Abstract
Background:
Gastric cancer (GC) peritoneal carcinomatosis is fatal. Delay in detection of peritoneal metastases contributes to high mortality; highlighting the need to develop biomarkers that can help identify patients at high risk for peritoneal recurrence (PR) or metastasis (PM).
Methods:
We performed a systematic discovery and validation for the identification of PR-prediction and PM-detection biomarkers by analyzing expression profiling datasets from 249 GC patients, followed by analysis of 426 patients from 3 cohorts for clinical validation.
Results:
Genomewide expression profiling identified a 12-gene panel for robust prediction of PR in GC patients (AUC=0.95), which was successfully validated in a second dataset (AUC=0.86). Examination of 216 specimens from a training cohort allowed us to establish a 6-gene based risk prediction model (AUC=0.72; 95%CI: 0.66-0.78), which was subsequently validated in an independent cohort of 111 GC patients (AUC=0.76; 95%CI: 0.67-0.83). In both cohorts, combining tumor morphology and depth of invasion further improved the predictive accuracy of the prediction model (AUC=0.84). Thereafter, we evaluated the performance of the identical 6-gene panel for its ability to detect PM by analyzing 210 GC specimens (prior 111 patients plus additional 99 cases), which discriminated patients with and without PM (AUC=0.72). Finally, our biomarker panel was also remarkably effective for identifying peritoneal micrometastasis (AUC=0.72), and its diagnostic accuracy was significantly enhanced when depth of invasion was included in the model (AUC=0.85).
Conclusions:
Our novel transcriptomic signature for risk-stratification and identification of high-risk patients with peritoneal carcinomatosis might serve as an important clinical decision-making in GC patients.
Keywords: Gastric cancer, peritoneal metastasis, peritoneal recurrence, biomarker, HIPEC
INTRODUCTION
Gastric cancer (GC) is the 2nd leading cause of cancer-related deaths worldwide (1). The majority of GC presents at an advanced stage, and consequently a significant number of patients are diagnosed with a metastatic disease at initial presentation or experience disease relapse even after adjuvant treatment. Peritoneal dissemination is the most common cause for cancer recurrence and distant metastasis in GC, resulting in poor prognosis with a dismal median survival duration of ~4 months (2,3). Although poor response to conventional systemic chemotherapy and suboptimal therapeutic options are primarily responsible for the poor outcomes in GC patients with peritoneal carcinomatosis, the lack of clinically robust diagnostic modalities to detect presence of peritoneal metastasis remains one of the most critical hurdles in improving disease outcomes in this malignancy.
The presence of intraperitoneal cancer cells in GC patients is intimately associated with a frequent peritoneal relapse (PR) or metastasis (PM) (4). The American Joint Committee on Cancer/International Union Against Cancer Classification 8th edition currently employs washing cytology for detection of malignant cells and a more accurate tumor staging, and cytology-positive tumors even without a visible metastatic tumor (P0/Cy1) are considered as advanced cancers with distant metastasis (5). The identification of intraperitoneal cancer cells is imperative because the treatment strategy for these patients is completely different, and surgery is typically not indicated in patients with peritoneal metastasis. However, cytological examination of peritoneal lavage fluid is clinically challenging due to its low sensitivity, inter-observer variability, and its limited ability to discriminate well-differentiated cancer cells from normal mesothelial cells (6-8). Moreover, the surgical procedure to obtain the specimens is invasive and performed under general anesthesia. Likewise, while imaging modalities such as computed tomography (CT) and positron emission tomography, are also commonly used for pre-treatment staging and therapeutic monitoring; they also suffer from poor sensitivity for detecting PM in patients with GC (9).
Patients with peritoneal carcinomatosis are usually diagnosed overtly with the presence of ascites or other gastrointestinal symptoms limiting their compliance to systemic treatment. A recent ACTS-GC trial demonstrated that adjuvant chemotherapy decreases the burden of disease relapse in patients with probable micrometastasis in the peritoneal cavity (2). Previous studies investigating the role of cytoreductive surgery with hyperthermic intraperitoneal chemotherapy (HIPEC), an emerging treatment in patients with peritoneal carcinomatosis, have highlighted the importance of minimal extent of peritoneal dissemination of cancer cells and complete cytoreduction for improving the long-term survival in GC (10-12). These results emphasize the clinical significance for detecting metastasized tumors at a lower burden state and indicated that early detection of peritoneal dissemination is imperative for improving treatment outcomes. Collectively, this highlights an important gap in knowledge and the importance of developing clinically relevant biomarkers for the identification of GC patients with high risk of PR or PM.
In this regard, over the last decade, several studies have analyzed clinical specimens from patients with peritoneal carcinomatosis in order to identify tissue-derived epigenomic and transcriptomic markers (13,14), metabolomic analytes(15), and the analysis of exosomal RNAs in peritoneal lavage fluid (16-18). While findings from some of these studies have been encouraging, certain shortcoming including non-comprehensive biomarker discovery and validation approaches, inadequate patient cohort size and the lack of independent validation cohorts have stifled their translation into clinical settings. Herein, we addressed these limitations of the previous studies and performed a systematic genome-wide transcriptomic expression profiling in tissue specimens from patients with GC, followed by rigorous bioinformatic approaches to identify a panel of genes for predicting PR following surgery and adjuvant therapy. After the discovery of gene markers, we subsequently established a risk-prediction model, which was successfully applied for validating this biomarker panel for predicting PR in two independent cohorts of patients with GC. Considering that PR is often associated with the existence of metastases within the peritoneal cavity, we subsequently assessed the diagnostic performance of our gene panel for its ability to detect PM, by analyzing independent cohorts of patients where patients either had metastasis at initial diagnosis or those with washing cytology positive P0/Cy1 tumors. This systematic and comprehensive biomarker discovery and validation effort allowed us to identify a 6-gene biomarker panel and subsequent establishment of a risk-stratification model for the identification of high-risk patients with peritoneal carcinomatosis – which if identified in a timely manner could potentially facilitate appropriate clinical decision-making and improving the treatment outcomes with cytoreductive surgery with HIPEC in patients with gastric cancer.
MATERIALS AND METHODS
This study was performed in accordance with the REMARK (Reporting recommendations for tumor MARKer prognostic studies) guidelines (19).
Biomarker discovery and in silico validation in genome-wide expression profiling datasets
For the biomarker discovery phase of our study, we analyzed genome-wide expression profiling data from the two publicly available datasets (GSE15081 and GSE62254). Data were downloaded from the Gene Expression Omnibus (GEO) database. The GSE15081 cohort included 140 GC cases; of which 32 cases lacking information on disease recurrence were excluded. The GSE62254 dataset included expression profiling data from 300 GC cases; wherein, 159 cases including 108 patients with a stage 1 or 4 cancer, 39 cases with other routes of tumor relapse (e.g. hematogenous, lymphogenous, and local recurrence), and 12 cases without recurrence data were excluded. This led to the final selection of 141 patients, which included 113 patients who did not experience tumor relapse and 28 with PR. Finally, 108 and 141 cases from each dataset were included in the analysis, as summarized in Table S1.
