Abstract
Gut dysbiosis is commonly observed in patients with cirrhosis and chronic gastrointestinal disorders, however, its effect on anti-tumor immunity in the liver is largely unknown. Here we studied how the gut microbiome affects anti-tumor immunity in cholangiocarcinoma. Primary sclerosing cholangitis (PSC) or colitis, two known risk factors for cholangiocarcinoma, which promote tumor development in mice caused an accumulation of CXCR2+ polymorphonuclear myeloid-derived suppressor cells (PMN-MDSC). A decrease in gut barrier function observed in mice with PSC and colitis allowed gut derived bacteria and lipopolysaccharide (LPS) to appear in the liver and induced CXCL1 expression in hepatocytes through a TLR4-dependent mechanism and an accumulation of CXCR2+ PMN-MDSC. On the contrary, neomycin treatment blocked CXCL1 expression, PMN-MDSC accumulation and inhibited tumor growth even in the absence of liver disease or colitis. Our study demonstrates that the gut microbiome controls hepatocytes to form an immunosuppressive environment by increasing PMN-MDSC to promote liver cancer.
Keywords: Gut microbiota, hepatocytes, MDSC, cholangiocarcinoma
INTRODUCTION
The gut and the liver are anatomically and physiologically connected, and this “gut–liver axis” controls not only on the liver pathology but also intrahepatic and systemic immune responses. As such, the gut microbiome is an important modulator of anti-tumor immunity (1–3). The intestinal barrier is the first line defense to separate intestinal lumen microbes from host (4). Defects in the gut barrier function have been described in different liver diseases (5,6). An impairment of barrier function increases intestinal permeability and promotes transportation of microbial products even intact bacteria into portal circulation. Altered intestinal microbiome composition, known as “dysbiosis”, is associated with intestinal barrier dysfunction, both of which can be commonly observed in chronic gastrointestinal diseases such as inflammatory bowel diseases (IBD), primary sclerosing cholangitis (PSC) and cirrhosis (7–9), and are all risk factors for the development of cholangiocarcinoma(10). However, the knowledge of the influence of dysbiosis and intestinal barrier dysfunction on liver immunity and tumor development is limited.
Cholangiocarcinoma is the second most common primary hepatic malignancy (11). Most cholangiocarcinoma patients present with unresectable disease at the time of diagnosis and the prognosis is poor (12). PSC, a chronic liver disease characterized by progressive inflammation and scarring of the medium and large bile ducts of the liver or the extrahepatic bile tree, is recognized as an important risk factor for cholangiocarcinoma (13). Clinical data suggests that colitis, a chronic inflammatory colon disease, contributes to cholangiocarcinoma development (10,14,15). Intestinal dysbiosis has been described in patients with PSC (16) or colitis (17). Yet, the role of gut microbiome in cholangiocarcinoma progression is unknown.
Myeloid-derived immunosuppressive cells (MDSCs) are a heterogenous population of immature myeloid cells with the ability to suppress both adaptive and innate immune responses through multiple mechanisms (18). The presence of MDSCs in cancer patients and their tumor-promoting functions are well documented. MDSCs can be grouped into polymorphonuclear PMN-MDSCs and monocytic M-MDSCs (19). Chemotaxis is important for MDSC accumulation and differs among these two MDSC populations. Specifically, M-MDSC recruitment is often mediated by CCL2-CCR2, whereas the binding of chemokine CXCL1 to its receptor CXCR2 is important for PMN-MDSC accumulation (19). Extrahepatic diseases such as tumors of other organs can induce MDSC accumulation in the liver (20), and hepatic MDSCs are known to promote liver tumors (21). Recently, the involvement of MDSC in patients with cholangiocarcinoma has been described and it has been suggested that targeting PMN-MDSC improves the efficacy of checkpoint inhibitor therapy in murine cholangiocarcinoma models(22).
Here, we studied how the gut microbiome can affect MDSCs in the liver. Combining different mouse models for PSC, colitis and cholangiocarcinoma, we demonstrate that Gram-negative commensal gut bacteria control accumulation of hepatic MDSC through a TLR4/CXCL1/CXCR2 dependent mechanism and thereby suppress anti-tumor immunity in the liver.
RESULTS
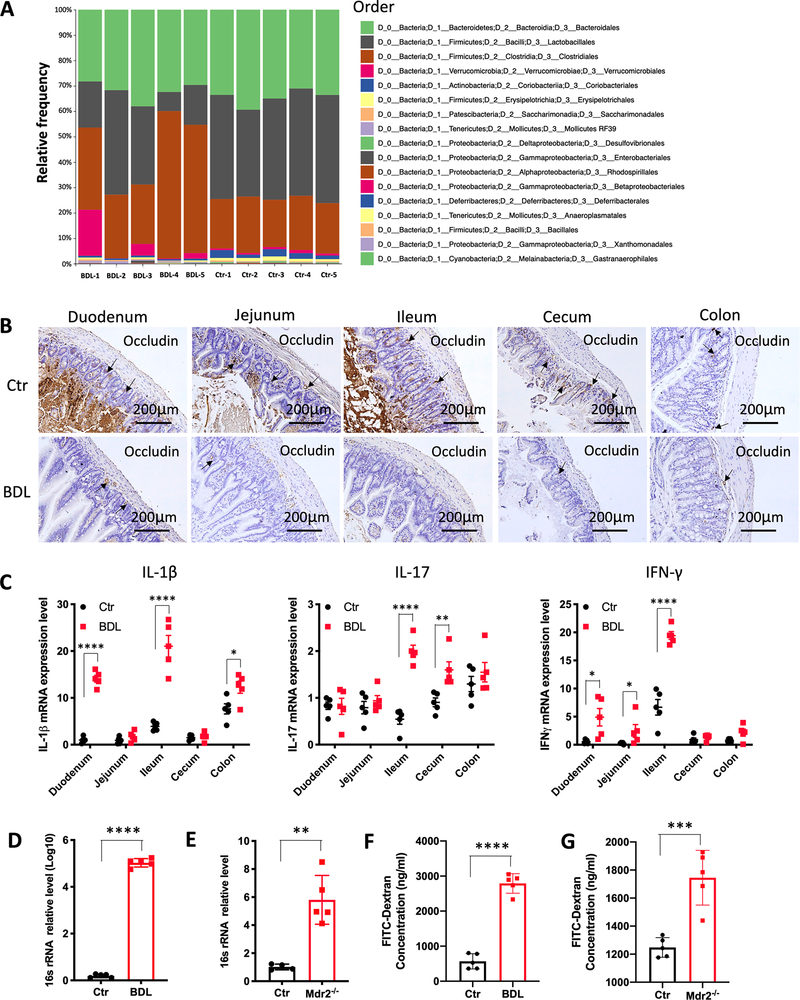
Impaired gastrointestinal barrier in PSC and colitis exposes the liver to the gut microbiome
Gut dysbiosis has been observed in patients with PSC (23). We examined two different well-established murine PSC models (24). A PSC-like state was either modeled in mice using bile duct ligation (BDL) (Supplementary Fig. S1A) or occurred spontaneously in Mdr2−/− mice (Supplementary Fig. S1B) (25) (26). As reported(27), sequencing of the gut microbiome revealed a different spectrum of commensal gut bacteria from mice after BDL (Fig. 1A). Profound changes of the gut commensals were observed at the class level as shown in Figure 1A. Especially notable, are the differences in abundances of Lactobacillales, Actinobacteria and Clostridiales. These taxa appear to be inversely related with Lactobacillales and Actinobacteria seen in higher abundance in the control samples and Clostridiales relatively more abundant after BDL (Fig. 1A and Supplementary Fig. S1C). Emerging evidence suggests that dysbiosis contributes to intestinal barrier dysfunction in PSC (28). Next, changes in the intestine were studied. Microscopic analysis revealed signs of chronic colitis with mucosal fibrosis and a reduction of goblet cells in mice after BDL (Supplementary Fig. S1D). Tight junction proteins (Occludin and ZO-1) were decreased in duodenum, jejunum, ileum, cecum of BDL mice, while no difference of Occludin and ZO-1 was observed in the colon (Fig. 1B, Supplementary Fig. S1E). Inflammatory cytokine mRNA levels (IL-1β, IL-17, IFN-γ, TGFβ, TNF-α, IL-10) were significantly elevated particularly in ileum tissue samples derived from mice after BDL (Fig. 1C, Supplementary Fig. S2A). These results indicated that PSC development led to inflammatory responses in the small bowel epithelium and an impairment of the gut barrier function. Next we studied the portal vein, which drains the blood from the intestine to the liver and thereby forms a physical connection between the liver and the gut microbiome. Indeed, higher bacterial 16sRNA levels were detected in portal vein blood in both PSC-like mouse models (Figs. 1D and 1E). We also observed a significant increase in serum levels of FITC-labelled dextran after oral administration further supporting an impaired intestinal barrier function in murine PSC-like models (Figs. 1F and 1G). Finally, we examined liver tissue for the presence of translocated bacteria. Plating liver tissue lysate revealed more bacterial colonies from mice with PSC-like lesions than from control mice (Supplementary Figs. S2B and S2C).
Figure 1.
PSC and colitis cause a leaky gut and bacterial translocation.
(A) BDL was performed in C57BL/6 mice. Two weeks later, stool samples from BDL and control (Ctr) mice were collected for 16s rRNA sequencing. n=5 for Ctr and BDL. Bar plots of the order levels in BDL and Ctr mice are shown. Relative abundance is plotted for each mouse.
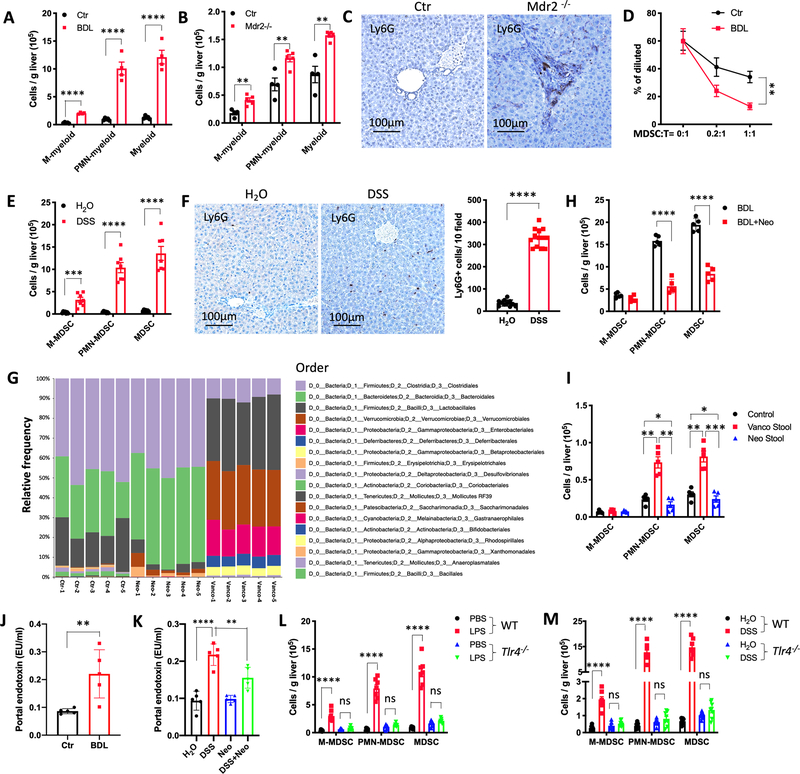
(B) BDL was performed in C57BL/6 mice. Two weeks later, duodenum, jejunum, ileum, cecum and colon were collected for immunochemistry (IHC). Representative IHC samples for Occludin are shown.
(C) BDL was performed in C57BL/6 mice. Two weeks later duodenum, jejunum, ileum, cecum and colon were collected for RT-PCR. The relative mRNA expression analysis for IL-1β, IL-17 and IFN-γ was performed. n=5 for Ctr and BDL. Data represent mean ± SEM. *p<0.05, **p<0.01, ****p<0.0001, two-way ANOVA.
(D) Quantitative real time PCR for relative 16s rRNA in portal vein blood of Ctr and BDL mice. n=5 for Ctr and BDL. Data represent mean ± SEM. ***p<0.001, Student’s t test.
(E) Quantitative real time PCR for relative 16s rRNA levels in portal vein blood of Ctr and Mdr2−/− mice. n=5 for Ctr and Mdr2-/−. Data represent mean ± SEM. **p<0.01, Student’s t test.
(F) Bile duct ligation (BDL) was performed in C57BL/6 mice. Two weeks later, BDL and control (Ctr) mice received 440mg/kg body weight FITC-Dextran by oral gavage. Four hours later, blood was collected. The concentration of FITC-Dextran was measured in blood. n=5 for Ctr and BDL. Data represent mean ± SEM. ****p<0.0001, Student’s t test.
(G) Ten-week old VBN/J control (Ctr) and Mdr2−/− mice received 440mg/kg body weight FITC-Dextran by oral gavage. Four hours later, blood was collected. The concentration of FITC-Dextran was measured. n=5 for Ctr and Mdr2-/−. Data represent mean ± SEM. ***p<0.001, Student’s t test.
Inflammatory bowel disease is well-known to have impaired intestinal barrier function, so we decided to extend our gut microbiome-liver studies to a well-established murine colitis model induced by dextran sulfate sodium (DSS)(29). Similar to mice with PSC like lesions, FITC-dextran concentrations in serum (Supplementary Fig. S2D) and bacterial 16sRNA in portal vein blood (Supplementary Fig. S2E) were higher in mice with colitis than in controls. Consistently, bacterial growth was detected in liver tissues and mesenteric lymph nodes (MLN) of DSS-colitis mice but not in liver tissues from control mice (Supplementary Figs. S2F and S2G). Different strains including Lactobacillus species were detected in the livers of mice with DSS induced colitis (Supplementary Fig. S2H). Bacterial translocation to liver was further confirmed using fluorescence in situ hybridization (FISH) using a probe specific for bacterial DNA (EUB338) (Supplementary Fig. S2I). These results demonstrate that both PSC and colitis cause impaired intestinal barrier function which permits microbes and their products to enter the portal circulation and enter the liver.
