Abstract
Pandrug-resistant (PDR) K. pneumoniae refractory to conventional treatment has been reported worldwide, causing a huge burden on the healthcare system, patient safety and the economy. K. pneumoniae is a prominent opportunistic pathogen causing hospital-acquired and community-acquired infections, but is rarely associated with infective endocarditis. Currently, there are sparse data guiding the optimal regimen when commonly used antibiotics fail, notably for the treatment of endocarditis infections. Here we report our experience in treating a 40-year-old female with PDR K. pneumoniae infection of cardiovascular implantable electronic device (CIED) and right-sided infective endocarditis. Initial susceptibility testing of the incriminated pathogen showed an apparent susceptibility to colistin but the prolonged course of colistin, gentamicin and meropenem did not resolve the infection. However, the synergistic combinations of aztreonam with ceftazidime-avibactam was able to overcome resistance and clear the infection rapidly. Genome sequencing showed that the PDR K. pneumoniae isolate belongs to the international high-risk clone ST14. The isolate harbored genes encoding NDM-1, OXA-48, CTX-M-14b, SHV-28 and OXA-1, explaining resistance to all β-lactams, including carbapenems. It carried the armA gene conferring resistance to all clinically important aminoglycosides and had alterations in GyrA, ParC and MgrB, explaining resistance to ciprofloxacin and colistin.
Subject terms: Microbiology, Antimicrobials, Bacteria, Clinical microbiology, Infectious-disease diagnostics, Microbial genetics, Pathogens
Introduction
Klebsiella pneumoniae is an important human pathogen responsible for a wide range of severe infections with an increasing scarcity of effective treatments1,2. The species is often associated with hospital-acquired bloodstream infections but is rarely a cause of infective endocarditis (IE) or cardiac implantable electronic device (CIED) infection3–5. Among gram-negative pathogens, K. pneumoniae has been only associated with 1.2% of native-valve IE and 4.1% of prosthetic-valve IE6. Currently, there is no clear evidence-based treatment guideline for K. pneumoniae causing IE7. Hence, most of the reported cases were treated according to the antibiotic susceptibilities of the cultured isolates with or without surgical intervention, which led to successful bacteraemia clearance ranging from 70 to 85% in susceptible strains that are not also hypervirulent (hvKP)3,6,8–10.
The emergence of co-resistance to β-lactams, aminoglycosides, quinolones, colistin and tigecycline in K. pneumoniae isolates poses a serious therapeutic challenge due to limited treatment options. In recent years, the incidences of pan-drug resistant (PDR) K. pneumoniae infections refractory to conventional treatment have been reported globally, causing a significant increase in long-term hospitalizations, morbidity and mortality11. Resistance to last-resort carbapenems in this species is mainly mediated by the production of β-lactamases, notably those belonging to the KPC, NDM, VIM and OXA-48-like type enzymes. In Saudi Arabia, K. pneumoniae is the most contributing organism to carbapenem resistance among all Enterobacterales, increasing from 0 to 33.3% in 10 years (2007–2016)12. Resistance to carbapenems in K. pneumoniae isolates from Saudi Arabia are mainly associated with the acquisition of OXA-48-like and NDM carbapenemases, although few recent studies have reported the detection of KPC carbapenemases in Klebsiella spp. isolates13,14.
Combination antibiotic therapy has been used as an option to treat patients with life-threatening PDR K. pneumoniae infections15,16. However, limited studies showed evidence-based combination antibiotic therapy to treat patients infected by PDR K. pneumoniae. This is particularly true for endocarditis, for which treatment options are already limited by the localization and characteristics of the infection. Most of the available information is driven from in vitro studies and just a few numbers of clinical in vivo studies17. The recent approach of therapeutic options such as the β-lactam-β-lactamase inhibitors ceftazidime-avibactam (CAZ/AVI) or meropenem-vaborbactam combinations showed potent inhibitors activity against class A and D carbapenemase producers (e.g. KPC, OXA-48-like, GES) but were ineffective against class B carbapenemase producers (e.g. NDM, VIM, IMP)18. Clinical trials and in vitro studies have demonstrated the activity of CAZ/AVI against ESBL-, AmpC-, KPC- and OXA-48-producing pathogens18–21. The combination of CAZ/AVI has been approved to be used as a therapeutic option to treat adults with complicated urinary tract infections, hospital-acquired pneumonia and other infections caused by MDR gram-negative pathogens22. Other studies have shown that CAZ-AVI plus aztreonam (ATM) can be an effective therapeutic combination against metallo-β-lactamases (MBLs)41. Here, we report the successful treatment of CIED and right-sided IE due to carbapenemase OXA-48- and NDM-producing K. pneumoniae strain ST14 using CAZ-AVI plus ATM only.
