Abstract
For patients with inflammatory bowel disease, cow’s milk allergy, and lactose intolerance, soymilk is a potential alternative to cow’s milk. In this study, we aimed to identify the effects of a soy protein-based low-protein diet on the body and organ weights and the gut microbiome of six-week-old mice fed a diet containing 20% (SP) or 5% (LP) soy protein for 14 days via 16S rRNA (V4) amplicon sequencing. Body weight gain (growth) and liver, spleen, and fat tissue weight were significantly suppressed by the LP diet. Operational taxonomic unit numbers and α-diversity were lower in the LP group than in the SP group. A principal coordinate analysis revealed differences in the gut microbiome compositions of SP and LP mice. The abundances of caecal Roseburia sp., Alistipes sp., and bacteria from the family Muribaculaceae were lower in the LP group than in the SP group. In contrast, the abundance of Desulfovibrionaceae, which is positively correlated with inflammation, was higher in the LP group than in the SP group. These results differed from the effects of a milk casein-based low-protein diet (reported previously). Based on these findings, we conclude that the undesirable effects of a low-protein diet and/or protein deficiency are related to changes in the gut microbiome composition and may differ depending on the kind of proteins used.
Keywords: Low-protein diet, Soy protein, Gut microbiome, Muribaculum, Desulfovibrionaceae, ICR mice
Graphical abstract
Highlights
-
•
Six-week-old ICR mice were fed a diet containing 20% (SP) or 5% (LP) soy protein for 14 days.
-
•
Body weight gain and liver, spleen, and fat tissue weight were significantly suppressed by the LP diet.
-
•
Caecal Roseburia sp., Alistipes sp., and bacteria from the family Muribaculaceae was lower in the LP.
-
•
Desulfovibrionaceae, which is positively correlated with inflammation, was higher in the LP group.
1. Introduction
Soybean is an important source of protein and has been used in traditionally processed and/or fermented foods in East Asia and other countries, such as in tofu, tempeh, natto, miso, and soy sauce. For patients with inflammatory bowel disease (IBD) and individuals with cow-milk allergy and/or lactose intolerance, soymilk is a potential alternative to cow’s milk (Heyman, 2006). β-Conglycinin (BC; 7S) and glycinin (11S) are the major protein molecules in soybean that constitute ~20%–40% of the total protein content of soymilk (Krishnan and Nelson, 2011). Although BC may induce soybean allergy, the protein and its hydrolysate reportedly possess antioxidant, antibacterial, immunomodulatory, and hypocholesterolemic properties (Ferreira et al., 2010; Xiang et al., 2016; Wang et al., 2017).
The gut microbiome of mammals including human and laboratory animals have been found to contain hundreds of species and ~11 log cells/g of bacteria. These microorganisms and their metabolites are considered to have important effects on the host’s health (Shanahan et al., 2021). For example, there are several reports on the correlation between typical indigenous bacterial groups and conditions such as obesity, diabetes, ageing, immune system-related diseases, IBD, and Alzheimer’s disease (Umirah et al., 2021; Ragonnaud and Biragyn, 2021). Dietary fibres and other food components such as lipids and proteins can change the composition of the gut microbiome rapidly and drastically (David et al., 2014; Wilson et al., 2020). Although most of the proteins are degraded and/or adsorbed in the stomach and small intestine, a proportion of these proteins reach the colon and caecum and are metabolised into various compounds, including amines, ammonia, phenols, H2S, and indoles (An et al., 2014).
As protein intake is important for protein synthesis in the muscles, various protein-rich functional foods are available for different age groups (from infants to the elderly). They are used to enhance the growth of infants and children (Uauy et al., 2015) and prevent sarcopenia in the elderly (Granic et al., 2020) and are used by individuals involved in sports and exercise training (Håvard et al., 2019). Additionally, a low-protein diet and/or protein deficiency reduce immune activity (Miyazaki et al., 2018). On the other hand, there are reports about the beneficial effect of a low-protein diet on inflammation related to the liver and gut (Guo et al., 2018). It may be the case that these effects of protein concentration in the diet are related to changes in the gut microbiome (Shanahan et al., 2021).
A study on the influence of a milk casein (MC)-based low-protein diet on the gut microbiome of mice revealed a decrease in the abundance of Bacteroides and Clostridium. (Masuoka et al., 2020). However, a different gut microbiome composition was reported in mice fed MC or soy protein (Xia et al., 2020). Therefore, we sought to clarify the effects of a soy protein-based low-protein diet on the body and organ weights and the gut microbiome of mice via 16S rRNA (V4) amplicon sequencing.
2. Materials and methods
2.1. Animal care
Animal experiments were performed in accordance with the ‘Fundamental Guidelines for Proper Conduct of Animal Experiments and Related Activities in Academic Research Institutions’ under the jurisdiction of the Ministry of Education, Culture, Sports, Science and Technology, Japan. The study protocol was approved by the Animal Experiment Committee of Tokyo University of Marine Science and Technology (Approval No. H31-5).
