Abstract
ACE2 and TMPRSS2 are key players on SARS-CoV-2 entry into host cells. However, it is still unclear whether expression levels of these factors could reflect disease severity. Here, a case–control study was conducted with 213 SARS-CoV-2 positive individuals where cases were defined as COVID-19 patients with respiratory distress requiring oxygen support (N = 38) and controls were those with mild to moderate symptoms of the disease who did not need oxygen therapy along the entire clinical course (N = 175). ACE2 and TMPRSS2 mRNA levels were evaluated in nasopharyngeal swab samples by RT-qPCR and logistic regression analyzes were applied to estimate associations with respiratory outcomes. ACE2 and TMPRSS2 levels positively correlated with age, which was also strongly associated with respiratory distress. Increased nasopharyngeal ACE2 levels showed a protective effect against this outcome (adjOR = 0.30; 95% CI 0.09–0.91), while TMPRSS2/ACE2 ratio was associated with risk (adjOR = 4.28; 95% CI 1.36–13.48). On stepwise regression, TMPRSS2/ACE2 ratio outperformed ACE2 to model COVID-19 severity. When nasopharyngeal swabs were compared to bronchoalveolar lavages in an independent cohort of COVID-19 patients under mechanical ventilation, similar expression levels of these genes were observed. These data suggest nasopharyngeal TMPRSS2/ACE2 as a promising candidate for further prediction models on COVID-19.
Subject terms: Virology, SARS-CoV-2, Viral infection, Gene expression, Infectious diseases, Respiratory tract diseases, Epidemiology, Transcription
Introduction
In late December 2019, SARS-CoV-2 has emerged as the etiologic agent of a novel human respiratory disease known as coronavirus disease 2019 (COVID-19)1. Most COVID-19 patients show mild to moderate symptoms of common respiratory viral infections such as dry cough, headache, and fever. However, some infected individuals may develop intense respiratory distress, pulmonary infiltrates, and secondary bacterial pneumonia2. This scenario can evolve to an even more critical condition of respiratory failure, septic shock and multi-organ disfunction2. Risk factors such as age, sex (male), hypertension, obesity, diabetes and smoking habit have been identified by their association with hospitalization or death due to COVID-19 on previous epidemiological studies3–5.
The angiotensin-converting enzyme 2 (ACE2) has been described as the entry receptor for SARS-CoV-2 and the transmembrane serine protease 2 (TMPRSS2) as an important priming enzyme required during this process6. Co-expression of ACE2 and TMPRSS2 genes has been detected by single-cell RNA-sequencing analyses on goblet secretory cells (nasal mucosa), type-2 pneumocytes (lungs), and absorptive enterocytes (small intestine), characterizing potential initial target sites for SARS-CoV-2 replication in humans7. However, data from ACE2 and TMPRSS2 expression on different airway tract sites of SARS-CoV-2 infected individuals is still sparse and have been raising divergent hypothesis about their potential impact on COVID-19 susceptibility.
Recent data found that expression levels of ACE2 were lower in the nasal epithelium of children under 10 years old, when compared to higher age groups, suggesting that age related ACE2 expression could help to explain lower susceptibility to SARS-CoV-2 among young people8. Moreover, evidence support that higher ACE2 expression levels could be expected on individuals with hypertension and upon angiotensin receptor blocker therapy9,10. Higher levels of ACE2 have also been found on the respiratory tract of smokers, supporting a risk effect hypothesis for greater ACE2 expression during SARS-CoV-2 infection11. On the other hand, protective effect against acute lung injury/inflammation has already been described for ACE2 and the angiotensin II type 2 receptor (AT2) in mice12. ACE2 deficiency has also been linked to exacerbation of adipose tissue inflammation upon high calorie diet induced obesity in mice, reinforcing its potential anti-inflammatory effects13.
Thus, the role of ACE2 expression levels on COVID-19 susceptibility still remains to be determined. Despite the relevance of TMPRSS2 on SARS-CoV-2 entry, poor data is available to understand how its expression is affected by different factors and whether it could be linked to SARS-CoV-2 severity. To further address these questions, here we describe the expression levels of ACE2 and TMPRSS2 at the upper respiratory tract from SARS-CoV-2 positive individuals and investigate their association with respiratory distress.
Methods
Subjects and study design
A case–control study was carried out including 213 individuals with SARS-CoV-2 infection confirmed by RT-qPCR in nasopharyngeal samples. Patients were enrolled at the Center for COVID-19 diagnosis from the Federal University of Rio de Janeiro (UFRJ) and at Hospital Naval Marcílio Dias (HNMD). The case group was enrolled at HNMD and included 38 individuals that showed intense respiratory distress defined here by their need of oxygen therapy after clinical evaluation (22 received oxygen by nasal cannula, 7 by noninvasive mechanical ventilation and 9 by intubation). The control group included 175 individuals that attended to UFRJ for COVID-19 diagnosis and presented only mild symptoms of COVID-19 along the entire disease course. Frequencies of males and females were paired between cases and controls to avoid confounding.
