Abstract
Since discovered in Hubei, China in December 2019, Corona Virus Disease 2019 named COVID-19 has lasted more than one year, and the number of new confirmed cases and confirmed deaths is still at a high level. COVID-19 is an infectious disease caused by SARS-CoV-2. Although RT-PCR is considered the gold standard for detection of COVID-19, CT plays an important role in the diagnosis and evaluation of the therapeutic effect of COVID-19. Diagnosis and localization of COVID-19 on CT images using deep learning can provide quantitative auxiliary information for doctors. This article proposes a novel network with multi-receptive field attention module to diagnose COVID-19 on CT images. This attention module includes three parts, a pyramid convolution module (PCM), a multi-receptive field spatial attention block (SAB), and a multi-receptive field channel attention block (CAB). The PCM can improve the diagnostic ability of the network for lesions of different sizes and shapes. The role of SAB and CAB is to focus the features extracted from the network on the lesion area to improve the ability of COVID-19 discrimination and localization. We verify the effectiveness of the proposed method on two datasets. The accuracy rate of 97.12%, specificity of 96.89%, and sensitivity of 97.21% are achieved by the proposed network on DTDB dataset provided by the Beijing Ditan Hospital Capital Medical University. Compared with other state-of-the-art attention modules, the proposed method achieves better result. As for the public COVID-19 SARS-CoV-2 dataset, 95.16% for accuracy, 95.6% for F1-score and 99.01% for AUC are obtained. The proposed network can effectively assist doctors in the diagnosis of COVID-19 CT images.
Keywords: Auxiliary diagnosis, Attention, COVID-19, Deep learning
1. Introduction
COVID-19 is a highly infectious viral pneumonia caused by 2019-nCoV (also known as SARS-CoV-2) [1], [2]. Since first discovered in Wuhan, Hubei, it has been found in 212 countries around the world. On March 11, 2020 local time, the Director-General of the World Health Organization Dr Tedros Adhanom Ghebreyesus announced that according to the assessment, the WHO believed that the current new crown pneumonia epidemic can be called a pandemic. As of March 1, 2021, COVID-19 has more than 110 million confirmed cases and more than 2.5 million confirmed deaths [3]. With the coming winter, there has been sharp and exponential rises in coronavirus infections across many countries in the world. It grows even faster with severe new variants of coronavirus and has caused huge human and economic losses. Early detection, isolation and treatment are the most effective means to prevent COVID-19 [4].
Reverse transcription polymerase chain reaction (RT-PCR) is considered to be the gold standard for SARS-CoV-2 diagnosis, which is pertain to nucleic acid test [5]. The average test time takes 2–3 h. Because it directly detects the viral nucleic acid in the collected specimens and has strong specificity and relatively high sensitivity, it has become the main detection method at present [6], [7], [8]. It can only detect whether the patient is infected with COVID-19, but cannot describe the specific infection. It is necessary to use common medical imaging for aided diagnosis. The widely used imaging tools include Computer Tomography (CT) and X-Ray. CT images are widely used in medical image processing, such as the treatment of lung-related diseases [9], [10]. CT images also can be used to classify COVID-19 and localize lesions. A large number of studies have shown that CT has an important role in the study of COVID-19 [11], [12], [13].
Deep learning has achieved the best results in image classification, image segmentation as well as medical image processing. For example, [14], [15], [16], [17] have achieved satisfactory results for medical image classification, detection and segmentation for lung cancer. It is of great significance to apply deep learning to COVID-19 lesion discrimination and localization.
In this paper, the deep learning method is used for the auxiliary diagnosis and localization of COVID-19 on CT images, and a new attention module is proposed. The attention mechanism includes modules for extracting multi-receptive field features, spatial attention blocks and channel attention blocks. Experiments show that this method has achieved significant results in the diagnosis and location of COVID-19 lesions. The contribution points of our method are as follows:
-
1)
Pyramid convolution module (PCM) is proposed to extract the features of multi-receptive field. So that the network can extract the features of different sizes of infection area, which improves the overall network diagnosis and positioning effect.
-
2)
The multi-receptive field spatial attention block (SAB) and the multi-receptive field channel attention block (CAB) are designed in the proposed network. The channel and spatial attention modules focus the extracted features on the lesion area to improve the accuracy of diagnosis and localization.
