Abstract
The autophagy-lysosomal degradation system has an important role in maintaining liver homeostasis by removing unnecessary intracellular components. Impaired autophagy has been linked to nonalcoholic fatty liver disease (NAFLD), which includes hepatitis, steatosis, fibrosis, and cirrhosis. Thus, gaining an understanding of the mechanisms that regulate autophagy and how autophagy contributes to the development and progression of NAFLD has become the focus of recent studies. Autophagy regulation has been thought to be primarily regulated by cytoplasmic processes; however, recent studies have shown that microRNAs (miRNAs) and transcription factors (TFs) also act as key regulators of autophagy by targeting autophagy-related genes. In this review, we summarize the miRNAs and TFs that regulate the autophagy pathway in NAFLD. We further focus on the transcriptional and posttranscriptional regulation of autophagy and discuss the complex regulatory networks involving these regulators in autophagy. Finally, we highlight the potential of targeting miRNAs and TFs involved in the regulation of autophagy for the treatment of NAFLD.
Subject terms: Transcription, Non-coding RNAs
Fatty liver disease: the interplay of RNAs and regulatory proteins
Small molecules of ribonucleic acid called microRNAs (miRNAs) and gene-regulating proteins called transcription factors (TFs) control the process of autophagy, which removes unnecessary components of cells and is impaired in nonalcoholic fatty liver disease (NAFLD). This condition is common in people who are obese or overweight. Autophagy is important for maintaining healthy cells and tissues, and is disrupted in several diseases. Researchers led by Chang Hwa Jung at Korea Food Research Institute in Wanju-gun, South Korea, review the interplay of miRNAs and TFs in regulating the activity of genes involved in autophagy and specifically its impairment in NAFLD. Complex regulatory networks being identified offer new insights into autophagy. The authors discuss the potential for developing new therapies for NAFLD using drugs that will interact with the miRNAs and TFs that control autophagy.
Introduction
Autophagy is important for maintaining intracellular protein homeostasis and for organelle quality control. In the liver, autophagy helps maintain metabolic homeostasis and, consequently, lipid balance, insulin sensitivity, and hepatocyte resistance to injuries, such as oxidative stress and inflammation1–5. Accumulating evidence indicates that autophagy is highly relevant to the pathogenesis of nonalcoholic fatty liver disease (NAFLD), including hepatitis and fibrosis, suggesting that modulating autophagy is a potential strategy for the treatment of NAFLD. However, the exact pathophysiological role of and the regulatory mechanisms underlying autophagy in NAFLD remain to be elucidated.
The regulatory network of transcriptional and posttranscriptional factors has recently attracted attention because of its roles in biological processes and in the development of various diseases6–8. Autophagy was initially considered a pathway exclusively regulated by cytoplasmic processes; however, over the past decade, a number of microRNAs (miRNAs) and transcription factors (TFs) have been found to control autophagy through the regulation of autophagy-related genes9,10. Several miRNAs and TFs that regulate autophagy pathways have been reported to play roles in NAFLD, but the interplay between miRNAs and TFs for the regulation of autophagy is not fully understood. Here, we review the interplay between miRNAs and TFs in regulating autophagy and suggest potential target networks for treating NAFLD.
The autophagy process
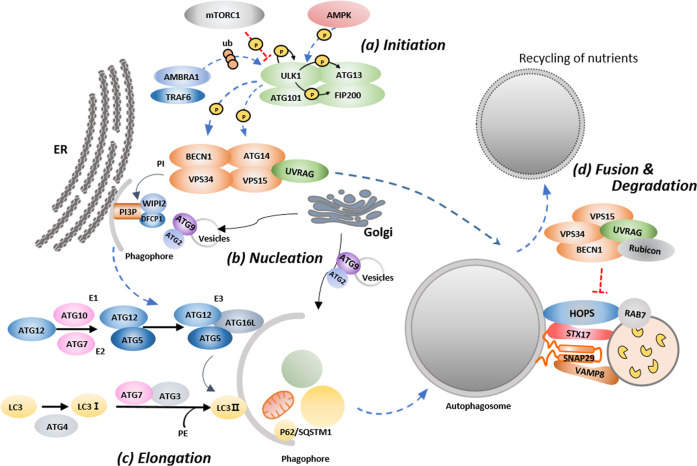
As shown in Fig. 1, the induction of autophagy begins with the Unc-51-like autophagy activating kinase 1 (ULK1) complex formed by the interaction of ULK1 with focal adhesion kinase family-interacting protein of 200 kDa (FIP200), autophagy-related protein (ATG)13, and ATG10111–13. Under starved conditions, ULK1 is autophosphorylated and activated; then, ULK1 phosphorylates ATG13 and FIP200 to induce autophagy14,15. In contrast, under nutrient-rich conditions, the activated mechanistic target of rapamycin (mTOR) inactivates ULK1 and ATG13 through phosphorylation16. AMP-activated protein kinase (AMPK) has also been reported to activate autophagy through the inhibition of mTORC1 and to directly phosphorylate several ATG proteins, including ULK1, Beclin1 (BECN1), and phosphatidylinositol 3-kinase complex III (VPS34)17–19. In the early stages of autophagy, activating molecule in Beclin1-regulated autophagy protein 1 (AMBRA1), an autophagy regulatory protein, interacts with the E3 ubiquitinase tumor necrosis factor receptor-associated factor 6 and mediates K63-linked polyubiquitination of ULK1, which enhances ULK1 kinase activity20,21.
Fig. 1. Overview of the autophagy pathway.
a When autophagy is induced by various stress factors, the Unc-51-like autophagy activating kinase (ULK) complex is activated and initiates autophagy. b Activated ULK triggers phosphatidylinositol 3-kinase complex III (VPS34) to form the phagophore at omegasomes. c The phagophore is elongated by two ubiquitin-like conjugation systems to form an autophagosome. The first system involves conjugation of autophagy-related protein (ATG)12 to ATG5 in a reaction that requires the E1-like enzyme ATG7 and E2-like enzyme ATG10. The second system involves the conjugation of C-type lectin 3 (LC3) to phosphatidylethanolamine (PE). d The mature autophagosome or amphisome binds a lysosome, and the autophagic cargo is degraded by acidic hydrolases in the lysosome and is recycled in the cell as nutrients.
Phagophores (i.e., isolation membranes) are nucleated at the endoplasmic reticulum (ER)–mitochondria or the ER–plasma membrane contact site. The ATG14L complex is a key component for the nucleation stage. The ATG14L complex consists of VPS34, VPS15, and BECN1, and it is regulated by the kinase activities of ULK1 and 5′ adenosine monophosphate-activated protein kinase (AMPK)19,22–25. Activated VPS34 generates phosphatidylinositol 3-phosphate and initiates the formation of the preautophagosomal phagophore26–28. Another key protein, involved in the elongation of the phagophore, is ATG9A/B, which is the only known autophagy-related transmembrane protein that circulates between the trans-Golgi network and the late endosome through vesicular trafficking29. Upon autophagy induction, a portion of the ATG9A pool transiently localizes to autophagic membranes, and its intracellular cycling is positively regulated by both ULK1 and VPS3430–32.
Two ubiquitin-like conjugation systems, ATG12 and C-type lectin 3/gamma-aminobutyric acid receptor-associated protein (LC3/GABARAP; ATG8 in yeast), are involved in phagophore expansion33. In the first system, ATG12 is covalently conjugated to ATG5, depending on the E1-like enzyme ATG7 and the E2-like enzyme ATG10. Then, the resulting ATG12-ATG5 compound conjugates with ATG16L1 to form the ATG12-ATG5-ATG16L1 complex, which localizes to the outer membrane of the forming autophagosome34,35. The second system involves the conjugation of the LC3/GABARAP family proteins to a lipid molecule. LC3/GABARAP is processed at the C-terminal glycine by the cysteine protease ATG4. Then, LC3/GABARAP is activated by ATG7 and is conjugated by ATG3 to the amino group of phosphatidylethanolamine36–38. Finally, the ATG16L1-ATG5-ATG12 complex functions as a scaffold for LC3/GABARAP lipidation39.
One of the most important molecules in the maturation of the autophagosome and in the fusion with lysosomes is the small GTPase protein RAB740. Fusion with lysosomes is facilitated by membrane tethering factors such as the homotypic fusion and protein sorting (HOPS) complex and soluble N-ethylmaleimide-sensitive factor-activating protein (SNAP) receptor (SNARE) proteins. The HOPS complex interacts with syntaxin17 (STX17), which interacts with SNAP29 and vesicle-associated membrane protein 8 (VAMP8) to promote autophagosome–lysosome fusion41,42. UV resistance-associated gene (UVRAG) also promotes the fusion of autophagosomes and lysosomes by binding to VPS16, which is a subunit of the HOPS complex23,43. ATG14 also directly binds to STX17-SNAP29 to form a complex with VAMP8 on lysosomes, which further promotes autophagosome–lysosome fusion44.
