Abstract
Allium sect. Cepa (Amaryllidaceae) comprises economically important plants, yet resolving the phylogenetic relationships within the section has been difficult as nuclear and chloroplast-based phylogenetic trees have been incongruent. Until now, phylogenetic studies of the section have been based on a few genes. In this study, we sequenced the complete chloroplast genome (plastomes) of four central Asian species of sect. Cepa: Allium oschaninii, A. praemixtum, A. pskemense and A. galanthum. Their chloroplast (cp) genomes included 114 unique genes of which 80 coded proteins. Seven protein-coding genes were highly variable and therefore promising for future phylogenetic and phylogeographic studies. Our plastome-based phylogenetic tree of Allium sect. Cepa revealed two separate clades: one comprising the central Asian species A. oschaninii, A. praemixtum, and A. pskemense, and another comprising A. galanthum, A. altaicum, and two cultivated species, A. cepa and A. fistulosum. These findings contradict previously reported phylogenies that relied on ITS and morphology. Possible explanations for this discrepancy are related to interspecific hybridization of species ancestral to A. galanthum and A. cepa followed by chloroplast capture; however, this is impossible to prove without additional data. Our results suggest that the central Asian Allium species did not play a role in the domestication of the common onion. Among the chloroplast genes, rpoC2 was identified as a gene of choice in further phylogeographical studies of the genus Allium.
Keywords: Chloroplast genome, SNP, Phylogeny, Chloroplast capture
1. Introduction
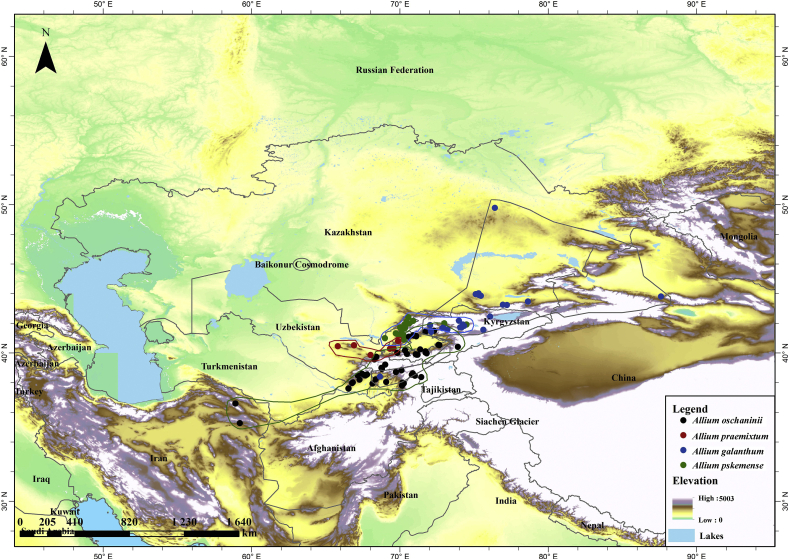
Allium sect. Cepa (Mill.) Prokh. (Amaryllidaceae) is a small group within the genus Allium L. that includes ten wild species and two economically important cultivated species, Allium cepa L. (common or bulb onion) and Allium fistulosum L. (bunching onion) (Fritsh and Friesen, 2002; Gurushidze et al., 2007). The wild species of this section occur naturally on dry rocky slopes in mountain areas in Asia. They are characterized by a long juvenile phase (3–10 years), are morphologically variable, and sometimes resemble A. cepa. Allium altaicum Pall., the most likely progenitor of A. fistulosum, grows in southern Siberia and Mongolia. Allium rhabdotum Stearn. occurs in Bhutan, Allium roylei Baker. in North-West India, Allium asarense R.M. Fritsch et Matin in Iran, and Allium farctum Wendelbo in Afghanistan and Pakistan. Allium vavilovii Popov et Vved. grows in the Kopetdag Range in Turkmenistan and northeastern Iran (Fritsh and Friesen, 2002). The ranges of the other species of sect. Cepa (Allium galanthum Kar. et Kir., Allium oschaninii O. Fedtsch., Allium praemixtum Vved. and Allium pskemense O.Fedtsch.) are within the Tien-Shan and Pamir-Alai mountain chains with isolated occurrences in northeastern Iran (A. oschaninii) and northeastern Kazakhstan (A. galanthum) (Fig. 1).
Fig. 1.
Distribution of central Asian species of Allium sect. Cepa.