-
3)
The proposed network can well locate the salient lesion area of COVID-19, which can provide auxiliary information for doctors to quantitative analysis for effective treatment.
The rest of this paper is organized as follows. Section 2 introduces some work related to our method. Section 3 describes our proposed method in detail. We introduce the datasets and report the experimental results in Section 4. Finally, we discuss and conclude the paper in Sections 5.
2. Application of deep learning in the diagnosis of COVID-19
Recently, deep learning has been widely used in many fields of computer vision and has achieved effective results, especially for medical image processing. Image classification is the basic task to use deep learning. Lecun et al. [18] first used convolutional neural networks for image classification. Many classic networks were proposed following, such as VGG [19], Inception [20], [21], [22], ResNet [23], DenseNet [24].
Attention mechanism was first widely used in the direction of natural language processing, and achieved the state-of-the-art results [25]. Computer vision has also introduced this idea, and many studies have achieved satisfactory results. Wang et al. [26] proposed Residual Attention Network (RAN) for image classification, which added an attention module in ResNet. The purpose was to obtain a larger receptive field as an attention map, which was more beneficial to classification. Hu et al. [27] proposed a general attention module SE-Net, which can learn the relationship between each channel and get the weight of different channels for multiply the original feature map. Inspired by this, Park et al. [28] proposed Convolutional Block Attention Module (CBAM). Compared with the SE-Net which only focused on the channel dimension, CBAM combined spatial attention and channel attention to guide features to focus on salient areas respectively. SE-Net and CBAM can be embedded in mainstream basic networks to improve the feature extraction capabilities of the network without significantly increasing the amount of calculation and parameters. In addition, Park et al. [29] also proposed Bottleneck Attention Module (BAM) to construct a hierarchical attention which is similar to a human perception procedure.
In recent years, deep learning has been used for medical image processing and have achieved satisfactory results. Due to the COVID-19 outbreak, many researchers have focused their research on the use of deep learning for study on COVID-19 [30], [31], [32], [33]. Wang et al. [34] proposed COVID-Net to screen suspected patients with coronavirus infection by identifying the obvious signs of the disease on chest X-rays. Compared with VGG-16 [20] and ResNet50 [24], the number of parameters and calculations of COVID-Net was greatly reduced, and the highest 93.3% for accuracy on the COVID-x dataset [34] was obtained. Ouchicha et al. [35] proposed a novel deep learning model named CVDNet to classify COVID-19 infection from normal and other pneumonia cases using chest X-ray images. The proposed model was constructed by using two parallel levels with different kernel sizes to capture local and global features of the inputs. Qian et al. [36] proposed a M3Lung-Sys to multi-class classification of pneumonia on CT images. Slice-level classification network and patient-level classification network were combined to classify CT images in multiple categories. Wang et al. [37] proposed a new joint learning framework to achieve accurate diagnosis of COVID-19 by effectively learning heterogeneous datasets with distributed differences. By redesigning the recently proposed COVID-Net [34], a strong backbone had been built in terms of network architecture and learning strategies. As well as, in order to improve the classification performance of each dataset, the contrast training target was adopted to enhance the domain invariance of semantic embedding. On SARS-CoV-2 dataset [38], the proposed method achieved 90.83% for accuracy, 90.87% for F1-Score and 96.24% for AUC. Harsh et al. [39] proposed a deep transfer learning algorithm to accelerate the detection of COVID-19 cases by using X-ray and CT-Scan images of the chest. On SARS-CoV-2 dataset [38], the proposed method achieved 94.04% for sensitivity and 95.86% for specificity.
Although the above methods have achieved good results, it does not propose a solution to the problem of COVID-19 infection area with different sizes and shapes. This paper proposes a novel attention module that involves multi-receptive field features to achieve more accurate representation for different lesions. The experimental results show that our method can get better results than other state-of-the-art methods and may provide auxiliary discrimination and localization for COVID-19 diagnosis.
3. Methods
Our method uses VGG16 as the backbone. The overall network is described in Fig. 1. There are five stages and there fully connection layers. Each stage includes two or three convolutional layers and a max pooling layer. Different from VGG16, we replace the convolution layer before each pooling layer with the proposed multi-receptive field attention module, which is shown in the yellow cubes in Fig. 1. The corresponding detailed architecture of the attention module is illustrated in the yellow dotted box. Following, we introduce the proposed attention in sub-sections.
