Abstract
Purpose:
KRASG12C is the most common KRAS mutation in primary lung adenocarcinoma (LUAD). Phase I clinical trials have demonstrated encouraging clinical activity of KRASG12C inhibitors in the metastatic setting. We investigated disease-free survival (DFS) and tumor genomic features in patients with surgically resected KRASG12C-mutant LUAD.
Experimental Design:
Patients who underwent resection of stage I-III LUAD and next-generation sequencing (NGS) were evaluated. Exclusion criteria were receipt of induction therapy, incomplete resection, and low-quality NGS. Mutations were classified as KRAS wild-type (KRASwt), G12C (KRASG12C), or non-G12C (KRASother). DFS was compared between groups using the log-rank test; factors associated with DFS were assessed using Cox regression. Mutual exclusivity and co-occurrence, tumor clonality, and mutational signatures were assessed.
Results:
In total, 604 patients were included: 374 KRASwt (62%), 95 KRASG12C (16%), and 135 KRASother (22%). Three-year DFS was not different between KRAS-mutant and KRASwt tumors. However, 3-year DFS was worse in patients with KRASG12C than KRASother tumors (log-rank p=0.029). KRASG12C tumors had more lymphovascular invasion (51% vs. 37%; p=0.032) and higher tumor mutation burden (median [interquartile range], 7.0 [5.3–10.8] vs. 6.1 [3.5–9.7]; p=0.021), compared with KRASother tumors. KRASG12C mutation was independently associated with worse DFS on multivariable analysis. Our DFS findings were externally validated in an independent The Cancer Genome Atlas cohort.
Conclusions:
KRASG12C mutations are associated with worse DFS after complete resection of stage I-III LUAD. These tumors harbor more-aggressive clinicopathologic and genomic features than other KRAS-mutant tumors. We identify a high-risk group for whom KRASG12C inhibitors may be investigated to improve survival.
Keywords: KRAS, G12C, lung adenocarcinoma, surgery, disease-free survival
Introduction
The KRAS gene encodes an oncoprotein involved in key signaling pathways for tumor growth and differentiation and is the most frequently mutated oncogene in all cancer types (1). Somatic mutations in KRAS are found in 25% to 33% of primary lung adenocarcinoma (LUAD) (2,3). In stage IV LUAD, patients with KRAS-mutant primary tumors have demonstrated poor overall survival (4) and less clinical benefit from standard-of-care systemic therapies, compared with KRASwt tumors (5).
During the last few decades, the KRAS protein has been viewed as undruggable, owing to the lack of deep pockets for direct small-molecule-inhibitor binding. Consequently, efforts to develop targeted therapies for KRAS-mutant tumors have focused on inhibition of downstream effector proteins in the MAPK pathway, such as BRAF, MEK, and ERK. However, inhibition of these downstream targets is often accompanied by on-target, nontumor toxicities, due to the inhibition of this signaling pathway in normal cells (6,7). As a result, this narrow therapeutic index has precluded the successful clinical development of agents targeting KRAS (5,8).
The majority of KRAS mutations in LUAD are single-base substitutions (SBSs) in codons 12 or 13—the most common being G12C, which occurs in 13% to 16% of LUAD (3,9). The recent discovery of direct KRASG12C inhibition by use of a trapping mechanism has led to promising preclinical and early-phase drug development (1,10–12). For example, in two phase I clinical trials of sotorasib and adagrasib (NCT03600883 and NCT03785249), encouraging clinical activity has been observed in patients with non-small cell lung cancer (NSCLC) (13–15).
Given the encouraging early results in the development of KRASG12C inhibitors, a greater understanding of the genomic complexity of KRASG12C-mutant tumors and the oncologic outcomes of patients with these tumors is needed, especially in the setting of curative-intent surgery for earlier-stage disease (16,17). To address this knowledge gap, we investigated and compared tumor genomic features and disease-free survival (DFS) in patients with surgically resected LUAD harboring mutations in KRASG12C, compared to KRASother and KRASwt tumors. The results of our study offer insight into the potential impact that KRASG12C inhibitors may have in patients with early-stage LUAD who undergo surgery as their first treatment.
Materials and Methods
Study Population and Data Collection
Following institutional review board approval at Memorial Sloan Kettering Cancer Center, patients who underwent complete (R0) surgical resection for pathologic stage I-III LUAD from February 2010 to December 2018 and had targeted next-generation sequencing (NGS; MSK-IMPACT) (18) performed on the primary tumor were identified. All patients provided written informed consent to participate in the institutional review board–approved protocol, and all studies were conducted in accordance with the ethical guidelines of the Declaration of Helsinki. Exclusion criteria were as follows: receipt of induction therapy, microscopic (R1) or macroscopic (R2) residual disease, and low-quality NGS (see CONSORT diagram, Supplementary Figure 1). Tumors were classified according to primary tumor KRAS mutation status as either wild-type (KRASwt), KRAS(G12C) mutation (KRASG12C), or other KRAS mutation (KRASother).
Prospectively collected demographic, imaging, staging (American Joint Committee on Cancer 8th edition), pathologic, genomic, recurrence, and follow-up data were reviewed. Predominant invasive LUAD histologic subtypes were classified as either lepidic, acinar, papillary, micropapillary, or solid (19). Follow-up was performed in accordance with National Comprehensive Cancer Network guidelines (20). Metachronous lesions were distinguished from recurrences using Martini and Melamed criteria (21), with confirmation of clonal relatedness using genomic data, as previously reported by our group (22).
MSK-IMPACT Sequencing
Tumor genomic profiling was performed and analyzed as previously described (18). The sequencing breadth of the IMPACT panel has increased over time, resulting in 8, 190, and 406 patients sequenced with 341-, 410-, and 468-gene panels, respectively. Tumor mutation burden (TMB) was defined as the total number of nonsynonymous single-nucleotide insertion or deletion mutations per megabase and was normalized by panel size by dividing the total number of mutations by the length of the coding region captured by each panel (0.98, 1.06, and 1.22 Mb in the 341-, 410-, and 468-gene panels, respectively). Fraction of genome altered (FGA) was computed from the output of the FACETS (Fraction and Allele-specific Copy number Estimates from Tumor Sequencing) algorithm, which provides accurate, purity- and ploidy-corrected, integer DNA copy number calls from sequenced samples (23). FGA is defined as the fraction of the genome that differs from the major integer copy number, which represents the integer total copy number spanning the largest portion of the genome.
