Here, we identified a novel circRNA, namely, circPIKfyve, that can act as a key regulator of the innate immune response in teleost fish. circPIKfyve acts as a molecular sponge by competitive adsorbing of miR-21-3p, thereby increasing the abundance of MAVS and activating the downstream NF-κB/IRF3 pathway to enhance the antiviral response. In addition, this study was the first to find that QKI protein is involved in regulating the formation of circPIKfyve in fish.
KEYWORDS: antiviral responses, circular RNA, MAVS, ceRNA, microRNA
ABSTRACT
Circular RNAs (circRNAs) are a class of widespread and diverse covalently closed circular endogenous RNAs that exert crucial functions in regulating gene expression in mammals. However, the function and regulation mechanism of circRNAs in lower vertebrates are still unknown. Here, we discovered a novel circRNA derived from PIKfyve, named circPIKfyve, that is related to the antiviral responses in teleost fish. The results showed that circPIKfyve plays essential roles in host antiviral immunity and inhibition of SCRV replication. Moreover, we also found that the antiviral effect inhibited by miR-21-3p could be reversed with the addition of circPIKfyve. Our data revealed that circPIKfyve is a competitive endogenous RNA (ceRNA) of MAVS by sponging miR-21-3p, leading to the activation of the NF-κB/IRF3 pathway, which then enhanced the innate antiviral responses. In addition, we found that the RNA binding protein QKI is involved in the formation and regulation of circPIKfyve. Our results provide strong evidence that circRNAs play a regulatory role in antiviral immune responses in teleost fish.
IMPORTANCE Here, we identified a novel circRNA, namely, circPIKfyve, that can act as a key regulator of the innate immune response in teleost fish. circPIKfyve acts as a molecular sponge by competitive adsorbing of miR-21-3p, thereby increasing the abundance of MAVS and activating the downstream NF-κB/IRF3 pathway to enhance the antiviral response. In addition, this study was the first to find that QKI protein is involved in regulating the formation of circPIKfyve in fish. The overall results of this study suggest that circPIKfyve plays an active regulatory role in the antiviral immune response of teleost fish.
INTRODUCTION
Innate immunity is the host’s initial defense system against pathogen invasion. Upon infection, viral nucleic acids are regarded as pathogen-associated molecular patterns (PAMPs), which are detected by pattern recognition receptors (PRRs) in host cells. To date, three classes of PRRs are involved in virus-specific component detection: Toll-like receptors (TLRs), retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs), and nucleotide oligomerization domain-like receptors (NLRs) (1, 2). Among them, RLR, the main PRR of RNA viruses that triggers antiviral responses, can trigger a series of signal cascades by recruiting the key adapter mitochondrial antiviral signaling protein (MAVS), thereby coordinating the production of type I interferons (IFN) and proinflammatory cytokines, ultimately suppressing viral replication (3–5). However, the excessive activation of MAVS-mediated antiviral responses will disrupt immune homeostasis. Thus, the signaling pathway should be precisely regulated to ensure that the invading viruses are eliminated while also avoiding excessive immune responses.
MAVS is also known as IFN-β promoter stimulator 1 (IPS-1), virus-induced signaling adaptor (VISA), and CARD adaptor-inducing IFN-β (Cardif) (5). The MAVS-mediated antiviral signaling pathway was initiated after RIG-I and MDA5 sense RNA viruses. This initiation of signaling drives interactions between the RLRs, MAVS, and corresponding proteins and organelles that mediate the activation of NF-κB and interferon regulatory factor 3/7 (IRF3/7), respectively (5, 6). In MAVS-deficient mice, the activation of IRF3 and NF-κB mediated by RIG-1 and MDA5 is impaired due to the absence of MAVS; therefore, these mice exhibit sensitivity to RNA viruses (6). Additionally, increasing evidence has shown that MAVS is also implicated in some other signaling, such as apoptosis and metabolic functions (7, 8). Similar to that in mammals, MAVS in teleost fish, which are lower vertebrates, also plays a key role in the host’s antiviral immune response (9). However, the related regulatory networks involved in MAVS-mediated IFN responses in lower vertebrates remain unknown.
Noncoding RNAs (ncRNAs) represent the majority of transcripts in cells. Circular RNAs (circRNAs) are a widespread form of ncRNAs in animal cells, which were originally discovered in the 1990s and were thought to be by-products of pre-mRNA missplicing (10). However, recent studies have shown that they are back-splicing products regulated by specific cis elements or trans factors (11–13). According to reports, the production of approximately one-third of these circRNAs is dynamically regulated by the RNA binding protein quaking (QKI). QKI binds to the recognition elements in the introns on both sides of the exons, causing the exons to approach each other and promote the biogenesis of circRNA (14, 15). Unlike linear RNA, circRNAs produced by the special splicing method have neither 5′–3′ polarity nor a polyadenylation tail, so they are more resistant to RNase R and could exist stably in the cytoplasm (16). Until now, circRNAs have been found to participate in various biological processes, including proliferation, invasion, and metastasis (17, 18). Emerging evidence suggests that circRNAs can act as microRNA (miRNA) sponges to regulate gene transcription and expression and can also interact or be translated with disease-related RNA binding proteins (RBPs) (19–21). Therefore, they quickly became a research hot spot in the field of RNA.
miRNAs are a class of short-chain ncRNAs composed of 18 to 25 nucleotides (22). miRNAs can act as a posttranscriptional regulator of target mRNA and participate in the regulation of various biological processes by inhibiting mRNA translation or promoting mRNA degradation, including development, apoptosis, proliferation, differentiation, and immune responses (23, 24). However, the upstream regulators of miRNAs are poorly understood. Recently, long noncoding RNAs (lncRNA), mRNAs, and pseudogenes have been hypothesized to have the ability to communicate and regulate each other through competitive combination of miRNA response elements (MREs) (25–28), which proves the new mechanism of gene regulation. It has been reported that circRNAs can also act as competitive endogenous RNA (ceRNA) to isolate miRNAs from their target genes (29, 30). For example, CIRS-7 was reported as the sponge of miRNA (31). Increasing evidence has shown that miRNA can bind to circRNAs and participate in the regulation of multiple physiological processes, such as development, immune responses, tumorigenesis, apoptosis, and viral infection (29–32). Although circRNAs function as an miRNA sponge has been studied in mammals, the circRNA-miRNA regulatory mechanism remains poorly understood in lower vertebrates, such as teleost fish.
As a representative population of lower vertebrates, fish is an important link in the evolution of invertebrates to vertebrates. Similar to higher vertebrates, it has a complete immune system. When a pathogen invades a host, it triggers multiple signal cascades in response to pathogen invasion, so it is considered an excellent biological model for immunological research (33). Growing evidence indicates that rhabdoviruses are one of the most significant viral pathogens in fish, causing severe hemorrhagic septicemia in freshwater and marine fish. Siniperca chuatsi rhabdovirus (SCRV), a single-stranded negative-sense RNA virus (34), is one of the main rhabdoviruses causing mass fish deaths, and its infection is characterized by the rapid occurrence of proinflammatory cytokine storms, which cause hemorrhagic sepsis and organ failure, leading to a large number of fish deaths (35). Recently, we conducted an in-depth study of the natural antiviral immunity in the teleost fish Miichthys miiuy (miiuy croaker), especially the function of miRNA in the immune response. It is still important to study the role of ncRNAs in antiviral immunity, which can help improve the level of diagnosis and treatment of fish diseases under viral infection.
In the present study, we found a novel immune-related circRNA in teleost fish, termed circPIKfyve, and we found that circPIKfyve could act as the endogenous competing RNA of miR-21-3p to promote the expression of MAVS, thereby modulating SCRV-induced antiviral immune responses. Meanwhile, we found that QKI is involved in regulating the production of circRNA in teleost fish for the first time. Our data not only elucidated the biological mechanism of the circRNA-miRNA-mRNA axis in the antiviral immune responses of fish but also provided new insights for the study of immune regulation in lower vertebrates.
RESULTS
Characterization of circPIKfyve involved in antiviral immunity.
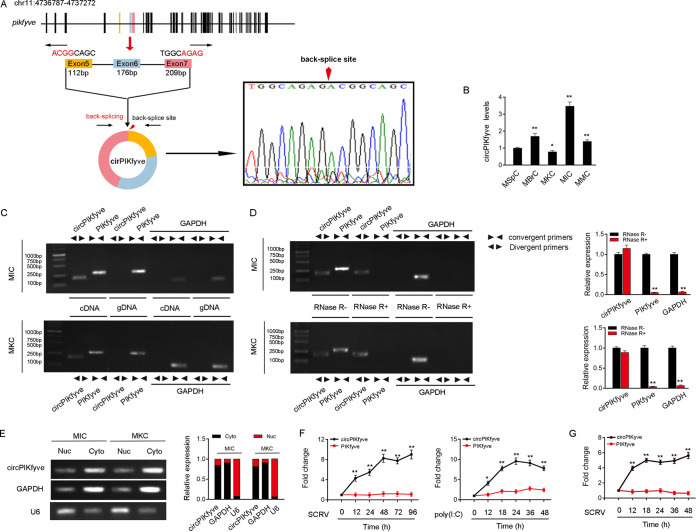
Several circRNAs are involved in the antiviral immune responses of mammals (10), but the role of circRNAs in the immune responses in lower vertebrates remains unclear. We used transcriptome sequencing (RNA-seq) data to compare the expression levels of circRNA after the infection of miiuy croaker with SCRV (GenBank accession no. PRJNA685924) and found that the expression of circPIKfyve was significantly upregulated after SCRV infection. Therefore, we focused on this gene for research. The bioinformatics prediction results showed that circPIKfyve is located in miiuy croaker chromosome 11 and consists of the head-to-tail splicing of exon 5 and exon 7, and the spliced mature sequence length is 490 bp (Fig. 1A, left). To determine whether the head-to-tail splicing is the result of trans-splicing or the genome rearrangement, we used several universal circRNA detection methods. We first designed divergent primers to amplify circPIKfyve. The result of Sanger sequencing confirmed the head-to-tail splicing in the reverse transcription-PCR (RT-PCR) product of circPIKfyve (Fig. 1A, right). We then evaluated the expression levels of circPIKfyve in untreated M. miiuy kidney cells (MKC), miiuy croaker muscle cells (MMC), miiuy croaker brain cells (MBrC), miiuy croaker intestine cells (MIC), and miiuy croaker spleen cells (MspC) (Fig. 1B). Among these cell lines, MIC and MKC showed the highest expression and the lowest expression of circPIKfyve, respectively. Therefore, we selected both MIC and MKC lines to investigate the function and regulatory mechanism of circPIKfyve. We used convergent primers to amplify the PIKfyve gene and divergent primers to amplify circPIKfyve. cDNA and genomic DNA (gDNA) were extracted separately from MKC and MIC and subjected to RT-PCR and agarose gel electrophoresis assays. The results shown in Fig. 1C indicated that circPIKfyve was amplified from cDNA by only divergent primers (an expected 178-bp fragment), whereas no amplification product was observed from gDNA. As the stability is considered one of the most crucial characteristics of circRNAs, we employed RNase R to confirm the cyclization of circPIKfyve. The results of RT-PCR and agarose gel electrophoresis showed that circPIKfyve, rather than linear PIKfyve or glyceraldehyde-3-phosphate dehydrogenase (GAPDH), resisted digestion by RNase R (Fig. 1D). In addition, we detected the subcellular location of circPIKfyve and found that circPIKfyve was mainly localized in the cytoplasm (Fig. 1E). These results suggested that circPIKfyve is a stable circRNA that is expressed and distributed mainly in the cytoplasm.
FIG 1.
