If combined with immune therapeutics, latency-reversing agents (LRAs) have the potential to reduce the size of the reservoir sufficiently that an engineered immune response can control the virus in the absence of antiretroviral therapy. We have identified a new class of LRAs that do not induce T-cell activation and that are able to potentiate, rather than inhibit, CD8+ T and NK cell cytotoxic effector functions.
KEYWORDS: HIV cure, HIV-1, HIV latency reversion, SIV, in vivo study
ABSTRACT
An ability to activate latent human immunodeficiency virus type 1 (HIV-1) expression could benefit many HIV cure strategies; however, most latency-reversing agents (LRAs) have proven disappointing. We evaluated AKT/mTOR activators as a potential new class of LRAs. Two glycogen synthase kinase-3 inhibitors (GSK-3i’s), SB-216763 and tideglusib (the latter already in phase II clinical trials), that activate AKT/mTOR signaling were tested. These GSK-3i’s reactivated latent HIV-1 present in blood samples from aviremic individuals on antiretroviral therapy (ART) in the absence of T cell activation, release of inflammatory cytokines, cell toxicity, or impaired effector function of cytotoxic T lymphocytes or NK cells. However, when administered in vivo to simian immunodeficiency virus (SIV)-infected rhesus macaques on suppressive ART, tideglusib exhibited poor pharmacodynamic properties and resulted in no clear evidence of significant SIV latency reversal. Whether alternative pharmacological formulations or combinations of this drug with other classes of LRAs will lead to an effective in vivo latency-reversing strategy remains to be determined.
IMPORTANCE If combined with immune therapeutics, latency-reversing agents (LRAs) have the potential to reduce the size of the reservoir sufficiently that an engineered immune response can control the virus in the absence of antiretroviral therapy. We have identified a new class of LRAs that do not induce T-cell activation and that are able to potentiate, rather than inhibit, CD8+ T and NK cell cytotoxic effector functions. This new class of LRAs corresponds to inhibitors of glycogen synthase kinase-3. In this work, we have also studied the effects of one member of this drug class, tideglusib, in SIV-infected rhesus monkeys. When tested in vivo, however, tideglusib showed unfavorable pharmacokinetic properties, which resulted in lack of SIV latency reversal. The disconnect between our ex vivo and in vivo results highlights the importance of developing novel LRAs with pharmacological properties that allow systemic drug delivery in relevant anatomical compartments harboring latent reservoirs.
INTRODUCTION
Despite its success, antiretroviral therapy (ART) is not curative; treatment must be continued for life. Human immunodeficiency virus (HIV) persists during ART in long-lived, latently infected CD4+ T cells. These cells typically express little or no viral protein unless activated, and thus, they escape immune recognition and clearance. One proposed strategy to eliminate or reduce the size of the latent reservoir involves initial activation of latent virus with small-molecule compounds termed latency-reversing agents (LRAs) (1). Since levels of virus production may not be sufficient to elicit a viral cytopathic effect, it may be necessary to actively promote immune clearance of these reactivated reservoir cells (2). Since only a small fraction of the cells are activated with each exposure to LRAs (3), it will be necessary to identify LRAs that can be safely administered in a repetitive manner. Unfortunately, most LRAs have proven ineffective or too toxic for serial use in vivo.
LRAs alone are unlikely to promote sharp decreases in the size of the latent reservoir needed to achieve a sustained viral remission in the absence of ART. LRAs will likely need to be combined with immune-based interventions, including vaccines or broadly neutralizing antibodies (bNAbs) (4). Combination strategies that both reactivate latent HIV and at the same time support elimination of those cells by immune-mediated approaches could reduce and control the latent reservoir (5). One key to this combination strategy is the identification of an effective LRA that can be safely administered multiple times and that does not compromise natural killer (NK) cell and cytotoxic T lymphocyte (CTL) action (6, 7). The ultimate aim would be to combine reservoir reduction with immune interventions in a manner that allows discontinuation of ART without viral rebound.
One approach to efficiently reactivate latent HIV in vitro in CD4+ T cells involves activation of the T-cell receptor (TCR) signaling cascade. Stimulation of the TCR ultimately results in the activation of several transcription factors, including, among others, the nuclear factor of activated T-cells (NFAT) and the nuclear factor κB (NF-κB), that in turn dynamically control transcriptional activity of the HIV-long terminal repeat region (LTR) (8). Furthermore, the multiple downstream phosphorylation events that follow TCR engagement induce an overall change in metabolic state of the cell as it moves from a resting catabolic state to one of growth and proliferation (9). This metabolic reprogramming has as its key regulators AKT (protein kinase B [PKB]) and the mammalian target of rapamycin (mTOR) (10). AKT and mTOR are both serine/threonine protein kinases involved in the regulation many cellular processes other than metabolism, such as cell growth, survival, proliferation, and apoptosis (11). mTOR participates in two multiprotein complexes, mTORC1 and mTORC2, as determined by association with different adapter and scaffolding proteins (12). When mTORC2 becomes activated (e.g., during TCR engagement), it phosphorylates AKT on S473, thus activating this kinase (13). Phosphorylated AKT, in turn, promotes NF-κB activity by phosphorylating and activating IκB kinase (IKK), leading in turn to IκBα degradation and translocation of NF-κB p50/p65 heterodimers, where they engage cognate κB enhancers (14, 15). Not surprisingly, replication of HIV-1 is tightly connected to AKT/mTOR functionalities (16); in fact, pharmacological inhibition of mTOR represses viral replication in vitro (17, 18).
Given the ability of AKT to induce NF-κB and promote, rather than impair, cell survival (19), we sought to identify new molecular targets within the AKT-regulated glycolytic pathway that induce NF-κB activation without inducing toxicity or overt cell activation. We focused on glycogen synthase-kinase 3 (GSK-3) as key regulator of the mTOR pathway. GSK-3 is a serine/threonine kinase that exists in two isoforms, α and β, that are structurally similar but not functionally identical (20). GSK-3 is constitutively active in quiescent (nonactivated) T cells and negatively regulates the mTOR pathway by introducing an inhibitory phosphorylation in Rictor, an accessory protein of mTORC2 (21), and stimulating the tuberous sclerosis 1-2 complex (TSC1-TSC2), a negative regulator of mTORC1 (22).
In the past decade, inhibiting GSK-3 activity via the introduction of small molecules has become an important strategy for treating neurodegenerative and psychiatric disorders (23, 24), mostly because of their ability to reduce tau phosphorylation. Interestingly, one GSK-3 inhibitor (GSK-3i), tideglusib, has already been proven safe for treatment in humans in a phase IIb human trial for treatment of congenital myotonic dystrophy (25), and it is also being explored as a treatment for autism (clinicaltrials.gov identification no. NCT02586935). Data from clinical studies have shown tideglusib to be generally safe and well tolerated in humans and also shown effects consistent with penetration into human brain following oral administration (26, 27).
In this work, we explored the potential of GSK-3i’s as LRAs in CD4+ T cells isolated from HIV-positive individuals on suppressive ART, while also testing their toxicity, T cell-activating properties, and effects on CTLs and NK cells. We went on to test the potential latency reversal effects of tideglusib in vivo in simian immunodeficiency virus (SIV)-infected rhesus macaques (RM) on suppressive ART.
RESULTS
GSK-3i’s induce nontoxic, noninflammatory latency reversion in CD4+ T cells isolated from blood of HIV-infected individuals on suppressive ART.
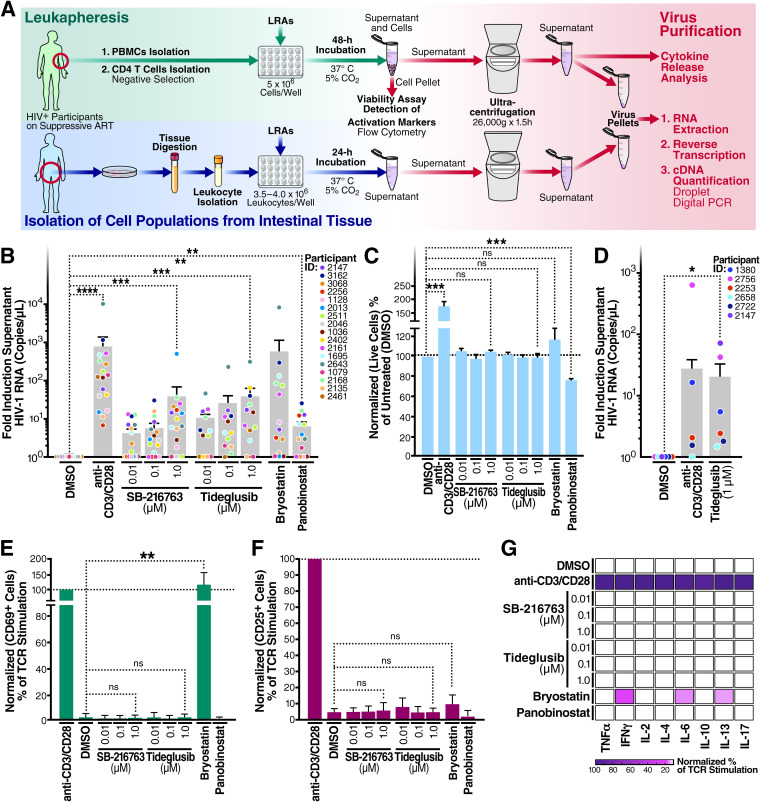
In Fig. 1A, we provide a schematic overview for our studies aimed at analyzing latency reversal in cells from 17 HIV-infected individuals on ART exposed to various candidate LRAs, including tideglusib and SB-216763 (GSK-3i’s). To test whether inhibition of GSK-3 leads to HIV latency reversal ex vivo, 5 × 106 total CD4+ T cells isolated from aviremic HIV-infected individuals on ART (n = 17) (Fig. 1B and Table 1) were stimulated with either (i) tideglusib, a GSK-3β inhibitor and activator of the AKT/mTOR pathway, (ii) SB-216763, an inhibitor of both GSK-3α and -β and activator of the AKT/mTOR pathway (28) (used here as a tool compound), (iii) anti-CD3/CD28-coated beads, to mimic TCR activation (used here as positive control for highly effective latency reversion), (iv) two previously studied LRAs (bryostatin and panobinostat), or (v) dimethyl sulfoxide (DMSO), a diluent control. After 48 h of stimulation, virions released into the culture supernatants were collected, and HIV RNA was quantified with droplet digital PCR (ddPCR) (Fig. 1B). For specific viral RNA quantification, we employed a primer/probe set that detects the 3′ sequence common to all completed HIV-1 RNAs (29).
FIG 1.
Inhibition of GSK-3 reverses latency in blood and gut tissue from HIV-positive individuals on ART studied ex vivo. (A) Schematic representation of the workflow followed for the analysis of latency reversion in blood and GALT cultures measured as HIV-1 virion levels (i.e., numbers of HIV-1 RNA copies per microliter) in the culture supernatant of blood or GALT samples. (B) Blood CD4+ T cells treated for 48 h with a single LRA or anti-CD3/CD28-coated beads. (C) Normalized percentage of live CD4+ T cells compared to untreated controls. (D) GALT samples were treated for 24 h with 1 μM tideglusib or anti-CD3/CD28 coated beads. (E and F) LRA-stimulated CD4+ T cells used for panels B and C were tested for surface expression of CD69 and CD25 activation markers. Lymphocytes were gated on single cells > live > CD3+ and CD69+ or CD25+. (G) After virus purification, culture supernatants of treated CD4+ T cells were tested for cytokine release using a proinflammatory panel including tumor necrosis factor alpha (TNF-α), IFN-γ, IL-2, IL-4, IL-6, IL-10, IL-13, and IL-17. Latency-reversal data are presented as fold induction relative to the untreated control (DMSO). Unless specified in the figure, treatments were done at the following concentrations/volumes: DMSO control, 0.01% (vol/vol) final volume; anti-CD3/CD28 coated beads, 125 μl (25 μl per 1 × 106 cells); bryostatin, 10 nM; panobinostat, 50 nM. Statistical significance was calculated from the HIV-1 RNA copy number values using a ratio paired t test compared with the DMSO control (*, P < 0.05; **, P < 0.001; ***, P < 0.0001; ****, P < 0.00001; ns, not significant). Error bars represent standard errors of the means.
TABLE 1.
Characteristics of HIV-1-positive study participantsa
| Participant ID | Age | Sex | Ethnicity | CD4 count (cells/μl) | ART regimen | ART start date (yr) |
|---|---|---|---|---|---|---|
| 2147 | 59 | Male | Asian | 597 | RPV/TDF/FTC | 1995 |
| 3162 | 54 | Male | White | 734 | RTV, DRV, ABC/TCV/3TC | 2000 |
| 3068 | 62 | Male | White | 452 | ABC/3TC, ETV, RGV | 2000 |
| 2256 | 60 | Male | White | 315 | EFV/TDF/FTC, RGV | 2001 |
| 1128 | 66 | Male | African American | 480 | 3TC, EFV, RGV | 2008 |
| 2013 | 68 | Male | White | 715 | ABC/TCV/3TC | 1996 |
| 2511 | 48 | Male | White | 334 | EFV/TDF/FTC, RGV | 2010 |
| 2046 | 52 | Male | Pacific Islander | 580 | ECV, EFV/TDF/FTC | 1995 |
| 1036 | 48 | Male | African American | 410 | EGV/TAF/FTC/COBI | 2008 |
| 2402 | 40 | Male | African American | 765 | RTV, DRV, RPV/TDF/FTC | 2006 |
| 1602 | 39 | Male | Native American | 459 | RPV/TDF/FTC | 2011 |
| 2643 | 55 | Male | White | 322 | ABC/TCV/3TC | 2017 |
| 1695 | 39 | Female | Mixed | 556 | FTC/TDF, ATV, RTV | 2010 |
| 2161 | 68 | Male | White | 687 | 3TC, RTV, DRV, TCV | 2005 |
| 1079 | 66 | Male | Latino | 481 | ETV, FTC/TAF | 2004 |
| 2135 | 60 | Male | White | 328 | RPV/TAF/FTC, RGV | 2006 |
| 2168 | 57 | Male | African American | 272 | ABC/TCV/3TC | 2000 |
| 2461 | 63 | Male | White | 496 | ABC/TCV/3TC | 1999 |
| 1380 | 60 | Female | African American | 760 | RPV/TAF/FTC | 2011 |
| 2658 | 39 | Male | Mixed | 614 | EFV/TDF/FTC | 2011 |
| 2722 | 56 | Male | Latino | 987 | FTC/TAF | 2016 |
| 2253 | 67 | Male | White | 643 | ABC/TCV/3TC | 2006 |
| 2756 | 45 | Male | Mixed | 799 | ABC/3TC, RTV, DRV | 2008 |
RPV, rilpivirine; TDF, tenofovir; FTC, emtricitabine; DRV, darunavir; ABC, abacavir; TCV, dolutegravir; 3TC, lamivudine; ETV, etravirine; RGV, raltegravir; EFV, efavirenz; ECV, entecavir; EGV, elvitegravir; TAF, tenofovir alafenamide; COBI, cobicistat; ATV, atazanavir; RTV, ritanovir.
Quantitating viral RNA in released virions may serve as a better proxy for the functional, infectious reservoir than cell-associated viral RNA, since many defective proviruses can transcribe viral RNA but not assemble into virions that successfully bud. Anti-CD3/anti-CD28 antibodies, the positive control in these studies, induced a 769.5-fold increase in supernatant HIV RNA compared to the DMSO diluent control. Treatment with 10 nM bryostatin induced a 574.4-fold increase in supernatant viral RNA, while treatments with 1 μM SB-216763 and 1 μM tideglusib caused a 38.6- and 38.3-fold change, respectively, over untreated controls (approximately 5% of the anti-CD3/anti-CD28 response). When the data were reanalyzed and expressed as median values of fold induction of released virion RNA, treatment with 1 μM tideglusib resulted in the release of 2-fold more virus-associated HIV-1 RNA than bryostatin (median values: 1 μM tideglusib, 10.3; bryostatin-1, 5.086), suggesting that different donors responded more uniformly to tideglusib than to treatment with bryostatin.
