Summary
Serine-arginine (SR) protein kinases (SRPKs) regulate the functions of the SR-rich splicing factors by phosphorylating multiple serines within their C-terminal arginine-serine-rich domains. Dysregulation of these phosphorylation events has been implicated in many diseases, suggesting SRPKs are potential therapeutic targets. In particular, aberrant SRPK1 expression alters the balances of proangiogenic (VEGF165) and antiangiogenic (VEGF165b) splicing isoforms of the key angiogenesis factor, vascular endothelial growth factor (VEGF), through the phosphorylation of prototypic SR protein SRSF1. Here, we report a protein-protein interaction (PPI) inhibitor of SRPKs, docking blocker of SRPK1 (DBS1), that specifically blocks a conserved substrate docking groove unique to SRPKs. DBS1 is a cell-permeable inhibitor that effectively inhibits the binding and phosphorylation of SRSF1 and subsequently switches VEGF splicing from the proangiogenic to the antiangiogenic isoform. Our findings thus provide a new direction for the development of SRPK inhibitors through targeting a unique PPI site to combat angiogenic diseases.
Subject areas: Biochemistry, Cell Biology, Structural Biology
Graphical abstract

Highlights
-
•
Structure-based design of a protein-protein interaction inhibitor of SRPK
-
•
PPI inhibitor of SRPK inhibits angiogenesis by switching VEGF splicing
-
•
SRPK-specific substrate docking groove serves as promising drug target site
-
•
Combination of PPI and ATP-competitive inhibitors of SRPK enhances antiangiogenesis
Biochemistry; Cell Biology; Structural Biology
Introduction
Pre-mRNA splicing in higher eukaryotic cells requires many auxiliary factors and involves a complex series of protein-protein, protein-RNA, and RNA-RNA interactions (Black, 2003). One essential family of splicing factors is serine-arginine (SR) proteins. SR proteins are characterized by the presence of multiple contiguous stretches of arginine-serine (RS) dipeptides, termed as RS domains, at their C-termini. The RS domains undergo reversible phosphorylation, and their states affect multiple facets of SR protein activity, including spliceosome assembly, splice site selection, mRNA export, and translation (reviewed in (Graveley, 2000; Shepard and Hertel, 2009). The prototypic SR protein, SRSF1, is a proto-oncogene that is upregulated in many forms of cancer (Das et al., 2012; Das and Krainer, 2014; Karni et al., 2007). Studies have shown that SRSF1 plays a critical role in regulating the splicing of the vascular endothelial growth factor A (VEGF-A, hereafter referred to as VEGF) and subsequently angiogenesis, a cancer hallmark essential for tumors to grow beyond a critical size or metastasize to another organ (Gammons et al., 2014; Mavrou and Oltean, 2016; Oltean et al., 2012).
VEGF is a multifunctional cytokine that plays critical regulatory roles in both pathological and physiological angiogenesis by binding and activating two receptors VEGFR-1 and VEGFR-2 on endothelial cells and activating their downstream signal transduction pathways (Dvorak, 2002; Robinson and Stringer, 2001). VEGF is implicated in a vast majority of solid human cancers, neovascular eye diseases, atherosclerosis, and arthritis, making it a desired therapeutic target. VEGF can be alternatively spliced to form multiple isoforms (Perrot-Applanat, 2012). Two major families of VEGF isoforms, the proangiogenic VEGFxxx and antiangiogenic VEGFxxxb isoforms, are generated differentially through the selection of the proximal splice site (PSS) or the distal splice site in the terminal exon 8 (Bates et al., 2013). Among the isoforms, VEGF165 is the predominant regulator of both physiological and pathological angiogenesis in humans. VEGF165 and its antagonist, VEGF165b, differ in the six C-terminal amino acid sequences (Pritchard-Jones et al., 2007; Varey et al., 2008). VEGF165b exhibits antiangiogenic potency and blocks angiogenesis in vivo (Cebe Suarez et al., 2006). A splicing switch from VEGF165b to the proangiogenic VEGF165 isoform promotes cell growth and migration. Mechanistically, SRSF1 phosphorylation by SR protein kinase (SRPK) 1, the most well-studied member of the SRPK family, promotes PSS usage and contributes to the augmented expression of VEGF165 in several forms of cancer (Merdzhanova et al., 2010; Nowak et al., 2008). Knockdown of SRPK1 or inhibition of SRPK1-mediated SRSF1 phosphorylation switches the balance to increase the levels of VEGF165b and subsequently inhibits angiogenesis and tumor growth in vivo (Gammons et al., 2013b; Nowak et al., 2010).
SRPKs and CDC2-like kinases (CLKs) are two major kinase families that phosphorylate SR proteins and play pivotal roles in the regulation of their trafficking and function during splicing (Giannakouros et al., 2011). While SRPKs selectively phosphorylate only RS dipeptides, CLKs are capable of phosphorylating both RS and proline-serine dipeptides. This difference in substrate specificities allows the two kinase families to fine-tune the extent of phosphorylation of SR proteins and regulate their functions in a coordinated manner. For instance, SRPK1 has been shown to initiate the phosphorylation of SR proteins in the cytoplasm to promote their nuclear import, whereas nuclear CLK1 further phosphorylates the SR proteins to relocate them from the nuclear speckles to the splicing sites (Colwill et al., 1996; Ngo et al., 2005). Because the two kinase families play critical roles at different regulatory points of pre-mRNA splicing, their normal expression and functions are critical for the well-being of cells (Dominguez et al., 2016; Nikas et al., 2020).
In particular, SRPK1 transduces EGF signaling through an Akt-SRPK-SR protein axis to regulate alternative splicing (Zhou et al., 2012). Abnormal expression of SRPK1 has been implicated in multiple types of solid tumors, including breast, pancreatic, colon, ovarian, and hepatocellular carcinomas (Bullock and Oltean, 2017; Gong et al., 2016; Hayes et al., 2007). In addition, SRPK1 is overexpressed in cancer cell samples of adult T cell leukemia and chronic myelogenous leukemia (Hishizawa et al., 2005). In line with these findings, Wang et al. reported that both overexpression and suppression of SRPK1 lead to constitutive activation of the AKT pathway through the disruption of pleckstrin-homology-domain-leucine-rich-repeat-protein-phosphatase-mediated dephosphorylation (Wang et al., 2014). This indicates that SRPK1 has the potential to function as either an oncogene or tumor-suppressor gene (Nikas et al., 2020). These findings together make SRPK1 an attractive alternative therapeutic target for treating angiogenic pathologies and cancers (Patel et al., 2019).
Several small-molecular inhibitors that target the ATP-binding clefts of SRPKs, including the isonicotinamide compound SRPIN340 and its derivatives, have been reported to date (Batson et al., 2017; Fukuhara et al., 2006; Gammons et al., 2013b). Among these inhibitors, SPHINX31 is the most potent one and inhibits SRPK1 at a nanomolar range (Batson et al., 2017). However, despite it binding to SRPK2 and CLK1 with affinities 50-fold lower than that of SRPK1, it is expected to exhibit some off-target activity against the two kinases. Given the unique regulatory roles of SRPKs and CLKs in splicing, it is important to identify specific inhibitors that could distinguish the two families of kinase to precisely inhibit abnormal splicing events to combat diseases. In addition, specific targeting of SRPKs might have therapeutic usage against infectious diseases such as hepatitis B virus and hepatitis C virus infections (Daub et al., 2002; Karakama et al., 2010). We and collaborators recently reported a highly potent irreversible SRPK inhibitor that covalently conjugates with Tyr227 of the spacer helix αS1 that is located immediately next to the ATP-binding cleft of SRPK1 (Hatcher et al., 2018). The exploitation of the unique SRPK spacer in inhibitor development improves both potency and selectivity. Yet, similar to all current SRPK-specific inhibitors, potential drawbacks still exist. First, these inhibitors must compete with intracellular ATP level that is in the millimolar range, which usually lowers the cellular potency of ATP-competitive kinase inhibitors. More importantly, a high possibility exists that drug-resistance-conferring mutation(s) develops at the ATP-binding clefts of SRPKs, such as the mutations of the “gatekeeper” residues that have been observed in many clinically approved ATP-competitive inhibitors (Ricci et al., 2002; Whittaker et al., 2010). Therefore, an alternative approach is preferred to target SRPKs.
Our previous works have shown that SRPK1 binds to SRSF1 with high affinity (Kd ~ 50 nM) and phosphorylates its C-terminal RS domain at approximately 12–14 sites, where the first eight serines are phosphorylated in a processive manner (Aubol et al., 2003; Velazquez-Dones et al., 2005). Crystal structures of SRPK1 in a complex with SRSF1 and a substrate peptide derived from the yeast SR-like protein Npl3p reveal that a docking groove that is distal to its active site confers to the high-affinity substrate binding and regulates the phosphorylation mode by interacting favorably with the arginines in alternating positions of the substrate peptides (Ngo et al., 2005, 2008). Alanine substitutions of four critical residues within the docking groove of SRPK1 significantly reduce SRSF1 binding and adversely affect phosphorylation and subcellular localization of the SR protein. In addition, we revealed that these critical docking groove residues are conserved among SRPKs and are responsible for the interaction and regulation of phosphorylation of other SR and SR-like substrates, including SRSF3 and AcinusS (Liang et al., 2014; Long et al., 2019). Therefore, we hypothesize that inhibitors blocking the docking groove can serve as specific therapeutic agents against SRPK functions.
