Abstract
High-throughput, high-content imaging technologies and multiplex slide-scanning have become widely used. Advantages of these approaches include the ability to archive digital copies of slides, review slides as teams using virtual microscopy software, and standardize analytical approaches. Barriers to implementation include both the cost and hardware and software inflexibility of dedicated slide-scanning devices. Here, we describe a simple method that allows any microscope to be used for slide-scanning. The only requirements are that the microscope be equipped with a motorized filter turret or wheels (for multi-channel fluorescence) and a motorized xyz stage. This example uses MetaMorph software, but the same principles can be used with any microscope control software that includes a few standard functions and allows programming of simple command routines, or journals. The series of journals that implement the method perform key functions, including assistance in defining an unlimited number of regions of interest and imaging parameters. Fully-automated acquisition is rapid, taking less than 3 hours to image 50 2.5mm regions of interest in four channels. Following acquisition, images can be easily stitched and displayed using open source or commercial image processing and virtual microscope applications.
Keywords: slide-scanning, fluorescence microscopy, digital pathology, tissue microarray, automated image acquisition
INTRODUCTION
As with all research, analysis of microscopic images is increasingly quantitative. These assessments rely on digital imaging using cameras with broad, linear dynamic ranges. However, manual methods of observation only capture selected fields within large specimens, thereby limiting the ability of other observers to evaluate the entire specimen and creating the potential for sampling bias. One way to overcome this is complete tissue, or slide, imaging. This is particularly advantageous for fluorescent samples, e.g., immunofluorescent stained cells and tissues, in which repeated viewing causes photobleaching and specimens degrade over time. Slide scanning also allows i) review on any computer without need for a microscope; ii) display of whole-slide images in group meetings, for both analysis and education of new investigators; iii) simple image annotation; iv) complete post-acquisition analysis, and v) sharing of unfiltered, raw data between investigators at different sites. Automated high-content imaging is also essential for multiplex technologies that image the same slides over many cycles staining (Goltsev et al., 2018; J.-R. Lin et al., 2018; Lin, Fallahi-Sichani, & Sorger, 2015; McKinley et al., 2017; Patel & Rodig, 2020; Rashid et al., 2019).
Most open-source and commercial microscope control packages now include a slide scanning module that allows users to capture a large area region of interest (ROI) as many smaller fields (tiles). The individual images are then stitched together to create a single image. After processing, this composite image can be saved in formats that allow rapid, network-based viewing using virtual microscopy software that allows simple slide navigation and rapid transition between low magnification and high magnifications. As a result of the power of such systems, slide scanning has become widely used.
Unfortunately, slide scanning modules within acquisition software are most often configured to image a single ROI after which a new ROI can be defined manually. This limits applicability of these tools to image tissue microarrays (TMAs) and similar technologies that enable the staining of many samples on a single slide (Dhir, 2008; Robertson, Savage, Reis-Filho, & Isacke, 2008). One could overcome this limitation by imaging the entire slide, but this would greatly increase the time required for data collection and create enormous data files that would be both difficult to stitch and view.
Multiple manufacturers have circumvented this challenge by developing automated slide scanning systems that only image regions of slides where specimens are present. These turnkey systems work well. However, opportunities for hardware customization are limited, and, due to the cost of the hardware, systems are typically available only within core facilities. Other shortcomings of some turnkey systems include frequent use of objectives with long working distances and low numerical apertures (NAs) to provide increased depth of field. This simplifies focusing at the cost of reduced resolution and extended exposure times, often leaving slides almost completely bleached. Turnkey systems may also use proprietary file formats that require conversion prior to analysis using other programs.
We sought to develop an automated slide scanning system that could easily be assembled within individual labs using only a simple fluorescent microscope as a cost-effective solution. We required that the solution i) be assembled using hardware already available in many labs; ii) allow for automated scanning of multiple ROIs; iii) create files of manageable sizes using standard formats, iv) include customizable focusing and image acquisition routines; v) produce high-quality images with relatively-short imaging times; and vi) be easily customized for specific purposes.
Focusing was a significant challenge. One option would be to use an alogorithm that defines the tissue plane based on measurements at key points. Systems that use a laser to determine slide position in the z axis might also be used. We were not satisfied with the results of either of these, likely because specimens within microarrays composed of formalin-fixed, paraffin-embedded tissues tend to vary in thickness and, in many cases, fold onto themselves or the slide. We considered software focusing at each tile, but this would extend the time required for imaging 10-fold. We therefore created journals that focus at user-defined intervals. These focus only if tissue is present and include checks to eliminate errors caused by debris or other artifacts. Basic Protocol 4 includes a more detailed explanation of the process.
This example uses MetaMorph software, but the journals could be translated for use in other programs, including MicroManager (Edelstein et al., 2014), that have internal command programming capabilities. This should allow application of the journals and conceptual solutions they describe to multiple imaging modalities and microscopes, objectives, stages, light sources, filters, and cameras produced by a wide range of vendors. For example, we use these journals with minor modifications for transmitted light imaging.
The journals work together and are accessed by a unified taskbar. The annotated code for these journals is provided (Supplemental Software Code S1-S43). Supplemental Movie 1 includes a short presentation of the procedures.
STRATEGIC PLANNING
Although specific hardware is described below, our aim was to develop an automated slide scanning system using existing equipment. Thus, the specific hardware used here should be considered only an example. Any wide-field epifluorescence microscope with either a motorized filter turret or other automated way to change wavelengths, e.g., wavelength-selectable light source and filter wheels, can be used.
A motorized stage is required; the high resolution (<0.5 μm xy repeatability) and linear encoders for the stage and focus motor used here allow precise positioning in all 3 axes. We have used a 4 position slide holder because 4 slides can be easily imaged overnight. However, systems that hold different numbers of slides can also be used. Care must be taken to ensure that slides are in register in the xy plane and are aligned with the camera focal plane, i.e., with limited z variance, across slides.
The system described here is equipped with a 5X objective for preliminary scans and a 20X objective for tiled image acquisition. Preliminary experiments showed that, for our purposes, overview scans using the 5X objective to image a nuclear stain worked well. For final imaging, the high NA 20X objective used here balances the competing demands of high resolution and manageable file sizes when imaging human intestinal biopsies and mouse intestinal cross sections, i.e., ~3 mm tissue cores. For other types of specimens, higher or lower magnification objectives may provide advantages. The only limitation is that the working distance be large enough to allow the stage to travel the entire z range needed without touching the slides. Use of immersion objectives is possible but may be difficult due to the challenges of maintaining sufficient immersion fluid.
Cameras with a wide field-of-view, high resolution, and excellent sensitivity are preferred. CMOS cameras are ideal for this purpose. The combination of camera, coupler, and objective magnification used here allows capture of ~48% of the illuminated area, which represents the majority of the area with uniform illumination, in each frame. The 20X objective (1.36 pixels/rAiry) results in near-Nyquist sampling. A 1.25X camera coupler would increase sampling, but it would also reduce each image to ~38% of the illuminated area and, therefore, require greater numbers of frames, extend imaging time, and increase file size. If greater sampling is needed, however, different cameras and couplers can easily be used. Although not essential for the protocols described here, the system is also equipped with a second camera port that houses a color CCD camera for transmitted light imaging, e.g., of hematoxylin and eosin-stained slides. The camera and coupler used for transmitted light result in individual frames sized similarly to those of the fluorescence imaging configuration.
This protocol and accompanying journals are written for MetaMorph 7.10. The journals also direct post-acquisition processing and stitching using MetaMorph. Open-source software, e.g., ImageJ/FIJI and ASHLAR can also be used (Muhlich, Russell, & Chen, 2017; Schindelin et al., 2012). An advantage of using ASHLAR for stitching is that it can be used for alignment and subtraction of images collected during multiplex imaging and staining. ASHLAR also creates the pyramidal files (Besson et al., 2019) that are often required for image display.
The complete system was designed to image tissue microarrays following fluorescent immunostaining. Standard approaches for creating (Kallioniemi, Wagner, Kononen, & Sauter, 2001; Kampf, Olsson, Ryberg, Sjostedt, & Ponten, 2012; Schweizer, Schumacher, & Rubin, 2004) and staining (Kampf et al., 2012; Lim et al., 2018; Lin, Fallahi-Sichani, Chen, & Sorger, 2016; J. R. Lin et al., 2018; Parra et al., 2017) tissue microarrays, including tissue processing, embedding, sectioning, blocking, and antigen retrieval are well-described. We have found that pre-bleaching with 4.5% H2O2 and 24 mM NaOH under bright light effectively reduces autofluorescence of formalin-fixed, paraffin-embedded tissues (J. R. Lin et al., 2018). We generally use SlowFade Diamond or ProLong Diamond (Invitrogen) antifade mountants. SlowFade, which is glycerol-based, is preferred if it will be necessary to remove the coverslips, e.g., for multiplex analysis. In this case, it may be helpful to reversibly secure the coverslips using nail polish. ProLong, which hardens, is preferred for archival storage but does not allow simple coverslip removal.
BASIC PROTOCOL 1 HARDWARE AND SOFTWARE CONFIGURATION
These journals are designed to work well with a wide variety of hardware configurations. The critical components include xyz motorized stage, automated filter changing, and high quality camera. This configuration is based on the hardware available in our lab. We used MetaMorph as the core software because it was already available in our lab. Thus, we were able to develop this system without purchasing new hardware or software. For those without MetaMorph, the conceptual approaches employed to solve the challenges of TMA imaging can be translated to other programs.
Materials
Upright or inverted wide-field microscope
The system used here is based on a Leica DM4000 fluorescent microscope, which has a motorized filter turret.
Motorized stage
The system described is equipped with a Ludl BioPrecision2 xy stage with linear encoders. The microscope is also equipped with a Ludl focus motor with linear encoder for z axis control. Both are controlled by a Ludl MAC 6000.
Objectives
This system is equipped with a Leica HC PL FLUOTAR 5X 0.15NA objective for preliminary scans and HCX PL FLUOTAR 20X 0.70NA objective for image acquisition.
Filters
The system described here is equipped with Chroma ET filter sets 49000, 49002, 49008, and 49009. Other filter sets can be used for imaging different fluorochromes. Ideally, filter cubes should be installed with the cube for the ubiquitous label, e.g., the nuclear stain, in position 1 followed by the cube for the fine focus channel in position 2 in order to minimize filter turret movement time.
Camera
Cameras with a wide-field-of-view, high resolution, and excellent sensitivity are preferred. CMOS cameras are ideal for this purpose. This system is equipped with an ORCA-Flash 4.0 LT+ CMOS camera (Hamamatsu) via a 1X coupler.
Antivibration table (optional)
Vibration isolation is not required, but is preferred, depending on the location in which the microscope is housed. The system used here is mounted on a TMC antivibration table with air pressure-based dry damping.
Software
Implementation requires MetaMorph 7.10 or greater (Molecular Devices) and the journals encoded within the file AutoscanJSuite.jzp.
Protocol Steps
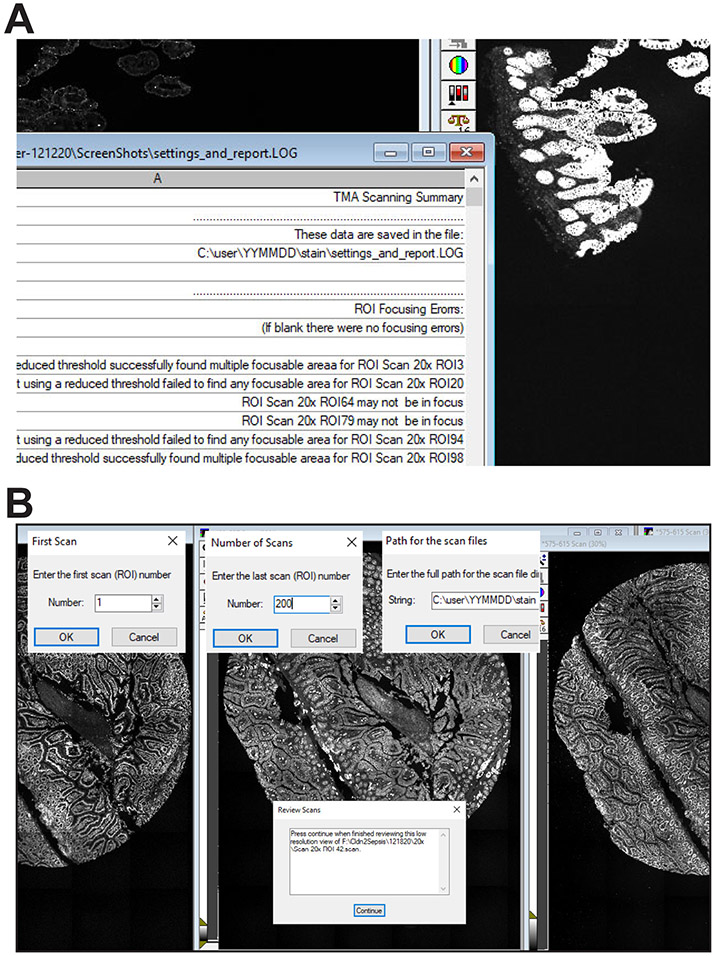
Open the MetaMorph Administrator to install and configure the hardware.
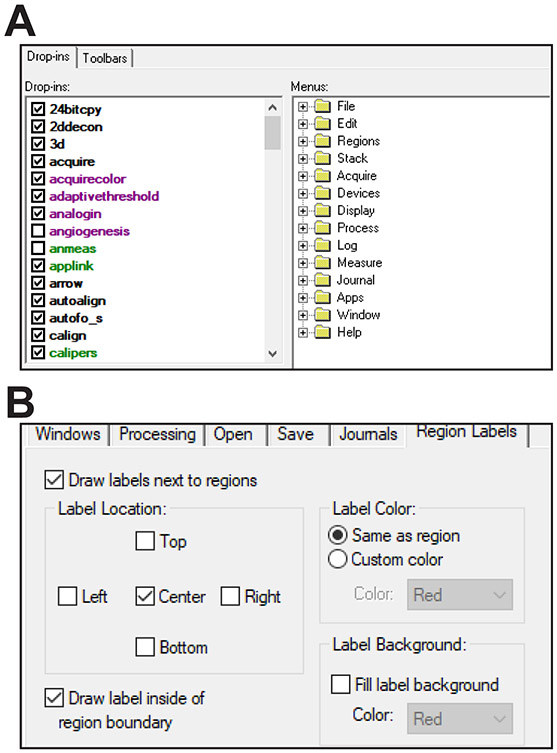
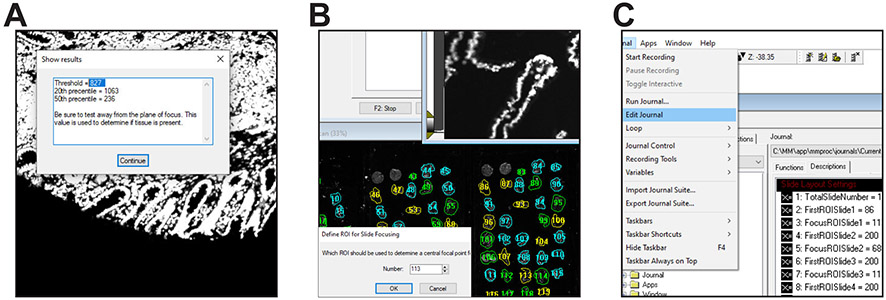
Enable the drop-ins aquire, adaptivethreshold, autofocus, measgrid, segimage, scanslide, and stitch (Fig. 1A). If any are not enabled, MetaMorph will generate an error message when the journal requiring the drop-in is run.