To assess the ability of tideglusib to activate latent proviruses from tissues, we conducted ex vivo stimulation assays using gut-associated lymphoid tissue (GALT) cultures (Fig. 1D). Cultures of GALT isolated from HIV-positive individuals on suppressive ART were treated with either anti-CD3/CD28-coated beads, 1 μM tideglusib, or DMSO diluent control. Of note, GALT cultures are extremely delicate and show decline in cell viability even if untreated for more than 1 day. For this reason, latency reversal in the GALT cultures was conducted for only 24 h. Supernatants of samples treated with tideglusib contained approximately 20 times more virus than untreated samples, suggesting that tideglusib is effective as an LRA in tissues, although these results must be considered preliminary given the small sample size.
Analysis of CD4+ T-cell viability using the CellTiter-Blue assay (Fig. 1C) confirmed by live/dead flow cytometric staining (data not shown) revealed that treatment with GSK-3 inhibitors or bryostatin did not alter cell viability. In comparison, treatment with panobinostat induced approximately a 30% decline in CD4+ T-cell viability within 48 h after treatment initiation.
To assess potential changes in cellular activation, CD4+ T cells were stimulated for 48 h with the LRAs. SB-216763 and tideglusib did not enhance expression of the CD69 or CD25 activation markers over the DMSO control (Fig. 1E and F). Conversely, activation of cells with bryostatin markedly increased CD69 surface expression (Fig. 1E). When cytokine production was assessed, the GSK-3i’s did not stimulate cytokine production in contrast to strong induction of cytokines by anti-CD3/anti-CD28 and more modest effects by bryostatin (Fig. 1G).
Together, these findings suggest that tideglusib and SB-216763 act as latency-reversing agents effectively promoting HIV virion production in culture of CD4+ T cells from aviremic individuals on suppressive ART. Latency reversal is achieved without changes in cell viability or induction of T cell activation and proinflammatory cytokine production. All of these favorable properties increased our interest in this class of LRA.
Inhibition of GSK-3 induces activation of several components of the AKT/mTOR pathway.
GSK-3 is constitutively active in resting CD4+ T cells (30) and naturally prevents activation of AKT/mTOR by (i) phosphorylating and inhibiting Rictor in the mTORC2 complex (21) and (ii) activating TSC2, thereby inhibiting the mTORC1 complex (31).
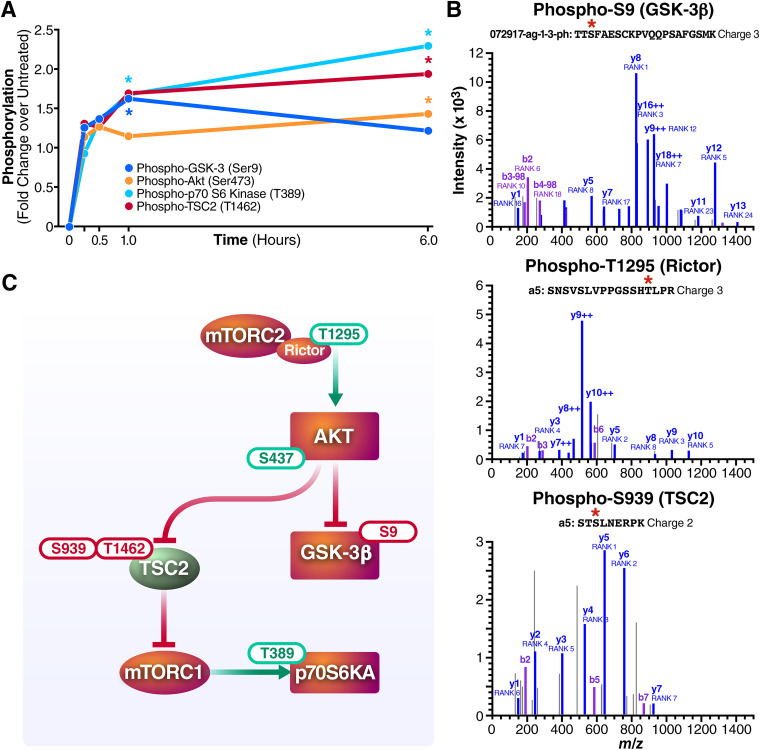
To explore how the AKT/mTOR pathway is modulated by the GSK-3i’s, we employed NanoString direct hybridization-based digital counting technology. NanoString allows both quantification of phosphorylation of a broad panel of proteins and assessment of expression of 700 unique genes at the mRNA level. CD4+ T cells from HIV-positive individuals on ART (n = 5) were treated with 1 μM tideglusib for 0 to 6 h with sample harvesting at 0, 15, 30, 60, and 360 min after stimulation. As early as 15 min after treatment, phosphorylation levels of phospho-GSK-3β, AKT, p70 S6 kinase (p70S6K), and TSC2 were detected (Fig. 2A). At 1 h after treatment, phospho-S9 on GSK-3β, phospho-T389 on p70S6K, and phospho-T1426 on TSC2 showed high levels of phosphorylation. Phosphorylation levels of p70S6K and TSC2 continued to increase in the following 6 h after treatment initiation, while levels of phospho-S9 on GSK-3β appeared to slowly decline. Detection of all these phosphorylations, particularly phosphorylation of p70S6K on T389, suggested activation of the mTOR pathway (32).
FIG 2.
Tideglusib activates the AKT/mTOR pathway in CD4+ T as detected by NanoString and whole-cell phosphoproteomics analysis. Tideglusib influences activation status of specific components of the AKT/mTOR pathway. (A) NanoString analysis of CD4+ T cells isolated from five HIV-infected individuals on suppressive ART treated with 1 μM tideglusib for up to 6 h. The differential expression of phospho-GSK3β (dark blue), phospho-AKT (orange), phospho-p70 S6 kinase (light blue), and phospho-TSC2 (red) is represented as linear fold change versus the untreated control (DMSO). Stars indicate points with P values of ≤0.05. Each data point is derived from 5 biological replicates (n = 5). (B) MS/MS spectra identifying GSK-3β S9 (TTS*FAESCKPVQQPSAFGSMK), TSC2 S939 (STS*LNERPK), and Rictor T1295 (SNSVSLVPPGSSHT*LPR). Phosphorylation was annotated by the Skyline spectrum builder, and y fragment ions are indicated in purple and blue, respectively. Fragment ion charge states are 1+ unless indicated otherwise. (C) Schematic representation of the phosphorylation sites detected by NanoString and phosphoproteomics on the respective protein/protein complexes.
To confirm the NanoString results and obtain further insights into how inhibition of GSK-3 acts as an LRA in CD4+ T cells, we performed an unbiased phosphoproteomics analysis of CD4+ T cells stimulated with tideglusib for 15 min (Fig. 2B). Analysis of the phosphospectra confirmed phosphorylation of GSK-3β on S9 as well as phosphorylation of Rictor on T1295 and TSC2 on S939. Together with phospho-T1426 on TSC2, phospho-S939 is another target of phosphorylation mediated by AKT (11); these two residues, once phosphorylated, prevent TSC2 from inhibiting mTORC1, consequently allowing mTORC1-mediated activation of p70S6K. Phosphorylation of T1295 on Rictor is an uncharacterized phosphorylation previously shown to be present in TCR-stimulated CD4+ T cells (33). Interestingly, GO enrichment analysis of phosphoproteomics data confirmed activation of components downstream of the TCR signaling cascade (Fig. 3C). Figure 2C shows a graphical summary of the phospho-sites detected in the NanoString and phosphoproteomics analysis, together with predicted protein-protein interactions. These results demonstrate that treatment of CD4+ T cells with tideglusib “disinhibits” the AKT/mTOR pathway, leading to activation of downstream substrates like p70S6 kinase.
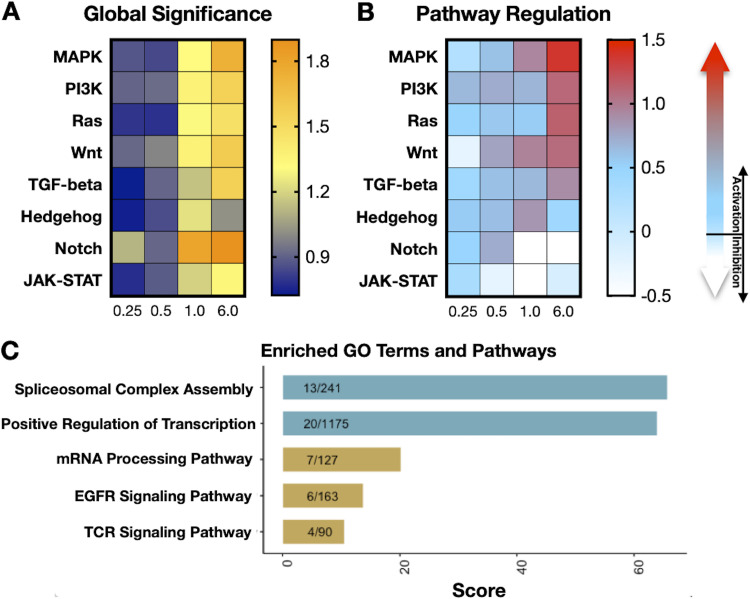
FIG 3.
Pathway regulation as induced by treatment with tideglusib in CD4+ T cells from HIV-positive donors on ART. (A) Heat map displaying global significance statistics and (B) pathway regulation obtained with the NanoString software following analysis of 700 RNA transcripts isolated from CD4+ T cells treated with 1 μM tideglusib for 0.25 to 6 h. A detailed list of the transcripts associated with each pathway is reported in the manufacturer’s protocol of the solid tumor assay (NanoString Technologies). The “global significance” of the differential expression of the analyzed RNA transcripts compared to the untreated control for each time point after treatment is reported with a color-coded score: a high score (orange) indicates transcripts that exhibit highly significant differential expression, while a low score (blue) indicates less significant differential expression. Regulation of selected pathways is reported with a color-coded score: light blue to red indicates pathway activation; light blue to white indicates pathway inhibition. (C) Tideglusib activates genes involved in splicing and positive regulation of transcription. A set of 142 gene names was extracted from the differential analysis of 1 μM tideglusib versus untreated phosphoproteomics results using cutoffs of log2 fold change of >1 or <−1 and a P value of <0.05. The gene set was then submitted to Enrichr for enrichment analysis. Tabular enrichment results obtained were downloaded into WikiPathways and Gene Ontology:Biological Process, and selected terms were plotted using R based on relevance, uniqueness, and combined score.
Tideglusib induces NF-κB binding with target DNA sequences and reverses latency in an NF-κB- and mTOR-dependent manner.
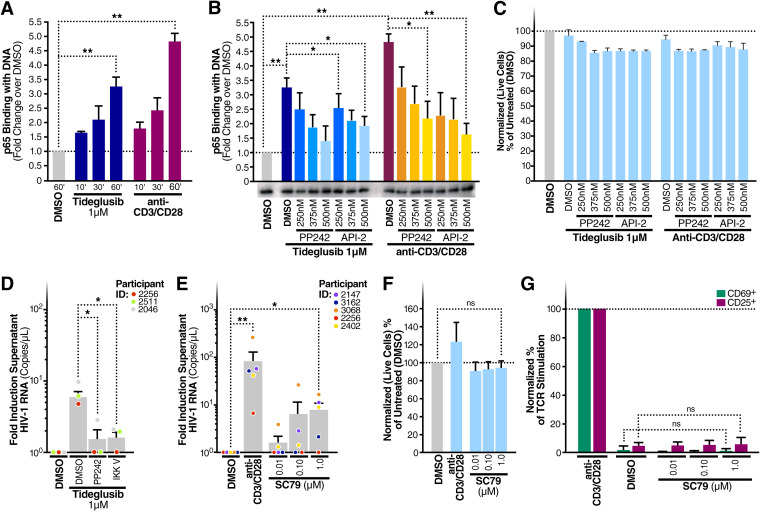
Because NF-κB plays an important role in HIV-LTR activation (34) and because AKT can induce NF-κB (14, 15, 35), we studied the effects of treating CD4+ T cells with 1 μM tideglusib for 10, 30, or 60 min, assessing the ability of the resultant nuclear extracts to bind to target κB DNA enhancer sites. As early as 10 min after incubation with tideglusib, we detected binding of p65 with target DNA sequences (Fig. 4A). The signal increased at the 30- and 60-min time points. To determine whether NF-κB activation by tideglusib involved activation of the AKT/mTOR pathway, CD4+ T cells were treated for 1 h with either tideglusib alone or increasing concentrations of PP242 (an mTOR inhibitor) or API-2 (a specific AKT inhibitor). As shown in Fig. 4B, inhibition of mTOR or AKT reduced the amount of p65 bound to target κB enhancers. These results link tideglusib-mediated inhibition of GSK-3β with activation of NF-κB, involving activation of the AKT and mTOR pathway.
FIG 4.
AKT and mTOR modulate tideglusib-mediated NF-κB activation in CD4+ T cells. (A) CD4+ T cells were purified from blood of three HIV-negative individuals and treated for the indicated times with either 1 μΜ tideglusib, anti-CD3/CD28, or a control DMSO solution. (B) Blood CD4+ T cells treated for 1 h with either 1 μΜ tideglusib, anti-CD3/CD28 coated beads, and increasing concentrations of the mTOR inhibitor PP242 or the AKT inhibitor API-2. Nuclear extracts were prepared from these cells (A and B), and p65 binding to target dsDNA was quantitated by enzyme-linked immunosorbent assay (ELISA). Extracts were also immunoblotted for TATA-binding protein (TBP) to test equivalence of protein loading. (C) CellTiter-Blue assays were used to assess potential changes in cell viability in CD4+ T cells treated as for panel B. (D) Fold induction of virion RNA in three HIV-infected individuals on ART treated with tideglusib (1 μM) and DMSO, PP242, or IKK-V, an IKK inhibitor. (E) Fold induction of virion RNA in five HIV-infected individuals on ART treated with SC79 (0.01 to 1 μM) or anti-CD3/CD28-coated beads or left untreated (DMSO control). (F) Normalized percentage of live CD4+ T cells compared to untreated controls. (G) LRA-stimulated CD4+ T cells were tested for surface expression of CD69 and CD25 activation markers. Lymphocytes were gated on single cells > live > CD3+ and CD69+ or CD25+. Unless otherwise specified in the figure, treatments were done at the following concentrations/volumes: DMSO control, 0.01% (vol/vol) final volume; anti-CD3/CD28-coated beads, 125 μl (25 μl per 1 × 106 cells). Data are means and SEM; statistical analysis employed paired two-tailed Student’s t test. *, P < 0.05; **, P < 0.01; ns, not significant.
We next investigated whether reactivation of latent virus by tideglusib was dependent on mTOR or NF-κB. A total of 5 × 106 CD4+ T cells isolated from aviremic HIV-infected individuals on ART were treated with tideglusib either alone or in the presence of either PP242 or IKK-V, a salicylamide-based inhibitor of IKK-2 that blocks NF-κB activation. As shown in Fig. 4D, inhibition of either mTOR or NF-κB strongly impaired the latency-reversing effects of tideglusib, reducing the amount of supernatant HIV RNA by approximately 80%. Of note, CD4+ T cells from one of the four donors analyzed (participant ID [PID], 2461) showed a lower response to mTOR or NF-κB inhibition (data not shown). The ability of mTOR inhibitors to prevent HIV latency reversion has been shown before (18, 36); here, we show that activation of mTOR (e.g., via GSK-3 inhibition) leads to latency reversal. In fact, specific activation of AKT (which leads to mTORC-1 activation [37]) with the small-molecule compound SC79 also resulted in enhanced release of virions in culture supernatants of CD4+ T cells from aviremic individuals on ART (Fig. 4E). As observed with GSK-3i’s, treatment with SC79 did not impair CD4+ T cell viability (Fig. 4F), nor did this agent induce expression of the cell activation markers CD69 and CD25 (Fig. 4G).
