Abstract
Background and Aims
Modern sugarcane cultivars (Saccharum spp.) are high polyploids, aneuploids (2n = ~12x = ~120) derived from interspecific hybridizations between the domesticated sweet species Saccharum officinarum and the wild species S. spontaneum.
Methods
To analyse the architecture and origin of such a complex genome, we analysed the sequences of all 12 hom(oe)ologous haplotypes (BAC clones) from two distinct genomic regions of a typical modern cultivar, as well as the corresponding sequence in Miscanthus sinense and Sorghum bicolor, and monitored their distribution among representatives of the Saccharum genus.
Key Results
The diversity observed among haplotypes suggested the existence of three founding genomes (A, B, C) in modern cultivars, which diverged between 0.8 and 1.3 Mya. Two genomes (A, B) were contributed by S. officinarum; these were also found in its wild presumed ancestor S. robustum, and one genome (C) was contributed by S. spontaneum. These results suggest that S. officinarum and S. robustum are derived from interspecific hybridization between two unknown ancestors (A and B genomes). The A genome contributed most haplotypes (nine or ten) while the B and C genomes contributed one or two haplotypes in the regions analysed of this typical modern cultivar. Interspecific hybridizations likely involved accessions or gametes with distinct ploidy levels and/or were followed by a series of backcrosses with the A genome. The three founding genomes were found in all S. barberi, S. sinense and modern cultivars analysed. None of the analysed accessions contained only the A genome or the B genome, suggesting that representatives of these founding genomes remain to be discovered.
Conclusions
This evolutionary model, which combines interspecificity and high polyploidy, can explain the variable chromosome pairing affinity observed in Saccharum. It represents a major revision of the understanding of Saccharum diversity.
Keywords: Saccharum, sugarcane, polyploidy, hybridization, founding ancestral genome, diversity
INTRODUCTION
Interspecific hybridization, sometimes accompanied by polyploidization, is an important evolutionary process in plants and is associated with the domestication and/or diversification of some major crops [e.g. banana (Simmonds 1962; Perrier et al., 2011), citrus (Wu et al., 2014), date palm (Flowers et al., 2019), rice (Santos et al., 2019) and wheat (McFadden and Sears, 1946)].
Polyploids are generally divided into two categories: autopolyploids, which formed within a single species, and allopolyploids, which resulted from hybridization between two or more species. Autopolyploids are typically characterized by random association among homologous chromosomes during meiosis, leading to polysomic segregation, whereas allopolyploids have sets of homoeologous chromosomes that do not typically pair, leading to disomic segregation (Doyle and Egan, 2010). A continuum in the parental divergence of polyploids yields many intermediate situations (Stebbins, 1950; Barker et al., 2016).
Polyploidy and recurrent interspecific hybridizations complicate the reconstruction of phylogenetic relationships between genera and species, particularly in higher-order polyploids, which may have a complex history of multiple allo- and/or autopolyploidization events (Fortune et al., 2007; Tennessen et al., 2014; Triplett et al., 2014).
Sugarcane belongs to Saccharum sensu stricto, a genus composed exclusively of higher-order polyploid (>4x) species. Despite its huge economic importance, the origin of sugarcane and the evolutionary history and taxonomy of the genus Saccharum (Poaceae; Andropogoneae) and its species are largely unresolved (Hodkinson et al., 2002; Welker et al., 2015). Several close genera (Erianthus section Ripidium/Tripidium, Miscanthus section Diandra, Narenga, Sclerostachya) that can occasionally hybridize with Saccharum have been proposed by some authors to be involved in the origin of Saccharum and are referred to as the ‘Saccharum complex’ by breeders (reviewed by Daniels and Roach, 1987, Grivet et al., 2006). However, molecular data, although limited so far, do not support an important direct contribution of these genera to Saccharum but suggest a monophyletic origin of this genus (Grivet et al., 2004).
The subdivision of the genus Saccharum is a matter of debate (Irvine, 1999; Evans and Joshi, 2016), but a subdivision into six species is generally used by sugarcane technologists (Daniels and Roach, 1987; Grivet et al., 2006). Among them, two species are wild (S. robustum and S. spontaneum); they are well differentiated but for both species the taxonomic limit and evolutionary history have been a matter of controversy (reviewed by Daniels and Roach, 1987). Saccharum spontaneum (2n = 40–128) is a highly polymorphic species with an extensive distribution from Africa to Southeast Asia. It generally has thin stalks with no or very low sugar content. Saccharum robustum (2n = 60, 80 and up to 200) is most likely native to Southeast Asia, southeast to Sulawesi, and has long thick stalks with little or no sugar. Four ‘species’ exist only in cultivation (S. officinarum, S. barberi, S. sinense and S. edule) and are considered by some authors to be horticultural groups. The most popular scenario for sugarcane domestication, among sugarcane specialists, was first established by Brandes (1956). In this scenario, sugarcane originated in New Guinea from wild S. robustum by human selection possibly as much as 8000 years ago, and resulted in a series of clones accumulating sugar in the stalks identified by botanists as S. officinarum (2n = 80). These cultivars were transported by humans to the Asian continent, where they hybridized with local forms of the wild species S. spontaneum, giving rise to a new series of cultivars better adapted to subtropical environments and to the emergence of sugar manufacturing (Daniels and Daniels, 1976). They are called S. barberi for cultivars from India (2n = 81–124) and S. sinense for cultivars from China (2n = 116–120). The interspecific origin of these two groups of formerly cultivated sugarcane was demonstrated by molecular cytogenetics (D’Hont et al., 2002). Saccharum edule (2n = 60–122) is cultivated for its edible aborted inflorescence in subsistence gardens from New Guinea to Fiji and is believed to correspond to natural mutant clones from S. robustum (Grivet et al., 2006).
The origin of modern cultivars is well documented. They are all derived from a few interspecific hybridization events performed a century ago by breeders between the formerly cultivated groups S. officinarum and S. barberi and the wild S. spontaneum followed by backcrossing with S. officinarum (Arceneaux, 1968; Daniels and Roach, 1987). They are all high polyploids and aneuploids, with around 120 chromosomes, and molecular cytogenetics studies have highlighted that 75–85 % of their chromosomes originated from S. officinarum and 15–25 % from S. spontaneum, including some chromosomes derived from interspecific recombinations (D’Hont et al., 1996; Cuadrado et al., 2004; Piperidis et al., 2010; Huang et al., 2020; Piperidis and D’Hont, 2020). Recently, a reference sequence assembly of one mosaic basic genome of sugarcane has been produced (Garsmeur et al., 2018) as well as an assembly of a tetraploid S. spontaneum (Zhang et al., 2018). However, because of its extreme genome complexity, an assembly of the polyploid genome of a cultivar has not been obtained yet.
The objectives of the present study were to gain insight into the origin and architecture of the complex genome of modern sugarcane cultivars. For this, we analysed the sequences of all 12 hom(oe)ologous haplotypes (12 BAC clones) from two distinct genomic regions of a typical modern sugarcane cultivar. This allowed us to differentiate three groups of haplotypes. To investigate the origin of the haplotypes, we exploited sequence data from accessions representative of the diversity of Saccharum. We showed that two groups of haplotypes were contributed by S. officinarum and one group by S. spontaneum. These results suggested that three founding genomes were involved in the origin of the Saccharum genus and modern sugarcane cultivars.
MATERIALS AND METHODS
BAC sequencing and annotation
Ten BAC clones from the sugarcane cultivar R570 library developed by Tomkins et al. (1999) and identified by Jannoo et al. (2007) as corresponding to hom(oe)ologous chromosome segments bearing the Adh1 gene were sequenced. Mate-pair libraries of ten BAC clones were produced and sequenced using the 454 method (FLX Titanium, Roche) and assembled with Newbler (Roche). Sequences were submitted to the EMBL database under the following accession numbers (BAC clone names in parentheses; Sh, Saccharum hybrid): HG531786 (Sh102M23), HG531788 (Sh111P05), HG531792 (Sh172H13), HG531793 (Sh182G15), HG531794 (Sh186P07), HG531797 (Sh192N12), HG531798 (Sh206M17), HG531799 (Sh209M19), HG531802 (Sh242M02) and HG531804 (Sh245F09). Two additional hom(oe)ologous BAC clones, Sh051L01 and Sh265O22 (accession numbers AM403006 and AM403007), were previously sequenced using the Sanger method (Jannoo et al., 2007).
Twelve BACs corresponding to hom(oe)ologous chromosome segments bearing the Rpa1 gene were identified and sequenced by de Setta et al. (2014). BAC sequences are available from GenBank under accession numbers KF184657 to KF184973.
