Abstract
Stress-associated proteins (SAPs) are known as response factors to multiple abiotic and biotic stresses in plants. However, the potential physiological and molecular functions of SAPs remain largely unclear. Castor bean (Ricinus communis L.) is one of the most economically valuable non-edible woody oilseed crops, able to be widely cultivated in marginal lands worldwide because of its broad adaptive capacity to soil and climate conditions. Whether SAPs in castor bean plays a key role in adapting diverse soil conditions and stresses remains unknown. In this study, we used the castor bean genome to identify and characterize nine castor bean SAP genes (RcSAP). Structural analysis showed that castor bean SAP gene structures and functional domain types vary greatly, differing in intron number, protein sequence, and functional domain type. Notably, the AN1-C2H2–C2H2 zinc finger domain within RcSAP9 has not been often observed in other plant families. High throughput RNA-seq data showed that castor bean SAP gene profiles varied among different tissues. In addition, castor bean SAP gene expression varied in response to different stresses, including salt, drought, heat, cold and ABA and MeJA, suggesting that the transcriptional regulation of castor bean SAP genes might operate independently of each other, and at least partially independent from ABA and MeJA signal pathways. Cis-element analyses for each castor bean SAP gene showed that no common cis-elements are shared across the nine castor bean SAP genes. Castor bean SAPs were localized to different regions of cells, including the cytoplasm, nucleus, and cytomembrane. This study provides a comprehensive profile of castor bean SAP genes that advances our understanding of their potential physiological and molecular functions in regulating growth and development and their responses to different abiotic stresses.
Keywords: Castor bean, Genomic characterization, Stress-associated proteins, Expression profile
1. Introduction
Stress-associated proteins (SAPs) participate in the plant responses to multiple abiotic and biotic stresses (Mukhopadhyay et al., 2004; Tyagi et al., 2014; Gao et al., 2016). High-throughput genome sequencing technology has led to the identification of SAPs in a range of plants, including Arabidopsis thaliana (L.) Heynh., Oryza sativa L. (Vij and Tyagi, 2006), Zea mays L. (Giri et al., 2013), Gossypium hirsutum L. (Gao et al., 2016), Solanum lycopersicum L. (Solanke et al., 2009), Populus euphratica Oliv. (Jia et al., 2016), Aeluropus littoralis (Gouan) Parl. (Zouari et al., 2007), Lobularia maritime (L.) Desv.(Ben et al., 2018), Sorghum bicolor (L.) Moench (Wang et al., 2013b). and Medicago truncatula Gaertn. (Zhou et al., 2018). However, the roles that SAPs play in response to different stresses remain poorly understood.
SAP family members are classified into five types based on the presence of A20, AN1 and/orC2H2 zinc finger domains (Dixit et al., 2018). These types include the A20-AN1-type, which contains one A20 domain and one type I AN1 domain; the AN1-type, which contains only a single type I AN1 domain; the AN1-AN1-types, which contains two type II AN1 domains, the AN1-AN1-C2H2–C2H2-type, which contains two type II AN1 domains and two C2H2 domains; and the AN1-AN1-C2H2-type, which contains two type II AN1 domains and a single C2H2 domain. The most common SAP type is the A20-AN1, whereas the AN1-AN1-C2H2-type has only been identified in Arabidopsis (Vij and Tyagi, 2006). SAP structural domain types may provide insight into SAP function, as domain types have been shown to be related to stress-specific transcriptional variation (Jia et al., 2016).
Previous studies have shown that SAP gene expression is induced by multiple biotic and abiotic stresses. In rice, the expression of OsSAP1 is induced by various stresses, including cold, salt, drought, water, heavy metal, wounding, and fungal infection (Mukhopadhyay et al., 2004; Tyagi et al., 2014). In P. euphratica, the gene expression of all SAPs increases in response to drought, salt, and heat stresses (Jia et al., 2016). In several cases, different SAP genes are simultaneously expressed in response to the same stresses (Ströher et al., 2009). Furthermore, SAP gene expression patterns vary in response to different stresses (Ben et al., 2018). Interestingly, AtSAPs that belong to the A20-AN1 SAP type do not respond to drought, genotoxic, oxidative, or wounding stresses (Ströher et al., 2009). In contrast, gene expression of the only AN1-AN1-type SAP in Arabidopsis, AtSAP12, is induced by all biotic and abiotic stresses (Ströher et al., 2009).
Functional studies have shown that SAPs confer stress tolerance to plants. Overexpression of OsSAP1 confers resistance to cold, saline and osmotic stresses in rice and pathogen resistance in tobacco (Dansana et al., 2014; Tyagi et al., 2014). Overexpression of OsSAP8, OsSAP9 and OsSAP11 increased plant tolerance to salt, drought and temperature stresses (Huang et al., 2008; Kanneganti and Gupta, 2008; Giri et al., 2011). The overexpression of AtSAP5 or AtSAP10 in Arabidopsis both improve plant resistance to heat (Dixit and Dhankher, 2011; Kim et al., 2015). Overexpression of MusaSAP1 in Musa × paradisiaca L. and the heterologously expressed SbSAP14 in rice provide stronger salt and oxidative tolerance (Sreedharan et al., 2012; Wang et al., 2013b). The overexpression of LmSAP in tobacco and yeast (Saccharomyces cervisiae) also improve toxic metal resistance (Ben et al., 2018). Recent studies have also shown that SAPs may function in coordination with hormone signals such as abscisic acid (ABA) (Zhang et al., 2016; Kang et al., 2013, 2017; Sharma et al., 2015). Intriguingly, the overexpression of AlSAP in rice not only confers stress resistance, but also raises yields of transgenic rice grown in unfavorable environments (Ben et al., 2012). Studies have revealed that SAPs may interact with diverse proteins by their zinc finger domains (Yin et al., 2017). However, to date no evidence indicates that SAP proteins directly bind promoter regions of genes to activate gene expression, suggesting that SAPs do not function as transcription factors, and may play various roles throughout the cell.
Castor bean (Ricinus communis L.), an economically important non-edible oilseed crop, has a strong tolerance to drought and salinity. Its seed oils contain a considerable amount ricinoleic acid (18 carbon hydroxyled fatty acid, over 85% of seed total oils), which has industrial uses, including in aviation oil, lubricants, paints, and biodiesel feedstock (Qiu et al., 2010; Wang et al., 2013a). Because of its economic importance, the castor bean has been widely cultivated in the marginal lands of many areas (Qiu et al., 2010; Xu et al., 2016). Investigating the physiological and molecular basis of adaptations to various environmental conditions is an essential prerequisite to creating stress-tolerant castor bean varieties. In this study we examined whether castor bean SAPs mediate plant responses and adaptation to diverse soil conditions and abiotic stresses. For this purpose, we identified nine castor bean SAP genes, investigated their expression profiles in response to various stresses, and identified their subcellular localization. This study provides fundamental and comprehensive information that will be useful in understanding the physiological and molecular basis of castor bean adaptation to various environments.
2. Materials and methods
2.1. Identification of SAPs in castor bean and other Euphorbiaceae species
The protein sequences of Arabidopsis A20/AN1-type zinc finger protein were obtained from National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov/). These sequences were used as queries to search candidate SAP family members in the castor bean genome database Phytozome (http://www.phytozome.net/) based on algorithms of the basic local alignment search tool (BLASTP). The same strategy was used to identify candidate SAP proteins from Jatropha curcas L., Manihot esculenta Crantz. and Hevea brasiliensis (Willd. ex A. Juss.) Muell. Arg., but the genome sequences of three species were acquired from NCBI. Each predicted SAP protein sequence was accepted by the A20/AN1 domain search using the SMART (http://smart.embl-heidelberg.de/) and Pfam (http://www.sanger.ac.uk/Software/Pfam/) databases, and protein sequences that contained the A20 or AN1 domains were ultimately regarded as SAP family members. Predictions of subcellular localization, isoelectric points (pIs) and theoretical molecular weight (MW) were analyzed using ProtComp 9.0 (http://linux1.softberry.com) and ProtParam (http://web.expasy.org/protparam).