Fig. 1.
Architecture of proposed network. (For interpretation of the references to color in this legend, the reader is referred to the web version of this article.)
3.1. Multi-receptive field attention module
The architecture of multi-receptive field attention module is shown in the yellow dotted box in Fig. 1. Assuming that the given input feature map is , the multi-receptive field feature map sets are obtained by passing it through a PCM. Then is sent to two attention blocks separately. Firstly, are summed and then sent to the SAB to obtain the spatial attention weight (). Secondly, are concatenated and then sent to the CAB to obtain the channel attention weight . Finally, the and are respectively multiplied with the feature map obtained by concatenating and to obtain the final feature map . The process can be expressed as the following (1), (2), (3):
| (1) |
| (2) |
| (3) |
where ⊗ is element-wise multiplication. is obtained feature map after the attention module. The following will introduce the blocks of PCM, SAB and CAB. Fig. 2 shows the detailed of the three blocks.
Fig. 2.
Architecture of PCM, SAB and CAB. (a) is the structure of PCM, (b) is the structure of SAB, (c) is the structure of CAB. As illustrated, the outputs of PCM are sent to both SAB and CAB which make the attention blocks contain multi-receptive field information.
3.1.1. . PCM
The PCM aims to transfer multi-receptive field information to the attention blocks. The detailed structure is shown in Fig. 2(a). The input feature map is first passed through a convolutional layer to adjust the number of channels to reduce the number of parameters. The number of channels of the kernel can be set freely. Then the convolutional layers with different kernel sizes is passed through to obtain the multi-receptive field feature map sets . The number of channels of kernels is the same as the number of input channels, and the size of kernels is related to the size of the input feature maps, as shown in Table 1.
Table 1.
The size and number of kernels for different input feature maps (based on VGG16).
| Input feature maps | Number of kernels | Kernel sizes | Channel |
|---|---|---|---|
| 4 | 3, 9, 15, 21 | 64 | |
| 4 | 3, 7, 11, 15 | 128 | |
| 4 | 3, 5, 7, 9 | 256 | |
| 4 | 1, 3, 5, 7 | 512 | |
| 4 | 1, 3, 5, 7 | 512 |
3.1.2. . SAB
The SAB uses the multi-receptive field feature map sets obtained from PCM to extract the spatial attention weight. The architecture is shown in Fig. 2(b). First, element-wise addition is performed on the multi-receptive field feature map sets to obtain the output . Then is passed to a convolutional layer with kernel size for refining features. An average pooling layer in the channel dimension is used to obtain feature map . After a 3 × 3 convolutional layer, the spatial attention weight can be obtained through a sigmoid on . The process can be shown in Eq. 4.
| (4) |
where denotes the sigmoid function. represents convolution with kernel size . is average pooling down-sampling operation.
3.1.3. . CAB
Similar to SAB, the CAB uses the obtained from PCM to extract the channel attention weight. The structure of CAB is shown in Fig. 2(c). Multi-receptive field feature map sets is concatenated to get . Then is through an average pooling layer in spatial dimension to obtain . Next,a 1 × 1 convolutional layer is used to change the number of channels and new feature map is obtained. Finally passing a sigmoid function, the channel attention weight is obtained. The process can be shown in Eq. (5).
| (5) |
where denotes the sigmoid function. represents convolution with kernel size . means average pooling down-sampling operation.
3.2. Loss function
The final result is described in Eq. (3). With the proposed attention module, the network can focus more on the lesion area and finally extract more representative features, which greatly improves the classification effect of the network.
The loss function used in this paper is the cross-entropy loss, which is commonly used in binary classification, as following:
| (6) |
Where y is the ground truth label of an input CT image, is the prediction results from the network.
4. Experiment
We verify the validity of our proposed method on two dataset, our own COVID-19 dataset DTDB and a public datasets SARS-CoV-2 [38].
DTDB is provided by the Beijing Ditan Hospital Capital Medical University, including 40 patients infected with COVID-19 in different periods and 40 people without COVID-19 infection. We extracted 3650 slices containing COVID-19 infection and 3600 slices not containing COVID-19. The dataset was divided into 5800 images for training, 1450 images for validation by using 5-fold cross-validation. We used basic networks with different attention modules to conduct experiments.