For all BAC clone sequences, the structure (exon–intron) and putative function of genes were automatically predicted using the GNPAnnot Community Annotation System (Guignon et al., 2012) available on the SouthGreen bioinformatics platform (https://www.southgreen.fr/). Gene predictions were manually curated using Artemis software as described in Garsmeur et al. (2011). Genes were numbered according to Jannoo et al. (2007) and de Setta et al. (2014) for the Adh1 and Rpa1 regions, respectively. Large transposable elements (TEs) were annotated in the Adh1 region as described in Garsmeur et al. (2011) and for the Rpa1 region the annotation from de Setta et al. (2014) was updated.
Identification of Miscanthus and Sorghum orthologous regions
Miscanthus sinensis and Sorghum bicolor orthologous regions were identified through BLASTN alignments of CDS sequences of all genes identified in the sugarcane BAC clones (16 and 6 genes for Adh1 and Rpa1 regions, respectively) onto scaffolds of a preliminary genome assembly of Miscanthus sinensis and Sorghum bicolor genomes (assembly v3.0.1, available at https://phytozome-next.jgi.doe.gov/). BLASTN hits were filtered with an e-value threshold of 1e−10, and for each region one Sorghum chromosome segment and two Miscanthus paralogous regions were identified. Genes were annotated as described above for sugarcane. The available Sorghum annotation (https://phytozome-next.jgi.doe.gov/) was compared with the de novo annotations to help improvement of manual curation with Artemis software.
Global sequence comparisons
Sugarcane hom(oe)ologous BAC sequences and the Miscanthus and Sorghum orthologous regions were aligned using BLASTN. All alignments were inspected with Artemis Comparison Tools (Carver et al., 2005).
Phylogenetic analyses
Hom(eo)ologous gene sequences, including exons and introns, were aligned with MAFFT (Katoh et al., 2009). Maximum-likelihood phylogenetic trees for genes shared by most haplotypes were constructed using PhyML with the GTR evolution model and the SH-like aLRT branch support with 1000 bootstrap replicates (Guindon et al., 2010).
Additional phylogenetic trees were constructed using the genomic segments shared between all hom(oe)ologous BAC sequences (from gene 6 to gene 7.5 for the Adh1 region and from gene 1 to gene 5 for the Rpa1 region) with an alignment-free method based on k-mer analysis using the AAF software (Fan et al., 2015): hom(eo)ologous genomic segments were split into k-mers of 30 bp and homopolymeric k-mers were discarded. A pairwise distance matrix representing the number of k-mers that differed between hom(oe)ologous segments was used to reconstruct phylogenetic relationships.
Trees were visualized with Seaview (Gouy et al., 2010) or FigTree v1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/).
Divergence times
The number of substitutions per synonymous site (Ks) was calculated between all hom(eo)ologous gene pairs belonging to three groups of haplotypes (A, B and C). Protein sequences were aligned with Clustal W (Larkin et al., 2007) and PAL2NAL (Suyama et al., 2006) was used to reconstruct the multiple codon alignment based on the corresponding aligned protein sequences. The Ks values were calculated with the Nei–Gojobori method implemented in PAML (Yang, 2007). This process was performed using a script available at http://github.com/tanghaibao/bio-pipeline/tree/master/synonymous_calculation/. Divergence times were estimated using the formula T = average Ks/(2 × 6.5 × 10–9) (Gaut et al., 1996).
Sequence data from accessions representative of Saccharum species and relatives
Two types of sequence data were used: whole-genome sequence (WGS) and targeted sequence capture data (Supplementary Data Table S1). Illumina paired-ends WGS data were available for two modern cultivars (including R570), 65 Saccharum spontaneum accessions and one S. officinarum accession (Garsmeur et al., 2018; Zhang et al., 2018). They represent on average a coverage of 7x of the total genome. In addition, pre-publication access to paired-end WGS data, obtained with the Illumina NovaSeq S4 platform, for 16 Saccharum accessions (one modern cultivar, three S. barberi, two S. officinarum, one S. robustum and nine S. spontaneum) was provided by the Joint Genome Institute (JGI). They represent on average a coverage of 13x of the total genome.
Targeted sequence data were available for 304 Saccharum accessions and relatives (Yang et al., 2019). This included sequences that mapped to 5914 sites of the Adh1 region but none that mapped to the Rpa1 region. We produced a second set of targeted capture sequences for 36 accessions (6 modern cultivars, 10 S. officinarum, 15 S. spontaneum, 2 S. barberi, 1 S. edule and 2 Miscanthus accessions). Sequencing libraries were built with 1.5 µg of DNA by accession using a protocol adapted from Kircher et al. (2012) and Meyer and Kircher (2010). DNAs were sheared to obtain an average of 300 bp on a Bioruptor® Standard (Catalogue No. UCD-200, Diagenode, Woburn, MA). Equal amounts of 16 genomic libraries were pooled to obtain at least 500 ng of DNA. Sequence capture by hybridization was performed on each library pool according to the manufacturer’s protocol for the myBaits® target capture kits (v3.02) with the custom oligonucleotide library designed by Arbor Biosciences. The regions targeted corresponded to 40 000 exons from sugarcane gene models annotated on the R570 reference sequence (Garsmeur et al., 2018), including 17 476 sites from the two regions analysed in this study (Adh1 and Rpa1).
Single-nucleotide polymorphism identification
Two subsets of read data were extracted from the WGS Illumina reads. The first one corresponded to reads that have a common k-mer (of size 20) with the BUSCO gene sets (Seppey et al., 2019) present in the monoploid sugarcane reference sequence of Garsmeur et al. (2018). The second subset corresponded to reads that have a common k-mer (of size 20) with one of the gene exons of the Adh1 or Rpa1 region. In silico Illumina reads were generated from the R570 BAC sequences with the tool art_illumina (Huang et al., 2012). For the targeted capture sequences, all the reads were used. All sequences were mapped on the monoploid sugarcane reference sequence of Garsmeur et al. (2018).
Single-nucleotide polymorphisms (SNPs) were identified as described by Garsmeur et al. (2018) (https://github.com/SouthGreenPlatform/VcfHunter/). This pipeline includes the mapping of sequence data onto the reference genome and identification and quality filtration of SNPs. For each accession and at each of the sites analysed, genotypes were determined if sequencing depth was at least 30. We coded genotypes as heterozygous if the variant occurred at least twice and at ≥4 % frequency. We coded genotypes as homozygous if no polymorphism was observed or if the variant occurred a single time and to a frequency of <1 % (which we considered to be a potential sequencing error). Ambiguous cases that did not meet these criteria were coded as missing data. Note that a few SNPs detected in the R570 BACs were not detected in the R570 WGS data. They may represent sequencing errors, technical artefacts or small variations between the sequenced accessions that represent material that has been vegetatively propagated for many years.
Multivariate analysis
Factor analyses of distances table (AFTDs) were performed for the three distinct sets of data. For accessions for which WGS data were available, the SNPs detected in the BUSCO gene set were used. For the other accessions, SNPs detected in the targeted sequence data sets were used. AFTDs were performed with the DARwin program (Perrier and Jacquemoud-Collet, 2006), using dissimilarity matrices calculated on SNPs with an in-house program, vcf2dis.1.0.py. (https://github.com/SouthGreenPlatform/VcfHunter/blob/master/README.md)
SNP origin analyses
The origin, S. officinarum/S. robustum versus S. spontaneum, of the three groups of haplotypes/BACs (A, B and C) was determined using two complementary analyses. In the first analysis, SNPs specific to each of the three haplotype groups (A, B and C) were identified and their origin was inferred based on their presence/absence in the two germplasm pools (S. officinarum/S. robustum versus S. spontaneum). In the second analysis, SNPs specific to the two germplasm pools were identified and their distribution in the three haplotype groups (A, B and C) was analysed. These analyses were performed with two in-house python scripts (vcf2origin_AFB.py and vcf2origin_BFA.py respectively; https://github.com/SouthGreenPlatform/sugarcane-origins/blob/master/vcf2origin_AFB.py).
