Abstract
Purpose:
Cancer associated fibroblasts (CAFs) are an important component of the tumor microenvironment (TME), but a systematic investigation of their molecular characteristics and clinical relevance are lacking. Here, we sought to compare CAFs across multiple cancer types to identify critical molecular pathways activated in CAF subtypes, which may contribute to clinical outcome, disease progression, and immunotherapy resistance.
Experimental Design:
We performed integrated analysis of CAFs from melanoma (MEL), head and neck squamous cell carcinoma (HNSC), and lung cancer (LC), and identified the molecular characteristics that are distinctly active in each CAF subtype. Gene signatures for individual CAF subtypes were identified and used to study the association of subtype abundance with clinical outcome and immunotherapy resistance.
Results:
We identified six CAF subtypes (pan-CAFs) shared across cancer types and uncovered the molecular characteristics and genetic pathways distinguishing them. Interestingly, these CAF subtypes express distinct immunosuppressive factors, such as CXCL12 and CXLC14, and stem-cell promoting factor IL-6. Additionally, we identified novel transcriptional drivers (MEF2C, TWIST1, NR1H3, RELB, and FOXM1) key to CAF heterogeneity. Furthermore, we showed that CAF subtypes were associated with different clinical outcomes and uncovered key molecular pathways that could activate or suppress cancer progression or were involved in resistance to anti-PD1 or anti-PD-L1 immunotherapy.
Conclusions:
Our study identifies the molecular characteristics of CAF subtypes shared across several cancer types, implicates cancer types that may benefit from CAF subtype targeted therapies, and identifies specific CAF subtypes associated with immunotherapy resistance.
Introduction
The tumor microenvironment (TME) is a complex ecosystem comprised of various cell types including malignant and stromal cells (1). Cancer associated fibroblasts (CAFs) is considered one of the most abundant stromal cells and are observed in almost all solid tumor types (2,3). A body of research suggests that CAFs are implicated in an array of pro-tumorigenic biological processes including, but not limited to, tumor cell invasion, cancer stem-cell renewal, chemo-resistance, and immune-cell evasion (4-6). However, in some cancer types, other studies suggest that CAFs can exert a tumor suppressive effect in the TME (7-9). The collection of these research endeavors embodies the notion that CAFs are a heterogenous population of cells and, to ensure precise targeting of tumor promoting CAFs, we need to better characterize CAF heterogeneity through the identification of specific markers.
In recent years, single-cell technology has been extensively applied to dissect cellular heterogeneity of solid tumors (10). Specifically, single-cell RNA-sequencing (scRNA-seq) has uncovered the malignant and/or stromal cellular heterogeneity across an array of cancer types including melanoma (MEL) (11), head and neck squamous cell carcinoma (HNSC) (12), lung cancer (LC) (13), medulloblastoma (14), pancreatic ductal adenocarcinoma (15), and glioma (16). As a result of this high-throughput analysis, the transcriptomic characteristics of CAFs have been described in several cancer types. Whether and how CAF subsets across different cancer types share similar gene expression profiles and molecular functions, however, are less clear. Furthermore, the clinical association of CAF subtypes and their relationship to immunotherapy resistance remains to be established. Here, we postulate that integrated scRNA-seq analysis of CAFs can provide a more consistent characterization of CAF subtypes, novel insights into their biological properties, and signaling pathways they may activate in TMEs, which in turn may influence clinical outcome and immunotherapy resistance.
Towards these, we have characterized and compared CAFs from three cancer types, MEL, HNSC and LC, and revealed the molecular and biological characteristics of CAF subtypes in solid tumors. Using the TCGA database, we further address the valuable prognostic utility of signature genes for these CAF subsets, providing new insights into how cancers residing in specific tissue types may benefit from CAF subtype targeted therapy. In distinct cancer types, we found that different CAF subtypes may activate their own signature molecular pathways to modulate the TME and resistance to immunotherapy.
Materials and Methods
Datasets
Single cell RNA-seq datasets containing CAFs from MEL (11) and HNSC (12) tumors were downloaded from the gene expression omnibus (GEO; GSE72056 and GSE103322, respectively), while dataset describing LC (13) tumors was from https://doi.org/10.1038/s41591-018-0096-5. We chose these datasets because they were among the first studies characterizing CAFs using scRNA-seq. TCGA bulk RNA-seq datasets with normalized read counts and FPKMs (fragment per kilobase of transcript per million mapped reads) were downloaded from the UCSC Xena platform (17).
CAF scRNA-seq analysis
MEL, HNSC, and LC CAFs were extracted from the full datasets based on the cell type annotation provided by the authors, but we also performed clustering analysis of all the cells to confirm the distinction of CAFs from other cell types. CAFs from each of the three cancers were then analyzed separately using Seurat (v3.1.5) (18), with the cluster numbers determined by the FindClusters function, using resolutions 0.05 ~ 1.0. The results were visualized using the uniform manifold approximation and projection for dimension reduction (UMAP) (19). The final resolutions (MEL, 0.5; HNSC, 0.3; LC, 0.15) were selected to closely match the CAF-clusters described in the original papers (11-13). The FindMarkers function was used to perform differential gene expression analysis with a Wilcoxon rank sum test to define marker genes for each CAF cluster, compared to all other cell types. The marker genes were subject to Gene Ontology (GO) analysis using the ClueGO (20) plugg-in via Cytoscape (21) (v3.7.1) with GO-Biological Process, KEGG-KEGG, and REACTOME-reactions as the ontologies and pathways. The enrichment for a GO term or pathway was considered significant at p < 0.05.
To compare and identify CAF sub-populations across cancer types, CAFs from MEL, HNSC and LC datasets were integrated using canonical correlation analysis (CCA) via Seurat (18). The final clustering resolution was selected so that each identified cluster had at least 10 marker genes (false discovery rate (FDR) < 0.05) with > 2-fold higher in expression compared to all other clusters. A bi-clustering heatmap was used to examine the specificity of the selected markers for all CAF subtypes.
Spatial distribution of pan-CAF subtypes analysis
The gene expression matrix of the integrated CAF scRNA-seq data, with the pan-CAF clustering labels, was uploaded to the CIBERSORTx (22) web server to generate a gene expression profile (GEP), using default settings except that the numbers of minimum and maximum genes for the GEP were set to 50 and 150, respectively. With this GEP and default parameters, CIBERSORTx estimated the pan-CAF abundances in bulk RNA-seq samples that were derived from three to six regional biopsies of tumors from patients (one tumor per patient) with non-small cell lung cancer (23). These RNA-seq data was downloaded from the GEO (GSE112996).
Transcription factor analysis
Using the transcription factor (TF) database described by Lambert et al (24), we identified which of the CAF subtype marker genes were TFs. To identify gene regulatory networks, target genes of pan-CAF specific TFs were identified and extracted from SCENIC (25) (v1.1.2.2). We then evaluated if the target genes of the identified TFs were enriched in the same pan-CAF subtypes.
Survival analysis using TCGA data
Using the MEL, HSNC and LC scRNA-seq data, we identified pan-CAF subtype specific gene signatures by considering both CAF and non-CAF cells. For each of the three cancer types, we performed differential gene expression analysis between cells belong to a particular pan-CAF subtype and all the other cells via the FindMarkers function in Seurat (18). Genes that were expressed significantly higher (adjusted p < 0.05 and log2[fold change] > 0.25) in the selected pan-CAF subtype than the other cells in all three cancer types and were also identified as pan-CAF subtype markers in our integrated clustering analysis were used as the pan-CAF specific signature genes.
