Abstract
In the present review, the authors shed light on the SARS-CoV-2 impact, persistence, and monitoring in the soil environment. With this purpose, several aspects have been deepened: i) viruses in soil ecosystems; ii) direct and indirect impact on the soil before and after the pandemic, and iii) methods for quantification of viruses and SARS-CoV-2 monitoring in soil. Viruses are present in soil (i.e. up to 417 × 107 viruses per g TS−1 in wetlands) and can affect the behavior and ecology of other life forms (e.g. bacteria), which are remarkably important for maintaining environmental equilibrium. Also, SARS-CoV-2 can be found in soil (i.e. up to 550 copies·g−1). Considering that the SARS-CoV-2 is very recent, poor knowledge is available in the literature on persistence in the soil and reference has been made to coronaviruses and other families of viruses. For instance, the survival of enveloped viruses (e.g. SARS-CoV) can reach 90 days in soils with 10% of moisture content at ambient. In such a context, the possible spread of the SARS-CoV-2 in the soil was evaluated by analyzing the possible contamination routes.
Keywords: COVID-19, Coronavirus, Viral abundance, Human viruses, Soil environment, Virus monitoring
Graphical abstract
1. Introduction
The recent COVID-19 pandemic caused by SARS-CoV-2 has demonstrated how viruses or pathogenic microorganisms can generate epidemic diseases and subsequently cause socio-economic and environmental damage (Atar and Atar, 2020; Mofijur et al., 2021; Razzaq et al., 2020). SARS-CoV-2 is a viral strain of the coronaviruses (CoVs), which are named after their shape, as the spike proteins present on their surface resemble a crown, or corona in Latin (Acter et al., 2020). CoVs are positive single-stranded RNA viruses characterized by small size (i.e. 60–220 nm in diameter) and detected in many animals, and therefore, a CoV can reach humans causing potential epidemic outbreaks, as previously occurred with MERS-CoV and SARS-CoV (Lee and Hsueh, 2020; Prompetchara et al., 2020; Su et al., 2016).
To date (April 30th, 2021), SARS-CoV-2 has been detected in almost all countries of the world (i.e. 223) by infecting more than 150.1 million people and causing almost 3.2 million deaths (WHO). These data underscore COVID-19 rapid and vast spread, due to its high reproduction number (R0) and long incubation period, factors that led to an easy and wide transmission of the infection (Cao et al., 2020; Liu et al., 2020). Also, the spread of SARS-CoV-2 variants, i.e. B.1.1.7, 501Y.V2, P.1 and B.1.617 by the United Kingdom, South Africa, Brazil and India, respectively, raises worries due to their alleged facility of transmission and wide mutations in the spike protein (Abdool Karim and de Oliveira, 2021; Moelling, 2021; Wang et al., 2021).
The main ways of virus spreads are aerosols and droplets, but it is well known that transmission can also occur via the fecal-oral route (Chen et al., 2020; Ding and Liang, 2020). Therefore, all the environmental compartments (i.e. water, air and soil) on which the virus can act should be necessarily monitored in order to control the spread of the infection (Cela-Dablanca et al., 2021; Núñez-Delgado, 2020a). However, the interactions between viruses and soil have received less attention compared to the other compartments, thus neglecting a possible contagion route. On the contrary, the relationship between water and wastewater, and the virus spread through the air particles, have been extensively studied (Anand et al., 2021b). Indeed, works published up to April 30th, 2021 and available on the Scopus database accounted for 1,524, 692 and just 36 for air, water and soil compartments, respectively (the keywords used were “SARS-CoV-2” and “air”, “SARS-CoV-2” and “water”, and “SARS-CoV-2” and “soil”, respectively).
Specifically, the possible interactions that can occur between wastewater and soil have received little investigation, but the interplay between these two matrices is continuous and frequent (Núñez-Delgado, 2020a). It is worth recalling that wastewater can be employed for secondary uses such as field irrigation, as well as the sewage sludge derived from wastewater treatment plants can be used as fertilizer in agricultural activities (Lamastra et al., 2018; Martínez-Puchol et al., 2020). This type of activity could allow the migration of the virus from wastewater and/or sewage sludge to the ground due to the fact that SARS-CoV-2 was found and proven to be present in these mentioned matrices (Balboa et al., 2020; Núñez-Delgado, 2020b). In addition, a source of interaction between the soil and SARS-CoV-2 can be due to the incorrect disposal of urban and hospital waste (Iyer et al., 2021), such as the protective devices used for the prevention of the virus (e.g. gloves and masks) (Rahman et al., 2020; Zand and Heir, 2020a).
Hence, this review paper is aimed at illustrating the possible impacts and consequences that the COVID-19 pandemic could have on the soil ecosystem. The manuscript is aimed at analyzing: i) the soil ecosystem and its health status before and after the pandemic; ii) the persistence of SARS-CoV-2 and other viruses in the soil; iii) the methods for SARS-CoV-2 detection and quantification in soil and iv) the monitoring and management of soil impacted by viruses.
2. Viruses in soil
During the COVID-19 pandemic, different basic essential parameters (e.g. national economics, public health) are assessed (Hsiang et al., 2020; Rundle et al., 2020; Siche, 2020). However, among all the environmental concerns of the current situation, soil health can be predominant for living organisms. Indeed, soil plays an important role in the decomposition degree of organic compounds, maintaining the biogeochemical cycle (Nannipieri et al., 2017). Also, the soil is one of the greatest reservoirs of microorganisms (Breitbart and Rohwer, 2005; Rohwer et al., 2009), which are responsible for a series of environmental chemistry reactions (Douglas, 2006). However, viruses (e.g. bacteriophages) can affect the bacterial population, causing detrimental effects on soil quality (Srinivasiah et al., 2013) (see section 2.2).
Viruses found in the soil environment can have an impact on economy and production likely due to an infectious cycle that helps the gene transformation process (Breitbart and Rohwer, 2005; Jain, 2003; Jones et al., 2007). In addition, specific viruses such as SARS-CoV-2 can be transmitted to the soil through infected water bodies, by means of sewage sludge and irrigation water, deeply affecting soil health, and with potential to eventually suffer adsorption-desorption phenomena in function of various parameters (e.g. pH, temperature, surface charges, moisture content) (see section 2.3).
2.1. Soil structure
The soil is not a unique and indistinct system, but is referred to individual ecosystems offering several habitats (e.g. rhizosphere, drilosphere), which jointly describe the whole and porous structure of this tridimensional entity that covers a high proportion of the world (George et al., 2019; Reich, 2014). The soil can be defined as a heterogeneous structure composed of different phases (i.e. solid, liquid and gaseous), whose aliquots and specific elements can be remarkably varied along with space and over time (Rabot et al., 2018). The content of organic matter (OM), i.e. mostly a mixture of partially or totally decomposed plants and animals, also characterizes the soil composition.
In such context, and regarding microbiota, the complexity of the soil ecosystems is meaningful to be understood, due to the fact that all the interactions between the host and virus or pathogens can occur in the soil through contact with host cells (Emerson, 2019; Munson-Mcgee et al., 2018), which leads to virus replication and progeny release (Iwanami et al., 2020). In addition, the physical-chemical demand of soil ecosystems considerably affected the virus-host interactions in the soil, allowing virus mutation due to the host resistance phenomena over the years (Sime-Ngando, 2014).
Several extraction methods (e.g. dispersed soil, aqueous two-phase partitioning) evaluating the bacterial number expressed as average per unit of inorganic mass are reported in studies on soil microbiology to cope with soil microheterogeneity, returning results fluctuating with various orders of magnitude (i.e. from 1 × 108 to 1 × 1010) (Insam, 2001). In addition, recent analytical methods focusing on the soil virome (i.e. the assembly of viruses) led to examine the virus role (e.g. coronaviruses) in the soil microbial communities (Gundy et al., 2009; Srinivasiah et al., 2008). Notwithstanding, the majority of studies are limited to the topsoil (i.e. up to 10 cm), and therefore, future attention should be addressed to increase the knowledge of viruses in deeper layers of soils within the overall biosphere (Williamson et al., 2017). Indeed, previous studies showed that viruses can percolate into soil reaching depths well below 10 m (De Serres et al., 1999; Keswick et al., 1984).
2.2. Viral ecology
A virus is not able to absorb and save energy, or even enabled out of their host target. Hence, a virus is not considered as an independent living organism in biology but as an infectious agent encompassed by a protein capsid, and is distinguished by internal (e.g. diameter, genetic material) and relational (e.g. host, other objects) characteristics (Regenmortel, 2000).
2.2.1. Bacteriophages
Bacteriophages (phages) are the main viruses in the aquatic ecosystems, and this statement should not be easily extended to soil environments due to the scarcity of bacterial biomasses in the bulk soils, which instead are abundant in the rhizosphere (Buée et al., 2009). Bacteriophages are classified into 21 morphotypes, being part of 13 virus families and almost 140 bacterial genera (Kimura et al., 2008). Phages also co-exist with bacteria by showing a predator-prey relationship in a sort of “arms race” explained by the theory of “coevolution”, as proposed by Alexander (1981). However, the coevolution can be affected by lytic and lysogenic phenomena.
Lytic phages quickly start a prolific cycle, where the phage genome is replicated and descendant phages are released via bacterial lysis (Feiner et al., 2015). Moderate phages start a lysogenic cycle when the phage genome is merged into the bacterial chromosome as prophages, which are concomitantly replicated with the bacterial host chromosome during its reproduction, and can enter a lytic cycle (Miller and Day, 2009). Pseudolysogeny is a precarious state where the phage genome is not able to start a lytic or lysogenic, mostly occurring under limited nutrient conditions (Feiner et al., 2015). Hence, lysogeny can be seen as the persistence approach by viruses at poor host abundance and activity, as also occurs in bulk soils (Trubl et al., 2018).
2.2.2. Virus abundance in soil
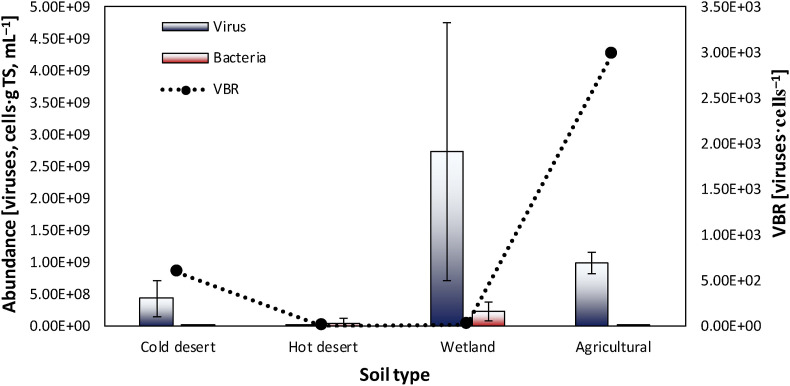
Viruses are notably abundant in various soils and are characterized by exhibiting more limited variability compared to host bacteria in the function of environmental circumstances. Indeed, viruses abound in soils with high OM and moisture content compared to dried and desert soils (Srinivasiah et al., 2008). In cold deserts, virus copiousness was composed of 23–64 × 107 viruses per g of soil total solids (TS−1) (Fig. 1 ), which was lower compared to wetlands and agricultural soils (i.e. 87–417 × 107 viruses per g TS−1, Fig. 1) (Williamson et al, 2005, 2007). On the contrary, the bacterial abundance shows a soil population considerably mutable in copiousness, ranging from 0.035 to 338 × 106 cells·g mL−1 (Fig. 1), which was 10,000-fold greater in wetlands compared to cold deserts (Fig. 1) (Williamson et al, 2005, 2007).
Fig. 1.
Viral (viruses·g TS−1) and bacterial (cells·mL−1) abundance in various soil types (i.e. cold and hot deserts, wetlands and agricultural soils). The values are the means and standard deviations of the reported data for each soil. The virus to bacteria ratio (VBR) was calculated assuming a conversion factor of 1 g mL−1 for bacterial abundance (Srinivasiah et al., 2008). Data were taken from Williamson et al. (2017, 2007, 2005).
Furthermore, pH and temperature may also play a significant role in viral and bacterial abundance in soil (Qu et al., 2020). Williamson et al. (2017) reported a reduction of virus copiousness in soil with extreme pH values (Table 1 ), thus affecting the virus persistence in soil. A substantial difference can be also seen by comparing the viral abundance in cold (Fig. 1) and hot deserts (i.e. 2.2 × 103–1 × 107 viruses per·g TS−1, Fig. 1), with lower temperatures probably promoting the virus persistence (Williamson et al., 2017). This should be seen as an important aspect, being real the possibility of reviving frozen pathogenic microorganisms such as the endospores of Siberian anthrax which can remain in the Permafrost for many years (Steffan et al., 2020).
Table 1.