Statistical and Genomic Data Analysis
The primary endpoint—DFS—was defined as the time from surgery to first recurrence or death from any cause; patients were otherwise censored at the time of last follow-up. DFS was estimated using the Kaplan-Meier method and compared between groups using the log-rank test. Median follow-up was estimated using the reverse Kaplan-Meier method. Patterns of recurrence were assessed using the cumulative incidence of locoregional recurrence (CIR-LR) and distant recurrence (CIR-DR) in the KRASG12C and KRASother groups. Patients with both locoregional and distant recurrence were included in the distant recurrence analysis. Clinicopathologic and genomic variables (including specific genes, pathways, TMB, and FGA) were compared between groups using Fisher’s exact test for categorical factors and the Wilcoxon rank-sum test for continuous factors. A total of two Cox regression models were used to quantify the association between clinicopathologic or genomic factors and DFS—(1) within the overall cohort and (2) within only the KRASG12C group—using hazard ratios (HRs) and 95% confidence intervals (CIs). Both regression analyses used identical lists of variables, with the exception of the KRAS-mutation group, which was included in only the Cox regression for the overall cohort. Multivariable models for all factors were adjusted for pathologic stage. To quantify the associations between specific genes and DFS, an additional univariable analysis was performed using all genes with an alteration frequency >8% after false-discovery rate (FDR) correction.
Analysis of specific somatic alterations was performed using OncoKB (24) to remove variants of unknown significance. For the analysis of co-occurrence and mutual exclusivity, we assessed all genes known to be drivers in LUAD (24). In total, 121 genes were identified at the intersection of the a priori pathway templates and the MSK-IMPACT panel. A pathway was considered to be altered in a tumor if at least one gene within the corresponding pathway template was altered. Mutual exclusivity and co-occurrence of alterations in genes and oncogenic cell signaling pathways were assessed using Fisher’s exact test, and p values were adjusted to correct for multiple comparisons using FDR correction.
Differences in primary tumor clonality were investigated between groups using the cancer cell fraction, as calculated by FACETS. Clonality assessment was able to be performed on 72 patients in the KRASG12C group and 95 patients in the KRASother group. Primary tumor clonality was defined as a cancer cell fraction >0.8, as in prior reports (25,26). Variant allele frequency is the fraction of sequence reads that contain a specific DNA variant, divided by the overall coverage at that locus.
Mutational signatures were computed using the most recent version of the SBS signatures defined in the Catalogue of Somatic Mutations in Cancer database for somatic mutation signature analysis (27). Tumors with ≥13.8 mutations/Mb were evaluated for the KRASG12C (n=67) and KRASother (n=83) groups, in accordance with the previously published threshold (28). Tumors were considered to have a detectable signature if the mean signature value was >0.1, as previously described (27).
Analyses were conducted using Stata 15.0 (StataCorp, College Station, TX) and R 3.5.1 (R Core Team, Vienna, Austria). Statistical tests were two-sided, and p<0.05 was considered statistically significant.
External Validation
External validation of the relationship between KRASG12C mutation and DFS was performed using The Cancer Genome Atlas (TCGA) LUAD data set (N=476) (20). Patients with pathologic stage I-III LUAD who did not receive induction therapy were included. Tumors were classified according to KRAS-mutation status. DFS was estimated using the Kaplan-Meier method and was compared between groups using the log-rank test.
Results
Clinicopathologic Characteristics
A total of 604 patients met the inclusion criteria (Table 1). KRAS mutation status was as follows: 374 KRASwt (62%), 95 KRASG12C (16%), and 135 KRASother (22%). The median age at resection was 68 years (interquartile range [IQR], 62–74), and two-thirds of patients were women (n=402 [67%]). Most patients (77%) had a history of smoking, with a median of 27 pack-years (IQR, 15–45). Patients with KRAS-mutant LUAD were more commonly smokers, compared with KRASwt LUAD. Sublobar resection was performed in 214 patients (35%). A majority of patients had pathologic stage I LUAD (n=447 [74%]); 95 patients (16%) had stage II LUAD, and 62 patients (10%) had stage III LUAD. Of note, patients with KRASG12C and KRASother tumors were more likely to have high (≥50%) programmed death-ligand 1 (PD-L1) expression (n=35/95 [37%] and n=47/135 [35%], respectively), compared with patients with KRASwt tumors (n=94/374 [25%]) (p=0.013). Adjuvant therapy was administered to 116 patients (19%) and was not statistically significantly different between groups. Regimens included chemotherapy only (n=95 [82%]), chemoradiation (n=18 [16%]), radiotherapy only (n=2 [1.7%]), and immunotherapy (n=1 [0.8%]). Median follow-up was 2.51 years (IQR, 1.74–3.30).
Table 1.
Comparison of clinicopathologic characteristics between the KRASwt, KRASG12C, and KRASother groups
| Characteristic | Total Cohort (n=604) | KRASwt (n=374) | KRASG12C (n=95) | KRASother (n=135) |
|---|---|---|---|---|
| Age at resection, years | 68 (62–74) | 68 (62–74) | 69 (64–75) | 67 (61–73) |
| Sex | ||||
| Male | 202 (33) | 130 (35) | 34 (36) | 38 (28) |
| Female | 402 (67) | 244 (65) | 61 (64) | 97 (72) |
| Smoking status | ||||
| Never | 138 (23) | 125 (33) | 4 (4.2) | 9 (6.7) |
| Ever | 466 (77) | 249 (67) | 91 (96) | 126 (93) |
| Pack-years (n=465) | 27 (15–45) | 10 (0–30) | 27 (16–43) | 30 (14–46) |
| FEV1 (n=592) | 94 (83–106) | 95 (84–108) | 90 (77–104) | 94 (83–105) |
| DLCO (n=586) | 84 (69–97) | 87 (72–99) | 79 (64–92) | 80 (66–95) |
| SUVmax (n=537) | 3.7 (1.9–7.2) | 3.4 (1.8–6.7) | 4.1 (2.3–8.1) | 4.0 (2.1–7.5) |
| Operative approach | ||||
| VATS | 521 (86) | 324 (87) | 79 (83) | 118 (87) |
| Open | 83 (14) | 50 (13) | 16 (17) | 17 (13) |
| Operative procedure | ||||
| Lobectomy or pneumonectomy | 390 (65) | 245 (66) | 61 (64) | 84 (62) |
| Sublobar | 214 (35) | 129 (34) | 34 (36) | 51 (38) |
| Pathologic tumor size, cm | 1.8 (1.2–2.8) | 1.8 (1.2–2.8) | 1.8 (1.3–2.7) | 2.0 (1.4–3.1) |
| LVI (n=599) | 248 (41) | 150 (41) | 49 (52) | 49 (36) |
| VPI | 109 (18) | 70 (19) | 20 (21) | 19 (14) |
| STAS (n=544) | 337 (62) | 183 (55) | 68 (75) | 86 (70) |
| Histologic subtype | ||||
| Lepidic | 88 (15) | 59 (16) | 10 (11) | 19 (14) |
| Acinar | 368 (61) | 218 (58) | 64 (67) | 86 (64) |
| Papillary | 43 (7.1) | 28 (7.5) | 4 (4.2) | 11 (8.1) |
| Micropapillary | 37 (6.1) | 25 (6.7) | 5 (5.3) | 7 (5.2) |
| Solid | 68 (11) | 44 (12) | 12 (13) | 12 (8.9) |
| pN status (n=603) | ||||
| N0 | 506 (84) | 311 (83) | 75 (79) | 120 (89) |
| N1 or N2 | 97 (16) | 62 (17) | 20 (21) | 15 (11) |
| pStage | ||||
| I | 447 (74) | 278 (74) | 66 (69) | 103 (76) |
| II | 95 (16) | 61 (16) | 16 (17) | 18 (13) |
| III | 62 (10) | 35 (9.4) | 13 (14) | 14 (10) |
| PD-L1 status | ||||
| None (<1%) | 343 (57) | 233 (62) | 45 (47) | 65 (48) |
| Low (1–49%) | 85 (14) | 47 (13) | 15 (16) | 23 (17) |
| High (≥50%) | 176 (29) | 94 (25) | 35 (37) | 47 (35) |
| Adjuvant therapy | 116 (19) | 68 (18) | 21 (22) | 27 (20) |
Data are no. (%) or median (interquartile range). DLCO, diffusing capacity of the lungs for carbon monoxide; FEV1, forced expiratory volume in 1 second; LVI, lymphovascular invasion; PD-L1, programmed death-ligand 1; STAS, spread through air spaces; SUVmax; maximum standardized uptake value; VPI, visceral pleural invasion; wt, wild-type.
