ABSTRACT
Assessing genetic relatedness of Plasmodium falciparum genotypes is a key component of antimalarial efficacy trials. Previous methods have focused on determining a priori definitions of the level of genetic similarity sufficient to classify two infections as sharing the same strain. However, factors such as mixed-strain infections, allelic suppression, imprecise typing methods, and heterozygosity complicate comparisons of apicomplexan genotypes. Here, we introduce a novel method for nonparametric statistical testing of relatedness for P. falciparum parasites. First, the background distribution of genetic distance between unrelated strains is computed. Second, a threshold genetic distance is computed from this empiric distribution of distances to demarcate genetic distances that are unlikely to have arisen by chance. Third, the genetic distance between paired samples is computed, and paired samples with genetic distances below the threshold are classified as related. The method is designed to work with any arbitrary genetic distance definition. We validated this procedure using two independent approaches to calculating genetic distance. We assessed the concordance of the novel nonparametric classification with a gold-standard Bayesian approach for 175 pairs of recurrent P. falciparum episodes from previously published malaria efficacy trials with microsatellite data from five studies in Guinea and Angola. The novel nonparametric approach was 98% sensitive and 84–89% specific in correctly identifying related genotypes compared with a gold-standard Bayesian algorithm. The approach provides a unified and systematic method to statistically assess relatedness of P. falciparum parasites using arbitrary genetic distance methodologies.
INTRODUCTION
Advances in genotyping technologies have opened new opportunities for understanding the molecular epidemiology of apicomplexan parasites, which include the Plasmodium malaria parasites as well as other clinically important parasites such as Cyclospora cayetanensis. Broadly, parasite genotyping is often performed with one of two major objectives. One objective is to obtain insights into broader, higher-level population trends. This type of analysis may help investigators differentiate populations originating from different geographic regions, or discriminate between populations derived from different hosts.1,2 Alternatively, finer “forensic-level” discrimination of infections may be required, where individual patient samples are compared with each other to determine whether they contain the same genotype. This level of discrimination applies in contexts where the investigator wishes to determine if a group of patients obtained their infections from precisely the same time point of exposure,3,4 or to understand whether a person who was infected in the past and was treated is now infected again with precisely the same parasites as before.5
An important example of where highly granular “forensic-level” discrimination of genotypes is required occurs in the context of antimalarial efficacy trials, which is arguably the most common motive to assess genetic similarity in Plasmodium falciparum. In these trials, patients with malaria infection are treated and monitored to evaluate clearance of infection. Episodes of recurrent parasitemia during the later periods of follow-up can be due either to new infections with unrelated strains or recrudescence of the original parasite. To distinguish between these two cases, parasites in patient blood samples from day 0 (D0) and the day of failure (DOF) are genotyped, and their genetic signatures are compared. Participants with matching D0–DOF genotypes are classified as recrudescences, and mismatches are counted as new infections. In this example, population-level discrimination is insufficient as there exists the requirement to distinguish between infections caused by parasites of the same population, propagating in the same geographic location (i.e., intrapopulation discrimination is needed). Indeed, the requirement for this level of discrimination also applies to other apicomplexan contexts; a similar level of intrapopulation discrimination is required when genotyping the foodborne parasite C. cayetanensis for the purposes of identifying case patients with genetically related infections, indicative of a common-source exposure to the parasite.4,6
Importantly, defining a match between two genotypes is not straightforward. Complex infections with more than one strain, allelic suppression, imprecise and insensitive typing methods, the presence of heterozygosity, and lack of diversity are all factors that complicate comparisons of genotypes.4,5,7–11
In the context of comparing paired samples from malaria drug efficacy trials, most studies genotype parasites by measuring the fragment lengths of three well-characterized length-polymorphic markers: msp1, msp2, and glurp. Less frequently, other methods used include neutral length polymorphic markers (microsatellites), single nucleotide–based bar codes, and (recently) targeted amplicon sequencing. Traditionally, the accepted methodology for comparing msp1/msp2/glurp genotyping data was to require a match at all three markers to classify a D0–DOF pair as a recrudescence.5 Recent simulation studies have shown, however, that a more relaxed criterion requiring a match at two of the three markers is less biased in settings with mixed-strain infections and high likelihood of missing data.12 A Bayesian algorithm for analysis of microsatellite data has been used that calculates the posterior probability of recrudescence based on the likelihood of the observed number of shared alleles, given the allelic frequencies.13 This algorithm was shown to outperform more naive counting-based methods that define a recrudescence based on a threshold number of matching loci.10
With the increased use of targeted amplicon sequencing and other novel genotyping methods, new algorithms have been described to distinguish between related and unrelated infections while accommodating these new types of genotyping data.4,11 These algorithms invariably calculate some measurement of distance with the goal of defining the relationships between each genotype in the test population. However, little attention has been paid to the selection of appropriate cutoff values to define what level of genetic similarity is evidence of a close or disparate genetic relationship. Unfortunately, frameworks for defining a rational and unbiased distance cutoff to facilitate a binary classification of paired genotypes (i.e., related or unrelated) are lacking. Consequently, these cutoffs are often selected empirically and without any clear basis, despite the major negative impacts that inappropriate cutoff selection can have on the final interpretation of results. Ideally, an unbiased system for selecting a rational distance cutoff for a binary classification would be generalizable to any/all available distance statistics. This feature would afford investigators the flexibility to select a statistic that suits their specific needs while retaining a uniform approach for cutoff selection.