Clinical cohorts for biomarker validation
A total of 426 patients with GC were enrolled for the training and validation of biomarkers for PR-prediction and PM-detection, which were drawn from three independent cohorts (Table S2). For the initial selection of patients within the training and validation cohorts for the development of PR-prediction biomarkers, we enrolled patients from two independent institutions; the training cohort comprised of patients enrolled at the University of Ulsan and Asan Medical Center, Seoul, Korea, and the validation cohort patients were enrolled at the Ajou University, Suwon, Korea. The patients in both cohorts received curative surgery for biopsy-proven stage 1 to 3 primary GC between 2008 and 2014. Patients who received neoadjuvant treatment and those in whom the cancers developed in the remnant stomach following previous partial gastrectomy were excluded. A total of 216 and 111 patients were included for the training and validation of PR-prediction biomarkers.
Furthermore, we also performed an additional analysis for evaluating the performance of our biomarker panel for detecting the presence of PM in GC patients. For these analyses, we analyzed specimens from 210 patients who received gastrectomy for the biopsy-proven stage 1 to 4 primary GC (called as PM evaluation cohort) including prior 111 patients in the validation cohort and additional 99 patients for inclusion of stage 4 GC cases from the Ajou University, Suwon, Korea and the Nagoya University, Nagoya, Japan. In all these cohorts, recurrence or disease progression was assessed by a laboratory test, endoscopy, and abdominopelvic CT on a regular basis, as recommended by the gastric cancer treatment guidelines (20).
Tissues were obtained from a representative malignant lesion in the surgically resected stomach specimen during the operation, snap-frozen in liquid nitrogen, and then stored at −80-°C. All surgical specimens were processed and examined according to the guidelines of the Japanese Gastric Cancer Association (21). The diagnosis of carcinoma was based on the modified Vienna classification (22), and the histological type was determined according to the World Health Organization classification (23). The depth of tumor invasion (T stage) and lymph node metastasis (N stage) were determined according to the 7th edition of the American Joint Committee on Cancer (24).
All procedures were conducted in accordance with the Helsinki Declaration. A written informed consent was obtained from all participants. This study was approved by the Institutional Review Boards of all participating institutions.
RNA extraction and gene expression analysis
Total RNA was isolated from fresh frozen surgical tissues using the RNeasy mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Real-time quantitative polymerase chain reaction (RT-qPCR) was performed using the SensiFAST™ probe Lo-ROX Kit (Bioline, London, UK) and the QuantStudio 6 Flex Real Time PCR System (Applied Biosystems, Foster City, CA). To ensure the reproducibility of the assays, we undertook several approaches including appropriate control templates, exclusion of any specimen with questionable RNA quality, and use of multi-replicates performed at different time points. Gene expression levels were evaluated with Applied Biosystems QuantStudio 6 Flex Real Time PCR System Software. The relative abundance of target genes was assessed and corrected to the expression level of beta-actin as an internal control using the 2−ΔCt method; ΔCt refers to the difference of Ct values between the gene of interest and beta-actin. Values were further transformed into the log2 data. The PCR primers used are described in Table S3.
Statistical analysis
Statistical analyses were performed using R, version 3.6.3, and MedCalc Statistical Software, version 19.2 (MedCalc Software Ltd, Ostend, Belgium). Wilcoxon rank-sum and Bonferroni tests were used to compare gene expression levels between PR and non-recurrence (NR) groups in the discovery phase. Random forest classifications with 10-fold cross-validation and synthetic minority over-sampling (SMOTE) approaches for final gene candidates were used to reassess the performance of candidate gene panels for biomarker discovery. In the in-silico validation phase, the performance of the risk model derived from candidate genes was assessed through logistic regression analysis with group dichotomization based on the median expression value of each gene. In the clinical validation phase, a gene-based risk score model was built by logistic regression through a backward elimination model. A high- or low-risk group was defined according to Youden’s index for recurrence prediction, and the median value of risk score for metastasis detection. Performance was evaluated using the receiver operating characteristic (ROC) curves and area under the curve (AUC) values. Regarding survival prediction, logistic regression and Cox regression analyses were used. Peritoneal recurrence free survival (pRFS) was assessed using the Kaplan-Meier method, and the pRFS was defined as the time from the surgery to the time of confirmed PR or death from any cause. Patients were censored at 5 years for pRFS if they were recurrence free and alive at 5 years following surgery. Patients who were lost to follow-up without any evidence of recurrence before 5 years were censored at the date of the last clinic visit. Cox proportional hazards models and bi-logistic regression were used for univariate and multivariable analyses, and outcomes were reported as hazard ratios and 95% confidence intervals (CIs). Biomarkers constituting gene expression levels were compared with Mann-Whitney U test. A threshold for statistical significance was set at P < 0.05.
RESULTS
Genome-wide gene expression profiling identified a 12-gene panel for predicting peritoneal recurrence in gastric cancer
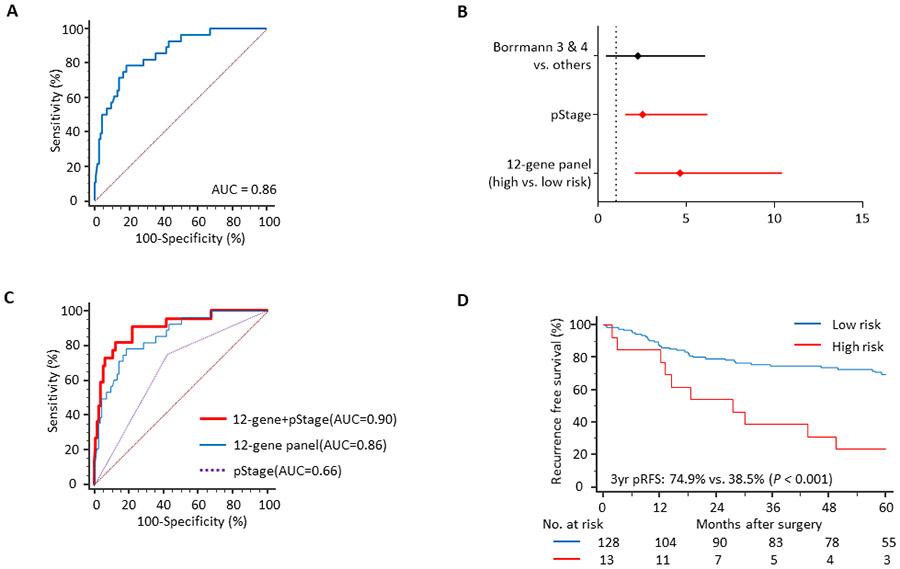
The overall workflow of this study is illustrated in Supplementary Figure S1. We undertook a systematic, comprehensive and an unbiased biomarker discovery effort by analyzing genome-wide expression profiling data from two publicly available datasets of patients with GC. The GSE15081 dataset was selected for the initial biomarker discovery, which included 33 patients with PR and 75 without any recurrence (NR). Differential gene expression (Wilcoxon rank-sum test, P < 0.01 and Bonferroni test, P < 0.05) and correlation analysis (< 0.5) led us to identify a panel of 13 genes that were differentially expressed between patients with PR vs. NR. Among this panel, the information regarding one of the genes was not present in the second public dataset (GSE62254), which was used for the in-silico validation of our discovery cohort findings. Accordingly, we finalized a panel of 12 genes, which included: ZBTB1, CHCHD3, KLHL41, POPDC2, LTBP3, CAVIN2 (SDPR), STT3B, TXNDC16, PHYHD1, KCNJ6, SLITRK6, and LMBR1. We developed a logistic regression model to predict peritoneal recurrence in GC patients, which exhibited an AUC of 0.95 (95% CI: 0.89-0.98, P < 0.001). Thereafter, the predictive accuracy of this 12-gene panel was validated in a second dataset (GSE62254), which once again confirmed the robustness of our biomarker discovery effort, as evidenced by the resulting AUC of 0.86 (95% CI: 0.79-0.91, sensitivity 79.2%, specificity 81.4%, positive predictive value [PPV] 69.1%, negative predictive value [NPV] 91.3%, positive likelihood ratio [PLR] 4.16, negative likelihood ratio [NLR] 0.26, and P < 0.001; Figure 1A). As illustrated in Table 1, our in-depth biomarker discovery effort utilized random forest classifications with 10-fold cross-validation and SMOTE modules to reassess the performance of 12 candidate genes and allowed us to assure that a 12-gene panel exhibited a robust performance in predicting PR in two independent genome-wide expression profiling datasets of GC patients. Random forest importance measures in the discovery dataset were described in Table S4.