Persistence (T90) of different enveloped (i.e. CoV and H1N1) and non-enveloped (i.e. Adenovirus, Enterovirus and Orthorevirus) virus causing respiratory diseases (e.g., pneumonia) reported for various matrices (i.e. water, tap water, biosolid and soil) in the function of the temperature (i.e. ambient, 4, 37 and 50 °C) and pH values. T90 is the time (d) needed for 1 log10 unit reduction.
| Viruses | Characteristics |
T90 [d] |
References | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Envelope | Genome type | Genome size [bases] | Diameter [nm] | Isoelectric point | 4 °C | Ambient | 37 °C | 50 °C | pH | Matrices | |||
| CoVa |  |
Enveloped | +ve sense single strand RNA | 30,000 | 60–220 | 6.24 | 28–588 | 1.6–59 | 6–9 | 15 min | 1b | Wastewater, tap water | (Ahmed et al., 2020b; Bivins et al., 2020; Cascella et al., 2020; Franklin and Bevins, 2020; Kumar et al., 2020c; Mohapatra et al., 2020) |
| H1N1 |  |
Enveloped | Single RNA | 13,500 | 80–120 | 6.50–7.00 | 200 | – | – | – | – | Water | (Dublineau et al., 2011; Michen and Graule, 2010) |
| Adenovirus |  |
Non-enveloped | Double strand RNA |
26,000–48,000 | 90–100 | 4.50 | 9–51 | 4.3–214 | – | – | 14–98c | Biosolids (i.e. manure, sludge) | (Magri et al., 2015; Michen and Graule, 2010; Wei et al., 2009) |
| Enterovirus |  |
Non-enveloped | +ve sense single strand RNA | 7200–8500 | 25–30 | 3.80–3.90 | – | 14 | – | – | – | Soil | (Michen and Graule, 2010; Pourcher et al., 2007) |
| Orthoreovirus |  |
Non-enveloped | Double strand RNA |
23,500 | 70–85 | 4.00–5.50 | 807 | 66–151 | – | – | 14–98c | Biosolid (i.e. sludge) | (Magri et al., 2015; Michen and Graule, 2010) |
a = SARS-CoV, SARS-CoV-2, MERS-CoV; b = pH of 2–3 and 11–12; c = pH of 9.
2.3. Human viruses in soil environment
2.3.1. Enveloped viruses
A CoV refers to an extended group of enveloped viruses (e.g. SARS-CoV-1, MERS-CoV, SARS-CoV-2) with a +ssRNA and crown-like heads on their egg-shaped surfaces (La Rosa et al., 2020) with diameters comprised between 60 and 220 nm (Table 1) (Cascella et al., 2020), which can influence the virus mobility in the soil environment. The genome sequence of SARS-CoV-2 was positively correlated with SARS-CoV-1 (i.e. 82%) (Chan et al., 2020), which was listed with MERS-CoV as a pathogenic agent with high mortality by causing severe pneumonia and respiratory failure in an infected subject (Qu et al., 2020). CoV can be spread via direct contact between humans (i.e. symptomatic or asymptomatic people) through the inhalation of the breathed virus in droplets (e.g. coughs or sneezes) or with infected surfaces (e.g. skin-to-skin, objects) (Qu et al., 2020). However, other sources of contamination such as the environmental compartments (e.g. soil) should be clarified.
Skeletal similarities between SARS-CoV and SARS-CoV-2 would suggest the possible use of already found data for SARS-CoV and its surrogates, in order to suppose the environmental fate of SARS-CoV-2 (Kumar et al., 2020c). The enveloped viruses differ from non-enveloped viruses due to genome, structure, replication, pathogenicity and persistence (Wigginton and Boehm, 2020). This aspect can be significant to determine the interactions (e.g., hydrophobic) that can allow the adsorption of CoV onto solid particles.
The time needed to reduce 90% the initial viral infectivity (T90) was reported to reach almost 40 d for SARS-CoV-2 in wastewater (Table 1) (Mohapatra et al., 2020). In addition, T90 of human CoV was comprised between 200 and 400 d at 4 °C in different water matrices (Table 1) (Kumar et al., 2020c), and can reach a value of 588 d in filtered tap water (Table 1), as reported by Franklin and Bevins (2020). The endurance of enveloped viruses such as the influenza virus (H1N1) in the water was determined as almost 200 days at 4 °C (Table 1) (Dublineau et al., 2011). Gutiérrez and Buchy (2012) reported that avian influenza (H5N1) could not maintain the viral load in sandy topsoil, but could remain in soil-based compost, thus indicating that different soil properties considerably influence the survival of the virus.
Although the World Health Organization (WHO) suggested that there was no evidence regarding the persistence of SARS-CoV-2 in wastewater or drinking water (Baldovin et al., 2021; Kitajima et al., 2020), the viral load of enveloped viruses such as CoV could be maintained for a prolonged time in the soil environment in the function of different parameters such as the temperature, moisture content, pH, OM, sunlight radiation, and occurrence of clays and nutrients (see section 2.5) (Anand et al., 2021a). For example, Bivins et al. (2020) recently evaluated that the increase of temperature from ambient to 50 °C can drastically reduce the T90 to 15 min (Table 1). Likewise, the survival of enteric viruses such as SARS-CoV can be decreased in dried soils (i.e. 15–25 d) compared to soils with 10% of moisture content (i.e. 60–90 d) at ambient temperature (i.e. 20 °C) (Bosch et al., 2006). In addition, the experiments performed on frozen and thawed samples could affect the microbial community involved, thus contributing to the reduction of the CoV particles in the investigated matrix (Bivins et al., 2020).
2.3.2. Non-enveloped viruses and other pathogenic microorganisms
Adenovirus, enterovirus, and orthoreovirus are non-enveloped viruses that can cause respiratory tract disease (Michen and Graule, 2010). These non-enveloped viruses are characterized by diameter size of 25–100 nm (Table 1) and could have a great potential to infect a large variety of environmental compartments such as the soil, being resistant to hostile conditions and water treatments (Kumar et al., 2020c).
For example, Adenovirus arrives in the soil environment after sewage sludge amendment (Horswell et al., 2010), and are subsequently sorbed by the soil particles through electrostatic interactions due to its isoelectric point (Table 1), as suggested by Wong et al. (2013). Adenovirus showed a T90 value comprised between almost 9 and 51 d at 4 °C (Table 1) depending on the biosolid considered (i.e. dairy and swine manure, respectively) (Wei et al., 2009). Also, in this case, an increase of temperature can allow a reduction of T90 to approximately 4 d (Table 1) (Wei et al., 2009). For Enterovirus, i.e. a virus of the Enterovirus genome, the survival in soil was estimated at approximately 14 d (Table 1) (Pourcher et al., 2007). Moreover, Orthoreovirus are reported to have a unique impact on mangrove soil (e.g. in river virome) (Jin et al., 2019), with a T90 ranging between 4 and 807 d (Table 1) depending on temperature, pH and ammonia nitrogen concentrations (Magri et al., 2015).
In addition, pathogenic soil bacteria or fungi can quickly infiltrate humans via cutaneous wounds, ingestion of infected foods or soils, and inhalation of respiratory droplets (Steffan et al., 2020). Yersinia pestis and Legionella spp. are the causative agents of pneumonic diseases with a high mortality rate (Baumgardner, 2012; Lynteris, 2017). For example, Legionella spp. can persist for months (i.e. 90–300 d) in a potting soil kept at temperatures comprised between −20 and 35 °C. Although bacterial diseases are not listed as highly contagious (Steffan et al., 2020), these aspects should also be considered in future policies to prevent new pandemics.
2.3.3. Fate of CoV in soil and crosstalk between ecosystems
The soil could be a viral sink acting as a secondary source for the spread of SARS-CoV-2 over an extended period (D. Zhang et al., 2020), likely due to inappropriate sanitization actions that can allow soil contamination by viruses (Foladori et al., 2020). In addition, the detection of a not negligible viral load of the human excrements has increased the concerns regarding the potential spread of SARS-CoV-2 through the soil and other environmental compartments within the ecosystems (e.g. plant, animal, ground-water) (Patel et al., 2020). Indeed, Wang et al. (2020) showed a viral load of human feces around 2.6·104 copies·mL−1. Afterwards, SARS-CoV-2 can reach wastewater treatment plants (WWTPs) via sewer systems, with various virus dilutions in the function of the degree of affectation of the pandemic (Foladori et al., 2020), population number, and other parameters such as temperature and travel time (Hart and Halden, 2020).
Ahmed et al. (2020) firstly showed the presence of SARS-CoV-2 in untreated wastewater ranging from 1.9 to 12 copies·100 mL−1. Zaneti et al. (2021) recently conducted a quantitative microbial risk assessment of COVID-19 in wastewater obtaining a viral load comprised between 1.03 × 102 and 1.31 × 104 copies mL−1. The disinfection of wastewater is strongly recommended by the WHO in order to avoid any virus discharge in the environment (Collivignarelli et al., 2020). However, Bogler et al. (2020) reported a partial removal of SARS-CoVs in conventional WWTPs, thus questioning the efficiency of some disinfection treatments. Indeed, 20% of soil samples collected nearby to the hospital receiving COVID-19 subjects and wastewater plant in Wuhan, recently resulted positive to SARS-CoV-2 RNA with an abundance comprised between 205 and 550 copies·g−1 (Wiktorczyk-Kapischke et al., 2021).
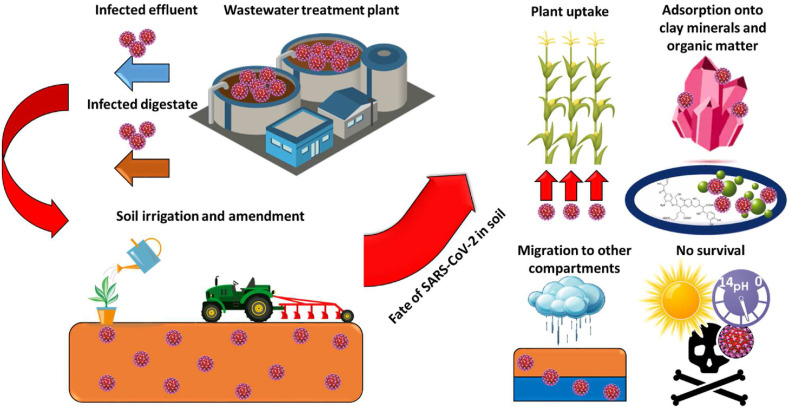
Hence, a substantial screening should be conducted on the wastewater effluents and sewage sludge before their application in soils to prevent the COVID-19 migration to other environmental compartments (Núñez-Delgado, 2020a). The use of infected wastewater and sewage sludge for soil irrigation and fertilization, respectively, could have a significant impact on the virus spread due to the groundwater contamination by SARS-CoV-2, and its potential uptake in crops (Fig. 2 ) (Usman et al., 2020), thus entering in the human food chain. A plant can use phosphorus incorporated by viruses after infecting and lysing microbial cells (Kuzyakov and Mason-Jones, 2018). This phenomenon can have a more pronounced harmful effect when untreated wastewater is improperly introduced into the environment due to the persistence of SARS-CoV-2 RNA (Ahmed et al., 2020b). Otherwise, the plant viral internalization can be reduced by the colloid fraction of soil (e.g. OM) (Mancuso et al., 2021). For example, Badawy et al. (1990) revealed that an enterovirus such as coxsackie virus can remain viable in crops for about 3–5 weeks.
Fig. 2.
Possible scenarios regarding the fate of SARS-CoV-2 in soil. SARS-CoV-2 can arrive in soil due to the discharge of infected effluent and digestate after an improper wastewater treatment. Afterwards, the virus can be taken by plants requiring the phosphorus, adsorbed onto clay minerals and organic substances due to electrostatic and hydrophobic interactions, respectively. Otherwise, SARS-CoV-2 can migrate from soil to other environmental compartments due to the reduction of ionic strength during rains. The virus survival can be limited by sunlight radiation, high temperature, acidic pH and the presence of pollutants.
Evapotranspiration phenomena can also accelerate the shift of the virus from soil or plants to the air moisture affecting the disease transmission (Lian et al., 2020). However, the viral particles could be strongly sorbed by soil due to envelope hydrophobicity of the SARS-CoV-2 (Fig. 2), affecting the survival (Table 1) and potential transmission of the virus (Mohapatra et al., 2020).
SARS-CoV-2 contains a further lipid membrane enveloping the capsid protein (Kumar et al., 2020b), probably impacting virus adsorption onto a solid fraction of soil (Fig. 2). A virus can be adsorbed by the soil through electron donor-acceptor (e.g. π-π) and hydrophobic (e.g. Van der Waals) interactions depending on the characteristics of the virus (e.g. lipophilicity, presence of functional groups) and soil properties (e.g. OM content, pH, ionic strength) (Betancourt et al., 2019; Bianco et al., 2021). Ye et al. (2016) recently simulated the SARS and MERS-CoV partitioning in wastewater, obtaining virus adsorption of 26% by the solid phase. The adsorption of enveloped viruses (e.g. SARS-CoV-2) can occur through hydrophobic interactions onto negatively charged soil particles with a sufficient equilibrium time generating hetero-aggregates (Katz et al., 2018). The occurrence of clay minerals can positively affect the enveloped virus adsorption onto soil particles with the increase of cation exchange capacity (CEC) (Kimura et al., 2008; Nasser, 2002). In addition, the presence of aluminum oxides in the soil can enhance the virus adsorption onto soil particles due to the combination of stronger electrostatic attraction and hydrophobic interactions (Attinti et al., 2010).