Roles of autophagy in NAFLD
NAFLD is caused by excessive hepatic fat accumulation that is not caused by significant alcohol consumption45. NAFLD is an umbrella term for diseases that includes simple hepatic steatosis, nonalcoholic steatohepatitis (NASH), and cirrhosis. Other than weight loss, there is currently no effective therapy for NAFLD. Studies are currently focused on understanding the molecular mechanisms underlying NAFLD to identify new therapeutic targets, and these studies have revealed a diverse repertoire of potential targets.
In obesity, the disruption of autophagy in the liver and in extrahepatic organs contributes to excessive lipid accumulation in the liver. Singh et al. were the first to suggest that autophagy might be a potential target for the treatment of NAFLD. In their study, ATG5 knockdown or pharmacological autophagy inhibition significantly increased the cellular triglyceride content46. This group suggested that autophagy has essential roles in lipolysis (lipophagy), wherein intracellular lipid droplets are degraded through a self-degradative pathway to alleviate hepatic inflammation and injury. In mouse livers lacking rubicon-like autophagy enhancer (RUBCNL)/Pacer, which promotes autolysosome formation in the late stages of autophagy, autophagic flux is impaired, resulting in lipid accumulation and liver fibrosis47. In contrast, enhancing autophagy by overexpressing Atg7 alleviated hepatic steatosis in ob/ob mice and in high-fat diet (HFD)-fed mice48.
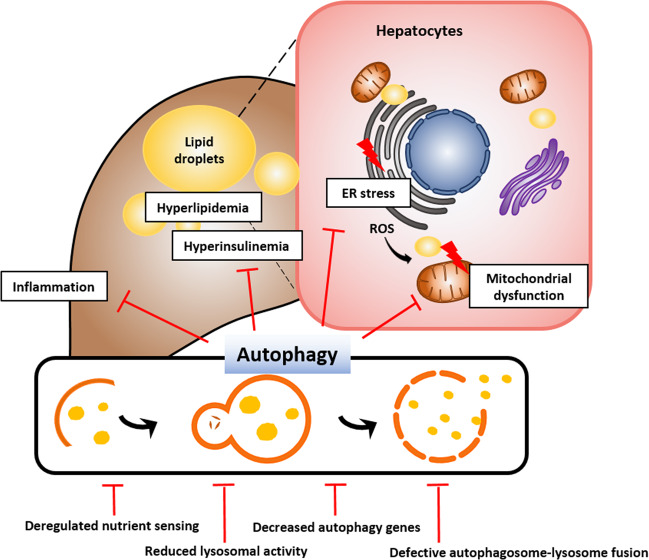
Hepatic lipotoxicity is a result of excess accumulation of harmful lipids through the dysregulation of the lipid milieu or intracellular lipid composition. Importantly, lipotoxicity is intimately associated with chronic inflammation and hepatocyte death, which are characteristics of NAFLD/NASH49. Lipotoxicity can also contribute to ER stress, mitochondrial dysfunction, and impaired autophagic flux (Fig. 2). For example, lipotoxicity-associated elevation in cytosolic calcium levels interferes with the fusion of autophagosomes and lysosomes, and treatment with calcium channel blockers restores autophagic flux and suppresses obesity-induced accumulation of protein aggregates50. Intake of a HFD stimulates ER stress, which can induce an inflammatory response. Suppressed autophagy in the liver can further worsen ER stress and inflammation. These findings indicate that a decrease in hepatic autophagy correlates with hepatic inflammation in NAFLD. In line with this finding, autophagy can protect the liver from lipotoxicity in NAFLD. Sequestosome 1 (SQSTM1 or p62)-mediated ULK1 activation induces autophagy and plays a hepatoprotective role through the degradation of Kelch-like ECH-associated protein 1and the activation of nuclear factor erythroid 2-related factor 2 under saturated fatty acid-induced lipotoxic conditions51.
Fig. 2. The role of autophagy in nonalcoholic fatty liver disease (NAFLD).
Excessive nutrients induce the enlargement of adipose tissue, which increases free fatty acids by promoting lipolysis and inflammatory adipokine secretion. This process leads to lipid accumulation and inflammatory response in the liver, resulting in endoplasmic reticulum (ER) stress and mitochondrial dysfunction. Autophagy, which can inhibit inflammation and promote the degradation of lipid droplets, has the potential as a novel therapeutic target in NAFLD.
Various studies have revealed some of the molecular mechanisms involved in the regulation of autophagy in NAFLD. Autophagy activity has been seen to decrease as an effect of the reduced expression of autophagy-related genes such as Atg7, Ulk1, and Atg948,52. Impaired autophagic flux caused by a decrease in lysosomal activity has been shown to be critical for the degradation of autophagosomes. Autophagic degradation is impaired owing to reduced lysosomal acidification and lysosomal proteolytic activity, such as those facilitated by hepatic cathepsin B and L53–55. Defects in autophagosome–lysosome fusion also cause autophagy disruption in NAFLD. Changes in intracellular lipid content can affect the overall activity of intracellular proteolytic pathways by inhibiting autophagosome–lysosome fusion56. Rubicon is a Beclin1-binding negative regulator of autophagosome–lysosome fusion. Rubicon is posttranscriptionally upregulated in HepG2 cells treated with saturated fatty acids and in the liver of HFD-fed mice54. In a recent chemokine study, osteopontin, a chemokine-like phosphorylated glycoprotein, promoted NAFLD progression by impairing autophagy57. Osteopontin secreted from fatty liver inhibits autophagosome–lysosome fusion by binding the integrins αVβ3 and αVβ5, and autophagic flux is recovered when αVβ3 and αVβ5 are blocked using specific antibodies. Another mechanism of autophagy regulation involves the deregulation of nutrient sensing. A representative example is the regulation of autophagy by mTORC1 through the downregulation of the ULK1-ATG13-FIP200 and VPS34-ATG14-BECN1 complexes. In fatty liver disease, mTORC1 is aberrantly activated and inhibits autophagy; it also contributes to the regulation of de novo lipogenesis through the upregulation of TFs such as sterol regulatory element-binding protein 158. In contrast, AMPK negatively regulates mTORC1 and induces autophagy through ULK1 or BECN1 phosphorylation59,60. Most studies show that AMPK activity is reduced in the fatty liver and that the activation of AMPK using genetic or pharmacological approaches reprograms lipid metabolism, thereby reducing hepatic steatosis61. The regulation of autophagy through AMPK also contributes to a reduction in hepatic steatosis to some extent.
MiRNAs that target autophagy-related genes in NAFLD
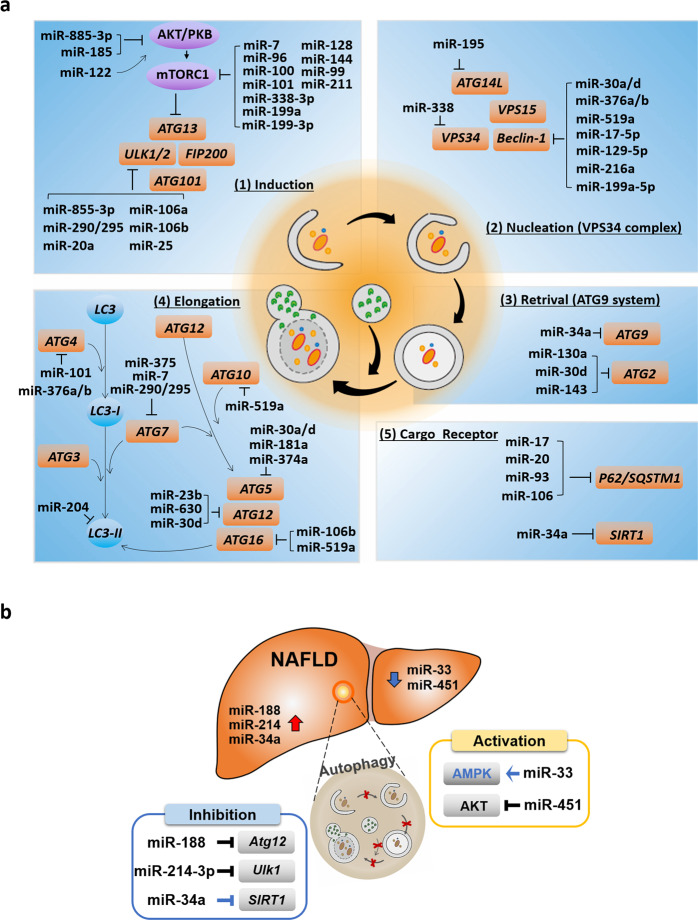
miRNAs are noncoding RNAs consisting of approximately 22 nucleotides62. They play roles in the posttranscriptional regulation of gene expression63. Approximately 60% of mammalian genes are known to be regulated by miRNAs64. Recent studies have reported that miRNAs also play roles in the regulation of autophagy; specifically, they regulate the expression of several key proteins in the various stages of the autophagy pathway. Therefore, we summarize representative miRNAs involved in the regulation of each stage of the autophagy pathway and discuss these mechanisms in the context of NAFLD (Fig. 3a).
