Abstract
The sphingolipid ceramide-1-phosphate (C1P) directly binds to and activates group IVA cytosolic phospholipase A2 (cPLA2α) to stimulate the production of eicosanoids. Because eicosanoids are important in wound healing, we examined the repair of skin wounds in knockout (KO) mice lacking cPLA2α and in knock-in (KI) mice in which endogenous cPLA2α was replaced with a mutant form having an ablated C1P interaction site. Wound closure rate was not affected in the KO or KI mice, but wound maturation was enhanced in the KI mice compared to that in wild-type controls. Wounds in KI mice displayed increased infiltration of dermal fibroblasts into the wound environment, increased wound tensile strength, and a higher ratio of type I:type III collagen. In vitro, primary dermal fibroblasts (pDFs) from KI mice showed substantially increased collagen deposition and migration velocity compared to pDFs from wild-type and KO mice. Additionally, KI mice showed an altered eicosanoid profile of reduced proinflammatory prostaglandins (PGE2 and TXB2) and an increased abundance of certain hydroxyeicosatetraenoic acid (HETE) species. Specifically, an increase in 5-HETE enhanced dermal fibroblast migration and collagen deposition. This gain-of-function role for the mutant cPLA2α was also linked to the relocalization of cPLA2α and 5-HETE biosynthetic enzymes to the cytoplasm and cytoplasmic vesicles. These findings demonstrate the regulation of key wound-healing mechanisms in vivo by a defined protein-lipid interaction and provide insights into the roles that cPLA2α and eicosanoids play in orchestrating wound repair.
INTRODUCTION
Wound healing is a dynamic process involving four main phases during which various types of cells and signaling molecules interact to complete the healing process in a timely and orderly fashion. The four distinct, yet overlapping, phases that make up the healing process for skin wounds, but are generally shared by repair processes in other tissues, are hemostasis, inflammation, proliferation, and remodeling (1, 2). During hemostasis, clotting factors work in tandem with blood vessel constriction to decrease bleeding (3). As the hemostasis phase ends, the wound transitions into the inflammatory phase, which is characterized by several hallmark events: vasodilation, increased vascular permeability, and neutrophil and macrophage infiltration into the wound site (4–7). After the wound has been cleared of any foreign debris and is properly prepped by the immune cells, followed by emigration of macrophages and inflammation resolution, the proliferation phase begins, at which point fibroblasts and keratinocytes migrate into the wound, proliferate, and initiate wound closure. Additionally, during this phase, angiogenesis occurs (8–10). The final phase of the healing process is remodeling, which involves the formation of new epithelial layers as well as increased deposition and crosslinking of collagen and the removal of excess extracellular matrix by fibroblast-derived matrix metalloproteinases (11–14). After completion of these processes, healed skin wounds have approximately 80% of the original tissue strength (15–17).
Here we focused on the role of eicosanoids in wound healing. Eicosanoids are oxidized derivatives of arachidonic acid (AA), eicosapentaenoic acid (EPA), and docosahexaenoic acid (DHA); they include prostaglandins, leukotrienes, hydroxyeicosatetraenoic acids (HETEs), prostacyclins, and thromboxanes, as well as lipid mediators such as Resolvins and Maresins, which are derived from 3-polyunsaturated fatty acids (PUFAs) (18–20). These bioactive lipids have emerged as key players in the normal wound-healing process. Indeed, a manifestation of impaired wound healing is the development of an imbalance between pro- and antiinflammatory lipids such as the arachidonic acid (AA)-derived eicosanoids (21). For example, prostaglandin E2 (PGE2) has been implicated in the regulation cell migration and meandering during the healing process of acute wounds (22). Leukotrienes are another class of eicosanoid that play a major role in the wound healing process, with leukotriene B4 (LTB4) being a prime example. In wound healing, the primary role of LTB4 is to recruit neutrophils to areas containing damaged tissue(23), one of the initial phases of wound healing.
Along with the healing of acute wounds, bioactive lipids reportedly play important roles in the impaired healing of chronic wounds. For example, the excess inflammation associated with the impaired healing of pressure ulcers correlates with high amounts of pro-inflammatory eicosanoids, such as leukotriene D4 (LTD4), leukotriene E4 (LTE4), and PGE2(21). Blocking eicosanoid production by cyclooxygenase 2 (COX-2) reduces inflammation and improves the healing of pressure ulcers in mice(24). Localized excess of PGE2 has also been linked to delayed wound healing in LIGHT −/− mice, which exhibit prolonged inflammation due to the lack tumor necrosis factor superfamily member 14, and to the inhibition of human fibroblast function and migration (18, 25). Furthermore, PGE1, an anti-inflammatory prostaglandin opposes the pro-inflammatory action of PGE2, is FDA-approved to treat human wounds(26). Clinical reports indicate that the bioactive fraction in platelet-rich plasma for wound therapy is a lipid component and that nutritional formulations containing pro-resolution lipids decrease the progression of pressure ulcers(27). Together, these findings support key and distinct roles for eicosanoids in the wound healing process and in its dysregulation or stalling. However, therapeutic treatments to modulate these pathways in the clinical setting to improve healing outcomes remain limited.
The synthesis of eicosanoids begins with an initial rate-limiting step – the generation of AA by a phospholipase A2 (PLA2) (28, 29). One of the major PLA2s involved in this initial step is group IVA cytosolic PLA2 (cPLA2α). Previously, our laboratory demonstrated that the translocation of cPLA2α to intracellular membranes, as well as AA release and production of eicosanoids, depended upon the sphingolipid ceramide-1-phosphate (C1P) interacting with the N-terminal lipid-binding C2 domain of cPLA2α at residue Arg59 and possibly at Arg57 and Lys58. (30–33) Later studies suggested that both Arg61 and His62 were also key residues for this interaction and for the cPLA2α translocation to membranes (34). Our laboratory previously reported that C1P derived from ceramide kinase (CERK) negatively regulates the migration of murine embryonic fibroblasts. Furthermore, C1P levels increase during the inflammatory phase, but are followed by a concomitant decrease during the proliferation and/or remodeling stage in human wound healing using the subcutaneuous, polytetrafluoroethylene (PTFE)-insertion model (35–37).
Based on the preceding findings, we hypothesized that the C1P-cPLA2α interaction regulates wound repair and regeneration. In this study, we tested this premise using a novel genetic mouse model, and we found that this lipid-protein interaction negatively regulated the later phases of wound healing. Thus, this study shows a defined lipid-protein interaction regulating a biological phenotype in an in vivo mammalian model, and our data suggest a previously unrecognized mechanistic paradigm for cPLA2α in shifting cell function depending on the association of the enzyme with specific lipid mediators.
RESULTS
Ablation of the C1P-cPLA2α interaction does not affect closure rate of acute wounds
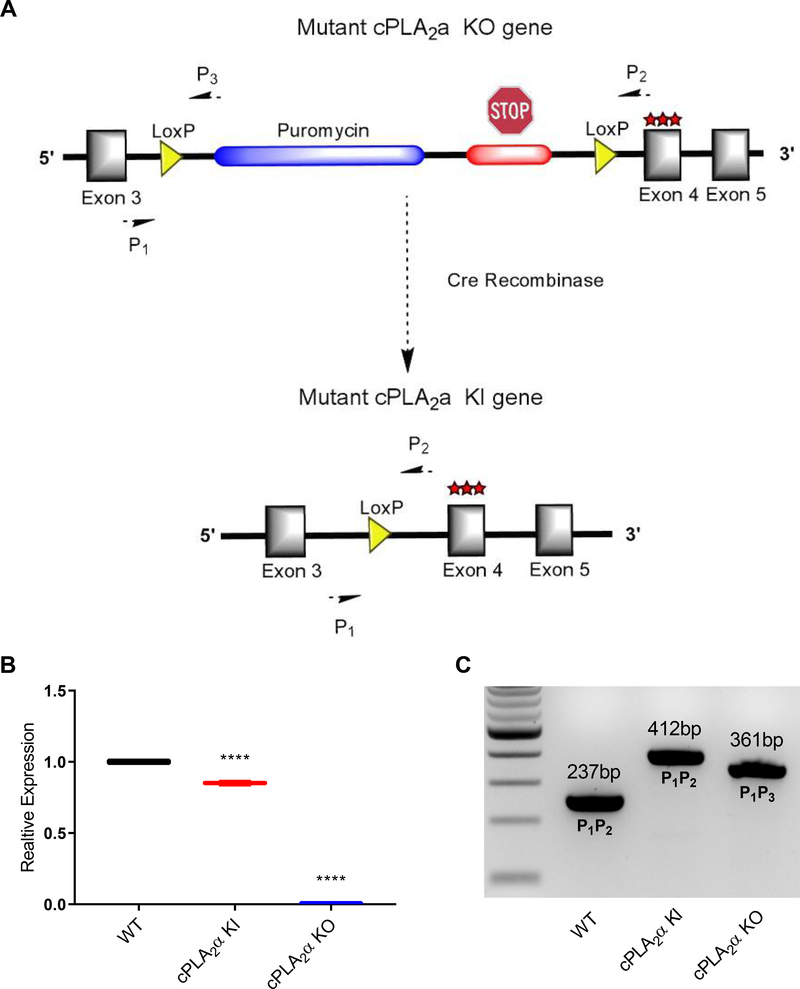
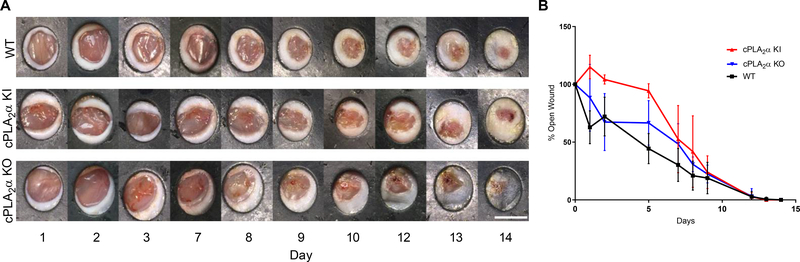
In order to study the role of the C1P-cPLA2α interaction in vivo, we generated both cPLA2α knockout (KO) mice that lacked cPLA2α in all tissues and cPLA2α knock-in (KI) mice that lacked the C1P interaction site in cPLA2α in all tissues (Fig. 1A). This mouse model was designed using a construct with an inserted cassette containing puromycin resistance, a premature stop codon flanked by loxP sites, and three mutated amino acids (R57A, K58A, R59A) in the endogenous cPLA2 locus (Fig. 1A). This design enabled the generation of cPLA2α KO mice producing the truncated protein in all tissues and cPLA2α KI mice by the addition of cytomegalovirus (CMV)-driven expression of CRE recombinase. KOs and KIs were confirmed by evaluating cPLA2α mRNA expression and genotyping (Fig. 1, B and C). Using these new genetic models, we examined acute wound healing in wild-type, cPLA2α KI, and cPLA2α KO mice using a punch-biopsy model as reported by Wang and co-workers (38). We observed no significant differences in the wound closure rate between the wild-type, cPLA2α KI, and cPLA2α KO mice (Fig. 2, A and B).
Fig. 1. cPLA2α KI and KO mice.
(A) Schematic representation of the cassette inserted into intron 3 of the endogenous cPLA2a locus. The cassette contains a puromycin resistance gene followed by a premature stop codon flanked by loxP sites. Insertion of the cassette causes cPLA2α KO. The cassette also contains a triple mutation at the C1P binding site (R57A, K58A, R59A) encoded in exon 4. CRE-mediated recombination excises the puromycin resistance gene and premature stop codon, generating KI mice that produce cPLA2α with the mutated C1P binding site instead of wild-type cPLA2α. (B) RNAs from primary dermal fibroblasts from WT, KI, and KO mice were converted to cDNA and used for quantitative PCR analysis using primers specific to cPLA2a. Samples were compared using ANOVA followed by Tukey’s post-hoc test. Data shown are means ± SD, n = 3–6 mice per genotype, ****P< 0.0001. (C) Genotyping of WT, KI, and KO mice using end point multiplex PCR using primers specific for the WT, KI, and KO alleles.
Fig. 2. Ablation of the C1P-cPLA2α interaction does not affect the wound closure rate in vivo.
(A) Images of acute wounds over 14 days in WT, cPLA2α KI, and cPLA2α KO mice using a previously described stinted excisional wound model (38). Scale bar, 6mm. (B) Wound closure rate was calculated as percent-opened wound for WT, KI, and KO mice over course of 14 days. Samples were compared using ANOVA followed by Tukey’s post-hoc test. Data shown are means ± SD, n = 4–8 mice per genotype.
Ablation of the C1P-cPLA2α interaction increases fibroblast migration and accelerates wound maturation
The lack of a significant effect on the rate of in vivo wound closure was not entirely unexpected, because the closure rate for acute wounds in mice depends more on contraction-based mechanisms than it does on cell proliferation–based mechanisms, in contrast to humans (39–41). We next examined the wounds for other healing processes that are similar in mice and humans, such as chemotactic responses, fibroblast migration, and wound maturation and remodeling (42, 43). Histological evaluations of healed wounds were performed in wild-type, cPLA2α KI, and cPLA2α KO mice. Masson’s trichrome stain of healed wounds demonstrated a marked increase in cell infiltration into the healed wounds of cPLA2α KI mice compared to both wild-type and cPLA2α KO mice (Fig. 3A).
