Abstract
Purpose:
Hexokinase II (HK2) protein expression is elevated in glioblastoma (GBM), and we have shown that HK2 could serve as an effective therapeutic target for GBM. Here, we interrogated compounds that target HK2 effectively and restrict tumor growth in cell lines, patient-derived glioma stem cells (GSCs), and mouse models of GBM.
Experimental Design:
We performed a screen using a set of 15 drugs that were predicted to inhibit the HK2-associated gene signature. We next determined the EC50 of the compounds by treating glioma cell lines and GSCs. Selected compounds showing significant impact in vitro were used to treat mice and examine their effect on survival and tumor characteristics. The effect of compounds on the metabolic activity in glioma cells was also assessed in vitro.
Results:
This screen identified the azole class of antifungals as inhibitors of tumor metabolism. Among the compounds tested, ketoconazole and posaconazole displayed the greatest inhibitory effect on GBM both in vitro and in vivo. Treatment of mice bearing GBM with ketoconazole and posaconazole increased their survival, reduced tumor cell proliferation, and decreased tumor metabolism. In addition, treatment with azoles resulted in increased proportion of apoptotic cells.
Conclusions:
Overall, we provide evidence that azoles exert their effect by targeting genes and pathways regulated by HK2. These findings shed light on the action of azoles in GBM. Combined with existing literature and preclinical results, these data support the value of repurposing azoles in GBM clinical trials.
Introduction
Cancer cells exhibit a key distinguishing feature compared with normal cells in how they generate ATP. Cancer cells generate more than 50% of their ATP from glycolysis rather than oxidative phosphorylation (OXPHOS), even under high oxygen conditions. This process is referred to as the “Warburg effect” or aerobic glycolysis (1). This metabolic remodeling seen in cancer cells results in alteration of synthesis and utilization of metabolites. It was originally thought that solid tumors benefit from the Warburg effect through enhanced glycolytic flux and faster ATP production per glucose molecule, ensuring sufficient supply of ATP for rapidly proliferating tumor cells, in particular under hypoxic conditions (2). However, metabolic reprogramming is utilized even under sufficient supply of oxygen and nutrients, suggesting that there are other advantages of metabolic reprogramming for cancer cell survival (1).
Glioblastoma (GBM) is the most common primary brain tumor in adults, displaying high therapeutic resistance and poor median survival of 12–18 months (3). To date, there has been no advance in targeted therapies for GBMs. Our group and others have shown that metabolic reprogramming plays an important role in tumor progression in GBM. We have shown that the glycolytic enzyme, hexokinase II (HK2), plays an important role in glucose flux into glycolysis or the pentose phosphate pathway (PPP) and its transcription is tightly regulated in part by HIF1α, glucose, cAMP, insulin, glucagon, and p53 (4). Specifically, we have also shown that downregulation of HK2 inhibits aerobic glycolysis, while promoting oxidative phosphorylation (OXPHOS), as demonstrated by increased O2 consumption, decreased extracellular lactate, and increased expression of genes involved in OXPHOS (5). Furthermore, we have shown that decreased HK2 expression results in sensitization of tumor cells to radiation and chemotherapy, concomitant with increased survival of GBM mouse models. We have proven that the function of HK2 is independent of its isoform HK1 (5). Collectively, this evidence supports that HK2 is a key driver of metabolic regulation in GBMs as it regulates tumor growth and proliferation and is in part responsible for resistance to chemoradiation.
Currently, no direct inhibitors of HK2 have been tested in human patients with GBM due to the inability of the drugs to cross the blood–brain barrier (BBB) or concern for their toxicity in patients. For example, 3-bromopyruvate (3-BrPA) is a glycolytic inhibitor that targets energy metabolism (6) and is an inhibitor of hexokinase (7, 8). Preclinical reports have demonstrated that 3-BrPA inhibits tumor growth upon intra-arterial or intratumoral administration. However, tissue autoradiography of rats infused with 14C-3-BrPA showed strong 14C signal in various organs except in the brain, suggesting that 3-BrPA does not cross the BBB (7). In addition, because of the highly reactive alkylating nature of 3-BrPA, its inhibitory effect is not only restricted to HK2 and is likely to be the reason for its toxic systemic effects.
Another widely studied hexokinase inhibitor is 2-deoxy-D-glucose (2-DG) that inhibits hexokinase by competing with glucose and accumulates inside the cells, which leads to inhibition of the glycolytic flux, depletion of ATP, cell-cycle arrest, and ultimately, cell death (9). Treatment with 2-DG in combination with 2 chemotherapeutic drugs, adriamycin and paclitaxel, proved to be effective in vivo by increasing the cytotoxic effects in xenograft mouse models of human osteosarcoma and non–small cell lung cancer (10). Treatment with 2-DG resulted in sensitization of GBM cells to treatment with histone deacetylase (HDAC) inhibitors (10). However, administration of 2-DG to patients with GBM at doses that were sufficient to limit glucose metabolism in cancer cells demonstrated signs of toxicity to the brain (11).
Given the limited progress in the field of therapeutics for GBM, there is a pressing need to expand the available treatment options through translation of preclinical discoveries to clinical trials. One possible means to expedite initiation of GBM clinical trials is through drug repurposing, by examining previously established drugs with a known track record of safety in humans. We hypothesized that identifying gene networks associated with HK2 and their selective targeting can reveal novel therapeutic options for GBM by inhibiting glycolysis, promoting OXPHOS, and reducing proliferation. Using a systems biology approach, we established the gene signature and pathways that are affected upon downregulation of HK2 to identify novel therapeutic vulner-abilities. Using a drug screen targeting potential HK2-regulated gene expression networks, we identified the azole class of antifungals as inhibitors. We examined the effect of azoles in vitro and in vivo on GBM metabolism and their potency as inhibitors of tumor growth.
Materials and Methods
Cell culture
Fetal normal human astrocytes (NHAs) immortalized with hTERT were purchased from ABM. U87, T98G, U251, and U118 cell lines were purchased from ATCC. Cells were grown in DMEM and 10% FBS. Normal neural stem cells (NSCs) were purchased from Thermo Fisher Scientific. Glioma stem cells (GSC; GSC8–18, GSC7–2, GBM8, and GSC30) were derived from human operative samples as described previously and cultured in GSC media (12). All cells were subjected to short tandem repeat (STR) profiling and confirmed Mycoplasma negative by PCR testing.
Viability assays and EC50 analysis
Cell viability assays were performed using the CellTiter-Glo Luminescent Cell Viability Assay (Promega) as per manufacturer’s protocol. Raw luminescent values were inputted into PRISM 7.0. Drug concentrations were log10 transformed and values normalized to percent viability with respect to vehicle-treated cells. The EC50values for each chemical were calculated using a log inhibitor versus normalized response with variable slope using a least-squares fit model.
HK2 knockdown
U87, T98G, GSC30, and GSC7–2 cells were treated with pooled HK2 siRNA (FlexiTube siRNA SI00004060, Qiagen) or AllStars negative siRNA control (SI03650318, Qiagen), together with the Qiagen siRNA transfection reagent (HiPerFect Transfection Reagent catalog No. 301705, Qiagen) as per manufacturer’s instructions. Final concentration of siRNA in all conditions was 5 nmol/L. HK2 siRNA-treated or scramble siRNA control–treated cells were collected for Western blot analysis (see below) or RNA isolation using the Qiagen RNeasy Extraction Kit (Qiagen, catalog No. 74104).