KRAS Mutation Status, Recurrence Patterns, and Survival
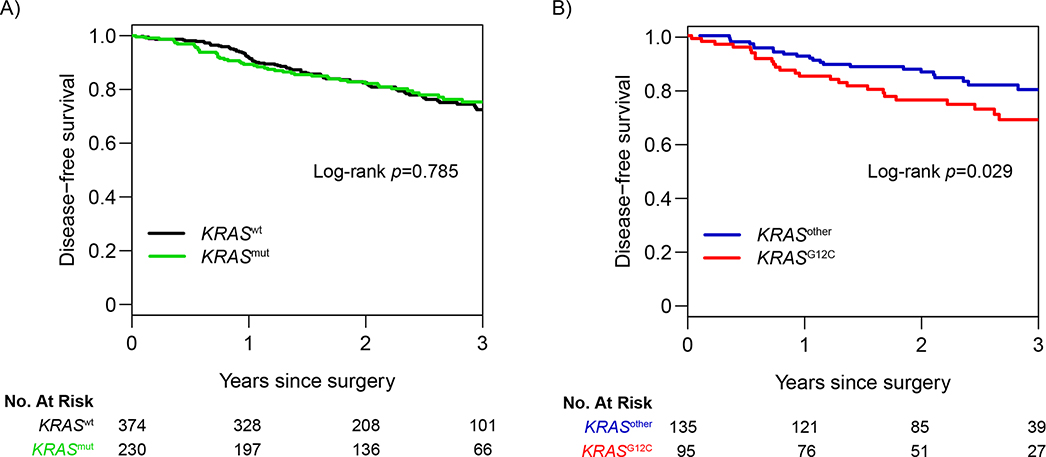
The specific types of KRAS alterations and associated frequencies are listed in Supplementary Table 1. The KRASother group predominantly comprised patients with G12D (n=33/135 [24%]), G12V (n=32/135 [24%]), and G12A (n=23/135 [17%]) mutations. Three-year DFS for the overall cohort was 73.6% (95% CI, 69.5%−78.0%). DFS was not statistically significant different between patients with any KRAS mutation (3-year DFS, 75.3% [95% CI, 69.2%−82.0%]) and patients with KRASwt LUAD (3-year DFS, 72.5% [95% CI, 67.1%−78.3%]), with a hazard ratio (HR) of 0.95 (95% CI, 0.68–1.34) (p=0.785; Figure 1A). However, among patients with KRAS-mutant tumors (n=230), DFS was substantially worse for patients with KRASG12C mutations (3-year DFS, 68.7% [95% CI, 58.9%−80.3%]) than for those with KRASother mutations (3-year DFS, 80.0% [95% CI, 72.4%−88.3%]), with an HR of 1.82 (95% CI, 1.06–3.15) (p=0.029; Figure 1B).
Figure 1. Association between KRAS mutation status and disease-free survival in the study cohort (N=604).
Three-year disease-free survival for (A) all KRAS-mutant (KRASmut) tumors versus KRAS wild-type (KRASwt) tumors and for (B) KRASG12C tumors versus all other KRAS-mutant tumors (KRASother).
The CIR-LR was higher in the KRASG12C group (3-year CIR-LR, 8.0% [95% CI, 2.8%−16.8%]) than in the KRASother group (3-year CIR-LR, 2.3% [95% CI, 0.6%−6.1%) but did not reach statistical significance (Gray’s p=0.119; Supplementary Figure 2A). The subtle observation of higher CIR-LR appeared to occur past the 2-year mark after surgery. Similarly, the CIR-DR was higher in the KRASG12C group (3-year CIR-DR, 20.8% [95% CI, 12.6%−30.4%]) than in the KRASother group (3-year CIR-DR, 13.0% [95% CI, 7.2%−20.7%]) but again did not reach statistical significance (Gray’s p=0.070; Supplementary Figure 2B). The observed divergence, however, appeared to occur earlier in the follow-up period. Overall, at 3 years after surgery, the CIR-DR was more than double the CIR-LR across both groups.
Clinicopathologic Factors Associated with DFS
On univariable analysis (N=604), the following factors were associated with DFS: diffusing capacity of the lungs for carbon monoxide (DLCO), primary tumor maximum standardized uptake value, open (thoracotomy) resection, pathologic tumor size, lymphovascular invasion (LVI), visceral pleural invasion (VPI), spread through air spaces (STAS), micropapillary or solid histologic subtype, pathologic node positivity, pathologic stage II or III LUAD, KRASG12C mutation, TMB, and FGA (p<0.1; Supplementary Table 2). On multivariable analysis, after adjustment for pathologic stage, KRASG12C mutation was independently associated with worse DFS (HR, 1.84 [95% CI, 1.01–3.36]; p=0.046) (Table 2). In addition, DLCO (HR, 0.99 [95% CI, 0.98–1.00]; p=0.031), LVI (HR, 2.36 [95% CI, 1.43–3.91]; p=0.001), VPI (HR, 1.66 [95% CI, 1.10–2.51]; p=0.015), STAS (HR, 1.81 [95% CI, 1.02–3.23]; p=0.044), and pathologic stage II or III (vs. stage I; HR, 1.80 [95% CI, 1.18–2.76]; p=0.007) were also independently associated with worse DFS.
