Abstract
Introduction
The COVID-19 pandemic has proven to be an unprecedented challenge to worldwide health, and strategies to mitigate the spread and severity of COVID-19 infection are urgently needed. Emerging evidence suggests that the composition of the gut microbiome and modification of microbial ecology via probiotics can affect susceptibility to a wide range of infections, including respiratory tract infections. In this study, we aim to evaluate the effects of the probiotic Lactobacillus rhamnosus GG (LGG) versus placebo on COVID-19 infection status and the gut microbiome in subjects with a household contact who has tested positive for COVID-19.
Methods and analysis
In this double-blinded, randomised, placebo-controlled trial, we will randomise 1132 subjects having a household contact who has recently (≤7 days) tested positive for COVID-19 to daily oral LGG or placebo for 28 days. We hypothesise that taking LGG as a probiotic will protect against COVID-19 infection and reduce the severity of disease in those who become infected (primary endpoint: decreased symptoms), and will be associated with beneficial changes in the composition of the gut microbiome. Stool samples and nasal swabs will be collected to evaluate the microbiome by 16S rRNA sequencing and the presence of SARS-CoV-2 by PCR, respectively. We will also conduct multivariate analysis of demographic, behavioural, temporal, and other variables that may predict development of symptoms and other outcomes.
Ethics and dissemination
This trial is conducted under a Food and Drug Administration Investigational New Drug for LGG, has received ethics approval by the institutional review board of Duke University and enrolment has begun. We plan to disseminate the results in peer-reviewed journals and at national and international conferences.
Trial registration number
Keywords: COVID-19, gastroenterology, infectious diseases, public health, microbiology, clinical trials
Strengths and limitations of this study.
This is the first randomised controlled trial evaluating the effect of the probiotic Lactobacillus rhamnosus GG (LGG) in preventing COVID-19 transmission and symptom development in exposed household contacts.
Even if we do not meet our primary endpoint, our microbiome studies and analytical approach will allow us to identify aspects of microbiota composition that may identify risk of COVID-19 infection; predict if some but not others may respond to LGG and shape our fundamental understanding of the effect of COVID-19 on microbial ecology.
In the interest of preserving social distancing, study procedures are entirely remote, thus consent and questionnaires are online, and study product and sample collection materials are mailed directly to subjects’ homes with all samples self-collected and returned via mail.
This remote model allows us to enrol patients across the USA, even though we are based out of a single institution.
Introduction
The SARS-CoV-2 (COVID-19) pandemic has significantly altered global public health, with over 141 million cases and over 3 million deaths worldwide as of 19 April 2021. Strategies are urgently needed to mitigate the spread and severity of COVID-19 infection. In several recent studies, COVID-19 severity has been associated with increased levels of inflammatory cytokines,1–3 presenting a potential target for intervention.
One potential method is via optimisation of the gut microbiome. The human body is a unique ecosystem made up of human and microbial cells; in fact, microbial cells outnumber human cells,4 and these microbes (microbiota) play a critical role in human health and disease.5 Manipulation of the gut microbiota through probiotics (live bacteria that are typically formulated in capsules for ingestion) has been shown to modulate the immune system and improve infectious outcomes, and is already well known to modulate the human inflammatory response, immune system, and infectious risk and outcomes.6 Emerging evidence suggests that the gut microbiota may likewise affect the risk of COVID-19 transmission and influence the severity and duration of symptoms7; therefore, modulation of gut microbiota through probiotic administration is a promising strategy for prophylaxis against and mitigation of COVID-19.