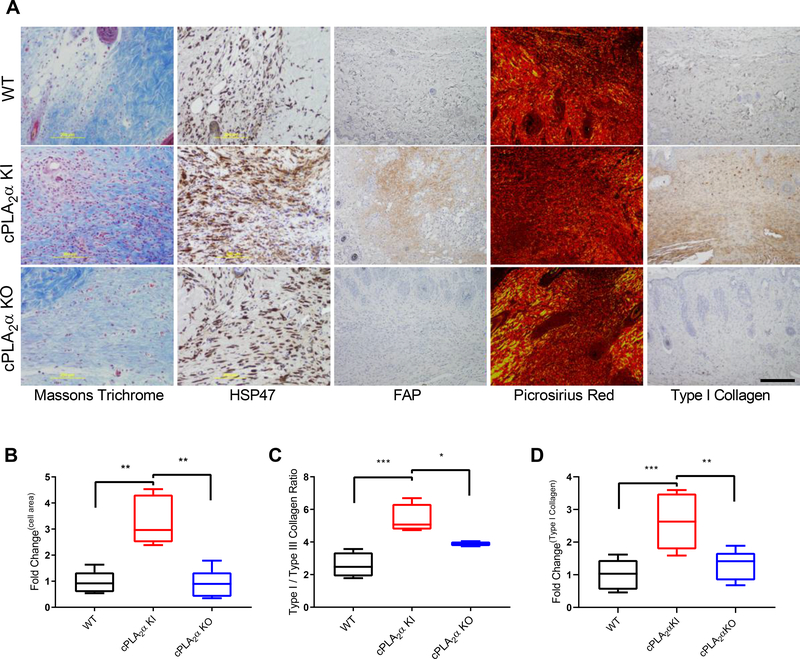
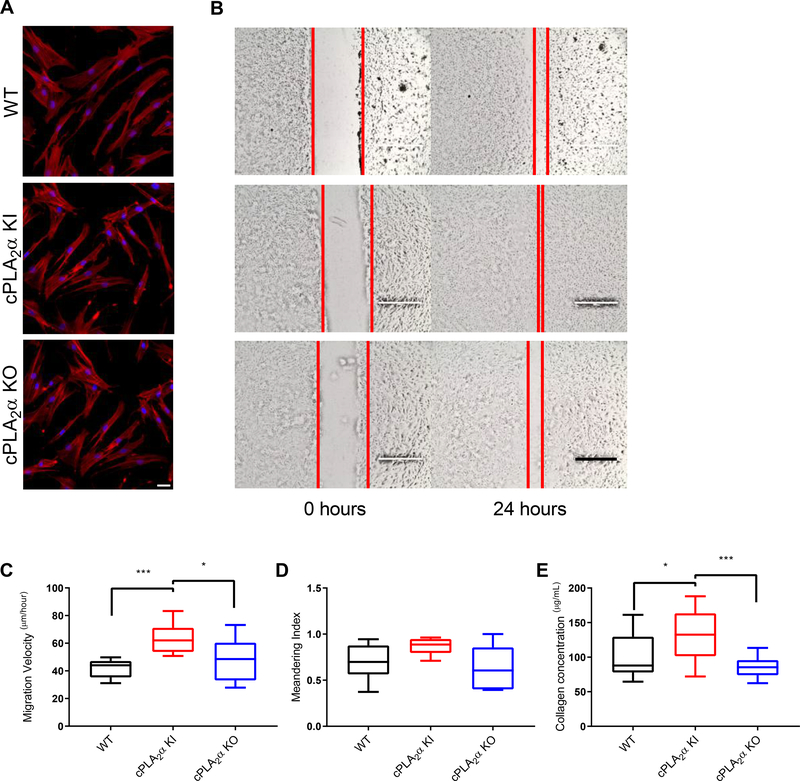
Fig. 3. Fibroblast infiltration and collagen deposition are increased in wounds in KI mice.
(A) Representative image showing Masson’s trichrome stain, HSP47 distribution, FAP distribution, Picrosirius red staining, and Type I collagen distribution in wound sections from WT, cPLA2α KI, and cPLA2α KO mice 10 days after excisional wounding. Scale bar, 200μm. (B) Graphical representation of FAP+ cells per field. The values are presented relative to that for WT, which was set to 1. (C) Graphical representation of the ratio of type I to type III collagen estimated from polarized light imaging of Picrosirius red –stained wound sections. (D) Graphical representation of the surface area that was positive for type I collagen in wound sections. The values are presented relative to that for WT, which was set to 1. Samples were compared using ANOVA followed by Tukey’s post-hoc test. Data shown are means ± SD, n = 4–7 mice per genotype, *P< 0.05, **P< 0.01, ***P< 0.001, ****P< 0.0001.
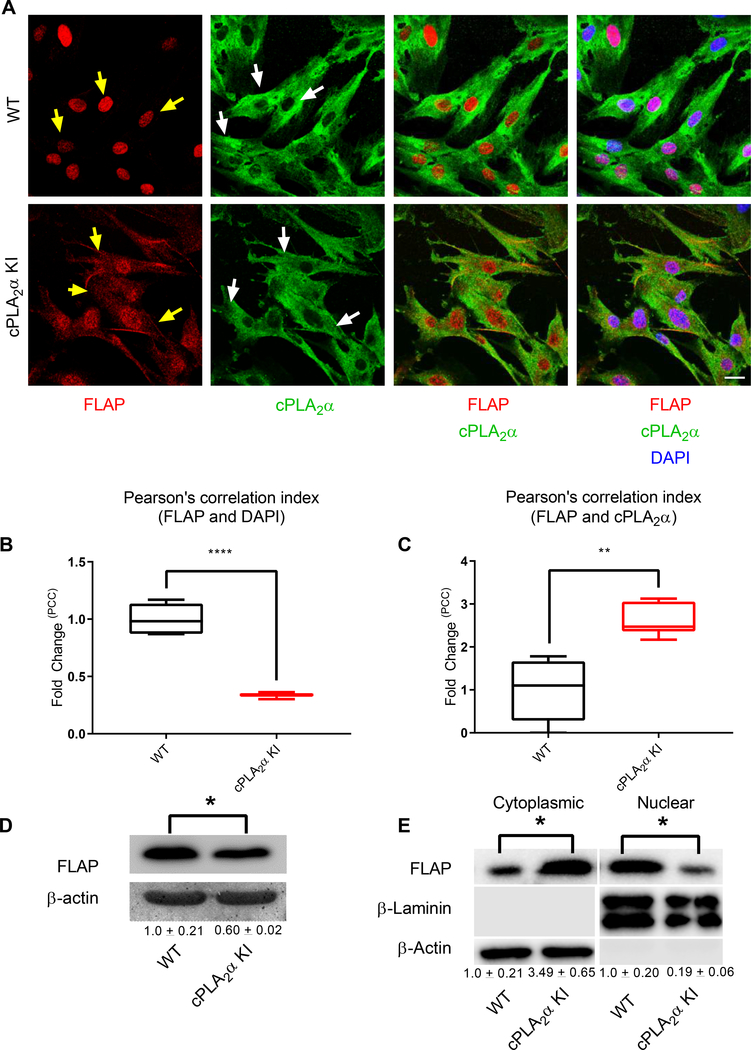
By day 10, the wound is normally expected to be undergoing both the proliferation and remodeling phases of wound healing, which involves the infiltration and proliferation of dermal fibroblasts (2, 44). In order to determine the cell type(s) infiltrating into the healed wounds of cPLA2α KI mice, we analyzed the immunohistochemical fibroblast marker fibroblast activation protein (FAP) and the collagen chaperone heat shock protein 47 (HSP47) in sections of wounded tissue. The healed wounds of cPLA2α KI mice contained increased numbers of dermal fibroblasts (Fig. 3, A and B). These data indicate that the C1P-cPLA2α interaction acts as a negative regulator of fibroblast migration into the wound environment.
To determine whether the increased numbers of dermal fibroblasts observed in the wounds of the cPLA2α KI mice affected wound repair and maturation negatively or positively, we examined collagen deposition by Picrosirius red staining, which under polarized light can reflect the amounts of Type I and Type III collagen (45). Using this method, a marked increase in the ratio of type I to type III collagen in the healed wounds of cPLA2α KI mice was observed (Fig. 3, A and C). Because the Picrosirius red methodology may have limited accuracy and reflect collagen density and packing rather than collagen composition, we confirmed our results by immunostaining wound sections for type I collagen. Significant increases in type I collagen were observed in wounds of cPLA2α KI mice compared to those of wild-type and cPLA2α KO mice (Fig. 3, A and D), which correlates with enhanced wound maturation (46, 47). Thus, our data demonstrate that the loss of C1P-cPLA2α interaction positively impacted acute wound healing.
Ablation of the C1P-cPLA2α interaction enhances wound remodeling and tensile strength
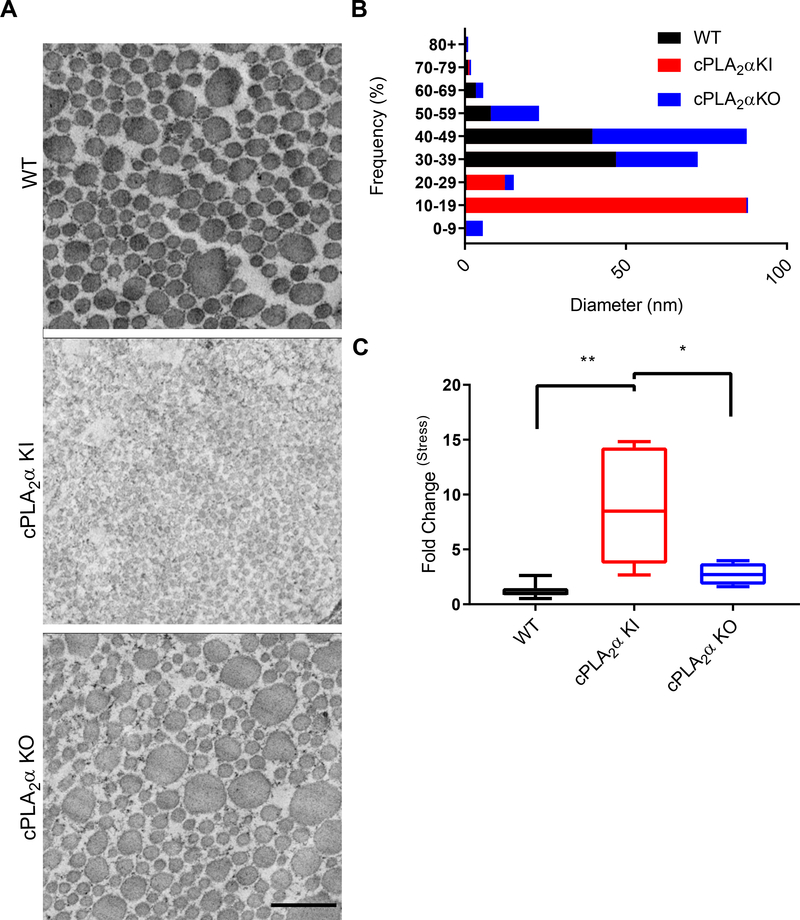
To determine whether the loss of the C1P-cPLA2α interaction affected the orderly deposition and cross-linking of collagen fibrils, we used electron microscopy to examine collagen fiber diameters in wounds from wild-type, cPLA2α KI, and cPLA2α KO mice (Fig. 4, A and B). Whereas the diameter of collagen fibers ranged from 30 to 49 nm in wounds of both cPLA2α KO and wild-type mice, it was reduced to a range of 10 to 19 nm in wounds from cPLA2α KI mice.
Fig. 4. Healed wounds from KI mice have thinner collagen fibers and increased tensile strength.
(A) Transmission electron microscopy (TEM) showing collagen fibers in stinted excisional wounds from WT, cPLA2α KI, and cPLA2α KO mice after 10 days of healing. Scale bar, 500 nm. (B) Graphical representations of collagen fiber diameter measured from TEM images. (C) Quantification of wound tensile strength in 2 cm incisional wounds from WT, KI, and KO mice 10 days after wounding. The values are presented relative to that for WT, which was set to 1. Samples were compared using ANOVA followed by Tukey’s post-hoc test. Data shown are means ± SD, n = 2–3 mice per group for TEM, n = 4–7 mice per group for wound tensile strength, *P< 0.05, **P< 0.01.
Because the tensile strength of healed wound tissue depends on type I collagen deposition and collagen fiber diameter, with many small fibers being stronger than fewer large fibers (48–50), we next examined the wound tensile strength in wild-type, cPLA2α KI, and cPLA2α KO mice. Healed wounds of cPLA2α KI mice exhibited significantly increased tensile strength compared to wounds from wild-type and cPLA2α KO mice (Fig. 4C). Taken together, these data indicate that genetic ablation of the C1P-cPLA2α interaction enhanced maturation, remodeling, and repair of acute wounds in mice.
