Abstract
Two microglial TAM receptor tyrosine kinases - Axl and Mer - have been linked to Alzheimer’s disease, but their roles in disease have not been tested experimentally. We find that in Alzheimer’s disease and its mouse models, induced expression of Axl and Mer in amyloid plaque-associated microglia was coupled to induced plaque decoration by the TAM ligand Gas6 and its co-ligand phosphatidylserine. In the APP/PS1 mouse model of Alzheimer’s disease, genetic ablation of Axl and Mer resulted in microglia that were unable to normally detect, respond to, organize, or phagocytose amyloid beta plaques. These major deficits notwithstanding, TAM-deficient APP/PS1 mice developed fewer dense-core plaques than APP/PS1 mice with normal microglia. Our findings reveal that the TAM system is an essential mediator of microglial recognition and engulfment of amyloid plaques, and that TAM-driven microglial phagocytosis does not inhibit, but rather promotes, dense-core plaque development.
The TAM receptor tyrosine kinases (RTKs) Axl and Mer (gene name Mertk) exert two key functions in macrophages and other immune sentinels1, 2. First, they are required for the phagocytosis of apoptotic cells (ACs)3, 4, 5. Mer, which is expressed by all phagocytic macrophages, is especially important for this process3. Second, both receptors, but most prominently Axl, function as negative feedback inhibitors of Toll-like and cytokine receptor signaling in dendritic cells and macrophages1, 2, 6. We have previously shown that these two functions are TAM-controlled in microglia, the tissue macrophages of the central nervous system (CNS)7. As genetic analyses have revealed that many of the genes associated with increased risk for Alzheimer’s disease (AD) and other neurodegenerative disorders are expressed in the CNS predominantly or exclusively by microglia, these macrophages have attracted increasing interest in the context of neurodegeneration8.
Human and mouse microglia normally express high levels of Mer and low levels of Axl7, 9, and we have shown that these RTKs play a critical role in microglial phagocytosis of the ACs that are generated during adult neurogenesis7. The third TAM receptor – Tyro3 – is not expressed by microglia7, 9, but is prominently expressed by neurons10. We and others have also shown that Axl gene expression is markedly up-regulated in microglia and other macrophages by inflammatory stimuli5, and that Axl up-regulation also occurs when macrophages are activated during the course of infection, tissue trauma, or disease3, 7. Correspondingly, RNA-seq analyses of laser-captured plaque-associated tissue from human AD, and single cell RNA-seq analyses of ‘disease-associated microglia’ (‘DAM’) in the 5xFAD mouse model of AD, have both documented up-regulation of microglial Axl mRNA in late-stage disease11, 12. The Axl tyrosine kinase is activated by Axl ectodomain binding of the ligand Gas6, which is in turn bound by the co-ligand phosphatidylserine13. Subsequent to Axl activation, metalloproteases cleave the Axl ectodomain from the cell surface5, and elevated cerebrospinal fluid levels of the soluble ectodomain (sAxl), in complex with Gas6, are predictive of disease development and progression in human AD14, 15. All of these observations have led to the classification of Axl as an ‘AD-associated gene’12. This notwithstanding, the roles that Axl and Mer might play in AD have not been studied.
We therefore examined the function of microglial TAM receptors and their ligands in AD and its mouse models. We studied hemizygous APP/PS1 mice, which carry an amyloidogenic ‘Swedish’ mutation of the human amyloid precursor protein (APP) gene and a pathogenic mutation of the human presenilin 1 (PSEN1) gene16, 17, both as transgenes driven by the mouse prion protein promoter. In humans, these mutations result in familial AD, due to elevated proteolytic production of amyloid beta (Aβ) peptides from the amino terminus of APP. We also examined hemizygous APP41 mice, which express a ‘Swedish’ + ‘London’ (V717I) mutant human APP under the control of the Thy-1 promoter18. Finally, we examined TAM regulation in human AD. We found that the full TAM system, including its universal ligand Gas6 and essential co-ligand phosphatidylserine, was engaged concomitantly with the deposition of Aβ plaques in the brain. Using single cell RNA sequencing, we observed that Axl−/−Mertk−/− microglia display a blunted transcriptomic response to Aβ plaques in APP/PS1 mice. Using single-cell live two-photon imaging of compound mutant mice, we found that Axl−/−Mertk−/− microglia are also unable to normally detect, respond to, organize, or phagocytose plaques. Remarkably, we observed that TAM-deficient APP/PS1 mice have fewer dense-core plaques than their TAM-expressing counterparts. We propose a model to account for these results.
RESULTS
Engagement of TAM receptors during disease
We detected pronounced elevation of microglial Axl protein in APP/PS1 mice (Fig. 1). This induction was tightly plaque-associated, and was not seen prior to the deposition of Aβ plaques. We identified microglia as ramified, multi-polar cells expressing Iba1 (ionized calcium-binding adaptor molecule 1), or green fluorescent protein (GFP) in Cx3cr1-eGFP mice7. (Perivascular macrophages are also GFP+ in these mice, but are readily distinguished from microglia. Iba1 and Cx3cr1 are also expressed by peripheral myeloid cells9 that may enter the brain and assume microglia-like properties in neurodegenerative disease19.) Microglia near to but not touching plaques, identified with the 6E10 antibody to the amino terminus of human APP, were Axl− in aged APP/PS1 mice, while those contacting plaques were strongly Axl+ (Fig. 1a). Importantly, Axl protein up-regulation is also seen in plaque-associated microglia in human AD20. Quantification of Axl fluorescence in plaque-associated versus non-plaque-associated microglia - PAM and NPAM, respectively - over the course of disease in APP/PS1 cortex revealed a modest Axl increase in NPAM between 7 and 12 months (mo) (Fig. 1b). However, robust up-regulation only occurred in plaque-associated cells (Fig. 1b). We did not detect significant plaque accumulation until ~5 mo in APP/PS1 mice, and did not measure microglial Axl up-regulation prior to this age (Fig. 1b; Extended Data Fig. 1a). Axl is not detectable by immunohistochemistry in healthy wild-type microglia7 (Extended Data Fig. 1a). At 9.5 mo, Axl levels were ~25-fold higher in PAM versus NPAM (Fig. 1b). We detected a similar Axl up-regulation in PAM in aged APP41 mice, in which Axl was also frequently concentrated at the center of plaques (Extended Data Fig. 1b). Since Axl activation is always accompanied by cleavage of the Axl ectodomain5, this plaque-deposited Axl is almost certainly sAxl.
Fig. 1. TAM receptor up-regulation is strictly plaque-associated.
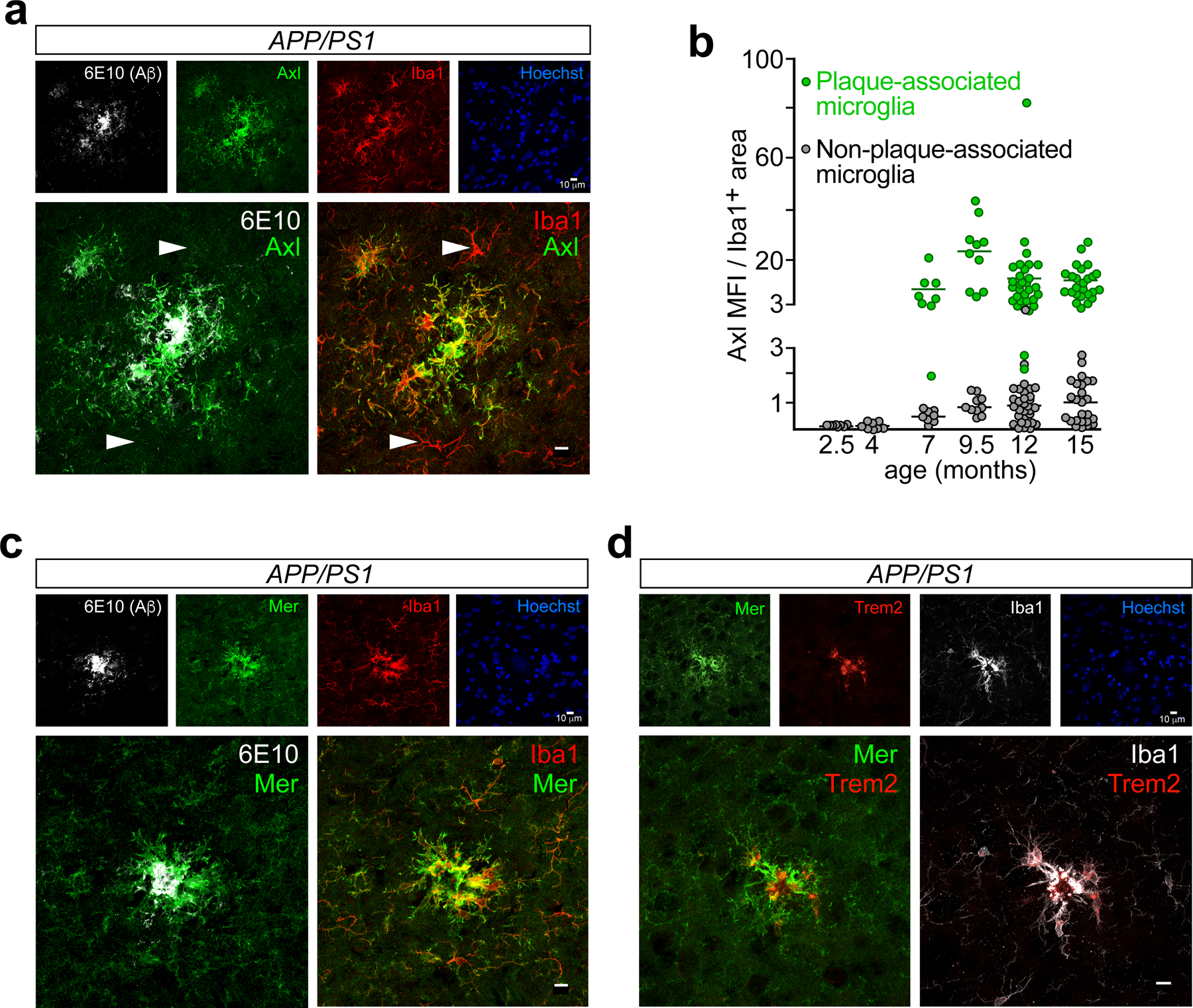
(a) Axl protein (green) in 9.5 mo APP/PS1 mice is up-regulated in microglia (Iba1, red) that are in contact with Aβ plaques (6E10, white), but not in microglia not contacting plaques (arrowheads in lower panels). Iba1 expression is also up-regulated in plaque-associated microglia. Hoechst stains nuclei. (b) Quantification of Axl mean fluorescence intensity (MFI) in cortical Iba1+ microglia in APP/PS1 mice over time reveals marked Axl up-regulation co-incident with the first deposition of plaques at ~5 mo, only in plaque-associated Iba1+ cells (green points). (c) Expression of Mer protein (green) in 9.5 mo APP/PS1 mice is seen in all cortical microglia (Iba1, red), but is further up-regulated in microglia that invest Aβ plaques (6E10, white). (d) The same cortical microglia that up-regulate Mer (green) and Iba1 (white) protein expression in 9.5 mo APP/PS1 mice also up-regulate expression of Trem2 (red). Lower panels in a, c, and d are enlargements of upper panels. a, c, and d contain representative images from N ≥ 3 sections per mouse from n = 3 mice from at least three independent replicates. Data in b were measured from N ≥ 3 images per mouse from n = 3 and 5 mice for 2.5 – 9.5 mo and 12 and 15 mo timepoints, respectively. All scale bars: 10μm.
In contrast to Axl, Mer is abundant in all normal human and mouse microglia7, 9. Although modest reductions in Mertk mRNA have been reported for plaque-associated microglia in APP/PS1 mice21, Mer protein expression was up-regulated in PAM in aged APP/PS1 (Fig. 1c) and APP41 (Extended Data Fig. 1c) mice. In the 9.5 mo APP/PS1 brain, we quantified a 3.5-fold elevation in Mer in PAM versus NPAM (Extended Data Fig. 1d). Axl+Merhi microglia were the same plaque-associated cells that also up-regulated Trem2 (Fig. 1d; Extended Data Fig. 1e), mutations in which increase the risk of developing human AD22. Although up-regulated Axl, Mer, and Trem2 were always present in the same cells, Trem2 and the TAMs appeared to occupy distinct membrane domains (Fig. 1d). Most Iba1+Axl+MerhiTrem2+ microglia in both APP/PS1 and APP41 mice were negative for TMEM119, a marker of homeostatic microglia23, although 1–2 cells at the very center of plaques were occasionally TMEM119+ (Extended Data Fig. 1f, g). Plaque-associated astrocytes display reactive astrocytosis in AD and its mouse models, but GFAP+ and S100b+ reactive astrocytes expressed no detectable Mer (Extended Data Fig. 1h and Ref. 7). Together, these results demonstrate that the concerted expression of Axl, Mer, and Trem2 is specific to plaque-associated microglia.
Engagement of TAM ligand and co-ligand during disease
Expression of the obligate Axl and pan-TAM ligand Gas613 was also strongly up-regulated at plaques in aged AD mice (Fig. 2a). This up-regulation was missed in earlier single cell RNA-seq analyses of mouse AD models because microglial expression of Gas6 mRNA, which is already high in normal microglia9, does not change over the course of disease in these models11. We found that the presence of plaque-associated Gas6 in APP/PS1 mice was tied to Axl up-regulation, and to an unusual feature of Gas6-Axl interaction: it has been shown that most of the Axl on cells in wild-type (WT) mouse tissues is constitutively bound by Gas6, and that expression of Gas6 protein in these tissues is Axl-dependent5. Gas6 expression is lost in Axl−/− but not Mertk−/− tissues, even though Gas6 mRNA levels are unchanged between Axl−/−, Mertk−/−, and WT tissues5. We therefore conclude that Gas6 protein is undetectable in non-diseased brains because these brains express little or no Axl (Extended Data Fig. 1a); and further, that acquired Axl in plaque-associated microglia leads to acquired plaque-localized Gas6. Consistent with this conclusion, plaque-associated Gas6 was sharply reduced in APP/PS1Axl−/− and eliminated entirely in APP/PS1Axl−/−Mertk−/− mice (Fig. 2a, quantified in Fig. 2b upper).
Fig. 2. Gas6 and phosphatidylserine decorate all Aβ plaques.
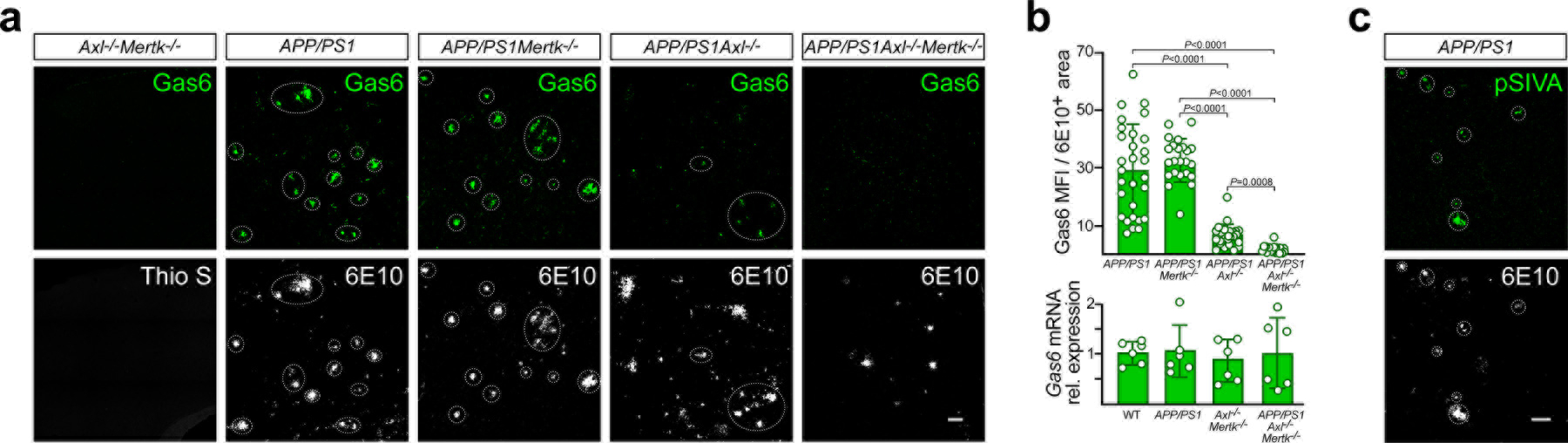
(a) There is no Gas6 protein (green) and are no plaques (Thio S, white, since 6E10 is human-specific) in the cortex of 12 mo WT (not shown) or Axl−/−Mertk−/− mice (left panels), but abundant Gas6 specifically associated with Aβ plaques (6E10, white) in APP/PS1 mice (second panels). Plaque-associated Gas6 is unchanged in APP/PS1Mertk−/− mice (third panels), severely reduced in APP/PS1Axl−/− mice (fourth panels), and eliminated entirely in APP/PS1Axl−/−Mertk−/− mice (fifth panels). Dotted ovals mark Gas6/6E10 coincidence. (b) (Upper graph) Quantification of results in a expressed as Gas6 MFI per 6E10+ area. (Lower graph) There is no change in the expression of cortical Gas6 mRNA between the indicated genotypes (n = 6 mice per group). (c) Externalization of PtdSer (detected with pSIVA, green, upper panel) in the cortex of 15 mo APP/PS1 mice is specific to 6E10+ Aβ plaques (lower panel). a and c contain representative images from N ≥ 3 sections from n = 4 and 3 mice of each genotype, respectively, from 3 and 2 independent replicates. a is quantified in b (upper). Data in b and all subsequent panels in the paper are represented as mean ±1 STD. Scale bars: 50μm. Kruskal Wallis test followed by Dunn’s multiple comparison test (b).
Amyloid plaques did not develop, and Gas6 did not appear, in aged Axl−/−Mertk−/− mice without the APP/PS1 allele (Fig. 2a), and Gas6 mRNA levels were constant across WT, APP/PS1, Axl−/−Mertk−/−, and APP/PS1Axl−/−Mertk−/− cortices at 12 mo (Fig. 2b lower). We also detected expression of Gas6 surrounding plaques in sections of human AD brain at BRAAK stage 6, but not in age-matched controls (Extended Data Fig. 2a), indicating that acquired Gas6 expression also marks plaques in human AD. Gas6 activates both Axl and Mer, but Mer can also be activated by the related ligand Protein S (Pros1)13. Although antibodies for high-resolution immunohistochemical detection of Pros1 in mouse are not currently available, microglia express abundant Pros1 mRNA9 and deep proteomic profiling of the 5xFAD hippocampus has revealed that elevated Pros1 is a novel biomarker for disease severity in this model24. It is therefore very possible that plaques are also decorated with Pros1.