Soy protein (SP: Fujipro F) and BC products were obtained from Fuji Oil (Izumisano, Japan). The Fujipro F contained 21% β-conglycinin (7S), 41% glycinin (11S), and 38% of other lipoproteins (Kawaguchi et al., 2018). Twelve 5-week-old male ICR mice (25 ± 2g) were purchased (Japan SLC Inc., Tokyo, Japan) and housed in metal wire cages (three mice per cage) at 22 ± 2 °C with a 12-h dark/light cycle. The mice were acclimatised to a semi-purified powder diet containing 20% (w/w) MC (Table 1) and were provided distilled water for drinking ad libitum. After 7 days, the mice were divided into two groups (n = 6, in each group)—standard protein (SP) and low protein (LP)—and were fed a diet containing either 20% (SP) or 5% (LP) soy protein for 14 days. During the feeding days 11–13, the defaecation frequency and faecal weight were measured.
Table 1.
Composition of test diets (g/100 g).
| SP | LP | |
|---|---|---|
| Soy-protein | 20.0 | 5.0 |
| DL-Methionine | 0.3 | 0.3 |
| Corn starch | 15.0 | 30.0 |
| Sucrose | 50.0 | 50.0 |
| Cellulose | 5.0 | 5.0 |
| Corn oil | 5.0 | 5.0 |
| Vitamin mix (AIN-76) | 1.0 | 1.0 |
| Mineral mix (AIN-76) | 3.5 | 3.5 |
| Choline bitartrate | 0.2 | 0.2 |
2.2. Plasma lipid and glucose levels
At the experimental endpoint, the mice were exsanguinated via the abdominal vein under anaesthesia using isoflurane (Fujifilm Wako Pure Chemical Corp., Osaka, Japan), and the liver, kidneys, spleen, and epididymal fat pads were removed and weighed. After ligation with yarn, the caecum was excised and placed on ice until microbial analysis was performed. Plasma triacylglyceride (TG), total cholesterol (TC), and glucose levels were determined using commercial kits following manufacturers’ instructions (Triglyceride E-Test Wako, Total Cholesterol E-Test Wako, Glucose CII-Test Wako, respectively; Fujifilm Wako Pure Chemical Corp.).
2.3. Analysis of caecal microbiota using the MiSeq system
The caecal contents were diluted using 99 vol (~5 mL) of phosphate-buffered saline (Nissui Pharmaceutical Co. Ltd., Tokyo, Japan), and the bacterial cell count was determined via dielectrophoretic impedance measurements (Hirota et al., 2014) using a bacterial counter (PHC Ltd., Tokyo, Japan).
16S rDNA (V4) amplicon sequencing was performed by Fasmac Co., Ltd. (Atsugi, Japan) as described in a previous report (Xia et al., 2020). Briefly, DNA was extracted from the caecal content using the MPure bacterial DNA extraction kit (MP Bio Japan, Tokyo, Japan). A DNA library was prepared using a two-step PCR (Sinclair et al., 2015). Next, the V4 region was amplified via a 23-cycle PCR using the following primers: forward 515f. And reverse 806r. Individual DNA fragments were tagged via an 8-cycle PCR using the individual mouse primers. DNA libraries were multiplexed and loaded onto an Illumina MiSeq system (Illumina, San Diego, CA, USA). Reads with a mismatched sequence at the start region were filtered using the FASTX Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/); poor-quality reads (below 20) and those shorter than 40 base pairs were omitted using Sickle (https://github.com/ucdavis-bioinformatics/sickle). Short-listed reads were merged using the paired-end merge script FLASH (http://ccb.jhu.edu/software/FLASH/), and 240–260 base pair reads were selected. Chimeras in the selected reads were identified and omitted using the QIIME2 bioinformatics pipeline (https://qiime2.org/). Sequences were clustered into operational taxonomic units (OTUs), with a 97% identity cut-off, using the QIIME2 workflow script and SILVA database (https://www.arb-silva.de/).
2.4. Statistical analysis
The results of body and organ weights and the alpha-diversity indices (diversity within a microbiome) are expressed as the mean values ± standard error of the mean. Data were subjected to an analysis of variance and the Student’s t-test using a statistical software package (Excel Statistic Ver. 6, Japan). A p-value < 0.05 was considered statistically significant. The alpha-diversity of the gut microbiome for the mice was determined using the Shannon-Wiener (H′) and Simpson’s diversity (D) indices (Kim et al., 2017). The beta diversity (distance between groups based on the differences in the OTUs present in each group) was assessed using unweighted and weighted UniFrac (Lozupone and Knight, 2005) and expressed using a principal coordinate analysis (PCoA) (Bunyavanich et al., 2016).
3. Results
3.1. Body, faecal, and organ weights and plasma lipid and glucose levels
The body weight of the mice at the start (0 day) was ~33 g; the mice fed the SP diet gained 10.5 g of body weight per 14 days, whereas mice fed LP gained only 3.9 g (p < 0.01, Table 2). The faecal and caecal content weights tended to be high in mice that were fed the LP diet, though the difference was not significant. Compared with mice fed the SP diet, the weights of the liver and spleen were lower in mice fed the LP diet (p < 0.01 and 0.05, respectively). The weight of the epididymal fat pad was 40% lower in the LP group (p < 0.05). Plasma TG and TC levels also tended to be lower in the LP group, though the difference was not significant. Additionally, plasma glucose levels were 20% lower in mice that were fed the LP diet (p < 0.01).
Table 2.