Expression profiles and characterization of circPIKfyve. (A) We confirmed the head-to-tail splicing of circPIKfyve in the circPIKfyve RT-PCR product by Sanger sequencing. (B) Relative expression of circPIKfyve in indicated cell lines was determined by qPCR. (C) RT-PCR validated the existence of circPIKfyve in MIC and MKC cell lines. circPIKfyve was amplified by divergent primers in cDNA but not gDNA. GAPDH was used as a negative control. (D) The expression of circPIKfyve and linear PIKfyve mRNA in both MIC and MKC lines was detected by RT-PCR assay followed by nucleic acid electrophoresis or qPCR assay in the presence or absence of RNase R. (E) circPIKfyve was mainly localized in the cytoplasm, and we used GAPDH as the cytoplasmic marker and U6 snRNA as the nuclear marker. RNA isolated from the nucleus (Nuc) and cytoplasm (Cyto) was used to analyze the expression of circPIKfyve by nucleic acid electrophoresis (up) and qPCR (down); n = 3. (F) qPCR for the abundance of circPIKfyve and linear PIKfyve mRNA in spleen tissues treated with SCRV (MOI, 5) and poly(I·C) at the indicated time points, respectively. (G) qPCR analysis of circPIKfyve and linear PIKfyve mRNA in MKC treated with SCRV (MOI, 5) at the indicated time points. All data represented the means ± SE from three independent triplicate experiments. *, P < 0.05; **, P < 0.01.
To further confirm the reliability of RNA-seq data, we treated miiuy croaker with SCRV and poly(I·C) and sampled spleen tissues at different times to extract RNA. We then quantitatively analyzed the expression level of miiuy croaker circPIKfyve by quantitative real-time PCR (qPCR). Considering that circRNAs were produced by linear RNA splicing, the expression levels of linear PIKfyve and circPIKfyve were also detected. The qPCR results confirmed that circPIKfyve was significantly upregulated under SCRV and poly(I·C) stimulation compared with linear PIKfyve (Fig. 1F). In addition, SCRV-treated MKC further confirmed the significant expression of circPIKfyve (Fig. 1G). In conclusion, circPIKfyve is present in the cytoplasm and can participate in innate immune responses induced by SCRV.
circPIKfyve enhances host antiviral innate immunity.
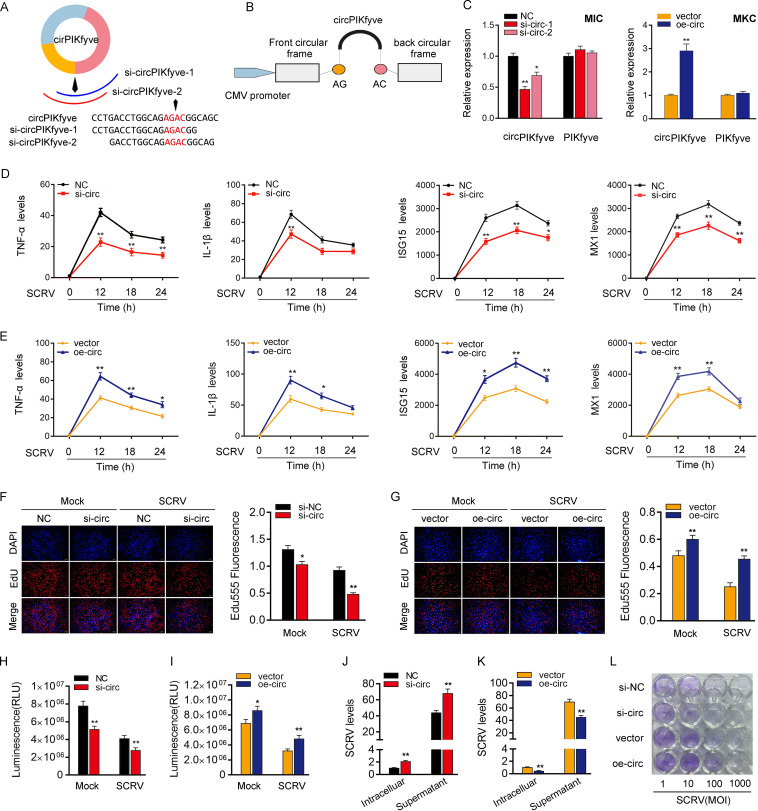
Small interfering RNAs (siRNAs) were designed against circPIKfyve, and the overexpression plasmid of circPIKfyve was constructed to detect the biological function of circPIKfyve (Fig. 2A and B). As expected, two siRNAs (si-circPIKfyve-1 and si-circPIKfyve-2) obviously decreased the expression level of circPIKfyve but did not affect the expression level of linear PIKfyve mRNA in MIC. In addition, si-circPIKfyve-1 can induce higher inhibitory efficiency. Thus, we chose si-circPIKfyve-1(si-circ) for the subsequent experiment (Fig. 2C). Meanwhile, the circPIKfyve overexpression plasmid was also successfully constructed, as it significantly increased the expression levels of circPIKfyve rather than linear PIKfyve mRNA in MKC cells (Fig. 2C). Considering that IFN-stimulated genes (ISGs) are important antiviral effectors, we investigated the role of circPIKfyve in regulating the expression of ISGs and inflammatory cytokines. As shown in Fig. 2D, upon SCRV infection, knockdown of circPIKfyve could significantly inhibit the expression levels of inflammatory cytokines and antiviral genes, such as those for tumor necrosis factor alpha (TNF-α), interleukin-1β (IL-1β), ISG15, and Mx1. In contrast, the overexpression of circPIKfyve (oe-circ) increased the expression levels of these genes under SCRV treatment (Fig. 2E). We conducted 5-ethynyl-2′-deoxyuridine (EdU) assays to examine the cell proliferation in MIC and MKC to further explore the function of circPIKfyve in antiviral innate immunity. The results showed the percentages of EdU-positive cells were considerably decreased with circPIKfyve knockdown (Fig. 2F) but greatly increased with circPIKfyve overexpression, suggesting circPIKfyve promotes proliferation in miiuy croaker cell lines (Fig. 2G). At the same time, when we investigated the effect of circPIKfyve on cell viability, we found that knockdown of circPIKfyve reduced cell viability and overexpression of circPIKfyve increased cell viability (Fig. 2H and I). In addition, we examined the effect of circPIKfyve on SCRV replication to explore the biological significance of circPIKfyve in SCRV-induced host cells. We found that circPIKfyve overexpression inhibited SCRV replication, whereas circPIKfyve knockdown promoted SCRV replication by detecting SCRV RNA (Fig. 2J and K). Additionally, SCRV replication was also monitored by the appearance of cytopathic effect (CPE) in epithelioma papulosum cyprini cells (EPC). As shown in Fig. 2L, the CPE is stronger when circPIKfyve was silenced, whereas overexpression of circPIKfyve attenuated the cytopathic effect. These results demonstrated that circPIKfyve can enhance SCRV replication. In summary, these data indicate that circPIKfyve, as a positive regulator, plays a crucial role in the regulation of antiviral immunity as well as cell proliferation and viability.
FIG 2.
circPIKfyve promotes antiviral innate immunity. (A and B) Schematic diagrams of siRNAs (A) and oe-circ structure (B). (C, left) qPCR analysis of circPIKfyve and linear PIKfyve mRNA in MIC treated with siRNAs. (Right) qPCR analysis of circPIKfyve and linear PIKfyve mRNA in MKC stably overexpressing circPIKfyve. (D and E) qPCR assays were performed to determine the expression levels of TNF-α, IL-1β, and ISG15 or MX1 in MIC transfected with si-circPIKfyve-1 (si-circ) or negative control (NC) (D) and MKC transfected with circPIKfyve overexpression plasmid (oe-circ) or control vector (vector) (E). (F and G) Cell proliferation was assessed by EdU assays in MIC transfected with si-circ or NC (F) and MKC transfected with oe-circ or vector (G). DAPI, 4′,6-diamidino-2-phenylindole. (H and I) Effect of circPIKfyve on cell viability after SCRV infection. MIC and MKC were transfected with si-circ (H) and oe-circ (I) for 48 h and then treated with SCRV for 24 h. Cell viability assays were used. RLU, relative light units. (J and K) circPIKfyve suppresses SCRV replication. MIC and MKC were transfected with NC or si-circ (J) and vector or oe-circ plasmid (K) for 48 h and then infected with SCRV. The qPCR analysis was conducted for intracellular and supernatant SCRV RNA expression. (L) EPC seeded in 48-well plates overnight were treated with cultural supernatants at the dose indicated for 48 h. The cell monolayers then were fixed with 4% paraformaldehyde and stained with 1% crystal violet. si-NC, si-circ, vector, or oe-circ was used. All data represent the means ± SE from three independent triplicate experiments. *, P < 0.05; **, P < 0.01.
circPIKfyve is able to regulate miR-21-3p expression and activity.
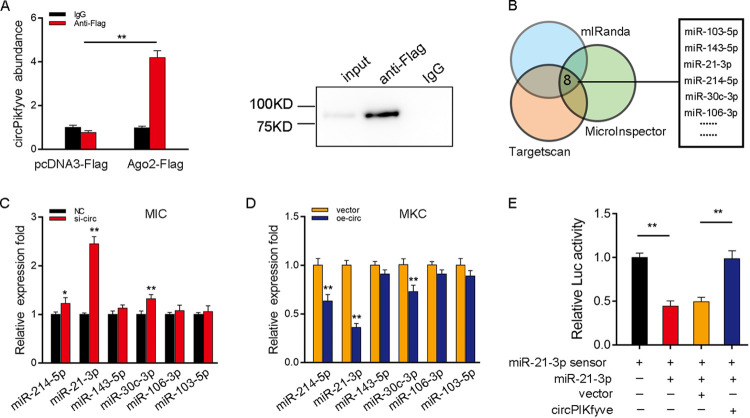
We examined the ability of circPIKfyve to bind to miRNAs in order to determine whether circPIKfyve can function as an miRNA sponge. To this end, we transfected Ago2-flag or pcDNA3.1-flag into MIC to conduct RNA immunoprecipitation (RIP) for Argonaute (Ago2). The results showed that endogenous circPIKfyve could be pulled down by Ago2-flag, and transfected Ago2-flag plasmid could increase the expression of Ago2 (Fig. 3A, left). Then we detected the Ago2 protein enrichment of input, anti-Flag, and IgG groups by Western blotting (Fig. 3A, right), and all of these results indicated that circPIKfyve has a binding site with miRNA. To further search for miRNAs combined with circPIKfyve, we first used miRNA target prediction tools, including TargetScan, miRanda, and MicroInspector, for prediction, and we selected 6 candidate miRNAs for further verification (Fig. 3B). Afterwards, we compared the expression levels of these candidate miRNAs in MIC transfected with si-circ or negative control and MKC transfected with circPIKfyve overexpression plasmid (oe-circ) or control vector. Among the six candidate miRNAs, miR-21-3p expression was significantly enhanced in response to circPIKfyve inhibition compared with other candidate miRNAs (Fig. 3C), whereas miR-21-3p expression was significantly reduced when circPIKfyve was overexpressed (Fig. 3D). To further consolidate the direct binding of miR-21-3p and circPIKfyve, we constructed the miR-21-3p sensor to detect whether circPIKfyve affects the activity of miR-21-3p. We transfected with the miR-21-3p sensor, along with miR-21-3p, control vector, or circPIKfyve overexpression plasmid, into EPC. The decreased luciferase activity induced by miR-21-3p was recovered when cotransfected with circPIKfyve overexpression plasmid, suggesting that circPIKfyve specifically sponged miR-21-3p, thereby preventing it from inhibiting luciferase activity (Fig. 3E). Taken together, these results suggested that circPIKfyve could regulate miR-21-3p expression and circPIKfyve functions as an miR-21-3p sponge.
FIG 3.
circPIKfyve regulates miR-21-3p expression and activity. (A) The Ago2-RIP assay for the amount of circPIKfyve in MIC transfected with Ago2-flag or pcDNA3.1-flag. We also detected the experimental group’s Western blot of Ago2 protein in input, anti-Flag, and IgG. (B) A schematic illustration showing overlapping of the target miRNAs of circPIKfyve predicted by TargetScan, miRanda, and MicroInspector. (C and D) Relative expression of candidate miRNAs in MIC and MKC transfected with si-circ (C) and oe-circ (D), respectively. (E) circPIKfyve reduces miR-21-3p activity. The relative luciferase activity was analyzed in MKC cotransfected with mimics, circPIKfyve overexpression plasmid, and control vector, together with miR-21-3p sensor. All data represent the means ± SE from three independent triplicate experiments. *, P < 0.05; **, P < 0.01.
cicrPIKfyve functions as an miRNA sponge of miR-21-3p.
