Abstract
Introduction
The consensus molecular subtypes (CMS) demonstrated prognostic value in metastatic colorectal cancer (mCRC). Similarly, a prognostic impact was suggested for the pre-consensus CRCAssigner (CRCA) classifier in early stages. The potential predictive role of these classifiers with regard to the choice of the first-line therapy has not been established. We investigated the prognostic and predictive impact of CMS and CRCA subtypes among mCRC patients treated in the TRIBE2 study.
Methods
Among 679 randomized patients, 426 and 428 (63%) samples were profiled according to CMS and CRCA classifications, respectively. The prognostic and predictive impact of both CMS and CRCA subtypes was investigated with univariate and multivariate analyses for progression-free survival (PFS), PFS 2 (PFS2), and overall survival (OS).
Results
Significant associations of CMS and CRCA subtypes with PFS, PFS2, and OS were demonstrated; the CMS classifier confirmed its independent prognostic value in the multivariable model (P value for PFS/PFS2/OS = 0.01/0.07/0.08). The effect of treatment intensification was independent of CMS subtypes (P value for interaction for PFS/PFS2/OS = 0.88/0.75/0.55). A significant interaction effect between CRCA subtypes and treatment arm was demonstrated in PFS (P = 0.02), PFS2 (P = 0.01), and OS (P = 0.008). The benefit of FOLFOXIRI seemed more relevant in the stem-like (PFS, hazard ratio = 0.60; P = 0.03) and mixed subtypes (hazard ratio = 0.44; P = 0.002). These findings were confirmed in a subgroup of patients of the previous TRIBE study.
Conclusions
We confirmed the independent prognostic role of CMS classification in mCRC independently of RAS/BRAF status. CRCA classification may help identifying subgroups of patients who may derive more benefit from FOLFOXIRI/bevacizumab.
Key words: consensus molecular subtypes, CRCAssigner, FOLFOXIRI/bevacizumab, metastatic colorectal cancer, biomarkers
Highlights
-
•
CMS subtypes have a prognostic impact in metastatic colorectal cancer.
-
•
The benefit of FOLFOXIRI/bevacizumab over doublets/bevacizumab is independent of CMS subtypes.
-
•
The stem-like CRCA subtype may derive higher benefit from the intensification of the upfront chemotherapy backbone.
Introduction
Colorectal cancer (CRC) is highly heterogeneous with a variable tumor microenvironment and response to cytotoxic drugs and targeted agents.1,2 In the last 7 years, six distinct classifications based on gene expression profiles have been developed and validated to better characterize the biology of colorectal tumors beyond genomic biomarkers commonly used in daily practice.3, 4, 5, 6, 7, 8 One of these classifications, the CRCAssigner (CRCA), identified five clinically relevant CRC subtypes—enterocyte, goblet-like, inflammatory, transit-amplifying (TA), and stem-like—with different expressions of genes involved in the WNT pathway, distinct cell types of the normal colon crypt,9 and which are associated with patient prognosis in early-stage disease.
In a previous study led by Sadanandam et al.,3 the CRCA subtypes revealed a prognostic impact in early-stage resected CRC tumors being the stem-like subtype characterized by poor overall survival (OS), but potential higher sensitivity to adjuvant 5-fluorouracil, leucovorin, irinotecan (FOLFIRI). Subsequent analyses confirmed the poor prognosis of the stem-like subtype in stage III patients and showed a lack of benefit from the addition of oxaliplatin to adjuvant 5-fluoruracil.10 The possible prognostic role of CRCA subtypes has never been assessed in clinical trials in the metastatic setting.
Subsequently, the six distinct classification systems (including CRCA) were merged into four transcriptome-based consensus molecular subtypes (CMS): CMS1: microsatellite instability (MSI) immune; CMS2: canonical; CMS3: metabolic; CMS4: mesenchymal.11
In the metastatic setting, the prognostic impact of CMS was retrospectively shown in three first-line randomized trials (AGITG MAX, CALGB 80405, FIRE-3). The immune CMS1 subtype was associated with the worst outcome and the epithelial CMS2 subtype with the best survival.12, 13, 14 Analyses of CALGB 80405 and FIRE-3 were carried out in the (K)RAS wild-type population of these studies, whereas the AGITG MAX included both RAS wild-type and mutated samples.
Although the main objectives of the Consensus classification were to provide a framework to capture the intrinsic heterogeneity of CRC and to drive informed drug development, its prognostic and predictive impact on existing therapies was also investigated. While FIRE-3 and CALGB 80405 studies provided inconsistent results with regard to the benefit from cetuximab versus bevacizumab,13,14 the AGITG MAX trial suggested increased benefit from bevacizumab in the CMS2 and CMS3 subtypes.12
Here we took advantage of the NanoCRC assay,15 recently developed to assess both CRCA and CMS subtypes using a single test with a restricted number of genes, to investigate the predictive role of CRCA and CMS subtypes with regard to the relative added value of the intensification of the upfront chemotherapy backbone from conventional doublets to the triplet FOLFOXIRI in combination with bevacizumab, as upfront treatment of metastatic CRC (mCRC). The identification of clinical and molecular factors able to identify patients more likely to derive higher benefit from the triplet is therefore crucial to optimize the cost/benefit balance of this therapeutic option.
We hypothesized that due to their poor prognosis in the early setting, stem-like/mesenchymal subtypes3,11 could derive a higher benefit from FOLFOXIRI/bevacizumab compared with doublets/bevacizumab.
Methods
Patients
TRIBE2 is a phase III, open-label, randomized, multicenter trial involving previously untreated mCRC patients. Patients received in a 1 : 1 ratio modified FOLFOX6 (mFOLFOX6) plus bevacizumab followed by FOLFIRI plus bevacizumab after disease progression or FOLFOXIRI plus bevacizumab followed by the reintroduction of the same agents after the evidence of disease progression.
