Abstract
The continued global pandemic of coronavirus disease 2019 (COVID-19) poses a serious threat to global public health and social stability and it has become a serious global public health problem. Unfortunately, existing diagnostic and therapeutic approaches for the prevention and control of COVID-19 have many shortcomings. In recent years, the emerging CRISPR/Cas technology can complement the problems of traditional methods. Biological tools based on CRISPR/Cas systems have been widely used in biomedicine. In particular, they are advantageous in pathogen detection, clinical antiviral therapy, drug, and vaccine development. Therefore, CRISPR/Cas technology may have great potential for application in the prevention and control of COVID-19 and emerging infectious diseases in the future. This article summarizes the existing applications of CRISPR/Cas technology in infectious diseases with the aim of providing effective strategies for the prevention and control of COVID-19 and other emerging infectious diseases in the future.
Keywords: CRISPR/Cas, SARS-CoV-2, pathogen detection, clinical therapy, drug and vaccine development
Introduction
Coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) first broke out in Wuhan, China, and rapidly grew into a global pandemic, seriously endangering human health and social development. By February 23, 2021, SARS-CoV-2 had become a global pandemic in 180 countries and regions, with the cumulative number of infections exceeding 110 million and the cumulative number of deaths exceeding 2.48 million. More importantly, with the ongoing mutation of SARS-CoV-2 and the coming winter and spring, these numbers may continue to increase (Grubaugh et al., 2020; Korber et al., 2020). In response to the SARS-CoV-2 pandemic, it is urgent to strengthen pathogen detection capacity and accelerate the development of vaccines and specific antiviral drugs. At present, the traditional nucleic acid detection method, mainly quantitative real-time reverse transcription-polymerase chain reaction (qRT-PCR), is recommended by WHO and some medical departments to detect pathogens. However, some researchers found that the results are unstable, and the sensitivity is even as low as 42.10%, and the sample processing requirements are very strict (Bustin and Nolan, 2020; Tesija Kuna et al., 2021). More importantly, the therapeutic effect of antiviral drugs has not achieved the expected effect, and symptomatic treatment is still the main treatment at present (Khan et al., 2020). Vaccine development is still in the clinical evaluation stage (Folegatti et al., 2020; Gao Q. et al., 2020; Tu et al., 2020), it will take some time before it can be applied, and even the mutation of the virus may make the vaccine less effective than expected (Becerra-Flores and Cardozo, 2020; Li Q. et al., 2020). Moreover, the vaccine may not be able to meet the huge global demand.
CRISPR (clustered regularly interspaced short palindromic repeats) consists of a gene encoding a Cas-related protein and the CRISPR array, which is a repetitive sequence within the prokaryotic genome, an immune weapon produced by the struggle between bacteria and phages throughout the history of evolution (Bondy-Denomy et al., 2013; Koonin et al., 2017). Therefore, bacteria can protect themselves by using the CRISPR/Cas system to eliminate foreign invading phage genes. Since 1987, CRISPR/Cas was discovered by Ishino in E. coli, scientists have been developing several biotechnologies based on this basic principle (Ishino et al., 1987). The development and application of the CRISPR/Cas system have given new impetus to the development of life sciences and biotechnology (Knott and Doudna, 2018). Especially in medical science, CRISPR/Cas has made remarkable achievements in gene editing, nucleic acid detection, functional gene screening, and so on (Zhou et al., 2014; Kurata et al., 2018; Pickar-Oliver and Gersbach, 2019; Wang et al., 2019; Yan et al., 2019). Also, in recent years, scientists have used the CRISPR/Cas system broadly in the prevention and control of infectious diseases. In the article, we reviewed recent advances regarding the application of the CRISPR/Cas system in the prevention and control of COVID-19 and other infectious diseases, such as in molecular diagnostics, infectious disease treatment, the antiviral drug, and vaccine development. And provide effective strategies for the prevention and control of COVID-19 and other future emerging infectious diseases.
Application of CRISPR/Cas in Pathogen Detection
Inadequacy of Existing Pathogen Detection
Pathogen detection is crucial for the prevention and control of infectious diseases. Early identification of pathogens enables identification of the source of infection and timely quarantine, so that appropriate anti-infective protocols can be developed to improve patient prognosis, especially for severe emerging infectious diseases such as COVID-19. However, the current detection of SARS-CoV-2 is mainly by qRT-PCR, which is time-consuming and dependent on specialized equipment and operators, which may not be available in developing countries or remote rural hospitals, and is not suitable for field testing (Corman et al., 2020; Lu et al., 2020). Besides, limited sensitivity may prevent the detection of asymptomatic infected persons, or some patients during the time-window of viral replication, who are likely to contribute to further transmission without medical treatment (Li Y. et al., 2020; Younes et al., 2020; Yu et al., 2020). overcome the shortcomings of qRT-PCR technology, some experts have suggested using a combined antibody assay to detect SARS-CoV-2 (Li Z. T. et al., 2020). Although enzyme‐linked immunosorbent assay (ELISA) has been successfully used to detect SARS-CoV-2, specific antibodies need to be produced after more than 7 days of infection with the virus (Lin et al., 2020; Lisboa Bastos et al., 2020; Wang Q. et al., 2020). In addition, antibody detection can produce cross-reactivity, and people with poor immunity may produce fewer antibodies, which can affect test results (Liu W. et al., 2020). Furthermore, isothermal amplification detection technology has a great prospect of application because of its simple operation and can be applied to field detection. However, the current isothermal amplification technology has its inherent defects in sensitivity, specificity, and anti-interference. Therefore, highly sensitive and specific, field-applicable pathogen detection is necessary for the detection of emerging infectious diseases ( Table 1 ).
Table 1.
Available diagnostic methods for COVID-19.
| Test | Detection type | Positive rate | Time | Characteristics | Limitations | References |
|---|---|---|---|---|---|---|
| qRT-PCR | nucleic acid detection | 38% | >2h | Currently the most common detection method, simple quantitative detection. | Time-consuming, poor sensitivity | (Liu R. et al., 2020) |
| ELISA | Immunological detection | 80.4% | >2h | Fast, easy to operate, low sample volumes | Cross-reactivity, Antibodies are produced about 7-10 days after infection. | (Liu W. et al., 2020) |
| RT-LAMP | nucleic acid detection | 92.9% | 40–60 min | Highly sensitive, fast, and convenient field testing | Prone to false positives | (Baek et al., 2020) |
| CRISPR-based detection | nucleic acid detection | 96% | 30-60min | Highly sensitive, high specificity, fast and convenient for field testing., low-cost | Not yet widespread and in clinical trials | (Patchsung et al., 2020) |
| ddPCR | nucleic acid detection | 40% | NA | Absolute quantification, high sensitivity | Expensive equipment, poor accessibility, and complexity of the operation | (Suo et al., 2020) |
| CT Scan | Radiological Screening | NA | NA | Check for disease progression, easy access, rapid detection | Unable to identify the pathogen of infection, clinical assistant tests | (Dong et al., 2020) |
Application of CRISPR/Cas-Based Systems for Rapid, Accurate Point-of-Care Diagnostics
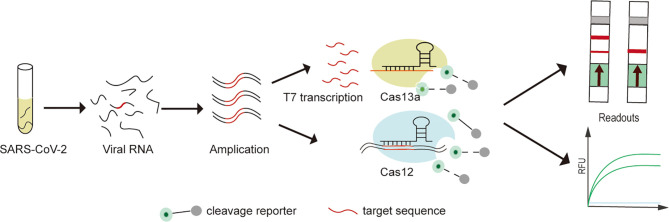
In recent years, emerging nucleic acid detection technologies based on CRISPR/Cas developments have opened up new opportunities for pathogen detection. SHERLOCK (Cas13a), DETECTR (Cas12a), CDetection (Cas12b) and Cas14-DETECTR, which have successfully performed rapid, highly sensitive and accurate detection of a variety of pathogens (Gootenberg et al., 2017; Chen et al., 2018a; Chen et al., 2018b; Harrington et al., 2018; Teng et al., 2019) ( Figure 1 ). Compared to traditional assays, CRISPR/Cas system has advantages in terms of rapidity, low cost, portability, ease of operation and extension, while maintaining high sensitivity and specificity (Zhou et al., 2018). In addition, the low sample quality requirements and high interference resistance mean that rapid sample pre-processing such as HUDSON can be combined to enable nucleic acid extraction on site without relying on specialized equipment (Gootenberg et al., 2017; Wang M. et al., 2020). In combination with lateral flow test strip technology for visualizing point-of-care diagnostics with the naked eye (Dai et al., 2020; Van Dongen et al., 2020). Fluorescence readout also can achieve point-of-care diagnostics with portable fluorescence collectors or colorimetric analysis (Yuan et al., 2020; Cheng et al., 2021).