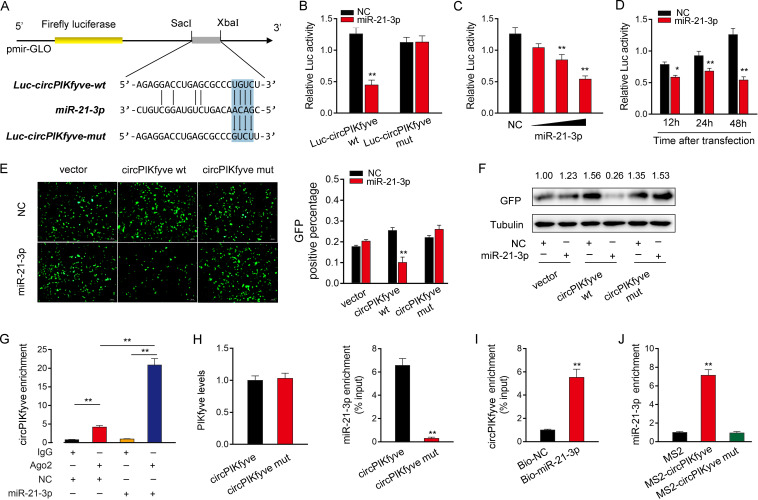
To investigate whether circPIKfyve could interact with miR-21-3p, we analyzed the sequences of circPIKfyve and found that circPIKfyve contained a binding site of miR-21-3p (Fig. 4A). Next, we constructed a luciferase plasmid of circPIKfyve (Luc-circPIKfyve-wt) and the mutated type with miR-21-3p binding sites mutated (Luc-circPIKfyve-mut) (Fig. 4A). Luciferase assays revealed that miR-21-3p can suppress the luciferase activity of the wild-type circPIKfyve luciferase plasmid, but it had no effect on the mutated type (Fig. 4B). Subsequently, we confirmed that miR-21-3p mimics inhibited luciferase activity in a dose- and time-dependent way (Fig. 4C and D). In addition, we inserted the wild or a mutated type of circPIKfyve into the mVenus-C1 vector and examined whether cotransfection with miR-21-3p could suppress the levels of green fluorescent protein (GFP). As shown in Fig. 4E and F, the results revealed that miR-21-3p could significantly inhibit the levels of GFP, which suggested that a direct interaction exists between circPIKfyve and miR-21-3p.
FIG 4.
circPIKfyve functions as a miRNA sponge of miR-21-3p. (A) Schematic illustration of circPIKfyve-wt and circPIKfyve-mut sequence cloned into pmirGLO luciferase reporter vectors. (B) The relative luciferase activities were detected in EPC after cotransfection with circPIKfyve-wt or circPIKfyve-mut and miR-21-3p mimic or NC. (C and D) The concentration gradient (C) and time gradient (D) experiments of miR-21-3p mimics were conducted. (E and F) circPIKfyve downregulated GFP expression. EPC were cotransfected with empty vector, circPIKfyve-wt or circPIKfyve-mut and miR-21-3p mimics, or NC. The fluorescence intensity and GFP expression were evaluated with an enzyme-labeling instrument and Western blotting, respectively. (G) The Ago2-RIP assay was executed in MIC after transfection with miR-21-3p mimics and NC, followed by qPCR to detect cirPIKfyve expression levels. (H and I) RNA pulldown assay was executed in MIC, followed by qPCR to detect the enrichment of miR-21-3p and circPIKfyve. (J) The MS2-RIP assay was executed in MIC after transfection with pcDNA3.1-MS2, pcDNA3.1-MS2-circPIKfyve, and pcDNA3.1-MS2-circPIKfyve-mut, followed by qPCR to detect the enrichment of miR-21-3p. All data represent the means ± SE from three independent triplicate experiments. *, P < 0.05; **, P < 0.01.
Given that miRNAs regulate target gene expression by binding to Ago2, we further tested the ability of circPIKfyve to bind to miR-21-3p. To this end, RIP assays were performed in MIC by cotransfecting Ago2-flag and miR-21-3p. The results from qPCR analysis indicated that circPIKfyve and miR-21-3p were efficiently pulled down by Ago2-flag (Fig. 4G). To further confirm the direct interaction between circPIKfyve and miR-21-3p, we conducted RNA pulldown assays with biotin-labeled circPIKfyve probe or biotin-labeled miR-21-3p, respectively. First, we detected the level of the PIKfyve mRNA after transfection with biotin-labeled circPIKfyve and circPIKfyve-mut and found there was no significant difference in the level of PIKfyve between the two groups (Fig. 4H, left). The results from qPCR analysis revealed that miR-21-3p could be pulled down by biotin-labeled circPIKfyve but not circPIKfyve-mut (Fig. 4H, right). Additionally, biotin-labeled miR-21-3p captured more circPIKfyve than the negative control (Fig. 4I). Furthermore, RIP assays were performed to test the direct interaction between circPIKfyve and miR-21-3p. To construct plasmids that can produce circPIKfyve identified by the MS2 protein, we cloned an MS2-12X fragment into pcDNA3.1, pcDNA3.1-circPIKfyve, and pcDNA3.1-circPIKfyve-mut plasmids. We also constructed a GFP and MS2 gene fusion expression vector to produce a GFP-MS2 fusion protein that can bind with the MS2-12X fragment and be identified using an anti-GFP antibody. Hence, miRNAs that interact with circPIKfyve can be pulled down by the GFP-MS2-circPIKfyve compounds. The results from qPCR assays showed that the pcDNA3.1-circPIKfyve RIP is significantly enriched for miR-21-3p compared with pcDNA3.1-circPIKfyve-mut or empty vector (Fig. 4J). Collectively, these data demonstrated that circPIKfyve can function as a sponge to bind to miR-21-3p.
miR-21-3p inhibits antiviral responses by targeting MAVS.
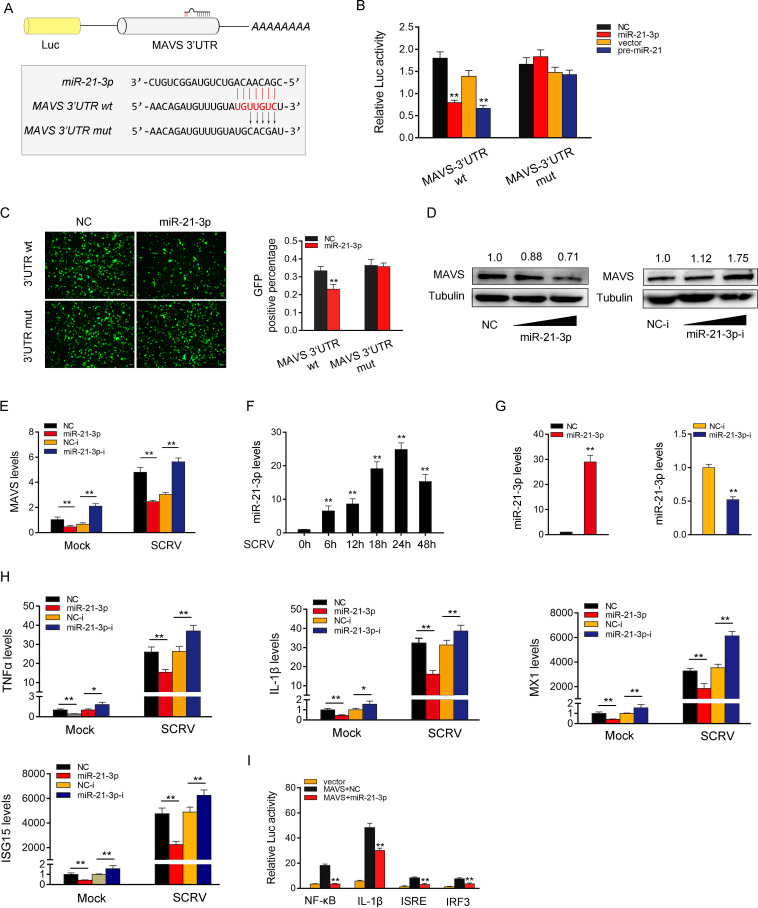
It is well known that miRNAs can posttranscriptionally regulate the expression of target mRNAs by binding to their 3′untranslated regions (3′-UTR). To this end, we predicted possible target genes of miR-21-3p through miRNA prediction programs. Among the candidate target genes, MAVS has been reported to be involved in innate antiviral responses (9). Therefore, we selected MAVS as the target gene of miR-21-3p for further study. In order to obtain direct evidence of miRNAs targeting the MAVS gene, we constructed MAVS-3′UTR wild-type and mutant-type reporter plasmids (Fig. 5A). We observed that miR-21-3p mimics and pre-miR-21 markedly inhibited the luciferase activity when cotransfected with the wild-type MAVS-3′UTR, whereas the MAVS-3′UTR mutant type led to a complete abrogation of the negative effect (Fig. 5B). To further confirm this result, we cloned the MAVS-3′UTR into the mVenus-C1 vector and explored the function of miR-21-3p on the expression of GFP. As shown in Fig. 5C, miR-21-3p mimics can downregulate GFP gene expression, and no change in fluorescence intensity was observed in EPC transfected with the mutant MAVS-3'UTR. Subsequently, we also investigated whether miR-21-3p regulates MAVS at the posttranscriptional level. As shown in Fig. 5D, miR-21-3p mimics reduced the expression level of MAVS in a dose-dependent manner. On the contrary, miR-21-3p inhibitor significantly increased the expression levels of MAVS in a dose-dependent manner. Meanwhile, further studies have shown that miR-21-3p affect the stability of MAVS mRNA expression (Fig. 5E). These results indicate that MAVS is a direct target of miR-21-3p, which regulates MAVS expression at the posttranscriptional level.
FIG 5.
miR-21-3p inhibits antiviral responses by targeting MAVS. (A) Schematic illustration of MAVS-3′-UTR wt and MAVS-3′UTR mut sequence cloned into luciferase reporter vectors. (B) The relative luciferase activities were detected in EPC after cotransfection with MAVS-3′-UTR wt or MAVS-3′UTR mut and mimics, pre-miR-21 plasmid, or control. (C) EPC were cotransfected with mVenus-MAVS-3′UTR wt or mutated mVenus-MAVS-3′UTR mut, together with NC and miR-21-3p. At 48 h posttransfection, the fluorescence intensity was evaluated by Varioskan LUX (Thermo). (D) Relative protein levels of MAVS were evaluated by Western blotting in MIC after cotransfection with the miR-21-3p mimics or inhibitors. (E) mRNA level of MAVS was evaluated by qPCR under SCRV. (F) The expression of miR-21-3p under different SCRV stimulation times by qPCR. (G) The effect of miR-21-3p mimics and inhibitors on endogenous miR-21-3p expression. MKC were transfected with NC or miR-21-3p (left) and NC-i and miR-21-3p-i (right) for 48 h, and then miR-21-3p expression was determined by qPCR. (H) MKC were transfected with NC, miR-21-3p, NC-i, or miR-21-3p-i. After 48 h posttransfection, the MIC were treated with SCRV for 24 h. The expression levels of TNF-α, IL-1β, Mx1, and ISG15 were analyzed by qPCR. (I) MKC were transfected with NC or miR-21-3p, together with MAVS expression plasmid, phRL-TK Renilla luciferase plasmid, and luciferase reporter genes. The luciferase activity was measured and normalized to Renilla luciferase activity. All data represent the means ± SE from three independent triplicate experiments. *, P < 0.05; **, P < 0.01.
We also detected the expression of miR-21-3p under different SCRV stimulation times by qPCR and found that miR-21-3p was significantly increased under SCRV stimulation (Fig. 5F). We measured the effects of synthetic miR-21-3p mimics and miR-21-3p inhibitors on the expression of miR-21-3p to explore the functional role of miR-21-3p in host antiviral responses. As expected, miR-21-3p mimics enhanced miR-21-3p expression sharply, whereas miR-21-3p inhibitors decreased miR-21-3p expression (Fig. 5G). The effect of miR-21-3p and SCRV on the expression patterns of the indicated genes were evaluated. The results showed that certain inflammatory cytokines and antiviral genes, including TNF-α, IL-1β, Mx1, and ISG15, were significantly decreased by the introduction of miR-21-3p mimics. On the contrary, the inhibition of endogenous miR-21-3p significantly elevated the expression of those genes compared with transfection of control inhibitors (Fig. 5H). Next, considering that miR-21-3p targets MAVS and regulates its expression, we attempted to investigate whether miR-21-3p affects MAVS-mediated activation of NF-κB and IRF3 signaling. The results showed that miR-21-3p mimics could inhibit the activation of NF-κB, IL-1β, ISRE, and IRF3 luciferases reporters (Fig. 5I). Collectively, these results indicate that MAVS is a direct target of miR-21-3p, and miR-21-3p participates in the regulation of antiviral responses by posttranscriptionally modulating the expression of MAVS.
circPIKfyve acts as an miR-21-3p sponge to enhance MAVS expression.