Clinical information from COVID-19 patients undergoing oxygen therapy (cases) was assessed from medical records and nasopharyngeal swab samples were collected at the time of hospital admission. For mild COVID-19 individuals (controls), information was obtained through a questionnaire application by qualified medical specialists. Exclusion criteria consisted of cancer, autoimmune diseases and/or pregnancy. No smokers were observed among enrolled participants. The present study was performed in accordance with relevant guidelines and regulations and approved by institutional ethical review boards from UFRJ and HNMD (protocol numbers 30161620.0.0000.5257 and 32382820.3.0000.5256 respectively), with written informed consents obtained from all participants.
Nucleic acid extraction and cDNA synthesis
Total nucleic acid extractions from nasopharyngeal swab samples were performed using the automated Maxwell System platform (Promega). 25 μl of total RNA was then submitted to cDNA synthesis with High-Capacity cDNA Reverse Transcription Kits (Thermo Fisher Scientific) and stored under − 20 °C.
Quantitative PCR
Expression levels of ACE2, TMPRSS2, and B2M (as a reference gene) were evaluated by quantitative PCR (qPCR) using GoTaq Probe qPCR System (Promega). The specific set of primers and probes (Integrated DNA Technologies) predesigned for exon-exon junctions were used: Hs.PT.58.27645939, Hs.PT.58.39738666 and, Hs.PT.39a.22214847. Reactions were performed in a final volume of 10 µL on a 7500 Real-Time PCR System (Thermo Fisher Scientific) following manufacturer’s recommendations. Genomic DNA amplification was not observed at no-reverse transcriptase controls. For relative quantification of expression levels, the fluorescence accumulation data of real-time RT-PCR reactions of each sample were used for fitting four parameters sigmoid curves (cycle × Rn) to represent each amplification curve using the library qPCR for the R statistical package version 3.0.1. The cycle of quantification (Cq) was determined for each amplification by the maximum of the second derivative of the fitted sigmoid curve. The efficiency of each amplification reaction was calculated as the ratio between the fluorescence (Rn) of the cycle of quantification (Cq) and the fluorescence of the cycle immediately preceding that (Cq-1). For each gene, efficiency was estimated by the mean of all the efficiencies for each amplification reaction for that gene. Once we had the average efficiency of the target (ET) and housekeeping (EH) genes with the quantification cycles (CqT and CqH) for all amplification reactions, we used the following equation to calculate the normalized expression values (Rq) of each target gene for each sample: Rq = ETΔCqT/EHΔCqH14. Relative expression results were log-transformed and represented in graphs as mean ± standard error.
Expression patterns in bronchoalveolar lavage
In addition to the main epidemiological study, an independent case-only cohort of 45 COVID-19 patients under mechanical ventilation was recruited to investigate whether ACE2 and TMPRSS2 expression patterns in lower respiratory tract could reflect those observed in the nasopharynx during severe/critical COVID-19. From the 45 samples, 11 were bronchoalveolar lavages (BAL) and 34 were swabs. They were all obtained from distinct individuals of two reference hospitals at the city of Belo Horizonte, also located at Brazilian Southeast region. Nasopharyngeal swabs samples were taken as baseline for comparisons by non-paired statistical analysis. All participants declared written informed consent approved by the institutional ethical review board (CAAE 32224420.3.0000.0008 and CAAE 31462820.3.0000.5149).
Statistical analyses
In the evaluation of the sociodemographic, clinical, and laboratory features between cases and controls, Wilcoxon rank-sum test were used for continuous numerical variables to test the hypothesis that different groups were drawn from the same distribution or distributions with the same median. Likewise, for categorical nominal variables, Fisher’s exact tests were used to evaluate frequencies among the different groups and test the hypothesis of independence. Kendall’s rank correlation coefficient analyses were estimated for continuous numerical variables. Correlation plots were generated with “ggstatsplot” and “grid” packages. Comparisons of ACE2 and TMPRSS2 expression levels between the different groups of interest were performed by contrasts obtained after both bi- and multivariate-linear models fitted by ordinary least square regressions. Normality of continuous data was assessed by Shapiro–Wilk test. A stepwise analysis was conducted to select covariates for multiple logistic regression models using the packages “tidyverse”, “caret”, “leaps” and, “MASS”15,16. Association between expression levels and respiratory distress requiring oxygen therapy was performed by crude and adjusted logistic regression models. This model was applied to investigate a role for ACE2 and TMPRSS2 expression levels as predictors of respiratory distress (binary outcome). All statistical analyses were performed on R (version 4.0.2).