An SNP was considered specific to one group of haplotypes/BACs (A, B or C) if (1) data were available for at least one haplotype for each group of haplotypes , and (2) the SNP was only found in one haplotype group. An SNP was used for origin analysis if (1) it was present in at least two accessions representative of one of the two germplasm pools and absent in all accessions of the other pool, and (2) if sequence data were available at its position for at least five S. officinarum/S. robustum accessions and five S. spontaneum accessions, among the accessions selected as representative of these germplasm pools. For the Rpa1 region, since data were available for a lower number of accessions, only three S. officinarum/S. robustum were required. The same criteria were used when considering SNPs from the two germplasm pools (S. officinarum/S. robustum versus S. spontaneum) and examining their presence among the haplotypes/BACs. For the Adh1 region, 45 S. officinarum, 8 S. robustum and 175 S. spontaneum accessions were used as representative of these species. For the Rpa1 region, 13 S. officinarum, 1 S. robustum and 84 S. spontaneum were used as representative of these species (Supplementary Data Table S1). The position on the monoploid sugarcane reference sequence (Garsmeur et al., 2018) of a set of 31 832 SNPs identified as specific to S. officinarum versus S. spontaneum is shown in Supplementary Data Table S2.
RESULTS
Comparison of two sets of 12 hom(oe)ologous haplotypes from modern cultivar R570
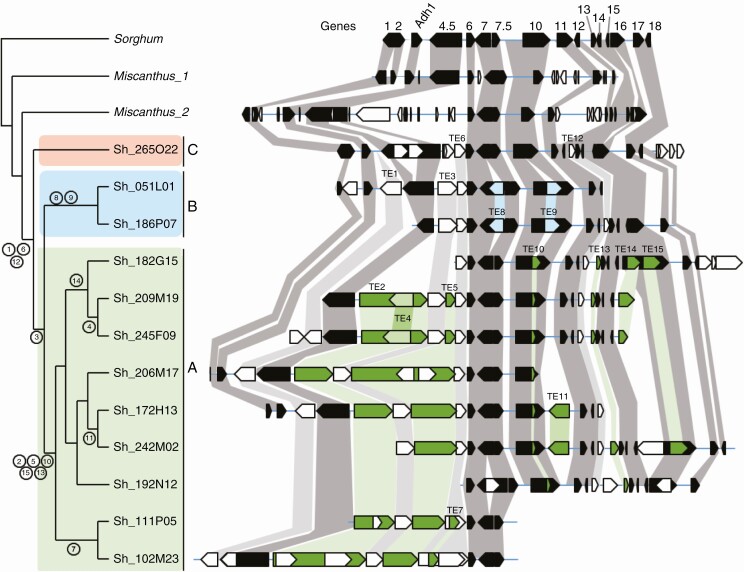
Two hom(oe)ologous sets of haplotypes from cultivar R570 were analysed. The first set corresponded to a region bearing the Adh1 gene located on sugarcane chromosome 1 (Garsmeur et al., 2018) and syntenic to Sorghum chromosome 1. This set contained 12 haplotypes, represented by 12 BAC clones. Sixteen genes with their corresponding allelic versions were annotated (Fig. 1, Supplementary Data Tables S3 and S4). The sequence that overlapped between pairs of hom(oe)ologous haplotypes varied from 35 to 113 kb, representing between 3 and 13 genes. One region of ~25 kb was shared by all hom(oe)ologous haplotypes and included three genes (genes 6–7.5).
Fig. 1.
Comparison of the 12 sugarcane hom(oe)ologous haplotypes (BACs) of the Adh1 region together and with Sorghum and Miscanthus orthologues. Genes are represented by black boxes and collinear genes are connected in dark grey. TEs are represented by white boxes; collinear TEs are connected in light grey or, when conserved within haplotype group A or B, in green or blue, respectively. Phylogenetic relationships among haplotypes are represented on the left with haplotypes from groups A, B and C highlighted in green, blue and red, respectively, and with circles positioning TE insertion times.
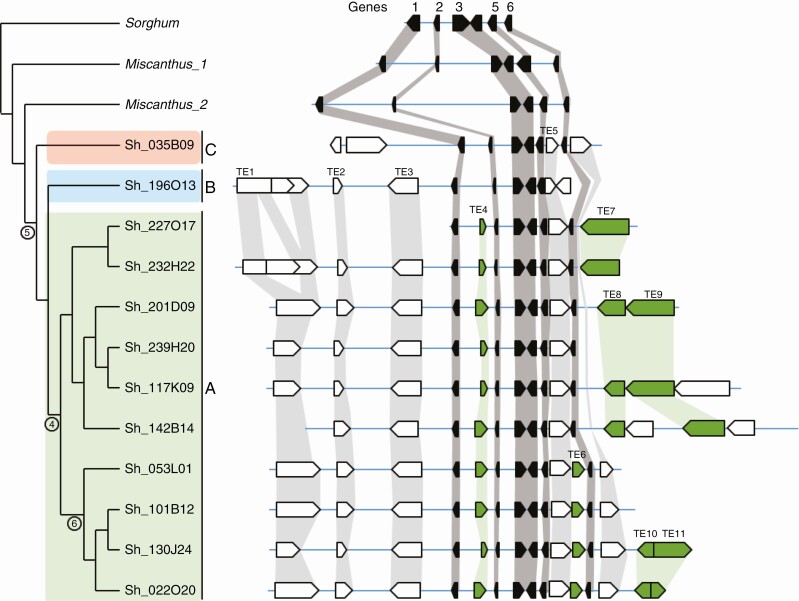
The second set corresponded to a region bearing the Rpa1 gene located on sugarcane chromosome 3 (Garsmeur et al., 2018) and syntenic to Sorghum chromosome 4. This set contained 12 haplotypes, represented by 12 BAC clones. Six genes with their corresponding allelic versions were annotated (Fig. 2, Supplementary Data Tables S3 and S4). The region shared by the 12 hom(oe)ologous haplotypes included five genes and represented around 35 kb (genes 1–5).
Fig. 2.
Comparison of the 12 sugarcane hom(oe)ologous haplotypes (BACs) of the Rpa1 region together and with Sorghum and Miscanthus orthologues. Genes are represented by black boxes and collinear genes are connected in dark grey. TEs are represented by white boxes; collinear TEs are connected in light grey or, when conserved within haplotype group A, in green. Phylogenetic relationships among haplotypes are represented on the left with haplotypes from groups A, B and C highlighted in green, blue and red, respectively and with circles positioning TE insertion times.
For both regions, the gene content and relative order were strictly conserved among all hom(oe)ologous sugarcane haplotypes. The percentage of nucleotide sequence identity was very high between all pairs of sugarcane hom(oe)ologous genes, with an average of 99.2 % for exons (ranging from 97.3 to 100 %) and 95.6 % for introns (ranging from 80.3 to 100 %).
The two regions were compared with their Sorghum and Miscanthus orthologous sequences. The gene content and order were also strictly conserved between sugarcane and the Sorghum orthologous sequences and the two ortho-paralogous M. sinensis sequences (Figs 1 and 2). Both sugarcane regions displayed a high level of nucleotide sequence conservation with Miscanthus and Sorghum, with an average of 96.5 and 93.4 % for exons and 89.7 and 75.8 % for introns, respectively.
Large TEs were annotated in the Adh1 and Rpa1 regions, representing distinct classes of TEs [long terminal repeat (LTR) retrotransposons, non-LTR retrotransposons and DNA transposons] (Figs 1 and 2, Supplementary Data Table S4). Several TE insertion sites were conserved across hom(oe)ologous haplotypes in both regions.
