Abstract
Background:
HIV proviral sequencing overcomes the limit of plasma viral load requirement by detecting all the “archived mutations”, but the clinical relevance remains to be evaluated.
Methods:
We included 25 participants with available proviral sequences (both intact and defective sequences available) and utilized the genotypic sensitivity score (GSS) to evaluate the level of resistance in their provirus and plasma virus. Defective sequences were further categorized as sequences with and without hypermutations. Personalized GSS score (pGSS) and total GSS score (tGSS) were calculated to evaluate the level of resistance to a whole panel of ARTs and to certain ART that a participant was using. The rate of sequences with drug resistance mutations (DRMs) within each sequence compartment (intact, defective and plasma viral sequences) was calculated for each participant.
Results:
Defective proviral sequences harbored more DRMs than other sequence compartments, with a median DRM rate of 0.25 compared to intact sequences (0.0, P=0.014) and plasma sequences (0.095, P=0.30). Defective sequences with hypermutations were the major source of DRMs, with a median DRM rate of 1.0 compared to defective sequences without hypermutations (0.042, P<0.001). Certain Apolipoprotein B Editing Complex 3 (APOBEC3)-related DRMs including reverse transcriptase gene mutations M184I, E138K, M230I, G190E and protease gene mutations M46I, D30N were enriched in hypermutated sequences but not in intact sequences or plasma sequences. All the hypermutated sequences had premature stop codons due to APOBEC3.
Conclusions:
Proviral sequencing may overestimate DRMs as a result of hypermutations. Removing hypermutated sequences is essential in the interpretation of proviral drug resistance testing.
Keywords: drug resistance mutations, HIV, provirus, hypermutation, antiretroviral therapy
Summary:
HIV proviral sequencing may overcall drug resistance mutations due to hypermutation.
Introduction
Human immunodeficiency virus (HIV) genotypic resistance testing has been recommended for antiretroviral therapy (ART) initiation, failure and modification [1], as transmitted or acquired drug resistance mutations (DRMs) represent important risk factors for virological failure [2–4]. Most commercially available tests rely on plasma HIV RNA sequencing utilizing Sanger sequencing methods to determine DRMs in reverse transcriptase (RT), protease (PR) and integrase (IN) genes. However, this method requires the plasma RNA copy number to be at least 500–1000 copies/ml and can only detect the major viral quasispecies in peripheral blood. A large body of evidence has shown that low-frequency minority quasispecies contribute to virological failure, but current RNA genotype tests are not capable of detecting them [5]. Due to these limitations of HIV RNA-based tests, HIV DNA-based test has been gaining popularity in recent years [6]. The DNA-based tests are able to identify the archived DRMs that are otherwise not readily detected in plasma sample due to low viral load; even additional DRMs that have never appeared in plasma can be detected in some cases [6, 7]. However, the clinical significance of those newly identified DRMs from DNA-based tests remain unclear, especially as the majority of proviral DNA are defective [1, 8]. In this current study, we aim to investigate the distribution of DRMs in different compartments of HIV proviral DNA and the clinical significance of those archived DRMs.
Methods
Participants
Study participants came from previously described multiple AIDS Clinical Trials Group (ACTG) studies that included a treatment interruption component (A371, A5068, A5197, A5170, and A5024) [9] and most of them were recruited in between 2000 and 2010. Only participants with pre-treatment interruption intact and defective proviral sequences, and post-treatment interruption plasma sequences available were included in this current study.
Sequence analyses
Proviral sequences from each participant were obtained using next-generation single-genome sequencing (NG-SGS) as described in our prior publication [10]. Briefly DNA was extracted from peripheral blood mononuclear cells (PBMCs) and limiting dilution proviral application was performed followed by nested PCR amplification. PCR amplicons were then subjected to Illumina sequencing system and sequence was assembled using the UltraCycler v1.0 system. Assembled DNA sequences were then subjected to an automated system to determine proviral DNA intactness [11]. To determine whether a sequence contains hypermutations, we used both Hypermut 2.0 and the original Hypermut program for hypermutations [12] from Los Alamos HIV sequence database. In the current study, we aligned all proviral and plasma sequences to HXB2 and extracted pol sequences for hypermutation and DRMs analysis.