In addition to ligands and receptors, TAM signaling requires binding of the phospholipid phosphatidylserine (PtdSer) to the amino-terminal ‘Gla’ domains of Gas6 and Pros13, 13. For example, a ‘Gla-less’ truncation mutant of Gas6 binds Axl with the same affinity as the full-length ligand, but is incapable of activating the receptor13. With respect to TAM receptor interaction, PtdSer is generally externalized on the surface of a cell – often an AC - that is apposed to a TAM-expressing cell1, 3. Although PtdSer is a component of every plasma membrane, in all normal cells it is enzymatically confined to the inner leaflet of the membrane bilayer3. Translocation of PtdSer to the plasma membrane surface is carried out by a set of PtdSer scramblases3 during apoptosis, and in response to Ca2+ entry, viral infection, and disease stressors3. During AC phagocytosis, Gas6 and/or Pros1 ‘bridge’ PtdSer that is externalized on the AC surface to TAM receptors expressed on the surface of microglia and other phagocytes3. To detect externalization of PtdSer in APP/PS1 mice, we stereotaxically injected their cortices with pSIVA, a ‘polarity-sensitive indicator for viability and apoptosis’ that fluoresces strongly only when bound to PtdSer25. The needle track of this injection (where cells were damaged) was strongly pSIVA+, but radiating out from the injection site we detected coincident pSIVA labeling of PtdSer on all 6E10+ Aβ plaques (Fig. 2c; Extended Data Fig. 2b). No pSIVA puncta were labeled outside the needle track after injection of WT brains (Extended Data Fig. 2c). Externalized plaque-associated PtdSer may mark the dystrophic neurites that are present in plaques and their immediate surroundings. Super-resolution microscopy suggested that Gas6 does indeed bridge PtdSer+ Aβ plaques to TAM+ microglia in the aged APP/PS1 brain (Extended Data Fig. 2d). Together, these results demonstrate that all of the essential components of the TAM system – the receptors, their ligand Gas6, and the essential co-factor PtdSer – are specifically up-regulated on and around Aβ plaques in AD.
TAM regulation of the microglial transcriptome in AD
To obtain a global perspective on how TAM signaling influences gene expression in PAM and NPAM, we performed comparative single cell RNA sequencing (scRNA-seq) of CD45+ cells sorted (Extended Data Fig. 3a) from plaque-burdened cortices at ~18mo, when plaques were abundant, using a 10X Genomics platform. Several prior RNA sequencing analyses have been performed with human and mouse microglia in neurodegenerative settings11, 21, 26, and these provide data for comparison with our results. We performed high-throughput single-cell transcriptomics of a total of 14,628 CD45+ immune cells from APP/PS1 and APP/PS1Axl−/−Mertk−/− cortices, and detected nine immune clusters by applying Seurat analyses27 (Extended Data Fig. 3b). Annotation using 18 marker genes (Extended Data Fig. 3c) allowed us to associate these clusters with specific cell types, of which microglia comprised the largest cluster. Also present in the CD45+ sort were NK and T cells, neutrophils, monocytes, and non-microglial macrophages, and smaller populations of B cells, oligodendrocytes, endothelial cells, and proliferating cells of various lineages (Extended Data Fig. 3b).
In a second clustering round, we identified seven microglial clusters, and numbered and staged these in the APP/PS1 and APP/PS1Axl−/−Mertk−/− replicates (Fig. 3a). Clusters were numbered based on transcriptomic state, with the largest cluster 0 (c0, Fig. 3a) representing the homeostatic microglial ground state. Mertk mRNA was distributed across all clusters, whereas Axl mRNA was largely restricted to clusters 4–6 (Extended Data Fig. 3d). Prominently expressed mRNAs in cluster 0 included Tmem119, Cx3cr1, P2ry12, and Maf (Fig. 3b, Extended Data Fig. 3e, f).
Fig. 3. TAM regulation of the microglial transcriptome in disease.
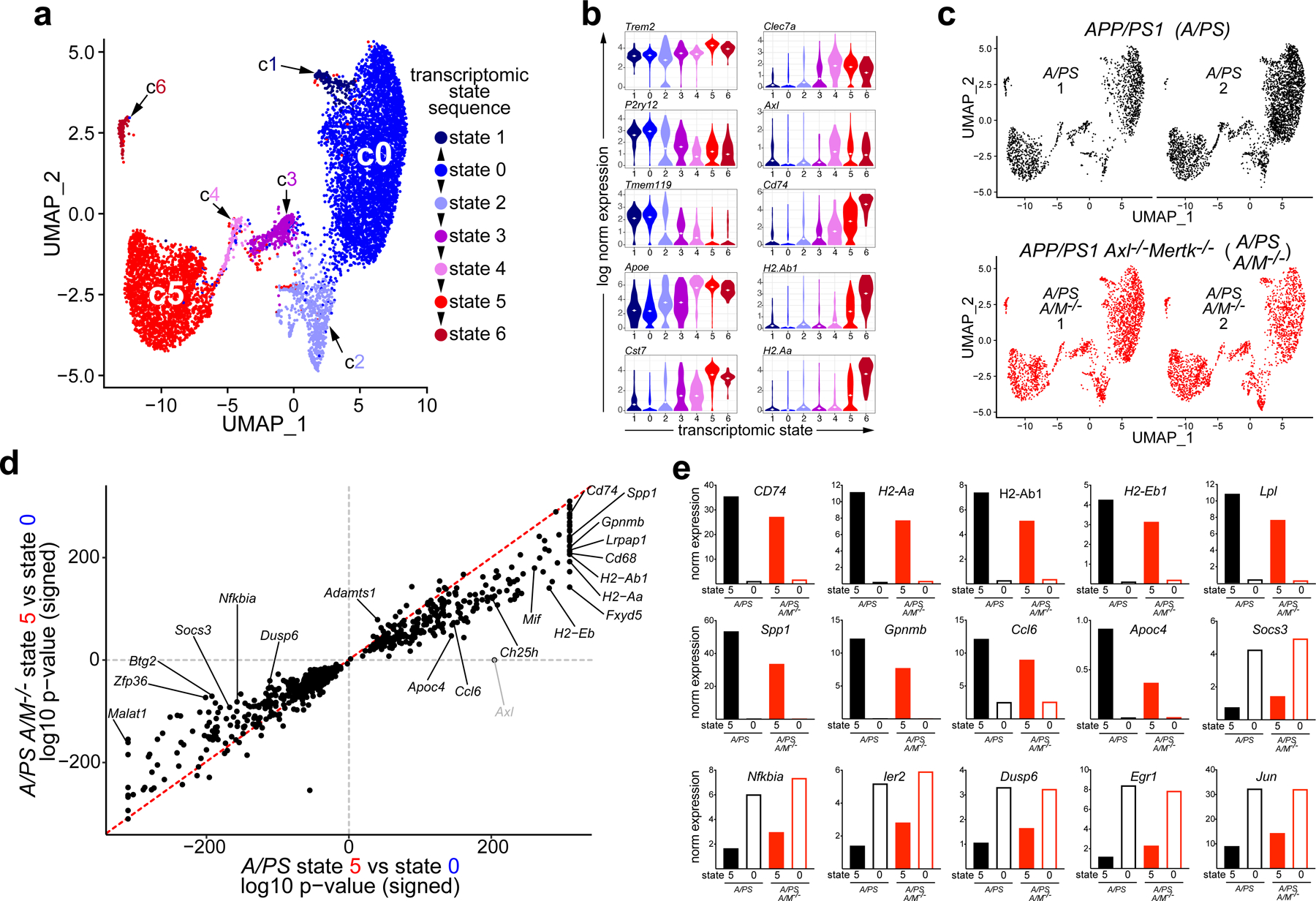
(a) Seven microglial cell clusters (c) defining seven transcriptomic states in combined APP/PS1 and APP/PS1 Axl−/−Mertk−/− microglia at 18 mo, displayed by uniform manifold approximation and projection (UMAP). c0 and c5 comprise major homeostatic (NPAM) and activated (PAM) transcriptomic states, respectively. (b) Violin plots for expression distribution of the indicated genes for the indicated microglial transcriptomic states, as defined in a. Dotted lines indicate mean. (c) UMAP for individual biological replicates of APP/PS1 (A/PS) and APP/PS1 Axl−/−Mertk−/− (A/PS A/M−/−) microglia yields the same cluster number and mapping for all replicates. (d) Scatter plot for signed log10 adjusted p-value of shared differentially expressed (DE) genes between state 5 vs state 0 in A/PS and A/PS A/M−/− microglia, respectively. Log10 adjusted p-value was signed by the up/down-regulation of the gene. Grey dot indicates where Axl would have been, since it was only differentially expressed in A/PS. (e) Normalized mean microglial expression of the indicated genes in transcriptomic state 0 versus 5, in A/PS (black) versus A/PS A/M−/− (red) microglia. DE analysis was performed by Seurat function “FindAllMarkers” and “FindMarkers” with default Wilcoxon Rank Sum test and logFC > 0.25 on pooled biological replicates. Genes with Bonferroni adjusted p-value < 0.05 were considered to be significant.
Cluster 1 microglia appear to represent a small one-way radiation from cluster 0, in which mRNAs indicative of immune stimulation, including Cst7, Axl, and type I interferon response genes (e.g., Ifitm3), were up-regulated (Fig. 3a, b). All remaining transcriptomic development from cluster 0 proceeded in order from clusters 2 through 6, with most microglia stopping at a fully activated cluster 5 (Fig. 3a, b; Extended Data Fig. 3e). By tracking microglial marker expression (Fig. 3b, Extended Data Fig. 3e, f), we determined that clusters 2, 3, and 4 represented cell sets in sequential transition states between the predominant clusters 0 and 5. These predominant clusters were largely equivalent to the homeostatic and ‘DAM’ cells, respectively, defined for 5xFAD microglia11 (Extended Data Fig. 3f). Although cluster 2 microglia exhibit a transcriptomic phenotype distinct from cluster 0, with diminished expression of Tmem119 and P2ry12 and increased expression of Apoe and Clec7a, among others (Fig. 3b), these cells mostly fell into the homeostatic cluster of Keren-Shaul et al.11 (Extended Data Fig. 3f). Similarly, cells in clusters 3 and 4 significantly but not entirely resembled those of their 5xFAD DAM stage 1, while cells in cluster 5 resembled those of DAM stage 211 (Extended Data Fig. 3f). RNAs abundant in cluster 5 cells included Cst7, Lpl, Trem2, and Csf1 (Extended Data Fig. 3e). The small cluster 6 appears to represent a further activation of cluster 5 in which microglia are primed for antigen presentation, in that cells in this cluster expressed elevated levels of mRNAs for the MHC class II trafficking adaptor CD74 and for multiple MHC class II proteins (Fig. 3b). This is of interest given evidence for T cell regulation of neuroinflammation in AD28.
Three features of microglial transcriptomic evolution in APP/PS1 versus APP/PS1Axl−/−Mertk−/− mice were apparent. First, we found that the same seven clusters were equivalently represented in the APP/PS1 and APP/PS1Axl−/−Mertk−/− replicates (Fig. 3c), indicating that Axl and Mer expression does not influence the evolution of microglial transcriptomic states in APP/PS1 mice. This is in contrast to the mutation of Trem2 in 5xFAD mice, which blocks the transition of cells to a maximally activated ‘DAM’ stage 2 state11. Importantly, one of the mRNAs prominent in DAM stage 2 is Axl, whose full microglial expression has been found to be Trem2-dependent11. While Trem2 protein is higher in PAM versus NPAM (Fig. 1d), Trem2 mRNA is only modestly increased in state 5 versus state 0 microglia (Fig. 3b), and expression of this mRNA is unaffected by mutation of Axl and Mertk.
Second, we observed that both the statistical significance and magnitude of induction of many mRNAs up-regulated in cluster 5 versus cluster 0 microglia was blunted upon mutation of Axl and Mertk (Fig. 3d, e, Extended Data Fig. 3g). This effect were seen for many DAM signature genes, including Spp1, Cd74, and Gpnmb. It was also apparent for genes involved in lipid metabolism (Lpl, Apoc4, Ch25h, Apoe) and MHC class II antigen presentation (H2-Aa, H2-Ab1, H2-Eb1, and Cd74), and for the ‘plaque-forming’ gene Ccl629. Although RNA for the phagocytosis regulator SIRPα was reduced in both transcriptomic states 5 and 0 in APP/PS1Axl−/−Mertk−/− microglia (Extended Data Fig. 3g), the principal phagocytic mediators effected by Axl and Mertk mutation were Axl and Mer themselves. (The Mertk mutation deletes an essential exon in the kinase domain and produces a protein null, but results in the synthesis of a non-functional mRNA that is detectable by PCR, RNase protection, and RNA-seq5, 30.) Conversely, we found that both the statistical significance (Fig. 3d) and magnitude (Fig. 3e, Extended Data Fig. 3g) of down-regulation of many genes reduced in cluster 5 relative to cluster 0 was dampened in APP/PS1Axl−/−Mertk−/− relative to APP/PS1 microglia. These included the homeostatic genes Malat1 and Nfkbia (Fig. 3d,e, Extended Data Fig. 3g).
Finally, we did not observe large-scale up-regulation of cytokine or chemokine mRNAs in state 5 versus state 0 microglia (Extended Data Fig. 3h), or in total mRNA purified from whole 12 mo cortex (Extended Data Fig. 3i), as a consequence of the combined loss of both microglial TAMs. Thus, it is unlikely that the phenotypes documented below result from major changes in APP/PS1 neuroinflammation consequent to Axl and Mertk mutation.
Microglial TAM activity at plaques
To determine if the TAM system perturbations documented above are of significance to microglial interaction with plaques, we used live two-photon imaging in APP/PS1 mice at 16 mo. We analyzed APP/PS1 hemizygotes that carried a single Cx3cr1GFP allele31 to label microglia, and then crossed these mice with both WT and Axl−/−Mertk−/− mutants. We labeled Aβ plaques using intraperitoneal injection of Methoxy-X0432 (MX04), a fluorescent amyloid dye, and then live-imaged 4–7 plaque-containing volumes per mouse in layers 1/2 of somatosensory cortex for ~90 min, using modifications of two-photon methods described previously7, 33. Representative time-lapse recordings of these analyses are shown in Supplementary Videos 1 and 2.
Quantification of microglial behavior allowed us to establish multiple differences in plaque recognition and interaction between WT and Axl−/−Mertk−/− microglia. First, we observed many fewer microglia encapsulating Aβ plaques in Cx3cr1GFPAPP/PS1Axl−/−Mertk−/− versus Cx3cr1GFPAPP/PS1 mice (Fig. 4a). Most plaques were enveloped by tightly bound and morphologically activated microglia in the latter (Fig. 4a, left), whereas plaques were often unattended by microglia in Cx3cr1GFPAPP/PS1Axl−/−Mertk−/− mice (Fig. 4a, right). Surface building reconstruction and quantification of imaging volumes (Fig. 4b) yielded a flat distribution of microglia cell body distances from plaques in Cx3cr1GFPAPP/PS1Axl−/−Mertk−/− mice, but showed a strong bias for microglia to be in contact with plaques in Cx3cr1GFPAPP/PS1 mice (Fig. 4c). This difference was independent of plaque size, although larger plaques were bound by more microglia in Cx3cr1GFPAPP/PS1 mice (Fig. 4c; Extended Data Fig. 4a).
Fig. 4. Microglia use Axl and Mer to detect, engage, and react to Aβ plaques.
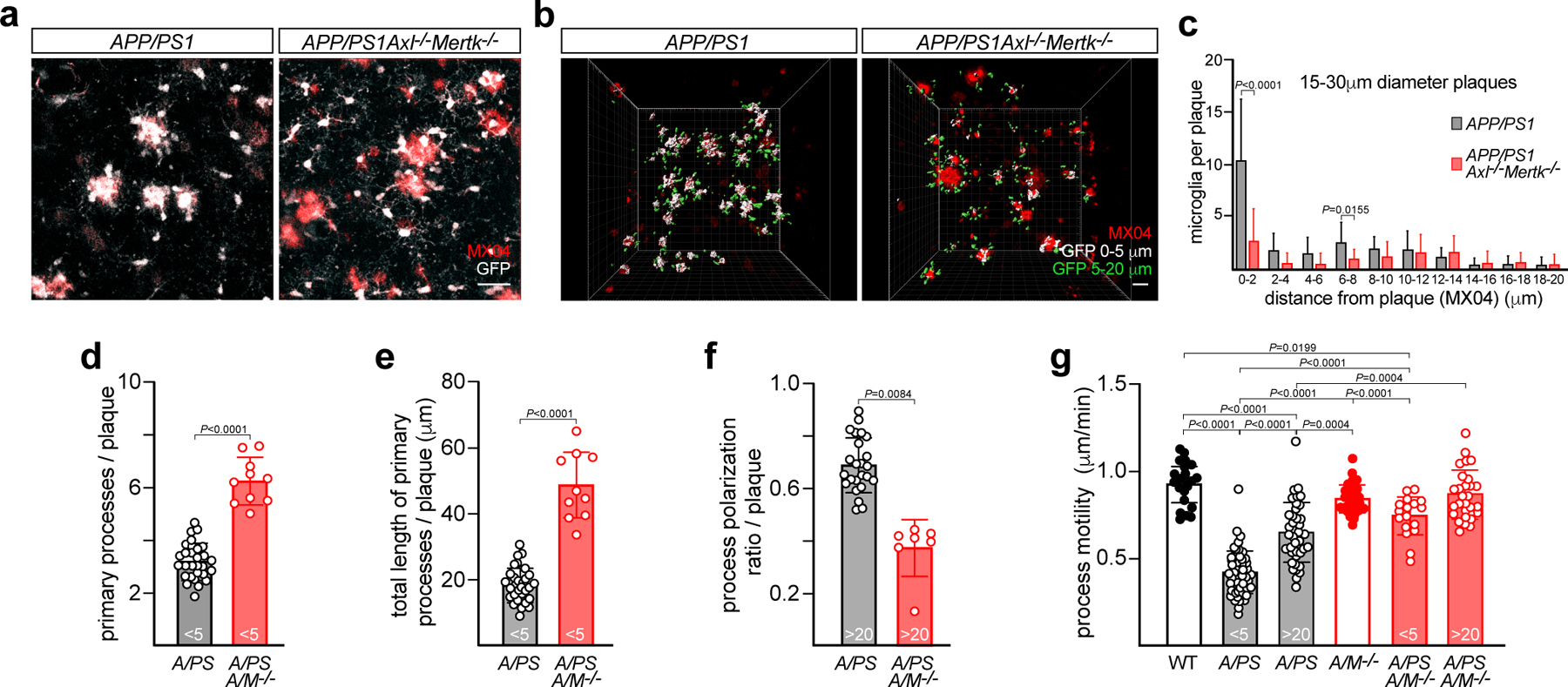
(a) Representative video stills from two-photon imaging of microglia (GFP signal, white) and amyloid plaques (MX04 signal, red) in 16 mo APP/PS1 and APP/PS1Axl−/−Mertk−/− cortex. (b) Imaris surface builds of microglial volumes at 0–5μm (white) and 5–20μm (green) from the edge of nearest plaque (red) in APP/PS1 and APP/PS1Axl−/−Mertk−/− cortex, representative of image volumes in cortices of n = 3 APP/PS1 and 5 APP/PS1 Axl−/−Mertk−/− mice. Scale bars: 30μm. (c) Distribution of distances of microglial cell body centroids from the edge of Aβ plaques in APP/PS1 and APP/PS1Axl−/−Mertk−/− cortex. 22 and 37 plaques were investigated from 3 and 4 mice for APP/PS1 and APP/PS1Axl−/−Mertk−/−, respectively. (d) Primary microglial processes per nearest plaque for microglia <5μm from plaques (PAM) in APP/PS1 (A/PS) and APP/PS1Axl−/−Mertk−/− (A/PS A/M−/−) cortex. (e) Summed length of primary microglial processes per nearest plaque for PAM in APP/PS1 and APP/PS1Axl−/−Mertk−/− cortex. (f) Process polarization to nearest plaque (ratio of summed length of primary processes oriented toward plaque to summed length of all primary processes) for NPAM (microglia >20μm from plaques) in APP/PS1 and APP/PS1Axl−/−Mertk−/− cortex. (g) Process motility for cortical PAM, NPAM, and non-diseased microglia in mice of the indicated genotypes. Data points are from 7–29 representative plaques from n = 3 mice from both genotypes (d–f), and 18–52 microglia from 2 (WT and Axl−/−Mertk−/−) and 3 (APP/PS1 and APP/PS1Axl−/−Mertk−/−) mice (g). Data represented as mean ±1 STD. Mann-Whitney test (d-f) and Kruskal Wallis test followed by Dunn’s multiple comparison test (g).
