Abstract
Background
Extensins are plant cell wall hydroxyproline-rich glycoproteins known to be involved in cell wall reinforcement in higher plants, and in defence against pathogen attacks. The ability of extensins to form intra- and intermolecular cross-links is directly related to their role in cell wall reinforcement. Formation of such cross-links requires appropriate glycosylation and structural conformation of the glycoprotein.
Scope
Although the role of cell wall components in plant defence has drawn increasing interest over recent years, relatively little focus has been dedicated to extensins. Nevertheless, new insights were recently provided regarding the structure and the role of extensins and their glycosylation in plant–microbe interactions, stimulating an interesting debate from fellow cell wall community experts. We have previously revealed a distinct distribution of extensin epitopes in Arabidopsis thaliana wild-type roots and in mutants impaired in extensin arabinosylation, in response to elicitation with flagellin 22. That study was recently debated in a Commentary by Tan and Mort (Tan L, Mort A. 2020. Extensins at the front line of plant defence. A commentary on: ‘Extensin arabinosylation is involved in root response to elicitors and limits oomycete colonization’. Annals of Botany 125: vii–viii) and several points regarding our results were discussed. As a response, we herein clarify the points raised by Tan and Mort, and update the possible epitope structure recognized by the anti-extensin monoclonal antibodies. We also provide additional data showing differential distribution of LM1 extensin epitopes in roots between a mutant defective in PEROXIDASES 33 and 34 and the wild type, similarly to previous observations from the rra2 mutant defective in extensin arabinosylation. We propose these two peroxidases as potential candidates to specifically catalyse the cross-linking of extensins within the cell wall.
Conclusions
Extensins play a major role within the cell wall to ensure root protection. The cross-linking of extensins, which requires correct glycosylation and specific peroxidases, is most likely to result in modulation of cell wall architecture that allows enhanced protection of root cells against invading pathogens. Study of the relationship between extensin glycosylation and their cross-linking is a very promising approach to further understand how the cell wall influences root immunity.
Keywords: Arabinosylation, cell wall, cross-linking, defence, extensin, immunocytochemistry, monoclonal antibodies, peroxidase
INTRODUCTION
Extensins are cell wall hydroxyproline-rich glycoproteins reported to be involved in many physiological processes in plants, including defence against pathogens. Extensins have been described to fortify the plant cell wall through the cross-linking of tyrosine residues, thus providing enhanced mechanical protection against pathogen invasion. However, such a cross-linking requires correct glycosylation of the protein backbone (Chen et al., 2015). This glycosylation consists of transfer of a sugar galactose onto a serine residue, and several arabinose residues onto hydroxyprolines (see glycan structure in figure S1 in Castilleux et al., 2020).
In a recent study, we investigated the impact of altered glycosylation of extensins on the plant defence response (Castilleux et al. 2020). Using an approach involving evaluation of mutants of Arabidopsis thaliana impaired in extensin glycosylation, microscopy and the employment of anti-extensin monoclonal antibodies LM1, JIM11, JIM12 and JIM20, we revealed that in response to elicitation with the bacterial peptide flagellin 22, the distribution of extensin epitopes in root cell walls differed between the mutants and the wild-type (WT). We also showed that in the rra2 and xeg113 mutants, both impaired in extensin arabinosylation, root colonization by the soil-borne pathogen Phytophthora parasitica was enhanced in comparison to the WT. These results highlighted, for the first time, a link between extensin glycosylation and plant defence, kindling interest and a very constructive Commentary from Tan and Mort (2020). Several points and questions were raised and will be addressed and discussed herein.
