Abstract
The species composition of plant communities is determined by a number of factors, including current environmental conditions as well as biogeographical and evolutionary history. Despite evidence that plant diversity decreases and species relatedness increases along latitudinal and environmental gradients (e.g., low temperatures), it remains unclear whether these same patterns occur along elevational gradients, especially in the subtropical mountainous areas harboring rich biodiversity. In this study, we explored the pattern of phylogenetic relatedness of woody angiosperm assemblages and examined the effects of temperature variables on the phylogenetic relatedness among angiosperm woody plants using generalized linear model in subtropical forest communities along a broad elevational gradient in the Dulong Valley of Yunnan Province, China. Our results showed that woody angiosperm species in local forest plots tend to be more phylogenetically related at higher elevations and in areas with lower temperatures. Additionally, winter average temperature, rather than mean annual temperature, is a major predictor of the pattern of increasing phylogenetic relatedness with increasing elevation. This finding is consistent with the prediction of ‘Tropical Niche Conservatism’ hypothesis, which highlights the role of niche constraints in driving phylogenetic community assembly along an elevational gradient.
Keywords: Dulong Valley, Ecological tolerance, Habitat filtering, Niche conservatism, Woody plants
1. Introduction
Plant communities are assembled through interactions between both environmental and evolutionary processes (Qian and Sandel, 2017; Qian et al., 2018). Community assemblages can only comprise species whose environmental tolerances allow them to maintain a population (Cardillo, 2011) under both the abiotic and biotic conditions of the local community (Fraterrigo et al., 2014; Qian et al., 2019a; Ricklefs, 1987; Willis et al., 2010). However, the traits that confer the ability to survive in a local environment are also constrained by evolutionary history (Donoghue, 2008). For example, cold and drought tolerance in plants are widely considered to be phylogenetically conserved (Wiens et al., 2010; Condamine et al., 2012; Crisp and Cook, 2012; Hawkins et al., 2014). Understanding how these environmental and evolutionary factors interact to influence plant community assemblage is a major challenge.
The ‘Tropical Niche Conservatism’ hypothesis (TNC) posits that phylogenetic relationships have a significant effect on community assembly along an environmental gradient (Wiens and Donoghue, 2004; Hettwer et al., 2012). This hypothesis also proposes that most angiosperm families originated and initially diversified in tropical climates (Latham and Ricklefs, 1993; Romdal et al., 2013). However, with the onset of global cooling in the Eocene, most tropical lineages were extirpated from the tropics, although some acquired adaptations enabling them to shift into the spreading temperate climates (Graham, 2011; Hawkins et al., 2014). Evolutionary innovations related to cold tolerance are thought to have only occurred in a few clades due to the conserved nature of ecological traits (e.g., cold tolerance) and the relative rarity of evolutionary events which confer novel functions (Latham and Ricklefs, 1993; Wiens and Donoghue, 2004; Donoghue, 2008). Due to the comparative rarity of cold-adapted lineages, there are a limited number of species which can fill niches in temperate environments (Ricklefs, 2006). The overall effect of this phenomenon is that the phylogenetic relatedness of species within an environment tends to increase as the temperature of that environment decreases, which has been demonstrated in several empirical studies (e.g., Giehl and Jarenkow, 2012; Qian et al., 2013, 2015; 2016; Qian and Sandel, 2017).
Climatic variation along elevational gradients is an excellent natural laboratory for examining how temperature influences the composition of different species assemblages in local communities (Qian et al., 2014; Rahbek et al., 2019). Species diversity along elevational gradients mirror species diversity along latitudinal gradient, and the underlying mechanisms responsible for these patterns are thought to be the same (Vetaas and Grytnes, 2002). Accordingly, some temperature-related variables, including mean annual temperature and minimum temperature of the coldest month, change in similar ways along elevational and latitudinal gradients (Qian et al., 2014; Muellner-Riehl et al., 2019). For example, minimum annual temperature is generally thought to significantly affect the distribution of species towards higher elevations and latitudes (Mittelbach et al., 2007; Sanders et al., 2007; Qian and Ricklefs, 2011). However, over a given distance, elevational gradients of temperature are generally steeper than latitudinal gradients of temperature. For example, the thermal gradient of a 100-km change in latitude is typically equivalent to that of a 100-m change in elevation (Jump et al., 2009; Stephenson and Das, 2011). Consequently, examining species richness along an elevational gradient is a more straightforward approach to disentangling the influences of temperature factors on phylogenetic relatedness (Körner, 2007).
The relatedness of species in local communities has been the subject of many studies, but the results of previous studies have been inconsistent. In tropical Asia and South America, woody angiosperm species are more distantly related to each other along elevational gradients (Culmsee and Leuschner, 2013; González-Caro et al., 2014; Qian, 2018). However, in temperate regions of Asia, woody angiosperm species are more closely related to each other along elevational gradients (Qian et al., 2014, 2019b; Chun and Lee, 2018), which is consistent with predictions of the TNC hypothesis. Because nearly all studies associated with elevational gradients have been limited to tropical and temperate areas, little information is known about subtropical regions. Therefore, more studies are needed to draw a general pattern on phylogenetic relatedness along elevational gradients.