2.2. Chromosome locations, gene duplication and phylogenetic analysis
The 10castor bean genetic linkage groups as 10 pseudochromosomes are derived from previously published work (Yu et al., 2019). The chromosomal locations of castor bean SAP genes were mapped on pseudochromosomes by MapInspect (http://www.plantbreeding.wur.nl/UK/softwaremapinspect.html). Gene tandem duplication events were checked from the online website PTGBase (Yu et al., 2015) (http://ocri-genomics.org/PTGBase/) based on the scaffold location information of genes. Multiple alignments of full-length SAP amino acid sequences from castor bean, A. thaliana, J. curcas, M. esculenta and H. brasiliensis were carried out using Clustal W (version 2.0) with default settings. Subsequently, the Neighbor-Joining (NJ) tree was constructed in MEGA 7.0 with 1000 bootstrap replicates (Kumar et al., 2016), and the genetic distance among SAPs were calculated based on the p-distance algorithm.
2.3. Gene structure and cis-actingelement analyses
The CDS and exon-intron structures of predicted castor bean SAP genes were analyzed using the Gene Structure Display Server (GSDS) (Hu et al., 2015) (http://gsds.cbi.pku.edu.cn/). The promoter regions (2 kb upstream of the initiation codon) of castor bean SAP genes were obtained from the database Phytozome, and the cis-regulatory elements were identified using the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
2.4. Gene expression analysis in various tissues
Castor bean SAP gene expression levels were retrieved from two data sets. One data set contained castor bean SAP gene expression levels from leaf, root, developing seed 1 (at 15 days of pollination), developing seed 2 (at 35 days of pollination) and endosperm (at 40 days of pollination) (Xu et al., 2013). The second data set (accession number ERA047687) contained castor bean SAP gene expression levels from the developing endosperm at the early and late stages, leaf, and developing male flower (Brown et al., 2012). Castor bean SAP gene expression levels were measured in transcripts per million reads (TPM) and fragments per kb per million reads (FPKM). The heat map of castor bean SAP gene expression levels was constructed using Multi-Experiment View (MEV) cluster software.
2.5. Plant material and treatments
Castor bean (ZB306 cultivar) seeds were sterilized in 3% H2O2 for 12 h, and then germinated in Petri plates containing several pieces of moistened filter paper at 30 °C. Each germinated seed was sown into one pot (10 cm in diameters and in height) and cultivated at 30 °C. Seedlings with two true leaves were treated with different abiotic stresses, including salt, drought, heat, cold, ABA (Sigma–Aldrich, Shanghai, China) and MeJA (Sigma–Aldrich, Shanghai, China). For salt and drought stresses, the pots with seedlings were transported into hydroponic boxes (40 cm in length, 30 cm in width and 15 cm in height) with either 300 mM NaCl or 20% PEG 6000 solutions poured over the surfaces of the pots (Wang et al., 2018). For heat stress treatment, the seedlings in pots were placed in a light growth incubator at 50 °C. For cold stress treatment, seedlings were kept at 4 °C. For phytohormone treatments, the entire seedling was sprayed with either 100 μM ABA or 100 μM MeJA. Each treatment was repeated at least three times across 15 seedlings. After treatment, the two true leaves of each seedling were harvested at five points in time (0, 1, 3, 6 and 12 h after treatment) and were rapidly frozen in liquid nitrogen.
2.6. RNA extraction and qRT-PCR
Equal amounts of two true leaves from each seedling were mixed in one sample for RNA extraction. Total RNA was extracted using the E. Z.N.A.™ HP Plant RNA Kit (OMEGA bio-tek) following the manufacturer's protocol. First-strand cDNA synthesis was performed using the all-in-one first-strand cDNA synthesis SuperMix (Transgen) with each reverse transcription reaction containing 100 ng total of RNA. Real-time quantitative PCR (qRT-PCR) was carried out on a Bio-Rad CFX Manager system (BIO-RAD, Berkeley, CA, USA), using TransStart Tip Green qPCR SuperMix (Transgen) qRT-PCR amplification was conducted following the manufacturer's recommendations: 95 °C for 2 min, followed by 40 cycles of amplification at 95 °C for 30 s, 60 °C for 15 s, and 72 °C for 10 s. Each gene at each point in time across the six treatments was amplified in triplicate and three independent biological replications were conducted. The primer sequences that were used in this research are listed in Table S1. The relative expression levels of genes were calculated with the 2−ΔΔCT algorithm. RcACTIN2 (see supplementary file 1) was used as an internal control reference gene to normalize relative expression levels (Xu et al., 2016).
2.7. Cloning castor bean SAP genes and transient expression system
Total RNA from roots, leaves, and immature seeds of castor bean (ZB306) was isolated and cDNA was synthesized as described above. cDNA from these three tissues were mixed equally and used to clone the full-length coding regions of nine castor bean SAP genes. The primer sequences used in the research are listed in Supplementary Table S1.
The coding sequences of nine castor bean SAPs (see the supplementary file 1) were inserted into the pSuper-1300 vector (derivative vector of pCAMBIA 1300) via a one-step cloning strategy (one-step cloning kit, Vazyme) to produce recombinant pSuper-RcSAPs-GFP. For subcellular localization, the pSuper-RcSAPs-GFP was transformed into four-week-old leaves of Nicotiana benthamiana Domin. using the Agrobacterium-mediated system following the protocol described by Chatre et al. (2005). N. benthamiana epidermal cells were scanned by confocal microscope (Fluoview fv1000, OLYMPUS) three days post-infection.
3. Results
3.1. Identification and phylogenetic analysis of castor bean SAP genes
To identify SAP genes in castor bean, 14 SAP protein sequences from Arabidopsis were used as queries to search the Phyozome database using BLASTp. In total, only nine putative SAP genes were identified in castor bean. These nine putative castor bean SAP genes were named based on their A20/AN1 domains and their location in the castor bean genome (Table 1). The lengths and MW of castor bean SAP genes varied greatly. Eight of nine castor bean SAP genes were distributed on chromosomes 4, 5, 6 and 7. RcSAP2 was localized to the unassembled scaffold 30055 (Supplementary Figure S1).
Table 1.
Characteristics of SAP genes in castor bean.
| Gene | Zinc Finger | Scaffold Location | CDS(bp) | Intron | Aa | pI | MW(kD) |
|---|---|---|---|---|---|---|---|
| RcSAP1 | A20–AN1 | 29625:66111-67350(+) | 501 | 0 | 166 | 8.26 | 18.56 |
| RcSAP2 | A20–AN1 | 30055:65043-66139(+) | 519 | 0 | 172 | 8.78 | 18.68 |
| RcSAP3 | A20–AN1 | 30147:748055-749409(+) | 501 | 1 | 166 | 9.02 | 17.98 |
| RcSAP4 | A20–AN1 | 30169:1052539–1055066(−) | 519 | 0 | 172 | 8.28 | 18.63 |
| RcSAP5 | AN1 | 29820:17148-17805(+) | 375 | 1 | 124 | 8.92 | 13.49 |
| RcSAP6 | AN1 | 30147:745169-745881(+) | 603 | 1 | 124 | 9.50 | 22.52 |
| RcSAP7 | AN1 | 30152:504246-505463(−) | 423 | 0 | 140 | 9.16 | 15.25 |
| RcSAP8 | AN1–AN1 | 29822:422676-423613(+) | 579 | 1 | 192 | 9.02 | 21.12 |
| RcSAP9 | AN1–C2H2–C2H2 | 29761:60693-61552(−) | 789 | 1 | 265 | 8.75 | 29.55 |
Note: (+): the gene orientation is forward, (−): the gene orientation is reverse. Aa denotes amino acid residues. CDS, pI and MW denote the coding sequence, isoelectric points and theoretical molecular weight, respectively.