Taken together, these results demonstrate that inhibition of GSK-3β with tideglusib leads to activation of NF-κB in an AKT/mTOR-dependent manner and that activation of both NF-κB and mTOR is required for tideglusib-mediated latency reversal. Based on the results of these collective studies, we provide a schematic summary of the proposed mechanism of action of tideglusib on the mTOR/AKT/NF-κB pathways in CD4+ T cells (Fig. 5).
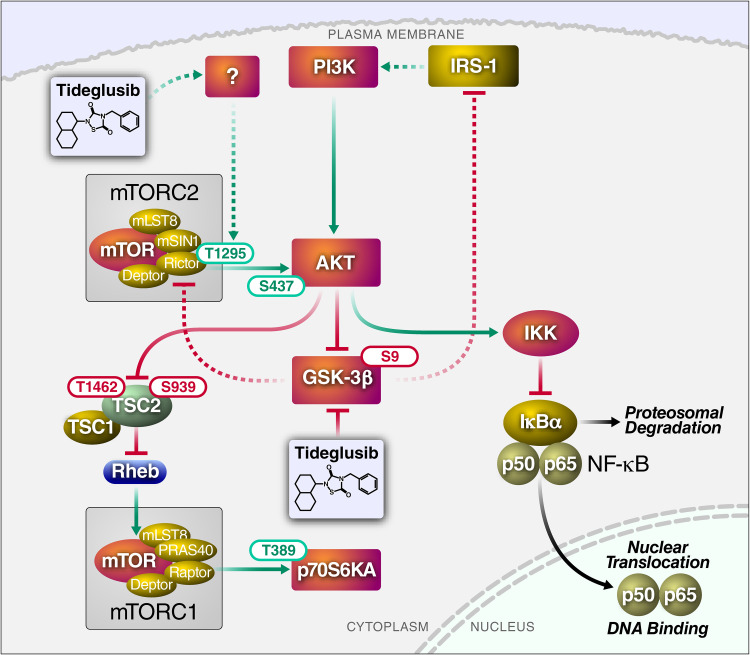
FIG 5.
Schematic summary of the pathway induced by the GSK-3i tideglusib, leading to activation of AKT/mTOR signaling. All phosphorylation events reported here were experimentally confirmed by NanoString analysis or whole-cell phosphoproteomics. Solid arrows/lines indicate interaction previously reported in the literature; dotted arrows/lines represent proposed interactions. Green arrows/lines indicate activation; red arrows/lines indicate inhibition (note that in many circumstances, a negative regulatory phosphorylation blocks the inhibitory effect of a protein, leading to activation). The indicated phosphorylation sites represent activating (green) or inhibitory (red) phosphorylation events.
Inhibition of GSK-3 does not affect CD4+ T-cell metabolism.
T cell receptor ligation causes a switch in cellular metabolism in T cells from oxidative phosphorylation to glycolysis involving the activation of multiple signaling pathways (38, 39). These signals converge on AKT, inducing phosphorylation of the glucose transporter-1 (GLUT1) and leading to an increase in its expression on the plasma membrane (40).
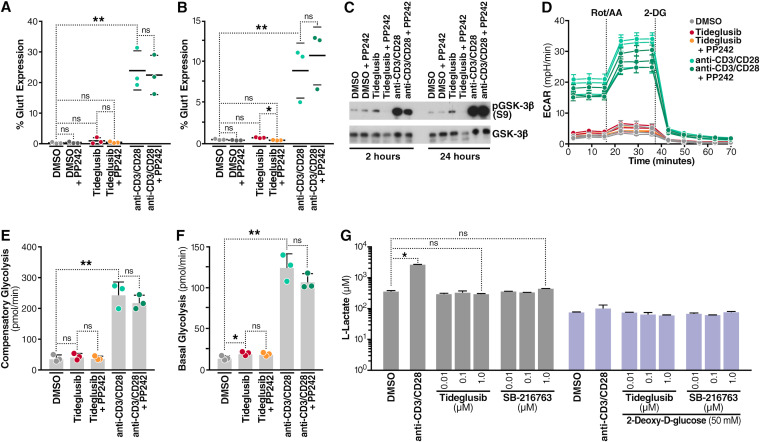
In view of the ability of tideglusib to activate the AKT/mTOR pathway, we investigated whether treatment with tideglusib resulted in major changes in the metabolism of resting CD4+ T cells. First, we measured plasma membrane expression of GLUT1 in CD4+ T cells isolated from blood of three HIV-negative individuals treated for 2 or 24 h with either 1 μM tideglusib or anti-CD3/CD28-coated beads, with or without the mTOR inhibitor PP242. As shown in Fig. 6A and B, tideglusib did not induce a detectable increase in GLUT1 surface expression compared to the untreated control at either 2 h (Fig. 6A) or 24 h (Fig. 6B). In contrast, cells activated with anti-CD3/CD28-coated beads exhibited a marked increase in plasma membrane expression of GLUT1 at both time points (as previously shown [41]). Compared to stimulation with anti-CD3/CD28-coated beads, addition of the mTOR inhibitor did not significantly alter expression of GLUT1. Biological activity of the GSK-3β inhibitor used in the analysis shown in Fig. 6A and B was confirmed in Western blots (Fig. 6C) by demonstrating an increase in phospho-S9 on GSK-3β.
FIG 6.
Inhibition of GSK-3 does not increase glycolytic metabolism in CD4+ T cells. Flow-cytometric assessment of surface expression of GLUT1 analysis in CD4+ T cells isolated from the blood of HIV-negative individuals after either 2 h (A) or 24 h (B) treatment with DMSO control, 1 μM tideglusib, or anti-CD3/CD28-coated beads, in the presence or absence of 250 nM PP242. (C) Extracts from cells used in panels A and B probed for phosphoS9-GSK-3β by immunoblotting. (D) Assessment of CD4+ T cell glycolytic profile using a Seahorse XF analyzer to determine OCR (oxygen consumption rate [data not shown]) and ECAR (extracellular acidification rate) measurement, 24 h after treatment as for panels A and B. (E) Measurement of compensatory glycolysis as derived from panel D. (F) Measurement of basal glycolysis as calculated from panel D. (G) Quantitation of l-lactate production measured in the supernatant of CD4+ T cell cultures, 24 h after treatment with the indicated GSK-3 inhibitors or anti-CD3/CD28, with or without 2-DG. All data represent 3 biological replicates except for data in panels D, E, and F, which represent 6 biological replicates. Data are means and SEM; statistical analysis was performed using a paired two-tailed Student's t test. *, P < 0.05; **, P < 0.01; ns, not significant. AA/Rot, antimycin A/rotenone; mpH/min, milli-pH units per minute.
To study the effect of tideglusib on glycolytic metabolism in CD4+ T cells, we utilized Seahorse XF technology. CD4+ T cells were treated for 24 h under the same conditions as in Fig. 6B and then subjected to Seahorse analysis. We measured extracellular acidification rate (ECAR) obtained during a standard glycolytic rate assay, including treatments with a combination of actinomycin A (AA) and rotenone (Rot), which are inhibitors of complex III and complex I, respectively (Fig. 6D). Response to Rot/AA did not result in an increase in compensatory glycolysis (Fig. 6E), indicating that tideglusib did not prime the cells to use glycolysis when ATP production from oxidative phosphorylation was artificially impaired. Measurement of basal glycolysis, on the other hand, showed a statistically relevant (P < 0.05) difference in basal glycolysis between untreated and tideglusib-treated samples (Fig. 6F). However, given the small change (i.e., the mean value for the DMSO-treated sample was 11.88 pmol/min, while that for the tideglusib-treated sample was 17.43 pmol/min), we concluded that treatment with tideglusib produced only a small effect on basal CD4+ T cell glycolysis. Furthermore, we could not detect increased medium acidification in cultures of CD4+ T cells treated with tideglusib or SB-216763, as shown in a colorimetric cell-based assay (Fig. 6G): 24 h after treatment, neither SB-216763 nor tideglusib induced increase of l-lactate concentrations in cell culture medium, while incubation with anti-CD3/anti-CD28-coated beads, as expected, caused a 10-fold increase in l-lactate production. As expected, treatments performed in the presence of the glycolysis inhibitor 2-deoxy-d-glucose (2-DG) dramatically reduced l-lactate production in all treated and untreated samples. Together, these findings indicate that inhibition of GSK-3β with tideglusib did not produce marked changes in glucose metabolism in human CD4+ T cells.
Tideglusib preserves viability and cytotoxic effector functions of CD8+ T and NK cells.
In the context of the “shock and kill” cure strategy, after successful reactivation of latent provirus, infected CD4+ T cells will likely require clearance by immune effector cells. Several previous studies, however, have shown that treatment with protein kinase C (PKC) agonists or histone deacetylase (HDAC) inhibitors impairs the cytotoxic effector functions of CD8+ T and NK cells (42–45). Accordingly, we have studied the effects of tideglusib on the viability and cytotoxic functions of CD8+ T and NK cells isolated from HIV-negative donors. Peripheral blood mononuclear cell (PBMC) cultures were treated for 48 h with 1 μM tideglusib or with two different LRAs: the PKC agonist bryostatin (10 nM) or the HDAC inhibitor panobinostat (50 nM). Cells were then either stimulated (with phorbol myristate acetate [PMA]/ionomycin or in the presence of K562 cells), to mimic in vivo activation, or left untreated. Viability and cytotoxic effector functions were measured by flow cytometry in both stimulated and nonstimulated CD8+ T and NK cell populations. As shown in Fig. 7A and D, tideglusib did not reduce viability of either CD8+ T or NK cells and, after activation, preserved full cytotoxic effector functions, as can be seen by the high levels of gamma interferon (IFN-γ) production and CD107a surface expression (Fig. 7B, C, E, and F). As shown in Fig. 7B and C, tideglusib in fact slightly enhanced production of IFN-γ and surface expression of the degranulation marker CD107a. A similar effect was noted in NK cells treated with tideglusib (Fig. 7E and F). On the other hand, as shown in Fig. 7A and D, panobinostat reduced the number of viable CD8+ T and NK cells to less than 40% (viability gates and gating-strategy are shown in Fig. S1 to S3 in the supplemental material). Given its high toxicity, IFN-γ production and expression of CD107a in panobinostat-treated samples were not included in the additional analyses. A full panel of flow data for CD8+ T and NK cells treated with panobinostat can be found in Fig. S2 (CD8+ T cells) and Fig. S4 (NK cells). Of note, the very few CD8+ T cells that survived panobinostat treatment showed high IFN-γ production and elevated surface expression of CD107a (Fig. S2). Treatment with bryostatin, while not as toxic as panobinostat, caused a substantial decrease in IFN-γ production in CD8+ T cells (Fig. 7B) and reduction of CD107a surface expression in NK cells (Fig. 7F).
FIG 7.
Treatment with tideglusib does not affect cytotoxic effector functions of CD8+ T, NK cells, or HIV-specific CTLs from HIV-positive individuals on ART. PBMCs isolated from five independent HIV-negative donors were cultured in the presence of the indicated LRAs orleft untreated (DMSO control) for 48 h. Treated PBMCs were then either left unstimulated or activated in the presence of PMA/ionomycin (for CD8+ T cells in panels A, B, and C) or K562 cells (for NK cells in panels D, E, and F). (A) Assessment of cell death in treated or treated and stimulated cells. Percent viability determined by viable cells relative to DMSO or DMSO with PMA/ionomycin is presented. (B and E) Fold change in IFN-γ-producing cells following treatment with tideglusib or bryostatin in the presence of PMA/ionomycin stimulation for CTLs (B) and NK cells (E). (C and F) Fold change in CD107a+ cells following treatment with tideglusib or bryostatin in the presence of PMA/ionomycin stimulation; data are for CTLs (C) and NK cells (F). The gating strategy is presented in Fig. S1 to S4. (D) Assessment of cell death in treated or treated and stimulated cells. Percent viability determined by viable cells relative to DMSO or DMSO plus K562 cells is presented. (G) Representative flow plots depicting infected CD4+ T-cell elimination (ICE) of gated HIVSF162-infected CD4+ T cells after a 1-h incubation in fresh medium alone (left column) or with negatively selected CD8+ T cells that had been initially stimulated for 6 days with HIVSF162-infected autologous CD4+ T-cell targets, without (middle) or with (right) tideglusib (1 μM). Data for a representative LTNP/EC (top row) and three participants (progressors) are shown (clinical data for the three study participants are reported in Table 4). Total percentages of HIV-infected (p24+) cells in each plot were determined as the sum of the percentages of the upper quadrants. ICE values (red) were calculated as follows: [(percent p24 expression of infected targets only − percent p24 expression of infected targets mixed with day 6 cells)/percent p24 expression of infected targets only] × 100. (H) Summary data of HIV-specific CD8+ T cell cytotoxic responses, measured by net GrB substrate fluorescence in infected targets (background fluorescence in cultures of effectors coincubated with uninfected targets has been subtracted) and ICE for 3 LTNP/EC (red circles) and 3 participants (progressors), which include CD8+ T cells derived from untreated (blue circles) or tideglusib-treated (red circles) cultures. Horizontal lines designate median values. Statistical significance was calculated using a ratio paired t test compared with each stimulated/DMSO control (*, P < 0.05; **, P < 0.005; ***, P < 0.0005; ns, not significant). Values are means and standard deviations.
Tideglusib was also nontoxic for CD8+ T cells isolated from HIV-positive individuals; in fact, production of granzyme B (GrB) and killing of autologous infected CD4+ T cells (infected-CD4+-T-cell elimination [ICE]) were not impaired by prolonged (i.e., 6 days) treatment with tideglusib (Fig. 7G and H). GrB production and ICE in HIV-specific CD8+ T cells from three long-term nonprogressor/elite controller (LTNP/EC) individuals were also measured. CD8+ T cells from LTNP/EC typically mediate greater lysis of infected targets than cells from progressors (46) and were therefore used here as positive controls. Overall, the results suggest that in contrast to many of the latency-reversing agents, tideglusib preserves and supports the cytotoxic effector functions of CD8+ T and NK cells and maintains the ability of CTLs to perform antigen-specific killing of target cells.
Oral and intravenous administration of tideglusib in SIV-infected rhesus macaques on ART, while well tolerated, does not significantly reactivate latent-provirus expression.
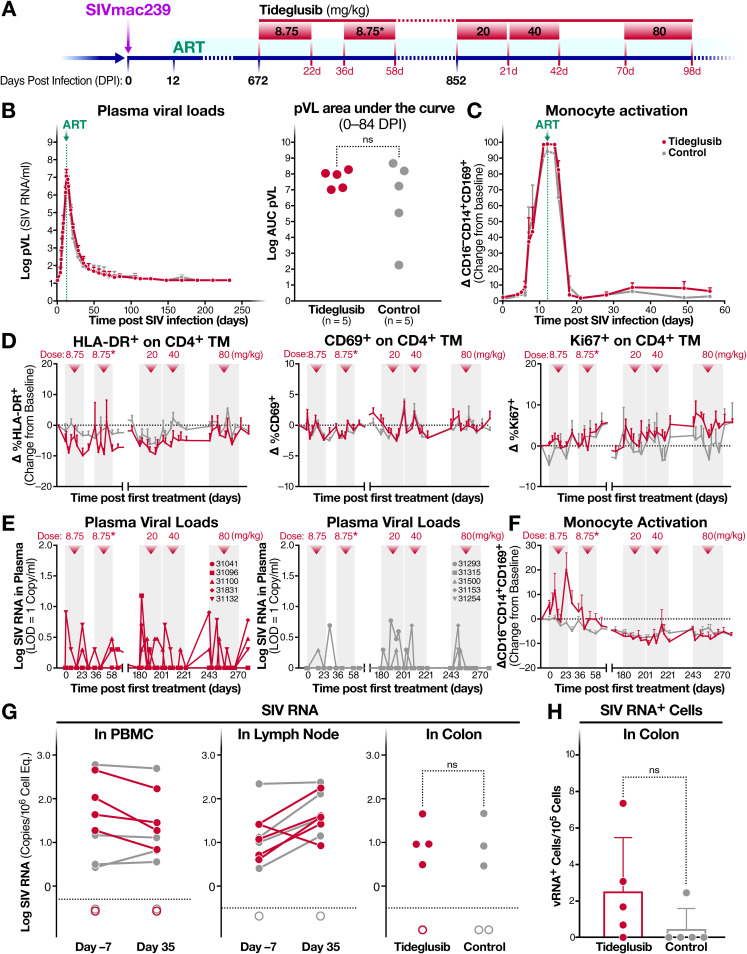
Of the drugs we have tested targeting the AKT/mTOR/GSK-3 signaling pathways, only tideglusib is currently in clinical trials. We therefore decided to test the latency-reversing effects of tideglusib in vivo, in 12 SIV-infected and ART-treated Indian-origin RM. Figure 8A summarizes the treatment timeline. We initially confirmed that tideglusib could induce SIV latency reversal ex vivo by stimulating CD4+ T cells isolated from aviremic SIV-infected RM on ART (n = 3) with tideglusib for 48 h. Cells from all three RM exhibited an increase in cell-associated viral RNA following tideglusib exposure (Fig. 9). Subsequently, we inoculated 12 RM intravenously with SIVmac239X and treated them with ART starting 12 days postinfection. All but one RM achieved stable full virologic suppression to <15 SIV RNA copies/ml plasma within 100 days postinfection. The nonsuppressed RM and an RM that was lost to study due to an unrelated illness were excluded from further analyses. The remaining 10 animals were assigned to two treatment groups and balanced for plasma viremia through 84 days postinfection (dpi) (Fig. 8B). Levels of monocyte activation were also similar between groups (Fig. 8C). Of note, monocyte activation, as shown by the interferon-triggered upregulation of CD169 (47, 48), increases with SIV plasma viral loads (pVL) during acute infection and declines after ART treatment (49), demonstrating that CD169 can be an effective marker for the presence of circulating SIV.