Figure 1.
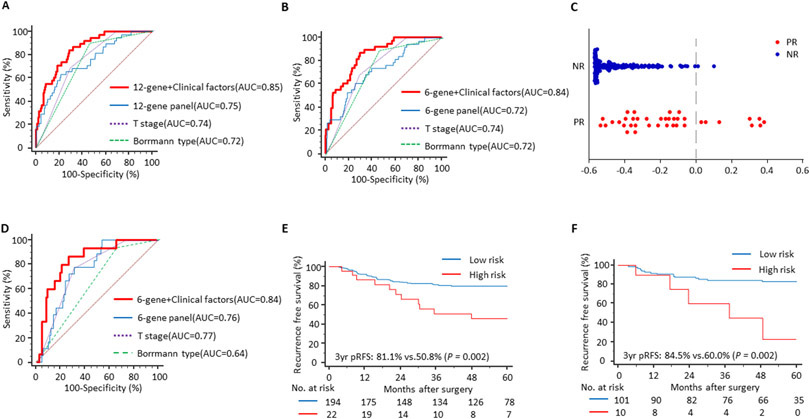
Performance of a 12-gene panel to predict peritoneal recurrence in a genomewide expression profiling dataset (GSE62254). (A) An ROC curve illustrating the performance of the gene panel, (B) A multivariate analysis, depicting that the gene panel is an independent prognostic factor for peritoneal recurrence free survival along with pathologic tumor stage (pStage), (C) Improvement in the performance of the biomarker panel to predict peritoneal recurrence when combined with pStage, and (D) The Kaplan-Meier curve showing the significant survival difference between two risk groups derived from the biomarker.
Table 1.
Summary of the biomarker performance to predict peritoneal recurrence and to detect peritoneal metastasis
| Performance to predict peritoneal recurrence | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| AUC | 95% CI | Accuracy | Sensitivity | Specificity | PPV | NPV | PLR | NLR | |
| Discovery (GSE15081) | |||||||||
| Original analysis | 0.95 | 0.89–0.98 | 0.90 | 0.94 | 0.88 | 0.87 | 0.91 | 7.83 | 0.07 |
| Random forest with SMOTE | 0.92 | N/A | 0.83 | 0.69 | 0.89 | 0.78 | 0.90 | 6.27 | 0.35 |
| In-silico validation (GSE62254) | |||||||||
| Original analysis | 0.86 | 0.79-0.91 | 0.86 | 0.79 | 0.81 | 0.69 | 0.91 | 4.16 | 0.26 |
| Random forest with SMOTE | 0.77 | N/A | 0.77 | 0.58 | 0.82 | 0.47 | 0.89 | 3.22 | 0.51 |
| Analysis with clinical samples | |||||||||
| Clinical training cohort | |||||||||
| 12-gene panel | 0.75 | 0.68-0.80 | 0.84 | 0.63 | 0.79 | 0.78 | 0.85 | 3.00 | 0.47 |
| 12 genes plus clinical factors* | 0.85 | 0.79-0.89 | 0.85 | 0.84 | 0.71 | 0.67 | 0.87 | 2.90 | 0.23 |
| 6-gene panel | 0.72 | 0.66-0.78 | 0.86 | 0.61 | 0.75 | 0.89 | 0.86 | 2.44 | 0.52 |
| 6 genes plus clinical factors* | 0.84 | 0.79-0.89 | 0.83 | 0.87 | 0.70 | 0.75 | 0.86 | 2.90 | 0.19 |
| Clinical validation cohort | |||||||||
| 6-gene panel | 0.76 | 0.67-0.83 | 0.82 | 0.78 | 0.69 | 0.87 | 0.83 | 2.52 | 0.32 |
| 6 genes plus clinical factors* | 0.84 | 0.75-0.90 | 0.76 | 0.87 | 0.73 | 0.76 | 0.86 | 3.22 | 0.18 |
| Performance to detect peritoneal metastasis | |||||||||
| AUC | 95% CI | Accuracy | Sensitivity | Specificity | PPV | NPV | PLR | NLR | |
| All peritoneal metastasis group | |||||||||
| 6-gene panel | 0.72 | 0.66-0.78 | 0.85 | 0.84 | 0.52 | 0.67 | 0.76 | 1.75 | 0.31 |
| 6 genes plus T stage | 0.86 | 0.80-0.90 | 0.85 | 0.81 | 0.80 | 0.67 | 0.86 | 4.05 | 0.24 |
| P0/Cy1 tumor group | |||||||||
| 6-gene panel | 0.72 | 0.69-0.81 | 0.91 | 0.88 | 0.48 | 0.67 | 0.91 | 1.69 | 0.25 |
| 6 genes plus T stage | 0.85 | 0.79-0.90 | 0.91 | 0.88 | 0.76 | 0.67 | 0.92 | 3.67 | 0.16 |
Abbreviations: AUC area under curve; CI confidence interval; PPV positive predictive value; NPV negative predictive value; PLR positive likelihood ratio; NLR negative likelihood ratio; SMOTE synthetic minority over-sampling technique; P0/Cy1: cytology positive gastric cancer without gross peritoneal metastasis
Clinical factors refer to depth of tumor invasion (T stage) and Borrmann type in gross appearance.
