Abstract
Background
Microbiome manipulation could enhance heat tolerance and help corals survive the pressures of ocean warming. We conducted coral microbiome transplantation (CMT) experiments using the reef-building corals, Pocillopora and Porites, and investigated whether this technique can benefit coral heat resistance while modifying the bacterial microbiome. Initially, heat-tolerant donors were identified in the wild. We then used fresh homogenates made from coral donor tissues to inoculate conspecific, heat-susceptible recipients and documented their bleaching responses and microbiomes by 16S rRNA gene metabarcoding.
Results
Recipients of both coral species bleached at lower rates compared to the control group when exposed to short-term heat stress (34 °C). One hundred twelve (Pocillopora sp.) and sixteen (Porites sp.) donor-specific bacterial species were identified in the microbiomes of recipients indicating transmission of bacteria. The amplicon sequence variants of the majority of these transmitted bacteria belonged to known, putatively symbiotic bacterial taxa of corals and were linked to the observed beneficial effect on the coral stress response. Microbiome dynamics in our experiments support the notion that microbiome community evenness and dominance of one or few bacterial species, rather than host-species identity, were drivers for microbiome stability in a holobiont context.
Conclusions
Our results suggest that coral recipients likely favor the uptake of putative bacterial symbionts, recommending to include these taxonomic groups in future coral probiotics screening efforts. Our study suggests a scenario where these donor-specific bacterial symbionts might have been more efficient in supporting the recipients to resist heat stress compared to the native symbionts present in the control group. These findings urgently call for further experimental investigation of the mechanisms of action underlying the beneficial effect of CMT and for field-based long-term studies testing the persistence of the effect.
Video abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s40168-021-01053-6.
Keywords: Microbiome transplantation, Marine microbiomes, Climate change, Microbiome flexibility, Thermal tolerance, Beneficial bacteria, 16S rRNA gene, Coral bleaching, Assisted evolution
Introduction
Reef-building corals are subjected to heat stress due to ocean warming. Frequent heatwaves induce coral bleaching, the disruption of the symbiosis between the coral host and its dinoflagellate symbionts. With ongoing ocean warming, coral bleaching events that entail high mortality have increased over the last decades and are expected to intensify [1], which calls for interventions that can enhance coral resilience. One such concept is “assisted evolution,” encompassing selective breeding of corals and the manipulation of coral-associated microbiome communities, like dinoflagellate symbionts and bacteria [2]. Multi-generational coral breeding studies are aiming to select for heat-tolerant offspring, but will require larger time scales [3]. However, manipulation of the fast-evolving microbiome could have substantial effects on much shorter time scales [4, 5]. Corals are associated with a diversity of microbes, such as dinoflagellates, other protists, fungi, bacteria, archaea, and viruses [6]. While we know that coral holobiont functioning relies on the supply of fixed carbon and several essential amino acids by dinoflagellate symbionts [7, 8], the roles of other holobiont members are still widely elusive [9]. Among all those other holobiont members, coral-associated bacterial communities (in the following “the microbiome”) have been studied and characterized for the past two decades [10–14] and we learned that they likely support diverse metabolic processes of the holobiont [8, 15–17]. Additionally, the coral microbiome can benefit the host through provision of vitamins, antioxidants, and antimicrobials, hence protecting against stressors and pathogens [8, 18]. Microbiome communities are coral species-specific, but can differ across space, time, and respond to environmental drivers, while certain parts of the microbiome are suggested to constitute a stable “core” community [6, 13, 14, 19–22]. Presumably, the flexibility of microbiome communities should allow manipulation by, e.g., administration of probiotics with the goal of promoting holobiont resilience [4]. Probiotic treatments have already proven to be an effective tool to tweak host health and performance in agriculture, insect model organisms, and human medicine. For instance, inoculations of crop plants with beneficial bacterial consortia have been performed to increase crop yields or to ward off plant pathogens [23]. Heat tolerance of the pea aphid, an insect model organism, was successfully enhanced through inoculation with a heat-tolerant strain of its obligate bacterial symbiont [24]. Particularly, approaches of human gut microbiome “transplantation” (i.e., fecal microbiome transplantations a.k.a. FMTs) have emerged as successful therapies relying on transmission of living, beneficial microbiomes from a healthy human donor to a symptomatic patient. Most prominently, FMTs are being employed as a treatment for several gastrointestinal conditions [25].
Microbiome manipulation for corals is still is in its infancy. Nonetheless, pioneering studies have demonstrated feasibility by showing that microbiome communities can be shaped through inoculation with cultured bacterial isolates or phages [26–29], while several have already taken the first steps of testing the probiotic potential of these inoculations, i.e., monitoring whether manipulation treatments are accompanied with improvements of coral health and resistance particularly under heat, pathogen, and pollutant stress [26, 30–32]. To further advance coral microbiome manipulation techniques, we set out to assess the effects of a field-based coral microbiome transplantation (CMT) procedure which intends to inoculate heat-sensitive corals with donor microbiomes using fresh tissue homogenates produced from heat-tolerant conspecific donor corals. This CMT strategy bypasses time-consuming culturing and screening for beneficial bacteria from healthy donors and importantly enables the transmission of the “unculturable” microbiome fraction. A cautious selection of healthy donors is crucial in order to minimize the undesired transmission of pathogens or pollutant agents during the procedure. We tested CMT for two cosmopolitan reef-building corals, Pocillopora sp. and Porites sp., from the Andaman Sea in Thailand. First, we assessed heat stress tolerance by employing short-term heat stress assays (sensu Oliver and Palumbi et al. [33] and Voolstra et al. [34]) in order to identify suitable donors and recipients in wild coral populations. We focused on high variability habitats that likely host corals of higher heat stress tolerance, while hypothesizing that corals from sheltered reefs of near-optimal reef conditions would display heat stress sensitivity [35, 36]. 16S rRNA gene metabarcoding was performed throughout the CMT procedure teaching us new lessons of bacterial uptake and microbiome flexibility. Most importantly, the reassessment of heat tolerance after inoculation indicated a beneficial effect of the CMT treatment. These results call for investigations to further explore its underlying mechanism of action of the observed beneficial effect and whether the CMT method has the potential to be developed towards a feasible probiotic intervention supporting coral health during heatwave events.
Results
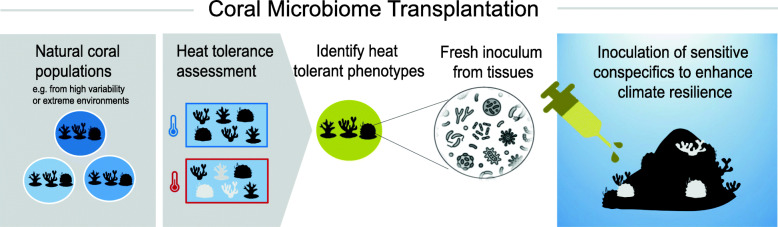
We present the results from two coral microbiome transplantation experiments with the underlying concept of using a fresh tissue homogenate of heat-tolerant donors from the wild, containing living microbiome communities, for the inoculation of heat-sensitive conspecifics to enhance their resilience under ocean warming (Fig. 1).
Fig. 1.
Coral microbiome transplantation. A field-based probiotic strategy to support coral heat tolerance during global ocean warming. Over the past years, microbiome transplantation has already been successfully employed as a clinical therapy for the treatment of several human gastrointestinal disorders. Applied to corals, this technique aims to expose heat-sensitive corals to bacterial communities of heat-resilient conspecific donor corals. Such donors can likely be found in reefs of environmental variability or extremes. CMT has several benefits. It bypasses time-consuming culturing and screening effort of bacterial isolates, which is required for the production of lab-cultured probiotics. Most importantly, this strategy enables the transmission of the “unculturable” fraction of the microbiome. Furthermore, reintroduction of CMT recipient corals will not clash with ethical considerations, since donor colonies can be locally sourced from reef habitats at the location of the application. As such, this approach can become a feasible, local management strategy for coral reefs. As ocean warming is progressing rapidly, an expeditious strategy like the CMT could represent a powerful probiotic intervention for corals
Heat tolerance assessment of corals in the wild
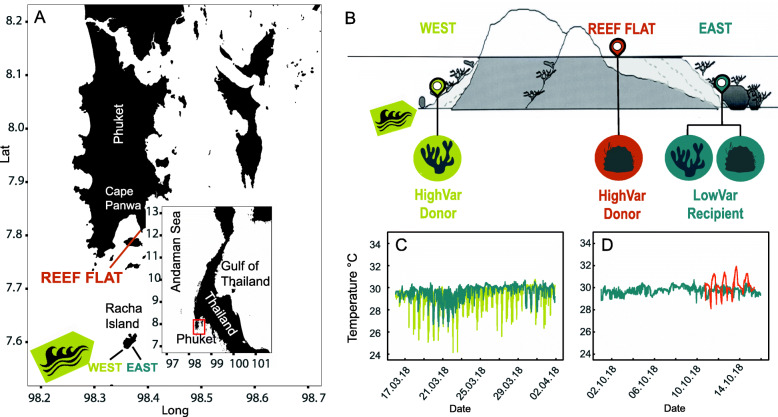
First, to identify suitable donor corals, we assessed heat stress responses of two coral species, Pocillopora and Porites, by screening fragments from wild colonies living in high and low variability reef sites, “LowVar” and “HighVar,” in the Andaman Sea (Fig. 2a-d). We used short-term heat stress assays (Fig. S1 A-B) and measured coral response variables such as the bleaching score of the coral tissues and photosynthetic efficiency of dinoflagellate symbionts to gain insight into stress condition before and after the heat exposure (Dataset S1). Corals of both species originating from the “HighVar” sites had a higher bleaching resistance compared to their “LowVar” conspecifics, as indicated by the overall decreased ∆-bleaching score of “LowVar” fragments under heat exposure (effect sizes ~ −1.0 and −1.1, Fig. 3a, b). In contrast, no significant changes were documented for “HighVar” corals under elevated temperature in the heat stress assays (effects sizes ~ −0.1 and −0.3). In our assays, this differential heat tolerance, “LowVar” vs. “HighVar” was slightly more pronounced for Porites than for Pocillopora. Porites from the “LowVar” site bleached significantly (p < 0.001), whereas their “HighVar” conspecifics exhibited no significant response under heat exposure (Tables S1-2). In contrast, the effect of heat exposure on pocilloporid corals from the “LowVar” site marginally failed to be statistically significant (effect size ~ −1.0, p = 0.051). Yet, the effect of heat exposure on “LowVar” corals was larger compared to the minor effect on “HighVar” conspecifics (effect size ~ −0.4), which hardly showed a bleaching score decline under heat exposure. Furthermore, only Pocillopora fragments showed a differential response between the two sites of origin based on photosynthetic efficiency (Fig. S2 A-B). Here, photosynthetic efficiency decreased in “LowVar” corals under heat exposure (p = 0.045, Tables S1-2), while it did not change for “HighVar” corals. Photosynthetic efficiency of Porites fragments from both sites significantly decreased under heat exposure (p = 0.001), showing no difference in the stress response of the dinoflagellate symbionts. Based on these outcomes, corals from the “LowVar” site were designated to be recipients, while the more heat-tolerant “HighVar” corals were used as microbiome donors in the CMT experiments.