The primary endpoint was the progression-free survival 2 (PFS2), defined as the time from randomization until the second evidence of disease progression or death. Secondary endpoints included PFS, defined as the time from randomization to the evidence of disease progression or death, and OS, defined as the time from randomization to death or last follow-up. All treatment drugs were administered up to eight cycles, followed by 5-fluorouracil plus bevacizumab until disease progression, unacceptable adverse events, or consent withdrawal.16
mRNA expression analyses
Formalin-fixed paraffin-embedded (FFPE) chemotherapy-naive samples were reviewed by two pathologists (G.F. and C.U.; Azienda Ospedaliero-Universitaria Pisana, Pisa, Italy). Samples with at least 20% of tumor content were selected; manual macrodissection of tumor-enriched areas was carried out from four 5-μm unstained slides. RNA was extracted with the RNeasy FFPE kit (Qiagen, Hilden, Germany) as per manufacturer's instructions. CRCA and CMS subtypes were identified using previously validated custom CRC subtype-based assays for nCounter platform (NanoString Technologies, Seattle, WA) and expression analyses (NanoCRCA17,18 and NanoCMS assays15). NanoCMS subtypes were identified using the rankCMS classifier.15
CRCAssigner R package (https://github.com/syspremed/CRCAssigner) was used to assign samples into CRCA subtypes with maximum Pearson correlation between each sample expression profile and CRCA-38 genes Prediction Analysis of Microarray (PAM) centroid.17 CMS subtypes were predicted using the rankCMS R package (https://github.com/syspremed/rankCMS), which is based on ranking of genes between the sample expression profile and the signature.15
Statistical analysis and ethical issues
Chi-square test, Fisher exact test, or Kruskall–Wallis test, were used as appropriate to compare clinical and molecular baseline characteristics of the gene expression population with those of the TRIBE2 study population, and of different CRCA and CMS subtypes. Survival curves of PFS, PFS2, and OS were assessed with the Kaplan–Meier method and compared by means of the log-rank test. Hazard ratios (HRs) and 95% confidence intervals were estimated with a Cox proportional hazard model. Multivariable models, including those covariates significant in univariate analysis (P ≤ 0.10), were built for PFS, PFS2, and OS.
Subgroup analyses of FOLFOXIRI/bevacizumab versus doublet/bevacizumab for PFS, PFS2, and OS were carried out using an interaction test to determine the impact of treatment effect in different CMS and CRCA subtypes.
The SAS version 9.4 software (SAS institute, Inc., Cary, NC) was used for statistical analyses. All patients' data were recorded in electronic case report forms and were reviewed by medical monitors. All patients provided written informed consent and the study was conducted in accordance with the Declaration of Helsinki. The data cut-off for the present analysis was 30 July 2019. TRIBE2 is registered at Clinicaltrials.gov NCT02339116.
Results
A total of 520 samples (77%) out of 679 patients randomized in the TRIBE2 study were available for gene expression analysis. Ninety-two samples were excluded due to failure of RNA extraction procedures or low RNA quality. Up to 428 (63%) and 426 (63%) samples were successfully profiled according to CRCA and CMS classifications, respectively (Supplementary Figure S1, available at https://doi.org/10.1016/j.esmoop.2021.100073). Overall, profiled samples originated from primary tumors.
The clinical and molecular characteristics of patients in the gene expression population did not significantly differ from those of the TRIBE2 study population except for a higher percentage of patients with primary tumor resected at baseline (61% versus 51%, P < 0.001) as a consequence of the higher availability of adequate tissue samples in these patients (Supplementary Table S1, available at https://doi.org/10.1016/j.esmoop.2021.100073).