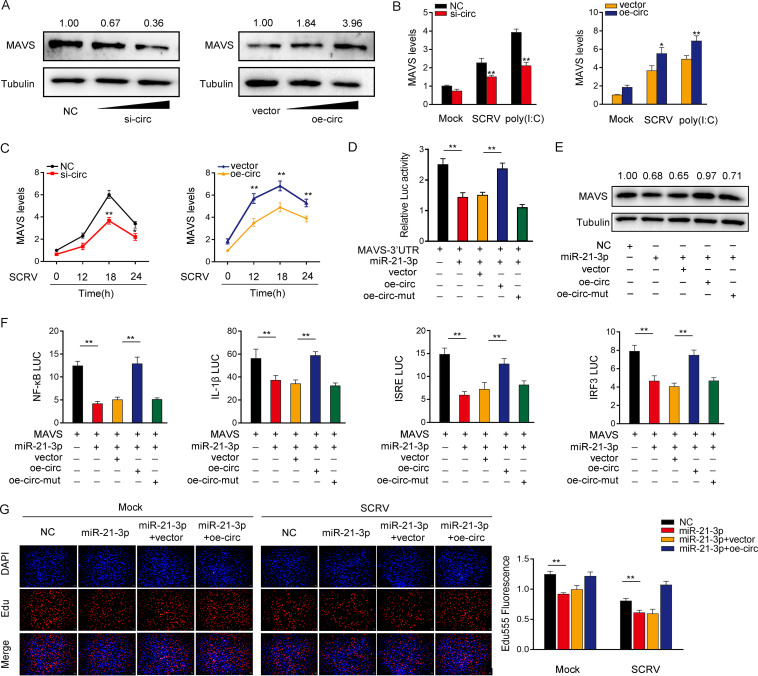
Given that circPIKfyve can interact with miR-21-3p and miR-21-3p targets MAVS and regulates its expression, we determined whether circPIKfyve can regulate MAVS. As shown in Fig. 6A, circPIKfyve knockdown significantly reduced the expression of MAVS protein, whereas the overexpression of circPIKfyve increased the expression of MAVS protein. Furthermore, the qPCR results showed that knockdown reduced the expression levels of MAVS in cells treated with SCRV or poly(I·C). In contrast, circPIKfyve overexpression increased the expression of MAVS at the mRNA level (Fig. 6B). Furthermore, overexpression of circPIKfyve increased the expression levels of MAVS in cells treated with SCRV, whereas knockdown of circPIKfyve reduced MAVS expression (Fig. 6C). We then determined whether circPIKfyve regulates MAVS expression through miR-21-3p. To this end, we cotransfected cells with the MAVS-3′UTR together with miR-21-3p, circPIKfyve overexpression plasmid, and circPIKfyve mutant plasmid (oe-circ-mut). The results showed that circPIKfyve could counteract the inhibitory effect of miR-21-3p on the MAVS-3′UTR (Fig. 6D). Strikingly, circPIKfyve could also counteract that effect of miR-21-3p on MAVS expression levels (Fig. 6E). These results demonstrated that circPIKfyve regulates MAVS expression through miR-21-3p. Given that miR-21-3p and MAVS participate in the regulation of NF-κB, IL-1β, ISRE, and IRF3 luciferase reporters, we examined the functional role of circPIKfyve in regulating these reporters. The results showed that circPIKfyve could counteract the negative effect of miR-21-3p on the luciferase activities of NF-κB, IL-1β, ISRE, and IRF3 luciferase reporters (Fig. 6F). Moreover, we attempted to explore the effect of the circPIKfyve/miR-21-3p regulatory loop on cell proliferation. The results indicated that circPIKfyve overexpression could counteract the negative effect of miR-21-3p on cell proliferation upon SCRV infection (Fig. 6G). Collectively, these data suggested that circPIKfyve is a ceRNA for miR-21-3p to regulate the expression of MAVS.
FIG 6.
circPIKfyve acts as a sponge of miR-21-3p to enhance MAVS expression. (A and B) Relative mRNA and protein levels of MAVS in MIC or MKC after cotransfection with NC, si-circ, vector, or oe-circ by Western blotting (A) and qPCR assays (B). (C) Relative mRNA levels of MAVS in MIC or MKC after cotransfection with NC, si-circ, vector, or oe-circ by qPCR assay. (D) The relative luciferase activities were detected in EPC after cotransfection with MAVS-3′UTR luciferase reporter vector, NC, mimics, oe-circ, or oe-circ-mut. (E) Western blot assays were performed in MKC after cotransfection with MAVS overexpression plasmid, NC, miR-21-3p mimics, oe-circ, or oe-circ-mut. (F) oe-circ counteracts the negative effect of miR-21-3p. Relative luciferase activities were detected in MIC after cotransfection with MAVS expression plasmid, pRL-TK Renilla luciferase plasmid, luciferase reporters, NC, mimics, or oe-circ. (G) Cell proliferation was assessed by EdU assays in MKC after cotransfection with NC, mimics, or oe-circ. All data represent the means ± SE from three independent triplicate experiments. *, P < 0.05; **, P < 0.01.
circPIKfyve is regulated by the RNA-binding protein QKI.
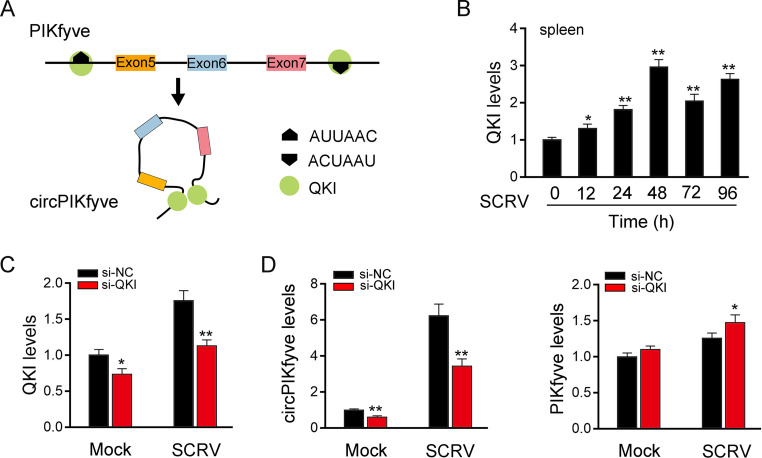
We then investigated which factors regulated circPIKfyve overexpression. According to reports, protein factors such as QKI, RNA binding motif protein 20 (RBM20), and the RNA editing enzyme ADAR contribute to the biogenesis of circRNAs in higher vertebrates (14, 36). However, in fish, the protein factors that affect the biogenesis of circRNA are unknown. Therefore, we analyzed the PIKfyve mRNA sequence in which the circPIKfyve sequence was located, as shown in Fig. 7A. We found that the intron upstream of exon 5 of PIKfyve contains the QKI binding site ACUAAU (14), and it was found that the downstream intron of PIKfyve exon 7 also contains the QKI binding site AUUAAC (37). This suggests that QKI is involved in the biogenesis of circPIKfyve. We focus on QKI and find that it is upregulated in SCRV-stimulated spleen (Fig. 7B). In addition, the expression levels of QKI and circPIKfyve in Fig. 7B and 1F decreased at 72 h and increased slightly at 96 h. We think that this is a dynamic equilibrium relationship. When the expression of QKI reaches the highest level 48 h after SCRV infection, QKI protein will accumulate in the cells with the passage of time. With the peak of QKI protein, the level of QKI mRNA will correspondingly decrease. With the passage of time, after the decrease of QKI, the expression of QKI protein will decrease. In short, we think that the relationship between the increase and decrease is a dynamic balance. To further investigate the function of QKI in circPIKfyve biogenesis, we silenced QKI and then detected the level of endogenous QKI expression. As shown in Fig. 7C, knockdown of QK1 can significantly downregulate the expression of QKI upon SCRV infection. On this basis, the results show that the expression of circPIKfyve is downregulated, while the expression level of linear PIKfyve is not affected after silencing of QKI in MIC (Fig. 7D). These results suggested that QKI is involved in the biosynthesis of circPIKfyve.
FIG 7.
circPIKfyve is regulated by the RNA-binding protein QKI. (A) Schematic diagram of QKI promoting circPIKfyve formation. (B) qPCR analysis of QKI mRNA in spleen cells treated with SCRV (MOI, 5) at the indicated time points. (C) qPCR analysis of QKI mRNA in MIC treated with si-QKI. (D) si-QKI regulates the mRNA expression of circPIKfyve upon SCRV infection. MIC were transfected with si-NC or si-QKI for 48 h and then treated with SCRV. The expression of circPIKfyve and linear circPIKfyve was analyzed by qPCR assays. All data represent the means ± SE from three independent triplicate experiments. *, P < 0.05; **, P < 0.01.
The ceRNA network of regulating MAVS is widely found in teleost fish.
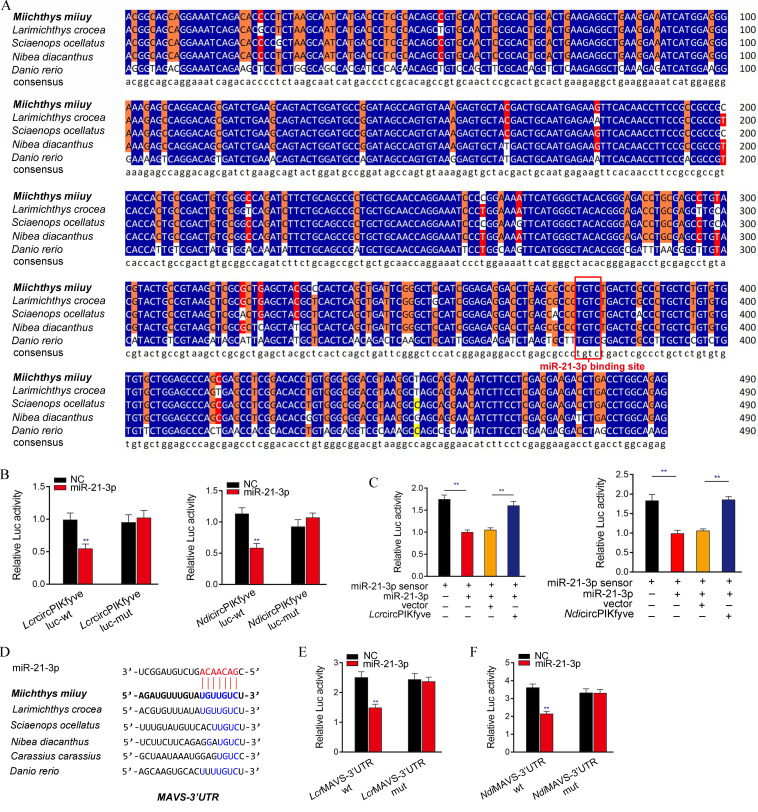
We performed the sequence alignment of circPIKfyve from different teleost fish to illustrate the generality of our findings. Interestingly, circPIKfyve displays high conservation in different fish species. In addition, the binding sites of miR-21-3p in circPIKfyve also showed high conservation (Fig. 8A). Therefore, we speculate that miR-21-3p interacts with circPIKfyve in different fish species. To verify this hypothesis, the circPIKfyve sequences of Larimichthys croceas and Nibea diacanthus were cloned into the pmirGLO vector and their mutated forms, with miR-21-3p binding sites mutated. Significantly, the luciferase assays revealed that miR-21-3p can suppress the luciferase activity of wild-type circPIKfyve luciferase plasmid in both fish species, but it had no effect on the mutated forms (Fig. 8B). Subsequent experiments were used to detect whether N. diacanthus and L. crocea circPIKfyve would affect the activity of miR-21-3p. As shown in Fig. 8C, the results showed that circPIKfyve in both types of fish could offset the inhibitory effect of miR-21-3p on the miR-21-3p sensor. These results suggested that circPIKfyve acts as a molecular sponge of miR-21-3p in other species. In addition, we also analyzed the binding sites of miR-21-3p and MAVS in other species (Fig. 8D). We also verified the finding that miR-21-3p targets the MAVS-3′UTR across species. Luciferase constructs were generated by cloning the MAVS-3′UTR of L. crocea and N. diacanthus into the pmirGLO vector in the mutant devoid of the miR-21-3p binding site as controls. Strikingly, miR-21-3p mimics were sufficient to decrease luciferase activities when cotransfected with the wild type of the MAVS-3′UTR of both fish species; however, it showed no effect on the luciferase activity of cells transfected with their mutant types (Fig. 8E and F). Taken together, these results showed that circPIKfyve could act as an endogenous RNA sponge to interact with miR-21-3p in different teleost fish (Fig. 9), which suggested that circPIKfyve contains strongly conserved elements among species, which is very important for preserving its function.