Fig. 2.
Collection sites and environmental properties. a Coral sites at Racha Island and Panwa reef flat in the Andaman Sea south of Phuket Island in Thailand (red rectangle). The impact of large-amplitude internal waves creates high variability habitats on the western shore of Racha Island (light green arrow). At the shallow reef flat in Panwa, high variability and extreme conditions are linked to diurnal solar and tidal variation. b Three distinct reef sites were selected: a high variability west shore site of Racha Island (“HighVar,” 15 m depth, light green); a high variability reef flat in Phuket Island, Panwa (“HighVar,” 0–2 m depth, orange); and a sheltered low variability reef site of stable environmental conditions at Racha Island east shore (“LowVar,” 15 m, teal). Corals from “HighVar” environments were designated as microbiome donors, whereas corals from the “LowVar” site were used as recipients during microbiome transplantation experiments. c, d In situ temperature profiles show the temperature history of corals prior to experiments. Strong fluctuations of temperature were measured at the “HighVar” west shore site (light green in c) and reef flat (orange in d), while comparably stable conditions are shown for the “LowVar” east shore site (teal in c and d). Branching coral, Pocillopora sp.; massive coral, Porites sp.
Fig. 3.
Coral bleaching responses during heat tolerance assessments before and after the coral microbiome transplantation experiments. a, b Effects of temperature (“29 °C” vs. “34 °C”) on the bleaching score of corals from sites of low and high environmental variability (“LowVar” and “HighVar”) are compared. c, d Next, data shows that the handling procedure during inoculation had no impact on the bleaching score of corals (“I”, inoculation group vs. “C”, sterile-filtered seawater (FSW) control group). Subsequently, e, f the temperature effects on the bleaching scores of the recipient group and the FSW control group are shown. Plots visualize ∆-bleaching score data (i.e., the difference of tissue color intensity at end–start of each experimental part). Swarm plots (left side plot) show raw data points and Cumming estimation plots (right) depict the effect sizes as the mean differences between the treatment groups using Cohen’s d and a 95% confidence interval. Significant differences are indicated by connecting lines (p < 0.001***, < 0.01**, < 0.05* from generalized linear/linear mixed effect models). Vertical error bars, 95% CI; N, individuals per treatment group; branching coral, Pocillopora; massive coral, Porites; light green, “HighVar” west shore corals; orange, “HighVar” reef flat corals; teal, “LowVar” east shore corals; colored circles represent the donor inoculum used: light green, “HighVar” Pocillopora donor; orange, “HighVar” Porites donor
Heat tolerance assessment of recipients after inoculation
The two CMT experiments, one for each coral species, consisted of two parts respectively (Fig. S1 C-D): first, the inoculation procedure, in which the treatment group received a donor-inoculum (“I”: recipients) and a control group received an inoculation treatment of sterile-filtered seawater (“C”: FSW control group); second, the reassessment of heat tolerance, where both groups were exposed to either a heat treatment of 34 °C or ambient of 29 °C resulting in four treatment groups, i.e., “I x 29 °C,” “I x 34 °C,” “C x 29 °C,” and “C x 34 °C.” The mere inoculation procedure did not entail any changes in the bleaching score and the photosynthetic efficiency (Fig. 3c,d and S2 C-D, Tables S1-2). However, it subsequently had a beneficial effect on the bleaching resistance of recipients when exposed to heat, as the pocilloporid “I” recipients did not significantly bleach under heat exposure (effect size ~ −0.5), but corals in the FSW control group bleached measurably (effect size ~ −1.6, p = 0.002, Fig. 3e). Porites “I” recipients were not entirely immune to bleaching, as the effect of heat exposure was significant and the bleaching score slightly reduced (effect size ~ −0.6, p = 0.004, Fig. 3f). Yet, in comparison, the stress response of the FSW control group “C” was far more severe (effect size ~ −1.5, p < 0.001), suggesting a beneficial effect of CMT on Porites recipients.
Measurements of photosynthetic efficiency did not reflect the beneficial effect of CMT in the same manner as shown by the bleaching score (Fig. S2 E-F). Over the course of time, the heat exposure led to a significant decrease of photosynthetic efficiency in both groups, “I x 34 °C” and “C x 34 °C.” However, at the end of the heat stress assays, there was no significant difference in photosynthetic efficiency levels between these two groups (Tables S1-2).
Microbiome data overview
To sequence DNA for microbiome analysis, coral and seawater samples were collected at three time points during the two CMT experiments, at the “start” and “end” of inoculation and at the “end” of heat tolerance reassessment (Fig. S1 C-D). α- and β-diversity analyses of the amplicon data rely on a rarefied data set containing 7177 amplicon sequence variants (ASVs) over 210 samples with 4000 reads per sample (Fig. S3). Further analyses used a filtered data set (i.e., “filt-10” data), where low abundant ASVs were removed from the original data resulting in 4604 ASVs over 293 samples and 2,335,885 reads in total (more details in Supplementary Materials and Methods; Dataset S2). Overall, the samples clustered by experiment, reflecting two different microbiome communities of the two coral species (ppermanova < 0.001). Source seawater tank communities were remarkably distant to both coral microbiomes, whereas seawater communities of experimental tanks were closer to the respective coral microbiomes (Fig. S4). This suggests that coral microbiome communities influence their immediate microbial surrounding to a certain degree rather than the opposite. Corals contained a high number of unique ASVs (~1500–2300) which made up 42% and 65% of all coral sequence reads in Pocillopora sp. and Porites sp., respectively. Comparably, very few ASVs (~70–400) were shared with the seawater microbiomes (Fig. S5).
Coral microbiome compositions
Pocilloporid microbiomes were mainly composed of Proteobacteria, Bacteroidetes, and a smaller proportion of Chloroflexi and Actinobacteria. Additionally, donors and inoculum were associated with Cyanobacteria and an unclassified taxon. Within these microbiomes, three bacterial species were strongly dominant, i.e., Candidatus Amoebophilus sp. (up to 90%), Alteromonas sp. (up to 62%), and a species of Rhodobacteraceae (up to 40%) (Fig. S6 A). Proteobacteria prevailed in the Porites microbiomes and some individuals had a large proportion of Tenericutes, Bacteroidetes, and Epsilonbacteraeota. Endozoicomonas sp. was the solely dominant bacterial species associated with Porites corals (up to 99%). In few individuals, Alteromonas sp. and a species of Entomoplasmatales were prevalent, but comparably less abundant (Fig. S6 B).
Microbiome differences between the donors and recipients at the experiment start
Both CMT experiments started with rather similar α-diversity metrics of the donor, inoculum, and recipient microbiomes (Fig. S7). Only Porites recipients initially had a slightly higher richness and significantly lower evenness compared to the donor and inoculum microbiomes (p = 0.058 and 0.045, respectively, Table S3), due to the dominance of Endozoicomonas. However, several compositional differences (β-diversity) were significant between the three sample groups at the experiment start, i.e., inoculum, donor samples from field collection, and recipient samples (ppermanova = 0.001 for both coral species, Fig. S8). Most notably, the β-diversity distance between the “donor+inoculum” microbiome community to the recipients’ community was particularly large in Porites (both ppairwise permanova < 0.01, Tables S4-5) and marginal in Pocillopora, where the “donor+inoculum” community was comparably less distant from the recipients.
When focusing on the microbiomes at the experiment start (based on “filt-10” data), we find that 146 ASVs were exclusively found in the pocilloporid inoculum, which represent potentially new donor bacteria, not yet present in the recipients’ microbiome prior to CMT (Fig. S9 A). Thirty ASVs were exclusively found in the Porites-donor inoculum (Fig. S9 B) when compared to the sequenced entirety of the recipients’ microbiomes. Interestingly, in Pocillopora, five Peredibacter-like species (phylogenetic order: Bdellovibrionales) and one Halobacteriovorax sp. were most abundant among these exclusive inoculum-specific ASVs, contributing to ~3% of the entire sequenced inoculum bacterial community. Other ASVs (0.1–0.3%) were an α-proteobacterium (Dstr-E11), three unclassified bacteria (uncultured Mollicutes), six Rhodobacteraceae, and five Alteromonadaceae. In Porites, six species of Endozoicomonas sp. were most prominent contributing to 3% of the entire inoculum community. To be considered, the “filt-10” data set represents the pocilloporid inoculum with 904,749 reads and Porites inoculum with only half of that (Fig. S9), which could be related to a sequencing bias or reflect a biological characteristic.
Bacterial densities during inoculation
Inoculations were performed in semi-enclosed micro-environments (Fig. S10), where bacterial densities were enriched by addition of the respective inoculum made of homogenized donor coral tissues. Bacterial cell counts showed that pocilloporid recipients were incubated in a bacterial density of 3.4 × 105 cells ml−1 inside the enclosures and with 1.5 × 104 cells ml−1 once the tubes were removed. Porites recipients were exposed to bacterial densities of 1.9 × 106 (day 1), 1.8 × 106 (day 2), and 1.2 × 106 (day 3) cells ml−1 inside the tubes and 1.7 × 105, 1.6 × 105, and 1.1 × 105 cells ml−1, respectively, when tubes were removed. Due to logistic limitations in the field, we could not perform a dosage test ahead of the experiments. To further develop the CMT method, more testing in this regard should be considered. Follow-up experiments are recommended to adjust bacterial cell densities of inocula prior to inoculation, which will require longer experimental time frames, laboratory space, and coral tissue material, but will allow for measuring dosage-dependent effects.