Table 2.
Multivariable analysis for disease-free survival for the total cohort and the KRASG12C group
| Group/Variablea | HRa | 95% CI | p |
|---|---|---|---|
| Total cohort (N=604) | |||
| Primary tumor SUVmax | 1.03 | 1.00–1.07 | 0.065 |
| DLCO | 0.99 | 0.98–1.00 | 0.031 |
| LVI | 2.36 | 1.43–3.91 | 0.001 |
| VPI | 1.66 | 1.10–2.51 | 0.015 |
| STAS | 1.81 | 1.02–3.23 | 0.044 |
| pStage | |||
| I | Ref | — | — |
| II or III | 1.80 | 1.18–2.76 | 0.007 |
| KRAS mutation status | |||
| Non-G12C | Ref | — | — |
| Wild-type | 1.45 | 0.85–2.47 | 0.2 |
| G12C | 1.84 | 1.01–3.36 | 0.046 |
| KRASG12C group (n=95) | |||
| LVI | 9.57 | 2.20–41.54 | 0.003 |
| VPI | 2.25 | 1.03–4.94 | 0.042 |
| Histologic subtype | |||
| Lepidic, acinar, or papillary | Ref | — | — |
| Micropapillary or solid | 2.15 | 0.95–4.87 | 0.067 |
CI, confidence interval; DLCO, diffusing capacity of the lungs for carbon monoxide; HR, hazard ratio; LVI, lymphovascular invasion; STAS, spread through air spaces; SUVmax, maximum standardized uptake value; VPI, visceral pleural invasion
Multivariable models for all factors were adjusted for pathologic stage.
Factors prognostic for DFS were then investigated within the KRASG12C group (n=95). On univariable analysis, age at resection, pack-years, DLCO, primary tumor maximum standardized uptake value, primary tumor size, LVI, VPI, STAS, micropapillary or solid subtype, pathologic node positivity, pathologic stage II or III, and TMB were associated with DFS (p<0.1; Supplementary Table 3). On multivariable analysis, primary tumor LVI (HR, 9.57 [95% CI, 2.20–41.54]; p=0.003) and VPI (HR, 2.25 [95% CI, 1.03–4.94]; p=0.042) were independently associated with DFS (Table 2).
Clinicopathologic and Genomic Differences Between Patients with KRASG12C and KRASother Tumors
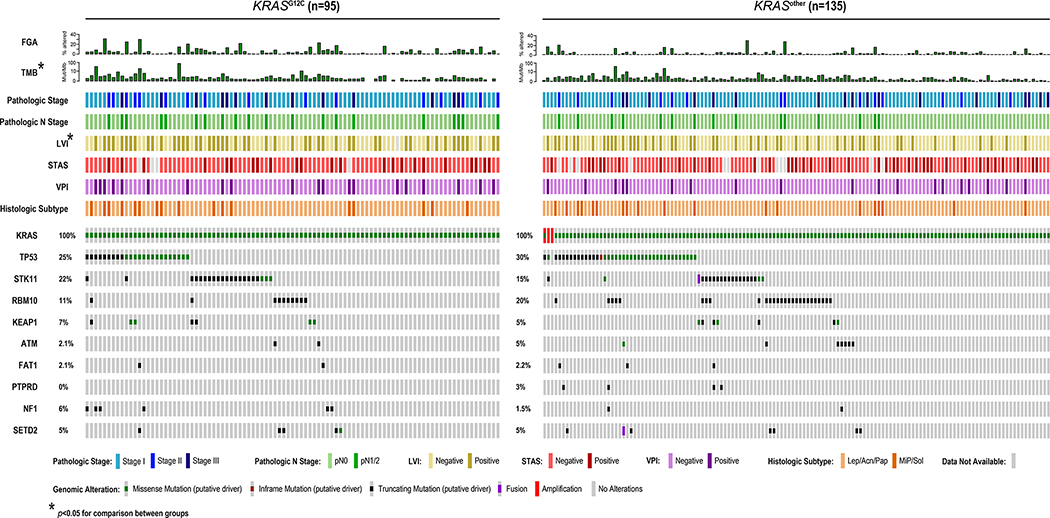
Clinicopathologic and genomic factors were then compared between patients with KRASG12C and KRASother tumors (Figure 2). LVI (51% vs. 37%; p=0.032) and pathologic lymph node metastasis (21% vs. 12%; p=0.059) were more common in the KRASG12C group. KRASG12C tumors were also found to have higher TMB (median [IQR], 7.0 [5.3–10.8] vs. 6.1 [3.5–9.7]; p=0.021) and FGA (x100; median [IQR], 3.8 [0.4–8.9] vs. 1.5 [0.2–7.3]; p=0.053).
Figure 2. Clinicopathologic and genomic features of KRAS-mutant tumors.
Comparison of clinicopathologic variables, genomic factors, and specific genes between the KRASG12C and other KRAS mutation (KRASother) groups. All genes with an alteration frequency >8% are shown. *p<0.05 for comparison between groups using Fisher’s exact test for categorical factors and the Wilcoxon rank-sum test for continuous factors. Acn, acinar; FGA, fraction of genome altered; Lep, lepidic; LVI, lymphovascular invasion; MiP, micropapillary; Pap, papillary; Sol, solid; STAS, spread through air spaces; TMB, tumor mutation burden; VPI, visceral pleural invasion.
The ten most commonly altered genes were compared between groups (Figure 2). Although no differences reached statistical significance, KRASG12C tumors had more STK11 and NF1 mutations than KRASother tumors. Conversely, KRASother tumors were nearly twice as likely to have a truncating mutation in RBM10, an RNA-binding protein and splicing regulator (29).
Genes Associated with DFS
To determine whether the differences in alteration frequencies described above were also prognostic for DFS, an additional univariable analysis was performed using the same list of genes (Supplementary Table 4). In the overall cohort, two genes (TP53: HR, 1.65 [95% CI, 1.18–2.31]; Q=0.024; and RBM10: HR, 0.43 [95% CI, 0.22–0.84]; Q=0.024) were associated with DFS after FDR correction, a finding that was previously reported by our group (30). In the KRASG12C group, no genes were statistically significantly associated with DFS after FDR correction.