Phylogenetic relationships among hom(oe)ologous haplotypes distinguished three groups of haplotypes
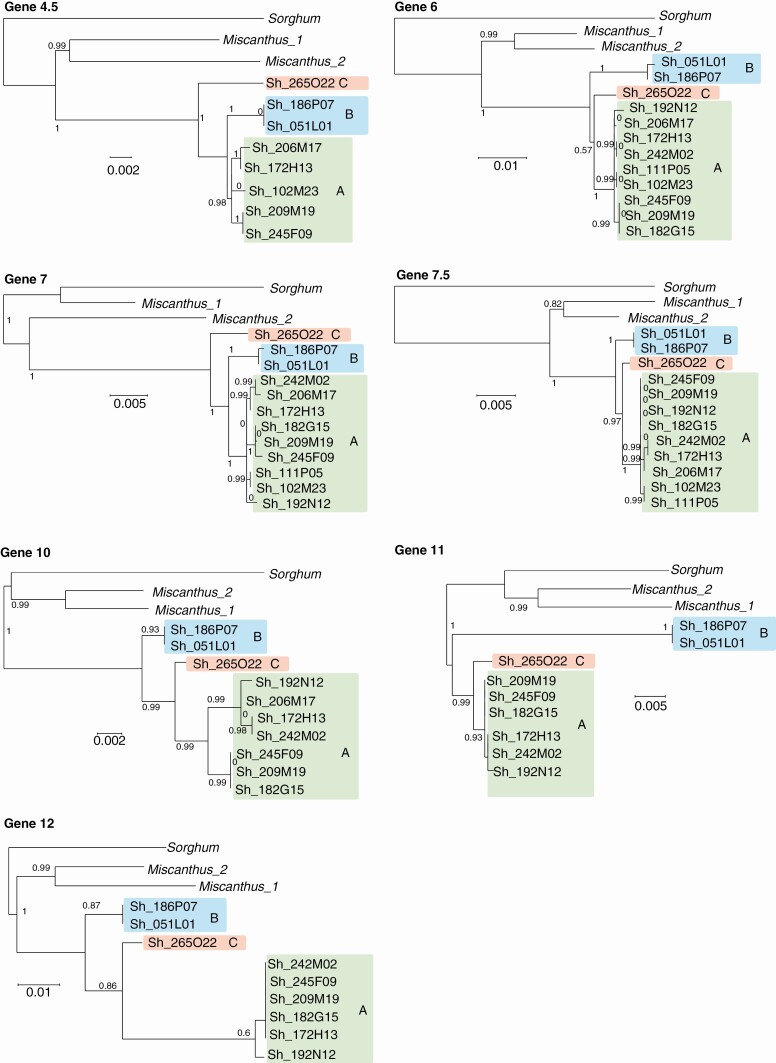
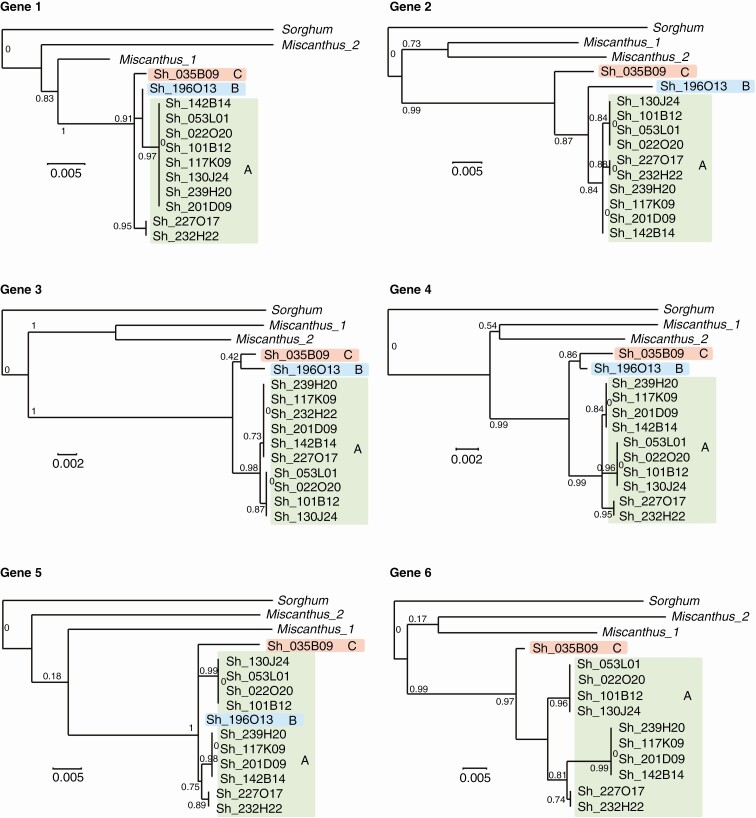
Phylogenetic relationships between hom(eo)ologous haplotypes were analysed based on (1) genomic regions shared by all haplotypes comprising genes 6–7.5 for the Adh1 region and genes 1–5 for the Rpa1 region (Figs 1 and 2) and (2) hom(oe)ologous copies of individual genes, including exons and introns (Figs 3 and 4).
Fig. 3.
Phylogenetic relationships between hom(oe)ologous genes in the Adh1 region. BACs belonging to haplotype groups A, B and C are highlighted in green, blue and red, respectively. Bootstrap values are indicated. Bar scales correspond to branch lengths.
Fig. 4.
Phylogenetic relationships between hom(oe)ologous genes in the Rpa1 region. BACs belonging to haplotype groups A, B and C are highlighted in green, blue and red, respectively. Bootstrap values are indicated. Bar scales correspond to branch lengths.
For the Adh1 region, both analyses revealed three groups of haplotypes, with a major group (named A) that included nine haplotypes (Sh182G15, Sh209M19, Sh245F09, Sh206M17, Sh172H13, Sh242M02, Sh192N12, Sh111P05 and Sh102M23), a second group (named B) that included two haplotypes (Sh051L01 and Sh186P07) and a third group (named C) with a unique haplotype (Sh265O22).
For the Rpa1 region, both analyses also revealed three groups of haplotypes, in accordance with the result of de Setta et al. (2014), with a major group (A) that included ten haplotypes (Sh227O17, Sh232H22, Sh201D09, Sh239H20, Sh117K09, Sh142B14, Sh053L01, Sh101B12, Sh130J24 and Sh022O20) and two groups (B and C) each with a single haplotype (Sh196O13 and Sh035B09, respectively). One exception was observed for gene 1 in haplotypes Sh227O17 and Sh232H22, which grouped separately from the other A haplotypes, suggesting that recombination may have occurred in these two haplotypes.
The two M. sinensis paralogues grouped generally together and always apart from the group of Saccharum hom(oe)ologous haplotypes.
TE insertion site conservation among hom(oe)ologous haplotypes reinforced the presence of three groups of haplotypes
In the Adh1 region, 15 TE insertion sites were conserved across two to nine hom(oe)ologous haplotypes (Fig. 1, Supplementary Data Table S4). Two TEs were shared only by the two haplotypes of group B (TE 8 and TE 9), nine TEs were conserved only across some or all available sequences for haplotypes of group A (TE 2, 4, 5, 7, 10, 11, 13, 14 and 15) and one TE (TE 3) was shared by haplotypes of groups A and B. The TE 1, TE 6 and TE 12 insertion sites were conserved across all haplotypes overlapping the corresponding regions. For TE 1, a 15-bp sequence corresponding to the short direct repeats of this TE was found in BAC Sh265O22. This suggested that TE 1 was present on the haplotype Sh265O22 but was removed by illegitimate recombination (Ma et al., 2004). The structure (complete versus fragment) was conserved among all other shared TEs, with two exceptions: TE 5 was found as a solo LTR in haplotypes Sh209M19 and Sh245F09, indicating that unequal homologous recombination between the two LTRs of the complete TE 5 occurred; TE 6 was found fragmented on haplotypes Sh111P05 and Sh102M23.
In the Rpa1 region, in the interval that overlapped for the three groups of haplotypes, TE 4 was found on all haplotypes from group A, TE 6 on several haplotypes of group A and one TE (TE 5) was conserved on all haplotypes (Fig. 2).
Globally, the pattern of TE insertions among hom(oe)ologous haplotypes reinforced the distinction between the three groups of haplotypes, especially for the Adh1 region, and made it possible to position the TE insertion events on the phylogenetic tree (Figs 1 and 2).
Chronology of divergence between Saccharum homoeologous haplotype groups and with Miscanthus paralogous orthologues
Divergence times were estimated from synonymous substitution rates (Ks) for each gene of both the Adh1 and the Rpa1 R570 region. On average, divergence times within homologous haplotypes of group A and within haplotypes of group B was low (0.05–0.34 Mya). Divergence time between haplotypes from groups A and B, from groups B and C and from groups A and C were estimated to be 0.84, 1.23 and 1.29 Mya, respectively (Table 1).
Table 1.
Divergence time between the three groups of haplotypes (A, B and C) and between Saccharum and Miscanthus
| K s | Mya | |||
|---|---|---|---|---|
| Adh1 | Rpa1 | Mean | ||
| Saccharum A–A | 0.0043 | 0.0048 | 0.0044 | 0.34 |
| Saccharum B–B | 0.0007 | – | – | 0.05 |
| Saccharum A–B | 0.0112 | 0.0102 | 0.0109 | 0.84 |
| Saccharum A–C | 0.0168 | 0.0166 | 0.0168 | 1.29 |
| Saccharum B–C | 0.0173 | 0.0129 | 0.0160 | 1.23 |
| Saccharum–Miscanthus | 0.0646 | 0.0874 | 0.0740 | 5.69 |
| Miscanthus 1– Miscanthus 2 | 0.0480 | 0.0510 | 0.0513 | 3.94 |
Divergence time between Saccharum and Miscanthus lineages was estimated to be 5.7 Mya and the whole-genome duplication in Miscanthus to be 3.9 Mya.