Here, we provide a generalized statistical framework for distinguishing between related and unrelated infections (i.e., in a binary manner) that is nonparametric and marker independent. We apply this framework to distances calculated using two approaches for real-world microsatellite datasets from P. falciparum antimalarial efficacy trials, and use this framework to select a cutoff that results in a comparable level of performance despite the choice of genetic distance measurement. In doing so, we provide investigators with a tool that facilitates the rational and unbiased selection of a cutoff to facilitate the binary classification of paired genotypes that is applicable in any context where a measurement of genetic distance is employed.
METHODS
We developed a three-step process for distinguishing between related and unrelated infections using a microsatellite dataset generated for P. falciparum as an example. For the first step, the genetic distance between all possible pairs of D0–D0 samples is calculated using any method for characterizing genetic distance (dissimilarity), as chosen by the investigator. In the second step, a threshold of genetic distance is calculated as the empiric lower fifth percentile of the observed D0–D0 pairwise distances. In the third step, pairwise genetic distances between paired D0–DOF samples are calculated, and any paired D0–DOF samples with a genetic distance below the defined threshold are then considered to be recrudescences (i.e., infections caused by the same parasites). For these samples, the likelihood of observing the genetic distance by chance is less than 5%, corresponding to a statistical test with a false-positive rate (α) of 5%. As part of this validation study, we used two different, independent genetic distance methods to assess the robustness of this approach.
Data collection.
We obtained previously collected and publicly available microsatellite genotyping data from five studies with 13 trial sites14–18 (Table 1). Seven microsatellite loci were included: TA1, Poly-α, PfPK2, TA109, TA2490, C2M34, and C3M69. Depending on the trial, data from 6 to 7 loci were available. Data represented the lengths of the tandem repeat loci, as measured using capillary electrophoresis.
Table 1.
Datasets included in nonparametric classification of recurrent parasitemias
| Study | Reference | Study sites | D0 samples | DOF samples | D0–D0 pairs | D0–DOF pairs |
|---|---|---|---|---|---|---|
| Angola 2013 | 14 | 2 | 50 | 25 | 721 | 25 |
| Angola 2015 | 15 | 3 | 137 | 35 | 3,976 | 35 |
| Angola 2017 | 16 | 3 | 101 | 38 | 1766 | 38 |
| Angola 2019 | 18 | 3 | 167 | 54 | 5,055 | 54 |
| Guinea 2016 | 17 | 2 | 46 | 23 | 531 | 23 |
| Total | 13 | 501 | 175 | 12,049 | 175 |
D0 = day 0; DOF = day of failure.
Data analysis.
We calculated the pairwise distance between each pair of D0–D0 samples from each trial, limiting pairwise comparisons to samples collected from the same trial site. The first genetic distance method (method A) used a simple match-counting algorithm. In this approach, the genetic distance Dij between samples i and j was defined as the proportion of loci with at least one matching allele. A matching allele was defined as two alleles differing by less than two base pairs, the typical assumed error rate of microsatellite fragment length measurement using capillary electrophoresis. For each pairwise comparison, loci for which one of the samples was missing data were excluded from both the numerator and the denominator.
The second distance calculation method used here (method B) was a heuristic algorithm for the calculation of a distance between sexual parasites.11 This method constitutes an algorithm based on propositional logic, set theory, and frequentist probabilities, and has been applied previously to multi-locus sequence-typing datasets generated for the apicomplexan parasite C. cayetanensis, and the nematode Strongyloides,3 and is therefore an appropriate measure of distance for use in the present comparison.
