Highlights
-
•
We estimated the hippocampal subfields volumes of 37 SCD and 40 controls using 7T MRI.
-
•
The SCD group presented significantly smaller volumes in the DG+CA2+CA3 bilaterally.
-
•
CA1 bilaterally and left ErC also presented a non-significant trend towards reduction.
-
•
Future studies are necessary to clarify the potential impact on cognitive performance.
Keywords: Sickle cell disease, Hippocampal segmentation, UHF MRI, 7T MRI, Brain imaging
Abstract
Sickle cell disease (SCD) is an inherited hemoglobinopathy that causes organ dysfunction, including cerebral vasculopathy and neurological complications. Hippocampal segmentation with newer and advanced 7 Tesla (7T) MRI protocols has revealed atrophy in specific subregions in other neurodegenerative and neuroinflammatory diseases, however, there is limited evidence of hippocampal involvement in SCD. Thus, we explored whether SCD may be also associated with abnormalities in hippocampal subregions. We conducted 7T MRI imaging in individuals with SCD, including the HbSS, HbSC and HbS/beta thalassemia genotypes (n = 53), and healthy race and age-matched controls (n = 47), using a customized head coil. Both T1- and T2-weighted images were used for automatic segmentation of the hippocampal subfields. Individuals with SCD had, on average, significantly smaller volume of the region including the Dentate Gyrus and Cornu Ammonis (CA) 2 and 3 as compared to the control group. Other hippocampal subregions also showed a trend towards smaller volumes in the SCD group. These findings support and extend previous reports of reduced volume in the temporal lobe in SCD patients. Further studies are necessary to investigate the mechanisms that lead to structural changes in the hippocampus subfields and their relationship with cognitive performance in SCD patients.
1. Introduction
Sickle cell disease (SCD) consists of a group of heterogeneous syndromes that share the inheritance of a mutated sickle hemoglobin (hemoglobin S, or HbS) (ter Maaten and Arogundade, 2010). SCD is one of the most common genetic disorders, with more than 5 million newborns carrying the mutated gene and more than 300,000 individuals being born with the disease yearly (Piel et al., 2013). The most predominant and severe type of SCD is caused by the homozygous inheritance of HbS (HbSS disease, or sickle cell anemia) (Davies and Gilmore, 2003). SCD may also originate from co-inheritance of HbS with other hemoglobin mutations, such as hemoglobin C (HbSC) or β-thalassemia (HbSβ-thalassemia) (Davies and Oni, 1997). β-thalassemia leads to absent (β0) or reduced (β+) synthesis of the beta-globin chains, and the coinheritance of HbS with β-thalassemia can be further categorized into two forms: HbSβ+ thalassemia and HbSβ0 thalassemia (Galanello and Origa, 2010). The genotypes HbSC and HbSβ+ tend to result in milder phenotypes, while HbSβ0 thalassemia may be as severe as HbSS disease (Serjeant, 1997).
Neurological complications of SCD include increased risk of stroke, silent cerebral infarction, and cognitive impairment, and are more common in individuals with HbSS disease (Dowling et al., 2010, Mackin et al., 2014); pediatric stroke happens almost exclusively in individuals with HbSS. HbSS individuals also tend to display lower cognitive performance when compared with individuals with HbSC (Jorgensen et al., 2017), possibly because anemia is less pronounced in HbSC. However, little research has been done on the impact of HbSC on the brain in adults.
Reduced cortical and subcortical volume has been previously reported in individuals with SCD when compared with healthy controls. Kirk et al (Kirk et al., 2009) showed cortical thinning in multiple brain regions in patients with SCD ranging from 5 to 21 years-old. They hypothesized that cortical thinning could be related with impaired cerebral perfusion. Mackin et al. (Mackin et al., 2014) reported reduced cortical thickness in the temporal lobe in adults with SCD, while Kawadler et al. (Kawadler et al., 2013) found significantly reduced volume in the hippocampus of SCD pediatric patients with silent cerebral infarction. Overall, evidence of the impact of SCD on brain regions remains limited, particularly in adults.
The hippocampus is a brain structure situated in the medial temporal lobe comprised of the Cornu Ammonis (CA) – which is further divided into three conventional histological subregions, CA1, CA2, and CA3 – the Dentate Gyrus (DG), and the Subiculum (Sub) (Flores et al., 2020). Additionally, the hippocampal formation includes the Entorhinal Cortex (ErC) (Witter et al., 2017). The hippocampal involvement in higher cognitive processing has been well investigated, especially as pertains the hippocampal primary role in short-term and episodic long-term memory and learning (Amaral and Witter, 1989). Hippocampal subfields are functionally interconnected, however functionalities are known to vary among the different subfields. For example, DG is suggested to be a preprocessor of incoming information, preparing it for subsequent processing in CA3, and potentially mediating learning, memory, and spatial encoding (Jonas and Lisman,). ErC, on the other hand, serves as the major input–output structure and mediates hippocampal connectivity with cortical regions (Witter et al.,). Cells in the DG receive excitatory input from ErC and send excitatory output to the CA3 region via the mossy fibers (Jonas and Lisman,). Also, CA1 and CA3 are believed to contribute to episodic memory processing (Hunsaker, 2008, Zheng et al., 2018). CA2 is the smallest subfield but studies found that lesions in this area can lead to abnormal social behavior by impairing social recognition memory (Stevenson and Caldwell, 2014).
As pathology within the hippocampus is increasingly linked to several neurological and neuropsychiatric diseases such as Alzheimer’s disease, epilepsy, and depression (Mueller et al., 2013, Mueller et al., 2007, Wisse et al., 2014, Small et al., 2011, de Flores et al., 2015), interest in the subfields of the hippocampus has grown. Subregions of the hippocampus differ in their neuronal electrophysiological (Milior et al., 2016), therefore, different stimuli can have different effects on each subfield depending on the disease (West et al., 2004). For instance, studies on humanized SCD mice and ischemic mice reported significant cognitive impairment that is associated with degeneration in the hippocampal subfields CA1, CA2, and CA3 (Olsson et al., 2003, Wang et al., 2016), while in Alzheimer’s disease, CA1 degeneration is considered an early biomarker (de Flores et al., 2015).