Supplementary Table S2, available at https://doi.org/10.1016/j.esmoop.2021.100073, shows the relative overlapping of CRCA and CMS classifications: CMS2 samples are mainly included in the enterocyte- and TA-CRCA subtypes, and 97% of stem-like samples are classified as CMS4. The CMS3 samples when profiled with CRCA are mostly represented by enterocyte and goblet-like subtypes.
Prognostic and predictive role of CRCA subtypes
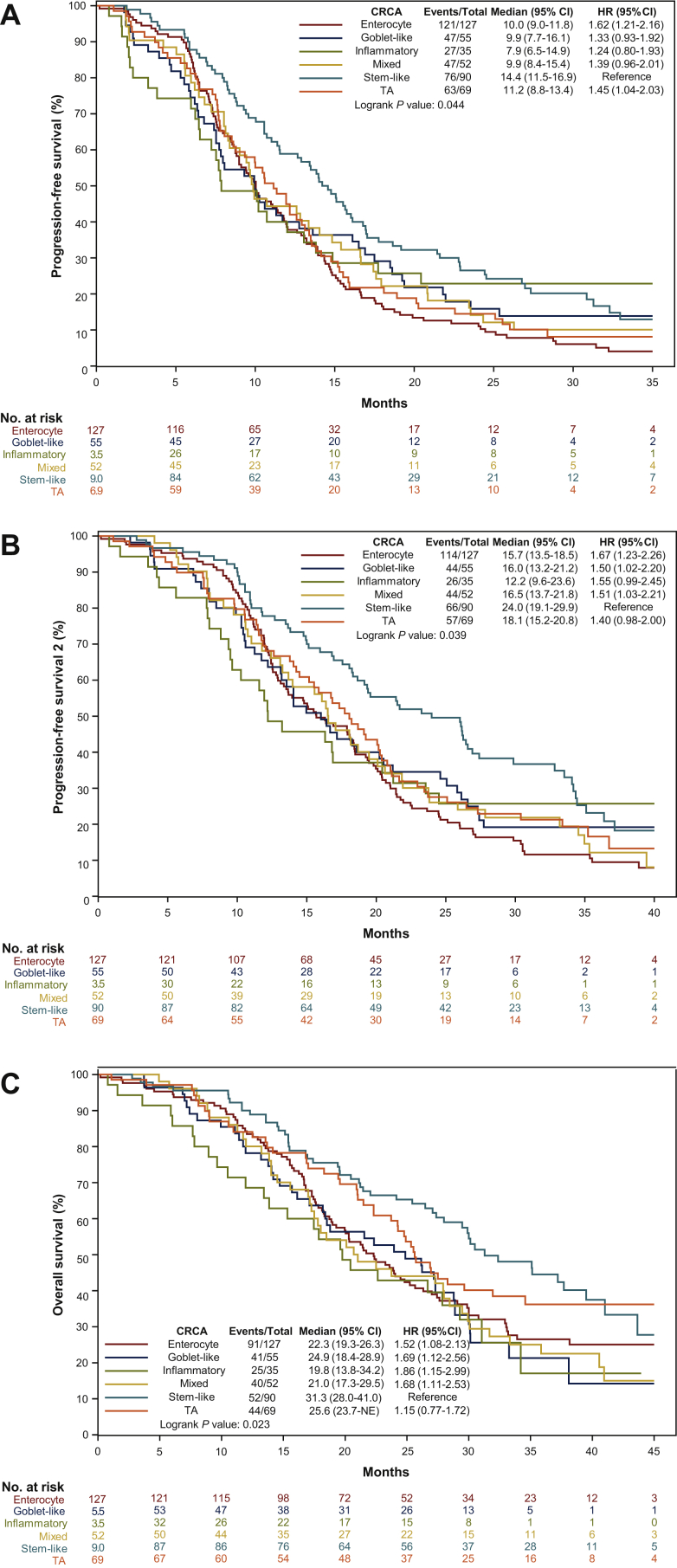
The distribution of CRCA subtypes was: 30% enterocyte, 13% goblet-like, 21% stem-like, 8% inflammatory, 16% TA, and 12% mixed. Baseline characteristics according to CRCA subtypes are summarized in Table 1. The proportion of synchronous and metachronous tumors differed according to CRCA subtypes (P = 0.004), with a higher proportion of metachronous tumors in TA subtype. The proportion of left-sided primary tumors was higher in enterocyte and TA subtypes, while right-sided tumors were prevalent in goblet-like and inflammatory subtypes (P < 0.001). RAS and BRAF wild-type tumors were more frequent in enterocyte, stem-like, and TA subtypes (P < 0.001), while a higher percentage (17%) of MSI-high samples were classified as inflammatory (P = 0.005). CRCA subtypes were significantly associated with PFS (P = 0.04), PFS2 (P = 0.04), and OS (P = 0.02). The stem-like subtype showed the best survival results, while the inflammatory subtype had the worst outcome (Figure 1A-C).
Table 1.
Clinical and molecular characteristics of TRIBE2 CRCAssigner patients
| Characteristics No. (%) |
Enterocyte (N = 127) | Goblet-like (N = 55) | Inflammatory (N = 35) | Stem-like (N = 90) | Transit-amplifying (N = 69) | Mixed (N = 52) | P |
|---|---|---|---|---|---|---|---|
| Median age (range), years | 60 (34-75) | 60 (34-75) | 59 (43-70) | 60 (29-74) | 62 (39-75) | 63 (34-74) | 0.54 |
| Sex | 0.43 | ||||||
| Male | 77 (61) | 29 (53) | 16 (46) | 48 (53) | 44 (64) | 31 (60) | |
| Female | 50 (39) | 26 (47) | 19 (54) | 42 (47) | 25 (36) | 21 (40) | |
| ECOG PS | 0.01 | ||||||
| 0 | 104 (82) | 49 (89) | 30 (86) | 81 (90) | 69 (100) | 45 (87) | |
| 1-2 | 23 (18) | 6 (11) | 5 (14) | 9 (10) | 0 | 7 (13) | |
| Synchronous metastases | 0.004 | ||||||
| Yes | 114 (90) | 53 (96) | 32 (91) | 85 (94) | 53 (77) | 47 (90) | |
| No | 13 (10) | 2 (3) | 3 (9) | 5 (6) | 16 (23) | 5 (10) | |
| Prior adjuvant chemotherapy | 0.11 | ||||||
| Yes | 0 | 0 | 0 | 1 (1) | 3 (4) | 1 (2) | |
| No | 127 (100) | 55 (100) | 35 (100) | 89 (99) | 66 (96) | 51 (98) | |
| Primary tumor site | <0.001 | ||||||
| Right | 37 (29) | 34 (62) | 20 (57) | 40 (44) | 21 (30) | 28 (54) | |
| Left or rectum | 90 (71) | 21 (38) | 15 (43) | 50 (56) | 48 (70) | 24 (46) | |
| Liver only disease | 0.20 | ||||||
| Yes | 33 (26) | 11 (20) | 12 (34) | 30 (33) | 26 (38) | 12 (23) | |
| No | 94 (74) | 44 (80) | 23 (66) | 60 (67) | 43 (62) | 39 (75) | |
| Missing data | 0 | 0 | 0 | 0 | 0 | 1 (2) | |
| Resected primary tumor | <0.001 | ||||||
| Yes | 32 (25) | 28 (51) | 29 (83) | 85 (94) | 49 (71) | 40 (77) | |
| No | 95 (75) | 27 (49) | 6 (17) | 5 (6) | 20 (29) | 12 (23) | |
| Molecular status | <0.001 | ||||||
| RAS mutated | 85 (67) | 35 (63) | 20 (57) | 58 (64) | 40 (58) | 36 (69) | |
| BRAF mutated | 7 (5) | 12 (22) | 10 (29) | 6 (7) | 1 (1) | 9 (17) | |
| RAS and BRAF wild-type | 30 (24) | 6 (11) | 4 (11) | 24 (27) | 26 (38) | 6 (12) | |
| Missing data | 5 (4) | 2 (4) | 1 (3) | 2 (2) | 2 (3) | 1 (2) | |
| Microsatellite status | 0.005 | ||||||
| MSS/pMMR | 119 (94) | 53 (96) | 27 (77) | 87 (97) | 65 (94) | 47 (90) | |
| MSI-high/dMMR | 6 (5) | 2 (4) | 6 (17) | 1 (1) | 2 (3) | 2 (4) | |
| Missing data | 2 (1) | 0 | 2 (6) | 2 (2) | 2 (3) | 3 (6) | |
| Treatment arm | 0.70 | ||||||
| Doublet/bev | 64 (50) | 32 (58) | 16 (46) | 46 (51) | 34 (49) | 31 (60) | |
| Triplet/bev | 63 (50) | 23 (42) | 19 (54) | 44 (49) | 35 (51) | 21 (40) |
bev, bevacizumab; CRC, colorectal cancer; dMMR, deficient mismatch repair; ECOG PS, Eastern Cooperative Oncology Group performance status; MSI-high, high microsatellite instability; MSS, microsatellite stable; pMMR, proficient mismatch repair.