Microbiome community changes observed after inoculation
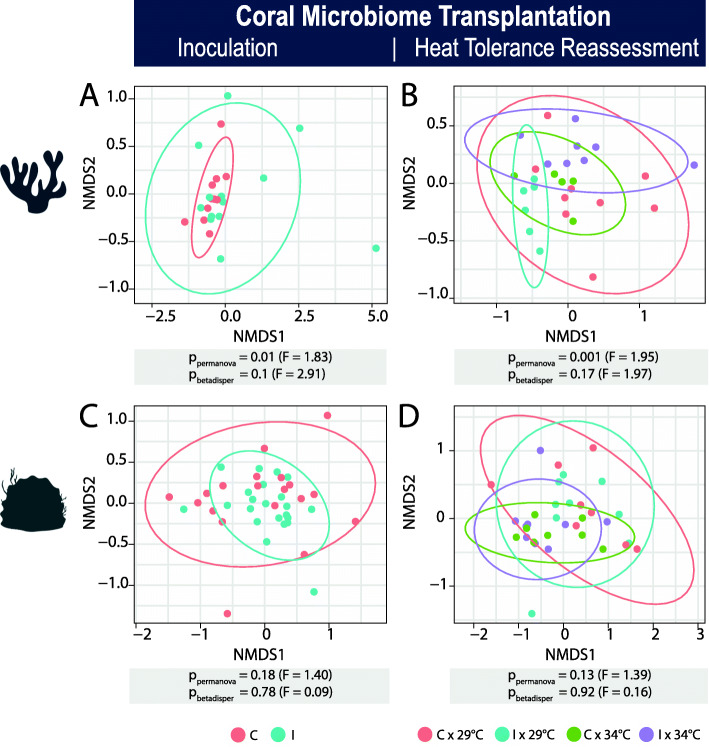
Significant reshaping of the recipients’ microbiome communities in response to CMT inoculation was demonstrated for Pocillopora, but not for Porites (Fig. 4). Microbiome diversity was slightly lower for the pocilloporid “I” recipients (Fig. S7, Fig. S11 B), and community structure was significantly changed after inoculation in comparison to the FSW control group (ppermanova = 0.01, Fig. 4a, Table S4). Subsequent heat exposure did not further affect α-diversity, but the microbiome community was changed in “I” recipients (ppermanova < 0.001, Fig. 4b). Notably, the differences between the “I” recipients and the FSW control group persisted after the heat stress assay (all ppairwise permanova < 0.05, Fig. 4b). In contrast, the microbiomes of Porites “I” recipients did not undergo any significant changes measured by α- and β-diversity in response to CMT (Fig. 4c, Fig. S7, Tables S 4-5). Even after heat exposure, Porites microbiomes of both groups remained without significant community rearrangement (Fig. 4d). Notably, community richness and diversity increased in both Porites groups “I” and “C” under heat exposure, but this effect was not significant (Fig. S7 D-F, Fig. S10 J-L, Table S3).
Fig. 4.
β-diversity of the coral microbiome communities throughout the coral microbiome transplantation experiments. The effects of the inoculation procedure (“I” vs. “C”) and the effects of subsequent heat exposure (“29 °C” vs. “34 °C”) on microbiome communities of “I” recipients and the “C” (FSW sterile-filtered seawater) control group are shown for a, b Pocillopora sp. and c, d Porites sp. Non-metric Multidimensional Scaling (nMDS) plots show microbiome communities based on Bray-Curtis dissimilarities. Group differences based on dissimilarities and dispersion were tested using PERMANOVA and BETADISPER analysis and p and F values are reported
Potentially transmitted bacterial species between the inoculum and recipients
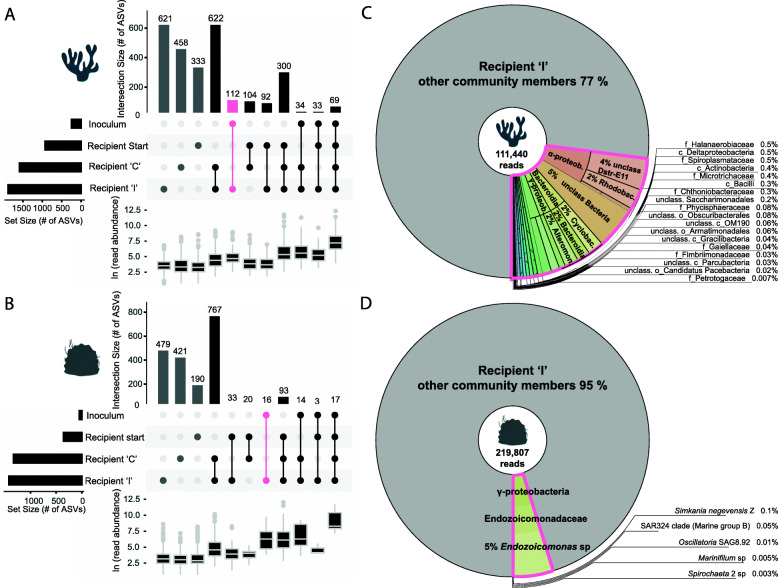
We identified 112 ASVs as exclusively shared by the microbiomes of the pocilloporid inoculum and the “I” recipient group, thus suggesting these bacterial species were potentially transferred by the CMT procedure (Fig. 5a). Sixteen of such ASVs were identified in the Porites experiment (Fig. 5b). The proportion of these donor-specific bacteria in “I” recipients was larger in Pocillopora compared to Porites (relative abundance of 23% vs. 5%, respectively; Fig. 5c, d). We found members of the Dstr-E11 group, unclassified Mollicutes, Rhodobacterales, Alteromonas sp. and other Alteromonadales, and Candidatus Amoebophilus sp. and other Cytophagales, as well as Bdellovibrionales occurring at lower abundances among the transmitted ASVs in Pocillopora (Table 1, Dataset S3). In Porites, donor-specific Endozoicomonas sp. ASVs constituted the majority of the transmitted bacterial community.
Fig. 5.
Transmission of bacteria in the coral microbiome transplantation experiments. UpSetR analyses identified amplicon sequence variants (ASVs) representing bacteria that were potentially transmitted from the inoculum to the recipient corals during the CMT treatment in a Pocillopora sp. and b Porites sp. The transmitted bacterial community, consisting of donor-specific bacteria, is represented by the exclusive overlap between the “inoculum” and “I” recipient microbiomes (marked pink). Set sizes are indicated by horizontal bars and unique and intersection group sizes are indicated by vertical bars. The inoculum set had no unique ASVs. A boxplot depicts the ln-transformed read counts per set, as an indicator of abundance. Krona plots for c Pocillopora sp. and d Porites sp. highlight the transmitted bacterial community within the total microbiomes of “I” recipients, showing relative abundances and phylogenetic classification at order, family, and species levels (SILVA database bootstrap > 80)
Table 1.
Transmission of bacteria in the coral microbiome transplantation experiments. A subset of the 112 transmitted bacterial candidates is shown for Pocillopora sp. This subset consists of amplicon sequence variants (ASVs) within the most abundant bacterial orders that contain >200 reads. For Porites sp., all 16 transmitted bacterial variants are shown. Bacterial orders [and families] are listed together with the number of ASVs within these taxonomic groups and their relative abundances within the “I” recipient microbiome community. For each bacterial order, up to three most abundant ASVs are listed with their lowest taxonomic classification (SILVA v 132) and nearest relative information from GenBank (NCBI)
| Pocillopora “I” recipient microbiomes | |||
| Order [families] | Relative abundance | No. of ASVs | Lowest taxonomic classification (SILVA) | sources and nearest relatives [% similarity; GenBank accession#] |
| Dstr-E11 [unclassified] | 3.73 | 1 | Unclassified Dstr-E11 | Montastraea franksi (coral) [99.75%; GU118824.1], Diploria strigosa (coral) [99.75%; GU118205.1] |
|
Bacteria [unclassified] Rhodobacterales [Rhodobacteraceae] |
3.57 | 1 | Unclassified Bacterium | Mollicutes bacterium, ascidians in eel pond [85.55%; EF137401.2], Anemonia viridis [86.43%; KC862086.1] |
| 2.43 | 6 | Unclassified Rhodobacteraceae | Lophelia pertusa (cold water coral) [97.52%; FJ041447.1], Stylophora pistillata (coral) [96.26%; KC669141.1] | |
| Unclassified Rhodobacteraceae | Tropicbacter sp. from marine sponge [100%; KP412857.1, MH818487.1] | |||
| Unclassified Rhodobacteraceae | Litopenaeus vannamei gut (shrimp) [99.5%; MK589151.1], diseased Montastraea faveolata (coral) [99.5%; FJ203299.1] | |||
| Clostridiales [Lachnospiraceae, Ruminococcaceae] | 2.11 | 13 | Unclassified Family XIII | endolithic bacteria from marine intertidal outcrop [99.51%; KT977254.1], healthy Montastraea faveolata (coral) [97.79%; FJ203646.1] |
| Anaerococcus sp. | Mouse cecum [100%; KY672406.1] | |||
| Ruminococcus torques group | Wastewater treatment system [99.75%; LR637623.1] | |||
| Cytophagales [Cyclobacteriaceae, Spirosomaceae, Amoebophilaceae] | 1.96 | 9 | Unclassified Cyclobacteriaceae | Montastraea faveolata, Montastraea franksi (corals) [100%; GU118859.1, GU118562.1] |
| Reichenbachiella sp. | Dinoflagellate culture [96.93%; KJ754644.1], Gulf of Mexico oil surface [95.27%; KF786696.1] | |||
| Candidatus Amoebophilus sp. | Astrangia poculata, Cladocora caespitosa, Galaxea fascicularis (corals) [95.52-95.70%; MK175907.1, KU354136.1, JQ235892.1] | |||
| Alteromonadales [Alteromonadaceae] | 1.63 | 8 | Alteromonas sp. |Favia sp. (coral mucus layer adjacent to black band mat) [100%; EF433118.1] |
| Two unclassified Alteromonadaceae | Alteromonas sp. from Mariana Trench [99.77%; MH010314.1], Xestospongia muta (sponge) [99.77%; CP031010.1], Polyhydroxyalkanoates producing bacteria [99.77%; KU521388.1] | |||
| Bacteria [unclassified] | 1.21 | 1 | Unclassified bacteria | Mollicutes bacterium from ascidians in eel pond [85.32%; EF137401.2], Anemonia viridis [86.18%; KC862086.1] |
| Flavobacteriales [Flavobacteriaceae, Weeksellaceae] | 0.74 | 7 | Tenacibaculum sp. | Flexibacter aurantiacus subsp. Copepodarum [99.06%; AB681014.1], Tenacibaculum sp. from biofilms that induce metamorphosis of marine polychaete [98.82%; MG819702.1] |
| Chryseobacterium sp. | Candidatus Amoebinatus massiliae [100%; AY204874.1], Chryseobacterium sp. in goldfish gut [99.76%; MN935216.1] | |||
| Tenacibaculum sp. | biofilms that induce metamorphosis of marine polychaete [99.29%; MG819702.1], shrimp gut [99.06%; KF342728.1] | |||
| Halanaerobiales [Halanaerobiaceae] | 0.55 | 1 | Halanaerobium sp. | Montastraea faveolata (healthy and diseased coral) [97.64–100%; FJ202891.1, JQ516458.1, FJ203280.1] |
| Chitinophagales [Chitinophagaceae] | 0.48 | 4 | Sediminibacterium sp. | tropical urban freshwater [100%; KX968184.1], ureolytic biocementation (carbonate precipitation) in soil [97.64%; MN656428.1] |
| Unclassified Chitinophagaceae | Ciona intestinalis (ascidian) [97.64%; F799383.1], shrimp gut [96.94%; KP947105.1] | |||
| Unclassified Chitinophagaceae | eutrophic freshwater lake [98.01%; EU273038.1], anaerobic digester [95.75%; MN157568.1] | |||
| Entomoplasmatales [Spiroplasmataceae] | 0.46 | 1 | Spiroplasma sp. | Arthropod-symbiotic Spiroplasma from shrimp [89.07 %; KR349130.1, KY115222.1] |
| Microtrichales [Microtrichaceae] | 0.36 | 4 | Unclassified Microtrichaceae Sva0996 marine group | Actinobacterium in Poecillastra compressa (deep sea sponge) [95.58%; KF597097.1], Fe-rich hydrothermal sediments [91.15%; FJ905720.1] |
| Unclassified Microtrichaceae Sva0996 marine group | Actinobacterium in coral reef sediment [99.75%; JN874654.1], Porites lutea (coral) [98.27%; KP303904.1] | |||
| IMCC26207 (Microtrichaceae) | Wastewater treatment system [100%; LR634799.1], sediments, lake water [99.75%; MF689304.1, KX367772.1] | |||
| Chthoniobacterales [Chthoniobacteraceae] | 0.26 | 1 | Candidatus Udaeobacter sp. | Mountain forest and soil at CO2 spring [100%; MG716938.1, HF952262.1] |
| Saccharimonadales [unclassified] | 0.24 | 1 | Unclassified Saccharimonadales | Soil [96.05%; JQ367084.2], planktonic bacteria [91.36%; Q472788.1] |