Recent advances in MRI techniques, such as 7 Tesla (T) MRI, have improved the spatial resolution and signal-to-noise ratio of MRI images (Krishnamurthy et al., 2019). These improvements enable more accurate volumetric analysis of the hippocampal subfields in in vivo studies and increase the performance of automatic segmentation methods (Knierim, 2015, Yushkevich et al., 2015, Boccardi et al., 2011, Iglesias et al., 2015, Giuliano et al., 2017). Manual segmentation of hippocampal subregions is time-consuming and labor intensive, limiting data collection and processing in large studies (Wisse et al., 2014, Iglesias et al., 2015). To compensate for this shortcoming, automated segmentation methods have been developed (Yushkevich et al., 2015, Iglesias et al., 2015, Yang et al., 2020).
This study aims to investigate how SCD impacts the volumes of hippocampal subfields. We used 7T MRI and an innovative radiofrequency (RF) coil to acquire high-resolution images of the hippocampal formation from a cohort of SCD participants and controls (N = 100). 7T tailored preprocessing and automatic segmentation software (Yushkevich et al., 2015) were used to compute the volumes of the hippocampal subfields. Based on previous evidence of atrophy in the hippocampus of SCD patients, our study aimed at verifying whole hippocampal volume reductions as well as exploring volume changes of the subfields. To the best of our knowledge, our study is the first to analyze SCD hippocampal subregions with high resolution 7T MRI, and to include both severe and milder genotypes of SCD.
2. Methods
2.1. Participants
Patients with SCD were recruited from the University of Pittsburgh Medical Center (UPMC) Adult Sickle Cell Program outpatient clinic under the University of Pittsburgh IRB protocols PRO12040139 and PRO08110422. Data were collected from 53 patients and 47 controls as part of a longitudinal study of neuroradiological biomarkers in SCD (ClinicalTrials.gov Identifier: NCT02946905). All patients with HbSS, HbSC and HbSβ thalassemia older than 18 years old and able to provide informed consent were informed about the study by staff members during their routine clinic visit, and offered entry into the study if they were in steady-state SCD (i.e. two weeks from an acute illness, including vaso-occlusive pain episodes). Eligibility criteria also included: 1) English-speaking; and 2) currently receiving routine follow-up care at the UPMC Adult Sickle Cell Program. Exclusion criteria included: 1) pregnancy, as determined by a positive urine human chorionic gonadotropin test at the time of the of the MRI; 2) acute medical problems including, but not limited to, acute vaso-occlusive episodes. Healthy controls, included in ClinicalTrials.gov identifier NCT02946905, were recruited from the community via brochures, and from the University of Pittsburgh Clinical and Translational Science Institute Pitt + Me Registry if older than 18 and able to provide informed consent. Healthy controls were age, race, and sex matched to the participants with SCD. Both patients with SCD and controls were excluded if pregnant or lactating, had any medical condition that could result in neurocognitive or brain dysfunction (other than those resulting from SCD), including diabetes mellitus, coronary artery disease, peripheral vascular disease, and other causes of cerebral vasculitis such as Systemic Lupus Erythematosus (SLE), or contraindications to MRI scanning such as electronic implants, magnetically-activated implants, tattoos above the shoulders, or brain implants.
2.2. MRI acquisition
MRI images were acquired using a 7T human MRI scanner (Siemens Magnetom, Germany) with a customized radiofrequency (RF) head coil with 16 transmit channels and 32 receive channels (Ibrahim et al., 2013), which produces homogenous images and whole brain coverage at 7T (Smagula et al., 2018, Krishnamurthy et al., 2019, Santini et al., 2018a, Santini et al., 2021; Ibrahim et al., 2017). T1-weighted (T1w) Magnetization Prepared RApid Gradient Echo (MPRAGE) was acquired with 0.75 mm isotropic resolution and the following parameters: TE/TI/TR = 2.17/1200/3000 ms, bandwidth = 391 Hz/Px, acceleration factor (Grappa) = 2, time of acquisition = 5:02 min. T2-weighted (T2w) images were acquired with high resolution in the plane perpendicular to the main axis of the hippocampus, slightly slanted from coronal acquisition. The sequence utilized was 2D turbo spin echo (TSE) with the following parameters: resolution = 0.375 × 0.375 × 1.5 mm3, TE/TR = 61/10060 ms, bandwidth = 264, acceleration factor (Grappa) = 2, time of acquisition = 3:32 min. Gradient echo (GRE) sequence – utilized as a secondary contrast for the calculations of the intracranial volume masks – was acquired with the following parameters: TE/TR = 8.16/24 ms, resolution 0.375 × 0.375 × 0.75 mm, acceleration factor 2, time of acquisition 8:20 min.
2.3. Image preprocessing and hippocampus segmentation
Both T1w and T2w images were denoised using an optimal forward and inverse variance-stabilizing transformation (VST), specifically designed for the Rice distribution (Foi, 2011). The forward VST converts the Rician heteroscedastic noise into a homoscedastic one (approximately Gaussian distributed), and thus the block-matching 4D (BM4D) denoising algorithm (Maggioni et al., 2013) was applied, followed by the inverse VST, to obtain the denoised images (Santini et al., 2018b). The images were then bias corrected with the SPM12 package using 30 mm of cutoff for the full width at half maximum (against 60 mm of the default) to account for the increased dielectric artifacts at higher field strength (Ibrahim et al., 2001). Fig. 1 shows the preprocessing methods applied in a representative T2w image from this study. The bias correction and denoising methods improved homogeneity and mitigated noise in the images.
Fig. 1.
Preprocessing of the T2-weighted images in a representative SCD patient. In a), raw image from the 7T scanner. In b), bias corrected image. In c), bias corrected and denoised image.