In this study, by using a structure-based approach, we developed an SRPK-specific peptide inhibitor that targets the unique SRPK docking groove directly. This inhibitor blocked PPI between SRPK1 and SRSF1 and is cell permeable. It could switch the splicing of VEGF from the proangiogenic isoform to the antiangiogenic isoform and block angiogenesis both in vitro and in vivo. More importantly, we evidenced that our inhibitor could be used together with the ATP-competitive SRPK inhibitor to inhibit angiogenesis with greater effectiveness. Our findings provide a new opportunity for cotargeting the ATP- and substrate-binding sites of SRPKs to combat angiogenic pathologies.
Result
Design of a non-ATP-competitive peptide inhibitor targeting the docking groove of SRPK1
Our previous works have identified an acidic SRPK-specific substrate docking groove in SRPK1 that is formed with the MAPK insert and a loop connecting helices αF and αG. We elucidated that this docking groove is critical for binding basic residue-rich substrates and regulates the mechanism of processive phosphorylation of SR proteins (Ngo et al., 2005, 2008). Mutations of four docking groove residues that are critical for substrate binding result in aberrant phosphorylation and subcellular localization of SRSF1. As this docking groove is conserved in SRPKs but absent in all other kinases, we hypothesized that inhibitor targeting this docking groove could block the binding and phosphorylation SRSF1 by SRPK1. Generally, the typical lack of natural ligands for PPIs as the starting template for inhibitor development and the relatively large surface area of PPI interface usually make it challenging to screen for drug-like small-molecule compounds that bind effectively enough to block the PPI (Buchwald, 2010; Sperandio et al., 2010). Therefore, we developed peptide inhibitors of SRPK1 based on the structural information of the kinase docking groove and the primary structures of substrates that bind to it. Basic residue-rich peptides based on the structures of SRSF1 and AcinusS were generated, and their inhibitory activities were tested. However, these peptides showed no or little inhibitory effects on the interaction of SRPK1 and SRSF1 at the tested concentrations (data not shown).
We have previously solved the crystal structure of SRPK1 bound to a 9-mer peptide (R-R-R-E-R-S-P-T-R) derived from the substrate Npl3p of the yeast ortholog Sky1p (PDB ID: 1WBP) (Ngo et al., 2005). This peptide, like the RS domain of SRSF1, binds to the docking groove with a high degree of charge and conformation complementarity. Therefore, we explored whether this Npl3p peptide serves as a better inhibitor of SRPK1 by synthesizing a 7-mer mimic R-E-R-A-R-T-R. The two N-terminal arginines were truncated owing to the lack of electron density in the complex structure, and the serine residue was mutated to alanine to prevent phosphorylation. The proline residue, which makes no contact with SRPK1 and orients toward the solvent, was mutated to arginine for enhancing the electrostatic interaction between the peptide and the acidic docking groove.
Isothermal titration calorimetry (ITC) experiments determined that the 7-mer peptide binds SRPK1 with a dissociation constant (Kd) of 112 ± 9.9 μM (Figure 1A). Next, we performed GST pull-down and kinase assays to evaluate whether the 7-mer could inhibit the interaction between SRPK1 and SRSF1. Our results revealed that although the 7-mer directly bound to SRPK1, it inhibited the binding and phosphorylation of SRSF1 by SRPK1 only at the highest concentrations tested (Figures 1D and 1G).
Figure 1.
20-mer peptide inhibitor (DBS1) binds SRPK1 directly and inhibits its phosphorylation of SRSF1
(A–C) Isothermal titration calorimetry studies of the binding of SRPK1 to peptides of different lengths. The raw thermograms (top panels) and the binding isotherms fitted to a single-site model (bottom panel) of (A) 7-mer, (B) 14-mer, and (C) 20-mer binding to SRPK1. (A) 7-mer binds to SRPK1 with a Kd value of 112 ± 9.5 μM. (B) 14-mer binds to SRPK1 with a Kd value of 7.85 ± 1.22 μM. (C) 20-mer binds to SRPK1 with a Kd value of 5.55 ± 1.12 μM. The reported errors correspond to the standard deviation (S.D.) of the fit. Each experiment was repeated at least three times with consistent results obtained, and only representative graphs are shown.
(D–F) In vitro GST pull-down assays of GST-tagged SRSF1 with SRPK1ΔNS3 in the presence of different peptide inhibitors. Samples were separated by SDS-PAGE and stained by Coomassie Blue. The top panels show the pull-down results and the bottom panels show the input. (C) The binding of GST-SRSF1 and SRPK ΔNS3 was blocked by the 7-mer at concentration of 400 μM or higher. (D) 14-mer and (E) 20-mer inhibited the binding of GST-SRSF1 and SRPK1ΔNS3 in dose-dependent manner.
(G–I) GST-SRSF1 was phosphorylated by SRPK1 in the presence of different concentrations of peptide inhibitors using 50 μM [32P] ATP. Radiolabeled protein bands corresponding to the phosphorylated GST-SRSF1 were quantified by using a phosphor imager and the Image Quant software. (G) 7-mer did not significantly inhibit the phosphorylation of SRSF1. (H) Top panel: 14-mer inhibited SRPK1-medated SRSF1 phosphorylation in a dose-dependent manner. Bottom panel: The band intensity of phosphorylated SRSF1 was plotted as a function of inhibitor concentration. The IC50 value represents the concentration of peptide inhibitor that reduced the phosphorylation of SRSF1 by 50% when compared with the no-peptide treatment control group. 14-mer inhibited SRPK1-mediated SRSF1 phosphorylation at an IC50 value of 91.18 μM. (I) 20-mer inhibited SRPK1-mediated SRSF1 phosphorylation at an IC50 value 10.77 μM. Data are expressed as mean ± S.D. for at least three independent experiments.
See also Figure S1.
To further guide our peptide inhibitor design, we solved a high-resolution crystal structure of an active fragment of SRPK1 (SRPK1ΔNS3) in complex with the 7-mer by soaking the peptide in apo-SRPK1 crystals. The overlay of our new structure with that of SRPK1 in complex with the Npl3p peptide reveals that SRPK1 adopts the same conformation in both models, and the 7-mer peptide also docks at the docking groove of SRPK1 (Figure 2A). However, we only observed the electron density of five amino acids (R3–R7), suggesting that the two N-terminal residues remain flexible. In addition, the peptide adopts a different conformation and shifts toward the right side of the docking groove compared with the Npl3p peptide in 1WBP and the N-terminal RS domain in the crystal structure of SRPK1 in complex with SRSF1 (PDB ID:3BEG) (Figure 2B).
Figure 2.
Complex crystal structure of SRPK1:7-mer peptide
(A) Overall structure of 7-mer in complex with SRPK1. 7-mer is colored in yellow and docks at the docking groove of SRPK1. Inset: Four SRPK1 residues that are previously identified to play critical roles in substrate binding and phosphorylation are shown in dark blue.
(B) Overlaid of the SRPK1: 7-mer and the structure of SRPK1 in complex with a 9-mer Npl3p peptide (PDB ID: 1WBP). The two structures were superimposed using the α carbons of the kinases. The 7-mer and 9-mer peptides are colored in yellow and light magenta, respectively. Comparison of the positions of peptides reveals that 7-mer shifts to the right side of the docking groove.
(C) Overlaid of the SRPK1:7-mer and the apo-SRPK1 (PDB ID: 1WAK) structures. The two structures were superimposed using the α carbons of the kinases. SRPK1:7-mer complex and apo-SRPK1 are colored light blue and wheat respectively. Upon binding of the 7-mer, the sidechains of H554 and W606 of SRPK1 flip outward. R3 of 7-mer interacts with the sidechains of D564 and E571 of SRPK1 through electrostatic interactions (magenta). R7 of 7-mer hydrogen bonds with the backbone carbonyl of L550 and the hydroxyl of Y616 of SRPK1 (gray).
See also Table S1.
Owing to the shift, the side chain of H554, compared with the apo structure of SRPK1, adopts a different rotamer to accommodate the peptide. The remaining peptides interact with the docking groove favorably through charge complementarity as observed in 1WBP and 3BEG. The carboxylates of residues D564 and E571 from helix αG of SRPK1 interact with R3 through electrostatic interactions. In addition, the guanidinium group of the arginine R7 forms hydrogen bonds with the backbone carbonyl of L550 and the phenolic hydroxyl group of Y616. The insertion of the R7 sidechain displaces the sidechain of W606 and pushes it away from the groove compared with the apo kinase (Figure 2C). The same change in the rotamer of W606 is observed in the models of 1WBP and 3BEG, suggesting that W606 might function as a gate of the substrate docking groove and must be flipped out for substrate binding.