FIG 8.
Effect of oral tideglusib on SIV viral reactivation in vivo. (A) Schematic showing timeline of rhesus macaque (RM) tideglusib treatment, including SIVmac239X infection, initiation of ART at 12 days postinfection (dpi), and oral tideglusib treatment phases (8.75 mg/kg for 22 days, starting at 672 dpi; 8.75 mg/kg for 22 days, starting at 708 dpi; 20 mg/kg for 21 days, starting at 852 dpi; 40 mg/kg for 21 days, starting at 873 dpi; and 80 mg/kg for 28 days, starting at 915 dpi). (B) Mean (and SEM) SIVmac239 plasma viral load profiles during initial SIV infection and ART suppression (left) and log area under the curve of SIV plasma viral loads for 0 to 84 dpi (right) for tideglusib-treated RM (red, n = 5) and control RM (gray, n = 5). (C) Mean (and SEM) change from baseline in percent CD169 on CD16− CD14+ classical monocytes in blood following SIV infection and ART suppression for tideglusib-treated RM (red, n = 5) and control RM (gray, n = 5). (D) Mean (and SEM) change from baseline in percent HLA-DR and CD69 (activation) and percent Ki67 (proliferation) on CD4+ memory T cells in blood during treatment periods (pale gray) of tideglusib-treated RM (red, n = 5) and control RM (gray, n = 5). (E) Individual SIV plasma viral load profiles of tideglusib-treated RM (left) and control RM (right) during treatment phases. (F) Mean (and SEM) change from baseline in percent CD169 on CD16− CD14+ classical monocytes in blood of tideglusib-treated RM (red, n = 5) and control RM (gray, n = 5) during treatment periods. (G) Cell-associated SIV RNA in peripheral blood mononuclear cells, lymph node cells, and colons of tideglusib-treated RM (red) and control RM (gray) at 7 days before or 35 days after the start of the 20-mg/kg-treatment period. Measurements below the limits of detection (dotted line) are depicted with unfilled circles. (H) Quantification of SIV RNA-positive cells by RNAscope in the colon of tideglusib-treated RM (red) and control RM (gray) on day 14 of the 40-mg/kg-treatment period. *, treatment with clinical product AMO-02. ns, not significant.
FIG 9.
Treatment with tideglusib reverses latency in blood CD4+ T cells from SIV-positive RM on ART studied ex vivo. Blood CD4+ T cells, isolated from three RM on suppressive ART, were either treated for 48 h with 1 μM tideglusib, treated with 10 nM PMA plus 500 nM ionomycin (PMA/I), or left untreated (DMSO control). Data are means and SEM.
After 96 weeks of antiviral suppression, to establish a stable latent viral reservoir, half of the animals (n = 5) were orally administered escalating doses of tideglusib, while the other half (n = 5) received vehicle control or no treatment. During the first two tideglusib dosing cycles, RM received the neat micronized drug substance followed by a clinical product (AMO-02) which is formulated to enhance bioavailability. Due to a limited supply of AMO-02, the neat drug substance was used for all subsequent dosing. During treatment, the animals were assessed for markers of CD4 memory T cell activation (HLA-DR and CD69) and proliferation (Ki67) in blood. As shown in Fig. 8D, tideglusib did not induce, even at the highest dose administered (80 mg/kg), activation or proliferation of CD4 memory T cells, consistent with ex vivo experiments. Similarly, tideglusib treatment had no effect on overall lymphocyte, naive, or memory subsets (central, transitional, and effector) of CD4 and CD8+ T cells and had no measurable effect on NK cell proliferation (data not shown).
In order to assess reversal of SIV latency during tideglusib treatment, we tested for an increase in SIV pVL (Fig. 8E) and evidence of monocyte activation (Fig. 8F). During the initial treatment period, oral administration of tideglusib caused no significant increase in plasma viremia. There was a 2.6-fold increase in the odds of detecting low-level viral blips in the tideglusib-treated animals, but this increase in low-level blipping did not reach statistical significance compared to the vehicle-treated animals (P value = 0.17).
We also examined CD169 expression on classical monocytes as a parallel marker of circulating SIV and innate immune activation. During the initial treatment phase, there was a marked increase in monocyte activation in the tideglusib-treated RM relative to control monkeys but no corresponding increase in pVL. This is not necessarily unexpected, as increases in plasma viremia may be small in magnitude and transitory but lead to type I interferon production, giving rise to CD169 expression.
In order to determine the effect of in vivo tideglusib treatment on SIV viral expression by SIV-infected cells, we performed qPCR quantification of cell-associated viral RNA (Fig. 8G) and DNA in PBMCs, lymph nodes, and colons of treated and untreated animals. As reported in Fig. 8G, levels of cell-associated SIV RNA in PBMCs, lymph nodes, and colons did not statistically differ in the treated and untreated RM. RNAscope analyses showed no statistically significant increase in the number of SIV RNA+ cells in colon biopsy tissues from tideglusib-treated versus control RM, although several tideglusib-treated animals trended to increased levels of cell-associated SIV RNA expression (Fig. 8H).
Next, we conducted pharmacokinetic analysis of heparinized samples collected after oral dosing. As presented in Table 2, plasma drug levels proved quite variable and didnot consistently increase with increasing oral dosing of the drug. Drug levels often fell below the level of detection (5 ng/ml). However, when we screened for effects on metabolism and liver function, there was a marked increase in liver alanine and aspartate transaminases, particularly at the higher doses (data not shown). These changes are consistent with effective absorption and enterohepatic circulation as described for oral administration of tideglusib to humans (26, 27). Other blood chemistry levels were normal. These findings suggest that the drug is effectively absorbed and delivered to the liver. However, because circulating plasma concentrations of tideglusib are highly variable, it is unclear whether the drug is effectively delivered to the relevant target tissues like peripheral lymph nodes. It is possible that plasma levels are restricted as a consequence of the enterohepatic cycle of absorption and clearance.
TABLE 2.
Tideglusib drug levels in plasma during the oral dosing phasea
| Dose (mg/kg/day) | Level (ng/ml) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Tideglusib |
Control |
||||||||||
| 31041 | 31096 | 31100 | 31831 | 31098 | 31132 | 31293 | 31315 | 31500 | 31153 | 31254 | |
| 8.75 | <5 | 10.0 | 9.0 | 11.3 | 4.9 | <5 | NA | NA | NA | NA | NA |
| 8.75b | <5 | <5 | <5 | <5 | <5 | <5 | NA | NA | NA | NA | NA |
| 20 | 36.2 | 14.6 | 33.8 | <5 | <5 | 20.3 | <5 | <5 | <5 | <5 | <5 |
| 40 | 18.27 | <5 | <5 | <5 | <5 | <5 | <5 | <5 | <5 | <5 | <5 |
| 80 | 54.7 | 21.1 | 30.8 | <5 | 10.9 | 7.4 | <5 | <5 | <5 | <5 | <5 |
Samples were collected at trough following each cycle of daily oral dosing with tideglusib at the indicated doses. Animals are identified by five-digit numbers. The limit of detection was 5 ng/ml. NA, not assayed.
AMO-02.
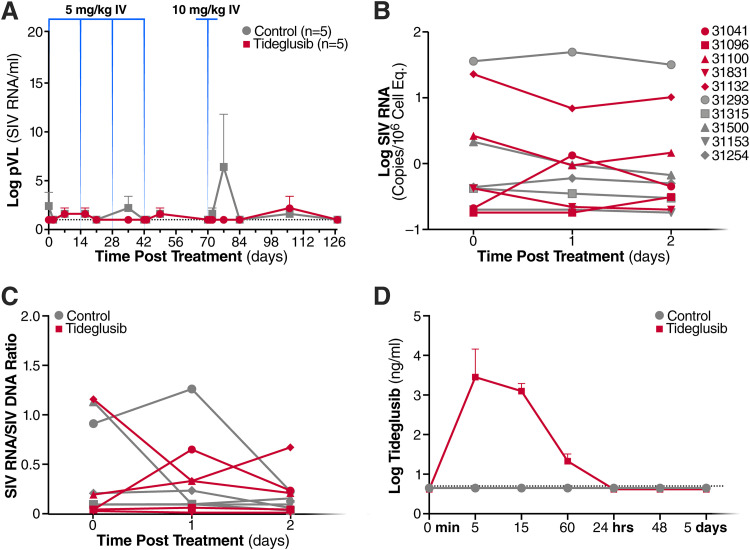
Low plasma bioavailability of tideglusib measured after oral administration prompted us to test whether intravenous administration would produce higher drug concentrations in plasma and result in detectable SIV latency-reversal. After a “washout” period of approximately 7 weeks, the animals received 4 biweekly doses of tideglusib administered intravenously at 5 mg/kg, followed by a single dose at 10 mg/kg 4 weeks later. Control animals were injected with vehicle control (100% polyethylene glycol 400 [PEG 400]). Plasma samples and PBMCs were collected for analysis of pVL and cell-associated SIV RNA, respectively, at 24 and 48 h following the first intravenous (i.v.) dose. In PBMC samples, change in SIV RNA/DNA ratio was also measured. As shown in Fig. 10, intravenous treatment of tideglusib did not result in detectable changes in SIV pVL (Fig. 10A), cell-associated viral RNA (Fig. 10B), or SIV RNA/DNA ratio (Fig. 10C). Furthermore, as indicated by measurements of plasma drug concentration (Fig. 10D), intravenous administration resulted in drug levels about 1,000-fold higher than during oral administration (Table 3). However, tideglusib was rapidly cleared from the circulation (with a peak between 5 and 15 min after injection) and was not detectable after 24 h. Taken together, these results suggest that tideglusib, as currently formulated, is not an effective latency-reversing agent for use in vivo in SIV-infected RM on ART.
FIG 10.
Effect of intravenous tideglusib on SIV viral reactivation in vivo. (A) Mean (and SEM) SIV plasma viral load profiles of RM following intravenous administration of tideglusib or vehicle control. Tideglusib was administered biweekly at 5 mg/kg at 1,027, 1,041, 1,055, and 1,069 dpi followed by a 10-mg/kg dose at 1,097 dpi. (B) Cell-associated SIV RNA (in log copies/106 cell equivalents) and (C) ratio of cell-associated SIV RNA to SIV DNA in PBMCs at 0, 1, and 2 days following a 5-mg/kg intravenous infusion of tideglusib or vehicle control at 1,027 dpi. (D) Mean (and SEM) tideglusib drug levels in plasma following a 5-mg/kg intravenous infusion of tideglusib or vehicle control at 1,027 dpi. Limit of detection, 5 ng/ml.
TABLE 3.
Tideglusib drug levels in plasma following intravenous dosinga
| Time after tideglusib infusion | Level (ng/ml) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Tideglusib |
Control |
||||||||||
| 31041 | 31096 | 31100 | 31831 | 31098 | 31132 | 31293 | 31315 | 31500 | 31153 | 31254 | |
| 0 min | <5 | <5 | <5 | <5 | <5 | <5 | <5 | <5 | <5 | <5 | <5 |
| 5 min | 17,719 | 17,749 | <5 | 14,675 | 14,693 | 9,536 | <5 | <5 | <5 | <5 | <5 |
| 15 min | 2,875 | 1,998 | 227 | 1,380 | 1,587 | 1,740 | <5 | <5 | <5 | <5 | <5 |
| 60 min | 40 | 32 | <5 | 32 | 25 | 26 | <5 | <5 | <5 | <5 | <5 |
| 24 h | <5 | <5 | <5 | <5 | <5 | <5 | <5 | <5 | <5 | <5 | <5 |
| 48 h | <5 | <5 | <5 | <5 | <5 | <5 | <5 | <5 | <5 | <5 | <5 |
| 7 days | <5 | <5 | <5 | <5 | <5 | <5 | <5 | <5 | <5 | <5 | <5 |
Samples were collected at the indicated times following i.v. administration of tideglusib at 5 mg/kg. Animals are identified by five-digit numbers. The limit of detection was 5 ng/ml.
DISCUSSION
All current approaches for reducing or eliminating the latent HIV reservoir would be propelled by latency-reversing agents that potently and safely reactivate latent proviruses within reservoir cells under conditions that lead to their recognition and destruction by immune effector cells. The first generation of LRAs studied (HDAC inhibitors and PKC activators) has fallen well short of the desired mark chiefly because of problems with toxicity, potency, and undesirable levels of T cell activation associated with proinflammatory cytokine production. Many of these LRAs also impair the function of the cytotoxic immune effectors (CTLs and NK cells) that likely will be needed to kill the reactivated reservoir cells (43, 44, 50). Further complicating the use of LRAs, after a single dose, these agents reactivate only a small fraction of the latent reservoir (3), and, even when LRAs can induce high levels of virus reactivation in vivo, as reported in a recent study on AZD5582, a mimetic of the second mitochondrion-derived activator of caspases (SMAC-mimetic) (51), this does not result in reduction in the total or replication-competent viral reservoir. Of note, in another study, treatment of SHIV-infected and ART-suppressed RM with AZD5582 resulted in no measurable latency reversion (52).
Serial administration of the LRA will almost certainly be required, which reemphasizes the need for safe and nontoxic agents. An ideal LRA will be (i) potent, (ii) nontoxic, (iii) able to be repeatedly dosed, (iv) non-cell activating, and (v) unable to impair the cytotoxic effector function of CTLs and NK cells.
In a search of new LRAs exhibiting many or all of these desirable properties, we focused on GSK-3 inhibitors. These compounds effectively reverse HIV latency ex vivo in CD4+ T cells isolated from HIV-positive individuals on suppressive ART, and they do so without inducing T cell activation. They also exhibit little or no toxicity. One member of the GSK-3i family, tideglusib, has been shown to be safe in humans with treatment regimens involving repeated dosing over many months (26, 27). The GSK-3i’s also do not promote T cell activation or production of proinflammatory cytokines. Finally, these agents do not impair the cytotoxic effector functions of CTLs and NK cells. Indeed, these agents can induce a small, but statistically significant, enhancement of IFN-γ production and CD107a surface expression in both activated CD8+ T and NK cells (Fig. 7A to F). In terms of potency, these agents consistently induced a 35- to 40-fold increase in production of virion RNA compared to the diluent control. This response was ∼5% of that obtained with anti-CD3/CD28 coated beads, which represents an extremely strong LRA that produces high-level T cell activation and proinflammatory cytokine release (Fig. 1B).