Statistically significant P values are reported in italics.
Figure 1.
(A) Progression-free survival, (B) progression-free survival 2, and (C) overall survival curves according to CRCA subtypes.
CI, confidence interval; CRCA, CRCAssigner; HR, hazard ratio; TA, transit-amplifying.
After adjustment for sex, Eastern Cooperative Oncology Group performance status (ECOG PS), primary tumor site, liver-only disease, primary tumor resection, mutational status, MSI status, and treatment arm, the prognostic impact of CRCA subtypes was not retained (Supplementary Table S3, available at https://doi.org/10.1016/j.esmoop.2021.100073).
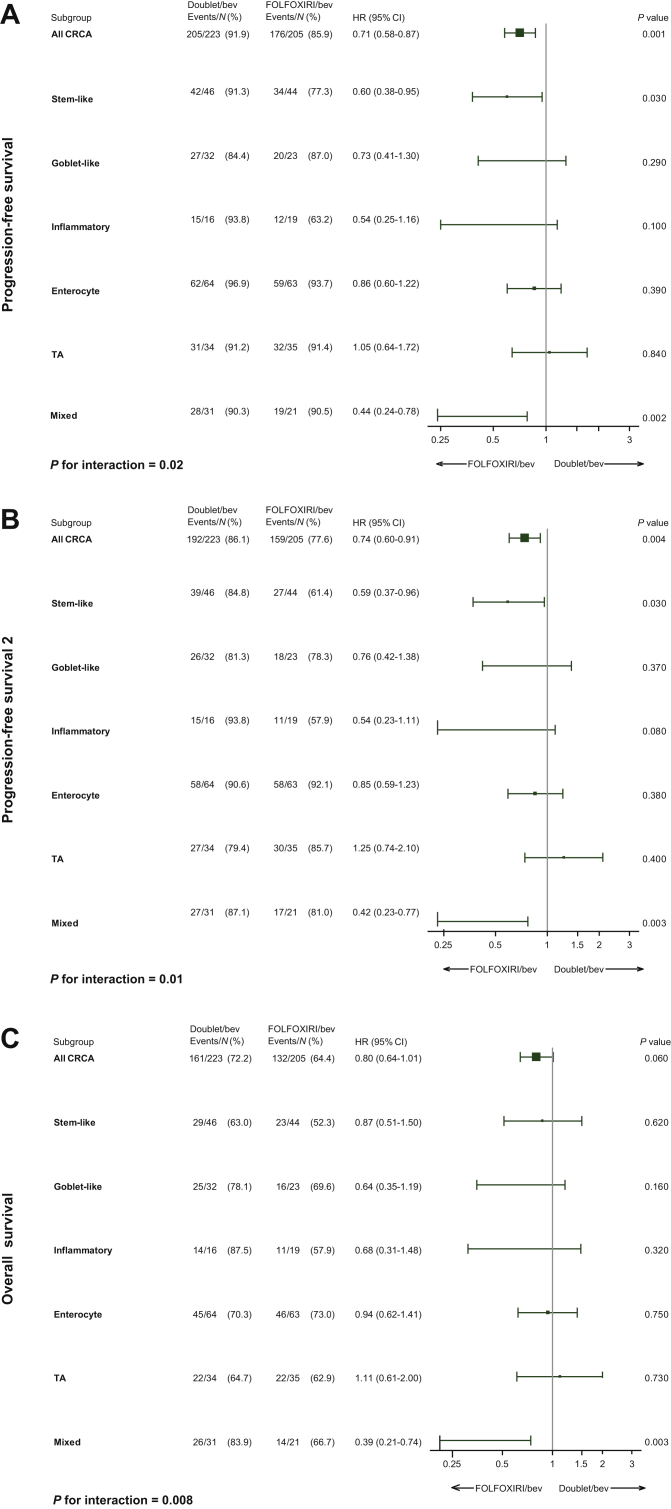
A significant interaction effect between CRCA subtypes and treatment arm was demonstrated for PFS (P = 0.02), PFS2 (P = 0.01), and OS (P = 0.008). The benefit from upfront FOLFOXIRI plus bevacizumab was more relevant in the stem-like (PFS, HR = 0.60, P = 0.03; PFS2, HR = 0.59, P = 0.03), and mixed subtypes (PFS, HR = 0.44, P = 0.002; PFS2, HR = 0.42, P = 0.003; OS, HR = 0.39, P = 0.003) (Figure 2A-C).
Figure 2.
Forest plots, predictive value of CRCA subtypes in terms of (A) progression-free survival, (B) progression-free survival 2, and (C) overall survival.
bev, bevacizumab; CI, confident interval; CRCA, CRCAssigner; HR, hazard ratio; TA, transit-amplifying.