The expression of these pan-CAF specific signature genes was extracted from the normalized FPKM values in the TCGA RNA-seq datasets of 31 cancer types. The FPKMs for each gene in a pan-CAF gene signature were used to compute Z-scores for each tumor sample. These Z-scores were then averaged to yield a single value, representing the overall expression of the signature genes in a tumor sample, with greater value meaning higher expression. For a cancer type, survival analysis was performed by dividing patients into quartiles based on the Z-scores. Quadrant 1 (tumors with the highest Z-score) were compared to Quadrant 4 (tumors with the lowest Z-score) for a CAF subtype. A log-rank and hazard ratio (HR) test were used to determine statistical significance between the two patient groups. Benjamini & Hochberg method was used to adjust for multiple comparisons for the log-rank p-values using the p.adjst function in R studio to calculate FDR. Cancer types with an FDR < 0.05 were used for further downstream analyses. For a more robust survival analysis, a univariate cox proportional hazards model was also performed with CAF subtype specific Z-score as a continuous variable. Furthermore, via STATA/IC 15.1, multivariate analysis was performed with CAF subtype specific z-scores while adjusting for age and gender as a continuous and dichotomous variable, respectively.
Gene set enrichment analysis
Of the cancer types showing a significant survival difference (FDR < 0.05) among tumors with high and low expression of signature genes for a pan-CAF subtype, gene set enrichment analysis (GSEA) (26) was performed. Specifically, differential expression between tumors in Quadrant 1 and 4 was determined using DESeq2 (version 1.26.0) (27). The genes were ranked by multiplying the -log2-transformation of the p-value with the sign of the log2(fold change). Gene sets in the REACTOME database were analyzed, with significant set to FDR < 0.001. To reduce redundancy and group enriched terms with overlapping genes, GSEA result was visualized and interpreted using the enrichment map (28) plug-in for cytoscape (21) (v3.7.1).
Association with immunotherapy resistance
We analyzed the transcriptome data in metastatic bladder and kidney metastatic tumors resistant to anti-PD-L1 immunotherapy (http://research-pub.gene.com/IMvigor210CoreBiologies) (29) and melanoma tumors resistant to anti-PD-1 immunotherapy (GEO: GSE78220) (30). The marker genes for individual pan-CAF subtypes were utilized as sets by the GSEA (26) to determine their significant enrichment between the therapeutically resistant or sensitive groups (FDR < 0.05), as described above.
Results
Molecular characterization of CAF heterogeneity for individual cancer types
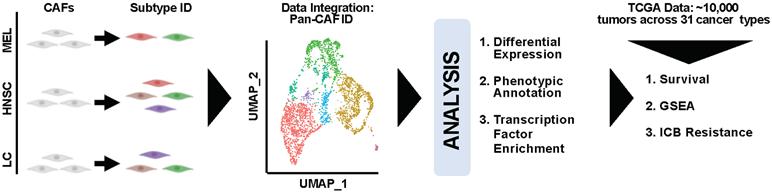
High throughput technology (e.g., scRNA-seq) offers a novel, robust, and systematic way to characterize CAF heterogeneity by molecular profiles. Thus, we obtained scRNA-seq data containing CAFs from MEL (11), HNSC (12), and LC (13) tumors, extracted the CAFs, and subclustered them to study CAF heterogeneity within each cancer type (Fig. 1). Next, we performed integrative and comparative analysis of the CAFs to identify pan cancer CAF (pan-CAF) subtypes and their corresponding molecular features (Fig. 1). Finally, using over 10,000 RNA-seq samples describing 31 cancer types, the association of pan-CAF specific gene signatures to clinical outcome, TME signaling pathways, or resistance to immune checkpoint blockade (ICB) was assessed (Fig. 1).
Figure 1. Schematic illustration of our experimental design and analytic approaches.
To start, we first performed CAF-focused sub-clustering analysis using Seurat (18) to confirm that we retrieved the correct cells and that our method could reproduce the clustering results in the original studies (11-13). As detailed in Supplementary Figure 1, although our CAF-focused analysis generated more consistently annotated CAF subclusters than the original work, partially due to the usage of same method and similar markers, it largely matched the previous CAF classifications but with some differences, for example, we distinguished melanoma CAFs from one to two subclusters.
Identification and molecular characterization of six pan-CAF subtypes
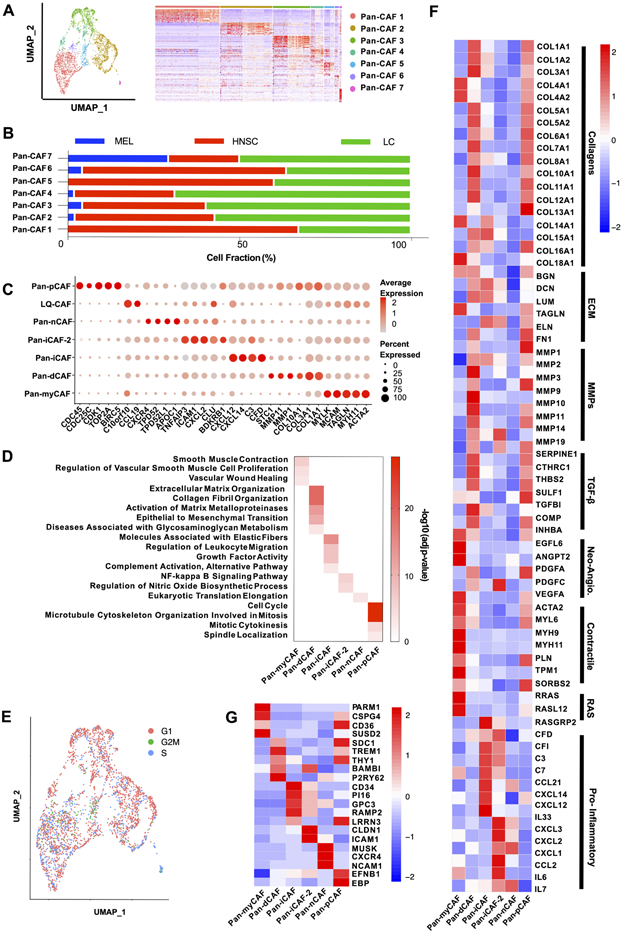
Although the CAF analysis of individual cancers (Supplementary Figure 1) shows common CAF subtypes across cancers, it remains unclear to what extent CAFs from different cancer types are related. To address this more precisely, we needed to perform integrated data analysis to remove batch effects and align gene expression among datasets. Therefore, we integrated CAFs from the three cancer types using Seurat (18). High-resolution clustering of the integrated data identified seven CAF subtypes (Fig. 2A), referred as pan-CAF 1-7. These pan-CAF subtypes were present across the three cancer types but at different abundances (Fig. 2B), and their relationship to the CAF subtypes identified from above study of individual cancers was analyzed (Supplementary Table 1). The result demonstrates the value of integrated analysis because such a fine separation was only feasible after cells from more than one cancer type were pulled together. Pan-CAF 1 comprised of 1007 cells (53.8% of all CAFs, red) (Fig. 2A) and was classified as pan-myCAFs, based on elevated expression of activated fibroblast markers (ACTA2) and smooth muscle cell markers (MYH11, MCAM, TAGLN, and MYLK) (Fig. 2C). GO analysis of its marker genes showed enrichments of gene sets related to smooth muscle contraction and vascular wound healing (Fig. 2D). Pan-CAF 2 comprised of 815 cells (28.3%, brown) (Fig. 2A) and was classified as pan-dCAFs, based on elevated expression of genes coding for collagen (COL1A1, COL3A1, etc.) and genes associated with GO terms of extracellular matrix (ECM) remodeling (Fig. 2D). Pan-CAF 3, comprised of 588 cells (20.5%, dark green), was characterized as pan-iCAFs based on high expression of CFD, C3, CXCL14, and CXCL12 (Fig. 2C) and GO term enrichment for inflammation (Fig. 2D). Pan-CAF 4, comprised of 202 cells (7.0%, light green) (Fig. 2A), was classified as pan-iCAFs-2 because these cells had high and unique expression of genes related to inflammation, including CXCL2 (Fig. 2C). Furthermore, GO analysis of pan-iCAF-2 marker genes found enrichment for NF-kB signaling pathway (Fig. 2D). Pan-CAF 5 comprised of 158 cells (5.5%, blue) (Fig. 2A) and was categorized as normal fibroblast (pan-nCAF) due to the enrichment of its marker genes for homeostasis (Fig. 2C). Pan-CAF 6 comprised of 69 cells (2.4%) and represented a low quality cellular cluster (LQ-CAF) without detectable marker genes, and thus were left out of further analysis (Fig. 2A). Pan-CAF 7, comprised of 34 cells (1.2%) (Fig. 2A, pink), was classified as pan-pCAF because of elevated expression of genes related to cell cycle (BIRC5, TOP2A, etc.) (Fig. 2C), and enrichment of GO terms associated with cellular proliferation (Fig. 2D). In addition, Seurat’s cell cycle analysis also showed that this cluster were enriched for genes related to the S phase of cell cycle (Fig. 2E). Note that we tested and validated the robustness of our classification of seven pan-CAFs by subsampling 75-90% of cells for clustering analysis (Supplementary Table 2).