Therefore, the migration of coronaviruses from soil to other environmental compartments should be limited (Kumar et al., 2020a), but their survival in soil could be guaranteed due to moisture content and low temperatures (section 2.4) (Mohan et al., 2021). However, the occurrence of acidic soil, i.e. with a pH lower than 7, can inactivate enveloped viral particles (Fig. 2) (Gutiérrez and Buchy, 2012). In addition, sunlight radiation can allow the inactivation of the SARS-CoV-2 virus (Fig. 2) by decreasing the T90 to a value lower than 10 min in the function of different parameters (i.e. latitude, season and hour) (Herman et al., 2020). Likewise, the presence of high concentrations of contaminants in environmental matrices (e.g. soil) due to the improper discharge of polluted wastewaters (Papirio et al., 2014) could significantly affect the T90 of SARS-CoV-2 (Foladori et al., 2020). For example, Touret et al. (2020) recently identified 15 drugs as inhibitors for SARS-CoV-2 in vitro replication. On the other hand, rainfall provides a higher flow rate in the soil column, thus increasing the viral migration via saturated soil pores, probably due to the decrease of soil ionic strength (Fig. 2) (Kumar et al., 2020c; Lama et al., 2020). Also, the migration of enveloped viruses (e.g. SARS-CoV-2) to other compartments can be enhanced by excessive soil tillage, which was reported to decrease OM content in soil of approximately 40% (Zhang et al., 2020).
3. Direct and indirect impact on the soil before and after pandemic
The advent of the COVID-19 in the world determined the closure of several secondary and tertiary sector activities (e.g. non-essential factories, tourism) as well as the reduction of traffic mobility due to the lockdown measures enforced by national policies, thus improving the air quality and reducing the dispersion of pollutants in soil or sediments (Table 2 ) (Berman and Ebisu, 2020; He et al., 2020; Tobías et al., 2020). The quarantine actions also allowed the cleaning of recreation areas (e.g. parks, seaside) due to the decrease of human activities in the mentioned spaces (Table 2) (SanJuan-Reyes et al., 2021). In addition, a reduction of the generated amount of solid waste (i.e. 20–30%) was observed during the global pandemic (Klemeš et al., 2020; Ragazzi et al., 2020). All this results in a beneficial effect on soil health status (Table 2), which is also supported by the reduction of SARS-CoV-2 abundance to zero, evaluated both in the middle and low-risk periods in soils sampled after the adopted stringent measures (Zhao et al., 2020).
Table 2.
Impact of COVID-19 on the soil and other environmental compartment status, human and other living organism health, waste management and economy.
| COVID-19 impact | Soil status | Other environmental compartment status | Human health | Other living organism health | Waste handling | Economy |
|---|---|---|---|---|---|---|
| Closure of work activities | ++ | +++ | + | + | + | +++ |
| Reduction of traffic mobility | +++ | ++ | ++ | ++ | + | +++ |
| Lockdown measures | ++ | ++ | +++ | ++ | ++ | +++ |
| Medical and hospital wastes | +++ | ++ | ++ | + | +++ | + |
| Personal protective equipment | +++ | ++ | +++ | +++ | +++ | +++ |
+ Less intensive; ++ Moderately intensive; +++ Very intensive.
In addition, the production of medical wastes (i.e. infected and uninfected) significantly raised after the pandemic started (e.g. up to 370% in Hubei province), probably increasing the potential transmission risk of SARS-CoV-2 (Adelodun et al., 2020; Klemeš et al., 2020) due to the improper hospital waste handling taking place in some cases (Table 2) (Yu et al., 2020). Therefore, several nations replaced the policies for recycling with waste landfilling or incineration to prevent the COVID-19 spread through the environment (Zambrano-Monserrate et al., 2020). The temperatures involved in a waste incinerator (i.e. >850 °C) as well as the thermophilic conditions which can occur during aerobic landfilling (i.e. up to 75 °C) can be considered as sanitization strategies to tackle the SARS-CoV-2 in wastes (Di Maria et al., 2020). Chlorine-based disinfection can be alternatively applied for the treatment of infected wastes before proper waste handling (Pandey et al., 2021), and is being extensively applied in WWTPs in disinfection tanks before the discharge of the treated wastewaters to a receiving water body (section 2.5). The major purpose of this way to proceed is the removal of residual pathogenic microorganisms in the waste for secure disposal (Cots et al., 2020). However, the increase of hospital or medical wastes led to the delay of municipal waste management activities (Table 2), which could negatively affect the soil status posing risks for the environment and human health (Kulkarni and Anantharama, 2020; Rahman et al., 2020).
Alongside the hospital and medical waste issue, personal protective equipment is being extensively used by citizens to cope with SARS-CoV-2 spread, such as face masks (e.g. surgical, KN95) (Feng et al., 2020), hand sanitizers (e.g. polyethylene bottles) and single-use plastic gloves (SanJuan-Reyes et al., 2021; Zand and Heir, 2020a), which are made of recalcitrant and durable materials, thus posing a threat for the soil status. For example, the average generation of health protection wastes considerably raised from 3.64 to 27.32 kg d−1 every 1000 inhabitants in Wuhan during the pandemic. This emerging type of waste would increase the amount of plastics yearly estimated to go in the sea and ocean (i.e. up to 12 million tons) (Patrício Silva et al., 2021). Moreover, face masks can be considered as potential microplastic sources in the environment, which could be easily swallowed by living organisms, thus entering the food chain and affecting human health (Aragaw, 2020). Hence, national policies could consider to provide health care waste collection bins in islands to cope with this issue, by raising awareness of the people via social media in order to perform a proper waste delivery (Zand and Heir, 2020b).
4. Methods for quantification of viruses and SARS-CoV-2 monitoring in soil
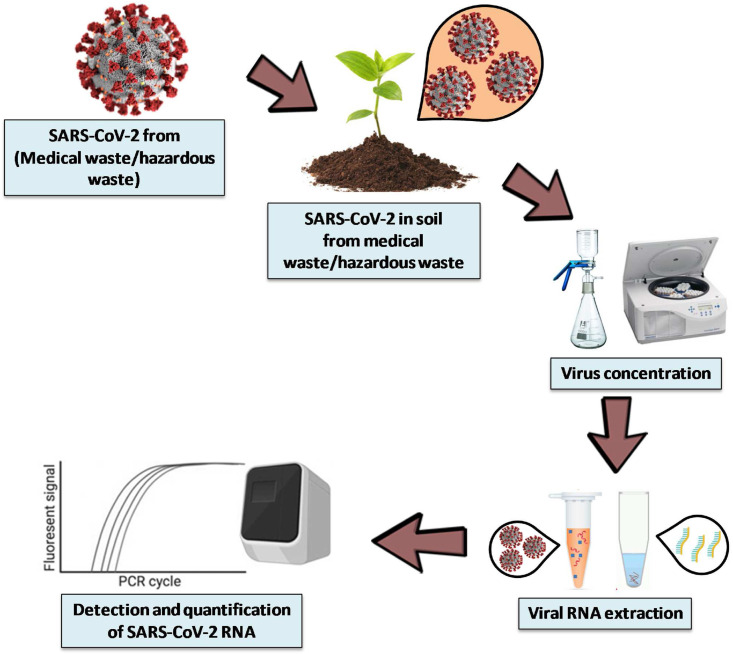
Soil chemical processes are of crucial importance for long-term sustainability of soils and the overall environment. With that in mind and knowing that it affects to both living organisms and all other components in that environmental compartment, this section mainly focuses on methods for virus elution during the extraction process, virus concentration techniques, detection, and quantification (Fig. 3 and Table 3 ).
Fig. 3.
A sequence of operations for SARS-CoV-2 detection and quantification in soil.
Table 3.
Summary of studies reporting methods for detection of viruses in soils, sediments, biosolids, sewage sludges and waters.
| S. no. | Sample type | Virus type | Extraction method | Detection method | References |
|---|---|---|---|---|---|
| 1 | Soil, sewage, biosolids | Bacteriophages | Elution method | Calculation of plaque forming units | Guzmán et al. (2007) |
| 2 | Coastal seawater | Poliovirus | Adsorption and elution with negatively charged membrane | Cell culture RT-PCR and direct RT-PCR | Katayama et al. (2002) |
| 3 | Sewage sludge | HAD (Human adenovirus), HAV (Hepatitis A virus), PV (Poliovirus), RV (Rotavirus) |
Elution and organic-based extraction protocol (Phenol/trizol/Silica based) | PCR, RT-PCR and nested PCR | Schlindwein et al. (2009) |
| 4 | Sludge | Enterovirus | Elution-concentration procedures | Agar overlay plaque formation technique | Albert and Schwartzbrod (1991) |
| 5 | Mercenaria Mercenaria (Hardshell clams) | Poliovirus and HAV | Extraction-concentration method | – | Alouini and Sobsey (1995) |
| 6 | Sludge | Enterovirus | Viral-Elution method | Cell culture and RT-PCR | Monpoeho et al. (2001) |
| 7 | Coastal sediment | Enteroviruses and Rotavirus | Elution and concentration | BGM cell cultures and indirect immunofluorescence | Jofre et al. (1989) |
| 8 | Agricultural soils | Bacteriophages | Elution method | VLP count by epifluorescence microscopy (EFM), and direct counts by transmission electron microscopy (TEM) |
Williamson et al. (2003) |
| 9 | Sewage | Human astroviruses (HAstV) | Adsorption-elution method | RT-PCR and quantitative real-time (qPCR) | Guimarães et al. (2008) |
| 10 | Freshwater, oligotrophic mountain lake, oligomesotrophic lake, eutrophic lake, domestic sewage | Virioplankton | Polyethylene glycol (PEG) and ultracentrifugation | TEM and EFM | Colombet et al. (2007) |
| 11 | Shellfish: oysters (Saccostrea forskali), cockles (Anadara nodifera) and mussels (Perna viridis), | Rotavirus | Adsorption-twice elution-extraction method | RT-nested PCR | Kittigul et al. (2015) |
| 12 | Potable water | Poliovirus I | Adsorption-elution method | – | Katzenelson et al. (1976) |
| 13 | Freshwater | Human rotavirus, Simian rotavirus and Poliovirus | PEG 6000 precipitation method | – | Lewis and Metcalf (1988) |
| 14 | Sewage | Group A rotaviruses | Adsorption- elution/ultrafiltration method | Ultracentrifugation-based method | Fumian et al. (2010) |
| 15 | Soil | SARS-CoV-2 | RNA extraction (NaCl and PEG 6000) | RT-qPCR | Zhang et al. (2020) |
| 16 | Soil | Bacteriophages | Negatively charged HA membranes, PEG and ultracentrifugation (UF) | Random amplified polymorphic DNA (RAPD), TEM |
Dias et al. (2020) |
| 17 | Freshwater beaches | Human adenovirus (HAdV), Human enterovirus (HEnV), and Human norovirus genogroups I/II | Cation-coated filtration method | qPCR | Lee et al. (2014) |
| 18 | Stool | Enterovirus, Adenovirus | Cell culture | PCR and hydridization | Allard et al. (1992) |
| 19 | Sediment | Poliovirus and Adenovirus | Elution method | PCR and qPCR | Miura et al. (2009) |
| 20 | Groundwater | Enteroviruses, reoviruses, HAV and Norwalk virus | Celite elution procedure | RT-PCR | Fout et al. (2003) |
4.1. Methods for virus elution
The first step to be followed for the quantification of viruses in soil matrices is the elution protocol (Fig. 3). Inhibitory compounds that can essentially affect the molecular biology-based techniques employed for virus detection are naturally existing in environmental samples such as soil (Guzmán et al., 2007). Thus, the elution method is a relatively low-cost approach based on the use of inorganic chemicals to minimize the abovementioned issue (Katayama et al., 2002).
Albert and Schwartzbrod (1991) developed a protocol for the elution process of virus in 3% beef extract media at alkaline pH solution (Table 3). Grabow et al. (1991) reported an elution technique using glycine and saline followed by a Freon insert of beef extract media. Furthermore, Alouini and Sobsey (1995) developed the virus elution protocol with 7% beef extract media. Using these protocols, Monpoeho et al. (2001) obtained up to 35% recovery of enteroviruses in bio-solid waste (Table 3). Likewise, Jofre et al. (1989) reported a recovery of enteroviruses up to 28%. Alouini and Sobsey (1995) reported poliovirus and Human Hepatitis A virus (HAV) recoveries of about 53% and 22%, respectively (Table 3).
Other studies used four different elution buffers for phages in soil samples, specifically 1% potassium citrate, 250 mM glycine buffer, 10% beef extract, and 10 mM sodium pyrophosphate, and found 29% viable phages recovery using glycine and beef extract buffers (Table 3) (Williamson et al., 2003). However, Williamson et al. (2003) found a higher rate of recovery of viruses, reaching up to 65% (Table 3).