Fig. 3. Schematic representation of core autophagy-related proteins and their regulation by miRNAs.
a Summary of miRNAs that target key proteins in different stages of autophagy. b miRNAs regulating autophagy-related genes in NAFLD. Blue indicates that the correlation between the miRNA-targeted protein and autophagy is not clearly demonstrated in NAFLD.
In the initial stages of autophagy, the ULK1/2 complex is directly controlled by mTORC1. A number of miRNAs targeting mTORC1 have been reported in various cell and tissue types. miR-7 induces autophagy through the negative regulation of mTORC165. In contrast, miR-199a, miR-338, miR-96, miR-100, miR-101, miR-128, miR-144, miR-99, and miR-211 have been suggested as potential positive regulators of autophagy through their direct inhibition of mTORC166–72. Several miRNAs have been reported to directly target the ULK1/2 complex. miR-885-3p was found to directly target ULK2, whereas the activation of miR-106a/b and miR-20a was found to inhibit autophagy through the downregulation of ULK1 expression73. In glucose-deficient melanoma cells, the miR-290/295 cluster code inhibited ULK1 expression, thereby inhibiting autophagy and endowing tumor cells with a survival advantage74. miR-25 in breast cancer cells also targets ULK1 and inhibits autophagy75. miRNAs also regulate autophagosome nucleation stage autophagy genes, such as VPS34, BECN1, and ATG14L. miR-30a/d, miR-376a/b, miR-519a, miR-17-5p, miR-129-5p, miR-199a-5p, and miR-216a inhibit BECN1 expression76–81. miR-338 inhibits the expression of VPS3482, and miR-195 inhibits ATG14-mediated autophagy83. Therefore, the suppression of these miRNAs promotes autophagy. Atg9 is the only transmembrane protein associated with the Golgi complex in the autophagy pathway, and it is targeted by miR-34a in the negative regulation of autophagy84. In addition, miR-130a, miR-30d, and miR-143 suppress ATG2 expression85–87.
The ubiquitin-like conjugation system for vesicle expansion is composed of several proteins, such as ATG7, ATG10A, ATG3, ATG12, ATG5, ATG16L1, and LC3. miR-375, miR-17, and miR-290/295 target ATG7, resulting in decreased ATG7 expression, thereby attenuating autophagy74,88,89. miR-30a, miR-181a, miR-374a, and miR-30d are direct regulators of ATG582,85,90,91. Moreover, miR-30d, miR-23b, and miR-630 are potential regulators of ATG1278,85,92. miR-106b and 519a target ATG16L1 expression in intestinal epithelial cells78,93. LC3B levels and autophagy are diminished by miR-20494,95. The miR-101 and miR376 family members miR-376a and miR-376b can negatively control ATG4 expression68,81,96. miR-519a has been shown to modulate E2-like enzyme ATG10A levels; however, the role of this interaction on autophagy has not yet been investigated78.
Several autophagy receptors have been discovered, including SQSTM1/p62, neighbor of breast cancer type 1 susceptibility protein gene 1 protein (NBR1), calcium-binding and coiled-coil domain-containing protein 2 (NDP52), optineurin (OPTN), and B-cell lymphoma 2/adenovirus E1B 19 kDa protein-interacting protein 3-like (NIX)97–101. Because these receptors are delivered to the autophagosome along with cellular cargo, autophagy receptor degradation is commonly used as a marker for autophagic degradation102. Members of a family of miRNAs that share a common seed sequence, including miR-17, miR-20, miR-93, and miR-106, were identified as direct regulators of SQSTM1/p62103,104.
Several studies have shown that changes in miRNA profiles are associated with a number of liver diseases. The expression profiles and signatures of these miRNAs can be used to distinguish between liver diseases with different etiologies and stages, from NAFLD to hepatocellular carcinoma (HCC)105. Recently, autophagy-related miRNAs were found to play important roles in NAFLD. We present a summary of the current knowledge on the regulation of autophagy by miRNAs and how miRNAs modify major autophagic effector proteins in NAFLD in Fig. 3b. Several studies have shown that autophagy regulation-related miRNAs, such as miR-214-3p, miR-188, and miR-34a, are expressed at high levels in NAFLD patients or in mouse models52,106. In our previous study, we observed that miR-214-3p has an inhibitory role in autophagy and is upregulated in HFD-induced fatty liver52. Therefore, an anti-mimic of miR-214-3p alleviated NAFLD by activating autophagy and depended on the upregulation of Ulk1. miR-188, known to target and suppress Atg12, was reported to be upregulated in the liver of obese mice107. According to Liu et al., the negative effects of miR-188 on the lipid and glucose metabolism-mediated autophagy pathway can be reversed by targeting Atg12. miR-34a is significantly upregulated in the liver of tissue with NAFLD106. miR-34a, a direct inhibitor of nicotinamide adenine dinucleotide-dependent protein deacetylase sirtuin 1 (SIRT1), is a potential biomarker of NAFLD108. Although the role of this interaction in the regulation of autophagy has not been observed, SIRT1 can have a direct role in autophagy regulation and affect the expression of autophagy-associated genes, including MAP1LC3A (LC3II), ATG5, and ATG7109,110. Stacchiotti et al. showed that therapeutic melatonin plays a role in NAFLD through the downregulation of miR-34a-5p and the activation of autophagy; however, they did not investigate the specific autophagy-related genes that were directly regulated by miR-34a111.
miRNAs can also directly or indirectly activate autophagy. miR-33 and miR-451 can activate autophagy, and these miRNAs are expressed at low levels in NAFLD112–114. The specific role of miR-33 in direct autophagy regulation in NAFLD remains unknown. However, Ghareghani et al. found that miR-33a and miR-33b activated autophagy by enhancing the activation of AMPK in HFD-fed mice112. miR-451 levels were reduced in the liver tissue of the obese mice, and miR-451 was found to be a negative regulator of migration inhibitory factor (MIF)115. Thus, Tang et al. found that MIF was upregulated in mice fed an HFD because the miR-451 level was reduced. Their results also indicated that MIF was reduced by miR-451 mimic transfection, which induced the inactivation of protein kinase B (Akt) and activation of LC3II. The same results were indicated upon MIF knockout. In contrast, increased Akt and reduced LC3II expression were induced by miR-451 silencing. In addition, Hur et al.113 showed that the downregulation of miR-451 in a NAFLD mouse model inhibited the production of fatty acid-induced proinflammatory cytokines through the AMPK/AKT pathway. Important miRNAs that can prevent or alleviate NAFLD can be modulated through autophagy activation.
TFs that target autophagy-related genes in NAFLD
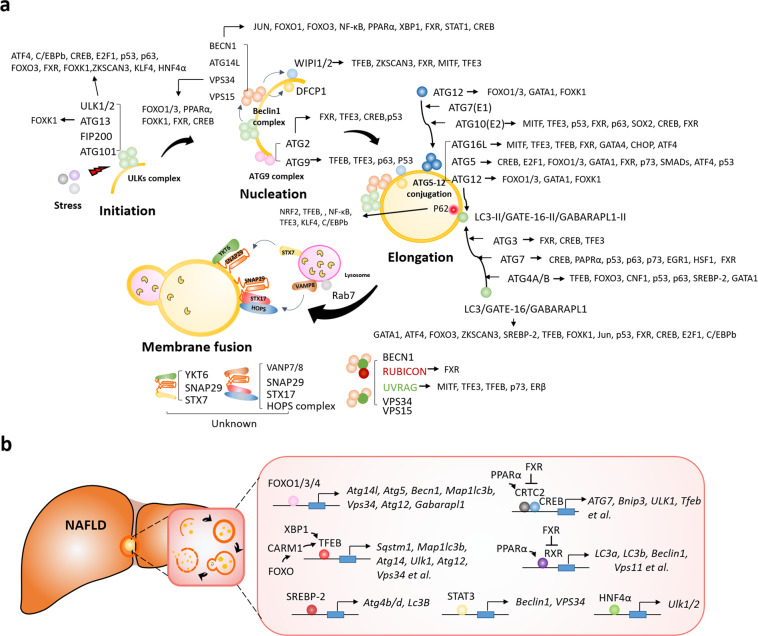
Currently, approximately 32 TFs involved in the regulation of autophagy-related genes have been reported; the most representative TFs are transcription factor EB (TFEB), cAMP response element-binding protein (CREB), forkhead box O proteins (FOXOs), farnesoid X receptor (FXR), and peroxisome proliferator-activated receptor alpha (PPARα) (Fig. 4a). We present a summary of the most representative TFs associated with autophagy-related genes in NAFLD in Fig. 4b.