In this study, we investigated the pattern of phylogenetic relatedness among woody angiosperms within subtropical forest assemblages along an elevational gradient in the Dulong Valley of Yunnan Province, China. Because temperature is one of the major environmental factors governing species distributions in local plant assemblages (Li et al., 2017), we explored the relationship of phylogenetic relatedness to both temperature and elevation variation. Specifically, we addressed two main questions: 1) Does phylogenetic relatedness of woody angiosperm assemblages increase with increasing elevation? 2) What environmental factor (e.g., mean annual temperature, winter average temperature) is the dominant determinant of phylogenetic relatedness in local communities of woody angiosperms?
2. Materials and methods
2.1. Study area and locations
The Dulong Valley is located at the border of Myanmar and China, and extends roughly 20 km west into Myanmar (He, 1998). This area has 2816 native seed plant species which belong to 778 genera and 171 families. Of these, 132 species are endemic to the valley (Li et al., 2007, 2011). This high degree of endemism and species richness is likely the result of interactions between the climatic, topographic and geologic variation of the site (Chaplin, 2005).
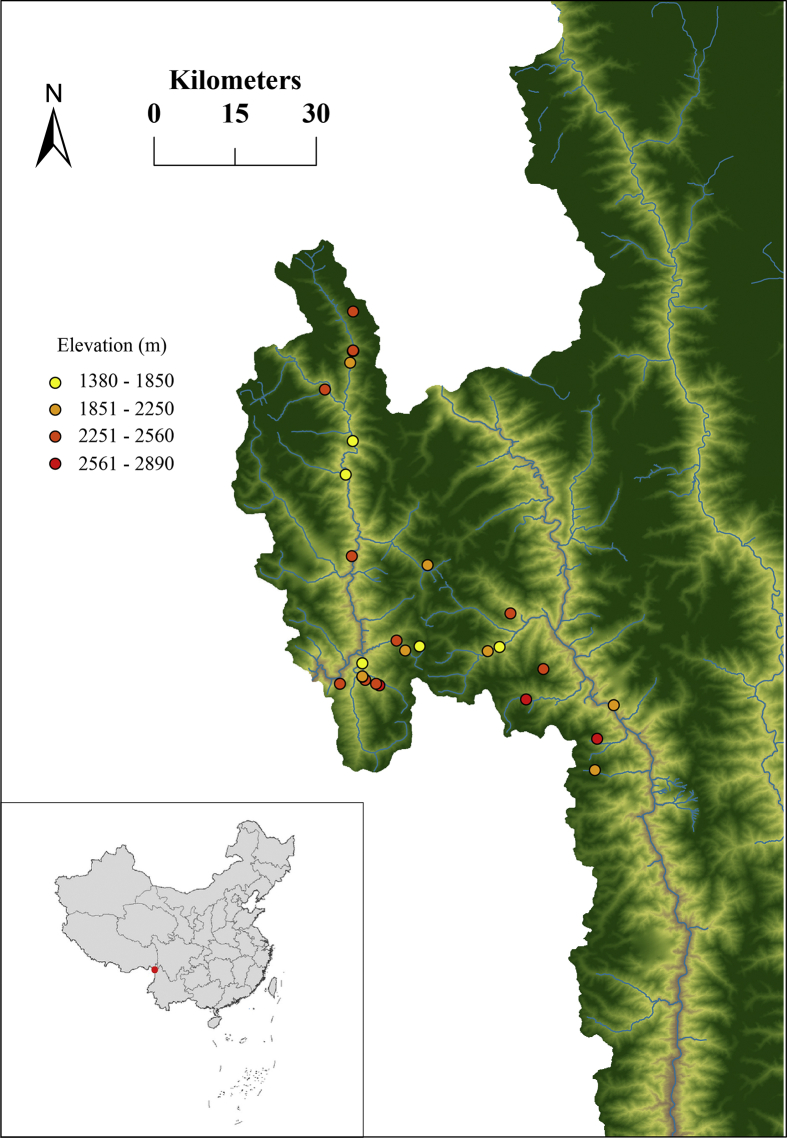
We obtained data on species composition for woody angiosperms in local assemblages from a recently published monograph (Wang et al., 2013). We identified woody angiosperms at 28 forest plots along an elevational gradient in the Dulong Valley. The plots extend from 1300 to 2900 m above sea level (Fig. 1). Each plot is 400 m2 with a homogeneous habitat. All woody angiosperms were identified to species or morphospecies (Wang et al., 2013). Botanical nomenclature was standardized according to Flora of China (http://www.efloras.org/). Dominant species, elevation, longitude and latitude for each forest plot used in this study are provided in Appendix S1.
Fig. 1.
Map showing locations of the forest plots used in the Dulong Valley of China.