As previously shown, both mTOR and NF-κB play a pivotal role regulating HIV-latency (18, 36, 53), and reactivation of quiescent proviruses involves induction of both signaling pathways. Indeed, these pathways may be at least partially intertwined. By disinhibiting the activity of the mTORC2 complex, tideglusib promotes NF-κB induction occurring in part through the activation of AKT by the mTORC2 complex (Fig. 4E). However, other pathways leading to NF-κB activation may also be in play, since neither the mTOR nor AKT inhibitors fully blocked NF-κB activation (Fig. 4B). The activation of multiple pathways by tideglusib is further supported by the NanoString results showing activation of mitogen-activated protein kinase (MAPK), phosphatidylinositol 3-kinase (PI3K), Ras, and Wnt signaling (Fig. 3B). In the Gene Ontology (GO) enrichment analysis, tideglusib also quickly altered spliceosome complex activity, overall transcription activation, and mRNA processing, plus stimulation of TCR and epidermal growth factor receptor signaling (Fig. 3C). Unraveling the detailed mechanism of action of GSK-3 inhibitors in CD4+ T cells could facilitate the identification of new agents that can synergize with tideglusib while retaining the desirable properties of an LRA. Indeed, it seems likely that combinations of LRAs will be required to overcome the multiplicity of blocks underlying HIV latency (50). However, these combinations need to retain the desirable properties of LRAs. One interesting approach might be to combine a GSK-3 inhibitor with disulfiram, which also activates AKT and has been proven safe for treatment in humans (54).
Despite the ability of GSK-3 inhibitors to induce activation of AKT, which is known to regulate GLUT1 surface expression and support activation of glycolysis (55), we could not detect induction of glycolysis after treatment of cells with tideglusib (Fig. 6). The extracellular signal-related kinase (ERK) (56), JAK-STAT (57), MAPK (58), and hexokinase II (59) are all directly connected with regulation of glucose metabolism in T cells. These pathways cooperate with AKT and mTOR to regulate metabolic reprograming of T cells upon TCR engagement. However, the magnitude of regulation of T-cell metabolism by these pathways may vary depending on several factors (60). The decreased activation of JAK-STAT detected in the NanoString studies (Fig. 3B), for example, might at least partially explain why tideglusib does not produce a metabolic shift in CD4+ T cells. Importantly, the lack of induction of glycolysis upon treatment with tideglusib may represent an additional favorable characteristic of this LRA, as proliferation and activity of CTLs was shown to be suppressed by lactate secreted by glycolytic CD4+ T cells (61).
The favorable ex vivo properties of tideglusib prompted us to test whether this agent functions as an effective LRA in vivo. For this testing, we chose the RM model. Briefly, RM were infected with SIV and then suppressed with ART. These animals were subsequently treated with tideglusib, first orally and subsequently intravenously. While tideglusib proved safe and nontoxic for all the treated animals, we were unable to detect a statistically significant increase in plasma viremia or cell-associated SIV RNA in animals treated with this drug compared to the untreated controls.
When tideglusib was administered orally, plasma levels proved highly variable, complicating interpretation of the experimental results. We did observe that at the highest oral dose (80 mg/kg), plasma drug levels of tideglusib ranged between 7.43 ng/ml and 54.7 ng/ml (or 0.16 μM) and that increases in hepatic alanine and aspartate transaminases occurred, indicative of drug exposure in the liver. During the 5-mg/kg intravenous treatment phase, plasma drug levels peaked quickly, reaching over 10,000 ng/ml, but rapidly declined within 60 min and were not detectable after 24 h. In our ex vivo experiments, HIV latency reversal in CD4+ T cells was observed after exposure to tideglusib at concentrations of 1 μM for 24 to 48 h. Accordingly, the limiting pharmacokinetic/pharmacodynamic (PK/PD) properties of tideglusib as currently formulated suggests that we were not able to deliver or sustain sufficiently high quantities of this drug in RM to induce reversal of SIV latency, particularly at doses which produce acceptable tolerable effects in the liver. Indeed, the clinical product AMO-02, currently in phase 2/3 trials (Clinical Trials no. NCT02858908) (25), was designed to enhance bioavailability by means of granules within the formulation, benefiting from a micronized drug substance and a bioavailability-enhancing nonionic surfactant. The formulation also includes diluents and stabilizers. Unfortunately, AMO-02 was not available to us for long-term dosing in RM.
While our ex vivo studies of the GSK-3 inhibitors highlight their many distinctly desirable properties as latency-reversing agents, in vivo effects of tideglusib, the prototypical GSK-3 inhibitor, in RM receiving ART proved disappointing. Different nontoxic formulations of these GSK-3 inhibitors, optimized for effective delivery to systemic lymphoid tissues at pharmacologically relevant levels, are needed to establish whether this class of AKT/mTOR activators can serve as effective LRAs in vivo.
MATERIALS AND METHODS
Study participants.
HIV-infected individuals were enrolled at the Zuckerberg San Francisco General Hospital based on the criteria of suppressive ART and undetectable plasma HIV-1 RNA levels (<50 copies per ml) for a minimum of 6 months. Leukapheresis was performed through the UCSF SCOPE cohort for the collection of >1 billion peripheral blood mononuclear cells (PBMCs). Characteristics of study participants are presented in Table 1. Characteristics of study participants shown in Fig. 7G and H are shown in Table 4.
TABLE 4.
| Participant | Date HIV+ (yr) | CD4 count (cells/mm3) | HIV RNA (copies/ml) | HLA Ab | HLA Bb |
|---|---|---|---|---|---|
| LTNP/EC-1 | 1982 | 1,662 | <50 | 32 | 27, 44 |
| LTNP/EC-2 | 1985 | 1,060 | <50 | 1, 31 | 8, 57 |
| LTNP/EC-3 | 1991 | 649 | <50 | 2, 3 | 7, 57 |
| Progressor-1 | 1995 | 558 | 81,182 | 1 | 52, 57 |
| Progressor-2 | 1989 | 289 | 23,151 | 2, 3 | 7, 14 |
| Progressor-3 | 1987 | 519 | <50a | 3 | 40, 57 |
Progressor-3 was on antiretroviral treatment.
The last two columns depict the HLA class I A and B alleles (haplotype) of each individual, which do not contain units and are typically shown as numerals. One number is shown for homozygosity and two numbers separated by a comma are shown for heterozygosity at a given locus.
Isolation and culture of peripheral blood mononuclear cells, CD4+ T cells, and gut-associated lymphoid tissue cells.
PBMCs were purified from whole blood or continuous-flow centrifugation leukapheresis products via density centrifugation on a Ficoll-Hypaque gradient. Subsequently, resting CD4+ T cells were enriched by negative depletion with an EasySepHuman CD4+ T cell isolation kit (STEMCELL). Enriched cells were cultured in RPMI medium supplemented with 10% fetal bovine serum, 1,000 U/ml penicillin, and 1 mg/ml streptomycin. PBMCs and CD4+ T cells from HIV-positive individuals were cultured and treated in the presence of 5 μM saquinavir to prevent spreading infection. For gut-associated lymphoid tissue (GALT) cells, to remove the epithelium and epithelial cells, 30 tissue biopsy tissue pieces were incubated in prewarmed buffer containing 10 mM dithiothreitol (DTT), 5 mM EDTA, 10 mM HEPES, and 5% fetal bovine serum (FBS) for 20 min at 37°C under continuous rotation and vortexed, and the supernatant was aspirated. Tissue pieces were then incubated for a second time in the same prewarmed buffer for a further 20 min at 37°C under continuous rotation. The sample was again vortexed and then rinsed for 20 min at 37°C under continuous rotation with RPMI, 10 mM HEPES, and 5% FBS. To then isolate lymphocytes from the lamina propria, a second, prewarmed buffer containing RPMI, 10 mM HEPES, 7.5 μg/ml DNase, and 5% FBS was applied to the tissue pieces, and these were again incubated for 20 min at 37°C under continuous rotation. The samples were then vortexed and aspirated numerous times with a blunt 20-gauge needle until tissue was visibly broken down. The cells were then rinsed twice with RPMI, 10 mM HEPES, and 5% FBS, passed through a 70-μm cell strainer, pelleted, and stained as described below for fluorescence-activated sorting. GALT cells from HIV-positive individuals were cultured and treated in the presence of 20 ng/ml enfuvirtide.
Drug treatments.
Treatment with anti-CD3/CD28-coated beads was done at 25 μl per 1 × 106 cells (for a bead-to-cell ratio of 1:1). Bryostatin (Sigma) was used at 10 nM; panobinostat (Sigma) was used at 50 nM; SB-216763 (Tocris Bioscience) was used at 10, 100, and 1,000 nM; tideglusib (Selleckchem) was used at 0.01, 0.1, and 1 μM; SC79 (Tocris Bioscience) was used at 0.01, 0.1, and 1 μM; PP242 (Tocris Bioscience) was used at 250 nM; IKK-V (Millipore) was used at 25 nM; 2-DG (Sigma-Aldrich) was used at 50 mM; and API-2 (Tocris Bioscience) was used at 250 nM.
Latency-reversing agent treatment conditions and measurement of virus-associated RNA.
Five million resting CD4+ T lymphocytes were plated and either left unstimulated (medium alone plus DMSO), stimulated with anti-CD3/CD28-coated beads, or incubated with LRAs for 48 h. The final DMSO percentage was never higher than 0.01% (vol/vol) for all treatments. After treatment, cells were collected and spun down at 1,000 rpm for 10 min in order to separate cells and supernatant: the cells were then used for assessment of viability and analysis of activation markers via flow cytometry. Supernatants were ultracentrifuged at 26,000 × g for 1.5 h at 4°C to purify released viruses. Pelleted viruses were resuspended in RTL lysis buffer (Qiagen), and RNA was extracted with an RNeasy minikit (Qiagen) according to the manufacturer’s protocol. The extracted RNA was then reverse transcribed with a Superscript III one-step RT-PCR system (Life Technologies), using the same primers as for ddPCR analysis. Reaction mixes contained 15 μl of a PCR mix containing reaction mix, Superscript III, primers (900 nM final concentration), and 10 μl purified RNA.
Preamplification was carried out using the following steps: reverse transcription at 50°C for 30 min, denaturation at 95°C for 2 min, 10 to 15 cycles of amplification (94°C 15 s, 55°C 30 s, and 68°C 30 s), and a final amplification at 68°C for 5 min on a GeneAmp PCR system 9700 (Thermo Fisher). Subsequently, ddPCR was applied to quantify preamplified cDNA. Each 25-μl ddPCR mix comprised the ddPCR probe supermix (no dUTP), 900 nM primers, 250 nM probe, and 8 μl cDNA. The following cycling conditions were used: 10 min at 95°C, 40 cycles each consisting of 30 s denaturation at 94°C followed by 59.4°C extension for 60 s, and a final 10 min at 98°C. Reaction mixes were loaded into the Bio-Rad QX-100 emulsification device, and droplets were formed following the manufacturer’s instructions. Then, samples were transferred to a 96-well reaction plate, and wells were sealed with a preheated Eppendorf 96-well heat sealer for 10 s, as recommended by Bio-Rad. Finally, samples were amplified on a Bio-Rad C1000 thermocycler and analyzed using a Bio-Rad QX100 ddPCR reader. Primers and probe used for HIV-1 RNA reverse transcription and ddPCR analysis were chosen as previously described (62): forward primer (5′→3′), CAGATGCTGCATATAAGCAGCTG (nucleotides 9501 to 9523); reverse primer (5′→3′), TTTTTTTTTTTTTTTTTTTTTTTTGAAGCAC [position 9629 to poly(A)]; and probe (5′→3′), 6-carboxyfluorescein (FAM)-CCTGTACTGGGTCTCTCTGG-MGB (positions 9531 to 9550).
Analysis of CD4+ T cell viability.
A 25-μl sample from each well/treatment was incubated with CellTiter-Blue reagent (Promega) for 3 to 4 h at 37°C. The assay is based on the ability of living cells to convert a redox dye (resazurin) into a fluorescent end product (resorufin). Colorimetric analysis of treated cells was done with a Perkin Elmer EnSpire 2300 multimode plate reader set to detect fluorescence (excitation, 579 nm; emission, 584 nm).
Cytokine release assay.
Supernatant was collected from the treated cell cultures described above and stored at −80°C for later analysis. Supernatant cytokine levels were determined using a V-PLEX proinflammatory panel 1 (human) kit (no. K15049D-1) and a V-PLEX IL-17A Gen. B (human) (no. K151WMD) from Meso Scale Discovery, following the manufacturer’s protocol.
RNA and protein expression profiling with NanoString.
Quantitative RNA and protein expression data were generated using the nCounter Vantage 3D solid tumor assay and the nCounter SPRINT profiler (NanoString Technologies), comprising 770 RNA and 30 protein targets, including positive and negative controls. One to three million negatively selected primary CD4+ T cells purified from five HIV-positive individuals on suppressive ART (PIDs, 2135, 2168, 1079, 1128, and 2511), as described above, were treated with 1 μM tideglusib or left untreated (DMSO control). Samples were then collected at 0.25, 0.5, 1, and 6 h, and RNA and protein lysates were prepared according to the manufacturer’s instructions. For RNA samples, an additional RNA purification step (Qiagen miRNeasy minikit) was necessary to ensure an appropriate sample input for the nCounter assay. A total of at least 30 ng RNA was used for each sample. Otherwise, samples were processed according to the manufacturer’s instructions. RNA and protein expression values were normalized and analyzed using nSolver Analysis Software 4.0 and the add-on Advanced Analysis Software 2.0.115 (NanoString Technologies). Normalization genes for each sample were automatically selected by the software based on the geNorm algorithm (63). Biological replicates were then grouped according to treatment, and the differential expression of each analyte type (RNA or protein target) was determined in comparison to the untreated condition by considering interdonor differences as confounding variables. Based on the differential expression of each gene, predefined gene sets by NanoString, representing different pathways included in this assay, were analyzed by calculating global significance scores for each gene set within each treatment, as follows:
where ti is the t statistic from the ith pathway gene. The directed global significance statistic is similar to the global significance statistic, but rather than measuring the tendency of a pathway to have differentially expressed genes, it measures the tendency to have over- or underexpressed genes. It is calculated similarly to the undirected global significance score, but it takes the sign of the t statistics into account: directed global significance = sign(U)|U|1/2, where
and where sign(U) equals −1 if U is negative and 1 if U is positive.
Phosphoproteomics.
Cells were lysed in a buffer containing 8 M urea, 100 mM Tris (pH 8.0), 150 mM NaCl, and protease inhibitors (Roche Complete tablet). Lysates were reduced with 4 mM TCEP [Tris(2-carboxyethyl)phosphine hydrochloride] for 30 min at room temperature and alkylated with 10 mM iodoacetamide for 30 min at room temperature in the dark. Samples were diluted 1:4 in 100 mM Tris (pH 8.0) to reduce urea concentration to 2 M and digested with trypsin (1:100 enzyme-to-substrate ratio) overnight at 37°C. Peptides were desalted with SepPak C18 solid-phase extraction columns (Waters) according the manufacturer’s specifications and lyophilized to dryness. Phosphopeptides were purified using an immobilized metal affinity chromatography approach. Samples were analyzed on a Thermo Scientific LTQ Orbitrap Elite mass spectrometry (MS) system equipped with an Easy nLC-1000 high-performance liquid chromatograph (HPLC) and autosampler system that is capable of maintaining back pressures of up to 10,000 lb/in2 for high-resolution chromatographic separations. The HPLC interfaces with the MS system via a nanoelectrospray source. Samples were injected onto a C18 reverse-phase capillary column (75-μm inner diameter by 25-cm length; packed with 1.9-μm C18 particles). Peptides were then separated by an organic gradient from 5% to 30% acetonitrile (ACN) in 0.1% formic acid over 180 min at a flow rate of 300 nl/min. The MS continuously collected spectra in a data-dependent fashion over the entire gradient. Raw mass spectrometry data were analyzed using the MaxQuant software package (version 1.3.0.5) (64).
