Abstract
Deep learning is becoming increasingly popular and available to new users, particularly in the medical field. Deep learning image segmentation, outcome analysis, and generators rely on presentation of Digital Imaging and Communications in Medicine (DICOM) images, and often radiation therapy (RT) structures as masks. While the technology to convert DICOM images and RT-Structures into other data types exists, no purpose-built Python module for converting NumPyarrays into RT-Structures exists. The two most popular deep learning libraries, Tensorflow and PyTorch, are both implemented within Python, and we believe a set of tools built in Python for manipulating DICOM images and RT-Structures would be useful and could save medical researchers large amounts of time and effort during the pre-processing and prediction steps. Our module provides intuitive methods for rapid data curation of RT-Structure files by identifying unique region of interest (ROI) names, ROI structure locations, and allowing multiple ROI names to represent the same structure. It is also capable of converting DICOM images and RT-Structures into NumPy arrays and SimpleITK Images, the most commonly used formats for image analysis and inputs into deep learning architectures, and radiomic feature calculations. Furthermore, the tool provides a simple method for creating a DICOM RTStructure from predicted NumPy arrays, which are commonly the output of semantic segmentation deep learning models. Accessing DicomRTTool via the public Github project invites open collaboration, while the deployment of our module in PyPi ensures painless distribution and installation. We believe our tool will be increasingly useful as deep learning in medicine progresses.
Introduction
Deep learning has become increasingly popular in the medical community, particularly for semantic segmentation1–13. As medical professionals begin to explore the creation of deep learning architectures, the number of people who need to process medical images to create both input for deep learning networks and radiation therapy (RT) structures from predictions will continue to grow. The Python programming language14 has become the most widely used programming language worldwide (http://pypl.github.io/PYPL.html), and is the base for the two most popular deep learning libraries, Tensorflow15 and PyTorch16. While technology (e.g., PlastiMatch17) to convert Digital Imaging and Communications in Medicine (DICOM) images and RT-Structures into other common data types (.nii, .nrrd, etc.) exists, there is currently no single purpose-built python module for converting prediction arrays back into DICOM RT-Structures. We believe the distribution of simple DICOM tools, created in Python, could save medical researchers large amounts of time during the pre-processing and prediction steps of deep learning. Here we describe our program, DicomRTTool, which is designed to alleviate several of the most time-consuming aspects of data preparation by quickly identifying unique region of interest (ROI) names, where image sets with all ROIs are located, and converting DICOM images and RT-Structures into NumPy arrays18 and SimpleITK images19. It also allows for the conversion of prediction NumPy arrays to DICOM RT-Structures.
Materials and Methods
To demonstrate our program’s capabilities we have created a Juypter notebook, Supplemental Material, which walks through the capabilities of our program: 1) Getting Data, 2) Reading Dicom and RT-Structure, 3) Saving as nifty, 4) Saving/loading NumPy, 5) Calculating radiomics, and 6) Predictions to RT-Structures, on a publically available brain tumor data20.
Installation
The DicomRTTool software is available through both Github (https://github.com/brianmanderson/Dicom_RT_and_Images_to_Mask) and PyPi. The list of module requirements is present in the requirements.txt file and are automatically installed when using pip.
pip install DicomRTTool
This module is compatible with Python3.x versions and above, on Windows and Linux machines.
Data preparation
Leveraging SimpleITK’s ability to rapidly identify series instance unique identifiers (UIDs), our model is able to separate and group DICOM images and RT-Structure files based on UIDs and referenced series instance UIDs, respectively, even when they are not present within the same folder. The module leverages threading and multi-processing, reducing time bottlenecks even when walking through multiple nested structures and patients.
Loading DICOM Images and RT-Structures
The DicomReaderWriter is built as a Python Object class, allowing the module to be initialized once and used multiple times. To specify the behavior of the DicomReaderWriter, several arguments can be passed. We maintain an updated description of arguments on the GitHub Wiki page (https://github.com/brianmanderson/Dicom_RT_and_Images_to_Mask/wiki) and recommend readers to inspect the latest version.
When RT-Structures are found, the list of ROIs present within the structure are automatically added to the module by reading the StructureSetROISequence Dicom tag. This allows for the user to investigate ROI structures without tediously reloading structures. All ROI names are automatically put into lowercase. Following TG-263 guidelines, capitalization should not be used to distinguish ROIs. Please note that this is only for loading masks and will not affect the ROI names present in the RT-Structures in any way.
Upon completion, the module will have a list of indexes, each associated with a unique UID for the image and RT-Structures. By default the module starts with index=0, but the user can set a desired index at any time using .set_index(index).
Conversion of DICOM to SimpleITK images and NumPy
The user can load images for a desired index easily using .get_images (). Images are generated via SimpleITK’s ImageSeriesReader().
