Abstract
Insect systemic immune responses to bacterial infections have been mainly studied using microinjections, whereby the microbe is directly injected into the hemocoel. While this methodology has been instrumental in defining immune signaling pathways and enzymatic cascades in the hemolymph, it remains unclear whether and to what extent the contribution of systemic immune defenses to host microbial resistance varies if bacteria invade the hemolymph after crossing the midgut epithelium subsequent to an oral infection. Here, we address this question using the pathogenic Serratia marcescens (Sm) DB11 strain to establish systemic infections of the malaria vector Anopheles gambiae, either by septic Sm injections or by midgut crossing after feeding on Sm. Using functional genetic studies by RNAi, we report that the two humoral immune factors, thioester-containing protein 1 and C-type lectin 4, which play key roles in defense against Gram-negative bacterial infections, are essential for defense against systemic Sm infections established through injection, but they become dispensable when Sm infects the hemolymph following oral infection. Similar results were observed for the mosquito Rel2 pathway. Surprisingly, blocking phagocytosis by cytochalasin D treatment did not affect mosquito susceptibility to Sm infections established through either route. Transcriptomic analysis of mosquito midguts and abdomens by RNA-seq revealed that the transcriptional response in these tissues is more pronounced in response to feeding on Sm. Functional classification of differentially expressed transcripts identified metabolic genes as the most represented class in response to both routes of infection, while immune genes were poorly regulated in both routes. We also report that Sm oral infections are associated with significant downregulation of several immune genes belonging to different families, specifically the clip-domain serine protease family. In sum, our findings reveal that the route of infection not only alters the contribution of key immunity genes to host antimicrobial defense but is also associated with different transcriptional responses in midguts and abdomens, possibly reflecting different adaptive strategies of the host.
Keywords: Anopheles gambiae, Mosquito innate immunity, Complement-like protein, C-type lectin, Serratia marcescens, Oral infections
Introduction
Insects deploy several humoral and cellular innate immune effector mechanisms to clear bacterial infections. While antimicrobial peptides (AMPs) [1, 2], melanization [3, 4], phagocytosis [5, 6], and complement-mediated attack [7, 8] are often described as the main players in different contexts, several knowledge gaps remain as to their regulation, specificity, and relative contribution to microbial clearance. This is further complicated by the fact that the vast majority of bacterial challenges in model insects have been established through an artificial route, by pricking the cuticle to introduce the microbes directly into the hemocoel [9, 10, 11, 12]. While this route of infection has allowed the dissection of systemic antimicrobial immune responses at different levels, it is associated with 2 major pitfalls: first, microbes are often introduced at large numbers to trigger pathogenesis, which might blur the readouts from distinct effector programs due to saturation effects; low-dose infections are most likely the norm in field conditions. For instance, it was recently shown that when a low dose of Staphylococcus aureus is injected into Drosophila, the melanization response but not hemocytes or Toll effectors plays a significant role in resisting the infection, whereas at higher doses, the role of hemocytes becomes predominant over that of melanization [13]. Second, this route of infection may not allow sufficient priming of the systemic response whether humoral or cellular. For instance, in the malaria vector Anopheles gambiae, invasion of the midgut epithelium by Plasmodium ookinetes triggers the release of a hemocyte differentiation factor, constituted of a lipoxin/lipocalin complex, into the hemolymph, which induces immune priming, preparing the host for a subsequent challenge. Lipocalin is produced by the abdominal wall, possibly in response to unknown signals originating from the invaded midgut [14]. Also, Plasmodium midgut invasion triggers the nitration of the basal surface of the midgut epithelium, which upon contact with hemocytes induces the release of hemocyte-derived microvesicles that activate the complement-mediated attack against invading parasites, through unknown factors they deliver [15]. These studies inform that midgut invasion seems to trigger different forms of innate immune priming which might not occur if this route is bypassed.
A large number of functional genetic studies in A. gambiae identified several immunity genes with roles in systemic antibacterial defense [11, 16, 17, 18, 19, 20, 21, 22, 23]. However, since bacterial challenges in these studies were performed by cuticle pricking, it remains unclear whether these genes significantly contribute to immune defense against systemic infections established through the oral route (i.e., after midgut invasion). This is particularly important since a previous study in Drosophila revealed that the virulent Serratia marcescens (Sm) Db11 strain is resistant to the Imd-mediated immune response during septic infections but is susceptible to the local Imd response in the gut after oral infections [24]. Sm is a Gram-negative bacterium with a broad host range including plants, vertebrates, and invertebrates [25], and an opportunistic pathogen to vertebrates [26, 27] and invertebrates [24, 28]. Its ability to efficiently colonize the midguts of insects [24, 29] and to invade the midgut epithelium reaching into the hemolymph [24] makes it an attractive microbe to address whether the route of infection alters the contribution of key immunity genes to systemic immune responses. Furthermore, Sm is one of the bacterial species identified frequently as a member of the microbiota in lab- and field-collected A. gambiae mosquitoes [30, 31, 32], which makes it more relevant to studying host-parasite interactions in this important malaria vector. Certain isolates of Sm compromised Plasmodium development when introduced into the midgut through a blood or sugar meal, most likely due to certain virulence factors released by the bacteria [30, 31]. However, the physiological relevance of Serratia symbiosis in insects remains poorly characterized. Here, we chose A. gambiae C-type lectin 4 (CTL4) and thioester-containing protein 1 (TEP1) which exhibit prominent roles in defense against systemic Gram-negative bacterial infections [11, 16, 19, 21] to determine whether the contribution of immune genes to mosquito resistance to Sm infections varies with the route of infection (oral vs. injection). TEP1 and CTL4 are required for the clearance of E. coli systemic infections [11, 16, 19, 21]; however, the fact that E. coli is not pathogenic to mosquitoes and that susceptibility studies require the injection of large numbers of bacteria (approximately 150,000 CFUs [21]) into the hemolymph raise legitimate questions concerning the significance of the immune contribution of these genes using this bacterial infection model and route of infection. To clarify this situation, we used the virulent Sm DB11 bacterial strain that kills mosquitoes at much lower CFUs than E. coli, to assess the true contribution of CTL4 and TEP1 to antibacterial defense. We also used RNA-seq analysis to determine whether the different routes of infection are associated with distinct transcriptional responses in the midguts and abdomens of infected mosquitoes.
Materials and Methods
A. gambiae Rearing
All experiments were performed with adult female A. gambiae G3 strain mosquitoes. Mosquitoes were maintained at 27 (±1)°C and 75 (±5)% humidity with 12-h day-night cycle. Larvae were reared in 752 cm2 plastic pans at a density of approximately 150 larvae per pan and given tropical fish food. Freshly emerged adult mosquitoes were collected from larval pans using a vacuum collector and maintained on 10% sucrose and given BALB/c mice blood (mice were anesthetized with ketamine) for egg laying.
Double-Stranded RNA Synthesis and Gene Silencing by RNA Interference
Double-stranded RNA (dsRNA) synthesis was performed using the T7 RiboMax Express Large Scale RNA production system (Promega) according to the manufacturer's instructions, and dsRNAs were purified as previously described [33]. Primers used for dsRNA production are listed in online suppl. Table 1; see www.karger.com/doi/10.1159/000511401 for all online suppl. material. In vivo gene silencing was performed as previously described [33]. In brief, mosquitoes were microinjected with 69 nL of 4 μg/μL solution of gene-specific dsRNA and allowed to recover for 3–4 days before proceeding with Sm infections. The efficiency of gene silencing by RNAi for TEP1 and CTL4 was quantified by Western blot in hemolymph extracts of naive mosquitoes at 3 days after dsRNA injection, as previously described [21], using the following dilution of primary antibodies: rabbit αTEP1 (1:1,000) and rabbit αCTL4 (1:1,000). Rabbit αSRPN3 (1:1,000) was used to control for loading. The silencing efficiency of Rel1 and Rel2 was determined by qRT-PCR in naïve mosquitoes at 3 days after dsRNA injection.
RNA Extraction and Real-Time PCR
Total RNA was isolated from whole mosquitoes at the indicated time points using the TRIzol reagent (Invitrogen) according to the manufacturer's instructions, and contaminant genomic DNA was removed by DNase I treatment. First-strand cDNA was produced from 1 μg of total RNA using the iScript cDNA synthesis kit as described by the manufacturer (Bio-Rad). qRT-PCR was performed in a CFX96 real-time detection system (Bio-Rad) using the SYBR Green JumpStartTM Taq ReadyMix (Sigma-Aldrich) according to the manufacturer's instructions. Relative gene expression was normalized relative to the mosquito gene encoding the ribosomal protein S7 and calculated using the comparative CT method after checking for the efficiency of target amplification. The primers used in qRT-PCR are listed in online suppl. Table 1.
Mosquito Infections with Sm and Survival Assays
Mosquito oral infections with Sm were performed by allowing mosquitoes to feed continuously on a sugar solution containing Sm that was prepared as follows. DsRed-expressing, gentamycin-resistant Sm strain DB11 [24] cultured exponentially at 37°C was washed with PBS and then diluted in a sterile 3% sucrose solution to a final OD600 = 1. Mosquitoes that fed on Sm-containing sugar pads were sorted out at 24 h after feeding on Sm with the help of a food colorant added to the sugar solution and used in subsequent experiments. Mosquitoes were maintained on Sm-containing sugar pads for the duration of the experiment. To determine whether our mosquito colony naturally contains Sm, DNA was extracted from a pool of 10 midguts dissected from either sugar-fed adult female mosquitoes or mosquitoes that have been feeding on the Sm DB11 strain for 24 h, using DNeasy Blood and Tissue Kit (Qiagen). A 100 ng of extracted DNA per sample was used to amplify a 175-bp amplicon of the LuxS gene involved in quorum sensing, using Sm LuxS-specific primers, For, 5′-TGCCTGGAAAGCGGCGATGG-3′, and Rev, 5′-CGCCAGCTCGTCGTTGTGGT-3′ [34], according to the following program (45 s at 95°C; 60 s at 66°C; 60 s at 72°C) for 33 cycles. As internal control, we PCR amplified a 298-bp amplicon of the gene encoding A. gambiae ribosomal protein S7 (Ag_S7) using primers, For, 5′-AGAACCAGCAGACCACCATC-3′, and Rev, 5′-GCTGCAAACTTCGGCTATTC-3′, according to the following program (45 s at 95°C; 60 s at 60°C; 60 s at 72°C) for 33 cycles. Amplicons were separated on a 1.2% agarose gel, stained with ethidium bromide, and analyzed on ChemiDoc MP (Bio-Rad).
Septic infections with Sm were performed by the intrathoracic microinjection of dsRNA-treated mosquitoes with a suspension of DsRed-expressing gentamycin-resistant Sm strain DB11 in PBS (OD600 = 0.0005). Mosquitoes treated with dsRNA specific to the β-galactosidase gene (dsLacZ) served as control. Mosquito survival was scored over a period of 8–10 days after Sm septic or oral infections. The Kaplan-Meier survival test was used to calculate the percent survival. Statistical significance of the observed differences was calculated using the log-rank test. Experiments were repeated at least 3 times using different mosquito and bacterial batches. At least 50 mosquitoes were utilized per sample per experiment.
Scoring Sm CFUs in Infected Mosquitoes
To determine Sm CFUs in whole mosquitoes following septic injections, batches of 8 mosquitoes each per genotype were grinded using a micropestle in 400-μL Luria Bertani (LB) broth at 24 h after Sm injections. The homogenate was serially diluted in the LB medium. After overnight culturing at 37°C on LB agar supplemented with gentamycin, CFUs were scored under a fluorescence stereomicroscope.
To determine Sm CFUs in the hemolymph, the hemolymph was collected 72 h after oral infections with Sm by perfusion as follows. In brief, mosquitoes were perfused with 5 μL of PBS injected into the thorax using a Nanoject II (Drumond Scientific) nanoinjector, and the perfused hemolymph was collected from a small incision made in the 3rd abdominal segment. Hemolymph was collected in sterile ice-cold PBS from batches of 4 or 5 mosquitoes each, serially diluted, and then plated on LB agar with the appropriate antibiotic. CFUs were scored after culturing overnight at 37°C on LB agar supplemented with gentamycin. Statistical significance was calculated using the Mann-Whitney test in GraphPad Prism software (version 6.0). Medians were considered significantly different if p < 0.05.
For the bacterial fitness experiment, dsCTL4 and dsLacZ (control) mosquitoes were injected with Sm prepared from a fresh bacterial culture (OD600 = 0.0005) or Sm collected by hemolymph perfusion from wild-type mosquitoes that have been feeding on Sm for 24 h. Mosquitoes injected with hemolymph perfusate received 4, 207, and 331 Sm CFUs in the 3 independent biological experiments performed. Sm proliferation in injected mosquitoes was scored by homogenizing batches of 8 whole mosquitoes each in the LB medium at 24 h after Sm injection. The homogenate was serially diluted in the LB medium. CFUs were scored under a fluorescence stereomicroscope after culturing overnight at 37°C on LB agar supplemented with gentamycin.
