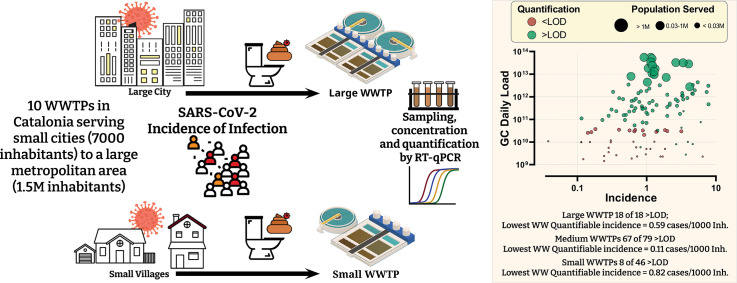
Abstract
Wastewater based epidemiology was employed to track the spread of SARS-CoV-2 within the sewershed areas of 10 wastewater treatment plants (WWTPs) in Catalonia, Spain. A total of 185 WWTPs inflow samples were collected over the period consisting of both the first wave (mid-March to June) and the second wave (July to November). Concentrations of SARS-CoV-2 RNA (N1 and N2 assays) were quantified in these wastewaters as well as those of Human adenoviruses (HAdV) and JC polyomavirus (JCPyV), as indicators of human faecal contamination. SARS-CoV-2 N gene daily loads strongly correlated with the number of cases diagnosed one week after sampling i.e. wastewater levels were a good predictor of cases to be diagnosed in the immediate future. The conditions present at small WWTPs relative to larger WWTPs influence the ability to follow the pandemic. Small WWTPs (<24,000 inhabitants) had lower median loads of SARS-CoV-2 despite similar incidence of infection within the municipalities served by the different WWTP (but not lower loads of HAdV and JCPyV). The lowest incidence resulting in quantifiable SARS-CoV-2 concentration in wastewater differed between WWTP sizes, being 0.11 and 0.82 cases/1000 inhabitants for the large and small sized WWTP respectively.
Keywords: Wastewater-based epidemiology, SARS-CoV-2, Pandemic surveillance, Small large WWTPs, Human adenoviruses, JC polyomavirus
Graphical abstract
1. Introduction
The COVID-19 pandemic, caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), went from a local issue in China to a truly global threat to human health, wellbeing and the economy, all in a matter of a few months. As of the end of November 2020, the disease has caused over 64 M infections and 1.4 M deaths and has dictated the agenda of governments for the past months (WHO, 2020).
The dominant impact the pandemic has had on everyday life has also resulted in a scientific and technological scramble for solutions, amongst which has been the application of wastewater-based epidemiology (WBE) to track the distribution and magnitude of infections amongst the population generating the wastewater (WW) (Medema et al., 2020a, Medema et al., 2020b; Mallapaty, 2020; Lesté-Lasserre, 2020; Ahmed et al., 2020a; Randazzo et al., 2020). While COVID-19 is mainly a respiratory illness, SARS-CoV-2 can also infect intestinal cells and it is shed in faeces in moderate quantities (between 102 and 107 GC/g) (Cheung et al., 2020; Wölfel et al., 2020; Zheng et al., 2020). This has prompted suggestions to apply WBE principles in tracking the spread of the disease (Bivins et al., 2020; Daughton, 2020; Hata and Honda, 2020), with the first reports of detection and quantification of the virus in wastewater (WW) in April 2020 (Ahmed et al., 2020a; Lodder and de Roda Husman, 2020; Medema et al., 2020b).
The theoretical scientific basis for tracking the virus in WW has been previously described (see Ahmed et al., 2020a; Wu et al., 2020), however, a number of practical uncertainties still persist. While these issues hinder the application of WBE for accurately measuring the actual number of infected individuals, the potential advantages – that an approximation affords to public health monitoring – are too valuable to dismiss. Being able to obtain an estimate of the degree of active cases within a specific community is paramount to controlling the spread and obtaining this information through WBE is cheaper and less laborious than carrying out diagnostic screening of individuals at a massive scale. At the initial stages of the pandemic, the health authorities in most countries struggled to cope with the sudden influx of testing requirements. As a consequence of this, the number of individual nasopharyngeal or oral swabs carried out during the first wave of the pandemic to determine if suspected cases were in fact positive had to be prioritised. Thus, the real degree of infection (actual prevalence) amongst a community could not be established. This was also later confirmed by seroprevalence studies in numerous countries which showed that a much larger portion of the populations under study were positive to SARS-CoV-2 antibodies, indicating that they have been or are infected with the virus (Eckerle and Meyer, 2020; Pollán et al., 2020). In clear contrast, the WBE approach bypasses the limitations of individual testing since it provides integrated information of the whole community. A few WW samples can provide the same population-level information as thousands of individual tests. Clearly WBE is not mean to replace individual diagnoses, but rather provide complementary information to health authorities and decision makers about i) the prevalence of infection, also over time, within specific geographic areas and ii) the identification of potential hot-spots while taking up a negligible quantity of medical resources and consumables.
While the potential benefits are clear, a number of uncertainties of the WBE monitoring of SARS-CoV-2 persist. These more variable parameters include: i) the large variability in both the faecal viral load of infected individuals and their shedding duration; ii) the proportion of the total infected individuals that shed the virus in faeces; and iii) the degradation of the viral RNA in WW and during storage of collected samples. The combination of these parameters is expected to differ substantially with the conditions present at WW treatment plant (WWTP) of different sizes, in terms of number of people they serve. That is, while the WW being received cannot be logically influenced directly by a WWTP at its inflow, the conditions present in the drainage area of small and big WWTPs are different and are hypothesised to affect the ability to employ WBE tracking of the virus within their respective communities.
Herein, 10 WWTPs of different sizes (ranging from 7 k to 1.5 M inhabitants (inh.) served) have been monitored for the presence of SARS-CoV-2 and the aims are threefold: (i) to demonstrate the quantitative relationship between the number of COVID-19 cases and the SARS-CoV-2 RNA detected in wastewater over the first and second waves of the pandemic; (ii) to shed light on the minimum number of COVID-19 cases within the sewershed area of a WWTP needed in order to have quantifiable SARS-CoV-2 RNA in WW; (iii) to contribute to a better understanding of population size effects on monitoring efforts. Additionally, Human adenoviruses (HAdV) and JC polyomavirus (JCPyV) have also been quantified in the WW samples. These viruses are commonly used as human faecal indicators, as they end up in sewage through gastrointestinal and urinary tract shedding (Bofill-Mas, 2016; Bofill-Mas et al., 2006; Bofill-Mas et al., 2000; Rusiñol and Girones, 2017), being persistent in wastewater samples worldwide and not prone to seasonal variations nor to variations from other environmental conditions (Rusiñol et al., 2014). Thus, their inclusion serves as both a quality control as well as to identify any dilution or other undesired mixing of WW. All this information is expected to aid the application of WBE in decision making by better defining the expectations and limits of the approach to policy makers.
2. Materials and methods
2.1. Wastewater samples – viral quantification
A total of 185 influent 24 h time-proportional composite WW samples from mid-March till early-November 2020 from 10 WWTPs, of varying sizes, in the Region of Catalonia (Spain) were collected. All samples were collected as a 24-h composite using the autosamplers available at the WW facilities. The monitored WWTPs (Table 1 ) serve 26% of the Catalan population (2.0 M people of a total of 7.6 M), including more than half of the City of Barcelona, one of the most COVID-19 affected areas in Spain with almost 300,000 cases by the end of November. All samples (250 mL) were collected at the studied WWTPs using gloves and sterile containers, transported at 4 °C and delivered to the laboratory at The University of Barcelona and stored at −80 °C until analysis.