Genotypic sensitivity score calculation
We used genotypic sensitivity score (GSS) to evaluate the level of HIV drug resistance, as published in prior studies [13, 14]. GSS was derived from Stanford HIV Database [15]. Resistance was graded to five levels: susceptible (score 1), potential low-level resistance (score 0.75), low-level resistance (score 0.5), intermediate (score 0.25) and high-level resistance (score 0) [13]. For each participant, personal GSS (pGSS) was calculated by adding the GSS score of each ART they used. Maximal pGSS in this study is three. For some participants, four ARTs instead of three were used. In this case, the smaller GSS from the two of the same-class ARTs was used. For example, for the regimen stavudine+ didanosine+ indinavir+ nelfinavir, we compared the GSSs for indinavir and nelfinavir and selected the smaller score to represent GSS for protease inhibitor class in this regimen. We also calculated total GSS (tGSS), including GSS for a panel of 15 ARTs (Supplementary Materials). Maximal tGSS in this study was 15. We did not include GSS for integrase strand transfer inhibitors (INSTI) since none of the participants received INSTIs.
Statistical analyses
We used non-parametric rank-based analysis with median and interquartile ranges (IQR) to compare pGSS, tGSS and DRM rates. Chi-square test or Fisher’s exact test was used to compare categorical variables and Dunn’s test, a non-parametric test, was used to account for multiple comparisons between different groups. P<0.05 was deemed statistically significant. STATA 13.1 was used for statistical analyses (StataCorp, College Station, Texas).
Results
Baseline characteristics
In this study, we included 25 participants and baseline characteristics were summarized in Table 1. The majority of participants were male and aged 40–50 years. Median CD4 cell count at time of ATI was 886 (IQR 784, 980) cells/mm3. Each participant had an average of 9 intact proviral sequences, 22 total defective sequences and 41 plasma sequences. All participants were infected with subtype B virus.
Table 1.
Baseline Characteristics (n=25).
| Age, median years (IQR) | 41 (38, 47) |
| Male sex, n (%) | 20 (80.0) |
| PTC, n (%) | 10 (40.0) |
| ART regimen, n | |
| ABC+ d4T+ 3TC+ APV/r | 8 |
| ABC+ 3TC+ NVP | 1 |
| ABC+ AZT+ EFV | 1 |
| AZT+ 3TC+ PI/r (NFV or ATV) | 3 |
| AZT+ 3TC+ NNRTI (EFV or NVP) | 2 |
| d4T+ 3TC+ NNRTI (EFV or NVP) | 3 |
| d4T+ 3TC+ PI/r (NFV or IDV) | 3 |
| d4T+ TDF+ EFV | 1 |
| d4T+ ddI+ EFV | 1 |
| d4T+ ddI+ IDV+ NFV | 1 |
| 3TC+ EFV+ IDV | 1 |
IQR, interquartile range; PTC, post-treatment controller; ART, antiretroviral therapy; ABC, abacavir; d4T, stavudine; 3TC, lamivudine; NNRTI, non-nucleos(t)ide reverse transcriptase inhibitor; APV, amprenavir; r, boosted by ritonavir; NVP, nevirapine; AZT, zidovudine; EFV, efavirenz; NFV, nelfinavir; ATV, atazanavir; IDV, indinavir; TDF, tenofovir disoproxil fumarate; ddI, didanosine.