Over last three decades, several hypotheses have been proposed regarding the phylogeny of sect. Cepa, the role of central Asia in the evolution in this group and the domestication of A. cepa. The central Asian species A. oschaninii was for some time treated as the most ancestral species of the cultivated onion (Wendelbo, 1971; Hanelt, 1985, 1990), but was shown to have different heterochromatic banding patterns and severe crossing barriers with A. cepa (Vosa, 1976; van Raamsdonk et al., 1992). In contrast, A. cepa and A. vavilovii (morphologically similar to A. oschaninii) have been successfully hybridized (van Raamsdonk et al., 1992). A. vavilovii resembles A. oschaninii in having a bubble-like hollow stem but its leaves are completely flat and falcate. Fritsch et al. (2001) proposed that the closest relatives of the common onion are four species with a bubble-like swelling in the lower part of the hollow scape (A. oschaninii, A. praemixtum, A. asarense and A. vavilovii). Based on morphological and distributional evidence, Fritsch and Friesen (2002) later excluded A. oschaninii and A. praemixtum from consideration as the closest relatives of A. cepa. Due to differences in morphology, A. galanthum has never been considered a close relative of A. cepa.
Determining the closest wild relative to the common onion is impossible without using molecular markers that have a phylogenetic signal. Unfortunately, the molecular phylogeny of sect. Cepa lags behind its morphological and karyological characterization.
Plastomes are a reliable source of information for inferring phylogeny and evolutionary history due to the absence of recombination and their low mutation rate (Jansen et al., 2012; Moore et al., 2010; Shaw et al., 2014); however, few studies have used plastomes analyze the phylogeny of sect. Cepa (Havey, 1992; Lilly and Havey, 2001; van Raamsdonk et al., 2003). The only study based on selected chloroplast sequences (van Raamsdonk et al., 2003) used only three cpDNA fragments (trnL-F, rps16, rbcL). This low coverage of existing cpDNA variation may explain the observed disagreement between phylogenetic trees based on cpDNA and nrDNA (ITS region) (Friesen and Klaas, 1998; Gurushidze et al., 2007), including the placement of A. pskemense in sect. Rhizirideum and A. roylei in an intermediate position between sections Cepa and Schoenoprasum, while both species were located within sect. Cepa based on nuclear DNA analysis (van Raamsdonk et al., 2003). To resolve this issue, we analyzed 16 sequenced plastomes of seven species from sect. Cepa, focusing on the central Asian species whose phylogenetic position in sect. Cepa is the most unclear and poorly understood. We characterized plastomes of the central Asian species by assessing the arrangement and variation of their genes, and used these plastome sequences to construct a phylogeny of Allium sect. Cepa.
2. Material and methods
2.1. Taxon sampling and DNA extraction
Samples of the central Asian species of Allium were collected during botanical expeditions to central Asia from 2014 to 2019 (Table 1). The sampled leaves were dried in silica-gel upon collecting. Total DNA was isolated by the CTAB protocol (Doyle and Doyle, 1987) from 1 g of well-dried leaves.
Table 1.
Source of new chloroplast genomes used in this study and location of voucher specimens.
| No | Species name | Location | Voucher |
|---|---|---|---|
| 1 | Allium galanthum Kar. & Kir | Almaty province, Kazakhstan, h = 741 m, N44.0119, E75.31301 | ZD1108(KUN) |
| 2 | A. galanthum Kar. & Kir | Jalalabad province, Kyrgyzstan, h = 1093 m, N41.6737, E72.8623 | ZD0932(KUN) |