Probiotics have been shown to improve a wide variety of infectious outcomes, including sepsis, ventilatory-associated pneumonia (VAP) and lower respiratory tract infections.6 8 A recent large trial in Nature reported that full-term healthy infants randomised to a synbiotic Lactobacillus intervention had a 40% reduction in sepsis or death from 9.0% (placebo) to 5.4% (Lactobacillus) (p<0.001),9 10 including a 34% reduction in lower respiratory tract infections with Lactobacillus (p=0.002). Likewise, a recent Cochrane meta-analysis of >3000 healthy subjects found that acute upper respiratory tract infections were decreased by 47% (p<0.001) with probiotics versus placebo; infection duration and antibiotic prescriptions were also decreased.11 These findings of decreased severity of illness are further supported by a second meta-analysis of >3000 healthy subjects who showed shorter illness episodes and fewer numbers of days absent from day care/school/work with probiotics versus placebo.12 In patients already hospitalised or in an intensive care unit, a recent National Institutes of Health-funded study of Lactobacillus rhamnosus GG (LGG) prophylaxis against VAP showed a 50% reduction with LGG versus placebo (40.0% vs 19.1%; p=0.007).13 Similarly, a meta-analysis by our group of ~3000 intensive care unit patients showed a 26% reduction in VAP and 20% reduction in overall infections with probiotics versus placebo, particularly in patients with a greater acuity of illness.14
Our group’s work, along with that of others, has shown that these improvements in clinical outcomes may be mediated by the effects of probiotics on the immune system and intestinal/lung barrier function. Our experimental pneumonia and infection studies have shown probiotics such as LGG can improve intestinal homeostasis, increase regulatory T-cells, normalise protective mucin production and decrease pro-inflammatory cytokines.15–18 Implications include improved survival from pneumonia, decreased markers of the systemic inflammatory response and reduced histopathological signs of lung injury in mice receiving LGG versus placebo.16 These protections may be related to increases in regulatory T-cells,16 which play a key immunoregulatory role in improving pneumonia outcomes by reducing lung injury through attenuation of excessive inflammatory injury19 resulting from infections, such as is thought to occur with COVID-19. Reports from other groups demonstrate specific protection against viral respiratory tract infections such as H1N1 influenza infection via modulation of antiviral gene expression,20 increased expression of toll-like receptors, and reduced secretion of inflammatory cytokines and chemokines21 in mice receiving Lactobacillus. These clinical and laboratory reports suggest a potent immunomodulatory role for Lactobacillus probiotic therapies in preventing or attenuating COVID-19. We chose to study LGG in particular given its success in the previous in vivo studies and clinical trials discussed above.
To better understand how LGG may affect the risk of COVID-19 transmission, we are conducting a randomised clinical trial of LGG versus placebo in COVID-19-exposed household contacts (EHCs). We have chosen to focus on this population given the high risk of infection, which has been estimated at 10%–20% in recent reports22–24; here, we conservatively anticipate that 10.5% of these individuals will develop symptomatic COVID-19 in the absence of LGG intervention. Longitudinal collection of stool and nasal samples will allow us to evaluate trends in the microbiome and monitor COVID-19 infection status, respectively. Most clinical trials employing probiotics focus on clinical outcomes but lack quantitative microbiome data. However, LGG has been associated with changes in microbiota composition in mouse studies,25 26 and human studies of other Lactobacillus spp have demonstrated that probiotic administration significantly alters the gut microbiota27 28; therefore, further studies such as ours are needed to elucidate its effects of LGG specifically on the human microbiome. In the interest of social distancing, we are employing a novel approach to conduct the study entirely remotely; this also allows us to recruit participants nationally across the USA. Overall, this study has the potential to significantly impact our approach to combating the COVID-19 pandemic in the USA and worldwide and contribute to the design of future probiotic therapies for COVID-19 and other infections.
Methods and analysis
Study design
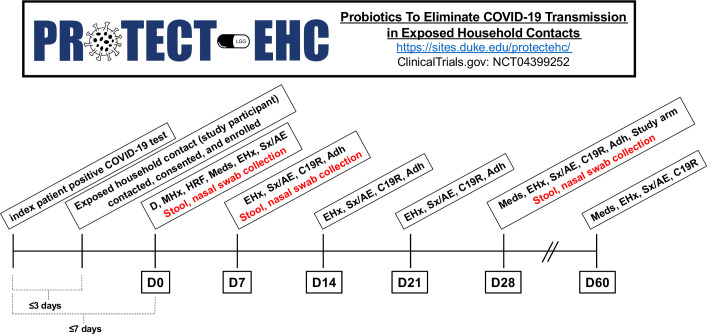
This is a double-blinded, randomised, placebo-controlled trial conducted at Duke University Hospital, an academic medical centre in Durham, North Carolina. This trial has been developed according to the Standard Protocol Items: Recommendations for Intervention Trials 2013 statement.29 Enrolment began in June 2020 and is expected to last until May 2022. We are actively recruiting other sites; interested parties should contact PROTECT-EHC@duke.edu, Paul.Wischmeyer@duke.edu or Anthony.Sung@duke.edu to discuss potential collaboration. Figure 1 outlines the study design and timeline.
Figure 1.
Study design and timeline. Adh, adherence to treatment; C19R, COVID-19-related event reporting; D, demographics; EHx, exposure history; HRF, household risk factors; LGG, Lactobacillus rhamnosus GG; Meds, medications; MHx, medical history; Sx/AE, symptoms/adverse events.
Eligibility
Inclusion criteria
Age ≥1 year (as children<1 year may not be able to take oral probiotics).
Household contact of someone diagnosed with COVID-19.
Willingness to stop taking other probiotics or to not take any other probiotic while on LGG/placebo (taking a probiotic at the time of screening will not be considered a reason for exclusion; however, subjects will be asked to stop taking their probiotic if they enrol on the study).