Figure 1.
Experimental protocol for SARS-CoV-2 detection on CRISPR/Cas detection platform. (1) The extracted SARS-CoV-2 viral RNA is first pre-amplified (RT-RPA and RT-LAMP) into double-stranded DNA (dsDNA). (2) For Cas13a-SHERLOCK assay, dsDNA is first T7 transcribed into single-stranded RNA (ssRNA), followed by activation of the Cas13a cleavage reporter. (3) For Cas12a or Cas12b assays, direct detection of pre-amplified dsDNA enables activation of the Cas12a or Cas12b cleavage reporter. (4) Visualization of CRISPR/Cas assay results by fluorescence and lateral flow readouts.
CRISPR/Cas based detection of SARS-CoV-2 has been actively researched by scientists since the COVID-19 outbreak ( Table 2 ). For example, James P. Broughton can successfully detect SARS-COV-2 within 30-40 minutes using the CRISPR-Cas12a-based method with a limit of detection (LoD) of up to 10 copies/µL, and the combination of lateral flow strip technology enables visible detection results in the field (Broughton et al., 2020b). Similarly, based on the CRISPR/Cas13a method, the SHERLOCK platform has been successfully validated in 534 clinical samples with a LoD of 42 copies/µL and 100% sensitivity for fluorescence readout and 97% for lateral-flow detection (Patchsung et al., 2020). Feng Zhang’s team developed the SARS-CoV-2 detection platform STOPCovid.v2 based on AapCas12b, which enables point-of-care diagnostics to be performed quickly and easily within 1 hour, without relying on specialized equipment. Also, it has obtained the FDA (US Food and Drug Administration) emergency use right of SARS-CoV-2 detection (Guglielmi, 2020; Joung et al., 2020). Besides, researchers using the Cas12a-based opvCRISPR platform were able to visualize fluorescence detection of SARS-CoV-2 with the naked eye in 45 min (Wang et al., 2021). It is worth noting that CRISPR/Cas technology allows not only rapid detection but also massively multiplexed nucleic acid detection. For example, Cheri M. Ackerman developed Combinatorial Arrayed Reactions for Multiplexed Evaluation of Nucleic acids (CARMEN) based on CRISPR/Cas13, which allows for the simultaneous differentiation of 169 viruses associated with humans, as well as the multiple identifications of various subtypes and even drug resistance mutations, thereby increasing detection rates while reducing detection costs (Ackerman et al., 2020).
Table 2.
Summary of the CRISPR/Cas platform for SARS-CoV-2 detection.
| Detection platforms | CRISPR/Cassystem | Limit of detection | Time | Characteristics | References |
|---|---|---|---|---|---|
| CREST | Cas13a | 10copies/μL | 30-50min | Combined with RT-PCR technology, CRISPR detection is stable. However, there may be drawbacks to field detection. | (Rauch et al., 2020) |
| SHERLOCK | Cas13a | 42 copies per reaction | 70min | Combination of RT-RPA amplification and lateral flow assay for rapid field testing. | (Patchsung et al., 2020) |
| SHINE | Cas13a | 10copies/μL | 50min | Improved sample pre-processing (HUDSON) combined with a one-step SHERLOCK for fast field inspection. | (Arizti-Sanz et al., 2020) |
| DETECTR | Cas12a | 10copies/μL | 45min | Combined with LAMP to solve the problem of unstable RPA amplification enables faster and more sensitive detection in the field. | (Broughton et al., 2020a) |
| STOPCovid | Cas12b | 100 copy per reaction | <60min | Integrated LAMP and Cas12b detection enables one-step detection, making it easier and faster to operate. | (Joung et al., 2020) |
| CARMEN | Cas13a | attomolar | NA | Enables large-scale pathogen detection and simultaneous identification of 165 human-associated viruses. | (Ackerman et al., 2020) |
Moreover, CRISPR/Cas technology has the unique ability to detect single nucleotide polymorphisms (SNP) compared to other above-mentioned detection technologies. Compared with conventional sequencing for SNP detection, CRISPR/Cas technology is simpler, faster, and cheaper, and more importantly, can detect low-frequency mutations. Faced with SARS-CoV-2 mutations such as the D614G and N501Y locus mutation. These mutations may increase the infectivity of SARS-COV-2 and even pose a significant threat to existing therapies and vaccines (Li Q. et al., 2020; Yurkovetskiy et al., 2020). So the identification of mutant strains based on high-specificity CRISPR/Cas technology has great significance. Although no researchers have yet performed CRISPR/Cas-based detection of the SARS-COV-2 mutation site, some scientists have successfully used Cas13a to detect the HBV drug resistance site rt204 (Gootenberg et al., 2017; Wang S. et al., 2020).