Results
Clinical characteristics of the study population
To investigate whether nasopharyngeal levels of ACE2 and TMPRSS2 are associated with respiratory distress susceptibility on COVID-19 patients, 213 SARS-CoV-2 RT-qPCR positive individuals were enrolled in this case–control study. Cases were defined as COVID-19 patients who required oxygen support over the course of SARS-CoV-2 infection while controls were COVID-19 patients that presented only mild symptoms of the disease. The mean age was significantly higher among cases as compared to controls (54 ± 24 vs 39 ± 13 years; p = 3.48 × 10–8). As expected by the study design, gender distributions were similar between the groups (Table 1). Most common symptoms in the present cohort were cough (70.42%), fever (63.85%), headache (61.50%), anosmia (57.28%) and myalgia (55.87%). Six out of the 13 symptoms evaluated showed significant association with disease severity. Anosmia and headache were significantly more frequent among controls while dyspnea and fatigue were more frequent among cases. All cases used at least one pharmacological intervention against SARS-CoV-2, while about 55% of the control group did so. Six therapeutic strategies (hydroxychloroquine, ceftriaxone, moxifloxacin/ciprofloxacin, levofloxacin, ondansetron, enoxaparin and corticoid use) were found more frequently among cases, though this probably reflects prescription after outcome occurrence (Table 1).
Table 1.
Epidemiological and demographic description of 213 individuals enrolled on the case–control study.
| Variable | Cases | Controls | p-value** | ||
|---|---|---|---|---|---|
| Total | Mean (SD) | Total | Mean (SD) | ||
| Age | 38 | 54 (± 24) | 175 | 39 (± 13) | 3.48 × 10–8 |
| Total | N (%) | Total | N (%) | p-value* | |
|---|---|---|---|---|---|
| Sex | 38 | 175 | 0.8435 | ||
| Female | 26 (68.42) | 114 (65.14) | |||
| Male | 12 (31.58) | 61 (34.85) | |||
| Symptoms | 38 | 175 | |||
| Fever | 30 (78.95) | 106 (60.57) | 0.0511 | ||
| Cough | 28 (73.68) | 122 (69.71) | 0.7718 | ||
| Sneeze | 5 (13.15) | 69 (39.42) | 0.0038 | ||
| Dyspnea | 31 (81.58) | 48 (27.49) | 1.21 × 10–9 | ||
| Coryza/Nasal Congestion | 7 (18.42) | 69 (39.42) | 0.0236 | ||
| Headache | 10 (26.32) | 121 (69.14) | 2.21 × 10–6 | ||
| Nausea | 4 (10.53) | 54 (30.86) | 0.0187 | ||
| Vomit | 2 (5.26) | 13 (7.43) | 0.9019 | ||
| Diarrhea | 6 (15.79) | 58 (33.14) | 0.0549 | ||
| Hash | 0 | 11 (07.97) | - | ||
| Myalgia | 16 (42.11) | 103 (58.86) | 0.0882 | ||
| Anosmia | 6 (15.79) | 116 (66.29) | 3.34 × 10–8 | ||
| Fatigue | 10 (26.32) | 11 (7.97) | 5.0 × 10–4 | ||
| Treatment | 37 | 149 | |||
| Any Treatment | 37 (100) | 82 (55.03) | 8.62 × 10–8 | ||
| Dipyrone/Paracetamol | 33 (89.19) | 120 (80.54) | 0.0532 | ||
| Loratadine | 1 (3.03) | 5 (3.36) | 0.1188 | ||
| Azithromycin | 22 (56.41) | 56 (37.58) | 0.0051 | ||
| Amoxicillin/Clavulanate | 10 (27.03) | 18 (12.08) | 0.0079 | ||
| Hydroxychloroquine | 12 (32.43) | 6 (4.03) | 4.22 × 10–8 | ||
| Ivermectin | 6 (16.22) | 22 (14.77) | 0.1183 | ||
| Nitazoxanide | 1 (2.70) | 3 (2.01) | 0.1172 | ||
| Ceftriaxone | 12 (32.43) | 1 (0.67) | 1.68 × 10–12 | ||
| Moxifloxacin/Ciprofloxacin | 8 (21.22) | 1 (0.67) | 2.60 × 10–8 | ||
| Levofloxacin | 16 (43.24) | 3 (2.01) | 1.30 × 10–14 | ||
| Ondansetron | 4 (10.81) | 1 (0.67) | 2.15 × 10–4 | ||
| Enoxaparin | 10 (27.03) | 1 (0.67) | 2.15 × 10–10 | ||
| Corticoid | 15 (40.54) | 1 (0.67) | 8.66 × 10–16 | ||
| Comorbidities | 38 | 157 | |||
| Any Comorbidity | 27 (71.05) | 71 (45.22) | 0.001 | ||
| Systemic Arterial Hypertension | 19 (50.00) | 22 (14.02) | 3.36 × 10–7 | ||
| Asthma | 3 (7.89) | 22 (14.02) | 0.1529 | ||
| Rhinitis | 1 (2.63) | 6 (3.82) | 0.2415 | ||
| Hypothyroidism | 2 (5.26) | 10 (6.37) | 0.2493 | ||
| Obesity | 13 (34.21) | 3 (1.91) | 1.46 × 10–11 | ||
| Diabetes | 10 (26.32) | 9 (5.73) | 1.25 × 10–4 | ||
| Hypercholesterolemia | 3 (7.89) | 1 (0.64) | 0.0034 | ||
| Anxiety/Depression | 2 (5.26) | 3 (1.91) | 0.1210 | ||
| Chronic Therapy | 28 | 129 | |||
| Any Chronic Therapy | 23 (82.14) | 61 (47.29) | 0.0065 | ||
| Angiotensin Receptor Blockers | 11 (39.29) | 11 (08.53) | 1.20 × 10–4 | ||
| ACE Inhibitors | 2 (7.14) | 3 (2.33) | 0.4208 | ||
| Serotonin Selective Reuptake Inhibitor | 2 (7.14) | 7 (5.43) | 0.9392 | ||
| β2 Adrenergic Agonists | 2 (7.14) | 6 (4.65) | 0.8627 | ||
| Thiazide Diuretics | 7 (25.00) | 11 (8.53) | 0.0462 | ||
| Intranasal Corticoids | 1 (3.57) | 6 (4.65) | 0.9690 | ||
| β Blockers | 4 (14.29) | 5 (3.88) | 0.0996 | ||
| Benzodiazepines | 2 (07.14) | 2 (1.55) | 0.2349 | ||
| Calcium Channel Blockers | 3 (10.71) | 3 (2.33) | 0.1106 | ||
| Statins | 4 (14.29) | 4 (3.10) | 0.0510 | ||
| Levothyroxine | 1 (3.57) | 8 (6.29) | 0.8631 | ||
| Metformin | 7 (25.00) | 9 (6.98) | 0.0169 |
*p-values for Fisher’s Exact test. ** p-values for Wilcoxon Signed Rank test. Significance after Bonferroni adjustment for multiple comparisons (α = 0.001) is represented in bold.
