ABSTRACT
Zoonotic knowlesi malaria has replaced human malaria as the most prevalent malaria disease in Malaysia. The persistence of knowlesi malaria in high-risk transmission areas or hotspots can be discouraging to existing malaria elimination efforts. In this study, retrospective data of laboratory-confirmed knowlesi malaria cases were obtained from the Sarawak Health Department to investigate the spatiotemporal patterns and clustering of knowlesi malaria in the state of Sarawak from 2008 to 2017. Purely spatial, purely temporal, and spatiotemporal analyses were performed using SaTScan software to define clustering of knowlesi malaria incidence. Purely spatial and spatiotemporal analyses indicated most likely clusters of knowlesi malaria in the northern region of Sarawak, along the Sarawak–Kalimantan border, and the inner central region of Sarawak between 2008 and 2017. Temporal cluster was detected between September 2016 and December 2017. This study provides evidence of the existence of statistically significant Plasmodium knowlesi malaria clusters in Sarawak, Malaysia. The analysis approach applied in this study showed potential in establishing surveillance and risk management system for knowlesi malaria control as Malaysia approaches human malaria elimination.
INTRODUCTION
Malaysia is aiming to eliminate human malaria by 2020. With zero indigenous human malaria cases in 2018, Malaysia has been on track to achieving this.1 Although the prospect for human malaria elimination in Malaysia is optimistic, this coincided with a disheartening increase in zoonotic knowlesi malaria in Malaysia. From 2013 to 2017, knowlesi malaria accounted for 11,380 of the 16,500 malaria cases reported in Malaysia.2 The bulk of the knowlesi malaria cases equivalent to 87% (9,902 cases) originated from Sabah and Sarawak, with an average incidence rate (IR) of 0.302 per 1,000 population.2
Beginning with the discovery of a large focus of natural Plasmodium knowlesi infections in humans in Kapit, Sarawak, Malaysian Borneo,3 knowlesi malaria has now been increasingly reported in Malaysia, primarily in Malaysian Borneo, and is found throughout Southeast Asia.4 With long-tailed and pig-tailed macaques as the monkey host, Anopheles latens have been found to be one of the main vectors of P. knowlesi transmission in Sarawak.5,6
Although current antimalarials remain efficacious, P. knowlesi infection can be severe and fatal.7 With the increasing knowlesi malaria incidence, it would be worthwhile to identify the spatiotemporal distribution of knowlesi malaria longitudinally. Current geospatial analysis helps to study epidemiology of a disease spatially and temporally, identifying high-risk areas, transmission changes, and peak transmission periods. This in turn provides a better understanding of knowlesi malaria transmission that can assist in devising effective control policy, and to determine exactly where and when to implement interventions.8
Longitudinal spatiotemporal patterns of P. knowlesi transmission have only been described in Peninsular Malaysia,9 at a regional scale and in parts of Sabah.10,11 Because malaria transmission can be markedly heterogeneous spatially, even at the scale of a village,12–14 and interventions should be precisely targeted, this study aimed to investigate the spatiotemporal patterns of knowlesi malaria in Sarawak, one of the states with the highest number of P. knowlesi infections, from 2008 to 2017.
METHODS
Study area.
Sarawak, East Malaysia, is situated at the northwest coast of Borneo, located close to the Equator between latitude 0°50′ and 5°N and between longitude 109°36′ and 115°40′E.15 It shares its northeastern border with Sabah, another state in East Malaysia; its northern borders with Brunei; and its southern and western borders with Kalimantan, Indonesia. It has an area of about 124,450 km2, with an estimated population of 2.81 million in 2019.16 Its land mass is only slightly smaller than that of the whole of Peninsular Malaysia (132,100 km2).17
Sarawak is made up of 11 divisions, which are divided into 31 districts (Supplemental Figure 1). Topographically, the coastal lowlands and plains are found along the west coast of Sarawak from the north to the south, which extends eastward into a large region of undulating hills that stretches into the mountainous region along the Sarawak–Kalimantan border.15
Ethical approval.
Ethics approval for this study was obtained from the Medical Research and Ethics Committee of the Ministry of Health, Malaysia, and registered with the National Medical Research Register, the Ministry of Health, Malaysia (NMRR ID: 17–344–34773).
Data collection.
Retrospective data of laboratory-confirmed P. knowlesi/P. malariae malaria cases for 2008–2017 were obtained from the Sarawak Health Department. These cases were diagnosed and confirmed via microscopy. Although all microscopically identified Plasmodium malariae or P. knowlesi cases in Sarawak were categorized as P. knowlesi/P. malariae cases,18 most cases were definitely P. knowlesi, as there was no indigenous case of P. malariae in Sarawak based on molecular detection of 887 samples from 2000 to 2011.19
In Sarawak, it is compulsory for all laboratory-confirmed cases of malaria to be notified to the nearest district health offices. Malaria case data were entered into the Online Malaria Cases Registration System which can be accessed by district health offices, state health offices, and the Ministry of Health, Malaysia. Geographical coordinates were geocoded from the residential addresses of the reported indigenous knowlesi malaria cases. Only 98% of the case locations were able to be geocoded and used for subsequent analysis. Population data of Sarawak state and each district were projected each year based on the 2010 National Census.20
Temporal pattern and spatial distribution of knowlesi malaria.