In order to further evaluate the clinical significance of our recurrence prediction biomarkers, we next validated the prognostic significance of this 12-gene panel in a gene expression profiling dataset (GSE62254). Among the three factors which were significant in a univariate analysis (the 12-gene panel, stage, and Borrmann type), we noted that the pathologic tumor stage and the gene expression panel were the only variables that emerged as significant predictors of pRFS in a multivariate cox proportional hazard model (Figure 1B). When we combined these two factors together, this led to further improvement in the recurrence prediction potential of the gene panel and yielded an AUC of 0.90 (vs. an AUC of 0.86 with the gene panel alone; Figure 1C). Finally, when we analyzed the pRFS in this cohort, the Kaplan-Meier analysis revealed that based upon our recurrence prediction model, patients categorized within the high-risk group had a significantly worse prognosis vs. those in the low-risk group (3-year pRFS: 74.9% vs. 38.5%, P < 0.001; Figure 1D). Taken together, our data highlight the potential importance of 12-gene panel in predicting PR in patients with GC.
A clinical training phase allowed establishment of a 6-gene based risk model for predicting recurrence in patients with gastric cancer
Following successful discovery and in-silico validation of our gene panel, we next undertook an effort to further refine our biomarker panel and established a risk-prediction model by training our biomarkers in a clinical training cohort of 216 GC patients, by performing RT-qPCR assays for each of the 12 genes. Gene expression levels were measured in all samples and we developed two independent prediction models through logistic regression and the backwards elimination approach. The 12-gene panel derived from a single-step based regression yielded an AUC value of 0.75 (95% CI: 0.68-0.80, sensitivity 63.2%, specificity 78.7%, PPV 78.2%, NPV 84.8%, PLR 3.00, NLR 0.47, P < 0.001), which was quite comparable to the AUC value of 0.72 (95% CI: 0.66-0.78, sensitivity 60.5%, specificity 75.3%, PPV 88.6%, NPV 86.2%, PLR 2.44, NLR 0.52, P < 0.001) for the reduced, 6-gene panel derived from the stepwise regression model (Table 1).
In the training cohort of patients, Cox proportional hazard analysis demonstrated that infiltrative tumor morphology (Borrmann type 3 and 4; HR 4.02, 95% CI: 1.75-9.24, P = 0.001), deeper invasion of tumor cells (advanced T stage; HR 1.91, 95% CI: 1.03-3.52, P = 0.039), and our 6-gene panel (HR 2.05, 95% CI: 1.01-4.16, P = 0.048), representing the patients within the high-risk group associated with a significantly shorter pRFS (Table 2). In view of these findings, we next questioned whether a combination of our gene panels along with these clinical factors might further improve the performance of both recurrence prediction models. Indeed, when we performed these analysis, we noted that the combination of our 12-gene panel together with the tumor morphology and T stage further improved the predictive performance (AUC = 0.85; 95% CI: 0.79-0.89, sensitivity: 84.2%, specificity: 71.3%, PPV 67.4%, NPV 86.8%, PLR 2.90, NLR 0.23, P < 0.001 vs. an AUC of 0.75 for the gene panel alone; Figure 2A). Furthermore, the predictive performance of this model was almost identical to the one that included the 6-gene panel (AUC= 0.84, 95% CI: 0.79-0.89, sensitivity: 86.8%, specificity: 70.2%, PPV 75.2%, NPV 85.6%, PLR 2.90, NLR 0.19, P < 0.001 vs. an AUC of 0.72 with the gene panel alone; Figure 2B). Through these analyses, we ensured that the integration of the gene panel together with the clinical prognostic indicators led to a significantly improved overall performance in predicting PR using both the 12 and 6-gene panels.
Table 2.
Cox regression analysis of prognostic factors for peritoneal recurrence free survival
| Clinical training cohort | Clinical validation cohort | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Variables | Univariate analysis | Multivariate analysis | Univariate analysis | Multivariate analysis | ||||||||
| HR | 95% CI | P-value | HR | 95% CI | P-value | HR | 95% CI | P-value | HR | 95% CI | P-value | |
| Differentiation | 4.36 | 1.73-10.98 | 0.002 | 2.23 | 0.84-5.91 | 0.108 | 2.75 | 0.81-9.37 | 0.105 | |||
| Tumor size | 1.13 | 1.04-1.20 | 0.003 | 1.02 | 1.01-1.03 | 0.003 | 1.01 | 0.99-1.02 | 0.091 | |||
| Borrmann type | 6.22 | 2.80-13.81 | <0.001 | 4.02 | 1.75-9.24 | 0.001 | 5.44 | 1.27-23.36 | 0.023 | 4.25 | 0.98-18.53 | 0.054 |
| Depth of invasion | 3.17 | 1.80-5.57 | <0.001 | 1.91 | 1.03-3.52 | 0.039 | 6.77 | 2.47-18.55 | <0.001 | 3.58 | 1.16-11.08 | 0.027 |
| 6-Gene panel (high- vs. low-risk) | 2.78 | 1.42-5.40 | 0.003 | 2.05 | 1.01-4.16 | 0.048 | 4.42 | 1.60-12.19 | 0.004 | 4.15 | 1.31-13.20 | 0.016 |
Figure 2.
Performance of the biomarker to predict peritoneal recurrence in clinical cohorts. In the training cohort, (A) a risk prediction model that included the 12-gene biomarker panel along with clinical factors (T stage and Borrmann type) was established; (B) The performance of a reduced 6-gene biomarker panel along with the same clinical variables. (C) A scatter plot illustrating the performance of the 6-gene biomarker to discriminate cases with peritoneal recurrence (PR) from non-recurrence group (NR). (D) The identical risk prediction formula derived from the training phase was validated in an independent cohort, and the ROC curves for the 6-gene biomarker and combined gene panel were produced. The biomarker consistently stratified the prognosis in (E) the training cohort and (F) the validation cohort in the Kaplan-Meier curves.
In order to prioritize and select a clinically relevant gene panel and considering the equivalent predictive potential of both gene panels, we selected the 6-gene panel to establish a risk-stratification model to predict PR in GC patients, as illustrated in Figure 2C. Based on the individual coefficient and constants derived from the logistic regression model of 6-gene panel, we developed a risk prediction formula as follows; 0.89290×ZBTB1+0.50046×CAVIN2−0.44275×CHCHD3−0.25294×LTBP3−0.16871×SLITRK6−0.14124×STT3B+0.23925. Of interest, the expression of two of the genes, ZBTB1 and CAVIN2, was significantly upregulated in tumors with PR in the training cohort (P = 0.041 and 0.004, respectively; Supplementary Figure S2A). We assessed the performance of this 6-gene panel for survival prediction and it yielded an AUC of 0.70 (95% CI: 0.63-0.77, P = 0.034) in logistic regression and 0.70 (95% CI: 0.63-0.76, P = 0.034) in Cox regression analysis.
