of VCP by a member of a novel human protein methyltransferase family. Nat Commun. 2012;3:1038. [DOI] [PubMed]
- 132.Cloutier P, Lavallee-Adam M, Faubert D, Blanchette M, Coulombe B. Methylation of the DNA/RNA-binding protein Kin17 by METTL22 affects its association with chromatin. J Proteom. 2014;100:115–24. doi: 10.1016/j.jprot.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 133.Hatanaka Y, Tsusaka T, Shimizu N, Morita K, Suzuki T, Machida S, et al. Histone H3 methylated at arginine 17 is essential for reprogramming the paternal genome in zygotes. Cell Rep. 2017;20:2756–65. doi: 10.1016/j.celrep.2017.08.088. [DOI] [PubMed] [Google Scholar]
- 134.Wu TP, Wang T, Seetin MG, Lai Y, Zhu S, Lin K, et al. DNA methylation on N(6)-adenine in mammalian embryonic stem cells. Nature. 2016;532:329–33. doi: 10.1038/nature17640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Aik W, Scotti JS, Choi H, Gong L, Demetriades M, Schofield CJ, et al. Structure of human RNA N(6)-methyladenine demethylase ALKBH5 provides insights into its mechanisms of nucleic acid recognition and demethylation. Nucleic Acids Res. 2014;42:4741–54. doi: 10.1093/nar/gku085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Liu F, Clark W, Luo G, Wang X, Fu Y, Wei J, et al. ALKBH1-Mediated tRNA Demethylation Regulates Translation. Cell. 2016;167:1897. doi: 10.1016/j.cell.2016.11.045. [DOI] [PubMed] [Google Scholar]
- 137.Westbye MP, Feyzi E, Aas PA, Vagbo CB, Talstad VA, Kavli B, et al. Human AlkB homolog 1 is a mitochondrial protein that demethylates 3-methylcytosine in DNA and RNA. J Biol Chem. 2008;283:25046–56. doi: 10.1074/jbc.M803776200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Haag S, Sloan KE, Ranjan N, Warda AS, Kretschmer J, Blessing C, et al. NSUN3 and ABH1 modify the wobble position of mt-tRNAMet to expand codon recognition in mitochondrial translation. EMBO J. 2016;35:2104–19. doi: 10.15252/embj.201694885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ringvoll J, Nordstrand LM, Vagbo CB, Talstad V, Reite K, Aas PA, et al. Repair deficient mice reveal mABH2 as the primary oxidative demethylase for repairing 1meA and 3meC lesions in DNA. EMBO J. 2006;25:2189–98. doi: 10.1038/sj.emboj.7601109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Fu D, Samson LD. Direct repair of 3,N-4-ethenocytosine by the human ALKBH2 dioxygenase is blocked by the AAG/MPG glycosylase. DNA Repair. 2012;11:46–52. doi: 10.1016/j.dnarep.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.You CJ, Wang PC, Nay SL, Wang JS, Dai XX, O’Connor TR, et al. Roles of Aag, Alkbh2, and Alkbh3 in the repair of carboxymethylated and ethylated thymidine lesions. ACS Chem Biol. 2016;11:1332–8. doi: 10.1021/acschembio.6b00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bian K, Lenz SAP, Tang Q, Chen FY, Qi R, Jost M, et al. DNA repair enzymes ALKBH2, ALKBH3, and AlkB oxidize 5-methylcytosine to 5-hydroxymethylcytosine, 5-formylcytosine and 5-carboxylcytosine in vitro. Nucleic Acids Res. 2019;47:5522–9. doi: 10.1093/nar/gkz395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chen ZJ, Qi MJ, Shen B, Luo GZ, Wu YM, Li JX, et al. Transfer RNA demethylase ALKBH3 promotes cancer progression via induction of tRNA-derived small RNAs. Nucleic