Access to email/internet to complete electronic consent (e-consent) via REDCap.
Exclusion criteria
Symptoms of COVID-19 at enrolment, including fever, respiratory symptoms (eg, cough, dyspnoea), gastrointestinal symptoms, anosmia, ageusia.
>7 days since original patient (index patient) associated with household contact had first positive COVID-19 test.
Taking hydroxychloroquine or remdesivir for any reason (as this would have the potential to decrease the expected rate of COVID-19 in this population and affect our power and sample size calculations).
Enrolled in a COVID-19 prophylaxis study (as this would have the potential to decrease the expected rate of COVID-19 in this population and affect our power and sample size calculations).
Any medical condition that would prevent taking oral probiotics or increase risks associated with probiotics including but not limited to inability to swallow/aspiration risk and no other methods of delivery (eg, no gastrostomy/jejunostomy tube), increased infection risk due to immunosuppression (eg, due to chronic immunosuppressive medication, prior organ or haematopoietic stem cell transplant, known neutropenia with absolute neutrophil count <500 cells/µL, HIV with CD4 <200 cells/µL), increased infection risk due to endovascular risk factors (eg, rheumatic heart disease, congenital heart defect, mechanical heart valves, endocarditis, endovascular grafts, permanent endovascular devices), increased infection risk due to mucosal incompetence (eg, gastro-oesophageal or intestinal injury including active bleeding, surgery of the oesophagus, stomach, small or large bowel, liver, gallbladder, hepatobiliary tree, spleen, or pancreas within 72 hours, suspected or documented ischaemic gut, severe acute pancreatitis).
Unable to read and follow directions in English or Spanish (as this study is being done remotely, any subject who cannot consent on their own/needs a witness to help them consent will be excluded).
Prisoners and institutionalised individuals (as the definition of ‘household contact’ would have a very different meaning in this setting).
Recruitment, screening and consent
Potential subjects may be approached in two ways. First, study coordinators identify Duke Health patients newly diagnosed with COVID-19 through the Duke Epic dashboard and call them to obtain their assent to contact their EHCs; following assent, we then call those EHCs (potential subjects). For COVID-19-positive patients who are unable to provide assent (eg, intubated with COVID-19) and for whom we are unable to contact their legal representative, we have a waiver to approach emergency contacts listed in the electronic medical record to identify household contacts. Second, potential subjects may hear about our study through flyers, social media platforms (https://www.facebook.com/protectehc/), or our dedicated study website (https://sites.duke.edu/protectehc/) and reach out to us directly.
Once identified, potential subjects complete an online self-screening form in REDCap; if eligible, they continue to the e-consent form in a second REDCap survey. They are given ample time to review the consent, which provides information about the purpose of the research, methods, potential risks and benefits, subject concerns and other study-related matters, though the consent notes an exclusion criterion of >7 days since the original patient (index patient) was diagnosed with COVID-19. Subjects <18 years may participate if their parent or legal guardian provides permission/consent, including willingness to administer LGG/placebo if necessary. Subjects aged 6–12 years need to be informed that they are participating in the study and subjects aged 12–18 years additionally need to provide assent.
Randomisation
After consent/assent, subjects are randomised using a permuted block randomisation technique (to ensure rolling balance between treatment arms) to receive either (1) LGG or (2) placebo in a 1:1 ratio. Both subjects and study coordinators are blinded to the intervention; our statistician generates the randomisation key and only the pharmacist dispensing the study product has access to this key. Product is dispensed via Federal Express overnight delivery. Both LGG and placebo come in indistinguishable foil packaging and as indistinguishable capsules to maintain blinding. Subjects may be unblinded if deemed medically necessary by their provider and the study principal investigator (PI).
Trial intervention
Subjects will take LGG or placebo once daily for 28 days starting from receipt of the study package. LGG, made by Culturelle (an i-Health and DSM subsidiary), comes in capsules each containing 10 billion colony-forming units of LGG (ATCC 53103). Placebo, also made by Culturelle, is made of microcrystalline cellulose, a common food additive used as a bulking agent in food preparation and vitamin supplements, that comes in capsules that contain 325 mg of microcrystalline cellulose. Subjects aged ≥5 years will be instructed to take two capsules per day, ideally taken together at the same time. Subjects aged <5 years will be instructed to take one capsule per day. Both LGG and placebo come as encapsulated powders; capsules may be opened and powder mixed with food or drink (eg, adding to water or apple sauce), with the exception of hot beverages that may inactivate LGG. Patients who develop symptoms of COVID-19 and/or are diagnosed with COVID-19 are instructed to consult with their primary care physician and continue taking the study product unless otherwise directed.