As expected, comorbidities were more prevalent in cases, with systemic arterial hypertension, obesity and diabetes showing up as major chronic risk factors for supplemental oxygen intervention during SARS-CoV-2 infection in the present cohort. Although chronic use of medications was similarly distributed among cases and controls, angiotensin receptor blockers were found more frequently among cases, suggesting a possible association of this class of drugs with COVID-19 severity (Table 1). As angiotensin receptor blockers are broadly used to control systemic arterial hypertension, we further investigated whether its association with oxygen therapy requirement during COVID-19 could be a consequence of the strong association found for systemic arterial hypertension per se. Logistic regression analysis was then performed on a sub cohort including all participants with hypertension (41 individuals), and no association between angiotensin receptor blockers and respiratory distress was found (Supplementary Table 1).
ACE2 and TMPRSS2 transcription levels
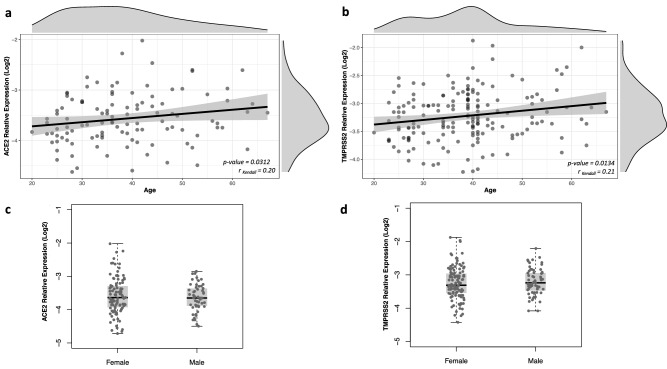
Transcription levels were determined in nasopharyngeal samples collected for disease diagnosis and, therefore, all samples were obtained in the course of infection. Expression of both genes was positively correlated to age (r = 0.20; p = 0.03 and r = 0.21; p = 0.01 for ACE2 and TMPRSS2, respectively), while no association with sex was observed (Fig. 1). Expression levels were not associated with any comorbidity or chronic use of medications (Supplementary Fig. 1 and Supplementary Fig. 1).
Figure 1.
ACE2 and TMPRSS2 expression in nasopharyngeal samples from SARS-CoV-2 positive individuals according to age and sex. Relative expression of both genes was determined by RT-qPCR using B2M gene as reference and results are shown in log-scale (base 2). The correlation between ACE2 and TMPRSS2 expression and age (years) was determined by Kendall rank correlation coefficient and density plots for each variable are depicted externally (a,b). Boxes represent mean and interquartile range, and whiskers represent upper and lower limit. Linear regression models were applied for comparisons of ACE2 (c) and TMPRSS2 (d) expression between male and female subjects.
Association between ACE2 and TMPRSS2 transcription levels and COVID-19 severity
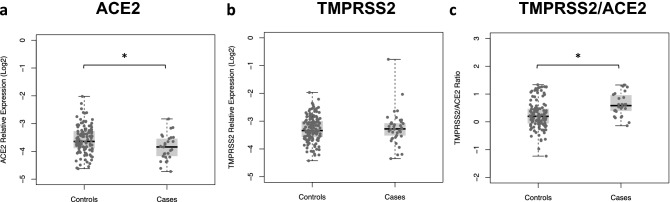
Transcription levels of ACE2 were significantly lower among cases than in controls (mean values of − 3.80 and − 3.60, respectively), while similar levels of TMPRSS2 were observed in both groups (Fig. 2a,b). Considering that functional impact of TMPRSS2 on SARS-CoV-2 entry is dependent on the presence of the receptor ACE2, we hypothesize that TMPRSS2 effect could be better explored in relation to ACE2 expression. TMPRSS2/ACE2 ratios were then determined for each individual and a linear regression analysis showed significantly higher ratios among cases compared to controls (mean values of 0.63 and 0.33, respectively) (Fig. 2c). Next, a stepwise search combined with logistic regression analysis was carried out to define the best covariates to be included in multiple models and then control for confounding. Results showed a better performance for the model including age, systemic arterial hypertension, diabetes and obesity as predictive variables for respiratory distress during COVID-19 (Supplementary Table 2).
