Herpesviruses establish lifelong quiescent infections in specific cells in the body and reactivate to produce infectious virus only when precise signals induce them to do so. The signals that induce herpesvirus reactivation are often studied only in one particular cell type infected with the virus.
KEYWORDS: MHV68, cytokines, gammaherpesvirus, reactivation
ABSTRACT
Gammaherpesviruses such as Epstein-Barr virus (EBV), Kaposi’s sarcoma-associated virus (KSHV), and murine gammaherpesvirus 68 (MHV68) establish latent infection in B cells, macrophages, and nonlymphoid cells and can induce both lymphoid and nonlymphoid cancers. Research on these viruses has relied heavily on immortalized B cell and endothelial cell lines. Therefore, we know very little about the cell type-specific regulation of virus infection. We have previously shown that treatment of MHV68-infected macrophages with the cytokine interleukin-4 (IL-4) or challenge of MHV68-infected mice with an IL-4-inducing parasite leads to virus reactivation. However, we do not know if all latent reservoirs of the virus, including B cells, reactivate the virus in response to IL-4. Here, we used an in vivo approach to address the question of whether all latently infected cell types reactivate MHV68 in response to a particular stimulus. We found that IL-4 receptor expression on macrophages was required for IL-4 to induce virus reactivation but that it was dispensable on B cells. We further demonstrated that the transcription factor STAT6, which is downstream of the IL-4 receptor and binds the virus gene 50 N4/N5 promoter in macrophages, did not bind to the virus gene 50 N4/N5 promoter in B cells. These data suggest that stimuli that promote herpesvirus reactivation may affect latent virus only in particular cell types but not in others.
IMPORTANCE Herpesviruses establish lifelong quiescent infections in specific cells in the body and reactivate to produce infectious virus only when precise signals induce them to do so. The signals that induce herpesvirus reactivation are often studied only in one particular cell type infected with the virus. However, herpesviruses establish latency in multiple cell types in their hosts. Using murine gammaherpesvirus 68 (MHV68) and conditional knockout mice, we examined the cell type specificity of a particular reactivation signal, interleukin-4 (IL-4). We found that IL-4 induced herpesvirus reactivation only from macrophages but not from B cells. This work indicates that regulation of virus latency and reactivation is cell type specific. This has important implications for therapies aimed at either promoting or inhibiting reactivation for the control or elimination of chronic viral infections.
INTRODUCTION
Herpesviruses establish chronic infections in their hosts. However, these viruses do not persistently replicate. Instead, they establish quiescent infections, termed latency. Latency is characterized by limited viral gene expression and no viral progeny production. Latent infections periodically reactivate and reenter the lytic cycle to produce infectious virus. Latency and reactivation of herpesviruses are tightly controlled by the host immune system, reflecting the coevolution of these viruses with their hosts. The development of drugs and vaccines to target herpesvirus, as well as efforts aimed at developing herpesviruses as vaccine vectors and oncolytic agents, requires that we understand the mechanisms that govern host control of herpesvirus latency.
The human gammaherpesviruses Epstein-Barr virus (EBV) and Kaposi’s sarcoma-associated herpesvirus (KSHV) infect humans only, and the cell culture systems available to study these viruses are limited to a few specific cell types. Pathogenesis and host-pathogen interactions of gammaherpesviruses can be examined using a close genetic relative of EBV and KSHV, murine gammaherpesvirus 68 (MHV68). MHV68 establishes latency in macrophages and B cells and leads to lymphomas and tumors in immunocompromised mice, similar to EBV and KSHV in humans (1–6).
Because herpesvirus infections are ubiquitous, we previously examined the effects of coinfection on herpesvirus latency and reactivation. We showed that helminth parasites induce herpesvirus reactivation from latency. We identified the host cytokine interleukin-4 (IL-4) and the transcription factor signal transducer and activator of transcription 6 (STAT6) that are required for parasite infection-driven reactivation of MHV68 in mice (7). We determined that IL-4 receptor (IL-4R) signaling through STAT6 and binding of STAT6 to a viral promoter induce MHV68 lytic gene expression. In vitro, treatment of bone marrow-derived macrophages (BMDMs) with IL-4 increases virus replication. These data indicate that IL-4 can induce herpesvirus replication in macrophages in vitro, but it does not tell us which types of cells are responding to IL-4 in vivo.
Other host cytokines both positively and negatively regulate gammaherpesvirus latency in a cell type-specific manner. Interferon gamma (IFN-γ), for example, suppresses MHV68 reactivation only in the peritoneum in macrophages but not in the spleen in B cells (8, 9). Similarly, IFN-γ suppresses KSHV lytic replication in only certain cell types (10, 11). Other cytokines, including type I interferon and IL-21, also differentially impact MHV68 replication and latency in epithelial cells, macrophages, and B cells (12, 13). Understanding these cell type-specific mechanisms will be critical for developing therapies for chronic viral infection that either enforce latency to prevent reactivation or promote reactivation to purge latent reservoirs and clear the virus.
In our previous work, we detected robust IL-4-induced reactivation of MHV68 from peritoneal cells but not from the spleen, perhaps indicating that IL-4 promotes MHV68 reactivation in a tissue and cell type-specific manner. To determine whether IL-4 promotes gammaherpesvirus reactivation in a cell type-specific manner, we tested the requirements of IL-4R signaling in macrophages and B cells. We found that treatment of a latently infected B cell line with IL-4 did not lead to virus reactivation. Because of the limitations with in vitro systems, we examined the necessity of IL-4R signaling in macrophages and B cells in vivo. Using IL-4R-floxed mice crossed with either macrophage- or B cell-specific cre recombinase drivers, we determined the reactivation of MHV68 following treatment with IL-4/anti-IFN-γ in mice lacking IL-4R either only on macrophages or only on B cells. We further examined the binding of STAT6 to a viral promoter of the latent-to-lytic switch gene (RTA [open reading frame 50 {ORF50}]). Our results indicate that IL-4 induces herpesvirus reactivation and STAT6 binding to the ORF50 promoter in a cell type-specific manner.
RESULTS
IL-4 treatment increases viral replication in macrophages but does not reactivate virus from a latently infected B cell line.
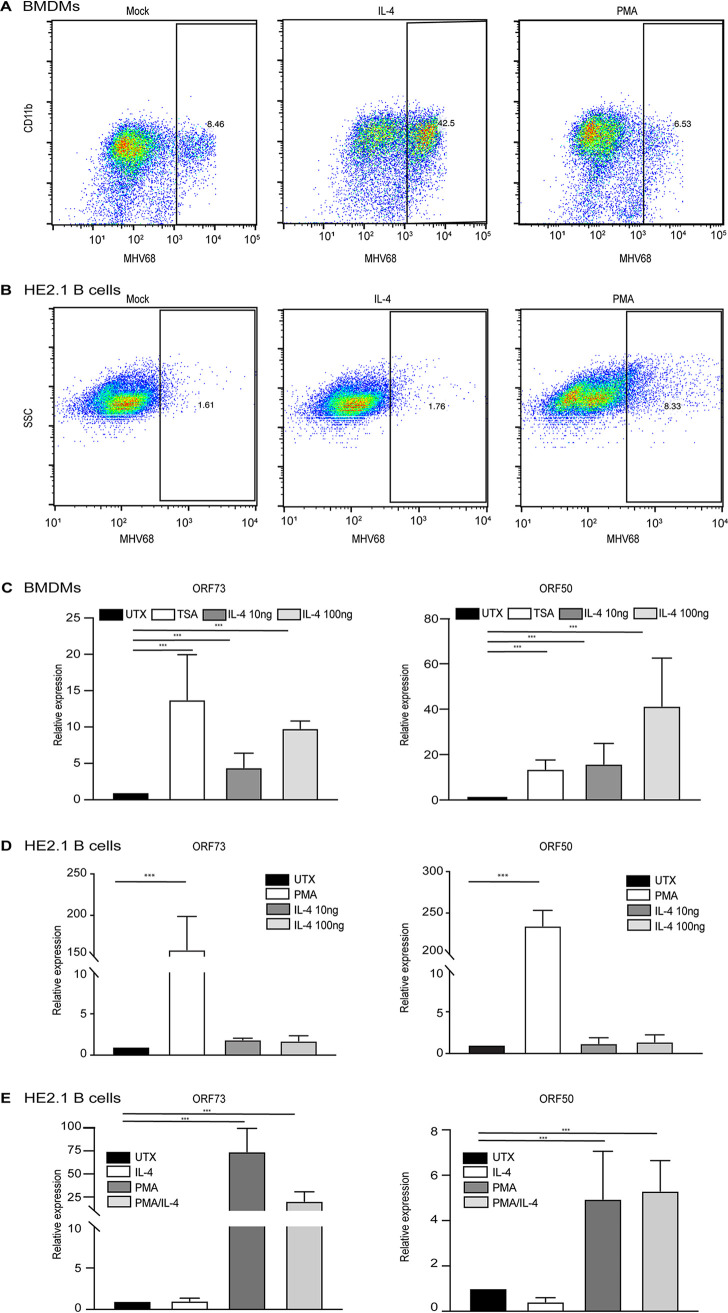
We first examined whether IL-4 reactivated MHV68 in vitro from two different cell types, macrophages and B cells, which harbor latent virus in vivo. In previous studies, we found that IL-4 treatment of bone marrow-derived macrophages (BMDMs) increased the replication of MHV68 (7). To confirm this, we treated BMDMs with IL-4 for 16 h before infection with MHV68. As expected, IL-4 treatment increased the number of cells that express MHV68 lytic proteins, as measured by a flow cytometry assay described previously (7), compared to untreated macrophages (Fig. 1A). To examine whether IL-4 has an effect on B cells similar to that on macrophages, we took advantage of a B cell line (HE2.1) that harbors reactivation-competent latent MHV68 (14). We treated the HE2.1 cells with IL-4 or phorbol 12-myristate 13-acetate (PMA), a positive control that induces virus reactivation in B cells but not macrophages (14, 15), and then examined the expression of MHV68 lytic proteins by flow cytometry. While PMA induced viral reactivation from B cells, as indicated by the increase in MHV68-positive cells, IL-4 treatment had no effect on MHV68 protein expression in the HE2.1 B cells (Fig. 1B). We further measured virus-specific gene expression after IL-4 treatment in macrophages and B cells. In accordance with our flow cytometry data, IL-4 treatment increased the expression of open reading frame 50 (Orf50) and Orf73 in infected macrophages to a level comparable to that with trichostatin A (TSA), a positive control that induces robust reactivation of MHV68 in macrophages (Fig. 1C) (15). Although PMA treatment increased the gene expression of Orf50 and Orf73 in HE2.1 B cells, we did not detect a significant increase in viral gene expression in this cell line after IL-4 treatment (Fig. 1D). We examined whether IL-4 could augment PMA treatment in HE2.1 B cells but found no additive effect of IL-4 plus PMA on virus gene expression (Fig. 1E). These data suggest that IL-4-induced reactivation of MHV68 is cell type specific. However, there are caveats with the use of cell lines, and de novo infection of primary B cells with MHV68 in tissue culture is limited. Another caveat of the in vitro experiments is that infection of macrophages with MHV68 in vitro replicates some aspects of latency, but it is not a true latent model, and lytic replication does occur (15). Therefore, we examined IL-4-induced reactivation from specific cell types in vivo.