In order to validate the predictive impact of CRCA subtypes, we profiled 120 chemotherapy-naive samples from patients enrolled in the previous TRIBE19 study of FOLFOXIRI plus bevacizumab versus FOLFIRI plus bevacizumab as initial therapy of unresectable mCRC.
The highest benefit from FOLFOXIRI/bevacizumab compared with FOLFIRI/bevacizumab was confirmed in the stem-like and mixed CRCA subtypes (PFS, stem-like HR = 0.43, P = 0.04; mixed HR = 0.38, P = 0.05). Due to the limited numbers of this cohort, no formal interaction between CRCA subtypes and treatment arm was evident (Supplementary Figure S2A and B, available at https://doi.org/10.1016/j.esmoop.2021.100073).
Prognostic and predictive role of CMS subtypes
The distribution of CMS subtypes was: 1% CMS1, 33% CMS2, 14% CMS3, and 52% CMS4. Baseline characteristics according to CMS subtypes are summarized in Table 2.
Table 2.
Clinical and molecular characteristics of TRIBE2 CMS predicted patients
| Characteristics No. (%) |
CMS1 (N = 4) | CMS2 (N = 142) | CMS3 (N = 58) | CMS4 (N = 222) | P |
|---|---|---|---|---|---|
| Median age (range), years | 56 (48-66) | 61 (35-75) | 61 (34-74) | 60 (29-75) | 0.65 |
| Sex | 0.42 | ||||
| Male | 2 (50) | 88 (62) | 35 (60) | 119 (54) | |
| Female | 2 (50) | 54 (38) | 23 (40) | 103 (46) | |
| ECOG PS | 0.26 | ||||
| 0 | 4 (100) | 128 (90) | 47 (81) | 197 (89) | |
| 1-2 | 0 | 14 (10) | 11 (19) | 25 (11) | |
| Synchronous metastases | 0.64 | ||||
| Yes | 4 (100) | 124 (87) | 53 (91) | 201 (91) | |
| No | 0 | 18 (13) | 5 (9) | 21 (9) | |
| Prior adjuvant chemotherapy | 0.62 | ||||
| Yes | 0 | 1 (<1) | 0 | 4 (2) | |
| No | 4 (100) | 141 (99) | 58 (100) | 218 (98) | |
| Primary tumor site | <0.001 | ||||
| Right | 2 (50) | 40 (28) | 26 (44) | 111 (50) | |
| Left or rectum | 2 (50) | 102 (72) | 32 (55) | 111 (50) | |
| Liver only disease | 0.29 | ||||
| Yes | 2 (50) | 46 (32) | 12 (21) | 63 (28) | |
| No | 2 (50) | 95 (67) | 46 (79) | 159 (72) | |
| Missing data | 0 | 1 (<1) | 0 | 0 | |
| Resected primary tumor | <0.001 | ||||
| Yes | 2 (50) | 62 (44) | 8 (14) | 191 (86) | |
| No | 2 (50) | 80 (56) | 50 (86) | 31 (14) | |
| Molecular status | <0.001 | ||||
| RAS mutated | 2 (50) | 96 (68) | 38 (66) | 136 (61) | |
| BRAF mutated | 1 (25) | 1 (<1) | 7 (12) | 36 (16) | |
| RAS and BRAF wild-type | 1 (25) | 41 (29) | 8 (14) | 46 (21) | |
| Missing data | 0 | 4 (3) | 5 (8) | 4 (2) | |
| Microsatellite status | 0.08 | ||||
| MSS/pMMR | 3 (75) | 132 (93) | 57 (98) | 203 (91) | |
| MSI-high/dMMR | 1 (25) | 4 (3) | 1 (2) | 13 (6) | |
| Missing data | 0 | 5 (4) | 0 | 6 (3) | |
| Treatment arm | 0.20 | ||||
| Doublet/bev | 3 (75) | 72 (51) | 24 (41) | 123 (55) | |
| Triplet/bev | 1 (25) | 70 (49) | 34 (59) | 99 (45) |
bev, bevacizumab; CMS, consensus molecular subtypes; dMMR, deficient mismatch repair; ECOG PS, Eastern Cooperative Oncology Group performance status; MSI-high, high microsatellite instability; MSS, microsatellite stable; pMMR, proficient mismatch repair.
Statistically significant P values are reported in italics.
The proportion of right- and left-sided tumors differed according to CMS subgroups (P < 0.001). CMS2 predominantly comprised left-sided primary tumors whereas other subtypes contained approximately equal proportions of right- and left-sided tumors. CMS subtypes differed also with regard to the distribution of RAS and BRAF mutations, with RAS and BRAF wild-type tumors mainly comprised in the CMS2 subtype (P < 0.001). Primary resected samples were mainly represented in CMS4 samples (86%), while the lowest percentage (14%) was reported in CMS3 (P < 0.001).