The risk-prediction model was successful in stratifying GC patients with and without peritoneal recurrence in an independent validation cohort
Following development of the 6-gene based risk prediction model, we evaluated its performance in an independent cohort of 111 patients within the validation cohort using the same risk prediction formula as the training cohort. It was quite reassuring to observe that our biomarker panel successfully discriminated patients that experienced PR from those without recurrence, with a corresponding AUC value of 0.76 (95% CI: 0.67-0.83, sensitivity: 77.8%, specificity: 68.8%, PPV 86.8%, NPV 83.2%, PLR 2.52, NLR 0.32, P = 0.006; Table 1). In line with our training phase data, a Cox regression analysis in the patients within this validation cohort once again demonstrated that the depth of tumor invasion (HR 3.58, 95% CI: 1.16-11.08, P = 0.027), tumor morphology (HR 4.25, 95% CI: 0.98-18.53, P = 0.054) and the 6-gene panel (HR 4.15, 95% CI: 1.31-13.20, P = 0.016), were significant prognostic indicators for predicting PR in GC patients (Table 2). Likewise, the combination of the tumor T stage and morphology further augmented the predictive performance of the gene panel (AUC = 0.84, 95% CI: 0.75-0.90, sensitivity: 86.7%, specificity 73.3%, PPV 76.1%, NPV 85.8%, PLR 3.22, NLR 0.18, P < 0.001; Figure 2D). As was the case in the training cohort patients, expression levels of the ZBTB1 and CAVIN2, were also significantly upregulated in tumors with PR (P = 0.023 and 0.048, respectively; Supplementary Figure S2B). However, the performance of risk prediction formula to predict survival was not significant. Collectively, the independent validation of the recurrence prediction model demonstrated the robustness of our biomarker in predicting PR in patients with GC.
The gene biomarker panel robustly predicted recurrence free survival of GC patients in the training and validation cohorts
Given that PR is almost always accompanied with poor survival in GC patients, we next evaluated the prognostic potential of our recurrence prediction biomarker panel in the training and validation cohort of patients. All patients were dichotomized into high and low-risk groups for PR according to the cutoff threshold values derived from the prediction model, followed by Kaplan-Meier analyses to determine the prognostic significance of our gene panel. In the training cohort, pRFS in the high-risk group patients was significantly worse compared to those in the low-risk group (3-year pRFS: 81.1% vs. 50.8%, P = 0.002; Figure 2E). Similarly, when we evaluated this hypothesis in an independent validation cohort of patients with GC, we observed significant differences in survival outcomes between the high- and low-risk groups (3-year pRFS: 84.5% vs. 60.0%, P = 0.002; Figure 2F). These results highlighted that in addition to the ability of our biomarker panel to predict PR, these biomarkers are also clinically significant in predicting the prognosis of GC patients.
The 6-gene biomarker panel robustly identifies presence of peritoneal metastasis at disease diagnosis in GC patients
Considering that PR is often a manifestation of metastases within the peritoneal cavity, we assessed the potential of our 6-gene panel for its ability to detect PM in patients with GC. For these analyses, we examined 210 clinical specimens from patients within the PM evaluation cohort, which included 31 specimens from patients accompanied with PM at the time of surgery. It was very encouraging to observe that our biomarker panel successfully distinguished GC patients with PM from those without distant metastasis, as evidenced by a robust AUC value of 0.72 (95% CI: 0.66-0.78, sensitivity 83.9%, specificity 52.0%, PPV 66.8%, NPV 75.8%, PLR 1.75, NLR 0.31, P < 0.001; Table 1).
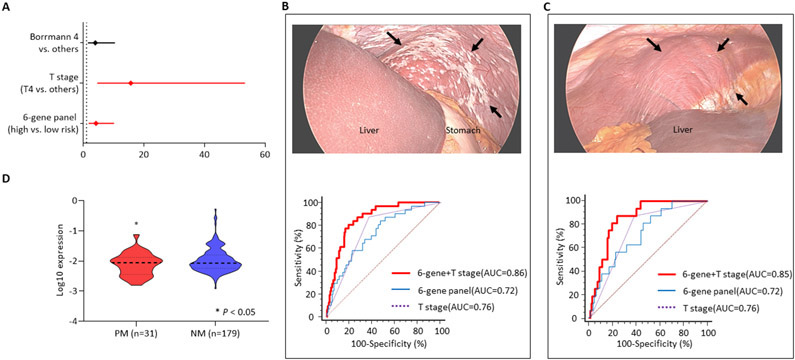
Multivariate logistic regression analysis revealed that the 6-gene panel and the depth of tumor invasion were the only significant indicators for PM (HR 4.47, 95% CI: 1.72-11.62, P = 0.002; HR 17.03, 95% CI: 4.88-59.42, P < 0.001, respectively; Figure 3A). Accordingly, we next evaluated the diagnostic accuracy of this combination panel, which clearly demonstrated a significant gain in diagnostic accuracy (AUC=0.86, 95% CI: 0.80-0.90, sensitivity 80.6%, specificity 80.4%, PPV 67.2%, NPV 86.4%, PLR 4.05, NLR 0.24, P < 0.001; Figure 3B) vs. the gene panel and T-stage individually. Collectively, these findings allowed us to better appreciate the clinical significance of our biomarker panel in not only its potential for predicting PR in surgically-resectable cases, but also its ability to detect PM in GC patients who are otherwise not candidates for surgical treatments.
Figure 3.
The 6-gene biomarker’s performance to detect peritoneal metastasis in the peritoneal metastasis evaluation cohort. (A) A forest plot demonstrating that the gene panel was a significant predictor for peritoneal metastasis (PM) of gastric cancer along with depth of tumor invasion (T stage) in multivariate logistic regression. (B) the 6-gene biomarker discriminated patients with PM from those without metastasis. The combination of T stage improved the performance of the gene panel (arrows indicate grossly disseminated tumor deposits on peritoneum). (C) The biomarker successfully identified peritoneal micro-metastasis of gastric cancer detected by washing cytology as well (no visible or minimal cancer is seen). (D) A violin plot exhibited a significant difference in gene expression of ZBTB1.
The gene biomarker panel successfully identified GC patients with peritoneal micrometastasis
Following the identification that our gene biomarker could successfully detect PM in GC patients, we next asked whether this biomarker panel can also identify patients with cytology positive P0/Cy1 tumors. These tumors are unfortunately often missed with the currently used diagnostic modalities in the clinic, but these patients are ideal candidates for receiving the newer therapies that can quite effectively treat peritoneal carcinomatosis and improve the overall survival outcomes in this subset of GC patients. Among 31 patients with PM from the PM evaluation cohort, 16 patients with micrometastasis were selected and the potential of the gene biomarker panel was evaluated. Our analysis of patients with P0/Cy1 cancers revealed that the diagnostic potential of our biomarker was quite remarkable (AUC=0.72, 95% CI: 0.69-0.81, sensitivity 87.5%, specificity 48.0%, PPV 67.2%, NPV 90.8%, PLR 1.69, NLR 0.25, P = 0.034), which was significantly improved when we combined it together with T-stage information from these patients (AUC=0.85, 95% CI: 0.79-0.90, sensitivity 87.5%, specificity 76.0%, PPV 67.2%, NPV 92.4%, PLR 3.67, NLR 0.16, P < 0.001; Figure 3C). In support of our previous findings, among the six genes constituting the biomarker, the expression levels of ZBTB1 were significantly higher in patients with PM (P = 0.036; Figure 3D). Taken together, our systematic and comprehensive biomarker discovery and validation effort allowed us to identify and validate a novel gene panel that could potentially predict peritoneal recurrence, as well as simultaneously allow detection of peritoneal metastasis – which are key features for improving the overall management of patients with GC and improving their overall survival.
