Abstract
Background & Aims:
We have previously identified tissue methylated DNA markers (MDMs) associated with pancreatic ductal adenocarcinoma (PDAC). In this case-control study, we aimed to assess the diagnostic performance of plasma MDMs for PDAC.
Methods:
Thirteen MDMs (GRIN2D, CD1D, ZNF781, FER1L4, RYR2, CLEC11A, AK055957, LRRC4, GH05J042948, HOXA1, PRKCB, SHISA9, and NTRK3) were identified based on selection criteria applied to results of prior tissue experiments and assays were optimized in plasma. Next, 340 plasma samples (170 PDAC cases, 170 controls) were assayed using target enrichment long-probe quantitative amplified signal method. Initially, 120 advanced stage PDAC cases and 120 healthy controls were used to train a prediction algorithm at 97.5% specificity using random forest (rForest) modeling. Subsequently, the locked algorithm derived from the training set was applied to an independent blinded test set of 50 early stage PDAC cases and 50 controls. Finally, data from all 340 patients were combined, and cross-validated.
Results:
The cross-validated AUC for the training set was 0.93 (0.89−0.96) for the MDM panel alone, 0.91 (95% CI: 0.87–0.96) for CA 19–9 alone, and 0.99 (0.98−1) for the combined MDM-CA 19–9 panel. In the test set of early stage PDAC, the AUC for MDMs alone was 0.84 (0.76–0.92), CA19–9 alone was 0.87 (0.79–0.94), and combined MDM-CA 19–9 panel was 0.90 (0.84–0.97) significantly better compared to either MDMs alone or CA19–9 alone (p=0.0382 and 0.0490, respectively). At a pre-set specificity of 97.5%, the sensitivity for the combined panel in the test set was 80% (28–99%) for stage 1 disease and 82% (68–92%) for stage 2 disease. Using the combined data sets, the cross-validated AUC was 0.9 (0.86−0.94) for the MDM panel alone and 0.89 for CA 19–9 alone (0.84–0.93) vs 0.97 (0.94−0.99) for the combined MDM-CA 19–9 panel, p≤0.0001 Overall, cross-validated sensitivity of MDM-CA 19–9 panel was 92% (83–98%) with an observed specificity of 92% at the preset specificity of 97.5%.
Conclusions:
Plasma MDMs in combination with CA 19–9 detect PDAC with significantly higher accuracy compared to either biomarker individually.
Keywords: Pancreatic Neoplasms/prevention and control, liquid biopsy, DNA methylation, Plasma/analysis, biomarkers
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is a leading cause of cancer-associated mortality in the United States, with a 5-year survival of 8% in all stages combined, one of the lowest among the major cancer killers.(1) Detecting PDAC at an early localized stage is associated with a 4-fold higher 5-year survival, which is further improved for sub-centimeter, pre-symptomatic tumors.(1, 2) Unfortunately, the vast majority of PDAC cases continue to be detected at a late stage.(3) Although the longer survival in early-stage PDAC is apparent both in the general population and high-risk groups, there is currently no effective screening paradigm for this highly lethal disease.(4, 5) A biomarker that reliably detects PDAC at early stages could potentially improve disease outcomes. Early stage PDAC has longer survival compared to late stage. Detecting more early stage disease will potentially improve the 5-year survival in this disease by identifying patients at a stage when they are likely to benefit the most from neoadjuvant therapy and surgery. Clinically available blood biomarkers, such as carbohydrate antigen 19–9 (CA 19–9), are unreliable for detecting early-stage PDAC, may be normal in advanced disease in subjects who do not express Lewis blood group antigens, and can be falsely elevated in with the setting of biliary obstruction and in patients with inflammatory pancreatitis.(6, 7)
Because of the limitations of CA 19–9 and other circulating proteins, more recent approaches to non-invasive biomarker development have focused on circulating exfoliated cancer cells or cancer cell products, such as DNA.(8, 9) This “liquid biopsy” approach alone and in combination with established protein biomarkers is anticipated to enhance early stage tumor detection with improved sensitivity and specificity. However, DNA mutations are heterogeneous among neoplasms of the same type in different persons or among clones of tumor cells in the same individual.(10) In PDAC, the combination of mutations in circulating tumor DNA (ctDNA) and protein biomarkers has been shown to detect early stage disease with 64% sensitivity;(11) however, potentially hundreds of additional genomic positions assayed in combination with protein markers might be required to improve upon this performance.(9) Methylated DNA markers (MDMs) appear to be more broadly informative than DNA mutations; as few as 4 MDMs are present in nearly all colorectal cancers and polyps.(12) Further, MDMs may be specific to tumors arising in different organs.(13, 14)
Using a comprehensive unbiased methylome sequencing approach, our laboratory has previously identified novel MDMs that accurately detect cancers throughout the gastrointestinal tract, including those of colorectal, esophageal, gastric, and hepatocellular origin.(15–18) Using this discovery approach followed by multistep validation of candidate markers, we have also previously identified MDMs in tissues that are associated with pancreatic cancer and precancer in independent tissues, pancreatic juice, and pancreatic cyst fluid.(19, 20) Plasma assay performance for these pancreatic neoplasia-associated MDMs has not yet been reported. In the present case-control study, we tested the hypothesis that MDMs would improve performance of CA 19–9 for PDAC detection.
Materials and Methods
Study Design
In this two-phase case-control study using archival plasma samples, we assessed the accuracy of MDMs alone and in combination with CA 19–9 to discriminate advanced stage PDAC cases from age- and sex-balanced control patients. The resulting multi-marker panel was then locked down and blindly tested in a second phase on an independent set of stage I and II PDAC cases and controls. This study design was adopted to optimally use the limited pool of archival samples from early-stage PDAC cases. Early stage PDAC samples are scant since the vast majority of patients present with advanced disease.(3) Despite access to a fairly large institutional biorepository, early stage PDAC samples are both scarce and highly sought after; we aimed to design the study to ensure that early stage PDAC samples are used judiciously. Data on blood CEA levels were not clinically available in the vast majority of PDAC patients included in this study. Since CA 19–9 is a more commonly used biomarker in clinical practice, and given sample volume restrictions and the primary aim of testing the plasma MDM panel we limited the number of additional biomarkers tested to CA 19–9 alone. The study was reviewed and approved by the Mayo Clinic Institutional Review Board (IRB). All participants in this study provided informed written consent to research under IRB protocol #354–06. The Mayo Clinic IRB is in compliance with the requirements of FDA regulations 21 CFR Parts 50 and 56 and HHS regulations 45 CFR 46, which are guided by the Belmont Report. In addition, the Mayo Clinic IRB complies with ICH guideline on Good Clinical Practices where they are consistent with FDA and HHS regulations.