Figure 2. Marker genes and biological processes for pan-CAF subtypes.
(A) UMAP (left) depicting subtypes and heatmap (right) depicting marker genes associated with pan-CAF subtypes. Pan-myCAF, myofibroblast-like CAFs; pan-dCAF, desmoplastic CAFs; pan-iCAF and pan-iCAF-2, inflammatory-like CAFs; pan-nCAF, normal myofibroblasts; pan-pCAF, proliferating CAFs. (B) Fraction of CAF subtypes in individual cancer types. (C) Dot plot showing expression of marker genes for pan-CAF subtypes. (D) Enriched gene sets for pan-CAF subtypes. (E) UMAP depicting cell cycle states of pan-CAF subtypes. (F) Heatmap of expression of genes associated with selected functions. (G) Heatmap of expression of genes encoding cell-surface proteins.
Next, gene sets curated by Qian et al (31) that described key CAF-related biological processes were used to further annotate the phenotypic and functional heterogeneity of the pan-CAFs. The analysis showed that pan-myCAFs expressed a distinct set of collagens (COL4A1, COL4A2, etc.), genes involved in angiogenesis (EGFL6, ANGPT2, and PDGFA), smooth muscle-related contractile genes, and RAS superfamily members (Fig. 2F). Pan-dCAFs expressed a different set of collagens (COL1A1, COL7A1, COL10A1, etc.), MMPs, TGFB-signaling molecules and ECM related genes (Fig. 2F). Similarly, we found that pan-pCAFs had a similar gene expression pattern with pan-dCAFs, suggesting that pan-pCAFs are proliferating pan-dCAFs (Fig. 2F). Lastly, both pan-iCAFs and pan-iCAFs-2 had low expression of collagen genes but high expression of pro-inflammatory molecules (Fig. 2F). Specifically, pan-iCAFs highly expressed pro-inflammatory and immunosuppressive factors IL-33, CXCL12, and CXCL14, while pan-iCAFs-2 highly expressed CXCL1-3, CCL2, IL-6, and IL-7 (Fig. 2F). These results highlight the similarity and difference among pan-CAF subtypes, as well as relevant cytokines and immune factors that could be produced by each CAF subtype for mediating its specific interactions with other TME or tumor cells. Lastly, to provide a framework for future experimental identification and isolation of pan-CAF subtypes, we identified marker genes that produce proteins on the surface of cellular membranes using the cell surface protein atlas database (32) (Fig. 2G).
Collectively, these results describe the transcriptomic and molecular characteristics of CAF subtypes commonly observed across multiple cancer types. In addition, we provide a framework for the isolation of pan-CAF subtypes for functional assays by delineating cell surface specific marker genes.
Intra- and Inter-tumor CAF heterogeneity
An interesting question is the spatial distribution of pan-CAFs in tumors. To address this, we used CIBERSORTx to estimate the proportions of pan-CAF subtypes in samples used for bulk RNA-seq (22). Although spatial RNA-seq data were reported previously for MEL (33) and LC (34), we were only able to obtain the dataset from tumor samples of non-small-cell lung cancer (NSCLC) (23). The bulk RNA-seq data were from three to six regions (T1-T6) of twelve tumors in twelve patients. Using CIBERSORTx and our integrated pan-CAF scRNA-seq data, we found that in general the estimated pan-CAF abundances showed a large variation among tumors/patients but a relatively small variation in samples taken from different regions of the same tumors (Supplementary Fig. S2). However, some regional biopsies were enriched for specific pan-CAF subtypes. For example, for the samples from patient 160003805, biopsy T2 and T5 showed a relatively reduced proportion of pan-iCAF compared to T3 and T4 (Supplementary Fig. S2B). Overall, the relative abundance of pan-CAFs estimated for these bulk RNA-seq samples is consistent with what was seen in our LC scRNA-seq data (Fig. 2).
Pan-CAF subtype specific transcription factors and gene regulatory networks
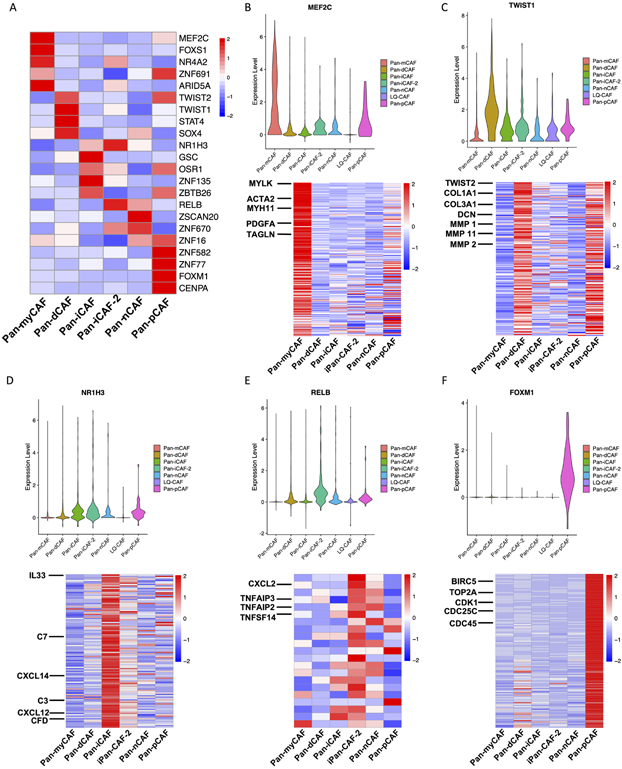
We next sought to identify transcription factors (TFs) and their targeted gene regulatory networks in order to better understand how pan-CAF subtypes are established and maintained genetically. For this, we applied the software SCENIC (25) to identify TFs and their targets that are highly active in one pan-CAF subtype vs others. We observed that pan-myCAF were enriched for the MEF2C (Fig. 3), which was previously suggested to regulate neo-angiogenesis (35). In addition, target genes for MEF2C such as MYLK, ACTA2, and others were all upregulated in pan-myCAFs (Fig. 3B). TWIST1 was among the TFs highest expressed in pan-dCAFs (Fig. 3), and it is a key factor for CAF transdifferentiation (36, 37). Furthermore, target genes for TWIST1 (TWIST2, COL1A1, MMP2, etc.) were all upregulated in pan-dCAFs (Fig. 3C). Pan-iCAFs and pan-iCAFs-2 showed high expression of NR1H3 (Fig. 3), a key regulator known to control transcriptional programs associated with inflammation (38). Target genes for NR1H3 (IL-33, CXCL14, CXCL12, etc.) were all upregulated in pan-iCAFs (Fig. 3D). In addition to NR1H3, pan-iCAFs-2 showed high expression of NF-kB subunit RELB, a known co-factor involved in promoting inflammatory transcriptional programs (38, 39). Furthermore, the key RELB target pro-inflammatory genes (CXCL2, TNFAIP3, etc.) were upregulated in Pan-iCAFs-2 (Fig. 3E). Lastly, pan-pCAFs showed high expression of FOXM1, which is implicated in cellular proliferation (40). In addition, known cycle regulatory target genes of FOXM1 (BIRC5, CDK1, etc.) were upregulated in pan-pCAFs (Fig. 3F). Collectively, these results identify key TFs driving or maintaining the gene expression programs in identified pan-CAF subtypes, providing further insights into the gene regulatory networks underlying CAF heterogeneity.