2.2. Phylogeny and community phylogenetic structure
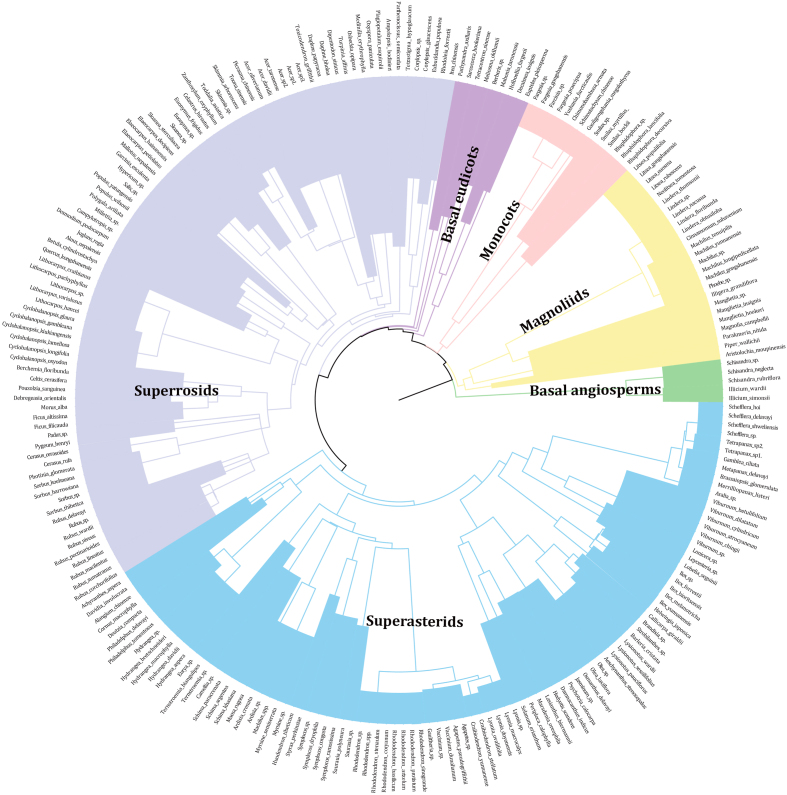
We used a recently published mega-phylogeny “GBOTB” as the basis to generate a phylogeny for the species present in this area. GBOTB was constructed using 79,881 taxa in GenBank and a backbone provided by Open Tree of Life (Smith and Brown, 2018). This is the most comprehensive and up-to-date time-calibrated species-level mega-phylogeny for seed plants. Genera and species present in the study area but absent from GBOTB were added to their respective families (in the case of genera) and genera (in the case of species) using the Phylomatic and BLADJ approaches (Webb et al., 2008) implemented in the V.PhyloMaker software (Jin and Qian, 2019). V.PhyloMaker places any missing species at the basal node within a given genus, and any missing genera at the basal node within their respective families. We pruned the phylogeny to retain only species present in our data (Fig. 2).
Fig. 2.
Phylogenies of woody angiosperms species in forest communities in the Dulong Valley of China.
We used two metrics to quantify phylogenetic relatedness among species for each forest plot: net relatedness index (NRI; Webb, 2000) and phylogenetic species variability (PSV; Helmus et al., 2007). Both metrics measure the degree of phylogenetic relatedness among species within a community, and both have been commonly used in earlier studies of phylogenetic community assembly (e.g., Li et al., 2015; Lu et al., 2018; Qian et al., 2019b). NRI measures the standardized effect size of the mean pairwise distance (MPD), which is used to estimate the mean phylogenetic relatedness among taxa within a community. NRI is defined as: −1 × (MPDobserved − MPDrandomized)/(sdMPDrandomized) (Webb, 2000), where MPDobserved is the observed MPD, MPDrandomized is the expected MPD of the randomized communities (n = 1000), and sdMPDrandomized is the standard deviation of the MPD for the randomized communities. A negative NRI value indicates that the observed MPD is greater than expected by chance (i.e., taxa are more distantly related than expected) and that phylogenetic evenness or overdispersion of taxa is present. In contrast, a positive NRI value indicates that the observed MPD is lower than what would be expected by chance (i.e., taxa are more closely related than expected by chance) and that phylogenetic clustering has taken place.
PSV is defined as: (ntrC − ΣC)/n(n − 1) (Helmus et al., 2007), where n is the number of taxa, C is a covariance matrix that represents the correlation of phylogenetic structure, trC is the sum of the diagonal elements of C, ΣC is the sum of all elements in C. PSV varies from one, which indicates that taxa assemblages are unrelated (phylogenetic overdispersion or evenness), to zero, which indicates that taxa assemblages are perfectly related (phylogenetic clustering). Therefore, regions with high PSV values represent phylogenetic overdispersion, whereas regions with low PSV values indicate phylogenetic clustering. This metric is independent of species richness (Savage and Cavender-Bares, 2012).
We assessed the significance of phylogenetic metrics for the forest plots by comparing their observed value to a null distribution where the species names were randomly shuffled across the tips of the phylogeny 999 times. Then, a two-tailed test was applied to judge whether the observed value was significantly lower or higher than the null model. Observed values that fell into the top or bottom 2.5% of the distribution for each forest plot were considered significant (Santos et al., 2010; Qian et al., 2013; Zhang and Zhang, 2017).