US Environmental Protection Agency (US EPA) proposed a protocol for elution to determine viral particles from soil samples, by using an adsorption process in an AlCl3 synthetic solution with acid pH. Researchers that used the US EPA protocol for the determination of different viruses, obtained a recovery of rotavirus, adenovirus, and both hepatitis A virus and Poliovirus of 90, 20 and 100%, respectively (Table 3) (Schlindwein et al., 2009).
Hence, several elution methods are available in the literature with various recovery efficiencies in the function of the physical-chemical properties of each investigated virus (e.g. specific density, morphology) and membrane attachment patterns (Table 3) (Lewis and Metcalf, 1988). On the other hand, the ultracentrifugation (UF)-based method can result in a greater virus recovery compared to the adsorption-elution method (Table 3) (Fumian et al., 2010). The filters utilized in the adsorption-elution approach can be obstructed by the high amount of detritus present in a soil sample, thus decreasing the recovery rate (Table 3) (Guimarães et al., 2008).
4.2. Virus concentration techniques
A concentration technique is generally employed before the quantification of viral particles in soil samples (Fig. 3). Colombet and Sime-Ngando (2012) determined virus particles from infected samples of humans, animals, and plants using different concentration methods. The UF method is commonly used to concentrate viruses (Table 3) (Ammersbach and Bienzle, 2011; Fumian et al., 2010; Nordgren et al., 2009), but the poly-ethylene glycol (PEG) method can be utilized for the determination of virus particles by inducing interaction of crystallinity between DNA during the precipitating and concentrating process (Table 3) (Colombet et al., 2007; Kittigul et al., 2015; Shieh et al., 1999).
Katzenelson et al. (1976) reported a recovery of 75% of poliovirus from soil and sediment samples using the PEG virus concentrated method (Table 3). Similarly, 70% of recovery of rotavirus can be obtained employing the PEG precipitation method at neutral pH, 15% concentration of PEG, and 3% beef extract-sodium nitrate eluent (Table 3) (Lewis and Metcalf, 1988). Both methods were extensively discussed in detail by Peyret (2015). However, Dias et al. (2020) proposed a modified protocol design for concentrating viral particles through UF and PEG techniques in soil samples (Table 3). Briefly, viral particles are taken after reinitializing and subsequently filtered through the diafiltration system. Afterwards, this retentate is re-concentrated through UF and PEG precipitation. Thus, in a UF method, viral pellets are re-concentrated and incubated under agitation with 3 h duration and SM buffer mixtures. In the case of the PEG method, a mixture solution (i.e. PEG 8000 and NaCl) is added to viral ultra-filtered retentates to final concentrations. However, Yamamoto et al. (1970) also reported that a molecular weight of PEG of 6000 can be effective in precipitating macromolecules. Indeed, Zhang et al. (2020) recently investigated the presence of SARS-CoV-2 in soil samples by precipitation with NaCl (0.3 mol L−1) and PEG 6000 (10%) overnight (Table 3).
Humic acid (HA) negatively charged membrane is another method for concentrating viral particles (Table 3) (Lee et al., 2014). In this method, the viral particle is concentrated by adsorption phenomena, where MgCl2 is used as a key agent in the viral suspension sample. Due to adsorption phenomena, the virus gets into the membrane and then the concentrated viral particles are collected for further analysis.
Dias et al. (2020) also determined viruses in soil samples through HA membranes (Table 3). Firstly, 50 mL of suspension sample is added with MgCl2 25 mM in a HA membrane having 47 mm diameter, and 0.45 μm pore size for viral concentration. Afterwards, the final viral suspension is filtered and centrifuged through a DNA concentrator to get the total volume of 500 μL. In the UF method, the morphology found by transmission electron microscopy (TEM) images showed different viral morphotypes containing larger viral particles. Dias et al. (2020) concluded from TEM images, that a greater number of viral particles can be observed in the HA membrane as compared to the other abovementioned methods. Indeed, Dias et al. (2020) showed in a metagenome sequencing employing these methods, 24 genera by using UF and HA membrane methods, and in the case of the PEG method, no genus was detected. Williamson et al. (2017) also observed virus particles from TEM images, which were Podoviridae, Myoviridae, Siphoviridae and other non-tailed filamentous structure virus particles.
In addition, Williamson et al. (2005) found different morphologies of viral particles in different soils such as wetland, agricultural, and forested. An amount of 56 and 80% of viral spherical particle structures and tailed phages was reported in agricultural, and other soils, respectively (Williamson et al., 2005). Similar viruses were observed by different researchers in an aquatic environment (Ackermann, 2001; Weinbauer, 2004; Wommack and Colwell, 2000). Swanson et al. (2009) found 70 and 5% viral spherical particle, and tailed phages, respectively, in an agricultural soil sample (Table 3). To be noted that, from TEM images, these researchers reported several non-tailed in addition to the tailed phages.
Hence, the HA membrane method represents a promising approach for obtaining metagenomic data of virus particles from soil samples compared to UF and PEG. Thus, the HA membrane can be considered the most suitable concentration method due to the low required background material in the electron micrographs, and also for recovering various morphotypes and viruses.
4.3. Virus detection
In general, after elution and concentration of viral particles, virus detection methods (e.g. cell culture, molecular techniques) are used for quantification of infectious viruses (Table 3). This procedure evaluates metagenomics after extracting the entire DNA or RNA from soil samples (Fig. 3) using for instance a soil kit (i.e. RNeasy® PowerSoil®), as previously reported by Zhang et al. (2020). Comparing to the isolation method, the latter frequently focuses only on a single virus. The DNA is subsequently broken up into several minute pieces and sequenced. The resulting sequence is examined to build the virus genomes of the sample (Trubl et al., 2020).
Fong and Lipp (2005) described virus detection by cell culture method, and samples were initially inoculated with different cell lines such as Madin-Darby bovine, pig, and buffalo green monkey kidney, CaCo-2, RD, A549, MA104, and FRhK-4. Staggemeier et al. (2011) reviewed different techniques for the detection of viral communities in soils and sediments. The virus detection (Table 3) and quantification techniques include most-probable number assays (MPNs), plaque assays (PAs), Polymerase Chain Reaction (PCR), TEM, epifluorescence microscopy (EfM), and flow cytometry (FC).
Allard et al. (1992) described viral detection through molecular techniques and Polymerase Chain Reaction (PCR), which has several advantages such as high detection limit and good accuracy in very small amounts and is much faster (Table 3) (Griffin et al., 2003; Lee and Jeong, 2004). However, depending on elution protocol and substances present in the soil samples, inhibition phenomena can occur (section 4.1). For example, PCR inhibition was reported in the elution process performed in the beef extract media (Lewis et al., 1985). Miura et al. (2009) reported that HA could hamper PCR detection if present on the soil samples (Table 3). Other researchers utilized hyphenated PCR methods such as Real-Time PCR (qPCR), Nested PCR, Multiplex PCR and Randomly Amplified Polymorphic DNA-PCR (RAPD-PCR) for detection of viral particles on soil samples. Rajtar et al. (2008) used the qPCR technique for detection of DNA of enteric viruses followed by quantification using fluorescence emission. Donaldson (2002) showed a greater detection sensitivity via qPCR than conventional PCR. These methods are commonly applied for the detection and quantification of SARS-CoV-2 in human-deriving samples (Table 4 ). Ehlers et al. (2005) reported good sensitivity through nested PCR due to its design of two pair primers. Le Guyader et al. (1994) detected rotavirus, and enterovirus in sediment samples using nested PCR. However, Katayama et al. (2002) reported a high probability of contamination through nested PCR (Table 3). Fout et al. (2003) developed a multiplex PCR for detection of enteric viruses in environmental samples and they concluded that multiplex PCR techniques need further investigation towards optimization and reaction conditions of PCR (Table 3). Winget and Wommack (2008) detected DNA fragments in viral-containing soil samples using random PCR amplification of polymorphic (RAPD-PCR) with a single decamer primer. This technique provides a banding pattern that can be used as a proxy fingerprint for the underlying complexity of the original DNA template. They found 105 and 104 viruses per reaction in the soil samples. Srinivasiah et al. (2013) developed RAPD-PCR, which has high accuracy for the detection of soil viral communities. They demonstrated RAPD-PCR fingerprinting is an inexpensive, high-throughput of viral community dynamics within environmental samples.
Table 4.
Methods applied for the detection and quantification of SARS-CoV-2 in human-deriving samples.
| Type of study | Assay method | Analyte | Concentration range | Tested samples | Reference |
|---|---|---|---|---|---|
| Research article | RT-LAMP | RNA | 80–500 copies·mL−1 | Nasopharyngeal swab | Huang et al. (2020) |
| Rapid communication | RT-PCR assay | RNA | 276 copies·reaction−1 | Nasopharyngeal swab | Pfefferle et al. (2020) |
| Research article | Antibody-based detection | RNA | – | Nasopharyngeal swab | Liu et al. (2020) |
| Protocol | CRISPR-based approach | RNA | 10–100 copies·μL−1 | Nasopharyngeal or oropharyngeal swabs | Zhang et al. (2021) |
| Research letter | CT-SCAN | – | – | – | Fang et al. (2020) |
Suttle (2007) described the protocol of PA method for detection of infected viruses in environmental samples, whose form a clearing (plaque) on a lawn of host cells on solid media. Suttle (2007) also described the MPN method mainly on microplates, which counted viable virus. This method detects the infected virus by the number of dilutions with cell lysis in microplates or well plates. Børsheim et al. (1990) described another technique for virus particles in environmental samples through TEM analysis, which provides data on both the abundance and morphology of virus-like particles. Suttle et al. (1990) used the EfM technique to estimate the virus particles in sediment and marine water samples. This method determined virus particles with a 4’, 6-diamidino-2-phenylindole fluorescent dye after virus concentration by different methods. Different fluorescent dyes, such as YO-PRO, SYBR Green and SYBR Gold were also used for the same techniques (Hennes and Suttle, 1995; Noble and Fuhrman, 1998; Winget et al., 2005). However, EfM has virus diversity limitations. Likewise, Brussaard (2004) described the FC method for further detection of virus diversity and quantify subpopulation of viral communities in environmental samples with a change in dye fluorescence frequency.
Some of the advanced technologies and soil management strategies currently available (and/or even other to be developed) are now required to restore soil quality due to the fact that the COVID-19 pandemic has advanced in an accelerated manner, but studies on that topic are still in a rough phase (see section 4.4). With this in mind, focusing specifically on virus detection/quantification, sequence-specific DNA probes designated with a fluorescent colorant could be produced to specifically target virome, with their copiousness analyzed by qPCR. This method can be suitable to quantify specific viruses (e.g. SARS-CoV-2) in the environment, but not quickly illustrative of soil native viruses (Trubl et al., 2020).
4.4. SARS-CoV-2 monitoring in soil and data management
The management of soil quality is undoubtedly required to reduce the urgent issue of environmentally spread diseases from pathogenic microorganisms (Qian et al., 2020). Although no works regarding the presence of SARS-CoV-2 in soil-related liquid samples have been performed (Conde-Cid et al., 2021), enveloped viruses could persist in soil matrices due to their structure and environmental conditions (Section 2.5). Therefore, SARS-CoV-2 should be rapidly monitored in the soil to prevent its transmission to humans by promoting effective detection strategies.
Conde-Cid et al. (2020) recently proposed both i) in-situ monitoring and ii) in-situ sampling for performing lab analyses. i) In-situ monitoring could be carried out by installing specific devices, which should be sterilized (e.g. chemicals, UV lights) before employment (Conde-Cid et al., 2020). Both flat and slope areas can be sampled through piezometers allowing the water vertical-flow from long distances, or otherwise tensiometers and vacuum pumps (Conde-Cid et al., 2020). Thus, the samples can be subsequently analyzed via rapid tests specifically for the detection of SARS-CoV-2 (Krüttgen et al., 2021), in order to evaluate the degree of mobilization of the virus onward the groundwater. ii) in-situ sampling for performing lab analyses can be conducted using sanitized stainless steel cores or Edelman-type probes to preserve or not the tridimensional structural integrity of the soil, respectively (Conde-Cid et al., 2020). Afterwards, all samples have to be stored on ice and conveyed to the laboratory for the extraction and quantification methods (see section 4.3) (Randazzo et al., 2020).
Data obtained from the monitoring of SARS-CoV-2 in soil samples could be stored in an online database for soil management. For example, smart web-based geospatial decision support systems (S-DSSs) can be used as a suitable tool for this purpose (Terribile et al., 2015). S-DSSs can combine soil databases with high-performance computing (HPC) to assess the spread of COVID-19 on a large scale with high-resolution detail (e.g. 20 m Sentinel) (Lal et al., 2020). In such context, several issues related to fragmentation of soil policy, detached soil administration for environmental and agricultural subjects, and rough and subdivided geospatial acquaintance regarding soil processes and characteristics, could be overcome.