CRISPR/Cas in Clinical Antiviral Therapy
Application of Cas9 Gene-Editing to Clinical Antiviral Therapy
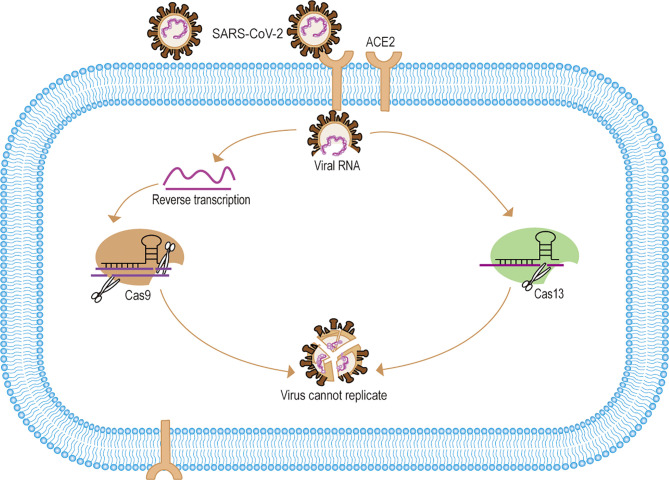
In the face of emerging infectious diseases such as SARS, MERS, and COVID-19, existing antiviral drugs are often ineffective. However, developing an effective antiviral drug is an arduous process. Therefore, clinical treatment is mainly supportive and to prevent complications, but in the absence of specific antivirals, this approach may result in a higher case fatality rate. The scientific community has been searching for ways to eliminate viruses more quickly and accurately, which could have significant implications for early source prevention and control of emerging infectious diseases. In recent years, experts using CRISPR/Cas9 gene-editing technology to treat many clinically refractory diseases. For one thing, the researchers used CRISPR/Cas9 targeting virus-specific sequences for gene editing to directly eliminate the virus to treat the disease. For example, The Edward M. Kennedy in the cell-free system trial used Cas9 to specifically target the HPV E6E7 gene to eliminate HPV infection to treat cervical cancer (Kennedy et al., 2014). Zhen has validated in animal studies that Cas9-targeted elimination of covalently closed circular DNA (cccDNA) is effective in the treatment of hepatitis B (Zhen et al., 2015). In addition, scientists also successfully used Cas9 to accurately eliminate Cytomegalovirus, Epstein-Barr virus infections in vitro (Ebina et al., 2013; Hu et al., 2014; Huo and Hu, 2019; Chen S. J. et al., 2020). For another thing, some researchers have used Cas9 to engineer T cells with the ability to treat specific diseases and thus indirectly eliminate viral infections (Zhao et al., 2018; Stadtmauer et al., 2020). For example, Cas9 was developed to improve CAR-T (Chimeric Antigen Receptor T-Cell Immunotherapy) therapy to treat incurable cancers such as B-cell leukemias and lymphomas (Brentjens et al., 2011; Porter et al., 2016; Eyquem et al., 2017; Labanieh et al., 2018). Moreover, many in vivo trial studies show Cas9 gene editing modifies CD4+ T cells to be effective in treating HIV infection, which may provide a new approach to refractory infectious diseases (Ebina et al., 2013; Liu et al., 2017; Hultquist et al., 2019; Xiao et al., 2019). With SARS-CoV-2 and other difficult to treat emerging infectious disease, CRISPR/Cas9 gene editing is likely to be one of the best treatments in the future (Grifoni et al., 2020) ( Figure 2 ). In addition, with ongoing research and development, Cas12a and Cas14 also hold great potential for gene editing to treat infectious diseases in the future (Gao Z. L. et al., 2020).
Figure 2.
Mechanisms of CRISPR/Cas systematic elimination of SARS-CoV-2. (1) SARS-CoV-2 targets the ACE2 receptor to release viral RNA into cells. (2) After reverse transcription of the viral RNA to form dsDNA, the pre-designed sgRNA-Cas9 identified and specifically cleaved the viral DNA. (3) For Cas13 the system can directly identify and eliminate viral RNA. (4) The CRISPR/Cas system eliminates the SARS-CoV-2 gene, thus making the viral RNA unable to replicate and the viral gene unable to be expressed, and eventually making SARS-CoV-2 unable to replicate.
Cas13 Specifically Targets RNA Viruses for Degradation
Compared to Cas9, gene editing of Cas13-targeted RNA may offer a new approach to RNA virus therapy (Freije et al., 2019). LshCas13a system specifically targeted E6E7mRNA of HPV has a positive effect on inhibiting human cervical cancer in vivo testing (Chen Y. et al., 2020). Besides, Hao Li used Cas13a cleavage of the Dengue virus NS3 gene to efficiently inhibits viral replication in vivo testing (Li H. et al., 2020). Interestingly, Timothy R. Abbott used Cas13d to develop the PAC-MAN platform (prophylactic antiviral CRISPR in human cells) for viral inhibition that can effectively degrade RNA from SARS-CoV-2 sequences and live influenza A virus (IAV) in human lung epithelial cells. Notably, this approach provides a novel treatment strategy for SARS-CoV-2 and influenza virus, among others (Abbott et al., 2020; Nguyen et al., 2020) ( Figure 2 ).
Application of CRISPR/Cas Systems in Vaccine and Drug Development
CRISPR/Cas9 Assisted Drug Development and Target Screening
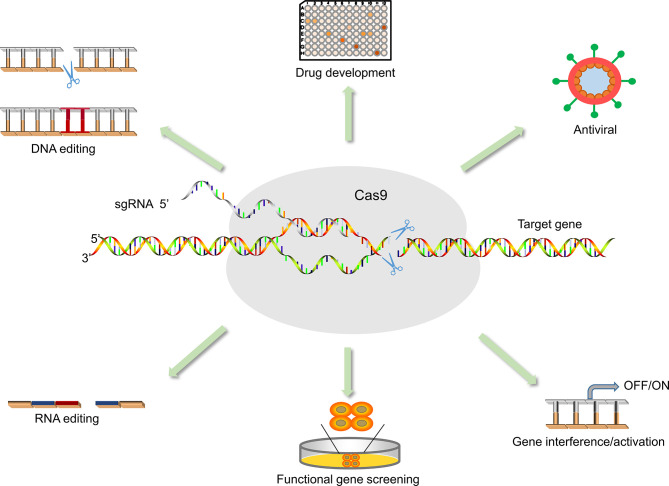
The rapid development of effective antiviral drugs is also essential for the treatment of COVID-19 ( Table 3 ). However, existing antiviral drugs are often ineffective in the treatment of emerging infectious diseases, mainly due to the lack of effective targets for the treatment of emerging viruses. Unfortunately, the main reason for the failure of antiviral drug development is also the difficulty in identifying effective targets, which makes development more difficult, expensive, and time-consuming. Moreover, host factors associated with viral action are critical to the success of treatment and drug development (Li B. et al., 2020; Wei et al., 2020). Therefore, how to identify functional targets more quickly and accurately is one of the challenges plaguing drug development. Although RNAi is undoubtedly a widely recognized and powerful method, it may produce misleading results due to off-target effects. As a result, the off-target effect may affect many functionally important genes (Settleman et al., 2018; Haley and Roudnicky, 2020). The emergence and development of CRISPR/Cas technology can more easily and quickly find potential drug targets to inhibit the replication of viruses, and provide safer and more feasible strategies for the treatment and prevention of diseases (Behan et al., 2019) ( Figure 3 ). For instance, Flint used CRISPR/Cas technology to screen the genome-wide library and found that N-acetylglucosamine-1-phosphate transferase subunits alpha and beta (GNPTAB) were potential targets for the anti-Ebola virus (Flint et al., 2019). Interestingly, researchers on the bat cells of genome-wide screening explore the target of widespread antiviral treatment, after screening detects MTHFD1 targets is a potential target for broad-spectrum antiviral drugs. MTHFD1 inhibitor caprolactone broad antiviral activities against the Zika virus, mumps virus, and importantly, SARS-CoV-2 (Anderson et al., 2020). Furthermore, CRISPR/Cas screening can reveal host genes that regulate SARS-CoV-2 infection, such as SRSF protein kinases 1 and 2, ACE2, and HMGB1 (Heaton et al., 2020; Hoffmann et al., 2020; Wang R. et al., 2020; Wei et al., 2020). This provides insight into the signaling pathways underlying viral action. Although these articles have not been peer-reviewed, these ideas provide good insights and thoughts for exploring responses to emerging infectious diseases such as SARS-COV-2.
Table 3.
Summary of current antiviral drugs or approaches to treat COVID-19.
| Type | Candidate therapies | Mechanism of action | Clinical efficacy | References |
|---|---|---|---|---|
| Antiviral | Remdesivir | Inhibition of viral RNA replication | Efficacy is controversial, Under clinical trial for SARS-CoV-2 | (Grein et al., 2020; Wang Y. et al., 2020) |
| Antiviral | Lopinavir-Ritonavir | Viral Protease Inhibitors, inhibits 3CLpro | Under clinical trial for SARS-CoV-2 | (Cao et al., 2020) |
| Antiviral | Hydroxy-chloroquine, Chloroquine | Increasing endosomal pH prevents membrane fusion between virus and host cells | Under clinical trial for SARS-CoV-2 | (Gautret et al., 2020; Pereira, 2020) |
| Antiviral | Favipiravir | RNA polymerase inhibitors | Under clinical trial for SARS-CoV-2 | (Cai et al., 2020; Du and Chen, 2020) |
| Antiviral | PAC-MAN method based on CRISPR-Cas13d, CRISPR-Cas9 | Specific degradation of SARS-COV-2 | Although not in clinical trials, its unique advantages provide therapeutic strategies for refractory, poorly treated viruses | (Abbott et al., 2020) |
| Symptomatic therapeutic | Bevacizumab, SARS-CoV-2-Specific Neutralizing Antibodies | Attenuating the Inflammatory Response | Under clinical trial for SARS-CoV-2 | (Thickett et al., 2001; Zhou and Zhao, 2020) |
Figure 3.