FIG 1.
IL-4 treatment increases viral replication in macrophages but does not reactivate virus from a latently infected B cell line. (A) Bone marrow-derived macrophages (BMDMs) were treated with 100 ng/ml IL-4 and 20 ng/ml PMA for 16 h in culture medium and then infected with MHV68 at an MOI of 5. Twenty-four hours after infection, cells were fixed, and cells expressing lytic viral proteins were determined by flow cytometry. (B) HE2.1 B cells were treated with 100 ng/ml IL-4 and 20 ng/ml PMA for 38 h, cells were then fixed, and cells expressing lytic viral proteins were determined by flow cytometry. SSC, side scatter. (C) BMDMs were treated with 130 nM TSA or different concentrations of IL-4 for 16 h and then infected with MHV68 at an MOI of 5. Transcripts of the virus genes Orf50 and Orf73 were determined 12 h after infection. Expression was normalized to the expression of the glyceraldehyde-3-phosphate dehydrogenase gene (Gapdh). Data are from three independent experiments. (D) HE2.1 B cells were treated with 20 ng/ml PMA or different concentrations of IL-4 for 24 h. RT-qPCR was conducted to assess the expression of the virus genes Orf50 and Orf73. Expression was normalized to the expression of Gapdh. Data are from three independent experiments. (E and F) HE2.1 B cells were treated with 20 ng/ml PMA, 100 ng/ml IL-4, or PMA plus IL-4 for 24 h. RT-qPCR was conducted to assess the expression of the virus genes Orf50 and Orf73. Expression was normalized to the expression of Gapdh. Data are from three independent experiments. UTX, untreated.
IL-4 receptor expression is required for MHV68 reactivation from macrophages.
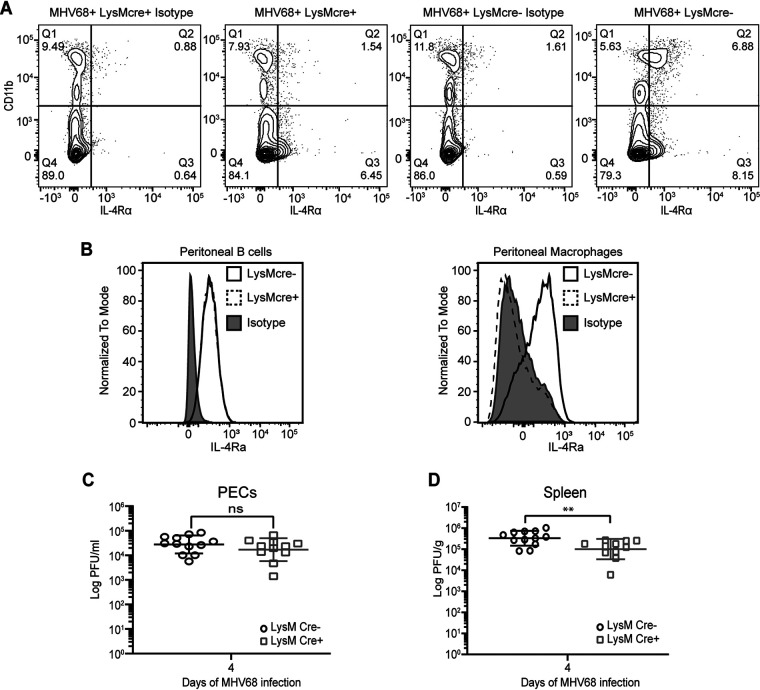
To determine whether IL-4-induced viral reactivation in vivo requires IL-4 receptor (IL-4R) on macrophages, we generated mice that are deficient in IL-4R specifically on myeloid cells, including macrophages. We crossed mice homozygous for loxP-flanked Il4rα genes, Il4rαflox/flox (Il4rαf/f) (16), with mice that express cre recombinase under the control of the lysozyme M (LysM) promoter (LysMcre). Gene knockout efficiency in Il4rαf/f × LysMcre-positive (Il4rαf/f × LysMcre+) mice was demonstrated by the absence of detectable levels of IL-4R expression in macrophages from peritoneal exudate cells (PECs) with and without virus infection (Fig. 2A and B). B cell expression of IL-4R in the peritoneum was normal in the Il4rαf/f × LysMcre+ mice (Fig. 2B).
FIG 2.
IL-4R signaling is not required for MHV68 replication. (A) Il4rαf/f × LysMcre+ and Il4rαf/f × LysMcre− mice were intraperitoneally infected with 106 PFU wild-type MHV68. Peritoneal lavage fluid was collected at 37 days post-infection, and flow cytometry was performed to detect IL-4R on peritoneal cells. Cells that have high expression levels of CD11b are macrophages, and CD11b-low or -negative cells include B cells. Contour plots are representative of data from one experiment. (B) Peritoneal exudate cells (PECs) were collected from uninfected Il4rαf/f × LysMcre+ and Il4rαf/f × LysMcre− mice, and flow cytometry was used to detect the IL-4 receptor on peritoneal B cells and macrophages. Representative histograms for two independent experiments are shown. (C and D) Il4rαf/f × LysMcre+ and Il4rαf/f × LysMcre− mice were infected by intraperitoneal injection with 106 PFU of MHV68. PECs (C) and whole spleens (D) were collected at day 4 post-infection, and the virus titer was determined by a plaque assay. Bars represent means ± standard deviations (SD). Each dot represents an individual mouse. ns, not significant; **, P < 0.01 (by a t test).
Before examining the role of IL-4 signaling in macrophages during latency, we first tested whether IL-4 signaling was required for the control of acute MHV68 replication. We compared virus replication in Il4rαf/f × LysMcre-positive and Il4rαf/f × LysMcre-negative (Il4rαf/f × LysMcre−) mice. After infecting mice with MHV68, PECs and spleen samples were collected on day 4 post-infection. Viral titers were determined by plaque assay. The Il4rαf/f × LysMcre-positive mice had levels of viral replication similar to those of Il4rαf/f × LysMcre-negative mice in the PECs and a small decrease in viral replication in spleen cells, indicating that IL-4 signaling has no or only a small effect on MHV68 acute replication (Fig. 2C and D).
We previously showed that injection with long-lasting IL-4 complexes (IL-4c) combined with blocking antibody to interferon gamma (IFN-γ) (IL-4c/anti-IFN-γ) mimicked intestinal helminth infection to induce MHV68 reactivation. While virus reactivation induced by parasite infection is highly variable, reactivation induced by IL-4c/anti-IFN-γ is more robust, as demonstrated by both bioluminescence imaging and an ex vivo reactivation assay (7). Therefore, we chose to use IL-4c/anti-IFN-γ to examine cell type-specific reactivation in vivo.