| 3 | A. pskemense B. Fedtsch | Parkent, Tashkent, Uzbekistan, h = 989 m, N 41.3268, E69.7272 | FM201902(TASH) |
| 4 | A. pskemense B. Fedtsch | Pskem, Tashkent, Uzbekistan. h = 1336 m, N 41.924956, E70.316807 | F201901(TASH) |
| 5 | A. oschaninii O. Fedtsch | Anzob Pass, east side of the Varzob River, Tajikistan, h = 1850 m, N38.994222, E68.7654 | ZD0708(KUN) |
| 6 | A. oschaninii O. Fedtsch | Sukh, Fergana province, Uzbekistan, h = 1171 m, N40.0263 E71.1334 | ZD0346(KUN) |
| 7 | A. oschaninii O. Fedtsch | Imomota, Andijan province, Uzbekistan, h = 795 m, N40.548118, E72.607991 | FKZ022(TASH) |
| 8 | A. praemixtum Vved | Jizzakh province, Uzbekistan, h = 900 m, N40.5236, E66.9111 | KT201903(TASH) |
2.2. Plastome sequencing, assembly and annotation
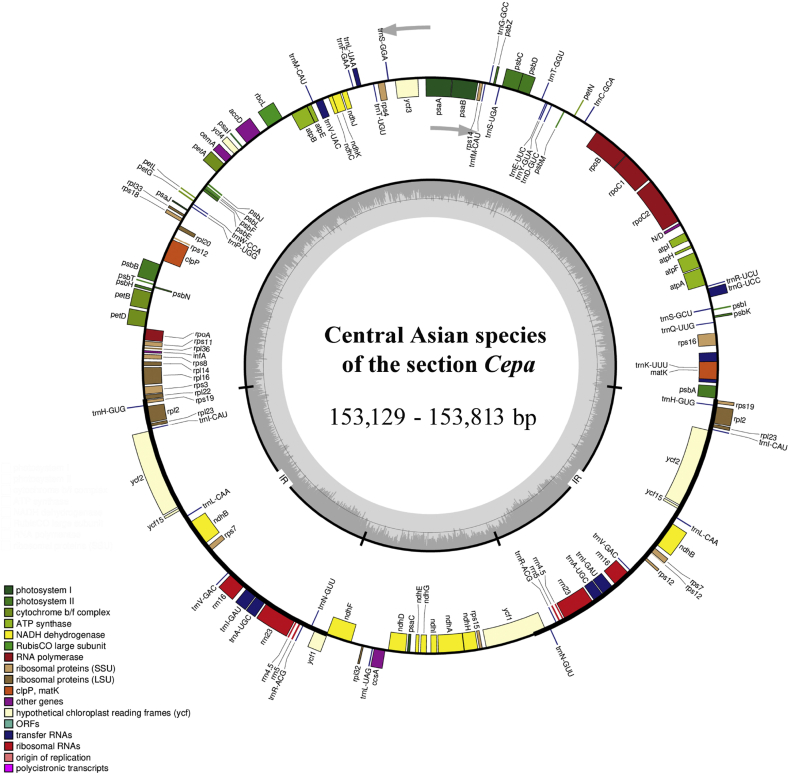
Libraries were constructed from 150 bp pair-end reads with insert sizes of 350 bp using the Genomic DNA Sample Prep Kit (Illumina), according to the manufacturer's protocol, and then sequenced on Illumina HiSeq 4000 system at Beijing Novogene Bioinformatics Technology Co., Ltd, Beijing, China. The plastid genomes were assembled with A. fistulosum as a reference (NC_040222, Yusupov et al., 2019) using software NovoPlasty v.3.8.3 (Dierckxsens et al., 2017). In accordance with the reference, start and stop codons and intron/exon boundaries for protein-coding genes were checked manually in Geneious v.10.0.2 (Kearse et al., 2012). The physical map of the circular cp genomes (Fig. 2) was created using the OGDRAW program (Greiner et al., 2019).
Fig. 2.
General plastome map of central Asian species of Allium sect. Cepa.
2.3. Assessment of gene variability in plastome comparisons
Single nucleotide polymorphisms (SNP) were assessed for 80 protein-coding genes used in phylogenetic analyses by Geneious v.10.0.2 (Kearse et al., 2012). SNP variability was assessed at several levels: with and without the outgroup, for two species clusters corresponding to two geographic regions, and within the two species, A. oschaninii and A. galanthum.
2.4. Phylogenetic analysis
The reconstruction of the phylogeny of sect. Cepa utilized 17 samples in total, including newly collected samples, all available plastomes of sect. Cepa in the NCBI database, plus one outgroup (Allium sativum L.). Eight of these plastomes were annotated and uploaded to the NCBI database (Table 1, Table 2).
Table 2.
Characteristics and GenBank accessions of plastomes of Allium sect. Cepa.
| Species | Total |
Large single copy (LSC) |
Small single copy (SSC) |
Inverted repeats (IRs) |
GenBank accessions | |
|---|---|---|---|---|---|---|
| Length (bp) | G + C (%) | Length (bp) | Length (bp) | Length (bp) | ||
| A. pskemense | 153,788 | 36.70 | 82,720 | 18,034 | 26,517 | NC_044411 |
| A. pskemense | 153,813 | 36.80 | 82,747 | 18,032 | 26,517 | MT300496 |
| A. oschaninii | 153,580 | 36.80 | 82,521 | 18,031 | 26,514 | NC_044470 |
| A. oschaninii | 153,643 | 36.80 | 82,549 | 17,984 | 26,555 | MT300495 |
| A. oschaninii | 153,581 | 36.80 | 82,522 | 18,031 | 26,514 | MT300494 |
| A. praemixtum | 153,226 | 36.80 | 82,162 | 18,042 | 26,511 | NC_044412 |
| A. fistulosum | 153,164 | 36.80 | 82,237 | 17,907 | 26,510 | NC_040222 |
| A. fistulosum | 153,162 | 36.80 | 82,235 | 17,907 | 26,510 | MK335927 |
| A. alataicum | 153,129 | 36.70 | 82,196 | 17,913 | 26,510 | NC_040972 |
| A. galanthum | 153,227 | 36.90 | 82,408 | 17,887 | 26,466 | MT300496 |
| A. galanthum | 153,222 | 36.90 | 82,402 | 17,888 | 26,466 | MT300497 |
| A. cepa_1 | 153,529 | 36.80 | 82,698 | 17,931 | 26,450 | KM088013 |
| A. cepa_2 | 153,568 | 36.80 | 82,738 | 17,930 | 26,450 | KM088015 |
| A. cepa_3 | 153,538 | 36.80 | 82,694 | 17,922 | 26,461 | KF728080 |
