SUMMARY
Low protein diets promote metabolic health in rodents and humans, and the benefits of low protein diets are recapitulated by specifically reducing dietary levels of the three branched-chain amino acids (BCAAs), leucine, isoleucine, and valine. Here, we demonstrate that each BCAA has distinct metabolic effects. A low isoleucine diet reprograms liver and adipose metabolism, increasing hepatic insulin sensitivity and ketogenesis, and increasing energy expenditure, activating the FGF21-UCP1 axis. Reducing valine induces similar but more modest metabolic effects, while these effects are absent with low leucine. Reducing isoleucine or valine rapidly restores metabolic health to diet-induced obese mice. Finally, we demonstrate that variation in dietary isoleucine levels helps explain body mass index differences in humans. Our results reveal isoleucine as a key regulator of metabolic health and the adverse metabolic response to dietary BCAAs, and suggest reducing dietary isoleucine as a new approach to treating and preventing obesity and diabetes.
Keywords: isoleucine, valine, branched-chain amino acids, insulin resistance, obesity, diabetes, body composition, adiposity, mice, human, body mass index
eTOC blurb
Yu & Richardson et al. find that restriction of dietary isoleucine or valine promotes metabolic health in mice, and that restriction of dietary isoleucine is required for the metabolic benefits of a low protein diet. Further, higher dietary isoleucine levels are associated with increased BMI in humans.
Graphical Abstract
INTRODUCTION
Obesity is a growing problem in the United States and worldwide. Over two-thirds of the population of the United States is overweight or obese (Ogden et al., 2014), and the prevalence of these conditions in most countries is rising rapidly. Obesity is a risk factor for many other conditions, including diabetes, cardiovascular disease, cancer, and Alzheimer’s disease (1999; Barbagallo and Dominguez, 2014; Giovannucci et al., 2010). While reducing caloric intake by dieting is an obvious solution to mediate this epidemic, long-term sustainable weight loss through calorie restriction has proven unsustainable for most individuals.
Recently, a growing body of literature has demonstrated that ‘a calorie is not just a calorie.’ Prospective and retrospective cohort analyses suggest that consumption of dietary protein, particularly from animal sources, increases the risk of diabetes, insulin resistance, cancer, and is associated with a higher overall risk of mortality (Akter et al., 2020; Huang et al., 2020; Lagiou et al., 2007; Levine et al., 2014; Sluijs et al., 2010). A short-term randomized control trial found that individuals consuming a low protein (LP) diet lost weight without calorie restriction; a LP diet also reduced adiposity and lowered fasting blood glucose (Fontana et al., 2016). Multiple studies in rodents have similarly found that LP diets promote leanness, glycemic control, and even increase lifespan, while high protein diets do the opposite (Fontana et al., 2016; Laeger et al., 2014; Maida et al., 2016; Solon-Biet et al., 2014; Solon-Biet et al., 2015).
We speculated that the beneficial effects of an LP diet might result from the reduced consumption of specific dietary amino acids (AAs). The three branched-chain amino acids (BCAAs; leucine/Leu, isoleucine/Ile, and valine/Val) have long been implicated in the etiology of type 2 diabetes; blood levels of BCAAs are positively correlated with insulin resistance, obesity, and diabetes in both humans and rodents (Batch et al., 2013; Connelly et al., 2017; Felig et al., 1969; Lotta et al., 2016; Newgard et al., 2009). Additionally, a number of interventions that reduce obesity and improve metabolic health in humans lower plasma levels of BCAAs (Magkos et al., 2013; Zheng et al., 2016). Essential AA levels are lower in plant-based vs. animal-based sources (Boye et al., 2012), and humans consuming vegan or pescatarian diets have lower BCAA levels than meat eaters (Elshorbagy et al., 2017; Schmidt et al., 2016). Finally, plasma BCAAs are specifically reduced in mice and humans fed a LP diet (Fontana et al., 2016; Solon-Biet et al., 2014).
Specifically restricting dietary BCAAs recapitulates many of the metabolic effects of an LP diet, improves lifelong metabolic health in mice and extends male lifespan when begun young, while BCAA supplementation promotes hyperphagia, obesity, and shortens lifespan (Fontana et al., 2016; Richardson et al., 2021; Solon-Biet et al., 2019). Further, specifically restricting dietary BCAAs restores metabolic health to Western diet-induced obese (DIO) mice, rapidly normalizing weight, adiposity, and glycemic control without caloric restriction (Cummings et al., 2018). A low BCAA diet similarly slows the accumulation of fat and preserves insulin sensitivity in a rat model of obesity (White et al., 2016). Collectively, these data demonstrate a key role for dietary BCAAs in metabolic health.
While most research to date has investigated the physiological roles of the BCAAs in combination, evidence is now emerging that the BCAAs may have distinct effects on molecular processes, metabolism, and health. The three BCAAs differentially activate the protein kinase mTORC1 (mechanistic Target Of Rapamycin Complex 1), a key regulator of many metabolic processes (Kennedy and Lamming, 2016; Sheen et al., 2011; Wolfson et al., 2016), and are involved in the posttranslational modification of distinct proteins (He et al., 2018). While catabolism of each BCAA shares several steps, the intermediate and final products of each BCAA are distinct. For example, 3-hydroxy-isobutyrate is a Val-specific catabolite that regulates trans-endothelial fatty acid transport (Jang et al., 2016). Furthermore, a recent study found that blood levels of Leu and Val correlated with decreased mortality, while blood levels of Ile correlated with increased mortality (Deelen et al., 2019).
Here, we tested the hypothesis that each BCAA has distinct contributions to the beneficial effects of a LP diet. We find that restriction of Ile is both necessary and sufficient for the beneficial metabolic effects of a LP diet. In contrast, the restriction of Val improves metabolic health to a lesser degree, but is not necessary for the effects of a LP diet. While Ile is an agonist of mTORC1 activity, reduced hepatic activity of mTORC1 does not mediate the effects of a Low Ile diet. Similarly, the effects of reduced Ile are not mediated by the other major AA sensing kinase GCN2 (general control nonderepressible 2). Instead, we find that reducing dietary Ile reprograms hepatic metabolism, and induces expression of the insulin-sensitizing and energy-balancing hormone FGF21 (fibroblast growth factor 21), activating the FGF21-UCP1 (uncoupling protein 1) axis and promoting adipose tissue beiging and energy expenditure. Reduction of dietary Ile or Val is sufficient to restore metabolic health to DIO mice, even as these animals continue to consume an otherwise unhealthy “Western” diet. Surprisingly, restriction of Leu has no beneficial metabolic effects in lean or DIO mice, and does not contribute to the beneficial effects of a LP diet. Finally, we find that the consumption of dietary protein with increased Ile content is associated with increased body mass index in humans. In conclusion, these results demonstrate that dietary Ile is a critical regulator of metabolic health and of the response to dietary BCAAs, and suggest that interventions based on decreasing dietary levels of Ile may be a translatable way to promote and restore metabolic health.
RESULTS
Reducing dietary Ile recapitulates many metabolic effects of BCAA estriction
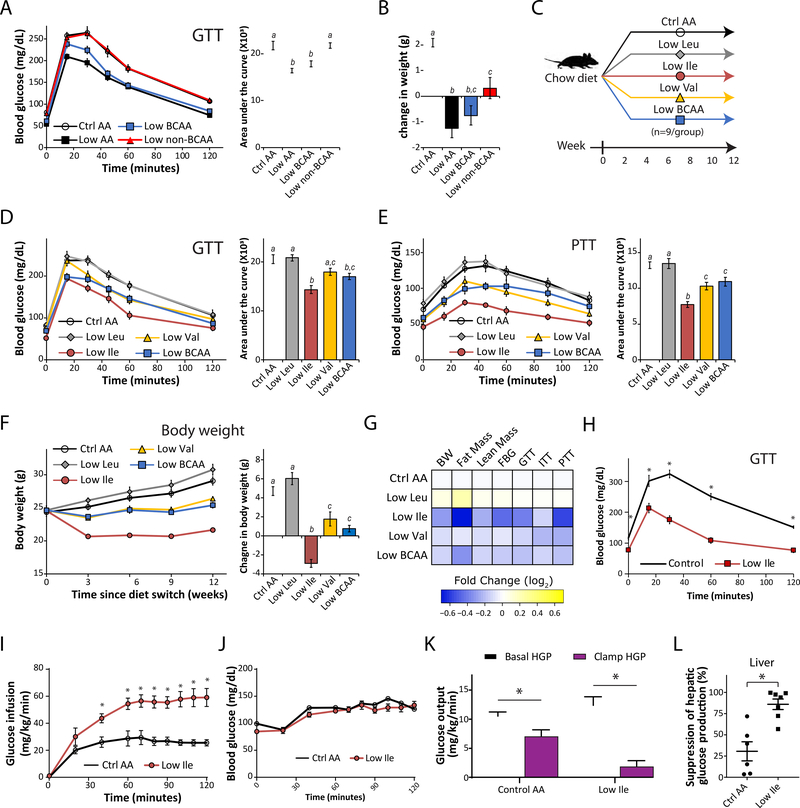
To examine the specific role of dietary BCAAs on metabolic health, we utilized an AA-defined Control (Ctrl AA) diet containing all twenty common AAs; the diet composition reflects that of a natural chow diet in which 21% of calories are derived from protein (Fontana et al., 2016). We created a diet series based on this diet, in which either all AAs (Low AA), the three BCAAs (Low BCAA), or the other essential AAs (Low non-BCAA) were reduced by 67%. All four of these diets are isocaloric, with identical levels of fat, and in the case of the latter two diets, non-essential AAs were increased to keep the calories derived from AAs constant (Table S1). Mice fed a Low AA or Low BCAA diet had significantly improved glucose tolerance and blunted weight gain vs. mice fed a Ctrl AA diet, while restriction of the other essential AAs had no effect on glucose tolerance and less impact on weight (Figures 1A and 1B). These results demonstrate the unique contribution of the BCAAs to the metabolic effects of a LP diet.
Figure 1. Dietary restriction of Ile or Val, but not Leu, improves metabolic health.
(A) Glucose tolerance of mice after 3 weeks on a Ctrl AA diet, a Low AA diet, a Low BCAA diet, or a Low non-BCAA diet (restricted in all essential AAs except for BCAAs). (B) Change in body weight of mice after 3 weeks, n=16–18/group. (C) Experimental scheme. (D and E) Glucose tolerance (D) and pyruvate tolerance (E) of mice after 3 and 5 weeks on the indicated diets, respectively. (F) Body weight and change in body weight of mice after 12 weeks. (D-F) n = 8–9/group. *p<0.05, Tukey-Kramer test following ANOVA, means with different letters are statistically different. (G) Heatmap of the metabolic effects of each diet; FBG = fasting blood glucose. (H) Glucose tolerance of mice after 3 weeks on a Ctrl AA or a Low Ile diet (n=12–13/group; *p<0.05, Sidak’s test post 2-way RM ANOVA). (I-L) Glucose infusion rate (I), blood glucose level (J), basal and clamp hepatic glucose production (K), and insulin responsiveness (L), were determined during a hyperinsulinemic-euglycemic clamp in mice maintained on a Ctrl AA diet or a Low-Ile diet for 3 weeks (n=6–7/group; *p<0.05, Student’s t-test). Data represented as mean ± SEM.
To determine the role of each individual BCAA, we designed a new diet series, in which the level of each individual BCAA – leucine (Low Leu), isoleucine (Low Ile), or valine (Low Val) – or all three BCAAs (Low BCAA) was reduced by 67%; all diets were again isocaloric with identical levels of fat, and the percentage of calories derived from AAs was kept constant by proportionally adjusting the amount of non-essential AAs (Figure 1C and Table S1). Mice fed either the Low Ile or Low BCAA diets for 3 weeks had significantly improved glucose tolerance compared to mice fed the Ctrl AA diet (Figure 1D); mice fed a Low Val diet also exhibited a trend towards improved glucose tolerance (p=0.06). In contrast, consumption of a Low Leu diet had no effect on glucose tolerance (Figure 1D). Mice fed the Low Ile, Low Val, or Low BCAA diets likewise had improved pyruvate tolerance but responded normally to insulin, indicating improved suppression of gluconeogenesis (Figures 1E and S1A); Low Leu fed mice were indistinguishable from Ctrl AA-fed mice. We determined fasting and glucose-stimulated glucose and insulin levels; while stimulated glucose levels were lower in mice fed Low Ile, Low Val, and Low BCAA diets, fasting and stimulated insulin levels were not significantly different between diet groups (Figure S1B).
LP and Low BCAA diets increase food intake while promoting leanness (Cummings et al., 2018; Fontana et al., 2016; Kobayashi et al., 2006). Mice fed a Low Ile or a Low BCAA diet had increased food consumption when normalized by body weight, yet both groups gained less weight, fat mass, and lean mass than Ctrl AA-fed mice (Figures 1F and S1C–S1H). Intriguingly, Low Ile-fed mice maintained the lowest weight of all groups throughout the study, while Low Val-fed animals also gained less weight (Figure 1F). While consumption of a Low Ile or Low BCAA diet reduced fat accretion (Figure S1G), mice fed a Low Ile, Low Val, or Low BCAA diet gained significantly less lean mass than Ctrl AA-fed mice (Figure S1H). The weight, fat mass, and lean mass of Low Leu-fed mice were similar to those of Ctrl AA-fed mice, with a trend for these mice to weigh more with increased adiposity. At the end of three months, Low Ile fed mice had the lowest adiposity, and were leaner than all other groups (Figure S1I).
The absence of metabolic benefits in Low Leu-fed mice was surprising, as Leu restriction is reported to improve metabolic health (Lees et al., 2017). While our AA defined diet is based on the AA profile of a commercially available natural diet (Fontana et al., 2016), casein-based rodent diets typically have a lower starting level of Leu than found in our Control AA diet. We wondered if the 67% restriction in the Low Leu diet did not restrict Leu sufficiently to induce metabolic benefits. We designed a new diet, ExLow Leu (Table S1), in which Leu was restricted by 87%; this mimics a 67% restriction of Leu in a casein-based diet. Even this severe restriction of Leu did not increase food consumption (Figure S1J–S1L), nor did it result in significant changes in fat mass or lean mass, although overall body weight was slightly reduced (Figure S1M). The ExLow Leu-fed mice also did not have improved glucose tolerance relative to Control AA-fed mice (Figure S1N–S1O).
Reducing either dietary Ile or Val alone is sufficient to recapitulate the beneficial physiological effects of reducing all three BCAAs or dietary protein (Figure 1G). Reducing Ile in particular robustly promotes glycemic control and leanness, while reducing Val has less dramatic but consistently beneficial effects. In contrast, reduction of Leu alone does not produce metabolic benefits, and may even be modestly deleterious.