Application of CRISPR/Cas9 in medical sciences. The application of Cas9 is not only for antiviral but also for functional gene screening, drug development, animal model vector construction, and signaling pathway research.
Application of CRISPR/Cas9 in Vaccine Development
The prevention and control of outbreaks and epidemics of COVID-19 and other emerging infectious diseases require not only specific antiviral drugs but also vaccines. Vaccination is one of the most economical and effective public health interventions for the prevention and control of infectious diseases and one of the effective ways to prevent the large-scale spread of infectious diseases. However, the development of a vaccine is a long, complex process, and expensive. How to develop specific vaccines quickly and easily is a difficult problem for scientists. In response to emerging infectious diseases, CRISPR/Cas9 could boost progress in vaccine development. For example, Mustafa Ozan Atasoy developed and applied a highly efficient and rapid NHEJ-CRISPR/Cas9 (non-homologous end-joining, NHEJ) and Cre–Lox-mediated genome-editing approach for simultaneous deletion of virulence factors and insertion of antigens into the infectious laryngotracheitis virus to generate recombinant, multivalent, and safer vaccine vectors (Atasoy et al., 2019). Homology-directed repair (HDR) CRISPR/Cas9 and erythrocyte binding be used for the rapid generation of recombinant turkey herpesvirus-vectored avian influenza virus vaccines (Chang et al., 2019). In addition to Manuel used the CRISPR/Cas9 gene-editing system to produce recombinant African swine fever virus (ASFVs) (Borca et al., 2018). And the recombinant ASFV has great significance for the development of the live attenuated vaccine. Significantly, the researchers generated mouse models expressing human angiotensin-converting enzyme II (hACE2) using the Cas9 knock-in technique, which was used to study the transmission and pathogenesis of SARS-CoV-2 and to provide a useful tool for evaluating COVID-19 vaccines and therapeutic agents (Sun et al., 2020) ( Figure 3 ).
Limitations and Improvements of the CRISPR/Cas System
Despite the many advantages of the emerging CRISPR/Cas system mentioned above, it also has some limitations. For example, Cas9 may have limited specificity of the recognition site during gene editing, which may cause a high off-target effect and eventually lead to permanent damage to the genome, such as cancer (Fu et al., 2013). In addition, Cas12 and Cas9 also need to recognize protospacer adjacent motif (PAM) sequences before activation, and PAM sequences may limit targeting and affect editing efficiency and flexibility (Mojica et al., 2009; Marraffini and Sontheimer, 2010). More importantly, the transport vector of the CRISPR/Cas system is also a major problem (Komor et al., 2017; Glass et al., 2018). Currently, viruses are mainly used as transport vectors, which may cause potential harm to the human body. In addition, the human body may be immune to sgRNA and Cas proteins, thus affecting the proper role of the CRISPR/Cas system (Kim et al., 2018). Beyond that, it is noteworthy that the CRISPR/Cas system may have implications for medical ethics (Brokowski and Adli, 2019).
As the CRISPR/Cas system has been deeply researched, some limitations have been addressed. Jennifer Doudna’s team used the anti-CRISPR protein AcrllA4 to reduce the incidence of off-target effects by a factor of four, without any disruption to gene editing (Chen et al., 2017; Harrington et al., 2017; Jiang et al., 2019). In terms of optimizing the delivery carrier of the CRISPR/Cas system, scientists have made gradual progress in using nanocarriers to replace virus carriers (Lee et al., 2017). Another approach is to use smaller Cas proteins such as Cas14 (Harrington et al., 2018). Even more exciting is that although the CRISPR/Cas system needs to recognize specific PAM sequences, recent SCIENCE reports that using near-PAMless engineered can almost eliminate the limitations of PAM. Thus, high precision targeting can be realized in the application of genome editing, while further reducing off-target effect (Nishimasu et al., 2018).
Conclusions
Although the clinical application of the CRISPR/Cas system is still in its infancy, its emergence provides many possibilities in the biomedical field. We should be more supportive and inclusive of emerging technologies. With continued in-depth research and clinical validation, we believe that the application of CRISPR/Cas system will play an irreplaceable role in the prevention and control of emerging infectious diseases in the future.
Author Contributions
RD and GD designed the study. RD and SC wrote the paper. JL, MY, YJ, HY, MC, and SC provided writing and revision suggestions. All authors contributed to the article and approved the submitted version.
Funding
The work was supported by the National Science and Technology Specific Projects (2018ZX10301407), Hunan Provincial Key Laboratory of Clinical Epidemiology (2020ZNDXLCL003), and The Key Scientific Research Projects in Colleges and Universities of Henan Province (20A330004). Henan Province University Science and Technology Innovation Talent Projects (17HASTIT045).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Abbott T. R., Dhamdhere G., Liu Y. X., Lin X. Q., Goudy L., Zeng L. P., et al. (2020). Development of CRISPR as an Antiviral Strategy to Combat SARS-Cov-2 and Influenza. Cell 181, 865–86+. 10.1016/j.cell.2020.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackerman C. M., Myhrvold C., Thakku S. G., Freije C. A., Metsky H. C., Yang D. K., et al. (2020). Massively Multiplexed Nucleic Acid Detection With Cas13. Nature 582, 277–282. 10.1038/s41586-020-2279-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D. E., Cui J., Ye Q., Huang B., Zu W., Gong J., et al. (2020). Orthogonal Genome-Wide Screenings in Bat Cells Identify MTHFD1 as a Target of Broad Antiviral Therapy. bioRxiv 2020.2003.2029.014209. 10.1101/2020.03.29.014209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arizti-Sanz J., Freije C. A., Stanton A. C., Petros B. A., Boehm C. K., Siddiqui S., et al. (2020). Streamlined Inactivation, Amplification, and Cas13-Based Detection of SARS-Cov-2. Nat. Commun. 11, 5921. 10.1101/2020.05.28.119131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atasoy M. O., Rohaim M. A., Munir M. (2019). Simultaneous Deletion of Virulence Factors and Insertion of Antigens Into the Infectious Laryngotracheitis Virus Using NHEJ-CRISPR/Cas9 and Cre-Lox System for Construction of a Stable Vaccine Vector. Vaccines 7 (4), 207. 10.3390/vaccines7040207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek Y. H., Um J., Antigua K. J. C., Park J. H., Kim Y., Oh S., et al. (2020). Development of a Reverse Transcription-Loop-Mediated Isothermal Amplification as a Rapid Early-Detection Method for Novel SARS-Cov-2. Emerg. Microbes Infect. 9, 998–1007. 10.1080/22221751.2020.1756698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becerra-Flores M., Cardozo T. (2020). SARS-Cov-2 Viral Spike G614 Mutation Exhibits Higher Case Fatality Rate. Int. J. Clin. Pract. 74 (8), e13525. 10.1111/ijcp.13525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behan F. M., Iorio F., Picco G., Goncalves E., Beaver C. M., Migliardi G., et al. (2019). Prioritization of Cancer Therapeutic Targets Using CRISPR-Cas9 Screens. Nature 568, 511–51+. 10.1038/s41586-019-1103-9 [DOI] [PubMed] [Google Scholar]