The SimpleITKImageHandle can easily be saved as a .nii or .nii.gz file using SimpleITK’s built-in WriteImage(image=image, fileName=filename.nii). The SimpleITKImageHandle is beneficial in that it maintains spacing information, direction, orientation, and origin. If NumPy arrays are desired, the user can transform an Image to a NumPy array through simpleITK.GetArrayFromImage(image_handle).
Identification of ROIs present within folders
The DicomReaderWriter can easily identify all the unique ROI names present within a set of data. This function can be beneficial for quickly identifying variant ROI names and building an associations file.
Locating RT-Structures by ROIs
If the user would like to identify where an individual ROI name is located, the DicomReaderWriter compiles a list of RT-Structures which contain a specific ROI name, using DicomReaderWriter.where_is_ROI(RoiName).
Conversion of DICOM RT-Structures to NumPy mask and SimpleITK images from RT-Structure
To create a binary mask of the structure, potentially for deep learning training, the user will need to specify a list of desired contour names. The module allows for variations of names with an associations dictionary, this allows the user to specify that multiple names (i.e ‘liver_bma’, ‘liver_final’) can all correspond to the same mask.
Conversion of prediction to DICOM-RT
NumPy prediction arrays from deep learning algorithms can easily be converted back into DICOM RT-Structures. The module generates a new RT-Structure file based on a template file if an RT-Structure is not already present. Contours are generated from the binary mask using a marching-squares method within Scikit-image21.
Generated contours for test cases including: creation of a circle, rectangle, and ‘target’ were reviewed in four systems: RayStation22, vv-Slicer23, Velocity24, and 3DSlicer25, Figure 1. Raystation, vv-Slicer, and Velocity were able to display all generated test case contours without any issues.
Figure 1:
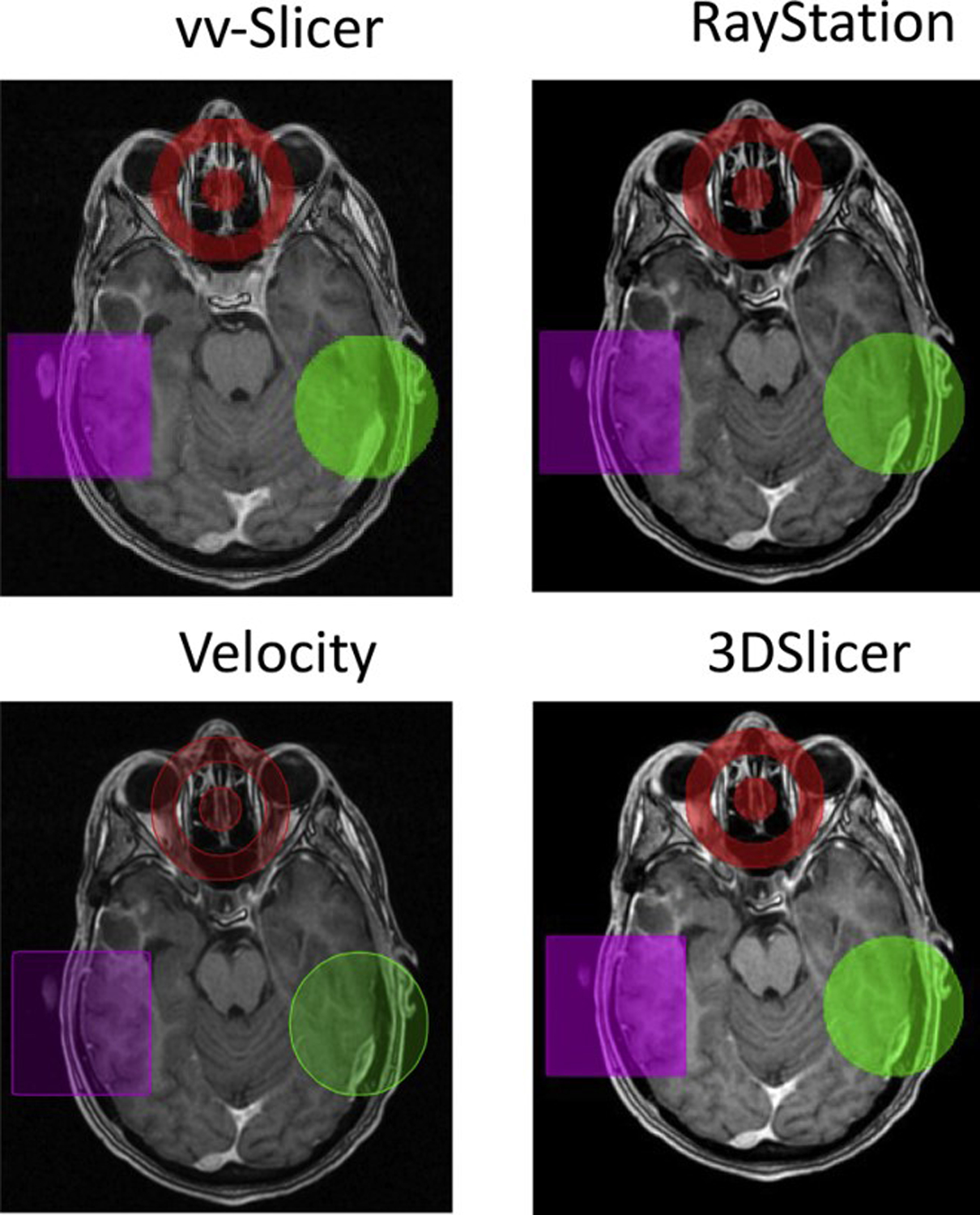
Examples of generated ‘circle’, ‘rectangle’, and ‘target’ in vv-slicer, Raystation, Velocity, and 3DSlicer.