Reducing dietary Ile increases hepatic insulin sensitivity
As reducing dietary Ile had the most pronounced effects on glucose and pyruvate tolerance, we fed an additional cohort of mice either a Ctrl AA or Low Ile diet, again observing robustly improved glucose tolerance in Low Ile fed mice (Figure 1H). We then used a hyperinsulinemic-euglycemic clamp to measure insulin sensitivity (Figures 1I–1L). Briefly, mice received a constant infusion of insulin, and radiolabeled glucose was co-infused as needed to maintain euglycemia. To achieve euglycemia, Low Ile fed mice required a significantly higher glucose infusion rate than Ctrl AA fed mice (Figures 1I and 1J). Calculating whole-body glucose uptake and hepatic glucose production, based on the fraction of labeled glucose in the circulation, revealed that Low Ile-fed mice had significantly improved hepatic insulin sensitivity (Figures 1K and 1L).
Reduction of dietary Ile is necessary for the beneficial metabolic effects of a low protein diet
We next decided to determine if reduced dietary levels of an individual BCAA, or of all three BCAAs, are required for a LP diet to promote metabolic health. To test this, we utilized an addback paradigm, in which we performed metabolic phenotyping on mice consuming a Ctrl diet, a Low AA diet, or a Low AA diet in which leucine (Low AA + Leu), isoleucine (Low AA + Ile), valine (Low AA + Val), or all three BCAAs (Low AA + BCAAs) were “added back” to Ctrl AA diet levels (Figure 2A). These diets were all isocaloric with equal levels of fat (Table S2).
Figure 2. Replenishing Ile significantly attenuates the metabolic effects of LP.
(A) Experimental scheme. (B) Average food consumption (normalized by body weight) during the 3rd week on the indicated diets. (C-E) Glucose tolerance (C), insulin tolerance (D), and pyruvate tolerance (E) of mice after 3, 4, or 5 weeks on the indicated diets, respectively. (F) Body weight and change in body weight of mice after 12 weeks. (B-F) n=4 cages of 3 animals each/group (B) or n=12/group (C-F); * = p<0.05, Sidak’s test post ANOVA, comparing all groups to the Ctrl AA and Low AA diet groups. (G) Heatmap of the metabolic effects of each diet. Data represented as mean ± SEM.
In agreement with our previous results and the satiating effect of protein, mice fed the Low AA diet ate significantly more than Ctrl AA diet-fed mice when normalized by body weight (Figures 2B and S2A–S2B). While addback of all three BCAAs blocked this effect, addback of each individual BCAA did not (Figures 2B and S2A–S2B). Mice consuming the Low AA diet for 3 weeks had significantly improved glucose tolerance relative to mice fed a Ctrl AA diet (Figure 2C). Addback of Ile, or of all three BCAAs, blunted this effect; in contrast, addback of Leu or Val had no significant effect on glucose tolerance (Figure 2C). Low AA diet-fed mice also had significantly improved sensitivity to insulin, an effect that was blunted by addback of either Ile, Val, or all three BCAAs, but not by addback of Leu (Figure 2D). Addback of Ile alone or of all three BCAAs blunted the effects of a LP diet on pyruvate tolerance, while addback of Leu or Val had no significant effect (Figure 2E).
Despite increased food consumption (Figure 2B), mice fed a Low AA diet weighed significantly less than Ctrl AA diet fed animals throughout the course of the study (Figure 2F), with reduced accretion of both fat mass and lean mass (Figures S2C–S2D). Addback of Ile alone or of all three BCAAs blunted the effect of a Low AA diet on body weight and lean mass, with addback of all three BCAAs also completely blocking the effect of a Low AA diet on fat mass (Figures 2F and S2C–S2D). In contrast, addback of Val had no effect on weight or body composition, while addback of Leu promoted the loss of both fat and lean mass (Figures 2F and S2C–S2D).
We used metabolic chambers to evaluate energy balance, analyzing food consumption, spontaneous activity, and energy expenditure. As expected, a Low AA diet increased food consumption, respiratory exchange ratio (RER), and energy expenditure (Figures S2E–S2L). Consistent with the effects of the individual BCAAs on body weight and composition, addback of either Ile or all three BCAAs blunted the effects of a Low AA diet on energy balance and fuel source utilization (Figures S2E–S2L). In contrast, addback of Val had minimal effects on these parameters, while addback of Leu resulted in increased food consumption and energy expenditure (Figures S2E–S2L).
As shown in Figure 2G, repletion of BCAAs or Ile alone blocks the ability of a Low AA diet to promote metabolic health, while repletion of Val partially blocks the ability of a Low AA diet to improve glucose homeostasis, but not body composition. In contrast, repletion of Leu potentiates the effects of a Low AA diet. In combination with the results of Figure 1, these results clearly demonstrate that reduced dietary levels of Ile are both sufficient to recapitulate the beneficial metabolic effects of a LP diet, and necessary for a LP diet to maximally promote metabolic health.
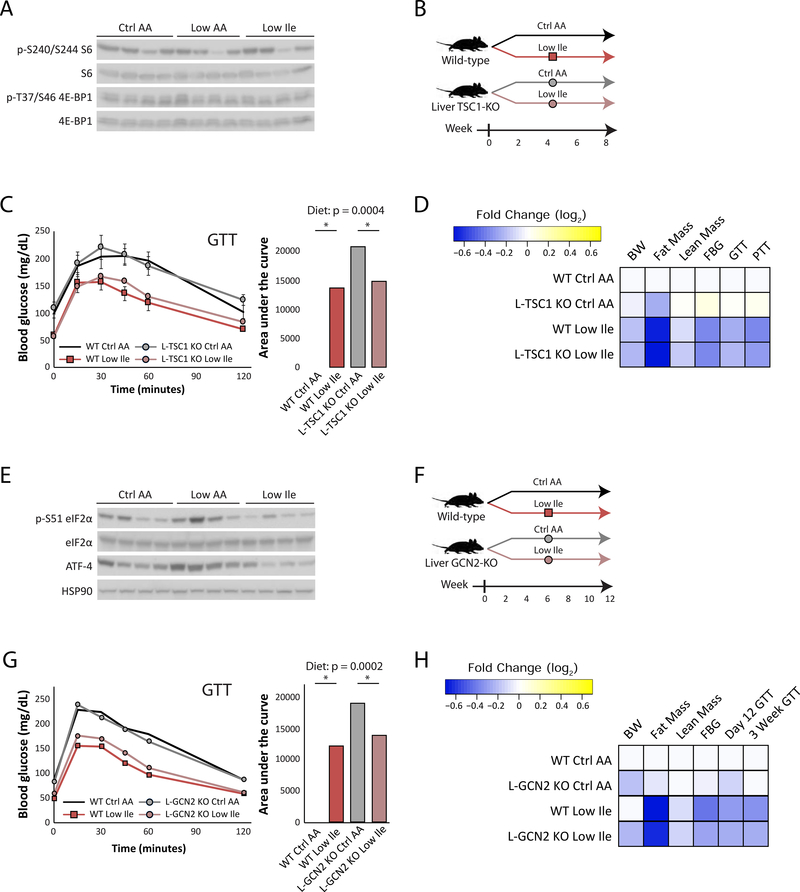
Hepatic mTORC1 and GCN2 are not required for the metabolic effects of a reduced Ile diet
While Leu is widely known to stimulate mTORC1, it has recently become apparent that Ile and Val are also potent mTORC1 agonists (Appuhamy et al., 2012; Arriola Apelo et al., 2014; Dyachok et al., 2016; Ikeda et al., 2017; Zhou et al., 2018). Indeed, AA sensing regulates stem cell proliferation, gut homeostasis, lifespan and mTORC1 in flies via Sestrin, which encodes a homologue of the mammalian Sestrin proteins which regulate BCAA sensing by mTORC1 (Chantranupong et al., 2014; Lu et al., 2021; Saxton et al., 2016). We were therefore surprised that Low AA and Low Ile feeding did not alter the phosphorylation of S6, a downstream readout of mTORC1, and 4E-BP1, a mTORC1 substrate, in the liver (Figure 3A). We decided to directly test if hepatic mTORC1 was involved in the metabolic effects of a Low Ile diet.
Figure 3. The metabolic effects of Ile restriction are independent of hepatic mTORC1 and GCN2 activity.
(A) Western blot analyses of mTORC1 signaling in the liver of fasted mice after 3 weeks of feeding the indicated diets. (B) Experimental scheme. (C) Glucose tolerance in WT and L-TSC1 KO mice fed the diets for 3 weeks (n=7–8/group; for AUC, statistics for the overall effects of genotype, diet, and the interaction represent the p value from a two-way ANOVA, *p<0.05, from a Sidak’s post-test examining the effect of parameters identified as significant in the two-way ANOVA). (D) Heatmap depiction of the metabolic effects of the indicated diets in WT and L-TSC1 KO mice. (E) Western blot analyses of GCN2 signaling in the liver of fasted mice after 3 weeks. (F) Experimental scheme. (G) Glucose tolerance in WT and L-GCN2 KO mice fed the indicated diets for 3 weeks (n=6–7/group; for AUC, statistics for the overall effects of genotype, diet, and the interaction represent the p value from a two-way ANOVA, *p<0.05, from a Sidak’s post-test examining the effect of parameters identified as significant in the 2-way ANOVA). (H) Heatmap of the metabolic effects of each diet. Data represented as mean ± SEM.
We utilized a mouse model of constitutive hepatic mTORC1 activity, in which Tsc1, a negative regulator of mTORC1, is deleted specifically in the liver (L-TSC1 KO) (Harputlugil et al., 2014; Sengupta et al., 2010; Yu et al., 2018) (Figure S3A). We fed L-TSC1 KO mice and their wild-type littermates (WT) either Ctrl AA or Low Ile diets (Figure 3B). Consumption of the Low Ile diet improved glucose tolerance equally well in L-TSC1 KO mice and their WT littermates (Figure 3C). Hepatic insulin sensitivity, assessed as suppression of gluconeogenesis during a pyruvate tolerance test, was also improved by Low Ile diet similarly in both genotypes (Figure S3B). A Low Ile diet also induced similar effects on the body weight, fat mass and lean mass of both WT and L-TSC1 KO mice (Figures S3C–S3E). Given the similar metabolic phenotypes of WT and L-TSC1 KO mice fed a Low Ile diet (Figure 3D), we conclude that suppression of hepatic mTORC1 activity is not required for the metabolic effects of a Low Ile diet.
The other major AA sensing kinase is GCN2, which is activated by uncharged tRNAs, and is essential for the adaptation to AA deprived diets (Dong et al., 2000; Wek et al., 1995; Xiao et al., 2011; Xiao et al., 2014; Zhang et al., 2002). We observed increased phosphorylation of the GCN2 substrate eIF2α and increased expression of ATF4, a downstream transcription factor, in mice fed a Low AA diet, but surprisingly not in mice fed a Low Ile diet (Figure 3E). We therefore decided to test if hepatic GCN2 signaling was involved in the metabolic effects of a Low Ile diet.
We placed mice lacking hepatic Gcn2 (L-GCN2 KO) and their wild-type littermates (WT) on Ctrl AA or Low Ile diets (Figure 3F). As GCN2 has been shown to be essential for the initial adaptation to a LP diet (Laeger et al., 2016), we performed a GTT after only twelve days (Figure S3F) as well as after three weeks of diet feeding (Figure 3G). At each time point we found that a Low Ile diet improved glucose tolerance equally well in WT and L-GCN2 KO mice. Mice fed a Low Ile diet had reduced body weight, fat mass, and lean mass, regardless of genotype (Figures S3G–S3I). Given both the western blotting data and the similar metabolic phenotypes of WT and L-GCN2 KO mice fed a Low Ile diet (Figure 3H), we conclude that a Low Ile diet does not promote metabolic health via activation of hepatic GCN2.
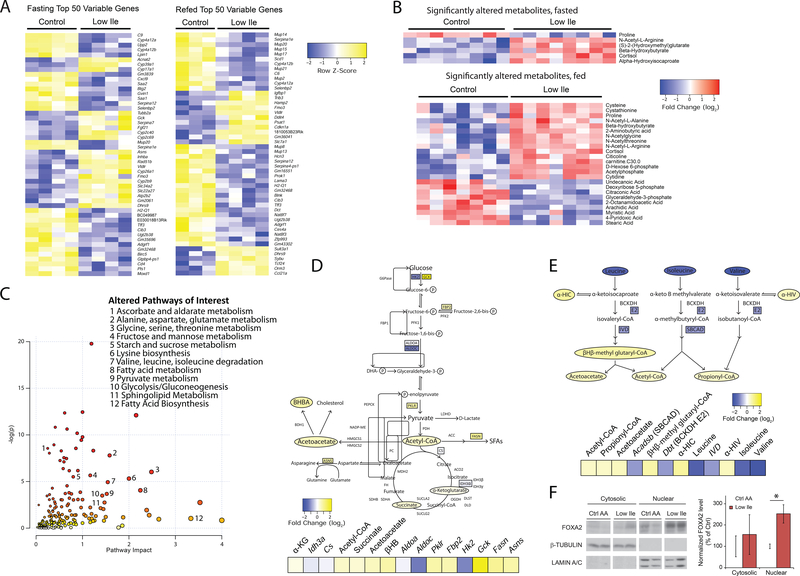
Reduction of dietary Ile reprograms hepatic metabolism
We next performed transcriptional profiling and metabolomics analysis on the livers of mice fed either Ctrl AA or Low Ile diets for three weeks, sacrificed either after an overnight fast or following refeeding. We observed a significant effect of a reduced Ile diet on hepatic gene expression, with 184 genes changing significantly in fasted mice and 255 genes changing significantly in refed mice (Figure 4A and Table S3). Intriguingly, the proteins encoded by the genes most significantly upregulated and downregulated in fasted mice by a Low Ile diet were enriched for Ile content relative to unchanged genes (Figure S4A). Performing targeted metabolomics, we identified 6 significantly altered metabolites in fasted mice and 24 significantly altered metabolites in refed mice (Figure 4B).
Figure 4. Ile restriction reprograms hepatic metabolism.
(A) Heatmaps of top 50 variable genes in transcriptomic analysis of Control or Low Ile-fed mouse livers. (B) Heatmaps of all significantly altered metabolites in targeted metabolite analysis of Control or Low Ile-fed mouse livers. In (A) and (B), mice were either fasted overnight or fasted overnight, then refed for 3 hours. (C) Representation of pathways altered in Low Ile-fed liver based on transcriptomic and metabolomics data. Metabolic pathways of interest are highlighted. (D) Drawn pathways in central metabolism altered in Low Ile feeding. (E) Drawn pathways in BCAA degradation altered in Low Ile feeding. Genes and metabolites colored by log2-fold change in transcriptomic and metabolomics data. Data in (C), (D), and (E) is integrated from significant changes in transcriptomic and metabolomic data gathered in the fasted state. (F) Representative cropped western blots and quantification of nuclear and cytosolic FoxA2 in the liver of fasted mice after 3 weeks of feeding the diets (n=6/group; *p<0.05, Sidak’s test post ANOVA). Data represented as mean ± SEM.