| A. cepa_4 | 153,586 | 36.80 | 82,719 | 17,931 | 26,468 | MK335926 |
| A. cepa_5 | 153,440 | 36.80 | 82,577 | 17,927 | 26,468 | KM088014 |
Multiple-sequence alignments were performed with MAFFT software (Katoh et al., 2002). Phylogenetic trees were reconstructed using Maximum likelihood (ML) and Bayesian inference (BI). For ML we employed RAxML-HPC BlackBox v.8.1.24 software (Stamatakis et al., 2006) with 1000 bootstrap replicates, and for BI we used MrBayes v.3.2.6 (Ronquist et al., 2003) with 1,000,000 generations with random trees sampled every 200 generations. In the latter analysis, after discarding the first 25% trees as burn-in, a 50% majority-rule consensus tree was constructed from the remaining trees to estimate posterior probabilities (PP). For analyses, a model of nucleotide substitution was selected based on the Akaike Information Criterion (AIC) using MrModelTest 2 (Nylander, 2004).
3. Results
3.1. Characterization of the plastomes of central Asian species of Allium sect. Cepa
The total length of the complete chloroplast genomes within sect. Cepa species ranged from 153,129 to 153,813 bp. The small single copy (SSC), large single copy (LSC) and inverted repeats (IR) regions of cp genomes ranged from 17,887 to 18,042 bp, 82,162 to 82,747 bp and 26,450 to 26,555 bp, respectively. For the central Asian species A. oschaninii, A. pskemense, and A. praemixtum, the SSC regions ranged from 17,984 to 18,042 bp (IR: 26,511–26,555 bp) for four species (A. cepa, A. galanthum, A. fistulosum, A. altaicum); the SSC regions ranged from 17,887 to 17,931 bp (IR: 26,450–26,510 bp). The complete chloroplast genomes of A. oschaninii from Uzbekistan (NC_044470 and MT300495) and Tajikistan (MT300494) differed in size due to the presence of indels (Table 1, Table 2).
The chloroplast genome of species of sect. Cepa encodes 134 genes, including 80 protein-coding genes, 30 tRNA genes, four rRNA genes, and 20 duplicated genes (Fig. 2; Table 3).
Table 3.
List of genes identified in plastomes of Allium sect. Cepa.
| Category of Genes | Gene group | Gene name |
|---|---|---|
| Self-replication | Ribosomal RNA genes | rrn4.5 × 2, rrn5 × 2, rrn16 × 2, rrn23 × 2 |
| Transfer RNA genes | trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnG-GCC, trnG-UCCa, trnH-GUG × 2, trnK-UUUa, trnL-UAAa, trnL-UAG, trnM-CAU, trnP-UGG, trnQ-UUG, trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-UGU, trnT-GGU, trnV-UACa, trnY-GUA, trnW-CCA, trnfM-CAU, trnA-UGCa × 2, trnI-CAU × 2, trnI-GAUa × 2, trnL-CAA × 2, trnN-GUU × 2, trnR-ACG × 2, trnV-GAC × 2 | |
| Ribosomal protein (small subunit) | rps2, rps3, rps4, rps7 × 2, rps8, rps11, rps12b × 2, rps14, rps15, rps16a, rps18, rps19 | |
| Ribosomal protein (large subunit) | rpl2a × 2, rpl14, rpl16a, rpl20, rpl22, rpl23 × 2, rpl32, rpl33, rpl36 | |
| RNA polymerase | rpoA, rpoB, rpoC1a, rpoC2 | |
| Translational initiation factor | infA | |
| Genes for photosynthesis | Subunits of photosystem I | psaA, psaB, psaC, psaI, psaJ, ycf3b, ycf4 |
| Subunits of photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | |
| Subunits of cytochrome | petA, petBa, petDa, petG, petL, petN | |
| Subunits of ATP synthase | atpA, atpB, atpE, atpFa, atpH, atpI | |
| Large subunit of Rubisco | rbcL | |
| Subunits of NADH dehydrogenase | ndhAa, ndhBa × 2, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | |
| Other genes | Maturase | matK |
| Envelope membrane protein | cemA | |
| Subunit of acetyl-CoA | accD | |
| Synthesis gene | ccsA | |
| ATP-dependent protease | clpPb | |
| Component of TIC complex | ycf1 × 2 | |
| Genes of unknown function | Conserved open reading frames | ycf2 × 2, ycf15 × 2 |
×2: Two gene copies in the IR regions.