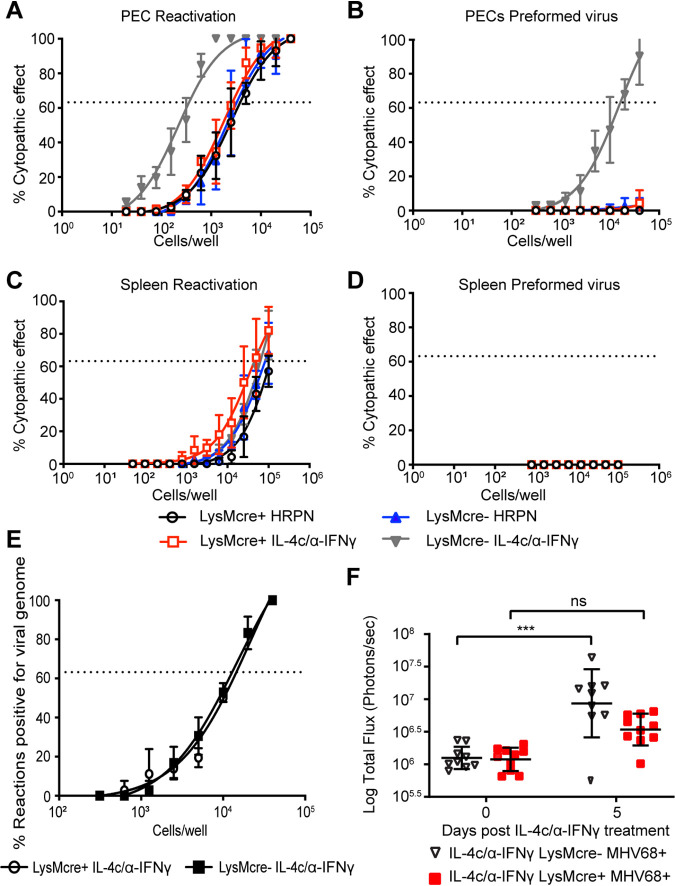
To determine if IL-4R expression is required on macrophages for virus reactivation in vivo, we infected Il4rαf/f × LysMcre-positive and -negative mice with MHV68. To induce reactivation from latency, we injected mice with either an isotype control (HRPN) or IL-4c/anti-IFN-γ one month after infection. Seven days after treatment, peritoneal cells, which harbor latent virus primarily in macrophages and B cells (4, 17), were collected for a limiting-dilution assay (LDA) to quantitate virus reactivation. In this ex vivo reactivation assay, explanted cells are plated on a mouse embryonic fibroblast (MEF) monolayer in 96-well plates in a limiting-dilution fashion. Cytopathic effect (CPE) is detected after 3 weeks, and the frequency of reactivating cells is determined using Poisson’s distribution (18). We can distinguish ex vivo reactivating virus from in vivo preformed virus by plating both live explanted cells and lysed cells. The lysed samples (termed disrupted) induce cytopathic effect of the MEF monolayers if they contain virus that reactivated in vivo prior to the collection of samples. The live explanted cells induce cytopathic effect on the MEF monolayers when virus reactivates during ex vivo culture. Similar to our previous findings, when Il4rαf/f × LysMcre− mice are injected with IL-4c/anti-IFN-γ, we observed substantial virus reactivation from latency compared with isotype-injected mice (HRPN) (Fig. 3A). Moreover, we detected preformed virus in the Il4rαf/f × LysMcre− mice injected with IL-4c/anti-IFN-γ (Fig. 3B), indicating that virus reactivated in vivo prior to collection. In contrast, when Il4rαf/f × LysMcre+ mice, which lack IL-4R on macrophages, were injected with IL-4c/anti-IFN-γ, there was no increase in virus reactivation ex vivo and no preformed virus (Fig. 3A and B). These data indicate that macrophage expression of IL-4R is required for IL-4c/anti-IFN-γ-induced virus reactivation in PECs. We also measured virus reactivation from splenocytes, which harbor latent virus primarily in B cells (19), and detected no significant increase in virus reactivation or preformed virus from either LysMcre+ or LysMcre− mice injected with IL-4c/anti-IFN-γ (Fig. 3C and D).
FIG 3.
IL-4R expression is required for MHV68 reactivation from macrophages. (A to D) Il4rαf/f × LysMcre+ and Il4rαf/f × LysMcre− mice were intraperitoneally infected with 106 PFU wild-type MHV68. Mice received the isotype control (HRPN) or both IL-4c and anti-IFN-γ at 28 days post-infection. Another dose of IL-4c was administered 2 days after the first treatment, and all mice were then euthanized at 5 days post-treatment. Peritoneal exudate cells (PECs) (A and B) and splenocytes (C and D) were processed into single-cell suspensions. (A and C) Reactivation frequencies were determined by ex vivo plating of serially diluted PECs and splenocytes on a MEF monolayer. (B and D) The presence of preformed virus in PECs and splenocytes was determined by disrupting the cell suspensions and plating serially diluted samples on MEF monolayers. Cytopathic effect was scored at 3 weeks post-plating. Groups of 3 to 5 mice were pooled for each experiment. Data represent the averages from 3 independent experiments. The error bars represent standard errors of the means. (E) Peritoneal exudate cells were processed into single-cell suspensions and subjected to nested PCR analysis to assess the frequency of cells harboring viral genomes. Groups of 3 to 5 mice were pooled for each infection and analysis. The data are pooled from 3 independent experiments. (F) Il4rαf/f × LysMcre+ and Il4rαf/f × LysMcre− mice were intraperitoneally infected with 106 PFU MHV68-M3FL. Mice received anti-IFN-γ and IL-4c after 40 days of infection. IL-4c was administered 2 days after the first treatment, and total flux (photons per second) from the abdominal region was then quantitated at day 5 post-treatment using an Ivis bioluminescence imager. Bars represent means ± SD. Each dot represents an individual mouse. The data are pooled from two independent experiments. ns, not significant; ***, P < 0.001.
Defective reactivation could be due to a failure of MHV68 to establish or maintain latency in IL-4R-deficient macrophages. To test this, the frequency of virus-positive peritoneal cells during chronic infection was quantitated by limiting-dilution nested PCR targeting MHV68 Orf72 (18). Equivalent numbers of viral genomes were detected in Il4rαf/f × LysMcre+ and Il4rαf/f × LysMcre− mice (Fig. 3E), indicating that latency was established at equivalent levels in Il4rαf/f × LysMcre+ and Il4rαf/f × LysMcre− mice.
To further confirm our LDA reactivation results, Il4rαf/f × LysMcre+ and Il4rαf/f × LysMcre− mice were infected with luciferase-tagged MHV68, MHV68-M3FL (20), and injected with IL-4c/anti-IFN-γ. Mice were imaged 5 days after injection in accordance with our previously published results (7). As expected, LysMcre− mice that received IL-4c/anti-IFN-γ displayed increased reactivation, whereas LysMcre+ mice did not, verifying that IL-4-induced reactivation in vivo requires IL-4R expression on macrophages (Fig. 3F). Collectively, these data suggest that macrophage expression of IL-4 receptor is required for IL-4c/anti-IFN-γ-induced virus reactivation in vivo.
IL-4R expression on macrophages is not required for cyclosporine-induced reactivation.
We next wanted to determine if the loss of IL-4R on macrophages generally inhibited virus reactivation. To test if Il4rαf/f × LysMcre mice exhibited reactivation to a stimulus unrelated to IL-4, we administered cyclosporine (CA) intraperitoneally. Cyclosporine is an immunosuppressive drug that reactivates MHV68 (20, 21). It targets multiple pathways in T cells that inhibit activation and proliferation. It is believed that cyclosporine increases the reactivation of MHV68 because T cells, especially CD8+ T cells, and the production of IFN-γ are important for the control of MHV68 (22, 23). Reactivation induced by cyclosporine should occur through IL-4 receptor-independent pathways.
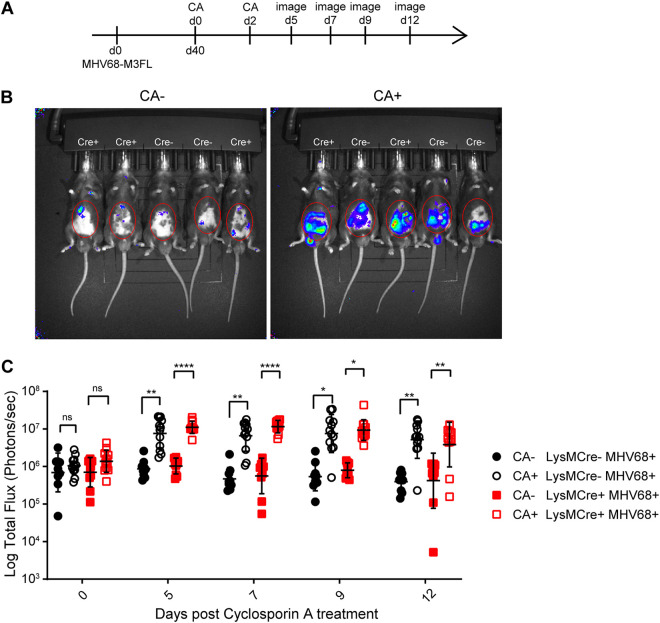
Il4rαf/f × LysMcre+ and Il4rαf/f × LysMcre− mice were infected with MHV68-M3FL to track virus reactivation in vivo. Infected mice were injected with cyclosporine on day 40 and day 42 of infection (Fig. 4A). The mice were imaged at several time points to capture the peak of reactivation, which occurred on day 7 after cyclosporine treatment (Fig. 4B and C). We detected reactivation from both LysMcre+ and LysMcre− mice after cyclosporine treatment (Fig. 4C), indicating that the lack of the IL-4 receptor in macrophages does not globally inhibit the in vivo reactivation of MHV68 in response to cyclosporine.