Given the alternate binding position of the 7-mer peptide, we speculated that the low inhibitory activity observed is due to the bound peptide “sliding out” of the docking groove during the assay, as “sliding” of the bound peptide/motif of the substrate along the docking groove is an intrinsic property that makes processive phosphorylation possible.
We therefore attempted to improve its inhibitory activity with proximity effect by generating 14-mer and 20-mer peptide ligands that resemble a dimer and trimer of the 7-mer peptide, respectively, to increase its local concentration and minimize the loss of inhibitory activity owing to sliding. ITC studies showed that both the 14-mer and the 20-mer peptides bound with significantly higher affinities to SRPK1 with Kd values of 7.9 ± 1.2 and 5.6 ± 1.1 μM (Figures 1B and 1C), respectively. We next tested the inhibitory effects of the peptides by using GST pull-down and kinase reaction assays. Our results revealed that both the 14-mer and the 20-mer effectively blocked the interaction between SRPK1 and SRSF1 in a dose-dependent manner and inhibited SRPK1 activity with IC50 values of 90 and 11 μM, respectively (Figures 1E, 1H, 1F, and 1I). Considering the improved binding affinity and inhibitory activity of the 20-mer peptide, we selected it as our inhibitor candidate and hereinafter referred to it as Docking Blocker of SRPK1 (DBS1). To demonstrate that DBS1 inhibits SRPK1 activity in a non-ATP-competitive manner, we tested the inhibitory activity of DBS1 in the presence of increased ATP concentrations. The ATP-competitive SRPK inhibitor SRPIN340 was used as a control (Fukuhara et al., 2006). The results revealed that the inhibitory activity of SRPIN340, but not DBS1, was reduced when excess ATP was present (Figure S1A). We next assayed the SRPK1-mediated SRSF1 phosphorylation at different SRSF1 or ATP concentrations in the presence of increasing amount of DBS1. Lineweaver-Burk plots of these assays revealed that DBS1 inhibited the kinase activity of SRPK1 in a competitive manner against the substrate SRSF1, but not ATP, with a Ki value of 2.12 μM (Figure S1B). Together, our results confirm that DBS1 is a new class of non-ATP-competitive SRPK inhibitor.
DBS1 inhibits SRPK docking groove selectively
To confirm whether DBS1 exerted its inhibitory effect through blocking of the docking groove, both ITC and microscale thermophoresis (MST) experiments were performed to compare the binding of DBS1 with SRPK1ΔNS3 and a docking groove mutant (SRPK1ΔNS3(DM)) in which four residues (D548, D564, E571, and K615) critical for substrate binding are mutated to alanines (Figure 3A) (Ngo et al., 2005). Our results revealed that the mutations abolished the binding of DBS1, confirming the inhibitor specificity for the SRPK1 docking groove (Figures 3B and 3C). The Kd value of DBS1 obtained through MST measurements is 3.2 ± 1.5 μM, which is in line with the ITC result (Figure 3D).
Figure 3.
DBS1 blocks the SRPK1 docking groove using its arginines
(A) Schematic diagram of the organization of SRPK1ΔNS3 and its docking groove mutant protein. Four residues at the docking groove that are critical for substrate interaction are mutated to alanine.
(B) In vitro GST pull-down assays of GST-tagged SRSF1 with SRPK1ΔNS3 or SRPK1ΔNS3_DM. Samples were separated by SDS-PAGE and stained by Coomassie Blue. Mutations at the docking groove abolish the interaction with SRSF1.
(C) ITC studies of the binding of DBS1 to SRPK1ΔNS3_DM. No binding between SRPK1ΔNS3_DM and DBS1 was detected. Experiment was repeated at least three times and only the representative data are shown.
(D) MicroScale Thermophoresis (MST) analysis to measure the binding affinity between SRPK1 and DBS1. A Kd of 3.15 ± 1.5 μM was determined for the interaction between DBS1 and SRPK1ΔNS3 using standard data analysis with MO. Affinity Analysis Software (5 s MST-on time for evaluation), while the interaction between SRPK1_DM with DBS1 showed no binding. Values are means ± S.D. for three independent experiments.
(E) The sequences of DBS1 and mutant peptides. Arginine at different locations of DBS1 (bolded) were substituted by alanine and the mutants are termed M1, M2, M3, and M4, respectively.
(F) In vitro GST pull-down assays of GST-tagged SRSF1 with SRPK1ΔNS3 in the presence of different concentrations of DBS1 or DBS1 mutants. Samples were separated by SDS-PAGE and stained by Coomassie Blue. Left panel: Input samples containing 500 nM GST-SRSF1 and SRPK1 in the presence of different concentrations of DBS1 or DBS1 mutants were shown. Right panel: DBS1 effectively blocked the interaction between SRPK1 and SRSF1 in a dose-dependent manner. M2 showed no inhibitory effect while the inhibitory activities of the other mutants were partially impaired.
(G) In vitro radioactive kinase assays in the presence of DBS1 or DBS1 mutants. GST-SRSF1 was phosphorylated by SRPK1 in the presence of different concentrations of DBS1 or mutants using 50 μM [2P] ATP. Radiolabeled GST-SRSF1 protein bands were scanned using a phosphor imager. DBS1 inhibited SRSF1 phosphorylation in a dose-dependent manner. In comparison, M2 mutant totally lost inhibitory effect and the other three mutants exhibited impaired inhibitory effects. Each experiment was performed three times independently, only the representative results are shown.
See also Figure S2.
Although the docking groove is unique to the SRPK family, the four critical residues are conserved among all family members (Liang et al., 2014). We next tested whether DBS1 could inhibit the highly homologous SRPK2 as well as two other SRSF1 kinases CLK1 and Akt/PKB that do not contain the docking groove (Figure S2A) (Blaustein et al., 2005; Colwill et al., 1996). DBS1 inhibited both SRPKs but not CLK1 or Akt/PKB, further confirming it specifically targets the SRPK docking groove. Interestingly, DBS1 appeared to be selective against SRPK1 over SRPK2, suggesting that targeting the docking groove may provide a new route to develop member-specific SRPK inhibitors, which will be discussed later.
The selectivity of DBS1 against SRPK was further evaluated by screening its inhibitory activity against 140 different kinases by using the kinase inhibitor profiling service provided by the International Center for Kinase Profiling at the MRC Protein Phosphorylation Unit. DBS1 showed high selectivity and did not significantly inhibit other protein kinases (Figure S2B). Interestingly, DBS1 did not inhibit SRPK1 in this screen, likely because a free RS-rich peptide instead of SRSF1 was used as the substrate in the phosphorylation assay. This observation matches our results that DBS1 targets the distal substrate docking groove instead of the ATP-binding or substrate-binding sites near the active site.
Arginines in the central region of DBS1 are critical for inhibition
The docking groove of SRPK1 preferentially interacts with basic residues on its substrates. To examine whether all arginines of DBS1 are necessary for its inhibitory activity and to identify the particular residues, if any, that play more important roles, we generated arginine-to-alanine mutations at four different regions of DBS1. These mutants are termed as DBS1 M1, M2, M3, and M4, and their inhibitory activities were tested using GST pull-down and kinase assays. We observed that compared with DBS1, all four mutants exhibited reduced inhibitory activities. In particular, DBS1 M2, which contains mutations in the midregion (i.e. R7, R8, and R10), failed to inhibit the interaction and phosphorylation of SRSF1 through SRPK1 (Figures 3E–3G). These results suggested that all arginines are required for the inhibitory activity of DBS1, and three arginines in the midregion play the most critical roles.
DBS1 is cell-penetrating
DBS1 is rich in arginines that mimic cell-penetrating peptides (CPPs) (Schmidt et al., 2010). To test whether DBS1 could enter cells without the aid of delivery reagents, we conjugated the peptide with fluorescein 5′-carboxylic acid (5-FAM) and transduced it into the metastatic melanoma cell line A375 and the cervical cancer cell line HeLa. Confocal microscopy showed that DBS1 alone was able to penetrate into the cells. However, the use of pyrenebutyrate, a delivery agent that enhances cellular uptake of basic peptides, further enhanced the cellular uptake of DBS1 (Takeuchi et al., 2006). By contrast, a 5-FAM-conjugated 20-mer control peptide with irrelevant amino acid sequence (LNGHEDAQAFPTRIVYLSKM) could not enter the cells without the aid of pyrenebutyrate (Figure 4). These findings indicate that the arginine-rich sequence of DBS1 might have conferred its cell-penetrating property to overcome the cell membrane permeability issue. Nevertheless, to ensure that the cellular delivery of DBS1 is efficient and consistent, we used pyrenebutyrate in the following experiments.