Body, organ and faecal weights, and levels of plasma lipids and glucose of tested mice.
| SP | LP | |
|---|---|---|
| Body weight (g) | ||
| Initial | 32.9 ± 0.8 | 32.8 ± 0.7 |
| 14 days feeding | 43.4 ± 1.7 | 36.5 ± 0.7∗∗ |
| Gain per 14 days | 10.5 ± 0.9 | 3.8 ± 0.6∗∗ |
| Defecation | ||
| Frequency (n/day/mouse) | 31 ± 2 | 26 ± 2 |
| Weight (g/day/mouse) | 0.51 ± 0.09 | 0.65 ± 0.04 |
| Organ weights (g) | ||
| Liver | 2.380 ± 0.125 | 1.771 ± 0.059∗∗ |
| Kidneys | 0.650 ± 0.041 | 0.563 ± 0.021 |
| Spleen | 0.150 ± 0.011 | 0.125 ± 0.007∗ |
| Epididymal fat pads | 1.916 ± 0.221 | 1.211 ± 0.122∗ |
| Caecum (Net) | 0.280 ± 0.031 | 0.356 ± 0.034 |
| Caecal content | 0.206 ± 0.033 | 0.272 ± 0.038 |
| Plasma lipids and glucose (mg/100 mL) | ||
| Triacylglyceride | 89.6 ± 14.7 | 63.0 ± 7.2 |
| Total-cholesterol | 230 ± 24 | 185 ± 13 |
| Glucose | 326 ± 13 | 262 ± 17∗∗ |
Values indicate the mean and SEM (n = 6 for body and organ weights, n = 4 for plasma lipids and glucose). ∗and ∗∗ indicate significant differences relative to the SP group when analysed by the Student’s t-test (∗p < 0.05, ∗∗p < 0.01).
3.2. Direct cell counts and alpha and beta diversity of the caecal microbiome
The total bacterial cell count in the caecal content of the tested mice was 11.3–11.4 log cell/g (Fig. 1A). In the 16S rDNA (V4) amplicon sequencing, the total read numbers obtained from mice fed the SP and LP diets were 99000 and 91000, respectively (Fig. 1B). Compared with the SP group, the OTU number in mice fed the LP diet was 20% lower (p < 0.05, Fig. 1C). The alpha-diversity Shannon-Weaver H′ values and the Simpson’s index (D) in mice fed LP diet were significantly lower than ones in mice fed SP diet (p < 0.05, Fig. 1D and E). The PCoA of the OTUs revealed that the SP and LP groups had different gut microbiome compositions (Fig. 1F and G).
Fig. 1.
Total bacterial count (A), total read number (B), operational taxonomic units (OTU) number (C), Shannon index (D), Simpson’s index (E) and principal coordinate analysis (PCoA; F and G) of the OTUs in the caecal microbiome of the mice fed a diet containing 20% (SP) or 5% soy protein (LP).
3.3. Microbiome composition
3.3.1. Phylum level
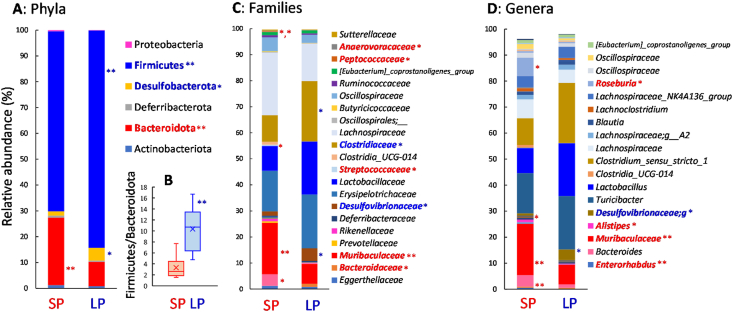
The predominant bacterial phylum in the tested mice was Firmicutes, and its abundance was higher in mice fed the LP diet (84%) than in those fed the SP diet (70%) (p < 0.01, Fig. 2A). In contrast, the abundance of the second-most dominant phylum, Bacteroidota, was higher in the SP (26%) than in the LP (9.5%) group (p < 0.01). The ratio of the Firmicutes to Bacteroidota (F/B) was three times higher in mice fed the LP diet (p < 0.01, Fig. 2B). The phylum Desulfobacterota was also dominant in mice fed the LP (4.9%) rather than the SP (1.7%) diet (p < 0.05). The abundance of Actinobacteria tended to be higher in the SP group (1.7%) than in the LP group (0.8%), though the difference was not significant.
Fig. 2.
Composition of the caecal microbiome at the phylum (A), family (C), and genus (D) levels in the mice fed a diet containing 20% (SP) or 5% (LP) soy protein. (B) The ratio of Firmicutes to Bacteroidota. ∗ and ∗∗ indicate significant differences between the groups determined by the Student’s t-test (∗p < 0.05, ∗∗p < 0.01).