Select Journal>Import Journal Suite from the dropdown menu within MetaMorph and choose the file AutoscanJSuite.jzp. Then select the import location, e.g., MM\App\mmproc\Journals\. The journals will automatically be updated to call one another according to the folder you choose.
Select Edit > Preferences, click the Region Labels tab, and click the boxes Draw labels next to regions, Center, Draw labels inside of region boundary, Draw labels with respect to the image zoom, and Label Color: Same as region (Fig. 1B). Leave the Fill label background box unchecked.
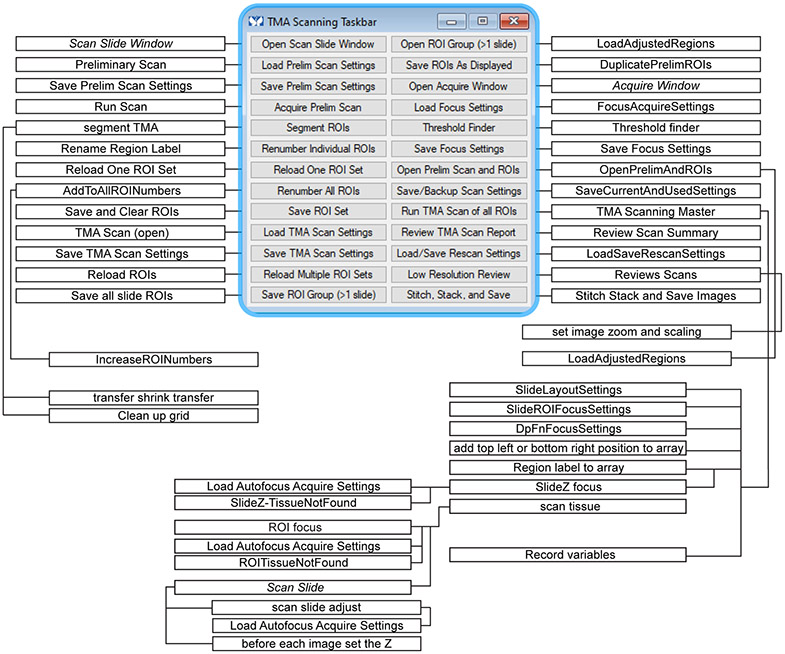
Within MetaMorph select Journal>Taskbars>Load Taskbar, navigate to the folder with these journals, and select TMA Taskbar.JTB. The taskbar (Fig. 2) and a flow chart showing the journals called by each button as well as secondary journals called by the primary journals is provided along with the text code of each journal (Supplemental Software Code S1-S43). Variables used are listed in Tables 1 and 2.
Autofocus acquisition parameter are stored in .AST files located in the subdirectory C:\MM\app\TMA\acquisitions\. There is one file for each channel to be used. These define channel, exposure time, binning (if needed), and other settings used for identifying the presence of tissue and autofocusing (e.g., autofocusing360_440.AST).
Define illumination settings. The journals provided expect illumination settings named with excitation and emission wavelengths, i.e., “360_440”, “488_527”, “575_615” and “637_684” Other names can be used as long as the journals calling these settings are appropriately modified.
Figure 1. Software configuration.
A. Use the MetaMorph Administrator to load required dropins. B. It can be helpful to define placement of ROI labels (these will be used later).
Figure 2. Taskbar and journals.
Flowchart showing relationship between taskbar buttons and journals. Items that are commands, rather than journals, are indicated in italics.
Table 1.
Journal-defined custom variables
| AcquireSettingFile | used in Load Autofocus Acquire Settings to define the file to be loaded |
| AcquireSettings | binary (Y/N) variable used in TMA Scanning Master to confirm that focus acquire settings have been set and saved correctly |
| ActiveImage | binary (Y/N) variable used in TMA Scanning Master to confirm that Stage list array.tif is open and active when using an existing stage list array file |
| AugmentRegionNumber | defined in AddToAllROINumbers and used in IncreaseROINumbers to indicate the constant value to be added when renumbering a set of ROIs |
| BottomRightX | used in Region label to array (or Region label to array (existing stage list array)) and used in SlideZ focus to define location of ROI designated for use in determining the central focal plane of each slide |
| BottomRightY | used in Region label to array (or Region label to array (existing stage list array)) and used in SlideZ focus to define location of ROI designated for use in determining the central focal plane of each slide |
| column | used in Segment TMA to define the grid |
| CurrentDpZ | used in scan slide adjust to track the most recent deep focus result within an ROI |
| CurrentSlide | used in SlideZ focus, scan tissue, and ROI focus and determined based on FirstROI variable |
| CurrentX, CurrentY | used in SlideZ focus and ROIZ focus to track stage position |
| Difference | used in Threshold finder to report the calculated threshold for the current image |
| Directory | defined by the Scan Slide variable ScanSlide.Directory and used in scan tissue |
| DpFocCh | defined in TMA Scanning Master and used in scan slide adjust to encode the channel, i.e. illumination settings, used for deep focusing |
| DpFocChk | used in scan slide adjust as counter to determine when a deep focus should be performed within an ROI |
| DpFocExp | defined in scan slide adjust to record value for entry into the summary log |
| DpFocInt | defined in TMA Scanning Master and used in scan tissue and scan slide adjust to indicate number of tiles imaged before deep focusing within an ROI |
| DpFocRange | defined in TMA Scanning Master and used in scan slide adjust to indicate the range, in microns, to be scanned when deep focusing within an ROI |
| DpStepSz | defined in TMA Scanning Master and used in scan slide adjust to indicate the size, in microns, of steps used when deep focusing within an ROI |
| DpFocVar | defined in TMA Scanning Master and used in scan slide adjust to indicate the size, in microns, of the acceptable difference in z between a newly identified focal point and the previous deep focal point when deep focusing |
| DpThreshold | defined in TMA Scanning Master and used in scan slide adjust to indicate the threshold used to determine if tissue is present when deep focusing |
| EndTime | defined in TMA Scanning Master and used in Record variables. |
| ErrorCode | Used in ROI focus and ROITissueNotFound. |
| FirstROINumber | used in Segment TMA to define the grid |
| FirstROIPos1 | binary (Y/N) variable used in TMA Scanning Master to ask if the first ROI on slide 1 is numbered 1 |
| FirstROISlide1, FirstROISlide2, FirstROISlide3, FirstROISlide4 | defined in TMA Scanning Master and used in SlideZ focus, scan tissue, and ROI focus to indicate the number of the first ROI on each slide. |
| FirstSubZ | numerical variable defined in ROI focus and used in scan slide adjust to tag the first tile within an ROI and to track the degree of discordance between previous and new focus positions |
| FnFocCh | defined in TMA Scanning Master and used in scan slide adjust to record the channel, i.e. illumination settings, used for fine focusing within an ROI |
| FnFocChk | introduced in scan tissue and used in scan slide adjust as a counter to determine when a fine focus should be performed within an ROI |
| FnFocExp | defined in scan slide adjust for entry into the summary log |
| FnFocInt | introduced in TMA Scanning Master and used in scan tissue and scan slide adjust to indicate the number of tiles imaged before fine focusing is performed within an ROI |
| FnFocRange | introduced in TMA Scanning Master and used in scan slide adjust to indicate the range, in microns, to be scanned when fine focusing within an ROI |
| FnFocStepSz | introduced in TMA Scanning Master and used in scan slide adjust to indicate the size, in microns, of steps used when fine focusing within an ROI. |
| FnFocVar | introduced in TMA Scanning Master and used in scan slide adjust; the size, in microns, of the acceptable difference in z between a newly identified focal point and the previous fine focal point when fine focusing within an ROI |
| FnThreshold | introduced in TMA Scanning Master and used in scan slide adjust to indicate the threshold used to determine if tissue is present in scan slide adjust |
| Focus | binary (Y/N) variable used in TMA Scanning Master to ask if z-origin is set and correct objective is in place |
| FocusChannel | defined by user in TMA Scanning Master and FocusAcquireSettings, used in Load Autofocus Acquire Settings, SlideZ Focus, ROI focus, and scan slide adjust to pass current focus channel information between journals |
| FocusROISlide1, FocusROISlide2, FocusROISlide3, FocusROISlide4 | introduced in TMA Scanning Master and used in Region label to array (or Region label to array (existing stage list array)) to indicate the number of the ROI to be used in determining the central focal plane of each slide |
| Height | used in SlideZ focus and ROI focus to direct stage y movements within the ROI |
| Homed | binary (Y/N) variable used in TMA Scanning Master to ask if stage is calibrated. |
| HowFarToMoveX | used in SlideZ focus and ROI focus to direct stage x movements within the ROI |
| HowFarToMoveY | used in SlideZ focus and ROI focus to direct stage y movements within the ROI |
| InitialZ | defined in TMA Scanning Master and used in SlideZ focus to record stage z position at start of TMA Scanning Master |
| Label | used in Region label to array and Region label to array (existing stage list array) to record the numerical value of the region label |
| LoadSave | used in LoadSaveRescanSettings to indicate whether settings are being loaded or saved |
| Mag | used in TMA Scanning Master, Review Scans, and Stich, Stack and Save Images to record the magnification setting used during TMA scanning |
| message | used in TMA Scanning Master, SlideZ focus, ROI focus, and Record variables to create text for entry into the summary log |
| moveXY | used in SlideZ focus and ROI focus to control how far in x and y directions the stage moves when focusing; the default value of 4 moves the stage 1/4 of the total width or height of the ROI |
| Name | used in scan tissue as string variable defined by Image.StageLabel for creating scan file name |
| NewArray | binary (Y/N) variable used in TMA Scanning Master to ask if a stage list array file needs to be created |
| NewDirectory | used in Stitch Stack and Save Images to define \stitched_stacks directory |
| NumberofScans | used in Review Scans and Stitch Stack and Save Images to calculate file names |
| Path | used in Review Scans and Stitch Stack and Save Images to calculate file names |
| Pattern | integer string used in SlideZ focus and ROI focus to move the xy stage within the ROI when determining focus points |
| Position | integer variable that increments for each plane in the stage list array file in add top left or bottom right position to array and Region label to array (or Region label to array (existing stage list array)) after being set to 1 in TMA Scanning Master |
| ReducedDpThreshold | defined in DpFnFocusSettings or in response to prompts and used as lower threshold in scan slide adjust |
| ReducedFnThreshold | defined in DpFnFocusSettings or in response to prompts and used as lower threshold in scan slide adjust |
| ReducedROIZCount | defined and used in ROITissueNotFound to track the number of times a focal point has been determined |
| ReducedROIZThreshold | defined in SlideROIFocusSettings or in response to prompts and used as lower threshold in ROITissueNotFound |
| RegionColumn | used in transfer shrink transfer TMA to number ROIs |
| RegionRow | used in transfer shrink transfer TMA to number ROIs |
| RepeatSlideROIZCount | defined and used in SlideZ-TissueNotFound to track the number of times a focal point has been determined |
| RepeatTotalZThreshold | defined in SlideROIFocusSettings or in response to prompts and used as lower threshold in SlideZ-TissueNotFound |
| ReviseThreshold | defined in ROITissueNotFound and used scan slide adjust to indicate that reduced deep and fine focus thresholds should be used when identifying tissue |
| ROI | used in Review Scans and Stitch Stack and Save Images to calculate file names |
| ROIDirectory | used in OpenPrelimAndROIs and LoadAdjustedRegions to track the directory where the preliminary scan and ROIs are stored. |
| ROIgroup | used in Save all slide ROIs to determine which ROI group is being saved |
| ROISetCount | used in Reload ROIs to count the sets being loaded |
| ROISetLoad | used in Reload One ROI Set to determine which ROI set should be loaded |
| ROISetNumber | used in Save and clear ROIs to determine which ROI set is being saved |
| ROISetsTotal | used in Reload ROIst o determine how many ROI sets will be loaded |
| ROIZ | used in ROI focus to record z origin of each ROI |
| ROIZ1, ROIZ2, ROIZ3 | temporary variables used in ROI focus to record focal points |
| ROIZcount | used in ROI focus to track the number of times a focal point has been determined |
| ROIZFocCh | defined in TMA Scanning Master and used in ROI focus to indicate the channel, i.e. illumination settings, used to determine the origin focal plane for each ROI |
| ROIZFocExp | defined in ROI focus to retains value for entry into the summary log |
| ROIZFocRange | defined in TMA Scanning Master and used in ROI focus to indicate the range, in microns, to be scanned in determining the origin focal plane for each ROI |
| ROIFocSettings | binary (Y/N) variable used in TMA Scanning Master to ask if the default (hard coded) settings will be used for ROI focus |
| ROIZFocStepSz | defined in TMA Scanning Master and used in ROI focus to indicate the size, in microns, of steps used when determining the origin focal plane for each ROI |
| ROIZFocVar | defined in TMA Scanning Master and used in ROI focus to indicate the size, in microns, of the acceptable difference in z between two identified focal points when determining the origin focal plane for each ROI |
| ROIZThreshold | defined in TMA Scanning Master and used in ROI focus to indicate the threshold used to determine if tissue is present in ROI focus. |
| row | used in Segment TMA to define the grid |
| ScanName | used in Review Scans and Stitch Stack and Save Images to store temporary file names |
| ScanslideCheck | used in TMA Scanning Master to query user about Scan Slide settings |
| ScanslideFocSettings | used in TMA Scanning Master to query user about Scan Slide settings |
| ScanSlideSettingFile | used in SaveCurrentAndUsedSettings to record the Scan Slide save directory |
| ScanSettings | binary (Y/N) variable used in TMA Scanning Master to confirm that settings have been entered correctly |
| SkipConfigurationQuestions | used in TMA Scanning Master to bypass configuration questions |
| slide | used in segment TMA to record the number of the slide for which ROIs are being defined |
| Slide1Z, Slide2Z, Slide3Z, Slide4Z | introduced in TMA Scanning Master and used in SlideZ focus and ROI focus to record of central focal plane for each slide. |
| SlideFocSettings | binary (Y/N) variable used in TMA Scanning Master to ask if the default (hard coded) slide and ROI focus settings will be used |
| SlideLayout | binary (Y/N) variable used in TMA Scanning Master to ask if the default (hard coded) slide layout will be used |
| SlideROIZ1 | introduced in TMA Scanning Master and used in SlideZ focus to record focal points determined within SlideZ focus |
| SlideROIZ2, SlideROIZ3 | used in SlideZ focus to record focal points determined within SlideZ focus |
| SlideROIZcount | used in SlideZ focus to track the number of times a focal point has been determined |
| SlideZ | introduced in TMA Scanning Master and used in SlideZ focus as temporary variable to record central focal plane determined for slide (based on designated ROI) |
| SlideZFocExp | defined in SlideZ Focus for entry into the summary log |
| SSfocus | binary (Y/N) variable used in TMA Scanning Master to ask if focusing journals are used within Scan Slide |
| Stage position | used in add top left or bottom right position to array |
| StartImage | defined in segment TMA and used in transfer shrink transfer TMA journals |
| StartingScan | used in Review Scans and Stitch Stack and Save Images to calculate file names |
| StartTime | defined in TMA Scanning Master and used in Record variables |
| SubZ | used in scan slide adjust and before each image set the Z to track the most recent fine focus result within an ROI |
| SuitePath | used in Load Autofocus Acquire Settings to detrermine where the journals and acquire settings are stored |
| tempdir | used in TMA Scanning Master and Review Scan Summary to hold the path and file name for the Edge List Log file |
| TissuePresent | used in SlideZ focus and ROI focus to indicate that, based on scaling and threshold variables, tissue is present in the field imaged |
| TissuePresentDp | used in scan slide adjust to determine if tissue is present, based on scaling and threshold variables, before deep focusing. |
| TissuePresentFn | used in scan slide adjust to determine if tissue is present, based on scaling and threshold variables, before fine focusing |
| TopLeftX | used in Region label to array (or Region label to array (existing stage list array)) and SlideZ focus to define location of ROI designated for use in determining the central focal plane of each slide |
| TopLeftY | used in Region label to array (or Region label to array (existing stage list array)) and SlideZ focus to define location of ROI designated for use in determining the central focal plane of each slide |
| TotalPlanes | introduced in TMA Scanning Master and used in Region label to array existing stage list array) to indicate number of planes in existing stage list array file |
| TotalSlideNumber | used in TMA Scanning Master to indicate number of slides on stage |
| TotalZFocCh | introduced in TMA Scanning Master and used in SlideZ focus to indicate the channel, i.e. illumination settings, used to determine the central focal plane of each slide |
| TotalZFocStepSz | introduced in TMA Scanning Master and used in SlideZ focus to indicate the size, in microns, of steps used when determining the central focal plane of each slide |
| TotalZFocVar | introduced in TMA Scanning Master and used in SlideZ focus to indicate the size, in microns, of the acceptable difference in z between two identified focal points when determining the central focal plane of each slide |
| TotalZRange | introduced in TMA Scanning Master and used in SlideZ focus to indicate the range, in microns, to be scanned in determining the central focal plane of each slide |
| TotalZThreshold | introduced in TMA Scanning Master and used in SlideZ focus to indicate the threshold used to determine if tissue is present in SlideZ focus |
| Width | used in SlideZ focus and ROI focus to direct stage x movements within the ROI |
| Xrange | used in SlideZ focus and ROI focus to direct stage x movements within the ROI |
| Yrange | used in SlideZ focus and ROI focus to direct stage y movements within the ROI |
Table 2.