Defective proviral sequences harbor most of DRMs
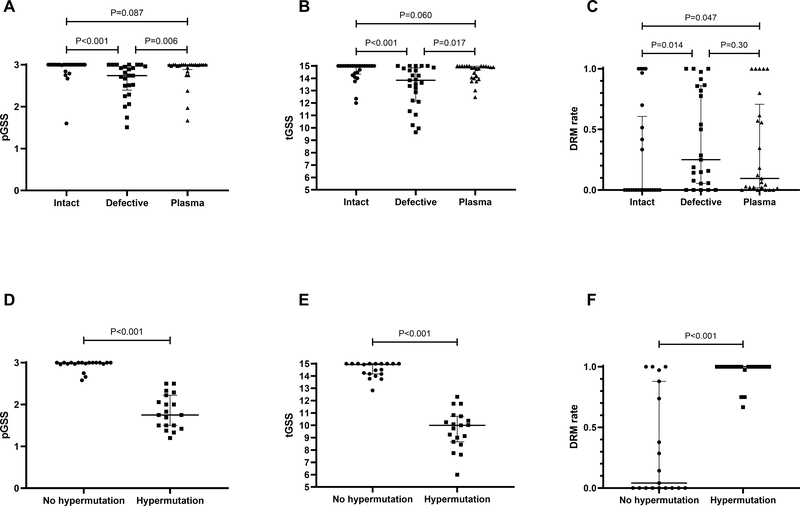
We further evaluated DRM distributions in different compartments of proviral DNA and plasma sequences. As shown in Figure 1A, the median pGSS of intact sequences was 3.00 (IQR 3.00–3.00), representing a very favorable susceptibility profile. Similarly, the median pGSS of plasma sequences was also 3.00 (IQR 2.93–3.00, P=0.09 compared to pGSS from intact sequences). In contrast, the pGSS of defective proviral sequences were significantly lower than those of either intact proviruses (median intact vs defective provirus pGSS: 3.00 vs 2.74, P<0.001) or plasma sequences (median plasma vs defective provirus pGSS: 3.00 vs 2.74, P=0.006).
Figure 1.
HIV sensitivity scores among different viral sequence compartment. (A-C) Personalized GSS, total GSS and DRM rates among intact proviral sequences, defective proviral sequences and plasma sequences. (D-E) Personalized GSS, total GSS and DRM rates between defective sequences with and without hypermutations.
The total GSS (tGSS) and the rate of DRMs followed the same patterns of distribution among intact sequences, defective sequences, and plasma sequences. Compared to intact proviral sequences, defective proviral sequences had significantly lower tGSS scores (15.00 vs 13.84, P<0.001, Figure 1B) and plasma sequences had modestly lower tGSS scores (15.00 vs 14.85, P=0.060). Similarly, the median rate of DRMs were 0, 0.25 (P=0.014 compared to intact sequences) and 0.095 (P=0.30compared to intact sequences) respectively in intact proviral, defective proviral and plasma sequences (Figure 1C). This result remained the same when stratified by NC and PTC status (data not shown).
We further stratified plasma viral sequences based on time points (pre-ART and post-ATI) and did not demonstrate significant differences in pGSS, tGSS and DRM rates (Supplementary Figure S1).
Defective sequences with hypermutations are the major contributor of DRMs in proviral sequences
Given prior evidence that APOBEC3 may generate DRMs even without exposure to ART [16], we evaluated the impact of hypermutation on DRMs in different sequence compartments. In 19 participants who had defective sequences with and without hypermutation, non-hypermutation defective sequences are associated with higher pGSS (P<0.001, Figure 1D), higher tGSS (P<0.001, Figure 1E) and lower DRM rates (P<0.001, Figure 1F).