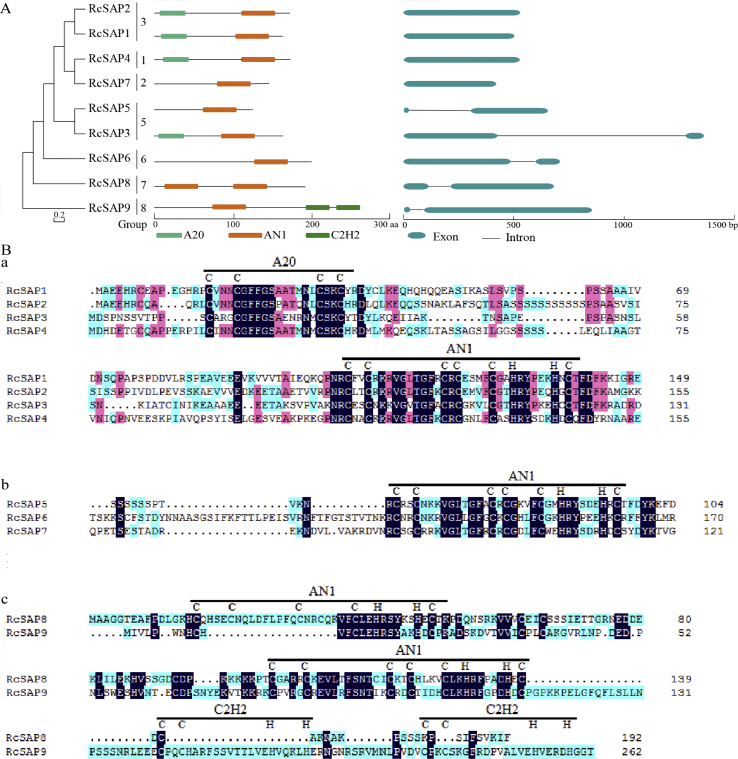
The structural features of castor bean SAP genes were analyzed using GSDS. Four castor bean SAP genes (RcSAP1, 2,4 and 7) had no introns and five castor bean SAP genes (RcSAP3,5,6, 8 and 9) had one intron (Fig. 1A and Table 1). According to the functional domain analysis by SMART and Pfam databases (Table 1 and Fig. 1A and B), RcSAP1-4 contained one A20 domain and one AN1 domain (type I, Cx2Cx9–12Cx1–2Cx4Cx2Hx5HxC consensus sequence) at their N terminus and C terminus respectively, RcSAP5-7 only had a single AN1 domain (type I) and RcSAP8 contained two AN1 domains (type II, Cx4Cx9–12Cx1–2Cx4Cx2Hx5HxC consensus sequence). Furthermore, RcSAP9 possessed a single AN1 domain (type II) with an additional paired C2H2 domain.
Fig. 1.
Identification of functional domains and structural divergence of RcSAP genes in castor bean. (A) Distribution of conserved domains (A20, AN1 and C2H2) of each castor bean SAP gene and their exon/intron organization on genome. (B) Alignment of protein sequences of RcSAPs. (a) Gene structure and distribution of A20 and AN1 domains in the A20-AN1-type; (b) gene structure and distribution of AN1 domain in the AN-type; (c) gene structure and distribution of AN1 and C2H2 domains the AN1-C2H2–C2H2 type.
Our search for SAP genes in the genomes of spurge family species (same family as castor bean) identified 11 SAP genes from J. curcas, 16 from M. esculenta and 11 from H. brasiliensis (Table 2). Functional domain analysis revealed that nine JcSAPs, 13 MeSAPs and seven HbSAPs were A20-AN1-type SAPs. Only one JcSAP and one MeSAP belonged to the two-AN1 type SAP. One MeSAP and three HbSAPs belonged to the single AN1 type SAP. The single AN1 type is absent in JcSAPs. One JcSAP and one MeSAP belonged to the SAP type with two AN1 and paired C2H2. This SAP type was absent in H. brasiliensis. Similar to the RcSAP9 in castor bean, only a single HbSAP gene of the AN1-AN1-C2H2 type was identified, suggesting the AN1-AN1-C2H2 type is rare in these plants. Of the nine castor bean SAPs, only four belong to the A20-AN1 type. Compared to the other spurge family species examined (J. curcas, M. esculenta and H. brasiliensis), there are fewer SAP genes in castor bean, and of these, fewer A20-AN1 type SAPs.
Table 2.
Comparison of SAP family members in R.communis, A. thaliana, J. curcas, M. esculenta and H. brasiliensis.
| Zinc finger domain | R. communis | A. thaliana | J. curcas | M. esculenta | H. brasiliensis |
|---|---|---|---|---|---|
| A20-AN1 | RcSAP2 | AtSAP1 | JCGZ_20731 | Manes.02G032000 | KAF2313176 |
| RcSAP4 | AtSAP2 | JCGZ_07939 | Manes.01G071400 | KAF2298958 | |
| RcSAP3 | AtSAP3 | JCGZ_25898 | Manes.06G166800 | KAF2300798 | |
| RcSAP1 | AtSAP4 | JCGZ_23668 | Manes.11G160700 | KAF2319417 | |
| AtSAP5 | JCGZ_16739 | Manes.S080300 | KAF2319447 | ||
| AtSAP6 | JCGZ_26211 | Manes.14G001000 | KAF2320699 | ||
| AtSAP7 | JCGZ_09438 | Manes.08G023800 | KAF2293600 | ||
| AtSAP8 | JCGZ_21704 | Manes.07G004500 | |||
| AtSAP9 | JCGZ_16738 | Manes.01G008400 | |||
| AtSAP10 | Manes.01G017400 | ||||
| Manes.07G004400 | |||||
| Manes.14G032700 | |||||
| Manes.10G145700 | |||||
| AN1 | RcSAP5 | AtSAP14 | Manes.01G008700 | KAF2321630 | |
| RcSAP6 | KAF2289734 | ||||
| RcSAP7 | KAF2316176 | ||||
| AN1-AN1 | RcSAP8 | AtSAP12 | JCGZ_09661 | Manes.15G180600 | |
| AN1-AN1-C2H2 | AtSAP13 | ||||
| AN1-C2H2–C2H2 | RcSAP9 | KAF2307917 | |||
| AN1-AN1-C2H2–C2H2 | AtSAP11 | JCGZ_21077 | Manes.09G023500 |
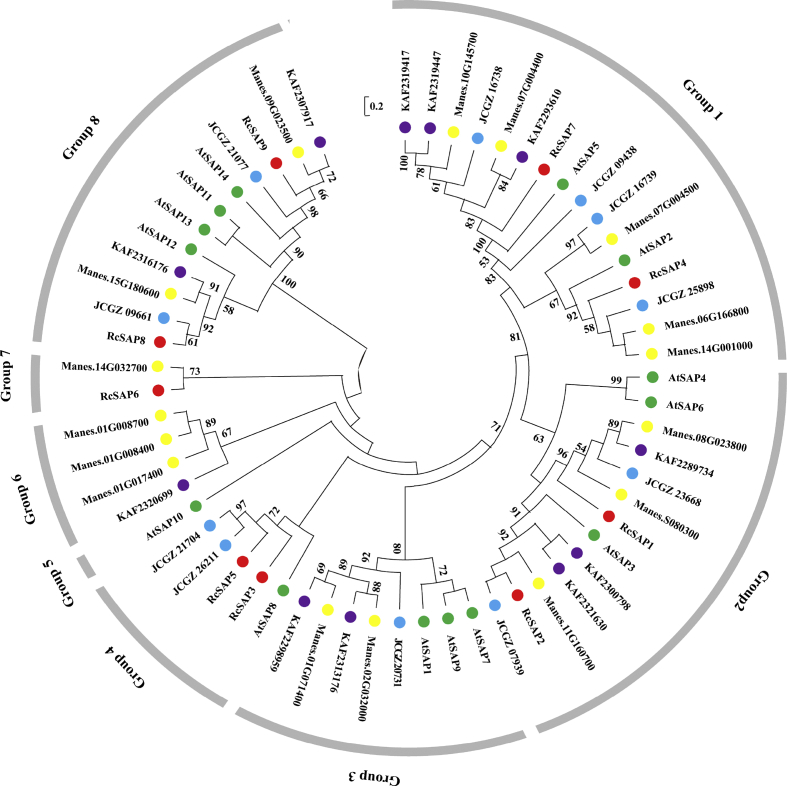
To determine the phylogenetic relationships between castor bean SAP genes, phylogenetic analysis was conducted with protein sequences of castor bean SAPs, A. thaliana SAPs, J. curcas SAPs, M. esculenta SAPs and H. brasiliensis SAPs. Phylogenetic analysis revealed eight distinct clusters of SAPs. Castor bean SAPs were absent from clusters 3, 5 and 6 (Fig. 2). Most SAP genes that shared the same functional domains were not clustered. These results suggest that outside of conserved zinc finger domains the amino acid sequences of SAP proteins are not phylogenetically conserved.