FIG 4.
Absence of IL-4R expression on macrophages does not impair cyclosporine-induced reactivation. (A) Timeline of treatments with cyclosporine (CA) and imaging to measure CA-induced MHV68-M3FL reactivation in mice. Total flux (photons per second) was measured using an Ivis bioluminescence imager. (B) Representative images of Il4rαf/f × LysMcre+ and Il4rαf/f × LysMcre− mice on day 7 after CA treatment. The red circles mark the areas used for total flux measurements. (C) Quantitation of total flux. The data shown are the results obtained from a pool of two independent experiments. Bars represent means ± SD. Each dot represents an individual mouse. ns, not significant; *, P < 0.05; **, P < 0.01; ****, P < 0.001.
The absence of IL-4 receptor on B cells does not affect IL-4c/anti-IFN-γ-induced MHV68 reactivation.
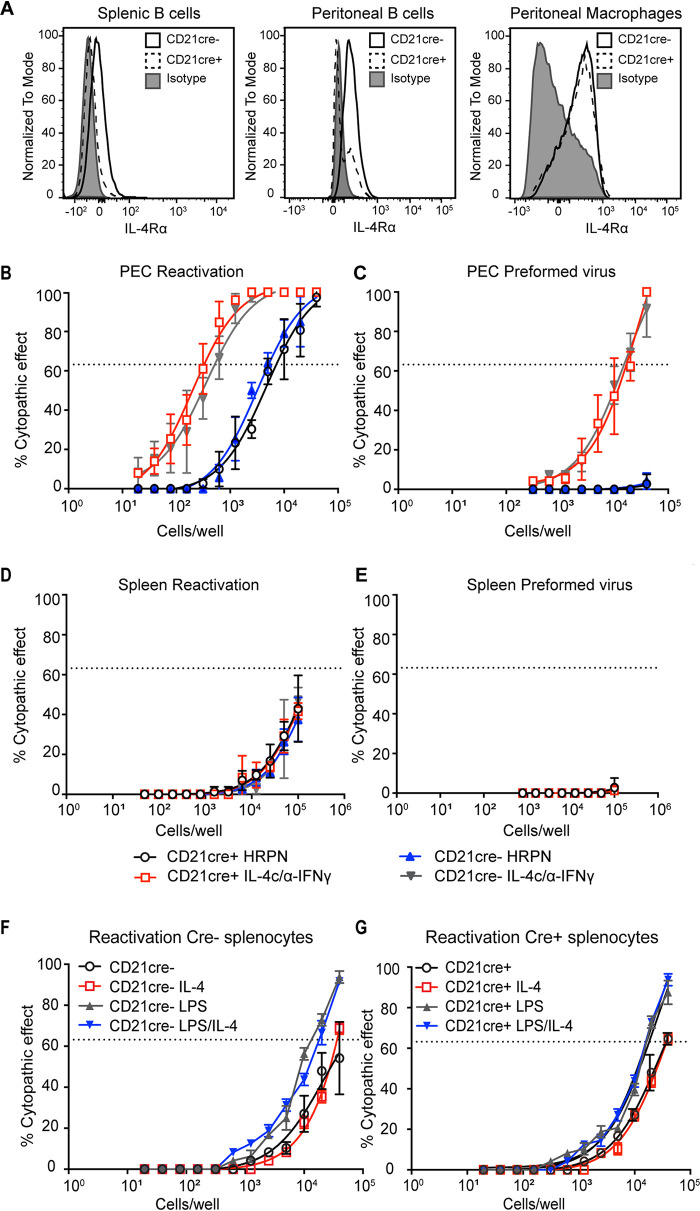
Since MHV68 also establishes latency in B cells, we next evaluated MHV68 reactivation frequency in mice lacking IL-4Rα on B cells. We crossed Il4rαf/f mice with Cd21-cre-expressing mice to delete B cell expression of IL-4Rα. We used Cd21-cre rather than the more commonly used Cd19-cre because both IL-4Rα and Cd19 are on the same mouse chromosome. CD21 is expressed on mature B cells in the peritoneum and the spleen, which are latency reservoirs of MHV68 (24–26). As shown in Fig. 5A, we used flow cytometry to confirm that cre recombinase successfully deleted the IL-4 receptor on B cells in the spleen and peritoneum of Il4rαf/f × Cd21-cre+ mice (Fig. 5A). IL-4R expression is intact on macrophages in Il4rαf/f × Cd21-cre+ mice (Fig. 5A).
FIG 5.
IL-4R expression is not required for MHV68 reactivation from B cells. (A) Whole spleen or peritoneal exudate cells (PECs) were collected from uninfected Il4rαf/f × Cd21-cre+ and Il4rαf/f × Cd21-cre− mice, and flow cytometry was used to detect IL-4R on B cells and macrophages. Representative histograms from two independent experiments are shown. (B to E) Il4rαf/f × Cd21-cre+ and Il4rαf/f × Cd21-cre− mice were intraperitoneally infected with 106 PFU wild-type MHV68. Mice received an isotype control (HRPN) or both IL-4c and anti-IFN-γ at 28 days post-infection. Another dose of IL-4c was administered 2 days after the first treatment, and mice were then euthanized at 5 days post-treatment. Splenocytes (D and E) and PECs (B and C) were processed into single-cell suspensions. (B and D) Reactivation frequencies were determined by plating serially diluted PECs and splenocytes on a MEF monolayer. (C and E) The presence of preformed virus in PECs and splenocytes was determined by disrupting the cell suspensions and plating serially diluted samples on MEF monolayers. Cytopathic effect was scored at 3 weeks post-plating. Groups of 3 to 5 mice were pooled for each infection and analysis. The data are pooled from 3 independent experiments. The error bars represent standard errors of the means. (F and G) Il4rαf/f × Cd21-cre+ and Il4rαf/f × Cd21-cre− mice were intraperitoneally infected with 106 PFU wild-type MHV68. Sixteen days after infection, spleen was processed into single-cell suspensions. Reactivation frequencies of Cre− (F) and Cre+ (G) splenocytes were determined by plating serially diluted splenocytes on a MEF monolayer with medium, LPS, IL-4, or IL-4 plus LPS. Groups of 3 mice were pooled for each infection and analysis. The data are pooled from 2 independent experiments.
We assayed the frequency of viral reactivation from Il4rαf/f × Cd21-cre+ and Il4rαf/f × Cd21-cre− mice with and without IL-4c/anti-IFN-γ in the peritoneum and the spleen. As expected, injection of latently infected Il4rαf/f × Cd21-cre− mice with IL-4c/anti-IFN-γ induced virus reactivation from explanted peritoneal cells (Fig. 5B) and induced virus reactivation in vivo, as detected by the presence of preformed virus (Fig. 5C). Similar to Cd21-cre− mice, Il4rαf/f × Cd21-cre+ mice injected with IL-4c/anti-IFN-γ also reactivated the virus to levels comparable to those in cre recombinase-negative mice (Fig. 5B and C). In summary, our results demonstrate that the absence of IL-4R on B cells in the peritoneum did not dampen MHV68 reactivation upon IL-4c/anti-IFN-γ treatment.
We also examined virus reactivation from the spleen following IL-4c/anti-IFN-γ treatment in vivo. We did not detect an increase in virus reactivation from the spleen after IL-4c/anti-IFN-γ treatment from either group of mice (Fig. 5D and E), indicating that IL-4c/anti-IFN-γ does not stimulate splenic reactivation of MHV68.
We previously reported that two signals, IL-4 and blockade of IFN-γ, are required to reactivate MHV68 (7). However, other work indicates that IFN-γ suppresses virus reactivation in macrophages but not B cells (8, 27). This0 raises the possibility that our second signal, anti-IFN-γ, is insufficient to overcome the blockade of virus reactivation in B cells. We examined whether lipopolysaccharide (LPS) stimulation of explanted cells, which has been previously shown to reactivate MHV68 from B cells (28), could synergize with IL-4 to induce MHV68 reactivation from B cells. We measured the virus reactivation frequency of Il4rαf/f × Cd21-cre+ and Il4rαf/f × Cd21-cre− splenocytes by stimulating cells at 16 days post-infection with LPS, IL-4, or LPS plus IL-4. LPS induced a small increase in virus reactivation, as expected, but we detected no increase in virus reactivation in splenocytes treated with IL-4 after explant. Splenocytes treated with both IL-4 and LPS had reactivation equivalent to that in LPS-only samples, indicating that even with LPS stimulation, IL-4 is unable to enhance reactivation in B cells (Fig. 5F and G).