Results
Case record, diagnostic and antibiotic treatment
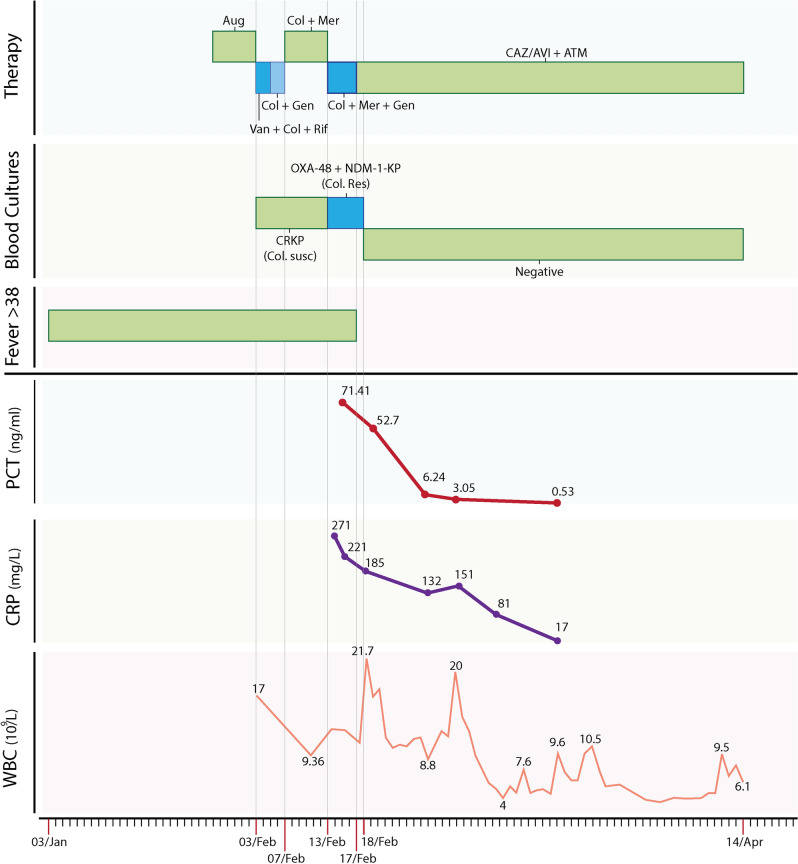
The patient is a 40-year-old female with known rheumatic heart disease since childhood who had a bioprosthetic mitral valve replacement in 2015. In July 2017, the patient had the mechanical valve replaced due to valve thrombosis and an insertion of a dual-chamber pacemaker to treat arrhythmia. In October 2017, she developed prosthetic mitral valve endocarditis due to Enterococcus faecalis that was cleared by six weeks of treatment with a combination of ampicillin and ceftriaxone. In January 2019, the patient had another infective endocarditis affecting the tricuspid valve, and the pacemaker leads due to a carbapenem-resistant K. pneumoniae, for which she was managed during ten days in two different hospitals before being transferred to our specialized cardiac center following several cardiac arrests from septic pulmonary emboli (Fig. 1). The antibiotic susceptibility performed in the transferring hospital suggested apparent susceptibility to gentamicin and colistin, and so, the patient was put on colistin (2.5 million IU twice daily) and gentamicin (7 mg/kg once daily) intravenously (IV), to which 500 mg meropenem twice daily due to acute kidney injury was added at day one. However, repeated blood cultures showed persistent bacteremia with a K. pneumoniae strain resistant to all tested antimicrobials, including those used for treatment (Table 1). A synergy of antibiotic combinations showed that only CAZ/AVI with ATM was an effective option for treatment (Table 1). Accordingly, the therapy was changed on day five to CAZ/AVI (2.5 gm every 8 h) and ATM (2 gm every eight hours) IV. Her fever resolved, and bacteremia cleared right after starting this combination, while inflammatory markers and creatinine levels improved over a couple of weeks. At 42 days of therapy, the pacer device and the lead tip were replaced. The culture from the tip of the lead did not grow any bacteria, and consequently, the antibiotic treatment was stopped at day 50 (Fig. 1). Although the source of the bacteremia was not found, the follow up at six months after discharge showed that the patient was completely healthy and asymptomatic.
Figure 1.
Patient timeline for antimicrobial exposure, fever pattern, duration of persistence of infection in blood cultures with trends of important laboratory investigations. Important dates marked from left to right at the bottom represented the start of symptoms (03/Jan), first hospital admission (03/Feb), transfer to the second hospital (07/Feb), transfer to our cardiac centre (13/Feb), the start of the therapy CAZ/AVI plus ATM combination (17/Feb), date of the first negative culture (18/Feb) and date of discharge from hospital (14/Apr). Aug augmentin, Van vancomycin, Col colistin, Rif rifampicin, Gen gentamicin, Mer meropenem, CAZ/AVI ceftazidime/avibactam, ATM aztreonam, CRKP carbapenem-resistant Klebsiella pneumoniae, OXA-48 oxacillinase, NDM New Delhi Metallo-β-lactamase, Susc susceptible; Res resistant; PCT procalcitonin; CRP C-reactive protein; WBC white blood cells.
Table 1.
Antibiotic resistance in K. pneumoniae SA-KpST14.
| Antimicrobial category | Antimicrobial agents | MIC (μg/ml) VITEK II | Interpretation (CLSI breakpoints) | Genes associated with resistance |
|---|---|---|---|---|
| β-lactams | Ampicillin | ≥ 32 | R | blaOXA-1, blaSHV-28, blaNDM-1, blaOXA-48, blaCTX-M-14b |
| Amoxicillin/Clavulanic acid | ≥ 32 | R | ||
| Piperacillin/Tazobactam | ≥ 128 | R | ||
| Ceftazidime/Avibactam | > 256* | R | ||
| Cefoxitin | ≥ 64 | R | ||
| Ceftazidime | ≥ 64 | R | ||
| Cefepime | ≥ 64 | R | ||
| Cefalotin | ≥ 64 | R | ||
| Ceftriaxone | ≥ 64 | R | ||
| Imipenem | > 32* | R | ||
| Meropenem | ≥ 16 | R | ||
| Aztreonam | > 256* | R | ||
| Fluoroquinolones | Ciprofloxacin | ≥ 4 | R | aac(6′)-Ib-cr, gyrA (S83Y, D87G). parC (S80I) |
| Aminoglycosides | Amikacin | ≥ 64 | R | aac(6′)-Ib-cr, aph(3′)-Ib, aph(6)-Id, dfrA12, armA, aadA2, strAB, aph(3′)-VI |
| Gentamicin | ≥ 16 | R | ||
|
Trimethoprim/ Sulfamethoxazole |
Trimethoprim/ Sulfamethoxazole |
≥ 4/76 | R | dfrA12 |
| Polymyxin | Colistin | ≥ 64** | R | IS5 disruption of mgrB gene |
| Tetracycline | Tigecycline | ≥ 256* | R | oqxAB§, acrAB§ |
| Fosfomycin | Fosfomycin | ≥ 1024* | R | fosA |
| Others | Nitrofurantoin | 128 | R | |
| Name of the antibiotic combination (ETEST) | Result of synergy test | |||
| Meropenem + Tigecycline | No zone | |||
| Meropenem + Ertapenem | No zone | |||
| Meropenem + Fosfomycin | No zone | |||
| Meropenem + Gentamicin | Very small zone of inhibition | |||
| Ceftazidime/Avibactam + Aztreonam | A large zone of Inhibition | |||
* ETEST method.