Discussion and Conclusion
Python is becoming increasingly popular and widely adopted by new users, and serves as the base for two of the most used deep learning libraries16,26. Our module offers a simple way of curating and converting patient DICOM and RT-Structure data into NumPy arrays and SimpleITK images, with a range of parameters to benefit each use case. This is particularly important within Radiation Oncology, which largely relies on DICOM RT-structures. Likewise, conversion of predictions back to DICOM RT-Structures is necessary for any deep learning segmentation task. Moreover, while we have constructed our module with deep learning explicitly in mind as a use-case, our module may also be adapted for other quantitative imaging applications, such as radiomic feature extraction27,28.
Being available on GitHub invites open collaboration, and deployment via PyPi ensures easy distribution. We believe this work will provide a useful tool for all medical researchers, novice or expert, involved with deep learning.
Supplementary Material
Acknowledgements:
The authors would like to acknowledge the scientific editors in the Research Medical Library at The University of Texas MD Anderson Cancer Center and funding from the Society of Interventional Radiology Foundation Allied Scientist Grant, the NIH (NCI R01CA235564), and the Helen Black Image Guided Fund. K.A. Wahid is supported by a training fellowship from The University of Texas Health Science Center at Houston Center for Clinical and Translational Sciences (TL1TR003169).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Research data are stored in an institutional repository and will be shared upon request to the corresponding author. Statistical analysis for this work was performed by Brian M Anderson.
Conflicts of Interest, Brian Anderson: None
Conflicts of Interest, Kareem Wahid: None
Conflicts of Interest, Kristy Brock: Dr. Brock reports grants from National Institutes of Health R01CA235564, during the conduct of the study; grants from RaySearch Laboratories, outside the submitted work.
References
- 1.Akkus Z, Galimzianova A, Hoogi A, Rubin DL, Erickson BJ. Deep Learning for Brain MRI Segmentation: State of the Art and Future Directions. J Digit Imaging. 2017;30(4):449–459. 10.1007/s10278-017-9983-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.To MNN, Vu DQ, Turkbey B, Choyke PL, Kwak JT. Deep dense multi-path neural network for prostate segmentation in magnetic resonance imaging. Int J Comput Assist Radiol Surg. 2018;13(11):1687–1696. 10.1007/s11548-018-1841-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hai J, Qiao K, Chen J, et al. Fully convolutional densenet with multiscale context for automated breast tumor segmentation. J Healthc Eng. 2019;2019. 10.1155/2019/8415485 [DOI] [PMC free article] [PubMed]
- 4.Roth HR, Shen C, Oda H, et al. A Multi-Scale Pyramid of 3D Fully Convolutional Networks for Abdominal Multi-Organ Segmentation. Accessed October 4, 2018. https://arxiv.org/pdf/1806.02237.pdf
- 5.Rigaud B, Anderson BM, Yu ZH, et al. Automatic Segmentation Using Deep Learning to Enable Online Dose Optimization During Adaptive Radiation Therapy of Cervical Cancer. Int J Radiat Oncol Biol Phys. 2021;109(4):1096–1110. 10.1016/j.ijrobp.2020.10.038 [DOI] [PubMed] [Google Scholar]
- 6.Chen L-C, Papandreou G, Kokkinos I, Murphy K, Yuille AL. SEMANTIC IMAGE SEGMENTATION WITH DEEP CON-VOLUTIONAL NETS AND FULLY CONNECTED CRFS. Accessed April 11, 2018. https://arxiv.org/pdf/1412.7062.pdf [DOI] [PubMed]
- 7.Wang G, Zuluaga MA, Li W, et al. DeepIGeoS: A Deep Interactive Geodesic Framework for Medical Image Segmentation. IEEE Trans Pattern Anal Mach Intell. Published online 2018:1–14. 10.1109/TPAMI.2018.2840695 [DOI] [PMC free article] [PubMed]