Mutual Exclusivity and Co-occurrence Patterns
Mutual exclusivity and co-occurrence patterns of individual genes and oncogenic pathways were then explored between groups. In the overall cohort, STK11 and KEAP1 were significantly co-occurrent, as previously described (31), whereas any KRAS mutation was predictably mutually exclusive with other RAS pathway genes (e.g., EGFR, BRAF, MET) (FDR-p<0.05; Supplementary Figure 3A). Interestingly, no genes were found to be statistically significantly co-occurrent with KRASG12C mutation or KRASother mutation. Among KRASG12C tumors, TP53 and NF1 mutations were observed to be significantly co-occurrent (p=0.03; Supplementary Figure 3B). Finally, among KRASother tumors, STK11 and KEAP1 were again found to statistically significantly co-occur (p=0.007; Supplementary Figure 3C).
Clonality Patterns and Somatic Mutation Signatures of KRASG12C and KRASother Tumors
Next, differences in primary tumor clonality were explored between the KRASG12C and KRASother groups. KRAS mutation was found to be a clonal event in 90% of KRASG12C tumors (n=65/72) and 91% (n=86/95) of KRASother tumors (Supplementary Figure 4). This confirms that KRAS mutations are an early, truncal alteration in the evolution of LUAD tumors.
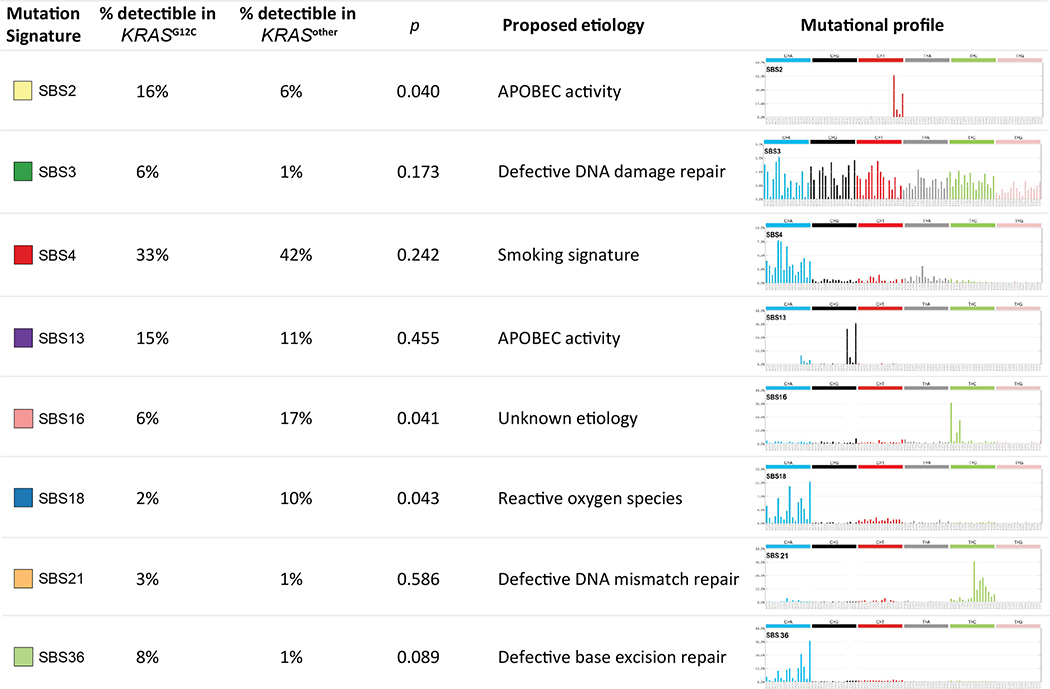
Detectable somatic mutation signatures were then investigated between the KRASG12C and KRASother groups (Figure 3). Overall, the smoking signature (SBS4) was most commonly detectable in both groups (33% vs. 42%, respectively; p=0.24). Interestingly, KRASG12C tumors had a statistically significantly higher prevalence of the SBS2 signature (attributed to APOBEC activity(27)), compared with KRASother tumors (16% vs. 6%; p=0.04). Conversely, KRASother tumors had higher rates of SBS16 (17% vs. 6%; p=0.041) and SBS18 (10% vs. 2%; p=0.043), compared with KRASG12C tumors.
Figure 3. Analysis of somatic mutational signatures in KRAS-mutant tumors.
Comparison of the relative frequencies of 8 detectible mutational signatures between the KRASG12C and other KRAS mutation (KRASother) groups, and proposed etiologies for these signatures.
External Validation of DFS Association
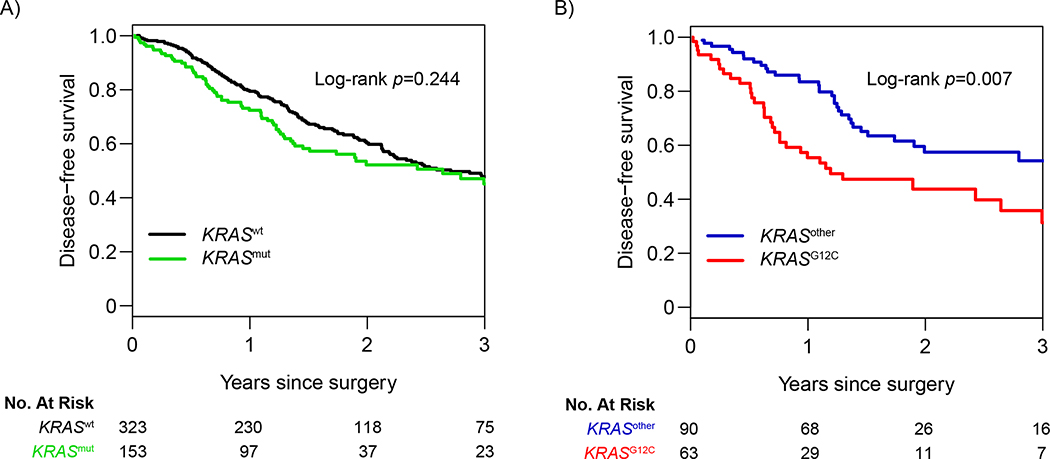
To externally validate our findings, we queried a TCGA data set for all patients with pathologic stage I-III LUAD who did not undergo induction therapy; a total of 476 patients were identified. KRAS mutation status was as follows: 323 KRASwt (68%), 63 KRASG12C (13%), and 90 KRASother (19%) (Supplementary Table 5). No difference was noted in 3-year DFS between patients with any KRAS mutation and patients with KRASwt LUAD (HR, 1.18 [95% CI, 0.89–1.56]; p=0.244; Figure 4A). However, among patients with KRAS-mutant tumors (n=153), patients with KRASG12C mutation were again found to have significantly worse 3-year DFS than patients with KRASother mutation (31.3% versus 54.2%, respectively; HR, 1.88 [95% CI, 1.18–3.00]; p=0.007; Figure 4B), confirming the relationship between KRASG12C mutation and DFS in our study.