| Bacteria [unclassified] | 0.22 | 1 | Unclassified Bacteria| Mollicutes bacterium from ascidians in eel pond [85.55%; EF137401.2], Anemonia viridis [86.43%; KC862086.1] |
| Bdellovibrionales [Bdellovibrionaceae] | 0.21 | 4 | Peredibacter sp. | biofilm on a copper-based antifouling paint, seawater [99.76%; JN594639.1, MH121376.1], Favia sp. (healthy and diseased coral) [98.12%; GU472125.1] |
| Peredibacter sp. | sediments [96.71%; KC925129.1], Ciona intestinalis (ascidian) [96.05%; KF799703.1], crab [96.01%; KC917599.1] | |||
| Peredibacter sp. | seawater [100%; MH121376.1], Favia sp. (healthy and diseased coral) [98.19%; GU472125] | |||
| Desulfobacterales [Desulfobacteraceae] | 0.19 | 1 | Desulfatitalea sp. | Montastraea faveolata (coral) [100%; JQ516448.1] |
| Betaproteobacteriales [Burkholderiaceae] | 0.19 | 4 | Burkholderia-Caballeronia-Paraburkholderia | human blood [99.77%; AB374482.1], Bradyrhizobium sp. from beech tree roots [99.53%; KX023689.1] |
| Ralstonia sp. | Ralstonia pickettii from soil, pinapple roots, other plants [100%; MT322968.1, LR797737.1, MT341804.1] | |||
| Bordetella sp. | Burkholderia bacterium from lupins and grass roots [99.77%; JN590346.1, LC031362.1], Bordetella sp. from water and sediment of abandoned uranium mine [99.77%; KF441609.1] | |||
| Porites “I” recipient microbiomes | |||
| Order [families included] | Relative abundance | No. of ASVs | Lowest taxonomic classification (SILVA) | sources and nearest relatives [% similarity; GenBank accession#] |
| Oceanospirillales [Endozoicomonadaceae] | 4.64 | 9 | Endozoicomonas sp. | Porites lutea (healthy coral) [99.77%; KF179706.1, KF179699.1], Porites compressa, Porites lobata [99.53%; FJ930621.1] |
| Endozoicomonas sp. | Porites lutea (healthy coral) [100%; KF180095.1, KF180125.1] | |||
| Kistimonas sp. | Neofibularia nolitangere (sponge) [97.44%, EU816849.1], Alcyonium gracillimum (soft coral) [96.97%; JF925015.1] | |||
| Chlamydiales [Simkaniaceae] | 0.10 | 1 | Simkania negevensis Z | endolithic bacteria from marine intertidal outcrop [94.4 2%; KT979567.1], sediments from Mariana trench [94.64%; MG580090.1] |
| SAR324 Marine group B [class:Deltaproteobacteria] | 0.05 | 2 | SAR324 clade (Marine group B) HF0200_14D13 | Bacterioplankton [89.05-89.30%; MG875850.1, JN232995.1] |
| SAR324 clade (Marine group B) HF0200_14D13 | Bacterioplankton [89.30-89.55%; MG875850.1, JN232995.1] | |||
| Nostocales Incertae Sedis | 0.01 | 1 | Oscillatoria SAG8.92 | endolithic bacteria from marine intertidal outcrop [99.26%; KT973113.1], Oculina patagonica and bleached Muricea elongata (corals) [99.02%; KU936867.1, DQ917838.1] |
| Cellvibrionales [Cellvibrionaceae] | 0.01 | 1 | Candidatus Endobugula sp. | corals in fish farm effluent [96.28%; GQ413096.1], Candidatus Endobugula glebosa in Bugula simplex (bryozoan) [96.28%; AY532642.1] |
| Bacteroidales [Marinifilaceae] | 0.01 | 1 | Marinifilum sp. | Montastraea faveolata (coral) [97.64%; FJ202823.1], corals in fish farm effluent [97.41%; GQ413742.1] |
| Spirochaetales [Spirochaetaceae] | 0.003 | 1 | Spirochaeta 2 sp. | Siderastrea stellata (bleached coral) [95.34%; JF835682.1], Millepora sp. (coral) [95.57%; HQ288601.1] |
Discussion
Probiotics could become a supporting intervention to alleviate the coral reef crisis by helping corals to endure increasing temperature stress through administration of beneficial bacteria [4]. In this regard, our experiments combine a field-based and application-focused perspective with the aim to develop probiotics. We tested a coral microbiome transplantation procedure for corals (a.k.a., "CMT", Fig. 1) using two locally and globally important reef-building coral species from the Thai Andaman Sea, Pocillopora sp. and Porites sp., showing that treatment with a CMT inoculum among conspecifics partially reduced the bleaching response of recipients, as assessed in a short-term heat stress test. We also assessed responses of the recipient microbiomes, which were distinct between the two coral species. In both coral species however, several donor-specific and typically dominant bacterial taxa (i.e., coral-resident bacteria or putative symbionts) were potentially transmitted through the CMT treatment and may be suspected of helping to establish a more heat-resistant recipient coral phenotype.
Recipients showed reduced bleaching responses under heat stress after inoculation
The first hurdle of performing the CMT was the inoculation procedure which required maintenance of recipients in small volumes of water with limited flow and enriched particle and bacterial loads [37]. This may represent challenging conditions for corals that are most commonly found in clear seawater with regular flow dynamics [38]. Our experiments showed that the procedure did not harm neither of the recipients. Here, bacterial densities of inoculation were all in the range of 105–107 of bacterial cells/mL in bulk and were slightly higher for Porites than for Pocillopora.
Next and most importantly, heat exposure following the inoculation treatment revealed a higher bleaching resistance in the recipient group compared to the control group that received a sterile-filtered seawater inoculum (i.e., a cell-free control treatment), indicating either a mitigation of stress or a delay of the onset of bleaching. This compares well with observations by Rosado et al. [30] where stress responses were strongly reduced after inoculation with putatively beneficial marine bacterial isolates, but not fully eliminated. Interestingly, the CMT did not benefit photosynthetic resilience, which is in line with the insight that bleaching susceptibility might not be necessarily linked to declines of symbiont performance [39]. Altogether, outcomes indicate that a reduction of stress on the host side of the holobiont was facilitated by the CMT treatment, preventing recipients from bleaching at the same high rates as the cell-free control group.
Coral-specific bacterial symbiont taxa were transmitted during CMT
Our inoculations had a beneficial effect on recipients' heat resistance, but did not trigger any large-scale restructuring of the microbiome in neither of the two coral species. The question remains whether large microbiome community changes are required for achieving an alternation of the phenotype. Studies have reported both cases, microbiome differences without measurable signs of phenotype changes or the contrary [40–43]. Indeed, substantial reshaping of the microbiome can remain silent when phylogenetically distinct, but functionally redundant bacteria take over niches [44]. On the other hand, we cannot exclude that already small-scale manipulation of the coral microbiome may already lead to phenotypic effects. Our sequencing data captured such small-scale changes that were linked with the reduced coral bleaching response observed, by identifying putative CMT-transmitted bacteria. Most of these donor-specific bacterial species, found in the microbiomes of recipients after inoculation, were members of several typical coral-resident taxa, whose nearest relatives have previously been found in many other coral microbiomes (see references in Table 1). Notably, several of these bacterial species were members of strongly dominant taxa of our coral species’ microbiomes, i.e., a Candidatus Ampoebophilus sp. and Alteromonas sp. in Pocillopora and Endozoicomonas sp. in Porites; among which Candidatus Ampoebophilus is suggested to have co-evolved with the coral holobiont [13] and Endozoicomonas is the most prominent candidate coral symbiont known to the coral microbiome research community [45, 46]. These transmitted bacteria originated from donor corals that are naturally coping well with higher environmental stress levels in high variability habitats [36]. Thus, their essential microbiome members may be well adapted to living in a stress-challenged holobiont and are likely to have evolved optimized traits to perform holobiont services more efficiently. Our findings suggest that these bacterial taxa (ASVs) identified by our analyses might more likely integrate into coral microbiomes than other more elusive bacterial species that were also inoculated as part of the CMT inoculum. Based on these results, we underline the importance of future research to elucidate dynamics and interactions of these potential coral symbionts with the host, specifically considering their potentially beneficial effects on coral holobiont health and stress tolerance [4].
Our list of transmitted donor-specific bacteria represents some of the potentially new species that were incorporated into recipient tissues or into the mucus after inoculation. Cells that loosely attach to the corals were excluded through thorough rinsing of the samples with sterile-filtered seawater. However, this list merely represents a list of “candidates,” which the 16S rRNA gene marker, the sequencing effort, and our analysis have been able to capture, while it is important to consider that the use of another marker gene region and a deeper sequencing effort could deliver a more accurate picture [47]. In future microbiome manipulation studies like ours, employment of deep sequencing, the use of full-length 16S rRNA sequences, or metabarcoding at bacterial strain level will very likely prove to be more efficient to capture the processes of bacterial transmission and microbiome community shift [48].