Ablation of the C1P-cPLA2α interaction enhances the migration of primary dermal fibroblasts
Because the enhanced wound maturation phenotype was linked to enhanced fibroblast infiltration and deposition of Type I collagen, we next determined whether these features were recapitulated in vitro with primary dermal fibroblast (pDFs) and whether the cells exhibited phenotypes consistent with mediating the observed in vivo phenotypes. We observed no phenotypic differences in cell morphology or actin fiber distribution in pDFs from the three genotypes (Fig. 5A). However, pDFs from cPLA2α KI mice exhibited significantly increased migration velocity compared to pDFs from wild-type and cPLA2α KO mice (Fig. 5, B and C). Additionally, we examined the movement patterns of pDFs from wild-type, cPLA2α KI, and cPLA2α KO mice using a meandering assay, which calculated the meandering index (measurement of displacement of the cell from the point of origin at the end point of the experimental run and dividing that distance by the track length) (35). We observed no significant difference in meandering between the genotypes (Fig. 5D). Lastly, collagen deposition in pDFs from cPLA2α KI mice was significantly increased compared to pDFs from cPLA2α KO and wild-type mice (Fig. 5E). The data indicate that the C1P-cPLA2α interaction negatively regulated pDF migration and collagen deposition, but did not play a substantial role in the migration polarity of pDFs.
Fig. 5. Loss of the C1P-cPLA2α interaction causes an increase in migration velocity and collagen deposition in primary dermal fibroblasts.
(A) Actin distribution in primary dermal fibroblasts (pDFs) collected from WT, cPLA2α KI, and cPLA2α KO mice. Scale bar, 40μm. (B) Monolayers of pDFs from WT, KI, and KO mice were scratch-wounded and followed for 24 hours. Red lines indicate the borders of the scratch wound. Scale bar, 200μm. (C) Migration velocity of pDFs from WT, KI, and KO mice after scratch-wounding. (D) Quantification of cell meandering in scratch-wounded pDFs from WT, KI, and KO mice. (E) Collagen deposition of pDFs from WT, KI, and KO mice. Samples were analyzed for type I collagen levels using an ELISA-based assay for soluble type I collagen. Data were compared using ANOVA followed by Tukey’s post-hoc test. Data shown are means ± SD, n = 6–12 cell isolates per genotype (5–6 mice per genotype were utilized to generate the cell isolates) for migration and meandering assays, n = 15–16 cell isolates per genotype (8–10 mice per genotype were used to generate the cell isolates) for collagen deposition assay, *P< 0.05, **P< 0.01, ***P< 0.001, ****P< 0.0001.
Ablation of the C1P-cPLA2α interaction alters the lipid profile in primary dermal fibroblasts
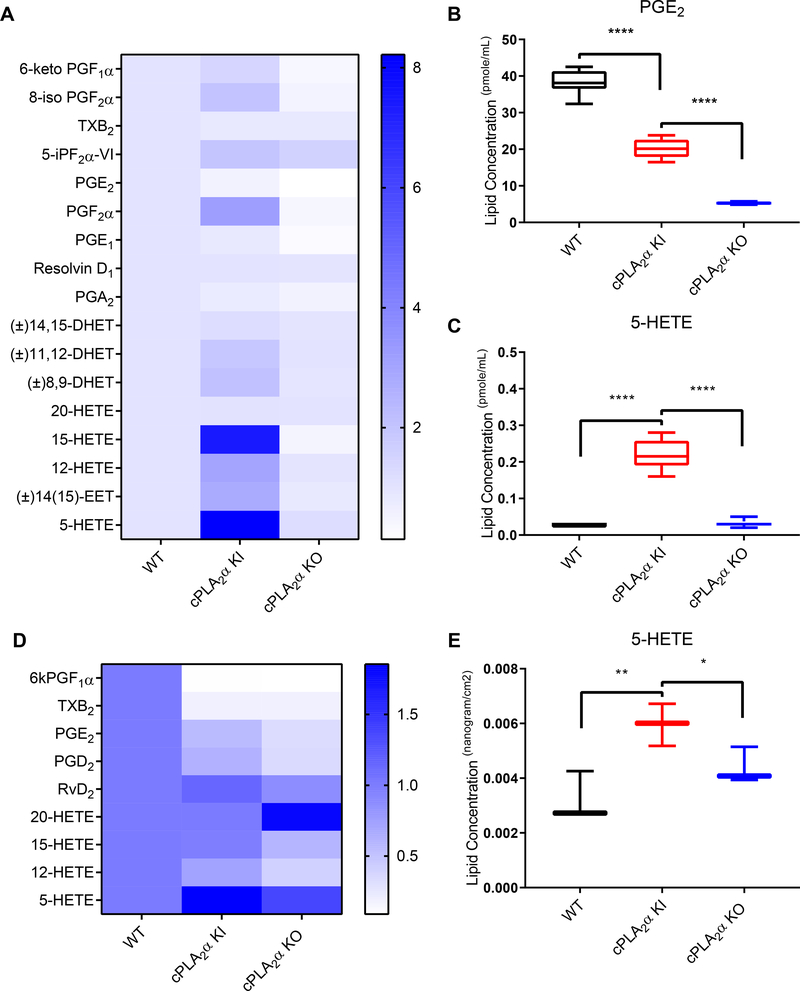
Because of the known involvement of eicosanoids in regulating fibroblast migration (22, 23, 35, 51, 52), we performed a targeted lipidomics screen on pDFs that had been subjected to scratch-wounding utilizing ultra performance liquid chromatography (UPLC) electrospray ionization coupled to tandem mass spectrometry (ESI-MS/MS). We observed significant decreases upon wounding in several AA-derived eicosanoids, including PGE2, in pDFs with the ablated C1P-cPLA2α interaction compared to wounded pDFs from wild-type mice (Fig. 6, A and B, table S1). Similar findings were observed in wounded pDFs from cPLA2α KO mice compared to wounded pDFs from wild-type mice (Fig. 6, A and B, table S1). Significant increases in the amounts of several HETES, including 5-HETE, were observed in cPLA2α KI pDFs when wounded compared to both wild-type and cPLA2α KO pDFs (Fig. 6, A and C, table S1). These data agreed with lipid profiles from wound tissue regarding the significant increase in 5-HETE and the decrease in PGE2 in acute wounds of cPLA2α KI mice at 10 days after wounding as compared to wound tissue from wild-type and cPLA2α KO mice (Fig. 6, D and E; table S2). These data support the hypothesis that the C1P-cPLA2α interaction was both a positive and negative regulator of eicosanoid production pathways.
Fig. 6. Genetic ablation of the C1P-cPLA2α interaction alters eicosanoid synthesis in primary dermal fibroblasts and healed excisional wounds.
(A) Complete lipid profile of pDFs from WT, cPLA2α KI, and cPLA2α KO mice shown as fold change in comparison to WT pDFs. (B) Quantification of PGE2 in pDFs from WT, KI, and KO mice. (C) Quantification of 5-HETE in pDFs from WT, KI, and KO mice. (D) Partial lipid profiles from excisional wounds in WT, KI, and KO mice. (E) Quantification of 5-HETE in excisional wounds from WT, KI, and KO mice. Samples were compared using ANOVA followed by Tukey’s post-hoc test. Data shown are means ± SD, n = n = 6–8 cell isolates per genotype (4–5 mice per genotype were used to generate the cell isolates) for (A–C), n = 3 mice (one wound per mouse) per genotype in (D, E). *P< 0.05, **P< 0.01, ***P< 0.001, ****P< 0.0001.
To determine whether the altered eicosanoid profile was in fact due to the mutant cPLA2α and not due to the induction of a different PLA2 pathway, we also performed lipid profiling of pDFs treated with the selective cPLA2α inhibitor pyrrophenone. Pyrrophenone blocked the increased 5-HETE abundance observed in cPLA2α KI pDFs without significantly affecting HETE levels in wild-type and cPLA2α KO pDFs (table S3). Thus, the increase in 5-HETE abundance observed in cPLA2α KI pDFs was due to the ablation of the C1P-cPLA2α interaction and not due to an artifactual adaptation leading to activation of a compensatory PLA2 system such as Ca2+-independent PLA2 (iPLA2).
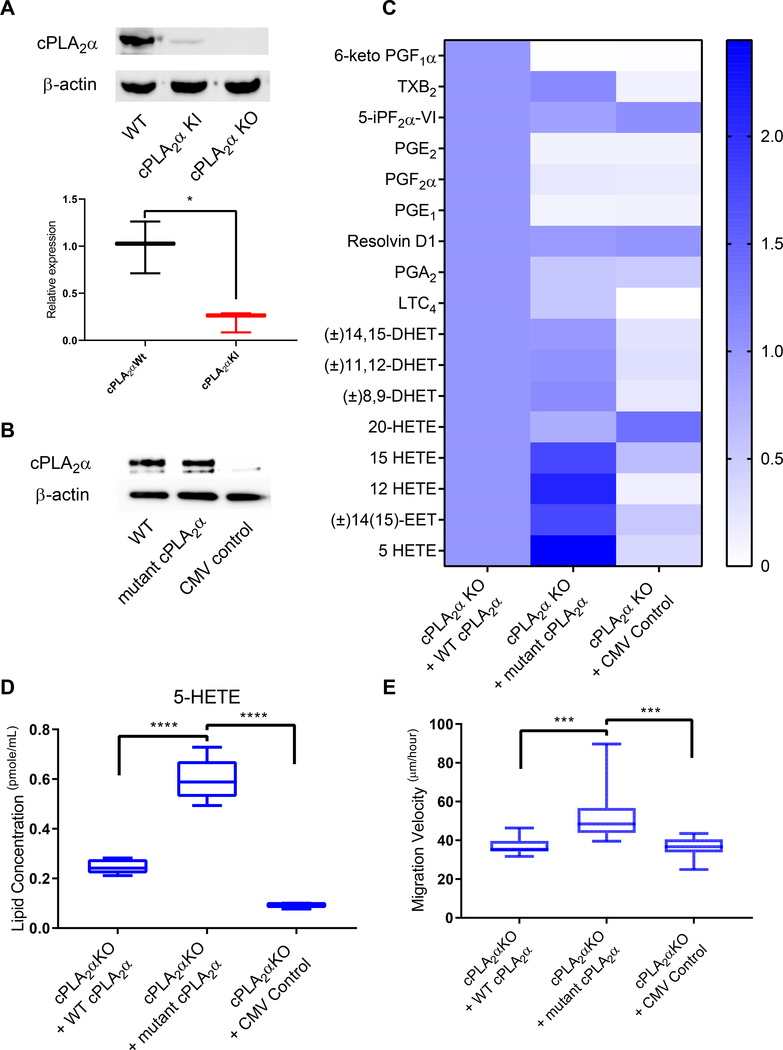
Unexpectedly, we found that the abundance of the mutant cPLA2α protein in cPLA2α KI pDFs was highly reduced compared to the amount of cPLA2α in wild-type pDFs (Fig. 7A), whereas mRNA abundance was only mildly, though significantly, reduced by 15% (Fig. 1B). To determine whether the altered eicosanoid profile was due to the ablation of the C1P-cPLA2α interaction and induction of a “gain of function” for the enzyme versus reduced protein abundance, we ectopically-expressed wild-type or mutant cPLA2α (R57A, K58A, R59A) in cPLA2α KO pDFs (Fig. 7B). Expression of mutant cPLA2α in cPLA2α KO pDFs induced the same eicosanoid profile as that observed in cPLA2α KI pDFs with even greater enhancement of 5-HETE production (Fig. 7, C and D). The expression of wild-type cPLA2α in cPLA2α KO pDFs rescued the eicosanoid profile to that of wild-type pDFs (Fig. 7C and D). Mutant cPLA2α expression also rescued the migration phenotype of cPLA2α KO pDFs (Fig. 7E). These data confirm that the C1P-cPLA2α interaction not only a positive regulated a large number of inflammatory eicosanoids (Fig. 6, A and B; Fig. 7, C and D; table S1), but also acted as a negative regulator of the production of 5-lipoxygenase–derived lipid mediators such as 5-HETE (Fig. 6, A and C; Fig. 7, C and D; table S1). Our data strongly suggest that ablation of the C1P-cPLA2α interaction leads to an increase in the enzymatic activity of cPLA2α, thus stimulating 5-HETE biosynthesis in pDFs.
Fig. 7. Expression of mutant cPLA2α reproduces the KI phenotype in KO pDFs.
(A) Immunoblotting and quantification of cPLA2α in pDFs from WT, cPLA2α KI, and cPLA2α KO mice. β-actin is a loading control. n = 3 cell isolates per genotype (3–4 mice per genotype were utilized to generate the cell isolates). (B) Immunoblotting for cPLA2α in cPLA2α KO pDFs in which an adenoviral (CMV) expression system (30) was used to drive expression of WT cPLA2α or C1P-binding mutant cPLA2α. Empty vector is a negative control. Data is representative of n = 3 independent experiments using cells isolated from different cPLA2α KO mice. (C) Lipid profiles of WT and KI pDFs expressing WT or mutant cPLA2α from the CMV vector. (D) Quantification of 5-HETE in KO pDFs expressing WT or mutant cPLA2α from the CMV vector. For (C) and (D), n = 3 (empty CMV control) and n = 9 (WT and mutant cPLA2α) cell isolates from 5 cPLA2α KO mice. (E) Migration velocity in KO pDFs expressing WT or mutant cPLA2α from the CMV vector. n = 12 cell isolates per genotype (6–9 mice per genotype were used to generate the cell isolates). Samples were compared using ANOVA followed by Tukey’s post-hoc test. Data shown are means ± SD*P< 0.05, **P< 0.01, ***P< 0.001, ****P< 0.0001.