Microarray and pathway analysis
RNA was extracted from U87 and GSC30 cells treated with siRNA pool against HK2 or control siRNA using the RNeasy Mini Kit (Qiagen, catalog No. 74104) following the manufacturer’s protocol. Biotinylated cDNA were prepared according to the standard Affymetrix protocol from 10 μg total RNA (Expression Analysis Technical Manual, 2001, Affymetrix). Following fragmentation, 10 μg of cRNA was hybridized for 16 hours at 45°C on Human Gene 2.0 ST Array. GeneChips were washed and stained in the Affymetrix Fluidics Station 400. GeneChips were then scanned at The Centre for Applied Genomics (TCAG), SickKids Hospital Toronto, Canada. The data were normalized with Affymetrix Expression Console Software using RMA analysis and log2 transformed. In summary, we collected RNA from biological duplicates of U87 siRNA control, U87 HK2 siRNA, GSC30 siRNA control, and GSC30 HK2 siRNA. Differentially expressed genes were then identified through comparison of gene expression profiles in HK2 knockdown cells with siRNA control cells. Significance was estimated using the Wilcoxon rank-sum test, with a Benjamini–Hochberg corrected P <0.05 and a minimum fold change of 1.5. Pathway analysis was performed using Gene Set Enrichment Analysis (GSEA v3.0 under JAVA 8.0 environment) from Broad Institute (Cambridge, MA). In short, a rank file (.rnk) was generated from P values and fold change from microarray analysis comparing control siRNA with HK2 siRNA-treated samples. The rank file was analyzed by GSEA preranked method using gene sets (Human_GOBP_All-Pathways_no_GO_iea_March _01_2018_symbol.gmt) compiled by Bader Lab (http://download.baderlab.org/EM_Genesets/March_01_2018/Human/symbol/Human_GOBP_AllPathways_no_GO_iea_March_01_2018_symbol.gmt) and parameters set to 2,000 gene-set permutations, gene sets size between 10 and 200. An enrichment map (version 3.0.0 of Enrichment Map software) was generated in Cytoscape (version 3.6.0) using enriched gene sets with FDR < 0.01 and the Jacaard coefficient set to 0.25 for connecting edges. The pathways were then clustered by the Markov Cluster Algorithm in the AutoAnnotate APP (v1.2) in Cytoscape.
Connectivity map analysis
Top 200 upregulated and top 200 downregulated genes from our HK2 siRNA microarray experiment were used to query connectivity map (C-Map) database (build02; https://portals.broadinstitute.org/cmap/; ref. 13). Compounds with a negative enrichment score and a P value less than 0.05 were recorded as potential therapeutic agents for cells expressing high HK2. Please refer to Supplementary Table S2 for the list of compounds used in the study and purchasing information.
Yeast fitness defect drug screen
We performed a yeast fitness defect drug screen on both homozygous and heterozygous HXK2 deletion strains to examine the sensitivity of yeast lacking the equivalent to human HK2 gene to our hit compounds following the method described previously (14).
Western blotting
Cell pellets were lysed in PLC lysis buffer containing protease and phosphatase inhibitors (Roche Inc.). Protein lysates were quantified using the bicinchoninic acid (BCA) assay (Pierce Chemical Co.). Thirty micrograms of total cell lysate protein was resolved on 10% SDS-PAGE gels and subjected to electrophoresis. Proteins were transferred to polyvinylidene difluoride membranes using a semi-dry transfer apparatus (Bio-Rad). Membranes were blocked in 5% BSA in PBS-T for 1 hour and probed for various proteins overnight in 5% nonfat milk or 5% BSA in TBS, 0.5% Tween-20 (TBS-T) or PBS, and 0.5% Tween-20 (PBS-T). Membranes were next washed 3 times for 5 minutes in TBS-T and incubated with horseradish peroxidase–conjugated secondary antibodies specific for the primary antibodies (Bio-Rad Laboratories, Inc.). Binding was detected using Chemiluminescence Reagent Plus (PerkinElmer Inc.). Antibodies were used at the following dilutions: Hexokinase I (1:1,000; Cell Signaling Technology; catalog No. C35C4), and HK2 (1:1,000; Cell Signaling Technology; catalog No. 2106).
Lactate production assay and hexokinase activity assay
Equal numbers of cells were plated in 96-well plates and the media were collected at 24- or 48-hour time points to analyze lactate concentration in triplicates using a colorimetric kit following the manufacturer’s instructions (catalog No. 120001100A; Eton Bioscience Inc.). The absorbance was estimated at 490 nm using a plate reader. Hexokinase activity assay kit was purchased from BioVision (catalog No. K789–100), and the assay was performed following manufacturer’s instructions.
13C glucose labeling, metabolite extraction, and gas chromatography/mass spectrometry measurements
For 13C labeling experiments, semiconfluent cells in a 10-cm dish were cultured in glucose-free DMEM (Sigma) with the compounds listed in Supplementary Table S2 and a 25 mmol/L 1:1 mixture of (U-13C6)glucose and (1-13C])glucose (Cambridge Isotope Laboratories) overnight. Cells were quenched with 1 mL ice-cold methanol and collected with a cell scraper. Four volumes of chloroform were added, and the cells were lightly vortexed and chilled in ice for 30 minutes for deproteinization. After centrifugation, the aqueous phase was collected and evaporated at room temperature. Dried polar metabolites were dissolved in 50 μL of 2% methoxyamine hydrochloride in pyridine (Pierce), sonicated for 30 minutes, and held at 37°C for 3 hours. After dissolution and reaction, 90-μL MBTSTFA + 1% TBDMCS (Pierce) was added and samples were incubated at 55°C for 1 hour. Gas chromatography/mass spectrometry (GC/MS) analysis was performed using an Agilent 6890 GC equipped with a 30-m DB-35MS capillary column connected to an Agilent 5975B MS operating under electron impact (EI) ionization at 70 eV. The identity and values of metabolites in glycolysis and OXPHOS were quantified and measured at the SickKids SPARC BioCentre in Toronto, Canada.
Animal xenograft studies
All animal procedures were carried out according to animal use protocols approved ethically by Institutional Animal Care Committee under the guidelines of the Canadian Council on Animal Care. Twenty 4- to 6-week-old NOD-SCID spontaneous male mice (NOD-SCID-Prkdcscid) were randomly separated into 2 groups and received intracranial injections of 5 × 104 GSC 8–18 cells or 1 × 105 GSC 7–2 or U87 cells. Cells were resuspended in 10 μL of PBS. At 10 days postinjection, the mice were randomized into vehicle, ketoconazole, or posaconazole treatments (25 mg/kg). Mice were sacrificed at signs of neurologic duress, followed by extraction of their brains and fixation in 4% paraformaldehyde.
IHC
Paraffin-embedded blocks were cut into 5-μm sections. Slides were processed as follows: dewaxed in xylene followed by rehydration in a standard alcohol series. Antigen retrieval was performed by pressure cooking for 20 minutes in citrate buffer (pH 6.0), followed by blocking of endogenous peroxidase activity in 3% H2O2. The primary antibodies were added and incubated overnight at 4°C. Antibodies were detected using a secondary HRP–labeled mouse or rabbit antibody detection system (Dako EnVision+ System-HRP catalog No. k4401, catalog No. k4403) followed by addition of 3,3′-diaminobenzidine (DAB) chromogen (Vector Laboratories) for visualization. Sections were counterstained with hematoxylin (Thermo Fisher Scientific Inc.) and slides were dehydrated sequentially in 70%, 80%, and 100% ethanol, followed by xylene. Slides were coverslipped and mounted in Permount (Thermo Fisher Scientific Inc.). Ki67 antibody (Dako, catalog No. F7268) was used at a 1:100 dilution. HK2 rabbit mAb (2876, Cell Signaling Technology) was used at a 1:300 dilution. TUNEL assay was performed using the DeadEnd Colorimetric Apoptosis Detection System (Promega). All images were captured on an Olympus IX73 fluorescent microscope system and analyzed using CellSens Dimension software.