2.3. Environmental data
We used mean annual temperature (MAT) and winter average temperature (WAT) to quantify environmental conditions in each plot. These variables were selected due to their common usage in testing the relationship between environmental factors and phylogenetic relatedness along elevational gradients (e.g., Culmsee and Leuschner, 2013; Qian, 2014; Chun and Lee, 2018). Temperature for this study was collected using iButton data loggers (DS1923 #F5) every 30 min from August 2018 to August 2019 at 9 sites, which were installed along a vertical interval of 200 m. We used generalized additive model in order to obtain unbiased estimates for the slopes of the temperature in cases where the relationship between temperature and elevation was not linear. Elevation data were used as an independent variable with the coordinates of the plot as a smooth factor to account for spatial autocorrelation to obtain the interpolated MAT and WAT for each plot.
2.4. Statistical analyses
We used a generalized linear model to assess the effect of temperature variables on phylogenetic relatedness among the angiosperm species of each forest plot. We included phylogenetic relatedness of each plot as the response variable and the MAT and WAT as one of the independent variables (NRI or PSV = β0 + β1MAT + β2WAT + ε). The MAT and WAT variables were all standardized by log 10 transformation to facilitate interpretation and comparisons among parameters. We determined which temperature variable better described phylogenetic metrics based on the Akaike information criterion (AIC) model selection, with smaller AIC scores indicating the more concise and accurate model (Symonds and Moussalli, 2011). All phylogenetic metrics and model analyses were implemented in “picante v.1.8.1”, “mgcv v.1.8e31”, and “psych v.1.9.12.31” packages (Kembel et al., 2010; Wood, 2017; Revelle, 2018) of R v.3.5.2 (R Development Core Team, 2018).
3. Results
A total of 236 woody angiosperms species from 135 genera and 69 families were identified in the 28 forest plots. Species richness varied greatly across plots, from 12 to 40.
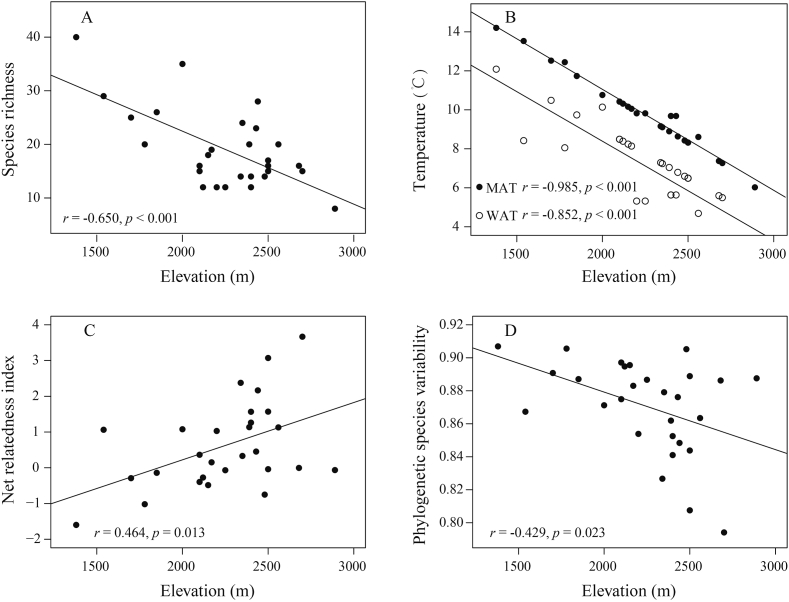
We found that elevation had a significant negative correlation with the species richness of woody angiosperms (r = −0.650, p < 0.001) (Fig. 3A). Additionally, elevation was negatively correlated with both mean annual temperature (r = −0.985, p < 0.001) and winter average temperature (r = −0.852, p < 0.001) (Fig. 3B). NRI increased with increasing elevation (r = 0.464, p = 0.013) (Fig. 3C), while PSV was negatively correlated with elevation (r = −0.429, p = 0.023) (Fig. 3D). Although these correlations are moderate, they indicate that species in local forest assemblages in the Dulong Valley are phylogenetically related at higher elevations. The AIC scores indicate the best model for describing phylogenetic relatedness include WAT, regardless of whether it was measured using NRI or PSV (Table 1).
Fig. 3.
Relationships of elevation with the number of species (A), mean annual temperature (MAT) and winter average temperature (WAT) (B), net relatedness index (C) and phylogenetic species variability (D) along an elevation gradient in forest communities in the Dulong Valley.
Table 1.
Generalized linear models for the correlation of phylogenetic relatedness with temperature factors along an elevation gradient in forest communities in the Dulong Valley.
| Variable | Net relatedness index (NRI) |
Phylogenetic species variability (PSV) |
||
|---|---|---|---|---|
| Coef. | AIC | Coef. | AIC | |
| MAT | −0.42∗ | 89.29 | 0.40∗ | −119.30 |
| WAT | −0.51∗ | 87.23 | 0.48∗ | −121.59 |
MAT = Mean annual temperature; WAT = Winter average temperature; Coef. = Regression coefficient; AIC = Akaike information criterion; ∗ = Significant correlations (P < 0.05).