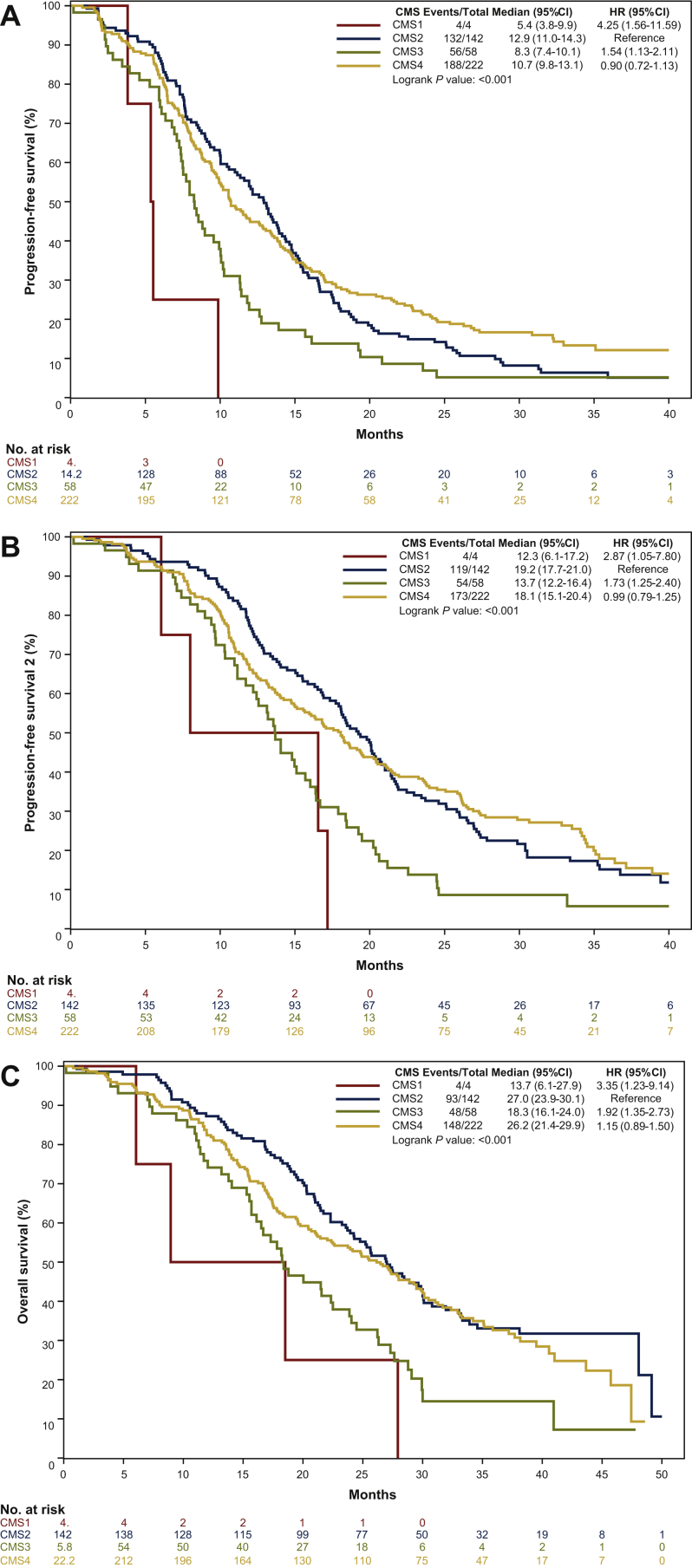
CMS subtypes were significantly associated with PFS (P < 0.001), PFS2 (P < 0.001), and OS (P < 0.001): CMS2 and CMS4 consistently showed better outcome than CMS1 and CMS3 (Figure 3A-C).
Figure 3.
(A) Progression-free survival, (B) progression-free survival 2 and (C) overall survival curves according to CMS.
CI, confident interval; CMS, consensus molecular subtypes; HR, hazard ratio.
In the multivariable model including sex, ECOG PS, primary tumor site, liver-only disease, primary tumor resection, RAS/BRAF mutational status, MSI status, and treatment arm as covariates, the prognostic impact of CMS subtypes remained significant in terms of PFS (P = 0.01), PFS2 (P = 0.07), and OS (P = 0.08) (Supplementary Table S4, available at https://doi.org/10.1016/j.esmoop.2021.100073).
No significant interaction effect between CMS subtypes and treatment arm was reported (P for interaction for PFS = 0.88, PFS2 = 0.75, OS = 0.55) (Supplementary Figure S3A-C, available at https://doi.org/10.1016/j.esmoop.2021.100073).
Discussion
Since their early development, transcriptome-based classifications have been regarded as intriguing tools able to provide accurate information about tumor biology and to unveil the functional implications of genetic alterations. Gaining a deeper knowledge about CRC heterogeneity has numerous potential implications for clinical research, including the possibility to drive drug development in specific subgroups, based on their specific features. To this end, the CMS classification marked a step forward in this field to overcome inconsistencies while strengthening similarities among pre-consensus classifications.11 Nevertheless, in order to find an immediate application for the CMS classification in clinical practice, different studies assessed the predictive impact of gene expression subtypes with regard to the efficacy of drugs used for the treatment of CRC, in particular antiangiogenic and anti-epidermal growth factor receptor agents.13,14 However, no reproducible association between CMS subtypes and the efficacy of targeted agents was found, as somehow expected based on the lack of unique target or pathway dependency of each subgroup and the biological diversity within subtypes. Oppositely, prognostic associations in the early and/or metastatic setting were found for both CMS and pre-consensus classifications including CRCA subtypes.12, 13, 14,20
We therefore investigated whether gene expression subtypes could predict the benefit from two treatment strategies, rather than from individual targeted agents, with the hypothesis that an intensified upfront treatment could be especially useful for subtypes with poor prognosis. In order to focus on a homogeneous series, we selected only chemotherapy-naive primary tumors for our analysis.
Based on our results, a heterogeneous effect of treatment intensification is evident among different CRCA subtypes. In particular, the stem-like and the mixed subgroups seem to derive higher benefit from the triplet. The possible predictive effect of CRCA classification regarding the role of the intensification of the chemotherapy backbone has never been assessed in mCRC patients. In the original publication by Sadanandam et al.,3 the stem-like subtype was associated with the worst outcome in early-stage CRC patients and a possible benefit from adjuvant chemotherapy was observed. In the same setting the poor prognosis of the stem-like subtype was confirmed in the NSABP C07 trial, but no interaction with the addition of oxaliplatin to 5-fluorouracil was observed.10 By a biological perspective, WNT signaling and stemness pathways are up-regulated in the stem-like subtype with high expression of myoepithelial and mesenchymal genes and low expression of differentiation markers.3 These features are associated with a proliferative advantage for cancer cells translating into an aggressive behavior that may be probably more efficaciously counteracted by a more intense upfront chemotherapy (FOLFOXIRI). In order to verify whether this association was due to a higher sensitivity of stem-like tumors to the triplet rather than to the addition of irinotecan to mFOLFOX6/bevacizumab, we analyzed a subgroup of samples with available material from the previous TRIBE study, where the addition of oxaliplatin to upfront FOLFIRI/bevacizumab was investigated.19 Consistent results were reported, thus corroborating the hypothesis that the upfront use of the triplet may be able to reverse the poor prognosis of these tumors. Similarly, we may speculate that the synergic effect of the three cytotoxics may better contrast the intrinsic heterogeneity of tumors belonging to the mixed subtype.