Fig. 2.
Phylogenetic analysis of SAP family members in Ricinus communis, Arabidopsis thaliana, Jatropha curcas, Manihot esculenta and Hevea brasiliensis. The Neighbor-Joining (NJ) tree was constructed based on protein sequences including castor bean (indicated by red dots), Arabidopsis (indicated by green dots), J. curcas (indicated by green dots), M. esculenta (indicated by green dots) and H. brasiliensis (indicated by purple dots). The bootstrap values (≥50) are shown on the root of each branch.
3.2. Expression patterns of castor bean SAP genes in various tissues
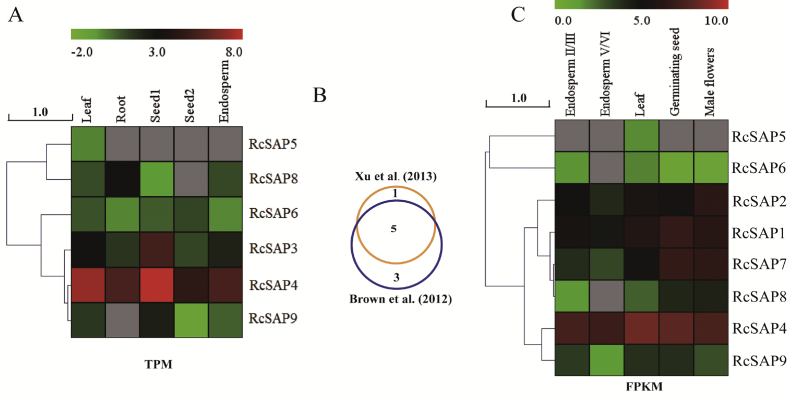
To investigate the expression patterns of castor bean SAP genes in different tissues under normal conditions, expression data were obtained from Xu et al. (2013) and Brown et al. (2012). Six (RcSAP3, 4, 5, 6, 8 and 9) and eight (except for RcSAP3) of nine castor bean SAP genes were identified from the two data sets, respectively (Fig. 3). Five transcripts (RcSAP4, 5, 6, 8 and 9) were detected in both data sets. These analyses showed that RcSAP3, 4 and 6 were expressed in all tested tissues, including in the leaf, root, developing seed1 (at the early stage of seed development), seed2 (at the middle stage of seed development) and developing endosperm. The expression levels of most castor bean SAP genes seem low, except for RcSAP4, which was relatively highly expressed across all tested tissues. According to data from Xu et al. (2013), RcSAP5 was expressed in the leaf and not in other tested tissues; RcSAP8 was not expressed in developing seed2; and RcSAP9 was not expressed in the root tissue. According to data from Brown et al. (2012), most castor bean SAP genes exhibited similar expression patterns among different tissues. For example, RcSAP5 was specifically expressed in the leaf; and RcSAP4 was relatively highly expressed across all tested tissues. We discovered that RcSAP1, 2 and 7 exhibited different patterns in the two data sets. Most castor bean SAP genes were expressed in various tissues, although their expression was not synchronous and was relatively low.
Fig. 3.
The heat maps indicating the expression patterns of RcSAP genes in various tissues. (A) the expression patterns of RcSAP genes among different tissues including leaf, root, developing seeds at the different stages (seed 1 and seed 2) and endosperm were obtained based on the dataset of the Tag-seq generated by Xu et al. (2013); (B) The overlapped RcSAP genes between two datasets. (C) The expression patterns of RcSAP genes among leaf, germinating seed, developing endosperm at the different stages (II/III, V/VI), and male flower tissues were obtained from the dataset of the RNA-seq generated by Brown et al. (2012). Expression values are log2-transformed.
3.3. Expression patterns of castor bean SAP genes in response to various stresses
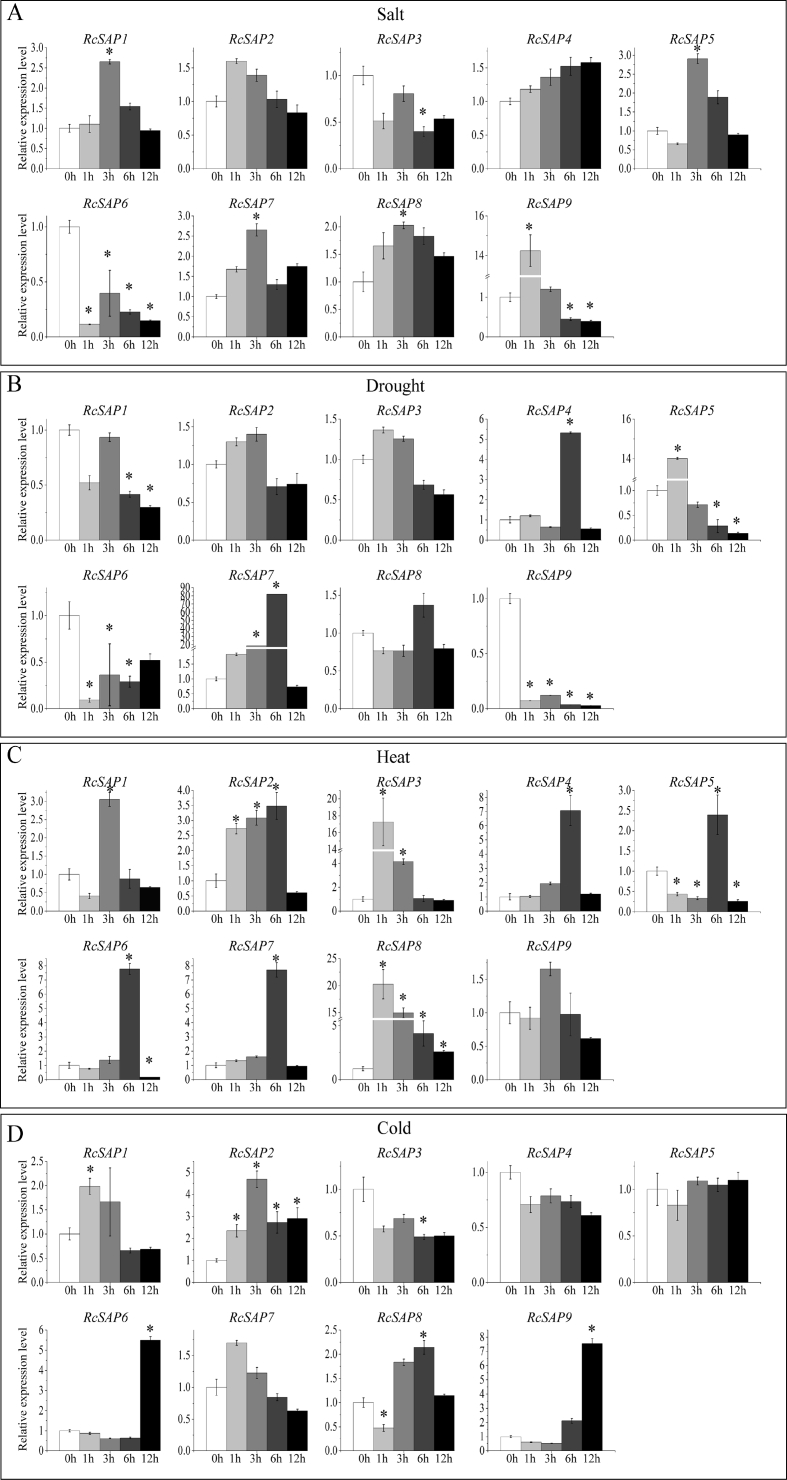
To investigate the potential function of castor bean SAP genes in response to stresses, we examined the gene expression of castor bean SAP genes in leaves treated with salt, drought, heat, and cold stresses (Fig. 4). Under salt stress, four castor bean SAP genes (RcSAP1, 5, 7 and 9) were significantly induced (the relative fold change ≥ 2) with the highest expression in RcSAP9 (14.24 folds) at 1 h of salt treatment (Fig. 4A). Under drought stress, three castor bean SAP genes (RcSAP4, 5 and 7) were significantly induced. Of these, the transcript peak of RcSAP7 reached over 80-fold at 6 h of drought treatment. The expression of RcSAP9 was apparently repressed, and the other SAP genes did not show significant differences in expression in response to drought treatment (Fig. 4B). Under heat stress, eight castor bean SAP genes (all except RcSAP9) were significantly induced with the highest expression in RcSAP8 (~20 fold) at 1 h of heat stress (Fig. 4C). Under cold stress, five castor bean SAP genes (RcSAP1, 2, 6, 8 and 9) were highly induced. The transcript peak of RcSAP9 was nearly eight-fold within 12 h of cold treatment. Other castor bean SAP genes did not show significant differences in transcript levels under cold treatment (Fig. 4D). When we compared the expression patterns of castor bean SAP genes under salt, drought, heat and cold treatments, we found that each SAP gene was strongly induced by at least one stress treatment, but that no SAP gene was highly induced by all four stress treatments. Of all castor bean SAP genes, RcSAP1, 5 and 7 were significantly induced under three stress treatments, and RcSAP3 was significantly induced by only heat treatment.