Similarly, there were obvious differences in microglial morphology and motility (Fig. 4a, d–g). Cx3cr1GFPAPP/PS1Axl−/−Mertk−/− microglia often displayed a ramified ‘resting’ configuration that was similar to that seen in WT mice (Fig. 4a, right; Supplementary Video 1). In contrast, Cx3cr1GFPAPP/PS1 microglia displayed an activated ‘amoeboid’ morphology, with larger cell bodies and fewer, shorter processes (Fig. 4a, left; Supplementary Video 1). Cx3cr1GFPAPP/PS1Axl−/−Mertk−/− PAM elaborated significantly more (Fig. 4d, Extended Data Fig. 4b) and longer (Fig. 4e, Extended Data Fig. 4c) primary processes than Cx3cr1GFPAPP/PS1 PAM.
Processes from microglia positioned >20μm from plaques (NPAM) were strongly oriented toward plaques in the Cx3cr1GFPAPP/PS1 cortex, but showed no orientation bias in the Cx3cr1GFPAPP/PS1Axl−/−Mertk−/− cortex (Fig. 4f, Extended Data Fig. 4d). In addition, microglial process motility – a reduction in which is indicative of microglial activation34 - was significantly reduced in APP/PS1 mice relative to WT (Fig. 4g). In contrast, process motility was only modestly lower than WT for APP/PS1Axl−/−Mertk−/− PAM (microglia <5 μm from the nearest plaque) (Fig. 4g), and was not statistically different from WT for APP/PS1Axl−/−Mertk−/− NPAM (Fig. 4g). Finally, AD microglia normally proliferate near plaques. Mutation of Trem2 in the 5xFAD mouse model blunts this microgliosis ~2-fold35, 36, and we measured a similarly blunted proliferation of microglia, primarily those in close proximity to plaques, in the cortex of Cx3cr1GFPAPP/PS1Axl−/−Mertk−/− relative to Cx3cr1GFPAPP/PS1 mice (Extended Data Fig. 4e). Thus, by all of the above measures, TAM-deficient microglia were strikingly deficient in their ability to detect, bind, and respond to amyloid plaques.
TAM-dependent phagocytosis and shaping of plaques
Microglia are vigorous phagocytes that exhibit an absolute TAM receptor requirement for AC engulfment7. Since TAM-mediated phagocytosis of ACs is entirely dependent on PtdSer externalization on the AC surface3, 13, and since all Aβ plaques are decorated with both PtdSer (Fig. 2c) and Gas6 (Fig. 2a, b), we used two-photon imaging to measure engulfment of MX04-labeled amyloid plaque material into GFP+ microglia that were either WT or Axl−/−Mertk−/−. We quantified internalized MX04 signal both per normalized volume and per GFP volume. These measurements revealed a clear deficit: we quantified ~10-fold lower levels of engulfed MX04-labelled plaque material inside of Axl−/−Mertk−/− as compared to WT microglia by both measures (Fig. 5a, b). Note that internalized amyloid material, which may be routed to acidic lysosomes, occupies a remarkable 9% of WT microglial volume in APP/PS1 mice (Fig. 5b). These data indicate that microglia cannot effectively phagocytose amyloid material without TAM receptors.
Fig. 5. TAM-deficient microglia can neither phagocytose nor organize plaques.
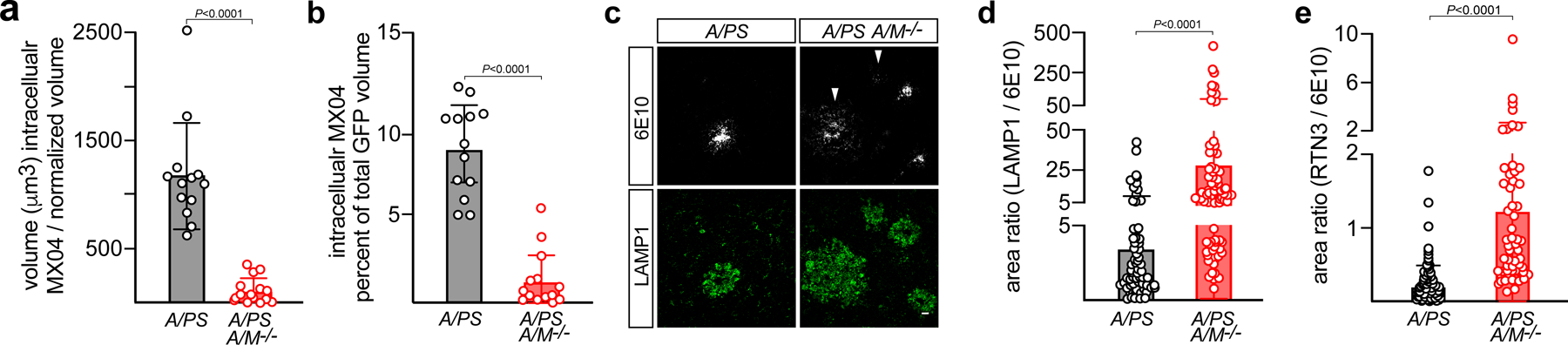
(a, b) MX04-labeled Aβ plaque material engulfed within GFP+ microglia, imaged in vivo, in 16 mo. APP/PS1 (A/PS) versus APP/PS1Axl−/−Mertk−/− (A/PS A/M−/−) cortex, normalized to imaging volume (a) and the volume of GFP+ cells (b). (c) Representative images of the halo of LAMP1+ dystrophic membranes (green, lower panels) that surround 6E10+ plaques in 12 mo APP/PS1 (A/PS; left) versus APP/PS1Axl−/−Mertk−/− (A/PS A/M−/−; right) cortex. Arrowheads mark weakly-staining, diffuse 6E10+ plaques, which are more common in the APP/PS1Axl−/−Mertk−/− brain (see also Extended Data Fig. 5). Scale bar: 10μm. (d) Quantification of the ratio of LAMP1+ area to 6E10+ plaque area across all plaque sizes, both dense-core and diffuse. (e) Quantification of ratio RTN3+ area to 6E10+ plaque area across all plaque sizes, both dense-core and diffuse. Data are 13–15 volumetric images from n= 3 and 4 mice for APP/PS1 and APP/PS1Axl−/−Mertk−/−, respectively (a–b). Data are 94–113 plaques (d) and 56–95 plaques (e) investigated from N ≥ 3 sections per mouse from n = 3 mice of each genotype. Mann-Whitney test (a, b, d, e). Data represented as mean ±1 STD.
Amyloid plaques are typically surrounded by a halo of dystrophic neuronal membranes37. This halo is shaped by microglial phagocytic activity37, and is marked by the expression of several proteins, including the autophagic and endo-lysosomal vesicular marker LAMP138 (examples in Fig. 5c). When we measured the area ratio of LAMP1+ membranes to 6E10+ plaques (examples in Extended Data Fig. 5), we quantified a 10-fold increase in this ratio in APP/PS1Axl−/−Mertk−/− versus APP/PS1 mice at 12 mo (Fig. 5d). This difference was primarily driven by an expansion of LAMP1 around weakly-staining, diffuse plaques (arrowheads in Fig. 5c), but was also seen for dense-core plaques (Extended Data Fig. 6a, b). A second protein that displays a similar but more circumscribed distribution in dystrophic membranes is the endoplasmic reticulum protein reticulon-3 (RTN3)39. When we performed an area ratio analysis for RTN3 surrounding 6E10-labeled plaques, we again measured a large increase in this ratio in APP/PS1Axl−/−Mertk−/− mice (Fig. 5e). This difference was also driven by a large expansion of RTN3 around weakly-staining, diffuse plaques, but was also seen for dense-core plaques (Extended Data Fig. 6c, d). Inspection of 6E10+ profiles suggested that diffuse, poorly-organized 6E10+ plaques were more common in APP/PS1Axl−/−Mertk−/− than APP/PS1 mice (Extended Data Fig. 5), and quantification confirmed this impression (Extended Data Fig. 6e).
We found that these multiple plaque perturbations were accompanied a ~2-fold increase, relative to APP/PS1, in the deposition of Aβ within and surrounding blood vessels in the APP/PS1Axl−/−Mertk−/− brain, indicative of cerebral amyloid angiopathy (CAA)40 (Extended Data Fig. 6f, g). This increased CAA, which is a common feature of AD40, is of interest given that a similar increase has been seen when microglia are depleted in 5xFAD mice29. Finally, and in keeping with the phagocytic deficits documented above, we always detected uncleared cCasp3+ apoptotic debris surrounding 6E10+ plaques in aged APP/PS1Axl−/−Mertk−/− mice, but never in APP/PS1 mice (Extended Data Fig. 6h).
Dense-core plaque burden in the absence of TAM signaling
Microglial phagocytosis has heretofore been thought to inhibit the growth of plaques8. Thus, a prediction from the relative inability of APP/PS1Axl−/−Mertk−/− microglia to engulf Aβ material is that aged TAM-deficient AD mice should display a much higher plaque burden than AD mice with WT microglia. Our initial indication that this was not the case came from the APP/PS1Axl−/−Mertk−/− two-photon volumes we recorded, most of which contained fewer MX04-labeled plaques than APP/PS1 volumes with WT microglia (examples in Fig. 6a). To measure plaque burden in these populations, we used semi-automated quantification of Thioflavin S (Thio S)-stained dense-core plaques in whole-brain sections to assess the effect of Axl and Mer deletion on dense-core plaque accumulation, size, and number across disease development. We also quantified dense-core plaques in APP/PS1Axl−/− and APP/PS1Mertk−/− single compound mutants at 12 mo. Representative Thio S-stained sagittal sections of APP/PS1 and APP/PS1Axl−/−Mertk−/− brains at 12 mo are shown in Fig. 6b.
Fig. 6. TAM-driven microglial phagocytosis favors dense-core plaque formation.
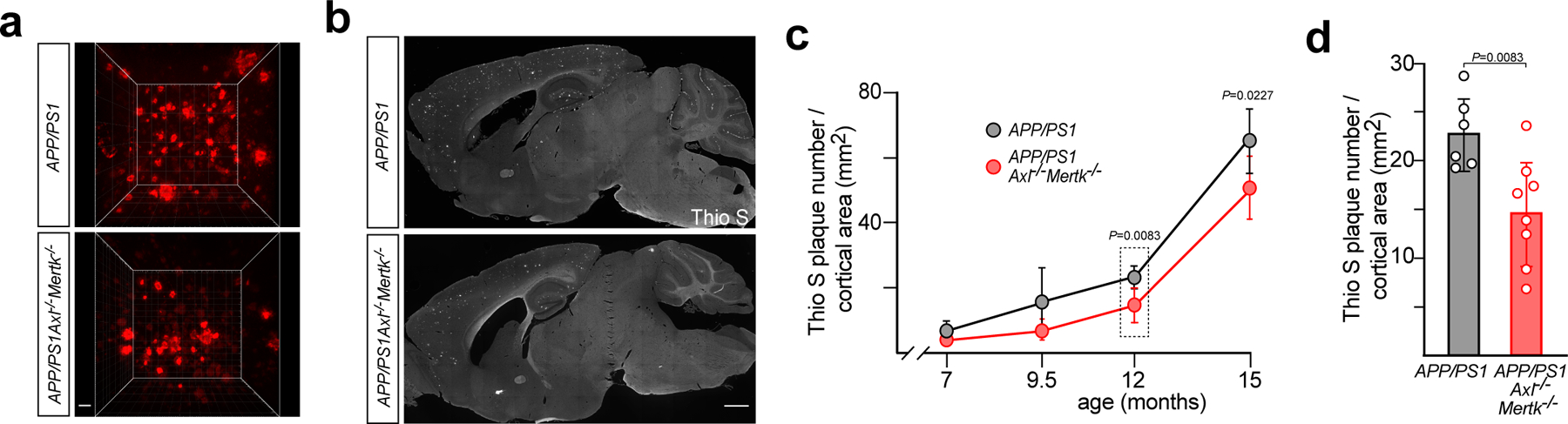
(a) Representative two-photon imaging volumes (X and Y 350μm, Z 300μm) of MX04-labeled plaques in 16 mo APP/PS1 and APP/PS1Axl−/−Mertk−/− cortex, as observed in image volumes in cortices of n = 3 (APP/PS1) and 5 (APP/PS1 Axl−/−Mertk−/−) mice. Scale bar: 30μm. (b) Representative sagittal sections from 12 mo APP/PS1 and APP/PS1Axl−/−Mertk−/− brains from at least 3 independent replicates, stained for dense-core plaques with thioflavin S (Thio S), quantified in d and Extended Data Fig. 7a, b. Scale bar: 1mm. (c) Quantification (see Methods) of Thio S-labeled dense-core Aβ plaque density, for cross-sectional plaque areas of all sizes, in APP/PS1 (gray) and APP/PS1Axl−/−Mertk−/− (red) cortex over time. Boxed data at 12 mo are detailed in d. n=4–8 per genotype per time point. (d) Thio S-labeled plaque density in cortex (of APP/PS1 (gray) and APP/PS1Axl−/−Mertk−/− (red) mice at 12 mo. Data points represent plaque density in n= 6 APP/PS1 and 8 APP/PS1Axl−/−Mertk−/− mice of the indicated genotypes averaged from N = 5 cortical sections of each brain. Student’s t-test (c, d). Data represented as mean ±1 STD.
Quantification revealed that, from initial plaque appearance to profuse deposition, APP/PS1Axl−/−Mertk−/− cortices displayed many fewer, not many more, dense-core plaques than APP/PS1 cortices (Fig. 6c). At 12 mo, APP/PS1Axl−/−Mertk−/− mice showed a 35% reduction in cortical Thio S+ plaque density relative to APP/PS1 (Fig. 6d). This reduction was seen across all plaque sizes, but was most pronounced for smaller plaques (Extended Data Fig. 7a). A similar reduction was seen in the hippocampus (Extended Data Fig. 7b). Importantly, these reductions in dense-core plaque density were not due to any change in the production of Aβ peptide, since soluble Aβ levels in the cortex and hippocampus at 4 and 12 mo were not significantly different between APP/PS1 and APP/PS1Axl−/−Mertk−/− mice (Extended Data Fig. 7c). Similarly, no change was apparent between these genotypes in cortical expression of the amyloid precursor protein (APP) (Extended Data Fig. 7d).
When we quantified dense-core plaque burden in the cortex and hippocampus of single compound mutants at 12 mo, we found that nearly all of the effect observed in the double mutants was due to mutation of Mer alone. APP/PS1Mertk−/− mice displayed reductions in dense-core plaque burden that largely phenocopied those seen in APP/PS1Axl−/−Mertk−/− mice (Extended Data Fig. 7e), whereas APP/PS1Axl−/− mice, while exhibiting a trend toward reduced plaque density in the hippocampus, displayed cortical plaque burdens that were equivalent to APP/PS1 (Extended Data Fig. 7f). Thus, while Axl is up-regulated in plaque-associated microglia, most of the phagocytosis-dependent construction of dense-core plaques is performed by Mer, the receptor that is constitutively present in microglia. Together, these data demonstrate that TAM-mediated phagocytosis promotes the deposition of dense-core Aβ plaques (Extended Data Fig. 7g).
Consistent with the findings of equivalent APP and Aβ levels in APP/PS1 and APP/PS1Axl−/−Mertk−/− mice, we detected equivalent reductions in an excitatory synaptogenesis index – the physically paired expression of the pre- and post-synaptic markers vGlut1 and PSD9541 – between these genotypes and WT mice within CA1 of the 15 mo hippocampus (Extended Data Fig. 8a). When we performed a fear conditioning assay assessing cognitive deficits in WT, Axl−/−Mertk−/−, APP/PS1, APP/PS1Axl−/−Mertk−/− mice, we detected a statistically significant deficit in fear acquisition in APP/PS1Axl−/−Mertk−/− relative to APP/PS1 mice (Extended Data Fig. 8b, c), but no significant difference in contextual fear memory between these genotypes (Extended Data Fig. 8d).
DISCUSSION
Our results lead to two principal conclusions. First, the TAM system is required for microglial recognition of, response to, and phagocytosis of Aβ plaques. And second, TAM-mediated microglial phagocytosis of Aβ material does not inhibit, but rather promotes, the formation of dense-core plaques.
The first of these conclusions is consistent with prior knowledge as to TAM action in microglia and other macrophages3, 5, 7. All Aβ plaques are decorated with externalized PtdSer, an essential TAM co-ligand13, and this PtdSer is bound to the amino-terminal ‘Gla’ domain of Gas613. The carboxy-terminal ‘SHBG’ domain of this ligand1, 13 is then bound to Axl and Mer on microglia. In this arrangement, the TAM ligand serves as a bridge between microglia and an Aβ plaque in the same way that TAM ligands bridge macrophages to PtdSer-expressing ACs during engulfment1, 3, 4, 5, 7, 13. In the absence of Axl and Mer, microglia display a diminished transcriptomic response to plaques, are compromised in their attachment to plaques, do not re-orient their processes toward plaques, and are blunted in their proliferative, process extension, and process motility responses to plaques. Most strikingly, Axl−/−Mertk−/− microglia cannot effectively phagocytose Aβ plaque material.
There are some similarities between these phenotypes and those seen when Trem2−/− mice are crossed into amyloidogenic AD mouse models22, 35, 36, 42, 43, 44 – but in general the TAM phenotypes are far stronger. The closest similarities are seen with respect to microglial proliferation and plaque binding. Microglial proliferation around plaques is reduced ~2-fold in the Trem2−/− compound mutants36, 42 and also ~2-fold for Axl−/−Mertk−/− compound mutants. Similarly, the number of microglia bound per plaque is reduced 2–4-fold in the Trem2−/− compound mutants35, 36, 42, 43 and ~5-fold in the Axl−/−Mertk−/− compound mutants. Loss of Trem2 from the APP/PS1–21 line results in a ~3.5-fold increase in the volume of LAMP1+ membranes surrounding ‘filamentous’ plaques and a ~1.5-fold increase for compact plaques in late-stage disease44, whereas these numbers for the APP/PS1Axl−/−Mertk−/− mice are ~8-fold and ~4-fold at a comparable stage of disease. Since dystrophic neurites arise around plaques devoid of microglial coverage37, the paucity of microglial plaque association seen in APP/PS1Axl−/−Mertk−/− mice may result in significant collateral axonal damage.