The yearly IR was calculated based on the number of reported cases per 1,000 population at state and district levels. For monthly temporal analysis, the knowlesi malaria cases were aggregated by month based on their date of diagnosis. Periodogram analysis was conducted by fast Fourier transform of the time series case data of knowlesi malaria using stats package spec.pgram() function in R version 3.5.3. Fast Fourier transform decomposes the time series data into sinusoidal signals at different frequencies, enabling identification of important periodicities such as seasonal and cyclical patterns. The time series data were further decomposed into trend, seasonal, and irregular components using locally estimated scatterplot smoothing21 through stl() function in R. This decomposition procedure was based on additive model:
| (1) |
where Yt is the observed time series data, Tt is the trend component which is the upward or downward shift in a dataset over time, St is the seasonal component which represents the repetitive pattern over time, and It is the irregular component which is the residual after trend and seasonal components have been removed. The Ljung–Box test was applied to examine the autocorrelation in the irregular component of the model. Under the null hypothesis in which autocorrelation is absent, the applied model is considered fit to adequately decompose time series data into trend and seasonal information. Cox–Stuart and Friedman tests were performed using tsutils package in R to assess the presence of trend and seasonality of the data. Also, spatial distribution of knowlesi malaria was analyzed using ArcGIS version 10.3 to illustrate digital maps of yearly and annualized IRs of knowlesi malaria. The IR reported in each district was grouped into categories of 1 IR intervals.
Spatiotemporal scan statistical analysis.
Purely spatial, purely temporal, and spatiotemporal analyses were carried out using SaTScan version 9.6 to identify spatial and temporal clustering of knowlesi malaria cases over the 10-year study period. A discrete Poisson-based model was applied in all scan statistical analyses of knowlesi malaria clusters.22 This model was selected under the assumption that the occurrence of cases follows a Poisson distribution based on known underlying population at risk. Maximum scanning window size was set at 50% of population at risk. Scanning process involved the comparison of distributions between observed cases and expected cases inside and outside a cluster under the null hypothesis of random distribution according to spatial, temporal, or spatiotemporal aspects. The observed cases represent the actual number of cases reported within the specified area, whereas the expected cases in each area were computed in proportion to its population size. The significance of each cluster was examined at a 95% confidence level through Monte–Carlo simulation, and significant clusters (P < 0.05) were reported. In purely spatial and spatiotemporal analyses, the potential candidate clusters are indicated in circular windows imposed by the software. For each observation, a cluster with the highest log likelihood ratio (LLR) is considered the most likely cluster (ranked no. 1), indicating that it was least likely to have occurred by chance. By contrast, other significant clusters were recognized as secondary clusters and ranked based on the LLR value.
RESULTS
Plasmodium knowlesi cases from 2008 to 2017.
Knowlesi malaria IRs in the whole of Sarawak spiked in 2009, 2012, 2015, and 2017 (Supplemental Figure 2). This appeared to be a cyclical phenomenon which occurred every 2–3 years. Whenever a spike in cases was recorded in that year, the number of cases fell in the following year. Periodogram analysis detected two important periodic patterns at 30 months (2.5 years), which was in accordance with the mentioned cyclical phenomenon and 12-month seasonality (Supplemental Table 1). The time series case data were decomposed into trend and seasonal components with lack of significant autocorrelation in the irregular components (P = 0.17). In general, there was an increasing trend of knowlesi malaria in Sarawak as demonstrated by the Cox–Stuart test (P < 0.01) (Supplemental Figure 3).
Yearly, knowlesi malaria cases peaked in October (seasonal index = 34.17, IQR = 14.64), which then mostly fell quite rapidly in the following months (Supplemental Figures 3 and 4). Relatively, the lowest number of cases were reported in February (seasonal index = −23.35, IQR = 15.69) based on the seasonality of the dataset. The Friedman test further supported the presence of a stable seasonal pattern of knowlesi malaria (P < 0.01). The greatest number of cases recorded by month was in October 2012 (183 cases), followed by November 2017 (165 cases) and October 2014 (137 cases). March 2008 recorded the lowest number of cases (15 cases), followed by February 2010 (16 cases).
Knowlesi malaria IRs in districts of Sarawak from 2008 to 2017.