Data collection
Using electronic questionnaires, data on demographics, medical history, household risks and infection details of index patient are collected upon enrolment; and data on medications, adherence, COVID-19 exposures, symptoms, adverse events (AEs)-related and COVID-19-related events are collected throughout the study (table 1). Subjects are instructed to document the date and time of the index patient’s positive COVID-19, date of symptom onset of the index patient and whether the index patient was hospitalised at testing. If available in the medical record, we will also document the COVID-19 strain of the index patient. Additionally, we request permission to access subject medical records to confirm events (eg, if a study subject is diagnosed with COVID-19, admitted, intubated, etc).
Table 1.
Study assessments and timing
| Domain | Assessment | Time point | Rationale |
| Questionnaires | Demographics | D0 | To be used in multivariate analysis |
| Medical history | D0 | To be used in multivariate analysis | |
| Household risk factors | D0 | To be used in multivariate analysis | |
| Medications | D0, D28, D60 | To be used in multivariate analysis | |
| Exposure history | D0, D7, D14, D21, D28, D60 | To be used in multivariate analysis | |
| Symptoms/adverse events | D0, D7, D14, D21, D28, D60 | Monitor safety | |
| COVID-19-related event reporting | D7, D14, D21, D28, D60 | To be used in multivariate analysis | |
| Adherence to treatment | D7, D14, D21, D28 | To be used in multivariate analysis | |
| Study arm (patients’ self-assessment of to which arm they were randomised) | D28 | To be used in multivariate analysis | |
| Microbiota | Stool collection (OMNIgene-GUT kit) | D0, D7, D28 | Gut microbiota analysis |
| Nasal swab collection | D0, D7, D28 | COVID-19 infection status |
Subjects self-collect and send nasal swabs and stool samples at day 0 (baseline, before starting probiotic), day 7 and day 28. They are provided with ORAcollect-RNA and OMNIgene-GUT collection kits for nasal and stool samples, respectively. Both kits contain an RNA preservative which maintains the integrity of the sample for up to 60 days at ambient temperatures. The sample tubes are mailed back to study coordinators in the provided prepaid return packaging and frozen at −80°C for batch analyses.
To ensure data security, electronic records of subject data will be maintained using a dedicated Microsoft Access Database, which is housed in an encrypted and password-protected Duke Cancer Institute (DCI) file server. Completed case report forms and demographic information will be stored and updated in REDCap. Access to electronic databases will be limited to study staff and clinical staff supporting the subject’s care. The DCI and/or Duke Medicine will manage the security and viability of the information technology infrastructure.
Clinical outcomes
Primary endpoint
The primary endpoint of this study is the incidence of symptoms of COVID-19, including: fever, chills, headache, muscle aches, runny nose, sore throat, cough, shortness of breath, nausea or vomiting, diarrhoea, stomach upset or pain, excessive bloating or gas, constipation, loss of sense of smell, loss of sense of taste, rash, painful toes or other symptoms related to COVID-19 diagnosis.
Secondary endpoints
Secondary endpoints include: laboratory-confirmed COVID-19 (all based on medically dictated, clinical testing and electronic medical record review); research laboratory-confirmed COVID-19 (all based on research testing of nasal swab and stool samples); asymptomatic clinical laboratory-confirmed COVID-19; asymptomatic research laboratory-confirmed COVID-19; symptomatic clinical laboratory-confirmed COVID-19; symptomatic research laboratory-confirmed COVID-19; complications of COVID-19 (eg, need for hospitalisation, intubation, mortality); types of symptoms and duration of symptoms. In patients who develop COVID-19, we will review medical records and analyse other clinical variables such as inflammatory markers. We will look at the incidence of these events through day 28 and also through day 60.
Analysis of microbiome data with our novel bioinformatics tools and software will additionally allow us to evaluate the interaction between COVID-19, LGG and the microbiome, specifically:
The impact of LGG on intestinal bacterial diversity (ie, comparing longitudinal changes in subjects who do not develop COVID-19 and receive LGG versus subjects who do not develop COVID-19 and receive placebo.
If there are baseline (D0) differences in the microbiome of subjects who develop COVID-19 versus those who do not (ie, a microbiome signature that may be protective against COVID-19).
If there are changes in the microbiome during infection in subjects receiving placebo who develop COVID-19 versus those who do not (ie, the effect of COVID-19 on the microbiome).
If there are longitudinal changes between subjects receiving LGG who develop COVID-19 versus those who do not (ie, how LGG may impact the microbiome to protect against COVID-19).
How these results are affected by demographic (eg, age, gender), behavioural (eg, shared bed, hours spent together), temporal and other factors.