For the antimalarial efficacy data, we plotted the distribution of pairwise D0–D0 distance for all sites combined and calculated the empiric 5% quintile (D0.05) for each method. We then calculated the pairwise distance between all paired D0–DOF samples from recurrent parasitemias. We classified recrudescences as participants for which Dij < D0.05.
We used a previously developed Bayesian algorithm13 to calculate the posterior probability of recrudescence for each D0–DOF pair. We next calculated the concordance between recrudescence calls for D0–DOF pairs using the Dij < D0.05 nonparametric classifier and pairs for which the Bayesian posterior probability of recrudescence was > 0.95.
RESULTS
A total of 175 pairs of D0–DOF samples were available from the previously generated microsatellite data14–17 (Table 1). We calculated a total of 10,286 D0–D0 pairwise distances, ranging from 514 to 3,976 per study.
Method A.
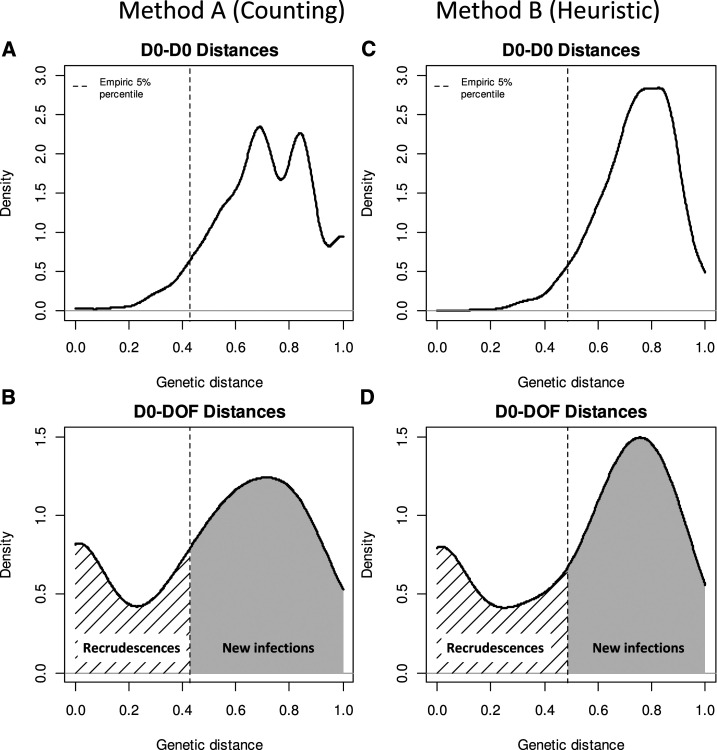
The distribution of pairwise distances using the match-counting method for D0–D0 samples was right-skewed with a median genetic distance of 0.71 (5/7 markers different) (Figure 1A). Overall, 95% of all genetic distances were greater than 0.43 for the real-world datasets (i.e., D(A)0.05 = 0.43). For genotyping data from seven loci, this corresponds to a threshold of < 3 unmatched loci to classify a set of D0–DOF samples as likely containing a shared strain and thus showing evidence of recrudescence.
Figure 1.
Distribution of pairwise genetic distance for all day 0–day 0 (D0) pairs, with empiric bottom 5% threshold denoted (A and C), and distribution distance between paired D0–day of failure (DOF) samples (B and D), using two different, independent definitions of genetic distance.
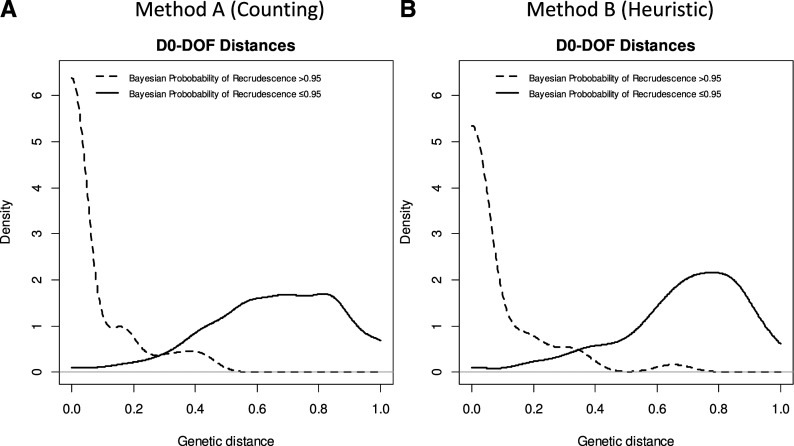
The distribution of genetic distances from the paired D0–DOF samples was bimodal (Figure 1B). After applying the threshold derived from the background D0–D0 pairwise distances of D(A)0.05 = 0.43, 55/175 (31%) of all D0–DOF observed pairwise distances were below the threshold, showing statistical evidence of recrudescence. Most recurrent parasitemias classified as recrudescences by the nonparametric classification had Bayesian posterior probabilities of recrudescence ≥ 0.95 (Figure 2A). The sensitivity and specificity of the nonparametric classifier was 98% and 89%, respectively, using the Bayesian classification as the gold standard (Table 2).