IL-4 does not regulate N4/N5 promoter activity of Orf50 in HE2.1 B cells.
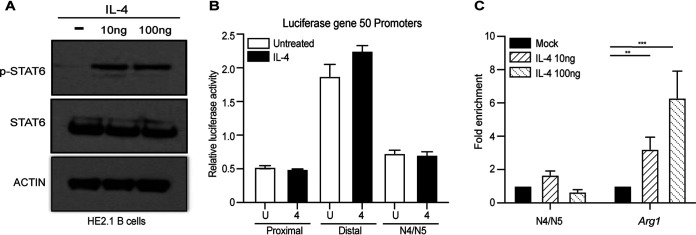
We showed previously in macrophages that phosphorylated STAT6 directly binds to a viral promoter, termed N4/N5, of Orf50, which is the latent-to-lytic switch gene (7, 29). The activation of Orf50 promotes viral replication in vitro in macrophages and virus reactivation in vivo (30–34). Despite the fact that B cells in the germinal centers respond to IL-4 produced by T follicular cells (35), one possibility is that IL-4 stimulation of B cells does not activate STAT6. We determined if IL-4 signaling was intact in HE2.1 B cells by measuring the phosphorylation of STAT6. We treated the B cells with a low dose and a high dose of IL-4 and probed for the phosphorylation of STAT6. As shown in Fig. 6A, both doses of IL-4 treatment induced the phosphorylation of STAT6, indicating that the HE2.1 B cells indeed respond to IL-4.
FIG 6.
STAT6 does not bind the N4/N5 promoter of RTA/Orf50 in HE2.1 B cells. (A) HE2.1 B cells were treated with different concentrations of IL-4 for 24 h. Total and phosphorylated STAT6 proteins were determined by Western blotting. (B) A20 B cells were transfected with vectors expressing luciferase under the control of three different RTA/Orf50 promoters, termed proximal, distal, and N4/N5. A dual-luciferase reporter assay was performed 24 h after treatment with IL-4. (C) HE2.1 B cells were treated with different concentrations of IL-4 for 24 h. DNA bound to STAT6 was immunoprecipitated, and quantitative PCR (qPCR) was performed for the N4/N5 promoter region or arginase-1 (Arg1). The percentage of the input after normalization to the immunoglobulin G control was calculated for both N4/N5 and Arg1. Data are from three independent experiments. Error bars are standard errors of the means. P values were calculated by one-way ANOVA. **, P < 0.01; ****, P < 0.001.
We previously demonstrated that IL-4 treatment of BMDMs could increase promoter activity and induce STAT6 binding to the N4/N5 promoter of Orf50. Therefore, we tested whether IL-4 could increase the promoter activity of three different promoters in A20 B cells. IL-4 did not increase luciferase expression from any of the three Orf50 promoters, suggesting that IL-4 does not increase the promoter activity of any of the Orf50 promoters (Fig. 6B). Finally, we determined whether IL-4 induced STAT6 binding to the N4/N5 promoter in HE2.1 B cells. Using STAT6 antibody-based chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR), we examined STAT6 binding to the N4/N5 promoter in HE2.1 B cells after IL-4 treatment. Compared to increased binding to the Arg1 promoter, which is activated by STAT6 signaling, STAT6 did not bind to the N4/N5 promoter of viral Orf50 upon IL-4 stimulation of HE2.1 B cells (Fig. 6C). Taken together, these data suggest that even though IL-4 activates STAT6 in B cells, IL-4 does not activate Orf50 promoters and STAT6 does not bind to the N4/N5 promoter of Orf50 in a latently infected B cell line.
DISCUSSION
We found that the expression of IL-4 receptor on macrophages was required for efficient MHV68 reactivation from latency in vivo upon the administration of IL-4c plus a blocking antibody to IFN-γ. Intriguingly, IL-4 receptor deficiency on B cells did not impair virus reactivation in response to IL-4c combined with anti-IFN-γ treatment. We also revealed that even though IL-4 treatment of both macrophages and B cells induces the phosphorylation of STAT6, phosphorylated STAT6 does not bind to the N4/N5 promoter of Orf50 in a B cell line latently infected with MHV68. Thus, our study suggests that different latently infected cell populations respond differentially to reactivation signals.
For gammaherpesviruses, reactivation from latency is controlled by one or more master regulators of transcription. The expression of these latent-to-lytic switch genes is silenced during latency, and reactivation signals turn on the expression of these genes. When expressed, these transactivator proteins induce the expression of lytic viral genes, leading to the production of virus. In the case of MHV68, this master regulator is ORF50 (also called RTA) (36). We demonstrated previously that IL-4 treatment of macrophages leads to the induction of ORF50 expression and the activation of one of the Orf50 promoters, N4/N5. We found that the host transcription factor downstream of the IL-4R, STAT6, binds to the N4/N5 promoter of Orf50 in macrophages (7). In this report, we found that even though IL-4 induces the phosphorylation of STAT6 in B cells, IL-4 treatment did not enhance the promoter activity of the Orf50 promoters. Moreover, STAT6 did not bind to the Orf50 N4/N5 promoter. These data suggest that this promoter is inaccessible to STAT6 binding in B cells and may indicate that the Orf50 N4/N5 promoter is differentially accessible to transcription factor binding in B cells and macrophages.
Interestingly, there are multiple Orf50 promoters for MHV68, termed proximal, distal, N3, and N4/N5 (29). It is not currently known why MHV68 Orf50 utilizes multiple different promoters or what the purposes of these various promoters are. It is possible that the different Orf50 promoters have different functions, depending on the cell type. One way to regulate the ability of these promoters to respond to various stimuli in different cells is to regulate the chromatin accessibility of the viral genome in macrophages and B cells. This differential chromosome accessibility may allow the virus to have distinct responses to various reactivation stimuli. The differential binding of STAT6 to the Orf50 N4/N5 promoter suggests that the MHV68 genome displays different chromosome structure states depending on the cell type.
MHV68 establishes latent infection in germinal center B cells and maintains long-term latency in memory B cells (25, 37). T follicular cells make IL-4 as part of the germinal center reaction to promote class switching (35). Therefore, even though germinal center and memory B cells that harbor latent virus encounter IL-4, they do not induce virus reactivation in response to IL-4 signals. This raises the question of whether the virus in latently infected B cells silences the Orf50 N4/N5 promoter or preferentially uses different Orf50 promoters to regulate the latent-to-lytic switch.
The closely related human gammaherpesvirus KSHV also has 4 different Orf50 promoters that induce the expression of 6 different RTA transcripts (38). There are also other herpesvirus open reading frames that are controlled by multiple promoters that produce alternative transcripts (39–41). One possibility is that these different promoters respond to different acute, latent, and reactivation signals. Another possibility is that the different isoforms of RTA produced have different transactivation potentials for various KSHV promoters. Both of these possibilities reflect the intricate regulation of herpesvirus gene expression and illustrate the multiple ways in which the virus has evolved to regulate latency and reactivation.
During the lifetime of the host, the host and the latent virus will experience many bystander infections. We hypothesize that there are conditions that are favorable for virus reactivation and conditions that are unfavorable. Similarly, virus may reactivate from one tissue or cell type but not from another tissue or cell type, depending on the signals received. The nature of chronic latent infection dictates that herpesviruses evolve exquisite sensitivity to the host environment.
MATERIALS AND METHODS
Animals, infections, and treatment.
All mice used in this study were from the C57BL/6 background and housed in a specific-pathogen-free facility at the University of Texas (UT) Southwestern Medical Center. Mice were maintained and used under a protocol approved by the UT Southwestern Medical Center Institutional Animal Care and Use Committee (IACUC). To generate mice conditionally deficient for IL4r, IL4rαf/f mice (16) were crossed to lysozyme M-cre recombinase-expressing mice or CD21-cre recombinase (42)-expressing mice for selective disruption of IL4rα in different cell types in vivo. Mice were used for infections at between 6 and 12 weeks of age, and all experiments used littermate controls. Mixed groups of both males and females were used in all experiments. MHV68 (WUSM strain) and MHV68-M3FL (20) were diluted in D10 (Dulbecco’s modified Eagle medium [DMEM] with 10% fetal bovine serum [FBS]) and administered intraperitoneally (106 PFU).
In vivo MHV68 reactivation.