**Broth Micro Dilution method.
§No genetic evidence was found to infer overexpression.
Genome sequence analysis
Genome sequencing showed that the carbapenem-resistant K. pneumoniae SA-KpST14 isolate harbored five different plasmids (Table 2). In silico analyses identified the strain as sequence type (ST)14 and detected genes explaining resistance to β-lactams, aminoglycosides, quinolones, phenicols and fosfomycin, as shown in Table 1. Otherwise, the genetic disruption of the mgrB regulator by insertion sequence IS5 explained resistance to colistin.
Table 2.
Genetic elements sizes and replicon types of plasmids of SA-KpST14 isolate and the presence of antimicrobial resistance genes.
| Named | Size | GC (%) | Plasmid Type | Antimicrobial resistance gene(s) | Accession No |
|---|---|---|---|---|---|
| SA-KpST14 | 5,378,785 bp | 57 | – | blaOXA-1, blaSHV-28, aac(6′)-Ib-cr, catB, fosA | CP071279 |
| pSA-KpST14-NDM-1 | 269, 329 bp | 46 | IncHI1B | blaNDM-1, aadA2, aph(3′)-VI, armA, mph(E), msr(E), sul1, dfrA12 | CP071280 |
| pSA-KpST14-OXA48-2 | 68, 932 bp | 51 | IncM1 | blaOXA-48, blaCTX-M-14b, aph(3′')-Ib, aph(6)-Id | CP071281 |
| pSA-KpST14-3 | 166,565 bp | 50 | IncFIB | – | CP071282 |
| pSA-KpST14-4 | 20,912 bp | 53 | IncR | – | CP071283 |
| KpST14-5 | 2,095 bp | 44 | – | – | CP071284 |
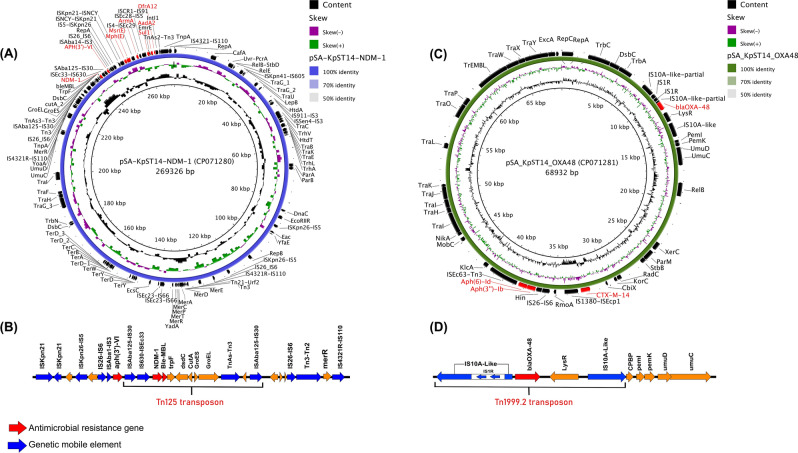
Resistance to last-resort carbapenems was associated with the presence of blaNDM-1 and blaOXA-48 genes that were located on two different plasmids (Fig. 2). The blaNDM-1, embedded in transposon Tn125 (11,192 bp) was located on an IncHI1B replicon-type plasmid (pSA-KpST14-NDM-1, 269, 329 bp) which also harbored resistance to aminoglycosides (aadA2, aph(3′)-VI and armA) , macrolides (mph(E) and msr(E)), sulfonamides (sul1) and trimethoprim (dfrA12) (Fig. 2A). The genetic structure of the Tn125 transposon was composed of the insertion sequence ISEc33-IS630 upstream the blaNDM-1 gene (New Delhi Metallo-beta-lactamase 1) and the bleMBL gene (bleomycin resistance protein), trpF (phosphoribosyl-anthranilate isomerase), dsdc (D-serine deaminase activator), cutA (divalent-cation tolerance protein), ATP-dependent chaperonin GroEL–GroES and incomplete TnAs3-Tn3 downstream (Fig. 2B). On the other hand, the blaOXA-48 carbapenemase was embedded in a classical Tn1999.2 transposon (5,639 bp) on an IncM1 plasmid (pSA-KpST14-OXA48-2, 68, 932 bp) with also carried the blaCTX-M-14b and aph(3′')-Ib genes (Fig. 2C). The genetic structure of Tn1999.2 transposon was as described composed of the lysR transcriptional regulator, blaOXA-48 (oxacillinase), flanked by two copies of insertion sequences IS10A-like in the opposite orientation and thus forming the IS10A-like -blaOXA-48-LysR- IS10A-like element. The IS10A-like is belonge to IS4 family which is 99.77% identity with IS10A (Accession number AF078527) and 99.62% identity with IS1999 (Accession number : AF133697).The insertion sequence IS10A-like located upstream of blaOXA-48 was truncated by the IS1 insertion (Fig. 2D).