Commensal Gram-negative gut bacteria cause accumulation of hepatic MDSC through a TLR4-dependent mechanism
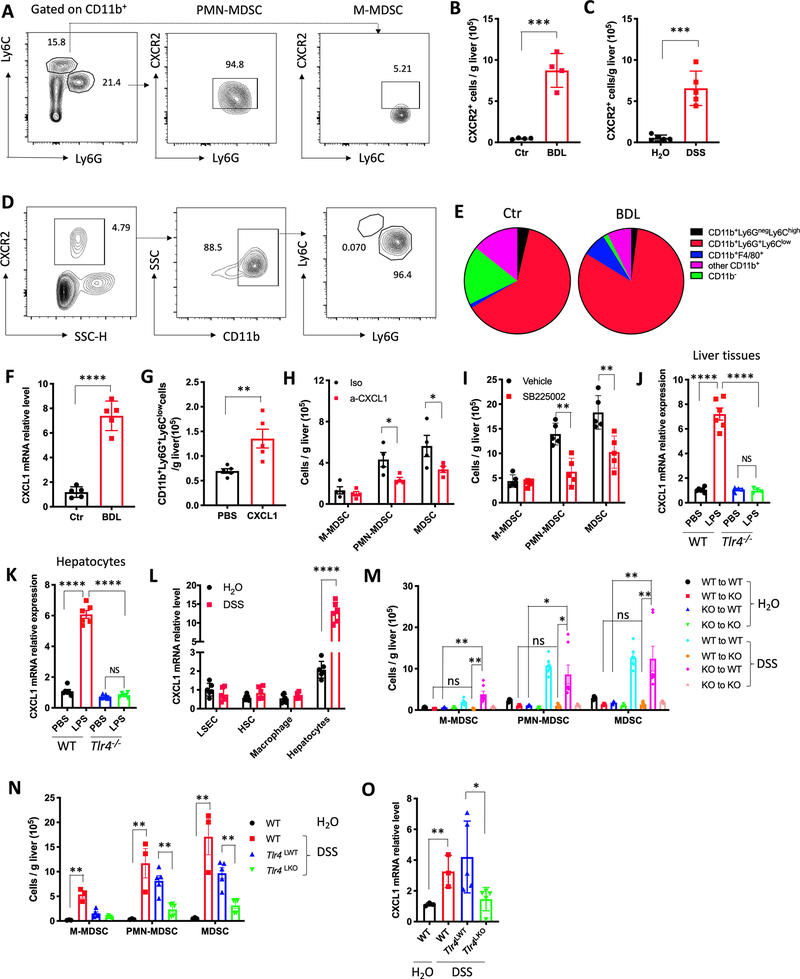
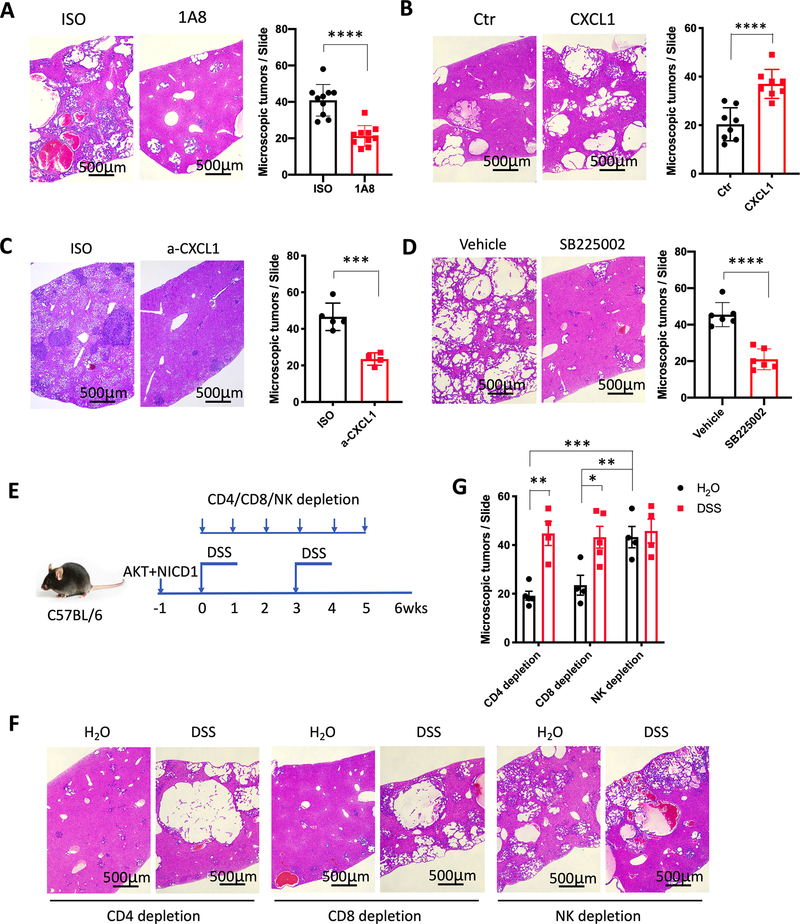
Next, we studied the immune cell subsets in livers of mice with PSC-like lesions. BDL caused a robust accumulation of hepatic CD11b+Gr-1+ myeloid cells compared to other immune cells (Fig. 2A and Supplementary Figs. S3A and S3B). CD11b+Gr-1+ myeloid cells consisted mainly of CD11b+Ly6G+Ly6Clow polymorphonuclear myeloid cells and to a lesser extent of CD11b+Ly6GnegLy6Chigh monocytic cells. In Mdr2−/− mice, the increase of hepatic CD11b+Ly6G+Ly6Clow cells was also found by flow cytometry (Fig. 2B and Supplementary Fig. S3C). Corroborating these findings, immunohistochemical analysis showed an increase of Ly6G+ cells in the livers of Mdr2−/− mice (Fig. 2C). To assess whether the PSC-like lesions induced myeloid cells exhibited immunosuppressive function, we isolated hepatic CD11b+Gr1+ myeloid cells and tested their ability to suppress CD8+ T cell proliferation in vitro. Indeed, CD8+ T cell proliferation was inhibited by BDL-PSC-recruited hepatic myeloid cells (Fig. 2D), proving that these CD11b+Gr-1+ cells were MDSCs. In addition, fewer CD69+CD8+ and Granzyme B+CD8+ hepatic T cells were seen in mice with PSC-like lesions (Supplementary Figs. S3D and S3E). PMN-MDSC depletion using 1A8 antibody significantly enhanced the frequency of TNF-α+CD8+T cells, IFN-γ+CD8+ T cells, and CD69+CD8+ T cells in liver tissues (Supplementary Fig. S3F). Finally, we studied livers from mice with DSS-induced colitis and observed similar results (Figs. 2E and 2F and Supplementary Fig. S3G). Accumulation of MDSC in mice after BDL ligation and Mdr2−/− mice was not limited to the liver and was also observed in spleen, gut and lung (Supplementary Figs. S3H and S3I), and in mice after BDL ligation increased bacterial 16sRNA in lung tissues were found (Supplementary Fig. S3J).
Figure 2.
LPS/TLR4 induces MDSC accumulation in the liver.
(A) Two weeks after BDL, the absolute number of hepatic M-myeloid, PMN-myeloid and total myeloid were determined in BDL and control (Ctr) mice. n=4 for Ctr and BDL. Data represent mean ± SEM. **** p<0.0001, two-way ANOVA.
(B) The absolute numbers of hepatic M-myeloid, PMN-myeloid and total myeloid were determined in FVBN/J control (Ctr) and Mdr2−/− mice at the age of 10 week. n=4 for Ctr, 5 for Mdr2-/−. Data represent mean ± SEM. ** p<0.01, two-way ANOVA.
(C) Immunochemistry of Ly6G in liver of Ctr or Mdr2−/− mice at the age of 10 week.
(D) Inhibition of T cell proliferation by myeloid cells was assessed by flow cytometry of T cells in co-culture experiments. Hepatic CD11b+Gr-1+ myeloid cells were purified from control (Ctr) or BDL mice. Splenic T cells (T) were isolated from normal C57BL/6 mice. T cells were labeled with CFSE and activated using anti-CD3ε/ anti-CD28. Myeloid cells and T cells were co-cultured at different ratios. The percentage of diluted CFSE after 72 hours co-culture was measured by FACS. Data represent mean ± SEM. ** p<0.01, two-way ANOVA.
(E) The absolute numbers of hepatic M-MDSC, PMN-MDSC and total MDSC were determined in C57BL/6 mice after 1 cycle of DSS treatment (2.5% DSS in drinking water for 1 week, followed by regular water for 2 weeks). n=6 for H2O, 7 for DSS. Data represent mean ± SEM. **** p<0.0001, two-way ANOVA.
(F) Immunochemistry of Ly6G in H2O or DSS treated liver tissues. Ly6G+ cells in each field were counted by a blinded investigator. n=11 for H2O, 13 for DSS. Data represent mean ± SEM. **** p<0.0001, Student’s t test.
(G) C57BL/6 mice were treated with H2O (Ctr), Vancomycin (Vanco), or Neomycin for 3 weeks before stool samples were collected for 16s rRNA sequencing. n=5 for Ctr and BDL. Bar plots of the order levels in BDL and Ctr mice are shown. Relative abundance is plotted for each mouse.
(H) C57BL/6 mice received Neomycin for 2 weeks prior to BDL (BDL+Neo). Two weeks after BDL, the absolute numbers of hepatic M-MDSC, PMN-MDSC and MDSC were determined. n=5 for BDL and BDL+Neo. Data represent mean ± SEM. **** p<0.0001, two-way ANOVA.
(I) C57BL/6 germ free mice were colonized with stool samples from mice treated with vancomycin (Vanco Stool) or neomycin (Neo Stool) for 3 weeks by oral gavage. Two weeks later, mice were sacrificed and the absolute numbers of hepatic M-MDSC, PMN-MDSC and MDSC were determined. n=5 for Control, Vanco stool, and Neo stool. Data represent mean ± SEM. *p<0.05, **p<0.01, ***p<0.001, two-way ANOVA.
(J) Bile duct ligation (BDL) was performed in C57BL/6 mice. Two weeks later, the concentration of endotoxin in portal vein was detected. n=5 for Ctr and BDL. Data represent mean ± SEM. **p<0.01, Student’s t test.
(K) C57BL/6 mice received H2O, DSS, Neo, and DSS+Neo for 7 days. The concentration of endotoxin in portal vein was detected. n=5 for H2O, DSS, Neo, and DSS+Neo. Data represent mean ± SEM. **p<0.01, ***p<0.001, one-way ANOVA.
(L) PBS or 2.5mg/kg LPS was i.p. injected in Tlr4−/− or C57BL/6 wildtype (WT) mice. Three days later, the absolute numbers of M-MDSC, PMN-MDSC and total MDSC were determined. Data represent mean ± SEM. n=6 for PBS and 7 for LPS in WT mice, n=6 for PBS and LPS in Tlr4−/− mice. ns, no significant. ****p<0.0001, two-way ANOVA.
(M) WT or Tlr4−/− C57BL/6 mice were treated with H2O or 2.5 % DSS for 1 cycle. The absolute numbers of M-MDSC, PMN-MDSC and total MDSC were determined. Data represent mean ± SEM. n=7 for H2O and DSS in WT mice, n=7 for PBS and 8 for LPS in Tlr4−/− mice. ns, no significant. ****p<0.0001, two-way ANOVA.
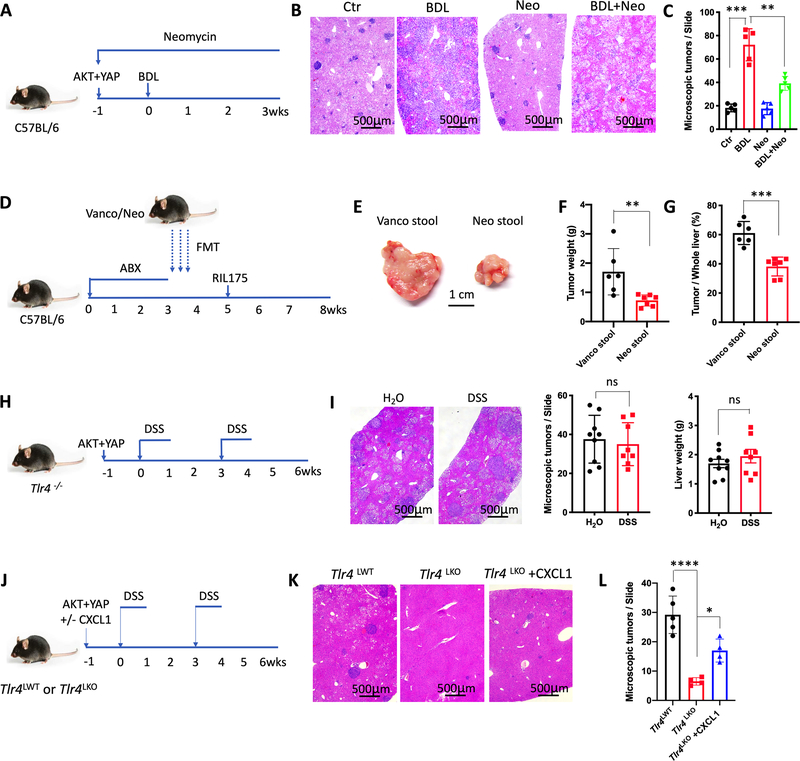
Next, we studied the effects of commensal gut bacteria on the accumulation of hepatic MDSC using neomycin and vancomycin treatment to selectively target Gram-negative and Gram-positive bacteria, respectively. As expected, vancomycin and neomycin treatment altered commensal gut bacteria (Fig. 2G), and neomycin treatment impaired the accumulation of hepatic PMN-MDSC in BDL mice (Fig. 2H and Supplementary Fig. S3K). Since PMN-MDSC were the major subset, total MDSC were also reduced after neomycin treatment (Fig. 2H and Supplementary Fig. S3K). Similar results were obtained in neomycin treated mice with DSS-induced colitis (Supplementary Figs. S3L and S3M), demonstrating that Gram-negative bacteria depletion by neomycin prevented MDSC accumulation in the liver. To study the immediate effects of neomycin or vancomycin-treated gut microbiome on hepatic MDSC, fecal microbiota transplantation (FMT) was performed. Germ-free (GF) mice were fed with cecum stool derived from either neomycin-treated specific pathogen free (SPF) mice (Neo-Stool, most Gram-positive bacteria left), or vancomycin-treated SPF mice (Vanco-Stool, most Gram-negative bacteria left) (Supplementary Fig. S3N). Oral gavage with Vanco-Stool significantly increased hepatic PMN-MDSC, while Neo-Stool had no effect (Fig. 2I and Supplementary Fig. S3O). Portal LPS concentration in GF mice were higher in mice after FMT with stools samples derived from mice treated with vancomycin than with neomycin (Supplementary Fig. S3P). These results indicated that Gram-negative bacteria promote MDSC accumulation in the liver.