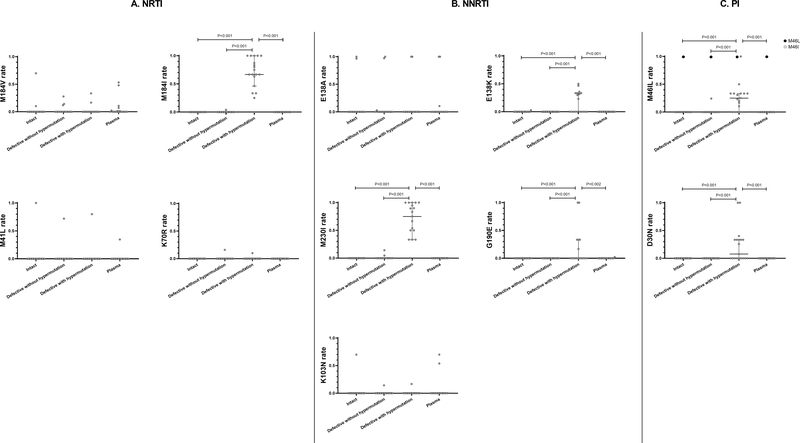
We next evaluated major DRMs distribution in different proviral compartments. For NRTI-related DRMs (Figure 2A), the most prevalent DRM was M184V/I in this group while thymidine analog resistance mutations (TAMs) were rare. M184V was evenly distributed in intact, defective without hypermutation, defective with hypermutation and plasma sequences (median rates 0.0 in four compartments). In comparison, M184I was enriched in defective sequences with hypermutations (median rate 0.67) while other compartments had median rates of 0.0 (P<0.001 with Dunn’s test). This is explained by APOBEC3-related G-to-A mutation leading to codon changes (ATG [methionine] to ATA [isoleucine]) at amino acid position 184. TAM-1 and TAM-2 were rare in this cohort and did not differ among different proviral and plasma compartments (Figure 2A). NNRTI-related DRMs were more prevalent in this cohort. As seen in Figure 2B, M230I and G190E were enriched in proviral sequences with hypermutation. Of note, one participant had G190E mutations both in the defective proviral and plasma sequences but not in intact HIV proviral sequences. However, those plasma sequences did not contain hypermutations based on the Hypermut program analysis. E138K, a DRM that can be selected by RPV as well as caused by APOBEC3, was preferably enriched in sequences with hypermutation (P<0.01 compared to all other sequence types, Figure 2B); in comparison, E138A was evenly distributed in four sequence compartments. K103N, a common NNRTI-associated DRM selected by NVP and EFV, was not common in this cohort and was evenly distributed in different compartments.
Figure 2.
Distribution of certain DRMs among intact sequences, defective sequences without hypermutations, defective sequences with hypermutations, and plasma sequences. (A) NRTI-related DRMs. (B) NNRTI-related DRMs. (C) PI-related DRMs. Only P value <0.05 was shown in this figure.
PI-related DRMs were less prevalent in this cohort. D30N and M46I were enriched in hypermutated sequences, while M46L was distributed evenly (Figure 2C). Of note, one participant had one plasma sequence containing M46I, but this plasma sequence did not contain hypermutations; the hypermutated sequences from this participant did not contain M46I mutations.
Hypermutated sequences with DRMs contain premature stop codons due to APOBEC3
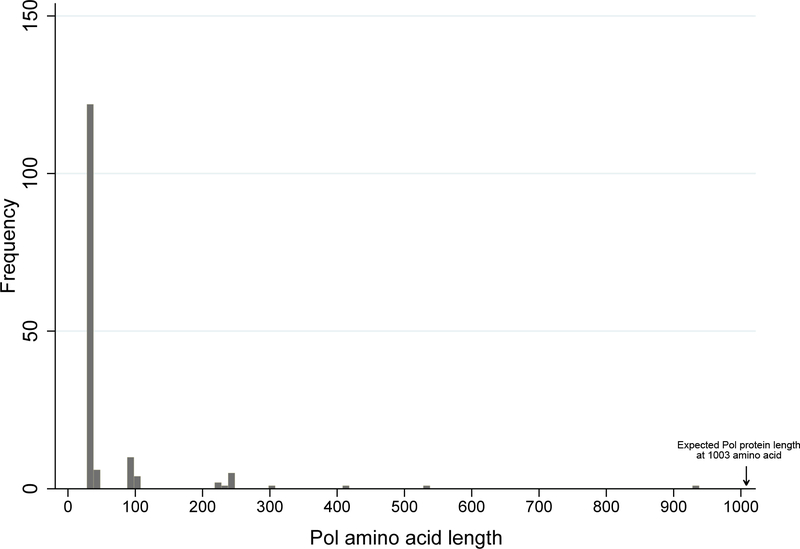
Among 163 defective pol gene sequences with hypermutations, only 4 (2.5%) could produce full-length Pol protein (1003 amino acid, Figure 3). None of those 4 sequences contained APOBEC3-derived DRMs. The majority of premature stop codon was derived from Tryptophan (TGG) to stop codon (TAA/TAG/TGA) mutations and 120 sequences (73.6%) had stop codons at pol amino acid position 34 due to W34Stop mutation (Figure 3).