Figure 3. TFs and regulatory gene programs associated with pan-CAF subtypes.
(A) Heatmap of gene expression of transcription factors. (B) Violin plot depicting the MEF2C expression (top) and heatmap depicting MEF2C target genes upregulated in pan-myCAFs. (C) Violin plot depicting the TWIST1 expression (top) and heatmap depicting TWIST1 target genes upregulated in pan-dCAFs. (D) Violin plot depicting the NR1H3 expression (top) and heatmap depicting NR1H3 target genes upregulated in pan-iCAFs. (E) Violin plot depicting the RELB expression (top) and heatmap depicting RELB target genes upregulated in pan-iCAF-2. (F) Violin plot depicting the FOXM1 expression (top) and heatmap depicting FOXM1 target genes upregulated in pan-pCAF.
Pan-cancer assessment of the prognostic utility of pan-CAF subtype-specific gene signatures
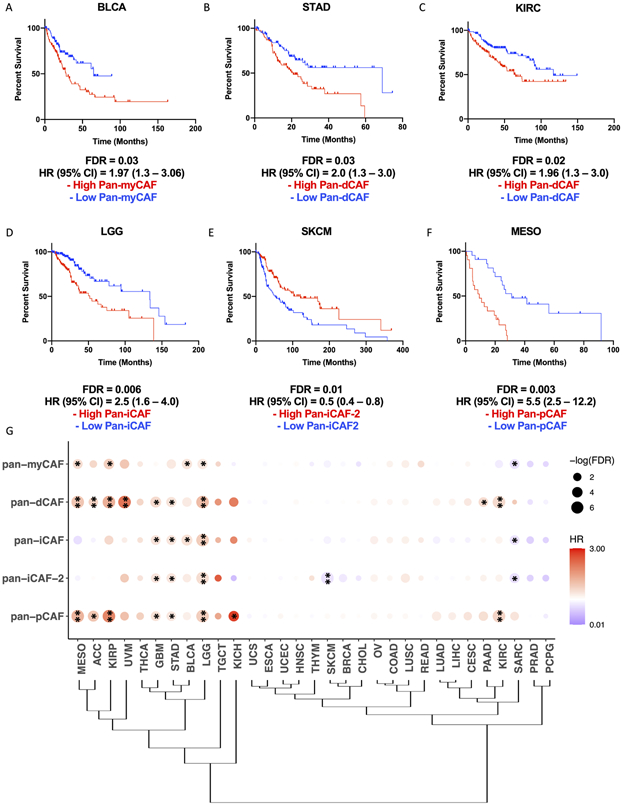
Previous studies suggest that CAFs can exert an anti-tumorigenic effect in the TME can lead to disease exacerbation in distinct cancer types (7-9). We wonder if this is related to CAF subtypes present differently in various cancers. Thus, we sought to assess how cancer type and tissue of origin may shape the CAF landscape of tumors and the pro- and anti-tumorigenic properties of identified pan-CAF subtypes. To address this, we curated pan-CAF subtype specific gene signatures and used them to estimate pan-CAF abundances across 31 cancer types available through the TCGA, using higher expression of the signature genes as a proxy for greater abundance of a pan-CAF subtype. Because the pan-CAF gene markers (Fig. 2) were defined to best distinguish CAF subtypes, we refined them by considering gene expression levels in tumor, immune, and other stroma cells to obtain more specific signature genes (see Methods and Supplementary Table S3). With them, we assessed the prognostic utility of our curated pan-CAF subtypes. We found that the expression of pan-myCAFs signature genes was highest in cholangiocarcinoma (CHOL) and lowest in kidney chromophobe (KICH) and kidney renal papillary cell carcinoma (KIRP) (Supplementary Fig. 3A). A high expression of pan-myCAFs signature genes was correlated with a poor prognosis in bladder urothelial carcinoma (BLCA) (Fig. 4A) and KIRP (Supplementary Fig. 3A), suggesting that pan-myCAFs may have a pro-tumorigenic effect in the BLCA and KIRP TMEs. The expression of pan-dCAFs signature genes was highest in CHOL and breast cancer (BRCA) and lowest in KICH (Supplementary Fig. 3B). In addition, high expression of pan-dCAF gene signature was correlated with a poor prognosis in stomach adenocarcinoma (STAD) (Fig. 4B), kidney renal clear cell carcinoma (KIRC) (Fig. 4C), uveal melanoma (UVM), KIRP, mesothelioma (MESO), and low grade glioma (LGG) cancer types (Supplementary Fig. 4B-E). The expression of pan-iCAF signature genes was highest in uterine carcinosarcoma (USC) and lowest in KICH (Supplementary Fig. 3C). In addition, high expression of pan-iCAFs signature genes was correlated with a poor prognosis in LGG (Fig. 4D). Furthermore, the expression of pan-iCAFs-2 genes was highest in KIRC and lowest in KICH (Supplementary Fig .3D). In addition, high expression of the iCAFs-2 genes was correlated with a poor prognosis in LGG (Supplementary Fig. 4F), while high expression of iCAFs-2 signature genes was correlated with a favorable clinical outcome in skin cutaneous melanoma (SKCM) (Fig. 4E). The expression of pan-pCAF signature genes was highest in CHOL and lowest in LGG (Supplementary Fig. 3E). High expression of the pan-pCAF genes was correlated with a poor prognosis for several cancer types including in MESO (Fig. 4F), KIRC, KIRP, and LGG (Supplementary Fig. 4G-I). To better assess the prognostic utility of our curated pan-CAF subtype gene signatures, we performed multivariate cox proportional hazard modeling while adjusting for age and gender. The results support a significant association of high expression of pan-CAF subtype specific signature genes with clinical outcomes for each of the cancer types described above (Supplementary Table S4).
Figure 4. Specific pan-CAF subtypes linked to clinical outcome in distinct cancer types.
Kaplan-Meier plots depicting the survival differences among patients with high and low pan-myCAF in BLCA (A), pan-dCAF in KIRC (B) and STAD (C), pan-iCAF in LGG (D), and pan-iCAF-2 in SKCM (E), and pan-pCAF in MESO (F). (G) Clustering of tumor types by the hazard ratios (HRs) for the six pan-CAF gene signatures. Dot plot in which the size is related to statistical significance and the color indicates hazard ratio (red = hazard ratio above one [poor prognosis] and blue = hazard ratio below one [favorable prognosis]). * FDR < 0.05, ** FDR < 0.01.