There is no consensus among research groups as to the effect of Trem2 deletion on Aβ plaque burden in amyloidogenic AD models22, 42, with different groups reporting either marginal increases or decreases, or no changes, in plaque density (measured with Thio S, 6E10, or amyloid dyes) that vary with the model employed, age analyzed, and brain region assessed. In contrast, APP/PS1Axl−/−Mertk−/− mice display a consistent reduction in dense-core plaque density, in both the cortex and the hippocampus, across the full course of disease in APP/PS1 mice. Most strikingly, Trem2−/− microglia exhibit a modest ~2-fold reduction in their ability to phagocytose MX04-labeled Aβ material36 and display normal phagocytosis of ACs in in vitro assays35, whereas Axl−/−Mertk−/− microglia exhibit a 10-fold deficit in their ability to phagocytose MX04-labeled Aβ material in vivo, and are incapable of any AC phagocytosis in vitro7. Together, these results argue that Mer and Axl are the main receptors that microglia use to detect, engage, and engulf amyloid plaques. Indeed, it is possible that some of phenotypes observed in Trem2−/− mice are in part TAM-mediated, since scRNA-seq analyses have indicated that up-regulation of Axl mRNA in the 5xFAD model is Trem2-dependent11.
The second of our principal conclusions is contrary to expectation. How does a 10-fold reduction in phagocytic capacity, coupled with a 2-fold reduction in microglial numbers and a 5-fold reduction in plaque binding, result in 35% fewer dense-core plaques? This finding should be considered in light of two sets of prior analyses in 5xFAD mice29, 45, 46. First, using two-photon imaging methods similar to ours, Baik and colleagues found that microglial deposition of previously-phagocytosed Aβ material, via exocytosis or microglial death, was essential for the growth of dense-core plaques45. An acidic environment, as is found in the lysosomes into which engulfed Aβ material is deposited47, is known to promote the formation of densely packed, protease-resistant Aβ fibrils45, 48. As noted above, we find that internalized Aβ material accounts for nearly 10% of WT microglial volume in APP/PS1 mice, and studies in another amyloidogenic model have reported even larger Aβ-laden lysosomal volumes within these cells29. This suggests that once routed to lysosomes, Aβ fibrils are compacted into dense-core material that is ‘indigestible’.
A second key observation is provided by studies in which microglia were killed by pharmacological inhibition of the CSF1 receptor29, 46. These studies demonstrated that plaques never appear in the 5xFAD brain when microglia are killed, except in the limited regions where microglia are spared from death29. Together, these findings and our results suggest that dense-core Aβ plaques do not form spontaneously, but are instead constructed from loosely-organized Aβ material by phagocytic microglia, and that TAM receptors are key components of the molecular machinery through which this is achieved. They are consistent with the hypothesis that dense-core plaques may represent a macrophage-mediated confinement mechanism, perhaps analogous to the granulomas of tuberculosis and other infections49, which limits the dissemination of toxic pre-plaque Aβ oligomers throughout the brain. This may in part explain why agents that disaggregate dense-core plaques but do little to alter the production of Aβ peptides have largely failed as AD therapeutics8, 50.
Methods
Mice.
C57BL/6J wild-type mice were obtained from The Jackson Laboratory. Mice were typically group-housed at approximately 22 degrees Celsius and provided with bedding and nesting material. All animals were maintained on a 12 hr light/dark cycle and given ad libitum access to standard rodent chow and water. The Axl−/30, Mertk−/−30, Axl−/−Mertk−/−30, and Cx3cr1GFP/+31 strains have been described previously. B6.Cg-Tg(APPSwePSEN1dE9) hemizygous mice16, 17 (APP/PS1) (JAX number: 005864) were crossed with Axl−/−, Mertk−/− and/or Axl−/−Mertk−/− lines to generate APP/PS1 Axl−/−, APP/PS1Mertk−/−, and APP/PS1Axl−/−Mertk−/− mice. For two-photon microscopy, APP/PS1 mice were crossed with Cx3cr1GFP/GFP or Cx3cr1GFP/GFP Axl−/−Mertk−/− strains. Only female APP/PS1Cx3cr1GFP/+ WT or Axl−/−Mertk−/− and healthy littermates were used in two-photon studies to avoid potential gender biases in pathology and microglial responses. APP41 mice, which express a ‘Swedish’ + ‘London’ (V717I) mutant human APP under the Thy-1 promoter 18, 51, 52, were a kind gift of Drs. Kuo-Fen Lee and Jiqing Xu. All lines have been backcrossed for >10 generations to and maintained on a C57BL/6 background. All animal procedures were conducted in compliance with the ethical guidelines and according to protocols approved by the Salk Institute Animal Care and Use Committee (Protocol No. 17–00009). Mice of both genders were randomly allocated to experimental groups unless otherwise noted.
Reagents and antibodies.
Antibodies used were as follows: anti-Axl (R&D AF854, 1:50), anti-Mer (eBioscience DS5MMER and R&D AF591, 1:200), anti-mouse Gas6 (R&D AF986, 1:50), anti-human Gas6 (R&D AF885, 1:25), anti-beta-amyloid, clone 6E10 (Biolegend 803001, 1:500), anti-Iba1 (Wako 019-19741 and Novus NB100-1028, 1:250 and 1:200, respectively), anti-GFAP (Dako z-334, 1:500), cleaved Casp3 (Cell Signaling 9661, 1:200), anti-CD68 (BioRad MCA1957, 1:200), anti-RTN3 (EMD Millipore ABN1723, 1:500), anti-Tmem119 (Abcam AB209064, 1:200), anti-LAMP1 (BD Biosciences, Cat# 553792, clone 1D4B, 1:200), and anti-Trem2 (R&D systems, AF1729 and BAF 1729, 1:200 and 1:50 respectively), anti-CD31 (R&D, AF3628, 1:50), anti-laminin (Sigma, L-9393, 1:500). For flow cytometry, anti-mouse CD16/32 (Biolegend 101301, 1:100), anti-CD45-PE, 2D1 (Invitrogen 12-9459-42, 1:25), anti-CD11b-FITC, M1/70 (Invitrogen 11-0112-85, 1:25). Anti-vGlut1 (Millipore, AB5905), and anti-PSD95 (Life Technologies, 51-6900) were kind gifts of the Nicola Allen lab. Secondary antibodies for immunohistochemistry were all used at 1:500 and were fluorophore-conjugated anti-rat (712- 545- 153 or 712- 165- 153 from Jackson ImmunoResearch), anti-goat (A-11056 from Life Technologies, or 705-166-147 from Jackson ImmunoResearch), anti-rabbit (A-11071 or A-21206 from Life Technologies), anti-sheep (A21098 from Thermo Fisher Scientific), anti-mouse (A-11029 from Life Technologies, 715-166-150 or 715- 176- 150 from Jackson ImmunoResearch), and streptavidin 555 (Invitrogen S21381). For immunoblotting, anti-6E10 (see above, 1:1000) and anti-GAPDH (Millipore MAB374, 1:10000) as loading control. For Li-Cor detection, IRDye 680RG IgG secondary antibodies of the corresponding species were used. Antibodies used in the quantification of Aβ42 peptides by ELISA were obtained as a kind gift from Dr. Marc Mercken from Janssen Pharmaceuticals: JRF AB042/26 for Aβ42 and detection antibody JRF/AbN/25, unlabeled. Key reagents and their usage were as follows: Thio S (Acros organics 213150250); Hoechst 33258 in a 1% solution of bisbenzimide in water (Sigma Aldrich B-2883); TrueBlack® Lipofuscin Autofluorescence Quencher (Biotium #23007). For biotinylation of detection antibody JRF/AbN/25 (EZ link Sulfo-NHS-LC Biotin, Thermo Fisher, Cat #21217) and subsequent dialysis (Slide-A-lyzer dialysis cassette, Pierce 0728, prod # 66415). Synthetic human Aβ1–42 peptides for ELISA standards (Anaspec Cat # 20276). pSIVA-IANBD™ was included in the pSIVA apoptotic detection kit (Novus Bio NBP2-29382) originally described in Kim et al.25. For in vivo amyloid plaque labeling, MX04 (Tocris 4920) was prepared and used as previously described32, following the manufacturer’s instructions. Stock MX04 was first dissolved in 100% DMSO then injected i.p. at a final concentration of 10 mg/kg in 10% DMSO, 45% propylene glycol, 45% PBS, pH 7.5) 24 h prior to imaging.
Immunohistochemistry and histochemistry.
Routine perfusion procedures were performed7. Briefly, P30 and adult mice (2.5 −16 mo) were anesthetized with a final concentration of 100mg/kg Ketamine/10mg/kg Xylazine in accordance with IACUC guidelines. Mice were then transcardially perfused with 20U/ml heparin in PBS followed by freshly prepared 4% PFA in PBS. Both hemispheres were post-fixed in 4% PFA in PBS overnight at 4°C and subsequently infiltrated in 30% sucrose in PBS for 1 day at 4°C and flash frozen in TBS tissue freezing medium. The left hemisphere was sagittally cryo-sectioned at 15μm, air-dried overnight and subsequently processed for staining. Antigen retrieval with citrate buffer was performed if needed by heating brain sections to 80°C for 3 min in pre-warmed citrate buffer and cooling to room temperature. Non-specific binding was blocked by 1h incubation in blocking buffer (PBS containing 0.3% Triton-X100, 0.1% Tween-20, 5% donkey serum and 2% IgG-free BSA). Sections were incubated overnight at 4°C with primary antibody diluted in blocking buffer, then washed in PBS 0.1% Tween-20, and incubated for 2 h at 22–24 °C in the dark with fluorophore-coupled secondary antibodies and subsequently in Hoechst for 5 min diluted in blocking buffer. Sections were washed, sealed with Fluoromount-G (SouthernBiotech) and stored at 4°C.
Cortical and hippocampal plaque density was quantified by staining with the fluorescent amyloid dye thioflavin S (Thio S). Thio S binds to the β pleated sheets of dense-core senile amyloid plaques, and provides for more reliable semi-automated quantification of plaques than immunostaining with 6E10. A serial set of 15μm sagittal sections (5 sections/mouse) were obtained 0.25mm-0.85mm from the midline, each spaced 0.15mm apart. Plaque abundance was quantified by histochemistry with Thio S staining using published protocols. Image analysis of the number of cortical and hippocampal Thio S+ plaque was done blind with respect to genotypes and in a semi-automated fashion with set intensity thresholds for all conditions, using the ‘analyze particles’ function on ImageJ or FIJI software. For evaluating pSIVA-injected APP/PS1 and WT mice, a serial set of 15μm sagittal sections were obtained between 1mm-2mm from the midline (0.5mm before and after the corresponding pSIVA injection sites, 15 sections/mouse) from both injected and un-injected control hemispheres. Serially cut sections from both hemispheres and genotypes (APP/PS1 vs WT) were further immunostained with 6E10 antibody and evaluated for co-localization of fluorescence emitted from pSIVA and amyloid.
Human postmortem AD brain sections.
Paraffin-embedded thin sections of postmortem human brain tissues were obtained from the UCSD Alzheimer’s Disease Research Center (ADRC), courtesy of Dr. Robert Rissman. The postmortem tissues used in this project were collected from the UCSD ADRC neuropathology core. All participants consented to brain donation at the time of enrollment in the ADRC. Three age-matched pairs of AD BRAAK stage 6 and clinically normal brain sections were evaluated. Sections underwent standard deparaffinization and permeabilization with .3% Triton-X100 before treated for 5min with 1X TrueBlack in 70% Ethanol. Subsequent immunostaining for human Gas6 and Aβ plaques using the antibodies detailed above was done with the standard immunofluorescence protocol detailed above without detergents in the buffers.
Isolation and purification of brain immune single cells for sequencing.
Mature APP/PS1 or APP/PS1 Axl−/−Mertk−/− mice (18 mo) were transcardially perfused with ice cold D-PBS containing Ca2+ and Mg2+, and brains were promptly dissected out and placed on pre-chilled petri dishes on ice to extract cortices. Cortices from two mice were pooled for each sample and two biological samples per genotype were included in our study. All steps were carried out on ice, unless otherwise specified. Pooled cortices were minced with a razor blade, suspended in 5ml D-PBS in 15ml tubes for tissue chunks to settle before removal of the supernatant. Single cell suspensions were prepared following a modified version of the Neural Tissue Dissociation kit from Miltenyi Biotec (130-094-802). Briefly, for each sample, an enzyme mixture 1 containing 50μl enzyme P in 1910μl buffer Z supplemented with 0.2μg DNase (Sigma) and an enzyme mixture 2 containing 30μl Buffer Y and 15μl of enzyme A were prepared and these enzyme mixtures were kept on ice before usage. The settled tissue pellet was resuspended in mixture 1 and horizontally agitated at room temperature for 15 min. Thirty μl of enzyme mixture 2 was added before mechanical dissociation on ice using a fire-polished Pasteur pipette. The cell suspension was further incubated at room temperature with constant rotation for 10min, after which the remaining mixture 2 was added. Samples were subjected to a second round of mechanical dissociation on ice with fire-polished Pasteur pipettes with decreasing diameter until no observable tissue pieces remained. The resulting cell suspensions were filtered using 70μm cell strainers, which were subsequently washed with 10ml D-PBS supplemented with 0.5% BSA. The cells were centrifuged at 300g for 5min at +4°C and pellets were suspended in 10ml 30% isotonic Percoll (GE Healthcare 17-0891-01) diluted in 1XHBSS and then centrifuged again at 700g for 15min at +4°C with minimum acceleration and braking. The bottom 5ml of the gradient containing microglia was collected, resuspended and passed through a 70μm cell strainer. The cell suspension was washed in ice-cold 1XHBSS for a total volume of 40ml and pelleted at 300g for 10min at +4°C.
Flow Cytometry and Fluorescence Activated Cell Sorting (FACS).
Dissociated cells were resuspended in 300 μl of FACS buffer (2% FBS and 1mM EDTA in D-PBS, sterile). Fluorescence labeling procedures were then carried out on ice. Fc-receptors were blocked by addition of anti-CD16/32 antibody (1:100 dilution, Biolegend) for 15 min followed by the addition of labelled antibodies: anti-CD45-PE (1:25, Biolegend), anti-CD11b-FITC (1:25, Biolegend) and Hoechst 33258 (1:1000) for 1 hour. Finally, samples were washed twice with FACS buffer and immediately taken to the Salk Institute Flow Cytometry core facility (samples were kept on ice from this point onwards, or chilled at 4C while undergoing FACS purification).
FACS purification was carried out on a BD FACS Aria Fusion sorter with 1x PBS for sheath fluid. For high viability, concentrated cells suitable for downstream 10X Genomics analysis, an 85-μm nozzle was used with sheath pressure set to 45PSI. Live cells were gated first (Hoechst dye negative), followed by exclusion of debris using forward and side scatter pulse area parameters (FSC-A and SSC-A), exclusion of aggregates using forward and side scatter pulse width parameters (FSC-W and SSC-W), before finally gating on CD45+ cells to be isolated. Cells were purified using a 1-drop single cell sort mode (for counting accuracy). These were directly deposited into a 1.5 ml Eppendorf tube without additional buffer to yield a sufficient concentration that permitted direct loading onto the 10X chip.
Single cell sequencing.
Single cell RNA Sequencing was conducted on FACS sorted brain immune cells following the user guide on the Next GEM single cell 3’ v3.1 protocol. Briefly, single cell suspensions of approximately 14,000–16,000 cells from each sample was directly loaded onto microfluidic chip with barcoded beads to generate Gel Bead-in-Emulsions (GEMs) using 10X Genomics Chromium Single Cell Controller. Reverse transcription of GEMs for first strand cDNA synthesis and cDNA amplification were carried out according to 10X Next GEM single cell 3’ v3.1 protocol. Following indexed scRNA-Seq library construction, the final library size distribution was determined using TapeStation (Agilent) and the concentration was measured by a Qubit fluorometer (ThermoFisher). The libraries were pooled in equal molar ratio, quantified by qPCR, and sequenced on Illumina NextSeq500 at 28 cycles for Read 1, 8 cycles for i7 index and 91 cycles for Read 2 at an average sequencing read depth of 33k-41k reads per cell.
Analysis of single cell RNA-seq.
Raw sequencing data were aligned to the reference built using Ensembl primary assembly and annotation (release-93) that included both protein coding genes and polymorphic pseudogenes by cell ranger pipeline (v3.1.0). The resulting filtered gene expression matrix was further analyzed by “Seurat” (3.2.1.9002)53. Cells that had less than 10% of reads mapped to mitochondria genes and 200–10000 expressed genes were used in the analysis. Samples were integrated by Seurat standard workflow “FindIntegrationAnchors” and “IntegrateData” functions with default settings. After manually examining the Principle Component (PC) elbow plot, the top 25 PCs of the integrated data were used for clustering analysis and Uniform Manifold Approximation and Projection (UMAP) analysis with default settings.
Doublet scores were calculated by R package “scds” (v1.2.3)54 using the raw count matrix as input. The top 5% cells ranked by hybrid scores and clusters enriched (>50%) with doublets were removed from the downstream analysis. Data was re-scaled and re-clustered after doublet removal. The expression patterns of a large group of well-known immune markers (Extended Data Fig. 3b) were carefully examined to annotate the clusters. A second round of analysis was performed on microglia to further reveal the relationship of cells within the population. The top 20 PCs were used for clustering and UMAP analysis. One cluster that co-expressed both microglia markers (P2ry12 and Fcrls) and T/NK markers (Cd3g and Nkg7) was regarded as doublets and was filtered out. Clustering resolution was set at 0.2 because the clusters highly correlated with the UMAP topologies. Differential Expression (DE) analysis was performed by Seurat function “FindAllMarkers” and “FindMarkers” with default Wilcoxon Rank Sum test and logFC > 0.25 on pooled biological replicates. Genes with Bonferroni adjusted p-value < 0.05 were considered to be significant. When plotted, the adjusted p-value was log10 transformed. Each adjusted p-value was added an extremely small number (1e-310) to avoid infinite values before transformation. Then the value was signed by the up-/down-regulation of the gene.
Supplemental Table 7 containing Trem2 positive expression from different stages of disease progression from Keren-Shaul et al.11 was used as validation dataset. Common symbols between this study and our top 10 DE genes ranked by logFC from each microglia cluster were used for heatmap plotting. Average expression values from both datasets were offset by 1 and log2 transformed and z-scaled.
RT-qPCR.
RNA from snap-frozen cortex or hippocampal tissue was isolated with TRIzol (Thermo Fisher Scientific 15596026) according to the manufacturer’s instructions. The RT Transcriptor First Strand cDNA Synthesis Kit (Roche) with anchored oligo(dT) primers (Roche) was used for reverse transcription. Quantitative PCR was run in a 384-well plate format on a QuantStudio Q5. Oligonucleotide primer sequences were ordered from Integrated DNA Technologies IDT and reconstituted in house. Primer sequences used were: Gas6 forward primer 5’−3’ AACTGGCTGAACGGGGAAG; and reverse primer 5’−3’ CTTCCCAGGTGGTTTCCGT. TNFα forward primer 5’−3’ GCCACCACGCTCTTCTGTCT and reverse primer 5’−3’ CAGCTGCTCCTCCACTTG GT. IFNα4 forward primer 5’−3’ CCCACAGCCCAGAGAGTGAC and reverse primer 5’−3’ GCCCTCTTGTTCCCGAGGT. Il-6 forward primer AGACAAAGCCAGAGTCCTTCAGA and reverse primer 5’−3’ GCCACTCCTTCTGTGACTCCA. Il-1β forward primer 5’−3’ CCTCTCCAGCCAAGCTTCC reverse primer 5’−3’ CTCATCAGGACAGCCCAGGT. IFNγ forward primer CAATCAGGCCATCAGCAACA and reverse primer 5’−3’ AACAGCTGGTGGACCAC CG. Relative expression of genes of interest was normalized to Gapdh (primer pairs: forward sequence 5’−3’ AGGTCGGTGTGAACGGATTTG; reverse sequence 5’−3’ GGGGTCGTTGATGGCAAC).