DISCUSSION
Accumulating evidence in recent years have allowed an improved understanding of the various molecular subtypes in gastric cancer (GC) (25,26). As a consequence, a recent study reported that a subtype of peritoneal GC cells has a unique gene expression, mutational and copy number alteration profile (27). In spite of exhaustive efforts at unveiling the genomic and epigenomic landscape of GC, much of this molecular knowledge has had a very little impact on the clinical management of disease in patients with GC. In particular, there is paucity of data on molecular biomarkers that can predict peritoneal recurrence (PR) and can identify peritoneal metastasis (PM) in GC patients – which has important implications for the improved disease management and treatment decision-making in this malignancy. In this study, we undertook a systematic and comprehensive biomarker discovery and validation effort, which led us to identify a 6-gene biomarker panel which predicted PR in patients with GC. Furthermore, we established a risk-stratification model that robustly identified high-risk patients with peritoneal carcinomatosis – which if identified in a timely manner could potentially facilitate appropriate clinical decision-making and improving the treatment outcomes in patients with GC.
In our study, we used primary tissue specimen to ascertain the high-risk patients for peritoneal carcinomatosis. In patients with a localized disease, surgery remains the preferred treatment choice, and it provides adequate specimen for biomarker identification such as our PR predicting gene panel as well as pathological analysis. In those with PM, gastrectomy is not indicated (28). Instead, tissues should be obtained with endoscopy, which is a safe procedure, to confirm the diagnosis and to guide the optimal treatment in all patients with GC. The clinical approaches to obtain analytical specimens from these patients should be safe and accessible, and not levy extra risk of invasiveness, because post-procedural complication can delay the commencement of an optimal therapy and eventually lead to treatment failure. From this perspective, a biomarker that can be measured in the primary tumor tissue is a suitable option for risk stratification of peritoneal carcinomatosis, regardless of the tumor stage (Supplementary Figure S3).
Our study revealed that the 3-year pRFS was 78.0% and 82.8% in the training and validation cohorts, which is slightly higher than the 3-year RFS rates of 72~74% observed in patients with stage 2 and 3 GC in previous studies (2,29). As our study included more than 10% of stage 1 patients with excellent prognosis in both cohorts and survival analysis was assessed only based on peritoneal recurrence, prognosis outcomes seemed a bit more favorable than the general outcomes reported in gastric cancer.
Gastric cancer with Borrmann type 3 and 4 on gross appearance is typically characterized by aggressive tumor infiltration. In particular, with regards to type 4 tumors, they are frequently associated with peritoneal recurrence or metastasis. Our results are in line with findings from previous studies and this could support the objectivity of clinical cohorts in this study (3,30,31).
From a functional viewpoint, various genes in our biomarker panel have been shown to be bonafide candidates involved in cancer pathogenesis. For instance, Zinc finger and BTB domain containing 1 (ZBTB1) is an important regulator of translesion DNA synthesis in damaged DNA (32). Recently, ZBTB1 was found to be one of the most commonly mutated genes in microsatellite instability (MSI) type GC (33). Likewise, epithelial-mesenchymal transition (EMT) is a key step for cancer cells to metastasize (34) and Caveolae Associated Protein 2 (CAVIN2) is involved in the transforming growth factor β (TGF-β) pathway which drives EMT in GC (35). Similarly, Coiled-Coil-Helix-Coiled-Coil-Helix Domain Containing 3 (CHCHD3), which is involved in mitochondrial cristae structure and organization, is associated with PD-1 activity in CD8+ T cell and serglycan which promotes EMT and tumor cell aggressiveness (36,37).
To date, the optimal treatment of peritoneal carcinomatosis from GC remains unclear. Although systemic or intraperitoneal chemotherapy improves overall survival compared with symptomatic care, the prognosis for this malignancy remains unsatisfactory (38). Cytoreductive surgery with HIPEC, are considered a safe and effective treatment for various cancer types (39,40), including GC. Previous studies reported promising results that this comprehensive treatment might provide a long-term survival even without recurrence in selected patients with limited metastasis as well as better prognosis than chemotherapy alone (41,42). Notably, our biomarker panel was effective in discriminating patients with P0/Cy1 tumor as well as those with gross carcinomatosis, and might serve as an important modality for identifying patients that are eligible for HIPEC treatment.
We would like to acknowledge some of the potential limitations of our study. First, despite using multiple clinical cohorts in this study, our study had a retrospective design. Second, we were unable to validate the performance of our biomarkers for its diagnostic potential for PM in endoscopic biopsy tissues. To overcome these limitations, a prospective multi-institutional study is required.
In conclusion, through a systematic and comprehensive discovery and validation effort, we have developed a novel gene expression biomarker panel that has a potential to predict peritoneal recurrence and simultaneously distinguish patients with peritoneal metastasis in patients with gastric cancer. We established a risk-stratification model for the identification of high-risk patients with peritoneal carcinomatosis – which when diagnosed in a timely manner might potentially facilitate appropriate clinical decision-making and improving the treatment outcomes in patients with gastric cancer.
Supplementary Material
TRANSLATIONAL RELEVANCE.
Peritoneal metastasis of gastric cancer is a fatal disease mainly due to delayed detection. The limited accuracy of current diagnostic modalities highlights the need of the biomarker that can identify patients at high risk of peritoneal recurrence or metastasis to improve prognosis of this malignancy. In this study, based on a systematic and comprehensive biomarker discovery and validation with multiple clinical cohorts, we developed a 6-gene based biomarker panel for identification of high-risk patients with peritoneal carcinomatosis. This biomarker panel was remarkably effective for detecting peritoneal micro-metastasis, which are ideal candidates for the new treatment, as well as predicting recurrence in peritoneal cavity. Moreover, its accuracy was significantly enhanced when combined with tumor depth of invasion. This novel transcriptomic signature could serve as an important clinical decision-making and potentially facilitate appropriate treatment in patients with gastric cancer.
ACKNOWLEDGMENTS
The present work was supported by the grants CA72851, CA181572, and CA187956 from the National Cancer Institute, National Institutes of Health, and a pilot grant from the Stupid Strong Foundation to A. Goel. We would like to thank Satoshi Nishiwada, Tatsuhiko Kakisaka, Yuma Wada, Yasuyuki Okada, and Divya Sahu for helping with their important insights into experiments and data analysis. The biospecimen and data used in this study were provided by Asan Bio-Resource Center, Korea Biobank Network 2019-05(184) and the Ajou Human Bio-Resource Bank (AHBB), a member of the National Biobank of Korea, which is supported by the Ministry of Health and Welfare.