4. Discussion
In the present study, we used two metrics (NRI and PSV) to examine the pattern of phylogenetic relatedness among woody angiosperm species and evaluated the effects of temperature variables on their phylogenetic relatedness in the subtropical forest plots along a broad gradient of elevation in the Dulong Valley. Both metrics measure phylogenetic relatedness as the standardized effect size, and PSV accounts for differences in species richness (Webb, 2000; Helmus et al., 2007; Savage and Cavender-Bares, 2012). Although patterns derived from NRI and PSV are generally congruent, the elevational gradient of phylogenetic relatedness derived from NRI is smoother than that derived from PSV (Fig. 3C and D).
Our data revealed that woody angiosperm assemblages showed greater relatedness to each other (phylogenetic clustering) at higher elevations in the Dulong Valley. This correlation between elevation and phylogenetic relatedness is consistent with the findings of previous studies on phylogenetic relatedness (using NRI to quantify phylogenetic relatedness) for plant assemblages along elevational gradients (Li et al., 2014; Qian et al., 2014, 2019b; Chun and Lee, 2018), and supports the predictions of the TNC hypothesis for phylogenetic community assemblages along temperature gradients. Phylogenetic clustering is commonly considered to result from environmental filtering (Webb et al., 2002; Bryant et al., 2008), because ecological traits are generally phylogenetically conserved and thus more closely related species are expected to be more ecologically similar (Webb et al., 2002). In our study, woody angiosperm species in forest plots had increased phylogenetic clustering as elevation increased and temperature decreased along a gradient in the Dulong Valley, suggesting that environmental filtering likely had a significant impact on the sorting of plants among plots (Cavender-Bares et al., 2009). However, our finding that phylogenetic relatedness increases with increasing elevation (and thus with decreasing temperature) is contrary to patterns that have been identified when studying the same phenomenon in tropical environments. For example, previous studies have found that in South American tropical tree communities, phylogenetic clustering occurs at high temperatures (low elevation), whereas phylogenetic overdispersion occurs at low temperatures (high elevations) (González-Caro et al., 2014; Qian, 2018). Similarly, Culmsee and Leuschner (2013) reported that in Malesia tropical environments phylogenetic relatedness of woody angiosperm assemblages was even at high elevations. Taken together, these studies indicate that in local forest communities that exist along a tropical elevational gradient there is an inverse relationship between the phylogenetic relatedness of woody angiosperm species and elevation. Qian and Ricklefs (2016) developed the ‘niche convergence’ or ‘Out of the Tropics’ hypothesis to explain why increasing elevation tends to result in the decline of phylogenetic relatedness among woody angiosperm species in tropical regions. This hypothesis posits that although most angiosperm clades originated in tropical or tropical-like conditions, some rapidly spread to colder environments, where they have evolved relatively slowly compared to their counterparts in the tropics. Furthermore, ecological tolerance for plant species across elevations in the tropics diversified among distantly related lineages through niche convergence (Jablonski et al., 2006). Therefore, taxa at higher elevations in the tropics tend to be more phylogenetically overdispersed.
Our study found that WAT, rather than MAT, described phylogenetic relatedness of woody angiosperm assemblages most robustly, although MAT varied more than WAT across the forest plots in the Dulong Valley (Table 1; Fig. 3B). One previous study has also indicated that WAT plays a more significant role than MAT in constructing phylogenetic relatedness of community assemblies along an elevational gradient (e.g., Qian, 2018). The TNC hypothesis explains how WAT shapes the impact of elevation of phylogenetic relatedness. Specifically, relatively few species have acquired adaptations to harsh environments (Latham and Ricklefs, 1993) and these traits are phylogenetically conserved (Donoghue, 2008). The result of this limitation is that phylogenetic relatedness tends to increase as WAT decreases across an elevational gradient (Qian et al., 2007; Wang et al., 2011).
In conclusion, our study showed that woody angiosperm species in local forest plots are more likely to be more closely related to each other at higher elevations and in areas of lower temperatures. This pattern of increasing phylogenetic relatedness with increasing elevation appears to be driven by winter average temperature, rather than mean annual temperature. Our findings on the impact of phylogenetic relatedness along an elevational gradient in a subtropical region are consistent with the ‘Tropical Niche Conservatism’ model, although our findings are distinct from those of studies in the tropics.
Author contributions
R. Li and J. Yue conceived and wrote the paper. J. Yue collected and analyzed the data.
Declaration of Competing Interest
None.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (grant nos. 31770228, 31570212, 31370243), the Belt and Road Project of West Light Foundation of the Chinese Academy of Sciences, Yunnan Science and Technology Innovation Team Program (grant no. 2019HC015), the Second Tibetan Plateau Scientific Expedition and Research Program (grant no. 2019QZKK0502), and the Biodiversity Survey and Assessment Project of the Ministry of Ecology and Environment, The People’s Republic of China, China (grant no. 2019HJ2096001006).
Footnotes
Peer review under responsibility of Editorial Office of Plant Diversity.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2020.08.003.