Patients Studied
For this single center study, we used archival plasma samples obtained from the Mayo Clinic SPORE in Pancreatic Cancer (P50 CA102701, PI: Gloria Petersen). Cases included adult patients with treatment-naïve biopsy-proven PDAC. Cancer staging was done according to the AJCC 8th edition.(21) Controls were patients recruited from primary care clinics at Mayo Clinic during routine wellness check visits, and matched to cases by sex- and age (+5 years) Those with a prior history of cancer (except non melanoma skin cancer) were excluded. Patients with known primary cancer outside of the pancreas within the last 5 years prior to plasma collection or a documented cancer outside of the pancreas within 3 years after plasma collection (not including basal cell or squamous cell skin cancers), and those with history of prior therapeutic radiation to the upper abdomen, prior history of transplant, or receiving chemotherapy class drugs in the 5 years prior to plasma collection were excluded. Mayo Clinic electronic health records were reviewed to abstract clinicopathological and demographic data for all study subjects using pre-designed study forms. The presence or absence signs and symptoms of abdominal or back pain, fatigue, unintentional weight loss, anorexia and jaundice at the time of index cancer diagnosis were ascertained by chart review of clinical notes performed by a board-certified gastroenterologist (S.M.). Source clinical note documents were from encounters with gastroenterology, pancreatic surgery or medical oncology. This was at the time point when patients were enrolled into the pancreas biorepository (IRB protocol #354–06) and the blood specimens, used in this study, were collected. Data from the study forms were subsequently entered in the electronic study database, and the final dataset was verified by a single expert reviewer (SM) to ensure accurate case-control phenotyping.
Marker selection
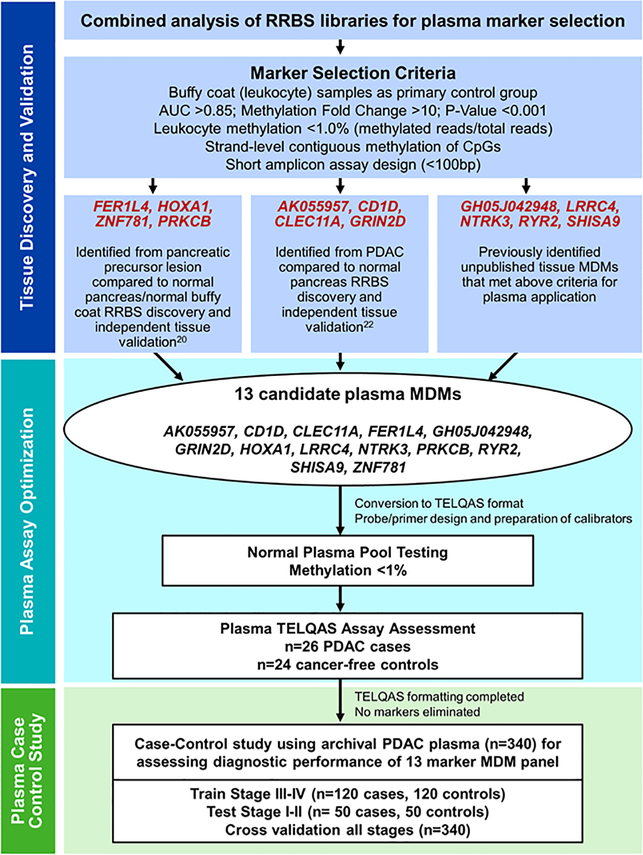
Thirteen MDMs were chosen for the PDAC plasma case-control study based on prior work. Gene annotations and genomic coordinates are listed in Supplemental Table 1. These hypermethylated candidates originated from several earlier tissue discovery and validation studies performed by our group. The first was a reduced representation bisulfite sequencing (RRBS) study using tissue biopsies from PDAC patients and comparing them to normal pancreas samples.(22) The second study also utilized RRBS but also included tissues from advanced and early stage precursor lesions (PanIN-3, 2, and 1).(20) For the current study, we extensively analyzed combined RRBS libraries using selection criteria and techniques to target candidates specifically for plasma application (Figure 1). In plasma, the normal background is predominantly cell free DNA derived from leukocytes, not epithelial cells. Hence, we utilized the non-cancer buffy coat sample cohort included in our sequencing results as the primary control group for comparing to PDAC and precursor groups. Four genes that were previously identified and tested in the PDAC study (GRIN2D, CD1D, CLEC11A, and AK055957) and similarly four from the precursor study (ZNF781, PRKCB, FER1L4, HOXA1) met current criteria and were chosen for plasma testing (Figure 1). Additionally, five novel genes were identified and subsequently validated in independent tissues (unpublished data): RYR2, LRRC4, GH05J042948, SHISA9, and NTRK3. This combined panel of 13 candidate MDMs was then optimized for plasma TELQAS (target enrichment with long probe quantitative amplified signal) assays in a sample set of 26 PDAC cases across all stages and 24 control plasmas from cancer-free subjects (Figure 1); all 13 MDM TELQAS designs passed this step and were carried forward.
Figure 1:
Marker selection algorithm for identifying best candidate plasma MDMs from tissue RRBS libraries and subsequent plasma assay optimization steps prior to training and testing in case-control study.
Biospecimen collection and molecular assay techniques
As part of the ongoing Mayo Clinic SPORE in Pancreatic Cancer biospecimen archiving protocol, blood from each subject was collected in a K2 EDTA Vacutainer® (BD, Franklin Lakes, NJ). Blood was processed according to standard operating procedures of the Mayo Clinic’s Biospecimens Accessioning and Processing core laboratory. Within 4 hours, the tubes were centrifuged at 1500xG (10 min), plasma removed and centrifuged a second time, aliquoted in 2mL cryotubes, and stored at −80C without any intervening thawing. For the purpose of this study, 4 ml of plasma from each subject was retrieved. Starting with 3.8 ml of plasma for the MDM assay to account for losses during handling and transfer, we used a final volume of 3mL for the extraction. The cell-free DNA (cfDNA) was purified and bisulfite converted from 3mL of plasma using a proprietary automated silica bead method (Exact Sciences, Madison, WI). A non-human DNA spike-in was used to control for processing aberrations. For all samples, 3 mL of plasma was initially subjected to Proteinase K treatment followed by lysis with detergent and chaotropic reagents. Silica coated binding beads and lysis buffer containing isopropyl alcohol were added to each sample for DNA capture and DNA precipitation. All samples were subjected to multiple rounds of washing on the Hamilton STARlet liquid handling system (Hamilton Company, Reno, NV) and binding beads were dried prior to DNA sample elution in elution buffer. Samples were then bisulfite converted as previously described(23) with the use of the Hamilton STARlet liquid handling system. Briefly, samples were initially denatured with sodium hydroxide. Ammonium bisulfite was added to each sample for deamination. Samples were subsequently bound to silica coated binding beads and subjected to multiple rounds of washing prior to desulphonation. Sample washing was repeated, and purified samples were eluted in elution buffer. 200uL of plasma was used to measure CA 19–9.
MDMs were assayed from the extracted DNA using TELQAS, a highly sensitive multiplexed format(18) that is a modification to the FDA-approved quantitative allele-specific realtime target and signal amplification assay.(24) TELQAS oligonucleotides (forward invasive primer, reverse primer, flap probe) were designed to 5’-cytosine-phosphate-guanine-3’(CpG) motifs within each of the 13 DMRs (IDT, Coralville, IA) as well as B3GALT6 (methylated reference gene) and RASSF1 (zebrafish spike-in processing control). Ten μL of bisulfite-treated DNA was used in triplex format—FAM, HEX, Quasar® 670 (Hologic, Marlborough, MA)—in which two markers plus the B3GALT6 reference gene were amplified and quantified. TELQAS reactions were performed on an Applied Biosystems™ 7500 Fast Dx Real-Time PCR Instrument (Applied Biosystems, Foster City, CA).