Districts with consistent IRs of more than 1.5 throughout the years were Julau, Pakan, Song, Kapit, and Kanowit, and Belaga and Lawas just to a slightly lesser extent. These districts are in the inner central region of Sarawak, and along the Sarawak–Kalimantan border. Song, Pakan, and Julau consistently registered the highest IR (Supplemental Figures 5 and 6).
The lowest IR (< 0.10) was recorded at the southern tip of Sarawak (Kuching and Samarahan divisions) and along the west coast in the central region (Mukah division) (Supplemental Figures 5 and 6). Besides, districts with few cases reported over the years (IR < 0.5) were Meradong, Sarikei, Sibu, Bintulu, Limbang, Miri, and Sri Aman. Districts where cases had not been reported from 2008 to 2017 were Asajaya and Matu (Supplemental Figures 5 and 6).
However, albeit the very low IR (0.01–0.20), districts which have begun to demonstrate an increase in IRs include Sri Aman (starting 2011), Bau (starting 2015), and Kuching (starting 2011). None of the districts demonstrated any decreasing trend in knowlesi malaria cases (Supplemental Figure 6).
An annualized IR for the period of 2008–2017 of all districts (Supplemental Figure 7) also reflected a similar picture as described earlier. The annualized IR in all districts was < 4. All districts located in the eastern side of the northern and central regions bordering Kalimantan had annualized IRs of 1–< 4, with the highest in Julau and Song. The rest of the districts had an annualized IR of < 1 and were located along the west coast of Sarawak.
Purely spatial and temporal analyses of knowlesi malaria cases in Sarawak from 2008 to 2017.
Spatial analysis indicated that the distribution of P. knowlesi infections was not random in Sarawak (Supplemental Table 2). It was also evident that the identified clusters involved the same districts every year. Four to five spatial clusters were present yearly. Each spatial cluster encompassed 2–10 districts. However, between 2013–2014 and 2017, there were more localized clusters (7–13 clusters) (Supplemental Figure 8).
Ten statistically significant clusters for high incidence of P. knowlesi infection during the period of 2008–2017 were determined (Supplemental Figure 8 and Table 2). The four relatively larger and most likely clusters for high incidence of P. knowlesi cases occurred in the eastern side of Sarawak, along the Sarawak–Kalimantan border. The most likely cluster (ranked no. 1) was centered at Julau, encompassing areas of Kanowit, Pakan, Saratok, Betong, Lubok Antu, and Song districts (radius = 46.33 km, relative risk [RR] = 16.16, LLR = 3,324.18, P < 0.001), with 1935 observed cases and 155.03 expected cases, whereas cluster 2 was centered at Kapit, encompassing areas of Belaga and Tatau (radius = 81.53 km, RR = 11.43, LLR = 2153.65, P < 0.001). The third cluster which was also the largest encompassed the areas of Marudi, northern to northeastern Belaga, southern parts of Limbang and Lawas, and parts of North Kalimantan (radius = 142.92 km, RR = 10.37, LLR = 1875.56, P < 0.001). There was also one cluster in northern Lawas (cluster 5) and another centered in the northern part of the Sarikei district (cluster 6). The rest of the smaller clusters were distributed along the northeast–southwest axis in the central region of Sarawak and one at northern Limbang (Supplemental Figure 8).
Basically, the likely clusters covered areas of all districts in the northern and central regions of Sarawak, except for areas at the west coast, that is, Miri, western part of Bintulu, Mukah, Dalat, Matu, Daru, Sibu, and Meradong. No cluster was detected in the southern parts of Sarawak.
Temporal analysis of the 10-year period indicated that the distribution of P. knowlesi infection was not random in Sarawak. The analysis identified one statistically significant 16-month period of high incidence of P. knowlesi infection, which happened between September 2016 and December 2017 (RR = 1.63, LLR = 136.54, P < 0.001), with 1,597 observed cases and 1,064 expected cases (Supplemental Table 3).
Spatiotemporal analysis of knowlesi malaria cases in Sarawak from 2008 to 2017.