- Bondy-Denomy J., Pawluk A., Maxwell K. L., Davidson A. R. (2013). Bacteriophage Genes That Inactivate the CRISPR/Cas Bacterial Immune System. Nature 493, 429–U181. 10.1038/nature11723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borca M. V., Holinka L. G., Berggren K. A., Gladue D. P. (2018). CRISPR-Cas9, a Tool to Efficiently Increase the Development of Recombinant African Swine Fever Viruses. Sci. Rep. 8 (1), 3154. 10.1038/s41598-018-21575-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brentjens R. J., Riviere I., Park J. H., Davila M. L., Wang X. Y., Stefanski J., et al. (2011). Safety and Persistence of Adoptively Transferred Autologous CD19-Targeted T Cells in Patients With Relapsed or Chemotherapy Refractory B-Cell Leukemias. Blood 118, 4817–4828. 10.1182/blood-2011-04-348540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brokowski C., Adli M. (2019). CRISPR Ethics: Moral Considerations for Applications of a Powerful Tool. J. Mol. Biol. 431, 88–101. 10.1016/j.jmb.2018.05.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton J. P., Deng X., Yu G., Fasching C. L., Servellita V., Singh J., et al. (2020. a). CRISPR–Cas12-Based Detection of SARS-Cov-2. Nat. Biotechnol. 38, 870–874. 10.1038/s41587-020-0513-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton J. P., Deng X. D., Yu G. X., Fasching C. L., Servellita V., Singh J., et al. (2020. b). CRISPR-Cas12-Based Detection of SARS-Cov-2. Nat. Biotechnol. 38, 870–U854. 10.1038/s41587-020-0513-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin S. A., Nolan T. (2020). RT-Qpcr Testing of SARS-Cov-2: A Primer. Int. J. Mol. Sci. 21 (8), 3004. 10.3390/ijms21083004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q., Yang M., Liu D., Chen J., Shu D., Xia J., et al. (2020). Experimental Treatment With Favipiravir for COVID-19: An Open-Label Control Study. Eng. (Beijing) 6, 1192–1198. 10.1016/j.eng.2020.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G., et al. (2020). A Trial of Lopinavir-Ritonavir in Adults Hospitalized With Severe Covid-19. N Engl. J. Med. 382, 1787–1799. 10.1056/NEJMoa2001282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang P. X., Ameen F., Sealy J. E., Sadeyen J. R., Bhat S., Li Y. Q., et al. (2019). Application of HDR-CRISPR/Cas9 and Erythrocyte Binding for Rapid Generation of Recombinant Turkey Herpesvirus-Vectored Avian Influenza Virus Vaccines. Vaccines 7 (4), 192. 10.3390/vaccines7040192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. S., Dagdas Y. S., Kleinstiver B. P., Welch M. M., Sousa A. A., Harrington L. B., et al. (2017). Enhanced Proofreading Governs CRISPR-Cas9 Targeting Accuracy. Nature 550, 407–410. 10.1038/nature24268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng M., Xiong E., Tian T., Zhu D., Ju H. Q., Zhou X. (2021). A CRISPR-Driven Colorimetric Code Platform for Highly Accurate Telomerase Activity Assay. Biosens. Bioelectron 172, 112749. 10.1016/j.bios.2020.112749 [DOI] [PubMed] [Google Scholar]
- Chen Y., Jiang H., Wang T., He D., Tian R., Cui Z., et al. (2020). In Vitro and in Vivo Growth Inhibition of Human Cervical Cancer Cells Via Human Papillomavirus E6/E7 Mrnas’ Cleavage by CRISPR/Cas13a System. Antiviral Res. 178, 104794. 10.1016/j.antiviral.2020.104794 [DOI] [PubMed] [Google Scholar]
- Chen J. S., Ma E., Harrington L. B., Da Costa M., Tian X., Palefsky J. M., et al. (2018. a). CRISPR-Cas12a Target Binding Unleashes Indiscriminate Single-Stranded Dnase Activity. Science 360, 436–439. 10.1126/science.aar6245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. S., Ma E. B., Harrington L. B., Da Costa M., Tian X. R., Palefsky J. M., et al. (2018. b). CRISPR-Cas12a Target Binding Unleashes Indiscriminate Single-Stranded Dnase Activity. Science 360, 436–43+. 10.1126/science.aar6245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. J., Wang S. C., Chen Y. C. (2020). Antiviral Agents as Therapeutic Strategies Against Cytomegalovirus Infections. Viruses-Basel 12 (1), 21. 10.3390/v12010021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V. M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D. K., et al. (2020). Detection of 2019 Novel Coronavirus, (2019-Ncov) by Real-Time RT-PCR. Euro Surveill. 25 (3), 2000045. 10.2807/1560-7917.ES.2020.25.3.2000045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y., Wu Y., Liu G., Gooding J. J. (2020). CRISPR Mediated Biosensing Toward Understanding Cellular Biology and Point-of-Care Diagnosis. Angew. Chem. Int. Ed. Engl. 59, 20754–20766. 10.1002/anie.202005398 [DOI] [PubMed] [Google Scholar]
- Dong X., Cao Y. Y., Lu X. X., Zhang J. J., Du H., Yan Y. Q., et al. (2020). Eleven Faces of Coronavirus Disease 2019. Allergy 75, 1699–1709. 10.1111/all.14289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y. X., Chen X. P. (2020). Favipiravir: Pharmacokinetics and Concerns About Clinical Trials for 2019-Ncov Infection. Clin. Pharmacol. Ther. 108, 242–247. 10.1002/cpt.1844 [DOI] [PubMed] [Google Scholar]
- Ebina H., Misawa N., Kanemura Y., Koyanagi Y. (2013). Harnessing the CRISPR/Cas9 System to Disrupt Latent HIV-1 Provirus. Sci. Rep. 3, 2510. 10.1038/srep02510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyquem J., Mansilla-Soto J., Giavridis T., Van Der Stegen S. J., Hamieh M., Cunanan K. M., et al. (2017). Targeting a CAR to the TRAC Locus With CRISPR/Cas9 Enhances Tumour Rejection. Nature 543, 113–117. 10.1038/nature21405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint M., Chatterjee P., Lin D. L., Mcmullan L. K., Shrivastava-Ranjan P., Bergeron E., et al. (2019). A Genome-Wide CRISPR Screen Identifies N-Acetylglucosamine-1-Phosphate Transferase as a Potential Antiviral Target for Ebola Virus. Nat. Commun. 10 (1), 285. 10.1038/s41467-018-08135-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folegatti P. M., Ewer K. J., Aley P. K., Angus B., Becker S., Belij-Rammerstorfer S., et al. (2020). Safety and Immunogenicity of the Chadox1 Ncov-19 Vaccine Against SARS-Cov-2: A Preliminary Report of a Phase 1/2, Single-Blind, Randomised Controlled Trial. Lancet 396, 467–478. 10.1016/S0140-6736(20)31604-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freije C. A., Myhrvold C., Boehm C. K., Lin A. E., Welch N. L., Carter A., et al. (2019). Programmable Inhibition and Detection of RNA Viruses Using Cas13. Mol. Cell 76 (5), 826–837.e11. 