CA 19–9 was quantitated from plasma samples using the MILLIPLEX® Map Kit (MilliporeSigma, Burlington, MA) on the Luminex® MAGPIX® analyzer (MilliporeSigma). Plasma samples were diluted 1:6 using the Serum Matrix provided in the kit as the diluent. Only CA 19–9 Antibody-Immobilized Magnetic Beads were used in the immunoassay. The assay was completed using the protocol supplied with the kit reagents. Quantitative results for each sample were generated from the median fluorescence intensity signals using the Luminex® xPONENT® software. CA 19–9 values obtained using this method was compared to serum CA 19–9 values where available from the Mayo Clinic Immunochemical Core Laboratory that used the protocol recommended by the ELISA kit manufacturer (Cobas/Roche, Basel, Switzerland).
Statistical analysis
Training in advanced stage PDAC and testing in early stage PDAC (Model-Validation #1)
The panel of 13 MDMs (with and without CA 19–9) was used to train a prediction algorithm for PDAC (Y/N) using random forest (rForest) modeling with 500 trees.(25) Briefly, each recursive partition tree of the forest is trained by taking a bootstrap random sample of the data. Each node or branch point of any given tree is determined by selecting, from a random subset of the markers, the most discriminant marker between cases and controls at that point of the tree. As a tree can have multiple nodes, a new random subset of the marker panel is taken at each node. This selection of markers at each node of the tree is done in order to “de-correlate” predictions amongst the trees within the forest.(26) rForest has been shown to provide superior generalizability and predictive accuracy in test data sets compared to logistic regression with minimal concerns of overfitting.(26) The training set consisted of 120 control patients and 120 late stage PDAC patients. The average predicted probability of PDAC across the 500 individual trees was then used to classify a patient as PDAC (Y/N) using a cut-off value that had 97.5% specificity in the training set. The trained model and predefined cut-off were then applied to a test set of control patients (N=50) and early stage PDAC patients (N=50). The training and testing of the model were carried out independently by two statisticians (DWM and WRB, respectively) who were blinded to the results in the other phase. The accuracy of the prediction model within the blinded test set was summarized as sensitivity, specificity, and area under the receiver operator characteristic curve (AUC) with corresponding 95% confidence intervals. Comparisons of AUCs between sub-models was done using the methods of DeLong et al.(27). McNemar’s test was used to compare sensitivity between sub-models at matched specificities.
In-silico cross-validation all stages combined (Model-Validation #2)
For the second validation, data from all 340 patients were combined and the prediction model was refit using rForest modeling with 500 trees. The refit prediction model was cross-validated by first randomly splitting the entire data set 2:1 (training: testing) 500 times. For each random split, the trained prediction model was used to predict PDAC (Y/N) within the test set. The sensitivity, specificity, and concordance across each of the 500 iterations were summarized as an average with corresponding 95% confidence intervals for the cross-validated diagnostic performance of the panel of markers. The effect of clinical characteristics on prediction accuracy of the trained models were assessed by comparing stratified AUCs across levels of the covariates as suggest by Janes et al. (28)
Sample Size Considerations
Samples sizes for the training and testing sets were determined a priori to ensure a) the half-width of the 95% confidence bounds for the estimated specificity (set a prior at 97.5%) and sensitivity (assumed at 90%) of the panel of markers would be no larger than ±10% and ±15% for each phase, respectively, and b) 90% power to detect an area under the ROC curve (AUC) of 0.85 or higher relative to a null AUC of 0.7.
Results
Patient Characteristics
A total of 340 patients (170 cases and 170 controls) were included in this study. Patients were divided into a training set comprising 120 advanced stage PDAC (Stage 3 and 4) cases and 120 controls and a test set with 50 early stage PDAC (Stage 1 and 2) cases and 50 controls. Within both sets, cases and controls were balanced on age and sex. Demographic characteristics of the study population are summarized in Table 1. There were 34 cases and 24 controls with a history of Type 2 diabetes mellitus. Accurate assessment of new-onset diabetes mellitus was not feasible due to lack of a prior blood glucose value for the majority of these study subjects. For those with a documented date of diabetes diagnosis in controls, 13/18 (72%) were diagnosed at age 50 or older and for cases, 26/32 (81%) were diagnosed at age 50 or older (p=0.4945).
Table 1:
Demographic and clinical characteristics of the study population
| Cases |
Controls |
||||||
|---|---|---|---|---|---|---|---|
| Training (N=120) | Testing (N=50) | Overall (N=170) | Training (N=120) | Testing (N=50) | Overall (N=170) | Overall P value | |
| Age | 0.066 | ||||||
| Median (Q1, Q3) | 64.9 (56.6, 71.3) | 72.3 (66.0, 78.0) | 66.6 (60.5, 73.9) | 63.0 (55.4, 70.2) | 69.9 (60.8, 75.4) | 64.7 (56.9, 71.6) | |
| Sex | 0.745 | ||||||
| Female | 64 (53.3%) | 17 (34.0%) | 81 (47.6%) | 67 (55.8%) | 17 (34.0%) | 84 (49.4%) | |
| Race | 0.009 | ||||||
| Black | 1 (0.8%) | 0 (0.0%) | 1 (0.6%) | 2 (1.7%) | 1 (2.0%) | 3 (1.8%) | |