No predictive impact of CMS subgroups was shown. Indeed, while the expected partial overlap between the stem-like and the CMS4 subtypes was found, and the 97% of stem-like samples were actually classified as CMS4, the CMS4 subgroup was highly heterogeneous, including not only samples classified as stem-like (40%), but also inflammatory (15%), enterocyte (12%), TA (6%), goblet-like (12%), and mixed (16%) subtypes.
The prognostic role of this classification in the metastatic setting was confirmed in a patients' population deeply different from those of other randomized trials analyzed so far. In particular, consistent with other experiences including previously untreated mCRC patients (FIRE-3, CALGB 80405, and AGITG MAX), patients with CMS3 tumors had the worst outcome when compared with those with CMS2 and CMS4 ones. However, differently from the FIRE3 and the CALGB 80405 studies,13,14 our trial mostly enrolled patients with RAS or BRAF mutant tumors, and differently from the MAX trial,12 only patients fit enough to receive a combination of chemotherapy were eligible.
Despite some technical limitations, we reproduced the partial overlapping between CMS and CRCA subtypes which is consistent with previous evidence15: in fact the majority of CMS2 samples were subtyped as enterocyte and TA according to the CRCA classifier, while almost all stem-like tumors were grouped as CMS4.
We acknowledge some limitations. Firstly, this is a retrospective analysis of a prospective trial. A significant proportion of patients with primary tumor not resected are missing due to insufficient tissue for analysis. Although this is a recurrent caveat of correlative studies, it is possible that patients with widespread metastatic disease at diagnosis, a known poor prognostic feature, are underrepresented. This may have contributed to the low prevalence of CMS1 subtype identified in our study. Furthermore, the CMS subtypes were identified using a reduced set of 38 genes previously selected from an early disease dataset included in the consensus analysis.10,18
The genes representing the CMS1 subtype may require further optimization when NanoCMS is applied to a metastatic population. Our small-panel CMS classifier also detected only two mixed/unclassified samples, whereas these were around 13% in the original CMS study. It is likely that those mixed/unclassified samples may have been assigned by NanoCMS to the CMS4 group.
In the meanwhile, Morris et al.21 developed and validated a new CMS classifier based on Nanostring Technology using 99 genes, which demonstrated good accuracy in classifying CMS subtypes and with prognostic relevance in mCRC. Hence, the research field in CRC gene signatures is in continuous evolution: the NanoCMS assay, taking advantage of a restricted number of genes, could be easily applicable in clinical practice, but requires further validation especially in the metastatic setting. Lastly, a possible intrinsic limitation of gene expression classifiers is the high genotypic and phenotypic intratumor heterogeneity of mCRC.22 Overall, the difficulty in the reproducibility of the calling of different subtypes due to these technical limitations should be recognized.
Despite these limitations, subtype-related features, the partial overlapping between the two classifiers, and the CMS prognostic value were consistent with previous findings.
In conclusion, the application of gene expression signatures not only to clinical practice but also to clinical studies' design is not immediate. Information provided by different classifications is not totally overlapping and may affect patients' management at different levels. In particular, the potential predictive impact of CRCA subtypes with regard to the efficacy of FOLFOXIRI/bevacizumab compared with doublets/bevacizumab is worthy of further investigation in order to optimize the cost/benefit balance of this therapeutic option. More data from clinical trials, and especially from RAS mutant patients' populations, are needed to throw light on the potential usefulness of gene expression signatures in daily practice.
Acknowledgements
The authors are grateful to all participating patients and their families and to the investigators from the participating Italian centers. BB, CA, AB, GZ, AF, and CC acknowledge ARCO Foundation for their support. EF, AS, GN, and BB acknowledge the National Institute for Health Research Biomedical Research Centre at The Royal Marsden and the Institute of Cancer Research.
Funding
This work was supported by ARCO Foundation, Pisa, Italy (no grant number). EF and AS acknowledge Cancer Research UK for PhD funding for EF through the Institute of Cancer Research/Royal Marsden Hospital (no grant number). The project was also funded by Bando Ricerca Salute Regione Toscana 2018 (IN BILICO) and by University of Pisa, Progetti di Ricerca di Ateneo (PRA) 2020-2021 (no grant number).
Disclosure
AS received research funding from Bristol-Myers Squibb, Merck KGaA, and Pierre Fabre. Patents: (i) ‘Colorectal cancer classification with differential prognosis and personalized therapeutic responses’ (patent number PCT/IB2013/060416); (ii) ‘Prognostic and treatment response predictive method’ [European (EP) patent application number: 18792565.6], and (iii) ‘Patient classification and prognostic method’ (international patent application number: PCT/EP2019/053845). CC is a consultant/advisory board member for Roche, Amgen, Bayer, Merck Serono, Servier. AF is a consultant/advisory board member for Bayer, Roche, Amgen, Eli-Lilly, Merck Serono, Sanofi, Servier. All remaining authors have declared no conflicts of interest.