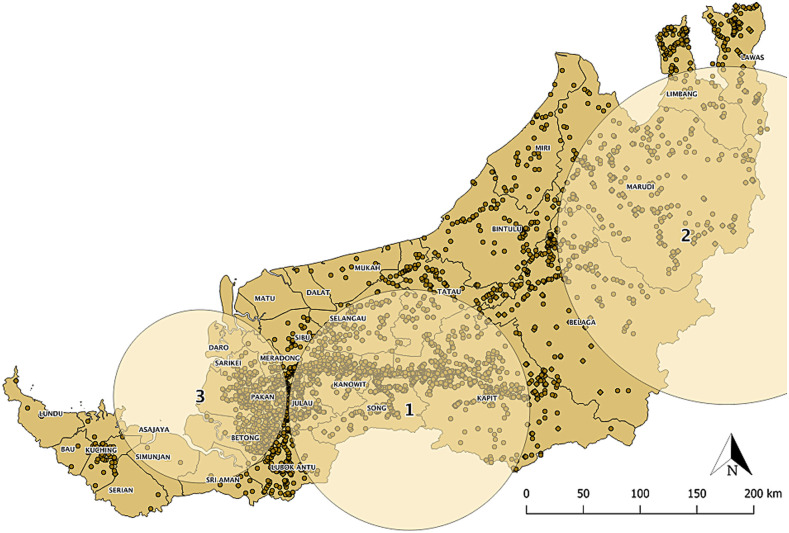
Three statistically significant clusters for high incidence of P. knowlesi infection were identified through spatiotemporal scan statistics (Figure 1, Supplemental Table 4). The most likely cluster was distributed throughout Song and Kanowit, and their adjacent districts (radius = 107.49 km, RR = 11.14, LLR = 2870.98, P < 0.001) during December 2012 to November 2017, with 2095 observed cases and 247.1 expected cases (Figure 1).
Figure 1.
Spatiotemporal clusters of knowlesi malaria from 2008 to 2017. Each dot on the map represents a case. These clusters were identified in time frames of December 2012–November 2017 (cluster 1), August 2012–July 2017 (cluster 2), and October 2017–December 2017 (cluster 3). This figure appears in color at www.ajtmh.org.
During the same period, albeit a shorter time frame (August 2012–July 2017), a secondary cluster was also identified in the northeastern part of Sarawak (radius = 150.88, RR = 10.53, LLR = 1,234.05, P < 0.001). Also, there was an emergence of a third cluster (radius = 77.34, RR = 3.84, LLR = 26.3, P < 0.001) in the southwestern coast of Sarawak for the three-month period (October 2017–December 2017). These clusters demonstrated that knowlesi malaria transmission had been stable and continuous for the past years in these districts.
DISCUSSION
Collectively, all data analyses indicated districts in the northern region of Sarawak, along the Sarawak–Kalimantan border, and the inner central region of Sarawak to be high-risk areas for knowlesi malaria between 2008 and 2017. Analyses also showed that knowlesi malaria transmission was stable in these clusters throughout the period. Similarly, a spatiotemporal study on malaria cases in Betong district, Sarawak, demonstrated a high malaria IR in Betong and Spaoh subdistricts. These hotspots were mainly concentrated in the northeastern to eastern parts of Betong district which are close to the borders of Pakan and Lubok Antu.23 The transmission clusters detected throughout the years were mostly situated in the inner and eastern regions of Sarawak, with undulating hills, or are mountainous. This may suggest geographical elevation as an associating factor of knowlesi malaria cases in Sarawak, as also indicated in Sabah.24,25
Ooi et al.18 predicted knowlesi malaria incidence to increase in Sarawak in the coming decades. In fact, the forecasted IR of 0.44 (per 1,000 population) for 2020 had already “come true” in 2017 (Supplemental Figure 2). There seemed to be a cyclical rise in knowlesi malaria cases in Sarawak, occurring every 2–3 years. The underlying reason for this phenomenon is not known. Also, it was noted that more localized clusters were observed in 2013–2014 and 2017 than in other years. Increase in the number of spatial clusters could be contributed by heightened diagnostic capacity, changes in vector abundance and behaviors, as well as frequent human activities such as deforestation or forest-related recreations. However, could the cyclical patterns of knowlesi malaria incidences, and the emergence of a higher number of larger knowlesi malaria spatial clusters (Supplemental Figure 8), predict a high knowlesi malaria incidence in the coming year? A similar study with a longer period of time should be able to confirm this. There was a decrease in P. knowlesi cases between 2015 and 2016 followed by a significant increase in cases in 2017. Similar to the situation in Sabah, researchers then believed that this could be due to the change in vector behaviors and ecological adaptations attributed to changing rainfall and weather patterns associated with the El Nino phenomenon.26,27 Furthermore, it was found that knowlesi malaria cases typically peaked in October. To some extent, this has been attributed to the fruit harvesting season between July and October annually.23 The macaques and humans converged in the same farms, orchards, and forest fringes for the fruits. With suitable vectors present, these became ideal grounds for knowlesi malaria transmission.28
None of the districts demonstrated any decreasing trend in knowlesi malaria cases. However, measures should be put in place to ensure areas with low number of cases (Samarahan, Mukah, and Kuching divisions; districts Meradong, Sarikei, Sibu, Bintulu, Miri, and Sri Aman) or no cases (Asajaya and Matu) remain at low risk, and to progress toward risk free. Besides, it would be interesting to examine the reason for some areas (e.g., Meradong) to have low incidence of knowlesi malaria despite being adjacent to districts with a high number of cases. Inversely, further local investigations can be carried out to understand the specific risk factors that led some districts to have a much higher transmission than their neighbors. These factors could be the presence of vectors, the environment, geography, or human-related activities.29
However, in Sarawak, there are limited studies associating human risk factors, improved diagnostic capacity, heightened awareness of knowlesi malaria, change in factors related to vectors’ and macaques’ behavior or habitat, and deforestation, with the increase in knowlesi malaria incidence.18 Nonetheless, a study at Betong district found males, adults, farmers, and the aboriginal Iban ethnics to have higher risk of contracting malaria.23 This is also corroborated by a study in Sabah, which demonstrated that among others, adult male, plantation work, sleeping outdoors, clearing vegetation, and having long grass around the house pose an increased risk for P. knowlesi infection.24,28 Indeed, more risk factors of knowlesi malaria are known in Sabah. The proportion of forest within a certain distance from a village or household, isolated households in patches of forest or plantation, deforestation, agricultural practices, and historical forest loss were found to be associated with the incidence of P. knowlesi infections.24,25 Besides, a recent molecular-based study reported that subpopulation of P. knowlesi associated with long-tailed macaque host was predominantly present at Betong, Sarikei, and Kapit divisions, whereas subpopulation associated with pig-tailed macaque was more common in Kanowit.30 These locations are within clusters of high incidences as identified in this study, and this warrants the importance of continuous molecular-based surveillance to understand the association among parasite subpopulations, distribution of macaque species, and clustering of cases.