Figure 4. Association between KRAS-mutation status and disease-free survival in The Cancer Genome Atlas cohort (N=476).
Three-year disease-free survival for (A) all KRAS-mutant (KRASmut) tumors versus KRAS wild-type (KRASwt) tumors and for (B) KRASG12C tumors versus all other KRAS-mutant tumors (KRASother).
Discussion
The recent development of KRASG12C inhibitors and their promising early results in phase I clinical trials have necessitated the genomic characterization and examination of long-term oncologic outcomes in patients with surgically resected KRASG12C-mutant tumors. We have shown that, compared with KRASother mutations, KRASG12C primary tumor mutations portended worse DFS in both our institutional data set as well as an external TCGA data set. KRASG12C mutation was also independently associated with worse DFS in our cohort. KRASG12C tumors appeared to contain a greater proportion of aggressive pathologic features (LVI, positive lymph nodes) and genomic perturbations (higher TMB and FGA) than KRASother tumors. Through extensive genomic characterization, we discovered that no common oncogenes or tumor suppressors were co-occurrent or mutually exclusive with KRASG12C tumors. However, in the overall cohort, STK11 and KEAP1 were significantly co-occurrent, a finding supported by studies in the metastatic setting (32). Finally, we have shown that the vast majority of both KRASG12C and KRASother tumors harbor clonal populations of KRAS-mutant cells, confirming that acquisition of this alteration is an early event in the mutagenesis of these tumors.
KRAS somatic mutations have been shown to be associated with decreased survival in prior studies, with 5-year overall survival ranging from 22% to 30% in patients with KRAS-mutant NSCLC (4,32). Although this association with poor prognosis is well documented for advanced disease, the influence of KRAS mutation on survival in patients with early-stage disease remains poorly characterized. In a smaller series (N=179), Nadal and colleagues found overall KRAS mutation was associated with worse DFS (log-rank p=0.006) and overall survival (log-rank p=0.046) (17). However, this cohort included all stages of LUAD, with 20% of KRAS-mutant tumors (n=21) being stage III or IV in this study. When the analysis was repeated for only patients with stage I disease (n=121), the survival difference substantially diminished (log-rank p=0.049). In the two larger, early-stage cohorts in our study (study cohort, N=604; TCGA external validation cohort, N=476), 3-year DFS was not statistically significantly different between patients with KRAS-mutant and KRASwt tumors, indicating there was no prognostic significance for the overall KRAS mutation population.
Targeting KRASG12C—the most prevalent of the KRAS alterations, present in up to half of KRAS-mutant tumors (33)—has led to encouraging tumor responses in phase I clinical trials (13,34). In the Nadal study, the 2-year DFS for patients with KRASG12C tumors was 42.9%, compared with approximately 65.0% for patients with other KRAS-mutant tumors. However, the KRASG12C group comprised only 35 patients, with an unreported stage distribution. In the present study, 3-year DFS for patients with KRASG12C tumors was also substantially worse than that for patients with KRASother tumors in our institutional cohort (68.7% vs 80.0%, respectively). Additionally, analysis of the TCGA database yielded similar results, with a 3-year DFS of 31.3% versus 54.2%, respectively, in this external validation cohort. Interestingly, the observed survival differences do not appear to be linked to comutation patterns, as no genes were significantly co-occurrent with KRASG12C alteration on genomic analysis, and likewise no other genes were prognostic for DFS within this group. One possible explanation for this survival detriment may involve the lack of immunogenicity in KRASG12C tumors. A recent study by Aredo and colleagues found that these tumors were significantly associated with low (1%−49%) PD-L1 expression (35). In the present study, KRASG12C-mutant tumors with high PD-L1 expression (≥50%)—although these were in the minority—were significantly more common in the KRASG12C group than in the KRASwt group (37% vs. 23%; p=0.013). On the basis of preclinical data, KRASG12C inhibitors enhance antitumor immunity, which may be helpful for eradicating micrometastatic disease (9). The ability to target this alteration in a newly defined high-risk population—whether in the adjuvant or neoadjuvant setting—has major therapeutic implications.
Numerous studies have shown that KRAS mutation is an early event in lung tumorigenesis (36–38). In a recent analysis of NSCLC tumor evolution, primary tumors with alterations in any of four genes (TP53, KEAP1, STK11, and EGFR) were shown to have a higher proportion of clonal tumor cell populations, signifying that these genes play a role in tumor initiation (36). In a separate study involving multi-region sequencing, KRAS mutations were present in both minimally invasive adenocarcinoma or adenocarcinoma in situ and paired invasive LUAD, suggesting that KRAS mutation is an early mutagenic event and an indicator of malignant transition (37). Furthermore, in the landmark TRACERx NSCLC tumor evolution study, 88% of samples with KRAS mutations were deemed to be clonal populations (38). In our study, 90% of overall KRAS-mutant tumors, as well as 90% of KRASG12C-mutant tumors, were found to harbor clonal cancer cell populations. This knowledge, coupled with the increased risk of recurrence in patients with KRASG12C tumors in this study, provides encouraging evidence to support the use of KRASG12C inhibitors for preventing or delaying tumor relapse in patients with early-stage LUAD.
Recent large-scale analyses have identified numerous somatic mutational signatures across the spectrum of cancer types (27). Although somatic mutational signature analysis most commonly relies on whole-genome or whole-exome sequencing data, our group has previously shown the feasibility of using MSK-IMPACT (28). In the present study, KRASG12C tumors were associated with high activity of the putative APOBEC mutational signature (SBS2), whereas KRASother tumors were associated with the putative reactive oxygen species signature (SBS18). Loss-of-function alterations in APOBEC-related genes lead to DNA hypermutation and inaccurate RNA editing and are associated with tumorigenesis as well as drug resistance (19,39,40). Furthermore, a recent analysis of immune-response-related mutational signatures in NSCLC showed that high TMB combined with APOBEC-related mutational signatures was predictive of response to immunotherapy (41), revealing an intriguing new patient population that may derive benefit from these agents.
Limitations of our study include a low number of death events in our early-stage cohort such that overall survival was unable to be explored as an outcome. Another limitation is that our findings require further validation from international data sets such as those from Asia, where the incidence of KRAS mutations in LUAD is lower, which would provide useful information regarding the implications of KRASG12C mutation and inhibition in a geographically diverse population. Despite these limitations, this externally validated analysis from the largest data set of patients with resected KRASG12C LUAD provides evidence to support the investigation of sotorasib, adagrasib (MRTX849), or other KRASG12C inhibitors in the adjuvant setting, with the goal of improving DFS.