Metaboanalyst was used to coordinate the RNA-Seq and metabolomics data, and identify pathways altered in response to Low Ile feeding (Figure 4C). We found alterations in several central metabolic processes, including carbohydrate metabolism and glycolysis, as well as BCAA degradation and fatty acid biosynthesis. In fasted mice we observed that a Low Ile diet increased glycolytic gene expression, Acetyl-CoA, and the TCA cycle intermediates succinate and α-ketoglutarate (Figure 4D). Additionally, we observed decreased levels of the other two BCAAs (Leu and Val) and increased levels of their catabolic intermediates, β-Hydroxy β-methylglutaryl-CoA (HMG-CoA) and propionyl-CoA (Figure 4E). This likely reflects altered BCAA catabolism, as blood levels of the BCAAs are not decreased in Low-Ile fed mice and liver levels of Leu and Val trend higher in the livers of Low Ile mice following feeding (Figure S4B and Table S3). We also observed elevated levels of hepatic β-Hydroxybutyrate in both the fasted and fed state, suggestive of increased ketogenesis, perhaps in response to increased acetoacetate and acetyl-CoA (Figure 4B). Expression of fatty acid synthase (Fasn) was significantly upregulated, suggesting that lipogenesis may be increased. Together, our analyses indicate profound changes in liver metabolism upon Low Ile feeding.
To gain insight into potential transcriptional regulators of the response to a Low Ile diet, we utilized MAGIC, a recently developed and experimentally validated tool for transcription factor enrichment analysis (Roopra, 2020). We identified 13 significantly enriched transcription factors predicted as drivers of the observed gene expression changes (Figure S4C). Intriguingly, one of these transcription factors was FOXA2, which was previously identified as a regulator of hepatic insulin sensitivity and ketogenesis (Wolfrum et al., 2004). FOXA2 activity is regulated by localization; cytoplasmic FOXA2 is inactive, while nuclear FOXA2 is active (Wolfrum et al., 2004). Consistent with the MAGIC prediction of active FOXA2, we found that nuclear levels of FOXA2 were significantly increased in the livers of Low Ile-fed mice (Figure 4F).
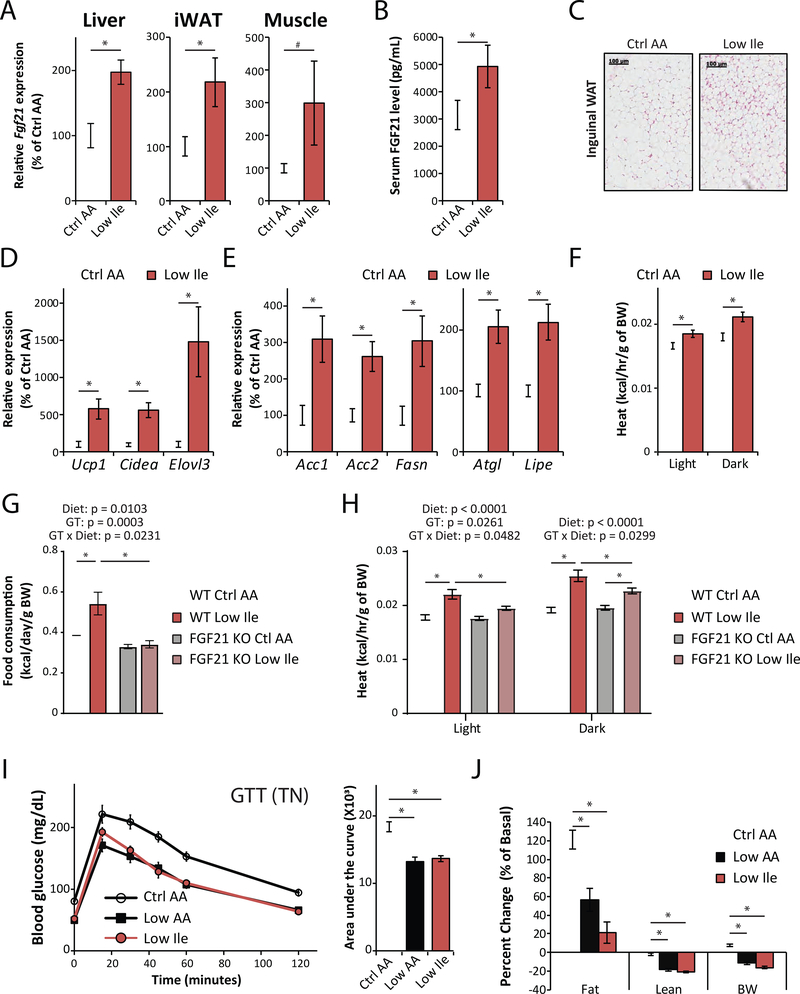
Reducing dietary Ile induces the FGF21-UCP1 axis and promotes energy expenditure
The FOXA2 target gene upregulated the most by a Low Ile diet, and one of the top 50 upregulated genes, is Fgf21 (Figures 4A and S4D). FGF21 is a hormone that promotes hepatic insulin sensitivity and regulates energy homeostasis (Fisher and Maratos-Flier, 2016; Xu et al., 2009), and is induced by a variety of stresses, including a LP diet (Fontana et al., 2016; Laeger et al., 2014). Although we observed no increase in plasma FGF21 in lean mice fed a reduced BCAA diet (Fontana et al., 2016), we subsequently observed that reducing dietary BCAAs acutely induces FGF21 in DIO mice (Cummings et al., 2018), and elevated FGF21 is also observed in diabetic humans fed a reduced BCAA diet (Karusheva et al., 2019). FGF21 promotes hepatic ketogenesis (Fisher et al., 2017; Inagaki et al., 2007; Potthoff et al., 2009), and we observed increased levels of increased levels of ketones (β-Hydroxybutyrate, BHB) in Low Ile-fed mice (Figure 4B and Table S3).
A Low Ile diet increased expression of Fgf21 mRNA in the liver, inguinal WAT, and skeletal muscle (Figure 5A). In agreement with this result, mice fed a Low Ile diet had increased levels of FGF21 in the plasma (Figure 5B); in contrast, a Low Leu or Low Val diet did not increase plasma FGF21 (Figure S4E). In agreement with a model in which a Low Ile diet induces ketogenesis via FGF21, we observed increased blood levels of BHB in Low Ile and Low BCAA-fed mice, but not in mice fed Low Leu or Low Val diets (Figure S4F).
Figure 5. Reducing dietary Ile induces the FGF21-UCP1 axis and promotes energy expenditure.
(A) Fgf21 expression in the liver, inguinal white adipose tissue (iWAT) and muscle of Ctrl AA or Low Ile fed mice after 3 weeks of diet, n=8/group. (B) Plasma FGF21 level in Ctrl AA or Low Ile fed mice after 3 weeks of diet, n=5–8/group. (C) Representative images of H&E stained iWAT of Ctrl AA or Low Ile fed mice. Scale bar=100 μm. n=5–6/group. (D and E) Expression of thermogenic genes (D), lipolytic and lipogenic genes (E) in iWAT of Ctrl AA or Low Ile fed mice after 3 weeks on the indicated diets. n=8/group. (A-E) *p<0.05, t-test. (F) Energy expenditure (Heat) in mice after 6 weeks on the diets as assessed by metabolic chambers. n=5–8/group. *p<0.05, Dunnett’s test post ANOVA, considering data from all groups shown in Figure S4L. (G-H) Food consumption per gram of body weight (G) measured after 3 weeks of diet, and energy expenditure (H) measured after 12 weeks of diet to wild-type and FGF21 KO mice (n=6–9/group; the overall effect of genotype (GT), diet, and the interaction represent the p-value from a 2-way ANOVA conducted separately for the light and dark cycles; *p<0.05, Sidak’s test post 2-way ANOVA). (I and J) Glucose tolerance (I) and change in body composition (J) of mice housed at thermoneutral (TN) temperature (28.5°C) (n=12/group; *p<0.05, Sidak’s test post ANOVA). Data represented as mean ± SEM.
FGF21 drives energy expenditure by promoting the beiging of WAT; we observed multilocular beige adipocytes in the inguinal WAT (iWAT) of mice fed a reduced Low Ile diet (Figure 5C), as well as in the iWAT of mice fed either a Low Val or Low BCAA diets (Figure S4G). In contrast, beiging was not observed in the iWAT of mice fed a Low Leu diet, or in the gonadal WAT of mice on any diet (Figure S4G). Low Ile-fed mice showed transcriptional changes consistent with beiging in iWAT;. we found increased expression of Ucp1 as well as two other key thermogenic genes, cell death-inducing DNA fragmentation factor alpha-like effector A (Cidea) and elongation of very long chain fatty acids protein 3 (Elovl3) (Figure 5D). Beiging of iWAT is characterized by induction of a futile cycle of lipolysis and lipogenesis (Mottillo et al., 2014), and expression of several key lipogenic genes, including acetyl-coenzyme A carboxylase 1 and 2 (Acc1 and Acc2) and Fasn, and the key lipolytic genes adipose triglyceride lipase (Atgl) and hormone-sensitive lipase (Lipe), was increased in iWAT of Low Ile-fed mice (Figure 5E).
We measured energy balance and assessed fuel source utilization in mice fed diets in which each individual BCAA or all three BCAAs were reduced (Figures 5F and S4H–S4O). While there was no difference in activity between groups, food consumption was increased in mice fed a Low Ile or Low Val diet (Figures S4H–S4J, and S4O). Energy expenditure normalized to body weight was increased in mice fed Low Ile, Low Val, or Low BCAA diets (Figures 5F and S4L). The respiratory exchange ratio (RER), which reflects substrate utilization, was increased at night in mice consuming Low Val or Low BCAA diets, suggesting increased carbohydrate utilization (Figure S4N).
We tested if FGF21 was necessary for the effects of a Low Ile diet by feeding Ctrl AA or Low Ile diets to wild-type (WT) and whole-body Fgf21 knockout mice (FGF21 KO), and examining the metabolic response. Food consumption showed a significant interaction between diet and genotype, with food consumption increasing in Low Ile-fed WT mice but not in Low Ile-fed FGF21 KO mice (Figures 5G and S5A–S5B). Despite the genotype-dependent effect of a Low Ile diet on food consumption, the weight, fat, and lean mass of WT and FGF21 KO mice responded similarly to a Low Ile diet (Figure S5C). Using metabolic chambers, we analyzed how energy balance was altered by the Low Ile diet in both genotypes (Figures 5H and S5D–S5F). A Low Ile diet induced a robust increase in energy expenditure normalized to body weight in WT mice, but that this effect was significantly blunted in FGF21 KO mice, with a significant interaction between diet and genotype during both the light and dark cycles (Figures 5H). However, we found no effect of genotype on the ability of a Low Ile diet to improve glucose tolerance (Figure S5G). We conclude that some, but not all, of the metabolic effects of a Low Ile diet are dependent on FGF21.
Given the lower adiposity of Low AA and Low Ile-fed mice, we examined the possibility that these mice were cold-stressed, as room temperature is below the thermoneutral temperature of mice (Ganeshan and Chawla, 2017), by housing mice fed at thermoneutrality. The metabolic impact on Low Ile and Low AA diets at thermoneutrality was virtually identical to that at room temperature, and both diets improved glucose tolerance and body composition regardless of housing temperature (Figures 5I–J and S5H–S5I). Housing temperature therefore does not influence the overall metabolic response to dietary Ile.
Benefits of restricting dietary Ile are amplified in the context of an unhealthy Western diet
Specifically reducing dietary BCAAs is sufficient to restore metabolic health to DIO mice, even while they continue to consume a high-fat, high-sucrose Western diet (WD) (Cummings et al., 2018). We examined the metabolic impact of reducing levels of the individual BCAAs in mice preconditioned with a WD for 12 weeks (Figure 6A). Briefly, the level of each individual BCAA or all three BCAAs was reduced by 67% in the context of a WD; all of the WDs were isocaloric with identical levels of fat, and the percentage of calories derived from AAs was kept constant by proportionally adjusting the amount of non-essential AAs (Table S4).
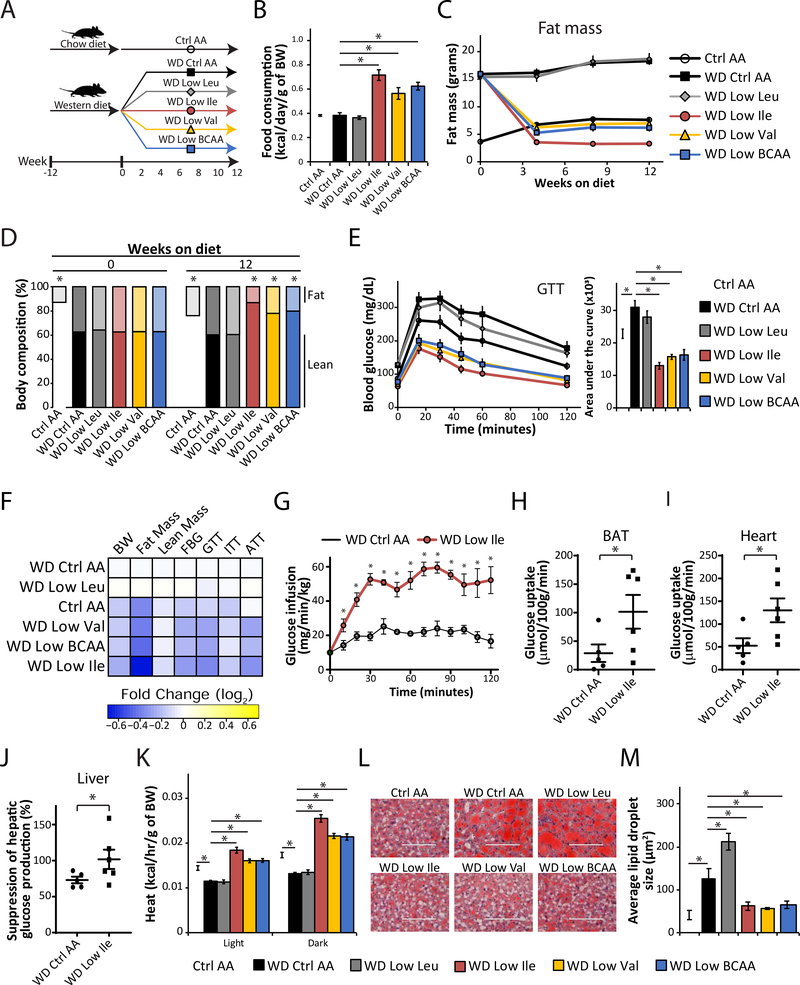
Figure 6. Restricting dietary Ile or Val improves metabolic health of DIO mice.