Anopheles latens, which is the main vector for P. knowlesi in Kapit, Sarawak, can be found abundantly in forest and farm.5,6 Nonetheless, it must be admitted that although An. latens is the main vector for P. knowlesi in Kapit,6 it may not be the main vector in other districts. Therefore, there could be other knowlesi malaria vectors in other areas, such as An. pujutensis, An. macarthuri, An. introlatus,6 and An. balabacensis,31 of the Leucosphyrus group, which may have behaviors different from those of the current known vector, contributing to the varying epidemiology of knowlesi malaria in Sarawak. Thus, the ecology and the vectors’ behaviors in relation to knowlesi malaria transmission patterns in other districts need to be studied too.
Malaria interventions should be locally tailored. In this case, the use of spatiotemporal analyses enables us to understand the transmissions in different geographical areas. Unfortunately, conventional human malaria control measures, including insecticide-treated bed nets, indoor residual spraying, and mass drug therapy, have limited efficacies against knowlesi malaria. These control measures do not block transmissions among the macaque hosts, and are inadequate against P. knowlesi vectors which bite outdoors and do not enter dwellings to feed and rest.32,33 Therefore, new approaches are needed to curb P. knowlesi transmission to humans. This may be achieved through specific vector control activities using local surveillance data, increasing awareness of knowlesi malaria, and improving diagnostics and treatment.26,29 With knowlesi malaria transmission which occurs in forested areas or non-domestic settings, target interventions onto a highly populated area are not encouraged. Targeted approach onto individuals who are of high risk may be more appropriate.14 Provisions of insecticide-treated protective items, such as hammocks, tarpaulins, bed nets, or uniforms, to individuals who spent the night outdoors in high-risk areas may also be considered, although their efficacies require evaluation.34 In Aceh, it was suggested that the District Health Office could collaborate with other relevant government departments and local stakeholders (farm owners, logging site supervisors, and farming companies) to engage forest workers in malaria elimination activities. The authors are also in the opinion that migration and mobility surveillance system should be linked to the malaria surveillance system for use across subdistricts, districts, and province.34 The gist of it is building strong networks to fight against knowlesi malaria.29
The aforementioned point on building networks to control knowlesi malaria appropriately applies to Sarawak. Some of the knowlesi malaria clusters in Sarawak encompassed the areas of Sabah, Brunei, and Kalimantan. The areas with high risks for knowlesi malaria in Sarawak are along the Sarawak–Kalimantan border, and knowlesi malaria is prevalent in these regions. These cross-border knowlesi malaria clusters add an extra layer to the complexity of knowlesi malaria monitoring and control. However, it is extremely crucial that interstate and international stakeholders come together and coordinate malaria surveillance and control. Failing to acknowledge malaria clusters in the adjacent state or country will not help malaria control efforts of one’s district, state, or country.8 Lessons learnt from the many cross-border malaria studies have also shown how positive actions taken by a nation for malaria control or the lack of it can greatly influence malaria transmission in its neighbor.8,29,35
One factor which is not considered in this study is the asymptomatic reservoir of knowlesi malaria. Application of PCR detection in routine surveillance of malaria is uncommon, which could allow asymptomatic cases to be undetected due to low sensitivity of microscopy in diagnosing low-density malaria infection. Hence, current data are not sufficient to represent the statewide prevalence of knowlesi malaria in Sarawak. Asymptomatic knowlesi malaria has been reported from Sabah,36 Sarawak, in the Belaga district,37 and Betong division,38 and from other Southeast Asian countries.39–42 These infections may contribute to sustained transmission or outbreaks,43 and they are potentially affected by geographical factors as well as sociodemography.44 Active case detection of asymptomatic and submicroscopic malaria is essential to establish an accurate picture of malaria burden and prevent misinformed interventions.37
With the eventual elimination of human malarias in this region, malaria control programs will now need to focus on zoonotic malarias, especially P. knowlesi and Plasmodium cynomolgi infections,45–47 which are prevalent in Malaysia. Therefore, knowlesi malaria stratification by time and space is a relevant tool that can cater the shift in focus of national malaria control programs. The analyses allow observation of high-risk areas to be made not only at a regional level but also with higher resolution, down to the local village or household levels, thereby giving the flexibility to articulate interventions based on local needs.29,48 Furthermore, a robust spatial and temporal cluster detection with reliably high statistical power in this study is made possible because of the availability of large amount of case data recorded over 10 years. This analytical method will be helpful in providing an understanding on when and where to concentrate public health resources with constant update from surveillance.