Figure 4.
DBS1 is a cell-permeable inhibitor
Internalization of the FAM tagged DBS1 and control peptide into A375 (left two panels) and HeLa cells (right two panels). The peptides were conjugated with fluorescein 5′-carboxylic acid (5-FAM) at the N-termini and imaged using confocal laser scanning microscopy (CLSM). Top panels: Cells without pyrenebutyrate treatment were incubated with 50 μM peptides overnight only. Bottom panels: Cells were first incubated with 50 μM peptides overnight, followed by treatment with 50 μM pyrenebytyrate for 15 min on the second day. Cells were washed three times before images were taken. DBS1 alone permeated into the cell. In contrast, the control peptide was hardly detected inside the cell. The efficiency of peptide internalization increased notably with the aid of pyrenebutyrate. The scale bar represents 30 μm.
Next, we tested the inhibitory effect of DBS1 on SR protein phosphorylation in cells. A375 or HeLa cells were treated with different concentrations of DBS1 with the aid of pyrenebutyrate. Western blot analysis with anti-phospho-SR protein antibodies revealed that DBS1 inhibited SR protein phosphorylation in treated cells (Figures 5A and 5B).
Figure 5.
DBS1 switches the VEGF pre-mRNA splicing from VEGF165 to VEGF165b dose-dependently
(A and B) Effects of DBS1 treatment on the phosphorylation of SR proteins. Total cell lysates from A375 or HeLa cells treated with the indicated concentrations of DBS1 was subjected to western blotting probed with the anti-phospho-SR protein antibody mAb1H4. (A) Top panel: The phosphorylation levels of multiple SR proteins were reduced upon DBS1 treatment in A375 cells. Bottom panel: The ratios of phospho-SR proteins versus tubulin were analyzed. (B) The phosphorylation levels of phospho-SR proteins were downregulated upon DBS1 treatment in HeLa cells.
(C and D) DBS1 switched the expression of VEGF isoforms in A375 cells in dose-dependent manner. Expression levels of VEGF165 mRNA or protein in DBS1-treated cells were analyzed by RT-PCR or western blotting. (C) Top panel: Expression levels of VEGF165 decreased in A375 cells upon treatment with DBS1. Bottom panel: The ratios of VEGF165 mRNA or protein versus GAPDH or tubulin, respectively, were quantified. (D) DBS1 increased VEGF165b expression. Top panel: Both mRNA and protein levels of VEGF165b increased upon DBS1 treatment in A375 cells. Bottom panel: The ratios of expression levels of VEGF165b relative to the reference were analyzed.
(E and F) Treatment of HeLa cells with DBS1 altered the expression of VEGF isoforms. (E) DBS1 decreased VEGF165 expression in HeLa cells. (F) The expression of VEGF165b significantly increased upon DBS1 treatment. Values are means ± S.D. for three independent experiments. ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.005.
DBS1 alters VEGF splicing from proangiogenic to antiangiogenic isoform
VEGF splicing is tightly regulated with SRPK1-mediated SRSF1 phosphorylation. Knockdown or inhibition of SRPK1 with different inhibitors switches VEGF splicing to produce the antiangiogenic VEGF165b isoform (Gammons et al., 2013a; Mavrou and Oltean, 2016). Because DBS1 inhibits SRSF1 phosphorylation through SRPK1 in vitro and the phosphorylation of endogenous SR proteins in cells, we tested the effect of DBS1 on VEGF splicing.
A375 or HeLa cells were treated with varying DBS1 dosages. We observed a clear dose-dependent shift in splicing to the VEGF165b isoform in both cell lines. Consistent with the shift in mRNA levels, treatment of 25 μM DBS1 in A375 cells also significantly suppressed and increased the protein expression of VEGF165 and VEGF165b, respectively (Figures 5C and 5D). Similar results were observed in HeLa cells. The protein expression level of VEGF165 decreased on DBS1 treatment, whereas that of VEGF165b increased significantly at 25 μM or higher concentrations (Figures 5E and 5F).
DBS1 suppresses angiogenesis
The balance of the VEGF165 and VEGF165b expressions dictate angiogenesis. On the basis of the observed effects of DBS1 on the switching of VEGF splicing, we examined whether DBS1 could attenuate angiogenesis through an in vitro tube formation assay that models the reorganization stage of angiogenesis (DeCicco-Skinner et al., 2014). In brief, trypsinized human umbilical vein endothelial cells (HUVECs) were cultivated using conditioned media (CM) collected from A375 or HeLa cells treated with different concentrations of DBS1 or an irrelevant control peptide derived from the RRM2 of SRSF1 fused with a CPP (GRKKRRQRRRPQSWQDLKDH), a sequence derived from the transactivator of transcription (TAT) protein of HIV-1. Pull-down and kinase activity assays were performed to confirm that the control peptide did not inhibit the binding and phosphorylation of SRSF1 through SRPK1 (Figure S3). HUVECs cultured in the endothelial cell growth medium with DBS1 served as a negative control to discount any antiangiogenic effect of DBS1 on HUVECs directly. After the HUVECs reached appropriate density, they were added on top of a growth-factor-reduced basement membrane matrix (Matrigel). We first measured the cytotoxicity of both DBS1 and the control peptide by using MTT. Our results illustrated that they have no significant cytotoxicity on both cell lines at the concentrations we tested (Figure S4A).
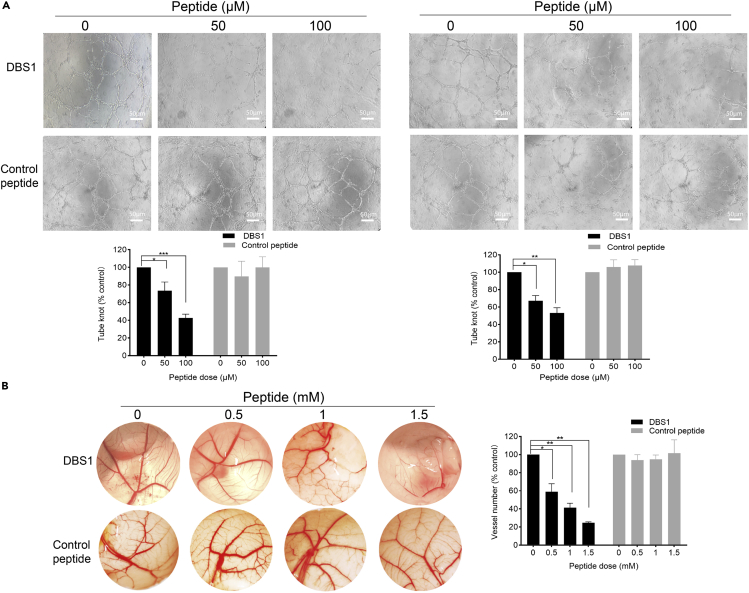
The antiangiogenic activity of DBS1 was determined by counting the branch points of the formed capillary-like tubes. Our results demonstrated that when HUVEC cells were treated with CM collected from DBS1-treated A375 or HeLa cells, the formation of capillary-like structures and branch points reduced significantly in a dose-dependent manner (Figure 6A). By contrast, HUVECs treated with either CM collected from the control peptide treated cells or DBS1 directly showed no effect on tube formation (Figures 6A and S4B). These results indicate that DBS1 is noncytotoxic to HUVECs and could potently inhibit angiogenesis.
Figure 6.
DBS1 inhibits angiogenesis
(A) DBS1 dose-dependently inhibited endothelial cell tube formation on Matrigel. Top panel: Human umbilical vein endothelial cells (HUVECs) were detached and resuspended in Media 199. The cells (3 × 104 cells/well) together with 100μL of conditioned media (CM) collected from DBS1 or control peptide treated A375 (Left panel) or HeLa cells (Right panel) for 24 h were added to 96-well cell culture plates coated with growth factor reduced Matrigel. After 6 h incubation at 37°C, the capillary-like tubes formed by HUVECs in each well were pictured using an inverted microscope with 10x magnification from three different viewing fields. Representative images are shown. The scale bar represents 50 μm. Bottom panel: Relative tube formation ability was calculated by comparing the tube knots formed in each sample with those of the no treatment control. DBS1 treatment suppressed angiogenesis in a dose-dependent manner while the control peptide exhibited no effect on tube formation. Assays were performed in triplicate with consistent results. Values are means ± S.D. for independent experiments.
(B) DBS1 suppressed angiogenesis in chick embryo chorioallantoic membrane (CAM). Left panel: CAMs were photographed 6 days after the application of either PBS or 100μL of 0, 0.5, 1, and 1.5 mM DBS1 (top panel) or control peptide (bottom panel). Right panel: Vessel number was quantified by ImageJ and quantified. Values are means ± S.D. for three independent experiments. ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.005.
See also Figures S3 and S4.