Figure 3.
Distribution of the projected Pol protein length (number of amino acid) derived from hypermutated proviral sequences until the first stop codon.
Discussion
In this study, we demonstrate that defective HIV proviral sequences harbor higher rates of DRMs, specifically in hypermutated proviral sequences. Certain DRMs related to APOBEC3 are preferentially enriched in sequences with hypermutations. These results suggest that proviral HIV DNA genotyping results may overestimate the frequency of certain DRMs, especially when hypermutated sequences are not removed. There has been debates regarding whether DRMs from hypermutated proviral sequences contribute to clinically significant drug resistance. Hypermutated sequences are usually replication-defective due to alteration of start codons and premature stop codons [8]. In vitro studies demonstrate that recombination can rescue those lethal and sublethal mutations caused by APOBEC3 [16, 17]. However, in patient-derived proviral and plasma sequences, hypermutated proviruses do not produce viable HIV virions [18]. Even in those having DRMs in hypermutated proviral sequences, their plasma sequences do not harbor DRMs [19]. In a study by Delviks-Frankenberry and colleagues, they demonstrate that recombination between hypermutated proviral sequences and intact sequences only contributes to 3.9 × 10−5 mutations/base pair/replication cycle, comparable to HIV mutation rates generated by error-prone reverse transcription; in addition, the chance of co-packaging of a hypermutated sequence and an intact sequence into one single virion is low [20]. These studies are consistent with our findings that even if certain DRMs (e.g. M184I, E183K, M230I, G190E for reverse transcriptase and D30N, M46I for protease) are enriched in hypermutated sequences, they are highly unlikely to contribute to clinical DRMs in plasma virus.
This study has the following clinical implications. Clinicians should use caution when interpreting HIV proviral DNA sequencing data, as those DRMs from hypermutation are highly unlikely to contribute to clinical drug resistance. For clinical and research laboratories performing HIV proviral sequencing, the results highlight the importance of bioinformatically removing hypermutated sequences from the analysis. Some proviral DNA sequencing platform, e.g. GenoSure Archive ™, excluded hypermutated sequences, which helps reduce false positive rates [21].
There are several limitations in our study. We have a small sample size and participants in this study were enrolled in early 2000s. None of the participants were receiving newer NRTIs, including tenofovir disoproxil fumarate or tenofovir alafenamide, new protease inhibitors (e.g. darunavir) or INSTIs, which limits our ability to evaluate DRMs against modern ARTs. In addition, the utility of proviral sequences/archived mutations may be even less useful in the settings of newer ARTs with higher genetic barriers (especially new protease inhibitors and INSTIs including dolutegraivr and bictegravir). All participants were infected with subtype B virus and thus we were unable to evaluate the association of HIV subtypes and DRMs distribution [22]. Each participant only had plasma viral sequences available at one or two time points when they were viremic. This limited our ability to fully rule out the possibility that DRMs from hypermutations can contribute to mature, infectious virions [23]. Given limited sample size and DRMs in this cohort, we were unable to evaluate other DRMs with regard to non-hypermutation sequences. In addition, we are only able to obtain CD4+ T cell from peripheral blood, while the majority of HIV provirus resides in difficult-to-reach anatomical sites including gut-associated lymphoid tissue [24]. Finally, we were unable to evaluate the selective pressure of autologous anti-HIV immunoglobulin G on rebound virus post-ATI, which may skew the DRM rate and GSS in rebound virus [25].