Mutations in KRASG12C are the most common KRAS mutations in lung cancer and are independently prognostic for poor DFS after complete resection of stage I-III LUAD. We have shown that the vast majority of KRASG12C tumors harbor clonal populations of KRASG12C-mutant cells, and these tumors appear to harbor more genomic perturbations and aggressive clinicopathologic features than other KRAS-mutant tumors. Additionally, KRASG12C mutation was not found to be co-occurrent with actionable alterations in other common LUAD driver genes. Our findings provide evidence supporting the investigation of KRASG12C inhibitors in the adjuvant setting in this vulnerable patient population.
Supplementary Material
Statement of Translational Relevance:
KRASG12C is the most prevalent of the KRAS alterations in primary lung adenocarcinoma (LUAD), present in up to half of cases. The association between KRAS somatic mutations and decreased survival is well-documented in metastatic LUAD. However, the influence of KRAS mutation on survival in early-stage disease remains poorly characterized. The recent development of KRASG12C inhibitors and their promising early results in phase I clinical trials necessitates the genomic characterization and examination of long-term oncologic outcomes in patients with surgically resected KRASG12C-mutant tumors. We have shown that, compared with KRASother mutations, KRASG12C primary tumor mutations portended worse disease-free survival in both our institutional data set and an external TCGA data set. KRASG12C tumors contain a greater proportion of aggressive pathologic features (LVI, positive lymph nodes) and genomic perturbations (higher TMB and FGA) than KRASother tumors. We identify a high-risk group for whom KRASG12C inhibitors may be investigated to improve survival.
Acknowledgments
David B. Sewell of the Department of Surgery, Memorial Sloan Kettering Cancer Center, provided editorial assistance. This study was supported by the National Cancer Institute (R01CA217169 and R01CA240472 to D.R.J., R01CA236615 to P.S.A., T32CA009501 to J.G.C., P30 CA008748 to Memorial Sloan Kettering Cancer Center), Hamilton Family Foundation (to D.R.J.), and Department of Defense (LC160212 to P.S.A.). This study was also supported by a Conquer Cancer Young Investigator Award (to Y.R.M.-G.), supported by Charles M. Baum and Carol A. Baum. Any opinions, findings, and conclusions expressed in this material are those of the authors and do not necessarily reflect those of the American Society of Clinical Oncology, Conquer Cancer, or Dr. Charles M. Baum and Carol A. Baum. Y.R.M.-G. has received training through an institutional K30 grant (CTSA UL1TR00457).
Conflicts of Interest: Yonina R. Murciano-Goroff has received travel, accommodations, and expenses from AstraZeneca. Sandra Misale consulted for Boehringer-Ingelheim. Matthew J. Bott serves as a consultant for AstraZeneca. Prasad S. Adusumilli has received research funding from ATARA Biotherapeutics and Acea Biosciences, has served on the Scientific Advisory Board or as consultant to ATARA Biotherapeutics, Bayer, Carisma Therapeutics, Imugene, and Takeda Therapeutics, and has patents, royalties, and intellectual property on mesothelin-targeted CARs and other T-cell therapies, method for detection of cancer cells using virus, and pending patent applications on T-cell therapies. Daniela Molena is a consultant for Johnson & Johnson, Urogen, Intuitive, and Boston Scientific. Gaetano Rocco has financial relationships with Scanlan. Rona Yaeger has received research funding from Array BioPharma/Pfizer and Boehringer Ingelheim and has consulted for Array BioPharma/Pfizer, Mirati Therapeutics, and Natera. Gregory J. Riely has received research funding from Novartis, Genentech, Millennium Pharmaceuticals, GlaxoSmithKline, Pfizer, Infinity Pharmaceuticals, ARIAD Pharmaceuticals, has a patent application submitted covering pulsatile use of erlotinib to treat or prevent brain metastases, and has received travel expenses from Merck Sharp & Dohme. James M. Isbell is a consultant for Genentech and has an equity interest in LumaCyte LLC. Bob T. Li has served as an uncompensated advisor and consultant to Amgen, Genentech, Boehringer Ingelheim, Lilly, AstraZeneca, Daiichi Sankyo, and has received consulting fees from Guardant Health and Hengrui Therapeutics. He has received research grants to his institution from Amgen, Genentech, AstraZeneca, Daiichi Sankyo, Lilly, Illumina, GRAIL, Guardant Health, Hengrui Therapeutics, MORE Health and Bolt Therapeutics. He has received academic travel support from Resolution Bioscience, MORE Health, and Jiangsu Hengrui Medicine. He is an inventor on two institutional patents at MSK and has intellectual property rights as a book author at Karger Publishers. David R. Jones serves as a consultant for AstraZeneca and on a Clinical Trial Steering Committee for Merck. All other authors have no potential conflicts to disclose.