Data were matched to the Swiss-Prot human reference sequence database. MaxQuant was configured to generate and search against a reverse sequence database for false-discovery-rate calculations. Variable modifications were allowed for methionine oxidation, protein N-terminus acetylation, and serine, threonine, or tyrosine phosphorylation. A fixed modification was indicated for cysteine carbamidomethylation. Full trypsin specificity was required. The first search was performed with a mass accuracy of ±20 ppm, and the main search was performed with a mass accuracy of ±6 ppm. A maximum of 5 modifications were allowed per peptide. A maximum of 2 missed cleavages were allowed. The maximum charge allowed was 7+. Individual peptide mass tolerances were allowed. For MS/MS matching, a mass tolerance of 0.5 Da was allowed and the top 6 peaks per 100 Da were analyzed. MS/MS matching was allowed for higher-charge states and water and ammonia loss events.
Data were searched against a concatenated database containing all sequences in both forward and reverse directions, with reverse hits indicating the false discovery rate of identifications. The data were filtered to obtain a peptide, protein, and site-level false discovery rate of 0.01. The minimum peptide length was 7 amino acids. Results were matched between runs, with a time window of 2 min for technical duplicates. The MaxQuant-analyzed data were subsequently analyzed using an in-house computational pipeline for statistical analysis of relative quantification with fixed and/or mixed effect models, implemented in the MSstats Bioconductor package (version 3.3.10). Contaminants, decoy hits, and peptides not containing phosphorylated residues were removed, and all samples were normalized by median-centering the log2-transformed MS1 intensity distributions. Then, the MSstats group Comparison function was run with the following options: no interaction terms for missing values, no interference, unequal intensity feature variance, and restricted technical and biological scope of replication.
Analysis of NF-κB (p65) binding with target DNA sequences.
To determine the specific NF-κB p65 DNA binding activity of treated CD4+ T cells, an aliquot of the same nuclear extracts used for the p65/TBP Western blots were assayed with a transcription factor assay kit (Abcam catalogue no. ab133112). Following the manufacturer’s instructions sequentially, nuclear extracts were incubated in the 96-well plate coated with double-strand DNA NF-κB oligonucleotides. The wells were washed and then incubated with primary anti-p65 antibody, which was then detected by incubation with horseradish peroxidase (HRP)-conjugated secondary antibody. Developing reagents were added, and absorbance was read at 450 nm. The level of absorbance indicates the DNA binding activity. For each donor experiment, binding specificity controls included an NF-κB competitor double-strand DNA (dsDNA) control well and a nonspecific binding control well. The assay was performed on three different donor samples.
GO enrichment analysis of phosphoproteomics data.
A set of 142 unique gene names was extracted from the differential analysis of 1 μM tideglusib versus untreated phosphoproteomics results using cutoffs of log2 fold change of >1 or <−1 and P values of ≤0.05 and translated from UniProt identifiers to HGNC (HUGO Gene Nomenclature Committee) symbols. The gene set was submitted to Enrichr for enrichment analysis against dozens of public ontologies and annotated gene sets. Tabular enrichments results were downloaded for WikiPathways and Gene Ontology:Biological Process, and selected terms were plotted using R based on relevance, uniqueness, and combined score. The combined score is the product of −log P value and Z-score.
Analysis of basal and compensatory glycolysis.
ECAR was determined using the Seahorse SF glycolytic rate assay kit (Agilent; 103344-100) and a Seahorse XF96 extracellular flux analyzer (Seahorse Bioscience). The instrument was a gift from the S.D. Bechtel, Jr. Foundation to the Gladstone Institutes. In brief, resting CD4+ T cells (2 × 105 per well) were seeded onto Cell-Tak-coated wells (354240; Corning) in buffered Seahorse XF RPMI medium (Agilent; 103576-100) supplemented with 10 mM glucose, 2 mM l-glutamine, 1 mM pyruvate, and 5 mM HEPES (pH 7.4). Measurements were obtained under basal conditions and after the addition of 0.5 μM rotenone/antimycin A (103015-100; Seahorse Bioscience) and 2-deoxy-d-glucose (2-DG) at a final well concentration of 50 mM (Agilent Technologies). Basal glycolysis and proton efflux rate (PER) were calculated according to reference 65. Lactate measurements were detected using the Cayman l-lactate assay kit (Cayman Chemical; 70010). In brief, 24 h posttreatment, supernatant from 200,000 primary resting and activated CD4+ T cells was harvested and measured according to the manufacturer’s recommendation.
Western blot analysis.
For analysis of phosphor-GSK-3β, proteins were extracted from cells with radioimmunoprecipitation assay (RIPA) buffer (diluted from 10× stock solution; Sigma-Aldrich catalogue no. 20-188) with protease and phosphatase inhibitors (Roche) added according to the manufacturer’s instructions. Laemmli 2× buffer was added to prepare the SDS-PAGE samples. Whole-cell lysates were run on NuPage 4-to-12% gradient bis-Tris gels with loading of equal cell numbers per lane and transferred to polyvinylidene difluoride (PVDF) membranes (Bio-Rad). Membranes were blocked for 1 h in Tris-buffered saline with Tween (TBST) plus 5% bovine serum albumin (BSA), followed by primary antibody incubation overnight at 4°C, a 1-h wash period, secondary-antibody incubation (1 h at room temperature [RT], with anti-rabbit HRP-conjugated antibodies [SouthernBiotech] diluted 1:10,000 in TBST plus 5% BSA plus 2.5% milk), and a final 2-h wash period. Protein levels were assessed using the following primary antibodies: p-S9-GSK3β (CST no. 9323) and pan-GSK3β (CST no. 9315). Blots were developed using an enhanced chemiluminescence (ECL) Western blot detection kit (PerkinElmer) or Luminata Forte (Millipore).
For analysis of p65, nuclear extracts were prepared from purified CD4+ T cells using the NE-PER nuclear and cytoplasmic extraction reagent supplemented with protease inhibitor cocktail (Thermo Scientific no. 78833), as specified by the manufacturer’s protocol. Protein concentration of extracts was determined by Bio-Rad protein assay (Bio-Rad no. 5000001). Nuclear extracts were resolved by electrophoresis on a 10% SDS-polyacrylamide gel using 15 μg of protein per condition and then transferred to PVDF membranes. Membranes were immunoblotted with primary antibodies for p65 (Cell Signaling Technology no. 8242) and TATA binding protein (Abcam no. ab63766); primary antibodies were diluted 1:1,000. Detection of primary antibodies was achieved by incubation with HRP-conjugated secondary antibody. Bands were visualized by chemiluminescent detection using Immobilon Forte Western HRP ECL substrate (Millipore catalogue no. WBLUF0100) followed by exposure to Hyperfilm ECL (GE/Amersham no. 45001508).
Lactate production analysis.
CD4+ T cells isolated from 3 HIV-negative donors were treated for 24 h at 37°C as indicated above either in normal RPMI or in medium with 50 mM 2-deoxy-d-glucose (2-DG). Subsequently, media of the treated samples were tested for production of l-lactated using a glycolysis cell-based assay kit (Cayman Chemical). This test measures the quantity of formazan, an artificial chromogenic dye that results from the reduction of tetrazolium salts in the presence of NADH in the culture medium. Colored formazan absorbs between 490 and 520 nm and can be detected by a plate reader. Since lactate dehydrogenase catalyzes the reaction between NAD+ and lactate, yielding pyruvate and NADH, which directly reduces the tetrazolium salt, the quantity of formazan produced is proportional to the quantity of l-lactate in the culture medium and is thus an indirect measurement of glycolysis. In order to determine specific l-lactate concentration, a standard curve was obtained analyzing absorbance of a known concentration of l-lactate as indicated in the manufacturer’s protocol.
Flow cytometry.
Freshly isolated PBMCs or CD4+ T cells were stained for viability, using the fixable viability dye Zombie violet (1:50; BioLegend) according to the manufacturer's recommendation. Surface staining was performed with the following antibodies at 1:100 dilutions in staining buffer for 20 min on ice: phycoerythrin (PE)-conjugated CD3-Cy7, allophycocyanin (APC)-conjugated CD4-Cy7, fluorescein isothiocyanate (FITC)-conjugated CD69, CD25-PE (all 1:100; BioLegend), and GLUT1-Ax647 (BD Bioscience, 566580). Cells were fixed in 1% paraformaldehyde (PFA)-containing PBS. All flow cytometry analyses were performed on the LSRII flow cytometer (BD Biosciences) using FlowJo software (v.10.4.2). Color compensations were performed using single-stained samples for each of the fluorochromes used. Data were analyzed using FlowJo software (V10.4.2; TreeStar, San Carlos, CA).
Immunophenotyping of RM T cells, NK cells, and monocytes was performed as previously described (66, 67) using whole blood stained with combinations of the following antibodies: anti-CD3 (SP34-2, APC-Cy7, and BUV395), anti-CD4 (L200, BV786, and BV510), anti-CD8alpha (SK1, BUV737), anti-CD11c (3.9, APC), anti-CD14 (M5E2, PE-Cy7, and FITC), anti-CD16 (3G8, BV650, and BV421), anti-CD20 (L27; Alexa Fluor 700, APC-Cy7, and APC-Fire), anti-CD28 (CD28.2, PE-Dazzle), anti-CD69 (FN50, TrueRed), anti-CD123 (7G3, TrueRed), anti-CD169 (7-239, PE), anti-CCR5 (3A9, APC), anti-CCR7 (15053, biotin), anti-HLA-DR (L243; BV510, PE-Dazzle, PE-ECD, and PE-Texas Red), anti-Ki67 (B56, FITC), and streptavidin (BV421 and BV605). Samples were analyzed on an LSRII flow cytometer (BD Biosciences), and data analysis was performed using FlowJo software (Tree Star).
T cell analyses were performed as previously described (68) by gating first on the small-lymphocyte population and then on the CD3+ T cell subsets before gating on CD4+ and CD8+ T cell subsets. Naive cells were defined as a contiguous population of CD28-intermediate, CD95-low, CCR7+ CCR5− cells, and memory cells were defined as CD95-high cells. Central memory cells were defined as CD28+ CCR5− CCR7+, transitional memory cells were defined as CD28+ CCR5+, and effector memory cells were defined as CD28− CCR5lo CCR7−. NK cell subsets were defined by gating on the small-lymphocyte population and then CD16 and CD56 expression on CD3− CD8+ NKG2A+ small lymphocytes as previously described (69). Monocyte populations were defined by CD14 and CD16 expression on HLA-DR+ CD20− CD3− small lymphocytes.
Assessment of CD8+ T cell cytotoxic effector functions.
After drug treatment, human PBMCs were cultured in complete RPMI in the presence of CD107a-BV650 (1:50; 328637; BioLegend) or CD107a-FITC antibodies (1:50; 555800; BD Bioscience) and PMA/ionomycin (50 ng/ml and 1 μM) for 3.5 h, with the addition of brefeldin A and monensin (eBioscience) for the last hour. Cells were subsequently stained for viability using the viability dye eFluor 506 (eBioscience) according to the provided protocol. Following surface marker staining with the antibodies (all at a 1:100 dilution) CD3-APC-H7 (560176; BD Biosciences), CD8-V450 (560347; BD Biosciences), CD45RA-APC (17–0458; eBioscience), and CCR7-PE-Cy7 (557648; BD Biosciences), cells were permeabilized and fixed using the Foxp3/transcription factor fixation/permeabilization concentrate and diluent kit (eBioscience). Intracellular staining was performed with IFN-γ–PerCP–Cy5.5 antibody (1:100; 560704; BD Bioscience) in Foxp3 Perm/Wash buffer for 1 h at 4°C. After washing and fixation of the cells in 1% paraformaldehyde–PBS, cells were analyzed by flow cytometry. Final data stem from having gated on CD8hi- and CD8lo-expressing T cells.
Assessment of NK cell cytotoxic effector functions.
Density centrifugation using a Ficoll-Hypaque gradient was performed to purify PBMCs from continuous-flow centrifugation leukapheresis products. Subsequently, cells were plated in 96-well U-bottom plates in complete RPMI, which was supplemented with 10 ng/ml recombinant interleukin 15 (IL-15) (R&D systems), treated with LRAs or diluent control (DMSO), and incubated at 37°C and 5% CO2 for 48 h.
To assay NK cell functionality, cells were analyzed by flow cytometry as previously described (70). Briefly, 1 × 106 cells/200 μl were stained with FITC-labeled anti-CD107 antibodies (1:50; BD Biosciences), mixed with 1 × 105 K562 cells, and incubated at 37°C and 5% CO2 for 1 h. Then, in order to enhance intracellular cytokine staining, protein transport was blocked with brefeldin A (Sigma-Aldrich) and Golgi Stop (BD Biosciences) according to the manufacturers’ instructions, and the cells were incubated at 37°C and 5% CO2 for another 5 h. Afterwards, cells were spun down (5 min, 330 × g, RT), the supernatant was removed, and the cells were stained with Zombie violet (1:50; BioLegend) according to the manufacturer protocol. Subsequently, cells were stained for surface proteins for 15 min at room temperature with the following antibodies: APC/Cy7–anti-CD3 (1:100; BioLegend), APC–anti-CD14 (1:100; BioLegend), APC–anti-CD19 (1:100; BioLegend), PE/Cy7–anti-CD56 (1:100; BD Biosciences), Alexa Fluor 700–anti-CD16 (1:100; BioLegend), PE–anti-CD7 (1:50; Thermo Fisher Scientific). Next, cells were permeabilized and fixed using the Foxp3/transcription factor fixation/permeabilization concentrate and diluent kit (eBioscience) and stained intracellularly with PerCP/Cy5–anti-IFN-γ (1:50; BD Bioscience) in Foxp3 Perm/Wash buffer for 1 h at 4°C. Finally, cells were washed and fixed in 1% paraformaldehyde–PBS and subjected to flow cytometry analysis on a LSRII flow cytometer (BD Biosciences).
Data were analyzed using FlowJo software (TreeStar). Samples were gated for singlets and for live, CD3-negative, CD14/CD19-negative, and CD7-positive cells as previously described (70). CD16-high CD56-dim, CD16-dim CD56-high cells were considered for analysis of IFN-γ and CD107a positivity. All gates were defined based on fluorescence-minus-one (FMO) samples.
Granzyme B cytotoxicity assay and infected-CD4+-T-cell elimination.
Day 6 effectors were prepared as previously described (46), but with the following modification. PBMCs were coincubated with HIVSF162-infected autologous CD4+ T cell targets (effector-to-target ratio [E:T], 25:1) with or without tideglusib at a final concentration of 1 μM in 96-well deep (1 ml)-well plates for 6 days. As before, cultured day 6 cells were then labeled with immunomagnetic beads (CD8+ T cell isolation kit II; Miltenyi Biotec) prior to negative selection of CD8+ T cells by magnetic automated cell sorting (46). Cytotoxic responses were measured against LIVE/DEAD fixable violet stain (Molecular Probes, Invitrogen Detection Technologies, Eugene, OR, USA)-labeled HIVSF162- infected or uninfected autologous CD4+ T cell targets in assays examining GrB target cell activity and infected-CD4+-T-cell elimination (ICE) as previously reported (46). Cells were analyzed on a FACSAria multilaser cytometer (Becton Dickinson) with FACSDiva software. Gates were drawn on labeled CD4+ T-cell targets, and 5,000 to 15,000 events were collected. Color compensations were performed using single-stained samples for each of the fluorochromes used. Data were analyzed using FlowJo software (TreeStar, San Carlos, CA).
Animals.