10.1016/j.molcel.2019.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Foden J. A., Khayter C., Maeder M. L., Reyon D., Joung J. K., et al. (2013). High-Frequency Off-Target Mutagenesis Induced by CRISPR-Cas Nucleases in Human Cells. Nat. Biotechnol. 31, 822–826. 10.1038/nbt.2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q., Bao L. L., Mao H. Y., Wang L., Xu K. W., Yang M. N., et al. (2020). Development of an Inactivated Vaccine Candidate for SARS-Cov-2. Science 369, 77–7+. 10.1126/science.abc1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z. L., Fan M. H., Das A. T., Herrera-Carrillo E., Berkhout B. (2020). Extinction of All Infectious HIV in Cell Culture by the CRISPR-Cas12a System With Only a Single Crrna. Nucleic Acids Res. 48, 5527–5539. 10.1093/nar/gkaa226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautret P., Lagier J. C., Parola P., Hoang V. T., Meddeb L., Mailhe M., et al. (2020). Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agents 56, 105949. 10.1016/j.ijantimicag.2020.105949 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Glass Z., Lee M., Li Y., Xu Q. (2018). Engineering the Delivery System for CRISPR-Based Genome Editing. Trends Biotechnol. 36, 173–185. 10.1016/j.tibtech.2017.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootenberg J. S., Abudayyeh O. O., Lee J. W., Essletzbichler P., Dy A. J., Joung J., et al. (2017). Nucleic Acid Detection With CRISPR-Cas13a/C2c2. Science 356, 438–43+. 10.1126/science.aam9321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grein J., Ohmagari N., Shin D., Diaz G., Asperges E., Castagna A., et al. (2020). Compassionate Use of Remdesivir for Patients With Severe Covid-19. New Engl. J. Of Med. 382, 2327–2336. 10.1056/NEJMoa2007016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Weiskopf D., Ramirez S. I., Mateus J., Dan J. M., Moderbacher C. R., et al. (2020). Targets of T Cell Responses to SARS-Cov-2 Coronavirus in Humans With COVID-19 Disease and Unexposed Individuals. Cell 181 (7), 1489–1501.e15. 10.1016/j.cell.2020.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubaugh N. D., Hanage W. P., Rasmussen A. L. (2020). Making Sense of Mutation: What D614G Means for the COVID-19 Pandemic Remains Unclear. Cell 182, 794–795. 10.1016/j.cell.2020.06.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielmi G. (2020). First CRISPR Test for the Coronavirus Approved in the United States. Nature. 10.1038/d41586-020-01402-9, https://www.nature.com/articles/d41586-020-01402-9 [DOI] [PubMed] [Google Scholar]
- Haley B., Roudnicky F. (2020). Functional Genomics for Cancer Drug Target Discovery. Cancer Cell 38, 31–43. 10.1016/j.ccell.2020.04.006 [DOI] [PubMed] [Google Scholar]
- Harrington L. B., Burstein D., Chen J. S., Paez-Espino D., Ma E., Witte I. P., et al. (2018). Programmed DNA Destruction by Miniature CRISPR-Cas14 Enzymes. Science 362, 839–842. 10.1126/science.aav4294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington L. B., Doxzen K. W., Ma E., Liu J. J., Knott G. J., Edraki A., et al. (2017). A Broad-Spectrum Inhibitor of CRISPR-Cas9. Cell 170 (6), 1224–1233.e15. 10.1016/j.cell.2017.07.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton B. E., Trimarco J. D., Hamele C. E., Harding A. T., Tata A., Zhu X., et al. (2020). SRSF Protein Kinases 1 and 2 are Essential Host Factors for Human Coronaviruses Including SARS-Cov-2. bioRxiv 2020.2008.2014.251207. 10.1101/2020.08.14.251207 [DOI] [Google Scholar]
- Hoffmann H.-H., Schneider W. M., Sánchez-Rivera F. J., Luna J. M., Ashbrook A. W., Soto-Feliciano Y. M., et al. (2020). Functional Interrogation of a SARS-Cov-2 Host Protein Interactome Identifies Unique and Shared Coronavirus Host Factors. Cell Host Microbe 29, 267–280.e265. 10.1101/2020.09.11.291716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Kaminski R., Yang F., Zhang Y., Cosentino L., Li F., et al. (2014). RNA-Directed Gene Editing Specifically Eradicates Latent and Prevents New HIV-1 Infection. Proc. Natl. Acad. Sci. U. S. A. 111, 11461–11466. 10.1073/pnas.1405186111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultquist J. F., Hiatt J., Schumann K., Mcgregor M. J., Roth T. L., Haas P., et al. (2019). CRISPR-Cas9 Genome Engineering of Primary CD4(+) T Cells for the Interrogation of HIV-Host Factor Interactions. Nat. Protoc. 14, 1–27. 10.1038/s41596-018-0069-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo H. F., Hu G. H. (2019). CRISPR/Cas9-Mediated LMP1 Knockout Inhibits Epstein-Barr Virus Infection and Nasopharyngeal Carcinoma Cell Growth. Infect. Agents And Cancer 14, 30. 10.1186/s13027-019-0246-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishino Y., Shinagawa H., Makino K., Amemura M., Nakata A. (1987). Nucleotide Sequence of the Iap Gene, Responsible for Alkaline Phosphatase Isozyme Conversion in Escherichia Coli, and Identification of the Gene Product. J. Bacteriol. 169, 5429–5433. 10.1128/JB.169.12.5429-5433.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F., Liu J. J., Osuna B. A., Xu M., Berry J. D., Rauch B. J., et al. (2019). Temperature-Responsive Competitive Inhibition of CRISPR-Cas9. Mol. Cell 73 (3), 601–610.e5. 10.1016/j.molcel.2018.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung J., Ladha A., Saito M., Kim N. G., Woolley A. E., Segel M., et al. (2020). Detection of SARS-Cov-2 With SHERLOCK One-Pot Testing. N Engl. J. Med. 383, 1492–1494. 10.1056/NEJMc2026172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy E. M., Kornepati A. V. R., Goldstein M., Bogerd H. P., Poling B. C., Whisnant A. W., et al. (2014). Inactivation of the Human Papillomavirus E6 or E7 Gene in Cervical Carcinoma Cells by Using a Bacterial CRISPR/Cas RNA-Guided Endonuclease. J. Of Virol. 88, 11965–11972. 10.1128/JVI.01879-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S., Siddique R., Shereen M. A., Ali A., Liu J., Bai Q., et al. (2020). Emergence of a Novel Coronavirus, Severe Acute Respiratory Syndrome Coronavirus 2: Biology and Therapeutic Options. J. Clin. Microbiol. 58 (5), e00187-20. 10.1128/JCM.01297-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Koo T., Jee H. G., Cho H. Y., Lee G., Lim D. G., et al. (2018). CRISPR Rnas Trigger Innate Immune Responses in Human Cells. Genome Res 8 (3), 367–373. 10.1101/gr.231936.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott G. J., Doudna J. A. (2018). CRISPR-Cas Guides the Future of Genetic Engineering. Science 361, 866–869. 10.1126/science.aat5011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor A. C., Badran A. H., Liu D. R. (2017). CRISPR-Based Technologies for the Manipulation of Eukaryotic Genomes. Cell 169, 559. 10.1016/j.cell.2017.04.005 [DOI] [PubMed] [Google Scholar]