Mosquito Treatment with Cytochalasin D
Cytochalasin D was dissolved in DMSO to make a 1 mg/mL stock solution from which a 62.5 μg/mL (120 μM) working solution in PBS was prepared. Each mosquito was injected with 69 nL of the working solution. Control groups included mosquitoes injected with PBS only and those injected with 6.25% DMSO in PBS. Mosquitoes injected with PBS, DMSO, or cytochalasin D were allowed to recover for 6 h before feeding on Sm (OD600 = 1) and for 24 h before injection with Sm (OD600 = 0.0005).
RNA Extraction, Library Preparation, and Sequencing
RNA was extracted from midguts and abdomens dissected from untreated control female mosquitoes (fed on 3% sugar solution) and from female mosquitoes treated by oral Sm feeding, Sm injection, and sterile PBS injection using a hybrid modified Trizol/RNeasy protocol (Qiagen). Untreated mosquitoes served as control for mosquitoes fed on Sm and PBS-injected mosquitoes as control for Sm-injected mosquitoes. By including these respective controls, we would be assessing transcriptional responses that are Sm specific in each route, which allows us to focus on genes regulated by Sm itself and not secondary to the infection procedure. The abdomen specimen refers to the whole abdomen excluding the gut in addition to the malpighian tubules and ovaries which were pulled out with the gut during dissection. Among treated mosquitoes, RNA was extracted from the indicated tissues at 6, 12, and 24 h after treatment. RNA quantification was performed using Qubit RNA HS Assay and quality check procedures via AATI Fragment Analyzer. QuantSeq 3′ mRNA-Seq Library Prep Kit (Lexogen) was used for construction of 3′ end RNA-seq libraries. Libraries were checked with the Qubit DNA Assay kit and AATI Fragment Analyzer again before pooling and sequencing. Illumina NextSeq 500 platform with standard protocol for 75-bp single-end read sequencing was utilized to sequence libraries at the Cornell Life Sciences Sequencing core facility. Three to six million reads were obtained per sample, which is equivalent approximately to a 20× or more coverage of the transcriptome. Quality control of raw reads was performed with FastQC, followed by trimming of the reads by BBMap (https://sourceforge.net/projects/bbmap/) and then mapping to the A. gambiae transcriptome (AgamP4.12) using Salmon [35].
Differential Gene Expression and Gene Ontology
Differential expression was analyzed on the transcript level using Bioconductor package DESeq2 [36]. A model with 2 categorical variables was fitted, 1 variable for the replicate and a second variable that contained a separate level for each of the 18 combinations of tissue (abdomen or midgut), time (6, 12, and 24 h), and treatment (Sm oral infection, Sm injection, and PBS injection), plus a level for the untreated control (only sugar fed) at time zero. Differential expression was analyzed by fitting a generalized linear model and testing for a significant difference in coefficients for treatment and control. This analysis was performed within each combination of time and tissue by comparing read counts between Sm oral infection and the untreated control, between PBS injection and the untreated control, and between Sm injection and PBS injection of the same time point. The transcript-specific p values for differential expression were adjusted for a false discovery rate, and only transcripts with a false discovery rate below 0.05 were labeled as differentially expressed. Genes with at least one differentially expressed transcript (DET) were labeled differentially expressed. To identify transcripts whose expression changes significantly and uniquely in response to oral feeding of Sm relative to untreated control and those whose expression changes significantly and uniquely in response to Sm injection relative to PBS injection, we took all transcripts that are differentially expressed in each treatment of interest (p value adjusted for false discovery rate <0.05) and removed all transcripts that showed differential expression in the same direction in any of the other comparisons, either according to false discovery rate-adjusted p value (p value <0.05) or according to fold change (fold change >1.5). The fold change criterion was included to be confident that the remaining transcripts are actually treatment specific. Enrichment of differentially expressed genes was tested for each of treatment-control comparison according to 4 classifications, namely, gene ontology terms from the molecular function and biological process ontology, KEGG pathways, and gene families. The enrichment tests used Wallenius noncentral hypergeometric distribution to account for transcript-length-dependent bias for detecting differential expression as implemented in the R package goseq[37]. The false discovery rate was calculated by selecting all groups from all 4 classifications that contain >1 significantly differentially expressed gene in any of the comparisons of differential expression and applying the Benjamini-Hochberg correction [38] to the enrichment p values of all these groups. Only terms with a false discovery rate below 0.05 were reported.
Results
Sm Invades the Hemolymph after Mosquito Oral Infection
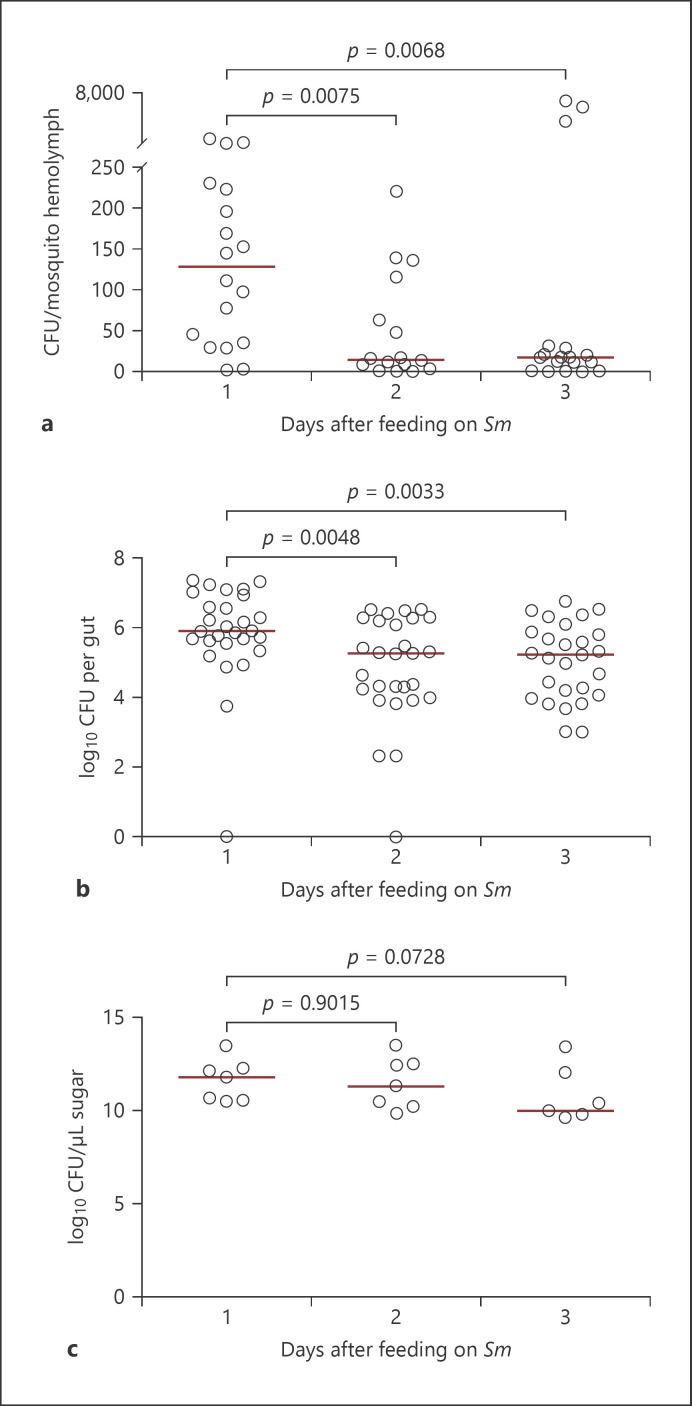
Sm efficiently colonizes the gut of A. gambiae mosquitoes after oral infection [29, 39]. It also colonizes the ovaries and is vertically transmitted to the progeny, which renders this bacterium an important tool for paratransgenic approaches that aim to generate mosquitoes resistant to Plasmodium infection [39]. Here, we monitored, during 3 consecutive days, Sm dynamics in the gut and hemolymph of mosquitoes feeding continuously on the Sm DB11 strain suspended in 3% sucrose solution. Sm DB11 is known to be virulent to insects, nematodes, and mice [24, 27, 28]. The results showed that Sm CFUs in the hemolymph are high on day 1 and then drop on days 2 and 3 after oral infection despite continuous feeding on Sm (Fig. 1a). Even though the highest numbers of Sm CFUs were detected on day 1, they were generally low, not exceeding 250 CFUs per mosquito. A similar trend was observed in the gut, whereby Sm CFUs dropped significantly by days 2 and 3 after oral infection; however, the guts generally contained much higher numbers (approximately 10,000-folds more) of Sm CFUs at all 3 days, relative to the hemolymph (Fig. 1b), suggesting that only few bacteria are present in the hemolymph at a given time despite the efficiency of Sm colonization of the gut. There were no significant differences in Sm CFUs in the sugar pads between all 3 days that could explain the significant drop observed in the gut CFUs at days 2 and 3 (Fig. 1c), indicating that Sm remains viable in the sugar solution for several days.
Fig. 1.
Serratia marcescens acquired through the oral route crosses the midgut epithelium into the hemolymph. a Hemolymph was collected by perfusion from batches of 5 mosquitoes each, at the indicated time points after feeding on Sm, and plated on LB agar containing the appropriate antibiotic. Each point on the scatter plot represents the mean CFU per batch per mosquito. Data were pooled from 5 independent biological experiments. b Guts of individual mosquitoes were dissected at the indicated time points after feeding on Sm, homogenized, and plated on LB agar containing the appropriate antibiotic. Data were pooled from 3 independent biological experiments. Each point on the scatter plot represents 1 midgut. c Bacterial counts were monitored in sugar pads harboring Sm (OD600 = 1) over a period of 3 days. Data shown are from 7 independent biological experiments. Each point on the scatter plot corresponds to CFU/μL of sugar solution in the pad per experiment. Medians are represented by red lines. Statistical analysis was performed using the Mann-Whitney test, and medians were considered significantly different if p < 0.05. Sm, Serratia marcescens; CFU, colony-forming unit.
The Route of Hemolymph Invasion by Sm Alters the Contribution of CTL4 and TEP1 to Bacterial Clearance
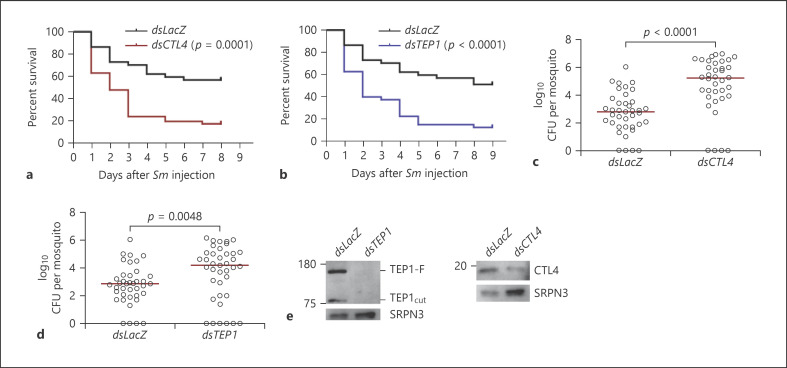
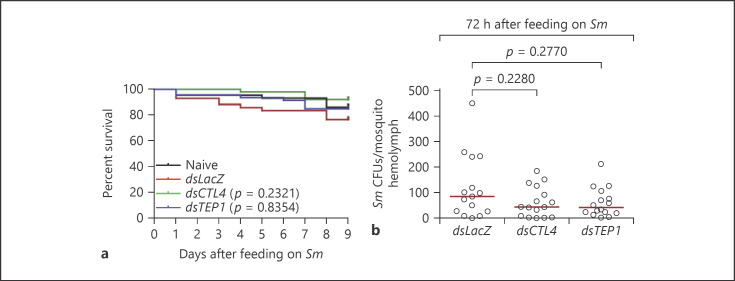
The fact that Sm invades the mosquito hemolymph in low numbers after oral infection, mimicking natural bacterial infections in the field, renders it an attractive model to address whether the route of hemolymph invasion (thoracic injection vs. crossing of the midgut) influences the contribution of key humoral immune factors to systemic antibacterial defense. To that purpose, we selected CTL4 and TEP1 as candidates due to their essential role in the systemic immune response against septic (i.e., through thoracic injection) Gram-negative bacterial infections [11, 19, 21, 40], specifically CTL4, which forms a heterodimeric complex with the lectin CTLMA2 that protects mosquitoes from septic E. coli, Pseudomonas, and Enterobacter cloacae infections [21]. First, we assessed the contribution of these genes to the susceptibility of A. gambiae adult female mosquitoes to septic Sm infections established through thoracic injection. Mosquitoes treated with gene-specific dsRNA for CTL4 and TEP1 were injected with an Sm suspension in PBS (OD600 = 0.0005) at day 3 after dsRNA administration. At this OD, the CFUs injected per mosquito ranged between 19 and 113. Both CTL4 and TEP1 knockdown (kd) compromised mosquito survival to injected Sm (Fig. 2a, b; online suppl. Fig. 1a, b) compared to LacZ kd control. Also, Sm proliferation in these genotypes was significantly higher than that in the control group (Fig. 2c, d). The Sm DB11 strain used herein is gentamycin resistant and expresses DsRed [24], which allows accurate measurement of CFUs in whole mosquito lysates, without interference from natural Sm strains whose presence seems to be sporadic and minor in our mosquitoes (online suppl. Fig. 2). This is further corroborated by a recent published work from our lab, in which the total OTUs belonging to the genus Serratia in the midguts of A. gambiae mosquitoes collected from our insectary over a 7-month period were about 12% [41]. Western blot analysis showed that both TEP1 and CTL4 were efficiently knocked down (Fig. 2e). Interestingly, when the same strain was used to establish systemic infections through oral feeding in CTL4 and TEP1 kd mosquitoes, no effect on survival was observed relative to the control group (Fig. 3a; online suppl. Fig. 3). We performed hemolymph perfusions at 72 h after oral infection to score the numbers of Sm that invaded the hemocoel in the different mosquito genotypes. Our data showed that Sm CFUs in the hemolymph of CTL4 and TEP1 kd mosquitoes were low and similar to those in the control (Fig. 3b); median values were 84.8, 43.6, and 42.2 for LacZ, CTL4, and TEP1 kd mosquitoes, respectively. This indicates that the immune function of these proteins becomes nonessential when Sm invades the hemolymph through the oral route. The differential contribution of CTL4 and TEP1 to immune defense against Sm in the 2 routes of infection is not attributed to differences in the numbers of Sm introduced into the hemolymph between both routes; 19 to 113 CFUs of Sm were injected into the mosquito hemolymph during septic infections, while the numbers of Sm that reached the hemolymph of wild-type mosquitoes at 24 h after oral infection ranged from 2 to 280 CFUs (Fig. 1a). Despite both numbers being relatively low, Sm proliferated dramatically in CTL4 and TEP1 kd mosquitoes at 24 h after Sm injection (Fig. 2c, d), while in oral infections, Sm CFUs in the hemolymph remained low in these genotypes, even at 72 h after feeding (Fig. 3b). A similar profile was noted in the LacZ kd controls of both treatments (injection vs. oral), suggesting that bacteria invading the hemolymph from the gut exhibit a more controlled proliferation compared to those injected directly into the hemolymph.