We next determined the portal blood concentration of LPS which is a major component of Gram-negative bacteria. BDL increased LPS concentration in portal vein blood (Fig. 2J). In accordance with results from prior studies (30) higher portal LPS concentrations were also seen in mice after DSS treatment (Fig. 2K), which was reversed by neomycin treatment (Fig. 2K). Furthermore, LPS i.p. challenge was sufficient to induce an accumulation of hepatic MDSC cells in wild type (WT) mice (Fig. 2L and Supplementary Fig. S3Q). Toll-like receptor 4 (TLR4) is the main receptor for LPS (31). Therefore, we studied its role in MDSC accumulation in more detail. Fig. 2L and Supplementary Fig. S3Q showed that TLR4 deficiency (Tlr4−/−) completely reversed the accumulation of hepatic CD11b+Gr1+ MDSC cells upon LPS challenge. Similarly, colitis-induced MDSC accumulation was abrogated in Tlr4−/− mice (Fig. 2M and Supplementary Fig. S3R). These data indicate that neomycin-sensitive Gram-negative bacteria control MDSC frequencies in the liver via LPS and TLR4 even in the absence of specific liver disease or colitis.
Hepatocytes mediate MDSC accumulation via LPS/TLR4/CXCL1
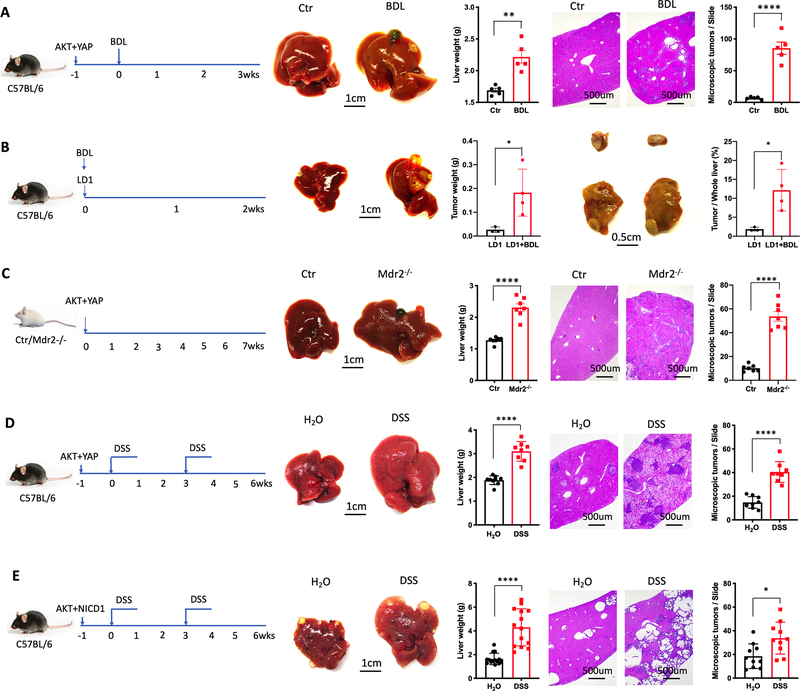
CXCR2 is a receptor crucial for neutrophil recruitment to inflammatory sites (32) and MDSC accumulation (33). In C57BL/6 mice, approximately 95% of CD11b+Ly6G+Ly6Clow polymorphonuclear myeloid cells expressed CXCR2, while approximately 5% CD11b+Ly6G−Ly6Chigh monocytic myeloid cells were CXCR2 positive (Fig. 3A). BDL and DSS treatment caused a significant increase of hepatic CXCR2+ cells (Figs. 3B and 3C and Supplementary Figs. S4A and S4B). PMN-MDSC accounted for around 85% of hepatic CXCR2+ leukocytes (Fig. 3D), and the percentage of PMN-MDSC in CXCR2+ cells increased after BDL and DSS treatment (Fig. 3E and Supplementary Fig. S4C). CXCL1 is the main ligand of CXCR2 in mice (34). Therefore, we studied CXCL1 expression in the liver of mice with PSC-like lesions and colitis. CXCL1 mRNA level in livers of BDL, Mdr2−/− and DSS-colitis mice increased significantly (Fig. 3F and Supplementary Figs. S4D and S4E) and CXCL1 over-expression in the liver led to an accumulation of hepatic PMN-MDSC (Fig. 3G and Supplementary Fig. S4F). As expected, CXCL1 antibody neutralization decreased hepatic PMN-MDSC as well as total MDSC (Fig. 3H and Supplementary Fig. S4G), showing that CXCL1 upregulation mediates accumulation of hepatic PMN-MDSC in mice with PSC-like lesion and colitis. Similar results were obtained when SB225002, a potent and selective CXCR2 inhibitor (35), was used in BDL-induced PSC-like lesions and DSS-induced colitis model (Fig. 3I and Supplementary Figs. S4H and S4I). Together, these data indicate that the CXCL1/CXCR2 axis regulates PMN-MDSC accumulation in the liver.
Figure 3.
Hepatocytes mediate MDSC accumulation via LPS/TLR4/CXCL1.
(A) Representative CXCR2 staining of hepatic M-MDSC and PMN-MDSC cells from three independent experiments.
(B) Absolute numbers of CXCR2+ cells in liver of control and BDL mice. Data represent mean ± SEM. n=4 for Ctr and BDL. ***p<0.001, Student’s t test.
(C) Absolute numbers of CXCR2+ cells in liver of H2O and DSS treated mice. Data represent mean ± SEM. n=5 for H2O and DSS. ***p<0.001, Student’s t test.
(D) Representative flow cytometry analysis of CXCR2+ infiltrating mononuclear cells in liver from three independent experiments.
(E) Composition of CXCR2+ infiltrating mononuclear cells in liver of control or BDL mice. Data represent pooled results from three experiments.
(F) CXCL1 mRNA expression levels were detected in liver tissues of control or BDL mice. n=5 for Ctr and BDL. Data represent mean ± SEM. ****p<0.0001, Student’s t test.
(G) Hydrodynamic injection of CXCL1 plasmid in C57BL/6 mice. The absolute numbers of PMN-MDSCs was determined 7 days later. n=5 for PBS and CXCL1. Data represent mean ± SEM. **p<0.01, Student’s t test.
(H) C57BL/6 mice were treated with 2.5% DSS for 7days. CXCL1 neutralization antibody or isotype control at 4 mg/kg was injected i.v. on day 5, 7 and 9. The mice were sacrificed at day 11. The absolute numbers of M-MDSC, PMN-MDSC and total MDSC were determined. Data represent mean ± SEM. n=4 for Iso and a-CXCL1. *p<0.05, two-way ANOVA.
(I) BDL was performed in C57BL/6 mice and the mice were treated with vehicle or SB225002 (10mg/kg, i.p. every other day). The absolute numbers of hepatic M-MDSC, PMN-MDSC and total MDSC was determined. n=5 for Vehicle and SB225002. Data represent mean ± SEM. ** p<0.01, two-way ANOVA.
(J) 2.5mg/kg LPS or saline was injected i.p. into Tlr4−/− or wild type (WT) mice. Three days later, CXCL1 mRNA levels in whole liver tissues were detected by real-time PCR. n=6 for each group. Data represent mean ± SEM. ns, no significant. ****p<0.0001, one-way ANOVA.
(K) Hepatocytes were isolated from Tlr4−/− or wild type (WT) mice. Then, hepatocytes were incubated with 100ng/ml LPS overnight. CXCL1 mRNA levels in hepatocytes was determined by real-time PCR. n=6 for each group. Data represent mean ± SEM. ns, no significant. ****p<0.0001, one-way ANOVA.
(L) C57BL/6 mice were treated with H2O or 2.5% DSS for 1cycle. Macrophage, LSEC, HSC and hepatocytes were isolated for RT-PCR to detect CXCL1 mRNA levels. n=5 for H2O and DSS. Data represent mean ± SEM. ****p<0.0001, two-way ANOVA.
(M) C57BL/6 mice (WT) or Tlr4−/− mice (KO) were lethally irradiated with 900 rad, followed by i.v. injection with 2×107 bone marrow cells from WT or KO mice. Six weeks later, the mice were treated with H2O or 2.5% DSS for 1cycle. Then, mice were sacrificed for M-MDSC, PMN-MDSC and total MDSC detection. n=3 for WT to WT+ H2O, WT to KO+ H2O, KO to WT+ H2O, KO to KO+ H2O, n=7 for WT to WT+ DSS, WT to KO+ DSS, KO to WT+ DSS, KO to KO+ DSS. Data represent mean ± SEM. ns, no significant. *p<0.05, **p<0.01, two-way ANOVA.
(N and O) C57BL/6 mice (WT) were treated with H2O or 2.5% DSS for 1cycle, Alb-Cre-; Tlr4 f/f (Tlr4LWT) and Alb-Cre+;Tlr4f/f (Tlr4LKO) mice were treated with 2.5% DSS for 1cycle. Then, mice were sacrificed for hepatic M-MDSC, PMN-MDSC and total MDSC detection (N) and CXCL1 mRNA levels in whole liver tissues (O). n=3 for WT+ H2O and WT+ DSS, n=5 for Tlr4LWT+DSS and Tlr4LKO+DSS. Data represent mean ± SEM. **p<0.01, two-way ANOVA.
Next, we studied the effect of LPS/TLR4 on CXCL1 expression in the liver. TLR4 deficiency abrogated the upregulation of CXCL1 mRNA expression both in whole liver tissue (Fig. 3J) and isolated hepatocytes (Fig. 3K) from mice subjected to LPS i.p. challenge, demonstrating that TLR4 on hepatocytes is essential for CXCL1 expression upon LPS challenge. Moreover, we found CXCL1 mRNA level significantly increased only in hepatocytes after DSS treatment but not in macrophages, liver sinusoidal endothelial cells (LSECs) or hepatic stellate cells (HSC) (Fig. 3L), indicating that hepatocytes are the main source of CXCL1 in this setting.
We next asked whether resident or bone marrow-derived/immune TLR4 is responsible for CXCL1 expression and PMN-MDSC accumulation in livers of TLR4-bone marrow chimeric mice. We first confirmed successful bone marrow transplantation by flow cytometry of CD45.1 and CD45.2 in peripheral blood (Supplementary Fig. S4J). No MDSC accumulation upon DSS treatment was seen in mice lacking Tlr4 in radioresistant cells (WT to KO), while MDSC significantly increased after DSS treatment in chimeric mice expressing Tlr4 in radioresistant cells (KO to WT) (Fig. 3M and Supplementary Fig. S4K). Next we tested mice lacking Tlr4 expression on hepatocytes (Tlr4LKO) by crossing Tlr4fl/fl mice with mice bearing an Albumin-Cre transgene (Alb-Cre) and found that DSS treatment could not induce hepatic MDSC accumulation in Tlr4LKO mice (Fig. 3N and Supplementary Fig. S4L). Similarly, hepatic CXCL1 mRNA expression was impaired in DSS treated Tlr4LKO mice (Fig. 3O). All these results demonstrated that TLR4 expression on hepatocytes was essential for LPS to induce CXCL1 expression in the liver.
PSC and colitis promote cholangiocarcinoma
PSC is an important risk factor for cholangiocarcinoma, and clinical data suggests that colitis was associated with cholangiocarcinoma (14). Using two well established murine cholangiocarcinoma models(36,37), we next tested whether our mouse models of PSC or colitis would promote cholangiocarcinoma. Plasmids encoding activated AKT and YAP or AKT and Notch1 were delivered by hydrodynamic injection, which resulted in formation of cholangiocarcinoma after 4 weeks (36,37). Microscopic analysis demonstrated tumor lesion with histological features of cholangiocarcinoma as previously described by others (Supplementary Figs. S5A and S5B)(38). Larger tumors were found in bile duct ligated mice upon AKT and YAP injection (Fig. 4A). In addition, we established cholangiocarcinoma cell line LD1 (Supplementary Fig. S5C). Copy number variation (CNV) sequencing of LD1 cell line indicated various mutations including AKT and YAP (Supplementary Table S1). Orthotopically implanted cholangiocarcinoma tumors grew faster in BDL-PSC mice (Fig. 4B). We also tested AKT and YAP delivery into Mdr2−/− mice and again noticed that PSC-like lesions promoted cholangiocarcinoma growth (Fig. 4C). Similar results were found in mice with colitis. More tumor burden was found in DSS treated mice upon hydrodynamic injection of either AKT and YAP (Fig. 4D) or AKT and Notch1 (Fig. 4E). Finally, accelerated cholangiocarcinoma growth was also observed in a second colitis mouse model (induced by 2,4,6-Trinitrobenzenesulfonic acid (TNBS) (Supplementary Fig. S5D) and in different mouse strains and different genders (Supplementary Figs. S5E and S5F). Together, these data indicate that both PSC and colitis promote cholangiocarcinoma growth in these mouse models.
Figure 4.
PSC and colitis promote cholangiocarcinoma.
(A) C57BL/6 mice were used to induce cholangiocarcinoma via hydrodynamic injection of AKT and YAP. One week later, BDL was performed on the mice. Mice were sacrificed 3 weeks after BDL. Representative liver images and H&E staining are shown. Microscopic tumors were counted. n=5 for Ctr and BDL. Data represent mean ± SEM. **p<0.01, **** p<0.0001, Student’s t test.