Figure 2.
Distribution of pairwise genetic distance for paired D0–day of failure (DOF) samples, stratifying by Bayesian posterior probability of recrudescence, using two different, independent definitions of genetic distance.
Table 2.
Comparison of nonparametric approach to gold-standard Bayesian approach
| Nonparametric threshold classification | |||||
|---|---|---|---|---|---|
| Method A | Method B | ||||
| Reinfection | Recrudescence | Reinfection | Recrudescence | ||
| Gold-standard Bayesian classification | Reinfection | 119 | 14 | 112 | 21 |
| Recrudescence | 1 | 41 | 1 | 41 | |
| Sensitivity | 98% | Sensitivity | 98% | ||
| Specificity | 89% | Specificity | 84% | ||
| PVP | 75% | PVP | 66% | ||
| PVN | 99% | PVN | 99% | ||
| Accuracy | 91% | Accuracy | 87% | ||
PVN = predictive value negative; PVP = predictive value positive;
Method B.
The results derived from using Barratt’s heuristic definition of genetic distance matched the results derived from method A. The pairwise genetic distance for unrelated D0–D0 samples was also unimodal and right-skewed, whereas paired D0–DOF samples were bimodal (Figures 1C and D). The derived threshold was calculated to be D(B)0.05 = 0.49. Comparing the classifications derived using method B under the threshold determination framework outlined here with the Bayesian classifications (our gold standard) yielded a sensitivity of 98% and a specificity of 84%.
DISCUSSION
Our findings show the potential for a nonparametric approach to distinguish recrudescent infections (i.e., those caused by the same strain) from new infections (i.e., those caused by a different strain) in the context of recurrent parasitemia caused by the malaria parasite P. falciparum. By characterizing the empiric distribution of pairwise genetic distances between D0–D0 samples using any arbitrary measure of distance, a background distribution of relatedness can be estimated. A threshold derived from this distribution can be calculated that identifies genetic distances unlikely to have arisen by chance. In the data from the settings we analyzed, even a threshold based on a simple genetic distance involving counting the number of matching loci (method A) has sufficient power to detect recrudescent infections (98% sensitivity and 89% specificity), when this framework was applied to select an appropriate cutoff distance, using a Bayesian algorithm as the gold standard. The Bayesian algorithm is a statistical approach that was specifically designed to estimate the likelihood of two complex samples containing a shared genotype.13 It uses a Markov chain Monte Carlo sampling strategy to jointly estimate the posterior probability of recrudescence as well as to infer hidden states, and takes several hours to run for a typical dataset. It has been independently evaluated and shown to identify recrudescences with high sensitivity and specificity.10 The concordance between the classifications obtained from the nonparametric method A, nonparametric method B, and more complex model-based approaches such as the Bayesian algorithm is evidence that each is likely capturing a high proportion of true cases of recrudescence. Moreover, the optimal threshold reported here (3/7 loci) to differentiate recrudescences from new infections using method A matches previous results from simulation studies, showing an intermediate number of matches as sufficient evidence to classify samples as containing a shared, recrudescing, strain.10 Finally, when using our framework to select an unbiased cutoff value, the similar performance displayed by two entirely different and independent methods for calculating genetic distance (method A and method B) suggests that this approach is robust to the choice of genetic distance definition.
In this study, our framework depended on having enough D0 genotypes to characterize the true background distribution of genetic distances in a population to allow accurate and precise determination of the key empiric 5% threshold. Investigators are therefore urged to genotype as many D0 samples from antimalarial efficacy trials as feasible. As more and more efficacy investigators genotype all D0 samples for analysis of molecular markers of resistance, and additional population-level molecular surveys take place, data on the frequencies of circulating alleles and genotypes are likely to increase. The optimal number of D0 samples that need to be genotyped will vary by factors such as transmission intensity, the diversity of the chosen markers, the prevalence of multi-strain infections, and the rate of treatment failure.