Figure 2.
ACE2 and TMPRSS2 expression in COVID-19 patients with (cases) and without (controls) respiratory distress. Relative expression of both genes was determined by RT-qPCR using B2M gene as reference and results are shown in log-scale (base 2). Boxes represent mean and interquartile range, and whiskers represent upper and lower limit. Linear regression models were applied for comparisons of ACE2 (a) and TMPRSS2 (b) expression levels and TMPRSS2/ACE2 ratio (c) between cases and controls. *p < 0.05.
Results of multiple models showed a protective effect for increased expression levels of ACE2 on nasopharynx either before or after adjustment for covariates (adjOR = 0.30; 95%CI = 0.09–0.91) while no significant association was found for TMPRSS2 (Table 2). Notably, a strong risk effect of TMPRSS2/ACE2 ratios on COVID-19 severity was found (adjOR = 4.28; 95%CI = 1.36–13.48) (Table 2). Since TMPRSS2/ACE2 fold change is based on transcription levels of both genes and a significant association was already found for ACE2 alone, a stepwise search combined with regression analyses was performed to compare nested models including both ACE2 and TMPRSS2/ACE2 ratio along with already selected non-genetic covariates. Results showed that a model including age, systemic arterial hypertension, diabetes, obesity and TMPRSS2/ACE2 minimized the Akaike Information Criteria (AIC = 97.11), suggesting that TMPRSS2/ACE2 ratio outperforms ACE2 expression predictive power to model COVID-19 respiratory outcome in this cohort (Supplementary Table 3).
Table 2.
Association between ACE2 and TMPRSS2 expression on nasopharynx and respiratory distress during COVID-19.
| Relative Log2 Expression Level | Cases mean (SD) | Controls mean (SD) | OR (95%CI) | adjOR (95%CI) * |
|---|---|---|---|---|
| ACE2 | − 3.85 (0.46) | − 3.60 (0.51) | 0.34 (0.14–0.83) | 0.30 (0.09–0.91) |
| Total = 172 | 27 | 145 | p-value = 0.0193 | p-value = 0.0318 |
| TMPRSS2 | − 3.24 (0.61) | − 3.23 (0.46) | 0.98 (0.47–2.03) | 1.05 (0.43–2.56) |
| Total = 210 | 37 | 173 | p-value = 0.9511 | p-value = 0.9068 |
| TMPRSS2/ACE2 ratio | 0.63 (0.42) | 0.33 (0.52) | 3.29 (1.37–7.88) | 4.28 (1.36–13.48) |
| Total = 171 | 26 | 145 | p-value = 0.0070 | p-value = 0.0131 |
*OR estimates adjusted for age, systemic arterial hypertension, diabetes and obesity. OR values represent an associated risk/protection according to an increase of 1 log of target’s expression relative to B2M gene. SD = standard deviation.
Expression of ACE2 and TMPRSS2 genes in lower respiratory tract
To further investigate whether results obtained from the nasopharynx might reflect transcription levels at lower respiratory tract from critically ill COVID-19 individuals, additional 34 nasopharyngeal swabs and 11 bronchoalveolar lavage (BAL) samples were obtained from an independent case-only cohort of individuals. Distribution of age and gender were similar between swab and BAL groups (Supplementary Table 4). Results of linear regression models showed similar transcription levels of ACE2 and TMPRSS2 in nasopharyngeal and BAL samples even after adjustment for the same covariates selected after stepwise regression (Fig. 3). No significant differences were found for TMPRSS2/ACE2 ratio whatsoever.
Figure 3.
ACE2 and TMPRSS2 expression levels on upper and lower respiratory tract of severe COVID-19 patients with respiratory failure. Relative expression levels of ACE2 (a) and TMPRSS2 (b) were determined by RT-qPCR on samples from nasopharyngeal swabs and bronchoalveolar lavages (BAL). TMPRSS2/ACE2 ratio were calculated for each individual and compared between swabs and BAL samples (c). Results are represented in log-scale (base 2) relative to B2M expression. Boxes represent mean and interquartile range, and whiskers represent upper and lower limit. Linear regression models were applied for comparisons between groups.
Discussion
Population-based studies on COVID-19 patients have been elucidating epidemiological factors contributing to disease severity such as age and chronic diseases (systemic arterial hypertension, diabetes and, obesity)2,5. Consistent with previous literature findings, age showed up as a major risk factor for the need of oxygen support during SARS-CoV-2 infection in the present study. Comorbidities such as systemic arterial hypertension, diabetes and obesity also appeared here as significant factors associated with respiratory distress.