Extraction of soluble Aβ and quantification of Aβ42 by ELISA
Briefly, whole cortices and hippocampi were swiftly dissected from freshly extracted brains on an ice-cold platform and ‘snap’ frozen in liquid nitrogen. Tissues were lysed on ice in RIPA buffer [50mM Tris-HCl, pH =8.0, 150mM NaCl, 0.1% SDS, 1% Triton-X 100, 0.5% deoxycholate] with protease and phosphatase inhibitor (Roche, Sigma) at 4:1 RIPA volume/brain wet weight. Homogenates were spun at 5000g first to deplete debris and then ultracentrifuged at 100,000 rpm at 4°C for 1 hour. Supernatants (containing soluble Aβ) from the cortical and hippocampal preparation were aliquoted and stored at −80°C until further use. Extracts from APP/PS1 and APP/PS1 Axl−/− Mertk−/− and their non-diseased littermates were prepared simultaneously and identically.
Standard sandwich ELISA measurements of soluble Aβ42 were performed based on published protocols55. Briefly, 96-well plates were pre-coated with anti-Aβ42 JRF AB042/26 capture antibody at a concentration of 1.5μg/ml 50ul per well overnight at 4°C. The following day, plates were blocked 150μl/well with 1% casein blocking buffer for 2 hours at room temperature. Plates were washed five times with PBS + 0.05%Tween-20. JRF/AbN/25 detection antibody against the N-terminus of Aβ had previously been biotinylated and dialyzed following manufacturer’s instructions. 25μl biotinylated detection antibody was diluted in blocking buffer was mixed with either 25μl of standards (synthetic human Aβ1–42 peptide) or 25μl extracellular media before loaded 50μl per well and in duplets. Following overnight incubation at 4°C, plates were rinsed and incubated in streptavidin-HRP at 50μl/well for 30min and developed in 1:1 mixture of Color Reagent A (H2O2) and Color Reagent B (Tetramethylbenzidine) (Cat # DY999). Plates were immediately measured on a TECAN Infinite® 200 PRO reader at 450nm.
pSIVA stereotaxic injections for in vivo PtdSer labeling.
Stereotactic injections were performed following previously published protocols 56. Briefly, thin wall glass pipettes were pulled on a Sutter Flaming/Brown puller and cut using sterile techniques resulting in tip diameters of 10–15μm. Adult APP/PS1 and WT control mice (15–16 mo) were anesthetized with isoflurane (4% for induction; 1%−2% during surgery). Mice were head-fixed in a computer-assisted stereotactic system with digital coordinate readout. Three sites were serially injected with pSIVA on the right hemisphere of each animal. Neocortical coordinates were (AP +1mm, ML+1.5mm, DV +1mm), (AP 0mm, ML+1.5mm, DV +1mm) and (AP −1mm, ML+1.5mm, DV +1mm) along the motor and somatosensory cortex around layer III/IV. First, craniotomy sites were marked and an electrical micro-drill with a fluted bit (0.5 mm tip diameter) was used to thin a 0.5–1 mm diameter part of the bone over the target injection site. Care was taken to ensure uninjured uplifting of the bone segment. Next, undiluted pSIVA dye was loaded into the glass pipette with sufficient volume and was gently lowered to the desired depth (using the DV coordinates). A total of 500μl pSIVA per injection site was injected slowly at 1nl/second over a period of 15–20min and with a 5-minute break halfway and at the end of each injection to avoid backflow before carefully retracting the injection pipette to the next target injection site (using AP and ML coordinates). At the end of the last injection, mice were sutured along the incision of the scalp, and given subcutaneous Buprenex SR (0.5mg/kg). Each mouse was allowed to recover before placement in their home cage for 1.5–2h before undergoing routine perfusion and processing of both injected right hemisphere and control left hemisphere (see previous section).
Surgery and animal preparation for in vivo two-photon imaging.
Surgical procedures closely followed established protocols 7, 33, 56. Briefly, mice were anesthetized with isoflurane (4–5% for induction; 1%−1.5% for maintenance), head-fixed with blunt ear bars, and kept at 36°C–37°C on a custom surgical bed (Thorlabs). Eyes were protected with vet ophthalmic ointment (Puralube). Depilator cream (Nair) was used to remove hair on top of the mouse’s head. The scalp was thoroughly cleansed and disinfected with a two-stage scrub of betadine and 70% ethanol. A scalp portion was surgically removed to expose frontal, parietal, and interparietal skull segments. Scalp edges were attached to the lateral sides of the skull using tissue compatible adhesive (3M Vetbond). A custom-machined metal plate was affixed to the skull with dental cement (Coltene Whaledent, cat. no. H00335). Ear bars were removed and the head was stabilized by clamping the skull-attached plate with a custom holder. An approximately 3mm diameter craniotomy was made over somatosensory cortex (center coordinates: AP −1.5mm, ML 1.5mm). A 1.5% agarose solution and coverslip were applied to the exposed tissue, and the coverslip was fixed to the skull with dental cement to control tissue motion. Imaging commenced immediately after optical window preparation (corresponding to 13–14h after intraperitoneal MX04 injection). Depth of anesthesia was monitored throughout the experiment and adjusted as needed to maintain a breath rate of approximately 55–65 breaths per minute. Animal temperature was maintained at 36°C–37°C and saline supplemented as needed to compensate for fluid loss.
Confocal microscopy.
One-photon laser scanning confocal images were acquired with a Zeiss LSM 710 Confocal microscope using Plan-Apochromat 20× 0.8-NA air-matched or 63× 1.4-NA oil objectives (laser lines: 405 nm, 488nm, 594nm and 633nm). Image size was 1024 × 1024 pixels. Stack thickness was typically 15μm for mouse brain sections and 5μm for postmortem paraffin-embedded sections. For synaptic quantification specifically, CA1 apical dendrites were imaged at 3μm thickness and z stack image were obtained (optical slice 0.29μm, 11 slices per 3μm stack). Exposure acquisition was set according to the WT or non-mutant samples in all experiments. Airyscan super-resolution images were acquired with a Zeiss LSM 880 Rear Port Laser Scanning Confocal and Airyscan FAST Microscope using 63x oil objective. Images were obtained and processed via the Zen Black and Zen Blue editions. Images for cortical and hippocampal plaque quantification were acquired with Olympus VS-120 Virtual Slide Scanning Microscope using a 10x objective.
Two-photon microscopy.
Live animal imaging was performed as previously described 7, 33, 56. Briefly, a Sutter Movable Objective Microscope (MOM) equipped with a pulsed femtosecond Ti:Sapphire laser (Chameleon Ultra II, Coherent) with two fluorescence detection channels was used for imaging (dichroic beamsplitter: FF520-Di02 (Semrock); blue emission filter: FF01-452/45 (Semrock); green emission filter: ET525/70M (Chroma); photomultiplier tubes: H7422-40 GaAsP (Hamamatsu)). Laser excitation wavelength was set to 830nm. Average laser power was <10-15mW at the tissue surface and adjusted with depth as needed to compensate for signal loss due to scattering and absorption. An Olympus 20× 1.0-NA water immersion objective was used for light delivery and collection. Z-stacks included up to 350 images, acquired at 1μm axial step size, used a 2-frame average, 512 × 512 pixel resolution, and 2.0x-10x zoom (corresponding to 350μm-72μm fields of view). Time-lapse recordings typically included 60–70 images/stack, acquired at 1.0–1.2 μm axial step size, used a 2-frame average, 60 stack repeats (corresponding to approximately 94 min total recording duration), 512 × 512 pixel resolution, and 3.3x-5x zoom (corresponding to 212 μm - 142 μm fields of view). Up to thirteen z-stacks, and four to seven time-lapse recordings were acquired per animal in layers 1 and 2 of the somatosensory cortex.
Imaging data analysis.
For fixed-brain thin sections, maximum intensity projection images of 212μm × 212μm (1024×1024 pixel resolution) were analyzed in FIJI. ~5–7 plaques per brain sections (3–5 sections per animal) were randomly chosen in the prefrontal cortex using the 6E10 channel. Mean fluorescent intensity (MFI) for Mer, Axl, and Gas6 expression analysis, was calculated as the quotient of integrated density divided by region of interest, for example, Iba1 area or 6E10 area, respectively, which were gated based on their intensity and applied for all images in each experiment. Plaque-associated and non-plaque associated Iba1 areas were classified manually by investigator blind with respect to genotypes. For quantification of dystrophic neurite area using LAMP1 or RTN3, area above set fluorescent thresholds for either LAMP1 or RTN3-marked neuritic dystrophy and 6E10 immuno-positive area were calculated and summed using the FIJI ‘analyze particle’ tool, and the ratio of summed area (LAMP1 or RTN3 area/6E10 area) was calculated on per plaque basis. For LAMP1 analysis for convenience of binary categorization, dense-core plaques were defined as plaques that contain a single or solid (usually bright) 6E10-positive core area ≥ 100μm2, and diffuse plaques were defined as the complement of these dense-core plaques that are characterized as those devoid of compact 6E10-positive area ≥ 100μm2. To assess excitatory synapse changes in the mouse hippocampus, co-localization of vGlut1 and PSD95 in the apical dendrite area of CA1 was analyzed using Imaris software (Bitplane). 3D z stack images were background subtracted and positive puncta of vGlut1 and PSD95 were selected and built as ‘spots’ by uniformly thresholding size and intensity across all sections and genotypes analyzed. PSD95 spots were then transformed into a distance vector using the ‘distance transform’ function, which was followed by a calculation of distance between vGlut1 spots and PSD95 spots. Puncta were considered colocalized if the distance between vGlut1 and PSD95 was ≤ 0.7μm. Number of colocalized puncta from each image was averaged for one section or for one animal and compared between experimental groups that contain 3 animals. ≥ 3 images per section, ≥ 3–5 sections per animal were analyzed.
For analyses of two-photon image stacks, Imaris software (version 9.1.2; Bitplane, Zurich, Switzerland) was used for three-dimensional reconstruction of GFP microglia and MX04-labeled dense-core plaques, and also for the analysis of (a) the distance of the centroid of GFP+ microglial cell bodies to the edge of a MX04+ mass, (b) microglial cell body volume, and (c) intracellular Aβ,from 1μm-step z-series stack images of both genotypes. Two types of ‘surfaces’, as digital representations that capture the volume of either a microglia cell body or a dense-core (not diffuse) plaque for the GFP channel and MX04 channel, respectively, were created under the same threshold for all stack images. For MX04 objects, a surface cutoff was set to be at the clear-cut border of the outlining of dense-core plaques, exclusive of surrounding diffuse material. Only dense-core plaques whose diameters fell between 10μm-40μm were included in the following analysis. For GFP surfaces, filters for “Surface grain size”, “Diameter of the largest sphere which fits into object”, “Seed points diameter”, “Quality for seed points” (e.g. to determine whether a structure belongs to the same or a neighboring cell), and volume cutoff were set to 0.3μm, 7.5μm, 3.75μm, 65a.u. and 65μm3, respectively, for capturing the structure of GFP microglial cell bodies in imaging volume of 142μm x142μm × 70μm for Fig. 4c and Extended Data Fig. 4a and a normalized imaging volume of 350μm × 350μm × 300μm in somatosensory cortex (512 × 512 pixel resolution). The same thresholds were applied to all images of both genotypes. Structures on edge and/or captured incompletely were manually deleted. Created surfaces were then used to determine distance of the centroid of individual GFP surfaces to the closest edge of MX04 objects with the build-in MATLAB function in Imaris.
For quantification of intracellular Aβ, a GFP ‘cell-body only’ channel was built with GFP surfaces as mask borders using the ‘set outside voxel to 0’ mask. MX04 and the ‘cell-body only’ GFP channel were then thresholded to capture the overlapping volume between MX04 and GFP channel using the ‘co-localization’ function in Imaris. The overlapping volume was then normalized to the total volume of GFP of the image as a representation of the percentage of GFP microglia cell body volume occupied by MX04 material. Primary process number, total length for PAM, and process polarization ratio for NPAM around dense-core plaques whose diameters fell between 10μm- 30μm were analyzed with FIJI based on maximum intensity projection stack images of 142µm x142μm × 60μm in volume. The following parameters were measured using volumetric analysis57, 58. For primary process number and total length, PAM were defined as those whose cell body centroids were 0–5μm away from the edge of the nearest MX04 edge 37. On average 3–5 microglia per plaque that were unambiguously distinct, showed clear process visibility, and were not between two plaques were selected. Primary process number and length from PAM were measured using the ‘free-hand’ line tool on FIJI. Z-stacks of these projected images were gone through to ensure processes continuity from the same cell across layers. Mainly distal primary processes were analyzed in this group of cells due to the fact that majority of their somata were next to or in direct contact with a plaque surface. For polarization index, NPAM were defined as those whose cell body centroids were 20–50μm from a dense-core MX04 edge and whose processes were clearly visible. Cells equidistant between two plaques were not included in the analysis. Calculation of this index was performed58 using the following criteria: processes oriented toward plaques were classified as those on the plaque side of a line drawn through the microglial cell body perpendicular to a line drawn to the nearest plaque. The polarization index for one microglia cell is the ratio of the summed length of all ‘toward’ primary processes divided by the summed length of all processes.
For structural dynamics of individual microglia processes, process velocity was assessed as previously described33, 59, by manually tracing process tips over time using the Manual Tracking plug-in on Fiji on Maximum Intensity Projected time lapse Videos, typically over 90-minute time spans that cover a FOV of 212μm × 212μm × 70μm in the somatosensory cortices of age-matched AD and healthy littermates of WT and Axl−/−Mertk−/− mice. As mentioned in the previous section, measurements were made for AD microglia whose cell body centroids were within 5μm from the edge of dense-core MeX04 edge as measured on projected images (PAM), and for those whose centroids were over 20μm from any plaque in the FOV (NPAM). Resting microglia were microglia from non-AD age-matched littermates. At least 2–3 and 4–7 processes were quantified from PAM and NPAM, respectively, to obtain an average process velocity per microglia.
For CAA quantification, CAA area percentage was defined as ‘6E10-positive vessel area’ by calculating the ratio of 6E10-positive area within CD31-positive area over total CD31 area in either somatosensory or prefrontal cortex of mice of each genotype. Measurements were averaged from images of respective brain areas from 15 sections (spanning >1.8mm3 of brain volume) per mouse.
Immunoblot analysis.
Tissues were snap frozen in liquid nitrogen. Frozen tissues were lysed using RIPA buffer and phosphatase protease inhibitor (Roche, Sigma) for 30min on ice. Supernatants were stored at −80°C after samples were spun down at 12,000 rpm for 5min. Immunoblotting was as previously published5. Briefly, equal amounts of protein (10μg) in 3XLaemmli buffer and 0.1M DTT were separated by electrophoresis through 4–12% Bis-Tris polyacrylamide gels (Novex, Life Technologies) and were transferred to PVDF membranes. Membranes were blocked with 1% casein block in PBS (BioRad 1610782), subsequently incubated with primary and secondary antibodies and washed with TBST (50mM Tris-HCl pH7.5, 0.15M NaCl and 0.25% Tween-20) between incubations. Blots were developed using an Odyssey Gel Imaging System (Li-Cor), and quantified using ImageStudio with Gapdh as loading control.
Contextual fear conditioning behavior test.
Fear acquisition and contextual memory tests were adapted from an established fear conditioning paradigm60. Briefly, mice were habituated for 5 minutes per animal for 5 consecutive days prior to the day of fear acquisition. One day prior to fear acquisition, mice were allowed to explore the conditioning context for 10min. All testing was performed between 0930 and 1430 h and all mice were acclimated to the testing room for at least one hour before the beginning of each behavioral assay. For fear acquisition (day 1), mice were evaluated for their abilities to learn the association between a context (conditional stimulus, C.S.) and an aversive mild electrical stimulus (unconditional stimulus, U.S.). Freezing behavior during and after three pairs of co-terminated stimuli of an auditory cue (30s, 90db, 2.5kHz) and an electric shock (0.5mA, 2s) was calculated as “% freezing”; i.e., the time the mouse was immobile during the time period in the presence of the paired context even when the aversive stimulus was absent. Freezing behavior was monitored and analyzed by an automated video-freeze software (Med Associates). Animals were returned to their home cage after the fear conditioning session. On day 2, 24 hours after fear acquisition, freezing behavior was measured in the original conditioning context for 3 minutes in order to test the memory retention of the learned association between C.S. and U.S. To avoid estrogen-related and other cognitive-irrelevant variations, fear conditioning was performed only on male mice in cohorts with matching ages and group caged ones.
Statistical Analyses and Reproducibility.
Numeric data analysis and statistics were performed with Microsoft Excel (ver15.36) and GraphPad Prism (version 8.0) software, except for Fig. 3 and Extended Data Fig. 3. All data in all figure panels of the paper are represented as mean ±1 standard deviation. In most cases, ‘‘n’’ is used to denote the number of mice per experimental group and ‘‘N’’ the number of brain sections/images/samples analyzed per mouse. Group sample sizes were chosen based on previous studies and/or power analysis. Experiments were performed with at least two independent replicates whenever applicable. No technical replicate samples across independent experiments were pooled in the datasets. Datasets displayed normal distribution and equal standard deviations unless indicated by unequal variance test. Statistical tests were always performed two-sided. Bonferroni adjusted P values were calculated for genes in Fig. 3 and Extended Data Fig 3. P values were calculated using Mann-Whitney test in Figs. 4d–f, 5a, b, d, e, and Extended Data Figs. 1d, 4b–d, 6a–e, g and 7a, c, d, 8c, d. Student’s t-test was used in Fig. 6c–d and Extended Data Fig 7b,e,f. For multiple group comparison, P values were calculated in Figs. 2b (upper and lower) and 4g, and Extended Data Fig 8a using Kruskal Wallis test followed by Dunn’s multiple comparison test. P values were calculated in Fig. 4c and Extended Data Figs. 4a and 4e using 2-way ANOVA with Sidak’s multiple comparison test. ‘‘n.s.’’ (not significant) indicates p > 0.05; significant P values are noted as exact values on graphs.
Extended Data
Extended Data Fig. 1. Expression of Axl, Mer, TMEM119, Trem2, and GFAP in plaque-burdened AD brains.