Fig. 4.
The expression patterns of RcSAP genes under various abiotic stresses based on qRT-PCR methods. (A), (B), (C) and (D) represent the expressional changes of each RcSAP gene in leaf tissues with treatment time under salt, drought, heat and cold treatments, respectively. ∗ denotes a significant difference (the fold change ≥ 2) compared with the control (at the 0 h). The bars represent means of relative expression level with standard error (SE), based on the three independent repetitions.
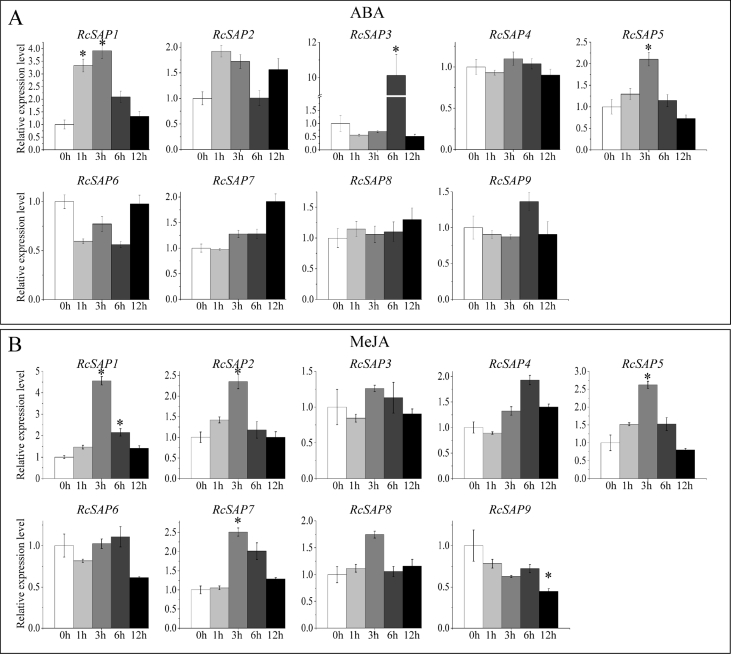
To test whether castor bean SAP gene expression is sensitive to ABA or JA signals, we examined castor bean SAP gene expression profiles after exogenous ABA and MeJA treatments (Fig. 5). We found that three castor bean SAP genes (RcSAP1, 3 and 5) were significantly induced by ABA treatment within 3 or 6 h (Fig. 5A). Similarly, four castor bean SAP genes (RcSAP1, 2, 5 and 7) were significantly induced by MeJA treatment within 3 h; expression of five castor bean SAP genes did not change in response to MeJA (Fig. 5B). Additionally, we noted that RcSAP1 and RcSAP5 were significantly induced by both ABA and MeJA signals.
Fig. 5.
The expression patterns of RcSAP genes in leaf tissues under ABA and MeJA treatments. Changes in gene expression of each RcSAP gene in leaf tissues with treatment time under ABA treatment (A) and MeJA treatment (B), respectively. The bars represent means of relative expression level with standard error (SE), based on the three independent repetitions. ∗ denotes a significant difference (the fold change ≥ 2) compared with the control (at the 0 h).
3.4. The identification of cis-acting elements in promoter regions of castor bean SAP genes
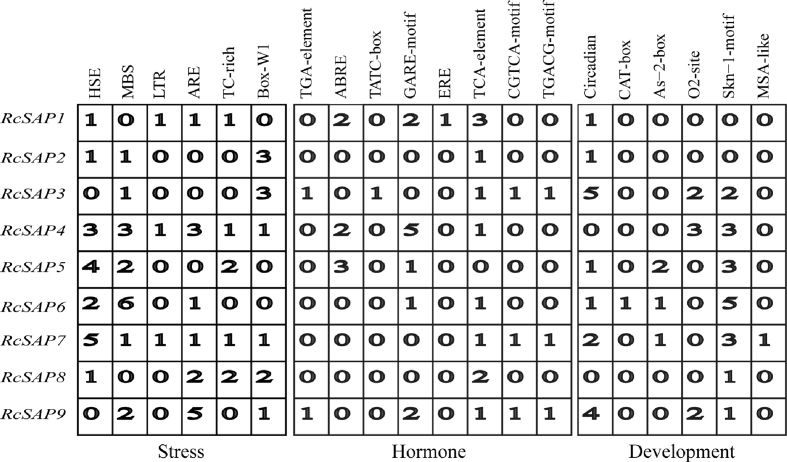
Cis-elements in promoter regions of genes are bound by transcription factor during transcriptional regulation. To investigate the mechanisms underlying castor bean SAP gene expression in response to stresses, we checked the −2000 bp of DNA sequence in promoter regions for each castor bean SAP gene. In total, we identified 20 cis-elements (see Fig. 6), including HSE, MBS, MYB, LTR, ARE and TC-rich repeats (in general, these cis-elements have been known to be functionally involved in regulating plant responses to diverse stresses), TGA, ABRE, TATC-box, GARE-motif, ERE, TCA-element, CGTCA-motif and TGACG-motif (these cis-elements have usually been considered to be binding sites with regulators responding to hormone signals), and Circadian, CAT-box, As-2-box, O2-site, Skn-1 motif and MSA-like (these cis-elements have been known to be functionally involved in binding with regulators that control plant growth and development). Promoter regions of most castor bean SAP genes harbor multiple cis-elements that are functionally involved in regulating plant responses to stresses. Seven of nine castor bean SAP genes harbor at least one or more HSE and MBS cis-elements, and six of nine castor bean SAP genes harbor one or more ARE and Box-W1 cis-elements in their promoter regions. In addition, we noted that only three castor bean SAP genes (RcSAP1, 3 and 9) have four or five kinds of cis-elements. Most castor bean SAP genes only have limited cis-elements that bind with hormone-responsive regulators. Similarly, only RcSAP6 and 7 harbor four kinds of cis-elements that bind with regulators involved in the regulation of plant growth and development. The Circadian and Skn-1 motif were distributed in the promoter regions of seven castor bean SAP genes. These results indicate that the promoter regions of castor bean SAP genes harbor various cis-elements, implying that these cis-elements may be bound by diverse factors during transcription regulation.
Fig. 6.
The identification of cis-acting elements in the promoters of RcSAPs. The numbers (from 1 to 6) indicates the number of each cis-element, and the 0 denotes absence. Stress box denotes the identified cis-acting elements functionally involved in responding to stresses, the hormone box denotes the identified cis-acting elements functionally involved in responding to hormone signals, and the development box denotes the identified cis-acting elements functionally involved in regulating plant development.