5. Conclusions
In the current COVID-19 pandemic, different basic essential parameters, mostly focusing on public health, are generally assessed. However, soil health, which plays a major role in bacterial growth, should be carefully evaluated to maintain environmental equilibrium. Indeed, these bacteria populations can be affected by viruses that are transmitted from infected water bodies such as untreated sewage, sewage sludge, and irrigation systems. This could deeply affect soil health likely due to a variety of phenomena, including strong adsorption-desorption of viruses such as SARS-CoV-2. In such a context, the improper treatment of wastewater would pose a threat to human and animal health. The viral load of enveloped viruses such as CoV could be maintained for a prolonged time in the soil environment in the function of different parameters such as the temperature, moisture content, pH, OM, sunlight radiation, and occurrence of clays and nutrients. Moreover, the characteristics of enveloped viruses (e.g. SARS-CoV-2) can influence the virus's mobility in the soil environment. Thus, the presence of SARS-CoV-2 could be directly monitored in soil matrices in-situ or with soil sampling for performing lab analyses after extraction and quantification methods (e.g. elution protocol, PEG, PCR). Therefore, future studies about SARS-CoV-2 should be especially aimed at focusing soil characteristics in order to cope with the COVID-19 pandemic and long-term sustainability.
Credit author statement
Uttpal Anand: Conceptualization and conceiving the original idea, Literature survey/mining, writing - the major original draft preparation, Manuscript structure. Francesco Bianco: Literature survey/mining, writing - the major original draft preparation, Figures/Illustrations and Tables preparation., Visualization, Response, Critical Review and Editing/Proofreading the manuscript, Retrieved and arranged the references. S. Suresh: Literature survey/mining, writing - the major original draft preparation, Visualization, Response, Critical Review and Editing/Proofreading the manuscript. Vijay Tripathi: Figures/Illustrations and Tables preparation. Avelino Núñez-Delgado: Visualization, Response, Critical Review and Editing/Proofreading the manuscript. Marco Race: Conceptualization and conceiving the original idea, Literature survey/mining, writing - the major original draft preparation, Figures/Illustrations and Tables preparation., Visualization, Response, Critical Review and Editing/Proofreading the manuscript, Manuscript structure.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Abdool Karim S.S., de Oliveira T. New SARS-CoV-2 variants — clinical, public health, and vaccine implications. N. Engl. J. Med. NEJMc2100362. 2021 doi: 10.1056/NEJMc2100362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackermann H.-W. Frequency of morphological phage descriptions in the year 2000. Arch. Virol. 2001;146:843–857. doi: 10.1007/s007050170120. [DOI] [PubMed] [Google Scholar]
- Acter T., Uddin N., Das J., Akhter A., Choudhury T.R., Kim S. Evolution of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) as coronavirus disease 2019 (COVID-19) pandemic: a global health emergency. Sci. Total Environ. 2020;730:138996. doi: 10.1016/j.scitotenv.2020.138996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adelodun B., Ajibade F.O., Ibrahim R.G., Bakare H.O., Choi K.S. Snowballing transmission of COVID-19 (SARS-CoV-2) through wastewater: any sustainable preventive measures to curtail the scourge in low-income countries? Sci. Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.140680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O'Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Bibby K., Haramoto E., Hewitt J., Huygens F., Gyawali P., Korajkic A., Riddell S., Sherchan S.P., Simpson S.L., Sirikanchana K., Symonds E.M., Verhagen R., Vasan S.S., Kitajima M., Bivins A. Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environ. Res. 2020 doi: 10.1016/j.envres.2020.110092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert M., Schwartzbrod L. Recovery of enterovirus from primary sludge using three elution concentration procedures. Water Sci. Technol. 1991;24:225–228. doi: 10.2166/wst.1991.0063. [DOI] [Google Scholar]
- Alexander M. Why microbial predators and parasites do not eliminate their prey and hosts. Annu. Rev. Microbiol. 1981 doi: 10.1146/annurev.mi.35.100181.000553. [DOI] [PubMed] [Google Scholar]
- Allard A., Albinsson B., Wadell G. Detection of adenoviruses in stools from healthy persons and patients with diarrhea by two-step polymerase chain reaction. J. Med. Virol. 1992;37:149–157. doi: 10.1002/jmv.1890370214. [DOI] [PubMed] [Google Scholar]
- Alouini S., Sobsey M.D. Evaluation of an extraction-precipitation method for recovering hepatitis A virus and poliovirus from hardshell clams (mercenaria mercenaria) Water Sci. Technol. 1995;31:465–469. doi: 10.1016/0273-1223(95)00313-C. [DOI] [Google Scholar]
- Ammersbach M., Bienzle D. Methods for assessing feline immunodeficiency virus infection, infectivity and purification. Vet. Immunol. Immunopathol. 2011;143:202–214. doi: 10.1016/j.vetimm.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Anand U., Adelodun B., Pivato A., Suresh S., Indari O., Jakhmola S., Jha H.C., Jha P.K., Tripathi V., Di Maria F. A review of the presence of SARS-CoV-2 RNA in wastewater and airborne particulates and its use for virus spreading surveillance. Environ. Res. 2021;196:110929. doi: 10.1016/j.envres.2021.110929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand U., Cabreros C., Mal J., Ballesteros F., Sillanpää M., Tripathi V., Bontempi E. Novel coronavirus disease 2019 (COVID-19) pandemic: from transmission to control with an interdisciplinary vision. Environ. Res. 2021;197:111126. doi: 10.1016/j.envres.2021.111126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragaw T.A. Surgical face masks as a potential source for microplastic pollution in the COVID-19 scenario. Mar. Pollut. Bull. 2020 doi: 10.1016/j.marpolbul.2020.111517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atar S., Atar I. An invited commentary on “The socio-economic implications of the coronavirus and COVID-19 pandemic: a review. Int. J. Surg. 2020;78:122. doi: 10.1016/j.ijsu.2020.04.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attinti R., Wei J., Kniel K., Thomas Sims J., Jin Y. Virus' (MS2, PdblX174, and Aichi) attachment on sand measured by atomic force microscopy and their transport through sand columns. Environ. Sci. Technol. 2010 doi: 10.1021/es903221p. [DOI] [PubMed] [Google Scholar]
- Badawy A.S., Rose J.B., Gerba C.P. Comparative survival of enteric viruses and coliphage on sewage irrigated grass. J. Environ. Sci. Heal. . Part A Environ. Sci. Eng. Toxicol. 1990;25:937–952. doi: 10.1080/10934529009375610. [DOI] [Google Scholar]
- Balboa S., Mauricio-Iglesias M., Rodriguez S., Martínez-Lamas L., Vasallo F.J., Regueiro B., Lema J.M. 2020. The Fate of SARS-COV-2 in Wastewater Treatment Plants Points Out the Sludge Line as a Suitable Spot for Incidence Monitoring. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldovin T., Amoruso I., Fonzo M., Buja A., Baldo V., Cocchio S., Bertoncello C. SARS-CoV-2 RNA detection and persistence in wastewater samples: an experimental network for COVID-19 environmental surveillance in Padua, Veneto Region (NE Italy) Sci. Total Environ. 2021;760:143329. doi: 10.1016/j.scitotenv.2020.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgardner D.J. Soil-related bacterial and fungal infections. J. Am. Board Fam. Med. 2012 doi: 10.3122/jabfm.2012.05.110226. [DOI] [PubMed] [Google Scholar]
- Berman J.D., Ebisu K. Changes in U.S. air pollution during the COVID-19 pandemic. Sci. Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.139864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt W.Q., Schijven J., Regnery J., Wing A., Morrison C.M., Drewes J.E., Gerba C.P. Variable non-linear removal of viruses during transport through a saturated soil column. J. Contam. Hydrol. 2019 doi: 10.1016/j.jconhyd.2019.04.002. [DOI] [PubMed] [Google Scholar]
- Bianco F., Race M., Papirio S., Oleszczuk P., Esposito G. The addition of biochar as a sustainable strategy for the remediation of PAH–contaminated sediments. Chemosphere. 2021;263:128274. doi: 10.1016/j.chemosphere.2020.128274. [DOI] [PubMed] [Google Scholar]
- Bivins A., Greaves J., Fischer R., Yinda K.C., Ahmed W., Kitajima M., Munster V.J., Bibby K. Persistence of SARS-CoV-2 in water and wastewater. Environ. Sci. Technol. Lett. 2020 doi: 10.1021/acs.estlett.0c00730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogler A., Packman A., Furman A., Gross A., Kushmaro A., Ronen A., Dagot C., Hill C., Vaizel-Ohayon D., Morgenroth E., Bertuzzo E., Wells G., Kiperwas H.R., Horn H., Negev I., Zucker I., Bar-Or I., Moran-Gilad J., Balcazar J.L., Bibby K., Elimelech M., Weisbrod N., Nir O., Sued O., Gillor O., Alvarez P.J., Crameri S., Arnon S., Walker S., Yaron S., Nguyen T.H., Berchenko Y., Hu Y., Ronen Z., Bar-Zeev E. Rethinking wastewater risks and monitoring in light of the COVID-19 pandemic. Nat. Sustain. 2020 doi: 10.1038/s41893-020-00605-2. [DOI] [Google Scholar]
- Børsheim K.Y., Bratbak G., Heldal M. Enumeration and biomass estimation of planktonic bacteria and viruses by transmission electron microscopy. Appl. Environ. Microbiol. 1990;56:352–356. doi: 10.1128/AEM.56.2.352-356.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch A., Pintó R.M., Abad F.X. Viruses in Foods. 2006. Survival and transport of enteric viruses in the environment. [DOI] [Google Scholar]
- Breitbart M., Rohwer F. Here a virus, there a virus, everywhere the same virus? Trends Microbiol. 2005;13:278–284. doi: 10.1016/j.tim.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Brussaard C.P.D. Optimization of procedures for counting viruses by flow cytometry. Appl. Environ. Microbiol. 2004;70:1506–1513. doi: 10.1128/AEM.70.3.1506-1513.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buée M., de Boer W., Martin F., van Overbeek L., Jurkevitch E. The rhizosphere zoo: an overview of plant-associated communities of microorganisms, including phages, bacteria, archaea, and fungi, and of some of their structuring factors. Plant Soil. 2009 doi: 10.1007/s11104-009-9991-3. [DOI] [Google Scholar]
- Cao Z., Zhang Q., Lu X., Pfeiffer D., Jia Z., Song H., Zeng D.D. 2020. Estimating the Effective Reproduction Number of the 2019-nCoV in China. medRxiv. [DOI] [Google Scholar]
- Cascella M., Rajnik M., Cuomo A., Dulebohn S.C., Di Napoli R. 2020. Features, Evaluation and Treatment Coronavirus (COVID-19) (StatPearls) [PubMed] [Google Scholar]
- Cela-Dablanca R., Santás-Miguel V., Fernández-Calviño D., Arias-Estévez M., Fernández-Sanjurjo M.J., Álvarez-Rodríguez E., Núñez-Delgado A. SARS-CoV-2 and other main pathogenic microorganisms in the environment: situation in Galicia and Spain. Environ. Res. 2021;197:111049. doi: 10.1016/j.envres.2021.111049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.W., Kok K.H., Zhu Z., Chu H., To K.K.W., Yuan S., Yuen K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microb. Infect. 2020 doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collivignarelli M.C., Collivignarelli C., Carnevale Miino M., Abbà A., Pedrazzani R., Bertanza G. SARS-CoV-2 in sewer systems and connected facilities. Process Saf. Environ. Protect. 2020 doi: 10.1016/j.psep.2020.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombet J., Robin A., Lavie L., Bettarel Y., Cauchie H.M., Sime-Ngando T. Virioplankton ‘pegylation’: use of PEG (polyethylene glycol) to concentrate and purify viruses in pelagic ecosystems. J. Microbiol. Methods. 2007;71:212–219. doi: 10.1016/j.mimet.2007.08.012. [DOI] [PubMed] [Google Scholar]
- Colombet J., Sime-Ngando T. Use of PEG, Polyethylene glycol, to characterize the diversity of environmental viruses. Microb. Ecol. 2012;58:728–736. [Google Scholar]
- Conde-Cid M., Arias-Estévez M., Núñez-Delgado A. SARS-CoV-2 and other pathogens could be determined in liquid samples from soils. Environ. Pollut. 2021 doi: 10.1016/j.envpol.2021.116445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde-Cid M., Arias-Estévez M., Núñez-Delgado A. How to study SARS-CoV-2 in soils? Environ. Res. 2020 doi: 10.1016/j.envres.2020.110464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cots J.M., Alós J., Bárcena M., Boleda X. Minimising the present and future plastic waste, energy and environmental footprints related to COVID-19. Renew. Sustain. Energy Rev. 2020 doi: 10.1016/j.rser.2020.109883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Serres G., Cromeans T.L., Levesque B., Brassard N., Barthe C., Dionne M., Prud’homme H., Paradis D., Shapiro C.N., Nainan O.V., Margolis H.S. Molecular confirmation of hepatitis A virus from well water: epidemiology and public health implications. J. Infect. Dis. 1999;179:37–43. doi: 10.1086/314565. [DOI] [PubMed] [Google Scholar]