3.3.2. Family level
As shown in Fig. 2C, in the case of families that belonged to the phylum Firmicutes, Lachnospiraceae (14%–24%), Erysipelotrichaceae (16%–21%), Clostriridiaceae (10%–23%), Lactibacillaceae (9.6%–20%), and Oscillospiraceae (3.1%–5.3%) were dominant. Among these dominant families, bacteria from the family Clostridiaceae in mice fed the LP diet were significantly higher than in mice fed the SP diet (p < 0.05). Bacteria from other dominant families besides Lachnospiraceae tended to be high in mice fed the LP diet. Certain Gram-positive cocci, such as Streptococcaceae (0.08%–0.46%) and Peptococcaceae (0.13%–0.30%) were higher in mice fed the SP diet than in those fed the LP diet (p < 0.05), although their abundance was not high. With regard to the phylum Bacteroidota, the dominant family was Muribaculuaceae (7.5%–20%), followed by Bacteroidaceae (1.2%–4.4%). Both the families in the SP group were 2.6 and 3.6 times higher than in the LP group (p < 0.05 and 0.01), respectively. Almost all of the Desulfobacteria OTUs, which were high in the LP group, were defined as Desulfovibrionaceae.
3.3.3. Genus level
In the case of genera that belonged to the family Lachnospiraceae, Roseburia (7.1%) and two Lachnospiraceae OTUs (4.4%–7.3%) were dominant in mice fed the SP diet (Fig. 2D). Among these genera, the abundance of Roseburia was low in mice fed the LP diet (0.1%, p < 0.05). The abundance of the genus Alistipes (family Rikenellaceae, phylum Bacteroidota) was higher in the SP group than in the LP group (p < 0.05), though it was not the dominant genus (1.0%). Enterorhabdus, which belong to the family Eggerthellaceae, and Actinobacteria were higher in mice fed SP (0.62%) rather than the LP diet (0.21%, p < 0.01).
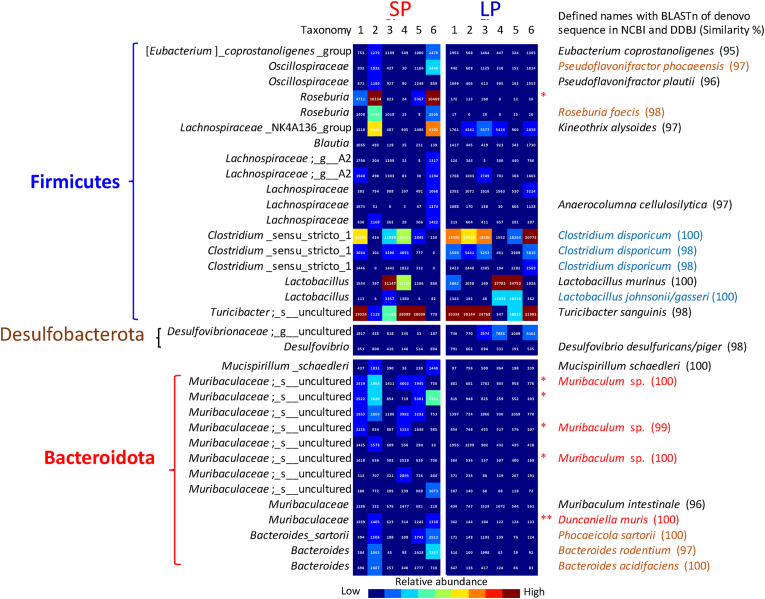
3.4. Dominant OTUs
The heat map of dominant OTUs with an abundance of 0.5% or more is shown in Fig. 3. Species names of the predominant and typical OTUs defined using BLASTn by the National Center for Biotechnology Information (NCBI, https://blast.ncbi.nlm.nih.gov/Blast.cgi) and DDBJ that displayed 95% or more similarity are also shown. In Firmicutes, the top four dominant OTUs, Turicibacter, Clostridium sensu stricto, Lactobacillus, and Lachnospiraceae NK4A136 group were defined as T. sanguinis-, C. disporicum-, L. murinus-, and Kineothrix alysoides-like bacteria, respectively. There was no significant difference for the dominant OTUs between the two diet groups, though C. disporicum-like bacteria tended to be high in mice fed an LP diet. The Roseburia OTU that showed high abundance in the SP group could not be defined, and the most similar bacteria that were identified using BLASTn was R. intestinalis (similarity = 91%).
Fig. 3.
Heat map showing the relative abundance of identified OTUs filtered by 0.5% counts of all identified OTUs in the caecal microbiome in mice fed a diet containing 20% (SP) or 5% (LP) soy protein. ∗ and ∗∗ indicate significant differences between the groups as determined by the Student’s t-test (∗p < 0.05, ∗∗p < 0.01).
In the case of OTUs from Bacteroidota, the three dominant Muribaculaceae OTUs that showed higher abundance in the SP group were defined as Muribaculum sp. like bacteria. The other Muribaculaceae OTU, which was also abundant in the SP group, was defined as Duncaniella muris. Although Bacteroidaceae abundance was significantly higher in mice fed an SP diet, the abundance of the dominant Bacteroidaceae OTUs identified as Phocaeicola sartorii, Bacteroides rodentium, and Bacteroides acidifaciens-like bacteria tended to be higher in mice fed an SP diet than in those fed an LP diet, but the difference was not significant.
4. Discussion
There are several reports about the suppression of weight gain in young mice and rats fed on MC- and soy protein-based low-protein diets (Chaumontet et al., 2019). Liver weight loss in mice fed a low-protein diet has also been reported (Pérez-Martí et al., 2017). Lower spleen weight in mice fed an LP diet can be regarded as a result of the effect of low protein intake on the immune system (Lewicki et al., 2014). The suppressive effects of a low-protein diet on fat tissue weight and the level of plasma lipids and glucose has been shown in rat models of type 2 diabetes (T2D) (Kitada et al., 2018).