(A) Experimental scheme. (B) Food consumption normalized by body weight during the 3rd week on the diets. (C – D) Fat mass (C), body composition (D), and glucose tolerance (E) of mice consuming the indicated diets. (B-E) n=5–6 cages of 2 animals each/group (B) or n = 10/group (C-E); (B, D, E) *p < 0.05, Dunnett’s test post ANOVA, each group compared to the WD Ctrl AA group. (F) Heatmap of the metabolic effects of each diet. (G-J) Glucose infusion rate (G), glucose uptake into BAT (H) and heart (I), and hepatic insulin responsiveness (J) was determined during a hyperinsulinemic-euglycemic clamp in mice preconditioned with a WD for 12 weeks and then maintained on either a WD Ctrl AA diet or a WD Low Ile diet for 3 weeks. Each symbol represents a single animal (n=5–6/group, 1-tailed Student’s t-test, *p<0.05). (K) Energy expenditure (Heat) as assessed by metabolic chamber (n =5–8/group). (L-M) Representative Oil-Red-O–stained sections of liver of mice after 12 weeks on the diets, scale bar=100 μm (L) and quantification of lipid droplet size (M) (n=4/group). (K-M) *p<0.05, Dunnett’s test post ANOVA, groups compared to the of WD Ctrl AA diet group. *p<0.05. Data represented as mean ± SEM.
Mice fed the WD Low Ile, WD Low Val, or WD Low BCAA diets ate more food than mice on the WD Control (Ctrl) AA diet (Figure 6B and Figure S6A–S6B). Despite eating more, mice fed these diets quickly lost excess weight and fat mass, with smaller changes in lean mass; the overall effect was reduced adiposity (Figures 6C–6D and S6C–S6D). The reduction in weight and adiposity was most pronounced in mice fed the WD Low Ile diet, which lost both lean mass and fat mass relative to Ctrl AA-fed mice never exposed to a WD. The weight and body composition of mice fed a WD Low Leu diet were indistinguishable from that of mice fed the WD Ctrl AA diet (Figures 6C–6D and S6C–S6D).
Mice fed the WD Ctrl AA and WD Low Leu diets were glucose intolerant and insulin resistant relative to Control AA-fed mice never exposed to a WD. However, mice fed either a WD Low Ile, WD Low Val, or WD Low BCAA diet had significantly improved glucose tolerance and insulin sensitivity (Figures 6E and S6E), and displayed significantly improved suppression of hepatic gluconeogenesis during an alanine tolerance test (Figure S6F). Overall, we found that reducing dietary Ile, Val, or all three BCAAs together restored the metabolic health of DIO mice consuming an otherwise WD, whereas restriction of Leu had no beneficial effects (Figure 6F).
We then used a hyperinsulinemic-euglycemic clamp to measure the insulin sensitivity of WD Ctrl AA and WD Low Ile-fed mice (Figures 6G–J and S6G–S6I). To achieve euglycemia, WD Low Ile fed mice required a significantly higher glucose infusion rate than WD Ctrl AA fed mice (Figure 6G). Glucose uptake rates into several key metabolic tissues, including brown adipose tissue (BAT), heart, iWAT, and skeletal muscle were substantially higher in Low Ile-fed mice (Figures 6H–I and S6G–H). Calculating whole-body glucose uptake and hepatic glucose production, based on the fraction of labeled glucose in the circulation, revealed that WD Low Ile-fed mice had significantly improved hepatic insulin sensitivity (Figures 6J and S6I).
The positive effects of WD Low Ile, WD Low Val, and WD Low BCAA diets on body composition and glycemic control were accompanied by increased energy expenditure (Figures 6K and S6J–K). While food consumption and respiratory exchange ratio were higher in mice fed either a WD Low Ile, WD Low Val, or WD Low BCAA diet (Figures S6L–S6O), changes in spontaneous activity were unremarkable, except that WD Low Ile fed mice had higher activity, which was statistically significant during the day (Figure S6P). Lastly, as WD feeding is well-known to induce hepatic steatosis, we examined the lipid accumulation in the livers of mice on these different diets. While mice consuming WD Ctrl AA and WD Low Leu diets show marked accumulation of lipid in their livers, mice fed either a WD Low Ile, WD Low Val, or WD Low BCAA diet had reduced hepatic lipid deposition and smaller lipid droplets, similar to levels found in Ctrl AA-fed mice never exposed to a WD (Figures 6L, 6M, and S6Q).
Increased Ile intake attenuates the beneficial effects of LP on obesity
As reducing dietary Ile is needed to exert the metabolic benefits of a LP diet in lean mice, we hypothesized that this might also be true in DIO mice. We tested this using a similar “addback” paradigm, feeding mice preconditioned with a naturally sourced WD either a WD Ctrl AA diet, a WD Low AA diet, or a Low AA diet in which leucine (WD Low AA + Leu), isoleucine (WD Low AA + Ile), valine (WD Low AA + Val), or all three BCAAs (WD Low AA + BCAAs) were “added back” to WD Ctrl AA diet levels (Figures 7A and S7A). These diets were all isocaloric with equal levels of fat (Table S5).
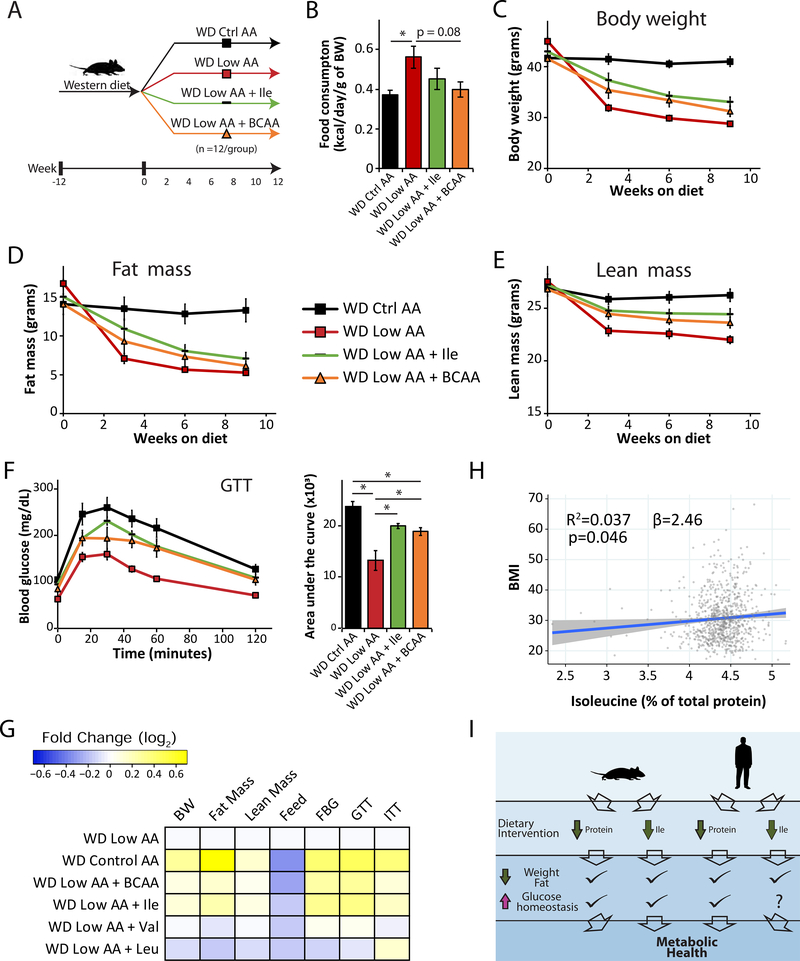
Figure 7. Increased Ile intake attenuates the beneficial effects of LP on obesity and is associated with higher BMI in human.
(A) Experimental scheme. (B) Average food consumption during the first month on the indicated diets normalized by body weight. (C–F) Body weight (C), fat mass (D), lean mass (E), and glucose tolerance (F) of mice consuming WD Ctrl AA, WD Low AA, WD Low AA + Ile, and WD Low AA + BCAA diets. (B-F) n=6 cages of 2 animals each/group (B) or n=11–12/group (C-F); *p<0.05, Sidak’s test post ANOVA; each group compared to the WD Ctrl AA and WD Low AA groups. (G) Heatmap of the metabolic effects of each diet. (H) Association between body mass index (BMI) and percent of total protein from Ile from the SHOW study (n=788, shaded area represents 95% CI). (I) Graphic summary of the study. Data represented as mean ± SEM.
Mice on the WD Low AA diet had significantly elevated food consumption that was repressed by addback of any of the three BCAAs, reaching statistical significance in the case of mice fed the WD Low AA + BCAA diet (Figures 7B and S7B). WD Low AA-fed mice had reduced body weight, fat mass and lean mass compared to WD Ctrl AA-fed mice, and addback of either Ile or BCAAs attenuated these effects on body weight and lean mass and blunted the effects on fat mass (Figures 7C–7E, S7C–S7E). Addback of Val had minimal effects on weight and body composition, while addback of Leu potentiated the effects of WD Low AA diet on lean mass (Figures S7F–S7H).
As expected, a WD Low AA diet improved glucose tolerance; addback of either Ile or all BCAAs blunted this effect, while addback of either Leu or Val did not (Figures 7F and S7I). Finally, a WD Low AA diet improved insulin sensitivity as assessed by ITT; addback of Ile or Leu blunted this improvement (Figure S7J). Overall, we find that reducing dietary Ile is required for the full metabolic benefits of a LP diet, and addback of dietary Ile, but not Val or Leu, is sufficient to make WD Low AA-fed mice metabolically similar to mice fed a WD Ctrl AA diet (Figure 7G).
Increased Ile intake is associated with higher BMI in humans
We next examined if the Ile level in the diet is associated with altered metabolic health in humans. Among a randomly selected, cross-sectional population-based sample of adults (>18 years old), we examined associations between estimated Ile intake relative to total protein and body mass index (kg/m2) determined from measured weight and height. After careful adjustment for confounding factors, we found that an increase in the intake of dietary Ile relative to total protein of a single percentage point – e.g., from 4% to 5% of protein – is associated with a 2.46 unit increase in BMI (p = 0.046) (Fig. 7H, Table S6). These results are consistent with our animal study findings and suggest that dietary intake of protein with a higher proportion of Ile increases BMI in a general population-based sample. Adjusting for the same factors, we found that the intake of Leu and Val were not significantly associated with BMI (Table S6). When considered in the context of our animal studies, these results suggest that dietary protein quality – and in particular, the proportion of Ile in dietary protein – may also be an important regulator of metabolic health in humans.
DISCUSSION
Recent studies have demonstrated that reducing dietary BCAAs promotes metabolic health in rodents. As the three BCAAs are normally consumed together (though in different quantities) and share very similar catabolic processes and fate, they are often studied in combination, rather than individually. Based on emerging evidence of distinct physiological and molecular roles for the individual BCAAs, we tested the hypothesis that dietary restriction of each of the three BCAAs might have distinct physiological and metabolic effects. Here, we demonstrate that in lean mice, dietary restriction of Ile recapitulates the effects of dietary BCAA and protein restriction, and is necessary for the full physiological and metabolic effects a LP diet. Further, restriction of dietary Ile reprograms hepatic and WAT metabolism, and is necessary and sufficient to restore metabolic health to diet-induced obese mice. Finally, dietary Ile levels correlate with BMI in humans, suggesting that these findings are directly relevant to human health.
Our studies here represent the first comprehensive dissection of the unique roles played by the three BCAAs in metabolic health and the response to a LP diet. Specifically reducing dietary Ile, and to a lesser degree Val, promotes glucose tolerance and reduces adiposity in lean mice, while restriction of Leu tended to have negative effects, with a trend towards increased adiposity. Restriction of dietary Ile or Val rapidly normalizes the weight and body composition of diet-induced obese mice, promoting fat mass loss as well as rapid improvements in glucose tolerance and insulin sensitivity, whereas Leu restriction has negligible effects in this context. Addback experiments demonstrated that dietary Ile is uniquely potent, as restriction of dietary Ile, but not Leu or Val, is required for the beneficial effects of a LP diet in both lean and DIO mice.
Multiple mechanisms likely underlie the metabolic benefits of a reduced Ile diet. We examined the contribution of several different potential physiological and molecular mechanisms, and tested the effects of a Low Ile diet in mouse models with alterations in mTOR, GCN2, or FGF21 signaling. The improved glucose homeostasis of Low Ile-fed mice is mediated in part by improved hepatic insulin sensitivity, but does not require suppression of hepatic mTORC1 signaling, activation of hepatic GCN2 signaling, or whole-body FGF21 signaling. Our results do not exclude the possibility that through action in non-hepatic tissues, mTOR or GCN2 play a role in the effects of a Low Ile diet. We have also not examined how mTOR or GCN2 signaling contributes to the effects of a LP diet; a LP diet reduces mTORC1 signaling in many tissues, and mTORC1 has been implicated in other studies as important for the effects of protein restriction (Harputlugil et al., 2014; Lamming et al., 2015; Solon-Biet et al., 2014).
FGF21 is induced in both humans and rodents in response to LP diets and plays a key role in the metabolic effects of such diets (Cummings et al., 2018; Fontana et al., 2016; Hill et al., 2018; Hill et al., 2019; Laeger et al., 2016; Laeger et al., 2014; Maida et al., 2016; Wanders et al., 2017). Here, we show that reduction of dietary Ile by 67% induces Fgf21 transcription in multiple tissues and raises blood levels of FGF21. Using mice lacking Fgf21, we show that FGF21 is required for a reduced Ile diet to increase food consumption, and plays a role in the increased energy expenditure of Low Ile-fed mice. Blood levels of FGF21 did not increase in Low Val-fed mice, which may explain in part the stronger metabolic impact of Ile restriction, and suggests that the effects of Val restriction are FGF21-independent.
Further research will be required to fully define the mechanisms by which dietary Ile regulates expression of Fgf21 and programs metabolism, as well as to determine if the action of FGF21 in the brain mediates the effect of dietary Ile on energy balance (Hill et al., 2019). Our results here hint that activation and nuclear localization of the transcription factor FOXA2 may contribute to the induction of Fgf21 by a reduced Ile diet. Understanding the role of FOXA2 in the regulation of hepatic Fgf21 expression by Ile, as well as clarifying the contribution of FGF21 from other tissues to the increased levels of FGF21 we find in Low Ile-fed mice, will be important to address in the future. Elucidating both the FGF21-dependent and –independent mechanisms by which a reduction in dietary Ile or Val promotes leanness without calorie restriction will likewise be a key area of future study.
An additional question is why reduction of Leu has distinct effects from reduction of Ile and Val. The reduction of hepatic levels of all three BCAAs, along with the increase in the levels of Leu catabolic intermediates and the end products of BCAA catabolism suggest that catabolism of Leu and Val is altered in Low Ile-fed mice. While it is tempting to speculate that the reduced levels of Leu and Val are responsible for the metabolic benefits of a Low Ile diet, this does not accord with the phenotypes we have observed. Instead of improving glucose tolerance and reducing adiposity, we find reducing Leu instead has a neutral to slightly negative effect on metabolic health. Further reduction of Leu, by up to 87%, likewise does not recapitulate the effects of Ile restriction. While a Low Val diet may improve some metabolic phenotypes via a reduction of skeletal muscle insulin resistance (Jang et al., 2016), the most dramatic effects of a Low Ile diet are on hepatic insulin sensitivity as well as reprogramming of WAT. It is therefore unlikely that the reduced levels of hepatic Leu and Val directly promote metabolic health.