Table 1.
Characteristics of the selected WWTPs organised according to the number of inhabitants served (the number of inhabitants registered within the municipalities cover by that WWTP).
| WWTP ID | Inhabitants served | WWTP Design capacity (Hab. Eq.) | Sampling interval (over 24 h) | Daily Volume (ML) | pH | Conductivity (μS/cm2) | BOD5 (mgO2/L) | COD (mgO2/L) | NH4—N (mgN/L) |
|---|---|---|---|---|---|---|---|---|---|
| A | 7090 | 27,000 | Every 20 min | 2.8 ± 1.2 | 7.46 ± 0.15 | 2365 ± 260 | 137 ± 68 | 350 ± 375 | 45 ± 8 |
| B | 8241 | 19,956 | Every hour | 1.5 ± 0.3 | 7.36 ± 0.24 | 1937 ± 179 | 190 ± 223 | 561 ± 329 | 55 ± 14 |
| C | 14,744 | 35,553 | Every hour | 2.1 ± 0.7 | 7.57 ± 0.27 | 3039 ± 653 | 143 ± 35 | 478 ± 174 | 42 ± 14 |
| D | 26,425 | 68,750 | Every 20 min | 7.4 ± 1.7 | 7.22 ± 0.20 | 3076 ± 199 | 136 ± 48 | 484 ± 78 | 40 ± 3 |
| E | 56,746 | 165,450 | Every hour | 23.1 ± 2.9 | 7.55 ± 0.16 | 2428 ± 155 | 291 ± 105 | 584 ± 304 | 60 ± 7 |
| F | 58,479 | 204,166 | Every hour | 17.4 ± 3.5 | 7.22 ± 0.24 | 2168 ± 267 | 331 ± 212 | 700 ± 339 | 44 ± 9 |
| G | 66,048 | 285,666 | Every hour | 26.6 ± 6.8 | 8.17 ± 0.21 | 2973 ± 502 | 69 ± 30 | 205 ± 74 | 10 ± 2 |
| H | 92,243 | 196,167 | Every hour | 22.1 ± 2.1 | 7.52 ± 0.18 | 1506 ± 106 | 195 ± 45 | 475 ± 101 | 44 ± 14 |
| I | 183,517 | 451,250 | Every hour | 24.9 ± 2.3 | 7.54 ± 0.15 | 1710 ± 212 | 390 ± 72 | 607 ± 100 | 42 ± 2 |
| J | 1,497,767 | 2,843,750 | Every hour | 341 ± 78.1 | 7.39 ± 0.13 | 2786 ± 215 | 364 ± 72 | 981 ± 175 | 42 ± 6 |
Mean values and standard deviations over the sampling period are shown. WWTP: wastewater treatment plant; ML: megalitres; BOD5: biological organic demand; COD: chemical oxygen demand; NH4-N: ammonium nitrogen.
As back up during analysis, 150 mL of each wastewater sample was stored. Concentration of the viral particles was performed by first removing debris by centrifuging 100 mL of sample at 4750 ×g for 30 min and processing the supernatant for concentration by ultrafiltration. Due to the severe shortage of filtration devices caused by the COVID-19 pandemic, concentration of viruses was initially achieved using Centricon® Plus-70 30 KDa (Millipore) and from 15th May onwards the automatic Concentrating Pipette (CP-Select™) with 150 KDa ultrafiltration tips (Innovaprep) was used. A recent study characterizing both Centricon® devices and the new CP-Select™, have shown that both concentration methods yield equivalent results (Forés et al., 2021). When using Centricon®, the viral particles present in a volume of 70 mL of supernatant were concentrated in 200–250 μL as previously described (Medema et al., 2020a). When using CP-Select™, between 80 and 90 mL of supernatant were filtered and eluted in a final volume of 240–300 μL. The concentration factors were thus between 280–350× for the Centricon® and between 266–375× for the CP-Select™. All samples were spiked with the bacteriophage MS2 as process control before any processing was carried out.
Viral DNA and RNA were co-extracted with QIAamp Viral RNA Mini Kit using the QIAcube automatic system (Qiagen). A total of 140 μL of sample concentrates were extracted into a final volume of 60 μL. To determine the genome copy numbers of SARS-CoV-2, the N1 and N2 assays targeting the viral nucleocapsid (US CDC, 2020) were used. Specific qPCR and RT-qPCR assays previously described were used to quantify MS2 bacteriophage (Pecson et al., 2009), HAdV (Bofill-Mas et al., 2006; Hernroth et al., 2002) and JCPyV (Pal et al., 2006) by using TaqMan™ Environmental Master Mix 2.0 and RNA UltraSense™ One-Step RT-qPCR System (Invitrogen) for DNA and RNA viruses respectively. Primers, probes and qPCR conditions are summarized in Table 1 of the supplementary material. The SARS-CoV-2 standard was constructed using the EURM-019 single stranded RNA fragments of SARS-CoV-2, provided by the European Commission Joint Research Centre. The other qPCR standards were prepared using synthetic gBlocks® Gene fragments (IDT) and serial dilutions were quantified with a Qubit® fluorometer (Thermo Fisher Scientific). All RT-qPCR were performed in quadruplicate and included non-template controls. To avoid contamination, qPCR preparation was performed in a clean laboratory and RNA/DNA extraction on a bench that is only used for that purpose. Template addition was done inside a PCR cabinet. The limits of detection (LOD) for SARS-CoV-2 assays were 1.4 × 103 GC/L (N1 assay) and 9.9 × 102 GC/L (N2 assay).
2.2. Ancillary data – daily infections & WWTP parameters
The number of daily new identified cases of infection in the corresponding sewersheds (i.e. municipalities served by a WWTP) of the sampled WWTP, as identified after individual PCR tests, is routinely compiled by the Catalan Health System and available on its open data platform (https://analisi.transparenciacatalunya.cat/resource). Data were handled through an in-house developed application available online on https://llegir-api-covid19.h2793818.stratoserver.net. In this database, each individual case is linked to a territorial code (at municipal level) and it is based on the residence address of the patient. For each of the sampled WWTPs, the source of WW overlaps very well to municipal levels. Thus, the daily summation of the number of cases in the municipalities which drain to specific WWTPs, can be considered as the number of new people shedding the virus in this WW. While it is known that not all carriers shed viral RNA, it was assumed that the ratio of shedders to non-shedders remained stable within the studied period and the differences in loads is mainly due to differences in number of individuals carrying the SARS-CoV-2 virus. For the specific case of the Besòs WWTP (WWTP-J) in Barcelona, the identification of new cases is slightly more complex, since the city of Barcelona is identified with a single municipal code but is served by two WWTPs, one of which is the Besòs WWTP which treats 65% of the city's WW. Thus, for the WWTP-J, the number of new cases in the drainage area was estimated as the sum of the cases in the relevant neighbouring municipalities plus 65% of the cases in the municipality of Barcelona. From these daily new cases, three estimates of the approximate number of people actively shedding the viral RNA in their excreta at any given time, was computed according to the equations below, where t = 0 represents the day when the WWTP sample was collected. The active cases in the 7 days following the day of sampling (Eq. (2)) was used as a way to check for correlation between the SARS-CoV-2 concentration in WW and the estimated number of upcoming COVID-19 cases assuming that individual tests often lag behind the start of the shedding period and sometimes behind symptoms.
| (1) |
| (2) |
| (3) |
Physicochemical and other parameters of the sampled WWTP were provided by the corresponding WWTP management companies, this included daily inflow volume of WW which was used to compute the daily load of genome copies (GC), BOD5, COD and other routine parameters.