Azole measurements in vivo
Posaconazole concentrations were determined by a high-performance liquid chromatography (HPLC)/MS-MS method. Briefly, posaconazole in mouse serum or xenograft tissue was extracted with methyl tert-butyl ether and the supernatant was dried. The extracts were reconstituted in methanol/water (4:1). The HPLC system consisted of a Shimadzu LC-20AD pump and a Shimadzu LC-20 AC autosampler (Shimadzu Corporation). The column used was a Phenomex hyperclone BDS C18 analytic column (50 × 2.0 mm, 5 μm). The binary mobile phase consisted of mobile phase A: 0.1% HCOOH in water mobile phase: B: 0.1% HCOOH in acetonitrile. The gradient conditions for the mobile phase are as follows: 0.0–1.0 minutes, 40%–100% B; 1.0–3.0 minutes, 100% B; 3.0–3.3 minutes, 100%–40% B; 3.3–6.3 minutes, 40% B. The flow rate was 0.5 mL/minute. The HPLC system was interfaced to an MDS Sciex triple quadrupole mass spectrometer (API 3200; Applied Biosystems) operating in the positive electrospray ionization mode. For multiple-reaction monitoring, the transitions monitored were m/z 701.5 to 683.5 for posaconazole. Data collection, peak integration, and calculation were performed using Applied Biosystems MDS Analyst 1.4.2 software.
Statistical analysis
Significance of alterations in mRNA expression from microarray data was estimated using the Wilcoxon rank-sum test, with a Benjamini–Hochberg corrected value of P < 0.05 and a minimum fold change of 1.5. In vitro experiments were performed in triplicates. Means and SDs or SEMs were computed. Student t test was used for pairwise comparison. Significance was defined at *, P < 0.05. Asterisks on graphs denote a significant difference (*, P < 0.05;**, P < 0.01;***, P < 0.001). In vivo survival was calculated using Kaplan–Meier analysis in Prism 6 (GraphPad), and statistical significance was measured using log-rank (Mantel–Cox) test.
Results
HK2 loss leads to distinct gene signature alterations
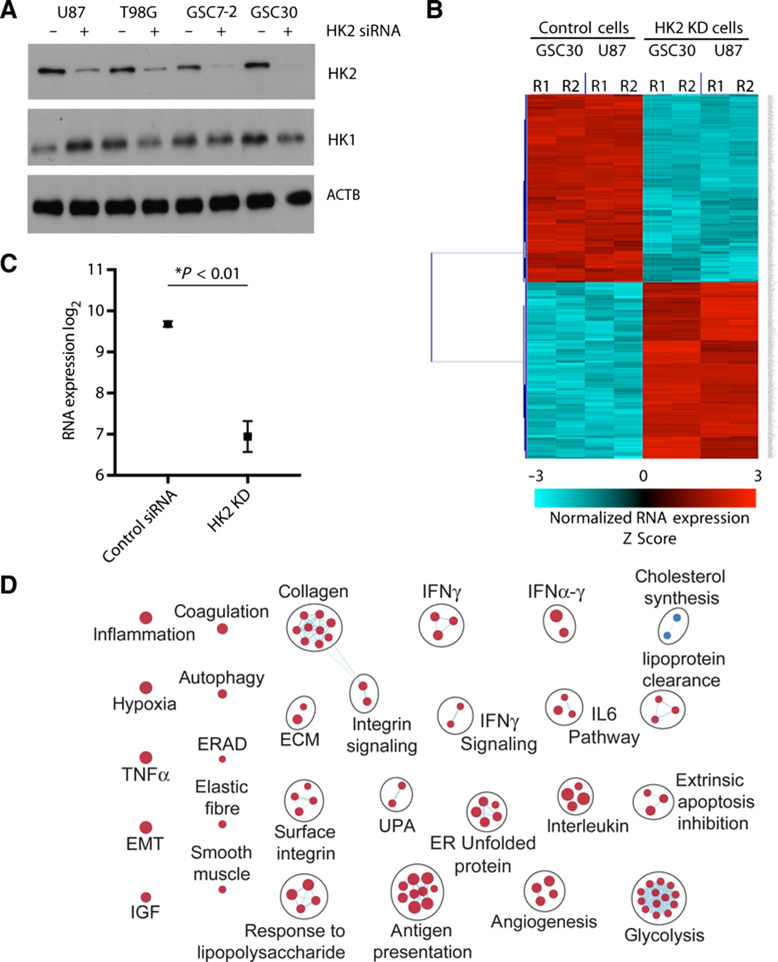
We have previously shown that loss of HK2 results in reduced GBM growth and increased sensitivity to radiation and alkylating therapies (5). On the basis of this, we postulated that genes and signaling pathways associated with HK2 knockdown would identify promising and targetable signaling pathways in GBM. To establish transcriptomic changes associated with HK2 expression, we knocked down HK2 expression using pooled siRNAs in GBM cell line U87, together with patient-derived GSCs (GSC30; Fig. 1A). Using Affymetrix Gene Expression 1.0 ST arrays, we compared the gene expression profiles of GBM cells treated with control siRNA with HK2-specific siRNAs. We next performed a Wilcoxon rank-sum test and identified 1,087 differentially expressed genes between control siRNA– and HK2 siRNA–treated cells (Fig. 1B). As expected, microarray results suggested a decrease in HK2 mRNA level that was further confirmed by RT-PCR analysis showing a 7-fold decrease in HK2 mRNA in 4 HK2 siRNA–treated cell lines relative to controls (Fig. 1C, corrected P < 0.05). Genes significantly downregulated with HK2 knockdown included well-characterized GBM relevant genes including MET, VEGFA, and VEGFC, whereas genes significantly upregulated with HK2 loss included genes typically associated with GBM stemness (SOX2, MEOX2) and the ERBB3 and ERBB4 receptors (data not shown). We next performed GSEA based on the list of transcripts downregulated or upregulated with loss of HK2 to identify molecular pathways associated with HK2 knockdown. An enrichment map was generated in Cytoscape and the pathways were clustered using the Markov Cluster Algorithm (Fig. 1D). Most notably the main cellular processes that were altered by knockdown of HK2 included glycolysis and angiogenesis. We also performed Ingenuity Pathway Analysis (IPA version 01–12) using our preranked list of genes that were differentially expressed. We identified the top ten canonical signaling pathways for both cell types and reported the negative log of the FDR. Several oncogenic signaling pathways were downregulated upon reduction of HK2 including the EGFR, KRAS, and MEK signaling pathways (Supplementary Fig. S1A). Oncogenic pathways associated with genes upregulated with HK2 silencing included Hedgehog, mTOR, and NOTCH signaling pathways (Supplementary Fig. S1B). These data demonstrated an intimate association between HK2 and regulation of key oncogenic pathways in GBMs.
Figure 1.
HK2 loss results in suppression of several oncogenic signaling pathways. A, Western blot analysis was performed 72 hours post-siRNA transfection to confirm HK2 knockdown in multiple GBM cell lines and GSC. Western blot analysis on HK1 was performed as a control to confirm HK2 siRNA specificity. B, Hierarchical clustering of 1,087 genes that were up- or downregulated with HK2 siRNA treatment greater than 1.5-fold based on microarray results. Microarray analysis was performed on 2 cell lines (U87, GSC30) with 2 biological replicates for each. R1 and R2 stand for biological replicates 1 and 2, respectively. C, RT-PCR analysis was performed to confirm HK2 mRNA decrease in HK2 siRNA–treated cells. This analysis was performed on 3 biological replicates and 3 technical replicates for each treatment condition indicated and data are presented as the average of decrease in 4 cell lines. D, Top 200 differentially expressed mRNAs with HK2 siRNA treatment in the U87 and GSC30 cell lines were subjected to pathway analysis using Cytoscape. Blue circles denote downregulated pathways, whereas red circles denote upregulated processes including hypoxia, metabolism, and blood vessel formation.