The metabolic phenotypes of Low Ile-fed mice, including improved glucose tolerance, may be due in part to increased catabolism of the BCAAs, particularly Leu and Val. Improved glucose tolerance is also observed in mice treated with BT2, an inhibitor of branched-chain α-keto acid dehydrogenase kinase (BCKDK), which results in increased BCAA catabolism (Zhou et al., 2019). One possible mechanism for this effect is that levels of the α-ketoacids of Leu and Val, which may increase due to the AA imbalance we have created in the diet, can inhibit BCKDK, which would increase BCAA catabolism. Increased levels of Acetyl-CoA and acetoacetate – which are in part Leu derived – would then be available to support increased ketogenesis and lipogenesis. In agreement with this idea, we observed elevated ketones only in the blood of Low Ile-fed mice, and not in the blood of mice fed diets in which only Leu or Val was reduced. Ketogenic diets can drive increased energy expenditure by activating BAT (Srivastava et al., 2013; Srivastava et al., 2012), and future research will be required to define how BCAA catabolism is altered in Low Ile-fed mice, and to elucidate the role of ketogenesis in the metabolic response to reduced dietary Ile.
Limitations of Study
Our study was conducted exclusively in males; while our recent work suggests that males and females have similar metabolic responses to BCAAs, we and others have identified sex-dependent differences in the response to BCAAs, including in the effect of these diets on FGF21 (Richardson et al., 2021; Solon-Biet et al., 2019). Furthermore, our study focused specifically on C57BL/6J mice, and others have shown that the response to other dietary interventions, such as calorie restriction, vary according to strain of mouse (Barrington et al., 2018; Liao et al., 2010; Mitchell et al., 2016). Future research should consider the effect of the individual AAs on the metabolic health of multiple genetic backgrounds and both sexes.
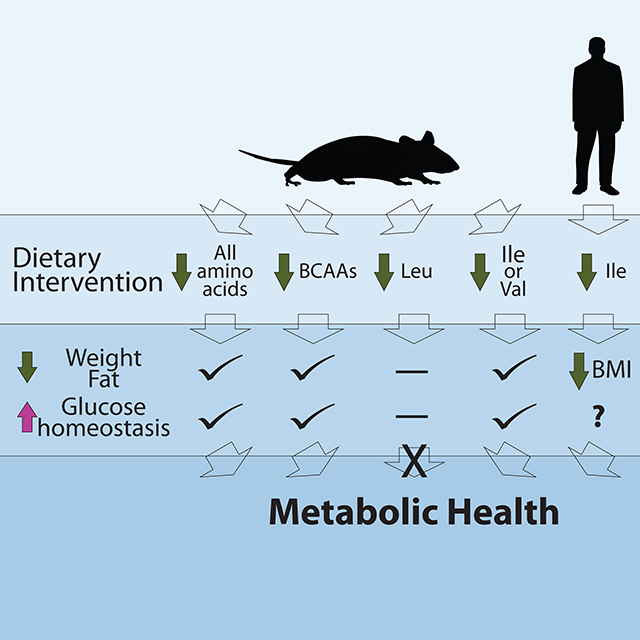
In summary, dietary Ile is a key mediator of metabolic health in mice and potentially in humans (Figure 7I). Specifically reducing dietary Ile reprograms hepatic and WAT metabolism, improves hepatic insulin sensitivity, increases hepatic ketogenesis and lipogenesis, and increases energy expenditure. We find that dietary levels of Ile are correlated with metabolic health in humans, and we link dietary levels of Ile to higher BMI. Critically, we show that reduction of dietary Ile corrects many metabolic abnormalities associated with obesity including adiposity, hyperglycemia, insulin resistance, and hepatic steatosis, restoring metabolic health to DIO mice. Our results demonstrate unique roles for each of the dietary BCAAs in metabolic health, highlights the critical importance of dietary levels of Ile for the metabolic health of both mice and humans, and define reduced Ile as critical to the metabolic effects of a LP diet. Finally, our results suggest that reducing dietary Ile may be a novel therapeutic and public health strategy to combat the twin epidemics of obesity and diabetes.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dudley W. Lamming (dlamming@medicine.wisc.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
The gene expression data generated during this study can be obtained from Gene Expression Omnibus (GEO) (GEO: GSE168588).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mouse information
All procedures were performed in conformance with institutional guidelines and were approved by the Institutional Animal Care and Use Committee of the William S. Middleton Memorial Veterans Hospital (Madison, WI, USA) or the University of Pennsylvania (Philadelphia, PA, USA). Except as noted otherwise, all the studies described here used male C57BL/6J mice purchased from The Jackson Laboratory (Bar Harbor, ME, USA) and all mice were acclimated to the animal research facility for at least one week before entering studies. To test the effects of individual BCAA restriction and low-protein diet repleted with individual BCAAs in the context of normal calorie, 10-week-old male mice were placed on control, individual BCAA restricted diets, or individual BCAA repleted low protein diets and maintained on their respective diets. To test the metabolic effects of individual BCAAs in the context of diet-induced obesity, 6-week-old male C57BL/6J mice were pre-conditioned with WD (TD.88137, Envigo, Madison, WI) for 12 weeks, and another group of mice were fed control chow diet (Purina 5001). Following 12-weeks of WD preconditioning, mice on WD diet were then switched to the amino acid defined Western diets described in Figures 6 and 7, while mice fed a chow diet were switched to a Control amino acid defined diet. To compare the metaboic effect of a Low Ile diet to that of of a Low AA diet, 10-week-old male mice were placed on either a Control AA, a Low AA, or a Low Ile diet for 3 weeks. All mice were maintained at a temperature of approximately 22°C, except for mice used to study the effect of themoneutral housing on the metabolic response to protein restriction and isoleucine restrction; for this study, 10-week-old male mice were placed on Control, Low AA or Low Ile diets, in a room maintained at approximately 28.5°C for 6 weeks.
To study if supppressed hepatic mTORC1 signaling is required for the effects of a Low Ile diet, male liver specific Tsc1 knockout mice were generated by crossing Albumin-Cre mice (Postic et al., 1999) from The Jackson Laboratory (003574) with mice expressing a conditional allele of Tsc1 from The Jackson Laboratory (005680), backcrossed to C57BL/6J mice at least three times, and genotyping was performed as previously described (Kwiatkowski et al., 2002). To study if the protein kinase GCN2 was required for the effects of a Low Ile diet, we crossed Albumin-Cre mice with mice expressing a conditional allele of Gcn2 (Maurin et al., 2005) from The Jackson Laboratory (008452) and genotyped male progeny as previously described.
To generate male FGF21 KO mice, we crossed CMV-Cre mice (Schwenk et al., 1995) from the Jackson Laboratory (006054) with mice expressing a floxed allele of FGF21 (Potthoff et al., 2009) from The Jackson Laboratory (022361), then crossed with C57BL/6J mice to remove CMV-Cre; genotyping was performed using PCR, primers can be found in the Key Resources Table. PCR Protocol: Stage 1, 95C°, 1:30; Stage 2 (x8) 95°C, 0:30, step down, 68°C, 1:00; Stage 3 (x28) 95°C, 0:30, 61.5°C, 0:30, 72°C, 1:00; Stage 4 72°C, 5:00, hold at 4°C. PCR was performed using DreamTaq DNA Polymerase (Thermo Scientific, EP0705) according to insert instructions.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-p-S6 ribosomal protein (Ser240/244) | Cell Signaling Technology | Cat# 2215; RRID: AB_331682 |
| Rabbit anti-S6 ribosomal protein | Cell Signaling Technology | Cat#2217; RRID: AB_331355 |
| Rabbit anti-4EBP1 | Cell Signaling Technology | Cat# 9644; RRID: AB_2097841 |
| Rabbit Anti-p-4E-BP1 (Thr37/46) | Cell Signaling Technology | Cat# 2855; RRID: AB_560835 |
| Rabbit anti-HSP90 | Cell Signaling Technology | Cat# 4877; RRID: AB_2233307 |
| Rabbit anti-p-eIF2α (Ser51) | Cell Signaling Technology | Cat# 9721; RRID: AB_330951 |
| Rabbit anti-eIF2α | Cell Signaling Technology | Cat# 9722; RRID: AB_2230924 |
| Rabbit anti-TSC1 | Cell Signaling Technology | Cat# 6935; RRID: AB_10860420 |
| Rabbit anti-ATF-4 | Cell Signaling Technology | Cat# 11815; RRID: AB_2616025 |
| Rabbit anti-β-Tubulin | Cell Signaling Technology | Cat# 2146; RRID: AB_2210545 |
| Mouse anti-FOXA2(HNF-3β) | Santa Cruz Biotechnology | Cat# sc-374376; RRID: AB_10989742 |
| Mouse anti-lamin A/C | Developmental Studies Hybridoma Bank | Cat# MANLAC2(10F8); RRID: AB_2618204 |
| Anti-rabbit IgG, HRP-linked Antibody | Cell Signaling Technology | Cat# 7074; RRID: AB_2099233 |
| Anti-mouse IgG, HRP-linked Antibody | Cell Signaling Technology | Cat# 7076; RRID: AB_330924 |
| Chemicals, peptides, and recombinant proteins | ||
| Human insulin | Eli Lilly | NDC 0002-8215-17 (Humulin R U-100) |
| TRI reagent | Sigma | T9494 |
| SYBR Green | ThermoFisher | 4309155 |
| Critical commercial assays | ||
| Ultra-sensitive mouse insulin ELISA | Crystal Chem | Cat# 90080 |
| Mouse/Rat FGF-21 Quantikine ELISA Kit | R&D Systems | Cat# MF2100 |
| Deposited data | ||
| Raw and analyzed data | This paper | GEO: GSE168588; Table S3 |
| Experimental models: organisms/strains | ||
| Mouse strain: C57BL/6J | The Jackson Laboratory | Cat# JAX:000664; RRID: IMSR_JAX:000664 |
| Mouse strain: Albumin-Cre | The Jackson Laboratory, (Postic et al., 1999) | Cat# JAX:003574; RRID: IMSR_JAX:003574 |
| Mouse strain:Tsc1loxP/loxP | The Jackson Laboratory, (Kwiatkowski et al., 2002) | Cat# JAX:005680; RRID: IMSR_JAX:005680 |
| Mouse strain: Gcn2loxP/loxP | The Jackson Laboratory, (Maurin et al., 2005) | Cat# JAX:008452; RRID: IMSR_JAX:008452 |
| Mouse strain:Fgf21loxP/loxP | The Jackson Laboratory, (Potthoff et al., 2009) | Cat# JAX:022361; RRID: IMSR_JAX:022361 |
| Mouse strain: CMV-Cre | The Jackson Laboratory, (Schwenk et al., 1995) | Cat# JAX:006054; RRID: IMSR_JAX:006054) |
| Mouse strain:C57BL/6J; Albumin-Cre Tsc1loxP/loxP | This paper | N/A |
| Mouse strain:C57BL/6J; Albumin-Cre Gcn2loxP/loxP | This paper | N/A |
| Mouse strain:C57BL/6J; Fgf21Δ/Δ | This paper | N/A |
| Oligonucleotides | ||
| Mouse TSC1 genotyping F: AGGAGGCCTCTTCTGCTACC | (Kwiatkowski et al., 2002) | F4536 |
| Mouse TSC1 genotyping R: CAGCTCCGACCATGAAGTG | (Kwiatkowski et al., 2002) | R4830 |
| Mouse GCN2 genotyping common F: TCTCCCAGCGGAATCCGCACATCG | The Jackson Laboratory | oIMR8796 https://www.jax.org/Protocol?stockNumber=008240&protocolID=23168 |
| Mouse GCN2 genotyping mutant R: TGCCACTGTCAGAATCTGAAGCAGG | The Jackson Laboratory | oIMR8791, https://www.jax.org/Protocol?stockNumber=008240&protocolID=23168 |
| Mouse GCN2 genotyping WT R: ATCCAGGCGTTGTAGTAGCGCACA | The Jackson Laboratory | oIMR8797, https://www.jax.org/Protocol?stockNumber=008240&protocolID=23168 |
| Mouse FGF21 genotyping WT F: ACCCCCTGAGCATGGTAGA | The Jackson Laboratory | 51425, https://www.jax.org/Protocol?stockNumber=033846&protocolID=37478 |
| Mouse FGF21 genotyping mutant F: CAGACCAAGGAGCACAGACC | The Jackson Laboratory | 51434, https://www.jax.org/Protocol?stockNumber=033846&protocolID=37478 |
| Mouse FGF21 genotyping common R: GCAGAGGCAAGTGATTTTGA | The Jackson Laboratory | 51435, https://www.jax.org/Protocol?stockNumber=033846&protocolID=37478 |
| Mouse Cre genotyping common F: GAACCTGATGGACATGTTCAGG | (Yu et al., 2019) | N/A |
| Mouse Cre genotyping common R: AGTGCGTTCGAACGCTAGAGCCTGT | (Yu et al., 2019) | N/A |
| Primers for RT-PCR: see Table S7 | This paper | N/A |
| Software and algorithms | ||
| Diet*Calc v1.5.0 | National Cancer Institute | https://epi.grants.cancer.gov/dhq2/dietcalc/ |
| edgeR package | (Robinson et al., 2010) | https://bioconductor.org/packages/release/bioc/html/edgeR.html |
| GraphPad Prism | GraphPad | http://www.graphpad.com/scientific-software/prism |
| Heatmapper | (Babicki et al., 2016) | http://www.heatmapper.ca/ |
| ImageJ | National Institutes of Health, (Schneider et al., 2012) | https://imagej.nih.gov/ij |
| MAVEN (version v.682) | (Clasquin et al., 2012) | http://genomics-pubs.princeton.edu/mzroll/index.php?show=index |
| MetaboAnalyst | (Chong et al., 2019) | https://www.metaboanalyst.ca/ |
| R (Version 3.5.3) | N/A | https://www.r-project.org/ |
| SAS (version 9.4) | SAS Institute, Inc. | https://www.sas.com/en_us/company-information.html |
| Other | ||
| Normal Chow | Purina | Cat# 5001 |
| Mouse diets (See Tables S1–S2 and S4–S5) | This paper | N/A |
All diets were obtained from Envigo, and diet compositions and item numbers are provided in Supplemental Tables 1–2 and 4–5. Mice were housed in a specific pathogen free (SPF) mouse facility with a 12:12 hour light/dark cycle and with free access to food and water. Animals were group housed in static microisolator cages, except when temporarily housed in a Columbus Instruments Oxymax/CLAMS metabolic chamber system. Group sizes are provided in the figure legends.