Origin of the three groups of haplotypes coexisting in modern cultivars
To determine the origin of the three distinct groups of R570 haplotypes, we exploited sequence data from accessions representative of the Saccharum species. These data were aligned to the R570 sugarcane monoploid reference sequence and SNPs were identified.
Because hybridization can occur between the different Saccharum species and because mislabelling of accessions is frequent in collections, we performed multivariate analyses for each of the three sets of sequence data (the WGS data and the two sets of targeted sequence data) to analyse the structure of the diversity within the Saccharum accessions and to select accessions representative of S. spontaneum, S. officinarum and S. robustum. For the data set from Yang et al. (2019), a preliminary multivariate analysis was performed to exclude accessions not belonging to the Saccharum genus. In this analysis, the first two axes clearly differentiated a large group of accessions labelled as Saccharum from scattered accessions that included accessions from closely related genera (Miscanthus, Erianthus, Sorghum) and several accessions probably mislabelled as Saccharum (Supplementary Data Fig. 1). These accessions were excluded and the remaining 272 Saccharum accessions were kept for further analysis.
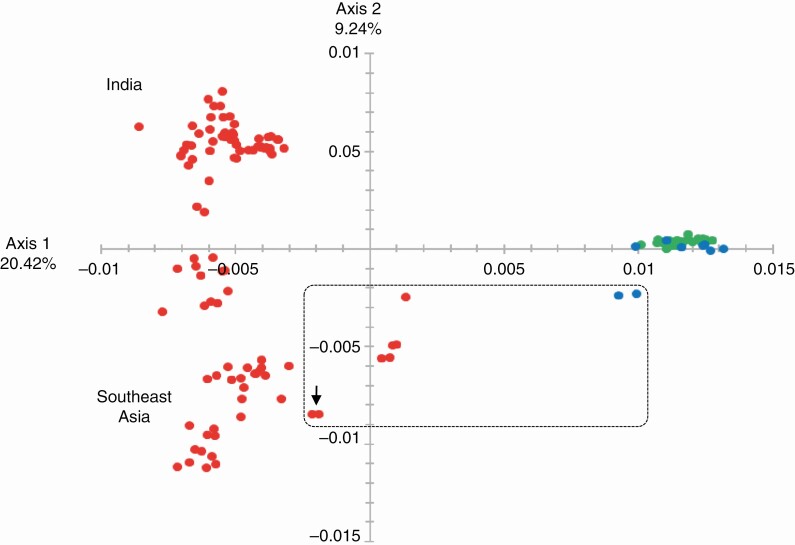
In the multivariate analyses performed for each of the three sets of sequence data only with the Saccharum accessions, the first axis separated S. officinarum and S. robustum accessions from S. spontaneum accessions. Accessions belonging to S. barberi, S. sinense and modern cultivars were in an intermediate position in accordance with their interspecific origin (Supplementary Data Figs S2, S3 and S4). Multivariate analyses were then performed without these hybrid accessions. The first axis clearly separated S. officinarum and S. robustum accessions from S. spontaneum accessions (Fig. 5 and Supplementary Data Figs S3b and S4). Saccharum spontaneum accessions formed a very large group, with the second axis largely separating accessions from India from accessions from Indonesia in the first axis of a multivariate analysis performed with targeted capture sequences of 142 accessions (Fig. 5). A few accessions labelled as S. robustum and S. spontaneum had intermediate positions. They could represent hybrids between these groups and thus were excluded from the set of accessions chosen as representatives of the three species for the selection of specific SNPs. Due to their close proximity in the multivariate analysis, S. robustum and S. officinarum accessions were further considered as one germplasm pool while S. spontaneum accessions were considered as a second germplasm pool for the selection of SNPs specific to each of these two germplasm pools.
Fig. 5.
First plane of a multivariate analysis separating accessions in two germplasm pools: one pool comprises S. officinarum (green) and S. robustum (blue) accessions, and one pool comprises S. spontaneum (red) accessions. Analysis is based on SNPs obtained from targeted capture sequences of 142 accessions (Yang et al., 2019). Accessions located between these two germplasm pools within the black rectangle may represent hybrids and were excluded for the selection of SNPs specific to these germplasm pools. Arrow points to accessions IN 84-088 and IN 84-089.
Two analyses were then performed with the selected accessions to study the origin of the three groups of R570 haplotypes/BACs (groups A, B and C). In the first analysis, we identified SNPs specific to each of the three groups of haplotypes/BACs in the Adh1 and Rpa1 regions (i.e. SNPs found only in all or some haplotypes from a single group). A total of 188 and 60 such SNPs were identified for the Adh1 and Rpa1 regions, respectively. Combining the two regions, 97 SNPs were found only in haplotype group A, 65 SNPs only in haplotype group B and 86 SNPs only in haplotype group C (Table 2 and Supplementary Data Tables S5 and S6). We then identified among the accessions selected as representative of S. robustum/S. officinarum and S. spontaneum which one had these SNPs. SNPs specific to a haplotype group in R570 but shared by both germplasm pools were interpreted as ancestral SNPs. All SNPs specific to haplotype groups A and B and present in only one germplasm pool were found only (with one exception) in the S. officinarum/S. robustum pool, suggesting that haplotypes A and B originated from this germplasm pool. This corresponded to 57 and 25 SNPs in the Adh1 and Rpa1 regions, respectively (Table 2). All SNPs specific to haplotype group C and found in only one germplasm pool were found only in the S. spontaneum pool, suggesting that haplotype C originated from this species. This corresponded to 28 and 16 SNPs in the Adh1 and Rpa1 regions, respectively (Table 2).
Table 2.
Distribution of SNPs specific to each of the three groups of haplotypes in representatives of the Saccharum species in regions Adh1 and Rpa1
| S. officinarum/S. robustum | S. spontaneum | All three species | Total | |
|---|---|---|---|---|
| Adh1 | ||||
| Group | ||||
| A | 29 | 0 | 44 | 73 |
| B | 28 | 1 | 19 | 48 |
| C | 0 | 27 | 40 | 67 |
| Total | 57 | 28 | 103 | 188 |
| Rpa1 | ||||
| Group | ||||
| A | 15 | 0 | 9 | 24 |
| B | 10 | 0 | 7 | 17 |
| C | 0 | 16 | 3 | 19 |
| Total | 25 | 16 | 19 | 60 |
In the second analysis, from the sequences of the Saccharum representatives that mapped to the Adh1 and Rpa1 regions, we identified SNPs that were specific to each germplasm pool (i.e. SNPs found only in some or all S. officinarum/S. robustum accessions versus SNPs found only in some S. spontaneum accessions). A total of 96 and 44 such SNPs were identified for the Adh1 and Rpa1 regions, respectively (Table 3 and Supplementary Data Tables S7 and S8). We then identified which of these SNPs were present in haplotype groups A, B and C. The SNPs found only in the S. officinarum/S. robustum accessions were exclusively (with one exception) found in R570 haplotype groups A and B while the SNPs found only in S. spontaneum accessions were exclusively found in R570 haplotype group C.
Table 3.
Distribution of SNPs specific to S. officinarum/S. robustum versus S. spontaneum in the three groups of haplotypes in regions Adh1 and Rpa1.
| Group A | Group B | Group A and B | Group C | Total | |
|---|---|---|---|---|---|
| Adh1 | |||||
| S. officinarum/S. robustum | 29 | 28 | 4 | 0 | 61 |
| S. Spontaneum | 0 | 1 | 0 | 27 | 28 |
| Total | 29 | 29 | 4 | 27 | 89 |
| Rpa1 | |||||
| S. officinarum/S. robustum | 15 | 10 | 2 | 0 | 25 |
| S. spontaneum | 0 | 0 | 0 | 16 | 16 |
| Total | 15 | 10 | 2 | 16 | 43 |
Both analyses clearly suggested that haplotype groups A and B originated from S. officinarum and S. robustum, while haplotype group C originated from S. spontaneum.
Distribution of SNPs specific to the three groups of haplotypes in Saccharum
The distribution of the 82 R570 SNPs specific to haplotype groups A and B that were found only in S. officinarum/S. robustum and of the 43 SNPs specific to group C found only in S. spontaneum was then analysed in the whole set of Saccharum accessions (Supplementary Data Table S1). We found SNPs specific to each of the three groups of haplotypes (A, B, C) in representatives of S. barberi, S. sinense and modern cultivars. These results are expected since they are hybrids between S. officinarum and S. spontaneum clones. SNPs specific to each of the three groups of haplotypes (A, B, C) were also found in most of the accessions from Yang et al. (2019) that these authors re-classified as hybrids. Many of these accessions probably corresponded to mislabelled accessions, as already suggested by Yang et al. (2019).