Journal-defined MetaMorph variables.
| Device.Focus.CurPos | MetaMorph device variable. |
| Device.Magnification.Setting | MetaMorph device variable. |
| Device.Stage.XPosition | MetaMorph device variable. |
| Device.Stage.YPosition | MetaMorph device variable. |
| DoesFileExist.ErrorCode | MetaMorph file variable. |
| Image.ActivePlane | MetaMorph image variable used in add top left or bottom right position to array and Region label to array (or Region label to array (existing stage list array)) to record the current plane (representing an ROI) of Stage list array.tif |
| Image.Annotation | MetaMorph image variable used in Region label to array (or Region label to array (existing stage list array)) used to store the text string for the Scan Slide directory name in each plane of Stage list array.tif Also used as designed in SlideZ focus, ROI focus, and scan slide adjust to record exposure times |
| Image.CameraX | MetaMorph image variable used in add top left or bottom right position to array, Region label to array (or Region label to array (existing stage list array)), scan tissue, and ROI focus to store bottom right ROI stage positions in each plane of Stage list array.tif |
| Image.CameraY | MetaMorph image variable used in add top left or bottom right position to array, Region label to array (or Region label to array (existing stage list array)), scan tissue, and ROI focus to store ROI bottom right stage positions in each plane of Stage list array.tif. |
| Image.FilePath | MetaMorph file variable. |
| Image.IllumSetting | MetaMorph image variable used in Region label to array (or Region label to array (existing stage list array)) and scan tissue to store the text string for the scan file name in each plane of Stage list array.tif |
| Image.NumPlanes | MetaMorph image variable. |
| Image.ScaleValueHigh | MetaMorph image variable. |
| Image.ScaleValueLow | MetaMorph image variable. |
| Image.StageLabel | MetaMorph image variable used in Region label to array (or Region label to array (existing stage list array)), scan tissue, and ROI focus to store the text string of the region label (the variable Label) in each plane of Stage list array.tif |
| Image.StageX | MetaMorph image variable used in add top left or bottom right position to array, Region label to array (or Region label to array (existing stage list array)), scan tissue, and ROI focus to store top left ROI stage positions in each plane of Stage list array.tif |
| Image.StageY | MetaMorph image variable used in add top left or bottom right position to array, Region label to array (or Region label to array (existing stage list array)), scan tissue, and ROI focus; to store top left ROI stage positions in each plane of Stage list array.tif |
| Journal.RunJournalLastDirectory | MetaMorph file variable |
| Region.Label | MetaMorph image/region variable |
| ScanSlide.BaseName | MetaMorph file variable |
| ScanSlide.Directory | MetaMorph file variable |
| ScanSlide.ScanUpperLeft.X; ScanSlide.ScanUpperLeft.y | MetaMorph programvariables |
| ScanSlide.ScanLowerRight.X ScanSlide.ScanLowerRight.Y | MetaMorph program variables |
| ScanSlide.Status.WaveNum | MetaMorph program variable |
| Stopwatches.Stopwatch1 | MetaMorph program variable |
| SuitePath | MetaMorph file variable |
Underlines indicate standard MetaMorph variables are used for non-standard purposes.
BASIC PROTOCOL 2 CREATE A PRELIMINARY SCAN
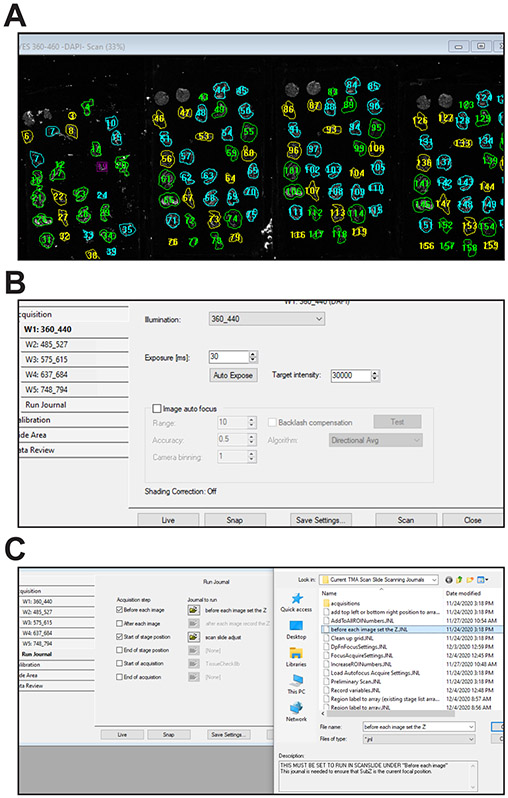
This protocol uses the scan slide module to create a preliminary low magnification scan of all slides to be scanned that will be used as an overview to map regions of interest (ROIs). This protocol and those that follow assume that four TMA slides are being imaged. Although the built-in Scan Slide module is used without modification in this protocol, taskbar buttons simplify loading and saving scanning parameters, including calibrations. These activate three simple journals, Preliminary Scan.JNL, Save Prelim Scan Settings.JNL, and Run Scan.JNL, that, respectively, load the preliminary scan settings file, save the revised settings file, and initiate the scan. For convenience, a taskbar button that opens the Scan Slide window is included.
Materials
Microscope, peripherals, and software configured as in Basic Protocol 1
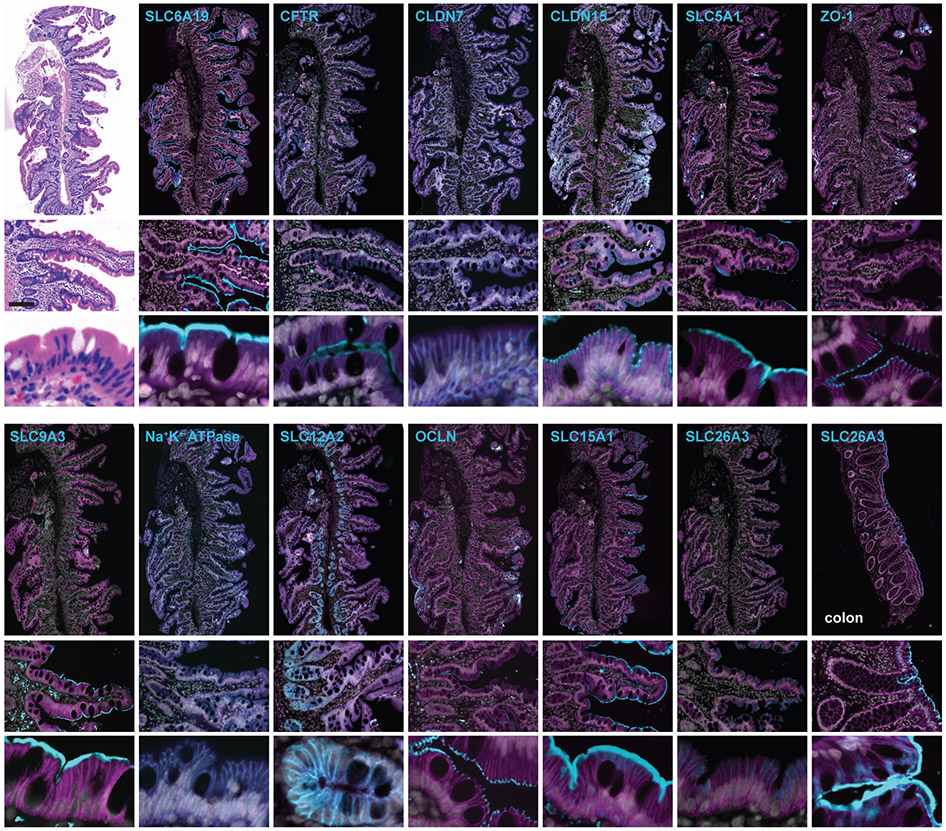
Stained slides (a list of primary antibodies used in these protocols can be found in Table 3)
Table 3.
Primary antibodies used.
| Antigen | Host | Clonality | Vendor | Catalog number | Lot number | Concentration | RRID |
|---|---|---|---|---|---|---|---|
| SLC6A19 | Rabbit | Polyclonal | Sigma | HPA043207 | R41029 | 3 μg/ml | AB_2678363 |
| CFTR | Rabbit | Polyclonal | Abcam | Ab59394 | GR139238 | 10 μg/ml | AB_940993 |
| CLDN7 | Rabbit | Monoclonal | Abcam | Ab207300 | GR251482-1 | 1.3 μg/ml | AB_2783812 |
| CLDN15 | Rabbit | Polyclonal | Thermo Fisher | 38-9200 | QJ228299 | 1.7 μg/ml | AB_2533391 |
| SLC5A1 | Rabbit | Polyclonal | Abcam | Ab14686 | GR140673-6 | 6 μg/ml | AB_301411 |
| ZO-1 | Rabbit | Polyclonal | Thermo Fisher | 61-7300 | 1452279A | 0.5 μg/ml | AB_138452 |
| SLC9A3 | Rabbit | Polyclonal | Abcam | Ab101817 | GR53426-4 | 1:100 | AB_10715387 |
| Na+-K+ ATPase | Rabbit | Polyclonal | Abcam | Ab7671 | GR161548-6 | 10 μg/ml | AB_306023 |
| SLC12A2 | Rabbit | Polyclonal | Abcam | Ab59791 | GR1790353-1 | 2 μg/ml | AB_944433) |
| OCLN | Rat | Monoclonal | J.R. Turner | Clone 5E5A6 | 5 μg/ml | AB_2819196 | |
| SLC15A1 | Rabbit | Polyclonal | D. Merlin | Merlin et al. Gastroenterol, 2001 | 1:100 | AB_2827376 | |
| SLC26A3 | Rabbit | Polyclonal | P.K. Dudeja | Gill et al. J Clin Invest, 2007 | 1:100 | AB_2827377 | |
| E-cadherin | Mouse | Monoclonal | Abcam | ab76055 | GR21603-5 | 1:300 | AB_1310159 |
Protocol Steps
-
Load slides onto the microscope stage. If the slides are going to be scanned twice or more, e.g., for background subtraction or multiplexing, be sure to push the slides tightly towards one corner of the holder, clamp them in place securely, and make a record of their positions.
If slides are stored at reduced temperature, it is important to allow them to completely equilibrate to room temperature before imaging. Slides should be cleaned with an ammonia-free solution, e.g., Sparkle, to minimize distortion and focusing errors caused by fingerprints and fine debris.
Anchoring is critical because any change in position will result in inaccurate ROI placement for additional scans.
-
Home the stage. Calibrate the xy control of the automated stage controller using the homing function under Devices > Stage > Move Stage to Absolute Position > Home Stage and then Set Origin.
If the stage does not have a homing function (or the homing function is difficult to use), a convenient and reproducible approach is to move the stage to the measurable limits of the encoders, e.g., by moving the stage beyond those limits and manually returning the stage to the first position that can be read accurately. This should be set to a fixed value, such as (0,0). The values are not critical, but the position must be reproducible.
A dedicated TMA Scanning Taskbar (Fig. 2) has been created to guide the user through the steps of the TMA scanning process. These are arranged in sequence, from top to bottom. If the taskbar has not already been loaded, do so using the Journal > Taskbars > Load Taskbar command. Load the TMA Taskbar.JTB file.
Open Scan Slide using the “Open Scan Slide Window” button at the top of the TMA Scanning Taskbar.
-
Load the settings file for collecting the 5X preliminary scan using the “Load Prelim Scan Settings” button on the TMA Scanning Taskbar. The settings file is located within the journal folder and named 5X TMA Prelim Scan.scansetting.
We use a 5X NA 0.15 objective for this preliminary scan. A 2.5X objective was tested but the resolution was insufficient for our ~2.5 mm diameter samples.