- Di Maria F., Beccaloni E., Bonadonna L., Cini C., Confalonieri E., La Rosa G., Milana M.R., Testai E., Scaini F. Minimization of spreading of SARS-CoV-2 via household waste produced by subjects affected by COVID-19 or in quarantine. Sci. Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.140803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias R.S., Abe A.E., Lima H.S., Silva L.C.F., de Paula S.O., da Silva C.C. Viral concentration methods for diversity studies in soil samples. Appl. Soil Ecol. 2020;155:103666. doi: 10.1016/j.apsoil.2020.103666. [DOI] [Google Scholar]
- Ding S., Liang T.J. Is SARS-CoV-2 also an enteric pathogen with potential fecal–oral transmission? A COVID-19 virological and clinical review. Gastroenterology. 2020;159:53–61. doi: 10.1053/j.gastro.2020.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson K. Detection, quantitation and identification of enteroviruses from surface waters and sponge tissue from the Florida Keys using real-time RT–PCR. Water Res. 2002;36:2505–2514. doi: 10.1016/S0043-1354(01)00479-1. [DOI] [PubMed] [Google Scholar]
- Douglas T. Viruses: making friends with old foes. Science (80-. ) 2006;312:873–875. doi: 10.1126/science.1123223. [DOI] [PubMed] [Google Scholar]
- Dublineau A., Batéjat C., Pinon A., Burguière A.M., Leclercq I., Manuguerra J.C. Persistence of the 2009 pandemic influenza a (H1N1) virus in water and on non-porous surface. PloS One. 2011 doi: 10.1371/journal.pone.0028043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers M.M., Grabow W.O.K., Pavlov D.N. Detection of enteroviruses in untreated and treated drinking water supplies in South Africa. Water Res. 2005;39:2253–2258. doi: 10.1016/j.watres.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Emerson J.B. Soil viruses: a new hope. mSystems. 2019;4 doi: 10.1128/mSystems.00120-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y., Zhang H., Xie J., Lin M., Ying L., Pang P., Ji W. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 2020;296:E115–E117. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feiner R., Argov T., Rabinovich L., Sigal N., Borovok I., Herskovits A.A. A new perspective on lysogeny: prophages as active regulatory switches of bacteria. Nat. Rev. Microbiol. 2015 doi: 10.1038/nrmicro3527. [DOI] [PubMed] [Google Scholar]
- Feng S., Shen C., Xia N., Song W., Fan M., Cowling B.J. Rational use of face masks in the COVID-19 pandemic. Lancet Respir. Med. 2020 doi: 10.1016/S2213-2600(20)30134-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foladori P., Cutrupi F., Segata N., Manara S., Pinto F., Malpei F., Bruni L., La Rosa G. SARS-CoV-2 from faeces to wastewater treatment: what do we know? A review. Sci. Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.140444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong T.-T., Lipp E.K. Enteric viruses of humans and animals in aquatic environments: health risks, detection, and potential water quality assessment tools. Microbiol. Mol. Biol. Rev. 2005;69:357–371. doi: 10.1128/MMBR.69.2.357-371.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fout G.S., Martinson B.C., Moyer M.W.N., Dahling D.R. A multiplex reverse transcription-PCR method for detection of human enteric viruses in groundwater. Appl. Environ. Microbiol. 2003;69:3158–3164. doi: 10.1128/AEM.69.6.3158-3164.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin A.B., Bevins S.N. Spillover of SARS-CoV-2 into novel wild hosts in North America: a conceptual model for perpetuation of the pathogen. Sci. Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.139358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumian T.M., Leite J.P.G., Castello A.A., Gaggero A., Caillou M.S.L. de, Miagostovich M.P. Detection of rotavirus A in sewage samples using multiplex qPCR and an evaluation of the ultracentrifugation and adsorption-elution methods for virus concentration. J. Virol. Methods. 2010;170:42–46. doi: 10.1016/j.jviromet.2010.08.017. [DOI] [PubMed] [Google Scholar]
- George P.B.L., Lallias D., Creer S., Seaton F.M., Kenny J.G., Eccles R.M., Griffiths R.I., Lebron I., Emmett B.A., Robinson D.A., Jones D.L. Divergent national-scale trends of microbial and animal biodiversity revealed across diverse temperate soil ecosystems. Nat. Commun. 2019 doi: 10.1038/s41467-019-09031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabow W.O.K., de Villiers J.C., Prinsloo N. An assessment of methods for the microbiological analysis of shellfish. Water Sci. Technol. 1991;24:413–416. doi: 10.2166/wst.1991.0101. [DOI] [Google Scholar]
- Griffin D.W., Donaldson K.A., Paul J.H., Rose J.B. Pathogenic human viruses in coastal waters. Clin. Microbiol. Rev. 2003 doi: 10.1128/CMR.16.1.129-143.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimarães F.R., Ferreira F.F.M., Vieira C.B., Fumian T.L.M., Shubo T., Leite J.P.G., Miagostovich M.P. Molecular detection of human astrovirus in an urban sewage treatment plant in Rio de Janeiro, Brazil. Mem. Inst. Oswaldo Cruz. 2008;103:819–823. doi: 10.1590/S0074-02762008000800013. [DOI] [PubMed] [Google Scholar]
- Gundy P.M., Gerba C.P., Pepper I.L. Survival of coronaviruses in water and wastewater. Food Environ. Virol. 2009 doi: 10.1007/s12560-008-9001-6. [DOI] [Google Scholar]
- Gutiérrez R.A., Buchy P. Contaminated soil and transmission of influenza virus (H5N1) Emerg. Infect. Dis. 2012;18:1530. doi: 10.3201/eid1809.120402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzmán C., Jofre J., Blanch A.R., Lucena F. Development of a feasible method to extract somatic coliphages from sludge, soil and treated biowaste. J. Virol. Methods. 2007;144:41–48. doi: 10.1016/j.jviromet.2007.03.017. [DOI] [PubMed] [Google Scholar]
- Hart O.E., Halden R.U. Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: feasibility, economy, opportunities and challenges. Sci. Total Environ. 2020:138875. doi: 10.1016/j.scitotenv.2020.138875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He G., Pan Y., Tanaka T. The short-term impacts of COVID-19 lockdown on urban air pollution in China. Nat. Sustain. 2020 doi: 10.1038/s41893-020-0581-y. [DOI] [Google Scholar]
- Hennes K.P., Suttle C.A. Direct counts of viruses in natural waters and laboratory cultures by epifluorescence microscopy. Limnol. Oceanogr. 1995;40:1050–1055. doi: 10.4319/lo.1995.40.6.1050. [DOI] [Google Scholar]
- Herman J., Biegel B., Huang L. Inactivation times from 290 to 315 nm UVB in sunlight for SARS coronaviruses CoV and CoV-2 using OMI satellite data for the sunlit Earth. Air Qual. Atmos. Heal. 2020 doi: 10.1007/s11869-020-00927-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horswell J., Hewitt J., Prosser J., Van Schaik A., Croucher D., MacDonald C., Burford P., Susarla P., Bickers P., Speir T. Mobility and survival of Salmonella Typhimurium and human adenovirus from spiked sewage sludge applied to soil columns. J. Appl. Microbiol. 2010 doi: 10.1111/j.1365-2672.2009.04416.x. [DOI] [PubMed] [Google Scholar]
- Hsiang S., Allen D., Annan-Phan S., Bell K., Bolliger I., Chong T., Druckenmiller H., Huang L.Y., Hultgren A., Krasovich E., Lau P., Lee J., Rolf E., Tseng J., Wu T. Publisher Correction: the effect of large-scale anti-contagion policies on the COVID-19 pandemic. Nature. 2020;585 doi: 10.1038/s41586-020-2691-0. E7–E7. [DOI] [PubMed] [Google Scholar]
- Huang W.E., Lim B., Hsu C.-C., Xiong D., Wu W., Yu Y., Jia H., Wang Y., Zeng Y., Ji M., Chang H., Zhang X., Wang H., Cui Z. RT-LAMP for rapid diagnosis of coronavirus SARS-CoV-2. Microb. Biotechnol. 2020;13:950–961. doi: 10.1111/1751-7915.13586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insam H. 2001. Developments in Soil Microbiology since the Mid 1960s. Geoderma. [DOI] [Google Scholar]
- Iwanami S., Kitagawa K., Ohashi H., Asai Y., Shionoya K., Saso W., Nishioka K., Inaba H., Nakaoka S., Wakita T., Diekmann O., Iwami S., Watashi K. Should a viral genome stay in the host cell or leave? A quantitative dynamics study of how hepatitis C virus deals with this dilemma. PLoS Biol. 2020 doi: 10.1371/journal.pbio.3000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer M., Tiwari S., Renu K., Pasha M.Y., Pandit S., Singh B., Raj N., Krothapalli S., Kwak H.J., Balasubramanian V., Jang S., Bin G., D K., Uttpal A., Narayanasamy A., Kinoshita M., Subramaniam M.D., Nachimuthu S.K., Roy A., Valsala Gopalakrishnan A., Ramakrishnan P., Cho S.G., Vellingiri B. Environmental survival of SARS-CoV-2 – a solid waste perspective. Environ. Res. 2021;197:111015. doi: 10.1016/j.envres.2021.111015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R. Horizontal gene transfer accelerates genome innovation and evolution. Mol. Biol. Evol. 2003;20:1598–1602. doi: 10.1093/molbev/msg154. [DOI] [PubMed] [Google Scholar]
- Jin M., Guo X., Zhang R., Qu W., Gao B., Zeng R. Diversities and potential biogeochemical impacts of mangrove soil viruses. Microbiome. 2019;7:58. doi: 10.1186/s40168-019-0675-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jofre J., Blasi M., Bosch A., Lucena F. Occurrence of bacteriophages infecting Bacteroides fragilis and other viruses in polluted marine sediments. Water Sci. Technol. 1989;21:15–19. doi: 10.2166/wst.1989.0072. [DOI] [Google Scholar]
- Jones J.B., Jackson L.E., Balogh B., Obradovic A., Iriarte F.B., Momol M.T. Bacteriophages for plant disease control. Annu. Rev. Phytopathol. 2007;45:245–262. doi: 10.1146/annurev.phyto.45.062806.094411. [DOI] [PubMed] [Google Scholar]
- Katayama H., Shimasaki A., Ohgaki S. Development of a virus concentration method and its application to detection of enterovirus and norwalk virus from coastal seawater. Appl. Environ. Microbiol. 2002;68:1033–1039. doi: 10.1128/aem.68.3.1033-1039.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz A., Peña S., Alimova A., Gottlieb P., Xu M., Block K.A. Heteroaggregation of an enveloped bacteriophage with colloidal sediments and effect on virus viability. Sci. Total Environ. 2018 doi: 10.1016/j.scitotenv.2018.04.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzenelson E., Fattal B., Hostovesky T. Organic flocculation: an efficient second-step concentration method for the detection of viruses in tap water. Appl. Environ. Microbiol. 1976;32:638–639. doi: 10.1128/aem.32.4.638-639.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keswick B.H., Gerba C.P., DuPont H.L., Rose J.B. Detection of enteric viruses in treated drinking water. Appl. Environ. Microbiol. 1984;47:1290–1294. doi: 10.1128/aem.47.6.1290-1294.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., Jia Z.J., Nakayama N., Asakawa S. Ecology of viruses in soils: past, present and future perspectives. Soil Sci. Plant Nutr. 2008 doi: 10.1111/j.1747-0765.2007.00197.x. [DOI] [Google Scholar]
- Kitajima M., Ahmed W., Bibby K., Carducci A., Gerba C.P., Hamilton K.A., Haramoto E., Rose J.B. SARS-CoV-2 in wastewater: state of the knowledge and research needs. Sci. Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.139076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittigul L., Singhaboot Y., Chavalitshewinkoon-Petmitr P., Pombubpa K., Hirunpetcharat C. A comparison of virus concentration methods for molecular detection and characterization of rotavirus in bivalve shellfish species. Food Microbiol. 2015;46:161–167. doi: 10.1016/j.fm.2014.07.020. [DOI] [PubMed] [Google Scholar]