Fig. 2.
TEP1 and CTL4 are required for defense against systemic Sm infections. a, b Survival assays of the indicated mosquito genotypes after injection with Sm (OD600 = 0.0005). One representative experiment is shown from at least 3 independent biological experiments. The Kaplan-Meier survival test was used to calculate the percent survival. Statistical significance of the observed differences was calculated using the log-rank test. c, d Bacterial proliferation assays conducted on the indicated mosquito genotypes injected with Sm (OD600 = 0.0005). Batches of 8 whole mosquitoes were grinded each in the LB medium at 24 h after infection, and CFUs were scored on LB plates supplemented with the appropriate antibiotic. Each circle on the scatter plot represents the mean CFU per mosquito per batch. Statistical analysis was performed using the Mann-Whitney test, and medians were considered significantly different if p < 0.05. Data shown are from 9 independent biological experiments. e Western blots showing the knockdown efficiencies of TEP1 and CTL4. αSRPN3 was used to control for loading. TEP1, thioester-containing protein 1; CTL4, C-type lectin 4; Sm, Serratia marcescens; CFU, colony-forming unit.
Fig. 3.
CTL4 and Tep1 are not required for defense against Sm oral infections. a Survival assays of the indicated mosquito genotypes after oral infection with Sm (OD600 = 1). One representative experiment is shown from at least 3 independent biological experiments. The Kaplan-Meier survival test was used to calculate the percent survival. Statistical significance of the observed differences was calculated using the log-rank test. b Bacterial proliferation assays conducted on the indicated mosquito genotypes after oral infection with Sm (OD600 = 1). Hemolymph was perfused from batches of 5 mosquitoes at 72 h after feeding on Sm, and CFUs were scored on LB plates supplemented with the appropriate antibiotic. Each circle on the scatter plot represents the mean CFU per mosquito per batch. Statistical analysis was performed using the Mann-Whitney test. Medians (black lines) were considered significantly different if p < 0.05. Data shown are from 3 independent biological experiments. TEP1, thioester-containing protein 1; CTL4, C-type lectin 4; Sm, Serratia marcescens; CFU, colony-forming unit.
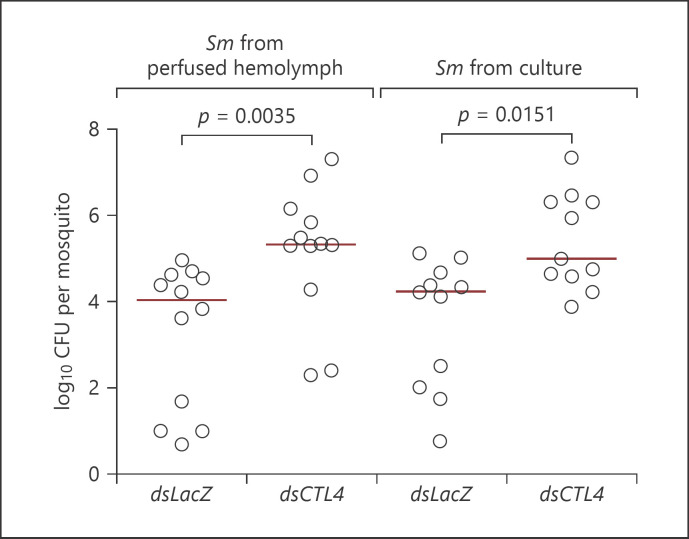
To try to explain the controlled proliferation of Sm in the hemolymph after oral feeding, we hypothesized that the process of midgut invasion might influence the fitness of bacterial cells possibly due to exposure to oxidants (reactive oxygen and nitrogen species) generated as part of the local epithelial immune response, which are known to damage bacterial cells (reviewed in [42]). For instance, oxidants generated by dual oxidase in the Drosophila gut limit microbial proliferation [43], and nitric oxide produced in the A. gambiae midgut by heme peroxidase 2 and NADPH oxidase 5 enhances Plasmodium cytotoxicity [44]. To determine if bacteria exhibit an altered fitness after crossing the midgut, the hemolymph was collected by perfusion from wild-type mosquitoes that have fed on Sm during 24 h, and bacterial cells in the perfusate were pelleted by centrifugation, washed, and injected into LacZ and CTL4 kd naïve mosquitoes. The same mosquito genotypes injected with Sm (OD600 = 0.0005) prepared from a fresh batch culture were used as control. Challenged mosquitoes were homogenized 24 h later to score Sm CFUs. The results show that bacteria prepared from hemolymph perfusates were able to proliferate to the same extent as those originating from a fresh culture, indicating that bacterial cells that cross the gut into the hemolymph do not seem to suffer from a reduced growth fitness (Fig. 4).
Fig. 4.
Invasion of the midgut epithelium does not alter the fitness of Sm. Bacterial proliferation assays conducted on the indicated mosquito genotypes after injection with Sm prepared from a fresh bacterial culture (OD600 = 0.0005) or collected from hemolymph perfusions of mosquitoes that have fed on Sm for 24 h. Batches of 7 whole mosquitoes were grinded in the LB medium at 24 h after infection, and CFUs were scored on LB plates supplemented with the appropriate antibiotic. Each point on the scatter plot represents the mean CFU per mosquito per batch. Statistical analysis was performed using the Mann-Whitney test. Medians (red lines) were considered significantly different if p < 0.05. Data shown are from 3 independent biological experiments. Sm, Serratia marcescens; CFU, colony-forming unit.
The Rel2 Pathway and Phagocytosis Are Dispensable for Defense against Sm Systemic Infections Established after Gut Invasion
The mosquito Rel2 pathway is involved in defense against systemic infections with Gram-negative and Gram-positive bacteria [45, 46], and also protects against P. falciparum ookinetes [45, 47, 48]. To determine the contribution of Rel2 to systemic defense against Sm that invade the hemolymph following an oral infection, LacZ (control), Rel2, and Rel1 kd mosquitoes were fed continuously on sugar pads containing a suspension of Sm at an OD600 = 1, and their survival was scored over 10 days after challenge. Neither Rel1 nor Rel2 silencing compromised mosquito survival to oral Sm infections (Fig. 5a; online suppl. Fig. 4a). In contrast, when systemic infection in these mosquito genotypes was established by injecting Sm into the hemocoel, Rel2 kd compromised mosquito survival (Fig. 5b; online suppl. Fig. 4b) and resistance (Fig. 5c) as noted from the enhanced bacterial proliferation relative to control. Hence, the Rel2 pathway contributes to immune defense against Sm systemic infections established through septic injury but not through feeding. The efficiency of Rel1 and Rel2 silencing in our hands is 44 and 50% (Fig. 5d), which is similar to what was reported previously for these genes [46, 49].
Fig. 5.
The Rel2 signaling pathway plays a role in mosquito tolerance and resistance against systemic but not oral Sm infections. Survival assays following Sm oral (OD600 = 1) (a) and systemic (OD600 = 0.0005) (b) infections in mosquitoes silenced for either Rel1 or Rel2. One representative experiment is shown from at least 3 independent biological experiments. The Kaplan-Meier survival test was used to calculate percent survival. Statistical significance of the observed differences was calculated using the log-rank test. c Bacterial proliferation assays conducted on Rel2 kd mosquitoes injected with Sm (OD600 = 0.0005). Batches of 8 whole mosquitoes were grinded in the LB medium at 24 h after infection, and CFUs were scored on LB plates supplemented with the appropriate antibiotic. Each point on the scatter plot represents the mean CFU per mosquito per batch. Statistical analysis was performed using Robust ANOVA in R. Data shown are from at least 3 independent biological experiments. d Transcript levels of Rel1 and Rel2 measured by qRT-PCR in whole female mosquitoes at 3 days following injection of their respective dsRNA. Error bars represent standard error of the mean of 2 biological repeats. Statistical analysis was done using Student's t test. Sm, Serratia marcescens; CFU, colony-forming unit.
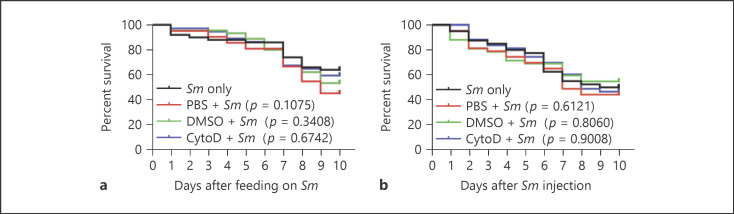
Phagocytosis is an important innate immune response that was shown to control host susceptibility to septic bacterial infections in A. gambiae [11, 50] and Drosophila [51, 52, 53, 54]. Additionally, blocking phagocytosis in Drosophila adults by cytochalasin D injection compromised the survival of the flies to oral Sm infections [24]. Based on these data, we hypothesized that the dispensable roles of CTL4, TEP1 and the Rel2 pathway in defense against Sm that gains access into the hemolymph after oral infection may be due to a primary role of phagocytosis in controlling host susceptibility through this route. To address this point, mosquitoes injected intrathoracically with 69 nL of 62.5 μg/mL solution (120 μM) of cytochalasin D in PBS were allowed to feed continuously on a sugar solution containing Sm at 6 h after cytochalasin D injection. Survival assays revealed that cytochalasin D treatment did not affect mosquito susceptibility to oral Sm infections (Fig. 6a; online suppl. Fig. 5a), despite the fact that the concentration of cytochalasin D used herein is higher than that which blocked phagocytosis in Drosophila adults [24] and A. gambiae cell lines [55]. Hence, our data suggest that, in the mosquito, phagocytosis may not play an essential role in controlling Sm that escapes into the hemolymph after oral infections. Also, cytochalasin D treatment did not compromise mosquito survival to Sm injection (Fig. 6b; online suppl. Fig. 5b), which was not surprising to us due to the primary immune defensive role of the humoral factors TEP1 and CTL4 in this infection route.
Fig. 6.
Blocking phagocytosis by cytochalasin D does not seem to significantly impact mosquito susceptibility to oral or systemic Sm injections. Survival of noninjected mosquitoes or mosquitoes preinjected with either PBS, DMSO, or cytochalasin D was monitored over a period of 10 days following oral (OD600 = 1) (a) or systemic (b) Sm infections. One representative experiment is shown from at least 3 independent biological experiments. The Kaplan-Meier survival test was used to calculate the percent survival. Statistical significance of the observed differences was calculated using the log-rank test. Sm, Serratia marcescens.