Hierarchical clustering of hazard ratios (HRs) for individual pan-CAF gene signatures indicates that TCGA tumors can be categorized into three groups. The first group consists of tumor types in which the majority of pan-CAF subtypes were strongly correlated with a poor prognosis: MESO, adrenocortical carcinoma (ACC), KIRP, UVM, thyroid carcinoma (THCA), glioblastoma (GBM), STAD, BLCA, LGG, testicular germ cell tumors (TGCT), and KICH (Fig. 4G). The second group consists of tumor types in which the majority of pan-CAF subtypes were moderately to not correlated with prognosis: uterine carcinosarcoma (UCS), esophageal carcinoma (ESCA), uterine corpus endometrial carcinoma (UCEC), HNSC, thymoma (THYM), SKCM, BRCA, CHOL, ovarian serous cystadenocarcinoma (OV), colon adenocarcinoma (COAD), lung squamous cell carcinoma (LUSC), rectum adenocarcinoma (READ), lung adenocarcinoma (LUAD), liver hepatocellular carcinoma (LIHC), cervical squamous cell carcinoma (CESC), pancreatic adenocarcinoma (PAAD), and KIRC (Fig. 4G). The third and final group consists of tumor types in which the majority of pan-CAF subtypes were correlated with a favorable prognosis: prostate adenocarcinoma (PRAD) and pheochromocytoma and paraganglioma (PCPG) (Fig. 4G). Collectively, these results indicate how cancer type and tissue of origin shape the CAF landscape and provide insight to which pan-CAF subtypes may contribute to a tumor promoting or tumor suppressive role in specific cancer types.
Association of Pan-CAF subtypes with specific molecular functions in tumors
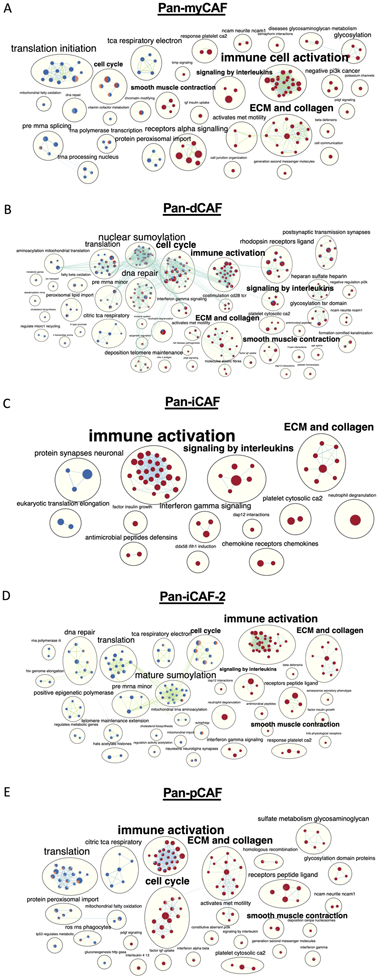
Since we observed that individual pan-CAF subtypes were correlated with a poor prognosis in specific cancer types, we sought to identify the molecular pathways that are deregulated in the tumors with greater abundance of the pan-CAF subtypes. For the cancer types in which a pan-CAF subtype was significantly associated with either poor or favor prognosis, we applied ranked gene set enrichment analysis (rGSEA). To better illustrate how functionally related gene sets were enriched (red color) or depleted (blue color) across multiple cancers, results were organized as a network (Fig. 5). This analysis revealed key and interesting similarities across cancer types among cancers with increased activities of pan-myCAFs, pan-dCAF, pan-iCAF, pan-iCAF-2, and pan-pCAF (Fig. 5). Specifically, tumors with increased abundances of these five pan-CAF subtypes showed a significant enrichment for active gene sets (nodes) associated with immune cell activation and ECM and collagen (Fig. 5), suggesting a likely common function of CAFs. The immune cell activation gene sets largely consisted of nodes related to innate immune response, while the ECM and collagen enrichment were mostly from nodes related to ECM remodeling (Fig. 5). However, when considered the total numbers of gene sets associated with immune cell activation, pan-iCAF and pan-iCAF-2 were linked to the highest number of gene sets (26 and 29, respectively) compared to pan-myCAF, pan-dCAF, and pan-pCAF (22, 22, and 23, respectively) (Fig. 5). Conversely, pan-iCAF and pan-iCAF-2 were associated with the lowest number of enriched gene sets (8 and 10, respectively) of ECM and collagen, compared to pan-myCAF, pan-dCAF, and pan-pCAF (14, 13, and 15, respectively) (Fig. 5 C, D). This result suggests that pan-iCAFs and pan-iCAFs-2 are potentially associated with a molecular microenvironment with enhanced immune cell activation while pan-myCAF, pan-dCAF, and pan-pCAF are associated with a molecular microenvironment that favors ECM remodeling.
Figure 5. Networks of REACTOME terms enriched or depleted in tumors with high expression of different pan-CAF signature genes.
Nodes are terms enriched (red circles) or depleted (blue circles) among genes expressed higher in tumors with increased expression of pan-CAF gene signatures, while edges link terms with overlapping genes. Connected nodes with similar functions are further summarized by a more generalized term using Enrichmentmap. In each node, the filled colors represented results from individual cancer types. (A) pan-myCAF results from BLCA and KIRP cancers. (B) pan-dCAF results for KIRC, KIRP, LGG, UVM, MESO, and STAD cancers. (C) pan-iCAF results for LGG. (D) pan-iCAF 2 results for LGG and SKCM cancers. (E) pan-pCAF for KIRC, KIRP, LGG, and MESO.
This analysis also revealed several other notable differences. Compared to tumors with a high proportion of pan-iCAF, pan-iCAF-2, or pan-pCAF, the tumors with a high proportion of pan-myCAF exhibited the highest number of gene sets (n = 4) associated with smooth muscle cell contraction (Fig. 5). In addition, compared to tumors with a high proportion of pan-myCAF, pan-dCAF, and pan-pCAF, tumors with a high proportion of pan-iCAF showed an increase in the number of gene sets associated with signaling by interleukins (Fig. 5), suggesting that pan-iCAFs are likely associated with a more inflammatory molecular microenvironment. Lastly, cancers with increased proportions of pan-dCAFs and pan-pCAFs showed positively enriched gene sets associated with cell cycle, suggesting that tumors with a high proportion of pan-dCAF and pan-pCAF are associated with a molecular microenvironment with an enhanced cell cycle potential (Fig. 5A, C).
Taken together, this data suggests that distinct pan-CAF subtypes reside in molecular TMEs that may provide favored or unfavored environments to facilitate or hinder tumor development across various cancer types, by activating or suppressing specific gene pathways, a hypothesis that need to be tested by future functional assays.