5-HETE drives the cPLA2α KI migration phenotype
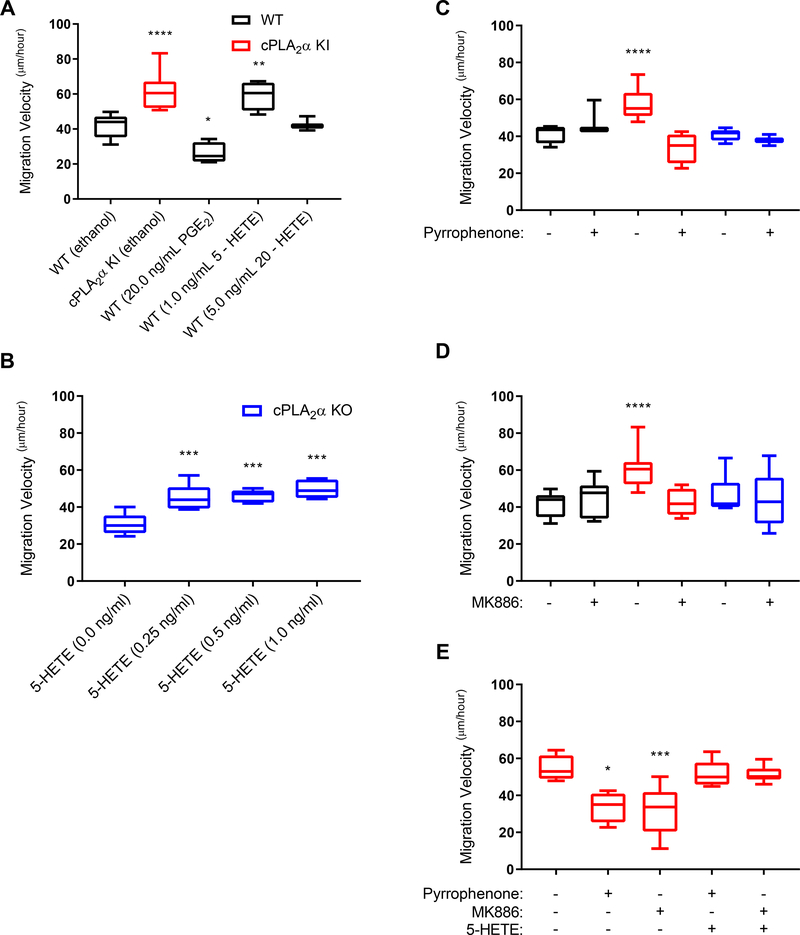
To determine whether the increased production of specific HETE species was responsible for the enhanced migration of cPLA2α KI pDFs, we exogenously applied specific eicosanoids and small molecule inhibitors of pertinent biosynthetic enzymes to cells. The addition of 5-HETE, but not 20-HETE, to wild-type pDFs recapitulated the increased migration velocity phenotype of cPLA2α KI pDFs (Fig. 8A), and it did so at a concentration only moderately above that measured in cPLA2α KI pDFs (Fig. 6, C, table S1). We hypothesized that the production of higher PGE2 levels in wild-type pDFs as compared to cPLA2α KO and cPLA2α KI pDFs necessitates higher levels of 5-HETE to increase the migration velocity because PGE2 is a known inhibitor of fibroblast migration (Fig. 8A). To test this idea, pDFs from cPLA2α KO mice, which produce low amounts of PGE2, were treated with various concentrations of 5-HETE. In contrast to wild-type pDFs, extremely low levels of 5-HETE enhanced the migration velocity of these pDFs to the same extent as that observed for cPLA2α KI pDFs (Fig. 8B). These data show that the loss of the C1P-cPLA2α interaction produced an optimal eicosanoid pattern for enhanced pDF migration by reducing the amounts of migration-suppressing eicosanoids, such as PGE2, while increasing the biosynthesis of pro-migratory lipid mediators like 5-HETE.
Fig. 8. Exogenous eicosanoids and small molecule inhibitors of cPLA2α and FLAP alter the migration velocity of dermal fibroblasts.
(A) Migration velocity of pDFs from WT and cPLA2α mice treated with ethanol or the indicated concentrations of PGE2, 5-HETE, or 20-HETE. (B) Migration velocity of pDFs from KO mice treated with increasing concentrations of 5-HETE. (C–E) Migration velocity of pDFs from WT, KI, and KO mice in the presence of pyrrophenone (C), MK886 (D), or the indicated combinations of pyrrophenone, MK886, and 5-HETE (E). Samples were compared using ANOVA followed by Tukey’s post-hoc test. Data shown are means ± SD, n = 3–10 cell isolates per genotype (3–8 mice per genotype were used to generate the cell isolates) for each treatment group. *P< 0.05, **P< 0.01, ***P< 0.001, ****P< 0.0001.
In control studies, pyrrophenone was again used to treat pDFs to show that the effects on migration velocity were due to the mutant cPLA2α and not activation of a different PLA2 pathway. Pyrrophenone blocked the enhanced migration velocity of cPLA2α KI pDFs but had no significant effect on wild-type and KO pDFs (Fig. 8C). The data support the finding that the enhanced migration velocity observed in the cPLA2α KI pDFs is due to a gain or shift in the enzymatic function of cPLA2α. To further confirm the role of 5-HETE in promoting pDF migration, we treated cells with MK886, an inhibitor of 5-lipoxygenase (5-LO)–activating protein (FLAP), to block 5-LO–dependent production of the biosynthetic precursor of 5-HETE. Whereas MK886 treatment had no significant effect on the migration velocity of wild-type and cPLA2α KO pDFs compared to untreated cells (Fig. 8D), it significantly decreased the migration velocity of cPLA2α KI pDFs to that of wild-type pDFs (Fig. 8D). Lastly, we treated cPLA2α KI pDFs with various combinations of pyrrophenone, MK886, and 5-HETE. 5-HETE restored the enhanced migration phenotype that was blocked by pyrrophenone and MK886 in cPLA2α KI pDFs (Fig. 8E). Together, the data support the conclusion that ablation of the C1P-cPLA2α interaction induced 5-HETE biosynthesis at the expense of PGE2 production, thus enhancing pDF migration.
cPLA2α KI dermal fibroblasts display altered colocalization of FLAP and cPLA2α
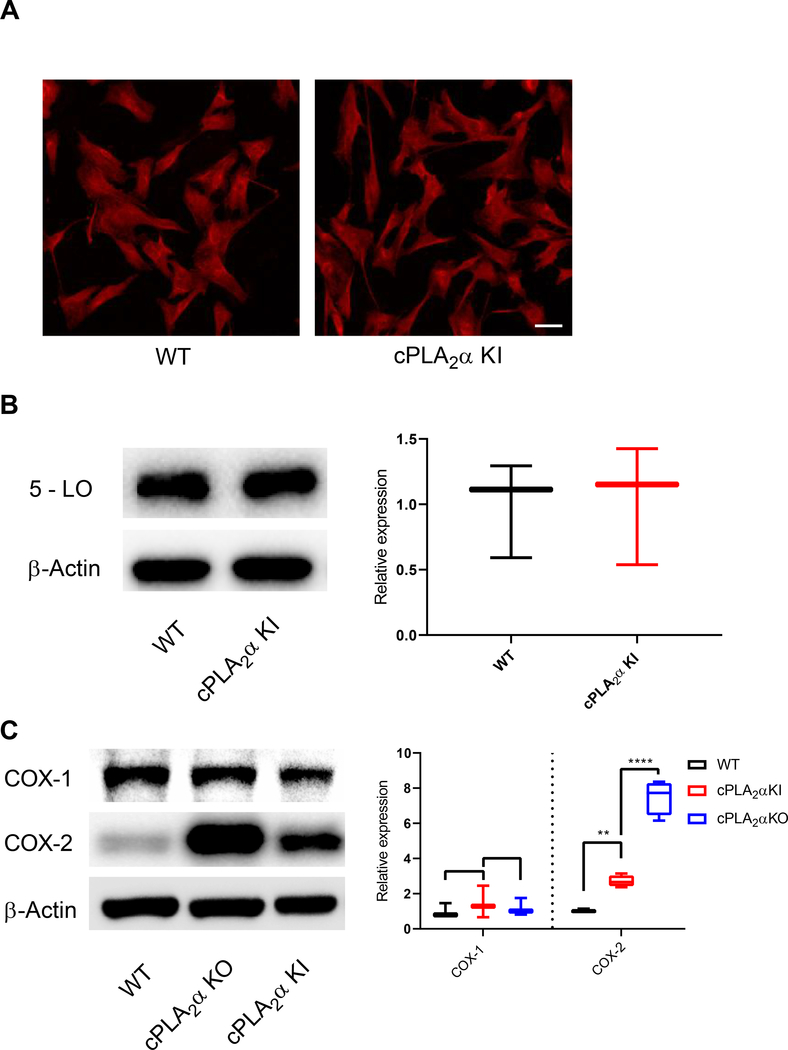
Previous work has demonstrated that both the abundance and localization of the biosynthetic enzymes play a key role in the production of both HETE and leukotriene (53) Thus, we began exploring the mechanism by which the mutant cPLA2α enzyme drove 5-HETE biosynthesis at the expense of inflammatory eicosanoids. Immunocytochemistry and confocal imaging revealed that both FLAP and cPLA2α were present in the cytoplasm in cPLA2α KI pDFs, in contrast to the strictly nuclear localization of FLAP and the Golgi and peri-nuclear localization of cPLA2α in wild-type pDFs (Fig. 9, A and B). Moreover, mutant cPLA2α and FLAP co-localized in the cytoplasm as vesiculated structures. cPLA2α KI pDFs, again, in contrast to their localization in wild-type pDFs (Fig. 9, A and C). Additionally, FLAP protein abundance was significantly reduced in the cPLA2α KI pDFs (0.60 ± 0.02) compared to wild-type pDFs (1.0 ± 0.21) (Fig. 9D). Immunoblotting of cytoplasmic and nuclear fractions from wild-type and cPLA2α KI pDFs confirmed the difference in localization for FLAP that was observed by confocal microscopy [cytoplasmic FLAP (normalized to β-actin): 1.0 ± 0.21 (wild-type) vs 3.49 ± 0.65 (cPLA2α KI); nuclear FLAP (normalized to β-laminin): 1.0 ± 0.20 (wild-type) vs 0.19 ± 0.06 (cPLA2α KI)] (Fig. 9E). To determine whether the abundance or localization of the biosynthetic enzymes involved in the secondary rate-limiting step in HETE and prostaglandin biosynthesis were affected, we examined the localization and abundance of 5-LO, COX-1, and COX-2. By confocal microscopy and Western blot analysis, we detected no significant differences in 5-LO localization or abundance between wild-type and cPLA2α KI pDFs (Fig. 10, A and B). Expression of COX-1 was not different between wild-type, cPLA2α KI, and cPLA2α KO pDFs, but expression of COX-2 increased in both cPLA2α KO and cPLA2α KI pDFs compared to wild-type cells (Fig. 10, C). These data demonstrate that the increase in 5-HETE biosynthesis in cPLA2α KI cells mutant likely arises from the cytoplasmic co-localization of FLAP with mutant form of cPLA2α that cannot bind to C1P rather than differential effects on the abundance and localization of the 5-LO and COX enzymes.
Fig. 9. Primary dermal fibroblasts from KI mice show altered localization of cPLA2α and FLAP.
(A) Immunofluorescence showing FLAP (nuclear and cytosolic, yellow arrows) and cPLA2α (Golgi/perinuclear and cytosolic, white arrows) in pDFs from WT and cPLA2α KI mice. Nuclei are labelled with DAPI. Scale bar, 40 μm. (B) Cytosolic localization of FLAP correlated to nuclear staining was quantified as fold-change in the Pearson’s Correlation Coefficient (PCC) in KI pDFs relative to that in WT pDFs. (C) C-localization of FLAP with cPLA2α was quantified as fold-change in the PCC in KI pDFs relative to that in WT pDFs. (D) Immunoblotting and quantification (numbers below blots) of FLAP in pDFs from WT and KI mice. (E) Immunoblotting and quantification of FLAP in nuclear and cytoplasmic fractions of WT and KI pDFs. Colocalization was determined using Pearson’s correlation index, samples were compared using unpaired students t-test with Welch’s correction. Data shown are means ± SD, n = 5–6 cell isolates per genotype (4–6 mice per genotype were used to generate the cell isolates) for confocal microscopy, n = 4 cell isolates per genotype (3 mice per genotype were used to generate the cell isolates) for immunoblot analysis, *P< 0.05, **P< 0.01, ***P< 0.001, ****P< 0.0001.
Fig. 10. Dermal fibroblasts from KI mice show no changes in the localization and abundance of 5-lipoxygenase.