HK2-associated gene signature identifies small-molecule inhibitors
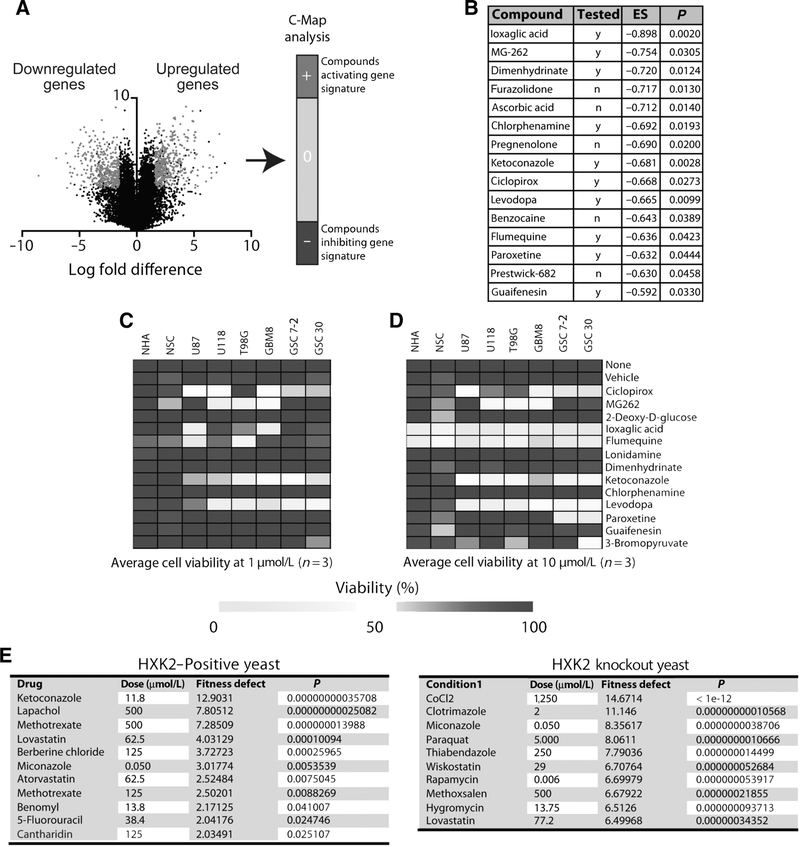
To date, an HK2-specific inhibitor does not exist and global HK2 inhibitors show toxicity and lack of specificity (11, 15). We hypothesized that genes downregulated by HK2 knockdown could provide the rationale for novel drug targets or pathways. We, therefore, performed C-Map analysis on the set of genes that were downregulated with HK2 knockdown in U87 and GSC30 cell lines (Fig. 2A; Supplementary Table S1). U87 (>2,296 citations in PubMed) cell lines are routinely employed for glioma research and provide valuable knowledge about this type of tumor. However, these cells are not perfect as their culture conditions over many decades have altered their genome and transcriptome. For this reason, we included GSCs (GSC30) derived from GBM patient samples to ensure consistency in outcome of our experiments.
Figure 2.
Gene signature associated with HK2 knockdown identified several small-molecule inhibitors that can target cells expressing high levels of HK2. A, Schematic of C-Map that was utilized for analysis of genes that were up- and downregulated with HK2 knockdown. B, List of top 15 drugs with a negative enrichment score from C-Map analysis that were predicted to inhibit gene signatures associated with HK2 knockdown. Cells were treated with the identified inhibitors at 1 μmol/L (C) and 10 μmol/L (D) and their viability was assessed using an ATP-cell titer GLO viability assay. The average viability of 3 independent experiments is shown and is depicted as a heatmap. E, Results from yeast drug screen on HXK2 (HK2 homolog) and HXK2 knockout (KO) identified ketoconazole as a potential inhibitor of HXK2, as it did not cause a fitness defect in HXK2 knockout yeast.
Our analysis identified 15 small molecules that were significant and could potentially downregulate HK2 and the transcripts that were downregulated with HK2 knockdown (Fig. 2B). Of these, 10 compounds were available for further testing on cells. We next performed an ATP-GLO viability assay on several GBM cell lines, including U87, T98G, U118, GBM8, GSC7–2, and GSC30, in comparison with normal NSCs and NHAs using the ten compounds and general glycolysis inhibitors such as lonidamine, 3BrPA, and 2-DG, at 1 μmol/L and 10 μmol/L concentrations (Fig. 2C and D). Levodopa, ciclopirox, and ketoconazole were found to specifically inhibit the growth of tumor cells but not NSCs and NHAs at both 1 μmol/L and 10 μmol/L. At 10 μmol/L, ioxiglic acid, MG-262, and flumequine were toxic to both normal and GBM cells (viability <70%, P < 0.05).
To independently test the role of azoles in HK2 activity, we performed a yeast fitness defect drug screen. Strikingly, HXK2-expressing yeast (the yeast homolog of HK2) was sensitive to ketoconazole as the top hit. In addition, HXK2 knockout yeast displayed no fitness defect when cultured with ketoconazole, supporting an indirect or direct interaction between ketoconazole and HXK2 (Fig. 2E).
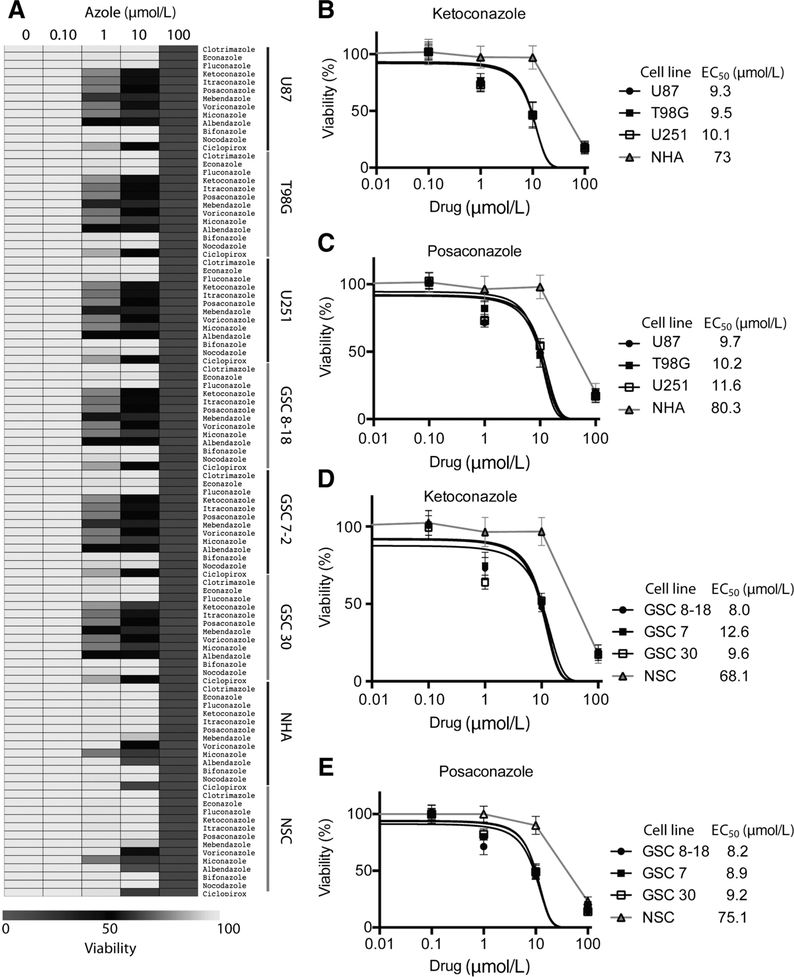
Azole drugs selectively target GBM cells but not normal cells
Our analysis of the predicted drug screen indicated that ketoconazole and ciclopirox were candidate drugs and given prior reports on the role of other antifungals in inhibiting brain tumor growth (16) or other cancers (17), we expanded our screen to test several additional azole compounds. We next tested the sensitivity of GBM cells lines, GSCs, NSCs, and NHAs, to clotrimazole, econazole, fluconazole, ketoconazole, itraconazole, posaconazole, mebendazole, voriconazole, miconazole, albendazole, bifonazole, and nocodazole at varying sub, low, and high micromolar doses (Fig. 3A). Ketoconazole and posaconazole were our top 2 hits displaying EC50 around 8–10 μmol/L in GBM cells but EC50 of >68 μmol/L in NHAs or NSCs (Figs. 3B–E). The EC50 for both ketoconazole and posaconazole in our study were well within the clinically achievable dose, as demonstrated by the clinical use of these drugs for other purposes (18).