There were two limitations pertaining to the method used for spatiotemporal analysis. First, SaTScan imposed a fixed circular scan window which moves across space and time setting to detect clusters, thus preventing potential irregular shaped clusters from being identified. Second, possible risk factors associated with case clustering were not assessed in this study. Hence, some improvement is needed to enhance the sensitivity of the spatiotemporal analyses, particularly in future analysis. This may include incorporating land use information,11 use of higher resolution data down to the village or household level to prevent erroneous missing out of high-risk areas, human movement data, locations of occupational risk groups, role of asymptomatic and undiagnosed cases, local vector population data, climatic data, and malaria transmission data from neighboring areas of Sarawak to assess risk at either side of the border.35
In conclusion, this study clearly highlighted the spatiotemporal epidemiology characteristics of knowlesi malaria as well as spatial and temporal clustering of the cases in Sarawak from 2008 to 2017. The findings and approach used in this study are especially helpful for the Malaysian health authority, particularly Sarawak Health Department, in enhancing malaria intervention strategies, high-risk area identification, effective resource distribution, as well as facilitating surveillance and risk management of vector-borne diseases.
Supplemental figures and tables
ACKNOWLEDGMENTS
We would like to thank the Director General of Health, the Ministry of Health, Malaysia, for granting permission to publish this study. Also, we would like to extend gratitude to the Sarawak Health Department for providing the knowlesi malaria disease data for the analysis.
Note: Supplemental figures and tables appear at www.ajtmh.org.
REFERENCES
- 1.World Health Organization , 2019. World Malaria Report 2019. Geneva, Switzerland: WHO. [Google Scholar]
- 2.Hussin N, Lim YAL, Goh PP, William T, Jelip J, Mudin RN, 2020. Updates on malaria incidence and profile in Malaysia from 2013 to 2017. Malar J 19: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh B, Lee KS, Matusop A, Radhakrishnan A, Shamsul SS, Cox-Singh J, Thomas A, Conway DJ, 2004. A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet 363: 1017–1024. [DOI] [PubMed] [Google Scholar]
- 4.Lee KS, Vythilingam I, 2013. Plasmodium knowlesi: Emergent human malaria in Southeast Asia. Lim YAL, Vythilingam I, eds. Parasites and Their Vectors. Vienna, Austria: Springer, 5–31. [Google Scholar]
- 5.Vythilingam I, Tan CH, Asmad M, Chan ST, Lee KS, Singh B, 2006. Natural transmission of Plasmodium knowlesi to humans by Anopheles latens in Sarawak, Malaysia. Trans R Soc Trop Med Hyg 100: 1087–1088. [DOI] [PubMed] [Google Scholar]
- 6.Tan CH, Vythilingam I, Matusop A, Chan ST, Singh B, 2008. Bionomics of Anopheles latens in Kapit, Sarawak, Malaysian Borneo in relation to the transmission of zoonotic simian malaria parasite Plasmodium knowlesi. Malar J 7: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Millar SB, Cox-Singh J, 2015. Human infections with Plasmodium knowlesi-zoonotic malaria. Clin Microbiol Infect 21: 640–648. [DOI] [PubMed] [Google Scholar]
- 8.Rejeki DSS, Fuad A, Widartono BS, Murhandarwati EEH, Kusnanto H, 2019. Spatiotemporal patterns of malaria at cross-boundaries area in Menoreh Hills, Java, Indonesia. Malar J 18: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Phang WK, Hamid MHA, Jelip J, Mudin RN, Chuang TW, Lau YL, Fong MY, 2020. Spatial and temporal analysis of Plasmodium knowlesi infection in Peninsular Malaysia, 2011 to 2018. Int J Environ Res Public Health 17: 9271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown R, Chua TH, Fornace K, Drakeley C, Vythilingam I, Ferguson HM, 2020. Human exposure to zoonotic malaria vectors in village, farm and forest habitats in Sabah, Malaysian Borneo. PLoS Negl Trop Dis 14: e0008617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fornace KM, et al. 2019. Local human movement patterns and land use impact exposure to zoonotic malaria in Malaysian Borneo. Elife 8: e47602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brooker S, Clarke S, Njagi JK, Polack S, Mugo B, Estambale B, Muchiri E, Magnussen P, Cox J, 2004. Spatial clustering of malaria and associated risk factors during an epidemic in a highland area of western Kenya. Trop Med Int Health 9: 757–766. [DOI] [PubMed] [Google Scholar]