(A) Immunofluorescence showing 5 - LO in pDFs from WT and cPLA2α KI mice. Scale bar, 40 μm. (B) Immunoblotting and quantification of 5 - LO in pDFs from WT and KI mice. (C) Immunoblotting and quantification of COX-1 and COX-2 in WT, KI, and KO pDFs. Data shown are means ± SD, n = 5–6 cell isolates per genotype (3–4 mice per genotype were used to generate the cell isolates) for confocal microscopy, n = 3 cell isolates per genotype (3 mice per genotype were used to generate the cell isolates) for immunoblot analysis, *P< 0.05, **P< 0.01, ***P< 0.001, ****P< 0.0001.
DISCUSSION
For the present study, we developed a genetic model to test whether the C1P-cPLA2α interaction plays an important role in the wound healing process. This new mouse model, which expresses cPLA2α with an ablated C1P interaction site instead of wild-type cPLA2α, was characterized by increased dermal fibroblast infiltration into the wound environment, enhanced progression through the wound healing stages as measured by increased Type I collagen deposition, reduced collagen fiber diameter, and enhanced wound strength. Altered eicosanoid profiles were also observed in pDFs and in the wounds themselves. Notably, the loss of inflammatory prostaglandins concomitant with an increase in 5-HETE increase was key for the altered fibroblast phenotypes. Our data support a role for the C1P-cPLA2α interaction during the inflammatory stage of wound healing as a negative regulator of the proliferation and remodeling stages. Indeed, C1P reportedly increases during the inflammatory stage of wound healing in human subjects, but then decreases during the proliferation and remodeling stages (54). Thus, blocking C1P production or the interaction of C1P with cPLA2α could be beneficial for “compressing” the inflammatory phase and accelerating the induction of the proliferation and remodeling phases. We therefore hypothesize that addition of exogenous 5-HETE or an inhibitor of the C1P-cPLA2α interaction may provide significant healing improvement. Such treatments may also be adaptable to chronic wounds, which fail to resolve and are stalled in the inflammatory stage due to continued association of cPLA2α with C1P. This observation is further supported by reports indicating that C1P generation by sphingomyelinase D from brown recluse spider venom causes ulcerated wounds in humans (55, 56).
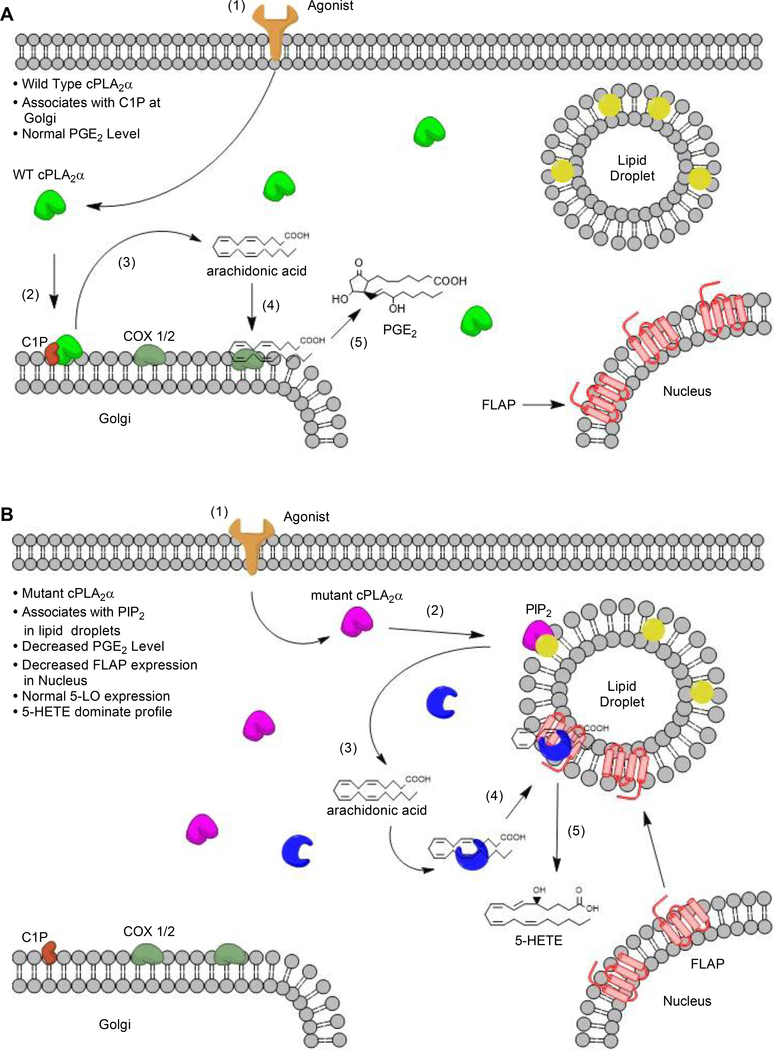
Our data, while confirming previous in vitro cell studies that loss of the C1P-cPLA2α interaction suppresses the production of certain proinflammatory prostaglandins, also revealed the unexpected finding that increased HETE biosynthesis is associated with the absence of C1P-mediarted regulation of cPLA2α but not to the absence of cPLA2α. The specific cPLA2α inhibitor pyrrophenone also mitigated the increased HETE biosynthesis and migration observed in pDFs from cPLA2α KI mice, but had no effect on cells from cPLA2α KO mice. These observations support a gain-of-function by the mutant enzyme rather than a developmental adaptation involving an increase in the biosynthesis of a separate PLA2 class. Notably, re-expression of mutant cPLA2α in cPLA2α KO cells induced the same lipid profile and migration phenotype. These observations support a model in which the inability to bind to C1P causes mutant cPLA2α to co-localize with FLAP and 5-HETE biosynthetic enzymes at vesicular structures in the cytoplasm instead of localizing with C1P at the Golgi apparatus and perinuclear membranes (Fig. 11, A and B).
Fig. 11. Schematic depiction of the proposed mechanism for C1P-mediated regulation of cPLA2α activity in wound repair.
(A) Under normal conditions, an inflammatory stimulus (for example, proinflammatory cytokines or mechanical trauma) initiates a signal cascade (1) that leads to the activation of cPLA2α and its translocation to intracellular membranes, such as the Golgi apparatus, where it binds to C1P (2). There, cPLA2α preferentially cleaves arachidonic acid–containing phospholipids at the sn-2 position, releasing free fatty acids (3). Free arachidonic acid can be converted by intracellular enzymes such as COX-1 and COX-2 (4) into PGE2 through a multi-step process (5). (B) When cPLA2α cannot bind to C1P, agonist-mediated signaling (1) leads to the activation of cPLA2α and its translocation to intracellular lipid droplets, where it binds to PIP2 (2). There, cPLA2α preferentially cleaves arachidonic acid–containing phospholipids, releasing free fatty acids (3). Free arachidonic acid binds to 5-LO (4), which interacts with FLAP at lipid droplets (4) instead of at the nucleus. FLAP-mediated activation of 5-LO enzymatic activity stimulates the production of 5-HETE, leading to a 5-HETE–dominated eicosanoid profile.
In further support of a gain-of-function mechanism for the cPLA2α C1P-binding mutant, the expression of additional eicosanoid biosynthetic enzymes, such as 5-LO and COX-1, were unaffected in the cPLA2α KO and cPLA2α KI pDFs in comparison to wild-type pDFs, which did not explain the loss of inflammatory prostaglandins and gain of HETEs. In contrast, COX-2 levels were increased in cPLA2α KO and cPLA2α KI pDFs, which may signify the activation of a compensatory mechanism to restore PGE2 levels in cPLA2α KO and cPLA2α KI pDFs. On the other hand, PGE2 or other COX-2–derived prostaglandins lost in the cPLA2α KO and cPLA2α KI pDFs may negatively feed-back on COX-2 expression. Regardless, the increased abundance of COX-2 does not explain the reduction in PGE2 observed in cPLA2α KI pDFs. Thus, the expression of enzymes controlling secondary rate-limiting steps in eicosanoid biosynthesis is unlikely to mediate the effects on eicosanoid biosynthesis and subsequent enhancement of the cellular migration in cPLA2α KI pDFs.
The use of lipidomics allowed for the detection and quantification of >40 eicosanoids, including different species of HETES. Whereas our rescue data (Fig. 8E) argue for 5-HETE as the bioactive eicosanoid driving the migration of pDFs, we cannot rule out the involvement of additional bioactive eicosanoids differentially produced in the KI pDFs that may contribute to both wound maturation in vivo and cellular migration in vitro. For example, 5-HETE can be further converted in cells and tissues to downstream metabolites, such as oxo- derivatives like 5-oxo-eicosatetraenoic acid (5-oxo-ETE), which is not included in our ESI-MS/MS protocol. Thus, the effects we observed may not be solely and directly attributable to 5-HETE. Indeed, the increases in 5-HETE in the cPLA2α KI pDFs are significant, but moderate, which may be suggestive of conversion of 5-HETE to an active downstream metabolite. Inhibition of both cPLA2α and FLAP in the KI pDFs would provide the same blockage of cPLA2α KI pDF migration in this metabolic paradigm. Alternatively, the moderate induction of 5-HETE in cPLA2α KI pDFs may reach the concentration threshold required to activate the cell surface receptor, and our data demonstrate that a lower concentration of 5-HETE is required when PGE2 is reduced. Furthermore, the increased amounts of 5-HETE maybe localized within the wound – to specific areas of the tissue with DF infiltration in vivo and near the pDFs subjected to mechanical trauma in vitro. This is not unprecedented, and our lipidomic analyses would fail to identify this possibility. Also of note, we cannot rule out an alternative intracellular signaling mechanism in the cPLA2α KI pDFs that may allow 5-HETE production in the cytoplasm to affect cellular migration differently than does the nuclear membrane production of 5-HETE in wild-type pDFs. Overall, future studies will focus on examining these possibilities as technological advancements in imaging mass spectrometric analysis continue and deuterated standards for 5-HETE metabolites become available.
Another remaining question is how the loss of the C1P-cPLA2α interaction shifts the enzyme to different cellular membranes. Indeed, cPLA2α is known to be regulated in multiple ways such as by Ca2+, phosphorylation, and interaction with bioactive lipids like PIP2 and C1P (21–28). Because C1P enhances cPLA2α partitioning to membranes in the presence of Ca2+, loss of the C1P interaction may shift cPLA2α to a more Ca2+-independent role akin to PIP2 regulation of the enzyme. Our previous work with both PIP2 and C1P activation of cPLA2α suggests this possibility because C1P inhibits the Ca2+-independent activation of cPLA2α by PIP2 in vitro, and C1P has higher affinity for the enzyme than does PIP2 (57). Because PIP2 is a Ca2+-independent allosteric activator of cPLA2α, the loss of C1P association may shift the enzyme to PIP2-enriched membranes, resulting in utilization of a different substrate pool linked to 5-HETE production by 5-LO (Fig. 10). Our data demonstrating co-localization of the cPLA2α C1P-binding mutant with FLAP, in contrast to the wild-type enzyme, supports this temporal shifting hypothesis, which intriguingly may be a key step in the transition from the inflammatory stage to the proliferation stage of wound healing. Ongoing studies are aimed at in-depth testing of this new hypothesis.
One conundrum is our finding that C1P association with cPLA2 inhibits pDF migration. Other laboratories have reported that C1P treatment induces cellular migration (58, 59). Our differential findings may be due to ceramide kinase–derived C1P inhibiting migration because of its localization to internal membranes with cPLA2α, whereas C1P generated at the plasma membrane or at other cellular localities enhances migration through association with factors such as annexins (60). Given that exogenous C1P treatment mainly increases the C1P content of the plasma membrane in various cell types, a pool of C1P at this site may drive of migration, whereas a pool at other sites may inhibit migration. An additional anabolic pathway for C1P generation does exist in mammalian cells, which may involve a sphingosine-1-phosphate (S1P)-acylase (61). Regardless, further study is required to elucidate the specific cellular roles of distinct C1P anabolic pathways based on their biosynthetic topology.
Another intriguing—but puzzling—observation is the difference between the localization of FLAP in cPLA2α KI pDFs compared to wild type pDFs. FLAP localized primarily to the nucleus in wild-type pDFs, in contrast to the cytoplasmic localization of FLAP in the cPLA2α KI cells. Currently, very little is known about FLAP translocation because it was initially considered to be a nuclear membrane protein. However, it has been reported that FLAP localizes to organelles such as lipid bodies (also called lipid droplets) (62). Lipid bodies play roles in many cellular processes, including lipid metabolism, membrane trafficking, and the regulation of inflammation. Indeed, phosphoinositides play key roles in lipid bodies by serving as signaling platforms for cellular trafficking and autophagy (63, 64), a finding that supports our hypothetical model of C1P-binding-mutant cPLA2α shifting to cytoplasmic localities such as the observed vesicular structures and lipid bodies by association with phosphoinositides and co-localizing with FLAP (Fig. 11). Such a concentration of enzymes as well as altered FLAP localization allows for greater colocalization with the mutant cPLA2α observed in our cPLA2α KI, which may explain the increase in HETE biosynthesis observed in cPLA2α KI cells.