| Caucasian | 104 (86.7%) | 50 (100.0%) | 154 (90.6%) | 116 (96.7%) | 48 (96.0%) | 164 (96.5%) | |
| Other | 15 (12.5%) | 0 (0.0%) | 15 (8.8%) | 2 (1.7%) | 1 (2.0%) | 3 (1.8%) | |
| Tobacco use | 0.021 | ||||||
| Current | 22 (18.5%) | 6 (12.0%) | 28 (16.6%) | 11 (9.2%) | 2 (4.0%) | 13 (7.7%) | |
| Former | 45 (37.8%) | 20 (40.0%) | 65 (38.5%) | 55 (46.2%) | 28 (56.0%) | 83 (49.1%) | |
| Never | 52 (43.7%) | 24 (48.0%) | 76 (45.0%) | 53 (44.5%) | 20 (40.0%) | 73 (43.2%) | |
| Alcohol use | 0.057 | ||||||
| Current | 56 (51.9%) | 34 (73.9%) | 90 (58.4%) | 81 (72.3%) | 29 (61.7%) | 110 (69.2%) | |
| Former | 23 (21.3%) | 6 (13.0%) | 29 (18.8%) | 7 (6.2%) | 9 (19.1%) | 16 (10.1%) | |
| None | 29 (26.9%) | 6 (13.0%) | 35 (22.7%) | 24 (21.4%) | 9 (19.1%) | 33 (20.8%) | |
| Diabetes mellitus | 0.002 | ||||||
| Type I | 2 (1.7%) | 0 (0.0%) | 2 (1.2%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | |
| Type II | 21 (17.6%) | 13 (26.0%) | 34 (20.1%) | 18 (15.0%) | 6 (12.2%) | 24 (14.2%) | |
| Type NS | 8 (6.7%) | 1 (2.0%) | 9 (5.3%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | |
| Biopsy within 14 days prior to blood collection | < 0.001 | ||||||
| No | 68 (56.7%) | 19 (38.0%) | 87 (51.2%) | 120 (100.0%) | 49 (98.0%) | 169 (99.4%) | |
| Yes Other | 16 (13.3%) | 5 (10.0%) | 21 (12.4%) | 0 (0.0%) | 1 (2.0%) | 1 (0.6%) | |
| Yes Pancreas | 36 (30.0%) | 26 (52.0%) | 62 (36.5%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | |
| History of chronic pancreatitis | 0.156 | ||||||
| Yes | 2 (1.7%) | 0 (0.0%) | 2 (1.2%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | |
| Pancreatic Surgery | < 0.001 | ||||||
| Yes | 17 (14.2%) | 29 (58.0%) | 46 (27.1%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | |
| Stage | < 0.001 | ||||||
| I | 0 (0.0%) | 5 (10.0%) | 5 (2.9%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | |
| II | 0 (0.0%) | 45 (90.0%) | 45 (26.5%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | |
| III | 60 (50.0%) | 0 (0.0%) | 60 (35.3%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | |
| IV | 60 (50.0%) | 0 (0.0%) | 60 (35.3%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | |
Training in advanced stage PDAC and testing in early stage PDAC (Model-Validation #1)
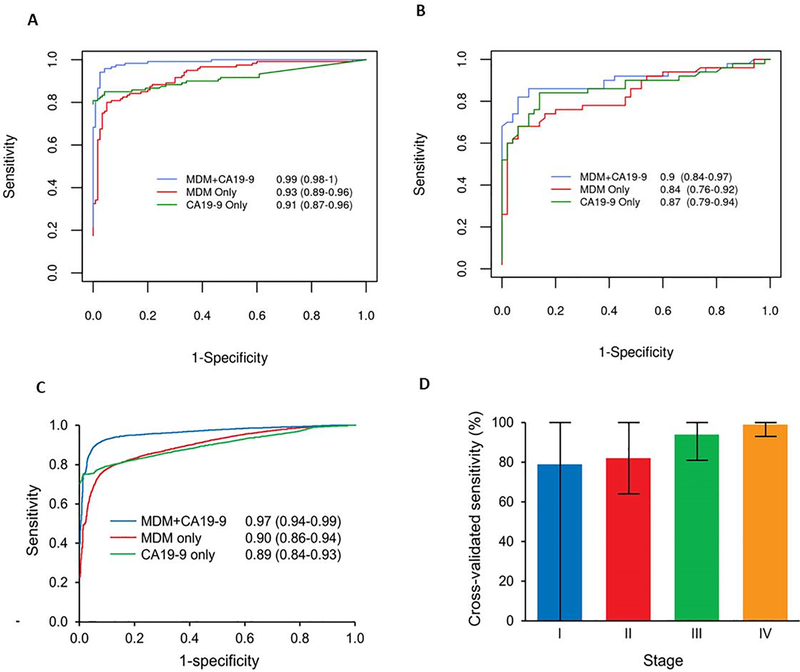
In the training set, individual MDM AUCs for differentiating cases from controls ranged from 0.75–0.86 (Supplemental Table 2). The cross-validated AUC within the training set for the MDM panel alone was 0.93 (95% CI: 0.89–0.96) as compared to CA 19–9 alone which was 0.91 (95% CI: 0.87–0.96) (p=0.4718) (Figure 2A). There proved to be significantly higher accuracy when the MDM panel and CA 19–9 were combined, with a combined cross-validated AUC of 0.99 (95% CI: 0.98–1.00); p<0.0004 compared with either component alone (Figure 2A). At a set specificity of 97.5% (95% CI: 93–99%), the cross-validated sensitivity of the MDM-CA 19–9 combination was 90% (95% CI: 83–95%) and 88% (95% CI: 81–93%) for CA-19 alone.
Figure 2:
A) Receiver operating curves for CA 19–9, MDM panel and combination of both in the training set of advanced stage PDAC (n=120) B) Receiver operating curves for CA 19–9, MDM panel and combination of both in the test set of early stage PDAC (n=50) C) Cross validated AUC demonstrating that the MDM-CA19–9 combination significantly enhances PDAC detection accuracy compared to either marker component alone D) Sensitivity of MDM-CA 19–9 combination across different PDAC stages at 92% specificity.
MDM cut-off values derived from the training set were applied to the test set comprising stage 1 and 2 PDAC cases and healthy controls. The AUC of the MDM panel alone was 0.84 (95% CI 0.76–0.92) and when combined with CA 19–9 was 0.90 (0.84–0.97), p=0.038 (Figure 2B). At the pre-set specificity cutoff of 97.5%, the observed sensitivity/specificity within the test set was 40% (26–55%)/98% (89–100%) for MDMs only, 70% (55–82%)/90% (78–97%) for CA19–9 only, and 82% (69–91%)/94% (83–99%) for the combination. At a matched specificity of 94% within the test set, the observed sensitivities were 68% (53–80%) for MDMs only, 68% (53–80%) for CA19–9 only, and 82% (69–91%) for the combination. There was no significant difference between the MDM only model and the CA19–9 only model (p=0.99, McNemar’s test). There was a statistically significantly higher sensitivity of the combined model in comparison to MDMs only (p=0.0455, McNemar’s test) and for CA19–9 only (p=0.0233, McNemar’s test). CA 19–9 at a cut-off of 35 IU/L detected a similar number of early stage cases but was falsely elevated in 17/50 (34%;95% CI: 21–49%) controls. At a higher cut-off of 55 IU/L CA 19–9 was falsely positive in only 3/50 (6%; 95% CI: 1–17%) controls at the cost of lower detection of only 30/45 (67%; 95% CI: 51–80%) Stage 2 cases.
In silico cross validation all stages combined (Model-Validation #2)
This analysis was done using the data on 170 cases and 170 controls. The cross-validated AUC was 0.9 (0.86−0.94) for the MDM panel alone and 0.89 for CA 19–9 alone (0.84–0.93); validating further the training and test set outcomes, the AUC was significantly higher at 0.97 (0.94−0.99) for the combined MDM-CA 19–9 panel, (p=<0.0001; Figure 2C).Using a preset cutoff of 97.5% in the trained model, the mean observed specificity was 92% (81–100%) and the mean sensitivity of the MDM-CA 19–9 combination was 92% (83–98%) overall. Detection rates were 79% in Stage 1, 82% in Stage 2, 94% in Stage 3, and 99% in Stage 4 PDAC (p=0.001, Figure 2D).