Coral microbiome dynamics: drivers of stability and flexibility
Microbiome analyses in this study taught us new valuable lessons about coral microbiome dynamics and flexibility (see current perspectives in [49]). Community dynamics in our experiments were most evident and community shifts were significant in the pocilloporid recipients, which responded with an increase of dispersion between individuals, despite the reputation of having a low microbiome flexibility and being coined as a microbiome regulator [14, 41]. In comparison, the community structure remained stable in Porites recipients, even though they had experienced a longer lasting and repeated inoculation procedure during CMT. We observed a dynamic Pocillopora and stable Porites microbiome in our study. The contrary has been so far reported in few other studies where pocilloporid microbiomes were the most inflexible [14, 41], which has been suspected to be a host species-specific characteristic. Interestingly, these inflexible microbiomes were strongly dominated by the bacterial taxon of Endozoicomonas. Our study demonstrates the contrast between a flexible pocilloporid microbiome against an inflexible and Endozoicomonas-dominated Porites microbiome, providing the insight that flexibility might be determined by the initial bacterial composition, rather than coral host species. In this regard, our results support the notion that community evenness and bacterial species dominance are drivers for microbiome stability in a holobiont context [50]. Additionally, our sequencing data captured a significant proportion of predatory bacterial taxa, members of Bdellovibrio and like organisms “BALOs” [51], in the flexible pocilloporid donor coral and the inoculum. Such micropredators can shape a bacterial community already at low abundances which compares to top-down predator effects in macroscopic ecosystems [52, 53]. In our experiments, species of Peredibacter sp. were transmitted to pocilloporid recipients and may have acted as the main drivers of the microbiome changes after inoculation, whereas BALOs were absent in the stable microbiomes of Porites. Noteworthy, BALOs constitute a suitable bacterial group to be considered for microbiome manipulation approaches as already implemented in aqua farming projects [54, 55]. Many BALOs are culturable and can protect corals from pathogenic and coral bleaching-associated Vibrios [29].
“Therapeutic agents” in a CMT
Our study comes from a bacterial microbiome-focused perspective hypothesizing that bacterial community changes and specific taxa can be meaningfully linked with the altered phenotype of recipients [56]. This mirrors the perspectives and developments of clinical fecal microbiome transplantation therapies, which has been mostly driven by the human gut microbiome research community and focusing on microbiome community changes that are associated with physiological responses of the recipients [25]. However, to this point, the question for the “therapeutic agents” of microbiome transplantation methods remains not fully resolved [57–60]. A beneficial effect of an inoculation may not solely stem from bacterial input, but other components of the inoculum should be considered.
In corals, a bacterial inoculum serving as an additional food source has been debated [28]. Our CMT inoculum, made from fresh coral tissues, could have indeed offered a food source in form of cell debris, microbial cells, and dissolved organic carbon that could have been ingested by the recipient. It is well known that a significant nutritious input can benefit resilience of corals over a time span of a month [61], but whether a smaller-scale input from an inoculation treatment (as the CMT) could have contributed enough energy reserves to sustain recipient corals through heat stress in our experiment remains unresolved here. Moreover, our current study design does not allow to disentangle the potential therapeutic efficacy of other inoculum components, which aside from bacteria, include biological agents, such as dinoflagellates, other protists, fungi, and phages [9], but also bioactive molecular matter, such as enzymes, signaling molecules, antimicrobials, or regulatory mRNAs [62]. We recommend for future experiments to work with multiple “control” treatments that use modifications and fractions of a CMT inoculum to address the question for the therapeutic agents of such inoculation treatments. Here, in particular, we recommend working with heat-killed inocula and self-inoculation treatments to test for the suspected nutritional benefit and sterile-filtered inocula which can reveal molecular effects stemming from the cell-free supernatant.
The next steps to further explore the CMT strategy
Our study encompasses experiments performed on a short-term schedule including 1–3 days of inoculation treatments and 1–2 days of acute heat stress exposure. To further develop this method for implementation in coral propagation efforts in a reef restoration setting [2], we emphasize on the importance of a meticulous selection screening for healthy donors to minimize the potential transmission of pathogenic and harmful bacteria or other adverse agents (e.g., environmental pollutants). The important next step will be to test recipients under gradually increasing temperature mimicking a natural bleaching event [36, 63] along with long-term monitoring that includes measuring their recovery potential [64]. Eventually, a reef-reintroduction experiment will be required. The persistence of coral microbiome changes and accompanying physiological effects after CMT or probiotic inoculations needs to be investigated. These effects will need to last for a duration of few weeks, since coral bleaching events typically occur locally due to “short-lived episodes of extreme heat” [1]. On a positive note, bacterial community differences after microbiome manipulation have lasted up to 7 days after treatment for the sea anemone Nematosella [65] and a donor footprint including health benefits has been successfully documented in human gut microbiome transplantation recipients for 1–2 years [66].
Conclusions
We draw the final conclusion from a microbiome-focused perspective. Approaches that investigate the probiotic potential of microbiome manipulation, such as several previous coral probiotic studies [30, 31, 67] and the CMT concept developed in our study (Fig. 1), are founded on the hypothesis that exposure of the holobiont to beneficial bacterial consortia can be applied to increase health and resilience. This could be a result of a shift towards a more efficient microbiome community composition through the proliferation of beneficial bacteria that were already present within the microbiome or the incorporation of “entirely new” taxa or strains that provide additional functions helping the holobiont cope with stress. Our 16S rRNA gene metabarcoding data suggests a possible scenario, which could have been the uptake of a specific “new” variant of a typical coral-resident or “symbiotic” bacterium that can provide holobiont services in a more efficient manner under stress conditions than the native “symbionts.” These results further suggest that such bacterial taxa should spcifically be included into the scope of coral probiotics developments. Our CMT experiments demonstrated that coral bleaching resistance can be positively influenced in a field-based setting. Hence, this strategy might reveal itself as a feasible approach to support coral heat resistance and find application to enhance efficiency of certain coral propagation and restoration efforts. If scaling up the method will not turn out to be feasible any soon, another perspective is that CMT can serve as an elegant manipulative tool which could help further advance the identification of probiotic bacterial species and strains, which then could be further developed into a probiotic inoculation treatment sensu [4, 30].
Materials and methods
Coral collection sites, taxa, and maintenance
Collection sites were located in the Andaman Sea, Thailand (Fig. 2a, see details in Supplementary Material and Methods). They were purposely chosen based on their local environmental differences and implications for coral heat tolerance [36, 68]. We refer to the collections sites as “LowVar” for sites of low temperature variability and “HighVar” for sites of high temperature variability (Fig. 2b). Fragments from visually healthy colonies of Pocillopora sp. (April 2018) and Porites sp. (November 2018) were collected. Both corals represent two distinct coral ecotypes (branching vs. massive morphology) and are abundant and ecologically significant coral species in Thailand [69]. All experiments were performed inside four 40 L tanks (Tables S6-7).
Heat tolerance assessment
We assessed coral heat tolerance by employing short-term heat tolerance assays to quantify stress responses in a high-throughput manner. We tested a suite of colonies from the different sites. Ten colonies from the “LowVar” and 9 from the “HighVar” site were screened in the genus Pocillopora, and 14 colonies from the “LowVar” and 12 from the “HighVar” site in the genus of Porites. Two fragments from each colony were randomly distributed among the two treatments, “34 °C” and “29 °C” (tanks N = 2). The “34 °C” treatment was established by ramping temperatures from 29 to 34 °C for 4 h, holding at 34 °C for 5 h (Pocillopora) or 6 h (Porites), and decreasing temperatures to ambient 29 °C within 4 h (Fig. S1 A-B). Afterwards, all corals were maintained at ambient temperature for 10–11 h until the next day. While Pocillopora corals were subjected to one 34 °C-heat peak, resulting in a full experiment of 24 h (see experiment schedule in Fig. S1 A), Porites corals were exposed twice under 34 °C-heat peak resulting in a heat tolerance assay of 72 h in total (Fig. S1 B).
Coral microbiome transplantation experiments
Based on the outcomes of the heat tolerance assessment, colonies from the “LowVar” site were designated to be recipients. For the coral microbiome transplantation (CMT) experiments, we collected four fragments per colony of Pocillopora (colony N = 5) and Porites (colony N = 10). Each experiment consisted of two parts: (i) the inoculation phase with two treatment groups, “I” (i.e., recipients of a CMT inoculation) and “C” (i.e., control group receiving a FSW inoculation), and (ii) the reassessment of heat tolerance resulting in four experimental groups, i.e., “I x 29 °C,” “I x 34 °C,” “C x 29 °C,” and “C x 34 °C.” We designated colonies from the “HighVar” sites to be donors and collected six Pocillopora fragments from the west shore of Racha Island (colony N = 3, two fragments per colony) and 18 Porites fragments from Panwa reef flat (colony N = 6, three fragments per colony). Inocula were prepared 2 h before each inoculation event by homogenizing the tissues of each donor fragment according to established protocols for coral pathogen transmission ([37], Supplementary Material and Methods and Fig. S10). Four fragments of each recipient colony were randomly distributed among the four experimental tanks to commence the inoculation phase. Inoculations were performed at 29 °C. Current pumps and aeration were interrupted and the seawater volume was reduced to 8 L (i.e., 6 cm water level). Next, PVC tubes (height 7 cm, ø 8 cm, volume 350 mL) were placed around each coral fragment to create a semi-enclosed microenvironment to which inoculation shots were added [70]. The control treatments consisted of shots of filtered seawater (FSW 0.2 μm) that were prepared without the addition of donor-tissue material. Pocilloporid recipients each received one inoculation shot over 24 h, while Porites recipients each received three shots repeated every 24 h over 3 days (Fig. S1 C-D). Corals were incubated with the inocula inside the tubes for 30 min (Pocillopora) or 2 h (Porites). Subsequently, PVC tubes were removed, water flow and aeration were switched back on, and tanks were filled up to 40 L, further diluting bacterial densities. After 24 h, a regular seawater exchange (50% twice a day) was continued. Differences between the two inoculation procedures stem from logistic limitations such as availability of coral material. Following the inoculation phase, heat stress tolerance was reassessed in both recipient groups. Heat tolerance assays were performed as described above with a minor modification of exposing Pocillopora recipients at 34 °C for 7 instead of 5 h.
Coral response variables
Stress response variables were measured for each fragment before and after each experimental part. First, a bleaching score was determined as a measure of dinoflagellate symbiont cell density in coral tissues. The same observer visually categorized individual fragments on the scale from “1” (bleached) to “6” (healthy), recording a minimum and maximum score (“Coral Health Chart,” Coral Watch, reefquest.org, [71]). Second, photosynthetic efficiency of symbionts was assessed in light-adapted fragments measuring effective quantum yield (Φ PSII = (Fm’ – F)/Fm’ = ΔF/Fm’, [72]) using a pulse amplitude-modulated fluorometer (Diving-PAM, Walz, Germany).
Microbiome sequencing
Coral and seawater samples were collected at three time points during the CMT experiments, “start” of inoculation, “end” of inoculation, and “end” of heat tolerance reassessment (Fig. S1 C-D). DNA was extracted following established protocols. The variable region V3–V4 of the 16S rRNA gene (357F [5′CCTACGGGAGGCAGCAG′3], 806R [5′GACTACHVGGGTWTCTAAT′3]) was amplified and sequenced [73], including quality control (QC) samples, i.e., PCR and DNA extraction kit blanks. Amplicon library preparation and sequencing were performed at the IKMB Sequencing Center (University of Kiel, Germany). Data were delivered by two Illumina runs and processed separately using QIIME2 v2019.7 integrating DADA2 [74]. Amplicon sequence variants (ASVs) from both runs were merged prior to classification with SILVA v132 [75]. Sequence reads from QC samples were used to identify contaminants and clean up the data (Dataset S4). Sequencing data are available in the NCBI Sequence Read Archive (SRA) under accession number PRJNA647757. A rarefied data set was generated after determination of a suitable subsampling depth using rarefaction tools as implemented in QIIME2. Rarefaction curves were plotted using function rarecurve (R package vegan v2.5-6). A filtered data set, “filt-10,” was created from the full data excluding rare ASVs with a total read abundance of <10. Details are provided in the Supplementary Materials and Methods and Dataset S5.