Appendix A. Supplementary data
The following is the supplementary data to this article:
Dominant species, elevation, longitude and latitude for the forest plots used in this study.
References
- Bryant J.A., Lamanna C., Morion H. Microbes on mountainsides: contrasting elevational patterns of bacterial and plant diversity. Proc. Natl. Acad. Sci. U. S. A. 2008;105:11505–11511. doi: 10.1073/pnas.0801920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardillo M. Phylogenetic structure of mammal assemblages at large geographical scales: linking phylogenetic community ecology with macroecology. Philos. Trans. R. Soc. B-Biol. Sci. 2011;366:2545–2553. doi: 10.1098/rstb.2011.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavender-Bares J., Kozak K.H., Fine P.V. The merging of community ecology and phylogenetic biology. Ecol. Lett. 2009;12:693–715. doi: 10.1111/j.1461-0248.2009.01314.x. [DOI] [PubMed] [Google Scholar]
- Chaplin G. Physical geography of the Gaoligong Shan area of southwest China in relation to biodiversity. Proc. Calif. Acad. Sci. 2005;56:527–556. [Google Scholar]
- Chun J.H., Lee C.B. Diversity patterns and phylogenetic structure of vascular plants along elevational gradients in a mountain ecosystem, South Korea. J. Mt. Sci. 2018;15:280–289. [Google Scholar]
- Condamine F.L., Sperling F.A., Wahlberg N.J. What causes latitudinal gradients in species diversity? Evolutionary processes and ecological constraints on swallowtail biodiversity. Ecol. Lett. 2012;15:267–277. doi: 10.1111/j.1461-0248.2011.01737.x. [DOI] [PubMed] [Google Scholar]
- Crisp M.D., Cook L.G. Phylogenetic niche conservatism: what are the underlying evolutionary and ecological causes? New Phytol. 2012;196:681–694. doi: 10.1111/j.1469-8137.2012.04298.x. [DOI] [PubMed] [Google Scholar]
- Culmsee H., Leuschner C. Consistent patterns of elevational change in tree taxonomic and phylogenetic diversity across Malesian mountain forests. J. Biogeogr. 2013;40:1997–2010. [Google Scholar]
- Donoghue M.J. A phylogenetic perspective on the distribution of plant diversity. Proc. Natl. Acad. Sci. U. S. A. 2008;105:11549–11555. doi: 10.1073/pnas.0801962105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraterrigo J.M., Wagner S., Warren R.J. Local-scale biotic interactions embedded in macroscale climate drivers suggest Eltonian noise hypothesis distribution patterns for an invasive grass. Ecol. Lett. 2014;17:1447–1454. doi: 10.1111/ele.12352. [DOI] [PubMed] [Google Scholar]
- Giehl E.L.H., Jarenkow J.A. Niche conservatism and the differences in species richness at the transition of tropical and subtropical climates in South America. Ecography. 2012;35:933–943. [Google Scholar]
- González-Caro S., Umana M.N., Alvarez E. Phylogenetic alpha and beta diversity in tropical tree assemblages along regional-scale environmental gradients in northwest South America. J. Plant Ecol. 2014;7:145–153. [Google Scholar]
- Graham A. The age and diversification of terrestrial New World ecosystem through Cretaceous and Cenozoic time. Am. J. Bot. 2011;98:336–351. doi: 10.3732/ajb.1000353. [DOI] [PubMed] [Google Scholar]
- Hawkins B.A., Rueda M., Rangel T.F. Community phylogenetics at the biogeographical scale: cold tolerance, niche conservatism and the structure of North American forests. J. Biogeogr. 2014;41:23–38. doi: 10.1111/jbi.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He D.M. Comprehensive study on the Dulong river area. Yunnan Geogr. Environ. Res. 1998;10:8–14. [Google Scholar]
- Helmus M.R., Bland T.J., Williams C.K. Phylogenetic measures of biodiversity. Am. Nat. 2007;169:E68–E83. doi: 10.1086/511334. [DOI] [PubMed] [Google Scholar]
- Hettwer G., Eduardo L., Jarenkow J.A. Niche conservatism and the differences in species richness at the transition of tropical and subtropical climates in South America. Ecography. 2012;35:933–943. [Google Scholar]
- Jablonski D., Roy K., Valentine J.W. Out of the tropics: evolutionary dynamics of the latitudinal diversity gradient. Science. 2006;314:102–106. doi: 10.1126/science.1130880. [DOI] [PubMed] [Google Scholar]
- Jin Y., Qian H. V.PhyloMaker: an R package that can generate very large phylogenies for vascular plants. Ecography. 2019;42:1353–1359. doi: 10.1016/j.pld.2022.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jump A.S., Matyas C., Penuelas J. The altitude-for-latitude disparity in the range retractions of woody species. Trends Ecol. Evol. 2009;24:694–701. doi: 10.1016/j.tree.2009.06.007. [DOI] [PubMed] [Google Scholar]
- Kembel S.W., Cowan P.D., Helmus M.R. Picante: R tools for integrating phylogenies and ecology. Bioinformatics. 2010;26:1463–1464. doi: 10.1093/bioinformatics/btq166. [DOI] [PubMed] [Google Scholar]