Prevalence of SARS-CoV-2 induced headache and anosmia has been found to vary considerably between cohorts17. Here, 61.5% of total participants reported headache and 57.28% showed olfactory impairment/dysfunction with a remarkably overrepresentation of these outcomes among controls. No significant differences were found for general chronic therapies between cases and controls. However, previous in silico findings warned for potentially higher ACE2 or TMPRSS2 levels during systemic arterial hypertension treatments18. Although we did not find any evidence of chronic therapy influencing ACE2 or TMPRSS2 expression (Supplementary Fig. 2), angiotensin receptor blockers were significantly more frequent among cases, suggesting an impact on COVID-19 severity. After a sub cohort analysis, the association was found to be driven by systemic arterial hypertension per se (Supplementary Table 1). Although preliminary, this supports recent findings that the use of this class of drugs is not associated with disease severity during SARS-CoV-2 infections19–21.
ACE2 has been described as the entry receptor for SARS-CoV-2 infection and TMPRSS2 as a major priming enzyme relevant to its fusion on the cell membrane6. Since the beginning of the pandemic, a lot of questions rely on how expression of these host factors could impact on COVID-19 pathogenesis. ACE2 and TMPRSS2 levels have previously been found lower in children than in young adults’ upper airways, raising the hypothesis that they could reflect age-related susceptibility to COVID-198,22. However, expression levels of these genes were not yet assessed in different age groups during infection. Here, in addition to the association with severity, age also appears to impact ACE2 and TMPRSS2 expression levels at the nasopharyngeal epithelium of SARS-CoV-2 infected individuals.
In the present study, we characterized the transcription levels of both genes in nasopharyngeal samples of COVID-19 patients and investigated their association with respiratory distress using a case–control study design. After logistic regression analysis, higher nasopharyngeal expression of ACE2 was significantly associated with protection against respiratory distress requiring supplemental oxygen during COVID-19. ACE2 protective effect may be supported by recent evidence of its “interferon stimulated gene” attributes, where higher levels of ACE2 transcripts could be a consequence of higher interferon activity induced by viral replication itself23 and then possibly acting as a hallmark of a more robust antiviral response7. Experimental data showing ACE2 upregulation on smokers suggests a risk effect for IFN induced overexpression of this gene in the lungs by IFN signaling activation11,22,24. However, contrasting with the pattern expected for the upper respiratory tract, exacerbation of IFN signaling in lungs could worsen coronavirus associated SARS by increasing inflammation and induce intense endothelial/alveolar damage25–27.
A risk effect for TMPRSS2 levels on COVID-19 severity could be expected once this enzyme triggers the viral S protein priming after its recognition by ACE2 entry receptor and is considered an important player host susceptibility to SARS-CoV-26. Moreover, TMPRSS2 expression was also found to be reshaped by smoking habits, a known risk factor for SARS22. However, since its functional activity on viral fusion depends on ACE2 mediated viral adsorption, an association could only be seen in this study when both targets were evaluated together. In fact, higher TMPRSS2/ACE2 ratios showed a prominent risk effect for respiratory distress requiring oxygen therapy during COVID-19 and comparisons between multiple models revealed that TMPRSS2/ACE2 ratio can be even more informative to model disease severity than ACE2 expression alone. These data suggest that further expression studies on SARS-CoV-2 pathogenesis should consider evaluating co-expression of both genes.
Finally, expression analysis from nasopharyngeal swab samples compared to BAL on an independent case-only cohort demonstrates similar expression patterns of ACE2, TMPRSS2 and TMPRSS2/ACE2 on upper and lower respiratory tracts of COVID-19 patients under mechanical ventilation. Recent work has shown that ACE2 and TMPRSS2 expression are significantly higher in nasal than in bronchial tissues in children and adults22. Our data suggests that, after viral colonization of both upper and lower respiratory tracts, ACE2 and TMPRSS2 assessed by nasopharyngeal swabs seems to reflect what is found on BAL samples. Then, expression levels of these genes could be further explored as potential biomarkers.
An important limitation of this study is that our data does not allow a clear interpretation of ACE2 and TMPRSS2 levels as predictors of disease severity, since case samples were obtained at the moment of hospital admission to initiate oxygen therapy. Prospective studies should then be carried out to better explore the prediction effect of these factors on COVID-19 severity. We also would like to address that, due to sample size limitations in the case group, we were unable to perform stratification analysis for different methods of oxygen therapy (nasal cannula, noninvasive mechanical ventilation and intubation). Then, we encourage future studies with greater sample sizes to do so in order to better define the contribution of ACE2 and TMPRRS2 expression levels on respiratory outcome severity.
Taken together, the present study shows an association between nasopharyngeal expression of ACE2 and TMPRSS2 genes and the need of oxygen therapy during COVID-19. Our data also supports that TMPRSS2 effect on modelling disease severity may be dependent on ACE2 levels, suggesting that the impact of this serine protease on COVID-19 might be better explored in combination to its partner ACE2.