The automatic segmentation of hippocampal subfields (ASHS) package (Yushkevich et al., 2015) and a 7T atlas from young adults’ data (Berron et al., 2017) were utilized to segment the hippocampal subfields. The images from the atlas were denoised using the same technique described above for the T1w and T2w images. The results of the segmentation were manually corrected when the fixes were obvious, such as removing pixels that do not overlap with the hippocampus, extending the segmentations until the boundaries of the hippocampal subfields, and removing cysts that were classified as hippocampal subregions (Berron et al., 2017). Fig. 2 shows an example where the manual fixes were applied; minor manual corrections were performed in most of the subjects using the software ITK-SNAP (Yushkevich et al., 2006). In 23 of the 100 subjects, either the automatic segmentation failed or the manual fixes were not obvious (mostly due to the lack of contrast caused by excessive motion artifact); these subjects were excluded from the analysis, yielding a final sample size of 77. Fig. 3 shows a T2-weighted acquisition, the hippocampal segmentation, and its 3D reconstruction in a representative SCD patient. The preprocessing techniques applied before segmentation with the ASHS package produced a smoother and more accurate segmentation of the hippocampal subfields. A summary of the methods is shown in Fig. 4.
Fig. 2.
Multiple slices of the original and manually corrected segmentation of the hippocampal subfields overlayed on the T2-weighted MR images. The “Raw segmentation” shows the output from the ASHS software. The “Corrected segmentation” shows the manual corrections applied to the segmentations. The corrected regions are indicated by the yellow arrows. The manual corrections were performed as follows: 1) pixels that are not connected to the rest of the segmentation were removed (a); 2) the closest label was extended in hippocampal regions where segmentation labels were missing, (b, d, and e); 3) cysts that were mislabeled as another hippocampal region were removed (c); and 4) segmentations that extended outside the hippocampus were removed (i). In the slices f and h, there were no manual corrections in the segmentations. The color code is the same as in Fig. 3. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 3.
Example of hippocampal subfields segmentation in a patient with SCD. a) coronal slice of the T2-weighted image; b, c) zoomed image showing details of the hippocampus structure, subject right and left, respectively; d, e) hippocampal subfields segmentations overlaying the T2-weighted image, subject right and left, respectively; f) 3D reconstruction of the hippocampal subfield segmentations. Abbreviations - cornu ammonis 1–3, CA1-3; dentate gyrus, DG; hippocampal tail, Tail; subiculum, Sub; entorhinal cortex, ErC.
Fig. 4.
Flowchart of the method implemented for the hippocampal subfields segmentation and analysis. T1w, T1-weighted contrast; T2w, T2-weighted contrast; GRE: gradient echo sequence.
Intracranial volume is often used as a covariate in brain volumetric analysis; intracranial masks were calculated with the SPM12 package using the MPRAGE and GRE acquisitions as channels for the segmentations after a co-registration. The segmentations for the white matter, gray matter, and cerebrospinal fluid were combined, and the missing pixels in the segmentation were filled using the MATLAB function imfill. Imperfections in the intracranial segmentations were manually corrected using the ITK-snap software (Yushkevich et al., 2006).
2.4. Statistical analysis
Sample characteristics were compared between SCD groups and controls using one-way ANOVA. The volumes of the hippocampal subfields were compared for patients and controls using analysis of covariance (ANCOVA), with sex, age, and intracranial volume as covariates. The hippocampal subregions DG, CA2, and CA3 were combined (DG + CA2 + CA3), as the small volumes of CA2 and CA3 could lead to inaccurate labeling. An analysis separating these regions is presented in the Supplementary Material (Table S1). Those measures that differed by SCD status (patients and controls) were considered for inclusion in multivariable models testing the associations of SCD status and neuroimaging measures. The p-value threshold for between-group differences was calculated based on the Bonferroni correction. The statistical analysis was performed using SPSS (version 26, IBM, Chicago, IL).
3. Results
3.1. Demographics and hemoglobin values of the participants
There was no statistically significant difference in the age or sex between the groups (Table 1). As expected, the hemoglobin levels were significantly different between groups, with the severe SCD group (HbSS and HbSβ0) having the lowest Hb level (9.10 ± 1.09 g/dL), the milder SCD group (HbSC and HbSβ+) having an intermediate level (11.25 ± 2.48 g/dL), and the controls having the highest Hb level (13.94 ± 1.55 g/dL) at the time of the assessment (Table 1).
Table 1.
Sample characteristics.
| SCD HbSS, HbSβ0 (severe) | SCD HbSC, HbSβ+ (milder) | Controls | F-statistic (p-value) | |
|---|---|---|---|---|
| N | 16 | 21 | 40 | – |
| Age, mean (SD) | 34.31(10.23) | 35.29(12.24) | 36.15(13.24) | 0.131 (0.88) |
| Sex, F/M | 7/9 | 9/12 | 16/24 | 0.41 (0.96) |
| Hb1, mean (SD) | 9.10 (1.09) | 11.25 (2.48) | 13.94 (1.55) | 43.5 (6.9E-13) * |
1 in g/dL
* p-value < 0.05.