Abbreviations:
- AUC
Area under curve
- CI
Confidence interval
- CT
Computed tomography
- GC
Gastric cancer
- HIPEC
Hyperthermic intraperitoneal chemotherapy
- PR
Peritoneal recurrence
- PM
Peritoneal metastasis
- ROC
Receiver operating characteristic
- pRFS
Peritoneal recurrence free survival
- RT-qPCR
Real-time quantitative reverse transcription polymerase chain reaction
- SMOTE
Synthetic minority over-sampling technique
- PPV
Positive predictive value
- NPV
Negative predictive value
- PLR
Positive likelihood ratio
- NLR
Negative likelihood ratio
Footnotes
Declarations of interest: None of the authors has any potential conflicts to disclose.
REFERENCES
- 1.Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin DM, Pineros M, et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer 2019;144(8):1941–53 doi 10.1002/ijc.31937. [DOI] [PubMed] [Google Scholar]
- 2.Sakuramoto S, Sasako M, Yamaguchi T, Kinoshita T, Fujii M, Nashimoto A, et al. Adjuvant chemotherapy for gastric cancer with S-1, an oral fluoropyrimidine. N Engl J Med 2007;357(18):1810–20 doi 10.1056/NEJMoa072252. [DOI] [PubMed] [Google Scholar]
- 3.Thomassen I, van Gestel YR, van Ramshorst B, Luyer MD, Bosscha K, Nienhuijs SW, et al. Peritoneal carcinomatosis of gastric origin: a population-based study on incidence, survival and risk factors. Int J Cancer 2014;134(3):622–8 doi 10.1002/ijc.28373. [DOI] [PubMed] [Google Scholar]
- 4.Kodera Y, Yamamura Y, Shimizu Y, Torii A, Hirai T, Yasui K, et al. Peritoneal washing cytology: prognostic value of positive findings in patients with gastric carcinoma undergoing a potentially curative resection. J Surg Oncol 1999;72(2):60–4; discussion 4-5 doi . [DOI] [PubMed] [Google Scholar]
- 5.Amin MB, Greene FL, Edge SB, Compton CC, Gershenwald JE, Brookland RK, et al. The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more "personalized" approach to cancer staging. CA Cancer J Clin 2017;67(2):93–9 doi 10.3322/caac.21388. [DOI] [PubMed] [Google Scholar]
- 6.Abe S, Yoshimura H, Tabara H, Tachibana M, Monden N, Nakamura T, et al. Curative resection of gastric cancer: limitation of peritoneal lavage cytology in predicting the outcome. J Surg Oncol 1995;59(4):226–9 doi 10.1002/jso.2930590405. [DOI] [PubMed] [Google Scholar]
- 7.Kodera Y, Nakanishi H, Yamamura Y, Shimizu Y, Torii A, Hirai T, et al. Prognostic value and clinical implications of disseminated cancer cells in the peritoneal cavity detected by reverse transcriptase-polymerase chain reaction and cytology. Int J Cancer 1998;79(4):429–33 doi . [DOI] [PubMed] [Google Scholar]
- 8.Schofield K, D'Aquila T, Rimm DL. The cell adhesion molecule, E-cadherin, distinguishes mesothelial cells from carcinoma cells in fluids. Cancer 1997;81(5):293–8. [PubMed] [Google Scholar]
- 9.Kim SJ, Kim HH, Kim YH, Hwang SH, Lee HS, Park DJ, et al. Peritoneal metastasis: detection with 16- or 64-detector row CT in patients undergoing surgery for gastric cancer. Radiology 2009;253(2):407–15 doi 10.1148/radiol.2532082272. [DOI] [PubMed] [Google Scholar]
- 10.Mielko J, Rawicz-Pruszynski K, Skorzewska M, Cisel B, Pikula A, Kwietniewska M, et al. Conversion Surgery with HIPEC for Peritoneal Oligometastatic Gastric Cancer. Cancers (Basel) 2019;11(11) doi 10.3390/cancers11111715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yonemura Y, Kawamura T, Bandou E, Takahashi S, Sawa T, Matsuki N. Treatment of peritoneal dissemination from gastric cancer by peritonectomy and chemohyperthermic peritoneal perfusion. Br J Surg 2005;92(3):370–5 doi 10.1002/bjs.4695. [DOI] [PubMed] [Google Scholar]
- 12.Bonnot PE, Piessen G, Kepenekian V, Decullier E, Pocard M, Meunier B, et al. Cytoreductive Surgery With or Without Hyperthermic Intraperitoneal Chemotherapy for Gastric Cancer With Peritoneal Metastases (CYTO-CHIP study): A Propensity Score Analysis. J Clin Oncol 2019;37(23):2028–40 doi 10.1200/JCO.18.01688. [DOI] [PubMed] [Google Scholar]
- 13.Kanda M, Shimizu D, Tanaka H, Tanaka C, Kobayashi D, Hayashi M, et al. Significance of SYT8 For the Detection, Prediction, and Treatment of Peritoneal Metastasis From Gastric Cancer. Ann Surg 2018;267(3):495–503 doi 10.1097/SLA.0000000000002096. [DOI] [PubMed] [Google Scholar]
- 14.Sawaki K, Kanda M, Miwa T, Umeda S, Tanaka H, Tanaka C, et al. Troponin I2 as a Specific Biomarker for Prediction of Peritoneal Metastasis in Gastric Cancer. Ann Surg Oncol 2018;25(7):2083–90 doi 10.1245/s10434-018-6480-z. [DOI] [PubMed] [Google Scholar]
- 15.Kaji S, Irino T, Kusuhara M, Makuuchi R, Yamakawa Y, Tokunaga M, et al. Metabolomic profiling of gastric cancer tissues identified potential biomarkers for predicting peritoneal recurrence. Gastric Cancer 2020. doi 10.1007/s10120-020-01065-5. [DOI] [PubMed] [Google Scholar]
- 16.Hiraki M, Kitajima Y, Koga Y, Tanaka T, Nakamura J, Hashiguchi K, et al. Aberrant gene methylation is a biomarker for the detection of cancer cells in peritoneal wash samples from advanced gastric cancer patients. Ann Surg Oncol 2011;18(10):3013–9 doi 10.1245/s10434-011-1636-0. [DOI] [PubMed] [Google Scholar]
- 17.Nakanishi K, Kanda M, Umeda S, Tanaka C, Kobayashi D, Hayashi M, et al. The levels of SYT13 and CEA mRNAs in peritoneal lavages predict the peritoneal recurrence of gastric cancer. Gastric Cancer 2019;22(6):1143–52 doi 10.1007/s10120-019-00967-3. [DOI] [PubMed] [Google Scholar]
- 18.Tokuhisa M, Ichikawa Y, Kosaka N, Ochiya T, Yashiro M, Hirakawa K, et al. Exosomal miRNAs from Peritoneum Lavage Fluid as Potential Prognostic Biomarkers of Peritoneal Metastasis in Gastric Cancer. PLoS One 2015;10(7):e0130472 doi 10.1371/journal.pone.0130472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM, et al. Reporting recommendations for tumor marker prognostic studies (REMARK). J Natl Cancer Inst 2005;97(16):1180–4 doi 10.1093/jnci/dji237. [DOI] [PubMed] [Google Scholar]