SARS-CoV-2 [38] dataset is a publicly available COVID-19 CT dataset, containing 1252 CT scans that are positive for SARS-CoV-2 infection (COVID-19) and 1230 CT scans for patients non-infected by SARS-CoV-2, 2482 CT scans in total. The dataset was divided into 1986 images for training, 496 images for validation by using 5-fold cross-validation. We compared with different state-of-the-art methods to verify the effectiveness of our method.
The proposed network was implemented in Pytorch [40]. All the COVID-19 CT images in our experiments had been resized to . In order to reduce the influence of overfitting caused by limited datasets, we employed several data augmentation operations, including random rotation and random flipping (up-down or left-right in x-y planes). The Adam optimizer was employed for training our model in an end-to-end manner with an initial learning rate of 0.001 and betas of (0.9, 0.999). The learning rate decayed by 0.1 every 30 epochs. The batch size was set to 16 on an NVIDIA GeForce GTX 1070 GPU with 8 GB memory.
On DTDB dataset and SARS-CoV-2 dataset, training on the training set took about 3.6 h and 2.5 h respectively. As for testing a CT slice, it costed an average of 0.058 s on an NVIDIA GeForce GTX 1070 GPU with 8 GB memory.
4.1. Evaluation criteria
The metrics employed to quantitatively evaluate classification are accuracy, sensitivity, specificity and F1-score.
Accuracy represents the accuracy rate of classification results, as shown in Eq. (7):
| (7) |
The sensitivity and specificity measure the classifier's ability to identify positive samples and negative samples respectively:
| (8) |
| (9) |
F1 score is a measure of classification problems. It is a harmonic average of the precision and recall, with a maximum of 1 and a minimum of 0, as show in Eq. (12).
| (10) |
| (11) |
| (12) |
4.2. Ablation studies
In this section, we performed ablation experiments to verify the effectiveness of the two attention blocks. We used VGG16 as the backbone. The input image was resized to 224 × 224. The kernels information of the PCM was shown in Table 1. The experimental results were shown in Table 2.
Table 2.
Experimental results of ablation experiments (best in bold).
| Description | Accuracy (%) | Specificity (%) | Sensitivity (%) |
|---|---|---|---|
| VGG16 [20] | 93.02 | 94.32 | 91.25 |
| VGG16 + SAB | 94.76 | 96.35 | 93.32 |
| VGG16 + CAB | 96.02 | 96.83 | 95.13 |
| VGG16 + SAB + CAB | 97.12 | 96.89 | 97.21 |
From Table 2, we can see that both SAB and CAB can improve the diagnostic effect of the network. The accuracy improved by 1.74% and 2.03% respectively. When SAB and CAB were combined, the accuracy was further improved, which was 4.10% higher than the basic network. Fig. 3 shows the ROC curve and the confusion matrix of the ablation studies.
Fig. 3.
The ROC curve (left) and confusion matrix of ablation studies (right).
4.3. Experimental results
For DTDB COVID-19 dataset, we compared with three widely used image classification attention modules, including Residual Attention Network (RAN) [26], SE-net [27], CBAM [28], Table 3 summarized the experimental results.
Table 3.
COVID-19 diagnostic results on DTDB COVID-19 dataset.(best in bold).
It can be seen from Table 3 that our attention modules had a better effect than other commonly used attention modules. For accuracy, our method was 2.36% higher than RAN, 3.06% higher than SE-Net, and 2.85% higher than CBAM. For sensitivity and specificity, our method was (2.36%, 2.11%), (1.54%, 4.65%), (0.16%, 5.45%) higher than RAN, SE-Net, and CBAM respectively. Experimental results proved that the attention module we proposed can better recognize COVID-19 in CT images. Fig. 4 shows the ROC curve and the confusion matrix of the experimental results.
Fig. 4.
The ROC curve (left) and the confusion matrix of different attention modules (right).
On SARS-CoV-2 dataset [38], we compared with baseline and two state-of-the-art methods in [37], [39]. Table 4 summarized the experimental results.
Table 4.
COVID-19 diagnostic results on SARS-CoV-2 dataset. (best in bold).
Experimental results illustrated that the proposed network achieved better result than other methods. Comparing with backbone, the proposed network improved the accuracy of 8.29%, F1-score of 8.14% and AUC of 6.85% respectively. As for [37], we achieved 4.33%, 4.73% and 2.77% improvements in accuracy, F1-score and AUC respectively. Compared to [39], the proposed method reported better results without transfer learning. The experimental results illustrated the effectiveness of the proposed method for COVID-19 discrimination.