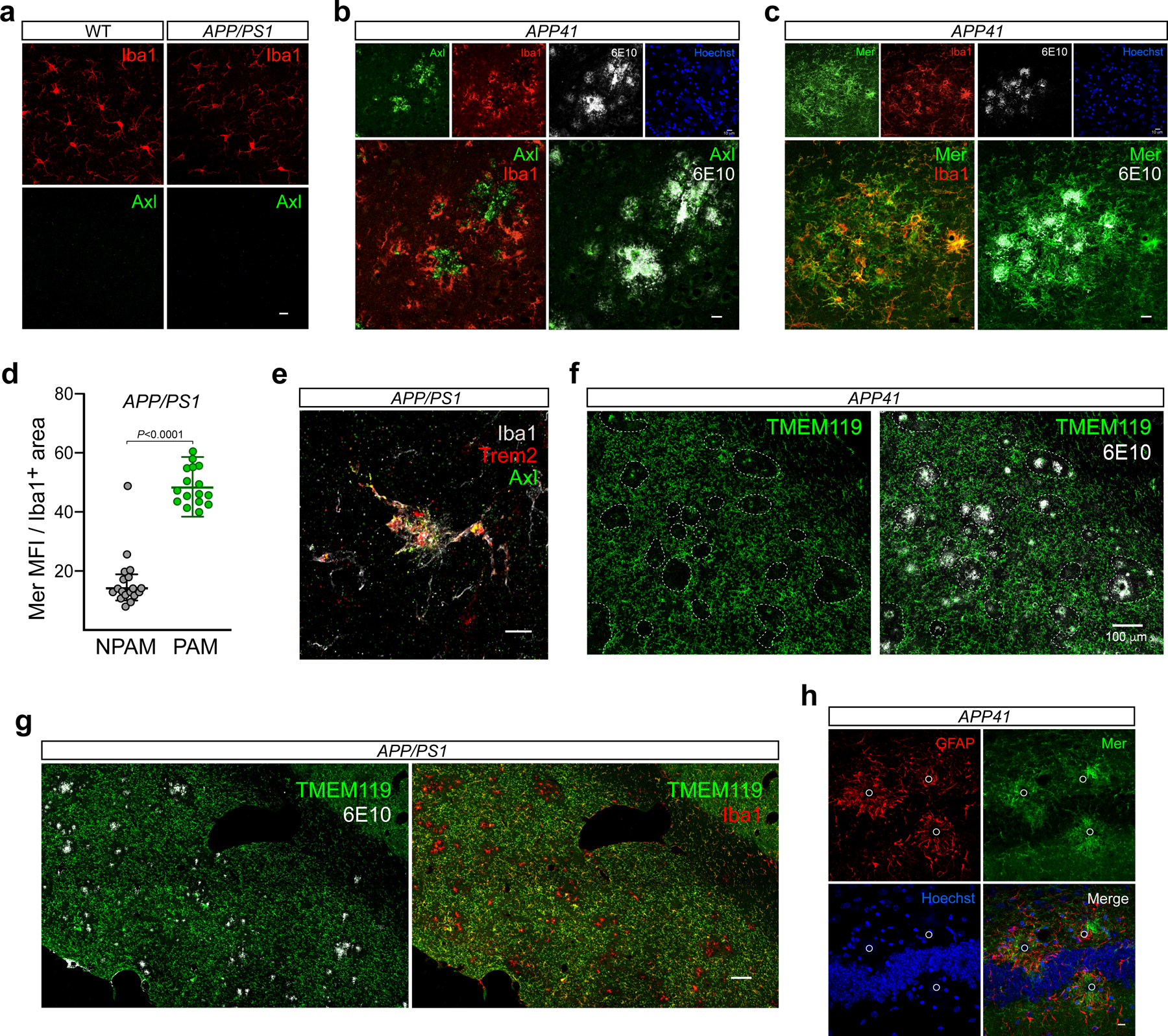
(a) Axl protein (green, lower panels) is undetectable in WT (left) and APP/PS1 (right) microglia (Iba1, red, upper panels) in the 4mo cortex, prior to the appearance of plaques in APP/PS1 mice. n = 3 per genotype. (b) Axl protein (green) in 15 mo APP41 mice (n = 3) is up-regulated in cortical microglia (Iba1, red) contacting Aβ plaques (6E10, white), and is also often concentrated in plaque centers, consistent with strong prior activation of Axl and subsequent cleavage of its ectodomain. (As is seen for other RTKs, robust activation of Axl results in nearly complete cleavage of the ectodomain from the cell surface.) Hoechst 33258 stains nuclei. (c) Expression of Mer protein (green) in 15 mo APP41 mice is seen in all cortical microglia (Iba1, red), but is further up-regulated in microglia that invest Aβ plaques (6E10, white). (d) Quantification of Mer up-regulation in Iba1+ plaque-associated microglia (PAM) versus non-plaque-associated microglia (NPAM) in APP/PS1 mice at 9.5 months. (e) Axl (green) and Trem2 (red) are up-regulated in the same Iba1+ (white) microglia cell in the 9.5 mo APP/PS1 cortex. (f) Expression of the homeostatic microglial marker TMEM119 (green) is lost in cortical microglia that surround plaques (6E10, white) in 15 mo APP41 mice, except for an occasional 1–2 cells at the center of plaques. (g) This same TMEM119 down-regulation is seen in 15 mo APP/PS1 mice. TMEM119− microglia surrounding 6E10+ plaques are strongly Iba1+. (h) The up-regulated Mer expression (green) seen in 15 mo APP41 mice is not in GFAP+ reactive astrocytes (red). (Activated S100b+ astrocytes are also negative for Mer expression by IHC1). Circles mark the position of Aβ plaques. Representative images obtained from immunostaining of N ≥ 3 sections from n ≡ 3 mice of each genotype. Scale bars: 10μm (a–c, e, h), 100μm (f, g). Mann-Whitney test (d). For all supplementary figure panels, data are represented as mean ±1 STD.
Extended Data Fig. 2. Gas6 and PtdSer decoration of Aβ plaques.
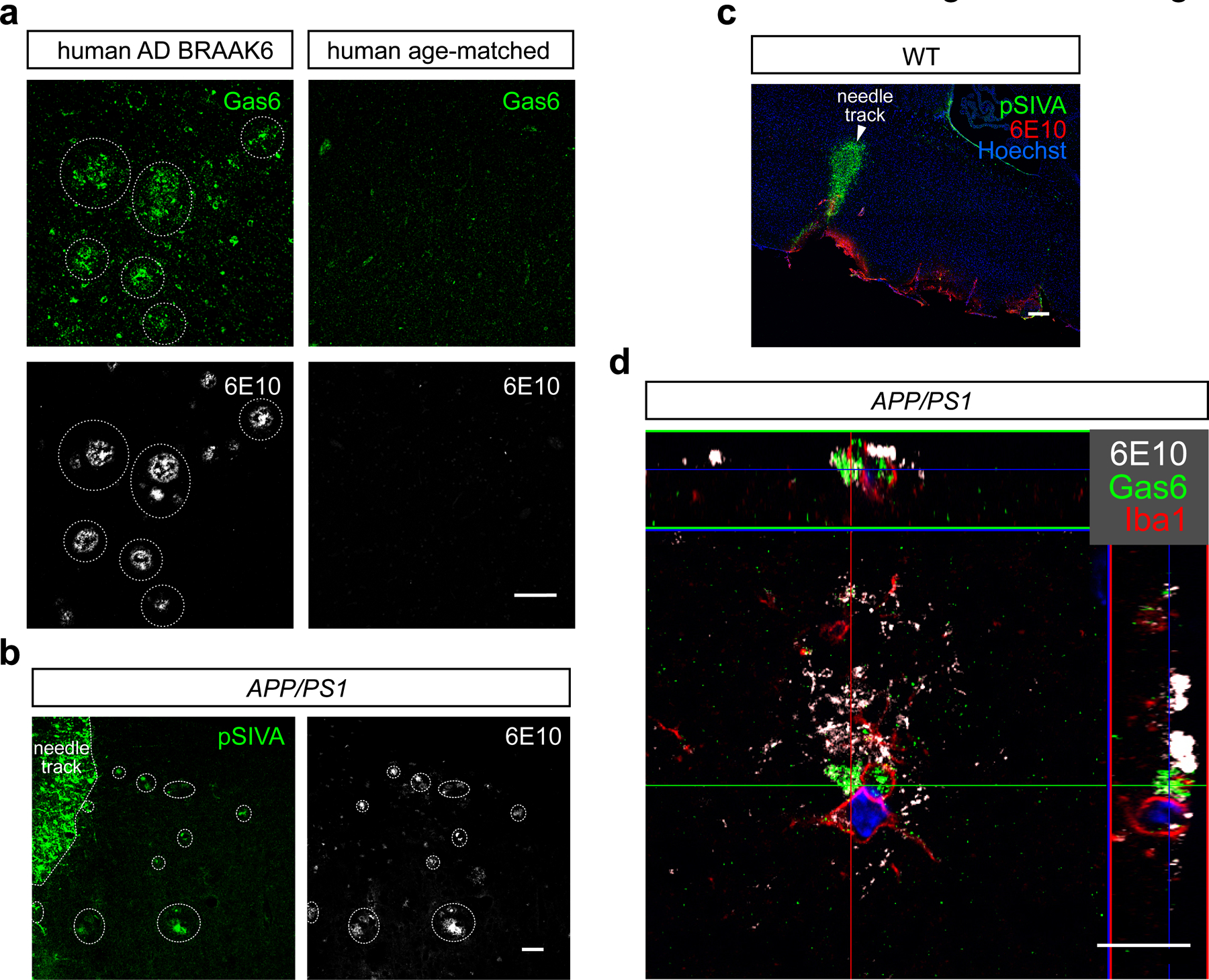
(a) Gas6 protein (green) decorates 6E10+ plaques (white) on sections of postmortem cortex from human patients with advanced (BRAAK stage 6) AD (left panels), but is not evident in the plaque-free cortex of cognitively normal age-matched controls (right panels). Representative images obtained from immunostaining of ≥3 sections from 3 individuals of each condition. (b) Visualization of externalized PtdSer in 15 mo APP/PS1 cortex following stereotaxic injection of pSIVA (green, left panel). pSIVA binds to externalized PtdSer in the needle track of the injection, where cells are damaged and undergoing apoptosis, and also to the PtdSer associated with all 6E10+ amyloid plaques adjacent to the injection site (white, right panel.) (c) A similar pSIVA injection in 15 mo WT mice labels only the needle track of the injection, since there are no plaques in these mice. (d) Airyscan super-resolution image of the juxtaposition of plaque-associated microglia (Iba1, red), Gas6 (green), and Aβ plaque (6E10, white) in 12 mo APP/PS1 mice (n = 5). Scale bars: 100μm (a), 50μm (b), 200μm (c), 10μm (d). n = 3 and 2 for APP/PS1 and WT control, respectively (b, c) from two independent experiments.
Extended Data Fig. 3. Transcriptomics of APP/PS1 and APP/PS1Axl−/−Mertk−/− microglia as quantified by single cell RNA-seq.
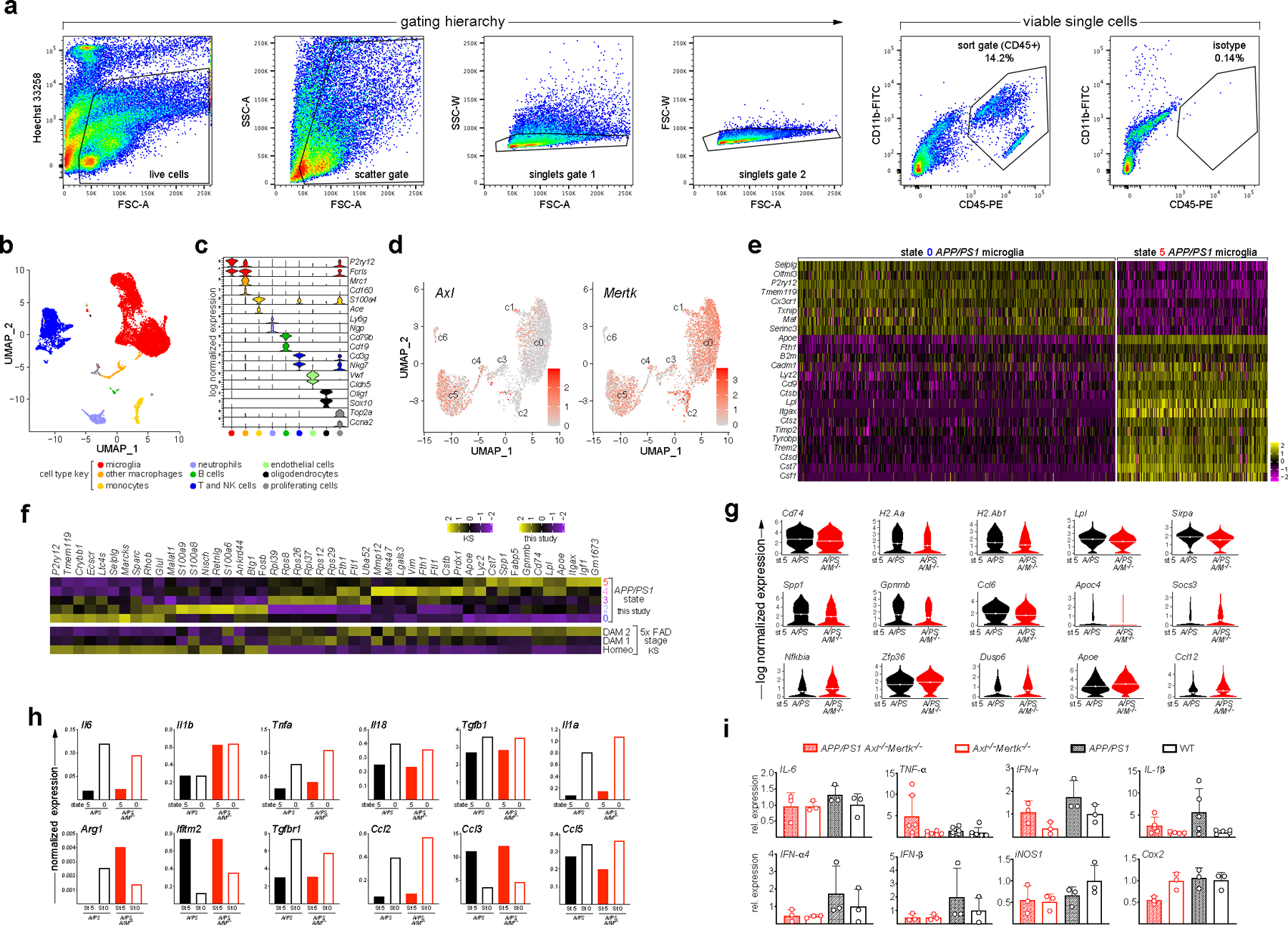
(a) Sorting scheme for isolation of CD45+ single cells. FSC and SSC, forward and side scatter, respectively. A, area; W, width. (b) Uniform manifold approximation and projection (UMAP) clustering of CD45+ cells sorted from 18 mo APP/PS1 (A/PS) and APP/PS1Axl−/−Mertk−/− (A/PS A/M−/−) cortices (combined) and annotated using the 18 marker genes in b. (c) Violin plots of population log-transformed normalized expression of the indicated genes in the indicated cell types. Cell type key applies to a and b. (d) Log-transformed normalized expression of Axl (left) and Mertk (right) mRNA in APP/PS1 cells within the microglial clusters defined in Fig. 3a. (e) Heat map of the scaled expression levels of the indicated genes in individual cells within transcriptomic state (cluster) 0 and state 5 microglia in the APP/PS1 cortex at 18 mo. (f) Comparative composite heat maps for the indicated genes across stages and transcriptomic states in 5xFAD and APP/PS1 mice, as quantified in this study (top five rows) and in Keren-Shaul et al.2 (bottom three rows), respectively. Values shown as z-scaled log-transformed normalized average of each group. (g) Violin plots of the log-transformed normalized expression distribution of the indicated genes at transcriptomic state 5 in A/PS (black) versus A/PS A/M−/− (red) microglia, as determined by single cell RNA-seq (scRNA-seq). Dotted lines indicate mean. (h) Mean expression level of the indicated cytokine and chemokine genes in state 5 versus state 0 microglia in A/PS (black) versus A/PS A/M−/− (red) microglia at 18 mo, as determined by scRNA-seq. (i) Relative expression level of the indicated inflammatory regulator mRNAs in RNA isolated from total cortex of mice of the indicated genotypes at 12 mo, as determined by qRT-PCR. n = 3–6. Kruskal-Wallis test with Dunn’s multiple comparison test. Data are represented as mean ±1 STD.
Extended Data Fig. 4. APP/PS1Axl−/−Mertk−/− microglia are unresponsive to Aβ plaques.
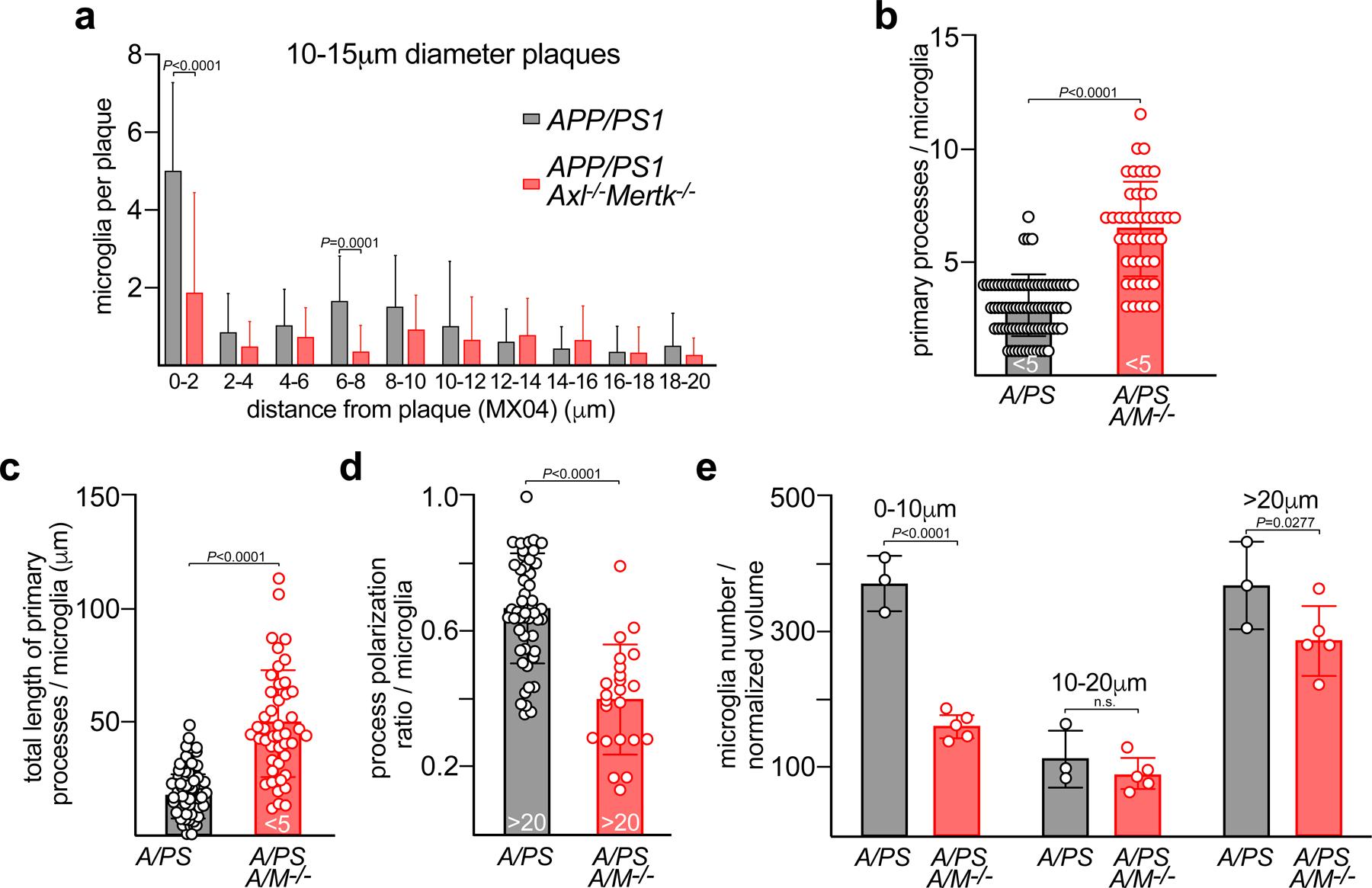
(a) Distribution of distance of microglial cell body centroids, in 2μm bins, from the edge of MX04-labeled Aβ plaques with diameters of 10–15μm in APP/PS1 (gray) and APP/PS1Axl−/−Mertk−/− (red) cortex. Values obtained for 56 and 23 plaques from 3 and 4 mice for APP/PS1 and APP/PS1Axl−/−Mertk−/−, respectively. (b) Number of imaged GFP+ primary processes per PAM (microglia <5μm from plaques) in APP/PS1 (gray, A/PS) and APP/PS1Axl−/−Mertk−/− (red, A/PS A/M−/−) cortex. (c) Summed length of primary microglial processes per PAM in APP/PS1 (gray) and APP/PS1Axl−/−Mertk−/− (red) cortex. (d) Process polarization ratio to nearest plaque per NPAM (microglia >20μm from plaques; see Materials and Methods) in APP/PS1 (gray) and APP/PS1Axl−/−Mertk−/− (red) cortex. (e) Quantification of microglial cell density in the cortex of 16 mo APP/PS1 (gray) and APP/PS1Axl−/−Mertk−/− (red) mice for microglia 0–10 μm, 10–20 μm, and >20 μm from the edge of the nearest plaque. Data points are from 45–129 cells (PAM) investing 10–29 plaques (b, c), and 21–49 cells peripheral to 7–24 plaques (d) from n = 3 mice per genotype (b–d). Points in e represent 3–5 imaging volumes from 3 APP/PS and 4 APP/PS1Axl−/−Mertk−/− mice. Two-way ANOVA with Sidak’s multiple comparison test (a, e) and Mann-Whitney’s test (b-d). Data are represented as mean ±1 STD.