3.2. Neuroimaging characteristics of patients with SCD vs. Controls
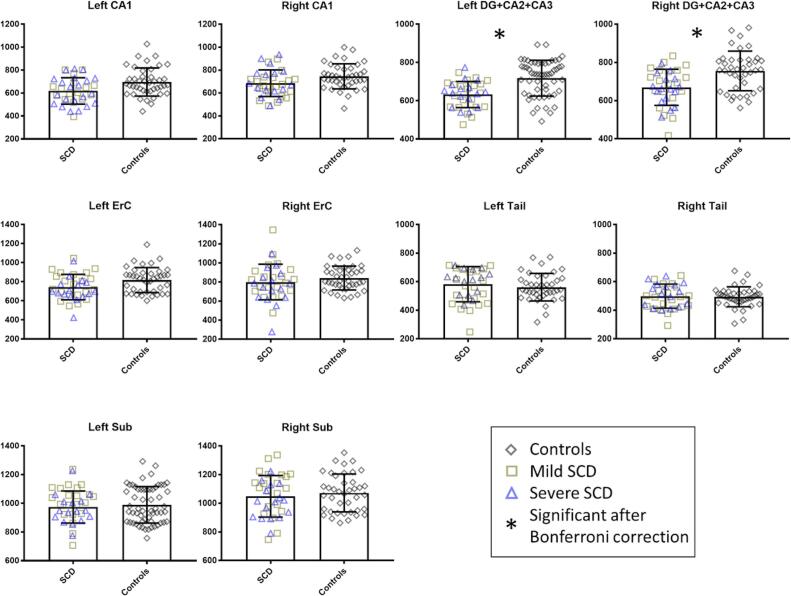
Results of the comparison of the hippocampal subfields volumes between SCD patients and controls are tabulated in Table 2. Fig. 5 shows the bar plots with the individual volumetric data points for all the subjects included in this study. Significant differences between the hippocampal volumes of the SCD and control groups were observed bilaterally in the region DG + CA2 + CA3: − 11.55% (F = 20.79, p-value = 0.020 × 10-3) in the left hippocampus, and − 11.36% (F = 14.09, p-value = 0.350 × 10−3) in the right hippocampus. There was also a trend towards a reduction of the left CA1 (− 10.52%, F = 5.69, p-value 0.020), right CA1 (−8.02%, F = 4.02, p-value 0.049), and left ErC (−9.03%, F = 5.68, p-value 0.020), which was not statistically significant after Bonferroni correction. The other hippocampal subregions, including the right ErC, bilateral Tail, and bilateral Sub, did not significantly differ between SCD and control groups. The whole left (−7.29%, F = 7.29, p-value 0.009) and right hippocampus (−6.57%, F = 4.93, p-value 0.030) showed a trend that did not remain statistically significant after Bonferroni correction. Between-group differences in the region DG + CA2 + CA3 were marginally attenuated after adjustment for intracranial volume (from 11.55% to 10.10% for the left hemisphere, and from 11.36% to 9.63% for the right hemisphere), and minimally attenuated with the inclusion of age and sex in the model, as shown in Table 3. No significant differences were found between the severe and milder SCD genotypes.
Table 2.
Neuroimaging characteristics of hippocampal subfields in subjects with sickle cell disease and matched controls.
| SCD (severe and milder) – Mean (SD) | Controls – Mean (SD) | Percentage difference SCD vs controls4 | F-statistic (p-valuec) | |
|---|---|---|---|---|
| Intracranial volume1 | 1,306,040 (114740) | 1,336,886 (130234) | −2.31% | 2.813 (0.097786)a |
| Total hippocampal volume1 | 4635.69 (541.56) | 4980.17 (557.54) | −6.92% | 6.400 (0.013601)b |
| Asymmetry Ratio2 | 0.03655 (0.02280) | 0.03385 (0.02242) | 7.97% | 0.475 (0.492749) b |
| Left Hemisphere | ||||
| Cornu Ammonis 11 | 619.02 (115.08) | 691.82 (122.65) | −10.52% | 5.69 (0.019689) b |
| DG + CA2 + CA31, 3 | 632.49 (70.68) | 715.05 (92.82) | −11.55% | 20.79 (0.000020)b* |
| Subiculum1 | 979.63 (116.29) | 999.65 (133.50) | −2.00% | 0.02 (0.899994) b |
| Entorhinal cortex1 | 743.54 (133.47) | 817.36 (131.31) | −9.03% | 5.68 (0.019773) b |
| Tail1, 3 | 583.22 (123.34) | 561.99 (96.57) | 3.78% | 1.34 (0.251466) b |
| Hippocampus1 | 2231.14 (250.92) | 2406.52 (281.49) | −7.29% | 7.29 (0.008650) b |
| Right Hemisphere | ||||
| Cornu Ammonis 11 | 685.96 (116.09) | 745.73 (108.93) | −8.02% | 4.02 (0.048854) b |
| DG + CA2 + CA33 | 669.81 (94.72) | 755.69 (103.89) | −11.36% | 14.09 (0.000350)b* |
| Subiculum1 | 1048.77 (145.29) | 1072.22 (131.94) | −2.19% | 0.01 (0.918181) b |
| Entorhinal cortex1 | 799.98 (186.52) | 841.61 (124.96) | −4.95% | 0.81 (0.371777) b |
| Tail1, 3 | 499.13 (84.39) | 494.83 (69.95) | 0.87% | 0.46 (0.501763) b |
| Hippocampus1 | 2404.55 (300.00) | 2573.64 (287.13) | −6.57% | 4.93 (0.029504) b |
*Significantly different after Bonferroni correction (p-value threshold: 0.05 / 10 subfields / 2 groups = 0.0025 for significance).
in mm3.
Defined as (right hippocampus - left hippocampus)/(right hippocampus + left hippocampus)*100.
Defined in the reference (Wisse et al., 2014).
Given 95% confidence interval.
Corrected for age and sex.
Corrected for age, sex, and intracranial volume.
uncorrected p-value.
Fig. 5.
Hippocampal subfield volume comparison between patients and controls. There was a significant volume difference between the patient and the control groups in the region DG + CA2 + CA3 bilaterally. Abbreviations - cornu ammonis 1–3, CA1-3; dentate gyrus, DG; entorhinal cortex, ErC; hippocampal Tail, Tail; subiculum, Sub. The error bars represent the standard deviation of the data. Controls (n = 40), Milder SCD (n = 21), Severe SCD (n = 16).
Table 3.
Multivariable Regression Models of Sickle Cell Disease Status predicting DG + CA2 + CA3 volume: beta coefficient and percentage change before and after adjustment for intracranial volume, age, and sex.
| Beta Coefficient (95% Confidence Intervals) in mm3, Percentage change, unadjusted p-value |
|||
|---|---|---|---|
| Unadjusted model | Model 1: adjusted for Intracranial volumes | Model 2: further adjusted for Age and Sex | |
| DG + CA2 + CA3, Right | 85.88 (40.62–131.13), 11.36%, 0.000312 | 72.76 (33.68–111.84), 9.63%, 0.000399 | 69.07 (32.39–105.75), 9.14%, 0.000350 |
| DG + CA2 + CA3, Left | 82.56 (44.87–120.24), 11.55%, 0.000040 | 72.25 (39.03–105.46), 10.10%, 0.000045 | 71.97 (40.51–103.43), 10.06%, 0.000020 |
4. Discussion
In this study, 7T MRI and innovative radiofrequency coil developments allowed the acquisition of high-resolution and high-quality images of the hippocampus. Preprocessing techniques such as denoising and bias correction further improved the image quality. Manual quality assurance and correction refined the automatic hippocampal segmentation method. The use of the GRE sequence with the T1-weighted MPRAGE also improved the estimation of the intracranial volume masks, although manual corrections were still necessary.