The two S. robustum (IN 84-076 and IS 76-184) that we excluded as representative of S. robustum based on the multivariate analysis displayed one SNP specific to group C in addition to SNPs specific to groups A and B, which may indicate a hybrid status. The few S. spontaneum clones that we excluded as representative of S. spontaneum displayed SNPs specific to groups A and B in addition to SNPs specific to group C, suggesting a hybrid status. Two of them (IN 84-088 and IN 84-089 in Fig. 5) positioned very close in the multivariate analysis to the other S. spontaneum accessions from Southeast Asia (Indonesia). These two accessions carried only SNPs specific to groups B and C except for one SNP that was specific to group A (in region Adh1). However, data from one of the two BACs/haplotypes of group B was missing at this position; thus the SNP could also have been present on the missing BAC/haplotype, invalidating this position as specific for group A. In addition, this SNP was found in every S. robustum, S. sinense and S. barberi accession and modern cultivar and in 24 out of the 28 S. officinarum accessions. This could indicate that this SNP is an ancestral SNP common to haplotypes of group A and B but not C, rather than specific to group A. These results suggested that these two accessions (IN 84-089 and IN 84-088) could be hybrids between the B and C ancestral founding genomes.
DISCUSSION
We analysed 12 hom(oe)ologous haplotypes for two genomic regions in the genome of a typical modern sugarcane cultivar (R570). These regions belong to chromosomes 1 and 3, for which 12 chromosome copies were revealed with chromosome-specific oligo probes by FISH in cultivar R570 (Piperidis and D’Hont, 2020). This number is in the range of chromosome copy numbers expected for a modern cultivar (Piperidis and D’Hont, 2020). Our results revealed for both regions the existence of three groups of haplotypes, with a major group (A) being present in nine or ten copies and two minor groups (B, C) being present in one or two copies. Two wild species are known in the Saccharum genus, S. spontaneum and S. robustum. The sweet domesticated canes, S. officinarum, are thought to have been domesticated from S. robustum (Brandes, 1956). SNPs specific to haplotypes A and B were found in representative accessions of S. robustum and S. officinarum but not in S. spontaneum. Conversely, SNPs specific for haplotype C were found in representative accessions of S. spontaneum but not in representatives from S. robustum and S. officinarum. The three groups of haplotypes were estimated to have diverged some 0.8–1.3 Mya. These results suggested that three founding genomes were involved in the origin of the Saccharum genus and modern sugarcane cultivars. The observed divergence time between the three groups of haplotypes is in the range of previous estimates of the divergence between the S. officinarum and S. spontaneum lineages (0.7–3.5 Mya) (Jannoo et al., 2007; Vilela et al., 2017; Yang et al., 2017).
The detection of specific SNPs from two groups of haplotypes (A and B) in the genomes of both S. officinarum and S. robustum is consistent with the common view that S. officinarum has been domesticated from S. robustum (Brandes, 1956). Furthermore, our study clearly demonstrated that the foundation of S. robustum, and hence S. officinarum, is heterogeneous and that interspecific hybridization or allopolyploidization occurred in the evolutionary history of this taxon in addition to autopolyploidy. The much higher proportion of haplotype A compared with haplotype B observed in both studied regions of a modern cultivar suggests that the hybridization events involved transmission of unreduced gametes or hybridizations between autopolyploids with different ploidy levels and/or were followed by backcrosses with the A founder genome.
Recently, Zhang et al., 2019 contradicted the general assumption that S. officinarum was domesticated from S. robustum based on the divergence time of 0.38 Mya they estimated between assembled haplotypes from these two species. This divergence time is close to the one we obtained between haplotypes within group A (0.34 Mya). The presence of two founder genomes in the origin of these species, revealed by our study, may have complicated the interpretation of Zhang et al. (2019).
Specific SNPs from group C were found only in S. spontaneum. This species is highly polymorphic, with a large distribution range from Africa to Southeast Asia that overlaps with S. robustum, from Kalimantan Island to Papua New Guinea (Grivet et al., 2004). The high diversity of this species is illustrated by the results of the multivariate analysis, which showed a large group with two main subgroups, one from India and one from East Asia (Fig. 5). Specific SNPs from group C but not from groups A and B were found in clones from these two S. spontaneum subgroups. Several clones identified as S. spontaneum in collections were found in intermediate positions between these S. spontaneum groups and the S. robustum/S. officinarum pool. These clones may be natural hybrids between these species since they displayed SNPs specific to groups A, B and C. These clones were found in the multivariate analysis mainly between the S. officinarum/S. robustum pool and the S. spontaneum accessions from East Asia (Indonesia). This can be explained by the fact that their distribution overlaps in these regions (Grivet et al., 2006). Some of these S. spontaneum clones, based on pairwise genetic distance with modern cultivars, were suggested by Yang et al. (2019) to be the ones mainly involved in the origin of modern cultivars. Our analysis suggested instead that their genetic proximity with modern cultivars is linked to their hybrid status with S. officinarum or S. robustum as they bear A and B alleles specific to these species.
We found SNPs specific to each of the three groups of haplotypes (A, B, C) in representatives of S. barberi and S. sinense, in accordance with their proposed natural interspecific hybrid origin between S. officinarum and S. spontaneum (Brandes, 1956; Daniels and Roach, 1987; D’Hont et al., 2002). The three groups of SNPs were also found in all modern cultivars tested, which are all derived from a few interspecific hybridizations made by breeders a century ago between a few S. officinarum, S. spontaneum and S. barberi clones. These interspecific hybridizations were followed by backcrosses with S. officinarum to recover good agronomic performance. This process resulted in the reduction of the proportion of S. spontaneum chromosomes, which was estimated based on molecular cytogenetics to represent between 15 and 25 % of the chromosomes in modern cultivars (D’Hont et al., 1996; Cuadrado et al., 2004; Piperidis et al., 2010; Huang et al., 2020; Piperidis and D’Hont, 2020). More recently this proportion was shown to vary from one to four copies depending on the hom(oe)ology group in the few cultivars analysed (Piperidis and D’Hont, 2020). The proportion of haplotypes A/B (11 haplotypes in the two regions) originating from S. officinarum versus haplotype C from S. spontaneum (one haplotype) observed in the two regions studied fits in this range.
This evolutionary model, implicating autopolyploid and allopolyploid/interspecific hybridization events, contradicts the assumption that S. officinarum has an autopolyploid origin (Garsmeur et al., 2011; Vilela et al., 2017; Yang et al., 2017) and could explain the variable chromosome pairing affinity observed in Saccharum (Jannoo et al., 2007). A few genetic maps have been built and all of them are partial. However, they revealed some variation in paring affinity. Some preferential pairing has been observed in S. robustum and S. officinarum (Mudge et al., 1996; Edmé et al., 2006; Aitken et al., 2007) but not in S. spontaneum (Al-Janabi et al., 1993; da Silva et al., 1995; Alwala, 2008). This observation could suggest that S. spontaneum accessions are autopolyploids (from genome founder C) with polysomic pairing. This autopolyploidy was verified recently for one tetraploid clone, AP85-441 (haploid of SES 208), from which a genome sequence was assembled (Zhang et al., 2018). For S. robustum and S. officinarum, the observed pairing behaviour is compatible with a mix of allo- and autopolyploid origins with the coexistence of two groups of homologues (from genome founders A and B) resulting in preferential pairing (A versus B) but each pairing being in a polysomic manner. In a modern cultivar such as R570, based on our data and Piperidis and D’Hont (2020), it is probably not rare that haplotypes B and C are present in two copies. This could explain the strong preferential pairing that we have observed for some S. spontaneum and some S. officinarum chromosomes (Grivet et al., 1996; Hoarau et al., 2001; Jannoo et al., 2004), while the other A haplotypes largely display polysomic inheritance. Occasional recombination between homoeologues may result in mosaic chromosome structures complicating the chromosome pairing picture observed (Jannoo et al., 2004).