4.4. Infection visualization
Above we quantitatively analyzed the experimental results of our network and showed the effectiveness of our method. Then we used the Grad-Cam [41] method for visualization and localization, which can verify the reliability of our method.
We randomly selected several test CT images and located the lesion area with Grad-CAM median value greater than 0.7. Fig. 5 shows the results.
Fig. 5.
The visualization results of Grad-CAM. Row 1 is the visualization results and row 2 are the corresponding original CT images.
As can be seen from the Fig. 5, our attention focused on the area where the positive samples were located. Our method can better locate the distinguishable COVID-19 lesion parts of CT images to improve the classification ability.
4.5. Lesion localization and quantitative analysis
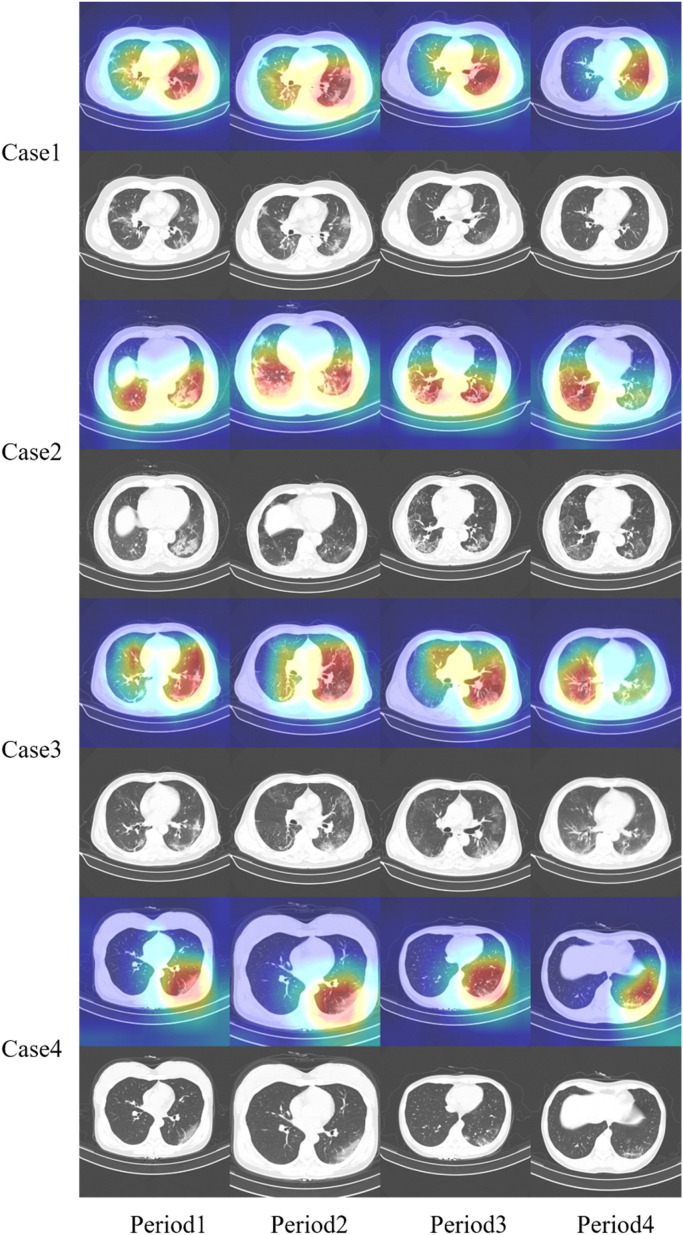
DTDB dataset contains some COVID-19 patients with CT cases at different periods. We used Grad-CAM to locate the lesion area of the COVID-19 patients from infection to cure on CT images. In a case of COVID-19 patients, Fig. 6 visualized CT slices with the largest area of feature map response greater than 0.7 and positive diagnosis of COVID-19. It showed the progress with treatment periods for 4 patients.
Fig. 6.
COVID-19 lesion localization in different periods of patients.