Adverse events
This is a minimal risk study that involves the use of a commercially available dietary supplement, and we do not anticipate any AEs related to the study product beyond the potential for bloating or excessive gas if taken in excess (beyond recommended dosing). However, because subjects have been exposed to COVID-19 prior to enrolment, they are at risk of developing COVID-19. Therefore, we will monitor AEs and serious AEs (SAEs). Subjects are asked to fill out a symptom questionnaire at each time point (table 1) and are instructed to contact a study team member if any side effects occur; permission to reach out to emergency contacts has been requested in the case subjects are not responsive. The PIs of this study will continuously monitor the conduct, data and safety of this study, including monitoring and tabulating AEs. AEs will be assessed according to the Common Terminology Criteria for Adverse Events (CTCAE) V.4.0; if CTCAE grading does not exist for an AE, severity will be graded as mild (1), moderate (2), severe (3), potentially life-threatening (4) or fatal (5). Additionally, all AEs are graded based on likelihood of being related to the study intervention. All SAEs will be reported to the Duke University Health System (DUHS) Institutional Review Board (IRB) within 24 hours for potentially life-threatening events and within 5 business days for non-potentially life-threatening events. If an unexpected frequency of grade III or IV events occurs, appropriate actions will be taken, including protocol amendment, dose de-escalation or potentially closure of the study. Additionally, we will conduct an interim analysis when COVID-19 symptom data are available from 538 patients (269 per arm); we will stop the trial at this time if the LGG arm has a higher incidence of COVID-19 symptoms than the placebo arm.
Statistical analysis
Power and sample size considerations
With 1076 subjects (538 per arm), the Χ2 test with one-sided alpha=5% has 80% power to detect a 40% reduction in the rate of COVID-19 from 10.5% (attack rate in household contacts based on Centers for Disease Control and Prevention reports)22 to 6.3%. The 40% relative reduction is estimated from data showing 30%–50% reduction in respiratory infections with LGG.9 10 30 31 As above, we will conduct an interim analysis when COVID-19 symptom data are available from 538 patients (269 per arm) and will stop the trial if the LGG arm has a higher incidence of COVID-19 symptoms than the placebo arm (a simulation study shows that the impact of the interim analysis is negligible in overall alpha level and power). Using two-sample t-tests to investigate the difference in microbiome compositional profiles between the LGG and placebo arms, enrolment of 1076 subjects (538 per arm) also achieves about 90% power at 5% alpha level to detect a difference in the alpha diversity equal to 1/5 of the SD within arms; for example, assuming that Shannon Diversity Index (SI) in the controls has mean at about 3 and SD about 0.5, then we are able to detect a difference in the mean SI for the LGG arm if it is <2.9 or >3.1. Because subjects may withdraw from the study early or be lost to follow-up, we will account for up to 5% attrition and plan to enrol 1132 subjects (566 per arm).
Data and statistical analysis
The primary endpoint of this study is the incidence of symptoms of COVID-19 in the LGG versus placebo arm after 28 days. Incidence rates will be compared by Χ2 test, using intention-to-treat methodology.
Secondary endpoints include clinical laboratory-confirmed COVID-19 (all based on clinical testing and electronic medical record review), research laboratory-confirmed COVID-19 (all based on research testing of nasal swab and stool samples), asymptomatic clinical laboratory-confirmed COVID-19, asymptomatic research laboratory-confirmed COVID-19, symptomatic clinical laboratory-confirmed COVID-19, symptomatic research laboratory-confirmed COVID-19, complications of COVID-19 (eg, need for hospitalisation, intubation, mortality), types of symptoms and duration of symptoms. We will look at the incidence of these events through day 28 and through day 60. We will also conduct multivariate analysis using the logistic regression method to adjust for demographic, behavioural (eg, shared bed, hours spent together), temporal, and other variables that may predict development of symptoms and other outcomes.
We will additionally compare the impact of LGG versus placebo on microbiome diversity, as well as the impact of COVID-19 on the microbiome and the impact of the microbiome on development of COVID-19. For microbiome analyses, stool swabs will be analysed using PCR and 16S rRNA sequencing as we have previously described.32 Briefly, we will sequence 16S rRNA using the Illumina HiSeq platform and analyse the data using the Qiime script package with parallel processing. Sequencing data will be de-noised and clustered using USEARCH and aligned to the 16S rRNA gene, using the align.seqs.py wrapper with the PyNAST algorithm and Greengenes reference alignment. Based on these results, we will calculate diversity (SI and Chao1) and construct phylogenetic trees using computational analysis software. SI will be compared using unpaired two-sided Student’s t-tests with a more stringent cut-off of 0.0125 given multiple comparisons by the Bonferroni correction for four time periods of independent comparisons. Changes in specific bacterial families of interest will be compared using a two-sided Student’s t-test, with normality confirmed by D’Agostino and Pearson omnibus test with p≤0.05. All other comparisons will be done using two-sided Mann-Whitney tests. The R packages vegan,33 phyloseq,34 APE,35 randomForest36 and arules37 will be used for identification of associative patterns between taxa most associated with LGG versus placebo or COVID-19 versus no COVID-19 and other metadata. Comparisons may be made between intraindividual samples (eg, D0 (baseline) vs D7 or D0 (baseline) vs D28) as well as between arms (eg, average diversity at D7 of subjects receiving LGG vs subjects receiving placebo; change in diversity (D0 vs D28) of subjects who develop COVID-19 vs subjects who do not develop COVID-19).