Supplementary Information
Acknowledgements
The authors thank all participants enrolled in this study. This work was supported by Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ) [grant numbers E-26/202.791/2019, E-26/010.002278/2019 and, E-26/210.179/2020 to C.C.C.; E-26/010.002434/2019 and E-26/210.178/2020 to A.T.; E-26/010.000168/20 to S.P.C.B and 202.922/2018 to R.S.A.], Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPQ) [312688/2017-2 and 439119/2018-9 to R.S.A. and 310627/2018-4 to R.P.S.], RedeVírus/MCTI [FINEP 01.20.0029.000462/20, CNPq 404096/2020-4], FAPEMIG [APQ-00475-20 to R.P.S], MEC/CAPES 118 [14/2020 - 23072.211119/2020-10] and FINEP [0494/20 01.20.0026.00 and UFMG-NB3 1139/20 to R.S.A ]. A.D.R is recipient of a post-doctoral fellowship from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES).
Author contributions
A.D.R., C.C.C, S.P.C.B., A.T. and R.S.A. conceived the study. I.C.L., H.T.S., D.S.F., R.M.G., R.E.A., G.G.R., S.N.F., J.S.O., T.M.P.P.C., R.P.S, R.S.A. and COVID-19 UFRJ Workgroup were responsible for sample collection and SARS-CoV-2 molecular diagnosis. A.D.R., J.L.F.A, T.B.A., C.A.V., J.M.A., S.N.F., J.S.O, H.J.A and S.P.C.B. conducted the experiments. A.D.R., M.R.A. and C.C.C. analyzed the data. I.C.L., H.T.S., D.S.F., O.C.F.J, R.M.G., R.E.A., G.G.R., M.M.T. and T.M.P.P.C. were responsible for subject’s recruitment and clinical follow-up. C.C.C, A.T, S.P.C.B, R.S.A financed the study. A.D.R. and C.C.C. wrote the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
A list of authors and their affiliations appears at the end of the paper.
Contributor Information
Cynthia Chester Cardoso, Email: cynthiac@biologia.ufrj.br.
COVID-19 UFRJ Workgroup:
Alice Laschuk Herlinger, Aliny dos Santos Carvalho, André Felipe Andrade dos Santos, Anna Carla Pinto Castiñeiras, Bianca Isabelle Barreto Teixeira, Bianca Ortiz da Silva, Bruno Clarkson, Bruno Eduardo Dematté, Camila Nacif, Camille Victória Leal Correia de Silva, Carolina Moreira Voloch, Caroline Macedo Nascimento, Carolyne Lalucha Alves L. da Graça, Cassia Cristina Alves Gonçalves, Cíntia Policarpo, Diana Mariani, Ekaterini Simões Goudouri, Elaine Sobral da Costa, Elisangela Costa da Silva, Enrico Bruno Riscarolli, Érica Ramos dos Santos Nascimento, Fabio Hecht Castro Medeiros, Fábio Luís Lima Monteiro, Fernanda Leitão dos Santos, Fernando Luz de Castro, Filipe Romero Rebello Moreira, Francine Bittencourt Schiffler, Gabriela Bergiante Kraychete, Gabriele Silveira da Cunha, Gisely Novaes Borges da Cunha, Guilherme Sant’Anna de Lira, Gustavo Peixoto Duarte da Silva, Harrison James Westgarth, Helena D.’Anunciação de Oliveira, Helena Keito Toma, Huang Ling Fang, Inês Corrêa Gonçalves, Ingrid Camelo da Silva, Isabela Labarba Carvalho de Almeida, Joissy Aprigio de Oliveira, Juliana Cazarin de Menezes, Juliana Tiemi Sato Fortuna, Karyne Ferreira Monteiro, Kissyla Harley Della Pascoa França, Laura Zalcberg Renault, Lendel Correia da Costa, Leticia Averbug Correa, Liane de Jesus Ribeiro, Lídia Theodoro Boullosa, Liliane Tavares de Faria Cavalcante, Luana dos Santos Costa, Lucas Matos Millioni, Luciana Jesus da Costa, Luiza Mendonça Higa, Marcela dos Santos Durães, Marcelo Amaral de Souza, Marcelo Calado de Paula Tôrres, Mariana Freire Campos, Mariana Quinto, Mariane Talon de Menezes, Marisa Souza Correia, Mateus Rodrigues de Queiroz, Matheus Augusto Calvano Cosentino, Mayla Gabryele Miranda de Melo, Mirela D’arc Ferreira da Costa, Pedro Henrique Costa da Paz, Raissa Mirella dos Santos Cunha da Costa, Raquel Fernandes Coelho, Richard Araujo Maia, Rodrigo de Moraes Brindeiro, Romina Carvalho Ferreira, Sérgio Machado Lisboa, Thamiris dos Santos Miranda, Victor Akira Ota, Victoria Cortes Bastos, and Viviane Guimarães Gomes
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-88944-8.