2.3. Statistical analysis & evaluation of viral recovery
MS2 recovery percentages were calculated as described in Forés et al., 2021. Briefly, viral recovery percentages were calculated according to experimental values obtained by spiking samples with MS2 viral stocks and using as input viral concentration the direct quantification of the viral stock added. To evaluate whether there were statistically significant differences between viral recoveries and between physicochemical parameters reported for each wastewater treatment facility we run Kruskal–Wallis test using SPSS software ver. 25.0 (IBM, Armonk, NY, USA). Results were considered to be significant with a p < 0.05. Pearson's correlation tests were run to evaluate the potential associations between mean viral concentrations and the maximum incidences reported and between viral concentrations and wastewater physicochemical parameters (BOD, COD and NH4-H).
3. Results and discussion
3.1. Differences between WWTP and viral recoveries
The overall mean viral recovery of the applied procedure using MS2 as a process control was 30.19% (with a 95% interval of confidence between 27.10 and 33.29%). Ultrafiltration with centrifugal ultrafilters that have been recently used for the concentration of SARS-CoV-2 from wastewater, reported similar results with MS2 as the process control (Forés et al., 2021; Medema et al., 2020a, Medema et al., 2020b). Although the physicochemical parameters between WWTP were significantly different (see boxplots SI Fig. 1), no significant differences were observed amongst the viral recoveries (p-value = 0.069).
3.2. SARS-CoV-2 loads over the first and second waves
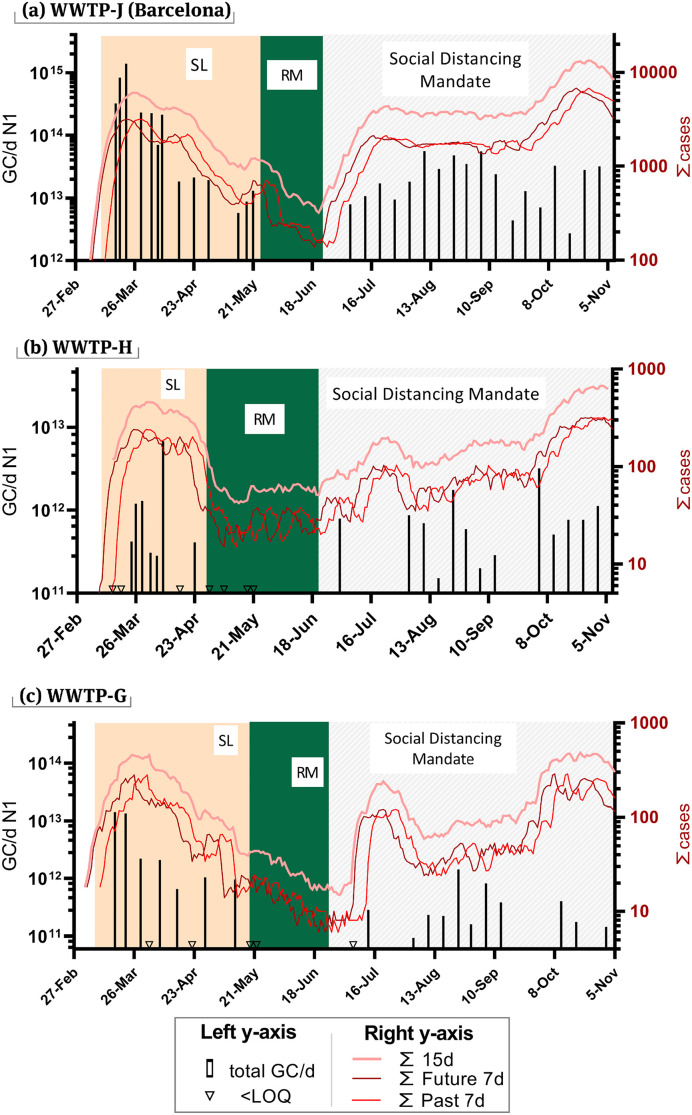
The WWTPs – WWTP-J (population served: 1,497,767), WWTP-H (population served: 93,796) and WWTP-G (population served: 66,048) were monitored for SARS-CoV-2 abundance (N1 and N2 assay) starting from mid-March and results till the start of November. Fig. 1 shows the SARS-CoV-2 quantification (N1 assay) and the estimates of cases as per Eqs. (2), (3). Background coloured areas indicate the different containment measures imposed on the respective populations by health authorities. In all three cities, N1 concentrations in WW matched well to estimates of infection prevalence, with high values preceding the upcoming high infection incidences, in most cases. The anticipation is also seeing through the correlations shown in Fig. 2 .
Fig. 1.
Prevalence of infection estimates and WW measured concentrations for SARS-CoV-2 (N1 assay) (a) WWTP-J in Barcelona, (b) WWTP-H and (c) WWTP-G. Values below LOQ are marked with an inverted triangle on the x-axis. WW values are expressed as daily loads. The sum of cases shows in shades of red were calculated using Eq. (2) (bright red), Eq. (3) (crimson-red), Eq. (4) (pastel red). Both y-axis are in log10 scale. The shading represents the enforced measures at the time: SL = strict lockdown; RM = reduce mobility.
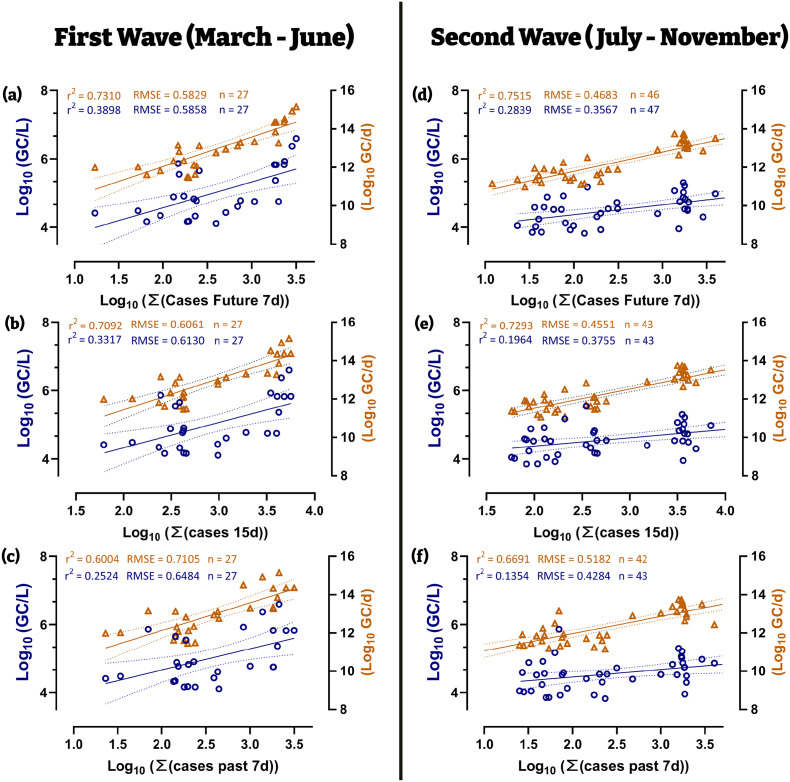
Fig. 2.
For WWTPs (WWTP- J, -G, -H): Linear correlations of the log converted values of viral concentration and daily viral loads with estimates for number of cases during the first wave (left half) and second wave (right half). (a–c and d–f follow Eqs. (2), (3)). Dotted line represents the 95% C.I. Difference in n between series is due to the daily flow at the WWTP inlet was not available on and daily GC loads could not be computed.