Perfect collinearity and a very high level of gene structural conservation among hom(oe)ologous sugarcane chromosomes were observed, with an average divergence in coding sequence of <1 % and all alleles being predicted to be functional (with one exception). The striking conservation of hom(oe)ologous genes that we observed confirmed and extended our previous results on two hom(oe)ologous BACs from the Adh1 region and seven hom(oe)ologous haplotypes from a region carrying the brown rust resistance (Bru1) gene (Jannoo et al., 2007; Garsmeur et al., 2011). One reason for this strikingly high level of gene conservation among hom(oe)ologous sugarcane haplotypes may be the relatively young age of the polyploidization event in Saccharum species, which may have given little time for the paralogous chromosomes to differentiate from each other. Another reason may be the high polyploidy and mixed allo- and autopolyploidy, with autopolyploidy restraining divergence through pairing and recombination between homologues and polysomic inheritance. The maintenance of a broad set of functional hom(oe)ologues could be involved in the remarkable productivity and phenotypic plasticity of sugarcane.
The comparison of the two sugarcane regions with Sorghum (2n = 2x = 20) orthologous regions and with the two ortho-paralogous Miscanthus (2n = 4x = 38) regions showed high gene synteny conservation. For both regions, gene phylogenetic analyses did not support the assumption that the allopolyploid event that arose around 3–4 Mya in the Miscanthus lineage after its divergence with Sorghum was shared with the Saccharum lineage (Kim et al., 2014), in agreement with Welker et al. (2015), Vilela et al. (2017) and Zhang et al. (2018). Our results suggested that, after its divergence from the Miscanthus lineage, the Saccharum lineage differentiated in a few sublineages (A, B, C, and possibly others) that further underwent auto- and/or allopolyploid events leading to the present day higher-order polyploids (>4x). No extant diploid representatives of these lineages are known, presumably having become extinct. In addition, no pure representatives of the A or B lineages were found in our sample. Further investigation should be made in particular within S. robustum, for which we analysed only a few accessions, but these were described to display important phenotypic variation that led some authors to separate them in several distinct taxonomic groups (Daniels and Roach, 1987).
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Figure S1: first plane of a multivariate analysis based on SNPs obtained from targeted capture sequences of 304 accessions. Figure S2: first plane of a multivariate analysis based on SNPs obtained from targeted capture sequences of 272 accessions. Figure S3: first plane of a multivariate analysis based on SNPs obtained from targeted capture sequences of 33 accessions. Figure S4: first plane of a multivariate analysis based on SNPs from WGS data from 83 accessions. Table S1: information on the sequence data used. Table S2: position on the sugarcane reference sequence of Garsmeur et al. (2018) of 31 832 SNPs identified as specific to S. officinarum or S. spontaneum based on targeted capture sequences from 35 S. officinarum and 91 S. spontaneum accessions from Yang et al. (2019) Table S3: genes annotated in the Adh1 and Rpa1 regions. Table S4: information on TEs conserved among hom(oe)ologous chromosome haplotypes in the Adh1 region Table S5: distribution of the SNPs specific to each of the three groups of haplotypes/BACs in the Adh1 region in representatives of Saccharum diversity. Table S6: distribution of the SNPs specific to each of the three groups of haplotypes/BACs in the Rpa1 region and in representatives of Saccharum diversity. Table S7: distribution of the SNPs specific to S. officinarum/S. robustum versus S. spontaneum in the haplotypes/BACs of the Adh1 region. Table S8: distribution of the SNPs specific to S. officinarum/S. robustum versus S. spontaneum in the haplotypes/BACs of the Rpa1 region.
ACKNOWLEDGEMENTS
We thank the South Green Bioinformatics platform (http://southgreen.cirad.fr) for providing a high-performance cluster, and the CIRAD Regional genotyping technology platform (GPTRG), Dr Dan S. Rokhsar, Dr Stephen Moose and Dr Anna Hale for sharing resources. We thank the JGI and collaborators for prepublication access to sugarcane diversity data for this study
FUNDING
This work was supported by the Centre de Coopération Internationale en Recherche Agronomique pour le Développement (CIRAD) and the International Consortium for Sugarcane Biotechnology (ICSB). The sequencing conducted by the US Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, is supported by the Office of Science of the US Department of Energy under Contract No. DE-AC02-05CH11231 (Community Science Program, CSP-504319).
LITERATURE CITED
- Aitken KS, Jackson PA, McIntyre CL. 2007. Construction of a genetic linkage map for Saccharum officinarum incorporating both simplex and duplex markers to increase genome coverage. Genome 50: 742–756. [DOI] [PubMed] [Google Scholar]
- al-Janabi SM, Honeycutt RJ, McClelland M, Sobral BW. 1993. A genetic linkage map of Saccharum spontaneum L. ‘SES 208’. Genetics 134: 1249–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alwala S, Kimbeng CA, Veremis JC, Gravois KA. 2008. Linkage mapping and genome analysis in a Saccharum interspecific cross using AFLP, SRAP and TRAP markers. Euphytica 164: 37–51. [Google Scholar]
- Arceneaux G. 1968. Breeding sugarcane varieties for the Northern Caribbean. In: Proceedings of the International Society of Sugar Cane Technologists 13: 1034–1046.
- Barker MS, Arrigo N, Baniaga AE, Li Z, Levin DA. 2016. On the relative abundance of autopolyploids and allopolyploids. New Phytologist 210: 391–398. [DOI] [PubMed] [Google Scholar]
- Brandes E. 1956. Origin, dispersal and use in breeding of the Melanesian garden sugarcane and their derivatives, Saccharum officinarum L. Proceedings of the International Society of Sugar Cane Technologists 9: 709–750. [Google Scholar]
- Carver TJ, Rutherford KM, Berriman M, Rajandream MA, Barrell BG, Parkhill J. 2005. ACT: the Artemis Comparison Tool. Bioinformatics 21: 3422–3423. [DOI] [PubMed] [Google Scholar]
- Cuadrado A, Acevedo R, Moreno Díaz de la Espina S, Jouve N, de la Torre C. 2004. Genome remodelling in three modern S. officinarum × S. spontaneum sugarcane cultivars. Journal of Experimental Botany 55: 847–854. [DOI] [PubMed] [Google Scholar]
- Daniels J, Daniels C. 1976. Buddhism, sugar and sugarcane. Sugarcane Breeders’ Newsletter 38: 35–60. [Google Scholar]
- Daniels J, Roach BT. 1987. Taxonomy and evolution. In: Heinz DJ, ed. Sugarcane Improvement through Breeding. Amsterdam: Elsevier, 7–84. [Google Scholar]
- D’Hont A, Grivet L, Feldmann P, Rao S, Berding N, Glaszmann JC. 1996. Characterisation of the double genome structure of modern sugarcane cultivars (Saccharum spp.) by molecular cytogenetics. Molecular & General Genetics 250: 405–413. [DOI] [PubMed] [Google Scholar]
- D’Hont A, Paulet F, Glaszmann JC. 2002. Oligoclonal interspecific origin of ‘North Indian’ and ‘Chinese’ sugarcanes. Chromosome Research 10: 253–262. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Egan AN. 2010. Dating the origins of polyploidy events. New Phytologist 186: 73–85. [DOI] [PubMed] [Google Scholar]
- Edmé SJ, Glynn NG, Comstock JC. 2006. Genetic segregation of microsatellite markers in Saccharum officinarum and S. spontaneum. Heredity 97: 366–375. [DOI] [PubMed] [Google Scholar]
- Evans DL, Joshi SV. 2016. Complete chloroplast genomes of Saccharum spontaneum, Saccharum officinarum and Miscanthus floridulus (Panicoideae: Andropogoneae) reveal the plastid view on sugarcane origins. Systematics and Biodiversity 14: 548–571. [Google Scholar]
- Fan H, Ives AR, Surget-Groba Y, Cannon CH. 2015. An assembly and alignment-free method of phylogeny reconstruction from next-generation sequencing data. BMC Genomics 16: 522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers JM, Hazzouri KM, Gros-Balthazard M, et al. 2019. Cross-species hybridization and the origin of North African date palms. Proceedings of the National Academy of Sciences of the USA 116: 1651–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortune PM, Schierenbeck KA, Ainouche AK, Jacquemin J, Wendel JF, Ainouche ML. 2007. Evolutionary dynamics of Waxy and the origin of hexaploid Spartina species (Poaceae). Molecular Phylogenetics and Evolution 43: 1040–1055. [DOI] [PubMed] [Google Scholar]
- Garsmeur O, Charron C, Bocs S, et al. 2011. High homologous gene conservation despite extreme autopolyploid redundancy in sugarcane. New Phytologist 189: 629–642. [DOI] [PubMed] [Google Scholar]