- 13.Gaudart J, et al. 2006. Space-time clustering of childhood malaria at the household level: a dynamic cohort in a Mali village. BMC Public Health 6: 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stresman G, Bousema T, Cook J, 2019. Malaria hotspots: is there epidemiological evidence for fine-scale spatial targeting of interventions? Trends Parasitol 35: 822–834. [DOI] [PubMed] [Google Scholar]
- 15.Sarawak Government , 2020. The Geography of Sarawak. Available at: https://www.sarawak.gov.my/web/home/article_view/159/176/. Accessed March 31, 2020. [Google Scholar]
- 16.Department of Statistics Malaysia , 2020. Malaysia @ a Glance. Available at: https://www.dosm.gov.my/v1/index.php?r=column/cone&menu_id=clJnWTlTbWFHdmUwbmtSTE1EQStFZz09. Accessed March 31, 2020. [Google Scholar]
- 17.Forestry Department Peninsular Malaysia , 2019. Forestry Statistics. Available at: https://www.forestry.gov.my/en/2016-06-07-02-53-46/2016-06-07-03-12-29. Accessed December 21, 2019. [Google Scholar]
- 18.Ooi CH, Bujang MA, Tg Abu Bakar Sidik TMI, Ngui R, Lim YA, 2017. Over two decades of Plasmodium knowlesi infections in Sarawak: trend and forecast. Acta Trop 176: 83–90. [DOI] [PubMed] [Google Scholar]
- 19.Singh B, Daneshvar C, 2013. Human infections and detection of Plasmodium knowlesi. Clin Microbiol Rev 26: 165–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Department of Statistics Malaysia , 2011. Population Distribution and Basic Demographic Characteristics 2010. Putrajaya, Malaysia: Department of Statistics Malaysia. [Google Scholar]
- 21.Cleveland R, Cleveland WS, McRae J, Terpenning IJ, 1990. STL: a seasonal-trend decomposition procedure based on loess. J Off Stat 6: 3–33. [Google Scholar]
- 22.Kulldorff M, 1997. A spatial scan statistic. Commun Stat Theory Methods 26: 1481–1496. [Google Scholar]
- 23.Jawahir M, Ikhwan R, Mutalip H, Hasim H, Che Dom N, 2019. Spatio-temporal distribution of malaria in Betong, Sarawak, Malaysia: a five years study. Serangga 24: 104–118. [Google Scholar]
- 24.Brock PM, Fornace KM, Grigg MJ, Anstey NM, William T, Cox J, Drakeley CJ, Ferguson HM, Kao RR, 2019. Predictive analysis across spatial scales links zoonotic malaria to deforestation. Proc Biol Sci 286: 20182351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fornace KM, Abidin TR, Alexander N, Brock P, Grigg MJ, Murphy A, William T, Menon J, Drakeley CJ, Cox J, 2016. Association between landscape factors and spatial patterns of Plasmodium knowlesi infections in Sabah, Malaysia. Emerg Infect Dis 22: 201–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cooper DJ, et al. 2020. Plasmodium knowlesi malaria in Sabah, Malaysia, 2015–2017: ongoing increase in incidence despite near-elimination of the human-only Plasmodium species. Clin Infect Dis 70: 361–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Solomon T, Loha E, Deressa W, Gari T, Lindtjorn B, 2019. Spatiotemporal clustering of malaria in southern-central Ethiopia: a community-based cohort study. PLoS One 14: e0222986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grigg MJ, et al. 2017. Individual-level factors associated with the risk of acquiring human Plasmodium knowlesi malaria in Malaysia: a case-control study. Lancet Planet Health 1: e97–e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Canelas T, Castillo-Salgado C, Ribeiro H, 2018. Analyzing the local epidemiological profile of malaria transmission in the Brazilian Amazon between 2010 and 2015. PLoS Curr 10: ecurrents.outbreaks.8f23fe5f0c2052bfaaa648e6931e4e1a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Divis PCS, Hu TH, Kadir KA, Mohammad DSA, Hii KC, Daneshvar C, Conway DJ, Singh B, 2020. Efficient surveillance of Plasmodium knowlesi genetic subpopulations, Malaysian Borneo, 2000–2018. Emerg Infect Dis 26: 1392–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ang XDJ, Kadir KA, Mohamad DSA, Matusop AA, Divis PCS, Yaman K, Singh B, 2020. New vectors in northern Sarawak, Malaysian Borneo, for the zoonotic malaria parasite, Plasmodium knowlesi. Parasites Vectors 13: 472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davidson G, Chua TH, Cook A, Speldewinde P, Weinstein P, 2019. Defining the ecological and