Statistical analysis
∆-values of coral response variables (end–start of each experimental part) were used for analyses. Effects of (i) temperature treatments (“34 °C” vs. “29 °C”) within the site of origin (“HighVar,” “LowVar”), (ii) inoculation (“I” vs. “C”), and (iii) subsequent temperature treatment of inoculated recipient corals (“34 °C” vs. “29 °C”) were evaluated using dabestR v0.2.3 6 [76] and linear mixed effect models (nlme v4 3.1-148 and lme4 v1.1-23 package). Where applicable, coral colony genotype was used as a random factor.
Microbiome α- and β-diversity analyses were performed on the rarefied data using phyloseq v1.32.0 and vegan v2.5-6 in R. We compared (i) coral and seawater microbiomes pooled across time points (“Pocillopora,” “Porites,” “seawater tank,” “seawater source”), (ii) coral groups at experiment “start” (“donor,” “inoculum,” “recipient start group”), (iii) at the “end” of inoculation, and (iv) at the “end” of heat tolerance reassessment. α-diversity metrics were analyzed by dabestR v0.2.3, ANOVA or Kruskal-Wallis, and generalized linear or linear mixed effect models where suitable. Dissimilarities (Bray-Curtis) and homogeneity of variances were tested by PERMANOVA with 9999 permutations using adonis2 and by PERMDISP using betadisper function, respectively. Pairwise tests followed respectively, pairwise.perm.manova or Tukey HSD test through betadisper. The “filt-10” data set was used to create stacked bar plots at the bacterial species level (showing the most dominant species: relative abundance >10%). UpSetR (v1.4.0 [77];) analyses were performed with “filt-10” data to (i) characterize shared bacterial taxa in the coral and seawater microbiome, (ii) identify exclusively unique taxa in the inoculum, and (iii) capture bacteria uniquely shared by the inoculum and the recipients after inoculation.
Supplementary Information
Additional file 1. Supplementary materials.
Additional file 2: Dataset S1. Analysis table of coral response variables. ‘LowVar’ site = Racha Island east shore, ‘HighVar’ site (Pocillopora) = Racha Island west shore, ‘HighVar’ site (Porites) = Panwa reef flat; heat tolerance (HT) assessment treatments: ‘34 °C’ and ‘29 °C’; Inoculation treatments: ‘I’ = inoculation; ‘C’ = sterile-filtered seawater (FSW) control group; n = replicate fragment numbers; mean difference = mean difference between start and end of a treatment; SD = standard deviation; SE = standard error; CI = confidence interval.
Additional file 3: Dataset S2. Analysis tables of microbiome data. Count tables are provided for (A) ‘filt-10’ data and (B) rarefied data (subsampled to 4 000 reads). Tables enclose read abundance counts per amplicon sequence variant (ASV), experiment metadata (i.e., treatment groups), and SILVA classification. TYPE = sample type; POC = Pocillopora; POR = Porites; SW = seawater. Timepoints include: t1 = start, t2 = end of inoculation, t3 = timepoint of water collection from the source tank during heat tolerance (HT) reassessment, t4 = end of HT reassessment. I = inoculation; C = FSW control group; H = heat exposure treatment 34 °C; A = ambient temperature treatment 29 °C.
Additional file 4: Dataset S3. Full table of potentially transmitted bacteria in the (A) Pocillopora and (B) Porites experiment. Amplicon sequence variants (ASVs) exclusively shared between the inoculum and the ‘I’ recipient’ group after inoculation are provided each with their respective SILVA based taxonomy and sequence. Read abundances for each potentially transmitted ASV show its occurrence within the different treatment groups. ASVs are marked in ‘green’, when also detected in the donor samples. Those, also detected in seawater samples, are marked in ‘blue’. ASVs are marked in red, when detected in the ‘I’ recipient group at the end of heat tolerance reassessment (i.e., ASVs that persisted within the recipients’ microbiomes until the very end of the experiment).
Additional file 5: Dataset S4. Quality control samples and clean-up of microbiome data. Tables show host-origin and contaminant amplicon sequence variant (ASV) sequences that were removed from the microbiome data set prior to downstream analyses. (A) Table shows ASV sequences of host-origin as matched with GenBank (NCBI). (B) Table shows ASVs identified as contaminants using DNA Extraction Kit and PCR blank samples. Stacked bar charts show bacterial community compositions of (C) extraction kit blank samples and (D) PCR blank samples. Additionally, scoring tables for contaminant ASVs are shown.
Additional file 6: Dataset S5. Protocol of raw read processing using QIIME2 V2019.7.
Acknowledgements
We acknowledge the team of the research unit Marine Ecology at Phuket Marine Biological Center for field logistics and support during the set up of coral experimental facilities. We thank M. Heckwolf, K. Bimson, L. Niewendieck, M. Suwareh, and V. Conrad for field assistance. We thank A. Hethke, I. Clefsen, and N. Pohl for their assistance in the laboratories; F. Wendt for advice regarding aquarium equipment; T. Bayer for his recommendations during the data analysis and manuscript preparation; and C.R. Voolstra for advice during the manuscript preparation.
Permissions
Research in Thailand was conducted under the permit of the National Research Council of Thailand (NRCT # 0002/632), and corals were collected under the collection and export permissions of CITES (export # AC.0510.6/0017 and # AC.0510.6/022; import # DE E-04829/18 and # DE E-01510/19).
Abbreviations
- CMT
Coral microbiome manipulation
- FSW
Sterile-filtered seawater
- ASV
Amplicon sequence variant
- QC
Quality control
- nMDS
Non-metric Multidimensional Scaling
- PERMANOVA
PERMutational multivariate ANalysis Of VAriance
- BETADISPER
BETA-diversity analysis of multivariate homogeneity of group DISPERsions (variances)
Authors’ contributions
AR conceived the study. AR and TD designed the experiment. TD and AR conducted coral experiments. AR, MW, LP, and TR performed coral collection. TD, RS, and AR performed lab work. TD, AR, and MW performed the data analysis and visualization. AR, TD, and MW wrote and edited the manuscript. UHH edited the manuscript. Field facilities were provided by the PMBC team and molecular laboratory space by the “Marine Symbioses” group at GEOMAR. The authors read and approved the final manuscript.
Funding
This research was funded by the DFG (German National Science Foundation) excellence initiative “Future Ocean” (# CP1782) awarded to AR and additional support from the DFG research center “Origin and Function of Metaorganisms” (# CRC1182-TP B1) awarded to UHH. Open Access funding enabled and organized by Projekt DEAL.
Availability of data and materials
Sequence data generated in this study are available under NCBI BioProject ID PRJNA647757. Source data underlying analyses are provided with the Supplementary Dataset files, i.e., analysis tables for phenotypic response variables are provided in Dataset S1; ASV count tables including metadata, corresponding taxonomic classification, and read sequences are presented in Dataset S2; and quality control samples and decontamination process of the data are presented in Dataset S4. The raw read processing protocol using QIIME v2019.7 including summary statistics is provided in Dataset S5.
Declarations
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hughes TP, Anderson KD, Connolly SR, Heron SF, Kerry JT, Lough JM, et al. Spatial and temporal patterns of mass bleaching of corals in the Anthropocene. Science (80-83) 2018;359:80–83. doi: 10.1126/science.aan8048. [DOI] [PubMed] [Google Scholar]
- 2.van Oppen MJH, Oliver JK, Putnam HM, Gates RD. Building coral reef resilience through assisted evolution. Proc Natl Acad Sci. 2015;112(8):2307–2313. doi: 10.1073/pnas.1422301112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan WY, Hoffmann AA, Oppen MJH. Hybridization as a conservation management tool. Conserv Lett. 2019;12:conl.12652. doi:10.1111/conl.12652.