-
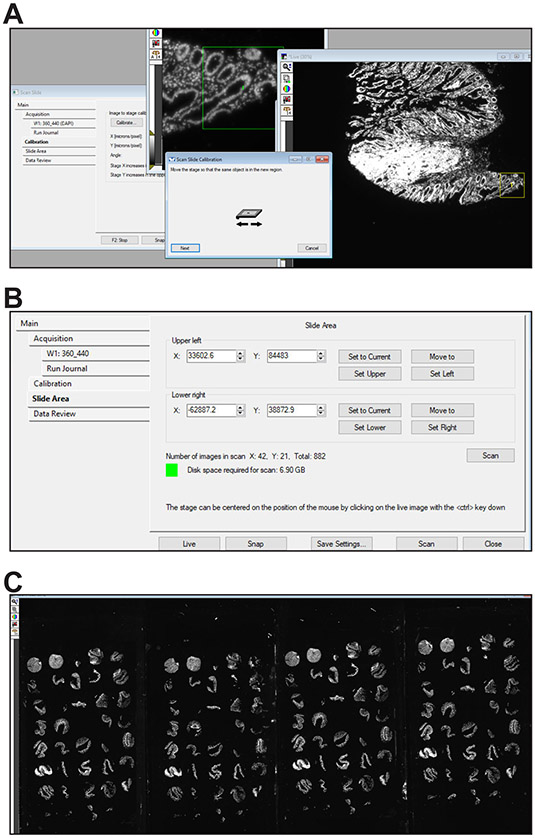
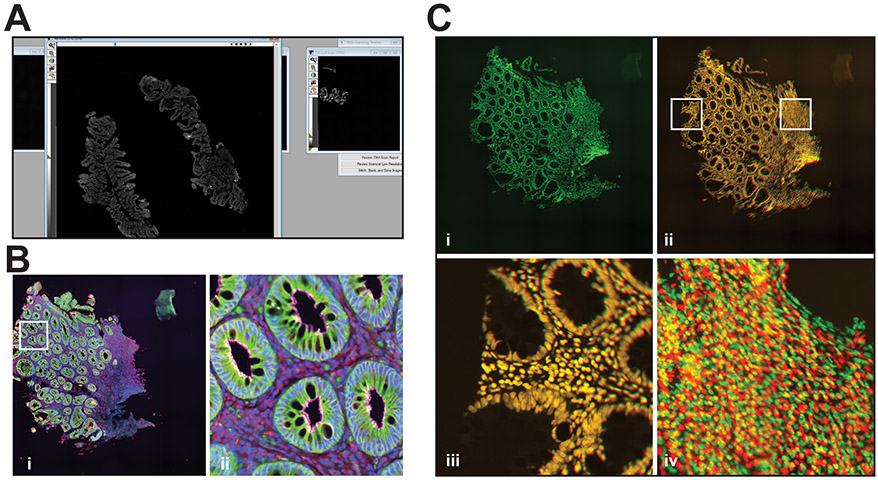
If the objective and stage have not been recently calibrated, click the Calibration tab within the Scan Slide window, select the objective to be used to make the preliminary scan, e.g., 5X, and follow the calibration procedure (Fig. 3A).
Calibration also calculates the pixel size. This calibration must be performed separately for each objective or magnification used.
-
Set the top left and bottom right corner positions for the rectangular region that will be scanned in the Slide Area tab within the Scan Slide window. This region must include all tissues to be scanned (Fig. 3B).
For this scan, it is useful to extend the corners slightly beyond the anticipated area needed to avoid having to repeat this scan.
Set the imaging parameters, e.g., channel and exposure time, in Scan Slide. Exposure should be set to the shortest interval that allows reliable tissue detection. DAPI or Hoechst dye-stained nuclei are ideal for this process. Only one channel is needed for the preliminary scan.
Set the save folder. The save folder will contain all the preliminary scan images and ROI files. For example, C:\Data\user\YYMMDD\stain.
Press the “Save Prelim Scan Settings” button in the taskbar. This saves the current settings over the previous 5X TMA Prelim Scan.scansetting file. This must be done because several other functions refer to the save location save in the settings file (Fig. 2).
Set the z-origin. Find tissue near the center of the scanned area and focus the image. Then, from the menus select Devices > Focus and choose the option to Set Origin. The z position should now be 0.
-
Start the scan the “Acquire Prelim Scan” button on the taskbar.
Watching the beginning of the scan in order to assess whether exposure, tile alignment, and region selection are correct can be helpful. At this point the scan can easily be aborted (using the Esc key) and re-initiated after correcting the errant settings.
At completion, a low-resolution stitched image of the scanned area that includes all tissues to be scanned should be displayed (Fig. 3C, see Table 4 for troubleshooting).
Figure 3. Preliminary scan.
A. Stage calibration. B. Slide scan window showing configuration before initiating preliminary scan. C. Representative low magnification preliminary scan of 4 slides stained with Hoechst 33342.
Table 4.
Troubleshooting.
| Problem | Possible Reason | Solution |
|---|---|---|
| The tiles are not aligned or are the incorrect size. | Objective magnification or calibration were set incorrectly in Scan Slide. | Repeat the Scan Slide calibration for the appropriate objective. If the 5X objective is recalibrated, repeat the preliminary scan. If the 20X objective is recalibrated it may be possible to use the existing ROIs after re-alignment. |
| The preliminary scan does not include all tissues. | The corners of the scanning area were set incorrectly. | Rescan after resetting the area scanned within the 5X preliminary scan Scan Slide settings file. |
| The journal cannot find a referenced journal and returns errors. | (i) The journal files are not in the correct folders. (ii) The journal files have been renamed. |
(i) Take care to correctly modify the journals during set up so that each calls the correct folder for the referenced journal. (ii) Do not rename the files unless the journals are modified to reflect the new names. |
| All images from one slide are out of focus. | (i) Tissues on the slide are outside of the defined focus range. (ii) The central focal place was determined incorrectly. (iii) The slide holder is not parallel to the camera. (iv) The slide is not seated in the holder. |
(i) Select a larger z-range for determining the initial central focal plane of each slide. (ii) Choose a different Focus ROI for that slide. (iii) Correct z alignment of the stage as described in the Equipment Setup section. (iv) Confirm that slides are correctly positioned. |
| All images from one area of a slide out of focus. | (i) Tissues are outside of the focus range. (ii) The slide is not firmly seated in the holder. |
(i) Re-check the focus range across the slide and select a larger z-range for focusing within each slide. If the range is large, realign the slide and/or stage. (ii) Confirm that slides are correctly positioned. |
| The wrong areas were scanned. | (i) The ROI boxes were not correctly aligned. (ii) Objective magnification or calibration were set incorrectly in Scan Slide. (iii) ROIs were not positioned for the objective used for final scans. |
(i) Reset the ROI positions. If the overview scan and ROIs are acceptable and the slide(s) has not been moved, restart the scan by pressing the Run TMA Scan taskbar button. (ii) Repeat the Scan Slide calibration and re-start either with the preliminary scan or the TMA scan. (iii) ROI realignment for the higher magnification objective was not performed properly. |
| Entire slides or unclustered ROIs are out of focus at all tile positions and are reported in the summary log. | (i) If the log indicates that no focus was achieved on the ROI, the threshold may be too high. (ii) The focus threshold may have been set too low and non-tissue signals interfered with the autofocusing. |
(i) Use the “Threshold Finder” button to find a better for these regions or empirically reduce the threshold value. (ii) Use the “Threshold Finder” button to find a better for these regions or empirically increase the threshold value. |
| Only the center tiles within an ROI are in focus. | Focus adjustments were not made during these ROI scans because the threshold for fine focusing was set too high. | Use the “Threshold Finder” button to find a better threshold or reduce the threshold empirically. |
| Some or all ROIs show focus drift unrelated to the region’s center. | (i) Refocusing is occurring on areas without tissue. (ii) Refocusing erratic because it is occurring over too large of a z-range. (iii) The tissue loses focus between refocusing steps because the tissue is not flat, e.g., sections are wrinkled. |
(i) Increase the value of the threshold for identifying tissue and, possibly, decrease the z-range and interval of gross focus rechecks. (ii) Decrease the z-range of fine focusing and gross focus rechecks and increase the frequency of fine focusing steps. (iii) Use more frequent focus intervals. Rechecking the fine focus on every tile may be necessary. |
| Despite frequent focusing intervals, fine focusing does not find the focal point. | Focal points are identified, but they are outside the acceptable variation between fine focus points. | Increase the acceptable variation between fine focus points. |
| ROIs are not identified correctly after pressing the Segment ROIs taskbar button | (i) The mask was made incorrectly. (ii) The grid was not precisely positioned. |
(i) Incorrect values can cause the Adaptive Threshold image to be white over non-tissue areas or black over tissue. Open the journal editor and change the values within the LowPass or Adaptive Threshold functions within segment TMA. (ii) Close all images (except for the low-resolution preliminary scan) and clear all ROIs. Then press the “Segment ROIs” taskbar button and place the grid more precisely. Be sure not to include bright areas that are not tissue, e.g., the coverslip edge. |
| Saved images names do not match ROI names. | (i)The wrong Stage List Array.tif file was used. (ii) The wrong ROIs were loaded |
(i) Open the preliminary scan (from the tab in Scan Slide) and manually reload the adjusted ROIs from the preliminary scan directory. In sequence, press “Open Scan Slide Window,” “Load TMA Scan Settings,” “Save TMA Scan Settings,” “Final Checks,” and “Run TMA Scan” buttons. Be sure to replay Yes to the query “Do you need to create a new stage list array?” prompt. (ii) Reload the correct set of ROIs by selecting the Correct set from Regions > Load Regions |
BASIC PROTOCOL 3 SELECT, SAVE, AND POSITION ROIS
ROIs can be selected manually. However, this can be a time-consuming process. For example, defining and labeling 200 ROIs, e.g, four slides with 50 positions in each tissue microarray, can take 30 - 60 minutes. The journals within this protocol create ROIs using a computer -assisted, semi-automated process.
In basic protocol 3, the journal segment TMA.JNL is triggered by a taskbar button. This applies a low pass filter and adaptive thresholding using user-defined variables, creates a binary image, and the calls the Measure Grid module, which generates a grid whose dimensions are defined by responses to prompts. After manually repositioning the grid, the user runs transfer shrink transfer TMA.JNL from the Measure Grid module. This outlines the tissue at each grid position, numbers the ROIs, and transfers the ROIs to the initial image. Finally, transfer shrink transfer TMA.JNL runs Clean up grid.JNL to close the intermediate images created during the process. ROIs can then be verified manually.
If software-generated ROIs need to be revised, new regions will need to be drawn manually. Rename Region Label.JNL allows manual renumbering of individual ROIs. If an ROI set is being reused, as in scanning multiple sections from the same TMA block, AddToAllROINumbers.JNL, which calls IncreaseROINumbers.JNL, can be used to renumber the entire ROI set. Save and clear ROIs.JNL, triggered by a taskbar button, saves the ROI set and assigns it to a specific slide number. Reload One ROI Set.JNL and Reload ROIs.JNL, both activated via the taskbar, reload one or all ROIs, respectively. Save all slide ROIs.JNL saves multiple sets of ROIs as a single group. LoadAdjustedRegions.JNL reloads a ROI group. Load TMA Scan Settings.JNL and TMA Scan Settings.JNL load and save Scan Slide settings for final acquisition, respectively.
Materials
Preliminary low magnification scan generated in Basic Protocol 2
Protocol Steps
-
Segment the image. Press the “Segment ROIs” button on the TMA Scanning Taskbar. This will apply a low pass filter and segmentation to the low-resolution preliminary scan image. This procedure takes advantage of the adaptive threshold function.
Optimization of the minimum and maximum image widths and intensity above local background setting is important for this to work well. These can be edited within the segment TMA.JNL file using the MetaMorph journal editor. The minimum and maximum object sizes can be determined using the region function. Gray levels over background will vary with exposure and can be determined based on the intensity of bright and dim tissues and bright and dim areas without tissue. The ROIs will be numbered consecutively from left to right starting with the top row and continuing to the bottom.
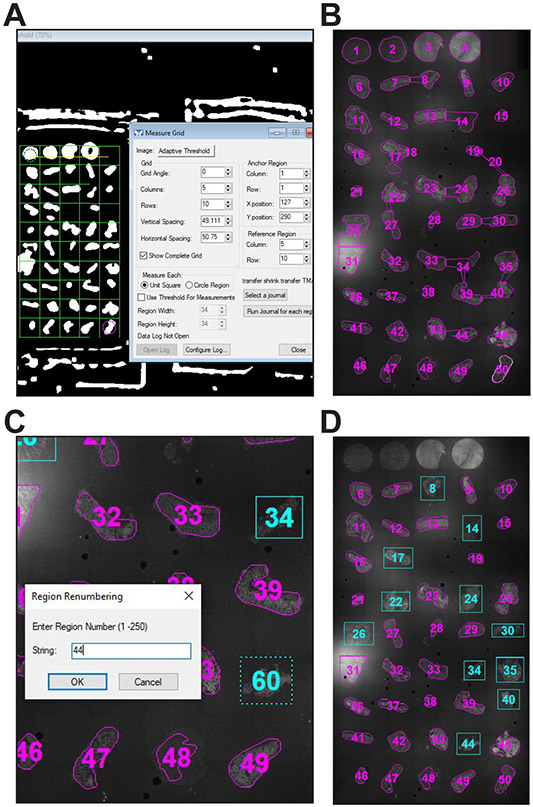
The process initiated by the “Segment ROIs” button will open a measurement grid within the segmented image (Fig. 4A). The numbers of columns and rows in the grid and numbering sequence for the ROIs (below) are defined by user responses to prompts just prior to grid placement. Adjust the position of the grid for automated ROI determination. The cyan circle (anchor) will be in the top left cell. Drag the cyan circle over the tissue in the top left corner of the TMA. Now drag the magenta circle over the tissue in the bottom right corner. If necessary, the angle of the grid can be adjusted using the angle option in the Measure Grid window or by dragging the yellow angle line on the image. Each tissue fragment should be in a separate cell within the grid.
Once the grid has been positioned, press the “Run Journal for Each Region” in the Measure Grid window. The transfer shrink transfer TMA journal should already be selected. Each tissue fragment will be traced as a ROI (Fig. 4B).
- Revise ROIs as needed. Delete any ROIs that are either too large or too small or do not enclose the area over the corresponding tissue fragment. ROIs over regions without tissue, e.g., an empty position in the TMA, or tissues that do not need to be imaged, e.g., orientation controls, can also be deleted.
-
Use the rectangular region tool to draw new ROIs until there is an ROI over each tissue to be imaged.The ROIs should be drawn tightly around each tissue, as Scan Slide includes some extra space around the perimeter of each ROI.
-
The new ROIs will be numbered consecutively from the last ROI created by the journal (Fig. 4C). They should be renamed using the “Renumber individual ROIs” button on the taskbar.Maintaining the naming sequence is essential for accurate assignment of images to specific tissues within each TMA. Ideally, the new names, i.e., numbers, should correspond with the tissue position within the TMA.It is most convenient to number empty positions within the TMA. There do not need to be regions corresponding to these numbers, e.g., positions 1 through 5 (Fig. 4D), but this approach will simplify assignment of images to specific tissues in downstream applications.
-
-
Save the ROIs using the “Save ROI Set” button on the taskbar. The user is prompted for the set number, which usually corresponds to the number of the slide position on the stage. The ROIs are then cleared so that the next set can be defined. The journals allow up to four separate ROI sets.
If scanning multiple tissues on different slides, the range of ROI numbers used for each slide must be recorded so that the images can be assigned correctly.
-
If scanning multiple sections from the same TMA block, e.g., with different stains, it is possible to reuse the same ROIs.
If taking this approach, it is critical that stain intensities in all channels allow use of the same exposures for each channel across the slides.- Use the “Reload one ROI set” button on the taskbar to load the previous ROI set. The prompts will ask which ROI set, i.e., slide number, to reload.