- 8.Trebeschi S, van Griethuysen JJM, Lambregts DMJ, et al. Deep Learning for Fully-Automated Localization and Segmentation of Rectal Cancer on Multiparametric MR. Sci Rep. 2017;7(1):5301. 10.1038/s41598-017-05728-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang D, Xu D, Zhou SK, et al. Automatic Liver Segmentation Using an Adversarial Image-to-Image Network. Accessed June 1, 2018. https://arxiv.org/pdf/1707.08037.pdf
- 10.Liu Z, Li X, Luo P, Loy CC, Tang X. Deep Learning Markov Random Field for Semantic Segmentation. 2016;1:415–423. 10.1007/978-3-319-46723-8 [DOI] [PubMed] [Google Scholar]
- 11.Anderson BM, Lin EY, Cardenas CE, et al. Automated Contouring of Contrast and Noncontrast Computed Tomography Liver Images With Fully Convolutional Networks. Advancesradonc. Published online 2020. 10.1016/j.adro.2020.04.023 [DOI] [PMC free article] [PubMed]
- 12.Seo H, Huang C, Bassenne M, Xiao R, Xing L. Modified U-Net (mU-Net) with Incorporation of Object-Dependent High Level Features for Improved Liver and Liver-Tumor Segmentation in CT Images. IEEE Trans Med Imaging. 2020;39(5):1316–1325. 10.1109/TMI.2019.2948320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X, Chen H, Qi X, Dou Q, Fu C-W, Heng P-A. H-DenseUNet: Hybrid Densely Connected UNet for Liver and Tumor Segmentation from CT Volumes. IEEE Trans Med Imaging. Published online June 11, 2018:1–1. 10.1109/TMI.2018.2845918 [DOI] [PubMed]
- 14.Van Rossum G, Drake FL. Python 3 Reference Manual. CreateSpace; 2009. [Google Scholar]
- 15.Abadi M, Agarwal A, Barham P, et al. TensorFlow: Large-Scale Machine Learning on Heterogeneous Distributed Systems. Accessed July 28, 2020. www.tensorflow.org.
- 16.Paszke A, Gross S, Massa F, et al. PyTorch: An Imperative Style, High-Performance Deep Learning Library.; 2019.
- 17.Zaffino P, Raudaschl P, Fritscher K, Sharp GC, Spadea MF. Technical Note: Plastimatch mabs, an open source tool for automatic image segmentation. Med Phys. 2016;43(9):5155–5160. 10.1118/1.4961121 [DOI] [PubMed] [Google Scholar]
- 18.Harris CR, Millman KJ, van der Walt SJ, et al. Array programming with NumPy. Nature. 2020;585(7825):357–362. 10.1038/s41586-020-2649-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beare R, Lowekamp B, Yaniv Z. Image segmentation, registration and characterization in R with simpleITK. J Stat Softw. 2018;86(1):1–35. 10.18637/jss.v086.i08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang Y, Lafata K, Sun W, et al. An investigation of machine learning methods in deltaradiomics feature analysis. PLoS One. 2019;14(12). 10.1371/journal.pone.0226348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Der Walt S, Schönberger JL, Nunez-Iglesias J, et al. Scikit-image: Image processing in python. PeerJ. 2014;2014(1). 10.7717/peerj.453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bodensteiner D RayStation: External beam treatment planning system. Med Dosim. 2018;43(2):168–176. 10.1016/j.meddos.2018.02.013 [DOI] [PubMed] [Google Scholar]
- 23.Seroul Pierre and Sarrut David.; 2008. Accessed January 11, 2021. http://www.creatis.insa-lyon.fr/rio/vv
- 24.Velocity | Varian. Accessed January 11, 2021. https://www.varian.com/products/interventional-solutions/velocity
- 25.3D Slicer. Accessed January 11, 2021. https://www.slicer.org/
- 26.Abadi M, Agarwal A, Barham P, et al. TensorFlow: Large-Scale Machine Learning on Heterogeneous Distributed Systems. Accessed December 10, 2017. https://static.googleusercontent.com/media/research.google.com/en//pubs/archive/45166.pdf
- 27.Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images are more than pictures, they are data. Radiology. 2016;278(2):563–577. 10.1148/radiol.2015151169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Griethuysen JJM, Fedorov A, Parmar C, et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77(21):e104–e107. 10.1158/0008-5472.CAN-17-0339 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