Sequencing of the 16S rRNA amplicon revealed that among the typical bacterial families in the gut of mice fed an LP diet, Clostridiaceae and Desulfovibrionaceae showed high abundance, and Muribaculaceae showed low abundance (Fig. 2C). Additionally, the abundance of the genus Roseburia was significantly lower in the LP group than in the SP group (Fig. 2D). These results are not in agreement with those from a previous study on mice maintained on MC (Masuoka et al., 2020). The variation may be due to the different effects exerted by soybean and MC on the gut microbiome. For example, the abundance of the families Bacteroidaceae and Muribaculaceae was lower and higher, respectively, in mice fed 20% soy protein than in mice fed 20% MC (Xia et al., 2020).
The dominant genus in the family Clostridiaceae in the LP diet-fed group was identified as C. disporicum (Fig. 3). Though an increase in the abundance of C. disporicum in murine caecum fed fermented loofah and some spices (cumin and coriander) has been reported (Shikano et al., 2019; Xia et al., 2019), the abundance decreased on a diet containing egg white, some edible algae, and algal polysaccharides (Takei et al., 2019; Takei et al., 2020; Xia et al., 2020). Although functional reports of C. disporicum are limited, the epimerase activity of this species, which produces ursodeoxycholic acid (UDCA) from chenodeoxycholic acid (CDCA) was recently reported (Tawtep et al., 2017). This might be correlated with low liver and fat tissue weights observed with the LP diet (Table 2).
Recently, bacteria from the family Desulfovibrionaceae have been identified as harmful. For example, there are reports about a positive relationship between endotoxin, which induces inflammation, and the abundance of bacteria from the family Desulfovibrionaceae (Zhai et al., 2019; Chen et al., 2020). An increase in gut Desulfovibrionaceae in rats fed a diet containing beef and sucrose stimulated oxidative stress (Hecke et al., 2019). In contrast, the prevalence of the bacteria from this family was decreased by the use of some dietary fibres such as inulin and fucoidan, antioxidants such as rutin, quercetin, and resveratrol, and food materials rich in dietary fibres and antioxidants such as edible brown algae and wasabi (Zhao et al., 2017; Guo et al., 2018; Li et al., 2019; Takei et al., 2019; Thomaz et al., 2020). The relationship between dietary protein deficiency, liver injury, oxidative stresses, and amelioration of that with the use of antioxidants has been reported (Maiti and Chatterjee, 2000; Nkosi et al., 2006).
Muribaculum, which had a low abundance in mice fed an LP diet, is a dominant genus in the murine gut (Lagkouvardos et al., 2016). Its abundance increases when mice are fed dietary fibre- and protein-rich foods such as rice bran and cyanobacteria, and this increase exerts an ameliorative effect on obesity and T2D in model mice (Shibayama et al., 2019; Yuan et al., 2020; Taniguchi et al., 2021). Furthermore, studies have suggested a correlation of the predominantly commensal Muribaculum sp. with the host immune system and metabolism, particularly carbohydrate metabolism (Lagkouvardos et al., 2016; Graham et al., 2018; Chun et al., 2020).
Multiple reports on the butyrate-producing species of Roseburia, which also showed low abundance in the LP group, have indicated their role as a marker of host health that can be correlated with its beneficial properties (Tamanai-Shacoori et al., 2017). For example, the relationship between Roseburia abundance and amelioration of signs of colitis, IBD, obesity, T2D, and neuronal system conditions, as well as immunomodulation, have been reported (Imharan et al., 2017; Gurung et al., 2019; Luo et al., 2019; Zheng et al., 2019; Nishiwaki et al., 2020). Although the ameliorative effect of a low-protein diet on gut inflammation has been described earlier (Guo et al., 2018), soy protein and its hydrolysate have antioxidant, antibacterial, immunomodulatory, and hypocholesterolemic properties (Ferreira et al., 2010; Xiang et al., 2016; Wang et al., 2017).
The results of our study regarding the effect of a low-protein diet on the gut microbiome have important differences relative to previous reports about mice maintained using MC (Masuoka et al., 2020). In our study, we observed that a low-protein diet was undesirable for growth, the gut microbiome, and its environment when mice were maintained using soy protein. The bacteria that showed an increase or decrease in prevalence in the gut of mice kept on a low-protein diet in this study can be regarded as soy protein susceptible gut indigenous bacteria (SIB). It has been shown that certain food materials and their SIBs have a synergistic effect on the host (Wilck et al., 2017). Isolation of the SIBs using a proper culturing method may help clarify the relationship between proteins and the gut microbiome.