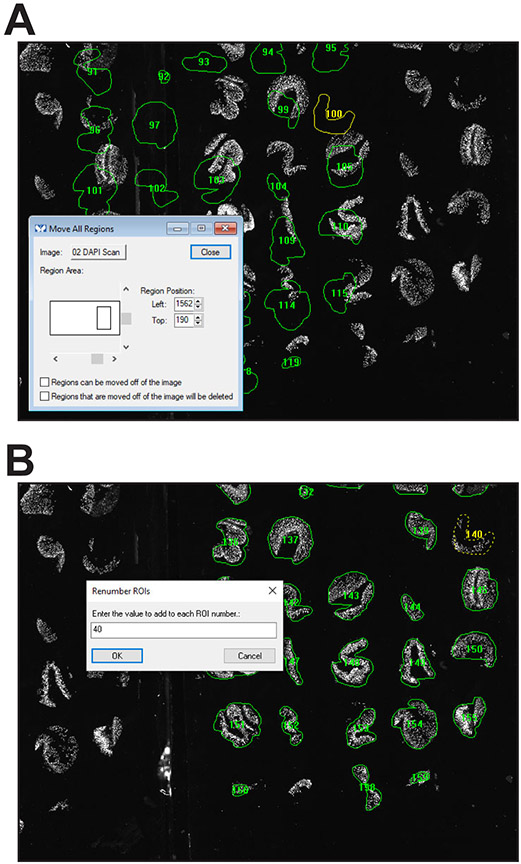
- Select Region > Move All Regions from the dropdown menu and use this tool to reposition the ROIs over tissues on the next slide (Fig. 5A).
- Press the “Renumber all ROIs” button on the taskbar. The prompts will ask what value should be added to the number of each ROI. For example, if each slide has 50 positions, i.e., tissue cores; the cores on the first slide were numbered 1 through 50; and the cores on the second slide needs be numbered 51 through 100, indicate that ROI numbers should be increased by 50 and begin with number 51 (Fig. 5B).
- After renumbering, Press the “Save ROI Set” button on the taskbar. The prompts will ask which slide corresponds to the ROIs being saved.
More commonly, different TMAs that have been labeled with the same stains will be imaged. In this case, repeat the process for each slide until all tissues to be imaged have been defined by ROIs. If, for example, the TMAs being imaged have 50 cores in each, the best approach is to assign the ROIs 1-50, 51-100, 101-150, and 151-200 for each of four slides. A prompt asks for the number to be assigned to the upper left grid position just prior to grid placement by the journal.
Press the “Open Scan Slide Window” button on the TMA Scanning Taskbar.
Press the “Load Prelim Scan Settings” button on the taskbar.
Press the “Reload Multiple ROI sets” button on the taskbar.
If desired, the entire group shown can now be saved by pressing the Press the “Save ROI Group (>1 slide)” button on the taskbar. Click the “original” radio button in response to the the prompt.
Press the “Load TMA Scan Settings” button on the taskbar.
Select the objective to be used for final imaging, e.g. 20X.
If the objective has not been calibrated, click the Calibration tab within the Scan Slide window, select the objective to be used to for final imaging, e.g., 20X, and follow the calibration procedure. Press the “Save TMA Scan Settings” button on the taskbar.
- Regions placed on the overview 5X scan can be slightly displaced at 20X due to differences between objectives and stitching of the low-resolution preliminary scan view. Fortunately, this misalignment, if present, is typically small, uniform across all slides, and easily corrected.
- Select a ROI surrounding a core with an easily recognizable, asymmetric tissue profile.
- On the Scan Slide Data Review tab, click the button to move the stage to the center of the ROI.
- Using the 20X objective, view the imaged region on screen. It can also help to look through the oculars, which typically provide a larger field of view.
- Assess ROI location relative to desired ROI to be imaged. To correct misalignment, select Region > Move All Regions from the dropdown menu and use this tool to adjust ROI placement.
-
Send the stage to the new center of the ROI using the button in the Scan Slide Data Review tab. Then send the stage to the upper left and lower right corners of the ROI to ensure that the tissue is completely enclosed within the region.It can be useful to watch through the oculars as the stage moves between the upper left and lower right corners. If taking this approach, be sure to reset the camera selector when finished.
- Correct misalignment again using the Move All Regions function until the center of the ROI corresponds to the tissue center.
- Repeat the process using several other ROIs across the slides until no corrections are needed.
Press the “Save ROI Group (>1 slide)” button on the taskbar. Click the “adjusted” radio button in response to the the prompt.If needed, the original or adjusted ROI groups can be reloaded by pressing the “Open ROI Group (>1 slide)” button on the taskbar.
-
Save an image of the regions superimposed on the preliminary scan by pressing the “Save ROIs as Displayed” button on the task bar. The file will be saved in the directory selected in the preliminary scan settings file.
This image can be useful if opened in a separate window, i.e., in a different program, for reference.
Figure 4. Placing ROIs.
A. Pressing the “Segment ROIs” button on the taskbar will display an adaptively thresholded image along with a measurement grid. B. ROIs are drawn automatically after the grid is positioned and the “Run Journal for each region” button is pressed. C. Some ROIs, e.g., 8, 24, and 34, include excess area and others, e.g., 20, have outlined artifacts. After placing new ROIs (shown in cyan), the new ROIs are renumbered to maintatin the sequence. D. Representative final ROI set (for slide 1).
Figure 5. Reusing an ROI set.
A. If two slides with sections derived from the same TMA block are imaged simultaneously, ROIs from the first slide can be reloaded and repositioned over the second slide. B. After placement, the entire second ROI set can be easily renumbered.
BASIC PROTOCOL 4 DETERMINE AND SET AUTOFOCUS PARAMETERS
Acquisition parameters include the settings used for detecting tissue and autofocusing. Once determined, these values can typically be used across multiple slides and slide sets. Therefore, this process does not generally need to be repeated for each scan. Some trial and error, depending on the microscope, sample, and stain, can improve scan quality and efficiency.
There are four separate autofocusing steps during acquisition. Although the channels used for each of these focusing steps can be selected individually, the ubiquitous stain, e.g., the nuclear stain, is typically used for the first three. Because the same nuclear stain is used for all tissues, optimized settings can often be used routinely across many specimens. Fine focusing on nuclei is generally insufficient due to the presence of contrast throughout the their thickness. Instead, the marker used for fine focusing should be selected on the basis of the other stains being imaged. For example, E-cadherin or γ-actin stains work well for tissues stained for epithelial membrane proteins. In this channel, the threshold setting is used to determine whether there is sufficient signal for focusing. The settings used for fine focusing can often be applied to other stains of similar intensity in the same wavelength channel.
The focusing step first determines the central focal plane of each slide using a designated ROI for each slide and a large z range. The second step uses the first as a starting point and checks focus over a narrower range to determine the z origin to be used within each ROI. The first and second steps operate similarly. In each case, the system collects an image at the center of the ROI and determines if tissue is present using a user-defined threshold variable. Software focusing is only activated if tissue is present; this test for the presence of tissue saves time and improves accuracy by eliminating focusing on areas with insufficient tissue, where articfacts could result in focusing errors. The stage then moves to another site within the region and repeats the process. Successful focusing is defined as two focal planes within a user-defined distance of one another. If, after checking 10 positions, the focusing has not been successful, the process is repeated using a lower threshold for tissue detection.
The third, deep, focusing step is triggered at the first tile within each ROI and at user-defined intervals thereafter. The journal first checks if tissue is present using either a higher or lower threshold, depending on whether a lower threshold was required during the second focusing step. If tissue is present, focusing is performed over a smaller range than those used for the first two steps. The journal then checks for tissue in the channel specified for fine focusing, again using one of two thresholds. If tissue is present, fine focusing is performed over an even narrower range. To be considered successful, tissue must be identified in both channels and, in both cases, focus must be within user-defined distances of the previous focal planes. If the process is successful, the deep and fine interval counters are reset. If not, the process is repeated at the next tile using an extended deep focusing range.
Fine focusing alone is set to be triggered more frequently than deep focusing and fine focusing together. If tissue is identified and fine focus is within the user-defined distances of the previous fine focus, the fine focus counter is reset. If the new focal plane exceeds acceptable variance from the previous fine focus deep and fine focusing are triggered at the next tile.
The values assigned to variables defined in this protocol will determine the accuracy of focusing during final TMA scanning. Three journals are used; FocusAcquireSettings.JNL, Threshold finder.JNL, and Save Acquire Settings.JNL. FocusAcquireSettings.JNL prompts the user for the channel to be defined and loads the corresponding settings. Threshold finder.JNL scales the image to the 20th and 50th percentile intensities and determines the difference between these so that that value, the threshold, can be entered later. Two thresholds can be determined for each channel; the second, lower, value is used to assess difference between 10th and 50th percentile intensities when tissue is not identified using the first threshold.
Values determined here can be entered manually when running the scan, or, for convenience, coded into the journals SlideLayoutSettings.JNL, SlideROIFocusSettings.JNL, and DpFnFocusSettings.JNL.
Materials
Stained slides
Preliminary low magnification scan generated in Basic Protocol 2
ROIs generated in Basic Protocol 3
Protocol Steps
Select the objective that will be used for final imaging, e.g., 20X.
- Define autofocusing acquisition parameters.
- Press the “Open Acquire Window” button on the taskbar.
- Press the “Load Focus Settings” button on the taskbar and select a channel that will be used for focusing. This will load the file corresponding to the channel selected, e.g., autofocusing360_440.AST for the nuclear stain from the \acquisitions subfolder within the folder that contains the TMA journals.
- If the Scan Slide window is not already open, open it using the “Open Scan Slide Window button.”
- Click on an ROI with an average staining intensity and move the stage to the center of the ROI using the Move to Region Center button in the Scan Slide Data Review tab.
- Press the “Show Live” button within the Acquire window.
- Focus on the tissue.
- Press the “Acquire” button within the Acquire window (not the Scan Slide window).
- Adjust the exposure time. Ideally, the exposure time should be short, i.e. less than 20 milliseconds, and allow the brightest pixels within tissue to be clearly distinguished from areas of the slide without tissue. In typical use, this means that the brightest pixels within nuclei are at least 10 - 20 % of the maximum intensity value for the camera, e.g., 1500 – 3000 for a 14 bit camera.
- Save the revised acquire settings file using the “Save Focus Settings” button on the taskbar.
- Identify thresholds to determine whether tissue is present and there is sufficient signal for focusing.
- Press the “Threshold Finder” button on the taskbar. This will return the intensity of the pixel at the 20th percentile (from bright to dim), the intensity of the pixel at the 50th percentile, and the difference between these values. The difference is the most important value to record (Fig. 6A).
- Press the “Show Live” button, defocus the tissue image by ~150 μm, and press the “Acquire” button. Then press the “Threshold Finder” button again. Record the difference between the 20th and the 50th percentile.
- Repeat the process using tissues with lower staining intensities to determine the range of differences between the 20th and the 50th percentile when tissue is present.
- Repeat the process using areas of the slides with the greatest background fluorescence, e.g., having brightly-fluorescent debris or other defect that fluoresces. Determine the range of differences between the 20th and the 50th percentile when tissue is not present.
- Identify a threshold value above which tissue is reliably present and below which tissue is not present. Accuracy in this step is important, as this is how the program determines whether sufficient tissue for focusing is present.
- Determine a second threshold value, typically 3 to 10 times smaller than the first, to be used if the first threshold is too high to allow detection of some tissues.
- Repeat the process for each channel used for focusing by the four focusing steps within the imaging process.
-
The threshold values determined for the channels used to determine the central focal plane of each slide, to determine the position used as the z origin within each ROI, for deep focusing within an ROI, and for fine focusing within an ROI should be assigned to the variables TotalZThreshold, ReducedTotalZThreshold, ROIZThreshold, ReducedROIZThreshold, DpThreshold, ReducedDpThreshold, FnThreshold, and ReducedFnThresholdThe same channel, e.g., a nuclear stain, is typically used for determining each slide’s central focal plane, ROI z origins, and deep focus within ROIs. In this case, the same threshold and reduced threshold values may be used for variables related to each step.
- Determine the z origin, z range, and ROI used to determine the central focal plane of each slide and the z range used for determining the z origin (central focal plane) within individual ROIs.
- For each slide, focus manually on the top left and bottom right ROIs and record the z position for each focal point.
- Calculate the mean and range of the two z positions.
- Identify an ROI with a focal plane close to the mean (Fig. 6B). This tissue should be stained brightly with the marker to be used for determining the central focal plane of the slide.
- Repeat for each slide.
- Determine the maximum z range across all slides, the midpoint of that range, and the maximum z range within any one slide. A z range across all slides of ~300 μm or less is acceptable. Similarly, the maximum z range within any one slide should be less than ~100 μm. If either of these exceeds those values, the camera and stage should be realigned.
- Adjust the focus to the midpoint across all slides and use the stage control window to set that as z = 0.
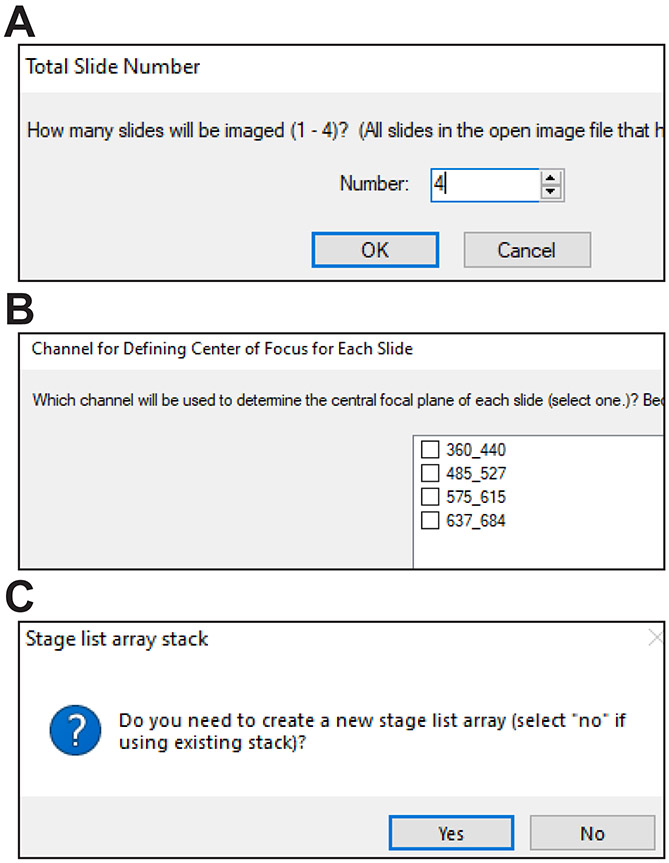
- Record the number of slides, first ROI number of each slide, and ROI used for focusing within each slide. These values should be assigned to the variables TotalSlideNumber,FirstROISlide1, FirstROISlide2, FirstROISlide3, FirstROISlide4; FocusROISlide1, FocusROISlide2, FocusROISlide3, and FocusROISlide4, respectively.
- To reduce the number of manual entries required to initiate a scan, the values of these variables may be encoded within SlideLayoutSettings.JNL.
- Define additional focusing variables.
- In order to prevent errors caused by focusing on bright debris or other artifacts, the first two focusing algorithms focus at multiple sites. When two focal points are within the z distances defined by the variables TotalZFocVar or ROIZFocVar, for determining the central focal plane for each slide and the z origin of each ROI, respectively, they are considered to be in agreement. The mean of these is then used as the central focal plane or origin. Because the value determined for the central focal plane of each slide does not need to be precise, TotalZFocVar is typically set between 5 and 10 μm. ROIZFocVar can be set to between 25% and 100% of TotalZFocVar.