We next used chick embryo chorioallantoic membrane (CAM) assay to validate the antiangiogenic effect of DBS1 in vivo (Lokman et al., 2012; Ribatti, 2017). Different dosages of DBS1 or control peptide (130, 260, and 390 μg) were applied to the CAM of an 8-day-old chicken embryo, and vascularization in the area of application was examined 6 days later. Our results revealed that DBS1, but not control peptide, significantly inhibited vascularization (Figure 6B). Together, our results clearly demonstrated that DBS1 is an antiangiogenic inhibitor.
Cotreatment of DBS1 with ATP-competitive inhibitor of SRPK1 inhibits angiogenesis more effectively
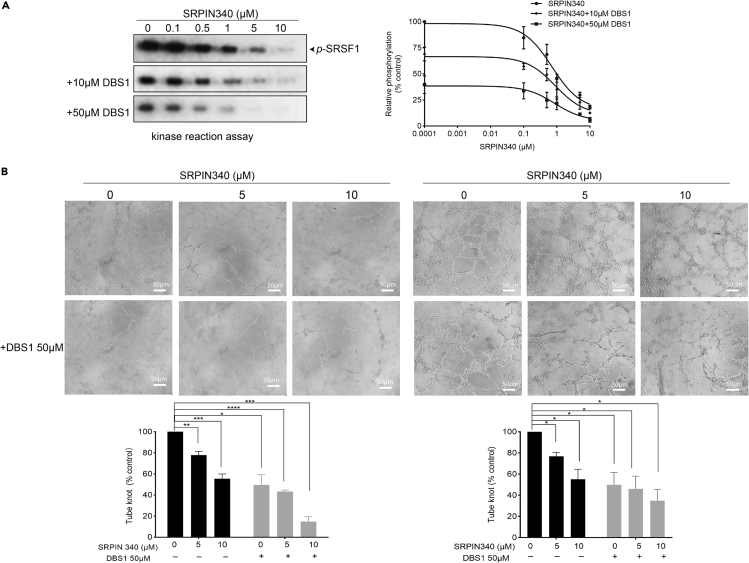
Our findings thus far indicate that the inhibition of the PPI of SRPK1 and its substrate lead to antiangiogenic activity. Studies have demonstrated that ATP-competitive inhibitors of SRPK1 are also antiangiogenic (Gammons et al., 2013b). Therefore, we tested whether combining DBS1 and SRPIN340 inhibits angiogenesis more effectively (Fukuhara et al., 2006). The first-generation SRPK1 inhibitor SRPIN340 was selected instead of other more potent inhibitors so that any significant improvement in inhibitory activity could be easily observed. We first tested whether combining the two inhibitors induces cytotoxicity to either A375 or HeLa cells. MTT assays revealed that cotreatment with 10 μM SRPIN340 and 50 μM DBS1 did not affect the viability of either A375 or HeLa cells (Figure S5A). In addition, HUVECs were directly treated with the combination of SRPIN340 and DBS1 to examine the effect of inhibitors on tube formation. We found no difference in tube formation when compared with the control (Figure S5B), indicating that cotreatment exerted neither toxicity nor antiangiogenic effects on HUVECs directly. Moreover, we performed an in vitro kinase assay by using SRPIN340 alone or in a mixture that contained a fixed amount of DBS1 (10 μM or 50 μM) and varying amounts of SRPIN340. We found that the cotreatment further suppressed SRSF1 phosphorylation, suggesting that dual inhibition may inhibit SRPK1 more potently (Figure 7A).
Figure 7.
Cotreatment of SRPIN340 and DBS1 suppresses angiogenesis more potently
(A) Left Panel: SRSF1 was phosphorylated by SRPK1 with 50 μM [32P] ATP in the presence of various concentrations of SRPIN340 alone, or with the addition of 10 μM or 50 μM DBS1. Radiolabeled SRSF1 protein bands corresponding to the phosphorylated SRSF1 were scanned. Right panel: The band intensity of phosphorylated SRSF1 was plotted as a function of SRPIN340 concentration. SRPIN340 inhibited SRPK1-mediated SRSF1 phosphorylation dose-dependently (Fukuhara et al., 2006). Combination of SRPIN340 and 50 μM DBS1 inhibited SRSF1 phosphorylation more effectively. Error bars represent the standard deviation for triplicate measurements.
(B) A375 or HeLa cells were treated with different concentrations of SRPIN340 alone or with the addition of 50 μM DBS1 for 24 h. The corresponding CM were then collected and subjected for tube formation assay. Top panel: The HUVECs (3 × 104 cells/well) together with 100 μL of CM collected from A375 (left panel) or HeLa cells (right panel) were added to 96-well cell culture plates coated with growth factor reduced Matrigel. Capillary-like tubes formed by HUVECs in each well were pictured using an inverted microscope with 10x magnification from three different viewing fields. Representative images are shown. The scale bar represents 50 μm. Bottom panel: Relative tube formation ability was quantified by comparing the number of tube knots of each sample with those of the no treatment control. Assays were performed in triplicate. Values are means ± S.D. for three independent experiments. ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.005.
See also Figure S5.
A tube formation assay was then performed as described earlier in the presence of SRPKIN340 only or the combination of SRPIN340 and DBS1. CM collected from either A375 or HeLa culture cotreated with SRPIN340 and DBS1 exhibited significantly lower angiogenic activity than with SRPIN340 treatment alone. In particular, cotreatment of A375 or HeLa cells with 10 μM SRPIN340 and 50 μM DBS1 suppressed tube formation nearly by 85% and 65% (Figure 7B), respectively. These results indicated that cotreatment of cancer cell lines with ATP-competitive inhibitor and PPI inhibitor of SRPK1 could inhibit angiogenesis more effectively.
Discussion
Over the past decade, accumulated evidence has indicated that SRPK1 plays important roles in multiple hallmarks of cancer, including angiogenesis and metastasis (Amin et al., 2011; Chang et al., 2015; Lin et al., 2014; van Roosmalen et al., 2015). SRPK1 is overexpressed in many forms of cancer and is correlated with advanced tumor staging and poor prognosis (Nikas et al., 2020). Downregulation or inhibition of SRPK1 in preclinical models usually results in tumor growth reduction, implicating SRPK1 as a potential cancer drug target (Mavrou et al., 2015). SRPK1 influences many oncogenic processes across diverse cancer forms. Among these processes, the role of SRPK1 in angiogenic pathogenesis has been the most elaborated. In particular, SRPK1-mediated phosphorylation of SRSF1 serves as a key pathogenic mechanism through alteration of VEGF pre-mRNA splicing to produce its proangiogenic isoform and to promote angiogenesis (Oltean et al., 2012). Although several ATP-competitive inhibitors of SRPKs have been developed, concerns regarding their abilities to compete with a high concentration of cellular ATP and the high potential of occurrence of drug resistance-conferring mutation(s) remain unsolved. We therefore aimed to identify an inhibitor that targets the region outside the ATP-binding site of SRPK1. Currently, most non-ATP-competitive kinase inhibitors are often peptide inhibitors derived from their native substrates, inhibitor proteins, or other binding partners. For example, a 20-mer peptide inhibitor derived from the heat-stable inhibitor protein of PKA targeting a substrate-binding region near the active site serves as a potent competitive inhibitor of PKA (Dalton and Dewey, 2006). A series of pseudosubstrate peptide inhibitors designed on the basis of an optimal peptide substrate sequence of AKT1 inhibits the kinase in a competitive manner with Ki as low as 0.1 μM (Luo et al., 2004). However, no inhibitor protein for SRPKs has been identified thus far, and owing to a lack of structural information on how the substrate binds near the active site, developing inhibitors for SRPK1 by using similar strategies is difficult. Molecular dynamic simulations and structural studies have demonstrated that the RS domain of SRSF1 is highly dynamic and gets phosphorylated in a processive manner, suggesting that the RS region most likely binds near the active site in a transient manner.
To overcome these limitations, we targeted a unique SRPK-specific docking groove that is distal to the active site. This docking groove is critical for the binding and phosphorylation of different substrates by SRPKs, so its blockage would lead to PPI inhibition. PPI interfaces are promising drug-targeting sites. However, PPI interfaces are usually large and lack deep pockets for inhibitors to specifically bind to. In addition, the presence of noncontiguous binding sites makes targeting PPIs much challenging. However, for SRPK1, the docking groove is a deep concave pocket that serves as an ideal inhibitor-targeting site. We first solved a new crystal structure of SRPK1 in complex with a 7-mer peptide inhibitor at the docking groove. This peptide was designed based on previous structural information obtained from SRPK1, substrate complex structure, but it exhibits surprisingly low inhibitory activity in vitro. The crystal structure revealed that the inhibitor shifts to an alternate binding position within the docking groove compared with previous SRPK1 complex structures. This finding suggests that the interaction at the docking groove is likely dynamic and explains the low activity of the 7-mer inhibitor. Inspired by this observation, we rationally designed DBS1, which serves as a PPI inhibitor for SRPK1. We demonstrated that DBS1 specifically binds to the docking groove, interferes with the binding of SRSF1, and inhibits its phosphorylation. This altered the expression of VEGF165 and VEGF165b, resulting in the inhibition of angiogenesis both in vitro and in vivo.