Compared with controls, our analysis shows that the region DG + CA2 + CA3 is the most affected hippocampal subregion in the SCD group, an effect observed with a mean difference of −11.55% on the left hemisphere and −11.36% on the right hemisphere. After adjusting for age, sex, and intracranial volume, the differences were marginally reduced, indicating that SCD is the major contributor to the overall difference. Moreover, the hippocampal subregions CA1 bilaterally, left ErC, and hippocampus bilaterally also showed a trend towards smaller volumes in the SCD group, but the differences were not statistically significant after Bonferroni correction.
These results are consistent with findings in pediatric populations with SCD. Kawadler et al. (2013) reported a volume difference of −6.74% in the right hippocampus and −10.26% in the left hippocampus in pediatric HbSS subjects with silent cerebral infarction when compared with healthy controls. If confirmed in future studies, our results indicate that the impact of SCD on hippocampal morphology reported in pediatric patients persists in adulthood. Future studies should assess the impact of these patterns of significant subregional hippocampal atrophy on cognitive decline.
Understanding how hippocampal subregion volumes differ in SCD may identify biomarkers of disease severity and cognition. For instance, our analysis found that the total hippocampal volume was not significantly different between patients and controls, whereas the DG + CA2 + CA3 subregion grouping showed a consistent volumetric difference between the two groups. This hippocampal region is believed to mediate learning, memory, and spatial encoding (Jonas and Lisman,), and abnormalities in this area could be associated with deficits in these cognitive domains in SCD.
The hippocampus is particularly vulnerable to hypoxia and inflammation (Iampietro et al., 2014), which are common pathogenic mechanisms in SCD. Mouse models of SCD reveal that hippocampal pathology is associated with cognitive deficits (Wang et al., 2016). Interestingly, we did not observe an association between the volume of hippocampal subregions and SCD genotype, despite evidence that individuals with milder genotypes tend to have better cognitive functioning (Jorgensen et al., 2017, Schatz, 2002). It is possible that compensatory mechanisms such as increased hippocampal activation and connectivity (Case et al., 2017, Hämäläinen et al., 2007) may confound the effect of hippocampal atrophy on cognition.
One limitation of this study is that hippocampal subregions may be inaccurately labeled as the adjacent subregions. In particular, CA2 and CA3 subregions are small and their borders do not present clear landmarks by MRI; therefore, they are more prone to mislabeling errors. On the other hand, the segmentation of DG is the most accurate (Yushkevich et al., 2015), since it follows well defined landmarks in the T2-weighted image. Nevertheless, only histological data could precisely identify all specific subregions. To reduce the impact of mislabeling, we combined CA2 and CA3 with DG, a common strategy (Yushkevich et al., 2015), and presented an exploratory analysis of these regions separately in the Supplementary Material. Additionally, the tail label is based on external landmarks (Flores et al., 2020) and may have limited meaningfulness, although some studies have found associations between the tail volume and clinical outcomes (Sozinova et al., 2008, Nogovitsyn et al., 2020). To further enhance the precision of our measurements, we excluded 23 subjects whose scans presented excessive motion artifacts, mostly due to artifacts in the T2w TSE acquisition. Future work in RF coil design, MRI sequence development, and image processing may lead to a higher inclusion rate.
In summary, we found that between-group differences in the volumes of hippocampal subfields followed a distinct spatial location, specifically more pronounced in DG + CA2 + CA3, bilaterally, as compared to other subregions. These associations were attenuated after adjustment for intracranial volume and demographics but remained significant. Further studies will be necessary to clarify the mechanisms that led to volume reduction in the hippocampal subfields and elucidate their significance as an imaging biomarker for cognitive deficits in individuals with SCD.
CRediT authorship contribution statement
Tales Santini: Software, Conceptualization, Methodology, Formal analysis, Writing - original draft, Writing - review & editing. Minseok Koo: Software, Writing - original draft, Writing - review & editing. Nadim Farhat: Methodology, Writing - review & editing. Vinicius P. Campos: Software, Writing - review & editing. Salem Alkhateeb: Methodology, Writing - review & editing. Marcelo A.C. Vieira: Software, Writing - review & editing. Meryl A. Butters: Methodology, Writing - review & editing. Caterina Rosano: Methodology, Formal analysis, Writing - original draft, Writing - review & editing. Howard J. Aizenstein: Conceptualization, Writing - review & editing. Joseph Mettenburg: Conceptualization, Writing - review & editing. Enrico M. Novelli: Conceptualization, Methodology, Writing - original draft, Supervision, Project administration, Funding acquisition, Writing - review & editing. Tamer S. Ibrahim: Conceptualization, Methodology, Resources, Writing - original draft, Supervision, Project administration, Funding acquisition, Writing - review & editing.
Acknowledgements
This work was supported by the National Institutes of Health under award numbers: R01HL127107, R01MH111265, R01AG063525, and T32MH119168. The first author was partially supported by CAPES Foundation, Ministry of Education of Brazil, under the award number 13385/13-5. This research was also supported in part by the University of Pittsburgh Center for Research Computing (CRC) through the resources provided.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nicl.2021.102655.
Contributor Information
Enrico M. Novelli, Email: noveex@upmc.edu.
Tamer S. Ibrahim, Email: tibrahim@pitt.edu.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- ter Maaten JC, Arogundade FA. Sickle cell disease. Comprehensive Clinical Nephrology: Elsevier; 2010. pp. 596–608.