From the CT images of patients with COVID-19, Fig. 6 illustrated obviously that the area of the lesion increased faster in the early stage. With the advancement of treatment for some periods, the lesion area decreased significantly, such as the 2, 4, 6, 8 rows in Fig. 6. The qualitative visualization results of the proposed method had the same trend and the high response region of the localization approached the real lesion region, shown in the 1, 3, 5, 7 rows in Fig. 6.
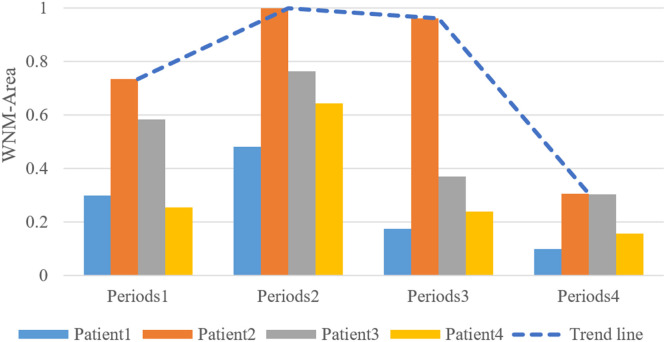
In order to illustrate our conclusion more intuitively, we quantitatively analyzed the weighted normalized maximum area of the cases in different periods of the 4 patients in Fig. 6. is described in Eq. (13).
| (13) |
where represents the feature map maximum response region area of the i-th patient j-th period, and represents the obtained probability output from the network for the corresponding slice with COVID-19 positive. represents normalization operation.
We used combined diagram to quantitatively analyze the weighted normalized maximum area (WNM-Area) in Fig. 7. It also reflects a basically consistent infection changing trend that the infection of coronavirus has been serious at early periods and then tend to decrease along with effective treatment. The proposed method can provide a quantitative analysis of infection area in different treatment periods and assist doctors to evaluate the progress of COVID-19 treatment effectively.
Fig. 7.
Combined diagram for variation of weighted normalized maximum area in different periods. The blue dotted tend line represents the change trend of the WNM-Area of the patients over time.
5. Conclusions
This paper presents a convolutional neural network structure for the diagnosis of COVID-19 on CT images. The structure of the network is based on VGG16. The designed attention modules are added in each stage before pooling layer. It is composed of pyramid convolution module (PCM), multi-receptive field spatial attention block (SAB) and multi-receptive field channel attention block (CAB). PCM can improve the diagnosis results of different size for lesion regions. Better classification accuracy is achieved by using SAB and CAB which can make features extracted by the network focus on lesion areas. Experiments performed on our DTDB COVID-19 dataset achieved 97.12% for accuracy, 96.89% for specificity, and 97.21% for sensitivity. Compared with other commonly used attention modules, our methods get better results. As for public COVID-19 SARS-CoV-2 dataset, more satisfactory results are obtained compared with the state-of-the-art methods. The quantitative and visualization results indicate that the features extracted by the proposed network focus on the localization of lesions regions, which is more helpful in COVID-19 diagnosis. The same results are achieved for lesion localization. By locating the lesion area of the same patient in different periods, it can assist doctors to determine the effectiveness of the treatment. The proposed method provides quantitative diagnostic information for doctors, as well as auxiliary lesion location information. Our method has the space for further optimization and also can be transformed to other medical diseases auxiliary diagnosis.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.WHOrganization, Who Director-General’s Opening Remarks at the Media Briefing On Covid-19-11 March 2020, 2020. 〈https://www.who.int/dg/speeches/detail/who-director-generals-opening-remarks-at-the-media-briefing-on-covid-19-11-march-2020〉.
- 2.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Tan W. A novel coronavirus from patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johns Hopkins University, COVID-19 Dashboard by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU), 2020. 〈https://coronavirus.jhu.edu/map.html〉.