Extended Data Fig. 5. Expansive areas of plaque-associated dystrophic LAMP1+ membrane and poorly compacted plaques in the APP/PS1Axl−/−Mertk−/− brain.
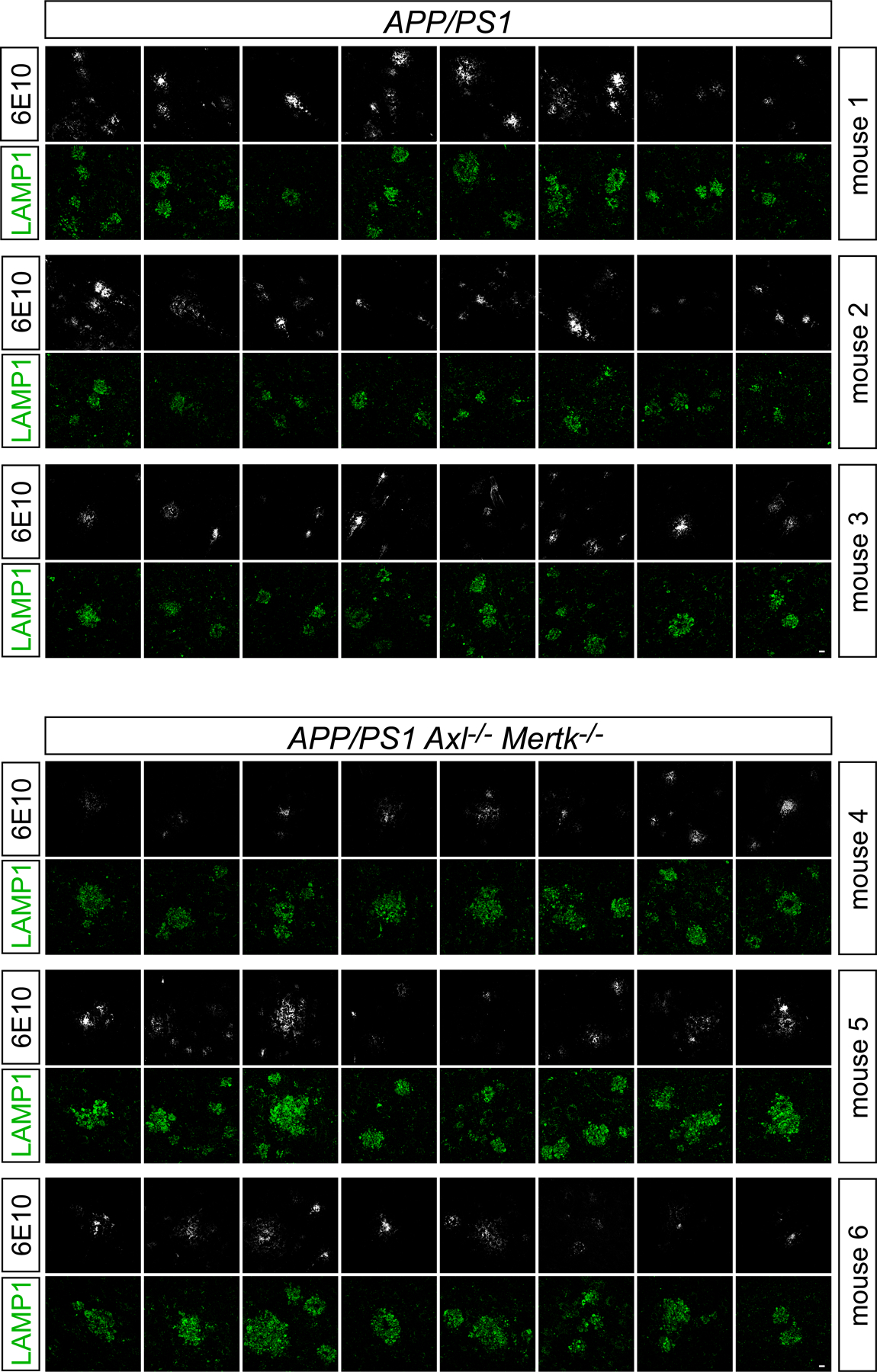
A montage of 24 paired sections in both APP/PS1 (top six rows) and APP/PS1Axl−/−Mertk−/− (bottom six rows) cortex, each stained with antibodies to both LAMP1 (green) and 6E10 (white). Each 6E10 image is paired with (from the same section as) the LAMP image immediately below. This montage, which is a subset of the images used to generate the data in Fig. 5d, is composed of images taken from three different mice of each genotype at 12 months. Note that: (a) 6E10+ Aβ plaques are in general more compact and brightly stained in APP/PS1 mice and more diffuse and weakly stained in APP/PS1Axl−/−Mertk−/− mice; and (b) the area occupied by LAMP1+ membrane is in general much larger in APP/PS1Axl−/−Mertk−/− mice. Scale bars: 10μm.
Extended Data Fig. 6. Accumulation of LAMP1+ dystrophic membrane and apoptotic cell debris in the APP/PS1Axl−/−Mertk−/− brain.
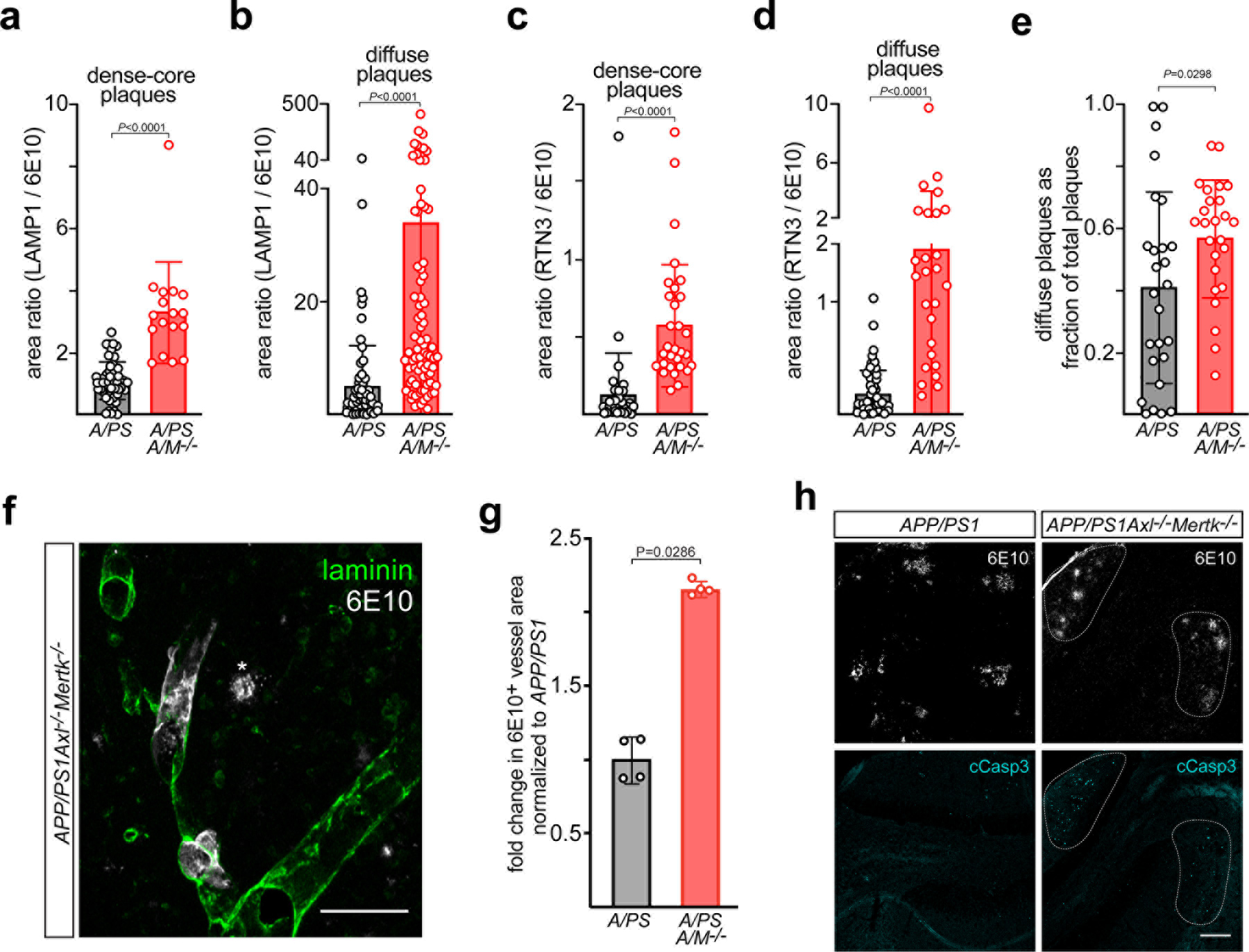
(a) Quantification of LAMP1/6E10 area ratio as in Fig. 5d, but only for dense-core plaques (plaques with solid 6E10+ cores with areas > 100μm2). (b) Quantification of LAMP1/6E10 area ratio as in Fig. 5d, but only for diffuse plaques (plaques without solid 6E10+ cores with areas > 100μm2). (c) Quantification of RTN3/6E10 area ratio as in Fig. 5e, but only for dense-core plaques. (d) Quantification of RTN3/6E10 area ratio as in Fig. 5e, but only for diffuse plaques. (e) Quantification of the density of diffuse plaques (defined as above) expressed as a fraction of total plaques in the cortex of mice of the indicated genotypes at 12 mo. Data represent diffuse plaques quantified from N = 4–5 sections from n= 6 mice per group. (f) Representative example of cerebral amyloid angiopathy (CAA) in the cortex of a 15 mo APP/PS1Axl−/−Mertk−/− mouse. 6E10+ Aβ material (white) is evident within laminin+ blood vessels (green). Asterisk marks an Aβ plaque in the parenchyma. (g) Quantification (see Methods) of CAA in the somatosensory cortex of 15 mo APP/PS1Axl−/−Mertk−/− mice (A/PS A/M−/−) relative to APP/PS1 mice (A/PS). n=4/group and measurements were averaged from N ≥ 15 sections (spanning ≥ 1.8mm3 of brain volume) per mouse. (h) cCasp3+ apoptotic debris (cyan, lower panels) accumulates around 6E10+ Aβ plaques (upper panels) in the APP/PS1Axl−/−Mertk−/− (right panels) but not the APP/PS1 (left panels) hippocampus at 12 mo. Images are representative of n = 3 mice per genotype from three independent experiments. Scale bars: 100μm. Data are 18–47 (a), 67–78 (b), 30–43 (c) and 26–52 (d) plaques investigated from N ≥ 3 sections per mouse from n = 3 mice of each genotype from at least 3 independent replicates. Mann-Whitney test (a–e, g). Data are represented as mean ±1 STD.
Extended Data Fig. 7. TAM (Mer) signaling promotes dense-core Aβ plaque accumulation with functional consequences.
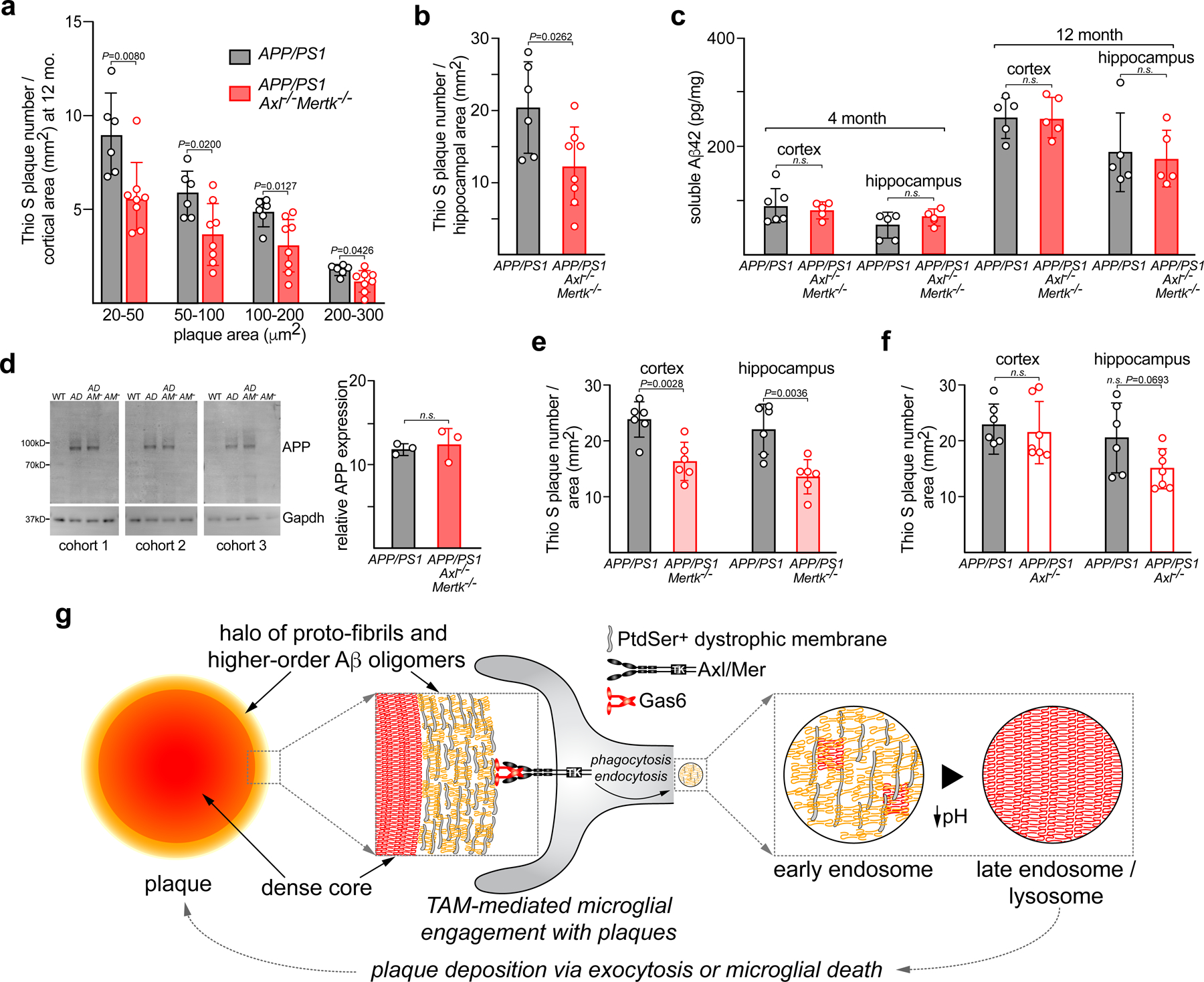
(a) Thio S plaque density in APP/PS1 (gray) versus APP/PS1Axl−/−Mertk−/− (red) cortex for plaques of the indicated size at 12 months. (b) Thio S+ plaque density (all plaque sizes) in APP/PS1 (gray) versus APP/PS1Axl−/−Mertk−/− (red) hippocampus at 12 months. (c) Soluble Aβ42 levels quantified in APP/PS1 (gray) versus APP/PS1Axl−/−Mertk−/− (red) cortex and hippocampus at 4 and 12 mo, as indicated. n= 5–6 per genotype. (d) Quantitative LI-COR western blot measurement of APP protein levels in the 12 mo cortex of 3 cohorts of mice (4 genotypes each cohort) of the indicated genotypes demonstrates no change in APP expression in APP/PS1 mice upon mutation of Axl and Mertk. Blots left and quantification right. (e) ThioS+ plaque density (all plaque sizes) in APP/PS1 (gray) versus APP/PS1Mertk−/− (pink) cortex and hippocampus at 12 months. (f) Thio S+ plaque density (all plaque sizes) in APP/PS1 (gray) versus APP/PS1Axl−/− (white) cortex and hippocampus at 12 months. Data points represent plaque density in n= 6–8 mice of the indicated genotypes averaged from N ≥ 5 cortical sections for each brain. Mann-Whitney test (a, c, d, i, j) and Student’s t-test (b, e, f). Data are represented as mean ±1 STD. (g) TAM-mediated microglial recognition, phagocytosis, and consolidation of Aβ plaques. Microglial Axl and Mer are bridged to the PtdSer-rich dystrophic membranes of plaques via TAM ligands, whose amino-terminal and carboxy-terminal domains bind PtdSer and Axl/Mer, respectively3, 26. Gas6 is shown, but a role for the Mer ligand Pros126 is not excluded. Engagement of the PtdSer-TAM ligand-TAM receptor complex activates the TAM tyrosine kinases (TK), which drives phagocytosis of forming plaque material. Internalized phagocytic cargo is eventually transferred to lysosomes, whose acidic interiors promote the aggregation of large, insoluble Aβ fibrils. Exocytosis or microglial death then delivers this aggregated material to growing dense-core plaques.
Extended Data Fig. 8. Functional consequences of TAM deletion in APP/PS1 mice.