3.5. Subcellular localization of castor bean SAP genes
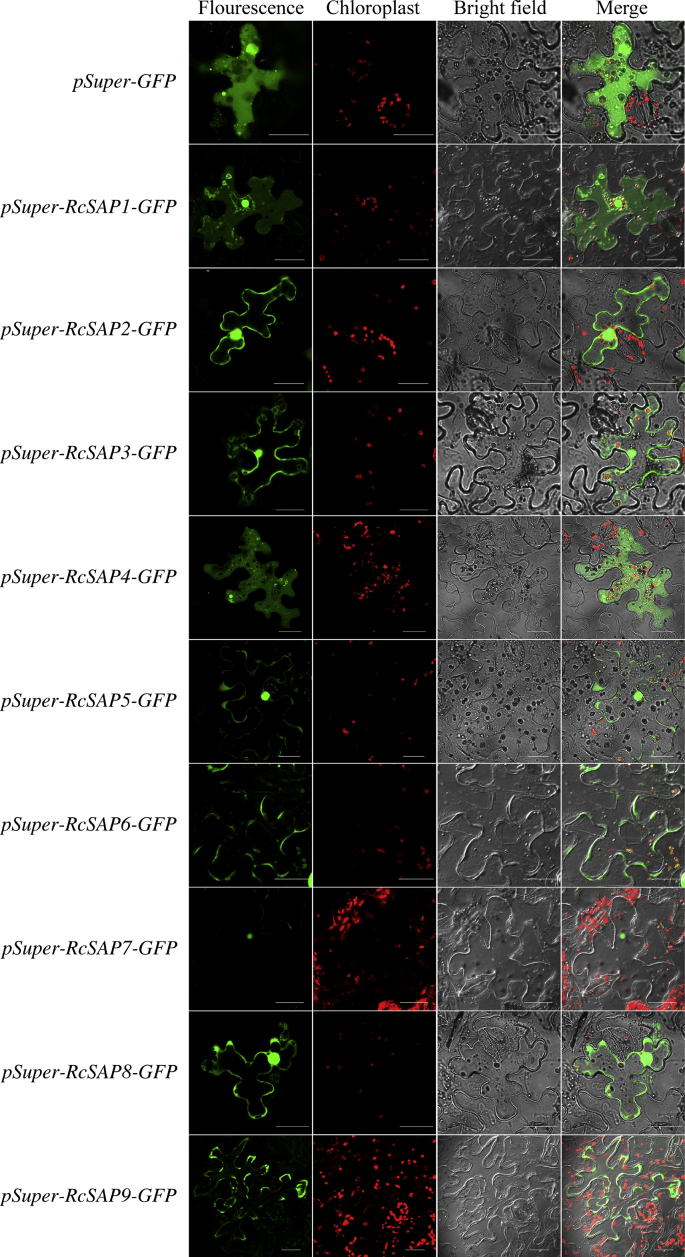
To investigate where castor bean SAP genes function within cells, we performed subcellular localization experiments for nine castor bean SAP genes by expressing pSuper-RcSAPs-GFP fusion proteins in tobacco leaf epidermal cells. Translated castor bean SAP genes were localized in different subcellular organelles (Fig. 7). RcSAP1 was detected in both the cytoplasm and nucleus; RcSAP2 was observed in the nucleus and the cytomembrane; RcSAP3, 5 and 8 were observed in the cytoplasm, nucleus and the cytomembrane; RcSAP4 was exclusively localized in cytoplasm; and RcSAP7 was observed primarily in the nucleus; RcSAP6 and 9 were exclusively localized in the cytomembrane. These findings clearly suggest that each castor bean SAP may have a unique function.
Fig. 7.
The subcellular localization of nine RcSAP genes. Nine RcSAPs were expressed with the fluorescent GFP label in transgenic tobacco leaves. The green fluorescence represents the expression of RcSAPs and the red fluorescence represents the auto-fluorescence of chloroplasts in tobacco leaf, and the “merge” represents their overlaps under confocal microscope; “bright field” indicates microscopy without fluorescence. Bar = 30 μm.
4. Discussion
In this study, we identified and characterized nine castor bean SAP genes at the genome level and provided a comprehensive gene expression profile that may reveal potential functions of SAPs in response to abiotic stresses in castor bean. There are fewer SAP genes in castor bean (nine SAPs) than in previously examined plants (e.g., maize has 11 SAPs, Arabidopsis has 14, M. truncatula has 17, P. euphratica has 18 and G. hirsutum has 37) (Gao et al., 2016; Jia et al., 2016; Zhou et al., 2018). Furthermore, castor bean has fewer SAPs than three species in the Euphorbiaceae family (J. curcas has 11 SAPs, M. esculenta has 16 and H. brasiliensis has 11). The smaller number of SAP genes might be related to the smaller genome size and the lower level of genome duplication during the evolution of castor bean. The castor bean genome (352 Mb) is smaller than those of J. curcas (379.5 Mb), M. esculenta (582.25 Mb) and H. brasiliensis (1.47 Gb), and castor bean, which has not undergone a recent species-specific whole-genome duplication event, has a lower level of genome duplication during its evolution (Yu et al., 2019; Tang et al., 2016).
We also found that the SAP gene types differ between castor bean and previously examined plant species. Specifically, the A20-AN1 type SAP is common in many plant species. J. curcas, M. esculenta and H. brasiliensis have nine, 13 and seven A20-AN1-type SAPs, respectively, whereas castor bean has only four A20-AN1-type SAPs. Similarly, A20-AN1-type SAPs predominate in other plants, including Arabidopsis, M. truncatula, P. euphratica and G. hirsutum (Gao et al., 2016; Jia et al., 2016; Zhou et al., 2018). However, the number of A20-AN1-type SAPs is variable in plant species. Phylogenetic analysis of Arabidopsis and four spurge species (castor bean, Jatropha curcass, M. esculenta and H. brasiliensis) revealed that the amino acid sequences of SAPs are not conserved outside of their shared functional domains (A20, AN1 and C2H2). Therefore, it remains unclear whether the structures of SAPs are associated with environmental tolerance in castor bean.
The expression profiles of castor bean SAP genes in various tissues and in response to multiple stresses reflect their potential functions. Expression analysis of castor bean SAP genes based on high throughput RNA-seq data of various tissues revealed that most castor bean SAP genes were expressed at low levels under normal conditions. RcSAP4 was relatively at relatively higher levels, implying that RcSAP4 might function in maintaining plant growth and seed development. We also found that castor bean SAP gene transcript levels differed slightly between the data sets of Xu et al. (2013) and Brown et al. (2012). This difference may be explained by different sequencing strategies and depths.
Previous studies have reported SAP genes could be rapidly and strongly induced by various abiotic stresses, including drought, salt, heat, and cold (Giri et al., 2013; Gao et al., 2016). In this study, we found that each castor bean SAP gene could be significantly induced by at least one stress treatment (i.e., salt, drought, heat and cold), but no single castor bean SAP gene was significantly induced by all four stress treatments. This indicates that not every RcSAP can be strongly induced in response to a given stress. Although a given RcSAP (excluding RcSAP3) can often be significantly induced by two or three stresses, expression patterns of these SAP genes varies in response to different abiotic stresses. These results suggest that each castor bean SAP gene might function differently when it is expressed in response to different stresses.
When we compared the expression patterns of A20-AN1 and AN1 castor bean SAP genes, we found that SAP genes of the same type did not exhibit similar expression patterns, suggesting that transcriptional regulation of castor bean SAP genes of the same type might vary in response to stresses. Previous studies have shown that two desert P. euphratica SAPs that belong to the AN1-AN1 type (PeuSAP16 and 1) are significantly induced by cold, salt, heat, and drought (Jia et al., 2016). However, in our study, the one AN1-AN1-type SAP in castor bean (RcSAP8) was not highly induced by salt or drought stresses. In Arabidopsis, AtSAP12, which is an AN1-AN1-type SAP, is not highly induced by cold or drought stresses (Ströher et al., 2009). In rice, OsSAP17, another AN1-AN1-type SAP, is strongly induced by salt and drought stresses, but not by cold treatment (Vij and Tyagi, 2006). These studies suggest that transcriptional regulation of SAP genes and the functions of SAPs in stress response may vary across plant species.