METHOD DETAILS
In vivo Procedures
Average food consumption during the first month of diet feeding was measured weekly by calculating the difference in food weight between the food put into the cage and that remaining at the end of each week. Food consumption was normalized to weight and lean mass determined at the same time food consumption was measured. For glucose and pyruvate/alanine tolerance tests, food was withheld overnight from ad libitum fed animals for 16 hours, and mice were then injected intraperitoneally with glucose (1 g/kg) or pyruvate (2 g/kg) or alanine (2 g/Kg) as described previously (Bellantuono et al., 2020; Yu et al., 2018). Insulin tolerance tests were performed by fasting mice for 4 hours starting at lights on, and then injecting insulin (0.75U/kg) intraperitoneally. Glucose stimulated insulin secretion (GSIS) assays were performed by recording glucose and collecting blood from overnight (16-hour) fasted mice before and 15min after an intraperitoneal injection of 1g/kg of glucose. Glucose measurements were taken using a Bayer Contour blood glucose meter and test strips. Body composition was assessed using an EchoMRI Body Composition Analyzer (EchoMRI, Houston, TX, USA) according to the manufacturer’s procedures. To assess metabolic physiology (O2, CO2, RER, and food consumption) and spontaneous activity, mice were placed into a Columbus Instruments Oxymax/CLAMS metabolic chamber system (Columbus Instruments, Columbus, OH, USA) and acclimated for approximately 24 hours prior to data collection, and data from a continuous 24-hour period was then selected for analysis as previously described (Yu et al., 2018).
To compare the similarities and differences between the in vivo metabolic phonotypes of mice fed each diet, we calculated the ratio of the value for each measurement in the individual diet group to the desired control group, and used the log transformed ratio to generate a summary heatmap using Heatmapper (Babicki et al., 2016). Values used were the final values of body weight, fat mass, and lean mass; food consumption; fasting blood glucose (FBG); and the AUCs for the GTT, ITT and ATT or PTT assays.
Hyperinsulinemic-Euglycemic Clamp
All hyperinsulinemic-euglycemic clamp studies were performed in mice at the Penn Diabetes Research Center Rodent Metabolic Phenotyping Core (University of Pennsylvania); mice were sourced directly from The Jackson Laboratory and diets were obtained from Envigo (Madison, WI).
Hyperinsulinemic-euglycemic clamp studies on lean mice were performed as previously described (Lamming et al., 2012) with some modifications. Indwelling jugular vein catheters were surgically implanted in the mice for infusion 5 days prior to the clamp study day. Mice were fasted for 5 hours prior to initiation of clamp and acclimated to the plastic restrainers for cut tail sampling. A [3-3H] glucose infusion was primed (5-μCi) and continuously infused for a 90 min equilibration period (0.05 μCi/min). Baseline measurements were determined in blood samples collected at −10 and 0 min (relative to the start of the clamp) for analysis of glucose, [3-3H] glucose specific activity and basal insulin. The clamp was started at t = 0 min with a continuous infusion of human insulin (1.25 mU/Kg/min; Novolin Regular Insulin) and glucose (D20 mixed with [3-3H] glucose 0.03 μCi/μL) was infused at variable glucose infusion rate (GIR) to maintain euglycemia. The mixing of D20 with [3-3H] glucose is required to maintain the specific activity constant during the clamp period. Blood samples were taken at t=80–120 min for the measurement of [3-3H] glucose specific activity and clamped insulin levels. 10 μCi of 2-[14C] deoxyglucose ([14C]2DG) was administered as an i.v. bolus 45 min (t=75 min) before the end of the study. After the final blood sample, animals were injected with a bolus of pentobarbital, and quadricep muscle and epididymal and brown adipose tissue were collected and frozen in liquid nitrogen and stored at −20°C for subsequent analysis.
Hyperinsulinemic-euglycemic clamp studies of obese, Western diet fed animals were performed as follows. Indwelling jugular vein and carotid artery catheters were surgically implanted in the mice for infusion 7 days prior to the clamp study day as previously described (Ayala et al., 2006). Mice were fasted for 5 hours prior to initiation of clamp and acclimated to the containers (plastic bowl with alpha dry). Jugular vein and arterial line are connected to the dual swivel 2 hours prior to the clamp initiation. A [3-3H] glucose infusion was primed (1.5 μCi) and continuously infused for a 90 min equilibration period (0.075 μCi/min). Baseline measurements were determined in arterial blood samples collected at −10 and 0 min (relative to the start of the clamp) for analysis of glucose, [3-3H] glucose specific activity, basal insulin and free fatty acids. The clamp was started at t = 0 min with a primed-continuous infusion of human insulin (2.5 mU/Kg/min; Novolin Regular Insulin), a donor blood infusion at 4.5 uL/min to prevent a 5% drop in the hematocrit, and glucose (D50 mixed with [3-3H] glucose 0.05 μCi/uL) was infused at variable glucose infusion rate (GIR) to maintain euglycemia. The mixing of D50 with [3-3H] glucose is required to maintain the specific activity constant during the clamp period. Arterial blood samples were taken at t=80–120 min for the measurement of [3-3H] glucose specific activity, clamped insulin and free fatty acid levels. 120 minutes after initiation of clamp, 14C-2DG (12μCi) is injected and arterial blood samples obtained at 2, 5, 10, 15 and 25 min to determine Rg, an index of tissue-specific glucose uptake in various tissues. After the final blood sample, animals were injected with a bolus of pentobarbital, and tissues were collected and frozen in liquid nitrogen and stored at −80°C for subsequent analysis.
Processing of samples and calculations: Radioactivity of [3-3H] glucose, [14C]2DG and [14C]2DG-6-phosphate were determined as previously described (Ayala et al., 2007). The glucose turnover rate (total Ra; mg/kg/min) was calculated as the rate of tracer infusion (dpm/min) divided by the corrected plasma glucose specific activity (dpm/mg) per kg body weight of the mouse. Glucose appearance (Ra) and disappearance (Rd) rates were determined using steady-state equations and endogenous glucose production (Ra) was determined by subtracting the GIR from total Ra. Tissue specific glucose disposal (Rg; μmol/100g tissue/min) was calculated as previously described (Ayala et al., 2007). The plasma insulin concentration was determined by mouse ELISA kit (Biomarker core, University of Pennsylvania).
Transcriptional profiling
10-week-old male C57BL/6J mice from The Jackson Laboratory were placed and maintained on Ctrl AA, Low AA and Low Ile diets for 3 weeks before being sacrificed in fasted (16-hour fast) or refed (3h-refeeding following 16-hour fast) state. Liver tissues were harvested and snap frozen in liquid nitrogen.
Transcriptional profiling: RNA was extracted from liver as previously described (Cummings et al., 2018). Concentration and purity of RNA was initially determined using a Nanodrop 2000c, and then submitted to the University of Wisconsin-Madison Biotechnology Center Gene Expression Center & DNA Sequencing Facility. RNA quality was assayed using an Agilent RNA NanoChip, and stranded mRNA libraries with polyA enrichment were prepared as described in the Illumina TruSeq Stranded mRNA Sample Preparation Guide Rev. E. To remove any genes that exhibited no or a very low number of mapped reads, only genes that had more than 1 count per million in at least 4 samples across all treatments were retained for further analysis. This resulted in a total of 14211 unique genes. Raw counts were normalized using the trimmed mean of M values (TMM normalization) (Robinson and Oshlack, 2010). Differential gene expression was modeled using the edgeR package (Robinson et al., 2010) in R (version 3.5.3) and pairwise comparisons were conducted between Fasted Control or Refed Ctrl AA and either Low AA or Low Ile. To control for type I errors, the Benjamini Hochberg adjusted p-value was used (5% FDR). We tested for over-representation of gene ontology (GO) terms or KEGG pathways in each comparison, using the limma package (Ritchie et al., 2015).
Metabolomics study
Metabolite extraction: To extract metabolites from the liver, snap-frozen liver samples were ground at liquid nitrogen temperature with a Cryomill (Retsch, Newtown, PA). The resulting liver powder (~20 mg) was weighed and then metabolites were extracted by adding −20°C acetonitrile:methanol:water (40:40:20 v/v) extraction solution, vortexed, and centrifuged at 16,000 x g for 10 min at 4°C. The volume of the extraction solution (μL) was 40x the weight of liver (mg) to make an extract of 25 mg tissue per mL solvent. The supernatant was collected for liquid chromatography-mass spectrometry (LC-MS) analysis.
Metabolite measurement by LC-MS: A quadrupole-orbitrap mass spectrometer (Q Exactive, Thermo Fisher Scientific, San Jose, CA) operating in negative or positive ion mode was coupled to hydrophilic interaction chromatography via electrospray ionization and used to scan from m/z 70 to 1000 at 1 Hz and 75,000 resolution. LC separation was on a XBridge BEH Amide column (2.1 mm x 150 mm, 2.5 μm particle size, 130 Å pore size; Waters, Milford, MA) using a gradient of solvent A (20 mM ammonium acetate, 20 mM ammonium hydroxide in 95:5 water: acetonitrile, pH 9.45) and solvent B (acetonitrile). Flow rate was 150 μL/min. The LC gradient was: 0 min, 85% B; 2 min, 85% B; 3 min, 80% B; 5 min, 80% B; 6 min, 75% B; 7 min, 75% B; 8 min, 70% B; 9 min, 70% B; 10 min, 50% B; 12 min, 50% B; 13 min, 25% B; 16 min, 25% B; 18 min, 0% B; 23 min, 0% B; 24 min, 85% B; 30 min, 85% B. Autosampler temperature was 5°C, and injection volume was 3 μL. For quantification of circulating amino acid concentrations, authentic standard amino acids were ran together to obtain standard curves. Data were analyzed using the MAVEN software (Clasquin et al., 2012) v.682 (http://genomics-pubs.princeton.edu/mzroll/index.php?show=download).
Metabolomics analysis: Before metabolomic analysis, a series of processing steps were applied to the entire data set to supply us with the most instructive and stable metabolites for further analysis. First, metabolites were normalized using a log base 2 transformation. Second, only metabolites with a signal-to-noise ratio (signal-to-noise ratio = mean/sample standard deviation) greater than or equal to 15 were kept for analysis. Third, metabolites that were missing from 15% or more of all samples were removed. To detect significantly differentially expressed metabolites between treatment groups, an empirical Bayes moderated linear model was fitted to each metabolite (Smyth, 2004). P values for each comparison were adjusted using the Benjamini–Hochberg procedure using a false discovery rate (FDR) of 20%. Pathway analysis was conducted using the online tool MetaboAnalyst (Chong et al., 2019). Using the Pathway Analysis function, we entered a list of significantly altered metabolites using the human metabolome database (HMDB) IDs from the linear model (p<0.05).
Assays and Kits
Unless noted otherwise, all assays were performed using samples from overnight fasted mice. Plasma insulin was quantified using a ultra-sensitive mouse insulin ELISA kit (90080), from Crystal Chem (Elk Grove Village, IL, USA). Blood FGF21 levels were assayed by a mouse/rat FGF-21 quantikine ELISA kit (MF2100) from R&D Systems (Minneapolis, MN, USA).
Histology
Unless noted otherwise, tissues were harvested from mice on the indicated diets following overnight (16-hr) fasting. Samples of brown adipose tissue (BAT), white adipose tissue (WAT), liver, and skin were isolated following euthanasia. Adipose tissue including epididymal white adipose tissue (eWAT), inguinal white adipose tissue (iWAT) and brown adipose tissue (BAT) was fixed in 4% paraformaldehyde overnight, sectioned and H&E stained by the UWCCC Experimental Pathology Laboratory. Liver was embedded in OCT, and then cryosectioned and Oil-Red-O stained by the UWCCC Experimental Pathology Laboratory. Liver, BAT and WAT sections were imaged using an EVOS microscope as previously described (Cummings et al., 2018). Skin was isolated from the abdomen and back of mice, paraformaldehyde-fixed (4%) overnight and then paraffin-embedded for evaluation (Kasza et al., 2014). For quantification of lipid droplet size in liver, 6 independent fields were obtained for tissue from each mouse by investigators blinded to the treatment group, and then quantified using NIH ImageJ.
Quantitative PCR
RNA was extracted from liver or adipose tissue using Tri-reagent (Sigma-Aldrich, St. Louis, MO, USA). 1 μg of RNA was used to generate cDNA using Superscript III (Invitrogen, Carlsbad, CA, USA). Oligo dT primers and primers for real-time PCR were obtained from Integrated DNA Technologies (Coralville, IA, USA). Reactions were run on an Applied Biosystems StepOne Plus machine (Applied Biosystems, Foster City, CA, USA) with Sybr Green PCR Master Mix (Invitrogen, Carlsbad, CA, USA). Actin was used to normalize the results from gene-specific reactions. Primer sequences are given in Table S7.
Immunoblotting
Unless noted otherwise, tissues including liver, muscle and adipose were harvested from mice on the indicated diets following overnight (16-hr) fasting. Tissue samples were lysed in cold RIPA buffer supplemented with phosphatase and protease inhibitor cocktail tablets. Tissues were lysed as previously described (Baar et al., 2016) using a FastPrep 24 (M.P. Biomedicals, Santa Ana, CA, USA) with bead-beating tubes (16466–042) from (VWR, Radnor, PA, USA) and zirconium ceramic oxide bulk beads (15340159) from (Thermo Fisher Scientific, Waltham, MA, USA) and then centrifuged at 13,300 rpm for 10 min and supernatant was subsequently collected. A NEPER nuclear and cytoplasmic extraction kit (78833) from (Thermo Fisher Scientific, Waltham, MA, USA) was used to separate nuclear and cytoplasmic proteins according to manufacturer instructions. Protein concentration was determined by Bradford (Pierce Biotechnology, Waltham, MA, USA). 20–30 μg protein was separated by SDS–PAGE (sodium dodecyl sulfate–polyacrylamide gel electrophoresis) on 8%, 10%, or 16% resolving gels (Thermo Fisher Scientific, Waltham, MA, USA) and transferred to PVDF membrane (EMD Millipore, Burlington, MA, USA). S6 (2217), HSP90 (4877), pS204/244 S6 (2215), p-T37/S46 4E-BP1 (2855), 4E-BP1 (9644), pS51-eIF2α (9721), eIF2α (9722), TSC1 (6935), β-Tubulin (2146), ATF-4 (11815), HRP-linked anti-mouse (7076) and HRP-linked anti-rabbit (7074) antibodies were purchased from Cell Signaling Technologies. FOXA2 antibody (sc-374376) was purchased from Santa Cruz Biotechnology. Lamin A/C antibody was obtained from the Developmental Studies Hybridoma Bank at the University of Iowa by Morris, G.E. (DSHB Hybridoma Product MANLAC2(10F8)). Imaging was performed using a GE ImageQuant LAS 4000 imaging station (GE Healthcare, Chicago, IL, USA). Quantification was performed by densitometry using NIH ImageJ software (Schneider et al., 2012).
SHOW study
We analyzed the association between dietary isoleucine intake and body mass indexes (BMI) in a sample of 2016–2017 Survey of Health of Wisconsin (SHOW) participants. SHOW is an ongoing population-based health examination survey of non-institutionalized residents of Wisconsin.
Detailed survey methods have been previously described (Nieto et al., 2010). Survey components relevant to the current analysis included an in-home interview accompanied by measurements of weight and height, and a self-administered questionnaire including the National Cancer Institute’s Diet History Questionnaire from which specific dietary intake variables were derived. The study population included 788 individuals that completed all parts of the survey. Demographic characteristics of the population are described in Table S6. All study protocols were approved by the University of Wisconsin Health Sciences Institutional Review Board, and all participants provided written informed consent as part of the initial home visit.