- 4.Peixoto RS, Rosado PM, Leite DC, Rosado AS, Bourne DG. Beneficial microorganisms for corals (BMC): proposed mechanisms for coral health and resilience. Front Microbiol. 2017;1–16. doi:10.3389/fmicb.2017.00341. [DOI] [PMC free article] [PubMed]
- 5.Epstein HE, Smith HA, Torda G, van Oppen MJH. Microbiome engineering: enhancing climate resilience in corals. Front Ecol Environ. 2019;17(2):100–108. doi: 10.1002/fee.2001. [DOI] [Google Scholar]
- 6.Bourne DG, Morrow KM, Webster NS. Insights into the coral microbiome: underpinning the health and resilience of reef ecosystems. Annu Rev Microbiol. 2016;70(1):317–340. doi: 10.1146/annurev-micro-102215-095440. [DOI] [PubMed] [Google Scholar]
- 7.Muscatine L, Porter JW. Reef corals: mutualistic symbioses adapted to nutrient-poor environments. Bioscience. 1977;27(7):454–460. doi: 10.2307/1297526. [DOI] [Google Scholar]
- 8.Robbins SJ, Singleton CM, Chan CX, Messer LF, Geers AU, Ying H, et al. A genomic view of the reef-building coral Porites lutea and its microbial symbionts. Nat Microbiol. 2019;4(12):2090–2100. doi: 10.1038/s41564-019-0532-4. [DOI] [PubMed] [Google Scholar]
- 9.van Oppen MJH, Blackall LL. Coral microbiome dynamics, functions and design in a changing world. Nat Rev Microbiol. 2019;1. 10.1038/s41579-019-0223-4. [DOI] [PubMed]
- 10.Rohwer F, Seguritan V, Azam F, Knowlton N. Diversity and distribution of coral-associated bacteria. Mar Ecol Prog Ser. 2002;243:1–10. doi: 10.3354/meps243001. [DOI] [Google Scholar]
- 11.Sunagawa S, Woodley CM, Medina M. Threatened corals provide underexplored microbial habitats. PLoS One. 2010;5(3):e9554. doi: 10.1371/journal.pone.0009554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McDevitt-Irwin JM, Baum JK, Garren M, Vega Thurber RL. Responses of coral-associated bacterial communities to local and global stressors. Front Mar Sci. 2017;4:262. doi: 10.3389/fmars.2017.00262. [DOI] [Google Scholar]
- 13.Pollock FJ, McMinds R, Smith S, Bourne DG, Willis BL, Medina M, Thurber RV, Zaneveld JR. Coral-associated bacteria demonstrate phylosymbiosis and cophylogeny. Nat Commun. 2018;9(1):1–13. doi: 10.1038/s41467-018-07275-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ziegler M, Grupstra CGBB, Barreto MM, Eaton M, BaOmar J, Zubier K, et al. Coral bacterial community structure responds to environmental change in a host-specific manner. Nat Commun. 2019;10(1):3092. doi: 10.1038/s41467-019-10969-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neave MJ, Michell CT, Apprill A, Voolstra CR. Endozoicomonas genomes reveal functional adaptation and plasticity in bacterial strains symbiotically associated with diverse marine hosts. Sci Rep. 2017;7(1). 10.1038/srep40579. [DOI] [PMC free article] [PubMed]
- 16.Thurber RV, Willner-Hall D, Rodriguez-Mueller B, Desnues C, Edwards RA, Angly F, Dinsdale E, Kelly L, Rohwer F. Metagenomic analysis of stressed coral holobionts. Environ Microbiol. 2009;11(8):2148–2163. doi: 10.1111/j.1462-2920.2009.01935.x. [DOI] [PubMed] [Google Scholar]
- 17.Wegley L, Edwards R, Rodriguez-Brito B, Liu H, Rohwer F. Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environ Microbiol. 2007;9(11):2707–2719. doi: 10.1111/j.1462-2920.2007.01383.x. [DOI] [PubMed] [Google Scholar]
- 18.Krediet CJ, Ritchie KB, Paul VJ, Teplitski M. Coral-associated micro-organisms and their roles in promoting coral health and thwarting diseases. Proc R Soc B Biol Sci. 2013;280(1755):20122328. doi: 10.1098/rspb.2012.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ziegler M, Roik A, Porter A, Zubier K, Mudarris MS, Ormond R, Voolstra CR. Coral microbial community dynamics in response to anthropogenic impacts near a major city in the central Red Sea. Mar Pollut Bull. 2016;105(2):629–640. doi: 10.1016/j.marpolbul.2015.12.045. [DOI] [PubMed] [Google Scholar]
- 20.Hernandez-Agreda A, Leggat W, Bongaerts P, Herrera C, Ainsworth TD. Rethinking the coral microbiome: simplicity exists within a diverse microbial biosphere. MBio. 2018;9(5):e00812–e00818. doi: 10.1128/mBio.00812-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chu ND, Vollmer SV. Caribbean corals house shared and host-specific microbial symbionts over time and space. Environ Microbiol Rep. 2016;8(4):493–500. doi: 10.1111/1758-2229.12412. [DOI] [PubMed] [Google Scholar]
- 22.Röthig T, Bravo H, Corley A, Prigge T-L, Chung A, Yu V, McIlroy SE, Bulling M, Sweet M, Baker DM. Environmental flexibility in Oulastrea crispata in a highly urbanised environment: a microbial perspective. Coral Reefs. 2020;39(3):649–662. doi: 10.1007/s00338-020-01938-2. [DOI] [Google Scholar]
- 23.Redman RS, Kim YO, Woodward CJDA, Greer C, Espino L, Doty SL, et al. Increased fitness of rice plants to abiotic stress via habitat adapted symbiosis: a strategy for mitigating impacts of climate change. PLoS One. 2011;6:1–10. doi: 10.1371/journal.pone.0014823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moran NA, Yun Y. Experimental replacement of an obligate insect symbiont. Proc Natl Acad Sci. 2015;112(7):2093–2096. doi: 10.1073/pnas.1420037112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Borody TJ, Khoruts A. Fecal microbiota transplantation and emerging applications. Nat Rev Gastroenterol Hepatol. 2012;9(2):88–96. doi: 10.1038/nrgastro.2011.244. [DOI] [PubMed] [Google Scholar]
- 26.Cohen Y, Joseph Pollock F, Rosenberg E, Bourne DG. Phage therapy treatment of the coral pathogen Vibrio coralliilyticus. Microbiologyopen. 2013;2(1):64–74. doi: 10.1002/mbo3.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Efrony R, Loya Y, Bacharach E, Rosenberg E. Phage therapy of coral disease. Coral Reefs. 2007;26(1):7–13. doi: 10.1007/s00338-006-0170-1. [DOI] [Google Scholar]
- 28.Damjanovic K, van Oppen MJH, Menéndez P, Blackall LL. Experimental inoculation of coral recruits with marine bacteria indicates scope for microbiome manipulation in Acropora tenuis and Platygyra daedalea. Front Microbiol. 2019;1702. doi:10.3389/fmicb.2019.01702. [DOI] [PMC free article] [PubMed]
- 29.Welsh RM, Rosales SM, Zaneveld JR, Payet JP, McMinds R, Hubbs SL, et al. Alien vs. predator: bacterial challenge alters coral microbiomes unless controlled by Halobacteriovorax predators. PeerJ. 2017;5:e3315. doi: 10.7717/peerj.3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosado PM, Leite DCA, Duarte GAS, Chaloub RM, Jospin G, Nunes da Rocha U, et al. Marine probiotics: increasing coral resistance to bleaching through microbiome manipulation. ISME J. 2018;13(4):921–936. doi: 10.1038/s41396-018-0323-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fragoso Ados Santos H, Duarte GAS, Rachid CTDC, Chaloub RM, Calderon EN, Marangoni LFDB, et al. Impact of oil spills on coral reefs can be reduced by bioremediation using probiotic microbiota. Sci Rep. 2015;5:1–11. doi: 10.1038/srep18268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jacquemot L, Bettarel Y, Monjol J, Corre E, Halary S, Desnues C, et al. Therapeutic potential of a new jumbo phage that infects Vibrio coralliilyticus, a widespread coral pathogen. Front Microbiol. 2018;2501. doi:10.3389/fmicb.2018.02501. [DOI] [PMC free article] [PubMed]
- 33.Oliver TA, Palumbi SR. Do fluctuating temperature environments elevate coral thermal tolerance? Coral Reefs. 2011;30(2):429–440. doi: 10.1007/s00338-011-0721-y. [DOI] [Google Scholar]
- 34.Voolstra CR, Buitrago-López C, Perna G, Cárdenas A, Hume BCC, Rädecker N, Barshis DJ. Standardized short-term acute heat stress assays resolve historical differences in coral thermotolerance across microhabitat reef sites. Glob Chang Biol. 2020;26(8):4328–4343. doi: 10.1111/gcb.15148. [DOI] [PubMed] [Google Scholar]
- 35.Camp EF, Schoepf V, Mumby PJ, Hardtke LA, Rodolfo-Metalpa R, Smith DJ, et al. The future of coral reefs subject to rapid climate change: lessons from natural extreme environments. Front Mar Sci. 2018;4. doi:10.3389/fmars.2018.00004.
- 36.Buerger P, Schmidt GM, Wall M, Held C, Richter C. Temperature tolerance of the coral Porites lutea exposed to simulated large amplitude internal waves (LAIW) J Exp Mar Bio Ecol. 2015;471:232–239. doi: 10.1016/j.jembe.2015.06.014. [DOI] [Google Scholar]
- 37.Gignoux-Wolfsohn SA, Aronson FM, Vollmer SV. Complex interactions between potentially pathogenic, opportunistic, and resident bacteria emerge during infection on a reef-building coral. FEMS Microbiol Ecol. 2017;93:1–10. doi: 10.1093/femsec/fix080. [DOI] [PubMed] [Google Scholar]
- 38.Kleypas JA, McManu JW, Mene LAB. Environmental limits to coral reef development: where do we draw the line? Am Zool. 1999;39(1):146–159. doi: 10.1093/icb/39.1.146. [DOI] [Google Scholar]
- 39.Gardner SG, Raina JB, Nitschke MR, Nielsen DA, Stat M, Motti CA, Ralph PJ, Petrou K. A multi-trait systems approach reveals a response cascade to bleaching in corals. BMC Biol. 2017;15(1):117. doi: 10.1186/s12915-017-0459-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shaver EC, Shantz AA, McMinds R, Burkepile DE, Vega Thurber RL, Silliman BR. Effects of predation and nutrient enrichment on the success and microbiome of a foundational coral. Ecology. 2017;98(3):830–839. doi: 10.1002/ecy.1709. [DOI] [PubMed] [Google Scholar]
- 41.Pogoreutz C, Rädecker N, Cárdenas A, Gärdes A, Wild C, Voolstra CR. Dominance of Endozoicomonas bacteria throughout coral bleaching and mortality suggests structural inflexibility of the Pocillopora verrucosa microbiome. Ecol Evol. 2018;8(4):2240–2252. doi: 10.1002/ece3.3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bourne DG, Iida Y, Uthicke S, Smith-Keune C. Changes in coral-associated microbial communities during a bleaching event. ISME J. 2008;2(4):350–363. doi: 10.1038/ismej.2007.112. [DOI] [PubMed] [Google Scholar]
- 43.McCauley M, Jackson CR, Goulet TL. Microbiomes of Caribbean octocorals vary over time but are resistant to environmental change. Front Microbiol. 2020;11:1272. doi: 10.3389/fmicb.2020.01272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burke C, Steinberg P, Rusch D, Kjelleberg S, Thomas T. Bacterial community assembly based on functional genes rather than species. Proc Natl Acad Sci U S A. 2011;108(34):14288–14293. doi: 10.1073/pnas.1101591108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bayer T, Neave MJ, Alsheikh-Hussain A, Aranda M, Yum LK, Mincer T, Hughen K, Apprill A, Voolstra CR. The microbiome of the Red Sea coral Stylophora pistillata is dominated by tissue-associated Endozoicomonas bacteria. Appl Environ Microbiol. 2013;79(15):4759–4762. doi: 10.1128/AEM.00695-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neave MJ, Apprill A, Ferrier-Pagès C, Voolstra CR. Diversity and function of prevalent symbiotic marine bacteria in the genus Endozoicomonas. Applied Microbiology and Biotechnology. 2016;100(19):8315–8324. doi: 10.1007/s00253-016-7777-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Knight R, Vrbanac A, Taylor BC, Aksenov A, Callewaert C, Debelius J, Gonzalez A, Kosciolek T, McCall LI, McDonald D, Melnik AV, Morton JT, Navas J, Quinn RA, Sanders JG, Swafford AD, Thompson LR, Tripathi A, Xu ZZ, Zaneveld JR, Zhu Q, Caporaso JG, Dorrestein PC. Best practices for analysing microbiomes. Nat Rev Microbiol. 2018;16(7):410–422. doi: 10.1038/s41579-018-0029-9. [DOI] [PubMed] [Google Scholar]