Figure 3.
Azole drugs selectively target GBM cells but not normal cells in vitro. A, Several azoles were used at varying doses (0–100 μmol/L) to assess viability of GBM cells and normal cells and the results are depicted as a heat-map. EC50 curves for ketoconazole (B) and posaconazole (C) in GBM cell lines and nonimmortalized NHAs. EC50 curves for ketoconazole (D) and posaconazole (E) in GSCs and fetal NSCs. All analyses were performed in triplicates and error bars represent SD of the data mean.
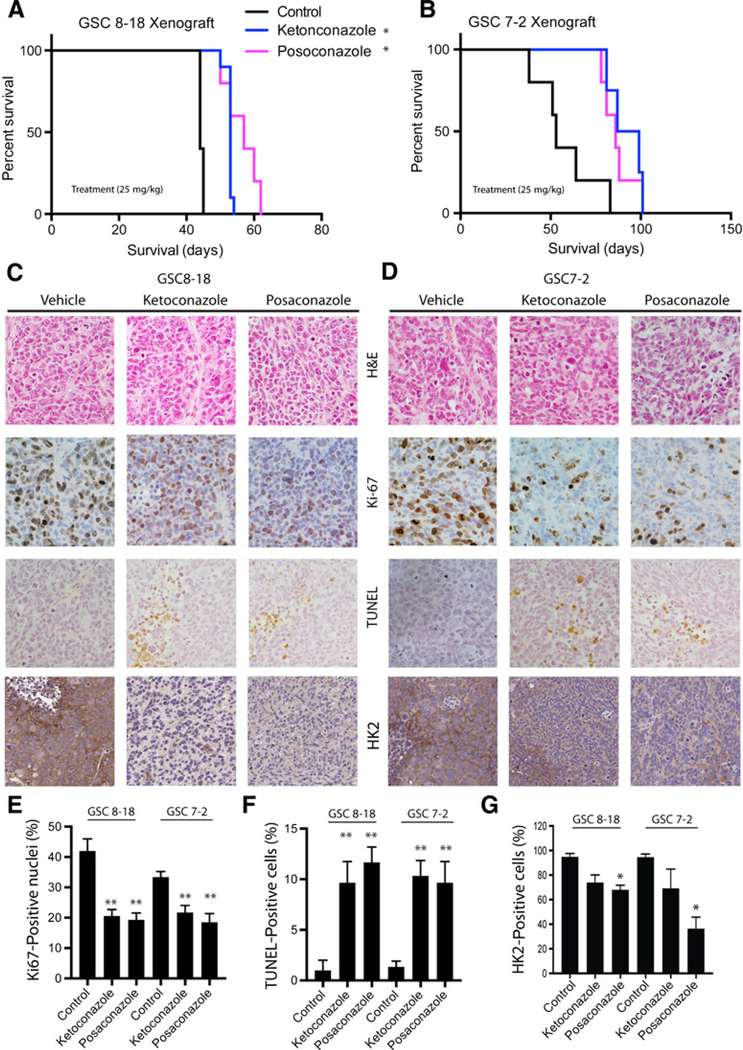
Ketoconazole and posaconazole inhibit GBM growth in several xenograft models
We next evaluated the in vivo inhibitory effect of ketoconazole and posaconazole in intracranial GBM mouse models. Mice were treated with 25 mg/kg of ketoconazole or posaconazole for 2 weeks, 1 week following intracranial injection of either GSC 8–18 or GSC 7–2 cells in NOD-SCID mice. The vehicle-treated GSC 8–18 xenograft model had an average life expectancy of 44 ± 2 days and was statistically significantly increased by either ketoconazole (54 ± 4 days, P < 0.05) or posaconazole (56 ± 4 days, P < 0.05; Fig. 4A). Likewise, vehicle-treated GSC 7–2 xenograft model displayed an average life expectancy of 57 ±16 days that was statistically significantly increased by either ketoconazole (84 ± 20 days, P < 0.05) or posaconazole (87 ± 8 days, P < 0.05; Fig. 4B). The histopathologic analysis of untreated and treated GSC tumors indicated that treatment with ketoconazole or posaconazole caused significant reduction in proliferation, as determined by Ki-67 staining (Figs. 4C–E) and significant induction of apoptosis, as evaluated by TUNEL staining in both xenograft models (Fig. 4C–F). Importantly, treatment of mice with both ketoconazole and posaconazole reduced expression of HK2 within the tumors, as determined by IHC analysis of HK2 protein level in the tumor sections (Fig. 4G). Similar prolonged survival of xenografts, together with reduced proliferation and increased apoptosis of tumor cells were observed in the U87 model (Supplementary Fig. S2A–S2C). Ketoconazole is known to cross the BBB; however, although posaconazole is predicted to cross the BBB, this has not been shown previously. Here, we confirmed that posaconazole crosses the BBB using mass spectrometry and detected the compound in both normal brain tissue and tumor in intracranial GBM xenografts (Supplementary Fig. S2D).
Figure 4.
Ketoconazole and posaconazole inhibit GBM growth in xenograft mouse models. Kaplan–Meier survival analysis of vehicle-treated, ketoconazole-treated (25 mg/kg), and posaconazole-treated (25 mg/kg) mouse GBM models intracranially injected with GSC 8–18 (A) and GSC7–2 (B) cells. C and D, Histopathologic analysis of GBMs extracted from mice treated with azoles by hematoxylin and eosin (H&E), TUNEL, Ki-67, and HK2 staining. E, Quantification of Ki-67–positive cells in azole-treated and vehicle-treated GBM xenografts as an indication of proliferating cells indicated a decrease in proportion of proliferating cells in treated tumors. F, Quantification of TUNEL-positive cells in treated and untreated GBM xenograft models as a measure for proportion of apoptotic cells suggested that treatment with both ketoconazole and posaconazole increased the percentage of apoptotic cells in tumors. G, Quantification of HK2 to determine the effect of azoles of HK2 protein level in tumor sections. All experiments were performed with a minimum of 3 biological replicates and 3 technical replicates. Asterisks on graphs denote a significant difference (*, P < 0.05, **, P < 0.01).