An underlying assumption of the proposed approach is that the parasite population is well mixed and homogenous at a given geographic sampling site as the approach is designed to facilitate intrapopulation-level discrimination. Unimodal distributions of the D0–D0 background genetic distance are evidence of homogenous mixing. Conversely, a non-unimodal distribution of D0–D0 background genetic distance might suggest possible subpopulations or importation of parasites from an outside population, complicating analysis of genetic relatedness. A multimodal distribution of distances could also indicate a dataset biased toward certain genotypes, which could be ameliorated by normalizing the number of each genotype in a population to the same frequency before distance calculation for cutoff selection. Alternatively, a bimodal or multimodal distribution resulting from distinct genetic subpopulations could indicate separation of populations by a geographic barrier preventing uniform intermixing (correctable by comparing only types from the same site), or a reproductive barrier. The possible existence of a “cryptic” reproductive barrier is supported if the specimens being compared share their geographic and/or host origin, yet a bimodal or multimodal distribution is still observed, even after normalization of genotypes as described earlier. In this case, our statistical framework could still be used for cutoff selection, although users are urged to consider the lower fifth percentile of only the intrapopulation distribution of distances (i.e., the distribution of distances only under the first, leftmost peak of the entire distribution), and not the entire distribution—and only after normalizing genotypes to the same frequency to control for sampling bias.
When performing malaria drug efficacy trials, investigators are urged to critically analyze genotyping data for proper interpretation of the final efficacy results. Datasets in which genetic distances do not show a wide-enough distribution are unlikely to yield valid recrudescence versus new infection classifications. In these cases, investigators should not attempt differentiation with invalid data and could consider using different genetic markers, as their panel might lack sufficient diversity to provide a wide distribution. Whole genome sequencing of a select number of samples or analysis of previously available genomes can help identify the most informative markers in a given geographic area, and can be performed in advance of the trial itself. However, many laboratories in endemic settings currently do not have capacity for capillary electrophoresis or sequencing. In these cases, investigators can consider partnering with capacity-building approaches such as the U.S. President’s Malaria Initiative–Supported Antimalarial Resistance Monitoring in Africa Network.19
Investigators should strive to use genotyping methods that have enough discriminatory power to yield wide genetic distance distributions. Equally, investigators should attempt to measure the analytic sensitivity of the genotyping methods used, for example using mixtures of laboratory strains. No amount of statistical wizardry is able to overcome the biases introduced by insensitive detection of minority strains, and investigators should ensure that the genotyping methods are sufficiently sensitive to detect low-frequency minority strains.20 Regardless of the genotyping method and classification algorithms used, a certain number of false positive and false negatives are bound to occur, and study reports should be transparent in describing the methodologies and should systematically publish all genotyping data. Investigators should clearly state how uncertainties in the classification approach were estimated and accounted for in the analysis, for example using Bayesian statistical approaches9 or sensitivity analyses.
To conclude, a generalized, nonparametric, marker-independent statistical framework for distinguishing between related and unrelated apicomplexan infections is described and validated using a real-world P. falciparum genotyping dataset. We demonstrate how this method can be applied to any measure of genetic distance to facilitate rational and unbiased cutoff distance selection when a binary classification of “related” and “unrelated” genotypes is desired. Although the present study applied this framework to microsatellite data collected for P. falciparum, it should be broadly applicable to any measure of genetic distance and for other apicomplexan parasites where binary classification of genotypes is the desired outcome.