- Piel F.B., Patil A.P., Howes R.E., Nyangiri O.A., Gething P.W., Dewi M., Temperley W.H., Williams T.N., Weatherall D.J., Hay S.I. Global epidemiology of sickle haemoglobin in neonates: a contemporary geostatistical model-based map and population estimates. Lancet. 2013;381(9861):142–151. doi: 10.1016/S0140-6736(12)61229-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies S.C., Gilmore A. The role of hydroxyurea in the management of sickle cell disease. Blood Rev. 2003;17(2):99–109. doi: 10.1016/s0268-960x(02)00074-7. [DOI] [PubMed] [Google Scholar]
- Davies S.C., Oni L. Fortnightly review: management of patients with sickle cell disease. BMJ. 1997;315(7109):656–660. doi: 10.1136/bmj.315.7109.656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanello R., Origa R. Beta-thalassemia. Orphanet J. Rare Dis. 2010;5(1):11. doi: 10.1186/1750-1172-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serjeant G.R. Sickle-cell disease. Lancet. 1997;350(9079):725–730. doi: 10.1016/S0140-6736(97)07330-3. [DOI] [PubMed] [Google Scholar]
- Dowling MM, Quinn CT, Rogers ZR, Buchanan GR. Acute silent cerebral infarction in children with sickle cell anemia. Pediatr Blood Cancer. 2010;54(3):461-4. Epub 2009/10/09. doi: 10.1002/pbc.22242. PubMed PMID: 19813251; PubMed Central PMCID: PMCPMC2807470. [DOI] [PMC free article] [PubMed]
- Mackin RS, Insel P, Truran D, Vichinsky EP, Neumayr LD, Armstrong FD, et al. Neuroimaging abnormalities in adults with sickle cell anemia: associations with cognition. Neurology. 2014;82(10):835-41. Epub 2014/02/14. doi: 10.1212/WNL.0000000000000188. PubMed PMID: 24523480; PubMed Central PMCID: PMCPMC3959758. [DOI] [PMC free article] [PubMed]
- Jorgensen D.R., Metti A., Butters M.A., Mettenburg J.M., Rosano C., Novelli E.M. Disease severity and slower psychomotor speed in adults with sickle cell disease. Blood Adv. 2017;1(21):1790–1795. doi: 10.1182/bloodadvances.2017008219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk GR, Haynes MR, Palasis S, Brown C, Burns TG, McCormick M, et al. Regionally specific cortical thinning in children with sickle cell disease. Cerebral Cortex. 2009;19(7):1549-56. [DOI] [PubMed]
- Kawadler J.M., Clayden J.D., Kirkham F.J., Cox T.C., Saunders D.E., Clark C.A. Subcortical and cerebellar volumetric deficits in paediatric sickle cell anaemia. Br. J. Haematol. 2013;163(3):373–376. doi: 10.1111/bjh.12496. [DOI] [PubMed] [Google Scholar]
- de Flores R, Berron D, Ding SL, Ittyerah R, Pluta JB, Xie L, et al. Characterization of hippocampal subfields using ex vivo MRI and histology data: Lessons for in vivo segmentation. Hippocampus. 2020;30(6):545-64. Epub 2019/11/02. doi: 10.1002/hipo.23172. PubMed PMID: 31675165; PubMed Central PMCID: PMCPMC7192771. [DOI] [PMC free article] [PubMed]
- Witter M.P., Doan T.P., Jacobsen B., Nilssen E.S., Ohara S. Architecture of the entorhinal cortex a review of entorhinal anatomy in rodents with some comparative notes. Front. Syst. Neurosci. 2017;11:46. doi: 10.3389/fnsys.2017.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D.G. Amaral M.P. Witter The three-dimensional organization of the hippocampal formation: A review of anatomical data Neuroscience. 31 3 1989 571 91 10.1016/0306-4522(89)90424-7. [DOI] [PubMed]
- Jonas P, Lisman J. Structure, function, and plasticity of hippocampal dentate gyrus microcircuits. Front Neural Circuits. 2014;8:107. Epub 2014/10/14. doi: 10.3389/fncir.2014.00107. PubMed PMID: 25309334; PubMed Central PMCID: PMCPMC4159971. [DOI] [PMC free article] [PubMed]
- Witter MP, Doan TP, Jacobsen B, Nilssen ES, Ohara S. Architecture of the Entorhinal Cortex A Review of Entorhinal Anatomy in Rodents with Some Comparative Notes. Front Syst Neurosci. 2017;11:46. Epub 2017/07/14. doi: 10.3389/fnsys.2017.00046. PubMed PMID: 28701931; PubMed Central PMCID: PMCPMC5488372. [DOI] [PMC free article] [PubMed]
- Hunsaker MR, Lee B, Kesner RP. Evaluating the temporal context of episodic memory: the role of CA3 and CA1. Behav Brain Res. 2008;188(2):310-5. Epub 2008/01/08. doi: 10.1016/j.bbr.2007.11.015. PubMed PMID: 18178264; PubMed Central PMCID: PMCPMC2675273. [DOI] [PMC free article] [PubMed]
- Zheng F, Cui D, Zhang L, Zhang S, Zhao Y, Liu X, et al. The Volume of Hippocampal Subfields in Relation to Decline of Memory Recall Across the Adult Lifespan. Front Aging Neurosci. 2018;10:320. Epub 2018/10/27. doi: 10.3389/fnagi.2018.00320. PubMed PMID: 30364081; PubMed Central PMCID: PMCPMC6191512. [DOI] [PMC free article] [PubMed]
- Stevenson E.L., Caldwell H.K. Lesions to the CA 2 region of the hippocampus impair social memory in mice. Eur. J. Neurosci. 2014;40(9):3294–3301. doi: 10.1111/ejn.12689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller S., Yushkevich P., Wang L., Van Leemput K., Mezher A., Iglesias J.E. Collaboration for a systematic comparison of different techniques to measure subfield volumes: announcement and first results. Alzheimer's & Dementia J. Alzheimer's Assoc. 2013;9(4):P51. [Google Scholar]