FIG 8.
ceRNA network of regulating MAVS is widely found in teleost fish. (A) Sequence alignment of circPIKfyve from various teleost fish species. (B) The relative luciferase activities were detected in EPC after cotransfection with LcrcircPIKfyve-wt or LcrcircPIKfyve-mut and mimics or NC (left) and EPC after cotransfection with NdicircPIKfyve-wt or NdicircPIKfyve-mut and mimics or NC (right). (C) LcrcircPIKfyve and NdicircPIKfyve reduce miR-21-3p activity. EPC were transfected with LcrcircPIKfyve or NdicircPIKfyve expression plasmid, mimics, and control vector, together with the miR-21-3p sensor. The luciferase activity was analyzed and normalized to Renilla luciferase activity. (D) Sequence alignment of MAVS containing miR-21-3p binding site among different teleost species. (E and F) miR-21-3p regulating the luciferase activity of MAVS-3′UTR is examined in N. diacanthus and L. crocea. All data represent the means ± SE from three independent triplicate experiments. **, P < 0.01.
FIG 9.
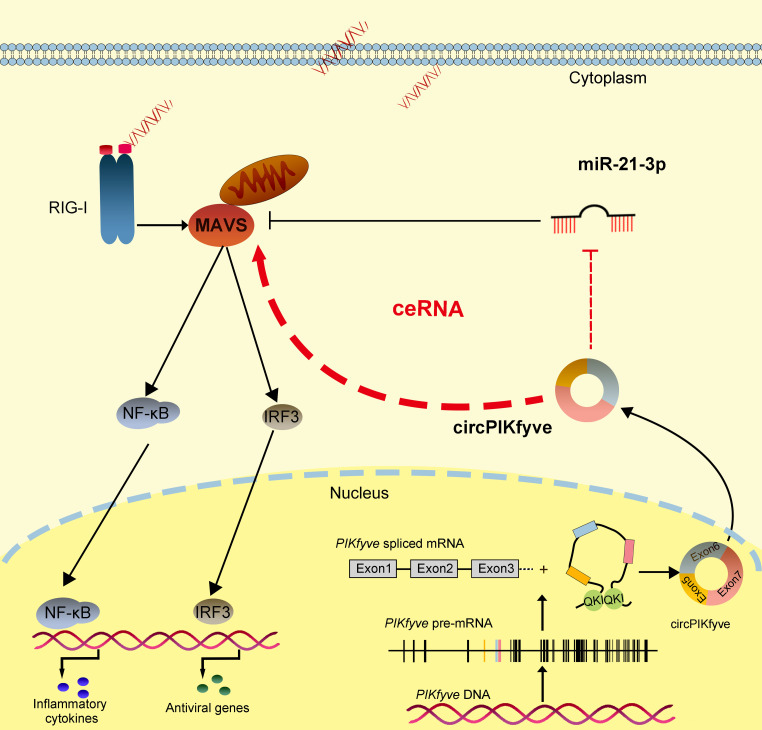
Schematic diagram of the mechanism underlying circPIKfyve as a ceRNA for miR-21-3p to regulate MAVS expression. miR-21-3p targets MAVS and represses MAVS-mediated antiviral responses, thereby regulating viral replication. circPIKfyve acts as a molecular sponge regulating miR-21-3p to enhance MAVS expression, thereby maintaining the stable antiviral responses and ensuring appropriate inflammatory responses.
DISCUSSION
In recent decades, the high fatality rate of aquatic organisms caused by viral infection has seriously restricted the development of aquaculture (35). The innate immune response of teleost plays a fundamental and central defensive role in viral infection (38). In recent years, with the development of immunology research of higher vertebrates, a new type of ncRNA-circRNA has been found to have excellent functional properties in immune responses (39). Similarly, we also found a large number of circRNAs in muiiy croaker. In this study, we discovered a new circRNA, circPIKfyve, that can participate in the regulation of innate immune responses. To this end, we constructed a circRNA-miRNA-mRNA regulatory network by regulating the expression of three genes, circPIKfyve, miR-21-3p, and MAVS, and then regulating signal pathways related to cellular immunity and viral pathogenesis. Our data also confirmed that miR-21-3p is a negative regulator of innate immune responses, which weakens the antiviral immune responses by targeting MAVS, thereby promoting virus replication. Furthermore, miR-21-3p also has been shown to be closely related to the antiviral immune response. In mammals, miR-21-3p can inhibit the production of type I IFN by targeting a variety of immune-related genes, thereby inhibiting the host's antiviral immune response (40). For example, miR-21-3p can target two important components, MyD88 and IRAK1 (41), in the TLR signaling pathway that leads to type I IFN production. However, fish and mammals are not exactly the same, and fish are specific in many ways, such as the presence of TLR4 not being found in most fish (42), in addition to TLR22 being unique to fish (43). These cases all indicate some differences between fish and mammals. We think this may be a survival strategy for the virus to escape the host's antiviral immune responses. circPIKfyve upregulates the expression of the immune-related gene MAVS by adsorbing miR-21-3p and enhances the antiviral signal pathway in order to reply to the escape mechanism of the virus.
MAVS is considered the key mediator of innate antiviral immune responses. When the host recognizes viral infection, RLRs, including RIG-I and MDA5, interact with MAVS to trigger the innate immune responses (6, 7). Most viruses have developed strategies to inhibit the activation of the RLR pathway to evade the host's antiviral immune responses (4). For example, hepatitis C virus (HCV) uses the protease NS3/4A to cleave the TM region of MAVS, causing MAVS to dissociate from the mitochondrial inner membrane and inhibiting the production of IFN and antiviral genes to escape the host's innate immune responses (44). In addition, the antiviral responses can be regulated by targeting antiviral genes with ncRNAs. For example, miR-3570 can inhibit the production of antiviral genes and promote viral replication by targeting MAVS (38). The lncRNA MARL enhances the expression of MAVS by adsorbing miR-122 and maintains the stability of the viral immune responses (9). N6-methylated circRNA inhibits the activation of the RIG-1 signaling pathway to resist the innate immune responses (45). The present study demonstrated that two ncRNAs, including miR-21-3p and circPIKfyve, play an important regulatory role in the host antiviral responses after RNA virus infection in teleost. Further research found that miR-21-3p has a negative regulatory effect on the viral signaling pathway mediated by MAVS. In contrast, circPIKfyve has a positive regulatory effect on the viral signaling pathway mediated by MAVS. The data indicated that the two ncRNAs could coordinately regulate MAVS expression to modulate the antiviral signaling pathway.
In eukaryotic cells, protein-coding RNA (mRNA) occupies only about 2% of the genome (46), and the remaining transcripts are classified as ncRNAs. ncRNAs, including miRNAs, lncRNAs, and circRNAs, have regulatory roles in gene expression (47). In recent decades, the mechanism and function of miRNAs have been deeply studied. However, research on the function of circRNAs is just beginning. Thus, understanding the biogenesis of circRNAs is important. Increasing evidence indicates that circRNA cyclization can aid ring formation by the close proximity of circRNA splice sites, which is mediated by complementary base pairing of inverted repeats in the introns flanking the circRNA-forming exons (48). The simplest example of this model of biogenesis is the interaction between QKI and the flanking introns of the forming circRNAs. For example, QKI binds upstream and downstream of the circRNA-forming exons in SMARCA5 to promote circRNA formation (14). In this study, it was found that QKI was related to the formation of circPIKfyve, and knockdown of QKI inhibited the expression of circPIKfyve, which showed biological effects similar to those of circPIKfyve knockdown.
The ceRNA hypothesis suggests that RNA transcripts, including mRNAs, lncRNAs, pseudogenes, and circRNAs, can cross talk and regulate the expression of miRNA through competition to bind miRNA responses elements (MREs), thereby constructing a new complex posttranscriptional regulatory network and mechanism (25). circRNAs can act as miRNA sponges to regulate the expression of mRNA target genes. Accumulating reports indicate that the ceRNA mechanism is the primary approach through which circRNAs perform their biological functions. For example, the circ-UBE2D2 sponge mechanism has been proven to promote breast cancer progression by adsorbing miR-1236 and miR-1287 (49). The overexpression of circHIPK3 can effectively inhibit the invasion and metastasis of bladder cancer cells by targeting miR-558 (50). Additionally, circRNAs_0005075 acts as a sponge for miR-431 and inhibits the progression of gastric cancer by regulating P53 (51). Genes regulated by differentially expressed circRNAs are involved in a variety of cellular processes, including apoptosis pathways, JAK-STAT, Wnt, TLRs, and RLR signaling pathways, all of which are related to cellular immunity and viral pathogenesis (52). Therefore, the regulatory mechanism of circRNAs in innate immune responses should be studied. It was reported that ssc_circ_009380 regulates transmissible gastroenteritis virus-induced inflammatory responses by targeting interleukin-6 through adsorbing miR-22 (53); circPIKfyve was predicted to contain miR-21-3p MREs. Therefore, we speculate that circPIKfyve exerts antiviral immune responses by binding miR-21-3p. Subsequently, a series of experiments, including biotin-labeled probe pulldown experiments, dual-luciferase reporter gene experiments, and RIP experiments, confirmed that circPIKfyve can directly interact with miR-21-3p. All experiments proved that circPIKfyve has a typical ceRNA mechanism and can be used as a sponge to absorb miR-21-3p, which indirectly enhanced the expression of MAVS and promoted the antiviral immune responses. Our current results show that circPIKfyve can indirectly target MAVS through miRNA to promote the production of antiviral genes, thereby resisting viral invasion and replication. This further proves that circRNAs participate in the regulation of immune responses as a new immunomodulatory molecule.
In conclusion, we found a new ncRNA, circPIKfyve, that is involved in the regulation of innate antiviral immune responses in teleost fish. Among them, miR-21-3p plays a negative regulatory role in MAVS-mediated antiviral responses. On the contrary, circPIKfyve plays a positive role in MAVS-mediated antiviral responses. We further confirmed the mechanism of the two ncRNAs plays a role in regulating the innate antiviral immune responses. circPIKfyve can be used as the ceRNA of miR-21-3p to reduce its inhibitory effect on MAVS expression, thereby inhibiting the replication of the virus. In addition, we found that QKI can promote the formation of circPIKfyve, which is consistent with studies in mammals that found QKI promotes the formation of circRNAs (14). In addition, we found that the structure and function of circPIKfyve are highly conserved in different teleost fish. In summary, our research reveals that circRNAs are involved in the host-virus interaction mechanism, which provides new insights into the role of circRNA in host antiviral immunity.
MATERIALS AND METHODS
Sample and challenge.