(B) Intrahepatic injection using 3×105 LD1 cell line were performed on C57BL/6 mice, followed by BDL. Two weeks later, the mice were sacrificed. Representative liver images and tumor images were shown. Tumor weight were detected. The ratio of tumor weight in whole liver weight was measured. n=4 for LD1 and LD1+BDL. Data represent mean ± SEM. *p<0.05, Student’s t test.
(C) FVBN/J (Ctr) or Mdr2−/− mice at 10 weeks were used to induce cholangiocarcinoma via hydrodynamic injection of AKT and YAP. Mice were sacrificed 7 weeks after injection. Representative liver images and H&E staining are shown. Microscopic tumors were counted. n=6 for Ctr and Mdr2-/−. Data represent mean ± SEM. **** p<0.0001, Student’s t test.
(D) C57BL/6 mice were used to induce cholangiocarcinoma via hydrodynamic injection of AKT and YAP. One week later, the mice were treated with 2.5% DSS in drinking water for 1 week, followed by regular water for 2 weeks (1 cycle). Mice were sacrificed after 2 cycles DSS treatment. Representative liver images and H&E staining are shown. Microscopic tumors were counted. n=9 for H2O, 8 for DSS. Data represent mean ± SEM. **** p<0.0001, Student’s t test.
(E) C57BL/6 mice were used to induce cholangiocarcinoma via hydrodynamic injection of AKT and NICD1. One week later, the mice were treated with 2.5% DSS in drinking water for 1 week, followed by regular water for 2 weeks (1 cycle). Mice were sacrificed after 2 cycles DSS treatment. Representative liver images and H&E staining are shown. Microscopic tumors were counted. n=18 for H2O,18 for DSS. Data represent mean ± SEM. *p<0.05, **** p<0.0001, Student’s t test.
Targeting PMN-MDSC reduces cholangiocarcinoma
Next, we investigated the contribution of MDSC/CXCL1/CXCR2 axis to cholangiocarcinoma progression. PMN-MDSC depletion by 1A8 antibody (Supplementary Fig. S6A) significantly reduced cholangiocarcinoma growth in mice with colitis (Fig. 5A). Hepatic CXCL1 overexpression by hydrodynamic delivery promoted cholangiocarcinoma growth (Fig. 5B) and was accompanied by an increase of hepatic PMN-MDSC (Supplementary Fig. S6B). Consistently, CXCL1 neutralization reduced cholangiocarcinoma burden progression in DSS-colitis mice (Fig. 5C) and decreased hepatic PMN-MDSC (Supplementary Fig. S6C). Similar results were obtained when SB225002 was used to pharmacologically block CXCR2 (Fig. 5D and Supplementary Fig. S6D).
Figure 5.
CXCL1/CXCR2/MDSC/NK axis regulates liver cancer development.
(A) AKT+ NICD1 tumor bearing C57BL/6 mice with DSS-colitis were treated with isotype control (ISO) or anti-Ly6G antibody (1A8) (200 μg, i.p. every other day). Microscopic tumors were counted after 2 cycles of DSS treatment. Data represent mean ± SEM. n=10 for ISO an 1A8. **** p<0.0001, Student’s t test.
(B) CXCL1 overexpression in cholangiocarcinoma was induced by hydrodynamic injection using AKT+NICD1+CXCL1 (CXCL1). Hydrodynamic injection using AKT+NICD1 was used as control (Ctr). Microscopic tumors were counted after 7 weeks. Data represent mean ± SEM. n=8 for Ctr and CXCL1. ****p<0.0001, Student’s t test.
(C) C57BL/6 mice were used to induce cholangiocarcinoma via hydrodynamic injection of AKT and YAP. Mice were treated with CXCL1 neutralization antibody (a-CXCL1) or isotype (ISO) control (4 mg/kg, i.v.) and sacrificed after 2 cycles DSS treatment. The microscopic tumors were counted. Data represent mean ± SEM. n=5 for Iso, 4 for a-CXCL1. ***p<0.001, Student’s t test.
(D) Cholangiocarcinoma was induced via hydrodynamic injection of AKT+ NICD1, followed by 2 cycles DSS treatment. Mice were treated with vehicle or SB225002 (10mg/kg, i.p. every other day). Microscopic tumors were counted. Data represent mean ± SEM. n=6 for Vehicle and SB225002. **** p<0.0001, Student’s t test.
(E, F and G) Cholangiocarcinoma was induced via hydrodynamic injection of AKT+ NICD1. Mice were treated with anti-CD4, anti-CD8 or NK depletion antibodies and killed after 2 cycles DSS treatment. Microscopic tumors were counted. n=5 for CD8 depletion in DSS-treated mice, 4 for some other groups. Data represent mean ± SEM. *p<0.05, **p<0.01, ***p<0.001, two-way ANOVA.
MDSC have been reported to promote liver cancer through inhibiting NK or T cell function (21,39). Next, we asked if NK or T cells mediated the cholangiocarcinoma-promoting function of MDSC. NK, CD4+ T, or CD8+ T cells were depleted respectively in cholangiocarcinoma-bearing mice with or without colitis (Fig. 5E, Supplementary Fig. S6E). Tumor volume in mice after depletion of CD4+ or CD8+ T cells with and without DSS treatment was similar to what we had seen before depletion (Fig. 4E). However, DSS treatment had no further effect on already increased tumor load in mice depleted of NK cells (Figs. 5F and 5G, Supplementary Fig. S6F), suggesting that MDSC, which were accumulated in mice after DSS treatment, could no longer promote iCCA without suppressing NK cell function.
Gut microbiome directs hepatocytes to control cholangiocarcinoma
The specific role of the gut microbiome on MDSC in the liver and tumor growth was further investigated. Neomycin treatment was used to eliminate Gram-negative bacteria in the gut, which resulted in fewer cholangiocarcinoma in BDL mice (Figs. 6A–C and Supplementary Figs. S7A and S7B) and DSS-colitis mice (Supplementary Figs. S7C–S7E). To directly address the effect of gut microbiome on liver tumor growth, we repeated the fecal transplant studies in gut sterilized mice using antibiotic treatment and monitored tumor growth of intrahepatic injected RIL175 tumor cells (Fig. 6D) in the absence of colitis or PSC-like lesions. Higher tumor burden was found in mice colonized with stool from mice treated with vancomycin (Figs. 6E–6G), which was accompanied by a higher number of liver infiltrating PMN-MDSC (Supplementary Fig. S7F). Interestingly, in the absence of PSC-like lesions or colitis, gut sterilization using an antibiotic cocktail (ABX, 0.5g/L vancomycin, 0.5g/L neomycin, and 0.5g/L primaxin) did not reduce cholangiocarcinoma growth (Supplementary Figs. S7G and S7H). The lack of effect was likely due to the low PMN-MDSC numbers in the absence of a leaky gut.
Figure 6.
Gut microbiome directs hepatocytes to control cholangiocarcinoma.
(A, B and C) Cholangiocarcinoma was induced by hydrodynamic injection of AKT+YAP and mice were treated with Neomycin. One week later, BDL was performed. Control (Ctr), BDL, neomycin (Neo) and BDL+ neomycin (BDL+Neo) mice were sacrificed 3 weeks after BDL (A). Representative H&E staining of livers are shown (B). Microscopic tumors (C) were counted. n=5 for each group. Data represent mean ± SEM. ***p<0.001, one-way ANOVA.
(D, E, F and G) C57BL/6 mice were treated with an antibiotics cocktail (0.5g/L vancomycin, 0.5g/L neomycin, and 0.5g/L primaxin) for 3 weeks, followed by oral gavage of cecum stool samples derived from mice treated for 3 weeks with Vancomycin (Vanco Stool) or Neomycin (Neo Stool). Two weeks later (Week 5), intrahepatic injection of 3×105 RIL175 cells was performed and mice were sacrificed at week 8 (D). Representative tumors are shown (E). Tumor weight (F) and the ratio of tumor in whole liver (G) are shown. n=6 for Vanco Stool, 7 for Neo Stool. Data represent mean ± SEM. *p<0.05, Student’s t test.
(H and I) Tlr4−/− mice were used to induce cholangiocarcinoma via hydrodynamic injection of AKT and YAP, then treated with H2O or DSS for 2 cycles (H). The microscopic tumors and liver weight (I) were determined. Data represent mean ± SEM. n=9 for H2O, 8 for DSS. ns, no significant, Student’s t test.
(J, K and L). Alb-Cre+;Tlr4f/f (Tlr4LKO) or Alb-Cre-;Tlr4f/f (Tlr4LWT) mice were used to induce cholangiocarcinoma via hydrodynamic injection of AKT+YAP, or AKT+YAP plus or without CXCL1. Mice were sacrificed after 2 cycles DSS treatment (J). Representative H&E staining are shown (K). Microscopic tumors were counted (L). n=5 for Tlr4LWT, 4 for Tlr4LKO and Tlr4LKO+CXCL1. Data represent mean ± SEM. *p<0.05, ****p<0.0001, two-way ANOVA.
Our study demonstrated that TLR4 expression on hepatocytes was critical for PMN-MDSC hepatic accumulation in leaky gut. The role of TLR4 in cholangiocarcinoma progression was tested. DSS-colitis failed to accelerate tumor growth upon AKT and YAP injection in Tlr4−/− mice (Figs. 6H and 6I) and no change in hepatic MDSC levels were seen (Supplementary Fig. S7I).Next we studied the specific role of TLR4 expression on hepatocytes and how changes in the gut microbiome affect hepatic MDSC. As expected, tumor growth was dramatically suppressed in mice lacking Tlr4 expression on hepatocytes (Tlr4LKO) after DSS treatment and injection with AKT + YAP (Figs. 6J–6L, Supplementary Fig. S7J). In contrast, CXCL1 overexpression (Supplementary Fig. S7K) rescued impaired tumor growth in Tlr4LKO mice (Figs. 6J–6L, Supplementary Fig. S7J). These results indicated that TLR4 expression on hepatocytes was important for cholangiocarcinoma tumor growth in the context of leaky gut. Together, these data demonstrate that targeting Gram-negative bacteria inhibits cholangiocarcinoma through a hepatocyte selective TLR4 signaling pathway, which leads to hepatic PMN-MDSC accumulation via CXCL1/CXCR2.
The gut microbiome from patients with cirrhosis affects myeloid cells in the liver.
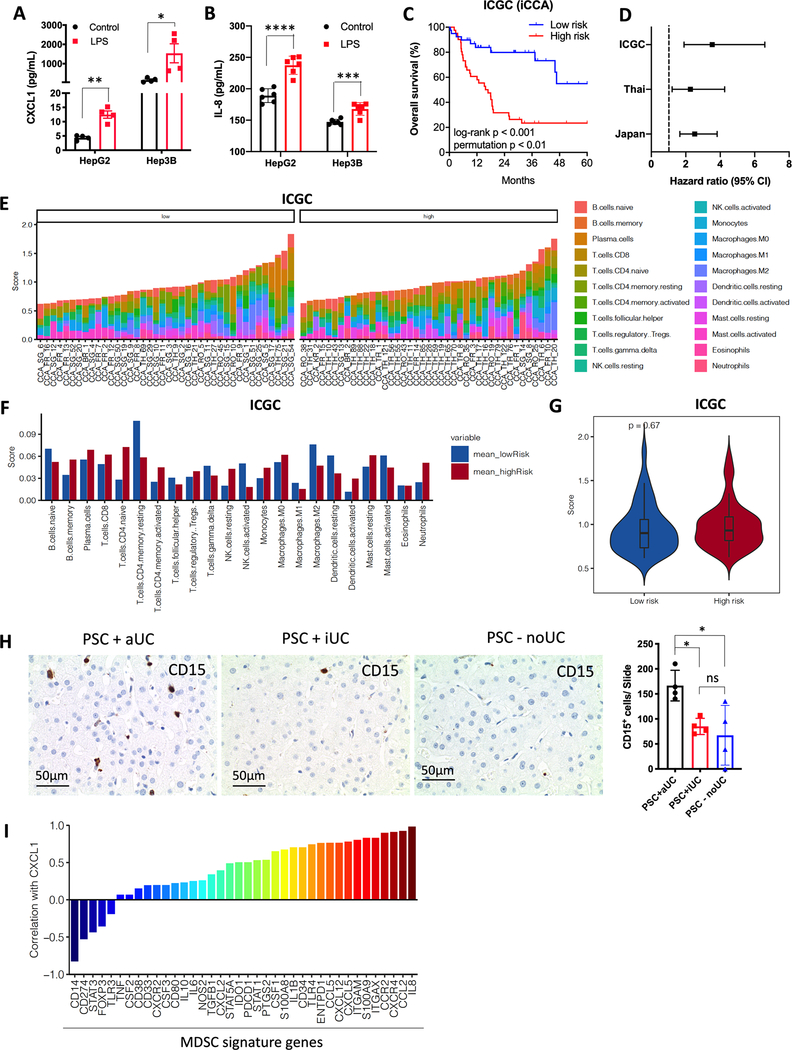
We conducted complimentary studies to assess the significance of our findings observed in animal models to the situation in patients. First, LPS was detected in liver samples from cirrhotic patients (Supplementary Fig. S8A) by immunohistochemistry. Next, the ability of LPS to induce the expression of CXCL1 and IL-8, an important CXCR2 ligand in humans, in human hepatic cell lines was tested. Similar to the observation in mice, LPS incubation induced a strong release of CXCL1 and IL-8 from both HepG2 and Hep3B cells (Figs. 7A and 7B). We also studied Tlr4 signature genes and patient outcome using bulk transcriptomic data from three cohorts of patients with iCCA (International Cancer Genome Consortium (ICGC)(40), Japan (https://www.ebi.ac.uk/ega/home) and Thailand(41)) (Supplementary Table S2). The Tlr4 pathway activity can be detected by using a panel of 153 Tlr4 signature genes (PathCards, pathway unification database) (Supplementary Table S3). BRB-Array analysis separated iCCA cases into two groups (low risk and high risk) according to Tlr4 gene signature expression levels (Supplementary Table S4). Patients with high Tlr4 gene signature expression displayed worse overall survival in all three cohorts (Fig. 7C, Supplementary Fig. S8B). The mean hazard ratio (95% CI) for the three cohorts were 3.54 (1.9–6.59, ICGC), 2.27 (1.21–4.26, Thai), and 2.52 (1.66–3.82, Japan) (Fig. 7D). We also applied CIBERSORT(42) to estimate the abundance of tumor infiltrating immune cell subsets in each tumor sample of the high-risk and low-risk groups of ICGC, Japan and Thailand cohorts based on the gene expression data (Fig. 7E, Supplementary Figs. S8C and S8D). Here, we did not find significant differences of each immune cell subset (Fig. 7F, Supplementary Figs. S8E and S8F) as well as the overall immune cell abundance (Fig. 7G, Supplementary Fig. S8G and S8H) in all three cohorts, suggesting that the high-risk and low-risk groups were not simply separated by the abundance of immune cells.