In summary, our study demonstrated that certain DRMs are preferentially enriched in hypermutated sequences and are unlikely to contribute to clinical drug resistance to HIV. Further studies with larger sample size, and multiple time points for plasma virus sampling would be warranted to validate our findings.
Supplementary Material
Acknowledgments
Funding
This work was supported by National Institutes of Health/National Institute of Allergy and Infectious Diseases grants (AI150396 to JZL), and the Harvard University Center for AIDS Research (5P30AI060354-08 to JZL).
Footnotes
Financial Disclosure
JZL has consulted for Abbvie and Jan Biotech.
This study results were in part presented at Conference on Retroviruses and Opportunistic Infections (CROI) 2021.
References
- 1.Panel on Antiretroviral Guidelines for Adults and Adolescents. Guidelines for the Use of Antiretroviral Agents in Adults and Adolescents with HIV. Department of Health and Human Services. In; 2020.
- 2.Lodi S, Gunthard HF, Gill J, Phillips AN, Dunn D, Vu Q, et al. Effectiveness of Transmitted Drug Resistance Testing Before Initiation of Antiretroviral Therapy in HIV-Positive Individuals. J Acquir Immune Defic Syndr 2019; 82(3):314–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gunthard HF, Calvez V, Paredes R, Pillay D, Shafer RW, Wensing AM, et al. Human Immunodeficiency Virus Drug Resistance: 2018 Recommendations of the International Antiviral Society-USA Panel. Clin Infect Dis 2019; 68(2):177–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Y, Gu L, Han Y, Xie J, Wang H, Lv W, et al. HIV-1 subtype B/B’ and baseline drug resistance mutation are associated with virologic failure: a multicenter cohort study in China. J Acquir Immune Defic Syndr 2015; 68(3):289–297. [DOI] [PubMed] [Google Scholar]
- 5.Li JZ, Paredes R, Ribaudo HJ, Svarovskaia ES, Metzner KJ, Kozal MJ, et al. Low-frequency HIV-1 drug resistance mutations and risk of NNRTI-based antiretroviral treatment failure: a systematic review and pooled analysis. JAMA 2011; 305(13):1327–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh H, Jones S, Vaamonde C, Wilkin T. Application of GenoSure Archive in Clinical Practice. Open Forum Infectious Diseases 2016; 3(Suppl_1). [Google Scholar]
- 7.Sotillo A, Sierra O, Martínez-Prats L, Gutiérrez F, Zurita S, Pulido F, et al. Analysis of drug resistance mutations in whole blood DNA from HIV-1 infected patients by single genome and ultradeep sequencing analysis. J Virol Methods 2018; 260:1–5. [DOI] [PubMed] [Google Scholar]
- 8.Ho YC, Shan L, Hosmane NN, Wang J, Laskey SB, Rosenbloom DI, et al. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 2013; 155(3):540–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Namazi G, Fajnzylber JM, Aga E, Bosch RJ, Acosta EP, Sharaf R, et al. The Control of HIV After Antiretroviral Medication Pause (CHAMP) Study: Posttreatment Controllers Identified From 14 Clinical Studies. J Infect Dis 2018; 218(12):1954–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharaf R, Lee GQ, Sun X, Etemad B, Aboukhater LM, Hu Z, et al. HIV-1 proviral landscapes distinguish posttreatment controllers from noncontrollers. J Clin Invest 2018; 128(9):4074–4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee GQ, Orlova-Fink N, Einkauf K, Chowdhury FZ, Sun X, Harrington S, et al. Clonal expansion of genome-intact HIV-1 in functionally polarized Th1 CD4+ T cells. The Journal of clinical investigation 2017; 127(7):2689–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rose PP, Korber BT. Detecting hypermutations in viral sequences with an emphasis on G --> A hypermutation. Bioinformatics 2000; 16(4):400–401. [DOI] [PubMed] [Google Scholar]