Acids Res. 2019;47:2533–45. doi: 10.1093/nar/gky1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Songe-Moller L, van den Born E, Leihne V, Vagbo CB, Kristoffersen T, Krokan HE, et al. Mammalian ALKBH8 possesses tRNA methyltransferase activity required for the biogenesis of multiple wobble uridine modifications implicated in translational decoding. Mol Cell Biol. 2010;30:1814–27. doi: 10.1128/MCB.01602-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Martin Carli JF, LeDuc CA, Zhang Y, Stratigopoulos G, Leibel RL. FTO mediates cell-autonomous effects on adipogenesis and adipocyte lipid content by regulating gene expression via 6mA DNA modifications. J Lipid Res. 2018;59:1446–60. doi: 10.1194/jlr.M085555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kazimierczyk M, Kasprowicz MK, Kasprzyk ME, Wrzesinski J. Human long noncoding RNA interactome: detection, characterization and function. Int J Mol Sci. 2020;21:1027. [DOI] [PMC free article] [PubMed]
- 148.Alvarez-Dominguez JR, Lodish HF. Emerging mechanisms of long noncoding RNA function during normal and malignant hematopoiesis. Blood. 2017;130:1965–75. doi: 10.1182/blood-2017-06-788695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Rodriguez-Malave NI, Rao DS. Long noncoding RNAs in hematopoietic malignancies. Brief Funct Genom. 2016;15:227–38. doi: 10.1093/bfgp/elv047. [DOI] [PubMed] [Google Scholar]
- 150.Zheng ZQ, Li ZX, Zhou GQ, Lin L, Zhang LL, Lv JW, et al. Long noncoding RNA FAM225A promotes nasopharyngeal carcinoma tumorigenesis and metastasis by acting as ceRNA to sponge miR-590-3p/miR-1275 and upregulate ITGB3. Cancer Res. 2019;79:4612–26. doi: 10.1158/0008-5472.CAN-19-0799. [DOI] [PubMed] [Google Scholar]
- 151.Zuo XL, Chen ZQ, Gao W, Zhang Y, Wang JG, Wang JF, et al. M6A-mediated upregulation of LINC00958 increases lipogenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. J Hematol Oncol. 2020;13:5. [DOI] [PMC free article] [PubMed]
- 152.Wang JR, Ding WM, Xu YK, Tao EF, Mo MJ, Xu W, et al. Long non-coding RNA RHPN1-AS1 promotes tumorigenesis and metastasis of ovarian cancer by acting as a ceRNA against miR-596 and upregulating LETM1. Aging. 2020;12:4558–72. doi: 10.18632/aging.102911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ban YY, Tan PQ, Cai J, Li JJ, Hu M, Zhou Y, et al. LNCAROD is stabilized by m6A methylation and promotes cancer progression via forming a ternary complex with HSPA1A and YBX1 in head and neck squamous cell carcinoma. Mol Oncol. 2020;14:1282–96. doi: 10.1002/1878-0261.12676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Jin D, Guo J, Wu Y, Du J, Yang L, Wang X, et al. m(6)A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1-miR-1914-3p-YAP axis to induce NSCLC drug resistance and metastasis. J Hematol Oncol. 2019;12:135. doi: 10.1186/s13045-019-0830-6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 155.Liu H, Xu Y, Yao B, Sui T, Lai L, Li Z. A novel N6-methyladenosine (m6A)-dependent fate decision for the lncRNA THOR. Cell Death Dis. 2020;11:613. doi: 10.1038/s41419-020-02833-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yoneda R, Ueda N, Uranishi K, Hirasaki M, Kurokawa R. Long noncoding RNA pncRNA-D reduces cyclin D1 gene expression and arrests cell cycle through RNA m(6)A modification. J Biol Chem. 2020;295:5626–39. doi: 10.1074/jbc.RA119.011556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Barros-Silva D, Lobo J, Guimaraes-Teixeira C, Carneiro I, Oliveira J, Martens-Uzunova ES, et al. VIRMA-dependent N6-methyladenosine modifications regulate the expression of long non-coding RNAs CCAT1 and CCAT2 in prostate cancer. Cancers. 2020;12:771. [DOI] [PMC free article] [PubMed]