Miiuy croaker (∼50 g) was obtained from Zhoushan Fisheries Research Institute, Zhejiang Province, China. Fish were acclimated in aerated seawater tanks at 25°C for 6 weeks before experiments. SCRV and poly(I·C) challenges were performed as follows. Briefly, fish were challenged with 200 μl SCRV at a multiplicity of infection (MOI) of 5 and 200 μl poly(I·C) (2.5 mg/ml) through intraperitoneal injection. As a comparison, 200 μl of physiological saline was used as a control. Afterwards, fish were sacrificed at different time points, and the spleen tissues were collected for RNA extraction. All animal experimental procedures were performed in accordance with the National Institutes of Health’s Guide for the Care and Use of Laboratory Animals (54), and the experimental protocols were approved by the Research Ethics Committee of Shanghai Ocean University (no. SHOU-DW-2018-047).
Cell culture and treatment.
M. miiuy spleen cells (MspC), M. miiuy kidney cells (MKC), M. miiuy muscle cells (MMC), M. miiuy brain cells (MBrC), and M. miiuy intestine cells (MIC) were cultured in L-15 medium (HyClone) supplemented with 15% fetal bovine serum (FBS; Gibco), 100 U/ml penicillin, and 100 μg/ml streptomycin at 26°C. Epithelioma papulosum cyprini cells (EPC) were maintained in medium 199 (Invitrogen) supplemented with 10% FBS, 100 U/ml penicillin, and 100 mg/ml streptomycin at 28°C in 5% CO2. For stimulation experiments, MKC and MIC were challenged with SCRV at an MOI of 5 and harvested at different times for RNA extraction.
Plasmid construction.
To construct the MAVS-3′UTR reporter vector, the 3′UTR region of the M. miiuy MAVS gene, as well as L. crocea and N. diacanthus MAVS-3′UTR, were amplified using PCR and cloned into pmirGLO luciferase reporter vector (Promega). Meanwhile, the sequences of M. miiuy MAVS 3′UTR were inserted into mVenus-C1 vector (Invitrogen), which included the sequence of enhanced GFP. To construct circPIKfyve overexpression vector, the full-length circPIKfyve cDNA was amplified by specific primer pairs and cloned into pLC5-ciR vector (Geneseed Biotech), which contained a front and back circular frame to promote RNA circularization. The circPIKfyve overexpression vectors of L. crocea and N. diacanthus were constructed by synthesizing the full-length circPIKfyve cDNA of L. crocea and N. diacanthus, respectively. The empty vector with no circPIKfyve sequence was used as a negative control. The mutated forms with point mutations in the miR-21-3p binding site were synthesized using Mut Express II fast mutagenesis kit V2 with specific primers (Table 1). An miR-21-3p sensor was created by inserting two consecutive miR-21-3p complementary sequences into psiCHECK vector (Promega). The correct construction of the plasmids was verified by Sanger sequencing and extracted through an EndoFree plasmid DNA miniprep kit (Tiangen Biotech). To build pcDNA3.1-MS2, the MS2-12X fragment was inserted into the BamHI and EcoRV restriction sites of the pcDNA3.1 vector, and then circPIKfyve was amplified and cloned into pcDNA3.1-MS2.
TABLE 1.
PCR primer information in this study
| Primer | Sequence (5′–3′) |
|---|---|
| MAVS-qRT-F | AGGCACCAACAATTCCAG |
| MAVS-qRT-R | ACGGAGCAGGCTTCACTT |
| TNF-α-qRT-F | GTTTGCTTGGTACTGGAATGG |
| TNF-α-qRT-R | TGTGGGATGATGATCTGGTTG |
| IL-1β-qRT-F | TACGATGGCTAATAACTCC |
| IL-1β-qRT-R | CATTGACAAAGTGCTCCA |
| MX1-qRT-F | GCTGCTTGTTTACTCCCA |
| MX1-qRT-R | ACCTGCATCATCTCCCTC |
| ISG15-qRT-F | TGAACGGACAGAAGACGC |
| ISG15-qRT-R | TGAGGAATACCTGCATGG |
| circPIKfyve-divergent-F | TACGCCCACTCAGCTGAT |
| circPIKfyve-divergent-R | TTTCCTGCTGCCGTCTCT |
| circPIKfyve-convergent-F | ACAGAACCCAAAATTGCC |
| circPIKfyve-convergent-R | AACATCCCCGTATCATCC |
| miR-21-3p-qRT-F | CGACAACAGTCTGTAGGCT |
| miR-21-3p-qRT-R | GGTCCAGTTTTTTTTTTTTTTTCAATG |
| miR-214-5p-qRT-F | GCCTGTCTACACTTGCTG |
| miR-214-5p-qRT-R | GTCCAGTTTTTTTTTTTTTTTGCAC |
| miR-143-5p-qRT-F | TGCAGTGCTGCATCTC |
| miR-143-5p-qRT-R | GTCCAGTTTTTTTTTTTTTTTCCAGA |
| miR-30c-3p-qRT-F | GCTGGGAGAGGGGTGT |
| miR-30c-3p-qRT-R | TCCAGTTTTTTTTTTTTTTTAGCGT |
| miR-103-5p-qRT-F | GAGCCTCTTTACAGTGCTG |
| miR-103-5p-qRT-R | GGTCCAGTTTTTTTTTTTTTTTCAAG |
| 5.8S rRNA-qRT-F | AACTCTTAGCGGTGGATCA |
| 5.8S rRNA-qRT-R | GTTTTTTTTTTTTTTTGCCGAGTG |
| GAPDH-qRT-F | ACCTTCACTCCTCCATCTT |
| GAPDH-qRT-R | AGGTCACAGACACGGTTG |
| U6-qRT-F | TGCGAGTAGCAGACCA |
| U6-qRT-R | CACGAGACCGAAACAC |
| circPIKfyve-F | CGGAATTCTAATACTTTCAGACGGCAGCAGGAAATCAG |
| circPIKfyve-R | CGGGATCCAGTTGTTCTTACCTCTGCCAGGTCAGGTCTTC |
| circPIKfyve-pmirGLO-F | CGAGCTCACGGCAGCAGGAAATCAG |
| circPIKfyve-pmirGLO-R | TGCTCTAGACTCTGCCAGGTCAGGTCTTC |
| circPIKfyve-pmirGLO-mut-F | TGAGCGCCCGTCTTGACTCGCCCTGCTCTGTGT |
| circPIKfyve-pmirGLO-mut-R | AGTCAAGACGGGCGCTCAGGTCCTCTCCGATGG |
| circPIKfyve-mVenus-F | TCAGATCTCGAGCTCAAGCTTACGGCAGCAGGAAATCAGAC |
| circPIKfyve-mVenus-R | CGGGCCCGCGGTACCGTCGACCTCTGCCAGGTCAGGTCTTCC |
| circPIKfyve-mVenus-mut-F | TGAGCGCCCGTCTTGACTCGCCCTGCTCTGTGT |
| circPIKfyve-mVenus-mut-R | AGTCAAGACGGGCGCTCAGGTCCTCTCCGATGG |
| miR-21-3p sensor-F | TCGAGGCTGTTGTCAGACATCCGACAGGCTGTTGTCAGACATCCGACAGGC |
| miR-21-3p sensor-R | GGCCGCCTGTCGGATGTCTGACAACAGCCTGTCGGATGTCTGACAACAGCC |
| Ago2-Flag-F | CCCAAGCTTGACAAAATGTATTCCTCTGC |
| Ago2-Flag-R | CGCGGATCCTTTCATCAGTGGGGTCTC |
| circPIKfyve-T7-F | TAATACGACTCACTATAGGGGCAGCCGCTGCTGCAACC |
| circPIKfyve-T7-R | AGAAGATCTGGCCGCACAGTC |
| pcDNA3.1-MS2-circPIKfyve-F | ACTATAGGGAGACCCAAGCTTGCAGCCGCTGCTGCAACC |
| pcDNA3.1-MS2-circPIKfyve-R | GCGGCCGTTACTAGTGGATCCAGAAGATCTGGCCGCACAGTC |
| pcDNA3.1-MS2-circPIKfyve-mut-F | TGAGCGCCCGTCTTGACTCGCCCTGCTCTGTGT |
| pcDNA3.1-MS2-circPIKfyve-mut-R | AGTCAAGACGGGCGCTCAGGTCCTCTCCGATGG |
| MAVS-HindIII-F | GACGATGACGACAAGAAGCTTTCGTCTGCCAAAGACAAACTGTA |
| MAVS-EcoRI-R | TGATGGATATCTGCAGAATTCCAGCCTCTGTCCTGTCTACTTCATG |
| MAVS-3′UTR-F | CTAGCTAGCCGTATGGTGCCTTATTG |
| MAVS-3′UTR-R | TGCTCTAGACAGCCTCTGTCCTGTCTACT |
| MAVS-3′UTR-mut-F | CACGAATGAAGATCAGACTGATGACTGTCTGCA |
| MAVS-3′UTR-mut-R | GTCTGATCTTCATTCGTGTACAAACATCTGTTATTATTATCATCATTATTATT |
| LcrMAVS-3′UTR-F | CCGCTCGAGACCCTCCAGACCTTTGAT |
| LcrMAVS-3′UTR-R | GTCGACGTCGACGCTCCTCGTTAATCCTCA |
| LcrMAVS-3′UTR-mut-F | ATCGCATTGAAGATCAGACTGATGACTCGTGC |
| LcrMAVS-3′UTR-mut-R | CTGATCTTCAATGCGATTATAAACACGTCTATTATTATCATCTTTATTATTG |
| NdiMAVS-3′UTR-F | AGAGGAACTTGGTTAGGTACCGGGGCTTTAAAACCTTTAAAGGA |
| NdiMAVS-3′UTR-R | CAGGTCGACTCTAGACTCGAGTCATTTATTTATAATTTCAGGTTTATTGATG |
| NdiMAVS-3′UTR-mut-F | AGACGCTACACGTGAAGATCAGACTGATGACTGTCATG |
| NdiMAVS-3′UTR-mut-R | CTTCACGTGTAGCGTCTATTATTATTATTATCATCATTATTATTAACA |
| LcrcircPIKfyve-F | CGGAATTCTAATACTTTCAGACGGCAGCAGGAAATCAG |
| LcrcircPIKfyve-R | CGGGATCCAGTTGTTCTTACCTCTGCCAGGTCAGGTCTTC |
| LcrcircPIKfyve-pmirGLO-F | CGAGCTCACGGCAGCAGGAAATCAG |
| LcrcircPIKfyve-pmirGLO-R | TGCTCTAGACTCTGCCAGGTCAGGTCTTC |
| LcrcircPIKfyve-pmirGLO-mut-F | TGAGCGCCCGTCTTGACTCGCCCTGCTCTGTGT |
| LcrcircPIKfyve-pmirGLO-mut-R | AGTCAAGACGGGCGCTCAGGTCCTCTCCGATGG |
| NdicircPIKfyve-F | CGGAATTCTAATACTTTCAGACGGCAGCAGGAAATCAG |
| NdicircPIKfyve-R | CGGGATCCAGTTGTTCTTACCTCTGCCAGGTCAGGTCTTC |
| NdicircPIKfyve-pmirGLO-F | CGAGCTCACGGCAGCAGGAAATCAG |
| NdicircPIKfyve-pmirGLO-R | TGCTCTAGACTCTGCCAGGTCAGGTCTTC |
| NdicircPIKfyve-pmirGLO-mut-F | TGAGCGCCCGTCTTGACTCGCCCTGCTCTGTGT |
| NdicircPIKfyve-pmirGLO-mut-R | AGTCAAGACGGGCGCTCAGGTCCTCTCCGATGG |
RNA oligoribonucleotides.
The miR-21-3p mimics are synthetic double-stranded RNAs (dsRNAs) with stimulating naturally occurring mature miRNAs. The miR-21-3p mimic sequence was 5′-CGACAACAGUCUGUAGGCUGUC-3′. The negative control mimic sequence was 5′-UUCUCCGAACGUGUCACGUTT-3′. miRNA inhibitors are synthetic single-stranded RNAs (ssRNAs) that sequester intracellular miRNAs and block their activity in the RNA interfering pathway. The miR-21-3p inhibitor sequence was 5′-GCCAGCUUACAGACUGCUGUUG-3′. The negative-control inhibitor sequence was 5′-CAGUACUUUUGUGUAGUACAA-3′. The RNA interference sequences for circPIKfyve are the following: si-circPIKfyve-1, 5′-CCUGACCUGGCAGAGACGGTT-3′; si-circPIKfyve-2, 5′-GACCUGGCAGAGACGGCAGTT-3′. The scrambled control RNA sequence was 5′-CCUGACCUGGCAGAGCACUTT-3′. The si-QKI sequence was 5′-GACUCACAGCCAAACAGUUTT-3′. The negative control mimic sequence was 5′-UUCUCCGAACGUGUCACGUTT-3′.