TRANSCRIPT ANALYSIS OF EXTENSIN ARABINOSYLATION GENES
The significance of our approach based on the p4h2, rra1, rra2 and xeg113 mutants, which are impaired in extensin glycosylation, was commented upon by Tan and Mort (2020), arguing that the transcript levels of the P4H and RRA isoforms were not shown. Although the transcript level does not always reflect the protein level, such an analysis would indeed have been relevant to address the compensation phenomenon that could occur, especially in the rra2 mutant which displayed the most interesting results. The RRA genes comprise three isoforms with RRA1 mainly expressed in the siliques, and RRA2 and RRA3 mostly expressed in roots (Egelund et al., 2007; Velasquez et al., 2015). Similarly, P4H2 is not the only P4H to be expressed in the root because this is also the case for P4H1, P4H5, P4H9, P4H10, P4H12 and P4H13 (Velasquez et al., 2011). Higher expression of RRA3 and P4H transcripts cannot therefore be excluded in the rra2 and p4h2 mutants respectively, potentially leading to a partial WT phenotype recovery and to reduction of the impact on the root response to elicitation. To our knowledge, the expression of genes involved in extensin glycosylation has not been analysed in response to pathogens or to pathogen-derived elicitors. Data regarding the expression of extensin genes in such a response remain poorly documented, although the EXTENSIN1 gene was previously shown to be overexpressed in A. thaliana seedlings upon infection with the bacterium Xanthomonas campestris (Merkouropoulos and Shirsat, 2003). Therefore, analysis of extensin gene transcripts, especially those related to extensin glycosylation, would help to better understand whether and how the gene expression is altered in response to pathogens or elicitors, thus providing further insights into interpretation of the results obtained with the anti-extensin immunolabelling and inoculation with P. parasitica.
IMMUNOLABELLING AND EXTENSIN EPITOPE RECOGNITION
Although extensin levels in situ were estimated through immunolabelling with anti-extensin antibodies, the format employed previously for presenting immunolabelling data in immuno-microscopy images, through detailed graphics and statistical analyses (Castilleux et al. 2020), was potentially over-complicated and weakened the evidence presented. We propose here in Table 1 an alternative summary of our data, emphasizing the major differences observed between the mutants and the WT for each antibody.
Table 1.
Overview of the immunolabelling results using the LM1, JIM11, JIM12 and JIM20 anti-extensin monoclonal antibodies on Flg22-elicited and non-elicited mutants and WT roots.
This summary is based on the results presented in Castilleux et al. (2020). The root fluorescence area was measured on the micrographs, after setting a certain intensity threshold defined according to the non-elicited wild-type condition and specific for each antibody. The fluorescence area was then related to the total root area observed in the micrographs to calculate the corresponding ratio (%) displayed here. Detailed graphs and statistical analyses are presented in figures S6–S9 of our previous paper (Castilleux et al., 2020).
The conclusions regarding the response to Flg22-elicitation are based on these statistical analyses. Each difference stated here is significant according to a Tukey’s multiple comparisons test at α = 5%, and with a P-value < 0.05. The terms ‘Similar signal’ and ‘No change of signal’ both mean that no significant difference was observed between non-elicited and elicited conditions. However, the term ‘No change in signal’ was specifically used for the mutants when the absence of differences between non-elicited and elicited conditions actually constituted a differential response to elicitation in comparison to WT.
Flg22: flagellin 22
Three main conclusions can be drawn: (1) a specific and unique distribution of epitopes associated with extensins was observed for each mutant impaired in extensin glycosylation; (2) the response to elicitation varied between mutants, and between the mutants and the WT; and (iii) the response to elicitation was epitope-specific.
Lack of knowledge related to the precise structure of the epitopes recognized by the anti-extensin antibodies weakens the significance of the immunolabelling approach, as pointed out by Tan and Mort (2020). In this regard, a very promising study was published by Ruprecht et al. (2017) in which a glycan array approach was employed to characterize the epitope of various anti-arabinogalactan proteins antibodies. This technique is a promising approach that can be applied to determine the precise structure of the epitopes recognized by the anti-extensin antibodies.