The intake of isoleucine and valine were estimated from the Diet History Questionnaire II (National Institutes of Health, 2010) using Diet*Calc software v 1.5.0 (National Cancer Institute, October 2012). The estimated levels of each AA are expressed as the percent (%) of total protein. The primary outcome was body mass index (kg/m2) derived from measured weight (kg) and height (m).
Multiple linear regression was performed in SAS 9.4 software (Cary, NC) with BMI as the outcome and percent of total protein from isoleucine or valine as the predictor of interest. In the model we also adjusted for age, gender, education, income, total caloric intake, and physical activity.
QUANTIFICATION AND STATISTICAL ANALYSIS
Unless otherwise stated, statistical analysis was conducted using Prism 7/8 (GraphPad Software). Tests involving repeated measurements were analyzed with two-way repeated-measures ANOVA, followed by a Tukey-Kramer or Dunnett’s post-hoc test as specified. All other comparisons of three or more means were analyzed by one-way or two-way ANOVA followed by a Dunnett’s,Tukey-Kramer, or Sidak’s post-hoc test as specified in the figure legends. Additional comparisons, if any, were corrected for multiple comparisons using the Bonferroni method. To calculate the percentage of isoleucine in each gene, we extracted amino acid sequences for SDE genes between the control and Low ILE groups in both the fasted and fed state using UniProt (UniProt, 2021) and calculated amino acid percentages using ProtrWeb (Xiao et al., 2015). To compare to unchanged genes, we extracted the same data from the 100 least altered (and non-significant) genes by fold-change.
Supplementary Material
Supplemental Table 3, related to Figure 4. Transcriptional profiling and metabolomics data. Genes, fold change of Control vs. Low Ile, p-value, and FDR adjusted p-value of data from RNA sequencing of fasted and fed livers; and Metabolites, fold change of Control vs. Low Ile, p-value, and FDR adjusted p-value of data from metabolite analysis of fasted and fed livers.
Highlights.
Reduced isoleucine or valine, but not leucine, promotes metabolic health in mice.
Reduced isoleucine is required for the metabolic benefits of a low protein diet.
The benefits of isoleucine restriction are mediated in part by FGF21.
Dietary levels of isoleucine are positively associated with BMI in humans.
ACKNOWLEDGEMENTS
We would like to thank Dr. Dawn Davis and Dr. Vincent Cryns for their valuable insights and comments. We thank Dr. Tina Herfel (Envigo) for assistance with diet formulation. The MANLAC2 (10F8) antibody was developed by G.E. Morris and was obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at The University of Iowa, Department of Biology, Iowa City, IA 52242. The work was supported in part by the NIH/NIA (AG056771, AG062328, and AG061635 to DWL), the NIH/NIGMS (GM113142 to CMA), the NIH/NIAMS (P30 AR066524 Pilot Award to IK), and the NIH/NIDDK (DP1DK113643 to JDR), by a Glenn Foundation Award for Research in the Biological Mechanisms of Aging to DWL, and startup funds from the UW-Madison School of Medicine and Public Health and Department of Medicine to DWL. Support for this research was provided by the UW-Madison Office of the Vice Chancellor for Research and Graduate Education with funding from the Wisconsin Alumni Research Foundation. The Survey of the Health of Wisconsin is funded by the Wisconsin Partnership Program. This research was conducted while DWL was an AFAR Research Grant recipient from the American Federation for Aging Research. DY was supported in part by a fellowship from the American Heart Association (17PRE33410983). NER was supported in part by a training grant from the UW Institute on Aging (NIA T32 AG000213). CLG was supported in part by a grant from Dalio Philanthropies and is supported by a Glenn Foundation for Medical Research Postdoctoral Fellowship in Aging Research. VF and MEM were supported in part by Research Supplements to Promote Diversity in Health-Related Research (R01 AG056771-01A1S1 and R01AG062328-03S1). HHP was supported in part by a NIA F31 predoctoral fellowship (AG066311). IK was supported in part by McArdle Departmental Funds. CJ was supported by the American Diabetes Association (1-17-PDF-076). Metabolomics work was supported by Diabetes Research Center Grant P30 DK019525. The UW Carbone Cancer Center (UWCCC) Experimental Pathology Laboratory is supported by P30 CA014520 from the NIH/NCI. Clamp studies were performed in the Rodent Metabolic Phenotyping Core of the University of Pennsylvania Diabetes Research Center (P30 DK19525); this award also supported in part the metabolomics analysis. This work was supported in part by the U.S. Department of Veterans Affairs (I01-BX004031), and this work was supported using facilities and resources from the William S. Middleton Memorial Veterans Hospital. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. This work does not represent the views of the Department of Veterans Affairs or the United States Government.
Footnotes
DECLARATION OF INTERESTS
DWL has received funding from, and is a scientific advisory board member of, Aeovian Pharmaceuticals, which seeks to develop novel, selective mTOR inhibitors for the treatment of various diseases. UW-Madison has applied for a patent based in part on the findings reported here, for which NER and DWL are inventors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- (1999). Diabetes mellitus: a major risk factor for cardiovascular disease. A joint editorial statement by the American Diabetes Association; The National Heart, Lung, and Blood Institute; The Juvenile Diabetes Foundation International; The National Institute of Diabetes and Digestive and Kidney Diseases; and The American Heart Association. Circulation 100, 1132–1133. [DOI] [PubMed] [Google Scholar]
- Akter S, Mizoue T, Nanri A, Goto A, Noda M, Sawada N, Yamaji T, Iwasaki M, Inoue M, Tsugane S, et al. (2020). Low carbohydrate diet and all cause and cause-specific mortality. Clin Nutr. [DOI] [PubMed] [Google Scholar]
- Appuhamy JA, Knoebel NA, Nayananjalie WA, Escobar J, and Hanigan MD (2012). Isoleucine and leucine independently regulate mTOR signaling and protein synthesis in MAC-T cells and bovine mammary tissue slices. J Nutr 142, 484–491. [DOI] [PubMed] [Google Scholar]
- Arriola Apelo SI, Singer LM, Lin XY, McGilliard ML, St-Pierre NR, and Hanigan MD (2014). Isoleucine, leucine, methionine, and threonine effects on mammalian target of rapamycin signaling in mammary tissue. J Dairy Sci 97, 1047–1056. [DOI] [PubMed] [Google Scholar]
- Ayala JE, Bracy DP, Julien BM, Rottman JN, Fueger PT, and Wasserman DH (2007). Chronic treatment with sildenafil improves energy balance and insulin action in high fat-fed conscious mice. Diabetes 56, 1025–1033. [DOI] [PubMed] [Google Scholar]
- Ayala JE, Bracy DP, McGuinness OP, and Wasserman DH (2006). Considerations in the design of hyperinsulinemic-euglycemic clamps in the conscious mouse. Diabetes 55, 390–397. [DOI] [PubMed] [Google Scholar]
- Baar EL, Carbajal KA, Ong IM, and Lamming DW (2016). Sex- and tissue-specific changes in mTOR signaling with age in C57BL/6J mice. Aging Cell 15, 155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babicki S, Arndt D, Marcu A, Liang Y, Grant JR, Maciejewski A, and Wishart DS (2016). Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res 44, W147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbagallo M, and Dominguez LJ (2014). Type 2 diabetes mellitus and Alzheimer’s disease. World J Diabetes 5, 889–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrington WT, Wulfridge P, Wells AE, Rojas CM, Howe SYF, Perry A, Hua K, Pellizzon MA, Hansen KD, Voy BH, et al. (2018). Improving Metabolic Health Through Precision Dietetics in Mice. Genetics 208, 399–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batch BC, Shah SH, Newgard CB, Turer CB, Haynes C, Bain JR, Muehlbauer M, Patel MJ, Stevens RD, Appel LJ, et al. (2013). Branched chain amino acids are novel biomarkers for discrimination of metabolic wellness. Metabolism 62, 961–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellantuono I, de Cabo R, Ehninger D, Di Germanio C, Lawrie A, Miller J, Mitchell SJ, Navas-Enamorado I, Potter PK, Tchkonia T, et al. (2020). A toolbox for the longitudinal assessment of healthspan in aging mice. Nature protocols 15, 540–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boye J, Wijesinha-Bettoni R, and Burlingame B (2012). Protein quality evaluation twenty years after the introduction of the protein digestibility corrected amino acid score method. Br J Nutr 108 Suppl 2, S183–211. [DOI] [PubMed] [Google Scholar]
- Chantranupong L, Wolfson RL, Orozco JM, Saxton RA, Scaria SM, Bar-Peled L, Spooner E, Isasa M, Gygi SP, and Sabatini DM (2014). The Sestrins interact with GATOR2 to negatively regulate the amino-acid-sensing pathway upstream of mTORC1. Cell reports 9, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J, Yamamoto M, and Xia J (2019). MetaboAnalystR 2.0: From Raw Spectra to Biological Insights. Metabolites 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clasquin MF, Melamud E, and Rabinowitz JD (2012). LC-MS data processing with MAVEN: a metabolomic analysis and visualization engine. Curr Protoc Bioinformatics Chapter 14, Unit14 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly MA, Wolak-Dinsmore J, and Dullaart RPF (2017). Branched Chain Amino Acids Are Associated with Insulin Resistance Independent of Leptin and Adiponectin in Subjects with Varying Degrees of Glucose Tolerance. Metab Syndr Relat Disord 15, 183–186. [DOI] [PubMed] [Google Scholar]
- Cummings NE, Williams EM, Kasza I, Konon EN, Schaid MD, Schmidt BA, Poudel C, Sherman DS, Yu D, Arriola Apelo SI, et al. (2018). Restoration of metabolic health by decreased consumption of branched-chain amino acids. The Journal of physiology 596, 623–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deelen J, Kettunen J, Fischer K, van der Spek A, Trompet S, Kastenmuller G, Boyd A, Zierer J, van den Akker EB, Ala-Korpela M, et al. (2019). A metabolic profile of all-cause mortality risk identified in an observational study of 44,168 individuals. Nature communications 10, 3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Qiu H, Garcia-Barrio M, Anderson J, and Hinnebusch AG (2000). Uncharged tRNA Activates GCN2 by Displacing the Protein Kinase Moiety from a Bipartite tRNA-Binding Domain. Molecular Cell 6, 269–279. [DOI] [PubMed] [Google Scholar]
- Dyachok J, Earnest S, Iturraran EN, Cobb MH, and Ross EM (2016). Amino Acids Regulate mTORC1 by an Obligate Two-step Mechanism. J Biol Chem 291, 22414–22426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshorbagy A, Jerneren F, Basta M, Basta C, Turner C, Khaled M, and Refsum H (2017). Amino acid changes during transition to a vegan diet supplemented with fish in healthy humans. Eur J Nutr 56, 1953–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felig P, Marliss E, and Cahill GF Jr. (1969). Plasma amino acid levels and insulin secretion in obesity. N Engl J Med 281, 811–816. [DOI] [PubMed] [Google Scholar]
- Fisher FM, Kim M, Doridot L, Cunniff JC, Parker TS, Levine DM, Hellerstein MK, Hudgins LC, Maratos-Flier E, and Herman MA (2017). A critical role for ChREBP-mediated FGF21 secretion in hepatic fructose metabolism. Molecular metabolism 6, 14–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher FM, and Maratos-Flier E (2016). Understanding the Physiology of FGF21. Annual review of physiology 78, 223–241. [DOI] [PubMed] [Google Scholar]
- Fontana L, Cummings NE, Arriola Apelo SI, Neuman JC, Kasza I, Schmidt BA, Cava E, Spelta F, Tosti V, Syed FA, et al. (2016). Decreased Consumption of Branched-Chain Amino Acids Improves Metabolic Health. Cell reports 16, 520–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganeshan K, and Chawla A (2017). Warming the mouse to model human diseases. Nature reviews. Endocrinology 13, 458–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannucci E, Harlan DM, Archer MC, Bergenstal RM, Gapstur SM, Habel LA, Pollak M, Regensteiner JG, and Yee D (2010). Diabetes and cancer: a consensus report. Diabetes Care 33, 1674–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harputlugil E, Hine C, Vargas D, Robertson L, Manning BD, and Mitchell JR (2014). The TSC complex is required for the benefits of dietary protein restriction on stress resistance in vivo. Cell reports 8, 1160–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XD, Gong W, Zhang JN, Nie J, Yao CF, Guo FS, Lin Y, Wu XH, Li F, Li J, et al. (2018). Sensing and Transmitting Intracellular Amino Acid Signals through Reversible Lysine Aminoacylations. Cell Metab 27, 151–166 e156. [DOI] [PubMed] [Google Scholar]
- Hill CM, Berthoud HR, Munzberg H, and Morrison CD (2018). Homeostatic sensing of dietary protein restriction: A case for FGF21. Front Neuroendocrinol 51, 125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill CM, Laeger T, Dehner M, Albarado DC, Clarke B, Wanders D, Burke SJ, Collier JJ, Qualls-Creekmore E, Solon-Biet SM, et al. (2019). FGF21 Signals Protein Status to the Brain and Adaptively Regulates Food Choice and Metabolism. Cell reports 27, 2934–2947 e2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Liao LM, Weinstein SJ, Sinha R, Graubard BI, and Albanes D (2020). Association Between Plant and Animal Protein Intake and Overall and Cause-Specific Mortality. JAMA Intern Med 180, 1173–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Kinoshita M, Kayama H, Nagamori S, Kongpracha P, Umemoto E, Okumura R, Kurakawa T, Murakami M, Mikami N, et al. (2017). Slc3a2 Mediates Branched-Chain Amino-Acid-Dependent Maintenance of Regulatory T Cells. Cell reports 21, 1824–1838. [DOI] [PubMed] [Google Scholar]
- Inagaki T, Dutchak P, Zhao G, Ding X, Gautron L, Parameswara V, Li Y, Goetz R, Mohammadi M, Esser V, et al. (2007). Endocrine regulation of the fasting response by PPARalpha-mediated induction of fibroblast growth factor 21. Cell Metab 5, 415–425. [DOI] [PubMed] [Google Scholar]
- Jang C, Oh SF, Wada S, Rowe GC, Liu L, Chan MC, Rhee J, Hoshino A, Kim B, Ibrahim A, et al. (2016). A branched-chain amino acid metabolite drives vascular fatty acid transport and causes insulin resistance. Nat Med 22, 421–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karusheva Y, Koessler T, Strassburger K, Markgraf D, Mastrototaro L, Jelenik T, Simon M-C, Pesta D, Zaharia O-P, Bódis K, et al. (2019). Short-term dietary reduction of branched-chain amino acids reduces meal-induced insulin secretion and modifies microbiome composition in type 2 diabetes: a randomized controlled crossover trial. The American Journal of Clinical Nutrition. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasza I, Suh Y, Wollny D, Clark RJ, Roopra A, Colman RJ, MacDougald OA, Shedd TA, Nelson DW, Yen MI, et al. (2014). Syndecan-1 is required to maintain intradermal fat and prevent cold stress. PLoS Genet 10, e1004514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy BK, and Lamming DW (2016). The Mechanistic Target of Rapamycin: The Grand ConducTOR of Metabolism and Aging. Cell Metab 23, 990–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Kato H, Hirabayashi Y, Murakami H, and Suzuki H (2006). Modulations of muscle protein metabolism by branched-chain amino acids in normal and muscle-atrophying rats. J Nutr 136, 234s–236s. [DOI] [PubMed] [Google Scholar]