The highest concentration of SARS-CoV-2 in WW was obtained for WWTP-J on the 22nd of March (3.92 × 106 GC/L), corresponding to a daily total load of 1.4 × 1015 GC entering the WWTP (Fig. 1a). This value matched well to the infection peak for the first wave of the pandemic in Barcelona, which reported the highest number of new daily reported infections just 4 days later (26th of March, 556 new daily cases). The largest 7-day cumulative sum (Eq. (2)) of daily cases for WWTP-J was recorded on the 28th of March. From these results, an early warning of approximately 6 days could be envisaged from WW data at the start of the pandemic (first wave), being especially relevant considering the limited clinical testing capacity at the time that probably delayed diagnosis. While the second wave of the pandemic (July–onwards) showed higher recorded cases, the concentration of N1 in WW was lower than that recorded in the first wave, suggesting that a larger number of cases were not diagnosed and registered during the first wave while acknowledging that other factors could be the cause.
The first wave peak for WWTP-G (Fig. 1c) most probably occurred sometime before the 17th of March, the day when first WW sample was collected. The city of WWTP-G was identified early on as one of the three main COVID-19 hotspots in Catalonia and was put under lockdown on the 12th of March, a few days before the rest of the country. The first two WW samples from WWTP-G showed the highest concentrations of N1 recorded while the number of daily new cases was not registered until the 26th of March and the weekly cumulative sum peaked on the 1st of April. These infection estimates, however, were most probably due to delays in diagnosis. In fact, the first reported cases diagnosed by PCR in this city dated on the 10th of March (18 positive cases) and health authorities put the city under lockdown only 2 days with the assumption that there were a large number of hidden cases.
WWTP-H (Fig. 1b), while only 30 km away from WWTP-G, suffered a less intense outbreak. The city showed very similar number of infections to the hotspot city (WWTP-G) during the first wave despite having a larger population (40% more inhabitants). In WWTP-H, the highest N1 concentrations in WW was recorded on the 8th of April while the peak of daily new cases was reported on the 29th of March and the peak for the cumulative 7 day sum was two days later (31st of March). Less clear is the N1 peak observed a week later (8th of April) and the lack of detection in the following week (16th April) since no dilution by rainfall applies. Besides, the concentration of other viral indicators (HAdV and JCPyV) in this sample was similar to preceding and following dates (SI Table 2).
The efficacy of the lockdown measures implemented nationwide are well reflected in the results from WW monitoring. Spain went into a nationwide lockdown on the 14th of March although some hotspot areas were locked down earlier (such as the city served by WWTP-G). This lockdown was organised in four phases of incremental restrictions, being the phase 0 the most restrictive and phase 4 the least. During the lockdown (orange shading in Fig. 1a–c), the concentration of SARS-CoV-2 in sewage progressively decreased together with daily new cases and estimates of infection prevalence. The lockdown in Spain was highly restrictive and generally well enforced, allowing only the bare essential movements. SARS-CoV-2 concentrations in sewage were reduced to below LOD in subsequent phases (Fig. 1a–c green shading) agreeing with the low number of reported cases during this period. The following phases, which mandated social distancing, mask wearing and limited some social activities while allowing travel between regions (Fig. 1a–c grey shading) quickly resulted in an increase of N1 concentrations above the LOD. Similar observations were reported in France after the lockdown and where concentrations of SARS-CoV-2 in sewage increased by 1.5 orders of magnitude after relaxing the restrictions (Trottier et al., 2020). The anticipation of the infection peak through WW is not as evident for the second wave, however the correlations done in Fig. 2 still show the best fit with the number of cases for the following 7 days of sampling. Overall, the health authorities and population were better prepared for the second wave and the lag in identifying cases was probably shorter and hence not as evident as for the first wave in Fig. 1.
The WW concentrations of SARS-CoV-2 RNA measured herein are well within the range of reported values in other WBE SARS-CoV-2 studies. Medema et al. (2020b) reported first wave maximum concentration of 7.9 × 105 GC/L (N1) in the Tilburg, Netherlands on the 16th of March (compare to. 3.9 × 106 GC/L recorded on the 22nd of March in Barcelona which saw a higher incidence of infection than the Netherlands). Analysis of SARS-CoV-2 in wastewater collected from other Spanish WWTPs located in areas with similar COVID-19 incidence during April (1 case per 1000 inhabitants) reported concentrations (N1 and N2 assays) ranging between 105 and 106 GC/L Randazzo et al. (2020). In Germany, WW concentrations reported for April (Westhaus et al., 2021) were up to 2 orders of magnitude lower than values reported herein despite having similar reported incidence rates (0.5–1.8 cases per 1000 inhabitants). Germany as a whole suffered a more contained COVID-19 outbreak during the first wave and the mismatch could be due to a higher detection rate of COVID-19 cases in Germany relative to Spain.
During the first wave in Catalonia, PCR tests were less readily available giving estimates that under-reported cases (Russell et al., 2020). In fact, it has been documented that the median viral loads in clinical samples over the first phase of the outbreak were higher as compared to the following period (Jacot et al., 2020). The higher WW loads during the first wave reinforces the idea that the number of real infections in the first wave was higher than the second wave, despite the fact that more cases were diagnosed during the second wave i.e. the percentage of cases identified out of the total cases was higher in the second wave.
3.3. Relationship between SARS-CoV-2 loads and COVID-19 incidence
Fig. 2 shows the linear correlation between either the log10 converted values of absolute concentration (in GC/L, blue dots, left y-axis) or total viral load (in GC/d, orange dots, right y-axis) and the estimation of COVID-19 prevalence for all three cities in the studied period (March to September). The best fit, in terms of the highest Pearson correlation coefficient and lowest RSME is seen from the regression of log daily viral loads (Fig. 2a & d-orange) against the summation of cases in the 7-days that followed WW sampling (Eq. (3)). That is the viral loads in WW are a better indicator of the cases that will be diagnosed within 1 week after sampling thus affording a level of anticipation of future values. The worst fit was observed with estimates of viral incidence that summed the number of new cases in the 7-days that preceded the sampling (Fig. 2c & f). Comparing the fit parameters, for the same estimation of cases, between waves, shows that the second wave data were better than that of the first wave. This is probably a result of health authorities being able to diagnose a larger proportion of the real number of cases during the second wave as opposed to the first in March–June.