- Körner C. The use of ‘altitude’ in ecological research. Trends Ecol. Evol. 2007;22:569–574. doi: 10.1016/j.tree.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Latham R.E., Ricklefs R.E. Global patterns of tree species richness in moist forests: energy-diversity theory does not account for variation in species richness. Oikos. 1993;67:325–333. [Google Scholar]
- Li H., Li X.H., Yang J. Distribution pattern and ecological reasons of the richness of the Gaoligong Mountains endemic seed plants. J. West China For. Sci. 2017;46:S58–S65. [Google Scholar]
- Li R., Dao Z.L., Ji Y.H. A floristic study on the seed plants of the northern Gaoligong Mountains in western Yunnan, China. Acta Bot. Yunnanica. 2007;29:601–615. [Google Scholar]
- Li R., Dao Z.L., Li H. Seed plant species diversity and conservation in the northern Gaoligong Mountains in western Yunnan, China. Mt. Res. Dev. 2011;31:160–165. [Google Scholar]
- Li R., Kraft N.J.B., Yu H. Seed plant phylogenetic diversity and species richness in conservation planning within a global biodiversity hotspot in eastern Asia. Conserv. Biol. 2015;29:1552–1562. doi: 10.1111/cobi.12586. [DOI] [PubMed] [Google Scholar]
- Li X.H., Zhu X.X., Niu Y. Phylogenetic clustering and overdispersion for alpine plants along elevational gradient in the Hengduan Mountains Region, southwest China. J. Systemat. Evol. 2014;52:280–288. [Google Scholar]
- Lu L.M., Mao L.F., Yang T. Evolutionary history of the angiosperm flora of China. Nature. 2018;554:234–238. doi: 10.1038/nature25485. [DOI] [PubMed] [Google Scholar]
- Mittelbach G.G., Schemske D.W., Cornell H.V. Evolution and the latitudinal diversity gradient: speciation, extinction and biogeography. Ecol. Lett. 2007;10:315–331. doi: 10.1111/j.1461-0248.2007.01020.x. [DOI] [PubMed] [Google Scholar]
- Muellner-Riehl A., Schnitzler J., Kissling W.D. Origins of global mountain plant diversity: testing the mountain-geobiodiversity hypothesis. J. Biogeogr. 2019;46:2826–2838. [Google Scholar]
- Qian H. Contrasting relationships between clade age and temperature along latitudinal versus elevational gradients for woody angiosperms in forests of South America. J. Veg. Sci. 2014;25:1208–1215. [Google Scholar]
- Qian H. Climatic correlates of phylogenetic relatedness of woody angiosperms in forest communities along a tropic elevational gradient in South America. J. Plant Ecol. 2018:394–400. [Google Scholar]
- Qian H., Deng T., Jin Y. Phylogenetic dispersion and diversity in regional assemblages of seed plants in China. Proc. Natl. Acad. Sci. U. S. A. 2019;116:23192–23201. doi: 10.1073/pnas.1822153116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian H., Field R., Zhang J.L. Phylogenetic structure and ecological and evolutionary determinants of species richness for angiosperm trees in forest communities in China. J. Biogeogr. 2016;43:603–615. [Google Scholar]
- Qian H., Hao Z.Q., Zhang J. Phylogenetic structure and phylogenetic diversity of angiosperm assemblages in forests along an elevational gradient in Changbaishan, China. J. Plant Ecol. 2014;7:154–165. [Google Scholar]
- Qian H., Ricklefs R.E. Latitude, tree species diversity and the metabolic theory of ecology. Global Ecol. Biogeogr. 2011;20:362–365. [Google Scholar]
- Qian H., Ricklefs R.E. Out of the tropical lowlands: latitude versus elevation. Trends Ecol. Evol. 2016;31:738–741. doi: 10.1016/j.tree.2016.07.012. [DOI] [PubMed] [Google Scholar]
- Qian H., Sandel B. Phylogenetic structure of regional angiosperm assemblages across latitudinal and climatic gradients in North America. Global Ecol. Biogeogr. 2017;26:1258–1269. [Google Scholar]
- Qian H., Sandel B., Deng T. Geophysical, evolutionary and ecological processes interact to drive phylogenetic dispersion in angiosperm assemblages along the longest elevational gradient in the world. Bot. J. Linn. Soc. 2019;190:333–344. [Google Scholar]
- Qian H., Wang X.H., Wang S.L. Environmental determinants of amphibian and reptile species richness in China. Ecography. 2007;30:471–482. [Google Scholar]
- Qian H., Wiens J.J., Zhang J. Evolutionary and ecological causes of species richness patterns in North American angiosperm trees. Ecography. 2015;38:241–250. [Google Scholar]
- Qian H., Zhang J., Hawkins B.A. Mean family age of angiosperm tree communities and its climatic correlates along elevational and latitudinal gradients in eastern North America. J. Biogeogr. 2018;45:259–268. [Google Scholar]
- Qian H., Zhang Y.J., Zhang J. Latitudinal gradients in phylogenetic relatedness of angiosperm trees in North America. Global Ecol. Biogeogr. 2013;22:1183–1191. [Google Scholar]