Cell transfection.
Transient transfection of cells with miRNA mimic, miRNA inhibitor, or siRNA was performed in 24-well plates using Lipofectamine RNAiMAX (Invitrogen), and cells were transfected with DNA plasmids using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instructions. For functional analyses, the overexpression plasmid (500 ng per well) or control vector (500 ng per well) and miRNA mimic (100 nM), miRNA inhibitor (100 nM), or siRNA (100 nM) were transfected into cells in culture medium and then harvested for further detection. For luciferase experiments, miRNA mimic (100 nM) or miRNA inhibitor (100 nM) and pmirGLO (500 ng per well) containing the wild or mutated MAVS 3′UTR plasmid were transfected into cells.
RNA extract and quantitative real-time PCR.
For the isolation and purification of both cytoplasmic and nuclear RNA from MIC, the Cytoplasmic & Nuclear RNA purification kit was used according to the manufacturer’s instructions (Norgen Biotek). Total RNA was isolated with TRIzol reagent (Invitrogen), and the cDNA was synthesized using the FastQuant RT kit (Tiangen), which includes DNase treatment of RNA to eliminate genomic contamination. The expression patterns of each gene were performed by using SYBR Premix Ex Taq (TaKaRa). The small RNA was extracted by using an miRcute miRNA isolation kit (Tiangen), and the miRcute miRNA FirstStrand cDNA synthesis kit (Tiangen) was applied to the reverse transcription of miRNAs. The expression analysis of miR-21-3p was executed by using the miRcute miRNA qPCR detection kit (Tiangen). Real-time PCR was performed in an Applied Biosystems QuantStudio 3 (Thermo Fisher Scientific). GAPDH and 5.8S rRNA were employed as endogenous controls for mRNA and miRNA, respectively. Primer sequences are displayed in Table S1 in the supplemental material.
Luciferase reporter assay.
Wild-type circPIKfyve and the mutant devoid of the miR-21-3p binding site were cotransfected with miR-21-3p mimics into EPC. At 48 h posttransfection, reporter luciferase activities were measured using the dual-luciferase reporter assay system (Promega). To determine the functional regulation of circPIKfyve, cells were cotransfected with MAVS overexpression plasmid or circPIKfyve overexpression plasmid, together with NF-κB, IL-1β, ISRE, and IRF3 luciferase reporter gene plasmids, phRL-TK Renilla luciferase plasmid, and either miR-21-3p mimic or negative controls. At 48 h posttransfection, the cells were lysed for reporter activity using the dual-luciferase reporter assay system (Promega). miR-21-3p sensor was cotransfected with miR-21-3p mimics or circPIKfyve overexpression plasmid. At 48 h posttransfection, the cells were lysed for reporter activity. All the luciferase activity values were achieved against the renilla luciferase control. Transfection of each construct was performed in triplicate in each assay. Ratios of renilla luciferase readings to firefly luciferase readings were taken for each experiment, and triplicates were averaged.
Western blotting.
Cellular lysates were generated by using 1× SDS-PAGE loading buffer. Proteins were extracted from cells and measured with the bicinchoninic acid protein assay kit (Vazyme), subjected to SDS-PAGE (10%) gel, and transferred to polyvinylidene difluoride (Millipore) membranes by semidry blotting (Bio-Rad Trans Blot turbo system). The membranes were blocked with 5% bovine serum albumin (BSA). Protein was blotted with different antibodies. The antibody against MAVS was diluted 1:500 (Abcam), anti-Flag and anti-tubulin monoclonal antibody were diluted 1:2,000 (Sigma), and horseradish peroxidase (HRP)-conjugated anti-rabbit IgG or anti-mouse IgG (Abbkine) was diluted 1:5,000. The results were representative of three independent experiments. The immunoreactive proteins were detected by using WesternBright ECL (Advansta). Digital imaging was performed with a cold charge-coupled device camera.
RNase R treatment.
The RNAs (10 μg) from MIC and MKC were treated with RNase R (3 U/μg; Epicenter) and incubated for 30 min at 37°C. The treated RNAs then were reverse transcribed with divergent primer or convergent primer and detected by qPCR and RT-PCR assay, followed by nucleic acid electrophoresis.
Nucleic acid electrophoresis.
The cDNA and gDNA PCR products were investigated using 2% agarose gel electrophoresis with Tris-acetate-EDTA (TAE) running buffer. DNA was separated by electrophoresis at 100 V for 30 min. The DNA marker was Super DNA marker (100 to 10,000 bp) (CWBIO). The bands were examined by UV irradiation.
RNA pulldown assay.
circPIKfyve and circPIKfyve-mut, with miR-21-3p binding sites mutated, were transcribed in vitro. The two transcripts were biotin labeled with the T7 RNA polymerase and biotin RNA labeling mix (Roche), treated with RNase-free DNase I, and purified with the RNeasy minikit (Qiagen). The whole-cell lysates from MIC (∼1.0 × 107) were incubated with purified biotinylated transcripts for 1 h at 25°C. The complexes were isolated by streptavidin agarose beads (Invitrogen). RNA was extracted from the remaining beads, and qPCR was used to evaluate the expression levels of miRNAs.
To conduct pulldown assays with biotinylated miRNA, MIC were harvested at 48 h after transfection and then incubated on ice for 30 min in lysis buffer (20 mM Tris, pH 7.5, 200 mM NaCl, 2.5 mM MgCl2, 1 mM dithiothreitol, 60 U/ml Superase-In, 0.05% Igepal, protease inhibitors). The lysates were precleared by centrifugation for 5 min, and 50 μl of the sample was aliquoted for input. The remaining lysates were incubated with M-280 streptavidin magnetic beads (Sigma). To prevent nonspecific binding of RNA and protein complexes, the beads were coated with RNase-free BSA and yeast tRNA (both from Sigma). The beads were incubated for 4 h at 4°C and washed twice with ice-cold lysis buffer, three times with the low-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.0, and 150 mM NaCl), and once with the high-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.0, and 500 mM NaCl) (39). RNA was extracted from the remaining beads with TRIzol reagent (Invitrogen) and evaluated by qPCR.
RIP.
RIP experiments were performed by using the Magna RIP RNA-binding protein immunoprecipitation kit (Millipore) by following the manufacturer’s protocol. The Ago-RIP assay was conducted in MIC (∼2.0 × 107)-transfected Ago2-flag or pcDNA3.1-flag and miR-21-3p mimics or control mimics. After 48 h of transfection, the cell extract was incubated with magnetic beads conjugated with IgG and anti-Flag antibody (Sigma). RNA was extracted from the remaining beads, and qPCR was used to evaluate the expression levels of circPIKfyve.
The MS2-RIP assay was also conducted in MIC (∼2.0 × 107) transfected with pcDNA3.1-MS2, pcDNA3.1-MS2-circPIKfyve, pcDNA3.1-MS2-circPIKfyve-mut, or pMS2-GFP (Addgene). To construct plasmids that could produce circPIKfyve identified by the MS2 protein, an MS2-12X fragment was cloned into pcDNA3.1, pcDNA3.1-circPIKfyve, and mutated circPIKfyve plasmid. Furthermore, a GFP and MS2 gene fusion expression plasmid was constructed to produce a GFP-MS2 fusion protein that could bind with the MS2-12X fragment and be identified using an anti-GFP antibody (Abcam). After 48 h transfection, MIC were used in RIP assays via the Magna RIP RNA-binding protein immunoprecipitation kit (Millipore) and an anti-GFP antibody by following the manufacturer’s protocol. RNA was extracted from the remaining beads, and qPCR was used to evaluate the expression levels of miRNAs (55, 56).
EdU cell proliferation assay.
The EdU assay was performed to assess the proliferation of cells by using BeyoClick EdU cell proliferation kit with Alexa Fluor 555 (Beyotime) by following the manufacturer’s instructions. The EdU cell lines were photographed and counted under a Leica DMiL8_fluorescence microscope and evaluated by Thermo Scientific Varioskan LUX. These experiments were repeated three times.
Cell viability.
Cell viability was measured at 48 h after transfection in MKC and MIC with CellTiter-Glo luminescent cell viability assays (Promega) according to the manufacturer’s instructions.
Virus yield quantification.
Miiuy croaker MIC and MKC were transfected with oligonucleotides and plasmids and then infected with SCRV (MOI, 5). A volume of 0.1 ml of the cultural supernatant was then serially diluted on the monolayer of EPC, and EPC were seeded into 48-well plates 1 day before measurement. The cell monolayer was washed with PBS, fixed with 4% paraformaldehyde, and stained with 1% crystal violet.
Statistical analysis.
Data are expressed as the means ± standard errors (SE) from at least three independent triplicate experiments. Student's t test was used to evaluate the data. The relative gene expression data were acquired using the 2−ΔΔCT method, and comparisons between groups were analyzed by one-way analysis of variance (ANOVA) followed by Duncan’s multiple-comparison tests (57). A P value of <0.05 was considered significant.
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China (31822057) and the National Key Research and Development Project (2018YFD0900503).
We have no conflict of interest to declare.