Abdomen and Midgut Transcriptional Responses after Sm Oral and Septic Infections
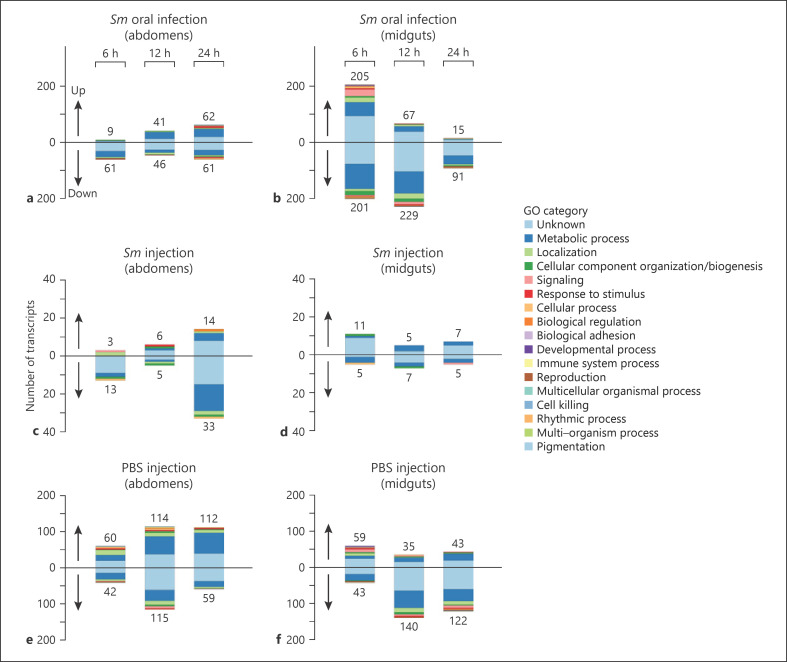
The fact that CTL4- and TEP1-mediated systemic immune responses did not provide resistance to Sm invading the hemolymph from the gut prompted us to monitor whether oral infection primes the tissue-specific expression of an immune gene repertoire in the fat body or midgut that could explain the dispensable roles of CTL4 and TEP1 in this route of infection. To that purpose, abdomens (excluding gut, malpighian tubules, and ovaries) and midguts (excluding hind- and foreguts) were dissected from wild-type mosquitoes at 6, 12, and 24 h after feeding on a 3% sucrose solution containing Sm Db11 (OD600 = 1) or after injection with an Sm suspension in PBS (OD600 = 0.0005), and transcriptional responses were monitored by RNA-seq. Mosquitoes fed on 3% sucrose solution or injected with sterile PBS were used as controls for Sm oral infection and Sm injection, respectively. Three independent biological experiments were performed. All DETs were determined according to a false discovery rate of 0.05 (online suppl. Table 2). In the abdomens, 70, 87, and 123 DETs were identified, respectively, at 6, 12, and 24 h after Sm oral infection with respect to untreated controls (i.e., only sugar fed), whereas 16, 11, and 47 DETs were identified at the respective time points after Sm injection with respect to PBS injection only (Fig. 7a, c), indicating that oral infection triggers more profound transcriptional changes in the abdomen than injection. In midguts, 406, 296, and 106 DETs were identified at 6, 12, and 24 h after Sm oral infection with respect to untreated controls, whereas 16, 12, and 12 DETs were identified at the respective time points after Sm injection (Fig. 7b, d), again indicating that oral infection had a greater influence on the midgut transcriptome than injection, which is rather expected since in the context of direct injection into the hemocoel, Sm is unlikely to invade the gut epithelium from the basal side. The low numbers of DETs in abdomens and midguts of Sm-injected mosquitoes are also likely due to the fact that PBS injection itself regulated a large number of transcripts in these tissues at all 3 time points (Fig. 7e, f). Of note, although the PBS solution used in these treatments is sterile, bacteria attached to the mosquito cuticle can still be introduced into the hemolymph due to the wounding process.
Fig. 7.
Number of DETs per treatment, colored according to top-level gene ontology terms within the “biological process” ontology. a, b DETs in abdomens and midguts, respectively, of mosquitoes fed on Sm. c, d DETs in abdomens and midguts, respectively, of mosquitoes injected with Sm. e, f DETs in abdomens and midguts, respectively, of mosquitoes injected with PBS. For this plot, each transcript was assigned to its top-level gene ontology term within the “biological process” ontology. Whenever a transcript was associated with >1 top-level term, the least common term was chosen. DETs, differentially expressed transcripts; Sm, Serratia marcescens.
Functional classification of all DETs in abdomens and midguts from all treatments revealed that metabolic genes are the most represented class followed by those involved in cellular localization (Fig. 7; online suppl. Table 2). Surprisingly, immunity genes were underrepresented in both abdomens and midguts from all treatments suggesting that oral and septic infections with Sm have little effect on the transcriptome of immunity genes. When comparing the DETs in midguts of mosquitoes injected with Sm to those of mosquitoes fed on Sm, a small overlap was observed (online suppl. Fig. 6a). The same was noted for abdomens (online suppl. Fig. 6b), indicating that different physiological responses are triggered in response to the different routes of Sm infection. To determine whether hemolymph infection following Sm injection or midgut crossing triggers route-specific unique responses to Sm in abdomens, we identified the transcripts whose expression changes significantly in response to Sm injection (Sm_inj) relative to PBS-injected (PBS_inj) control but not in response to any other treatment (i.e., Sm oral feeding [Sm_of] vs. untreated control [UC] or Sm_inj vs. UC or PBS vs. UC) and transcripts whose expression changes significantly in response to Sm_of relative to UC but not in response to any other treatment (i.e., Sm_inj vs. PBS_inj or Sm_inj vs. UC or PBS_inj vs. UC). In abdomens, where physiological responses are expected to be more relevant to hemolymph infection with Sm due to the presence of the fat body and sessile hemocytes, only one transcript, vacuolar protein sorting 60 (Vps 60; AGAP005100), showed significant change in expression unique to Sm_inj versus PBS_inj (online suppl. Table 3). Vps proteins are involved in the formation of multivesicular bodies which play important roles in the endocytic degradation of proteins and also in the formation of exosomes [56], which are small extracellular vesicles that mediate intercellular communication to regulate several biological processes including tissue repair [57]. The upregulation of Vps60 in abdomens may reflect enhanced investment in repair processes in response to Sm infection. Tissue repair and regeneration processes are crucial for host tolerance to infection [58]. Alternatively, this upregulation may indicate an increase in the protein secretory capacity of the fat body in response to immune activation and infection, which in Drosophila was associated with enhanced tolerance to infection [59]. On the other hand, 3 transcripts showed a downregulated expression profile in abdomens unique to Sm_of versus UC (online suppl. Table 4) including, CCR4-NOT (CNOT) transcription subunit complex 3 (AGAP009030), very long-chain enoyl-reductase (AGAP010714), and UPF0518 (AGAP011705). CNOT is a large multi-subunit RNA deadenylase, composed of catalytic and noncatalytic subunits that is conserved in eukaryotes and plays key roles in mRNA degradation and turnover, hence controlling the rate of protein expression [60]. It also plays an effector role in miRNA-mediated gene silencing [61]. As such, CNOT is involved in regulating several physiological processes in the cell including cell death, autophagy, immunity, inflammation, and differentiation to mention a few [60, 62]. It is tempting to speculate that the downregulation of CNOT3, a noncatalytic subunit essential for CNOT activity [63], in abdomens may increase the stability of mRNAs involved in immunity, tissue repair, or stress response which might favor host tolerance to systemic infection established after feeding on Sm. The very long-chain enoyl-reductase is involved in the synthesis of sphingolipids and glycerophospholipids [64], and its downregulation may indicate a shift in lipid metabolism, whereas UPF0518 has no known function. GO enrichment analysis of the route-unique transcripts listed in online suppl. Tables 3 and 4 suggests that the global physiological response associated with the oral route includes most of that associated with Sm injection (except 4 genes), in addition to other specific functions (online suppl. Tables 5, 6). These oral route-specific responses are mainly attributed to the midgut (33 out of 36 genes) and are enriched in biological processes related mainly to protein translation, protein folding, protein modification, DNA damage repair, and cell cycle regulation among others (online suppl. Table 5). These responses could reflect the pathology induced by Sm to the midgut epithelium.
A Wallenius noncentral hypergeometric distribution was used to test for the enrichment of GO terms, KEGG pathways, and gene families in the total set of differentially regulated genes in abdomens and midguts of mosquitoes fed on or injected with Sm, relative to UC and PBS-injected control, respectively. The results identified 13 unique terms (4 gene families, 5 KEGG pathways, and 4 GO terms) that were significantly overrepresented, the majority of which were associated with functions related to protein translation, processing, and export, followed by terms related to metabolic processes, in particular oxidative phosphorylation (OXPHOS), and 1 associated with immunity (online suppl. Table 7). Concerning immunity, only the clip-domain serine protease family (CLIPs) was significantly overrepresented in the midgut of mosquitoes at 12 h after Sm oral infection (Fig. 8; online suppl. Table 7). In total, 10 CLIPs were downregulated in this treatment including CLIPC4, CLIPB4, CLIPB1, CLIPC9, CLIPB13, CLIPA8, CLIPA4, CLIPA6, CLIPA1, and CLIPA7. CLIPs are key components of serine protease cascades that regulate important insect immune responses specifically melanization and Toll pathway activation [3, 65, 66]. Among the enriched CLIPs, CLIPB4 and CLIPC9, both catalytic clips, are involved in the melanization of P. berghei ookinetes in refractory mosquito backgrounds [67, 68], while CLIPA8 and CLIPA7 are noncatalytic CLIPs that act as positive and negative regulators of Plasmodium melanization, respectively [67]. The melanization response to fungal infections requires CLIPA8 [69], while both CLIPA8 and CLIPC9 play an essential role in the melanization response to bacterial infections [68, 70, 71]. CLIPA1, CLIPA4, CLIPA6, and CLIPB1 do not seem to be involved in Plasmodium melanization [67], whereas the roles of CLIPC4 and CLIPB13 in the melanization response remain to be elucidated. The downregulation of this significant number of CLIP genes suggests that Sm oral infection may suppress the melanization response regulated by several of these CLIPs. Of note, the differential regulation of CLIPs in the midgut is most likely attributed to hemocytes attached to the midgut surface and not to the midgut epithelium per se, as insect CLIPs are mainly expressed in hemocytes and fat body cells [65, 66]. Indeed, several of the overrepresented CLIPs in our study including CLIPA7, CLIPA8, CLIPB1, CLIPB4, CLIPB13, CLIPC4, and CLIPC9 were among the genes identified in transcriptomic studies of mosquito hemocytes [72, 73]. Also, PPO6 which is hemocyte specific [73, 74] was among the DETs identified in mosquito midguts in response to Sm oral infections (online suppl. Table 2), further indicating that some of the immunity genes identified in the midgut transcriptome are attributed to midgut-attached hemocytes rather than to the midgut epithelium. Indeed, there is evidence that contact between midgut epithelial cells and the gut microbiota which occurs during Plasmodium midgut invasion initiates systemic immune priming by triggering hemocyte differentiation and their attraction to the midgut surface where they present antimicrobial activities including complement activation [14, 15, 75]. Of note, the KEGG pathway enrichment analysis identified 12 genes involved in OXPHOS that are all downregulated in the midgut after feeding on Sm, suggesting that midgut infection may be triggering a shift in the gut metabolic program. As for the abdomens, only the FOXO signaling pathway is enriched after Sm injections but not feeding (online suppl. Table 7). In Drosophila, the FOXO transcription factor activates AMP production under nutritional stress independent of Toll and Imd pathways [76]. FOXO is also required for Drosophila to survive oral infections with Sm[77]. Whether FOXO signaling plays a similar role in mosquito immunity against bacterial septic injections and oral infections remains to be elucidated.
Fig. 8.
Heatmap of estimated log-fold changes of all CLIP genes. Log-fold changes were estimated for each comparison indicated by column labels. The column labels start with symbol for control (UC, unchallenged; iPBS, PBS injection), followed by a symbol for the tissue (a, abdomen; g, gut). The symbols for the treatments are OF (oral feeding) and iSm (injection of Serratia marcescens). The timing of treatment is indicated in hours (h). The row dendrogram shows a hierarchical clustering of the dissimilarities between CLIP genes in their log-fold change patterns among all comparisons shown in the plot. CLIP, clip-domain serine proteases.
In addition to CLIPs, few other genes belonging to distinct immune gene families, though not overrepresented, were also downregulated after Sm oral but not septic infections (online suppl. Table 2). These include, Eiger, GNBPB1, the scavenger receptors SCRB5, SCRB7, and SCRB9, PPO6, TEP2, and CTLMA1. Eiger was downregulated in abdomens at 24 h after Sm oral infections. It is a TNF ortholog which, in Drosophila, is also expressed in the fat body [78] and plays an important role in regulating melanization, AMP expression, and immune defense against extracellular pathogens [78, 79]. GNBPB1 was also downregulated in abdomens at 24 h after Sm oral infections, and it was previously shown to contribute to anti-Plasmodium immunity [22]. Two members of the scavenger receptor gene family, SCRB5 and SCRB7, were downregulated in abdomens at 12 and 6 h after Sm oral infections, respectively, while SCRB9 was downregulated in the midgut at 6 h after infection. The role of these receptors in mosquito immunity has not been investigated, but members of this family are involved in the phagocytic uptake of bacteria in Drosophila [80, 81, 82]. PPO6, TEP2, and CTLMA1 were also downregulated in the midgut in response to Sm oral infection. PPO6, a phenoloxidase expressed in mosquito adults, is involved in the melanization reaction to P. berghei ookinetes, bacteria, and fungi [33, 69, 70]. While the roles of TEP2 and CTLMA1 in immune defense are unknown, certain members of the TEP and CTL families are key players in the mosquito antimicrobial defense [19, 21, 33, 83, 84, 85]. On the other hand, only 4 immunity genes were upregulated after Sm oral infections; galectin 5 and cecropin A were upregulated in abdomens, whereas CTL6 and lysozyme C7 (LYSC7) were upregulated in the midgut at the indicated time points (online suppl. Table 2). Whether these genes are involved in controlling Sm proliferation in the hemolymph following oral infection will require further investigations. Altogether, these results suggest that Sm invasion of the hemolymph following oral infection is seemingly associated with transcriptional suppression of several immune genes involved in different facets of the humoral and cellular immune response.