- In order to prevent gross focusing errors, the deep and fine focusing algorithms compare the newly-determined focal plane to the previously-determined focal plane. If the difference between the new deep focus position and the previous position exceeds the value of DpFocVar, the new focus position is rejected and deep focusing is attempted on the next tile. If the difference between the new fine focus position and the previous position exceeds the value of FnFocVar but less than twice the value of FnFocVar, the new focus position is rejected and fine focusing is attempted on the next tile. If the difference between the new fine focus position and the previous position is more than twice FnFocVar, fine focusing is repeated with a 3-fold expanded range. Depending on the flatness of the specimen being imaged and planar alignment of the slide with the camera, these values can be set to between 1 and 3 μm, with DpFocVar being greater than or equal to FnFocVar.
- The MetaMorph “Adjust Focus” command is used to determine focus in each step. This command relies on values for Range, Step Size, and Number of Steps; only two of these need to be specified, the third is calculated. The values for TotalZRange, TotalZFocStepSz, ROIZRange, ROIZFocStepSz, DpFocRange, DpStepSz, FnFocRange, and FnFocStepSz must be specified.
- The variable TotalZRange should be 25% to 50% greater than the maximum z range across all slides. For example, if the maximum z range is 300 μm, TotalZRange should be set to 450. Step Size should be less than 5% of the range and, in practice, should be less than 15 μm. Thus, if TotalZRange is 450, value 15 is reasonable for TotalZFocStepSz. Using larger and smaller than necessary values for TotalZRange and TotalZFocStepSz, respectively, has minimal impact on total scanning time since the process only occurs once for each slide. TotalZFocCh should be ubiquitously present within tissues, photostable, and with a high signal:noise ratio. A nuclear stain is ideal. TotalZThreshold corresponds to the threshold for that channel, as determined above. These values can be encoded in the SlideROIFocusSettings.JNL.
- Focusing time is reduced by determining the z origin within each ROI over a limited range. That range is defined by the variable ROIZFocRange, which should be 25% to 50% greater than the maximum focus range within a single slide, but should not be more than 150 μm. As above, the algorithm focuses at multiple sites within each ROI. The z origin for each ROI is determined two focal points, which must be within ROIZFocVar of one another. If two focal points within ROIZFocVar cannot be identified, this is recorded as a possible focusing error and displayed in the settings and report.LOG file, which is saved in the same folder as the images and displayed at the end of the scanning process. Using larger and smaller than necessary values for ROIZFocRange and ROIZFocStepSz, respectively, has substantial impact on total scanning time since the process is repeated once for each ROI. ROIZFocCh defines the channel used to determe the z origin. ROIZThreshold and ReducedROIZThreshold are used to identify tissue. If the same channel that is used to define the central focal plane of each slide is used for determining the z origin within ROIs, each ROIZ variable can be set to the same value as corresponding SlideZ variable. The settings can, however, also be defined independently. As above, these values can be entered in response to prompts or encoded in SlideROIFocusSettings.JNL.
-
The ranges for deep and fine focusing are defined by DpFocRange and FnFocRange, respectively. Typical values are 5 – 10 μm and 1 – 3 μm, respectively. DpStepSz and FnStepSz are generally 1 – 3 μm and 0.2 – 0.6 μm, respectively. DpFocCh, DpThreshold, FnFocCh, FnThreshold, are determined as described above. These values are defined by the user and can be entered in response to prompts or encoded in DpFnFocusSettings.JNL.Using larger values than necessary can dramatically increase scanning time. These values are influenced by objective magnification and NA.
-
The number of frames, i.e., tiles, collected before deep or fine focusing is defined by the variables DpFocInt and FnFocInt, respectively. The settings chosen depend on the flatness of the tissue within each ROI. Values of 20 and 5, respectively, often work well. Some trial and error may allow these to be increased further. Typically, DpFocInt should be a multiple of FnFocInt. These values can be encoded in DpFnFocusSettings.JNL.Using smaller values than necessary can increase scanning time.
To minimize the number of manual entries required when initiating a scan, the values of the variables above may be encoded within SlideROIFocusSettings.JNL and DpFnFocusSettings.JNL (Fig. 6C).
Figure 6. Setting focusing parameters.
A. Defining the threshold is critical. The journals use this parameter to determine whether tissue is present. B. ROIs to be used to determine the central focal point of each slide can be indicated in response to prompts when running TMA scans. C. Alternatively, ROIs that define each slide can be encoded in the SlideLayoutSettings journal.
BASIC PROTOCOL 5 ACQUIRE TILED IMAGES
This is the central protocol that scans the slides using a series of journals. In preparation, the preliminary scan and ROIs are loaded by OpenPrelimAndROIs.JNL. After defining channels and exposure times and saving the settings using Save TMA Scan Settings.JNL, TMA Scanning Master.JNL is activated by the “Run TMA Scan of all ROIs” button on the taskbar. After manual or automated entry of the settings, the latter using SlideLayoutSettings.JNL, SlideROIFocusSettings.JNL, and DpFnFocusSettings.JNL, and confirmation of other settings, TMA Scanning Master.JNL calls add top left or bottom right position to array.JNL to move to and record the top left and bottom right corner of each ROI to a plane in the file Stage list array.TIF (this step is omitted if an existing Stage list array.TIF file is being reused).
TMA Scanning Master.JNL then calls Region label to array.JNL, or, if an existing Stage list array.TIF file is being reused, Region label to array (existing stage list array).JNL. Either of these journals steps through the planes of Stage list array.TIF and, for each ROI designated as the defining the central focal plane of a slide, moves to that region and runs SlideZ focus.JNL. SlideZ focus.JNL attempts to find and focus on tissue in nine regions. If a consensus focus cannot be found, SlideZ-tissue not found.JNL runs and attempts to find and focus on tissue using a reduced threshold. Consensus focus is defined as two focal points within TotalZFocVar of one another. If found, it is recorded as the central focal plane for that slide, e.g., Slide1Z for slide 1. If neither SlideZ focus.JNL nor SlideZ-tissue not found.JNL find consensus focus, an error is logged.
TMA Scanning Master.JNL then calls Scan tissue.JNL, which again cycles through each plane of Stage list array.TIF. Scan tissue.JNL generates a file name from the ROI label, determines the current slide number, and enters the coordinates of the ROI corners from Stage list array.TIF into the Scan Slide module before calling ROI focus.JNL. In turn, ROI focus.JNL moves the stage to the z plane defined for the current slide, sets the channel for focusing, loads the acquire settings for the focusing channel into acquire, and attempts to find and focus on tissue in nine regions, using commands similar to those in SlideZ focus.JNL. If a consensus focus, defined using variables that parallel those in SlideZ focus.JNL, cannot be found, ROITissueNotFound.JNL makes a second attempts using a reduced threshold, similar to SlideZ-tissue not found.JNL. If ROITissueNotFound.JNL runs, its success or failure in finding consensus focus is logged and reported when Record variables.JNL runs after the last ROI is acquired.
Scan tissue.JNL then sets the variable FirstSubZ to 1 and the deep and fine focus counters to one greater than the designated interval before running Scan Slide. If scan slide adjust.JNL is set to run at the start of each stage position within the ROI, the journal checks if deep focus is needed. If deep focus is needed, the channel is set the deep focus channel, z to the value determined by ROI focus.JNL or ROITissueNotFound.JNL. An image is acquired and used to check for tissue using the standard threshold or, if ROITissueNotFound.JNL ran, the reduced threshold. If tissue is not found, the journal stops and looks for tissue again at the next stage position. If tissue is found, deep focusing is performed over the distance specified or, if this is the first tile in the ROI, i.e., when FirstSubZ = 1, twice that value. After changing to the fine focus channel, scan slide adjust.JNL checks for tissue using the standard threshold or the reduced threshold. If tissue is not found in the fine focus channel, the journal stops and the counters are set so that fine focusing is triggered at the next stage position. Otherwise, fine focusing is performed. For both deep and fine focusing, the new focus position (z) is compared to the previous value and, if the new value exceeds the acceptable variation, the new focus is rejected and the counters are set to focus at the next stage position. Depending on the magnitude of difference between the new and previous z values, FirstSubZ may be reset to a value of 1 or greater to increase the range over which focus is checked at the next tile position. Conversely, FistSubZ is set to 0 if focusing is successful.
Fine focus is activated if deep focusing has occurred or if the FnFocChk counter equals FnFocInt. In either case, the channel is set to to the fine focus channel and an image is aquired to to determine if tissue is present based on the threshold or reduced threshold (depending on whether ROITissueNotFound.JNL ran). If tissue is present, focusing is performed using the defined variables for range, step size, and acceptable variation from the previous position. If it is within acceptable variation of the previous focus position this becomes the new focus position, the fine focus counter is reset, and FirstSubZ is set to 0. If either tissue is not present or not the new focus position is beyond the accepted range, fine focusing is triggered at the next stage position. Depending on the magnitude of discordance between new and previous focus positions, FirstSubZ is reset and deep focusing may be activated at the next stage position.
Finally, before each image set the Z.JNL must be set to run before each image. This journal moves the stage to the most recently determined fine focus z at wavelength 1 of each set. This is critical, because the MetaMoprh Scan Slide module is programmed to return to the initial z position (when Scan Slide was started) before acquiring each tile.
Materials
System configured as in Basic Protocol 1
Preliminary low magnification scan generated in Basic Protocol 2
ROIs generated in Basic Protocol 3
Files created and settings determined in Basic Protocol 4
Stained slides
Protocol Steps
- Adjust the Scan Slide settings.
- Press the “Open Scan Slide Window” button at the top of the taskbar.
- Press the “Open Prelim Scan and ROIs” button. Click the “adjusted” radio button in response to the the prompt (Fig. 7A).
- Press the “Load TMA Scan Settings” (or “Load/Save Rescan Settings”) button on the taskbar.
- In Scan Slide, select the number of required wavelengths, illumination settings (e.g., filter cubes), and exposures in each channel for the tiled images (Fig. 7B).
-
When scanning tissue labeled by multiple fluorochromes, the focal point of one stain may be displaced from that of the other fluorochromes, which may also be displaced from each other. This can be addressed using the offset function in Scan Slide.Note that the entered z offset is from the previous channel based on the order of channel selection in the wavelengths tab.
- Click the Scan Slide Journals tab and set before each image set the Z.JNL to run Before each image (Fig. 7C). Also set scan slide adjust.JNL to run at Start of stage position.
- Set shading correction if that function is being used.
- Confirm the save file path and name. Each set of scans should be in a separate sub-folder within the parent folder for all the scans to be made of the TMA slides. For example, C:\Data\user\YYMMDD\stain. Scan files will be imaged with the magnification used and ROI number, e.g., “Scan 20X ROI 1” for ROI 1.
- Press the “Save/Backup Scan Settings” button. This will overwrite the TMA scan current settings.scansetting file within the journals folder and save a copy of the file in the folder where the images will be saved.
If variable values encoded in SlideLayoutSettings.JNL, SlideROIFocusSettings.JNL, and DpFnFocusSettings.JNL are being used, confirm that they are correct.
Set the focus to the center z point across all slides. This was set to 0 in Basic Protocol 4.
Press the “Run TMA Scan of all ROIs” button on the taskbar.
A prompt will ask “Are you certain that everything is correctly configured?” Those who are experienced in using the procedures, are using the variables encoded in SlideLayoutSettings.JNL, SlideROIFocusSettings.JNL, and DpFnFocusSettings.JNL, and are confident that all other settings are correct can respond affirmatively. This will skip to the prompt asking if a new Stage list array.TIF needs to be created.
- If the response to the previous is negative, several sets of prompts will be triggered.
- The first set asks about general configuration.
- Has the stage has been calibrated?
- What magnifcation will be used? This is followed by a request for verification that the correct objective is in place and that z is set to a central focal point for all slides/ROIs.
- The user is then asked to confirm that acquire settings files been created for each channel used for autofocusing.
- Finally, the journal asks if the correct TMA Scan Settings been saved and loaded in the Scan Slide window.
- The second set of prompts asks if the slide layout and focus settings are set correctly. In each case, the user is presented with the default variable values (encoded in SlideLayoutSettings.JNL, SlideROIFocusSettings.JNL, and DpFnFocusSettings.JNL) and asked if these are correct. In each case, a No response triggers queries for the value of each variable. The default values are shown in the response boxes as guides.
- A prompt will ask if a new Stage list array.TIF needs to be created (Fig. 8).
- If a new aray does need to be created, the user is asked to confirm that the preliminary scan with adjusted ROIs is loaded as the active window.
- If the user presses Continue, the stage will move to the coordinate positions (opposite corners) of each ROI. These values will be saved in a plane of the file Stage list array.TIF, which has one plane for each ROI. The file will then be saved in the location specified in Scan Slide.
- The user is instructed to press Cancel and end the process if the image and ROIs are not loaded correctly.
-
If a new aray does not need to be created, the program requests confirmation that Stage list array.TIF is open and selected as the active image.If the slides are being reimaged, e.g., after an additional round of staining, and their coordinate positions, based on stage xy calibration, are nearly identical, it may be possible to re-use the stage list array. This saves the time required for the preliminary scan and for the stage to move to the corners of each ROI.
The journal will now run without any further user input.
The journals will direct system to determine the central focal plane of each slide before moving to the first ROI on the first slide. The z origin for that ROI is then determined and Scan Slide triggered to collect tiled images of the ROI.
-
If Scan slide adjust.JNL is set to run, deep and fine focusing are performed regularly using the variables set above.
It can be helpful to watch acquisition of the first ROI or two to ensure that the process is running correctly.
After ROI 1 is fully imaged, the stage moves to the next ROI and repeats the process. ROIs are imaged by physical position and may not be scanned in the numbered order.
After the last ROI, a summary log text file lists the values of imaging variables and provides a report of success of failure when slide or ROI focusing needed to be performed with a lower threshold.
The summary file can be reopened and displayed by pressing the “Review TMA Scan Report” button on the taskbar.
Figure 7. Final scan settings.
A. Set of four slides with ROIs after pressing the Open Prelim Scan and ROIs button on the taskbar. B. Scan slide configuration for multi-wavelength (channel) imaging. C. If the focusing journals are being used, they should be selected on the Run Journal tab of the Scan slide window.
Figure 8. TMA scan.
A. Prompt asking about slide layout. This and other slide layout values can be encoded in journals to simplify scan initiation. B. Prompt asking which wavelength will be used for slide focusing. C. Prompt asking if a new stage list array file should be created.
BASIC PROTOCOL 6 REVIEW THE SCANS
When scanning ~200 ROIs, it is common for a few to have errors in ROI placement, focusing, or other problems. Correcting mispositioned ROIs and revising focusing parameters can overcome these errors. In most cases, errors are readily recognized with low-resolution stitching. The procedure below allows rapid review of collected images. ROIs can then be reimaged, as necessary, before moving on to further processing and analysis.
This protocol uses Review Scans.JNL to quickly step through low resolution stitched images. The journal set image zoom and scaling.JNL defines the display variables.