Figure 7.
TLR4 gene signatures are associate with poor survival of cholangiocarcinoma patients.
(A and B) Human HepG2 or Hep3B cell line was stimulated with 100ng/ml LPS overnight. CXCL1 (A) and IL-8 (B) concentrations in supernatant were determined by ELISA. Data represent mean ± SEM. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, one-way ANOVA.
(C) Total of 81 intrahepatic cholangiocarcinoma (iCCA) cases were divided into two groups (low-risk and high-risk) according to the expression of Tlr4 activation-associated genes. Log-rank (Mantel-Cox) test was performed to detect overall survival of the two groups. log-rank p value and permutation p value are provided.
(D) Hazard ratio with 95% confidence interval (CI) for the three cohorts ICGC, Thai, and Japan are shown.
(E, F and G) CIBERSORT was applied to estimate the abundance of different immune cells based on TLR4 gene expression level. The abundance of different immune cell in each iCCA patient are shown (E). The abundance of different immune cell in low-risk and high-risk iCCA patient are shown (F). The overall abundance of immune cells in low-risk and high-risk groups are shown (G).
(H) Immunochemistry of CD15 in liver tissues of patients with PSC+aUC, PSC+iUC, or PSC-noUC. The sum of CD15+ cells from 10 slides was calculated. Data represent mean ± SEM. *p<0.05, one-way ANOVA.
(I) The correlation of CXCL1 expression with MDSC signature gene was analyzed using the published GEO DataSets (GSE118373).
Next, we conducted two different studies to investigate a link between the gut microbiome and MDSC in patients. The level of hepatic myeloid cells in PSC patients was measured. We used a cohort of PSC patients with active ulcerative colitis (PSC+aUC), inactive ulcerative colitis (PSC+iUC) or without ulcerative colitis (PSC-noUC) (Supplementary Table S5). Myeloid cells in liver sections were detected by immunohistochemistry using CD15 staining. The presence of CD15+ cells in PSC with active colitis was significantly higher than in those PSC patients with inactive colitis or PSC patients without colitis (Fig. 7H), suggesting that active colitis in patients may increase the number of intrahepatic myeloid cells. In addition, we interrogated a gene expression data set derived from patients with PSC (GSE118373), and found that CXCL1 expression was positively correlated with most of the MDSC signature genes (43) (Fig. 7I and Supplementary Table S6) (44).
Finally, we decided to extend our studies beyond cholangiocarcinoma and PSC and asked whether stool samples derived from other patients with gut dysbiosis may affect intrahepatic MDSC. We performed fecal microbiota transplantation (FMT) into germ free mice using stool samples from patients with liver cirrhosis due to alcoholic hepatitis or healthy control (Supplementary Figs. S9A and S9B, Supplementary Tables S7 and S8). FMT from cirrhosis patients into germ-free mice increased (Myeloperoxidase) MPO+ myeloid cells and Ly6G mRNA expression level in the liver of mice (Supplementary Figs. S9C to S9E). An increase of CXCL1 expression was also observed but this change did not reach statistical significance (Supplementary Fig. S9F). In summary, a set of different human studies support our murine studies and demonstrate that the gut microbiome can induce myeloid cells accumulation in the liver, which are associated with worse clinical outcome in cholangiocarcinoma.
DISCUSSION
The gut microbiome has profound effects on systemic immune responses and it has been recognized as an important regulator of anti-tumor immunity (1,3). The portal vein, which drains blood from the small and large intestine delivers large amounts of commensal gut bacteria derived products to the liver (45). There are several studies suggesting that gut commensal bacteria promote development and growth of hepatocellular carcinoma (HCC) (46,47). Most of these studies focused on how the gut microbiome promotes carcinogenesis and malignant transformation of HCC. In contrast, there is only very limited knowledge on how the gut microbiome controls anti-tumor immunity in the liver and primary liver cancer. Here we studied how the gut microbiome may affect anti-tumor immunity in cholangiocarcinoma, the second most common type of liver cancer. We studied how two different well-known risk factors for cholangiocarcinoma, PSC and colitis, promote the development of cholangiocarcinoma and demonstrate that gut derived commensal bacteria are being exposed to the liver and cause an accumulation of immunosuppressive CXCR2+ PMN-MDSC in the liver ultimately promoting the growth of cholangiocarcinoma. We show that Gram-negative commensal bacteria induce TLR4-dependent CXCL1 production from hepatocytes which leads to an accumulation of CXCR2+PMN-MDSC. Interestingly, this mechanism can also be found in the absence of PSC or colitis using stool samples from patients with liver cirrhosis or mice treated with vancomycin. Thus, our study suggests that gut commensal bacteria can direct hepatocytes to form a tumor-promoting environment by recruiting PMN-MDSC.
Studying cholangiocarcinoma and risk factors for biliary cancer using animal models is complicated by the fact that cholangiocarcinomas (CCA) are classically sub-divided into three groups depending on the anatomical site of origin: intrahepatic CCA (iCCA), perihilar CCA (pCCA) and distal CCA (dCCA) (48). Currently, there is no ideal mouse model available for extrahepatic cholangiocarcinoma (49), so we decided to use two different iCCA models, which have previously been established by others. A recent meta-analysis identified inflammatory bowel disease and cirrhosis among others as risk factors for iCCA and extrahepatic CCA with an odds ratio of up to 15 (50). Primary sclerosing cholangitis is another well-known risk factor for iCCA (51,52). In the absence of a well-characterized and highly reproducible PSC animal model, in which mice will develop fibrous-obliterative cholangitis of the intra- and extrahepatic bile ducts in association with inflammation of the gut with predominant right-sided colitis and the development of cholangiocellular carcinoma (24), we decided to combine well established animal models for colitis and PSC with models for cholangiocarcinoma. As expected, both colitis and PSC promoted growth of cholangiocarcinoma significantly.
The contribution of MDSC to colitis associated tumorigenesis is well established (53,54) and gut derived commensal bacteria have emerged as important factors during initiation and promotion of colon cancer (55,56). Hepatic macrophages play a central role in the pathogenesis of chronic liver injury (57). Resident macrophages (Kupffer cells) recognize pathogen-associated molecular patterns and can promote the formation of the inflammasome (58). NLRP3 inflammasome activation was also described in Mdr2-associated cholestasis (59). We noticed a specific accumulation of PMN-MDSC in mice with colitis, PSC and upon fecal transplant of stool samples derived from vancomycin treated mice. PMN-MDCS were CXCR2+ in contrast to CD11b+F4/80+ macrophages or CD11b+Ly6GnegLy6Chigh M-MDSC. This promoted us to study CXCL1 and TLR4 expression in the liver. Unexpectedly, TLR4 expression, which has previously been described to also be expressed by hepatocytes (60,61), induced CXCL1 expression and an accumulation of PMN-MDSC much more than TLR4 on M-MDSC and macrophages.
Our studies show a TLR4 dependent accumulation of hepatic PMN-MDSC by Gram-negative commensal bacteria, but we did not investigate the role of other TLRs and therefore can only conclude that TLR4 is necessary but not necessarily sufficient for the responses observed and we cannot exclude other components in the microenvironment such as stroma or paracrine signaling may also contributed to enhanced tumor growth.
While we were able to demonstrate that commensal bacteria can be found in the liver of mice with colitis, and that transfer of stool samples can induce PMN-MDSC, it is important to note that it is not clear whether intact bacteria or bacteria derived components are necessary to induce CXCL1 expression in the liver.
It has previously been shown that PSC results in gut dysbiosis in patients (16,23), which prompted us to study in more detail the underlying mechanisms of how bile duct changes may cause an increase of LPS in portal vein blood leading to an accumulation of MDSC. It has been suggested that bile acid changes in bile acid composition found in PSC shape the gut microbiota (16). Here we show that PSC-dependent dysbiosis caused local gut barrier dysfunction associated with local inflammatory responses ultimately leading to a leaky gut and the ability of Gram-negative bacteria (Gram-negative bacteria derived products) to transmigrate in the portal vein blood. These observations are supported by recent findings from Nakamoto et al. (28) who demonstrated that Klebsiella pneumonia in the microbiota of patients with PSC disrupts the epithelial barrier to initiate bacterial translocation and liver inflammatory responses. Studies in Mdr2−/− mice indicated that intestinal dysbiosis amplifies the hepatic Nlrp3-mediated innate immune response (59) and activation of γ/δ T cells (62).
Although largely generated from PSC or colitis models, the finding that Gram-negative bacteria induce hepatic MDSC accumulation has more general application. Our results demonstrate that colonization of Gram-negative bacteria enriched stool in germ free mice is sufficient to increase hepatic MDSC, showing that the MDSC regulating mechanism is independent of PSC or colitis condition. Only hepatic PMN-MDSC, but not M-MDSC, increased in Gram-negative bacteria colonized germ-free mice, which could be explained by the different receptors on the two subsets (19,33). MDSCs have broad immunosuppressive functions and inhibit both adaptive and innate immune responses (18). Their tumor-promoting function is general and has been reported in many kinds of tumors (18). Indeed, our results show that the increased hepatic MDSCs not only promote cholangiocarcinoma, but also promote the growth of orthotopic RIL175 HCC tumor, suggest that our finding applies to all liver tumor types. Here we show that MDSC exert their immunosuppressive function leading to accelerated tumor growth, but it is possible that other components in the tumor microenvironment such as stroma or paracrine signaling may also promote tumor growth (63).
Dysbiosis and increased gut permeability are commonly presented in chronic gastrointestinal diseases (64). Increased blood LPS has been reported in patients with different chronic liver disease such as nonalcoholic fatty live disease and cirrhosis(65). In addition, patients with liver cirrhosis often present bacterial translocation (66). Our study suggests that gut microbiome induced hepatic MDSC may be a common contributor to liver tumor development in the context of chronic gastrointestinal diseases.
It has been demonstrated that gut microbiota promoted diethylnitrosamine (DEN)-hepatotoxin carbon tetrachloride (CCl4) induced HCC in a TLR4 dependent manner (67), indicating the essential role of Gram-negative bacteria/LPS/TLR4 in HCC promotion. In addition, Gram negative bacteria have also been shown to be crucial in infection and inflammation (68,69). Indeed, in cholangiocarcinoma, we used two PSC-like models (BDL and Mdr2−/−), two colitis models (DSS and TNBS), three cholangiocarcinoma models (AKT/YAP, AKT/Notch and intrahepatic injection of LD1) in three mouse strains and both genders, and demonstrated that PSC and colitis accelerate cholangiocarcinoma progression in mice. More importantly, Gram-negative bacteria depletion by neomycin treatment inhibited cholangiocarcinoma progression and hepatic PMN-MDSC accumulation. Colonization of Gram-negative bacteria into antibiotics cocktail (ABX)-treated mice promoted RIL175 HCC tumor growth. Therefore, Gram-negative bacteria is a potential therapeutic target for cholangiocarcinoma.
Our studies suggest that TLR4 expression on hepatocytes promotes growth of iCCA via recruiting PMN-MDSC. Interestingly, in TLR4−/− mice colitis had no effect on tumor growth and was similar to what was seen in TLR4 wildtype mice without colitis, suggesting that TLR4 signaling may also control tumor growth through MDSC independent pathways and TLR4’s role in cholangiocarcinoma may even be more complex. As a matter of fact, both pro-and anti-tumor functions have been reported and activation of TLR4 on dendritic cells has been found to improve antigen presentation and lead to better activation of cytotoxic T cells (70), whereas the opposite has been seen in (DEN)-hepatotoxin carbon tetrachloride (CCl4) induced HCC (67).
The principle of our finding also can be applied to humans. We detected LPS in livers of patients with liver cirrhosis, and LPS induces human hepatocyte cells to produce CXCL1. More CD15+ myeloid cells were found in PSC patients with active colitis compared to PSC patients with inactive colitis or without colitis. Of note these samples are extremely rare and we had to restrict our analysis to patients with primary sclerosing cholangitis. Complementary to that we studied three different cohorts of cholangiocarcinoma patients from Japan, Thailand and from the International Cancer Genome Consortium. Although this data lacks complete clinical data, we were able to show that a Tlr4 gene signature gene expression was associated with worse survival of cholangiocarcinoma patients.
Our understanding of how the gut microbiome controls immunological mechanism in cancer is largely derived from findings observed in mice. We are aware that animal models can never completely mimic the situation in a patient. Both PSC and CCA models used in this study have limitations: CCA induced by hydrodynamic injections arise in hepatocytes and not cholangiocytes (71). BDL as well as Mdr2−/− mice cause PSC-like lesions, which can only resemble certain aspects of PSC seen in patients (72). However, experiments done using human cells and gene expression data from patient cohorts described here clearly support our main conclusions derived from experiments in mice and only future clinical trials will answer how we can translate findings described here into therapeutic applications(73).
In summary, our study shows that Gram-negative bacteria/LPS controls hepatocytes to form an immunosuppressive microenvironment by inducing CXCR2+ PMN-MDSC accumulation through TLR4-dependent CXCL1 production, thus promote liver tumor growth (Supplementary Figure S10). The finding is not limited to PSC or colitis promoting cholangiocarcinoma, but also applies to other liver tumors with increased liver exposure to gut microbiome. Our study also suggests gut bacteria as a target for prevention and treatment of liver cancer with underlying chronic gastrointestinal conditions.