Patient and public involvement
Household contacts exposed to COVID-19 were involved in the development and conduct of this clinical trial protocol.
Discussion
COVID-19 is a unique and novel challenge that does not yet have a vaccine, treatment or cure. Among the multitude of strategies under development (testing, vaccination, antivirals, immunomodulatory agents, apps), little is known about the potential for probiotics and the microbiome to impact COVID-19 transmission. Because probiotics are known to have protective effects in other infectious settings (including upper and lower respiratory tract infections, VAP, sepsis and death),6 38 we are conducting the first double-blinded, randomised, placebo-controlled trial to evaluate the effect of the probiotic LGG on development of symptomatic COVID-19 in EHCs. Microbiome sampling will further allow us to evaluate interaction between COVID-19 infection and clinical outcomes, LGG and the microbiome; specifically, impact of LGG on the microbiome on COVID-19 infection, symptomatology and clinical complications; differences in baseline microbiome predicting risk of COVID-19 infection (ie, protective microbiome signature); effect of COVID-19 infection on changes in microbiome; impact of LGG on microbiome in EHC at high risk of COVID-19 and how results are affected by covariates.
The remote design of the study allows us to preserve social distancing; in addition, it allows us to recruit nationally while keeping study costs low. However, this does present the limitation of dependence on self-collection of samples. Encouragingly, our extensive experience with self-collection of stool39 and studies such as the American Gut Project have shown that remotely conducted studies relying on self-collection and mailing of stool samples can yield high-quality data.40 While there is no way to self-collect and mail peripheral blood (which otherwise would have significantly strengthened the study), for subjects enrolled at participating academic medical centres, there is the potential to access peripheral blood samples collected as part of COVID-19 biorepository studies that can be later analysed and combined with this data. Finally, although this trial relies on self-report of symptoms, we have requested permission to access subjects’ medical records to confirm COVID-19 diagnoses and other medical events; for participants outside of the DUHS, we will have the subject request copies of appropriate medical records from their doctor/institution.
Our trial has the potential to have a significant and readily implementable impact on the COVID-19 pandemic in the USA and worldwide. With the wide range of interventions receiving attention for potential therapeutic use in COVID-19, very little attention has been devoted to understanding the role of our microbial ecosystem, which is perhaps what most fundamentally makes us human (as we are >50% cellularly microbial).4 This project presents a unique opportunity to demonstrate that our symbiotic microbes can be valuable partners in the fight against infectious diseases. Insights gleaned from this trial will inform understanding of the relationships between the microbiome and COVID-19 and allow for potential identification of clinically relevant microbiome targets to mitigate the spread of the COVID-19 pandemic. The results of this trial could fundamentally transform the care of COVID-19, as well as reshape our scientific understanding and approach to maintaining health in the face of infectious threats (while avoiding the development of the super-pathogens facilitated by traditional antibiotics). Further, these data have the opportunity to demonstrate that probiotics may serve as a safe, low-cost, commercially available and rapidly deployable intervention against other pandemic disease (ie, influenza, new SARS virus). Throughout millennia, the microbiota has evolved alongside humans as critical symbionts essential to our survival; now, it may prove to be a critical ally in the fight against COVID-19’s devastating threat to mankind.
Ethics and dissemination
This trial is conducted under a Food and Drug Administration Investigational New Drug for LGG and has been approved by the IRB of Duke University. This protocol was designed and will be conducted and reported in accordance with the International Conference on Harmonization Harmonized Tripartite Guidelines for Good Clinical Practice, the Declaration of Helsinki, and applicable federal, state and local regulations.
We plan to disseminate the results in peer-reviewed journals and at national and international conferences.
Supplementary Material
Footnotes
Twitter: @paul_wischmeyer
ADS and PEW contributed equally.
Contributors: ADS, PEW, LB, ML, DJ, S-HJ and AZ designed the study and the protocol submission. HT and PEW wrote the manuscript. ADS and PEW revised the manuscript and approved the final version.
Funding: This work is supported by a grant from the Duke University Microbiome Center. iHealth is donating LGG and placebo for the trial.