5. Conclusion
In this study, we characterised the effects of a low-protein diet in mice that were administered soy protein, and measured the body and organ weights, and analysed the gut microbiome of six-week-old mice fed a diet containing 20% (SP) or 5% (LP) soy protein for 14 days via 16S rRNA (V4) amplicon sequencing. The body weight gain (growth), and the liver, spleen, and fat tissue weights were significantly suppressed in mice on the LP diet. Further, the OTU number and α-diversity were lower in the LP group. A PCoA revealed that SP and LP fed mice had different gut microbiome compositions. The caecal Roseburia sp., Alistipes sp., and bacteria from the family Muribaculaceae in the LP group were lower than that in the SP group. In contrast, the abundance of bacteria from the family Desulfovibrionaceae was high in the LP group. These results differ from the effects of a low-protein diet maintained using MC. We concluded that the undesirable effects of a low-protein diet and/or protein deficiency are related to changes in the gut microbiome composition and that these effects may differ depending on the kinds of proteins used.
CRediT authorship contribution statement
Saori Nakamura: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Data curation, Writing – original draft, Visualization. Takashi Kuda: Conceptualization, Methodology, Validation, Formal analysis, Resources, Data curation, Writing – review & editing, Visualization, Supervision, Project administration. Yuko Midorikawa: Investigation. Hajime Takahashi: Conceptualization, Methodology, Supervision. Bon Kimura: Conceptualization, Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was partially supported by the Fuji Foundation for Protein Research, Tokyo, Japan. We would like to thank Editage (www.editage.com) for English language editing.
References
- An C., Kuda T., Yazaki T. Caecal fermentation, putrefaction and microbiotas in rats fed milk casein, soy protein or fish meal. Appl. Microbiol. Bioteshnol. 2014;98:2779–2787. doi: 10.1007/s00253-013-5271-5. [DOI] [PubMed] [Google Scholar]
- Bunyavanich S., Shen N., Grishin A. Early-life gut microbiome composition and milk allergy resolution. J. Allergy Clin. Immunol. 2016;138:1122–1130. doi: 10.1016/j.jaci.2016.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumontet C., Azzout-Marniche D., Blais A. Low-protein and methionine, high-starch diets increase energy intake and expenditure, increase FGF21, decrease IGF-1, and have little effect on adiposity in mice. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2019;316:R486–R501. doi: 10.1152/ajpregu.00316.2018. [DOI] [PubMed] [Google Scholar]
- Chen Y.H., Wang Y.C., Chiu C.C. Housing condition-associated changes in gut microbiota further affect the host response to diet-induced nonalcoholic fatty liver. J. Nutr. Biochem. 2020;79:108362. doi: 10.1016/j.jnutbio.2020.108362. [DOI] [PubMed] [Google Scholar]
- Chun Y.W., Gwak H.J., Moon S. Functional dynamics of bacterial species in the mouse gut microbiome revealed by metagenomic and metatranscriptomic analyses. PloS One. 2020;15 doi: 10.1371/journal.pone.0227886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David L.A., Maurice C.F., Carmody R.N. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira E.S., Silva M.A., Demonte A. β-Conglycinin (7S) and glycinin (11S) exert a hypocholesterolemic effect comparable to that of fenofibratein rats fed a high-cholesterol diet. J. Funct. Foods. 2010;2:275–283. [Google Scholar]
- Graham D.B., Luo C., O’Connell D.J. Antigen discovery and specification of immunodominance hierarchies for MHCII-restricted epitopes. Nat. Med. 2018;24:1762–1772. doi: 10.1038/s41591-018-0203-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granic A., Mendonça N. Effects of dietary patterns and low protein intake on sarcopenia risk in the very old: the Newcastle 85+ study. Clin. Nutr. 2020;39:166–173. doi: 10.1016/j.clnu.2019.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J.H., Wang H.Y., Zhang Q. The low protein diet affects the nonspecific inflammatory response of middle-aged and old mice through mTOR. Eur. Rev. Med. Pharmacol. Sci. 2018;22:7551–7561. doi: 10.26355/eurrev_201811_16297. [DOI] [PubMed] [Google Scholar]
- Guo X., Tang R., Yang S. Rutin and its combination with inulin attenuate gut dysbiosis, the inflammatory status and endoplasmic reticulum stress in paneth cells of obese mice induced by high-fat diet. Front. Microbiol. 2018;9:2651. doi: 10.3389/fmicb.2018.02651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurung M., Li Z., You H. Role of gut microbiota in type 2 diabetes pathophysiology. EBio Med. 2019;51:102590. doi: 10.1016/j.ebiom.2019.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecke T.V., De Vrieze J., Boon N. Combined consumption of beef-based cooked mince and sucrose stimulates oxidative stress, cardiac hypertrophy, and colonic outgrowth of Desulfovibrionaceae in Rats. Mol. Nutr. Food Res. 2019;64 doi: 10.1002/mnfr.201800962. [DOI] [PubMed] [Google Scholar]
- Heyman M.B. Lactose intolerance in infants, children, and adolescents. Pediat. 2006;118:1270–1286. doi: 10.1542/peds.2006-1721. [DOI] [PubMed] [Google Scholar]
- Hirota K., Inagaki R., Hamada R. Evaluation of a rapid oral bacteria quantification system using dielectrophoresis and the impedance measurement. Biocontrol Sci. 2014;19:45–49. doi: 10.4265/bio.19.45. [DOI] [PubMed] [Google Scholar]
- Imharan F., Vila A.V., Bonder M.J. Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease. Gut. 2017;67:108–119. doi: 10.1136/gutjnl-2016-312135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi T., Kita R., Shinyashiki N. Physical properties of tofu gel probed by water translational/rotational dynamics. Food Hydrocolloids. 2018;77:474–481. [Google Scholar]