- 4.Taipale J., Romer P., Linnarsson S. Population-scale testing can suppress the spread of COVID-19. medRxiv Prepr. 2020 doi: 10.1101/2020.04.27.20078329. [DOI] [Google Scholar]
- 5.Smyrlaki I., Ekman M., Vondracek M., Papanicoloau N., Lentini A., Aarum J., Reinius B. Massive and rapid COVID-19 testing is feasible by extraction-free SARS-CoV-2 RT-qPCR. Nat. Commun. 2020;(2020) doi: 10.1038/s41467-020-18611-5. 11.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wynes M.W., Sholl L.M., Dietel M., Schuuring E., Tsao M.S., Yatabe Y., Hirsch F.R. An international interpretation study using the ALK IHC antibody D5F3 and a sensitive detection kit demonstrates high concordance between ALK IHC and ALK FISH and between evaluators. J. Thorac. Oncol. 2014;9(5):631–638. doi: 10.1097/JTO.0000000000000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang C., Wen T., Shi F.J., Zeng X.Y., Jiao Y.J. Rapid detection of IgM antibodies against the SARS-CoV-2 virus via colloidal gold nanoparticle-based lateral-flow assay. ACS Omega. 2020 doi: 10.1021/acsomega.0c01554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chinese Society of Laboratory Medicine Expert consensus on nucleic acid detection of 2019-novel coronavirus (2019-ncov) Natl. Med. J. China. 2020;100(00) E003-E003. [Google Scholar]
- 9.de Barry O., Cabral D., Kahn J.E., Vidal F., Carlier R.Y., El Hajjam M. 18-FDG pseudotumoral lesion with quick flowering to a typical lung CT COVID-19. Radiol. Case Rep. 2020;15(10):1813–1816. doi: 10.1016/j.radcr.2020.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lorensen W.E., Cline H.E. Marching cubes: a high resolution 3D surface construction algorithm. ACM SIGGRAPH Comput. Graph. 1987;21(4):163–169. [Google Scholar]
- 11.Lei J., Li J., Li X., Qi X. CT imaging of the 2019 novel coronavirus (2019-nCoV) pneumonia. Radiology. 2020;295(1):18. doi: 10.1148/radiol.2020200236. 18-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.J. Zhao, Y. Zhang, X. He, P. Xie, COVID-CT-Dataset: A CT Scan Dataset about COVID-19, 2020. https://arxiv. org/pdf/2003.13865 v1.pdf.
- 13.Zheng B., Liu Y., Zhu Y., Yu F., Jiang T., Yang D., Xu T. MSD-Net: multi-scale discriminative network for COVID-19 lung infection segmentation on CT. IEEE Access. 2020;8:185786–185795. doi: 10.1109/ACCESS.2020.3027738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O. Ronneberger, P. Fischer, T. Brox, U-net: convolutional networks for biomedical image segmentation, in: Proceedings of the International Conference on Medical image computing and computer-assisted intervention, Springer, Cham, 2015 October. pp. 234–241.
- 15.Kashf D.W.A., Okasha A.N., Sahyoun N.A., El-Rabi R.E., Abu-Naser S.S. Predicting DNA lung cancer using artificial neural network. Int. J. Acad. Pedagog. Res. (IJAPR) 2018;2(10):6–13. [Google Scholar]
- 16.Zheng J., Yang D., Zhu Y., Gu W., Zheng B., Bai C., Shi W. Pulmonary nodule risk classification in adenocarcinoma from CT images using deep CNN with scale transfer module. IET Image Process. 2020;14(8):1481–1489. [Google Scholar]
- 17.Lakshmanaprabu S.K., Mohanty S.N., Shankar K., Arunkumar N., Ramirez G. Optimal deep learning model for classification of lung cancer on CT images. Future Gener. Comput. Syst. 2019;92:374–382. [Google Scholar]
- 18.LeCun Y., Bottou L., Bengio Y., Haffner P. Gradient-based learning applied to document recognition. Proc. IEEE. 1998;86(11):2278–2324. [Google Scholar]
- 19.Simonyan K., Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv Prepr. arXiv. 2014;1409:1556. [Google Scholar]
- 20.C. Szegedy, W. Liu, Y. Jia, P. Sermanet, S. Reed, D. Anguelov, A. Rabinovich, Going deeper with convolutions, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2015. pp. 1–9.
- 21.Ioffe S., Szegedy C. Batch normalization: accelerating deep network training by reducing internal covariate shift. arXiv Prepr. arXiv. 2015;1502:03167. [Google Scholar]
- 22.C. Szegedy, V. Vanhoucke, S. Ioffe, J. Shlens, Z. Wojna, Rethinking the inception architecture for computer vision, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2016. pp. 2818–2826.
- 23.K. He, X. Zhang, S. Ren, J. Sun, Deep residual learning for image recognition, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2016. pp. 770–778.
- 24.G. Huang, Z. Liu, L. Van Der Maaten, K.Q. Weinberger, Densely connected convolutional networks, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2017. pp. 4700–4708.