- Mueller S.G., Stables L., Du A.T., Schuff N., Truran D., Cashdollar N., Weiner M.W. Measurement of hippocampal subfields and age-related changes with high resolution MRI at 4 T. Neurobiol. Aging. 2007;28(5):719–726. doi: 10.1016/j.neurobiolaging.2006.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisse L.E.M., Biessels G.J., Heringa S.M., Kuijf H.J., Koek D.(H.).L., Luijten P.R., Geerlings M.I. Hippocampal subfield volumes at 7T in early Alzheimer's disease and normal aging. Neurobiol. Aging. 2014;35(9):2039–2045. doi: 10.1016/j.neurobiolaging.2014.02.021. [DOI] [PubMed] [Google Scholar]
- Small S.A., Schobel S.A., Buxton R.B., Witter M.P., Barnes C.A. A pathophysiological framework of hippocampal dysfunction in ageing and disease. Nat. Rev. Neurosci. 2011;12(10):585–601. doi: 10.1038/nrn3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Flores R., La Joie R., Landeau B., Perrotin A., Mézenge F., de La Sayette V., Eustache F., Desgranges B., Chételat G. Effects of age and Alzheimer's disease on hippocampal subfields: comparison between manual and FreeSurfer volumetry. Hum. Brain Mapp. 2015;36(2):463–474. doi: 10.1002/hbm.22640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- G. Milior M.A. Di Castro L.P. Sciarria S. Garofalo I. Branchi D. Ragozzino C. Limatola L. Maggi 6 1 2016 10.1038/srep38242. [DOI] [PMC free article] [PubMed]
- West MJ, Kawas CH, Stewart WF, Rudow GL, Troncoso JC. Hippocampal neurons in pre-clinical Alzheimer's disease. Neurobiology of aging. 2004;25(9):1205-12. Epub 2004/08/18. doi: 10.1016/j.neurobiolaging.2003.12.005. PubMed PMID: 15312966. [DOI] [PubMed]
- Olsson T., Wieloch T., Smith M.-L. Brain damage in a mouse model of global cerebral ischemia. Brain Res. 2003;982(2):260–269. doi: 10.1016/s0006-8993(03)03014-2. [DOI] [PubMed] [Google Scholar]
- Wang L, Almeida LEF, de Souza Batista CM, Khaibullina A, Xu N, Albani S, et al. Cognitive and behavior deficits in sickle cell mice are associated with profound neuropathologic changes in hippocampus and cerebellum. Neurobiol Dis. 2016;85:60-72. Epub 2015/10/16. doi: 10.1016/j.nbd.2015.10.004. PubMed PMID: 26462816; PubMed Central PMCID: PMCPMC4688201. [DOI] [PMC free article] [PubMed]
- de Flores R., La Joie R., Chételat G. Structural imaging of hippocampal subfields in healthy aging and Alzheimer’s disease. Neuroscience. 2015;309:29–50. doi: 10.1016/j.neuroscience.2015.08.033. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy N, Santini T, Wood S, Kim J, Zhao T, Aizenstein HJ, et al. Computational and experimental evaluation of the Tic-Tac-Toe RF coil for 7 Tesla MRI. PloS one. 2019;14(1):e0209663. Epub 2019/01/11. doi: 10.1371/journal.pone.0209663. PubMed PMID: 30629618. [DOI] [PMC free article] [PubMed]
- Knierim J.J. The hippocampus. Curr. Biol. 2015;25(23):R1116–R1121. doi: 10.1016/j.cub.2015.10.049. [DOI] [PubMed] [Google Scholar]
- Yushkevich P.A., Pluta J.B., Wang H., Xie L., Ding S.-L., Gertje E.C., Mancuso L., Kliot D., Das S.R., Wolk D.A. Automated volumetry and regional thickness analysis of hippocampal subfields and medial temporal cortical structures in mild cognitive impairment. Hum. Brain Mapp. 2015;36(1):258–287. doi: 10.1002/hbm.22627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccardi M., Ganzola R., Bocchetta M., Pievani M., Redolfi A., Bartzokis G., Camicioli R., Csernansky J.G., de Leon M.J., deToledo-Morrell L., Killiany R.J., Lehéricy S., Pantel J., Pruessner J.C., Soininen H., Watson C., Duchesne S., Jack Jr C.R., Frisoni G.B., Ashford J.W., Rosen A., Adamson M., Bayley P., Sabri O., Furst A., Black S.E., Weiner M. Survey of protocols for the manual segmentation of the hippocampus: preparatory steps towards a joint EADC-ADNI harmonized protocol. J. Alzheimer's Dis. 2011;26(s3):61–75. doi: 10.3233/JAD-2011-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias J.E., Augustinack J.C., Nguyen K., Player C.M., Player A., Wright M., Roy N., Frosch M.P., McKee A.C., Wald L.L., Fischl B., Van Leemput K. A computational atlas of the hippocampal formation using ex vivo, ultra-high resolution MRI: application to adaptive segmentation of in vivo MRI. Neuroimage. 2015;115:117–137. doi: 10.1016/j.neuroimage.2015.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliano A., Donatelli G., Cosottini M., Tosetti M., Retico A., Fantacci M.E. Hippocampal subfields at ultra high field MRI: a n overview of segmentation and measurement methods. Hippocampus. 2017;27(5):481–494. doi: 10.1002/hipo.22717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Zhuang X., Mishra V., Sreenivasan K., Cordes D. A multi-scale convolutional neural network based automated hippocampal subfield segmentation toolbox. NeuroImage. 2020;218:116947. doi: 10.1016/j.neuroimage.2020.116947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim T., Zhao Y., Krishnamurthy N., Raval S., Zhao T., Wood S. International Society of Magnetic Resonance in Medicine; Salt Lake City; Utah: 2013. 20-To-8 Channel Tx Array with 32-Channel Adjustable Receive-Only Insert for 7T Head Imaging; p. 4408. [Google Scholar]
- Ibrahim T ST, Raval S, Krishnamurthy N, Wood S, Kim J, Zhao Y, Wu X, Yacoub E, Aizenstein H, Zhao T. Towards Homogenous 7T Neuro Imaging: Findings and Comparisons between 7T TTT and NOVA RF Coil Systems. In Proc of the 25th International Society of Magnetic Resonance in Medicine Annual Meeting; Honolulu, Hawaii, USA2017.