Fig. 4. Transcription factors (TFs) that target autophagy-related genes in nonalcoholic fatty liver.
a Summary of TFs targeting autophagy-related genes in the different stages of autophagy progression. b Transcription factors directly involved in regulating autophagy-related genes in NAFLD.
TFEB acts as a master regulator of lysosomal biogenesis, autophagy, lysosomal exocytosis, lipid catabolism, energy metabolism, and the immune response116. TFEB is a member of the microphthalmia/transcription factor E (MIT/TFE) family of TFs, and it recognizes and binds to the E-box sequence (CANNTG)117. For efficient DNA binding, TFEB dimerizes with itself or with other MIT/TFE family members, such as IGHM Enhancer 3 (TFE3), transcription factor EC (TFEC), and microphthalmia-associated TF (MITF)118. TFEB regulates the expression of key genes in autophagy, such as BECN1, WIPI1 (which encodes WD repeat domain phosphoinositide-interacting protein 1), GABARAP, ATG16 L, ATG5, and UVRAG119. TFEB activity is regulated by phosphorylation (at S122, S142, and S211) by mTORC1; phosphorylated TFEB is sequestered into the cytoplasm, and the induction of transcription of the target gene is suppressed120,121. In contrast, when mTORC1 activity is inhibited during nutrient deficiency, TFEB is dephosphorylated and rapidly migrates to the nucleus, where it binds the promoter of the target gene. Similarly, mTORC1 also inhibits the nuclear localization of TFE3 and MITF, effectively inhibiting the transcriptional induction of autophagy genes122. In HFD-fed mice that overexpress TFEB, lipid accumulation in the liver was reduced, and metabolic syndrome was attenuated during a 24-hour starvation period123. In line with the role of TFEB in energy control, modest restriction of calorie consumption and regular exercise may be promising approaches for treating early-stage NAFLD, whereas therapeutic agents that target TFEB may be promising for preventing the development of NAFLD into NASH.
FOXO1, which has an important role in both glucose and lipid metabolism, promotes lipid droplet breakdown through transcriptional activation of hepatic autophagy by inducing a number of autophagy-related genes, including Atg5, Atg14, Vps34, Atg12, and Gabarap1124,125. Under nutrient starvation conditions, AMPK phosphorylates FOXO3 in the nucleus, and FOXO3 inhibits the transcription of S-phase kinase-associated protein 2 (SKP2), which is an SKP1, Cullins, F-box protein (SCF) E3 ubiquitin ligase complex critical for the degradation of coactivator-associated arginine methyltransferase 1 (CARM1). Stabilized CARM1 promotes histone H3 Arg17 demethylation and binds TFEB as a coactivator, thereby promoting the expression of autophagy-related genes126. In addition, spliced X-box binding protein 1 (sXBP1), the key TF that promotes the adaptive unfolded protein response, regulates autophagy-related genes by occupying the −743 to −523 site of the TFEB promoter in the liver under fasting conditions127. However, evidence showing that these mechanisms can be targeted for NAFLD therapy is still insufficient.
The fasting transcriptional activator CREB upregulates autophagy genes, including ATG7 and ULK1, by recruiting CREB-regulated transcription coactivator 2 (CRTC2) in hepatocytes. On the other hand, FXR transrepresses autophagy-related genes by disrupting the CREP-CRTC2 complex, suggesting that the FXR-CREB axis is a key physiological switch for regulating autophagy128. The CREB–CRTC2 complex upregulates the expression of TFEB, in addition to autophagy-related genes. Similar to the way CRTC2 competes with FXR for binding CREB in proximal autophagy-related genes, PPARα competes with FXR for binding the DR1 response element in the promoters of these genes, and thus, PPARα acts as a ligand-dependent transactivator129,130.
SREBP-2, a major transcriptional regulator of cholesterol metabolism, targets lipid metabolic processes, and it also directly activates autophagy-related genes during cellular sterol depletion in the liver131. Signal transducer and activator of transcription 3 (STAT3) also regulates autophagy-related genes. Interleukin (IL)-17A inhibits autophagy by activating STAT3 in hepatic fibrosis; in turn, STAT3 downregulates the expression of BECN1 and VPS34, which are involved in the development of hepatic fibrosis132. On the other hand, in methionine/choline-deficient diet-induced NASH, autophagy-related proteins such as BECN1 and SQSTM1/p62 are upregulated, but autophagic flux is impaired by hypoxia-inducible factor-1 alpha (HIF-1α), which induces liver steatosis and inflammation133. It has recently been reported that hepatocyte nuclear factor 4 alpha (HNF4α), which is essential for hepatocyte differentiation and has an important role in liver function, also has an important role in autophagy. Hnf4α expression was reduced in the fatty liver of mice fed an HFD for a prolonged period and activated autophagy by directly regulating Ulk1 expression in the liver52.
The interplay of miRNAs and TFs in autophagy regulation in NAFLD
miRNAs and TFs often play coordinating roles in the regulation of various cellular processes via a complex signal transduction network in the liver134,135. For example, in human HCC cells, miR-223 and FOXO3a modulate doxorubicin-induced cytoprotective autophagy, contributing to chemoresistance. However, miR-223 overexpression suppresses Foxo3a-modulated autophagy, which enhances doxorubicin sensitivity in a mouse xenograft model of HCC, suggesting that this miRNA/TF axis is an important mechanism for drug resistance development in HCC136,137. TFEB-mediated transactivation is also regulated by miR-30-5p, which suppresses TFEB-dependent downstream gene expression by binding to coordinated lysosomal expression and regulation element, leading to the inhibition of lysosomal biogenesis and autophagy in mouse liver138.
Accumulating evidence shows that miR-34a is involved in NAFLD, and miR-34a expression is increased in NASH patients and in obese or diabetic mice108,139,140. miR-34a promotes hepatic steatosis through the suppression of various TFs, such as HNF4α141, PPARα, and SIRT1, which promote the expression of autophagy-related genes142–144. These observations suggest that the miR-34a/TF axis may inhibit NAFLD progression through transcriptional regulation of autophagy52,109,130,145. Interestingly, miR-34a is directly activated by nuclear receptor liver X receptor-α, a ligand-dependent TFr involved in hepatic cholesterol metabolism. miR-34a also inhibits Atg4B and Rab8b, which regulate autophagic flux, leading to the progression of hepatic steatosis146,147. Considering the role of LXR in cholesterol homeostasis and that increased hepatic free cholesterol is associated with the development of NASH from NAFL in obese mice, cross-talk between TFs, miR-34a, and autophagy may be important for controlling NASH development.
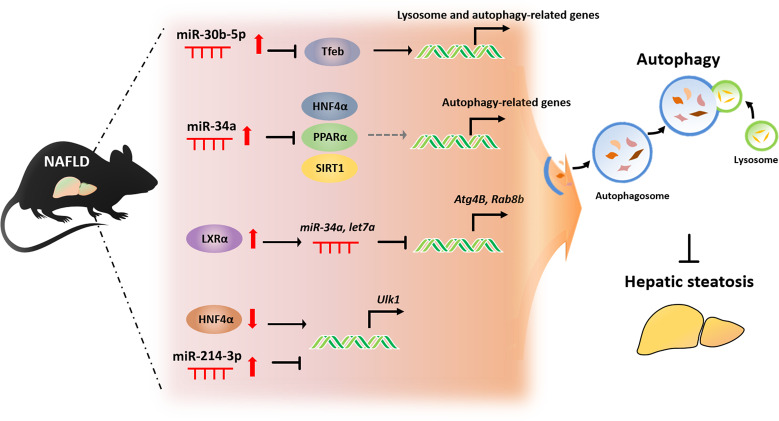
Recently, we reported certain miRNAs and TFs that regulate autophagy in the development of HFD-induced fatty liver. As shown in Fig. 5, we found that miR-214-3p and HNF4α modulated Ulk1 expression and autophagy in hepatocytes52. Our results indicate that autophagy in the fatty liver was attenuated only when mice were fed a 45% HFD for a prolonged period, which led to a significant reduction in the expression of autophagy-related genes, such as Ulk1. This downregulation of autophagy was caused by increased miR-214-3p and decreased HNF4α levels in hepatocytes. miR-214-3p negatively regulates Ulk1 expression through direct binding of the 3´-UTR sequence of Ulk1, and HNF4α induces autophagy by directly binding Ulk1, promoting its transcription. Thus, both miR-214-3p and HNF4α act as regulatory factors of Ulk1 expression. Although the inhibition of miR-214-3p in the fatty liver appears to restore HNF4α expression, miR-214-3p does not directly regulate HNF4α, suggesting that miR-214-3p and HNF4α independently regulate Ulk1 expression. The interplay between miR-214-3p and HNF4α and their involvement in the regulation of autophagy in the fatty liver are summarized in Fig. 5. Taken together, we propose that miR-214-3p and HNF4α are potential targets for NAFLD therapy.