Co-variate analyses
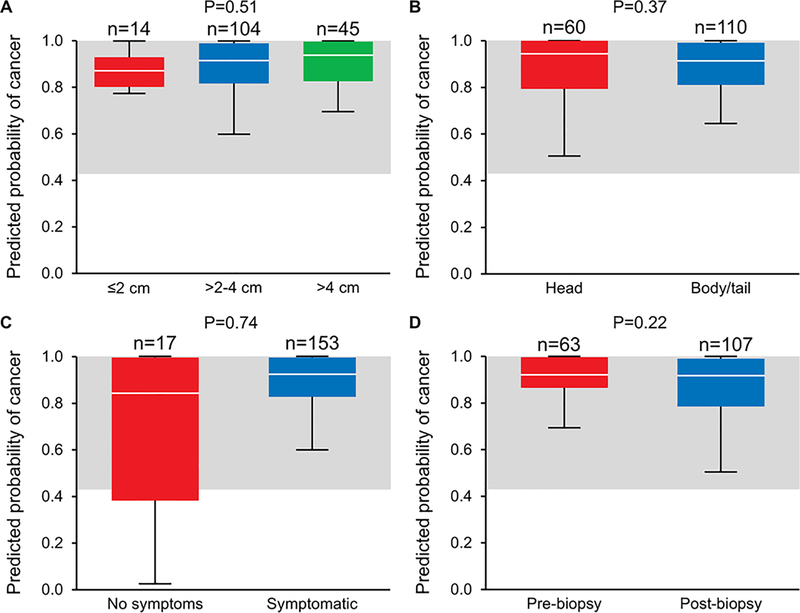
The AUC for the MDM panel was not significantly affected by age, sex, race, or presence of diabetes (Table 2). An analysis of predicted probability of cancer using the cross-validated model in cases showed no significant differences based on tumor size (less than or greater than 4 cm), tumor location (head vs body/tail), temporal relationship of plasma collection with biopsy, or presence or absence of symptoms at the time of PDAC diagnosis for both the MDM-CA 19–9 combination (Figure 3 A-D) and MDMs alone (Supplemental Figure 1).
Table 2:
Analysis of potential confounding variables on the overall accuracy (estimated AUC with corresponding 95% CI) of the MDMs rForest model
| Covariate | Grouping of Covariate | P value | |
|---|---|---|---|
| No AUC (95% CI) |
Yes AUC (95% CI) |
||
| Age>65 years | 0.92 (0.87–0.96) | 0.88 (0.83–0.93) | 0.3092 |
| Male | 0.87 (0.81–0.92) | 0.93 (0.9–0.97) | 0.0532 |
| White Race | 0.78 (0.57–0.99) | 0.91 (0.87–0.94) | 0.2468 |
| Ever Smoker | 0.92 (0.87–0.96) | 0.89 (0.84–0.93) | 0.3744 |
| Alcohol use | 0.89 (0.81–0.97) | 0.91 (0.87–0.94) | 0.7774 |
| History of Diabetes mellitus | 0.91 (0.87–0.94) | 0.86 (0.78–0.95) | 0.3779 |
Figure 3:
Box plot of the predicted probability of cancer from the cross-validated MDMs + CA 19–9 rForest model demonstrating no significant differences in model performance based on A) Tumor size, B) Tumor location, C) Presence of symptoms at the time of diagnosis, D) Timing of biopsy in relationship to blood draw. The Gray shaded area indicates probability values exceeding the 92nd percentile in controls with group sizes indicated above each box plot.
CA 19–9 correlation between study and clinical test methods
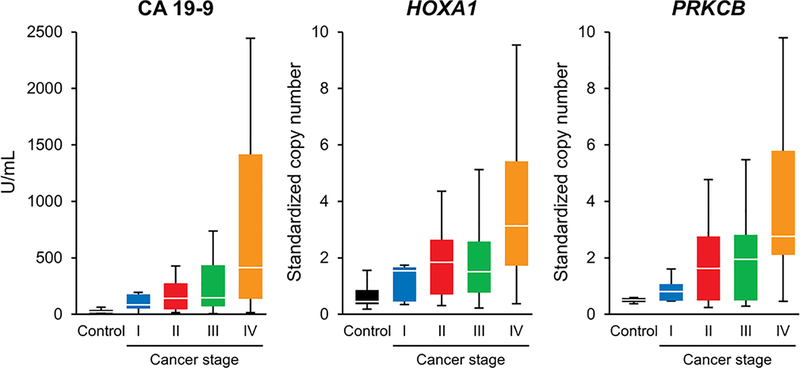
Of the 340 samples analyzed in this study, Mayo Clinic Immunochemical Core Laboratory tested pretreatment CA 19–9 results were available on 119 (15 (9%) controls and 104 (61%) case) samples. The median time of collection is 5 days after pathology confirmed diagnosis (25th percentile=3 days prior to diagnosis, 75th percentile=14 days post diagnosis). A comparison of CA 19–9 values derived using the study protocol and those obtained by the clinical test yielded excellent correlation (Pearson correlation coefficient = 0.946, 95% CI (0.922, 0.962), Lin’s concordance correlation coefficient = 0.935, 95% CI (0.908, 0.954)) (Supplemental Figure 2). Similar to plasma CA 19–9 values which are known to increase with advancing tumor stage, several of the plasma MDMs demonstrated increased signal with stage advancement (Figure 4 and Supplemental Figure 3).
Figure 4:
Distributions in plasma from controls and PDAC patients by stage of disease of CA 19–9 values and standardized copy numbers of selected MDMs, HOXA1 and PRKCB.
Discussion
In this case-control study using archival plasma samples, we demonstrate that a panel of plasma MDMs in combination with CA 19–9 detects PDAC across all stages with moderate to high accuracy. These findings were confirmed by both training-test set comparison and in silico cross-validation. To the best of our knowledge, this is the largest study reporting outcomes for a diagnostic DNA methylation biomarker panel in PDAC.(29) Plasma MDM marker measurements were not significantly influenced by tumor size, location and presence or absence of symptoms, indicating feasibility of clinical application for detecting PDAC at early pre-symptomatic stages. Moreover, several plasma MDMs identified in this study demonstrate incremental circulating levels with advancing PDAC stage, a property that will need to be explored in future studies aimed at monitoring disease progression and response to treatment.
The search for novel circulating biomarkers that can either outperform or complement CA 19–9 has spanned several decades without any circulating biomarker surviving the rigor of validation. With ongoing advances in molecular analysis techniques over the past few years there are some emerging novel biomarkers that have high reported accuracy for PDAC diagnosis. By reprogramming cells derived from advanced PDAC tissue and subsequently performing proteomic analysis of conditioned medium from precursor PanINs cultured as organoids, Kim et al., identified 107 human proteins specific to advanced stage PanINs.(30) In a large study aimed at discovery and multi-stage validation of these candidate markers using human PDAC plasma across all stages, the authors reported sensitivity of 87% for the combination of THBS2 and CA 19–9 at 98% specificity.(30) In another recent study, a panel of eight protein markers including CA 19–9 and DNA mutations at 1,933 distinct genomic positions was evaluated in eight cancer types, including PDAC, and found to demonstrate >99% specificity at a median sensitivity of 70%;(9) however, these results have not been validated in independent patient samples.