For IL-4c/anti-IFN-γ-induced reactivation, both anti-IFN-γ and long-lasting IL-4c were injected intraperitoneally on day 28 of MHV68 infection, and a second dose of IL-4c was injected on day 30 as described previously by Reese et al. (7). Five hundred micrograms of anti-IFN-γ (clone R4-6A2; BioXCell) or an isotype control (clone HRPN; BioXCell) was diluted in phosphate-buffered saline (PBS). Five micrograms of IL-4 (catalog no. AF-214-14; Peprotech) and 25 μg of anti-IL-4 (clone 11B11; BioXCell) were complexed prior to intraperitoneal injection into mice. For cyclosporine-induced reactivation, 50 mg/kg of body weight of cyclosporine (lots 098M4084V and 095M4084V; Sigma) with corn oil as a carrier was injected intraperitoneally into the infected mice on the days indicated in the figures. Mice were imaged on day 0 before cyclosporine injection and on days 5, 7, 9, and 12 following treatment.
Cell cultures and infections.
Murine fibroblast (3T12; ATCC CCL-164) cells were cultured in DMEM (Corning) supplemented with 5% fetal bovine serum (Biowest), 2 mM l-glutamine (Gibco), 1% HEPES (Corning), 10 U/ml penicillin, and 10 μg/ml streptomycin sulfate (Corning) (complete DMEM). Cells were cultured at 37°C with 5% CO2. A20 B cell lines were maintained in a solution containing RPMI 1640 (Gibco), 10% fetal bovine serum, 2 mM l-glutamine, 100 U/ml penicillin, 100 mg/ml streptomycin, and 50 μM β-mercaptoethanol. A20-HE2.1 B cell lines (14) (kindly provided by Laurie Krug) were maintained in a solution containing RPMI 1640 (Gibco), 10% fetal bovine serum, 2 mM l-glutamine, 100 U/ml penicillin, 100 mg/ml streptomycin, 50 μM β-mercaptoethanol, and 300 μg/ml G418. Mouse embryonic fibroblasts (MEFs) were obtained from C57BL/6 mouse embryos. BMDMs were harvested from C57BL/6J mice and differentiated for 7 days with 10% CMG-14 supernatants (43) in complete DMEM (10% fetal bovine serum, 1% HEPES, 2 mM l-glutamine). On day 7, 5 × 105 cells per well were plated in 6-well plates, pretreated with IL-4 (Peprotech) for 16 h, infected with MHV68 for 1 h, washed with PBS, and cultured in medium containing cytokines.
Bone marrow-derived macrophage cultures and infections.
Bone marrow was harvested from C57BL/6J mice and differentiated for 7 days with 10% CMG-14 supernatants in complete DMEM (10% fetal calf serum [FCS], 1% HEPES, 2 mM l-glutamine). At day 7, 1.5 × 105 cells were plated per well in 24-well plates, rested for 2 to 3 days, pretreated with IL-4 (Peprotech) at 10 ng/ml unless otherwise noted, infected with MHV68 (multiplicity of infection [MOI] of 5 for 1 h), washed, and resuspended in medium containing cytokines. Cells were harvested for fluorescence-activated cell sorter (FACS) analysis 24 h after infection. Infected macrophages were fixed with 4% formaldehyde, blocked with 10% mouse serum and 1% Fc block (CD16/32; BioLegend), and then stained with polyclonal rabbit antibody to MHV68 (7, 44, 45) (1:1,000), followed by secondary goat anti-rabbit Alexa Fluor 647 (Invitrogen).
Determination of MHV68 viral titers.
3T12 cells were seeded in 6-well plates at 3 × 105 cells/well. Homogenized tissue or viral stock supernatants were serially diluted in D5 medium (DMEM, 5% FBS, 2 mM l-glutamine, 1 mM HEPES) and added to 3T12 cell monolayers for 1 h at 37°C. Samples were overlaid with 1% methylcellulose in DMEM with 5% FBS. Plates were incubated at 37°C for 7 days and then stained with crystal violet (0.2% crystal violet in 20% ethanol) to visualize plaques.
Ex vivo limiting-dilution assay for reactivation and persistent replication.
To determine the frequency of cells harboring latent virus capable of reactivation ex vivo, single-cell suspensions from peritoneal exudate cells (PECs) and spleens were plated in 2-fold serial dilutions on MEF monolayers (maintained in DMEM with 10% FBS) (18). Cytopathic effect (CPE) was scored 3 weeks after plating. To distinguish preformed infectious virus from virus that reactivates ex vivo from live cells upon explantation, parallel samples were mechanically disrupted using 0.5-mm silica beads on the Precellys 24 tissue homogenizer (Bertin Technologies) to kill the cells but keep any infectious virus intact and then plated on the monolayer of MEFs to release preformed virus. The 63.2% Poisson distribution horizontal line represents the frequency at which one reactivation event is likely to occur per number of plated cells. For ex vivo treatment of latently infected splenic cells, whole spleen was harvested from infected mice on day 16. Single-cell suspensions from spleens were stimulated with medium alone, 1 μg/ml LPS, 100 ng/ml IL-4, or 1 μg/ml LPS plus 100 ng/ml IL-4 and plated in 2-fold serial dilutions on MEF monolayers. CPE was scored 3 weeks after plating.
Limiting-dilution PCR to detect viral genomes.
To compare the frequencies of cells harboring viral genomes, single-cell suspensions from PECs and spleens were assayed by nested PCR (5). In brief, cells were serially diluted in a solution containing uninfected 3T12 cells to maintain a total number of 104 cells per PCR and lysed overnight at 56°C with proteinase K. Two rounds of PCR were performed using six dilutions per sample with 12 reactions per dilution with primers specific for the MHV68 ORF72 gene. A plasmid containing ORF72 was included at 0.1, 1.0, and 10 copies as positive controls. Products were visualized on a 1.5% agarose gel. Data points for limiting-dilution PCR (LD-PCR) represent the means and the standard errors of the means for three replicate experiments. Frequencies of viral genome-carrying cells were obtained by calculating the cell density at which 63.2% of the wells were positive based on the Poisson distribution.
Bioluminescence imaging of mice using MHV68-M3FL.
Mice were infected with 106 PFU per mouse of MHV68-M3FL. The mice were anesthetized with isoflurane and imaged after intraperitoneal injection of 150 mg/kg of d-luciferin (Gold Biotechnology). Bioluminescence images of MHV68-M3FL-infected mice were collected on the Ivis Lumina series III system (Perkin Elmer), and data collection and analysis were done with Living Image software (Caliper Life Sciences). Mice were exposed for 3 min, for 3 consecutive pictures. Total luminescence was calculated by measuring the total flux (photons per second) in a region of interest (ROI) for each mouse. One ROI size was used for each image, to accommodate the size of the mice. The pictures with peak luminescence were selected as final data.
Flow cytometry.
Whole spleens were processed into a single-cell suspension and strained. Peritoneal exudate cells were collected by peritoneal lavage and washed with PBS. Any samples with blood were lysed with ACK lysis buffer (Lonza BioWhittaker). Fc receptors were blocked on cells with anti-CD16/32 (clone 2.4G2; Tonbo). Cells were stained with BV605-anti-CD19 (clone 1D3; BD Biosciences), allophycocyanin (APC)-Cy7-anti-CD19 (1D3; BD Biosciences), peridinin chlorophyll protein (PerCP)-Cy5.5-anti-F4/80 (BM8.1; Tonbo), APC-Cy7-anti-F4/80 (BM8; BioLegend), fluorescein isothiocyanate (FITC)-anti-CD11b (M1/70; BioLegend), phycoerythrin (PE)-Cy7-anti-CD11b (M1/70; Tonbo), PE-anti-IL-4Rα/CD124 (I015F8; BioLegend), and Violet Fluor 450-anti-CD3 (17A2; Tonbo). Rat PE-anti-IgG2b (LTF-2; Tonbo) was used as the isotype control for CD124. Cells were suspended in FACS buffer (PBS with 1% FBS) and then analyzed on an LSR II flow cytometer. Data were analyzed using FlowJo software (TreeStar, Inc., San Carols, CA).
Luciferase reporter assays.
A20 B cells were electroporated via a Nucleofector kit (Lonza) according to the manufacturer’s instructions. Two micrograms of pGL4.10 reporter plasmids containing proximal, distal, or N4/N5 promoters (7, 29) was cotransfected with pGL4-73 Renilla for normalization. pGL4.10-Luc was used as a negative control. After transfection, 100 ng/ml IL-4 was added to the wells. All cells were collected at 24 h post-transfection and lysed. Luciferase activity was measured using the Dual-Glo luciferase assay system (catalog no. E2920; Promega). All transfections were repeated in triplicate.
Reverse transcription-qPCR (RT-qPCR).
RNA was isolated using an RNeasy minikit (Qiagen), and cDNA was prepared using a SuperScript Vilo cDNA synthesis kit (Invitrogen) according to the manufacturer’s instructions. Quantitative PCR was performed using PowerUp SYBR green master mix (Applied Biosystems) in a Quant Studio 7 Flex real-time PCR system using the primers listed in Table 1. qPCR was performed on three biological replicates, and the fold change was calculated by the ΔΔCT method.
TABLE 1.