A total of 12 purpose-bred, male rhesus macaques (RM, Macaca mulatta) of Indian genetic background were used for this work. Animals were housed at the Oregon National Primate Research Center (ONPRC) under the oversight of the Institutional Animal Care and Use Committee, following the standards prescribed by the United States National Institutes of Health Guide for the Care and Use of Laboratory Animals. These RM were specific pathogen free, defined as being free of cercopithecine herpesvirus 1, d-type simian retrovirus, simian T-lymphotrophic virus type 1, rhesus rhadinovirus, and Mycobacterium tuberculosis. RM were intravenously inoculated with 2 TZM-bl assay focus-forming units of RM PBMC-expanded SIVmac239X (71) before starting daily subcutaneous injections of ART (consisting of 5.1 mg kg−1 day−1 tenofovir disoproxil (TDF), 40 mg kg−1 day−1 emtricitabine (FTC), and 2.5 mg kg−1 day−1 dolutegravir (DTG) in a solution containing 15% (vol/vol) Kleptose at pH 4.2 as previously described (72) at 12 dpi). Before the tideglusib treatment period, one RM was lost to study due to an unrelated illness; a second RM never fully suppressed SIV (>1,000 copies/ml plasma after >600 days of ART) and was excluded from later analysis.
The remaining 10 animals were randomized into two treatment groups (5 per group) such that the areas under the curves of pVL from 0 to 84 dpi were similar between the groups. For oral administration, 2 formulations of tideglusib (AMO Pharma Ltd.) were administered daily, mixed with food. This includes (i) a neat micronized drug substance and (ii) a clinical product formulated as a powder for oral suspension (AMO-02). The neat drug substance was administered for 21 to 28 days starting 672 dpi at 8.75 mg/kg, 852 dpi at 20 mg/kg, 873 dpi at 40 mg/kg, and 915 dpi at 80 mg/kg. AMO-02 or placebo was administered for 22 days starting 708 dpi at 8.75 mg/kg. For intravenous administration, the neat drug substance was formulated in PEG 400 and administered at 5 mg/kg at 1,027, 1,041, 1,055, 1,069, and 1,097 dpi.
SIV quantification.
SIV RNA levels in plasma were measured as described previously using a quantitative real-time/digital RT-PCR assay targeting SIV Gag, with a threshold limit of detection of 15 copies/ml (standard) or 1 copy/ml (ultrasensitive) (49). Cell-associated SIV RNA and DNA levels in tissues and cells were measured using nested quantitative hybrid real-time/digital RT-PCR and PCR assays targeting SIV Gag following protocols described previously (49).
SIV RNA in situ hybridization and image analysis.
RNAscope was performed on formaldehyde-fixed, paraffin-embedded tissue sections (5 μm) according to a previously published protocol (73) with the following minor modifications: heat-induced epitope retrieval was performed by boiling slides in 1× target retrieval solution (322000; ACD) for 30 min., followed by incubation at 40°C with a 1:10 dilution of protease III (322337; ACD) in 1× PBS for 20 min. Slides were incubated with the target probe SIVmac239X (312811; ACD) for 2 h at 40°C, and amplification was performed with RNAscope 2.5 HD detection kits (322360; ACD) according to the manufacturer’s instructions, with 0.5× wash buffer (310091; ACD) used between steps. The resulting signal was detected with Warp Red chromogen (WR806M; Biocare Medical). Slides were counterstained with Churukian-Allison-Tacha (CAT) hematoxylin (CATHE-GL; Biocare Medical) and mounted with Clearmount (17885-15; EMS) until dry. Coverslips were applied using Permount (SP15-100; Fisher Scientific), and slide were scanned at ×40 magnification on an Aperio AT2 (Leica Biosystems). RNAscope images were analyzed for the total number of viral-RNA-positive cells/105 total cells (quantitative) using the CytoNuclear module (v1.6) within Halo software (v2.3.2089.27; Indica Labs). Module settings to distinguish positive versus negative cells were established on concomitantly run control slides containing acutely SIV-infected and SIV-uninfected samples.
Serum chemistry.
Serum chemistry analysis was performed by the ONPRC Division of Comparative Medicine, Department of Clinical Pathology, using the Hariba ABX Petra 400 clinical chemistry system (Kyoto, Japan), according to the manufacturer’s instructions.
Assessment of tideglusib plasma concentration.
Tideglusib analysis was adapted from unpublished methods provided by AMO Pharma Ltd. and as previously described (74). A concentrated stock solution of internal standard CCG-50014 (Cayman Chemical) was prepared in DMSO at 10 mg/ml before being further diluted in methanol to a final working concentration of 1 μg/ml. Tideglusib micronized drug substance was prepared in methanol at 1 mg/ml. Standard curves were prepared by spiking naive sodium heparin RM plasma at 0, 5, 10, 50, 100, 500, and 1,000 ng/ml of tideglusib from methanol stocks and serial dilution. Next, 150 μl of spiked standards and samples was aliquoted into 1.5 ml Eppendorf tubes, spiked with 10 ng of internal standard, pretreated with 150 μl of 1:1 isopropanol-water, touch vortexed, and finally centrifuged at setting 4 for 1 min to clarify in a benchtop centrifuge. Pretreated samples were then loaded onto SLE+ 400-μl cartridges (Biotage, Charlotte, NC) and, after a 5-min loading step, eluted three times using gravity with 400 μl of dichloromethane, waiting 5 min between each elution, into glass 13- by 100-mm tubes. Elution was finished with a brief high-positive-pressure push for 1 to 2 s at 20 lb/in2. Samples were dried in a Turbovap (Biotage, Charlotte, NC) at 1 liter/min nitrogen at 25°C for 15 min. Samples were reconstituted in 80 μl of 80% acetonitrile/water and then filtered using 0.2-μm spin filters. Samples above the highest curve point were diluted 1:10 or 1:50 using naive RM plasma and reprepared.
Samples were analyzed using a 5500 QTRAP hybrid triple-quadrupole linear ion trap mass spectrometer (Applied Biosystems, Carlsbad, CA) with electrospray ionization (ESI) in positive mode. The mass spectrometer was interfaced to a Shimadzu (Columbia, MD) SIL-20AC XR auto-sampler followed by 2 LC-20AD XR LC pumps. Analytes were separated on an Atlantis dC18 50-mm-long by 2.1-mm-inner-diameter column held at 40°C in a Shimadzu CTO-20AC column oven using an isocratic mobile phase consisting of 1 mM ammonium fluoride and acetonitrile at a 20:80 ratio delivered at a variable flow rate. The initial flow rate was 0.3 ml/min and held for 1 min, then ramped to 1 ml/min over 0.1 min, held for 2.4 min, and ramped back down to the original 0.3 ml/min in 0.1 min, and re-equilibrated for 1.4 min. HPLC flow was directed to the source from 0.5 to 3.7 min; during the other times, the flow was directed to waste. The source parameters were as follows: curtain gas, 40; IonSpray voltage, 4000; source and gas temperature, 400; ion source gas 1, 35; ion source gas 2, 25; and collision-associated dissociation gas, −2. The multiple-reaction monitoring (MRM) transitions were optimized using infusion of each compound and included the following quantifier MRMs: tideglusib, m/z = 335.1→91 (parameters: declustering potential (DP), 50; entrance potential (EP), 10; collision energy (CE), 40; collision cell exit potential (CXP), 25), and CCG_50014, m/z = 317.2→109 (parameters: DP, 75; EP, 10; CE, 40; CXP, 25). Qualifier MRM transitions were tideglusib, m/z = 335.1→65, and CCG_50014, m/z = 317.2→95. Data were acquired using Analyst 1.7.3 (AB Sciex, Ontario, Canada) and analyzed using Multiquant 3.0.1 (AB Sciex, Ontario, Canada).
Statistical analysis.
Comparisons was carried out groupwise and calculated using a ratio paired Student's t test (comparing populations derived from the same donor). Unless indicated otherwise, data are presented as means and standard errors of the means (SEM).
For the analysis of viral blips in monkeys, we used a mixed-effects logistic model that estimated how the odds of blips (values > 1) change as a function of (i) the animal the data were measured in, (ii) the day the observation was made, (iii) whether the animal was treated, and (iv) the interaction between day of observation and treatment. These fits were made using the glmer function, part of the lmerTest in the R package.
ACKNOWLEDGMENTS
We thank the study participants, the UCSF-SCOPE team, Marielle Cavrois and Nandhini Raman (Gladstone Flow Cytometry Core), Alex Pico and Reuben Thomas (Gladstone Bioinformatics Core), the Center for Advanced Technology (UCSF), the Quantitative Molecular Diagnostics Core of the AIDS and Cancer Virus Program, Frederick National Laboratory for Cancer Research, the Bioanalytical Shared Resource/Pharmacokinetics Core (OHSU), John C. W. Carroll for graphics arts, and Robin Givens for administrative support.
This work was supported by the amfAR Institute for HIV Cure Research, the University of California San Francisco-Gladstone Institute of Virology & Immunology Center for AIDS Research, the James B. Pendleton Charitable Trust, the Oregon National Primate Research Center (ONPRC) NIH grant award P51OD011092, and the following grants from the National Institutes of Health/National Institute of Allergy and Infectious Diseases: P30 AI027763, P01 AI131374, R01 AI127219, R01GM11790 (to S.K.P.), R01MH112457 (to S.K.P.), R01AI147777 (to N.R.R.), and R01939 (to M.O.). This work was also supported in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN261200800001E (to J.D.L.).
A.G., R.S., A.A.O., and W.C.G. designed the experiments; A.G., R.S., W.B., B.V.-M., P.A.H., M.M., H.S.S., S.A.M., D.R., L.E.H., J.R.J., T.A.P., Z.W.G., F.H., M.N., J.D.L., J.D.E., and E.H. performed experiments or sample analysis; A.G., R.S., W.B., B.V.-M., P.A.H., S.A.M., and W.C.G. analyzed the data. A.G. and W.C.G drafted the manuscript, and M.O., S.K.P., T.J.H., N.J.K., E.V., N.R.R., S.G.D., L.J.P., M.S., J.D.L., B.K., A.A.O., and W.C.G. revised the manuscript.
Michael Snape and Bernard Kiernan are employees and shareholders in AMO Pharma Ltd. and as such receive salaries from this company as well as having ownership of a financial interest in the products the company is developing.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Deeks SG. 2012. HIV: shock and kill. Nature 487:439–440. 10.1038/487439a. [DOI] [PubMed] [Google Scholar]
- 2.Rasmussen TA, Tolstrup M, Sogaard OS. 2016. Reversal of latency as part of a cure for HIV-1. Trends Microbiol 24:90–97. 10.1016/j.tim.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 3.Battivelli E, Dahabieh MS, Abdel-Mohsen M, Svensson JP, Tojal Da Silva I, Cohn LB, Gramatica A, Deeks S, Greene WC, Pillai SK, Verdin E. 2018. Distinct chromatin functional states correlate with HIV latency reactivation in infected primary CD4(+) T cells. Elife 7:e34655. 10.7554/eLife.34655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barouch DH, Deeks SG. 2014. Immunologic strategies for HIV-1 remission and eradication. Science 345:169–174. 10.1126/science.1255512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwarzer R, Gramatica A, Greene WC. 2020. Reduce and control: a combinatorial strategy for achieving sustained HIV remissions in the absence of antiretroviral therapy. Viruses 12:188. 10.3390/v12020188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ward AR, Mota TM, Jones RB. 2020. Immunological approaches to HIV cure. Semin Immunol 2020:101412. 10.1016/j.smim.2020.101412. [DOI] [PubMed] [Google Scholar]
- 7.Jones RB, Walker BD. 2016. HIV-specific CD8+ T cells and HIV eradication. J Clin Invest 126:455–463. 10.1172/JCI80566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siliciano RF, Greene WC. 2011. HIV latency. Cold Spring Harb Perspect Med 1:a007096. 10.1101/cshperspect.a007096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Almeida L, Lochner M, Berod L, Sparwasser T. 2016. Metabolic pathways in T cell activation and lineage differentiation. Semin Immunol 28:514–524. 10.1016/j.smim.2016.10.009. [DOI] [PubMed] [Google Scholar]
- 10.Buck MD, O'Sullivan D, Pearce EL. 2015. T cell metabolism drives immunity. J Exp Med 212:1345–1360. 10.1084/jem.20151159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manning BD, Toker A. 2017. AKT/PKB signaling: navigating the network. Cell 169:381–405. 10.1016/j.cell.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aylett CH, Sauer E, Imseng S, Boehringer D, Hall MN, Ban N, Maier T. 2016. Architecture of human mTOR complex 1. Science 351:48–52. 10.1126/science.aaa3870. [DOI] [PubMed] [Google Scholar]
- 13.Bridges D, Saltiel AR. 2015. Phosphoinositides: key modulators of energy metabolism. Biochim Biophys Acta 1851:857–866. 10.1016/j.bbalip.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dan HC, Cooper MJ, Cogswell PC, Duncan JA, Ting JP, Baldwin AS. 2008. Akt-dependent regulation of NF-{kappa}B is controlled by mTOR and Raptor in association with IKK. Genes Dev 22:1490–1500. 10.1101/gad.1662308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dan HC, Ebbs A, Pasparakis M, Van Dyke T, Basseres DS, Baldwin AS. 2014. Akt-dependent activation of mTORC1 complex involves phosphorylation of mTOR (mammalian target of rapamycin) by IkappaB kinase alpha (IKKalpha). J Biol Chem 289:25227–25240. 10.1074/jbc.M114.554881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taylor HE, Calantone N, Lichon D, Hudson H, Clerc I, Campbell EM, D'Aquila RT. 2020. mTOR overcomes multiple metabolic restrictions to enable HIV-1 reverse transcription and intracellular transport. Cell Rep 31:107810. 10.1016/j.celrep.2020.107810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roy J, Paquette JS, Fortin JF, Tremblay MJ. 2002. The immunosuppressant rapamycin represses human immunodeficiency virus type 1 replication. Antimicrob Agents Chemother 46:3447–3455. 10.1128/aac.46.11.3447-3455.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Besnard E, Hakre S, Kampmann M, Lim HW, Hosmane NN, Martin A, Bassik MC, Verschueren E, Battivelli E, Chan J, Svensson JP, Gramatica A, Conrad RJ, Ott M, Greene WC, Krogan NJ, Siliciano RF, Weissman JS, Verdin E. 2016. The mTOR complex controls HIV latency. Cell Host Microbe 20:785–797. 10.1016/j.chom.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kitagishi Y, Kobayashi M, Kikuta K, Matsuda S. 2012. Roles of PI3K/AKT/GSK3/mTOR pathway in cell signaling of mental illnesses. Depress Res Treat 2012:752563. 10.1155/2012/752563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cohen P, Frame S. 2001. The renaissance of GSK3. Nat Rev Mol Cell Biol 2:769–776. 10.1038/35096075. [DOI] [PubMed] [Google Scholar]
- 21.Chen C-H, Shaikenov T, Peterson TR, Aimbetov R, Bissenbaev AK, Lee S-W, Wu J, Lin H-K, Sarbassov DD. 2011. ER stress inhibits mTORC2 and Akt signaling through GSK-3beta-mediated phosphorylation of rictor. Sci Signal 4:ra10. 10.1126/scisignal.2001731. [DOI] [PubMed] [Google Scholar]
- 22.Inoki K, Ouyang H, Zhu T, Lindvall C, Wang Y, Zhang X, Yang Q, Bennett C, Harada Y, Stankunas K, Wang CY, He X, MacDougald OA, You M, Williams BO, Guan KL. 2006. TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell 126:955–968. 10.1016/j.cell.2006.06.055. [DOI] [PubMed] [Google Scholar]