Ketoconazole and posaconazole reduce glycolytic metabolism in GBM cells
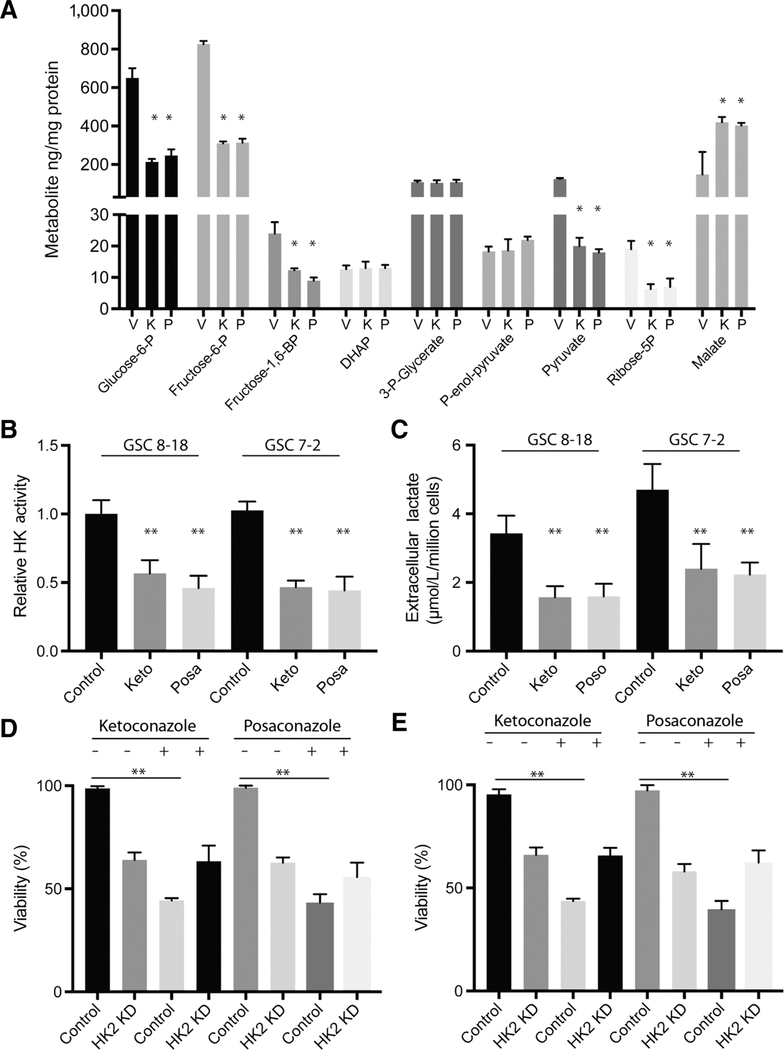
We next explored whether antitumor growth effects of ketoconazole and posaconazole observed in vitro and in vivo were due to inhibitory effects on tumor metabolism. We performed C13 glucose–labeled flux analysis using GC/MS on GSC 8–18 cells treated with vehicle (DMSO), ketoconazole, or posaconazole for 24 hours. This 24-hour time point was selected because viability did not decrease with these treatment conditions. Compared with the vehicle-treated cells, we observed a significant reduction in several intermediate metabolites of glycolysis including glucose-6-phosphate, fructose-6-phosphate, fructose-1,6-bisphosphate, ribose-5-phosphate, and pyruvate, in addition to an increase in malate, which is an intermediate product in the Krebs cycle (Fig. 5A). Similar C13 glucose–labeling results were observed in U87 cells treated with ketoconazole or posaconazole (Supplementary Fig. S3A). Further supporting an antiglycolytic effect, ketoconazole- and posaconazole-treated GSC 8–18 and GSC 7–2 cells showed a significant reduction in total hexokinase activity (Fig. 5B) along with a decrease in extracellular lactate production (Fig. 5C). Ketoconazole- and posaconazole-treated U87 cells also showed reduced lactate and hexokinase activity (Supplementary Fig. S3). Knockdown of HK2 by siRNA treatment of GSC 8–18 and GSC 7–2 cells reduced the cell viability compared with control siRNA–treated cells (Fig. 5D and E). Likewise, GSC 8–18 and GSC 7–2 cells treated with ketoconazole or posaconazole displayed reduced viability compared with cells not treated with drugs but treated with control siRNA (Fig. 5D and E). Strikingly, HK2 knockdown in GSC 8–18 and GSC 7–2 cells showed no additive effect when combined with ketoconazole or posaconazole treatment, confirming that HK2 protein expression is required for the function of ketoconazole and posaconazole (Fig. 5D and E). Similar results were observed in U87 cells (Supplementary Fig. S3).
Figure 5.
Ketoconazole and posaconazole reduce metabolism in GBM cells. A, 13C glucose–labeled flux analysis in GSC 8–18 cells treated with vehicle (DMSO; V), ketoconazole (K), and posaconazole (P) for 24 hours and quantification of glycolytic intermediates. B and C, Quantification of relative hexokinase (HK) activity and extracellular lactate production in GSC 8–18 cells treated with ketoconazole (Keto) or posaconazole (Posa), compared with vehicle-treated cells (V) at 24 hours. Alamar blue cell viability analysis on drug-treated and vehicle-treated GSC 8–18 (D) and GSC 7–2 (E) cells transfected with HK2 siRNA or scrambled siRNA at 72 hours. All experiments were performed with a minimum of 3 biological replicates and 3 technical replicates. Asterisks on graphs denote a significant difference (*, P < 0.05, **, P < 0.01). KD, knockdown.
Discussion
Despite varying genetic backgrounds, solid tumors including GBM rely on metabolic reprogramming to sustain anabolic processes for lipid, protein, and nucleotide production to support their growth and survival (5, 19). Our group and others have shown that HK2 is a promising therapeutic target in GBM as it is a driver of tumor metabolism, growth, invasion, and angiogenesis. Furthermore, there is preferential HK2 expression in GBM compared with normal brain making it a good therapeutic target (5, 20). We have also previously shown that targeting HK2 not only reduces tumor growth in vivo, it promotes radio- and chemo-sensitivity in GBM. Similar results have been found in several other tumors including cervical cancer, lung cancer, and hepatocellular cancer (5, 21, 22). Thus, targeting HK2 is acompelling and attractive therapeutic strategy.
Currently, HK2 is not clinically actionable as there are no specific HK2 inhibitors, and global HK inhibitors exhibit high cytotoxicity in normal tissues including the brain, which normally expresses HK1 (23). Given the lack of HK2-specific inhibitors, we employed a systems biology approach to first determine the gene expression signatures that correspond with high HK2 expression. Using gene expression analysis, we identified several hundred genes downregulated with the loss of HK2 and pathway analysis of the downregulated genes demonstrated enrichment of several oncogenic signaling pathways and biological processes known to be effector pathways of HK2 and glycolysis including MAPK/ERK, VEGF, and mTOR signaling pathways (5, 22, 24). We utilized C-Map analysis on HK2-associated gene signatures and identified small-molecule inhibitors that are predicted to mimic loss of HK2 and genes downregulated by loss of HK2. From our initial screen, we identified ketoconazole, an antifungal agent, which is known to cross the BBB in mice (25). Ketoconazole has previously been shown to decrease GBM growth while given simultaneously with temozolomide (26). Mechanistically, ketoconazole is a strong inhibitor of 5-lipoxygenase, thromboxane synthase (27, 28), and the drug efflux pump at the BBB (28). Further expansion of our screen to other azole compounds identified posaconazole as a top inhibitor of GBM growth in vitro. We observed EC50 values ranging from 8–12 μmol/L in GBM cell lines and GSCs, which are clinically achievable based on the human dosing of ketoconazole and posaconazole for other diseases (29). At the same time, the EC50 values in NSC and NHA, both of which are negative for HK2 expression, were 6 to 10 times higher than tumor cells, providing a critical therapeutic window for targeting tumor while avoiding normal brain cells. In addition to ketoconazole and posaconazole, we found that itraconazole, levodopa, and mebendazole conferred significant growth-inhibitory effect on glioma cells. Posaconazole is the next-generation variant of itraconazole and therefore, we focused our analysis on posaconazole. Levodopa (dopamine) has been shown to exert growth-inhibitory effect on GSCs and induce differentiation of NSCs (30). Mebendazole has been shown to confer survival benefits in 2 preclinical mouse models of GBM (16) and is now being tested in a phase I clinical trial for management of pediatric gliomas (NCT01837862).