3.4. Inferences from WWTPs of different sizes
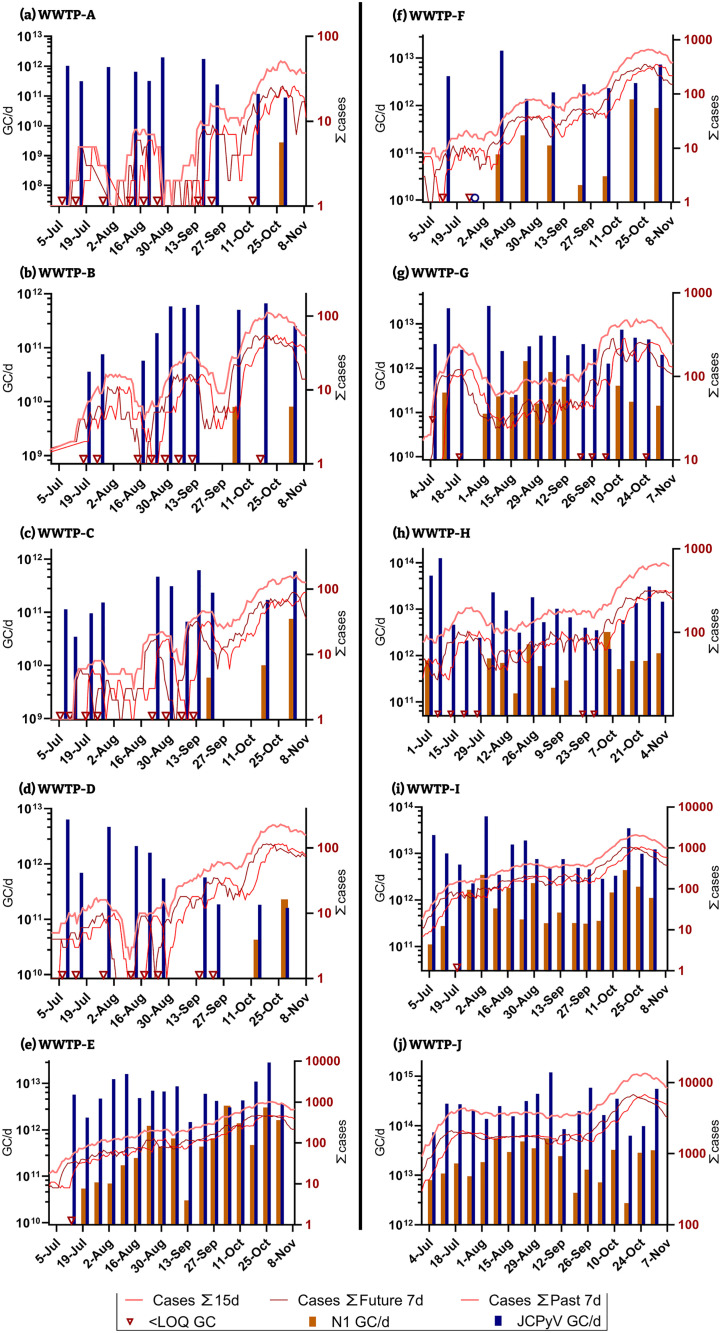
From the beginning of the second wave (second peak of COVID-19 infections), a total of 10 WWTPs, the three already shown, and an additional seven were added to the monitoring campaign. This new dataset included a wider range of WWTPs sizes (the largest of which are WWTP-J and WWTP-I (Table 1) and different geographical areas in Catalonia. The large and medium sized WWTPs showed a similar increasing trend in the concentrations of SARS-CoV-2 (N1 assay) from July to November coinciding with the progressive rise of reported cases during this period i.e. the so called second wave) (Fig. 3 ). Significant levels of human faecal contamination, by means of JCPyV quantification, were persistently detected over the sampling period in all tested facilities, indicating that the non-detected or low SARS-CoV-2 values were not due to the presence of PCR inhibitory materials in the WW. The recovery of the bacteriophage MS2 also confirmed that there was no failure in the analytical process (mean calculated recovery values were 40.50 ± 22.88%).
Fig. 3.
Estimated number of active infections (lines, right y-axis) and WW loads of SARS-CoV-2 (N1 assay), orange bar plot left y-axis) and JCPyV (blue bar plot left y-axis) viruses for the second wave at all 10 WWTPs.
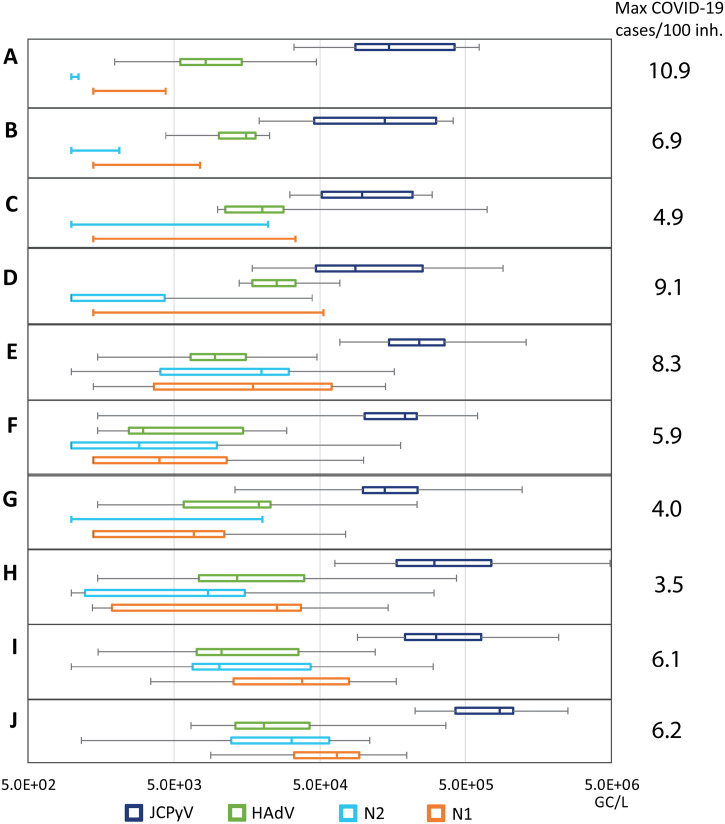
Besides the incidence level, concentrations of SARS-CoV-2 (N1 and N2) were consistently higher in larger facilities compared with smallest plants (Fig. 4 ). This trend was not observed for the human faecal indicators analysed (HAdV and JCPyV), showing no differences according the size of the population served. Like SARS-CoV-2, some infections of the HAdV virus cause gastrointestinal and respiratory diseases (Rusiñol and Girones, 2017). Respiratory adenoviruses are detected in nasopharyngeal swabs at lower mean viral loads (5.0 × 103 GC/mL) than SARS-CoV-2 (6.0 × 106 GC/mL) (Jacot et al., 2020). Although the level of excretion in faeces is relatively similar between the viruses (log10 7–10 GC/g) (reviewed by Rusiñol and Girones, 2017 and Medema et al., 2020a, Medema et al., 2020b), the detection of SARS-CoV-2 RNA in stool samples is limited to a shorter duration of shedding (from 1 to 33 days; (Zheng et al., 2020)) compared to HAdV (up to 192 day after infection (Lion et al., 2010)). Polyomaviruses, and specially JCPyV, are also persistently excreted by infected individuals with or without symptoms of a disease, sometimes for years, and in this case via the urinary tract (Bofill-Mas, 2016; Moens et al., 2013). The excreted viral particles found in wastewater will thus originate from faeces, urine and also oral fluids and their persistence in this complex water matrix will determine the chance of detection. It has been calculated that 90% of the viable SARS-CoV-2 (T90) are inactivated in 1.5 days in wastewater at room temperature (Bivins et al., 2020) whereas, using qPCR and DNAse treatment to quantify structured viral particles, the T90 of HAdV is 60.9 d and that of JCPyV is 63.9 d (Bofill-Mas et al., 2006). Previous studies on small WWTP (approximately 2100 inhabitants) have validated the use of both faecal markers, JCPyV and HAdV, as they are not affected by day-to-day variations (Mayer et al., 2016). HAdV and JCPyV have been described to occur at much more similar concentrations in WW (Bofill-Mas et al., 2006; Rusiñol and Girones, 2017) than in the current study in which JCPyV concentrations are higher. A decrease in HAdV concentrations were observed since March 2020 (SI Table 2) thus it might be that lockdown avoided community transmission of HAdV while the transmission of JCPyV, that has been described to occur within families, could have not been affected.
Fig. 4.