Discussion
In all organisms, the vast majority of microbial infections are established through initial interactions between microbes and host at barrier epithelia. There is growing evidence, in both invertebrates and vertebrates, that the route of infection determines the adaptive strategies of the host in terms of the nature of immune responses engaged to deal with the insult [86, 87]. In Drosophila, oral and systemic infections with Pseudomonas entomophila triggered the evolution of resistance in fly populations that was infection-route specific [86]. Also, oral infection of Drosophila with different RNA viruses revealed different patterns of virus clearance and immune priming compared to systemic injections [87]. In a similar context, Anopheles coluzzii oral infections with O'nyong nyong arbovirus shared little overlap in transcriptional responses with intrathoracic injections [88]. Route-specific immune responses have been also described in mammals. For instance, the intranasal administration of the vaccinia virus to mice triggered a stronger adaptive response in magnitude and diversity compared to local intradermal injections [89]. In another study, infection of mice with Brucella melitensis through 3 different routes, intradermally, intraperitoneally, and intranasally, revealed route-specific contributions of the 3 lymphoid populations, CD4+ T cells, B cells, and γδ+ T cells [90]. Interestingly, the authors also showed that the type IV secretion system which is required for Brucella persistence in the lungs after intranasal infections does not seem to promote persistence in the skin after intradermal infections, suggesting that the route of infection influences not only the physiology of the immune response but also the contribution of certain virulence factors to microbial persistence. In this work, we used the Sm DB11 strain as a model mosquito pathogen to score the contribution of 2 key humoral antibacterial factors, TEP1 and CTL4, to immune defense against hemolymph infections established either through injection or midgut invasion after oral feeding, in adult A. gambiae female mosquitoes.
Sm is detected in the mosquito hemolymph 1 day after oral infection, but the numbers drop significantly during the following 2 days concomitant with a reduction in Sm CFUs in the midguts. This reduction is not due to increased bacterial death in the sugar pads used to feed mosquitoes since bacterial CFUs in the pads did not change significantly during this period. The reduction in Sm hemolymph CFUs in days 2 and 3 could be explained by immune defenses active at the level of the midgut epithelium that may restrict the numbers of bacteria that successfully invade the midgut into the hemolymph [24, 29]. It may also reflect a reduction in mosquito feeding due to chronic infection of the gut by Sm. In fact, the gustatory receptor gene Gr9 was shown to be associated with Sm infection phenotype of A. gambiae midguts, and silencing this gene increased Sm colonization of the midgut, indicating that Sm infection may trigger a behavioral feeding response [29]. The composition of the gut microbiota in Drosophila also influences its foraging behavior [91]. We did not address whether blood feeding would influence the dynamics of Sm invasion of the hemolymph; however, we expect that it would be more difficult for Sm to cross the midgut during blood feeding since the peritrophic matrix [92] and the dityrosine network produced by a peroxidase dual oxidase system [93] will likely restrict midgut permeability to microbes.
We found that CTL4 and TEP1 are required for mosquito resistance to Sm infections of the hemolymph established following injection but not oral infection. The latter route may trigger systemic immune priming by gut epithelia creating redundancy in bacterial defense among different arms of the immune response. The numbers of Sm that gain access to the hemolymph after oral infection are likely to be small as inferred from our hemolymph perfusion assays. In Drosophila, melanization was shown to be essential for immune defense against septic infections with a small dose of S. aureus[13]. Our attempts to measure PO activity following oral Sm infections were not conclusive, as some trials showed activation while others did not (data not shown). There are 2 plausible explanations for this inconsistency: first, hemolymph invasion after oral infection is likely to occur in waves and not at one single time point which makes it difficult to pinpoint the optimal time point for measuring hemolymph PO activity. Second, small numbers of bacteria are most likely reaching the hemolymph in each wave, as inferred from the small numbers of Sm CFUs scored in the hemolymph after oral infections (Fig. 1), which might not trigger a measurable PO activity. This is in contrast to bacterial injections where the time of hemolymph infection and the dose of introduced bacteria can be optimized to trigger a measurable PO response [70, 94]. However, the fact that silencing TEP1, a key upstream regulator of the mosquito melanization response [69, 70, 95], did not alter mosquito susceptibility to Sm oral infection and that PPO6 and several CLIPs were downregulated after feeding on Sm suggest that melanization may not play an essential role in this route of infection. We observed that bacteria invading the hemolymph from the gut remained at low numbers, as compared to those injected directly into the hemolymph, even in control (dsLacZ) mosquitoes. It remains unclear whether this is attributed to a longer generation time (i.e., reduced cell division rate) associated with bacteria that cross the midgut into the hemolymph but not with those directly injected into the hemolymph. Reduced proliferation is expected to benefit bacterial persistence since the release of cell wall components, such as peptidoglycan, during bacterial cell division would activate PGRPs leading to Imd pathway activation in fat body cells [20, 96, 97, 98]. However, it is worth noting that the mosquito complement-like system, which plays a key role in antibacterial immunity, may not be sensitive to bacterial proliferation since TEP1 was shown to efficiently bind E. coli bioparticles [40, 95]. Another possibility is that exposure to the midgut triggers some alterations of the outer membrane of bacteria, resulting in changes in susceptibility to immune effectors [99]. We showed that bacteria that invade the hemolymph from the midgut proliferate efficiently after extraction and injection into the hemolymph of control or dsCTL4 mosquitoes, indicating that they have not lost fitness. These results suggest that the combination of the midgut invasion process and the exposure to the hostile hemolymph environment may impose a certain stress on the bacteria associated with a reduced proliferation rate. This stress may have been relieved through the process of extraction and washing before the cells are injected into another mosquito.
Rel2 silencing did not affect mosquito survival to oral Sm infections, suggesting that the Rel2/Imd pathway may be either nonessential for defense against Sm invasion of the hemolymph following an oral infection or that the pathway is not activated through this route. In Drosophila, Sm was sensitive to the local Imd response in the gut but failed to activate the systemic response in the fat body after crossing the gut epithelium into the hemolymph [24]. Currently, it is not possible to accurately score the activation of the mosquito Rel2 pathway due to the absence of a specific gene expression signature associated with this pathway. However, in our RNA-seq analysis, several immunity genes that are known to be at least partially regulated by Rel2, such as TEP1, APL1, several CLIPs, and FBNs among others [45, 47], were not upregulated neither in the midgut nor in the abdomens after oral Sm infection, suggesting that the Imd pathway may not be activated through this route. Phagocytosis is an essential determinant of Drosophila susceptibility to Sm oral infection [24]. However, this does not seem to be the case in A. gambiae since the treatment of mosquitoes with cytochalasin D did not alter their susceptibility to oral Sm infection. This result may suggest either the existence of functional redundancy among different branches of the immune response in this route of infection or that phagocytosis may not be essential when small numbers of bacteria are present in the hemolymph. Indeed, a recent study in Drosophila revealed that hemocyte-deficient flies did not succumb to a low-dose S. aureus infection, but a high dose did compromise their survival [13].
Our RNA-seq analysis identified a limited number of DETs in the midguts after Sm oral infection, specifically at the late 24-h time point. This agrees with a previous microarray-based study that compared the transcriptional responses in the guts of antibiotic-treated mosquitoes at 3 days after Sm oral infection with those of antibiotic-treated uninfected mosquitoes [29]. Another microarray-based study in A. gambiae that compared the gut transcriptomes of antibiotic-treated and untreated mosquitoes also identified a limited set of differentially expressed genes [100]. On the other hand, oral infections in Drosophila trigger dramatic changes in the gut transcriptome [101, 102]. This discrepancy can be explained by the different nature of the food source of both species; while Drosophila feeds mainly on fermented and rotten fruits rich in yeast and bacteria [103, 104], A. gambiae mosquitoes feed mainly on human blood which is sterile. It should also be noted that in oral infections, the number of DETs increased in the abdomens with time, whereas the inverse was observed in midguts. This DETs pattern in abdomens may be due to the continuous crossing of Sm from the gut into the hemolymph triggering physiological responses in the fat body and/or hemocytes attached to it or due to signaling between the gut epithelium on the one hand and the fat body and hemocytes on the other. Interorgan communication has been mainly studied in Drosophila whereby pathogen-infected intestinal cells signal to hemocytes, which in turn regulate intestinal regeneration [105, 106]. There is also evidence for signaling between the gut and fat body in Drosophila to regulate energy homeostasis [107, 108]. Our RNA-seq analysis also revealed that the response to Sm oral infections is more pronounced in the midgut, and it becomes even more pronounced when the analysis is restricted to transcripts that uniquely respond to feeding on Sm. Most of these transcripts are associated with biological processes related to protein translation, cell cycle, and DNA repair, which may not be surprising since Sm infection of the Drosophila gut was shown to trigger significant damage to the gut epithelium that alters cell morphology and physiology [109]. Epithelial damage of the gut and enhancement of gut physiological responses associated with stress, cell renewal, and proliferation have been also observed in Drosophila intestinal infections with Erwinia carotovora [101]. Only 4 genes showed an expression pattern unique to Sm injection suggesting that most of the transcriptional response is triggered by the wounding process per se. This was not surprising since a previous study by Dimopoulos et al. [110] showed that the predominant transcriptional responses triggered by septic and sterile injury in the refractory L3–5 mosquito strain were shared, suggesting that most of these responses are attributed to injury and/or wound healing; injury-specific transcriptional responses were dominated by functional groups pertaining to carbohydrate metabolism, whereas septic infection was dominated by immunity genes. Interestingly, a separate study showed that wounding of A. gambiae mosquitoes by the injection of water or dsRNA triggers the killing of P. falciparum parasites in a TEP1-dependent manner [111]. To better understand the relationship between wounding and immune defense to Plasmodium, the authors performed a genome-wide analysis of the transcriptional response to wounding in adult A. gambiae mosquitoes and identified 53 genes with statistically significant regulation that were enriched mainly in genes involved in proteolysis-related processes including several CLIP genes. Wounding also triggered the expression of several immunity genes with known anti-Plasmodium roles such as TEP1, LRIM1, APL1C, and FBN9 among others [112]. The fact that wounding triggers the expression of several mosquito immunity genes explains most likely why no immunity genes showed an expression pattern unique to Sm_inj in our study. The complex physiological responses triggered by wounding in other insects (reviewed in [113]) lend further support to our conclusion. In Drosophila, for instance, where the wound healing process is best characterized, cellular responses mediated by hemocytes, epithelial cells, and fat body cells act in concert with humoral factors including hemolectin and fondue to seal the wound, clear tissues debris, and initiate soft clot formation that becomes eventually melanized by the action of crystal cell-derived phenoloxidase [114, 115, 116, 117, 118, 119, 120, 121, 122]. Interestingly, fat body cells were also shown to secrete antimicrobial peptides locally to protect from wound infection [115].
Our transcriptomic analysis revealed that metabolic genes are the most represented functional class of all DETs in abdomens and midguts from all treatments. Knowing that metabolism is at the core of all biological processes, this result comes as no surprise. There is currently ample evidence in mammals, specifically from studies in mice, that cellular metabolism shapes the activation and differentiation of myeloid and lymphoid immune cells during infection [123, 124]. This relation has been particularly addressed in macrophages, whereby proinflammatory macrophages of the M1 type exhibit a metabolic shift to aerobic glycolysis associated with the production of nitric oxide, reactive oxygen species, and prostaglandins, whereas M2 macrophages exhibit a shift towards OXPHOS and increased dependency on a complete Kreb's cycle fueled by glucose, fatty acids, and glutamine (reviewed in [125]). Our knowledge of immunometabolism in mosquitoes is fragmented, with emerging evidence pointing toward a cross-talk between metabolic genes or metabolic signaling pathways and immune defense processes. In A. stephensi, P. falciparum infection was shown to induce the expression of insulin-like peptides that favor parasite development by suppressing the NF-κB signaling pathway in the midgut and by triggering metabolic shifts in this tissue independent of NF-κB [126]. Human insulin ingested by mosquitoes during blood feeding was also shown to enhance Plasmodium development by inhibiting NF-κB-dependent immune responses [127]. This reciprocal effect between immunity and insulin signaling has been also reported in Drosophila [128]. Another example of cross-talk between immunity and metabolism in mosquitoes is the finding that Lipophorin, a multifunctional carrier involved in lipid transport and metabolism, and its receptor are upregulated in Aedes aegypti mosquitoes following infection with Gram-positive bacteria and fungi in a Toll/Rel1-dependent manner [129]. In A. gambiae, apolipophorin-II/I was shown to control TEP1 expression during systemic infections with E. coli and Beauveria bassiana [130]. Metabolic decisions may also influence the outcome of mosquito infection with microbes independent of immunity, and this has been mainly studied in the context of Plasmodium infections. For instance, the susceptibility of refractory and susceptible strains of A. gambiae to infection with P. berghei was shown to be influenced by broad metabolic differences between these strains, whereby the refractory strain exhibits rapid utilization of lipids, impaired mitochondrial respiration, and increased glycolytic activity leading to higher ROS production that is toxic to malaria parasites [131]. In a more recent study, Lampe et al. [132] elegantly showed that the timely expression of blood-meal-inducible miR-276 finely regulates the rate of amino acid catabolism, terminating the investment in reproductive processes and providing excess resources for the sporogonic development of P. falciparum. In a similar context, it would be interesting to determine whether metabolic shifts induced by blood feeding would influence mosquito resistance to bacterial and fungal infections and through what mechanisms. It was interesting to note that genes involved in OXPHOS were overrepresented in our KEGG pathway enrichment analysis, and all were downregulated in the midgut after feeding on Sm. This metabolic shift away from OXPHOS (a catabolic process) may reflect increased dependency on anabolic processes such as aerobic glycolysis that would be required to promote midgut tissue repair in response to the damage triggered by Sm intestinal infection [24]. Tissue repair processes are known to be anabolic in nature and contribute to host tolerance to infection [124, 133]. Our transcriptomic analysis also revealed that the abdomen transcriptome was substantially larger in Sm oral infections relative to injections, at all 3 time points (compare Fig. 7a, c), despite the fact that injections resulted eventually in higher loads of Sm in the hemolymph. While these results may reflect different adaptive strategies of the host in response to different routes of infection with the same microbe, they possibly pinpoint also to a potential key role of the midgut epithelium in priming immune and non-immune physiological responses in the fat body and hemocytes that should act in concert to control hemolymph infections.