- Kwiatkowski DJ, Zhang H, Bandura JL, Heiberger KM, Glogauer M, el-Hashemite N, and Onda H (2002). A mouse model of TSC1 reveals sex-dependent lethality from liver hemangiomas, and up-regulation of p70S6 kinase activity in Tsc1 null cells. Hum Mol Genet 11, 525–534. [DOI] [PubMed] [Google Scholar]
- Laeger T, Albarado DC, Burke SJ, Trosclair L, Hedgepeth JW, Berthoud HR, Gettys TW, Collier JJ, Munzberg H, and Morrison CD (2016). Metabolic Responses to Dietary Protein Restriction Require an Increase in FGF21 that Is Delayed by the Absence of GCN2. Cell reports 16, 707–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laeger T, Henagan TM, Albarado DC, Redman LM, Bray GA, Noland RC, Munzberg H, Hutson SM, Gettys TW, Schwartz MW, et al. (2014). FGF21 is an endocrine signal of protein restriction. J Clin Invest 124, 3913–3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagiou P, Sandin S, Weiderpass E, Lagiou A, Mucci L, Trichopoulos D, and Adami HO (2007). Low carbohydrate-high protein diet and mortality in a cohort of Swedish women. J Intern Med 261, 366–374. [DOI] [PubMed] [Google Scholar]
- Lamming DW, Cummings NE, Rastelli AL, Gao F, Cava E, Bertozzi B, Spelta F, Pili R, and Fontana L (2015). Restriction of dietary protein decreases mTORC1 in tumors and somatic tissues of a tumor-bearing mouse xenograft model. Oncotarget 6, 31233–31240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamming DW, Ye L, Katajisto P, Goncalves MD, Saitoh M, Stevens DM, Davis JG, Salmon AB, Richardson A, Ahima RS, et al. (2012). Rapamycin-induced insulin resistance is mediated by mTORC2 loss and uncoupled from longevity. Science 335, 1638–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees EK, Banks R, Cook C, Hill S, Morrice N, Grant L, Mody N, and Delibegovic M (2017). Direct comparison of methionine restriction with leucine restriction on the metabolic health of C57BL/6J mice. Scientific reports 7, 9977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine ME, Suarez JA, Brandhorst S, Balasubramanian P, Cheng CW, Madia F, Fontana L, Mirisola MG, Guevara-Aguirre J, Wan J, et al. (2014). Low protein intake is associated with a major reduction in IGF-1, cancer, and overall mortality in the 65 and younger but not older population. Cell Metab 19, 407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao CY, Rikke BA, Johnson TE, Diaz V, and Nelson JF (2010). Genetic variation in the murine lifespan response to dietary restriction: from life extension to life shortening. Aging Cell 9, 92–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotta LA, Scott RA, Sharp SJ, Burgess S, Luan J, Tillin T, Schmidt AF, Imamura F, Stewart ID, Perry JR, et al. (2016). Genetic Predisposition to an Impaired Metabolism of the Branched-Chain Amino Acids and Risk of Type 2 Diabetes: A Mendelian Randomisation Analysis. PLoS Med 13, e1002179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Temp U, Müller-Hartmann A, Esser J, Grönke S, and Partridge L (2021). Sestrin is a key regulator of stem cell function and lifespan in response to dietary amino acids. Nature Aging 1, 60–72. [DOI] [PubMed] [Google Scholar]
- Magkos F, Bradley D, Schweitzer GG, Finck BN, Eagon JC, Ilkayeva O, Newgard CB, and Klein S (2013). Effect of Roux-en-Y gastric bypass and laparoscopic adjustable gastric banding on branched-chain amino acid metabolism. Diabetes 62, 2757–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maida A, Zota A, Sjoberg KA, Schumacher J, Sijmonsma TP, Pfenninger A, Christensen MM, Gantert T, Fuhrmeister J, Rothermel U, et al. (2016). A liver stress-endocrine nexus promotes metabolic integrity during dietary protein dilution. J Clin Invest 126, 3263–3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurin AC, Jousse C, Averous J, Parry L, Bruhat A, Cherasse Y, Zeng H, Zhang Y, Harding HP, Ron D, et al. (2005). The GCN2 kinase biases feeding behavior to maintain amino acid homeostasis in omnivores. Cell Metab 1, 273–277. [DOI] [PubMed] [Google Scholar]
- Mitchell SJ, Madrigal-Matute J, Scheibye-Knudsen M, Fang E, Aon M, González-Reyes JA, Cortassa S, Kaushik S, Gonzalez-Freire M, Patel B, et al. (2016). Effects of Sex, Strain, and Energy Intake on Hallmarks of Aging in Mice. Cell Metab 23, 1093–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottillo EP, Balasubramanian P, Lee YH, Weng C, Kershaw EE, and Granneman JG (2014). Coupling of lipolysis and de novo lipogenesis in brown, beige, and white adipose tissues during chronic beta3-adrenergic receptor activation. J Lipid Res 55, 2276–2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Cancer Institute, E.a.G.R.P. (October 2012). Diet*Calc Analysis Program. [Google Scholar]
- National Institutes of Health, E.a.G.R.P. (2010). Diet History Questionnaire. [Google Scholar]
- Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, Haqq AM, Shah SH, Arlotto M, Slentz CA, et al. (2009). A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab 9, 311–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto FJ, Peppard PE, Engelman CD, McElroy JA, Galvao LW, Friedman EM, Bersch AJ, and Malecki KC (2010). The Survey of the Health of Wisconsin (SHOW), a novel infrastructure for population health research: rationale and methods. BMC Public Health 10, 785–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden CL, Carroll MD, Kit BK, and Flegal KM (2014). Prevalence of childhood and adult obesity in the United States, 2011–2012. JAMA 311, 806–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postic C, Shiota M, Niswender KD, Jetton TL, Chen Y, Moates JM, Shelton KD, Lindner J, Cherrington AD, and Magnuson MA (1999). Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J Biol Chem 274, 305–315. [DOI] [PubMed] [Google Scholar]
- Potthoff MJ, Inagaki T, Satapati S, Ding X, He T, Goetz R, Mohammadi M, Finck BN, Mangelsdorf DJ, Kliewer SA, et al. (2009). FGF21 induces PGC-1alpha and regulates carbohydrate and fatty acid metabolism during the adaptive starvation response. Proc Natl Acad Sci U S A 106, 10853–10858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson NE, Konon EN, Schuster HS, Mitchell AT, Boyle C, Rodgers AC, Finke M, Haider LR, Yu D, Flores V, et al. (2021). Lifelong restriction of dietary branched-chain amino acids has sex-specific benefits for frailty and life span in mice. Nature Aging 1, 73–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, and Oshlack A (2010). A scaling normalization method for differential expression analysis of RNA-seq data. Genome biology 11, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roopra A (2020). MAGIC: A tool for predicting transcription factors and cofactors driving gene sets using ENCODE data. PLoS Comput Biol 16, e1007800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton RA, Knockenhauer KE, Wolfson RL, Chantranupong L, Pacold ME, Wang T, Schwartz TU, and Sabatini DM (2016). Structural basis for leucine sensing by the Sestrin2-mTORC1 pathway. Science 351, 53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt JA, Rinaldi S, Scalbert A, Ferrari P, Achaintre D, Gunter MJ, Appleby PN, Key TJ, and Travis RC (2016). Plasma concentrations and intakes of amino acids in male meat-eaters, fish-eaters, vegetarians and vegans: a cross-sectional analysis in the EPIC-Oxford cohort. Eur J Clin Nutr 70, 306–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nature methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenk F, Baron U, and Rajewsky K (1995). A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res 23, 5080–5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Peterson TR, Laplante M, Oh S, and Sabatini DM (2010). mTORC1 controls fasting-induced ketogenesis and its modulation by ageing. Nature 468, 1100–1104. [DOI] [PubMed] [Google Scholar]
- Sheen JH, Zoncu R, Kim D, and Sabatini DM (2011). Defective regulation of autophagy upon leucine deprivation reveals a targetable liability of human melanoma cells in vitro and in vivo. Cancer Cell 19, 613–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluijs I, Beulens JW, van der AD, Spijkerman AM, Grobbee DE, and van der Schouw YT (2010). Dietary intake of total, animal, and vegetable protein and risk of type 2 diabetes in the European Prospective Investigation into Cancer and Nutrition (EPIC)-NL study. Diabetes Care 33, 43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK (2004). Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Statistical applications in genetics and molecular biology 3, Article3. [DOI] [PubMed] [Google Scholar]
- Solon-Biet SM, Cogger VC, Pulpitel T, Wahl D, Clark X, Bagley E, Gregoriou GC, Senior AM, Wang QP, Brandon AE, et al. (2019). Branched chain amino acids impact health and lifespan indirectly via amino acid balance and appetite control. Nat Metab 1, 532–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solon-Biet SM, McMahon AC, Ballard JW, Ruohonen K, Wu LE, Cogger VC, Warren A, Huang X, Pichaud N, Melvin RG, et al. (2014). The ratio of macronutrients, not caloric intake, dictates cardiometabolic health, aging, and longevity in ad libitum-fed mice. Cell Metab 19, 418–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solon-Biet SM, Mitchell SJ, Coogan SC, Cogger VC, Gokarn R, McMahon AC, Raubenheimer D, de Cabo R, Simpson SJ, and Le Couteur DG (2015). Dietary Protein to Carbohydrate Ratio and Caloric Restriction: Comparing Metabolic Outcomes in Mice. Cell reports 11, 1529–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S, Baxa U, Niu G, Chen X, and Veech RL (2013). A ketogenic diet increases brown adipose tissue mitochondrial proteins and UCP1 levels in mice. IUBMB Life 65, 58–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S, Kashiwaya Y, King MT, Baxa U, Tam J, Niu G, Chen X, Clarke K, and Veech RL (2012). Mitochondrial biogenesis and increased uncoupling protein 1 in brown adipose tissue of mice fed a ketone ester diet. FASEB J 26, 2351–2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt, C. (2021). UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res 49, D480–D489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanders D, Forney LA, Stone KP, Burk DH, Pierse A, and Gettys TW (2017). FGF21 Mediates the Thermogenic and Insulin-Sensitizing Effects of Dietary Methionine Restriction but Not Its Effects on Hepatic Lipid Metabolism. Diabetes 66, 858–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek SA, Zhu S, and Wek RC (1995). The histidyl-tRNA synthetase-related sequence in the eIF-2 alpha protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Molecular and Cellular Biology 15, 4497–4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White PJ, Lapworth AL, An J, Wang L, McGarrah RW, Stevens RD, Ilkayeva O, George T, Muehlbauer MJ, Bain JR, et al. (2016). Branched-chain amino acid restriction in Zucker-fatty rats improves muscle insulin sensitivity by enhancing efficiency of fatty acid oxidation and acyl-glycine export. Molecular metabolism 5, 538–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfrum C, Asilmaz E, Luca E, Friedman JM, and Stoffel M (2004). Foxa2 regulates lipid metabolism and ketogenesis in the liver during fasting and in diabetes. Nature 432, 1027–1032. [DOI] [PubMed] [Google Scholar]
- Wolfson RL, Chantranupong L, Saxton RA, Shen K, Scaria SM, Cantor JR, and Sabatini DM (2016). Sestrin2 is a leucine sensor for the mTORC1 pathway. Science 351, 43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F, Huang Z, Li H, Yu J, Wang C, Chen S, Meng Q, Cheng Y, Gao X, Li J, et al. (2011). Leucine Deprivation Increases Hepatic Insulin Sensitivity via GCN2/mTOR/S6K1 and AMPK Pathways. Diabetes 60, 746–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F, Yu J, Guo Y, Deng J, Li K, Du Y, Chen S, Zhu J, Sheng H, and Guo F (2014). Effects of individual branched-chain amino acids deprivation on insulin sensitivity and glucose metabolism in mice. Metabolism 63, 841–850. [DOI] [PubMed] [Google Scholar]
- Xiao N, Cao DS, Zhu MF, and Xu QS (2015). protr/ProtrWeb: R package and web server for generating various numerical representation schemes of protein sequences. Bioinformatics 31, 1857–1859. [DOI] [PubMed] [Google Scholar]
- Xu J, Lloyd DJ, Hale C, Stanislaus S, Chen M, Sivits G, Vonderfecht S, Hecht R, Li YS, Lindberg RA, et al. (2009). Fibroblast growth factor 21 reverses hepatic steatosis, increases energy expenditure, and improves insulin sensitivity in diet-induced obese mice. Diabetes 58, 250–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Yang SE, Miller BR, Wisinski JA, Sherman DS, Brinkman JA, Tomasiewicz JL, Cummings NE, Kimple ME, Cryns VL, et al. (2018). Short-term methionine deprivation improves metabolic health via sexually dimorphic, mTORC1-independent mechanisms. FASEB J 32, 3471–3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, McGrath BC, Reinert J, Olsen DS, Lei L, Gill S, Wek SA, Vattem KM, Wek RC, Kimball SR, et al. (2002). The GCN2 eIF2alpha kinase is required for adaptation to amino acid deprivation in mice. Mol Cell Biol 22, 6681–6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Ceglarek U, Huang T, Li L, Rood J, Ryan DH, Bray GA, Sacks FM, Schwarzfuchs D, Thiery J, et al. (2016). Weight-loss diets and 2-y changes in circulating amino acids in 2 randomized intervention trials. Am J Clin Nutr 103, 505–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Shao J, Wu CY, Shu L, Dong W, Liu Y, Chen M, Wynn RM, Wang J, Wang J, et al. (2019). Targeting BCAA Catabolism to Treat Obesity-Associated Insulin Resistance. Diabetes 68, 1730–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhou Z, Peng J, and Loor JJ (2018). Methionine and valine activate the mammalian target of rapamycin complex 1 pathway through heterodimeric amino acid taste receptor (TAS1R1/TAS1R3) and intracellular Ca(2+) in bovine mammary epithelial cells. J Dairy Sci 101, 11354–11363. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 3, related to Figure 4. Transcriptional profiling and metabolomics data. Genes, fold change of Control vs. Low Ile, p-value, and FDR adjusted p-value of data from RNA sequencing of fasted and fed livers; and Metabolites, fold change of Control vs. Low Ile, p-value, and FDR adjusted p-value of data from metabolite analysis of fasted and fed livers.
Data Availability Statement
The gene expression data generated during this study can be obtained from Gene Expression Omnibus (GEO) (GEO: GSE168588).