METHODS
Animal studies
C57BL/6 and BALB/c mice were purchased from Charles River. Mdr2−/− (Stock NO: 002539), FVBN/J (Stock NO: 001800), B6 CD45.1 (JAX NO: 002014), C57BL/10 (Stock NO: 000665), Albumin-Cre (Stock NO: 003574), Tlr4fl (Stock NO: 024872) were purchased from Jackson laboratory. Tlr4−/− mice were kindly provided by Dr. Giorgio Trinchieri (Cancer and inflammation program, NIH). Mice were randomly divided into 5 mice per cage. After one week, the mice were used for experiments. All experiments were conducted according to local institution guidelines and approved by the Animal Care and Use Committee of the National Institutes of Health, Bethesda, USA. Hydrodynamic injection was used to induce intrahepatic cholangiocarcinoma (74). For NICD+AKT cholangiocarcinoma, 20μg NICD, 4μg AKT and 1μg hyperactive sleeping beauty 2 (HSB2) transposase plasmids were diluted into 1.6ml PBS, and injected into tail vein within 5–7 seconds(36). For YAP+AKT cholangiocarcinoma, 30μg YAP, 20μg AKT and 2μg HSB2 were injected hydrodynamically(37). Mice were treated with indicated dose of Dextran Sulfate Sodium (DSS) for 7 days, and regular water for 14 days (1 cycle). Fresh DSS water was replaced every other day. For intrahepatic LD1 cholangiocarcinoma model, 3×105 LD1 cholangiocarcinoma cells were injected into liver. Two weeks later, the mice were sacrificed for further detection. Mice were treated with 2.5 mg/kg LPS (i.p., L2880, Sigma), 200 μg anti-CD4 (i.p. once a week, clone GK1.5, BioxCell), 200 μg anti-CD8 (i.p. once a week, clone 2.43, BioxCell), or 600 μg anti-NK (i.v. every other day, clone PK136, BioxCell) for depletion, 200 μg anti-Ly6G (i.p. every other day, clone 1A8, BioxCell), 4 mg/kg CXCL1 neutralization antibody (i.v. every other day, MAB453, R&D Systems) for neutralization, 10 mg/kg CXCR2 inhibitor (i.p. every other day, SB225002, Tocris Bioscience) for inhibition. At the end of experiments, mice were sacrificed to collect organs for RNA isolation, flow cytometry and histological analysis. All tumor measurements were performed by a blinded investigator.
Cell lines
Three human HCC cell lines HepG2, Hep3B and Huh7, one murine HCC cell line RIL-175(75), one murine B-cell lymphoma A20, one murine cholangiocarcinoma cell line LD1 were used in this study. HepG2, Hep3B, Huh7, A20 were purchased from ATCC. RIL-175 has been described previously(20). LD1 cholangiocarcinoma cell line in C57BL/6 background was established in our lab and is derived from a mouse after hydrodynamic injection with AKT and YAP plasmids. The sequencing data of LD1 was uploaded as SRA submission SUB8582522, BioProject accession PRJNA679802. All cell lines were used less than 12 passages before experiments were conducted.
Flow cytometry
For surface markers staining, cells were stained with antibodies for 15 mins at 4°C, followed by washing with flow cytometry buffer. For intracellular staining, cells were stained with Foxp3/transcription factor staining buffer (eBioscience) according to manufacturer’s instructions. Antibodies used in this study were as follows: anti-CD19-PerCP/Cy5.5 (clone eBio1D3, eBioscience), anti-CD3-FITC (clone 17A2, BD Pharmigen), anti-CD4-Alexa Fluor 700 (clone GK1.5, Biolegend), anti-CD8-Pacific Blue (clone 53–6.7, Biolegend), anti-TCRb-BV510 (clone H57–587, Biolegend), PBS57/CD1d-tetramer-APC (NIH core facility), anti-CD11b-Pacific Blue (clone M1/70, Biolegend), anti-Gr1-PerCP/Cy5.5 (clone RB6–8C5, Biolegend), anti-Ly6G-Alexa Fluor 700 (clone 1A8, Biolegend), anti-Ly6C-APC (cloneHK1.4, Biolegend), anti-F4/80-FITC (clone BM8, Biolegend), anti-CXCR2-PE (clone SA044G4, Biolegend), anti-CD69-Pacific blue (clone H1.2F3, Biolegend), anti-Granzyme B-FITC (clone GB11, Biolegend), anti-TNF-α-PE (clone MP6-XT22, Biolegend), anti-IFN-γ-APC (clone XMG1.2, Biolegend).The immune cell subsets were identified by markers: B cells: CD3−CD19+; Hepatic CD4+ T cells: CD3highCD4+; CD8+ T cells: CD3+CD8+; NKT cells: TCRb+CD1d-Tetramer+, PMN-MDSC: CD11b+Ly6G+Ly6Clow; M-MDSC: CD11b+Ly6G−Ly6Chigh; Macrophage: CD11b+F4/80+. The absolute number of immune cells calculated by multiplying frequency by the total live cells, then divided by liver weight.
Microscopic tumor analysis
Liver tissues were fixed in formalin overnight, followed by fixation in 70% alcohol. Three liver tissues with similar size from the same part of the liver were fixed on 1 slide. After Hematoxylin and eosin (H&E) staining, microscopic tumors in each slide were calculated.
Endotoxin assay
Portal vein blood was collected in endotoxin free tubes. After centrifugation at 10,000g for 5 mins, the plasma was used to detect endotoxin concentration. ToxinSensor™ chromogenic LAL endotoxin assay kit (L00250, GenScript) was used according to the manufacturer’s instructions. All determinations were performed in duplicates.
Gut permeability assay
Mice were administrated with FITC-dextran (4KDa, Sigma) at the dose of 440mg/ kg body weight 4 hours after fasting. After 4 hours of the administration, blood was collected by cardiac puncture. The concentration of FITC in serum was detected by fluorescence plate reader at excitation 485nm/emission 528nm. Serum from mice not administrated with FITC-dextran was used as control.
Bacteria translocation
Liver tissues were aseptically harvested from BDL-induced PSC mice, Mdr2−/− mice and normal C57BL/6 mice treated with or without DSS. The cell suspension of liver was plated on LB medium without any antibiotics, then incubated at 37°C for 24 hours. Colony Formation Units (CFU) were counted and calculated as per milligram of tissue. For identification of bacteria, the liver from DSS-treated or control mice was homogenized and plated onto 5% sheep blood, chocolate, MacConkey agar, and CDC anaerobic agar (Remel, Inc.) and Fastidious Broth (Hardy Diagnostics), then incubated at 35°C in 5% CO2 or in anaerobic conditions, as appropriate. Pure subculture isolates were identified by MALDI-TOF mass spectrometry (BioTyper; Bruker, Billerica, MA).
Bacteria colonization
C57BL/6 mice in SPF animal room were treated with vancomycin (0.5g/L) or neomycin (0.5g/L) for 3 weeks. Then, the cecum stool from 5 mice were collected into 10ml anaerobic sterile glycerol. Germ free mice in C57BL/6 background were performed oral gavage with 200ul cecum stool solution in anaerobic sterile glycerol. Two weeks later, the germ-free mice were sacrificed for further detection. Stool samples from two cirrhotic patients and one healthy donor were used for fecal transplantation in germ-free mice. Mice were garaged with 100 ul stool sample (1 g stool dissolved in 30 ml Luria-Bertani (LB) medium containing 15% glycerol under anaerobic conditions) at week 5–6 and repeated two weeks later. Two weeks after the second gavage, mice were placed on the control liquid diet for 15 days as described (76).
16s rRNA sequencing
DNA extraction and amplification were performed using Eppendorf liquid handling robots. The V4 region of the 16S rDNA gene (515F-806R) was sequenced for 10 samples for Fig 1A, and 15 samples for Fig 2G; generating paired-end, overlapping reads on the Illumina MiSeq platform (77). The demultiplexed paired end fastq files were pre-processed and analyzed using QIIME 2 version 2–2020.2 (https://qiime2.org) (78). The DADA2 algorithm (79), implemented in QIIME2, was used for error modelling and filtering the raw fastq files. Post denoising and chimera removal; a total of 573,739 sequences was retained for 10 samples with an average of 57,373 sequences per sample for Fig 1A, a total of 835,408 sequences was retained for 15 samples with an average of 55,693 sequences per sample for Fig 2G. Taxonomic classification was performed using the QIIME2 feature-classifier (https://github.com/qiime2/q2-feature-classifier) plugin trained on the Silva 132 database (80). The Alpha and Beta-diversity analyses were performed using the diversity plugin (https://github.com/qiime2/q2-diversity) at a rarefied sampling depth of 50000 for Fig 1A, and at a rarefied sampling depth of 49700 for Fig 2G. The Bioproject accession number of the 16s rRNA sequencing for mouse stools is PRJNA680370.
DNA extraction and 16s rDNA sequencing for human samples (Supplementary Fig. S9A) were performed as described(76). Raw 16s rRNA sequencing reads of human stool samples can be found in the NCBI SRA associated with Bioproject PRJNA517994 (Cirrhosis) and PRJNA525701 (Healthy), under the following BioSample IDs: SAMN11083186 (Healthy), SAMN10856936 (Cirrhosis #1) and SAMN10856982 (Cirrhosis #2).
Human cohort
Patient cohorts have been described (76). Patients with cirrhosis due to alcoholic hepatitis were enrolled from the InTeam Consortium (ClinicalTrials.gov identifier number: NCT02075918). Inclusion and exclusion criteria have been published(76). Liver biopsies were done if clinically indicated as part of routine clinical care for diagnostic purposes. The baseline characteristics are shown in Supplementary Table S7. The protocol was approved by the Ethics Committee of each participating center and patients were enrolled after written informed consent was obtained from each patient.
Fluorescence in situ hybridization (FISH)
Bacteria in liver tissues was visualized using FISH. Liver tissues were fixed in 4% paraformaldehyde overnight at 4°C, followed by washing with PBS and dehydrated in 15% sucrose overnight at 4°C. Then, the samples were embedded in OCT compound. The slides with frozen samples will be applied to probes at the concentration of 2 pmol/ul in prewarmed hybridization buffer (900 mM NaCl, 20 mM Tris pH 7.5, 0.01% SDS, 20% formamide). Next, the slides will be incubated at 46°C in a humid chamber for 2 hours and washed at 48°C for 15 minutes in wash buffer (215 mM NaCl, 20 mM Tris pH 7.5, 5 mM EDTA). Then, the slides will be dipped in water, then in 100% ethanol, air-dried, and coverslips were mounted using ProLongGold antifade reagent (Life Technologies). At last, slides were analyzed using confocal microscopy. The probe used to detect bacteria in liver is EUB338-Alexa 488: all bacteria 16S rRNA GCTGCCTCCCGTAGGAGT(81).
CXCL1 overexpression in mice
To perform CXCL1 overexpression, 20μg CXCL1 overexpression plasmid (Cat no: MG50150-UT, Sino Biological) was hydrodynamically injected into mice. In tumor free mice, the mice were sacrificed for further determination 7 days later. In tumor bearing mice, the mice were sacrificed for further determination 7 weeks later.
Quantitative reverse transcription PCR (RT-qPCR)
Total RNA from whole liver tissues, hepatocytes, liver sinusoidal endothelial cell, macrophages and hepatic stellate cells was isolated using RNeasy Mini Kit (Cat. NO. 74104, Qiagen) according to manufacturer’s instructions. cDNA synthesis was performed using iScript™ cDNA synthesis kit (Cat. NO. 170–8891, BIO-RAD). For germ-free mice colonized with human stool samples, RNA was extracted from liver tissues using TRIzol reagent (Cat NO. 15596018, Thermo Fisher) and cDNA was generated with High Capacity cDNA Reverse Transcription kit (Cat. NO. 4368814, Thermo Fisher). RT-PCR was performed using iQ SYBR Green Supermix (Cat. NO. 1708882, BIO-RAD). The primers were used for RT-PCR: CXCL1: Forward, 5’-CTT GAA GGT GTT GCC CTC AG-3’, Reverse, 5’-AAG GGA GCT TCA GGG TCA AG-3’; Ly6G: Forward, 5’- ACA CAA CTA CCT GCC CCT TC-3’, Reverse, 5’- CAG ATG GGA AGG CAG AGA TT-3’. GAPDH: Forward, 5’-CCT GCA CCA CCA ACT GCT TA-3’, Reverse 5’-TCA TGA GCC CTT CCA CAA TG-3’. The relative expression levels of CXCL1 were determined by 2-^^CT methods. GAPDH expression level was used as control.
Western Blot
Whole cell lysates were obtained by adding ice-cold lysis buffer (Mammalian Protein Extract Reagent, Thermo Fisher Scientific) containing protease inhibitors (Halt Protease and Phosphatase Inhibitor, Thermo Scientific). The lysed cells were then centrifuged at 12,000 rpm for 20 minutes to remove cellular debris. Protein concentration of supernatant was determined by the Pierce BCA Protein Assay (Thermo Fisher Scientific). Proteins were then resolved by SDS-PAGE electrophoresis and transferred to PVDF membranes (BioRad). Membranes were incubated with primary antibodies at 4°C overnight in 2% nonfat milk in TBS-Tween. The primary antibody dilution was 1:1000 unless otherwise indicated. The following primary antibodies were used for immunoblotting analysis: CK-7 (ab181598) and CK-19 (ab52625) from Abcam (Cambridge, MA), β-Actin (4970S) from Cell Signaling Technology (Danvers, MA). After overnight incubation, membranes were washed in TBS-Tween and then corresponding horseradish peroxidase conjugated secondary antibodies were added to membrane at a concentration of 1:2000 and incubated for 1 hour at room temperature. Immunoblots were visualized with enhanced chemiluminescence reagents (Clarith/Clarity Max ECL, BioRad).