- 48.Callahan BJ, Wong J, Heiner C, Oh S, Theriot CM, Gulati AS, McGill SK, Dougherty MK. High-throughput amplicon sequencing of the full-length 16S rRNA gene with single-nucleotide resolution. Nucleic Acids Res. 2019;47(18):e103. doi: 10.1093/nar/gkz569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Voolstra CR, Ziegler M. Adapting with microbial help: microbiome flexibility facilitates rapid responses to environmental change. BioEssays. 2020;:2000004. doi:10.1002/bies.202000004. [DOI] [PubMed]
- 50.De Roy K, Marzorati M, Negroni A, Thas O, Balloi A, Fava F, et al. Environmental conditions and community evenness determine the outcome of biological invasion. Nat Commun. 2013;4(1):1–5. doi: 10.1038/ncomms2392. [DOI] [PubMed] [Google Scholar]
- 51.Davidov Y, Jurkevitch E. Diversity and evolution of Bdellovibrio-and-like organisms (BALOs), reclassification of Bacteriovorax starrii as Peredibacter starrii gen. nov., comb. nov., and description of the Bacteriovorax-Peredibacter clade as Bacteriovoracaceae fam. nov. Int J Syst Evol Microbiol. 2004;54(5):1439–1452. doi: 10.1099/ijs.0.02978-0. [DOI] [PubMed] [Google Scholar]
- 52.Johnke J, Cohen Y, de Leeuw M, Kushmaro A, Jurkevitch E, Chatzinotas A. Multiple micro-predators controlling bacterial communities in the environment. Curr Opin Biotechnol. 2014;27:185–190. doi: 10.1016/j.copbio.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 53.Chen H, Laws EA, Martin JL, Berhane T-K, Gulig PA, Williams HN. Relative contributions of Halobacteriovorax and bacteriophage to bacterial cell death under various environmental conditions. MBio. 2018;9(4). 10.1128/mBio.01202-18. [DOI] [PMC free article] [PubMed]
- 54.Cao H, Wang H, Yu J, An J, Chen J. Encapsulated Bdellovibrio powder as a potential bio-disinfectant against whiteleg shrimp-pathogenic Vibrios. Microorganisms. 2019;7(8):244. doi: 10.3390/microorganisms7080244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Richards GP, Fay JP, Dickens KA, Parent MA, Soroka DS, Boyd EF. Predatory bacteria as natural modulators of Vibrio parahaemolyticus and Vibrio vulnificus in seawater and oysters. Appl Environ Microbiol. 2012;78(20):7455–7466. doi: 10.1128/AEM.01594-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reshef L, Koren O, Loya Y, Zilber-Rosenberg I, Rosenberg E. The coral probiotic hypothesis. Environ Microbiol. 2006;8(12):2068–2073. doi: 10.1111/j.1462-2920.2006.01148.x. [DOI] [PubMed] [Google Scholar]
- 57.Ott SJ, Waetzig GH, Rehman A, Moltzau-Anderson J, Bharti R, Grasis JA, et al. Efficacy of sterile fecal filtrate transfer for treating patients with Clostridium difficile infection. Gastroenterology. 2017;152:799–811. doi: 10.1053/j.gastro.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 58.Leonardi I, Paramsothy S, Doron I, Semon A, Kaakoush NO, Clemente JC, et al. Fungal trans-kingdom dynamics linked to responsiveness to fecal microbiota transplantation (FMT) therapy in ulcerative colitis. Cell Host Microbe. 2020;27:823–829. doi: 10.1016/j.chom.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Draper LA, Ryan FJ, Smith MK, Jalanka J, Mattila E, Arkkila PA, Ross RP, Satokari R, Hill C. Long-term colonisation with donor bacteriophages following successful faecal microbial transplantation. Microbiome. 2018;6(1):220. doi: 10.1186/s40168-018-0598-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bojanova DP, Bordenstein SR. Fecal transplants: what is being transferred? PLoS Biol. 2016;14(7):e1002503. doi: 10.1371/journal.pbio.1002503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aichelman HE, Townsend JE, Courtney TA, Baumann JH, Davies SW, Castillo KD. Heterotrophy mitigates the response of the temperate coral Oculina arbuscula to temperature stress. Ecol Evol. 2016;6(18):6758–6769. doi: 10.1002/ece3.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Castro-Oliveira L, Silva MO, Carvalho RDO, Anchiêta A, Benevides LJ, Oliveira CJF, et al. Omics of probiotic bacteria: which features to seek? Genet Mol Res. 2020;19:gmr18599. doi: 10.4238/gmr18599. [DOI] [Google Scholar]
- 63.Grottoli AG, Warner ME, Levas SJ, Aschaffenburg MD, Schoepf V, Mcginley M, et al. The cumulative impact of annual coral bleaching can turn some coral species winners into losers. Glob Chang Biol. 2014;20(12):3823–3833. doi: 10.1111/gcb.12658. [DOI] [PubMed] [Google Scholar]
- 64.Schoepf V, Grottoli AG, Levas SJ, Aschaffenburg MD, Baumann JH, Matsui Y, Warner ME. Annual coral bleaching and the long-term recovery capacity of coral. Proc R Soc B Biol Sci. 2015;282(1819):20151887. doi: 10.1098/rspb.2015.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Domin H, Zurita-Gutiérrez YH, Scotti M, Buttlar J, Humeida UH, Fraune S. Predicted bacterial interactions affect in vivo microbial colonization dynamics in Nematostella. Front Microbiol. 2018;9:728. doi: 10.3389/fmicb.2018.00728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Goloshchapov OV, Olekhnovich EI, Sidorenko SV, Moiseev IS, Kucher MA, Fedorov DE, Pavlenko AV, Manolov AI, Gostev VV, Veselovsky VA, Klimina KM, Kostryukova ES, Bakin EA, Shvetcov AN, Gumbatova ED, Klementeva RV, Shcherbakov AA, Gorchakova MV, Egozcue JJ, Pawlowsky-Glahn V, Suvorova MA, Chukhlovin AB, Govorun VM, Ilina EN, Afanasyev BV. Long-term impact of fecal transplantation in healthy volunteers. BMC Microbiol. 2019;19(1):312. doi: 10.1186/s12866-019-1689-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Morgans CA, Hung JY, Bourne DG, Quigley KM. Symbiodiniaceae probiotics for use in bleaching recovery. Restor Ecol. 2019;:rec.13069. doi:10.1111/rec.13069.
- 68.Brown B, Dunne RP, Somerfield PJ, Edwards AJ, Simons WJF, Phongsuwan N, et al. Long-term impacts of rising sea temperature and sea level on shallow water coral communities over a ~40 year period. Sci Rep. 2019;9:1–12. doi: 10.1038/s41598-019-45188-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brown B, Phongsuwan N. Delayed mortality in bleached massive corals on intertidal reef flats around Phuket, Andaman Sea, Thailand. Phuket Mar Biol Cent Res Bull. 2012;48:43–48. [Google Scholar]
- 70.Fuess LE, Pinzón CJH, Weil E, Grinshpon RD, Mydlarz LD. Life or death: disease-tolerant coral species activate autophagy following immune challenge. Proc R Soc B Biol Sci. 2017;284(1856):20170771. doi: 10.1098/rspb.2017.0771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Siebeck UE, Marshall NJ, Klüter A, Hoegh-Guldberg O. Monitoring coral bleaching using a colour reference card. Coral Reefs. 2006;25(3):453–460. doi: 10.1007/s00338-006-0123-8. [DOI] [Google Scholar]
- 72.Genty B, Briantais JM, Baker NR. The relationship between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochim Biophys Acta - Gen Subj. 1989;990(1):87–92. doi: 10.1016/S0304-4165(89)80016-9. [DOI] [Google Scholar]
- 73.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79(17):5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Callahan BJ, McMurdie PJ, Holmes SP. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 2017;11(12):2639–2643. doi: 10.1038/ismej.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41(Database issue):D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ho J, Tumkaya T, Aryal S, Choi H, Claridge-Chang A. Moving beyond P values: data analysis with estimation graphics. Nature Methods. 2019;16(7):565–566. doi: 10.1038/s41592-019-0470-3. [DOI] [PubMed] [Google Scholar]
- 77.Conway JR, Lex A, Gehlenborg N. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics. 2017;33(18):2938–2940. doi: 10.1093/bioinformatics/btx364. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Supplementary materials.
Additional file 2: Dataset S1. Analysis table of coral response variables. ‘LowVar’ site = Racha Island east shore, ‘HighVar’ site (Pocillopora) = Racha Island west shore, ‘HighVar’ site (Porites) = Panwa reef flat; heat tolerance (HT) assessment treatments: ‘34 °C’ and ‘29 °C’; Inoculation treatments: ‘I’ = inoculation; ‘C’ = sterile-filtered seawater (FSW) control group; n = replicate fragment numbers; mean difference = mean difference between start and end of a treatment; SD = standard deviation; SE = standard error; CI = confidence interval.
Additional file 3: Dataset S2. Analysis tables of microbiome data. Count tables are provided for (A) ‘filt-10’ data and (B) rarefied data (subsampled to 4 000 reads). Tables enclose read abundance counts per amplicon sequence variant (ASV), experiment metadata (i.e., treatment groups), and SILVA classification. TYPE = sample type; POC = Pocillopora; POR = Porites; SW = seawater. Timepoints include: t1 = start, t2 = end of inoculation, t3 = timepoint of water collection from the source tank during heat tolerance (HT) reassessment, t4 = end of HT reassessment. I = inoculation; C = FSW control group; H = heat exposure treatment 34 °C; A = ambient temperature treatment 29 °C.
Additional file 4: Dataset S3. Full table of potentially transmitted bacteria in the (A) Pocillopora and (B) Porites experiment. Amplicon sequence variants (ASVs) exclusively shared between the inoculum and the ‘I’ recipient’ group after inoculation are provided each with their respective SILVA based taxonomy and sequence. Read abundances for each potentially transmitted ASV show its occurrence within the different treatment groups. ASVs are marked in ‘green’, when also detected in the donor samples. Those, also detected in seawater samples, are marked in ‘blue’. ASVs are marked in red, when detected in the ‘I’ recipient group at the end of heat tolerance reassessment (i.e., ASVs that persisted within the recipients’ microbiomes until the very end of the experiment).
Additional file 5: Dataset S4. Quality control samples and clean-up of microbiome data. Tables show host-origin and contaminant amplicon sequence variant (ASV) sequences that were removed from the microbiome data set prior to downstream analyses. (A) Table shows ASV sequences of host-origin as matched with GenBank (NCBI). (B) Table shows ASVs identified as contaminants using DNA Extraction Kit and PCR blank samples. Stacked bar charts show bacterial community compositions of (C) extraction kit blank samples and (D) PCR blank samples. Additionally, scoring tables for contaminant ASVs are shown.
Additional file 6: Dataset S5. Protocol of raw read processing using QIIME2 V2019.7.
Data Availability Statement
Sequence data generated in this study are available under NCBI BioProject ID PRJNA647757. Source data underlying analyses are provided with the Supplementary Dataset files, i.e., analysis tables for phenotypic response variables are provided in Dataset S1; ASV count tables including metadata, corresponding taxonomic classification, and read sequences are presented in Dataset S2; and quality control samples and decontamination process of the data are presented in Dataset S4. The raw read processing protocol using QIIME v2019.7 including summary statistics is provided in Dataset S5.