Fig. 5. The interplay of miRNAs and transcription factors (TFs) in regulating autophagy in the fatty liver.
miR-30b-5p and miR-34a downregulate autophagy-related gene expression by directly inhibiting TFs, such as transcription factor EB (TFEB), Hnf4α, peroxisome proliferator-activated receptor alpha (PPARα), and NAD-dependent protein deacetylase sirtuin 1 (SIRT1). Liver X receptor-α (LXRα) transcriptionally activates miR-34a and let7a, which directly target the 3’UTR of Atg4B and Rab8b. miR-214-3p and HNF4α reciprocally regulate Ulk1 expression.
Concluding remarks
Studies on the molecular regulation of autophagy mainly focus on how autophagy-related proteins bind and function in the cytoplasm. However, as described in this review, there has been growing interest in understanding the roles of miRNAs and TFs that influence the autophagy pathway. Transcriptional regulation of autophagy may be associated with posttranslational regulation to coordinate the fine-tuning of autophagic flux, especially in cells under chronic stress. In addition, the transcriptional induction of autophagy-related genes may prevent the depletion of the corresponding proteins under stress conditions, as the degradation of autophagy-related proteins is enhanced under normal conditions. Thus, it is important to identify novel co-regulatory networks of miRNAs and TFs that contribute to autophagy, characterize these networks within the context of autophagy-related proteins, and determine how these networks are perturbed in NAFLD. Given the studies that demonstrate the benefits of targeting autophagy for treating NAFLD, transcriptional regulation of autophagy is expected to have similar benefits for the treatment of NAFLD. The effects of the currently available NAFLD drugs on autophagy are not yet known; however, we believe that selective induction of autophagy will be a useful therapeutic strategy for NAFLD.
Acknowledgements
This work was supported by the Main Research Program of the Korea Food Research Institute, which is funded by the Ministry of Science, ICT, & Future Planning of the Republic of Korea.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lemasters JJ. Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res. 2005;8:3–5. doi: 10.1089/rej.2005.8.3. [DOI] [PubMed] [Google Scholar]
- 2.Ogata M, et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol. Cell. Biol. 2006;26:9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scherz-Shouval R, Elazar Z. ROS, mitochondria and the regulation of autophagy. Trends Cell Biol. 2007;17:422–427. doi: 10.1016/j.tcb.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 4.Ryter SW, Cloonan SM, Choi AM. Autophagy: a critical regulator of cellular metabolism and homeostasis. Mol. Cells. 2013;36:7–16. doi: 10.1007/s10059-013-0140-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Russell RC, Yuan H-X, Guan K-L. Autophagy regulation by nutrient signaling. Cell Res. 2014;24:42–57. doi: 10.1038/cr.2013.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinez NJ, Walhout AJ. The interplay between transcription factors and microRNAs in genome-scale regulatory networks. Bioessays. 2009;31:435–445. doi: 10.1002/bies.200800212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Le TD, et al. Inferring microRNA and transcription factor regulatory networks in heterogeneous data. BMC Bioinformatics. 2013;14:92. doi: 10.1186/1471-2105-14-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang XM, Guo L, Chi MH, Sun HM, Chen XW. Identification of active miRNA and transcription factor regulatory pathways in human obesity-related inflammation. BMC Bioinformatics. 2015;16:76. doi: 10.1186/s12859-015-0512-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Füllgrabe J, Klionsky DJ, Joseph B. The return of the nucleus: transcriptional and epigenetic control of autophagy. Nat. Rev. Mol. Cell Biol. 2014;15:65–74. doi: 10.1038/nrm3716. [DOI] [PubMed] [Google Scholar]
- 10.Di Malta C, Cinque L, Settembre C. Transcriptional regulation of autophagy: mechanisms and diseases. Front. Cell Dev. Biol. 2019;7:114. doi: 10.3389/fcell.2019.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hara T, et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ganley IG, et al. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J. Biol. Chem. 2009;284:12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mercer CA, Kaliappan A, Dennis PB. A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy. Autophagy. 2009;5:649–662. doi: 10.4161/auto.5.5.8249. [DOI] [PubMed] [Google Scholar]
- 14.Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr. Opin. Cell Biol. 2010;22:132–139. doi: 10.1016/j.ceb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 15.Lazarus MB, Novotny CJ, Shokat KM. Structure of the human autophagy initiating kinase ULK1 in complex with potent inhibitors. ACS Chem. Biol. 2015;10:257–261. doi: 10.1021/cb500835z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jung CH, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimura N, et al. A possible linkage between AMP‐activated protein kinase (AMPK) and mammalian target of rapamycin (mTOR) signalling pathway. Genes Cells. 2003;8:65–79. doi: 10.1046/j.1365-2443.2003.00615.x. [DOI] [PubMed] [Google Scholar]
- 18.Kim J, Kundu M, Viollet B, Guan K-L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim J, et al. Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell. 2013;152:290–303. doi: 10.1016/j.cell.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fimia GM, et al. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–1125. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- 21.Nazio F, et al. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat. Cell Biol. 2013;15:406–416. doi: 10.1038/ncb2708. [DOI] [PubMed] [Google Scholar]
- 22.Furuya N, Yu J, Byfield M, Pattingre S, Levine B. The evolutionarily conserved domain of Beclin 1 is required for Vps34 binding, autophagy and tumor suppressor function. Autophagy. 2005;1:46–52. doi: 10.4161/auto.1.1.1542. [DOI] [PubMed] [Google Scholar]
- 23.Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol. Biol. Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan Y, Flinn RJ, Wu H, Schnur RS, Backer JM. hVps15, but not Ca2+/CaM, is required for the activity and regulation of hVps34 in mammalian cells. Biochem. J. 2009;417:747–755. doi: 10.1042/BJ20081865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Russell RC, et al. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat. Cell Biol. 2013;15:741–750. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Axe EL, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burman C, Ktistakis NT. Regulation of autophagy by phosphatidylinositol 3-phosphate. FEBS Lett. 2010;584:1302–1312. doi: 10.1016/j.febslet.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 28.Polson HE, et al. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy. 2010;6:506–522. doi: 10.4161/auto.6.4.11863. [DOI] [PubMed] [Google Scholar]
- 29.Orsi A, et al. Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Mol. Biol. Cell. 2012;23:1860–1873. doi: 10.1091/mbc.E11-09-0746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Young ARJ, et al. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J. Cell Sci. 2006;119:3888–3900. doi: 10.1242/jcs.03172. [DOI] [PubMed] [Google Scholar]
- 31.Webber JL, Tooze SA. Coordinated regulation of autophagy by p38α MAPK through mAtg9 and p38IP. EMBO J. 2010;29:27–40. doi: 10.1038/emboj.2009.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mack HI, Zheng B, Asara JM, Thomas SM. AMPK-dependent phosphorylation of ULK1 regulates ATG9 localization. Autophagy. 2012;8:1197–1214. doi: 10.4161/auto.20586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat. Rev. Mol. Cell Biol. 2001;2:211–216. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- 34.Mizushima N, et al. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J. Cell Sci. 2003;116:1679–1688. doi: 10.1242/jcs.00381. [DOI] [PubMed] [Google Scholar]