- Klemeš J.J., Fan Y. Van, Tan R.R., Jiang P. Minimising the present and future plastic waste, energy and environmental footprints related to COVID-19. Renew. Sustain. Energy Rev. 2020 doi: 10.1016/j.rser.2020.109883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüttgen A., Cornelissen C.G., Dreher M., Hornef M.W., Imöhl M., Kleines M. Comparison of the SARS-CoV-2 Rapid antigen test to the real star Sars-CoV-2 RT PCR kit. J. Virol. Methods. 2021 doi: 10.1016/j.jviromet.2020.114024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni B.N., Anantharama V. Repercussions of COVID-19 pandemic on municipal solid waste management: challenges and opportunities. Sci. Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.140693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Mazumder P., Mohapatra S., Kumar Thakur A., Dhangar K., Taki K., Mukherjee S., Kumar Patel A., Bhattacharya P., Mohapatra P., Rinklebe J., Kitajima M., Hai F.I., Khursheed A., Furumai H., Sonne C., Kuroda K. A chronicle of SARS-CoV-2: seasonality, environmental fate, transport, inactivation, and antiviral drug resistance. J. Hazard Mater. 2020;124043 doi: 10.1016/j.jhazmat.2020.124043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Mohapatra S., Mazumder P., Singh A., Honda R., Lin C., Kumari R., Goswami R., Jha P.K., Vithanage M., Kuroda K. Making waves perspectives of modelling and monitoring of SARS-CoV-2 in aquatic environment for COVID-19 pandemic. Curr. Pollut. Reports. 2020 doi: 10.1007/s40726-020-00161-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Thakur A.K., Mazumder P., Kuroda K., Mohapatra S., Rinklebe J., Ramanathan A., Cetecioglu Z., Jain S., Tyagi V.K., Gikas P., Chakraborty S., Tahmidul Islam M., Ahmad A., Shah A.V., Patel A.K., Watanabe T., Vithanage M., Bibby K., Kitajima M., Bhattacharya P. Frontier review on the propensity and repercussion of SARS-CoV-2 migration to aquatic environment. J. Hazard. Mater. Lett. 2020;1:100001. doi: 10.1016/j.hazl.2020.100001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzyakov Y., Mason-jones K. Nano-scale undead drivers of microbial life, biogeochemical turnover and ecosystem functions. Soil Biol. Biochem. 2018 [Google Scholar]
- La Rosa G., Bonadonna L., Lucentini L., Kenmoe S., Suffredini E. Coronavirus in water environments: occurrence, persistence and concentration methods - a scoping review. Water Res. 2020 doi: 10.1016/j.watres.2020.115899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal R., Brevik E.C., Dawson L., Field D., Glaser B., Hartemink A.E., Hatano R., Lascelles B., Monger C., Scholten T., Singh B.R., Spiegel H., Terribile F., Basile A., Zhang Y., Horn R., Kosaki T., Sánchez L.B.R. Managing soils for recovering from the covid-19 pandemic. Soil Syst. 2020 doi: 10.3390/soilsystems4030046. [DOI] [Google Scholar]
- Lama G.F.C., Errico A., Francalanci S., Solari L., Preti F., Chirico G.B. Evaluation of flow resistance models based on field experiments in a partly vegetated reclamation channel. Geosciences. 2020;10:47. doi: 10.3390/geosciences10020047. [DOI] [Google Scholar]
- Lamastra L., Suciu N.A., Trevisan M. Sewage sludge for sustainable agriculture: contaminants' contents and potential use as fertilizer. Chem. Biol. Technol. Agric. 2018;5:10. doi: 10.1186/s40538-018-0122-3. [DOI] [Google Scholar]
- Le Guyader F., Dubois E., Menard D., Pommepuy M. Detection of hepatitis A virus, rotavirus, and enterovirus in naturally contaminated shellfish and sediment by reverse transcription-seminested PCR. Appl. Environ. Microbiol. 1994;60:3665–3671. doi: 10.1128/AEM.60.10.3665-3671.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.S., Lee C., Marion J., Wang Q., Saif L., Lee J. Occurrence of human enteric viruses at freshwater beaches during swimming season and its link to water inflow. Sci. Total Environ. 2014;472:757–766. doi: 10.1016/j.scitotenv.2013.11.088. [DOI] [PubMed] [Google Scholar]
- Lee H.K., Jeong Y.S. Comparison of total culturable virus assay and multiplex integrated cell culture-PCR for reliability of waterborne virus detection. Appl. Environ. Microbiol. 2004;70:3632–3636. doi: 10.1128/AEM.70.6.3632-3636.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P.I., Hsueh P.R. Emerging threats from zoonotic coronaviruses-from SARS and MERS to 2019-nCoV. J. Microbiol. Immunol. Infect. 2020;53:365–367. doi: 10.1016/j.jmii.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis G.D., Loutit M.W., Austin F.J. A method for detecting human enteroviruses in aquatic sediments. J. Virol. Methods. 1985;10:153–162. doi: 10.1016/0166-0934(85)90101-6. [DOI] [PubMed] [Google Scholar]
- Lewis G.D., Metcalf T.G. Polyethylene glycol precipitation for recovery of pathogenic viruses, including hepatitis A virus and human rotavirus, from oyster, water, and sediment samples. Appl. Environ. Microbiol. 1988;54:1983–1988. doi: 10.1128/aem.54.8.1983-1988.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian X., Piao S., Li L.Z.X., Li Y., Huntingford C., Ciais P., Cescatti A., Janssens I.A., Peñuelas J., Buermann W., Chen A., Li X., Myneni R.B., Wang X., Wang Y., Yang Y., Zeng Z., Zhang Y., McVicar T.R. Summer soil drying exacerbated by earlier spring greening of northern vegetation. Sci. Adv. 2020 doi: 10.1126/sciadv.aax0255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Liu L., Kou G., Zheng Y., Ding Y., Ni W., Wang Q., Tan L., Wu W., Tang S., Xiong Z., Zheng S. Evaluation of nucleocapsid and spike protein-based enzyme-linked immunosorbent assays for detecting antibodies against SARS-CoV-2. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.00461-20. e00461-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Gayle A.A., Wilder-Smith A., Rocklöv J. The reproductive number of COVID-19 is higher compared to SARS coronavirus. J. Trav. Med. 2020;27 doi: 10.1093/jtm/taaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynteris C. A “suitable soil”: plague's urban breeding grounds at the dawn of the third pandemic. Med. Hist. 2017 doi: 10.1017/mdh.2017.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magri M.E., Fidjeland J., Jönsson H., Albihn A., Vinnerås B. Inactivation of adenovirus, reovirus and bacteriophages in fecal sludge by pH and ammonia. Sci. Total Environ. 2015;520:213–221. doi: 10.1016/j.scitotenv.2015.03.035. [DOI] [PubMed] [Google Scholar]
- Mancuso G., Perulli G.D., Lavrnic S., Morandi B., Toscano A. Sars-cov-2 from urban to rural water environment: occurrence, persistence, fate, and influence on agriculture irrigation. A review. Water (Switzerland) 2021;13:764. doi: 10.3390/w13060764. [DOI] [Google Scholar]
- Martínez-Puchol S., Rusiñol M., Fernández-Cassi X., Timoneda N., Itarte M., Andrés C., Antón A., Abril J.F., Girones R., Bofill-Mas S. Characterisation of the sewage virome: comparison of NGS tools and occurrence of significant pathogens. Sci. Total Environ. 2020;713:136604. doi: 10.1016/j.scitotenv.2020.136604. [DOI] [PubMed] [Google Scholar]
- Michen B., Graule T. Isoelectric points of viruses. J. Appl. Microbiol. 2010 doi: 10.1111/j.1365-2672.2010.04663.x. [DOI] [PubMed] [Google Scholar]
- Miller R.V., Day M.J. Bacteriophage Ecology. 2009. Contribution of lysogeny, pseudolysogeny, and starvation to phage ecology. [DOI] [Google Scholar]
- Miura T., Masago Y., Chan Y.-M., Imai T., Omura T. Detection of bacteria and enteric viruses from river and estuarine sediment. J. Water Environ. Technol. 2009;7:307–316. doi: 10.2965/jwet.2009.307. [DOI] [Google Scholar]
- Moelling K. Within-host and between-host evolution in SARS-CoV-2—new variant's source. Viruses. 2021;13:751. doi: 10.3390/v13050751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mofijur M., Fattah I.M.R., Alam M.A., Islam A.B.M.S., Ong H.C., Rahman S.M.A., Najafi G., Ahmed S.F., Uddin M.A., Mahlia T.M.I. Impact of COVID-19 on the social, economic, environmental and energy domains: lessons learnt from a global pandemic. Sustain. Prod. Consum. 2021;26:343–359. doi: 10.1016/j.spc.2020.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan S.V., Hemalatha M., Kopperi H., Ranjith I., Kumar A.K. SARS-CoV-2 in environmental perspective: occurrence, persistence, surveillance, inactivation and challenges. Chem. Eng. J. 2021;405:126893. doi: 10.1016/j.cej.2020.126893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra S., Gayathri Menon N., Mohapatra G., Pisharody L., Pattnaik A., Menon N. Gowri, Bhukya P.L., Srivastava M., Singh M., Barman M.K., Gin K.Y.-H., Mukherji S. The novel SARS-CoV-2 pandemic: possible environmental transmission, detection, persistence and fate during wastewater and water treatment. Sci. Total Environ. 2020;142746 doi: 10.1016/j.scitotenv.2020.142746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monpoeho S., Maul A., Mignotte-Cadiergues B., Schwartzbrod L., Billaudel S., Ferré V. Best viral elution method available for quantification of enteroviruses in sludge by both cell culture and reverse transcription-PCR. Appl. Environ. Microbiol. 2001;67:2484–2488. doi: 10.1128/AEM.67.6.2484-2488.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munson-Mcgee J.H., Peng S., Dewerff S., Stepanauskas R., Whitaker R.J., Weitz J.S., Young M.J. A virus or more in (nearly) every cell: ubiquitous networks of virus-host interactions in extreme environments. ISME J. 2018 doi: 10.1038/s41396-018-0071-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nannipieri P., Ascher J., Ceccherini M.T., Landi L., Pietramellara G., Renella G. Microbial diversity and soil functions. Eur. J. Soil Sci. 2017;68:12–26. doi: 10.1111/ejss.4_12398. [DOI] [Google Scholar]
- Nasser A. Contribution of microbial activity to virus reduction in saturated soil. Water Res. 2002;36:2589–2595. doi: 10.1016/S0043-1354(01)00461-4. [DOI] [PubMed] [Google Scholar]
- Noble R., Fuhrman J. Use of SYBR Green I for rapid epifluorescence counts of marine viruses and bacteria. Aquat. Microb. Ecol. 1998;14:113–118. doi: 10.3354/ame014113. [DOI] [Google Scholar]
- Nordgren J., Matussek A., Mattsson A., Svensson L., Lindgren P.-E. Prevalence of norovirus and factors influencing virus concentrations during one year in a full-scale wastewater treatment plant. Water Res. 2009;43:1117–1125. doi: 10.1016/j.watres.2008.11.053. [DOI] [PubMed] [Google Scholar]
- Núñez-Delgado A. What do we know about the SARS-CoV-2 coronavirus in the environment? Sci. Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.138647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Núñez-Delgado A. SARS-CoV-2 in soils. Environ. Res. 2020;190:110045. doi: 10.1016/j.envres.2020.110045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey D., Verma S., Verma P., Mahanty B., Dutta K., Daverey A., Arunachalam K. SARS-CoV-2 in wastewater: challenges for developing countries. Int. J. Hyg Environ. Health. 2021 doi: 10.1016/j.ijheh.2020.113634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papirio S., Zou G., Ylinen A., Di Capua F., Pirozzi F., Puhakka J.A. Effect of arsenic on nitrification of simulated mining water. Bioresour. Technol. 2014;164:149–154. doi: 10.1016/j.biortech.2014.04.072. [DOI] [PubMed] [Google Scholar]
- Patel M., Chaubey A.K., Pittman C.U., Mlsna T., Mohan D. Coronavirus (SARS-CoV-2) in the environment: occurrence, persistence, analysis in aquatic systems and possible management. Sci. Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.142698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrício Silva A.L., Prata J.C., Walker T.R., Duarte A.C., Ouyang W., Barcelò D., Rocha-Santos T. Increased plastic pollution due to COVID-19 pandemic: challenges and recommendations. Chem. Eng. J. 2021 doi: 10.1016/j.cej.2020.126683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyret H. A protocol for the gentle purification of virus-like particles produced in plants. J. Virol. Methods. 2015;225:59–63. doi: 10.1016/j.jviromet.2015.09.005. [DOI] [PubMed] [Google Scholar]
- Pfefferle S., Reucher S., Nörz D., Lütgehetmann M. Evaluation of a quantitative RT-PCR assay for the detection of the emerging coronavirus SARS-CoV-2 using a high throughput system. Euro Surveill. 2020;25:2000152. doi: 10.2807/1560-7917.ES.2020.25.9.2000152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourcher A.M., Françoise P.B., Virginie F., Agnieszka G., Vasilica S., Gérard M. Survival of faecal indicators and enteroviruses in soil after land-spreading of municipal sewage sludge. Appl. Soil Ecol. 2007 doi: 10.1016/j.apsoil.2006.10.005. [DOI] [Google Scholar]
- Prompetchara E., Ketloy C., Palaga T. Immune responses in COVID-19 and potential vaccines: lessons learned from SARS and MERS epidemic. Asian Pac. J. Allergy Immunol. 2020 doi: 10.12932/AP-200220-0772. [DOI] [PubMed] [Google Scholar]
- Qian H., Zhang Q., Lu T., Peijnenburg W.J.G.M., Penuelas J., Zhu Y.-G. Lessons learned from COVID-19 on potentially pathogenic soil microorganisms. Soil Ecol. Lett. 2020 doi: 10.1007/s42832-020-0068-9. [DOI] [Google Scholar]
- Qu G., Li X., Hu L., Jiang G. An imperative need for research on the role of environmental factors in transmission of novel coronavirus (COVID-19) Environ. Sci. Technol. 2020 doi: 10.1021/acs.est.0c01102. [DOI] [PubMed] [Google Scholar]
- Rabot E., Wiesmeier M., Schlüter S., Vogel H.-J. Soil structure as an indicator of soil functions: a review. Geoderma. 2018;314:122–137. doi: 10.1016/j.geoderma.2017.11.009. [DOI] [Google Scholar]