Materials
Image sets acquired in Basic Protocol 5
Protocol Steps
Low resolution preview images of all ROIs can be reviewed relatively quickly by pressing the “Low Resolution Review” button on the taskbar.
Respond to user prompts for ROI numbers, scan file locations, and magnification used during imaging (Fig. 9A).
Determine whether the displayed scan requires reimaging (Fig. 9B) and which scanning parameters should be modified (see Table 4 for troubleshooting).
Press “Continue” to view the next scan.
Figure 9. Post-acquisition review.
A. Summary report shown at the completion of scanning. B. Low power review of scans is simplified through a journal that directs successive presentation of each stitched ROI.
BASIC PROTOCOL 7 REIMAGE ROIS AS NEEDED
After determing ROIs to be reimaged in Basic Protocol 6, variables are modified as needed and rescanning is activated using the journals described previously.
Materials
Stained slides
Preliminary low magnification scan generated in Basic Protocol 2
ROIs generated in Basic Protocol 3
Files created and settings determined in Basic Protocol 4
Protocol Steps
- Rescan ROIs using the taskbar tools
- Create a sub-folder in the image folder, e.g., C:\Data\user\YYMMDD\stain\rescans.
- Press the “Open Scan Slide Window” button on the taskbar.
-
Press the “Load/Save Rescan Settings” button on the taskbar and select “load” in response to the prompt.If using the same acquisition settings as in the original scan, it may be easiest to load the original TMA scan settings (using the button on the taskbar) as a starting point.
-
Change the save location to the rescans subfolder created above. Press the “Load/Save Rescan Settings” button on the taskbar and select “save” in response to the prompt.If the original TMA scans were used as a starting point, the new configuration will be saved over the previous rescan settings file.
- Open preliminary scan.
- If a new preliminary scan is needed, return the Basic Protocol 2 and follow the subsequent protocols.
- If reusing the previous preliminary scan, press the “Open Scan Slide Window” button followed by the “Load Prelim Scan Settings” button. Then Press “Open Prelim Scan and ROIs” and select “do not load ROIs” in response to the prompt.
Press the “Open Scan Slide Window” button (if needed) and the “Load/Save Rescan Settings” if the rescan settings are not loaded.
- Create ROIs
- Typically, relatively few ROIs need to be rescanned. If this is the case, use the region tools to draw a ROI around each tissue that needs to be rescanned. Press the “Renumber individual ROIs” button on the taskbar to name these correctly.
- If there are many regions to be rescanned, load either the ROIs for one slide or all slides by pressing either “Reload One ROI Set” or “Open ROI Group (>1 slide)” button on the taskbar, respectively.
Verify positions of all ROIs as in Basic Protocol 3.
Press the “Save ROI Group (>1 slide)” button on the taskbar to save the modified ROIs in the rescan folder. Click the “rescan” radio button in response to the the prompt.
Adjust other scan settings, as needed, and press the “Load/Save Rescan Settings” button on the taskbar and select “save” in response to the prompt.
From the ROIs present, select one on each slide as the focusing ROI. Either edit the SlideLayoutSettings.JNL journal or enter the slide layout settings, including the new FocusROIs for each slide, in response to the prompts while running the scan (below). If using this approach, the previous values for FirstROI on each slide can be used as long as they bracket the ROIs to be rescanned.
Press the “Run TMA Scan of all ROIs” button on the taskbar.
Review the summary log.
Review the rescan images using Basic Protocol 6.
BASIC PROTOCOL 8 STITCH, STACK, AND ASSEMBLE IMAGES
MetaMorph can be used for post-acquisition processing and stitching, but open-source software, e.g., ImageJ/FIJI and ASHLAR can also be effective.(Muhlich et al., 2017; Schindelin et al., 2012). ASHLAR is particularly advantageous when subtracting background or aligning multiple cycles of images and also creates pyramidal OME-TIFF files (Besson et al., 2019) that are necessary for responsive virtual microscopy as part of the stitching process. All of these stitching methods create images that that are compatible with open-source software such as ImageJ/FIJI, GiMP, and QuPath (Bankhead et al., 2017). We use an OMERO Plus server with PathViewer (Glencoe Software) for image display, but a variety of open-source virtual microscopy solutions are available (Bankhead et al., 2017). Open-source options for OMERO servers are also available (Allan et al., 2012).
Materials
Image files generated in the basic protocol
MetaMorph or ASHLAR software
Protocol Steps
To keep the scanning microscope free for acquisition, images can be transferred to an, offline computer, e.g. a workstation, for assembly into composite stitched images.
- Use MetaMorph or ASHLAR to stitch the images. ASHLAR should be used if aligning multiple sets of images from the same tissues.
- Stitching with MetaMorph:
- Press the “Stitch, Stack, and Save” button on the taskbar. The stitched images will be saved in a subfolder stitched-stacks within the image folder.
- Most virtual microscope applications, including OMERO Plus PathViewer, require pyramidal OME-TIFF image files. These can be created from the multi-wavelength image stack using the Java-based open source Bio-Formats program (Linkert et al., 2010) with bioformats2raw or raw2ometiff programs (Glencoe Software).
- Stitching with ASHLAR:
- ASHLAR is open source software under continual refinement and is available for download from GitHub. ASHLAR runs in Python and has no graphical user interface. Some fundamental knowledge of Python is therefore required for its use. Unlike MetaMorph, ASHLAR can align several staining cycles of the same tissue based on a reference channel from each cycle. The data must be described by a .scan file and the reference channels must all be the same number (the default reference channel is 0). This feature makes ASHLAR suitable for multiplexing and background subtraction experiments. The output is a series of stitched TIFF files ready for analysis.
-
Determine the necessary parameters required to stitch the tiled images. Tiled images collected in MetaMorph are listed in the reverse order from the ASHLAR default. Command line options (--flip-x and --flip-y) have been integrated into ASHLAR to correct for this. If there are sensor noise issues in the corners of tiles, this can be offset by applying a Gaussian filter (--filter-sigma SIGMA). It is recommended that you begin with a filter of 1 and increase as needed. When run from the home directory of a scanning project with multiple subdirectories containing image files, the following command line would stitch two cycles of staining in the same way, aligning by the default channel (0) and saving them to the subdirectory stitched/ashlar –flip-x –flip-y --filter-sigma 1 \c1\f\Scan 20X ROI1.scan \c1\b\Scan 20X ROI1.scan \c2\f\Scan 20X ROI1.scan \c2\b\Scan 20X ROI1.scan --o \stitched\
- Write a Python script to run ASHLAR on all the scans you need to stitch. The process of stitching is time intensive, especially if multiple imaging cycles are involved. To save user interface time, write a looping script to execute ASHLAR on each .scan file, each of which represents an ROI. Some basic knowledge of Python programming is required.
- (optional) ASHLAR has built in functionality to output pyramidal OME-TIFF image files.
BASIC PROTOCOL 9 REPEAT SCANNING FOR MULITPLEX IMMUNOSTAINING OR BACKGROUND SUBTRACTION
Multiplex imaging has become a powerful adjunct to immunofluorescent staining of tissues. This approach allows cycles of staining and de-staining, with imaging at each step. As a result, tissues and individual cells can be labeled with 20 or more stains. Nuclear or other staining can then be used to align individual image sets with one another. A simpler approach, in which slides are stained for nuclei, imaged, stained for other markers (and nuclei), and imaged again can also be used to digitally subtract tissue autofluorescence from acquired images. When combined, these approaches enable powerful downstream analyses that include treating the data as if it were obtained using a flow cytometer and unsupervised cluster analyses. Many excellent articles have been written about staining, de-staining or quenching, image registration, and downstream analysis (Goltsev et al., 2018; Kallioniemi et al., 2001; Kampf et al., 2012; Lin et al., 2016; Lin et al., 2015; J. R. Lin et al., 2018; McKinley et al., 2017; Muhlich et al., 2017; Parra et al., 2017; Preibisch, Saalfeld, & Tomancak, 2009), and these topics are not discussed here.
Materials
Stained slides
Preliminary low magnification scan generated in Support Protocol 2
ROIs generated in Support Protocol 3
Settings determined in Support Protocol 4
Protocol Steps
Remove coverslips.
Strip antibodies or inactivate fluorophores (Bolognesi et al., 2017; Du et al., 2019; Gendusa, Scalia, Buscone, & Cattoretti, 2014; J. R. Lin et al., 2018).
Re-stain slides.
-
Recoverslip and reload the slides.
If tissue has moved or the slides are not reloaded in register with the original placement it may not be possible to re-use the ROIs and Stage list array.tif file.
Press the “Open Scan Slide Window” button at the top of the taskbar.
Press the “Load Prelim Scan Settings” buttons on the taskbar.
- If the slides are reloaded in near perfect register, the previous preliminary scan may be re-used after being validated.
- Load the previous preliminary scan using “Open Prelim Scan and ROIs” button the Scan Slide Data Review tab.
- Correct ROI positions as in Basic Protocol 3.
-
Change the Scan Slide save directory to the new directory and press “Save Prelim Scan Settings” to create a new 5X TMA Prelim Scan.scansetting file.This overwrite the previous preliminary scan settings file.
-
After confirming that all ROIs are correctly positioned, press the “Save ROI Group (>1 slide)” button.The program will overwrite the previous ROI file. If necessary, this file, which is found in the preliminary scan folder, can be renamed before saving the new file.
- If a new preliminary scan is needed, change the Scan Slide save directory to the new directory, adjust the region to be scanned, press “Save Prelim Scan Settings” to a create a new 5X TMA Prelim Scan.scansetting file.
- Create a new preliminary scan as in Basic Protocol 2
- Load previous ROIs, generally one slide at a time using the “Reload One ROI Set” button or create new ROIs as in Basic Protocol 3.
-
After confirming that all ROIs are correctly positioned, press the “Save ROI Group (>1 slide)” button.The program will overwrite the previous ROI file. If necessary, this file, which is found in the preliminary scan folder, can be renamed before saving the new file.
If necessary, adjust focusing parameters as in Basic Protocol 4.
Press the “Load TMA Scan Settings” button.
Change the Scan Slide save directory to the new directory and adjust all acquistioin settings as in Basic Protocol 5.
Press the “Save TMA Scan Settings” button.
Press the “Run TMA Scan of all ROIs” button.
COMMENTARY
BACKGROUND INFORMATION
Complete tissue, or slide imaging allows for archiving digital copies of slides and making it possible for other observers to evaluate the whole specimen, using virtual microscopy software. Multiple manufacturers have developed automated slide scanning systems that work well but are, in general, only available in core facilities. Here, we sought to develop an automated slide scanning system that could easily be assembled within individual labs using existing resources and is applicable to many different microscopes, including upright and inverted wide-field and confocal microscopes from many manufacturers. This protocol allows for automated and customizable scanning of multiple regions of interest within relatively short imaging times with output files being images in standard formats and manageable sizes.
The method described here can be widely applied to transmitted light and fluorescence imaging using wide-field, confocal, or other, e.g. super resolution, light microscopes. It will be particularly useful for investigators using advanced preparation techniques, e.g., immunofluorescent staining, to analyze specimen collections, e.g., tissue microarrays, as well as high-content and other multiplex imaging approaches. As described, the protocol is designed for multi-wavelength fluorescence, wide-field imaging of TMAs. It is suitable for multiplex staining approaches, in which antibodies are stripped or fluorochromes inactivated between imaging cycles, thereby allowing analysis of 20 or more markers in individual cells (Bolognesi et al., 2017; Gerdes et al., 2013; Goltsev et al., 2018; Lin et al., 2016; J. R. Lin et al., 2018; McKinley et al., 2017; Patel & Rodig, 2020). Methods for preparation and staining of TMAs are discussed in detail elsewhere (Bolognesi et al., 2017; Lin et al., 2016) as are approaches to multiplex staining and analysis (Goltsev et al., 2018; Li, Germain, & Gerner, 2017; Lin et al., 2016; McKinley et al., 2017; Patel & Rodig, 2020). With multiplexing, this approach using TMAs is suited to quantitative analysis of multiple samples on a single slide, as it simplifies standardization of staining and imaging parameters across samples within each slide. This approach also allows inclusion of internal controls and replicate samples on each slide.
CRITICAL PARAMETERS
Configuration of the microscope with the appropriate filter cube(s), camera(s), and a motorized stage is a pre-requisite to this protocol. Control of the automated filters and motorized stage must be integrated with the microscope control software.
Camera/stage xy alignment and field-of-view selection are critical. The camera must be aligned with the xy movements of the stage. This can be accomplished by tracking an identifiable object along the edge of the field-of-view and making incremental adjustments to the camera’s rotation until the object glides gently along the edge of the field-of-view with minimal drift. The camera’s position, once adjusted, must be made secure. In addition, focus should not be lost at the corners. This is generally not a problem with high quality, planar objectives but should be verified. Light fall-off at the field edges can also be a challenge. Minor defects can be corrected by field size selection and use of shading correction, but larger problems are best resolved by adjusting the hardware. The Scan Slide module staggers tiles slightly to reduce the impact of image defects at the corners when the tiles are stitched. ASHLAR (Muhlich et al., 2017) can also apply a light Gaussian filter during stitching to compensate for limited image distortion and light fall-off at the edges of each tile.
Proper stage z alignment is essential. The first concern is flatness, the more level the stage is the more rapidly scanning will progress and the easier stitching will be. A perfectly flat stage would minimize the need for repeated focusing when collecting tiles, which would dramatically accelerate scanning. We have found that a four-slide stage with a range of up to 300 μm over the four 75mm x 25mm slides is within tolerance; smaller ranges are even better. We generally allow a 50% buffer, e.g., a 450 μm range, when setting the range for determining the central focal plane of each slide.
Stage xy alignment when replacing slides is crucial. This primarily applies to application in which multiple scans of the same slides are collected, e.g., for background subtraction and multiplex imaging. Many slide holders allow some extra space that could interfere with precise placement. Pushing the slides towards a corner, e.g., top right, of the holder and securing them with clips eliminates this problem.
TROUBLESHOOTING
When scanning is successful the focus will be clear in each and the seams between tiles will be imperceptible (Fig. 10A, B). When scanning is only moderately successful a stitched image can be skewed (Fig. 10C) and if it is completely unsuccessful it will not be possible to stitch together the tiles, particularly if using MetaMorph for stitching. Detailed troubleshooting advice is provided in Table 4.
Figure 10. Stitching.
A. High resolution stitched images can be created and assembled into image stacks automatically. B. Four color overlay of imaged tissue at low power (i) and an enlarged view of the boxed area (ii). C. Single-channel stitched image of nuclei collected after correct calibration of Scan Slide (i). When this (green) is combined with a second image (red) acquired with improperly calibrated Scan Slide settings (ii), a few areas align (left box, enlarged in iii) but most do not (right box, enlarged in iv).
UNDERSTANDING RESULTS
This system has allowed us to scan numerous tissue microarrays from experimental mice (Graham et al., 2019; Kuo et al., 2019; Nalle et al., 2019; Raju et al., 2020) and human biopsies (Turner, 2020). Over 5,000 scanned images have been compiled into the publicly accessible Atlas of Intestinal Transport (https://www.jrturnerlab.com/atlasofintestinaltransport; Fig. 11A). Each image was saved in pyramidal OME-TIFF format, allowing OMERO Plus and PathViewer to act as a virtual microscope, replete with threshold adjustments for each channel (Fig. 11B).