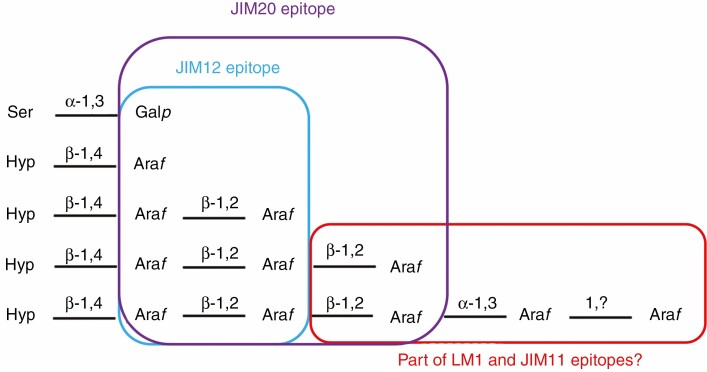
Interestingly, our immunolabelling data may have already provided some insight into the nature of these epitopes, as the presence/absence of binding could be related to the state of extensin glycosylation in the different mutants. Based on these results, we hypothesized that the JIM12 epitope would contain only the galactose on the serine residue and the two first arabinose residues on the hydroxyprolines, while part of the LM1, JIM11 and JIM20 epitopes would comprise at least one of the arabinoses missing in the xeg113 mutant. With a similar reasoning, it was also recently shown that the JIM20 epitope is likely to comprise at least one of the three first arabinoses in the extensin glycan but not the four arabinose added by ExAD (Beuder et al. 2020). Taking these results together, we propose an update of the probable epitope structure recognized by these antibodies (Fig. 1). Hence, a study in which the anti-extensin antibodies would be used on the different mutants impaired in extensin glycosylation (sgt1, hpat1-2-3, rra1-2-3, xeg113 and exad mutants) could be of interest to fully decipher the epitope structure recognized, thus allowing a more precise analysis of all the results obtained so far with anti-extensin antibodies (results summarized in Castilleux et al., 2018).
Fig. 1.
Possible epitope structure recognized by the anti-extensin monoclonal antibodies (mAbs) LM1, JIM11, JIM12 and JIM20. Proposal for the epitope structure recognized by the LM1, JIM11, JIM12 and JIM20 anti-extensin mAbs, based on immunocytochemical observations and immunoblots performed on Arabidopsis thaliana mutants impaired in extensin arabinosylation and wild-type roots (Beuder et al., 2020; Castilleux et al., 2020). The JIM12 epitope would comprise part or the entire structure containing the two first arabinoses and the galactose from the extensin glycan moiety. The JIM20 epitope would contain part or the entire structure containing the three first arabinoses and the galactose from the extensin glycan moiety. The LM1 and JIM11 epitopes may include the third arabinose and/or following arabinose residues on the extensin glycan moiety. Ser: serine. Hyp: hydroxyproline. Galp: galactopyranose. Araf: arabinofuranose.This figure is an update of figure 7 published in Castilleux et al. (2020).