- Garsmeur O, Droc G, Antonise R, et al. 2018. A mosaic monoploid reference sequence for the highly complex genome of sugarcane. Nature Communications 9: 2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut BS, Morton BR, McCaig BC, Clegg MT. 1996. Substitution rate comparisons between grasses and palms: synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL. Proceedings of the National Academy of Sciences of the USA 93: 10274–10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Molecular Biology and Evolution 27: 221–224. [DOI] [PubMed] [Google Scholar]
- Grivet L, D’Hont A, Roques D, Feldmann P, Lanaud C, Glaszmann JC. 1996. RFLP mapping in cultivated sugarcane (Saccharum spp.): genome organization in a highly polyploid and aneuploid interspecific hybrid. Genetics 142: 987–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivet L, Daniels C, Glaszmann JC, D’Hont A. 2004. A review of recent molecular genetics evidence for sugarcane evolution and domestication. Ethnobotany Research and Applications 2: 9–17. [Google Scholar]
- Grivet L, Glaszmann J-C, D’Hont A. 2006. Molecular Evidence of Sugarcane Evolution and Domestication In: Darwin’s Harvest: new approaches to the origins, evolution, and conservation of crops. Columbia University Press, 49–66. [Google Scholar]
- Guignon V, Droc G, Alaux M, et al. 2012. Chado controller: advanced annotation management with a community annotation system. Bioinformatics 28: 1054–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology 59: 307–321. [DOI] [PubMed] [Google Scholar]
- Hoarau J-Y, Offmann B, D’Hont A, et al. 2001. Genetic dissection of a modern sugarcane cultivar (Saccharum spp.). I. Genome mapping with AFLP markers. Theoretical and Applied Genetics 103: 84–97. [DOI] [PubMed] [Google Scholar]
- Hodkinson TR, Chase MW, Lledó MD, Salamin N, Renvoize SA. 2002. Phylogenetics of Miscanthus, Saccharum and related genera (Saccharinae, Andropogoneae, Poaceae) based on DNA sequences from ITS nuclear ribosomal DNA and plastid trnLintron and trnL-F intergenic spacers. Journal of Plant Research 115: 381–392. [DOI] [PubMed] [Google Scholar]
- Huang W, Li L, Myers JR, Marth GT. 2012. ART: a next-generation sequencing read simulator. Bioinformatics 28: 593–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Chen H, Han J, et al. 2020. Species-specific abundant retrotransposons elucidate the genomic composition of modern sugarcane cultivars. Chromosoma 129: 45–55. [DOI] [PubMed] [Google Scholar]
- Irvine JE. 1999. Saccharum species as horticultural classes. Theoretical and Applied Genetics 98: 186–194. [Google Scholar]
- Jannoo N, Grivet L, David J, D’Hont A, Glaszmann JC. 2004. Differential chromosome pairing affinities at meiosis in polyploid sugarcane revealed by molecular markers. Heredity 93: 460–467. [DOI] [PubMed] [Google Scholar]
- Jannoo N, Grivet L, Chantret N, et al. 2007. Orthologous comparison in a gene-rich region among grasses reveals stability in the sugarcane polyploid genome. Plant Journal 50: 574–585. [DOI] [PubMed] [Google Scholar]
- Katoh K, Asimenos G, Toh H. 2009. Multiple alignment of DNA sequences with MAFFT. In: Posada D, ed. Methods in molecular biology. Bioinformatics for DNA sequence analysis. Totowa, NJ: Humana Press, 39–64. [DOI] [PubMed] [Google Scholar]
- Kim C, Wang X, Lee TH, Jakob K, Lee GJ, Paterson AH. 2014. Comparative analysis of Miscanthus and Saccharum reveals a shared whole-genome duplication but different evolutionary fates. Plant Cell 26: 2420–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Sawyer S, Meyer M. 2012. Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Research 40: e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- Ma J, Devos KM, Bennetzen JL. 2004. Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Research 14: 860–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden ES, Sears ER. 1946. The origin of Triticum spelta and its free-threshing hexaploid relatives. Journal of Heredity 37: 81 107. [DOI] [PubMed] [Google Scholar]
- Meyer M, Kircher M. 2010. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harbor Protocols 2010: pdb.prot5448. [DOI] [PubMed] [Google Scholar]
- Mudge J, Andersen WR, Kehrer RL, Fairbanks DJ. 1996. A RAPD genetic map of Saccharum officinarum. Crop Science 36: cropsci1996.0011183X003600050046x. [Google Scholar]
- Perrier X, Jacquemoud-Collet JP. 2006. DARwin software, CIRAD, Montpellier. http://DARwin.cirad.fr (29 January 2021, date last accessed). [Google Scholar]
- Perrier X, De Langhe E, Donohue M, et al. 2011. Multidisciplinary perspectives on banana (Musa spp.) domestication. Proceedings of the National Academy of Sciences of the USA 108: 11311–11318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperidis G, Piperidis N, D’Hont A. 2010. Molecular cytogenetic investigation of chromosome composition and transmission in sugarcane. Molecular Genetics and Genomics 284: 65–73. [DOI] [PubMed] [Google Scholar]
- Piperidis N, D’Hont A. 2020. Sugarcane genome architecture decrypted with chromosome-specific oligo probes. The Plant Journal 103: 2039–2051. [DOI] [PubMed] [Google Scholar]
- Santos JD, Chebotarov D, McNally KL, et al. 2019. Fine scale genomic signals of admixture and alien introgression among Asian rice landraces. Genome Biology and Evolution 11: 1358–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seppey M, Manni M, Zdobnov EM. 2019. BUSCO: assessing genome assembly and annotation completeness. Methods in Molecular Biology 1962: 227–245. [DOI] [PubMed] [Google Scholar]
- de Setta N, Monteiro-Vitorello CB, Metcalfe CJ, et al. 2014. Building the sugarcane genome for biotechnology and identifying evolutionary trends. BMC Genomics 15: 540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva J, Honeycutt RJ, Burnquist W, et al. 1995. Saccharum spontaneum L. ‘SES 208’ genetic linkage map combining RFLP- and PCR-based markers. Molecular Breeding 1: 165–179. [Google Scholar]
- Stebbins GL. 1950. Variation and evolution in plants. New York: Columbia University Press. [Google Scholar]
- Simmonds NW. The Evolution of the Bananas 101–131 (Longman, 1962). [Google Scholar]
- Suyama M, Torrents D, Bork P. 2006. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Research 34: W609–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen JA, Govindarajulu R, Ashman TL, Liston A. 2014. Evolutionary origins and dynamics of octoploid strawberry subgenomes revealed by dense targeted capture linkage maps. Genome Biology and Evolution 6: 3295–3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomkins JP, Yu Y, Miller-Smith H, Frisch DA, Woo SS, Wing RA. 1999. A bacterial artificial chromosome library for sugarcane. Theoretical and Applied Genetics 99: 419–424. [DOI] [PubMed] [Google Scholar]
- Triplett JK, Clark LG, Fisher AE, Wen J. 2014. Independent allopolyploidization events preceded speciation in the temperate and tropical woody bamboos. New Phytologist 204: 66–73. [DOI] [PubMed] [Google Scholar]
- Vilela MM, Del Bem LE, Van Sluys MA, et al. 2017. Analysis of three sugarcane homo/homeologous regions suggests independent polyploidization events of Saccharum officinarum and Saccharum spontaneum. Genome Biology and Evolution 9: 266–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welker CAD, Souza-Chies TT, Longhi-Wagner HM, Peichoto MC, McKain MR, Kellogg EA. 2015. Phylogenetic analysis of Saccharum s.l. (Poaceae; Andropogoneae), with emphasis on the circumscription of the South American species. American Journal of Botany 102: 248–263. [DOI] [PubMed] [Google Scholar]
- Wu GA, Prochnik S, Jenkins J, et al. 2014. Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nature Biotechnology 32: 656–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Song J, You Q, Paudel DR, Zhang J, Wang J. 2017. Mining sequence variations in representative polyploid sugarcane germplasm accessions. BMC Genomics 18: 594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Song J, Todd J, et al. 2019. Target enrichment sequencing of 307 germplasm accessions identified ancestry of ancient and modern hybrids and signatures of adaptation and selection in sugarcane (Saccharum spp.), a ‘sweet’ crop with ‘bitter’ genomes. Plant Biotechnology Journal 17: 488–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution 24: 1586–1591. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhang X, Tang H, et al. 2018. Allele-defined genome of the autopolyploid sugarcane Saccharum spontaneum L. Nature Genetics 50: 1565–1573. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhang Q, Li L, et al. 2019. Recent polyploidization events in three Saccharum founding species. Plant Biotechnology Journal 17: 264–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.