Concentrations of SARS-CoV-2 (N1 and N2 assays), HAdV and JCPyV in different sized WWTP for the period July and November 2020. Midline shows the median, edges of the box plot show the 2nd and 3rd quartiles and the whiskers show the minimum and maximum value measured. Maximum incidences per 1000 inh. reported over the period July–November are shown next to the figure.
During the first weeks of the second wave, no SARS-CoV-2 RNA was detected in the smallest WW facilities (WWTP-A, —B, —C and —D), it was not until mid-September, when the infection rate relative to population was higher than ever (including the first wave), that SARS-CoV-2 was detected and quantified in their WW samples. In terms of population served these four WWTP are the smallest of the WWTP studied herein and all serve less than 27,000 inhabitants (Table 1). Since the concentration of HAdV and JCPyV in these WWTPs were similar to those measured in the other WWTPs, the lack of detection of SARS-CoV-2 in WW was not caused by dilution of WW from other sources such as industry or storm water, nor by failures in the analytical process. This substantiates our working hypothesis that small WWTPs are not as informative to sample as their larger counterparts since a higher proportion of infection in needed within the sewershed in order to produce quantifiable WW results. A summary of the results from the sampling carried out during the second wave are shown in Fig. 4. The JCPyV and HAdV viruses were quantifiable in most samples independently of WWTPs size and the concentration was very similar in between WWTPs, this was in contrast with the N1 and N2 assays. Correlation analyses were performed between the mean concentrations and the maximum incidences reported. Bigger WW facilities presented in mean values higher incidences (Pearson's correlation coefficient p-value = 0.85) (see Fig. 2 Supplementary material). Other authors have observed positive correlation between the SARS-CoV-2 concentrations and the active cases reported even where low COVID-19 prevalence was reported (D'Aoust et al., 2021; Medema et al., 2020a, Medema et al., 2020b). Although SARS-CoV-2 RNA can be detected in WW from small facilities when only few clinical cases are reported (Gonzalez et al., 2020), sporadic detection would not allow consistent interpretation.
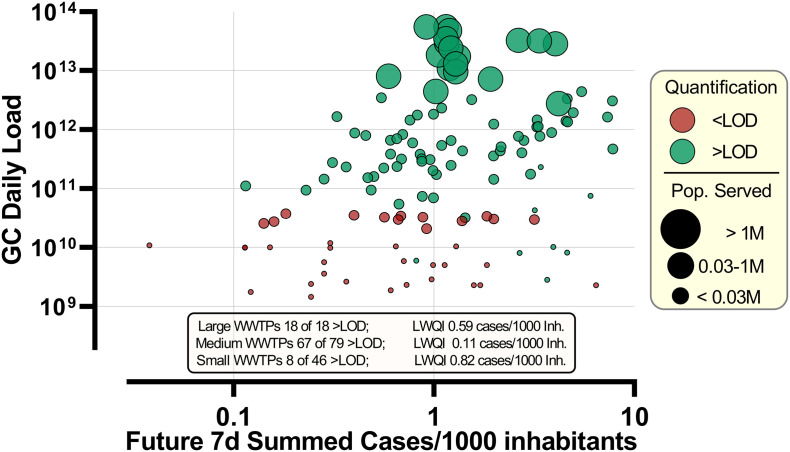
The difference in prevalence of infection and WWTP size is also visualised in Fig. 5 . While all but one sampling dates recorded a prevalence rate higher than 0.1 cases per 1000 inhabitants, small WWTPs were more likely to have WW levels lower than the LOD. The samples from the large WWTP-J were quantifiable (i.e. above LOD) 100% of the time, the lowest WW quantifiable incidence (LWQI) for this plant was recorded at 0.59 cases per 1000 inhabitants. A lower incidence was never recorded thus it is highly possible that WW level would remain positive even at lower incidences. A stark difference in the LWQI is seen between the small (LWQI = 0.82 cases/1000 inh.) and medium (LQWI = 0.11 cases/1000 inh.) sized WWTPs.
Fig. 5.
Bubble Plot of the second wave showing the summed future 7d cases per 1000 inhabitants against the total daily N1 GC loads measured. Tick mark diameters are organised in three groups by size depending on the number of people being served by the WWTP. Red dots indicate measured concentrations below LOD and indicate only the maximum daily load possible. LWQI = lowest WW quantifiable incidence i.e. the lowest incidence of infection which resulted in a quantifiable concentration of N1 GC in WW.
In other words, while the analytical LOD in terms of GC/L are the same amongst samples from different WWTPs, the LOD in terms of prevalence of infection (i.e. incidence of shedding relative to total number of inhabitants) is higher in small cities. It is well documented that obtaining a representative WW sample from a small population is more challenging than for large populations (Aymerich et al., 2017; Medema et al., 2020a, Medema et al., 2020b; Ort et al., 2010). Putting incidence between WWTPs into perspective, and incidence rate of 0.7 cases/1000 inh. is only between 5 and 19 cases for the four smallest WWTPs (WWTP A-D) but over 1000 in the largest WWTP. The variability with such small numbers is high; for example, a portion of the infected individuals might never shed the virus in faeces throughout the course of their infection, some might be residing in other municipalities while being registered in a different one. Another factor is the high variability in viral shedding both in terms of loads and duration which would be even more extreme with such low numbers of active cases since they would not average out. One has also to bear in mind that toilet use and flushing occurs in discrete events of a short duration. A composite WW sample that is collected through pulses, tens of minutes apart, may very well miss a relevant discharge from a shedding individual. The sewer residence time, sewage fluid dynamics and composition are also expected to be influential on the sampling variability and hence utility of using WBE to monitor small areas with a low number of infected individuals in absolute terms.
Sampling the septic tanks of cruise ships and aeroplanes for SARS-CoV-2 RNA has already been carried out and positive samples were identified (Ahmed et al., 2020b). While cruise ships and aeroplanes might seem superficially similar to very small cities or neighbourhoods, the conditions for WW are very different. In aeroplanes, for instance, WW is often not diluted during flushing since the toilets are vacuum driven and greywater entry, and hence dilution, is limited to hand washing. Septic tanks on an aeroplane also allows for a better homogenisation of WW resulting in sampling strategy being less critical and grab samples being sufficiently representative. When it comes to the large WWTP-J, 0.7 cases per 1000 inhabitants corresponds to 1048 cases in absolute terms. In this case, even with the same sampling uncertainties, the probability of sampling during a relevant discharge event is much higher and the distribution of discharge is more homogeneous over time. While it is more challenging to sample small WWTPs with low incidence of infection, it is not excluded that if the practical limitations of representative sampling and homogenisation are overcome, small WWTPs could also be as useful to sample as their larger counterparts.
The three largest incidence levels which had WW values at or below LOD were reported at, 6.4 cases per 1000 inhabitants (WWTP-B, 52 cases), 3.2 cases/1000 inh. (WWTP-G, 211 cases) and 2.0 cases/1000 inh. (WWTP-F, 116 cases). Hart and Halden (2020) modelled a range of minimum infection rates needed in order to surpass the LOD. Depending on a range on numerous parameters they approximate that the upper bounds are 0.00005% and the lower bounds at 0.88%. While our results fall within this range, they are at the lower end i.e. higher prevalence of infection is need in order to be able to measure WW concentrations above LOD. The concentration and loads for small plants are rather variable, as an example, on the 29th of October 2020 an incidence of 0.324% (23 cases) was recorded in the area served by WWTP-A. This gave a concentration of 4.39 × 103 GC/L (daily flow = 1163 m3) while a virtually identical incidence of 0.329% (87 cases) in WWTP-D gave a concentration of 5.31 × 104 GC/L (daily flow =4421 m3), note that WWTP-D serves 3.7× more people than WWTP-A.