- Koonin E. V., Makarova K. S., Zhang F. (2017). Diversity, Classification and Evolution of CRISPR-Cas Systems. Curr. Opin. Microbiol. 37, 67–78. 10.1016/j.mib.2017.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W. M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., et al. (2020). Tracking Changes in SARS-Cov-2 Spike: Evidence That D614G Increases Infectivity of the COVID-19 Virus. Cell 182, 812–81+. 10.1016/j.cell.2020.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurata M., Yammoto K., Moriarity B. S., Kitagwa M., Largaespada D. A. (2018). CRISPR/Cas9 Library Screening for Drug Target Discovery. J. Hum. Genet. 63 (2), 179–186. 10.1038/s10038-017-0376-9 [DOI] [PubMed] [Google Scholar]
- Labanieh L., Majzner R. G., Mackall C. L. (2018). Programming CAR-T Cells to Kill Cancer. Nat. Biomed. Eng. 2, 377–391. 10.1038/s41551-018-0235-9 [DOI] [PubMed] [Google Scholar]
- Lee K., Conboy M., Park H. M., Jiang F., Kim H. J., Dewitt M. A., et al. (2017). Nanoparticle Delivery of Cas9 Ribonucleoprotein and Donor DNA in Vivo Induces Homology-Directed DNA Repair. Nat. BioMed. Eng. 1, 889–901. 10.1038/s41551-017-0137-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Clohisey S. M., Chia B. S., Wang B., Cui A., Eisenhaure T., et al. (2020). Genome-Wide CRISPR Screen Identifies Host Dependency Factors for Influenza a Virus Infection. Nat. Commun. 11, 164. 10.1038/s41467-019-13965-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Wang S., Dong X., Li Q., Li M., Li J., et al. (2020). CRISPR-Cas13a Cleavage of Dengue Virus NS3 Gene Efficiently Inhibits Viral Replication. Mol. Ther. Nucleic Acids 19, 1460–1469. 10.1016/j.omtn.2020.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Wu J., Nie J., Zhang L., Hao H., Liu S., et al. (2020). The Impact of Mutations in SARS-Cov-2 Spike on Viral Infectivity and Antigenicity. Cell 182 1284-1294, e1289. 10.1016/j.cell.2020.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Yao L., Li J., Chen L., Song Y., Cai Z., et al. (2020). Stability Issues of RT-PCR Testing of SARS-Cov-2 for Hospitalized Patients. J. Med. Virol. 92 (7), 903–908. 10.1002/jmv.25786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z. T., Yi Y. X., Luo X. M., Xiong N., Liu Y., Li S. Q., et al. (2020). Development and Clinical Application of a Rapid Igm-Igg Combined Antibody Test for SARS-Cov-2 Infection Diagnosis. J. Of Med. Virol. 92, 1518–1524. 10.1002/jmv.25727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D. C., Liu L., Zhang M. X., Hu Y. L., Yang Q. T., Guo J. B., et al. (2020). Evaluations of the Serological Test in the Diagnosis of 2019 Novel Coronavirus (SARS-Cov-2) Infections During the COVID-19 Outbreak. Eur. J. Of Clin. Microbiol. Infect. Diseases 39, 2271–2277. 10.1007/s10096-020-03978-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisboa Bastos M., Tavaziva G., Abidi S. K., Campbell J. R., Haraoui L. P., Johnston J. C., et al. (2020). Diagnostic accuracy of serological tests for covid-19: systematic review and meta-analysis. BMJ 370, m2516. 10.1136/bmj.m2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Chen S., Jin X., Wang Q., Yang K., Li C., et al. (2017). Genome Editing of the HIV Co-Receptors CCR5 and CXCR4 by CRISPR-Cas9 Protects CD4(+) T Cells From HIV-1 Infection. Cell Biosci. 7, 47. 10.1186/s13578-017-0174-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R., Han H., Liu F., Lv Z., Wu K., Liu Y., et al. (2020). Positive Rate of RT-PCR Detection of SARS-Cov-2 Infection in 4880 Cases From One Hospital in Wuhan, China, From Jan to Feb 2020. Clin. Chim. Acta 505, 172–175. 10.1016/j.cca.2020.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Liu L., Kou G., Zheng Y., Ding Y., Ni W., et al. (2020). Evaluation of Nucleocapsid and Spike Protein-Based Enzyme-Linked Immunosorbent Assays for Detecting Antibodies Against SARS-Cov-2. J. Clin. Microbiol. 58 (6), e00461-20. 10.1128/JCM.00461-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Wang L., Sakthivel S. K., Whitaker B., Murray J., Kamili S., et al. (2020). US CDC Real-Time Reverse Transcription PCR Panel for Detection of Severe Acute Respiratory Syndrome Coronavirus 2. Emerg. Infect. Dis. 26 (8), 1654–1665. 10.3201/eid2608.201246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini L. A., Sontheimer E. J. (2010). Self Versus Non-Self Discrimination During CRISPR RNA-Directed Immunity. Nature 463, 568–571. 10.1038/nature08703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica F. J. M., Diez-Villasenor C., Garcia-Martinez J., Almendros C. (2009). Short Motif Sequences Determine the Targets of the Prokaryotic CRISPR Defence System. Microbiol. (Reading) 155, 733–740. 10.1099/mic.0.023960-0 [DOI] [PubMed] [Google Scholar]
- Nguyen T. M., Zhang Y., Pandolfi P. P. (2020). Virus Against Virus: A Potential Treatment for 2019-Ncov (SARS-Cov-2) and Other RNA Viruses. Cell Res. 30, 189–190. 10.1038/s41422-020-0290-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H., Shi X., Ishiguro S., Gao L., Hirano S., Okazaki S., et al. (2018). Engineered CRISPR-Cas9 Nuclease With Expanded Targeting Space. Science 361, 1259–1262. 10.1126/science.aas9129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patchsung M., Jantarug K., Pattama A., Aphicho K., Suraritdechachai S., Meesawat P., et al. (2020). Clinical Validation of a Cas13-Based Assay for the Detection of SARS-Cov-2 RNA. Nat. Biomed. Engineering 4, 1140–1149. 10.1038/s41551-020-00603-x [DOI] [PubMed] [Google Scholar]
- Pereira B. B. (2020). Challenges and Cares to Promote Rational Use of Chloroquine and Hydroxychloroquine in the Management of Coronavirus Disease 2019 (COVID-19) Pandemic: A Timely Review. J. Of Toxicol. And Environ. Health Part B Critical Rev. 23, 177–181. 10.1080/10937404.2020.1752340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickar-Oliver A., Gersbach C. A. (2019). The Next Generation of CRISPR-Cas Technologies and Applications. Nat. Rev. Mol. Cell Biol. 20 (8), 490–507. 10.1038/s41580-019-0131-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter D. L., Levine B. L., Kalos M. (2016). Chimeric Antigen Receptor-Modified T Cells in Chronic Lymphoid Leukemia (Vol 365 2011). New Engl. J. Of Med. 374, 998–998, pg 725. 10.1056/NEJMx160005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch J. N., Valois E., Solley S. C., Braig F., Lach R. S., Audouard M., et al. (2020). A Scalable, Easy-to-Deploy, Protocol for Cas13-Based Detection of SARS-Cov-2 Genetic Material. J Clin Microbiol 59, e02402–20. 10.1101/2020.04.20.052159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settleman J., Sawyers C. L., Hunter T. (2018). Challenges in Validating Candidate Therapeutic Targets in Cancer. Elife 7, e32402. 10.7554/eLife.32402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtmauer E. A., Fraietta J. A., Davis M. M., Cohen A. D., Weber K. L., Lancaster E., et al. (2020). CRISPR-Engineered T Cells in Patients With Refractory Cancer. Science 367, 1001–100+. 