Figure 2.
Sequence representation of the two carbapenemase-encoding plasmids carried by the SA-KpST14 isolate. (A) Genetic structure of pSA-KpST14-NDM-1 plasmid, (B) Gene composition of the bla-NDM-1-bearing Tn125 transposon, (C) Genetic structure of pSA-KpST14-OXA48-2 plasmid, (D) Gene composition of the blaOXA-48 -bearing Tn1999.2 transposon. The circular map was generated with the Blast Ring Image Generator (BRIG)59 software and the schematic diagram of the genetic structure was generated with Easyfig60.
Virulence factors
Genome sequences showed that the K. pneumoniae SA-KpST14 strain lacked the capsule regulator gene rmpA/rmpA2 and the siderophore aerobactin factors that are characteristic of invasive strains. However, the strain possessed several other iron acquisitions and siderophore genes, including the enterobactin (Ent), yersiniabactin (Ybt) and salmochelin (Sal) genes. Of these, the ybt locus was identified within the integrative and conjugative element-5 (ICEKp-5) as previously described23–25. Capsular type of SA-KpST14 was determined based on the K-locus's gene content, which corresponded to KL2 (99.73%) and known allelic type wzc2 wzi2. The LPS O antigen was determined by sequence identity to wzm and wzt genes, which corresponded to O locus O1v1 (99.99%). Otherwise, the isolate harbored other intrinsic virulence factors previously associated with the adherence, biofilm formation, secretion system and efflux pump factors listed in Table 3.
Table 3.
Virulence factors in K. pneumoniae SA-KpST14.
| Virulence factor | Category | Related genes | Function |
|---|---|---|---|
| Adherence | Type I fimbriae | fimA-K | Adhering to human mucosal or epithelial surfaces |
| E. coli common pilus | ecpRABCDE | Cell adherence and biofilm formation26 | |
| Biofilm formation | Type 3 fimbriae | mrkABCDFHI | Promotes mucous adherence, tissue colonisation, and biofilm formation26 |
| Iron uptake | Enterobactin | entABCDEF, | Enterobactin promotes bacterial growth around blood vessels 27 |
| fepABCDG | |||
| fes | |||
| ybdA | |||
| Yersiniabactin | ybtAEPQSTUX | Most common virulence genes associated with human K. pneumoniae infection 23–25 | |
| fyuA | |||
| irp12 | |||
| Salmochelin | iroE | iroE is coded for protein with hydrolytic activity to degrade salmochelins and enterobactin to release iron 27,28 | |
| Secretion system | T6SS | tssABCDFGHIJKLM | Bacterial Competition, Cell invasion, Type-1 fimbriae expression, in vivo Colonization, and to puncture target cells and deliver lethal effectors29–31 |
| Immune evasion | K2 capsule | manBC | Evading the host immune system 32 |
| wcaJ | |||
| galf | |||
| gnd | |||
| ugd | |||
| wza, wzi | |||
| cpsACP | |||
| Serum resistance | LPS | glf | Essential structural component and immunodominant molecules of the outer membrane 33,34 |
| wbbMNO | |||
| wzm wzt |
Discussion
Right-sided IE accounts for 5–10% of all IE cases, and more than 50% of these cases are due to intravenous drug use6. The presence of an implantable endovascular device imposes a higher risk of developing gram-negative endocarditis along with hospital stay, history of invasive procedures and other risk factors for developing gram-negative bacteremia in general8. The weaker adhesion ability of gram-negative bacteria (GNB) has been attributed to the low prevalence of CIED and IE caused by non-HACEK GNB35. Presentation can be associated with septic emboli to the lungs as in the presented case6. Liver abscess and urinary tract infection are the most common source when bacteremia is present36. Mortality rates vary depending on the pathogen, virulence factors, complications and the valve involved, reaching up to 49% in one article6,37. Urgent surgical interventions as a first-choice treatment is recommended for uncontrolled infection to prevent complications, including heart failure and embolic events35,38.
In Enterobacterales, PDR is observed among carbapenemase-producing bacteria, especially among K. pneumoniae, as it can easily acquire mobile genetic elements through horizontal gene transfer39. The risk of acquiring carbapenemase-producing bacteria increases in a patient with prior surgery, extended hospital stays and the presence of wounds40.
Latest Infectious Diseases Society of America (IDSA) guidelines on the treatment of carbapenem-resistant enterobacterales (CREs) in general recommended the use of CAZ/AVI with ATM or cefiderocol monotherapy for MBL-producers (e.g., NDM, VIM or IMP) and CAZ/AVI monotherapy or cefiderocol monotherapies for OXA-48-like producers41. While second-line options included tigecycline, eravacycline, colistin and fosfomycin in limited indications and they recommended against combinations of antimicrobials when a β-lactam is susceptible41.
However, the guideline did not focus on cases of CIED or IE but rather gave a recommendation for infections outside the urinary tract in general, and no recommendations were given for conditions where two or more resistance genes are detected within the same species41.