- Smagula S.F., Karim H.T., Rangarajan A., Santos F.P., Wood S.C., Santini T., Jakicic J.M., Reynolds C.F., Cameron J.L., Vallejo A.N., Butters M.A., Rosano C., Ibrahim T.S., Erickson K.I., Aizenstein H.J. Association of hippocampal substructure resting-state functional connectivity with memory performance in older adults. Am. J. Geriatric Psychiatry. 2018;26(6):690–699. doi: 10.1016/j.jagp.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santini T, Zhao Y, Wood S, Krishnamurthy N, Kim J, Farhat N, et al. In-vivo and numerical analysis of the eigenmodes produced by a multi-level Tic-Tac-Toe head transmit array for 7 Tesla MRI. PloS one. 2018;13(11):e0206127. doi: 10.1371/journal.pone.0206127. PubMed PMID: 30481187; PubMed Central PMCID: PMCPMC6258503 products in development to declare. This does not alter our adherence to PLOS ONE policies on sharing data and materials. [DOI] [PMC free article] [PubMed]
- Santini T, Wood S, Krishnamurthy N, Martins T, Aizenstein HJ, Ibrahim TS. Improved 7 Tesla transmit field homogeneity with reduced electromagnetic power deposition using coupled Tic Tac Toe antennas. Scientific reports. 2021;11(1):3370. Epub 2021/02/11. doi: 10.1038/s41598-020-79807-9. PubMed PMID: 33564013; PubMed Central PMCID: PMCPMC7873125. [DOI] [PMC free article] [PubMed]
- Foi A. Noise estimation and removal in MR imaging: The variance-stabilization approach. 2011 IEEE International symposium on biomedical imaging: from nano to macro: IEEE; 2011. pp. 1809–1814.
- Maggioni M., Katkovnik V., Egiazarian K., Foi A. Nonlocal transform-domain filter for volumetric data denoising and reconstruction. IEEE Trans. Image Process. 2013;22(1):119–133. doi: 10.1109/TIP.2012.2210725. [DOI] [PubMed] [Google Scholar]
- Santini T, Brito F, Wood S, Martins T, Mettenburgh J, Aizenstein H, et al. Noise mitigation from high-resolution 7T MRI images. In Proc of the 26th International Society of Magnetic Resonance in Medicine Annual Meeting; Paris, France2018.
- Ibrahim T.S., Lee R., Abduljalil A.M., Baertlein B.A., Robitaille P.-M. Dielectric resonances and B 1 field inhomogeneity in UHFMRI: computational analysis and experimental findings. Magn. Resonance Imag. 2001;19(2):219–226. doi: 10.1016/s0730-725x(01)00300-9. [DOI] [PubMed] [Google Scholar]
- Berron D., Vieweg P., Hochkeppler A., Pluta J.B., Ding S.-L., Maass A., Luther A., Xie L., Das S.R., Wolk D.A., Wolbers T., Yushkevich P.A., Düzel E., Wisse L.E.M. A protocol for manual segmentation of medial temporal lobe subregions in 7 Tesla MRI. NeuroImage Clin. 2017;15:466–482. doi: 10.1016/j.nicl.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich P.A., Piven J., Hazlett H.C., Smith R.G., Ho S., Gee J.C., Gerig G. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage. 2006;31(3):1116–1128. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]
- Iampietro M, Giovannetti T, Tarazi R. Hypoxia and inflammation in children with sickle cell disease: implications for hippocampal functioning and episodic memory. Neuropsychol Rev. 2014;24(2):252-65. Epub 2014/04/20. doi: 10.1007/s11065-014-9259-4. PubMed PMID: 24744195. [DOI] [PubMed]
- Jorgensen DR, Metti A, Butters MA, Mettenburg JM, Rosano C, Novelli EM. Disease severity and slower psychomotor speed in adults with sickle cell disease. Blood Adv. 2017;1(21):1790-5. Epub 2018/01/04. doi: 10.1182/bloodadvances.2017008219. PubMed PMID: 29296825; PubMed Central PMCID: PMCPMC5728099 interests. [DOI] [PMC free article] [PubMed]
- Schatz J, Finke RL, Kellett JM, Kramer JH. Cognitive functioning in children with sickle cell disease: a meta-analysis. J Pediatr Psychol. 2002;27(8):739-48. Epub 2002/10/31. doi: 10.1093/jpepsy/27.8.739. PubMed PMID: 12403864. [DOI] [PubMed]
- Case M, Zhang H, Mundahl J, Datta Y, Nelson S, Gupta K, et al. Characterization of functional brain activity and connectivity using EEG and fMRI in patients with sickle cell disease. NeuroImage Clinical. 2017;14:1-17. Epub 2017/01/25. doi: 10.1016/j.nicl.2016.12.024. PubMed PMID: 28116239; PubMed Central PMCID: PMCPMC5226854. [DOI] [PMC free article] [PubMed]
- Hamalainen A, Pihlajamaki M, Tanila H, Hanninen T, Niskanen E, Tervo S, et al. Increased fMRI responses during encoding in mild cognitive impairment. Neurobiology of aging. 2007;28(12):1889-903. Epub 2006/09/26. doi: 10.1016/j.neurobiolaging.2006.08.008. PubMed PMID: 16997428. [DOI] [PubMed]
- Yushkevich PA, Amaral RS, Augustinack JC, Bender AR, Bernstein JD, Boccardi M, et al. Quantitative comparison of 21 protocols for labeling hippocampal subfields and parahippocampal subregions in in vivo MRI: towards a harmonized segmentation protocol. Neuroimage. 2015;111:526-41. Epub 2015/01/18. doi: 10.1016/j.neuroimage.2015.01.004. PubMed PMID: 25596463; PubMed Central PMCID: PMCPMC4387011. [DOI] [PMC free article] [PubMed]
- Sozinova E.V., Kozlovskiy S.A., Vartanov A.V., Skvortsova V.B., Pirogov Y.A., Anisimov N.V., Kupriyanov D.A. The role of hippocampal parts in verbal memory and activation processes. Int. J. Psychophysiol. 2008;69(3):312. doi: 10.1016/j.ijpsycho.2008.05.328. [DOI] [Google Scholar]
- Nogovitsyn N, Muller M, Souza R, Hassel S, Arnott SR, Davis AD, et al. Hippocampal tail volume as a predictive biomarker of antidepressant treatment outcomes in patients with major depressive disorder: a CAN-BIND report. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2020;45(2):283-91. Epub 2019/10/15. doi: 10.1038/s41386-019-0542-1. PubMed PMID: 31610545; PubMed Central PMCID: PMCPMC6901577. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.