- 13.Boyd MA, Moore CL, Molina J-M, Wood R, Madero JS, Wolff M, et al. Baseline HIV-1 resistance, virological outcomes, and emergent resistance in the SECOND-LINE trial: an exploratory analysis. The Lancet HIV 2015; 2(2):e42–e51. [DOI] [PubMed] [Google Scholar]
- 14.Pironti A, Pfeifer N, Kaiser R, Walter H, Lengauer T. Improved therapy-success prediction with GSS estimated from clinical HIV-1 sequences. J Int AIDS Soc 2014; 17(4 Suppl 3):19743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu TF, Shafer RW. Web resources for HIV type 1 genotypic-resistance test interpretation. Clin Infect Dis 2006; 42(11):1608–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fourati S, Malet I, Lambert S, Soulie C, Wirden M, Flandre P, et al. E138K and M184I mutations in HIV-1 reverse transcriptase coemerge as a result of APOBEC3 editing in the absence of drug exposure. AIDS 2012; 26(13):1619–1624. [DOI] [PubMed] [Google Scholar]
- 17.Mulder LC, Harari A, Simon V. Cytidine deamination induced HIV-1 drug resistance. Proc Natl Acad Sci U S A 2008; 105(14):5501–5506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kieffer TL, Kwon P, Nettles RE, Han Y, Ray SC, Siliciano RF. G-->A hypermutation in protease and reverse transcriptase regions of human immunodeficiency virus type 1 residing in resting CD4+ T cells in vivo. J Virol 2005; 79(3):1975–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neogi U, Shet A, Sahoo PN, Bontell I, Ekstrand ML, Banerjea AC, et al. Human APOBEC3G-mediated hypermutation is associated with antiretroviral therapy failure in HIV-1 subtype C-infected individuals. J Int AIDS Soc 2013; 16:18472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delviks-Frankenberry KA, Nikolaitchik OA, Burdick RC, Gorelick RJ, Keele BF, Hu WS, et al. Minimal Contribution of APOBEC3-Induced G-to-A Hypermutation to HIV-1 Recombination and Genetic Variation. PLoS Pathog 2016; 12(5):e1005646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andreatta K, Willkom M, Martin R, Chang S, Wei L, Liu H, et al. Switching to bictegravir/emtricitabine/tenofovir alafenamide maintained HIV-1 RNA suppression in participants with archived antiretroviral resistance including M184V/I. J Antimicrob Chemother 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smit E, White E, Clark D, Churchill D, Zhang H, Collins S, et al. An association between K65R and HIV-1 subtype C viruses in patients treated with multiple NRTIs. J Antimicrob Chemother 2017; 72(7):2075–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hernandez MM, Fahrny A, Jayaprakash A, Gers-Huber G, Dillon-White M, Audigé A, et al. Impact of Suboptimal APOBEC3G Neutralization on the Emergence of HIV Drug Resistance in Humanized Mice. J Virol 2020; 94(5):e01543–01519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Estes JD, Kityo C, Ssali F, Swainson L, Makamdop KN, Del Prete GQ, et al. Defining total-body AIDS-virus burden with implications for curative strategies. Nat Med 2017; 23(11):1271–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bertagnolli LN, Varriale J, Sweet S, Brockhurst J, Simonetti FR, White J, et al. Autologous IgG antibodies block outgrowth of a substantial but variable fraction of viruses in the latent reservoir for HIV-1. Proc Natl Acad Sci U S A 2020; 117(50):32066–32077. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.