Therapeutic options used in the literature with successful results for carbapenemase co-producing (NDM-1 and OXA-48 like) K. pneumoniae includes the use of ATM in combination with CAZ/AVI as this has shown promising results in NDM producing Enterobacterales in-vitro and in-vivo. It is thought that this is due to the efficacy of ATM against MBLs in general with the addition of the effect of the avibactam component in CAZ/AVI on extended-spectrum β-lactamases, and ambler class A, and D carbapenmases, which are often co-produced by some strains22,42. The presence of serine β-lactamases along with NDM-1 gene was detected in up to 30% in one study for which the combination of CAZ/AVI and ATM has shown synergy in vitro and in vivo43. Several studies have proposed this effect42,44–47, and other β-lactamase inhibitors (Clavulanate and Tazobactam) has also been tested with ATM showing variable degrees of successful results48. It is worth mentioning that an in-vitro study was comparing the synergy of CAZ/AVI plus ATM with Meropenem-Vaborbactam plus ATM in NDM-1 non-OXA-48 like co-producer E. coli and K. pneumoniae strains which showed similar synergy against these CREs44. A single product formulation of aztreonam-avibactam is currently in phase III clinical trial, which will address many issues with using a two-drug combination like susceptibility testing and epidemiologic surveillance data43. Clinicians should be aware that resistance to CAZ/AVI in K. pneumoniae may emerge while on treatment, and meropenem susceptibility may be restored as previously reported by mutations in the omega loop of blaKPC in the carbapenemase-producing strain, which may require testing MICs every time a phenotypic or genotypic alteration occur49.
In the presented case, we started the CAZ/AVI combination with ATM on the 4th day of hospital admission, which resulted in rapid clearance of bacteremia (in one day). The decision to start this combination was guided by the available literature at that time (February 2019) and synergy testing using gradient diffusion strips showing positive synergy results. The fact that the patient avoided the indicated open heart surgery for valve replacement just by using this combination proves that it is an effective antimicrobial combination in similar cases.
Materials and methods
Bacterial isolate and antimicrobial susceptibility testing
The K. pneumoniae strain, namely SA-KpST14, was recovered from a 40-year-old female with rheumatic heart disease (RHD) at Prince Sultan Military Medical City, Riyadh, Saudi Arabia. The initial antimicrobial susceptibility testing was performed using the VITEK‑2 system (BioMerieux, Brussels, Belgium). Etest (bioMérieux, Durham, NC) was used to determine the MICs for aztreonam, imipenem, ceftazidime/avibactam and synergetic activities of antibiotic combinations listed in Table 1. Antimicrobial MIC interpretations were in accordance to CLSI guidelines. A zone of hope for antimicrobial activity was defined based on the definition of synergy (1 plus 1 equals more than 2); hence, a zone of hope is defined as (0 plus 0 equals more than 1)50. MICs of colistin were confirmed by broth dilution methods done according to CLSI guidelines. Carba-R test using GeneXpert system (Cepheid, USA) was initially used to detect the presence of carbapenemase-resistance genes.
Complete genome sequencing
Genomic DNA (gDNA) of the K. pneumoniae SA-KpST14 strain was extracted from an overnight culture on LB agar using the QIAamp DNA Mini Kit (QIAgen, Germany) according to the manufacturer instructions. The quality and purity of the extracted DNA were checked using the Nanodrop 2000 spectrophotometer (Thermofisher, USA) and Qubit 3.0 Fluorometer with the dsDNA HS (High sensitivity) kit (Thermofisher, USA). Short reads sequences were generated on the Illumina MiSeq platform using the Nextera-XT library preparation kit (Illumina, San Diego, CA). Long reads sequencing with the Oxford Nanopore Technology (ONT) were generated using the ligation sequencing kit SQK-LSK109 (Oxford Nanopore Technologies, Ltd., UK) on the MinION sequencer (Oxford Nanopore Technologies, Ltd., UK).
Hybrid genome assembly and annotation
The Oxford Nanopore MinION and Illumina MiSeq reads were assembled with Unicycler (version 0.4.8)51 or following the EToKi pipeline (Enterobase Tool Kit) (version 1.0)52 using the default settings. The hybrid genome assembly generated six contigs with an average sequencing coverage depth of 60×. The NCBI Prokaryotic Genome Annotation Pipeline was used for the annotation of the SA-KpST14 chromosome and plasmid sequences53. The genome of SA-KpST14 consisted of one 5,378,785 bp chromosome and five plasmids designated pSA_KpST14-NDM-1 (269,329 bp, 46% GC content), pSA_KpST14-OXA48-2 (68,932 bp, 51% GC content), pSA_KpST14-3 (166,565 bp, 50% GC content), pSA_KpST14-4 (20.912 bp, 53% GC content), and pSA_KpST14-5 (2,095 bp, 44% GC content) (Table 2).
Identification of antibiotic resistance, virulence genes and plasmid replicon typing
Identification of antibiotic resistance genes and virulence factors were determined with ABRicate (https://github.com/tseemann/abricate) (version 0.9.8) using the ResFinder (version 2.1)54, Comprehensive Antimicrobial Resistance (CARD)55, virulence factors (VFDB)56 and Kaptive (version 0.7.3)57 databases. Basic plasmid characteristics were determined using the PlasmidFinder (version 1.3)58 software. Plasmid maps were drawn using the Blast Ring Image Generator (BRIG)59 software and Easyfig60.
Ethical approval
This study was approved by the Cardiac Research Department of Prince Sultan Cardiac Center in Riyadh, Saudi Arabia (Reference number R21004). We confirm that all research in this study was performed in accordance with the relevant guidelines and regulations after obtaining informed consent for conducting and publishing this study. All rules and regulations of ICH-GCP and the Declaration of Helsinki were followed.