REFERENCES
- 1.Galal L, Hamidović A, Dardé ML, Mercier M, 2019. Diversity of Toxoplasma gondii strains at the global level and its determinants. Food Waterborne Parasitol 15: e00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ajzenberg D, Bañuls AL, Su C, Dumètre A, Demar M, Carme B, Dardé ML, 2004. Genetic diversity, clonality and sexuality in Toxoplasma gondii. Int J Parasitol 34: 1185–1196. [DOI] [PubMed] [Google Scholar]
- 3.Barratt JLN, Sapp SGH, 2020. Machine learning-based analyses support the existence of species complexes for Strongyloides fuelleborni and Strongyloides stercoralis. Parasitology 147: 1184–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nascimento FS, et al. 2020. Evaluation of an ensemble-based distance statistic for clustering MLST datasets using epidemiologically defined clusters of cyclosporiasis. Epidemiol Infect 148: e172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Health Organization , 2008. Methods and Techniques for Clinical Trials on Antimalarial Drug Efficacy: Genotyping to Identify Parasite Populations. Geneva, Switzerland: WHO. [Google Scholar]
- 6.Hlavsa MC, et al. 2017. Using molecular characterization to support investigations of aquatic facility-associated outbreaks of cryptosporidiosis - Alabama, Arizona, and Ohio, 2016. MMWR Morb Mortal Wkly Rep 66: 493–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Juliano JJ, Gadalla N, Sutherland CJ, Meshnick SR, 2010. The perils of PCR: can we accurately ‘correct’ antimalarial trials? Trends Parasitol 26: 119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greenhouse B, Myrick A, Dokomajilar C, Woo JM, Carlson EJ, Rosenthal PJ, Dorsey G, 2006. Validation of microsatellite markers for use in genotyping polyclonal Plasmodium falciparum infections. Am Soc Trop Med Hyg 75: 836–842. [PMC free article] [PubMed] [Google Scholar]
- 9.Felger I, Snounou G, Hastings I, Moehrle JJ, Beck HP, 2020. PCR correction strategies for malaria drug trials: updates and clarifications. Lancet Infect Dis 20: e20–e25. [DOI] [PubMed] [Google Scholar]
- 10.Jones S, Plucinski M, Kay K, Hodel EM, Hastings IM, 2020. A computer modelling approach to evaluate the accuracy of microsatellite markers for classification of recurrent infections during routine monitoring of antimalarial drug efficacy Antimicrob Agents Chemother 64: e01517–e01519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barratt JLN, Park S, Nascimento FS, Hofstetter J, Plucinski M, Casillas S, Bradbury RS, Arrowood MJ, Qvarnstrom Y, Talundzic E, 2019. Genotyping genetically heterogeneous Cyclospora cayetanensis infections to complement epidemiological case linkage. Parasitology 146: 1275–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones S, Kay K, Hodel EM, Chy S, Mbituyumuremyi A, Uwimana A, Menard D, Felger I, Hastings I, 2019. Improving methods for analyzing antimalarial drug efficacy trials: molecular correction based on length-polymorphic markers msp-1, msp-2, and glurp. Antimicrob Agents Chemother 63: e00590–e00619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Plucinski MM, Morton L, Bushman M, Dimbu PR, Udhayakumar V, 2015. Robust algorithm for systematic classification of malaria late treatment failures as recrudescence or reinfection using microsatellite genotyping. Antimicrob Agents Chemother 59: 6096–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plucinski MM, 2015. Efficacy of artemether-lumefantrine and dihydroartemisinin-piperaquine for treatment of uncomplicated malaria in children in zaire and uíge provinces, Angola. Antimicrob Agents Chemother 59: 437–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plucinski MM, et al. 2017. Efficacy of artemether–lumefantrine, artesunate–amodiaquine, and dihydroartemisinin–piperaquine for treatment of uncomplicated Plasmodium falciparum malaria in Angola, 2015. Malar J 16: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davlantes E, Dimbu PR, Ferreira CM, Joao MF, Pode D, Félix J, 2018. Efficacy and safety of artemether–lumefantrine, artesunate–amodiaquine, and dihydroartemisinin–piperaquine for the treatment of uncomplicated Plasmodium falciparum malaria in three provinces in Angola, 2017. Malar J 17: 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beavogui AH, et al. 2020. Efficacy and safety of artesunate–amodiaquine and artemether–lumefantrine and prevalence of molecular markers associated with resistance, Guinea: an open-label two-arm randomised controlled trial. Malar J 19: 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dimbu PR, et al. 2020. Continued low efficacy of artemether-lumefantrine in Angola, 2019. Antimicrob Agents Chemother 65: e01949-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halsey ES, et al. 2017. Capacity development through the US President’s malaria initiative–supported antimalarial resistance monitoring in Africa network. Emerg Infect Dis 23: S53–S56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Messerli C, Hofmann NE, Beck HP, Felger I, 2017. Critical evaluation of molecular monitoring in malaria drug efficacy trials and pitfalls of length-polymorphic markers. Antimicrob Agents Chemother 61: e01500–e01516. [DOI] [PMC free article] [PubMed] [Google Scholar]