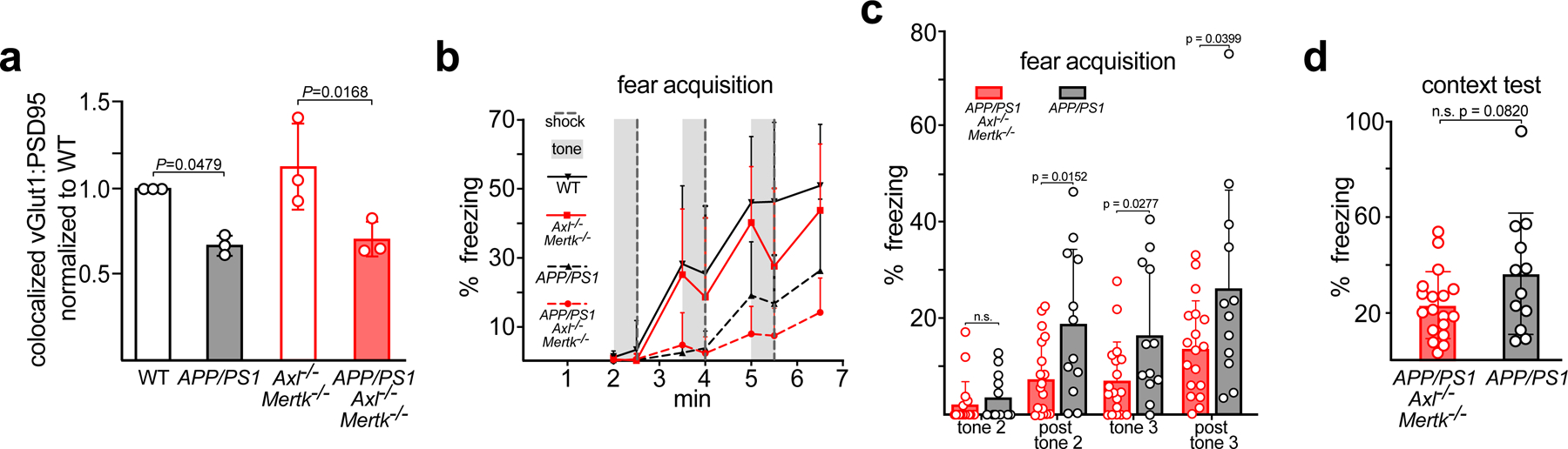
(a) Quantification of the 3D colocalization of the excitatory pre- and post-synaptic markers vGlut1 and PSD95 in the 15mo hippocampus (CA1) (see Methods), as an index of synaptic connectivity. The previously documented decrease in co-localization of these markers in APP/PS1 mice is not altered by the combined mutation of Axl and Mertk. Stack size is 85 × 85 ×3 μm3 per image, averaged 3 images per CA1 section across 3–5 sections per mouse. Data points represent synaptic density index in n = 3 mice of the indicated genotypes. (b) Acquisition of association between a 30 s auditory tone and a subsequent co-terminal 2 s 0.5 mA foot shock, expressed as percent time immobile (% freezing) during the indicated intervals, over three successive trial intervals in 15 mo mice of the indicated genotypes (see Methods). (c) Data in b plotted for the indicated genotypes with the indicated statistical significance per interval. (d) Contextual fear memory as assayed by percent of a 3 minute interval in which mice of the indicated genotypes were immobile (% freezing) when returned to the same testing cage 24 h after the fear acquisition trials of h (see Methods). A cohort of group-housed male mice (n = 12–20/group) were used in the behavioral assay. Data points in h represent the mean % freezing of each group in the interval duration immediately prior to the point. Each data point in d is the % freezing of one animal in the duration of the testing period. Kruskal Wallis test followed by Dunn’s multiple comparison test (a) and Mann-Whitney test (c, d). Data are represented as mean ±1 STD.
Supplementary Material
Left, example two-photon time-lapse recording showing microglia (GFP signal, white) and amyloid plaques (MX04 signal, red) in somatosensory cortical layer 1 of an anesthetized APP/PS1Cx3cr1GFP/+ mouse. Each image is a maximum-intensity projection of a 70 μm-thick z-stack (1.2μm axial step size) recorded every ~93s between 100μm and 30μm depth below the pia. Right, example two-photon time-lapse recording showing microglia (GFP signal, white) and amyloid plaques (MX04 signal, red) in somatosensory cortical layer 1 of an age-and gender-matched anesthetized APP/PS1Axl−/−Mertk−/−Cx3cr1GFP/+ mouse. Each image is a maximum-intensity projection of a 70μm-thick z-stack (1.2μm axial step size) recorded every ~93s between 94μm and 24μm depth below the pia. Elapsed time is indicated in the upper right corner. Amyloid plaques are closely encapsulated by highly polarized microglia in APP/PS1 but not APP/PS1Axl−/−Mertk−/− mice. Additionally, microglia near plaques typically show fewer and less elaborate processes, higher process polarization, and reduced process motility in APP/PS1 as compared to APP/PS1Axl−/−Mertk−/− mice. Supplementary videos 1 and 2 were prepared using Fiji and custom-written scripts in Igor Pro (version 6.04; WaveMetrics). Scale bar, 30 μm.
Left, example two-photon time-lapse recording showing microglia (GFP signal, white) and amyloid plaques (MX04 signal, red) in somatosensory cortical layers 1/2 of an anesthetized APP/PS1Cx3cr1GFP/+ mouse. Each image is a maximum-intensity projection of a 70μm-thick z-stack (1.2μm axial step size) recorded every ~93s between 148μm and 78μm depth below the pia. Right, example two-photon time-lapse recording showing microglia (GFP signal, white) and amyloid plaques (MX04 signal, red) in somatosensory cortical layers 1/2 of an age-and gender-matched anesthetized APP/PS1Axl−/−Mertk−/−Cx3cr1GFP/+ mouse. Each image is a maximum-intensity projection of a 70μm-thick z-stack (1.2μm axial step size) recorded every ~93s between 135μm and 65μm depth below the pia. Elapsed time is indicated in the upper right corner. Amyloid plaques are closely encapsulated by highly polarized microglia in APP/PS1 but not APP/PS1Axl−/−Mertk−/− mice. Additionally, microglia near plaques typically show fewer and less elaborate processes, higher process polarization, and reduced process motility in APP/PS1 as compared to APP/PS1Axl−/−Mertk−/− mice. Scale bar, 30μm.
Acknowledgements
We thank J. Hash for technical assistance, M. Mercken (Janssen Pharmaceuticals) for Aβ42 antibodies, R. Rissman and the Alzheimer’s Disease Research Center (ADRC) at UCSD for AD brain sections, M. Shokhirev for advice on bioinformatic analyses, S. Parylak and F. Gage for advice on fear conditioning assays, and members of the Lemke lab and the Nomis Center for discussions. Supported by grants from the US National Institutes of Health (RF1 AG060748 and R01 AI101400 to G.L., DP2 NS083038, R01 NS108034, and U01 NS103522 to A.N., P30 AG062429 to the UCSD ADRC, and P30 CA014195 and S10-OD023689 to the Salk Institute), the Cure Alzheimer’s Fund and the Coins for Alzheimer’s Research Trust (to G.L.), and the Leona M. and Harry B. Helmsley Charitable Trust (to the Salk Institute); by Goeddel’s Chancellor’s, Marguerite Vogt, and H.A and Mary K. Chapman Charitable Trust graduate fellowships (to Y.H.), and by Anderson, Nomis, and Sweden-America Foundation postdoctoral fellowships (to K.E.H.).
Footnotes
Competing interests: The authors declare no competing interests.
Data Availability.
We declare that the main data supporting the findings of this study are available within the article and its Extended Data Information files. Sequencing data were deposited at Gene Expression Omnibus with the accession number GSE160523. Additional supporting raw data are available from the authors upon request.
References
- 1.Lemke G Biology of the TAM receptors. Cold Spring Harbor Perspectives 5(11), 10.1101/cshperspect.a009076. (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu Q & Lemke G Homeostatic regulation of the immune system by receptor tyrosine kinases of the Tyro 3 family. Science 293, 306–311 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Lemke G How macrophages deal with death. Nat Rev Immunol 19, 539–549 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scott RS et al. Phagocytosis and clearance of apoptotic cells is mediated by MER. Nature 411, 207–211 (2001). [DOI] [PubMed] [Google Scholar]
- 5.Zagórska A, Través PG, Lew ED, Dransfield I & Lemke G Diversification of TAM receptor tyrosine kinase function. Nat Immunol 15, 920–928 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rothlin CV, Ghosh S, Zuniga EI, Oldstone MB & Lemke G TAM receptors are pleiotropic inhibitors of the innate immune response. Cell 131, 1124–1136 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Fourgeaud L et al. TAM receptors regulate multiple features of microglial physiology. Nature 532, 240–244 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Long JM & Holtzman DM Alzheimer Disease: An Update on Pathobiology and Treatment Strategies. Cell 179, 312–339 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.ImmGen C Open-source ImmGen: mononuclear phagocytes. Nat Immunol 17, 741 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Lai C & Lemke G An extended family of protein-tyrosine kinase genes differentially expressed in the vertebrate nervous system. Neuron 6, 691–704 (1991). [DOI] [PubMed] [Google Scholar]
- 11.Keren-Shaul H et al. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell 169, 1276–1290 e1217 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Yin Z et al. Immune hyperreactivity of Abeta plaque-associated microglia in Alzheimer’s disease. Neurobiology of aging 55, 115–122 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Lew ED et al. Differential TAM receptor-ligand-phospholipid interactions delimit differential TAM bioactivities. eLife 3:e03385 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mattsson N et al. CSF protein biomarkers predicting longitudinal reduction of CSF beta-amyloid42 in cognitively healthy elders. Translational psychiatry 3, e293 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sainaghi PP et al. Growth Arrest Specific 6 Concentration is Increased in the Cerebrospinal Fluid of Patients with Alzheimer’s Disease. Journal of Alzheimer’s disease : JAD 55, 59–65 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Jankowsky JL et al. Mutant presenilins specifically elevate the levels of the 42 residue beta-amyloid peptide in vivo: evidence for augmentation of a 42-specific gamma secretase. Hum Mol Genet 13, 159–170 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Jankowsky JL et al. Co-expression of multiple transgenes in mouse CNS: a comparison of strategies. Biomol Eng 17, 157–165 (2001). [DOI] [PubMed] [Google Scholar]
- 18.Rockenstein E, Mallory M, Mante M, Sisk A & Masliaha E Early formation of mature amyloid-beta protein deposits in a mutant APP transgenic model depends on levels of Abeta(1–42). J Neurosci Res 66, 573–582 (2001). [DOI] [PubMed] [Google Scholar]
- 19.Labzin LI, Heneka MT & Latz E Innate Immunity and Neurodegeneration. Annu Rev Med 69, 437–449 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Roy ER et al. Type I interferon response drives neuroinflammation and synapse loss in Alzheimer disease. J Clin Invest 130, 1912–1930 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krasemann S et al. The TREM2-APOE Pathway Drives the Transcriptional Phenotype of Dysfunctional Microglia in Neurodegenerative Diseases. Immunity 47, 566–581 e569 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ulland TK & Colonna M TREM2 - a key player in microglial biology and Alzheimer disease. Nat Rev Neurol 14, 667–675 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Bennett ML et al. New tools for studying microglia in the mouse and human CNS. Proc Natl Acad Sci U S A 113, E1738–1746 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim DK et al. Deep proteome profiling of the hippocampus in the 5XFAD mouse model reveals biological process alterations and a novel biomarker of Alzheimer’s disease. Exp Mol Med 51, 1–17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim YE, Chen J, Langen R & Chan JR Monitoring apoptosis and neuronal degeneration by real-time detection of phosphatidylserine externalization using a polarity-sensitive indicator of viability and apoptosis. Nat Protoc 5, 1396–1405 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Friedman BA et al. Diverse Brain Myeloid Expression Profiles Reveal Distinct Microglial Activation States and Aspects of Alzheimer’s Disease Not Evident in Mouse Models. Cell Rep 22, 832–847 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Satija R, Farrell JA, Gennert D, Schier AF & Regev A Spatial reconstruction of single-cell gene expression data. Nat Biotechnol 33, 495–502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gonzalez H & Pacheco R T-cell-mediated regulation of neuroinflammation involved in neurodegenerative diseases. J Neuroinflammation 11, 201 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spangenberg E et al. Sustained microglial depletion with CSF1R inhibitor impairs parenchymal plaque development in an Alzheimer’s disease model. Nature communications 10, 3758 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu Q et al. Tyro-3 family receptors are essential regulators of mammalian spermatogenesis. Nature 398, 723–728 (1999). [DOI] [PubMed] [Google Scholar]
- 31.Jung S et al. Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol Cell Biol 20, 4106–4114 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klunk WE et al. Imaging Abeta plaques in living transgenic mice with multiphoton microscopy and methoxy-X04, a systemically administered Congo red derivative. J Neuropathol Exp Neurol 61, 797–805 (2002). [DOI] [PubMed] [Google Scholar]
- 33.Nimmerjahn A, Kirchhoff F & Helmchen F Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science 308, 1314–1318 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Nimmerjahn A Two-photon imaging of microglia in the mouse cortex in vivo. Cold Spring Harb Protoc 2012 (2012) 10.1101/pdb.prot069294. [DOI] [PubMed] [Google Scholar]
- 35.Wang Y et al. TREM2 lipid sensing sustains the microglial response in an Alzheimer’s disease model. Cell 160, 1061–1071 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y et al. TREM2-mediated early microglial response limits diffusion and toxicity of amyloid plaques. J Exp Med 213, 667–675 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Condello C, Yuan P, Schain A & Grutzendler J Microglia constitute a barrier that prevents neurotoxic protofibrillar Abeta42 hotspots around plaques. Nature communications 6, 6176 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hassiotis S et al. Lysosomal LAMP1 immunoreactivity exists in both diffuse and neuritic amyloid plaques in the human hippocampus. Eur J Neurosci 47, 1043–1053 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Hu X et al. Transgenic mice overexpressing reticulon 3 develop neuritic abnormalities. EMBO J 26, 2755–2767 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Greenberg SM et al. Cerebral amyloid angiopathy and Alzheimer disease - one peptide, two pathways. Nat Rev Neurol 16, 30–42 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blanco-Suarez E, Liu TF, Kopelevich A & Allen NJ Astrocyte-Secreted Chordin-like 1 Drives Synapse Maturation and Limits Plasticity by Increasing Synaptic GluA2 AMPA Receptors. Neuron 100, 1116–1132 e1113 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jay TR et al. Disease Progression-Dependent Effects of TREM2 Deficiency in a Mouse Model of Alzheimer’s Disease. J Neurosci 37, 637–647 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jay TR et al. TREM2 deficiency eliminates TREM2+ inflammatory macrophages and ameliorates pathology in Alzheimer’s disease mouse models. J Exp Med 212, 287–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yuan P et al. TREM2 Haplodeficiency in Mice and Humans Impairs the Microglia Barrier Function Leading to Decreased Amyloid Compaction and Severe Axonal Dystrophy. Neuron 90, 724–739 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baik SH, Kang S, Son SM & Mook-Jung I Microglia contributes to plaque growth by cell death due to uptake of amyloid beta in the brain of Alzheimer’s disease mouse model. Glia 64, 2274–2290 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Sosna J et al. Early long-term administration of the CSF1R inhibitor PLX3397 ablates microglia and reduces accumulation of intraneuronal amyloid, neuritic plaque deposition and pre-fibrillar oligomers in 5XFAD mouse model of Alzheimer’s disease. Mol Neurodegener 13, 11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fu H et al. Complement component C3 and complement receptor type 3 contribute to the phagocytosis and clearance of fibrillar Abeta by microglia. Glia 60, 993–1003 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu X et al. Amyloid seeds formed by cellular uptake, concentration, and aggregation of the amyloid-beta peptide. Proc Natl Acad Sci U S A 106, 20324–20329 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cambier CJ, Falkow S & Ramakrishnan L Host evasion and exploitation schemes of Mycobacterium tuberculosis. Cell 159, 1497–1509 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Panza F, Lozupone M, Logroscino G & Imbimbo BP A critical appraisal of amyloid-beta-targeting therapies for Alzheimer disease. Nat Rev Neurol 15, 73–88 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Hutter-Paier B et al. The ACAT inhibitor CP-113,818 markedly reduces amyloid pathology in a mouse model of Alzheimer’s disease. Neuron 44, 227–238 (2004). [DOI] [PubMed] [Google Scholar]
- 52.Imbimbo BP et al. CHF5074, a novel gamma-secretase modulator, attenuates brain beta-amyloid pathology and learning deficit in a mouse model of Alzheimer’s disease. Br J Pharmacol 156, 982–993 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stuart T et al. Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902 e1821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bais AS & Kostka D scds: computational annotation of doublets in single-cell RNA sequencing data. Bioinformatics 36, 1150–1158 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schmidt SD, Mazzella MJ, Nixon RA & Mathews PM Abeta measurement by enzyme-linked immunosorbent assay. Methods Mol Biol 849, 507–527 (2012). [DOI] [PubMed] [Google Scholar]
- 56.Tufail Y et al. Phosphatidylserine Exposure Controls Viral Innate Immune Responses by Microglia. Neuron 93, 574–586 e578 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koenigsknecht-Talboo J et al. Rapid microglial response around amyloid pathology after systemic anti-Abeta antibody administration in PDAPP mice. J Neurosci 28, 14156–14164 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bolmont T et al. Dynamics of the microglial/amyloid interaction indicate a role in plaque maintenance. J Neurosci 28, 4283–4292 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bernier LP et al. Nanoscale Surveillance of the Brain by Microglia via cAMP-Regulated Filopodia. Cell Rep 27, 2895–2908 e2894 (2019). [DOI] [PubMed] [Google Scholar]
- 60.Kitamura T et al. Island cells control temporal association memory. Science 343, 896–901 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Left, example two-photon time-lapse recording showing microglia (GFP signal, white) and amyloid plaques (MX04 signal, red) in somatosensory cortical layer 1 of an anesthetized APP/PS1Cx3cr1GFP/+ mouse. Each image is a maximum-intensity projection of a 70 μm-thick z-stack (1.2μm axial step size) recorded every ~93s between 100μm and 30μm depth below the pia. Right, example two-photon time-lapse recording showing microglia (GFP signal, white) and amyloid plaques (MX04 signal, red) in somatosensory cortical layer 1 of an age-and gender-matched anesthetized APP/PS1Axl−/−Mertk−/−Cx3cr1GFP/+ mouse. Each image is a maximum-intensity projection of a 70μm-thick z-stack (1.2μm axial step size) recorded every ~93s between 94μm and 24μm depth below the pia. Elapsed time is indicated in the upper right corner. Amyloid plaques are closely encapsulated by highly polarized microglia in APP/PS1 but not APP/PS1Axl−/−Mertk−/− mice. Additionally, microglia near plaques typically show fewer and less elaborate processes, higher process polarization, and reduced process motility in APP/PS1 as compared to APP/PS1Axl−/−Mertk−/− mice. Supplementary videos 1 and 2 were prepared using Fiji and custom-written scripts in Igor Pro (version 6.04; WaveMetrics). Scale bar, 30 μm.
Left, example two-photon time-lapse recording showing microglia (GFP signal, white) and amyloid plaques (MX04 signal, red) in somatosensory cortical layers 1/2 of an anesthetized APP/PS1Cx3cr1GFP/+ mouse. Each image is a maximum-intensity projection of a 70μm-thick z-stack (1.2μm axial step size) recorded every ~93s between 148μm and 78μm depth below the pia. Right, example two-photon time-lapse recording showing microglia (GFP signal, white) and amyloid plaques (MX04 signal, red) in somatosensory cortical layers 1/2 of an age-and gender-matched anesthetized APP/PS1Axl−/−Mertk−/−Cx3cr1GFP/+ mouse. Each image is a maximum-intensity projection of a 70μm-thick z-stack (1.2μm axial step size) recorded every ~93s between 135μm and 65μm depth below the pia. Elapsed time is indicated in the upper right corner. Amyloid plaques are closely encapsulated by highly polarized microglia in APP/PS1 but not APP/PS1Axl−/−Mertk−/− mice. Additionally, microglia near plaques typically show fewer and less elaborate processes, higher process polarization, and reduced process motility in APP/PS1 as compared to APP/PS1Axl−/−Mertk−/− mice. Scale bar, 30μm.
Data Availability Statement
We declare that the main data supporting the findings of this study are available within the article and its Extended Data Information files. Sequencing data were deposited at Gene Expression Omnibus with the accession number GSE160523. Additional supporting raw data are available from the authors upon request.