Informed consent
Informed written consent was taken from the patient to conduct and publish this study and no personal data will be disclosed or breached beyond the principal investigator.
Acknowledgements
MFA acknowledges funding from King Abdullah International Medical Research Center (RC17/027 and RC18/374) as part of the funding program to monitor antimicrobial resistance and develop antimicrobial agents.
Author contributions
M.F.A., M.A., M.D. designed the study. M.F.A., L.O., B.A., M.A.A., A.A.A. and M.D. did the majority of the experimental work, including antimicrobial susceptibility testing, library preparation and WGS, and bioinformatic analysis. M.A., Y.S.A., A.A. and M.B. treated the patient as part of a multidisciplinary team, collected and wrote the patient’s clinical data. M.F.A., M.A. and M.D. wrote the manuscript. All authors provided critical feedback and helped shape the clinical research, analysis and manuscript.
Data availability
The complete genome sequence of K. pneumoniae isolate SA_KpST14 has been deposited in GenBank under accession no. CP071279 for the chromosome, CP071280 for pSA_KpST14-NDM-1, CP071281 for pSA_KpST14-OXA48-2, CP071282 for pSA_KpST14-3, CP071283 for pSA_KpST14-4, CP071284 for pSA_KpST14-5. These sequences are part of BioProject no. PRJNA705688.
Competing interets
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Majed F. Alghoribi and Moayad Alqurashi.
References
- 1.Wyres KL, Lam MMC, Holt KE. Population genomics of Klebsiella pneumoniae. Nat. Rev. Microbiol. 2020;18:344–359. doi: 10.1038/s41579-019-0315-1. [DOI] [PubMed] [Google Scholar]
- 2.Paczosa MK, Mecsas J. Klebsiella pneumoniae: Going on the offense with a strong defense. Microbiol. Mol. Biol. Rev. 2016;80:629–661. doi: 10.1128/MMBR.00078-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benenson S, et al. Carbapenem-resistant Klebsiella pneumoniae endocarditis in a young adult Successful treatment with gentamicin and colistin. Int. J. Infect. Dis. 2009;13:e295–e298. doi: 10.1016/j.ijid.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 4.Raymond T, Wiesen J, Rehm S, Auron M. Carbapenem-resistant klebsiella pneumoniae prosthetic valve endocarditis: A feared combination of technology and emerging pathogens. Infect. Dis. Clin. Pract. 2014;22:113–115. doi: 10.1097/IPC.0b013e318287c881. [DOI] [Google Scholar]
- 5.Brouqui P, Raoult D. Endocarditis due to rare and fastidious bacteria. Clin. Microbiol. Rev. 2001;14:177–207. doi: 10.1128/CMR.14.1.177-207.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riangwiwat T, Dworkin J. Tricuspid valve infective endocarditis due to Klebsiella pneumoniae in intravenous drug user Hawaii. J. Med. Public Health. 2019;78:98–102. [PMC free article] [PubMed] [Google Scholar]
- 7.Habib G, et al. 2015 ESC guidelines for the management of infective endocarditis. Eur. Heart J. 2015;36:3075–3123. doi: 10.1093/eurheartj/ehv319. [DOI] [PubMed] [Google Scholar]
- 8.Iacovelli A, et al. A challenging case of carbapenemase-producing Klebsiella pneumoniae septic thrombophlebitis and right mural endocarditis successfully treated with ceftazidime/avibactam. Infection. 2018;46:721–724. doi: 10.1007/s15010-018-1166-9. [DOI] [PubMed] [Google Scholar]
- 9.Chaari A, et al. Efficacy of tigecycline-colistin combination in the treatment of carbapenem-resistant Klebsiella pneumoniae endocarditis. J. Glob. Antimicrob. Resist. 2015;3:214–216. doi: 10.1016/j.jgar.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 10.Zimhony O, et al. Endocarditis caused by extended-spectrum-β-lactamase-producing Klebsiella pneumoniae: Emergence of resistance to ciprofloxacin and piperacillin-tazobactam during treatment despite initial susceptibility. Antimicrob. Agents Chemother. 2006;50:3179–3182. doi: 10.1128/AAC.00218-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karakonstantis S, Kritsotakis EI, Gikas A. Pandrug-resistant gram-negative bacteria: A systematic review of current epidemiology, prognosis and treatment options. J. Antimicrob. Chemother. 2020;75:271–282. doi: 10.1093/jac/dkz401. [DOI] [PubMed] [Google Scholar]
- 12.Balkhy HH, et al. Ten-year resistance trends in pathogens causing healthcare-associated infections; reflection of infection control interventions at a multi-hospital healthcare system in Saudi Arabia, 2007–2016. Antimicrob. Resist. Infect. Control. 2020;9:21. doi: 10.1186/s13756-020-0678-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alghoribi MF, et al. Genomic analysis of the first KPC-producing Klebsiella pneumoniae isolated from a patient in Riyadh: A new public health concern in Saudi Arabia. J. Infect. Public Health. 2020;13:647–650. doi: 10.1016/j.jiph.2020.01.003. [DOI] [PubMed] [Google Scholar]
- 14.Hala S, et al. First report of Klebsiella quasipneumoniae harboring bla KPC-2 in Saudi Arabia. Antimicrob. Resist. Infect. Control. 2019;8:2. doi: 10.1186/s13756-019-0653-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wyres KL, et al. Distinct evolutionary dynamics of horizontal gene transfer in drug resistant and virulent clones of Klebsiella pneumoniae. PLoS Genet. 2019;15:2. doi: 10.1371/journal.pgen.1008114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kofteridis DP, et al. Treatment pattern, prognostic factors, and outcome in patients with infection due to pan-drug-resistant gram-negative bacteria. Eur. J. Clin. Microbiol. Infect. Dis. 2020;39:965–970. doi: 10.1007/s10096-019-03784-9. [DOI] [PubMed] [Google Scholar]