Aberrant DNA methylation is an early event in carcinogenesis and appears to be more broadly informative than most protein and gene mutation biomarkers for early detection of cancer.(12) Further, recent advances in technology have resulted in improved analytical sensitivity for MDMs when assayed from plasma.(18) The recent development and widespread availability of high throughput assay methods that can detect minute amounts of circulating tumor DNA has transformed this field.(18, 31) In a recent study, methylation-specific PCR for blood-based DNA methylation targeting 28 genes in 95 patients with PDAC yielded only moderate accuracy with a specificity of 83% and sensitivity of 76%.(32) Genes targeted in that panel were selected based on a literature search for previously published biomarkers, which is unlike the unbiased next-generation methylome tissue discovery approach that yielded the MDM panel tested in our study. Moreover, the TELQAS assay technique utilized in the present study has demonstrated the requisite analytical sensitivity to meaningfully advance ctDNA detection from plasma.(33) Overall, the role of MDMs in PDAC is at a nascent stage and continues to evolve. Collectively, there is early evidence for further developing MDMs for multiple clinical applications in PDAC management.
A central assumption with most approaches to liquid biopsy is that molecular aberrations present in tumors enter the circulation and can be targeted as detection markers.(8) However, the target signal in plasma is typically present in minute quantities relative to the vast pool of background circulating nucleic acid which is derived from leukocytes and other normal non-dysplastic cells, and it is critical that targeted markers be discriminated from this normal background. The candidate markers selected for this study were derived from a tissue-based discovery effort based on their uniquely high presence in PDAC relative to normal pancreas and normal leukocytes, a pattern that leads to the highest likelihood of efficacy in a blood-based application.(20, 22) Using a similar approach with several other cancer types, we have demonstrated encouraging results for early tumor detection in plasma applications.(16, 18, 34) Furthermore, several plasma MDMs included in this panel were selected from tissue MDMs that not only distinguish PDAC from normal tissue but also discriminate between advanced precursor lesions (PanIN-3) and earlier stage precursors (PanIN-1 and 2).(20) The progression from low-grade to high-grade precursor lesions in PDAC spans several decades and this provides a window of opportunity for early detection.
Longitudinal surveillance efforts in high-risk individuals can detect PDAC at early stages and a large prospective study recently reported excellent outcome of 85% 3-year survival in surveillance-detected PDAC compared to a 5-year survival rate of less than 10% in the general population.(4) Several patients who underwent a major pancreatic resection were found to not have histologically confirmed PDAC or high-grade dysplasia.(4) This poses significant risk to patients in such surveillance programs, given the morbidity involved with pancreatic resections.(35) A biomarker that can detect advanced dysplasia and early invasive cancer can potentially transform the screening paradigm in high-risk individuals by refining case selection for surgical resection. There are several other high-risk cohorts, such as subjects with a genetic predisposition to PDAC, new-onset diabetes mellitus, chronic pancreatitis, and pancreatic cystic neoplasms that would potentially benefit from a high-risk screening algorithm.(36–38) However, given the relatively low event-rate of PDAC in the general population, average risk screening has not been feasible to date. Rather, an approach that combines clinical risk prediction models and an accurate biomarker has been proposed as a path to early detection.(39–41) We have recently validated a cyst fluid MDM panel for detecting pancreatic cancer and advanced neoplasia in pancreatic cystic lesions(20) and used pancreatic juice MDMs for discriminating PDAC from subjects with chronic pancreatitis.(19, 22) Future studies aimed at assessing combined performance of plasma MDMs with cyst fluid and pancreatic juice MDMs may further enhance diagnostic accuracy in certain high-risk patient subpopulations.
While PDAC is the second leading cancer killer in the U.S., its relatively low prevalence has thwarted efforts to cost-effectively perform single organ screening for this tumor in the general population. However, one could envision a future screening paradigm that simultaneously targets multiple cancers from a single medium based on a rationale that leverages their cumulative prevalence.(8) MDMs evaluated in the present study could be included in a multicancer screening test that includes PDAC among its targets. The MDM panel used in this study has potential cross-reactivity with other cancers. Based on our tissue RRBS data and in-silico analyses, approximately 75% of the MDMs in this panel show strong cross-reactivity with DNA from primary tissues of colon and lung adenocarcinoma. We have not yet tested for cross-reactivity with these cancers in plasma samples. The potential for cross-reactivity with other cancers would need to be discussed with patients prior to use. A positive test result would prompt diagnostic evaluation initially directed at pancreatic cancer. If none were found, additional diagnostic testing might be required. While this is anticipated to be an acceptable trade-off when used in a population at high risk for pancreatic cancer, the same would not apply to the average risk population. In an average risk population, a broad diagnostic survey test (e.g. PET-CT) has been used to evaluate positive results of a prototype liquid biopsy test that is intended to detect multiple cancers.(31) Alternatively, other investigators have shown highly comparable specificity for pancreas compared to other cancers using DNA methylation as assayed by whole genome bisulfite sequencing.(42) This approach has been reported to provide tissue of origin for positive test results but is a more expensive and cumbersome compared to the TELQAS assays used in our study. A final clinical test in an average risk population may require a combination of these approaches.
Our study has several limitations. First, the small number of Stage 1 cases limits precision of our predictions for very early PDACs. Despite that, the sensitivity and specificity of the panel remained relatively high in a large subset of Stage 2 PDAC. Furthermore, high discrimination for PDAC by the MDM-CA 19–9 combination remained consistent on rigorous cross-validation. There are two separate modeling steps performed in this study. The first model was developed entirely on a training set of Stage III/IV PDAC patients and then applied to a test set of Stage I/II patients. For the second model, the entire set of Stage I through Stage IV was combined and cross-validated by randomly splitting the cases into two-third training and one-third testing groups. Second, the relatively small volume and collection method of plasma used may have influenced DNA yield and downstream molecular detection thresholds. The optimal volume of plasma remains to be determined, and optimization of collection methods aimed at improving plasma tumor DNA integrity may further augment results in future studies.(43) A stabilized blood sample that optimizes DNA yield and quality is a key pre-analytical requirement for any liquid biopsy approach. Recently it has been demonstrated that proprietary stability buffers are superior for preservation of cells and cell-free DNA (cfDNA) in whole blood when compared to samples collected in K2 ETDA. Prospective blood collection using these more optimal preservation methods may further improve the diagnostic accuracy of our MDM panel. Third, the case blood samples were obtained from patients being clinically evaluated at Mayo Clinic for suspected PDAC and the presence or absence of symptoms at the time of PDAC diagnosis were determined by a retrospective chart review, so these results cannot be immediately applied to a prospective screening paradigm. Interestingly, although only a small subset of cases in this study were incidentally detected PDAC, MDM intensity was not affected by presence or absence of disease-specific symptoms in the case group. This suggests the likely ability of these plasma MDMs to detect asymptomatic disease, which is a key property for any biomarker designed to be an effective screening tool. Finally, our study did not include a control group of patients with chronic pancreatitis and other risk factors for pancreatic cancer. The present study included 170 healthy controls which did not permit an exhaustive analysis to exclude potential cross-reactivity with benign conditions that lead to increased cell turnover. A more comprehensive specificity study is planned for the MDM panel used in the present study, as we have done previously with stool DNA candidate markers.(44) This is a critical step that we intend to address in future studies prior to marker elimination and establishing and locking diagnostic cut-offs comparing both normal healthy and disease controls to cases with PDAC.