Figure 11. Representative Data.
A. Searchable and exportable database displaying a few of over 5,000 images available at jrturnerlab.com/database-viewer/atlas-of-intestinal-transport. Clicking on a thumbnail opens the corresponding pyramidal OME-TIFF file on the OMERO Plus server using PathViewer. B. Display of human duodenal biopsy stained for SLC6A19 (Na+-dependent neutral amino acid cotransporter AT1; cyan), E-cadherin (magenta), ZO-1 (yellow), and nuclei (Hoechst 33342; blue). Colors and scaling were applied using the user-modifiable viewing options control box (left).
As an example, a duodenal biopsy from an individual human subject is shown when stained by hematoxylin and eosin or using 12 different immunostains to detect transport and barrier proteins (Fig. 12). In this case serial sections were stained. However, similar results can be achieved with multiplex, or cyclic, immunofluorescent staining.(Du et al., 2019; Lin et al., 2016; J. R. Lin et al., 2018; McKinley et al., 2017) Regardless of the staining methods used, the protocol described here allows most labs within a simple wide-field fluorescent microscope to generate images of a quality comparable or superior to those obtained using turnkey scanning systems.
Figure 12. Representative Data.
Serial sections of a duodenal biopsy from a healthy, 26-year-old, female subject were stained by hematoxylin and eosin or with 12 different immunostains. This was one of 50 tissue cores in a single TMA. After imaging and post-acquisition processing using this protocol, pyramidal OME-TIFF images were displayed. Each file (~400 GB) is shown in full (top images, bar = 250 μm), or zoomed in on successively smaller areas (middle images, bar = 100 μm and bottom images, bar = 10 μm). SLC6A19 (Na+-dependent neutral amino acid cotransporter AT1; B0AT1), CFTR (cystic fibrosis transmembrane conductance regulator), CLDN7 (claudin-7), CLDN15 (claudin-15), SLC5A1 (Na+-dependent glucose cotransporter 1; SGLT1), ZO-1 (zonula occludens-1), SLC9A3 (Na+-H+ exchanger 3; NHE3), Na+-K+ ATPase, SLC12A2 (Na+-K+-Cl- transporter 1; NKCC1), OCLN (occludin), and SLC15A1 (H+-peptide cotransporter 1; HPEPT1(Merlin et al., 2001)) are all expressed at appropriate positions along the crypt-villus and basolateral-apical polarity axes. SLC26A3 (downregulated in adenoma; DRA(Gill et al., 2007)), is poorly-expressed in duodenum; but detection and apical expression within surface epithelial are confirmed in proximal colon (bottom right image set). Antibodies used are listed in Table 1.
TIME CONSIDERATIONS
Time required for installation of the hardware and software can vary from minutes to hours, depending on the hardware configuration. Creation of the preliminary scan takes 5 – 10 minutes per slide. The semi-automated ROI selection process reduces the time required for ROI assignments to ~5 minutes per slide. Defining other imaging parameters can be very quick, particularly if these have been established previously.
Acquisition of tiled images is the longest part of the process. The time required is dependent on the number and size of ROIs being scanned, imaging parameters, size of the camera capture area, speed of the motorized filter cube turret, pace of the automated stage, and number of channels imaged. For example, our configuration is optimized for fluorochromes with spectral characteristics of DAPI/Hoechst 33342, fluorescein/Cy2/Alexafluor 488, Texas red/Alexafluor 594, and Cy5/Alexafluor 647. In general, one slide with 50 2.5 mm diameter tissue cores can be imaged in 4 channels at 20X in less than 3 hours. Some groups have reported successful use of longer wavelength dyes, e.g., Cy5.5/Alexafluor750 using Chroma filter set 49007, as a fifth channel. This would require the illumination system to excite and the camera to detect the dye and would extend imaging time.
The time required for filter cube changes is a substantial part of the imaging time. This could be largely eliminated through use of higher intensity, wavelength-selectable light sources, e.g., LEDs or lasers, multi-bandpass dichroic mirror, and individual emission filters on a fast filter wheel; a multi-bandpass emission filter is not recommended as it will result in considerably increased background due to the broad spectrum autofluorescence of paraffin-embedded tissues. We have found that this configuration can reduce imaging time by more nearly 50%, i.e., to less than 2 hours for a 50 TMA slide imaged in four channels. Although stitching is processor intensive, but most PCs can stitch 40-50 ROIs in less than 1 hour.
Supplementary Material
SIGNIFICANCE STATEMENT.
Complete tissue, or slide, imaging simplifies microscopic evaluation of large specimens, allows virtual microscopy review by observers at different, and digitally archives slides for future review. Commercial slide-scanning systems are available, but their cost typically restricts availability to core facilities. Here, we demonstrate an easily deployable, automated slide-scanning system using a simple wide-field fluorescence microscope and widely-available, commercial microscope control software. After semi-automated detection of individual tissue microarray samples, the system independently acquires tiled images of the regions defined by each tissue sample. Review, stitching, and channel compilation are also automated. The system is compatible with microscopes, stages, light sources, and cameras from a wide range of manufacturers.
ACKNOWLEDGEMENTS
This work was supported by the National Institute of Diabetes Digestive and Kidney Disease (R01DK61931, R01DK068271, R24DK099803) and the Harvard Digestive Disease Center (P30DK034854).
We thank Jeremy Hermann, Lora MDM Ong, Roman Pavlyuk, and Sunil Yeruva, for their contributions to early stages of this work and Prof. Didier Merlin (Georgia State University) for generously providing anti-HPEPT1 antibody.
Footnotes
Conflicts of interest
JRT is a cofounder of and hold shares in Thelium Therapeutics. NRG is an employee of Molecular Devices. ER and CA are employees of Glencoe Software.
INTERNET RESOURCES
Image data are available at https://www.jrturnerlab.com/database-viewer/atlas-of-intestinal-transport. A journal suite will be made available for download at jrturnerlab.com and a public repository, e.g., Github.
LITERATURE CITED
- Allan C, Burel JM, Moore J, Blackburn C, Linkert M, Loynton S, … Swedlow JR (2012). OMERO: flexible, model-driven data management for experimental biology. Nat Methods, 9(3), 245–253. doi: 10.1038/nmeth.1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankhead P, Loughrey MB, Fernandez JA, Dombrowski Y, McArt DG, Dunne PD, … Hamilton PW (2017). QuPath: Open source software for digital pathology image analysis. Sci Rep, 7(1), 16878. doi: 10.1038/s41598-017-17204-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besson S, Leigh R, Linkert M, Allan C, Burel JM, Carroll M, … Swedlow JR (2019). Bringing Open Data to Whole Slide Imaging. Digit Pathol (2019), 2019, 3–10. doi: 10.1007/978-3-030-23937-4_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolognesi MM, Manzoni M, Scalia CR, Zannella S, Bosisio FM, Faretta M, & Cattoretti G (2017). Multiplex Staining by Sequential Immunostaining and Antibody Removal on Routine Tissue Sections. Journal of Histochemistry and Cytochemistry, 65(8), 431–444. doi: 10.1369/0022155417719419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhir R (2008). Tissue microarrays: an overview. Methods in Molecular Biology, 441, 91–103. doi: 10.1007/978-1-60327-047-2_6 [DOI] [PubMed] [Google Scholar]
- Du Z, Lin JR, Rashid R, Maliga Z, Wang S, Aster JC, … Santagata S (2019). Qualifying antibodies for image-based immune profiling and multiplexed tissue imaging. Nat Protoc, 14(10), 2900–2930. doi: 10.1038/s41596-019-0206-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein AD, Tsuchida MA, Amodaj N, Pinkard H, Vale RD, & Stuurman N (2014). Advanced methods of microscope control using muManager software. J Biol Methods, 1(2). doi: 10.14440/jbm.2014.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendusa R, Scalia CR, Buscone S, & Cattoretti G (2014). Elution of High-affinity (>10-9 KD) Antibodies from Tissue Sections: Clues to the Molecular Mechanism and Use in Sequential Immunostaining. Journal of Histochemistry and Cytochemistry, 62(7), 519–531. doi: 10.1369/0022155414536732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes MJ, Sevinsky CJ, Sood A, Adak S, Bello MO, Bordwell A, … Ginty F (2013). Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue. Proc Natl Acad Sci U S A, 110(29), 11982–11987. doi: 10.1073/pnas.1300136110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill RK, Borthakur A, Hodges K, Turner JR, Clayburgh DR, Saksena S, … Dudeja PK (2007). Mechanism underlying inhibition of intestinal apical Cl/OH exchange following infection with enteropathogenic E. coli. Journal of Clinical Investigation, 117(2), 428–437. doi: 10.1172/JCI29625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goltsev Y, Samusik N, Kennedy-Darling J, Bhate S, Hale M, Vazquez G, … Nolan GP (2018). Deep Profiling of Mouse Splenic Architecture with CODEX Multiplexed Imaging. Cell, 174(4), 968–981 e915. doi: 10.1016/j.cell.2018.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham WV, He W, Marchiando AM, Zha J, Singh G, Li HS, … Turner JR (2019). Intracellular MLCK1 diversion reverses barrier loss to restore mucosal homeostasis. Nature Medicine, 25(4), 690–700. doi: 10.1038/s41591-019-0393-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallioniemi OP, Wagner U, Kononen J, & Sauter G (2001). Tissue microarray technology for high-throughput molecular profiling of cancer. Human Molecular Genetics, 10(7), 657–662. doi: 10.1093/hmg/10.7.657 [DOI] [PubMed] [Google Scholar]
- Kampf C, Olsson I, Ryberg U, Sjostedt E, & Ponten F (2012). Production of tissue microarrays, immunohistochemistry staining and digitalization within the human protein atlas. J Vis Exp(63). doi: 10.3791/3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo WT, Shen L, Zuo L, Shashikanth N, Ong M, Wu L, … Turner JR (2019). Inflammation-induced Occludin Downregulation Limits Epithelial Apoptosis by Suppressing Caspase-3 Expression. Gastroenterology, 157(5), 1323–1337. doi: 10.1053/j.gastro.2019.07.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Germain RN, & Gerner MY (2017). Multiplex, quantitative cellular analysis in large tissue volumes with clearing-enhanced 3D microscopy (Ce3D). Proc Natl Acad Sci U S A, 114(35), E7321–E7330. doi: 10.1073/pnas.1708981114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JCT, Yeong JPS, Lim CJ, Ong CCH, Wong SC, Chew VSP, … Iqbal J (2018). An automated staining protocol for seven-colour immunofluorescence of human tissue sections for diagnostic and prognostic use. Pathology, 50(3), 333–341. doi: 10.1016/j.pathol.2017.11.087 [DOI] [PubMed] [Google Scholar]
- Lin J-R, Izar B, Wang S, Yapp C, Mei S, Shah P, … Sorger PK (2018). A simple open-source method for highly multiplexed imaging of single cells in tissues and tumours. bioRxiv 151738. doi: 10.1101/151738 [DOI] [Google Scholar]
- Lin JR, Fallahi-Sichani M, Chen JY, & Sorger PK (2016). Cyclic Immunofluorescence (CycIF), A Highly Multiplexed Method for Single-cell Imaging. Curr Protoc Chem Biol, 8(4), 251–264. doi: 10.1002/cpch.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JR, Fallahi-Sichani M, & Sorger PK (2015). Highly multiplexed imaging of single cells using a high-throughput cyclic immunofluorescence method. Nat Commun, 6, 8390. doi: 10.1038/ncomms9390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JR, Izar B, Wang S, Yapp C, Mei S, Shah PM, … Sorger PK (2018). Highly multiplexed immunofluorescence imaging of human tissues and tumors using t-CyCIF and conventional optical microscopes. Elife, 7. doi: 10.7554/eLife.31657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkert M, Rueden CT, Allan C, Burel JM, Moore W, Patterson A, … Swedlow JR (2010). Metadata matters: access to image data in the real world. J Cell Biol, 189(5), 777–782. doi: 10.1083/jcb.201004104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley ET, Sui Y, Al-Kofahi Y, Millis BA, Tyska MJ, Roland JT, … Coffey RJ (2017). Optimized multiplex immunofluorescence single-cell analysis reveals tuft cell heterogeneity. JCI Insight, 2(11). doi: 10.1172/jci.insight.93487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlin D, Si-Tahar M, Sitaraman SV, Eastburn K, Williams I, Liu X, … Madara JL (2001). Colonic epithelial hPepT1 expression occurs in inflammatory bowel disease: transport of bacterial peptides influences expression of MHC class 1 molecules. Gastroenterology, 120(7), 1666–1679. Retrieved from http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=11375948 [DOI] [PubMed] [Google Scholar]
- Muhlich J, Russell DP, & Chen Y-A (2017). Alignment by Simultaneous Harmonization of Layer/Adjacency Registration. Github, github.com/sorgerlab/ashlar. [Google Scholar]
- Nalle SC, Zuo L, Ong M, Singh G, Worthylake AM, Choi W, … Turner JR (2019). Graft-versus-host disease propagation depends on increased intestinal epithelial tight junction permeability. Journal of Clinical Investigation, 129(2), 902–914. doi: 10.1172/JCI98554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra ER, Uraoka N, Jiang M, Cook P, Gibbons D, Forget MA, … Rodriguez-Canales J (2017). Validation of multiplex immunofluorescence panels using multispectral microscopy for immune-profiling of formalin-fixed and paraffin-embedded human tumor tissues. Sci Rep, 7(1), 13380. doi: 10.1038/s41598-017-13942-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SS, & Rodig SJ (2020). Overview of Tissue Imaging Methods. Methods in Molecular Biology, 2055, 455–465. doi: 10.1007/978-1-4939-9773-2_21 [DOI] [PubMed] [Google Scholar]
- Preibisch S, Saalfeld S, & Tomancak P (2009). Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics, 25(11), 1463–1465. doi: 10.1093/bioinformatics/btp184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raju P, Shashikanth N, Tsai PY, Pongkorpsakol P, Chanez-Parades S, Steinhagen PR, … Turner JR (2020). Inactivation of paracellular cation-selective claudin-2 channels attenuates immune-mediated experimental colitis in mice. J Clin Invest. doi: 10.1172/JCI138697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid R, Gaglia G, Chen YA, Lin JR, Du Z, Maliga Z, … Santagata S (2019). Highly multiplexed immunofluorescence images and single-cell data of immune markers in tonsil and lung cancer. Sci Data, 6(1), 323. doi: 10.1038/s41597-019-0332-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson D, Savage K, Reis-Filho JS, & Isacke CM (2008). Multiple immunofluorescence labelling of formalin-fixed paraffin-embedded (FFPE) tissue. BMC Cell Biol, 9, 13. doi: 10.1186/1471-2121-9-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, … Cardona A (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods, 9(7), 676–682. doi: 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer MS, Schumacher L, & Rubin MA (2004). Constructing tissue microarrays for research use. Curr Protoc Hum Genet , Chapter 10, Unit 10 17. doi: 10.1002/0471142905.hg1007s39 [DOI] [PubMed] [Google Scholar]
- Turner JR (2020). Atlas of Intestinal Transport. https://www.jrturnerlab.com/database-viewer/atlas-of-intestinal-transport. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.