Primers used for RT-qPCR
| Primer | Sequence |
|---|---|
| Gapdh forward | GGGTGTGAACCACGAGAAATA |
| Gapdh reverse | GTCATGAGCCCTTCCACAAT |
| Orf50 forward | AGAAACCCACAGCTCGCACTT |
| Orf50 reverse | CAATATGCTGGACAGGCGTATC |
| Orf73 forward | CCAGAAGCTTGTGTACTTGTGGAT |
| Orf73 reverse | AAATACCACAGCAGCGTAGAAGGT |
| Arg1 forward | GCACAACTCACGTACAGACA |
| Arg1 reverse | TGAGGCATTGTTCAGACTTCC |
| N4/N5 forward | GCCGTCCCTTATCTACAGTCA |
| N4/N5 reverse | CTATCATGGGGGCCAGGC |
Immunoblot analysis.
The total protein lysate was harvested in radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, and 25 mM Tris with a protease inhibitor cocktail). Protein concentrations were determined using a Bradford assay (Bio-Rad). Proteins were separated on 4 to 12% Bis-Tris plus gels (Thermo Fisher Scientific) and transferred to a nitrocellulose membrane. Antibodies against STAT6 (Santa Cruz Biotechnology, Dallas, TX), tyrosine 641-phosphorylated STAT6 (Santa Cruz Biotechnology, Dallas, TX), and actin (1:5,000) (catalog no. A2228; Sigma) were detected by the use of secondary donkey anti-rabbit (1:5,000) (catalog no. 711-035-152; Jackson Immuno Research Laboratory) and goat anti-mouse peroxidase (1:5,000) (catalog no. 115-035-174; Jackson Immuno Research Laboratory) by immunoblot analysis with the Luminata Forte Western horseradish peroxidase (HRP) substrate (Millipore).
Chromatin immunoprecipitation.
Cells from a 10-cm dish were cross-linked in 1% formaldehyde in PBS for 10 min at room temperature and quenched in 0.125 M glycine for 10 min. The cross-linked cell suspension was then spun down at 800 × g at 4°C for 5 min. The cell pellet was washed twice with cold PBS and lysed with 1 ml ChIP lysis buffer (50 mM HEPES [pH 7.9], 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100). Nuclei were collected by centrifugation at 1,000 × g for 10 min at 4°C and resuspended in 0.5 ml of ChIP shearing buffer. Samples were then sonicated until DNA was fragmented to an average distribution of about 200 to 300 bp using a Bioruptor (Diagenode) for a total of 30 cycles (30 s on and 30 s off). The sonicated nuclear lysate was spun at 15,000 × g for 5 min at 4°C, and the pellet was discarded. The sonicated nuclear lysate was incubated with 10 μg rabbit monoclonal anti-Stat6 (catalog no. sc-374021) or rabbit IgG overnight. Twenty microliters of mixed protein A and G Dynabeads (Thermo Fisher) was added, and tubes were rotated for 2 h at 4°C. Beads were washed with a low-salt immune complex (20 mM Tris [pH 8.0], 1% NP-40, 2 mM EDTA, 150 mM NaCl, 0.1% SDS, 0.5% deoxycholic acid), a high-salt immune complex (20 mM Tris [pH 8.0], 1% NP-40, 2 mM EDTA, 500 mM NaCl, 0.1% SDS, 0.5% deoxycholic acid), a lithium chloride immune complex (10 mM Tris [pH 8.0], 0.25 M LiCl, 1% NP-40, 1% deoxycholic acid, 1 mM EDTA), and Tris-EDTA twice. After washing, complexes were eluted off the beads using elution buffer (1% SDS and 100 mM NaHCO3) at 65°C for 30 min. Protein-DNA complexes were de-cross-linked using de-cross-linking buffer (50 mM Tris-HCl [pH 6.8], 500 mM NaCl, 5 mM EDTA, 0.5 mg/ml proteinase K) at 60°C overnight. DNA was then purified using the Zymo ChIP DNA clean and concentrator kit (Zymo Research) and used for qPCR analysis using the primers listed in Table 1.
Statistical analysis.
Data were analyzed with Prism 7 software (GraphPad Software, San Diego, CA). In all graphs, significant values are indicated with asterisks (*, P < 0.05; **, P < 0.01; ***, P < 0.001), which were determined by one-way analysis of variation (ANOVA) with multiple comparisons (for multiple samples) and Student’s t test (for two samples), as indicated in the figure legends.
ACKNOWLEDGMENTS
We thank the UTSW Flow Cytometry Core, the UTSW Animal Resource Center, the UTSW IACUC for use of their facilities, the Ivan D’Orso lab for use of their sonicator, and Laurie Krug for providing HE2.1 B cells.
This work was supported by the NIH (1R01AI130020-01A1), the American Heart Association (17SDG33670071), CPRIT (RP200118), and the Pew Scholars Program. C.Z. is supported by the Creigh Weyer Foundation.
G.W., C.Z., and T.A.R. designed experiments. G.W. and C.Z. performed experiments and analyzed data. G.W., C.Z., and T.A.R. wrote and edited the manuscript. T.A.R. secured funding. T.C., L.T., and A.L. assisted with experiments.
REFERENCES
- 1.Barton E, Mandal P, Speck SH. 2011. Pathogenesis and host control of gammaherpesviruses: lessons from the mouse. Annu Rev Immunol 29:351–397. doi: 10.1146/annurev-immunol-072710-081639. [DOI] [PubMed] [Google Scholar]
- 2.Tarakanova VL, Suarez F, Tibbetts SA, Jacoby MA, Weck KE, Hess JL, Speck SH, Virgin HW, IV.. 2005. Murine gammaherpesvirus 68 infection is associated with lymphoproliferative disease and lymphoma in BALB β2 microglobulin-deficient mice. J Virol 79:14668–14679. doi: 10.1128/JVI.79.23.14668-14679.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee KS, Groshong SD, Cool CD, Kleinschmidt-DeMasters BK, van Dyk LF. 2009. Murine gammaherpesvirus 68 infection of IFNγ unresponsive mice: a small animal model for gammaherpesvirus-associated B-cell lymphoproliferative disease. Cancer Res 69:5481–5489. doi: 10.1158/0008-5472.CAN-09-0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weck KE, Kim SS, Virgin HW, IV, Speck SH. 1999. Macrophages are the major reservoir of latent murine gammaherpesvirus 68 in peritoneal cells. J Virol 73:3273–3283. doi: 10.1128/JVI.73.4.3273-3283.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weck KE, Kim SS, Virgin HW, IV, Speck SH. 1999. B cells regulate murine gammaherpesvirus 68 latency. J Virol 73:4651–4661. doi: 10.1128/JVI.73.6.4651-4661.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flaño E, Husain SM, Sample JT, Woodland DL, Blackman MA. 2000. Latent murine γ-herpesvirus infection is established in activated B cells, dendritic cells, and macrophages. J Immunol 165:1074–1081. doi: 10.4049/jimmunol.165.2.1074. [DOI] [PubMed] [Google Scholar]
- 7.Reese TA, Wakeman BS, Choi HS, Hufford MM, Huang SC, Zhang X, Buck MD, Jezewski A, Kambal A, Liu CY, Goel G, Murray PJ, Xavier RJ, Kaplan MH, Renne R, Speck SH, Artyomov MN, Pearce EJ, Virgin HW. 2014. Helminth infection reactivates latent γ-herpesvirus via cytokine competition at a viral promoter. Science 345:573–577. doi: 10.1126/science.1254517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steed A, Buch T, Waisman A, Virgin HW, IV.. 2007. Gamma interferon blocks gammaherpesvirus reactivation from latency in a cell type-specific manner. J Virol 81:6134–6140. doi: 10.1128/JVI.00108-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steed AL, Barton ES, Tibbetts SA, Popkin DL, Lutzke ML, Rochford R, Virgin HW, IV.. 2006. Gamma interferon blocks gammaherpesvirus reactivation from latency. J Virol 80:192–200. doi: 10.1128/JVI.80.1.192-200.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang J, Renne R, Dittmer D, Ganem D. 2000. Inflammatory cytokines and the reactivation of Kaposi’s sarcoma-associated herpesvirus lytic replication. Virology 266:17–25. doi: 10.1006/viro.1999.0077. [DOI] [PubMed] [Google Scholar]
- 11.Park M-K, Cho H, Roh SW, Kim S-J, Myoung J. 2019. Cell type-specific interferon-γ-mediated antagonism of KSHV lytic replication. Sci Rep 9:2372. doi: 10.1038/s41598-019-38870-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan CSE, Lawler C, May JS, Belz GT, Stevenson PG. 2016. Type I interferons direct gammaherpesvirus host colonization. PLoS Pathog 12:e1005654. doi: 10.1371/journal.ppat.1005654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collins CM, Speck SH. 2015. Interleukin 21 signaling in B cells is required for efficient establishment of murine gammaherpesvirus latency. PLoS Pathog 11:e1004831. doi: 10.1371/journal.ppat.1004831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forrest JC, Speck SH. 2008. Establishment of B-cell lines latently infected with reactivation-competent murine gammaherpesvirus 68 provides evidence for viral alteration of a DNA damage-signaling cascade. J Virol 82:7688–7699. doi: 10.1128/JVI.02689-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodwin MM, Molleston JM, Canny S, El Hassan MA, Willert EK, Bremner R, Virgin HW. 2010. Histone deacetylases and the nuclear receptor corepressor regulate lytic-latent switch gene 50 in murine gammaherpesvirus 68-infected macrophages. J Virol 84:12039–12047. doi: 10.1128/JVI.00396-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herbert DR, Hölscher C, Mohrs M, Arendse B, Schwegmann A, Radwanska M, Leeto M, Kirsch R, Hall P, Mossmann H, Claussen B, Förster I, Brombacher F. 2004. Alternative macrophage activation is essential for survival during schistosomiasis and downmodulates T helper 1 responses and immunopathology. Immunity 20:623–635. doi: 10.1016/S1074-7613(04)00107-4. [DOI] [PubMed] [Google Scholar]