- Kim B., Shin J., Guevarra R.B. Deciphering diversity indices for a better understanding of microbial communities. J. Microbiol. Biotechnol. 2017;27:2089–2093. doi: 10.4014/jmb.1709.09027. [DOI] [PubMed] [Google Scholar]
- Kitada M., Ogura Y., Suzuki T. A low-protein diet exerts a beneficial effect on diabetic status and prevents diabetic nephropathy in Wistar fatty rats, an animal model of type 2 diabetes and obesity. Nutr. Metab. 2018;15:20. doi: 10.1186/s12986-018-0255-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan H.B., Nelson R.L. Proteomic analysis of high protein soybean (Glycine max) accessions demonstrates the contribution of novel glycinin subunits. J. Agric. Food Chem. 2011;59:2432–2439. doi: 10.1021/jf104330n. [DOI] [PubMed] [Google Scholar]
- Lagkouvardos I., Pukall R., Abt B. The Mouse Intestinal Bacterial Collection (miBC) provides host-specific insight into cultured diversity and functional potential of the gut microbiota. Nature Microbiol. 2016;1:16131. doi: 10.1038/nmicrobiol.2016.131. [DOI] [PubMed] [Google Scholar]
- Lewicki S., Lewicka A., Kalicki B. The influence of vitamin B12 supplementation on the level of white blood cells and lymphocytes phenotype in rats fed a low-protein diet. Cent. Eur. J. Immunol. 2014;39:419–425. doi: 10.5114/ceji.2014.47723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Li J., Mao G., Yan L. Effect of the sulfation pattern of sea cucumber-derived fucoidanoligosaccharides on modulating metabolic syndromes and gut microbiotadysbiosis caused by HFD in mice. J. Foof Funct. 2019;55:193–210. [Google Scholar]
- Lozupone C., Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W., Shen Z., Deng M. Roseburia intestinalis supernatant ameliorates colitis induced in mice by regulating the immune response. Mol. Med. Rep. 2019;20:1007–1016. doi: 10.3892/mmr.2019.10327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti S., Chatterjee A.K. Differential response of cellular antioxidant mechanism of liver and kidney to arsenic exposure and its relation to dietary protein deficiency. Environ. Toxicol. Pharmacol. 2000;8:227–235. doi: 10.1016/s1382-6689(00)00046-6. [DOI] [PubMed] [Google Scholar]
- Masuoka H., Suda W., Tomitsula E. The influences of low protein diet on the intestinal microbiota of mice. Sci. Rep. 2020;10:17077. doi: 10.1038/s41598-020-74122-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki A., Kandasamy S., Michael H. Protein deficiency reduces efficacy of oral attenuated human rotavirus vaccine in a human infant fecal microbiota transplanted gnotobiotic pig model. Vaccine. 2018;8:6270–6281. doi: 10.1016/j.vaccine.2018.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiwaki H., Ito M., Ishida T. Meta-analysis of gut dysbiosis in Parkinson’s disease. Mov. Disord. 2020;35:1626–1635. doi: 10.1002/mds.28119. [DOI] [PubMed] [Google Scholar]
- Nkosi C.Z., Opoku A.R., Terblanche S.E. Antioxidative effects of pumpkin seed (Cucurbita pepo) protein isolate in CCl4-Induced liver injury in low-protein fed rats. Phytother Res. 2006;20:935–940. doi: 10.1002/ptr.1977. [DOI] [PubMed] [Google Scholar]
- Pérez-Martí A., Garcia-Guasch M., Tresserra-Rimbau A. A low-protein diet induces body weight loss and browning of subcutaneous white adipose tissue through enhanced expression of hepatic fibroblast growth factor 21 (FGF21) Mol. Nutr. Food Res. 2017;61:1600725. doi: 10.1002/mnfr.201600725. [DOI] [PubMed] [Google Scholar]
- Ragonnaud E., Biragyn A. Gut microbiota as the key controllers of “healthy” aging of elderly people. Immun. Adeing. 2021;18:1401. doi: 10.1186/s12979-020-00213-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanahan F., Ghosh T.S., O’Toole P.W. The healthy microbiome—what is the definition of a healthy gut microbiome? Gatroenterol. 2021;160:483–494. doi: 10.1053/j.gastro.2020.09.057. [DOI] [PubMed] [Google Scholar]
- Shibayama J., Goto M., Kuda T. Effect of rice bran fermented with Schharomyces cerevisiae and Lactobacillus plantarum on gut microbiome of mice fed high-sucrose diet. Benef. Microbes. 2019;10:811–821. doi: 10.3920/BM2019.0072. [DOI] [PubMed] [Google Scholar]
- Shikano A., Kuda T., Shibayama J. Effects of Lactobacillus plantarum Uruma-SU4 fermented green loofah on lasma lipid levels and gut microbiome of high-fat diet fed mice. Food Res. Int. 2019;121:817–824. doi: 10.1016/j.foodres.2018.12.065. [DOI] [PubMed] [Google Scholar]
- Sinclair L., Osman O.A., Bertilsson S. Microbial community composition and diversity via 16S rRNA gene amplicons: evaluating the Illumina platform. PloS One. 2015;10 doi: 10.1371/journal.pone.0116955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei M., Kuda T., Fukunaga M. Effects of edible algae on caecal microbiomes of ICR mice fed a high-sucrose and low–dietary fibre diet. J. Appl. Phycol. 2019;31:3969–3978. [Google Scholar]
- Takei M.N., Kuda T., Taniguchi M. Detection and isolation of low molecular weight alginate- and laminaran susceptible gut indigenous bacteria from ICR mice. Carbohydr. Polym. 2020;238:116205. doi: 10.1016/j.carbpol.2020.116205. [DOI] [PubMed] [Google Scholar]
- Tamanai-Shacoori Z., Smida I., Bousarghin L. Roseburia spp.: a marker of health? Future Microbiol. 2017;12:157–170. doi: 10.2217/fmb-2016-0130. [DOI] [PubMed] [Google Scholar]
- Taniguchi, M., Kuda, T., Takei, M., et al., Effects of fermented Aphanizomenon flos-aquae on the caecal microbiome of mice fed a high-sucrose and low-dietary fibre diet. J. Appl. Phycol. 33, 397-407.