Association of Pan-CAF subtypes with anti-PD1 and anti-PD-L1 resistance
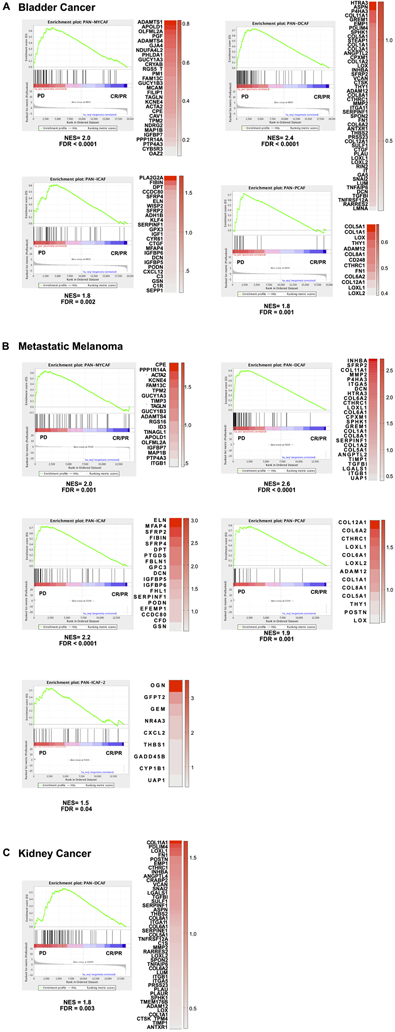
Therapeutic antibodies targeting programmed death-ligand 1 (PD-L1) or programmed death-1 (PD-1) can induce a robust and durable response among patients diagnosed with metastatic urothelial cancer (29) and metastatic melanoma (30). Since we observed specific pan-CAF subtypes express immunosuppressive inflammatory factors (e.g., CXCL12, CXCL14), we studied if pan-CAF subtypes are associated with the effectiveness of immune targeting therapy. For this, we obtained two public datasets that describe the transcriptomic features of metastatic urothelial tumors treated with anti-PD-L1 (29) and metastatic melanoma tumors treated with anti-PD-1 (30). We used our curated pan-CAF subtype specific signature genes to perform GSEA analysis between patients with progressive disease (PD) and patients with complete response (CR) or partial response (PR) upon either anti-PD-L1 or anti-PD-1 treatment. The result showed a positive and significant enrichment of gene sets associated with pan-myCAF, pan-dCAF, pan-iCAF, and pan-pCAF among metastatic bladder cancer patients who experienced PD when receiving anti-PD-L1 (Fig. 6A). Similarly, we found a positive and significant enrichment for gene sets associated with pan-myCAF, pan-dCAF, pan-iCAF, pan-pCAF, and pan-iCAF-2 in metastatic melanoma patients who experienced PD while receiving anti-PD-1 (Fig. 6B). However, we only observed a positive and significant enrichment of gene sets associated with pan-dCAF among patients diagnosed with metastatic kidney cancer who experienced PD when treated with anti-PD-L1 (Fig. 6C). Collectively, these results suggest that specific pan-CAFs are potentially associated with ICB resistance in metastatic bladder, melanoma, and kidney cancer.
Figure 6. Pan-CAF subtypes in anti-PD1 resistant tumors.
(A) Gene set enrichment analysis for bladder tumors treated with anti-PD1 showing significant enrichment of pan-myCAF (left), pan-dCAF (left-middle), and pan-iCAF (right-middle), and pan-pCAF (right) gene signature in the progressive disease (PD) patients compared to complete response (CR) and partial response (PR) patients. (B) Gene set enrichment analysis for metastatic melanoma tumors treated with anti-PD1 treatment showing significant enrichment of pan-myCAF (left), pan-dCAF (left-middle), pan-iCAF (right-middle), pan-pCAF (right), and pan-iCAF-2 (left-bottom) gene signature in the PD patients compared to CR and PR patients. (C) Gene set enrichment analysis for kidney tumors treated with anti-PD1 treatment showing significant enrichment of pan-dCAF gene signature in the PD patients compared to CR and PR patients. The heatmaps in each panel shows the expression difference of the leading edge genes between PD and CR/PR tumors.
Discussion
We provide genetic features for six pan-CAF subtypes that are commonly observed across cancer types, including a myofibroblast-, desmoplastic-, two inflammatory-, normal-, and a proliferating-like CAF. Using the TCGA database, we study the clinical relevance for individual pan-CAF subtypes and link pan-CAF subtypes with underlying gene expression programs that can play critical roles in tumor biology, clinical outcome, and ICB resistance.
A few previous studies have investigated the similarities of CAF heterogeneity across cancer types. A recent scRNA-seq study profiled CAFs from LC, colorectal cancer, and ovarian cancer (31). Similar to their findings, we observe a myCAF population that is characterized by high expression of smooth muscle related contractive genes and activated fibroblast markers (i.e., ACTA2). However, we also observed that pan-myCAF highly express genes related to angiogenesis (31). This difference is important and consistent with a previous study in pancreatic ductal adenocarcinoma (PDAC) models that linked ACTA2+ CAFs to angiogenesis (8). Furthermore, a scRNA-seq study profiling stroma cells from triple negative breast cancer (TNBC) (41) and breast cancer (42) characterize myCAFs as having elevated expression of activated fibroblast markers, suggesting that pan-myCAF might be conserved in breast cancer. In addition, our findings suggest that BLCA and KIRP may benefit the most from pan-myCAF targeted therapy. The poor clinical outcome observed among patients diagnosed with these cancer types might be linked to the activation of underlying pathways associated with smooth muscle cell contraction and innate immune cell activation. Innate immune cell activation is associated with establishing an immunosuppressive microenvironment which may also elucidate a potential explanation as to why pan-myCAFs are linked to ICB resistance in metastatic bladder and melanoma cancer.
Our pan-cancer characterization of CAF heterogeneity led to the discovery of two CAF subtypes involved in ECM remodeling: pan-dCAF and pan-pCAF. Pan-dCAF showed elevated expression of TWIST1, suggesting that epithelial-mesenchymal-transition (EMT) may be required for the transdifferentiation of this highly invasive CAF phenotype (43). Consistently with this, previous studies have shown that depletion of TWIST1 in CAFs derived from both ESCC and STAD led to reduced tumor cancer cell migration and invasion (36, 37). Indeed, our pan-cancer TCGA analysis showed that pan-dCAFs were correlated with a poor prognosis in STAD, further highlighting the clinical relevance of pan-dCAFs in this stomach cancer. In addition, we found that pan-dCAFs highly express STC-1, which - in studies using CAFs derived from colorectal cancer - has been implicated in promoting tumor cell intravasation and metastases (44). Interestingly, we observed that pan-dCAFs also highly express VEGFA and PDGFC soluble factors, suggesting this CAF subset may contribute to the establishment of a pro-angiogenic microenvironment essential for cancer cell invasion and metastases (45). Lastly, our findings suggest that STAD, KIRC, UVM, KIRP, MESO, and LGG are cancer types that might benefit the most from potential therapeutic intervention targeting pan-dCAFs. Importantly, positive enrichment of cell cycle gene sets and an expansion in the number of gene sets associated with the ECM and collagen annotation was observed in cancer types that had a poor clinical outcome associated with pan-dCAFs, suggesting key molecular pathways that pan-dCAFs modulate in the TME. Lastly, pan-dCAFs were enriched in metastatic melanoma, bladder, and kidney tumors resistant to ICB therapy. A review of the leading edge genes driving enrichment of pan-dCAFs in these ICB resistant tumors pointed to the potential important factors, TGFB1 and genes related to collagen. Indeed, inhibition of TGFB1 and downstream signaling pathways has been shown to significantly improve response to ICB therapeutic intervention (29). Lastly, the production of collagens in the TME might provide a physical barrier that impairs adaptive immune cell trafficking to sites of neo-antigen presentation.
In our pan-cancer characterization of CAFs, we identified a novel subtype defined as pan-pCAF. This pan-CAF phenotype displays gene expression patterns similar to what was observed in pan-dCAFs; however, a fundamental distinction is that pan-pCAF had upregulation of gene programs associated with cell cycle. Single cell RNA-seq analysis of CAFs derived from a mouse breast cancer model also identified a pCAF, suggesting that this molecular subtype might be conserved across species (46). In addition, we observe a positive and significant enrichment of pan-pCAFs in metastatic bladder and melanoma tumors resistant to ICB therapy, and, similar to pan-dCAF, genes related to collagen were primarily driving enrichment. Overall, the identification of pan-pCAF subtype merits further study into the different contexts of how this subtype is derived and why the proliferation of specifically the dCAF is favored for tumor development and pathogenesis.