- 17.Rekow MM, Darrah EJ, Mboko WP, Lange PT, Tarakanova VL. 2016. Gammaherpesvirus targets peritoneal B-1 B cells for long-term latency. Virology 492:140–144. doi: 10.1016/j.virol.2016.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weck KE, Barkon ML, Yoo LI, Speck SH, Virgin HW, IV.. 1996. Mature B cells are required for acute splenic infection, but not for establishment of latency, by murine gammaherpesvirus 68. J Virol 70:6775–6780. doi: 10.1128/JVI.70.10.6775-6780.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sunil-Chandra NP, Efstathiou S, Nash AA. 1992. Murine gammaherpesvirus 68 establishes a latent infection in mouse B lymphocytes in vivo. J Gen Virol 73:3275–3279. doi: 10.1099/0022-1317-73-12-3275. [DOI] [PubMed] [Google Scholar]
- 20.Hwang S, Wu T-T, Tong LM, Kim KS, Martinez-Guzman D, Colantonio AD, Uittenbogaart CH, Sun R. 2008. Persistent gammaherpesvirus replication and dynamic interaction with the host in vivo. J Virol 82:12498–12509. doi: 10.1128/JVI.01152-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sunil-Chandra NP, Efstathiou S, Nash AA. 1994. The effect of acyclovir on the acute and latent murine gammaherpesvirus-68 infection of mice. Antivir Chem Chemother 5:290–296. doi: 10.1177/095632029400500502. [DOI] [Google Scholar]
- 22.Ehtisham S, Sunil-Chandra NP, Nash AA. 1993. Pathogenesis of murine gammaherpesvirus infection in mice deficient in CD4 and CD8 T cells. J Virol 67:5247–5252. doi: 10.1128/JVI.67.9.5247-5252.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Braaten DC, Sparks-Thissen RL, Kreher S, Speck SH, Virgin HW, IV.. 2005. An optimized CD8+ T-cell response controls productive and latent gammaherpesvirus infection. J Virol 79:2573–2583. doi: 10.1128/JVI.79.4.2573-2583.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahearn JM, Fischer MB, Croix D, Goerg S, Ma M, Xia J, Zhou X, Howard RG, Rothstein TL, Carroll MC. 1996. Disruption of the Cr2 locus results in a reduction in B-1a cells and in an impaired B cell response to T-dependent antigen. Immunity 4:251–262. doi: 10.1016/S1074-7613(00)80433-1. [DOI] [PubMed] [Google Scholar]
- 25.Collins CM, Boss JM, Speck SH. 2009. Identification of infected B-cell populations by using a recombinant murine gammaherpesvirus 68 expressing a fluorescent protein. J Virol 83:6484–6493. doi: 10.1128/JVI.00297-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fischer MB, Goerg S, Shen L, Prodeus AP, Goodnow CC, Kelsoe G, Carroll MC. 1998. Dependence of germinal center B cells on expression of CD21/CD35 for survival. Science 280:582–585. doi: 10.1126/science.280.5363.582. [DOI] [PubMed] [Google Scholar]
- 27.Goodwin MM, Canny S, Steed A, Virgin HW. 2010. Murine gammaherpesvirus 68 has evolved gamma interferon and Stat1-repressible promoters for the lytic switch gene 50. J Virol 84:3711–3717. doi: 10.1128/JVI.02099-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gargano LM, Forrest JC, Speck SH. 2009. Signaling through Toll-like receptors induces murine gammaherpesvirus 68 reactivation in vivo. J Virol 83:1474–1482. doi: 10.1128/JVI.01717-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wakeman BS, Johnson LS, Paden CR, Gray KS, Virgin HW, Speck SH. 2014. Identification of alternative transcripts encoding the essential murine gammaherpesvirus lytic transactivator RTA. J Virol 88:5474–5490. doi: 10.1128/JVI.03110-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu T-T, Usherwood EJ, Stewart JP, Nash AA, Sun R. 2000. Rta of murine gammaherpesvirus 68 reactivates the complete lytic cycle from latency. J Virol 74:3659–3667. doi: 10.1128/jvi.74.8.3659-3667.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu T-T, Tong L, Rickabaugh T, Speck S, Sun R. 2001. Function of Rta is essential for lytic replication of murine gammaherpesvirus 68. J Virol 75:9262–9273. doi: 10.1128/JVI.75.19.9262-9273.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song MJ, Hwang S, Wong WH, Wu T-T, Lee S, Liao H-I, Sun R. 2005. Identification of viral genes essential for replication of murine γ-herpesvirus 68 using signature-tagged mutagenesis. Proc Natl Acad Sci U S A 102:3805–3810. doi: 10.1073/pnas.0404521102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moorman NJ, Lin CY, Speck SH. 2004. Identification of candidate gammaherpesvirus 68 genes required for virus replication by signature-tagged transposon mutagenesis. J Virol 78:10282–10290. doi: 10.1128/JVI.78.19.10282-10290.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pavlova IV, Virgin HW, IV, Speck SH. 2003. Disruption of gammaherpesvirus 68 gene 50 demonstrates that Rta is essential for virus replication. J Virol 77:5731–5739. doi: 10.1128/jvi.77.10.5731-5739.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vinuesa CG, Linterman MA, Goodnow CC, Randall KL. 2010. T cells and follicular dendritic cells in germinal center B‐cell formation and selection. Immunol Rev 237:72–89. doi: 10.1111/j.1600-065X.2010.00937.x. [DOI] [PubMed] [Google Scholar]
- 36.Speck SH, Ganem D. 2010. Viral latency and its regulation: lessons from the γ-herpesviruses. Cell Host Microbe 8:100–115. doi: 10.1016/j.chom.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Flaño E, Kim I-J, Woodland DL, Blackman MA. 2002. γ-Herpesvirus latency is preferentially maintained in splenic germinal center and memory B cells. J Exp Med 196:1363–1372. doi: 10.1084/jem.20020890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wakeman BS, Izumiya Y, Speck SH. 2017. Identification of novel Kaposi’s sarcoma-associated herpesvirus Orf50 transcripts: discovery of new RTA isoforms with variable transactivation potential. J Virol 91:e01434-16. doi: 10.1128/JVI.01434-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Minarovits J. 2006. Epigenotypes of latent herpesvirus genomes. Curr Top Microbiol Immunol 310:61–80. doi: 10.1007/3-540-31181-5_5. [DOI] [PubMed] [Google Scholar]
- 40.Lieberman PM. 2013. Keeping it quiet: chromatin control of gammaherpesvirus latency. Nat Rev Microbiol 11:863–875. doi: 10.1038/nrmicro3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O’Grady T, Feswick A, Hoffman BA, Wang Y, Medina EM, Kara M, van Dyk LF, Flemington EK, Tibbetts SA. 2019. Genome-wide transcript structure resolution reveals abundant alternate isoform usage from murine gammaherpesvirus 68. Cell Rep 27:3988–4002.e5. doi: 10.1016/j.celrep.2019.05.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kraus M, Alimzhanov MB, Rajewsky N, Rajewsky K. 2004. Survival of resting mature B lymphocytes depends on BCR signaling via the Igα/β heterodimer. Cell 117:787–800. doi: 10.1016/j.cell.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 43.Takeshita S, Kaji K, Kudo A. 2000. Identification and characterization of the new osteoclast progenitor with macrophage phenotypes being able to differentiate into mature osteoclasts. J Bone Miner Res 15:1477–1488. doi: 10.1359/jbmr.2000.15.8.1477. [DOI] [PubMed] [Google Scholar]
- 44.Weck KE, Canto AJD, Gould JD, O'Guin AK, Roth KA, Saffitz JE, Speck SH, Virgin HW. 1997. Murine γ-herpesvirus 68 causes severe large-vessel arteritis in mice lacking interferon-γ responsiveness: a new model for virus-induced vascular disease. Nat Med 3:1346–1353. [DOI] [PubMed] [Google Scholar]
- 45.Tarakanova VL, Molleston JM, Goodwin M, Virgin HW. 2010. MHV68 complement regulatory protein facilitates MHV68 replication in primary macrophages in a complement independent manner. Virology 396:323–328. doi: 10.1016/j.virol.2009.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]