With one intron.
With two introns.
3.2. Variation in protein-coding genes
Plastome protein-coding genes of 17 Allium species contained a total of 2,239 SNPs. Within sect. Cepa, however, without including A. sativum, the number of SNPs was substantially lower (1,482 SNPs). Comparison of six plastomes representing three central Asian species (A. oschaninii, A. praemixtum and A. pskemense) revealed 451 SNPs, 10 plastomes representing four species (wild A. altaicum and A. galanthum, and cultivated A. cepa and A. fistulosum) revealed 290 SNPs. The number of SNPs in three plastomes of A. oschaninii collected in different areas and two of plastomes of A. galanthum collected in different areas was 52 and 18, respectively (Table 4).
Table 4.
Number of SNPs in plastome protein-coding genes in different clades of Allium sect. Cepa.
| Data type | Total number of SNPs | Number of SNPs |
||||||
|---|---|---|---|---|---|---|---|---|
| matK | rpoC2 | ycf1 | ndhF | rpoB | accD | ycf2 | ||
| Section Cepa plus one outgroup | 2239 | 79 | 101 | 255 | 93 | 62 | 35 | 30 |
| Section Cepa | 1482 | 51 | 60 | 179 | 67 | 42 | 31 | 19 |
| Central Asian species (A. praemixtum, A. oschaninii, A. pskemense) | 451 | 25 | 31 | 75 | 24 | 23 | 13 | 7 |
| Northeast Asian species (A. galanthum, A. altaicum) plus A. fistulosum and A. cepa | 290 | 14 | 14 | 68 | 22 | 14 | 15 | 4 |
| A. oschaninii | 52 | 7 | 3 | 8 | 2 | 2 | 2 | 0 |
| A. galanthum | 18 | 0 | 0 | 5 | 2 | 4 | 1 | 1 |
Of the 80 protein-coding genes examined, seven (accD, ycf2, ycf1, rpoC2, ndhF, rpoB, matK) were highly variable (number of SNPs ≥ 30). The ycf1 gene was the most variable. The ranking of gene variability was unchanged when the outgroup species was included. Several genes showed a high level of variability in two intra-specific variability assessments (two plastomes of A. galanthum and three plastomes of A. oschaninii): ycf1 (A. galanthum – 8 SNPs and A. oschaninii – 5 SNPs), matK (A. oschaninii – 7 SNPs), rpoB (A. galanthum – 4 SNPs) and rpoC2 (A. oschaninii – 3 SNPs) (Table 4).
3.3. Phylogenetic analysis
The best fit model for genome regions (80 protein-coding genes, whole, SSC, IRs, LSC) was the generalized time reversible plus gamma model (GTR + G), whereas for single protein-coding genes the best model was GTR.
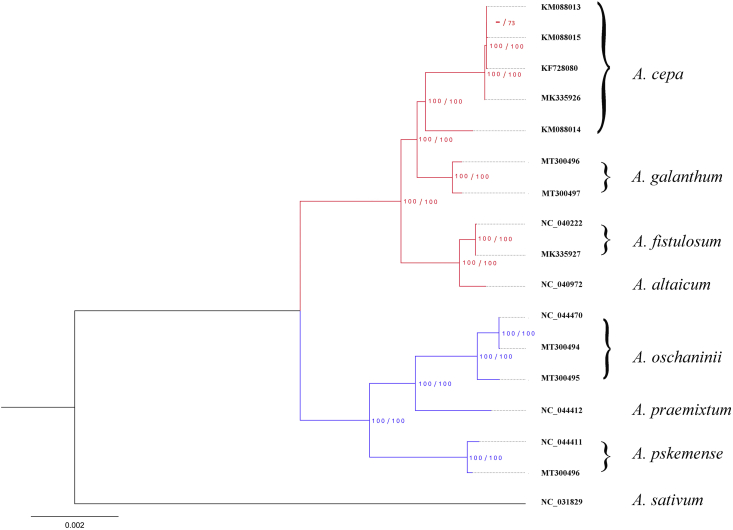
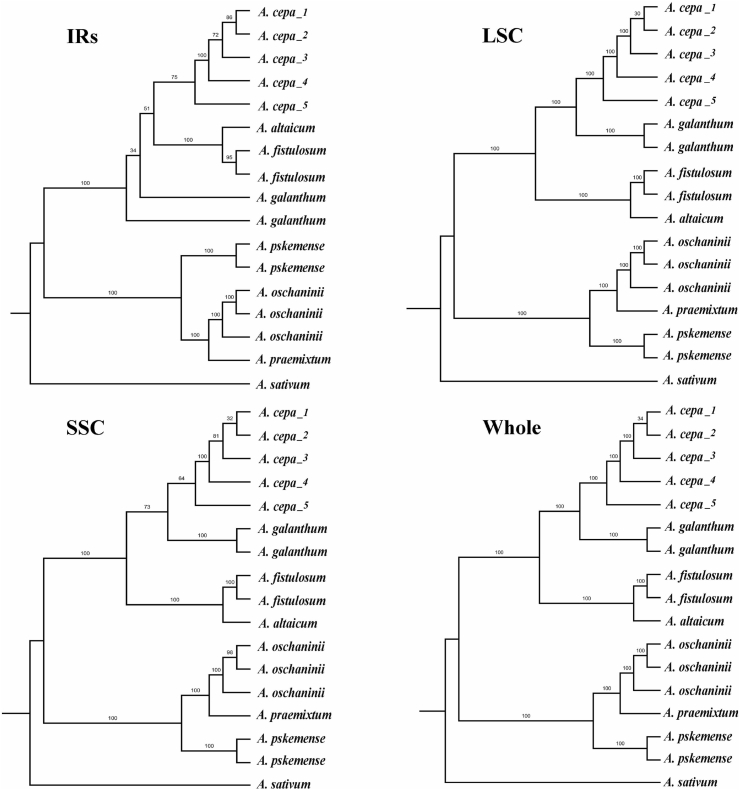
The phylogenetic trees for 17 plastomes, SSC and LSC constructed using Maximum likelihood (ML) and Bayesian inference (BI) had the same topology with the tree based on 80 protein-coding genes. In all trees produced there were two major clades with 100% support in both ML and BI, one comprising A. praemixtum, A. oschaninii, and A. pskemense, and another comprising A. altaicum, A. galanthum, and two cultivated species A. fistulosum and A. cepa (Fig. 3, Fig. 5).