In conclusion, this study demonstrates that a panel of plasma MDMs can detect PDAC across all stages and enhances significantly the diagnostic yield of the existing clinically used biomarker CA 19–9. The next study in this continuum will need to focus on marker elimination in independent samples with a goal to remove the MDMs that are only minimally contributing to the panel’s diagnostic performance and set a specific MDM panel algorithm with diagnostic cutoffs, include an incremental number of early stage PDAC cases and include control patients with underlying pancreatic diseases and risk factors for PDAC. A panel of circulating tumor markers like the ones described in this study, once validated prospectively, could potentially be used for early detection in groups at high risk for PDAC. These MDMs also have potential for incorporation into a future universal cancer detection blood test. Further optimization and prospective validation studies are indicated to assess this promising approach to early detection of PDAC.
Supplementary Material
Statement of translational relevance.
Pancreatic cancer is a leading cause of cancer related deaths. Carbohydrate antigen 19–9 (CA 19–9), a widely used blood biomarker for pancreatic cancer, may miss cancers especially at early-stage disease. We have previously identified methylated DNA markers (MDMs) in pancreatic tissues that are strongly associated with pancreatic cancer and advanced precancer. This study describes a panel of MDMs in plasma that accurately discriminate pancreatic cancer cases from cancer-free controls. The AUC for the combined MDM-CA 19–9 panel training set was 0.99 (0.98−1). In the test set of early stage PDAC, the AUC for MDMs alone was 0.84 (0.76–0.92), CA19–9 alone was 0.87 (0.79–0.94), and combined MDM-CA 19–9 panel was 0.90 (0.84–0.97) significantly better compared to either MDMs alone or CA19–9 alone (p=0.0382 and 0.0490, respectively). Overall, cross-validated cancer sensitivity of MDMs and CA 19–9 combined was 92% (83–98%) at a specificity of 92% (81–100%). Further optimization and prospective validation studies are indicated to assess this promising new diagnostic approach. These MDMs also have potential for being studied for universal cancer screening and for pancreas cancer detection in high-risk individuals.
Acknowledgements:
The authors dedicate this work to the memory of Dr. David A. Ahlquist (1951–2020), who inspired this work and played key roles in study concept and design, analysis and interpretation of data, critical review of the manuscript for important intellectual content, and obtained funding.
Funding was provided by the Carol M. Gatton Foundation (to Dr Ahlquist). Exact Sciences (Madison, WI) provided blinded assays and critical assay reagents. Dr. Majumder was supported by a career enhancement award funded by Mayo Clinic SPORE in Pancreatic Cancer (P50 CA102701). This work was partially supported by R37 CA214679 to Dr. Kisiel, and P50 CA 102701 and U01 CA210138 to Dr. Petersen. The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
Abbreviations:
- PDAC
Pancreatic ductal adenocarcinoma
- MDM
Methylated DNA marker
- CA 19–9
Carbohydrate antigen 19–9
- cfDNA
cell-free DNA
- RRBS
Reduced representation bisulfite sequencing
- TELQAS
Target enrichment long-probe quantitative amplified signal
Footnotes
Conflict of Interest Statement and Disclosures: Mayo Clinic and Exact Sciences have an intellectual property development agreement. Drs. Majumder & Kisiel and Messrs. Taylor & Mahoney are listed as inventors under this agreement and could share potential future royalties paid to Mayo Clinic. Drs. Lidgard and Allawi are Exact Sciences employees. The other authors of this manuscript have no conflicts of interest to declare. The abstract summarizing preliminary results from this study were presented at Digestive Disease Week 2019 in San Diego, CA.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA: a cancer journal for clinicians. 2018;68:7–30. [DOI] [PubMed] [Google Scholar]
- 2.Ishikawa O, Ohigashi H, Imaoka S, Nakaizumi A, Uehara H, Kitamura T, et al. Minute carcinoma of the pancreas measuring 1 cm or less in diameter--collective review of Japanese case reports. Hepato-gastroenterology. 1999;46:8–15. [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA: a cancer journal for clinicians. 2019;69:7–34. [DOI] [PubMed] [Google Scholar]
- 4.Canto MI, Almario JA, Schulick RD, Yeo CJ, Klein A, Blackford A, et al. Risk of Neoplastic Progression in Individuals at High Risk for Pancreatic Cancer Undergoing Long-term Surveillance. Gastroenterology. 2018;155:740–51.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Owens DK, Davidson KW, Krist AH, Barry MJ, Cabana M, Caughey AB, et al. Screening for Pancreatic Cancer: US Preventive Services Task Force Reaffirmation Recommendation Statement. Jama. 2019;322:438–44. [DOI] [PubMed] [Google Scholar]
- 6.Tempero MA, Uchida E, Takasaki H, Burnett DA, Steplewski Z, Pour PM. Relationship of carbohydrate antigen 19–9 and Lewis antigens in pancreatic cancer. Cancer research. 1987;47:5501–3. [PubMed] [Google Scholar]
- 7.Locker GY, Hamilton S, Harris J, Jessup JM, Kemeny N, Macdonald JS, et al. ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2006;24:5313–27. [DOI] [PubMed] [Google Scholar]
- 8.Ahlquist DA. Universal cancer screening: revolutionary, rational, and realizable. NPJ precision oncology. 2018;2:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen JD, Li L, Wang Y, Thoburn C, Afsari B, Danilova L, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science (New York, NY). 2018;359:926–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kulemann B, Rösch S, Seifert S, Timme S, Bronsert P, Seifert G, et al. Pancreatic cancer: Circulating Tumor Cells and Primary Tumors show Heterogeneous KRAS Mutations. Sci Rep. 2017;7:4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen JD, Javed AA, Thoburn C, Wong F, Tie J, Gibbs P, et al. Combined circulating tumor DNA and protein biomarker-based liquid biopsy for the earlier detection of pancreatic cancers. Proceedings of the National Academy of Sciences of the United States of America. 2017;114:10202–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zou H, Allawi H, Cao X, Domanico M, Harrington J, Taylor WR, et al. Quantification of methylated markers with a multiplex methylation-specific technology. Clinical chemistry. 2012;58:375–83. [DOI] [PubMed] [Google Scholar]
- 13.Gai W, Ji L, Lam WKJ, Sun K, Jiang P, Chan AWH, et al. Liver- and Colon-Specific DNA Methylation Markers in Plasma for Investigation of Colorectal Cancers with or without Liver Metastases. Clinical chemistry. 2018;64:1239–49. [DOI] [PubMed] [Google Scholar]
- 14.Kisiel JB, Wu CW, Taylor WR, Allawi HT, Yab TC, Foote PH, et al. Multi-Site Gastrointestinal Cancer Detection by Stool DNA. Gastroenterology. 2018;154:S–95. [Google Scholar]
- 15.Taylor WR, Kisiel JB, Yab TC, Mahoney DW, Smyrk TC, Boardman LA, et al. Discovery of Novel DNA Methylation Markers for the Detection of Colorectal Neoplasia: Selection by Methylome-Wide Analysis. Gastroenterology. 2014;146:S–30. [Google Scholar]