Interestingly, positive samples have been detected when reported COVID-19 cases reached 0.05–0.10 cases per 1000 inhabitants (0.005–0.010%) (Hata et al., 2020). The same authors calculated that SARS-CoV-2 was detectable if one in 100,000 persons shed 109 GC/g of faeces, which has been later reported to occur only in a low percentage of the population (≈10%) (Endo et al., 2020). The presence of a patient with such a high load may be possible, because shedding rates are highly variable amongst individuals (reviewed by (Medema et al., 2020a, Medema et al., 2020b) but the presence of this “superexcretor” in the smallest WWTP will be less probable. One should also keep in mind that such comparisons between different WWTPs and, especially, between different countries are error-prone since they might have different ratios of identified cases to total real cases, the latter of which can only be approximated by epidemiological models.
Overall, the higher incidence prevalence required within a population served by small WWTPS does not mean that these plants should not be monitored but the limitations in terms of sensitivity should be kept in mind, and great care should be taken when comparing WWTPs of difference size. Alternative sampling strategies could also be employed such as primary sludge sampling. Promising results have been reported (Balboa et al., 2020; Peccia et al., 2020). Limitations with this approach include that sampling proportionally at a city or neighbourhood level (as opposed to sampling from WWTP that are designed to precipitate the sludge) is not practical and issues with homogenisation of the sample are expected to increase uncertainty.
4. Conclusions
Applying WBE to track the spread of COVID-19 within the sewershed areas served by small WWTPs (<24,000 inhabitants) is more challenging. Higher prevalence of infection amongst the population is required in these areas in order to have WW level of SARS-CoV-2 (N1 and N2 assays) above the LOD.
-
▪
Wastewater SARS-CoV-2 GC (N1 assay) in terms of total loads is a much better predictor of prevalence of infection than SARS-CoV-2 GC concentration.
-
▪
In all 10 of the monitored WWTPs, WW loads are congruent with estimations of active cases within the sewershed.
-
▪
Concentrations of SARS-CoV-2 (N1 assay) in WW showed good correlations with the number of cases that would be diagnosed from 0 to 7 days after sampling.
-
▪
When lack of SARS-CoV-2 RNA detection, HAdV and JCPyV surveillance can help understanding dilution effects or failures in the analytical process.
-
▪
The lowest incidence of infection which resulted in a quantifiable concentration of SARS-CoV-2 in WW for medium sized WWTPs was 0.11 cases/1000 inh. while for small WWTPs this was 0.82 cases/1000 inh.
CRediT authorship contribution statement
Rusiñol M.- sampling, sample analysis, data gathering, data analysis and writing.
Zammit I.- sampling, data gathering, data analysis and writing.
Itarte M: sample analysis and reviewing.
Eva Forés: sample analysis, process controls production.
Martínez-Puchol S: sample analysis and reviewing.
Girones R: conceptualization and reviewing.
Borrego C.M.: conceptualization, sampling, formal analysis, reviewing.
Corominas LL: conceptualization, sampling, formal analysis, reviewing.
Bofill-Mas S: conceptualization, sampling, sample analysis, formal analysis, reviewing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was partially funded by the projects PCI2019-103643, RTI2018-097346-B-I00, and AGL2017-86797-C2-1-R funded by MCIN/AEI/10.13039/501100011033/ FEDER “A way of making Europe”, by the European Union. S. Bofill-Mas is a Serra-Hunter fellow at the University of Barcelona. This study is performed with partial support from the Water Research Institute of the University of Barcelona. IDAEA-CSIC is a Centre of Excellence Severo Ochoa (Spanish Ministry of Science and Innovation, ProjectCEX2018-000794-S). ICRA authors acknowledge the support from the Economy and Knowledge Department of the Catalan Government through Consolidated Research Group (ICRA-ENV 2017 SGR 1124). ICRA is part of the CERCA program. We would like to give a big thanks to our families and colleagues, for their support and help behind the scenes.
Editor: Damia Barcelo
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2021.147463.
Appendix A. Supplementary data
Supplementary material
References
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O'Brien J.W.…Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Angel N., Bibby K., Bivins A., Dierens L.…Mueller J.F. Detection of SARS-CoV-2 RNA in commercial passenger aircraft and cruise ship wastewater: a surveillance tool for assessing the presence of COVID-19 infected travellers. J. Travel Med. 2020;27(5) doi: 10.1093/jtm/taaa116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aymerich I., Acuña V., Ort C., Rodríguez-Roda I., Corominas L. Fate of organic microcontaminants in wastewater treatment and river systems: an uncertainty assessment in view of sampling strategy, and compound consumption rate and degradability. Water Res. 2017;125:152–161. doi: 10.1016/j.watres.2017.08.011. [DOI] [PubMed] [Google Scholar]
- Balboa S., Mauricio-Iglesias M., Rodríguez S., Martínez-Lamas L., Vasallo F.J., Regueiro B., Lema J.M. The fate of SARS-CoV-2 in wastewater treatment plants points out the sludge line as a suitable spot for incidence monitoring. medRxiv. 2020 doi: 10.1101/2020.05.25.20112706. 2020.2005.2025.20112706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivins A., Greaves J., Fischer R., Yinda K.C., Ahmed W., Kitajima M.…Bibby K. Environmental Science & Technology Letters; 2020. Persistence of SARS-CoV-2 in Water and Wastewater. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bofill-Mas S., editor. Polyomavirus - Global Water Pathogen Project Part 3 Viruses. Michigan State University; E. Lansing, MI, UNESCO: 2016. [Google Scholar]
- Bofill-Mas S., Pina S., Girones R. Documenting the epidemiologic patterns of polyomaviruses in human populations by studying their presence in urban sewage. Appl. Environ. Microbiol. 2000;66(1):238–245. doi: 10.1128/aem.66.1.238-245.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bofill-Mas S., Albinana-Gimenez N., Clemente-Casares P., Hundesa A., Rodriguez-Manzano J., Allard A.…Girones R. Quantification and stability of human adenoviruses and Polyomavirus JCPyV in wastewater matrices. Appl. Environ. Microbiol. 2006;72(12):7894–7896. doi: 10.1128/aem.00965-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung K.S., Hung I.F.N., Chan P.P.Y., Lung K.C., Tso E., Liu R.…Leung W.K. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from a Hong Kong cohort: systematic review and meta-analysis. Gastroenterology. 2020;159(1):81–95. doi: 10.1053/j.gastro.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Aoust P.M., Mercier E., Montpetit D., Jia J.J., Alexandrov I., Neault N., Baig A.T., Mayne J., Zhang X., Alain T., Langlois M.A., Servos M.R., MacKenzie M., Figeys D., MacKenzie A.E., Graber T.E., Delatolla R. Quantitative analysis of SARS-CoV-2 RNA from wastewater solids in communities with low COVID-19 incidence and prevalence. Water Res. 2021;188 doi: 10.1016/j.watres.2020.116560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughton C.G. Wastewater surveillance for population-wide Covid-19: the present and future. Sci. Total Environ. 2020;736 doi: 10.1016/j.scitotenv.2020.139631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckerle I., Meyer B. SARS-CoV-2 seroprevalence in COVID-19 hotspots. Lancet. 