- 17.Sader HS, Castanheira M, Shortridge D, Mendes RE, Flamm RK. Antimicrobial activity of ceftazidime-avibactam tested against multidrug-resistant enterobacteriaceae and Pseudomonas aeruginosa isolates from US Medical Centers, 2013 to 2016. Antimicrob. Agents Chemother. 2017;61:2. doi: 10.1128/AAC.01045-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chew KL, et al. Aztreonam-avibactam combination restores susceptibility of aztreonam in dual-carbapenemase-producing enterobacteriaceae. Antimicrob. Agents Chemother. 2018;62:2. doi: 10.1128/AAC.00414-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alraddadi BM, et al. Efficacy of ceftazidime-avibactam in the treatment of infections due to carbapenem-resistant enterobacteriaceae. BMC Infect. Dis. 2019;19:772. doi: 10.1186/s12879-019-4409-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Temkin E, et al. Ceftazidime-avibactam as salvage therapy for infections caused by carbapenem-resistant organisms. Antimicrob. Agents Chemother. 2017;61:2. doi: 10.1128/AAC.01964-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fiore M, et al. Ceftazidime-avibactam combination therapy compared to ceftazidime-avibactam monotherapy for the treatment of severe infections due to carbapenem-resistant pathogens: A systematic review and network meta-analysis. Antibiotics. 2020;9:388. doi: 10.3390/antibiotics9070388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shirley M. Ceftazidime-avibactam: A review in the treatment of serious gram-negative bacterial infections. Drugs. 2018;78:675–692. doi: 10.1007/s40265-018-0902-x. [DOI] [PubMed] [Google Scholar]
- 23.Lam MMC, et al. Population genomics of hypervirulent Klebsiella pneumoniae clonal-group 23 reveals early emergence and rapid global dissemination. Nat. Commun. 2018;9:1–10. doi: 10.1038/s41467-017-02088-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lam MMC, et al. Genetic diversity, mobilisation and spread of the yersiniabactin-encoding mobile element ICEKp in klebsiella pneumoniae populations. Microb. Genom. 2018;4:2. doi: 10.1099/mgen.0.000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farzand R, et al. A virulence associated siderophore importer reduces antimicrobial susceptibility of Klebsiella pneumoniae. Front. Microbiol. 2021;12:52. doi: 10.3389/fmicb.2021.607512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alcántar-Curiel MD, et al. Multi-functional analysis of Klebsiella Pneumoniae fimbrial types in adherence and biofilm formation. Virulence. 2013;4:129–138. doi: 10.4161/viru.22974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bachman MA, Lenio S, Schmidt L, Oyler JE, Weiser JN. Interaction of lipocalin 2, transferrin, and siderophores determines the replicative niche of Klebsiella pneumoniae during pneumonia. MBio. 2012;3:2. doi: 10.1128/mBio.00224-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu M, Valdebenito M, Winkelmann G, Hantke K. Functions of the siderophore esterases IroD and IroE in iron-salmochelin utilization. Microbiology. 2005;151:2363–2372. doi: 10.1099/mic.0.27888-0. [DOI] [PubMed] [Google Scholar]
- 29.Alteri CJ, Mobley HLT. The versatile type VI secretion system. Microbiol. Spectr. 2016;4:2. doi: 10.1128/microbiolspec.VMBF-0026-2015. [DOI] [Google Scholar]
- 30.Hsieh PF, Lu YR, Lin TL, Lai LY, Wang JT. Klebsiella pneumoniae type VI secretion system contributes to bacterial competition, cell invasion, type-1 fimbriae expression, and in vivo colonization. J. Infect. Dis. 2019;219:637–647. doi: 10.1093/infdis/jiy534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barbosa VAA, Lery LMS. Insights into Klebsiella pneumoniae type VI secretion system transcriptional regulation. BMC Genom. 2019;20:2. doi: 10.1186/s12864-019-5885-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fodah RA, et al. Correlation of Klebsiella pneumoniae comparative genetic analyses with virulence profiles in a murine respiratory disease model. PLoS ONE. 2014;9:e107394. doi: 10.1371/journal.pone.0107394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kelly SD, et al. Klebsiella pneumoniae O1 and O2ac antigens provide prototypes for an unusual strategy for polysaccharide antigen diversification. J. Biol. Chem. 2019;294:10863–10876. doi: 10.1074/jbc.RA119.008969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clarke BR, et al. A bifunctional O-antigen polymerase structure reveals a new glycosyltransferase family. Nat. Chem. Biol. 2020;16:450–457. doi: 10.1038/s41589-020-0494-0. [DOI] [PubMed] [Google Scholar]