evolutionary drivers of Plasmodium knowlesi transmission within a multi-scale framework. Malar J 18: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shearer FM, et al. 2016. Estimating geographical variation in the risk of zoonotic Plasmodium knowlesi infection in countries eliminating malaria. PLoS Negl Trop Dis 10: e0004915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herdiana H, et al. 2016. Malaria risk factor assessment using active and passive surveillance data from Aceh Besar, Indonesia, a low endemic, malaria elimination setting with Plasmodium knowlesi, Plasmodium vivax, and Plasmodium falciparum. Malar J 15: 468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mercado CEG, et al. 2019. Spatiotemporal epidemiology, environmental correlates, and demography of malaria in Tak Province, Thailand (2012–2015). Malar J 18: 240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fornace KM, Nuin NA, Betson M, Grigg MJ, William T, Anstey NM, Yeo TW, Cox J, Ying LT, Drakeley CJ, 2016. Asymptomatic and submicroscopic carriage of Plasmodium knowlesi malaria in household and community members of clinical cases in Sabah, Malaysia. J Infect Dis 213: 784–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiram AI, Ooi CH, Rubio JM, Hisam S, Karnan G, Sukor NM, Artic MM, Ismail NP, Alias NW, 2019. Evidence of asymptomatic submicroscopic malaria in low transmission areas in Belaga district, Kapit division, Sarawak, Malaysia. Malar J 18: 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siner A, Liew ST, Kadir KA, Mohamad DSA, Thomas FK, Zulkarnaen M, Singh B, 2017. Absence of Plasmodium inui and Plasmodium cynomolgi, but detection of Plasmodium knowlesi and Plasmodium vivax infections in asymptomatic humans in the Betong division of Sarawak, Malaysian Borneo. Malar J 16: 417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Imwong M, et al. 2019. Asymptomatic natural human infections with the simian malaria parasites Plasmodium cynomolgi and Plasmodium knowlesi. J Infect Dis 219: 695–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van den Eede P, Vythilingam I, Ngo DT, Nguyen VH, Le XH, D’Alessandro U, Erhart A, 2010. Plasmodium knowlesi malaria in Vietnam: some clarifications. Malar J 9: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lubis IND, Wijaya H, Lubis M, Lubis CP, Divis PCS, Beshir KB, Sutherland CJ, 2017. Contribution of Plasmodium knowlesi to multispecies human malaria infections in North Sumatera, Indonesia. J Infect Dis 215: 1148–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Herdiana H, et al. 2018. Two clusters of Plasmodium knowlesi cases in a malaria elimination area, Sabang Municipality, Aceh, Indonesia. Malar J 17: 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Okell LC, Bousema T, Griffin JT, Ouedraogo AL, Ghani AC, Drakeley CJ, 2012. Factors determining the occurrence of submicroscopic malaria infections and their relevance for control. Nat Commun 3: 1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ouedraogo M, Samadoulougou S, Rouamba T, Hien H, Sawadogo JEM, Tinto H, Alegana VA, Speybroeck N, Kirakoya-Samadoulougou F, 2018. Spatial distribution and determinants of asymptomatic malaria risk among children under 5 years in 24 districts in Burkina Faso. Malar J 17: 460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raja TN, Hu TH, Kadir KA, Mohamad DSA, Rosli N, Wong LL, Hii KC, Simon Divis PC, Singh B, 2020. Naturally acquired human Plasmodium cynomolgi and P. knowlesi infections, Malaysian Borneo. Emerg Infect Dis 26: 1801–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grignard L, Shah S, Chua TH, William T, Drakeley CJ, Fornace KM, 2019. Natural human infections with Plasmodium cynomolgi and other malaria species in an elimination setting in Sabah, Malaysia. J Infect Dis 220: 1946–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ta TH, Hisam S, Lanza M, Jiram AI, Ismail N, Rubio JM, 2014. First case of a naturally acquired human infection with Plasmodium cynomolgi. Malar J 13: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.DePina AJ, Andrade AJB, Dia AK, Moreira AL, Furtado UD, Baptista H, Faye O, Seck I, Niang EHA, 2019. Spatiotemporal characterisation and risk factor analysis of malaria outbreak in Cabo Verde in 2017. Trop Med Health 47: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.