ABA and JA are critical signals that regulate plant responses to environmental stresses (Kang et al., 2017). Several studies have reported that most SAPs are rapidly induced by ABA or JA treatment (Gao et al., 2016; Jia et al., 2016), indicating that the transcription of SAP genesis closely linked with hormone signals. Here, we found that only three castor bean SAP genes (RcSAP1, 3 and 5) were significantly induced by exogenous ABA treatment or four SAPs (RcSAP1, 2, 5 and 7) were significantly induced by exogenous MeJA treatment within 12 h post-treatments. However, exogenous ABA and MeJA treatments elicited different expression patterns from castor bean SAP genes than did abiotic stresses. These results suggest that SAP gene expression in castor bean may be regulated independently from ABA or JA signal pathways, or at least partially independent. This possibility indicates that the regulatory mechanisms of gene transcription for most castor bean SAP genes are complex.
Cis-acting elements within the promoter region of genes determine whether a given gene is activated or repressed during plant growth and development or in response to environmental stresses. Here, we identified 20 important and known cis-acting elements functionally involved in regulating gene expression in response to diverse stresses, hormone signals, and the maintenance of plant growth and development. Furthermore, when we compared cis-acting elements in promoter regions of SAP genes of various plant species (castor bean, P. euphratica and, G. hirsutum, Cucumis sativus var. sativus L. and Glycine max (L.) Merr.) (Gao et al., 2016; Jia et al., 2016; Lai et al., 2020; Zhang et al., 2019), we found that the presence of the cis-acting elements was partially conserved among different plants. These results imply castor bean SAP genes are functionally diverse and their transcriptional regulation is complex. Notably, the promoter regions of most castor bean SAP genes contain multiple cis-acting elements functionally involved in regulating plant responses to stresses. The transcriptional activation of most castor bean SAP genes in response to different stresses can be explained, at least partially, by specific cis-acting elements in their promoter regions. For example, eight of the nine castor bean SAP genes were strongly induced by heat stress; correspondingly, the promoter regions of these genes contain cis-acting heat shock elements (HSE) that are bound by transcription factors that regulate plant responses to heat. Most castor bean SAP genes are not induced by ABA or MeJA signals, because the promoter regions of most castor bean SAP genes lack the cis-elements bound by either ABA-responsive transcription factors (ABRE), or MeJA-responsive transcription factors (CGTCA-motif and TGACG-motif).
The potential function of proteins can be determined by their subcellular localization. To our knowledge, the subcellular locations of only a few SAPs (Arabidopsis AtSAP5, AtSAP9, AtSAP10 and AtSAP13; rice OsSAP11) have been determined (Dixit and Dhankher, 2011; Giri et al., 2011; Kang et al., 2011; Dixit et al., 2018). In this study, we found that castor bean SAPs were localized in different subcellular organelles. In combination with our findings that SAP gene structure, expression profiles, and promoter regions vary, these findings suggest that SAP genes in castor bean are involved in maintaining plant growth and development and play important roles in regulating plant responses to various abiotic stresses.
5. Conclusions
In this study, we identified nine castor bean SAP genes and characterized their gene structures and functional domains. Expression patterns of castor bean SAP genes was tissue-specific and varied in response to diverse abiotic stresses, suggesting that transcriptional regulation of these genes is complex. Our finding that gene expression for only three castor bean SAP genes was induced by ABA and/or MeJA, suggest that regulation of these genes may operate independently, or at least partially independently, of ABA and MeJA signal pathways. Cis-element analyses and subcellular localization of each castor bean SAP gene add to our understanding of the physiological and molecular functions of SAP genes involved in regulating growth and development and response to different abiotic stresses.
Author contributions
Z. Wang, S. Chen and A. Liu conceived and designed the experiments; Z. Wang and J. Kuang performed the experiments; J. Kuang and B. Han prepared plant materials; Z. Wang and A. Liu wrote the paper. All authors reviewed and approved the final manuscript.
Declaration of Competing Interest
The authors declare no conflict of interest.
Acknowledgements
This research was funded by National Natural Science Foundation of China (31661143002, 31771839, 31701123and 31501034), Yunnan Applied Basic Research Projects (2016FB060 and 2016FB040). We thank Prof. Piaotao Wang (from the State Key Laboratory of Cotton Biology, College of Life Sciences, Henan University) for giving us the pSuper 1300 vector.
Footnotes
Peer review under responsibility of Editorial Office of Plant Diversity.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2020.07.010.
Contributor Information
Zaiqing Wang, Email: wangzaiqing@mail.kib.ac.cn.
Jingge Kuang, Email: kjg1106@163.com.
Bing Han, Email: hanbing@mail.kib.ac.cn.
Suiyun Chen, Email: chensuiyun@ynu.edu.cn.
Aizhong Liu, Email: liuaizhong@mail.kib.ac.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Ben S.R., Fabre D., Mieulet D. Expression of the Aeluropus littoralis AlSAP gene in rice confers broad tolerance to abiotic stresses through maintenance of photosynthesis. Plant Cell Environ. 2012;35:626–643. doi: 10.1111/j.1365-3040.2011.02441.x. [DOI] [PubMed] [Google Scholar]
- Ben S.R., Farhat K.A., Ben H.N. The LmSAP gene isolated from the halotolerant Lobularia maritima improves salt and ionic tolerance in transgenic tobacco lines. Funct. Plant Biol. 2018;45:378–390. doi: 10.1071/FP17202. [DOI] [PubMed] [Google Scholar]
- Brown A.P., Kroon J.T., Swarbreck D. Tissue-specific whole transcriptome sequencing in castor, directed at understanding triacylglycerol lipid biosynthetic pathways. PLoS One. 2012;7 doi: 10.1371/journal.pone.0030100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatre L., Brandizzi F., Hocquellet A. Sec22 and Memb11 are v-SNAREs of the anterograde endoplasmic reticulum - Golgi pathway in tobacco leaf epidermal cells. Plant Physiol. 2005;139:1244–1254. doi: 10.1104/pp.105.067447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dansana P.K., Kothari K.S., Vij S. OsiSAP1 overexpression improves water-deficit stress tolerance in transgenic rice by affecting expression of endogenous stress-related genes. Plant Cell Rep. 2014;33:1425–1440. doi: 10.1007/s00299-014-1626-3. [DOI] [PubMed] [Google Scholar]
- Dixit A., Tomar P., Vaine E. A stress-associated protein, AtSAP13, from Arabidopsis thaliana provides tolerance to multiple abiotic stresses. Plant Cell Environ. 2018;41:1171–1185. doi: 10.1111/pce.13103. [DOI] [PubMed] [Google Scholar]
- Dixit A.R., Dhankher O.P. A novel stress-associated protein 'AtSAP10' from Arabidopsis thaliana confers tolerance to nickel, manganese, zinc, and high temperature stress. PLoS One. 2011;6 doi: 10.1371/journal.pone.0020921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W., Long L., Tian X. Genome-wide identification and expression analysis of stress-associated proteins (SAPs) containing A20/AN1 zinc finger in cotton. Mol. Genet. Genom. 2016;291:2199–2213. doi: 10.1007/s00438-016-1252-6. [DOI] [PubMed] [Google Scholar]
- Giri J., Dansana P.K., Kothari K.S. SAPs as novel regulators of abiotic stress response in plants. Bioessays. 2013;35:639–648. doi: 10.1002/bies.201200181. [DOI] [PubMed] [Google Scholar]