- Tawtep S., Fukiya, Lee J. Isolation of six novel 7-oxo- or urso-type secondary bile acid-producing bacteria from rat cecal contents. J. Biosci. Bioeng. 2017;124:514–522. doi: 10.1016/j.jbiosc.2017.06.002. [DOI] [PubMed] [Google Scholar]
- Tomaz F.S., Tomsett K.I., Panchel S.K. Wasabi supplementation alters the composition of the gut microbiota of diet-induced obese rats. J. Food Funct. 2020;67:103868. [Google Scholar]
- Uauy R., Kurpad A., Tano-Debrah K. Role of protein and amino acids in infant and young child nutrition: protein and amino acid needs and relationship with child growth. J. Nutr. Sci. Vitaminol. 2015;61:S192–S194. doi: 10.3177/jnsv.61.S192. [DOI] [PubMed] [Google Scholar]
- Umirah F., Neoh C.F., Ramasamy K. Differential gut microbiota composition between type 2 diabetes mellitus patients and healthy controls: a systematic review. Diabetes Res. Clin. Pract. 2021 doi: 10.1016/j.diabres.2021.108689. (in press) [DOI] [PubMed] [Google Scholar]
- Wang Y., Wang Z., Handa C.L., Xu J. Effects of ultrasound pre-treatment on the structure of β-conglycinin and glycinin and the antioxidant activity of their hydrolysates. Food Chem. 2017;218:165–172. doi: 10.1016/j.foodchem.2016.09.069. [DOI] [PubMed] [Google Scholar]
- Wilck N., Matus M.G., Keamy S.M. Salt-responsive gut commensal modulates TH17 axis and disease. Nature. 2017;551:585–589. doi: 10.1038/nature24628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A.S., Koller K.R., Ramaboli M.C. Diet and the human gut microbiome: an international review. Dig. Dis. Sci. 2020;65:723–740. doi: 10.1007/s10620-020-06112-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y., Fukunaga M., Kuda T. Detection and isolation of protein susceptible indigenous bacteria affected by dietary milk-casein, albumen and soy-protein in the caecum of ICR mice. Int. J. Biol. Macromol. 2020;144:813–820. doi: 10.1016/j.ijbiomac.2019.09.159. [DOI] [PubMed] [Google Scholar]
- Xia Y., Kuda T., Toyama A. Detection and isolation of bacteria affected by dietary cumin, coriander, turmeric, and red chili pepper in the caecum of ICR mice. J. Funct. Foods. 2019;61:103467. [Google Scholar]
- Xiang N., Lyu Y., Zhu X., Bhunia A.K., Narsimhan G. Methodology for identification of pore forming antimicrobial peptides from soy protein subunits β-conglycinin and glycinin. Peptides. 2016;85:27–40. doi: 10.1016/j.peptides.2016.09.004. [DOI] [PubMed] [Google Scholar]
- Yuan Y., Zhou J., Zheng Y. Beneficial effects of polysaccharide-rich extracts from Apocynum venetumleaves on hypoglycemic and gut microbiota in type 2 diabetic mice. Biomed. Pharmacother. 2020;127:110182. doi: 10.1016/j.biopha.2020.110182. [DOI] [PubMed] [Google Scholar]
- Zhai S., Qin S., Li L. Dietary butyrate suppresses inflammation through modulating gut microbiota in high-fat diet-fed mice. FEMS Microbiol. Lett. 2019;336:fnz153. doi: 10.1093/femsle/fnz153. [DOI] [PubMed] [Google Scholar]
- Zhao L., Zhang Q., Ma W. A combination of quercetin and resveratrol reduces obesity in high-fat diet-fed rats by modulation of gut microbiota. Food Funct. 2017;8:4644–4656. doi: 10.1039/c7fo01383c. [DOI] [PubMed] [Google Scholar]
- Zheng Q., Li D., He Y. Discrepant gut microbiota markers for the classification of obesity-related metabolic abnormalities. Sci. Rep. 2019;9:13424. doi: 10.1038/s41598-019-49462-w. [DOI] [PMC free article] [PubMed] [Google Scholar]