Given its specificity against the SRPK docking groove, DBS1 also inhibited the homologous SRPK2 which docking groove is conserved. Conversely, it did not inhibit other SRSF1 kinases such as CLK1, CLK2 (based on the phosphorylation screen), and Akt/PKB. It also failed to inhibit the panel of kinases tested. Although some kinases appear to be activated instead, this is likely owing to a charge or buffer effect caused by the high concentration of DBS1 being used. Alternatively, the presence of a threonine in DBS1 might also lead to off-target phosphorylation by the tested kinases. Nonetheless, these results revealed that DBS1 is an SRPK-specific inhibitor.
Because of the non-ATP-competitive nature of DBS1, it can be coadministered with other SRPK inhibitors that target the ATP-binding clefts. The observation that cotreatment with DBS1 and SRPIN340 inhibited angiogenesis with improved effectiveness provides a new direction for developing antiangiogenic treatment. Such a strategy will likely achieve better selectivity, lower the inhibitor dosage, and reduce cytotoxic and side effects.
SRPK inhibitors that have been reported thus far are specific to the family, but they failed to distinguish between different family members, that is, SRPK1, SRPK2, and so on. Here we showed that DBS1 is more selective toward SRPK1 compared with SRPK2. Therefore, our strategy to target the docking groove of SRPK might provide a new solution to overcoming this caveat. We recently demonstrated a residue that is located at the core of the docking groove, L568 in SRPK1 and H601 in SRPK2, plays regulatory role in the processive phosphorylation mechanism of SR proteins. SRPK1 processively phosphorylates SRSF1 at approximately eight sites, whereas SRPK2 does so with a lower processivity of approximately five sites. When H601 of SRPK2 is converted to a leucine residue as in SRPK1, the processivity increases to eight sites, underscoring the importance of this core residue (Long et al., 2019). We hypothesize that such increase in processivity is likely because histidine residue can adopt different protonation states. Therefore, although docking groove residues critical for substrate binding are conserved among all SRPKs, differences in the amino acid identity within the core of the groove could provide a new opportunity to develop member-specific inhibitors for the SRPK family.
We chose peptides instead of small molecules as the docking-groove blocker because small molecules could not effectively block the large PPI interfaces. Furthermore, peptides consist of natural amino acids or closely related analogs, are generally nontoxic, and rarely undergo drug-drug interactions. Although peptide inhibitors generally could not permeate the cellular membrane, the identification and application of CPPs, such as the TAT tag in our control peptide, has mostly solved the problem (Futaki et al., 2001). Many CPPs are basic, and it is believed that their electrostatic interactions with the negatively charged phospholipids and proteoglycans on the cellular membrane initiate the cellular entry. Further investigations revealed that the guanidinium group of arginine makes it a more effective residue than lysine in facilitating the uptake (Bechara and Sagan, 2013). Therefore, the arginine-rich design of DBS1 not only makes it effective to compete with the RS domain of SRSF1 at the SRPK1 docking groove but also confers it self-cell-permeability characteristic without further modifications.
Here, we demonstrated that a peptidyl inhibitor that targets the SRPK-specific docking groove could block the interaction between SRPK1 and SRSF1 and subsequently inhibit angiogenesis, providing evidence that the interference of the docking groove is a feasible and promising direction for designing highly specific SRPK PPI inhibitors. Although effective delivery of peptide inhibitors orally remains challenging, advances in nasal, pulmonary, and other noninvasive delivery approaches or the usage of nanocarriers could provide solutions in future. Optimization of peptides is comparatively simpler and more diversified than using small molecules. Further structural modifications of DBS1 such as D-amino acid or unnatural amino acid substitutions, N-terminal acetylation and C-terminal amidation, PEGylation, and lipidation could help enhance the stability and delivery of the inhibitor and develop it into a potent drug candidate.
Limitations of the study
Our study has identified a peptide inhibitor DBS1 that blocks the protein-protein interaction between SRPK1 and SRSF1. While it specifically targets SRPKs and appears to be more selective against SRPK1 than SRPK2, further work is still required to optimize it into an SRPK-family-member-specific inhibitor. In addition, DBS1 is still an early inhibitor whose inhibitory activity remains to be improved. Fortunately, given its peptidyl nature, rational modifications to improve its activity and stability is highly feasible.
Despite SRPKs serving as promising antiangiogenic drug targets, several inhibitors have been developed, but so far no comprehensive splicing analysis has been performed to investigate the effects of SRPK inhibition at the transcriptome-wide scale. It would be of interest how inhibition of specific members of the SRPK family affects splicing on a global level.
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Jacky Chi Ki Ngo (jackyngo@cuhk.edu.hk)
Materials availability
Materials are available upon reasonable request, but we may require a payment and/or a completed Materials Transfer Agreement if there is potential for commercial application.
Data and code availability
All relevant data are available from the Lead Contact upon request. The coordinates of SRPK1:7-mer complex have been deposited in the Protein DataBank under the accession number 7DD1.
Methods
All methods can be found in the accompanying transparent methods supplemental file.
Acknowledgments
We thank Professor Ricky Ngok Shun Wong and Dr. Patrick Ying Kit Yue (Hong Kong Baptist University) for providing technical advice on the tube-formation assays. We also thank Professor Knud Jørgen Jensen (University of Copenhagen) for helpful discussion. This work was supported by the General Research Fund grant from the Hong Kong Research Grants Council (Grant number 14122319); the Chinese University of Hong Kong direct grant scheme (Grant numbers 133136856 and BL18592); CUHK School of Life Sciences Seed Fund and Impact Fund Scheme; and the Innovation and Technology Fund (funding support to the State Key Laboratory for Agrobiotechnology, CUHK) of the Hong Kong SAR, China, to J.C.K.N.
Author contributions
Conceptualization, Q.L., C.Z., and J.C.K.N.; Crystallography, Q.L., C.Z., and K.W.Y.Y.; Inhibitor Design, Q.L., C.Z., Y.Z., J.X. and J.C.K.N.; Biochemical and Cellular Experiments, Q.L., C.Z., H.L., S.W.C.W., and B.S.W.M.; CAM Assays, C.C., Q.X., and S.X.; Writing – Original Draft, Q.L., and C.Z.; Review and Editing, J.X., S.X., and J.C.K.N.
Declaration of interests
The authors declare no competing interests.
Published: May 21, 2021
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2021.102423.