Fig. 3.
Phylogenetic tree based on 80 protein-coding genes of 16 plastomes of Allium sect. Cepa plus an outgroup plastome (A. sativum). In the phylogenetic tree Maximum Likelihood bootstrap proportion values and Bayesian posterior probabilities are shown next to the nodes, and a dash (−) indicates support at a node < 50%. Scale bar represents the expected substitutions per site.
Fig. 5.
Phylogenetic trees based on whole genomes, IRs, LSC and SSC regions. Maximum likelihood values are given above nodes.
The phylogenetic trees based on the most variable seven protein-coding genes (matK, rpoC2, ycf1, ndhF, rpoB, accD, ycf2, and all seven combined) were highly similar. They all showed the two major clades described above with minor differences in sub-clade patterns (Fig. 4). The tree for rpoC2 was congruent with the tree for complete chloroplast genomes.
Fig. 4.
Seven phylogenetic trees based on a single protein-coding gene each plus tree from the combination of all seven genes. Maximum likelihood values are given next to nodes.
4. Discussion
4.1. Variability in protein-coding genes
Among the genes comprising the plastomes of sect. Cepa, several were exceptional in the extent of their variability (matK, rpoC2, ycf1, ndhF, rpoB, accD, ycf2) with matK, rpoC2 and ycf1 also showing substantial variation at the intra-specific level. Among these highly variable genes, rpoC2 produced exactly the same tree topology with the tree for complete chloroplast genomes, making it a priority gene for further phylogenetic studies in sect. Cepa (Fig. 4).
4.2. Phylogenetic analysis
The plastome-based phylogenetic tree agrees well with the tree produced by Havey (1992) from cpDNA RFLPs in the distant position of A. oschaninii and A. pskemense, which were placed at the root of the clade of sect. Cepa. In comparison, the plastome-based phylogenetic tree differed from the tree van Raamsdonk et al. (2003) obtained using three cpDNA fragments (trnL-F, rps16 and rbcL). In the latter tree A. oschaninii was sister to A. fistulosum, A. altaicum, and A. galanthum and distant from A. pskemense. This discrepancy is due to the limited number of loci of cpDNA fragments used by van Raamsdonk et al. (2003). As we show here, there is always some incongruence among the phylogenetic relationships based on particular genes. This incongruence disappears, however, with an increase in the number of genes used (Fig. 3, Fig. 5).