References
- 1.Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 2013;503(7477):548–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prior IA, Lewis PD, Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res 2012;72(10):2457–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson ML, Sima CS, Chaft J, Paik PK, Pao W, Kris MG, et al. Association of KRAS and EGFR mutations with survival in patients with advanced lung adenocarcinomas. Cancer 2013;119(2):356–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Janne PA, van den Heuvel MM, Barlesi F, Cobo M, Mazieres J, Crino L, et al. Selumetinib plus docetaxel compared with docetaxel alone and progression-free survival in patients with KRAS-mutant advanced non-small cell lung cancer: The SELECT-1 randomized clinical trial. Jama-J Am Med Assoc 2017;317(18):1844–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Welsh SJ, Corrie PG. Management of BRAF and MEK inhibitor toxicities in patients with metastatic melanoma. Ther Adv Med Oncol 2015;7(2):122–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fakih M, Vincent M. Adverse events associated with anti-EGFR therapies for the treatment of metastatic colorectal cancer. Curr Oncol 2010;17:S18–S30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Janne PA, Shaw AT, Pereira JR, Jeannin G, Vansteenkiste J, Barrios C, et al. Selumetinib plus docetaxel for KRAS-mutant advanced non-small-cell lung cancer: a randomised, multicentre, placebo-controlled, phase 2 study. Lancet Oncol 2013;14(1):38–47. [DOI] [PubMed] [Google Scholar]
- 9.Canon J, Rex K, Saiki AY, Mohr C, Cooke K, Bagal D, et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 2019;575(7781):217–23. [DOI] [PubMed] [Google Scholar]
- 10.Patricelli MP, Janes MR, Li LS, Hansen R, Peters U, Kessler LV, et al. Selective inhibition of oncogenic KRAS output with small molecules targeting the inactive state. Cancer Discov 2016;6(3):316–29. [DOI] [PubMed] [Google Scholar]
- 11.Janes MR, Zhang JC, Li LS, Hansen R, Peters U, Guo X, et al. Targeting KRAS mutant cancers with a covalent G12C-specific inhibitor. Cell 2018;172(3):578–89. [DOI] [PubMed] [Google Scholar]
- 12.Lito P, Solomon M, Li LS, Hansen R, Rosen N. Allele-specific inhibitors inactivate mutant KRAS G12C by a trapping mechanism. Science 2016;351(6273):604–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xue JY, Zhao YL, Aronowitz J, Mai TT, Vides A, Qeriqi B, et al. Rapid non-uniform adaptation to conformation-specific KRAS(G12C) inhibition. Nature 2020;577(7790):421–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janne P PK, Ou I, Rybkin I, Johnson M. A phase 1 clinical trial evaluating the pharmacokinetics (PK), safety, and clinical activity of MRTX849, a mutant-selective small molecule KRAS G12C inhibitor, in advanced solid tumors. AACR-NCI-EORTC International Conference on Molecular Targets. Boston, MA. Available at https://www.mirati.com/wp-content/uploads/2019/10/AACR-NCI-EORTC-Clinical-Data-Presentation_Janne_October-2019-1-1.pdf. Accessed November 23, 2020. [Google Scholar]
- 15.Hong DS FM, and Strickler JH et al. , Li BT. KRAS(G12C) inhibition in advanced solid tumors. N Engl J Med 2019. [in press]. [Google Scholar]
- 16.Jeanson A, Tomasini P, Souquet-Bressand M, Brandone N, Boucekine M, Grangeon M, et al. Efficacy of immune checkpoint inhibitors in kras-mutant non-small cell lung cancer (NSCLC). J Thorac Oncol 2019;14(6):1095–101. [DOI] [PubMed] [Google Scholar]
- 17.Nadal E, Chen G, Prensner JR, Shiratsuchi H, Sam C, Zhao L, et al. KRAS-G12C mutation is associated with poor outcome in surgically resected lung adenocarcinoma. J Thorac Oncol 2014;9(10):1513–22. [DOI] [PubMed] [Google Scholar]
- 18.Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn 2015;17(3):251–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caso R, Sanchez-Vega F, Tan KS, Mastrogiacomo B, Zhou J, Jones GD, et al. The underlying tumor genomics of predominant histologic subtypes in lung adenocarcinoma. J Thorac Oncol 2020. [Online ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.The Cancer Genome Atlas: Lung Adenocarcinoma. Available at https://www.nccn.org/professionals/physican_gls/pdf/nscl.pdf. Accessed June 9, 2020.
- 21.Martini N, Melamed MR. Multiple primary lung cancers. J Thorac Cardiovasc Surg 1975;70(4):606–12. [PubMed] [Google Scholar]
- 22.Chang JC, Alex D, Bott M, Tan KS, Seshan V, Golden A, et al. Comprehensive next-generation sequencing unambiguously distinguishes separate primary lung carcinomas from intrapulmonary metastases: comparison with standard histopathologic approach. Clin Cancer Res 2019;25(23):7113–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen R, Seshan VE. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res 2016;44(16):e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, et al. OncoKB: A precision oncology knowledge base. JCO Precis Oncol 2017;2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Razavi P, Li BT, Brown DN, Jung B, Hubbell E, Shen R, et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat Med 2019;25(12):1928–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol 2012;30(5):413–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alexandrov LB, Kim J, Haradhvala NJ, Huang MN, Ng AWT, Wu Y, et al. The repertoire of mutational signatures in human cancer. Nature 2020;578(7793):94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 2017;23(6):703–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Serrano P, Hammond JA, Geralt M, Wuthrich K. Splicing site recognition by synergy of three domains in splicing factor RBM10. Biochemistry 2018;57(10):1563–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones GD, Brandt WS, Shen R, et al. A genomic-pathologic annotated risk model to predict recurrence in early-stage lung adenocarcinoma: a prospective observational cohort study. JAMA Surg 2020. [Online ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Papillon-Cavanagh S, Doshi P, Dobrin R, Szustakowski J, Walsh AM. STK11 and KEAP1 mutations as prognostic biomarkers in an observational real-world lung adenocarcinoma cohort. ESMO Open 2020;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arbour KC, Jordan E, Kim HR, Dienstag J, Yu HA, Sanchez-Vega F, et al. Effects of co-occurring genomic alterations on outcomes in patients with KRAS-mutant non-small cell lung cancer. Clin Cancer Res 2018;24(2):334–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roman M, Baraibar I, Lopez I, Nadal E, Rolfo C, Vicent S, et al. KRAS oncogene in non-small cell lung cancer: clinical perspectives on the treatment of an old target. Mol Cancer 2018;17(1):33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hong DS, Fakih MG, Strickler JH, Desai J, Durm GA, Shapiro GI, et al. KRAS(G12C) inhibition with sotorasib in advanced solid tumors. N Engl J Med 2020;383(13):1207–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aredo JV, Padda SK, Kunder CA, Han SS, Neal JW, Shrager JB, et al. Impact of KRAS mutation subtype and concurrent pathogenic mutations on non-small cell lung cancer outcomes. Lung Cancer 2019;133:144–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Testa U, Castelli G, Pelosi E. Lung cancers: molecular characterization, clonal heterogeneity and evolution, and cancer stem cells. Cancers (Basel) 2018;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Izumchenko E, Chang X, Brait M, Fertig E, Kagohara LT, Bedi A, et al. Targeted sequencing reveals clonal genetic changes in the progression of early lung neoplasms and paired circulating DNA. Nat Commun 2015;6:8258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jamal-Hanjani M, Wilson GA, McGranahan N, Birkbak NJ, Watkins TBK, Veeriah S, et al. Tracking the evolution of non-small-cell lung cancer. N Engl J Med 2017;376(22):2109–21. [DOI] [PubMed] [Google Scholar]
- 39.Swanton C, McGranahan N, Starrett GJ, Harris RS. APOBEC enzymes: mutagenic fuel for cancer evolution and heterogeneity. Cancer Discov 2015;5(7):704–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salter JD, Bennett RP, Smith HC. The APOBEC protein family: united by structure, divergent in function. Trends Biochem Sci 2016;41(7):578–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen H, Chong W, Teng C, Yao Y, Wang X, Li X. The immune response-related mutational signatures and driver genes in non-small-cell lung cancer. Cancer Sci 2019;110(8):2348–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.