Pan-cancer characterization of CAFs led to the discovery of two inflammatory CAF subtypes: pan-iCAF and pan-iCAF-2. Importantly, both pan-iCAF and pan-iCAF-2 highly expressed genes involved in complement cascade activation. A previous study showed that complement component 5 (C5) indirectly contributes to suppressed T-cell activity in the TME by acting as a chemoattractant for immunosuppressive myeloid cells (47). The identification of the expression of CAF-subtype specific complement factors merits further research into the role of these factors in the TME. We observed that pan-iCAF specifically expressed CXCL12 and CXCL14 (48,49). Studies in prostate cancer suggest that CAF-derived CXCL12 and CXCL14 promote macrophage differentiation and M2 polarization (48,49), thus suggesting that pan-iCAFs may have an immunosuppressive role in the TME. Our TCGA analysis identified LGG as a cancer type that might benefit from pan-iCAF targeted therapy. Patients with high pan-iCAFs might be linked to a poor clinical outcome due to an enrichment of gene sets and pathways related to interleukin signaling, which is linked to the requirement and activation of innate immune cells and the establishment of an immunosuppressive microenvironment. Furthermore, our observed enrichment of pan-iCAF in ICB resistant metastatic bladder and melanoma tumors further highlights the immunosuppressive roles of pan-iCAFs. Among the core enriched genes for pan-iCAF in ICB resistant tumors we identify DCN, CFD, C3, and CXCL12; however, the role of these factors in ICB resistance is largely unknown and warrants further investigation.
For pan-iCAF-2, we observed specific expression of CCL2. In LIHC, it has been shown that CAFs contributes to an immunosuppressive microenvironment by recruiting circulating MDSCs through the secretion of CCL2 (50). Furthermore, pan-iCAF-2 specifically expresses IL-6. Previous studies have shown that CAF-derived IL-6 is implicated in cancer stem cell self-renewal in breast cancer (5), drug resistance in breast cancer (5) and non-small cell lung cancer (NSCLC) (51) and CAF-activation (52). In addition, we identified LGG as a cancer type that might benefit from pan-iCAF-2 targeted therapies. Lastly, we observed enrichment of pan-iCAF-2 in ICB resistant metastatic melanoma tumors. Among the core enriched genes for pan-iCAF-2 in ICB resistant tumors we identified CXCL2, however, its role in ICB resistance is not well studied.
There are a few limitations of our study worth noting. First of all, our work is computational and omic nature, thus it will be important to validate the CAF markers from our findings to identify and characterize CAF subtypes in solid tumor tissues using experimental methods such as fluorescence-activated cell sorting (FACS), immunohistochemistry, and/or immunofluorescence. Secondly, although we have tried our best to achieve the robustness of our clustering analysis of CAFs in the three cancer types, we envision that our clustering results can be further improved and refined with larger datasets (more cells and more tumor types) and additional software. Thirdly, in our TCGA analysis we relied on the expression of signature genes to assess the prognostic utility of individual pan-CAFs across cancer types. While this makes the result more clinically relevant (since it is relatively easy to test the expression of a few marker genes), the findings can be improved with other orthogonal approaches for determining cell population abundance, such as FACS. In relation to this, it would also be interesting to determine the spatial distribution of CAF subtypes, using technology such as spatial transcriptomics.
In conclusion, we have comprehensively and systematically characterized six-distinct CAF subtypes commonly observed across multiple cancer types (pan-CAFs). Our work suggests specific cancer types that might benefit from pan-CAF subtype specific targeted therapy, a finding that can be experimentally tested in the future. Importantly, we identify several pan-CAF subtypes that are related to ICB resistance which can be evaluated in future functional assays. As the increasing widely usage of single cell technologies, we anticipate that more datasets including CAFs across an array of other cancer types will become available. Ultimately, our current study provides a framework for future studies to characterize and compare CAF heterogeneity across cancer types and research their functional and clinical roles.
Supplementary Material
Translational Relevance.
Cancer associated fibroblasts (CAFs) can support or suppress tumor growth. However, marker genes and molecular mechanisms that distinguish tumor-promoting from -suppressive CAFs is unclear. In addition, it is not fully understood how tissue of tumor origin affects the functions of CAFs in distinct tumor microenvironments. Here, at single cell resolution, we identify and describe the molecular characteristics of six CAF subtypes across cancer types. Using the TCGA resource, we demonstrate that these CAF subtypes are strongly associated with a poor or favorable clinical outcome in distinct cancer types. Importantly, we also study the underlying molecular mechanisms that may link these CAF subtypes to tumor progression and resistance to immune checkpoint blockade. Taken together, our work provides substantial evidence for the prognostic utility associated with CAF subtype specific gene signatures, and identify key molecular pathways associated with individual CAF subtypes that may contribute to tumor progression and immunotherapy resistance.
Acknowledgements
This work is supported by NIH/NCI (R01CA211336 to D Zheng and R01CA175495 to X Zang), and US/DOD (BC190403 to X Zang), PM Galbo is supported by the NIH/National Center for Advancing Translational Science (NCATS) Einstein-Montefiore CTSA training grant (TL1 TR002557).
Footnotes
Conflict of Interest: None to declare
Data and Code Availability
All the scRNA-seq and bulk RNA-seq data were from previously publications or public domains. The code for the analysis is available at https://github.com/bioinfoDZ/panCAF.
References
- 1.Whiteside. The tumor microenvironment and its role in promoting tumor growth. Oncogene 2008; 27: 5904–5912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen X and Song E. Turning foes to friends: targeting cancer-associated fibroblasts. Nat Rev Drug Discov 2019;18:99–115. [DOI] [PubMed] [Google Scholar]
- 3.Liu T, Han C, Wang S, Fang P, Ma Z, Xu L, et al. Cancer-associated fibroblasts: an emerging target of anti-cancer immunotherapy. J Hematol Oncol 2019;12:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang X, Jin X, Malladi S, Zou Y, Wen YH, Brogi E, et al. Selection of bone metastasis seeds by mesenchymal signals in the primary tumor stroma. Cell 2014;154:1060–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Su S, Chen J, Yao H, Liu J, Yu S, Lao L, et al. CD10+ GPR77+ cancer-associated fibroblasts promote cancer formation and chemoresistance by sustaining cancer stemness. Cell 2018;172:841–856. [DOI] [PubMed] [Google Scholar]
- 6.Costa A, Kieffer Y, Scholer-Dahirel A, Pelon F, Bourachot B, Cardon M, et al. Fibroblast heterogeneity and immunosuppressive environment in human breast cancer. Cancer Cell 2018;33:463–479. [DOI] [PubMed] [Google Scholar]
- 7.Ozdemi BC, Pentcheva-Hoan T, Carstens JL, Zheng X, Wu CC, Simpson TR, et al. Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppressive and accelerates pancreas cancer with reduced survival. Cancer Cell 2014;25:719–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rhim AD, Oberstein PE, Thomas DJ, Mirek ET, Palermo CF, Sastra SA, et al. Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer Cell 2014;25:735–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brechbul HM, Finlay-Schultz J, Yamamoto TM, Gillen AE, Cittelly DM, Tan AC, et al. Fibroblast subtypes regulate responsiveness of luminal breast cancer to estrogen. Clin Cancer Res 2017;23: 1710–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ren X, Kang B, Zhang Z. Understanding tumor ecosystems by single-cell sequencing: promises and limitations. Genome Biol 2018; 19: 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tirosh I, Izar B, Prakadan SM, Wadsworth MH, Treacy D, Trombetta JJ. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 2016;352:189–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Puram SV, Tirosh I, Parikh AS, Patel AP, Yizhak K, Gillespie S, et al. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell 2017;171:1611–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lambrechts D, Wauters E, Boeckx B, Aibar S, Nittner D, Burton O, et al. Phenotype molding stromal cells in the lung tumor microenvironment. Nat Med 2018;24:1277–1289. [DOI] [PubMed] [Google Scholar]