CROSS-LINKING OF EXTENSINS AND PEROXIDASES
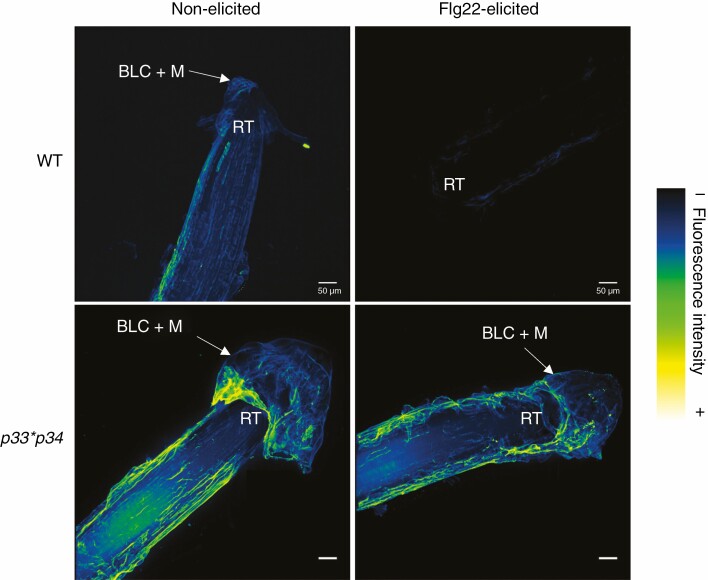
In addition to a different extensin epitope distribution in response to elicitation, we also showed that zoospores of the soilborne pathogen P. parasitica accumulated more abundantly over the root tissues of the xeg113 and rra2 mutants as compared to the WT (Castilleux et al. 2020). We hypothesized that the defect of extensin glycosylation in the mutants leads to a reduced rate of cross-linking of extensins, an altered plant cell wall architecture and, ultimately, a weakened root defence (Castilleux et al., 2020). However, although this speculation appeared to be shared by Tan and Mort (2020), they highlighted that evidence is missing regarding mechanisms by which extensin glycosylation is involved in plant–microbe interactions. We agree that it would have been a major support to our hypothesis to show whether the degree of extensin cross-linkings was actually altered in the mutants. However, such an analysis in vivo is not straightforward for several reasons. So far, the link between extensin glycosylation and the cross-linking has been predicted in silico (Velasquez et al., 2015) and shown in vitro (Chen et al., 2015), but not yet in vivo. One approach may be the application of immunoblot with anti-extensin antibodies to reveal a potential shift in molecular weight in the proteins or polypeptides that bind to the antibodies, as was recently conducted in Arabidopsis anthers (Jacobowitz et al., 2019). In this way, the alteration of extensin cross-linking could be examined in the xeg113 and rra2 mutants. Alternatively, focus can be given to peroxidases that potentially catalyse the cross-linking of extensins. Jacobowitz et al. (2019) recently identified the PEROXIDASES 9 and 40 as being able to catalyse the cross-linking of extensins in A. thaliana. As a preliminary approach, here we have investigated two other peroxidases in Arabidopsis, namely PEROXIDASES 33 and 34, that appeared to be promising candidates in catalysing this cross-linking (Passardi et al., 2006; Daudi et al., 2012). We performed immunolabelling using the anti-extensin antibody LM1 on roots of the double mutant p33*p34. In this double mutant, extensin epitopes were detected along the root, with a stronger signal when compared to the WT (Fig. 2). Interestingly, when roots were elicited with flagellin 22, nearly no signal was observed in the WT, whereas in the p33*p34 mutant the LM1 antibody was still binding similarly to the non-elicited condition. This suggests that the mutation of the genes encoding PEROXIDASES 33 and 34 impacts detection of the extensin LM1 epitope (and possibly the formation of extensin cross-links) in response to elicitation with flagellin 22. This result is particularly intriguing because the same response had been observed in the rra2 mutant (Castilleux et al., 2020) and supports the hypothesis that arabinosylation plays its central role in plant–microbe interactions by allowing the necessary cross-linking of extensins to occur within the plant cell wall.
Fig. 2.
Distribution of the LM1 extensin epitope in Arabidopsis thaliana p33*p34 double mutant and WT root tips. Immunolabelling was performed on 9-d-old roots using the anti-extensin monoclonal antibody LM1. Roots were either elicited with 1 μm Flg22 or not elicited. Images are 3-D reconstructions of 1-μm section stacks and were obtained with a Leica SP2 inverted confocal laser scanning microscope (λ excitation, 488 nm; λ emission, 507–550 nm). For each condition, five plant root tips were observed. Immunolabelling was performed as described in Castilleux et al. (2020). Scale bars = 50 μm. RT, root tip; BLC + M, border-like cells and mucilage. Flg22: flagellin 22.
CONCLUSION
Extensins appear to play an important role in root defence, with arabinosylation essential for their correct function, probably through a controlled cross-linking catalysed by specific cell wall peroxidases. Identifying the exact mechanism by which plant cell wall extensins contribute to root protection in response to invading pathogens remains a major current challenge. Nevertheless, these recent studies provide a novel and promising angle of investigation for exploring cell wall involvement in root–microbe interactions.