- 35.Otomo C, Metlagel Z, Takaesu G, Otomo T. Structure of the human ATG12~ ATG5 conjugate required for LC3 lipidation in autophagy. Nat. Struct. Mol. Biol. 2013;20:59. doi: 10.1038/nsmb.2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanida I, Tanida-Miyake E, Ueno T, Kominami E. The human homolog of Saccharomyces cerevisiae Apg7p is a Protein-activating enzyme for multiple substrates including human Apg12p, GATE-16, GABARAP, and MAP-LC3. J. Biol. Chem. 2001;276:1701–1706. doi: 10.1074/jbc.C000752200. [DOI] [PubMed] [Google Scholar]
- 37.Tanida I, Tanida-Miyake E, Komatsu M, Ueno T, Kominami E. Human Apg3p/Aut1p homologue is an authentic E2 enzyme for multiple substrates, GATE-16, GABARAP, and MAP-LC3, and facilitates the conjugation of hApg12p to hApg5p. J. Biol. Chem. 2002;277:13739–13744. doi: 10.1074/jbc.M200385200. [DOI] [PubMed] [Google Scholar]
- 38.Geng J, Klionsky DJ. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. EMBO Rep. 2008;9:859–864. doi: 10.1038/embor.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fujita N, et al. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol. Biol. Cell. 2008;19:2092–2100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jäger S, et al. Role for Rab7 in maturation of late autophagic vacuoles. J. Cell Sci. 2004;117:4837–4848. doi: 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- 41.Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012;151:1256–1269. doi: 10.1016/j.cell.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 42.Jiang P, et al. The HOPS complex mediates autophagosome-lysosome fusion through interaction with syntaxin 17. Mol. Biol. Cell. 2014;25:1327–1337. doi: 10.1091/mbc.E13-08-0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liang C, et al. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat. Cell Biol. 2008;10:776–787. doi: 10.1038/ncb1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu R, Zhi X, Zhong Q. ATG14 controls SNARE-mediated autophagosome fusion with a lysosome. Autophagy. 2015;11:847–849. doi: 10.1080/15548627.2015.1037549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chalasani N, et al. The diagnosis and management of non-alcoholic fatty liver disease: practice Guideline by the American Association for the Study of Liver Diseases, American College of Gastroenterology, and the American Gastroenterological Association. Hepatology. 2012;55:2005–2023. doi: 10.1002/hep.25762. [DOI] [PubMed] [Google Scholar]
- 46.Singh R, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheng X, Sun Q. RUBCNL/Pacer and RUBCN/Rubicon in regulation of autolysosome formation and lipid metabolism. Autophagy. 2019;15:1120–1121. doi: 10.1080/15548627.2019.1596500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang L, Li P, Fu S, Calay ES, Hotamisligil GS. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 2010;11:467–478. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Masarone M, et al. Role of oxidative stress in pathophysiology of nonalcoholic fatty liver disease. Oxid. Med. Cell. Longev. 2018;2018:9547613. doi: 10.1155/2018/9547613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park HW, et al. Pharmacological correction of obesity-induced autophagy arrest using calcium channel blockers. Nat. Commun. 2014;5:4834. doi: 10.1038/ncomms5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee DH, et al. SQSTM1/p62 activates NFE2L2/NRF2 via ULK1-mediated autophagic KEAP1 degradation and protects mouse liver from lipotoxicity. Autophagy. 2020;16:1949–1973. doi: 10.1080/15548627.2020.1712108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee D. H. et al. Mir214-3p and Hnf4a/Hnf4α reciprocally regulate Ulk1 expression and autophagy in nonalcoholic hepatic steatosis. Autophagy10.1080/15548627.2020.1827779, 1–17 (2020). [DOI] [PMC free article] [PubMed]
- 53.Fukuo Y, et al. Abnormality of autophagic function and cathepsin expression in the liver from patients with non-alcoholic fatty liver disease. Hepatol. Res. 2014;44:1026–1036. doi: 10.1111/hepr.12282. [DOI] [PubMed] [Google Scholar]
- 54.Tanaka S, et al. Rubicon inhibits autophagy and accelerates hepatocyte apoptosis and lipid accumulation in nonalcoholic fatty liver disease in mice. Hepatology. 2016;64:1994–2014. doi: 10.1002/hep.28820. [DOI] [PubMed] [Google Scholar]
- 55.Inami Y, et al. Hepatic steatosis inhibits autophagic proteolysis via impairment of autophagosomal acidification and cathepsin expression. Biochem. Biophys. Res. Commun. 2011;412:618–625. doi: 10.1016/j.bbrc.2011.08.012. [DOI] [PubMed] [Google Scholar]
- 56.Koga H, Kaushik S, Cuervo AM. Altered lipid content inhibits autophagic vesicular fusion. FASEB J. 2010;24:3052–3065. doi: 10.1096/fj.09-144519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tang M, et al. Osteopontin acts as a negative regulator of autophagy accelerating lipid accumulation during the development of nonalcoholic fatty liver disease. Artif. Cells Nanomed. Biotechnol. 2020;48:159–168. doi: 10.1080/21691401.2019.1699822. [DOI] [PubMed] [Google Scholar]
- 58.Han J, Wang Y. mTORC1 signaling in hepatic lipid metabolism. Protein Cell. 2018;9:145–151. doi: 10.1007/s13238-017-0409-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hoegg MB, Robbins SM, McGhee JD. Characterization of the C. elegans erlin homologue. BMC Cell Biol. 2012;13:2. doi: 10.1186/1471-2121-13-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang D, et al. AMPK regulates autophagy by phosphorylating BECN1 at threonine 388. Autophagy. 2016;12:1447–1459. doi: 10.1080/15548627.2016.1185576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Garcia D, et al. Genetic liver-specific AMPK activation protects against diet-induced obesity and NAFLD. Cell Rep. 2019;26:192–208. doi: 10.1016/j.celrep.2018.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 63.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 64.Chiang HR, et al. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev. 2010;24:992–1009. doi: 10.1101/gad.1884710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang Y, Liu J, Liu C, Naji A, Stoffers DA. MicroRNA-7 regulates the mTOR pathway and proliferation in adult pancreatic beta-cells. Diabetes. 2013;62:887–895. doi: 10.2337/db12-0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ozturk DG, et al. MITF-MIR211 axis is a novel autophagy amplifier system during cellular stress. Autophagy. 2019;15:375–390. doi: 10.1080/15548627.2018.1531197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang Z, Yuan B, Fu F, Huang S, Yang Z. Hemoglobin enhances miRNA-144 expression and autophagic activation mediated inflammation of microglia via mTOR pathway. Sci. Rep. 2017;7:11861. doi: 10.1038/s41598-017-12067-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Frankel LB, et al. microRNA-101 is a potent inhibitor of autophagy. EMBO J. 2011;30:4628–4641. doi: 10.1038/emboj.2011.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ge YY, et al. MicroRNA-100 promotes the autophagy of hepatocellular carcinoma cells by inhibiting the expression of mTOR and IGF-1R. Oncotarget. 2014;5:6218–6228. doi: 10.18632/oncotarget.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu P, et al. Inhibition of MicroRNA-96 ameliorates cognitive impairment and inactivation autophagy following chronic cerebral hypoperfusion in the rat. Cell. Physiol. Biochem. 2018;49:78–86. doi: 10.1159/000492844. [DOI] [PubMed] [Google Scholar]
- 71.Han J, et al. miR-338-3p confers 5-fluorouracil resistance in p53 mutant colon cancer cells by targeting the mammalian target of rapamycin. Exp. Cell Res. 2017;360:328–336. doi: 10.1016/j.yexcr.2017.09.023. [DOI] [PubMed] [Google Scholar]
- 72.Fornari F, et al. MiR-199a-3p regulates mTOR and c-Met to influence the doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res. 2010;70:5184–5193. doi: 10.1158/0008-5472.CAN-10-0145. [DOI] [PubMed] [Google Scholar]
- 73.Wu H, et al. MiR-20a and miR-106b negatively regulate autophagy induced by leucine deprivation via suppression of ULK1 expression in C2C12 myoblasts. Cell. Signal. 2012;24:2179–2186. doi: 10.1016/j.cellsig.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 74.Chen Y, Liersch R, Detmar M. The miR-290-295 cluster suppresses autophagic cell death of melanoma cells. Sci. Rep. 2012;2:808. doi: 10.1038/srep00808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang Z, et al. MicroRNA-25 regulates chemoresistance-associated autophagy in breast cancer cells, a process modulated by the natural autophagy inducer isoliquiritigenin. Oncotarget. 2014;5:7013–7026. doi: 10.18632/oncotarget.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang XL, Shi HJ, Lin SQ, Ba MC, Cui SZ. MicroRNA-216a enhances the radiosensitivity of pancreatic cancer cells by inhibiting beclin-1-mediated autophagy. Oncol. Rep. 2015;34:1557–1564. doi: 10.3892/or.2015.4078. [DOI] [PubMed] [Google Scholar]