REFERENCES
- 1.Kawasaki T, Kawai T, Akira S. 2011. Recognition of nucleic acids by pattern recognition receptors and its relevance in autoimmunity. Immunol Rev 243:61–73. doi: 10.1111/j.1600-065X.2011.01048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kawai T, Akira S. 2006. Innate immune recognition of viral infection. Nat Immunol 7:131–137. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- 3.Yoneyama M, Fujita T. 2009. RNA recognition and signal transduction by RIG-I-like receptors. Immunol Rev 227:54–65. doi: 10.1111/j.1600-065X.2008.00727.x. [DOI] [PubMed] [Google Scholar]
- 4.Yoneyama M, Fujita T. 2007. RIG-I family RNA helicases: cytoplasmic sensor for antiviral innate immunity. Cytokine Growth Factor Rev 18:545–551. doi: 10.1016/j.cytogfr.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 5.LeBerre M, Lamoureux A, Louise Y, Lauret E, Boudinot P, Brémont M. 2009. Mitochondrial antiviral signaling protein plays a major role in induction of the fish innate immune responses against RNA and DNA viruses. J Virol 83:7815–7827. doi: 10.1128/JVI.00404-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumar H, Kawai T, Kato H, Sato S, Takahashi K, Coban C, Yamamoto M, Uematsu S, Ishii K, Takeuchi O, Akira S. 2006. Essential role of IPS-1 in innate immune responses against RNA viruses. J Exp Med 203:1795–1803. doi: 10.1084/jem.20060792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guan K, Zheng Z, Song T, He X, Xu C, Zhang Y, Ma S, Wang Y, Xu Q, Cao Y, Li J, Yang X, Ge X, Wei C, Zhong H. 2013. MAVS regulates apoptotic cell death by decreasing K48-linked ubiquitination of voltage-dependent anion channel 1. Mol Cell Biol 33:3137–3149. doi: 10.1128/MCB.00030-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chao CC, Gutiérrez-Vázquez C, Rothhammer V, Mayo L, Wheeler MA, Tjon EC, Zandee SEJ, Blain M, de Lima KA, Takenaka MC, Avila-Pacheco J, Hewson P, Liu L, Sanmarco LM, Borucki DM, Lipof GZ, Trauger SA, Clish CB, Antel JP, Prat A, Quintana FJ. 2019. Metabolic control of astrocyte pathogenic activity via cPLA2-MAVS. Cell 179:1483–1498. doi: 10.1016/j.cell.2019.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chu Q, Xu T, Zheng W, Chang R, Zhang L. 2020. Long noncoding RNA MARL regulates antiviral responses through suppression miR-122-dependent MAVS downregulation in lower vertebrates. PLoS Pathog 16:e1008670. doi: 10.1371/journal.ppat.1008670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, Le Noble F, Rajewsky N. 2013. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 11.Cocquerelle C, Mascrez B, Hétuin D, Bailleul B. 1993. Mis-splicing yields circular RNA molecules. FASEB J 7:155–160. doi: 10.1096/fasebj.7.1.7678559. [DOI] [PubMed] [Google Scholar]
- 12.Chen LL, Yang L. 2015. Regulation of circRNAs biogenesis. RNA Biol 12:381–388. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang XO, Dong R, Zhang Y, Zhang JL, Luo Z, Zhang J, Chen JJ, Yang L. 2016. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res 26:1277–1287. doi: 10.1101/gr.202895.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips C, Roslan S, Schreiber A, Gregory P, Goodall G. 2015. The RNA binding protein quaking regulates formation of circRNAs. Cell 160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 15.Teplova M, Hafner M, Teplov D, Essig K, Tuschl T, Patel D. 2013. Structure-function studies of STAR family Quaking proteins bound to their in vivo RNA target sites. Genes Dev 27:928–940. doi: 10.1101/gad.216531.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeck WR, Sharpless NE. 2014. Detecting and characterizing circular RNAs. Nat Biotechnol 32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. 2013. Cell-type specific features of circular RNA expression. PLoS Genet 9:e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salzman J. 2016. Circular RNA expression: its potential regulation and function. Trends Genet 32:309–316. doi: 10.1016/j.tig.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rong D, Sun H, Li Z, Liu S, Dong C, Fu K, Tang W, Cao H. 2017. An emerging function of circRNA-miRNAs-mRNA axis in human diseases. Oncotarget 8:73271–73281. doi: 10.18632/oncotarget.19154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang M, Huang N, Yang X, Luo J, Yan S, Xiao F, Chen W, Gao X, Zhao K, Zhou H, Li Z, Ming L, Xie B, Zhang N. 2018. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene 37:1805–1814. doi: 10.1038/s41388-017-0019-9. [DOI] [PubMed] [Google Scholar]
- 21.Huang A, Zheng H, Wu Z, Chen M, Huang Y. 2020. Circular RNA-protein interactions: functions, mechanisms, and identification. Theranostic 10:3503–3517. doi: 10.7150/thno.42174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hatfield SD, Shcherbata HR, Fischer KA, Nakahara K, Carthew RW, Ruohola-Baker H. 2005. Stem cell division is regulated by the microRNA pathway. Nature 435:974–978. doi: 10.1038/nature03816. [DOI] [PubMed] [Google Scholar]
- 23.Yoshino H, Seki N, Itesako T, Chiyomaru T, Nakagawa M, Enokida H. 2013. Aberrant expression of microRNAs in bladder cancer. Nat Rev Urol 10:396–404. doi: 10.1038/nrurol.2013.113. [DOI] [PubMed] [Google Scholar]
- 24.Catto JW, Alcaraz A, Bjartell AS, De Vere White R, Evans CP, Fussel S, Hamdy FC, Kallioniemi O, Mengual L, Schlomm T, Visakorpi T. 2011. MicroRNA in prostate, bladder, and kidney cancer: a systematic review. Eur Urol 59:671–681. doi: 10.1016/j.eururo.2011.01.044. [DOI] [PubMed] [Google Scholar]
- 25.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. 2011. A ceRNA hypothesis: the Rosetta stone of a hidden RNA language? Cell 146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang W, Du WW, Li X, Yee AJ, Yang BB. 2016. Foxo3 activity promoted by non-coding effects of circular RNA and Foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene 35:3919–3931. doi: 10.1038/onc.2015.460. [DOI] [PubMed] [Google Scholar]
- 27.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. 2013. Natural RNA circles function as efficient microRNA sponges. Nature 495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 28.Wang K, Liu F, Zhou LY, Long B, Yuan SM, Wang Y, Liu CY, Sun T, Zhang XJ, Li PF. 2014. The long noncoding RNA CHRF regulates cardiac hypertrophy by targeting miR-489. Circ Res 114:1377–1388. doi: 10.1161/CIRCRESAHA.114.302476. [DOI] [PubMed] [Google Scholar]
- 29.Guo LL, Song CH, Wang P, Dai LP, Zhang JY, Wang KJ. 2015. Competing endogenous RNA networks and gastric cancer. World J Gastroenterol 21:11680–11687. doi: 10.3748/wjg.v21.i41.11680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qi X, Lin Y, Chen J, Shen B. 2020. Decoding competing endogenous RNA networks for cancer biomarker discovery. Brief Bioinform 21:441–457. doi: 10.1093/bib/bbz006. [DOI] [PubMed] [Google Scholar]
- 31.Hansen TB, Kjems J, Damgaard CK. 2013. Circular RNA and miR-7 in cancer. Cancer Res 73:5609–5612. doi: 10.1158/0008-5472.CAN-13-1568. [DOI] [PubMed] [Google Scholar]
- 32.Liu M, Luo C, Dong J, Guo J, Luo Q, Ye C, Guo Z. 2020. CircRNAs_103809 suppresses the proliferation and metastasis of breast cancer cells by sponging microRNA-532-3p (miR-532-3p). Front Genet 11:485. doi: 10.3389/fgene.2020.00485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu T, Chu Q, Cui J, Huo R. 2018. MicroRNA-216a inhibits NF-κB-mediated inflammatory cytokine production in teleost fish by modulating p65. Infect Immun 86:e00256-18. doi: 10.1128/IAI.00256-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tao JJ, Gui JF, Zhang QY. 2007. Isolation and characterization of a rhabdovirus from co-infection of two viruses in mandarin fish. Aquaculture 262:1–9. doi: 10.1016/j.aquaculture.2006.09.030. [DOI] [Google Scholar]
- 35.Zhang Q, Gui JF. 2015. Virus genomes and virus-host interactions in aquaculture animals. Sci China Life Sci 58:156–169. doi: 10.1007/s11427-015-4802-y. [DOI] [PubMed] [Google Scholar]
- 36.Khan MA, Reckman YJ, Aufiero S, van den Hoogenhof MM, van der Made I, Beqqali A, Koolbergen DR, Rasmussen TB, van der Velden J, Creemers EE, Pinto YM. 2016. RBM20 regulates circular RNA production from the titin gene. Circ Res 119:996–1003. doi: 10.1161/CIRCRESAHA.116.309568. [DOI] [PubMed] [Google Scholar]
- 37.Zong FY, Fu X, Wei WJ, Luo YG, Heiner M, Cao LJ, Fang Z, Fang R, Lu D, Ji H, Hui J. 2014. The RNA-binding protein QKI suppresses cancer-associated aberrant splicing. PLoS Genet 10:e1004289. doi: 10.1371/journal.pgen.1004289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu T, Chu Q, Cui J, Bi D. 2017. Inducible microRNA-3570 feedback inhibits the RIG-I-dependent innate immune responses to rhabdovirus in teleost fish by targeting MAVS/IPS-1. J Virol 92:e01594-17. doi: 10.1128/JVI.01594-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi J, Hu N, Mo L, Zeng Z, Sun J, Hu Y. 2018. Deep RNA sequencing reveals a repertoire of human fibroblast circular RNAs associated with cellular responses to herpes simplex virus 1 infection. Cell Physiol Biochem 47:2031–2045. doi: 10.1159/000491471. [DOI] [PubMed] [Google Scholar]
- 40.Shi J, Feng P, Gu T. 2020. MicroRNA-21-3p modulates FGF2 to facilitate influenza A virus H5N1 replication by refraining type I interferon response. Biosci Rep 40:BSR20200158. doi: 10.1042/BSR20200158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Y, Chen J, Wang H, Shi J, Wu K, Liu S, Liu Y, Wu J. 2013. HCV-induced miR-21 contributes to evasion of host immune system by targeting MyD88 and IRAK1. PLoS Pathog 9:e1003248. doi: 10.1371/journal.ppat.1003248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alexander R, Tom G, Hans-Martin S. 2010. Toll-like receptor signaling in bony fish. Vet Immunol Immunopathol 134:139–150. doi: 10.1016/j.vetimm.2009.09.021. [DOI] [PubMed] [Google Scholar]
- 43.Matsuo A, Oshiumi H, Tsujita T, Mitani H, Kasai H, Yoshimizu M, Matsumoto M, Seya T. 2008. Teleost TLR22 recognizes RNA duplex to induce IFN and protect cells from birnaviruses. J Immunol 181:3474–3485. doi: 10.4049/jimmunol.181.5.3474. [DOI] [PubMed] [Google Scholar]
- 44.Ferreira AR, Magalhães AC, Camões F, Gouveia A, Vieira M, Kagan JC, Ribeiro D. 2016. Hepatitis C virus NS3-4A inhibits the peroxisomal MAVS-dependent antiviral signaling responses. J Cell Mol Med 20:750–757. doi: 10.1111/jcmm.12801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen YG, Chen R, Ahmad S, Verma R, Kasturi SP, Amaya L, Broughton JP, Kim J, Cadena C, Pulendran B, Hur S, Chang HY. 2019. N6-methyladenosine modification controls circular RNA immunity. Mol Cell 76:96–109. doi: 10.1016/j.molcel.2019.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann Y, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, International Human Genome Sequencing Consortium. 2001. Initial sequencing and analysis of the human genome. Nature 409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 47.Qu Z, Adelson D. 2012. Evolutionary conservation and functional roles of ncRNA. Front Genet 3:205. doi: 10.3389/fgene.2012.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dragomir M, Calin G. 2018. Circular RNAs in cancer—lessons learned from microRNAs. Frontiers Oncol 8:179. doi: 10.3389/fonc.2018.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Y, Li J, Du C, Zhang L, Zhang Y, Zhang J, Wang L. 2019. Upregulated circular RNA circ-UBE2D2 predicts poor prognosis and promotes breast cancer progression by sponging miR-1236 and miR-1287. Transl Oncol 12:1305–1313. doi: 10.1016/j.tranon.2019.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li Y, Zheng F, Xiao X, Xie F, Tao D, Huang C, Liu D, Wang M, Wang L, Zeng F, Jiang G. 2017. CircHIPK3 sponges miR-558 to suppress heparinase expression in bladder cancer cells. EMBO Rep 18:1646–1659. doi: 10.15252/embr.201643581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu J, Chen Z, Song Y, Zhu Y, Dou G, Shen X, Zhou Y, Jiang H, Li J, Peng Y. 2020. CircRNAs_0005075 suppresses carcinogenesis via regulating miR-431/p53/epithelial-mesenchymal transition axis in gastric cancer. Cell Biochem Funct 38:932–942. doi: 10.1002/cbf.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Awan FM, Yang BB, Naz A, Hanif A, Ikram A, Obaid A, Malik A, Janjua HA, Ali A, Sharif S. 2020. The emerging role and significance of circular RNAs in viral infections and antiviral immune responses: possible implication as theranostic agents. RNA Biol 18:1–15. doi: 10.1080/15476286.2020.1790198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ma X, Zhao X, Zhang Z, Guo J, Guan LJ, Li J, Mi M, Huang Y, Tong D. 2018. Differentially expressed non-coding RNAs induced by transmissible gastroenteritis virus potentially regulate inflammation and NF-κB pathway in porcine intestinal epithelial cell line. BMC Genomics 19:747. doi: 10.1186/s12864-018-5128-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed. National Academies Press, Washington, DC. [Google Scholar]
- 55.Wu XS, Wang F, Li HF, Hu YP, Jiang L, Zhang F, Li ML, Wang XA, Jin YP, Zhang YJ, Lu W, Wu WG, Shu YJ, Weng H, Cao Y, Bao RF, Liang HB, Wang Z, Zhang YC, Gong W, Zheng L, Sun SH, Liu YB. 2017. Lnc RNA-PAGBC acts as a micro RNA sponge and promotes gallbladder tumorigenesis. EMBO Rep 18:1837–1853. doi: 10.15252/embr.201744147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fu T, Ji K, Jin L, Zhang J, Wu X, Ji X, Fan B, Jia Z, Wang A, Liu J, Bu Z, Ji J. 2021. ASB16-AS1 up-regulated and phosphorylated TRIM37 to activate NF-κB pathway and promote proliferation, stemness, and cisplatin resistance of gastric cancer. Gastric Cancer 24:45–55. doi: 10.1007/s10120-020-01096-y. [DOI] [PubMed] [Google Scholar]
- 57.Livak KJ, Schmittgen T. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]