- 35.Falcone M, et al. risk factors and outcomes of endocarditis due to non-HACEK gram-negative bacilli: Data from the prospective multicenter italian endocarditis study cohort. Antimicrob. Agents Chemother. 2018;62:1–11. doi: 10.1128/AAC.02208-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hassan SA, Akhtar A, Falah NU, Khan M. An unusual case of Klebsiella pneumoniae endocarditis. Cureus. 2020;12:e6999. doi: 10.7759/cureus.6999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bekker T, Govind A, Weber DM. A case of polymicrobial, gram-negative pulmonic valve endocarditis. Case Rep. Infect. Dis. 2019;2019:6439390. doi: 10.1155/2019/6439390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saitto G, et al. Infectious aortic root pseudoaneurysm after bentall procedure: To treat or not to treat by redo operation? Aorta. 2019;7:90–92. doi: 10.1055/s-0039-1694013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wyres KL, et al. Distinct evolutionary dynamics of horizontal gene transfer in drug resistant and virulent clones of Klebsiella pneumoniae. PLoS Genet. 2019;15:e1008114. doi: 10.1371/journal.pgen.1008114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baron SA, et al. Successful treatment and digestive decolonisation of a patient with osteitis caused by a carbapenemase-producing Klebsiella pneumoniae isolate harbouring both NDM-1 and OXA-48 enzymes. J. Glob. Antimicrob. Resist. 2019;18:225–229. doi: 10.1016/j.jgar.2019.06.001. [DOI] [PubMed] [Google Scholar]
- 41.Tamma, P. D. et al. Infectious Diseases Society of America Guidance on the Treatment of Extended-Spectrum β-lactamase Producing Enterobacterales (ESBL-E), Carbapenem-Resistant Enterobacterales (CRE), and Pseudomonas aeruginosa with Difficult-to-Treat Resistance (DTR-P. aerug. Clin. Infect. Dis.21287, 1–15 (2020). [DOI] [PubMed]
- 42.Benchetrit L, Mathy V, Armand-Lefevre L, Bouadma L, Timsit J-F. Successful treatment of septic shock due to NDM-1-producing Klebsiella pneumoniae using ceftazidime/avibactam combined with aztreonam in solid organ transplant recipients: report of two cases. Int. J. Antimicrob. Agents. 2020;55:105842. doi: 10.1016/j.ijantimicag.2019.10.023. [DOI] [PubMed] [Google Scholar]
- 43.Tan X, et al. Therapeutic options for metallo-β-lactamase-producing enterobacterales. Infect. Drug Resist. 2021;14:125–142. doi: 10.2147/IDR.S246174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Biagi M, et al. Searching for the optimal treatment for metallo- and serine-β-lactamase producing enterobacteriaceae: Aztreonam in combination with ceftazidime-avibactam or meropenem-vaborbactam. Antimicrob. Agents Chemother. 2019;63:2. doi: 10.1128/AAC.01426-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marshall S, et al. Can ceftazidime-avibactam and aztreonam overcome β-lactam resistance conferred by metallo-β-lactamases in enterobacteriaceae? Antimicrob. Agents Chemother. 2017;61:1–9. doi: 10.1128/AAC.02243-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shah PJ, Tran T, Emelogu F, Tariq F. Aztreonam, ceftazidime/avibactam, and colistin combination for the management of carbapenemase-producing klebsiella pneumoniae bacteremia: A case Report. J. Pharm. Pract. 2019 doi: 10.1177/0897190019882262. [DOI] [PubMed] [Google Scholar]
- 47.Shaw E, et al. Clinical outcomes after combination treatment with ceftazidime/avibactam and aztreonam for NDM-1/OXA-48/CTX-M-15-producing Klebsiella pneumoniae infection. J. Antimicrob. Chemother. 2018;73:1104–1106. doi: 10.1093/jac/dkx496. [DOI] [PubMed] [Google Scholar]
- 48.Emeraud C, et al. Aztreonam plus clavulanate, tazobactam, or avibactam for treatment of infections caused by metallo-β-lactamase-producing gram-negative bacteria. Antimicrob. Agents Chemother. 2019;63:1–7. doi: 10.1128/AAC.00010-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shields, R. K. et al. Emergence of Ceftazidime-Avibactam Resistance and Restoration of Carbapenem Susceptibility in Klebsiella pneumoniae Carbapenemase-Producing K pneumoniae: A Case Report and Review of Literature. Open forum Infect. Dis.4, ofx101 (2017). [DOI] [PMC free article] [PubMed]
- 50.Avery LM, Nicolau DP. Assessing the in vitro activity of ceftazidime/avibactam and aztreonam among carbapenemase-producing Enterobacteriaceae: Defining the zone of hope. Int. J. Antimicrob. Agents. 2018;52:688–691. doi: 10.1016/j.ijantimicag.2018.07.011. [DOI] [PubMed] [Google Scholar]
- 51.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou Z, Alikhan NF, Mohamed K, Fan Y, Achtman M. The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res. 2020;30:138–152. doi: 10.1101/gr.251678.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haft DH, et al. RefSeq: An update on prokaryotic genome annotation and curation. Nucleic Acids Res. 2018;46:D851–D860. doi: 10.1093/nar/gkx1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zankari E, et al. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jia B, et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017;45:D566–D573. doi: 10.1093/nar/gkw1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen L, Zheng D, Liu B, Yang J, Jin Q. VFDB 2016: Hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res. 2016;44:D694–D697. doi: 10.1093/nar/gkv1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wyres KL, et al. Identification of Klebsiella capsule synthesis loci from whole genome data. Microb. genomics. 2016;2:e000102. doi: 10.1099/mgen.0.000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Carattoli A, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011;12:2. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sullivan MJ, Petty NK, Beatson SA. Easyfig: A genome comparison visualizer. Bioinformatics. 2011;27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The complete genome sequence of K. pneumoniae isolate SA_KpST14 has been deposited in GenBank under accession no. CP071279 for the chromosome, CP071280 for pSA_KpST14-NDM-1, CP071281 for pSA_KpST14-OXA48-2, CP071282 for pSA_KpST14-3, CP071283 for pSA_KpST14-4, CP071284 for pSA_KpST14-5. These sequences are part of BioProject no. PRJNA705688.