Immunohistochemistry
Immunohistochemistry was carried out using 5μm paraffinembedded sections of mouse or human liver tissues using the universal animal IHC kit (Lot: KT4418A, Innovex) according to the manufactureŕs instructions. The primary antibody of anti-CK19 (Abcam, ab52625), anti-HNF-4α (Santa Cruz, sc-374229), anti-Ly6G (clone 1A8, BioxCell for Figs. 2D and 2F, BD Biosciences for supplementary Fig. 2G), anti-MPO (Biocare Medical, Catalog Number: PP023AA), anti-CD15 (DAKO, M3631), anti-Lipopolysaccharide Core, mAb WN1 222–5 (HycultBiotech, Cat#HM6011; RRID:AB_2750644) was used. ImageJ software was used to count the positive staining cells.
ELISA
Total of 5X105 human hepatocyte cell line HepG2 or Hep3B was seeded into 6-well plate with 100ng/ml LPS (L2880, Sigma) challenge overnight. CXCL1 and IL-8 concentration in supernatant were determined using Human GRO alpha ELISA Kit (Invitrogen, REF: BMS2122) and human IL-8 ELISA Kit (Invitrogen, Catalog #: KHC0081) respectively according to the manufactureŕs instructions.
T cell proliferation assay
Hepatic MDSC were isolated using myeloid-derived suppressor cell isolation kit (Order no: 130–094-538, Miltenyi) according to the manufacturer’s instructions. T cells from spleen were labeled with CFSE and stimulated using T cell activation/expansion kit (Order no: 130–093-627, Miltenyi). Total of 1X105 CFSE-labeled splenic T cells were seeded into 96-well plate, and co-cultured with MDSC with different ratios (MDSC:T=0:1, 02:1, 1:1). Seventy-two hours later, diluted CFSE+ CD8+ T cells were measured as proliferated T cells using flow cytometry.
Isolation of lymphocytes from gut
Lymphocytes isolation from gut was performed as previously described (82). Briefly, tissue segments were incubated in 30 ml of extraction media (30 ml RPMI + 93 μl 5% (w/v) dithiothreitol (DTT) + 60 μl 0.5 M EDTA + 500 μl fetal bovine serum (FBS)) for 15 min at 37 °C. Minced tissues to 25 ml of digestion media (25 ml RPMI + 12.5 mg dispase + 37.5 mg collagenase II + 300 μl FBS), and stir at 500 rpm for 30 min at 37 °C. Filter digested tissue through a 100 μm cell strainer into a 50 ml tube. Rinse the strainer with 20 ml of RPMI containing 10% FBS. Centrifuge the filtered solution at 500 x g for 10 min at 4 °C. Resuspend pellet in 1 ml of RPMI containing 10% FBS. The cells are now ready for flow cytometry analysis.
Isolation of hepatocyte, liver sinusoidal endothelial cell, macrophage and hepatic stellate cell
Previously reported protocol was followed to isolate the four types of cells from the same liver (83,84). Mice were euthanized with CO2. Then, the liver was perfused with 5 mM HEPES and 0.5 mM EDTA in HBSS at 37°C for 5 mins, followed by perfusion with 0.05% collagenase IV (Sigma, C5138) in HBSS supplemented with 5 mM HEPES and 0.5 mM CaCl2 at 37°C for 5 mins. The livers were excised and homogenized, and then passed through 70-μm filter. The suspension was centrifuged at 50g for 3 mins to separate hepatocytes (pellet) from non-parenchymal cells (NPC, supernatant). Hepatocytes were further enriched by magnetic bead depletion of anti-CD45 and anti-CD146 to deplete most immune cells and endothelial cells respectively. The NPC-fraction was then submitted to a 15% Optiprep density gradient, and then centrifuged at 1500 g for 25 min at room temperature. The well-defined interface of cells was carefully collected, and then centrifuged at 500 g for 5 mins at 4°C. The NPCs were submitted to cell staining and cell sorting. Antibodies used for staining were as follows: anti-CD146-APC (clone ME-9F1, Biolegend), anti-Tie2-PE (clone TEK4, Biolegend), anti-F4/80-FITC (clone BM8, Biolegend), anti-CD3-PE/Cy7 (clone 17A2, Biolegend). LSECs were sorted as CD146+Tie2high F4/80− CD11b− CD3−, Macrophage were sorted as F4/80+CD11b+CD146− Tie2− CD3− cells, hepatic stellate cells were negatively sorted as CD3− CD146− Tie2− F4/80− CD11b− cells. Cell sorting was carried out with a FACS Aria II (BD Biosciences). All experiments were performed with 95% yield of purity for each subset.
Bone marrow transplantation (BMT)
CD45.1 WT C57BL/6 mice and CD45.2 Tlr4−/− mice were lethally irradiated with 900 rad. Four to six hours later, 2×107 bone marrow cells were intravenously injected into the irradiated mice. All experiments were performed 6 weeks after BMT. Successful BMT was confirmed flow cytometry of CD45.1 and CD45.2 in peripheral blood.
Survival analysis of cholangiocarcinoma patients
To indicate the potential association of Tlr4-related genes and patient outcomes, we used bulk transcriptomic data from three cohorts, i.e., International Cancer Genome Consortium (ICGC), Japan and Thailand. Survival information was available for 115 patients of ICGC cohort, 162 patients of Japan cohort, and 85 patients of Thai cohort. To perform survival analysis based on 153 Tlr4-related gene signatures (Supplementary Table S2), we applied BRB-Array Tools (version 4.6.0) (85) developed by Biometric Research Branch of National Cancer Institute. Kaplan-Meier curves were provided for two risk groups obtained by 10-fold cross validation. We performed 100 times of permutation for the log-rank test. Both log-rank p value and permutation p value were provided to indicate the statistical significance.
Statistical analysis
The sample sizes for animal studies were guided by previous murine studies in our laboratory. Statistical analysis was carried out using GraphPad Prism 8 (GraphPad Software). The significant differences between groups were calculated by Student’s unpaired t test, one-way, or two-way ANOVA (Tukey’s and Bonferroni’s multiple comparison test). Log-rank (Mantel-Cox) test were used to determine significance between survival curves. p<0.05 was considered as statistically significant.
Supplementary Material
STATEMENT OF SIGNIFICANCE.
Myeloid derived suppressor cells have been shown to be induced by tumors and suppress anti-tumor immunity. Here we show that the gut microbiome can control accumulation of myeloid derived suppressor cells in the liver in the context of a benign liver disease or colitis.
Acknowledgments
We thank Ascharya Balaji from NCI/NIH for analyzing and quantifying the 16s rRNA data.
Financial support:
T.F.G., X.W.W and G.T. were supported by the Intramural Research Program of the NIH, NCI (ZIA BC 011345, ZO1 BC01087). T.L. and S.W. were supported by the Deutsche Forschungsgemeinschaft (LO-1676/4-1, SFB/TR209 project B08, SFB/TR57 project Q1 and WA-4610/1-1). This study was supported in part by services provided by P30 DK120515. AK acknowledges funding support from the National Institute of Allergy and Infectious Diseases (R01AI132389, R21AI130800).
Conflict of interest disclosure statement:
B.S. has been consulting for Ferring Research Institute, Intercept Pharmaceuticals, HOST Therabiomics, Mabwell Therapeutics and Patara Pharmaceuticals. B.S.’s institution UC San Diego has received grant support from BiomX, NGM Biopharmaceuticals, CymaBay Therapeutics and Synlogic Operating Company.
References
- 1.Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 2018;359(6371):97–103 doi 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillere R, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science (New York, NY) 2018;359(6371):91–7 doi 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 3.Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science (New York, NY) 2018;359(6371):104–8 doi 10.1126/science.aao3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peterson LW, Artis D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol 2014;14(3):141–53 doi 10.1038/nri3608. [DOI] [PubMed] [Google Scholar]
- 5.Szabo G Gut-liver axis in alcoholic liver disease. Gastroenterology 2015;148(1):30–6 doi 10.1053/j.gastro.2014.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abu-Shanab A, Quigley EM. The role of the gut microbiota in nonalcoholic fatty liver disease. Nature reviews Gastroenterology & hepatology 2010;7(12):691–701 doi 10.1038/nrgastro.2010.172. [DOI] [PubMed] [Google Scholar]
- 7.Tamboli CP, Neut C, Desreumaux P, Colombel JF. Dysbiosis in inflammatory bowel disease. Gut 2004;53(1):1–4 doi 10.1136/gut.53.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lemoinne S, Kemgang A, Ben Belkacem K, Straube M, Jegou S, Corpechot C, et al. Fungi participate in the dysbiosis of gut microbiota in patients with primary sclerosing cholangitis. Gut 2020;69(1):92–102 doi 10.1136/gutjnl-2018-317791. [DOI] [PubMed] [Google Scholar]
- 9.Bajaj JS, Heuman DM, Hylemon PB, Sanyal AJ, White MB, Monteith P, et al. Altered profile of human gut microbiome is associated with cirrhosis and its complications. J Hepatol 2014;60(5):940–7 doi 10.1016/j.jhep.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tyson GL, El-Serag HB. Risk factors for cholangiocarcinoma. Hepatology 2011;54(1):173–84 doi 10.1002/hep.24351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rizvi S, Khan SA, Hallemeier CL, Kelley RK, Gores GJ. Cholangiocarcinoma - evolving concepts and therapeutic strategies. Nature reviews Clinical oncology 2018;15(2):95–111 doi 10.1038/nrclinonc.2017.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Razumilava N, Gores GJ. Cholangiocarcinoma. Lancet 2014;383(9935):2168–79 doi 10.1016/S0140-6736(13)61903-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lazaridis KN, LaRusso NF. Primary Sclerosing Cholangitis. The New England journal of medicine 2016;375(12):1161–70 doi 10.1056/NEJMra1506330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shaib YH, El-Serag HB, Davila JA, Morgan R, McGlynn KA. Risk factors of intrahepatic cholangiocarcinoma in the United States: a case-control study. Gastroenterology 2005;128(3):620–6. [DOI] [PubMed] [Google Scholar]
- 15.Welzel TM, Graubard BI, El-Serag HB, Shaib YH, Hsing AW, Davila JA, et al. Risk factors for intrahepatic and extrahepatic cholangiocarcinoma in the United States: a population-based case-control study. Clin Gastroenterol Hepatol 2007;5(10):1221–8 doi 10.1016/j.cgh.2007.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Little R, Wine E, Kamath BM, Griffiths AM, Ricciuto A. Gut microbiome in primary sclerosing cholangitis: A review. World journal of gastroenterology 2020;26(21):2768–80 doi 10.3748/wjg.v26.i21.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manichanh C, Borruel N, Casellas F, Guarner F. The gut microbiota in IBD. Nature reviews Gastroenterology & hepatology 2012;9(10):599–608 doi 10.1038/nrgastro.2012.152. [DOI] [PubMed] [Google Scholar]
- 18.Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol 2009;9(3):162–74 doi 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eggert T, Wolter K, Ji J, Ma C, Yevsa T, Klotz S, et al. Distinct Functions of Senescence-Associated Immune Responses in Liver Tumor Surveillance and Tumor Progression. Cancer Cell 2016;30(4):533–47 doi 10.1016/j.ccell.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kapanadze T, Gamrekelashvili J, Ma C, Chan C, Zhao F, Hewitt S, et al. Regulation of accumulation and function of myeloid derived suppressor cells in different murine models of hepatocellular carcinoma. J Hepatol 2013;59(5):1007–13 doi 10.1016/j.jhep.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoechst B, Ormandy LA, Ballmaier M, Lehner F, Kruger C, Manns MP, et al. A new population of myeloid-derived suppressor cells in hepatocellular carcinoma patients induces CD4(+)CD25(+)Foxp3(+) T cells. Gastroenterology 2008;135(1):234–43 doi 10.1053/j.gastro.2008.03.020. [DOI] [PubMed] [Google Scholar]
- 22.Loeuillard E, Yang J, Buckarma E, Wang J, Liu Y, Conboy C, et al. Targeting tumor-associated macrophages and granulocytic myeloid-derived suppressor cells augments PD-1 blockade in cholangiocarcinoma. J Clin Invest 2020;130(10):5380–96 doi 10.1172/jci137110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sabino J, Vieira-Silva S, Machiels K, Joossens M, Falony G, Ballet V, et al. Primary sclerosing cholangitis is characterised by intestinal dysbiosis independent from IBD. Gut 2016;65(10):1681–9 doi 10.1136/gutjnl-2015-311004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fickert P, Pollheimer MJ, Beuers U, Lackner C, Hirschfield G, Housset C, et al. Characterization of animal models for primary sclerosing cholangitis (PSC). J Hepatol 2014;60(6):1290–303 doi 10.1016/j.jhep.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tag CG, Sauer-Lehnen S, Weiskirchen S, Borkham-Kamphorst E, Tolba RH, Tacke F, et al. Bile duct ligation in mice: induction of inflammatory liver injury and fibrosis by obstructive cholestasis. J Vis Exp 2015(96) doi 10.3791/52438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smit JJ, Schinkel AH, Oude Elferink RP, Groen AK, Wagenaar E, van Deemter L, et al. Homozygous disruption of the murine mdr2 P-glycoprotein gene leads to a complete absence of phospholipid from bile and to liver disease. Cell 1993;75(3):451–62 doi 10.1016/0092-8674(93)90380-9. [DOI] [PubMed] [Google Scholar]
- 27.Cabrera-Rubio R, Patterson AM, Cotter PD, Beraza N. Cholestasis induced by bile duct ligation promotes changes in the intestinal microbiome in mice. Scientific reports 2019;9(1):12324 doi 10.1038/s41598-019-48784-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakamoto N, Sasaki N, Aoki R, Miyamoto K, Suda W, Teratani T, et al. Gut pathobionts underlie intestinal barrier dysfunction and liver T helper 17 cell immune response in primary sclerosing cholangitis. Nat Microbiol 2019;4(3):492–503 doi 10.1038/s41564-018-0333-1. [DOI] [PubMed] [Google Scholar]