Supplemental information
References
- Amin E.M., Oltean S., Hua J., Gammons M.V., Hamdollah-Zadeh M., Welsh G.I., Cheung M.K., Ni L., Kase S., Rennel E.S. WT1 mutants reveal SRPK1 to be a downstream angiogenesis target by altering VEGF splicing. Cancer Cell. 2011;20:768–780. doi: 10.1016/j.ccr.2011.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubol B.E., Chakrabarti S., Ngo J., Shaffer e., Nolen B., Fu X.-D., Ghosh G., Adams J.A. Processive phosphorylation of alternative splicing factor/splicing factor 2. Proc. Natl. Acad. Sci. U S A. 2003;100:12601–12606. doi: 10.1073/pnas.1635129100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D.O., Mavrou A., Qiu Y., Carter J.G., Hamdollah-Zadeh M., Barratt S., Gammons M.V., Millar A.B., Salmon A.H., Oltean S. Detection of VEGF-A(xxx)b isoforms in human tissues. PLoS One. 2013;8:e68399. doi: 10.1371/journal.pone.0068399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batson J., Toop H.D., Redondo C., Babaei-Jadidi R., Chaikuad A., Wearmouth S.F., Gibbons B., Allen C., Tallant C., Zhang J. Development of potent, selective SRPK1 inhibitors as potential topical therapeutics for neovascular eye disease. ACS Chem. Biol. 2017;12:825–832. doi: 10.1021/acschembio.6b01048. [DOI] [PubMed] [Google Scholar]
- Bechara C., Sagan S. Cell-penetrating peptides: 20 years later, where do we stand? FEBS Lett. 2013;587:1693–1702. doi: 10.1016/j.febslet.2013.04.031. [DOI] [PubMed] [Google Scholar]
- Black D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- Blaustein M., Pelisch F., Tanos T., Munoz M.J., Wengier D., Quadrana L., Sanford J.R., Muschietti J.P., Kornblihtt A.R., Caceres J.F. Concerted regulation of nuclear and cytoplasmic activities of SR proteins by AKT. Nat. Struct. Mol. Biol. 2005;12:1037–1044. doi: 10.1038/nsmb1020. [DOI] [PubMed] [Google Scholar]
- Buchwald P. Small-molecule protein-protein interaction inhibitors: therapeutic potential in light of molecular size, chemical space, and ligand binding efficiency considerations. IUBMB Life. 2010;62:724–731. doi: 10.1002/iub.383. [DOI] [PubMed] [Google Scholar]
- Bullock N., Oltean S. The many faces of SRPK1. J. Pathol. 2017;241:437–440. doi: 10.1002/path.4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebe Suarez S., Pieren M., Cariolato L., Arn S., Hoffmann U., Bogucki A., Manlius C., Wood J., Ballmer-Hofer K. A VEGF-A splice variant defective for heparan sulfate and neuropilin-1 binding shows attenuated signaling through VEGFR-2. Cell Mol. Life Sci. 2006;63:2067–2077. doi: 10.1007/s00018-006-6254-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y., Wu Q., Tian T., Li L., Guo X., Feng Z., Zhou J., Zhang L., Zhou S., Feng G. The influence of SRPK1 on glioma apoptosis, metastasis, and angiogenesis through the PI3K/Akt signaling pathway under normoxia. Tumour Biol. 2015;36:6083–6093. doi: 10.1007/s13277-015-3289-2. [DOI] [PubMed] [Google Scholar]
- Colwill K., Pawson T., Andrews B., Prasad J., Manley J.L., Bell J.C., Duncan P.I. The Clk/Sty protein kinase phosphorylates SR splicing factors and regulates their intranuclear distribution. EMBO J. 1996;15:265–275. [PMC free article] [PubMed] [Google Scholar]
- Dalton G.D., Dewey W.L. Protein kinase inhibitor peptide (PKI): a family of endogenous neuropeptides that modulate neuronal cAMP-dependent protein kinase function. Neuropeptides. 2006;40:23–34. doi: 10.1016/j.npep.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Das S., Anczukow O., Akerman M., Krainer A.R. Oncogenic splicing factor SRSF1 is a critical transcriptional target of MYC. Cell Rep. 2012;1:110–117. doi: 10.1016/j.celrep.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S., Krainer A.R. Emerging functions of SRSF1, splicing factor and oncoprotein, in RNA metabolism and cancer. Mol. Cancer Res. 2014;12:1195–1204. doi: 10.1158/1541-7786.MCR-14-0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daub H., Blencke S., Habenberger P., Kurtenbach A., Dennenmoser J., Wissing J., Ullrich A., Cotten M. Identification of SRPK1 and SRPK2 as the major cellular protein kinases phosphorylating hepatitis B virus core protein. J. Virol. 2002;76:8124–8137. doi: 10.1128/JVI.76.16.8124-8137.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCicco-Skinner K.L., Henry G.H., Cataisson C., Tabib T., Gwilliam J.C., Watson N.J., Bullwinkle E.M., Falkenburg L., O’Neill R.C., Morin A. Endothelial cell tube formation assay for the in vitro study of angiogenesis. J. Vis. Exp. 2014;91:1–8. doi: 10.3791/51312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez D., Tsai Y.H., Weatheritt R., Wang Y., Blencowe B.J., Wang Z. An extensive program of periodic alternative splicing linked to cell cycle progression. Elife. 2016;5:e10288. doi: 10.7554/eLife.10288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak H.F. Vascular permeability factor/vascular endothelial growth factor: a critical cytokine in tumor angiogenesis and a potential target for diagnosis and therapy. J. Clin. Oncol. 2002;20:4368–4380. doi: 10.1200/JCO.2002.10.088. [DOI] [PubMed] [Google Scholar]
- Fukuhara T., Hosoya T., Shimizu S., Sumi K., Oshiro T., Yoshinaka Y., Suzuki M., Yamamoto N., Herzenberg L.A., Hagiwara M. Utilization of host SR protein kinases and RNA-splicing machinery during viral replication. Proc. Natl. Acad. Sci. U S A. 2006;103:11329–11333. doi: 10.1073/pnas.0604616103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futaki S., Suzuki T., Ohashi W., Yagami T., Tanaka S., Ueda K., Sugiura Y. Arginine-rich peptides. J. Biol. Chem. 2001;276:5836–5840. doi: 10.1074/jbc.M007540200. [DOI] [PubMed] [Google Scholar]
- Gammons M.V., Dick A.D., Harper S.J., Bates D.O. SRPK1 inhibition modulates VEGF splicing to reduce pathological neovascularization in a rat model of retinopathy of prematurity. Invest. Ophthalmol. Vis. Sci. 2013;54:5797–5806. doi: 10.1167/iovs.13-11634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammons M.V., Fedorov O., Ivison D., Du C., Clark T., Hopkins C., Hagiwara M., Dick A.D., Cox R., Harper S.J. Topical antiangiogenic SRPK1 inhibitors reduce choroidal neovascularization in rodent models of exudative AMD. Invest. Ophthalmol. Vis. Sci. 2013;54:6052–6062. doi: 10.1167/iovs.13-12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammons M.V., Lucas R., Dean R., Coupland S.E., Oltean S., Bates D.O. Targeting SRPK1 to control VEGF-mediated tumour angiogenesis in metastatic melanoma. Br. J. Cancer. 2014;111:477–485. doi: 10.1038/bjc.2014.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannakouros T., Nikolakaki E., Mylonis I., Georgatsou E. Serine-arginine protein kinases: a small protein kinase family with a large cellular presence. FEBS J. 2011;278:570–586. doi: 10.1111/j.1742-4658.2010.07987.x. [DOI] [PubMed] [Google Scholar]
- Gong L., Song J., Lin X., Wei F., Zhang C., Wang Z., Zhu J., Wu S., Chen Y., Liang J. Serine-arginine protein kinase 1 promotes a cancer stem cell-like phenotype through activation of Wnt/beta-catenin signalling in NSCLC. J. Pathol. 2016;240:184–196. doi: 10.1002/path.4767. [DOI] [PubMed] [Google Scholar]
- Graveley B.R. Sorting out the complexity of SR protein functions. RNA. 2000;6:1197–1211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatcher J.M., Wu G., Zeng C., Zhu J., Meng F., Patel S., Wang W., Ficarro S.B., Leggett A.L., Powell C.E. SRPKIN-1: a covalent SRPK1/2 inhibitor that potently converts VEGF from pro-angiogenic to anti-angiogenic isoform. Cell Chem. Biol. 2018;25:460–470 e466. doi: 10.1016/j.chembiol.2018.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes G.M., Carrigan P.E., Miller L.J. Serine-arginine protein kinase 1 overexpression is associated with tumorigenic imbalance in mitogen-activated protein kinase pathways in breast, colonic, and pancreatic carcinomas. Cancer Res. 2007;67:2072–2080. doi: 10.1158/0008-5472.CAN-06-2969. [DOI] [PubMed] [Google Scholar]
- Hishizawa M., Imada K., Sakai T., Ueda M., Hori T., Uchiyama T. Serological identification of adult T-cell leukaemia-associated antigens. Br. J. Haematol. 2005;130:382–390. doi: 10.1111/j.1365-2141.2005.05619.x. [DOI] [PubMed] [Google Scholar]
- Karakama Y., Sakamoto N., Itsui Y., Nakagawa M., Tasaka-Fujita M., Nishimura-Sakurai Y., Kakinuma S., Oooka M., Azuma S., Tsuchiya K. Inhibition of hepatitis C virus replication by a specific inhibitor of serine-arginine-rich protein kinase. Antimicrob. Agents Chemother. 2010;54:3179–3186. doi: 10.1128/AAC.00113-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karni R., de Stanchina E., Lowe S.W., Sinha R., Mu D., Krainer A.R. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat. Struct. Mol. Biol. 2007;14:185–193. doi: 10.1038/nsmb1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang N., Zeng C., Tao K.P., Sou W.H., Hsia H.P., Qu D., Lau S.N., Ngo J.C. Primary structural features of SR-like protein acinusS govern the phosphorylation mechanism by SRPK2. Biochem. J. 2014;459:181–191. doi: 10.1042/BJ20131091. [DOI] [PubMed] [Google Scholar]