2020;396(10250):514–515. doi: 10.1016/S0140-6736(20)31482-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo A., Abbott S., Kucharski A., Funk S. Estimating the overdispersion in COVID-19 transmission using outbreak sizes outside China [version 3; peer review: 2 approved] Wellcome Open Res. 2020;5(67) doi: 10.12688/wellcomeopenres.15842.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forés E., Bofill-Mas S., Itarte M., Martínez-Puchol S., Hundesa A., Calvo M.…Rusiñol M. 2021. Evaluation of Two Rapid Ultrafiltration-based Methods for SARS-CoV-2 Concentration From Wastewater. (under review) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez R., Curtis K., Bivins A., Bibby K., Weir M.H., Yetka K., Thompson H., Keeling D., Mitchell J., Gonzalez D. COVID-19 surveillance in Southeastern Virginia using wastewater-based epidemiology. Water Res. 2020;186 doi: 10.1016/j.watres.2020.116296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart O.E., Halden R.U. Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: feasibility, economy, opportunities and challenges. Sci. Total Environ. 2020;730 doi: 10.1016/j.scitotenv.2020.138875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata A., Honda R. Potential sensitivity of wastewater monitoring for SARS-CoV-2: comparison with norovirus cases. Environ. Sci. Technol. 2020;54(11):6451–6452. doi: 10.1021/acs.est.0c02271. [DOI] [PubMed] [Google Scholar]
- Hata A., Honda R., Hara-Yamamura H., Meuchi Y. Detection of SARS-CoV-2 in wastewater in Japan by multiple molecular assays-implication for wastewater-based epidemiology (WBE) medRxiv. 2020;2020.2006.2009.20126417 doi: 10.1101/2020.06.09.20126417. [DOI] [Google Scholar]
- Hernroth B.E., Conden-Hansson A.C., Rehnstam-Holm A.S., Girones R., Allard A.K. Environmental factors influencing human viral pathogens and their potential indicator organisms in the blue mussel, Mytilus edulis: the first Scandinavian report. Appl. Environ. Microbiol. 2002;68:4523–4533. doi: 10.1128/AEM.68.9.4523-4533.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacot D., Greub G., Jaton K., Opota O. Viral load of SARS-CoV-2 across patients and compared to other respiratory viruses. Microbes Infect. 2020 doi: 10.1016/j.micinf.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesté-Lasserre C. Coronavirus found in Paris sewage points to early warning system. Science. 2020;80 doi: 10.1126/science.abc3799. [DOI] [Google Scholar]
- Lion T., Kosulin K., Landlinger C., Rauch M., Preuner S., Jugovic D.…Matthes-Martin S. Monitoring of adenovirus load in stool by real-time PCR permits early detection of impending invasive infection in patients after allogeneic stem cell transplantation. Leukemia. 2010;24(4):706–714. doi: 10.1038/leu.2010.4. [DOI] [PubMed] [Google Scholar]
- Lodder W., de Roda Husman A.M. SARS-CoV-2 in wastewater: potential health risk, but also data source. Lancet Gastroenterol. Hepatol. 2020;5(6):533–534. doi: 10.1016/S2468-1253(20)30087-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallapaty S. How sewage could reveal true scale of coronavirus outbreak. Nature. 2020;580:176–177. doi: 10.1038/d41586-020-00973-x. [DOI] [PubMed] [Google Scholar]
- Mayer R.E., Bofill-Mas S., Egle L., Reischer G.H., Schade M., Fernandez-Cassi X.…Farnleitner A.H. Occurrence of human-associated Bacteroidetes genetic source tracking markers in raw and treated wastewater of municipal and domestic origin and comparison to standard and alternative indicators of faecal pollution. Water Res. 2016;90:265–276. doi: 10.1016/j.watres.2015.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020;7(7):511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Medema G., Been F., Heijnen L., Petterson S. Implementation of environmental surveillance for SARS-CoV-2 virus to support public health decisions: opportunities and challenges. Curr. Opin. Environ. Sci. Health. 2020;17:49–71. doi: 10.1016/j.coesh.2020.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens U., Van Ghelue M., Song X., Ehlers B. Serological cross-reactivity between human polyomaviruses. Rev. Med. Virol. 2013;23(4):250–264. doi: 10.1002/rmv.1747. [DOI] [PubMed] [Google Scholar]
- Ort C., Lawrence M.G., Reungoat J., Mueller J.F. Sampling for PPCPs in wastewater systems: comparison of different sampling modes and optimization strategies. Environ. Sci. Technol. 2010;44(16):6289–6296. doi: 10.1021/es100778d. [DOI] [PubMed] [Google Scholar]
- Pal A., Sirota L., Maudru T., Peden K., Lewis A.M., Jr. Real-time, quantitative PCR assays for the detection of virus-specific DNA in samples with mixed populations of polyomaviruses. J. Virol. Methods. 2006;135:32–42. doi: 10.1016/j.jviromet.2006.01.018. [DOI] [PubMed] [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A.…Omer S.B. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020;38(10):1164–1167. doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecson B.M., Martin L.V., Kohn T. Quantitative PCR for determining the infectivity of bacteriophage MS2 upon inactivation by heat, UV-B radiation, and singlet oxygen: advantages and limitations of an enzymatic treatment to reduce false-positive results. Appl. Environ. Microbiol. 2009;75(17):5544–5554. doi: 10.1128/AEM.00425-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollán M., Pérez-Gómez B., Pastor-Barriuso R., Oteo J., Hernán M.A., Pérez-Olmeda M.…Vázquez de la Villa A. Prevalence of SARS-CoV-2 in Spain (ENE-COVID): a nationwide, population-based seroepidemiological study. Lancet. 2020;396(10250):535–544. doi: 10.1016/S0140-6736(20)31483-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusiñol M., Girones R. In: Global Water Pathogens Project. Rose J.B., Jiménez-Cisneros B., editors. UNESCO, Michigan Michigan State University; E. Lansing, MI: 2017. Summary of excreted and waterborne viruses. [Google Scholar]
- Rusiñol M., Fernandez-Cassi X., Hundesa A., Vieira C., Kern A., Eriksson I.…Girones R. Application of human and animal viral microbial source tracking tools in fresh and marine waters from five different geographical areas. Water Res. 2014;59:119–129. doi: 10.1016/j.watres.2014.04.013. [DOI] [PubMed] [Google Scholar]
- Russell T.W., Hellewell J., Abbott S., Jarvis C., van Zandvoort K., Group C.N.W.…Kucharski A. Centre for Mathematical Modeling of Infectious Diseases Repository; 2020. Using a Delay-adjusted Case Fatality Ratio to Estimate Under-reporting. [Google Scholar]
- Trottier J., Darques R., Ait Mouheb N., Partiot E., Bakhache W., Deffieu M.S., Gaudin R. Post-lockdown detection of SARS-CoV-2 RNA in the wastewater of Montpellier, France. One Health. 2020;10 doi: 10.1016/j.onehlt.2020.100157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- US CDC . 2020. 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel - Instructions for Use. Atlanta, GA, USA. [Google Scholar]
- Westhaus S., Weber F.-A., Schiwy S., Linnemann V., Brinkmann M., Widera M.…Ciesek S. Detection of SARS-CoV-2 in raw and treated wastewater in Germany – suitability for COVID-19 surveillance and potential transmission risks. Sci. Total Environ. 2021;751 doi: 10.1016/j.scitotenv.2020.141750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO WHO coronavirus (COVID-19) dashboard. 2020. https://covid19.who.int
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A.…Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wu F., Zhang J., Xiao A., Gu X., Lee W.L., Armas F.…Alm E.J. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. mSystems. 2020;5(4):e00614–e00620. doi: 10.1128/mSystems.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S., Fan J., Yu F., Feng B., Lou B., Zou Q.…Liang T. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January–March 2020: retrospective cohort study. BMJ. 2020;369:m1443. doi: 10.1136/bmj.m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material