In conclusion, we provide evidence using gene silencing and transcriptomic analysis that the dynamics of immune defense to bacterial hemolymph infections through the midgut are different from those of hemolymph infections established by septic injections. The key difference between both routes is that the first involves the gut as a natural route towards establishing systemic infection while the second utilizes the more artificial or “naturally less common” wounding process to do so. Being at the front line of microbial defense, it is not surprising that the midgut epithelium, in addition to its classical evolutionary conserved role in local immune defense through its physical impermeability and chemical defenses, also plays an important role in priming physiological responses in distant organs that provide the host with better resistance and tolerance in case the microbe succeeds in crossing this barrier to establish a systemic infection. The nature of these protective physiological responses and how they are primed by the midgut epithelium remain largely unknown.
Statement of Ethics
This study was carried out according to the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (Bethesda, USA). Animal protocol was approved by the Institutional Animal Care and Use Committee (IACUC) of the American University of Beirut (permit number 17-10-451). The IACUC functions in compliance with the Public Health Service Policy on the Humane Care and Use of Laboratory Animals (USA) and adopts the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health.
Conflict of Interest Statement
The authors have no conflicts of interest to declare.
Funding Sources
This work was supported by AUB Kamal A. Shair CRSL Research award 103599 (to M.A.O.), the National Institutes of Health, the National Institute for Allergy and Infectious Disease, grant R01 AI140760 (to M.A.O.), the New York State Department of Agriculture and Markets (C00235GG) (to N.B.), NSF Grants IOS1656118 and IOS1653021 (to N.B.), and NIH R21AI153934 (to N.B.). The funding sources had no role in the preparation of data or the manuscript.
Author Contributions
M.A.O. and A.S.D. conceived and designed the study. A.S.D. performed all functional genetic studies. M.A.O. and N.B. designed the transcriptomic analysis. N.B. and X.Y. performed the RNA-seq library construction and sequencing. M.A.O., H.D., N.B., and X.Y. analyzed the RNA-seq data. M.A.O. wrote the manuscript. All authors critically reviewed and edited the manuscript.
Supplementary Material
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Acknowledgements
We thank Kamal A. Shair Central Research Science Laboratory for providing free access to their equipment. We also thank Domonique Ferrandon (Strasbourg University) for providing the Serratia marcescens strain DB11.
References
- 1.Bulet P, Stöcklin R. Insect antimicrobial peptides: structures, properties and gene regulation. Protein Pept Lett. 2005;12((1)):3–11. doi: 10.2174/0929866053406011. [DOI] [PubMed] [Google Scholar]
- 2.Hanson MA, Dostálová A, Ceroni C, Poidevin M, Kondo S, Lemaitre B. Synergy and remarkable specificity of antimicrobial peptides in vivo using a systematic knockout approach. Elife. 2019;8:e44341. doi: 10.7554/eLife.44341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakhleh J, El Moussawi L, Osta MA. The melanization response in insect immunity. In: Ligoxygakis P, editor. Insect immunity. Vol.52. Amsterdam, Netherlands: Elsevier; 2017. pp. p. 2–20. [Google Scholar]
- 4.Nappi A, Poirié M, Carton Y. The role of melanization and cytotoxic by-products in the cellular immune responses of Drosophila against parasitic wasps. Adv Parasitol. 2009;70:99–121. doi: 10.1016/S0065-308X(09)70004-1. [DOI] [PubMed] [Google Scholar]
- 5.King JG, Hillyer JF. Infection-induced interaction between the mosquito circulatory and immune systems. PLoS Pathog. 2012;8((11)):e1003058. doi: 10.1371/journal.ppat.1003058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wood W, Martin P. Macrophage functions in tissue patterning and disease: new insights from the fly. Dev Cell. 2017;40((3)):221–33. doi: 10.1016/j.devcel.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dostalova A, Rommelaere S, Poidevin M, Lemaitre B. Thioester-containing proteins regulate the Toll pathway and play a role in Drosophila defence against microbial pathogens and parasitoid wasps. BMC Biol. 2017;15((1)):79. doi: 10.1186/s12915-017-0408-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Povelones M, Osta MA, Christophides GK. The complement system of malaria vector mosquitoes. In: Raikhel AS, editor. Progress in mosquito research. Vol.51. Amsterdam, Netherlands: Elsevier; 2016. pp. p. 223–42. [Google Scholar]
- 9.Blandin S, Moita LF, Köcher T, Wilm M, Kafatos FC, Levashina EA. Reverse genetics in the mosquito Anopheles gambiae: targeted disruption of the defensin gene. EMBO Rep. 2002;3((9)):852–6. doi: 10.1093/embo-reports/kvf180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hillyer JF, Schmidt SL, Christensen BM. Rapid phagocytosis and melanization of bacteria and Plasmodium sporozoites by hemocytes of the mosquito Aedes aegypti. J Parasitol. 2003;89((1)):62–9. doi: 10.1645/0022-3395(2003)089[0062:RPAMOB]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 11.Moita LF, Wang-Sattler R, Michel K, Zimmermann T, Blandin S, Levashina EA, et al. In vivo identification of novel regulators and conserved pathways of phagocytosis in A. gambiae. Immunity. 2005;23((1)):65–73. doi: 10.1016/j.immuni.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 12.Vodovar N, Acosta C, Lemaitre B, Boccard F. Drosophila: a polyvalent model to decipher host-pathogen interactions. Trends Microbiol. 2004;12((5)):235–42. doi: 10.1016/j.tim.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 13.Dudzic JP, Hanson MA, Iatsenko I, Kondo S, Lemaitre B. More than black or white: melanization and Toll share regulatory serine proteases in Drosophila. Cell Rep. 2019;27((4)):1050–61.e3. doi: 10.1016/j.celrep.2019.03.101. [DOI] [PubMed] [Google Scholar]
- 14.Ramirez JL, de Almeida Oliveira G, Calvo E, Dalli J, Colas RA, Serhan CN, et al. A mosquito lipoxin/lipocalin complex mediates innate immune priming in Anopheles gambiae. Nat Commun. 2015;6:7403. doi: 10.1038/ncomms8403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castillo JC, Ferreira ABB, Trisnadi N, Barillas-Mury C. Activation of mosquito complement antiplasmodial response requires cellular immunity. Sci Immunol. 2017;2((7)):eaal1505. doi: 10.1126/sciimmunol.aal1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dong Y, Aguilar R, Xi Z, Warr E, Mongin E, Dimopoulos G. Anopheles gambiae immune responses to human and rodent Plasmodium parasite species. PLoS Pathog. 2006;2((6)):e52. doi: 10.1371/journal.ppat.0020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong Y, Dimopoulos G. Anopheles fibrinogen-related proteins provide expanded pattern recognition capacity against bacteria and malaria parasites. J Biol Chem. 2009;284((15)):9835–44. doi: 10.1074/jbc.M807084200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong Y, Taylor HE, Dimopoulos G. AgDscam, a hypervariable immunoglobulin domain-containing receptor of the Anopheles gambiae innate immune system. PLoS Biol. 2006;4((7)):e229. doi: 10.1371/journal.pbio.0040229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levashina EA, Moita LF, Blandin S, Vriend G, Lagueux M, Kafatos FC. Conserved role of a complement-like protein in phagocytosis revealed by dsRNA knockout in cultured cells of the mosquito, Anopheles gambiae. Cell. 2001;104((5)):709–18. doi: 10.1016/s0092-8674(01)00267-7. [DOI] [PubMed] [Google Scholar]
- 20.Meister S, Agianian B, Turlure F, Relógio A, Morlais I, Kafatos FC, et al. Anopheles gambiae PGRPLC-mediated defense against bacteria modulates infections with malaria parasites. PLoS Pathog. 2009;5((8)):e1000542. doi: 10.1371/journal.ppat.1000542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schnitger AK, Yassine H, Kafatos FC, Osta MA. Two C-type lectins cooperate to defend Anopheles gambiae against Gram-negative bacteria. J Biol Chem. 2009;284((26)):17616–24. doi: 10.1074/jbc.M808298200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warr E, Das S, Dong Y, Dimopoulos G. The Gram-negative bacteria-binding protein gene family: its role in the innate immune system of Anopheles gambiae and in anti-Plasmodium defence. Insect Mol Biol. 2008;17((1)):39–51. doi: 10.1111/j.1365-2583.2008.00778.x. [DOI] [PubMed] [Google Scholar]
- 23.Sigle LT, Hillyer JF. Eater and draper are involved in the periostial haemocyte immune response in the mosquito Anopheles gambiae. Insect Mol Biol. 2018;27((4)):429–38. doi: 10.1111/imb.12383. [DOI] [PubMed] [Google Scholar]
- 24.Nehme NT, Liégeois S, Kele B, Giammarinaro P, Pradel E, Hoffmann JA, et al. A model of bacterial intestinal infections in Drosophila melanogaster. PLoS Pathog. 2007;3((11)):e173. doi: 10.1371/journal.ppat.0030173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grimont PA, Grimont F. The genus Serratia. Annu Rev Microbiol. 1978;32:221–48. doi: 10.1146/annurev.mi.32.100178.001253. [DOI] [PubMed] [Google Scholar]
- 26.Hejazi A, Keane CT, Falkiner FR. The use of RAPD-PCR as a typing method for Serratia marcescens. J Med Microbiol. 1997;46((11)):913–9. doi: 10.1099/00222615-46-11-913. [DOI] [PubMed] [Google Scholar]
- 27.Falkiner CL, Chauvet S, Andrès E, Aurouze M, Vallet I, Michel GP, et al. Virulence factors of the human opportunistic pathogen Serratia marcescens identified by in vivo screening. EMBO J. 2003;22((7)):1451–60. doi: 10.1093/emboj/cdg159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pradel E, Zhang Y, Pujol N, Matsuyama T, Bargmann CI, Ewbank JJ. Detection and avoidance of a natural product from the pathogenic bacterium Serratia marcescens by Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2007;104((7)):2295–300. doi: 10.1073/pnas.0610281104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stathopoulos S, Neafsey DE, Lawniczak MK, Muskavitch MA, Christophides GK. Genetic dissection of Anopheles gambiae gut epithelial responses to Serratia marcescens. PLoS Pathog. 2014;10((3)):e1003897. doi: 10.1371/journal.ppat.1003897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bahia AC, Dong Y, Blumberg BJ, Mlambo G, Tripathi A, BenMarzouk-Hidalgo OJ, et al. Exploring Anopheles gut bacteria for Plasmodium blocking activity. Environ Microbiol. 2014;16((9)):2980–94. doi: 10.1111/1462-2920.12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bando H, Okado K, Guelbeogo WM, Badolo A, Aonuma H, Nelson B, et al. Intra-specific diversity of Serratia marcescens in Anopheles mosquito midgut defines Plasmodium transmission capacity. Sci Rep. 2013;3:1641. doi: 10.1038/srep01641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tchioffo MT, Boissière A, Abate L, Nsango SE, Bayibéki AN, Awono-Ambéné PH, et al. Dynamics of bacterial community composition in the malaria mosquito's epithelia. Front Microbiol. 2015;6:1500. doi: 10.3389/fmicb.2015.01500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blandin S, Shiao SH, Moita LF, Janse CJ, Waters AP, Kafatos FC, et al. Complement-like protein TEP1 is a determinant of vectorial capacity in the malaria vector Anopheles gambiae. Cell. 2004;116((5)):661–70. doi: 10.1016/s0092-8674(04)00173-4. [DOI] [PubMed] [Google Scholar]
- 34.Joyner J, Wanless D, Sinigalliano CD, Lipp EK. Use of quantitative real-time PCR for direct detection of Serratia marcescens in marine and other aquatic environments. Appl Environ Microbiol. 2014;80((5)):1679–83. doi: 10.1128/AEM.02755-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14((4)):417–9. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15((12)):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11((2)):R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc, B. 1995;57((1)):289–300. [Google Scholar]