- R Development Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2018. R: A Language and Environment for Statistical Computing. [Google Scholar]
- Rahbek C., Borregaard M.K., Antonelli A. Building mountain biodiversity: geological and evolutionary process. Science. 2019;365:1114–1119. doi: 10.1126/science.aax0151. [DOI] [PubMed] [Google Scholar]
- Revelle W. Northwestern University; Evanston, Illinois, USA: 2018. Psych: Procedures for Personality and Psychological Research. [Google Scholar]
- Ricklefs R.E. Community diversity: relative roles of local and regional processes. Science. 1987;235:167–171. doi: 10.1126/science.235.4785.167. [DOI] [PubMed] [Google Scholar]
- Ricklefs R.E. Evolutionary diversification and the origin of the diversity-environment relationship. Ecology. 2006;87:S3–S13. doi: 10.1890/0012-9658(2006)87[3:edatoo]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Romdal T.S., Araujo M.B., Rahbek C. Life on a tropical planet: niche conservatism and the global diversity gradient. Global Ecol. Biogeogr. 2013;22:344–350. [Google Scholar]
- Sanders N.J., Lessard J.P., Fitzpatrick M.C. Temperature, but not productivity or geometry, predicts elevational diversity gradients in ants across spatial grains. Global Ecol. Biogeogr. 2007;16:640–649. [Google Scholar]
- Santos B.A., Arroyo-Rodríguez V., Moreno C.E. Edge-related loss of tree phylogenetic diversity in the severely fragmented Brazilian Atlantic forest. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage J.A., Cavender-Bares J. Habitat specialization and the role of trait lability in structuring diverse willow (genus Salix) communities. Ecology. 2012;93:S138–S150. [Google Scholar]
- Smith S.A., Brown J.W. Constructing a broadly inclusive seed plant phylogeny. Am. J. Bot. 2018;105:302–314. doi: 10.1002/ajb2.1019. [DOI] [PubMed] [Google Scholar]
- Stephenson N.L., Das A.J. Common on “change in climatic water balance drive downhill shifts in plant species' optimum elevations”. Science. 2011;334:177. doi: 10.1126/science.1205740. [DOI] [PubMed] [Google Scholar]
- Symonds M.R.E., Moussalli A. A brief guide to model selection, multimodel inference and model averaging in behavioural ecology using Akaike's information criterion. Behav. Ecol. Sociobiol. 2011;65:13–21. [Google Scholar]
- Vetaas O.R., Grytnes J.A. Distribution of vascular plant species richness and endemic richness along the Himalayan elevation gradient in Nepal. Global Ecol. Biogeogr. 2002;11:291–301. [Google Scholar]
- Wang C.Y., He Z.R., Peng C.M. Science Press; Beijing: 2013. Vegetation and Plant Research in Dulongjiang River (Upper Irrawaddy River) Watershed and Adjacent Area. [Google Scholar]
- Wang Z., Fang J., Tang Z. Patterns, determinants and models of woody plant diversity in China. Proc. R Soc. B Biol Sci. 2011;278:2122–2132. doi: 10.1098/rspb.2010.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb C.O. Exploring the phylogenetic structure of ecological communities: an example for rain forest trees. Am. Nat. 2000;156:145–155. doi: 10.1086/303378. [DOI] [PubMed] [Google Scholar]
- Webb C.O., Ackerly D.D., Kembel S.W. Phylocom: software for the analysis of phylogenetic community structure and trait evolution. Bioinformatics. 2008;24:2098–2100. doi: 10.1093/bioinformatics/btn358. [DOI] [PubMed] [Google Scholar]
- Webb C.O., Ackerly D.D., McPeek M.A. Phylogenies and community ecology. Annu. Rev. Ecol. Systemat. 2002;33:475–505. [Google Scholar]
- Wiens J.J., Ackerly D.D., Allen A.P. Niche conservatism as an emerging principle in ecology and conservation biology. Ecol. Lett. 2010;13:1310–1324. doi: 10.1111/j.1461-0248.2010.01515.x. [DOI] [PubMed] [Google Scholar]
- Wiens J.J., Donoghue M.J. Historical biogeography, ecology and species richness. Trends Ecol. Evol. 2004;19:639–644. doi: 10.1016/j.tree.2004.09.011. [DOI] [PubMed] [Google Scholar]
- Willis C.G., Halina M., Lehman C. Phylogenetic community structure in Minnesota oak savanna is influenced by spatial extent and environmental variation. Ecography. 2010;33:565–577. [Google Scholar]
- Wood S.N. second ed. Chapman and Hall/CRC Press; 2017. Generalized Additive Models: an Introduction with R. [Google Scholar]
- Zhang H.X., Zhang M.L. Spatial patterns of species diversity and phylogenetic structure of plant communities in the Tianshan Mountains, arid central Asia. Front. Plant Sci. 2017;8:2134. doi: 10.3389/fpls.2017.02134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dominant species, elevation, longitude and latitude for the forest plots used in this study.