One of the more important outcomes of this study is the demonstration of a specific role for a defined lipid-protein interaction in vivo. Genetic ablation of this lipid-protein interaction identified specific mechanistic roles for the interaction in vivo. Whereas many lipid-protein interactions have been biophysically and cellularly defined, such as that between diacylglycerol (DAG) and protein kinase C (PKC), the findings have not been directly confirmed in vivo. In the present study, we have bolstered a decade of enzymology, biophysical assays, and cellular analyses to show that the C1P-cPLA2α interaction is a major regulator of cPLA2α function in vivo, both positively to drive prostaglandin formation and negatively to inhibit the formation of HETEs, through a previously unknown mechanism of shifting the subcellular localization and/or activity of the enzyme to alter the eicosanoid profile. These findings open the door to examination of the temporal shift in cPLA2α function in cells in response to various inflammatory agonists in many physiologic contexts. For example, neutrophils and macrophages may shift their lipid profile to switch from inflammation to recruitment of more pro-resolution cell types in timing- and agonist-dependent ways. Notably, there is precedent for a shift between pro-inflammatory and pro-resolution phenotypes that are regulated by cPLA2α-derived lipids in many physiological processes and cell types (65–67). In conclusion, our study shows that the C1P-cPLA2α interaction plays an important role in the wound healing process. Our findings show the importance of lipids, and more specifically eicosanoids, in the wound healing process and is likely to provide the foundation for further research in the development of drugs that increase the healing rate of wounds and the strength of healed wounds.
Methods and Materials
Western blotting and quantification
Western blot analysis and quantification was performed as described previously (68–70) using 20 μg of protein from each extract. Briefly, western immunoblots were quantitated by densitometry using Bio-Rad image Lab software and dividing the gene of interest by the loading control for each sample. Monoclonal anti-cPLA2α (Santa Cruz Biotech 4–4B-3C); 1:200), rabbit polyclonal anti- cPLA2α (Abcam ab135825); 1:200), Mouse monoclonal ant-COX-1 (abcam 695); 1:1000), Rabbit monoclonal anti-COX-2 (abcam ab15191); 1:1000), Mouse monoclonal anti-5-LO (Santa Cruz Biotech sc-515821); 1:500), and rabbit polyclonal anti-FLAP (Abcam ab85227); 1:800), was used to identify the protein of interest. Anti β-actin antibody (1:5000) (Cell Signaling Technology) was used as a normalizing factor.
Cytoplasmic and Nuclear fractionation extraction
Nuclear extracts were prepared using the NE-PER nuclear and cytoplasmic extraction kit (Thermo-Fisher, Cat. no.78833) according to manufacturer’s instructions.
RNA Isolation, Reverse Transcription-PCR, and Quantitative PCR
RNA isolation and qRT-PCR were undertaken as previously described (71). Premixed primer-probe sets, and TaqMan Universal PCR master mix were purchased from Applied Biosystems, and the cDNA was amplified using CFX Connect real-time PCR detection system (Bio-Rad). Gene expression levels were calculated by the ΔΔCt method, normalized to GAPDH expression, and presented as fold of mean values ± SD.
cPLA2α KO and KI mouse generation
To generate KO mice, 129/Sv ES cells were electroporated with linearized version of the targeting vector shown in Fig. 1A containing multiple loxP sites flanking a cassette containing a puromycin resistance gene followed by a premature stop codon and triple mutation at the C1P binding site, R57A, K58A, R59A. Following electroporation G418- and puromycin-resistant clones containing our desired homologous recombination event were identified via PCR and DNA sequencing.
To generate KI mice, Cre recombinase treatment of ES cells was used to eliminate the cassette containing the premature stop codon, which allowed for the translation of our triple mutation at the C1P binding site, R57A, K58A, R59A. Target ES clones were transfected using pCMV-Cre and analyzed via PCR and sequencing for confirmation of CRE-mediated recombination between the two loxP sites.
PCR-based identification and genotyping of WT, KI, and KO mice
Genotyping of WT, KI, and KO mice was performed by first collecting genomic DNA using an Accustart II genotyping kit followed by polymerase chain reaction (PCR) using the following primers: PLA2 58534–58556 (P1), 5’-TGAGGGTCGTGCTGTAGAGTTAG-3’; PLA2 58780–58757 (P2), 5’-TGCCAGATGTGAACTTACTTCCAG-3’; and SV40 2590–2567 (P3) 5’-CTGCATTCTAGTTGTGGTTTGTCC-3’. 50ng of genomic DNA was used for each reaction along with the following primer concentrations, KI: 0.2 μmol/L PLA2 58534–58556 (P1) 0.2 μmol/L PLA2 58780–58757 (P2) and, cPLA2α KO: 0.2 μmol/L PLA2 58534–58556 (P1) 0.1 μmol/L PLA2 58780–58757 (P2), and 0.1 μmol/L SV40 2590–2567 (P3), the following reaction cycles were repeated 33 times, 94°C for 1 min, 59°C for 1 min, and 72°C for 2 min. Reaction products expected are as follows WT: 237bp, KI: 412bp, and KO:361bp and were examined using a 2% agarose gel.
Isolation of mouse dermal fibroblasts
pDFs were isolated and cultured from 10-week-old WT, KI, and KO C57/Blk6 mice as previously described (72).
Soluble collagen assay
pDFs obtained from WT, KI, and KO mice were plated at a density of 5.0 × 103/well in 96 well plates in high glucose DMEM supplemented with 20% FBS and 2% penicillin/streptomycin and allowed to adhere and grow at standard incubation conditions for 24 h. Collagen deposition was then measured using a soluble collagen assay (Cell BioLabs) following manufacturers protocol.
Excisional biopsy punch
6mm excisional wounds were generated in 10-week-old WT, KI, and KO C57/Blk6 males as described Silicone stints were then place around the wound and a combination of 6 sutures and glue were used to hold said stints in place. Wounds were dressed using Tegaderm (3M Medical) and imaged over the course of 14 days (73). Wound images were analyzed using the Fiji image J bundle.
Histology
6 mm excisional wounds were generated as previously described (73). After 10 days, wounds were prepared for histological evaluation using the following procedure. Excised wounds were fixed by placing them in 4% paraformaldehyde for 24 hours, following fixation the wound was placed in a cassette that allowed for the dehydration of the tissue, followed by clearing of the tissue using xylene (Fisher brand), and finally imbedding the tissue in paraffin wax. Sections (5 μm) of the paraffin block were placed on clear glass slides for further treatment and staining. Staining and probing for Masson’s trichrome, Picrosirius red, Hsp47 (rabbit polyclonal anti-Hsp47 (Abcam ab109117); 1:4000), rabbit polyclonal anti-Fibroblast activating protein (Abcam ab53066); 1:200), and mouse polyclonal anti-type I collagen (Millipore AB765P); 1:1000) were performed by Histo-Scientific Research Laboratories Inc.
Cell count quantification
Masson’s trichrome stained images were opened in Fiji image J bundle, converted to 8bit images, and threshold processed using the auto threshold selction option. Once threshold was set images were processed using the imageJ watershed setting. Lastly, the newly generated images were analyzed using the analyze particles option in imageJ.
FAP and Type I Collagen quantification
FAP and Type I Collagen quantification was performed via the following process, images were loaded into Adobe photoshop and background was subtracted as follows, blue (#0000FF) and purple (#800080), and light blue (#ADD8E6). Following background subtraction, new images were loaded in Fiji image J bundle and converted into 8bit images. Thresholds for these images were set using the auto selection option in image J. Lastly, images were analyzed using the analyze particles tool in image J.
Picrosirius red Quantification
Picrosirius red quantification was performed via the following process, images were loaded into Adobe photoshop and background was subtracted as follows, black (#000000) and yellow (#FFFF00) for Type I or black (#000000) and red (#FF0000) for Type III collagen. Following background subtraction, new images were loaded in Fiji image J bundle and converted into 8bit images. Thresholds for these images were set using the auto selection option in image J. Lastly, images were analyzed using the analyze particles tool in image J.
Colocalization Quantification
Colocalization was determined using velocity software (quorum Technologies). Briefly, images were loaded into velocity software and two wave lengths were selected for analysis, 405 and 594 for DAPI to FLAP, 488 and 594 for cPLA2α to FLAP, and 594 for 5-LO localization. Velocity generated localization data as Pearson Correlation coefficients, at which point the WT samples were set to 1 and all other samples were normalized accordingly (74).
Immunocytochemistry
pDFs from WT, KI, and KO were seeded into 24 well plates that contained round cover slips at 3.5 × 104/well and fixed for 20 min in 4% paraformaldehyde, following 4% paraformaldehyde fixation cells were washed 3x in PBS and then treated with 100% ice cold methanol for 5 min. Following the fixation and methanol treatment cells were immunolabeled as previously described(75, 76) with the following antibodies: mouse monoclonal anti-cPLA2α (Santa Cruz Biotech 4–4B-3C); 1:50), rabbit polyclonal anti-FLAP (Abcam ab85227); 1:100), Mouse monoclonal anti-5-LO (Santa Cruz Biotech sc-515821); 1:50) and mouse monoclonal anti-calnexin (Abcam ab22595); 1:50). All secondary antibodies were obtained from Invitrogen Life Technologies (Grand Island, NY; Alexa™ Fluor) and used at a dilution of 1:500. DAPI counterstain was performed by the addition of NucBlue (Invitrogen) to each well for 15min immediately followed by 3x washes in PBS. Once all washes were completed 500uL of Ibidi mounting medium was applied to each well.
Wound tensile strength measurements
20 mm incisional wounds were generated in 10-week-old WT, KI, and KO C57/Blk6 males using a scalpel (Bard-Parker) to form a single sagittal incision along the line on the dorsum. Wounds were dressed using butterfly stitches and self-adhesive medical tape. Mice were monitored, and any mice that removed their dressing within 72 h were removed from the study. Wounds were allowed to heal for 10 days, and the wound was excised and trimmed into a bar bell shape, so that the midsection was 5 mm wide and the length was 20 mm. Once trimmed, samples were placed on a MTS Synergie 100 (MTS Corporation, Eden Prairie MN) and stretched to failure at a displacement rate of 10mm/minute as performed previously for wound tensile strength.
Scratch-induced mechanical trauma of fibroblasts
pDFs obtained from WT, KI, and KO mice were plated at a density of 2 × 106 on 10 cm tissue culture plates in high glucose DMEM supplemented with 20% FBS (Gibco) and 2% penicillin/streptomycin. Following the overnight incubation cells were rested in 2% FBS (Gibco) 2% penicillin/streptomycin (Bio Whittaker) high glucose DMEM (Gibco) for 2 hours, then the media was exchanged with 0% FBS 2% penicillin/ high glucose DMEM, and mechanical trauma was induced on the monolayer by performing scratched across the diameter of the plate in an asterisk pattern using 4 × 20 μl pipette tips on a multichannel micro pipettor. Media was taken for lipidomic analysis at 0 h and 2 h.
Eicosanoid preparation and analysis
Eicosanoids were extracted using a modified extraction process and analyzed by UPLC ESI-MS/MS as previously described (77). Briefly, Media from cells (4 mL) was combined with an IS mixture comprised of 10% methanol (400 μl) and glacial acetic acid (20 μl) before spiking with internal standard (20 μl) containing the following deuterated eicosanoids (2 pmol/μl, 40 pmol total) (All standards purchased from Cayman Chemicals): (d4) 6keto-Prostaglandin F1α, (d4) Prostaglandin F2α, (d4) Prostaglandin E2, (d4) Prostaglandin D2, (d8) 5-Hydroxyeicosa-tetranoic acid (5-HETE), (d8) 12-Hydroxyeicosa-tetranoic acid (12-HETE) (d8) 15-Hydroxyeicosa-tetranoic acid (15-HETE), (d6) 20-Hydroxyeicosa-tetranoic acid (20-HETE), (d11) 8,9 Epoxyeicosa-trienoic acid, (d8) 14,15 Epoxyeicosa-trienoic acid, (d8) Arachidonic acid, (d5) Eicosapentaenoic acid, (d5) Docosahexaenoic acid, (d4) Prostaglandin A2, (d4) Leukotriene B4, (d4) Leukotriene C4, (d4) Leukotriene D4, (d4) Leukotriene E4, (d5) 5(S),6(R)-Lipoxin A4, (d11) 5-iPF2α-VI, (d4) 8-iso Prostaglandin F2α, (d11) (±)14,15-DHET, (d11) (±)8,9-DHET, (d11) (±)11,12-DHET, (d4) Prostaglandin E1, (d4) Thromboxane B2, (d6) dihmo gamma linoleic acid, (d5) Resolvin D2, (d5) Resolvin D1, (d5) Maresin2, and (d5) Resolvin D3. Samples and vial rinses (5% MeOH; 2 ml) were applied to Strata-X SPE columns (Phenomenex), previously washed with methanol (2 ml) and then dH2O (2 ml). Eicosanoids eluted with isopropanol (2 ml), were dried in vacuuo and reconstituted in EtOH:dH2O (50:50;100 μl) prior to UPLC ESI-MS/MS analysis.