- 29.Chassaing B, Aitken JD, Malleshappa M, Vijay-Kumar M. Dextran sulfate sodium (DSS)-induced colitis in mice. Current protocols in immunology 2014;104:Unit 15.25. doi 10.1002/0471142735.im1525s104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gabele E, Dostert K, Hofmann C, Wiest R, Scholmerich J, Hellerbrand C, et al. DSS induced colitis increases portal LPS levels and enhances hepatic inflammation and fibrogenesis in experimental NASH. J Hepatol 2011;55(6):1391–9 doi 10.1016/j.jhep.2011.02.035. [DOI] [PubMed] [Google Scholar]
- 31.Hoshino K, Takeuchi O, Kawai T, Sanjo H, Ogawa T, Takeda Y, et al. Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the Lps gene product. Journal of immunology (Baltimore, Md : 1950) 1999;162(7):3749–52. [PubMed] [Google Scholar]
- 32.Cacalano G, Lee J, Kikly K, Ryan AM, Pitts-Meek S, Hultgren B, et al. Neutrophil and B cell expansion in mice that lack the murine IL-8 receptor homolog. Science (New York, NY) 1994;265(5172):682–4. [DOI] [PubMed] [Google Scholar]
- 33.Katoh H, Wang D, Daikoku T, Sun H, Dey SK, Dubois RN. CXCR2-expressing myeloid-derived suppressor cells are essential to promote colitis-associated tumorigenesis. Cancer Cell 2013;24(5):631–44 doi 10.1016/j.ccr.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Balkwill F Cancer and the chemokine network. Nature reviews Cancer 2004;4(7):540–50 doi 10.1038/nrc1388. [DOI] [PubMed] [Google Scholar]
- 35.Alves-Filho JC, Sonego F, Souto FO, Freitas A, Verri WA Jr., Auxiliadora-Martins M, et al. Interleukin-33 attenuates sepsis by enhancing neutrophil influx to the site of infection. Nature medicine 2010;16(6):708–12 doi 10.1038/nm.2156. [DOI] [PubMed] [Google Scholar]
- 36.Fan B, Malato Y, Calvisi DF, Naqvi S, Razumilava N, Ribback S, et al. Cholangiocarcinomas can originate from hepatocytes in mice. J Clin Invest 2012;122(8):2911–5 doi 10.1172/JCI63212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang S, Song X, Cao D, Xu Z, Fan B, Che L, et al. Pan-mTOR inhibitor MLN0128 is effective against intrahepatic cholangiocarcinoma in mice. Journal of hepatology 2017;67(6):1194–203 doi 10.1016/j.jhep.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seehawer M, Heinzmann F, D’Artista L, Harbig J, Roux PF, Hoenicke L, et al. Necroptosis microenvironment directs lineage commitment in liver cancer. Nature 2018;562(7725):69–75 doi 10.1038/s41586-018-0519-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoechst B, Voigtlaender T, Ormandy L, Gamrekelashvili J, Zhao F, Wedemeyer H, et al. Myeloid derived suppressor cells inhibit natural killer cells in patients with hepatocellular carcinoma via the NKp30 receptor. Hepatology (Baltimore, Md) 2009;50(3):799–807 doi 10.1002/hep.23054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chaisaingmongkol J, Budhu A, Dang H, Rabibhadana S, Pupacdi B, Kwon SM, et al. Common Molecular Subtypes Among Asian Hepatocellular Carcinoma and Cholangiocarcinoma. Cancer Cell 2017;32(1):57–70 e3 doi 10.1016/j.ccell.2017.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jusakul A, Cutcutache I, Yong CH, Lim JQ, Huang MN, Padmanabhan N, et al. Whole-Genome and Epigenomic Landscapes of Etiologically Distinct Subtypes of Cholangiocarcinoma. Cancer discovery 2017;7(10):1116–35 doi 10.1158/2159-8290.Cd-17-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12(5):453–7 doi 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murakami S, Shahbazian D, Surana R, Zhang W, Chen H, Graham GT, et al. Yes-associated protein mediates immune reprogramming in pancreatic ductal adenocarcinoma. Oncogene 2017;36(9):1232–44 doi 10.1038/onc.2016.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Govaere O, Cockell S, Van Haele M, Wouters J, Van Delm W, Van den Eynde K, et al. High-throughput sequencing identifies aetiology-dependent differences in ductular reaction in human chronic liver disease. J Pathol 2019;248(1):66–76 doi 10.1002/path.5228. [DOI] [PubMed] [Google Scholar]
- 45.Jenne CN, Kubes P. Immune surveillance by the liver. Nat Immunol 2013;14(10):996–1006 doi 10.1038/ni.2691. [DOI] [PubMed] [Google Scholar]
- 46.Schwabe RF, Greten TF. Gut microbiome in HCC - Mechanisms, diagnosis and therapy. Journal of hepatology 2020;72(2):230–8 doi 10.1016/j.jhep.2019.08.016. [DOI] [PubMed] [Google Scholar]
- 47.Yu LX, Schwabe RF. The gut microbiome and liver cancer: mechanisms and clinical translation. Nat Rev Gastroenterol Hepatol 2017;14(9):527–39 doi 10.1038/nrgastro.2017.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rizvi S, Gores GJ. Pathogenesis, diagnosis, and management of cholangiocarcinoma. Gastroenterology 2013;145(6):1215–29 doi 10.1053/j.gastro.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Loeuillard E, Fischbach SR, Gores GJ, Rizvi S. Animal models of cholangiocarcinoma. Biochim Biophys Acta Mol Basis Dis 2019;1865(5):982–92 doi 10.1016/j.bbadis.2018.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clements O, Eliahoo J, Kim JU, Taylor-Robinson SD, Khan SA. Risk factors for intrahepatic and extrahepatic cholangiocarcinoma: A systematic review and meta-analysis. J Hepatol 2020;72(1):95–103 doi 10.1016/j.jhep.2019.09.007. [DOI] [PubMed] [Google Scholar]
- 51.Karlsen TH, Folseraas T, Thorburn D, Vesterhus M. Primary sclerosing cholangitis - a comprehensive review. J Hepatol 2017;67(6):1298–323 doi 10.1016/j.jhep.2017.07.022. [DOI] [PubMed] [Google Scholar]
- 52.Fung BM, Tabibian JH. Cholangiocarcinoma in patients with primary sclerosing cholangitis. Curr Opin Gastroenterol 2020;36(2):77–84 doi 10.1097/MOG.0000000000000616. [DOI] [PubMed] [Google Scholar]
- 53.Ibrahim ML, Klement JD, Lu C, Redd PS, Xiao W, Yang D, et al. Myeloid-Derived Suppressor Cells Produce IL-10 to Elicit DNMT3b-Dependent IRF8 Silencing to Promote Colitis-Associated Colon Tumorigenesis. Cell Rep 2018;25(11):3036–46 e6 doi 10.1016/j.celrep.2018.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang T, Fan C, Yao A, Xu X, Zheng G, You Y, et al. The Adaptor Protein CARD9 Protects against Colon Cancer by Restricting Mycobiota-Mediated Expansion of Myeloid-Derived Suppressor Cells. Immunity 2018;49(3):504–14 e4 doi 10.1016/j.immuni.2018.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dejea CM, Fathi P, Craig JM, Boleij A, Taddese R, Geis AL, et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science (New York, NY) 2018;359(6375):592–7 doi 10.1126/science.aah3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science (New York, NY) 2017;358(6369):1443–8 doi 10.1126/science.aal5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krenkel O, Tacke F. Liver macrophages in tissue homeostasis and disease. Nat Rev Immunol 2017;17(5):306–21 doi 10.1038/nri.2017.11. [DOI] [PubMed] [Google Scholar]
- 58.Zannetti C, Roblot G, Charrier E, Ainouze M, Tout I, Briat F, et al. Characterization of the Inflammasome in Human Kupffer Cells in Response to Synthetic Agonists and Pathogens. Journal of immunology (Baltimore, Md : 1950) 2016;197(1):356–67 doi 10.4049/jimmunol.1502301. [DOI] [PubMed] [Google Scholar]
- 59.Liao L, Schneider KM, Galvez EJC, Frissen M, Marschall HU, Su H, et al. Intestinal dysbiosis augments liver disease progression via NLRP3 in a murine model of primary sclerosing cholangitis. Gut 2019;68(8):1477–92 doi 10.1136/gutjnl-2018-316670. [DOI] [PubMed] [Google Scholar]
- 60.Mencin A, Kluwe J, Schwabe RF. Toll-like receptors as targets in chronic liver diseases. Gut 2009;58(5):704–20 doi 10.1136/gut.2008.156307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schwabe RF, Seki E, Brenner DA. Toll-like receptor signaling in the liver. Gastroenterology 2006;130(6):1886–900 doi 10.1053/j.gastro.2006.01.038. [DOI] [PubMed] [Google Scholar]
- 62.Tedesco D, Thapa M, Chin CY, Ge Y, Gong M, Li J, et al. Alterations in Intestinal Microbiota Lead to Production of Interleukin 17 by Intrahepatic γδ T-Cell Receptor-Positive Cells and Pathogenesis of Cholestatic Liver Disease. Gastroenterology 2018;154(8):2178–93 doi 10.1053/j.gastro.2018.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Høgdall D, Lewinska M, Andersen JB. Desmoplastic Tumor Microenvironment and Immunotherapy in Cholangiocarcinoma. Trends Cancer 2018;4(3):239–55 doi 10.1016/j.trecan.2018.01.007. [DOI] [PubMed] [Google Scholar]
- 64.Carding S, Verbeke K, Vipond DT, Corfe BM, Owen LJ. Dysbiosis of the gut microbiota in disease. Microb Ecol Health Dis 2015;26:26191 doi 10.3402/mehd.v26.26191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Soares JB, Pimentel-Nunes P, Roncon-Albuquerque R, Leite-Moreira A. The role of lipopolysaccharide/toll-like receptor 4 signaling in chronic liver diseases. Hepatol Int 2010;4(4):659–72 doi 10.1007/s12072-010-9219-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cirera I, Bauer TM, Navasa M, Vila J, Grande L, Taura P, et al. Bacterial translocation of enteric organisms in patients with cirrhosis. Journal of hepatology 2001;34(1):32–7 doi 10.1016/s0168-8278(00)00013-1. [DOI] [PubMed] [Google Scholar]
- 67.Dapito DH, Mencin A, Gwak GY, Pradere JP, Jang MK, Mederacke I, et al. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell 2012;21(4):504–16 doi 10.1016/j.ccr.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Peleg AY, Hooper DC. Hospital-acquired infections due to gram-negative bacteria. The New England journal of medicine 2010;362(19):1804–13 doi 10.1056/NEJMra0904124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Aujla SJ, Chan YR, Zheng M, Fei M, Askew DJ, Pociask DA, et al. IL-22 mediates mucosal host defense against Gram-negative bacterial pneumonia. Nature medicine 2008;14(3):275–81 doi 10.1038/nm1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Apetoh L, Ghiringhelli F, Tesniere A, Obeid M, Ortiz C, Criollo A, et al. Toll-like receptor 4-dependent contribution of the immune system to anticancer chemotherapy and radiotherapy. Nature medicine 2007;13(9):1050–9 doi 10.1038/nm1622. [DOI] [PubMed] [Google Scholar]
- 71.Wang G, Wang Q, Liang N, Xue H, Yang T, Chen X, et al. Oncogenic driver genes and tumor microenvironment determine the type of liver cancer. Cell death & disease 2020;11(5):313 doi 10.1038/s41419-020-2509-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fickert P, Pollheimer MJ, Beuers U, Lackner C, Hirschfield G, Housset C, et al. Characterization of animal models for primary sclerosing cholangitis (PSC). Journal of hepatology 2014;60(6):1290–303 doi 10.1016/j.jhep.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Models for Immuno-oncology Research. Cancer Cell 2020;38(2):145–7 doi 10.1016/j.ccell.2020.07.010. [DOI] [PubMed] [Google Scholar]
- 74.Chen X, Calvisi DF. Hydrodynamic transfection for generation of novel mouse models for liver cancer research. Am J Pathol 2014;184(4):912–23 doi 10.1016/j.ajpath.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yu SJ, Ma C, Heinrich B, Brown ZJ, Sandhu M, Zhang Q, et al. Targeting the crosstalk between cytokine-induced killer cells and myeloid-derived suppressor cells in hepatocellular carcinoma. J Hepatol 2019;70(3):449–57 doi 10.1016/j.jhep.2018.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Duan Y, Llorente C, Lang S, Brandl K, Chu H, Jiang L, et al. Bacteriophage targeting of gut bacterium attenuates alcoholic liver disease. Nature 2019;575(7783):505–11 doi 10.1038/s41586-019-1742-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proceedings of the National Academy of Sciences of the United States of America 2011;108 Suppl 1(Suppl 1):4516–22 doi 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 2019;37(8):852–7 doi 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 2016;13(7):581–3 doi 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Donghi D, Pechlaner M, Finazzo C, Knobloch B, Sigel RK. The structural stabilization of the κ three-way junction by Mg(II) represents the first step in the folding of a group II intron. Nucleic Acids Res 2013;41(4):2489–504 doi 10.1093/nar/gks1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Manfredo Vieira S, Hiltensperger M, Kumar V, Zegarra-Ruiz D, Dehner C, Khan N, et al. Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science (New York, NY) 2018;359(6380):1156–61 doi 10.1126/science.aar7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Couter CJ, Surana NK. Isolation and Flow Cytometric Characterization of Murine Small Intestinal Lymphocytes. J Vis Exp 2016(111) doi 10.3791/54114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mohar I, Brempelis KJ, Murray SA, Ebrahimkhani MR, Crispe IN. Isolation of Non-parenchymal Cells from the Mouse Liver. Methods in molecular biology (Clifton, NJ) 2015;1325:3–17 doi 10.1007/978-1-4939-2815-6_1. [DOI] [PubMed] [Google Scholar]
- 84.Bigorgne AE, John B, Ebrahimkhani MR, Shimizu-Albergine M, Campbell JS, Crispe IN. TLR4-Dependent Secretion by Hepatic Stellate Cells of the Neutrophil-Chemoattractant CXCL1 Mediates Liver Response to Gut Microbiota. PLoS One 2016;11(3):e0151063 doi 10.1371/journal.pone.0151063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Simon R, Lam A, Li M-C, Ngan M, Menenzes S, Zhao Y. Analysis of gene expression data using BRB-array tools. Cancer Informatics 2007;3:11–7. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.