Contributor Information
A. Sadanandam, Email: anguraj.sadanandam@icr.ac.uk.
C. Cremolini, Email: chiaracremolini@gmail.com.
Supplementary data
References
- 1.Molinari C., Marisi G., Passardi A., Matteucci L., De Maio G., Ulivi P. Heterogeneity in colorectal cancer: a challenge for personalized medicine? Int J Mol Sci. 2018;19(12):3733. doi: 10.3390/ijms19123733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vogelstein B., Papadopoulos N., Velculescu V.E., Zhou S., Diaz L.A., Kinzler K.W. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sadanandam A., Lyssiotis C.A., Homicsko K. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat Med. 2013;19(5):619–625. doi: 10.1038/nm.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Sousa E Melo F., Wang X., Jansen M. Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nat Med. 2013;19(5):614–618. doi: 10.1038/nm.3174. [DOI] [PubMed] [Google Scholar]
- 5.Schlicker A., Beran G., Chresta C.M. Subtypes of primary colorectal tumors correlate with response to targeted treatment in colorectal cell lines. BMC Med Genomics. 2012;5:66. doi: 10.1186/1755-8794-5-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marisa L., de Reyniès A., Duval A. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med. 2013;10(5):e1001453. doi: 10.1371/journal.pmed.1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Budinska E., Popovici V., Tejpar S. Gene expression patterns unveil a new level of molecular heterogeneity in colorectal cancer. J Pathol. 2013;231(1):63–76. doi: 10.1002/path.4212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roepman P., Schlicker A., Tabernero J. Colorectal cancer intrinsic subtypes predict chemotherapy benefit, deficient mismatch repair and epithelial-to-mesenchymal transition. Int J Cancer. 2014;134(3):552–562. doi: 10.1002/ijc.28387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Humphries A., Wright N.A. Colonic crypt organization and tumorigenesis. Nat Rev Cancer. 2008;8(6):415–424. doi: 10.1038/nrc2392. [DOI] [PubMed] [Google Scholar]
- 10.Song N., Pogue-Geile K.L., Gavin P.G. Clinical outcome from oxaliplatin treatment in stage II/III colon cancer according to intrinsic subtypes. JAMA Oncol. 2016;2(9):1162–1169. doi: 10.1001/jamaoncol.2016.2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guinney J., Dienstmann R., Wang X. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21(11):1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mooi J.K., Wirapati P., Asher R. The prognostic impact of consensus molecular subtypes (CMS) and its predictive effects for bevacizumab benefit in metastatic colorectal cancer: molecular analysis of the AGITG MAX clinical trial. Ann Oncol. 2018;29(11):2240–2246. doi: 10.1093/annonc/mdy410. [DOI] [PubMed] [Google Scholar]
- 13.Lenz H.-J., Ou F.-S., Venook A.P. Impact of consensus molecular subtype on survival in patients with metastatic colorectal cancer: results from CALGB/SWOG 80405 (Alliance) J Clin Oncol. 2019;37(22):1876–1885. doi: 10.1200/JCO.18.02258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stintzing S., Wirapati P., Lenz H.-J. Consensus molecular subgroups (CMS) of colorectal cancer (CRC) and first-line efficacy of FOLFIRI plus cetuximab or bevacizumab in the FIRE3 (AIO KRK-0306) trial. Ann Oncol. 2019;30(11):1796–1803. doi: 10.1093/annonc/mdz387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fontana E., Ragulan C., Eason K. Validated nCounter platform to stratify colorectal cancer (CRC) into Consensus Molecular Subtypes (CMS) and CRCassigner subtypes in Asian population. Ann Oncol. 2017;28:x43. [Google Scholar]
- 16.Cremolini C., Antoniotti C., Rossini D. Upfront FOLFOXIRI plus bevacizumab and reintroduction after progression versus mFOLFOX6 plus bevacizumab followed by FOLFIRI plus bevacizumab in the treatment of patients with metastatic colorectal cancer (TRIBE2): a multicentre, open-label, phase 3, randomised, controlled trial. Lancet Oncol. 2020;21(4):497–507. doi: 10.1016/S1470-2045(19)30862-9. [DOI] [PubMed] [Google Scholar]
- 17.Ragulan C., Eason K., Fontana E. Analytical validation of multiplex biomarker assay to stratify colorectal cancer into molecular subtypes. Sci Rep. 2019;9(1):7665. doi: 10.1038/s41598-019-43492-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fontana E., Nyamundanda G., Cunningham D. Association between transit-amplifying signature and outcomes of patients treated with anti-epidermal growth factor receptor (EGFR) therapy in colorectal cancer. Ann Oncol. 2019;30:v201–v202. [Google Scholar]
- 19.Cremolini C., Loupakis F., Antoniotti C. FOLFOXIRI plus bevacizumab versus FOLFIRI plus bevacizumab as first-line treatment of patients with metastatic colorectal cancer: updated overall survival and molecular subgroup analyses of the open-label, phase 3 TRIBE study. Lancet Oncol. 2015;16(13):1306–1315. doi: 10.1016/S1470-2045(15)00122-9. [DOI] [PubMed] [Google Scholar]
- 20.Pogue-Geile K.L., Andre T., Song N. Association of colon cancer (CC) molecular signatures with prognosis and oxaliplatin prediction-benefit in the MOSAIC Trial (Multicenter International Study of Oxaliplatin/5FU-LV in the Adjuvant Treatment of Colon Cancer) J Clin Oncol. 2019;37(suppl 15):3503. [Google Scholar]
- 21.Morris J.S., Luthra R., Liu Y. Development and validation of a gene signature classifier for consensus molecular subtyping of colorectal carcinoma in a CLIA-certified setting. Clin Cancer Res. 2020;27:120–130. doi: 10.1158/1078-0432.CCR-20-2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dunne D., Alderdice M., O'Reilly P.G. Cancer-cell intrinsic gene expression signatures overcome intratumoural heterogeneity bias in colorectal cancer patient classification. Nat Commun. 2017;8:15657. doi: 10.1038/ncomms15657. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.