Analysis of Eicosanoids by UPLC ESI-MS/MS
Eicosanoids were separated using a Shimadzu Nexara X2 LC-30AD coupled to a SIL-30AC auto injector, coupled to a DGU-20A5R degassing unit in the following way as previously described by us(77). A 14 minute, reversed phase LC method utilizing an Acentis Express C18 column (150mm x 2.1mm, 2.7μm) was used to separate the eicosanoids at a 0.5 ml/min flow rate at 40°C as previously described by us(77). The column was equilibrated with 100% Solvent A [acetonitrile:water:formic acid (20:80:0.02, v/v/v)] for 5 min and then 10 μl of sample was injected. 100% Solvent A was used for the first two minutes of elution. Solvent B [acetonitrile:isopropanol:formic acid (20:80:0.02, v/v/v)] was increased in a linear gradient to 25% Solvent B at 3 min, to 30% at 6 min., to 55% at 6.1 min, to 70% at 10 min, and to 100% at 10.10 min. 100% Solvent B was held constant until 13.0 min, where it was decreased to 0% Solvent B and 100% Solvent A from 13.0 min to 13.1 min. From 13.1 min to 14.0 min. Solvent A was held constant at 100%.
Eicosanoids were analyzed via mass spec using an AB Sciex Triple Quad 5500 Mass Spectrometer as previously described by us(77). Q1 and Q3 were set to detect distinctive precursor and product ion pairs. Ions were fragmented in Q2 using N2 gas for collisionally induced dissociation. Analysis used multiple-reaction monitoring in negative-ion mode. Eicosanoids were monitored using precursor → product MRM pairs. The mass spectrometer parameters used were: Curtain Gas: 20; CAD: Medium; Ion Spray Voltage: −4500V; Temperature: 300°C; Gas 1: 40; Gas 2: 60; Declustering Potential, Collision Energy, and Cell Exit Potential vary per transition.
Migration analysis of fibroblasts
Cells were seeded into 24 well tissue culture plates at a density of 7.5 × 104 and allowed to grow to confluence. Once a confluent monolayer was achieved cells were placed in 2% FBS/2% penicillin/streptomycin, high glucose DMEM media and allowed to rest for 2-hours. After the 2-hour resting period, the media was exchanged with 0% FBS, high glucose DMEM media, and mechanical trauma was induced on the monolayer by performing scratched across the diameter of each well using a 20 μl pipette tip. Cells were observed using a live cell incubation chamber mounted on a Keyence BZ-X710 microscope, which took images every 3 min for 24hr. Migration Velocity and meandering index (measurement of displacement of the cell from the point of origin at the end point of the experimental run and dividing that distance by the track length) were calculated using the Keyence VW-9000 motion analysis software.
For exogenous addition of bioactive lipids and small molecule inhibitor studies, once a confluent monolayer was achieved, cells were placed in 2% FBS, High Glucose DMEM media containing either the addition of various eicosanoids at the following concentrations (20 ng/mL PGE2, 0.25ng/mL 5-HETE, 0.5ng/mL 5-HETE, 1.0 ng/mL 5-HETE, and 5.0 ng/mL 20-HETE) or MK886 (7.5 nM) (Cayman Chemical), pyrrophenone (50 nM) (Cayman Chemical), or a combination of MK886/pyrrophenone and 5-HETE (1.0 ng/ml) for two hours. The media was then exchanged with 0% FBS, high glucose DMEM media containing either the addition of various eicosanoids at the following concentrations (20 ng/mL PGE2, 0.25ng/mL 5-HETE, 0.5ng/mL 5-HETE, 1.0 ng/mL 5-HETE, and 5.0 ng/mL 20-HETE) or MK886 (7.5 nM) (Cayman Chemical), pyrrophenone (50 nM) (Cayman Chemical), or a combination of either MK886/pyrrophenone and 5-HETE (1.0 ng/ml). Following the media exchange, migration was analyzed as detailed in the above paragraph. Media was also taken for lipidomic analysis at 0 h and 2 h.
For adenoviral treatment of fibroblasts, cells were infected with adenovirus containing either GFP-cPLA2α (WT), GFP-cPLA2α (R57A/K58A/R59A), or an CMV-control virus (100, 100, and 50 multiplicity of infection, respectively) as described previously described by us (78). Following the media exchange, migration was analyzed as detailed in the above paragraphs. Media was again taken for lipidomic analysis at 0 h and 2 h.
Electron microscopy
6 mm excisional wounds were generated as previously described(73). After 10 days, mice were transcardially perfused with a phosphate buffered solution containing 5% glutaraldehyde and 4% paraformaldehyde (pH 7.3). Whole carcasses were post-fixed for 2 weeks in the same solution to ensure optimal tissue preservation. For all studies, 6 mm hole biopsies were taken of the wounded area processed for standard transmission electron microscopic analysis as previously described. For the EM analysis, a JEOL JEM 1400 transmission electron microscope equipped with a Gatan Ultrascan digital camera system was employed to collect a minimum of 4 electron micrographs per animal. These images were used to determine the collagen fiber diameter as described by Khorasani and colleague(50).
Statistical Analysis
Graphing and statistics were performed using Prism GraphPad (GraphPad Software, San Diego, CA) Data were analyzed using, unpaired students t-test with Welch’s correction, ANOVA followed by Tukey’s post-hoc test, or persons correlation coefficient were applicable. All data reported as mean ± standard deviation (SD) using a minimum of three n’s with experiments repeated on at least two separate occasions. p < 0.05 was considered statistically significant.
Ethical considerations
The mouse studies were undertaken under the supervision and approval of the VCU IACUC (Protocol# AM10089) and USF IACUC (Protocol# IS00004094 and IS00004110) following standards set by the Federal and State government. The animal assurance number for VCU is #D16–00180 (A3281–01). USF and VCU are fully accredited by AAALAC International as program #000434 and #000036 respectively.
Supplementary Material
Table S1. In vitro lipid profiles of mechanically injured primary dermal fibroblasts from WT, KI, and KO mice.
Table S2. In vivo lipid profiles of acute excisional wounds in WT, KI, and KO mice.
Table S3. The altered lipid profile in primary dermal fibroblasts from KI mice is cPLA2α-specific.
Acknowledgments:
We would like to thank Dr. John Ryan and his lab group in the Biology Department at VCU for use of his lab space to finish experiments integral to the completion of this manuscript.
Funding: This work was supported by research grants from the Veteran’s Administration (VA Merit Review, I BX001792 (CEC) and a Research Career Scientist Award, 13F-RCS-002 (CEC)); from the National Institutes of Health via HL125353 (CEC, REB, EHH), HD087198 (CEC), RR031535 (CEC), R01138495 (JJR), F32Al108088 (LAH). The contents of this manuscript do not represent the views of the Department of Veterans Affairs or the United States Government. Microscopy was performed at the VCU Microscopy Facility, supported, in part, by funding from NIH-NCI Cancer Center Support Grant P30 CA016059.
Data and materials availability: The mass spectrometric eicosanoid data are available at the NIH Common Fund’s National Metabolomics Data Repository (NMDR) website, the Metabolomics Workbench, https://www.metabolomicsworkbench.org, where it has been assigned Project IDs ST001252 and ST001254. The data can be accessed directly through the Project DOIs 10.21228/M87H7W and 10.21228/M87H7W. This database is supported by NIH grant U2C-DK119886. All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
Footnotes
Competing interests: The authors declare that they have no competing interests.
REFERENCES AND NOTES
- 1.Enoch S, and Price P. 2004. Pathophysiology-Of-Healing @ www.worldwidewounds.com..
- 2.Clark RAF 1988. Wound Repair. In The Molecular and Cellular Biology of Wound Repair Springer US, Boston, MA. 3–50. [Google Scholar]
- 3.Versteeg HH, Heemskerk JWM, Levi M, and Reitsma PH. 2013. New Fundamentals in Hemostasis. Physiol. Rev 93: 327–358. [DOI] [PubMed] [Google Scholar]
- 4.Kim M-H, Liu W, Borjesson DL, Curry F-RE, Miller LS, Cheung AL, Liu F-T, Isseroff RR, and Simon SI. 2008. Dynamics of neutrophil infiltration during cutaneous wound healing and infection using fluorescence imaging. J. Invest. Dermatol 128: 1812–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koh TJ, and DiPietro LA. 2011. Inflammation and wound healing: the role of the macrophage. Expert Rev. Mol. Med 13: e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diegelmann RF, Evans MC, and others. 2004. Wound healing: an overview of acute, fibrotic and delayed healing. Front Biosci 9: 283–9. [DOI] [PubMed] [Google Scholar]
- 7.Eming SA, Wynn TA, and Martin P. 2017. Inflammation and metabolism in tissue repair and regeneration. Science 356: 1026–1030. [DOI] [PubMed] [Google Scholar]
- 8.Broughton G, Janis JE, and Attinger CE. 2006. Wound healing: an overview. Plast. Reconstr. Surg 117: 1e-S–32e-S. [DOI] [PubMed] [Google Scholar]
- 9.Sivamani RK 2014. Eicosanoids and Keratinocytes in Wound Healing. Adv. wound care 3: 476–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.desJardins-Park HE, Foster DS, and Longaker MT. 2018. Fibroblasts and wound healing: an update. Regen. Med 13: 491–495. [DOI] [PubMed] [Google Scholar]
- 11.Goldberg GI, Wilhelm SM, Kronberger A, Bauer EA, Grant GA, and Eisen AZ. 1986. Human fibroblast collagenase. Complete primary structure and homology to an oncogene transformation-induced rat protein. J. Biol. Chem 261: 6600–5. [PubMed] [Google Scholar]
- 12.Löffek S, Schilling O, and Franzke CW. 2011. Biological role of matrix metalloproteinases: A critical balance. Eur. Respir. J 38: 191–208. [DOI] [PubMed] [Google Scholar]
- 13.Krzyszczyk P, Schloss R, Palmer A, and Berthiaume F. 2018. The Role of Macrophages in Acute and Chronic Wound Healing and Interventions to Promote Pro-wound Healing Phenotypes. Front. Physiol 9: 419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hesketh M, Sahin KB, West ZE, and Murray RZ. 2017. Macrophage Phenotypes Regulate Scar Formation and Chronic Wound Healing. Int. J. Mol. Sci 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ireton JE, Unger JG, and Rohrich RJ. 2013. The role of wound healing and its everyday application in plastic surgery: a practical perspective and systematic review. Plast. Reconstr. surgery. Glob. open 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodrigues M, Kosaric N, Bonham CA, and Gurtner GC. 2019. Wound Healing: A Cellular Perspective. Physiol. Rev 99: 665–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morris JL, Cross SJ, Lu Y, Kadler KE, Lu Y, Dallas SL, and Martin P. 2018. Live imaging of collagen deposition during skin development and repair in a collagen I – GFP fusion transgenic zebrafish line. Dev. Biol 441: 4–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dhall S, Wijesinghe DS, Karim ZA, Castro A, Vemana HP, Khasawneh FT, Chalfant CE, and Martins-Green M. 2015. Arachidonic acid-derived signaling lipids and functions in impaired healing. Wound Repair Regen 23: 644–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fischer R, Konkel A, Mehling H, Blossey K, Gapelyuk A, Wessel N, von Schacky C, Dechend R, Muller DN, Rothe M, Luft FC, Weylandt K, and Schunck W-H. 2014. Dietary omega-3 fatty acids modulate the eicosanoid profile in man primarily via the CYP-epoxygenase pathway. J. Lipid Res 55: 1150–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tapiero H, Ba GN, Couvreur P, and Tew KD. 2002. Polyunsaturated fatty acids (PUFA) and eicosanoids in human health and pathologies. Biomed. Pharmacother 56: 215–22. [DOI] [PubMed] [Google Scholar]
- 21.Dhall S, Wijesinghe DS, a Karim Z, Castro A, Vemana HP, Khasawneh FT, Chalfant CE, and Martins-Green M. 2015. Arachidonic acid-derived signaling lipids and functions in impaired healing. Wound Repair Regen 23: 644–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.White ES, Atrasz RG, Dickie EG, Aronoff DM, Stambolic V, Mak TW, Moore BB, and Peters-Golden M. 2005. Prostaglandin E(2) inhibits fibroblast migration by E-prostanoid 2 receptor-mediated increase in PTEN activity. Am. J. Respir. Cell Mol. Biol 32: 135–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo L, Tanaka R, Kanazawa S, Lu F, Hayashi A, Yokomizo T, and Mizuno H. 2017. A synthetic leukotriene B4 receptor type 2 agonist accelerates the cutaneous wound healing process in diabetic rats by indirect stimulation of fibroblasts and direct stimulation of keratinocytes. J. Diabetes Complications 31: 13–20. [DOI] [PubMed] [Google Scholar]
- 24.Romana-Souza B, dos Santos JS, Bandeira LG, and Monte-Alto-Costa A. 2016. Selective inhibition of COX-2 improves cutaneous wound healing of pressure ulcers in mice through reduction of iNOS expression. Life Sci 153: 82–92. [DOI] [PubMed] [Google Scholar]