Our results are important in light of a long-standing debate about the most probable ancestor of the domesticated species A. cepa. Although A. vavilovii is accepted as the closest wild relative of A. cepa based on nrDNA (Friesen and Klaas, 1998; Fritsch et al., 2001, Gurushidze et al., 2007), the immediate ancestor is still unknown. A. oschaninii, once considered to be closest to the ancestral species, is no longer considered as such based on the results of crossing experiments (van Raamsdonk and de Vries, 1992). Our analyses of Allium plastomes indicates that the closest wild relative of A. cepa is A. galanthum, which is consistent with the crossability dendrogram constructed by van Raamsdonk et al. (1992) (Fig. 3). In contrast, the latter crossability dendrogram corresponds poorly to the similarity of the nuclear DNA of these two species (e.g., van Raamsdonk et al., 2000; Gurushidze et al., 2007). Unfortunately, an appealing conclusion from the two species cpDNA similarity, that A. galanthum played a role in the domestication of the onion, several lines of evidence contradict this interpretation. First, A. cepa and A. galanthum differ in scape characteristics (fistulose vs. solid). Second, there are large differences in the nuclear DNA of the two species (Gurushidze et al., 2007). Most importantly, although crosses between A. cepa and A. galanthum produce F1 hybrids, the chromosomes of the F1 hybrids of A. galanthum are discriminated from chromosomes of A. cepa in the continuing backcrosses, in which A. galanthum acts as the cytoplasmic donor and A. cepa acts as the nucleus donor. The chromosome regions from A. galanthum decrease in number and completely disappear by B3 (Yamashita and Tashiro, 1999), indicating that A. galanthum and A. cepa do not share a common ancestor.
Allium galanthum was a member of the clade including, besides A. cepa, another cultivated species A. fistulosum and its accepted wild progenitor A. altaicum, while A. oshaninii, A. praemixtum and A. pskemense formed another clade. The phylogenetic position of three species native to Central Asian mountains (A. oschaninii, A. praemixtum, A. pskemense) contradicts previously reported ITS results (Fritsch et al., 2001; Gurushidze et al., 2007). Morphological characteristics of the three species also correspond to the phylogenetic tree of nrDNA. The possible reasons for this discrepancy are probably related to interspecific hybridization between species ancestral to A. galanthum and A. cepa followed by chloroplast capture, but this is impossible to prove without additional data.
A serious limitation of our study for understanding the evolution of Allium sect. Cepa is unavailability of chloroplast genomes of A. rhabdotum, A. roylei, A. farctum, A. asarense and A. vavilovii. Further progress is impeded by these gaps in our knowledge. Future efforts in reconstructing the evolution of sect. Cepa and domestication of the onion must be directed towards studying these five species.
5. Conclusions
In this study, we tested the hypothesis that central Asian species of Allium sect. Cepa (A. oschaninii, A. praemixtum, A. pskemense and A. galanthum) are the closest wild relatives of the cultivated A. cepa. Our results suggest that none of these species could have played a role in domestication of the common onion and therefore it is highly unlike that central Asia was the origin of A. cepa domestication. Our analysis also revealed that the plastid gene rpoC2 produced exactly the same tree topology as the phylogenetic tree based on 80 protein-coding genes, whole-genome plastomes, SSC and LSC regions, and therefore is a gene of choice in further phylogeographical and phylogenetic studies of the Allium.
Author contributions
ZY and TD carried out this research, ZY, DM, KT collected the material. HS and KT conceived and designed the research, SV, KT, FK, TD and HS discussed the results and revised the manuscript. TD and HS funded the research. All of the authors read and approved the manuscript.
Declaration of Competing Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgements
We are grateful to two anonymous reviewers for comments on the manuscript. The manuscript has greatly improved due to these invaluable comments. The authors in particular wish to thank Dr. David E. Boufford for editing the English. This study was supported by the Second Tibetan Plateau Scientific Expedition and Research (STEP) program (2019QZKK0502), the International Partnership Program of Chinese Academy of Sciences (151853KYSB20180009) and the Strategic Priority Research Program of Chinese Academy of Sciences (XDA20050203) to H.S., and the Youth Innovation Promotion Association of Chinese Academy of Sciences (2019382), the Young Academic and Technical Leader Raising Foundation of Yunnan Province (2019HB039), the Chinese Academy of Sciences “Light of West China” Program and the Biodiversity Survey, Monitoring and Assessment (2019HB2096001006) to T. D, and the the Belt and Road Project of West Light Foundation of the Chinese Academy of Sciences.
Footnotes
Peer review under responsibility of Editorial Office of Plant Diversity.
Contributor Information
Komiljon Tojibaev, Email: ktojibaev@mail.ru.
Hang Sun, Email: sunhang@mail.kib.ac.cn.