- 25.A. Vaswani, N. Shazeer, N. , Parmar, J. Uszkoreit, L. Jones, A.N. Gomez, I. Polosukhin, Attention is all you need, in: Advances in Neural Information Processing Systems, 2017. pp. 5998–6008.
- 26.F. Wang, M. Jiang, C. Qian, S. Yang, C. Li, H. Zhang, X. Tang, Residual attention network for image classification, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2017. pp. 3156–3164.
- 27.J. Hu, L. Shen, G. Sun, Squeeze-and-excitation networks, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2018. pp. 7132–7141.
- 28.S. Woo, J. Park, J.Y. Lee, Kweon So, CBAM: convolutional block attention module, in: Proceedings of the European Conference on Computer Vision (ECCV), 2018. pp. 3–19.
- 29.Park J., Woo S., Lee J.Y., Kweon I.S. BAM: Bottleneck attention module. arXiv Prepr. arXiv. 2018;1807:06514. [Google Scholar]
- 30.Elkorany A.S., Elsharkawy Z.F. COVIDetection-net: a tailored COVID-19 detection from chest radiography images using deep learning. Optik. 2021;231(3) doi: 10.1016/j.ijleo.2021.166405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oh Y., Park S., Ye J.C. Deep learning covid-19 features on CXR using limited training data sets. IEEE Trans. Med. Imaging. 2020;39(8):2688–2700. doi: 10.1109/TMI.2020.2993291. [DOI] [PubMed] [Google Scholar]
- 32.Sitaula C., Hossain M.B. Attention-based VGG-16 model for COVID-19 chest X-ray image classification. Appl. Intell. 2020:1–14. doi: 10.1007/s10489-020-02055-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hussain E., Hasan M., Rahman M.A., Lee I., Tamanna T., Parvez M.Z. CoroDet: a deep learning based classification for COVID-19 detection using chest X-ray images. Chaos Solitons Fractals. 2021;142 doi: 10.1016/j.chaos.2020.110495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang L., Lin Z.Q., Wong A. COVID-net: a tailored deep convolutional neural network design for detection of covid-19 cases from chest x-ray images. Sci. Rep. 2020;10(1):1–12. doi: 10.1038/s41598-020-76550-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ouchicha C., Ammor O., Meknassi M. CVDNet: a novel deep learning architecture for detection of coronavirus (Covid-19) from chest x-ray images. Chaos Solitons Fractals. 2020;140 doi: 10.1016/j.chaos.2020.110245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qian X., Fu H., Shi W., Chen T., Fu Y., Shan F., Xue X. M3 Lung-Sys: a deep learning system for multi-class lung pneumonia screening from CT imaging. IEEE J. Biomed. Health Inform. 2020;24(12):3539–3550. doi: 10.1109/JBHI.2020.3030853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Z., Liu Q., Dou Q. Contrastive cross-site learning with redesigned net for COVID-19 CT classification. IEEE J. Biomed. Health Inform. 2020;24(10):2806–2813. doi: 10.1109/JBHI.2020.3023246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soares E., Angelov P., Biaso S., Froes M.H., Abe D.K. Trends in emergency department visits and hospital admissions in health care systems in 5 states in the first months of the COVID-19 pandemic in the US. JAMA Intern. Med. 2020;180:1328–1333. doi: 10.1101/2020.04.24.20078584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Panwar H., Gupta P.K., Siddiqui M.K., Morales-Menendez R., Bhardwaj P., Singh V. A deep learning and grad-CAM based color visualization approach for fast detection of COVID-19 cases using chest X-ray and CT-Scan images. Chaos Solitons Fractals. 2020;140 doi: 10.1016/j.chaos.2020.110190. 110190.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.A. Paszke, S. Gross, F. Massa, A. Lerer, J. Bradbury, G. Chanan, T. Killeen, Z. Lin, N. Gimelshein, L. Antiga. Pytorch: An imperative style, high-performance deep learning library, in: Proceedings of the Advances in Neural Information Processing Systems, 2019, pp. 8026–8037.
- 41.Selvaraju R.R., Cogswell M., Das A., Vedantam R., Parikh D., Batra D. Grad-cam: visual explanations from deep networks via gradient-based localization. Int. J. Comput. Vis. 2020;128(2):336–359. [Google Scholar]