- 77.Zhao K, et al. Methylation of microRNA-129-5P modulates nucleus pulposus cell autophagy by targeting Beclin-1 in intervertebral disc degeneration. Oncotarget. 2017;8:86264–86276. doi: 10.18632/oncotarget.21137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huang Y, Guerrero-Preston R, Ratovitski EA. Phospho-DeltaNp63alpha-dependent regulation of autophagic signaling through transcription and micro-RNA modulation. Cell Cycle. 2012;11:1247–1259. doi: 10.4161/cc.11.6.19670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chatterjee A, Chattopadhyay D, Chakrabarti G. miR-17-5p downregulation contributes to paclitaxel resistance of lung cancer cells through altering beclin1 expression. PLoS ONE. 2014;9:e95716. doi: 10.1371/journal.pone.0095716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yu Y, et al. Targeting microRNA-30a-mediated autophagy enhances imatinib activity against human chronic myeloid leukemia cells. Leukemia. 2012;26:1752–1760. doi: 10.1038/leu.2012.65. [DOI] [PubMed] [Google Scholar]
- 81.Korkmaz G, le Sage C, Tekirdag KA, Agami R, Gozuacik D. miR-376b controls starvation and mTOR inhibition-related autophagy by targeting ATG4C and BECN1. Autophagy. 2012;8:165–176. doi: 10.4161/auto.8.2.18351. [DOI] [PubMed] [Google Scholar]
- 82.Su Z, Yang Z, Xu Y, Chen Y, Yu Q. MicroRNAs in apoptosis, autophagy and necroptosis. Oncotarget. 2015;6:8474–8490. doi: 10.18632/oncotarget.3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shi G, et al. Increased miR-195 aggravates neuropathic pain by inhibiting autophagy following peripheral nerve injury. Glia. 2013;61:504–512. doi: 10.1002/glia.22451. [DOI] [PubMed] [Google Scholar]
- 84.Yang J, et al. MiR-34 modulates Caenorhabditis elegans lifespan via repressing the autophagy gene atg9. Age (Dordr.) 2013;35:11–22. doi: 10.1007/s11357-011-9324-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang X, et al. mir-30d Regulates multiple genes in the autophagy pathway and impairs autophagy process in human cancer cells. Biochem. Biophys. Res. Commun. 2013;431:617–622. doi: 10.1016/j.bbrc.2012.12.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lin XT, et al. MicroRNA-143 targets ATG2B to inhibit autophagy and increase inflammatory responses in Crohn’s disease. Inflamm. Bowel Dis. 2018;24:781–791. doi: 10.1093/ibd/izx075. [DOI] [PubMed] [Google Scholar]
- 87.Kovaleva V, et al. miRNA-130a targets ATG2B and DICER1 to inhibit autophagy and trigger killing of chronic lymphocytic leukemia cells. Cancer Res. 2012;72:1763–1772. doi: 10.1158/0008-5472.CAN-11-3671. [DOI] [PubMed] [Google Scholar]
- 88.Chang Y, et al. miR-375 inhibits autophagy and reduces viability of hepatocellular carcinoma cells under hypoxic conditions. Gastroenterology. 2012;143:177–187. doi: 10.1053/j.gastro.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 89.Comincini S, et al. microRNA-17 regulates the expression of ATG7 and modulates the autophagy process, improving the sensitivity to temozolomide and low-dose ionizing radiation treatments in human glioblastoma cells. Cancer Biol. Ther. 2013;14:574–586. doi: 10.4161/cbt.24597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fu XT, et al. MicroRNA-30a suppresses autophagy-mediated anoikis resistance and metastasis in hepatocellular carcinoma. Cancer Lett. 2018;412:108–117. doi: 10.1016/j.canlet.2017.10.012. [DOI] [PubMed] [Google Scholar]
- 91.Tekirdag KA, Korkmaz G, Ozturk DG, Agami R, Gozuacik D. MIR181A regulates starvation- and rapamycin-induced autophagy through targeting of ATG5. Autophagy. 2013;9:374–385. doi: 10.4161/auto.23117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang P, et al. MicroRNA 23b regulates autophagy associated with radioresistance of pancreatic cancer cells. Gastroenterology. 2013;145:1133–1143. doi: 10.1053/j.gastro.2013.07.048. [DOI] [PubMed] [Google Scholar]
- 93.Zhai Z, Wu F, Chuang AY, Kwon JH. miR-106b fine tunes ATG16L1 expression and autophagic activity in intestinal epithelial HCT116 cells. Inflamm. Bowel Dis. 2013;19:2295–2301. doi: 10.1097/MIB.0b013e31829e71cf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xiao J, et al. MiR-204 regulates cardiomyocyte autophagy induced by ischemia-reperfusion through LC3-II. J. Biomed. Sci. 2011;18:35. doi: 10.1186/1423-0127-18-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mikhaylova O, et al. VHL-regulated MiR-204 suppresses tumor growth through inhibition of LC3B-mediated autophagy in renal clear cell carcinoma. Cancer Cell. 2012;21:532–546. doi: 10.1016/j.ccr.2012.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Korkmaz G. et al. MIR376A is a regulator of starvation-induced autophagy. PLoS ONE8, e82556 (2013). [DOI] [PMC free article] [PubMed]
- 97.Korac J, et al. Ubiquitin-independent function of optineurin in autophagic clearance of protein aggregates. J. Cell Sci. 2013;126:580–592. doi: 10.1242/jcs.114926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang J, Ney PA. Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ. 2009;16:939–946. doi: 10.1038/cdd.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.von Muhlinen N, et al. LC3C, bound selectively by a noncanonical LIR motif in NDP52, is required for antibacterial autophagy. Mol. Cell. 2012;48:329–342. doi: 10.1016/j.molcel.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lamark T, Kirkin V, Dikic I, Johansen T. NBR1 and p62 as cargo receptors for selective autophagy of ubiquitinated targets. Cell Cycle. 2009;8:1986–1990. doi: 10.4161/cc.8.13.8892. [DOI] [PubMed] [Google Scholar]
- 101.Bjorkoy G, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bjorkoy G, et al. Monitoring autophagic degradation of p62/SQSTM1. Methods Enzymol. 2009;452:181–197. doi: 10.1016/S0076-6879(08)03612-4. [DOI] [PubMed] [Google Scholar]
- 103.Meenhuis A, et al. MiR-17/20/93/106 promote hematopoietic cell expansion by targeting sequestosome 1-regulated pathways in mice. Blood. 2011;118:916–925. doi: 10.1182/blood-2011-02-336487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li H, et al. MicroRNA-17-5p contributes to osteoarthritis progression by binding p62/SQSTM1. Exp. Ther. Med. 2018;15:1789–1794. doi: 10.3892/etm.2017.5622. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 105.Kim KM, Kim SG. Autophagy and microRNA dysregulation in liver diseases. Arch. Pharm. Res. 2014;37:1097–1116. doi: 10.1007/s12272-014-0439-9. [DOI] [PubMed] [Google Scholar]
- 106.Xu Y, et al. A metabolic stress-inducible miR-34a-HNF4alpha pathway regulates lipid and lipoprotein metabolism. Nat. Commun. 2015;6:7466. doi: 10.1038/ncomms8466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liu Y, et al. miR-188 promotes liver steatosis and insulin resistance via the autophagy pathway. J. Endocrinol. 2020;245:411–423. doi: 10.1530/JOE-20-0033. [DOI] [PubMed] [Google Scholar]
- 108.Cermelli S, Ruggieri A, Marrero JA, Ioannou GN, Beretta L. Circulating microRNAs in patients with chronic hepatitis C and non-alcoholic fatty liver disease. PLoS ONE. 2011;6:e23937. doi: 10.1371/journal.pone.0023937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lee IH, et al. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc. Natl Acad. Sci. USA. 2008;105:3374–3379. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang Y., Zheng Z. J., Jia Y. J., Yang Y. L. & Xue Y. M. Role of p53/miR-155-5p/sirt1 loop in renal tubular injury of diabetic kidney disease. J. Transl. Med.16, 146 (2018). [DOI] [PMC free article] [PubMed]
- 111.Stacchiotti A. et al. Melatonin effects on non-alcoholic fatty liver disease are related to microRNA-34a-5p/Sirt1 axis and autophagy. Cells. 8, 1053 (2019). [DOI] [PMC free article] [PubMed]