References
- Dierckxsens N., Mardulyn P., Smits G. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017;45:e18. doi: 10.1093/nar/gkw955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J.J., Doyle J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987;19:11–15. [Google Scholar]
- Friesen N., Klaas M. Origin of some minor vegetatively propagated Allium crops studied with RAPD and GISH. Genet. Resour. Crop Evol. 1998;45:511–523. [Google Scholar]
- Fritsch R.M., Friesen N. Evolution, domestication and taxonomy. In: Rabinowitch H.D., Currah L., editors. Allium Crop Science: Recent Advances. CABI Publ; Wallingford: 2002. pp. 5–30. [Google Scholar]
- Fritsch R., Matin F., Klaas M. Allium vavilovii Popov et Vved. and a new Iranian species are the closest among the known relatives of the common onion A. cepa L. (Alliaceae) Genet. Resour. Crop Evol. 2001;48:401–408. [Google Scholar]
- Greiner S., Lehwark P., Bock R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019;47:W59–W64. doi: 10.1093/nar/gkz238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurushidze M., Mashayekhi S., Blattner F.R. Phylogenetic relationships of wild and cultivated species of Allium section Cepa inferred by nuclear rDNA ITS sequence analysis. Plant Syst. Evol. 2007;269:259–269. [Google Scholar]
- Hanelt P. Zur Taxonomie, Chorologie und Ökologie der Wildarten von Allium L. sect. Cepa (Mill.) Prokh. Flora. 1985;176:99–116. [Google Scholar]
- Hanelt P. Taxonomy, evolution and history. In: Rabinowitch H.D., Brewster J.L., editors. Onion and Allied Crops, Volume I. Botany, Physiology and Genetics. CRC press; Boca Raton, Florida: 1990. pp. 1–26. [Google Scholar]
- Havey M.J. Restriction enzyme analysis of the chloroplast and nuclear 45s ribosomal DNA of Allium sections Cepa and Phyllodolon (Alliaceae) Plant Syst. Evol. 1992;183:17–31. [Google Scholar]
- Jansen R.K., Ruhlman T.A. Genomics of Chloroplasts and Mitochondria. 2012. Plastid genomes of seed plants; pp. 103–126. [Google Scholar]
- Katoh K., Misawa K., Kuma K.I. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M., Moir R., Wilson A. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly J.W., Havey M.J. Sequence analysis of a chloroplast intergenic spacer for phylogenetic estimates in Allium section Cepa and a PCR-based polymorphism detecting mixtures of male-fertile and male-sterile cytoplasmic onion. Theor. Appl. Genet. 2001;102:78–82. [Google Scholar]
- Moore M.J., Soltis P.S., Bell C.D. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc. Natl. Acad. Sci. U. S. A. 2010;107:4623–4628. doi: 10.1073/pnas.0907801107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylander J.A., Ronquist F., Huelsenbeck J.P., Nieves-Aldrey J.L. Bayesian phylogenetic analysis of combined data. Syst. Biol. 2004;53:47–67. doi: 10.1080/10635150490264699. [DOI] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Shaw J., Shafer H.L., Leonard O.R. Chloroplast DNA sequence utility for the lowest phylogenetic and phylogeographic inferences in angiosperms: the tortoise and the hare IV. Am. J. Bot. 2014;101:1987–2004. doi: 10.3732/ajb.1400398. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analysis with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- van Raamsdonk L.W.D., de Vries T. Biosystematic studies in Allium L. Section cepa. Bot. J. Linn. Soc. 1992;109:131–143. [Google Scholar]
- van Raamsdonk L.W.D., Wiestma W.A., De Vries J.N. Crossing experiments in Allium L. Section Cepa. Bot. J. Linn. Soc. 1992;109:193–303. [Google Scholar]
- van Raamsdonk L.W.D., Vrielink-van Ginkel M., Kik C. Phylogeny reconstruction and hybrid analysis in Allium subgenus Rhizirideum. Theor. Appl. Genet. 2000;100:1000–1009. doi: 10.1007/s00122-003-1335-8. [DOI] [PubMed] [Google Scholar]
- van Raamsdonk L.W.D., Ensink W., van Heusden A.W. Biodiversity assessment based on cpDNA and crossability analysis in selected species of Allium subgenus Rhizirideum. Theor. Appl. Genet. 2003;107:1048–1058. doi: 10.1007/s00122-003-1335-8. [DOI] [PubMed] [Google Scholar]
- Vosa G.C. Heterochromatic patterns in Allium. I. The relationships between the species of the Cepa group and its allies. Heredity. 1976;36:383–392. [Google Scholar]
- Wendelbo P. Flora Iranica. K.H. Rechinger. Austria; 1971. Alliaceae; pp. 1–123. [Google Scholar]
- Yamashita K., Tashiro Y. Possibility of developing a male sterile line of shallot (Allium cepa L. Aggregatum Group) with cytoplasm from A. galanthum Kar. et Kir. J. Japan. Soc. Hort. Sci. 1999;68:256–262. [Google Scholar]
- Yusupov Z., Deng T., Liu C. The complete chloroplast genome of Allium fistulosum. Mitochondr. DNA B. 2019;4:489–490. [Google Scholar]