ACKNOWLEDGEMENTS
We are grateful to Professor Christophe Dunand and Dr Philippe Ranocha (University of Toulouse, France) for providing the double mutant p33*34 and for their very helpful expertise on peroxidases. Microscopy images were obtained on the Cell Imaging Platform of Normandy (PRIMACEN, http://www.primacen.fr) at the Faculty of Sciences, University of Rouen Normandie, Mont Saint Aignan, France. The peptide Flg 22 was synthesized and provided by Jérôme Leprince and Benjamin Lefranc (PRIMACEN).
FUNDING
This work was supported by La Région Normandie, la Fédération de Recherche ‘Normandie Vegetal’ (FED 4277) and le Centre Mondial de l’Innovation Roullier.
LITERATURE CITED
- Beuder S, Dorchak A, Bhide A, Rune Moeller S, Petersen BL, MacAlister CA. 2020. Exocyst mutants suppress pollen tube growth and cell wall structural defects of hydroxyproline O-arabinosyltransferase mutants. The Plant Journal 103: 1399–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castilleux R, Plancot B, Ropitaux M, et al. 2018. Cell wall extensins in root–microbe interactions and root secretions. Journal of Experimental Botany 69: 4235–4247. [DOI] [PubMed] [Google Scholar]
- Castilleux R, Plancot B, Gügi B, et al. 2020. Extensin arabinosylation is involved in root response to elicitors and limits oomycete colonization. Annals of Botany 125: 751–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Dong W, Tan L, Held MA, Kieliszewski MJ. 2015. Arabinosylation plays a crucial role in extensin cross-linking in vitro. Biochemistry Insights 8: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daudi A, Cheng Z, O’Brien JA, et al. 2012. The apoplastic oxidative burst peroxidase in Arabidopsis is a major component of pattern-triggered immunity. The Plant Cell 24: 275–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelund J, Obel N, Ulvskov P, et al. 2007. Molecular characterization of two Arabidopsis thaliana glycosyltransferase mutants, rra1 and rra2, which have a reduced residual arabinose content in a polymer tightly associated with the cellulosic wall residue. Plant Molecular Biology 64: 439–451. [DOI] [PubMed] [Google Scholar]
- Jacobowitz JR, Doyle WC, Weng JK. 2019. PRX9 and PRX40 are extensin peroxidases essential for maintaining tapetum and microspore cell wall integrity during Arabidopsis anther development. The Plant Cell 31: 848–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkouropoulos G, Shirsat AH. 2003. The unusual Arabidopsis extensin gene atExt1 is expressed throughout plant development and is induced by a variety of biotic and abiotic stresses. Planta 217: 356–366. [DOI] [PubMed] [Google Scholar]
- Passardi F, Tognolli M, De Meyer M, Penel C, Dunand C. 2006. Two cell wall associated peroxidases from Arabidopsis influence root elongation. Planta 223: 965–974. [DOI] [PubMed] [Google Scholar]
- Ruprecht C, Bartetzko MP, Senf D, et al. 2017. A synthetic glycan microarray enables epitope mapping of plant cell wall glycan-directed antibodies. Plant Physiology 175: 1094–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Mort A. 2020. Extensins at the front line of plant defence. A commentary on: ‘Extensin arabinosylation is involved in root response to elicitors and limits oomycete colonization’. Annals of Botany 125: vii–viii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasquez SM, Marzol E, Borassi C, et al. 2015. Low sugar is not always good: impact of specific O-glycan defects on tip growth in arabidopsis. Plant Physiology 168: 808–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasquez SM, Ricardi MM, Dorosz JG, et al. 2011. O-glycosylated cell wall proteins are essential in root hair growth. Science (New York, N.Y.) 332: 1401–1403. [DOI] [PubMed] [Google Scholar]