10.1126/science.aba7365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S. H., Chen Q., Gu H. J., Yang G., Wang Y. X., Huang X. Y., et al. (2020). A Mouse Model of SARS-Cov-2 Infection and Pathogenesis. Cell Host Microbe 28 (1), 124–133.e4. 10.1016/j.chom.2020.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo T., Liu X., Feng J., Guo M., Hu W., Guo D., et al. (2020). Ddpcr: A More Accurate Tool for SARS-Cov-2 Detection in Low Viral Load Specimens. Emerg. Microbes Infect. 9, 1259–1268. 10.1080/22221751.2020.1772678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng F., Guo L., Cui T., Wang X. G., Xu K., Gao Q., et al. (2019). Cdetection: CRISPR-Cas12b-Based DNA Detection With Sub-Attomolar Sensitivity and Single-Base Specificity. Genome Biol. 20, 132. 10.1186/s13059-019-1742-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesija Kuna A., Hanzek M., Vukasovic I., Nikolac Gabaj N., Vidranski V., Celap I., et al. (2021). Comparison of diagnostic accuracy for eight SARS-CoV-2 serological assays. Biochem Med (Zagreb) 31, 010708. 10.11613/BM.2021.010708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thickett D. R., Armstrong L., Christie S. J., Millar A. B. (2001). Vascular Endothelial Growth Factor May Contribute to Increased Vascular Permeability in Acute Respiratory Distress Syndrome. Am. J. Of Respir. And Crit. Care Med. 164, 1601–1605. 10.1164/ajrccm.164.9.2011071 [DOI] [PubMed] [Google Scholar]
- Tu Y. F., Chien C. S., Yarmishyn A. A., Lin Y. Y., Luo Y. H., Lin Y. T., et al. (2020). A Review of SARS-Cov-2 and the Ongoing Clinical Trials. Int. J. Mol. Sci. 21 (7), 2657. 10.3390/ijms21072657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dongen J. E., Berendsen J. T. W., Steenbergen R. D. M., Wolthuis R. M. F., Eijkel J. C. T., Segerink L. I. (2020). Point-of-Care CRISPR/Cas Nucleic Acid Detection: Recent Advances, Challenges and Opportunities. Biosens. Bioelectron 166, 112445. 10.1016/j.bios.2020.112445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Wang L., Zou X., Duan S., Li Z., Deng Z., et al. (2019). Advances in CRISPR-Cas Systems for RNA Targeting, Tracking and Editing. Biotechnol. Adv. 37, 708–729. 10.1016/j.biotechadv.2019.03.016 [DOI] [PubMed] [Google Scholar]
- Wang M., Zhang R., Li J. (2020). CRISPR/Cas Systems Redefine Nucleic Acid Detection: Principles and Methods. Biosens. Bioelectron 165, 112430. 10.1016/j.bios.2020.112430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Du Q., Guo B., Mu D., Lu X., Ma Q., et al. (2020). A Method To Prevent SARS-CoV-2 IgM False Positives in Gold Immunochromatography and Enzyme-Linked Immunosorbent Assays. J. Clin. Microbiol. 58(6), e00375-20. 10.1128/JCM.00375-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Qian C., Pang Y., Li M., Yang Y., Ma H., et al. (2021). Opvcrispr: One-Pot Visual RT-LAMP-CRISPR Platform for SARS-Cov-2 Detection. Biosens. Bioelectron 172, 112766. 10.1016/j.bios.2020.112766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Simoneau C. R., Kulsuptrakul J., Bouhaddou M., Travisano K., Hayashi J. M., et al. (2020). Functional Genomic Screens Identify Human Host Factors for SARS-Cov-2 and Common Cold Coronaviruses. Cell 184, 106–119 e114. 10.1101/2020.09.24.312298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Li H., Kou Z., Ren F., Jin Y., Yang L., et al. (2020). Highly Sensitive and Specific Detection of Hepatitis B Virus DNA and Drug Resistance. Clin. Microbiol. Infect. 29, S1198–743X(20)30226-3. 10.1016/j.cmi.2020.04.018. [DOI] [PubMed] [Google Scholar]
- Wang Y., Zhang D., Du G., Du R., Zhao J., Jin Y., et al. (2020). Remdesivir in Adults With Severe COVID-19: A Randomised, Double-Blind, Placebo-Controlled, Multicentre Trial. Lancet 395, 1569–1578. 10.1016/S0140-6736(20)31022-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J., Alfajaro M. M., Deweirdt P. C., Hanna R. E., Lu-Culligan W. J., Cai W. L., et al. (2020). Genome-Wide CRISPR Screens Reveal Host Factors Critical for SARS-Cov-2 Infection. Cell 184, 76–91 e13. 10.1016/j.cell.2020.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Q., Chen S., Wang Q., Liu Z., Liu S., Deng H., et al. (2019). CCR5 Editing by Staphylococcus Aureus Cas9 in Human Primary CD4(+) T Cells and Hematopoietic Stem/Progenitor Cells Promotes HIV-1 Resistance and CD4(+) T Cell Enrichment in Humanized Mice. Retrovirology 16, 15. 10.1186/s12977-019-0482-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan F., Wang W., Zhang J. (2019). CRISPR-Cas12 and Cas13: The Lesser Known Siblings of CRISPR-Cas9. Cell Biol. Toxicol. 35, 489–492. 10.1007/s10565-019-09489-1 [DOI] [PubMed] [Google Scholar]
- Younes N., Al-Sadeq D. W., Al-Jighefee H., Younes S., Al-Jamal O., Daas H. I., et al. (2020). Challenges in Laboratory Diagnosis of the Novel Coronavirus SARS-Cov-2. Viruses 12 (6), 582. 10.3390/v12060582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan C., Tian T., Sun J., Hu M., Wang X., Xiong E., et al. (2020). Universal and Naked-Eye Gene Detection Platform Based on the Clustered Regularly Interspaced Short Palindromic Repeats/Cas12a/13a System. Anal. Chem. 92, 4029–4037. 10.1021/acs.analchem.9b05597 [DOI] [PubMed] [Google Scholar]
- Yurkovetskiy L., Wang X., Pascal K. E., Tomkins-Tinch C., Nyalile T. P., Wang Y., et al. (2020). Structural and Functional Analysis of the D614G SARS-Cov-2 Spike Protein Variant. Cell 183, 739–751 e738. 10.1016/j.cell.2020.09.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Yan L., Wang N., Yang S., Wang L., Tang Y., et al. (2020). Quantitative Detection and Viral Load Analysis of SARS-Cov-2 in Infected Patients. Clin. Infect. Dis. 71, 793–798. 10.1093/cid/ciaa345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J. J., Lin Q. D., Song Y. P., Liu D. L. (2018). Universal Cars, Universal T Cells, and Universal CAR T Cells. J. Of Hematol. Oncol. 11 (1), 132. 10.1186/s13045-018-0677-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen S., Hua L., Liu Y. H., Gao L. C., Fu J., Wan D. Y., et al. (2015). Harnessing the Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR)/CRISPR-Associated Cas9 System to Disrupt the Hepatitis B Virus. Gene Ther. 22, 404–412. 10.1038/gt.2015.2 [DOI] [PubMed] [Google Scholar]
- Zhou L., Peng R., Zhang R., Li J. (2018). The Applications of CRISPR/Cas System in Molecular Detection. J. Cell Mol. Med. 22, 5807–5815. 10.1111/jcmm.13925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G. Y., Zhao Q. (2020). Perspectives on Therapeutic Neutralizing Antibodies Against the Novel Coronavirus SARS-Cov-2. Int. J. Of Biol. Sci. 16, 1718–1723. 10.7150/ijbs.45123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Zhu S., Cai C., Yuan P., Li C., Huang Y., et al. (2014). High-Throughput Screening of a CRISPR/Cas9 Library for Functional Genomics in Human Cells. Nature 509, 487–491. 10.1038/nature13166 [DOI] [PubMed] [Google Scholar]