Competing interests: iHealth has provided unrestricted gift funding donation to support probiotic research in COVID-19 to PEW and ADS.
Patient and public involvement: Patients and/or the public were involved in the design, or conduct, or reporting, or dissemination plans of this research. Refer to the Methods section for further details.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Li X, Xu S, Yu M, et al. Risk factors for severity and mortality in adult COVID-19 inpatients in Wuhan. J Allergy Clin Immunol 2020;146:110–8. 10.1016/j.jaci.2020.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen G, Wu D, Guo W, et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest 2020;130:2620–9. 10.1172/JCI137244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tang Y, Liu J, Zhang D, et al. Cytokine storm in COVID-19: the current evidence and treatment strategies. Front Immunol 2020;11:1708. 10.3389/fimmu.2020.01708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sender R, Fuchs S, Milo R. Are we really Vastly Outnumbered? revisiting the ratio of bacterial to host cells in humans. Cell 2016;164:337–40. 10.1016/j.cell.2016.01.013 [DOI] [PubMed] [Google Scholar]
- 5.Sonnenburg JL, Sonnenburg ED. Vulnerability of the industrialized microbiota. Science 2019;366:eaaw9255. 10.1126/science.aaw9255 [DOI] [PubMed] [Google Scholar]
- 6.Davison JM, Wischmeyer PE. Probiotic and synbiotic therapy in the critically ill: state of the art. Nutrition 2019;59:29–36. 10.1016/j.nut.2018.07.017 [DOI] [PubMed] [Google Scholar]
- 7.Walton GE, Gibson GR, Hunter KA. Mechanisms linking the human gut microbiome to prophylactic and treatment strategies for COVID-19. Br J Nutr 2020:1–9. 10.1017/S0007114520003980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wischmeyer PE, McDonald D, Knight R. Role of the microbiome, probiotics, and 'dysbiosis therapy' in critical illness. Curr Opin Crit Care 2016;22:347–53. 10.1097/MCC.0000000000000321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Panigrahi P, Parida S, Nanda NC, et al. A randomized synbiotic trial to prevent sepsis among infants in rural India. Nature 2017;548:407–12. 10.1038/nature23480 [DOI] [PubMed] [Google Scholar]
- 10.Tancredi DJ. Global health: probiotic prevents infections in newborns. Nature 2017;548:404–5. 10.1038/nature23540 [DOI] [PubMed] [Google Scholar]
- 11.Hao Q, Dong BR, Wu T, et al. Probiotics for preventing acute upper respiratory tract infections. Cochrane Database Syst Rev 2015;5. 10.1002/14651858.CD006895.pub3 [DOI] [PubMed] [Google Scholar]
- 12.King S, Glanville J, Sanders ME, et al. Effectiveness of probiotics on the duration of illness in healthy children and adults who develop common acute respiratory infectious conditions: a systematic review and meta-analysis. Br J Nutr 2014;112:41–54. 10.1017/S0007114514000075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morrow LE, Kollef MH, Casale TB. Probiotic prophylaxis of ventilator-associated pneumonia: a blinded, randomized, controlled trial. Am J Respir Crit Care Med 2010;182:1058–64. 10.1164/rccm.200912-1853OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manzanares W, Lemieux M, Langlois PL, et al. Probiotic and synbiotic therapy in critical illness: a systematic review and meta-analysis. Crit Care 2016;19:262. 10.1186/s13054-016-1434-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khailova L, Baird CH, Rush AA, et al. Lactobacillus rhamnosus GG treatment improves intestinal permeability and modulates inflammatory response and homeostasis of spleen and colon in experimental model of Pseudomonas aeruginosa pneumonia. Clin Nutr 2017;36:1549–57. 10.1016/j.clnu.2016.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khailova L, Baird CH, Rush AA, et al. Lactobacillus rhamnosus GG improves outcome in experimental Pseudomonas aeruginosa pneumonia: potential role of regulatory T cells. Shock 2013;40:496–503. 10.1097/SHK.0000000000000066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khailova L, Petrie B, Baird CH, et al. Lactobacillus rhamnosus GG and Bifidobacterium longum attenuate lung injury and inflammatory response in experimental sepsis. PLoS One 2014;9:e97861. 10.1371/journal.pone.0097861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khailova L, Frank DN, Dominguez JA, et al. Probiotic administration reduces mortality and improves intestinal epithelial homeostasis in experimental sepsis. Anesthesiology 2013;119:166–77. 10.1097/ALN.0b013e318291c2fc [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neill DR, Fernandes VE, Wisby L, et al. T regulatory cells control susceptibility to invasive pneumococcal pneumonia in mice. PLoS Pathog 2012;8:e1002660. 10.1371/journal.ppat.1002660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiso M, Takano R, Sakabe S, et al. Protective efficacy of orally administered, heat-killed Lactobacillus pentosus b240 against influenza A virus. Sci Rep 2013;3:1563. 10.1038/srep01563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanaka A, Seki M, Yamahira S, et al. Lactobacillus pentosus strain b240 suppresses pneumonia induced by Streptococcus pneumoniae in mice. Lett Appl Microbiol 2011;53:35–43. 10.1111/j.1472-765X.2011.03079.x [DOI] [PubMed] [Google Scholar]