Both ketoconazole and posaconazole compounds led to reduction in the number of proliferating tumor cells, while they increased the proportion of apoptotic cells and overall survival in GBM mouse models. The compounds exhibited direct antiglycolytic properties as evidenced by C13 glucose–labeling experiments. Ketoconazole and posaconazole treatments in vitro resulted in a significant reduction in several intermediate metabolites of glycolysis including glucose-6-phosphate, fructose-6-phosphate, fructose-1,6-bisphosphate, and pyruvate, consistent with an antiglycolytic property. Furthermore, we show that these azoles exert their tumor growth–inhibitory effect through modulation of HK2, as siRNA-mediated loss of HK2 in GBM cells notably rescued the tumor growth inhibition and antiglycolytic properties of both ketoconazole and posaconazole. Adding to these findings is the heterozygous yeast screen, where loss of HK2 yeast homolog, HXK2, resulted in loss of sensitivity to ketoconazole. Overall, these findings strongly argue that these 2 antifungal agents work best in tumor cells exhibiting high glycolysis and HK2 expression. Both ketoconazole and posaconazole have had long-standing clinical use in humans, through both oral and topical applications (29). There is clear evidence that ketoconazole crosses the BBB, and in this study, we show that both ketoconazole and posaconazole crosses the BBB; as confirmed by mass spectrometry results in intracranial GBM models.
Herein, we have established that ketoconazole and posaconazole display selective growth-inhibitory effects in GBM cells compared with nontumor cells. We observed significant in vivo survival benefits with both agents, including reduced proliferation and increased apoptosis in 3 independent xenograft mouse models. In addition, we established that azoles exert their antimetabolic action through inhibition of HK2 activity. We show that decrease in HK2 results in a distinct shift in gene expression and oncogenic pathway regulation. Future in-depth studies are required to uncover the mechanism through which ketoconazole and posaconazole target GBM cells and tumor metabolism. Overall, these results lend further mechanistic support to introduce azoles into practice for treatment of GBMs.
Supplementary Material
Supplemental Figure 1. Gene-set enrichment analysis (GSEA). Based on the list of top 200 differentially expressed genes with HK2 knockdown in the two cell lines (U87 and GSC30), a GSEA using the molecular signature database identified oncogenic signatures downregulated (A) or upregulated (B) with HK2 knockdown.
Supplemental Figure 3. Ketoconazole and Posaconazole reduce tumor metabolism in U87 cells.
(A) 13C-glucose labeled flux analysis in U87 cells treated with vehicle (DMSO) or Ketoconazole for 24 hours and quantification of glycolytic intermediates. (B-C) Quantification of relative hexokinase activity and extracellular lactate production (C) in U87 cells treated with Ketoconazole or Posaconazole, compared to vehicle treated cells at 24h. (D-E) Alamar blue cell viability analysis on Ketoconazole and Posaconazole treated U87 cells transfected with HK2 siRNA or scrambled siRNA for 72 hours. All experiments were performed with a minimum of 3 biological replicates and 3 technical replicates. Asterisks on graphs denote a significant difference (*p < 0.05, **p < 0.01, ***p < 0.001).
Supplemental Figure 2. Ketoconazole and Posaconazole inhibit GBM growth in a U87 xenograft model.
(A) Kaplan-Meier survival curve of vehicle-treated, Ketoconazole-treated (25mg/kg), and Posaconazole-treated (25mg/kg) U87 GBM xenograft models. (B) Immunohistochemical analysis of Ki67 was performed to determine tumor cell proliferation upon treatment with Ketoconazole and Posaconazole. (C) Ki-67 quantification was performed to estimate the proportion of proliferating tumor cells in drug versus control treated U87 mouse GBM models. (D). Detection of Posaconazole in U87 mouse tumor tissue was performed using mass spectrometry as described in ‘Materials and Methods’ section. Amount of Posaconazole in tumor (U87 and GSC8-18) was compared to untreated normal brain tissue (Control) and normalized per gram of tissue as baseline. The amount of compounds in normal brain, U87 and GSC8-18 derived tumor samples were normalized to the Control sample. Asterisks on graphs denote a significant difference (*p < 0.05).
Translational Relevance.
Through a systematic drug screening approach, we identified ketoconazole and posaconazole as inhibitors of hexokinase II (HK2). We have demonstrated that both drugs decrease tumor growth likely through blocking HK2 and affecting tumor metabolism. Prior studies suggested azoles as effective therapeutic compounds for gliomas and here, we have established the mechanism by which they influence glioblastoma (GBM) metabolism, oncogenic pathways regulated by HK2, and growth and progression of GBM. Two azoles, ketoconazole and posaconazole, displayed selective antitumoral properties in GBM cells and led to significant in vivo survival benefits, including reduced proliferation and increased apoptosis in 3 independent mouse models. In addition, we established that azoles cross the blood–brain barrier (BBB) and exert their antimetabolic action through inhibition of HK2 activity. We show that decrease in HK2 results in a distinct shift in gene expression and oncogenic pathway regulation. Overall, these results lend further mechanistic support to azoles being repurposed into practice through clinical trials for patients with GBM.