- 14.Hovestadt V, Smith KS, Bihannic L, Filbin MG, Shaw ML, Baumgartner A, et al. Resolving medulloblastoma cellular architecture by single-cell genomics. Nature 2019;572:74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peng J, Sun BF, Chen CY, Zhou JY, Chen YS, Chen H, et al. Single-cell RNA-seq highlights intratumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res 2019;29:777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Venteicher AS, Tirosh I, Hebert C, Yizhak K, Neftel C, Filbin MG, et al. Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq. Science 2017;355:eaai8478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldman MJ, Craft B, Hastie M, Repecka K, McDade F, Kamath A, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol 2020;38:675–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 2018;36:411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Becht E, Mclnnes L, Healy J, Dutertre CA, Kwok I, Ng L, et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol 2018;37:38–44. [DOI] [PubMed] [Google Scholar]
- 20.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009;25:1091–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newman AM, Steen CB, Liu CL, Gentles A, Chanudhuri AA, Scherer F, et al. Determining cell types abundance and expression form bulk tissues with digital cytometry. Nat Biotechnol 2019;37(7):773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jia Q, Wu W, Wang Y, Alexander PB, Sun C, Gong Z, et al. Local mutational diversity drives intratumoral immune heterogeneity in non-small cell lung cancer. Nat Commun 2018;9(1):5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M, et al. The human transcription factors. Cell 2018;172:650–665. [DOI] [PubMed] [Google Scholar]
- 25.Aibar S, Gonzale-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, et al. SCENIC: Single-cell regulatory network inference and clustering. Nat Methods 2018;14:1083–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005;102:15545–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data using DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One 2010;5:e13984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mariathasan S, Turley SJ, Nickles D, Castiglioni A, Yuen K, Wang Y, et al. TGFB attenuates tumor response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018;554(7693):544–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan S, et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell 2016;165(1):35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qian J, Olbrech S, Boeckx B, Vos H, Laoui D, Etlioglu E, et al. A pan-cancer blueprint of the heterogenous tumor microenvironment revealed by single-cell profiling. Cell Res 2020;30:745–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buasch-Fluck D, Hofmann A, Bock T, Frei AP, Cerciello F, Jacobs A, et al. A mass spectrometric-derived cell surface protein atlas. PLoS One 2015;10(3):e0121314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thrane K, Eriksson H, Maaskola J, Hansson J, Lundeberg J. Spatially resolved transcriptomics enables dissection of genetic heterogeneity in stage III cutaneous malignant melanoma. Cancer Res 2018;78(20):5970–5979. [DOI] [PubMed] [Google Scholar]
- 34.Joshi K, de Massy MR, Ismail M, Reading JL, Uddin I, Woolston A, et al. Spatial heterogeneity of the T cell receptor repertoire reflects the mutational landscape in lung cancer. Nat Med 2019;25(10):1549–15590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maiti D, Xu Z, Duh EJ. Vascular endothelial growth factor induces MEF2C and MEF2-dependent activity in endothelial cells. Invest Ophthalmol Vis Sci 2015;49:3640–3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yeo SY, Ha SY, Lee KW, Cui Y, Yang ZT, Xuan YH, et al. Twist1 is highly expressed in cancer-associated fibroblasts of esophageal squamous cell carcinoma with a prognostic significance. Oncotarget 2017;8:65265–65280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee KW, Yeo SY, Sung CO, Kim SH. Twist1 is a key regulator of cancer-associated fibroblasts. Cancer Res 2015;75:73–85. [DOI] [PubMed] [Google Scholar]
- 38.Chen L, Fukuda N, Shimizu S, Kobayashi H, Tanaka S, Nakamura Y, et al. Role of complement 3 in renein generation during the differentiation of mesenchymal stem cells to smooth muscle cells. Am J Physiol Cell Physiol 2020;18:C981–C990 [DOI] [PubMed] [Google Scholar]
- 39.Bren GD, Sloan NJ, Miyoshi H, Pennington KN, Pobst LJ, Paya CV. Transcription of the RelB gene is regulated by NF-kappaB. Oncogene 2001;20:7722–33 [DOI] [PubMed] [Google Scholar]
- 40.Liao GB, Li XZ, Zeng S, Liu C, Yang SM, Yang L, et al. Regulation of the master regulator FOXM1 in cancer. Cell Commun Signal 2018;16:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu SZ, Roden DL, Wang C, Holliday H, Harvey K, Cazet AS, et al. Stromal cell diversity associated with immune evasion in human triple-negative breast cancer. EMBO J 2020;13:e104063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kieffer Y, Hocin HR, Gentric G, Pelon F, Bernard C, Bourachot B, et al. Single-cell analysis reveals fibroblast cluster linked to immunotherapy resistance in cancer. Cancer Discov 2020;10:1330–1351. [DOI] [PubMed] [Google Scholar]
- 43.Yan J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell 2008;14:818–29. [DOI] [PubMed] [Google Scholar]
- 44.Pena C, Cespedes MV, Lindh MB, Kiflemariam S, Mezheyeuski A, Edqvist PH, et al. STC1 expression by cancer-associated fibroblasts drives metastasis of colorectal cancer. Cancer Res 2013;73:1287–97. [DOI] [PubMed] [Google Scholar]
- 45.O’Connel JT, Sugimoto H, Crooke VG, MacDonald BA, Mehta AI, LeBleu VS, et al. VEGF-A and tenascin-C produced by S100A4+ stromal cells are important for metastatic colonization. Proc Natl Acad Sci USA 2011;108:16002–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bartoschek M, Oskolkov N, Bocci M, Lovrot J, Larsson C, Sommarin M, et al. Spatially and functionally distinct subclasses of breast cancer-associated fibroblasts revealed by single cell RNA sequencing. Nat Commun 2018;9:5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Markiewski MM, DeAngelis RA, Benencia F, Ricklin-Lichtsteiner SK, Koutoulaki A, Gerard C, et al. Modulation of the antitumor immune response by complement. Nat Immunol 2008;9:1225–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Comito G, Giannoni E, Segura CP, Barcellos-de-Souza P, Raspollini MR, Raspollini MR, et al. Cancer-associated fibroblasts and M2-polarized macrophage synergize during prostate carcinoma progression. Oncogene 2014;33:2423–31. [DOI] [PubMed] [Google Scholar]
- 49.Augsten M, Sjoberg E, Frings O, Vorrink SU, Frijhoff J, Olsson E, et al. Cancer-associated fibroblasts expressing CXCL14 rely upon NOS1-derived nitric oxide signaling for their tumor-supporting properties. Cancer Res 2014;74:2999–3010. [DOI] [PubMed] [Google Scholar]
- 50.Yang X, Lin Y, Shi Y, Li B, Liu W, Yin W, et al. FAP promotes immunosuppression by cancer-associated fibroblasts in the tumor microenvironment via STAT3-CCL2 signaling. Cancer Res 2016;76:4124–35. [DOI] [PubMed] [Google Scholar]
- 51.Shintani Y, Fujiwara A, Kimura T, Kawamura T, Funaki S, Minami M, et al. Il-6 secreted from cancer-associated fibroblasts mediates chemoresistance in NSCLC by increasing epithelial-mesenchymal transition signaling. J Thorac Oncol 2016;11:1482–92. [DOI] [PubMed] [Google Scholar]
- 52.Albrengues J, Bertero T, Grasset E, Bonan S, Maiel M, et al. Epigenetic switch drives the conversion of fibroblasts into proinvasive cancer-associated fibroblasts. Nat Commun 2015;6:10204. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.