- 20.Japanese Gastric Cancer A Japanese gastric cancer treatment guidelines 2014 (ver. 4). Gastric Cancer 2017;20(1):1–19 doi 10.1007/s10120-016-0622-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Japanese Gastric Cancer A Japanese classification of gastric carcinoma: 3rd English edition. Gastric Cancer 2011;14(2):101–12 doi 10.1007/s10120-011-0041-5. [DOI] [PubMed] [Google Scholar]
- 22.Schlemper RJ, Riddell RH, Kato Y, Borchard F, Cooper HS, Dawsey SM, et al. The Vienna classification of gastrointestinal epithelial neoplasia. Gut 2000;47(2):251–5 doi 10.1136/gut.47.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flejou JF. [WHO Classification of digestive tumors: the fourth edition]. Ann Pathol 2011;31(5 Suppl):S27–31 doi 10.1016/j.annpat.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 24.Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol 2010;17(6):1471–4 doi 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 25.Cancer Genome Atlas Research N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513(7517):202–9 doi 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cristescu R, Lee J, Nebozhyn M, Kim KM, Ting JC, Wong SS, et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med 2015;21(5):449–56 doi 10.1038/nm.3850. [DOI] [PubMed] [Google Scholar]
- 27.Wang R, Song S, Harada K, Ghazanfari Amlashi F, Badgwell B, Pizzi MP, et al. Multiplex profiling of peritoneal metastases from gastric adenocarcinoma identified novel targets and molecular subtypes that predict treatment response. Gut 2020;69(1):18–31 doi 10.1136/gutjnl-2018-318070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fujitani K, Yang HK, Mizusawa J, Kim YW, Terashima M, Han SU, et al. Gastrectomy plus chemotherapy versus chemotherapy alone for advanced gastric cancer with a single non-curable factor (REGATTA): a phase 3, randomised controlled trial. Lancet Oncol 2016;17(3):309–18 doi 10.1016/S1470-2045(15)00553-7. [DOI] [PubMed] [Google Scholar]
- 29.Bang YJ, Kim YW, Yang HK, Chung HC, Park YK, Lee KH, et al. Adjuvant capecitabine and oxaliplatin for gastric cancer after D2 gastrectomy (CLASSIC): a phase 3 open-label, randomised controlled trial. Lancet 2012;379(9813):315–21 doi 10.1016/S0140-6736(11)61873-4. [DOI] [PubMed] [Google Scholar]
- 30.Lee JH, Son SY, Lee CM, Ahn SH, Park DJ, Kim HH. Factors predicting peritoneal recurrence in advanced gastric cancer: implication for adjuvant intraperitoneal chemotherapy. Gastric Cancer 2014;17(3):529–36 doi 10.1007/s10120-013-0306-2. [DOI] [PubMed] [Google Scholar]
- 31.Otsuji E, Yamaguchi T, Sawai K, Sakakura C, Okamoto K, Takahashi T. Regional lymph node metastasis as a predictor of peritoneal carcinomatosis in patients with Borrmann type IV gastric carcinoma. Am J Gastroenterol 1999;94(2):434–7 doi 10.1111/j.1572-0241.1999.873_b.x. [DOI] [PubMed] [Google Scholar]
- 32.Kim H, Dejsuphong D, Adelmant G, Ceccaldi R, Yang K, Marto JA, et al. Transcriptional repressor ZBTB1 promotes chromatin remodeling and translesion DNA synthesis. Mol Cell 2014;54(1):107–18 doi 10.1016/j.molcel.2014.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu J, McCleland M, Stawiski EW, Gnad F, Mayba O, Haverty PM, et al. Integrated exome and transcriptome sequencing reveals ZAK isoform usage in gastric cancer. Nat Commun 2014;5:3830 doi 10.1038/ncomms4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hugo H, Ackland ML, Blick T, Lawrence MG, Clements JA, Williams ED, et al. Epithelial--mesenchymal and mesenchymal--epithelial transitions in carcinoma progression. J Cell Physiol 2007;213(2):374–83 doi 10.1002/jcp.21223. [DOI] [PubMed] [Google Scholar]
- 35.Luo Y, Wu J, Wu Q, Li X, Wu J, Zhang J, et al. miR-577 Regulates TGF-beta Induced Cancer Progression through a SDPR-Modulated Positive-Feedback Loop with ERK-NF-kappaB in Gastric Cancer. Mol Ther 2019;27(6):1166–82 doi 10.1016/j.ymthe.2019.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogando J, Saez ME, Santos J, Nuevo-Tapioles C, Gut M, Esteve-Codina A, et al. PD-1 signaling affects cristae morphology and leads to mitochondrial dysfunction in human CD8(+) T lymphocytes. J Immunother Cancer 2019;7(1):151 doi 10.1186/s40425-019-0628-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manou D, Karamanos NK, Theocharis AD. Tumorigenic functions of serglycin: Regulatory roles in epithelial to mesenchymal transition and oncogenic signaling. Semin Cancer Biol 2020;62:108–15 doi 10.1016/j.semcancer.2019.07.004. [DOI] [PubMed] [Google Scholar]
- 38.Ishigami H, Fujiwara Y, Fukushima R, Nashimoto A, Yabusaki H, Imano M, et al. Phase III Trial Comparing Intraperitoneal and Intravenous Paclitaxel Plus S-1 Versus Cisplatin Plus S-1 in Patients With Gastric Cancer With Peritoneal Metastasis: PHOENIX-GC Trial. J Clin Oncol 2018;36(19):1922–9 doi 10.1200/JCO.2018.77.8613. [DOI] [PubMed] [Google Scholar]
- 39.Spiliotis J, Halkia E, Lianos E, Kalantzi N, Grivas A, Efstathiou E, et al. Cytoreductive surgery and HIPEC in recurrent epithelial ovarian cancer: a prospective randomized phase III study. Ann Surg Oncol 2015;22(5):1570–5 doi 10.1245/s10434-014-4157-9. [DOI] [PubMed] [Google Scholar]
- 40.Verwaal VJ, van Ruth S, de Bree E, van Sloothen GW, van Tinteren H, Boot H, et al. Randomized trial of cytoreduction and hyperthermic intraperitoneal chemotherapy versus systemic chemotherapy and palliative surgery in patients with peritoneal carcinomatosis of colorectal cancer. J Clin Oncol 2003;21(20):3737–43 doi 10.1200/JCO.2003.04.187. [DOI] [PubMed] [Google Scholar]
- 41.Coccolini F, Catena F, Glehen O, Yonemura Y, Sugarbaker PH, Piso P, et al. Complete versus incomplete cytoreduction in peritoneal carcinosis from gastric cancer, with consideration to PCI cut-off. Systematic review and meta-analysis. Eur J Surg Oncol 2015;41(7):911–9 doi 10.1016/j.ejso.2015.03.231. [DOI] [PubMed] [Google Scholar]
- 42.Passot G, Vaudoyer D, Villeneuve L, Kepenekian V, Beaujard AC, Bakrin N, et al. What made hyperthermic intraperitoneal chemotherapy an effective curative treatment for peritoneal surface malignancy: A 25-year experience with 1,125 procedures. J Surg Oncol 2016;113(7):796–803 doi 10.1002/jso.24248. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.