Acknowledgments
This work was supported by a CIHR Project Grant (awarded to G. Zadeh). S. Agnihotri received CIHR Fellowship award.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
References
- 1.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the warburg effect: the metabolic requirements of cell proliferation. Science 2009;324:1029–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mathupala SP, Rempel A, Pedersen PL. Glucose catabolism in cancer cells: identification and characterization of a marked activation response of the type II hexokinase gene to hypoxic conditions. J Biol Chem 2001;276: 43407–12. [DOI] [PubMed] [Google Scholar]
- 3.Ostrom QT, Gittleman H, Xu J, Kromer C, Wolinsky Y, Kruchko C, et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2009–2013. Neuro Oncol 2016;18:v1–v75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pedersen PL, Mathupala S, Rempel A, Geschwind JF, Ko YH. Mitochondrial bound type II hexokinase: a key player in the growth and survival of many cancers and an ideal prospect for therapeutic intervention. Biochim Biophys Acta 2002;1555:14–20. [DOI] [PubMed] [Google Scholar]
- 5.Vartanian A, Agnihotri S, Wilson MR, Burrell KE, Tonge PD, Alamsa hebpour A, et al. Targeting hexokinase 2 enhances response to radio-chemotherapy in glioblastoma. Oncotarget 2016;7:69518–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ko YH, Verhoeven HA, Lee MJ, Corbin DJ, Vogl TJ, Pedersen PL. A translational study “case report” on the small molecule “energy blocker” 3-bromopyruvate (3BP) as a potent anticancer agent: from bench side to bedside. J Bioenerg Biomembr 2012;44:163–70. [DOI] [PubMed] [Google Scholar]
- 7.Kunjithapatham R, Geschwind JFH, Rao PP, Boronina TN, Cole RN, Ganapathy-Kanniappan S. Systemic administration of 3-bromopyruvate reveals its interaction with serum proteins in a rat model. BMC Res Notes 2013;6:277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gandham SK, Talekar M, Singh A, Amiji MM. Inhibition of hexokinase-2 with targeted liposomal 3-bromopyruvate in an ovarian tumor spheroid model of aerobic glycolysis. Int J Nanomedicine 2015; 10:4405–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aft RL, Zhang FW, Gius D. Evaluation of 2-deoxy-D-glucose as a chemotherapeutic agent: mechanism of cell death. Br J Cancer 2002; 87:805–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maschek G, Savaraj N, Priebe W, Braunschweiger P, Hamilton K, Tidmarsh GF, et al. 2-Deoxy-D-glucose increases the efficacy of adriamycin and paclitaxel in human osteosarcoma and non-small cell lung cancers in vivo. Cancer Res 2004;64:31–4. [DOI] [PubMed] [Google Scholar]
- 11.Singh D, Banerji AK, Dwarakanath BS, Tripathi RP, Gupta JP, Mathew TL, et al. Optimizing cancer radiotherapy with 2-deoxy-d-glucose dose escalation studies in patients with glioblastoma multiforme. Strahlenther Onkol 2005;181:507–14. [DOI] [PubMed] [Google Scholar]
- 12.Bhat KPL, Balasubramaniyan V, Vaillant B, Ezhilarasan R, Hummelink K, Hollingsworth F, et al. Mesenchymal differentiation mediated by NF-κB promotes radiation resistance in glioblastoma. Cancer Cell 2013; 24:331–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, et al. The connectivity map: using. Science 2006;313:1929–35. [DOI] [PubMed] [Google Scholar]
- 14.Hillenmeyer ME, Fung E, Wildenhain J, Pierce SE, Hoon S, Lee W, et al. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science 2008;320:362–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shoshan MC. 3-bromopyruvate: targets and outcomes. J Bioenerg Biomembr 2012;44:7–15. [DOI] [PubMed] [Google Scholar]
- 16.Bai RY, Staedtke V, Aprhys CM, Gallia GL, Riggins GJ. Antiparasitic mebendazole shows survival benefit in 2 preclinical models of glioblastoma multiforme. Neuro Oncol 2011;13:974–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pantziarka P, Bouche G, Meheus L, Sukhatme V, Sukhatme VP. The repurposing drugs in oncology (ReDO) project. Ecancermedicalscience 2014;8:442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernardo VA, Cross SJ, Crews KR, Flynn PM, Hoffman JM, Knapp KM, et al. Posaconazole therapeutic drug monitoring in pediatric patients and young adults with cancer. Ann Pharmacother 2013;47:976–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agnihotri S, Zadeh G. Metabolic reprogramming in glioblastoma: the influence of cancer metabolism on epigenetics and unanswered questions. Neuro Oncol 2016;18:160–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agnihotri SK, Shen R, Li J, Gao X, Büeler H. Loss of PINK1 leads to metabolic deficits in adult neural stem cells and impedes differentiation of newborn neurons in the mouse hippocampus. FASEB J 2017;31: 2839–2853. [DOI] [PubMed] [Google Scholar]
- 21.Liu Y, Murray-Stewart T, Casero RA, Kagiampakis I, Jin L, Jiawen Z, et al. Targeting hexokinase 2 inhibition promotes radiosensitization in HPV16 E7-induced cervical cancer and suppresses tumor growth. Int J Oncol 2017;50:2011–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li M, Jin R, Wang W, Zhang T, Sang J, Li N, et al. STAT3 regulates glycolysis via targeting hexokinase 2 in hepatocellular carcinoma cells. Oncotarget 2017;8:24777–24784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolf A, Agnihotri S, Munoz D, Guha A. Developmental profile and regulation of the glycolytic enzyme hexokinase 2 in normal brain and glioblastoma multiforme. Neurobiol Dis 2011;44:84–91. [DOI] [PubMed] [Google Scholar]
- 24.Wolf A, Agnihotri S, Micallef J, Mukherjee J, Sabha N, Cairns R, et al. Hexokinase 2 is a key mediator of aerobic glycolysis and promotes tumor growth in human glioblastoma multiforme. J Exp Med 2011; 208:313–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iwen PC, Miller NG. Enhancement of ketoconazole penetration across the blood-brain barrier of mice by dimethyl sulfoxide. Antimicrob Agents Chemother 1986;30:617–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kast RE, Boockvar JA, Brüning A, Cappello F, Chang W-W, Cvek B, et al. A conceptually new treatment approach for relapsed glioblastoma: coordinated undermining of survival paths with nine repurposed drugs (CUSP9) by the international initiative for accelerated improvement of glioblastoma care. Oncotarget 2013;4:502–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nardone PA, Slotman GJ, Vezeridis MP. Ketoconazole: a thromboxane synthetase and 5-lipoxygenase inhibitor with antimetastatic activity in B16-F10 melanoma. J Surg Res 1988;44:425–9. [DOI] [PubMed] [Google Scholar]
- 28.Venishetty VK, Komuravelli R, Kuncha M, Sistla R, Diwan PV. Increased brain uptake of docetaxel and ketoconazole loaded folate-grafted solid lipid nanoparticles. Nanomedicine 2013;9:111–21. [DOI] [PubMed] [Google Scholar]
- 29.Sugar AM, Alsip SG, Galgiani JN, Graybill JR, Dismukes WE, Cloud GA, et al. Pharmacology and toxicity of high-dose ketoconazole. Antimicrob Agents Chemother 1987;31:1874–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dolma S, Selvadurai HJ, Lan X, Lee L, Kushida M, Voisin V, et al. Inhibition of dopamine receptor D4 impedes autophagic flux, proliferation, and survival of glioblastoma stem cells. Cancer Cell 2016;29:859–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Gene-set enrichment analysis (GSEA). Based on the list of top 200 differentially expressed genes with HK2 knockdown in the two cell lines (U87 and GSC30), a GSEA using the molecular signature database identified oncogenic signatures downregulated (A) or upregulated (B) with HK2 knockdown.
Supplemental Figure 3. Ketoconazole and Posaconazole reduce tumor metabolism in U87 cells.
(A) 13C-glucose labeled flux analysis in U87 cells treated with vehicle (DMSO) or Ketoconazole for 24 hours and quantification of glycolytic intermediates. (B-C) Quantification of relative hexokinase activity and extracellular lactate production (C) in U87 cells treated with Ketoconazole or Posaconazole, compared to vehicle treated cells at 24h. (D-E) Alamar blue cell viability analysis on Ketoconazole and Posaconazole treated U87 cells transfected with HK2 siRNA or scrambled siRNA for 72 hours. All experiments were performed with a minimum of 3 biological replicates and 3 technical replicates. Asterisks on graphs denote a significant difference (*p < 0.05, **p < 0.01, ***p < 0.001).
Supplemental Figure 2. Ketoconazole and Posaconazole inhibit GBM growth in a U87 xenograft model.
(A) Kaplan-Meier survival curve of vehicle-treated, Ketoconazole-treated (25mg/kg), and Posaconazole-treated (25mg/kg) U87 GBM xenograft models. (B) Immunohistochemical analysis of Ki67 was performed to determine tumor cell proliferation upon treatment with Ketoconazole and Posaconazole. (C) Ki-67 quantification was performed to estimate the proportion of proliferating tumor cells in drug versus control treated U87 mouse GBM models. (D). Detection of Posaconazole in U87 mouse tumor tissue was performed using mass spectrometry as described in ‘Materials and Methods’ section. Amount of Posaconazole in tumor (U87 and GSC8-18) was compared to untreated normal brain tissue (Control) and normalized per gram of tissue as baseline. The amount of compounds in normal brain, U87 and GSC8-18 derived tumor samples were normalized to the Control sample. Asterisks on graphs denote a significant difference (*p < 0.05).