References
- 1.Wu JT, Leung K, Leung GM. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet. 2020;395:689–697. doi: 10.1016/S0140-6736(20)30260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu Z, McGoogan JM. Characteristics of and Important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: Summary of a report of 72314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 3.Zheng Z, et al. Risk factors of critical & mortal COVID-19 cases: A systematic literature review and meta-analysis. J. Infect. 2020;81:e16–e25. doi: 10.1016/j.jinf.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang X, et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir. Med. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karagiannidis C, et al. Case characteristics, resource use, and outcomes of 10 021 patients with COVID-19 admitted to 920 German hospitals: an observational study. Lancet Respir. Med. 2020;8:853–862. doi: 10.1016/S2213-2600(20)30316-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffmann M, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ziegler CGK, et al. SARS-CoV-2 receptor ACE2 Is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181:1016–1035.e19. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bunyavanich S, Do A, Vicencio A. Nasal gene expression of angiotensin-converting enzyme 2 in children and adults. JAMA - J. Am. Med. Assoc. 2020;323:2427–2429. doi: 10.1001/jama.2020.8707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishiyama Y, et al. Upregulation of angiotensin-converting enzyme 2 after myocardial infarction by blockade of angiotensin II receptors. Hypertension. 2004;43:970–976. doi: 10.1161/01.HYP.0000124667.34652.1a. [DOI] [PubMed] [Google Scholar]
- 10.Wang X, et al. The effects of different angiotensin II type 1 receptor blockers on the regulation of the ACE-AngII-AT1 and ACE2-Ang(1–7)-Mas axes in pressure overload-induced cardiac remodeling in male mice. J. Mol. Cell. Cardiol. 2016;97:180–190. doi: 10.1016/j.yjmcc.2016.05.012. [DOI] [PubMed] [Google Scholar]
- 11.Smith JC, et al. Cigarette smoke exposure and inflammatory signaling increase the expression of the SARS-CoV-2 receptor ACE2 in the respiratory tract. Dev. Cell. 2020;53:514–529.e3. doi: 10.1016/j.devcel.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imai Y, et al. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005;436:112–116. doi: 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patel VB, et al. ACE2 deficiency worsens epicardial adipose tissue inflammation and cardiac dysfunction in response to diet-induced obesity. Diabetes. 2016;65:85–95. doi: 10.2337/db15-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Michael W. Pfaffl. Quantification strategies in real-time PCR. in A-Z of quantitative PCR (ed. Stephen A Bustin) 87–112 (International University Line, 2004).
- 15.Wickham H, et al. Welcome to the tidyverse. J. Open Source Softw. 2019;4:1686. doi: 10.21105/joss.01686. [DOI] [Google Scholar]
- 16.Venables, W. N. & Ripley, B. D. Modern Applied Statistics with S. (Springer New York, 2002). 10.1007/978-0-387-21706-2.
- 17.Zubair AS, et al. Neuropathogenesis and neurologic manifestations of the coronaviruses in the age of coronavirus disease 2019: A review. JAMA Neurol. 2020;77:1018–1027. doi: 10.1001/jamaneurol.2020.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saheb Sharif-Askari N, et al. Cardiovascular medications and regulation of COVID-19 receptors expression. Int. J. Cardiol. Hypertens. 2020;6:100034. doi: 10.1016/j.ijchy.2020.100034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Milne S, Yang CX, Timens W, Bossé Y, Sin DD. SARS-CoV-2 receptor ACE2 gene expression and RAAS inhibitors. Lancet Respir. Med. 2020;8:e50–e51. doi: 10.1016/S2213-2600(20)30224-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J, Wang X, Chen J, Zhang H, Deng A. Association of renin-angiotensin system inhibitors with severity or risk of death in patients with hypertension hospitalized for coronavirus disease 2019 (COVID-19) infection in Wuhan, China. JAMA Cardiol. 2020;5:825–830. doi: 10.1001/jamacardio.2020.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lam KW, et al. Continued in-hospital angiotensin-converting enzyme inhibitor and angiotensin II receptor blocker use in hypertensive COVID-19 patients is associated with positive clinical outcome. J. Infect. Dis. 2020;222:1256–1264. doi: 10.1093/infdis/jiaa447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saheb Sharif-Askari, N. et al. Airways Expression of SARS-CoV-2 Receptor, ACE2, and TMPRSS2 Is Lower in Children Than Adults and Increases with Smoking and COPD. Mol. Ther. - Methods Clin. Dev.18, 1–6 (2020). [DOI] [PMC free article] [PubMed]
- 23.Lieberman NAP, et al. In vivo antiviral host transcriptional response to SARS-CoV-2 by viral load, sex, and age. PLoS Biol. 2020;18:1–17. doi: 10.1371/journal.pbio.3000849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silver JS, et al. Inflammatory triggers associated with exacerbations of COPD orchestrate plasticity of group 2 innate lymphoid cells in the lungs. Nat. Immunol. 2016;17:626–635. doi: 10.1038/ni.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Channappanavar R, et al. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe. 2016;19:181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ackermann M, et al. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in covid-19. N. Engl. J. Med. 2020;383:120–128. doi: 10.1056/NEJMoa2015432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carsana L, et al. Pulmonary post-mortem findings in a series of COVID-19 cases from northern Italy: a two-centre descriptive study. Lancet Infect. Dis. 2020;20:1135–1140. doi: 10.1016/S1473-3099(20)30434-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.