- Ragazzi M., Rada E.C., Schiavon M. Municipal solid waste management during the SARS-COV-2 outbreak and lockdown ease: lessons from Italy. Sci. Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.141159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M.M., Bodrud-Doza M., Griffiths M.D., Mamun M.A. Biomedical waste amid COVID-19: perspectives from Bangladesh. Lancet Glob. Heal. 2020 doi: 10.1016/S2214-109X(20)30349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajtar B., Majek M., Polański Ł., Polz-Dacewicz M. Enteroviruses in water environment--a potential threat to public health. Ann. Agric. Environ. Med. 2008;15:199–203. [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razzaq A., Sharif A., Aziz N., Irfan M., Jermsittiparsert K. Asymmetric link between environmental pollution and COVID-19 in the top ten affected states of US: a novel estimations from quantile-on-quantile approach. Environ. Res. 2020;191:110189. doi: 10.1016/j.envres.2020.110189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regenmortel M.H.V. Biological Centrifugation. 2000. Introduction to the species concept in virus taxonomy. In virus taxonomy. Seventh report of the international committee on taxonomy of viruses. [Google Scholar]
- Reich P.B. The world-wide “fast-slow” plant economics spectrum: a traits manifesto. J. Ecol. 2014 doi: 10.1111/1365-2745.12211. [DOI] [Google Scholar]
- Rohwer F., Prangishvili D., Lindell D. Roles of viruses in the environment. Environ. Microbiol. 2009;11:2771–2774. doi: 10.1111/j.1462-2920.2009.02101.x. [DOI] [PubMed] [Google Scholar]
- Rundle A.G., Park Y., Herbstman J.B., Kinsey E.W., Wang Y.C. COVID‐19–Related school closings and risk of weight gain among children. Obesity. 2020;28:1008–1009. doi: 10.1002/oby.22813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SanJuan-Reyes S., Gómez-Oliván L.M., Islas-Flores H. COVID-19 in the environment. Chemosphere. 2021 doi: 10.1016/j.chemosphere.2020.127973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlindwein A., Simões C., Barardi C. Comparative study of two extraction methods for enteric virus recovery from sewage sludge by molecular methods. Mem. Inst. Oswaldo Cruz. 2009;104:576–579. doi: 10.1590/S0074-02762009000400007. [DOI] [PubMed] [Google Scholar]
- Shieh Y.-S.C., Calci K.R., Baric R.S. A method to detect low levels of enteric viruses in contaminated oysters. Appl. Environ. Microbiol. 1999;65:4709–4714. doi: 10.1128/aem.65.11.4709-4714.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siche R. What is the impact of COVID-19 disease on agriculture? Sci. Agropecu. 2020;11:3–6. doi: 10.17268/sci.agropecu.2020.01.00. [DOI] [Google Scholar]
- Sime-Ngando T. Environmental bacteriophages: viruses of microbes in aquatic ecosystems. Front. Microbiol. 2014 doi: 10.3389/fmicb.2014.00355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasiah S., Bhavsar J., Thapar K., Liles M., Schoenfeld T., Wommack K.E. Phages across the biosphere: contrasts of viruses in soil and aquatic environments. Res. Microbiol. 2008 doi: 10.1016/j.resmic.2008.04.010. [DOI] [PubMed] [Google Scholar]
- Srinivasiah S., Lovett J., Polson S., Bhavsar J., Ghosh D., Roy K., Fuhrmann J.J., Radosevich M., Wommack K.E. Direct assessment of viral diversity in soils by random PCR amplification of polymorphic DNA. Appl. Environ. Microbiol. 2013;79:5450–5457. doi: 10.1128/AEM.00268-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staggemeier R., Almeida S.E. de M., Spilki F.R. Methods OF virus detection IN soils and sediments. VIRUS Rev. Res. 2011;16 doi: 10.17525/vrr.v16i1-2.50. [DOI] [Google Scholar]
- Steffan J.J., Derby J.A., Brevik E.C. Soil pathogens that may potentially cause pandemics, including severe acute respiratory syndrome (SARS) coronaviruses. Curr. Opin. Environ. Sci. Heal. 2020 doi: 10.1016/j.coesh.2020.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S., Wong G., Shi W., Liu J., Lai A.C.K., Zhou J., Liu W., Bi Y., Gao G.F. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttle C.A. Marine viruses — major players in the global ecosystem. Nat. Rev. Microbiol. 2007;5:801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- Suttle C.A., Chan A.M., Cottrell M.T. Infection of phytoplankton by viruses and reduction of primary productivity. Nature. 1990;347:467–469. doi: 10.1038/347467a0. [DOI] [Google Scholar]
- Swanson M.M., Fraser G., Daniell T.J., Torrance L., Gregory P.J., Taliansky M. Viruses in soils: morphological diversity and abundance in the rhizosphere. Ann. Appl. Biol. 2009;155:51–60. doi: 10.1111/j.1744-7348.2009.00319.x. [DOI] [Google Scholar]
- Terribile F., Agrillo A., Bonfante A., Buscemi G., Colandrea M., D'Antonio A., De Mascellis R., De Michele C., Langella G., Manna P., Marotta L., Mileti F.A., Minieri L., Orefice N., Valentini S., Vingiani S., Basile A. A Web-based spatial decision supporting system for land management and soil conservation. Solid Earth. 2015 doi: 10.5194/se-6-903-2015. [DOI] [Google Scholar]
- Tobías A., Carnerero C., Reche C., Massagué J., Via M., Minguillón M.C., Alastuey A., Querol X. Changes in air quality during the lockdown in Barcelona (Spain) one month into the SARS-CoV-2 epidemic. Sci. Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.138540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touret F., Gilles M., Barral K., Nougairède A., van Helden J., Decroly E., de Lamballerie X., Coutard B. In vitro screening of a FDA approved chemical library reveals potential inhibitors of SARS-CoV-2 replication. Sci. Rep. 2020;10:13093. doi: 10.1038/s41598-020-70143-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trubl G., Hyman P., Roux S., Abedon S.T. Coming-of-Age characterization of soil viruses: a user's guide to virus isolation, detection within metagenomes, and viromics. Soil Syst. 2020;4:23. doi: 10.3390/soilsystems4020023. [DOI] [Google Scholar]
- Trubl G., Jang H. Bin, Roux S., Emerson J.B., Solonenko N., Vik D.R., Solden L., Ellenbogen J., Runyon A.T., Bolduc B., Woodcroft B.J., Saleska S.R., Tyson G.W., Wrighton K.C., Sullivan M.B., Rich V.I. 2018. Soil Viruses Are Underexplored Players in Ecosystem Carbon Processing. mSystems. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usman M., Farooq M., Hanna K. Existence of SARS-CoV-2 in wastewater: implications for its environmental transmission in developing communities. Environ. Sci. Technol. 2020 doi: 10.1021/acs.est.0c02777. [DOI] [PubMed] [Google Scholar]
- Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., Graham B.S., Mascola J.R., Chang J.Y., Yin M.T., Sobieszczyk M., Kyratsous C.A., Shapiro L., Sheng Z., Huang Y., Ho D.D. 2021. Antibody Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7. Nature 1–6. [DOI] [PubMed] [Google Scholar]
- Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA, J. Am. Med. Assoc. 2020 doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J., Jin Y., Sims T., Kniel K.E. Survival of human adenovirus 41 in land-applied manure and biosolids. Food Environ. Virol. 2009;1:148–154. doi: 10.1007/s12560-009-9021-x. [DOI] [Google Scholar]
- Weinbauer M.G. Ecology of prokaryotic viruses. FEMS Microbiol. Rev. 2004;28:127–181. doi: 10.1016/j.femsre.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Wigginton K.R., Boehm A.B. Environmental engineers and scientists have important roles to play in stemming outbreaks and pandemics caused by enveloped viruses. Environ. Sci. Technol. 2020 doi: 10.1021/acs.est.0c01476. [DOI] [PubMed] [Google Scholar]
- Wiktorczyk-Kapischke N., Grudlewska-Buda K., Wałecka-Zacharska E., Kwiecińska-Piróg J., Radtke L., Gospodarek-Komkowska E., Skowron K. SARS-CoV-2 in the environment—non-droplet spreading routes. Sci. Total Environ. 2021 doi: 10.1016/j.scitotenv.2021.145260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson K.E., Fuhrmann J.J., Wommack K.E., Radosevich M. Viruses in soil ecosystems: an unknown quantity within an unexplored territory. Annu. Rev. Virol. 2017 doi: 10.1146/annurev-virology-101416-041639. [DOI] [PubMed] [Google Scholar]
- Williamson K.E., Radosevich M., Smith D.W., Wommack K.E. Incidence of lysogeny within temperate and extreme soil environments. Environ. Microbiol. 2007 doi: 10.1111/j.1462-2920.2007.01374.x. [DOI] [PubMed] [Google Scholar]
- Williamson K.E., Radosevich M., Wommack K.E. Abundance and diversity of viruses in six Delaware soils. Appl. Environ. Microbiol. 2005 doi: 10.1128/AEM.71.6.3119-3125. 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson K.E., Wommack K.E., Radosevich M. Sampling natural viral communities from soil for culture-independent analyses. Appl. Environ. Microbiol. 2003;69:6628–6633. doi: 10.1128/aem.69.11.6628-6633.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winget D.M., Williamson K.E., Helton R.R., Wommack K.E. Tangential flow diafiltration: an improved technique for estimation of virioplankton production. Aquat. Microb. Ecol. 2005 doi: 10.3354/ame041221. [DOI] [Google Scholar]
- Winget D.M., Wommack K.E. Randomly amplified polymorphic DNA PCR as a tool for assessment of marine viral richness. Appl. Environ. Microbiol. 2008;74:2612–2618. doi: 10.1128/AEM.02829-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wommack K.E., Colwell R.R. Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 2000;64:69–114. doi: 10.1128/mmbr.64.1.69-114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K., Voice T.C., Xagoraraki I. 2013. Effect of Organic Carbon on Sorption of Human Adenovirus to Soil Particles and Laboratory Containers. Water Res. [DOI] [PubMed] [Google Scholar]
- Yamamoto K.R., Alberts B.M., Benzinger R., Lawhorne L., Treiber G. Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virology. 1970;40:734–744. doi: 10.1016/0042-6822(70)90218-7. [DOI] [PubMed] [Google Scholar]
- Ye Y., Ellenberg R.M., Graham K.E., Wigginton K.R. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ. Sci. Technol. 2016 doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]
- Yu H., Sun X., Solvang W.D., Zhao X. Reverse logistics network design for effective management of medical waste in epidemic outbreaks: insights from the coronavirus disease 2019 (COVID-19) outbreak in Wuhan (China) Int. J. Environ. Res. Publ. Health. 2020 doi: 10.3390/ijerph17051770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambrano-Monserrate M.A., Ruano M.A., Sanchez-Alcalde L. Indirect effects of COVID-19 on the environment. Sci. Total Environ. 2020 doi: 10.1016/j.scitotenv.2020.138813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zand A.D., Heir A.V. Emerging challenges in urban waste management in Tehran, Iran during the COVID-19 pandemic. Resour. Conserv. Recycl. 2020;162:105051. doi: 10.1016/j.resconrec.2020.105051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zand A.D., Heir A.V. Environmental impacts of new Coronavirus outbreak in Iran with an emphasis on waste management sector. J. Mater. Cycles Waste Manag. 2020 doi: 10.1007/s10163-020-01123-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaneti R.N., Girardi V., Spilki F.R., Mena K., Westphalen A.P.C., da Costa Colares E.R., Pozzebon A.G., Etchepare R.G. Quantitative microbial risk assessment of SARS-CoV-2 for workers in wastewater treatment plants. Sci. Total Environ. 2021;754:142163. doi: 10.1016/j.scitotenv.2020.142163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Yang Y., Huang X., Jiang J., Li M., Zhang X., Ling H., Li J., Liu Y., Li G., Li W., Yi C., Zhang T., Jiang Y., Xiong Y., Hu Z., Wang X., Deng S., Zhao P., Qu J., He Z., Wang X., Deng S., Zhao P., Qu J. SARS-CoV-2 spillover into hospital outdoor environments. medRxiv. 2020 doi: 10.1101/2020.05.12.20097105. 2020.05.12.20097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Niu L., Hu K., Hao J., Li F., Gao Z., Wang X. Influence of tillage, straw-returning and mineral fertilization on the stability and associated organic content of soil aggregates in the north China plain. Agronomy. 2020;10:951. doi: 10.3390/agronomy10070951. [DOI] [Google Scholar]
- Zhang W., Liu K., Zhang P., Cheng W., Li Linfei, Zhang F., Yu Z., Li Lifeng, Zhang X. CRISPR-based approaches for efficient and accurate detection of SARS-CoV-2. Lab. Med. 2021;52:116–121. doi: 10.1093/labmed/lmaa101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Atoni E., Du Y., Zhang H., Donde O., Huang D., Xiao S., Ma T., Shu Z., Yuan Z. 2020. First Study on Surveillance of SARS-CoV-2 RNA in Wastewater Systems and Related Environments in Wuhan: Post-lockdown. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]