- 39.Wang S, Dos-Santos ALA, Huang W, Liu KC, Oshaghi MA, Wei G, et al. Driving mosquito refractoriness to Plasmodium falciparum with engineered symbiotic bacteria. Science. 2017;357((6358)):1399–402. doi: 10.1126/science.aan5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yassine H, Kamareddine L, Chamat S, Christophides GK, Osta MA. A serine protease homolog negatively regulates TEP1 consumption in systemic infections of the malaria vector Anopheles gambiae. J Innate Immun. 2014;6((6)):806–18. doi: 10.1159/000363296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saab SA, Dohna HZ, Nilsson LKJ, Onorati P, Nakhleh J, Terenius O, et al. The environment and species affect gut bacteria composition in laboratory co-cultured Anopheles gambiae and Aedes albopictus mosquitoes. Sci Rep. 2020;10((1)):3352. doi: 10.1038/s41598-020-60075-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ezraty B, Gennaris A, Barras F, Collet JF. Oxidative stress, protein damage and repair in bacteria. Nat Rev Microbiol. 2017;15((7)):385–96. doi: 10.1038/nrmicro.2017.26. [DOI] [PubMed] [Google Scholar]
- 43.Ha EM, Oh CT, Bae YS, Lee WJ. A direct role for dual oxidase in Drosophila gut immunity. Science. 2005;310((5749)):847–50. doi: 10.1126/science.1117311. [DOI] [PubMed] [Google Scholar]
- 44.Oliveira Gde A, Lieberman J, Barillas-Mury C. Epithelial nitration by a peroxidase/NOX5 system mediates mosquito antiplasmodial immunity. Science. 2012;335((6070)):856–9. doi: 10.1126/science.1209678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dong Y, Das S, Cirimotich C, Souza-Neto JA, McLean KJ, Dimopoulos G. Engineered Anopheles immunity to Plasmodium infection. PLoS Pathog. 2011;7((12)):e1002458. doi: 10.1371/journal.ppat.1002458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meister S, Kanzok SM, Zheng XL, Luna C, Li TR, Hoa NT, et al. Immune signaling pathways regulating bacterial and malaria parasite infection of the mosquito Anopheles gambiae. Proc Natl Acad Sci U S A. 2005;102((32)):11420–5. doi: 10.1073/pnas.0504950102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garver LS, Dong Y, Dimopoulos G. Caspar controls resistance to Plasmodium falciparum in diverse anopheline species. PLoS Pathog. 2009;5((3)):e1000335. doi: 10.1371/journal.ppat.1000335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mitri C, Jacques JC, Thiery I, Riehle MM, Xu J, Bischoff E, et al. Fine pathogen discrimination within the APL1 gene family protects Anopheles gambiae against human and rodent malaria species. PLoS Pathog. 2009;5((9)):e1000576. doi: 10.1371/journal.ppat.1000576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garver LS, Bahia AC, Das S, Souza-Neto JA, Shiao J, Dong Y, et al. Anopheles Imd pathway factors and effectors in infection intensity-dependent anti-Plasmodium action. PLoS Pathog. 2012;8((6)):e1002737. doi: 10.1371/journal.ppat.1002737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sandiford SL, Dong Y, Pike A, Blumberg BJ, Bahia AC, Dimopoulos G. Cytoplasmic actin is an extracellular insect immune factor which is secreted upon immune challenge and mediates phagocytosis and direct killing of bacteria, and is a Plasmodium antagonist. PLoS Pathog. 2015;11((2)):e1004631. doi: 10.1371/journal.ppat.1004631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kocks C, Cho JH, Nehme N, Ulvila J, Pearson AM, Meister M, et al. Eater, a transmembrane protein mediating phagocytosis of bacterial pathogens in Drosophila. Cell. 2005;123((2)):335–46. doi: 10.1016/j.cell.2005.08.034. [DOI] [PubMed] [Google Scholar]
- 52.Kurucz E, Márkus R, Zsámboki J, Folkl-Medzihradszky K, Darula Z, Vilmos P, et al. Nimrod, a putative phagocytosis receptor with EGF repeats in Drosophila plasmatocytes. Curr Biol. 2007;17((7)):649–54. doi: 10.1016/j.cub.2007.02.041. [DOI] [PubMed] [Google Scholar]
- 53.Melcarne C, Ramond E, Dudzic J, Bretscher AJ, Kurucz É, Andó I, et al. Two Nimrod receptors, NimC1 and Eater, synergistically contribute to bacterial phagocytosis in Drosophila melanogaster. FEBS J. 2019;286((14)):2670–91. doi: 10.1111/febs.14857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shiratsuchi A, Mori T, Sakurai K, Nagaosa K, Sekimizu K, Lee BL, et al. Independent recognition of Staphylococcus aureus by two receptors for phagocytosis in Drosophila. J Biol Chem. 2012;287((26)):21663–72. doi: 10.1074/jbc.M111.333807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lombardo F, Ghani Y, Kafatos FC, Christophides GK. Comprehensive genetic dissection of the hemocyte immune response in the malaria mosquito Anopheles gambiae. PLoS Pathog. 2013;9((1)):e1003145. doi: 10.1371/journal.ppat.1003145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Piper RC, Katzmann DJ. Biogenesis and function of multivesicular bodies. Annu Rev Cell Dev Biol. 2007;23:519–47. doi: 10.1146/annurev.cellbio.23.090506.123319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Golchin A, Hosseinzadeh S, Ardeshirylajimi A. The exosomes released from different cell types and their effects in wound healing. J Cell Biochem. 2018;119((7)):5043–52. doi: 10.1002/jcb.26706. [DOI] [PubMed] [Google Scholar]
- 58.McCarville JL, Ayres JS. Disease tolerance: concept and mechanisms. Curr Opin Immunol. 2018;50:88–93. doi: 10.1016/j.coi.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Troha K, Im JH, Revah J, Lazzaro BP, Buchon N. Comparative transcriptomics reveals CrebA as a novel regulator of infection tolerance in D. melanogaster. PLoS Pathog. 2018;14((2)):e1006847. doi: 10.1371/journal.ppat.1006847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ukleja M, Valpuesta JM, Dziembowski A, Cuellar J. Beyond the known functions of the CCR4-NOT complex in gene expression regulatory mechanisms: new structural insights to unravel CCR4-NOT mRNA processing machinery. Bioessays. 2016;38((10)):1048–58. doi: 10.1002/bies.201600092. [DOI] [PubMed] [Google Scholar]
- 61.Chen Y, Boland A, Kuzuoğlu-Öztürk D, Bawankar P, Loh B, Chang CT, et al. A DDX6-CNOT1 complex and W-binding pockets in CNOT9 reveal direct links between miRNA target recognition and silencing. Mol Cell. 2014;54((5)):737–50. doi: 10.1016/j.molcel.2014.03.034. [DOI] [PubMed] [Google Scholar]
- 62.Yamaguchi T, Suzuki T, Sato T, Takahashi A, Watanabe H, Kadowaki A, et al. The CCR4-NOT deadenylase complex controls Atg7-dependent cell death and heart function. Sci Signal. 2018;11((516)):eaan3638. doi: 10.1126/scisignal.aan3638. [DOI] [PubMed] [Google Scholar]
- 63.Morita M, Oike Y, Nagashima T, Kadomatsu T, Tabata M, Suzuki T, et al. Obesity resistance and increased hepatic expression of catabolism-related mRNAs in Cnot3+/− mice. EMBO J. 2011;30((22)):4678–91. doi: 10.1038/emboj.2011.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Paul S, Gable K, Dunn TM. A six-membrane-spanning topology for yeast and Arabidopsis Tsc13p, the enoyl reductases of the microsomal fatty acid elongating system. J Biol Chem. 2007;282((26)):19237–46. doi: 10.1074/jbc.M701774200. [DOI] [PubMed] [Google Scholar]
- 65.Kanost MR, Jiang H. Clip-domain serine proteases as immune factors in insect hemolymph. Curr Opin Insect Sci. 2015;11:47–55. doi: 10.1016/j.cois.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Veillard F, Troxler L, Reichhart JM. Drosophila melanogaster clip-domain serine proteases: structure, function and regulation. Biochimie. 2016;122:255–69. doi: 10.1016/j.biochi.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 67.Volz J, Müller HM, Zdanowicz A, Kafatos FC, Osta MA. A genetic module regulates the melanization response of Anopheles to Plasmodium. Cell Microbiol. 2006;8((9)):1392–405. doi: 10.1111/j.1462-5822.2006.00718.x. [DOI] [PubMed] [Google Scholar]
- 68.Sousa GL, Bishnoi R, Baxter RH, Povelones M. The CLIP-domain serine protease CLIPC9 regulates melanization downstream of SPCLIP1, CLIPA8, and CLIPA28 in the malaria vector Anopheles gambiae. BioRxiv. 2020 doi: 10.1371/journal.ppat.1008985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yassine H, Kamareddine L, Osta MA. The mosquito melanization response is implicated in defense against the entomopathogenic fungus Beauveria bassiana. PLoS Pathog. 2012;8((11)):e1003029. doi: 10.1371/journal.ppat.1003029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.El Moussawi L, Nakhleh J, Kamareddine L, Osta MA. The mosquito melanization response requires hierarchical activation of non-catalytic clip domain serine protease homologs. PLoS Pathog. 2019;15((11)):e1008194. doi: 10.1371/journal.ppat.1008194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schnitger AK, Kafatos FC, Osta MA. The melanization reaction is not required for survival of Anopheles gambiae mosquitoes after bacterial infections. J Biol Chem. 2007;282((30)):21884–8. doi: 10.1074/jbc.M701635200. [DOI] [PubMed] [Google Scholar]
- 72.Pinto SB, Lombardo F, Koutsos AC, Waterhouse RM, McKay K, An C, et al. Discovery of Plasmodium modulators by genome-wide analysis of circulating hemocytes in Anopheles gambiae. Proc Natl Acad Sci U S A. 2009;106((50)):21270–5. doi: 10.1073/pnas.0909463106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Severo MS, Landry JJM, Lindquist RL, Goosmann C, Brinkmann V, Collier P, et al. Unbiased classification of mosquito blood cells by single-cell genomics and high-content imaging. Proc Natl Acad Sci U S A. 2018;115((32)):E7568–77. doi: 10.1073/pnas.1803062115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Muller HM, Dimopoulos G, Blass C, Kafatos FC. A hemocyte-like cell line established from the malaria vector Anopheles gambiae expresses six prophenoloxidase genes. J Biol Chem. 1999;274((17)):11727–35. doi: 10.1074/jbc.274.17.11727. [DOI] [PubMed] [Google Scholar]
- 75.Barletta ABF, Trisnadi N, Ramirez JL, Barillas-Mury C. Mosquito midgut prostaglandin release establishes systemic immune priming. iScience. 2019;19:54–62. doi: 10.1016/j.isci.2019.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Becker T, Loch G, Beyer M, Zinke I, Aschenbrenner AC, Carrera P, et al. FOXO-dependent regulation of innate immune homeostasis. Nature. 2010;463((7279)):369–73. doi: 10.1038/nature08698. [DOI] [PubMed] [Google Scholar]
- 77.Fink C, Hoffmann J, Knop M, Li Y, Isermann K, Roeder T. Intestinal FoxO signaling is required to survive oral infection in Drosophila. Mucosal Immunol. 2016;9((4)):927–36. doi: 10.1038/mi.2015.112. [DOI] [PubMed] [Google Scholar]
- 78.Mabery EM, Schneider DS. The Drosophila TNF ortholog eiger is required in the fat body for a robust immune response. J Innate Immun. 2010;2((4)):371–8. doi: 10.1159/000315050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schneider DS, Ayres JS, Brandt SM, Costa A, Dionne MS, Gordon MD, et al. Drosophila eiger mutants are sensitive to extracellular pathogens. PLoS Pathog. 2007;3((3)):e41. doi: 10.1371/journal.ppat.0030041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Guillou A, Troha K, Wang H, Franc NC, Buchon N. The Drosophila CD36 Homologue croquemort is required to maintain immune and gut homeostasis during development and aging. PLoS Pathog. 2016;12((10)):e1005961. doi: 10.1371/journal.ppat.1005961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ramet M, Pearson A, Manfruelli P, Li X, Koziel H, Gobel V, et al. Drosophila scavenger receptor CI is a pattern recognition receptor for bacteria. Immunity. 2001;15((6)):1027–38. doi: 10.1016/s1074-7613(01)00249-7. [DOI] [PubMed] [Google Scholar]
- 82.Stuart LM, Deng J, Silver JM, Takahashi K, Tseng AA, Hennessy EJ, et al. Response to Staphylococcus aureus requires CD36-mediated phagocytosis triggered by the COOH-terminal cytoplasmic domain. J Cell Biol. 2005;170((3)):477–85. doi: 10.1083/jcb.200501113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Osta MA, Christophides GK, Kafatos FC. Effects of mosquito genes on Plasmodium development. Science. 2004;303((5666)):2030–2. doi: 10.1126/science.1091789. [DOI] [PubMed] [Google Scholar]
- 84.Povelones M, Upton LM, Sala KA, Christophides GK. Structure-function analysis of the Anopheles gambiae LRIM1/APL1C complex and its interaction with complement C3-like protein TEP1. PLoS Pathog. 2011;7((4)):e1002023. doi: 10.1371/journal.ppat.1002023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Simoes ML, Mlambo G, Tripathi A, Dong Y, Dimopoulos G. Immune regulation of Plasmodium is Anopheles species specific and infection intensity dependent. MBio. 2017;8((5)) doi: 10.1128/mBio.01631-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Martins NE, Faria VG, Teixeira L, Magalhães S, Sucena É. Host adaptation is contingent upon the infection route taken by pathogens. PLoS Pathog. 2013;9((9)):e1003601. doi: 10.1371/journal.ppat.1003601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mondotte JA, Gausson V, Frangeul L, Blanc H, Lambrechts L, Saleh MC. Immune priming and clearance of orally acquired RNA viruses in Drosophila. Nat Microbiol. 2018;3((12)):1394–403. doi: 10.1038/s41564-018-0265-9. [DOI] [PubMed] [Google Scholar]