- 23.Eldar-Finkelman H, Martinez A. 2011. GSK-3 inhibitors: preclinical and clinical focus on CNS. Front Mol Neurosci 4:32. 10.3389/fnmol.2011.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martinez A, Gil C, Perez DI. 2011. Glycogen synthase kinase 3 inhibitors in the next horizon for Alzheimer's disease treatment. Int J Alzheimers Dis 2011:280502. 10.4061/2011/280502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horrigan J, Gomes TB, Snape M, Nikolenko N, McMorn A, Evans S, Yaroshinsky A, Della Pasqua O, Oosterholt S, Lochmuller H. 2020. A phase 2 study of AMO-02 (tideglusib) in congenital and childhood-onset myotonic dystrophy type 1 (DM1). Pediatr Neurol 112:84–93. 10.1016/j.pediatrneurol.2020.08.001. [DOI] [PubMed] [Google Scholar]
- 26.Tolosa E, Litvan I, Höglinger GU, Burn D, Lees A, Andrés MV, Gómez-Carrillo B, León T, del Ser T, TAUROS Investigators. 2014. A phase 2 trial of the GSK-3 inhibitor tideglusib in progressive supranuclear palsy. Mov Disord 29:470–478. 10.1002/mds.25824. [DOI] [PubMed] [Google Scholar]
- 27.Lovestone S, Boada M, Dubois B, Hull M, Rinne JO, Huppertz HJ, Calero M, Andres MV, Gomez-Carrillo B, Leon T, del Ser T, ARGO Investigators. 2015. A phase II trial of tideglusib in Alzheimer's disease. J Alzheimers Dis 45:75–88. 10.3233/JAD-141959. [DOI] [PubMed] [Google Scholar]
- 28.Cross DA, Culbert AA, Chalmers KA, Facci L, Skaper SD, Reith AD. 2001. Selective small-molecule inhibitors of glycogen synthase kinase-3 activity protect primary neurones from death. J Neurochem 77:94–102. 10.1046/j.1471-4159.2001.t01-1-00251.x. [DOI] [PubMed] [Google Scholar]
- 29.Bullen CK, Laird GM, Durand CM, Siliciano JD, Siliciano RF. 2014. New ex vivo approaches distinguish effective and ineffective single agents for reversing HIV-1 latency in vivo. Nat Med 20:425–429. 10.1038/nm.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wood JE, Schneider H, Rudd CE. 2006. TcR and TcR-CD28 engagement of protein kinase B (PKB/AKT) and glycogen synthase kinase-3 (GSK-3) operates independently of guanine nucleotide exchange factor VAV-1. J Biol Chem 281:32385–32394. 10.1074/jbc.M604878200. [DOI] [PubMed] [Google Scholar]
- 31.Medina M, Wandosell F. 2011. Deconstructing GSK-3: the fine regulation of its activity. Int J Alzheimers Dis 2011:479249. 10.4061/2011/479249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, Lim D, Peterson TR, Choi Y, Gray NS, Yaffe MB, Marto JA, Sabatini DM. 2011. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science 332:1317–1322. 10.1126/science.1199498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mayya V, Lundgren DH, Hwang SI, Rezaul K, Wu L, Eng JK, Rodionov V, Han DK. 2009. Quantitative phosphoproteomic analysis of T cell receptor signaling reveals system-wide modulation of protein-protein interactions. Sci Signal 2:ra46. 10.1126/scisignal.2000007. [DOI] [PubMed] [Google Scholar]
- 34.Ruelas DS, Greene WC. 2013. An integrated overview of HIV-1 latency. Cell 155:519–529. 10.1016/j.cell.2013.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dan HC, Antonia RJ, Baldwin AS. 2016. PI3K/Akt promotes feedforward mTORC2 activation through IKKalpha. Oncotarget 7:21064–21075. 10.18632/oncotarget.8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin S, Liao Q, Chen J, Zhang L, He Q, Zhu H, Zhang X, Xu J. 2018. TSC1 and DEPDC5 regulate HIV-1 latency through the mTOR signaling pathway. Emerg Microbes Infect 7:138. 10.1038/s41426-018-0139-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laplante M, Sabatini DM. 2009. mTOR signaling at a glance. J Cell Sci 122:3589–3594. 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Menk AV, Scharping NE, Moreci RS, Zeng X, Guy C, Salvatore S, Bae H, Xie J, Young HA, Wendell SG, Delgoffe GM. 2018. Early TCR signaling induces rapid aerobic glycolysis enabling distinct acute T cell effector functions. Cell Rep 22:1509–1521. 10.1016/j.celrep.2018.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Man K, Kallies A. 2015. Synchronizing transcriptional control of T cell metabolism and function. Nat Rev Immunol 15:574–584. 10.1038/nri3874. [DOI] [PubMed] [Google Scholar]
- 40.MacIver NJ, Michalek RD, Rathmell JC. 2013. Metabolic regulation of T lymphocytes. Annu Rev Immunol 31:259–283. 10.1146/annurev-immunol-032712-095956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frauwirth KA, Riley JL, Harris MH, Parry RV, Rathmell JC, Plas DR, Elstrom RL, June CH, Thompson CB. 2002. The CD28 signaling pathway regulates glucose metabolism. Immunity 16:769–777. 10.1016/s1074-7613(02)00323-0. [DOI] [PubMed] [Google Scholar]
- 42.Walker-Sperling VE, Pohlmeyer CW, Tarwater PM, Blankson JN. 2016. The effect of latency reversal agents on primary CD8+ T cells: implications for shock and kill strategies for human immunodeficiency virus eradication. EBioMedicine 8:217–229. 10.1016/j.ebiom.2016.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clutton G, Xu Y, Baldoni PL, Mollan KR, Kirchherr J, Newhard W, Cox K, Kuruc JD, Kashuba A, Barnard R, Archin N, Gay CL, Hudgens MG, Margolis DM, Goonetilleke N. 2016. The differential short- and long-term effects of HIV-1 latency-reversing agents on T cell function. Sci Rep 6:30749. 10.1038/srep30749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clutton GT, Jones RB. 2018. Diverse impacts of HIV latency-reversing agents on CD8+ T-cell function: implications for HIV cure. Front Immunol 9:1452. 10.3389/fimmu.2018.01452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mota TM, McCann CD, Danesh A, Huang SH, Magat DB, Ren Y, Leyre L, Bui TD, Rohwetter TM, Kovacs CM, Benko E, MacLaren L, Wimpelberg A, Cannon CM, Hardy WD, Safrit JT, Jones RB. 2020. Integrated assessment of viral transcription, antigen presentation, and CD8(+) T cell function reveals multiple limitations of class I-selective histone deacetylase inhibitors during HIV-1 latency reversal. J Virol 94:e01845-19. 10.1128/JVI.01845-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Migueles SA, Osborne CM, Royce C, Compton AA, Joshi RP, Weeks KA, Rood JE, Berkley AM, Sacha JB, Cogliano-Shutta NA, Lloyd M, Roby G, Kwan R, McLaughlin M, Stallings S, Rehm C, O'Shea MA, Mican J, Packard BZ, Komoriya A, Palmer S, Wiegand AP, Maldarelli F, Coffin JM, Mellors JW, Hallahan CW, Follman DA, Connors M. 2008. Lytic granule loading of CD8+ T cells is required for HIV-infected cell elimination associated with immune control. Immunity 29:1009–1021. 10.1016/j.immuni.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim WK, McGary CM, Holder GE, Filipowicz AR, Kim MM, Beydoun HA, Cai Y, Liu X, Sugimoto C, Kuroda MJ. 2015. Increased expression of CD169 on blood monocytes and its regulation by virus and CD8 T cells in macaque models of HIV infection and AIDS. AIDS Res Hum Retroviruses 31:696–706. 10.1089/AID.2015.0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van der Kuyl AC, van den Burg R, Zorgdrager F, Groot F, Berkhout B, Cornelissen M. 2007. Sialoadhesin (CD169) expression in CD14+ cells is upregulated early after HIV-1 infection and increases during disease progression. PLoS One 2:e257. 10.1371/journal.pone.0000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Okoye AA, Hansen SG, Vaidya M, Fukazawa Y, Park H, Duell DM, Lum R, Hughes CM, Ventura AB, Ainslie E, Ford JC, Morrow D, Gilbride RM, Legasse AW, Hesselgesser J, Geleziunas R, Li Y, Oswald K, Shoemaker R, Fast R, Bosche WJ, Borate BR, Edlefsen PT, Axthelm MK, Picker LJ, Lifson JD. 2018. Early antiretroviral therapy limits SIV reservoir establishment to delay or prevent post-treatment viral rebound. Nat Med 24:1430–1440. 10.1038/s41591-018-0130-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim Y, Anderson JL, Lewin SR. 2018. Getting the “kill” into “shock and kill”: strategies to eliminate latent HIV. Cell Host Microbe 23:14–26. 10.1016/j.chom.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nixon CC, Mavigner M, Sampey GC, Brooks AD, Spagnuolo RA, Irlbeck DM, Mattingly C, Ho PT, Schoof N, Cammon CG, Tharp GK, Kanke M, Wang Z, Cleary RA, Upadhyay AA, De C, Wills SR, Falcinelli SD, Galardi C, Walum H, Schramm NJ, Deutsch J, Lifson JD, Fennessey CM, Keele BF, Jean S, Maguire S, Liao B, Browne EP, Ferris RG, Brehm JH, Favre D, Vanderford TH, Bosinger SE, Jones CD, Routy JP, Archin NM, Margolis DM, Wahl A, Dunham RM, Silvestri G, Chahroudi A, Garcia JV. 2020. Systemic HIV and SIV latency reversal via non-canonical NF-kappaB signalling in vivo. Nature 578:160–165. 10.1038/s41586-020-1951-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dashti A, Waller C, Mavigner M, Schoof N, Bar KJ, Shaw GM, Vanderford TH, Liang S, Lifson JD, Dunham RM, Ferrari G, Tuyishime M, Lam CK, Nordstrom JL, Margolis DM, Silvestri G, Chahroudi A. 2020. SMAC mimetic plus triple-combination bispecific HIVxCD3 retargeting molecules in SHIV.C.CH505-infected, antiretroviral therapy-suppressed rhesus macaques. J Virol 94:e00793-20. 10.1128/JVI.00793-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Planas D, Zhang Y, Monteiro P, Goulet JP, Gosselin A, Grandvaux N, Hope TJ, Fassati A, Routy JP, Ancuta P. 2017. HIV-1 selectively targets gut-homing CCR6+CD4+ T cells via mTOR-dependent mechanisms. JCI Insight 2:93230. 10.1172/jci.insight.93230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Doyon G, Zerbato J, Mellors JW, Sluis-Cremer N. 2013. Disulfiram reactivates latent HIV-1 expression through depletion of the phosphatase and tensin homolog. AIDS 27:F7–F11. 10.1097/QAD.0b013e3283570620. [DOI] [PubMed] [Google Scholar]
- 55.Palmer CS, Duette GA, Wagner MCE, Henstridge DC, Saleh S, Pereira C, Zhou J, Simar D, Lewin SR, Ostrowski M, McCune JM, Crowe SM. 2017. Metabolically active CD4+ T cells expressing Glut1 and OX40 preferentially harbor HIV during in vitro infection. FEBS Lett 591:3319–3332. 10.1002/1873-3468.12843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marko AJ, Miller RA, Kelman A, Frauwirth KA. 2010. Induction of glucose metabolism in stimulated T lymphocytes is regulated by mitogen-activated protein kinase signaling. PLoS One 5:e15425. 10.1371/journal.pone.0015425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wofford JA, Wieman HL, Jacobs SR, Zhao Y, Rathmell JC. 2008. IL-7 promotes Glut1 trafficking and glucose uptake via STAT5-mediated activation of Akt to support T-cell survival. Blood 111:2101–2111. 10.1182/blood-2007-06-096297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tamas P, Hawley SA, Clarke RG, Mustard KJ, Green K, Hardie DG, Cantrell DA. 2006. Regulation of the energy sensor AMP-activated protein kinase by antigen receptor and Ca2+ in T lymphocytes. J Exp Med 203:1665–1670. 10.1084/jem.20052469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chehtane M, Khaled AR. 2010. Interleukin-7 mediates glucose utilization in lymphocytes through transcriptional regulation of the hexokinase II gene. Am J Physiol Cell Physiol 298:C1560–C1571. 10.1152/ajpcell.00506.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Palmer CS, Ostrowski M, Balderson B, Christian N, Crowe SM. 2015. Glucose metabolism regulates T cell activation, differentiation, and functions. Front Immunol 6:1. 10.3389/fimmu.2015.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Palmer CS, Cherry CL, Sada-Ovalle I, Singh A, Crowe SM. 2016. Glucose metabolism in T cells and monocytes: new perspectives in HIV pathogenesis. EBioMedicine 6:31–41. 10.1016/j.ebiom.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Laird GM, Bullen CK, Rosenbloom DI, Martin AR, Hill AL, Durand CM, Siliciano JD, Siliciano RF. 2015. Ex vivo analysis identifies effective HIV-1 latency-reversing drug combinations. J Clin Invest 125:1901–1912. 10.1172/JCI80142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:RESEARCH0034. 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cox J, Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26:1367–1372. 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 65.Mookerjee SA, Brand MD. 2015. Measurement and analysis of extracellular acid production to determine glycolytic rate. J Vis Expl 2015:e53464. 10.3791/53464:e53464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pitcher CJ, Hagen SI, Walker JM, Lum R, Mitchell BL, Maino VC, Axthelm MK, Picker LJ. 2002. Development and homeostasis of T cell memory in rhesus macaque. J Immunol 168:29–43. 10.4049/jimmunol.168.1.29. [DOI] [PubMed] [Google Scholar]
- 67.Picker LJ, Hagen SI, Lum R, Reed-Inderbitzin EF, Daly LM, Sylwester AW, Walker JM, Siess DC, Piatak M, Jr., Wang C, Allison DB, Maino VC, Lifson JD, Kodama T, Axthelm MK. 2004. Insufficient production and tissue delivery of CD4+ memory T cells in rapidly progressive simian immunodeficiency virus infection. J Exp Med 200:1299–1314. 10.1084/jem.20041049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Okoye AA, Rohankhedkar M, Abana C, Pattenn A, Reyes M, Pexton C, Lum R, Sylwester A, Planer SL, Legasse A, Park BS, Piatak M, Jr., Lifson JD, Axthelm MK, Picker LJ. 2012. Naive T cells are dispensable for memory CD4+ T cell homeostasis in progressive simian immunodeficiency virus infection. J Exp Med 209:641–651. 10.1084/jem.20112071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.DeGottardi MQ, Okoye AA, Vaidya M, Talla A, Konfe AL, Reyes MD, Clock JA, Duell DM, Legasse AW, Sabnis A, Park BS, Axthelm MK, Estes JD, Reiman KA, Sekaly RP, Picker LJ. 2016. Effect of anti-IL-15 administration on T cell and NK cell homeostasis in rhesus macaques. J Immunol 197:1183–1198. 10.4049/jimmunol.1600065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Milush JM, Lopez-Verges S, York VA, Deeks SG, Martin JN, Hecht FM, Lanier LL, Nixon DF. 2013. CD56negCD16(+) NK cells are activated mature NK cells with impaired effector function during HIV-1 infection. Retrovirology 10:158. 10.1186/1742-4690-10-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Del Prete GQ, Park H, Fennessey CM, Reid C, Lipkey L, Newman L, Oswald K, Kahl C, Piatak M, Jr., Quinones OA, Alvord WG, Smedley J, Estes JD, Lifson JD, Picker LJ, Keele BF. 2014. Molecularly tagged simian immunodeficiency virus SIVmac239 synthetic swarm for tracking independent infection events. J Virol 88:8077–8090. 10.1128/JVI.01026-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Del Prete GQ, Smedley J, Macallister R, Jones GS, Li B, Hattersley J, Zheng J, Piatak M, Keele BF, Hesselgesser J, Geleziunas R, Lifson JD. 2016. Short communication: Comparative evaluation of coformulated injectable combination antiretroviral therapy regimens in simian immunodeficiency virus-infected rhesus macaques. AIDS Res Hum Retroviruses 32:163–168. 10.1089/aid.2015.0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Deleage C, Wietgrefe SW, Del Prete G, Morcock DR, Hao X-P, Piatak JM, Bess J, Anderson JL, Perkey K, Reilly C, McCune JM, Haase AT, Lifson JD, Schacker TW, Estes JD. 2016. Defining HIV and SIV reservoirs in lymphoid tissues. Pathog Immun 1:68. 10.20411/pai.v1i1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Saini NK, Suresh PS, Lella M, Bhamidipati RK, Rajagopal S, Mullangi R. 2018. LC-MS/MS determination of tideglusib, a novel GSK-3beta inhibitor in mice plasma and its application to a pharmacokinetic study in mice. J Pharm Biomed Anal 148:100–107. 10.1016/j.jpba.2017.09.022. [DOI] [PubMed] [Google Scholar]