- 158.Hu XG, Peng WX, Zhou HX, Jiang JH, Zhou XC, Huang DS, et al. IGF2BP2 regulates DANCR by serving as an N6-methyladenosine reader. Cell Death Differ. 2020;27:1782–94. doi: 10.1038/s41418-019-0461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Chen S, Zhou L, Wang Y. ALKBH5-mediated m(6)A demethylation of lncRNA PVT1 plays an oncogenic role in osteosarcoma. Cancer Cell Int. 2020;20:34. doi: 10.1186/s12935-020-1105-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.He Y, Hu H, Wang Y, Yuan H, Lu Z, Wu P, et al. ALKBH5 inhibits pancreatic cancer motility by decreasing long non-coding RNA KCNK15-AS1 methylation. Cell Physiol Biochem. 2018;48:838–46. doi: 10.1159/000491915. [DOI] [PubMed] [Google Scholar]
- 161.Yan JJ, Huang XF, Zhang XJ, Chen ZJ, Ye C, Xiang WC, et al. LncRNA LINC00470 promotes the degradation of PTEN mRNA to facilitate malignant behavior in gastric cancer cells. Biochem Biophys Res Commun. 2020;521:887–93. doi: 10.1016/j.bbrc.2019.11.016. [DOI] [PubMed] [Google Scholar]
- 162.Sun T, Wu Z, Wang X, Wang Y, Hu X, Qin W, et al. LNC942 promoting METTL14-mediated m(6)A methylation in breast cancer cell proliferation and progression. Oncogene. 2020;39:5358–72. doi: 10.1038/s41388-020-1338-9. [DOI] [PubMed] [Google Scholar]
- 163.Lan T, Li H, Zhang DL, Xu L, Liu HL, Hao XY, et al. KIAA1429 contributes to liver cancer progression through N6-methyladenosine-dependent post-transcriptional modification of GATA3. Mol Cancer. 2019;18:186. [DOI] [PMC free article] [PubMed]
- 164.Liu B, Zhou J, Wang C, Chi Y, Wei Q, Fu Z, et al. LncRNA SOX2OT promotes temozolomide resistance by elevating SOX2 expression via ALKBH5-mediated epigenetic regulation in glioblastoma. Cell Death Dis. 2020;11:384. doi: 10.1038/s41419-020-2540-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wang X, Zhang J, Wang Y. Long noncoding RNA GAS5-AS1 suppresses growth and metastasis of cervical cancer by increasing GAS5 stability. Am J Transl Res. 2019;11:4909–21. [PMC free article] [PubMed] [Google Scholar]
- 166.Alarcon CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N-6-methyladenosine marks primary microRNAs for processing. Nature. 2015;519:482–5. doi: 10.1038/nature14281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Han J, Wang JZ, Yang X, Yu H, Zhou R, Lu HC, et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol Cancer. 2019;18:110. doi: 10.1186/s12943-019-1036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Peng W, Li J, Chen RR, Gu QO, Yang P, Qian WW, et al. Upregulated METTL3 promotes metastasis of colorectal Cancer via miR-1246/SPRED2/MAPK signaling pathway. J Exp Clin Cancer Res. 2019;38:393. [DOI] [PMC free article] [PubMed]
- 169.Wang HS, Deng QQ, Lv ZY, Ling YY, Hou X, Chen ZJ, et al. N6-methyladenosine induced miR-143-3p promotes the brain metastasis of lung cancer via regulation of VASH1. Mol Cancer. 2019;18:181. [DOI] [PMC free article] [PubMed] [Retracted]