- Lin J.C., Lin C.Y., Tarn W.Y., Li F.Y. Elevated SRPK1 lessens apoptosis in breast cancer cells through RBM4-regulated splicing events. RNA. 2014;20:1621–1631. doi: 10.1261/rna.045583.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokman N.A., Elder A.S., Ricciardelli C., Oehler M.K. Chick chorioallantoic membrane (CAM) assay as an in vivo model to study the effect of newly identified molecules on ovarian cancer invasion and metastasis. Int. J. Mol. Sci. 2012;13:9959–9970. doi: 10.3390/ijms13089959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Y., Sou W.H., Yung K.W.Y., Liu H., Wan S.W.C., Li Q., Zeng C., Law C.O.K., Chan G.H.C., Lau T.C.K. Distinct mechanisms govern the phosphorylation of different SR protein splicing factors. J. Biol. Chem. 2019;294:1312–1327. doi: 10.1074/jbc.RA118.003392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Smith R.A., Guan R., Liu X., Klinghofer V., Shen J., Hutchins C., Richardson P., Holzman T., Rosenberg S.H. Pseudosubstrate peptides inhibit Akt and induce cell growth inhibition. Biochemistry. 2004;43:1254–1263. doi: 10.1021/bi034515p. [DOI] [PubMed] [Google Scholar]
- Mavrou A., Brakspear K., Hamdollah-Zadeh M., Damodaran G., Babaei-Jadidi R., Oxley J., Gillatt D.A., Ladomery M.R., Harper S.J., Bates D.O. Serine-arginine protein kinase 1 (SRPK1) inhibition as a potential novel targeted therapeutic strategy in prostate cancer. Oncogene. 2015;34:4311–4319. doi: 10.1038/onc.2014.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrou A., Oltean S. SRPK1 inhibition in prostate cancer: a novel anti-angiogenic treatment through modulation of VEGF alternative splicing. Pharmacol. Res. 2016;107:276–281. doi: 10.1016/j.phrs.2016.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merdzhanova G., Gout S., Keramidas M., Edmond V., Coll J.L., Brambilla C., Brambilla E., Gazzeri S., Eymin B. The transcription factor E2F1 and the SR protein SC35 control the ratio of pro-angiogenic versus antiangiogenic isoforms of vascular endothelial growth factor-A to inhibit neovascularization in vivo. Oncogene. 2010;29:5392–5403. doi: 10.1038/onc.2010.281. [DOI] [PubMed] [Google Scholar]
- Ngo J.C., Chakrabarti S., Ding J.H., Velazquez-Dones A., Nolen B., Aubol B.E., Adams J.A., Fu X.D., Ghosh G. Interplay between SRPK and Clk/Sty kinases in phosphorylation of the splicing factor ASF/SF2 is regulated by a docking motif in ASF/SF2. Mol. Cell. 2005;20:77–89. doi: 10.1016/j.molcel.2005.08.025. [DOI] [PubMed] [Google Scholar]
- Ngo J.C., Giang K., Chakrabarti S., Ma C.T., Huynh N., Hagopian J.C., Dorrestein P.C., Fu X.D., Adams J.A., Ghosh G. A sliding docking interaction is essential for sequential and processive phosphorylation of an SR protein by SRPK1. Mol. Cell. 2008;29:563–576. doi: 10.1016/j.molcel.2007.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikas I.P., Themistocleous S.C., Paschou S.A., Tsamis K.I., Ryu H.S. Serine-arginine protein kinase 1 (SRPK1) as a prognostic factor and potential therapeutic target in cancer: current evidence and future perspectives. Cells Basel. 2020;9:19. doi: 10.3390/cells9010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak D.G., Amin E.M., Rennel E.S., Hoareau-Aveilla C., Gammons M., Damodoran G., Hagiwara M., Harper S.J., Woolard J., Ladomery M.R. Regulation of vascular endothelial growth factor (VEGF) splicing from pro-angiogenic to anti-angiogenic isoforms: a novel therapeutic strategy for angiogenesis. J. Biol. Chem. 2010;285:5532–5540. doi: 10.1074/jbc.M109.074930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak D.G., Woolard J., Amin E.M., Konopatskaya O., Saleem M.A., Churchill A.J., Ladomery M.R., Harper S.J., Bates D.O. Expression of pro- and anti-angiogenic isoforms of VEGF is differentially regulated by splicing and growth factors. J. Cell Sci. 2008;121:3487–3495. doi: 10.1242/jcs.016410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oltean S., Gammons M., Hulse R., Hamdollah-Zadeh M., Mavrou A., Donaldson L., Salmon A.H., Harper S.J., Ladomery M.R., Bates D.O. SRPK1 inhibition in vivo: modulation of VEGF splicing and potential treatment for multiple diseases. Biochem. Soc. Trans. 2012;40:831–835. doi: 10.1042/BST20120051. [DOI] [PubMed] [Google Scholar]
- Patel M., Sachidanandan M., Adnan M. Serine arginine protein kinase 1 (SRPK1): a moonlighting protein with theranostic ability in cancer prevention. Mol. Biol. Rep. 2019;46:1487–1497. doi: 10.1007/s11033-018-4545-5. [DOI] [PubMed] [Google Scholar]
- Perrot-Applanat M. VEGF isoforms. Cell Adh. Migr. 2012;6:526–527. doi: 10.4161/cam.23256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard-Jones R.O., Dunn D.B., Qiu Y., Varey A.H., Orlando A., Rigby H., Harper S.J., Bates D.O. Expression of VEGF(xxx)b, the inhibitory isoforms of VEGF, in malignant melanoma. Br. J. Cancer. 2007;97:223–230. doi: 10.1038/sj.bjc.6603839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribatti D. The chick embryo chorioallantoic membrane (CAM) assay. Reprod. Toxicol. 2017;70:97–101. doi: 10.1016/j.reprotox.2016.11.004. [DOI] [PubMed] [Google Scholar]
- Ricci C., Scappini B., Divoky V., Gatto S., Onida F., Verstovsek S., Kantarjian H.M., Beran M. Mutation in the ATP-binding pocket of the ABL kinase domain in an STI571resistant BCR/ABL-positive cell line. Cancer Res. 2002;62:5995–5998. [PubMed] [Google Scholar]
- Robinson C.J., Stringer S.E. The splice variants of vascular endothelial growth factor (VEGF) and their receptors. J. Cell Sci. 2001;114:853–865. doi: 10.1242/jcs.114.5.853. [DOI] [PubMed] [Google Scholar]
- Schmidt N., Mishra A., Lai G.H., Wong G.C. Arginine-rich cell-penetrating peptides. FEBS Lett. 2010;584:1806–1813. doi: 10.1016/j.febslet.2009.11.046. [DOI] [PubMed] [Google Scholar]
- Shepard P.J., Hertel K.J. The SR protein family. Genome Biol. 2009;10:242. doi: 10.1186/gb-2009-10-10-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperandio O., Reynes C.H., Camproux A.C., Villoutreix B.O. Rationalizing the chemical space of protein-protein interaction inhibitors. Drug Discov. Today. 2010;15:220–229. doi: 10.1016/j.drudis.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Takeuchi T., Kosuge M., Tadokoro A., Sugiura Y., Nishi M., Kawata M., Sakai N., Matile S., Futaki S. Direct and rapid cytosolic delivery using cell-penetrating peptides mediated by pyrenebutyrate. ACS Chem. Biol. 2006;1:299–303. doi: 10.1021/cb600127m. [DOI] [PubMed] [Google Scholar]
- van Roosmalen W., Le Devedec S.E., Golani O., Smid M., Pulyakhina I., Timmermans A.M., Look M.P., Zi D., Pont C., de Graauw M. Tumor cell migration screen identifies SRPK1 as breast cancer metastasis determinant. J. Clin. Invest. 2015;125:1648–1664. doi: 10.1172/JCI74440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varey A.H., Rennel E.S., Qiu Y., Bevan H.S., Perrin R.M., Raffy S., Dixon A.R., Paraskeva C., Zaccheo O., Hassan A.B. VEGF 165 b, an antiangiogenic VEGF-A isoform, binds and inhibits bevacizumab treatment in experimental colorectal carcinoma: balance of pro- and antiangiogenic VEGF-A isoforms has implications for therapy. Br. J. Cancer. 2008;98:1366–1379. doi: 10.1038/sj.bjc.6604308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez-Dones A., Hagopian J.C., Ma C.T., Zhong X.Y., Zhou H., Ghosh G., Fu X.D., Adams J.A. Mass spectrometric and kinetic analysis of ASF/SF2 phosphorylation by SRPK1 and Clk/Sty. J. Biol. Chem. 2005;280:41761–41768. doi: 10.1074/jbc.M504156200. [DOI] [PubMed] [Google Scholar]
- Wang P., Zhou Z., Hu A., Ponte de Albuquerque C., Zhou Y., Hong L., Sierecki E., Ajiro M., Kruhlak M., Harris C. Both decreased and increased SRPK1 levels promote cancer by interfering with PHLPP-mediated dephosphorylation of Akt. Mol. Cell. 2014;54:378–391. doi: 10.1016/j.molcel.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker S., Kirk R., Hayward R., Zambon A., Viros A., Cantarino N., Affolter A., Nourry A., Niculescu-Duvaz D., Springer C. Gatekeeper mutations mediate resistance to BRAF-targeted therapies. Cancer. 2010;2:35–41. doi: 10.1126/scitranslmed.3000758. [DOI] [PubMed] [Google Scholar]
- Zhou Z., Qiu J., Liu W., Zhou Y., Plocinik R.M., Li H., Hu Q., Ghosh G., Adams J.A., Rosenfeld M.G. The Akt-SRPK-SR axis constitutes a major pathway in transducing EGF signaling to regulate alternative splicing in the nucleus. Mol. Cell. 2012;47:422–433. doi: 10.1016/j.molcel.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are available from the Lead Contact upon request. The coordinates of SRPK1:7-mer complex have been deposited in the Protein DataBank under the accession number 7DD1.