- 22.Burke RM, Midgley CM, Dratch A, et al. Active Monitoring of persons exposed to patients with confirmed COVID-19 - United States, January-February 2020. MMWR Morb Mortal Wkly Rep 2020;69:245–6. 10.15585/mmwr.mm6909e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boulware DR, Pullen MF, Bangdiwala AS, et al. A randomized trial of hydroxychloroquine as postexposure prophylaxis for Covid-19. N Engl J Med 2020;383:517–25. 10.1056/NEJMoa2016638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Madewell ZJ, Yang Y, Longini IM, et al. Household transmission of SARS-CoV-2: a systematic review and meta-analysis of secondary attack rate. medRxiv 2020. 10.1101/2020.07.29.20164590. [Epub ahead of print: 31 Jul 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi C-W, Cheng M-Y, Yang X. Probiotic Lactobacillus rhamnosus GG promotes mouse gut microbiota Diversity and T ceCell Differentiation. Front Microbiol 2020;11:607735. 10.3389/fmicb.2020.607735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng Y-C, Liu J-R. Effect of Lactobacillus rhamnosus GG on energy metabolism, leptin resistance, and gut microbiota in mice with diet-induced obesity. Nutrients 2020;12:2557. 10.3390/nu12092557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu G, Chong H-X, Chung FY-L, et al. Lactobacillus plantarum DR7 modulated bowel movement and gut microbiota associated with dopamine and serotonin pathways in stressed adults. Int J Mol Sci 2020;21:4608. 10.3390/ijms21134608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang W-C, Pan C-H, Wei C-C, et al. Lactobacillus plantarum PS128 improves physiological adaptation and performance in triathletes through gut microbiota modulation. Nutrients 2020;12:2315. 10.3390/nu12082315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chan A-W, Tetzlaff JM, Altman DG, et al. Spirit 2013 statement: defining standard protocol items for clinical trials. Ann Intern Med 2013;158:200–7. 10.7326/0003-4819-158-3-201302050-00583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hao Q, Dong BR, Wu T. Probiotics for preventing acute upper respiratory tract infections. Cochrane Database Syst Rev 2015;2:CD006895. 10.1002/14651858.CD006895.pub3 [DOI] [PubMed] [Google Scholar]
- 31.Morrow LE, Casale T, Kollef M. Probiotic prophylaxis of ventilator-associated pneumonia. Chest 2009;136:36S. 10.1378/chest.136.4_MeetingAbstracts.36S-h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peled JU, Gomes ALC, Devlin SM, et al. Microbiota as predictor of mortality in allogeneic hematopoietic-cell transplantation. N Engl J Med 2020;382:822–34. 10.1056/NEJMoa1900623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oksanen J G-BF, Kindt R, Legendre P, et al. Vegan: community ecology package. R package version 2.0-9 2013.
- 34.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 2013;8:e61217. 10.1371/journal.pone.0061217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paradis E, Claude J, Strimmer K. Ape: analyses of phylogenetics and evolution in R language. Bioinformatics 2004;20:289–90. 10.1093/bioinformatics/btg412 [DOI] [PubMed] [Google Scholar]
- 36.Liaw AWM. Classification and regression by randomForest. R News 2002;2:18–22. [Google Scholar]
- 37.Hahsler M, Grün B, Hornik K. arules - A Computational Environment for Mining Association Rules and Frequent Item Sets. J Stat Softw 2005;14. 10.18637/jss.v014.i15 [DOI] [Google Scholar]
- 38.Wischmeyer PE. Are we creating survivors…or victims in critical care? delivering targeted nutrition to improve outcomes. Curr Opin Crit Care 2016;22:279–84. 10.1097/MCC.0000000000000332 [DOI] [PubMed] [Google Scholar]
- 39.Peled JU, Gomes ALC, Devlin SM, et al. Microbiota as predictor of mortality in allogeneic hematopoietic-cell transplantation. N Engl J Med 2020;382:822–34. 10.1056/NEJMoa1900623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McDonald D, Hyde E, Debelius JW, et al. American gut: an open platform for citizen science microbiome research. mSystems 2018;3:e00031–00018. 10.1128/mSystems.00031-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.