- 170.Yi DD, Wang R, Shi XB, Xu L, Yilihamu Y, Sang JF. METTL14 promotes the migration and invasion of breast cancer cells by modulating N6-methyladenosine and hsa-miR-146a-5p expression. Oncol Rep. 2020;43:1375–86. doi: 10.3892/or.2020.7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Klinge CM, Piell KM, Tooley CS, Rouchka EC. HNRNPA2/B1 is upregulated in endocrine-resistant LCC9 breast cancer cells and alters the miRNA transcriptome when overexpressed in MCF-7 cells. Sci Rep. 2019;9:9430. [DOI] [PMC free article] [PubMed]
- 172.Zhu H, Gan X, Jiang X, Diao S, Wu H, Hu J. ALKBH5 inhibited autophagy of epithelial ovarian cancer through miR-7 and BCL-2. J Exp Clin Cancer Res. 2019;38:163. doi: 10.1186/s13046-019-1159-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Chen T, Hao YJ, Zhang Y, Li MM, Wang M, Han WF, et al. m(6)A RNA methylation is regulated by MicroRNAs and promotes reprogramming to pluripotency. Cell Stem Cell. 2015;16:289–301. doi: 10.1016/j.stem.2015.01.016. [DOI] [PubMed] [Google Scholar]
- 174.Cai XL, Wang X, Cao C, Gao YE, Zhang SQ, Yang Z, et al. HBXIP-elevated methyltransferase METTL3 promotes the progression of breast cancer via inhibiting tumor suppressor let-7g. Cancer Lett. 2018;415:11–9. doi: 10.1016/j.canlet.2017.11.018. [DOI] [PubMed] [Google Scholar]
- 175.Cui X, Wang Z, Li J, Zhu J, Ren Z, Zhang D, et al. Cross talk between RNA N6-methyladenosine methyltransferase-like 3 and miR-186 regulates hepatoblastoma progression through Wnt/β-catenin signalling pathway. Cell Prolif. 2020;53:e12768. doi: 10.1111/cpr.12768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Shen XP, Ling X, Lu H, Zhou CX, Zhang JK, Yu Q. Low expression of microRNA-1266 promotes colorectal cancer progression via targeting FTO. Eur Rev Med Pharmacol Sci. 2018;22:8220–6. doi: 10.26355/eurrev_201812_16516. [DOI] [PubMed] [Google Scholar]
- 177.Xu C, Yuan B, He T, Ding B, Li S. Prognostic values of YTHDF1 regulated negatively by mir-3436 in Glioma. J Cell Mol Med 2020;24:7538–49. [DOI] [PMC free article] [PubMed]
- 178.Yang Z, Li J, Feng GX, Gao S, Wang Y, Zhang SQ, et al. MicroRNA-145 modulates N-6-methyladenosine levels by targeting the 3′-untranslated mRNA region of the N-6-methyladenosine binding YTH domain family 2 protein. J Biol Chem. 2017;292:3614–23. doi: 10.1074/jbc.M116.749689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017;27:626–41. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Di Timoteo G, Dattilo D, Centron-Broco A, Colantoni A, Guarnacci M, Rossi F, et al. Modulation of circRNA metabolism by m(6)A modification. Cell Rep. 2020;31:107641. doi: 10.1016/j.celrep.2020.107641. [DOI] [PubMed] [Google Scholar]
- 181.Park OH, Ha H, Lee Y, Boo SH, Kwon DH, Song HK, et al. Endoribonucleolytic cleavage of m(6)A-containing RNAs by RNase P/MRP complex. Mol Cell. 2019;74:494–507.e8. doi: 10.1016/j.molcel.2019.02.034. [DOI] [PubMed] [Google Scholar]
- 182.Chen RX, Chen X, Xia LP, Zhang JX, Pan ZZ, Ma XD, et al. N(6)-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat Commun. 2019;10:4695. doi: 10.1038/s41467-019-12651-2. [DOI] [PMC free article] [PubMed] [Google Scholar]