- 16.Anderson BW, Suh YS, Choi B, Lee HJ, Yab TC, Taylor WR, et al. Detection of Gastric Cancer with Novel Methylated DNA Markers: Discovery, Tissue Validation, and Pilot Testing in Plasma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2018;24:5724–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qin Y, Wu CW, Taylor WR, Sawas T, Burger KN, Mahoney DW, et al. Detection of Esophageal Cancer by Assay of Novel Methylated DNA Markers in Plasma. Gastroenterology. 2018;154:S–87. [Google Scholar]
- 18.Kisiel JB, Dukek BA, R VSRK, Ghoz HM, Yab TC, Berger CK, et al. Hepatocellular Carcinoma Detection by Plasma Methylated DNA: Discovery, Phase I Pilot, and Phase II Clinical Validation. Hepatology (Baltimore, Md). 2019;69:1180–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Majumder S, Raimondo M, Taylor WR, Yab TC, Berger CK, Dukek BA, et al. Methylated DNA in Pancreatic Juice Distinguishes Patients With Pancreatic Cancer From Controls. Clinical gastroenterology and hepatology : the official clinical practice journal of the American Gastroenterological Association. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Majumder S, Taylor WR, Yab TC, Berger CK, Dukek BA, Cao X, et al. Novel Methylated DNA Markers Discriminate Advanced Neoplasia in Pancreatic Cysts: Marker Discovery, Tissue Validation, and Cyst Fluid Testing. The American journal of gastroenterology. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chun YS, Pawlik TM, Vauthey JN. 8th Edition of the AJCC Cancer Staging Manual: Pancreas and Hepatobiliary Cancers. Annals of surgical oncology. 2018;25:845–7. [DOI] [PubMed] [Google Scholar]
- 22.Kisiel JB, Raimondo M, Taylor WR, Yab TC, Mahoney DW, Sun Z, et al. New DNA Methylation Markers for Pancreatic Cancer: Discovery, Tissue Validation, and Pilot Testing in Pancreatic Juice. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015;21:4473–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lidgard GP, Domanico MJ, Bruinsma JJ, Light J, Gagrat ZD, Oldham-Haltom RL, et al. Clinical performance of an automated stool DNA assay for detection of colorectal neoplasia. Clinical gastroenterology and hepatology : the official clinical practice journal of the American Gastroenterological Association. 2013;11:1313–8. [DOI] [PubMed] [Google Scholar]
- 24.Imperiale TF, Ransohoff DF, Itzkowitz SH, Levin TR, Lavin P, Lidgard GP, et al. Multitarget stool DNA testing for colorectal-cancer screening. The New England journal of medicine. 2014;370:1287–97. [DOI] [PubMed] [Google Scholar]
- 25.Breiman L Machine Learning (2001) 45: 5. 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 26.Couronné R, Probst P, Boulesteix AL. Random forest versus logistic regression: a large-scale benchmark experiment. BMC bioinformatics. 2018;19:270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–45. [PubMed] [Google Scholar]
- 28.Janes H, Longton G, Pepe M. Accommodating Covariates in ROC Analysis. The Stata journal. 2009;9:17–39. [PMC free article] [PubMed] [Google Scholar]
- 29.Henriksen SD, Madsen PH, Krarup H, Thorlacius-Ussing O. DNA Hypermethylation as a Blood-Based Marker for Pancreatic Cancer: A Literature Review. Pancreas. 2015;44:1036–45. [DOI] [PubMed] [Google Scholar]
- 30.Kim J, Bamlet WR, Oberg AL, Chaffee KG, Donahue G, Cao XJ, et al. Detection of early pancreatic ductal adenocarcinoma with thrombospondin-2 and CA19–9 blood markers. Science translational medicine. 2017;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lennon AM, Buchanan AH, Kinde I, Warren A, Honushefsky A, Cohain AT, et al. Feasibility of blood testing combined with PET-CT to screen for cancer and guide intervention. Science (New York, NY). 2020;369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henriksen SD, Madsen PH, Larsen AC, Johansen MB, Drewes AM, Pedersen IS, et al. Cell-free DNA promoter hypermethylation in plasma as a diagnostic marker for pancreatic adenocarcinoma. Clinical epigenetics. 2016;8:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allawi H T, Giakoumopoulos M, Flietner E, Oliphant A, Volkmann C, Aizenstein B, et al. Detection of lung cancer by assay of novel methylated DNA markers in plasma. Cancer research. 2017;77:712-. [Google Scholar]
- 34.Qin Y, Wu CW, Taylor WR, Sawas T, Burger KN, Mahoney DW, et al. Discovery, Validation, and Application of Novel Methylated DNA Markers for Detection of Esophageal Cancer in Plasma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2019;25:7396–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aslanian HR, Lee JH, Canto MI. AGA Clinical Practice Update on Pancreas Cancer Screening in High-Risk Individuals: Expert Review. Gastroenterology. 2020;159:358–62. [DOI] [PubMed] [Google Scholar]
- 36.Kirkegard J, Mortensen FV, Cronin-Fenton D. Chronic Pancreatitis and Pancreatic Cancer Risk: A Systematic Review and Meta-analysis. The American journal of gastroenterology. 2017;112:1366–72. [DOI] [PubMed] [Google Scholar]
- 37.Chari ST, Leibson CL, Rabe KG, Timmons LJ, Ransom J, de Andrade M, et al. Pancreatic cancer-associated diabetes mellitus: prevalence and temporal association with diagnosis of cancer. Gastroenterology. 2008;134:95–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu C, Hart SN, Polley EC, Gnanaolivu R, Shimelis H, Lee KY, et al. Association Between Inherited Germline Mutations in Cancer Predisposition Genes and Risk of Pancreatic Cancer. Jama. 2018;319:2401–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharma A, Kandlakunta H, Nagpal SJS, Feng Z, Hoos W, Petersen GM, et al. Model to Determine Risk of Pancreatic Cancer in Patients With New-Onset Diabetes. Gastroenterology. 2018;155:730–9.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abe T, Blackford AL, Tamura K, Ford M, McCormick P, Chuidian M, et al. Deleterious Germline Mutations Are a Risk Factor for Neoplastic Progression Among High-Risk Individuals Undergoing Pancreatic Surveillance. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2019:Jco1801512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Corral JE, Mareth KF, Riegert-Johnson DL, Das A, Wallace MB. Diagnostic Yield From Screening Asymptomatic Individuals at High Risk for Pancreatic Cancer: A Meta-analysis of Cohort Studies. Clinical gastroenterology and hepatology : the official clinical practice journal of the American Gastroenterological Association. 2019;17:41–53. [DOI] [PubMed] [Google Scholar]
- 42.Liu MC, Oxnard GR, Klein EA, Swanton C, Seiden MV, Liu MC, et al. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Annals of Oncology. 2020;31:745–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pottekat A, Allawi HT, Boragine GT, Kaiser MW, Sander T, Krueger C, et al. A comprehensive assessment of the impact of preanalytical variables on cell free DNA and circulating tumor cells in blood. Cancer research. 2018;78:4598. [Google Scholar]
- 44.Ahlquist DA TW, Yab TC, Devens ME, Mahoney DW, Boardman LA, Thibodeau SN, Zou H, Domanico MJ, Berger BM, Lidgard GP. Aberrantly methylated gene marker levels in stool: effect of demographic exposure, body mass, and other patient characteristics. . J Mol Biomark Diagn. 2012;3. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.