- 88.Carissimo G, Pain A, Belda E, Vernick KD. Highly focused transcriptional response of Anopheles coluzzii to O'nyong nyong arbovirus during the primary midgut infection. BMC Genomics. 2018;19((1)):526. doi: 10.1186/s12864-018-4918-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hughes LJ, Townsend MB, Gallardo-Romero N, Hutson CL, Patel N, Doty JB, et al. Magnitude and diversity of immune response to vaccinia virus is dependent on route of administration. Virology. 2020;544:55–63. doi: 10.1016/j.virol.2020.02.001. [DOI] [PubMed] [Google Scholar]
- 90.Demars A, Lison A, Machelart A, Van Vyve M, Potemberg G, Vanderwinden JM, et al. Route of infection strongly impacts the host-pathogen relationship. Front Immunol. 2019;10:1589. doi: 10.3389/fimmu.2019.01589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wong AC, Wang QP, Morimoto J, Senior AM, Lihoreau M, Neely GG, et al. Gut microbiota modifies olfactory-guided microbial preferences and foraging decisions in Drosophila. Curr Biol. 2017;27((15)):2397–404.e4. doi: 10.1016/j.cub.2017.07.022. [DOI] [PubMed] [Google Scholar]
- 92.Rodgers FH, Gendrin M, Wyer CAS, Christophides GK. Microbiota-induced peritrophic matrix regulates midgut homeostasis and prevents systemic infection of malaria vector mosquitoes. PLoS Pathog. 2017;13((5)):e1006391. doi: 10.1371/journal.ppat.1006391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kumar S, Molina-Cruz A, Gupta L, Rodrigues J, Barillas-Mury C. A peroxidase/dual oxidase system modulates midgut epithelial immunity in Anopheles gambiae. Science. 2010;327((5973)):1644–8. doi: 10.1126/science.1184008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nakhleh J, Christophides GK, Osta MA. The serine protease homolog CLIPA14 modulates the intensity of the immune response in the mosquito Anopheles gambiae. J Biol Chem. 2017;292((44)):18217–26. doi: 10.1074/jbc.M117.797787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Povelones M, Bhagavatula L, Yassine H, Tan LA, Upton LM, Osta MA, et al. The CLIP-domain serine protease homolog SPCLIP1 regulates complement recruitment to microbial surfaces in the malaria mosquito Anopheles gambiae. PLoS Pathog. 2013;9((9)):e1003623. doi: 10.1371/journal.ppat.1003623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Choe KM, Lee H, Anderson KV. Drosophila peptidoglycan recognition protein LC (PGRP-LC) acts as a signal-transducing innate immune receptor. Proc Natl Acad Sci U S A. 2005;102((4)):1122–6. doi: 10.1073/pnas.0404952102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Choe KM, Werner T, Stöven S, Hultmark D, Anderson KV. Requirement for a peptidoglycan recognition protein (PGRP) in relish activation and antibacterial immune responses in Drosophila. Science. 2002;296((5566)):359–62. doi: 10.1126/science.1070216. [DOI] [PubMed] [Google Scholar]
- 98.Iatsenko I, Kondo S, Mengin-Lecreulx D, Lemaitre B. PGRP-SD, an extracellular pattern-recognition receptor, enhances peptidoglycan-mediated activation of the Drosophila Imd pathway. Immunity. 2016;45((5)):1013–23. doi: 10.1016/j.immuni.2016.10.029. [DOI] [PubMed] [Google Scholar]
- 99.Cole JN, Nizet V. Bacterial evasion of host antimicrobial peptide defenses. Microbiol Spectr. 2016;4((1)):1–40. doi: 10.1128/microbiolspec.VMBF-0006-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dong Y, Manfredini F, Dimopoulos G. Implication of the mosquito midgut microbiota in the defense against malaria parasites. PLoS Pathog. 2009;5((5)):e1000423. doi: 10.1371/journal.ppat.1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Buchon N, Broderick NA, Poidevin M, Pradervand S, Lemaitre B. Drosophila intestinal response to bacterial infection: activation of host defense and stem cell proliferation. Cell Host Microbe. 2009;5((2)):200–11. doi: 10.1016/j.chom.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 102.Chakrabarti S, Liehl P, Buchon N, Lemaitre B. Infection-induced host translational blockage inhibits immune responses and epithelial renewal in the Drosophila gut. Cell Host Microbe. 2012;12((1)):60–70. doi: 10.1016/j.chom.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 103.Becher PG, Flick G, Rozpędowska E, Schmidt A, Hagman A, Lebreton S, et al. Yeast, not fruit volatiles mediate Drosophila melanogaster attraction, oviposition and development. Funct Ecol. 2012;26((4)):822–8. [Google Scholar]
- 104.Zhu J, Park KC, Baker TC. Identification of odors from overripe mango that attract vinegar flies, Drosophila melanogaster. J Chem Ecol. 2003;29((4)):899–909. doi: 10.1023/a:1022931816351. [DOI] [PubMed] [Google Scholar]
- 105.Ayyaz A, Li H, Jasper H. Haemocytes control stem cell activity in the Drosophila intestine. Nat Cell Biol. 2015;17((6)):736–48. doi: 10.1038/ncb3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chakrabarti S, Dudzic JP, Li X, Collas EJ, Boquete JP, Lemaitre B. Remote control of intestinal stem cell activity by haemocytes in Drosophila. PLoS Genet. 2016;12((5)):e1006089. doi: 10.1371/journal.pgen.1006089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Scopelliti A, Bauer C, Yu Y, Zhang T, Kruspig B, Murphy DJ, et al. A neuronal relay mediates a nutrient responsive gut/fat body axis regulating energy homeostasis in adult Drosophila. Cell Metab. 2019;29((2)):269–e10. doi: 10.1016/j.cmet.2018.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Song W, Cheng D, Hong S, Sappe B, Hu Y, Wei N, et al. Midgut-derived activin regulates glucagon-like action in the fat body and glycemic control. Cell Metab. 2017;25((2)):386–99. doi: 10.1016/j.cmet.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lee KZ, Lestradet M, Socha C, Schirmeier S, Schmitz A, Spenlé C, et al. Enterocyte purge and rapid recovery is a resilience reaction of the gut epithelium to pore-forming toxin attack. Cell Host Microbe. 2016;20((6)):716–30. doi: 10.1016/j.chom.2016.10.010. [DOI] [PubMed] [Google Scholar]
- 110.Dimopoulos G, Christophides GK, Meister S, Schultz J, White KP, Barillas-Mury C, et al. Genome expression analysis of Anopheles gambiae: responses to injury, bacterial challenge, and malaria infection. Proc Natl Acad Sci U S A. 2002;99((13)):8814–9. doi: 10.1073/pnas.092274999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nsango SE, Abate L, Thoma M, Pompon J, Fraiture M, Rademacher A, et al. Genetic clonality of Plasmodium falciparum affects the outcome of infection in Anopheles gambiae. Int J Parasitol. 2012;42((6)):589–95. doi: 10.1016/j.ijpara.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 112.Nsango SE, Pompon J, Xie T, Rademacher A, Fraiture M, Thoma M, et al. AP-1/Fos-TGase2 axis mediates wounding-induced Plasmodium falciparum killing in Anopheles gambiae. J Biol Chem. 2013;288((22)):16145–54. doi: 10.1074/jbc.M112.443267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Krautz R, Arefin B, Theopold U. Damage signals in the insect immune response. Front Plant Sci. 2014;5:342. doi: 10.3389/fpls.2014.00342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bidla G, Lindgren M, Theopold U, Dushay MS. Hemolymph coagulation and phenoloxidase in Drosophila larvae. Dev Comp Immunol. 2005;29((8)):669–79. doi: 10.1016/j.dci.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 115.Franz A, Wood W, Martin P. Fat body cells are motile and actively migrate to wounds to drive repair and prevent infection. Dev Cell. 2018;44((4)):460–70.e3. doi: 10.1016/j.devcel.2018.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Galko MJ, Krasnow MA. Cellular and genetic analysis of wound healing in Drosophila larvae. PLoS Biol. 2004;2((8)):E239. doi: 10.1371/journal.pbio.0020239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Goto A, Kadowaki T, Kitagawa Y. Drosophila hemolectin gene is expressed in embryonic and larval hemocytes and its knock down causes bleeding defects. Dev Biol. 2003;264((2)):582–91. doi: 10.1016/j.ydbio.2003.06.001. [DOI] [PubMed] [Google Scholar]
- 118.Karlsson C, Korayem AM, Scherfer C, Loseva O, Dushay MS, Theopold U. Proteomic analysis of the Drosophila larval hemolymph clot. J Biol Chem. 2004;279((50)):52033–41. doi: 10.1074/jbc.M408220200. [DOI] [PubMed] [Google Scholar]
- 119.Lesch C, Goto A, Lindgren M, Bidla G, Dushay MS, Theopold U. A role for hemolectin in coagulation and immunity in Drosophila melanogaster. Dev Comp Immunol. 2007;31((12)):1255–63. doi: 10.1016/j.dci.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 120.Losick VP, Fox DT, Spradling AC. Polyploidization and cell fusion contribute to wound healing in the adult Drosophila epithelium. Curr Biol. 2013;23((22)):2224–32. doi: 10.1016/j.cub.2013.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Scherfer C, Karlsson C, Loseva O, Bidla G, Goto A, Havemann J, et al. Isolation and characterization of hemolymph clotting factors in Drosophila melanogaster by a pullout method. Curr Biol. 2004;14((7)):625–9. doi: 10.1016/j.cub.2004.03.030. [DOI] [PubMed] [Google Scholar]
- 122.Scherfer C, Qazi MR, Takahashi K, Ueda R, Dushay MS, Theopold U, et al. The Toll immune-regulated Drosophila protein Fondue is involved in hemolymph clotting and puparium formation. Dev Biol. 2006;295((1)):156–63. doi: 10.1016/j.ydbio.2006.03.019. [DOI] [PubMed] [Google Scholar]
- 123.Jung J, Zeng H, Horng T. Metabolism as a guiding force for immunity. Nat Cell Biol. 2019;21((1)):85–93. doi: 10.1038/s41556-018-0217-x. [DOI] [PubMed] [Google Scholar]
- 124.Wang A, Luan HH, Medzhitov R. An evolutionary perspective on immunometabolism. Science. 2019;363((6423)):eaar3932. doi: 10.1126/science.aar3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.O'Neill LA, Pearce EJ. Immunometabolism governs dendritic cell and macrophage function. J Exp Med. 2016;213((1)):15–23. doi: 10.1084/jem.20151570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Pietri JE, Pakpour N, Napoli E, Song G, Pietri E, Potts R, et al. Two insulin-like peptides differentially regulate malaria parasite infection in the mosquito through effects on intermediary metabolism. Biochem J. 2016;473((20)):3487–503. doi: 10.1042/BCJ20160271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pakpour N, Corby-Harris V, Green GP, Smithers HM, Cheung KW, Riehle MA, et al. Ingested human insulin inhibits the mosquito NF-κB-dependent immune response to Plasmodium falciparum. Infect Immun. 2012;80((6)):2141–9. doi: 10.1128/IAI.00024-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Musselman LP, Fink JL, Grant AR, Gatto JA, Tuthill BF, 2nd, Baranski TJ. A complex relationship between immunity and metabolism in Drosophila diet-induced insulin resistance. Mol Cell Biol. 2018;38((2)):e00259-17. doi: 10.1128/MCB.00259-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cheon HM, Shin SW, Bian G, Park JH, Raikhel AS. Regulation of lipid metabolism genes, lipid carrier protein lipophorin, and its receptor during immune challenge in the mosquito Aedes aegypti. J Biol Chem. 2006;281((13)):8426–35. doi: 10.1074/jbc.M510957200. [DOI] [PubMed] [Google Scholar]
- 130.Kamareddine L, Nakhleh J, Osta MA. Functional interaction between apolipophorins and complement regulate the mosquito immune response to systemic infections. J Innate Immun. 2016;8((3)):314–26. doi: 10.1159/000443883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Oliveira JH, Gonçalves RL, Oliveira GA, Oliveira PL, Oliveira MF, Barillas-Mury C. Energy metabolism affects susceptibility of Anopheles gambiae mosquitoes to Plasmodium infection. Insect Biochem Mol Biol. 2011;41((6)):349–55. doi: 10.1016/j.ibmb.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lampe L, Jentzsch M, Kierszniowska S, Levashina EA. Metabolic balancing by miR-276 shapes the mosquito reproductive cycle and Plasmodium falciparum development. Nat Commun. 2019;10((1)):5634. doi: 10.1038/s41467-019-13627-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Eming SA, Wynn TA, Martin P. Inflammation and metabolism in tissue repair and regeneration. Science. 2017;356((6342)):1026–30. doi: 10.1126/science.aam7928. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data