- 25.White ES, Atrasz RG, Dickie EG, Aronoff DM, Stambolic V, Mak TW, Moore BB, and Peters-Golden M. 2005. Prostaglandin E2 Inhibits Fibroblast Migration by E-Prostanoid 2 Receptor–Mediated Increase in PTEN Activity. Am. J. Respir. Cell Mol. Biol 32: 135–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Co. OP 2010. Opalmon -Oral prostaglandin E1 derivative preparation. 2010: 2–6. [Google Scholar]
- 27.Hoeferlin LA, Huynh QK, Mietla JA, Sell SA, Tucker J, Chalfant CE, and Wijesinghe DS. 2015. The Lipid Portion of Activated Platelet-Rich Plasma Significantly Contributes to Its Wound Healing Properties. Adv. wound care 4: 100–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leslie CC 1997. Properties and regulation of cytosolic phospholipase A2. J. Biol. Chem 272: 16709–12. [DOI] [PubMed] [Google Scholar]
- 29.Clark JD, Schievella AR, Nalefski EA, and Lin LL. 1995. Cytosolic phospholipase A2. J. Lipid Mediat. Cell Signal. 12: 83–117. [DOI] [PubMed] [Google Scholar]
- 30.Lamour NF, Subramanian P, Wijesinghe DS, V Stahelin R, V Bonventre J, and Chalfant CE. 2009. Ceramide 1-Phosphate Is Required for the Translocation of Group IVA Cytosolic Phospholipase A2 and Prostaglandin Synthesis. J. Biol. Chem 284: 26897–26907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoeferlin LA, Wijesinghe DS, and Chalfant CE. 2013. The Role of Ceramide-1-Phosphate in Biological Functions. In Handbook of experimental pharmacology 153–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ward KE, Bhardwaj N, Vora M, Chalfant CE, Lu H, and Stahelin RV. 2013. The molecular basis of ceramide-1-phosphate recognition by C2 domains. J. Lipid Res 54: 636–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stahelin RV, Subramanian P, Vora M, Cho W, and Chalfant CE. 2007. Ceramide-1-phosphate Binds Group IVA Cytosolic Phospholipase a 2 via a Novel Site in the C2 Domain. J. Biol. Chem 282: 20467–20474. [DOI] [PubMed] [Google Scholar]
- 34.Stahelin RV, Subramanian P, Vora M, Cho W, and Chalfant CE. 2007. Ceramide-1-phosphate Binds Group IVA Cytosolic Phospholipase a 2 via a Novel Site in the C2 Domain. J. Biol. Chem 282: 20467–20474. [DOI] [PubMed] [Google Scholar]
- 35.Wijesinghe DS, Brentnall M, Mietla J. a., Hoeferlin L. a., Diegelmann RF, Boise LH, and Chalfant CE. 2014. Ceramide kinase is required for a normal eicosanoid response and the subsequent orderly migration of fibroblasts. J. Lipid Res 55: 1298–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alaish SM, Bettinger DA, Olutoye OO, Gould LJ, Yager DR, Davis A, Crossland MC, Diegelmann RF, and Kelman Cohen I. 1995. Comparison of the polyvinyl alcohol sponge and expanded polytetrafluoroethylene subcutaneous implants as models to evaluate wound healing potential in human beings. Wound Repair Regen 3: 292–298. [DOI] [PubMed] [Google Scholar]
- 37.Wijesinghe DS, Warncke UO, and Diegelmann RF. 2016. Human as the Ultimate Wound Healing Model: Strategies for Studies Investigating the Dermal Lipidome. Curr. Dermatol. Rep 5: 244–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang X, Ge J, Tredget EE, and Wu Y. 2013. The mouse excisional wound splinting model, including applications for stem cell transplantation. Nat. Protoc 8: 302–309. [DOI] [PubMed] [Google Scholar]
- 39.GRILLO HC, WATTS GT, and GROSS J. 1958. Studies in wound healing: I. Contraction and the wound contents. Ann. Surg 148: 145–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kennedy DF, and Cliff WJ. 1979. A systematic study of wound contraction in mammalian skin. Pathology 11: 207–22. [DOI] [PubMed] [Google Scholar]
- 41.McGrath MH, and Simon RH. 1983. Wound geometry and the kinetics of wound contraction. Plast. Reconstr. Surg 72: 66–73. [DOI] [PubMed] [Google Scholar]
- 42.Wong VW, Sorkin M, Glotzbach JP, Longaker MT, and Gurtner GC. 2011. Surgical Approaches to Create Murine Models of Human Wound Healing. J. Biomed. Biotechnol 2011: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Galiano RD, Michaels VJ, Dobryansky M, Levine JP, and Gurtner GC. 2004. Quantitative and reproducible murine model of excisional wound healing. Wound Repair Regen 12: 485–492. [DOI] [PubMed] [Google Scholar]
- 44.Midwood KS, Williams LV, and Schwarzbauer JE. 2004. Tissue repair and the dynamics of the extracellular matrix. Int. J. Biochem. Cell Biol 36: 1031–1037. [DOI] [PubMed] [Google Scholar]
- 45.Schmidt MJ, Tschoeke A, Noronha L, de Moraes RS, Mesquita RA, Grégio AMT, Alanis LRA, Ignácio SA, dos Santos JN, de Lima AAS, Luiz TS, Michels AC, Aguiar MCF, and Johann ACBR. 2016. Histochemical analysis of collagen fibers in giant cell fibroma and inflammatory fibrous hyperplasia. Acta Histochem 118: 451–455. [DOI] [PubMed] [Google Scholar]
- 46.Gonzalez A. C. de O., Costa TF, de A. Andrade Z, and Medrado ARAP. 2016. Wound healing - A literature review. An. Bras. Dermatol 91: 614–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lorenz HP, and Longaker MT. 2008. Wounds: Biology, Pathology, and Management. In Surgery Springer New York, New York, NY. 191–208. [Google Scholar]
- 48.Pickett BP, Burgess LP, Livermore GH, Tzikas TL, and Vossoughi J. 1996. Wound healing. Tensile strength vs healing time for wounds closed under tension. Arch. Otolaryngol. Head. Neck Surg 122: 565–8. [DOI] [PubMed] [Google Scholar]
- 49.Xue M, and Jackson CJ. 2015. Extracellular Matrix Reorganization During Wound Healing and Its Impact on Abnormal Scarring. Adv. wound care 4: 119–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khorasani H, Zheng Z, Nguyen C, Zara J, Zhang X, Wang J, Ting K, and Soo C. 2011. A quantitative approach to scar analysis. Am. J. Pathol 178: 621–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Romana-Souza B, dos Santos JS, Bandeira LG, and Monte-Alto-Costa A. 2016. Selective inhibition of COX-2 improves cutaneous wound healing of pressure ulcers in mice through reduction of iNOS expression. Life Sci 153: 82–92. [DOI] [PubMed] [Google Scholar]
- 52.Sivamani RK 2014. Eicosanoids and Keratinocytes in Wound Healing. Adv. wound care 3: 476–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rådmark O, Werz O, Steinhilber D, and Samuelsson B. 2015. 5-Lipoxygenase, a key enzyme for leukotriene biosynthesis in health and disease. Biochim. Biophys. Acta - Mol. Cell Biol. Lipids 1851: 331–339. [DOI] [PubMed] [Google Scholar]
- 54.Contaifer D, Carl DE, Warncke UO, Martin EJ, Mohammed BM, Van Tassell B, Brophy DF, Chalfant CE, and Wijesinghe DS. 2017. Unsupervised analysis of combined lipid and coagulation data reveals coagulopathy subtypes among dialysis patients. J. Lipid Res 58: 586–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hadanny A, Fishlev G, Bechor Y, Meir O, and Efrati S. 2016. Nonhealing Wounds Caused by Brown Spider Bites. Adv. Skin Wound Care 29: 560–566. [DOI] [PubMed] [Google Scholar]
- 56.DeLozier JB, Reaves L, King LEJ, and Rees RS. 1988. Brown recluse spider bites of the upper extremity. South. Med. J 81: 181–184. [DOI] [PubMed] [Google Scholar]
- 57.Subramanian P, Vora M, Gentile LB, V Stahelin R, and Chalfant CE. 2007. Anionic lipids activate group IVA cytosolic phospholipase A2 via distinct and separate mechanisms. J. Lipid Res 48: 2701–2708. [DOI] [PubMed] [Google Scholar]
- 58.Granado MH, Gangoiti P, Ouro A, Arana L, González M, Trueba M, and Gómez-Muñoz A. 2009. Ceramide 1-phosphate (C1P) promotes cell migration. Cell. Signal. 21: 405–412. [DOI] [PubMed] [Google Scholar]
- 59.Kim C, Liu R, Kucia M, and Ratajczak MZ. 2011. New Evidence That the Bioactive Lipid Ceramide-1-Phosphate (C1P) Is a Potent Chemoattractant for Mesenchymal Stromal Cells (MSC), Endothelial Progenitor Cells (EPCs) and Very Small Embryonic-Like Stem Cells (VSELs), Demonstrating Its Potential Involvemen…. Blood 118. [Google Scholar]
- 60.Hankins JL, Ward KE, Linton SS, Barth BM, V Stahelin R, Fox TE, and Kester M. 2013. Ceramide 1-phosphate mediates endothelial cell invasion via the annexin a2-p11 heterotetrameric protein complex. J. Biol. Chem 288: 19726–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Binoda C, Portoukalian J, Schmitt D, Rodriguez-Lafrasse C, and Ardail D. 2004. Subcellular compartmentalization of ceramide metabolism: MAM (mitochondria-associated membrane) and/or mitochondria? Biochem. J. 382: 527–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bozza PT, Magalhães KG, and Weller PF. 2009. Leukocyte lipid bodies - Biogenesis and functions in inflammation. Biochim. Biophys. Acta 1791: 540–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Skotland T, Hessvik NP, Sandvig K, and Llorente A. 2018. Exosomal lipid composition and the role of ether lipids and phosphoinositides in exosome biology. J. Lipid Res. jlr.R084343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wallroth A, and Haucke V. 2018. Phosphoinositide conversion in endocytosis and the endolysosomal system. J. Biol. Chem 293: 1526–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Serhan CN, Chiang N, and Van Dyke TE. 2008. Resolving inflammation: dual anti-inflammatory and pro-resolution lipid mediators. Nat. Rev. Immunol 8: 349–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Serhan CN 2010. Novel lipid mediators and resolution mechanisms in acute inflammation: to resolve or not? Am. J. Pathol 177: 1576–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schwab JM, and Serhan CN. 2006. Lipoxins and new lipid mediators in the resolution of inflammation. Curr. Opin. Pharmacol 6: 414–420. [DOI] [PubMed] [Google Scholar]
- 68.DeLigio JT, Lin G, Chalfant CE, and Park MA. 2017. Splice variants of cytosolic polyadenylation element–binding protein 2 (CPEB2) differentially regulate pathways linked to cancer metastasis. J. Biol. Chem 292: 17909–17918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vu NT, Park MA, Shultz MD, Bulut GB, Ladd AC, and Chalfant CE. 2016. Caspase-9b Interacts Directly with cIAP1 to Drive Agonist-Independent Activation of NF-kappaB and Lung Tumorigenesis. Cancer Res 76: 2977–2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shapiro BA, Vu NT, Shultz MD, Shultz JC, Mietla JA, Gouda MM, Yacoub A, Dent P, Fisher PB, Park MA, and Chalfant CE. 2016. Melanoma Differentiation-associated Gene 7/IL-24 Exerts Cytotoxic Effects by Altering the Alternative Splicing of Bcl-x Pre-mRNA via the SRC/PKCδ Signaling Axis. J. Biol. Chem 291: 21669–21681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shultz JC, Vu N, Shultz MD, Mba M-UU, Shapiro BA, and Chalfant CE. 2012. The Proto-oncogene PKCι regulates the alternative splicing of Bcl-x pre-mRNA. Mol. Cancer Res 10: 660–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Seluanov A, Vaidya A, and Gorbunova V. 2010. Establishing primary adult fibroblast cultures from rodents. J. Vis. Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang X, Ge J, Tredget EE, and Wu Y. 2013. The mouse excisional wound splinting model, including applications for stem cell transplantation. Nat. Protoc 8: 302–309. [DOI] [PubMed] [Google Scholar]
- 74.Dunn KW, Kamocka MM, and McDonald JH. 2011. A practical guide to evaluating colocalization in biological microscopy. Am. J. Physiol. Cell Physiol 300: C723–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Benusa SD, George NM, Sword BA, DeVries GH, and Dupree JL. 2017. Acute neuroinflammation induces AIS structural plasticity in a NOX2-dependent manner. J. Neuroinflammation 14: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vu NT, Park MA, Shultz JC, Goehe RW, Hoeferlin LA, Shultz MD, Smith SA, Lynch KW, and Chalfant CE. 2013. hnRNP U enhances caspase-9 splicing and is modulated by AKT-dependent phosphorylation of hnRNP L. J. Biol. Chem 288: 8575–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Simanshu DK, Kamlekar RK, Wijesinghe DS, Zou X, Zhai X, Mishra SK, Molotkovsky JG, Malinina L, Hinchcliffe EH, Chalfant CE, Brown RE, and Patel DJ. 2013. Non-vesicular trafficking by a ceramide-1-phosphate transfer protein regulates eicosanoids. Nature 500: 463–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lamour NF, and Chalfant CE. 2008. Ceramide kinase and the ceramide-1-phosphate/cPLA2alpha interaction as a therapeutic target. Curr. Drug Targets 9: 674–682. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. In vitro lipid profiles of mechanically injured primary dermal fibroblasts from WT, KI, and KO mice.
Table S2. In vivo lipid profiles of acute excisional wounds in WT, KI, and KO mice.
Table S3. The altered lipid profile in primary dermal fibroblasts from KI mice is cPLA2α-specific.