- 112.Ghareghani P, et al. Aerobic endurance training improves nonalcoholic fatty liver disease (NAFLD) features via miR-33 dependent autophagy induction in high fat diet fed mice. Obes. Res. Clin. Pract. 2018;12:80–89. doi: 10.1016/j.orcp.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 113.Hur W, et al. Downregulation of microRNA-451 in non-alcoholic steatohepatitis inhibits fatty acid-induced proinflammatory cytokine production through the AMPK/AKT pathway. Int. J. Biochem. Cell Biol. 2015;64:265–276. doi: 10.1016/j.biocel.2015.04.016. [DOI] [PubMed] [Google Scholar]
- 114.Zeng N, et al. MiR-451a attenuates free fatty acids-mediated hepatocyte steatosis by targeting the thyroid hormone responsive spot 14 gene. Mol. Cell Endocrinol. 2018;474:260–271. doi: 10.1016/j.mce.2018.03.016. [DOI] [PubMed] [Google Scholar]
- 115.Tang H, et al. Swimming prevents nonalcoholic fatty liver disease by reducing migration inhibitory factor through Akt suppression and autophagy activation. Am. J. Transl. Res. 2019;11:4315–4325. [PMC free article] [PubMed] [Google Scholar]
- 116.Napolitano G, Ballabio A. TFEB at a glance. J. Cell Sci. 2016;129:2475–2481. doi: 10.1242/jcs.146365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.LeBlanc JF, McLane KE, Parren PW, Burton DR, Ghazal P. Recognition properties of a sequence-specific DNA binding antibody. Biochemistry. 1998;37:6015–6022. doi: 10.1021/bi9730792. [DOI] [PubMed] [Google Scholar]
- 118.Brady OA, Martina JA, Puertollano R. Emerging roles for TFEB in the immune response and inflammation. Autophagy. 2018;14:181–189. doi: 10.1080/15548627.2017.1313943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Füllgrabe J, Ghislat G, Cho DH, Rubinsztein DC. Transcriptional regulation of mammalian autophagy at a glance. J. Cell Sci. 2016;129:3059–3066. doi: 10.1242/jcs.188920. [DOI] [PubMed] [Google Scholar]
- 120.Martina JA, Chen Y, Gucek M, Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 2012;8:903–914. doi: 10.4161/auto.19653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Puertollano R., Ferguson S. M., Brugarolas J. & Ballabio A. The complex relationship between TFEB transcription factor phosphorylation and subcellular localization. EMBO J.37, e98804 (2018). [DOI] [PMC free article] [PubMed]
- 122.Perera R. M., Di Malta C. & Ballabio A. MiT/TFE family of transcription factors, lysosomes, and cancer. Annu. Rev. Cancer Biol. 3, 203–222 (2019). [DOI] [PMC free article] [PubMed]
- 123.Settembre C, et al. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat. Cell Biol. 2013;15:647–658. doi: 10.1038/ncb2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Xiong X, Tao R, DePinho RA, Dong XC. The autophagy-related gene 14 (Atg14) is regulated by forkhead box O transcription factors and circadian rhythms and plays a critical role in hepatic autophagy and lipid metabolism. J. Biol. Chem. 2012;287:39107–39114. doi: 10.1074/jbc.M112.412569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Liu HY, et al. Hepatic autophagy is suppressed in the presence of insulin resistance and hyperinsulinemia: inhibition of FoxO1-dependent expression of key autophagy genes by insulin. J. Biol. Chem. 2009;284:31484–31492. doi: 10.1074/jbc.M109.033936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Shin HJ, et al. AMPK-SKP2-CARM1 signalling cascade in transcriptional regulation of autophagy. Nature. 2016;534:553–557. doi: 10.1038/nature18014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhang Z. et al. The unfolded protein response regulates hepatic autophagy by sXBP1-mediated activation of TFEB. Autophagy10.1080/15548627.2020.1788889, 1–15 (2020). [DOI] [PMC free article] [PubMed]
- 128.Seok S, et al. Transcriptional regulation of autophagy by an FXR-CREB axis. Nature. 2014;516:108–111. doi: 10.1038/nature13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lee JM. Transcriptional coordination of hepatic autophagy by nutrient-sensing nuclear receptor PPARα and FXR. Ann. Pediatr. Endocrinol. Metab. 2016;21:193–198. doi: 10.6065/apem.2016.21.4.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lee JM, et al. Nutrient-sensing nuclear receptors coordinate autophagy. Nature. 2014;516:112–115. doi: 10.1038/nature13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Seo YK, et al. Genome-wide localization of SREBP-2 in hepatic chromatin predicts a role in autophagy. Cell Metab. 2011;13:367–375. doi: 10.1016/j.cmet.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhang XW, et al. Antagonism of Interleukin-17A ameliorates experimental hepatic fibrosis by restoring the IL-10/STAT3-suppressed autophagy in hepatocytes. Oncotarget. 2017;8:9922–9934. doi: 10.18632/oncotarget.14266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wang X, et al. Macrophage-specific hypoxia-inducible factor-1α contributes to impaired autophagic flux in nonalcoholic steatohepatitis. Hepatology. 2019;69:545–563. doi: 10.1002/hep.30215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Shalgi R, Lieber D, Oren M, Pilpel Y. Global and local architecture of the mammalian microRNA-transcription factor regulatory network. PLoS Comput. Biol. 2007;3:e131. doi: 10.1371/journal.pcbi.0030131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhang HM, et al. Transcription factor and microRNA co-regulatory loops: important regulatory motifs in biological processes and diseases. Brief. Bioinformatics. 2015;16:45–58. doi: 10.1093/bib/bbt085. [DOI] [PubMed] [Google Scholar]
- 136.Zhou Y, et al. miR-223 overexpression inhibits doxorubicin-induced autophagy by targeting FOXO3a and reverses chemoresistance in hepatocellular carcinoma cells. Cell Death Dis. 2019;10:843. doi: 10.1038/s41419-019-2053-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Long C, Cen S, Zhong Z, Zhou C, Zhong G. FOXO3 is targeted by miR-223-3p and promotes osteogenic differentiation of bone marrow mesenchymal stem cells by enhancing autophagy. Hum. Cell. 2021;34:14–27. doi: 10.1007/s13577-020-00421-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Guo H. et al. Nuclear miR-30b-5p suppresses TFEB-mediated lysosomal biogenesis and autophagy. Cell Death Differ.10.1038/s41418-020-0602-4 (2020). [DOI] [PMC free article] [PubMed]
- 139.Pogribny IP, et al. Difference in expression of hepatic microRNAs miR-29c, miR-34a, miR-155, and miR-200b is associated with strain-specific susceptibility to dietary nonalcoholic steatohepatitis in mice. Lab Invest. 2010;90:1437–1446. doi: 10.1038/labinvest.2010.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Li S, et al. Differential expression of microRNAs in mouse liver under aberrant energy metabolic status. J. Lipid Res. 2009;50:1756–1765. doi: 10.1194/jlr.M800509-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Xu Y, et al. A metabolic stress-inducible miR-34a-HNF4α pathway regulates lipid and lipoprotein metabolism. Nat. Commun. 2015;6:7466. doi: 10.1038/ncomms8466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ding J, et al. Effect of miR-34a in regulating steatosis by targeting PPARα expression in nonalcoholic fatty liver disease. Sci. Rep. 2015;5:13729. doi: 10.1038/srep13729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Castro RE, et al. miR-34a/SIRT1/p53 is suppressed by ursodeoxycholic acid in the rat liver and activated by disease severity in human non-alcoholic fatty liver disease. J. Hepatol. 2013;58:119–125. doi: 10.1016/j.jhep.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 144.Jia N, et al. Amelioration of hepatic steatosis is associated with modulation of gut microbiota and suppression of hepatic miR-34a in Gynostemma pentaphylla (Thunb.) Makino treated mice. Nutr. Metab. (Lond.) 2018;15:86. doi: 10.1186/s12986-018-0323-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ou X, Lee MR, Huang X, Messina-Graham S, Broxmeyer HE. SIRT1 positively regulates autophagy and mitochondria function in embryonic stem cells under oxidative stress. Stem Cells. 2014;32:1183–1194. doi: 10.1002/stem.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kim YS, et al. LXRα activation inhibits autophagy and lipophagy in hepatocytes by dysregulating ATG4B and Rab-8B, reducing mitochondrial fuel oxidation. Hepatology. 2020;73:1307–1326. doi: 10.1002/hep.31423. [DOI] [PubMed] [Google Scholar]
- 147.Miller AM, et al. MiR-155 has a protective role in the development of non-alcoholic hepatosteatosis in mice. PLoS ONE. 2013;8:e72324. doi: 10.1371/journal.pone.0072324. [DOI] [PMC free article] [PubMed] [Google Scholar]