- Giri J., Vij S., Dansana P.K. Rice A20/AN1 zinc-finger containing stress-associated proteins (SAP1/11) and a receptor-like cytoplasmic kinase (OsRLCK253) interact via A20 zinc-finger and confer abiotic stress tolerance in transgenic Arabidopsis plants. New Phytol. 2011;191:721–732. doi: 10.1111/j.1469-8137.2011.03740.x. [DOI] [PubMed] [Google Scholar]
- Hu B., Jin J., Guo A.Y. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31:1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Wang M.M., Jiang Y. Expression analysis of rice A20/AN1-type zinc finger genes and characterization of ZFP177 that contributes to temperature stress tolerance. Gene. 2008;420:135–144. doi: 10.1016/j.gene.2008.05.019. [DOI] [PubMed] [Google Scholar]
- Jia H., Li J., Zhang J. Genome-wide survey and expression analysis of the stress-associated protein gene family in desert poplar, Populus euphratica. Tree Genet. Genomes. 2016;12:1–16. [Google Scholar]
- Kang M., Abdelmageed H., Lee S. AtMBP-1, an alternative translation product of LOS2, affects abscisic acid responses and is modulated by the E3 ubiquitin ligase AtSAP5. Plant J. 2013;76:481–493. doi: 10.1111/tpj.12312. [DOI] [PubMed] [Google Scholar]
- Kang M., Fokar M., Abdelmageed H. Arabidopsis SAP5 functions as a positive regulator of stress responses and exhibits E3 ubiquitin ligase activity. Plant Mol. Biol. 2011;75:451–466. doi: 10.1007/s11103-011-9748-2. [DOI] [PubMed] [Google Scholar]
- Kang M., Lee S., Abdelmageed H. Arabidopsis stress associated protein 9 mediates biotic and abiotic stress responsive ABA signaling via the proteasome pathway. Plant Cell Environ. 2017;40:702–716. doi: 10.1111/pce.12892. [DOI] [PubMed] [Google Scholar]
- Kanneganti V., Gupta A.K. Overexpression of OsiSAP8, a member of stress associated protein (SAP) gene family of rice confers tolerance to salt, drought and cold stress in transgenic tobacco and rice. Plant Mol. Biol. 2008;66:445–462. doi: 10.1007/s11103-007-9284-2. [DOI] [PubMed] [Google Scholar]
- Kim G.D., Cho Y.H., Yoo S.D. Regulatory functions of evolutionarily conserved AN1/A20-like Zinc finger family proteins in Arabidopsis stress responses under high temperature. Biochem. Biophys. Res. Commun. 2015;457:213–220. doi: 10.1016/j.bbrc.2014.12.090. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai W., Zhou Y., Pan R. Identification and expression analysis of stress-associated proteins (SAPs) containing A20/AN1 zinc finger in cucumber. Plants. 2020;9:400–415. doi: 10.3390/plants9030400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay A., Vij S., Tyagi A.K. Overexpression of a zinc-finger protein gene from rice confers tolerance to cold, dehydration, and salt stress in transgenic tobacco. Proc. Natl. Acad. Sci. USA. 2004;101:6309–6314. doi: 10.1073/pnas.0401572101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu L., Yang C., Tian B. Exploiting EST databases for the development and characterization of EST-SSR markers in castor bean (Ricinus communis L.) BMC Plant Biol. 2010;10:278. doi: 10.1186/1471-2229-10-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma G., Giri J., Tyagi A.K. Rice OsiSAP7 negatively regulates ABA stress signalling and imparts sensitivity to water-deficit stress in Arabidopsis. Plant Sci. 2015;237:80–92. doi: 10.1016/j.plantsci.2015.05.011. [DOI] [PubMed] [Google Scholar]
- Solanke A.U., Sharma M.K., Tyagi A.K. Characterization and phylogenetic analysis of environmental stress-responsive SAP gene family encoding A20/AN1 zinc finger proteins in tomato. Mol. Genet. Genom. 2009;282:153–164. doi: 10.1007/s00438-009-0455-5. [DOI] [PubMed] [Google Scholar]
- Sreedharan S., Shekhawat U.K., Ganapathi T.R. MusaSAP1, a A20/AN1 zinc finger gene from banana functions as a positive regulator in different stress responses. Plant Mol. Biol. 2012;80:503–517. doi: 10.1007/s11103-012-9964-4. [DOI] [PubMed] [Google Scholar]
- Ströher E., Wang X.J., Roloff N. Redox-dependent regulation of the stress-induced zinc-finger protein SAP12 in Arabidopsis thaliana. Mol. Plant. 2009;2:357–367. doi: 10.1093/mp/ssn084. [DOI] [PubMed] [Google Scholar]
- Tang C., Yang M., Fang Y. The rubber tree genome reveals new insights into rubber production and species adaptation. Native Plants. 2016;2:16073–16082. doi: 10.1038/nplants.2016.73. [DOI] [PubMed] [Google Scholar]
- Tyagi H., Jha S., Sharma M. Rice SAPs are responsive to multiple biotic stresses and overexpression of OsSAP1, an A20/AN1 zinc-finger protein, enhances the basal resistance against pathogen infection in tobacco. Plant Sci. 2014;225:68–76. doi: 10.1016/j.plantsci.2014.05.016. [DOI] [PubMed] [Google Scholar]
- Vij S., Tyagi A.K. Genome-wide analysis of the stress associated protein (SAP) gene family containing A20/AN1 zinc-finger(s) in rice and their phylogenetic relationship with Arabidopsis. Mol. Genet. Genom. 2006;276:565–575. doi: 10.1007/s00438-006-0165-1. [DOI] [PubMed] [Google Scholar]
- Wang C., Li G.R., Zhang Z.Y. Genetic diversity of castor bean (Ricinus communis L.) in Northeast China revealed by ISSR markers. Biochem. Systemat. Ecol. 2013;51:301–307. [Google Scholar]
- Wang Y., Zhang L., Zhang L. A novel stress-associated protein SbSAP14 from Sorghum bicolor confers tolerance to salt stress in transgenic rice. Mol. Breed. 2013;32:437–449. [Google Scholar]
- Wang Z., Yang C., Chen H. Multi-gene co-expression can improve comprehensive resistance to multiple abiotic stresses in Brassica napus L. Plant Sci. 2018;274:410–419. doi: 10.1016/j.plantsci.2018.06.014. [DOI] [PubMed] [Google Scholar]
- Xu W., Chen Z., Ahmed N. Genome-wide identification, evolutionary analysis, and stress responses of the GRAS gene family in castor beans. Int. J. Mol. Sci. 2016;17:1004. doi: 10.3390/ijms17071004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Li F., Ling L. Genome-wide survey and expression profiles of the AP2/ERF family in castor bean (Ricinus communis L.) BMC Genom. 2013;14:785. doi: 10.1186/1471-2164-14-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin M., Wang Y., Zhang L. The Arabidopsis Cys2/His2 zinc finger transcription factor ZAT18 is a positive regulator of plant tolerance to drought stress. J. Exp. Bot. 2017;68:2991–3005. doi: 10.1093/jxb/erx157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A., Li F., Xu W. Application of a high-resolution genetic map for chromosome-scale genome assembly and fine QTLs mapping of seed size and weight traits in castor bean. Sci. Rep. 2019;9:11950–11960. doi: 10.1038/s41598-019-48492-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Ke T., Tehrim S. PTGBase: an integrated database to study tandem duplicated genes in plants. Database. 2015;15:1–10. doi: 10.1093/database/bav017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Zheng W., Cao X. Genomic analysis of stress associated proteins in soybean and the role of GmSAP16 in abiotic stress responses in Arabidopsis and Soybean. Front. Plant Sci. 2019;10:1453–1471. doi: 10.3389/fpls.2019.01453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Lan H., Shao Q. An A20/AN1-type zinc finger protein modulates gibberellins and abscisic acid contents and increases sensitivity to abiotic stress in rice (Oryza sativa) J. Exp. Bot. 2016;67:315–326. doi: 10.1093/jxb/erv464. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Zeng L., Chen R. Genome-wide identification and characterization of stress-associated protein (SAP) gene family encoding A20/AN1 zinc-finger proteins in Medicago truncatula. Arch. Biol. Sci. 2018;70:87–98. [Google Scholar]
- Zouari N., Ben Saad R., Legavre T. Identification and sequencing of ESTs from the halophyte grass Aeluropus littoralis. Gene. 2007;404:61–69. doi: 10.1016/j.gene.2007.08.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.